Abstract
Two new species of Russulasubg.Heterophyllidia from Guangdong Province of China were described and illustrated based on morphological characters, and their identity supported by molecular phylogeny. R.luofuensis is morphologically characterized by a grayish yellow to brownish orange pileus center with a purplish gray to grayish magenta margin, a surface that is cracked and broken into small golden-brown patches, subglobose to broadly ellipsoid basidiospores with warts fused in short or long chains and a suprapellis composed of hyphal extremities with inflated, ellipsoid or globose cells and attenuated terminal cell. R.subbubalina is distinguished by the blanched almond to dark salmon pileus that is cracked with age, subglobose to broadly ellipsoid basidiospores with wart fused in short or long chains and frequently connected by line connections, a suprapellis with hyphal ends composed of inflated or ellipsoid cells and attenuated terminal cell, and pileocystidia that are mainly clavate and sometimes with round or ellipsoid appendage. The phylogenetic analyses based on ITS-nrLSU-mtSSU-TEF1 dataset were performed using maximum likelihood and Bayesian analysis. In terms of morphological features and molecular data, the former species belongs to subsect. Virescentinae, whereas the latter comes under subsect. Heterophyllinae.
Keywords: Luofu Mountain, new species, phylogeny, Russulaceae , taxonomy
Introduction
Russula Pers. is the largest genus of Russulaceae, estimated at least to contain 2000 species, which has resulted in many complex and multilevel classifications (Buyck et al. 2018; Adamčík et al. 2019; Wijayawardene et al. 2020). Recent molecular phylogenetic studies have indicated eight subgenera within the genus R.subg.Glutinosae Buyck & X.H. Wang, R.subg.Archaeae Buyck & V. Hofst., R.subg.Compactae (Fr.) Bon, R.subg.Crassotunicatae Buyck & V. Hofst., R.subg.Heterophyllidiae Romagnesi, R.subg.Malodorae Buyck & V. Hofst., R.subg.Brevipedum Buyck & V. Hofst., and R.subg.Russula (Buyck et al. 2018, 2020). The infrageneric classification system of Russula based on a multi-locus phylogenetic analysis was followed in this study. This genus is globally distributed and occurs across a wide range of habitats from Arctic tundra to tropical forests and forms ectomycorrhizal relationships with diverse plants (Knudsen and Borgen 1982; Buyck et al. 1996; Looney et al. 2018). Some species of Russula are famous edible fungi and are also commercially traded worldwide (Looney et al. 2018; Wang 2020). According to recent statistics on the resource diversity of Chinese macrofungi, there are 78 edible Russula species in China (Wu et al. 2019).
Guangdong Province is located in the southern coastal area of China, which is one of the Chinese provinces with tropical and subtropical climates. The climate can be divided into the middle subtropical, the southern subtropical, and the tropical climate zones, from north to south. The annual average temperature in Guangdong Province is 19–24 °C and the annual average rainfall is 1500–2000 mm. Abundant moisture, moderate to high temperatures, and variegated physiography support luxuriant and highly diversified plant growth. Broad-leaved evergreen forests, intermixed with coniferous and deciduous trees, cover much of the land. During the rainy season, the forest ecosystem can facilitate the fruiting of most ectomycorrhizal fungi, among which the members of Russula are very common. Recently, 16 new species and one epitype of Russula from Guangdong Province have been reported (Das et al. 2017; Zhang et al. 2017; Song et al. 2018a, b; Li et al. 2019; Yuan et al. 2019; Zhou et al. 2020). Obviously, Guangdong Province has become a hotspot in research on biodiversity of Chinese Russula, which makes it more vital for us to continue to explore it.
Northern hemisphere species within subg. Heterophyllidia are mainly characterized by the mostly medium to large basidiomata, equal lamellae, mild to strongly acrid taste, white or cream and rarely ochre spore print, basidiospores with inamyloid or partly amyloid suprahilar spot, mostly abundant gloeocystidia that are typically mucronate to obtuse-rounded, and absence of primordial hyphae. During a survey of the habitat diversity and geographic distribution of Russula in Guangdong Province, some interesting specimens of subg. Heterophyllidia were found that were different from known species. In this study, two new species from Guangdong Province are presented based on the morphological characters and molecular data.
Materials and methods
Morphological study
Fresh specimens were collected and photographed in Luofu Mountain Provincial Nature Reserve, Guangdong Province, South China. Collections were dried at 45–55 °C and deposited in the herbarium of the Research Institute of Tropical Forestry, Chinese Academy of Forestry (RITF). The macromorphological characters were described based on detailed notes and photographs. The color codes mostly refer to Kornerup and Wanscher (1981). The description templates and terminology of the micromorphological characters were taken from Adamčík et al. (2019). Estimates of spore ornamentation density from scanning electron microscopy pictures follow Adamčík and Marhold (2000). The hymenial cystidia density estimates refer to Buyck (1991). Experiments were performed on dried specimens using a ZEISS Imager M2 (Carl Zeiss AG; Germany). The basidiospores were observed and measured in Melzer’s reagent from a lateral view excluding ornamentation. After pretreatment of dried specimens in 5% potassium hydroxide (KOH), other micromorphological characters were identified and measured in Congo red. The coloring of the cystidia contents was observed in a sulfovanillin (SV) solution (Caboň et al. 2017). The pileipellis were examined in cresyl blue to verify the presence of ortho- or metachromatic reactions (Buyck 1989). The structure and ornamentation of the basidiospores were illustrated using a scanning electron microscopy (SEM-JEOL JSM-6510). Basidiospore measurements are presented as (Min–)AV-SD–AV–AV+SD(–Max), where Min is the minimum value, Max is the maximum value, AV is the average value, SD is the standard deviation, and Q represents the length/width ratio of the basidiospores.
Molecular study
The total genomic DNA was extracted from dry specimens following an improved CTAB protocol (Zhou and Liang 2011). We amplified and sequenced the following four loci with standard primer sets: 600 base pairs of the ITS region of rDNA using the primers ITS1 and ITS4 (White et al. 1990); 900 base pairs of the nuclear ribosome large subunit (nrLSU) using the primers LROR and LR5 (Vilgalys and Hester 1990); 600 base pairs of the ribosomal mitochondrial small subunit (mtSSU) with primers MS1 and MS2 (White et al. 1990); 900 base pairs of the translation elongation factor 1-alpha (TEF1) using primers EF1-F and EF1-R (Morehouse et al. 2003). Successful PCR products were subjected to automated DNA sequencing on an ABI 3730 DNA analyser using an ABI BigDye 3.1 terminator cycle sequencing kit (Shanghai Sangon Biological Engineering Technology and Services CO., Ltd, Shanghai, China). The newly generated sequences were submitted to GenBank database (Table 1).
Table 1.
GenBank accession numbers for sequences used in phylogenetic tree. The newly generated sequences are in bold.
| Taxon | Voucher | Location | ITS | nrLSU | mtSSU | TEF1 | Reference |
|---|---|---|---|---|---|---|---|
| R. aeruginea | AT2003017 | Sweden | DQ421999 | DQ421999 | ‒ | ‒ | Buyck et al. 2008 |
| R. albidogrisea | K15091234 | China | KY767807 | ‒ | ‒ | MN617847 | Das et al. 2017 |
| R. albidogrisea | RITF1871 | China | MW397095 | MW397128 | MW403841 | ‒ | Unpublished |
| R. amoena | SAV F–3147 | Slovakia | MT017544 | ‒ | MT417190 | MT417211 | Wisitrassameewong et al. 2020 |
| R. aureoviridis | H16082612 | China | KY767809 | ‒ | ‒ | MN617846 | Das et al. 2017 |
| R. aureoviridis | RITF4709 | China | MW646980 | MW646992 | MW647003 | MW650849 | This work |
| R. bella | SFC20170819-05 | South Korea | MT017552 | ‒ | MT196931 | MT199655 | Wisitrassameewong et al. 2020 |
| R. bubalina | K15052614 | China | MG018742 | ‒ | ‒ | ‒ | Li et al. 2019 |
| R. bubalina | RITF1863 | China | MW397097 | ‒ | MW403843 | ‒ | Unpublished |
| R. crustosa | BPL265 | USA | KT933966 | KT933826 | ‒ | ‒ | Looney et al. 2016 |
| R. cyanoxantha | FH 12-201 | Germany | KR364093 | KR364225 | ‒ | ‒ | De Crop et al. 2017 |
| R. cyanoxantha | RITF4682 | China | MW646981 | MW646993 | MW647004 | ‒ | This work |
| R. dinghuensis | GDGM45244 | China | KU863579 | ‒ | ‒ | MN617848 | Zhang et al. 2017 |
| R. dinghuensis | RITF5142 | China | MW646982 | MW646994 | MW647005 | ‒ | This work |
| R. grisea | UE2005.08.16-01 | Sweden | DQ422030 | DQ422030 | ‒ | ‒ | Buyck et al. 2008 |
| R. grisea | FH12234 | Germany | KT934006 | KT933867 | ‒ | ‒ | Looney et al. 2016 |
| R. heterophylla | UE20.08.2004-2 | Sweden | DQ422006 | DQ422006 | ‒ | ‒ | Buyck et al. 2008 |
| R. ilicis | 563IC52 | Europe | AY061682 | ‒ | ‒ | ‒ | Miller and Buyck 2002 |
| R. lakhanpalii | AG 17-1584 | India | MN262088 | ‒ | ‒ | ‒ | Ghosh et al. 2020 |
| R. lakhanpalii | RITF2600 | China | MW646983 | MW646992 | MW647006 | MW650850 | This work |
| R. lotus | RITF499 | China | MK860699 | MW397129 | MK860706 | ‒ | Song et al. 2019 |
| R. luofuensis | RITF4706 | China | MW646973 | MW646985 | MW646996 | MW650842 | This work |
| R. luofuensis | RITF4707 | China | MW646974 | MW646986 | MW646997 | MW650843 | This work |
| R. luofuensis | RITF4708 | China | MW646975 | MW646987 | MW646998 | MW650844 | This work |
| R. luofuensis | RITF4712 | China | MW646976 | MW646988 | MW646999 | MW650845 | This work |
| R. luofuensis | RITF4714 | China | MW646977 | MW646989 | MW647000 | MW650846 | This work |
| R.maguanensis | XHW4765 | China | MH724918 | MH714537 | ‒ | MH939983 | Wang et al. 2019 |
| R. mustelina | FH12226 | Germany | KT934005 | KT933866 | ‒ | ‒ | Looney et al. 2016 |
| R. orientipurpurea | SFC20170819-08 | South Korea | MT017550 | ‒ | MT196926 | MT199651 | Wisitrassameewong et al. 2020 |
| R. orientipurpurea | SFC20170725-37 | South Korea | MT017548 | ‒ | MT196927 | MT199652 | Wisitrassameewong et al. 2020 |
| R. pallidula | RITF2613 | China | MH027958 | MH027960 | MW403845 | MW650852 | Chen et al. 2019, This work |
| R. pallidula | RITF3331 | China | MH027959 | MH027961 | MW403846 | MW650853 | Chen et al. 2020, This work |
| R. parvovirescens | SDRM 6280 | USA | MK532789 | ‒ | ‒ | ‒ | Unpublished |
| R. phloginea | CNX530524068 | China | MK860701 | MK860704 | MK860708 | MK894877 | Song et al. 2019 |
| R. phloginea | CNX530524304 | China | MK860700 | MK860703 | MK860707 | MK894876 | Song et al. 2019 |
| R. prasina | HMAS 281232 | China | MH454351 | ‒ | ‒ | ‒ | Hyde et al. 2019 |
| R. pseudobubalina | GDGM70632 | China | MF433036 | ‒ | ‒ | ‒ | Li et al. 2019 |
| R. subbubalina | RITF4710 | China | MW646978 | MW646990 | MW647001 | MW650847 | This work |
| R. subbubalina | RITF4715 | China | MW646979 | MW646991 | MW647002 | MW650848 | This work |
| R. subpallidirosea | RITF4083 | China | MK860697 | MK860702 | MK860705 | MK894875 | Song et al. 2019 |
| R.substriata | XHW4766 | China | MH724921 | MH714540 | ‒ | MH939986 | Wang et al. 2019 |
| R. vesca | RITF5038 | China | MW646984 | ‒ | MW647007 | MW650851 | This work |
| R. vesca | BPL284 | USA | KT933978 | KT933839 | ‒ | ‒ | Looney et al. 2016 |
| R. virescens | HJB9989 | Belgium | DQ422014 | DQ422014 | ‒ | ‒ | Buyck et al. 2008 |
| R. viridicinnamomea | K15091418 | China | MK049972 | ‒ | ‒ | MN617850 | Yuan et al. 2019 |
| R. viridicinnamomea | RITF3324 | China | MW397098 | MW397130 | MW403847 | ‒ | Unpublished |
| R. viridirubrolimbata | HBAU 15011 | China | MT337526 | ‒ | ‒ | ‒ | Deng et al. 2020 |
| R. werneri | IB1997/0786 | Europe | DQ422021 | DQ422021 | ‒ | ‒ | Unpublished |
| R. xanthovirens | GDGM 71145 | China | MG786056 | ‒ | ‒ | ‒ | Song et al. 2018b |
Phylogenetic analysis
Species in the subg. Heterophyllidia with high similarity to our new species and partially representative species that are closely related to subsect. Heterophyllinae (Fr.) Jul. Schäff. and subsect. Virescentinae Singer were selected for phylogenetic analyses. Russulamaguanensis J. Wang, X.H. Wang, Buyck & T. Bau and R.substriata J. Wang, X.H. Wang, Buyck & T. Bau were used as outgroup. NCBI accession numbers and references of sequences used in the phylogenetic tree are listed in Table 1. Initial sequence alignment was performed using the online version MAFFT 7.0 (http://mafft.cbrc.jp/alignment/server/) with manual evaluations and adjustments in BioEdit when necessary to obtain reliable and reasonable results (Hall 1999). The final aligned result was submitted to TreeBASE (S27792). Maximum likelihood (ML) and Bayesian analysis (BA) were implemented for the phylogenetic analyses. The maximum likelihood was carried out by using RAxML-HPC2 on XSEDE (8.2.12) through the CIPRES Science Gateway (www.phylo.org). The ML analysis was executed by applying the rapid bootstrap algorithm with 1000 replicates to affirm the consistency of the results under the GAMMA model. Bootstrap support (BS) ≥70% on the final tree was regarded as significant. The BA was performed on XSEDE (MrBayes 3.2.7a) through the CIPRES Science Gateway (www.phylo.org) under the GTR model. Four independent Markov chains were run for a total of 50000000 generations, trees were sampled every 100 generations, and the first 25% of the trees were discarded as the burn-in phase of each analysis. The Bayesian posterior probability (PP) values were obtained from the 50% majority-rule consensus trees, and nodes with PP ≥0.95 were considered to be significantly supported.
Results
Phylogeny
Both the ML analysis and BA of combined ITS-nrLSU-mtSSU-TEF1 sequences dataset resulted in similar tree topologies, and only the ML tree is shown in Fig. 1. The posterior probabilities for the BA are also shown along the branches. The phylogenetic analyses confirmed that both subsect. Virescentinae and subsect. Heterophyllinae were a monophyletic group; each strongly supported by BS (100%) and PP (1). Additionally, the monophyly of the remaining 4 subsections of subg. Heterophyllidia was also significantly supported.
Figure 1.
Phylogenetic tree of based on the ITS-nrLS-mtSSU-TEF1 dataset. Species in the subg. Heterophyllidia with high similarity to our new species and partially representative species that are closely related to subsect. Heterophyllinae and subsect. Virescentinae were selected. Russulamaguanensis and R.substriata were used as outgroup. Bootstrap support (BS) ≥70% are shown. Bayesian Posterior Probabilities (PP) ≥0.95 are given. Infrageneric classification follows Buyck et al. (2018).
The samples of the two new species, R.luofuensis and R.subbubalina, formed each a strongly supported clade (BS 100%, PP 1.00) and were clearly distinct from known and sequenced species of the subg. Heterophyllidia. R.luofuensis clustered together with Chinese species R.albidogrisea J. W. Li & L. H. Qiu, which is sister to a clade comprising R.viridirubrolimbata J. Z. Ying, R.parvovirescens Buyck, D. Mitch. & Parrent and R.virescens (Schaeff.) Fr. with 99% bootstrap support and 1.00 posterior probabilities. Our second species, R.subbubalina clustered with Chinese species R.viridicinnamomea F. Yuan & Y. Song and formed a sister clade to two Chinese species (R.bubalina J.W. Li & L.H. Qiu and R.pseudobubalina J.W. Li & L.H. Qiu) with 99% bootstrap support and 1.00 posterior probabilities.
Taxonomy
Russula luofuensis
B. Chen & J. F. Liang sp. nov.
27D67B5D-5FAB-541A-ACD9-140BCA3DFB80
MB838836
Figure 2.
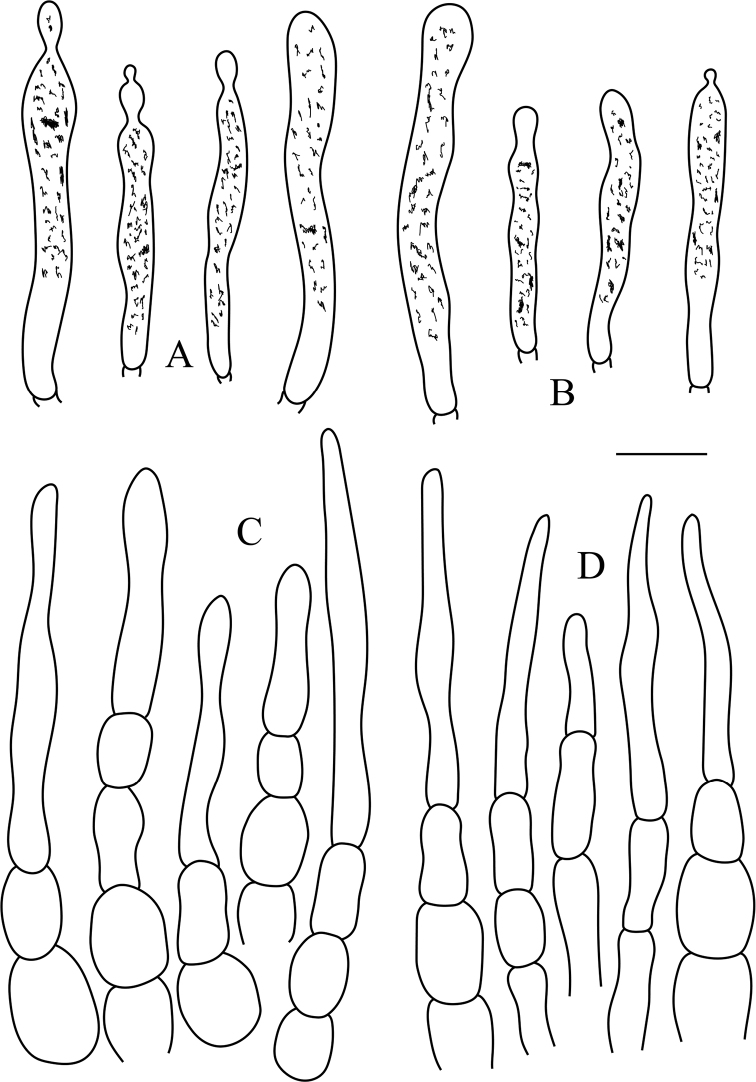
Fruiting bodies (A, B) and basidiospores (C, D) of Russulaluofuensis (RITF4708). Fruiting bodies (E, F) and basidiospores (G, H) of R.subbubalina (RITF 3715). Scale bars: 20 mm (A, B, E, F).
Figure 3.
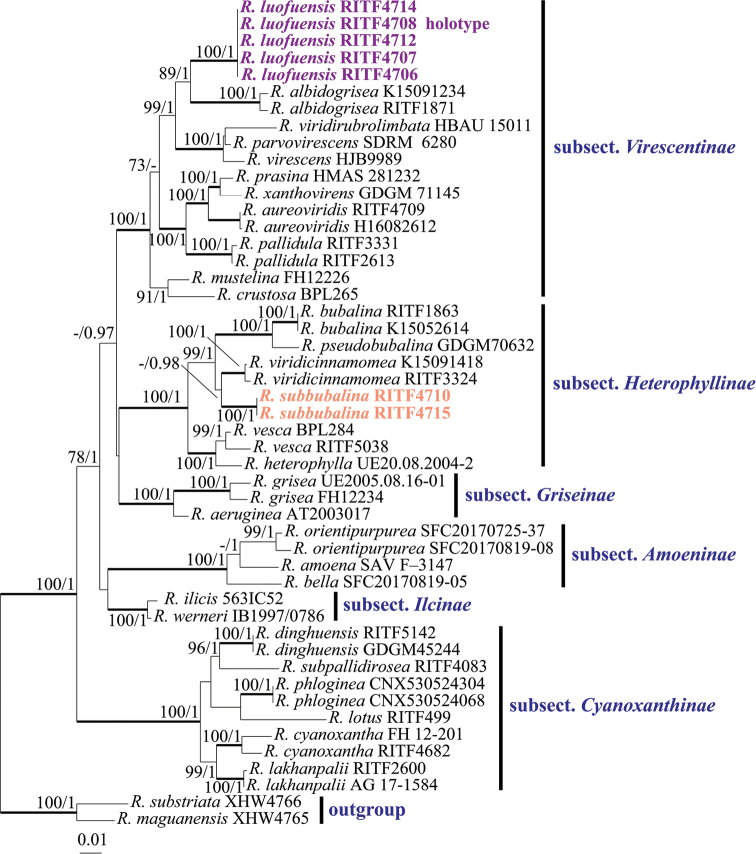
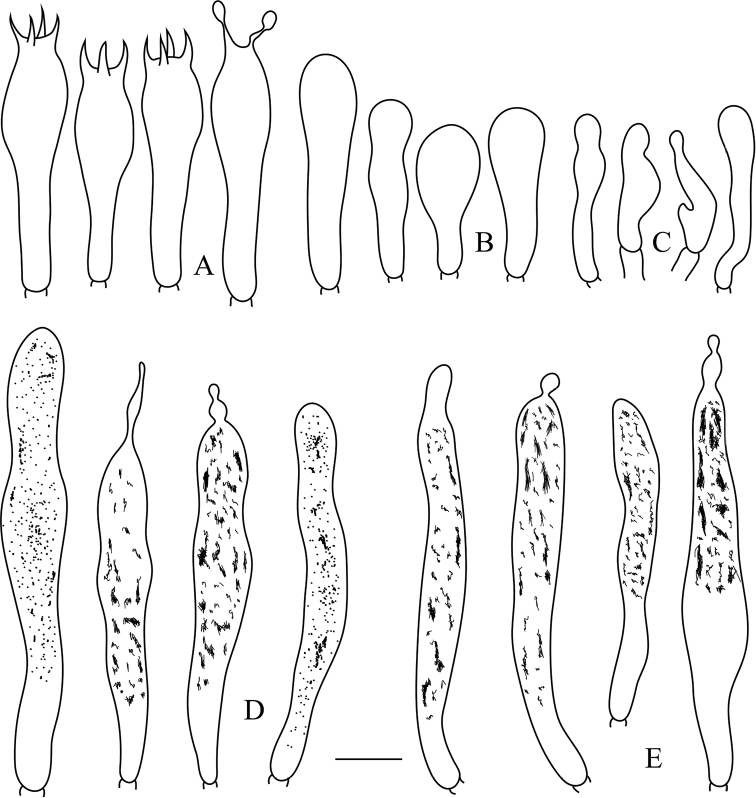
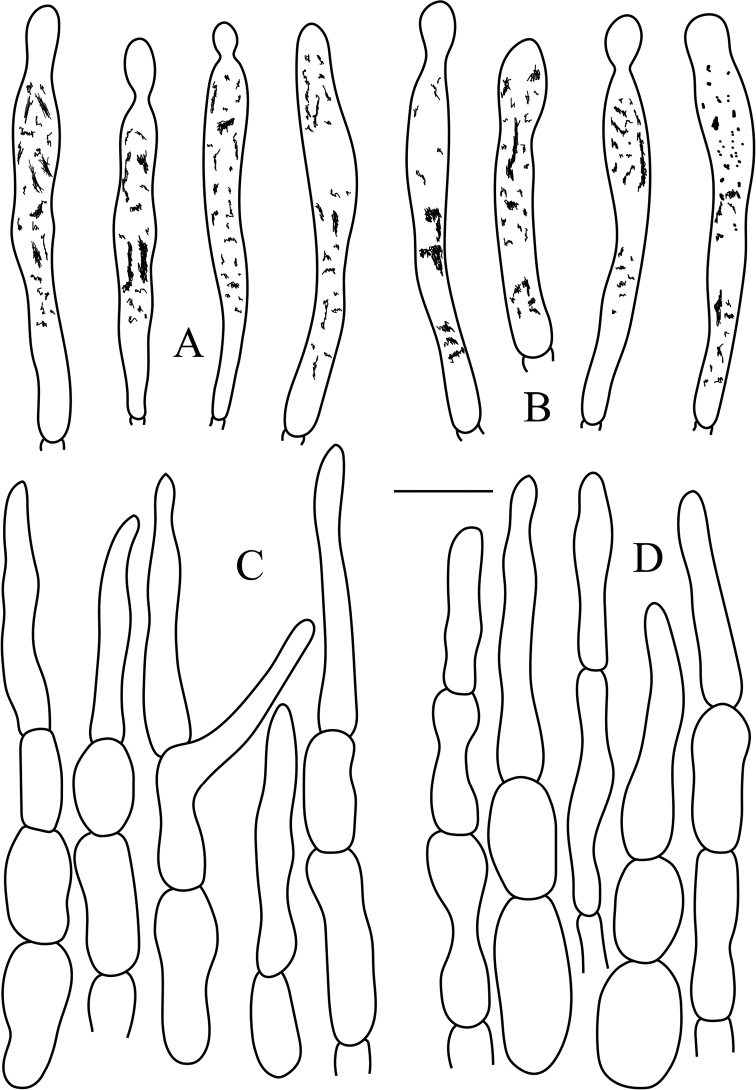
Russulaluofuensis (RITF 4708) A basidia B basidiola C marginal cells D hymenial gloeocystidia on lamellae sides E hymenial gloeocystidia on lamellae edges. Scale bar: 10 μm.
Figure 4.
Russulaluofuensis (RITF 4708) A pileocystidia near the pileus margin B pileocystidia near the pileus center C hyphal terminations near the pileus margin D hyphal terminations near the pileus center. Scale bar: 10 μm.
Diagnosis.
Basidiomata medium-sized to large; grayish yellow to brownish orange pileus center, purplish gray to grayish magenta towards the margin, surface cracking and broken into small golden-brown patches, peeling to 1/2 of the radius; subglobose to broadly ellipsoid basidiospores with warts fused in short or long chains; hymenial gloeocystidia mainly clavate; suprapellis composed of hyphal extremities with inflated, ellipsoid or globose cells and attenuated terminal cell; pileocystidia always one-celled, apically typically obtuse.
Holotype.
China. Guangdong Province, Huizhou City, Boluo County, Luofu Mountain Provincial Nature Reserve, 23°15'47.13"N, 114°3'45.42"E, 90 m asl., in mixed Fagaceae forests of Cyclobalanopsis and Castanopsis, 22 August 2020, leg. CB446 (RITF4708).
Etymology.
The species name refers to the type locality, Luofu Mountain Provincial Nature Reserve.
Description.
Basidiomata medium-sized to large; pileus 35–80 mm in diameter; initially hemispheric when young, applanate to convex, convex with a depressed center after mature; margin incurved, not cracked, striation short and inconspicuous; surface dry, glabrous, peeling to 1/2 of the radius, cracking and broken into small golden-brown patches, patches crowded towards the center, with smaller patches towards the margin; grayish yellow (4B5) to brownish orange (5C5) in the center, purplish gray (13B2) to grayish magenta (13B3) towards the margin. Lamellae adnate to subfree, 2–4 mm deep, 8–10 at 1 cm near the pileus margin, white (1A1) to cream; lamellulae absent; furcations occasional near the stipe; edge entire and concolor. Stipe 30–50 × 10–25 mm, cylindrical, slightly inflated towards the base, white (1A1), with yellowish (2A2) tinge at the base, and medulla initially stuffed becoming hollow. Context 2–3 mm thick in half of the pileus radius, white (1A1), unchanging when bruised, taste mild, odor inconspicuous. Spore print pale yellowish (2A2).
Basidiospores (5.0–)5.8–6.6–7.5(–8.6) × (4.5–)5.4–6.2–7.0(–8.0) μm, Q = (1.0–)1.02–1.08–1.14(–1.26), subglobose to broadly ellipsoid; ornamentation of medium-sized, moderately distant to dense [6–8(–9) in a 3 μm diameter circle] amyloid warts or spines, 0.3–0.6 μm high, locally reticulate, frequently fused in short or long chains [2–3(–4) in the circle], occasionally to frequently connected by line connections [1–2(–3) in the circle]; suprahilar spot medium-sized, amyloid. Basidia (35.0–)36.7–39.8–42.8(–45.5) × (9.0–)9.5–10.0–10.5(–11.2) μm, mostly 4-spored, some 2- and 3-spored, clavate; basidiola clavate or subcylindrical, ca. 6.5–11.5 μm wide. Hymenial gloeocystidia on lamellae sides dispersed to moderately numerous, ca. 600–900/mm2, (59.0)63.2–71.3–79.3(83.6) × (7.0)7.7–8.8–9.9(10.5) μm, clavate or narrowly clavate, apically mainly obtuse, occasionally acute, often with 3–10 μm long appendage, thin-walled; contents heteromorphous or granulose, mainly in the middle and upper part, turning reddish black in SV. Hymenial gloeocystidia on lamellae edges often smaller, (49.5–)56.2–64.3–72.4(–80.2) × (6.2–)7.3–8.3–9.4(–10.0) μm, clavate, or subcylindrical, sometimes fusiform, apically mainly obtuse, occasionally mucronate, sometimes with 3–6 μm long appendage thin-walled; contents heteromorphous, turning reddish black in SV. Marginal cells (15.2–)19.8–23.5–27.2(–30.6) × (3.5–)4.0–4.8–5.6(–7.0) μm, subcylindrical or clavate, often flexuous. Pileipellis orthochromatic in cresyl blue, not sharply delimited from the underlying context, 260–300 μm deep, two-layered; suprapellis 120–150 μm deep, hyphal endings composed of inflated, ellipsoid or globose cells with attenuated terminal cells; subpellis 120–160 μm deep, composed of repent, intricate, 2–6 μm wide hyphae. Hyphal terminations near the pileus margin typically unbranched, occasionally flexuous, thin-walled; terminal cells (9.2–)18.6–28.2–37.8(–50.8) × (3.2–)3.9–5.0–6.1(–8.2) μm, mainly narrowly lageniform, occasionally clavate or cylindrical, apically attenuated or constricted, occasionally obtuse; subterminal cells frequently shorter and wider, ca. 4–9 μm wide, typically unbranched. Hyphal terminations near the pileus center similar to those near the pileus margin; terminal cells (10.2–)18.4–27.4–36.4(–44.8) × (3.2–)3.6–4.7–5.8(–6.8) μm, mainly lageniform, occasionally fusiform or subcylindrical, apically attenuated or constricted; subterminal cells often shorter and wider, rarely branched, ca. 4–7 μm wide. Pileocystidia near the pileus margin always one-celled, (23.3–)27.9–35.0–42.2(–47.5) ×3.5–4.8–6.0(–8.3) μm, mainly clavate, occasionally subcylindrical or fusiform, apically typically obtuse, occasionally acute, often with round or ellipsoid, 3–6 μm long appendage, thin-walled; contents heteromorphous or granulose, turning reddish black in SV. Pileocystidia near the pileus center similar in size, always one-celled, (24.6–)27.2–34.8–42.5(–48.2) × 3.0–4.2–5.4(–6.8) μm, thin-walled, mainly clavate, occasionally fusiform, apically often obtuse or occasionally acute, occasionally with 2–4 μm long appendage, contents heteromorphous or granulose, turning reddish black in SV. Cystidioid hyphae In subpellis and context with heteromorphous contents, oleiferous hyphae in subpellis with refringent contents.
Additional specimens examined.
China. Guangdong Province, Huizhou City, Boluo County, Luofu Mountain Provincial Nature Reserve, 23°15'44.11"N, 114°3'16.77"E, 120 m asl., in mixed Fagaceae forests of Cyclobalanopsis and Castanopsis, 22 August 2020, leg. CB444 (RITF4706); ibid., 22 August 2020, leg. CB445 (RITF4707); ibid., 22 August 2020, leg. CB450 (RITF4712); ibid., 22 August 2020, leg. CB452 (RITF4714).
Notes.
The combination of morphological features and phylogenetic analysis place R.luofuensis in subsect. Virescentinae. Phylogenetically, our new species R.luofuensis is clustered with R.albidogrisea with 89% bootstrap support and 1.00 posterior probabilities, which is also from Guangdong Province of China. However, R.albidogrisea differs from R.luofuensis in having a white to grayish pileus with acute, even to slightly undulate margin, often smaller basidiospores [(5.1–)5.3–5.6–6.0(–6.4) × (4.6–)4.8–5.1–5.3(–5.6) μm], longer hymenial gloeocystidia on lamellae sides (35–50 × 5–11 μm) and hymenial gloeocystidia on lamellae edges (37–46 × 9–12 μm, Das et al. 2017).
Given cracking surface, R.viridirubrolimbata, R.parvovirescens, R.virescens and R.crustosa Peck of subsect. Virescentinae resemble R.luofuensis. However, R.viridirubrolimbata, originally described from China, can be distinguished by a light yellowish olive to yellowish olive pileus center with a pinkish red to light jasper red margin and absence of hymenial gloeocystidia on lamellae edges (Ying 1983; Deng et al. 2020). The American species R.parvovirescens possesses a greenish brown to metallic bluish green pileus with green patches (Buyck et al. 2006). Russulavirescens (originally reported from Europe) is distinct in its green to yellowish green pileus (Sarnari 1998). Russulaalbidogrisea, originally reported from North America, has a brownish-yellow, greenish or subolivaceous pileus with small spot-like areolae or pseudo-verrucae, shorter basidia [(29–)30–32–33.5(–35) × (7.5–)8–9.5–10.5(–11) μm] and absence of hymenial gloeocystidia on the lamellar edges (Adamčík et al. 2018).
Russula subbubalina
B. Chen & J. F. Liang sp. nov.
91CC489C-BA99-53AF-91C1-B4F7B6F73FF9
MB838837
Figure 5.
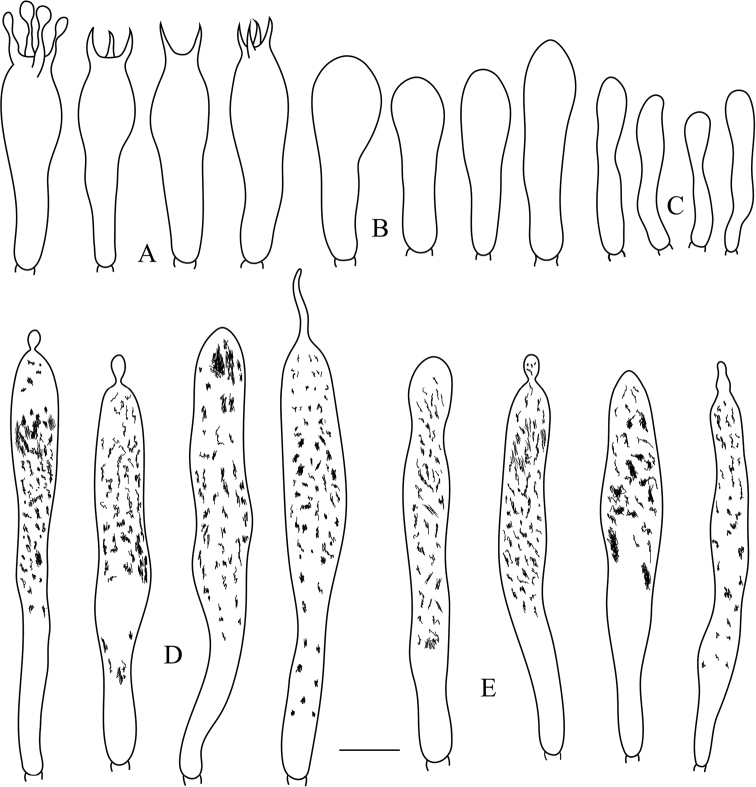
Russulasubbubalina (RITF 4710) A basidia B basidiola C marginal cells D hymenial gloeocystidia on lamellae sides E hymenial gloeocystidia on lamellae edges. Scale bar: 10 μm.
Figure 6.
Russulasubbubalina (RITF 4710) A pileocystidia near the pileus margin B pileocystidia near the pileus center C hyphal terminations near the pileus margin D hyphal terminations near the pileus center. Scale bar: 10 μm.
Diagnosis.
Basidiomata medium-sized to large; dark salmon pileus with rusty spots when young, blanched almond with a cracked margin after maturation, surface pruinose in some parts; adnate to slightly adnexed lamellae; subglobose to broadly ellipsoid basidiospores with warts fused in short or long chains and frequently connected by line connections; clavate or ellipsoid basidiola; hymenial gloeocystidia clavate or fusiform, apically mainly obtuse; suprapellis with hyphal ends composed of inflated or ellipsoid cells and attenuated terminal cell; pileocystidia mainly clavate, apically typically obtuse, sometimes with round or ellipsoid appendage.
Holotype.
China. Guangdong Province, Huizhou City, Boluo County, Luofu Mountain Provincial Nature Reserve, 23°15'43.80"N, 114°3'5.40"E, 220 m asl., in mixed Fagaceae forests of Cyclobalanopsis and Castanopsis, 22 August 2020, leg. CB448 (RITF4710).
Etymology.
Referred to its morphological resemblance to R.bubalina.
Description.
Basidiomata medium-sized to large; pileus 50–100 mm in diameter; initially hemispheric when young, applanate to convex, convex with a slightly depressed center after mature; margin incurved, cracked with age, striation short and inconspicuous; surface dry, glabrous, peeling to 1/4 of the radius, pruinose in some part; dark salmon with rusty spots when young, blanched almond after maturation, shallower at the margin. Lamellae adnate to slightly adnexed, 3–5 mm deep, 11–13 at 1 cm near the pileus margin, white (1A1) to cream; lamellulae sometimes present and irregular in length; furcations present especially near the stipe; edge entire and concolor. Stipe 30–55 × 5–15 mm, cylindrical, slightly inflated towards the base, white (1A1) to blanched almond, with rusty tinge towards the base, and medulla initially stuffed becoming hollow. Context 3–4 mm thick in half of the pileus radius, white (1A1), unchanging when bruised, taste mild, odor inconspicuous. Spore print white (1A1) to cream.
Basidiospores (5.2–)5.6–6.2–6.8(–7.2) × (4.5–)4.9–5.3–5.7(–6.2) μm, Q = (1.0–)1.08–1.17–1.25(–1.38), subglobose to broadly ellipsoid; ornamentation of relatively small, moderately distant to dense [6–8(–9) in a 3 μm diameter circle] amyloid warts or spines, 0.3–0.5 μm high, locally reticulate, fused in short or long chains [2–3(–4) in the circle], frequently connected by line connections [3–4(–5) in the circle]; suprahilar spot medium-sized, amyloid. Basidia (30.5–)31.7–34.8–37.8(–43.0) × (6.3–)7.5–8.1–8.8(–9.4) μm, mostly 4-spored, some 2- and 3-spored, clavate; basidiola clavate or ellipsoid, ca. 5.5–10 μm wide. Hymenial gloeocystidia on lamellae sides Moderately numerous, ca. 800–1000/mm2, (41.0)49.1–56.7–64.3(68.5) × (6.5)7.2–8.1–9.0(10.0) μm, clavate or fusiform, apically mainly obtuse, occasionally acute, sometimes with 4–10 μm long appendage, thin-walled; contents heteromorphous or granulose, turning reddish black in SV. Hymenial gloeocystidia on lamellae edges Often longer, (40.5–)52.6–63.0–73.5(–83.6) × (4.6–)6.7–8.1–9.6(–10.8) μm, mainly clavate, occasionally fusiform, apically typically obtuse, sometimes with 3–8 μm long appendage, thin-walled; contents heteromorphous-crystalline, turning reddish black in SV. Marginal cells (14.0–)19.0–23.4–27.7(–34.2) × (3.4–)3.7–4.5–5.3(–5.8) μm, clavate, lageniform or fusiform, often flexuous. Pileipellis Orthochromatic in cresyl blue, sharply delimited from the underlying context, 400–450 μm deep, two-layered; suprapellis180–200 μm deep, hyphal endings composed of inflated or ellipsoid cells with attenuated terminal cells; subpellis 240–260 μm deep, composed of horizontally oriented, relatively dense, intricate, 3–6 μm wide hyphae. Hyphal terminations near the pileus margin sometimes branched, occasionally flexuous, thin-walled; terminal cells (14.8)20.9–26.6–32.3(38.0) × 3.5–4.0–4.6(5.5) μm, mainly narrowly lageniform, occasionally cylindrical, apically attenuated or constricted; subterminal cells frequently shorter and wider ca. 3–8 μm wide, occasionally branched. Hyphal terminations near the pileus center similar to those near the pileus margin; terminal cells (14.3–)17.5–22.7–27.8(–33.7) × (3.4–)3.7–4.1–4.6(–5.0) μm, lageniform, clavate or cylindrical, apically attenuated or constricted, sometimes obtuse; subterminal cells often wider, rarely branched, ca. 4–8 μm wide. Pileocystidia near the pileus margin always one-celled, (27.9–)35.1–40.5–45.9(–48.9) × (3.8–)4.2–4.7–5.3(–5.7) μm, mainly clavate, occasionally fusiform, apically typically obtuse, sometimes with round or ellipsoid 2–6 μm long appendage, thin-walled; contents heteromorphous, turning reddish black in SV. Pileocystidia near the pileus center similar in shape, always one-celled, (23.7–)25.6–31.8–38.0(–46.0) × (3.3–)4.2–4.8–5.4(–6.0) μm, thin-walled, mainly clavate, occasionally fusiform or subcylindrical, apically typically obtuse, sometimes with 4–6 μm long appendage, contents granulose, turning reddish in SV. Cystidioid hyphae In subpellis and context with granulose contents, oleiferous hyphae frequent in subpellis with yellowish contents.
Additional specimens examined.
China. Guangdong Province, Huizhou City, Boluo County, Luofu Mountain Provincial Nature Reserve, 23°15'41.70"N, 114°3'5.21"E, 240 m asl., in mixed Fagaceae forests of Cyclobalanopsis and Castanopsis, 22 August 2020, leg. CB453 (RITF4710).
Notes.
Both morphology and phylogeny place R.subbubalina clearly in subsect. Heterophyllinae. In our phylogenetic tree, R.viridicinnamomea is the sister taxon to R.subbubalina but differs from it by the typically smaller basidiomata (30–50 μm), an emerald green-tinged buff pileus with undulate and tearing margin and longer hymenial gloeocystidia on the lamellae edges (36.5–63 × 4–12 μm, Yuan et al. 2019).
Morphologically, R.subbubalina may be confused in the field with two recently reported new species: R.bubalina and R.pseudobubalina also from Guangdong Province of China. However, R.bubalina has the typically smaller basidiomata (35–54 μm), a striate pileus margin and basidiospores with warty ornamentations not forming reticulum (Li et al. 2019), whereas R.pseudobubalina possesses the typically smaller basidiomata (31–46 μm), never forked lamellae, basidiospores with isolated warts, and often shorter hymenial gloeocystidia on the lamellae edges (23.4–37.8–65.5 × 6.2–8.3–10.0 μm, Li et al. 2019).
Supplementary Material
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31770657, 31570544 and 31900016), the Science and Technology Planning Project of Guangdong Province (No. 2019B121202005), and the Science and Technology Planning Project of Guangdong Forestry (No. 2019-03).
Citation
Chen B, Song J, Chen Y, Zhang J, Liang J (2021) Morphological and phylogenetic evidence for two new species of Russula subg. Heterophyllidia from Guangdong Province of China. MycoKeys 82: 139–157. https://doi.org/10.3897/mycokeys.82.64913
References
- Adamčík S, Jančovičová S, Buyck B. (2018) The Russulas described by Charles Horton Peck. Cryptogamie Mycologie 39(1): 3–108. 10.7872/crym/v39.iss1.2018.3 [DOI] [Google Scholar]
- Adamčík S, Looney B, Caboň M, Jančovičová B, Adamčíková K, Avis PG, Barajas M, Bhatt RP, Corrales A, Das K, Hampe F, Ghosh A, Gates G, Kälviäinen V, Khalid AN, Kiran M, Lange RD, Lee H, Lim YW, Kong A, Manz C, Ovrebo C, Saba M, Taipale T, Verbeken A, Wisitrassameewong K, Buyck B. (2019) The quest for a globally comprehensible Russula language. Fungal Diversity 99(1): 369–449. 10.1007/s13225-019-00437-2 [DOI] [Google Scholar]
- Adamčík S, Marhold K. (2000) Taxonomy of the Russulaxerampelina group. I. Morphometric study of the Russulaxerampelina group in Slovakia. Mycotaxon 76: 463–480. [Google Scholar]
- Buyck B. (1989) Valeur taxonomique du bleu de crésyl pour le genre Russula. Bulletin de la Société Mycologique de France 105: 1–6. [Google Scholar]
- Buyck B. (1991) The study of microscopic features in Russula 2. Sterile cells of the hymenium. Russulales News 1: 62–85. [Google Scholar]
- Buyck B, Hofstetter V, Eberhardt U, Verbeken A, Kauff F. (2008) Walking the thin line between Russula and Lactarius: the dilemma of Russulasubsect.Ochricompactae. Fungal Diversity 28: 15–40. [Google Scholar]
- Buyck B, Mitchell D, Parrent J. (2006) Russulaparvovirescens sp. nov., a common but ignored species in the eastern United States. Mycologia 98: 612–615. 10.1080/15572536.2006.11832664 [DOI] [PubMed] [Google Scholar]
- Buyck B, Thoen D, Watling R. (1996) Ectomycorrhizal fungi of the Guinea-Congo region. Proceedings of the Royal Society of Edinburgh 104B: 313–333. 10.1017/S0269727000006175 [DOI]
- Buyck B, Wang XH, Adamčíková K, Caboň M, Jančovičová S, Hofstetter V, Adamčík S. (2020) One step closer to unravelling the origin of Russula: subgenus Glutinosae subg. nov. Mycosphere 11: 285–304. 10.5943/mycosphere/11/1/6 [DOI] [Google Scholar]
- Buyck B, Zoller S, Hofstetter V. (2018) Walking the thin line… ten years later: the dilemma of above- versus below-ground features to support phylogenies in the Russulaceae (Basidiomycota). Fungal Diversity 89: 267–292. 10.1007/s13225-018-0397-5 [DOI] [Google Scholar]
- Caboň M, Eberhardt U, Looney B, Hampe F, Kolařík M, Jančovičová S, Verbeken A, Adamčík S. (2017) New insights in Russula subsect. Rubrinae: phylogeny and the quest for synapomorphic characters. Mycological Progress 16: 877–892. 10.1007/s11557-017-1322-0 [DOI] [Google Scholar]
- Chen B, Jiang XM, Song J, Liang JF, Wang SK, Lu JK. (2019) Morphological and phylogenetic analyses reveal the new species Russulapallidula from China. Sydowia 71: 1–10. 10.12905/0380.sydowia71-2019-0001 [DOI] [Google Scholar]
- Das K, Ghosh A, Chakraborty D, Li JW, Qiu LH, Baghela A, Halama M, Hembrom ME, Mehmood T, Parihar A, Pencakowski B, Bielecka M, Reczyńska K, Sasiela D, Singh U, Song Y, Świerkosz K, Szczęśniak K, Uniyal P, Zhang JB, Buyck B. (2017) Fungal Biodiversity Profiles 31–40. Cryptogamie Mycologie 38(3): 353–406. 10.7872/crym/v38.iss3.2017.353 [DOI] [Google Scholar]
- De Crop E, Nuytinck J, Van de Putte K, Wisitrassameewong K, Hackel J, Stubbe D, Hyde KD, Roy M, Halling RE, Moreau PA, Eberhardt U. (2017) A multi-gene phylogeny of Lactifluus (Basidiomycota, Russulales) translated into a new infrageneric classification of the genus. Persoonia 38: 5–80. 10.3767/003158517X693255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CY, Shi LY, Wang J, Xiang Z, Li SM, Li GJ, Yang H. (2020) Species diversity of Russulavirescens complex “qingtoujun” in southern China. Mycosystema 39: 1–23. [Google Scholar]
- Ghosh A, Das K, Bhatt RP, Hembrom ME. (2020) Two new species of Genus Russula from Western Himalaya with morphological details and phylogenetic estimations. Nova Hedwigia 111: 115–130. 10.1127/nova_hedwigia/2020/0588 [DOI] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, Soares Adriene MDS, Plautz Jr HL, Helio LP, Helen MPS, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai Y-C, Ji X-H, Tian X-M, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, do Nascimento BR, dos Santos JEF, de Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang JM, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC, Doilom M. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Knudsen H, Borgen T. (1982) Russulaceae in Greenland. In: Arctic and Alpine Mycology 1. University of Washington Press, Seattle and London, 1–559.
- Kornerup A, Wanscher JH. (1981) Taschenlexikon der Farben, 3rd edn. Muster-Schmidt Verlag, Göttingen.
- Li JW, Zheng JF, Song Y, Yuan F, Qiu LH. (2019) Three novel species of Russula from southern China based on morphological and molecular evidence. Phytotaxa 392(4): 264–276. 10.11646/phytotaxa.392.4.2 [DOI] [Google Scholar]
- Looney BP, Meidl P, Piatek MJ, Miettinen O, Martin FM, Matheny PB, Labbé JL. (2018) Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytologist 218: 54–65. 10.1111/nph.15001 [DOI] [PubMed] [Google Scholar]
- Looney BP, Ryberg M, Hampe F, Sánchez-García M, Matheny PB. (2016) Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Molecular Ecology 25: 630–647. 10.1111/mec.13506 [DOI] [PubMed] [Google Scholar]
- Miller SL, Buyck B. (2002) Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications. Mycological Research 106: 259–276. 10.1017/S0953756202005610 [DOI] [Google Scholar]
- Morehouse EA, James TY, Ganley ARD, Vilgalys R, Berger L, Murphy PJ, Longcore JE. (2003) Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Molecular Ecology 12: 395–403. 10.1046/j.1365-294X.2003.01732.x [DOI] [PubMed] [Google Scholar]
- Sarnari M. (1998) Monografia illustrate de genere Russula in Europa. Tomo Secondo. AMB, Centro Studi Micologici, Trento.
- Song J, Liang JF, Mehrabi-Koushki M, Krisai-Greilhuber I, Ali B, Bhat VK, Cerna-Mendoza A, Chen B, Chen ZX, Chu HL, Corazon-Guivin MA, Alves de Silva G, De Kesel A, Dima B, Dovana F, Farokhinejad R, Ferisin G, Guerrero-Abad JC, Guo T, Han LH, Ilyas S, Justo A, Khalid AN, Khodadadi-Pourarpanahi S, Li TH, Liu C, Lorenzini M, Lu JK, Mumtaz AS, Oehl F, Pan XY, Papp V, Qian W, Razaq A, Semwal KC, Tang LZ, Tian XL, Vallejos-Tapullima A, van der Merwe NA, Wang SK, Wang CQ, Yang RH, Yu F, Zapparoli G, Zhang M, Antonín V, Aptroot A, Aslan A, Banerjee A, Chatterjee S, Dirks AC, Ebrahimi L, Fotoujifar KB, Ghosta Y, Kalinina LB, Karahan D, Liu J, Maiti MK, Mookherjee A, Nath PS, Panja B, Saha J, Sevciková H, Voglmayr H, Yazici K, Haelewaters D. (2019) Fungal Systematics and Evolution: FUSE 5. Sydowia 71: 141–245. 10.12905/0380.sydowia71-2019-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Buyck B, Li JW, Yuan F, Zhang JB, Qiu LH. (2018a) Two novel and a forgotten Russula species in sect. Ingratae (Russulales) from Dinghushan Biosphere Reserve in southern China. Cryptogamie Mycologie 39(3): 341–357. 10.7872/crym/v39.iss3.2018.341 [DOI] [Google Scholar]
- Song Y, Li JW, Buyck B, Zheng JF, Qiu LH. (2018b) Russulaverrucospora sp. nov. and R.xanthovirens sp. nov., two novel species of Russula (Russulaceae) from southern China. Cryptogamie Mycologie 39(1): 129–142. 10.7872/crym/v39.iss1.2018.129 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Buyck B, Wang XH, Bau T. (2019) Visiting Russula (Russulaceae, Russulales) with samples from southwestern China finds one new subsection of R.subg.Heterophyllidia with two new species. Mycological Progress 18: 771–784. 10.1007/s11557-019-01487-1 [DOI] [Google Scholar]
- Wang XH. (2020) Taxonomic comments on edible species of Russulaceae. 554 Mycosystema 39(9): 1617–1639.
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk P, Kolaříková Z, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu JC, Wang Y, Tian FH, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, de A Santiago ALCM, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan J, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Kuhnert E, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Liu P, Cavender JC, Kang YQ, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Lateef AA, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Erdoğdu M. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Wisitrassameewong K, Park MS, Lee H, Ghosh A, Das K, Buyck B, Looney BP, Caboň M, Adamčík S, Kim C, Kim CS, Lim YW. (2020) Taxonomic revision of RussulasubsectionAmoeninae from South Korea. MycoKeys 75: 1–29. 10.3897/mycokeys.75.53673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhou L W, Yang ZL, Bau T, Li TH, Dai YC. (2019) Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species. Fungal Diversity 98: 1–76. 10.1007/s13225-019-00432-7 [DOI] [Google Scholar]
- Ying JZ. (1983) A study on Russulaviridi-rubrolimata sp. nov. and its related species of subsection Virescentinas. Acta Mycologica Sinica 2(1): 34–37. [Google Scholar]
- Yuan F, Song Y, Buyck B, Li JW, Qiu LH. (2019) Russulaviridicinnamomea F. Yuan & Y. Song, sp. nov. and R.pseudocatillus F. Yuan & Y. Song, sp. nov., two new species from southern China. Cryptogamie Mycologie 40(1): 45–56. 10.5252/cryptogamie-mycologie2019v40a4 [DOI] [Google Scholar]
- Zhang JB, Li JW, Li F, Qiu LH. (2017) Russuladinghuensis sp. nov. and R.subpallidirosea sp.nov., two new species from southern china supported by morphological and molecular evidence. Cryptogamie Mycologie 38(2): 191–203. 10.7872/crym/v38.iss2.2017.191 [DOI] [Google Scholar]
- Zhou LL, Liang JF. (2011) An improved protocol for extraction of DNA from macrofungi. Guangdong Forest Science Technology 27: 13–16. [Google Scholar]
- Zhou SY, Song Y, Chen KX, Li JW, Buyck B, Qiu LH. (2020) Three novel species of RussulaPers.subg.Compactae (Fr.) Bon from Dinghushan Biosphere Reserve in southern China. Cryptogamie Mycologie 41(14): 219–234. 10.5252/cryptogamie-mycologie2020v41a14 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.