Abstract
The coronavirus pandemic, which appeared in Wuhan, China, in December 2019, rapidly spread all over the world in only a few weeks. Faster testing techniques requiring less resources are key in managing the pandemic, either to enable larger scale testing or even just provide developing countries with limited resources, particularly in Africa, means to perform tests to manage the crisis. Here, we report an unprecedented, rapid, reagent-free and easy-to-use screening spectroscopic method for the detection of SARS-CoV-2 on RNA extracts. This method, validated on clinical samples collected from 280 patients with quantitative predictive scores on both positive and negative samples, is based on a multivariate analysis of FTIR spectra of RNA extracts. This technique, in agreement with RT-PCR, achieves 97.8% accuracy, 97% sensitivity and 98.3% specificity while reducing the testing time post RNA extraction from hours to minutes. Furthermore, this technique can be used in several laboratories with limited resources.
Subject terms: Biological techniques, Biophysics, Biotechnology, Medical research
Introduction
According to the World Health Organization (WHO), a pandemic is the worldwide spread of a new disease, characterized by a rapid propagation and high mortality rate. Transmitted by viruses, bacteria and other pathogens, it kills millions of people. Several pandemics are well-known in human history, from various plagues in the Middle Ages to the Spanish influenza pandemic in the last century, with 50 million deaths1 ascribed to H1N1 type virus2. It is worth noting that RNA viruses are particularly dangerous since they have high mutation rates, enhanced virulence and evolvability. They have been involved in most severe epidemics such as HIV3, HCV4, Ebola5, H1N6,7, etc. Most of these diseases resulted from an animal-to-human transmission, and a lack of accessible and rapid diagnostic tests generally hampered adequate health response and efficient management of the disease.
Presently, the world is experiencing an unprecedented health crisis, SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) referred to as a Covid-19 disease8–10. The first cases were reported in December 2019 in Wuhan, China; then it rapidly spread worldwide in only a few days. The fast spread of Covid-19 is mainly attributed to the mode of transmission of the virus and the high volume of business and tourism airline traffic. Moreover, emerging mutations known as Covid-19 variants (United Kingdom, Brazil and South Africa) increased transmissibility of the virus and improve its ability to escape the host immune system. The number of infected people is still increasing, with more than 103 million confirmed cases and more than 2.26 million confirmed deaths worldwide11. Beside the global health crisis, this pandemic has also triggered devastating social and economic impacts across the globe.
Even with significant medical resources in the developed world, most sophisticated healthcare systems are being overwhelmed by the magnitude of the pandemic. From limited healthcare workers to the lack of medical capacity, many developing countries are facing unprecedented health challenges in managing the Covid-19 situation12. The first confirmed Covid-19 cases in Africa were reported in Egypt (Feb 14, 2020), mostly imported from Europe13. Between February and March 2020, the pandemic spread rapidly, and most of the African countries reported several confirmed Covid-19 cases, with an increased rate of infections and an offset, time-shifted spread when compared to European neighbors confirming the origin of the pandemic13.
Currently, a new chapter in Covid-19 fight is open with the vaccine phase14–16. The COVID-19 vaccine race is launched in early 2021 and several countries worldwide are already undergoing massive vaccination programs. Indeed, since the identification of SARS-CoV-2 several vaccines have started to be rolled out. Some of them were made available in only few months for emergency reasons instead of years (> 10 years) and others are under clinical trials14–17. Even if the vaccines are expected to slow down human-to-human transmission, the rapid increase of virus mutations is still challenging for the efficacy of the vaccines and the overall management of the pandemic18. Moreover, little is known about the safety, immunity, protection and transmission level of vaccinated patients19,20. Therefore, rapid testing is still a critical cornerstone in the overall management of this pandemic, enabling healthcare to trace and contain the virus as well as to efficiently prepare the current vaccination phase.
Several diagnostic assays have been reported for SARS-Cov-2 detection21. Among them, virus isolation22, RNA quantification23, antigen detection and serological methods for detecting IgM and IgG are the most widely used methods in laboratory diagnosis and in virologic studies24. Very recently, new innovative technologies have been reported, with reliable efficiency in decreasing the overall time of the analysis and in pushing the limits of detection. Some of them are RT-PCR-like such as nano-PCR25, multiplex RT-PCR26, isothermal amplification27; and others rely on the integration of interesting CRISPR-based detection28–30.
Currently, diagnosis of SARS-Cov-2 virus is mainly based on the quantitative polymerase chain reaction (RT-PCR) for the detection of viral nucleic acids31,32. These methods have several limitations such as sample handling, requiring samples in the acute phase, and testing time, which ranges from 2 to 4 h for a simple PCR acquisition to more than 12 h for the overall processing and handling time. Moreover, this technique also requires the use of expensive kits that are mostly sourced from western and Chinese suppliers and cannot be guaranteed for African countries particularly during lockdown periods. In other words, the diagnosis of SARS-Cov-2 infected patients in African countries using the PCR technique is inadequate and requires, in several countries, more than 5 days to get the test results to the patients.
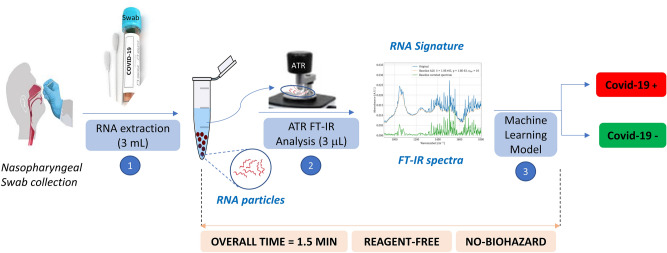
In line with these considerations, herein we report an unprecedented and highly accessible screening method for the detection of SARS-CoV-2 using extracted RNA samples. This method is based on a straightforward combination of infrared (IR) spectroscopy and machine learning (ML). Compared to RT-PCR, this method is faster (1.5 min vs 2–4 h post-RNA extraction), requires no reagents, and less biohazard waste is generated at the end of the test (Fig. 1).
Figure 1.
Sequential FT-IR-based assay for detection of SARS-CoV-2: sample collection, RNA extraction, FTIR analysis and then machine learning.
The use of IR spectroscopy for viral detection is relatively new33–35. Although the method has several advantages, it might require a sizeable number of samples to enhance its sensitivity, specificity and accuracy. In this work, we use, for the first time, machine learning to build classification models able to predict the patient's infection, based on the IR spectra of the extracted RNAs. To the best of our knowledge, this technology, based on a dual and complementary combination of FT-IR and machine learning on RNA samples, has not been reported yet, given the high diagnosis performance results achieved, suggests spectroscopy as a promising tool for viral diagnosis. We also used sparse classification techniques to improve the interpretability of models. To the best of our knowledge, this also the first time such technique is used on FTIR-based diagnosis techniques. We perform experiments on two sets of samples. A first set of 280 clinical samples is used to assess the sensitivity, specificity and accuracy. A second set of synthetic RNA samples is used to evaluate the limit of detection and further assess the selectivity against 15 other respiratory viruses.
Results
Clinical data
Research on spectroscopy based viral detection relies on different fields including microbiology, spectroscopy, data processing and machine learning. The efficiency of this method relies on the quality of the clinical specimens, the protocol used, hardware configuration, signal pre-processing techniques and the choice and tuning of statistical algorithms. We outline here the combination that achieves the highest performance. Other set-ups are detailed and discussed in Supplementary Information.
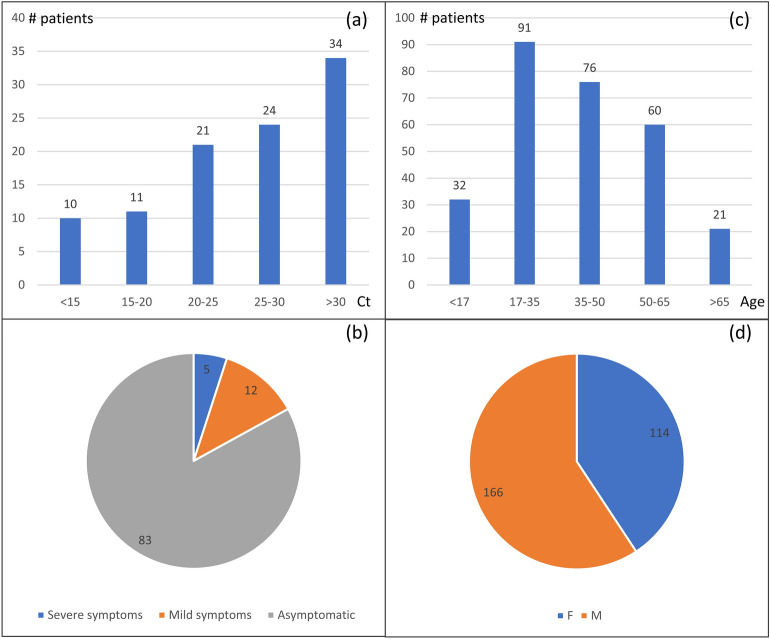
In this study, 280 RNA extracts from nasopharyngeal samples collected from 280 Moroccan patients are used to train, test, and validate our classification models (Fig. 2). These samples were collected from 100 SARS-CoV-2 PCR positive patients and 180 SARS-CoV-2 PCR negative patients with ages ranging from 11 to 67 years old. Cycle threshold (Ct) values of positive samples ranged from 11.7 to 34 with an average of 25.6 and a median of 26.1. 17 Covid-19 patients were experiencing symptoms while the remainder of the tested cohort was asymptomatic.
Figure 2.
Clinical data of Covid-19 tested patients. (a) Ct values distribution based on the number of samples (b) Distribution of symptomatic and asymptomatic positive patients, (c) Distribution of patients by age. (d) Distribution of patients by sex.
Nasopharyngeal samples specimens were collected using swabs. They were immediately inserted into sterile tubes containing 1–3 mL of viral transport media. We used extraction kits based on the magnetic beads method, followed by washing steps then elution. 100 μL of viral transport media were added to the preloaded kit, while the remaining purification process was fully automated by the extractor in Viral Mode. The samples output is of 50 µL. These protocols were based on the manufacturer’s recommendation. The real-time PCR assay was then performed using the Takyon Real-Time One-Step RT-PCR Master Mix and Eurogenetic kit for covid-19 E-gene (see Table 1, Supplementary Information). The RNA quality of the samples was determined by the optical method of absorbance in the ultraviolet. For nucleic acids, the three main wave numbers of interest are 260 nm, 280 nm and 230 nm. The ratio of the absorbance at 260 versus 280 nm (A260/280) is generally used to assess the purity of nucleic acids as well as the DNA vs RNA ratio. Absorbance at 280 nm is indicative of proteins content in the sample. Measurements at 230 nm were used to determine the amounts of other contaminants that may be present in the samples, such as guanidine thiocyanate and guanidine hydrochloride, common in nucleic acid purification kits (see Supplementary Information).
Table 1.
Comparison of predicting performance on the testing set using Logistic Regression.
| Spectral region (cm−1) | Spectra signal | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (× 100) | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (× 100) |
|---|---|---|---|---|---|---|---|---|---|
| Out-of-sample | In-sample | ||||||||
| 600 to 4500 | Raw | 95.7 | 94.1 | 96.7 | 98.4 | 100 | 100 | 100 | 100 |
| Baseline Corrected | 96.8 | 94.1 | 98.3 | 99.8 | 100 | 100 | 100 | 100 | |
| 1st Der | 96.8 | 97 | 98.3 | 99.9 | 100 | 100 | 100 | 100 | |
| 2nd Der | 97.8 | 97 | 98.3 | 99.1 | 100 | 100 | 100 | 100 | |
| 900 to 1800 | Raw | 83.1 | 70.5 | 86.5 | 89 | 92.1 | 82.3 | 100 | 94.3 |
| Baseline Corrected | 84.2 | 73.5 | 86.5 | 90.5 | 94.7 | 88.2 | 100 | 95.5 | |
| 1st Der | 93.6 | 94.1 | 92.4 | 97.7 | 100 | 100 | 100 | 100 | |
| 2nd Der | 94.7 | 94.1 | 95.4 | 97.2 | 100 | 100 | 100 | 100 | |
FTIR and machine learning
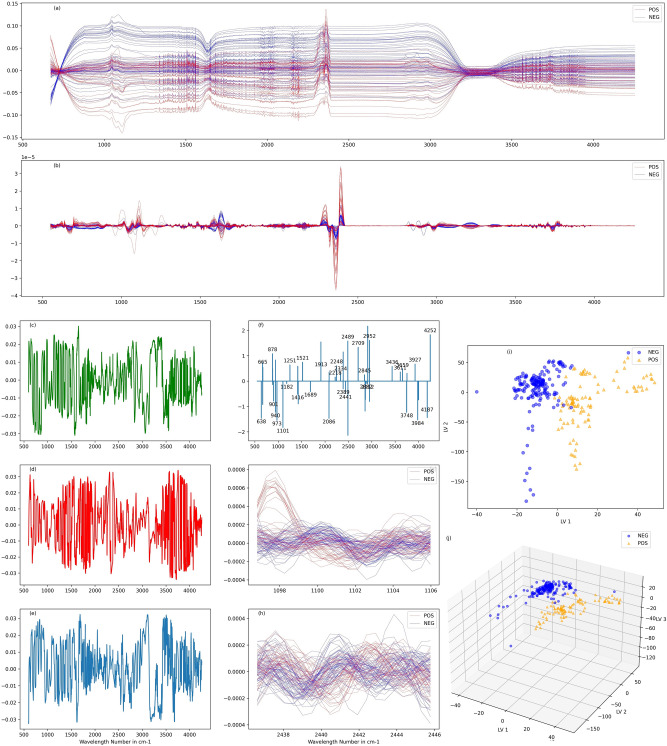
Primary analysis of the vibrational chemical bonds, shape and band intensity on the raw recorded spectra (Fig. 3a) did not give any useful information on the Covid-19 status of the patients, and no details can be extracted even after in-depth investigation of baseline corrected spectra. We then apply machine learning algorithms to the spectra and compare their out-of-sample performance on the testing set (Fig. 3).
Figure 3.
Detection of SARS-CoV-2 with multivariable analysis. (a) Raw Spectra (b) Sample of 2nd derivative of Savitzky–Golay smoothened spectra of positive and negative samples. (c, d, e) First three latent variables of PLS-DA. (f) Coefficients of variables selected by sparse classification algorithm of second derivative of raw spectra (g, h) Zooms on regions indicated by sparse classification. (i)Projection of the 280 spectra used according to the first two latent variables obtained. (j) Projection of the 280 spectra used according to the first three latent variables obtained.
We apply discrete second derivative of the raw spectra acquired (Fig. 3b). We center and normalize the data of the transformed spectra. We finally use machine learning algorithms to build classification models. We also evaluate other methods such as first derivative and found that best results are achieved using first and second derivative (see Supplementary Information). We report a sparse solution for sparse logistic regression in Fig. 3f, that stress out relevant wavenumbers in our classification method.
Given the measurement’s sampling interval in this region (~0.48 cm−1), a total of 8287 features (variables) were included for the 280 samples. Due to the unbalanced ratio between the number of samples (280) and the number of variables (~ 8200), we use dimension reduction techniques, namely principal component analysis (PCA), partial least square (PLS) and Sparse Classification36 and then use logistic regression for PCA and PLS and support vector machine (SVM) and kernel SVM for classification.
To improve interpretability, we also use the sparse classification approach. Classic methods such as PCA and PLS build principal components and latent variables using all available variables, which hinders their interpretability (Fig. 3c,d,e). Here we also build classification models using Mixed Integer Optimization based classification algorithms. These methods build a classification model using a relatively low number of variables, much lower than the number of the features of the spectra, to improve interpretability (Fig. 3f). Given a data matrix X (representing spectra in our case), a response vector Y (here positive or negative outcome), a loss function ℒ and a regularization function π, sparse classification algorithms build classification models by solving the following type of problem:
where γ is a non-negative parameter, k a positive integer and ||.||0 is the L0 norm indicating the number of non-zero variables in β. In this work we use a sigmoid function as a loss function, which is identical to logistic regression, and Tikhonov regularization. Regularization provides robustness to the model built vs noise. Tikhonov regularization is particularly efficient in addressing normal noise and has proven to be particularly efficient in spectroscopy applications37.
Dimension reduction methods project the data into a lower dimension space that is believed to allow a strong predictive power. Thus, they bolster the performance of the downstream binary classification methods. We use of cross-validation to tune the hyperparameters of these methods. For the performance evaluation, we use, as a standard practice, 67% of the samples for training and the remaining for testing. We randomly choose 185 samples for training and 95 for testing. In Supplementary Information, we also assess statistical significance by randomly shuffling 25 times the training/testing samples and report tests results in Table 1. All binary classification models are evaluated based on their ability to discriminate the outcome of interest. We present standard deviation for all reported metrics in Supplementary Information. We report AUC and its standard deviation for all methods in Supplementary Information. This metric captures the information about the model performance independently of the detection threshold selected. Moreover, we report the accuracy, sensitivity and specificity (see Supplementary Information for a full comparison of algorithms’ respective performance).
We observed that the choice of spectral region and data processing technique play a key role in the performance of the models. Indeed, expanding the spectral region from the classic bio fingerprint region of 1800–900 cm−1 to 600-4500 cm−1 boosts the accuracy by up to 12.6% (in the case of raw spectra). Although less important, there are also benefits using signal processing transformations. When 600–4500 cm−1 region is chosen, the improvement reported is 1.9%. The second derivative seems to offer the highest performance and improves the stability of the algorithms (see Supplementary Information). The choice of classification algorithm is less important (we provide a full discussion in Supplementary Information). The highest results were achieved using second derivative of captured spectra and the PLS-based methods and Logistic regression achieve an accuracy of 97.8%, a sensitivity of 97%, and specificity of 98.3%.
The biomarkers depicted by the sparse model (which results from an unsupervised method) are largely discussed in the FTIR spectroscopy literature, the most representative are reported in Table 2.
Table 2.
Tentative assignment of wavenumber markers used by sparse classification.
| Wave number (cm−1) | Tentative Assignments38–42 |
|---|---|
| 638, 665 | Guanine breathing mode |
| 878 | Out-of-plane vibrations of nucleobases |
| 901, 940 | Ribose phosphate backbone |
| 973 | C–O and C–C ribose |
| 1101 | Symmetric stretching P–O–C, RNA ν(C–O) ribose |
| 1182 | C–O and phosphate vibrations |
| 1251 | P = O, PO2−asym |
| 1416 | Stretching C–N, N–H and C–H deformation |
| 1521 | C nucleobase both in RNA ss or ds, amide II |
| 1689 | C6=O6 of G in ds, Amid I, C=O Guanine and N–H deformation, νasym(C2=O) vibration in RNA |
| 1913, 2086 | bands of second order |
| 2218, 2248, 2334, 2389, 2441, 2489, 2709, 2845, 2882, 2952 | Stretch C–H, N–H |
| 3611, 3436, 3659, 3748, 3927 | Stretch N–H asym and O–H asym |
Careful analysis of the maximum footprint spectral region of the RNAs spectra for the positive and negative samples of SARS-CoV-2 RNAs (Fig. 3f), indicates the presence of three main visible domains: one located at 600–1350 cm−1, the other at 1500–1700 cm−1 and at 2300–3900 cm−1. The first domain is attributed to the phosphate backbone vibrations (νP-O) with the 1000–1182 cm−1 region arising from symmetric stretching vibrations of PO2− and assigned to the νC-O stretching vibration of the phosphodiester and the ribose. In addition, the spectral region 1200–1300 cm−1 could be attributed to PO2− asymmetric stretching vibration of the RNA usually centered at the 1251 cm−1. The third 1500–1700 cm−1 region could be assigned to RNA nucleobases. Furthermore, this region overlaps with a series of biomarker bands usually ascribed to Amid I and II vibrations. The region 2400–3900 cm−1 is in line with the stretching vibrations of OH, NH, and CH groups. Taken together, these data are clearly in accordance with the RNA signature confirming the robustness of the FT-IR/machine learning dual coupling in virus detection and patient classification.
In order to assess the specificity of the technique towards other viruses and its limit of detection (LoD) we investigate a set of samples and analyze them using FTIR spectroscopy and machine learning.
Specificity towards other viruses
We use a total of 123 samples including 31 samples featuring the entire genome of SARS-CoV-2 with various concentrations as well as samples including the entire genome of each of human bocavirus 1, human coronavirus 229E, human coronavirus NL63, human coronavirus OC43, human enterovirus 68, human parainfluenza virus 1, human parainfluenza virus 4, rhinovirus 89, influenza A, influenza B, influenza H3N2measles, MERS coronavirus mumps and SARS-CoV-1 (2 samples each with 500 copies/μl concentration). We also performed experiments on42 PCR positive controls containing four individual non-infectious DNA plasmids coding for the RdRp gene, the E gene, the N gene, the RNAse P gene and 20 DEPC-treated water samples. The concentration of SARS-CoV-2 samples vary between 10,000 copies/μl and 0.5 copies/μl. A full description is provided in Table 3.
Table 3.
Synthetic RNA viruses used to assess the specificity. (a) Anti-sense strand, (b) sense strand.
| Name | Accession | Virus Type | Length |
|---|---|---|---|
| Twist synthetic Influenza H1N1 (2009) | NC_026431, NC_026431, NC_026431, NC_026431, NC_026431, NC_026431, NC_026431 | ssRNA (−)a | 13,158 |
| Twist synthetic Influenza H3N2 | NC_007366, NC_007367, NC_007368, NC_007369, NC_007370, NC_007371, NC_007372, NC_007373, | ssRNA (−) | 13,627 |
| Twist synthetic Influenza B | NC_002204, NC_002205, NC_002206, NC_002207, NC_002208, NC_002209, NC_002211 | ssRNA (−) | 14,452 |
| Twist synthetic human Bocavirus | MG953830.1 | ssDNA | 5164 |
| Twist synthetic human Enterovirus 68 | NC_038308.1 | ssRNA (+)b | 7367 |
| Twist synthetic human Rhinovirus 89 | NC_001617.1 | ssRNA (+) | 7152 |
| Twist synthetic Mumps virus | NC_002200.1 | ssRNA (−) | 15,384 |
| Twist synthetic human Parainfluenza virus 1 | NC_003461.1 | ssRNA (−) | 15,600 |
| Twist synthetic Measles virus | NC_001498.1 | ssRNA (−) | 15,894 |
| Twist synthetic human Parainfluenza virus 4 | NC_21928.1 | ssRNA (−) | 17,052 |
| Twist synthetic human Coronavirus 4 | NC_0022645.1 | ssRNA (+) | 27,317 |
| Twist synthetic human Coronavirus NL63 | NC_005831.2 | ssRNA (+) | 27,553 |
| Twist synthetic human Coronavirus OC43 | NC_006213.1 | ssRNA (+) | 30,741 |
| Twist synthetic human Coronavirus Tor 2 | NC_004718.3 | ssRNA (+) | 29,751 |
| Twist synthetic human Coronavirus 2c EMC/2012 | JX869059.2 | ssRNA (+) | 30,119 |
We recorded 64 spectra for each sample using the same parameters and equipment as those used for the clinical samples (Table S2) and then averaged the 64 spectra for each sample. We then apply the second derivative of the average spectra in the 1800–900 cm−1 region which resulted in 1867 variables. We first disregard the SARS-CoV-2 samples of less than 25 copies/μl to assess the specificity. 100% accuracy is achieved on the resulting set using sparse classification algorithm in-sample. The separation is achieved using only 13 variables out of the 1867 variables constituting the spectra in the 1800-900 cm−1 region. 2 pairs of these variables are adjacent and overlap in the spectra, which indicate that only 11 frequencies could be used to separate the set. The number of data points (109 spectra) is almost one order of magnitude lower than the number of dimensions used to classify. We enumerate the frequencies and provide a tentative assignment for the attribution of the specific wavenumbers (Table 4). Remarkably, we found that the set of variables used in this model and the set of variables used for clinical samples are disjoint, which indicates that the viral signature could be found in various variables settings. The variables used by the sparse algorithm for clinical samples can also be used to separate the positive synthetic samples from the synthetic negative ones.
Table 4.
Variables used to separate positives and negative samples with 100% accuracy.
| Wave number (cm−1) | Tentative assignments38–42 |
|---|---|
| 1038 | ν(C–O) ribose. symmetric stretching P–O–C |
| 1074 | νasym(PO2−) symmetric and ν (C-O) ribose |
| 1131 and 1172 | RNA ν (C=O). ribose |
| 1172 and 1174 | ν (C-O) and phosphate vibrations (non-H-bonded mode of C–OH) x |
| 1210 | νasym (PO2−) |
| 1640. 1670. 1673 | Amid I.νa (C2=O).νasym(C5=O).νasym (C6=O). can overlap with water (ν2) |
| 1760 | ν carbonyl stretching |
It is worth noting that the 11 frequencies used for the separation are in line with the RNA signature (Table 4). These frequencies can be classified into two spectral regions. The first one, 1038–1220 cm−1, is typically assigned to the fingerprint of the sugar and the phosphate backbone of the RNA. Indeed, 1038 and 1074 cm−1 bands could be assigned to the (C–O) ribose and P–O–C symmetric stretching at position 5 of the ribose. 1172 and 1174 cm−1 are ascribed to the single C–O bond vibrations of the ribose and phosphate as well as to the free hydroxy groups at 2’ position. 1210 cm−1 frequency is mainly ascribed to ν (PO2−) asymmetric vibration. The second region, 1640–1760 cm−1, can be attributed to in-plane vibrations of nucleobases related to double bonds in the 5 and 6 member rings heterocycles: C=C, C=N and C=O of purine (A. G) and pyrimidines (C. U). 1640–1673 frequencies are in line with vibrations of amid I, νa (C2=O), νa (C5=O) and νa (C6=O). Strong band at 1760 cm−1 is a typical marker of carbonyl groups.
To assess out-of-sample performance, we use Leave-one-out technique. We iteratively remove the spectrum for each sample of human bocavirus 1, human coronavirus 229E, human coronavirus NL63, human coronavirus OC43, human enterovirus 68, human parainfluenza virus 1, human parainfluenza virus 4, rhinovirus 89, influenza A, B and H3N2 measles, MERS coronavirus, mumps and SARS-COV-1 and train with remaining 108 spectra. The algorithm predicts correctly 25 out of the 30 samples presented (2 of each virus). The five samples that are predicted as false positives are one sample of each of influenza A and B, MERS, coronavirus OC43, and enterovirus 68. We also note that the training set is relatively limited regarding the variety of viruses. Undoubtedly, the out of sample specificity might be improved with a richer training set, i.e., by increasing the number of samples in the training set.
Limit of detection
The limit of detection (LoD) is a measure of the analytic sensitivity of the method, regarding the lowest copy number detected 95–100% of the time. Its quantification is highly dependent on the technique used, its accuracy, the kits’ specification and the component to be analyzed43. Regarding the quantification of LoD for SARS-CoV-2 detection, several units were reported such as copies/ml, copies/μl, copies/reaction volume, mol/l making the comparisons between the techniques difficult44,45. In our case we used a copies/μl as the correlation between Ct and the concentration (copy/μl) is easier to handle46.
For this study, we train sparse classification algorithm using the samples with concentration higher than 25 copies/μl and test on the remaining samples (Table 5). We achieve100% accuracy of all samples with concentrations as low as 10 copies/μl. In addition, we observe a weak detection with concentrations below 10 copies/μl on synthetic samples, which is consistent with the results obtained on clinical samples. Indeed, clinical samples with Ct of 31 and higher are correctly diagnosed. This level of Ct is equivalent to a number of copies close to 10 copies/μl44. It is worth noting that all clinical samples with a Ct higher or equal to 32, which corresponds to the limit of our RT-PCR set-up, are correctly diagnosed by our technique.
Table 5.
Detection limit: distribution of predictions on positive samples after training depending on samples’ concentration.
| Concentration (copies/ul) | Number of samples | Number of correct predictions |
|---|---|---|
| 25 | 3 | 3 |
| 10 | 3 | 3 |
| 5 | 3 | 1 |
| 3 | 3 | 0 |
| 0.5 | 5 | 0 |
Discussion
During the last centuries, the most severe and devastating pandemics, with high mortality rate, have been mainly ascribed to RNA viruses. Presently, the world is experiencing an unprecedented health crisis due to a new RNA virus, SARS-CoV-2 (Covid-19). This pandemic still has dramatic effects with devastating economic, social and health consequences. The diagnosis of SARS-CoV-2 and the control of its dissemination raised serious questions in developing countries. While no efficient treatment is now available, the mass diagnosis of populations during the next months is crucial to limit the spread of Covid-19. Many countries are still trying to scale up diagnostic screening tests to meet the high demand. Undoubtedly, there is an urgent need for an effective and appropriate diagnostic test for all populations to control the spread and minimize its devastating consequences.
To address these issues, we demonstrate in this work that the use of FT-IR spectroscopy on RNA extracts and machine learning is highly convenient and appropriate for the diagnosis of Covid-19 disease. Indeed, 280 symptomatic and asymptomatic-suspected Moroccan patients with specific clinical indications were tested for their Covid-19 status using swab samples. Among the samples of the 280 patients, 100 were Covid-19 positives and 180 negatives by quantitative RT–PCR assay used as a control. The extracted RNAs were then analyzed by ATR infrared spectroscopy and the obtained spectra were used to train and test a machine learning classification method. The machine learning was then applied and showed very promising results with 97.8% accuracy, 97% sensitivity and 98.3% specificity. The FTIR spectra indicates the presence of three main visible domains located at 600–1350 cm1, at 1500–1700 cm−1 and at 2300–3900 cm−1 clearly ascribed to RNA fingerprint, i.e., the phosphate backbone vibrations (νP-O), the νC-O stretching vibrations of the ribose sugar and the specific RNA nucleobases. The region 2400–3900 cm−1 is attributed to the stretching vibrations of OH, NH, and CH group.
To get further insight into the selectivity of this technique against other viruses, we used 123 samples including 31 samples featuring the entire genome of SARS-COV-2 as well as samples including the entire genome of human bocavirus, human coronaviruses (229E, NL63,OC43, MERS, SARS-CoV), enterovirus 68, human influenza viruses (A, B, H3N2), human parainfluenza viruses(1 and 4) and rhinovirus 89. We found that this technique is 100% in agreement with RT-PCR used a standard control, reaching high viral selectivity even with the high structural similarities of the RNA viruses. Furthermore, the limit of detection was assessed using both synthetic and clinical samples with different concentrations, from 0.5 to 25 copies/μL. The LoD was determined to be 10 copies/μL for both samples.
A remarkable work was just published by Martin and Coll47. This work uses ATR-FTIR on a saliva matrix instead of RNA extract. This report is highly interesting and complementary to the present work for comparison studies, i.e., saliva matrix versus RNA extract. Indeed, the performances observed from saliva are less than those observed with the RNA extracts in terms of sensitivity (97 vs 95%) and specificity (98.3% vs 89%), the accuracy for RNA extract is high 97.8% while it is not reported for the saliva. In other words, saliva swab is a complex medium containing complex cellular materials including phospholipids, viral and non-viral nucleic acids and proteins, etc. making the repeatability inter-patients difficult and the interpretability of spectra. The use of extracted viral RNA has several advantages in terms of purity (one major component) allowing clear signature, high correlation and interpretability of the FT-IR spectra.
Furthermore, the covid-19 diagnosis community is working on developing new techniques to increase testing capacity, overcome limitations (such as the use of reagents) and shorten the testing time. Each technique requires resources (equipment, reagents, etc.) that may not overlap creating thus the possibility to expand the testing capabilities. The multiplicity of testing techniques can be an answer to the testing challenge that World population is facing. The performances of FTIR-based technique are comparable to other techniques developed by the scientific community (Table 6).
Table 6.
Characteristics of different Sars-CoV-2 detection techniques. (a) Authors' estimates. (b) The matrix solution was prepared with α-cyano-hydroxy-cinnamic acid (CHCA) at 1% in acetonitrile/0.1% trifluoroacetic acid (1:1). c) RT-PCR Limit of Detection in copies/mL44.
| ATR-FTIR | CRISPR–Cas12-based22 | MALDI-MS48 | RT-PCR | |
|---|---|---|---|---|
| Test Ct range | 11 to 39 | 20 to 37 | 16 to 37 | – |
| Accuracy | 98% | 98% | 94% | – |
| Sensitivity | 97% | 95% | 95% | – |
| Specificity | 98% | 100% | 93% | – |
| LoD (in copies/μl) | 10 | 10 | NA | 10 to 511c |
| Sample-to-result time (min) | 21 | 30–40 | 25-30a | 120 |
| Consumables | RNA extraction kits | RNA extraction & Crisper kits | CHCAb | RNA extraction & PCR kits |
| Equipment | FTIR Spectrometer, RNA extractor | RT-Lamp, Lateral flow Strip, RNA extractor | Mass Spectrometer | PCR machine, RNA extractor |
The FTIR spectroscopy shows a promising potential for mass testing since it is reagent free, fast and provides strong predicting performances. The models developed in this study provide 97% sensitivity and could enable the use of FTIR as a tool that could filter samples going through PCR, which could expand the testing capabilities, especially in developing countries where the testing capacity is saturated. All laboratories performing RNA extraction (virtually all PCR testing centers) could benefit from this technique, which would cut post-RNA testing time from hours to minutes and could boost testing capacity whenever RNA extraction capacity is under-utilized relatively to PCR. This technology could also enable epidemiologic surveillance. It can be adapted not only in clinical laboratories and hospitals but also in several locations as well (airports, schools and universities, etc.) using a simple portable monitoring FT-IR device.
In summary, the combination of FT-IR and machine learning allows quantitative predictive agreement and classification of both positive and negative samples of SARS-CoV-2 in a faster manner compared to RT-PCR (1 to 1.5 min vs. 2–5 h) with 97.8% accuracy for the detection of this coronavirus in 280 patient samples. Compared to the RT-PCR requiring the enzymes and amplification kits, the FTIR spectroscopic method presented here is reagent-free and can be used in point of care with limited facilities. Finally, the use of RNA extracts and FTIR spectroscopy could also be considered in the diagnosis of other viruses.
Materials and methods
Samples collection and RNA extraction
Nasopharyngeal swab specimens were collected using only swabs with a synthetic tip. Swabs were immediately inserted into sterile tubes containing 1–3 ml of viral transport media. In this study, total RNA was extracted using four different nucleic acid extractors from different vendors Amplix (ZP01001), Molarray (MA-32T), Bioer (NPA-32P) and Genrui (v3-NE48/96). For all four methods, viral transport media was added to the preloaded Kit (Bioer Bsc86 Magabio Plus Virus DNA/RNA Purification Kit III, Genrui nucleic acid extraction kit NE-01A and 3150001 Amplix viral Nucleic acid extraction kit), while the remaining purification process was fully automated by corresponding extractor in Viral Mode. These protocols are all dedicated to viral RNA extraction and based on a magnetic beads method which ensure a good quality of total RNA after quantification. Extractions were performed according to the manufacturer's guidelines.
RT real time PCR
Real-time RT-PCR assay was performed using the Takyon Real-Time One-Step RT-PCR Master Mix and Eurogenetic kit for both E and RdRp SARS-CoV-2 genes as per the protocol below:
Each 25 μL reaction mixture contained 12.5 μL of 2X reaction buffer, 1 μL of forward and reverse primers at 10 mM, 0.5 μL of probe at 10 mM, 0.25 μL of RTenzyme, 0.5 μL RNase inhibitor and 5 μL of total extracted RNA with a concentration of 2 ng/ul or higher. Amplification was carried out in 96-well plates on QuantStudio1 machine. Thermocycling conditions consisted of 55 °C for 10 min for reverse transcription, followed by 95 °C for 3 min and then 45 cycles of 95 °C for 15 s and 58 °C for 30 s. Each run included one SARS-CoV-2 positive control and one template control. For a routine workflow, the E gene assay was carried out as the first-line screening indicator followed by confirmatory testing with the RdRp gene assay. This assay is setup with the same conditions as the E gene. Both E and RdRp screening assays were performed according to the manufacturer’s guidelines.
ATR-FTIR spectroscopy
We use ATR-FTIR spectroscopy (Attenuated Total Reflection Fourier Transform Infrared) to collect spectra. ATR-FTIR spectra were acquired using Jasco 4600 ATR-FTIR spectrometer with a deuterated lanthanum α-alanine doped triglycine sulphate (DLaTGS) pyroelectric detector. The detector is operated with temperature stabilization using electrical peltier temperature control. The spectrometer is paired with a high-intensity ceramic light source. We perform single reflection ATR using high-throughput monolithic diamond crystal and 64 spectra are averaged. We apply the torque-limiter pressure applicator for reproducible sample pressure contact for sample measurements. We use distilled water as solvent background. We spread 3 μL of each sample on the ATR crystal, ensuring that no air bubbles were trapped. We do not dry samples on the spectra to simplify sample manipulation and decrease the testing time, at the expense of having to deal with the absorption from water. After the acquisitions, we clean the crystal with ethanol (70% v/v) and dry it using paper towel (see Supplementary Information). We collect the 600–8000 cm−1 region with a spectral resolution of 2 cm−1. We use 600 cm−1 to 4500 cm−1 (Table 1) as a representative region in our study, with a special focus on the traditional 900 to 1800 region (Fig. 3), known as the RNA bio fingerprint region28.
Machine learning
We used mainly publicly available libraries on a commercial personal computer. PCA, PLS, Logistic regression, SVM, Kernel SVM and Discriminant analysis were all solved using “sklearn” v0.23 running on Python 3.7.3. We use “SubsetSelection” package on Julia language to perform sparse classification; warm-starting the algorithm with a good solution is key to reduce the computational time. We use the “signal” library for signal processing on Python 3.7.3. A quarter of the training data was used for cross-validation to tune the hyperparameters of the algorithms used. The computer used is has an Intel CORE i7-8750H CPU at 2.2 GHz with 16 GB RAM running on Windows 10.
Ethical and biosafety statement
In this work all methods were carried out in accordance with relevant guidelines and in line with the Moroccan’ regulations. All experimental protocols were approved by Institutional Ethic Committee UM6P-Anoual. Informed consent was obtained from all subjects and also, for subjects under 18, informed consent from a parent and/or legal guardian. No identity of any sample was related to the name of the patient or other information that could lead to personal identification. Specific patient information was limited to the present work (sex, age and symptoms). All samples used for RT-PCR were split for FT-IR analyses and used according to Laboratory protocols. All samples were deactivated before RNA extraction in a biosecurity Lab. All materials were cleaned before and after each experiment to prevent any source of contamination. All consumables and Covid waste are managed by a private company and incinerated according to the authorities’ guidelines.
Supplementary Information
Acknowledgements
This work is a part of technology development by team “Smart Spectra”. We would like to thank the X-Prize Fast Covid-19 Testing team for the acquisition of synthetic samples described in Table 4.
Author contributions
D. L. K. coordinated the present research, designed the analytical approach and analyzed data. A.F.G. contributed to the analysis of the data. D. B. supervised D.L. K. and A. F. G. on this work. S. L. and N. M. conceived and designed the study and the protocol of experiments, analyzed RNA samples and double-checked RT-PCR data and performed experiments. O. L. performed the calibration of the FTIR machine. N. A. contributes to design the study, to build the team, to tune the optical settings and to validate the optical spectrum. O. M. contributed to the analysis of spectra. R. B. headed the study at the chemistry/biology interface, analysis of spectra, writing the manuscript and supervision of O. L. J. F. headed the biological studies and supervised S. L. and N. M. All authors participate in writing, reading and correcting the manuscript and agreed to its contents. J. B. and J-L. G. provided clinical samples. F. Z. E. A. and F. J. participated in the RNA extraction, performed PCR tests and contributed to results analysis.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the authors.
Competing interests
The authors declare no competing interests. D.L.K, S.L., N.M., D.B., N.A, J.F. and R.B. are co-inventors of this technology.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95568-5.
References
- 1.Nickol ME, Kindrachuk J. A year of terror and a century of reflection: Perspectives on the great influenza pandemic of 1918–1919. BMC Infect. Dis. 2019;19:1–10. doi: 10.1186/s12879-019-3750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Morens, 1D. M. 918 Influenza: The Mother of All Pandemics. Emerg. Infect. Dis. 2006;12:15–22. doi: 10.3201/eid1209.05-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemelaar J. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 2013;66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Manns MP, Buti M, Gane E, Pawlotsky JM, Razavi H, Terrault N, Younossi Z. Hepatitis C virus infection. Nat. Rev. Dis. Prim. 2017 doi: 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 5.Fedson DS. What treating Ebola means for pandemic influenza. J. Public Health Policy. 2018;39:268–282. doi: 10.1057/s41271-018-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fineberg HV. Global health: Pandemic preparedness and response - Lessons from the H1N1 influenza of 2009. N. Engl. J. Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 7.Reperant LA, Osterhaus ADME. AIDS, Avian flu, SARS, MERS, Ebola, Zika… what next? Vaccine. 2017;35:4470–4474. doi: 10.1016/j.vaccine.2017.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascarella, G., Strumia, A., Piliego, C., Bruno, F., Del Buono, R., Costa, F., Scarlata & Agrò, S. F. E. COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med. 288, 192–206 (2020). [DOI] [PMC free article] [PubMed]
- 10.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin. Immunol. 2020 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO, No Title, p. WHO Coronavirus Disease (COVID-19) Dashboard, released Friday February 5th 2021.
- 12.Lone SA, Ahmad A. COVID-19 pandemic—An African perspective. Emerg. Microbes Infect. 2020;9:1300–1308. doi: 10.1080/22221751.2020.1775132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert G, Pullano G, Pinotti F, Valdano E, Poletto C, Boëlle PY, D’Ortenzio E, Yazdanpanah Y, Eholie SP, Altmann M, Gutierrez B, Kraemer MUG, Colizza V. Preparedness and vulnerability of African countries against importations of COVID-19: A modelling study. Lancet. 2020;395:871–877. doi: 10.1016/S0140-6736(20)30411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabortya S, Mallajosyulab V, Tato CM, Tan GS, Wang TT. SARS-CoV-2 vaccines in advanced clinical trials: Where do we stand? Adv. Drug Deliv. Rev. 2021 doi: 10.1016/j.addr.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 16.Sharma O, Sultan AA, Ding H, Triggle CR. A review of the progress and challenges of developing a vaccine for COVID-19. Front. Immunol. 2020;11:1–17. doi: 10.3389/fimmu.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Thanh T, Andreadakis Z, Kumar A, et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug. Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 18.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021 doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson EJ, Rouphael NG, Widge AT, et al. Safety and immunogenicity of SARS-CoV-2 mRNA- 1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voysey, M., Clemens, S. A. C, Madhi, S. A. et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet (2020) https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)32661-1/abstract. [DOI] [PMC free article] [PubMed]
- 21.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: The disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 22.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Bordi L, Piralla A, Lalle E, Giardina F, Colavita F, Tallarita M, Sberna G, Novazzi F, Meschi S, Castilletti C, Brisci A, Minnucci G, Vettamanzi V, Baldanti F, Capobianchi MR. Rapid and sensitive detection of SARS-CoV-2 RNA using the SimplexaTM COVID-19 direct assay. J. Clin. Virol. 2020;128:104416. doi: 10.1016/j.jcv.2020.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, Liu X, Chen Z, Huang H, Zhang F, Luo W, Niu X, Hu P, Wang L, Peng H, Huang Z, Feng L, Li F, Zhang F, Li F, Zhong N, Chen L. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheong J, Yu H, Lee CY, et al. Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat. Biomed. Eng. 2020;4:1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reijns MAM, Thompson L, Acosta JC, Black HA, Sanchez-Luque FJ, Diamond A, et al. A sensitive and affordable multiplex RT- qPCR assay for SARS-CoV-2 detection. PLoS Biol. 2020;18(12):e3001030. doi: 10.1371/journal.pbio.3001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, Stewart de Ramirez SA, Valera E, Cunningham BT, King WP, Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. PNAS. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nouri R, Tang Z, Dong M, Liu T, Kshirsagar A, Guan W. CRISPR-based detection of SARS-CoV-2: A review from sample to result. Biosens. Bioelectron. 2021 doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo L, Sun X, Wang X, Liang C, Jiang H, Gao Q, Qu B, Fang S, Mao Y, Chen Y, Feng G, Gu Q, Wang RR, Zhou Q, Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:4–7. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., Miao, X., treithorst, J. A., Granados, A., Sotomayor-Gonzalez, A., Zorn, K., Gopez, A., Hsu, E., Gu, W., Miller, S., Pan, C. Y., Guevara, H., Wadford, D. A., Chen, J. S. & Chiu, C. Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020). [DOI] [PMC free article] [PubMed]
- 31.Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Y, Chang L, Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30:1–14. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos MCD, Morais CLM, Nascimento YM, Araujo JMG, Lima KMG. Spectroscopy with computational analysis in virological studies: A decade (2006–2016) TrAC - Trends Anal. Chem. 2017;97:244–256. doi: 10.1016/j.trac.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandes, J. N., Dos Santos, L. M. B., Chouin-Carneiro, T., Pavan, M. G., G.Garcia, G. A., David, M. R., Beier, J. C., Dowell, F. E., Maciel-de-Freitas, R. & Sikulu-Lord, M. T. Rapid, noninvasive detection of Zika virus in Aedes aegypti mosquitoes by near-infrared spectroscopy. Sci. Adv. (2018). 10.1126/sciadv.aat0496. [DOI] [PMC free article] [PubMed]
- 35.Khan RS, Rehman IU. Spectroscopy as a tool for detection and monitoring of Coronavirus (COVID-19) Expert Rev Mol Diagn. 2020;20:647–649. doi: 10.1080/14737159.2020.1766968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertsimas D, Van Parys B. Sparse high-dimensional regression: Exact scalable algorithms and phase transitions. Ann. Stat. 2020;48:300–323. doi: 10.1214/18-AOS1804. [DOI] [Google Scholar]
- 37.Geinguenaud, F., Militello, V. & Arluison, V. in Methods in molecular biology (Clifton, N.J.), U. Walker, John M.(School of Life and Medical Sciences University of Hertfordshire Hatfield, Hertfordshire, Ed. (2020; 10.1007/978-1-0716-0278-2), vol. 2113, pp. 119–133. [DOI] [PubMed]
- 38.Geinguenaud F, Militello V, Arluison V. Application of FTIR spectroscopy to analyze RNA structure. In: Arluison V, Wien F, editors. RNA spectroscopy methods in molecular biology. Humana; 2020. [DOI] [PubMed] [Google Scholar]
- 39.Wood BR. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016;45:1980–1998. doi: 10.1039/C5CS00511F. [DOI] [PubMed] [Google Scholar]
- 40.Banyaya M, Sandbrinkb J, Strombergb R, Graslund A. Characterization of an RNA bulge structure by Fourier transform infrared spectroscopy. Biochem. Biophys. Res. Comm. 2004;324:634–639. doi: 10.1016/j.bbrc.2004.09.098. [DOI] [PubMed] [Google Scholar]
- 41.Movasaghi Z, Rehman S, Rehman UI. Fourier Transform Infrared (FTIR) spectroscopy of biological tissues. Appl. Spec. Rev. 2008;43:134–179. doi: 10.1080/05704920701829043. [DOI] [Google Scholar]
- 42.Dovbeshko GI, Gridina NY, Kruglova EB, Pashchuk OP. FTIR spectroscopy studies of nucleic acid damage. Talanta. 2000;53:233–246. doi: 10.1016/S0039-9140(00)00462-8. [DOI] [PubMed] [Google Scholar]
- 43.Parker, J., Fowler, N., Walmsley, M. L., Schmidt, T., Charrer, J., Kowaleski, J., Grimes, T., Hoyos, S. &Jack Chen, J. Analytical Sensitivity Comparison between Singleplex Real-Time PCR and a Multiplex PCR Platform for Detecting Respiratory Viruses. PloS ONE 10, e0143164, 1–9 (2015). [DOI] [PMC free article] [PubMed]
- 44.Fung B, Gopez A, Servellita V, Arevalo S, Ho C, Deucher A, Thornborrow E, Chiu C, Miller S. Direct comparison of SARS-CoV- 2 analytical limits of detection across seven molecular assays. J. Clin. Microbiol. 2020;58:e01535–e1620. doi: 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, Chen W-H, Yin X. Limits of detection of 6 approved RT–PCR kits for the novel SARS-Coronavirus-2 (SARS-CoV-2) Clinical Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barauna VG, Singh MN, Barbosa LL, Marcarini WD, Vassallo PF, Mill JG, Ribeiro-Rodrigues R, Campos LGG, Warnke PH, Martin FL. ultrarapid on-site detection of SARS-CoV-2 infection using simple ATR-FTIR spectroscopy and an analysis algorithm: High sensitivity and specificity. Anal. Chem. 2021 doi: 10.1021/acs.analchem.0c04608. [DOI] [PubMed] [Google Scholar]
- 48.Nachtigall FM, Pereira A, Trofymchuk OS, et al. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020;38:1168–1173. doi: 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the authors.