Abstract
Potent neutralizing SARS-CoV-2 antibodies often target the spike protein receptor-binding site (RBS), but the variability of RBS epitopes hampers broad neutralization of multiple sarbecoviruses and drifted viruses. Here, using humanized mice, we identified an RBS antibody with a germline VH gene that potently neutralized SARS-related coronaviruses, including SARS-CoV and SARS-CoV-2 variants. X-ray crystallography revealed coordinated recognition by the heavy chain of non-RBS conserved sites and the light chain of RBS with a binding angle mimicking the angiotensin-converting enzyme 2 (ACE2) receptor. The minimum footprints in the hypervariable region of RBS contributed to the breadth of neutralization, which was enhanced by immunoglobulin G3 (IgG3) class switching. The coordinated binding resulted in broad neutralization of SARS-CoV and emerging SARS-CoV-2 variants of concern. Low-dose therapeutic antibody treatment in hamsters reduced the virus titers and morbidity during SARS-CoV-2 challenge. The structural basis for broad neutralizing activity may inform the design of a broad spectrum of therapeutics and vaccines.
Keywords: SARS-CoV-2, variants of concern, SARS-CoV, neutralizing antibody, receptor-binding domain, IgG3, X-ray crystallography
Graphical abstract
Antigenic variability in SARS-related coronavirus decreases the likelihood of cross-neutralization by monoclonal antibodies. Onodera et al. identify a broad neutralizing SARS-CoV-2 antibody in humanized mice that targets a vulnerable site of SARS-CoV and SARS-CoV-2 variants. Structural analysis reveals that the broad neutralization is coordinated by the heavy and light chains and is enhanced by IgG3 class switching.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, triggered by the novel virus SARS-CoV-2 (CoV2), has caused more than 198 million infections and more than 4.2 million deaths worldwide as of August 3, 2021 (https://www.who.int/). Whereas host immune responses are considered to play both beneficial and detrimental roles for disease progression and outcome in the acute period, neutralizing antibodies that are maintained in the convalescent period protect against re-infection in animal models (Baum et al., 2020a; McMahan et al., 2021). Indeed, convalescent plasma and monoclonal antibody therapeutics received emergency use authorization by the U.S. Food and Drug Administration (Gottlieb et al., 2021; Weinreich et al., 2021), with more antibody seeds under preclinical and clinical development.
Major epitopes for CoV2-neutralizing antibodies reside in the receptor-binding domain (RBD) of spike (S) protein (Andreano et al., 2021; Piccoli et al., 2020). RBD epitopes are further classified into four classes based on the structure of antigen-antibody complex (Barnes et al., 2020; Yuan et al., 2021). Class 1 and class 2 epitopes overlap angiotensin-converting enzyme 2 (ACE2) binding sites (receptor-binding site [RBS]), thereby constituting the major target for highly potent neutralizing antibodies (Barnes et al., 2020). However, low conservation of the RBS sequence between CoV2 and other coronaviruses (CoVs) decreases the likelihood of cross-neutralization of multiple CoVs by RBS antibodies. Supporting this, only 1.2% (6 clones) of CoV2-neutralizing antibodies (482 clones) deposited in the coronavirus antibody database (CoV-AbDab) cross-neutralize SARS-CoV (CoV1) among sarbecoviruses (Raybould et al., 2021). CoV1 cross-neutralizing activity was also absent among highly potent CoV2-neutralizing antibodies that were selected from 453 neutralizing antibodies (Andreano et al., 2021). In addition, many RBS antibodies are vulnerable to CoV2 escape mutations under monotherapeutic usage (Baum et al., 2020b; Li et al., 2020; Weisblum et al., 2020) because of the antigenic drift of the RBS epitopes (Piccoli et al., 2020; Starr et al., 2021). Thus, similar to antibodies to other mutating RNA viruses, potent neutralizing activity and breadth to antigenically divergent viruses are incompatible characters in most CoV2-neutralizing antibodies.
Neutralizing RBS antibodies are frequently encoded by structurally related VH genes and share similar binding modes and footprints (Robbiani et al., 2020; Yuan et al., 2020b). Such convergent antibody responses in many individuals could increase the risk of generating antibody escape mutations because of limited diversity of humoral immune responses in the population. Indeed, CoV2 variants of concern (VOCs) in multiple lineages have emerged in several countries, including the United Kingdom (B.1.1.7; Alpha) (Volz et al., 2021), South Africa (B.1.351; Beta) (Tegally et al., 2021), and Brazil (P.1; Gamma) (Faria et al., 2021). All of these VOCs possess the N501Y mutation in RBD, probably contributing to the enhanced transmissibility by increased ACE2 binding (Andreano et al., 2020). Beta and Gamma VOCs share two additional RBD mutations in K417 and E484 that contribute to the antigenic drift and escape from therapeutic monoclonal antibodies and human serum antibodies from convalescent individuals or vaccinees (Garcia-Beltran et al., 2021; Supasa et al., 2021; Wang et al., 2021b; Wibmer et al., 2021; Zhou et al., 2021). Furthermore, circulation of SARS-related viruses in animal reservoirs pose continuing concerns as the next pandemic potential (Hu et al., 2017). At least two SARS-related CoVs have the ability to infect primary human airway cells (Menachery et al., 2015, 2016). Therefore, isolation of broad neutralizing antibodies against SARS-related CoVs in animal reservoirs and drifted CoV2 in humans advances the development of vaccines and therapeutics against CoVs.
In the present study, we have identified a highly potent CoV2-neutralizing antibody, named NT-193, with the breadth to CoV1 and resistance to CoV2 escape mutations. Structures of the antibody bound to monomeric RBD revealed the distinct binding mode of this RBS antibody, by which the antibody mimics the binding angle of ACE2. Furthermore, the sites of vulnerability in CoV2 and other SARS-related CoVs were unveiled by crystallography as the NT-193 target. CoV1 cross-neutralizing activity of this antibody depended highly on the avidity effect of an immunoglobulin (Ig) G subclass, and robust cross-neutralizing activity was achieved by IgG3 switching. Given the V gene usage of near germline, this distinct class of antibody is a promising target for broad neutralizing vaccines.
Results
NT-108 and NT-193 potently neutralize SARS-CoV-2 despite atypical VH usage
Here, we used TC-mAb mice, which stably maintain an engineered chromosome containing the loci of the entire human Ig heavy and kappa chains in a murine Ig knockout background (Satofuka et al., 2020). After sequential immunization with spike/RBD antigens from CoV2, CoV1, and MERS (Middle East respiratory syndrome)-CoV, B cells binding to the CoV2 RBD were sorted for single-cell culture as previously described (Figure 1 A) (Adachi et al., 2019). Among the high-affinity CoV2 RBD binders, we identified two potent neutralizing antibody clones by vesicular stomatitis virus (VSV)-based pseudovirus assay (Figure 1B). One clone (#108) was specific to the CoV2 RBD, but another clone (#193) was cross-reactive to CoV2/CoV1 RBDs. Both clones were of the human IgG1 subclass. Hereafter, the #108 and #193 clones are denoted as NT-108 and NT-193, respectively.
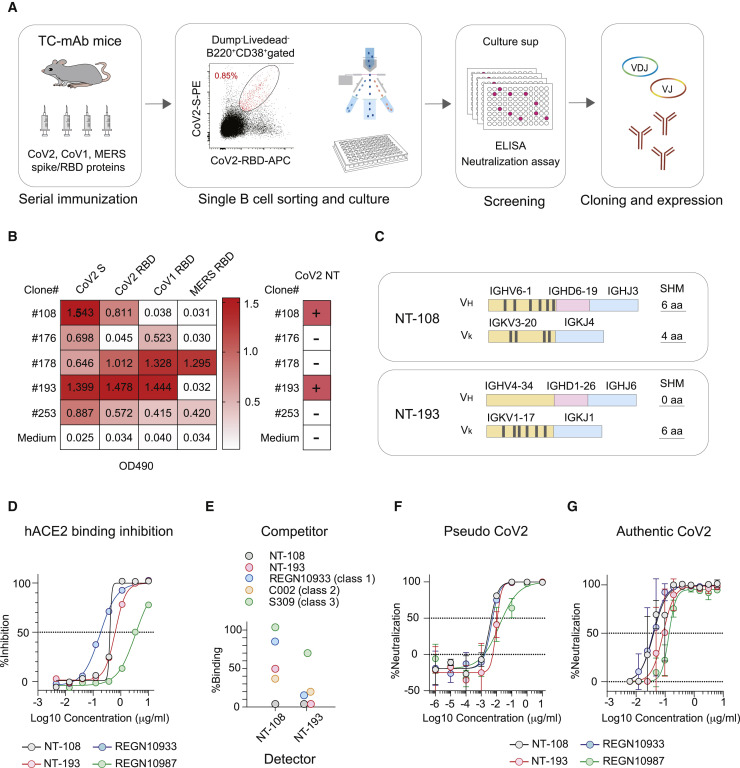
Figure 1.
NT-108/NT-193 antibodies potently neutralize SARS-CoV-2
(A) Schematic of the experimental workflow for isolating RBD monoclonal antibodies from TC-mAb mice. CoV2 spike (S) binding monoclonal antibodies were established by a single-cell culture approach from immune mice that express human antibodies.
(B) Binding of monoclonal antibodies in culture supernatant to the indicated S and RBD proteins was evaluated by ELISA. CoV2-neutralizing activity (CoV2 NT) in culture supernatant was assessed using a pseudovirus system. Two clones (#108, NT-108; #193, NT-193) exhibited neutralizing activity. The representative data from two independent experiments are shown.
(C) VH/Vk gene usage and somatic hypermutations in NT-108 and NT-193.
(D) Amounts of CoV2 RBD bound to hACE2-coated plates were assessed after preincubation with serially diluted NT-108/NT-193. The antibody-mediated inhibition on hACE2-RBD binding is plotted as the percentage.
(E) After masking RBD-bound plates with competitive antibodies of the indicated class, NT-108 and NT-193 was loaded to assess the binding to unmasked RBD.
(F and G) Neutralizing activity against CoV2 was assessed using pseudovirus (F) and authentic virus (G).
Values represent mean ± SD (F and G). The representative data from three independent experiments are shown (D–G). See also Figure S1.
Neutralizing antibodies are often encoded by convergent VH genes, such as IGHV3-53, IGHV3-66, IGHV3-30, IGHV1-2, and IGHV1-69 (Andreano et al., 2021; Robbiani et al., 2020; Yuan et al., 2020b). Unlike these stereotypic VH genes, NT-108 and NT-193 use the atypical IGHV6-1 and IGHV4-34 genes, respectively (Figure 1C). Among the 937 CoV2 RBD antibodies isolated from COVID-19 patients, only 3 IGHV6-1-encoded antibodies were deposited into CoV-AbDab (as of August 3, 2021) (Raybould et al., 2021). Likewise, IGHV4-34-encoded antibodies were rare but more frequently recovered from B cells of COVID-19 patients (Woodruff et al., 2020), as reflected in the deposition of 13 IGHV4-34-encoded antibodies in the database (Raybould et al., 2021). Antibodies encoded by IGHV4-34 genes, generally of the IgM isotype, are autoreactive to carbohydrates expressed on red blood cells and DNA (Bhat et al., 1993; Spellerberg et al., 1995). Despite the usage of this VH gene, NT-193 IgG1 did not develop detectable signals of HEp-2 binding under conditions in which the cells were brightly stained by two reference IgG1 antibodies (51F1 and 72H11) (Sakakibara et al., 2017) (Figure S1A). Moreover, polyreactivity to dsDNA, insulin, and LPS was weaker than that of the reference antibody under ELISA-based polyreactivity assay (Wardemann et al., 2003), as was the case for two CoV2-neutralizing antibodies in the clinical stage (Figure S1B). Thus, the autoreactivity and polyreactivity of NT-193 were negligible despite usage of the IGHV4-34 gene.
RBS epitopes can be subdivided into either class 1 and class 2 (Barnes et al., 2020) or RBS-A, RBS-B, and RBS-C (Yuan et al., 2021). In addition, there are non-RBS epitopes within RBD, denoted as either class 3 and class 4 or S309-proteoglycan and CR3022-cryptic sites. Antibodies against the class 4 epitope are generally less potent than those against other classes of epitopes (Yuan et al., 2020a); therefore, we depriotized the antibodies against this epitope class from our initial screening. Recombinant human IgG1/kappa antibodies were expressed based on the NT-108 and NT-193 VH/VL sequences. We chose REGN10933 (isolate from a humanized mouse) and REGN10987 (isolate from convalescent individuals) as the benchmark of clinical-stage antibodies (Baum et al., 2020b), because REGN10933 exhibits the most potent neutralization activity among clinical-stage antibodies in vitro (Rappazzo et al., 2021) and the antibody cocktail is effective for therapeutic usage (Baum et al., 2020a; Weinreich et al., 2021). In addition, a S309 antibody, another clinical-stage antibody against the class 3 epitope, was expressed as a representative of CoV1/CoV2 cross-neutralizing antibody (Pinto et al., 2020). Both NT-108 and NT-193 IgG1 inhibited RBD binding to human ACE2 (Figure 1D), suggesting their epitopes overlap RBS. We performed competitive ELISA in which the RBD class 1–3 epitopes were masked by a representative antibody of each class (Figure 1E). Strong competition by a class 2 antibody (C002) confirmed that both NT-108 and NT-193 are RBS antibodies against the class 2 epitope. Additional competition by a class 1 antibody (REGN10933) suggested that NT-193 recognizes an epitope encompassing class 1 and 2. Moreover, NT-108 and NT-193 likely recognize partially overlapping epitopes, because they compete with each other. Both antibodies were highly potent neutralizers of CoV2-pseudotyped and CoV2-authentic viruses, with a half-maximal inhibitory concentration (IC50) as potent as that for the REGN antibodies (IC50 < 100 ng/mL) (Figures 1F and 1G). Thus, we identified highly neutralizing RBS antibodies with unconventional VH gene usage.
NT-193-neutralizing activity depends on the IgG subclass
Only 24 ± 9 S trimers are present on the surfaces of individual CoV2 virions (Ke et al., 2020). Such a low density of RBS epitopes may interfere with bivalent binding by antibodies required for inter-RBS crosslinking, as previously observed in HIV-1 (Galimidi et al., 2015). For HIV-1, elevated neutralization was achieved by Env binding antibodies in the context of the IgG3 subclass, which bears a long and flexible hinge region (Richardson et al., 2019; Scharf et al., 2001). To assess the possible contribution of the IgG3-mediated avidity effect on CoV2 neutralization, we first compared the RBD binding affinities of NT-108 and NT-193 in the context of IgG1, IgG3, and Fab by surface plasmon resonance (SPR) analysis. Irrespective of the IgG subclasses, NT-193 bound to both CoV2 and CoV1 RBD at high affinity (apparent affinities of approximately 10−10∼−11 M) (Figure 2 A). However, the RBD binding affinities of NT-193 Fab dropped to less than 1/100-fold of NT-193 IgG antibodies (Figure 2A). The reduction of NT-108 affinity was modest (2- to 4-fold decrease) in the same condition, underscoring the large contribution of the avidity effect on the NT-193 IgG affinities. Therefore, the possible impacts of avidity effects on the breadth and neutralizing potency were assessed using NT-193.
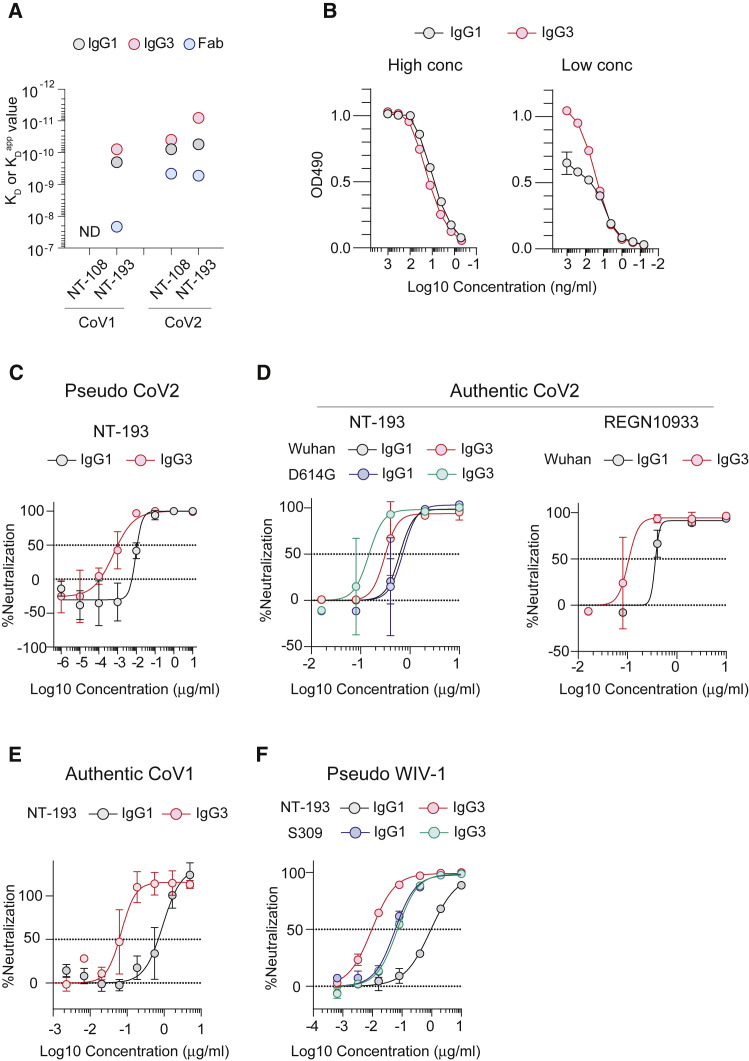
Figure 2.
NT-193 enhances CoV2- and CoV1-neutralizing activity by the IgG3 subclass
(A) RBD binding affinity of NT-108 and NT-193 was assessed by SPR using CoV1/CoV2 RBD immobilized on the sensor. The KD value for Fab and apparent KD (KDapp) value for IgG1 and IgG3 are shown. ND, not detected. The representative data from two independent experiments are shown.
(B) ELISA plates were coated with CoV2 RBD at high (2 μg/mL) or low (0.1 μg/mL) concentrations. The amounts of NT-193 bound to RBD in the IgG1 and IgG3 subclass are plotted at optical density (OD).
(C and D) CoV2-neutralizing activities of NT-193 and REGN10933 to Wuhan and D614G strains were determined by pseudovirus (C) and authentic virus (D).
(E) CoV1-neutralizing activities of IgG1 and IgG3 purified by the size exclusion fractionation were determined using authentic virus.
(F) WIV-1-neutralizing activities of the indicated antibodies were determined by pseudovirus.
Values represent mean ± SD (B–F). The representative data from three independent experiments are shown (B–F). See also Figure S2.
The avidity difference between the IgG1 and the IgG3 subclasses could be underestimated by extremely high-affinity binding under SPR analysis. Moreover, to mimic the low density of RBS epitopes on CoV2 virions, ELISA plates were coated at a non-saturating concentration and then NT-193 of the IgG1 and IgG3 subclasses were added to the plates to assess their binding avidity (Figure 2B). Subclass-dependent differences were not observed in the binding curve under the saturating concentration of RBD, but increased amounts of IgG3 antibodies bound to a non-saturating concentration of RBD compared with the IgG1 subclass. Furthermore, the IgG3-dependent enhancement correlated with a modest but noticeable increase in neutralizing activity against CoV2-pseudotyped and CoV2-authentic viruses (Figures 2C and 2D). The IgG3-dependent enhancement was further assessed using the D614G variant, which is considered to express S trimers more abundantly than the Wuhan strain by reduced shedding of the S1 (Zhang et al., 2020). The variant also expresses RBD with one-open conformation at elevated frequency (Weissman et al., 2021). The usage of this variant exaggerated the IgG3-dependent enhancement (Figure 2D), suggesting that the phenomenon depends on the density of S trimers and/or the frequencies of open conformation. The enhancement was also noticeable from REGN10933, whose epitope is masked in closed RBD conformation (Figure 2D).
A distinct feature of NT-193 is its cross-reactivity with CoV1 (Figures 1B and 2A). Although the cross-reactivity was not extended to other CoVs (MERS-CoV and four seasonal CoVs) (Figure S2), the breadth to CoV1 in sarbecoviruses is a meaningful and important character, given the circulation of zoonotic CoV1-related viruses, including WIV-1, which has the ability to infect human airway cells (Hu et al., 2017; Menachery et al., 2015, 2016). Therefore, the neutralizing activity of NT-193 against CoV1 and WIV-1 was examined in both IgG1 and IgG3 subclasses (Figures 2E and 2F). First, we observed that NT-193 IgG3 potently neutralized these CoV1-related viruses at low IC50 (<100 ng/mL). The potency and breadth of neutralization to the CoV1-related viruses and CoV2 are noteworthy, because the neutralizing activity against CoV1, CoV2, or both is generally weaker (IC50 > 100 ng/mL) in the previously isolated, cross-neutralizing antibodies (Brouwer et al., 2020; Hansen et al., 2020; Zost et al., 2020). Furthermore, potent cross-neutralizing activity was achieved by non-aggregated forms of the IgG3 subclass (Figure 2E). No IgG3-dependent enhancement of the neutralization activity in a class 3 RBD antibody (S309) suggests that the phenomenon is not common in RBD antibodies but depends on the epitopes. Indeed, the class 3 epitope can be recognized by the S309 antibody independently of open and closed conformation (Pinto et al., 2020), and the binding character is distinct from that of REGN10933 (class 1) with IgG3-dependent enhancement activity (Figure 2D). Given IgG3-dependent enhancement in NT-193 activity, this antibody may recognize the epitope that is exposed under open RBD conformation.
Structure of the NT-193 Fab-RBD complex reveals a distinct binding mode
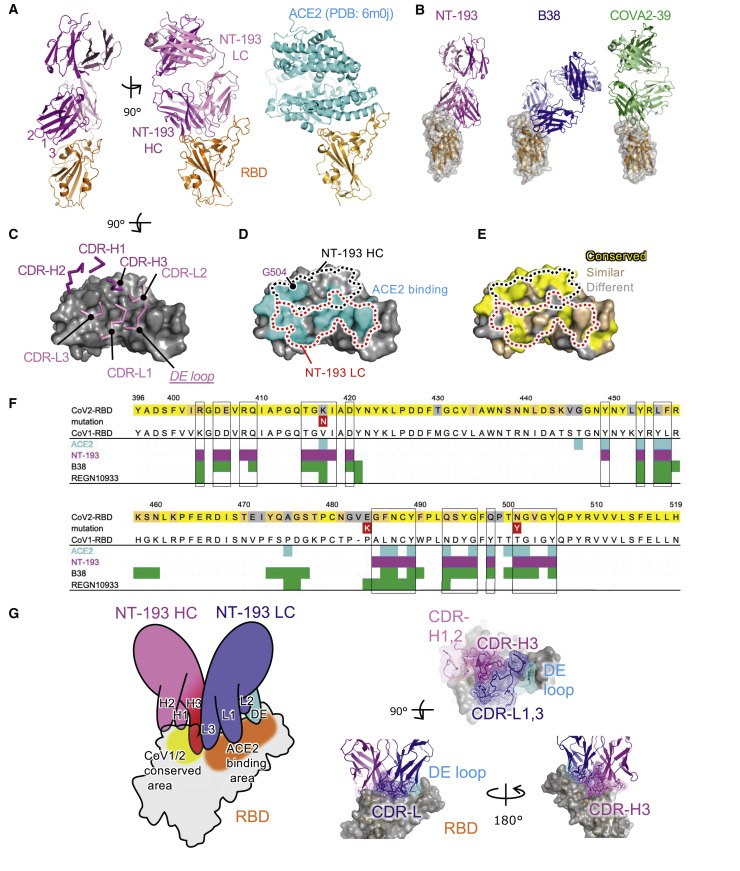
To gain the structural information on potent cross-neutralization activity by NT-193 and enhancement by the IgG3 subclass, we performed X-ray crystallographic analysis. Crystals of the complex of the RBD protein with NT-193 Fab were successfully obtained. The structure of the complex was determined at a resolution of 2.8 Å (Table S1). NT-193 recognizes the top areas of RBD, which widely overlap the binding site of ACE2 (Figure 3 A), but not those of S309 (class 3) and CR3022 (class 4) antibodies (Figure S3A). Class 1/RBS-A and class 2/RBS-B antibodies also recognize the top areas of RBD (Barnes et al., 2020; Yuan et al., 2021); however, the recognition mode of NT-193 is clearly distinct from the benchmark antibodies B38 (class 1) and COVA2-39 (class 2) (Figures 3B and S3B). The NT-193 antibody uses not only complementarity determining region (CDR)-1 and CDR-3 of the light chain (hereafter designated as CDR-Ls) but also the framework region, the DE loop, for recognition of the ACE2 binding site (Figures 3C and 3D; Table S2). Somatic hypermutations substituted six amino acids in the light chain, among which positions 31 and 66 with Ser to Gly mutations were located at the interface with RBD. These Ser to Gly substitutions are unlikely to cause steric hindrance or unsuitable electrostatic complementarity (Figures S4A and S4B), suggesting little, if any, contribution of somatic hypermutations on NT-193 binding. However, the protruding CDR-3 of the heavy chain (hereafter designated as CDR-Hs), together with only small portions of CDR-H1 and CDR-H2 of the heavy chain, are responsible for binding to the CoV1/CoV2 conserved area next to the ACE2 binding site (Figures 3E–3G, S4C, and S5). This site is included in the cryptic epitope that faces inward to the closed prefusion trimer, presumably restricting NT-193 access to RBD of a closed/down configuration. Given the predominance (∼55%) of one-open RBD trimers (Ke et al., 2020), this raises the possibility that a longer hinge in the IgG3 subclass may enhance inter-S-trimer cross-linking by the avidity effect for achieving higher neutralizing activity. Furthermore, these structural features result in an atypical binding mode that permits broad coverage of ACE2 binding sites and potent cross-reactivity by avidity effect with minimum footprints (Figures 3F, 3G, and S5).
Figure 3.
Structure of the NT-193 Fab-RBD complex reveals a distinct binding mode
(A) Overviews of the NT-193 complex at two angles are shown on the left (magenta, NT-193 heavy chain; pink, NT-193 light chain; orange, RBD; numbers 1–3 represent CDR-H1, CDR-H2, and CDR-H3, respectively). The structure of the ACE2-RBD complex is shown on the right (PDB: 6m0j). ACE2 is shown in cyan.
(B) Comparison of the NT-193 complex (colored the same as in A) with the representative class 1/RBS-A antibody complex, B38 (dark and light blue), and class 2/RBS-B antibody complex, COVA2-39 (dark and light green).
(C) CDR binding sites (magenta and pink, stick representations) are shown on the surface model of RBD (gray).
(D) Comparison between ACE2 and NT-193 binding sites on RBS. The binding areas for ACE2 are shown in cyan. The regions surrounded by dotted lines indicate the binding areas for light chains (red, LC) and heavy chains (black, HC).
(E) Amino acid sequence conservation of RBS between CoV1 and CoV2. Yellow, conserved; Light orange, similar; gray, non-conserved. The dotted lines are the same as in (D).
(F) Mapping of antibody binding residues on RBD. The residues that contact ACE2, antibody, or RBD within a distance of 4 Å are defined as binding residues. The RBD residues conserved with CoV1 are colored similarly to those in (E). The binding residues of RBD for ACE2 and antibody are mapped with color. The mutations in VOCs are shown (red).
(G) Recognition mode of NT-193 toward RBD. Left, schematic of the NT-193-RBD complex. The light chain of NT-193 uses CDR-L1/L3 (blue) and DE loop (cyan) for the recognition of RBD. CDR-H3 (magenta) and CDR-L3 dominantly bind to RBD. Right, cartoon models of CDR-Hs (magenta and pink), CDR-Ls (blue), and the DE loop (blue and cyan) of NT-193 are shown with a meshed surface.
See also Figures S3–S5 and Tables S1 and S2.
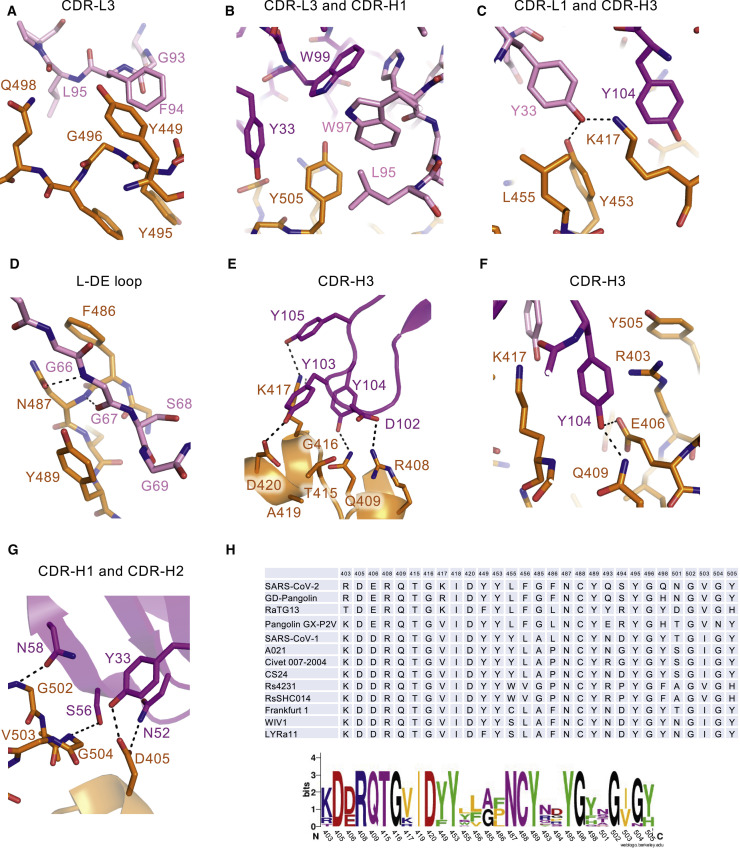
Looking in more detail at the binding interfaces of the CDR regions, F94 of CDR-L3 forms a π-π interaction with Y449 of RBD (Figure 4 A). Furthermore, Y505 of RBD is surrounded by a hydrophobic patch that includes W97 and L95 of CDR-L3, as well as Y33 of CDR-H1 (Figure 4B). In contrast, Y33 of CDR-L1 makes a hydrophobic interaction with L455 and polar interactions with K417 and Y453 (Figure 4C). The main chain of the DE loop of light chain forms hydrogen bonds with RBD (N487) and is sandwiched between Y489 and F486 (Figure 4D). Numerous hydrophobic interactions were observed in the CDR-H3 heavy chain, including those formed by three continuous tyrosines, namely, Y103, Y104, and Y105 (Figures 4E and 4F), which recognize the CoV1/2 conserved region. Y104 protrudes deeply to establish π-cation interactions with K417 and R403 of RBD and hydrogen bonds with E406 and Q409 (Figure 4F). However, N58 and S56 of CDR-H2 recognize the main-chain N atoms of G502 and G504 of the RBD loop (Figure 4G). In addition, N52 of CDR-H2 and Y33 of CDR-H1 make hydrogen bonds with D405 (Figure 4G). Finally, the CoV2 RBD epitopes recognized by the NT-193 light chain encompass most of the ACE2 binding site (the 595 Å2 surface area is buried), whereas the CDR-H3 of its heavy chain further mediates NT-193 anchoring into the CoV-1/2 conserved area, presumably relevant for the formation of the closed prefusion trimer, with the 498 Å2 surface area buried by heavy chain (Figure S4C). This binding mode is in stark contrast to the representative class 1 antibodies B38 and REGN10933 that mainly rely on heavy chain to cover ACE2 binding sites (Figures 3F and S5). Overall, the previously mentioned binding residues of RBD for interaction with NT-193 are relatively conserved and functionally maintained within clade 1 sarbecoviruses that include clade 1a (CoV1 related) and clade 1b (CoV2 related) (Figure 4H). With coordinated binding by heavy and light chains, NT-193 is capable of targeting these amino acid residues that are conserved among clade 1 sarbecoviruses, resulting in broad sarbecovirus-neutralizing activity (Figure 3G).
Figure 4.
Key footprints of NT-193 are relatively conserved in clade 1 sarbecoviruses
(A–G) Detailed recognition mode of CDR-L3 (A), CDR-L3 and CDR-H1 (B), CDR-L1 and CDR-H3 (C), DE loop of light chain (D), CDR-H3 (E and F), and CDR-H1 and CDR-H2 (G) of NT-193. The coloring is the same as in Figure 3. All conserved residues (see H) are included and labeled. The dotted lines are indicated as polar interactions.
(H) Top, amino acid sequence alignment of all NT-193 binding residues in clade 1 sarbecoviruses. Bottom, conservation of each residue was analyzed by WebLogo.
See also Figures S4 and S5 and Tables S1 and S2.
NT-193 is resistant to escape mutations of SARS-CoV-2
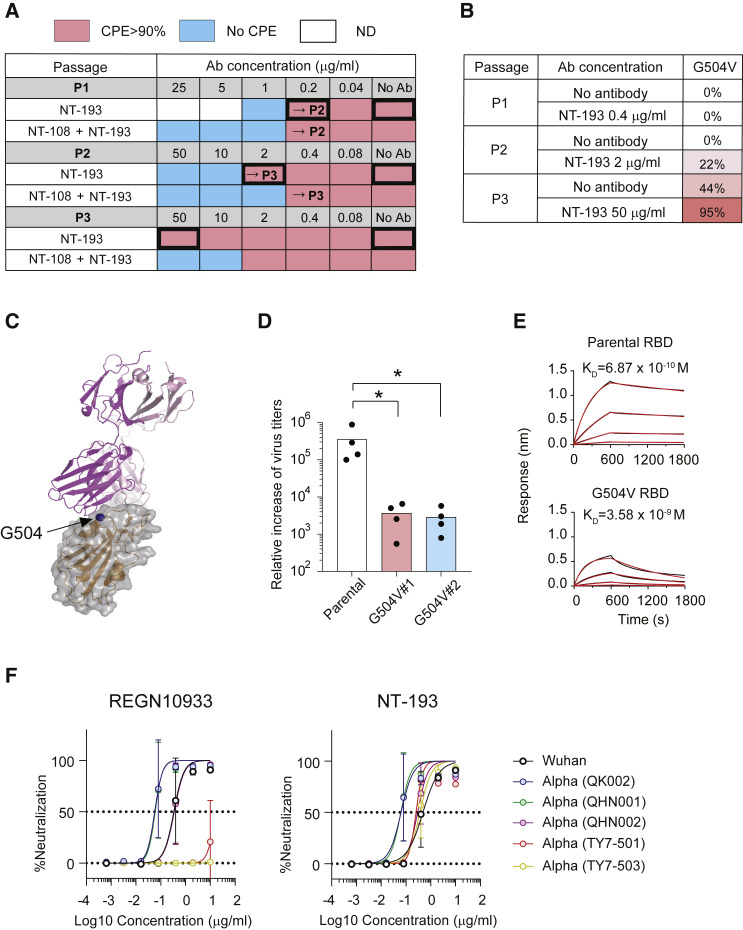
Potential resistance to neutralization escape was evaluated by escape mutation screening in vitro. In this assay, the viruses were passaged up to three times in the VeroE6/TMPRSS2 cell line in the presence of serially diluted IgG1 antibodies (Figure 5 A). After passaging three times, we observed cytopathic effects (CPEs) by the viruses that escaped from neutralization by NT-193 IgG1, but not by the NT-108/NT-193 cocktail that was set as the control for suppressing the emergence of escape variants (Figure 5A). The RBD mutations required for NT-193 escape were determined by viral RNA sequencing from CPE+ wells (Figure 5B). More than 90% of the sequenced viruses carried a single replacement mutation at position 504 within RBD in two separate experiments. The outcome of escape mutation screening largely depends on initial viral stock and culture condition; therefore, we repeated the experiments using a different viral stock and culture condition and examined whether the same escape mutations are reproducibly isolated (Figure S6A) (Kobayashi et al., 2008). We confirmed that the same G504V mutation emerged as a single RBD mutation using NT-193 IgG3 under a different screening condition. Whereas the amino acids next to G504 (N501, G502, and Y505) are directly interacting with ACE2 (Figure 3F), G504 is not in the ACE binding site and is located at the outer ledge of the NT-193 heavy-chain footprint on the CoV1/CoV2 conserved site (Figures 3D, 4G, and 5C), likely enabling escape from heavy-chain recognition. The possible impact of the G504V mutation on viral fitness was further assessed using G504V mutant clones, which were confirmed to remain resistant to NT-193 after cloning (Figure S6B). G504V mutants poorly replicated in VeroE6 cell lines (Figure 5D), and the defect can be at least partly explained by the reduced ACE2 affinity of G504V RBD relative to that of parental RBD (Figure 5E). Thus, the NT-193-escaping G504V mutation imposed a large cost on viral fitness, thereby losing the competitive advantage over circulating CoV2. This idea is supported by the rare frequency (<0.001%) of G504V mutation among CoV2 sequences in the GISAID database (as of August 3, 2021).
Figure 5.
NT-193 is resistant to escape mutations of SARS-CoV-2
(A) Passaging results of escape mutant screening are shown, with the qualitative percentage of CPE observed in each dilution (red, >90% CPE; blue, no CPE; white, not determined). Bold-bordered boxes indicate dilutions that were sequenced for identifying escape mutations.
(B) Identified escape mutations by deep sequencing of passaged virus. Viral RNAs were isolated at day 4 after infection from wells with the highest monoclonal antibody concentration among detectable CPE. RNA sequencing (RNA-seq) analysis was performed to identify changes relative to the input virus. Percentages of sequencing reads that contained the indicated mutant sequence are shown.
(C) Position of G504 is highlighted in dark blue.
(D) VeroE6 cells were infected with virus solution, including parental or G504 mutant viruses, and then the increase of virus titers from day 0 to day 3 was plotted. Each dot represents the number from the individual experiment, and the combined data from four independent experiments are shown. Statistical significance is indicated above the columns; ∗p < 0.05 (two-tailed Mann-Whitney test).
(E) Binding affinity between RBD (parental or G504 mutant) and ACE2 protein was determined by biolayer interferometry.
(F) Serially diluted REGN10933 and NT-193 were mixed with the indicated virus strains and then loaded onto VeroE6/TMPRSS2 cell lines. At day 3, the CPEs were visualized by crystal violet staining.
Values represent mean ± SD. The representative data from three independent experiments are shown (E and F). See also Figure S6.
CoV2 VOC have recently emerged with potential alternation in pathogenicity, transmissibility, and antigenicity. We thus evaluated the cross-neutralizing activity of NT-193 against these naturally emerged VOC strains (3 from Alpha and 2 from Gamma) (Figure 5F). The Gamma variants carry the E484K mutation, causing a 10- to 1,000-fold reduction in the neutralization activities of RBS antibodies, including REGN10933 and LY-CoV555, upon monotherapeutic usage (Wang et al., 2021b). In addition, the Gamma variants bear the K417 mutation that permits escape from REGN10933 and LY-COV016 (Baum et al., 2020b; Shi et al., 2020; Starr et al., 2021). Despite these mutations, NT-193 IgG1 neutralized both VOCs even at the lower IC50 required for neutralizing the original Wuhan strain, whereas REGN10933 reduced neutralizing activity to the Gamma variants below the detection limit (IC50 > 1 μg/mL) (Figure 5F). Increases in VOC-neutralizing activities have been previously observed in other antibodies, but the mechanisms remain to be addressed (Hoffmann et al., 2021; Dejnirattisai et al., 2021; Wang et al., 2021a, 2021b; Zhou et al., 2021). Although the Gamma variants remained susceptible by NT-193, the SPR analysis showed that NT-193 Fab bound the RBD variant with Gamma mutations (K417T/E484K/N501Y) at high (KD = 3.0 × 10−8 M) but reduced affinity compared with the original RBD (KD = 1.2 × 10−9 M), suggesting the hydrophobic K417-Y104 interaction somewhat contributes to NT-193 binding. These data underscore the resistance of NT-193 to emerging mutations that allow viral escape from previously isolated RBS antibodies in the clinical stage.
Prophylactic and therapeutic use of NT-193 protects against SARS-CoV-2 infection in hamsters
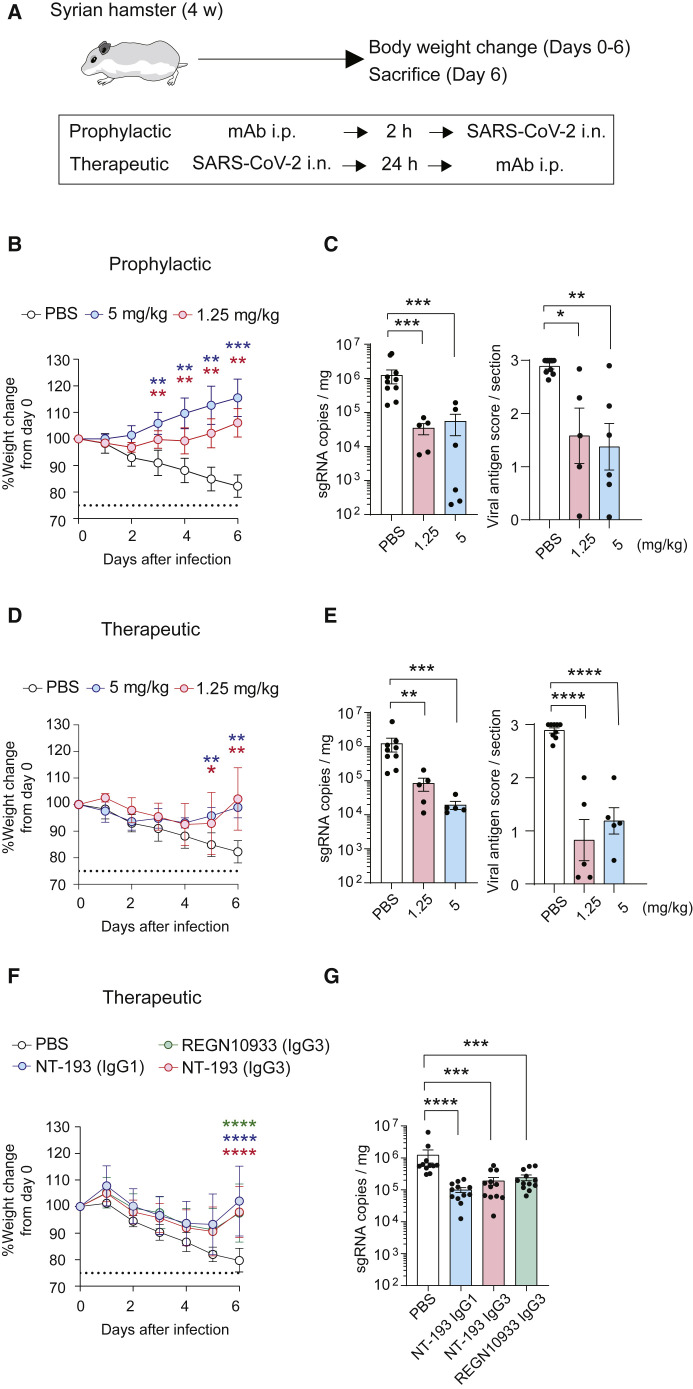
The Syrian hamster model has been used to assess prophylactic and therapeutic activities of several monoclonal antibodies against CoV2 infection (Baum et al., 2020a; Rogers et al., 2020). This model recapitulates non-lethal but severe forms of CoV2 infection with pneumonia and body weight loss of infected animals (Imai et al., 2020). Using this Syrian hamster model, we examined the prophylactic and therapeutic efficacy of NT-193 monotherapy. Previously isolated monoclonal antibodies provided prophylactic or therapeutic effects when given a >0.25–0.5 mg/kg dosage or a >4 mg/kg dosage, respectively (Andreano et al., 2021; Baum et al., 2020a; Rogers et al., 2020). In this study, the hamsters were treated with 5 or 1.25 mg/kg of IgG3 antibodies at 2 h before (prophylaxis) or 1 day after (therapy) intranasal challenge with CoV2 (Wuhan strain) (Figure 6 A). Prophylaxis with monotherapy prevented severe body weight loss in the hamsters until day 6 after infection (Figure 6B). The prophylactic effects were more pronounced in the 5 mg/kg group than in the 1.25 mg/kg group, and the effects became evident as early as 2 days after infection. The difference in body weight between treated and untreated groups further widened at later time points, resulting in reduced viral genomes and antigens in the lungs at day 6 after infection (Figure 6C). Similar to prophylaxis, all therapeutic treatments, including the groups at the 1.25 mg/kg dosage, prevented body weight loss in hamsters by day 6 after infection (Figure 6D) and reduced the viral genomes and antigens detected in the lungs (Figure 6E). Although therapeutic efficacy was slightly attenuated compared with prophylactic efficacy, the efficacy of NT-193 IgG3 at the 1.25 mg/kg dosage was comparable to that provided by REGN10933 IgG3 of the same dosage, which is one of the most potent neutralizing and protective antibodies to the original CoV2 strain (Figures 6F and 6G) (Baum et al., 2020b). In addition, despite better neutralizing activity of the IgG3 subclass in vitro, therapeutic efficacies of the IgG3 and IgG1 subclasses were equivalent under the challenge condition that induced more profound weight loss (<85% of initial body weight) than previous studies (Baum et al., 2020a; Imai et al., 2020; Rogers et al., 2020).
Figure 6.
Prophylactic and therapeutic use of NT-193 protects against SARS-CoV-2 infection in hamsters
(A) Schematic of the experimental design to assess prophylactic and therapeutic effects of NT-193. i.p., intraperitoneal; i.n., intranasal.
(B–E) Body weight changes of CoV2-challenged hamsters were monitored daily, followed by either prophylactic (B) or therapeutic (D) antibody injection at the indicated doses. The value represents mean ± SD. CoV2 viral loads in lung tissues of hamsters receiving either prophylactic (C) or therapeutic (E) monoclonal antibodies at the indicated doses are shown. Left, subgenomic RNA copies. Right, N antigen score.
(F and G) Hamsters were therapeutically injected by the indicated antibodies (1.25 mg/kg), and body weights (F) and subgenomic RNA copies (G) were monitored. Each dot represents the result from an individual animal. Dashed lines indicate the humane endpoint (75% weight change).
The combined data from more than two independent experiments (n = 5–12 per group) are shown. Statistical significance is indicated above the columns; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (two-tailed Mann-Whitney test).
Discussion
We showed here a RBS antibody that acquired neutralization potency and breadth for SARS-related coronaviruses, including emerging CoV2 VOCs. Several lines of evidence demonstrated that this antibody acquired broad neutralization activity by two strategies. The first strategy is coordinated antigen recognition by heavy and light chains. Cross-reactivity is mainly acquired through recognition of conserved sites by heavy chains, and neutralization potency is achieved through broad masking of the RBS by light chains. Such functional compartmentalization in heavy and light chains probably permits the breadth and potent neutralization by a single antibody clone, despite these characters being incompatible in most neutralizing RBS antibodies. X-ray crystallography revealed that contact residues for high-affinity binding by NT-193 are relatively conserved in clade 1b (CoV2 related) and clade 1a (CoV1 related) sarbecoviruses, probably because of the necessity for formation of a closed prefusion trimer and/or ACE2 binding. Such a conformational constraint likely makes the key footprints of NT-193 the vulnerable sites for emerging CoV2 variants and circulating CoV1-related viruses in animal reservoirs as the next pandemic potential (Hu et al., 2017).
The second strategy of NT-193 for acquiring robust cross-neutralization activity is the avidity effect by IgG3 switching. The avidity effect increased CoV1 neutralization activity solely by switching from IgG1 to IgG3, despite the similarity of apparent Fab affinities to RBD. We speculate that IgG3 switching potentiates neutralization activity of NT-193 through the long and flexible hinge of the IgG3 subclass, as is the case for HIV-1 antibodies (Richardson et al., 2019; Scharf et al., 2001). Supporting this, we observed that increased amounts of NT-193 IgG3 bound to RBD antigens that were sparsely coated on plates. In addition, NT-193 recognizes a cryptic epitope that is only accessible by antibody under the open configuration of RBD. Transition between open and closed configurations of RBD generates heterogenous S-trimer structures with different combinations of open and closed RBD conformations. During the steady state, CoV1 S trimers have higher frequencies of open configuration compared with CoV2 S trimers (Walls et al., 2019). In addition, the numbers of S trimers present on the individual virion are different between CoV2 (24 ± 9 per virion) (Ke et al., 2020) and CoV1 (65 per virion) (Beniac et al., 2006). Therefore, the difference in the open/closed RBD conformation and S-trimer density between CoV1 and CoV2 could affect inter- and/or intra-S-trimer cross-linking by NT-193 IgG3.
In vitro escape mutation screening identified G504 as another key footprint of NT-193. Recently, the in vitro evolution approach with random mutagenesis and selection by yeast display libraries was applied to generate an anti-SARS-related virus-neutralizing antibody, ADG-2 (Rappazzo et al., 2021). This engineered process elevated the potency and breadth against SARS-related virus neutralization, and coincidently, the binding mode of this antibody depended on G504. G504V mutation is infrequently detected (<0.001%) among CoV2 sequences deposited in the GISAID database. Together with attenuated ACE2 binding and replication of G504V mutants, G504 mutations likely lose the competitive advantage over circulating CoV2. The low-resolution cryoelectron microscopy (cryo-EM) structure showed that ADG-2 recognizes only a small portion of the ACE2 binding site. Therefore, although NT-193 and ADG-2 share G504 as the key footprint, it is important to stress that the NT-193 binding mode differs from that of ADG-2, including coordinated binding by heavy and light chains and overall ACE2 mimicry for the binding angle. Furthermore, NT-193 uses the near-germline heavy chain, making NT-193-like antibodies more attractive targets of vaccines, because poorly mutated antibodies can be elicited by immunization with less labor and time than heavily mutated antibodies. In addition, NT-193 may leave more room for potentiating the neutralizing activity by introducing additional affinity-enhancing mutations during affinity-based maturation of humoral immune responses.
NT-193 was originally isolated from humanized mice, but the NT-193-like antibodies sharing structural and functional characters are potentially elicited in COVID-19 patients by following reasons. First, IgG3 antibodies are elicited in severe COVID-19 patients during the acute phase (Amanat et al., 2020; Zohar et al., 2020). Second, the humoral response in severe patients is characterized by the robust extrafollicular responses that harbor IGVH4-34-encoded antibodies at elevated frequencies (Woodruff et al., 2020). Third, IGHV4-34-encoded monoclonal antibodies with CoV1/CoV2 cross-reactivity were recovered from COVID-19 patients and deposited in the database (Raybould et al., 2021). Even though IGHV4-34-encoded antibodies are present in extrafollicular response in COVID-19 patients, infrequent recovery of the similar antibodies from memory B cell repertoires suggests that IGHV4-34-encoded antibodies are less competitive than those with convergent VH genes that are frequently recovered from memory repertoires across multiple individuals (Andreano et al., 2021; Robbiani et al., 2020; Yuan et al., 2020b). Rational design of vaccine formulations and adjuvants may be required to overcome such possible roadblocks that prevent IGHV4-34-encoded antibodies from recruitment into memory B cell repertoires.
Neutralizing RBS antibodies are frequently encoded by convergent VH genes that share similar binding modes and footprints (Robbiani et al., 2020; Yuan et al., 2020b). Such focused antibody responses could increase the risk of generating antibody escape mutations because of limited diversity. Indeed, persistent infection in an immunocompromised COVID-19 patient generated several variants, including the one with the E484K mutation (Choi et al., 2020), allowing escape from convergent neutralizing antibodies (Gaebler et al., 2021). The emerging VOCs possess the E484K mutation (Tegally et al., 2021), raising the concern of viral escapes from vaccine-induced immunity and therapeutic antibodies. Therefore, countermeasures to boost resistance to escape mutations need to be prepared. We believe that NT-193 is one of the candidates possessing such activities via its distinct binding mode that is not used by convergent antibodies. Further analysis of NT-193 will guide the development of broad protective interventions against emerging CoV2 variants and SARS-related viruses with pandemic potential.
Limitations of the study
A limitation of this study is that there was no apparent difference in the therapeutic activities of NT-193 between the IgG1 and the IgG3 subclasses against the original CoV2 strain. This point needs to be further investigated using either CoV1 or WIV-1 for challenge, because more robust enhancement of neutralization was observed by using these viruses. In addition, it is important to assess the therapeutic activities under the VOC-challenged model. Optimal therapeutic protection by NT-193 in vivo could be regulated by the IgG Fc effector functions, in addition to the neutralization activity in vitro (Winkler et al., 2021), but this point remains to be clarified.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| REGN10933 | Hansen et al., 2020 | N/A |
| C002 | Robbiani et al., 2020 | N/A |
| REGN10987 | Hansen et al., 2020 | N/A |
| CR3022 | ter Meulen et al., 2006 | Accession#DQ168569.1, DQ168570.1 |
| S309 | Pinto et al., 2020 | N/A |
| 51F12 (anti-nuclear antibody) | Sakakibara et al., 2017 | N/A |
| 72H11 (anti-nuclear antibody) | Sakakibara et al., 2017 | N/A |
| Human IgG1 | Southern Biotech | Cat#0151K-01 |
| HPR-conjugated goat anti-human IgG | Southern Biotech | Cat#2040-05 |
| Hyper-immune rabbit serum raised against the GST-tagged N protein of SARS-CoV-2 | In-house | N/A |
| Histofine® Simple Stain MAX PO (R) | Nichirei Biosciences | Cat#424141 |
| Alexa Fluor 594 goat anti-Human IgG | Thermo Fisher Scientific | Cat#A-11014 |
| Anti-mouse FcγRII/III | In-house | 2.4G2 |
| Anti-mouse CD43, biotinylated | BD Biosciences | Cat#553269 |
| Anti-mouse CD90, biotinylated | Thermo Fisher Scientific | Cat#13-0903-85 |
| Anti-mouse CD3, biotinylated | BioLegend | Cat#100304 |
| Anti-mouse c-kit, biotinylated | BioLegend | Cat#105803 |
| Anti-mouse F4/80, biotinylated | BioLegend | Cat#123106 |
| Anti-mouse Gr-1, biotinylated | Thermo Fisher Scientific | Cat#13-5931-85 |
| Anti-mouse CD4, biotinylated | BioLegend | Cat#100404 |
| Anti-mouse CD8, biotinylated | BioLegend | Cat#100704 |
| Anti-mouse CD11b, biotinylated | BioLegend | Cat#101204 |
| Anti-mouse Ter119, biotinylated | BioLegend | Cat#116204 |
| Anti-mouse CD93, biotinylated | Thermo Fisher Scientific | Cat#13-5892-85 |
| Anti-mouse CD11c, biotinylated | BioLegend | Cat#117304 |
| Anti-mouse CD138, biotinylated | BD Biosciences | Cat#553713 |
| Anti-human IgD, biotinylated | BD Biosciences | Cat#555777 |
| Anti-mouse B220, BV786 conjugated | BioLegend | Cat#103246 |
| Anti-mouse CD38, AF700 conjugated | Thermo Fisher Scientific | Cat#56-0381-82 |
| Rabbit anti-SARS-CoV-2 N antibody | Nagata et al., 2021 | N/A |
| Bacterial and virus strains | ||
| h-CoV-19/Japan/TY-WK-521/2020 | National Institute of Infectious Diseases | EPI_ISL_408667 |
| hCoV-19/Japan/QK002/2020 | National Institute of Infectious Diseases | EPI_ISL_768526 |
| hCoV-19/Japan/QHN001/2020 | National Institute of Infectious Diseases | EPI_ISL_804007 |
| hCoV-19/Japan/QHN002/2020 | National Institute of Infectious Diseases | EPI_ISL_804008 |
| hCoV-19/Japan/TY7-501/2021 | National Institute of Infectious Diseases | EPI_ISL_833366 |
| hCoV-19/Japan/TY7-503/2021 | National Institute of Infectious Diseases | EPI_ISL_877769 |
| QH-329-037-p3d1 | National Institute of Infectious Diseases | DRA012137 |
| SARS coronavirus Frankfurt 1 | Thiel et al., 2003 | AY291315 |
| VSV pseudovirus bearing SARS-CoV-2 spike protein | Tani et al., 2021 | N/A |
| VSV pseudovirus bearing WIV-1 spike protein | In-house | N/A |
| Escherichia coli strain DH5alpha | Clontech | Cat#9057 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 RBD (amino acids: 331–529) | In-house | MN994467 |
| G504V RBD | In-house | N/A |
| SARS-CoV-2 spike protein (amino acids: 1-1213) | In-house | MN994467 |
| SARS-CoV-1 RBD (amino acids: 310-509) | In-house | EU371563 |
| SARS-CoV-1 spike protein (amino acids: 15-1195) | In-house | EU371563 |
| MERS-CoV RBD (amino acids: 367-606) | In-house | KF192507 |
| MERS-CoV spike protein (amino acids: 13-1296) | In-house | KF192507 |
| HCoV-NL63 spike protein (amino acids: 22-1293) | In-house | NC_005831 |
| HCoV-OC43 spike protein (amino acids: 12-1299) | In-house | NC_006213 |
| HCoV-229E spike protein (amino acids: 12-1109) | In-house | NC_002645 |
| HCoV-HKU1 spike protein (amino acids: 13-1293) | In-house | NC_006577 |
| Recombinant Human ACE2 | BioLegend | Cat#792006 |
| HiTrap Protein G HP Columns | Cytiva | Cat#17-0404-01 |
| StrepTactin Sepharose | iba | Cat#2-5030-010 |
| cOmplete His-Tag Purification Resin | Roche | Cat#0589682001 |
| Superose TM 6 Increase 10/300 GL | Cytiva | Cat#29-0915-96 |
| Superdex TM 200 Increase 10/300 GL | Cytiva | Cat#28-9909-44 |
| Superdex TM 200 10/300 GL | Cytiva | Cat#17517501 |
| Gel Filtration Standard | BioRad | Cat#1511901 |
| TALON® Metal Affinity Resin | Clontech | Cat#635504 |
| HisTALON Buffer Set | Clontech | Cat#635651 |
| Ni-NTA agarose | QIAGEN | Cat#30230 |
| Recombinant mouse IL-2 | Peprotech | Cat#212-12 |
| Recombinant mouse IL-4 | Peprotech | Cat#214-14 |
| Recombinant mouse IL-5 | Peprotech | Cat#215-15 |
| Fetal bovine serum | Biowest | Cat#S1780-500 |
| Fetal bovine serum | Hyclone | Cat#SH30396.03 |
| Normal goat serum | Jackson Immuno Research Laboratories | Cat#005-000-121 |
| DNA (Calf Thymus) | Worthington Biochemical Corporation | Cat#LS002105 |
| Insulin from bovine pancreas | Sigma-Aldrich | Cat#I5500 |
| Lipopolysaccharides from Escherichia coli O111:B4 | Sigma-Aldrich | Cat#L3024 |
| AddaVax | InvivoGen | Cat#vac-adx-10 |
| Freund’s Adjuvant, Incomplete | Sigma | Cat#F5506-10ML |
| iSeq 100 Reagent | Illumina | Cat#20021533 |
| Low glucose DMEM | Fujifilm Wako Pure Chemicals | Cat#041-29775 |
| DMEM (High Glucose) | Fujifilm Wako Pure Chemicals | Cat#044-29765 |
| RPMI | Fujifilm Wako Pure Chemicals | Cat#189-02025 |
| Opti-MEM I medium | Thermo Fisher Scientific | Cat#31985070 |
| NEAA | Thermo Fisher Scientific | Cat#11140-050 |
| HEPES | Thermo Fisher Scientific | Cat#15630-080 |
| HEPES | DOJINDO | Cat#340-01376 |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat#11360-070 |
| 2-mercapt ethanol | Thermo Fisher Scientific | Cat#21985-023 |
| Geneticin | Thermo Fisher Scientific | Cat#10131-027 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15140-122 |
| Poly-L-Lysine | Fujifilm Wako Pure Chemicals | Cat#333-30751 |
| Bovine serum albumin | Sigma-Aldrich | Cat#A2153 |
| Tween-20 | Fujifilm Wako Pure Chemicals | Cat#167-11515 |
| Triton X-100 | Nacalai Tesque | Cat#12969-25 |
| Can Get Signal #2 | TOYOBO | Cat#NKB-301 |
| OPD substrate | Sigma-Aldrich | Cat#P8287 |
| Formalin | Fujifilm Wako Pure Chemicals | Cat#062-01661 |
| Crystal violet solution | Sigma-Aldrich | Cat#V5265 |
| Polyethylene glycol 8,000 | Hampton Research | Cat#HR2-535 |
| Ethylene Glycol | Fujifilm Wako Pure Chemicals | Cat#058-00986 |
| 3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide | Nacalai Tesque | Cat#23547-34 |
| Tissue-Tek® Paraffin WaxII60 | Sakura Finetek Japan | Cat#7810 |
| Retrieval Solution pH6 | Nichirei Biosciences | Cat#415281 |
| 3,3′-diaminobenzidine | Sigma-Aldrich | Cat#D5637 |
| Hematoxylin cryst. (C.I. 75290) | Merck | Cat#1.04302.0025 |
| Multimount 480 | Matsunami Glass | Cat#FM48005 |
| Fluoro-KEEPER Antifade reagent with DAPI | Nacalai Tesque | Cat#12745-74 |
| TRIzol | Thermo Fisher Scientific | Cat#15596 |
| Streptavidin MicroBeads | Miltenyi Biotec | Cat#130-048-101 |
| Streptavidin-eFluor450 | Thermo Fisher Scientific | Cat#48-4317-82 |
| Streptavidin-HRP | Southern Biotech | Cat#7100-05 |
| Streptavidin-HRP | Thermo Fisher Scientific | Cat#434323 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| HBS-EP Buffer | Cytiva | Cat#BR100188 |
| HBS-EP+ 10X | Cytiva | Cat#BR100669 |
| Glycine 1.5 | Cytiva | Cat#BR100354 |
| NaOH 50 | Cytiva | Cat#BR100358 |
| Human-b2-microglobulin | In-house | NP_004039.1 |
| Biotinylated-human-b2-microglobulin | In-house | N/A |
| Critical commercial assays | ||
| RNeasy mini kit | QIAGEN | Cat#74104 |
| Direct-zol RNA Miniprep | Zymo research | Cat#R2052 |
| Gibson Assembly Master Mix | New England Biolabs | Cat#E2611 |
| SuperScript VILO Master Mix | Thermo Fisher Scientific | Cat#11755250 |
| Wizard SV Gel and PCR Clean-Up System | Promega | Cat#A9282 |
| Wizard Plus SV Minipreps DNA Purification Systems | Promega | Cat#A1460 |
| PureYield Plasmid Midiprep System | Promega | Cat#A2495 |
| R-Phycoerythrin Labeling Kit - NH2 | DOJINDO | Cat#LK23 |
| Allophycocyanin Labeling Kit - NH2 | DOJINDO | Cat#LK21 |
| QIAseq FX DNA Library kit | QIAGEN | Cat#180473 |
| Bright-Glo luciferase assay system | Promega | Cat#E2620 |
| Expi293 expression system | Thermo Fisher Scientific | Cat#A29133 |
| Cell counting kit 8 | Fujifilm Wako Pure Chemicals | Cat#343-07623 |
| QuantiTect Probe RT-PCR Kit | QIAGEN | Cat#204443 |
| Kallestad HEp-2 IFA kit | Bio-Rad | Cat#30471 |
| Biotin Capture Kit | Cytiva | Cat#28920233 |
| Amine Coupling Kit | Cytiva | Cat#BR100050 |
| Deposited data | ||
| Crystal structure of RBD-NT193 complex | This paper | PDB: 7E5O |
| NT-108 | This paper | Accession#MW619740, MW619741 |
| NT-193 | This paper | Accession#MW619742, MW619743 |
| BWA-mem version 0.7.13-r1126 | Li and Durbin, 2009 | https://github.com/lh3/bwa |
| VarScan version 2.4.3 | Koboldt et al., 2012 | http://varscan.sourceforge.net/using-varscan.html |
| Experimental models: Cell lines | ||
| VeroE6/TMPRSS2 cells | JCRB Cell Bank | JCRB1819 |
| VeroE6 cells | ATCC | CRL-1586 |
| NB21.2D9 | Kuraoka et al., 2016 | N/A |
| Experimental models: Organisms/strains | ||
| TC-mAb mouse | Satofuka et al., 2020. | N/A |
| Syrian hamster | Japan SLC | N/A |
| Oligonucleotides | ||
| CpG ODN1760 | Eurofins genomics | N/A |
| 5′- AAATTTTGGGGACCAGGAAC −3′ | Eurofins genomics | NIID_2019-nCOV_N_F2 |
| 5′- TGGCAGCTGTGTAGGTCAAC −3′ | Eurofins genomics | NIID_2019-nCOV_N_R2 |
| 5′- FAM- ATGTCGCGCATTGGCATGGA-BHQ −3′ | Eurofins genomics | NIID_2019-nCOV_N_P2 |
| ARTIC-N2 primer | Itokawa et al., 2020 | https://www.protocols.io/researchers/kentaro-itokawa |
| Recombinant DNA | ||
| Plasmid: pCMV-CoV2S-foldon-avi | This paper | N/A |
| Plasmid: pCAGGS-COV2RBD-avi | This paper | N/A |
| Plasmid: pCAGGS-COV2RBD_G504V | This paper | N/A |
| Plasmid: pMT-COV1_S | This paper | N/A |
| Plasmid: pMT-HCoV-NL63_S | This paper | N/A |
| Plasmid: pMT-HCoV-OC43_S | This paper | N/A |
| Plasmid: pMT-HCoV-229E_S | This paper | N/A |
| Plasmid: pMT-HCoV-HKU1_S | This paper | N/A |
| Plasmid: pHLsec-CoV2RBD | This paper | N/A |
| Plasmid: pHLsec-CoV1RBD | This paper | N/A |
| Plasmid: pHLsec-MERSRBD | This paper | N/A |
| Plasmid: phIgG1-NT193 | This paper | N/A |
| Plasmid: phIgG3-NT193 | This paper | N/A |
| Plasmid: phIgk-NT193 | This paper | N/A |
| Plasmid: phIgG1-NT108 | This paper | N/A |
| Plasmid: phIgk-NT108 | This paper | N/A |
| Plasmid: phIgG1-REGN10933 | This paper | N/A |
| Plasmid: phIgk-REGN10933 | This paper | N/A |
| Plasmid: phIgG1-REGN10987 | This paper | N/A |
| Plasmid: phIglambda-REGN10987 | This paper | N/A |
| Plasmid: phIgG1-S309 | This paper | N/A |
| Plasmid: phIgk-S309 | This paper | N/A |
| Plasmid: phIgG1-C002 | This paper | N/A |
| Plasmid: phIgk-C002 | This paper | N/A |
| Plasmid: pVRC-NT193Fab | This paper | N/A |
| Software and algorithms | ||
| Graphpad Prism 9 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| StepOne Software v2.3 | Applied Biosystems | https://www.thermofisher.com/jp/ja/home/technical-resources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html |
| FLOWJO X 10.7.2 | BD | https://www.bdbiosciences.com/en-in/products/software/flowjo-v10-software |
| Octet Data Analysis Software 11.1.0.4 | Sartorius | https://www.sartorius.com/en/products/protein-analysis/octet-systems-software |
| BIAevaluation version 4.1.1 | Cytiva | https://www.cytivalifesciences.com/en/gb/shop/protein-analysis/spr-label-free-analysis |
| T200 Evaluation version 1.0 | Cytiva | https://www.cytivalifesciences.com/en/gb/shop/protein-analysis/spr-label-free-analysis |
| FV10-ASW 4.2 Viewer | Olympus | N/A |
| Origin 2017 | OriginLab | https://www.originlab.com/ |
| UNICORN 6.3 | Cytiva | https://www.cytivalifesciences.com/en/gb/shop/chromatography |
| Imaging Software cellSens ver. 1.16 | Olympus | N/A |
| BWA-mem version 0.7.13-r1126 | Li and Durbin, 2009 | https://github.com/lh3/bwa |
| VarScan version 2.4.3 | Koboldt et al., 2012 | http://varscan.sourceforge.net/using-varscan.html |
| XDS | Kabsch, 2010 | https://xds.mr.mpg.de/ |
| CCP4 7.1 | Winn et al., 2011 | http://www.ccp4.ac.uk |
| PHENIX version 1.18 | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| The PyMol Molecular Graphics System | Schrödinger, LLC | https://pymol.org/ |
| PDBePISA | Krissinel and Henrick, 2007 | https://www.ebi.ac.uk/pdbe/pisa/ |
| Coot version 0.9.1 | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| MolProbity | Davis et al., 2007 | https://www.phenix-online.org/ |
| Other | ||
| StepOnePlus Real-Time PCR System | Applied Biosystems | https://www.thermofisher.com/order/catalog/product/4376598#/4376598 |
| iSeq 100 Sequencing System | Illumina | https://www.illumina.com/systems/sequencing-platforms/iseq/order-iseq-100.html |
| FACSAria III Cell Sorter | BD Biosciences | https://www.bdbiosciences.com/ja-jp/products/instruments/flow-cytometers/research-cell-sorters/bd-facsaria-iii |
| iMark microplate reader | Bio-Rad | https://www.bio-rad.com/ja-jp/product/imark-microplate-absorbance-reader?ID=58cca7aa-d943-4e32-9bea-0fe5d140fb9e |
| Epoch2 | Biotek | https://www.biotek.com/products/detection-microplate-readers/epoch-2-microplate-spectrophotometer/ |
| GloMax Navigator Microplate Luminometer | Promega | https://www.promega.com/products/microplate-readers-fluorometers-luminometers/microplate-luminometers/glomax-navigator-system/?catNum=GM2000&cs=y |
| FV1000 confocal laser scanning microscope | Olympus | https://www.olympus-lifescience.com/en/technology/museum/micro/2004/ |
| System Microscope BX53 | Olympus | https://www.olympus-lifescience.com/en/microscopes/upright/bx53f2/ |
| Digital microscope camera DP71 | Olympus | https://www.olympus-lifescience.com/en/technology/museum/micro/2006/ |
| TissueLyser LT instrument | QIAGEN | https://www.qiagen.com/jp/shop/pcr/tissuelyser-lt/ |
| ÄKTA pure protein purification system | Cytiva | https://www.cytivalifesciences.com/en/us/shop/chromatography/chromatography-systems/akta-pure-p-05844 |
| Biacore 3000 | Cytiva | https://www.cytivalifesciences.com/en/us/shop/biacore-3000-goldseal-p-02795 |
| Biacore T200 | Cytiva | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-free-analysis/systems/biacore-t200-p-05644 |
| Biolayer Interferometry | Sartorius | OctetRED96e system |
| LS Columns | Miltenyi Biotec | Cat#130-042-401 |
| Sensor chip CM5 Series S | Cytiva | Cat#29149603 |
| Streptavidin Biosensors | Sartorius | Cat#5020 |
| Microscope slide | Matsunami Glass | Cat#FRC-11 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yoshimasa Takahashi. (ytakahas@niid.go.jp).
Materials availability
TC-mAb mice are available from Tottori University or Trans Chromosomics, Inc. on request as availability permits. All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Mice
TC-mAb mice were established as previously described (Satofuka et al., 2020). In brief, an artificial chromosome containing human Ig heavy chain locus (hChr.14) and Ig kappa light chain locus (hCh.2) was generated. The mice carrying the artificial chromosome (IGHK-NAC) were mated with endogenous Ig-knockout strain. All mice were maintained under specific pathogen free conditions and female were used at 6 weeks of age. All work was performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the National Institute of Infectious Diseases, Japan.
Hamsters
Syrian hamsters were purchased from Japan SLC. All hamsters were maintained under specific pathogen free conditions and female were used at 1 month of age. All work was performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the National Institute of Infectious Diseases, Japan.
Method details
Recombinant S antigens
The human codon-optimized nucleotide sequence coding for the S protein of the CoV2 isolate (GenBank: MN994467) was synthesized commercially (Eurofins Genomics). The RBD (amino acids: 331–529), along with the signal peptide (amino acids 1–20; MIHSVFLLMFLLTPTESYVD) plus a histidine tag, was cloned into the mammalian expression vector pCAGGS. A soluble version of the S protein (amino acids 1–1213), including the T4 foldon trimerization domain, a histidine tag, and a strep-tag, was cloned into the mammalian expression vector pCMV. The protein sequence was modified to remove the polybasic cleavage site (RRAR to A), and two stabilizing mutations were also introduced (K986P and V987P; wild-type numbering) (Amanat et al., 2020). S proteins of CoV1 (GenBank: EU371563, amino acids 15–1195), MERS-CoV (GenBank: KF192507, amino acids 13–1296), HCoV-NL63 (GenBank: NC_005831, amino acids 22-1293), HCoV-OC43 (GenBank NC_006213, amino acids: 12-1299), HCoV-229E (GenBank: NC_002645, amino acids 12-1109), and HCoV-HKU1 (GenBank: NC_006577, amino acids 13–1293) were also cloned into the pMT vector, including a T4 foldon trimerization domain, a histidine tag, and a strep-tag. The RBDs of the S proteins of CoV1 (amino acids: 310-509) and MERS-CoV (amino acids: 367-606), along with a histidine-tag, were also cloned into the mammalian expression vector pHLsec. G504V RBD mutant of CoV2 was generated by site direct mutagenesis using primers with introduced relevant nucleotide substitutions and Gibson assembly (NEB) based on the RBD expression vector. Recombinant proteins were produced using a Drosophila expression system or Expi293F cells (Thermo Fisher Scientific). Supernatants from transfected cells were harvested on day 5 post-transfection and recombinant proteins were purified using Ni-NTA Agarose (QIAGEN) and/or StrepTactin Sepharose (Sigma-Aldrich). To generate S protein-fluorochrome probes, recombinant RBD and S proteins of CoV2 were conjugated with APC and PE (Dojindo).
Immunization and virus challenge
Mice were intraperitoneally primed with the recombinant CoV2 RBD protein (10 μg/body) in AddaVax adjuvant (InvivoGen), and then successively boosted with CoV1 S, CoV2 RBD, and MERS-CoV RBD proteins in incomplete Freund’s adjuvant (Sigma-Aldrich) plus CpG at three weeks intervals. Sera were collected at day 7 after final boosting to measure titers of antigen-specific human IgG antibodies. Syrian hamsters were anesthetized by i.p. injection of ketamine hydrochloride/xylazine and then i.n. infected with the CoV2 (hCoV-19/Japan/TY-WK-521/2020) virus at a dose of 104 TCID50 in a volume of 80 μl. The antibodies were diluted with PBS at 5 or 1.25 mg/kg of body weight and then administered by i.p. injection either 2 h before viral challenge for prophylactic treatment or 1 day after viral challenge for therapeutic treatment. All hamsters were monitored daily for survival and body weight loss until 6 days after infection. The humane endpoint was set as a weight loss of 25% relative to the initial body weight at the time of viral challenge (day 0).
ELISA and indirect immunofluorescence assay
ELISA plates were coated with either RBDs of CoV1, CoV2, or MERS-CoV; or S-trimer proteins of CoV1, CoV2, MERS-CoV, HCoV-NL63, HCoV-OC43, HCoV-229E, or HCoV-HKU1 at 2 μg/mL. In some experiments, the concentration of coating antigen was changed to 0.1 μg/mL. After blocking with PBS containing 1% BSA, serially-diluted sera, culture supernatants of single cells, or monoclonal antibodies were applied to the plates, and then incubated with goat anti-human IgG-HRP (Southern Biotech). HRP-activity was visualized with an OPD substrate (Sigma-Aldrich), and OD490 was measured using an iMark Microplate Reader (Bio-Rad). To examine the polyreactivity, ELISA plates were coated with either 10 μg/mL dsDNA of calf thymus (Worthington Biochemical Corporation), 5 μg/mL insulin from bovine pancreas, or 10 μg/mL LPS of E.coli O111:B4 (Sigma-Aldrich). For dsDNA, poly-L-lysine (Wako) was pre-coated at 10 μg/mL. After blocking, the plates were loaded with serially-diluted IgG antibodies, and incubated with goat anti-human IgG-HRP, followed by OPD substrate. Indirect immunofluorescence assay using HEp-2 cell line was performed using HEp-2 IFA kit according to the instruction (Bio-Rad). Primary IgG1 antibodies were added at 10 μg/mL in PBS containing 0.1% BSA and 0.05% Tween-20 and then the bound IgG1 antibodies were detected by 1:200-diluted Alexa594-goat anti-human IgG (H+L) (Thermo Fisher Scientific).
Flow cytometric analysis
Splenocytes of TC-mAb mice were pre-treated with anti-FcγRII/III monoclonal antibody and then incubated with biotinylated monoclonal antibodies against mouse CD43, CD90, CD3, c-kit, F4/80, Gr-1, CD4, CD8, CD11b, Ter119, CD93, CD11c, CD138, and human IgD. Class-switched memory B cells were enriched with a MACS system using streptavidin microbeads (Miltenyi Biotec). This was followed by staining with B220-BV786, CD38-AF700, PE-labeled CoV2-S, APC-labeled CoV2-RBD, streptavidin-efluor450, and DAPI. Stained cells were analyzed and sorted as single cells into 96-well plates using a FACSAria instrument (BD Biosciences).
Single cell culture
Feeder cells (NB21.2D9 cells) were seeded into 96-well plates at 1000 NB21.2D9 cells per well in B cell medium (BCM; RPMI-1640 containing 10% Hyclone FBS, 1 mM sodium pyruvate, 10 mM HEPES, MEM nonessential amino acid, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 55 μM 2-mercaptoethanol) 1 day before single B cell sorting. The next day, murine IL-4 (Peprotech: 2 ng/mL), IL-2 (Peprotech: 4 ng/mL), and IL-5 (Peprotech: 5 ng/mL) were added to the cultures, and mouse/human B cells were directly sorted into 96-well plates at one cell per well, and then co-cultured with a feeder line. After cultivation, culture supernatants were harvested and subjected to ELISA for mapping the epitopes of the secreted monoclonal antibodies. Cultured clonal B cells were also frozen for V(D)J sequence analysis and V(D)J gene recovery for producing recombinant monoclonal antibodies.
Generation of monoclonal antibodies
Recombinant monoclonal antibodies were prepared as described previously (Adachi et al., 2019; Tiller et al., 2008). In brief, VH/VL genes of selected B cell samples from single-cell cultures or published monoclonal antibodies were cloned into human IgG1 or IgG3 heavy chain, and kappa or lambda light chain expression vectors. Expression plasmids encoding the heavy and light chains of the Fab were also prepared. Pairs of heavy and light chain expression vectors were transfected into Expi293F cells using ExpiFectamine reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The supernatant was collected at 4 days post-transfection. The IgG and Fab antibodies were purified from the culture supernatant using a protein G column (Thermo Fisher Scientific) and a Talon resin affinity chromatography (Clontech), respectively, and then subjected to further analysis after dialysis against PBS. To remove soluble aggregates for further experiments, size exclusion chromatography was performed. After the centrifugation, the supernatant of either IgG1 or IgG3 was loaded onto a Superdex 200 10/300 GL column (Cytiva) at a flow rate of 0.5 mL/min using PBS as running buffer. Gel Filtration Standard (BioRad) was used according to the manufacturer’s instruction.
Inhibition of ACE2 binding
To assess antibody-mediated inhibition of CoV2 RBD binding to ACE2, recombinant human ACE2 (Biolegend) was coated onto 96-well plates (Thermo fisher scientific) and incubated overnight at 4°C. After blocking with PBS containing 1% BSA, a recombinant CoV2 RBD bearing an avitag was pre-incubated with serially diluted antibodies for 2 h at RT. The mixture was transferred to a well of an ACE2-coated plate and then incubated for overnight at 4°C. The binding of the CoV2 RBD to ACE2 was developed using streptavidin-HRP (Southern Biotech) and an OPD-substrate (Sigma-Aldrich). Absorbance at 490 nm was measured using an iMark Microplate Reader. The inhibition percentage was calculated as (OD490 of positive-control – OD490 of sample) / (OD490 of positive control – OD490 of negative-control) x 100. The half-maximal effective concentration (EC50) was determined using a nonlinear regression curve fit (GraphPad Prism).
Virus neutralization assay
For the pseudovirus-based neutralization assay, a VSV pseudovirus bearing CoV2- or WIV-1 S proteins was incubated with serially diluted antibodies for 1 h at 37°C or 4°C. The mixture was inoculated in VeroE6 or VeroE6/TMPRSS2 cells (JCRB1819, JCRB Cell Bank) seeded in 96-well solid white flat-bottom plates (Corning), and then incubated for 24 h at 37°C with 5% CO2. Luciferase activity in cultured cells was measured using the Bright-Glo luciferase assay system (Promega) with a GloMax Navigator Microplate Liminomater (Promega). The neutralization percentage was calculated as (viral control signals – sample signals) / (viral control signals – cell-only control signals) x 100. The half-maximal inhibitory concentration (IC50) was determined using a nonlinear regression curve fit (GraphPad Prism). For the authentic virus neutralization assay, a mixture of serially diluted antibodies and 100 TCID50 of the hCoV-19/Japan/TY-WK-521/2020 strain, variant strains (Alpha: hCoV-19/Japan/QK002/2020, hCoV-19/Japan/QHN001/2020, hCoV-19/Japan/QHN002/2020, Gamma: hCoV-19/Japan/TY7-501/2021, hCoV-19/Japan/TY7-503/2021, D614G: QH-329-037-p3d1), or CoV1 Frankfurt-1 strain was incubated at 37°C for 1 h (CoV2) or 4 h (CoV1) before being inoculated in VeroE6/TMPRSS2 cells seeded in 96-well flat-bottom plates (TPP). After culture for 5 days at 37°C with 5% CO2, the plates were fixed with 20% formalin (Fujifilm) and stained with crystal violet solution (Sigma-Aldrich). The absorbance at 595 nm was measured using an Epoch 2 instrument (Biotek). For SARS-CoV, survival ratio of VeroE6/TMPRSS2 cells was determined after 2-days culture using cell counting kit 8 (Fujifilm), and the absorbance at 450 nm was measured using an iMark Microplate Reader (Bio-Rad). The neutralization percentage was calculated as (sample signals – viral control signals) / (cell only control signals – viral control signals) x 100.
Surface plasmon resonance assay
The full-length and Fab fragments of NT-108 and NT-193 were dissolved in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Surfactant P20). SPR experiments were performed with a Biacore3000 or T200 (Cytiva). Streptavidin was immobilized on the CM5 sensor chip by amine-coupling using the standard amine coupling kit or immobilized on the CAP chip by the standard biotin capture kit, followed by the biotinylated CoV2 RBD. The biotinylated β2-microglobulin was used as a negative control protein. The NT-108 and NT-193 antibodies were injected over the immobilized RBD protein, at a flow rate of 30 μl/min. Flow cells were regenerated with 10 mM glycine pH 1.5. For the binding analysis of RBD variant to NT-193 Fab, the full-length IgG3 of NT-193 was immobilized on the CM5 chip by direct amine coupling and the original and variant RBDs were injected in similar way to the other orientation described above. The data were analyzed using BIA evaluation version 4.1.1, T200 evaluation version 1.0 and ORIGIN version 2017 software.
Biolayer interferometry
The kinetics of ACE2 binding to CoV2-RBD was determined using an OctetRED96e system (ForteBio) at 30°C with shaking at 1,000 rpm. Biotinylated RBDs were loaded at 5 μg/mL in 1 × kinetic buffer (0.1% BSA and 0.02% Tween-20 in PBS) for 600 s onto streptavidin biosensors (ForteBio) and incubated with serially diluted hACE2 (60, 20, 6.7, 2.2, 0.74, and 0 nM) (BioLegend) for 600 s, followed by immersion in 1 × kinetics buffer for 1,200 s of dissociation time. The binding curves were fitted in a 1:1 binding model and the association rate constant (kon), the dissociation rate constant (koff), and the dissociation constant (KD) values were calculated using the Octet Data Analysis software (ForteBio).
Crystallization, data collection, and structural determination
Purified CoV2 RBD and NT-193 Fab were buffer exchanged to 20 mM Tris pH8.0, 100 mM NaCl and mixed in the molar ratio of 1:1. Crystals of the RBD-NT-193 complex (7.5 mg/mL) were grown at 293K by the sitting drop vapor-diffusion method. The final crystallization condition was 0.1 M HEPES pH 7.5, 10% (w/v) PEG 8000, 8% (w/v) Ethylene glycol. The crystals were cryo-cooled in a liquid nitrogen and the X-ray diffraction experiment was performed at beamline BL-17A of Photon Factory (Tsukuba, Japan). The X-ray diffraction dataset was processed with XDS and scaled with Aimless in the CCP4 program package (Evans, 2006; Kabsch, 2010; Winn et al., 2011). The structure was solved by the molecular replacement method using the Phaser program in the PHENIX package with the RBD structure (PDB: 7jmp) as a search probe, and the structure refinement was carried out using phenix.refine and COOT (Emsley et al., 2010; Liebschner et al., 2019). The stereochemical properties of the structure were assessed using MolProbity (Davis et al., 2007). Figures were prepared with PyMOL (http://pymol.sourceforge.net). Intermolecular contact atoms were analyzed using PISA and CONTACT in the CCP4 program package (Krissinel and Henrick, 2007).
In vitro escape mutation selection
Antibody escape mutants were selected using two methods. For the first method, VeroE6/TMPRSS2 cells were seeded in 12-well plates (TPP), and a mixture of diluted antibodies and 1000 TCID50 of the CoV2 strain was added to the cells following 1 h culture at 37°C. After culturing for 4 days, the cells were monitored for CPE, and the culture supernatant from the well exhibiting evident CPE at the highest antibody concentration was used for the next round of selection and sequencing. For sequencing, total RNA was extracted from culture supernatants using TRIzol (Life Technologies) and RNeasy mini kit (QIAGEN). For the second method, we used VeroE6/TMPRSS2 cells and CoV2 strain that had been maintained independently of the first method. Virus-inoculated cells were recovered at day 3 post-infection and mixed with the diluted antibodies (0.6-50 μg/mL) in 24-well plates. After 1 h of incubation at RT, the mixtures of virus-infected cells and antibodies were added to cells that were suspended in 24-well plates. After 2 days of incubation at 37°C, CPE-positive wells were identified, and virus-infected cells were used for the next round of selection. After three rounds of selection, cells were infected with the viruses in the presence of diluted antibodies (0.25-8 μg/mL). CPE+ wells were subjected to RBD sequencing. The mutant viruses were purified through the limiting dilution method and then added to VeroE6 cells at serially diluted concentrations. After 3 days of incubation, the levels of CPEs were quantified using an MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) reagent. TCID50 was determined as the dilution that caused the half-maximal levels of CPEs according to the absorbance.
Next-generation sequencing and variant allele frequency analysis
Whole-genome amplification of CoV2 was carried out using a modified version of the ARTIC Network’s protocol for CoV2 genome sequencing (Itokawa et al., 2020). A next-generation sequencing (NGS) library was constructed using the QIAseq FX DNA Library kit (QIAGEN), and sequenced using an iSeq platform (Illumina). All short-read sequences were mapped to the CoV2 Wuhan-Hu-1 reference genome sequence (GenBank: MN908947.3) using the bwa mem algorithm (version 0.7.13-r1126) (Li and Durbin, 2009); next, variant allele frequency analysis was conducted using VarScan version 2.4.3 (Koboldt et al., 2012).
qRT-PCR analysis of viral RNA
Viral loads were determined by qRT-PCR to detect nucleocapsid genomic RNA of CoV2. Total RNA was extracted from lung homogenate of hamsters using the Direct-zol miniprep kit (Zymo580 research). Whole-lung homogenates were prepared using a TissueLyser LT instrument (QIAGEN). qRT-PCR reactions were performed in duplicate using the QuantiTect Probe RT-PCR Kit (QIAGEN), using the following cycling protocol: 50°C for 30 min and 95°C for 15 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. The primer and probe sequences used for qRT-PCR were as follows: forward primer: 5′- AAATTTTGGGGACCAGGAAC −3′, reverse primer: 5′- TGGCAGCTGTGTAGGTCAAC −3′, probe: 5′- FAM- ATGTCGCGCATTGGCATGGA-BHQ −3′. The standard curve was constructed using serial 10-fold dilutions of purified PCR products with a known copy number (from 10 to 106 copies/μL).
Histopathological analysis
Hamsters were anesthetized and perfused with 2 mL of 10% phosphate-buffered formalin, and the lungs were harvested and fixed. Fixed tissues were routinely embedded in paraffin and sectioned (3 μm). For immunohistochemistry, antigen retrieval of formalin-fixed hamsters tissue sections was performed by autoclaving at 121°C for 10 min in retrieval solution at pH 6.0 (Nichirei, Tokyo, Japan). CoV2 antigens were detected using a standard immunoperoxidase method, and a rabbit anti- CoV2 N antibody was used as the primary antibody (Nagata et al., 2021). Nuclei were counterstained with hematoxylin for 10 s. Scores were determined based on the percentage of N antigen-positive cells, as determined by immunohistochemistry in each group using the following scoring system: 0, no antigen positive cells; 1, antigen positive cells were occasionally observed in each cut sections (1-3 antigen-positive areas per section were observed in the high magnification); 2, scattered positive cells were observed (4-9 antigen-positive areas per section were observed in the high magnification); 3, many positive cells were diffusely and widely observed (more than 10 antigen-positive areas per section were observed in the low magnification). Mean scores from all lung sections (5-10 lung sections/animal) in each animal were calculated.
Study approval
Animal procedures were approved by the Animal Ethics Committee of the National Institute of Infectious Diseases, Japan, and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee. In vitro escape mutation screening experiments for CoV2 were performed at the Biosafety Level-3 facility of the Research Center for Zoonosis Control, Hokkaido University, and the National Institute of Infectious Diseases following the institutional guidelines.
Quantification and statistical analysis
Statistical analysis for virus titers in culture, body weight change and viral antigens in hamsters are described in the relevant figure legends. Each dot represents the numbers from individual experiment (virus titers) or the result from individual hamster. Values represent mean ± SD. Data were analyzed using Prism (GraphPad) by a two-tailed Mann-Whitney test. Significance was defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Further statistical details for each plot can be found in the corresponding figure legends.
Acknowledgments
We thank Akira Dosaka, Eriko Izumiyama, and Ryoko Itami at NIID for technical support. We also thank the beamline staff at Photon Factory and Swiss Light Source for supporting data collection. This work was supported by Japan Agency for Medical Research and Development grants JP19fk0108111 (to T.H. and Y.T.), JP20fk0108298 (to T.H., K.M., and Y.T.), JP20am0101093 (to K.M.), JP20ae0101047 (to K.M.), JP20fk0108251 (to H. Sawa), and JP20am0101124 (to Y.K.); by Ministry of Education, Culture, Sports, Science and Technology grants JPMXS0420100119 (to K.M.) and 20H05773 (to T.H.), by the Naito Foundation (to T.H.); by the Takeda Foundation (to K.M.); and by the Joint Usage/Research Center program of the Institute for Frontier Life and Medical Sciences, Kyoto University (to T.H.).
Author contributions
Conceptualization: K.M. and Y.T. Methodology: S.F., H.S., Y.K., M.O., and T.H. Investigation: T.O., S.K., Y. Adachi, S.M., A.S., T.N., T.I., T.T., Y. Anraku, K.Y., C.T., H.F., M.S., Y.O., N.S., N.I., N.N., T. Suzuki., J.S., T. Sekizuka, K.T., and L.S. Funding acquisition: Y.K., H. Sawa, T.H., K.M., and Y.T. Project administration: T.K., M.K., T. Suzuki, H. Ssawa, and T.H. Supervision: K.M. and Y.T. Writing – original draft: T.O., K.M., and Y.T. Writing – review & editing: A.S., T. Sekizuka, H. Satofuka, H. Sawa, Y.K., T.H., K.M., and Y.T.
Declaration of interests
A.S. is an employee of Shionogi & Co., Ltd. M.O. is a CEO, employee, and shareholder of Trans Chromosomics, Inc. These authors acknowledge a potential conflict of interest and attest that the work contained in this report is free of any bias that might be associated with the commercial goals of the company. T.O., Y. Adachi, M.O., T.H., K.M., and Y.T. declare that an intellectual property application has been filed using the data presented in this paper. The other authors declare that they have no competing interests.
Published: August 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.08.025.
Supplemental information
Data and code availability
Coordinates and structure factors have been deposited in the Protein Data Bank. Sequences data for NT-108 and NT-193 have been deposited in GenBank. All NGS reads were deposited in the DNA Data Bank of Japan. Accession numbers are listed in the Key Resources Table. Data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adachi Y., Tonouchi K., Nithichanon A., Kuraoka M., Watanabe A., Shinnakasu R., Asanuma H., Ainai A., Ohmi Y., Yamamoto T., et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat. Commun. 2019;10:3883. doi: 10.1038/s41467-019-11821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat N.M., Bieber M.M., Chapman C.J., Stevenson F.K., Teng N.N. Human antilipid A monoclonal antibodies bind to human B cells and the i antigen on cord red blood cells. J. Immunol. 1993;151:5011–5021. [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimidi R.P., Klein J.S., Politzer M.S., Bai S., Seaman M.S., Nussenzweig M.C., West A.P., Jr., Bjorkman P.J. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell. 2015;160:433–446. doi: 10.1016/j.cell.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokawa K., Sekizuka T., Hashino M., Tanaka R., Kuroda M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE. 2020;15:e0239403. doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Nakahara K., Seki T., Miki S., Kawauchi S., Suyama A., Wakasa-Morimoto C., Kodama M., Endoh T., Oosugi E., et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kuraoka M., Schmidt A.G., Nojima T., Feng F., Watanabe A., Kitamura D., Harrison S.C., Kepler T.B., Kelsoe G. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwata-Yoshikawa N., Sano K., Ainai A., Shiwa N., Shirakura M., Kishida N., Arita T., Suzuki Y., Harada T., et al. The peripheral T cell population is associated with pneumonia severity in cynomolgus monkeys experimentally infected with severe acute respiratory syndrome coronavirus 2. bioRxiv. 2021 doi: 10.1101/2021.2001.2007.425698. [DOI] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the Coronavirus Antibody Database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.I., Lambson B.E., Crowley A.R., Bashirova A., Scheepers C., Garrett N., Abdool Karim S., Mkhize N.N., Carrington M., Ackerman M.E., et al. IgG3 enhances neutralization potency and Fc effector function of an HIV V2-specific broadly neutralizing antibody. PLoS Pathog. 2019;15:e1008064. doi: 10.1371/journal.ppat.1008064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S., Arimori T., Yamashita K., Jinzai H., Motooka D., Nakamura S., Li S., Takeda K., Katayama J., El Hussien M.A., et al. Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus. Sci. Rep. 2017;7:16428. doi: 10.1038/s41598-017-16681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satofuka H., Abe S., Moriwaki T., Okada A., Kazuki K., Tanaka H., Yamazaki K., Hichiwa G., Morimoto K., Takayama H., et al. Efficient human-like antibody repertoire and hybridoma production in trans-chromosomic mice carrying megabase-sized human immunoglobulin loci. Research Square. 2020 doi: 10.21203/rs.3.rs-103241/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf O., Golding H., King L.R., Eller N., Frazier D., Golding B., Scott D.E. Immunoglobulin G3 from polyclonal human immunodeficiency virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J. Virol. 2001;75:6558–6565. doi: 10.1128/JVI.75.14.6558-6565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Spellerberg M., Chapman C., Hamblin T., Stevenson F. Dual recognition of lipid A and DNA by human antibodies encoded by the VH4-21 gene. A possible link between infection and lupus. Ann. N Y Acad. Sci. 1995;764:427–432. doi: 10.1111/j.1749-6632.1995.tb55858.x. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Kimura M., Tan L., Yoshida Y., Ozawa T., Kishi H., Fukushi S., Saijo M., Sano K., Suzuki T., et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein. Virol J. 2021;18:16. doi: 10.1186/s12985-021-01490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics Á., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. COVID-19 Genomics UK (COG-UK) consortium Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloniniyi O.K., Henry A.R., Darko S., et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373:eabh1766. doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., Nussenzweig M.C. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Wilson I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021;538:192–203. doi: 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell. 2020;183:1508–1519. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank. Sequences data for NT-108 and NT-193 have been deposited in GenBank. All NGS reads were deposited in the DNA Data Bank of Japan. Accession numbers are listed in the Key Resources Table. Data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.