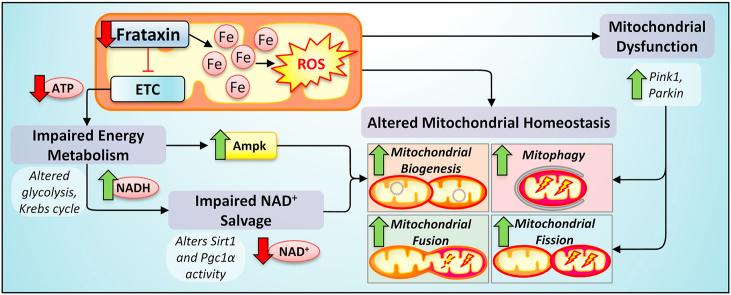
Abstract
Due to the high redox activity of the mitochondrion, this organelle can suffer oxidative stress. To manage energy demands while minimizing redox stress, mitochondrial homeostasis is maintained by the dynamic processes of mitochondrial biogenesis, mitochondrial network dynamics (fusion/fission), and mitochondrial clearance by mitophagy. Friedreich's ataxia (FA) is a mitochondrial disease resulting in a fatal hypertrophic cardiomyopathy due to the deficiency of the mitochondrial protein, frataxin. Our previous studies identified defective mitochondrial iron metabolism and oxidative stress potentiating cardiac pathology in FA. However, how these factors alter mitochondrial homeostasis remains uncharacterized in FA cardiomyopathy. This investigation examined the muscle creatine kinase conditional frataxin knockout mouse, which closely mimics FA cardiomyopathy, to dissect the mechanisms of dysfunctional mitochondrial homeostasis. Dysfunction of key mitochondrial homeostatic mechanisms were elucidated in the knockout hearts relative to wild-type littermates, namely: (1) mitochondrial proliferation with condensed cristae; (2) impaired NAD+ metabolism due to perturbations in Sirt1 activity and NAD+ salvage; (3) increased mitochondrial biogenesis, fusion and fission; and (4) mitochondrial accumulation of Pink1/Parkin with increased autophagic/mitophagic flux. Immunohistochemistry of FA patients' heart confirmed significantly enhanced expression of markers of mitochondrial biogenesis, fusion/fission and autophagy. These novel findings demonstrate cardiac frataxin-deficiency results in significant changes to metabolic mechanisms critical for mitochondrial homeostasis. This mechanistic dissection provides critical insight, offering the potential for maintaining mitochondrial homeostasis in FA and potentially other cardio-degenerative diseases by implementing innovative treatments targeting mitochondrial homeostasis and NAD+ metabolism.
Keywords: Iron loading, Iron, Mitochondria, Cardiomyopathy, Mitochondrial homeostasis
Graphical abstract
Highlights
-
•
Mitochondrial dysfunction due to frataxin-deficiency in the heart involved dysregulation of iron and redox biology.
-
•
Cardiac frataxin-deficiency caused mitochondrial proliferation with abnormal cristae structure.
-
•
Enhanced Pgc1α pathway upon frataxin-deficiency promoted mitochondrial biogenesis.
-
•
Increased mitochondrial fission and fusion potentiated proliferation of smaller, but elongated mitochondria.
-
•
Mitochondrial dysfunction upon frataxin-deficiency activated Pink1/Parkin signaling and increased mitophagic flux.
-
•
Impaired NAD + metabolism is due to perturbations in Sirt1 activity and NAD + salvage upon frataxin-deficiency.
Abbreviations
- Ampk
AMP-activated protein kinase
- BAF
Bafilomycin A1
- ddPCR
digital droplet PCR
- Drp1
dynamin-related protein 1
- EDS
energy dispersive X-ray spectroscopy
- FA
Friedreich's ataxia
- Fis1
mitochondrial fission protein 1
- FU
relative fluorescence unit
- Hsp90
heat shock protein 90
- H & E
hematoxylin and eosin
- i.p.
intraperitoneal injection
- ISC
iron-sulfur cluster
- KO
Knockout
- Lc3
microtubule-associated protein 1 light chain 3A
- LV
left ventricular
- Mff
mitochondrial fission factor
- Mfn1
Mitofusin 1
- mtDNA
mitochondrial DNA
- NAM
nicotinamide
- Nampt
nicotinamide phosphoribosyltransferase
- ND1
NADH dehydrogenase subunit 1
- NMN
nicotinamide mononucleotide
- Nmnat1
nicotinamide mononucleotide adenylyltransferase 1
- Nrf1/2
nuclear respiratory factors 1 and 2
- OD
optical density
- pAmpk
phosphorylated AMP-activated protein kinase
- Paris
Parkin-interacting substrate
- Pgc1α
peroxisome proliferation activator receptor γ -coactivator 1α
- PPARγ
peroxisome proliferation activator receptor γ
- SDS
sodium dodecyl sulfate
- TEM
transmission electron microscopy
- Tfam
mitochondrial transcription factor A
- WT
wild-type
- β2m
β-2-microglobulin
1. Introduction
Frataxin is a nuclear-encoded, mitochondrial protein [1] that is pivotal for mitochondrial iron metabolism [2], especially for iron-sulfur cluster (ISC) and heme synthesis [1,[3], [4], [5]]. Both ISCs and heme are vital for mitochondrial respiratory complexes, particularly, complex I-III [6]. Accordingly, frataxin-deficiency results in complex I-III inhibition [7]. In humans, the decreased expression of frataxin causes Friedreich's ataxia (FA), which is characterized by ataxia and a fatal hypertrophic cardiomyopathy [1,8]. Yet, despite reduced mitochondrial iron utilization, mitochondrial iron influx continues, causing mitochondrial iron-loading as non-ferritin-bound iron aggregates without a protective ferritin shell [3,4]. Considering that inappropriately bound iron is redox active and toxic [9,10], this could contribute to the redox dysregulation observed in the heart of FA patients [11].
Analogous pathology with mitochondrial iron loading and oxidative stress has also been reported by our laboratory examining the heart of the muscle creatine kinase conditional frataxin knockout mice (referred to herein as “frataxin KO mice”) [3,12]. These mice recapitulate the histological, biochemical and redox biology of FA cardiomyopathy [3,4,8,12,13]. In fact, our previous studies have demonstrated significant protein and glutathione oxidation associated with reactive oxygen species production in the heart, as well as marked perturbation in antioxidant defense mediated by the master regulator of the antioxidant response, nuclear factor erythroid 2-related factor-2 [12]. The oxidative damage to mitochondrial proteins and mitochondrial DNA (mtDNA) could lead to further mitochondrial dysfunction and mtDNA loss [14].
Damage to mtDNA severely affects mitochondrial function due to the 13 mtDNA-encoded proteins vital for the electron transport chain [15]. In FA patients, mtDNA is decreased in the heart and cerebellum [16]. In yeast FA models, increased mtDNA lesions and mtDNA loss were observed subsequent to iron-loading [14,17]. Therefore, mitochondrial redox active iron likely contributes to mtDNA degradation via reactive oxygen species generation. Mitochondrial dysfunction depletes ATP and NAD+ levels, and leads to the release of mitochondrial pro-apoptotic proteins [18,19]. Thus, the selective recycling of defective mitochondria by mitochondrial-specific autophagy (i.e., mitophagy) prevents these detrimental effects [18]. Mitophagy is mediated by the binding of the autophagosomal protein, microtubule-associated protein 1 light chain 3A (Lc3), to outer mitochondrial membrane adapters, such as Fundc1 [18]. Our previous studies using the frataxin KO mouse heart identified an accumulation of autophagic/mitophagic markers, including Lc3, p62 and Fundc1, suggesting the induction of autophagy/mitophagy and/or inhibition of autophagic flux [8].
Mitochondria constantly undergo fusion or fission, which is collectively known as “mitochondrial network dynamics” [20,21]. In cardiomyocytes and neurons, due to their high energy demand, mitochondrial dynamics plays significant roles in governing their viability [18,20,21]. As neural and cardio-degeneration are major pathologies of FA, the alterations in mitochondrial dynamics warrants investigation. Recently, two major regulators of mitochondrial dynamics and mitophagy were identified: Parkin, an E3 ubiquitin ligase; and Pink1, a Ser/Thr kinase that translocates to damaged mitochondria to recruit and activate Parkin [22]. Activated Parkin also translocates to the damaged mitochondria and poly-ubiquitinates outer mitochondrial membrane proteins, such as the mitochondrial fusion protein, mitofusin 1 (Mfn1). This leads to Mfn1 degradation, inhibition of mitochondrial fusion, and ensuing mitophagic degradation [22,23]. Conversely, mitochondrial fission is facilitated by the adapter-mediated targeting of dynamin-related protein 1 (Drp1) to mitochondrial constriction sites, where it constricts the outer mitochondrial membrane and causes mitochondrial fission [24].
For mitochondrial biogenesis to restore mitochondrial deficits, the peroxisome proliferation activator receptor (PPAR) γ-coactivator 1α (Pgc1α) is the major transcriptional regulator of mitochondrial biogenesis and energy metabolism [25,26]. In response to metabolic deficiency, Pgc1α is induced and enhances the transcriptional induction of nuclear respiratory factors (Nrf) 1 and 2, and the mitochondrial transcription factor A (Tfam) [[25], [26], [27]]. Nrf1/2 and Tfam then coordinate the transcription of nuclear DNA and mtDNA-encoded mitochondrial proteins, respectively [26,27].
The regulation of Pgc1α is highly complex, involving numerous transcriptional and post-translational mechanisms. These include: (1) transcriptional activation by 5′adenosine monophosphate-activated protein kinase (Ampk), or repression by the Parkin-interacting substrate (Paris) [[28], [29], [30]]; (2) phosphorylation by Ampk leading to Pgc1α activation [31]; (3) phosphorylation by glycogen synthase kinase 3β (Gsk3β) leading to Pgc1α nuclear proteasomal degradation [32,33]; and (4) deacetylation by the nuclear NAD+-dependent histone deacetylase, Sirt1, leading to enhanced Pgc1α activity [28,29].
The mechanisms governing mitochondrial homeostasis in the heart when challenged with severe mitochondrial dysfunction, iron-loading and redox stress due to frataxin-deficiency remain uncharacterized. Herein, we implemented the well-characterized frataxin KO mouse model that leads to deletion of frataxin in the heart and skeletal muscle [1,3,4,8,12,13,34]. This model closely mimics human FA cardiac pathophysiology, and herein, we identify pronounced mitochondrial proliferation in the heart of the KO versus their WT littermates. These mitochondria in the KO demonstrate abnormally condensed and ill-defined cristae associated with markers of mitochondrial dysfunction, e.g., increased Pink1 and Parkin. The mechanisms of mitochondrial proliferation entail increased mitochondrial biogenesis, increased mitochondrial fission, with perturbations in NAD+ metabolism being evident in the KO mice. Therefore, maintenance of intracellular NAD+, mitochondrial electron transport, and the related redox state of the cell, could represent a strategy for treating mitochondrial cardiomyopathy and the impaired cardiac function in FA.
2. Methods
2.1. Animals
The muscle creatine kinase-Cre conditional frataxin KO mice (homozygous for the loxP-flanked Frda exon 4 allele) and their WT (homozygous for the wild-type Frda allele) littermates were generated on a mixed C57Bl/6J and 129/Sv-ter background and maintained and genotyped, as described [8,12,34]. Both male and female mice were examined at either 4 weeks or 10 weeks of age. Of note, there were no statistically significant differences identified between the same genotype across genders. Approximately equal number of males and females in each genotype were used for all assessments. All representative western blots show a pair of WT and KO littermates of the same sex. Animal procedures were approved by the Animal Ethics Committee of The University of Sydney (AEC2018/1406).
To assess autophagic flux, 10 week WT and KO littermates (n = 6/genotype/group) were administered either saline (vehicle) or Bafilomycin A1 (BAF; 6 mg/kg; Cayman Chemical, Ann Arbor, MI) via i.p. injection. After 30 min, mice were then anesthetized using isoflurane and the left ventricular (LV) tissues were excised for protein analysis. Mice were randomly assigned to groups with similar gender and weight distribution.
2.2. Transmission electron microscopy (TEM)
Hearts of WT and KO mice were trans-cardiac perfused, washed and fixed using K buffer (2.5% glutaraldehyde, 4% paraformaldehyde in 0.1 mol/L phosphate buffer, pH 7.4). LV tissue was dissected, post-fixed with 1% osmium tetroxide and 1.5% potassium ferricyanide, and prepared according to standard TEM sample preparation protocols [35]. LV tissues were imaged using a JEOL JEM-1400 TEM equipped with an energy dispersive X-ray spectroscopy (EDS) system operated at 120 kV. EDS was performed at scanning TEM mode. TEM images were analyzed using ImageJ Fiji (NIH, Bethesda, MD, USA).
Appropriate blinding of investigators was utilized when assessing mitochondrial morphology. Measurements were taken from at least 3 images/mouse (6 WT and 6 KO per age group) and more than 100 mitochondria/image. The average number of mitochondria were measured by quantifying the number of mitochondria/image [36]. The mitochondria: myocyte area ratio was calculated by normalizing the total area occupied by the mitochondria/cardiomyocyte by the total area of each cardiomyocyte. The mitochondrial cross-sectional area was quantified from the cross-sectional area of each individual mitochondrion/image. The mitochondrial aspect ratio was calculated by measuring the longest axis divided by the shortest axis per mitochondrion, as performed previously [36].
2.3. Protein isolation, mitochondrial isolation, and western blot analysis
Whole hearts from WT and frataxin KO mice were trans-cardiac perfused with ice-cold PBS, then the LV tissues were excised and homogenized on ice in lysis buffer (150 mmol/L NaCl, 10 mmol/L Tris-HCl [pH 7.4], 0.5% (w/v) sodium dodecyl sulfate (SDS), 1 mmol/L EDTA, 40 μmol/L NaF, 1% (v/v) Triton X-100) containing a 1 x solution of PhosSTOP (Roche Diagnostics; Risch-Rotkreuz, Switzerland) and a 1x solution of protease inhibitor cocktail (Roche Diagnostics) using standard procedures [12]. To assess mitochondrial protein, the Mitochondrial Isolation Kit for Tissue (Thermo Fisher Scientific, Waltham, MA, USA) was implemented. Western blots were performed as previously described [12].
Primary antibodies were used at dilutions from 1/500–1/2000 and included Pgc1α (ab54481; Abcam, Cambridge, MA, USA), Nrf1 (ab34682; Abcam), Tfam (ab131607; Abcam), Ampk (ab32047; Abcam), pAmpk (Thr172; 2535S; Cell Signaling Technology, Danvers, MA, USA), Gsk3β (9832; Cell Signaling Technology), pGsk3β (Ser9; 9332; Cell Signaling Technology), Sirt1 (ab7343; Abcam), Paris (ab130867; Abcam), PPARγ (16643-1-AP; ProteinTech, Rosemont, IL, USA), anti-acetyl lysine (ab80178; Abcam), Mfn1 (13798-1-AP; ProteinTech), Opa1 (ab42364; Abcam), Drp1 (ab56788; Abcam), Fis1 (10956-1-AP; ProteinTech), Mff (ab81127; Abcam), Pink1 (NB100-493; Novus Biology, Centennial, CO, USA), Parkin (ab77924; Abcam), Lc3 (PD014; MBL International, Woburn, MA, USA), Fundc1 (ARP53280_P050; Aviva Systems Biology, San Diego, CA, USA), Sqstm1/p62 (ab56416; Abcam), Gapdh (2118S; Cell Signaling Technology), Tomm20 (H00009804-M01; Abnova, Walnut, CA, USA), and Hsp90 (ab13495; Abcam). Secondary antibodies used were goat anti-rabbit and goat anti-mouse (A0545 and A9917, respectively; 1/10,000; Sigma-Aldrich, St. Louis, MO, USA) conjugated with horseradish peroxidase.
Densitometric data were expressed as “Protein/Gapdh KO versus WT (a.u.)” or “Protein/Tomm20 KO versus WT (a.u.)”, where the expression of the protein of interest was first normalized to the protein-loading control, Gapdh or Tomm20, respectively, for all samples. All data were universally divided by the average WT value, thereby maintaining the coefficient of variation of these data.
2.4. Genomic DNA isolation and digital droplet PCR
Genomic DNA was extracted using the Wizard® SV Genomic DNA Purification System (Promega, Madison, WI, USA). Genomic DNA concentration was measured using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
To assess mtDNA copy number, the mtDNA gene, NADH-ubiquinone oxidoreductase chain 1 (ND1), was examined by digital droplet PCR (ddPCR) using an established protocol [15]. Briefly, 100 pg of genomic DNA was used in a 20 μL duplex reaction containing ddPCR Supermix (BioRad, Hercules, CA, USA), 5 U HaeIII restriction enzyme (NEB, Ipswich, MA, USA), and 1 μL of the nuclear or mtDNA probes. For the mtDNA probe, ND1 (FAM; Assay ID: dMmuCNS343824284; Bio-Rad) was implemented. For the nuclear genome probe, β-2-microglobulin (β2m; HEX; Assay ID: dMmuCNS576316288, Bio-Rad) was used. Standard ddPCR procedure was conducted following the manufacturer's instructions (Bio-Rad). To deduce mtDNA copies/copy of nuclear DNA in the heart, ND1 copy number were normalized to that of the single-copy nuclear gene, β2m.
2.5. RNA isolation and RT-qPCR
Total mRNA was isolated from the LV tissue of WT and frataxin KO mice according to previously described methods [12]. The cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). RT-qPCR was performed using a Roche LightCycler 480 PCR (Roche Diagnostics; Risch-Rotkreuz, Switzerland). Taqman probes (Thermo Fisher Scientific) targeting mouse Nrf1 (Mm00447996_m1), Tfam (Mm00447485_m1) and Gapdh (Mm99999915_g1) were used. Data were analyzed by the ΔΔCT method using Genex Software (MultiD Analyses, Göteborg Sweden). Analysis of gene expression fold change was conducted by an investigator who was blinded to mouse group identities.
2.6. Immunoprecipitation
Upon extensive perfusion of whole hearts from WT and frataxin KO mice with ice-cold PBS, LV tissue was excised and homogenized in 500 μL of ice-cold RIPA buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS and 1x solution of protease and phosphatase inhibitors (Roche Diagnostics)] using a Dounce glass homogenizer [12]. Immunoprecipitation was performed as described previously [12], but using Pgc1α antibody (5 μg; ab54481; Abcam).
2.7. Nuclear Sirt1 deacetylase activity
Sirt1 deacetylase activity was evaluated in the nuclear extract from PBS perfused LV tissues using the CycLex SIRT1/Sir2 Deacetylase Fluorometric Assay Kit (CycLex, Nagano, Japan), as previously described [37]. Briefly, the final reaction mixture (100 μL) contained 50 mmol/L Tris-HCl (pH 8.8), 4 mmol/L MgCl2, 0.5 mmol/L DTT, 0.25 mA/mL Lysyl endopeptidase, 1 μmol/L Trichostatin A, 200 μmol/L NAD+, and 5 μL of nuclear sample. After mixing thoroughly, samples were incubated for 10 min/RT and the fluorescence intensity (ex. 340 nm; em. 460 nm) was measured every 30 s for a total of 60 min immediately after the addition of the fluoro-substrate peptide (20 μmol/L final concentration) using a FLUOstar Optima Fluorometer (BMG Labtech, Cary, NC, USA) and normalized by the protein content using the Bradford protein assay. These results are reported as relative fluorescence unit (FU)/mg of protein. Assessment of enzymatic activity was conducted by an investigator who was blinded to mouse group identities.
2.8. Intracellular NAD+, NADH, NAM and NMN assessment
Whole hearts from 4 to 10 week WT and frataxin KO mice were comprehensively perfused with ice-cold PBS (pH 7.4) to remove excess blood prior to organ removal. Then, NAD+, NADH, NAM and NMN levels were extracted from LV homogenates, as previously described [38]. Briefly, 100 μL of homogenate was added to 20 μL of [D4]-NAM in 5% formic acid (v/v). Samples were then mixed with 300 μL of acetonitrile, vortexed for 15 s, centrifuged and then dried via a speed vacuum overnight at 37 °C. Samples were then reconstituted in 100 μL of 10 mmol/L ammonium acetate with 0.1% formic acid. Separation and quantitation of NAD+, NADH, NAM and NMN were performed using a TSQ Vantage bench-top mass spectrometer (ThermoFisher Scientific) operated in positive ion multiple reaction monitoring mode, as described previously [38]. The NAD+, NADH, NAM and NMN were separated on a Phenomenex NH2 column (150 mm × 2 mm x 3 μm; Phenomenex, Torrance, CA) using the same gradient and mobile phase, as described previously [38].
Briefly, the analytes were separated using a binary solvent gradient consisting of 5 mmol/L NH4OAc (pH 9.5) adjusted with ammonia (mobile phase “A”) and acetonitrile (mobile phase “B”) with a flow rate of 250 μL/min. The initial solvent composition at injection was 25% A, followed by a 2 min gradient to 45% “A” and a fast gradient ramp to 80% “A” (0.1 min), which was maintained for 5.9 min. Then, “A” was increased again to 95% (2 min), held for 13 min and then reverted to initial conditions (0.1 min) for equilibration, with a total run time of 30 min. The percent coefficient of variation ranged from 2.8 to 15.8%, with the limit of quantification being 0.1–0.2 pmol for NAD+, NADH, NAM and NMN [38]. Assessment of NAD+ metabolite levels were conducted by an investigator who was blinded to mouse group identities.
2.9. Nuclear NAD+ consumption assay
NAD+ consumption activity was examined using nuclear extracts from PBS perfused LV tissue, as previously described [37]. The final reaction mixture contained 10 mmol/L MgCl2, Triton X-100 (1%), and 20 μmol/L NAD+ in 50 mmol/L Tris buffer (pH 8.1). The plate was then incubated for 1 h/37 °C and the amount of NAD+ consumed was measured as described above. NAD+ consumption was normalized to protein concentration using the Bradford protein assay. Assessment of NAD+ consumption was conducted by an investigator who was blinded to mouse group identities. The results were expressed as μg NAD+ consumed/h/mg nuclear protein.
2.10. ATP assessment
Intracellular ATP levels were measured from harvested LV heart tissue from WT and frataxin KO mice that had been comprehensively perfused with ice-cold PBS. The assessment of ATP levels was performed using a luciferin-luciferase assay system (P1041 and E1500, Promega), as previously described [39]. In brief, tissue was homogenized in 250 μL of extraction solution (10 mmol/L KH2PO4, 4 mmol/L MgSO4, pH 7.4), heated for 98 °C/4 min, and placed on ice. Then, 50 μL of sample was added to a 100 μL reaction solution (50 mmol/L NaAsO2, 20 mmol/L MgSO4, pH 7.4, and 800 μg of luciferin/luciferase enzyme). Luminosity at 259 nm was measured with an extinction coefficient of 15,400 mol/L−1.cm−1 using a Turner Designs TD 20/20 luminometer (Stratec Biomedical Systems, Germany) [39]. ATP levels were normalized to protein concentration using the Bradford protein assay and reported as μg/mg protein. Analysis of ATP levels was conducted by an investigator who was blinded to mouse group identities.
2.11. Human heart immunohistochemistry
LV heart specimens from 5 male and 5 female FA patients were provided by Dr. Arnulf Koeppen (Albany Medical College, Albany, NY, USA). Control LV heart specimens were from 12 age- and gender-matched subjects and include both pre-mortem and post-mortem donors. The pre-mortem donor hearts were provided by Dr. Sean Lal (Sydney Heart Bank, University of Sydney), and were procured for heart transplantation, but not used for this purpose because of logistical reasons. For these samples, anatomical pathology reports confirmed normal histology. The post-mortem control hearts were obtained from the National Disease Research Interchange (Philadelphia, PA, USA). Examination of immunohistochemical staining for the proteins below revealed no significant difference in localization and intensity between pre- and post-mortem tissues. Therefore, the quantification of staining intensity was combined for assessment. Clinical details of the 10 FA patients and 12 controls, including their cause of death, are outlined in Table S1.
For immunohistochemistry, 5 μm paraffin sections were processed by standard methods [40]. Briefly, heat-induced epitope retrieval was conducted with Dako buffers (pH 6.1 or pH 9.0; Agilent Technologies, Santa Clara, CA, USA) using a decloaking chamber (Biocare Medical, Pacheco, CA, USA) for 125 °C/30 s. After a 3% hydrogen peroxide incubation for 5 min and incubation with protein block (Agilent Technologies) for 30 min, primary antibodies against PGC1α (1:750; ab191838; Abcam), MFN1 (1:250; 13798-1-AP; ProteinTech), DRP1 (1:750; ab56788; Abcam), FIS1 (1:500; 10956-1-AP; ProteinTech), and SQSTM1/P62 (1:1000; ab56416; Abcam) were prepared in Dako antibody diluent (Agilent Technologies) and used. Rabbit IgG and mouse IgG2a (Agilent Technologies) were also utilized as relevant isotype controls. Following incubation with Dako EnVision Peroxidase Polymer (anti-rabbit or anti-mouse), antigen signals were detected using Dako DAB+ Chromogen Substrate (Agilent Technologies). Slides were then counter-stained by hematoxylin staining and mounted in DPX (Sigma-Aldrich).
DAB staining was quantified using ImageJ Fiji software. For each image, DAB staining intensity was calculated using its optical density (OD = log [max intensity/mean intensity]) and normalized to the number of cardiomyocytes to correct for the extensive fibrotic lesions in the FA patient specimens. Between 9 and 11 images were captured per patient specimen and analyzed to deduce a representative OD/patient specimen. These values were then averaged across patient specimens to give a final mean DAB intensity for the control and FA patient groups. Hematoxylin and eosin (H & E) staining were utilized to assess general tissue architecture, as performed previously [8]. Human tissue handling procedures were approved by the Human Ethics Committee of The University of Sydney (HREC2012/2814, HREC2019/399).
2.12. Statistical analysis
Normality of data were assessed by Shapiro-Wilk test. Homogeneity of variance of datasets were assessed by the Bartlett test. Data with normal distribution was subjected to parametric Student's t-test (two-tailed), or one-way ANOVA with Tukey's post hoc test. Data were considered statistically significant when p < 0.05. Results are expressed as mean ± SEM. Statistical analyses were conducted using GraphPad Prism Ver. 8.2.1 (San Diego, CA, USA).
3. Results
3.1. Progressive proliferation of disorganized and dysfunctional mitochondria in the frataxin-deficient heart
Our previous studies comprehensively characterized the molecular alterations that result in mitochondrial iron-loading and oxidative damage that hinder mitochondrial function in the heart of the frataxin KO mouse model [1,3,4,8,12]. The processes of mitochondrial homeostasis, i.e., biogenesis, fission, fusion, and mitophagy, are essential for attenuating mitochondrial dysfunction [18], but these have not been comprehensively assessed in FA cardiomyopathy. Considering the iron-loading, redox stress, and mitochondrial damage observed in these mice [4,8,12,34], we hypothesized that mitochondrial homeostasis is markedly dysregulated and this warranted careful investigation.
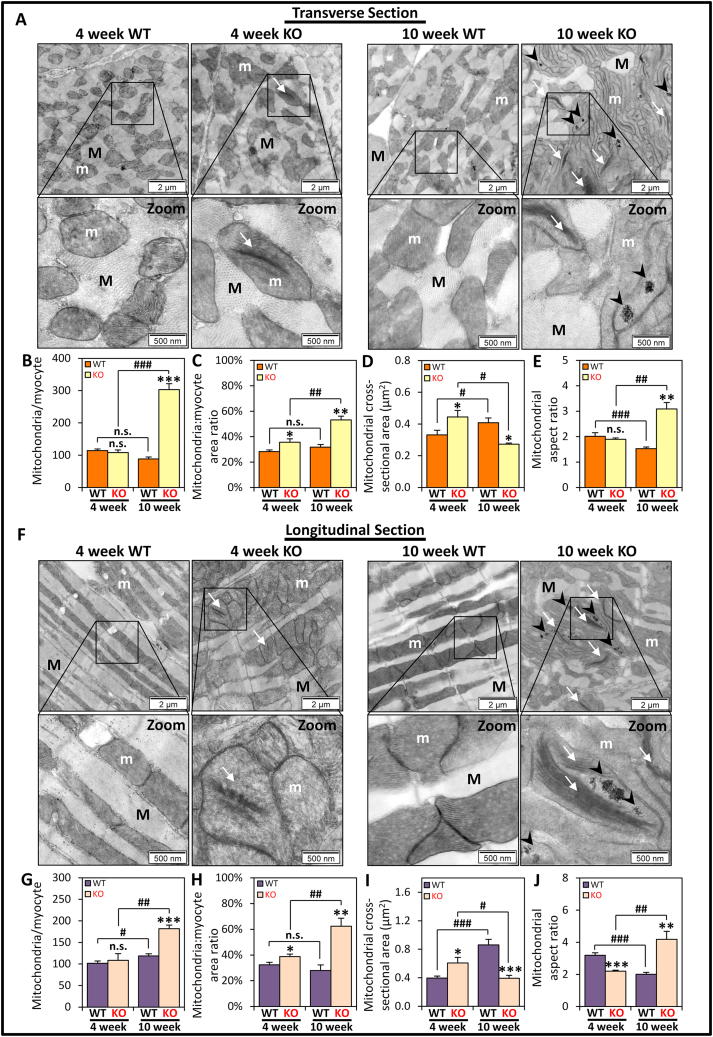
Alterations in mitochondrial homeostasis are well associated with mitochondrial morphological changes [18,20,21,26]. Thus, TEM was initially used to comprehensively examine cardiac mitochondrial morphology in frataxin KO mice versus WT littermates. In these studies, LV sections were utilized, as the KO heart demonstrates defective systolic function, fibrosis and iron-deposits within this region [8]. We quantified the alterations from transverse (Fig. 1A–E) and longitudinal (Fig. 1F–J) sections relative to the muscle fiber, to provide a spatial perspective of both anatomical planes of the cardiomyocyte and its mitochondria. The KO mice at 4 weeks of age were shown to exhibit no gross, histological, or functional cardiac defects relative to the WT, while pronounced cardiac pathology had developed by 10 weeks [3,4,8,12,13,34]. As such, 4 and 10 week mice were compared at the TEM level to assess cardiac disease progression.
Fig. 1.
TEM assessment demonstrates alterations of mitochondrial morphologies in LV sections of 4 and 10 week WT and KO littermates. Representative TEM micrographs showing 4 and 10 week WT and KO LV cardiomyocytes in: (A-E) transverse sections and (F-J) longitudinal sections. Mitochondria (m) were found within the myofiber (M) with condensed mitochondrial cristae (white arrows) in the KO at both ages, while iron-aggregates (black arrowheads) were only in 10 week KO mitochondria. Quantitative assessment of (B, G) the average number of mitochondria, (C, H) mitochondria: myocyte area ratio, (D, I) mitochondrial cross-sectional area, and (E, J) mitochondrial aspect ratio, between 4 and 10 week KO and WT mice. Data are mean ± SEM (n = 6 mice/genotype/age). One-way ANOVA with Tukey's post hoc tests were used. *, p < 0.05; **, p < 0.01; ***, p < 0.001, versus WT of respective age. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 versus 4 week mice. n.s., not significant. Scale bars in (A, F) are 2 μm, or 500 nm, as indicated.
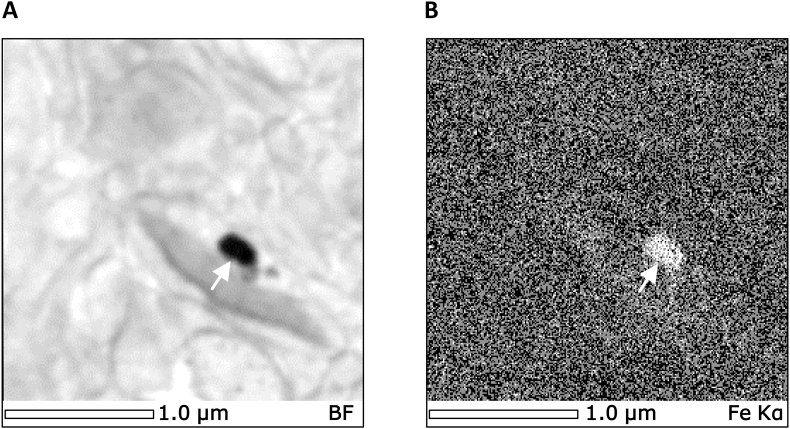
Even in asymptomatic 4 week KO mice, observable differences in mitochondrial morphology could be detected relative to WT littermates in transverse (Fig. 1A–E) and longitudinal (Fig. 1F–J) planes. An interesting feature of the mitochondria from the 4 week KO mice was the presence of condensed mitochondrial cristae that were absent in WTs (white arrows; Fig. 1A, F, left panels). This characteristic and a loss of cristae structure became more prevalent by 10 weeks of age in the KO animal (white arrows; Fig. 1A, F, right panels). Electron-dense accumulations (black arrowheads; Fig. 1A, F) consistent with elemental iron, as determined by energy dispersive X-ray spectroscopy (white arrow; Fig. 2), were only observed within the mitochondria of 10 week KOs, as previously reported [4,12,34]. We did not detect any iron aggregates outside of the mitochondria in either anatomical plane, which provides further evidence that the iron accumulation occurs within the mitochondria. Electron scatter analysis of these mitochondrial iron-aggregates did not result in any identifiable diffraction patterns (data not shown). These results suggested that the iron-aggregates are an amorphous composite. This conclusion is in good agreement with our previous biophysical and biochemical analysis using this same mouse model [4]. In these latter studies, mitochondrial iron accumulation was demonstrated to be a phosphorus, sulfur and iron-containing non-crystalline aggregate [4].
Fig. 2.
Electron-dense X-ray spectroscopy (EDS) analysis demonstrates iron accumulations within the LV mitochondria of 10 week frataxin KO mice. (A) Scanning transmission electron micrograph of the region subjected to EDS analysis. (B) Corresponding X-ray elemental map for iron Kα (6.4 keV) over the identical field shown in (A). The white arrows in (A) and (B) indicate electron-dense iron accumulations. Scale bar is 1 μm, as indicated.
In transverse sections, the spatial distribution of mitochondria in 4 week WT and KO cardiomyocytes appear predominantly as isolated organelles or small clusters distributed amongst the myofibers (Fig. 1A). However, in longitudinal sections, pronounced mitochondrial clustering was observed in the KOs, as opposed to being parallel to the myofibers in WTs (Fig. 1F). Furthermore, mitochondria in 10 week KO hearts were overly abundant in disorganized, entangled masses that displaced sarcomeric structures in both anatomical planes, which contrasts with 10 week WTs (Fig. 1A, F).
Detailed quantitative assessment of the differences in mitochondrial morphology in transverse and longitudinal sections confirmed significantly greater mitochondrial number per myocyte (mitochondria/myocyte) only in the 10 week KO in both planes versus the WT (Fig. 1B, G). The mitochondria: myocyte area ratio of 4 and 10 week KOs in both anatomical planes were significantly increased versus the respective WTs (Fig. 1C, H). This ratio only significantly increased with age in the KO, but not in the WT (Fig. 1C, H), which demonstrates the pathology was specific to the KO animal. The quantitative assessments of these two indicators, namely mitochondria/myocyte and mitochondria: myocyte area ratio (Fig. 1B, C, G, H), confirm mitochondrial proliferation in the frataxin KO mice, which could be the result of elevated mitochondrial biogenesis or increased mitochondrial fission.
The mitochondrial cross-sectional area was significantly increased in the 4 week KO versus WT in both anatomical planes, while a significant decrease occurred in the 10 week KOs (Fig. 1D, I). An increase in mitochondrial cross-sectional area could be due to increased mitochondrial biogenesis or mitochondrial fusion, while a decrease is likely due to increased mitochondrial fission. The mitochondrial aspect ratio was then used to measure mitochondrial shape by assessing the ratio between the lengths of the major axis relative to the minor axis of individual mitochondria (Fig. 1E, J) [36]. At 4 weeks of age, the mitochondrial aspect ratio was comparable between WT and KO mice in transverse sections (Fig. 1E), but was significantly decreased for the KO in longitudinal sections (Fig. 1J). This demonstrates a shortening of mitochondria in the 4 week KO mice, suggesting increased mitochondrial fission. In contrast, a significant increase in the mitochondrial aspect ratio was observed in both anatomical planes in the 10 week KO mice versus WTs (Fig. 1E, J), indicating mitochondrial elongation in the KOs potentially due to increased mitochondrial fusion.
Collectively, the comprehensive quantitative TEM assessment in Fig. 1 demonstrates pronounced alterations in cardiomyocyte mitochondrial morphology. These morphological changes suggest increased mitochondrial biogenesis and fission in both 4 and 10 week KO mice, while increased mitochondrial fusion likely occurs at 10 weeks of age.
3.2. Enhanced Pgc1α, Nrf1 and Tfam expression could promote mitochondrial biogenesis in the frataxin KO heart
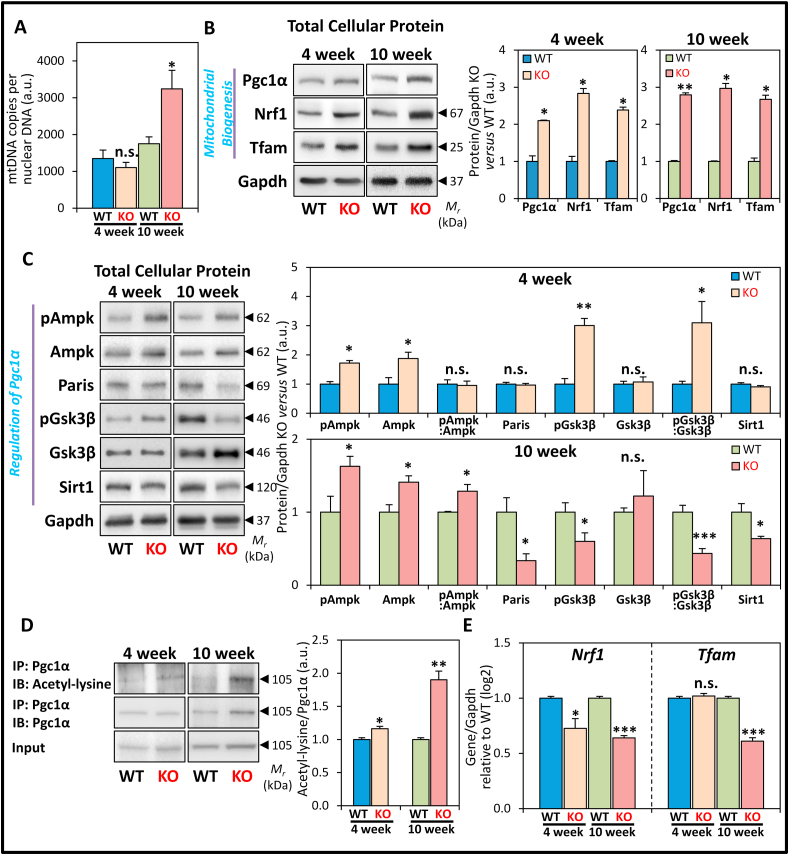
Our morphological assessment of LV mitochondrial number was supplemented by examining the copy number of the mtDNA gene, NADH dehydrogenase subunit 1 (ND1; Fig. 3A), which is outside of the mtDNA regions susceptible to deletion in disease states [15]. This was normalized to the single copy nuclear gene, β-2-microglobulin (β2m) [15], to deduce mtDNA copies/copy of nuclear DNA in the heart. The normalized ND1: β2m copy numbers were only significantly increased in 10 week KOs versus WT littermates (Fig. 3A). Therefore, the observed increase in mitochondrial number in 10 week KO mice (Fig. 1A, B, F, G) directly corresponds to the increased mtDNA copy number.
Fig. 3.
Expression of markers of mitochondrial biogenesis and proteins involving the regulation of Pgc1α in LV tissue of WT and KO littermates at 4 and 10 weeks. (A) Quantitation of mtDNA copies of ND1 versus the single copy nuclear DNA β2m gene. (B) Representative immunoblots and densitometry showing total protein expression of the mitochondrial biogenesis markers: Pgc1α, Nrf1, Tfam. (C) Representative immunoblots and densitometry showing total protein expression of the Pgc1α regulators: pAmpk, Ampk, Paris, pGsk3β, Gsk3β, and Sirt1. (D) Representative immunoblots and densitometry showing immunoprecipitation of Pgc1α with detection of acetyl-lysine and Pgc1α levels in the immunoprecipitate, and Pgc1α level in the input. (E) RT-qPCR analysis of Nrf1 and Tfam mRNA expression. Data are mean ± SEM (n = 4–6 mice/genotype/age). Unpaired two-tailed Student's t-tests were used. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT of respective age. a.u., arbitrary unit. n.s., not significant.
As hypothesized earlier, increased mitochondrial number, mitochondria: myocyte area ratio, and mitochondrial cross-sectional area could be due to enhanced mitochondrial biogenesis. This response could increase mitochondrial mass to attenuate mitochondrial deficits [26] that are observed in frataxin KO mice [1,3,4,8,12,13,34]. To assess this possibility, LV protein expression of the major transcriptional regulator of mitochondrial biogenesis, Pgc1α, and its downstream targets, Nrf1 and Tfam [26,27] were examined by western analysis (Fig. 3B). The expression of total cellular Pgc1α, Nrf1 and Tfam was significantly increased in the 4 and 10 week KOs versus WT littermates (Fig. 3B). The increase in these proteins at 4 weeks of age could reflect enhanced mitochondrial mass and play a role in the mechanisms causing the increased mitochondria: myocyte area ratio and mitochondrial cross-sectional area in the KO mice (Fig. 1C, D, H, I). The sustained activation of the Pgc1α-Nrf1-Tfam mitochondrial biogenesis pathway in the frataxin KO heart could explain the increased mitochondrial number and elevated mtDNA copy number by 10 weeks of age (Fig. 1, Fig. 3A).
3.3. The up-regulation of Pgc1α expression occurs by Ampk, Paris, and PPARγ regulation in the frataxin KO heart
Pgc1α is a key transcriptional regulator of mitochondrial biogenesis [25,26], with multiple, complex molecular mechanisms being involved in its regulation [[28], [29], [30], [31], [32], [33]]. To elucidate the regulatory mechanism responsible for increased Pgc1α protein levels, we first examined expression of the transcriptional regulators of Pgc1α, namely Ampk and Paris (Fig. 3C), which induce and repress Pgc1α expression, respectively [[28], [29], [30], [31]].
The activation of Ampk is governed by increased phosphorylation at Thr172 (pAmpk) relative to total Ampk level that then leads to increased Pgc1α levels [28,29]. In 4 week KO mice, while both the levels of pAmpk and total Ampk were significantly elevated versus WT mice, the pAmpk: Ampk ratio was not significantly altered (Fig. 3C). This observation suggested unchanged Ampk activity at this age. However, by 10 weeks of age, pAmpk was higher than total Ampk levels in the KOs, resulting in a significant increase in the pAmpk: Ampk ratio versus WT mice (Fig. 3C). Examining the expression of Paris, this was not significantly altered in the 4 week KO mice, although it was significantly decreased in the 10 week KOs versus WTs (Fig. 3C). Both Ampk activation and the decrease in Paris at 10 weeks may result in increased Pgc1α levels at this age [30].
Also relevant to the regulation of Pgc1α is PPARγ activation in the heart, which has been demonstrated to transcriptionally induces Pgc1α and Tfam expression [41]. Considering this, PPARγ protein expression was then investigated and demonstrated to be significantly increased in both 4 and 10 week KO hearts relative to their respective WT littermates (Fig. S1). Hence, the up-regulation of PPARγ protein could, in part, also explain the increased protein levels of Pgc1α and Tfam in the KO mice at both ages (Fig. 3B).
Collectively, Ampk activation, Paris down-regulation and PPARγ activation are known to transcriptionally up-regulate Pgc1α [29,30,41], and could explain the increased Pgc1α protein expression in the 4 and 10 week KO hearts (Fig. 3B).
3.4. The activity of Pgc1α is suppressed by Gsk3β and Sirt1 regulation in the frataxin KO heart at 10 weeks of age
To examine if the regulation of Pgc1α activity could also be altered, we then assessed the protein expression of Gsk3β and Sirt1 (Fig. 3C), which have been reported to post-translationally modify Pgc1α to regulate its cellular localization and activity [28,29,32,33]. Notably, the nuclear pool of Pgc1α has a short half-life [42], which is known to be regulated by Gsk3β-mediated phosphorylation of Pgc1α that results in the proteasomal degradation of nuclear Pgc1α [32,33].
Examination of Gsk3β expression in the KO mice showed that at 4 weeks of age, the level of the Ser9 phosphorylated Gsk3β (pGsk3β) was significantly elevated versus WTs (Fig. 3C). In contrast, the total Gsk3β protein level was unchanged, resulting in a significantly increased pGsk3β: Gsk3β ratio. The increase in this ratio indicates an inhibition of Gsk3β activity [43], and could therefore decrease nuclear Pgc1α degradation and increase its transcriptional activity [32]. In contrast at 10 weeks of age, Ser9 pGsk3β level was significantly decreased, while total Gsk3β level was marginally increased in the KO relative to the WT, resulting in a significant decrease in the pGsk3β: Gsk3β ratio (Fig. 3C). This indicates the activation of Gsk3β kinase activity in the 10 week KO mice, as we have reported previously [12], and this may result in proteasomal degradation of nuclear Pgc1α [32].
Studies then assessed Sirt1 protein expression (Fig. 3C), which deacetylates Pgc1α to increase Pgc1α transcriptional activity [28,29] and leads to Pgc1α nuclear translocation [33]. Sirt1 level was unchanged in the 4 week KO hearts versus WTs, while at 10 weeks of age its expression was significantly decreased in the KO (Fig. 3C). Taken together, at 10 weeks of age, Gsk3β activation could decrease Pgc1α nuclear levels, while decreased Sirt1 expression may lead to increased Pgc1α acetylation, and collectively, both these effects could suppress Pgc1α transcriptional activity.
3.5. Acetylation of Pgc1α that inhibits its activity is markedly increased in the 10 week KO heart
To ascertain if the transcriptional activity of Pgc1α was suppressed in the 10 week KO heart, the acetylation status of Pgc1α was then examined, which inversely governs its activity [26,27]. We analyzed acetyl-lysine levels of immunoprecipitated Pgc1α from LV lysates normalized to total Pgc1α. At 4 weeks of age, the Pgc1α acetyl-lysine/Pgc1α ratio was only slightly, but significantly increased in KOs versus WTs (Fig. 3D). In contrast, at 10 weeks of age, the Pgc1α acetyl-lysine/Pgc1α ratio was markedly and significantly increased in the KO relative to the WT (Fig. 3D).
The decreased Pgc1α transcriptional activity, as suggested from its increased acetylation (Fig. 3D), should lead to reduced mRNA levels of its downstream targets, Nrf1 and Tfam [26,27], particularly in 10 week KOs. In agreement with this hypothesis, RT-qPCR demonstrated Nrf1 mRNA levels were significantly decreased in 4 week KOs versus WTs, while Tfam mRNA was not significantly altered (Fig. 3E). By 10 weeks, both Nrf1 and Tfam mRNA levels were significantly decreased in the KO (Fig. 3E), which is consistent with the increased Pgc1α acetylation observed at this age (Fig. 3D). The seemingly paradoxical increase in Nrf1 and Tfam protein expression (Fig. 3B) despite decreased Nrf1 and Tfam transcript levels (Fig. 3E) must be considered with care due to: (1) the dynamic nature of both transcription and translation; (2) the half-lives of the proteins and mRNAs involved; and (3) the multiple mechanisms of feedback regulation that are known in the Pgc1α-Nrf1-Tfam pathway [25,44,45].
Considering this latter observation and caveat, increasing Tfam protein levels have been demonstrated to inhibit Nrf1 transcriptional activity and thereby decrease Tfam mRNA levels [45]. Therefore, the decreased Tfam mRNA levels at 10 weeks of age (Fig. 3E) could be mediated by this negative feedback mechanism due to increased Tfam protein expression (Fig. 3B). Moreover, Nrf1 transcription is also regulated by the antioxidant defense pathway mediated through the nuclear factor erythroid 2-related factor-2 [46]. We [12] and others [47] have recently demonstrated that this latter protein is markedly down-regulated in the frataxin-deficient mice, which could also explain the observed decrease in Nrf1 mRNA levels (Fig. 3E).
In summary, the studies in Fig. 3C–E demonstrate the regulation of Pgc1α expression and activity is complex and controlled by multiple molecular mechanisms. These mechanisms could collectively culminate in the increased Pgc1α expression, but also its inhibited transcriptional activity in the failing heart of the 10 week frataxin KO mouse.
3.6. Decreased Sirt1 activity and NAD+: NADH ratio is due to impaired NAD+ metabolism and salvage in the 10 week frataxin KO heart
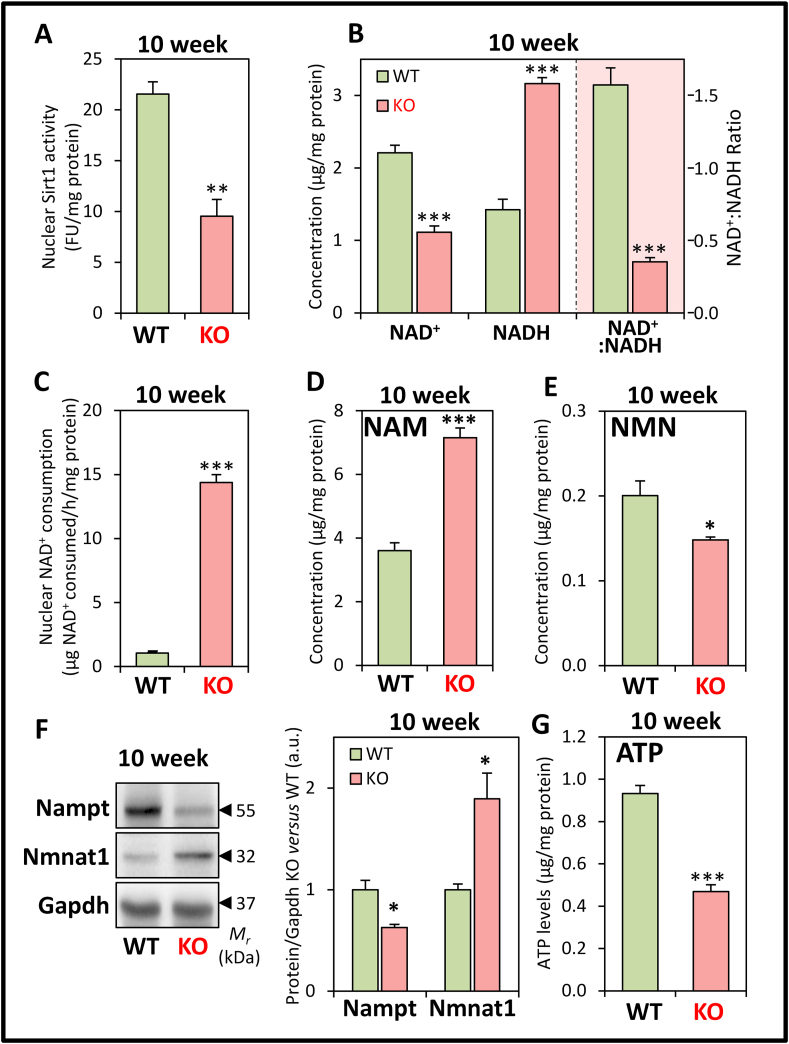
Considering the significant decrease in Sirt1 protein in the 10 week KO versus WT (Fig. 3C), we hypothesized that the activity of Sirt1 is also likely reduced at this age in the KO mice. To examine this, nuclear Sirt1 activity was assessed at 10 weeks, and was found to be significantly decreased in the KOs versus WTs (Fig. 4A). This finding corroborated the decrease in Sirt1 protein at this age (Fig. 3C), and could contribute to increased Pgc1α acetylation in the KO mice (Fig. 3D).
Fig. 4.
Examination of nuclear Sirt1 activity, NAD+/NADH levels, nuclear NAD+ consumption, NAD+ metabolites, NAD+ salvage pathway enzymes and ATP levels in the LV tissue of 10 week WT and KO littermates. (A) Nuclear Sirt1 activity. (B) NAD+ and NADH levels, and NAD+:NADH ratio. (C) nuclear NAD+ consumption. (D) NAM levels. (E) NMN levels. (F) Nampt and Nmnat1 protein expression. (G) ATP levels. All studies were performed in 10 week KO mice versus WT littermates. Data are mean ± SEM (n = 4–8 mice/genotype). Unpaired two-tailed Student's t-tests were used. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT littermates.
Since Sirt1 activity is NAD+-dependent [28,29], mechanistically this could indicate an NAD+ imbalance in the KO that may alter mitochondrial biogenesis [28]. Indeed, intracellular NAD+ was demonstrated to be significantly decreased, while NADH was significantly increased in the 10 week KOs versus WTs (Fig. 4B). These alterations resulted in a significantly decreased NAD+: NADH ratio (Fig. 4B). Of note, as found for the 10 week mice, a similar relative difference in NAD+, NADH, and the NAD+: NADH ratio between the WT and KO mice were also observed at 4 weeks of age (data not shown). The NAD+ deficiency in the frataxin KO heart at 10 weeks of age could be explained by the pronounced and significant increase in nuclear NAD+ consumption versus WT mice (Fig. 4C).
Increased nuclear NAD+ consumption may alter the metabolic balance of the NAD+ salvage pathway, which is responsible for >99% of NAD+ synthesis in the mouse heart [48]. Considering this, intracellular nicotinamide (NAM) was examined since it is a product of NAD+ consumption by enzymes, such as Sirt1 in the nucleus [19,28]. In 10 week KO mice, NAM was significantly increased versus WT littermates (Fig. 4D). This accumulation of NAM could further inhibit Sirt1 activity [19]. However, despite increased NAM, its direct downstream product in the NAD+ salvage pathway, NAM mononucleotide (NMN [19,28]), was significantly decreased in 10 week KOs (Fig. 4E). As NMN is then adenylylated to form NAD+, thereby completing the NAD+ salvage cycle [19,28], our finding suggests a dysregulation in the enzymes of this pathway in the 10 week KOs.
These imbalances in NAD+ metabolites in 10 week mice are corroborated by significant alterations in the expression of key enzymes of the NAD+ salvage pathway between the WT and KO mice (Fig. 4F). In fact, the significant decrease in NAM phosphoribosyltransferase (Nampt) expression observed in the KO (Fig. 4F) could raise NAM and decrease NMN levels, as this enzyme converts NAM to NMN [19,28]. A significant increase in the expression of nuclear-specific NMN adenylyltransferase 1 (Nmnat1), which converts NMN to NAD+ [19,28], was also observed in the KO (Fig. 4F), and this could further result in the observed decrease in NMN levels. Notably, the levels of cardiac Nampt have been demonstrated to directly correlate with NAD+ levels in the mouse heart [49], which supports our data on NAD+ levels (Fig. 4B). Together, these alterations in expression of key NAD+ metabolizing enzymes could mediate the observed NAM and NMN imbalance (Fig. 4D and E). This dysregulation in NAD+ homeostasis in the 10 week KOs is an indication of mitochondrial dysfunction [19], that is known to occur in these mice [3,4,8,34].
The pronounced mitochondrial defect induced by frataxin deletion was also demonstrated by the marked decrease in intracellular ATP in the LV of 10 week KO mice (Fig. 4G) that is probably due to inhibited mitochondrial respiration in these mice [50]. The ATP deficiency could also cause the increased Ampk phosphorylation in the 10 week KO mice (Fig. 3C), suggesting increased catabolism and autophagy [51] (examined below).
3.7. Increased expression of mitochondrial fusion and fission proteins in the frataxin KO heart
Given the observed mitochondrial abnormalities (Fig. 1) and bioenergetics-deficits (Fig. 4), changes in mitochondrial network dynamics could alleviate mitochondrial stress through complementation (via mitochondrial fusion), or compartmentalization (via mitochondrial fission), to restore mitochondrial function [24]. The observed increase in mitochondrial number (Fig. 1B, G), increased mitochondria: myocyte area ratio (Fig. 1C, H) and decreased mitochondrial cross-sectional area (in 10 week KO mice; Fig. 1D, I) may result from increased mitochondrial fission. On the other hand, increased mitochondrial elongation, as deduced from the mitochondrial aspect ratio in 10 week KOs (Fig. 1E, J), could be mechanistically explained by increased mitochondrial fusion. To assess this hypothesis, molecular markers of mitochondrial fission and fusion were then examined.
In mammals, fusion of the outer and inner mitochondrial membranes is facilitated by Mfn1/2, and Opa1, respectively [24]. Alternatively, mitochondrial fission compartmentalizes damaged mitochondrial components into daughter organelles that could be removed by mitophagy [24]. This mitochondrial fission process involves cytoplasmic Drp1 interacting with mitochondrial fission protein 1 (Fis1) and mitochondrial fission factor, Mff, thereby encircling the constriction site on the outer mitochondrial membrane [24]. A recent study has also indicated that Fis1 may directly bind Mfn1/2 to disrupt fusion machinery and promote mitochondrial fragmentation, independent of Drp1 [52].
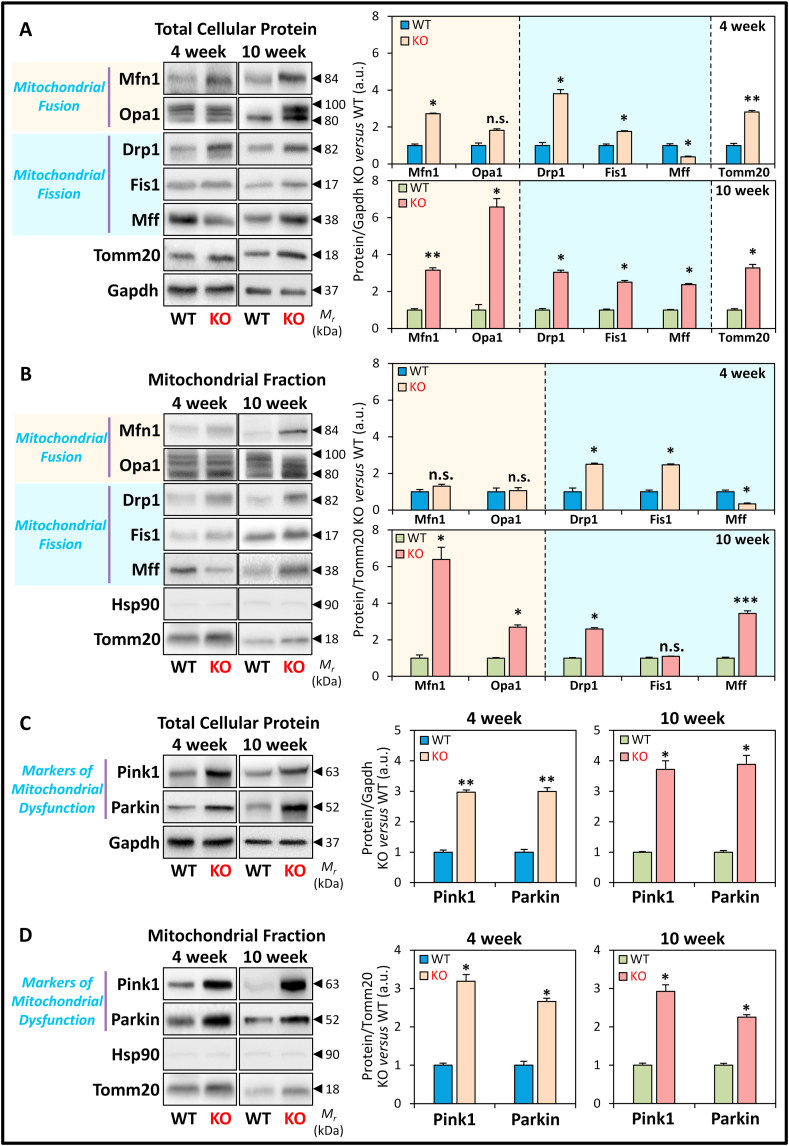
At 4 weeks of age, LV total protein expression of the mitochondrial fusion marker, Mfn1, was significantly increased in the KO hearts versus WTs (Fig. 5A). Opa1 appeared as multiple bands between 80 and 100 kDa [53] with its expression only being marginally (p > 0.05) increased in the 4 week KOs (Fig. 5A). The mitochondrial fission markers, Drp1, and to a lesser extent, Fis1, were significantly increased in 4 week KOs, whereas Mff was significantly decreased (Fig. 5A). The outer mitochondrial membrane protein and mitochondrial marker, Tomm20, was also significantly increased in the KO at 4 weeks (Fig. 5A), which corresponds to the increased mitochondrial cross-sectional area in the frataxin-deficient heart (Fig. 1D, I). These proteins were then examined in the mitochondrial fraction and normalized to Tomm20 expression (Fig. 5B).
Fig. 5.
Protein markers of mitochondrial fusion and fission, and mitochondrial dysfunction in the LV tissue of 4 and 10 week WT and KO littermates. (A) Representative immunoblots and densitometry showing total protein expression of mitochondrial fusion markers: Mfn and Opa1, and mitochondrial fission markers: Drp1, Fis1, and Mff. (B) Representative immunoblots and densitometry of the mitochondrial fraction assessing the markers of mitochondrial dynamics in (A). (C) Representative immunoblots and densitometry showing total protein expression of Pink1 and Parkin. (D) Representative immunoblots and densitometry of the mitochondrial fraction assessing Pink1 and Parkin expression. Gapdh was used as a loading control for total protein-loading in (A, C), while Hsp90 and Tomm20 were used as protein-loading controls for cytoplasmic and mitochondrial fractions in (B, D), respectively. Data are mean ± SEM (n = 4–6 mice/genotype/age). Unpaired two-tailed Student's t-tests were used. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT. a.u., arbitrary unit. n.s., not significant.
Of note, assessment of the mitochondrial fraction is crucial to detect translocation of cytoplasmic proteins to the mitochondrion (e.g., Drp1), and to normalize the mitochondrial protein expression to changes in mitochondrial mass. Therefore, the mitochondrial fraction provides a more sensitive assessment of mitochondrial protein levels than total cellular levels in the context of the quantitative alterations observed in mitochondrial morphology (Fig. 1). In the mitochondrial fraction, the expression of the fusion proteins, Mfn1 and Opa1, were not significantly altered in 4 week KOs versus WTs (Fig. 5B). However, at 4 weeks of age, the fission proteins, Drp1 and Fis1 were significantly increased in the mitochondrial fractions of the KOs versus WTs, despite normalization to increased Tomm20 levels, whereas Mff was significantly decreased in the KOs (Fig. 5B).
At 10 weeks of age, total protein expression of the mitochondrial fusion (Mfn1 and Opa1) and fission (Drp1, Fis1, and Mff) markers, as well as Tomm20, were all significantly increased in the KO (Fig. 5A). When normalized to Tomm20 expression in the mitochondrial fraction (Fig. 5B), the expression of the fusion proteins (Mfn1 and Opa1) was significantly increased in 10 week KOs versus WTs. On the other hand, only the mitochondrial fission proteins, Drp1 and Mff, but not Fis1, were significantly increased in the 10 week KOs relative to their WT littermates (Fig. 5B). Notably, the detection of cytosolic Drp1 in the mitochondrial fraction in both 4 and 10 week KOs is a robust indicator of mitochondrial fission [24] at both ages. Expression of the cytoplasmic marker, heat shock protein 90 (Hsp90), was very low in the mitochondrial fraction in the KO and WT in both age groups, indicating minimal cytoplasmic contamination (Fig. 5B).
Collectively, these findings demonstrate progressive alterations in expression of proteins that regulate mitochondrial dynamics in KO mice. In the mitochondrial fraction at 4 weeks, the expression of all 3 mitochondrial fission markers (i.e., Drp1, Fis1, Mff) was significantly altered in the KOs, while no significant change was observed for both fusion markers (i.e., Mfn1 and Opa1; Fig. 5B). In contrast, in the mitochondrial fraction at 10 weeks, both mitochondrial fusion (Mfn1 and Opa1) and fission (Drp1 and Mff) markers were significantly increased in KO mice relative to WTs.
Molecular data from mitochondrial fractions above are consistent with the ultrastructural alterations observed in Fig. 1. The early activation of mitochondrial biogenesis and fission markers (Fig. 3, Fig. 5) in the 4 week KO are consistent with the increased mitochondria: myocyte area ratio (Fig. 1C, H) and the observed decrease in the mitochondrial aspect ratio in the longitudinal plane (Fig. 1J). By 10 weeks of age, the progressive induction of mitochondrial fusion in the KO would result in mitochondrial elongation, as determined by the mitochondrial aspect ratio (Fig. 1E, J). This together with sustained mitochondrial fission in the 10 week KO would contribute to increased mitochondrial number in the KO (Fig. 1B, G), increased mitochondria: myocyte area ratio (Fig. 1C, H), and decreased mitochondrial cross-sectional area (Fig. 1D, I). These observations indicate that alterations in the expression of proteins involved in mitochondrial dynamics occur as early as 4 weeks of age, which may be regulated by sensors of mitochondrial dysfunction (e.g., Pink1 and Parkin) that affect mitochondrial turnover [23].
3.8. Increased Pink1 and Parkin protein in the frataxin KO heart indicates mitochondrial dysfunction
Pink1 and Parkin are sensors of mitochondrial dysfunction and regulators of mitochondrial dynamics, which make them markers of mitochondrial health, function, and quality control [23]. In addition, Pink1 and Parkin regulate mitochondrial-specific autophagy, known as mitophagy [23]. In response to decreased mitochondrial membrane potential and mitochondrial damage, Pink1 translocates to mitochondria, and recruits and activates Parkin through its phosphorylation [22,23]. Phosphorylated Parkin then targets Mfn1 for poly-ubiquitination and degradation to inhibit mitochondrial fusion [22,23]. Hence, expression and localization of Pink1 and Parkin are critical in regulating mitochondrial dynamics and biogenesis, and also facilitate mitophagic processing of damaged mitochondria [22,23]. As such, expression of these two proteins was examined (Fig. 5C and D).
Total LV protein expression of Pink1 and Parkin were significantly increased in the KO versus WT at 4 weeks and 10 weeks of age (Fig. 5C). Moreover, a robust and significant increase in Pink1 and Parkin levels normalized to Tomm20 were detected in mitochondrial fractions for both age groups (Fig. 5D). These results demonstrated mitochondrial localization of Pink1 and Parkin, suggesting mitochondrial dysfunction in frataxin KO hearts as early as 4 weeks of age. As a mechanistic consideration, Parkin can also indirectly regulate Pgc1α expression via coordinating proteasomal degradation of the Pgc1α transcriptional repressor, Paris [30]. The observed up-regulation of Parkin could decrease Paris levels (Fig. 3C), thereby causing the increased Pgc1α expression in 10 week KO mice (Fig. 3B).
3.9. Inhibition of autolysosomal degradation by bafilomycin A1 demonstrates enhanced autophagic and mitophagic flux in the frataxin KO heart
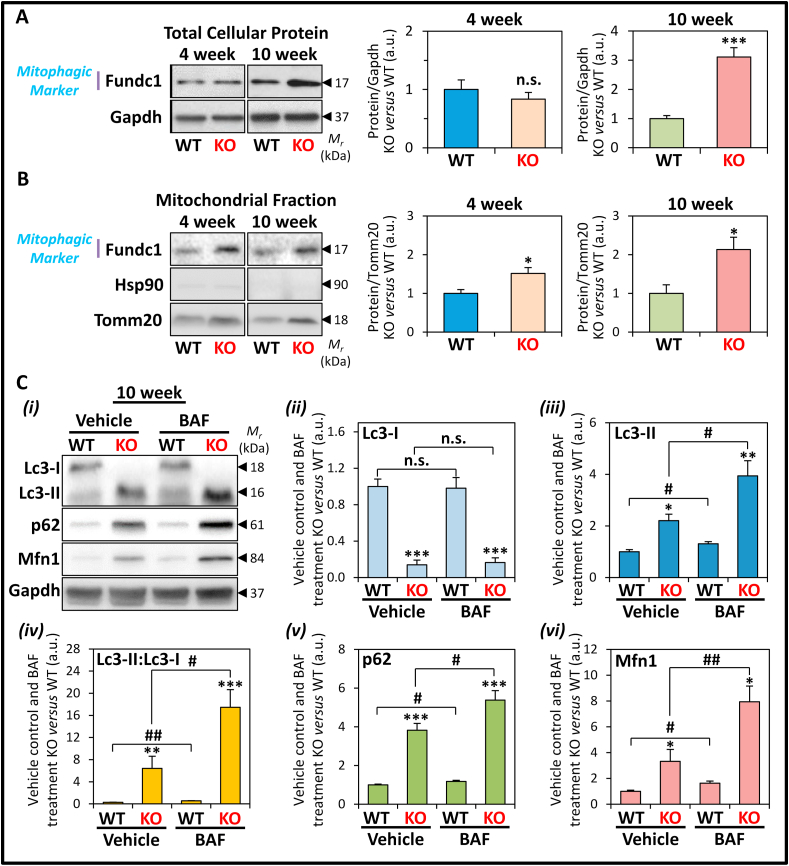
The mitochondrial localization of Pink1 and Parkin promotes mitophagy [22,23]. In support of this, and in agreement with our previous studies [8], total protein expression of the mitophagic marker, Fundc1, was found to be significantly increased at 10 weeks, but not at 4 weeks, in the frataxin KO mice versus WT littermates (Fig. 6A). However, the subsequent mitochondrial fractionation procedure demonstrated Fundc1 expression in mitochondria was observed to be significantly increased in both 4 and 10 week KO hearts (Fig. 6B).
Fig. 6.
Protein expression of the mitophagic marker, Fundc1, in 4 and 10 week WT and KO littermates, and assessment of autophagic/mitophagic flux upon Bafilomycin A1 (BAF) treatment in the LV tissue of 10 week frataxin KO mice versus WT littermates. (A, B) Representative immunoblots and densitometry showing Fundc1 expression in (A) total cellular protein, and (B) mitochondrial fractions, of 4 and 10 week KO versus WT littermates. Gapdh was used as a loading control for total protein-loading in (A), while Hsp90 and Tomm20 were used as protein-loading controls for cytoplasmic and mitochondrial fractions in (B), respectively. In (A, B) data are mean ± SEM (n = 4–6). Unpaired two-tailed Student's t-tests were used. (C) BAF inhibits autolysosomal degradation of autophagic components, i.e., Lc3-II and p62. Representative (i) immunoblots and (ii-vi) densitometry of (ii) Lc3-I, (iii) Lc3-II, (iv) Lc3-II:Lc3-I ratio, (v) p62, and (vi) Mfn1, in the heart from 10 week WT and KO littermates after treatment for 30 min with or without BAF (6 mg/kg; i.p.). Data are mean ± SEM (n = 6 mice/genotype). One-way ANOVA with Tukey's post hoc tests were used. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT. #, p < 0.05; ##, p < 0.01 versus vehicle control. a.u., arbitrary unit. n.s., not significant.
Considering the marked alterations in key proteins involved in mitochondrial dynamics (Fig. 5) and mitophagy (Fig. 6A and B), it was important to investigate the rate of mitochondrial elimination by autophagy. Specifically, the autophagic flux of WT and KO mice was assessed, as an indication of the heart's capacity to undergo autophagic degradation [54]. To assess this in vivo, 10 week mice were administered BAF (see Methods, Section 2.1), which inhibits autophagosome-lysosome fusion and thereby inhibits autolysosomal degradation of autophagic cargo [54]. Expression of the classical autophagic substrates, Lc3-II and p62, and mitophagic substrate, Mfn1, were then assessed to monitor autophagic and mitophagic flux in the LV of WT and KO mice after BAF-treatment (Fig. 6C).
In vehicle-treated mice, the WT expressed both Lc3-I (18 kDa) and Lc3-II (16 kDa), whereas in the KO, Lc3-II was the major form observed (Fig. 6Ci-iii). Considering our previous report of enhanced expression of autophagic initiation machinery [8], this finding of increased conversion of Lc3-I to Lc3-II in KO mice indicated stimulation of autophagosome formation and possibly an inhibition of Lc3-II degradation [54,55]. This alteration resulted in a significantly increased Lc3-II:Lc3-I ratio in vehicle-treated KO mice versus WT littermates (Fig. 6Civ) [8]. Following BAF treatment, a further significant increase in Lc3-II levels, without affecting Lc3-I expression, was observed in BAF-treated WT and KO mice versus the respective vehicle-treated mice (Fig. 6Ci-iii).
Notably, the effect of BAF on increasing Lc3-II expression was significantly more marked in the 10 week KO than the WT relative to their respective vehicle-treated counterparts (Fig. 6Ci, iii). Consequently, BAF treatment resulted in a slight, but significant increase in the Lc3-II:Lc3-I ratio in WT mice, while there was a marked and significant increase in this ratio in the KOs versus their vehicle-treated counterparts (Fig. 6Civ). Mechanistically, these results confirm BAF-mediated inhibition of autolysosomal Lc3-II degradation, thereby enabling an assessment of autophagic flux that measures the capacity of autolysosomal degradation in these mice [54]. Therefore, the autophagic flux is enhanced in the KO mice compared to the WT mice. However, the enhanced autophagic flux in the KO is likely exceeded by the increased autophagic initiation, thereby resulting in an accumulation of Lc3-II in the vehicle-treated KO versus WT mice.
Additional mechanistic dissection using BAF treatment was performed by examining the expression of the autophagic adapter and substrate, p62 [54], which was significantly elevated in vehicle-treated KOs versus WTs (Fig. 6Ci, v). This result again indicates increased autophagosome generation [54,55], but also possible inhibition of p62 degradation in the 10 week KO mice. Similarly to the Lc3-II:Lc3-I ratio, BAF treatment inhibited the autolysosomal degradation of p62, resulting in a slight, but significant increase in p62 levels in WT mice (Fig. 6Ci, v). In the KOs, BAF treatment also caused a marked and significant increase in p62 expression versus their vehicle-treated counterparts (Fig. 6Ci, v). Furthermore, BAF treatment resulted in an additional enhancement of Mfn1 expression in KO mice versus vehicle-treated littermates (Fig. 6Ci, vi), which suggests selective mitophagy also occurs via autolysosomal degradation and enhanced mitophagic flux.
Collectively, these observations in Fig. 6 suggest that: (1) Mfn1 is a substrate of autophagic degradation in the KO mice, which is consistent with enhanced mitophagy; (2) enhanced autophagic flux in the KO compared to the WT mice demonstrated that autophagic degradation is not inhibited in the KO mice; and (3) the accumulation of autophagic substrates in the KO mice is likely the result of an overwhelming induction of autophagic initiation, despite the already increased autophagic flux in these animals.
3.10. Alterations in mitochondrial biogenesis, fusion, fission, and autophagy in FA patients corroborate the studies in frataxin KO mice
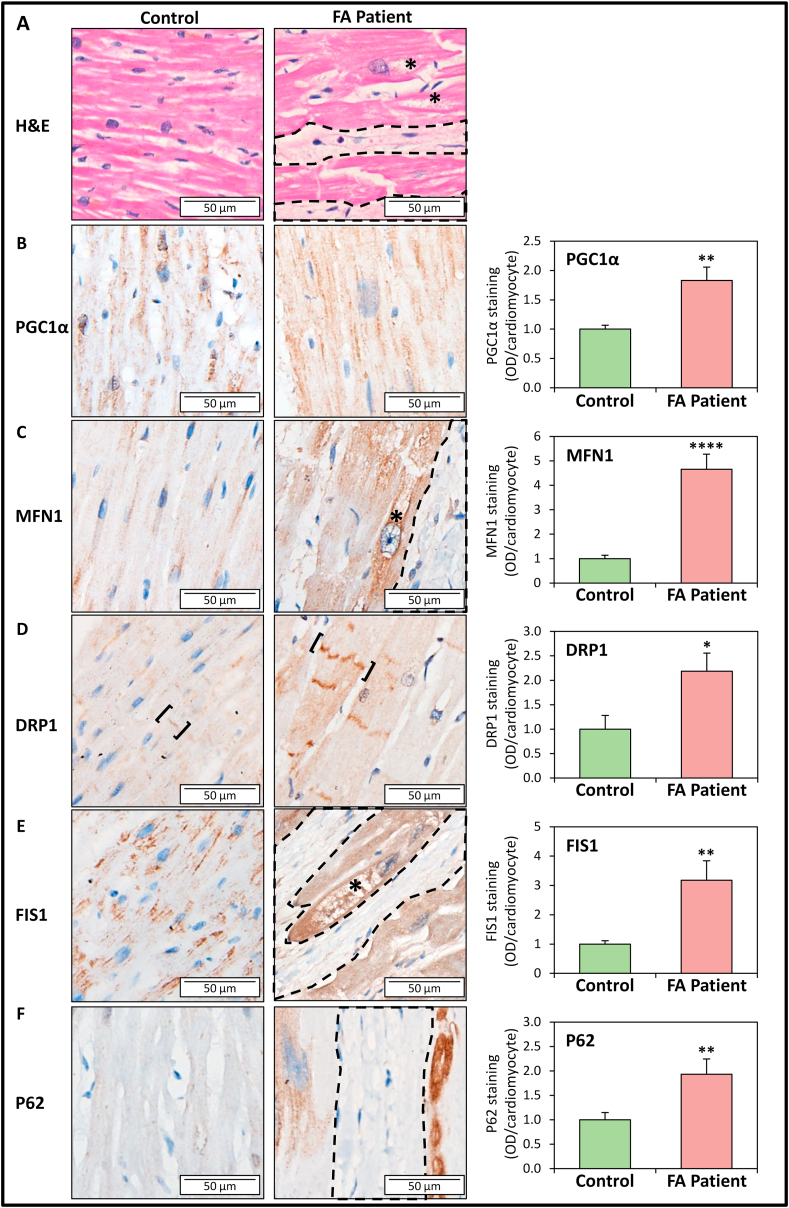
The dysregulation in mitochondrial homeostatic proteins observed in the frataxin KO heart prompted us to compare the levels and cellular localization of PGC1α, MFN1, DRP1, FIS1 and P62 in post-mortem LV sections of hearts from FA and age/gender-matched control patients (Table S1). In longitudinal sections of LV tissue from FA patients, pronounced fibrotic lesions (circled by dashed lines) and cardiomyocyte disarray and degeneration (asterisks) can be identified by H & E staining, which is absent in controls (Fig. 7A). Consistent with our previous investigation [8], intact cardiomyocytes of FA patients demonstrate cardiomyocyte hypertrophy, consistent with the cardiomyopathy observed in the frataxin KO mice [34].
Fig. 7.
Immunohistochemistry assessment of protein markers of mitochondrial biogenesis, fusion, fission, and autophagy in LV sections of FA patients relative to age- and gender-matched controls. Representative micrographs of staining for: (A) H & E, (B) PGC1α, (C) MFN1, (D) DRP1, (E) FIS1, and (F) P62 and their corresponding quantification in LV sections from FA patients and age- and gender-matched controls (see Table S1). Dash-encircled regions indicate areas of interstitial fibrosis; asterisks indicate areas of cardiomyocyte degeneration; and square-brackets indicate disorganized intercalated discs. Quantification of immunohistochemistry was performed using ImageJ Fiji with staining intensity represented as optical density (OD)/cardiomyocyte, where OD = log [max intensity/mean intensity]. Data are mean ± SEM (n = 10–12 specimens, 9–11 images were analyzed per specimen). Unpaired two-tailed Student's t-tests were used. *, p < 0.05; **, p < 0.01; ****, p < 0.0001 versus control. n.s., not significant. Scale bar: 50 μm.
In control patients, immunohistochemistry staining of the mitochondrial biogenesis marker PGC1α [29] was demonstrated to be expressed in the nucleus and cytosol (Fig. 7B). The mitochondrial fusion protein, MFN1, and mitochondrial fission proteins, DRP1 and FIS1, were observed in control patients primarily in the perinuclear and interfibrillar regions, which are typical locations of cardiomyocyte mitochondria (Fig. 7C–E). In contrast, in FA patients, immunostaining of PGC1α, MFN1, DRP1 and FIS1 were all significantly enhanced relative to controls (Fig. 7B–E). These results are consistent with the enhanced expression of these proteins in LV lysates from frataxin KO mice (Fig. 3, Fig. 5A, B), suggesting enhanced mitochondrial biogenesis, fusion and fission machinery in the FA patient hearts.
Interestingly, while PGC1α immunostaining was significantly increased in FA patients, it was mostly absent in the nucleus (Fig. 7B). This result could suggest PGC1α inactivation and/or increased nuclear PGC1α degradation, as observed in the frataxin KO mice (Fig. 3C and D). DRP1 expression was also detected in the intercalated discs of the control and this was particularly enhanced in FA patients (square-brackets; Fig. 7D). The intercalated discs in FA patients also appeared structurally disorganized and discontinuous, as reported previously [56]. For MFN1 and FIS1, their expression in FA patients was enhanced in the vacuolated, degenerating cardiomyocytes (asterisks; Fig. 7C, E), proximal to fibrotic lesions (encircled by dashed lines). Similarly, immunostaining of the autophagic substrate, P62, was predominantly expressed in degenerating cardiomyocytes surrounded by fibrotic lesions (encircled by dashed lines) in FA patients, while there was minimal staining in control sections (Fig. 7F).
Overall, these immunohistochemistry studies examining FA patients strongly support the analyses of the heart from the frataxin KO mice and confirm the significantly enhanced expression of markers of mitochondrial biogenesis, fusion/fission and autophagy.
4. Discussion
Defective mitochondrial iron metabolism and oxidative stress potentiate cardiac pathology invoked by the loss of frataxin [1,3,4,8,12], although how these factors alter mitochondrial homeostasis remains uncharacterized in FA cardiomyopathy. In FA patients, cardiomyopathy is the major cause of death with mitochondrial dysfunction being a major factor [3,4,8]. In the current study using the frataxin KO heart, TEM ultrastructural assessment demonstrated extensive proliferation of structurally abnormal mitochondria, with iron deposits being a distinct feature. Within the mitochondrial matrix, mitochondrial cristae condensation was also observed in the KO at 4 and 10 weeks of age. Notably, elongated and entangled mitochondria were observed in both anatomical planes of 10 week KO hearts, which markedly displaced cardiac myofibers. This contrasts the well-organized mitochondria arranged parallel to myofibers in the WT heart.
These mitochondrial ultrastructural alterations in frataxin-deficient cardiomyopathy could be attributed to two distinct regulatory mechanisms of Pgc1α, which is a major regulator of cardiac mitochondrial biogenesis and energy metabolism [25,26,57]. In terms of the first mechanism regulating Pgc1α, there was an early and sustained induction of Pgc1α-Nrf1-Tfam signaling in the KO mice from 4 weeks of age, when the mice showed no structural or functional cardiac defects [8], through to 10 weeks of age, when the mice developed cardiac hypertrophy and heart failure [8]. This sustained up-regulation of Pgc1α could contribute to progressive mitochondrial proliferation, as indicated by increased mitochondrial number, mitochondria: myocyte area ratio and mtDNA copy number.
The increased Pgc1α expression could also act to maintain adequate mitochondrial energy demand during the development of cardiac hypertrophy [57]. Notably, cardiac Pgc1α overexpression in mice has been demonstrated to result in a dilated cardiomyopathy with proliferation of enlarged mitochondria that displaced sarcomeric assemblies at the ultrastructural level [58]. This was morphologically comparable to the mitochondrial proliferation observed in the 10 week frataxin KO mice (Fig. 1A, F). However, increased Pgc1α expression was also implicated in the rapid onset of human heart failure due to mtDNA mutations [59]. Therefore, increased mitochondrial biogenesis despite mtDNA damage may be a maladaptive response that exacerbates heart failure [59].
Regarding the second mechanism regulating Pgc1α, there was an inhibition of Pgc1α activity as demonstrated by increased Pgc1α acetylation. This was most pronounced in the failing hearts of 10 week frataxin KO mice [8]. This inhibition of Pgc1α is prevalent in pathological cardiac hypertrophy and heart failure, where a metabolic switch mediated by Pgc1α inhibition shifts energy metabolism from fatty acid oxidation to reliance on glycolysis to facilitate efficient ATP production [57,60]. Of note, the failing adult heart and fetal heart both undergo a similar metabolic reprogramming towards glucose metabolism and the activation of a fetal gene program [60]. Considering this, we have previously identified fetal gene reactivation in the 10 week KOs [8], which also exhibited the distinctive elongated and curvilinear mitochondrial morphology that is characteristic of fetal cardiac mitochondria [61]. Moreover, the pronounced condensation of the mitochondrial cristae in the KO heart (Fig. 1A, F) is morphologically identical to those observed in the cardiac Pgc1α/β knockout mice [62]. Collectively, in the frataxin KO heart, a progressive stimulation of Pgc1α expression early in the disease process at 4 weeks is likely a major factor driving mitochondrial proliferation, while the subsequent inactivation of Pgc1α at 10 weeks could be responsible for the metabolic reprogramming that occurs as the heart progresses towards failure.
The regulation of the Pgc1α pathway is complex with many upstream regulators as well as feedback loops that govern their own expression [[29], [30], [31],45,63,64]. Herein, we demonstrated the progressive induction of Pgc1α expression could be mediated by: (1) increased Ampk activation that increases Pgc1α expression and activity [31,63], which could be potentiated by increased microRNA miR-144 expression [65] that we demonstrated to be up-regulated in 10 week KO mice [66]; (2) inhibition of the Pgc1α transcriptional repressor, Paris [30] at 10 weeks, likely due to the up-regulation of Parkin; (3) inhibition of Gsk3β activity at 4 weeks that could reduce nuclear Pgc1α degradation and increase Pgc1α activity [32]; and (4) elevated PPARγ expression that transcriptionally induces Pgc1α [41] (Fig. 8A). The subsequent inhibition of Pgc1α activity in the failing 10 week frataxin KO hearts is attributed to increased Pgc1α acetylation. This Pgc1α inhibition was in good agreement with the decreased expression and activity of the NAD+-dependent Sirt1 deacetylase [29], and also the decreased NAD+/NADH ratio at 10 weeks of age. Increased PPARγ expression also inhibits Sirt1 level and activity [67], which could further the Pgc1α inhibition in the frataxin KO heart. Our studies herein demonstrate for the first time in FA cardiomyopathy that decreased Sirt1 activity results in increased Pgc1α acetylation and its subsequent inactivation (Fig. 8A). Additionally, Pgc1α inactivation in the 10 week KO mice may be due to Gsk3β activation that causes nuclear Pgc1α degradation [29] and decreased expression and activity of Sirt1, which is required for Pgc1α nuclear translocation [33]. This finding in the KO mice is strongly corroborated in FA patient LV sections, where PGC1α immunostaining was overall enhanced, but its nuclear expression was substantially decreased.
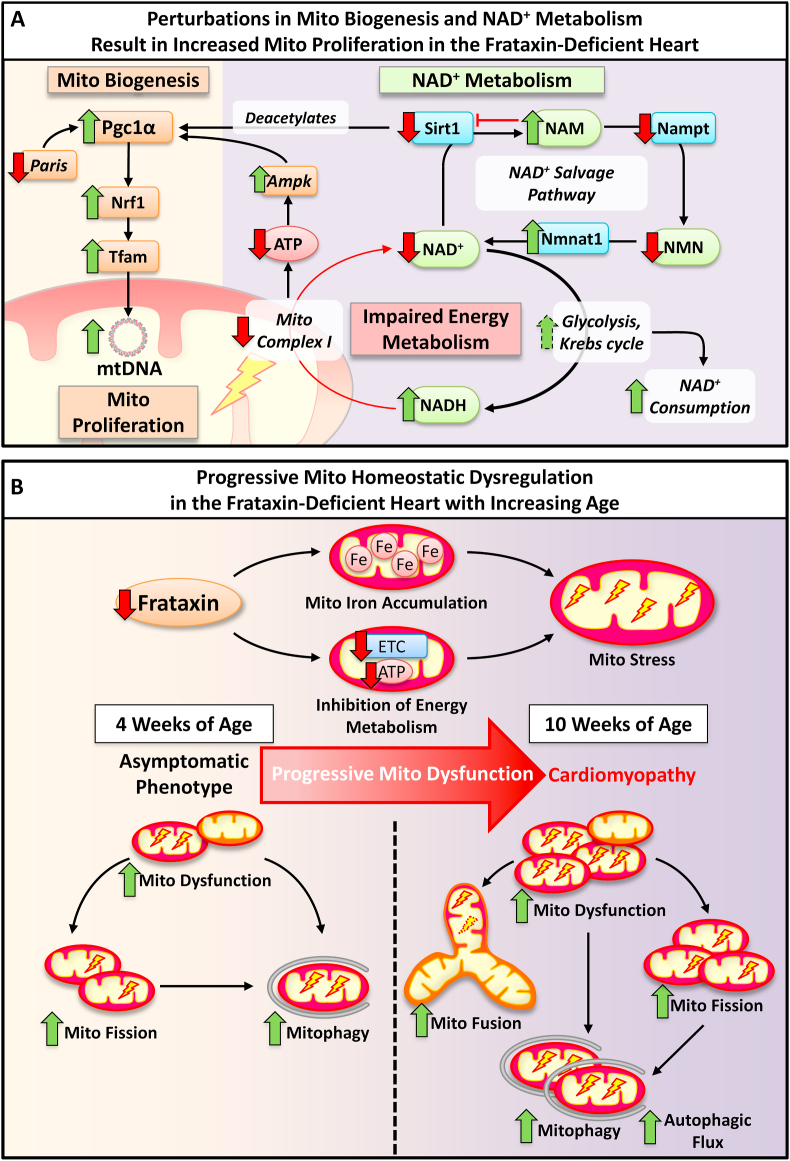
Fig. 8.
Schematic illustrating the effects of cardiac frataxin-deficiency on the development of mitochondrial (Mito) homeostatic dysregulation. (A) In 10 week-old frataxin KO mice hearts, enhanced mitochondrial proliferation was evident by the up-regulation of mitochondrial biogenesis (involving Pgc1α, Nrf1 and Tfam) and increased mtDNA. The increased Pgc1α protein could be potentiated by transcriptional regulation via increased Ampk and decreased Paris. Concurrently, there was also increased Pgc1α acetylation suggesting inhibition of the protein at 10 weeks, which could be due to decreased Sirt1 expression and activity caused by a feedback inhibition due to increased NAM. The NAD+ salvage pathway was also dysregulated with the inhibition of Nampt but up-regulation of Nmnat1, causing increased NAM and reduced NMN, respectively. Moreover, increased NAD+ consumption via increased glycolysis and Kreb cycle, together with decreased mitochondrial complex I activity, would result in a decreased NAD+: NADH ratio and impaired energy metabolism in 10 week KOs. (B) In FA, frataxin-deficiency progressively leads to LV mitochondrial iron-accumulation and inhibition of the electron transport chain that causes mitochondrial redox stress. Asymptomatic 4 week frataxin KO mice progressively develop a fatal, hypertrophic cardiomyopathy by 10 weeks. Our data in the frataxin-deficient heart identified progressive activation of: (1) mitochondrial dysfunction, involving the mitochondrial recruitment of Pink1 and Parkin observed from 4 week; (2) mitochondrial fission, involving mitochondrial translocation of Drp1 and increased Fis1 from 4 week; (3) mitochondrial fusion, involving enhanced Mfn1 and Opa1 expression that was significant at 10 weeks; and (4) mitophagy from 4 week, and increased autophagic flux at 10 week. Overall, there is progressive dysregulation of mitochondrial homeostasis in the frataxin-deficient heart that contributes to FA cardiac pathology.
Our current investigation demonstrated that the decreased Sirt1 activity in the frataxin-deficient heart was due to a decrease in the intracellular NAD+ pool. This NAD+ deficiency in FA cardiomyopathy is potentiated by a dysregulation in the NAD+ salvage pathway, which is responsible for >99% of NAD+ synthesis in the mouse heart [48]. Specifically, we identified a down-regulation of Nampt resulting in NAM accumulation that in turn inhibits Sirt1 activity [19], and an up-regulation of Nmnat1 contributing to a decline in NMN level, thereby disrupting the NAD+ salvage pathway in the frataxin-deficient heart (Fig. 8A).
Moreover, previous studies using the frataxin KO mouse also identified energy metabolic alterations that could contribute to the observed NAD+ deficiency, including: (1) increased glycolysis that increases NAD+ consumption; (2) increased expression of NAD+-reducing enzymes involved in the Krebs cycle, the catabolism of branched-chain amino acids and ketone bodies, and pyruvate decarboxylation; and (3) diminished mitochondrial complex I activity, which initiates the electron transport chain with NADH oxidation [50,68] (Fig. 8A). Collectively, NAD+ deficiency caused by impaired NAD+ salvage and dysregulated NAD+-dependent energy metabolism exacerbates the metabolic defects in frataxin-deficiency (Fig. 8A), which could play a major role in potentiating mitochondrial dysfunction, redox stress, and energy deficiency in FA.
In the frataxin KO mice, spatiotemporal alterations in mitochondrial fusion and fission markers fittingly corresponded with our quantitative ultrastructural assessment of mitochondrial morphology, and demonstrated a progressive mitochondrial dysfunction. In particular, there was an activation of mitochondrial fission from 4 weeks, with concurrent fission and fusion occurring by 10 weeks of age (Fig. 8B). Enhanced expression of both mitochondrial fusion and fission markers was also observed in the FA patient heart, thereby strongly supporting the results from the frataxin KO mice. Mechanistically, Drp1-mediated fission is induced by mitochondrial dysfunction in cardiac ischemia and reperfusion injury, and is a mediator of pathology [69]. Hence, mitochondrial dysfunction and bioenergetic deficits due to frataxin-deficiency could directly cause an early induction of Drp1-mediated mitochondrial fission and potentiate cardiomyopathy in FA (Fig. 8B). Moreover, increased Fis1 could also contribute to enhanced mitochondrial fragmentation in the KO mice independent of Drp1, via the inhibition of fusion machinery [52]. An early induction of mitochondrial fission due to frataxin-deficiency could also contribute to the fragmented mitochondrial network that was observed in in vitro models of FA [47,70]. Additionally, the oxidative stress-mediated increase in glutathione oxidation that we demonstrated in 10 week KOs [12], and that was also observed in other FA cardiac models [71], could stimulate mitochondrial fusion [72].
Cross-talk between mitochondrial fusion and fission in frataxin-deficiency may be mediated by the induction of Parkin, through its roles in both processes [69]. The early mitochondrial accumulation of Pink1, Parkin and Fundc1 at 4 weeks could be induced by Nrf1 [73], and indicates mitochondrial damage and mitophagic activation. Mitophagy was confirmed by the accumulation of Mfn1 following autolysosomal inhibition by BAF-treatment. Collectively, mitochondrial dysfunction caused by frataxin-deficiency could increase mitochondrial fission and mitochondrial recycling via the activation of Drp1 and Pink1-Parkin signaling. The subsequent induction of mitochondrial fusion potentially acts to increase mitochondrial capacity and cardiac energetics [74] (Fig. 8B). Thus, for the first time, our findings identify a significant role for mitochondrial dynamics in the development of FA cardiomyopathy.
Herein, using the well-established lysosomotropic agent, BAF [54], we confirmed increased autophagic and mitophagic flux in 10 week KOs, rather than an inhibition of autolysosomal degradation that we previously speculated [8]. Therefore, the heightened Lc3-II and p62 expression in vehicle-treated KO mice could be the result of chronic stress-induced autophagic initiation [8], but without equivocal autolysosomal degradation to prevent their accumulation. In FA patient LV sections, P62 was also observed in the degenerating cardiomyocytes proximal to fibrotic lesions, which is typical of the failing human heart [75].
Our previous studies examining the muscle creatine kinase conditional KO mouse demonstrated that while frataxin is deleted in the heart and skeletal muscle, only in the heart was significant molecular, cellular and whole organ pathology observed [12]. This is probably because frataxin is a mitochondrial protein that plays a key role in mitochondrial function and that the heart is markedly dependent on the aerobic mitochondrial respiration, with an almost exclusive dependence on aerobic metabolism [12]. In contrast, skeletal muscle mainly depends on anaerobic glycolysis, and as such, the effects of frataxin depletion are not marked in this tissue [12]. Thus, due to the lack of functional consequences, the deletion of frataxin in the skeletal muscle is unlikely to have significant bearing on the current results obtained examining the heart, where the major pathology is observed.
Our investigation has identified crucial bioenergetics deficits and molecules that could be targeted for novel therapeutic strategies for treatment of the cardiomyopathy in FA. For example, pharmacological targeting of Ampk could be a potential therapeutic strategy for FA via the cardio-protective effects mediated by: (1) enhancing Pgc1α-mediated mitochondrial biogenesis [76,77]; and (2) modulating autophagy or Pink1-mediated mitophagy to circumvent heart failure [78,79]. In terms of targeting mitochondrial network dynamics, the inhibition of mitochondrial fission [[80], [81], [82], [83]], or over-expression of mitochondrial fusion proteins [84], has been demonstrated to improve heart function following cardiac injury. Alternatively, NAD+ supplementation could potentially be an innovative therapeutic avenue for FA cardiomyopathy [13,68]. In particular, NAD+ precursor supplementation may be beneficial for FA treatment, as this could increase sirtuin activities [19]. Recent studies using an NAD+-donating agent or NMN modestly attenuated FA cardiomyopathy [13,68]. Therefore, the current investigation has identified a number of molecular alterations in the FA cardiomyopathy that could be crucial for rationalizing future treatment strategies for effective therapy.
In summary, frataxin-deficiency results in an accumulation of intramitochondrial redox active iron aggregates and inhibition of energy production that contributes to mitochondrial dysfunction [3,4,34,50]. In 4 week KO mice, as an early response to the bioenergetic deficits and mitochondrial dysfunction, mitochondrial biogenesis via Pgc1α activation, enhanced Drp1-mediated mitochondrial fission, and Pink1/Parkin signaling were observed (Fig. 8B). This concomitant activation of Pink1/Parkin would also act to suppress mitochondrial fusion to further promote fission and mitophagic recycling of damaged mitochondria [22,23]. By 10 weeks of age, the oxidative damage and metabolic deficits caused by frataxin-deficiency persist in the KO mice. As such, mitochondrial fusion is also activated as an additional rescue response, probably in an attempt to increase mitochondrial capacity and supplement cardiac bioenergetics [74]. Together with continual activation of mitochondrial biogenesis and fission that contribute to increased mitochondrial mass, there is continued Pink1/Parkin signaling and mitophagy, again as part of a frustrated attempt in mitochondrial quality control (Fig. 8B). Relevant to this analysis, it is also important to consider the heterogeneity of mitochondria within the cell [85,86], and especially in cardiomyocytes [87], in terms of redox state, membrane potential and mitochondrial network dynamics. Therefore, these dynamic and potentially competing processes of mitochondrial homeostasis could occur in parallel in distinct populations of cardiac mitochondria as an attempt to reconstitute mitochondrial energetics in the frataxin-deficient heart. However, mitochondrial dysfunction persists in the KO mice due to the genetic ablation of frataxin, demonstrating the metabolic defect could not be overcome.
Author contributions
M.L.H. Huang conceptualized the project. M.L.H. Huang, D.R. Richardson and S. Chiang designed research; M.L.H. Huang, D.R. Richardson and S. Chiang analyzed all presented data; M.L.H. Huang and S. Chiang performed research presented in Fig. 1, Fig. 2, Fig. 3, 5-7, and Supplementary Figure; N. Braidy performed research presented in Fig. 4; S. Maleki contributed reagents and analytical tools for IHC studies presented in Fig. 7; S. Lal contributed specimen samples for IHC studies; M.L.H. Huang, D.R. Richardson and S. Chiang wrote the paper; and all authors contributed to editing of the paper.
Funding
This work was supported by: National Health and Medical Research Council [Early Career Fellowship APP1074033 to M.L.H.H., Senior Principal Research Fellowships APP1062607 and APP1159596 to D.R.R.; and Project Grant #1002867 to D.R.R.], Muscular Dystrophy Association USA [Development Grant MDA417129 to M.L.H.H. and Research Grant to D.R.R.], University of Sydney [DVCR Fellowship and Early Career Researcher Grant to M.L.H.H.], an Australian Postgraduate Award to S.C., and Australian Research Council [Discovery Early Career Award DE170100628 to N.B.].
Declaration of competing interest
The authors of this article have stated explicitly that there are no conflict of interest in connection with this article.
Acknowledgments
The authors acknowledge support of the Australian Microscopy & Microanalysis Research Facility (University of Sydney, Australia), Dr. Arnulf H. Koeppen (Albany Medical College, NY, USA), who kindly provided FA patient heart samples, and Dr. Amy Anzovino who contributed to the Gsk3β western blots presented in Fig. 3.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102038.
Contributor Information
Des R. Richardson, Email: d.richardson@griffith.edu.au.
Michael L.-H. Huang, Email: huang.michaellh@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang M.L.H., Lane D.J.R., Richardson D.R. Mitochondrial mayhem: the mitochondrion as a modulator of iron metabolism and its role in disease. Antioxidants Redox Signal. 2011;15(12):3003–3019. doi: 10.1089/ars.2011.3921. [DOI] [PubMed] [Google Scholar]
- 2.Richardson D.R., Lane D.J., Becker E.M., Huang M.L.H., Whitnall M., Suryo Rahmanto Y., Sheftel A.D., Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U. S. A. 2010;107(24):10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang M.L.H., Becker E.M., Whitnall M., Rahmanto Y.S., Ponka P., Richardson D.R. Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant. Proc. Natl. Acad. Sci. U. S. A. 2009;106(38):16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitnall M., Rahmanto Y.S., Huang M.L.H., Saletta F., Lok H.C., Gutierrez L., Lazaro F.J., Fleming A.J., St Pierre T.G., Mikhael M.R., Ponka P., Richardson D.R. Identification of nonferritin mitochondrial iron deposits in a mouse model of Friedreich ataxia. Proc. Natl. Acad. Sci. U. S. A. 2012;109(50):20590–20595. doi: 10.1073/pnas.1215349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napier I., Ponka P., Richardson D.R. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105(5):1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 6.Kim H.J., Khalimonchuk O., Smith P.M., Winge D.R. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta. 2012;1823(9):1604–1616. doi: 10.1016/j.bbamcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17(2):215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 8.Huang M.L.H., Sivagurunathan S., Ting S., Jansson P.J., Austin C.J., Kelly M., Semsarian C., Zhang D., Richardson D.R. Molecular and functional alterations in a mouse cardiac model of Friedreich ataxia: activation of the integrated stress response, eIF2alpha phosphorylation, and the induction of downstream targets. Am. J. Pathol. 2013;183(3):745–757. doi: 10.1016/j.ajpath.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 9.Richardson D.R., Ponka P. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim. Biophys. Acta. 1997;1331(1):1–40. doi: 10.1016/s0304-4157(96)00014-7. [DOI] [PubMed] [Google Scholar]
- 10.Richardson D.R., Ponka P. Development of iron chelators to treat iron overload disease and their use as experimental tools to probe intracellular iron metabolism. Am. J. Hematol. 1998;58(4):299–305. doi: 10.1002/(sici)1096-8652. (199808)58:4<299::aid-ajh9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 11.Michael S., Petrocine S.V., Qian J., Lamarche J.B., Knutson M.D., Garrick M.D., Koeppen A.H. Iron and iron-responsive proteins in the cardiomyopathy of Friedreich's ataxia. Cerebellum. 2006;5(4):257–267. doi: 10.1080/14734220600913246. [DOI] [PubMed] [Google Scholar]
- 12.Anzovino A., Chiang S., Brown B.E., Hawkins C.L., Richardson D.R., Huang M.L.H. Molecular alterations in a mouse cardiac model of Friedreich Ataxia: an impaired Nrf2 response mediated via upregulation of Keap1 and activation of the Gsk3beta axis. Am. J. Pathol. 2017;187(12):2858–2875. doi: 10.1016/j.ajpath.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Chiang S., Kalinowski D.S., Dharmasivam M., Braidy N., Richardson D.R., Huang M.L.H. The potential of the novel NAD+ supplementing agent, SNH6, as a therapeutic strategy for the treatment of Friedreich's ataxia. Pharmacol. Res. 2020;155:104680. doi: 10.1016/j.phrs.2020.104680. [DOI] [PubMed] [Google Scholar]
- 14.Karthikeyan G., Lewis L.K., Resnick M.A. The mitochondrial protein frataxin prevents nuclear damage. Hum. Mol. Genet. 2002;11(11):1351–1362. doi: 10.1093/hmg/11.11.1351. [DOI] [PubMed] [Google Scholar]
- 15.Li B., Kaushik S., Kalinowski P., Kim B., Gershome C., Ching J., Poburko D. Droplet digital PCR shows the D-Loop to be an error prone locus for mitochondrial DNA copy number determination. Sci. Rep. 2018;8(1):11392. doi: 10.1038/s41598-018-29621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley J.L., Blake J.C., Chamberlain S., Thomas P.K., Cooper J.M., Schapira A.H. Clinical, biochemical and molecular genetic correlations in Friedreich's ataxia. Hum. Mol. Genet. 2000;9(2):275–282. doi: 10.1093/hmg/9.2.275. [DOI] [PubMed] [Google Scholar]
- 17.Wilson R.B., Roof D.M. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 1997;16(4):352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda Y., Shirakabe A., Brady C., Zablocki D., Ohishi M., Sadoshima J. Molecular mechanisms mediating mitochondrial dynamics and mitophagy and their functional roles in the cardiovascular system. J. Mol. Cell. Cardiol. 2015;78:116–122. doi: 10.1016/j.yjmcc.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane A.E., Sinclair D.A. Sirtuins and NAD(+) in the development and treatment of metabolic and cardiovascular diseases. Circ. Res. 2018;123(7):868–885. doi: 10.1161/circresaha.118.312498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank S. Dysregulation of mitochondrial fusion and fission: an emerging concept in neurodegeneration. Acta Neuropathol. 2006;111(2):93–100. doi: 10.1007/s00401-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Chan D.C. Critical dependence of neurons on mitochondrial dynamics. Curr. Opin. Cell Biol. 2006;18(4):453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scarffe L.A., Stevens D.A., Dawson V.L., Dawson T.M. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37(6):315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Spiegelman B.M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 26.Finck B.N., Kelly D.P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116(3):615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoli I., Alesci S., Blackman M.R., Su Y.A., Rennert O.M., Chrousos G.P. Mitochondria as key components of the stress response. Trends Endocrinol. Metab. 2007;18(5):190–198. doi: 10.1016/j.tem.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Canto C., Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harbor Symp. Quant. Biol. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011;93(4):884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin J.-H., Ko Han S., Kang H., Lee Y., Lee Y.-I., Pletinkova O., Troconso Juan C., Dawson Valina L., Dawson Ted M. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's Disease. Cell. 2011;144(5):689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U. S. A. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson B.L., Hock M.B., Ekholm-Reed S., Wohlschlegel J.A., Dev K.K., Kralli A., Reed S.I. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22(2):252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson R.M., Barger J.L., Edwards M.G., Braun K.H., O'Connor C.E., Prolla T.A., Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7(1):101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27(2):181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 35.Shami G., Cheng D., Henriquez J., Braet F. Assessment of different fixation protocols on the presence of membrane-bound vesicles in Caco-2 cells: a multidimensional view by means of correlative light and 3-D transmission electron microscopy. Micron. 2014;67:20–29. doi: 10.1016/j.micron.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Picard M., White K., Turnbull D.M. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J. Appl. Physiol. 2013;114(2):161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braidy N., Guillemin G.J., Mansour H., Chan-Ling T., Poljak A., Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PloS One. 2011;6(4) doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Bustamante S., Jayasena T., Richani D., Gilchrist R.B., Wu L.E., Sinclair D.A., Sachdev P.S., Braidy N. Quantifying the cellular NAD+ metabolome using a tandem liquid chromatography mass spectrometry approach. Metabolomics. 2017;14(1):15. doi: 10.1007/s11306-017-1310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braidy N., Poljak A., Grant R., Jayasena T., Mansour H., Chan-Ling T., Guillemin G.J., Smythe G., Sachdev P. Mapping NAD(+) metabolism in the brain of ageing Wistar rats: potential targets for influencing brain senescence. Biogerontology. 2014;15(2):177–198. doi: 10.1007/s10522-013-9489-5. [DOI] [PubMed] [Google Scholar]
- 40.Menezes S.V., Fouani L., Huang M.L.H., Geleta B., Maleki S., Richardson A., Richardson D.R., Kovacevic Z. The metastasis suppressor, NDRG1, attenuates oncogenic TGF-beta and NF-κB signaling to enhance membrane E-cadherin expression in pancreatic cancer cells. Carcinogenesis. 2019;40(6):805–818. doi: 10.1093/carcin/bgy178. [DOI] [PubMed] [Google Scholar]
- 41.Kalliora C., Kyriazis I.D., Oka S.-I., Lieu M.J., Yue Y., Area-Gomez E., Pol C.J., Tian Y., Mizushima W., Chin A., Scerbo D., Schulze P.C., Civelek M., Sadoshima J., Madesh M., Goldberg I.J., Drosatos K. Dual peroxisome-proliferator-activated-receptor-α/γ activation inhibits SIRT1-PGC1α axis and causes cardiac dysfunction. JCI Insight. 2019;5(17) doi: 10.1172/jci.insight.129556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trausch-Azar J., Leone T.C., Kelly D.P., Schwartz A.L. Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway. J. Biol. Chem. 2010;285(51):40192–40200. doi: 10.1074/jbc.M110.131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimes C.A., Jope R.S. The multifaceted roles of glycogen synthase kinase 3β in cellular signaling. Prog. Neurobiol. 2001;65(4):391–426. doi: 10.1016/S0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 44.Ramachandran B., Yu G., Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J. Biol. Chem. 2008;283(18):11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee E.J., Kang Y.C., Park W.-H., Jeong J.H., Pak Y.K. Negative transcriptional regulation of mitochondrial transcription factor A (TFAM) by nuclear TFAM. Biochem. Biophys. Res. Commun. 2014;450(1):166–171. doi: 10.1016/j.bbrc.2014.05.082. [DOI] [PubMed] [Google Scholar]
- 46.Piantadosi C.A., Carraway M.S., Babiker A., Suliman H.B. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008;103(11):1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.La Rosa P., Petrillo S., Turchi R., Berardinelli F., Schirinzi T., Vasco G., Lettieri-Barbato D., Fiorenza M.T., Bertini E.S., Aquilano K., Piemonte F. The Nrf2 induction prevents ferroptosis in Friedreich's Ataxia. Redox Biol. 2021;38:101791. doi: 10.1016/j.redox.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mori V., Amici A., Mazzola F., Di Stefano M., Conforti L., Magni G., Ruggieri S., Raffaelli N., Orsomando G. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PloS One. 2014;9(11) doi: 10.1371/journal.pone.0113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu C.P., Oka S., Shao D., Hariharan N., Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ. Res. 2009;105(5):481–491. doi: 10.1161/circresaha.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutak R., Xu X., Whitnall M., Kashem M.A., Vyoral D., Richardson D.R. Proteomic analysis of hearts from frataxin knockout mice: marked rearrangement of energy metabolism, a response to cellular stress and altered expression of proteins involved in cell structure, motility and metabolism. Proteomics. 2008;8(8):1731–1741. doi: 10.1002/pmic.200701049. [DOI] [PubMed] [Google Scholar]
- 51.Krishan S., Richardson D.R., Sahni S. Adenosine monophosphate–activated kinase and its key role in catabolism: structure, regulation, biological activity, and pharmacological activation. Mol. Pharmacol. 2015;87(3):363–377. doi: 10.1124/mol.114.095810. [DOI] [PubMed] [Google Scholar]
- 52.Yu R., Jin S.B., Lendahl U., Nistér M., Zhao J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 2019;38(8) doi: 10.15252/embj.201899748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178(5):749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., Adamopoulos I.E., Adeli K., Adolph T.E., Adornetto A., Aflaki E., Agam G., Agarwal A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2021;17(1):1–382. doi: 10.1080/15548627.2020.1797280. 1, fourth ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramirez R.L., Becker A.B., Mazurkiewicz J.E., Feustel P.J., Gelman B.B., Koeppen A.H. Pathology of intercalated discs in Friedreich cardiomyopathy. J. Am. Coll. Cardiol. 2015;66(15):1739–1740. doi: 10.1016/j.jacc.2015.06.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riehle C., Abel E.D. PGC-1 proteins and heart failure. Trends Cardiovasc. Med. 2012;22(4):98–105. doi: 10.1016/j.tcm.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehman J.J., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P. Peroxisome proliferator–activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106(7):847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sebastiani M., Giordano C., Nediani C., Travaglini C., Borchi E., Zani M., Feccia M., Mancini M., Petrozza V., Cossarizza A., Gallo P., Taylor R.W., d'Amati G. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J. Am. Coll. Cardiol. 2007;50(14):1362–1369. doi: 10.1016/j.jacc.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 60.Mettauer B., Zoll J., Garnier A., Ventura-Clapier R. Heart failure: a model of cardiac and skeletal muscle energetic failure. Pflügers Archiv. 2006;452(6):653–666. doi: 10.1007/s00424-006-0072-7. [DOI] [PubMed] [Google Scholar]
- 61.Kim H.D., Kim D.J., Lee I.J., Rah B.J., Sawa Y., Schaper J. Human fetal heart development after mid-term: morphometry and ultrastructural study. J. Mol. Cell. Cardiol. 1992;24(9):949–965. doi: 10.1016/0022-2828(92)91862-y. [DOI] [PubMed] [Google Scholar]
- 62.Lai L., Wang M., Martin O.J., Leone T.C., Vega R.B., Han X., Kelly D.P. A role for peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. J. Biol. Chem. 2014;289(4):2250–2259. doi: 10.1074/jbc.M113.523654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao L., Zhong W., Dong H., Guo W., Sun X., Zhang W., Yue R., Li T., Griffiths A., Ahmadi A.R., Sun Z., Song Z., Zhou Z. ATF4 activation promotes hepatic mitochondrial dysfunction by repressing NRF1-TFAM signalling in alcoholic steatohepatitis. Gut. 2020 doi: 10.1136/gutjnl-2020-321548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao L., Huang X., Xu M., Yang L., Hua F. MiR-144 protects the heart from hyperglycemia-induced injury by regulating mitochondrial biogenesis and cardiomyocyte apoptosis. FASEB J. 2020;34(2):2173–2197. doi: 10.1096/fj.201901838R. [DOI] [PubMed] [Google Scholar]
- 66.Chiang S., Huang M.L.H., Richardson D.R. Treatment of dilated cardiomyopathy in a mouse model of Friedreich's ataxia using N-acetylcysteine and identification of alterations in microRNA expression that could be involved in its pathogenesis, Pharmacol. Res. 2020;159:104994. doi: 10.1016/j.phrs.2020.104994. [DOI] [PubMed] [Google Scholar]
- 67.Han L., Zhou R., Niu J., McNutt M.A., Wang P., Tong T. SIRT1 is regulated by a PPARγ–SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res. 2010;38(21):7458–7471. doi: 10.1093/nar/gkq609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin A.S., Abraham D.M., Hershberger K.A., Bhatt D.P., Mao L., Cui H., Liu J., Liu X., Muehlbauer M.J., Grimsrud P.A., Locasale J.W., Payne R.M., Hirschey M.D. Nicotinamide mononucleotide requires SIRT3 to improve cardiac function and bioenergetics in a Friedreich's ataxia cardiomyopathy model. JCI Insight. 2017;2(14) doi: 10.1172/jci.insight.93885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dorn G.W., II Mitochondrial fission/fusion and cardiomyopathy. Curr. Opin. Genet. Dev. 2016;38:38–44. doi: 10.1016/j.gde.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du J., Zhou Y., Li Y., Xia J., Chen Y., Chen S., Wang X., Sun W., Wang T., Ren X., Wang X., An Y., Lu K., Hu W., Huang S., Li J., Tong X. Identification of Frataxin as a regulator of ferroptosis. Redox Biol. 2020;32:101483. doi: 10.1016/j.redox.2020.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Purroy R., Medina-Carbonero M., Ros J., Tamarit J. Frataxin-deficient cardiomyocytes present an altered thiol-redox state which targets actin and pyruvate dehydrogenase. Redox Biol. 2020;32:101520. doi: 10.1016/j.redox.2020.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thaher O., Wolf C., Dey P.N., Pouya A., Wüllner V., Tenzer S., Methner A. The thiol switch C684 in Mitofusin-2 mediates redox-induced alterations of mitochondrial shape and respiration. Neurochem. Int. 2018;117:167–173. doi: 10.1016/j.neuint.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Lu Y., Ding W., Wang B., Wang L., Kan H., Wang X., Wang D., Zhu L. Positive regulation of human PINK1 and Parkin gene expression by nuclear respiratory factor 1. Mitochondrion. 2020;51:22–29. doi: 10.1016/j.mito.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 74.Liesa M., Shirihai Orian S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metabol. 2013;17(4):491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinet W., Timmermans J.-P., De Meyer G.R.Y. Chapter Five - methods to assess autophagy in situ—transmission electron microscopy versus immunohistochemistry. In: Galluzzi L., Kroemer G., editors. Methods Enzymol. Academic Press; 2014. pp. 89–114. [DOI] [PubMed] [Google Scholar]
- 76.Yin M., van der Horst I.C., van Melle J.P., Qian C., van Gilst W.H., Sillje H.H., de Boer R.A. Metformin improves cardiac function in a nondiabetic rat model of post-MI heart failure. Am. J. Physiol. Heart Circ. Physiol. 2011;301(2):H459–H468. doi: 10.1152/ajpheart.00054.2011. [DOI] [PubMed] [Google Scholar]
- 77.Li X., Liu J., Lu Q., Ren D., Sun X., Rousselle T., Tan Y., Li J. AMPK: a therapeutic target of heart failure—not only metabolism regulation. Biosci. Rep. 2019;39(1) doi: 10.1042/bsr20181767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Wang Y., Zou M., Chen C., Chen Y., Xue R., Dong Y., Liu C. AMPK blunts chronic heart failure by inhibiting autophagy. Biosci. Rep. 2018;38(4) doi: 10.1042/bsr20170982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang B., Nie J., Wu L., Hu Y., Wen Z., Dong L., Zou M.H., Chen C., Wang D.W. AMPKα2 protects against the development of heart failure by enhancing mitophagy via PINK1 phosphorylation. Circ. Res. 2018;122(5):712–729. doi: 10.1161/circresaha.117.312317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu D., Dasgupta A., Chen K.-H., Neuber-Hess M., Patel J., Hurst T.E., Mewburn J.D., Lima P.D.A., Alizadeh E., Martin A., Wells M., Snieckus V., Archer S.L. Identification of novel dynamin-related protein 1 (Drp1) GTPase inhibitors: therapeutic potential of Drpitor1 and Drpitor1a in cancer and cardiac ischemia-reperfusion injury. FASEB J. 2020;34(1):1447–1464. doi: 10.1096/fj.201901467R. [DOI] [PubMed] [Google Scholar]
- 81.Sharp W.W., Fang Y.H., Han M., Zhang H.J., Hong Z., Banathy A., Morrow E., Ryan J.J., Archer S.L. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014;28(1):316–326. doi: 10.1096/fj.12-226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ong S.-B., Subrayan S., Lim S.Y., Yellon D.M., Davidson S.M., Hausenloy D.J. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121(18):2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 83.Givvimani S., Munjal C., Tyagi N., Sen U., Metreveli N., Tyagi S.C. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PloS One. 2012;7(3) doi: 10.1371/journal.pone.0032388. e32388-e32388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ong S.-B., Kalkhoran S.B., Cabrera-Fuentes H.A., Hausenloy Derek J. Mitochondrial fusion and fission proteins as novel therapeutic targets for treating cardiovascular disease. Eur. J. Pharmacol. 2015;763:104–114. doi: 10.1016/j.ejphar.2015.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wikstrom J.D., Katzman S.M., Mohamed H., Twig G., Graf S.A., Heart E., Molina A.J.A., Corkey B.E., de Vargas L.M., Danial N.N., Collins S., Shirihai O.S. β-cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes. 2007;56(10):2569–2578. doi: 10.2337/db06-0757. [DOI] [PubMed] [Google Scholar]
- 86.Kuznetsov A.V., Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int. J. Mol. Sci. 2009;10(4):1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu X., Thai P.N., Lu S., Pu J., Bers D.M. Intrafibrillar and perinuclear mitochondrial heterogeneity in adult cardiac myocytes. J. Mol. Cell. Cardiol. 2019;136:72–84. doi: 10.1016/j.yjmcc.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.