Abstract
Cancer is one of the leading causes of death worldwide producing estimated cost of $161.2 billion in the US in 2017 only. Early detection of cancer would not only reduce cancer mortality rates but also dramatically reduce healthcare costs given that the 17 million new cancer cases in 2018 are estimated to grow 27.5 million new cases by 2040. Analytical devices based upon paper substrates could provide effective, rapid, and extremely low cost alternatives for early cancer detection compared to existing testing methods. However, low concentrations of biomarkers in body fluids as well as the possible association of any given biomarker with multiple diseases remain as one of the greatest challenges to widespread adoption of these paper-based devices. However, recent advances have opened the possibility of detecting multiple biomarkers within the same device, which could be predictive of a patient’s condition with unprecedented cost-effectiveness. Accordingly, this review highlights the recent advancements in paper-based analytical devices with a multiplexing focus. The primary areas of interest include lateral flow assay and microfluidic paper-based assay formats, signal amplification approaches to enhance the sensitivity for a specific cancer type, along with current challenges and future outlook for the detection of multiple cancer biomarkers.
Keywords: Cancer detection, paper-based devices, multiplexing, biomarkers, signal amplification
Graphical Abstract
Paper-based analytical devices, including lateral flow and microfluidic devices, are adept at detecting multiple biomarkers. Growing interests in the multiplexing field in general indicate the potential for multiplex paper devices to improve the understanding of cancer patients’ condition. Here, the developments in the multiplex paper-based devices are reviewed, and the trends in the device configurations and signal amplification approaches are summarized.
1. Introduction
Cancer imposes a high toll on human life and economics. In 2018, 1 in 6 deaths across the globe were caused by cancer and approximately 70% of deaths from cancer occur in low- and middle-income countries.[1] Despite these overwhelming numbers, early detection, diagnosis and monitoring of cancer patients still play a key role in the reduction in mortality rates for cancer and the economic burden on the healthcare field.[2] Current clinical practices for early detection of cancer generally benefit from molecular level identification of cancer biomarkers by common analytical techniques such as enzyme linked immunosorbent assay (ELISA) or quantitative polymerase chain reaction (qPCR) using body fluids such as urine, blood, or serum. Organ-level investigations can also be performed by more complex methods such as mammography, colonoscopy, or computer tomography.[3] However, the need for special instruments (that can be expensive to purchase and operate) as well as personnel that require training to perform (sometimes lengthy) procedures also contribute to the extremely high overall healthcare costs for cancer diagnosis and the screening, especially for longitudinal monitoring of patients.[4] For instance, bladder cancer is ranked 9th in the world in frequency of diagnosis (~430,000 new cases diagnosed annually).[5] Moreover, on a per-case basis, bladder cancer is the costliest cancer type to diagnose and treat ($90,000-$200,000 per person)[6] due to required continuous monitoring via invasive, in-clinic procedures or “send-out lab tests”.[7] If cancer detection could be simplified to easy-to-use, sensitive solutions that remove the need for expensive analytical equipment and highly trained personnel, the costs could be reduced dramatically for this, and other types of cancer.
Paper-based analytical devices, both in the forms of lateral flow assays (LFAs) and microfluidic paper-based analytical devices (μPADs), have been proposed as low-cost alternatives to meet many of the current cancer diagnostic challenges.[8] Paper-based analytical devices are composed of inexpensive substrates that are designed to contain all of the processing steps necessary to detect a particular biomarker using a sample taken from a patient. Capillary-driven fluid flow along the porous paper sample provides for all fluid transport and separation processes on the device, requiring little technical expertise or additional equipment to obtain the test results. The paper substrates of existing paper-based devices provide new opportunities for chemical modification to advance the detection mechanisms for enhanced sensitivity. Further, ease of storage, transport, and disposal attributes make them a robust and user-friendly testing platform that solve many challenges that have so far hindered widespread use of biomarkers for cancer detection.
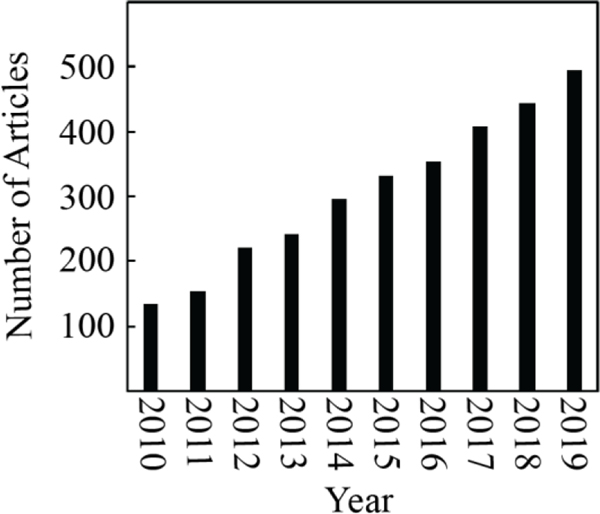
Despite these promising features of paper-based assays, their sensitivity and specificity still require improvement before they can achieve widespread adoption. This lack of sensitivity and specificity is directly related to the low abundance of cancer biomarkers present in body fluids, and even when present at high concentrations, cancer biomarkers are often indicative of other diseases or health conditions. Table 1 summarizes a list of cancer biomarkers highlighting their association with cancer and several other disease conditions. This problem of specificity is not limited to paper-based devices but also exists for laboratory-based analytical testing. Yet, where only one biomarker could potentially indicate multiple disease states, several biomarkers in tandem have a much higher probability of specifically indicating a particular disease. Accordingly, multiplexing approaches that utilize detection of multiple biomarkers in a single assay have been increasingly explored. These studies have benefitted from statistical analyses of genomic data to establish adequate panels of cancer biomarkers that are capable of diagnosing, with higher specificity, many cancer types such as pancreatic cancer[9], prostate cancer, ovarian cancer[10], gastric cancer[11] and recurrence of breast cancer.[10] Particularly, the number of publications within the past ten years in this arena reflects the growing interest in this field (Figure 1).
Table 1.
Common cancer biomarkers and their relationship with various diseases.
| Biomarker | Related Cancer Diagnosis | Related Benign Diagnosis | Ref |
|---|---|---|---|
| μRNA 21 | Breast, colon, lung, pancreas, prostate, and gastric cancer | Cardiac and pulmonary fibrosis, myocardial infarction | [14] |
| μRNA 155 | Breast, colon, lung cancer | Asthma, cystic fibrosis, | [14b, 15] |
| μRNA 424 | Non-small cell lung cancer, breast cancer | Fragile X-associated tremor/ataxia syndrome, amyotrophic lateral sclerosis | [16] |
| μRNA 210 | Breast cancer | Hypoxia, cardiovascular diseases | [17] |
| AFP | Liver, ovarian, testicular cancer | Pregnancy, hepatitis | [18] |
| CEA | Colorectal, gastrointestinal, ovarian, urinary tract, breast, lung, medullary thyroid cancers | Pancreatitis, cholecystitis, peptic ulcer disease, liver cirrhosis, hepatitis, inflammatory bowel disease, and benign extrahepatic biliary obstruction | [19] |
| NSE | Non-small cell lung cancer | Brain injury, spinal cord injury, stroke, cardiac arrest | [20] |
| PSA | Prostate cancer | Prostate hyperplasia, prostatitis | [21] |
| CA 125 | Ovarian | Ovarian cysts, benign ovarian tumors | [22] |
| CA 19–9 | Colorectal cancer, gastric cancer, pancreatic cancer, cholangiocarcinoma | Pancreatitis, cholecystitis, bronchiectasis, bronchiolitis, emphysema, idiopathic interstitial pneumonia, collagen disease associated pulmonary fibrosis, pleural effusion, tuberculosis, diabetes mellitus, cystic fibrosis, renal failure, autoimmune disorders, gastric ulcer, and benign ovarian cyst | [18b, 20] |
| CA 72–4 | Primarily gastrointestinal and gynecological cancers | Pancreatitis, liver cirrhosis, pneumonia, rheumatic illness, and ovarian cysts | [19] |
| CA 15–3 | Breast, lung, ovarian | Benign breast conditions, endometriosis, hepatitis | [23] |
| HCG | Testicular, trophoblastic disease, germ cell tumors, choriocarcinoma | Pregnancy, pregnancy-related disorders, gestational trophoblastic disease | [24] |
| Ratio of PG I/II | Gastric cancer | Superficial and atrophic gastritis | [25] |
| EPCAM | Breast, prostate, lung, colorectal, or non-small cell lung cancer | Congenital tufting enteropathy | [26] |
| GCP3 | Liver cancer and, squamous lung cancer, and non-small cell liver cancer | [27] | |
| CRP | Colorectal cancer, ovarian cancer, and prostate cancer | Arthritis, obesity, diabetes, cardiovascular diseases, and neurological diseases | [28] |
Figure 1.
Current trends in cancer diagnosis field for detecting multiple biomarkers. This data was generated by an advanced keyword-based searching approach on Web of Science with the following search terms: ((multiplex or panel or simultaneous) and (cancer or tumor) and biomarker)) covering the time frame between 2010 and 2019.
As a common goal of multiplex approaches, improvements in the diagnostic performance of screening tests for early-stage cancer diagnosis could provide both system-wide and patient level benefits. For example, a small change in the diagnostic performance of a screening test can have dramatic impacts on the overall cost per cancer detected, depending on the prevalence of the cancer type. An improvement in the discriminative capacity of a screening test, e.g., an increase in sensitivity from 90–95%, will reduce the cost per cancer detected by $230,000 for ovarian cancer when women at age 50–54 (~30 cases/100,000) are screened. However, this improvement will decrease the costs by only $7,900 for lung cancer when smokers are screened (~850 cases/100,000).[12] An enhancement in the diagnostic performance is also proven to reflect improvements in clinical outcomes. In this context, patients with early-stage ovarian cancer could be discriminated from non-cancer individuals with a high accuracy (receiver operating curve-ROC- area under the curve-AUC- of 0.9611 and positive predictive value of 88.9%) using a validated 4-biomarker panel (CA125, HE-4, E-CAD, and IL-6) as opposed to screening a single biomarker (ROC AUC of 0.8513 and positive predictive value of 65.2% for CA125).[13] At the patient level, the effects of the diagnostic performance could be more dramatic. The consequences of false negative results will be substantial for a fast-progressing cancer type with high mortality rates.[12] Moreover, false positive results can impact the patients with several side-effects of unnecessary treatments. Multiplex paper-based devices, in this regard, have been implementing novel signal enhancement strategies to improve the detection capacity for low levels of multiple biomarkers on a single paper device. However, the lack of further statistical analysis that combines the quantified levels of multiple biomarkers to provide a discriminative decision limits their current potential for cancer diagnosis. Accordingly, these devices will only be positioned appropriately for cancer diagnosis if the future studies evolve in this direction.
This review article aims to summarize this growing interest over the past ten years by highlighting recent advancements in paper devices for the detection of multiple cancer biomarkers. After a brief introduction on cancer biomarkers and their biorecognition mechanisms, advances in paper-based device configurations are discussed. Moreover, signal amplification strategies are highlighted for various types of cancer biomarkers that are integrated into multiplexed devices. Finally, advantages and current limitations for detection of multiple cancer biomarkers using paper-based devices are considered while touching upon what could be done to address some of these challenges.
2. Cancer Biomarkers and Current Challenges in Cancer Diagnosis
Some of the most widely studied cancer biomarkers are proteins.[29] Although most markers that are derived from proteomic evaluations are still in the discovery or translation stages, there are already some FDA-approved protein biomarkers used for clinical detection of cancer.[30] These proteins are produced by either cancer cells (or other cells that are interacting with cancer cells) and serve as a signature for cancer diagnosis. These protein markers are sometimes present in blood or urine, allowing for non-invasive sample collection. However, concentration of these markers can be extremely low, making their quantification very challenging to resolve. One way to overcome this problem is to follow protein enrichment procedures[31] to concentrate the amount of protein in samples, but this requires additional labor-intensive protocols and raises overall costs for cancer diagnosis. In addition, the presence of other proteins in body fluid may create background noise and cross-reactivity issues between proteins and the recognition component of the assay, thereby increasing the risk of a false positive signal. Therefore, the low abundance of cancer-specific protein biomarkers remains the greatest issue for achieving sensitive and specific protein detection and subsequent quantification. Enzymes are of particular interest as signatures for cancer diagnosis due to the fact that certain enzyme levels and their distribution in the serum is a reflection of the disease condition.[32] However, enzymes, like most other proteins, are often sensitive to the ambient testing conditions such as temperature, pH, or ionic strength and can lose their activity under extreme conditions. Therefore, enzyme activity-based biorecognition approaches could be significantly affected by the testing conditions leading to poor diagnostic performance if the optimum ambient conditions are not maintained.
Extracellular nucleic acids (circulating DNA/RNA markers) are another type of molecular biomarker for cancer diagnosis.[33] Circulating DNAs are defined as DNA fragments that are originated from tumor cells and are found circulating in the blood.[34] Even though the exact sources of circulating DNA and their transport into the bloodstream are not fully understood, a large fraction of these markers are assumed to form as a result of apoptosis.[35] Because these fragments have been shown at elevated levels in cancer patients’ circulation as opposed to healthy subjects, they are an attractive target for cancer diagnosis.[36] Another type of circulating nucleic acid markers is microRNA (miRNA). miRNAs are endogenous, single-stranded non-coding small RNA with length of ~22 nucleotides that are involved in regulating protein coding genes. Dysregulation of miRNAs can lead to overexpression of oncogenes or decreased expression of tumor suppressor genes that are associated with tumor development.[37] Therefore, differential expression of miRNAs could be a promising tool for cancer screening. Despite their short half-life and low abundance in body fluids, circulating nucleic acids can be quantified by DNA microarrays, northern blot, and quantitative real-time polymerase chain reaction (qRT-PCR). However, currently, these techniques rely upon instrumentation and trained personnel that increase the cost of detection and quantification.
Additionally, circulating tumor cells (CTCs) have been explored in the context of cancer detection. CTCs are rare cell subunits that are shed into circulation by a primary tumor to invade the blood stream and potentially initiate new tumor growth in other parts of the body.[38] Detection of CTCs from blood circulation and their quantification are of great interest as a useful biomarker for diagnosing, treatment management, and monitoring cancer patients.[39] However, CTCs are present at relatively low numbers in the circulation, for example 1–10 CTCs per mL of whole blood as opposed to millions of red blood cells in the circulation.[40] This makes their isolation and quantification a complex and labor intensive process. Similar to CTCs, cancer cell-excreted vesicles such as exosomes have received recent interests as a promising diagnostic tool.[41] Exosomes are membrane-bound vesicles with 30–150 μm in size and carry a range of cellular information including DNA, RNA, and proteins. They are found in different body fluids such as blood plasma, serum, urine, breast milk, and therefore, very attractive biomarkers for cancer diagnosis.[42] A number of different separation techniques have been applied to isolate exosomes from these body fluids including ultracentrifugation, precipitation and size exclusion chromatograph.[43] However, lipoprotein contamination in blood samples is still a major issue and needs to be addressed with the development of new purification methods for the exosome extraction.[44]
Regardless of biomarker type or measurement technique, single biomarker driven cancer diagnosis approaches also suffer from a lack of specificity toward a single cancer type. This is directly related to the substantial heterogeneity in cancer tissues and the biomarker’s track record as a signature for other diseases. In addition, clinical studies have already demonstrated strong evidence that a combination of biomarkers in a multiplex format would contain more useful information about the presence of disease, and even disease development (as compared to a single biomarker), significantly increasing diagnostic accuracy.[45] However, current detection strategies for multiple biomarkers require high-cost instrumentation and operation by skilled personnel. In this regard, paper-based analytical devices offer rapid, simple, low cost, and instrument-free detection approaches for low abundance cancer biomarkers. In the following section, the fundamentals of cancer biomarker biorecognition mechanisms and their implementation to paper-based analytical devices are briefly discussed.
3. Biorecognition and Detection Mechanisms of Paper-Based Devices
Lateral flow assays (LFAs) and microfluidic paper-based analytical devices (μPADs) are widely studied formats for paper-based analytical devices. The biomarker recognition mechanism used in these paper devices relies upon either label-based or label-free detection methods. Label-based assays mainly identify the biomarker from a complex mixture of body fluid (in which many proteins and small molecules are present along with the target biomarker) through highly specific interactions between the capture ligand and the biomarker. In the sandwich format, labeled biomarkers are separated from other constituents through their attachment to detection ligands that are localized on the paper surface. In this case, signals originating from labels are collected at the detection site, and signal intensity is directly proportional to the amount of biomarker that is captured on the paper. As an alternative method, a competitive format of biorecognition can be used whereby the biomarker competes with the label to be captured by the detection ligand. In this competitive format, the amount of captured label present on the paper is inversely related to the amount of biomarker.
Depending on the biomarker type, a number of different ligands are available for the biomarker recognition process. In this process, capture ligands are conjugated on to the surface of the label (especially in the sandwich format assays) to separate the biomarker from the liquid sample. Then, detection ligands (that are immobilized on the paper surface at the detection site) capture the labeled biomarker as the liquid sample flows across the paper. The most commonly used ligands are antibodies, which are known for their affinity to protein and enzyme biomarkers through non-covalent interactions. Based on their production method, antibodies can be classified as polyclonal or monoclonal antibodies. Polyclonal antibodies are produced by a combination of various B cell clones from animal sources. Therefore, they contain a heterogeneous population of antibodies that are specific to multiple different epitopes and possess batch-to-batch variations.[46] Due to this heterogeneity, diagnostic methods are usually faced with cross-reactivity issues and subsequent false positive results, especially when used for the recognition of multiple biomarkers.[47] Alternatively, monoclonal antibodies that are produced from a single cell line demonstrate much higher affinity towards a single epitope.[48] Further, monoclonal antibodies have minimal batch-to-batch variations. With the caveat that monoclonal antibodies are relatively expensive to produce, their enhanced specificity for a target biomarker makes them suitable for capturing/detection ligands for multiplex assays. There are also other non-antibody protein ligands that have been explored in the context of paper-based devices due to their cost-effective production and antibody mimicking properties.[49] For example, nanobodies are the smallest antibody fragments with a full biomarker or antigen-binding potential.[50] Due to their characteristic shape, nanobodies can bind structurally hidden binding domains in small cavities that may be inaccessible to conventional antibodies.[51] Additionally, non-antibody binding proteins such as affibodies, DARPins, and anticalins are usually engineered from a constant core protein with a number of biomarker recognition surfaces to mimicking the function of antibodies[52] and therefore, have a great promise to play a cost-effective role in paper-based devices.
Aptamers are another class of capturing ligands. Aptamers are short-chain peptides or single stranded oligonucleotides with high affinity to protein or nucleic acid-based biomarkers through non-covalent interaction or hybridization event, respectively. Oligonucleotide-based aptamers are produced by a process called systematic evolution of ligands by exponential enrichment (SELEX).[53] In this method, aptamers with a high affinity to a target biomarker are selected from a library of oligonucleotides. Then, non-binding aptamers are discarded, and the number of bound aptamers is expanded by a PCR cycle. The aptamer selection and expansion process is repeated for multiple rounds to enhance the enrichment of the oligonucleotide pool. The resulting collection of aptamers is highly specific for the target biomarker, and therefore, can potentially overcome the cross-reactivity challenges from which most antibody-based biorecognition approaches suffer. Additionally, due to this synthetic preparation method, selected aptamers are reproducible. Aptamers also exhibit higher stability at ambient conditions as compared to antibodies, making them potential candidates to replace or complement the biorecognition role of antibodies.[54]
In addition to capture/detection ligands, the selection of appropriate labels is another important consideration in the biomarker recognition and detection processes for paper-based devices. In this context, labels can be composed of functional nanoparticles (NPs) that have distinct optical, electrical, or magnetic properties (as well as surface chemistries) that allow for flexibility in the conjugation methods for different capture ligand types.[55] The most widely used nanoparticles in paper-based devices include gold nanoparticles (Au NPs), latex (polystyrene) beads, or quantum dots (QDs). Specifically, Au NPs are known for their unique optical properties that depend on their size and shape that provides easy-to-read visual output (color) once they are captured on the paper surface. This visual signal can usually be detected with the naked eye and provides semi-quantitative readouts for colorimetric measurements. In addition, the color intensity can be quantified by external imaging tools to help determine the biomarker amount. Similar to Au NPs, latex beads are also useful materials for color driven biomarker quantification and are cost effective alternatives to Au NPs. However, the color created by Au NPs or latex beads may be similar to the natural color of the sample, especially in whole blood samples that are applied to the paper device. In such cases, color-inducing labels that are featured with more discriminative color could be implemented to overcome this issue. Even though Au NPs and latex beads are capable of generating easy-to-read outputs, the visual signals are usually not sensitive for quantitative measurements. As an alternative to Au NPs and latex beads, QDs are nanoscale semi-conducting materials with superior fluorescence brightness and have a higher stability when compared to fluorescence dyes. When such materials are excited by a light source (usually in the UV range), they emit light at the longer wavelengths. The intensity of the emitted light can be correlated with biomarker concentration. Even though QDs generate fluorescence signals with high signal-to-noise ratio and sensitive biorecognition platforms, their bioconjugation with capture ligands can be a complex process that limits their wide implementation to the paper-based devices.[56]
Some of the aforementioned issues with low sensitivity of colored NPs and limited ligand bioconjugation capability of QDs have led to new innovations in paper-based devices and have opened up alternative routes for label-free detection approaches.[57] The biorecognition mechanism of these approaches is similar to the label-based detections in that ligands (capture and detection ligands) are used to capture the biomarker on the detection site. However, as opposed to the use of labels in order to detect a biomarker, label-free detection methods rely upon a change in the detection signal, either an electrical signal for electrochemical platforms or an optical signal for plasmon-based methods, that is realized as a result of the presence of the biomarker. The change in the detection signal could be either the case with a biomarker binding to a surface at the detection site (producing the electrical signal or a change in the light absorption feature), or the biomarker reacting with a special reagent (such as redox active material) and the resulting chemical product inducing a change in the electrical property at the detection site. This detected signal can be correlated with biomarker concentration for quantitative measurements.
So far, we have summarized the biorecognition elements and the components for detection mechanism for a typical paper-based device. In the following sections, we will focus on the implementation of these necessary elements to the multiplex form of paper-based devices. We will emphasize configuration aspects of the paper devices, as well as detection and signal amplification methods for sensitive and specific detection of multiple cancer biomarkers.
4. Multiplex Lateral Flow Assays
4.1. Operational and Configurational Aspects of Multiplex LFAs
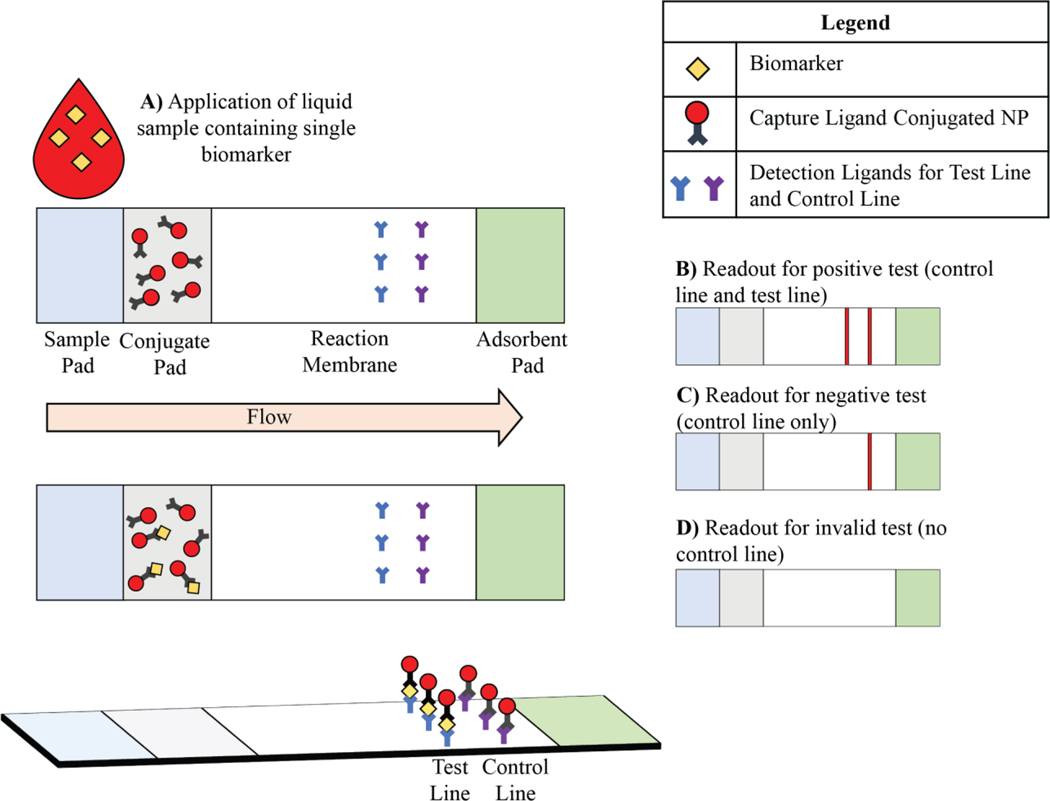
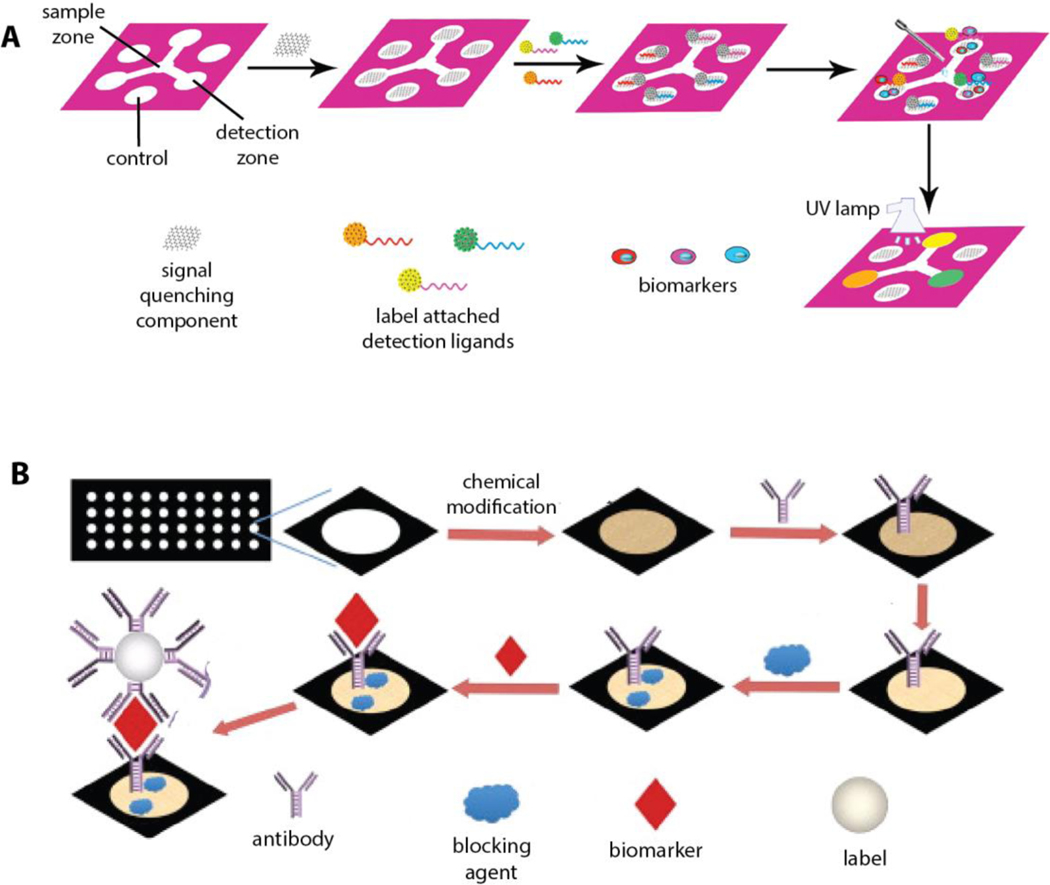
A typical LFA for detecting a single biomarker consists of four primary components: a sample pad, conjugate pad, reaction membrane, and adsorbent pad, each serving a distinct function on LFAs. These pads are oriented in this sequence with some degree of overlap in order to facilitate flow of the liquid sample through the device. The sample pad is where the liquid sample is applied to the paper device. The paper is pre-loaded with buffers, surfactants, blocking reagents and additives that help to account for any chemical variability present in the sample. Next is the conjugate pad that contains pre-deposited capture ligand-conjugated NPs; as the liquid sample flows from the sample pad to conjugate pad, the desired analyte attaches to the capture ligand-conjugated NPs. The liquid sample then meets the reaction membrane that includes detection ligands that are printed on the membrane in the form of two parallel lines. The first line (test line) contains detection ligands that are specific to the biomarker while the detection ligands on the second line (control line) can only capture NPs. Finally, the adsorbent pad acts as a sink to continuously collect the liquid from the reaction membrane and therefore, drives the flow of the sample through the device (Figure 2). These four pads are then set in a backing and encased in a cassette.
Figure 2.
Schematics illustration of a common sandwich format LFA configuration and its application on single biomarker detection.
The materials used for each of these paper components plays a key role in enabling the pad to function adequately. For example, the sample pad and adsorbent pad are often cellulose-based papers with a high capacity to adsorb liquid. Meanwhile, the conjugate pad is usually made from glass fibers that allows for NPs to flow across the reaction membrane as the liquid diffuses. Lastly, the reaction membrane is most commonly made from nitrocellulose; providing a high level of irreversible adsorption for detection ligands and helps the detection ligands remain attached on the membrane as the liquid moves across.[58]
The configuration of a typical LFA that is mentioned in this section could be modified so that multiple biomarkers can be detected on a single LFA and a number of emerging design strategies have already been explored for various applications.[59] The most commonly explored device design for multiple cancer biomarker detection has a single LFA device format with multiple spatially arranged detection sites on the reaction membrane. Detection ligands for each biomarker are immobilized either as multiple test lines or test dots. Depending on the label’s characteristics and the adequate detection method, the presence of each biomarker can be confirmed and further quantified.
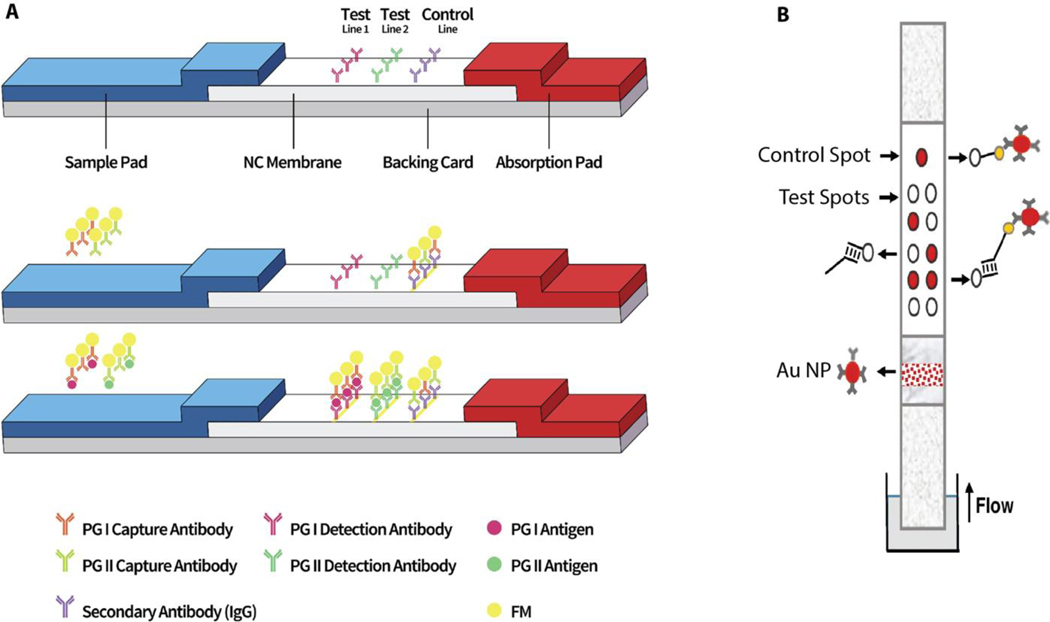
Multiplex LFAs in a multiple test line format have been explored for detection of biomarkers for a number of different cancer types. For example, Li et al. developed multiplex LFA using fluorescent latex beads to detect two gastric cancer biomarkers, pepsinogen I (PG I) and pepsinogen II (PG II).[60] Monoclonal antibodies for PG I and PG II were printed as two test lines on the reaction membrane to detect the biomarkers (Figure 3a). The fluorescence signals generated on the test lines were analyzed by a custom design analytical device to quantify PG I and PG II levels as well as the ratio of PG I to PG II which is accepted as another indication of gastric cancer.
Figure 3.
Schematic description of LFA configuration designs for multiplex cancer biomarker detection. a) PG I and PG II were captured at multiple test lines, Reproduced with permission.[60] Copyright 2019, Elsevier. b) Alleles-specific primers were captured at oligonucleotides containing multiple test dots. Reproduced with permission.[61] Copyright 2019, Springer.
Another LFA design for detection of multiple cancer biomarkers involves the immobilization of detection ligands as multiple test spots on a single device. Recently, Galaziou’s group designed an LFA strip with 10 detection spots that is capable of testing single nucleotide polymorphisms (SNP) that are related to breast cancer.[61] The device is configured such that each SNP is demonstrated by a pair of spots where left and right spots represent the presence of wild-type and mutated alleles, respectively. At the detection spots, complementary oligonucleotides to the specific primers were immobilized (Figure 3b). The device detected and quantified variations in five alleles using antibody-functionalized, Au NPs by using the visual signals. The color intensity of the detection spots was measured by a common flatbed scanner in order to determine the allelic fractions. The assay’s performance using clinical samples demonstrated that it could be used to assist in the diagnosis of breast cancer.
Although an LFA configuration with multiple test lines or dots can be useful for detecting multiple cancer biomarkers, the space allocated in the multiplexing detection area and the size required for the resolution of signal (e.g., lines/dots) ultimately limits the number of target biomarkers than can be detected. However, one way to overcome this problem would be to increase the size of the reaction membrane in order to fit more test lines/dots as needed. In this case, the operational length of the paper device becomes longer, and the time needed for the sample to diffuse across the device will necessarily be exponentially longer, limiting the speed of the assay performance.[62] Alternatively, more test lines/dots can be printed at a given detection area with smaller separation. However, this may cause issues with signal interference that originate from neighboring detection areas next to each other and will also subsequently affect the analysis for biomarker quantification. In this case, incorporation of multicolor labels to the device configuration that are specific to each target biomarker would be another approach to achieve multiple biomarker detection in the same space on an LFA.
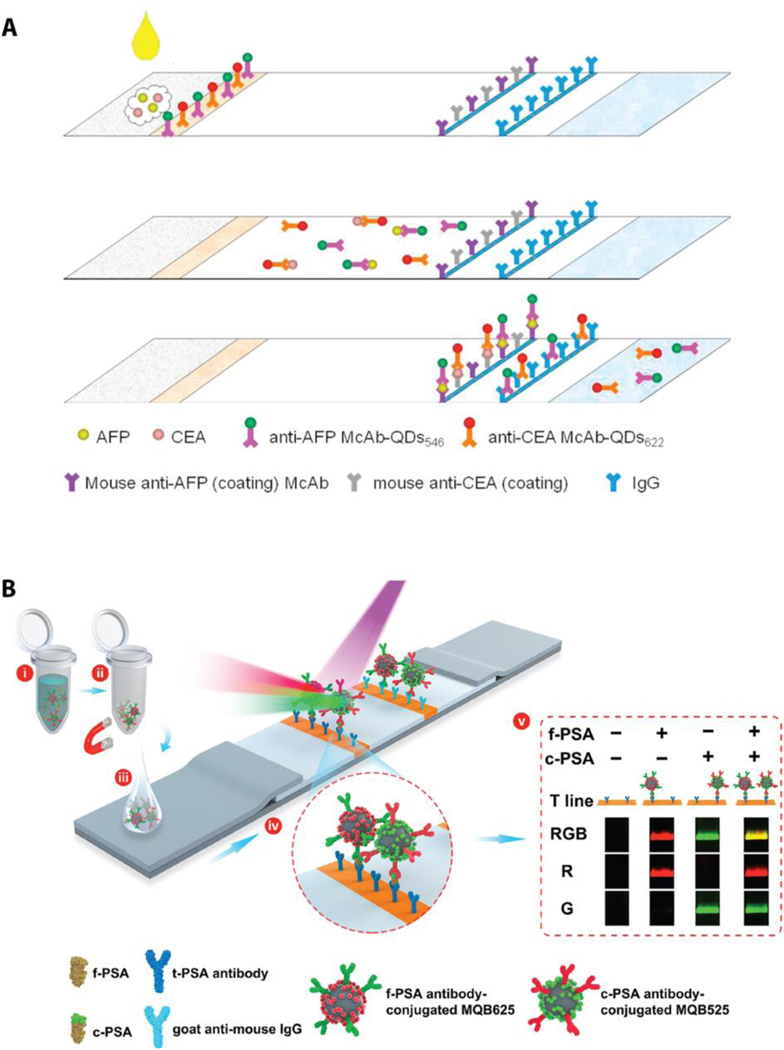
Wang et al. implemented a multicolor labeling approach to develop an LFA for multiplex detection of cancer biomarkers CEA and AFP.[63] Instead of creating multiple lines, detection ligands (mouse anti-AFP and mouse anti-CEA) were localized at the single test line (Figure 4a). Moreover, QDs of two different colors were used as the labeling material for the biomarkers. Specifically, QDs were selected from commercially available products that emit lights at 546 nm and 620 nm upon UV exposure. The LFA developed in this study was tested on human serum samples and demonstrated high specificity (94% AFP and 97% CEA) and high sensitivity (93% AFP and 87% CEA). While this LFA was limited by the number of antibodies that was immobilized on a single line, the results indicated the potential to detect multiple cancer biomarkers with multicolor QDs. In a similar approach, Rong et al. demonstrated a paper device with multiplex detection capacity for free PSA (f-PSA) and complex PSA (c-PSA) using multicolor magnetic quantum dot nanobeads (MQBs) on a single test line.[64] In this example, MQBs include super paramagnetic iron oxide at the core while quantum beads are attached to the polymer (PEI)-coated core via electrostatic interactions. MQBs with red color and green color were then conjugated with antibodies for f-PSA and c-PSA, respectively (Figure 4b). The magnetic features of these label components aid in the isolation and enrichment of biomarker-MQBs complexes from the liquid sample. Once the sample diffused on the paper device, red and green color tagged PSA complexes were captured by the mouse monoclonal t-PSA antibody that was spotted at the test line. A custom designed, smartphone-based optical device was used to record the optical signals of MQBs that were detected at the test line. The simultaneous detection of f-PSA and c-PSA in clinical serum samples were compared with the values that were obtained by a commercial immunoassay system. The results were consistent with the commercial system.[64]
Figure 4.
Schematic representation of multiplex LFA configurations with multicolor labels and biomarker detection at the single test line. a) AFP and CEA are labeled with multicolor QDs and captured at the single test line. Reproduced with permission.[63] Copyright 2015, Elsevier. b) Multicolor magnetic quantum nanobeads (MQBs) are used to label f-PSA and c-PSA in order to detect at the single test line. (i) Mixing biomarkers with MQBs, (ii) magnetic field induced biomarker separation, (iii) sample application on the LFA, (iv) capturing biomarkers on the antibody immobilized test line, (v) detection of biomarkers and interpretation of the resulting measurements. Reproduced with permission.[64] Copyright 2019, Elsevier.
After introducing configurational aspects of LFAs that are used for multiple cancer biomarker detection, we will now focus on specific examples of multiplex LFAs that are designed for detecting protein, nucleic acid and exosome-based biomarkers (Table 2).
Table 2.
Summary of LFAs for the multiplex detection of cancer biomarkers.
| Biomarkers | Sample | Configuration | Signal | Reporter | Limit of Detection | Analysis Time | Ref |
|---|---|---|---|---|---|---|---|
| CEA and CA15–3 | Serum | Multiple Test Lines | Fluorescence-quenching | QDs and QBs | CEA: 0.06 ng/mL CA15–33: 0.09 ng/mL | 2–3 min for visual, 15 min for fluorescent | [70] |
| CEA and NSE | Serum | Multiple Test Lines | Fluorescent | QDs and QBs | CEA: 0.0378 ng/mL NSE: 0.0378 ng/mL | < 15min | [65] |
| fPSA and cPSA | Serum | Single Test Line | Fluorescent | Multicolored QDs | fPSA: 0.009 ng/mL cPSA: 0.087 ng/mL | 60 min | [64] |
| AFP and CEA | Spiked serum | Single Test Line | Fluorescent | Multicolored QDs | AFP: 3ng/mL CEA 2ng/mL | 15 min | [63] |
| CEA and NSE | Serum | Multiple Test Lines | Magnetic | Magnetic Nanobeads | CEA: 0.045 ng/mL NSE: 0.094 ng/mL | N/A | [68] |
| CEA, and CYFRA: 21–1 | Serum | Multiple Test Lines | Fluorescent | QDs | CEA: 0.35 ng/mL CYFRA: 21–1: 0.16 ng/mL | 15 min | [67] |
| CEA and AFP | Serum and whole blood | Multiple Test Lines | Visual and chemiluminescent | Au NPs | AFP: 0.21 ng/mL CEA: 0.2 ng/mL | 30 min | [71] |
| PG I and PG II | Plasma | Multiple Test Lines | Fluorescent | Fluorescent Microsphere | PG I: 2.6 ng/mL PG II: 1.0 ng/mL | 15 min | [60] |
| SNPs | Spiked bovine serum | Multiple Test Spots | Visual | Au NPs | 5 fmol of extended primer (~17 pM) | 2 hr | [61] |
| miRNA (21, 155, 210) | Spiked serum | Multiple Test Lines | Colorimetric | Au NPs | μRNA 21: 0.073 nM μRNA 155: 0.061 nM μRNA210: 0.085 nM | 10 min | [75] |
| miRNA (210, 424) | Spiked serum | Multiple Test Lines | Colorimetric | Au NPs | 10 pmol (~2 μM) | 2–5 min | [76] |
| KS and BA associated DNAs | Buffer | Multiple Test Lines | SERS | Au NPs- | DNA (KS): 0.043 pM DNA (BA): 0.074 pM | 20 min | [80] |
| Synthetic biomarkers | Urine | Multiple Test Lines | Fluorescent | Au NPs | 0.1 pM | 30–60 min | [73] |
| Exosomes from healthy (CD63) and prostate cancer (CD44) | Spiked serum | Multiple Test Lines | Fluorescent | Fluorescent dye | Healthy (CD63): 0.6 ng/mL Prostate cancer (CD44): 1 ng/mL | <10 min | [84] |
4.2. Multiplex LFAs for Protein Biomarkers Detection
A number of signal enhancement and signal amplification approaches have been implemented to multiplex LFA designs in order to solve biomarker’s low abundance problem. For example, Xiao et al. developed an LFA capable of detecting NSE and CEA, which are biomarkers associated with lung cancer.[65] This device implemented a quantum beads (QBs)-based signal enhancement approach. Briefly, QBs incorporate many QDs into a single NP. Given that protein conjugation on a 5 nm QD is limited to only 2–5 proteins with about 100 kDa in size,[66] QBs can increase the quantity of antibody-bound QDs and enhanced the fluorescent signal. In addition, a simple custom designed handheld device known as a Handing system was used to detect the fluorescence signals. This study demonstrated that QBs had a greater sensitivity than QDs for the detection of CEA and NSE. When extended to clinical samples, Xiao et. al’s multiplex LFA detected CEA with a sensitivity of 99% and specificity of 97% while sensitivity and specificity for NSE were 97% and 100%, respectively.[65] In another work, Chen et al. studied a multiplex LFA that used fluorescent quantum dot-doped carboxylate-functionalized latex NPs to label a Cytokeratin-19 fragment (CYFRA 21–1) and CEA for early screening and prognosis of lung cancer.[67] This LFA utilizes the multiple test line format to spatially localize CYFRA 21–1 and CEA on the paper. Further, fluorescence signals collected from the test lines were analyzed with a portable fluorescence strip reader in order to quantify the amount of biomarker. The quantification results from clinical serum samples were comparable to those obtained via commercial immunoassay kit.
Another signal enhancement approach includes magnetic NPs as the label component of the paper device. Magnetic NPs exhibit a high signal-to-noise ratio on paper given that paper substrate has no magnetic background. Therefore, magnetic NPs-based LFAs provide considerable advantages over LFAs that generate optical-based signals for the detection of biomarkers. Using this approach, Lu et al. designed a multiplex LFA capable of detecting CEA and NSE using magnetic NPs.[68] Their custom-made magnetic NPs consisted of iron NPs with carboxyl surface modification to allow for antibody conjugation. The magnetic NPs that were 80 nm in size were found to be the optimum size for the assay performance. Once the assay was performed on this paper device, the magnetic signals on the test lines (one for CEA and one for NSE) and the control line were recorded by a magnetic assay reader. The magnetic signal intensities were correlated with the biomarker concentration using the standard curve that was generated from a plot of known biomarker concentration vs magnetic signal intensity. This paper device showed high specificity (97% for NSE and 95% for CEA) and sensitivity (87% for NSE and 93% for CEA) for the lung cancer biomarkers when tested with clinical serum samples.[68]
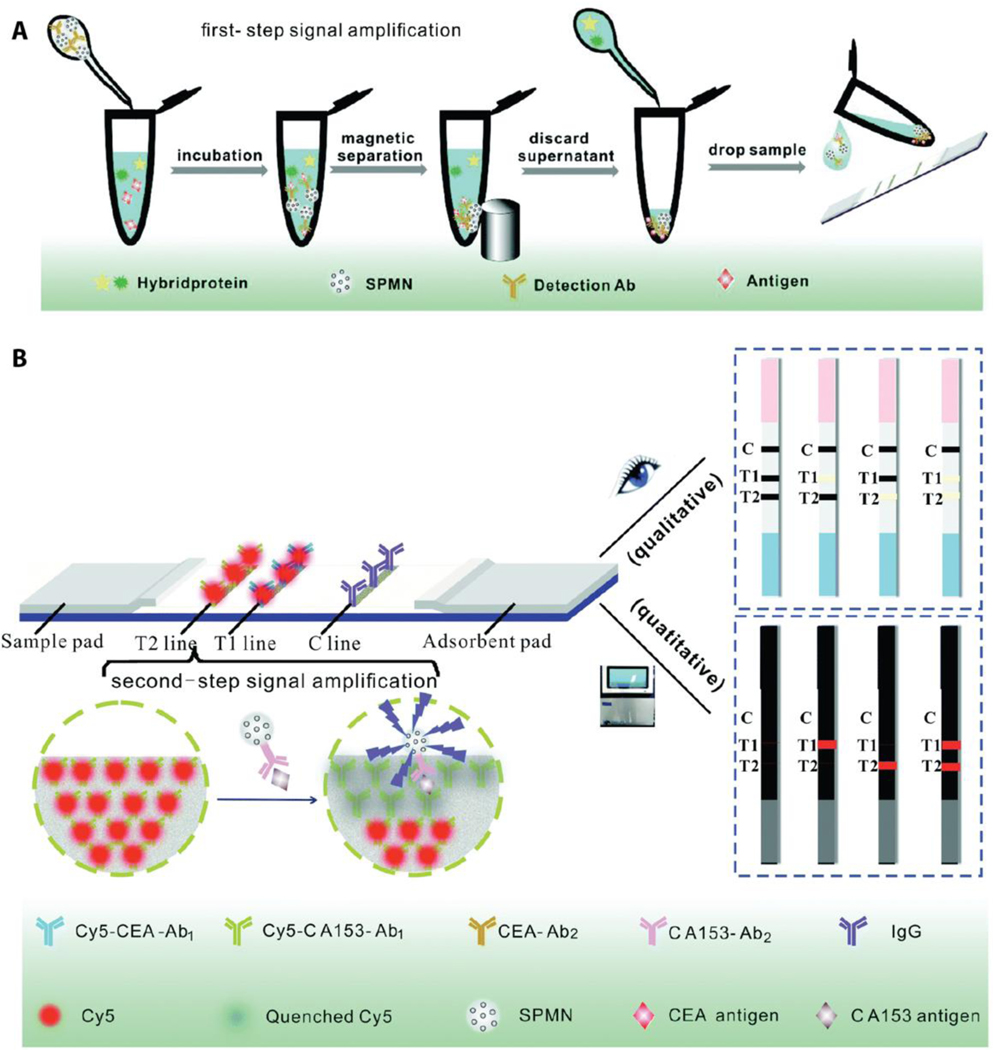
In addition to their feature of high signal-to-noise ratio as the label component for the multiplex LFAs, magnetic NPs have also been used for protein enrichment and separation from body fluids. In this case, magnetic NPs interact with the target biomarker to form complexes that can enhance the target biomarker concentration and detection sensitivity. If a magnetic NP-driven biomarker enrichment process is combined with a fluorophore-based detection method, the assay will suffer from an undesired fluorophore quenching problem. This is mainly due to the ability of magnetic NPs to quench various fluorophores with high efficiency.[69] Zhang et al. utilized this fluorescence quenching feature in order to create a multiplex assay to quantify CEA and CA153 for breast cancer diagnosis.[70] In this work, Cy5 molecules were utilized as a fluorescent label and detection antibodies conjugated with Cy5 molecules were immobilized on the two test lines. Additionally, iron oxide-based super paramagnetic nanoparticles (SPMN) were conjugated with the capture antibodies of CEA and CA153 to enable magnetic-field-induced biomarker enrichment and separation. SPMN-biomarker complexes that are aggregated in the test lines reduce the fluorescence intensity of Cy5 molecules, which can be then correlated with the amount of biomarker. The two-step-signal enhancement method (first via protein enrichment and second via quenching of fluorophore molecules) increases the sensitivity of this paper device with the limits of detection of 0.06 ng/mL and 0.09 U/mL for CEA and CA153, respectively. Moreover, SPMN-biomarker complexes form a visual signal on the paper device for rapid colorimetric qualitative readout. SPMNs also provide a 5-fold enhanced colorimetric response for the enriched biomarker when compared to the visual signal generated from the non-enriched sample. When tested with clinical serum samples, this dual-mode LFA demonstrated consistent clinical efficiency with the commercial diagnostic technologies in terms of sensitivity and specificity for the detection of both biomarkers for diagnosis of breast cancer.[70] Figure 5 represents a schematic demonstration of SPMN-induced biomarker enrichment from the body fluid as well as simultaneous qualitative and quantitative detection of CEA and CA153.
Figure 5.
Schematic illustration of the multiplex LFA with two step signal enhancement approach. a) Antibody conjugated super paramagnetic nanoparticles (SPMN) form complexes with target biomarkers of CEA and CA153 and these complexes are separated from the sample with an external magnet. b) Once SPMN-biomarker complexes are introduced to the multiplex LFA, captured CEA and CA153 results in loss in fluorescence signal of Cy5 due to SPMN induced fluorescence quenching. The aggregated SPMN also generate rapid visual color in the test line. Reproduced with permission.[70] Copyright 2017, Royal Society of Chemistry.
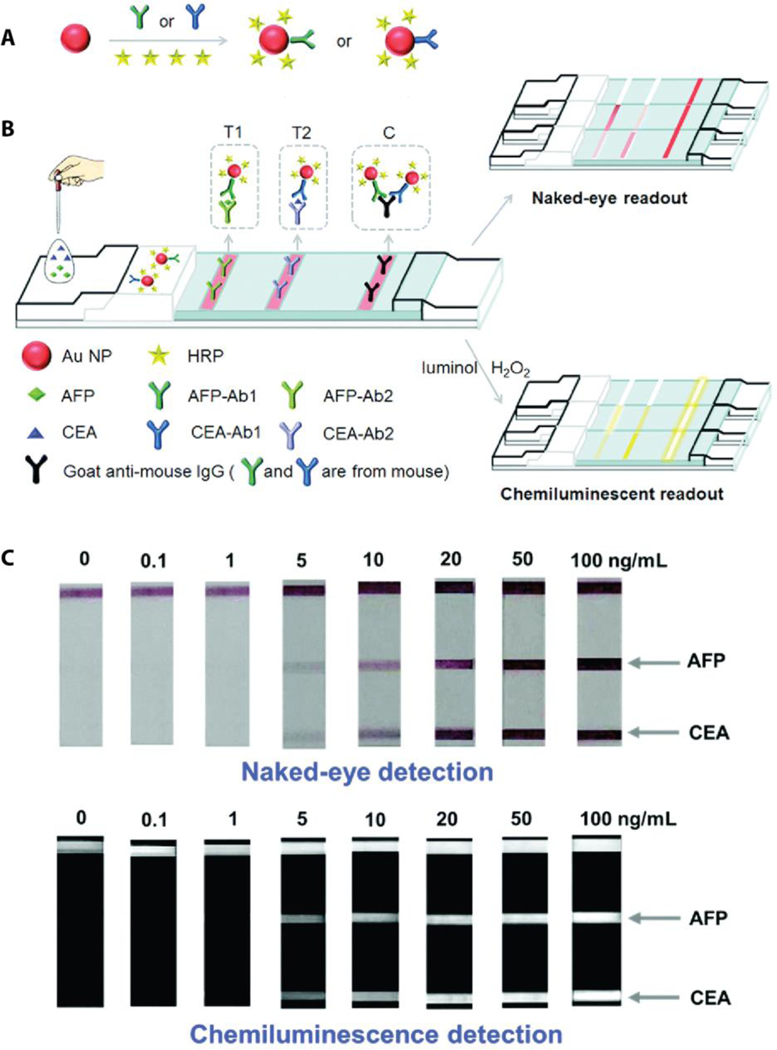
Another common approach for amplification of detection signal involves the collection of emitted light that is produced from a chemical reaction, also known as chemiluminescence. In a typical chemiluminescent LFA, capture antibodies are tagged with enzymes. Once the target biomarker-capture antibody complex is captured at the test line, enzymes are then used to create optical signals through production of light emitting components that are formed by enzyme-substrate interaction. Horseradish peroxidase (HRP) conjugated capture antibodies are commercially available enzyme-antibody conjugates that usually carry one or two HRP molecules on the antibody for a chemiluminescence reaction. In this regard, Chen et al. prepared a multiplex LFA that included Au NPs with antibody conjugated HRP for chemiluminescence detection of CEA and AFP, diagnostic biomarkers for malignant tumors.[71] The optical signal that was generated from HRP-luminol reaction in the presence of H2O2 was detected by a portable chemiluminescence analyzer. The most striking feature of this multiplex device is the large number of HRP molecules that are attached to a single Au NP (243 HRP per Au NP). This large number of HRP molecules accounts for the amplification of optical signal, producing a limit of detection of 0.2 ng/mL for CEA and 0.21 ng/mL for AFP. In addition to these low limits of detection, Au NPs create visual readouts at the test lines that can be seen with the naked eye indicating CEA and AFP at the level of 5 ng/mL (Figure 6).
Figure 6.
A multiplex LFA implements enzymatic signal amplification strategy and provides dual-readouts for sensitive detection of cancer biomarkers of CEA and AFP. a) The scheme of Au NP conjugation with the capture ligands (antibodies for CEA and AFP) and HRP molecules. b) Schematic representation of CEA and AFP detection by visual and chemiluminescent readouts. c) The results of simultaneous detection of CEA and AFP on the multiplex LFA demonstrate the detection limits for visual and chemiluminescence readouts. Reproduced with permission.[71] Copyright 2016, Royal Society of Chemistry.
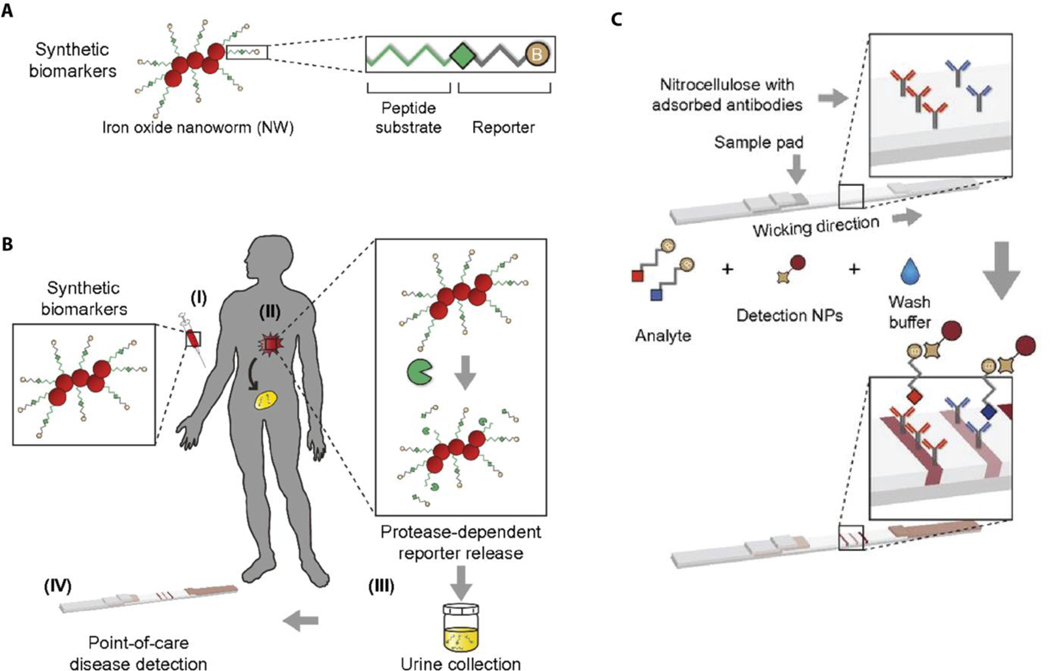
Rather than a direct investigation of biomarkers from samples of body fluid, an unconventional amplification approach would be to explore the products of an in vivo response of dysregulated activities that are subsequently cleared in body fluid.[72] Warren et al. proposed such an in-situ amplification technique to recognize multiple engineered synthetic cancer biomarkers to detect colorectal cancer from urine samples.[73] In this study, synthetic peptide-coated iron oxide nanoworms are administered to the circulation, which are engineered to accumulate in the tumor tissue. In the presence of matrix metalloproteinases (MMPs), which are over-expressed in solid tumors, the surface peptide layer of the synthetic biomarkers is cleaved, releasing biotinylated reporter peptides that will ultimately be eliminated in the patient’s urine. (Figure 7). In addition to these nanoworms, biotinylated free reporter peptides are injected intravenously, allowing for normalization of any variation in the urine. Once patient samples are applied to the multiplex LFA, both reporters and free reporters are labeled with streptavidin conjugated Au NPs and then captured at different test lines by reporter specific antibodies. Then, the paper device is scanned to quantify the relative colorimetric reporter intensities as opposed to the signal of free reporter signal. In order to determine the accuracy of this device, the rate of true positives (sensitivity) and false positives (1-specificity) are analyzed by receiver-operating characteristics curves. The results show that the multiplex LFA is highly accurate and discriminates colorectal cancer with an AUC of 0.90.
Figure 7.
Protease sensitive nanoworms probe the presence of tumor tissue in the body. a) Synthetic biomarkers include iron oxide NPs that are conjugated with protease specific peptide substrate and the peptide reporter. b) (I) Synthetic biomarkers are intravenously injected to the patient. (II) Nanoworms accumulate at the tumor site and release reporters through proteolytic cleavage of peptide substrate. (III) A patient’s urine sample is collected. (IV) The urine sample is applied to the multiplex LFA for the diagnosis. c) Detection antibodies are immobilized at the test lines and reporter analyte conjugated Au NPs aggregate at the test lines as the urine sample diffuses across the paper device. Reproduced with permission.[73] Copyright 2014, National Academy of Science.
4.3. Multiplex LFAs for Nucleic Acid Biomarkers Detection
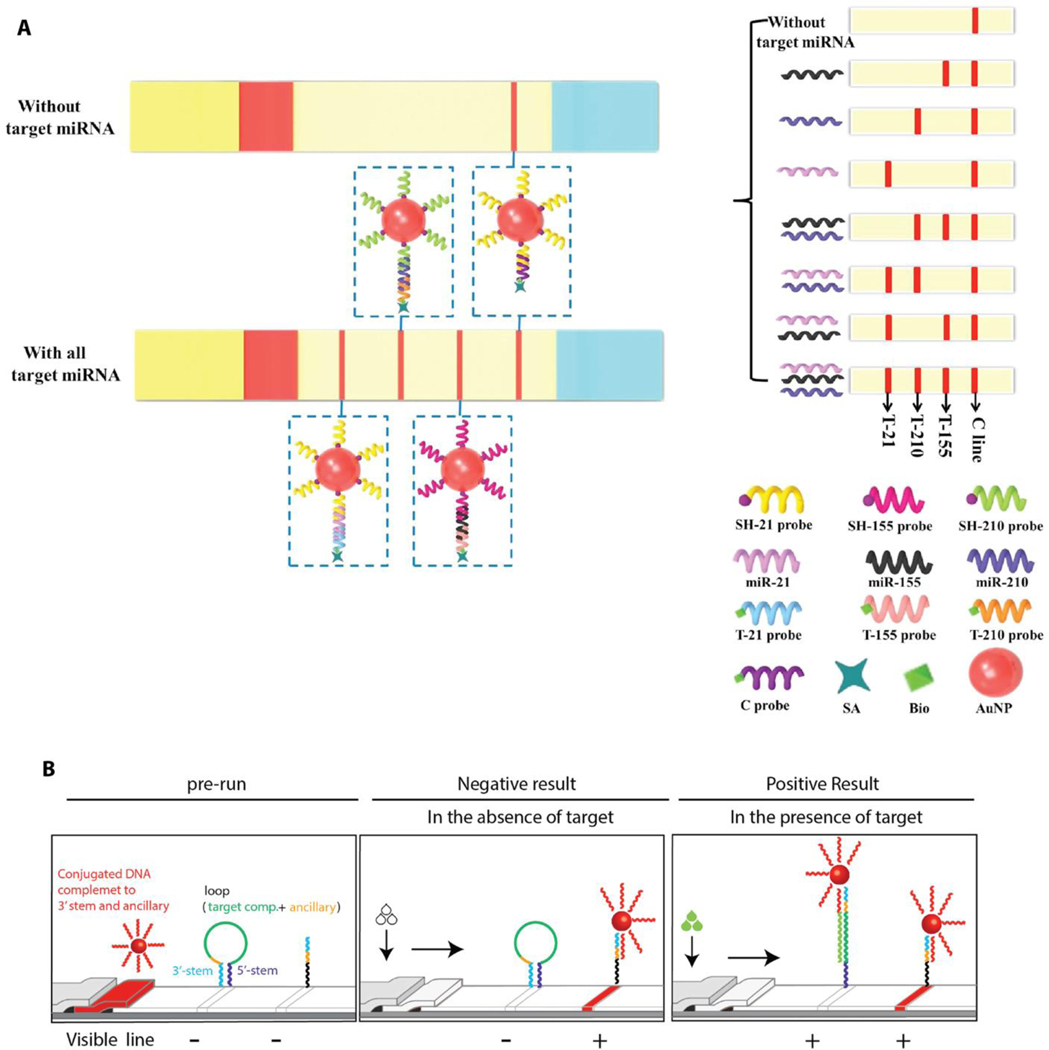
Similar to protein biomarkers, nucleic acid-based cancer biomarkers also suffer from a limited abundance in body fluid. This becomes a critical issue if the target biomarker is downregulated in the presence of cancer. Accordingly, recent studies have devoted considerable effort toward development of sensitive and specific methods for miRNA detection.[74] For example, Zheng et al. have developed an LFA for the visual detection of multiple miRNAs using a sandwich-type nucleic acid hybridization reaction, similar to sandwich immunoassays with antibodies.[75] Specifically, Au NPs were conjugated with thiol-modified single stranded DNA (ssDNA) in order to capture the target miRNAs from the liquid sample. The streptavidin-biotin modified detection ligands (ssDNAs) were immobilized on the test lines. Once the liquid sample including the target miRNAs (miRNA 155, 21, and 210) migrates across the paper device, the sandwich-type nucleic acid hybridization reaction results in ssDNA-miRNA-ssDNA/AuNP complexes that aggregate at the test lines to form visual signals (Figure 8a). Further quantification of the three miRNAs were performed by the intensity measurement of the test lines using ImageJ software. The resulting threshold for the visual detection of miRNA 155, 21, and 210 were found to be 0.01 nM, 0.01 nM, and 0.05 nM, respectively. In addition, the detection limits of these miRNAs were lower than the enzymatic-based signal amplification method. Javani et al. demonstrated a unique design of an LFA for detection of miRNA 210 and miRNA 424 where unmodified detection ligands (oligonucleotide molecular beacons) captured the target miRNAs on the test lines.[76] In typical nucleic acid-based LFAs, the detection ligands (ssDNAs) are usually modified with streptavidin-biotin in order to enhance their immobilization on the paper. This study demonstrated that the detection ligands (molecular beacons) can be immobilized on the paper without protein-based modification, reducing the overall cost of the paper device. These molecular beacons consisted of a single-stranded hair-pin DNA with characteristic stem and loop structure. The loop includes 18–30 base pairs that is complimentary to a specific target (miRNA 210 and miRNA 424 in this case) with the stem having 5–7 base pairs.[77] Molecular beacons were attached to the paper at the test lines from their stem, the 3′ and 5′ ends, and their loop structure was in a closed state in the absence of a target miRNA. In the presence of a target miRNA, the loop structure opened to hybridize the target to capture at the test line (Figure 8b). In addition to molecular beacon-based detection mechanism, this multiplex LFA incorporated DNA-conjugated, Au NPs to tag the target miRNAs. Thus, the color formation at the test line was associated with the Au NPs confirmed the presence of target miRNAs. Colorimetric signals were analyzed, and the limit of detection was found as 10 pmol for both miRNA 210 and miRNA 424.
Figure 8.
Schematic illustration of nucleic acid-based LFAs for multiple miRNA detection. a) Target miRNA-specific ssDNAs serve as the capturing and detecting ligands. Sandwich-type hybridization complexes (ssDNA-miRNA-ssDNA-Au NR) are formed at the test lines for colorimetric readout. Reproduced with permission.[75] Copyright 2018, Elsevier. b) Molecular level target miRNA recognition mechanism of molecular beacon-based LFA. Reproduced with permission.[76] Copyright 2017, Elsevier.
In addition to colorimetric methods for detection of cancer associated multiple nucleic acids, surface enhanced Raman scattering (SERS) technique has been also explored as an alternative signal enhancement approach on multiplex LFAs. This method involves the gain and loss of energy between an incident light and vibrational motions of a specific target molecule, also known as a SERS tag, on a SERS active label (Au NPs).[78] The resulting spectrum with narrow and sharp peaks is specific to the SERS tag and very sensitive to its surrounding environment. For example, certain peaks in this spectrum can change their position or intensity as a result of an attachment of another molecule on the surface of the SERS label such as a target biomarker.[79] Thus, the presence of a target molecule can be identified by monitoring the changes in this spectrum. In this context, Wang et al. has developed a multiplex LFA that can simultaneously detect dual nucleic acids using SERS-based detection method.[80] This study has focused on the differentiation of two diseases, Kaposi’s sarcoma (KS) and bacillary angiomatosis (BA), that exhibited similar clinical presentation.[81] In particular, Au NPs (SERS label) were functionalized with a SERS tag (malachite green isothiocyanate-MGITC) and were conjugated with DNA probes that were complementary from one end to the KS and BA associated DNA targets. Additionally, the DNA probes for KS and BA were immobilized on the two test lines on the paper device. Once the sample with target DNAs was introduced to the device, the target DNAs were hybridized from their one end with the DNA probes that were localized on the surface of Au NPs. Then, the Au NPs were localized at the test lines by hybridization of the target DNAs from their other end with the DNA probes on the paper. In order to quantify the target DNAs at each test line, the test lines were sequentially exposed to a laser light (632.8 nm), and the Raman peak intensity centered at 1617 cm−1 that was originated from the SERS tag (MGITC) was then monitored. The limit of detection values of target DNAs for KS and BA were estimated as 0.043 pM and 0.074 pM, respectively. These highly sensitive results were 10,000 fold less than the values that were obtained from a colorimetric detection method.[82]
4.4. Multiplex LFAs for Exosome Detection
Even though the detection of cancer cells-derived exosomes on LFAs has not been explored as broadly as other biomarker types such as proteins and nucleic acid biomarkers,[83] there was a recent study that demonstrated a multiplex LFA for detecting exosomes with multiple surface protein markers. Here, an isotachophoresis separation technique on a paper device was developed for a rapid isolation and identification of prostate cancer line-derived exosomes with multiple exosomal protein biomarkers.[84] Briefly, the isotachophoresis separation technique was based on an electrokinetic sample separation in a discontinuous electrolyte environment. Depending on the electrophoretic mobility of the components of a mixture, each component can be separated into pure detection zones. Specifically, the electrophoretic mobilities of target exosomes for prostate cancer cells and normal exosomes that were obtained from healthy donors were different and resulted in separation and enrichment of these exosomes at the test lines of the paper device. In order to create the discontinuous electrolyte environment, the sample loading section of this paper device, representing the cathode side of the device, was immersed in a pH 7.1 buffer solution while the other end of the paper was soaked in a pH 7.9 buffer solution (the anode side of the device). Additionally, monoclonal antibodies against CD63 and CD44 that were protein markers for normal and target exosomes, respectively were immobilized on two separate test lines of the paper device before the device assembly. When a mixture of red dye labeled target exosomes and green dye labeled normal exosomes was introduced to the paper, an electrical stimuli (equivalent to 150 V) was applied to the anode side of the device, and the cathode side was then set to ground. During this process, negatively charged target and normal exosomes were mobilized from the sample loading site towards the test lines and finally captured at the respective test lines. Then, the fluorescence signals that were associated with the target and normal exosomes were collected to determine the limits of detection as 1 ng/mL and 0.6 ng/mL, respectively. The comparison between the sensitivity of this device with the commercial ELISA assay demonstrated that the limit of detection for this paper device was 30-fold less than that of the commercial assay.
5. Multiplex Microfluidic Paper-Based Analytical Devices
Microfluidic paper-based analytical devices (μPADs) are slightly different than LFAs in terms of the device configuration. However, similar biorecognition elements such as labels, capture and detection ligands are still used for the development of analytical tools for cancer biomarkers detection. μPADs implement the principles of microfluidics, manipulation of small amount of fluids using channels, to facilitate small-scale, capillary-action to process the flow of fluids on paper. Rather than overlapping a series of pads like in LFAs, μPADs are created by modifying a single piece of paper (usually chromatography paper) with pore sizes ranging from 11–25 μm.[85] The paper modification involves the use of hydrophobic materials to form hydrophilic fluid flow paths (such as fluidic channels in a microfluidic device) on the paper. Thus, small amount of liquid sample (usually in the μL range) can be directed from an inlet toward desired locations on the paper via capillary action and this process takes place without the need for an external device to create a pressure drop (e.g., pumps). Collection of the sample in multiple separate detection zones allows for interaction of the sample only with target specific biorecognition components and therefore, multiple biomarkers can be identified at distinct locations on the paper.
A number of different patterning strategies have been applied to paper-based devices in order to generate hydrophobic barriers for the multiplex μPADs and to confine sample flow within the defined areas.[86] Each patterning method requires full penetration of hydrophobic material through paper substrate. Photolithography, the fabrication method for the first example of μPADs used for analytical measurements,[87] involves impregnating paper with photoresist (light-sensitive material) and then its polymerization by UV exposure. Once a patterned transparency mask is applied on the photoresist-coated paper during the UV exposure step, the photoresist that only interacts with the UV light is polymerized, and therefore becomes solidified. Uncured photoresist is then washed with organic solvent to form hydrophilic channels while cured photoresist defines the boundaries of the channels. Alternatively, wax-printing using wax-based ink in a commercially-available printer is used to print patterns on the paper, and then the wax ink melts through the paper upon application of heat.[88] Inkjet printing, on the other hand, focuses on printing pre-polymer ink on paper and then its polymerization by either heat or UV exposure.[89] Flexographic printing, another form of direct ink printing, applies multiple layers of hydrophobic ink through paper using a specialized printer at a much faster rate.[90] Even though its rapid production of hydrophobic patterns seems advantageous for a high-throughput fabrication process, flexographic printing requires a specialized printer and multiple printing steps for full penetration of the ink. Alternatively, paper-cutting and shaping can also be used to define flow boundaries without chemical treatment with waxes, polymers, or solvents.[91] However, these patterning approaches require high-cost cutting instruments that limit their wide use in the μPAD fabrication. Despite issues with its low resolution, wax printing remains the most common technique for patterning the hydrophobic barriers on paper substrate for multiplex μPAD fabrication.
The multiplex forms of μPADs that are described in this section are classified as two dimensional (2D μPADs) and three dimensional paper devices (3D μPADs) according to the number of patterned papers incorporated to the paper device. 2D μPADs have a simpler architecture and are fabricated on a single piece of filter paper. 3D μPADs are more complicated in that they utilize multiple layers of patterned papers. The layered structure of 3D μPADs reduces undesired contact between reagents and allows for control over multi-step operation. Even though the overall device designs of 2D μPADs and 3D μPADs are different, fabrication methods for hydrophobic barriers and thus creation of hydrophilic channels are similar. In the next section, we will discuss the details of configurational and operational characteristics of 2D and 3D μPADs and their application in multiple cancer biomarkers detection.
5.1. Multiplex 2D Microfluidic Paper-Based Analytical Devices (Multiplex 2D μPADs)
5.1.1. Configurational and Operational Aspects of Multiplex 2D μPADs
The configuration of 2D μPADs for multiplex detection of analytes can take two forms, namely branched or array formats. 2D μPADs with a branched configuration are usually designed around a centralized sample zone with multiple hydrophilic channels radiating outwards to detection zones (Figure 9a). Once the sample is applied to the device from the sample zone, it is directed to multiple detection zones through the branched patterns of hydrophilic channels. Depending on the assay, detection zones may be pre-loaded with target biomarker-specific reagents (capture and detection ligands and signal generating components) to give off a measurable signal upon sample introduction, or these reagents can alternatively be introduced to the detection zone in a sequential manner to generate the signal for biomarker identification. As an alternative to the branched architecture, an array of hydrophilic spots can be used for multiple biomarkers detection on the paper (Figure 9b). In this array type of configuration, the hydrophilic spots are generated by the previously mentioned patterning techniques. Each spot in the array is dedicated to capture and detect one type of target biomarker. Sequential addition of assay components (capture/detection ligands, labels) as well as the sample on the hydrophilic spot serves to generate the signal and quantify the amount of biomarker that is present in the sample. For example, sandwich immunoassay-type 2D μPADs start with capture antibody loading on the hydrophilic spot. A blocking agent, usually BSA, is then added to the spot in order to minimize non-specific protein adhesion on the paper. Next, the sample is added to capture the target biomarker and finally, detection antibody conjugated label is added to tag the captured biomarker. The signal that is generated by the label is collected and analyzed for correlating the signal intensity with the amount of biomarker in the sample.
Figure 9.
Schematic illustration of typical multiplex 2D μPADs configurations. a) A branched configuration with multiple hydrophilic channels confines the diffusion of liquid sample along the channels and allows for collecting the sample at the detection zones where each biomarker is detected separately. Reproduced with permission.[92] Copyright 2016, Elsevier. b) An array of hydrophilic spots is created for simultaneous detection of multiple cancer biomarkers within separate detection spots. In this configuration, each spot is specifically modified for detection of one type of biomarker. Reproduced with permission.[93] Copyright 2013, Elsevier.
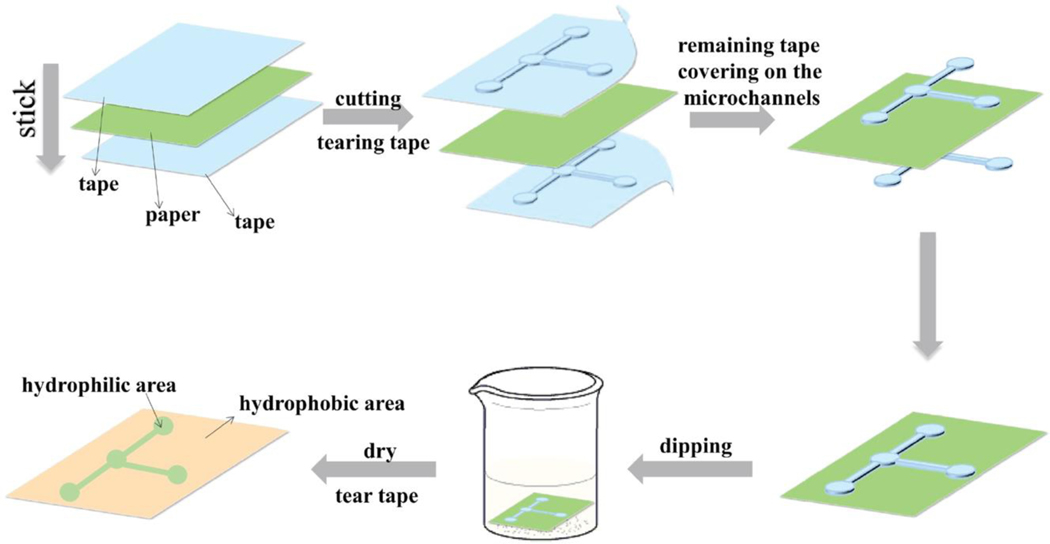
Innovations in the development of 2D μPADs provide the opportunity to reduce fabrication time and cost. In order to simplify the overall 2D μPAD fabrication process, for example, Guo et al. recently developed a novel 2D μPAD fabrication method in which a polystyrene solution (derived from recycled foam plastics) was used to create patterns of hydrophilic channels on the paper.[94] In this method, adhesive masks were applied on both sides of the paper. Then, patterns of channels and detection zones were cut by a cutting plotter. The cutting strength was carefully adjusted in a way that only adhesive masks were cut, and not the paper itself. The adhesive tape representing the patterns of channels and detection zones remained on the paper and the rest was removed. The hydrophobic material (recycled polystyrene) was then introduced to the paper by simply dipping the paper with the tape in polystyrene-chloroform solution and subsequently letting the paper dry at room temperature. Once the tape mask was removed, the sections covered by the mask were left as the patterns of multiple hydrophilic branches (Figure 10). This new technique reduced the fabrication time and cost by using recycled polymer as the hydrophobic patterning material and indicated a promising alternative to simplify fabrication of multiplex 2D μPADs.
Figure 10.
Schematic representation of a novel dipping method to fabricate branched configuration of a multiplex 2D μPAD by using recycled polystyrene as the hydrophobic material. Reproduced with permission.[94] Copyright 2019, Elsevier.
5.1.2. Multiplex 2D μPADs for Protein Biomarkers
Capture Ligand Related Sensitivity Enhancement Strategies
Increasing sensitivity of 2D μPADs for the detection of multiple protein biomarkers has been a primary focus in recent years. One common obstacle is maintaining antibody attachment to the cellulose-based paper devices. This becomes a major issue especially when the liquid sample or solutions of other reagents, such as detection antibody or washing buffer, are introduced after capture antibody immobilization on the paper. Notably, a previous study demonstrated that 34 to 42% of antibodies attached to the cellulose-based paper through non-specific adsorption can desorb from the paper upon washing with buffer solution,[95] which may eventually affect the sensitivity of the paper devices for detection of protein biomarkers. One possible solution to this problem could be the covalent immobilization of antibodies on the cellulose paper requiring additional steps for chemical modification on the paper. Various paper modification methods have been reported[96] but chitosan coating is the most commonly used paper modification approach for developing sensitive 2D μPADs.[94, 97] Chitosan, a cationic polysaccharide, interacts with anionic cellulose and can be readily crosslinked with glutaraldehyde where capture antibodies are covalently immobilized on the 2D μPAD. For example, Wang et al. adapted chitosan modified paper for detecting CEA, AFP, and CA-125 on an array format 2D μPAD.[97a] In this work, chitosan modification improved not only the stability of immobilized capture antibodies that are specific to each biomarker but also the wet-strength of the paper device. Alternatively, periodate oxidation of cellulose paper has been used for grafting capture antibodies on the paper. This approach relies on the creation of aldehyde groups on the cellulose fibers upon reacting with sodium periodate and utilization of these aldehyde groups in covalent attachment of capture antibodies. Ge et al. demonstrated covalent immobilization of capture antibodies on the periodate modified detection spots of an array format 2D μPAD for AFP, CA 153, and CA 125.[93] Even though periodate oxidation provided a simple route for paper modification, it required multiple washing steps to complete removal of unreacted sodium periodate that may eventually affect the wet-strength of the paper.
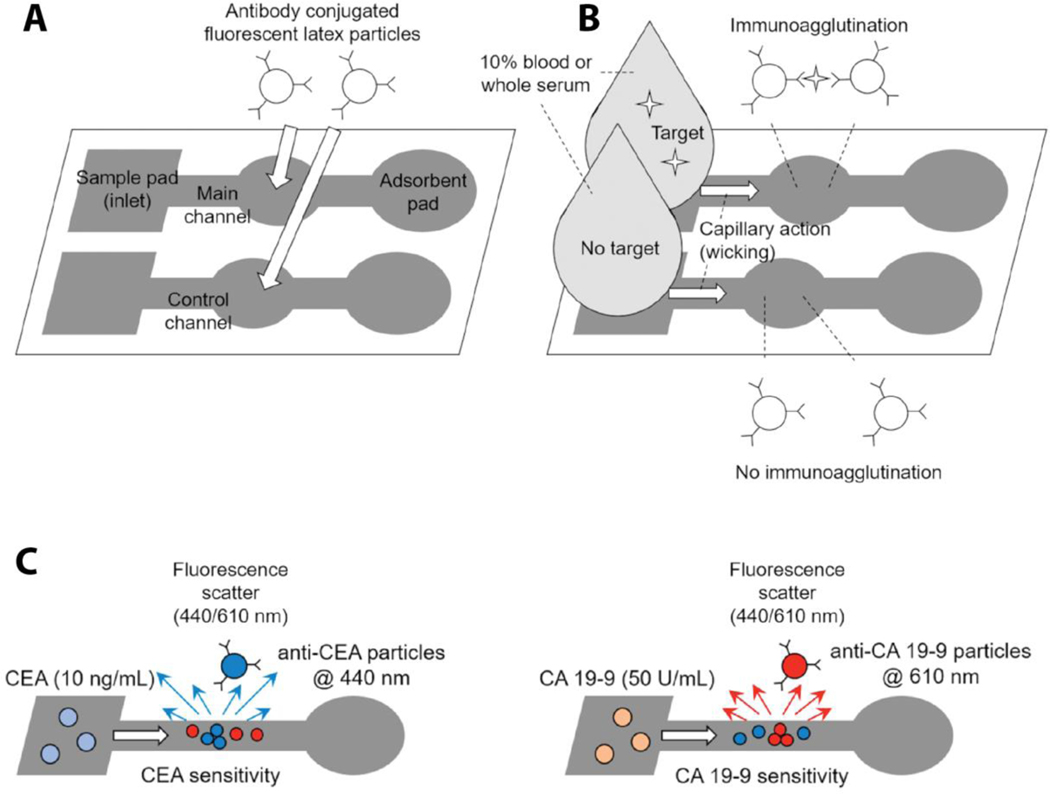
Rather than covalent attachment of capture antibodies on the modified paper, their immobilization on stationary platforms (that remain immobile upon liquid introduction) could be an alternative approach for securing capture antibodies on the paper device. Baynes and Yoon showed a novel multiplex 2D μPAD design for the detection of colorectal cancer related biomarkers (CEA and CA 19–9), in which capture antibodies were localized on the surface of submicron latex beads.[98] These beads were large enough to exhibit limited mobility on the paper. Therefore, capture antibodies remained in the main channel where beads were initially loaded (Figure 11). This study employed a fluorescence scattering immunoagglutination detection method in order to measure the change in the scattered light angles upon UV exposure that resulted from biomarker-capture antibody aggregation on the bead surface. The results demonstrated low detection limits for CEA (1 pg/mL) in blood and for CA 19–9 (1 U/mL) in serum (about 100 times lower than previously reported in human serum).[99]
Figure 11.
Schematic representation of the multiplex 2D μPAD with the strategy to minimize capture antibody desorption from the paper device. a) The overall outline for the 2D μPAD configuration. b) Summary of the biomarker capturing mechanism via immunoagglutination. Once the target biomarker containing sample reaches to the main channel, biomarker-antibody interaction induces aggregation of latex particles. c) Aggregated particles exhibit characteristic changes on the fluorescence light scattering angles at 440 nm and 610 nm under UV exposure for CEA and CA 19–9, respectively. The change in the scattered light is measured for determining the biomarker quantity. Reproduced with permission.[98] Copyright 2018, SAGE.
Label Related Sensitivity Enhancement Strategies
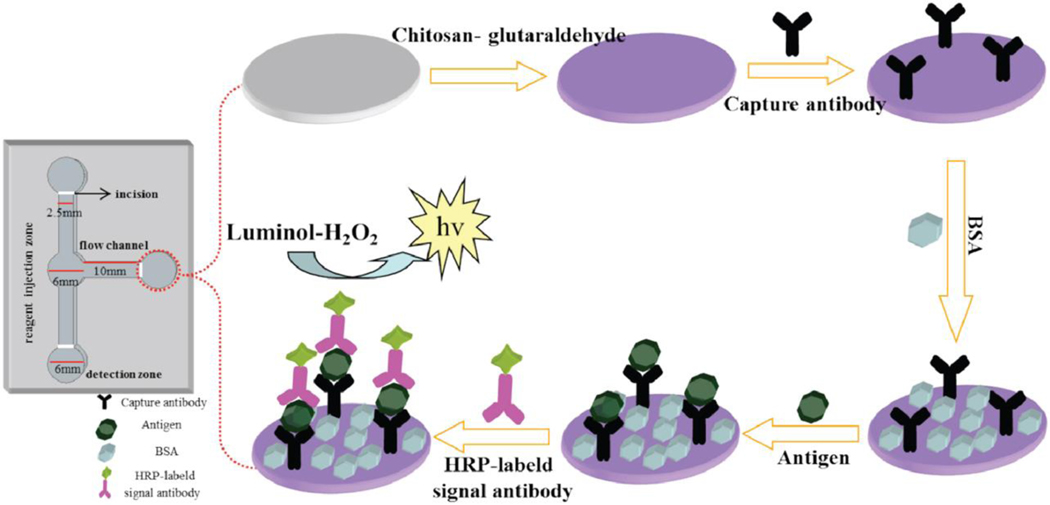
In addition to enhancing capture antibody attachment on the 2D μPADs, researchers have been exploring various signal enhancement approaches for the development of sensitive paper devices for detection of multiple protein biomarkers.[94, 97a, 100] The branched form 2D multiplex μPAD that was developed by Guo et al. included an enzymatic signal amplification strategy that is based on a chemiluminescent detection mechanism. Specifically, HRP conjugated detection antibodies for CEA, AFP, and PSA were incorporated into the assay design (Figure 12).[94] Once the sample was introduced to the paper, target biomarkers were captured in the three detection zones. Then, the solution of HRP conjugated detection antibodies was added to the detection zones to form antibody-biomarker complexes. Next, luminol and H2O2 were added to the detection zone that led to luminol oxidation and the resulting emitted light was detected by a chemiluminescence detector. The analytical performance of this device was capable of generating low detection limits for CEA, AFP, and PSA at 0.02 ng/mL, 2 ng/mL, and 0.3 ng/mL, respectively. Additionally, quantification of these biomarkers in the human serum spiked samples revealed comparable results with the standard ELISA method.[94]
Figure 12.
Schematic of the signal amplification strategy for sensitive detection of CEA, AFP, and PSA (antigen) on the three detection zones of a multiplex 2D μPAD. Reproduced with permission.[94] Copyright 2019, Elsevier.
An alternative method for improving the detection efficiency of 2D μPADs involves incorporation of a “turn on” mechanism for quenched fluorescence signals in the detection sites. Ge et al. developed an array format of a multiplex 2D μPAD where copper-mediated signal recovery of quenched QDs was used to improve the sensitivity of the assay.[93] The device was able to simultaneously detect four different cancer biomarkers of AFP, CA 125, CA 153, and CEA. Capture antibodies that were specific to these markers were immobilized in individually dedicated detection zones in the array format. Dithizone coordinated CdTe QDs were also localized in the detection zones. The coordination of dithizone at the surface of the CdTe QDs suppressed the fluorescent signal via fluorescent resonant energy transfer (FRET). In this sandwich immunoassay, detection antibodies that were labeled with copper oxide nanobodies released Cu2+ ions that activated the fluorescence of CdTe QDs in the detection sites. As a result of this quenching and “turn on” mechanism, the assay achieved high sensitivity with very low limits of detection (1.0 × 10−3 ng/mL for AFP, 2.0 × 10−3 ng/mL for CA 125, 1.0 × 10−4 ng/mL for CA 153, 5.0 × 10−3 ng/mL for CEA).
5.1.3. Multiplex 2D μPADs for Detection of Circulating Tumor Cells
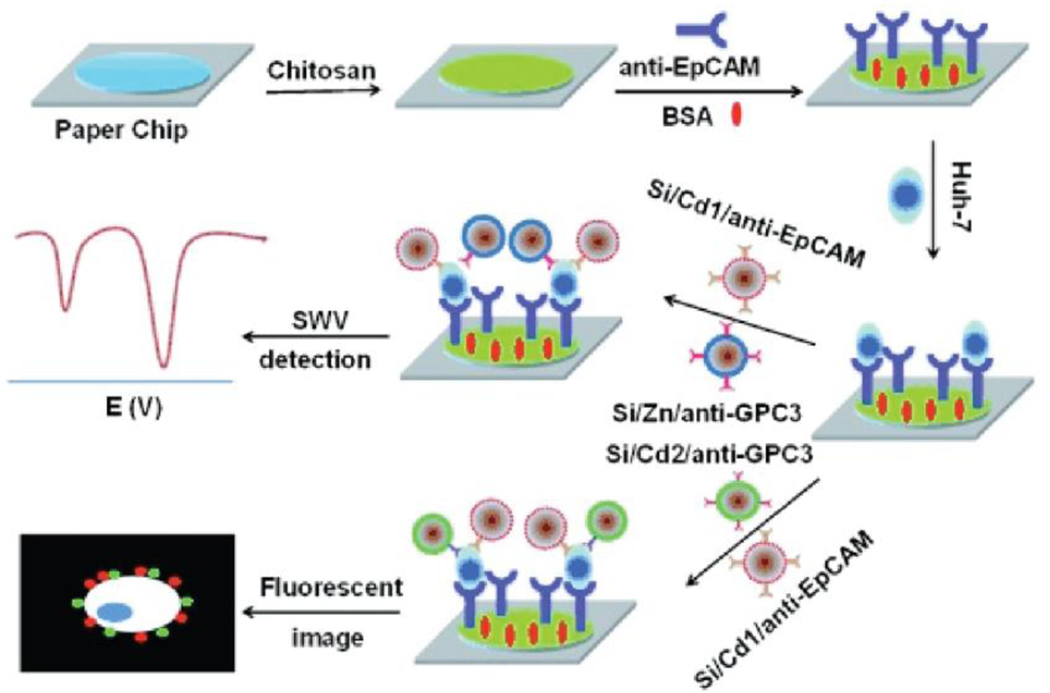
There are different approaches for employing a multiplexing strategy for detecting CTCs from the blood stream. For instance, CTCs can be captured through recognition of multiple biomarkers that are specific to a CTC type, followed by detection using paper-based devices. Accordingly, Wu et al. developed a 2D μPAD that can identify a model CTC, cell line of Huh7, by detecting cell surface biomarkers of EpCAM and GPC3.[97b] Figure 13 illustrates the CTCs detection mechanism that was used at the detection zone. Briefly, this study used silica NPs that contain multiple types of QDs in order to amplify electrochemical and fluorescent signals for the detection of cancer cells. Specifically, electrochemical and fluorescent signals of the device are collected through a screen printed carbon electrode. The μPAD was loaded with three types of silica NPs that are labeled with Cd1, Cd2, and Zn QDs to tag EpCAM and GPC3. The presence of surface proteins (EpCAM and GPC3) on the Huh7 cells were confirmed by the fluorescence signals of Cd1 and Cd2 loaded NPs. Additionally, the presence of these biomarkers were identified by the electrical signals that were enhanced by the incorporation of Cd1 and Zn to the silica NPs. Further, labeling silica NPs with multiple QDs was found to increase the sensitivity for EpCAM detection 2.3 times and the sensitivity for GPC3 2.8 times compared to the μPAD sensitivity that contains silica NPs with a single QD.[97b]
Figure 13.
Schematic illustration of the 2D μPAD for detection of multiple cell surface biomarkers for a model CTC, Huh 7. Reproduced with permission.[97b] Copyright 2014, Wiley.
The detection of multiple CTC types on a single device has also been attempted. Specifically, Liang et al. reported a 2D μPAD design to demonstrate fluorescence-based detection of three cancer cell types (adenocarcinoma cells (MCF-7), human acute promyelocytic leukemia cells (HL-60), and human chronic myelogenous leukemia cells (K562). The design included biorecognition of cancer cells by cell specific aptamers.[92] The aptamer probes were initially conjugated to QDs that were loaded on mesoporous silica NPs (MSNs/QDs) and produced different detection signals at different wavelengths. Thus, each aptamer probe could be differentiated by the color of MSNs/QDs that they were attached. Then, aptamer conjugated MSNs/QDs were localized at the detection zone where graphene oxide was previously loaded. The presence of graphene oxide resulted in quenching the fluorescence signals of aptamer conjugated MSNs/QDs. When the sample of cell mixture was introduced to the device, aptamer probes were released as a result of their interaction with corresponding target cancer cells leading to the emission of fluorescent signals. Quantification of this fluorescence signals revealed the limits of detection for MCF-7, HL-60, and K562 as 62 cells/mL, 70 cells/mL, and 65 cells/mL, respectively.
A summary of the research efforts for the development of 2D μPADs for detection of multiple cancer biomarkers is included in Table 3.
Table 3.
Summary of 2D μPADs for the multiplex detection of cancer biomarkers.
| Biomarkers | Sample | Configuration | Signal | Reporter | Limit of Detection | Analysis Time | Ref |
|---|---|---|---|---|---|---|---|
| AFP, CA 125, CEA | Serum | Array | Chemiluminescent | HRP | AFP: 25 ng/mL CA 125: 35 ng/mL CEA: 5 ng/mL | 3.5 min | [97a] |
| AFP, CA-125, CEA | Serum | Array | Chemiluminescent | HRP | AFP: N/A CA-125: N/A CEA: 0.05 ng/mL | 16 min | [100] |
| AFP, CEA, PSA | Serum | Branched | Chemiluminescent | HRP | AFP: 2 ng/mL CEA: 0.02 ng/mL PSA 0.03 ng/mL | 30 min | [94] |
| CA 19–9, CEA | Whole blood or serum | Branched | Fluorescent | Multicolored polystyrene particles | CA 19–9: 0.1 U/mL CEA: 1pg/mL | 1 min | [98] |
| EpCAM and GCP3 positive cells | Phosphate buffer solution | Single Detection Zone | Electrochemical and fluorescent | CdTe and ZnSe QDs on silica NPs | 10 cells/mL | 85 min | [97b] |
| K562 (leukemia), MCF-7 (breast cancer), HL-60 (leukemia) | Serum | Branched | Visual and fluorescent | QDs on silica NPs | K562: 65 cells/mL MCF-7: 62 cells/mL HL-60: 70 cells/mL | 30 min | [92] |
5.2. Multiplex 3D Microfluidic Paper-Based Analytical Devices (Multiplex 2D μPADs)
5.2.1. Configurational and Operational Aspects of Multiplex 3D μPADs
Recent advancements in μPADs offer unique opportunities to further develop paper devices that can manipulate the fluid flow not only in two dimension along the hydrophilic channels but also in the third (vertical) dimension. These devices exhibit key features such as integration of essential elements (working, reference and counter electrodes) for electrochemical-based detection methods as well as flexibility to combine multiple detection methods on a single device. Further, incorporation of adequate reagents/buffers at different layers on the 3D device simplifies the complex assay procedure that requires multiple and sequential reagent addition and washing steps.[101] In addition, manipulation of fluid diffusion between layers (from top to the bottom layer) brings the opportunity to apply a necessary filtration step that is not feasible on LFAs or 2D μPADs.
The assembly of 3D μPADs involves patterning of hydrophobic barriers on each individual paper layer if the device consists of a multilayered design. This is often performed through patterning strategies that were briefly described in the earlier section. Paper layers are then assembled by stacking these individual pieces on top of each other. Alternatively, patterning of hydrophobic barriers can be formed on a piece of paper, and the 3D device can be assembled by following carefully designed sequential bend-and-fold steps in an origami style. Figure 14 illustrates examples of multiplex 3D μPADs with layered and origami type device configurations. For both configuration designs, a binding clip is required to secure the necessary contact between the layers and to maintain the fluid flow in vertical direction. Any additional reagent, buffers, or chemical modification that are specific to the assay are typically added to their respective layer before the assembly process.
Figure 14.
Schematics of typical multiplex 3D μPADs configurations. a) Wax printed two paper sheets (paper A and paper B) were chemically modified to develop multiple working electrodes on paper A and counter and reference electrodes on paper B. b) Multiple detection zones were dedicated for electrochemical-based detection of four protein biomarkers. Reproduced with permission.[99] Copyright 2012, Elsevier. c) Optical images of the 3D μPAD that exhibits origami style device assembly. d) Description of assay reagents that are loaded on each layer and the flow direction of the sample once applied to the sample layer. Reproduced with permission.[102] Copyright 2020, Elsevier. e) The 3D μPAD including a rotational disks format. i) Three layers of rotating disks with electrodes integrated in the auxiliary and detection disks. ii) The assembly of the rotational 3D device. iii) The control mechanism of the “on/off’ states of upper and lower valves. Reproduced with permission.[103] Copyright 2018, Elsevier.
Multiplex assays using a typical layered 3D μPAD configuration apply a simple operational procedure. The sample is usually added to the sample layer, then moves onto the splitting layer and reaches to the test layer to perform the assay. Horizontal flow of the liquid sample splits the sample and carries the analytes within layers that hold various functions such as filtering, reserving necessary reagents for the assay, collecting the liquid in the waste and the detection zones. The splitting layer is important for multiplex analysis to allow for localization of the sample on multiple separate spots which is especially important in a limited space on a single device. Once the sample reaches the detection zone, the assay-specific detection signals are generated in the presence of target biomarkers. Finally, the device is unfolded for further quantitative or qualitative analysis using assay-specific detection method.
5.2.2. Multiplex 3D μPADs for Protein Biomarkers
Chemical Modification-Based Signal Enhancement Strategies
Current trends in identification and quantification of multiple protein biomarkers using 3D μPADs are mainly based on electrochemical detection methods. Modifications to the detection zones (working electrode) by incorporating different surface coatings on the paper or introducing functional nanomaterials to the detection system have been the foremost emphasis in the development of recent 3D μPADs for multiple protein biomarkers detection. For example, Wu et al. integrated graphene onto a chitosan-modified paper to enhance electrochemical signals for the detection of a panel of four cancer biomarkers (AFP, CEA, CA 125, CA 153).[104] Multiple detection zones (working electrodes) that were dedicated to each biomarker were functionalized with graphene oxide in order to accelerate the electron transfer, and then biomarkers were captured on the paper in a sandwich immunoassay. In this assay, HRP molecules that were chemically linked to detection antibodies through a synthetic polymer chain were then reacted with a substrate (O-phenylenediamine-H2O2 mixture). Finally, differential pulse voltammetric measurements were taken when the electroactive species (2,2′-diaminoazobenzene) were produced upon HRP-induced catalytic reaction. This particular device could reach limits of detection for these biomarkers ranging from 0.01 ng/mL – 0.05 ng/mL. A slightly modified 3D μPAD design was also created that could detect these biomarkers at a sub picogram per milliliter level. In this device, detection antibodies were tagged with silica NPs where multiple HRP molecules were conjugated. Incorporation of multiple HRP molecules to the detection system aided in increasing the sensitivity of this paper device that was reflected in the lower limits of detection for the target biomarkers (0.001 ng/mL AFP, 0.005 ng/mL CEA, 0.001 ng/mL CA 125, and 0.005 CA 153 ng/mL).[105] In addition to graphene modification, multi-walled carbon nanotubes (MWCNTs) were applied to the sensing component of an origami type multiplex 3D μPAD in order to produce signal enhancement in the detection of CA 125 and CEA.[106] The device utilized HRP- O-phenylenediamine-H2O2 electrochemical system for an immunoassay type biomarker recognition and its analytical performance was verified by testing with biomarkers spiked-PBS solutions. MWCNTs mediated enhancement of the electronic conductivity of the working electrode resulted in sensitive detection of CA 125 and CEA at limits of detection of 0.05 ng/mL and 0.001 U/mL, respectively. Once the device was tested using clinical samples, quantitation of biomarkers produced relative errors that were less than 5.8% for CA 125 and 6.5% for CEA when compared to reference values from a commercially available chemiluminescence method.
Modification of the working electrode with metal-based coatings on 3D μPADs can also be utilized to increase the surface area of the working electrode and consequently the amount of immobilized capture ligands. Li et al. demonstrated a multiplex 3D paper device with a nanoporous silver modified working electrode for enhancement of detection of two protein biomarkers (AFP and CEA).[107] Biomarker recognition was achieved by chitosan-coated, nanoporous Au NP labels that were loaded with large amount of metal ions such as Cu2+ and Pb2+. It was hypothesized that the presence of these metal ions on the NP labels played a key role in electrochemical signal enhancement. Additionally, the sensitivity of this device was found to be higher than the device with an unmodified electrode. When the assay’s performance was compared with the results that were obtained from a commercially available electrochemiluminescent method, this multiplex 3D μPAD could detect AFP and CEA with a relative error of 3.2%. In a similar signal enhancement approach, another electrochemical-based multiplex 3D μPAD was developed through the use of a cuboid silver coated working electrode.[108] In this 3D μPAD design, nanoporous silver-chitosan coated NPs were loaded with Cu2+ and Ag+ and were used to tag CA 125 and CA 199 on the working electrode via sandwich antibody conjugation. The electrochemical responses upon capturing biomarkers at the working electrode were investigated by cyclic voltammetry. The cuboid silver modified electrode produced much higher sensitivity for these biomarkers than unmodified working electrode and the limits of detection, 0.02 mU/mL and 0.04 mU/mL for CA 125 and CA 199, respectively, were much lower than the cutoff values used for the biomarkers in clinical diagnosis.[108]
The working electrode modification using Au, either in its NPs or nanorods (NRs) forms, is another attractive metal-based coating strategy for electrochemically active multiplex 3D μPADs. Li et al. designed an origami type 3D paper device with a working electrode where the electrode surface was coated with Au NPs for detection of multiple protein biomarkers (CEA and AFP).[109] Similar to previous examples of silver coated paper devices, addition of Au NPs lowered the resistance while increasing the conductivity of the electrode that resulted in improved sensitivity of the device. This 3D μPAD employed 3D graphene sheets to tag the biomarkers. These graphene sheets were previously loaded with dual redox probes (methylene blue and carboxyl ferrocene) before incorporating them into the electrode in order to trace the electrochemical changes upon target biomarker capture. The detection limits of this multiplex device were determined to be 0.5 pg/mL and 0.8 pg/mL for CEA and AFP, respectively. In the presence of other interfering agents such as CA 125, BSA and PSA, the device demonstrated minimum cross-reactivity with a relative error in the range of −3% ~ 3.5%. Similar to signal enhancement by Au NPs, Ma et al. demonstrated Au NRs-based signal enhancement on a 3D paper device capable of detecting two protein biomarkers (CA 125 and CEA) at the working electrode.[110] This work additionally applied a metal ion induced-electrochemical signal enhancement strategy by incorporating metal ions (Pb2+ and Cd2+) to BSA coated Au NP labels for the detection of biomarkers with a sandwich immunoreaction. The combination of metal ions on the labels and Au NRs coating on the paper surface created dual effects for improving the sensitivity of the multiplex device and the limits of detection for CEA and CA 125 were found to be 0.08 pg/mL and 0.06 mU/mL, respectively. Once this device was tested using clinical samples, the absolute error compared to a commercial electrochemiluminescent single-analyte test was less than 3.6%. In addition, minimal cross-reactivity between these two biomarkers overall reflected the clinical potential of this 3D μPAD design for detecting multiple protein biomarkers.
ZnO nanorods (ZNRs) are another metallic material of interest that have been explored in the development of sensitive electrochemical-based 3D μPADs for multiplex analysis of cancer biomarkers. For instance, due to high surface area of ZNRs, low tendency to agglomerate and their unique electron transportation properties, the working electrode of a paper device was modified with ZNRs to develop an origami type 3D μPAD for the multiplex detection of HCG, PSA, and CEA.[111] In this device, capture antibodies were immobilized on the ZNRs. The fundamental detection mechanism of this multiplex assay relied on the change in the electrical current once H2O2 was reduced by BSA-stabilized silver NPs that were attached to graphene oxide labels. BSA-stabilized silver NPs contributed not only catalyzing H2O2 but also aided in maintaining detection antibody-label conjugation for target biomarkers. Analysis of the resulting electrochemical signals upon introduction of biomarkers to the paper device revealed the limits of detection for HCG, PSA, and CEA to be 0.0007 mIU/mL, 0.35 pg/mL, and 0.33 pg/mL, respectively. In addition, quantification of these biomarkers from clinical serum samples showed the relative error as 4.8% when compared to a commercial electrochemiluminescent method.[111] A more complex immunoassay design of a 3D paper device including a photoactive matrix (ZNRs coupled with CdS QDs) with visible light absorbency and photoelectrochemical activity was created by Ge et al. for detecting CEA and AFP.[112] Briefly, CdS QDs were galvanostatically deposited on the ZNRs surface that were grown on the Au/Pd NPs coated paper electrode. CEA and AFP were tagged with nanoporous silver NP labels that were conjugated with detection antibodies and glucose oxidase (GOx). Upon biomarker capture and glucose introduction to the device, the large amount of GOx produced H2O2 aided in light-induced electrical signal enhancement. The overall sensing mechanism of this electrochemical device was based on visible light sensitization of CdS QDs at the working electrode that resulted in enhanced light absorption and improved the photocurrent. This photoelectrical activity was further enhanced by the production of H2O2, an electron donor to scavenge the holes in the valence band of ZNRs that were generated by their interaction with visible light-sensitized CdS QDs (Figure 15a). Substantially amplified photocurrent response was about 2.4 times greater than carbon nanotube modified working electrode and resulted in improved sensitivity for cancer biomarkers of CEA and AFP with the limits of detection of 0.3 pg/mL and 0.5 pg/mL, respectively. The multiplex device showed relative errors of less than 3.5% for human serum samples when compared with reference values that were obtained from a commercial electrochemiluminescent detection method.[112]
Figure 15.
a) Schematic illustration of the detection mechanism of the electrochemical-based multiplex 3DμPAD including a photoactive matrix. i) The working electrode was prepared by coating the paper with Au/Pd NPs. ii) ZNRs were grown on the modified electrode and coupled with CdS QDs. iii) Capture antibodies for CEA and AFP were immobilized on the ZNRs surface and the surface of the paper were blocked with BSA. iv) Nanoporous silver NPs-based labels including detection antibodies and GOx were introduced. v) Photocurrent activity under visible light (“on” or “off”) was recorded following the glucose addition to the electrode. Reproduced with permission.[112] Copyright 2015, Royal Society of Chemistry. b) Layered paper configuration for multi-parameter aptamer-based electrochemical detection of protein biomarkers (CEA and NSE). (1) sample inlet, (2) filtration sit, (3) counter electrode, (4) reference electrode, (5) detection zone, (6) working electrode. Reproduced with permission.[113] Copyright 2019, Elsevier.
Label Free Methods for Signal Enhancement Strategies
Even though the majority of electrochemical-based 3D paper devices implement sandwich type immunoassays as the biorecognition element, there are also examples of 3D paper devices that adapt label-free biorecognition using aptamers. For example, Wang et al. developed a multiplex 3D μPAD with a layered configuration in which simultaneous detection of lung cancer biomarkers of CEA and NSE was achieved by electrochemical aptamer-based sensing.[113] In this study, thiol modified DNA aptamers against CEA and NSE were synthesized. Two carbon working electrodes were formed on the bottom paper by modifying the electrode surface with amino functional graphene (NG)-Thionin (THI)-Au NPs (NH2-G/THI/Au NPs) and Prussian blue (PB)-poly (3,4-ethylenedioxythiophene) (PEDOT)-Au NPs (PB/PEDOT/Au NPs) that were the electroactive species in this assay design. Then, the solutions of CEA aptamers and NSE aptamers were dropped onto the surface of the NH2-G/THI/AuNPs and PB/PEDOT/AuNPs modified electrodes leading to covalent attachment between Au NPs and aptamers. A microfluidic channel patterned paper layer (filtration layer) was sandwiched between the bottom layer (with the working electrodes) and another paper layer on which counter and reference electrodes were created. Finally, a fourth paper layer, the sample layer that was printed according to the design pattern, was placed on top and the assembly of the 3D device was achieved by stacking these paper layers and securing their close contact with double sided tape (Figure 15b). Once the liquid sample was introduced to the sample layer (top paper), it diffused in the vertical direction and was filtered while moving through the layers. The electrical responses upon capturing CEA and NSE at the working electrodes were recorded by the differential pulse voltammetry. With the use of optimum Au NPs size (determined to be 15 nm in size), this 3D μPAD demonstrated the analytical performance at very low detection limits for CEA and NSE, 2 pg/mL and 10 pg/mL, respectively as compared to the clinical cutoff values of 5 ng/mL for CEA and 15 ng/mL for NSE.
5.2.3. Multiplex 3D μPADs for Nucleic Acid Biomarkers
Sample preparation for nucleic acid testing often involves extraction of nucleic acids from a cell lysed mixture (usually via a centrifuging process), amplification of the nucleic acids, and the detection procedure. The filtration feature of 3D paper devices offers unique opportunities to simplify this sample preparation process for multiple nucleic acid biomarkers in particular. The first example in this context for integration of small RNA extraction and amplification process on a paper device was shown by Deng et. al, for the detection of two cancer biomarkers that are associated with lung cancer (miRNA 155 and miRNA 21).[114] In this work, the extraction of nucleotides from cancer cell lysate was performed through a poly(ether sulfone) (PES) filtration set up where cell lysate was introduced to PES layers. Then, a mixture of amplification solution including a hairpin probe was added to the PES layers. An origami type, foldable and patterned paper (foldable detection chip) was also prepared by adding a DNA probe that is conjugated to QD labels to the hydrophilic spots on the top layer. Streptavidin-biotinylated capture probes for the target miRNAs were dispensed on the test spots at the second and third layers of the foldable paper device. Next, the PES layers were assembled with the folded paper by aligning the PES layers with the patterns of the folded and placed in a heating block for exponential amplification reaction (EXPAR). During the amplification process, target miRNAs diffused from the top layer and captured at the second and third layers due to their interaction with the corresponding DNA probes. Finally, the paper device was unfolded, and fluorescence intensities generated upon UV exposure at the detection spots were used for quantification of target miRNA amounts (Figure 16a). The unique design of this 3D μPAD eliminated the centrifuging-based nucleotides isolation step from cell lysates and provided a straightforward sample preparation process for miRNA extraction. Even though the amplification procedure required additional equipment in the form of a portable heating block, this process exhibited a shorter isothermal process (20 minutes) relative to other lengthy isothermal amplification strategies (over 6 hr).[115] This multiplex device produced optical signals from QD labels at two distinct wavelengths enabling detection of two RNA biomarkers on a single device. Also, this device achieved the sensitivity range from 3 × 105 copies to 3 × 108 copies. Further, this 3D μPAD showed an increase in optical signals from samples that were extracted from clinical tumor samples as opposed to samples that were extracted from healthy tissues.
Figure 16.
Novel 3D μPAD designs for the detection of nucleic acid biomarkers with fluorescence-based detection methods. a) Schematic illustration of the 3D μPAD configuration that is capable of DNA amplification on the paper device. b) Diagram of DNA amplification process on the 3D μPAD from lysing cells in a sample to filtration and finally fluorescent detection of the captured miRNAs. Reproduced with permission.[114] Copyright 2017, American Chemical Society. c) Schematic representation of the multiplex detection mechanism that used silver coating-based signal enhancement for the recovered fluorescent signal and generation of visual signal upon miRNA detection. Reproduced with permission.[116] Copyright 2017, Elsevier.
Another fluorescence-based detection of multiple miRNAs was explored by Liang et al. with the goal of reducing background optical signals and subsequent signal enhancement for detection of miRNA21 and miRNA210 on an origami type 3D μPAD.[116] The group used interconnected dense flower-like silver (FLS) layer deposition onto the paper surface at the detection zone to reduce the background fluorescence of the paper. The proposed signal generation mechanism was based on fluorescence signal recovery of quenched carbon dots (CDs) in the presence of miRNA biomarkers. Specifically, CDs labeled DNAs (DNA1-CD) were immobilized on the FLS coated paper and hybridized with CD quencher carrying DNA strand (DNA2-CeO2). Once the solution of target miRNAs was introduced to the detection zone, the fluorescence of DNA1-CD was recovered and measured by a spectrophotometer under excitation at 390 nm. In addition to enhanced fluorescence signal by the fluorescence recovery, the quenching component (DNA-CeO2), exhibited the potential for generating visual color when reacted with H2O2 (Figure 16b).
5.2.4. Multiplex 3D μPADs for Detection of Circulating Tumor Cells
There are only a few examples to date of 3D μPADs that have been developed for detection of CTCs. The general assay principle for such paper devices relies on an electrochemiluminescence detection method. For example, Wu et al. designed a multiplex 3D μPAD that used a macroporous paper with a particle retention size of 25 μm for the detection of four types of cancer cells: MCF-7, HL-60, K562, and CCRF-CEM.[117] This study used aptamer-mediated CTCs attached on the working electrode. In addition, coating the electrode surface with Au NPs increased the surface area for cell binding as well as facilitated the transfer of electrons to increase the sensitivity of the electrical signals. Moreover, cells were labeled with concavanalin-A-conjugated AuPd NPs to enable catalyzation of the disproportionation of H2O2 once introduced to the working electrode. The resulting O2 from this reaction then reacted with peroxydisulfate that was subsequently added to the electrode to produce electrochemiluminescence signals. The analytical performance of this device revealed that the limits of detection for MCF-7, HL-60, K562, and CCRF-CEM were 250 cells/mL, 236 cells/mL, 241 cells/mL, and 265 cells/mL, respectively.
A 3D μPAD with a better sensitivity for detection of smaller cell numbers was designed for electrochemiluminescent detection of multiple types of CTCs, specifically MCF-7, CCRF-CEM, HeLa, and K562 cells.[118] In this work, aptamer-based capture ligands were applied to the working electrode for CTC attachment. A Sudoku-like structure of this 3D device allowed for storage of assay reagents in different layers such as CTC labeling component, graphene QDs, and other reagents that are essential for the assay in different layers. This configuration aided in directing the fluid flow such that captured cells could interact with the signal generating reagents in a sequential manner. Further, the electrochemiluminescence signal amplification was achieved by incorporation of two efficient co-reactants (silver NPs and semicarbizide) that enabled dual mode signal enhancement. Addition of these dual enhancers created a strong electrochemiluminescence response, and resulted in much lower detection limits of CTCs; 38 cells/mL, 53 cells/mL, 67 cells/mL, and 42 cells/mL for MCF-7, CCRF-CEM, HeLa, and K562 cells, respectively. These detection limits were found to be one order of magnitude more sensitive than a previous study.[117]
A summary of multiplex analysis approaches using 3D μPADs for cancer biomarkers is listed in Table 4.
Table 4.
Summary of 3D μPADs for the multiplex detection of cancer biomarkers.
| Biomarkers | Sample | Configuration | Signal | Reporter | Limit of Detection | Analysis Time | Ref |
|---|---|---|---|---|---|---|---|
| CA125 CEA CA15–3 | Serum | Stacked | Photoelectrochemical | Carbon Dots | CA125: 3.6 mU/mL CA15–3: 3.8 mU/mL CEA: 1.8 pg/mL | 20 min | [119] |
| AFP CA125 CA199 CEA | Serum | Stacked | Electrochemiluminescent | Ru(bpy)32 | AFP: 0.15 ng/mL CA 125: 0.6 U/mL CA 199: 0.17 U/mL CEA: 0.5 ng/mL | 30 min | [99] |
| AFP CEA | Serum | Origami | Photoelectrochemical | Glucose oxidase on silver NPs | AFP: 0.5 pg/mL CEA: 0.3 pg/mL | 8 min | [112] |
| CA125 CEA | Serum | Stacked | Electrochemical | HRP | CA 125: 0.2 mU/mL CEA: 0.01ng/mL | CA 125: 2.5 min CEA: 3.3 min | [106] |
| CEA NSE | Serum | Stacked | Electrochemiluminescent | Au NPs | CEA: 2 pg/mL NSE: 10 pg/mL | 1 hr | [113] |
| AFP CEA | Serum | Origami | Electrochemical | Gu NPs | AFP: 0.8 pg/mL CEA: 0.5 pg/mL | 40 min | [120] |
| AFP CA153 CA199 CEA | Serum and whole blood | Origami | Chemiluminescent | Silver NPs | AFP: 25 ng/mL CA153: 0.4 U/mL CA199: 0.06 U/mL CEA: 0.02 ng/mL | 9.3 min | [101] |
| AFP CEA | Serum | Origami | Electrochemical | Nanoporous Au chitosan | AFP: 0.08 pg/mL CEA 0.06 pg/mL | 5.3 min | [107] |
| CA125 CA 199 | Serum | Origami | Electrochemical | Nanoporous silver chitosan | CA 125: 0.08 mU/mL CA 199: 0.1 mU/mL | 6 min | [108] |
| CEA HCG PSA | Serum | Origami | Electrochemical | Reduced graphene oxide silver NPs | CEA: 0.33 pg/mL HCG: 0.0007 mU/mL PSA: 0.35 pg/mL | 5 min | [111] |
| AFP CEA | Standard | Origami | Electrochemiluminescent | Au/Pd NPs | AFP: 0.82 fg/cell CEA: 24.69 fg/cell | 110 min | [121] |
| CA125 CEA | Serum | Origami | Electrochemiluminescent | Au-BSA NPs | CA 125: 0.06 mU/mL CEA: 0.08 pg/mL | 7 min | [110] |
| CA199 AFP CEA | Serum | Origami | Fluorescent | Fluorescein isothiocyanate | AFP: 0.03 ng/mL CA 199: 0.09 U/mL CEA: 0.05 ng/mL | 5 min | [102] |
| AFP CA125 CA153 CEA | Serum | Stacked | Electrochemical | HRP on silver NPs | AFP: 0.001 ng/mL CA125: 0.001 ng/mL CA153: 0.005 ng/mL CEA: 0.005 ng/mL | 4.2 min | [105] |
| AFP CA125 CA153 CEA | Standard solutions | Stacked | Electrochemical | HRP | AFP: 0.01 ng/mL CA125: 0.05 ng/mL CA153: 0.05 ng/mL CEA: 0.01 ng/mL | 4.2 min | [104] |
| miRNA21 Folate Receptor (FR) | Standard | Origami | Fluorescent | MnO2 sheets | miRNA 21: 0.0033 fM FR: 0.667 ng/mL | 1 hr | [122] |
| miRNA21 miRNA155 | NE buffer | Origami | Fluorescent | QDs | 3 × 10⁶ copies | 20 min | [114] |
| miRNA21 miRNA210 | Standard | Origami | Visual and fluorescent | Silver enhanced fluorophores | miRNA-21: 0.06 fM miRNA-210: 0.03 fM | 15 min | [116] |
| CCRF-CEM K562 HL-60 MCF-7 | Spiked Serum | Origami | Electrochemiluminescent | Au/Pd NPs | CCRF-CEM: 265 cells/mL K562: 241 cells/mL HL-60: 236 cells/mL MCF: 250 cells/mL | 5 min | [117] |
| CCRF-CEM HeLa K562 MCF-7 | Spiked Serum | Origami | Electrochemiluminescent | Semicarbazide and graphene QD enhanced silver NPs | CCRF-CEM: 53 cells/mL HeLa: 67 cells/mL K562: 42 cells/mL MCF-7: 38 cells/mL | 2 hr | [118] |
6. Summary and Outlook
Early detection, diagnosis and monitoring of cancer play a critical role in reducing mortality rates for cancer and overall healthcare costs. Paper-based analytical devices could serve as cost effective platforms for rapid and sensitive detection of multiple cancer biomarkers from body fluids. In this context, novel multiplex LFAs and μPADs have been developed to provide easy-to-use, sensitive diagnostic tools. These devices produce measurable signals for delivering both semi-quantitative and quantitative results. Given that cancer biomarkers are usually found at trace levels in body fluids, notable innovations for signal enhancement strategies are of the most important features of these devices.
The sensitivity of multiplex LFAs and μPADs can be directly linked to a number of key design features including incorporation of multiple labels in a single NP body, amplification of optical signals through enzyme driven chemical reactions, or chemical modifications on the paper to improve the electrical properties of the paper electrodes, each of which have been reviewed herein. Future devices will continue to optimize device layout and the exploration of new methods to achieve highly sensitive and specific indicators of the presence of extremely small amounts of biomarkers. One critical consideration moving forward is the possible cross-reactivity among multiple biomarkers as well as other proteins that are present in the sample. One way to approach to this problem is to create innovative sensing mechanisms that derive their inspiration from biomarker-specific biological processes.[123] For instance, leveraging enzymatic activity to measure biomarker levels is just one example for integrating catalytic reactions to such biorecognition mechanisms.[73, 124] Enzymes are known for their high affinity to a particular substrate while going through catalytic reactions. Therefore, enzyme specificity toward only one substrate could be utilized to reduce other biomarkers’ interference to the catalytic reaction. In addition, the implementation of new signal enhancement strategies that are uniquely programmed to amplify a signal upon this catalytic reaction and produce a pronounced visible color change could bring these devices closer to the goal of achieving point of care results without the need for analytical equipment.[124]
Even though technology-driven advencements in the detection of multiple biomarkers on paper-based devices would provide highly sensitive and specific quantitative outcomes in the future, additional key steps in the device design will need to be taken so that robust clinical decisions can be made for cancer diagnosis using these devices. Specifically, the current multiplex paper-based devices are lacking statistical evaluations of the quantified levels of multiple biomarkers. Future efforts in the field of multiplex paper-based devices should be devoted to broaden the scope of selected biomarkers that includes various clinically proven panels of biomarkers with high specificity towards a single cancer type. This will require collaboration between material scientists, chemists, clinicians and statisticians to successfully develop new sensing platforms with real clinical utility for cancer diagnosis.
From a broader perspective, widespread adoption of paper-based cancer diagnostics will also depend on overcoming current medical, societal and logistical impediments. In other words, even once technological advances in paper-based devices enable them to achieve high diagnostic performance, these barriers still stand in the way of bringing these technologies to patients at the point of care. Paper-based devices, as all other medical devices, must go through a strict regulatory approval process by the appropriate regulatory authority before being available on the market. The complex workflow of these regulatory processes for point of care devices and variations in these processes that change according to the regulatory authority make the commercialization and translation of the paper-based technologies non-trivial.[125] This is one of the reasons why there are limited numbers of paper-based devices for cancer diagnosis that are currently available in the market.[8a] Moreover, these devices need to be accepted by the public as effective diagnostic tools. As new and effective products become available in the future, healthcare providers will need to work on educating the public to raise the awareness on the significance, similar to regular mammogram or pap-smear tests for the screening purposes. Also, robust reimbursement strategies are required in the healthcare system so that these technologies can be accessible to wider patient population.[126] In addition to the aforementioned medical and societal challenges for paper-based diagnostic tools, there are still logistical issues in terms of transportation, storage, and managing supply chain of the paper-based diagnostics. Even though these problems are not always major issues in high-income countries, they still remain challenges in low- and middle-income countries, especially for the people who live in rural and resource-poor areas.
It may also be possible in the future to utilize not only body fluids for detection of disease-specific biomarkers, but also gasses like breath or body odor. For instance, numerous studies have suggested that canines have the ability to detect a number of diseases through the scents produced by patients with cancer[127] and even possibly COVID-19.[128] It is possible that detection devices (called electric or artificial noses) could be implemented instead for detection of these diseases.[129] One promising technology in this area is the increasing use of metal-organic frameworks (MOFs), which have pores that can be precisely tuned to have the desired shape, size, and surface chemistry to detect a specific chemical entity. In recent years, a number of research groups have shown how arrays of MOF can be used to make better electronic noses,[130] and could one day be used to detect diseases from subtle changes in body odor or breath.
Overall, recent innovations in the design and configuration of paper-based devices have been responsible for advancements in multiplexed analysis of cancer biomarkers and will continue to inspire future studies. By adopting novel biorecognition systems with signal enhancement strategies, the multiplex paper-based devices will deliver much more accurate and specific results in the future that will aid in early diagnosis of cancer and correspondingly timelier, life-saving treatments for patients. In particular, translating these early stage laboratory prototypes to affordable commercial products (similar to at-home use of pregnancy tests) will put diagnostic technologies in the hands of patients in a cost effective manner. Further, future paper-based devices will one day likely implement simple, portable readout systems in a uniform way. This will enable cancer diagnosis not in the clinics but also in resource-poor settings. Accordingly, smartphone-based readouts would offer low cost, simple and fast data generation and analysis. Given that less than 30% of the population in low-income countries have access to health services for cancer treatment, the ability to make cancer screening and monitoring readily available using portable devices would be transformative in healthcare areas that currently have no other options for diagnosis.
Acknowledgements
This work was supported by the NIH grant with the grant number 1R01 AR074285-01. B.Z. Ahlmark was supported by the Brackenridge Summer Research Fellowship from the Honors College at the University of Pittsburgh.
Biography
Dr. Nihan Yonet-Tanyeri received her PhD degree from the University of Illinois at Urbana-Champaign in 2011. Currently, she is a researcher at the University of Pittsburgh. Her research interests include biomaterials for biosensing and drug delivery applications.

Benjamin Z. Ahlmark is the recipient of the Brackenridge Summer Research Fellowship from the Honors College at the University of Pittsburgh. He is an undergraduate researcher with interests including biomaterials for biosensing and drug design and delivery.

Dr. Steven R. Little is currently the William Kepler Whiteford Professor and Chair of Chemical Engineering at the University of Pittsburgh. He is also the co-founder of Qrono Inc., a company which is the first to provide polymer-based custom design controlled release formulations. His research focuses on controlled release and pharmaceutical formulation as well as biomaterial and pharmaceutical excipient interactions, materials behavior ex vivo and in situ, new devices for detection of biomarkers, and the use of biomimetic design principles.

References
- [1].W. H. Organization in Cancer, Key Facts, URL: https://www.who.int/news-room/fact-sheets/detail/cancer, accessed: October, 2020. [Google Scholar]
- [2].a) DeSantis CE, Bray F, Ferlay J, Lortet-Tieulent J, Anderson BO and Jemal A, Cancer Epidemiology Biomarkers & Prevention 2015, 24, 1495–1506; [DOI] [PubMed] [Google Scholar]; b) Siegel R, DeSantis C. and Jemal A, Ca-a Cancer Journal for Clinicians 2014, 64, 104–117. [DOI] [PubMed] [Google Scholar]
- [3].Centers for Disease Control and Prevention in Screening Tests, URL: https://www.cdc.gov/cancer/dcpc/prevention/screening.htm, accessed: October, 2020. [Google Scholar]
- [4].Chen X, Gole J, Gore A, He Q, Lu M, Min J, Yuan Z, Yang X, Jiang Y, Zhang T, Suo C, Li X, Cheng L, Zhang Z, Niu H, Li Z, Xie Z, Shi H, Zhang X, Fan M, Wang X, Yang Y, Dang J, McConnell C, Zhang J, Wang J, Yu S, Ye W, Gao Y, Zhang K, Liu R. and Jin L, Nature Communications 2020, 11, 3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chou R, Selph SS, Buckley DI, Gustafson KS, Griffin JC, Grusing SE and Gore JL, Cancer 2016, 122, 842–851. [DOI] [PubMed] [Google Scholar]
- [6].Sievert KD, Amend B, Nagele U, Schilling D, Bedke J, Horstmann M, Hennenlotter J, Kruck S. and Stenzl A, World journal of urology 2009, 27, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yeung C, Dinh T. and Lee J, Pharmacoeconomics 2014, 32, 1093–1104. [DOI] [PubMed] [Google Scholar]
- [8].a) Mahmoudi T, de la Guardia M. and Baradaran B, TrAC Trends in Analytical Chemistry 2020, 125, 115842; [Google Scholar]; b) Ratajczak K. and Stobiecka M, Carbohydrate Polymers 2020, 229, 115463. [DOI] [PubMed] [Google Scholar]
- [9].Bunger S, Laubert T, Roblick UJ and Habermann JK, Journal of Cancer Research and Clinical Oncology 2011, 137, 375–389. [DOI] [PubMed] [Google Scholar]
- [10].Borrebaeck CAK, Nature Reviews Cancer 2017, 17, 199–204. [DOI] [PubMed] [Google Scholar]
- [11].Xiao H, Zhang Y, Kim Y, Kim S, Kim JJ, Kim KM, Yoshizawa J, Fan LY, Cao CX and Wong DTW, Scientific Reports 2016, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hartwell L, Mankoff D, Paulovich A, Ramsey S. and Swisher E, Nat Biotechnol 2006, 24, 905–908. [DOI] [PubMed] [Google Scholar]
- [13].Han C, Bellone S, Siegel ER, Altwerger G, Menderes G, Bonazzoli E, Egawa-Takata T, Pettinella F, Bianchi A, Riccio F, Zammataro L, Yadav G, Marto JA, Penet M-F, Levine DA, Drapkin R, Patel A, Litkouhi B, Ratner E, Silasi D-A, Huang GS, Azodi M, Schwartz PE and Santin AD, Gynecologic oncology 2018, 149, 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Kumarswamy R, Volkmann I. and Thum T, RNA Biology 2011, 8, 706–713; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC and Croce CM, Proceedings of the National Academy of Sciences 2006, 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mahesh G. and Biswas R, Journal of Interferon & Cytokine Research 2019, 39, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang F, Liang R, Tandon N, Matthews ER, Shrestha S, Yang J, Soibam B, Yang J. and Liu Y, Cellular and Molecular Life Sciences 2019, 76, 903–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guan Y, Song X, Sun W, Wang Y. and Liu B, Oxidative Medicine and Cellular Longevity 2019, 2019, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, Morgan TR, Kim HY, Lee WM, Bonkovsky HL and Dienstag JL, Gastroenterology 2010, 138, 493–502; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) American Association for Clinical Chemistry in Alpha-fetoprotein (AFP) Tumor Marker, URL: https://www.aacc.org/science-and-research/clinical-chemistry-trainee-council/trainee-council-in-english/pearls-of-laboratory-medicine/2013/tumor-markers-afp-and-hcg, accessed: October, 2020. [Google Scholar]
- [19].Kotzev A. and Draganov P, Gastrointestinal Tumors 2018, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haque A, Ray SK, Cox A. and Banik NL, Metabolic Brain Disease 2016, 31, 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Alberts AR, Schoots IG and Roobol MJ, International Journal of Urology 2015, 22, 524–532. [DOI] [PubMed] [Google Scholar]
- [22].Dochez V, Caillon H, Vaucel E, Dimet J, Winer N. and Ducarme G, Journal of Ovarian Research 2019, 12, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].a) Li X, Dai D, Chen B, Tang H, Xie X. and Wei W, Disease Markers 2018, 2018, 1–15; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) American Association for Clinical Chemistry in CA 15–3, URL: https://labtestsonline.org/tests/tumor-markers, accessed: October, 2020. [Google Scholar]
- [24].Stenman U-H, Tiitinen A, Alfthan H. and Valmu L, Human Reproduction Update 2006, 12, 769–784. [DOI] [PubMed] [Google Scholar]
- [25].Massarrat S. and Haj-Sheykholeslami A, Archives of Iranian Medicine 2016, 19, 137–140. [PubMed] [Google Scholar]
- [26].Keller L, Werner S. and Pantel K, Cell Stress 2019, 3, 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].a) Kolluri A. and Ho M, Frontiers in Oncology 2019, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Aviel-Ronen S, Lau SK, Pintilie M, Lau D, Liu N, Tsao MS and Jothy S, Modern Pathology 2008, 21, 817–825. [DOI] [PubMed] [Google Scholar]
- [28].a) Heikkila K, Ebrahim S. and Lawlor DA, Journal of Epidemiology & Community Health 2007, 61, 824–833; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li J, Jiao XD, Yuan ZF, Qiu HF and Guo RX, Medicine 2017, 96, 7; [Google Scholar]; c) Xu Q, Chen Y-J, Liu Z-Q, Chu L, Fang J-M, Zhang X. and Zhao H-X, Asian Journal of Andrology 2014, 16, 467; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Gupta SC, Kunnumakkara AB, Aggarwal S. and Aggarwal BB, Frontiers in Immunology 2018, 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Henry NL and Hayes DF, Molecular Oncology 2012, 6, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Füzéry AK, Levin J, Chan MM and Chan DW, Clinical proteomics 2013, 10, 13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ruhaak LR, Nguyen UT, Stroble C, Taylor SL, Taguchi A, Hanash SM, Lebrilla CB, Kim K. and Miyamoto S, Proteomics Clin Appl 2013, 7, 664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Duffy MJ, Clinical Cancer Research 1996, 2, 613. [PubMed] [Google Scholar]
- [33].a) Schwarzenbach H, Hoon DSB and Pantel K, Nature Reviews Cancer 2011, 11, 426–437; [DOI] [PubMed] [Google Scholar]; b) Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, Ali S, Cleator S, Kenny L, Stebbing J, Rutherford M, Sethi H, Boydell A, Swenerton R, Fernandez-Garcia D, Gleason KLT, Goddard K, Guttery DS, Assaf ZJ, Wu HT, Natarajan P, Moore DA, Primrose L, Dashner S, Tin AS, Balcioglu M, Srinivasan R, Shchegrova SV, Olson A, Hafez D, Billings P, Aleshin A, Rehman F, Toghill BJ, Hills A, Louie MC, Lin CJ, Zimmermann BG and Shaw JA, Clin Cancer Res 2019, 25, 4255–4263; [DOI] [PubMed] [Google Scholar]; c) Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, Goddard KS, Hastings RK, Luo J, Ogle O, Woodley L, Ali S, Stebbing J. and Coombes RC, Clin Cancer Res 2017, 23, 88–96; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Das J, Ivanov I, Sargent EH and Kelley SO, Journal of the American Chemical Society 2016, 138, 11009–11016. [DOI] [PubMed] [Google Scholar]
- [34].Fleischhacker M. and Schmidt B, Biochim Biophys Acta 2007, 1775, 181–232. [DOI] [PubMed] [Google Scholar]
- [35].Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD and Knippers R, Cancer Res 2001, 61, 1659–1665. [PubMed] [Google Scholar]
- [36].Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R. and Rosenfeld N, Nat Rev Cancer 2017, 17, 223–238. [DOI] [PubMed] [Google Scholar]
- [37].Wang H, Peng R, Wang J, Qin Z. and Xue L, Clinical Epigenetics 2018, 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].a) Paget S, Cancer Metastasis Rev 1989, 8, 98–101; [PubMed] [Google Scholar]; b) Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verástegui E. and Zlotnik A, Nature 2001, 410, 50–56. [DOI] [PubMed] [Google Scholar]
- [39].a) Balic M, Lin H, Williams A, Datar RH and Cote RJ, Expert Rev Mol Diagn 2012, 12, 303–312; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang C, Xia B-R, Jin W-L and Lou G, Cancer Cell International 2019, 19, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Joosse SA and Pantel K, Cancer Research 2013, 73, 8–11. [DOI] [PubMed] [Google Scholar]
- [41].Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y. and Zheng S, Signal Transduction and Targeted Therapy 2020, 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jayaseelan VP, Cancer Gene Therapy 2020, 27, 395–398. [DOI] [PubMed] [Google Scholar]
- [43].Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FWN, Das S, Driedonks TAP, Fernández-Messina L, Haderk F, Hill AF, Jones JC, Van Keuren-Jensen KR, Lai CP, Lässer C, di Liegro I, Lunavat TR, Lorenowicz MJ, Maas SLN, Mäger I, Mittelbrunn M, Momma S, Mukherjee K, Nawaz M, Pegtel DM, Pfaffl MW, Schiffelers RM, Tahara H, Théry C, Tosar JP, Wauben MHM, Witwer KW and Nolte-’t Hoen ENM, Journal of Extracellular Vesicles 2017, 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Simonsen JB, Circulation Research 2017, 121, 920–922. [DOI] [PubMed] [Google Scholar]
- [45].a) Hanash SM, Pitteri SJ and Faca VM, Nature 2008, 452, 571–579; [DOI] [PubMed] [Google Scholar]; b) Mirus JE, Zhang Y, Li CI, Lokshin AE, Prentice RL, Hingorani SR and Lampe PD, Clin Cancer Res 2015, 21, 1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stills in Chapter HF 11 - Polyclonal Antibody Production, Eds.: Suckow MA, Stevens KA and Wilson RP, Academic Press, Boston, 2012, pp. 259–274. [Google Scholar]
- [47].Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, Anagnostou V. and Rimm D, BioTechniques 2010, 48, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pasqualini R. and Arap W, Proceedings of the National Academy of Sciences of the United States of America 2004, 101, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].a) Pinto Torres JE, Goossens J, Ding J, Li Z, Lu S, Vertommen D, Naniima P, Chen R, Muyldermans S, Sterckx YGJ and Magez S, Scientific Reports 2018, 8, 9019; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Doerflinger SY, Tabatabai J, Schnitzler P, Farah C, Rameil S, Sander P, Koromyslova A. and Hansman GS, mSphere 2016, 1, e00219–00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang EY and Shah K, Frontiers in Oncology 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S. and Wyns L, Proceedings of the National Academy of Sciences of the United States of America 2006, 103, 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Binz HK, Amstutz P. and Plückthun A, Nature Biotechnology 2005, 23, 1257–1268. [DOI] [PubMed] [Google Scholar]
- [53].Stoltenburg R, Reinemann C. and Strehlitz B, Biomolecular Engineering 2007, 24, 381–403. [DOI] [PubMed] [Google Scholar]
- [54].Toh SY, Citartan M, Gopinath SCB and Tang T-H, Biosensors and Bioelectronics 2015, 64, 392–403. [DOI] [PubMed] [Google Scholar]
- [55].a) Jazayeri MH, Amani H, Pourfatollah AA, Pazoki-Toroudi H. and Sedighimoghaddam B, Sensing and Bio-Sensing Research 2016, 9, 17–22; [Google Scholar]; b) Zhang J, Liu B, Liu H, Zhang X. and Tan W, Nanomedicine (Lond) 2013, 8, 983–993; [DOI] [PubMed] [Google Scholar]; c) Francis JE, Mason D. and Lévy R, Beilstein journal of nanotechnology 2017, 8, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Foubert A, Beloglazova NV, Rajkovic A, Sas B, Madder A, Goryacheva IY and De Saeger S, TrAC Trends in Analytical Chemistry 2016, 83, 31–48. [Google Scholar]
- [57].Noviana E, McCord CP, Clark KM, Jang I. and Henry CS, Lab on a Chip 2020, 20, 9–34. [DOI] [PubMed] [Google Scholar]
- [58].Mansfield MA in The Use of Nitrocellulose Membranes in Lateral-Flow Assays, Eds.: Wong RC and Tse HY, Humana Press, Totowa, NJ, 2005, pp. 71–85. [Google Scholar]
- [59].a) Wu Y, Zhou Y, Leng Y, Lai W, Huang X. and Xiong Y, Biosensors and Bioelectronics 2020, 157, 112168; [DOI] [PubMed] [Google Scholar]; b) Li J. and Macdonald J, Biosensors and Bioelectronics 2016, 83, 177–192. [DOI] [PubMed] [Google Scholar]
- [60].Li KJ, Li XQ, Fan YL, Yang CH and Lv XF, Sensors and Actuators B-Chemical 2019, 286, 272–281. [Google Scholar]
- [61].Galaziou A, Christopoulos TK and Ioannou PC, Analytical and Bioanalytical Chemistry 2019, 411, 3769–3776. [DOI] [PubMed] [Google Scholar]
- [62].Washburn EW, Physical Review 1921, 17, 273–283. [Google Scholar]
- [63].Wang CY, Hou F. and Ma YC, Biosensors & Bioelectronics 2015, 68, 156–162. [DOI] [PubMed] [Google Scholar]
- [64].Rong Z, Bai ZK, Li JN, Tang H, Shen TY, Wang Q, Wang CW, Xiao R. and Wang SQ, Biosensors & Bioelectronics 2019, 145, 8. [DOI] [PubMed] [Google Scholar]
- [65].Xiao K, Wang K, Qin WJ, Hou YF, Lu WT, Xu H, Wo Y. and Cui DX, Talanta 2017, 164, 463–469. [DOI] [PubMed] [Google Scholar]
- [66].Chan WCW and Nie S, Science 1998, 281, 2016. [DOI] [PubMed] [Google Scholar]
- [67].Chen ZH, Liang RL, Guo XX, Liang JY, Deng QT, Li M, An TX, Liu TC and Wu YS, Biosensors & Bioelectronics 2017, 91, 60–65. [DOI] [PubMed] [Google Scholar]
- [68].Lu WT, Wang K, Xiao K, Qin WJ, Hou YF, Xu H, Yan XY, Chen YR, Cui DX and He JH, Scientific Reports 2017, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].a) Yu J, Yang L, Liang X, Dong T. and Liu H, Analyst 2015, 140, 4114–4120; [DOI] [PubMed] [Google Scholar]; b) Zhang Q, Su R, Wang J-P and Zhang Q, Journal of Nanoscience and Nanotechnology 2016, 16, 7427–7432. [Google Scholar]
- [70].Zhang B, Ma WJ, Li FX, Gao WC, Zhao Q, Peng WP, Piao JF, Wu XL, Wang HJ, Gong XQ and Chang J, Nanoscale 2017, 9, 18711–18722. [DOI] [PubMed] [Google Scholar]
- [71].Chen YP, Sun JS, Xianyu YL, Yin BF, Niu YJ, Wang SB, Cao FJ, Zhang XQ, Wang Y. and Jiang XY, Nanoscale 2016, 8, 15205–15212. [DOI] [PubMed] [Google Scholar]
- [72].Loynachan CN, Soleimany AP, Dudani JS, Lin Y, Najer A, Bekdemir A, Chen Q, Bhatia SN and Stevens MM, Nature Nanotechnology 2019, 14, 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Warren AD, Kwong GA, Wood DK, Lin KY and Bhatia SN, Proceedings of the National Academy of Sciences of the United States of America 2014, 111, 3671–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dave VP, Ngo TA, Pernestig A-K, Tilevik D, Kant K, Nguyen T, Wolff A. and Bang DD, Laboratory Investigation 2019, 99, 452–469. [DOI] [PubMed] [Google Scholar]
- [75].Zheng WL, Yao L, Teng J, Yan C, Qin PZ, Liu GD and Chen W, Sensors and Actuators B-Chemical 2018, 264, 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Javani A, Javadi-Zarnaghi F. and Rasaee MJ, Analytical Biochemistry 2017, 537, 99–105. [DOI] [PubMed] [Google Scholar]
- [77].Gao X, Xu H, Baloda M, Gurung AS, Xu L-P, Wang T, Zhang X. and Liu G, Biosensors and Bioelectronics 2014, 54, 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Raman CV and Krishnan KS, Nature 1928, 121, 501–502. [Google Scholar]
- [79].Fu X, Cheng Z, Yu J, Choo P, Chen L. and Choo J, Biosens Bioelectron 2016, 78, 530–537. [DOI] [PubMed] [Google Scholar]
- [80].Wang X, Choi N, Cheng Z, Ko J, Chen L. and Choo J, Analytical Chemistry 2017, 89, 1163–1169. [DOI] [PubMed] [Google Scholar]
- [81].Rosales CM, McLaughlin MD, Sata T, Katano H, Veno PA, de Las Casas LE and Miranda RN, AIDS patient care and STDs 2002, 16, 573–577. [DOI] [PubMed] [Google Scholar]
- [82].Mancuso M, Jiang L, Cesarman E. and Erickson D, Nanoscale 2013, 5, 1678–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].a) Chen C, Lin B-R, Hsu M-Y and Cheng C-M, Journal of visualized experiments : JoVE 2015, e52722-e52722; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheng N, Song Y, Shi Q, Du D, Liu D, Luo Y, Xu W. and Lin Y, Analytical Chemistry 2019, 91, 13986–13993; [DOI] [PubMed] [Google Scholar]; c) Lee J, Kim H, Heo Y, Yoo YK, Han SI, Kim C, Hur D, Kim H, Kang JY and Lee JH, Analyst 2020, 145, 157–164; [DOI] [PubMed] [Google Scholar]; d) Wu T, Yang Y, Cao Y, Huang Y, Xu L-P, Zhang X. and Wang S, Science China Chemistry 2018, 61, 1423–1429. [Google Scholar]
- [84].Guo S, Xu J, Estell AP, Ivory CF, Du D, Lin Y. and Dong W-J, Biosensors and Bioelectronics 2020, 164, 112292. [DOI] [PubMed] [Google Scholar]
- [85].Liana DD, Raguse B, Gooding JJ and Chow E, Sensors (Basel) 2012, 12, 11505–11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].a) Cate DM, Adkins JA, Mettakoonpitak J. and Henry CS, Analytical Chemistry 2015, 87, 19–41; [DOI] [PubMed] [Google Scholar]; b) Martinez AW, Phillips ST, Whitesides GM and Carrilho E, Analytical Chemistry 2010, 82, 3–10; [DOI] [PubMed] [Google Scholar]; c) Sher M, Zhuang R, Demirci U. and Asghar W, Expert Review of Molecular Diagnostics 2017, 17, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Martinez AW, Phillips ST, Butte MJ and Whitesides GM, Angew Chem Int Ed Engl 2007, 46, 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Carrilho E, Martinez AW and Whitesides GM, Anal Chem 2009, 81, 7091–7095. [DOI] [PubMed] [Google Scholar]
- [89].a) Li X, Tian J, Garnier G. and Shen W, Colloids and Surfaces B: Biointerfaces 2010, 76, 564–570; [DOI] [PubMed] [Google Scholar]; b) Yamada K, Takaki S, Komuro N, Suzuki K. and Citterio D, Analyst 2014, 139, 1637–1643. [DOI] [PubMed] [Google Scholar]
- [90].Olkkonen J, Lehtinen K. and Erho T, Anal Chem 2010, 82, 10246–10250. [DOI] [PubMed] [Google Scholar]
- [91].Chitnis G, Ding Z, Chang C-L, Savran CA and Ziaie B, Lab on a Chip 2011, 11, 1161–1165. [DOI] [PubMed] [Google Scholar]
- [92].Liang LL, Su M, Li L, Lan FF, Yang GX, Ge SG, Yu JH and Song XR, Sensors and Actuators B-Chemical 2016, 229, 347–354. [Google Scholar]
- [93].Ge SG, Ge L, Yan M, Song XR, Yu JH and Liu SS, Biosensors & Bioelectronics 2013, 43, 425–431. [DOI] [PubMed] [Google Scholar]
- [94].Guo XY, Guo YM, Liu W, Chen Y. and Chu WR, Spectrochimica Acta Part a-Molecular and Biomolecular Spectroscopy 2019, 223, 7. [DOI] [PubMed] [Google Scholar]
- [95].Jarujamrus P, Tian J, Li X, Siripinyanond A, Shiowatana J. and Shen W, Analyst 2012, 137, 2205–2210. [DOI] [PubMed] [Google Scholar]
- [96].Kong F. and Hu YF, Anal Bioanal Chem 2012, 403, 7–13. [DOI] [PubMed] [Google Scholar]
- [97].a) Wang SM, Ge L, Song XR, Yu JH, Ge SG, Huang JD and Zeng F, Biosensors & Bioelectronics 2012, 31, 212–218; [DOI] [PubMed] [Google Scholar]; b) Wu Y, Xue P, Hui KM and Kang Y, ChemElectroChem 2014, 1, 722–727. [Google Scholar]
- [98].Baynes C. and Yoon JY, Slas Technology 2018, 23, 30–43. [DOI] [PubMed] [Google Scholar]
- [99].Ge L, Yan JX, Song XR, Yan M, Ge SG and Yu JH, Biomaterials 2012, 33, 1024–1031. [DOI] [PubMed] [Google Scholar]
- [100].Wang J, Li W, Ban L, Du W, Feng X. and Liu B-F, Sensors and Actuators B: Chemical 2018, 254, 855–862. [Google Scholar]
- [101].Ge L, Wang S, Song X, Ge S. and Yu J, Lab on a Chip 2012, 12, 3150. [DOI] [PubMed] [Google Scholar]
- [102].Jiao YC, Du C, Zong LJ, Guo XY, Han YF, Zhang XP, Li L, Zhang CW, Ju Q, Liu JH, Yu HD and Huang W, Sensors and Actuators B-Chemical 2020, 306, 9. [Google Scholar]
- [103].Sun X, Li B, Tian C, Yu F, Zhou N, Zhan Y. and Chen L, Analytica Chimica Acta 2018, 1007, 33–39. [DOI] [PubMed] [Google Scholar]
- [104].Wu YF, Xue P, Hui KM and Rang YJ, Biosensors & Bioelectronics 2014, 52, 180–187. [DOI] [PubMed] [Google Scholar]
- [105].Wu YF, Xue P, Kang YJ and Hui KM, Analytical Chemistry 2013, 85, 8661–8668. [DOI] [PubMed] [Google Scholar]
- [106].Wang PP, Ge L, Yan M, Song XR, Ge SG and Yu JH, Biosensors & Bioelectronics 2012, 32, 238–243. [DOI] [PubMed] [Google Scholar]
- [107].Li WP, Li L, Li M, Yu JH, Ge SG, Yan M. and Song XR, Chemical Communications 2013, 49, 9540–9542. [DOI] [PubMed] [Google Scholar]
- [108].Li WP, Li L, Ge SG, Song XR, Ge L, Yan M. and Yu JH, Biosensors & Bioelectronics 2014, 56, 167–173. [DOI] [PubMed] [Google Scholar]
- [109].Li L, Li W, Yang H, Ma C, Yu J, Yan M. and Song X, Electrochimica Acta 2014, 120, 102–109. [Google Scholar]
- [110].Ma C, Li WP, Kong QK, Yang HM, Bian ZQ, Song XR, Yu JH and Yan M, Biosensors & Bioelectronics 2015, 63, 7–13. [DOI] [PubMed] [Google Scholar]
- [111].Sun GQ, Zhang LN, Zhang Y, Yang HM, Ma C, Ge SG, Yan M, Yu JH and Song XR, Biosensors & Bioelectronics 2015, 71, 30–36. [DOI] [PubMed] [Google Scholar]
- [112].Ge SG, Li WP, Yan M, Song XR and Yu JH, Journal of Materials Chemistry B 2015, 3, 2426–2432. [DOI] [PubMed] [Google Scholar]
- [113].Wang Y, Luo JP, Liu JT, Sun S, Xiong Y, Ma YY, Yan S, Yang Y, Yin HB and Cai XX, Biosensors & Bioelectronics 2019, 136, 84–90. [DOI] [PubMed] [Google Scholar]
- [114].Deng HP, Zhou XM, Liu QW, Li BF, Liu HX, Huang R. and Xing D, Acs Applied Materials & Interfaces 2017, 9, 41151–41158. [DOI] [PubMed] [Google Scholar]
- [115].Zhao W, Ali MM, Brook MA and Li Y, Angewandte Chemie International Edition 2008, 47, 6330–6337. [DOI] [PubMed] [Google Scholar]
- [116].Liang LL, Lan FF, Yin XM, Ge SG, Yu JH and Yan M, Biosensors & Bioelectronics 2017, 95, 181–188. [DOI] [PubMed] [Google Scholar]
- [117].Wu LD, Ma C, Ge L, Kong QK, Yan M, Ge SG and Yu JH, Biosensors & Bioelectronics 2015, 63, 450–457. [DOI] [PubMed] [Google Scholar]
- [118].Yang HM, Zhang Y, Li L, Zhang LN, Lan FF and Yu JH, Analytical Chemistry 2017, 89, 7511–7519. [DOI] [PubMed] [Google Scholar]
- [119].Zhang Y, Ge L, Li M, Yan M, Ge SG, Yu JH, Song XR and Cao BQ, Chemical Communications 2014, 50, 1417–1419. [DOI] [PubMed] [Google Scholar]
- [120].Li L, Li W, Yang H, Ma C, Yu J, Yan M. and Song X, Electrochimica Acta 2014, 120, 102–109. [Google Scholar]
- [121].Su M, Liu H, Ge S, Ren N, Ding L, Yu J. and Song X, RSC Advances 2016, 6, 16500–16506. [Google Scholar]
- [122].Tian T, Li L, Zhang Y, Liu HY, Zhang LN, Yan M. and Yu JH, Sensors and Actuators B-Chemical 2018, 261, 44–50. [Google Scholar]
- [123].Soleimany AP and Bhatia SN, Trends in Molecular Medicine 2020, 26, 450–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Acharya AP, Theisen KM, Correa A, Meyyappan T, Apfel A, Sun T, Tarin TV and Little SR, Advanced Healthcare Materials 2017, 6, 1700808. [DOI] [PubMed] [Google Scholar]
- [125].Yetisen AK, Akram MS, Lowe CR, Lab on a chip 2013, 13, 2210–2251. [DOI] [PubMed] [Google Scholar]
- [126].Quinn AD, Dixon D. and Meenan BJ, Crit Rev Clin Lab Sci 2016, 53, 1–12. [DOI] [PubMed] [Google Scholar]
- [127].a) Ehmann R, Boedeker E, Friedrich U, Sagert J, Walles T. and Friedel G, European Respiratory Journal 2011, 38, p2787; [DOI] [PubMed] [Google Scholar]; b) McCulloch M, Jezierski T, Broffman M, Hubbard A, Turner K. and Janecki T, Integrative Cancer Therapies 2006, 5, 30–39. [DOI] [PubMed] [Google Scholar]
- [128].a) Grandjean D, Sarkis R, Tourtier J-P, Julien-Lecocq C, Benard A, Roger V, Levesque E, Bernes-Luciani E, Maestracci B, Morvan P, Gully E, Berceau-Falancourt D, Pesce J-L, Lecomte B, Haufstater P, Herin G, Cabrera J, Muzzin Q, Gallet C, Bacqué H, Broc J-M, Thomas L, Lichaa A, Moujaes G, Saliba M, Kuhn A, Galey M, Berthail B, Lapeyre L, Méreau O, Matteï M-N, Foata A, Bey L, Philippe A-S, Abassi P, Pisani F, Delarbre M, Orsini J-M, Capelli A, Renault S, Bachir K, Kovinger A, Comas E, Stainmesse A, Etienne E, Voeltzel S, Mansouri S, Berceau-Falancourt M, Leva B, Faure F, Dami A, Costa MA, Tafanelli J-J, Luciani J-B, Casalot J-J, Charlet L, Ruau E, Issa M, Grenet C, Billy C. and Desquilbet L, bioRxiv 2020, 10.1101/2020.06.03.132134 [DOI] [Google Scholar]; b) Jendrny P, Schulz C, Twele F, Meller S, von Köckritz-Blickwede M, Osterhaus ADME, Ebbers J, Pilchová V, Pink I, Welte T, Manns MP, Fathi A, Ernst C, Addo MM, Schalke E. and Volk HA, BMC Infectious Diseases 2020, 20, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wilson AD and Baietto M, Sensors (Basel) 2009, 9, 5099–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].a) Yao M-S, Tang W-X, Wang G-E, Nath B. and Xu G, Advanced Materials 2016, 28, 5229–5234; [DOI] [PubMed] [Google Scholar]; b) Campbell MG, Liu SF, Swager TM and Dincă M, Journal of the American Chemical Society 2015, 137, 13780–13783; [DOI] [PubMed] [Google Scholar]; c) Ellern I, Venkatasubramanian A, Lee JH, Hesketh P, Stavila V, Robinson A. and Allendorf M, Micro & Nano Letters 2013, 8, 766–769; [Google Scholar]; d) Gustafson JA and Wilmer CE, Sensors and Actuators B: Chemical 2018, 267, 483–493; [Google Scholar]; e) Gustafson JA and Wilmer CE, The Journal of Physical Chemistry C 2017, 121, 6033–6038; [Google Scholar]; f) Zhang Y, Lan D, Wang Y, Cao H. and Jiang H, Physica E: Low-dimensional Systems and Nanostructures 2011, 43, 1219–1223. [Google Scholar]