Abstract
Inorganic polyphosphate (polyP) is produced by both bacteria and their eukaryotic hosts, and appears to play multiple important roles in the interactions between those organisms. However, the detailed mechanisms of how polyP synthesis is regulated in bacteria and how it influences both bacterial and host biology remain largely unexplored. In this review, we examine recent developments in the understanding of how bacteria regulate the synthesis of polyP, what roles polyP plays in controlling virulence in pathogenic bacteria, and the effects of polyP on the mammalian immune system, as well as progress on developing drugs that may be able to target bacterial polyP synthesis as novel means of treating infectious disease.
Keywords: Polyphosphate, Bacterial Survival, Pathogenicity, Immune Regulation
Why Study Polyphosphate?
Inorganic polyphosphate (polyP) is a universally conserved biomolecule composed of covalently linked units of phosphate monomers, as depicted in Figure 1A, with functions as widely varied as life itself. In bacteria, polyP has been linked to a myriad of functions, including energy storage, metabolic regulation, stress responses, viability, colonization, pathogenicity, virulence, mobility, and antibiotic resistance [1–14]. PolyP is involved in the movement of large magnetotactic bacteria in suboxic zones of the Black Sea, and has recently been investigated as a basis for a photo-microbial fuel cell [15, 16]. In eukaryotes, polyP plays roles in everything from oxidative and divalent cation stress responses in Saccharomyces cerevisiae to blood coagulation, macrophage differentiation, leukocyte proliferation, neutrophil recruitment, and platelet functions in mammalian systems [17–22], as well as playing a protective role in neuronal signaling by preventing glutamate excitotoxicity [23]. PolyP is important in archaea, as well. In Methanosarcina mazei, for example, polyP has been shown to accumulate under phosphate starvation conditions, acting to regulate the transcription of multiple phosphate metabolism and transport genes including those encoding the pstSCAB-phoU complex [24], while in several species of Sulfolobales, polyP is involved in archaeal motility, adhesion, and biofilm formation in [25].
Figure 1. The structure and regulation of polyP in bacteria.
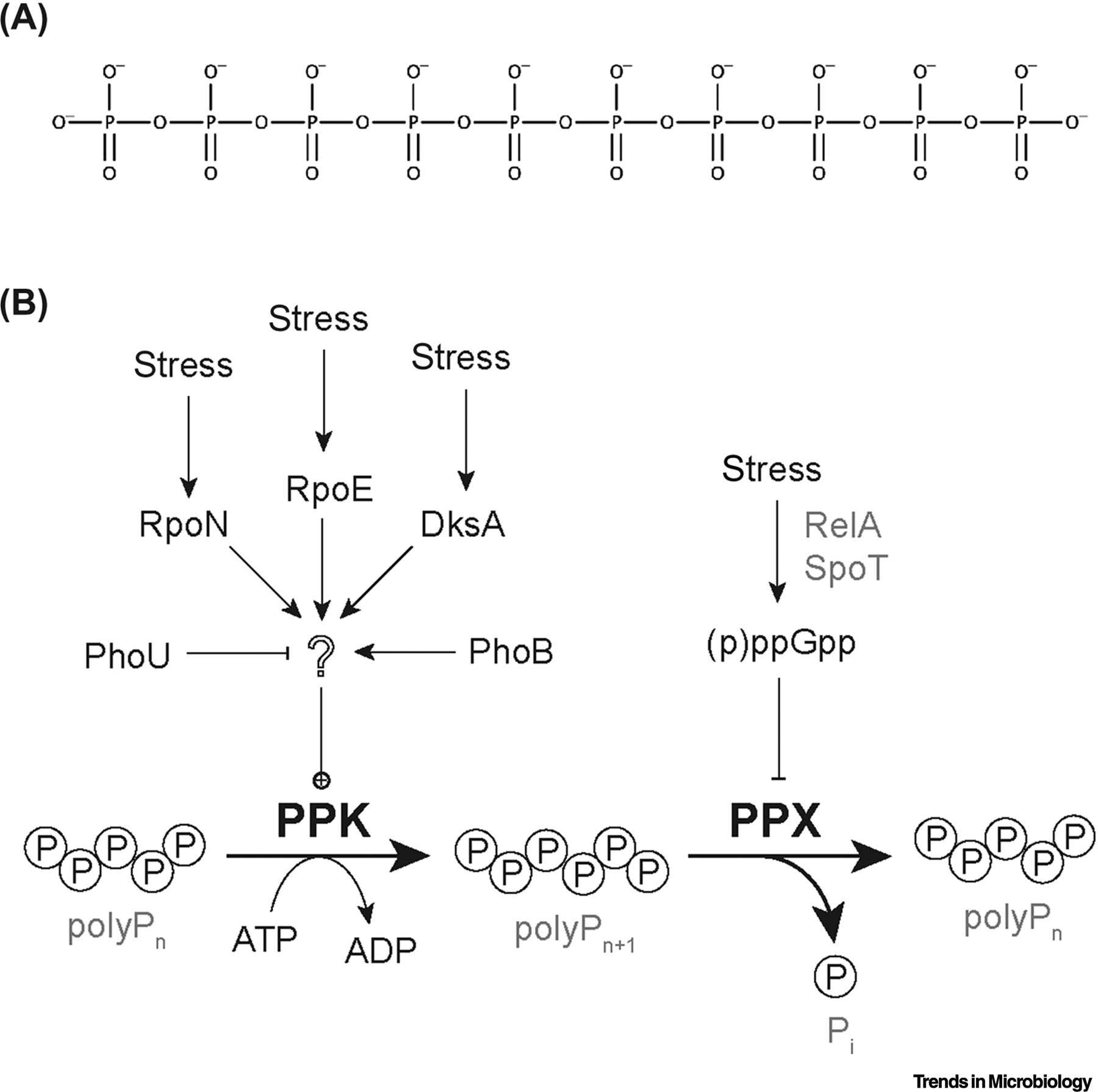
(A) The structure of polyP. A short polyP molecule (polyP10). Bacterial polyP can be up to 1000 phosphate units long, while eukaryotic polyP is usually substantially shorter. (B) The current model for polyP regulation in Escherichia coli. PolyP is synthesized by polyP kinase (PPK) and degraded by exo-polyPase (PPX). Starvation stress stimulates RelA and SpoT to synthesize (p)ppGpp, which inhibits PPX, but does not influence PPK activity and is not required for induction of polyP synthesis. Induction of polyP synthesis does depend on the transcription factors RpoN, RpoE, DksA, and PhoB, although none of these activate transcription of the ppk gene itself. PhoU is a negative regulator of both phosphate transport and polyP synthesis. None of the mechanism(s) by which these regulators affect PPK activity are currently known.
With such diverse functions, it is of little surprise that new studies are steadily discovering new and reimagined roles for polyP. It is the purpose of this review to discuss advances in the field of polyP biology over the past few years, emphasizing the importance of understanding polyP functions in host-microbe interactions and how polyP represents a promising, but complicated target for clinical applications.
Overturning Established Models of Polyphosphate Regulation
There are two unrelated kinases utilized by many bacteria to synthesize polyP: PPK1 or PPK (see Glossary) and PPK2 [1]. PPK1 predominately catalyzes the synthesis of polyP from ATP. PPK2 is different in that it can efficiently use GTP or ATP to synthesize polyP and is more efficient at hydrolyzing polyP to synthesize nucleotide triphosphates [26]. Some bacteria possess homologs of both PPK1 and PPK2; others possess only one, or in some cases, neither. While this review focuses primarily on polyP synthesis by PPK1, it is important to note that some species (such as Francisella tularensis) have both PPK1 and PPK2 homologs, and that most studies we highlight only focus on one of these enzymes. For brevity, and because most recent studies examine PPK1 functions, it is the intent of this review to focus largely on PPK1. For clarity, however, we will take care to indicate when PPK2 is also present in a system.
Many bacteria, including the model Gram-negative bacterium Escherichia coli (E. coli), which has a single PPK1 homolog (called simply PPK) [27], activate polyP synthesis in response to environmental stress [28], and the many roles of polyP in stress tolerance [4, 6, 8, 9, 11, 12, 29–36] are central to the function of polyP in host-microbe interactions. Nevertheless, the molecular mechanisms of polyP regulation in bacteria have been largely ignored since early studies in E. coli by Arthur Kornberg’s lab in the late 1990s [37, 38]. Those studies proposed a model in which the repression of polyP-degrading exopolyphosphatase (PPX) activity by the stringent response alarmone (p)ppGpp [39] was the key regulatory step in polyP synthesis [38, 40], and this idea was generally accepted until very recently [41]. However, we have recently demonstrated that induction of polyP synthesis in E. coli is, in fact, independent of (p)ppGpp synthesis [42], although it does depend on the presence of the RNA polymerase-binding stringent transcription factor DksA as well as on the alternative sigma factors RpoN and RpoE, which are best known for their roles in responding to nitrogen starvation and cell envelope disruption stresses, respectively [42, 43]. We do not currently know what gene or genes regulated by these transcription factors directly influence polyP levels, since ppk transcription is not activated under stress conditions in E. coli [43], although there does appear to be transcriptional control of ppk in some other bacteria in response to stress [44, 45]. Our lab has also recently published a study describing point mutations in E. coli PPK that strongly activate polyP synthesis in vivo without affecting the enzyme’s active site or in vitro activity [46] suggesting a role for some form of post-translational regulation of PPK activity, either by post-translational modification of PPK itself or by interaction with other proteins, but this remains to be tested. It has been known for some time that the phosphate transport regulators PhoU and PhoB regulate polyP accumulation in many bacteria [40, 47–49], but how they do so and whether they interact with other regulatory systems remains to be determined. In summary, as shown in Figure 1B, there is a great deal remaining to be discovered about the genes and proteins involved in controlling polyP accumulation, even in E. coli, which has a long history of polyP research.
Bacterial Polyphosphate and Lon Protease
One of the areas ripe for exploration is the relationship between bacterial polyP and Lon protease, one of the major protein-degrading enzymes of bacteria [50]. It is well established, at least in E. coli, that polyP interacts with Lon. The effects of this interaction, though, remain incompletely understood. Early on, Kornberg reported that polyP activates Lon-mediated degradation of a subset of ribosomal proteins, contributing to recovery from amino acid starvation [51–53]. Recently, the Konieczny lab has shown that polyP regulates DNA replication initiation by acting on Lon during the stringent stress response in E. coli [54]. In this report, the researchers showed both in vitro and in vivo that polyP activates Lon protease to target and degrade the essential replication initiation protein DnaA. DnaA inhibits replication initiation when bound to ATP and stimulates replication when bound to ADP [55, 56]. Gross and Konieczny demonstrated that polyP associates with DnaA-ADP, but not with DnaA-ATP, and that this association is necessary for the Lon protease to degrade DnaA. This results in depletion of DnaA-ADP, while enriching the DnaA-ATP repressor pool. However, polyP inhibits the Lon-dependent degradation of some other proteins, including the model protein α-casein [57] and, notably, the cell division inhibitor SulA [53]. This suggests a general Lon-dependent network by which polyP accumulation inhibits cell division. PolyP, Lon, DnaA, and ribosomal proteins are all very highly conserved among bacteria, and DnaA is a conserved target for Lon degradation [58]. Importantly, Lon is implicated in multiple polyP-dependent functions, some of which will be discussed below. The mechanism by which polyP affects Lon targeting and activity is currently unknown. Further deciphering of the global effect of polyP on Lon targeting and activity, and therefore on the bacterial proteome, is essential for understanding polyP biology and represents an exciting area for future research.
Polyphosphate in Pathogenic Bacteria
Much of the research in the field focuses on pathogenic bacteria and how polyP is involved in disease. PolyP production is required for virulence in many pathogens [2, 3, 8, 12–14, 35], and recent research has now begun to establish more detailed molecular mechanisms for these roles.
For example, one report found that polyP synthesis is essential for the virulence of Salmonella enterica serovar Typhimurium and its survival in Dictyostelium discoideum, a social amoeba used as a model for macrophage interactions [59]. The developmental cycle of D. discoideum progresses through three stages (aggregation, elevation, and culmination), which S. enterica sv. Typhimurium delays by inducing a starvation-like transcriptional response while selectively impairing expression of genes required for chemotaxis and aggregation [60–62]. S. enterica sv. Typhimurium, like E. coli, has only a single PPK1 homolog (called simply “PPK”) and lacks PPK2. The study by Varas et al found that Δppk S. enterica sv. Typhimurium were deficient in impairing developmental progress of D. discoideum [59]. Proteomic profiling of infected amoebae revealed that the WT strain triggered a robust response, including the expression of DNA repair enzymes, but the Δppk strain did not induce the expression of DNA repair enzymes. Importantly, while the WT strain was able to replicate and survive in the amoeba, the Δppk mutant was not, despite being internalized at significantly higher levels. This suggests that S. enterica sv. Typhimurium induces DNA damage through an unknown pathway that depends on continued bacterial survival, and that polyP synthesis is essential for this survival. What mechanisms are specifically regulated by polyP in S. enterica sv. Typhimurium and how those mechanisms allow it to evade host defenses remain to be explored; however, D. discoideum is clearly a powerful model system for deciphering these kinds of bacterial-eukaryotic interactions.
Another example of a newly-discovered role for polyP in bacterial pathogenesis is the recent report by Tang-Fichaux et al. showing that production of the DNA-damaging toxin colibactin by a variety of strains of E. coli is dependent on the presence of ppk [63]. The loss of polyP, either by deletion of ppk or by chemical inhibition of PPK activity with mesalamine (see The Discovery and Testing of Microbial Polyphosphate Kinase Inhibitors section below) decreased expression of the promoter driving the expression of colibactin synthesis genes, reducing the genotoxicity of E. coli.
There are many reports of polyP acting as a pro-virulence factor in bacteria. However, Rohlfing et al. tell a different story [64]. They investigated the role of PPK1 in Francisella tularensis, which possesses both PPK1 and PPK2. In F. tularensis major pathogenic elements are expressed from the Francisella Pathogenicity Island (FPI), which is regulated in a (p)ppGpp-dependent manner. Rohlfing et al. found that a Δppk1 mutant had higher expression of the FPI genes observed, suggesting that ppk expression actually antagonized Francisella pathogenicity. A Lon deletion strain (Δlon) also showed increased transcript levels of multiple virulence genes, though not to the level of the Δppk1 mutant. FPI expression in a Δlon Δppk strain was comparable to the Δppk1 single mutant. Western blots revealed that the level of an FPI protein was higher in the Δlon, Δppk1, and Δlon Δppk1 strains than in WT, further arguing that the expression of polyP represses the expression of FPI pathogenicity elements, likely through a Lon-dependent mechanism. These results emphasize the importance of understanding both the polyP-Lon relationship and the individual mechanisms utilized by different pathogenic species.
Regulation of virulence gene expression and protein stability are not the only roles polyP may play in pathogenic bacteria’s survival in a host. Roewe et al. have recently reported on the effect of bacterial polyP on innate host defense in an E. coli sepsis model in mice [34]. They found that polyP directly affects innate immune response in a chain-length dependent manner. Injecting long-chain polyP (lcPolyP, ~300–1000 phosphate units [31]), similar to that synthesized by bacterial PPK, into a host along with bacteria resulted in markedly increased mortality. High levels of polyP reduced the ability of neutrophils and macrophages to phagocytize bacteria while also reducing the expression of macrophage-attracting chemokines (such as CCL2 and CXCL10) and cytokines like INFβ. Moreover, lcPolyP bound to the surfaces of macrophages and was internalized, resulting in drastic alterations to gene expression and macrophage polarization from an M1 (pro-inflammatory) to an M2 (anti-inflammatory) phenotype. Interestingly, lcPolyP not only drove a M2 phenotypic response, but it also overrode the LPS-induced M1 response by enhancing M2 genes in LPS-activated macrophages while simultaneously antagonizing M1 genes, even to the extent of impairing the expression of iNOS genes and the secretion of NO2− into the supernatant. LcPolyP also altered the type I interferon response to LPS, resulting in a less responsive macrophage population. As a final blow to the innate response, lcPolyP interfered with antigen presentation by suppressing the expression of the Major Histocompatibility Complex (MHC) invariant chain as well as the important costimulatory proteins CD80 and CD86. Importantly, the reduction in the expression of the MHC-invariant chain was also seen in vivo, demonstrating interference by lcPolyP in the interplay between the innate and adaptive immune systems.
Supporting the idea that bacterial polyP plays an important role in modulating innate immune responses, Rijal et al. have recently reported that pathogenic Mycobacterium species, including M. tuberculosis and M. smegmatis (both of which possess PPK1 and PPK2, although in this study only PPK1 was examined) secrete polyP and that this secreted bacterial polyP increases survival of these bacteria after phagocytosis by either human macrophages or D. discoideum amoeba [65]. In these experiments, polyP inhibited both phagosome acidification and lysosome activity. Intriguingly, in D. discoideum, the putative polyP receptor GrlD was required for these effects, suggesting that eukaryotic cells may possess signaling pathways directly responsive to bacterial polyP. There is evidence that at least some other pro-inflammatory pathogenic bacteria can secrete or maintain substantial extracellular levels of polyP [66, 67] so, while important questions about physiological polyP concentration and sources during natural infections remain unresolved, the observation that bacterial-type polyP can repress the innate immune response is both important and informative for the study of polyP biology and suggests a molecular mechanism underlying the essentiality of polyP production for virulence in many pathogens.
Polyphosphate War: Clashing Functions in Host-Pathogen Interactions
PolyP is produced by both bacteria and eukaryotes. Not only does polyP play a role in bacterial survival, but it is also involved in host defense and repair mechanisms. For example, Suess et al. found that polyP plays multiple roles in wound healing and leukocyte biology [22]. They demonstrated that fibrocyte differentiation depended on platelet-derived polyP and that low concentrations of polyP (1 – 2 pM) promoted fibrocyte differentiation in peripheral blood mononuclear cell (PBMC) cultures, while higher (30 – 125 pM) appeared to decrease fibrocyte differentiation while concurrently increasing macrophage differentiation in the absence of serum. Interestingly, they also found that 0 – 10 pM polyP gradients acted as chemoattractants for neutrophils, suggesting a novel role for recruiting neutrophils to sites of tissue damage. Finally, they found that extracellular polyP played a role in proliferation, as concentrations of 100 μM or higher inhibited proliferation of PBMCs in vitro. In this study, Suess et al. convincingly demonstrated that polyP is highly involved in the innate response, playing roles in maturation of fibroblasts and macrophages while also acting as a chemoattractant for neutrophils into areas of inflammation.
The relationship between polyP and neutrophils takes on a new level of significance when considering neutrophilic responses to bacterial sepsis. In bacterial sepsis, neutrophils release traps composed of neutrophilic proteins (such as neutrophil elastase), DNA, and histones into the microvasculature in multiple tissues [68]. These Neutrophil Extracellular Traps (NETs) capture circulating bacteria while stimulating inflammation and coagulation in the surrounding tissue. While this serves as an effective mechanism to capture bacteria, the subsequent clotting and inflammation often cause significant tissue and organ damage. In 2017, McDonald et al. investigated the role of NETs, histone H4, and polyP in sepsis-induced intravascular coagulation [69]. To investigate the specific role of polyP in the neutrophilic response and to determine if H4 histone-driven polyP release from platelet granules drove coagulation, they treated septic mice with a monoclonal blocking antibody against polyP. While blocking polyP did not affect the quantity of NETs or platelets in the microvasculature of the liver in septic mice, it significantly reduced the amount of thrombin cleavage activity and subsequent clotting, suggesting that polyP is a crucial factor in the platelet-NET-coagulation response. Taken with Suess et al.’s more recent results, a more informative - yet complicated - picture of polyP’s role in the innate immune responses begins to emerge, where polyP both draws neutrophils to sites of infection to commence the innate response, while also driving the coagulation and inflammatory response in the area by regulating the activity of thrombin as well as the differentiation of macrophages and fibroblasts.
Given all of this, host polyP has become a target of great interest for potential medical treatments. One target of interest is inositol hexakisphosphate kinase I (IP6K1), which is known to regulate the levels of polyP produced by platelets in mice [70]. Recently, Hou et al. reported that impairing host IP6K1 substantially reduced the production of polyP from platelets, which resulted in enhanced host bacterial killing while reducing pulmonary neutrophil accumulation, thus minimizing tissue damage induced by highly active neutrophilic responses [71]. This reduction in lung damage was observed when mice were challenged with E. coli, Staphylococcus aureus, or purified LPS. The decrease in platelet-derived polyP resulted in fewer neutrophil-platelet aggregations (NPAs), which ultimately led to the reduction of neutrophil accumulation in the alveolar tissue. They found that by using an IP6K1-specific inhibitor they could induce the reduction of NPAs in both mice and a culture of human primary neutrophils and platelets, demonstrating a clinical significance to their work. This identification of a polyP-driven immune mechanism that can be altered to enhance bacterial clearance and reduce host damage demonstrates the importance of understanding polyP on a larger scale.
In general, the data we have discussed in this section demonstrates multiple roles for polyP in host immune responses, including recruiting neutrophils, controlling fibroblast and macrophage differentiation, and altering the local microenvironmental chemistry. How this interacts with the immunomodulatory effects of polyP discussed in the previous section remains to be determined. There is clearly a delicate and complicated relationship between host and pathogen polyP usage, which has been summarized in Figure 2 (Key Figure).
Figure 2. The PolyP Wars: The Struggle Between Host and Pathogen PolyP.
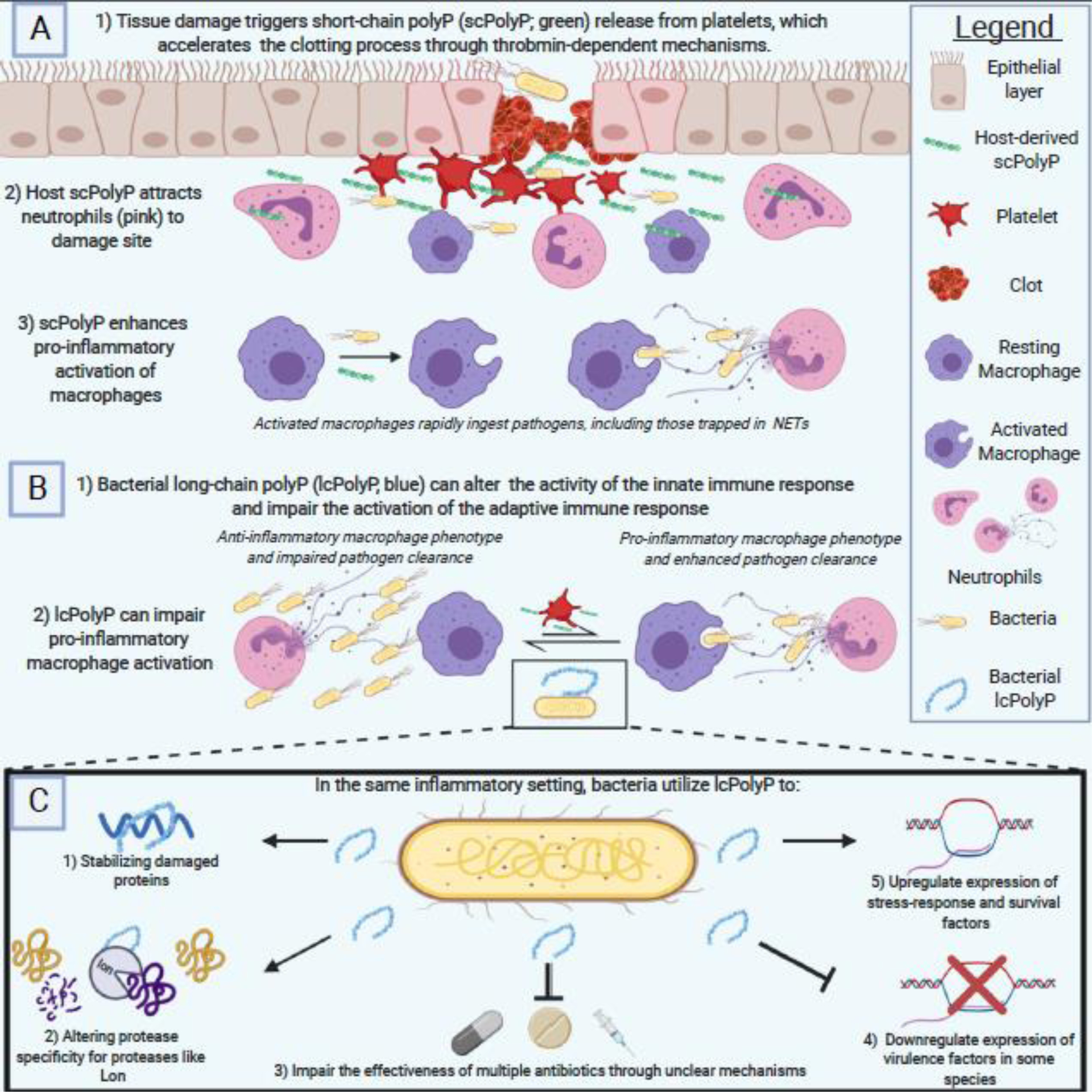
A) Host polyP utilization begins as soon as damage is detected, with platelets releasing short-chain polyP (scPolyP) to accelerate thrombin-dependent clotting and neutrophil recruitment. Host polyP also drives macrophage differentiation into the pro-inflammatory M1 phenotype that facilitates rapid phagocytosis and clearance of pathogens. B) Meanwhile, pathogenic bacteria utilize long-chain polyP (lcPolyP) to impair the host immune response, driving the anti-inflammatory M2 activation of macrophages while also impairing the expression of MHC Class II molecules to hamper the adaptive immune response. C) Internally, bacteria utilize lcPolyP for a wide variety of functions from stabilizing damaged proteins to regulating expression of crucial stress response and virulence factors. The two sides clash in an age-old contest using an ancient biomolecule as their weapon of choice. Figure created using BioRender.
Polyphosphate: Too Much of a Good Thing?
PolyP accumulation is not entirely beneficial for the host. As discussed above, it can lead to increased coagulation, heightened inflammation, and greater infiltration of neutrophils into tissue that may be damaged more by the innate response than by the bacteria that triggered it. Independent of bacterial triggers, however, polyP has been implicated in several disorders and diseases. In humans, for example, polyP has been linked to cancer-associated thrombosis (reviewed in [72]). Conversely, polyP has also been associated with beneficial results, including inhibiting metastasis and inducing apoptosis (reviewed in [73]). Indeed, polyP is often assigned counterproductive functions in the same system. For example, polyP accelerates the formation of amyloid fibrils in vitro, which are associated with a wide range of human disease such as Alzheimer’s disease, Parkinson’s disease, and dialysis-related amyloidosis [74], but polyP can also be neuroprotective, by preventing over excitement of neurons by glutamate signaling [23]. While the focus of this article is primarily on the importance of polyP in microbes and in host-microbe interaction, the literature on polyP biology is far more extensive and demonstrates the importance of approaching this ancient, universal molecule with an open mind.
The Discovery and Testing of Microbial Polyphosphate Kinase Inhibitors
The enzymology of polyP production in mammals is not well understood (although one recent paper suggests that the mitochondrial F1F0 ATPase synthesizes polyP [75]), but the well-characterized prokaryotic enzymes of microbial polyP metabolism [1] offer a unique opportunity to develop therapeutics targeting a multitude of infectious diseases. The idea that bacterial polyP metabolism could be a useful target for antimicrobial therapy is not new [76–79]. Several groups have recently reported substantial progress in this area and have identified a chemically diverse group of PPK1 inhibitors (Table 1), with exciting new data showing that bacterial polyP synthesis is targetable in vivo and that doing so may have useful anti-virulence effects. Only a small handful of studies have explored PPK2 as a target for inhibitors [77, 78, 80], and this remains an intriguing area for future work.
Table 1.
Structures and activities of PPK1 inhibitors
| Structure | Chemical ID | Target Species | In vitro PPK1 Inhibitiona | In Vivo Effects |
|---|---|---|---|---|
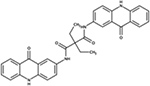
|
NSC618160 | P. aeruginosa | +++ | modestly reduced virulence in D. discoideum [81] |
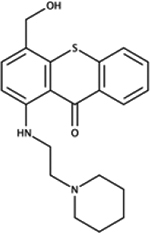
|
NSC166366 | P. aeruginosa | +++ | reduced virulence in D. discoideum [81] |
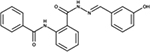
|
NSC205574 | P. aeruginosa | +++ | strongly reduced virulence in D. discoideum [81] |
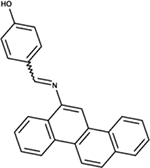
|
NSC141672 | P. aeruginosa | +++ | strongly reduced virulence in D. discoideum [81] |
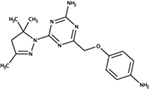
|
NSC696924 | P. aeruginosa | +++ | strongly reduced virulence in D. discoideum [81] |
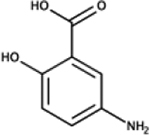
|
mesalamine (5-amino-salicylic acid) | E. coli | + | reduced polyP, mimics ppk phenotypes [87] |
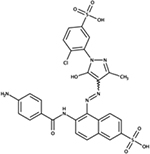
|
NSC75963 | E. coli | ND | mimics ppk phenotypes [85] |
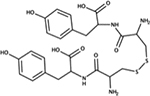
|
NSC333714 | E. coli | ND | mimics ppk phenotypes [85] |
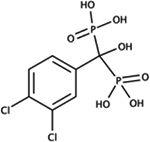
|
[(3,4‐dichlorophenyl)(hydroxy)phosphonatomethyl]phosphonate | E. coli | ++ | ND [80] |
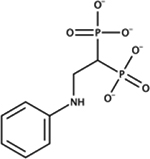
|
[2‐(phenylamino)‐1‐phosphonatoethyl]phosphonate | E. coli | ++ | ND [80] |
|
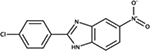
2-(4-chlorophenyl)-5-nitro-1H-benzo[d]imidazole | E. coli | + | mimics ppk phenotypes, treats UPEC infections [86] |
|
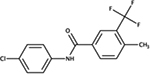
N-(4-chlorophenyl)-4-methyl-3-(trifluoromethyl)benzamide | E. coli | + | mimics ppk phenotypes, treats UPEC infections [86] |
+++ = IC50 < 10 μM, ++ = IC50 < 70 μM, + = IC50 > 100 μM, ND = not determined.
Using in silico modeling in combination with an in vivo screen of Pseudomonas aeruginosa virulence with D. discoideum as a host, the Chavez lab identified five compounds that potently inhibit PPK1 in vitro (IC50 < 10 μM) and also reduce bacterial virulence, mimicking the effect of a P. aeruginosa ppk1 knockout mutation [81], although this is complicated somewhat by the fact that D. discoideum is one of the few eukaryotes with a PPK1 homolog [82], and that polyP production by D. discoideum is involved in phagocytosis [83]. Usefully, however, the same compounds also inhibit E. coli PPK in vitro [84]. Another in silico screen for potential PPK inhibitors by the Bardaweel group identified two compounds that mimicked the effects of a ppk null mutation on both E. coli metabolism (as determined using the Biolog™ platform) and reduced total biofilm production [85], but they did not report the effect of these compounds on in vitro PPK activity. In contrast, an in vitro screen of bisphosphonic acid derivatives (a family of compounds which contains many clinically important enzyme inhibitors) by the Berlicki lab identified two compounds with IC50s for PPK1 of 50 – 60 μM, but did not test their effect on living bacteria [80]. Most recently, exciting new results from the Sun lab have identified two more PPK inhibitors, also from an initial in silico screen, that not only increase the stress sensitivity of uropathogenic E. coli (UPEC) under lab growth conditions, but also significantly reduce bacterial burden in an in vivo mouse model of UPEC infection [86]. Neither of these compounds are especially potent inhibitors of PPK activity in vitro (IC50 > 320 μM), but the fact that they are effective anti-virulence treatments in vivo is extremely encouraging. Surprisingly, none of the above studies directly measured the effect of inhibitors on bacterial polyP content, which will be an important control in future experiments.
By screening a library of FDA-approved drugs for inhibitory activity against E. coli PPK, the Jakob lab identified the front-line inflammatory bowel disease drug mesalamine (5-aminosalicylic acid) as a PPK inhibitor [87]. Although its inhibitory activity in vitro was also modest (increasing the Km of PPK for ATP by 4-fold at 1 mM mesalamine), at concentrations comparable to those used therapeutically, mesalamine significantly reduced polyP accumulation by cultures of E. coli, P. aeruginosa, and Vibrio cholerae, as well as in the gut microbiota of mesalamine-treated mice and humans. The mechanism(s) by which mesalamine reduces inflammation have been debated for many years [88], but these results may suggest that, in fact, we actually have been using bacterial polyP as a therapeutic target for quite some time. It remains to be seen, however, whether inhibiting polyP production will be a therapeutically useful strategy for dealing with other bacterial infections in humans, especially in light of the multiple roles of polyP in both host and microbe biology.
Concluding Remarks
Along with the renaissance of studies on the biology of polyP have come substantial improvements in the tools and assays for studying polyP in biological systems. These have been thoroughly reviewed recently [89], and include new and streamlined extraction and quantification techniques [90–93] as well as convenient biochemical methods for length determination and end-labeling of polyP [94, 95]. We expect that these and related technologies will be increasingly important as the community of researchers interested in polyP continues to grow.
The first description of polyP in living organisms was over a century ago [96], but our understanding of how it fits into cellular physiology has been slow in coming. There are no easy answers when studying polyP biology, but there is a bounty of questions to be explored (see Outstanding Questions), and a dynamic and growing community of researchers asking those questions. It’s an exciting time for polyP, and we are eager to see what new insights will be revealed about this ancient molecule.
Outstanding Questions Box.
What are the pathway(s) by which bacteria regulate polyP accumulation in response to different environmental stresses? How well-conserved are the regulatory mechanisms being identified in E. coli?
How does the interaction of polyP and Lon protease change the global proteome of bacteria under different conditions? How does this differ between species?
Is the anti-virulence effect of polyP unique to Francisella, or do other bacteria also use similar mechanisms?
What are the immunologically-relevant levels of short- and long-chain polyP in an active infection site? What is the balance between host and bacterial polyP during infection?
What receptors do eukaryotic cells, including neutrophils, use to sense and respond to polyP? Can they distinguish between short- and long-chain polyP, and if so, how?
How and under what circumstances do bacteria release polyP? Do they actively secrete polyP or is it released by cell lysis? Does this vary from species to species?
Can PPK inhibitors or other polyP-manipulating drugs be used to treat infections or modulate immune responses in a clinically useful way?
Highlights.
PolyP plays multiple roles in bacterial regulation, including controlling proteolysis by the Lon protease and regulating virulence factors, both positively and negatively
PolyP plays a major role in host repair by facilitating thrombin-driven coagulation and in defense mechanisms by recruiting neutrophils and driving M1 macrophage differentiation
Bacteria use polyP to manipulate host immune responses, impairing phagocytic cell functions, modulating inflammatory responses, and impairing antigen presentation
Dictyostelium discoideum is emerging as a powerful model system to investigate polyP biology
Multiple drugs influencing polyP levels in different organisms have recently been discovered
Glossary
- CCL2
C-C Motif Chemokine Ligand 2. One of several chemokine receptors utilized by monocytes and basophils to detect and direct migration towards areas with high C-C chemokine receptor 2 (CCR2) concentration.
- CD80
An immunoglobin expressed on antigen presenting cells that binds to a T cell’s CD28 receptor to provide essential costimulatory signals for activation. Closely related to CD86.
- CD86
An immunoglobin expressed on antigen presenting cells that binds to a T cell’s CD28 receptor to provide essential costimulatory signals for activation. Closely related to CD80.
- CXCL10
A pro-inflammatory cytokine that binds to CXCR3 on monocytes, Natural Killer cells, and T cells to stimulate pleiotropic effects related to antimicrobial activity.
- DksA
RNA polymerase-binding transcription factor involved in bacterial stringent response
- F1F0 ATPase
protein complex responsible for ATP synthesis in mitochondria
- FPI
Francisella Pathogenicity Island, genetic locus encoding multiple factors necessary for F. tularensis virulence
- histone H4
one of the five histones involved in DNA packaging. Its presence outside of the host cell triggers immune activity as it should only be inside healthy cells.
- homolog
a species-specific version of a gene or protein that is found among multiple species that share a common ancestor.
- IC50
concentration of an inhibitor which halves activity of the target
- INFβ
antiviral chemokine secreted by many immune cells. It stimulates macrophages and natural killer cells.
- iNOS
inducible Nitric Oxide Synthase; produces nitric oxide which acts to regulate the host immune response
- IP6K1
Canonically converts inositol hexakisphosphate to diphosphoinositol pentakisphosphate but has recently been shown to be involved in the production or regulation of mammalian polyP.
- Km
Michaelis-Menten constant; the concentration of substrate at which an enzyme acts at half its maximal velocity
- Lon
major bacterial protease involved in protein turnover and regulation
- LPS
lipopolysaccharide; strongly immuno-stimulatory outer membrane lipid of Gram-negative bacteria
- MHC
major histocompatibility complex; utilized by cells to present antigen fragments to T and B cells
- PBMC
the portion of blood cells containing the mononuclear lineages, which include the lymphocytes (T cells, B cells, NK cells) and monocytes.
- PhoB
bacterial transcription factor that positively regulates phosphate uptake
- PhoU
negative regulator of phosphate uptake in bacteria
- (p)ppGpp
guanosine penta- and tetraphosphate; second messengers that are global regulators of starvation stress response
- PPK1
or PPK, family of polyphosphate kinases, synthesizes polyP from ATP
- PPK2
family of polyphosphate kinases, synthesizes polyP from NTPs
- PPX
exopolyphosphatase; breaks down polyP to orthophosphate
- RelA
(p)ppGpp synthase
- RpoE
global regulator of bacterial cell envelope stress responses
- RpoN
global regulator of bacterial nitrogen starvation stress response
- RpoS
global regulator of bacterial general stress response
- SpoT
(p)ppGpp synthase/hydrolase
- stringent response
bacterial starvation stress response mediated by (p)ppGpp and DksA
- Western blot
technique for detecting and quantifying proteins using specific antibodies
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Achbergerova L and Nahalka J (2011) Polyphosphate--an ancient energy source and active metabolic regulator. Microb Cell Fact 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Candon HL et al. (2007) Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J Bacteriol 189 (22), 8099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha SB et al. (2012) Generation and envelope protein analysis of internalization defective Brucella abortus mutants in professional phagocytes, RAW 264.7. FEMS Immunol Med Microbiol 64 (2), 244–54. [DOI] [PubMed] [Google Scholar]
- 4.Kim KS et al. (2002) Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc Natl Acad Sci U S A 99 (11), 7675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornberg A et al. (1999) Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68, 89–125. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa N et al. (2000) Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J Bacteriol 182 (23), 6687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz-Severin J et al. (2015) Multiple antibiotic susceptibility of polyphosphate kinase mutants (ppk1 and ppk2) from Pseudomonas aeruginosa PAO1 as revealed by global phenotypic analysis. Biol Res 48, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng L et al. (2012) Polyphosphate kinase 1 is required for the pathogenesis process of meningitic Escherichia coli K1 (RS218). Future Microbiol 7 (3), 411–23. [DOI] [PubMed] [Google Scholar]
- 9.Rao NN et al. (2009) Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78, 605–47. [DOI] [PubMed] [Google Scholar]
- 10.Rao NN and Kornberg A (1996) Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol 178 (5), 1394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashid MH et al. (2000) Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182 (1), 225–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tunpiboonsak S et al. (2010) Role of a Burkholderia pseudomallei polyphosphate kinase in an oxidative stress response, motilities, and biofilm formation. J Microbiol 48 (1), 63–70. [DOI] [PubMed] [Google Scholar]
- 13.Jenal U and Hengge-Aronis R (2003) Regulation by proteolysis in bacterial cells. Curr Opin Microbiol 6 (2), 163–72. [DOI] [PubMed] [Google Scholar]
- 14.Zygmunt MS et al. (2006) Identification of Brucella melitensis 16M genes required for bacterial survival in the caprine host. Microbes Infect 8 (14–15), 2849–54. [DOI] [PubMed] [Google Scholar]
- 15.Schulz-Vogt HN et al. (2019) Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J 13 (5), 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai YC et al. (2017) Polyphosphate metabolism by purple non-sulfur bacteria and its possible application on photo-microbial fuel cell. J Biosci Bioeng 123 (6), 722–730. [DOI] [PubMed] [Google Scholar]
- 17.Trilisenko L et al. (2019) The Reduced Level of Inorganic Polyphosphate Mobilizes Antioxidant and Manganese-Resistance Systems in Saccharomyces cerevisiae. Cells 8 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travers RJ et al. (2015) Polyphosphate, platelets, and coagulation. Int J Lab Hematol 37 Suppl 1, 31–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrissey JH et al. (2012) Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood 119 (25), 5972–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puy C et al. (2016) Platelet-Derived Short-Chain Polyphosphates Enhance the Inactivation of Tissue Factor Pathway Inhibitor by Activated Coagulation Factor XI. PLoS One 11 (10), e0165172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SA et al. (2010) Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood 116 (20), 4353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suess PM et al. (2019) Extracellular Polyphosphate Promotes Macrophage and Fibrocyte Differentiation, Inhibits Leukocyte Proliferation, and Acts as a Chemotactic Agent for Neutrophils. J Immunol 203 (2), 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiolino M et al. (2019) Inorganic Polyphosphate Regulates AMPA and NMDA Receptors and Protects Against Glutamate Excitotoxicity via Activation of P2Y Receptors. J Neurosci 39 (31), 6038–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paula FS et al. (2019) The potential for polyphosphate metabolism in Archaea and anaerobic polyphosphate formation in Methanosarcina mazei. Sci Rep 9 (1), 17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recalde A et al. (2021) The Role of Polyphosphate in Motility, Adhesion, and Biofilm Formation in Sulfolobales. Microorganisms 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H et al. (2002) A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc Natl Acad Sci U S A 99 (26), 16678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn K and Kornberg A (1990) Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem 265 (20), 11734–9. [PubMed] [Google Scholar]
- 28.Albi T and Serrano A (2016) Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J Microbiol Biotechnol 32 (2), 27. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda A and Kornberg A (1997) Polyphosphate kinase as a nucleoside diphosphate kinase in Escherichia coli and Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 94 (2), 439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda A et al. (1999) Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc Natl Acad Sci U S A 96 (25), 14264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno SN and Docampo R (2013) Polyphosphate and its diverse functions in host cells and pathogens. PLoS Pathog 9 (5), e1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid MH and Kornberg A (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97 (9), 4885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rashid MH et al. (2000) Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97 (17), 9636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roewe J et al. (2020) Bacterial polyphosphates interfere with the innate host defense to infection. Nat Commun 11 (1), 4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srisanga K et al. (2019) Polyphosphate kinase 1 of Burkholderia pseudomallei controls quorum sensing, RpoS and host cell invasion. J Proteomics 194, 14–24. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari P et al. (2019) Inorganic polyphosphate accumulation suppresses the dormancy response and virulence in Mycobacterium tuberculosis. J Biol Chem 294 (28), 10819–10832. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ault-Riche D et al. (1998) Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol 180 (7), 1841–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuroda A et al. (1997) Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem 272 (34), 21240–3. [DOI] [PubMed] [Google Scholar]
- 39.Gourse RL et al. (2018) Transcriptional Responses to ppGpp and DksA. Annu Rev Microbiol 72, 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao NN et al. (1998) Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol 180 (8), 2186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Melderen L and Wood TK (2017) Commentary: What Is the Link between Stringent Response, Endoribonuclease Encoding Type II Toxin-Antitoxin Systems and Persistence? Front Microbiol 8, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray MJ (2019) Inorganic Polyphosphate Accumulation in Escherichia coli Is Regulated by DksA but Not by (p)ppGpp. J Bacteriol 201 (9), e00664–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray MJ (2020) Interactions between DksA and stress-responsive alternative sigma factors control inorganic polyphosphate accumulation in Escherichia coli. J Bacteriol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munevar NF et al. (2017) Differential regulation of polyphosphate genes in Pseudomonas aeruginosa. Mol Genet Genomics 292 (1), 105–116. [DOI] [PubMed] [Google Scholar]
- 45.Sanyal S et al. (2013) Polyphosphate kinase 1, a central node in the stress response network of Mycobacterium tuberculosis, connects the two-component systems MprAB and SenX3-RegX3 and the extracytoplasmic function sigma factor, sigma E. Microbiology (Reading) 159 (Pt 10), 2074–2086. [DOI] [PubMed] [Google Scholar]
- 46.Rudat AK et al. (2018) Mutations in Escherichia coli Polyphosphate Kinase That Lead to Dramatically Increased In Vivo Polyphosphate Levels. J Bacteriol 200 (6), e00697–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morohoshi T et al. (2002) Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl Environ Microbiol 68 (8), 4107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Almeida LG et al. (2015) phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl Environ Microbiol 81 (9), 3006–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grillo-Puertas M et al. (2016) PhoB activation in non-limiting phosphate condition by the maintenance of high polyphosphate levels in the stationary phase inhibits biofilm formation in Escherichia coli. Microbiology (Reading) 162 (6), 1000–1008. [DOI] [PubMed] [Google Scholar]
- 50.Cho Y et al. (2015) Individual and collective contributions of chaperoning and degradation to protein homeostasis in E. coli. Cell Rep 11 (2), 321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuroda A (2006) A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci Biotechnol Biochem 70 (2), 325–31. [DOI] [PubMed] [Google Scholar]
- 52.Kuroda A et al. (2001) Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293 (5530), 705–8. [DOI] [PubMed] [Google Scholar]
- 53.Nomura K et al. (2004) Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J Biol Chem 279 (33), 34406–10. [DOI] [PubMed] [Google Scholar]
- 54.Gross MH and Konieczny I (2020) Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli. Nucleic Acids Res 48 (10), 5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurokawa K et al. (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J 18 (23), 6642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speck C et al. (1999) ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J 18 (21), 6169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osbourne DO et al. (2014) Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered 5 (4), 264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J et al. (2019) Lon recognition of the replication initiator DnaA requires a bipartite degron. Mol Microbiol 111 (1), 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varas MA et al. (2018) Inorganic Polyphosphate Is Essential for Salmonella Typhimurium Virulence and Survival in Dictyostelium discoideum. Front Cell Infect Microbiol 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bozzaro S and Eichinger L (2011) The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets 12 (7), 942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loomis WF (1996) Genetic networks that regulate development in Dictyostelium cells. Microbiol Rev 60 (1), 135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sillo A et al. (2011) Salmonella typhimurium is pathogenic for Dictyostelium cells and subverts the starvation response. Cell Microbiol 13 (11), 1793–811. [DOI] [PubMed] [Google Scholar]
- 63.Tang-Fichaux M et al. (2020) The Polyphosphate Kinase of Escherichia coli Is Required for Full Production of the Genotoxin Colibactin. mSphere 5 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohlfing AE et al. (2018) Polyphosphate Kinase Antagonizes Virulence Gene Expression in Francisella tularensis. J Bacteriol 200 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rijal R et al. (2020) Polyphosphate is an extracellular signal that can facilitate bacterial survival in eukaryotic cells. Proceedings of the National Academy of Sciences, 202012009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neilands J and Kinnby B (2020) Porphyromonas gingivalis initiates coagulation and secretes polyphosphates - A mechanism for sustaining chronic inflammation? Microb Pathog, 104648. [DOI] [PubMed] [Google Scholar]
- 67.Noegel A and Gotschlich EC (1983) Isolation of a high molecular weight polyphosphate from Neisseria gonorrhoeae. J Exp Med 157 (6), 2049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brinkmann V et al. (2004) Neutrophil extracellular traps kill bacteria. Science 303 (5663), 1532–5. [DOI] [PubMed] [Google Scholar]
- 69.McDonald B et al. (2017) Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood 129 (10), 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh S et al. (2013) Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 122 (8), 1478–86. [DOI] [PubMed] [Google Scholar]
- 71.Hou Q et al. (2018) Inhibition of IP6K1 suppresses neutrophil-mediated pulmonary damage in bacterial pneumonia. Sci Transl Med 10 (435). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Almeida VH et al. (2019) Novel Aspects of Extracellular Vesicles as Mediators of Cancer-Associated Thrombosis. Cells 8 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kulakovskaya EV et al. (2018) Inorganic Polyphosphate and Cancer. Biochemistry (Mosc) 83 (8), 961–968. [DOI] [PubMed] [Google Scholar]
- 74.Zhang CM et al. (2019) Possible mechanisms of polyphosphate-induced amyloid fibril formation of beta2-microglobulin. Proc Natl Acad Sci U S A 116 (26), 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baev AY et al. (2020) Inorganic polyphosphate is produced and hydrolyzed in F0F1-ATP synthase of mammalian mitochondria. Biochem J 477 (8), 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kornberg A, Novel Antimicrobial Therapies, KORNBERG ARTHUR, US, 2002. [Google Scholar]
- 77.Shum KT et al. (2011) Aptamer-mediated inhibition of Mycobacterium tuberculosis polyphosphate kinase 2. Biochemistry 50 (15), 3261–71. [DOI] [PubMed] [Google Scholar]
- 78.Singh M et al. (2016) Establishing Virulence Associated Polyphosphate Kinase 2 as a drug target for Mycobacterium tuberculosis. Sci Rep 6, 26900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gautam LK et al. (2019) Bacterial Polyphosphate Kinases Revisited: Role in Pathogenesis and Therapeutic Potential. Curr Drug Targets 20 (3), 292–301. [DOI] [PubMed] [Google Scholar]
- 80.Burda-Grabowska M et al. (2019) Bisphosphonic acids and related compounds as inhibitors of nucleotide- and polyphosphate-processing enzymes: A PPK1 and PPK2 case study. Chem Biol Drug Des 93 (6), 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bravo-Toncio C et al. (2016) Dictyostelium discoideum as a surrogate host-microbe model for antivirulence screening in Pseudomonas aeruginosa PAO1. Int J Antimicrob Agents 47 (5), 403–9. [DOI] [PubMed] [Google Scholar]
- 82.Gomez-Garcia MR and Kornberg A (2004) Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc Natl Acad Sci U S A 101 (45), 15876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H et al. (2005) Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc Natl Acad Sci U S A 102 (8), 2731–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campos F et al. (2019) Fluorescence enzymatic assay for bacterial polyphosphate kinase 1 (PPK1) as a platform for screening antivirulence molecules. Infect Drug Resist 12, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bashatwah RM et al. (2018) Discovery of potent polyphosphate kinase 1 (PPK1) inhibitors using structure-based exploration of PPK1Pharmacophoric space coupled with docking analyses. J Mol Recognit 31 (10), e2726. [DOI] [PubMed] [Google Scholar]
- 86.Peng L et al. (2020) Discovery and antibacterial study of potential PPK1 inhibitors against uropathogenic E. coli. J Enzyme Inhib Med Chem 35 (1), 1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dahl JU et al. (2017) The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol 2, 16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hauso O et al. (2015) 5-Aminosalicylic acid, a specific drug for ulcerative colitis. Scand J Gastroenterol 50 (8), 933–41. [DOI] [PubMed] [Google Scholar]
- 89.Christ JJ et al. (2020) Methods for the Analysis of Polyphosphate in the Life Sciences. Anal Chem 92 (6), 4167–4176. [DOI] [PubMed] [Google Scholar]
- 90.Christ JJ and Blank LM (2018) Enzymatic quantification and length determination of polyphosphate down to a chain length of two. Anal Biochem 548, 82–90. [DOI] [PubMed] [Google Scholar]
- 91.Christ JJ and Blank LM (2018) Analytical polyphosphate extraction from Saccharomyces cerevisiae. Anal Biochem 563, 71–78. [DOI] [PubMed] [Google Scholar]
- 92.Pokhrel A et al. (2019) Assaying for Inorganic Polyphosphate in Bacteria. J Vis Exp (143). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dahl J-U et al. (2018) Extraction and Quantification of Polyphosphate (polyP) from Gram-negative Bacteria. Bioprotocol 8 (18), e3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith SA et al. (2018) DNA ladders can be used to size polyphosphate resolved by polyacrylamide gel electrophoresis. Electrophoresis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baker CJ et al. (2020) Diversification of polyphosphate end-labeling via bridging molecules. PLoS One 15 (8), e0237849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meyer A (1904) Orientierende Untersuchungen ueber Verbreitung, Morphologie, und Chemie des Volutins. Bot Zeit. 62, 113–152. [Google Scholar]


