Abstract
Nucleosome remodeling provides access to genomic DNA for recruitment of the transcriptional machinery to mediate gene expression. Aberrant function of nucleosome remodeling complexes has been correlated to human cancer, making them emerging therapeutic targets. The bromodomain PHD finger transcription factor, BPTF, is the largest member of the human nucleosome remodeling factor NURF. Over the last five years, BPTF has become increasingly identified as a pro-tumorigenic factor, prompting investigations into the molecular mechanisms associated with BPTF function. Despite a druggable bromodomain, small molecule discovery is at an early stage. Here we highlight recent investigations into the biology being discovered for BPTF, chemical biology approaches used to study its function, and small molecule inhibitors being designed as future chemical probes and therapeutics.
Keywords: BPTF, NURF, Bromodomain, Chemical Probe, Chemical Epigenetics
Graphical Abstract
Introduction
Epigenetics research focuses on the molecular mechanisms associated with the heritability of genetic information [1]. Chemical epigenetics is a subdiscipline of this field with a chief aim of developing chemical tools, probe molecules, and technological advances to illuminate these mechanisms at the molecular level, and in some cases with atomic-level precision [2]. Through this lens, we will provide an update on the current biology and chemical epigenetics approaches used to study an emerging epigenetic regulatory protein, the bromodomain and plant homeodomain (PHD) finger transcription factor, BPTF, while highlighting opportunities for innovation.
Bromodomain and PHD finger-containing proteins, such as BPTF, are two classes of epigenetic effector proteins, or “readers”. The PHD finger family is one of the largest classes of readers found in 291 human proteins [3]; however, drug discovery efforts targeting this domain have proven difficult with no chemical probes yet reported. Conversely, there are 46 human bromodomain-containing proteins. These bromodomains can be classified into eight structural families. Despite early efforts, significant chemical probes/drug candidates were not reported in the primary literature until 2010 when the first submicromolar inhibitors were reported for class II bromodomain and extraterminal (BET) proteins [4,5]. Chemical biologists and medicinal chemists have since made significant progress studying the molecular mechanisms of BET bromodomains and advancing therapeutic agents to clinical trials. Comparatively, translational studies of inhibitors for many non-BET bromodomain-containing proteins have lagged.
In this Current Opinion, we review the recent literature concerning the emerging biology and chemical biology of a class I non-BET bromodomain-containing protein, BPTF, the largest member of the nucleosome remodeling factor, NURF. We describe the role of BPTF in normal and pathophysiology, elaborate on new chemical biology tools, and discuss the first set of chemical inhibitors as important steps towards chemical probe development and novel therapeutics (Figure 1).
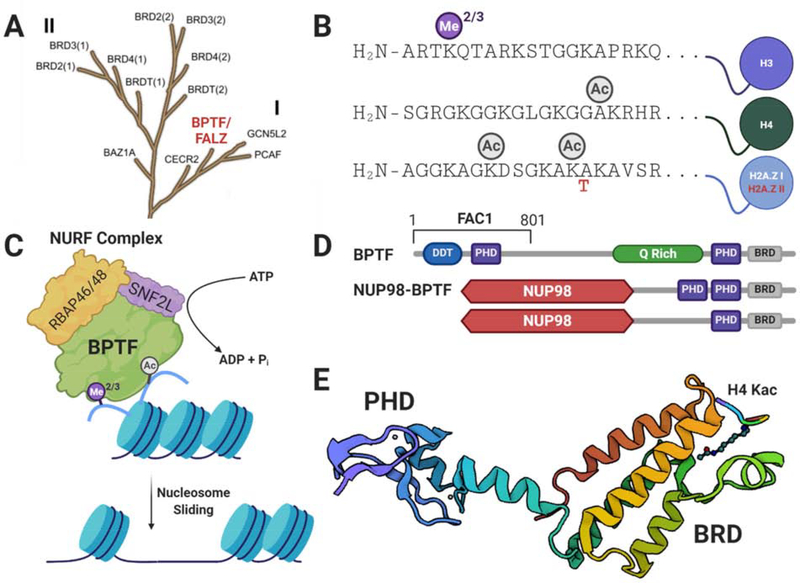
Figure 1: BPTF structure and function as an epigenetic reader protein.
A) Branch of the bromodomain phylogenetic tree representing the BET family (II) and family I, to which BPTF/FALZ belongs. (adapted with permission from Filippakopoulos et al. [14]) B) Modified Histone binding partners of BPTF’s PHD domain (H3 K4me2/3) and BRD (H4 K16ac and H2A.Z I/II K7ac, K13ac). C) Recruitment of NURF to chromatin through modified histone interactions facilitates cis nucleosome sliding. D) Domain diagram of BPTF with DNA binding homeobox and different transcription factor (DDT) domain, PHD domains, glutamine rich region (Q rich), and bromodomain (BRD) shown. The residues corresponding to the FAC1 truncation are outlined above (top). Also shown are the two domain diagrams of two reported NUP98-BPTF fusion proteins (middle, bottom). E) X-ray crystal structure of BPTF PHD-BRD in complex with acetylated H4 peptide shown in chainbow. PHD and bromodomains are annotated. PDB: 3QZV.
Discovery of BPTF
BPTF is expressed in several isoforms, including the full-length protein, a shorter N-terminal isoform, fetal Alzheimer’s clone 1 (FAC1), and oncogenic BPTF fusion proteins. FAC1 was identified first from amyloid plaques of Alzheimer’s patients [6] and later shown to regulate gene transcription through interaction with specific DNA sequences and the Myc-associated zinc finger protein ZF87/MAZ [7,8]. FAC1 shares the N-terminal 801 amino acids of BPTF and likely arises from an alternative splicing event [9]. BPTF is homologous to NURF301, the largest subunit of the Drosophila NURF complex which was isolated from embryo extracts the same year as the identification of FAC1.[10] The full length BPTF gene, also named FALZ, was identified through a cDNA database search for bromodomain motifs. BPTF contains a DDT DNA-association domain [11], two PHD fingers, a bromodomain, three LXXLL nuclear receptor binding motifs, and a glutamine-rich acidic region (Figure 1D) [9]. Apart from the C-terminal PHD and bromodomain, the functions of the additional domains remain largely uncharacterized. Additionally, an oncogenic fusion protein containing the c-terminal chromatin binding domains of BPTF, NUP98-BPTF, was recently identified from patient samples with acute megakaryoblastic leukemia (Figure 1D). BPTF-NUTM1 was also identified in acute lymphoblastic leukemia samples but is less well-characterized. [12,13].
BPTF Interactome and Functionality
BPTF serves as the largest subunit of NURF, the founding member of the ISWI family of ATP-dependent chromatin remodeling complexes [15]. Human NURF contains an ISWI ATPase domain, SNF2L, and WD-50 repeat containing protein RbAp46/48 (Figure 1C) [16]. In Drosophila, reconstitution of the ATPase and NURF301 was shown to be both sufficient and necessary for maintaining nucleosome remodeling activity. Deletion of the N-terminal 121 amino acids of NURF301 preserved complex formation but reduced activity [10]. A truncated NURF301 isoform lacking the PHD and Bromodomain also forms a NURF complex [17]. Given their difference in size, it remains unclear if either FAC1 or the BPTF fusion proteins form stable NURF complexes.
Once recruited to chromatin through interactions with modified histones or gene-specific transcription factors, NURF is responsible for sliding nucleosomes in cis to alter nucleosome position. The resulting changes in DNA accessibility allows for transcription factors to bind and regulate gene expression [15,18–20]. In mice, BPTF regulates genes and signaling pathways for early tissue development [21], and BPTF loss-of-function is embryonic lethal [22]. BPTF is essential for thymocyte maturation [23], immune homeostasis [24], melanocyte stem cell differentiation [25], and mammary gland development [26]. In flies., a NURF complex containing a BPTF isoform without the PHD and bromodomain leads to impaired spermatogenesis [17].
Chemical biologists have contributed to the understanding of BPTF PHD- and bromodomain-chromatin interactions using synthetic peptides and nucleosomes with post-translational modifications (PTMs) (Figure 1B). The C-terminal PHD domain was shown to directly interact with H3 K4me2/3 using synthetic H3 tail peptides [27] and in cells via co-immunoprecipitation [28]. Trimethylation of K4 on H3 is closely associated with the 5’ end of actively transcribed genes [29], whereas mono- and dimethylation are more broadly distributed across active genes [30,31] and are found at enhancers [32]. Although the PHD domain binds the H3 K4me3 (1–15) peptide robustly (Kd = 2.7 μM ) [28], Morrison et al. found that BPTF binding was prevented when the histone was incorporated in nucleosomes. Further investigations revealed the basic H3 tail associates with nucleosomal DNA which precludes binding. Additional modifications to H3 tails that change the electrostatic environment (e.g. lysine acetylation) are necessary to weaken the association with DNA and allow PHD domain binding [33]. These studies underscore the importance of characterizing histone interactions in their native environment.
In the context of BPTF bromodomain interactions, Ruthenburg et al. found that the bromodomain promiscuously interacts with several monoacetylated H4 peptides, including K12ac, K16ac, and K20ac. However, when these modifications were incorporated into nucleosomes containing an H3 K4me3 histone, the BPTF bromodomain showed enhanced specificity for the H4 K16ac mark. The interaction of BPTF’s C-terminal PHD domain with H3 K4me3 is ~100 fold stronger than the interaction between the bromodomain and acetylated H4. Tethering the PHD domain to H3 directs bromodomain binding to H4 K16ac to achieve specificity [34]. Both the PHD domain and bromodomain were shown to colocalize with H3 K4me3 and H4 K16ac near transcription start sites in chromatin immunoprecipitation experiments [27,34,35]. Mutations in either domain abrogated binding [34]. Furthermore, treatment of cells with bromodomain inhibitors reduces chromatin residence time [36] and BPTF-mediated transcription in reporter assays [37**].
Although H4 acetylation is considered the canonical interaction with BPTF, recent experiments showed the BPTF bromodomain can engage both isoforms of histone variant H2A.Z. Acetylation at K7 and K13 on synthetic peptides was the highest affinity modification pattern [38,39*] with similar affinity for BPTF as H4 K16ac. Although BPTF co-immunoprecipitates with nucleosomes containing H2A.Z, a direct interaction in a cellular context needs to be verified [35]. Beyond histone interactions, BPTF/FAC1 interacts with various transcription factors and nucleosome remodeling subunits which can be targeted for modulating BPTF function (Figure 2A) [23,40–45]. Searching for novel BPTF interactions continues to be an active area of research [46].
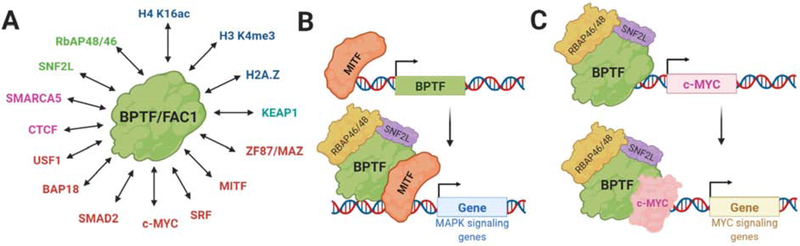
Figure 2: Molecular mechanisms associated with BPTF function.
A) Reported interactions of BPTF/FAC1. Histone proteins shown in blue, transcriptional regulators in red, antioxidant response proteins in cyan, chromatin regulators in magenta, and NURF subunits in green. B) Mechanistic role of MITF and BPTF in melanoma. MITF regulates the expression of BPTF and associates with NURF to co-regulate the expression of cell-cycle-regulating genes. C) BPTF drives c-Myc expression and binds to c-Myc to facilitate its transcriptional activity. Supplementary Table 1 is provided as a resource for additional direct BPTF genes in various disease states.
Emerging Oncogenic Roles of BPTF
BPTF was first implicated in disease by Buganim et al. through characterizing a translocation breakpoint in the BPTF gene at the chromosome 17q24.3 locus. BPTF was overexpressed in cells showing this translocation, and knockdown of BPTF slowed proliferation, indicating its role in driving malignancy. Further investigation showed amplification of BPTF in human tumors, most significantly in lung cancer and neuroblastomas [47*], the latter of which is closely correlated to 17q aberrations [48]. BPTF overexpression has now been verified in several cancers including non-small-cell lung cancer [49], hepatocellular carcinoma [50], and colorectal cancers [51]. BPTF’s most characterized protumorigenic role has been in melanoma, where BPTF overexpression predicts poor survival outcomes. BCL2, BCL-XL, and CCND2 were identified as key genes regulated by BPTF to have proliferative and antiapoptotic effects on cancer cells [52**]. In melanoma, BPTF expression is activated by microphthalmia-associated transcription factor (MITF), which binds at the BPTF promoter to facilitate transcription (Figure 2B) [53]. In a downstream signaling event, MITF associates with NURF affecting MAPK signaling to regulate cell cycle and survival genes [25].
A second emerging mechanism for BPTF and cancer is through an interaction with c-Myc, a transcription factor that is overexpressed in many human cancers [54*]. BPTF silencing impairs c-Myc recruitment to chromatin and reduces DNA accessibility at c-Myc target genes while also regulating Myc expression (Figure 2C) [55]. Through these mechanisms, BPTF silencing leads to reduced cell proliferation and replication stress [54*,56]. Therefore, disruption of the c-Myc-BPTF interaction is a potential strategy for the treatment of c-Myc driven tumors. In high grade gliomas, knockdown of the additional BPTF subunits did not have a significant effect on proliferation, suggesting additional function of BPTF outside of NURF [55,57]. Given the emerging role of BPTF in cancer, there is a significant unmet need for new synthetic inhibitors.
Chemical Biology Approaches to Study BPTF Function
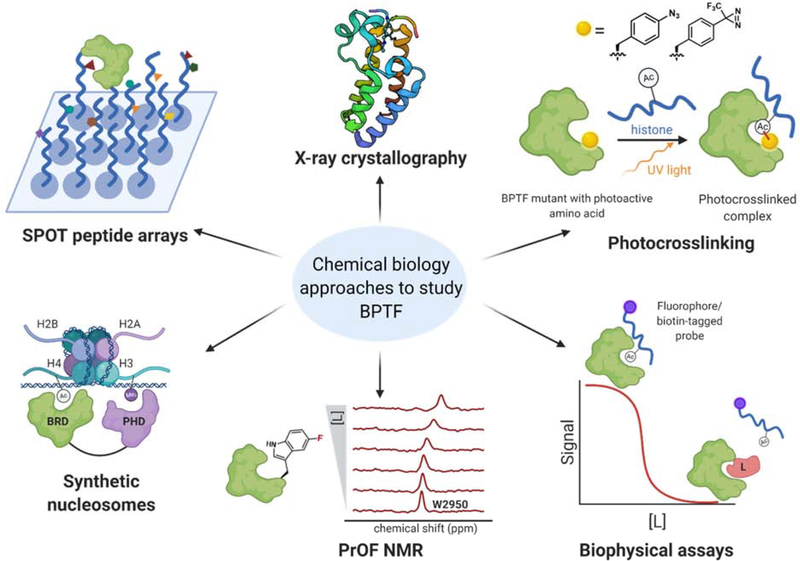
To enhance our understanding of BPTF function, a number of chemical biology approaches have been developed (Figure 3). In the context of deciphering native histone interactions, synthetic peptide arrays (SPOT blots) have been used with BPTF to panel diverse PTMs. This method involves synthesis of peptide arrays with PTMs on a cellulose membrane, followed by incubation with the protein of interest, and detection by western blotting or fluorescence imaging. Ruthenburg et al. used SPOT blotting to identify the BPTF bromodomain-H4 K16ac interaction [34]**. Filippakopolous et al. also used SPOT arrays to look at combinations of histone PTMs, and discovered that the BPTF bromodomain bound strongly with H3 pT3, K4ac, K9ac [58]. To profile the binding of multidomain proteins with modified histones, Mauser et al. developed a mixed peptide array screening tool [59]. An exhaustive set of modified histone peptides were mixed pairwise and incubated with a tandem PHD-bromodomain construct. This method confirmed the synergistic binding of BPTF with H4 K16ac and H3 K4me3. However, the weak affinity of histone peptides for bromodomains poses a challenge for SPOT array-based methods and highlights the need for orthogonal biophysical assays to validate these interactions.
Figure 3: Chemical biology tools developed to study BPTF interactions with peptides and small-molecule inhibitors.
Methods include SPOT peptide arrays, photocrosslinking, direct binding assays such as NMR and SPR, and competitive inhibition-based biophysical tools including AlphaScreen and fluorescence polarization. Synthetic nucleosomes provide a more biologically relevant platform and structural characterization has been enabled by X-ray crystallography (PDB 3QZT).
NMR is one of serval biophysical techniques employed for quantifying bromodomain and PHD domain-ligand interactions in vitro [28,60*]. To increase the speed of analysis, a protein-observed 19F NMR (PrOF NMR) method has been developed [61]. 19F-labelling of W2950 in the BPTF bromodomain binding site provides a reporter resonance which is responsive to changes in ligand binding. The change in chemical shift as a result of increasing ligand concentration is monitored to measure affinity. The fluorine substitution has been shown to induce minimal effects on the bromodomain structure and function [62*]. PrOF NMR is particularly suited for quantifying moderate-to-weak affinity binders of BPTF, such as acetylated histones [38,39*]. The resonances in these experiments are sufficiently resolved that two different proteins have been studied in the same NMR test tube [37**], as well as fluorinated multidomain proteins [63]. PrOF NMR may therefore be useful to study the tandem BPTF PHD-bromodomain interactions with native or synthetic ligands.
Affinity-based photocrosslinking has also been used to discover new binding partners for bromodomains [46]. Sudhamalla et al. used amber suppression to incorporate p-azido-L-phenylalanine (pAzF) in the binding site of bromodomains. This photosensitive amino acid can form a covalent bond with binding partners to capture transient interactions. Preliminary work with BPTF W2950AzF showed that the mutant underwent photocrosslinking with a tetraacetylated H4 peptide. This protein was subsequently used to validate a protein-protein interaction with acetylated H2A.Z [38]. Recent work optimized 4-(trifluoromethyldiazirinyl)-phenylalanine (tmdF) incorporation into BPTF and demonstrated that the W2950tmdF variant can crosslink with H4 proteins and endogenous histones [64*]. Further proteomic analysis has yet to be described.
While SPOT blotting, NMR, and photocrosslinking have played a key role in validating BPTF binding to PTMs, nucleosome-based approaches provide a more biologically relevant system to study protein-protein interactions. Ruthenburg et al. used synthetic mononucleosomes containing modified histones, prepared via expressed protein ligation [34**]. Using the tandem PHD-bromodomain, they found that the bivalent interaction conferred selectivity to the bromodomain binding with H4 K16ac which was not discernible through the SPOT arrays. To distinguish between intra- vs internucleosomal binding mechanisms, they constructed dinucleosomes using heteromeric DNA ligation of mononucleosomes. In this model, simultaneous engagement of histones across different nucleosomes was not observed, indicating that BPTF binds to chromatin via an intranucleosomal mode. To improve the throughput and sensitivity of nucleosome-based studies, Nguyen et al. designed DNA-barcoded nucleosome libraries [65]. This study validated previous findings of bivalent binding with H3 K4me3 and H4 K16ac and found that the previously unstudied pentaacetylated H4 Kac5 marks enhanced affinity by seven-fold. A high-throughput nucleosome remodeling assay profiled the nucleosome sliding activity of ISWI remodelers [66*]. This assay revealed that nucleosomes with H2A.Z have enhanced remodeling rates and that the nucleosome’s acidic patch is required for remodeling.
A number of other biophysical assays have been reported to study BPTF-ligand interactions such as isothermal calorimetry (ITC) [37**], surface plasmon resonance (SPR), AlphaScreen [68*], and fluorescence polarization [34**,69]. Further insight into the structural biology of the protein has been enabled by X-ray crystallography with histones and small-molecule inhibitors. Recently, Ycas et al. reported five of the first BPTF bromodomain-small molecule cocrystal structures, which provide key information for future structure-based drug design [68*]. BPTF-specific cell-based experiments such as a luciferase reporter assay [37**], ATAC-seq, and ChIP-seq [26] have also been optimized, providing a framework for studying the biological effects from BPTF inhibition.
Small Molecule Inhibitors of BPTF
Given the biological relevance of BPTF and its role in disease, there is a need to develop small-molecule inhibitors. The BPTF bromodomain remains the most extensively investigated. Vidler et al. used the computational program SiteMap to assess the druggability of human bromodomains [70]. BPTF was predicted to be highly druggable (Dscore 0.95 vs 0.93 for BRD4), along with other class I family members PCAF, GCN5L2, and CECR2. However, unlike BET bromodomains and other family I bromodomains, chemical probe development for BPTF is still in its infancy. Table 1 shows the small-molecule inhibitors reported for the BPTF bromodomain.
Table 1:
BPTF bromodomain inhibitors with in vitro affinity values and reported off-target effects. Functional groups acting as acetyl lysine mimics are shown in blue based on reported cocrystal structures.
| Inhibitor | BPTF in vitro affinity | Reported off-targets | References, PDB ID |
|---|---|---|---|
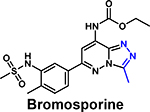
|
Kd = 1.8 μM (ITC) | pan-bromodomain inhibitor | [71] 5IGK (with BRD4) |
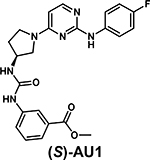
|
rac-AU1 Kd = 2.8 μM (ITC) | Kinases: TRKC Kd = 200 nM CDKL2 Kd = 260 nM (KINOMEscan) |
[26,37**,62*] |
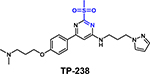
|
Kd = 120 nM (ITC) | CECR2 Kd = 10 nM (ITC) | [36,39*] 7KDZ |
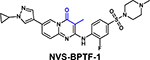
|
Kd = 3–71 nM (BROMOscan) | BRPF Kd = 37 nM CECR2 Kd = 66 nM GCN5L2 Kd = 62 nM PCAF Kd = 74 nM (BROMOscan) |
[72] |
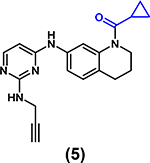
|
IC50 = 36 μM (AlphaScreen) | Not determined | [68*] 7KDW |
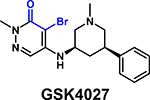
|
Kd = 1.7 μM (SPR) | PCAF/GCN5 Ki = 1.4 nM (BROMOscan) | [68*,76] 7K6R |
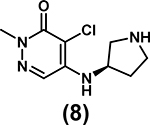
|
Kd = 6 μM (SPR) | Not determined | [68*] |
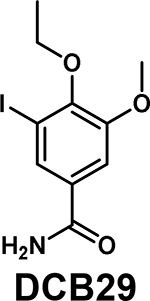
|
Kd = 17.9 μM (SPR) | Not reported | [73] |
|
Kd = 35.5 μM (Bio-layer Interferometry) | Not determined | [74] |
In the first report of a BPTF-small molecule ligand, bromosporine was identified as a pan-bromodomain inhibitor with nanomolar potency for BET bromodomains and a moderate affinity for BPTF (1.8 – 9 μM) [68*,71]. Subsequently, Urick et al. reported AU1 as a BPTF inhibitor (Kd = 2.8 μM), discovered from a library of 229 small molecules via a PrOF NMR screen simultaneously screening against fluorinated BPTF and BRD4 bromodomains [37**]. Although comparable in affinity to bromosporine, AU1 was the first selective inhibitor for BPTF over BET bromodomains. Using a BPTF-dependent luciferase reporter assay, AU1 was used to demonstrate the importance of bromodomain function on transcriptional activity. In Eph4 cells, AU1 treatment resulted in decreased proliferative capacity and G1 arrest of the cell cycle [26]. AU1 treatment also reduced c-Myc-DNA occupancy. Further SAR analysis led to the active (S)-enantiomer which was used in cell-based experiments, along with CRISPR/Cas9 BPTF depleted cells, to identify a BPTF-sensitive chronic myelogenous leukemia cell line, K562 [62*]. Despite the early use of AU1, limitations such as off-target kinase activity, low ligand efficiency, and stability, made AU1 a challenging inhibitor to develop and signified the need for new and more potent BPTF inhibitors.
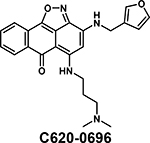
Several new inhibitors have been recently identified. TP-238 was reported as a CECR2/BPTF chemical probe by Takeda and the Structural Genomics Consortium (SGC), with 12-fold higher selectivity for CECR2 (Kd = 10 nM) [36]. TP-238 was used to validate the engagement of the BPTF bromodomain binding site by acetylated H2A.Z [39*]. Novartis and SGC reported NVS-BPTF-1 with in vitro binding affinity of 3–71 nM [72]. However, NVS-BPTF-1 is described as poorly soluble with inadequate ADME (absorption, distribution, metabolism, and excretion) properties for in vivo applications. Two moderate affinity BPTF binders, DCB29 [73] and C620–0696 [74] have also been reported (Kd = 17.9 and 35.5 μM, respectively). While an extensive analysis was not conducted, C620–0696 suppressed Myc protein levels in non-small-cell lung cancer cells.
Although most inhibitor development efforts are still in preliminary stages, they have enabled the cross-validation of a number of BPTF-specific biophysical methods. Ycas et al. used several of these compounds to optimize SPR and AlphaScreen assays and cross-validate PrOF NMR binding studies [68*]. A tetrahydroquinoline, discovered from a fragment screen [75], was investigated as a starting point for inhibitor development. Deconstructed fragments derived from GSK4027, a PCAF/GCN5 inhibitor [76], were also tested. Pyridazinone-based fragment 8 had a Kd of 6 μM and ligand efficiency of 0.45. The high ligand efficiency, compared to 0.22 for AU1, makes this scaffold a suitable lead for further inhibitor development.
Summary
New molecular mechanisms associated with BPTF function in human development and disease are emerging. Chemical biology has played a significant role in deciphering mechanisms through new tools and perturbing function through inhibitor development. While significant progress has been made for inhibiting the bromodomain as the most druggable domain, selectivity and ADME properties need to be improved to develop useful tool compounds for validating BPTF functional inhibition. In vivo studies using inhibitors have yet to be reported. One approach recently applied for class I bromodomains PCAF/GCN5, are proteolytically targeting chimeric molecules (PROTACs), which efficiently degrade these proteins [77*]. Importantly, PROTACs can recapitulate knock-down phenotypes in the absence of therapeutic effects from bromodomain inhibition. BPTF PROTACs are envisioned to be reported soon and may also lead to improved selectivity due to the formation of distinct ternary complexes. Relative to the bromodomain, the BPTF PHD domain remains a more challenging drug target. Despite the absence of inhibitors, mutagenesis of the PHD domain and use of a nonselective compound disulfiram have been used to support the role of the PHD domain of NUP98 fusions in transformed leukemia cells [60*]. More selective inhibitors would be a significant contribution to further elucidate the functional significance of the role of BPTF in oncogenic fusions and potential NURF-dependent and independent mechanisms. Given the increased number of reports associating BPTF in disease, and the growing number of BPTF small molecule inhibitors, we predict the next five years will produce exciting advances in our understanding of BPTF biology and new classes of BPTF chemical probes.
Supplementary Material
Acknowledgement
This work was supported by the National Institute of General Medical Sciences [R01GM121414-04], the National Institutes of Health Biotechnology Training Grant [5T32GM008347-23, N.M.O], and the University of Minnesota IEM Engineering in Medicine Doctoral Fellowship 2020 (H.Z) for financial support. We would also like to acknowledge BioRender, which was used to create all figures in this report.
Abbreviations
- ADME
absorption, distribution, metabolism, and excretion
- BET
bromodomain and extraterminal
- BPTF
bromodomain- and PHD finger-containing transcription factor
- BRD
bromodomain
- CECR2
cat eye syndrome chromosome region candidate 2
- DNL
DNA-barcoded nucleosome libraries
- FAC1
fetal Alzheimer’s clone 1
- ITC
isothermal calorimetry
- ISWI
imitation switch
- MAPK
mitogen-activated protein kinase
- MITF
microphthalmia-associated transcription factor
- NURF
nucleosome remodeling factor
- PHD
plant homeodomain
- PROTAC
proteolysis targeting chimera
- PTM
post translational modification
- pAzF
p-azido-L-phenylalanine
- SGC
Structural Genomics Consortium
- SNF2L
SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A, Member 1
- SPR
surface plasmon resonance
- tmdF
4-(trifluoromethyldiazirinyl)-phenylalanine
Footnotes
Declaration of Competing Interest
The authors declare that they have no competing interests influencing the work reported here.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A: An operational definition of epigenetics. Genes Dev 2009, 23:781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujimori DG, Conway SJ: Editorial overview: Chemical genetics and epigenetics. Curr Opin Chem Biol 2016, 33:vi–vii. [DOI] [PubMed] [Google Scholar]
- 3.Sbardella G: Methyl-Readers and Inhibitors. In Chemical Epigenetics. Edited by Mai A Springer International Publishing; 2019:339–399. [Google Scholar]
- 4.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. : Selective inhibition of BET bromodomains. Nature 2010, 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. : Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowser R, Giambrone A, Davies P: FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain. Dev Neurosci 1995, 17:20–37. [DOI] [PubMed] [Google Scholar]
- 7.Jordan-Sciutto KL, Dragich JM, Rhodes JL, Bowser R: Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator. J Biol Chem 1999, 274:35262–35268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan-Sciutto KL, Dragich JM, Caltagarone J, Hall DJ, Bowser R: Fetal Alz-50 clone 1 (FAC1) protein interacts with the myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry 2000, 39:3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones MH, Hamana N, Shimane M: Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics 2000, 63:35–39. [DOI] [PubMed] [Google Scholar]
- 10.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C: Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 2001, 8:531–543. [DOI] [PubMed] [Google Scholar]
- 11.Doerks T, Copley R, Bork P: DDT - A novel domain in different transcription and chromosome remodeling factors. Trends Biochem Sci 2001, 26:145–146. [DOI] [PubMed] [Google Scholar]
- 12.Roussy M, Bilodeau M, Jouan L, Tibout P, Laramée L, Lemyre E, Léveillé F, Tihy F, Cardin S, Sauvageau C, et al. : NUP98-BPTF gene fusion identified in primary refractory acute megakaryoblastic leukemia of infancy. Genes, Chromosom Cancer 2018, 57:311–319. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, et al. : The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017, 49:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, Knapp S: The bromodomain interaction module. FEBS Lett 2012, 586:2692–2704. [DOI] [PubMed] [Google Scholar]
- 15.Alkhatib SG, Landry JW: The nucleosome remodeling factor. FEBS Lett 2011, 585:3197–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R: Isolation of human NURF: A regulator of Engrailed gene expression. EMBO J 2003, 22:6089–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon SY, Xiao H, Wu C, Badenhorst P: Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet 2009, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwanbeck R, Xiao H, Wu C: Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem 2004, 279:39933–39941. [DOI] [PubMed] [Google Scholar]
- 19.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C: ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 1999, 97:833–842. [DOI] [PubMed] [Google Scholar]
- 20.Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C: Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell 1997, 1:141–150. [DOI] [PubMed] [Google Scholar]
- 21.Landry J, Sharov AA, Piao Y, Sharova L V., Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MSH, et al. : Essential role of chromatin remodeling protein bptf in early mouse embryos and embryonic stem cells. PLoS Genet 2008, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goller T, Vauti F, Ramasamy S, Arnold HH: Transcriptional regulator BPTF/FAC1 is essential for trophoblast differentiation during early mouse development. Mol Cell Biol 2008, 28:6819–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C: Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev 2011, 25:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu B, Wang Y, Wang C, Wang GG, Wu J, Wan YY: BPTF Is Essential for T Cell Homeostasis and Function. J Immunol 2016, 197:4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koludrovic D, Laurette P, Strub T, Keime C, Le Coz M, Coassolo S, Mengus G, Larue L, Davidson I: Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells. PLoS Genet 2015, 11:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frey WD, Chaudhry A, Slepicka PF, Ouellette AM, Kirberger SE, Pomerantz WCK, Hannon GJ, dos Santos CO: BPTF Maintains Chromatin Accessibility and the Self-Renewal Capacity of Mammary Gland Stem Cells. Stem Cell Reports 2017, 9:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. : A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442:86–90. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ: Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006, 442:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T: Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419:407–411. [DOI] [PubMed] [Google Scholar]
- 30.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T: Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 2004, 6:73–77. [DOI] [PubMed] [Google Scholar]
- 31.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K: High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129:823–837. [DOI] [PubMed] [Google Scholar]
- 32.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. : Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007, 39:311–318. [DOI] [PubMed] [Google Scholar]
- 33.Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, Musselman CA: The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. 2018, [DOI] [PMC free article] [PubMed]
- 34. Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. : Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 2011, 145:692–706. ** This study was notable as it showed for the first time a bivalent interaction with two epigenetic reader domains in the context of a doubly modified nucleosomes.
- 35.Kim K, Punj V, Choi J, Heo K, Kim JM, Laird PW, An W: Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenetics and Chromatin 2013, 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Structural Genomics Consortium. TP-238 A chemical probe for CECR2/BPTF bromodomains. https://www.thesgc.org/chemical-probes/TP-2382019,
- 37. Urick AK, Hawk LML, Cassel MK, Mishra NK, Liu S, Adhikari N, Zhang W, Dos Santos CO, Hall JL, Pomerantz WCK: Dual Screening of BPTF and Brd4 Using Protein-Observed Fluorine NMR Uncovers New Bromodomain Probe Molecules. ACS Chem Biol 2015, 10:2246–2256. ** Using a PrOF NMR dual screening method against a small molecule library, the authors uncovered the first small-molecule inhibitor selective for the bromodomain of BPTF over the first bromodomain of BRD4.
- 38.Perell GT, Mishra NK, Sudhamalla B, Ycas PD, Islam K, Pomerantz WCK: Specific Acetylation Patterns of H2A.Z Form Transient Interactions with the BPTF Bromodomain. Biochemistry 2017, 56:4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olson NM, Kroc S, Johnson JA, Zahid H, Ycas PD, Chan A, Kimbrough JR, Kalra P, Schönbrunn E, Pomerantz WCK: NMR Analyses of Acetylated H2A.Z Isoforms Identify Differential Binding Interactions with the Bromodomain of the NURF Nucleosome Remodeling Complex. Biochemistry 2020, 59:1871–1880. * The authors highlight a non-cannonical BPTF bromodomain interaction with a diacetylated motif on histone variant H2A.Z.
- 40.Li X, Wang S, Li Y, Deng C, Steiner LA, Xiao H, Wu C, Bungert J, Gallagher PG, Felsenfeld G, et al. : Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood 2011, 118:1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen J V., Hyman AA, Stunnenberg HG, et al. : Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142:967–980. [DOI] [PubMed] [Google Scholar]
- 42.Sun S, Zhong X, Wang C, Sun H, Wang S, Zhou T, Zou R, Lin L, Sun N, Sun G, et al. : BAP18 coactivates androgen receptor action and promotes prostate cancer progression. Nucleic Acids Res 2016, 44:8112–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, et al. : Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol 2012, 14:753–763. [DOI] [PubMed] [Google Scholar]
- 44.Qiu Z, Song C, Malakouti N, Murray D, Hariz A, Zimmerman M, Gygax D, Alhazmi A, Landry JW: Functional Interactions between NURF and Ctcf Regulate Gene Expression. Mol Cell Biol 2015, 35:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strachan GD, Morgan KL, Otis LL, Caltagarone J, Gittis A, Bowser R, Jordan-Sciutto KL: Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like ech-associated protein. Biochemistry 2004, 43:12113–12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudhamalla B, Dey D, Breski M, Nguyen T, Islam K: Site-specific azide-acetyllysine photochemistry on epigenetic readers for interactome profiling. Chem Sci 2017, 8:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buganim Y, Goldstein I, Lipson D, Milyavsky M, Polak-Charcon S, Mardoukh C, Solomon H, Kalo E, Madar S, Brosh R, et al. : A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PLoS One 2010, 5. * This work, which identified a translocation on chromosome arm 17q resulting in increased BPTF expression and cellular proliferation, was the first time BPTF was associated a malignant phenotype.
- 48.Bown N, Cotterill S, Łastowska M, O’Neill S, Pearson ADJ, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, et al. : Gain of Chromosome Arm 17q and Adverse Outcome in Patients with Neuroblastoma. N Engl J Med 1999, 340:1954–1961. [DOI] [PubMed] [Google Scholar]
- 49.Dai M, Lu J-J, Guo W, Yu W, Wang Q, Tang R, Tang Z, Xiao Y, Li Z, Sun W, et al. : BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget 2015, 6:33878–33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao S, Liu L, Fang M, Zhou X, Peng X, Long J, Lu X: BPTF Associated with EMT Indicates Negative Prognosis in Patients with Hepatocellular Carcinoma. Dig Dis Sci 2015, 60:910–918. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Kim MS, Yoo NJ, Lee SH: BPTF, a chromatin remodeling-related gene, exhibits frameshift mutations in gastric and colorectal cancers. Apmis 2016, 124:425–427. [DOI] [PubMed] [Google Scholar]
- 52. Dar AA, Nosrati M, Bezrookove V, De Semir D, Majid S, Thummala S, Sun V, Tong S, Leong SPL, Minor D, et al. : The role of BPTF in melanoma progression and in response to BRAF-targeted therapy. J Natl Cancer Inst 2015, 107:1–9. ** The authors validate the oncogenic role of BPTF in melanoma, identifying it as a novel target for anticancer therapies.
- 53.Dar AA, Majid S, Bezrookove V, Phan B, Ursu S, Nosrati M, De Semir D, Sagebiel RW, Miller JR, Debs R, et al. : BPTF transduces MITF-driven prosurvival signals in melanoma cells. Proc Natl Acad Sci U S A 2016, 113:6254–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Richart L, Carrillo-De Santa Pau E, Río-Machín A, De Andrés MP, Cigudosa JC, Lobo VJSA, Real FX: BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat Commun 2016, 7. * This report uncovers that BPTF interacts with c-Myc to drive transcription and tumor growth, providing insight into the molecular mechanisms of BPTF’s biological activity.
- 55.Green AL, Desisto J, Flannery P, Lemma R, Knox A, Lemieux M, Sanford B, Rourke RO, Ramkissoon S, Jones K, et al. : BPTF regulates growth of adult and pediatric high-grade glioma through the MYC pathway. Oncogene 2020, doi: 10.1038/s41388-019-1125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richart L, Real FX, Sanchez-Arevalo Lobo VJ: c-MYC partners with BPTF in human cancer. Mol Cell Oncol 2016, 3:e1152346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Y, Yuan F, Li Y, Wang G, Lin Z, Chen L: Bromodomain PHD-finger transcription factor promotes glioma progression and indicates poor prognosis. Oncol Rep 2019, 41:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. : Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mauser R, Kungulovski G, Meral D, Maisch D, Jeltsch A: Application of mixed peptide arrays to study combinatorial readout of chromatin modifications. Biochimie 2018, 146:14–19. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y, Guo Y, Gough SM, Zhang J, Vann KR, Li K, Cai L, Shi X, Aplan PD, Wang GG, et al. : Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion. Nat Commun 2020, 11:3339. * This is the first small-molecule inhibitor study of NUP98-BPTF fusion and its role in leukemogenesis. This work shows the importance of PHD-finger inhibition as a strategy for targeting acute myeloid leukemia.
- 61.Divakaran A, Kirberger SE, Pomerantz WCK: SAR by (Protein-Observed) 19 F NMR. Acc Chem Res 2019, 52:3407–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kirberger SE, Ycas PD, Johnson JA, Chen C, Ciccone MF, Woo RWL, Urick AK, Zahid H, Shi K, Aihara H, et al. : Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor. Org Biomol Chem 2019, 17:2020–2027. * In-cell experiments using the BPTF bromdomain inhibitor AU1 and CRISPR/Cas9 are described, identifying the chronic myelogenous leukemia K562 cell line to be sensitive to BPTF inhibition. This cell line has high Myc levels, making it is a useful model system for future studies.
- 63.Kalra P, McGraw L, Kimbrough JR, Pandey AK, Solberg J, Cui H, Divakaran A, John K, Hawkinson JE, Pomerantz WCK: Quantifying the Selectivity of Protein–Protein and Small Molecule Interactions with Fluorinated Tandem Bromodomain Reader Proteins. ACS Chem Biol 2020, doi: 10.1021/acschembio.0c00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wagner S, Sudhamalla B, Mannes P, Sappa S, Kavoosi S, Dey D, Wang S, Islam K: Engineering bromodomains with a photoactive amino acid by engaging ‘Privileged’ tRNA synthetases. Chem Commun 2020, 56:3641–3644. * An improved photocrosslinking method is described in this paper, which can be a useful chemical biology tool to discover new interacting partners of BPTF.
- 65.Nguyen UTT, Bittova L, Müller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW: Accelerated chromatin biochemistry using dnA-barcoded nucleosome libraries. Nat Methods 2014, 11:834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dann GP, Liszczak GP, Bagert JD, Müller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et al. : ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548:607–611. * In this work, the authors develop a DNA-barcoded library-based assay to measure remodelling rates of ISWI subunits. Their data highlights the significance of the nucelosome acidic patch in chromatin remodelling.
- 67.Clapier CR, Nightingale KP, Becker PB: A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res 2002, 30:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ycas PD, Zahid H, Chan A, Olson NM, Johnson JA, Talluri SK, Schonbrunn E, Pomerantz WCK: New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay development. Org Biomol Chem 2020, 18:5174–5182. * In this study, several of the first BPTF cocrystal structures in the primary literature, including with bromosporine, TP-238, GSK4027 and compound 5 are reported. This provides important structural information to address the key challenge of rational chemical probe design for BPTF.
- 69.Sudhamalla B, Dey D, Breski M, Islam K: Chemical Science epigenetic readers for interactome profiling. Chem Sci 2017, 8:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vidler LR, Brown N, Knapp S, Hoelder S: Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem 2012, 55:7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picaud S, Leonards K, Lambert J-P, Dovey O, Wells C, Fedorov O, Monteiro O, Fujisawa T, Wang C-Y, Lingard H, et al. : Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia. Sci Adv 2016, 2:e1600760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Structural Genomics Consortium. NVS-BPTF-1 A chemical probe for BPTF. https://www.thesgc.org/chemical-probes/NVS-BPTF-12019,
- 73.Zhang D, Han J, Lu W, Lian F, Wang J, Lu T, Tao H, Xiao S, Zhang F, Liu Y-C, et al. : Discovery of alkoxy benzamide derivatives as novel BPTF bromodomain inhibitors via structure-based virtual screening. Bioorg Chem 2019, 86:494–500. [DOI] [PubMed] [Google Scholar]
- 74.Xu J, Wang Q, Lai E, Leung H, Li Y, Fan X, Wu Q: Compound C620–0696, a new potent inhibitor targeting BPTF, the chromatin-remodeling factor in non-small-cell lung cancer. 2020, 14:60–67. [DOI] [PubMed] [Google Scholar]
- 75.Johnson JA, Nicolaou CA, Kirberger SE, Pandey AK, Hu H, Pomerantz WCK: Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains. ACS Med Chem Lett 2019, 10:1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humphreys PG, Bamborough P, Chung CW, Craggs PD, Gordon L, Grandi P, Hayhow TG, Hussain J, Jones KL, Lindon M, et al. : Discovery of a potent, cell penetrant, and selective p300/CBP-associated factor (PCAF)/general control nonderepressible 5 (GCN5) bromodomain chemical probe. J Med Chem 2017, 60:695–709. [DOI] [PubMed] [Google Scholar]
- 77. Bassi ZI, Fillmore MC, Miah AH, Chapman TD, Maller C, Roberts EJ, Davis LC, Lewis DE, Galwey NW, Waddington KE, et al. : Modulating PCAF/GCN5 immune cell function through a PROTAC approach. ACS Chem Biol 2018, doi: 10.1021/acschembio.8b00705. * This is the first report of a PCAF/GCN5 PROTAC, showing that the heterobifunctional targeting approach can be used to degrade family I bromodomains.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.