Abstract
Healthy bone homeostasis hinges upon a delicate balance and regulation of multiple processes that contribute to bone development and metabolism. While examining hematopoietic regulation by Tle4, we have uncovered a previously unappreciated role of Tle4 on bone calcification using a novel Tle4 null mouse model. Given the significance of osteoblasts in both hematopoiesis and bone development, this study investigated how loss of Tle4 affects osteoblast function. We used dynamic bone formation parameters and microCT to characterize the adverse effects of Tle4 loss on bone development. We further demonstrated loss of Tle4 impacts expression of several key osteoblastogenic genes, including Runx2, Oc, and Ap, pointing toward a potential novel mechanism for Tle4-dependent regulation of mammalian bone development in collaboration with the RUNX family members.
Keywords: Tle4, osteoblast, Runx2, bone mineralization, Tle4-Runx axis, bone calcification
Introduction
Normal bone development is a dynamic process that depends on the balance between bone formation and bone resorption. These two processes are largely mediated by osteoblasts and osteoclasts, respectively. An imbalance of these two forces results in various bone pathologies, including osteopetrosis and osteoporosis (Cohen, 2006; Kaul et al., 2015). Osteoblasts are derived from mesenchymal cells that are triggered by Wnt signaling toward osteoblastic differentiation (Hill et al., 2005; Houschyar et al., 2019). Various factors, including Wnt, BMP signaling, and Runx2, have been found to play roles, not only in normal osteoblast function, but also maturation and viability (Cohen, 2006; Kozhemyakina et al., 2015). Runx2 and Osterix regulate the differentiation of mesenchymal stem cells (MSCs) to osteoblastic lineages (Asada and Katayama, 2014). The loss of Runx2, a known interaction partner of Tle co-repressors, results in the absence of bone formation thought to be secondary to aberrant osteoblast differentiation in mice (Choi et al., 2001). Additionally, previous studies have shown that Runx2 expression induces osteoblastic differentiation of mouse stromal cells (Baniwal et al., 2012). Runx2 augments mesenchymal lineage proliferation while also assisting the commitment to osteoblasts by regulating a series of signaling pathways that include Wnt, FGF and PTH, as well as Dlx5 (Komori, 2019). Moreover, Runx2 enhances the expression of bone matrix protein genes including Col1a1, Spp1, Ibsp, Bglap2 and Fin1 (Komori, 2019). Smad and p38 MAPK signaling pathways regulate Runx2 promoting osteoblast and chondrocyte differentiation (Wu et al., 2016). Other key regulators of osteoblast differentiation include osteoproteregin, osterix, and alkaline phosphatase, and osteopontin. These factors are expressed at different stages of osteoblast differentiation and regulate osteoblast precursor fate decisions, bone metabolism, and osteoclast induction (Cohen, 2006).
The Groucho/TLE family of proteins are intimately involved in the regulation of various signaling pathways critical to cell fate and development, including receptor tyrosine kinase/Ras/MAPK, Notch, and Wnt signaling (Zhang and Dressler, 2013; Chodaparambil et al., 2014). The Groucho/TLE family have been extensively studied as corepressors of various binding partners, including the RUNX/AML family through the C-terminal VWRPY Groucho recruitment motif (Levanon et al., 1998; Chen and Courey, 2000). In leukemia, we have shown t(8;21) leukemic cell viability and growth are sensitive to TLE4 levels and that loss of the TLE homolog in zebrafish, Gro3, cooperates with AML1-ETO to create a myeloid leukemia phenotype (AML) (Dayyani et al., 2008). Having identified the tumor suppressor role of TLE4 in myeloid leukemias, we generated a novel Tle4 knockout mouse model to better understand its role in mammalian development (Sweetser et al., 2005; Dayyani et al., 2008; Wheat et al., 2014). In addition to various hematopoietic abnormalities, we unexpectedly found a severe bone development defect in these mice leading to severe runting and decreased bone mineralization (Wheat et al., 2014). Similar dual functions have been described for other regulators of bone development that also can function as tumor suppressor genes including FoxO members and ARF which drives bone remodeling and osteosarcoma development in mice through both p53 independent and dependent mechanisms (Rauch et al., 2010; Le et al., 2018; Ma et al., 2018; Schmitt-Ney, 2020).
Hematopoiesis and normal bone development are intimately connected (Bianco, 2011; Despars and St-Pierre, 2011). In concert with other resident bone tissue cells, osteoblasts create and protect a hospitable hematopoietic stem cell (HSC) microenvironment (Cohen, 2006; Yin and Li, 2006). Co-cultures of MSCs with various leukemic cell lines increase osteoblastic markers such as Runx2, Osx, Opn (Le et al., 2018). Initial studies of hematopoiesis in the bone marrow found many HSCs in close proximity to the inner bone endosteal area, which has triggered much inquiry into the specific relevance of osteoblasts in HSC maintenance and niche (Yin and Li, 2006; Garcia-Garcia et al., 2015). Osteoblasts communicate with HSCs through direct receptor-ligand interactions (e.g., Ang1/Tie2 and TPO/MPL), to support HSC adhesion and residence in the niche, including interactions between N-cadherin/β-catenin, and osteopontin/β1integrin (Yin and Li, 2006; Le et al., 2018). In addition, osteoblasts secrete factors including G-CSF, hepatocyte growth factor and osteopontin that regulate the pool size of the CD34+ progenitor population (Le et al., 2018). Osteoblasts regulate HSC migration in and outside of the bone marrow through CXCL12/CXCR4 and VCAM-1/VLA-4 (Le et al., 2018). The constitutive activation of β-catenin in osteoblasts and resultant expression of the Notch ligand Jagged-1 activates Notch signaling in HSC leading to the development of AML. This underscores the importance of normal regulation of mediators of osteoblast differentiation on normal hematopoiesis (Kode et al., 2014). Dicer1 or Ptpn11 deficient osteoprogenitor cells in mice display myelodysplastic syndrome and secondary acute myeloid leukemias, as well as juvenile myelomonocytic leukemia-like myeloproliferative neoplasms, respectively (Le et al., 2018). This is further demonstrated by studies that found HSC populations increased in parallel with expansion of osteoblasts due to parathyroid hormone treatment in mice (Calvi et al., 2003), while PTH activation enhances the migration of long-term repopulating HSCs (Even et al., 2021). Moreover, a previous study targeted ablation of osteoblasts in mice found that the loss of osteoblasts significantly reduced HSC and hematopoietic progenitor populations (Visnjic et al., 2004). Furthermore, animals deficient in Sca marker present bone abnormalities (Aguila and Rowe, 2005). Thus, osteoblast function intimately connects bone formation and hematopoiesis. Specifically Car/LepR+ CXCL12 expressing cells create a niche for HSCs cells while simultaneously give rise to osteoblasts (Galan-Diez and Kousteni, 2018). The similarity of Tle4 null mice to Runx2 null mice suggested the loss of Tle4 might either impair the function or the expression of Runx2. To better characterize the nature of the defect in bone development and maintenance in Tle4 null mice we have used Tle4 null and conditional Tle4 knockout mice and performed assays of osteoblast function and development in bone stromal cultures and mesenchymal bone marrow cell lines.
Materials and Methods
Generation of Tle4 Null and Conditional Tle4 Knockout Mice
For these experiments we used Tle4 null (T4KO) and Tle4 conditional knockout mice generated in our laboratory as previously described (Wheat et al., 2014). Briefly, conditional mice were constructed by targeting LoxP sites to flank exon 2 of Tle4 via homologous recombination using the 129S6/SvEvTac ES cell line (T4F). To generate T4KO, resultant mice were crossed with β-actin:Cre mice (gift of Dr. Susan Dymecki) to delete exon 2 in all tissues. Heterozygote mice were backcrossed to C57BL/6 background for over six generations and interbred to generate Tle4 null mice. For conditional knockout of Tle4, homozygous T4F mice containing Mx1-Cre (T4F cre) were used. Excision of Tle4 exon 2 was induced with three intraperitoneal injections of 15 mg/kg polyinosinic-polycytidylic acid (pIpC; Sigma) separated by 48 h. pIpC treatment induces interferon-γ signaling with activation of Cre expression and subsequent Cre recombinase excision of exon 2 of Tle4. This is predicted to cause a frameshift resulting in a premature stop codon and non-functional truncated Tle4 protein (Wheat et al., 2014). Demonstration of T4F knockout efficiency by pIpC was performed by PCR using primers mT4WTvFlpR 5′- GGAGACTTGGAAAACGCTGA-3′, mT4PcreF 5′- CAAAGGGCCCCAGAATCTT-3′and mT4PcreR 5′- CGACCGACTTGTAGCCATTT-3′. Mice were housed in a specific pathogen-free environment with a 12-h light/dark cycle, 30–70% relative humidity and approximately 70°F ambient temperature, in groups not surpassing four adult animals. Mice had ad libitum access to tap water and standard rodent chow (Prolab® RMH 3500, Scotts Distributing, Hudson, NH, United States). For analysis mice were euthanized by inhalation of 100% CO2. This study was carried out in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee.
Cell Culture, shRNA Construction, and Lentiviral Infection
ST2D cells (Generous gift of Dr. Baruch Frenkel) were generated by stably transforming mouse mesenchymal ST2 cells derived from bone marrow with a doxycycline-inducible Runx2 expression vector (Baniwal et al., 2012). ST2D cells were cultured in RPMI-1640 (Lonza, Walkersville, MD, United States) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO, United States) and 1% penicillin/streptomycin (Invitrogen, United States). Cells were maintained at 37°C and 5% CO2. When indicated, cells were also cultured with 350 ng/mL doxycycline (Sigma-Aldrich, United States) or DMSO (Sigma-Aldrich, United States). Non-targeting control (scr) and Tle4-specific shRNA constructs were developed using the lentiviral vector FUGW and delivered to cells via lentiviral delivery as previously described (Dayyani et al., 2008). The Tle4 shRNA (shTle4) used has the following target sequence: AGTGATGACAACTTGGTGG and a control scrambled shRNA (scr) CAGTCGCCATTAGTTCCAC. Infected cells were identified by GFP fluorescence detected using FACS LSRII or GFP-selected via cell sorting with FACS Aria (BD, United States).
Generation of Stromal Cultures
Stromal cultures were generated from bones of 1-week old T4WT or T4KO littermates as previously described (Mukherjee et al., 2008; Wheat et al., 2014). After harvesting femur and humeri, whole bones were crushed and plated on tissue culture plates with MEMα (Invitrogen, United States) supplemented with 20% FBS and 1% Penicillin/Streptomycin (Invitrogen, United States). After 3 days, non-adherent cells were removed and media was changed to osteogenic media containing 100 μM β-glycerophosphate, 2.84 μM ascorbic acid, and 10 nM dexamethasone. After 1 week in osteogenic media, stromal cultures were either lysed with Trizol (Invitrogen, United States) for RNA or stained for alkaline phosphatase activity (Sigma).
In osteoblast function experiments, ST2D cells were treated with Tle4-specific (T4KD) or scramble control shRNA (SCR) via lentiviral expression. One week after spinoculation, GFP + ST2D cells were selected using FACS Aria (BD, United States) and cultured in 6-well plates. Upon reaching confluence, ST2D cells were cultured in osteogenic media, with or without 350 ng/mL doxycycline. After 2 days, ST2D cultures were lysed with Trizol (Invitrogen, United States) for RNA. In a separate experiment RNA was harvested from ST2D cells cultured after stimulation with 350 ng/mL doxycycline at 24, 48, and 72 h.
Expression Analysis via qRT-PCR
RNA was harvested from whole bone lysate, stromal cultures, or ST2D cell culture using Trizol (Invitrogen, United States). Expression levels of select differentially expressed genes and others of interest were performed via qRT-PCR as previously described (Wheat et al., 2014). Briefly, the RNA was reversed transcribed using the M-MLV Reverse transcriptase kit (Invitrogen, United States), followed by the quantitative analysis using either SYBR Green system (Bio-Rad, United States) or predesigned TaqMan Gene Expression Assay (Applied Biosystems, United States). Primer sequences for SYBR Green assays are listed in Supplementary Table 1. Tle4 expression was assayed using the TaqMan Gene Expression Assay (Tle4: Mm01196934). The expression levels of genes of interest were normalized to the expression levels of the 18S housekeeping gene.
Mineral Apposition Rate Assay
For mineral apposition assay, 2-month old T4F and T4F cre littermates were irradiated with two doses of 450cGy and subsequently transplanted with 1 × 104 Lineage- c-Kit + Sca-1 + (LKS) cells from 2-month old wild-type C57BL/6 mice via tail vein injection. These transplanted wild-type LKS cells were isolated using a FACS Aria (BD, United States). Eight weeks after transplant, Tle4 excision was induced by three pIpC (Sigma) intraperitoneal injections at a dose of 15 mg/kg every 48 h. Four weeks after Tle4 excision, recipient mice were pulsed with 20 mg/kg calcein (Sigma- Aldrich, United States) via intraperitoneal injection. After 1 week, recipients were given 30 mg/kg demeclocycline (Sigma- Aldrich, United States) via intraperitoneal injection. Femurs were harvested 3 days after demeclocycline injection and fixed in 70% ethanol. Femurs were processed for resin embedding for mineral apposition rate analysis and for immunohistochemistry staining of Runx2 (PA5-86506, ThermoFisher Scientific, Waltham, MA, United States), Oc (MBS2003553, MyBiosource, San Diego, CA, United States) and β-catenin (ab6302, Abcam, Cambridge, United Kingdom) (Bouxsein et al., 2010).
MicroCT Bone Analysis
Microcomputed tomographic analysis (microCT) was performed on a subset of lumbar vertebrae. The femora were scanned at a resolution of 6 μm using a Scanco-35 microCT (Scanco United States, Inc., Southeastern, PA, United States). Each scan included a phantom containing air, saline and a bone reference material (1.18 g/cm3) for conversion of Houndsfield units to mineral density in g/cm3. Reconstruction of the individual projections to computed tomography volume data was performed using instrument software. Specimen-specific thresholds were determined by first selecting a volume of interest, generating the attenuation histogram, and determining the threshold that segments mineralized tissue from background. Properties determined included medullary bone mineral density (BMD), cortical bone thickness, AP distance, vertebral pedicle length, and trabecular bone volume fraction (trabecular bone volume to total volume ratio, BV/TV) (Bouxsein et al., 2010).
Statistics
Analyses used student’s unpaired t-test with Graphpad Prism for comparing two genotypes (Graph-pad Software, La Jolla, CA, United States). Data are presented as floating bars showing the minimum to maximum values or scatter plots, and values of p < 0.05 are considered statistically significant.
Results
Loss of Tle4 Leads to Defective Bone Development
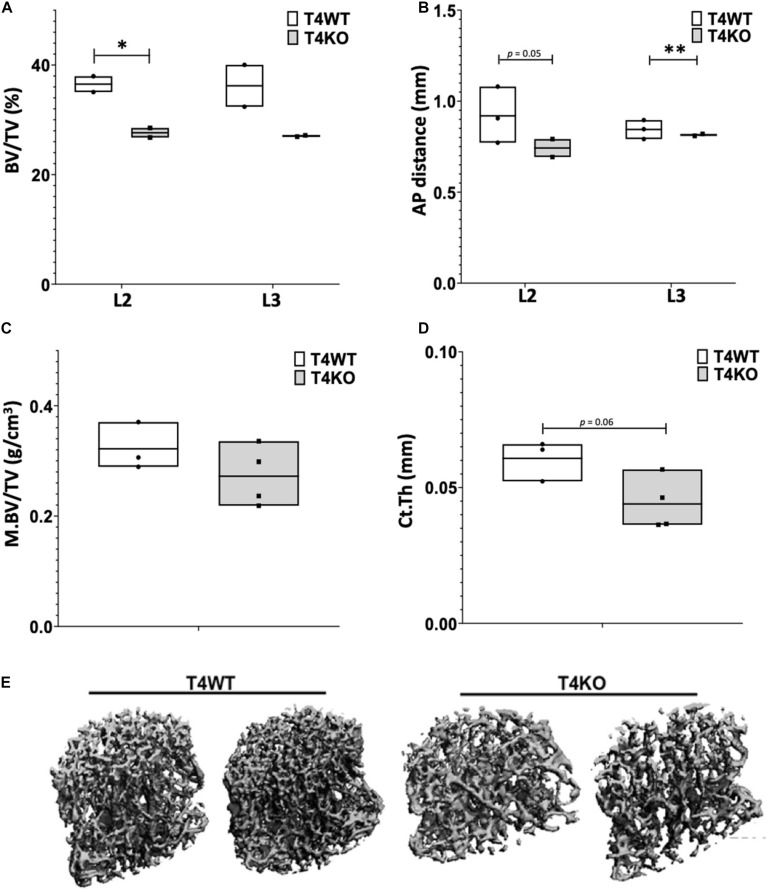
To assess bone formation in Tle4 null mice, we performed microCT analysis of lumbar vertebrae of 3-day old T4KO and wild-type (T4WT) littermates. T4KO mice exhibit decreased trabecular bone volume fraction (BV/TV) and AP distance measurements compared to T4WT counterparts, suggesting loss of Tle4 is associated with decreased bone density and vertebral pedicle length (Figures 1A,B). Additionally, femurs of 8 to 9-day-old T4KO and T4WT littermates were harvested for microCT analysis of medullary and cortical bone (Figures 1C,D). This analysis showed T4KO mice have reduced medullary bone density (M.BV/TV) and cortical bone thickness (Ct.Th). Moreover, microCT image reconstructions of trabecular bone in L3 vertebrae of 3 to 4-week old T4KO and T4WT littermates further illustrate decreased bone calcification and increased trabecular space in T4KO vertebrae compared to those of T4WT (Figure 1E). This is consistent with previous findings that loss of Tle4 is associated with deficits in vertebral and long bone formation.
FIGURE 1.
T4KO mice exhibit deficiencies in bone formation. MicroCT measurements of (A) BV/TV and (B) AP distance in L2 and L3 vertebrae of T4KO and T4WT littermates demonstrate decreased bone density and spinal canal diameters (n = 2 biologic duplicates per arm; ∗p < 0.05, ∗∗p < 0.01, Student t-test). Femurs collected from 9-day old T4KO and T4WT littermates were analyzed for (C) medullary bone density and (D) cortical thickness. Both measurements show decreased trend in T4KO vs. T4WT mice (n = 3–4 mice per arm, technical duplicates). (E) MicroCT image reconstructions of trabecular bone in L3 vertebrae of 3-to-4 weeks old T4WT and T4KO mice, illustrating decreased bone density and trabecular bone formation in Tle4 null mice (n = 2 biologic duplicates per arm).
Dynamic Bone Formation and Osteoblast Deficiencies in Conditional Tle4 Knockout Mouse
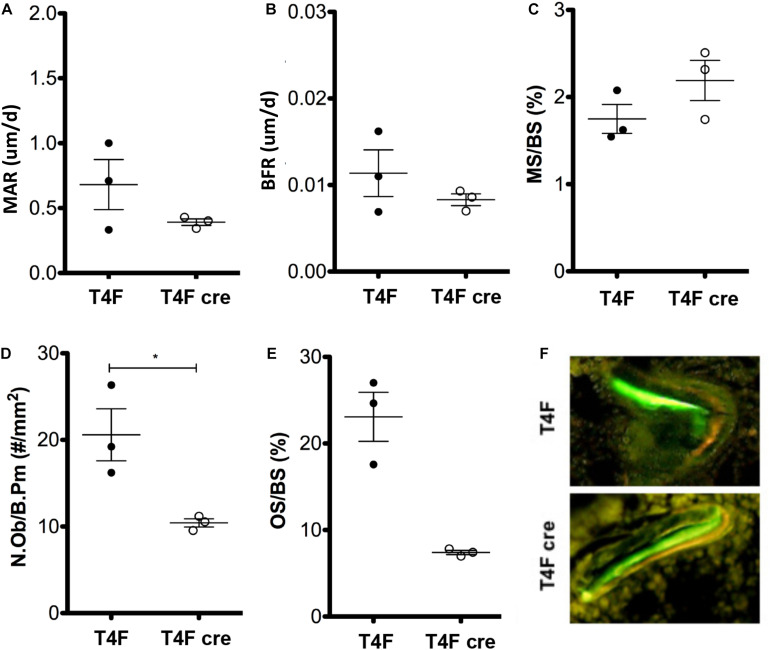
To further characterize the effects of Tle4 loss on bone development, we pursued a dynamic double-label mineral apposition rate (Calvi et al., 2003) assay using mice that have loxP target sites flanking exon 2 of Tle4 with or without Cre recombinase driven by the Mx1 promoter (T4F cre and T4F, respectively). In order to isolate the effects of Tle4 loss to bone marrow mesenchymal cells we replaced the bone marrow hematopoietic cells with wild type cells by bone marrow transplantation prior to knockout of Tle4 by pIpC in these Mx1-Cre expressing mice. The high degree of knockout efficiency of exon 2 of Tle4 in these conditional T4F-cre mice was demonstrated in harvested bone marrow of similarly pIpC treated T4F-cre compared to T4F mice lacking Mx1-cre (Supplementary Figure 1). The MAR assay revealed T4F cre mice have multiple lower dynamic parameters of bone formation (Figures 2A–F). While ratios of mineralizing to bone surface areas (MS/BS) were similar, MAR and bone formation rates (BFR) were lower in T4F cre mice compared to their control T4F counterparts (Figures 2A–C). Additionally, T4F cre mice had lower numbers of osteoblasts per given bone perimeter area (Figure 2D). Combined with lower osteoid to bone surface area ratios (Figure 2E), these results point toward an association between decreased bone formation and conditional loss of Tle4 in adult mice.
FIGURE 2.
MAR assay and trichrome staining results indicate contrasting effects of Tle4 loss on bone formation and calcification. T4F cre mice of 8-to-9 days old have decreased (A) MAR and (B) BFR rates, measurements of average bone calcification per osteoblast and total bone formation, respectively, while having similar (C) ratio of mineralizing to bone surface ratios. (D) T4F cre mice also had less N.Ob/B.Pm values, an estimate of the number of osteoblasts per area of bone surface. (E) Trichrome staining reveals T4F cre mice have significantly reduced osteoid compared to T4F controls (n = 3 mice per arm; *p < 0.05, Student t-test). (F) Representative image demonstrating decreased bone formation in T4F cre mice vs. T4F mice via MAR double-stain of calcein and demeclocycline.
Bone Defects Due to Tle4 Loss May Be Mediated Through Dysregulation of Canonical Regulators of Bone Development
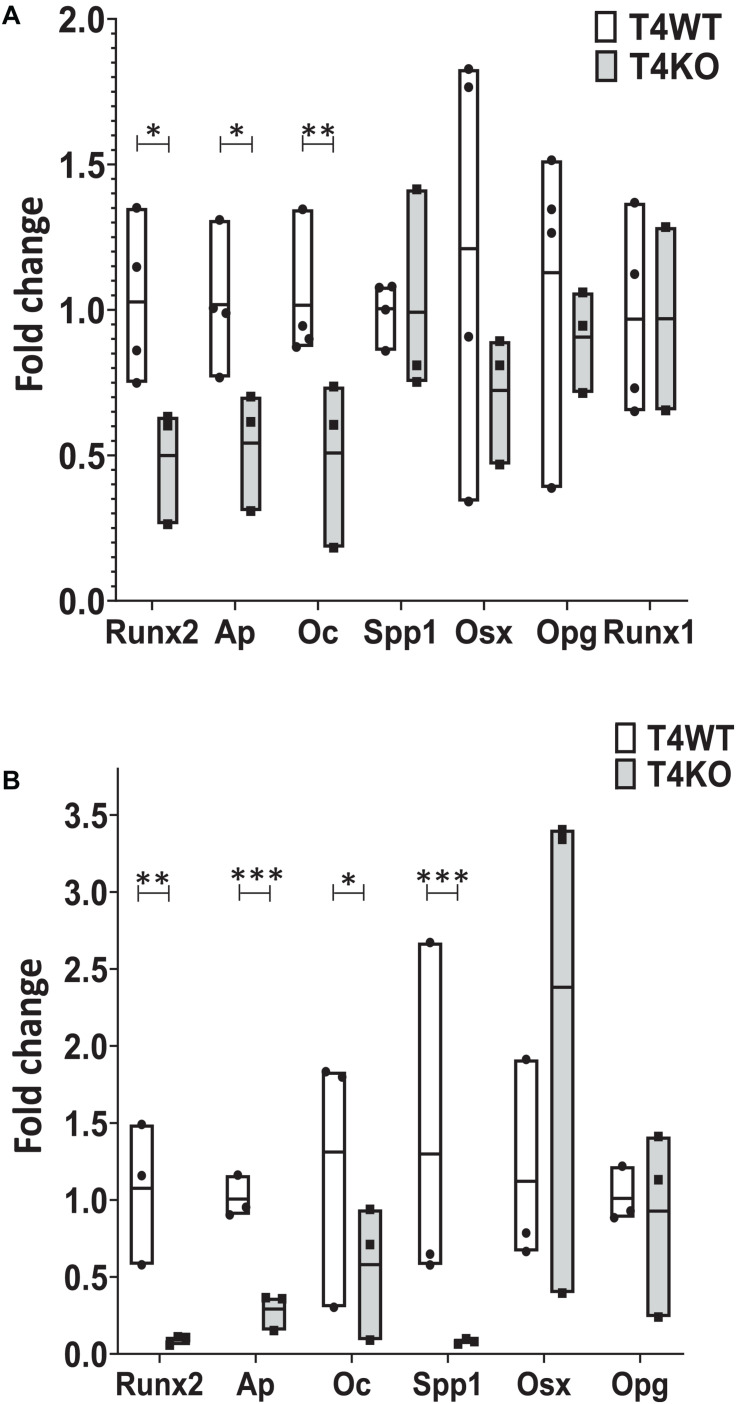
Given the defective bone and bone marrow phenotype seen in T4KO mice, we hypothesized that loss of Tle4 may affect osteoblast function and development. To assess this, we first harvested RNA from flushed whole bone lysates of 1-week-old T4KO and T4WT littermates. Expression analysis, using qPCR, revealed T4KO bone had significantly lower levels of Ap, Runx2, and Oc expression (Figure 3A). Ap is often used as a proxy for osteoblast function while Runx2 and Oc have previously been connected to osteoblast maturation and bone anabolic regulation (Jang et al., 2012). To minimize bone cell heterogeneity, crushed 1-week-old T4KO and T4WT femurs were cultured in osteogenic media to generate osteoblast stromal cultures. T4KO stromal cultures demonstrated significant decreases in osteoblast genes, including Runx2, Ap, Oc, and Spp1 (Figure 3B). Moreover, alkaline phosphatase staining qualitatively showed a trend toward decreased alkaline phosphatase activity in T4KO stromal cells (Supplementary Figure 2).
FIGURE 3.
Loss of Tle4 affects expression of osteoblast function and differentiation regulators. (A) qRT-PCR using RNA from 1-week old T4WT and T4KO flushed whole bone lysate shows T4KO has decreased expression of Ap, Runx2, and Oc (n = 6 biologic replicates, technical triplicates; *p < 0.05, **p < 0.01, Student t-test). (B) Stromal cultures from T4WT and T4KO were lysed for qRT-PCR analysis, which shows significantly reduced levels of Runx2, Ap, Oc, and Spp1 expression in T4KO compared to T4WT (n = 3 biologic replicates per arm, technical triplicates; *p < 0.05, **p < 0.01, ***p < 0.001, and Student t-test).
Our T4KO results revealed that absence of Tle4 was associated with significant reductions of Runx2, suggesting T4KO-associated bone abnormalities might be due to decreased Runx2 expression. To further elucidate the time course of Tle4-mediated effects on bone development factors through the Runx axis and considering the recent data on the Runx1 involvement in bone development (Tang et al., 2020), we assessed the expression levels of Runx1 in the 1-week-old T4KO and T4WT littermates, and found that the absence of Tle4 does not affect the expression levels of Runx1 (Figure 3A) suggesting the calcification defects in Tle4 KO mice are more likely explained by Runx2 inhibition.
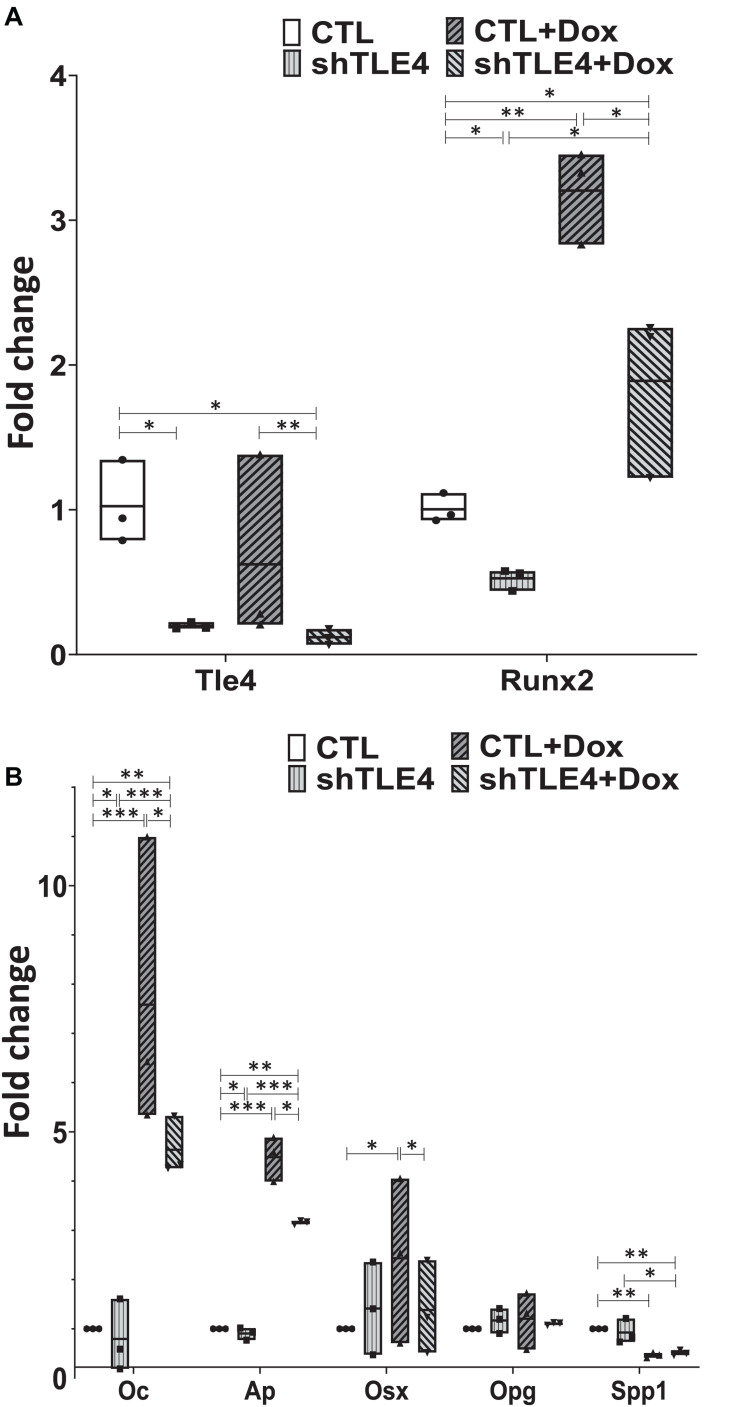
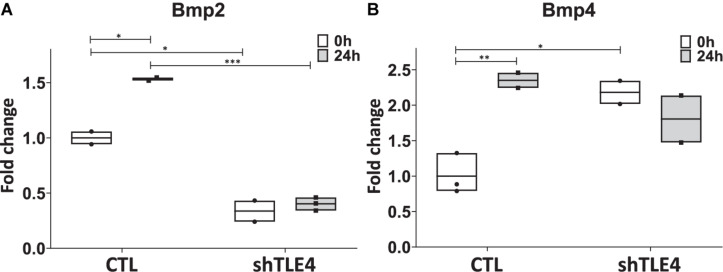
To better understand the mechanisms underlying Tle4 effect on bone development and the interplay with Runx2, we turn to an in vitro system using mouse mesenchymal ST2 cells stably transformed with a doxycycline-inducible Runx2 vector (ST2D) (Dayyani et al., 2008; Baniwal et al., 2012). In these ST2D cells the addition of doxycycline leads to an induction of Runx2 expression (Figure 4A). In this system we evaluated the effects of knocking down Tle4 expression via lentiviral delivery of Tle4-specific shRNA. Expression analysis using qPCR demonstrated over 80% reduction in Tle4 message via shRNA in both doxycycline and DMSO cultures and showed that ectopic expression of Runx2 in the absence of Tle4 shRNA did not significantly affect Tle4 expression levels (Figure 4A). In this system we demonstrated Tle4 knockdown reduced endogenous Runx2 expression levels by approximately 50% in ST2D cells cultured in control DMSO media. This result correlated nicely with that found in vivo. The addition of doxycycline significantly increased Runx2 expression levels. Tle4 knockdown via shRNA was not able to prevent this increase driven from a lentiviral promoter, though the increase was blunted, likely reflecting repression of Runx2 from the endogenous promoter. Induction of Runx2 by doxycycline resulted in a significant increase in the expression of Oc, Ap, and Osx after 48 h, and this increase was significantly decreased in Tle4 knockdown arms (Figure 4B). To further understand the relationship between Tle4 and Runx2, we queried expression levels of Bmp2 and Bmp4, given their previously described roles as regulators of Runx2 and normal skeletal development (Bandyopadhyay et al., 2006; Krishnan et al., 2006). We demonstrated that loss of Tle4 caused a significant reduction of Bmp2 expression at baseline and prevented upregulation in response to Runx2 induction. Interestingly, levels of Bmp4 were increased with Tle4 knockdown, but the upregulation seen with Runx2 expression was blocked (Figure 5).
FIGURE 4.
Tle4 knockdown in ST2D mouse mesenchymal cells demonstrates similarities in aberrant expression of osteoblast regulator genes seen in osteoblast cultures derived from T4KO mice. (A) qRT-PCR using RNA harvested from Tle4 knockdown (shTle4) and control (CTL) ST2D cells using scrambled shRNA confirm effective Tle4 knockdown and Runx2 induction after 48 hr of doxycycline induction which remains decreased compared to the scramble controls. (B) qRT-PCR experiments show expression of Ap and Oc are significantly reduced in Tle4 knockdown ST2D cells compared to control. Doxycycline-induced expression of Runx2 significantly increases expression of these genes compared to DMSO control, remaining however, in lower levels compared to scramble (n = 3 biologic replicates per arm, technical triplicates; *p < 0.05, **p < 0.01, ***p < 0.001, and Student t-test).
FIGURE 5.
Tle4 knockdown in ST2D cells cause aberrant levels of Bmp2 and Bmp4 expression in response to Runx2 expression. (A) Twenty-four hours after Runx2 induction, upregulation of Bmp2 in control (CTL) cells. Knockdown of Tle4 represses Bmp2 expression and blocks upregulation with Runx2 induction. (B) Induction of Runx2 also leads to an upregulation of Bmp4 in control cells. This upregulation is not seen in the presence of Tle4 knockdown (n = 3 biologic replicates per arm, technical triplicates; *p < 0.05, **p < 0.01, ***p < 0.001, and Student t-test).
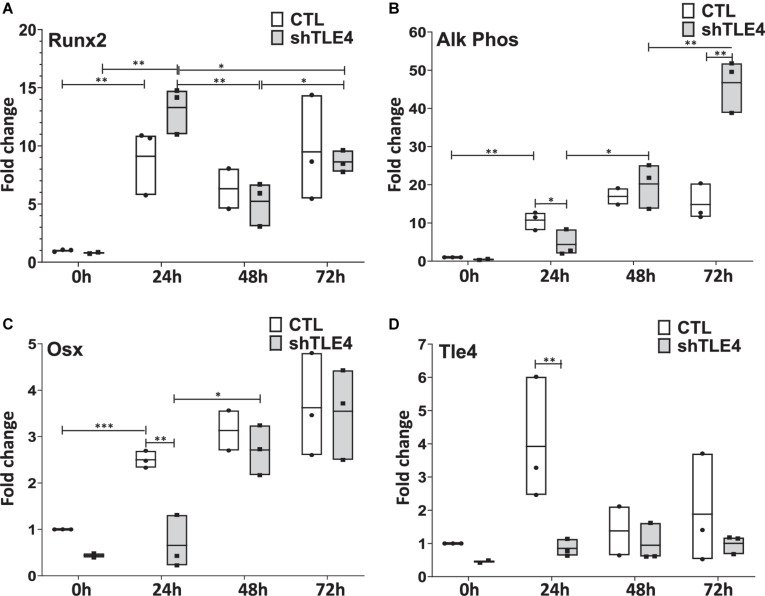
Extended time-course experiments demonstrate that Tle4 knockdown creates an initial surge of Runx2 expression by 24 h of knockdown compared to control, followed by decreased Runx2 levels at 48 and 72 h; consistent with the above observations (Figure 6). However, we found decreased expression levels of Runx2-mediated regulators of bone, including Alp and Osx at 24 h. This suggests the loss of Tle4 blunted the ability of Runx2 to upregulate Osx and Alp expression at early time points. By 72 h after Tle4 knockdown, the differential expression of these genes is lost, suggesting a time-delay of Alp and Osx induction due to Tle4 knockdown. These experiments support a role of Tle4 in regulating Runx2 and, subsequently, its target genes canonically associated with osteoblast differentiation and function.
FIGURE 6.
Time-course gene expression study demonstrates altered expression of Runx2 and its bone-related targets due to Tle4 knockdown (shTLE4) compared with control cells (CTL) in ST2D cells at 0, 24, 48, and 72 h. (A) ST2D cells that contain a doxycycline inducible Runx2 gene demonstrated maximal Runx2 induction after 24 h. (B) This is associated with an upregulation of alkaline phosphatase that was slightly delayed with Tle4 knockdown. (C) Tle4 knockdown also slows the induction of the osteoblast-specific transcription factor Osterix (Osx). (D) The knockdown of Tle4 achieved with a Tle4 specific shRNA was demonstrated at the different time points in this experiment. (0—knockdown to 37%, 24 h to 22%, 48 h to 69%, 72 h to 53%) (n = 3 biologic replicates per arm, technical triplicates; *p < 0.05, **p < 0.01, ***p < 0.001, and Student t-test).
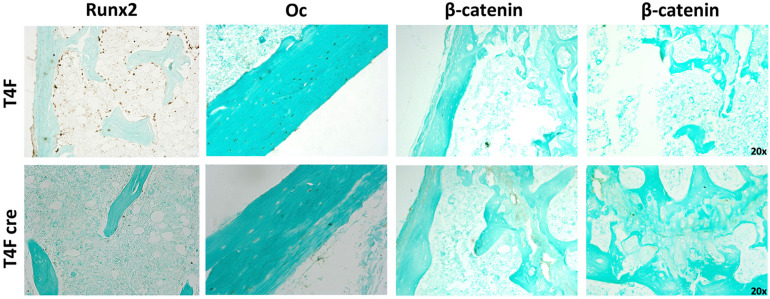
Immunohistochemistry using femurs from mice used in MAR assay revealed decreased amounts of Runx2 and Osteocalcin in T4F cre mice compared to control (Figure 7). In T4F mice, Runx2 positivity can be seen in numerous osteoid-lining cells, suggestive of Runx2-positive periosteal osteoblasts. The frequency and localization of these Runx2-positive cells dramatically drops in T4F cre mice. Similarly, osteocalcin positivity in cortical bone seen in T4F mice is not evident in the conditional Tle4 knockout mice. In addition to Runx2, β-catenin-mediated Wnt signaling has also been previously described as an important mediator of bone development and osteoblast differentiation (Hill et al., 2005; Cohen, 2006; Rodda and McMahon, 2006; Kook et al., 2015). Given previous reports implicating Tle4 as a negative regulator of Wnt signaling, we queried β-catenin levels via immunohistochemistry in bones of the T4F and T4F cre mice used in the above MAR assay as an exploratory objective. T4F cre femurs showed increased β-catenin signals in the epiphyseal and cortical areas. While T4F cre and T4KO mice display bone calcification and osteoblast function defects, loss of Tle4 in bone leads to increased β-catenin levels, which is consistent with previous reports implicating Tle4 as a repressor of Wnt signaling. However, using the above-described ST2D system, we were not able to find a Tle4 knockdown-dependent change in expression of canonical Wnt target gene expression (data not shown).
FIGURE 7.
T4F cre mice have significantly reduced levels of key regulators of bone development and osteoblast function. Immunohistochemistry assay using femurs harvested from T4F cre mice of 8-to-9 days old in MAR assay reveals decrease in Runx2 and Oc levels and increased β-catenin signals in the epiphyseal and cortical areas compared to T4F control counterparts (n = 3 mice per arm, representative images).
Discussion
The Groucho/TLE family of proteins has been extensively studied in Drosophila where it has been termed a master regulatory gene in development via its interaction with a number of important signaling pathways including Notch and Wnt and also can be recruited by transcription factors members of Hex, Runx, Nkx, Lef1/Tcf, Pax, Six and c-myc (Jennings and Ish-Horowicz, 2008; Agarwal et al., 2015). Our understanding of the roles of this protein family in vertebrate development is limited. The novel Tle4 null mouse provides valuable insight into the previously unappreciated roles of Tle4 in mammalian vertebrates regarding bone maturation, medullary hematopoiesis and HSPC maintenance. One of the striking abnormalities in the T4KO mice is a decreased calcification of the skeleton (Wheat et al., 2014). This impaired ossification is apparent in both membranous and endochondral bones by birth. The above phenotypes are more intense and progressive in an age-dependent manner in T4KO mice than in Grg5 null mice lacking a truncated member of the Groucho/TLE family (Wheat et al., 2014). Our earlier characterization of Tle4 KO mice suggested Tle4 also affects osteoclast function as demonstrated by an increase in osteoclasts by tartrate-resistance acid phosphatase (TRAP) staining (Wheat et al., 2014). In the current study microCT and MAR assay measurements indicate loss of Tle4 impaired bone formation, calcification, and osteoid production. The lethality of Tle4 null mice and the effects in hematopoietic cells and observed degradation of the bone marrow niche made it technically challenging to isolate the effect on osteoblasts in germline knockout mice. Conditional Tle4 mice transplanted with normal bone marrow hematopoietic cells served as a proxy for osteoblast-specific effects of Tle4 loss. Mx1-cre system has been demonstrated as one of the most commonly “deleter strain” in experimental hematology (Velasco-Hernandez et al., 2016). Previous work by Park et al. (2012) have demonstrated the contributory role of Mx1-expressing bone mesenchymal cells toward the generation of new osteoblasts responsible for new bone formation, supporting the use of the Mx1-Cre model in our experiments to examine the role of Tle4 loss in osteoblast function (Park et al., 2012). The concordance of observations seen in these models and studies strongly suggest that Tle4 may affect osteoblasts and other periosteal cells that are responsible for bone production and maintenance.
In the context of our previous work identifying hematopoietic defects in Tle4 null mice, we have attempted to determine whether the bone abnormalities due to Tle4 loss can be attributable to dysfunctional osteoblasts. We had shown Tle4 loss significantly impairs LSK differentiation into granulocyte, monocyte, macrophage progenitors and LSK self-renewal and adversely affects the integrity of bone marrow niche and stroma (Wheat et al., 2014). Osteoblasts are known to play a critical role in maintaining the bone marrow niche (Calvi et al., 2003; Asada et al., 2013; Fulzele et al., 2013; Even et al., 2021). In this current work qRT-PCR analysis using T4KO mouse samples from flushed whole bones and cultured stromal cells revealed significantly decreased expression of various transcription factors and regulators responsible for osteoblast function and differentiation, including Ap and Oc—both frequently used proxies for describing osteoblast function. Interestingly, osteocalcin is one of the main components of ground substance that, together with Type 1 collagen, constitute the bone matrix (Hill et al., 2005; Asada et al., 2013; Fulzele et al., 2013). Osx demonstrates a multifunctional role on osteoblast differentiation, growth and homeostasis, since its deletion in several time points postnatally in growing and adult bones causes defects in maturation, morphology and function of osteocytes (Zhou et al., 2010; Liu et al., 2020). Decreased Oc expression may lend insight into a physiologic basis for the Tle4 knockout-induced bone phenotype.
Wnt signaling has been described as a central mediator of bone formation (Hill et al., 2005; Cohen, 2006; Rodda and McMahon, 2006; Kook et al., 2015). Surprisingly, we observed defective bone formation and calcification in T4KO mice, in which Wnt signaling is expected to be activated especially given higher levels of β-catenin signal as determined by immunohistochemistry (Kronenberg, 2003; Chodaparambil et al., 2014). However, previous studies have shown that there is an intimate relationship between the timing of Wnt signaling and normal osteoblast differentiation; and thus, constitutive or increased Wnt signaling at an inappropriate stage of osteoblast development may be detrimental to normal bone growth (Rodda and McMahon, 2006; Janeway and Walkley, 2010). Alternatively, the detrimental effects of Tle4 loss on Runx2 activity might outweigh the effects of Wnt activation on osteoblast differentiation and calcification. Our results do not preclude the possibility that loss of Tle4 is responsible for the bone defects through other mechanisms and the potential effects of paracrine hormonal or Tle4 levels in other non-Mx1-expressing compartments.
The ST2D system provided insight into potential molecular mechanisms that may explain our findings, including not only Wnt signaling but also dysregulation of Bmp signaling and consequently Runx2 expression. Previous studies have demonstrated loss of function of Bmp2 and Bmp4 impair bone condensation and skeletal development (Bandyopadhyay et al., 2006; Krishnan et al., 2006; Wu et al., 2016). There is an interplay between BMP and RUNX2 in regulating osteoblast differentiation (Lowery and Rosen, 2018). BMP signaling is required for transcriptional activity of Runx2 and Runx2 enhances the sensitivity of cells to BMPs (Phimphilai et al., 2006).
Our experiments indicate there is a decrease in Runx2 expression in T4KO bone, T4KO stromal cells, and in ST2D cells with Tle4 knockdown in a time dependent manner. Previous studies have shown that TLE proteins are capable of interacting with Runx2, a critical regulator of bone development and maturation (McLarren et al., 2000; Choi et al., 2001; Kaul et al., 2015). The RUNX protein family is known to form co-repressor complexes with TLE proteins (Javed et al., 2000). Thus, the TLE proteins might affect both Runx2 expression as well as the activity of the Runx2 protein. The TLE effect on Runx2 expression could reflect interference of Runx2 transcriptional autoregulation (Drissi et al., 2000). In our studies, even if the experiments do not distinguish whether the blunting in the expression of these osteoblast genes with Tle4 knockdown is due to decreased Runx2 expression or decreased function of the Runx2 protein in the absence of Tle4, the differential expression levels of Runx2-mediated regulators of bone development are most likely a downstream effect consequent of the decreased endogenous Runx2 expression after Tle4 knockdown. Runx2 null mice demonstrate bone phenotypes similar, but more severe than our T4KO mouse: expiring at birth and completely missing skeletal and bone development, owing to defective osteoblast maturation (Komori et al., 1997; Otto et al., 1997; Okura et al., 2014). Previous studies have shown the importance of Runx2 in normal bone development, as Runx2 null mice lack bone ossification and osteoblast differentiation (Cohen, 2006). The similarity of Tle4 null mice to Runx2 null mice suggested the loss of Tle4 might either impair the function or the expression of Runx2. The less severe effect observed with Tle4 knockout could reflect complementary effects from the expression of other Tle family members along with Runx2 residual expression which thus induce the osteoblastic genes expression later during bone development in the Tle4 loss background, in contrast to the early in time severe effects of Tle4 loss in bone development. Additional studies are required to further characterize the interaction and potential regulatory role of Tle4 on Runx2 expression levels and function as it may relate to the defective bone development in the absence of Tle4. While direct functional interactions between RUNX and TLE have been described (Westendorf, 2006) and a possibility of the requirement of such a direct active interaction in the early osteoblastic development is suggested in our time dependent experiments, additional experiments may reveal further insight into whether TLE exerts a direct regulatory effect on RUNX transcription, stability or targets.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee.
Author Contributions
DS, TS, and ET: study conception and design, writing manuscript, and study supervision. TS, ET, CH, RY, CR, and PG: study conduct and data acquisition. TS, ET, CR, PG, and DS: methodology development and data analysis and interpretation. ET and DS: revising manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Baruch Frenkel and Jiali Yu for generous gift of ST2D cells and scientific discourse. We would also like to thank Nicolas Severe, Janaina Da Silva, and Marie Demay for guidance with mineral apposition rate assay and bone histology. We would also further like to thank Meredith Weglarz, Amy Galvin, and Maris Handley for assistance with flow cytometry and cell sorting. The authors would also like to thank Joanna Yeh, Yiyun Zhang, Matthew Jones, George Murphy, and Bob Varelas for their guidance, comments, and discourse. The authors gratefully acknowledge the support of the Malika Ray, Asok K. Ray, FRCS/(Edin) Initiative for Child Health.
Footnotes
Funding. This work was supported in part by the National Institutes of Health Grant R01 CA115772 (DS), Swim Across America (TS and DS), The Hill Family Fund for the Diagnosis and Management of Rare and Undiagnosed Diseases at Mass General (DS and ET), the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under Award Number R03HD099516 (PG), and The Malika Ray, Asok K. Ray, M.D., FRCS/(Edin) Initiative for Child Health Research (PG).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.671029/full#supplementary-material
References
- Agarwal M., Kumar P., Mathew S. J. (2015). The Grpucho/Transducin-like enhacer of split protein family in animal development. IUBMB Life 71: 1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguila H., Rowe D. (2005). Skeletal development, bone remodeling, and hematopoiesis. Immunol. Rev. 208 7–18. 10.1111/j.0105-2896.2005.00333.x [DOI] [PubMed] [Google Scholar]
- Asada N., Katayama Y. (2014). Regulation of helatopoiesis in endosteal microenvironments. Int. J. Hematol. 99 679–684. 10.1007/s12185-014-1583-1 [DOI] [PubMed] [Google Scholar]
- Asada N., Katayama Y., Sato M., Minagawa K., Wakahashi K., Kawano H., et al. (2013). Matrix-embedded osteocytes regulate mobilization of hematopoietic stem/progenitor cells. Cell Stem Cell 12 737–747. 10.1016/j.stem.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Tsuji K., Cox K., Harfe B. D., Rosen V., Tabin C. J. (2006). Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2:e216. 10.1371/journal.pgen.0020216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal S. K., Shah P. K., Shi Y., Haduong J. H., Declerck Y. A., Gabet Y., et al. (2012). Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells. Osteoporos. Int. 23 1399–1413. 10.1007/s00198-011-1728-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P. (2011). Bone and hematopoietic niche: atale of two stem cells. Blood 117 5281–5288. 10.1182/blood-2011-01-315069 [DOI] [PubMed] [Google Scholar]
- Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J., Muüller R. (2010). Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25 1468–1486. 10.1002/jbmr.141 [DOI] [PubMed] [Google Scholar]
- Calvi L., Adams G., Weibrecht K., Weber J. M., Olson D. P., Knight M. C., et al. (2003). Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425 841–845. 10.1038/nature02040 [DOI] [PubMed] [Google Scholar]
- Chen Q., Courey A. (2000). Groucho/TLE family proteins and transcriptional repression. Genes Chromosomes Cancer 249 1–16. 10.1016/s0378-1119(00)00161-x [DOI] [PubMed] [Google Scholar]
- Chodaparambil J. V., Pate K. T., Hepler M. R., Tsai B. P., Muthurajan U. M., Luger K., et al. (2014). Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 33 719–731. 10.1002/embj.201387188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Pratap J., Javed A., Zaidi S. K., Xing L., Balint E., et al. (2001). Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98 8650–8655. 10.1073/pnas.151236498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. M., Jr. (2006). The new bone biology: pathologic, molecular, and clinical correlates. Am. J. Med. Genet. A 140 2646–2706. 10.1002/ajmg.a.31368 [DOI] [PubMed] [Google Scholar]
- Dayyani F., Wang J., Yeh J. R., Ahn E. Y., Tobey E., Zhang D. E., et al. (2008). Loss of TLE1 and TLE4 from the del(9q) commonly deleted region in AML cooperates with AML1-ETO to affect myeloid cell proliferation and survival. Blood 111 4338–4347. 10.1182/blood-2007-07-103291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despars G., St-Pierre Y. (2011). Bidirectional interactions between bone metabolism and hematopoiesis. Exp. Hematol. 39 809–816. 10.1016/j.exphem.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Drissi H., Luc Q., Shakoori R., Chuva De Sousa Lopes S., Choi J. Y., Terry A., et al. (2000). Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184 341–350. [DOI] [PubMed] [Google Scholar]
- Even J., Yi L., Chang C. K., Rossi F. M. V. (2021). The parathyroid hormone-dependent activation of osteoblasts enhances hematopoietic stem cell migration and reduces their engraftment abilities. bioRxiv [Preprint]. 10.1101/2021.03.04.433901 [DOI] [Google Scholar]
- Fulzele K., Krause D. S., Panaroni C., Saini V., Barry K. J., Liu X., et al. (2013). Myelopoiesis is regulated by osteocytes through Gsalpha-dependent signaling. Blood 121 930–939. 10.1182/blood-2012-06-437160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Diez M., Kousteni S. (2018). A bone marrow niche-derived molecular switch between oestogenesis and hematopoiesis. Genes Dev. 32 324–326. 10.1101/gad.314013.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia A., de Castillejo C. L., Mendez-Ferrer S. (2015). BMSCs and hematopoiesis. Immunol. Lett. 168 129–135. 10.1016/j.imlet.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Hill T. P., Später D., Taketo M. M., Birchmeier W., Hartmann C. (2005). Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 8 727–738. 10.1016/j.devcel.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Houschyar K. S., Tapking C., Borelli M. R., Popp D., Duscher D., Maan Z. N., et al. (2019). Wnt pathway in bone repair and regeneration-what do we know so far. Front. Cell Dev. Biol. 6:170. 10.3389/fcell.2018.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway K. A., Walkley C. R. (2010). Modeling human osteosarcoma in the mouse: from bedside to bench. Bone 47 859–865. 10.1016/j.bone.2010.07.028 [DOI] [PubMed] [Google Scholar]
- Jang W. G., Kim E. J., Kim D. K., Ryoo H. M., Lee K. B., Kim S. H., et al. (2012). BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J. Biol. Chem. 287 905–915. 10.1074/jbc.m111.253187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A., Guo B., Hiebert S., Choi J. Y., Green J., Zhao S. C., et al. (2000). Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBFa/AML/PEBP2a) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113 2221–2231. 10.1242/jcs.113.12.2221 [DOI] [PubMed] [Google Scholar]
- Jennings B. H., Ish-Horowicz D. (2008). The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9:205. 10.1186/gb-2008-9-1-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul H., Hall B. K., Newby C., Ventikos Y. (2015). Synergistic activity of polarised osteoblasts inside condensations cause their differentiation. Sci. Rep. 5:11838. 10.1038/srep11838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode A., Manavalan J. S., Mosialou I., Bhagat G., Rathinam C. V., Luo N., et al. (2014). Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature 506 240–244. 10.1038/nature12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. (2019). Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 20:1694. 10.3390/ijms20071694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formationowing to maturational arrest od osteoblasts. Cell 89 755–764. 10.1016/s0092-8674(00)80258-5 [DOI] [PubMed] [Google Scholar]
- Kook S. H., Heo J. S., Lee J. C. (2015). Crucial roles of canonical Runx2-dependent pathway on Wnt1-induced osteoblastic differentiation of human periodontal ligament fibroblasts. Mol. Cell. Biochem. 402 213–223. 10.1007/s11010-015-2329-y [DOI] [PubMed] [Google Scholar]
- Kozhemyakina E., Lassar A., Zelzer E. (2015). A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 142 817–831. 10.1242/dev.105536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Bryant H. U., Macdougald O. A. (2006). Regulation of bone mass by Wnt signaling. J. Clin. Invest. 116 1202–1209. 10.1172/jci28551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H. (2003). Developmental regulation of the growth plate. Nature 423 332–335. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Le P. M., Andreeff M., Battula V. L. (2018). Osteogenic niche in the regulation of normal hematopoiesis and leukemogenesis. Haematologica 103 1945–1955. 10.3324/haematol.2018.197004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Goldstein R., Bernstein Y., Tang H., Goldenberg D., Stifani S., et al. (1998). Transcriptional repression by AML1 and FEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. U.S.A. 95 11590–11595. 10.1073/pnas.95.20.11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Li M., Wang S., Xiao Z., Xiong Y., Wang G. (2020). Recent advances of osterix transcription factor in osteoblast differentiation and bone formation. Front. Cell Dev. Biol. 8:601224. 10.3389/fcell.2020.601224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery J., Rosen V. (2018). The BMP pathway and its inhibitor in the skeleton. Physiol. Rev. 98 2431–2452. 10.1152/physrev.00028.2017 [DOI] [PubMed] [Google Scholar]
- Ma X., Liu Y., Liu Y., Alexandrov L. B., Edmonson M. N., Gawad C., et al. (2018). Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555 371–376. 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren K., Lo R., Grbavec D., Thirunavukkarasu K., Karsenty G., Stifani S. (2000). The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the Runt-related factor Cbfa1. J. Biol. Chem. 275 530–538. 10.1074/jbc.275.1.530 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Raje N., Schoonmaker J. A., Liu J. C., Hideshima T., Wein M. N., et al. (2008). Pharmacologic targeting of a stem/progenitor population in vivo is associated with enhanced bone regeneration in mice. J. Clin. Invest. 118 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura H., Sato S., Kishikawa S., Kaneto S., Nakashima T., Yoshiba N., et al. (2014). Runx2-I isoform contributes to fetal bone formation even in the absence of specific N-terminal amino acids. PLoS One 9:e108294. 10.1371/journal.pone.0108294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F., Thornell A., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., et al. (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell Stem Cell 89 765–771. 10.1016/s0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- Park D., Spencer J. A., Koh B. I., Kobayashi T., Fujisaki J., Clemens T. L., et al. (2012). Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10 259–272. 10.1016/j.stem.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phimphilai M., Zhao Z., Boules H., Roca H., Franceschi R. T. (2006). BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 21 637–646. 10.1359/jbmr.060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch D. A., Hurchla M. A., Harding J. C., Deng H., Shea L. K., Eagleton M. C., et al. (2010). The ARF tumor suppressor regulates bone remodeling and osteosarcoma development in mice. PLoS One 5:e15755. 10.1371/journal.pone.0015755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda S. J., McMahon A. P. (2006). Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133 3231–3244. 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- Schmitt-Ney M. (2020). The FOXO’s advantages of being a family: considerations on function and evolution. Cells 9:787. 10.3390/cells9030787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D. A., Peniket A. J., Haaland C., Blomberg A. A., Zhang Y., Zaidi S. T., et al. (2005). Delineation of the minimal commonly deleted segment and identification of candidate tumor-suppressor genes in del(9q) acute myeloid leukemia. Genes Chromosomes Cancer 44 279–291. 10.1002/gcc.20236 [DOI] [PubMed] [Google Scholar]
- Tang J., Xie J., Chen W., Tang C., Wu J., Wang Y., et al. (2020). Runt-related transcription factor 1 is required for murine osteoblast differentiation and bone formation. J. Biol. Chem. 295 11669–11681. 10.1074/jbc.ra119.007896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Sawen P., Bryder D., Cammenga J. (2016). Potential pitfalls of the Mx1-cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Rep. 7 11–18. 10.1016/j.stemcr.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnjic D., Kalajzic Z., Rowe D. W., Katavic V., Lorenzo J., Aguila H. L. (2004). Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood 103 3258–3264. 10.1182/blood-2003-11-4011 [DOI] [PubMed] [Google Scholar]
- Westendorf J. J. (2006). Transcriptional co-repressors of Runx2. J. Cell. Biochem. 98 54–64. 10.1002/jcb.20805 [DOI] [PubMed] [Google Scholar]
- Wheat J. C., Krause D. S., Shin T. H., Chen X., Wang J., Ding D., et al. (2014). The corepressor Tle4 is a novel regulator of murine hematopoiesis and bone development. PLoS One 9:e105557. 10.1371/journal.pone.0105557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Chen G., Li Y. P. (2016). TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Li L. (2006). The stem cell niches in bone. J. Clin. Invest. 116 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Dressler G. R. (2013). The Groucho protein Grg4 suppresses Smad7 to activate BMP signaling. Biochem. Biophys. Res. Commun. 440 454–459. 10.1016/j.bbrc.2013.09.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang Z., Feng J. Q., Dusevich V. M., Sinha K., Zhang H., et al. (2010). Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc. Natl. Acad. Sci. U.S.A. 107 12919–12924. 10.1073/pnas.0912855107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.