Abstract
To date, the overall response rate of PD-1 blockade remains unsatisfactory, partially due to limited understanding of tumor immune microenvironment (TIME). B-cell lymphoma 9 (BCL9), a key transcription co-activator of the Wnt pathway, is highly expressed in cancers. By genetic depletion and pharmacological inhibition of BCL9 in tumors, we found that BCL9 suppression reduced tumor growth, promoted CD8+ T cell tumor infiltration, and enhanced response to anti-PD-1 treatment in mouse colon cancer models. To determine the underlying mechanism of BCL9’s role in TIME regulation, single-cell RNA-seq was applied to reveal cellular landscape and transcription differences in the tumor immune microenvironment upon BCL9 inhibition. CD155-CD226 and CD155-CD96 checkpoints play key roles in cancer cell/CD8+ T cell interaction. BCL9 suppression induces phosphorylation of VAV1 in CD8+ T cells and increases GLI1 and PATCH expression to promote CD155 expression in cancer cells. In The Cancer Genome Atlas database analysis, we found that BCL9 expression is positively associated with CD155 and negatively associated with CD226 expression. BCL9 is also linked to adenomatous polyposis coli (APC) mutation involved in patient survival following anti-PD-1 treatment. This study points to cellular diversity within the tumor immune microenvironment affected by BCL9 inhibition and provides new insights into the role of BCL9 in regulating CD226 and CD96 checkpoints
Subject terms: Cancer microenvironment, Cancer genomics, Tumour immunology
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide.1 In the last few years, significant new insights into the molecular pathways underlying CRC have provided several new therapeutic options.2–4 However, despite the advances in chemotherapeutic and combined targeted treatment options, most patients with metastatic CRC still exhibit poor survival. As such, there is still an unmet need for more effective treatments.5,6 Recently, new therapeutic strategies that reinvigorate the immune response directed towards cancer cells were developed. Some of these have successfully paved their way to the clinic, for instance, immune checkpoint inhibitors directed against programmed cell death protein 1 (PD-1) or cytotoxic T lymphocyte-related protein 4 (CTLA-4) are effective against microsatellite-unstable CRC.7 Nevertheless, significant challenges remain, especially for microsatellite-stable CRC, which is characterized by a poor response to these checkpoint inhibitors.7
The Wnt signaling pathway is a tightly regulated and receptor-mediated transduction pathway that is involved in embryonic development and adult tissue homeostasis.8,9 It plays a prominent role in CRC with 80% of patient samples displaying adenomatous polyposis coli (APC) and β-catenin mutations.10 In canonical Wnt signaling, Frizzled receptor activation regulates the expression/intracellular localization of β-catenin (β-cat), which binds to co-activators such as B-cell lymphoma 9 (BCL9) and its homolog, B-cell lymphoma 9-like (B9L), thereby mediating Wnt transcription.11 We previously generated a potent and selective inhibitor targeting the interaction between BCL9 and β-cat-hsBCL9CT-24, which suppresses cancer cell growth and promotes intratumoral infiltration of cytotoxic T cells by reducing regulatory T cells (Treg).12
Wnt signaling further contributes to cancer maintenance by mediating immune evasion and resistance to immunotherapies.13 Tumors leverage two main Wnt-mediated mechanisms to subvert surveillance and cytotoxicity of the immune response. The first promotes the differentiation and activity of Treg, while the second minimizes the extent of CD8+ effector T cell (Teff) infiltration into the tumor microenvironment.14–16 Consequently, activation of β-cat results in T cell exclusion, resistance to immunotherapy, and shortened survival of colon cancer patients. Targeting β-cat to block Wnt signaling is a promising strategy to affect immunosurveillance and prevent tumor initiation and metastasis. Especially, elevated Wnt signaling has been implicated in immunosuppressive phenotypes, including insufficient tumor infiltration of immune cells, upon which clinical response to CTLA-4 and PD-1/programmed cell death-ligand 1 (PD-L1) immune checkpoint inhibitors (e.g., ipilimumab and pembrolizumab) is dependent.17–20 Recent reports have shown that combining Wnt inhibitors with immuno-oncology (IO) therapies may have synergistic effects in preventing cancer progression.12
CD226 is mainly expressed by monocytes, platelets, T cells, and natural killer (NK) cells,21–24 and is regarded as a costimulatory receptor. It has two extracellular Ig-like domains and an intracellular kinase domain that phosphorylates downstream signaling effector after binding to CD155 or CD112 (nectin-2).25 CD155 (or poliovirus receptor (PVR)/nectin-like molecule-5 or necl5) is a cell adhesion molecule that is commonly overexpressed in tumors associated with poor outcome.26 Cellular events induced by CD155 include promotion of cell adhesion,21 increase in cell migration, and reduced intrinsic cell contacts.27 Recently, CD155 was reported to also regulate the function of tumor infiltrating lymphocytes (TILs) by directly interacting with both stimulatory and inhibitory signaling pathways in T and NK cells. Indeed, CD155 competitively binds to the costimulatory receptor CD226,26 as well as the inhibitory receptors CD96 and TIGIT. Together, these molecules constitute a pathway that is analogous to that of CD28/CTLA-4.28 Particularly, after costimulatory signaling is established through CD226, NK cell adhesion and cytotoxicity as well as cytokine secretion are activated.29 In contrast, interaction of CD155 with TIGIT and CD96 contributes to a “cold tumor” phenotype that facilitates tumor escape and metastasis, leading to poor outcome.25 Overexpression of CD155 in tumor cells was also associated with a reduction in TILs and worse treatment outcome, presumably due to interactions between CD155 and the inhibitory receptors CD96 and TIGIT.30–32
In CRC, immune checkpoint inhibitors (ICIs) such as PD-1 and PD-L1 antibodies show poor response, suggesting that other immune checkpoints should be targeted to achieve clinical benefits. In a recent study, CD155 was reported to act as an immune checkpoint ligand for tumor and tumor-associated myeloid cells, thus representing a potential novel ICI target.33 Furthermore, it was reported that elevated expression of CD155 in human metastatic melanoma is associated with decreased sensitivity to anti-PD-1 immunotherapy.34 It, however, remains to be established whether a combination of PD-1 blockade and downstream activation of CD155-CD226 could create costimulatory cytotoxic signaling in CD8+ T cells, and whether this might exert a synergistic therapeutic effect.
In this study, we explored the involvement of Wnt signaling in the tumor immune microenvironment (TIME) by assessing how inactivation of BCL9, which acts as a necessary co-factor of β-cat, affects the activity of tumor infiltrating T cells. Specifically, we hypothesized that by inhibiting BCL9 activity, we could shift the balance of CD226 and CD96 checkpoints towards more cytotoxicity and thereby impede tumor growth.
Results
Depletion of Bcl9 inhibits tumor growth by modulating immune cell infiltration
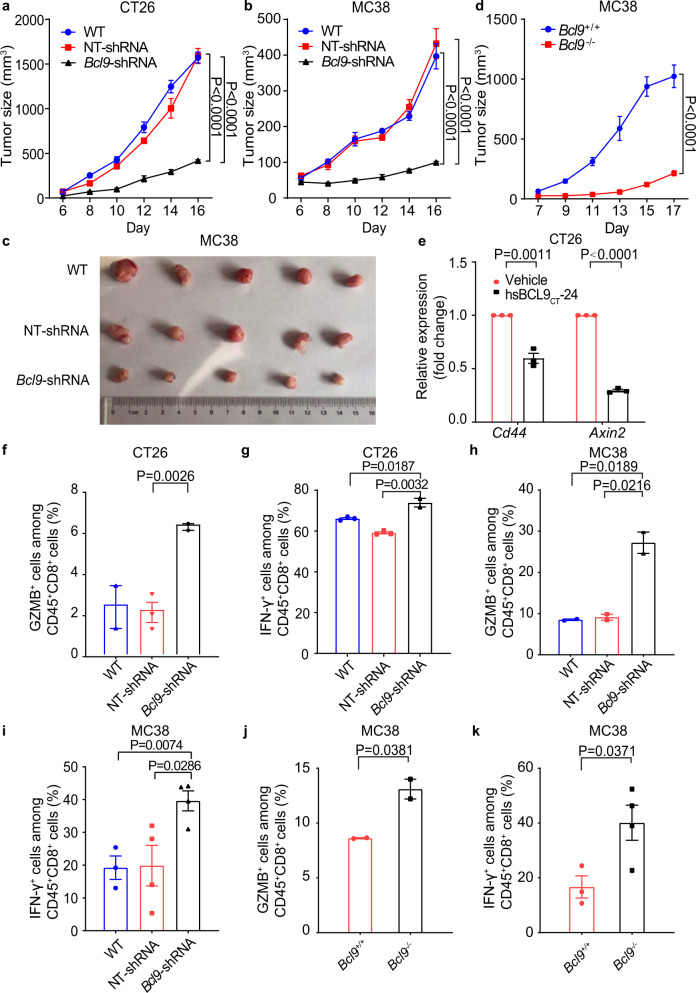
To characterize the function of BCL9 (the human gene name) during CRC growth, we designed a shRNA lentivirus plasmid vector pGIPZ to deplete Bcl9 (the mouse gene name) expression in murine CRC cell lines. Specifically, we depleted Bcl9 in the MC38 and CT26 cell lines (supplementary Fig. 1a, b), as these express high β-catenin levels and are characterized by Wnt/β-cat dependent growth.35–37 Tumor growth in mice subcutaneously bearing CT26 or MC38 cells infected with Bcl9-shRNA was significantly suppressed compared to wild-type (WT) mice: tumor growth inhibition (TGI) rate of 73.8% by day 16 in the CT26 model and 83.9% by day 16 in the MC38 model. Compared to NT-shRNA, CT26 or MC38 tumors infected with Bcl9-shRNA exhibited a TGI of 74.1% and 85.2% by day 16, respectively (Fig. 1a, b, and c).
Fig. 1.
BCL9 suppression promotes CD8+ T cells infiltration. a CT26 cells transduced with non-targeting (NT)-shRNA or Bcl9-shRNA were inoculated in BALB/c mice (n = 5 per cohort). b MC38 cells transduced with non-targeting (NT)-shRNA or Bcl9-shRNA were inoculated in C57BL/6 mice (n = 6 per cohort). c Image of tumor tissue from (c). d Tumor growth in Bcl9+/+ (wild-type) and Bcl9–/– (Bcl9 knockout) mice injected subcutaneously (s.c.) with MC38 cells. e qRT-PCR measurement of Cd44 and Axin2 expression in CT26 tumor tissue treated with hsBCL9CT-24 (i.p., 25 mg/kg) or vehicle. f Percentage of GZMB+ cells among CD45+CD8+ T cells in tumors from (a) was analyzed. g Percentage of IFN-γ+ cells among CD45+CD8+ T cells in tumors from (a) was analyzed. h Percentage of GZMB+ cells among CD45+CD8+ T cells in tumors from (b) was analyzed. i Percentage of IFN-γ+ cells among CD45+CD8+ T cells in tumors from (b) was analyzed. j Percentage of GZMB+ cells among CD45+CD8+ T cells in tumors from (c) was analyzed. k Percentage of IFN-γ+ cells among CD45+CD8+ T cells in tumors from (c) was analyzed. Results were denoted as means ± SEM for experiments performed in triplicate. Each experiment was repeated three times, and the statistical significance of differences between groups was determined by Two-way ANOVA or non-parametric test (One-tailed or Two-tailed). P < 0.05 means statistically significant.
Previously, we used the hsBCL9CT-24 inhibitor to block interaction between BCL9 and β-cat, and showed that tumor growth inhibition ratio was higher in immune competent than immune compromised mice, suggesting that Bcl9 expressed by immune cells also mediates the therapeutic effect of Bcl9 knockdown.12 We, therefore, also analyzed MC38 tumor growth in WT and Bcl9–/– full knockout mice. MC38 cells continued to grow progressively in WT mice, while growth was attenuated in Bcl9–/– mice (TGI of 80.5% on day17; Fig. 1d). Wnt downstream-signaling markers such as Cd44 and Axin2 were reduced in CT26 or MC38 tumors infected with Bcl9-shRNA (supplementary Fig. 1c, d), as well as in tumors treated with hsBCL9CT-24 (compared to vehicle-treated tumors; Fig. 1e), suggesting that BCL9 suppression exhibits robust anti-tumor effects by targeting oncogenic Wnt signaling.
To investigate whether Bcl9 depletion affects tumor immune infiltration, we characterized immune cells derived from CT26 and MC38 tumors implanted in immunocompetent mice by flow cytometry. In Bcl9-depleted CT26 tumors, the ratio of tumor-infiltrated Treg was significantly decreased (Supplementary Fig. 1e, while the proportions of cytotoxic granzyme B (GZMB)+CD8+ T cells, IFN-γ+ CD8+ T cells, and effector CD8+ T cells were increased (Fig. 1f, g, Supplementary Fig. 1f), implying that Bcl9 depletion inhibits immunosuppressive immune cells. In Bcl9-depleted MC38 tumors, cytotoxic CD8+ T cells were also significantly increased (Fig. 1h, i), while the ratio of tumor-infiltrated Treg cells were decreased (Supplementary Fig. 1g). Similarly, in Bcl9–/– mice injected s.c. with MC38 cells, the ratio of tumor-infiltrated Treg cells was significantly decreased (Supplementary Fig. 1h), and the proportion of cytotoxic CD8+ T cells was increased (Fig. 1j, k). Overall, it appears that Bcl9 depletion, either in tumor or stromal cells, not only reduces tumor growth, but also promotes infiltration of cytotoxic and effector CD8+ T cells.
Bcl9 depletion combined with PD-1 blockade improves the TIME
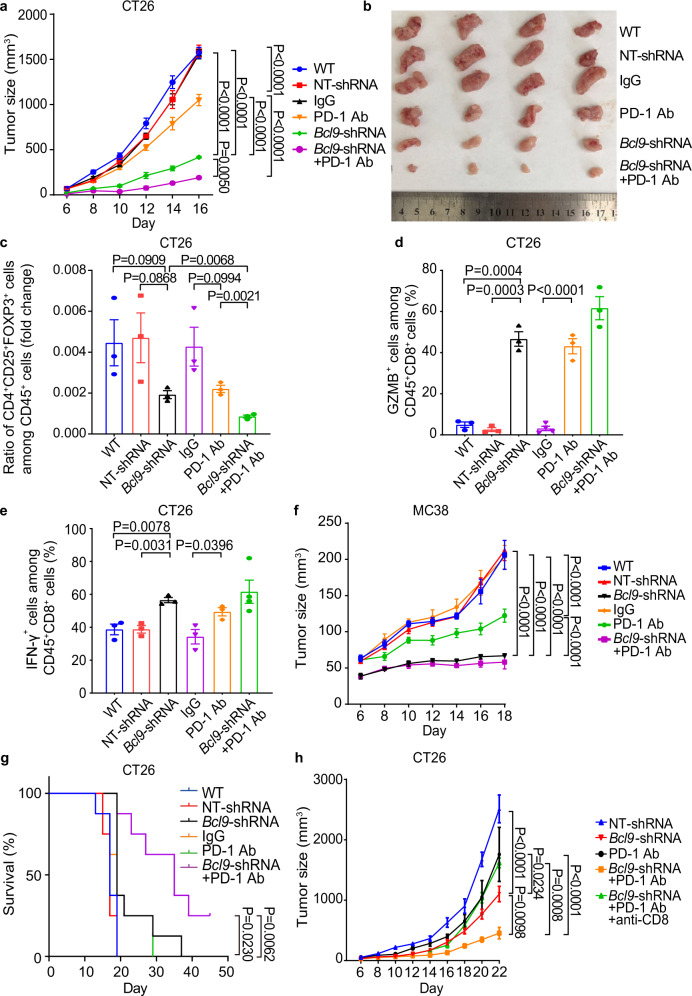
We then examined whether Bcl9 depletion has a synergistic effect with anti-PD-1 on tumor growth. Mice inoculated with Bcl9-shRNA infected in CT26 tumors were treated with a mouse anti-PD-1 monoclonal antibody. Tumor growth was significantly reduced in BCL9-depleted tumors treated with anti-PD-1 compared to anti-PD-1, with a TGI of 81.8% by day 16 (Fig. 2a, b). Flow cytometry of CT26 tumors revealed that anti-PD-1 treatment further increased the abundance of cytotoxic GZMB+CD8+ T cells and IFN-γ+ CD8+ T cells, but decreased the Treg cells (Fig. 2c, e). We also examined the combination of Bcl9 depletion and anti-PD-1 in MC38 tumors compared to NT-shRNA tumors and confirmed that depletion of Bcl9 decreases tumor size in response to anti-PD-1, with a TGI of 87.1% by day 18 (Fig. 2f). Finally, combination of Bcl9 depletion and PD-1 blockade also improved response and survival rates in the CT26 mouse model (Fig. 2g). Overall, these results suggest that depletion of Bcl9 combined with anti-PD-1 treatment can further increase T cell cytotoxicity and effector function in the TIME.
Fig. 2.
Inhibition of BCL9 enhances response to anti-PD-1 antibody in CRC models. a Combination treatment of Bcl9-shRNA and anti-PD-1 antibody resulted in almost complete regression of the CT26 tumor. BALB/c mice were either inoculated with CT26 cells via single flank implantation and treated with immunoglobulin G (IgG), anti-PD-1 antibody [twice-weekly (BIW)], or with CT26 cells that transduced with non-targeting (NT)-shRNA or Bcl9-shRNA. The mice were subsequently treated with anti-PD-1 antibody as indicated after tumor volume reached 40–60 mm3 (n = 5 per cohort). b Image of tumor tissue from (a). c Ratio of CD4+CD25+FOXP3+ cells among CD45+ cell populations in the tumors from (a). d Percentage of GZMB+ cells among CD45+CD8+ T cells in the tumor from (a) was analyzed. e Percentage of IFN-γ+ T cells among CD45+CD8+ T cells in the tumor from (a) was analyzed. f Mice inoculated with Bcl9-shRNA-transduced-MC38 cells and treated with anti-PD-1 antibody resulted in almost complete regression of the MC38 tumor. C57BL/6 mice were inoculated with MC38 cells via single flank implantation and treated with immunoglobulin G (IgG) or anti-PD-1 antibody (BIW), or with Bcl9-shRNA-transduced-MC38 cells and treated with anti-PD-1 antibody as indicated after tumor volume reached 40–60 mm3 (n = 5 per cohort). Ab, antibody. Results were denoted as means ± SEM for experiments performed in triplicate. g Kaplan–Meier analysis of survival end point (tumor size >2000 mm3, n = 8 per cohort). h Mice were inoculated with Bcl9-shRNA-transduced-CT26 cells used anti-PD-1 antibody resulted in almost complete regression of the CT26 tumor. BALB/c mice were inoculated with CT26 cells via single flank implantation and treated with anti-PD-1 antibody (BIW), the mice were inoculated with Bcl9-shRNA-transduced-CT26 cells used anti-CD8 antibody (intraperitoneal injection, i.p., 300 μg per mouse at days 12, 15, and 19 after tumor cell inoculation) plus anti-PD-1 antibody as indicated after tumor volume reached 40–60 mm3 (n = 5 per cohort). Each experiment was repeated three times, and the statistical significance of differences between groups was determined by Two-way ANOVA or non-parametric test (Two-tailed). P < 0.05 means statistically significant.
To directly assess the role of T cells in reducing tumor growth in Bcl9-shRNA tumors during anti-PD-1 treatment, we depleted CD8+ T cells using a CD8-neutralizing antibody and subsequently examined the effect of Bcl9-shRNA combined with anti-PD-1 on tumor growth. It was found that in CD8+ T cell-depleted tumor-bearing mice anti-PD-1 no longer exerted an additional inhibitory effect on tumor growth when combined with Bcl9 depletion (Fig. 2h). The data imply that Bcl9 depletion enhances anti-tumoral CD8+ T cell-mediated immune reactions, thereby amplifying the response to anti-PD-1 in CRC mouse models.
T-cell profiling by scRNA-seq after depletion or pharmacological inhibition of Bcl9
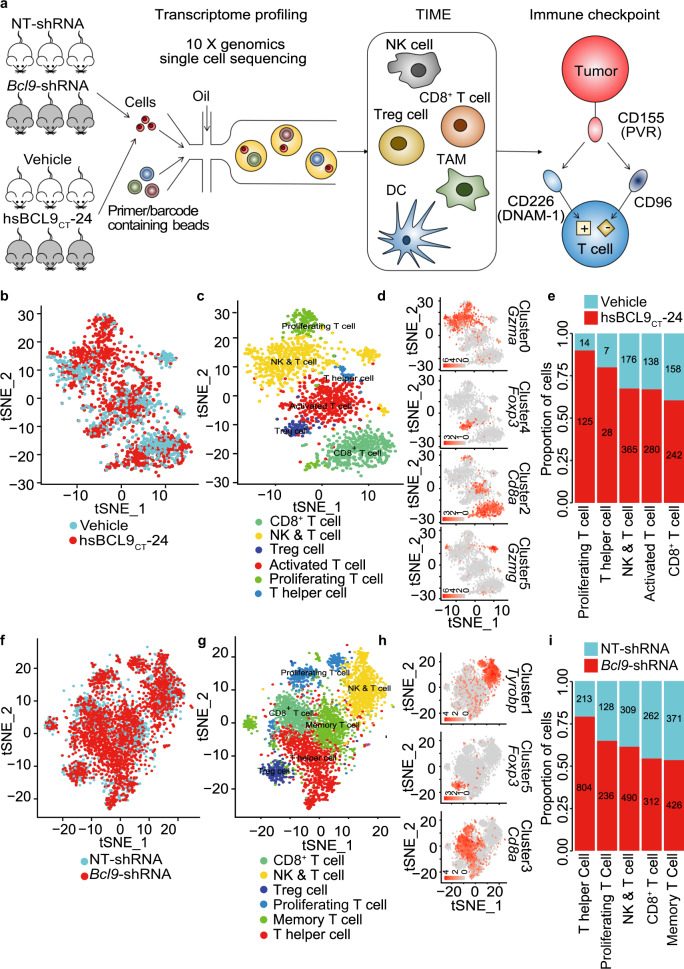
Next, we studied the transcriptional changes in the TIME after Bcl9 depletion and hsBCL9CT-24 treatment using single-cell RNA-seq (scRNA-seq) profiling. CT26 tumors infected with Bcl9-shRNA versus hsBCL9CT-24, or treated with hsBCL9CT-24 versus vehicle, were collected at day 14 and rapidly digested into a single-cell suspension for scRNA-seq using 10x Genomics (Fig. 3a, Supplementary Table 1). Following gene expression normalization for read depth and mitochondrial read count, we obtained high-quality expression data for 95,816 cells (Supplementary Fig. 2a, Supplementary Table 2). After graph-based clustering, 8 cell types were identified based on marker gene expression (Supplementary Fig. 2b, Supplementary Table 3), including cancer cells (n = 73,174) by Wnt10a, NK&T cells (n = 5374) by Cd3e, macrophages (n = 11,823) by C1qc, while classical (n = 11823) and plasmacytoid dendritic cells (n = 352) (cDCs and pDCs) were identified based on H2-Aa and Klk1b27, respectively (Supplementary Fig. 2b, Supplementary Table 3). Compared to NT-shRNA tumors, we observed increased proportions of T cells, cDCs, and pDCs in Bcl9-shRNA tumors (Supplementary Fig. 2c, d). Similar differences were observed in hsBCL9CT-24-treated tumors (Supplementary Fig. 2c, d).
Fig. 3.
Depletion of BCL9 changes T-cell cellular landscapes. a The workflow of scRNA sequencing. b tSNE plot of the tumor sample following treatment with vehicle or hsBCL9CT-24 (two groups), color-coded by their associated clusters. c Dot plot of the six clusters of T cells from (b). d tSNE plot of color-coded expression (gray to orange) of marker genes for the clusters from (c). e The proportion of CD8+ T and NK&T cells from 6 samples (vehicle and hsBCL9CT-24). f tSNE plot of the tumor sample that CT26 cells transduced with non-targeting (NT)-shRNA or Bcl9-shRNA (two groups), color-coded by their associated clusters. g Dot plot of the seven clusters of T cells from (f). h tSNE plot of color-coded expression (gray to orange) of marker genes for the clusters from (g). i The proportion of CD8+ T and NK&T cells from 6 samples (NT-shRNA and Bcl9-shRNA)
As infiltration of cytotoxic and effector CD8+ T cells increased after genetic depletion and pharmacological inhibition of BCL9, we investigated the cellular landscape associated with this T cell response in more detail using scRNA-seq data. We subclustered T cells using a similar approach as for the cell types (Supplementary Fig. 3a–d) and observed 6 T cell subclusters in CT26 tumors treated with hsBCL9CT-24 (Fig. 3b, c, Supplementary Fig. 3e, f). These included CD8+ T cells (marker Cd8a), Treg cells (marker Foxp3), and NK&T cells (marker: Klrb1a, Gzma, and Gzmg) (Fig. 3c, d, Supplementary Fig. 3e–g, k, l, Supplementary Table 3). Cell fraction analysis showed that CD8+ T cells and NK&T cells were increased after hsBCL9CT-24 treatment (Fig. 3e). In CT26 tumors infected with Bcl9-shRNA and NT-shRNA similar subclusters were identified (Fig. 3f, g, Supplementary Fig. 3h, i) including cytotoxic CD8+ T cells (marker Cd8a), Treg cells (marker Foxp3) and NK&T cells (marker Tyrobp and Klrb1a) (Fig. 3g, h, Supplementary Fig. 3h, j, m, n and o). Again, CD8+ T cells and NK&T cells were increased in BCL9-depleted tumors (Fig. 3i). Overall, these result indicate that BCL9 suppression drives a complex remodeling of infiltrating immune cells, including CD8+ T cells and NK&T cells.
Ligand–receptor interaction identifies the correlation between CD226-CD155 and BCL9
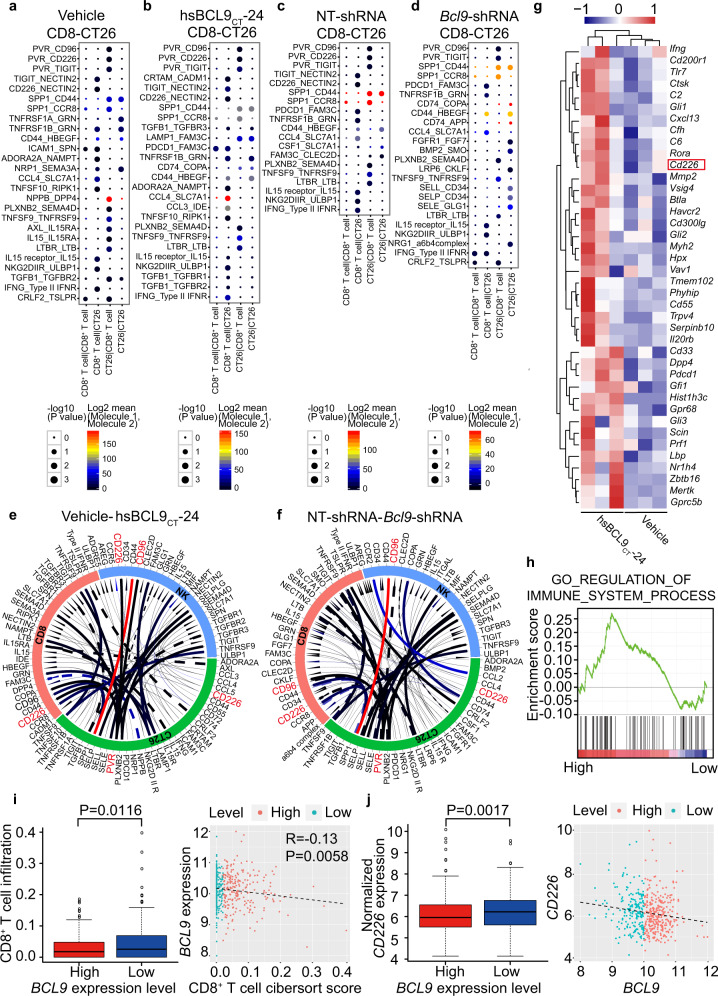
To identify potential interaction between CT26 tumor cells and infiltrating immune cells, we scored ligand–receptor interaction between cancer cells and CD8+ T cells by calculating average receptor and average ligand expression in each cell type (Fig. 4a–d). After computing scores for each tumor separately, we averaged them across each tumor model and treatment condition to identify conserved interactions (Fig. 4e, f). Many of the highest-scoring interactions are part of the chemokine family, including TGF-β, IL15, CCL4, and their receptors. Remarkably, CD155/PVR on cancer cells interacted with CD226 expressed in CD8+ T cells in all 4 treatment groups, suggesting that this interaction plays a key role in immune modulation.
Fig. 4.
BCL9 is associated with CD155-CD226 checkpoint in CT26-CD8 T+ cell interaction. a Interaction analysis between CD8+ T and CT26 cells in vehicle group. b Interaction analysis between CD8+ T and CT26 cells in hsBCL9CT-24 treated group. c Interaction analysis between CD8+ T and CT26 cells in NT-shRNA group. d Interaction analysis between CD8+ T and CT26 cells in Bcl9-shRNA group. e Circos plots of all of the putative ligand–receptor interactions in vehicle and hsBCL9CT-24 treated groups. The color of the line represents the average expression of the ligand–receptor pair in the pair of cells, and the thickness of the line represents the significance of the ligand–receptor enrichment in the pair of cells. The solid line represents the interaction of the hsbcl9CT-24 group, and the dotted line represents the interaction of the vehicle group. f Circos plots of all of the putative ligand–receptor interactions in NT-shRNA and Bcl9-shRNA groups. The solid line represents the interaction of Bcl9-shRNA group, and the dotted line represents the interaction of the NT-shRNA group. g Heatmap analysis of CT26 tumor pretreated with vehicle or hsBCL9CT-24 (i.p., 25 mg/kg). h GSEA analysis of CT26 tumor pretreated with vehicle or hsBCL9CT-24 (i.p., 25 mg/kg). i Scatter and boxplot analyses of CD8+ T cell infiltration associated with BCL9 expression level in TCGA. j Scatter and boxplot analyses of CD226 expression associated with BCL9 expression level in TCGA. P < 0.05 means statistically significant.
To further explore this interaction in Bcl9-deficient tumors, RNA-seq analyses were performed in CT26 tumor-bearing mice treated with hsBCL9CT-24. We found that Cd226 expression increased significantly compared to vehicle-treated mice (Fig. 4g). In a gene set enrichment analysis (GSEA) of differentially-expressed genes, hsBCL9CT-24-treated tumors showed positive correlation with immune regulation (Fig. 4h), consistent with our scRNA-seq findings. Interestingly, T cells were activated and cytotoxic, as indicated by Prf1 and Gzmb that were expressed in CD8+ T cells (Supplementary Fig. 4a–d).
Next, we explored the correlation between T cells and the Wnt signaling pathway using CIBERSORT (https://cibersort.stanford.edu) in CRC tumors profiled by The Cancer Genome Atlas (TCGA). We found that tumors with low expression of BCL9, CTNNB1, and TCF4 (i.e., tumors with low Wnt pathway activation) contained significantly more infiltrated CD8+ T cells (Supplementary Fig. 4e–g). In contrast, tumors expressing high levels of DVL1, AXIN1, and AXIN2 (i.e., high Wnt pathway activation) contained more Tregs (Supplementary Fig. 4h–j). These findings confirm that there is a correlation between BCL9 and aberrant Wnt pathway activation and CD8+ T cell suppression in Colon adenocarcinoma (COAD).
Finally, we evaluated the correlation between BCL9, CD226, and CD8+ T cell infiltration. The fraction of CD8+ T cells was significantly lower in COAD when BCL9 was high (Fig. 4i), while CD226 was negatively correlated with BCL9 (P < 0.05) (Fig. 4j). Additionally, the abundance of CD8+ T cells was increased in COAD when CD226 was high (P < 0.05; Supplementary Fig. 5a). Since BCL9 was negatively correlated with CD226 and CD8+ T cells in COAD, these findings suggest a correlation between BCL9 and CD226-positive CD8+ T cell infiltration in CRC.
Since expression of the immune checkpoints PD-1 and PD-L1 is correlated with infiltrating immune cells and Bcl9 depletion is synergistic with anti-PD-1 treatment in mouse tumor models, we studied the relationship among CD226, PD-1 (or PDCD1), and PD-L1 (or CD274) in CRC, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and triple-negative breast cancer (TNBC). Both PD-1 and PD-L1 were positively correlated with CD226 in LUAD, LUSC, TNBC and COAD (Supplementary Fig. 5b–i). Triple immunostaining of the murine model of CRC further revealed that CD226 was positive in CD8+ T cells, while CD155 was positive in CT26 cells (Supplementary Fig. 5j, k), consistent with the immunofluorescence staining data of the human ovarian cancer revealing that CD155 was co-stained with P53 for cancer cells marker (Supplementary Fig. 5i). Overall, it appears that CD226 and PD-1/PD-L1 are correlated with multiple cancer types, suggesting that CD226 activation may compliment anti-PD-L1/PD-1 immunotherapy by activating distinct T cell subsets with synergistic effects.
BCL9 associated APC mutation is negatively correlated with immunotherapy outcome
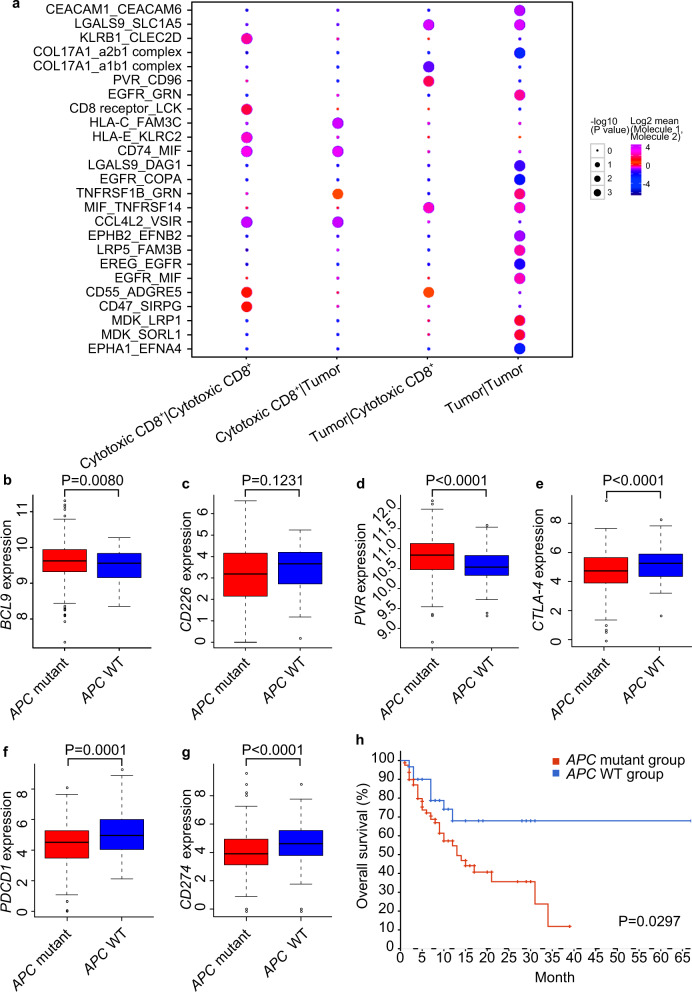
To further confirm that the interactions of CD155 with CD226 and CD96 play an important immunoregulatory role in CRC, we quantified cell–cell interaction in a human CRC scRNA-seq dataset. Thirteen patients were enrolled with histopathologically confirmed adenocarcinoma diagnosed by colonoscopy. All tumor tissues were obtained during stage I surgical resection and the samples were rapidly digested to a single-cell suspension and subjected to scRNA-seq. A similar approach as for the mouse CRC models was used to obtain clusters and identify cell types based on marker genes. When assessing cell type interactions between CD8+ T cells (marker gene: CD8A) and tumor cells (marker gene: EpCAM) (Fig. 5a), most of them showed interactions known to be mediating immunity, including that of CD155 with CD96 between CD8+ T cells and tumor cells.
Fig. 5.
BCL9 and CD155-CD226 are associated with APC mutation, which is correlated with patient survival after immune checkpoint inhibitor treatment. a Cell–cell interaction in scRNA-seq database for human CRC samples. b Boxplot analyses of BCL9 expression associated with APC mutation level in TCGA. c Boxplot analyses of CD226 expression associated with APC mutation level in TCGA. d Boxplot analyses of CD155/PVR expression associated with APC mutation level in TCGA. e Box plots showing expression differences of CTLA-4 between APC non-mutation and APC mutation tumors. f Box plots showing expression differences of PDCD1 (PD-L1) between APC non-mutation and APC mutation tumors. g Box plots showing expression differences of CD274 (PD-L1) between APC non-mutation and APC mutation tumors. h Correlation of APC mutation with immunotherapy outcome in CRC specimens. OS, overall survival. P < 0.05 means statistically significant.
In order to investigate the clinical significance of BCL9 and CD155-CD226 in ICI therapy response, we used a cohort of 110 CRC patients from cBioPortal database to analyze the association between gene mutation and overall survival (OS) after treatment.38 To further explore the relationship between APC mutation and marker genes, we employed the TCGA dataset, and found that APC mutation was positively correlated with BCL9 and CD155/PVR expression, but negatively correlated with CD226 in COAD in TCGA (Fig. 5b–d).39 However, expressions of CTLA-4, PD-L1, and PD-1 were significantly reduced in APC mutant compared to APC WT tumors (Fig. 5e–g).
Given the adverse role of mutated APC in the immune microenvironment and its effect on PD-1 expression, patients with APC mutation may not benefit from immunotherapy. We analyzed the association between APC mutation and immunotherapy outcome using a cohort of CRC patients treated with ICIs showing that they had significantly shorter OS time after immunotherapy than that of APC WT patients (Fig. 5h).
Depletion of BCL9 modulates CD226-CD155 signaling via phosphorylation of VAV1
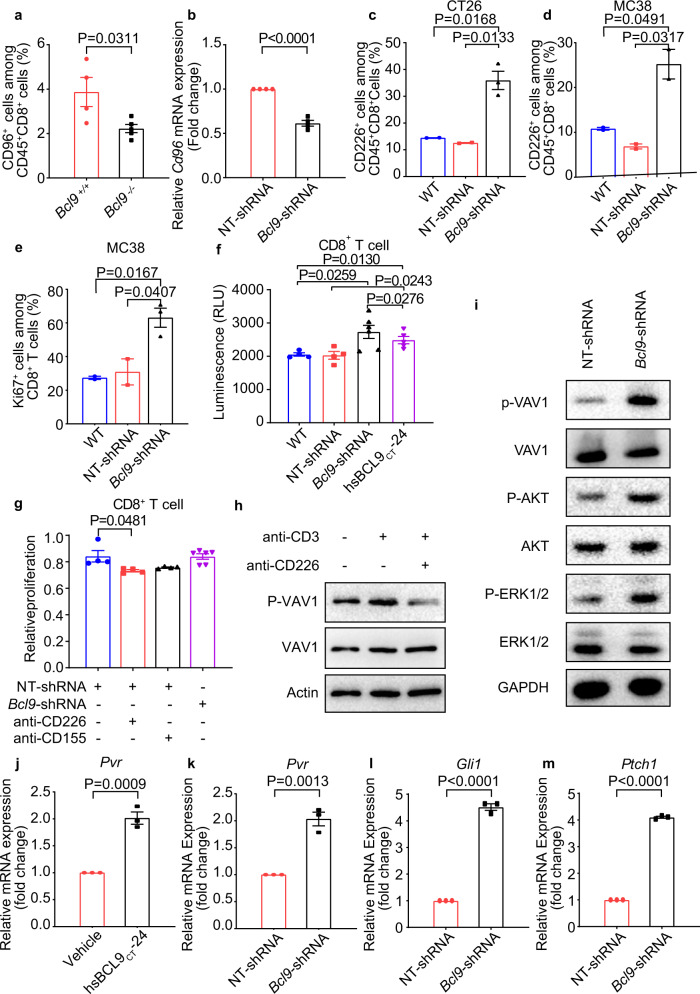
CD226 is a transmembrane receptor present on NK and CD8+ T cells. CD155 competitively binds to CD226 and CD96.26 The guanine nucleotide exchange factor VAV1 is required for the activation of T cells. Interestingly, upon T-cell activation, the costimulatory molecule CD226 was reported to be one of the most enriched VAV1 interactors.40 Significantly less CD96+CD8+ T cell infiltration was observed in Bcl9–/– mice bearing CT26 tumor cells suggesting that Bcl9 deficiency may promote T cell activation and potentially enhance responsiveness to PD-1 checkpoint blockade (Fig. 6a). Decreased mRNA expression of Cd96 is also found in Bcl9-shRNA treated CD8+ T cells compared with NT-shRNA (Fig. 6b), hsBCL9CT-24 and vehicle control (Supplementary Fig. 6a). In contrast, depletion of Bcl9 in CT26 tumor cells increased CD226+CD8+ T cell tumor infiltration accompanied by increased expression of Pvr/CD155 (Fig. 6c). In Bcl9-depleted MC38 tumors, CD226+CD8+ T cell tumor infiltration was also increased (Fig. 6d). These results suggest that Bcl9 suppression promotes CD226+CD8+ T cell tumor infiltration, but decreases that of CD96+CD8+ T cells. CD226 was reported to impact CD8+ T cell activity and function.41 In MC38 tumors, Bcl9 depletion and knockout both increased T cell proliferation indicted by staining of Ki67 (Fig. 6e, Supplementary Fig. 6b). Interestingly, Bcl9-shRNA or hsBCL9CT-24 treated CT26 tumor cells co-cultured with CD8+ T cells promoted the proliferation of CD8+ T cells in vitro (Fig. 6f). We then used CD226 and CD155 neutralizing antibodies in a CD8+ T cell proliferation assay. The proliferation of CD8+ T cells induced by Bcl9-shRNA was abolished by neutralizing antibodies against CD155 and CD226 (Fig. 6g, Supplementary Fig. 6c), suggesting that CD8+ T cell proliferation is dependent on CD226-CD155 signaling. Engagement of CD226 is known to elicit tyrosine phosphorylation of VAV1.40 Neutralizing antibodies against CD226 indeed decreased the activation of VAV1, as shown by its phosphorylation on Tyr174 (Fig. 6h), implying that engagement of CD226 induces tyrosine phosphorylation of VAV1. Treatment of hsBCL9CT-24 or Bcl9-shRNA in CD8+ T cells increased the level of tyrosine phosphorylation of VAV1, AKT, and ERK, while stimulation of CD8+ T cells increased the global quantities of tyrosine phosphorylation (Fig. 6i, Supplementary Fig. 6d). Our results indicate that suppression of Bcl9 promotes VAV1-AKT-ERK signaling cascades in CD8+ T cells.
Fig. 6.
Inhibition of BCL9 promotes CD155-CD226 checkpoint, which signals via VAV1 phosphorylation. a Percentage of CD96+ cells among CD45+CD8+ T cells from MC38 tumor tissue in BCL9+/+ and BCL9−/− mice was analyzed. b qRT-PCR measurement of Cd96 in CD8+ T cells treated with NT-shRNA or Bcl9-shRNA. c Percentage of CD226+ cells among CD45+CD8+ T cells from CT26 tumor tissue in BALB/c mice inoculated with wild-type (WT), NT-shRNA or Bcl9-shRNA-transduced-CT26 cells was analyzed. d Percentage of CD226+ cells among CD45+CD8+ T cells from MC38 tumor tissue in C57BL/6 mice inoculated with WT, NT-shRNA or Bcl9-shRNA-transduced-MC38 cells was analyzed. e Percentage of Ki67+ cells among CD45+CD8+ T cells from MC38 tumor tissue in C57BL/6 mice inoculated WT, NT-shRNA or Bcl9-shRNA-transduced-MC38 cells was analyzed. f Proliferation of CD8+ T cell co-cultured with CT26 were treated with Bcl9-shRNA or hsBCL9CT-24 and analyzed by Cell titer-Glo® Luminescent cell viability assay. g Mouse CD8+ T cells proliferation was measured after treatment with NT-shRNA or Bcl9-shRNA in the presence of anti-CD155 and anti-CD226 antibodies. h Immunoblots showing VAV1 and VAV1 phosphorylation on Tyr174 (p-VAV1) in equal amounts of total lysates from CD8+ T cells that were treated with or without CD3 antibody (Thermo, 16-0032-81, 0.25 μg) and/or anti-CD226 antibody (Thermo, 0.25 μg) for 5 min. i Immunoblots showing phosphorylated VAV1 (p-VAV1), phosphorylated ERK1/2 (p-ERK1/2), phosphorylated AKT (p-AKT) as well as total VAV1, AKT, and ERK1/2 in equal amounts of total lysates from CD8+ T cells treated with NT-shRNA or Bcl9-shRNA. j qRT-PCR measurement of CD155/Pvr in CT26 cells treated with hsBCL9CT-24 (5 μM) or vehicle for 24 h. k qRT-PCR measurement of CD155/Pvr expression in CT26 cells transduced with NT-shRNA or Bcl9-shRNA. l qRT-PCR measurement of Gli1 expression in CT26 cells treated with NT-shRNA or Bcl9-shRNA. m qRT-PCR measurement of Ptch1 expression in CT26 cells treated with NT-shRNA or Bcl9-shRNA. Results were denoted as means ± SEM for experiments performed in triplicate. Each experiment was repeated three times, and the statistical significance of differences between groups was determined by non-parametric test. P < 0.05 means statistically significant.
CD155, expressed on the surface of cancer cells, was shown to promote tumor invasiveness.42 Its upregulation in tumor environment–infiltrating myeloid cells restrained anti-tumor immunity by impairing anti-tumor T lymphocyte and NK cell function.43 hsBCL9CT-24 treatment increased mRNA expression of CD155/Pvr in CT26 cells (Fig. 6j), while Bcl9 depletion in CT26 similarly increased expression of CD155/Pvr (Fig. 6k). It was reported that sonic hedgehog activates CD155 gene expression.44 We thus examined whether key components of sonic hedgehog Gli1 (transcription factor) and Patch1 (receptor) were changed. Increased mRNA levels of Gli1 and Ptch1 were seen in hsBCL9CT-24 or Bcl9-shRNA treated CT26 tumor cells (Fig. 6l, m, Supplementary Fig. 6e, f), suggesting that BCL9 suppression activates CD155 expression via sonic hedgehog signaling.
Depletion of BCL9 inhibits Treg migration
The role of BCL9 in Treg cells was investigated using the Transwell migration method to simulate tumor infiltration by immune cells. In vitro migration of Treg cells was reduced in Bcl9-depleted CT26 cells compared with the NT-shRNA group (supplementary Fig. 7a). Likewise, Treg cells migration in Bcl9-depleted MC38 cells was also reduced (Supplementary Fig. 7b). In MC38 cells treated with hsBCL9CT-24 decreased migration of Tregs was observed (Supplementary Fig. 7c). When Ctnnb1 was depleted via shRNA in MC38 cells, Treg cell migration was also reduced, as expected (Supplementary Fig. 7d).
Next, we explored the molecular mechanisms underlying Treg cell migration by examining expression levels of known regulators, including Ccl4, Ccl22, and Tgf-β using real-time quantitative PCR. Bcl9 depletion significantly increased Ccl4 expression and significantly reduced Tgf-β expression levels in CT26 cells (Supplementary Fig. 7e, f).12 hsBCL9CT-24 and CCR4 antagonist C021 were employed to investigate changes in the ability of Tregs to infiltrate tumors. Both inhibitors significantly reduced the tumor infiltration ratio of Treg cells, but interestingly, it did not change the proportion of Treg cells in the spleen (supplementary Fig. 7g, h). This result suggests that Treg cell infiltration is mediated via CCR4 and BCL9/β-cat pathways.
BCL9 depletion induces transcriptional differences in CD8+ T cells
GSEA was performed to compare T cells in hsBCL9CT-24-treated vs. vehicle-exposed CT26 tumors. This revealed that pathways related to IL-2, CD4+ T cell memory formation and stimulation of CD8+ T were enriched in tumor-infiltrating T cells treated with hsBCL9CT-24 (Supplementary Fig. 8a, c). Tumor-infiltrating T cells in Bcl9-shRNA compared to NT-shRNA bearing tumors exhibited pathways enriched for TLR, FOXP3, and IL-2 (Supplementary Fig. 8b, d). We found a couple of pathways related to CD8+ T cells that are enriched in hsBCL9CT-24 and Bcl9-shRNA groups (Supplementary Fig. 9a–f).
Next, transcription factor activity underlying specific mouse CD8+ T-cell clusters was analyzed in the scRNA-seq data using SCENIC.45 This identified MYC, JUN, STAT1, JUNB, and FL1 as potential transcription factors of CD8+ T cells in Bcl9-shRNA but not NT-shRNA tumors (Supplementary Fig. 10a). It was found that MYC and STAT1 expression were linked to CD155-related signaling, while downregulation of Jun, Junb, and Fli1 were linked to CD226-related signaling, in which Jun and Junb were identified as CD226 gene promoters (Supplementary Fig. 10a). We then used SCENIC to analyze co-expression of transcription factors and their putative target genes. This identified Irf1, Junb, Jun, Stat1, and Elf1 as candidate transcription factors underlying gene expression differences in T cells (Supplementary Fig. 10b–f).
Discussion
In this paper, we uncovered that cytotoxic CD8+ T cell tumor infiltration was increased by pharmacological inhibition and genetic depletion of BCL9. We analyzed the TIME associated with this intervention at single-cell resolution. RNA sequencing technique was applied to characterize CD8+ T and NK&T cells and to study their roles in the TIME models. Cellular landscapes and transcriptional features were presented with T, NK, and tumor cells. Key cell clusters and signaling properties among CD8+ T and NK&T cells were described and CD155-CD226 was found to be a potential checkpoint regulated by BCL9. We found that CD155/PVR-CD226 signaling occurs through VAV1 phosphorylation and BCL9 inhibition promotes antibody-mediated PD-1 blockade via promoting cytotoxic CD8+ T cell tumor infiltration in mouse models. BCL9 suppression blocks VAV1 phosphorylation in CD8+ T cell as well as increases GLI1 and PATCH expression to promote CD155 expression in cancer cells (Supplementary Fig. 12). TCGA analysis revealed that BCL9 upregulation is associated with APC mutation.
We for the first time reveal BCL9 suppression results in altered immune infiltration. Enrichment of CD8+ T and NK&T cells in tumor is upon BCL9 genetic depletion and pharmacological inhibition. BCL9 suppression leads to the engagement of CD226 which in turn activates of VAV1 and promotes CD155 expression possibly via increasing mRNA levels of Gli1 and Patch. BCL9 depletion also significantly decreases CCL22 expression in tumor cells. Pharmacological inhibition of BCL9 reduces tumor infiltration by Treg cells due to inhibition of CCR4 expression. Inhibition of Wnt signaling may function through the CCL22-CCR4 axis in Treg cells tumor infiltration.46–48
This study is the first demonstration of using scRNA-seq to identify cell clusters (e.g., T cells, macrophages, fibroblasts, endothelial cells, dendritic cells, and granulocytes) in mouse models with Bcl9 genetic depletion and pharmacological inhibition. We present comprehensive cell distribution map in tumors with genetic depletion of BCL9 and pharmacological inhibition. Alterations in the proportions of T-cell subpopulations, including decreased percentages of CD8+ T cells, tumor-associated macrophages, cancer-associated fibroblasts, tumor endothelial cells, tumor-associated neutrophils, and dendritic cells, indicate a complex TIME in tumors regulated by BCL9. These findings introduce a new way to investigate the role of BCL9 in TIME. Clearly manipulation of cellular microenvironment is of value to dissect functional roles of immune cells of different type in the context of cancer therapeutic outcome evaluation and prognosis prediction.
Immune checkpoint blockade is effective in only a subset of CRC patients with microsatellite instability-high (MSI-H) tumors.49 In these patients, the TIME is dynamic during malignant progression. Recent studies have emphasized the role of the TIME in therapeutic resistance in cancer.50 Our scRNA-seq study sheds light on the composition of the TIME, which may facilitate the development of novel immunotherapy and enhance understanding about drug resistance in CRC. By identifying differences in immune cell subtypes (e.g., CD8+ T and NK&T cells) following BCL9 modulation and CD155-CD226 as a potential checkpoint, our data will certainly help CRC diagnosis and therapy.
BCL9 may be important mainly in APC mutated tumors.51–53 CRC patients with APCmut showed significantly shorter overall survival after ICI immunotherapy than that of APC wild-type patients. BCL9 may be negatively correlated with ICI treatment, suggesting inhibition of BCL9 may enhance the outcome of ICI immunotherapy in patients with APCmut.
There are a number of reports about BCL9 role in non-immune regulation in cancer development. Loss of BCL9/9l was reported to block colonic tumorigenesis and mutations.51 Bcl9 and Pygo were found to synergize downstream of Apc to effect intestinal neoplasia in Apcmin/+ mouse models.53 BCL9/9L-β-catenin signaling is associated with poor outcome and affects stemness maintenance in colorectal CRC.54 Deregulation of BCL9 is an important contributing factor to tumor progression in CRC.55 But there is no reports about the role of BCL9 in immune-oncology yet.
CD226 expression is not only associated with PD-1/PD-L1 in CRC, but also in triple-negative breast cancer, lung adenocarcinoma and lung carcinoma suggested by TCGA. It is positively correlated with APC mutation, which is an indicator of OS of CRC patients treated with ICIs.56 It appears that high infiltration of CD226+CD8+ T cells might be a survival marker associated with the outcome of ICI treatment in patients of different cancer types.
In the mouse scRNA-seq, we identified that both CD155-CD226 and CD155-CD96 interactions were significantly enriched between CD8+ T and CT26 cells while in the human scRNA-seq dataset, only CD155-CD96 was identified. This may be due to the difference of two immune systems and CD226 may plays different roles in the human vs. the mouse. Another possibility is that missing CD155-CD226 in human samples is an isolated case because the scRNA-seq data were generated among CRC patients with liver metastasis.
PVR, the ligand for TIGIT, is shared with CD226. TIGIT exerts immunosuppressive action by competing with CD226 for the same CD155 ligand. Blockage of CD226 completely abrogated the effect of TIGIT/PD-L1 in the tumor but did not impact IFN-γ-producing CD8+ T cell frequency in the tumor-draining lymph node,57 suggesting a unique interplay among CD226, TIGIT and PD-1 in the tumor microenvironment. However, the primary focus of this paper was to study co-stimulatory role of CD226 and co-inhibitory role CD96 in TIME of CRC, respectively.
To investigate the role of Wnt signaling in cancer cells, immune cells, and TIME, receptively, we used three different approaches, including genetic depletion, knockout and pharmacological inhibition. First, in the CT26/MC38 genetic depletion model, CT26 or MC38 cell was Bcl9 depleted, while the BALB/c and C57BL/6 mice are wild-type. This model shows how Bcl9 depleted cancer cell impacts on the wild-type CD8+ T cell tumor infiltration. Second, in the model of Bcl9 knockout model, the mice were subcutaneously implanted with wild-type MC38 cell. This model demonstrates how Bcl9 knockout immune cells were changed in wild-type TIME and how BCL9 driven Wnt signaling alters transcriptional profile of CD8+ T cells. Third, in the hsBCL9CT-24 treated CT26/MC38 model, hsBCL9CT-24, which is a BCL9 and β-cat inhibitor, suppressed transcription of Wnt signaling both in immune cells and cancer cells, without depleting or knockout BCL9 and β-cat protein.
In the CT26 microsatellite stable (MSS) tumor model, anti-PD-1 treatment showed a TGI of 33.88%. Tumor growth was significantly reduced in Bcl9-depleted tumors treated with anti-PD-1 antibody, compared to NT-shRNA tumors receiving anti-PD-1 alone, with a TGI of 81.8% by day 16 (Fig. 2a, b), while in the MC38 model with MSI, anti-PD-1 therapy displayed a TGI of 56.72%. We also observed in the same model that depletion of Bcl9 markedly decreased the tumor size in response to PD-1 antibody, with a TGI of 87.06% by day 18 (Fig. 2f). Our results show that Bcl9 depletion enhanced the therapeutic effect of anti-PD-1 antibody in both models irrespective of MSS or MSI.
Although our results suggest that targeting the BCL9-CD155-CD226 cascade might have therapeutic implication for CRC, additional preclinical studies are required to validate this hypothesis.
In summary, we found that BCL9 suppression reduced tumor growth, promoted CD8+ T cell tumor infiltration, and enhanced anti-PD-1 response. Single-cell RNA-seq revealed that CD226 and CD96 immune checkpoints plays an important role in TIME. Our analyses confirmed many important observations that were previously made by bulk sequencing, either in vitro or in animal models, and highlighted key areas for further studies of BCL9 in the tumor immune microenvironment. These findings would extend our understanding of BCL9 involvement in the tumorigenesis of CRC.
Materials and methods
Mouse xenograft model
CT26 Bcl9/NT-shRNA cancer cells were cultured as above and harvested for subcutaneously (s.c.) inoculation (4 × 105 cells in PBS) in the right flank region of BALB/c female mice (purchased from Charles River) at 6–8 weeks of age. MC38 Bcl9/NT-shRNA cancer cells were cultured as above and harvested for s.c. inoculation (1 × 106 cells in PBS) in the right flank region of C57BL/6 female mice (purchased from Charles River) at 6–8 weeks of age. BCL9–/– mice, on a C57BL/6 background and were obtained from Konrad Basler’s laboratory, Switzerland. MC38 cancer cells were cultured as above and harvested for s.c. inoculation (1 × 106 cells in PBS) in the right flank region of BCL9–/– or WT C57BL/6 female mice. Tumor volume was measured every other day (V = 0.5 ab,2 where a and b are the long and short diameters of the tumor, respectively). Tumor growth inhibition rate (TGI %) per dosing group was calculated according to the following formula: TGI% =[1 − (TVi − TV0)/(TVvi − TVv0)] × 100%, in which TVi represents the mean tumor volume of a dosing group on a specific day, TV0 is the mean tumor volume of a dosing group on day 0, TVvi is the mean tumor volume of vehicle group on a specific day, and TVv0 is the mean tumor volume of vehicle group on day 0. All the animal experiments and care protocols were approved by the Animal Care Committee of Fudan University and conformed to the Animal Management Rules of the National Health Commission of the People’s Republic of China.
In the CT26 model survival experiment, mice were grouped into six randomized cohorts (n = 8) and given IgG control, anti-PD-1 antibody [10 mg/kg, intraperitoneal (i.p.) injection, twice-weekly (BIW)], or a combination arm [Bcl9-shRNA + anti-PD-1 antibody] via i.p. injection. Tumor size of >2000 mm3 was set as the endpoint.
In the CT26 combination experiment (n = 5) and MC38 (n = 8) model, mice were grouped into six randomized cohorts and given IgG control, anti-PD-1 antibody (10 mg/kg, i.p., BIW), or a combination arm (Bcl9-shRNA + anti-PD-1 antibody) via i.p. injection.
For CD8 depletion, anti-CD8α (YTS169.4, BioXcell BE0117) was injected i.p. (300 μg per mouse) at days 12, 15, and 19 after tumor cell inoculation.
10x library preparation and sequencing
The single cell suspensions concentration was 700–1200 cells/μl to input and barcode with a 10x Chromium Controller (10x Genomics). Each barcoded cell sample’s RNA was reverse-transcribed to generate libraries with the reagents from 10x genomics Single Cell 5’ Gel Bead Kit according to the manufacturer’s instructions. Finally sequencing was performed with an Illumina Hiseq 3000 according to the manufacturer’s instructions (Illumina).
RNA isolation and RNA-Seq analysis
We obtained a total of 6 tumor samples of vehicle and hsBCL9CT-24 groups, from which we extracted RNA using the Tirol regent above. Samples were sequenced using the BGISEQ-500 platform at The Beijing Genomics Institute (BGI) for Genomics and Bioinformatics. Raw counts were then normalized to fragments per kilobase of transcript per million mapped reads (FPKM). Differential gene expression was performed based on the negative binomial distribution with the DEGSeq package using the default settings (Wald significance test). To identify enriched signaling pathways, we utilized GSEA and GO analysis.
Quantification and statistical analysis
PCA, t-SNE, and UMAP analysis
All cells that had the number of UMI sequences of low-quality were removed. From the remaining cells, the similarity and variability of cells were summarized by principle component analysis (PCA). As a result, the similarity between cells was observed by the aid of PCA. The expression trend of cell genes is proportional to the sample distance. For visualization, the dimensionality of each dataset was further reduced using either the Barnes-Hut t-Distributed Stochastic Neighbor embedding (t-SNE) or Uniform Manifold Approximation and Projection (UMAP) with Seurat functions Run-TSNE and Run-UMAP. The 7 principle components were summarized and visualized by tSNE (t-Distributed Stochastic Neighbor Embedding) using the default settings of the RunTSNE function. We reanalyzed cells from each of these seven cell types separately in order to identify subclusters. The cells were contrasted using the Seurat Find Markers function to identify marker genes for these subclusters.
TMB analysis
TCGA COAD patient mutation information and expression data was downloaded from BROAD GDAC Firehose (http://firebrowse.org/). We kept only the primary solid tumor patient samples for the following analysis. TMB score was calculated as follows: total number of truncating mutations * 2.0 + total number of non-truncating mutations * 1.0 Truncating mutations include nonsense, frame-shift deletion or insertion, and splice-site mutations. Non-truncating mutations include missense, in-frame deletion or insertion, and nonstop mutations.58
GSVA analysis
GSVA analysis of four group samples was shown as the average expression of each gene of the related cells by using the C2 KEGG pathway subclass data in the MsigDB database to obtain the GSVA score of each pathway. In these data, stimulation of CD8+ T signaling was enriched in tumor-infiltrating T cells in the hsBCL9CT-24 treated tumor compared with vehicle. TLR and IL2 stimulation signaling were enriched in tumor-infiltrating T cells in tumor bearing Bcl9-shRNA compared with NT-shRNA.
SCENIC analysis
The SCENIC analysis of four group samples were run using the 20-thousand motifs database for RcisTarget and GRNboost (SCENIC version 0.1.5). The input matrix was the normalized expression matrix, output from Seurat.
Statistical analysis
SPSS 22.0 for windows (Chicago, IL, USA) was used for data analysis, and statistical significance was determined using a t-test. Numerical data processing and statistical analysis were performed with GraphPad Prism 8; values are expressed as means ± SEM. The P values were then calculated using unpaired one/two-tailed Student-t tests. P < 0.05 was considered statistically significant. For analysis of TCGA dataset, A Kaplan–Meier curve was constructed to compare the overall and disease-free survival rates of two groups. Log-Rank P value and HR were calculated with SPSS 22.0. Correlation analysis of gene expression in tumor-infiltrating immune cells was conducted with TIMER. Non- parametric test was applied to small cohort.
Supplementary information
Supplementary information marked up changes
Acknowledgements
We thank Shuru Shen for assistance in sample preparation, FACS analysis, and animal studies. This study was partially supported by grants from National Natural Science Foundation of China (81872895 and 82073881 to D.Z.; 81872915, 82073904, and 82011530150 to M.-W.W.), Shanghai Municipal Education Commission (Shanghai Top-Level University Capacity Building Program DGF817029-04 to M.-W.W.), Shanghai Science and Technology Commission (18ZR1403900, 20430713600, and 18JC1413800 to D.Z.) and Fudan-SIMM Joint Research Fund (FU-SIMM20181010 to D.Z. and D.Y.).
Author contributions
D.Z. contributed the idea, oversaw the project, analyzed the data, and prepared the manuscript. M.F. collected tumor samples, analyzed the data, and edited the manuscript. Z.E.W. and S.M. performed bioinformatics analysis and participated in manuscript writing. Z.W. performed GSEA data analysis. Y.Z. and D.Y. prepared the samples for scRNA-seq. E.T. collected tumor samples and participated in western blot analysis. Y.Z. performed qPCR analysis. C.L. and F.H. collected tumor samples. H.L participated in western blot analysis. C.X. participated in data analysis. J.J., J.D., K.Y. and J.B.Q. edited the manuscript. M.-W.W. and D.L. advised the project, analyzed the data, and wrote the manuscript jointly with D.Z.
Data availability
Public Data Resources: The TCGA datasets, including COAD and READ, were downloaded from cBioPortal (http://www.cbioportal.org/). Human scRNA-seq data accession number: SUB9924819. All high-throughput data, flow cytometry data, and immunohistochemical data supporting the current study have been deposited in https://figshare.com/s/970ed37d44f9ac3489db. The custom code used in the manuscript is published at https://figshare.com/s/970ed37d44f9ac3489db.
Competing interests
The authors declare no competing interests.
Contributor Information
Diether Lambrechts, Email: diether.lambrechts@kuleuven.be.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Di Zhu, Email: zhudi@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-021-00730-0.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Halama N, et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell. 2016;29:587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Stintzing S, et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019;30:1796–1803. doi: 10.1093/annonc/mdz387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat. Rev. Clin. Oncol. 2020;17:11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner H, Chen C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br. J. Cancer. 2018;119:785–792. doi: 10.1038/s41416-018-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin-Acevedo JA, et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J. Hematol. Oncol. 2018;11:39. doi: 10.1186/s13045-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31:2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin PJ, Kinzler KW, Sparks A. B. beta-catenin mutations: insights into the APC pathway and the power of genetics. Cancer Res. 2016;76:5587–5589. doi: 10.1158/0008-5472.CAN-16-2387. [DOI] [PubMed] [Google Scholar]
- 11.Sampietro J, et al. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol. Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Feng M, et al. Pharmacological inhibition of beta-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating Treg cells. Sci. Adv. 2019;5:eaau5240. doi: 10.1126/sciadv.aau5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2019;29:44–65. doi: 10.1016/j.tcb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Tian T, Kalland KH, Ke X, Qu Y. Targeting Wnt/beta-catenin signaling for cancer immunotherapy. Trends Pharm. Sci. 2018;39:648–658. doi: 10.1016/j.tips.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Yang K, et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548:602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, et al. beta-catenin mediates tumor-induced immunosuppression by inhibiting cross-priming of CD8(+) T cells. J. Leukoc. Biol. 2014;95:179–190. doi: 10.1189/jlb.0613330. [DOI] [PubMed] [Google Scholar]
- 17.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 18.Gattinoni L, Ji Y, Restifo NP. Wnt/beta-catenin signaling in T-cell immunity and cancer immunotherapy. Clin. Cancer Res. 2010;16:4695–4701. doi: 10.1158/1078-0432.CCR-10-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert C, et al. Pembrolizumab versus Ipilimumab in advanced melanoma. N. Engl. J. Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 20.Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4:522–526. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reymond N, et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 2004;199:1331–1341. doi: 10.1084/jem.20032206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima H, et al. CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J. Biol. Chem. 2003;278:36748–36753. doi: 10.1074/jbc.M300702200. [DOI] [PubMed] [Google Scholar]
- 23.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 24.Shibuya A, et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573–581. doi: 10.1016/S1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, et al. Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5. Proc. Natl Acad. Sci. USA. 2019;116:988–996. doi: 10.1073/pnas.1815716116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Zheng Q, Xin N, Wang W, Zhao C. CD155, an onco-immunologic molecule in human tumors. Cancer Sci. 2017;108:1934–1938. doi: 10.1111/cas.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 28.Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 2017;276:112–120. doi: 10.1111/imr.12518. [DOI] [PubMed] [Google Scholar]
- 29.Bottino C, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 31.Deuss FA, Watson GM, Fu Z, Rossjohn J, Berry R. Structural basis for CD96 immune receptor recognition of nectin-like protein-5, CD155. Structure. 2019;27:219–228 e213. doi: 10.1016/j.str.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Husain B, et al. A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol. Cell Proteom. 2019;18:2310–2323. doi: 10.1074/mcp.TIR119.001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li XY, et al. CD155 loss enhances tumor suppression via combined host and tumor-intrinsic mechanisms. J. Clin. Invest. 2018;128:2613–2625. doi: 10.1172/JCI98769. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Lepletier A, et al. Tumor CD155 expression is associated with resistance to Anti-PD1 immunotherapy in metastatic melanoma. Clin. Cancer Res. 2020;26:3671–3681. doi: 10.1158/1078-0432.CCR-19-3925. [DOI] [PubMed] [Google Scholar]
- 35.Klose J, et al. Salinomycin inhibits metastatic colorectal cancer growth and interferes with Wnt/β-catenin signaling in CD133+ human colorectal cancer cells. BMC Cancer. 2016;16:896. doi: 10.1186/s12885-016-2879-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan G, et al. Novel role of STRAP in progression and metastasis of colorectal cancer through Wnt/¦Â-catenin signaling. Oncotarget. 2016;7:16023–16037. doi: 10.18632/oncotarget.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, et al. beta-Catenin inhibition shapes tumor immunity and synergizes with immunotherapy in colorectal cancer. Oncoimmunology. 2020;9:1809947. doi: 10.1080/2162402X.2020.1809947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoadley KA, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaud, G. et al. The costimulatory molecule CD226 signals through VAV1 to amplify TCR signals and promote IL-17 production by CD4(+) T cells. Sci Signal10.1126/scisignal.aar3083 (2018). [DOI] [PubMed]
- 41.Gilfillan S, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 2008;205:2965–2973. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molfetta, R. et al. CD155: A multi-functional molecule in tumor progression. Int. J. Mol Sci. 10.3390/ijms21030922 (2020). [DOI] [PMC free article] [PubMed]
- 43.Lanier LL. NK cell recognition. Annu Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 44.Solecki DJ, Gromeier M, Mueller S, Bernhardt G. n. & Wimmer, E. Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J. Biol. Chem. 2002;277:25697–25702. doi: 10.1074/jbc.M201378200. [DOI] [PubMed] [Google Scholar]
- 45.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gobert M, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res. 2009;69:2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- 47.Iellem A, et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 2001;194:847–853. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiyama D, et al. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl Acad. Sci. USA. 2013;110:17945–17950. doi: 10.1073/pnas.1316796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganesh K, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019;16:361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gay DM, et al. Loss of BCL9/9l suppresses Wnt driven tumourigenesis in models that recapitulate human cancer. Nat. Commun. 2019;10:723. doi: 10.1038/s41467-019-08586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deka J, et al. Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 2010;70:6619–6628. doi: 10.1158/0008-5472.CAN-10-0148. [DOI] [PubMed] [Google Scholar]
- 53.Mieszczanek J, van Tienen LM, Ibrahim AEK, Winton DJ, Bienz M. Bcl9 and Pygo synergise downstream of Apc to effect intestinal neoplasia in FAP mouse models. Nat. Commun. 2019;10:724. doi: 10.1038/s41467-018-08164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moor AE, et al. BCL9/9L-beta-catenin Signaling is Associated With Poor Outcome in Colorectal Cancer. EBioMedicine. 2015;2:1932–1943. doi: 10.1016/j.ebiom.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mani M, et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577–7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, L. & Shay, J. W. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. J. Natl Cancer Inst. 109, 10.1093/jnci/djw332 (2017). [DOI] [PMC free article] [PubMed]
- 57.Johnston RJ, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Li M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol. 2019;20:4. doi: 10.1186/s12865-018-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information marked up changes
Data Availability Statement
Public Data Resources: The TCGA datasets, including COAD and READ, were downloaded from cBioPortal (http://www.cbioportal.org/). Human scRNA-seq data accession number: SUB9924819. All high-throughput data, flow cytometry data, and immunohistochemical data supporting the current study have been deposited in https://figshare.com/s/970ed37d44f9ac3489db. The custom code used in the manuscript is published at https://figshare.com/s/970ed37d44f9ac3489db.