Abstract
Nanoscale coordination polymers are promising vehicles for anticancer drug delivery because their surface composition and particle size can be tuned to exploit the enhanced permeability and retention effect, and their reversible interaction with metal cations enables triggered drug release at the tumor site. Here we develop a novel nanoscale coordination polymer using the diblock copolymer poly(2-methacryloyloxyethyl phosphorylcholine)-block-poly(serinyl acrylate) (PMPC-b-PserA) and demonstrate its use for encapsulation of a hydrophobic drug and triggered drug release to induce breast cancer cell apoptosis in vitro. The zwitterionic PMPC block was inspired by the antifouling structure of cell membranes, and the PserA block by the amphoteric amino acids of proteins. The polymer was synthesized by reversible addition-fragmentation chain transfer polymerization, and a mixture of the polymer and FeCl3 self-assembled into nanoparticles via complexation of Fe3+ with PserA, with the hydrophilic PMPC block at the particle surface. At a 3:1 molar ratio of Fe3+ to serA, the hydrodynamic diameter of the particles was 22.2 nm. Curcumin, a natural water-insoluble polyphenol used to enhance the effects of chemotherapeutics, was encapsulated in the particles as an oil-in-water emulsion, with an encapsulation efficiency of 99.6% and a particle loading capacity of 32%. Triggered release of curcumin was achieved by adding deferoxamine, an FDA-approved Fe3+ chelating agent; curcumin release efficiency increased at higher deferoxamine concentrations and lower pH. Triggered release of curcumin induced apoptosis in human triple-negative breast cancer cells; cell viability decreased to 34.3% after 24 h of treatment with the curcumin-loaded nanoparticles and deferoxamine, versus >80% viability without deferoxamine to trigger drug release. The biocompatibility, tunable composition and size, high hydrophobic drug loading, and triggered release capability of this nanoscale coordination polymer make it well-suited for use in anticancer drug delivery.
Keywords: nanoscale coordination polymer, diblock zwitterionic copolymer, polymeric colloid, anticancer drug delivery, triggered release
Graphical Abstract
INTRODUCTION
Current drug delivery systems are often limited by poor drug bioavailability,1 lack of tissue selectivity,2 and uncontrolled drug release.3 Efforts to improve drug delivery are focused on new delivery vehicles,4 new targeting strategies,5 and new routes of administration.6 Nanoparticles such as polymeric micelles,7, 8 liposomes,9, 10 dendrimers,11, 12 and hydrogels13, 14 have emerged as promising drug delivery vehicles15 that enhance therapeutic efficacy by prolonging drug circulation,16 preventing side effects,17 improving bioavailability,18 and enabling tissue targeting.19, 20 Nanoscale coordination polymers (NCPs) are polymers that contain metal cation centers linked by ligands in the polymer and can undergo self-assembly into nanoparticles due to this coordination.21, 22 NCPs are advantageous for drug delivery due to their ease of formulation, tunable particle size and surface composition, and triggered drug release.23–25 Drug delivery via NCPs uses passive targeting such as the enhanced permeability and retention effect to target tumors: tumor tissues are disorganized and are dilated with pores, resulting in greater accumulation of nanoparticles in tumor tissues than in other tissues.26, 27 Stimuli-responsive drug release is achieved by destabilizing the metal-ligand interactions in the NCP particle core.28, 29
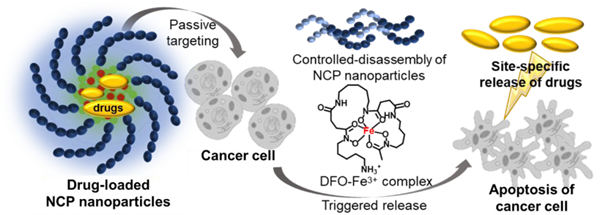
Here we present a novel NCP that forms a stable colloidal nanoparticle with tunable particle size, high hydrophobic drug loading, and triggered drug release, and demonstrate its use for triggered release of curcumin to induce apoptosis in triple-negative breast cancer cells in vitro. NCP nanoparticles were produced by coordination of Fe3+ and the diblock copolymer poly(2-methacryloyloxyethyl phosphorylcholine)-block-poly(serinyl acrylate) (PMPC-b-PserA). Serinyl acrylate (serA) is an amphoteric L-serine-based monomer that has metal-coordination capacity.30, 31 The PMPC block is a zwitterionic polymer that extends drug circulation time and reduces drug toxicity via its hydrophilicity.32, 33 PMPC-b-PserA was synthesized by reversible addition fragmentation chain transfer (RAFT) polymerization, which facilitates controls of molecular weight, composition, polydispersity, and block architecture.34 The resulting NCP nanoparticles possess a PMPC shell, are stable in solution, and can encapsulate drug molecules in the hydrophobic core. Drug release is triggered by an acidic pH environment (as in tumor tissues) and by addition of a chelating agent. We used the hydrophobic anticancer drug curcumin and MDA-MB-231, a triple-negative breast cancer cell line, as a model system to demonstrate the high loading of a hydrophobic drug, lack of cytotoxicity in the absence of curcumin, and triggered drug release by the NCP nanoparticles (Scheme 1).35, 36 Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione), the principal curcuminoid of turmeric, induces apoptosis in breast cancer cell lines37, 38 and has been used to enhance the effects of chemotherapy in cancer treatment.39 Curcumin release was triggered by reducing the solution pH and by adding deferoxamine (DFO), an FDA-approved chelating agent.40
Scheme 1.
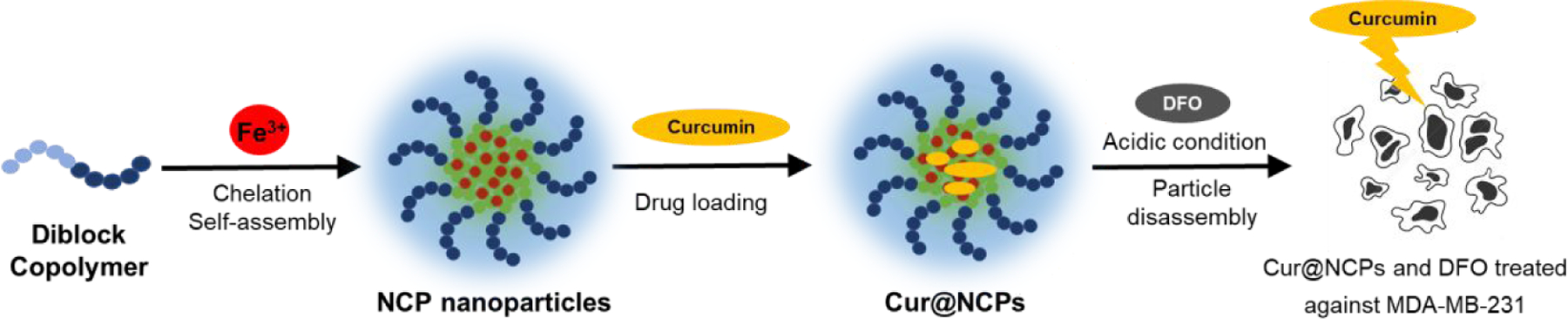
Schematic of the synthesis of curcumin-loaded nanoscale coordination polymers (Cur@NCPs) and their disassembly and release of curcumin, which induces the apoptosis of breast cancer cells (MDA-MB-231). The diblock copolymer PMPC-b-PserA self-assembles into NCP nanoparticles via complexation of Fe3+ and PserA. Cur@NCPs are produced by partitioning of curcumin into the hydrophobic particle core via oil-in-water emulsion. Addition of the chelating agent deferoxamine mesylate (DFO) in an acidic solution causes disassembly of the Cur@NCPs, curcumin release, and cell apoptosis.
MATERIALS AND METHODS
Materials
2-methacryloyloxyethyl phosphorylcholine (MPC, 97%), L-serine (≥99.5%), sodium phosphotungstate hydrate, sodium carbonate (≥99.5%), TWEEN 80, 4-cyano-4-(phenylcarbonothioylthio) pentanoic acid (CPD, 97%), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldric. HPLC-grade tetrahydrofuran and chloroform, acetone, deuterium oxide, and deferoxamine mesylate (DFO, 95%) were purchased from Acros Organics. 4,4’-azobis (4-cyanovaleric acid) (ACVA, 98%), curcumin (95%), acryloyl chloride (55%), and iron (III) chloride hexahydrate were purchased from Alfa Aesar. Nanosep centrifugal devices with Omega membrane (3K MWCO) were purchased from Pall Corporation. Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS), and (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) (MTT) assay kits were purchased from Thermo Fisher Scientific. Dialysis membranes (8K MWCO) were purchased from Spectrum Laboratories, Inc. Copper mesh was purchased from Ted Pella. Copper sulfate was purchased from Showa Co., Ltd. MDA-MB-231 breast cancer cells were purchased from the Bioresource Collection and Research Center in Taiwan. Deionized water with a minimum resistivity of 18.0 MΩ·m was used in experiments.
Preparation of serA monomer
L-serine (10.0 g, 95.2 mmol) was dissolved in deionized water (100 mL) at 90 °C, and basic cupric carbonate (11.8 g, 95.5 mmol) was added to the solution. The mixture was stirred for 10 min followed by filtering the insoluble residue. 60 mL of deionized water and 20 mL of acetone were added to wash the residue with the solution maintained in an ice bath. Acryloyl chloride (9.6 mL, 118.8 mmol) dissolved in 30 mL acetone was added dropwise to the solution. The pH of the mixture was maintained at 9.5 by simultaneously adding 2.0 M KOH(aq) and acryloyl chloride. The solution was removed from the ice bath and was stirred at room temperature for 12 h. The precipitated complex was removed by filtration and was washed with deionized water, methanol, and diethyl ether in series. The solid was dispersed in deionized water (200 mL) with addition of 8-quinolinol (32.0 g, 220.4 mmole) and chloroform solution (150 mL). The solution was stirred at room temperature for 12 h, and green precipitate in the chloroform phase was removed by filtration. To remove traces of 8-hydroxyquinoline, three extractions with chloroform through a separatory funnel were performed. The water layer was concentrated, and the white solid L-serinyl acrylate monomer (serA) was precipitated from tetrahydrofuran. The serA yield was 56%.
Synthesis of PMPC
Homopolymer PMPC was synthesized by RAFT polymerization with CPD as chain transfer agent, and ACVA as initiator. MPC (1 g, 3.4 mmole), CPD (18.9 mg, 0.068 mmole), and ACVA (3.8 mg, 0.0136 mmole) were dissolved in deionized water (1.411 mL) with methanol (1.976 mL) in a round-bottom flask. After purging with argon for 30 min to remove oxygen, the flask was heated at 70 °C for 3.5 h. The mixture was then quenched by liquid nitrogen to terminate the reaction, following by dialyzing against water with cellulose tubular membranes for 8 h with periodic bath changes to remove unreacted monomers. Pink solid PMPC was obtained after freeze drying, and served as a macro-chain transfer agent in the subsequent synthesis process. The PMPC yield was 76%.
Synthesis of PMPC-b-PserA
The diblock copolymer PMPC-b-PserA was synthesized by extending the macro-chain transfer agent, PMPC, by RAFT polymerization. Polymerization was performed in water with ACVA as the initiator. The reaction mixture was purged with argon for 30 min and was heated in a stirred oil bath at 70 °C. After reacting for 16 h, the mixture was immediately quenched with liquid nitrogen. To remove unreacted monomer, the solution was dialyzed against water using cellulose tubular membranes for 8 h with periodic bath changes. The solution was then freeze-dried to obtain PMPC-b-PserA. The PMPC-b-PserA yield was 68.4%.
Preparation of NCP nanoparticles
NCP nanoparticles were prepared by chelation of PserA and Fe3+ and subsequent self-assembly. The copolymer was dissolved in deionized water at 50 mg/mL, and FeCl3 was dissolved in deionized water at 0.7 M. The polymer, iron salt solution, and additional water were purged with argon for 30 min to remove oxygen. For 250 µL of synthesis, polymer solution, 14.3 µL of FeCl3, and 186.7 µL of additional water was mixed. The molar ratio of Fe3+ to serA was varied from 3 to 12 with a fixed polymer concentration of 10 mg/mL to identify the formulation of the most stable NCP nanoparticles. After mixing PMPC-b-PserA and FeCl3, the solution was incubated in a shaker at room temperature overnight to allow chelation and NCP nanoparticles formation. After NCP nanoparticles were formed, 10 µL of Na2CO3 (0.5M) was added to the mixture to induce precipitation of Fe-oxyhydroxides,41 and the mixture was filtered to remove the precipitate. Diluted NCP solution filtered through a 0.22-µm membrane filter was analyzed by UV-vis spectroscopy to confirm chelation of polymer and Fe3+, and the particle size was evaluated by DLS. Freeze-dried NCP nanoparticles were analyzed by FT-IR spectroscopy. NCPs were stained with sodium phosphotungstate hydrate and were freeze-dried before TEM imaging.42
Disassembly of NCP nanoparticles
NCP nanoparticle disassembly is critical for their use in triggered drug release. Disassembly of the synthesized NCP nanoparticles was quantified based on the chelated DFO-Fe3+ concentration at various DFO concentrations and at different pH values. DFO (40 mg/mL) was dissolved in water to prepare a stock solution. DFO stock solution (0, 3.28, 6.56 µL) was added to 10 µL of NCP solution and phosphate buffer at pH 5.5, 6.5, 7.4 to obtain a total volume of 50 µL. The mixtures were incubated and allowed to react in Nanosep centrifugal devices for 20 min. After the reaction, the mixtures were centrifuged at 9,000 rpm for 10 min followed by addition of 100 µL phosphate buffer and were centrifuged again three times to ensure that most of the chelated DFO-Fe3+ was separated. The DFO-Fe3+ concentration was measured using UV-vis spectroscopy (absorbance at 430 nm) based on a calibration curve (Figure S1A).
Characterization of polymers and nanoparticles
1H NMR spectra were recorded in D2O at 600 MHz on a Bruker NMR spectrometer to analyze polymer structures. Polymer molecular weight was analyzed by gel permeation chromatography (GPC). GPC measurements were performed using 0.15 M NaCl as the mobile phase, and polyethylene oxide calibration standards, operating at 0.6 mL/min at 40 °C with an A3000-single-pore GPC/SEC column (300 × 8 mm) equipped with a Viscotek RI detector (VE3580, Malvern). Attenuated total reflection-Fourier transform infrared (ATR FT-IR) analysis was performed with an IRSpirit spectrophotometer (Shimadzu, Japan) with Miracle Single Reflection ATR with the KBr pellet technique in a range from 3800–600 cm-1. UV-vis absorption spectra were recorded using a UV-1900 spectrophotometer (Shimadzu, Japan). Particle size was measured using DLS with a Malvern Zetasizer Nano-ZS instrument with a scattering angle of 90° at 25 °C. TEM was performed using a JEM-2000FXII TEM at a magnification of 10,000X and an accelerating voltage of 200 kV.
Preparation of curcumin-loaded NCP nanoparticles
Curcumin-loaded NCP nanoparticles were prepared using an oil-in-water emulsion method.43 The optimal formulation for emulsification was identified by varying the volumetric ratio of the dispersed phase (curcumin) and continuous phase (NCP nanoparticle solution).44 Curcumin stock solution was prepared by dissolving curcumin in CHCl3 at 4 mg/mL. Different amounts (100 µL, 125 µL, 150 µL, 175 µL, 200 µL) of the curcumin stock solution were added dropwise into a mixture of 200 µL NCP nanoparticle solution and 800 µL of deionized water under magnetic stirring at 300 rpm in the dark. The mixture was incubated and agitated overnight in an open atmosphere to allow CHCl3 to evaporate. The products were freeze-dried and the loading capacity and encapsulation efficiency were determined by adding 1 mL of acetone to the solid to dissolve unentrapped curcumin. (Free curcumin is highly soluble in acetone, while PMPC-modified NCPs are insoluble.) The dissolved curcumin was quantified by UV-vis spectroscopy at 420 nm using a calibration curve for curcumin in acetone (Figure S1B). The encapsulation efficiency and loading capacity were calculated as follows:
| (1) |
| (2) |
where C0 is the initial curcumin concentration (mg/mL), Cx is the unentrapped curcumin concentration (mg/mL), and Cp is the polymer concentration (mg/mL).
Precipitated curcumin-loaded NCP nanoparticles (Cur@NCPs) were purified by repetitive extraction with acetone. Purified Cur@NCPs were freeze-dried for further experiments. Cur@NCPs dissolved in water were characterized by UV-vis spectroscopy. For TEM imaging, Cur@NCPs were stained with sodium phosphotungstate hydrate and freeze-dried.
Triggered release of curcumin
Cur@NCPs and DFO were dissolved separately in deionized water at 10 mg/mL and 1 mg/mL, respectively. Based on a previous study,45 the ratio of DFO to incorporated Fe3+ was calculated to be 1 at a final Cur@NCPs concentration of 360 µg/mL. The buffer pH was adjusted to pH 5.5, 6.5, and 7.4. Cur@NCPs solution (360 µL), DFO solution (69 µL), and buffer (571 µL) were mixed for a final solution volume of 1 mL. The reactions were allowed to proceed for 30 min, and the products were measured by UV-vis spectroscopy. After the mixtures were freeze-dried, the powder was rinsed with 1.5 mL acetone to quantify the released curcumin from Cur@NCPs, since the polymer and chelated DFO are both insoluble in acetone. Dissolved curcumin was quantified by UV-vis spectroscopy at 420 nm based on a curcumin calibration curve (Figure S1B). A curcumin release profile was obtained by dialysis and was quantified using a curcumin calibration curve in 0.5% Tween 80 (Figure S2A-B).46
Cell culture
Triple-negative breast cancer cells (MDA-MB-231) were cultured in 10-mm diameter cell culture dishes in DMEM supplemented with 10% FBS, 4 mM L-glutamine, and 1 mM sodium pyruvate at 37°C and 5% CO2 for 2–3 days before viability experiments. The cells were subcultured every 2–3 days.
MTT assay
An MTT assay was used to analyze the effect of triggered curcumin release on MDA-MB-231 cell viability. MDA-MB-231 cells were plated in a 96-well plate at 6,000 cells per well and were incubated for 24 h. Various concentrations of curcumin were prepared with 0.1% DMSO solvent as a negative control; Cur@NCPs were dissolved at 10 mg/mL in DMEM without FBS. All samples were filtered with a 0.22-µm membrane filter to ensure sterilization. The growth medium was removed, and the cells were exposed to various concentrations of curcumin (0–100 µg/mL) and Cur@NCPs (0, 90, 150, 270, 360 µg/mL) with and without subsequent addition of DFO (estimated molar ratio of DFO to incorporated Fe3+: 1 and 1.5, respectively). After 24 h of co-culturing, the solution was removed and MTT (5 mg/mL in DMEM) was added and the samples were incubated at 37 °C for 3 h. The growth medium was removed and 100 µL of DMSO was added to each well to dissolve formazan crystals. Cell viability was measured using an ELISA reader (Synergy 2, BioTek, USA) based on optical density at 540 nm, and the cell viability was calculated with the following formula:
| (3) |
with the O.D.(s) =O.D. value of samples, O.D.(c) =O.D. value of the negative control group, and O.D.(b) =O.D. value of the blank. All samples are tested triplicate.
RESULTS AND DISCUSSION
Synthesis and characterization of PMPC-b-PserA and NCP nanoparticles
The diblock copolymer PMPC-b-PserA was synthesized using RAFT polymerization as illustrated in Scheme 2. First, PMPC was synthesized using CPD as the chain transfer agent and ACVA as the initiator, then PMPC-b-PserA was synthesized by chain-extending PMPC with serA. Analysis of polymer composition by 1H NMR (Figure S3) indicated that the degree of polymerization for MPC and serA were 55 and 25, respectively. Polymer molecular weight (Mn) and polydispersity indices (Đ) were characterized using GPC (Figure S4); the PMPC Mn was 8,312 Da with Đ=1.17, and the PMPC-b-PserA Mn was 13,081 Da with Đ=1.21, consistent with the 1H NMR results.
Scheme 2.
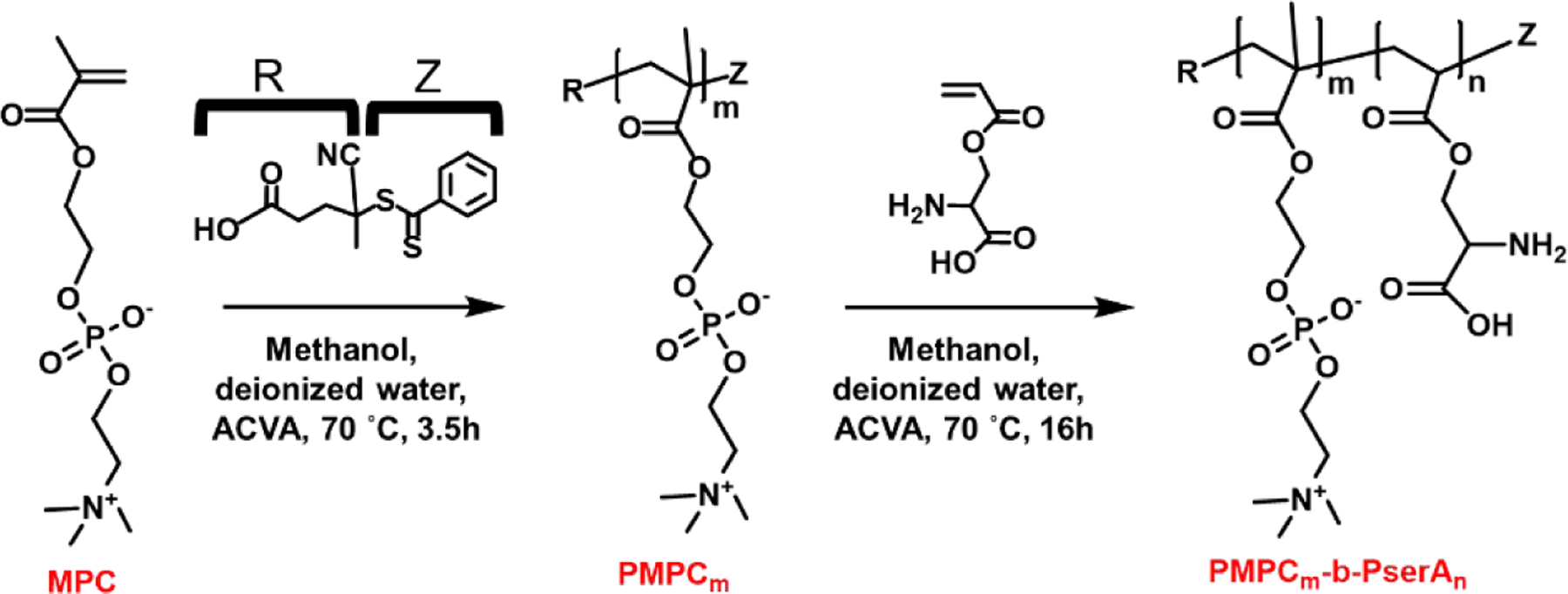
Synthesis of the diblock copolymer PMPC-b-PserA by RAFT polymerization. (m = 55, n = 25)
NCP nanoparticles were produced by complexation between Fe3+ and the PserA block of PMPC-b-PserA.47 The average hydrodynamic diameters of PMPC and PMPC-FeCl3 at a Fe3+ to MPC molar ratio of 3 were 4.2±0.6 nm and 3.17±0.6 nm, respectively, based on DLS measurements (Figure S5A), indicating that no particle formation occurred due to mixing of PMPC and FeCl3. The UV-vis spectra of PMPC and PMPC-FeCl3 (Figure S5B) were nearly identical, consistent with the DLS data. These results showed that PMPC does not complex with Fe3+. FT-IR spectroscopy was used to investigate Fe3+ chelation by the diblock copolymer. Figure 1A shows FT-IR spectra of PMPC-b-PserA and polymer-Fe3+ complexes. The complexes were formed by coordination bonding between PserA and Fe3+ (PMPC does not complex with Fe3+); therefore, the analyses focused on the pendant groups COO− and NH3+ in PserA. Prior to complexation, peaks were observed at 1717 and 1063 cm−1, attributed to C=O and C-N-C in PMPC, respectively. Peaks at 1220, 1063, and 785 cm−1 were attributed to phosphate groups in PMPC.48 Peaks at 1630, 1404, and 1158 cm−1 were assigned to asymmetric stretching (COO−), symmetric stretching (COO−), and stretching vibration of ester groups in PserA. The band at 1520 cm−1 could correspond to bending of amine groups, and the broad band in the 2700–3500 cm−1 range is associated with stretching of NH3+ (N-H) and OH−.49 The IR spectrum is consistent with previously reported data.31 After complexation, the IR spectrum of the NCP nanoparticles exhibited significant changes in COO− and NH3+ stretching. The intensity of the 1400 cm−1 absorption band of COO− stretch increased. The absorption band of N-H bending shifted from 1520 cm−1 to 1562 cm−1 and became stronger, and N-H stretching absorption bands (2700–3500 cm−1) appeared. These changes originate from the change in dipole moment when Fe3+ complexes with the pendant group of PserA.30, 49 UV-vis spectroscopy confirmed the formation of NCP nanoparticles. The spectrum of FeCl3 in deionized water shows a strong absorption peak at 292 nm (Figure 1B),50 and the NCP nanoparticle solution exhibits a charge transfer absorption centered around 300 nm, corresponding to the charge transfer transition between Fe3+ and deprotonated amino group.51 These results show that polymer-Fe3+ complexation occurs via PserA.
Figure 1.
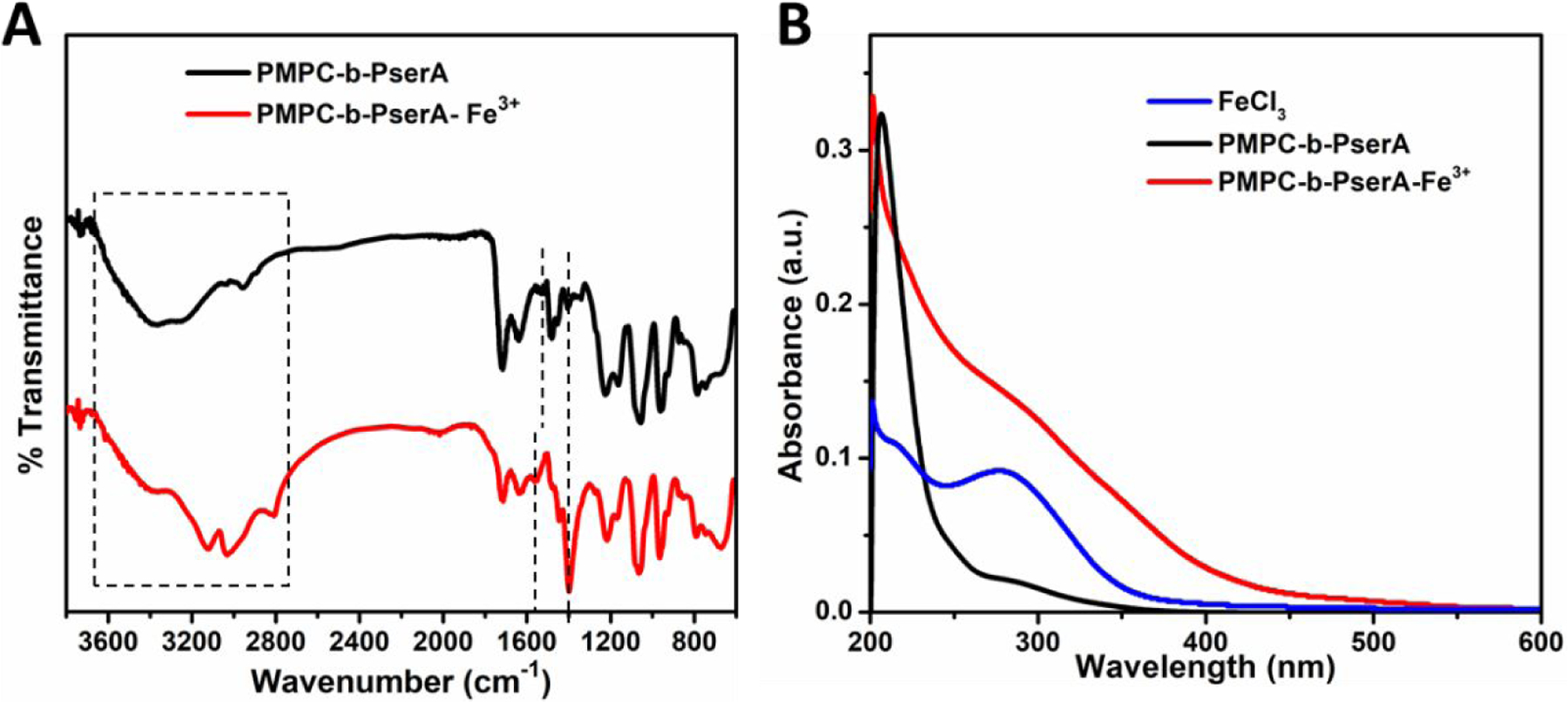
(A) IR spectra of PMPC-b-PserA and PMPC-b-PserA-Fe3+. (B) UV-vis spectra of deionized water solutions of FeCl3 (blue), PMPC-b-PserA (black), and a mixture of FeCl3 and PMPC-b-PserA (red).
NCP nanoparticles were produced by self-assembly via complexation of Fe3+ and the PserA block of PMPC-b-PserA (Scheme 3). The resulting particles exhibit a hydrophobic Fe-oxyhydroxides core with hydrophilic surfaces, PMPC.41 We optimized nanoparticle synthesis by titrating PMPC-b-PserA (10 mg/mL) with FeCl3 to obtain Fe3+/serA molar ratios of 3, 6, 9, and 12. The resulting particles were filtered through 0.22-µm filters and were characterized by UV-vis spectroscopy, DLS, and TEM. Figure 2A includes UV-vis spectra and photos of the resulting particle solutions. The absorbances of NCP nanoparticles with Fe3+/serA = 6 and 9 were significantly reduced, due to removal of precipitates by filtration. Significantly higher absorbance at Fe3+/serA = 3 and a homogenous solution appearance (inset in Figure 2A-a) indicated colloidally stable particles. At a Fe3+/serA ratio ≥ 6, the reaction resulted in immediate precipitation (inset in Figure 2A-b and 2A-c). At a Fe3+/serA ratio of 12, most of the particles precipitated and therefore were not included in further characterizations. DLS data (Figure 2B) showed that the average hydrodynamic diameters of PMPC-b-PserA and NCP nanoparticles (Fe3+/serA=3) were 9.7±1.5 and 22.2±5.6 nm, respectively, indicating particle formation after introduction of FeCl3. The average hydrodynamic diameters of the NCP particles increased to 65.1±21.2 and 111.4±28.3 nm when the Fe3+/serA ratio was increased to 6 and 9, respectively. At a Fe3+/serA ratio of 3, 95% of particle diameters fell within 15.7–32.7 nm with a median of 18.2 nm, which is suitable for using the enhanced permeability and retention effect to passively accumulate drugs in tumor tissues.26 TEM imaging showed that NCP nanoparticles at Fe3+/serA = 3 exhibit a spherical morphology (Figure 2C). Based on TEM, the NCP particle diameter varied from 16 to 25 nm with an average diameter of 20.5±1.9 nm (Figure 2D), similar to the DLS results. Together, these results confirm the formation of colloidal stable NCP nanoparticles via complexation of PMPC-b-PserA and Fe3+ with a Fe3+/serA molar ratio of 3.
Scheme 3.
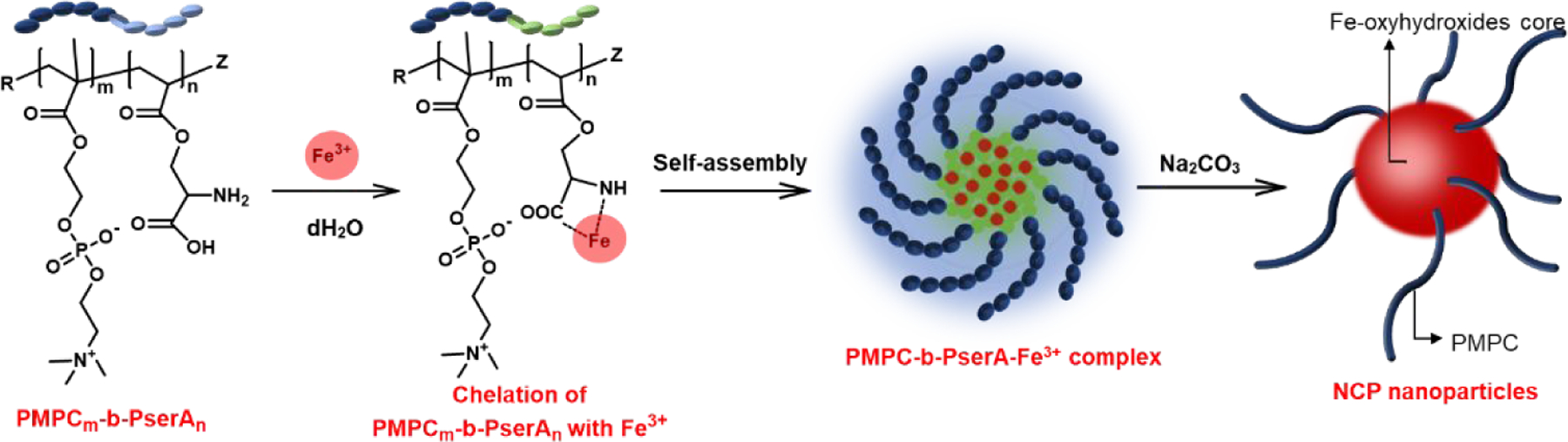
Self-assembly of PMPC-b-PserA into NCP nanoparticles in water via Fe3+ chelation. Addition of Na2CO3 induces formation of the Fe-oxyhydroxides core, with hydrophilic PMPC at the particle surface.
Figure 2. Characterization of NCP nanoparticle formation and size.
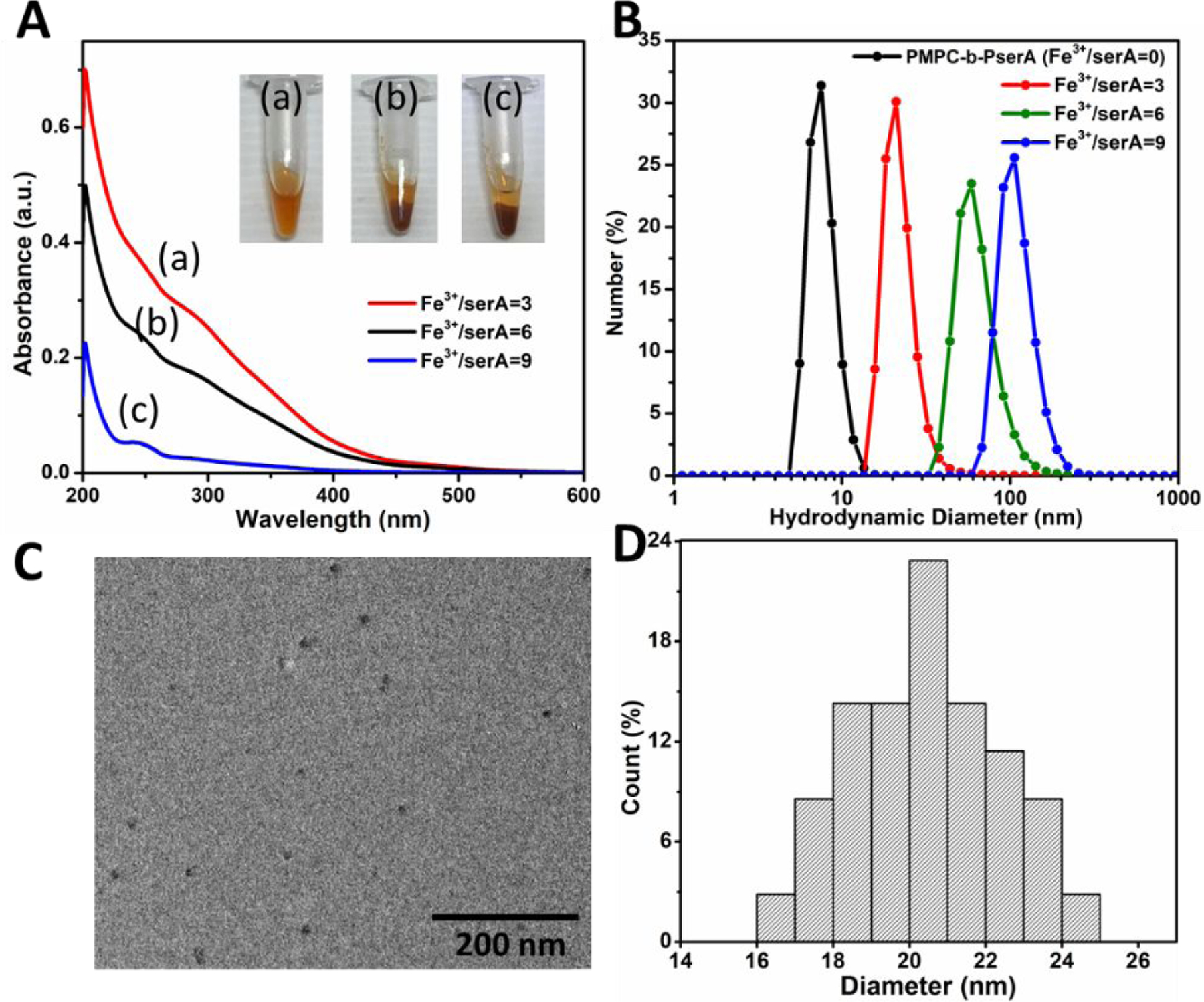
(A) UV-vis spectra of NCP nanoparticles at Fe3+ to serA molar ratios of 3 (a), 6 (b), and 9 (c). Inset: photographs of corresponding colloid solutions. (B) DLS analysis of the polymer and NCP nanoparticles showed that the average hydrodynamic diameter varied from 9.7±1.5 to 111.4±28.3 nm when the Fe3+/serA was increased from 0 to 9. (C) TEM image of NCP nanoparticles (Fe3+/serA = 3), showing a spherical particle morphology. (D) Size distribution of NCP nanoparticles (Fe3+/serA = 3); the particle diameter varied from 16 to 25 nm (average diameter, 20.5±1.9 nm).
Disassembly of NCP nanoparticles
NCP nanoparticle disassembly can be achieved by adding chelators to compete with metal cation complexation with the polymer.43, 52 Disassembly can be used to release drug molecules incorporated in the NCP nanoparticles. Deferoxamine, a clinically utilized metal chelator, and Fe3+ form a stable hexadentate structure with a high affinity constant of 1031 M-1.53 Figure S6 shows the UV-vis spectra of DFO, FeCl3, and DFO-Fe3+. The complex spectrum exhibited an absorption peak at 430 nm, in agreement with a previous report.54 Figure 3A shows the spectra of NCP nanoparticles and NCP nanoparticles with DFO. The particle spectrum shows a broad absorption between 200 and 450 nm which diminishes after DFO addition. Additionally, the spectrum of NCP nanoparticles with DFO shows a peak at 430 nm which corresponds to the DFO-Fe3+ complexes. Therefore, we conclude that the particles disassemble via DFO chelation of Fe3+.
Figure 3. Characterization of NCP nanoparticles with and without DFO.
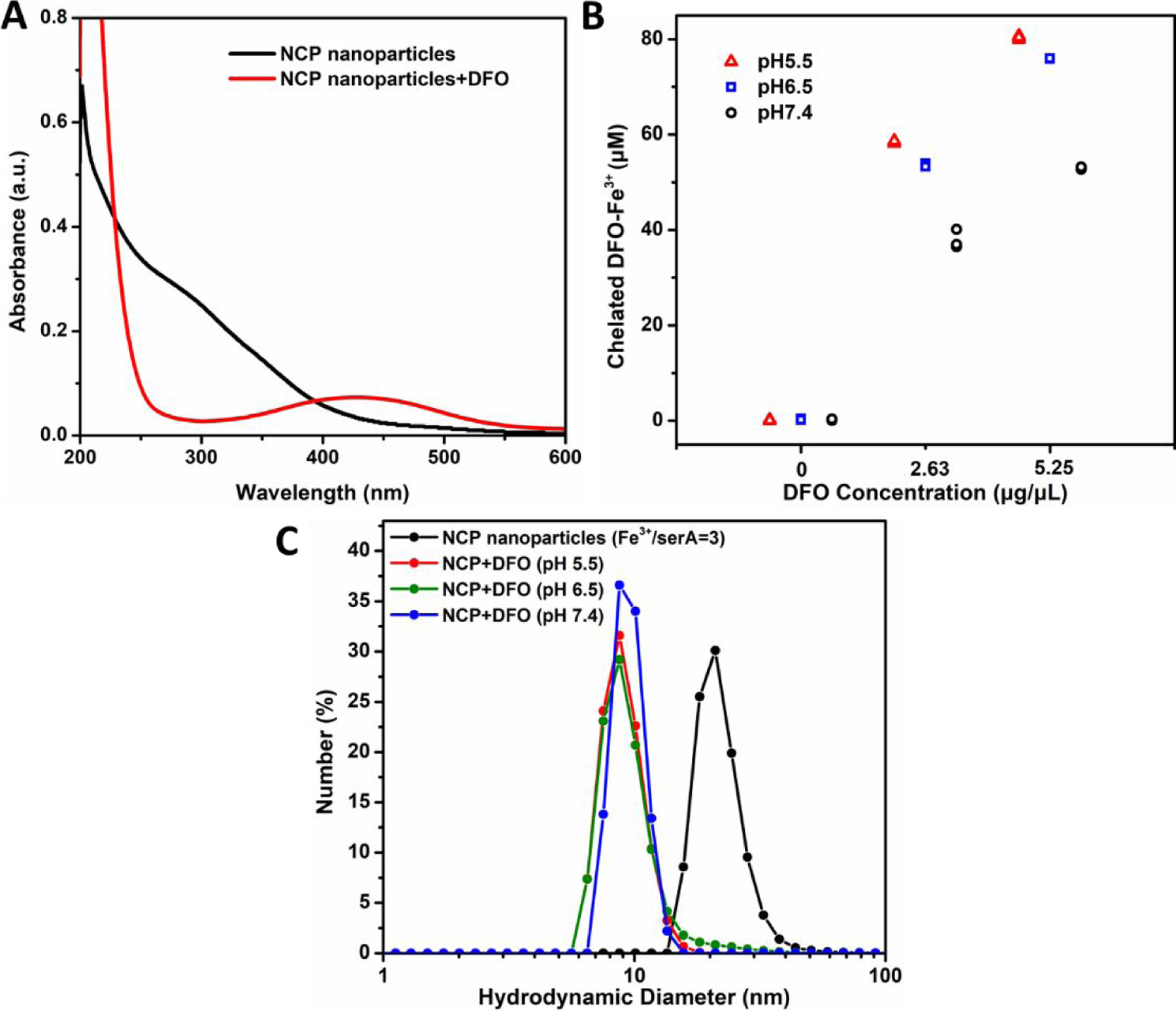
(A) UV-vis spectra of NCP nanoparticles (Fe3+/serA=3) with DFO (red) and without DFO (black). After DFO addition, the broad absorption of NCP nanoparticles between 200 and 450 nm diminished, and a peak at 430 nm appeared. (B) Concentrations of DFO-Fe3+ complexes for NCP nanoparticles treated with different amount of DFO at different solution pH values. (C) DLS analysis of the NCP nanoparticles (Fe3+/serA = 3) with and without DFO (2.6 µg/µL) at pH 5.5, 6.5, and 7.4.
The dissociation of Fe3+ and serA triggered by DFO can be modulated by changing the solution pH. PserA is an amphoteric polymer with carboxyl and amine groups, and the polymer charge is influenced by the solution pH.30, 31 NCP nanoparticle disassembly was evaluated by treating the particles with different amounts of DFO under different solution pH from 5.5 to 7.4. The resulting DFO-Fe3+ complexes were collected with Nanosep centrifugal devices and were quantitated by UV-vis spectroscopy using a calibration with DFO-Fe3+ standards. Higher DFO-Fe3+ complex concentration indicated greater particle disassembly. In Figure 3B, no disassembly was observed at 0 µg/µL DFO. At DFO concentrations of 2.6 and 5.2 µg/µL, the DFO-Fe3+ concentration increased to 37.9±2 and 52.9±0.3 µM, respectively, indicating particle disassembly. Lower solution pH also caused greater particle disassembly. Lower solution pH results in protonation of carboxylate groups, reducing interactions of PserA with Fe3+.31 The results (Figure 3C) demonstrate the size change in the disassembling process. The hydrodynamic diameter of NCP nanoparticles (at Fe3+/serA=3) was 22.2±5.6 nm before adding DFO, and ~9 nm after adding DFO at 2.6 µg/µL.
A possible mechanism for triggered release is illustrated in Scheme 4. At neutral pH, the pendant NH2 and COO− groups of PserA form complexes with Fe3+, resulting in a network of coordination bridges between PserA molecules and Fe3+. This complexation leads to a hydrophobic moiety by disrupting H-bonds between the amine groups of PserA and water molecules. The hydrophilic PMPC block drives self-assembly to form NCP nanoparticles.55 The addition of DFO leads to NCP nanoparticle disassembly because DFO replaces PserA in complexes with Fe3+. Under acidic conditions (such as in tumor tissue), both the amine and carboxyl groups of PserA are protonated and PserA becomes a cationic polymer, reducing its complexation with Fe3+ and allowing DFO to more effectively cause particle disassembly. It is concluded that more efficient NCP nanoparticles disassembly can be achieved by reducing the solution pH and increasing the amount of DFO. In tumor tissues, the disassembly of NCP may be achieved by the acidic environment of tumor tissue and possible interaction between the metallodrug delivery and transferrin receptor 1 (TfR1). 56, 57
Scheme 4.
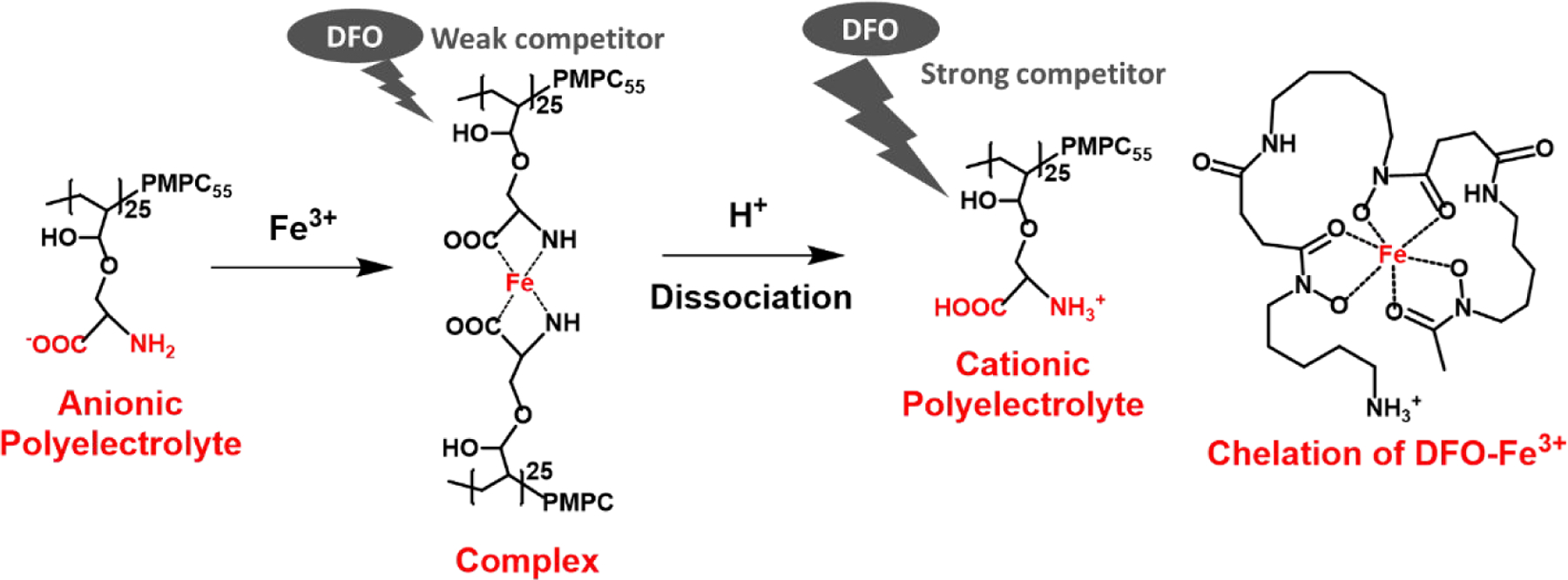
Mechanism of DFO- and proton-induced disassembly of PMPC-b-PserA NCP nanoparticles.
Triggered release of curcumin and induction of apoptosis in vitro
Curcumin was selected as the model drug for studying drug encapsulation and release by the NCP nanoparticles because of its anticancer activity and its low water solubility (11 ng/mL)58, which limits its utility.59 The NCP nanoparticle addresses this limitation by encapsulating curcumin dissolved in CHCl3 in a hydrophobic core via an oil-in-water emulsion method. Unincorporated curcumin was removed using acetone washes and was quantitated by UV-vis spectroscopy to calculate encapsulation efficiency and loading capacity (Figure 4A). The UV-vis spectrum of curcumin in acetone exhibits a distinct absorption at 420 nm. During emulsion formation, curcumin is incorporated via hydrophobic interactions between the polyphenol groups of curcumin and the hydrophobic particle core.60, 61 The resulting Cur@NCP particles appeared homogeneous and spherical by TEM (Figure 4B), with a particle diameter ranging 8 to 26 nm (18.2 nm average diameter) (Figure 4C). The oil-in-water emulsion method was optimized by varying the volume ratio of CHCl3 to deionized water (0.1, 0.125, 0.15, 0175, and 0.2). Encapsulation efficiency was estimated by normalizing the loaded curcumin to the total curcumin, and the loading capacity was calculated as the ratio of loaded curcumin to polymer. The encapsulation efficiency varied between 80% and 100% for all CHCl3 to deionized water ratios, and the loading capacity increased from 14% to 32% over the range of CHCl3 to deionized water ratios (Figure 4D). In contrast, a mixture of free PMPC-b-PserA polymer with curcumin showed a low encapsulation efficiency and loading capacity (14.4% and 3.6%, respectively), because the free polymer does not contain a hydrophobic core for curcumin partitioning. These results demonstrate that the NCP particle’s hydrophobic core can be utilized for drug loading. Cur@NCPs with a CHCl3 to deionized water ratio of 0.15 exhibited an encapsulation efficiency and loading capacity of 99.6% and 26%, respectively. This particle formulation was used to investigate triggered curcumin release in subsequent cell apoptosis experiments.
Figure 4. Characterization of Cur@NCPs.
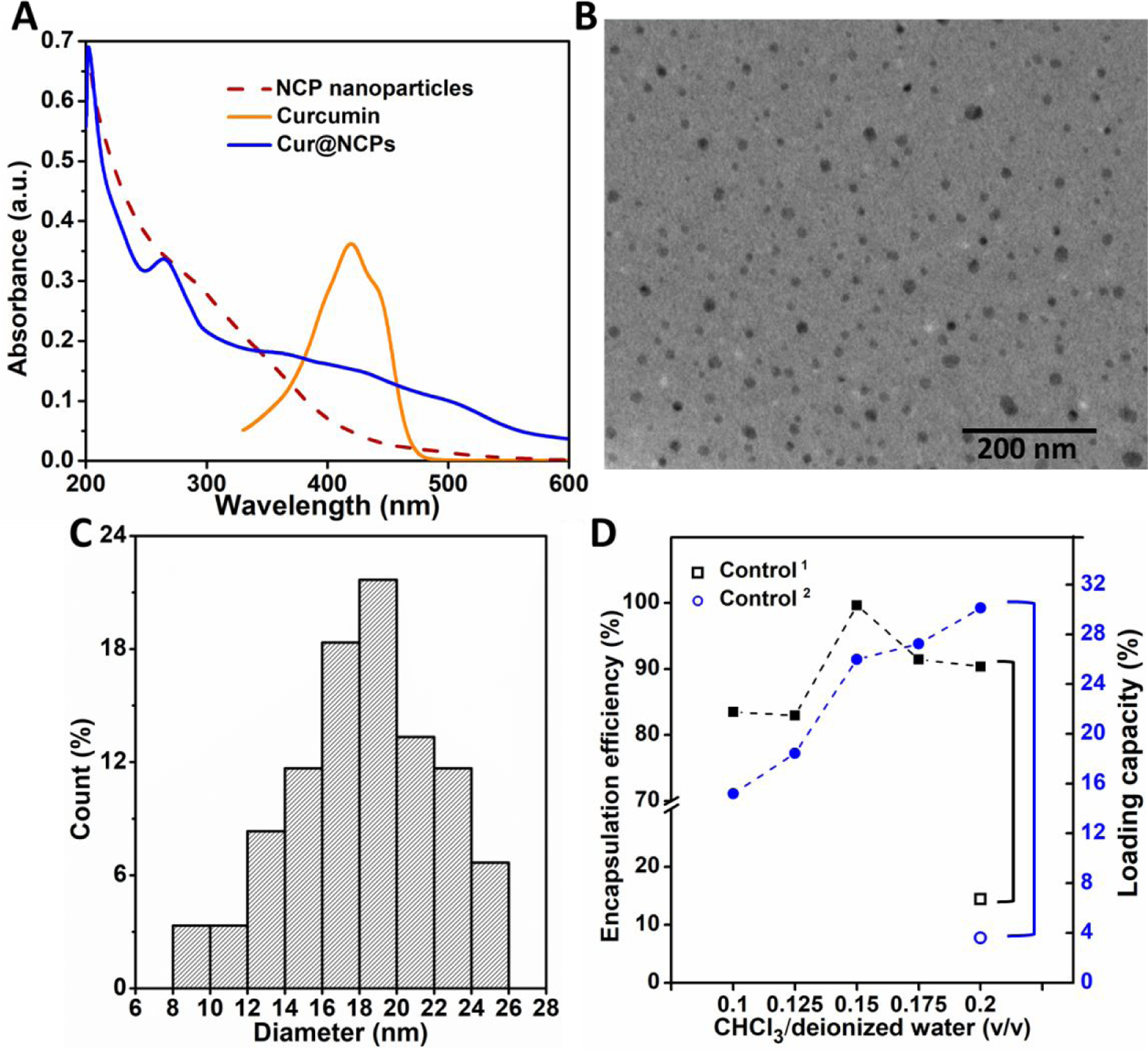
(A) UV-vis spectra of curcumin in acetone (orange) and NCP nanoparticles in deionized water before (red) and after (blue) entrapping curcumin. (B) TEM image of Cur@NCPs. (C) Size distribution of Cur@NCPs. (D) Encapsulation efficiency and loading capacity of Cur@NCPs using different volumetric ratios of CHCl3 to deionized water. Control1 and Control2 indicate encapsulation efficiency and loading capacity of a 1 mL PMPC-b-PserA solution (10mg/mL) without Fe3+ with addition of 200 µL curcumin in CHCl3 (4 mg/mL).
Triggered release of curcumin was demonstrated by treating Cur@NCPs with DFO for 30 min in phosphate buffer at pH 5.5, 6.5, and 7.4, as in the NCP nanoparticle disassembly experiments described in Figure 3. DFO treatment caused particle disassembly, indicated by a missing broad absorption between 250 and 450 nm which would indicate intact Cur@NCPs, and led to a new absorption band centered around 430 nm, indicating formation of DFO-Fe3+ complexes (Figure 5A). The results (Figure 5B) show DFO treatments release 3–12 µg curcumin, while Cur@NCPs alone without DFO released <1 µg curcumin. The amount of curcumin released in an acidic solution was more than two times higher, consistent with the previous NCP particle disassembly results.
Figure 5. Triggered release of curcumin.
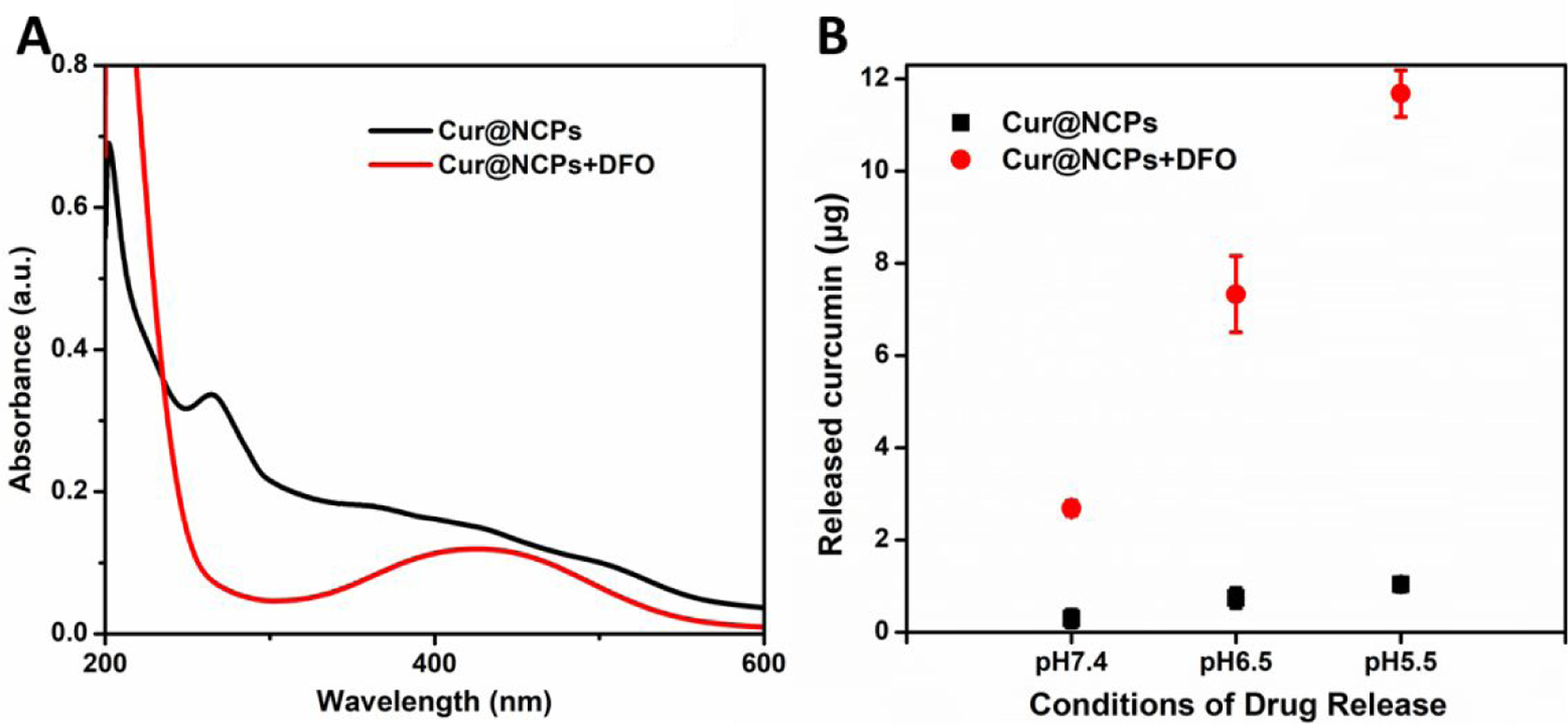
(A) UV-vis absorption spectra of Cur@NCPs with DFO (red) and without DFO (black). (B) Release of curcumin from Cur@NCPs with DFO (red) and without DFO (black) at different solution pH values, at an estimated molar ratio of DFO/Fe3+ of 1.
Curve fitting of the curcumin release data was performed using a first-order model; the results are shown in Figure S2 and are summarized in Table S2. The first-order model, dC/dt=-kC, where k is the first order rate constant, has been used previously to describe absorption or elimination of therapeutic agents. At 0 h, the Ct/C0 from both Cur@NCPs and Cur@NCPs+DFO are 1. At 0.5 h, C0.5 for the Cur@NCPs was 3.6, while Cur@NCPs+DFO was 13.2, which shows a significant difference in the initial curcumin release with and without DFO. From 0.5 to 12.5 h, the release rate constants from Cur@NCPs and Cur@NCPs+DFO were 0.17 and 0.26, respectively, indicating that curcumin release with addition of DFO is faster than without DFO.
Curcumin can induce cell apoptosis.37 We investigated whether triggered release of curcumin by the NCP nanoparticles could induce apoptosis of human triple-negative breast cancer cells (MDA-MB-231). Cell apoptosis and viability were estimated using an MTT assay. Cell viability after a 24 h treatment with different concentrations of curcumin alone decreased from 80% to 20% as the curcumin concentration increased from 10 to 100 µg/mL, consistent with previous studies.38 In the absence of curcumin, cell viabilities varied between 80% and 100% when the cells were treated with the polymer, FeCl3, DFO, DFO-Fe3+, NCP nanoparticles, and NCP nanoparticles+DFO. Cell viability was greater than 80% in all samples tested, well above the recommended 70% cutoff for cytotoxicity.62 Likewise, cells treated with Cur@NCPs at from 90 to 360 µg/mL without DFO showed 80–90% viability. When DFO was added at a 1:1 molar ratio with Fe3+, cell viability decreased to 64.9±5.1% and 41.9±4.4% at 270 and 360 µg/mL Cur@NCPs, respectively, indicating cell apoptosis due to curcumin release. When DFO was added at a 1.5 molar ratio with Fe3+, the cell viability decreased further to 57.8±5.6% and 34.3±12% at 270 and 360 µg/mL Cur@NCPs, respectively.
We hypothesize that curcumin release occurs inside the cells because curcumin solubility in water is extremely low (v), and the reduced cell viability caused by triggered curcumin release shown in Figure 6 would not likely have occurred if curcumin were released outside the cells. In addition, the NCP nanoparticles are coated with PMPC, which has been shown to penetrate cell membranes without cytotoxicity. Therefore, we hypothesized the particles might be internalized and the curcumin release might occur inside the cells.
Figure 6. Induction of breast cancer cell apoptosis in vitro.
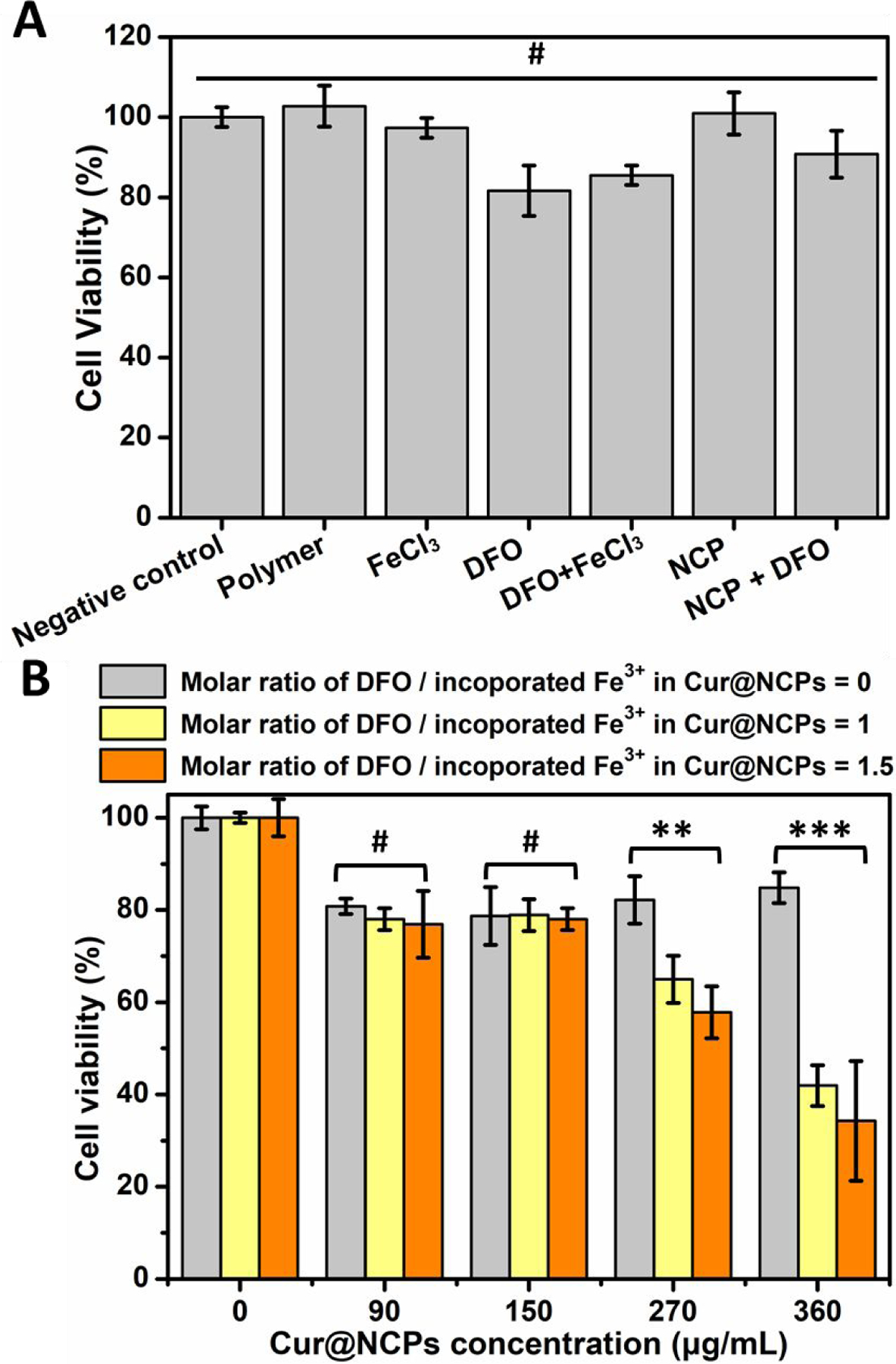
(A) Viability of triple-negative breast cancer cells treated polymer (360 µg/mL), FeCl3 (360 µg/mL), DFO (990 µg/mL), FeCl3 (360 µg/mL) with DFO (990 µg/mL), NCPs (360 µg/mL), and NCPs (360 µg/mL) with DFO (990 µg/mL). (B) Viability of breast cancer cells treated with Cur@NCPs at different concentrations, at DFO/Fe3+ molar ratios of 0, 1, and 1.5. **p<0.01, ***p<0.001, #no statistical significance (p>0.05).
CONCLUSIONS
PMPC-PserA block copolymers are attractive core-shell composite drug nanocarriers because of their biocompatibility, size tunability, and reversible chelation of metal ions. We developed PMPC-b-PserA-based nanoscale coordination polymers (NCPs) with high colloidal stability, high encapsulation efficiency of a hydrophobic drug, and triggered drug release for use as an anticancer drug carrier. The amphoteric NCP nanoparticles were synthesized via metal-ligand coordination between the pendant group of PserA and Fe3+, facilitating disassembly of NCP nanoparticles triggered by low pH (as found in tumor tissues) and competitive DFO chelation. The PMPC shells covering the metallic core endow the NCP nanoparticles with colloidal stability, biocompatibility, and the suitability for delivery into the blood. The tunable size of the NCP nanoparticles (~20 nm diameter for a Fe3+/serA molar ratio of 3) allows accumulation in tumors via the enhanced permeability and retention effect. We demonstrate the utility of the NCP nanoparticles for loading the hydrophobic drug curcumin with excellent encapsulation efficiency and release triggered by acidic conditions and addition of DFO in vitro. This NCP nanoparticle drug carrier system provides a promising platform for investigating the routes of cellular uptake and therapeutic efficacy of drug-loaded NCP nanoparticles in vivo.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the Ministry of Science and Technology (MOST 106–2622-E-008–009-CC2, 108–2221-E-008 −062 -MY3, 108–2628-E-008 −006 -MY3 and 109–3111-8–008-001) and Veterans General Hospitals and University System of Taiwan Joint Research Program (VGHUST109-V4–1-1) for financial support of this project. JL acknowledge the National Institute of Health (MH118160 and AG061383) for financial support of this project.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge on the ACS Publications website at DOI: https://pubs.acs.org/doi/10.1021/acsami.1c03191. Table of abbreviations. UV-vis analysis of DFO-Fe3+ complex, curcumin in acetone, and curcumin in phosphate buffer with 0.5% Tween 80. Drug release profiles of Cur@NCPs. Table of the release profile statistics. 1H NMR spectra of serA monomer, PMPC homopolymer, and PMPC-PserA block copolymer. GPC analysis of PMPC homopolymer and PMPC-PserA block copolymer. DLS and UV-vis analysis of PMPC and Fe3+. Cell viability tests.
The authors declare no competing financial interests.
REFERENCES
- (1).Feazell RP; Nakayama-Ratchford N; Dai H; Lippard SJ Soluble Single-Walled Carbon Nanotubes as Longboat Delivery Systems for Platinum(IV) Anticancer Drug Design. J. Am. Chem. Soc 2007, 129 (27), 8438–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lim CK; Singh A; Heo J; Kim D; Lee KE; Jeon H; Koh J; Kwon IC; Kim S Gadolinium-Coordinated Elastic Nanogels for Invivo Tumor Targeting and Imaging. Biomaterials 2013, 34 (28), 6846–6852. [DOI] [PubMed] [Google Scholar]
- (3).Liu D; Yang F; Xiong F; Gu N The Smart Drug Delivery System and Its Clinical Potential. Theranostics 2016, 6 (9), 1306–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Senapati S; Mahanta AK; Kumar S; Maiti P Controlled Drug Delivery Vehicles for Cancer Treatment and Their Performance. Signal Transduct. Target. Ther 2018, 3 (1), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Morales-Cruz M; Delgado Y; Castillo B; Figueroa CM; Molina AM; Torres A; Milián M; Griebenow K Smart Targeting to Improve Cancer Therapeutics. Drug Des. Devel. Ther 2019, 13, 3753–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jain KK Drug Delivery Systems - An Overview. Methods Mol. Biol 2008, 437 (216), 1–50. [DOI] [PubMed] [Google Scholar]
- (7).Jones M; Leroux J Polymeric Micelles - a New Generation of Colloidal Drug Carriers. Eur J Pharm Biopharm 1999, 48 (2), 101–111. [DOI] [PubMed] [Google Scholar]
- (8).Gupta R; Shea J; Scafe C; Shurlygina A; Rapoport N Polymeric Micelles and Nanoemulsions as Drug Carriers: Therapeutic Efficacy, Toxicity, and Drug Resistance. J. Control. Release 2015, 212, 70–77. [DOI] [PubMed] [Google Scholar]
- (9).Akbarzadeh A; Rezaei-sadabady R; Davaran S; Joo SW; Zarghami N Liposome : Classification , PrepNew Aspects of Liposomesaration , and Applications. Nanoscale Res. Lett 2013, 8 (102), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bibi S; Lattmann E; Mohammed AR; Perrie Y Trigger Release Liposome Systems: Local and Remote Controlled Delivery? J. Microencapsul 2012, 29 (3), 262–276. [DOI] [PubMed] [Google Scholar]
- (11).Kesharwani P; Jain K; Jain NK Dendrimer as Nanocarrier for Drug Delivery. Prog. Polym. Sci 2014, 39 (2), 268–307. [Google Scholar]
- (12).Cationic Poly‑L‑lysine Dendrimer Complexes DOX.Pdf [Google Scholar]
- (13).Hoare TR; Kohane DS Hydrogels in Drug Delivery: Progress and Challenges. Polymer (Guildf) 2008, 49 (8), 1993–2007. [Google Scholar]
- (14).Lin CC; Metters AT Hydrogels in Controlled Release Formulations: Network Design and Mathematical Modeling. Adv. Drug Deliv. Rev 2006, 58 (12–13), 1379–1408. [DOI] [PubMed] [Google Scholar]
- (15).Hossen S; Hossain MK; Basher MK; Mia MNH; Rahman MT; Uddin MJ Smart Nanocarrier-Based Drug Delivery Systems for Cancer Therapy and Toxicity Studies: A Review. J. Adv. Res 2019, 15, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li X; Yang Z; Yang K; Zhou Y; Chen X; Zhang Y; Wang F; Liu Y; Ren L Self-Assembled Polymeric Micellar Nanoparticles as Nanocarriers for Poorly Soluble Anticancer Drug Ethaselen. Nanoscale Res. Lett 2009, 4 (12), 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cabral H; Kataoka K Progress of Drug-Loaded Polymeric Micelles into Clinical Studies. J. Control. Release 2014, 190, 465–476. [DOI] [PubMed] [Google Scholar]
- (18).Aqil F; Munagala R; Jeyabalan J; Vadhanam MV Bioavailability of Phytochemicals and Its Enhancement by Drug Delivery Systems. Cancer Lett 2013, 334 (1), 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Grund S; Bauer M; Fischer D Polymers in Drug Delivery—State of the Art and Future Trends. Adv. Eng. Mater 2011, 13 (3), B61–B87. [Google Scholar]
- (20).Lee YH; Chang DS Fabrication, Characterization, and Biological Evaluation of Anti-HER2 Indocyanine Green-Doxorubicin-Encapsulated PEG-b-PLGA Copolymeric Nanoparticles for Targeted Photochemotherapy of Breast Cancer Cells. Sci Rep 2017, 7, 46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rieter WJ; Pott KM; Taylor KML; Lin W Nanoscale Coordination Polymers for Platinum-Based Anticancer Drug Delivery. J. Am. Chem. Soc 2008, 130 (35), 11584–11585. [DOI] [PubMed] [Google Scholar]
- (22).Solórzano R; Suárez-García S; Novio F; Lorenzo J; Alibés R; Busqué F; Ruiz-Molina D Nanoscale Coordination Polymers for Medicine and Sensors. Adv. Inorg. Chem 2020, 76, 3–31. [Google Scholar]
- (23).Liu D; Poon C; Lu K; He C; Lin W Self-Assembled Nanoscale Coordination Polymers with Trigger Release Properties for Effective Anticancer Therapy. Nat. Commun 2014, 5 (May), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Imaz I; Hernando J; Ruiz-Molina D; Maspoch D Metal-Organic Spheres as Functional Systems for Guest Encapsulation. Angew. Chemie - Int. Ed 2009, 48 (13), 2325–2329. [DOI] [PubMed] [Google Scholar]
- (25).Amorín-Ferré L; Busqué F; Bourdelande JL; Ruiz-Molina D; Hernando J; Novio F Encapsulation and Release Mechanisms in Coordination Polymer Nanoparticles. Chem. - A Eur. J 2013, 19 (51), 17508–17516. [DOI] [PubMed] [Google Scholar]
- (26).Maeda H; Wu J; Sawa T; Matsumura Y; Hori K Tumor Vascular Permeability and the EPR Effect in Macromolecular Therapeutics: A Review. J. Control. Release 2000, 65 (1–2), 271–284. [DOI] [PubMed] [Google Scholar]
- (27).Acharya S; Sahoo SK PLGA Nanoparticles Containing Various Anticancer Agents and Tumour Delivery by EPR Effect. Adv. Drug Deliv. Rev 2011, 63 (3), 170–183. [DOI] [PubMed] [Google Scholar]
- (28).Zhao J; Yang Y; Han X; Liang C; Liu J; Song X; Ge Z; Liu Z Redox-Sensitive Nanoscale Coordination Polymers for Drug Delivery and Cancer Theranostics. ACS Appl. Mater. Interfaces 2017, 9 (28), 23555–23563. [DOI] [PubMed] [Google Scholar]
- (29).Ghosh Chaudhuri R; Paria S Core/Shell Nanoparticles: Classes, Properties, Synthesis Mechanisms, Characterization, and Applications. Chem. Rev 2012, 112 (4), 2373–2433. [DOI] [PubMed] [Google Scholar]
- (30).Dual-Stimuli-Responsive L‑Serine-Based Zwitterionic UCST-Type Polymer with Tunable Thermosensitivity.Pdf [Google Scholar]
- (31).Luo CH; Sun XX; Wang F; Wei N; Luo FL Utilization of L-Serinyl Derivate to Preparing Triple Stimuli-Responsive Hydrogels for Controlled Drug Delivery. J. Polym. Res 2019, 26 (12), 1–9. [Google Scholar]
- (32).Goda T; Ishihara K; Miyahara Y Critical Update on 2-Methacryloyloxyethyl Phosphorylcholine (MPC) Polymer Science. J. Appl. Polym. Sci 2015, 132 (16), 1–10.25866416 [Google Scholar]
- (33).Ishihara K Revolutionary Advances in 2-Methacryloyloxyethyl Phosphorylcholine Polymers as Biomaterials. J. Biomed. Mater. Res. - Part A 2019, 107 (5), 933–943. [DOI] [PubMed] [Google Scholar]
- (34).Chiefari J; K. (Bill) Chong Y; Ercole F; Krstina J; Jeffery J; P. T. Le T; T. A. Mayadunne R; F. Meijs G; L. Moad C; Moad G; Rizzardo E; H. Thang S Living Free-Radical Polymerization by Reversible Addition−Fragmentation Chain Transfer: The RAFT Process. Macromolecules 1998, 31 (16), 5559–5562. [Google Scholar]
- (35).Chen Q; Men Y; Wang H; Chen R; Han X; Liu J Curcumin Inhibits Proliferation and Migration of A549 Lung Cancer Cells through Activation of ERK1/2 Pathway-Induced Autophagy. Nat. Prod. Commun 2019, 14 (6), 1–7. [Google Scholar]
- (36).Liu Q; Loo WTY; Sze SCW; Tong Y Curcumin Inhibits Cell Proliferation of MDA-MB-231 and BT-483 Breast Cancer Cells Mediated by down-Regulation of NFκB, CyclinD and MMP-1 Transcription. Phytomedicine 2009, 16 (10), 916–922. [DOI] [PubMed] [Google Scholar]
- (37).Choudhuri T; Pal S; Agwarwal ML; Das T; Sa G Curcumin Induces Apoptosis in Human Breast Cancer Cells through P53-Dependent Bax Induction. FEBS Lett 2002, 512 (1–3), 334–340. [DOI] [PubMed] [Google Scholar]
- (38).Chiu T-L; Su C-C Curcumin Inhibits Proliferation and Migration by Increasing the Bax to Bcl-2 Ratio and Decreasing NF-ΚBp65 Expression in Breast Cancer MDA-MB-231 Cells. Int J Mol Med 2009, 23 (4), 469–475. [DOI] [PubMed] [Google Scholar]
- (39).Tan BL; Norhaizan ME Curcumin Combination Chemotherapy: The Implication and Efficacy in Cancer. Molecules 2019, 24 (14), 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tam T; Leung-Toung R; Li W; Wang Y; Karimian K; Spino M Iron Chelator Research: Past, Present, and Future. Curr. Med. Chem 2005, 10 (12), 983–995. [DOI] [PubMed] [Google Scholar]
- (41).Dideriksen K; Baker JA; Stipp SLS Equilibrium Fe Isotope Fractionation between Inorganic Aqueous Fe(III) and the Siderophore Complex, Fe(III)-Desferrioxamine B. Earth Planet. Sci. Lett 2008, 269 (1–2), 280–290. [Google Scholar]
- (42).Jackson CL; Chanzy HD; Booy FP; Drake BJ; Tomalia DA; Bauer BJ; Amis EJ Visualization of Dendrimer Molecules by Transmission Electron Microscopy (TEM): Staining Methods and Cryo-TEM of Vitrified Solutions. Macromolecules 1998, 31 (18), 6259–6265. [Google Scholar]
- (43).Buwalda S; Nottelet B; Bethry A; Kok RJ; Sijbrandi N; Coudane J Reversibly Core-Crosslinked PEG-P(HPMA) Micelles: Platinum Coordination Chemistry for Competitive-Ligand-Regulated Drug Delivery. J. Colloid Interface Sci 2019, 535, 505–515. [DOI] [PubMed] [Google Scholar]
- (44).Ramazani F; Chen W; VanNostrum CF; Storm G; Kiessling F; Lammers T; Hennink WE; Kok RJ Strategies for Encapsulation of Small Hydrophilic and Amphiphilic Drugs in PLGA Microspheres: State-of-the-Art and Challenges. Int. J. Pharm 2016, 499 (1–2), 358–367. [DOI] [PubMed] [Google Scholar]
- (45).Hamilton JL; Kizhakkedathu JN Polymeric Nanocarriers for the Treatment of Systemic Iron Overload. Mol. Cell. Ther 2015, 3 (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zeng L; An L; Wu X Modeling Drug-Carrier Interaction in the Drug Release from Nanocarriers. J. Drug Deliv 2011, 2011, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ashmead HD Comparative Intestinal Absorption and Subsequent Metabolism of Metal Amino Acid Chelates and Inorganic Metal Salts 1991, 1 (1), 306–319. [Google Scholar]
- (48).Kuroda K; Miyoshi H; Fujii S; Hirai T; Takahara A; Nakao A; Iwasaki Y; Morigaki K; Ishihara K; Yusa SI Poly(Dimethylsiloxane) (PDMS) Surface Patterning by Biocompatible Photo-Crosslinking Block Copolymers. RSC Adv 2015, 5 (58), 46686–46693. [Google Scholar]
- (49).Ndokoye P; Ke J; Liu J; Zhao Q; Li X L-Cysteine-Modified Gold Nanostars for SERS-Based Copper Ions Detection in Aqueous Media. Langmuir 2014, 30 (44), 13491–13497. [DOI] [PubMed] [Google Scholar]
- (50).Loures CCA; Alcântara MAK; Filho HJI; Teixeira ACSC; Silva FT; Paiva TCB; Samanamud GRL Advanced Oxidative Degradation Processes: Fundamentals and Applications. Int. Rev. Chem. Eng 2013, 5 (2), 102. [Google Scholar]
- (51).Banerjee S; Maji T; Paira TK; Mandal TK Amino-Acid-Based Zwitterionic Polymer and Its Cu(II)-Induced Aggregation into Nanostructures: A Template for CuS and CuO Nanoparticles. Macromol. Rapid Commun 2013, 34 (18), 1480–1486. [DOI] [PubMed] [Google Scholar]
- (52).Muhammad F; Guo M; Qi W; Sun F; Wang A; Guo Y; Zhu G PH-Triggered Controlled Drug Release from Mesoporous Silica Nanoparticles via Intracelluar Dissolution of ZnO Nanolids. J. Am. Chem. Soc 2011, 133 (23), 8778–8781. [DOI] [PubMed] [Google Scholar]
- (53).Cheung W; Patel M; Ma Y; Chen Y; Xie Q; Lockard JV; Gao Y; He H π-Plasmon Absorption of Carbon Nanotubes for the Selective and Sensitive Detection of Fe3+ Ions. Chem. Sci 2016, 7 (8), 5192–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Alberti G; Emma G; Colleoni R; Nurchi VM; Pesavento M; Biesuz R Simple Solid-Phase Spectrophotometric Method for Free Iron(III) Determination. Arab. J. Chem 2019, 12 (4), 573–579. [Google Scholar]
- (55).Mane SR; Sathyan A; Shunmugam R Biomedical Applications of PH-Responsive Amphiphilic Polymer Nanoassemblies. ACS Appl. Nano Mater 2020, 3 (3), 2104–21117. [Google Scholar]
- (56).Huang WT; Larsson M; Lee YC; Liu DM; Chiou GY Dual Drug-Loaded Biofunctionalized Amphiphilic Chitosan Nanoparticles: Enhanced Synergy between Cisplatin and Demethoxycurcumin against Multidrug-Resistant Stem-like Lung Cancer Cells. Eur. J. Pharm. Biopharm 2016, 109, 165–173. [DOI] [PubMed] [Google Scholar]
- (57).Li H; Sun H; Qian ZM The Role of the Transferrin-Transferrin-Receptor System in Drug Delivery and Targeting. Trends Pharmacol. Sci 2002, 23 (5), 206–209. [DOI] [PubMed] [Google Scholar]
- (58).Song Z; Feng R; Sun M; Guo C; Gao Y; Li L; Zhai G Curcumin-Loaded PLGA-PEG-PLGA Triblock Copolymeric Micelles: Preparation, Pharmacokinetics and Distribution in Vivo. J. Colloid Interface Sci 2011, 354 (1), 116–123. [DOI] [PubMed] [Google Scholar]
- (59).Gupta SC; Patchva S; Aggarwal BB Therapeutic Roles of Curcumin: Lessons Learned from Clinical Trials. AAPS J 2013, 15 (1), 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Woolley GA; Deber CM Peptides in Membranes: Lipid‐induced Secondary Structure of Substance P. Biopolymers 1987, 26 (S0), S109–S121. [DOI] [PubMed] [Google Scholar]
- (61).Young JK; Graham WH; Beard DJ; Hicks RP The Use of UV-Visible Spectroscopy for the Determination of Hydrophobic Interactions between Neuropeptides and Membrane Model Systems. Biopolymers 1992, 32 (8), 1061–1064. [DOI] [PubMed] [Google Scholar]
- (62).International Organization for Standardization. (2009) Biological evaluation of medical devices — Part 5: Tests for in vitro cytotoxicity (ISO standard no. 10993–5:2009) Retrieved from https://www.iso.org/standard/36406.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


