Significance
In the search for druggable targets to treat TNBC, ERβ, an estrogen receptor and tumor suppressor, has been suggested because it is found in 30% of TNBCs. In the present study, we found that except for SP1, the other tethering partners of ERβ (Fos, Jun, Fra1) are down-regulated in TNBC, meaning that the tumor-suppressive functions of ERβ are severely compromised. However, RNA seq revealed other driver pathways such as CYPs involved in the synthesis of fatty acid epoxides and in the inactivation of calcitriol and retinoic acid. Thus, ERβ agonists are unlikely to be useful in the treatment of TNBC. Instead, there is the danger that at estrogen response elements, ERβ may be acting as ERα does, and increase proliferation.
Keywords: breast cancer, cytochrome P450, estrogen receptor beta, fatty acid oxidation
Abstract
To identify regulators of triple-negative breast cancer (TNBC), gene expression profiles of malignant parts of TNBC (mTNBC) and normal adjacent (nadj) parts of the same breasts have been compared. We are interested in the roles of estrogen receptor β (ERβ) and the cytochrome P450 family (CYPs) as drivers of TNBC. We examined by RNA sequencing the mTNBC and nadj parts of five women. We found more than a fivefold elevation in mTNBC of genes already known to be expressed in TNBC: BIRC5/survivin, Wnt-10A and -7B, matrix metalloproteinases (MMPs), chemokines, anterior gradient proteins, and lysophosphatidic acid receptor and the known basal characteristics of TNBC, sox10, ROPN1B, and Col9a3. There were two unexpected findings: 1) a strong induction of CYPs involved in activation of fatty acids (CYP4), and in inactivation of calcitriol (CYP24A1) and retinoic acid (CYP26A1); and 2) a marked down-regulation of FOS, FRA1, and JUN, known tethering partners of ERβ. ERβ is expressed in 20 to 30% of TNBCs and is being evaluated as a target for treating TNBC. We used ERβ+ TNBC patient-derived xenografts in mice and found that the ERβ agonist LY500703 had no effect on growth or proliferation. Expression of CYPs was confirmed by immunohistochemistry in formalin-fixed and paraffin-embedded (FFPE) TNBC. In TNBC cell lines, the CYP4Z1-catalyzed fatty acid metabolite 20-hydroxyeicosatetraenoic acid (20-HETE) increased proliferation, while calcitriol decreased proliferation but only after inhibition of CYP24A1. We conclude that CYP-mediated pathways can be drivers of TNBC but that ERβ is unlikely to be a tumor suppressor because the absence of its main tethering partners renders ERβ functionless on genes involved in proliferation and inflammation.
Triple-negative breast cancer (TNBC) is not estrogen or progesterone dependent but several laboratories have reported the presence of estrogen receptor β (ERβ) in 20 to 30% of TNBCs (1–6). Lobular cancer also expresses ERβ (7), and these observations have led to the suggestion that ERβ could be targeted in treatment of these cancers. Most studies to date have found that ERβ represses proliferation and migration in cancers (2, 8–12) but there are also studies showing that ERβ induces a more aggressive phenotype in TNBC cells (4, 13).
Before considering ERβ as a target for treatment, the role of ERβ in these cancers has to be determined. Since there is no ERα in TNBC, estrogen should stimulate ERβ. However, in one clinical study, estrogen was used to treat TNBC and was found to have no clinical benefit (6). TNBC (14) remains a disease with no effective therapeutic targets. There is an intensive search for the drivers of TNBC. ERβ has been reported to alter the extracellular matrix in breast cancer (5, 15–18) and to enhance the innate immune response (19, 20). This means that ERβ agonists acting on stromal, endothelial, and immune cells may be useful in treatment of TNBC without direct action on the cancer cells.
In addition to steroid and peptide hormones, breast cancer may be influenced by nonsteroidal ligands of nuclear receptors (metabolites of cholesterol, vitamin D, retinoic acid), and by metabolites of fatty acids which do not use nuclear receptors for their activity (21). Some possible drivers of TNBC have been suggested. These include the autotaxin–lysophosphatidic axis (22) and fatty acid epoxygenases (21, 23, 24). Certain CYPs (the cytochrome P450 family) have also been implicated in TNBC, particularly those involved in activating fatty acids (23), inactivating calcitriol (25), and inactivating retinoic acid (26). Of these, only CYP24A1 has been reported to be ERβ-regulated (27). CYP4Z1 is known to be overexpressed in human breast cancer (28, 29) and is associated with high-grade tumors and poor prognosis (30). Expression of CYP4Z1 in T47D cells caused an increase of 20-hydroxyeicosatetraenoic acid (20-HETE), which is thought to drive proliferation and invasiveness of breast cancer (23, 24, 31). As this manuscript was in preparation, a very detailed analysis of the role of fatty acid epoxides and CYPs which produce them in TNBC was published. The study clearly shows that fatty acid epoxides can be drivers of TNBC (23).
Although epidemiological studies suggest a role for vitamin D in prevention of breast cancer, and although in breast cancer cell lines it can suppress proliferation and invasiveness, clinical treatment with vitamin D has had no effect on proliferation or apoptosis in women with breast cancer (32).
For most of its transcriptional activity, ERβ does bind to estrogen response elements (EREs) on DNA but tethers to transcription factors like AP1 (Fos-Jun), Sp1, and nuclear factor κB (NFκB) without itself binding to DNA (33). Mice in which the DNA-binding domain of ERβ is removed from the gene show a mild phenotype (34) because ERβ utilizes tethering more often than it uses direct ERE binding (35).
One method of checking for the effect of treatment of ERβ agonists in TNBC is to use ERβ agonists in mice harboring TNBC patient-derived xenografts (PDXs). Another is to compare the gene expression profile in normal and malignant parts of cancers for evidence of ERβ-regulated driver genes. The use of TNBC cell lines can provide some information because they lack ERα, PR, and HER2, but these cell lines were derived from other types of breast cancer, not TNBC, and it cannot be assumed that their genome represents that of TNBC.
In the present study, we have used RNA sequencing (RNA-seq), TNBC PDXs, and immunohistochemistry of formalin-fixed and paraffin-embedded (FFPE) TNBC samples and have found no evidence that ERβ is a tumor suppressor in TNBC. However, we did identify CYPs as a pathway that could be driving TNBC. When TNBC cell lines are engineered to express ERβ, proliferation and invasiveness are reduced. This is why the finding that ERβ is expressed in more than 30% of patients with TNBC is puzzling. To assess whether ERβ is a tumor suppressor in TNBC, we used three approaches: 1) RNA-seq to identify estrogen-regulated genes that are differently expressed between malignant (m) and normal adjacent (nadj) parts of five TNBCs; 2) ERβ-positive TNBC PDXs to examine the effect of an ERβ agonist (LY500307); and 3) FFPE TNBC slides to confirm gene expression revealed by the RNA-seq. We found 1) LY500307 had no effect on proliferation in TNBC PDXs; 2) a marked down-regulation of ERβ-tethering partners FOS, FRA1, and JUN in RNA-seq; and 3) a marked elevation of genes known to be overexpressed in TNBC as well as several CYPs involved in synthesis of fatty acid metabolites and in the degradation of calcitriol and retinoic acid.
Results
RNA-Seq Data.
Several studies have compared differences between malignant and normal parts of TNBC. In the present study, RNA-seq revealed some marked differences between the malignant and nadj parts of TNBC (SI Appendix, Table S1). These include increased expression of genes already known to be increased in TNBC. Matrix metalloproteinases (MMPs), chemokines, lysophosphatidic acid receptors (LPARs), and, in keeping with the known basal characteristics of TNBC, MMP7, BIRC5/survivin, Wnt-10A and -7B, sox10, ROPN1B, and Col9a3. There was an 8.6-fold increase in expression of melanoma-inhibitory activity, a protein which promotes invasiveness in several cancers. Anterior-grade protein 2 (AGR2) was increased 6-fold in mTNBC. AGR2 was identified previously (36) as a marker of poor prognosis in TNBC. AGR2 and 3 are two related disulfide isomerases which appear to have different effects in breast cancer. AGR2 is an estrogen-regulated protooncogene associated with a more aggressive phenotype while AGR3 is androgen-regulated and associated with a more favorable prognosis. In the present study, both AGR2 and 3 were equally increased by 6-fold. The consequence of the coexpression of these two genes requires further investigation.
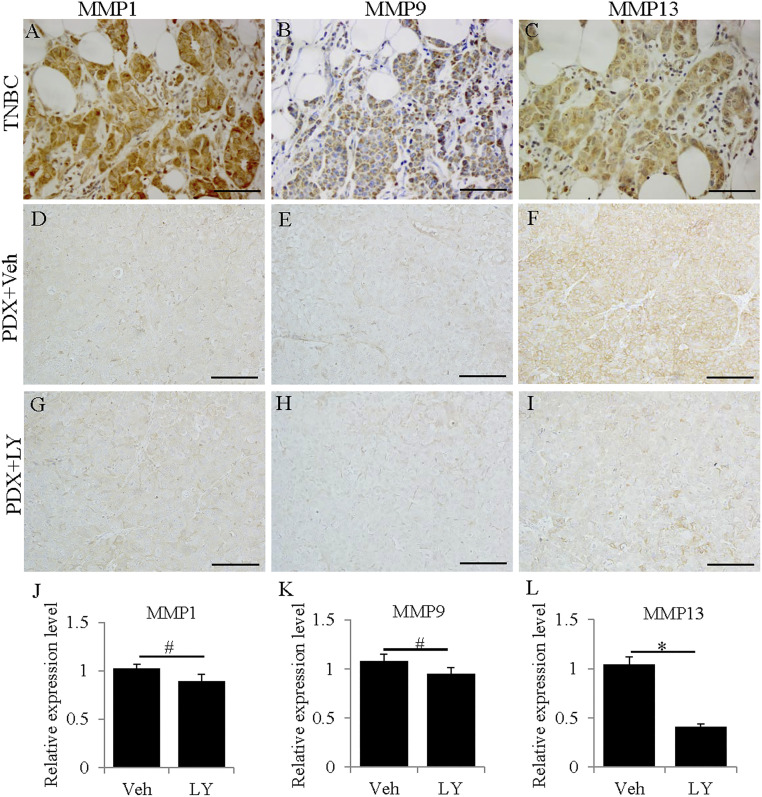
MMPs have several roles in cancer (37, 38). MMP1, 3, 7, 9, 10, 11, 12, and 14 are up-regulated in cancer (38). In the present study, MMP1, 3, 7, 9, 12, and 13 were all very highly expressed in mTNBC (Fig. 1 and Table 1). MMP2 and 27 were down-regulated. MMP1 (collagenase), MMP3 (stromolysin), and MMP9 (gelatinase) degrade the extracellular matrix to promote invasiveness and metastasis in cancer, while MMP13 is involved in tumor angiogenesis (39). The differential regulation of MMPs in mTNBC may indicate something about estrogen signaling in these samples. ERα is known to up-regulate (40) and ERβ to down-regulate MMPs 9 and 13 (41). The high expression of these two MMPs in TNBC suggests an absence of ERβ-suppressive function and may indicate an ERα-like action (activation at EREs) of ERβ. According to the RNA-seq, MMP2 and 27 were down-regulated in mTNBC. Since MMP2 and 27 are regulated by SP1 (42), their down-regulation may reflect an interaction of ERβ with SP1 (not significantly changed; Table 2) on the promotors of these genes.
Fig. 1.
Expression of MMPs in TNBCs and TNBC PDXs. MMP1, 9, and 13 were highly expressed in TNBC patient samples (A–C). MMP1 (D and G) and MMP9 (E and H) were weakly expressed in TNBC PDX samples and were not significantly changed by LY500307 treatment (J and K) (#P > 0.05). MMP13 (F and I) was highly expressed in TNBC PDXs and was significantly down-regulated by LY500307 treatment (L) (*P < 0.05). (J–L) Error bars represent standard deviations. (Scale bars, 100 μm.)
Table 1.
MMPs in TNBC
| MMPs | Log2 fold change (P value) |
| MMP1 | 8.7 (0.0007) |
| MMP3 | 7.1 (0.02) |
| MMP7 | 7.3 (0.001) |
| MMP9 | 4.2 (0.01) |
| MMP12 | 4.0 (0.08) |
| MMP13 | 9.0 (0.001) |
| MMP2 | −2.0 (0.02) |
| MMP27 | −3.8 (0.002) |
Table 2.
LPA pathway in TNBC
| LPA receptors | Log2 fold change (P value) |
| LPAR1(GPR26) | −2.5 (0.0008) |
| LPAR2 | 4.3 (0.008) |
| LPAR3 | 6.1 (0.0007) |
| ENPP2 | −4.5 (1.19E-10) |
| ENPP5 | 3.5 (0.001) |
MMP13 was 9-fold up-regulated in mTNBC. It is regulated by ETS translocation variant 4 (ETV4) (39, 43), a transcription factor whose overexpression in TNBC is an indicator of poor prognosis and increased metastasis. Its 5.9-fold up-regulation in the present TNBC analysis suggests that it could be the regulator of MMP13.
Han et al. (44) have reported that MMP7 is regulated by FOXc1 in TNBC and together with WNT-5A participates in the invasiveness of TNBC. In our mTNBC, neither FOXc1 nor WNT-5A was up-regulated. However, RUNX2, a transcription factor which regulates MMP7, was increased 2.5-fold.
Chemokines in TNBC.
As shown in Table 3, there was a marked increase in transcripts of several chemokines in mTNBC. There was no increase in the receptors of these chemokines which are expressed in monocytes and dendritic cells invading the tumor. One possible explanation for this is that monocytes and dendritic cells, which express the receptors, were not attracted to the tumor. As suggested by Gonzalez-Avila et al. (38), although the chemokines were released by the PDX, they were degraded by the high concentration of MMPs in the environment and were ineffective in recruiting immune cells to the tumor.
Table 3.
Chemokines and chemokine receptors in TNBC
| Log2 fold change (P value) | |
| Chemokines | |
| CXCL17 | 6.2 (0.008) |
| CXCL13 | 5.7 (0.0002) |
| CXCL9 | 3.3 (0.05) |
| CXCL10 | 7.2 (0.001) |
| CXCL11 | 5.2 (0.02) |
| CXCL14 | −5.0 (0.001) |
| CXCL12 | −4.2 (2.40E-06) |
| Receptors | |
| CXCR6 | Not changed |
| CXCR4 | Not changed |
| CXCR3 | Not changed |
LPAS in TNBC.
ENPPs (ectonucleotide pyrophosphatase/phosphodiesterases) are a family of enzymes which is responsible for the production of lysophosphatidic acid from lysophosphatidylcholine. In TNBC, ENPP2 was down-regulated but ENPP5 was up-regulated. LPA is a signaling molecule which binds to one of six lysophosphatidic acid receptors (LPAR1 to 6). LPAR2 and 3 were up-regulated by 4.3- and 6.1-fold, respectively (Table 2). Thus, LPA signaling could be a driver in TNBC.
Genes Previously Reported to Be Highly Expressed in TNBC That Were Not Up-Regulated in the Present Study.
GDF10 was reported by Zhou et al. (45) to be a repressor of proliferation and epithelial–mesenchymal transition in TNBC but it was down-regulated by 7.3-fold in the present study. NOTCH signaling has been called a hallmark of TNBC (46) but, in the present study, Notch 2 to 4 were all decreased in mTNBC as were NOTCH downstream genes (HES, HEY, CCND1, Notch-regulated ankyrin repeat protein).
Expression of FOS, Jun, and SP1 in TNBC.
One notable finding of this study is the marked reduction in expression of the ERβ-tethering partners Fos C, Fos B, JunB, and JunD (Table 4). The loss of tethering partners suggests that ERβ may not be functional as an antiinflammatory and antiproliferative factor and that it may instead be driving proliferation through its actions at EREs. This may explain the high expression (fourfold [log2]) of the ERα-inducible genes TFF1 and TFF3.
Table 4.
Changes in ERβ-tethering partners
| Transcript | Log2 fold change (P value) |
| Fos | −5.2 (3.7E-07) |
| Fos B | −7.0 (1.0E-14) |
| JunD | −1.8 (0.008) |
| JunB | −3.5 (5.3E-05) |
| Sp1 | NS |
| NFκBIE | 2.1 (0.02) |
NS, not significant.
Growth Curves of TNBC PDX Growth.
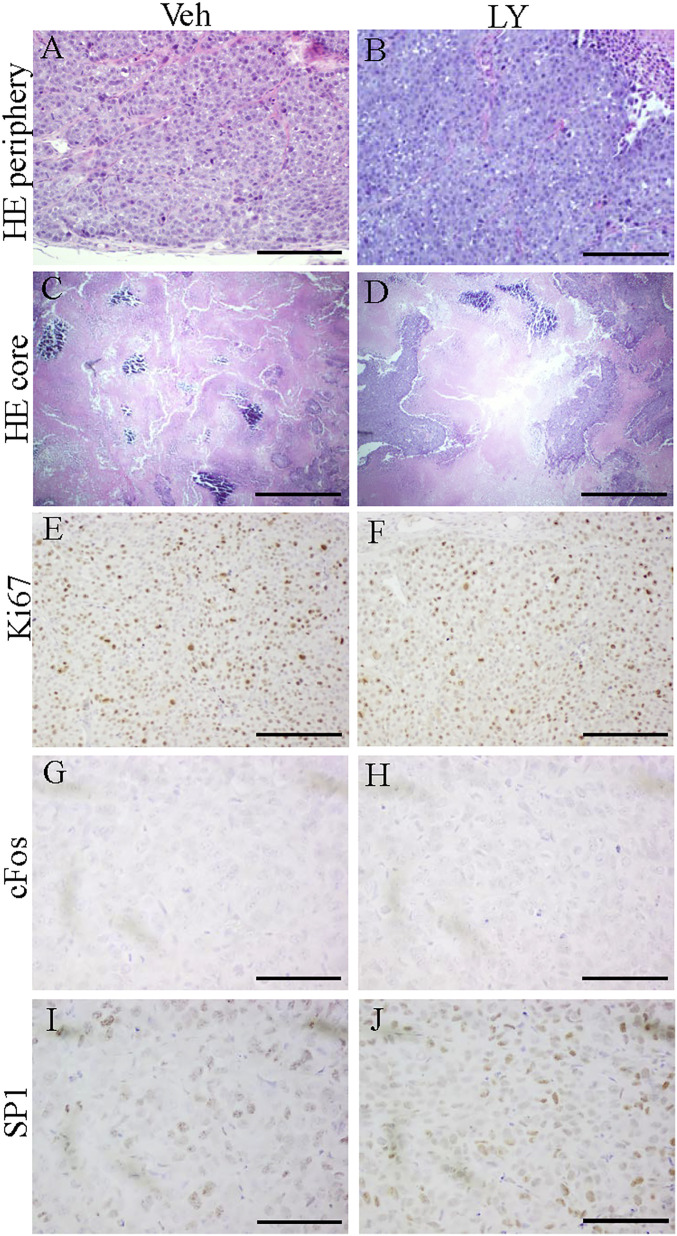
In both LY500307-treated and untreated mice, PDXs grew at similar rates. After 31 d, the tumors were excised and examined. The cancer cells in the xenografts were densely packed together and there was poor vascularization so there were large necrotic regions in the core of the tumors (Fig. 2 A–D). There was no difference in proliferation between the two groups as assessed by Ki67 staining (Fig. 2 E and F).
Fig. 2.
H&E staining and expression of Ki67, cFOS, and SP1 in TNBC PDXs. H&E staining of five PDXs in each treatment group demonstrated that cancer cells were densely packed together in peripheral zones (A and B) and that there were large necrotic regions in the core of the PDX tumors (C and D). Ki67 was highly expressed in both the Veh-treated group and LY500307-treated group (E and F). Expression of cFOS was not detected by immunohistochemistry staining (G and H). SP1 was highly expressed in Veh-treated or LY500307-treated PDX tumors (I and J). (Scale bars, 100 μm [A, B, and E–J] and 500 μm [C and D].)
In the ERβ+ xenografts, expression of Fos (Fig. 2 G and H), FRA1, JunB, JunD, and NFκB, which are tethering partners of ERβ, was reduced to below the level of detection by immunochemical staining and their expression was not changed by the ERβ agonist LY500307. SP1 was highly expressed in all of the PDXs (Fig. 2 I and J). These results indicate that genes regulated by ERβ tethering to SP1 should be ERβ-regulated in the PDX while those genes whose promoters respond to ERβ in association with AP1 or NFκB might not be influenced by the presence of the ERβ agonist. This latter class of genes includes those involved in proliferation and inflammation, which are normally down-regulated by ERβ.
CYP Change in TNBC.
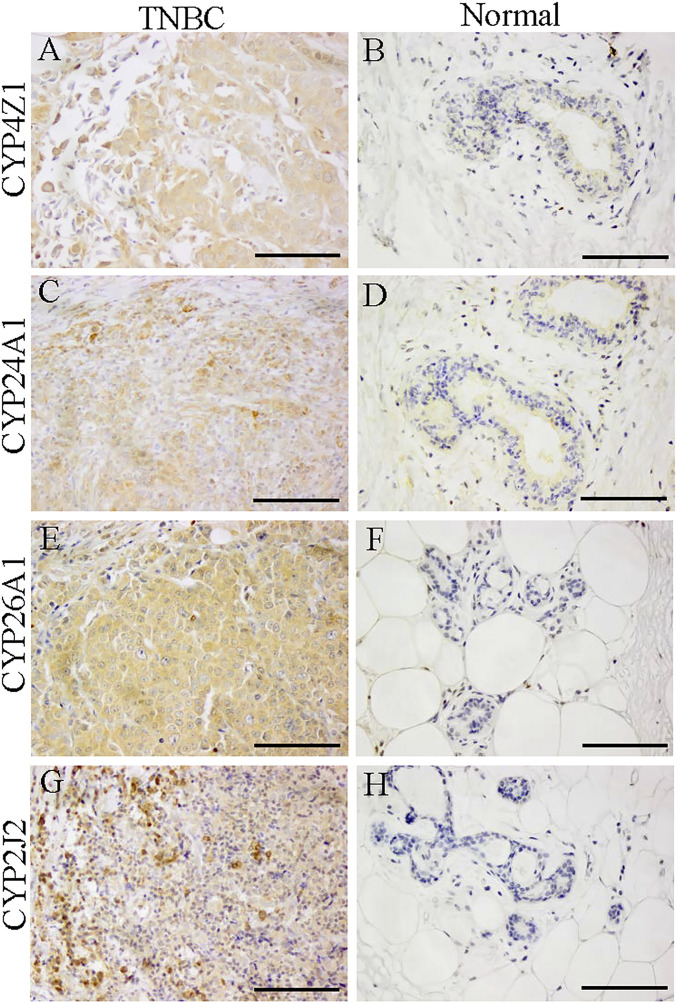
Transcripts of several members of the cytochrome P450 family were highly expressed in TNBC (Table 5). The functions of these CYPs are listed in Table 5. Immunohistochemistry revealed that these CYPs of families 4Z, 24A, 26A, and 2J2 were strongly expressed in all TNBC samples (Fig. 3), indicating that fatty acid metabolites and loss of calcitriol and retinoic acid may be involved in growth or progression of TNBC (23, 32).
Table 5.
CYPs expressed in TNBC
| CYPs | Log2 fold change | Function |
| 4F8 | 8.5 | Catalyzes hydroxylation of arachidonic acid, omega-2 and omega-3, PGH1 and PGH2, docosapentaenoic acid, PGI2, and epoxidation of docosahexaenoic acid and docosapentaenoic acid. |
| 4F11 | 3.2 | Catalyzes ω-hydroxylation of 1) short chain and medium chain to initiate β-oxidation and fatty acids and 2) leukotrienes and HETEs to initiate their inactivation. |
| 4F22 | 11 | The fatty acid ω-hydroxylase required for acylceramide synthesis. Its substrate is ultra–long-chain fatty acids. |
| 27B1 | 16 | Vitamin D 1-α hydroxylase. |
| 4Z1 | 36 | Catalyzes the production of 20-HETE from arachidonic acid, which stimulates growth and invasiveness of breast cancer cells. |
| 26A1 | 6.5 | Metabolizes and inactivates retinoic acid, terminating its prodifferentiative functions. |
| 24A1 | 6.7 | A 24-hydroxylase which inactivates calcitriol. |
| 2J2 | 7.2 | Catalyzes the conversion of endogenous polyunsaturated fatty acids to signaling molecules such as eicosatrienoic acid epoxides, which cause growth and invasiveness of MDA231 cells. |
Fig. 3.
Expression of CYPs in TNBC and normal breast. The FFPE slides were from 10 TNBCs and 10 normal breasts. Representative pictures are shown for staining with each antibody. Cyp4Z1 (A and B), Cyp24A1 (C and D), Cyp26A1 (E and F), and Cyp2J2 (G and H) were more highly expressed in TNBC samples than in normal breast. (Scale bars, 100 μm.)
CYPs in PDX and TNBC Cell Lines.
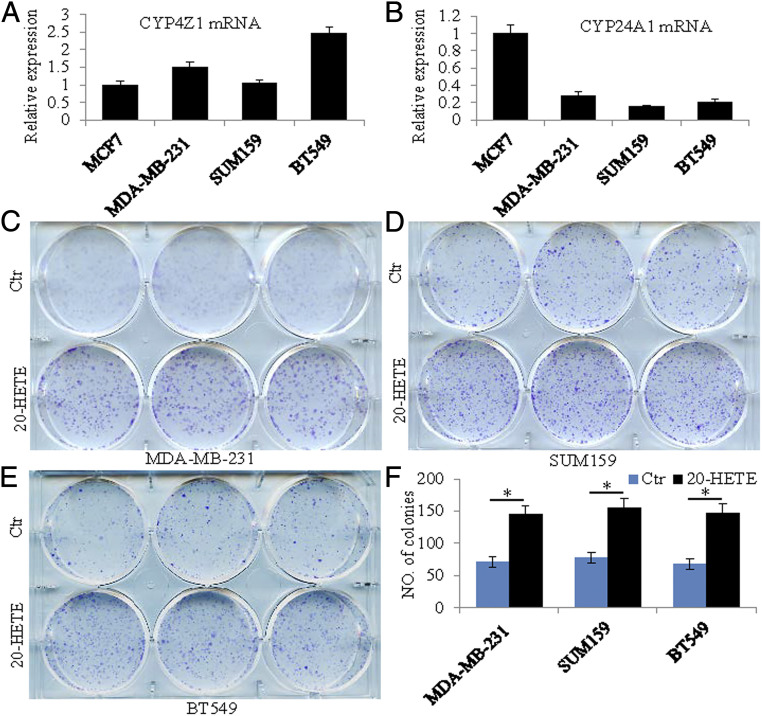
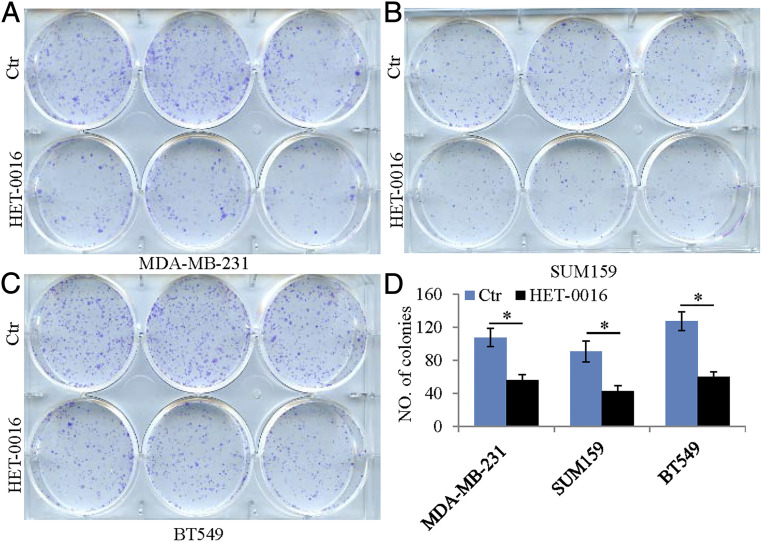
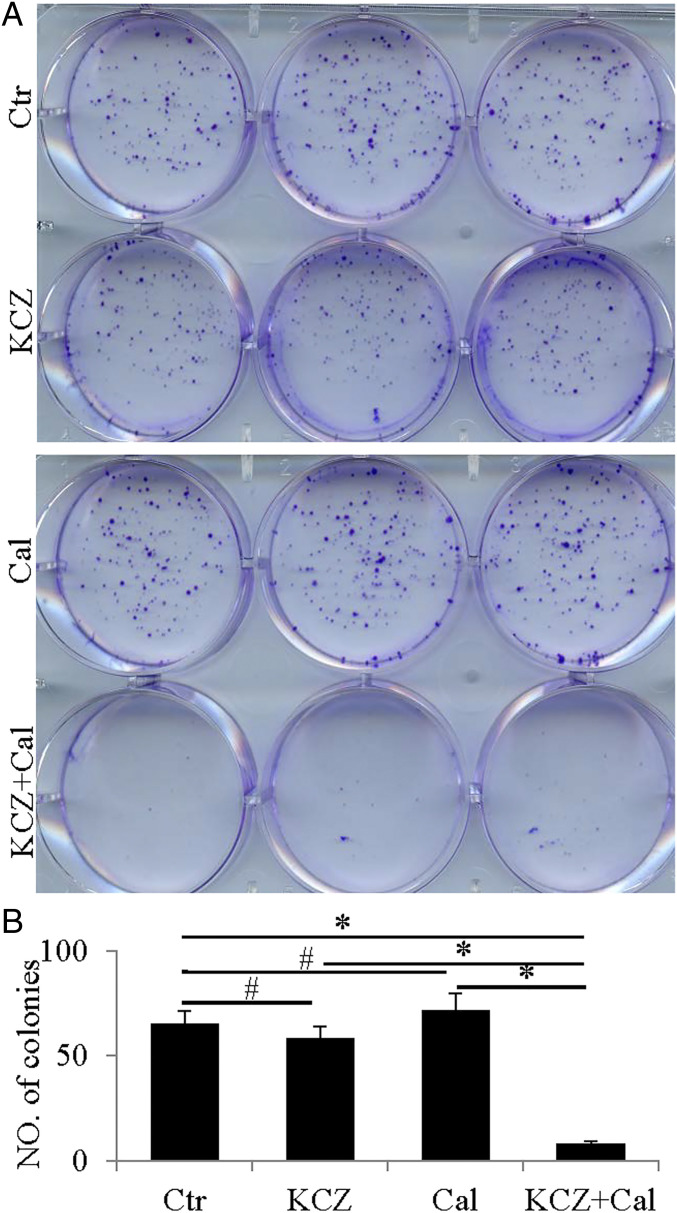
The PDXs expressed CYP24A1, 26A1, and 4Z1 but their level of expression was not changed by LY500703. These CYPs were also expressed in three TNBC cell lines, BT549, SUM159, and MDA-MB-231 (Fig. 4 A and B). To examine whether changes in expression or activity of these CYPs have effects on growth and migration of TNBC cell lines, we treated all three cell lines with 20-HETE (10 nM); HET-0016 (10 µM), an inhibitor of synthesis of 20-HETE; ketoconazole (20 μM), a general inhibitor of CYPs; calcitriol (10 nM); and the active hormone of vitamin D. We found that the receptor for 20-HETE, GPR 75, was increased 2.6-fold (log2) in TNBC over normal breast and in the cell lines, 20-HETE stimulated growth (Fig. 4 C–F), and HET-0016 reduced growth in all three cell lines (Fig. 5). Calcitriol 4-fold (log2) by itself did not affect growth but, in the presence of ketoconazole, it prevented growth in the TNBC cell line (Fig. 6). Thus, 20-HETE is a possible driver of TNBC and calcitriol is an inhibitor of growth when the activity of its degrading enzyme, CYP24A1, is blocked. In all three TNBC cell lines, 20-HETE stimulated cell migration and HET-0016 inhibited migration (SI Appendix, Figs. S1 and S2).
Fig. 4.
Expression of Cyp4Z1 and Cyp24A1 in breast cancer cell lines and the effect of 20-HETE on the growth of TNBC cells. qRT-PCR showed expression of Cyp4Z1 messenger RNA (mRNA) and Cyp24A1 mRNA in ERα-positive cells (MCF-7) and TNBC cells (MDA-MB-231, SUM159, and BT549) (A and B). 20-HETE treatment promoted growth in all three TNBC cells (C–E). Statistical analyses of the number of colonies in control (Ctr) or 20-HETE–treated TNBC cells (*P < 0.05) (F).
Fig. 5.
Role of HET-0016 in the growth of TNBC cells. HET-0016 treatment inhibited growth in all three TNBC cells (A–C). Statistical analyses of the number of colonies in control or HET-0016–treated TNBC cells (*P < 0.05) (D).
Fig. 6.
Effect of calcitriol (Cal) and ketoconazole (KCZ) on the growth of TNBC cells. Calcitriol treatment or ketoconazole treatment did not affect the growth of TNBC cells (A). Treatment with calcitriol and ketoconazole significantly reduced growth (A). Statistical analyses of the number of colonies in control, ketoconazole-, calcitriol-, or ketoconazole and calcitriol–treated TNBC cells (#P > 0.05, *P < 0.05) (B).
Discussion
Several studies have reported that ERβ is expressed in TNBC and the question has arisen as to whether ERβ ligands could be effective in the treatment of TNBC. We have used three cohorts of samples to address this question. 1) We grew ERβ-positive TNBC PDXs provided by the Jackson Laboratory in mice and treated half of the mice with the ERβ agonist LY500703. Before starting the study, we used immunohistochemistry to confirm the presence of ERβ in the PDX. After 31 d the tumors were excised and analyzed; 2) five TNBC frozen samples with a malignant area and an adjacent normal area from Imperial College London. These samples were used for RNA-seq; and 3) a set of FFPE slides from Imperial College London. Genes of interest obtained from the RNA-seq data were analyzed by immunohistochemistry in the PDX and the FFPE samples.
The PDX study revealed that although ERβ was expressed at the start and was maintained in the tumors at the end of the experiment, treatment of the PDX-bearing mice with the ERβ agonist had no effect on the growth or proliferation index of the tumors but did repress the expression of MMPs. RNA-seq revealed that Fos, Jun, Jun b, and Jun d were all very much reduced in mTNBC. SP1 and NFκB were unchanged but the NFκB inhibitor, NFκBIE, was induced in mTNBC. What these data suggest is that those genes whose regulation is dependent on the tethering of ERβ to AP1 or NFκB are not ERβ-regulated in TNBC, while those that are dependent on binding of ERβ to ERE or tethering to SP1 are responsive to ERβ ligands. What is not clear is whether, in the absence of its tethering factors, ERβ acts as ERα at EREs and causes proliferation.
Previous studies (27, 47) have shown that when the TNBC cell line MDA-MB-231 was engineered to express ERβ, all of the well-known ERβ-regulated genes (TGFRβ3, IGFBP4, Spink4) were induced and proliferation and invasiveness were inhibited. In the present study, the limited effect of ERβ expression in TNBC suggests caution in extrapolation from TNBC cell lines to human TNBC samples. The MDA-MB-231 cell line was isolated from a patient with invasive ductal carcinoma (IDC) which expressed ERα (48). All of the other TNBC cell lines were also derived from breast cancers that were not originally TNBC (49). These cell lines do have in common with TNBC the lack of expression of ER, PR, or HER2 but that does not make them true representatives of TNBC. This difference between the TNBC cell lines and TNBC in patient samples may explain why other pathways previously reported to be involved in TNBC cell lines (GPNMP, EGFR, Notch 1–3, mTOR, AKT 1 and 2, TGFβ, activin receptor, SMAD 7) were all down-regulated in RNA-seq of the TNBC patient samples.
MMPs are responsible for migration and metastasis in many cancers (27). From ERβ−/− mice, we know that, in the mammary gland, MMPs are ERβ–down-regulated genes (50). In TNBC, MMP1, 7, 9, 12, and 13 were highly expressed (increased over normal tissue by 415-, 158-, 30-, 15-, and 512- [linear] fold, respectively). The high expression in TNBC of MMPs indicates that down-regulation of these genes by ERβ requires tethering of ERβ to AP1 or NFκB.
Chemokines are key factors which recruit immune cells to cancers. Although they were very highly expressed in TNBC, none of their receptors, CXCRs, was increased and there were no increases in markers of B or T cells. One possible explanation for this dissociation between high chemokine expression and low recruitment of immune cells is inactivation of the chemokines by the high levels of MMPs in these cancers. The degradation of chemoattractant genes in TNBC may explain why markers of monocytes and macrophages (CD163), neutrophils (CD177), and T cells (TIMD4) are all very down-regulated in our mTNBC. There was increased expression of the chemokines CXCL10 (30-fold) and CXCL11 (150-fold). In addition, CXCL13 was increased 50-fold in TNBC. It has been reported that CXCR4 and CXCL10 overexpression predicts a favorable outcome in TNBC (51) but CXCR4 was not increased in TNBC in the present study.
Expression of CXCL12, a chemoattractant for lymphocytes, was reduced by 4.2-fold in mTNBC. Knockout of the receptors for CXCL12, namely CXCR4 and CXCR7, inhibits proliferation and invasiveness in the TNBC cell line MDA231 (49), and it has been suggested that the CXCR12 pathway could be used as a target for treatment of TNBC. In the present study, there was a marked loss of CXCL12 in mTNBC, suggesting that at least in this small sample (five TNBC patients), CXCL12 is not a driver.
Genes involved in the regulation of pH were very highly expressed in TNBC. Maintenance of an acidic environment of cancer is a well-characterized phenomenon which gives cancer cells a growth advantage over normal cells (52).
Some of the most unexpected changes revealed in the RNA-seq data were the remarkable elevation of expression in TNBC of CYPs involved in the activation of fatty acids and the inactivation of retinoic acid and calcitriol. Except for CYP24A1, whose expression is increased in MDA231 when these cells are engineered to express ERβ (27) and by treatment of TNBC cells with ERβ agonists, these CYPs are not known to be estrogen-regulated. Fatty acid metabolites formed through the action of CYPs are known to promote cancer and invasiveness while the inactivation of calcitriol and retinoic acid would be expected to remove the differentiative function of retinoic acid and the antiproliferative actions of calcitriol. Immunohistochemical analysis of the FFPE samples confirmed the strong expression of CYPs in mTNBC. The effects of fatty acid and vitamin D metabolism on proliferation were assessed in TNBC cell lines. The arachidonic acid metabolite 20-HETE stimulated growth and the 20-HETE inhibitor HET-0016 prevented this growth. Treatment of TNBC cells with calcitriol had no effect on growth but, in combination with the CYP inhibitor ketoconazole, calcitriol reduced cell growth, indicating that overexpression of CYP24A1 prevents the growth-inhibiting effects of calcitriol.
ERβ is expressed in both luminal and basal cells of the breast. The normal role of ERβ in the basal cells is restraining regulation of proliferation. This is clear from the phenotype of the mammary gland in ERβ knockout mice (50): When the ERβ gene is removed, there is increased proliferation of the basal cells. From the present studies, we conclude that, in TNBC, ERβ is relatively inactive as a transcription factor because of the low expression of its tethering partners AP1 and NFκB and that ERβ agonists may be of limited use in treatment of TNBC.
Materials and Methods
Hematoxylin and Eosin Staining.
Xenograft tumors were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and embedded in paraffin. Specimens were sectioned at 5 μm and mounted on glass slides. Slides were deparaffinized with xylene before hydrating through graded alcohols to water. Samples were stained with hematoxylin and eosin (H&E) followed by dehydration through graded alcohols to xylene prior to sealing with Permount.
All PDXs were completely sectioned and every 10th section was stained with hematoxylin. Thereafter, every 20th section was probed with antibodies.
Immunohistochemistry.
Paraffin-embedded sections were deparaffinized and heated in a LabVision PT module (Thermo Fisher Scientific) at 97 °C for 10 min to retrieve antigens. Sections were blocked with buffer composed of 50% (volume [vol]/vol) methanol and 3% (vol/vol) H2O2 and then transferred to PBS containing 3% (weight/vol) bovine serum albumin with 0.1% Nonidet P-40. This was followed by overnight incubation at 4 °C with primary antibodies: anti-MMP9 (1:50; Santa Cruz), anti-MMP1 (1:100; Abcam), anti-MMP14 (1:50; Abcam), anti-MMP13 (1:100; Abcam), anti-CYP24A1 (1:25; Abcam), anti-CUP4Z1 (1:100; LifeSpan Biosciences), anti-CYP26A1 (1:250; Abcam), anti-CYP2J2 (1:10; Abcam), anti-Ki67 (1:1,000; Abcam), anti–c-jun (1:100; Novus Biologicals), and anti-SP1 (1:100; Novus Biologicals). Slides were incubated with secondary antibody followed by a horseradish peroxidase polymer kit (Biocare Medical; GHP516) for 30 min. Finally, slides were developed with 3,3′-diaminobenzidine tetrahydrochloride and counterstained with hematoxylin.
TNBC samples were available from Imperial College London. Five samples were used for RNA-seq and 15 were used for immunohistochemistry. Three of the samples used for RNA-seq were primary grade 3 IDC and three were recurrent IDC.
Cell Lines and Reagents.
Human breast cancer cell lines MCF-7, MDA-MB-231, SUM159, and BT549 were maintained in our laboratory. MCF-7, SUM159, and BT549 were grown in Dulbecco’s modified Eagle’s medium (Gibco) and MDA-MB-231 cells were grown in RPMI-1640 (Gibco), and in all cases medium was supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Invitrogen). All cells were maintained at 37 °C with 5% CO2 in a humidified incubator. Ketoconazole, 20-HETE, HET-0016, and calcitriol were purchased from Sigma-Aldrich. The ERβ agonist LY500307 was developed by Eli Lilly.
qRT-PCR.
Total RNA was extracted from cells using an RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. RNA samples were reverse-transcribed using the High-Capacity cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The complementary DNA (cDNA) was amplified using a LightCycler 96 real-time PCR instrument (Roche). Reactions were prepared using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and universal cycling conditions (95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s). Reaction specificity was confirmed by melting curve analysis. Primers used were CYP24A1 forward: 5′-GCAGCCTAGTGCAGATTT-3′ and reverse: 5′-ATT-CACCCAGAACTGTTG-3′; and CYP4Z1 forward: 5′-CTTTCCAGATGGACGCTC-CTTACCT-3′ and reverse: 5′-GGCAAAATGCTGCCCAATGCAGTTC-3′. Glyceraldehyde 3-phosphate dehydrogenase was used to normalize expression.
Clonogenic Assay.
Cells were seeded in triplicate into 6-well plates (100 to 200 cells per well). The day after seeding, cells were cultured in the absence or presence of 10 nM 20-HETE or 10 μM HET-0016 as indicated in complete media for 10 to 14 d and drug medium was replaced every 3 to 4 d. Colonies formed were fixed with methanol and stained with 0.5% crystal violet. Cells were seeded in triplicate into 6-well plates (200 to 800 cells per well). The day after seeding, cells were cultured in the absence or presence of 10 nM calcitriol mixed with 20 μM ketoconazole from 72 to 96 h. Drugs were replaced with fresh medium at different time points. After incubation for 5 to 10 d, colonies formed were fixed with methanol and stained with 0.5% crystal violet.
Wound-Healing Cell-Migration Assay.
When breast cancer cells (MDA-MB-231, SUM159, and BT549) reached full confluence, the monolayer was scratched with a 1-mm micropipette tip. After washing, these cells were then treated with 10 nM 20-HETE for 48 h or 10 µM HET-0016 for 24 h. Images were captured by an inverted microscope (Nikon; Eclipse TS100).
Mouse Protocols at the Jackson Laboratory.
Drug efficacy was determined in the following PDX breast tumor models: TM00097 (BR1077) and TM00098 (BR1126). For each model, 21 6- to 8-wk-old female NOD.Cg-Prkdc<scid>Il2rg<tm1Wjl>/SzJ (NSG; JAX 5557) mice were used. Mice were implanted orthotopically into the inguinal mammary fat pad with tumor fragments to produce study-ready cohorts of P3 for PDX breast cancer model TM00098 (BR1126) or P4 of model TM00097 (BR1077).
Body weights and clinical observations were recorded once or twice weekly. Digital caliper measurements were initiated to determine tumor volume once or twice weekly when tumors became palpable. When the tumor volumes reached ∼70 to 120 mm3, mice were then randomly divided into the LY500307-treated group (LY) and vehicle-treated group (Veh). Both groups were dosed every 2 d for 14 doses (study days 0 through 26). Body weights, clinical observations, and digital caliper measurements were recorded twice weekly post dose initiation. Tumors were collected from all killed animals and cut into two pieces. One piece of each tumor was flash-frozen, and the other was fixed in buffered PFA. All mice were killed by CO2 asphyxiation on study day 27.
RNA-seq and analysis were done at the core facility for Bioinformatics and Expression Analysis, at Huddinge University Hospital, Karolinska Institute using Illumina TruSeq Stranded RNA assays and analyzed for gene expression analysis, looking at the gene sets C2 and hallmark from the Molecular Signatures Database (MSigDB). This gene set analysis was done for each comparison.
Supplementary Material
Acknowledgments
This study was supported by the Brockman Foundation, the Robert A. Welch Foundation (Grant E-0004), the Swedish Cancer Fund, and the Swedish Science Council.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104162118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Shanle E. K., et al., Research resource: Global identification of estrogen receptor β target genes in triple negative breast cancer cells. Mol. Endocrinol. 27, 1762–1775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shanle E. K., et al., Prognostic significance of full-length estrogen receptor beta expression in stage I–III triple negative breast cancer. Am. J. Transl. Res. 7, 1246–1259 (2015). [PMC free article] [PubMed] [Google Scholar]
- 3.Smart E., Hughes T., Smith L., Speirs V., Estrogen receptor β: Putting a positive into triple negative breast cancer? Horm. Mol. Biol. Clin. Investig. 16, 117–123 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hinsche O., Girgert R., Emons G., Gründker C., Estrogen receptor β selective agonists reduce invasiveness of triple-negative breast cancer cells. Int. J. Oncol. 46, 878–884 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Schüler-Toprak S., et al., Agonists and knockdown of estrogen receptor β differentially affect invasion of triple-negative breast cancer cells in vitro. BMC Cancer 16, 951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wisinski K. B., et al., Targeting estrogen receptor beta in a phase 2 study of high-dose estradiol in metastatic triple-negative breast cancer: A Wisconsin Oncology Network Study. Clin. Breast Cancer 16, 256–261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B., et al., Differential expression of estrogen receptor α, β1, and β2 in lobular and ductal breast cancer. Proc. Natl. Acad. Sci. U.S.A. 111, 1933–1938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramasamy K., Samayoa C., Krishnegowda N., Tekmal R. R., Therapeutic use of estrogen receptor β agonists in prevention and treatment of endocrine therapy resistant breast cancers: Observations from preclinical models. Prog. Mol. Biol. Transl. Sci. 151, 177–194 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Murphy L. C., Leygue E., The role of estrogen receptor-β in breast cancer. Semin. Reprod. Med. 30, 5–13 (2012). [DOI] [PubMed] [Google Scholar]
- 10.McNamara K. M., et al., The presence and impact of estrogen metabolism on the biology of triple-negative breast cancer. Breast Cancer Res. Treat. 161, 213–227 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Guo L., et al., Expression and prognostic value of estrogen receptor β in patients with triple-negative and triple-positive breast cancer. Exp. Ther. Med. 9, 2147–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua S. A., et al., Estrogen receptor beta protein in human breast cancer: Correlation with clinical tumor parameters. Cancer Res. 63, 2434–2439 (2003). [PMC free article] [PubMed] [Google Scholar]
- 13.Austin D., et al., Estrogen receptor-beta is a potential target for triple negative breast cancer treatment. Oncotarget 9, 33912–33930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litzenburger B. C., Brown P. H., Advances in preventive therapy for estrogen-receptor-negative breast cancer. Curr. Breast Cancer Rep. 6, 96–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg K., et al., Expression of estrogen receptor beta increases integrin alpha1 and integrin beta1 levels and enhances adhesion of breast cancer cells. J. Cell. Physiol. 222, 156–167 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Zalewski A., Cecchini E. L., Deroo B. J., Expression of extracellular matrix components is disrupted in the immature and adult estrogen receptor β-null mouse ovary. PLoS One 7, e29937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piperigkou Z., et al., Estrogen receptor beta modulates breast cancer cells functional properties, signaling and expression of matrix molecules. Matrix Biol. 56, 4–23 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kousidou O. Ch., et al., Estradiol-estrogen receptor: A key interplay of the expression of syndecan-2 and metalloproteinase-9 in breast cancer cells. Mol. Oncol. 2, 223–232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L., et al., Pharmacological activation of estrogen receptor beta augments innate immunity to suppress cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 115, E3673–E3681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham M., Gilkeson G., Estrogen receptors in immunity and autoimmunity. Clin. Rev. Allergy Immunol. 40, 66–73 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Mitra R., et al., CYP3A4 mediates growth of estrogen receptor-positive breast cancer cells in part by inducing nuclear translocation of phospho-Stat3 through biosynthesis of (±)-14,15-epoxyeicosatrienoic acid (EET). J. Biol. Chem. 286, 17543–17559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volden P. A., et al., Mammary adipose tissue-derived lysophospholipids promote estrogen receptor-negative mammary epithelial cell proliferation. Cancer Prev. Res. (Phila.) 9, 367–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apaya M. K., et al., Integrated omics-based pathway analyses uncover CYP epoxygenase-associated networks as theranostic targets for metastatic triple negative breast cancer. J. Exp. Clin. Cancer Res. 38, 187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J. G., et al., Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Res. 67, 6665–6674 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Flanagan L., et al., Efficacy of vitamin D compounds to modulate estrogen receptor negative breast cancer growth and invasion. J. Steroid Biochem. Mol. Biol. 84, 181–192 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Helvig C., Taimi M., Cameron D., Jones G., Petkovich M., Functional properties and substrate characterization of human CYP26A1, CYP26B1, and CYP26C1 expressed by recombinant baculovirus in insect cells. J. Pharmacol. Toxicol. Methods 64, 258–263 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Reese J. M., et al., ERβ-mediated induction of cystatins results in suppression of TGFβ signaling and inhibition of triple-negative breast cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 115, E9580–E9589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu W., et al., Increased expression of CYP4Z1 promotes tumor angiogenesis and growth in human breast cancer. Toxicol. Appl. Pharmacol. 264, 73–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rieger M. A., et al., Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Res. 64, 2357–2364 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Yang X., Hutter M., Goh W. W. B., Bureik M., CYP4Z1—A human cytochrome P450 enzyme that might hold the key to curing breast cancer. Curr. Pharm. Des. 23, 2060–2064 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Wei X., et al., Elevated 14,15-epoxyeicosatrienoic acid by increasing of cytochrome P450 2C8, 2C9 and 2J2 and decreasing of soluble epoxide hydrolase associated with aggressiveness of human breast cancer. BMC Cancer 14, 841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnan A. V., Swami S., Feldman D., The potential therapeutic benefits of vitamin D in the treatment of estrogen receptor positive breast cancer. Steroids 77, 1107–1112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kushner P. J., et al., Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74, 311–317 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Maneix L., et al., Estrogen receptor β exon 3-deleted mouse: The importance of non-ERE pathways in ERβ signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 5135–5140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao C., et al., Genome-wide mapping of estrogen receptor-beta-binding regions reveals extensive cross-talk with transcription factor activator protein-1. Cancer Res. 70, 5174–5183 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Salmans M. L., Zhao F., Andersen B., The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: A potential drug target and biomarker. Breast Cancer Res. 15, 204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shay G., Lynch C. C., Fingleton B., Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 44–46, 200–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Avila G., et al., Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit. Rev. Oncol. Hematol. 137, 57–83 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Dumortier M., et al., ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res. 20, 73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad N., Chen S., Wang W., Kapila S., 17β-Estradiol induces MMP-9 and MMP-13 in TMJ fibrochondrocytes via estrogen receptor α. J. Dent. Res. 97, 1023–1030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gustafsson J. A., Strom A., Warner M., Update on ERbeta. J. Steroid Biochem. Mol. Biol. 191, 105312 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Nalvarte I., et al., Estrogen receptor β controls MMP-19 expression in mouse ovaries during ovulation. Reproduction 151, 253–259 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Singsuksawat E., Thuwajit C., Charngkaew K., Thuwajit P., Increased ETV4 expression correlates with estrogen-enhanced proliferation and invasiveness of cholangiocarcinoma cells. Cancer Cell Int. 18, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han B., et al., FOXC1-induced non-canonical WNT5A-MMP7 signaling regulates invasiveness in triple-negative breast cancer. Oncogene 37, 1399–1408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou T., et al., GDF10 inhibits proliferation and epithelial-mesenchymal transition in triple-negative breast cancer via upregulation of Smad7. Aging (Albany NY) 11, 3298–3314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giuli M. V., Giuliani E., Screpanti I., Bellavia D., Checquolo S., Notch signaling activation as a hallmark for triple-negative breast cancer subtype. J. Oncol. 2019, 8707053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramaniam M., Pitel K. S., Bruinsma E. S., Monroe D. G., Hawse J. R., TIEG and estrogen modulate SOST expression in the murine skeleton. J. Cell. Physiol. 233, 3540–3551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chavez K. J., Garimella S. V., Lipkowitz S., Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 32, 35–48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M., et al., Impact of CXCR4 and CXCR7 knockout by CRISPR/Cas9 on the function of triple-negative breast cancer cells. OncoTargets Ther. 12, 3849–3858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner M., et al., Ventral prostate and mammary gland phenotype in mice with complete deletion of the ERβ gene. Proc. Natl. Acad. Sci. U.S.A. 117, 4902–4909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuan T., Li T., Yi C., Identification of CXCR4 and CXCL10 as potential predictive biomarkers in triple negative breast cancer (TNBC). Med. Sci. Monit. 26, e918281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lebelo M. T., Joubert A. M., Visagie M. H., Warburg effect and its role in tumourigenesis. Arch. Pharm. Res. 42, 833–847 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.