Graphical abstract
Abbreviations: CDF, cumulative distribution function; CGGA, Chinese Glioma Genome Atlas; CNA, copy number alternations; CNV, copy number variation; CSF-1, colony-stimulating factor-1; DMP, differentially methylated position; GBM, glioblastoma; GEO, Gene Expression Omnibus; GO, gene ontology; GSEA, gene set enrichment analysis; GSVA, gene set variation analysis; IGR, intergenic region; IHC, immunohistochemistry; IL, interleukin; KEGG, Kyoto Encyclopaedia of Genes and Genomes; PAM, partition around medoids; LGG, low grade glioma; PCA, principal component analysis; MMP-2, matrix metalloproteinase-2; MT1, MMP membrane type 1 matrix metalloprotease; TGF-β, tumor growth factor-β; TNFα, tumor necrosis factor α; SNP, single-nucleotide polymorphism; SNV, single-nucleotide variant; TAM, tumor associated macrophage; TCGA, The Cancer Genome Atlas; TIMP-2, tissue inhibitor of metalloproteinase-2; TLR2, toll-like receptor 2; TME, tumor microenvironment; TSS, transcription start site; WGCNA, weighted gene co-expression network analysis; pamr, prediction analysis for microarrays; SVM, Support Vector Machines; BBB, brain blood barrier; CHOL, Cholangiocarcinoma; OV, Ovarian serous cystadenocarcinoma; LIHC, Liver hepatocellular carcinoma; ESCA, Esophageal carcinoma; PAAD, Pancreatic adenocarcinoma; STAD, Stomach adenocarcinoma; COAD, Colon adenocarcinoma; KIRC, Kidney renal clear cell carcinoma; READ, Rectum adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; HNSC, Head and Neck squamous cell carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; LUSC, Lung squamous cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; KICH, Kidney Chromophobe; BRCA, Breast invasive carcinoma; THCA, Thyroid carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; SKCM, Skin Cutaneous Melanoma; BLCA, Bladder Urothelial Carcinoma; SARC, Sarcoma; THYM, Thymoma; LUAD, Lung adenocarcinoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; ACC, Adrenocortical carcinoma; PRAD, Prostate adenocarcinoma
Keywords: Macrophage, Glioma microenvironment, Machine learning, Immunotherapy, Prognostic model
Abstract
Background
Gliomas are one of the most common types of primary tumors in central nervous system. Previous studies have found that macrophages actively participate in tumor growth.
Methods
Weighted gene co-expression network analysis was used to identify meaningful macrophage-related gene genes for clustering. Pamr, SVM, and neural network were applied for validating clustering results. Somatic mutation and methylation were used for defining the features of identified clusters. Differentially expressed genes (DEGs) between the stratified groups after performing elastic regression and principal component analyses were used for the construction of MScores. The expression of macrophage-specific genes were evaluated in tumor microenvironment based on single cell sequencing analysis. A total of 2365 samples from 15 glioma datasets and 5842 pan-cancer samples were used for external validation of MScore.
Results
Macrophages were identified to be negatively associated with the survival of glioma patients. Twenty-six macrophage-specific DEGs obtained by elastic regression and PCA were highly expressed in macrophages at single-cell level. The prognostic value of MScores in glioma was validated by the active proinflammatory and metabolic profile of infiltrating microenvironment and response to immunotherapies of samples with this signature. MScores managed to stratify patient survival probabilities in 15 external glioma datasets and pan-cancer datasets, which predicted worse survival outcome. Sequencing data and immunohistochemistry of Xiangya glioma cohort confirmed the prognostic value of MScores. A prognostic model based on MScores demonstrated high accuracy rate.
Conclusion
Our findings strongly support a modulatory role of macrophages, especially M2 macrophages in glioma progression and warrants further experimental studies.
1. Background
Gliomas, a collective term referring to tumors originating from neuroglial cells, are categorized into astrocytoma, gangliocytoma, oligodendroglioma, ependymoma and others. They are one of the most common types of primary tumors in central nervous system [1]. They can be generally inferred by the WHO grades, with grade I being the most benign and grade IV (glioblastoma, GBM) the most malignant type. There are increasing molecular markers identified for prediction of patient survival rate, including mutational status, DNA methylation, and members of pathways involved in tumor suppression, proliferation and migration [2], [3]. As a result, WHO proposed an updated grading system for CNS tumors integrating molecular diagnosis [4].
Macrophages are phagocytic cells critical for host defense against endogenous or exogenous pathogens in innate immunity. They can develop from circulating monocytes or are seeded in various tissues during embryogenesis, the latter being named resident or tissue macrophages. Inflammatory stimuli are potent inducers of the activation of macrophages. Depending on the types of stimuli and cytokine profiles, it is proposed that macrophages can differentiate into the classically activated M1 or alternatively activated M2 type [5]. M1 macrophages are induced by stimuli such as lipopolysaccharide and interferon γ, and are capable of secreting pro-inflammatory cytokines. In contrast, M2 macrophages are activated by interleukin-4 (IL-4), -10, -13, colony-stimulating factor-1 (CSF-1) and tumor growth factor-β (TGF-β). They are connected to tissue remodelling, angiogenesis and allergy response. A plethora of studies focusing on tumor microenvironment (TME) have identified the importance of non-tumor cells in sustenance of tumor growth and response to immunotherapy [6], [7]. As a constitutive part of TME, tumor associated macrophages (TAMs) account for up to 50% of the total glioma noncancerous cell population, and are actively engaged in promoting tumor progression and metastasis [8]. The exact molecular profile of TAMs remains undetermined. It is emphasized that TAMs in glioma are in fact a heterogeneous group of cells consisting of microglia, infiltrating blood borne macrophages and monocytes coming through compromised brain blood barrier (BBB) [9], [10], [11]. BBB has been proposed to be responsible for the largely unsuccessful immunotherapies and other treatment modalities in gliomas. Macrophages have a natural ability to traverse the BBB, which macrophages could be loaded with anti-cancer agents [12]. Macrophages are also found to mediate drug resistance in glioma CSF-1 receptor inhibition immunotherapy by switching to M1 type with enhanced phagocytosis that promotes tumor cell death [13]. Moreover, it is demonstrated that in gliomas, the grade of malignancy is associated with the number of infiltrating myeloid cells, which consist of microglias and macrophages [14].
Macrophages have been shown to participate in tumor migration and invasion as well as angiogenic switch [15], [16]. In many solid tumors such as ovarian, lung, liver and kidney cancers, high levels of macrophages are associated with poor prognosis [17]. Similar finding was also noticed in gliomas [18]. In GBM with lower survival rate, microglias and macrophages upregulates their expression of CD45, TIE2 and CD163 and facilitate angiogenesis [19]. According to transcriptome profile, consensus clustering has identified three subtypes of GBMs including classical (CL), mesenchymal (MES), and proneural (PN) [20]. MES GBMs, being the subtype with less favourable outcome, and recurrent gliomas transitioning to MES both show higher levels of M2 macrophages than the non-MES GBM subtypes [20]. The role of M2 macrophages in the pathogenesis of gliomas remains unclear. Secretion of IL-10 through JAK2/STAT3 signaling by M2 macrophages participates in the glioma tumorigenesis [21]. M2 macrophages also release insulin-like growth factor-bind protein 1 (IGFBP1) and IL-6 to promote angiogenesis [22], [23]. Likewise, M1 macrophages have diverse roles in tumor microenvironment of gliomas. Suppressed M2 polarization and increased M1 macrophages have been reported to both inhibit tumor proliferation [24] and correlate with poor prognosis [25] simultaneously. Currently, M0/M1/M2 macrophages are currently seen as a continuous spectrum of states. The integrative analysis on these macrophage subgroups together would be promising.
Weighted gene co-expression network analysis (WGCNA) is a systems biology method for identification of gene correlation and is widely used in cancer research. In the present study, we employed WGCNA to identify meaningful macrophage-related gene modules in glioma patients. Genes within the identified module were extracted for clustering. Machine learning including prediction analysis for microarrays (pamr), Support Vector Machines (SVM), and neural network was used to validate the clustering results. Significant differentially expressed genes (DEGs) between the stratified groups after performing elastic regression and Principal component analyses (PCA) were used for the construction of risk scores, thereafter called MScores. Pan-cancer analysis was performed to further validate the prognostic value and define the function of MScores. We confirm that MScores could also predict immunotherapeutic efficiency. By using these multi-dimensional analyses, we have established the prognostic value of macrophages in glioma patients, with the special attention to the M2-like property.
2. Methods
2.1. Patient and cohort inclusion
This study collected 1991 diffuse glioma samples from two databases: The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA). For the TCGA cohort (672 glioma samples of combined LGG and GBM), the RNA-seq data and corresponding clinical information were retrieved from TCGA database (http://cancergenome.nih.gov/). Three CGGA validation cohorts were employed in this study, including two RNA-seq cohorts (CGGA325 and CGGA693) and a microarray cohort (CGGAarray). The RNA-seq and microarray data, clinical and survival information were downloaded from the CGGA database (http://www.cgga.org.cn). Single-cell expression matrices were obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) GSE138794. Eight scRNA sequencing samples were used for analysis. Samples from 15 patient cohorts diagnosed with gliomas were included in this study for external validation of MScores (Table S1). Pan-cancer data for functional annotation and validation of MScores were from the TCGA dataset.
2.2. WGCNA identifying macrophage related genes
The WGCNA package in R version 3.6.1 was used to perform WGCNA. The expression profile of top 5000 genes from TCGA cohort was applied as the input of WGCNA. The association between individual genes and macrophage densities was quantified by gene significance, and the correlation between module eigengenes and gene expression profiles was represented by module membership. A power of β = 3 automatically calculated by pickSoftThreshold function and a scale-free R2 = 0.85 automatically calculated by softConnectivity function were set as soft-threshold parameters to ensure a scale-free topology network and produce a TOM matrix. After recalculating module eigengenes, the module dendrogram depicting the relationship among the eigengenes and the macrophages was plotted using plotEigengeneNetworks function. A total of seven modules including red (33 genes), turquoise (1022 genes), yellow (291 genes), green (135 genes), blue (658 genes), brown (648 genes), and grey (2213 genes) were generated. The correlation between the turquoise module and xCell-defined M2 macrophages, M1 macrophages, macrophages, monocytes were 0.64, 0.68, 0.66, and 0.62, respectively, indicating a selective expression of the turquoise module in monocyte-derived macrophages. Given the remarkable correlation strength with the macrophage subgroups, turquoise module was deemed as macrophages-specific and was selected for further analysis. The correlation between module membership and gene significance was visualized using verboseScatterplot. Genes within the turquoise module were thus chosen for GO (gene ontology) and KEGG (Kyoto Encyclopaedia of Genes and Genomes) functional enrichment analyses.
2.3. Delineation and validation of immune subtypes
Univariate cox regression analysis was performed on the 1022 genes extracted from turquoise module to identify prognostic genes with the criteria of P < 0.001. Based on the 945 prognostic genes, we applied consensus clustering algorithm of partition around medoids (PAM) to identify robust clusters of TCGA patients [26]. The cumulative distribution function (CDF) and consensus heatmap were used to assess the optimal K value of 2. To validate the immune subtypes in three CGGA cohorts, we trained a pamr classifier in the discovery cohort to predict the immune subtypes for patients in the validation cohort (package pamr) based on the 434 intersected genes from TCGA and three CGGA cohorts. The clustering results were further validated by SVM and neural network.
2.4. Genomic alterations in immune subtypes
Somatic mutations and somatic copy number alternations (CNAs) were downloaded from the TCGA database as previously described [27], [28]. Genomic event enrichment was determined using GISTIC analysis. CNAs and altered peaks associated with the two clusters were obtained using GISTIC 2.0 analysis (https://gatk.broadinstitute.org). The R package TCGAbiolinks was used for downloading the WES-derived somatic mutation data acquired by Mutect2 [29]. The most differentially mutated genes were detected using Fisher’s exact test. The co-occurrence and mutually exclusive mutations were detected using CoMEt algorithm.
2.5. Genome-wide methylation array
The differentially methylated positions (DMPs) were stratified per genetic feature/chromosome and compared to the total number of 450 k probes associated with the respective genetic feature/chromosome. Fisher’s exact tests were used to calculate the probability that the number of DMPs of a specific genetic feature/chromosome was significantly different from the expected number. Using the numbers of hypermethylated versus the hypomethylated DMPs, a second Fisher’s exact test was performed to assess whether the distribution was significantly different in any genetic feature/chromosome. The threshold for statistical significance was set to a Bonferroni-adjusted α value of 0.05. R package ChAMP was used to analyze Illumina Infinium 450 k DNA methylation array data. β values were normalized using peak-based correction. Differential methylation probes and regions were identified using the limma package and Bumphunter algorithm [30]. Correlation between probe signals and gene expression levels was evaluated by Pearson correlation, and the same numbers of probes as in the true DMP set were randomly selected from all probes to construct 100 random sets. R package clusterProfiler was used for GO analyses of the DEGs and DMP-associated genes and Gene set enrichment analysis (GSEA).
2.6. Single cell sequencing
R package Seurat v3.1.2 was used to process the single-cell data expression matrix. The data were first normalized by ‘NormalizeData’. ‘FindVariableGenes’ was then used to identify 2000 highly variable genes. ‘FindIntegrationAnchors’ and ‘Integratedata’ were used to merge 8 GBM sample data sets as previously described [31]. After using ‘RunPCA’ to perform PCA, a K-nearest neighbor graph was constructed by the “FindNeighbors” function, and the ‘FindClusters’ function was used to alternately combine cells together with the highest resolution. Finally, ‘UMAP’ was used for visualization. In single cell sequencing analysis, the cut-off point was defined as the median value of gene expression levels.
2.7. Annotation of the immune infiltrating microenvironment
ESTIMATE was performed to evaluate the immune cell infiltration level (immune scores) and stromal content (stromal scores) for each sample. The enrichment levels of 64 immune signatures were quantified by the xCell algorithm [32]. The relative fraction of 22 immune cell types in tumor tissues were estimated using CIBERSORT algorithm [9]. Gene set variation analysis (GSVA) was performed to study GO pathways. Seven types of classified immune checkpoints signalling pathways were investigated from two previous published studies [33], [34].
2.8. Identification of an immune-related signature
Univariate Cox regression analysis was performed to determine the differentially expressed immune genes with prognostic significance with a p value <0.05 between subtypes. Elastic regression analysis and PCA were further used to calculate the MScores of patients. The extracted principal component 1 served as the signature score. The MScore of each patient after the prognostic value of gene signature score was obtained by the following calculation:
where i represented the expression of genes with HR > 1, and j the expression of genes with HR < 1.
2.9. Prediction of immunotherapy response
The IMvigor210 cohort, which is an urothelial carcinoma cohort treated with the anti‐PD‐L1 antibody atezolizumab was used for prediction of patient response to immunotherapy [35]. Based on the Creative Commons 3.0 License, complete expression data and clinical data were downloaded from http://research-pub.Gene.com/IMvigor210CoreBiologies. The GSE78220 cohort, a melanoma cohort receiving anti-PD-1 (pembrolizumab or nivolumab) immunotherapy, was also included for prediction of immunotherapy response [36]. Raw data were then normalized using the DEseq2 R package, and the count value was transformed into the TPM value.
2.10. Construction and validation of a prognostic model
Ultimately, nomogram is a form of visualized multi-factor regression analysis commonly used for cancer survival rate prediction. Variables selected for construction of the nomogram included the calculated prognostic scores, ages, pathological stages of glioma and mutation status. Univariate and multivariate regression analyses were also used to evaluate the prognostic value of these factors.
2.11. RNA sequencing of glioma samples
Tumor tissues from 48 glioma patients were collected for sequencing as previously described. Briefly, 1 μg RNA was used as input material for RNA sample preparations. DNA was sheared followed by sequencing library preparation using NEBNext Ultra RNA Library Prep Kit. PCR was then performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and the Index (X) Primer. After target region capture by biotin-labeled probes, the captured libraries were sequenced on an Illumina Hiseq platform to generate 125/150 bp paired-end reads. In-house perlscripts were used to process raw data (raw reads). Then reads containing adapter and ploy-N, and low-quality reads were removed to obtain clean data (clean reads). Reference genome and gene model annotation files were obtained from the genome website (http://genome.ucsc.edu). The reference genome index was built using Hisat2 v2.0.5 and paired-end clean reads were aligned to the reference genome. FeatureCounts v1.5.0-p3 was then used to count the read numbers mapped to each gene. TPM of each gene was calculated based on the gene length and reads count mapped to this gene.
2.12. Immunohistochemistry
Immunohistochemistry (IHC) staining was conducted as previously described [27], [28]. Paraffin-embedded tissues of 40 glioma samples (25 LGG samples and 15 GBM samples) with the corresponding sequencing data from the Xiangya Neurosurgey (XYNS) cohort were used for immunohistochemistry (IHC). The patient characteristics used for IHC were shown in Table S2. Sections were boiled in sodium citrate buffer (pH 6.0) for antigen retrieval, and endogenous HRP activity was blocked with 3% H2O2. Sections were washed with 10% phosphate buffered saline (PBS) and marked with PAP pen. After blockage with 5% BSA, sections of 4 μm thickness were incubated with primary antibody against CD163 (Rabbit, 1:500, Proteintech, China) under room temperature for 12 h. The target was detected using a two-step detection kit (PV-9000, ZSGB-Bio, China), which the sections were incubated with horse radish peroxidase conjugated secondary antibody and the antibody reaction was visualized using 3, 3′-diaminobenzidine (DAB) development. Sections were then counterstained with hematoxylin and scanned with Pannoramic Scanner (Pannoramic DESK, P-MIDI, P250, P1000, Hungary).
2.13. Statistical analysis
Kaplan-Meier curves with log-rank test were used to assess survival difference between groups. The univariate and multivariate Cox regression analyses were performed to detect the prognostic factors. Pearson correlation and distance correlation analyses were used to calculate correlation coefficients. Contingency tables were analyzed by χ2 contingency test. The OS and MScores were calculated using the R package survival and cutoff values determined. Based on the dichotomized MScores, patients were grouped as with high or low MScore in each data set, and the computational batch effect was reduced by the R package sva. Data were visualized using the R package ggplot2. OncoPrint was used to delineate the mutation landscape of TCGA by the maftools R package [37]. All survivorship curves were generated using R package survminer. Heatmaps were generated based on pheatmap. All statistical analyses were conducted using R software. P < 0.05 was considered statistically significant.
3. Results
3.1. Identification of macrophage density as a potential prognostic marker
The flow chart of our study design was shown in Fig. S1A. We sought to determine the prognostic value of macrophages in glioma by studying the macrophage-related genes using WGCNA. After stratifying patients by high and low median levels of Macrophages, M1 macrophages, M2 macrophages calculated by xCell algorithm, survival analysis revealed a clear distinction between the two subtypes (Fig. S1B). To evaluate the potential prognostic value of macrophages, we performed WGCNA with the expression profile of top 5000 genes from TCGA cohort as the input to search for macrophage-specific genes. A power β = 3 was selected as the soft threshold for a scale-free network construction. Seven modules were identified by clustering dendrogram (Fig. S2A). Given the close correlation strength with each macrophage subgroup (M2 macrophages, r = 0.64, p = 2e−79; M1 macrophages, r = 0.68, p = 1e−91; Macrophages, r = 0.66, p = 2e−86), turquoise module was selected for subsequent analyses (Fig. S2B and S2C). We investigated the correlation between the module membership and macrophage-related gene significance, which reached 0.83 and suggested that the expression levels of these macrophage-related genes were slightly influenced by other cells (Fig. S2D). GO functional enrichment analysis found that the genes were concentrated in pathways involving neutrophil chemotaxis and chemokine-mediated signalling (Fig. S2E). KEGG analysis showed that the genes were enriched in the cytokine-receptor interaction (Fig. S2F).
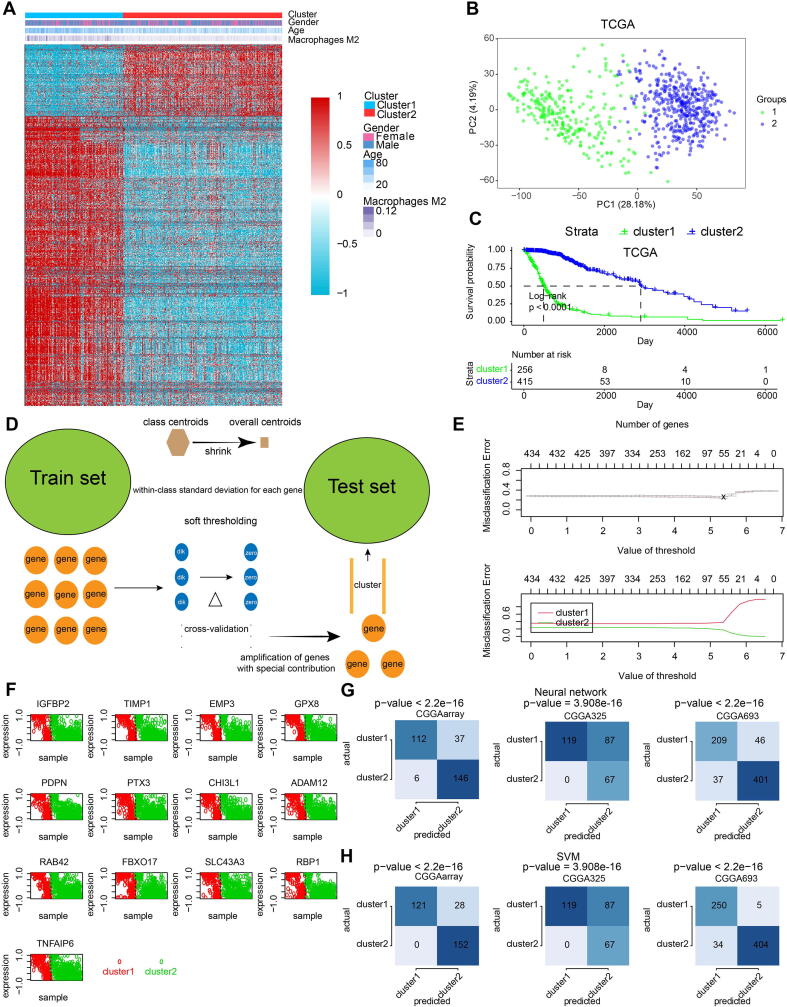
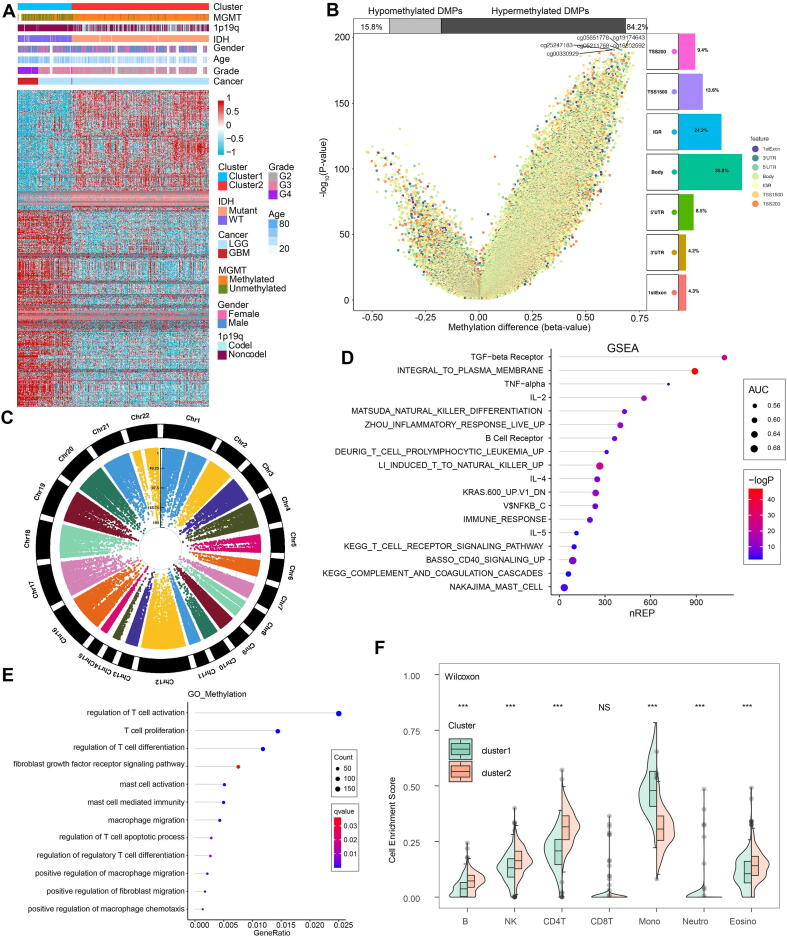
We subsequently extracted 945 genes from module turquoise by WGCNA for the subsequent generation of macrophage density. PAM was performed for glioma patients with the corresponding gene expression profiles in TCGA cohort (Fig. 1A). The optimal number of clusters was evaluated by ConsensusClusterPlus package (Fig. S3A). Clustering results were most stable when the number was set to two (K = 2). The delineated groups based on the 945 genes showed distinct patterns of clinical traits and macrophage levels (Fig. 1A). PCA managed to differentiate the samples from the TCGA dataset (Fig. 1B). Survival analyses of the two clusters confirmed an obviously lower survival probability curve for cluster 1 (Fig. 1C). Subsequently, combining the gene expression profiles from three CGGA cohorts, 434 genes were identified from these 954 genes to validate the clustering results by pamr (Fig. 1D). Optimal threshold values were selected (Fig. 1E). Thirteen genes identified by pamr, including IGFBP2, TIMP1, EMP3, PTX3, CHI3L1, PDPN, were identified as the ones with the strongest power to differentiate the samples (Fig. 1F). Notably, TIMP1 and EMP3 are associated with recruitment of macrophages to tumors [38], [39]. IGFBP2 and PDPN are also associated with M2 polarization [40], [41]. PTX3 has recently been reported to suppress the polarization of M2 macrophages. M2 macrophage-secreted CHI3L1 could promote gastric and breast cancer metastasis [42], and CHI3L1 modulates an immune suppressive microenvironment by reprogramming M2-like TAM in GBM [43]. TNFAIP6, RBP1, FBXO17, and GPX8 promote metastasis and invasiveness of tumor [44], [45], [46], [47]. SLC43A3 and ADAM12 have been revealed as the biomarker and therapeutic target in tumor patients [48], [49]. RAB42 has been proved to be prognostic marker in glioma [50]. Thus, these thirteen genes could be regarded as reliable discriminatory markers of macrophage signature from the biological point of view. Samples were then clustered into two groups with high or low death risk by pamr in three CGGA cohorts, respectively (Fig. S3B-D). Neural network and SVM were performed for validation of the clustering as well, which the contingency table showed the consistency in clustering results among pamr, SVM, and neural network (Fig. 1G, H). PCA also managed to differentiate the samples from three individual datasets (Fig. S3E-G). Survival analyses of the two clusters confirmed a lower survival probability curve for cluster 1 (Fig. S3H-J).
Fig. 1.
Construction and validation of M2 macrophage-related clusters based on machine learning. A, clustering dendrogram demonstrating good separation of the two clusters determined via PAM algorithm by traits. B, sample clustering by PCA in the TCGA dataset. C, Kaplan-Meier survival analysis of the two clusters. D, validation of clustering by pamr. E, selection of optimal threshold and exhibition of misclassification error. F, heatmap illustrating the differentiation power of 13 genes, red dots and green dots representing samples classified by genes. G, validation of clustering by neural network. H, validation of clustering by SVM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Clinical traits and TME characteristics of the macrophage-stratified groups
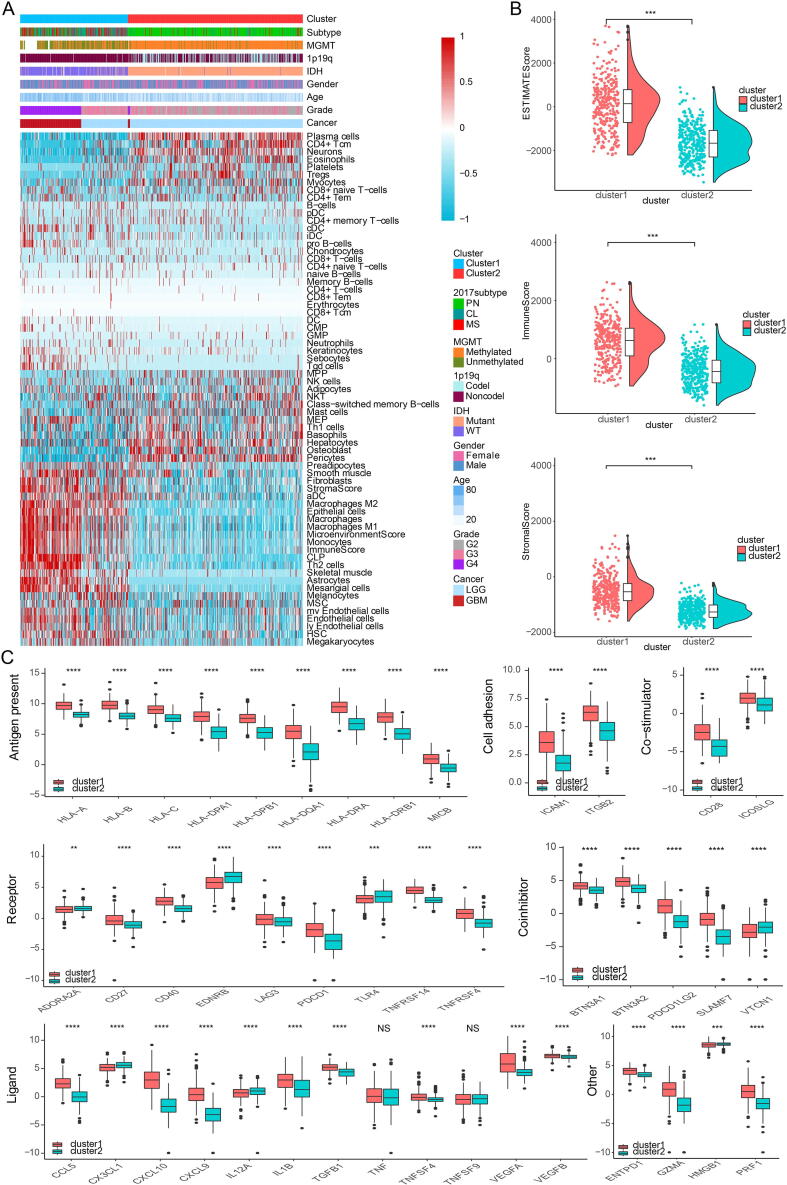
We then proceeded to investigate the TME characteristics of the two clusters. The expression difference of the levels of 64 cell types in two defined subtypes were investigated in TCGA (Fig. 2A). It was found that increased cells such as fibroblasts, macrophages and monocytes were related to cluster 1, i.e. worse survival probability. Moreover, CIBERSORT algorithm showed that the expression of several types of immune cells including M0/1/2 macrophages and neutrophils were higher in cluster 1 (Fig. S4A). The association between ESTIMATE scores of the immune infiltrating microenvironment, an indicator of the cancer biological behaviour, and clusters, as well as levels of immune cells was examined in TCGA cohort (Fig. 2B). ESTIMATEScores, ImuneScores and StromalScores were all higher in cluster 1 than in cluster 2 (Fig. 2B). Given that immune checkpoint molecules were critical in regulating tumor immunity [28], [51], we then compared the levels of several series of immune checkpoint molecules related to antigen presentation, cell surface receptor, coinhibition, ligand and cell adhesion between the two clusters. Immune checkpoint markers tended to be overexpressed in cluster 1 (Fig. 2C).
Fig. 2.
Characterization of the two clusters. A, dendrogram correlating the levels of 64 cell types and clusters in TCGA. B, ESTIMATEScores, ImuneScores and StromalScores of the two clusters in TCGA. C, molecule levels in the pathways involved in immune checkpoint pathways in TCGA.
The pathological gradings of glioma were also significantly different between clusters 1 and 2 (p < 2.2e−16), with a higher gradings in cluster 1 in TCGA (Fig. S4B). The proportions of samples with IDH mutation and chromosome 1p/19q codeletion in cluster 1 were higher than those in cluster 2 (Fig. S4B), also indicating a more malignant propensity in cluster 1. Results regarding the proportion of patients with MGMT promoter methylation were less universal, with data from the TCGA database showing the most significant difference while data from the other three databases statistically insignificant difference (Fig. S4B). The proportions of the four GBM subtypes in clusters 1 and 2 were significantly different in TCGA (p < 2.2e−16), showing that the more malignant CL and ME subtypes accounted for the majority of cluster 1 samples (Fig. S4B).
The expression differences of hypoxia pathways in two clusters were explored using GSVA. Investigated pathways included cell response regulation, hypoxia-induced intrinsic apoptosis, Hypoxia-Inducible Factor 1α (HIF1A) and others. These pathways were found to be more activated in cluster 1 in TCGA, suggesting a tendency for cell hypoxia, which is a universal marker for malignant tumor proliferation, in this group (Fig. S4C). We also interrogated the relationship between metabolic pathways, such as pyrimidine synthesis and sulfur metabolism, and subtypes. The metabolic pathways were overrepresented in cluster 1, proving a more active proliferation of glioma cells in these samples (Fig. S4D).
3.3. Macrophage-enriched group showed more malignant genomic features
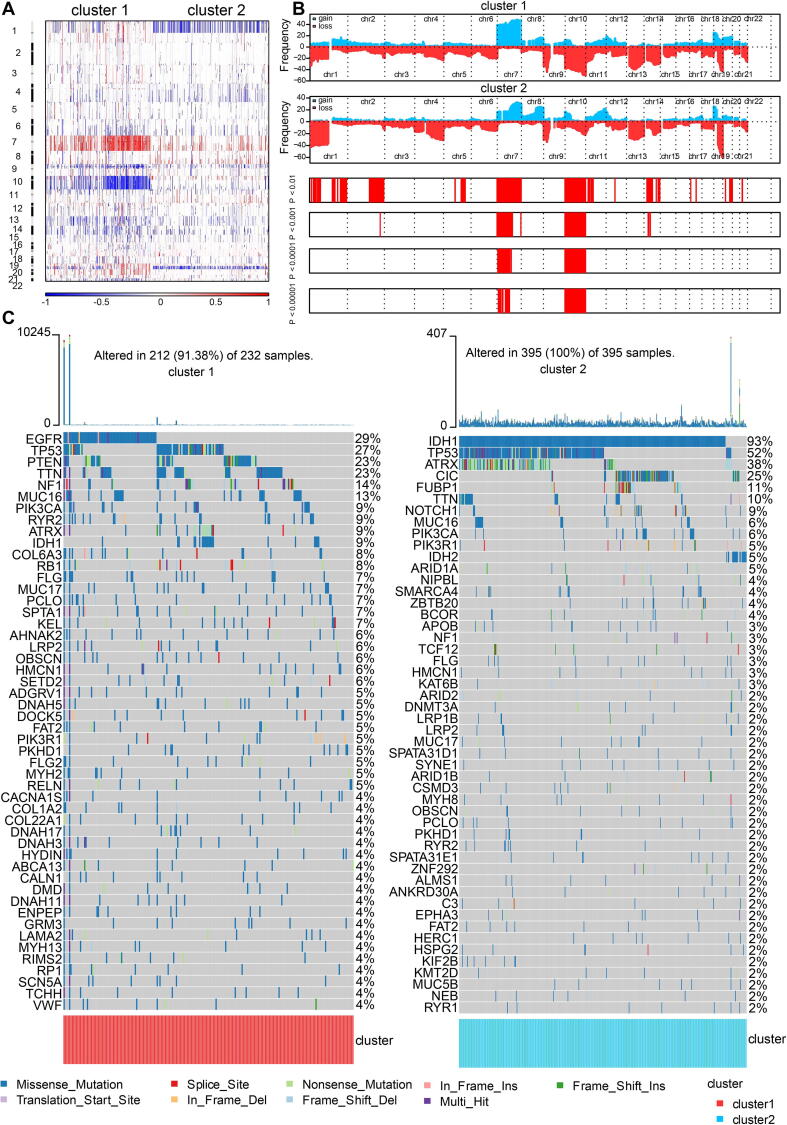
Somatic mutation analysis and copy number variation (CNV) were performed using the TCGA dataset to explore genomic traits of the two clusters (Table S3). A global CNV profile was obtained by comparing the two clusters (Fig. 3A, 3B, Table S4). According to somatic mutation analysis, mutations in EGFR (29%), TP53 (27%), PTEN (23%) and TTN (23%) were most highly enriched in cluster 1. In comparison, IDH1 (93%), TP53 (52%), ARTX (38%) and CIC (25%) mutations were enriched in cluster 2 (Fig. 3C). Missense mutation was the predominant gene alteration type in all these genes except for ATRX, in which frame-shifting deletion was the most common type.
Fig. 3.
Genomic features of the two clusters. A, global CNV profile of the two cluster. B, distribution of gain or loss of function mutation in the two clusters. C, list of the most frequently altered genes in clusters 1 and 2.
Different types of somatic mutations, including the single-nucleotide variant (SNV), single-nucleotide polymorphism (SNP), insertion, deletion and intergenic region (IGR), were analyzed using the R package maftools [52]. Silent, nonsense, missense, intronic, 5′ and 3′ UTR mutations were more common in cluster 1 than in cluster 2 (Fig. S5A). Among the detected SNVs, C > T appeared to be the most common mutation in cluster 1 (Fig. S5B). The T to A, C to T and C to A mutations occurred more frequently in cluster 1 than in cluster 2. While the frequencies of insertion and deletion were not statistically different between the two clusters, SNPs were significantly more common in cluster 1 (Fig. S5C). The top 17 most mutated cancer-related genes were listed in Fig. S5D. Common carcinogenic pathways were more active in cluster 1 (Fig. S5E). The strongest co-occurrent pairs of gene alteration were ATRX-TP53 and ATRX-IDH1, which was in accordance with previous reports [53], [54], [55]. It was suggested that acquisition of a second cancer-related gene alteration may dictate the development of certain tumor types, and that TP53, IDH1, ATRX are functionally linked [55], [56] (Fig. S5F). On the other hand, the most mutually exclusive pairs were CIC-TP53 and PTEN-IDH1.
3.4. DMPs in macrophage-enriched gliomas were associated with tumor progression
By adopting a Δbeta value >0.4 and a p value <0.05, a total of 8359 DMPs were identified, among which 8301 DMPs were upregulated and 58 DMPs were down-regulated. The distribution of identified DMPs in two clusters was exhibited by heatmap (Fig. 4A). Genomes of cluster 1 demonstrated an overall hypomethylation trend, with 84.2% of DMPs being hypomethylated (Fig. 4B). In terms of the location in relation to genes, DMPs were concentrated in IGRs and transcription start sites (TSSs). Focal analysis showed the distribution of identified DMPs in human chromosomes (Fig. 4C). GSEA of the DMP-associated genes showed that the hypermethylated genes with highly positive β differences have more essential contributions to tumor-associated biological processes such as TGFβ, tumor necrosis factor α (TNFα) and leukocytotrophic process (IL-2, Fig. 4D). GO enrichment analysis also revealed increased T cell activation, proliferation and differentiation activities (Fig. 4E). Enrichment scores of the main immune cell types in the two clusters were calculated and compared. Monocyte and neutrophil levels were higher in cluster 1 than in cluster 2, while B, NK and CD4+ T cell levels were higher in cluster 2 (Fig. 4F).
Fig. 4.
Methylation characteristics of the two clusters. A, clustering dendrogram by DMPs showing good separation of the two clusters by clinical and genetic traits. B, volcano plot of DMPs and their position in genes. C, Manhattan plot of the genome-wide DNA differential methylation. D, GSEA of the two clusters. E, GO functional enrichment analysis. F, comparison of enrichment scores of several immune cell types in the two clusters. *** p < 0.001.
3.5. Generation of MScore and its functional annotation
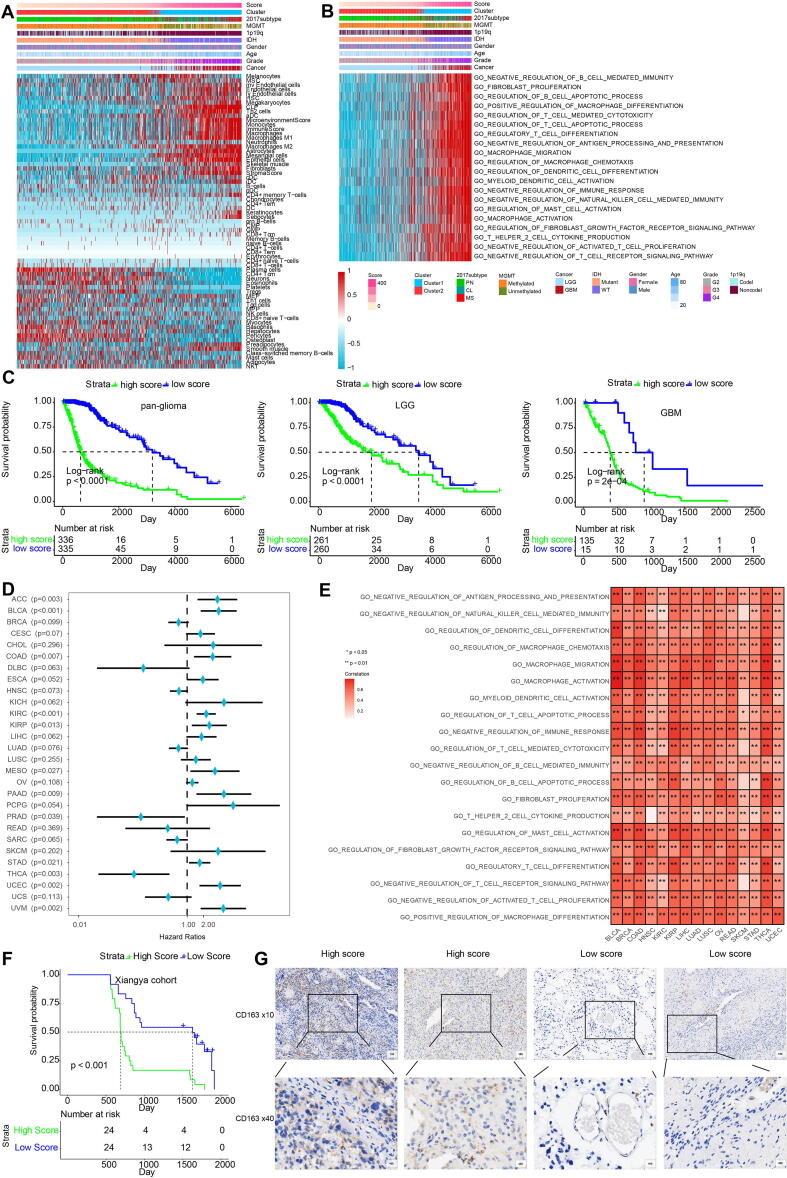
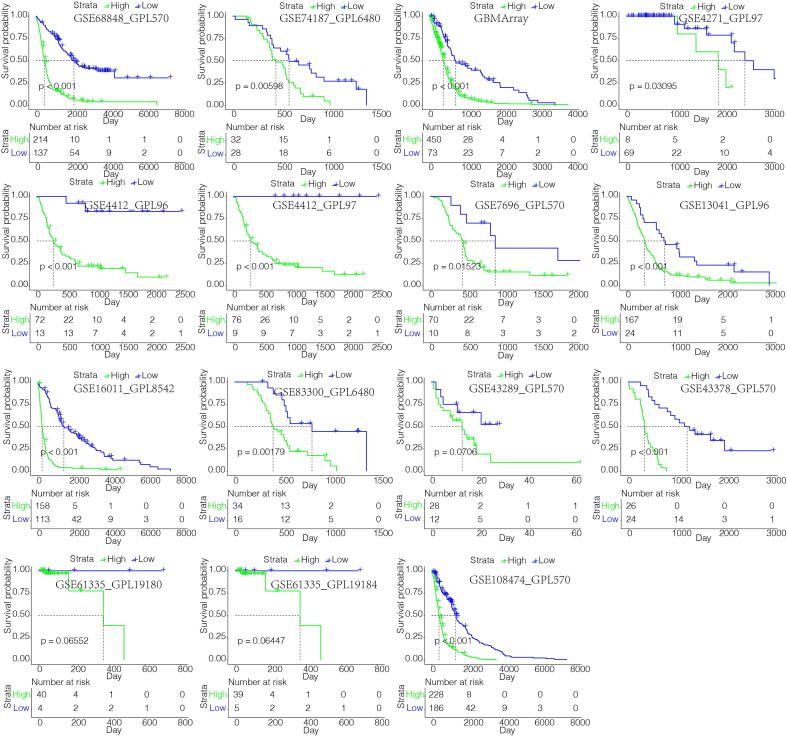
By performing elastic net regression analysis and PCA algorithm (Fig. S6A), 26 macrophage-related genes were derived from the 434 genes and their coefficients were obtained (Fig. S7A, Table S5), with the highest coefficients being TIMP1, EMP3, IGFBP2, PDPN and SSTR5. In accordance, regulation of TIMP1 and EMP3 in macrophages is associated with extracellular matrix remodeling and recruitment of macrophages to tumors [38], [39]. IGFBP2 and PDPN are also associated with M2 polarization [40], [41]. SSTR5 has been identified as the receptor controlling activation on macrophages [57]. The encoded protein interaction network was constructed using the STRING database (Fig. S7B) [58]. Of these genes, single cell RNA-sequencing results showed that PDPN, EMP3 and F2RL2 were the ones most highly expressed in macrophages (Fig. S7C). UMAP plot showed that most of these 26 genes were enriched in neoplastic cells and macrophages (Fig. S8). The macrophage-related gene signature was used to calculate MScores by PCA (Table S6). Sankey plot revealed a high consistency between macrophage-related clusters and MScores (Fig. S6B). The correlation of the expression levels of 64 cell types and MScores was then evaluated. There was a positive correlation between the scores and the levels of fibroblasts, macrophages and monocytes (Fig. 5A). In the TGCA dataset, survival analysis demonstrated a good separation of patients with different death risks by high and low MScores (Fig. 5C). The prognostic value of MScores was further validated in CGGAarray, CGGA325, and CGGA693 datasets (Fig. S6C). The separation was more significant in all or low-grade glioma (LGG) groups than in GBM group. Pathways related to macrophage activation and migration, dendritic cell differentiation and negative regulation of T cell proliferation were more active in the samples with higher scores (Fig. 5B). High MScores were associated with higher expression of immune infiltrating cells such as monocytes, neutrophils, and mast cells (S9A), as well as with several immune checkpoint molecules (Fig. S9B). In 15 external independent glioma datasets, MScores were proved to significantly stratify patients’ overall survival probabilities (Fig. 6). We then subclassified patients from TCGA dataset as LGG and GBM, and IDH-mutant and wildtype. Our results also confirmed the discrimination ability of MScores in these subgroups (Fig. S10A), as well as in astrocytoma, oligoastrocytoma and glioma patients from CGGA dataset (Fig. S10B). Furthermore, the prognostic value of MScores was explored in other types of cancers (Fig. S11), High- and low-MScores properly discriminated patients’ overall survival. They were detrimental in cancers such as pheochromocytoma and paraganglioma, kidney chromophobe and uveal melanoma, and protective in diffuse large B-cell lymphoma, prostate adenocarcinoma and thyroid cancer (Fig. 5D), which is partly in accordance with previous studies [59], while not compatible with others [60], [61]. The protective role of MScore in some cancers indicated the heterogeneity of the tumor microenvironment among different cancer types. GO enrichment functional analysis of MScores in several cancer types also showed that MScores were positively correlated with macrophage activation, fibroblast proliferation, regulation of mast cell activation, and regulatory T cell differentiation, which indicated an immune-suppressive microenvironment (Fig. 5E). In pan-cancer analysis, MScores were also correlated with higher expression of immune infiltrating cells (Fig. S9C). Finally, we evaluated MScores in our glioma patients, and found that high score patients had lower survival probabilities (p < 0.0001, Fig. 5F). IHC staining of CD163, a marker of M2 macrophage, showed that samples with high MScores had higher expression of CD163 (Fig. 5G).
Fig. 5.
Characterization of the MScore. A, dendrogram correlating the MScores and 64 cell types. B, GO functional enrichment analysis correlating different immune regulatory processes with MScores. C, survival analyses of MScores in pan-glioma, LGG and GBM groups from TCGA. D, Hazard ratios of MScores in different cancer types based on univariate Cox regression analysis. GBM, Glioblastoma multiforme; LGG, Brain Lower Grade Glioma; CHOL, Cholangiocarcinoma; OV, Ovarian serous cystadenocarcinoma; LIHC, Liver hepatocellular carcinoma; ESCA, Esophageal carcinoma; PAAD, Pancreatic adenocarcinoma; STAD, Stomach adenocarcinoma; COAD, Colon adenocarcinoma; KIRC, Kidney renal clear cell carcinoma; READ, Rectum adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; HNSC, Head and Neck squamous cell carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; LUSC, Lung squamous cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; KICH, Kidney Chromophobe; BRCA, Breast invasive carcinoma; THCA, Thyroid carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; SKCM, Skin Cutaneous Melanoma; BLCA, Bladder Urothelial Carcinoma; SARC, Sarcoma; THYM, Thymoma; LUAD, Lung adenocarcinoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; ACC, Adrenocortical carcinoma; PRAD, Prostate adenocarcinoma. E, GO enrich functional analysis of MScores in several cancer types. F, MScore discriminating survival probabilities in Xiangya glioma cohort. G, CD163 staining for 25 LGG samples and 15 GBM samples. IHC against CD163 molecule demonstrating the different M2 macrophage densities in the two MScore groups.
Fig. 6.
MScores discriminating survival probabilities in the majority of glioma cohorts.
3.6. Construction of a prognostic nomogram based on MScores
After establishing macrophage density as a suitable marker for survival prediction of gliomas, we further investigated its prediction efficiency by univariate and multivariate regression analysis. MScores were identified as a hazardous factor in TCGA and CGGA 693 cohorts (Fig. S12A, S12B). A prognostic nomogram was then developed by combing prognostic factors, including MScores, patient ages, tumor grades, IDH mutation, and chromosome 1p/19q codeletion (Fig. S12C). In TCGA dataset, predicted probabilities corresponded well with the actual one- to five-year overall survival rates of glioma patients (Fig. S12D). The Kaplan-Meier survival curve demonstrated a good discrimination of survival probabilities of the two clusters (p < 0.0001) (Fig. S12E). The ROC curve confirmed the discriminative ability of this nomogram (AUC = 0.802, Fig. S12F). The efficiency of the prognostic model was validated in CGGA 693 cohort. Predicted probabilities corresponded well with the actual four-year overall survival rates of glioma patients (Fig. S12G). The Kaplan-Meier survival curve demonstrated a good discrimination of survival probabilities of the two clusters (p < 0.0001) (Fig. S12H). The ROC curve confirmed the discriminative ability of this nomogram (AUC = 0.737, Fig. S12I).
3.7. Macrophage-stratified groups predicted response to immunotherapies
The potential immunogenic and tumorigenic features of MScore was explored. A series of immunogenic and tumorigenic factors were summarized (Table S7). Regarding the antigen presentation capacity, high MScore group exhibited higher level of leukocyte fraction, stromal fraction, lymphocyte infiltration signature score, and macrophage regulation, indicating the immune infiltration characteristics of high MScore group (Fig. S13A-D). Besides, high MScore group exhibited higher TCR diversity that was associated with tumor immunogenicity (Fig. S13E-F). High MScore group presented higher neoantigens, silent mutation rate, number of segments, fraction altered, aneuploidy score, all of which correlated with tumor malignancy (Fig. S13G-K). Notably, microsatellite instability (MSI), an indicator of better immunotherapy response, was found with lower level in high MScore group (Fig. S13L).
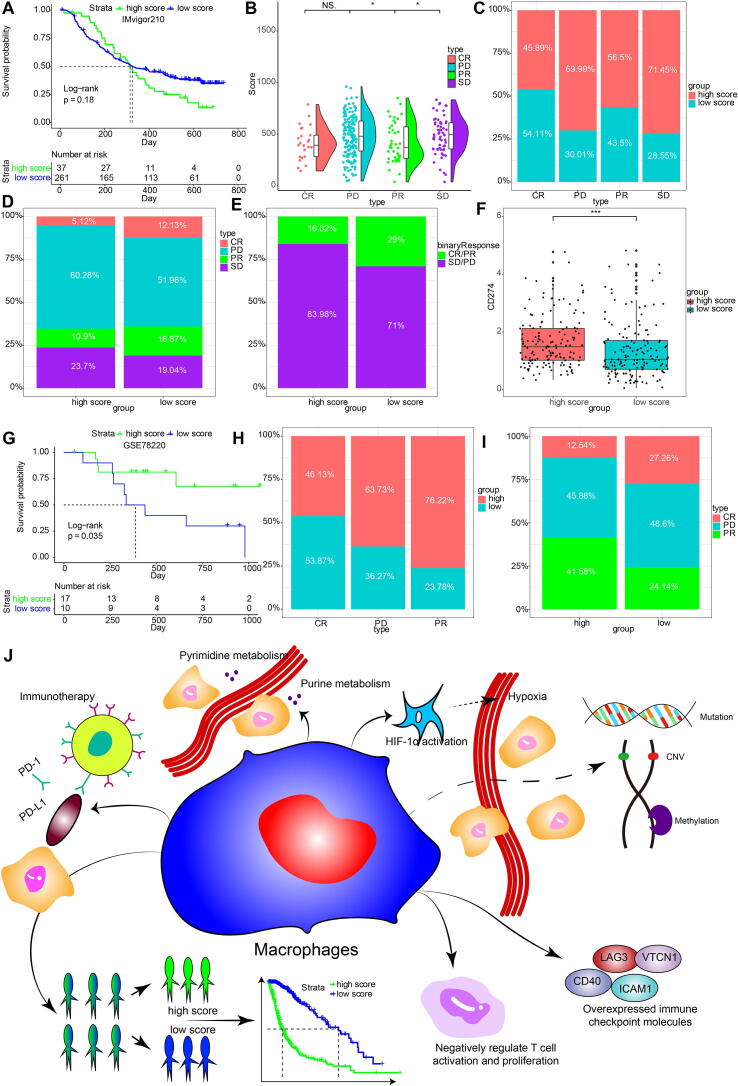
We evaluated whether MScores were able to predict therapeutic effects of immune blockade treatment in glioma patients. High and low MScores failed to stratify patients by survival probability from the IMvigor210 cohort (p = 0.18, Fig. 7A). Nevertheless, when further stratified the patients according to immunotherapeutic response types, the progressive disease, stable disease and partial response groups showed different MScores, which partial response and complete response correlated with lower MScore (Fig. 7B). The progressive disease group and the stable disease group showed a higher percentage of high MScore compared with the partial response group and the complete response group (Fig. 7C, D). We also grouped the therapeutic response in a binary mode, and found that the complete/partial response group had a higher percentage of high scores than the stable/progressive disease group (Fig. 7E). The levels of PD-L1 (CD274) were also higher in the high MScore group, indicating the immune escape in tumor microenvironment of high MScore group (Fig. 7F, p < 0.0001). The prognostic value of MScores was also tested in GSE78220, a melanoma cohort under PD-1 blockage treatment series. High MScore patients showed a more favourable therapeutic response and experienced prolonged survival (Fig. 7G-I).
Fig. 7.
Predictive value of MScore in immunotherapy response. A, Kaplan-Meier curve of high and low MScore groups in IMvigor210 cohort. B, rain-cloud plot showing MScores of CR, PR, PD and SD groups. CR, complete response; PR, partial response; PD, progressive disease; SD, stable disease. C, the bar chart showing proportions of high and low MScores. D, the bar chart showing proportions of CR/PR and SD/PD patients in high and low MScore groups. E, Constitution of the four therapeutic response types in high and low MScore groups. F. comparison of collective CD274 levels in the two MScore groups. G, Survival analysis of MScores in a melanoma cohort. H, Proportions of high and low MScores in different response groups. I, proportions of different response groups in high and low MScore groups. J, physiologic functions of M2 macrophages. CR, complete response; NS, not significant; PD, progressive disease; PR, partial; SD, stable disease. * p < 0.05; *** p < 0.001.
4. Discussion
In the current in silico study, we interrogated the reliability of macrophage density as a marker for glioma prognosis by using WGCNA for the first time. Genes derived from the module of WGCNA were used for glioma patient grouping. Machine learning including pamr, neural network, and SVM were applied for validating the clustering results based on macrophage. For the validation of the prognostic value of macrophages, we evaluated the comprehensive landscapes of tumor genomics, TME, metabolism, and hypoxia pathways of macrophage density-stratified glioma patients. A risk score based on the DEGs between macrophage-related clusters was generated by PCA. We then investigated its associated metabolic functions, biological functions, immunotherapeutic response and immune cell expression (Fig. 7J).
TME refers to the stromal cell, cytokine and chemokine niche that supports the tumor tissue. A previous study has found that glioma TAMs partly overlap with either M1 or M2 polarized macrophages [62]. They express surface markers such as CD163, CD204, CD206, and produce anti-inflammatory cytokines like IL-10 and TGF-β, i.e. they demonstrate classic features of M2 macrophages. In gliomas, M2 macrophages secrete IL-10 to activate JAK/STAT3 pathway, leading to tumor tissue growth [21].TGF-β promotes glioma cell migration by upregulation of integrin and matrix metalloproteinase-2 (MMP-2) as well as suppression of tissue inhibitor of metalloproteinase-2 (TIMP-2) [63]. CSF-1, a growth factor capable of promoting M2 polarization, is constitutively secreted by glioma cells in order to recruit microglia [64], [65]. Toll-like receptor 2 (TLR2) can induce M2 polarization in schistosomiasis and can be produced by macrophages under anti-inflammatory conditions in turn [65], [66]. Activation of TLR2 results in upregulation of membrane type 1 matrix metalloprotease (MT1-MMP) in microglia and subsequent glioma growth [67]. In summary, by assuming the anti-inflammatory M2 polarization, TAMs are able to facilitate tumor tissue growth and migration. Nevertheless, a recent multi-omic study finds that TAMs have both the canonical M1 and M2 profiling, showing the complex composition of TAMs [68]. Myeloid derived suppressor cells (MDSCs) are proposed as a group of potent immune suppressors that develop from myeloid cells in response to pathogenic stimuli [69]. They are categorized into two main groups, polymorphonuclear (PMN-) and monocytic (M-) MDSCs, the latter can differentiate into macrophages and dendritic cells. More importantly, MDSCs shows strong potency to differentiate into TAMs when homing to tumors [70]. We propose that the macrophage entity as defined by the xCell algorithm in the present study is very likely to encompass the M2 subgroup of TAMs and M-MDSCs.
The genomic alteration related to macrophage entity was first investigated. The IDH missense mutations confer better survival outcome in glioma patients. Nevertheless, LGGs carrying the IDH mutations are more prone to develop into secondary GBMs, especially when tertiary genetic alterations in oncogenes like PIK3CA and PDGFRA occur in the same patient [71]. The present study finds that the IDH1 missense mutations are overrepresented in the cluster 2 (93%) compared with the cluster 1 (9%), in accordance with previous findings that IDH mutations are more enriched in LGGs than in high grade ones [72]. Besides, TP53 (53% in cluster 2 vs 27% in cluster 1) is associated with low-grade gliomas and acts as a tumor suppressor [73]. ATRX mutation, an important indicator of telomere maintenance and benign glioma [74], is also overrepresnted in the cluster 2 (38%) compared with the cluster 1 (9%). In consistent with the previous findings, ATRX mutations are mutually associated with p53 mutation and IDH mutation but exclusive from 1p/19q codeletion in cluster 1, which is characterized by better prognosis [75]. Likewise, EGFR, which is the most enriched mutated gene in cluster 1 (29%) and whose alteration occurs in less than 2% of cluster 2 cases as identified by somatic mutation analysis, has been reported to be frequently activated in GBM [76]. PTEN, another oncogene in GBM, is also more frequently mutated in cluster 1 (23%) compared with cluster 2 (<2%).
We also explored the epigenetic modifications of glioma patients. DNA methylation abnormalities have been strongly associated with gliomas [77], [78]. CpG islands hypermethylation in 5′ promoter region is able to inhibit the transcription of tumor suppressor genes [79], while hypomethylation in turn opens up nucleosome and activates expression of oncogenes [80], [81]. Hypermethylation in the promoter region of MGMT, which encodes a DNA repair protein defending against mutagenesis, confers better prognosis of GBM patients treated with radiotherapy and temozolomide [82]. Indeed, gliomas patients in cluster 2 in our study show a hypermethylation trend. Previous studies have proved that IDH-mutant gliomas tend to be associated with hypermethylation, which is also consistent with our findings [83].
Considering the vital role of macrophage in tumor microenvironment, we tried to establish a robust relationship between macrophage signature and immune signatures. Hypoxia and hypermetabolism are two vital regulators of tumor microenvironment and determinants of tumor malignancy regarding their roles in apoptosis, autophagy, DNA damage, and immunosuppression [84], [85]. In accordance, patients in cluster 1 are closely associated with hypoxia pathways such as hypoxia inducible factor 1α signaling pathway and hypermetabolism pathways such as pyrimidine metabolism [86], purine metabolism [87] that have been proved to be associated with tumor progression and metastasis. Immune Score and Stromal Score serve to evaluate the infiltrating inflammatory cell density and stromal cell density within the tumor tissue [88]. They have been shown to correlate with worse clinical outcome in other types of tumors [89], [90]. Patients with higher MScores generally have higher ESTIMATE/Immune/Stromal Scores. Besides, higher MScores are associated with a series of tumor immunogenic factors. They also demonstrate an active antigen presentation profile and higher immune infiltration level. This suggests that tumor cells in these patients are in a more active state, and may elicit a more robust phagocytosis and cytotoxicity response by antigen presentation. Indeed, our analyses show that higher MScore in glioma also denotes higher inflammatory cell adhesion, trafficking, immune costimulatory and cytotoxicity activity, exemplified by the increased expression of ICAM1, CXCL9, CXCL10, CD27, BTN3A1, BTN3A2 and granzyme A molecules. On the contrary, the elevated PD1, PDCD1LG2 and LAG3 levels suggests that T cell anergy is activated to facilitate the glioma escaping tumor surveillance system. Meanwhile, the potent vascularization inducer VEGFA and the angiostatic molecule CXCL9 are both overexpressed in high MScore samples, with the levels of VEGFA are generally higher than those of CXCL9. Furthermore, M2 macrophage marker CD163 is more expressed in high MScore samples. Overall, our analyses demonstrate active pro- and anti-inflammatory processes, as well as angiogenesis and angiostasis in patients with higher MScores, indicating an ‘escalated battle’ between tumor progression and suppression in this group of patients that tilted toward tumor progression and the more M2-like property. This also denotes that glioma with higher MScores are populated by macrophages that encompass both the traditionally defined M1 and M2 phenotypes, which further emphasizes that M0/M1/M2 is a continuum instead of well delineated categories.
Moreover, CD11+MHC-II+ macrophages are found to be capable of removing anti-PD-1 antibodies from CD8+ T cells by Fc domain-Fcγ receptor interaction, thus neutralizing the therapeutic effect of PD-1 antibodies [91]. Whether the same mechanism applies to anti-PD-1L antibody remains to be determined. To support this theory, our analyses show that the high MScore group has lower MSI level and more frequent stable/progressive disease patients than responsive patients, representing a worse response to immunotherapies. Nevertheless, high MScore predicted increased survival and better immunotherapy response. The different characteristics of MScores in IMvigor cohort (urothelial carcinoma) and GSE78220 cohort (melanoma) suggested the heterogeneity in tumor microenvironment of different cancer types. Given that anti-PD1/PDL1 immunotherapy relies on T cell activity, MScore is presumably expected to exhibit better predictive value in TAMs targeting immunotherapies. There are indeed several macrophage-targeting immunotherapies on trial like DSP-0509 and Imiquimod. But after exhaustive searching, no publicly available dataset could be obtained to enable computational prediction of MScores. It would definitely be interesting to investigate the value of MScore in TAM-targeting immunotherapies in the future.
It should be noted that the macrophages in the present study are still a heterogeneous group of cells that share a panel of similar surface markers defined by cell type enrichment analysis tools. The general hazardous role of MScores indicated a more M2-like property. Considering the highly diverse and plastic nature of macrophages, further delineation and sub-classification of these macrophages may serve to unveil the underlying role of macrophages in glioma ontogeny, thus conferring more accurate prognostic prediction capabilities.
5. Conclusions
In conclusion, our in silico analyses have identified a macrophage gene signature consisting of 26 macrophage-specific genes, and established their prognostic value in glioma. Our findings strongly support a modulatory role of macrophages, especially M2 macrophages, in glioma progression. This conclusion warrants further experimental studies for validation.
Funding
This work was supported by the National Natural Science Foundation of China (NO. 82073893, 81402249, 81703622 and 81873635), China Postdoctoral Science Foundation (NO. 2018M633002), Hunan Provincial Natural Science Foundation of China (NO. 2018JJ3838, 2019JJ50963), Hunan Provincial Health Committee Foundation of China (C2019186). Xiangya Hospital Central South University Postdoctoral Foundation.
7. Availability of data and materials
All data used in this work can be acquired from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), the Cancer Genome Atlas (TCGA) datasets (https://xenabrowser.net/), the Chinese Glioma Genome Atlas (CGGA) datasets (http://www.cgga.org.cn/).
8. Author’s contributions
HZ, YBL, QC, WW, ZW, and ZL designed and drafted the manuscript;
HZ, QC, WW, and ZD wrote figure legends and revised the manuscript;
QC, HZ, ZW, ZD, and WW conducted data analysis;
QC, LZ, ZL, HC, and SF funded the study
All authors have read and approved the final manuscript.
CRediT authorship contribution statement
Hao Zhang: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation, Software, Validation, Writing - review & editing. Yue-Bei Luo: Data curation, Writing – original draft. Wan-Tao Wu: Writing - review & editing. Li-Yang Zhang: Funding acquisition. Ze-Yu Wang: Software, Validation. Zi-Yu Dai: Software, Validation. Song-Shan Feng: Funding acquisition. Hui Cao: Funding acquisition. Quan Cheng: Conceptualization, Methodology, Software, Visualization, Investigation, Supervision, Funding acquisition. Zhi-Xiong Liu: Visualization, Investigation, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.019.
Contributor Information
Quan Cheng, Email: chengquan@csu.edu.cn.
Zhixiong Liu, Email: zhixiongliu@csu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ostrom Q.T., Cioffi G., Gittleman H., Patil N., Waite K., Kruchko C. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludwig K., Kornblum H.I. Molecular markers in glioma. J Neurooncol. 2017;134:505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 5.Mills C.D., Kincaid K., Alt J.M., Heilman M.J., Hill A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 6.Rybstein M.D., Bravo-San Pedro J.M., Kroemer G., Galluzzi L. The autophagic network and cancer. Nat Cell Biol. 2018;20:243–251. doi: 10.1038/s41556-018-0042-2. [DOI] [PubMed] [Google Scholar]
- 7.Maman S., Witz I.P. A history of exploring cancer in context. Nat Rev Cancer. 2018;18:359–376. doi: 10.1038/s41568-018-0006-7. [DOI] [PubMed] [Google Scholar]
- 8.Watters J.J., Schartner J.M., Badie B. Microglia function in brain tumors. J Neurosci Res. 2005;81:447–455. doi: 10.1002/jnr.20485. [DOI] [PubMed] [Google Scholar]
- 9.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss N., Miller F., Cazaubon S., Couraud P.O. The blood-brain barrier in brain homeostasis and neurological diseases. BBA. 2009;1788:842–857. doi: 10.1016/j.bbamem.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Buonfiglioli A., Hambardzumyan D. Macrophages and microglia: the cerberus of glioblastoma. Acta Neuropathol Commun. 2021;9:54. doi: 10.1186/s40478-021-01156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek S.-K., Makkouk A.R., Krasieva T., Sun C.H., Madsen S.J., Hirschberg H. Photothermal treatment of glioma; an in vitro study of macrophage-mediated delivery of gold nanoshells. J Neurooncol. 2011;104:439–448. doi: 10.1007/s11060-010-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y., Bao J., Zhao Q., Zhou T., Sun X. A spatio-temporal model of macrophage-mediated drug resistance in glioma immunotherapy. Mol Cancer Ther. 2018;17:814–824. doi: 10.1158/1535-7163.MCT-17-0634. [DOI] [PubMed] [Google Scholar]
- 14.Brandenburg S., Blank A., Bungert A.D., Vajkoczy P. Distinction of microglia and macrophages in glioblastoma: close relatives, different tasks? Int J Mol Sci. 2020;22 doi: 10.3390/ijms22010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condeelis J., Pollard J.W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Zumsteg A., Christofori G. Corrupt policemen: inflammatory cells promote tumor angiogenesis. Curr Opin Oncol. 2009;21:60–70. doi: 10.1097/CCO.0b013e32831bed7e. [DOI] [PubMed] [Google Scholar]
- 17.Komohara Y., Jinushi M., Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komohara Y., Ohnishi K., Kuratsu J., Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216:15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 19.Blank A., Kremenetskaia I., Urbantat R.M., Acker G., Turkowski K., Radke J. Microglia/macrophages express alternative proangiogenic factors depending on granulocyte content in human glioblastoma. J Pathol. 2021;253:160–173. doi: 10.1002/path.5569. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32 doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi L., Yu H., Zhang Y., Zhao D., Lv P., Zhong Y. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget. 2016;7:71673–71685. doi: 10.18632/oncotarget.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nijaguna M.B., Patil V., Urbach S., Shwetha S.D., Sravani K., Hegde A.S. Glioblastoma-derived macrophage colony-stimulating factor (MCSF) induces microglial release of insulin-like growth factor-binding protein 1 (IGFBP1) to promote angiogenesis. J Biol Chem. 2015;290:23401–23415. doi: 10.1074/jbc.M115.664037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Xu Y., Sun J., Chen W., Zhao L., Ma C. M2-like tumor-associated macrophages drive vasculogenic mimicry through amplification of IL-6 expression in glioma cells. Oncotarget. 2017;8:819–832. doi: 10.18632/oncotarget.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue N., Zhou Q., Ji M., Jin J., Lai F., Chen J. Chlorogenic acid inhibits glioblastoma growth through repolarizating macrophage from M2 to M1 phenotype. Sci Rep. 2017;7:39011. doi: 10.1038/srep39011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C., Lee J., Choi S.A., Kim S.-K., Wang K.-C., Park S.-H. M1 macrophage recruitment correlates with worse outcome in SHH Medulloblastomas. BMC Cancer. 2018;18:535. doi: 10.1186/s12885-018-4457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010; 26: 1572-3. [DOI] [PMC free article] [PubMed]
- 27.Zhang H., Chen Z., Wang Z., Dai Z., Hu Z., Zhang X. Correlation between APOBEC3B expression and clinical characterization in lower-grade gliomas. Front Oncol. 2021;11:625838. doi: 10.3389/fonc.2021.625838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., He J., Dai Z., Wang Z., Liang X., He F. PDIA5 is correlated with immune infiltration and predicts poor prognosis in gliomas. Front Immunol. 2021;12:628966. doi: 10.3389/fimmu.2021.628966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive integration of single-cell data. Cell. 2019;177 doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Wang X., Chen X., Zhang Q., Hong J. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front Genet. 2020;11:363. doi: 10.3389/fgene.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., Zhang Q., Yu C., Cao Y., Zuo Y., Yang L. Immune cell infiltration-based signature for prognosis and immunogenomic analysis in breast cancer. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa026. [DOI] [PubMed] [Google Scholar]
- 36.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 38.Fan C, Zhang X, Zhang P, Zhao J, Shen H, Zhang Y, et al. LPS stimulation during HCV infection induces MMP/TIMP1 imbalance in macrophages. J Med Microbiol. 2020; 69: 759-66. [DOI] [PMC free article] [PubMed]
- 39.Chen Q., Jin J., Huang X., Wu F., Huang H., Zhan R. EMP3 mediates glioblastoma-associated macrophage infiltration to drive T cell exclusion. J Exp Clin Cancer Res. 2021;40:160. doi: 10.1186/s13046-021-01954-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L., Zhang X., Song Q., Liu L., Forbes E., Tian W. IGFBP2 promotes tumor progression by inducing alternative polarization of macrophages in pancreatic ductal adenocarcinoma through the STAT3 pathway. Cancer Lett. 2021;500:132–146. doi: 10.1016/j.canlet.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bieniasz-Krzywiec P., Martin-Perez R., Ehling M., Garcia-Caballero M., Pinioti S., Pretto S. Podoplanin-expressing macrophages promote lymphangiogenesis and lymphoinvasion in breast cancer. Cell Metab. 2019;30 doi: 10.1016/j.cmet.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Zhang S., Wang Q., Zhang X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol. 2017;10:36. doi: 10.1186/s13045-017-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen A., Jiang Y., Li Z., Wu L., Santiago U., Zou H. Chitinase-3-like-1 protein complexes modulate macrophage-mediated immune suppression in glioblastoma. J Clin Invest. 2021 doi: 10.1172/JCI147552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin S.B., Jang H.R., Xu R., Won J.Y., Yim H. Active PLK1-driven metastasis is amplified by TGF-beta signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2020;39:767–785. doi: 10.1038/s41388-019-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao L., Wang Q., Ren W., Zheng J., Li S., Dou Z. The RBP1-CKAP4 axis activates oncogenic autophagy and promotes cancer progression in oral squamous cell carcinoma. Cell Death Dis. 2020;11:488. doi: 10.1038/s41419-020-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suber T.L., Nikolli I., O’Brien M.E., Londino J., Zhao J., Chen K. FBXO17 promotes cell proliferation through activation of Akt in lung adenocarcinoma cells. Respir Res. 2018;19:206. doi: 10.1186/s12931-018-0910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khatib A., Solaimuthu B., Ben Yosef M., Abu Rmaileh A., Tanna M., Oren G. The glutathione peroxidase 8 (GPX8)/IL-6/STAT3 axis is essential in maintaining an aggressive breast cancer phenotype. Proc Natl Acad Sci U S A. 2020;117:21420–21431. doi: 10.1073/pnas.2010275117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mender I., Batten K., Peyton M., Vemula A., Cornelius C., Girard L. SLC43A3 is a biomarker of sensitivity to the telomeric DNA damage mediator 6-thio-2'-deoxyguanosine. Cancer Res. 2020;80:929–936. doi: 10.1158/0008-5472.CAN-19-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X., Xie X., Liu P., Yang L., Chen B., Song C. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating miR-34a. Oncogene. 2018;37:6316–6326. doi: 10.1038/s41388-018-0410-1. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G.-H., Zhong Q.-Y., Gou X.X., Fan E.X., Shuai Y., Wu M.N. Seven genes for the prognostic prediction in patients with glioma. Clin Transl Oncol. 2019;21:1327–1335. doi: 10.1007/s12094-019-02057-3. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H., Fan F., Yu Y., Wang Z., Liu F., Dai Z. Clinical characterization, genetic profiling, and immune infiltration of TOX in diffuse gliomas. J Transl Med. 2020;18:305. doi: 10.1186/s12967-020-02460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayakonda A., Lin D.-C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Network C.G.A.R., Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X.-Y., Gerges N., Korshunov A., Sabha N., Khuong-Quang D.-A., Fontebasso A.M. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 55.Xie Y., Tan Y., Yang C., Zhang X., Xu C., Qiao X. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol Med. 2019;16:784–796. doi: 10.20892/j.issn.2095-3941.2019.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ichimura K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012;29:131–139. doi: 10.1007/s10014-012-0090-4. [DOI] [PubMed] [Google Scholar]
- 57.Perez J., Viollet C., Doublier S., Videau C., Epelbaum J., Baud L. Somatostatin binds to murine macrophages through two distinct subsets of receptors. J Neuroimmunol. 2003;138:38–44. doi: 10.1016/s0165-5728(03)00094-8. [DOI] [PubMed] [Google Scholar]
- 58.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geng X., Chen C., Huang Y., Hou J. The prognostic value and potential mechanism of matrix metalloproteinases among prostate cancer. Int J Med Sci. 2020;17:1550–1560. doi: 10.7150/ijms.46780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie Z., Li X., He Y., Wu S., Wang S., Sun J. Immune cell confrontation in the papillary thyroid carcinoma microenvironment. Front Endocrinol (Lausanne) 2020;11:570604. doi: 10.3389/fendo.2020.570604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y.H., Cai K., Xu P.P., Wang L., Huang C.X., Fang Y. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther. 2021;6:10. doi: 10.1038/s41392-020-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szulzewsky F., Pelz A., Feng X., Synowitz M., Markovic D., Langmann T. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wick W., Platten M., Weller M. Glioma cell invasion: regulation of metalloproteinase activity by TGF-beta. J Neurooncol. 2001;53:177–185. doi: 10.1023/a:1012209518843. [DOI] [PubMed] [Google Scholar]
- 64.Park J.E., Dutta B., Tse S.W., Gupta N., Tan C.F., Low J.K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene. 2019;38:5158–5173. doi: 10.1038/s41388-019-0782-x. [DOI] [PubMed] [Google Scholar]
- 65.Coniglio S.J., Eugenin E., Dobrenis K., Stanley E.R., West B.L., Symons M.H. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong W., Huang F., Sun L., Yu A., Zhang X., Xu Y. Toll-like receptor-2 regulates macrophage polarization induced by excretory-secretory antigens from Schistosoma japonicum eggs and promotes liver pathology in murine schistosomiasis. PLoS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vinnakota K., Hu F., Ku M.-C., Georgieva P.B., Szulzewsky F., Pohlmann A. Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro Oncol. 2013;15:1457–1468. doi: 10.1093/neuonc/not115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnstone S.E., Reyes A., Qi Y., Adriaens C., Hegazi E., Pelka K. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell. 2020;182 doi: 10.1016/j.cell.2020.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabrilovich D.I., Bronte V., Chen S.H., Colombo M.P., Ochoa A., Ostrand-Rosenberg S. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425. doi: 10.1158/0008-5472.CAN-06-3037. author reply 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar V., Cheng P., Condamine T., Mony S., Languino L.R., McCaffrey J.C. CD45 phosphatase inhibits STAT3 transcription factor activity in myeloid cells and promotes tumor-associated macrophage differentiation. Immunity. 2016;44:303–315. doi: 10.1016/j.immuni.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wakimoto H., Tanaka S., Curry W.T., Loebel F., Zhao D., Tateishi K. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20:2898–2909. doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cancer Genome Atlas Research N, Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haase S., Garcia-Fabiani M.B., Carney S., Altshuler D., Núñez F.J., Méndez F.M. Mutant ATRX: uncovering a new therapeutic target for glioma. Expert Opin Ther Targets. 2018;22:599–613. doi: 10.1080/14728222.2018.1487953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karsy M., Guan J., Cohen A.L., Jensen R.L., Colman H. New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17:19. doi: 10.1007/s11910-017-0722-5. [DOI] [PubMed] [Google Scholar]
- 76.Network C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malta TM, de Souza CF, Sabedot TS, Silva TC, Mosella MS, Kalkanis SN, et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018; 20: 608-20. [DOI] [PMC free article] [PubMed]
- 78.Etcheverry A., Aubry M., de Tayrac M., Vauleon E., Boniface R., Guenot F. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esteller M., Cordon-Cardo C., Corn P.G., Meltzer S.J., Pohar K.S., Watkins D.N. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res. 2001;61:2816–2821. [PubMed] [Google Scholar]
- 80.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 82.Gorlia T., van den Bent M.J., Hegi M.E., Mirimanoff R.O., Weller M., Cairncross J.G. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981–22981/CE.3. Lancet Oncol. 2008;9:29–38. doi: 10.1016/S1470-2045(07)70384-4. [DOI] [PubMed] [Google Scholar]
- 83.Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jing X., Yang F., Shao C., Wei K., Xie M., Shen H. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18:157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siddiqui A., Ceppi P. A non-proliferative role of pyrimidine metabolism in cancer. Mol Metab. 2020;35:100962. doi: 10.1016/j.molmet.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front Immunol. 2018;9:1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ge P.L., Li S.F., Wang W.W., Li C.B., Fu Y.B., Feng Z.K. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging (Albany NY). 2020;12:5479–5499. doi: 10.18632/aging.102971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang H., Wu X., Chen Y. Stromal-immune score-based gene signature: a prognosis stratification tool in gastric cancer. Front Oncol. 2019;9:1212. doi: 10.3389/fonc.2019.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arlauckas S.P., Garris C.S., Kohler R.H., Kitaoka M., Cuccarese M.F., Yang K.S. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work can be acquired from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), the Cancer Genome Atlas (TCGA) datasets (https://xenabrowser.net/), the Chinese Glioma Genome Atlas (CGGA) datasets (http://www.cgga.org.cn/).