Graphical abstract

Keywords: DNA Replication, Flap endonuclease, DNA Polymerase δ, Single-molecule fluorescence, FRET, PIFE, Fluorescence enhancement, Fluorescence quenching
Abstract
Flap endonuclease 1 (FEN1) is an important component of the intricate molecular machinery for DNA replication and repair. FEN1 is a structure-specific 5′ nuclease that cleaves nascent single-stranded 5′ flaps during the maturation of Okazaki fragments. Here, we review our research primarily applying single-molecule fluorescence to resolve important mechanistic aspects of human FEN1 enzymatic reaction. The methodology presented in this review is aimed as a guide for tackling other biomolecular enzymatic reactions by fluorescence enhancement, quenching, and FRET and their combinations. Using these methods, we followed in real-time the structures of the substrate and product and 5′ flap cleavage during catalysis. We illustrate that FEN1 actively bends the substrate to verify its features and continues to mold it to induce a protein disorder-to-order transitioning that controls active site assembly. This mechanism suppresses off-target cleavage of non-cognate substrates and promotes their dissociation with an accuracy that was underestimated from bulk assays. We determined that product release in FEN1 after the 5′ flap release occurs in two steps; a brief binding to the bent nicked-product followed by longer binding to the unbent nicked-product before dissociation. Based on our cryo-electron microscopy structure of the human lagging strand replicase bound to FEN1, we propose how this two-step product release mechanism may regulate the final steps during the maturation of Okazaki fragments.
1. Introduction
The rectification of aberrant structures generated during DNA replication and repair is indispensable for maintaining genomic integrity. This crucial task is carried out by several families of structure-specific nucleases among which is the structure-specific 5′ nuclease family. The members of this family play an important role in the resolution of diverse range of aberrant DNA whose only common attribute is single-stranded/double-stranded (ss/ds) junctions: nicks, gaps, flaps, bubbles and four-way junctions. These toxic structures are highly common pathway intermediates in replication, repair, recombination, noncoding RNA removal and transcription termination in organisms spanning from bacteria to humans. How 5′ nucleases achieve their catalytic selectivitiy by relying mainly on the contours of their substrates is a thought-provoking question. Pertinent to this question is the observation that even minor variations in the substrate entice extreme catalytic variability. Thus, whereas sequence-based specificity explains part of the fidelity of DNA replication, key information is still missing about the mechanism controlling the precise accuracy by which the structure-based excision occurs.
5′ nucleases are highly conserved endo- and/or exo-nucleases that hydrolyze phosphodiester bonds situated 5′ end of ss/dsDNA junctions (Fig. 1A) [1], [2], [3], [4]. This unified site of cleavage of diverse DNA structures is mediated by sharply bending the DNA at the ss/dsDNA junctions to position the scissile phosphate near the metal ions of the active site (Fig. 1B). Furthermore, for some family members; catalysis has been shown to require changes in the protein conformation in order to assemble the active site [5], [6], [7], [8], [9], [10], [11], [12] and to position the scissile phosphate closer to the catalytic metal centers [6], [7], [10], [11], [12]. Different 5′ nuclease family members are proposed to utilize superfamily-conserved and unique structural features to interact with and interrogate the bent DNA conformer. Better understanding of the mechanism of 5′ nucleases would require the characterization of the conformational changes of both DNA and protein and the control mechanisms for the selection or the induced-fit that lead to an exquisite catalytic selectivity. Additionally, it is important to understand how the interactions of 5′ nucleases with partner proteins in various DNA repair pathways recruite the nuclease and influence its substrate recognition and cleavage.
Fig. 1.
The recognition and cleavage of the cognate substrate by FEN1. (A) Flap equilibration results from the complementarity of the downstream and upstream primers to the template generating an equilibrating double flap (DF) and single flap (SF). FEN1 recognizes the equilibrating DF substrate and incises the ss-dsDNA substrate one nucleotide into the junction thus generating a sealable nick. (B) The cap-helical gateway, where the active site and the 3′ flap binding pocket are located, is largely disordered before FEN1 binding to DNA (1UL1.pdb) [74], and becomes ordered upon DNA binding (5UM9.pdb) [67]. In the substrate complex, the DNA is bent at the junction by ∼100° angle and the 5′ flap is threaded into the cap-helical gateway.
Here, we present a timely review of the status of the field of single molecular investigations of an important 5′ nuclease namely flap endonuclease 1 (FEN1) using single-molecule Förster resonance energy transfer (SMFRET) and fluorescence modulations methods. Recently, this extensive single-molecule work together with previous ensemble kinetics and structural investigations elucidated the reaction pathway of FEN1 and the conformational dynamics of its associated DNA substrates. These key findings are instrumental for the understanding of the catalysis and regulation of FEN1 in many DNA transactions. Moreover, we review our recent cryo electron (Cryo-EM) structure of Pol δ–DNA–PCNA–FEN1 complex which has broad implications for lagging strand DNA synthesis. The present work offers a guideline to the use of single-molecule techniques to investigate similar molecular systems that can bend, modify and process DNA.
Besides, the Introduction and Conclusion sections, this review is organized according to the main checkpoints along the reaction pathway of FEN1 and the respective underlying experiments as follows:
1.1. Single molecule fluorescence techniques
Single-molecule fluorescence is becoming increasingly popular for investigating biomolecular reactions [13], [14], [15], [16]. Fluorescence detection can be performed under ambient conditions of aqueous media and room temperatures. The popularity of this technique increased by the improvement of the signal-to-background ratio due to the advent of photostable synthetic dyes such as Cy3 and Cy5, which can emit 105-106 photons before photobleaching [17]. Single-molecule techniques have several advantages including the revelation of population distributions masked by ensemble averaging, exploration of hidden heterogeneity and direct observation of dynamical state changes arising from photophysics and photochemistry, without the requirement for synchronization [18].
Based on the mode of sample illumination and emission collection, single-molecule fluorescence detection can be broadly divided into two main techniques; namely confocal and wide-field, that have been devised to minimize out-of-focus background from the surroundings [18]. Confocal microscopy applies pin-holes in the excitation and emission pathways to restrict the excitation volume below the diffraction-limit and perform point-by-point detection of the fluorescence bursts from the individual molecules diffusing in and out of the detection volume using photodiodes at high temporal resolution up to sub-millisecond [19], [20]. In wide-field microscopy, total-internal reflection (TIR) circumvents the background from solution by exciting the molecules at the surface [21], [22]. The temporal resolution is limited to few tens of milliseconds, however trajectories from hundreds of surface-immobilized molecules can be recorded per experiment [23], [24]. Recently, temporal resolutions on the order of milliseconds were achieved by applying stroboscopic excitation [25] and by using fast sCMOS sensors [26].
One of the widespread fluorescence detection methods in bioscience is SMFRET, which is a spectroscopic technique for measuring distances at the molecular level in the range of 1–10 nm [27]. SMFRET relies on the dipole–dipole coupling between two fluorophores with spectral overlap resulting in radiationless energy transfer between the two fluorophores [28]. The efficiency of energy transfer is directly proportional to the distance between the two fluorophores and may also be influenced by their orientation and the surrounding environment [29]. SMFRET is well-suited for real time recording of biomolecular reactions undergoing structural changes at the single-molecule level and determining the rates of the steps of biomolecular reactions [23]. However, SMFRET requires the choice of an optimized FRET pair, detectable FRET change, and stable acceptor.
Some fluorophores such as Cy3 and Cy5 are continuously interchanging between dim and bright states via cis–trans isomerization [30]. The proximity of proteins to such dyes results in distance-dependent enhancement of the fluorescence of the dye. The single-molecule method based on the protein-induced fluorescence enhancement is termed (SMPIFE) [31]. SMPIFE has the advantages of relying on a single fluorophore without the need for protein labeling and of being sensitive to distances within the range of 0–4 nm [32]. Both SMPIFE and SMFRET were employed to investigate the catalytic steps of archaeal FEN1 [33], as well as to detect the motion of helicases on RNA or DNA such as the translocation activity of human Retinoic acid inducible–gene I (RIG-I) on dsRNA [34] and DNA binding and translocation of E. coli Rep [35]. This combination was also used to study the filament formation dynamics of E. coli RecA on ssDNA [36], the repetitive looping of the 5′ ssDNA tail of the superfamily 1 bacterial helicase (PcrA) [37], the interactions between E. coli DNA polymerase I and carcinogenic DNA adducts [38] and the dynamics of the exonuclease proofreading activity in E. coli DNA polymerase III [39].
The foundation for the effect of proteins on isomerization and consequently dye fluorescence was primarily based on either steric hindrance [40] or contacts to specific residues of the protein with the dye with minimal role of the DNA-dye structure [41]. However, the fluorescence properties of DNA-coupled cyanine dyes can be influenced by several other variables including DNA sequence, position of the dye and DNA structure [42], [43]. In a recent study, we presented a new single-molecule technique based on protein-induced fluorescence quenching, which we refered to by SMPIFQ [44]. It is note-worthy to mention that none of the known fluorescence quenching mechanisms [45], [46], [47] can explain the PIFQ effect. The initial fluorescence state of the DNA–Dye complex can be influenced by several molecular interactions such as steric hindrance imposed by the DNA structure on the dye, electrostatic, hydrophobic, π-π stacking and hydrogen bonding [48], [49]. Using a comprehensive library of oligomers with various sequences, fluorophore positions and types, we illustrated that the DNA-dye structure dictates the initial state and fluorescence lifetime of the DNA-coupled Cyanine-dyes which in turn set the course for the fluorophore to experience either enhancement or quenching by protein interactions that disrupt the initial DNA-Dye structure. The relationship between the initial and final fluorescence states can provide a systematic experimental layout to obtain either PIFQ or PIFE as desired, which circumvents the present arbitrary nature for design and control in PIFE experiments [44].
1.2. FEN1 role in the maturation of Okazaki fragments
FEN1 plays a fundamental role in the maturation of Okazaki fragments. Synthesis of an Okazaki fragment is initiated by the addition of 8–12 nt RNA primer by the primase subunit of DNA polymerase alpha (Pol α) that is then extended by the Pol α-polymerase subunit into a 30–35 nucleotide RNA-DNA primer [50], [51], [52]. During the maturation of Okazaki fragments in lagging strand synthesis, short ssDNA or ssRNA 5′ flaps are generated by the limited strand displacement activity of the lagging strand replicase DNA polymerase delta (Pol δ) into the 5′ end of the previous Okazaki fragment [53], [54]. FEN1 recognizes and cleaves these flaps in joint with Pol δ by the coordination of proliferating cell nuclear antigen (PCNA) sliding clamp [55], [56], [57], [58], [59]. FEN1 hydrolyzes the phosphodiester bond one nucleotide into the 5′ end of ss/ds-DNA junctions (Fig. 1A) in order to generate a nick that can be subsequently ligated by DNA Ligase 1 (LIG1) [2], [60], [61], [62]. It is estimated that during the cell cycle of mammalian cells nearly 50 million Okazaki fragments need to be processed in order to generate contiguous lagging strands [63]. Moreover, studies showed correlation between FEN1 levels and tumor aggressiveness in cancerous cells in human [64] leading to the investigation of FEN1 as a potential therapeutic target in cancer [65], [66].
The substrate and product complexes of FEN1 provided detailed understanding of how FEN1 uses superfamily conserved and unique structural features to interact and recognize its substrate [6], [67], [68]. The 5′ flap substrate is formed of three strands, where the annealing of the 5′-flap and the 3′-flap strands to the template strand constitutes the down and upstream dsDNA regions, respectively (Fig. 1A). The main structural/functional features of FEN1 include the helical gateway topped by the helical cap for the selection of ss-5′-flap substrates, the hydrophobic wedge between the 3′ flap binding site and the cap-helical gateway, and the helix-2turn-helix (H2TH) motif that interacts with the downstream DNA (Fig. 1B). The binding sites for dsDNA on either side of the active site are spaced one helical turn apart thus imposing significant bending (∼100°) at the junction nick [6], [67]. DNA bending provides a scaffold that allows FEN1 to interact and recognize the various features of the junctions – fully paired nick junction, one nucleotide 3′ flap and 5′ flap – and discriminates against continuous dsDNA since FEN1 can’t bend dsDNA sharply. The 5′ nucleases conserved helical gateway in FEN1 has a unique helical cap that forms a narrow tunnel through which only ss-5′-flap with free end can thread [6], [69], [70], [71], [72] (Fig. 1B). The 5′ flap threading is required to guide the scissile phosphate into the active site. In the absence of DNA, the 3′ flap binding pocket and the cap-helical gateway that include catalytically-indispensable residues are largely disordered [67], [68]. However, on 3′ flap binding the cap-helical gateway and the 3′ flap binding pocket undergo a disorder-to-order transition thus proposing DNA-induced ordering of the cap-helical gateway as a mechanism that couples 3′ flap recognition and positioning of the nick junction and the 5′ flap with assembly of the active site and licensing of cleavage [6], [73], [74] (Fig. 1B).
Several single-molecule fluorescence studies from our group have been a cornerstone in connecting the pieces of information from static X-ray crystal structures and bulk methods to provide an unprecedented level of detail to the kinetics and mechanism of FEN1 reaction [44], [72], [75]. Recently, we solved the first structure of the human Pol δ–DNA–PCNA–FEN1 complex thus revealing various mechanistic aspects of the maturation of Okazaki fragments [76]. Collectively, these studies unraveled the underlying kinetic schemes for DNA bending, protein disorder-to-order transition, active-site assembly, and 5′ flap incision. In this review, we discuss the application of complementary single-molecule fluorescence techniques: PIFE, PIFQ and FRET to investigate an entire biomolecular reaction from the instance of encountering the substrate by the enzyme until the final step of product release exemplified by FEN1 on the ss-dsDNA junction substrate. In addition, we present the interactions between individual components of the Pol δ–DNA–PCNA–FEN1 complex obtained by Cryo-EM microscopy. The methodology presented in this review is aimed as a guide for tackling other biomolecular enzymatic by PIFE, PIFQ and FRET and their combinations.
2. Monitoring DNA bending and 5′ flap cleavage
2.1. Substrate labeling and experimental conditions
The double flap (DF) substrate discussed throughout this review has 6 nucleotide 5′ flap and a single nucleotide 3′ flap and is denoted as DF-6,1. The DF-6,1 substrate is classified according to the complementarity of the 3′ flap to the template into “equilibrated” (EQ DF-6,1), where the 3’ flap is complementary to the template (Fig. 1A), and “non-equilibrated” (NonEQ DF-6,1) with no complementarity. For the detection of the conformational changes by SMFRET, we select either flap- or internal-labeling scheme for these substrates. The respective labeling scheme will be denoted as a subscript after the name of the substrate. Both substrates have a biotin moiety for the immobilization via biotin/neutravidin linkage onto the surface of a functionalized glass coverslip. The imaging is performed on a custom-built objective-based TIRF setup [77] in buffer containing either CaCl2 or MgCl2 for DNA bending only or bending and 5’ flap cleavage, respectively [75].
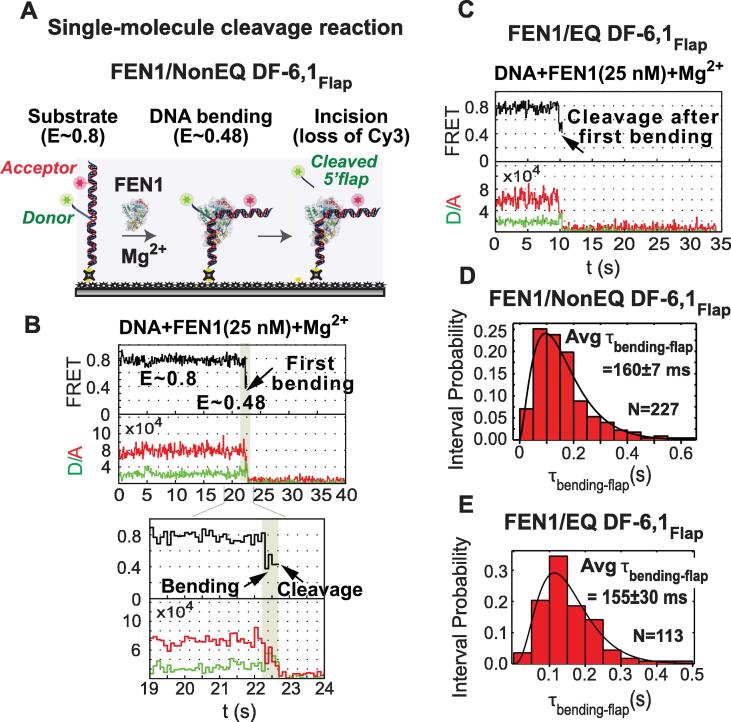
In the flap-labeling scheme (NonEQ DF-6,1Flap), the donor (Cy3) is attached via phosphoramidite linkage to the 5′ end of the 5′ flap and the acceptor (Alexa Fluor 647) is linked to a base upstream of the nick junction in the template strand (Fig. 2A) [71]. This scheme reports on DNA bending and 5′ flap threading in the presence of Ca2+. In the presence of Mg2+, it will additionally report on 5′ flap release indicated by the disappearance of the donor signal upon cleavage. However, the flap-labeling scheme does not provide any further information about FEN1 interaction with the nicked DNA product. The FRET efficiency time traces show that the substrate prior to FEN1 binding is in a single high FRET conformer. DNA bending and 5′ flap threading are manifested by the increase in distance between the 5′ flap and the upstream duplex leading to decrease in FRET efficiency (Fig. 2A, lower panel). The anti-correlated intensity fluctuations of the donor and acceptor emission signals in the SMFRET traces arise from the distance-dependent energy transfer from donor to acceptor. In the presence of Ca2+, FEN1 forms a stable complex with the bent DNA conformer.
Fig. 2.
DNA bending by FEN1 in the presence of Ca2+ for non-equilibrating (NonEQ) and equilibrating (EQ) DF substrates. (A) Flap-labeling scheme, the donor is positioned at the 5′ flap end and the acceptor is at the upstream of the nick. FEN1 bends the DNA thus increases the distance between the donor and acceptor and consequently decreases FRET (lower panel). (B) Internal labeling-scheme, the donor and acceptor are at the downstream and upstream of the nick, respectively. Upon DNA bending by FEN1, the distance between the two fluorophores decreases resulting in the increase of FRET (lower panel). The substrates used in A and B are NonEQ DF, where FEN1 forms a stable bent DNA complex. (C) Representative time trace of FEN1 bending EQ DF shows the formation of less stable bent DNA complex. The dwell times τbending and τunbending of the bent and unbent states are marked by purple and blue, respectively. (D) The histograms of the dwell times in the unbent and bent states were fitted to single-exponential decay functions to generate kbending (1/τunbending) and kunbending (1/τbending), respectively. The association rate constant (kon-bending) is calculated from the slope of the linear fit of kbending versus FEN1 concentration. The dissociation rate constant (koff-unbending) is the intercept of the y-axis of the linear fit to kunbending. Kd = koff-unbending/kon-bending. The error bars of kbending and kunbending represent the standard error of the exponential fit. The errors of the association and dissociation constants are the standard errors of the linear fit. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In the internal-labeling scheme (NonEQ DF-6,1Internal), both the donor (Cy3 replacing a base) and the acceptor (Alexa Fluor 647) linked to a base are in the template strand and positioned at the downstream and upstream of the nick, respectively (Fig. 2B). The internal-labeling scheme reports on DNA bending in the presence of Ca2+. It can also monitor the steps subsequent to 5′ flap release such as the binding to the nicked duplex product in the presence of Mg2+. In this scheme, DNA bending results in the increase of FRET due to the decrease in distance between the donor and acceptor (Fig. 2B, lower panel). The FRET efficiency time traces also show that the substrate prior to FEN1 binding is in a single conformation and forms a stable bent substrate complex with FEN1.
2.2. FEN1 actively bends the DNA with near diffusion-limited kinetics
Using SMFRET, we show that the substrate alone exists as a single FRET conformer and that FEN1 recognizes and actively bends the substrate into the distorted bent conformation (Fig. 2A and 2B) [72]. There is an ongoing debate of whether upon DNA damage recognition, the conformational distortion occurs via protein capturing of a particular conformer (conformational selection) or protein active molding of DNA into a distorted conformation. Throughout our experiments, we only observed a single conformer for the DF substrate. Also, we did not detect any other conformer even by increasing the temporal resolution to sub-millisecond by applying confocal microscopy on freely-diffusing substrate. Applying burst variance anaylsis (BVA) and photon-by-photon hidden Markov modeling (H2MM) methods, we only observed a single conformer [78]. On the other hand, we tried slowing down conformational changes in DNA by increasing divalent ion concentration and obtained the same outcome. Our molecular dynamics (MD) simulations showed that the most energetically favorable conformer of DF-6,1 is the extended form (∼165°). A steep energy barrier of ∼ 14 kcal/mole is required to break the base stacking in the extended conformer and bend the DNA [72]. There is a possibility that conformational transitions in DNA may exist at the nano or picosecond regime, which is far beyond the realm of the currently available temporal resolution for single-molecule detection.
Active DNA distortion has been also reported in other structure-specific DNA repair enzymes. Both BVA and photon-by-photon (H2MM) methods [78], and free MD simulations showed that the nick substrate behaves similarly to flap substrates and is actively bent by another member of the 5′ nucleases super-family, the human mismatch repair exonuclease 1 (EXO1) [72]. The human xeroderma pigmentosum group G (XPG) protein which is essential for efficient processing of unpaired (bubble) regions in nucleotide excision repair [79], [80], unstacks, and sculpts duplex DNA of bubble substrates into strongly bent structures [81]. In addition, human apurinic/apyrimidinic endonuclease 1 (APE1) which is critical for repairing oxidative DNA lesions in the base excision repair pathway [82], [83], performs DNA sculpting mechanism and protein-imposed structural adjustments of the DNA substrate to recognize and accommodate several diverse DNA substrates into its active site [84].
Other structure-specific DNA repair enzymes exploit conformational dynamics in their substrate recognition mechanisms. Human Holliday junction 5′ flap endonuclease (GEN1), a member of the 5′ nucleases super-family, was shown to capture one of the interchanging Holliday junction (HJ) conformers followed by active distortion and resolution of the HJ [85]. Similar mechanism was reported for other HJ resolvases, where conformational selection still continued during the initial enzyme binding [86]. DNA binding proteins such as Rad4/XPC in human exploits the conformational dynamics of their substrates for DNA mismatch recognition and binding [87]. The E. coli DNA polymerase I (Pol I) recognizes and binds to the gapped DNA substrate and polymerizes across the gap [88]. It was reported that gapped DNA substrate undergoes rapid interconversions between stacked and unstacked states, where the unstacked conformations exhibit increased fraying of 1–2 nt around the gap and adopt a more bent structure. The weaker base stacking in gapped structures increases the importance of backbone electrostatic repulsion and shielding from cations resulting in an average increase in bending around the gap. Thus, the bending of gapped structures could be interpreted by either the increased flexibility of unstacked structures or the transient unstacking events, or a combination of both [78]. This finding led to the proposal of a two-step mechanism for substrate-recognition by structure-specific DNA binding enzymes as Pol I, where the substrate is bound in an initial conformational selection step detecting the increased flexibility of the DNA followed by an ‘on-protein’ conformational search, in both the protein and DNA [89].
However, addressing the relevance of conformational sampling versus active distortion requires consideration of the higher order chromatin structure and the functioning of these nucleases within the context of other components in the biological reaction. For example, the recent structure of Pol δ bound to DNA and its processivity clamp PCNA showed that Pol δ interacts with the 5′ ssDNA template exiting the active site and bends it by 90° [76]. Furthermore, FEN1 binds PCNA right across the template strand [76] suggesting that it might be actively picking up a bent DNA from Pol δ.
Interestingly, the time traces show transitioning in the equilibrated junction (EQ DF-6,1Internal) in contrast to the non-equilibrated junction (Fig. 2C) [72], indicating the reduced stability of the bent conformer suggesting the dissociation of the 3′ flap from the 3′ flap-binding pocket and the 3′ flap pairing to the template strand before FEN1 can rebind to it. Nonetheless, both substrates are cleaved with equal activities as discussed below. The transitioning in the case of equilibrated junction allowed us to calculate the DNA bending association (kon-bending) and dissociation (koff-unbending) rate constants (Fig. 2D) [72]. The results show that FEN1 bends the DNA with near diffusion limited kinetics.
2.3. FEN1 always cleaves the 5′ flap after DNA bending
Replacing Ca2+ with the catalytically active Mg2+ in the flap labeling scheme allows for monitoring the reaction up to 5′ flap release (Fig. 3A) [72]. The Cy3 donor is generally stable for several minutes under our experimental conditions. In order to confirm that the loss of donor particles is due to 5′ flap cleavage and not due to donor photobleaching, we quantified the donor loss in the presence of Mg2+ in comparison to Ca2+. We found that in the case of Mg2+, the donor loss was much higher than that in Ca2+ and coincided with the entry of FEN1 into the flow cell. Also, the donor loss in presence of Mg2+ unlike Ca2+ was preceded by DNA bending confirmed by clear anti-correlation between the donor and acceptor intensities [72]. The FRET time trace of NonEQ DF-6,1Flap shows short DNA bending (E ≃ 0.48) dwell time, as indicated by the anti-correlated donor and acceptor intensities, prior to the incision concluded by a single-step loss of signal from both donor and acceptor due to the 5′ flap release (Fig. 3B) [72]. The change in fluorescence upon FEN1 binding is significantly distinguishable from noise and can be reliably assigned to either FRET or PIFQ, as described below [44]. Notably, every DNA bending event results in a successful cleavage reaction producing 5′ flap ssDNA and nicked dsDNA as observed also in EQ DF-6,1Flap (Fig. 3C) [72]. The probability density plot of the distributions of the dwell time before cleavage (τbending-Flap) of NonEQ and EQ DF-6,1 showed that both substrates are cleaved with similar kinetics (Fig. 3D & 3E) [72]; this is consistent with findings from bulk cleavage assays [75], [90], [91]. Hence, the 5′ flap release is instantaneous [55], [72], [91], the single turnover rate for catalysis (kSTO) can be determined from the inverse of the average dwell time prior to cleavage (kSTO = 1/Avg τbending-Flap). This kSTO includes DNA bending, protein ordering for active site assembly, chemistry and 5′ flap release. The kSTO obtained from SMFRET cleavage was slightly slower than that from bulk assays due to the lower reaction temperature in the single-molecule assays [72]. Both the diffusion-limited association rate and the 5′ flap incision from the first bending event demonstrate that FEN1 cleavage for the cognate substrate proceeds with diffusion-limited kinetics [72]. This is consistent with the reported diffusion-limited cleavage kinetics from bulk cleavage assays [55], [91].
Fig. 3.
FEN1 cleaves cognate substrate on the first encounter. (A) Schematic depicting DNA bending followed by the incision of the 5′ flap by FEN1 in the single-molecule experiment. (B) Representative time trace (upper panel) shows DNA bending and loss of donor upon the first encounter of FEN1 with NonEQ DF-6,1Flap. The zoomed-in view (lower panel) emphasizes the dwell time of the bent state before cleavage (τbending-Flap). (C) Representative time trace of the cleavage of EQ DF-6,1Flap from the first instance of bending. (D) The NonEQ DF-6,1Flap dwell times (τbending-Flap) of N events are fitted to a gamma distribution. The average dwell time (Avg τbending-Flap) is used to determine the single turnover catalytic constant kSTO as 1/Avg τbending-Flap. (E) The dwell time distribution of EQ DF-6,1Flap. The uncertainties in Avg τbending-Flap correspond to the standard error of the mean.
3. Defining features in the enzymatic reaction of FEN1
3.1. 5′ flap threading occurs after DNA bending
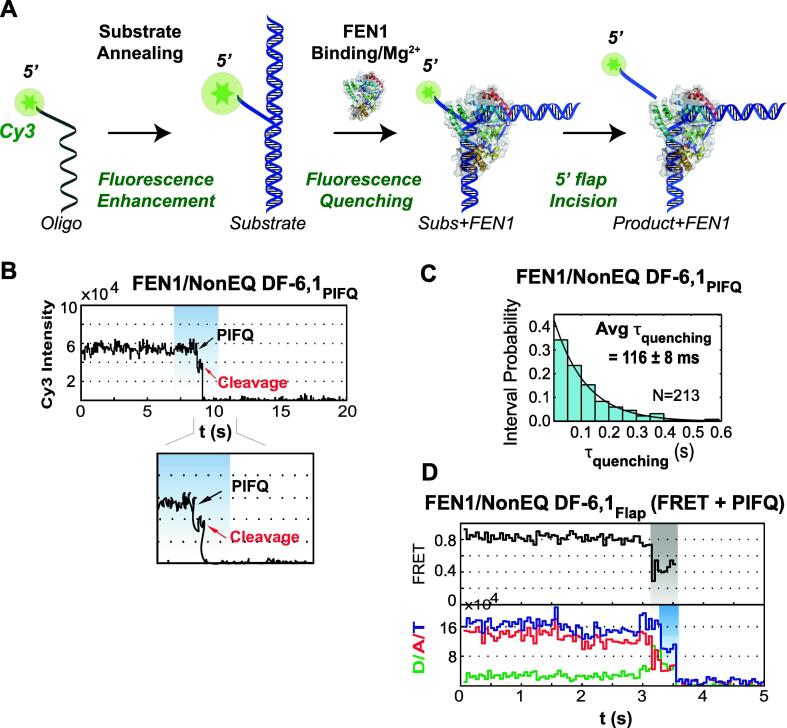
The timing of the 5′ flap threading relative to DNA bending has been under intensive investigation biochemically [55], [69], [70], [92], [93], [94] and structurally [6], [67], [68], [73], [74]. We pinned down the timing of 5′ flap threading through a series of SMPIFQ and SMFRET experiments. The hybridization of the 5′ flap oligo labeled at the 5′ end with Cy3 via phosphoramidite linkage (pCy3) with the template and upstream primers results in fluorescence enhancement [44]. The binding of FEN1 to the substrate quenches the fluorescence of pCy3 back to the level of the 5′ flap oligo prior to hybridization. Therefore, the PIFQ effect is preceded by nucleic-acid induced fluorescence enhancement while FEN1 binding modulates the fluorescence of pCy3 in the DF substrate. Therefore, in the presence of Mg2+, SMPIFQ would detect the quenching of pCy3 fluorescence followed by 5′ flap release (Fig. 4A) [44]. The SMPIFQ cleavage experiment was performed on non-equilibrating DF-6,1 labeled with pCy3 dye at the 5′ flap, termed NonEQ DF-6,1PIFQ [44]. The time trace showed quenching of pCy3 fluorescence upon FEN1 binding followed by complete loss upon the incision of the 5′ flap similar to the change observed in SMFRET (Fig. 4B) [44]. The average dwell time (τquenching) in SMPIFQ was ∼ 40 ms shorter when compared to that from SMFRET and its distribution exhibited a single-exponential decay indicating a single rate-limiting step in contrast to the rise and decay in the case of SMFRET (Fig. 4C cf. 3D) [44]. Also, it is worth-mentioning that the start points of FRET and PIFQ are distinguishable in the SMFRET experiment by following the change in FRET and the total fluorescence intensity of both the donor and acceptor, respectively (Fig. 4D) [44]. Two discrete features in the total fluorescence can be observed after the onset of the low FRET state in the FRET time trace (shaded-gray); the first is a constant total fluorescence intensity and the second is a drop in the total intensity resulting from donor quenching (shaded-blue) just before cleavage (Fig. 4D). The incidence of FRET change before PIFQ supports our conclusion that PIFQ reports on a step involving 5′ flap threading after DNA bending. This can be explained by the difference in the beginning of change between SMFRET and SMPIFQ, where in SMFRET the change starts on DNA bending and continues through the 5′ flap threading while in SMPIFQ, FEN1 induces the quenching of Cy3 fluorescence once the flap has been fully threaded and positioned into the active site. Our results suggest that FEN1 bends the DNA and requires an extra 40 ms before threading the 5′ flap. This result demonstrates that SMPIFQ can provide important mechanistic information on the 5′ flap threading in relation to DNA bending during cleavage thus complementing SMFRET.
Fig. 4.
5′ Flap threading occurs after DNA bending. (A) Schematic of the principle behind SMPIFQ experiment. Fluorescence enhancement by annealing the 5′ flap oligo labeled at 5′ end with Cy3 via phosphoramidite linkage (pCy3). The substrate is termed DF-6,1PIFQ. Fluorescence quenching upon FEN1 binding to the substrate is followed by 5′ flap release. (B) Representative time trace (top panel) showing the quenching of fluorescence of pCy3 followed by loss of signal upon interaction with FEN1. The zoom-in-view (lower panel) shows the dwell time of PIFQ before signal loss, termed (τquenching). (C) The probability distribution of τquenching is fitted to a single-exponential decay. The average dwell time (Avg τquenching) is the mean of the distribution. The uncertainty corresponds to the standard error of the mean. (D) Representative time trace shows both FRET (shaded-gray) and PIFQ (shaded-blue) demonstrated by the total fluorescence intensity (blue) during SMFRET cleavage, where pCy3 quenching before donor loss due to PIFQ results in the decrease of the total intensity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, the conformation and/or positioning of the bent DNA differ before and after the threading of 5′ flap. FRET of the bent DF-30,1Internal under conditions where 5′ flap threading is blocked was slightly, but consistently, different from that when the 5′ flap is threaded. This suggests that the substrate might not be fully distorted and/or properly positioned before 5′ flap threading. Also, the blocking of the 5′ flap threading markedly increases the dissociation rate constant of the bent DNA conformer (koff-unbending = 50 ± 0.02 s−1) [72]. These findings are consistent with the crystal structures of pre- and post-threaded FEN1:DNA complexes which showed that despite FEN1 inducing sharp DNA bending, the downstream DNA was far from the active site in the pre-threaded structure representing the FEN1–DNA complex before 5′ flap binding compared to that of the threaded structure, where the 5′ flap is threaded through the cap-helical gateway [68].
3.2. 3′ flap-induced protein ordering after DNA bending
The binding of the 3′ flap in the equilibrated DF substrate was proposed to drive protein ordering in the cap-helical gateway and the 3′ flap binding pocket [6], [73], [74]. Although DNA bending by FEN1 occurs at diffusion-limited rate, the conformational changes associated with the positioning of the 5′ flap scissile phosphodiester bond into the active site of FEN1 are essential and rate-limiting for the 5′ flap cleavage [91]. In vivo, the native equilibrated junction may exist either as a single 5′ flap which requires active modeling by FEN1 into a double 5′- and 3′-flap or as a DF with a readily accessible 3′ flap for FEN1 to bind. In order to study the effect of the 3′ flap on DNA bending, we made a single flap substrate termed SF-6,0 by removing the single unpaired 3′ flap in NonEQ DF-6,1. This substrate showed 34 fold decrease in FEN1 cleavage activity [55].
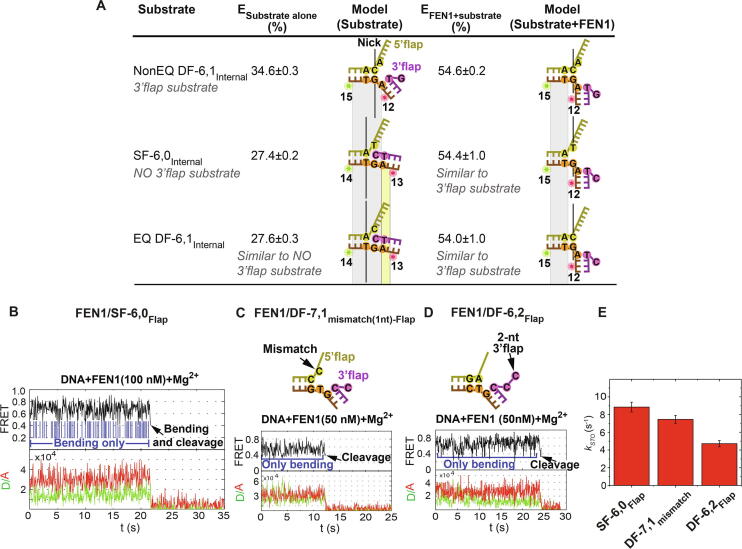
The next query was whether the equilibrated junction exists as a SF or DF. Using an internal-labeling scheme, we found the FRET states of EQ DF-6,1Internal and SF-6,0Internal to be similar (E ≃ 0.27) and less than that of NonEQ DF-6,1Internal (E ≃ 0.34) (Fig. 5A) [72]. This suggests that EQ DF exists as a SF with slightly less extended geometry than that of dsDNA (E ≃ 0.23). MD simulations also showed that EQ SF did not equilibrate to a DF [72]. However, FEN1 was reported to cleave both EQ and NonEQ DF substrates with equal activity [90] (Fig. 3D and 3E). This led us to the proposition that FEN1 actively generates a 3′ flap at the nick junction of the cognate EQ DF substrate. Therefore, we investigated the formation of the 3′ flap in EQ DF and SF junctions by measuring the FRET states of various nick-junction positions in NonEQ DF-6,1, EQ DF-6,1 and SF-6,0 substrates and also in presence of FEN1 [72].
Fig. 5.
3′ Flap-induced protein ordering after DNA bending. (A) FEN1 actively generates one nt 3′ flap thus inducing protein ordering. Comparison of the FRET states of internal-labeled substrates represented as percentages. Both EQ DF-6,1Internal and SF-6,0Internal have similar FRET which is less than that of NonEQ DF-6,1 suggesting that EQ DF exists as SF. The position of the nick relative to the donor and acceptor is shifted by one base pair compared to that in NonEQ DF-6,1Internal since EQ DF-6,1Internal substrate exists in a SF form. The bent conformers in EQ DF-6,1Internal and SF-6,0Internal exhibit the same FRET as that in NonEQ DF-6,1Internal indicating that the nick junction position must have shifted by one base pair in EQ DF-6,1Internal and SF-6,0Internal. Consequently, FEN1 can actively create a 3′ flap at the nick junction. (B,C,D) Cleavage accuracy of FEN1 upon encountering non-cognate substrates. SMFRET traces of the cleavage of FEN1 to SF-6,0Flap, FEN1 on DF-7,1mismatch(1nt)-Flap (a substrate that has one nucleotide gap instead of nick junction) and FEN1 on DF-6,2Flap (a substrate with 6 nt 5′ flap and 2 nt 3′ flap), respectively, showing multiple abortive DNA bending cycles before a successful cleavage event. (E) Bar chart of kSTO for the three non-cognate substrates in B-D.
The position of the nick relative to the donor and acceptor is shifted by one base pair compared to that in NonEQ DF-6,1Internal as EQ DF-6,1Internal substrate exists in a SF form (Fig. 5A). The bent conformers in EQ DF-6,1Internal and SF-6,0Internal exhibit the same FRET (E ≃ 0.54) as that in NonEQ DF-6,1Internal indicating that the nick junction must have shifted by one base pair in EQ DF-6,1Internal and SF-6,0Internal upon the formation of 3′ flap by FEN1. Consequently, FEN1 can actively create a 3′ flap in the non-cognate SF-6,0 substrate. This mechanism would explain the 1 nt shift of the cleavage site in SF versus DF substrates [55], [95].
The blocking of the 5′ flap threading via biotin/neutravidin impaired DNA bending to some extent and decreased the binding stability to a level comparable to that of SF-6,0. Thus, the ability of the 5′ flap to thread into the cap-helical gateway is required for the 3′ flap-induced protein ordering to form the stably and properly bent DNA conformer necessary for catalysis [69], [70], [71], [72]. Therefore, we deduce that FEN1 verifies the structural features of its cognate substrate by active DNA bending, 5′ flap threading and protein ordering through active formation of the 3′ flap. Drawbacks in any of these steps promote the dissociation of FEN1 from the non-cognate substrates through the decrease in the binding stability [72]. It is likely that the binding and selection of FEN1-substrate is a multistep process which starts by initial weakly bound bent DNA and progresses towards tight recognition intermediates [72], [91]. The majority of interactions in the initial phase may take place far from the active site and progress towards the engagement with the cleavage site in the DNA, thus enabling specific contacts between the active site and the scissile bond that license catalysis and reduce the conformational entropy. A similar mechanism was elegantly demonstrated by the time-resolved trapping of the reaction intermediates of human EXO1 thus revealing a sequence of successive interlocking conformational changes which guide the substrate into a metal-mediated inline-hydrolysis mechanism that catalyzes the cleavage [11].
3.3. Cleavage accuracy and missed-cleavage opportunities
SMFRET cleavage assays provide a direct measurement of the accuracy of FEN1 since they can unambiguously differentiate between DNA bending events that lead to catalysis versus those that don’t. Furthermore, by accessing the lifetime of the bent conformer without cleavage and kSTO of the cleavage events, key information on the mechanism of assembly of catalytically competent active site can be provided. Traces of single-molecule cleavage showed that non-cognate SF-6,0Flap underwent multiple cycles of DNA bending and unbending before a successful DNA bending event led to 5′ flap cleavage by chance (Fig. 5B) [72]. Although, kon-bending for SF-6,0 remained diffusion-limited similar to that of EQ DF-6,1, koff-unbending increased significantly to 23.3 ± 3.8 s−1, which is 7-fold faster than kSTO. Therefore, the bent conformer in SF-6,0 is destabilized to rates which are limiting for catalysis thus reducing the probability of assembling catalytically competent active sites [72]. We next cleaved two non-cognate substrates where koff-unbending is not rate-limiting for catalysis; a substrate that has one nucleotide gap instead of nick junction (DF-7,1mismatch(1nt)-Flap) (Fig. 5C) and DF with 6 nt 5′ flap and 2 nt 3′ flap (DF-6,2Flap) (Fig. 5D) [72]. Both substrates exhibited koff-unbending that was ∼ 13–15 fold slower than that of SF-6,0 and ∼ 3–4 fold longer than kSTO of the cognate substrate. Although, the lifetimes of the bent states were long enough to support catalysis in both non-cognate substrates (DF-7,1mismatch(1nt)-Flap and DF-6,2Flap), cleavage was not successful as evident from the multiple transitions. These transitions demonstrate that FEN1 bends both cognate and non-cognate substrates but selectively stabilizes the bent DNA intermediate and promotes assembly of catalytically-competent active site in the cognate substrates, while it destabilizes the bent DNA intermediate and reduces the probability of assembly of catalytically-competent active sites in non-cognate substrates.
The inability to detect abortive intermediary bent states in bulk assays leads to undermining of FEN1 accuracy and verification of the features of its cognate substrate in order to inhibit off-target cleavage. Also, bulk assays do not take into account the prior unsuccessful attempts in the cleavage of the non-cognate substrate resulting into lower estimation of the kSTO rate compared to the actual cleavage rate of the scissile phosphate bond. Finally, our results suggest a much higher accuracy of FEN1 inside the cell since it is less likely to access the same non-cognate substrate with high frequency.
Regardless of whether the lifetime of the bent conformer is limiting or not, the kSTO rates of FEN1 for the tested non-cognate substrates (Fig. 5E) [72] were similar and comparable to that of the cognate substrate. These results support that FEN1 likely has intrinsic mechanisms that block the probable formation of catalytically competent active sites with non-cognate substrates to inhibit off-target incision. This supports a scenario where the 3′ flap-induced protein ordering could act once per DNA bending event, thus locking the DNA into either catalytically-competent to be incised or -incompetent conformations that lead to DNA release from the bent conformation. Interestingly, a recent NMR spectroscopy study suggested that FEN1-DNA complex shows evidence of millisecond timescale motions in the arch region that may be required for DNA to enter the active site. Therefore, FEN1 local conformational flexibility which spans a range of dynamic timescales is crucial to reach the catalytically relevant ensemble [96]. It is possible that the initial binding of non-cognate substrates is positioned in a way that traps those substrates in a state that can’t promote subsequent interactions. This in turn will markedly reduce the probability of the protein conformational sampling to form catalytically competent protein-DNA interactions. In this scenario, FEN1 needs multiple DNA binding/bending trials until stochastically one DNA bending attempt positions the substrate in a state that can promote subsequent interactions.
There are major short-flap and minor long-flap pathways for processing OF. In the short-flap pathway, the flap length is restricted by the limited strand activity of Pol δ and the active hand-off mechanism between Pol δ and FEN1 (nick translation), where FEN1 cleaves the short flap (1–6 nt) and generates a nick sealable by LIG 1 [54], [97]. However, long flaps were also shown to exist both in vitro and in vivo [92], [98], [99]. In this minor pathway, the long-flap strand is bound by replication protein A (RPA), where Dna2 helicase/nuclease is recruited to the site while cleavage by FEN1 becomes inhibited [100], [101]. Dna2 displaces RPA [102] and progressively cleaves the flap to the point where Dna2 dissociates due to its lower affinity for the short flap [92] or FEN1 displaces Dna2 [103], [104], and the substrate becomes accessible again for FEN1. Interestingly, we showed that the miscleavage after DNA bending can also occur in the cognate substrate under the specific condition of increasing the 5′ flap length; kSTO however remains similar to that in DF-6,1 [75]. The miscleavage will result in the binding of RPA to the long 5′ flap and the inhibition of FEN1 cleavage activity [75]. We propose that miscleavage upon increasing the 5′ flap length is one of the mechanisms that may trigger the requirement of the long flap processing pathway to reduce the 5′ flap length and displace RPA before FEN1 can access and cleave the 5′ flap. This newly identified mechanism can only be resolved by following cleavage at the single-molecule level. We also investigated the behavior of FEN1 on short and long RNA flaps in EQ DF substrates. In this case, FEN1 showed ∼ 3-fold and ∼ 10-fold reduction in the stability of the bent complexes with the short and long RNA flaps, respectively, in comparison to the corresponding DNA substrates [75]. However, single-molecule cleavage assays exhibited slightly faster cleavage kinetics with RNA-flap substrates compared to the counterpart DNA substrates which is constituent with a previous study [105]. In conclusion, we found that although FEN1 exhibits reduced stability on RNA-flap substrates, its catalytic efficiency is not limited significantly by the higher dissociation rate [75].
4. Monitoring the kinetics of product release
4.1. Cleavage of the internal-labeled substrate
The catalytic constant (kcat) from steady state measurement is significantly slower than the kSTO determined by the SMFRET cleavage of DF-6,1Flap [55], [90], [91]. It has been shown by bulk cleavage kinetics that, following cleavage, the 5′ flap release is fast, whereas the nicked-duplex release limits the kcat [55], [91]. However, the kSTO rate 6.5 ± 1.2 s−1 from EQ DF-6,1Flap cleavage only reports on DNA bending, protein ordering for active site assembly, chemistry and 5′ flap release but not the nicked duplex release. Therefore, we performed the SMFRET cleavage of DF-6,1Internal as a complementary assay to monitor steps subsequent to 5′ flap release. This assay detects the time spent by EQ DF-6,1Internal substrate (E ≃ 0.3) upon FEN1 bending at high FRET (E ≃ 0.52) before 5′ flap release and FRET falls to (E ≃ 0.25) (Fig. 6A) [75]; a state that we assign to the unbent nicked-product. This assumption is supported by the observation that the mean FRET of histograms of DF-6,1Internal substrate is ∼ 0.05 higher than the corresponding nicked dsDNA. More importantly, the FEN1 concentration (250 nM) at which the cleavage reaction was performed is ∼ 50-fold higher than Kd-bending of DF-6,1Internal (4.8 nM) but well below Kd-bending of the nicked-product which was determined to be 580 ± 130 nM [75]. As FEN1 bends and cleaves the 5′ flap with diffusion-limited kinetics [72], this FEN1 concentration ensures a high fraction of cleavage within a single turnover without rebinding and bending of the nicked-product after its release. Therefore, in the unlikely event that DNA bending is not followed by 5′ flap cleavage, the FRET should change from 0.3 (unbent substrate) to 0.52 (bent substrate) and back to 0.3 (unbent substrate), whereas in the case of DNA bending followed by 5′ flap cleavage, the FRET should change from 0.3 (unbent substrate) to 0.52 (bent substrate) and back to 0.25 (unbent nicked product) [75].
Fig. 6.
Release kinetics of the nicked-product. (A) Schematic of the single-molecule cleavage experiment of EQ DF-6,1Internal. The zoom-in view of a representative cleavage trace shows three-states as fitted by vbFRET (0.3, 0.52 and 0.25) corresponding to the three DNA conformers, unbent EQ DF-6,1Internal, bent EQ DF-6,1Internal and unbent nicked-product, respectively. The dwell times of the bent state (τbending-Internal) is fitted to a gamma distribution. The average τbending-Internal is reported with the standard error of the mean. (B) The cleavage of nonEQ DF-6,1Internal was performed at 40 mM KCl concentration to promote FEN1 rebinding to the nicked-product. Representative trace shows the cleavage of NonEQ DF-6,1Internal and the three DNA conformers in A. The FRET state and the schematic of substrate/product conformer in each step are illustrated. After cleavage, FEN1 rebinds/bends the nicked-product as demonstrated by the rapid transitions between 0.25 (unbent product) and 0.52 (bent product). The dwell time distributions of FEN1 binding to the bent product (τbending-Internal) (red) and unbent product (τproduct-unbent) (blue) were fitted to gamma functions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.2. Promoting FEN1 rebinding to the nicked product
The dwell time of the EQ DF-6,1Internal in the bent state Avg τbending-Internal 270 ± 70 ms (Fig. 6A) is ∼ 100 ms longer than the time required for cleaving the 5′ flap (Fig. 3E). This established that FEN1 remains bound to the bent DNA conformer for an extended time after 5′ flap release. However, the corresponding kSTO 3.7 s−1 from Avg τbending-Internal (Fig. 6A) still does not agree with the much slower kcat 1.4 ± 0.1 s−1 from bulk assays. Therefore, we considered the possibility that FEN1 remains bound to the unbent nicked-product for some time before dissociating into solution [55], [91]. First, we measured FEN1 binding to the nicked-product substrate with internal-label under equilibrium conditions at 40 and 100 mM KCl concentrations (Kd-bending 62 ± 9 nM and 580 ± 130 nM), respectively [75]. Second, in order to test this hypothesis, we performed the same cleavage experiment under conditions that favor FEN1 rebinding to the nicked-product by lowering KCl salt concentration from 100 to 40 mM (Fig. 6B) [75]. Lower salt concentration increases the affinity of FEN1 for the nicked-product of DF-6,1 without affecting the kSTO of the reaction, i.e. Avg τbending-Flap is 155 ± 30 ms at 100 mM KCl compared to 180 ± 40 ms at 40 mM KCl for the EQ DF-6,1Flap substrate [75]. The cleavage experiment is performed under continuous flow, where acquitision starts before and continues after FEN1 is introduced into the flow cell. The onset time of the bending event (t ≃ 17 s) (Fig. 6B) marks the entry of FEN1 into the flow cell and encountering the substrate at near-diffusion limited kinetics. The expectation was that if FEN1 is still bound for some time to the unbent nicked-product after 5′ flap release, there will be a lag phase at low FRET (E ≃ 0.25), which reflects the duration at which FEN1 remains in this complex before it dissociates and another FEN1 rapidly binds and bends the released nicked-product to achieve high FRET (E ≃ 0.52).
At 40 mM KCl concentration, the increase in Avg τbending-Internal 570 ± 115 ms (Fig. 6B) compared to Avg τbending-Flap 180 ± 40 ms of the EQ DF-6,1 indicates that the dwell time of bent product (τproduct-bent) was extended by 390 ± 120 ms. Interestingly, we observed an elongated phase with unbent nicked-product (τproduct-unbent) at E ≃ 0.25 for a duration of 2100 ± 420 ms before FEN1 dissociation followed by re-binding/bending of the product and attaining high FRET bent state (E ≃ 0.52) (Fig. 6B).
These results can be interpreted such that product release by FEN1 after the 5′ flap release occurs in two steps: first, Avg τproduct-bent wherein FEN1 briefly holds the product in bent state for 390 ± 120 ms. Second, Avg τproduct-unbent wherein FEN1 remains bound to the unbent product for 2100 ± 420 ms before dissociating into solution. Therefore, the actual Avg τrelease can be the sum of these two dwell times, which yields a krelease of 0.40 ± 0.07 s−1 for the nicked-product. The resulting kcat of 0.37 ± 0.06 s−1 (1/(Avg τbending-Flap + Avg τrelease)) is in agreement with the rates determined by our bulk assays [75] and previous reports [55], [90]. Collectively, these results suggest that the release of the nicked DNA product, mainly from an unbent state, is the rate-limiting step for FEN1 reaction.
4.3. Monitoring of total FEN1 reaction by PIFE
However, the previous SMFRET experiment based on product rebinding did not provide a direct evidence that FEN1 remained bound to the unbent nicked-product during the lag phase. Thus, we applied PIFE by introducing a singly-labeled substrate where Cy3 is at position 10 in the downstream dsDNA of nonEQ DF-6,1. This dye location induced 35% enhancement upon protein binding through PIFE without relying on conformational changes (Fig. 7A) [75]. The SMPIFE experiments were performed under standard SMFRET cleavage conditions (250 nM FEN1, 100 mM KCl). In SMPIFE, the dwell time of the enhanced-fluorescence state (τPIFE) comprises the duration that FEN1 spends binding to the substrate, cleaving the 5′ flap, and any subsequent binding to the nicked-product (Fig. 7B) [75]. Based on the kinetics from SMFRET, if FEN1 released the product from a bent state immediately after cleavage, τPIFE should be relatively short, whereas a longer τPIFE would imply that FEN1 remained bound to the product after a productive binding/cleavage event.
Fig. 7.
Direct measurement of total FEN1 reaction by PIFE. (A) Singly-labeled nonEQ DF-6,1PIFE substrate with Cy3 positioned at the downstream duplex. Bulk time-resolved fluorescence lifetime measurements are depicted for the substrate (black) and in presence of 1 μM FEN1 showing 35% fluorescence enhancement (red). (B) Representative trace of SMPIFE cleavage experiment with nonEQ DF-6,1PIFE illustrating the respective substrate/product conformer. The enhanced-fluorescence state dwell time (τPIFE) distribution is fitted to gamma function. The means and standard errors of the means are reported. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fitting the dwell time distribution of τPIFE for cleavage events with gamma function (Fig. 7B) resulted in a lengthy Avg τPIFE of 2210 ± 500 ms, which translates into a rate of 0.45 ± 0.10 s−1. This rate is in good agreement with kcat observed in bulk assays as well as kcat from SMFRET experiments at lower salt [75]. Collectively, both SMFRET and SMPIFE experiments show FEN1 product release mechanism to have two steps, a fast unbending step and a relatively slow product release step from the unbent state. These experiments illustrate the power of single-molecule fluorescence assays in resolving intermediary steps inaccessible to bulk assays.
5. Kinetic timeline of FEN1 catalytic reaction at the single-molecule level
The kinetic parameters obtained from SMFRET on flap- and internal-labeled substrates as well as SMPIFE and SMPIFQ experiments enabled us to construct a comprehensive reaction mechanism of FEN1 on its substrate at the single-molecule level (Fig. 8). To recapitulate, FEN1 binds and actively bends its cognate substrate by diffusion-limited kinetics, threads the 5′ flap, molds/binds the 3′ flap thus inducing protein ordering and assembly of the active site, resulting in incision and prompt release of the 5′ flap. However, for the non-cognate substrates, FEN1 inhibits off-target incision by blocking or reducing the probability of formation of catalytically competent active site resulting in repetitive abortive bending events and dissociation of FEN1 from the bent state. The slow release of nicked-product occurs in two-steps indicating the relatively high affinity and stability of the FEN1-nicked-product complex. The product release rates (kproduct-bent) and (kproduct-unbent) are determined for nonEQ DF-6,1Internal at 40 mM KCl in presence of Mg2+. FEN1 can rebind/rebend the nicked product at diffusion-limited kinetics, however at a slower on-rate than the substrate. There is no significant change in kon-bending of the substrate at lower KCl concentration. Likewise, kSTO is similar for both EQ DF-6,1Flap and nonEQ DF-6,1Flap and is independent of KCl concentration. The kon and koff rates of the substrate and product are determined in presence of Ca2+. These findings demonstrate the merit of applying complementary single-molecule fluorescence techniques augmented with various labeling schemes to elucidate the details of an entire enzymatic reaction as exemplified by FEN1.
Fig. 8.
Kinetic timeline of FEN1 enzymatic reaction. FEN1 binds and bends DF substrate at diffusion-limited kinetics. kSTO starts from DNA bending, whereas kthreading initiates from 5′ flap threading till the 5′ flap incision and instantaneous release. FEN1 inhibits off-target cleavage of non-cognate substrates. The release of nicked-product occurs in two steps. FEN1 can rebind/rebend the nicked product at diffusion-limited kinetics, although at a slower on-rate than the substrate. The product release rates (kproduct-bent) and (kproduct-unbent) are determined for nonEQ DF-6,1Internal at 40 mM KCl in presence of Mg2+. kSTO is similar for both EQ DF-6,1Flap and nonEQ DF-6,1Flap and independent of KCl concentration [75]. The substrate and product kon and koff rates are determined in presence of Ca2+.
6. Potential roles of the two-step product release based on Pol δ–DNA–PCNA–FEN1 structure
The synthesis process of Okazaki fragments is initiated by Pol α that lacks proofreading activity [50]. Pol δ is a heterotetrameric protein complex composed of the catalytic subunit (p125) and three regulatory subunits (p50 or B-subunit, p66 and p12) required for the optimal activity of the holoenzyme [106]. The limited strand displacement activity of Pol δ during the invasion of the 5′ end of the previous unmatured Okazaki fragment helps in removing the Pol α-synthesized primer. The incorporation rate for each nucleotide successively decreases to 10–20% of that of the preceding nucleotide. Therefore, nascent flap acts as a progressive molecular brake on Pol δ [97]. The maturation of Okazaki fragments occurs by the joint action of Pol δ and FEN1, coordinated by the PCNA sliding clamp, where FEN1 cleaves the ssDNA 5′ flaps producing ligatable nicks. The iterative strand displacement and cleavage by Pol δ and FEN1 completely remove the RNA primer in a process termed nick translation [107], [108]. The Pol δ–DNA–PCNA–FEN1 complex sustains an efficient and processive nick translation during which the 1 nt flap is the main FEN1′s substrate.
Using Cryo-EM microscopy, we reported the first structure for human Pol δ–DNA–PCNA–FEN1 complex (Fig. 9A), hence revealing various mechanistic details of the maturation of Okazaki fragments [76]. Pol δ has three PCNA-interacting (PIP-box) motifs located on the p125, p66 and p12 subunits that can individually interact with PCNA [109], [110]. Pol δ is bound to one of the three PCNA monomers through the C-terminal domain (CTD) of the p125 subunit. The catalytic core of Pol δ, composed of N-terminal, palm, thumb and exonuclease domains, is situated on top of PCNA in an open configuration, while the regulatory subunits extend sideways (Fig. 9A). This arrangement permits PCNA to thread and stabilize the DNA exiting the catalytic cleft and to recruit FEN1 to one of the two unoccupied monomer in a toolbelt fashion. FEN1 preferentially binds the more exposed PIP-box on PCNA (Fig. 9A). The third PIP-box on PCNA is sterically hidden [76], suggesting that LIG1 would be recruited from solution and this requires movement of Pol δ out of the way or its dissociation. This mechanism is consistent with the distributive action of LIG1 during maturation of Okazaki fragments [97]. It was also reported that acute depletion of LIG1 in yeast permits nick translation to progress up to the dyad of the pre-assembled nucleosome, and the Okazaki fragment termini are enriched around known transcription factor binding sites [111]. Therefore, nucleosomes and DNA binding factors may trigger the stalling of Pol δ and its dissociation from PCNA and/or DNA to allow LIG1 to bind and seal the nick of a mature Okazaki fragment [76].
Fig. 9.
Potential roles of the two-step product release. (A) Cryo-EM structure of Pol δ-DNA-PCNA-FEN1 complex. (B) Proposed toolbelt model for Pol δ and FEN1 bound to PCNA processing an Okazaki fragment. (C) Potential interactions between FEN1 and Pol δ synthesizing an Okazaki fragment. FEN1 cap-helical gateway and the loop spanning residues 45–59 that includes the the 3′ flap binding pocket, shown as gold ribbons, are disordered in the Cryo-EM structure shown in A and may become ordered upon interaction with Pol δ after strand displacement during the invasion of the 5′ end of the previous Okazaki fragment. (D) Overlay of the three Cryo-EM models of different Pol δ-DNA-PCNA conformers with increasing tilting of PCNA relative to the polymerase. (E) Model of Pol δ-DNA-PCNA-FEN1 complex with PCNA corresponding to the conformer with highest tilting (Conf 3 shown in D). Tilting of PCNA results in the increased exposure of the vacant PIP-box site (PIP-3), marked by a purple ribbon corresponding to the p21 peptide bound to PCNA (PDB ID: 1AXC) [112]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Our Pol δ–DNA–PCNA–FEN1 complex structure showed the mutual positions of Pol δ, DNA, and PCNA to be analogous to those of the processive Pol δ holoenzyme and that FEN1 occupies the space between the N-terminal and palm domains of the p125 subunit (Fig. 9A). The resolution of FEN1 is lower than the rest of the complex, which is compatible with FEN being flexibly tethered to PCNA. FEN1 is situated across the template strand (Fig. 9A) and properly oriented to bind the downstream duplex DNA when Pol δ encounters the previously synthesized Okazaki fragment (Fig. 9B) [76]. Remarkably, the template strand was already at 90° bent angle similar to that required for FEN1 activity [6], [72]. Therefore, Pol δ may handoff a bent nick junction to FEN1 and aid in positioning the 5′ flap to thread into the cap-helical gateway (Fig. 9B). Notably, a pre-bent junction is not required for FEN1 activity since FEN1 can actively bend the nick junction in diffusion-limited kinetics [72], [75]. Nick translation takes place at 10-fold faster rates than the release of nicked-product by FEN1 [75], [91], [97]. Therefore, Pol δ and FEN1 must be actively handing off their products during nick translation. This is also suggested by our structure, which shows the proximity of FEN1 to the template strand and the potential interaction between FEN1, particularly the cap-helical gateway and loops that include the 3′ flap binding pocket, and Pol δ that may facilitate their products handoff during nick translation (Fig. 9C) [76].
It remains unclear how LIG1 would access the hidden third PIP-box during the nick-translation reaction. PCNA can adopt a tilting conformer which may expose the third PIP-box (Fig. 9D) [76]. It is also possible that during the nick translation reaction that Pol δ is tethered flexibly to PCNA when the DF is bound to FEN1. The tilting of PCNA and the flexible tethering of Pol δ may provide a window of opportunity for LIG1 to access the hidden third PIP-box (Fig. 9E) [76]. However, it remains unclear how Pol δ–PCNA–FEN1 would signal the binding of LIG1. As mentioned before, the nicked-product release by FEN1 occurs in two steps, where FEN1 binds briefly to the bent conformer followed by a lengthy binding to the extended conformer [75]. We propose that the bent conformer is more compatible with Pol δ binding, while the extended conformer sequesters the nick till Pol δ moves out of the way and LIG1 is recruited from solution. In this mechanism, LIG1 needs to bind PCNA before Pol δ rebinds the FEN1-bent nick product. We anticipate that LIG1 binding will sterically exclude Pol δ and break the continuity of its communication with FEN1 during the nick translation reaction.
7. Conclusion and future outlook
DNA replication and repair inside cells are built around an intricate set of molecular machineries that carry out these vital processes with the utmost performance and highest fidelity. FEN1 is at the heart of the process of the maturation of Okazaki fragments culminating into the synthesis of contiguous lagging strands. The roles of FEN1, Pol δ and PCNA sliding clamp are orchestrated by dynamic interactions within this protein complex, which may also have direct implications on the way LIG1 seals the nicks on the lagging strand. In this review, we presented our research work using single-molecule fluorescence to unveil the mechanistic aspects of FEN1 enzymatic reaction and our Cryo-EM structure of the reconstituted DNA–Pol δ–PCNA–FEN1 complex for maturation of Okazaki fragments.
Collectively, our work demonstrates the merit of using complementary single-molecule fluorescence techniques to elucidate the details of an entire enzymatic reaction as exemplified by FEN1. Also, we illustrate the remarkable selectivity of FEN1 for its cognate substrate through the verification of its key features, while promoting the dissociation of the non-cognate substrates to inhibit off-target incision by blocking or reducing the probable formation of catalytically competent active sites. We propose that different members of the 5′ nucleases family share similar DNA-bending-induced disorder-to-order transitioning but differ in the mechanisms that couple this transitioning with active site assembly.
Establishing a variety of fluorescence assays to monitor FEN1 along the path of substrate recognition, cleavage and product release will provide foundation for studying its interactions with PCNA, Pol δ, and LIG1 during the maturation of Okazaki fragments. By systematically adding PCNA, Pol δ, or LIG1 and tailoring the individual or combination of FRET, PIFE, and PIFQ assays to selectively visualize specific intermediary structures, it will be possible to follow the transfer of the flap substrate and nick product between FEN1 and Pol δ or the nick product between FEN1 and LIG1 in absence or presence of Pol δ. The tight integration of findings from SM assays with Cryo-EM structures and other structural techniques of intermediary steps during the maturation of Okazaki fragments will allow us to build reliable molecular movies of lagging strand DNA synthesis and maturation of Okazaki fragments.
In a wider perspective of the field, genetic and biochemical studies continue to unveil the roles that FEN1 plays in many DNA transactions including processing intermediates of Okazaki fragment maturation, long-patch base excision repair, telomere maintenance, and stalled replication fork rescue. The ongoing efforts for understanding the functional and mechanistic properties of FEN1 and other 5′ nucleases within the context of DNA replication and repair inside the cell may culminate in identifying potential therapeutic targets for human cancers, neuromuscular and neurodegenerative diseases.
In summary, performing enzymology while monitoring the conformation of the substrate at the single-molecule level is a challenging endeavor. This review showed how investment in the development of variety of assays using various fluorescence modulations methods (FRET, PIFE and PIFQ) were necessary to follow binding and distortion of the substrate and product along the path of catalysis. Investment in understanding and controling PIFE and PIFQ on demand [44] and their combination with FRET will be a key development to realize the full potential of single-molecule imaging in enzymology. Finally, devising and interpreting experiments aimed towards the achievement of comprehensive mechanistic understanding will benefit from their application to systems that are well-characterized biochemically and structurally.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This research was supported by King Abdullah University of Science and Technology through the KAUST Competitive Research Grants URF/1/3764‐01‐01 and URF/1/4036‐01‐01 to S.M.H.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Contributor Information
Mohamed A. Sobhy, Email: mohamed.sobhy@kaust.edu.sa.
Samir M. Hamdan, Email: samir.hamdan@kaust.edu.sa.
References
- 1.Tsutakawa S.E., Lafrance-Vanasse J., Tainer J.A. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 2014;19:95. doi: 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsutakawa S.E., Tainer J.A. Double strand binding-single strand incision mechanism for human flap endonuclease: implications for the superfamily. Mech Ageing Dev. 2012;133(4):195. doi: 10.1016/j.mad.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finger L.D., Atack J.M., Tsutakawa S., Classen S., Tainer J., Grasby J. The wonders of flap endonucleases: structure, function, mechanism and regulation. Sub-Cell Biochem. 2012;62:301. doi: 10.1007/978-94-007-4572-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Kao H.I., Bambara R.A. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 5.Sakurai S., Kitano K., Okada K., Hamada K., Morioka H., Hakoshima T. Preparation and crystallization of human flap endonuclease FEN-1 in complex with proliferating-cell nuclear antigen, PCNA. Acta Crystallogr D Biol Crystallogr. 2003;59(5):933. doi: 10.1107/s0907444903004815. [DOI] [PubMed] [Google Scholar]
- 6.Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145(2):198. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orans J., McSweeney E.A., Iyer R.R., Hast M.A., Hellinga H.W., Modrich P. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145(2):212. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devos J.M., Tomanicek S.J., Jones C.E., Nossal N.G., Mueser T.C. Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J Biol Chem. 2007;282(43):31713. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, Princz LN, Klugel MF, Habermann B, Pfander B, Biertumpfel C. Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage. eLife 2015, 4:e12256. [DOI] [PMC free article] [PubMed]
- 10.Liu Y., Freeman A.D., Declais A.C., Wilson T.J., Gartner A., Lilley D.M. Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA. Cell reports. 2015;13(11):2565. doi: 10.1016/j.celrep.2015.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y., Hellinga H.W., Beese L.S. Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I. Proc Natl Acad Sci USA. 2017;114(23):6010. doi: 10.1073/pnas.1704845114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Algasaier S.I., Exell J.C., Bennet I.A., Thompson M.J., Gotham V.J., Shaw S.J., Craggs T.D., Finger L.D., Grasby J.A. DNA and Protein Requirements for Substrate Conformational Changes Necessary for Human Flap Endonuclease-1-catalyzed Reaction. J Biol Chem. 2016;291(15):8258. doi: 10.1074/jbc.M115.698993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uphoff S., Sherratt D.J. Single-Molecule Analysis of Bacterial DNA Repair and Mutagenesis. Annu Rev Biophys. 2017;46:411. doi: 10.1146/annurev-biophys-070816-034106. [DOI] [PubMed] [Google Scholar]
- 15.Mohapatra S., Lin C.T., Feng X.A., Basu A., Ha T. Single-Molecule Analysis and Engineering of DNA Motors. Chem Rev. 2020;120(1):36. doi: 10.1021/acs.chemrev.9b00361. [DOI] [PubMed] [Google Scholar]
- 16.Ray S., Widom J.R., Walter N.G. Life under the Microscope: Single-Molecule Fluorescence Highlights the RNA World. Chem Rev. 2018;118(8):4120. doi: 10.1021/acs.chemrev.7b00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapanidis A.N., Weiss S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J Chem Phys. 2002;117(24):10953. [Google Scholar]
- 18.Moerner W.E., Fromm D.P. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev Sci Instrum. 2003;74(8):3597. [Google Scholar]
- 19.Deniz A.A., Dahan M., Grunwell J.R., Ha T., Faulhaber A.E., Chemla D.S. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc. Natl. Acad. Sci. U. S. A. 1999;96(7):3670. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapanidis A.N., Laurence T.A., Lee N.K., Margeat E., Kong X., Weiss S. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38(7):523. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 21.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89(1):141. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2(11):764. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 23.Walter N.G., Huang C.Y., Manzo A.J., Sobhy M.A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5(6):475. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25(1):78. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 25.Farooq S., Hohlbein J. Camera-based single-molecule FRET detection with improved time resolution. PCCP. 2015;17(41):27862. doi: 10.1039/c5cp04137f. [DOI] [PubMed] [Google Scholar]
- 26.Juette M.F., Terry D.S., Wasserman M.R., Altman R.B., Zhou Z., Zhao H. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13(4):341. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stryer L. Fluorescence Energy-Transfer as a Spectroscopic Ruler. Annu Rev Biochem. 1978;47:819. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 28.Selvin P.R., Ha T., Enderle T., Ogletree D.F., Chemla D.S., Weiss S. Fluorescence resonance energy transfer between a single donor and a single acceptor molecule. Biophys J. 1996;70(2):Wp302. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha T., Enderle T., Ogletree D.F., Chemla D.S., Selvin P.R., Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. U. S. A. 1996;93(13):6264. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia K., Wan Y., Xia A.D., Li S.Y., Gong F.B., Yang G.Q. Characterization of photoinduced isomerization and intersystem crossing of the cyanine dye Cy3. J Phys Chem A. 2007;111(9):1593. doi: 10.1021/jp067843i. [DOI] [PubMed] [Google Scholar]
- 31.Hwang H., Kim H., Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108(18):7414. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang H., Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem Soc Rev. 2014;43(4):1221. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craggs T.D., Hutton R.D., Brenlla A., White M.F., Penedo J.C. Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates. Nucleic Acids Res. 2014;42(3):1857. doi: 10.1093/nar/gkt1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myong S., Cui S., Cornish P.V., Kirchhofer A., Gack M.U., Jung J.U. Cytosolic viral sensor RIG-I Is a 5 '-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myong S., Rasnik I., Joo C., Lohman T.M., Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437(7063):1321. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 36.Joo C., McKinney S.A., Nakamura M., Rasnik I., Myong S., Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126(3):515. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 37.Park J., Myong S., Niedziela-Majka A., Lee K.S., Yu J., Lohman T.M. PcrA helicase dismantles reca filaments by reeling in DNA in uniform steps. Cell. 2010;142(4):544. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vrtis K.B., Markiewicz R.P., Romano L.J., Rueda D. Carcinogenic adducts induce distinct DNA polymerase binding orientations. Nucleic Acids Res. 2013;41(16):7843. doi: 10.1093/nar/gkt554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J., Jergic S., Jeon Y., Cho W.K., Lee R., Dixon N.E. Dynamics of Proofreading by the E. coli Pol III Replicase. Cell Chem Biol. 2018;25(1):57. doi: 10.1016/j.chembiol.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Gatzogiannis E., Chen Z.X., Wei L., Wombacher R., Kao Y.T., Yefremov G. Mapping protein-specific micro-environments in live cells by fluorescence lifetime imaging of a hybrid genetic-chemical molecular rotor tag. Chem Commun. 2012;48(69):8694. doi: 10.1039/c2cc33133k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stennett E.M.S., Ciuba M.A., Lin S., Levitus M. Demystifying PIFE: The Photophysics Behind the Protein-Induced Fluorescence Enhancement Phenomenon in Cy3. J Phys Chem Lett. 2015;6(10):1819. doi: 10.1021/acs.jpclett.5b00613. [DOI] [PubMed] [Google Scholar]
- 42.Lee W., von Hippel P.H., Marcus A.H. Internally labeled Cy3/Cy5 DNA constructs show greatly enhanced photo-stability in single-molecule FRET experiments. Nucleic Acids Res. 2014;42(9):5967. doi: 10.1093/nar/gku199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kretschy N., Sack M., Somoza M.M. Sequence-dependent fluorescence of Cy3-and Cy5-labeled double-stranded DNA. Bioconj Chem. 2016;27(3):840. doi: 10.1021/acs.bioconjchem.6b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid F., Raducanu V.S., Zaher M.S., Tehseen M., Habuchi S., Hamdan S.M. Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation. Nat Commun. 2019;10:2104. doi: 10.1038/s41467-019-10137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stennett E.M.S., Ciuba M.A., Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chem Soc Rev. 2014;43(4):1057. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 46.Berezin M.Y., Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem Rev. 2010;110(5):2641. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honda M., Park J., Pugh R.A., Ha T., Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35(5):694. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levitus M., Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q Rev Biophys. 2011;44(1):123. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 49.Spiriti J., Binder J.K., Levitus M., van der Vaart A. Cy3-DNA stacking interactions strongly depend on the identity of the terminal basepair. Biophys J. 2011;100(4):1049. doi: 10.1016/j.bpj.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perera R.L., Torella R., Klinge S., Kilkenny M.L., Maman J.D., Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. Elife. 2013;2 doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerbe L.K., Kuchta R.D. The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry. 2002;41(15):4891. doi: 10.1021/bi016030b. [DOI] [PubMed] [Google Scholar]
- 52.Okazaki R., Arisawa M., Sugino A. Slow Joining of Newly Replicated DNA Chains in DNA Polymerase I-Deficient Escherichia-Coli Mutants. Proc Natl Acad Sci U S A. 1971;68(12):2954. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgers P.M.J., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273(31):19756. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 54.Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18(22):2764. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finger L.D., Blanchard M.S., Theimer C.A., Sengerova B., Singh P., Chavez V. The 3 '-Flap Pocket of Human Flap Endonuclease 1 Is Critical for Substrate Binding and Catalysis. J Biol Chem. 2009;284(33):22184. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kao H.I., Henricksen L.A., Liu Y., Bambara R.A. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J Biol Chem. 2002;277(17):14379. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 57.Gomes X.V., Burgers P.M.J. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19(14):3811. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson E., Garg P., Burgers P.M.J. The pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem. 2004;279(3):1907. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 59.Garg P., Burgers P.M.J. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40(2):115. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 60.Harrington J.J., Lieber M.R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13(5):1235. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrington J.J., Lieber M.R. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8(11):1344. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 62.Balakrishnan L., Bambara R.A. Flap endonuclease 1. Annu Rev Biochem. 2013;82:119. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balakrishnan L., Bambara R.A. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286(9):6865. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng L., Jia J., Finger L.D., Guo Z., Zer C., Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39(3):781. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Pel D.M., Barrett I.J., Shimizu Y., Sajesh B.V., Guppy B.J., Pfeifer T. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9(1) doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward T.A., McHugh P.J., Durant S.T. Small molecule inhibitors uncover synthetic genetic interactions of human flap endonuclease 1 (FEN1) with DNA damage response genes. PLoS ONE. 2017;12(6) doi: 10.1371/journal.pone.0179278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsutakawa S.E., Thompson M.J., Arvai A.S., Neil A.J., Shaw S.J., Algasaier S.I. Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability. Nat Commun. 2017;8:15855. doi: 10.1038/ncomms15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H., Shi R., Han W., Cheng J., Xu X., Cheng K. Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1. Nucleic Acids Res. 2018;46(21):11315. doi: 10.1093/nar/gky911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gloor J.W., Balakrishnan L., Bambara R.A. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010;285(45):34922. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel N., Atack J.M., Finger L.D., Exell J.C., Thompson P., Tsutakawa S. Flap endonucleases pass 5'-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5'-ends. Nucleic Acids Res. 2012;40(10):4507. doi: 10.1093/nar/gks051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sobhy M.A., Joudeh L.I., Huang X., Takahashi M., Hamdan S.M. Sequential and multistep substrate interrogation provides the scaffold for specificity in human flap endonuclease 1. Cell Rep. 2013;3(6):1785. doi: 10.1016/j.celrep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Rashid F., Harris P.D., Zaher M.S., Sobhy M.A., Joudeh L.I., Yan C. Single-molecule FRET unveils induced-fit mechanism for substrate selectivity in flap endonuclease 1. Elife. 2017;6 doi: 10.7554/eLife.21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chapados B.R., Hosfield D.J., Han S., Qiu J., Yelent B., Shen B. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116(1):39. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 74.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24(4):683. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaher M.S., Rashid F., Song B., Joudeh L.I., Sobhy M.A., Tehseen M. Missed cleavage opportunities by FEN1 lead to Okazaki fragment maturation via the long-flap pathway. Nucleic Acids Res. 2018;46(6):2956. doi: 10.1093/nar/gky082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lancey C., Tehseen M., Raducanu V.S., Rashid F., Merino N., Ragan T.J. Structure of the processive human Pol delta holoenzyme. Nat Commun. 2020;11:1109. doi: 10.1038/s41467-020-14898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sobhy M.A., Elshenawy M.M., Takahashi M., Whitman B.H., Walter N.G., Hamdan S.M. Versatile single-molecule multi-color excitation and detection fluorescence setup for studying biomolecular dynamics. Rev Sci Instrum. 2011;82(11) doi: 10.1063/1.3657153. [DOI] [PubMed] [Google Scholar]
- 78.Harris P.D., Hamdan S.M., Habuchi S. Relative contributions of base stacking and electrostatic repulsion on DNA nicks and gaps. J Phys Chem B. 2020;124(47):10663. doi: 10.1021/acs.jpcb.0c06941. [DOI] [PubMed] [Google Scholar]
- 79.Odonovan A., Davies A.A., Moggs J.G., West S.C., Wood R.D. Xpg Endonuclease Makes the 3' Incision in Human DNA Nucleotide Excision-Repair. Nature. 1994;371(6496):432. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 80.Evans E., Fellows J., Coffer A., Wood R.D. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16(3):625. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsutakawa S.E., Sarker A.H., Ng C., Arvai A.S., Shin D.S., Shih B. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc Natl Acad Sci U S A. 2020;117(25):14127. doi: 10.1073/pnas.1921311117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M.X., Wilson D.M. Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Sign. 2014;20(4):678. doi: 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schermerhorn K.M., Delaney S. A chemical and kinetic perspective on base excision repair of DNA. Acc Chem Res. 2014;47(4):1238. doi: 10.1021/ar400275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoitsma N.W., Whitaker A.M., Beckwitt E.C., Jang S., Agarwal P.K., Houten B.V. AP-endonuclease 1 Sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop. Nucleic Acids Res. 2020;48(13):7345. doi: 10.1093/nar/gkaa496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sobhy M.A., Bralic A., Raducanu V.S., Takahashi M., Tehseen M., Rashid F. Resolution of the Holliday junction recombination intermediate by human GEN1 at the single-molecule level. Nucleic Acids Res. 2019;47(4):1935. doi: 10.1093/nar/gky1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou R.B., Yang O., Declais A.C., Jin H., Gwon G.H., Freeman A.D.J. Junction resolving enzymes use multivalency to keep the Holliday junction dynamic. Nat Chem Biol. 2019;15(3):269. doi: 10.1038/s41589-018-0209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chakraborty S., Steinbach P.J., Paul D., Mu H., Broyde S., Min J.H. Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex. Nucleic Acids Res. 2018;46(3):1240. doi: 10.1093/nar/gkx1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uyemura D., Lehman I.R. Biochemical characterization of mutant forms of DNA polymerase I from Escherichia coli. I. The polA12 mutation. J Biol Chem. 1976;251(13):4078. [PubMed] [Google Scholar]
- 89.Craggs T.D., Sustarsic M., Plochowietz A., Mosayebi M., Kaju H., Cuthbert A. Substrate conformational dynamics facilitate structure-specific recognition of gapped DNA by DNA polymerase. Nucleic Acids Res. 2019;47(20):10788. doi: 10.1093/nar/gkz797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tarantino M.E., Bilotti K., Huang J., Delaney S. Rate-determining Step of Flap Endonuclease 1 (FEN1) Reflects a Kinetic Bias against Long Flaps and Trinucleotide Repeat Sequences. J Biol Chem. 2015;290(34):21154. doi: 10.1074/jbc.M115.666438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song B., Hamdan S.M., Hingorani M.M. Positioning the 5'-flap junction in the active site controls the rate of flap endonuclease-1-catalyzed DNA cleavage. J Biol Chem. 2018;293(13):4792. doi: 10.1074/jbc.RA117.001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gloor J.W., Balakrishnan L., Campbell J.L., Bambara R.A. Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 2012;40(14):6774. doi: 10.1093/nar/gks388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finger L.D., Patel N., Beddows A., Ma L., Exell J.C., Jardine E. Observation of unpaired substrate DNA in the flap endonuclease-1 active site. Nucleic Acids Res. 2013;41(21):9839. doi: 10.1093/nar/gkt737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel N., Exell J.C., Jardine E., Ombler B., Finger L.D., Ciani B. Proline Scanning Mutagenesis Reveals a Role for the Flap Endonuclease-1 Helical Cap in Substrate Unpairing. J Biol Chem. 2013;288(47):34239. doi: 10.1074/jbc.M113.509489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thompson M.J., Gotham V.J.B., Ciani B., Grasby J.A. A conserved loop-wedge motif moderates reaction site search and recognition by FEN1. Nucleic Acids Res. 2018;46(15):7858. doi: 10.1093/nar/gky506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bennet I.A., Finger L.D., Baxter N.J., Ambrose B., Hounslow A.M., Thompson M.J. Regional conformational flexibility couples substrate specificity and scissile phosphate diester selectivity in human flap endonuclease 1. Nucleic Acids Res. 2018;46(11):5618. doi: 10.1093/nar/gky293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stodola J.L., Burgers P.M. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat Struct Mol Biol. 2016;23(5):402. doi: 10.1038/nsmb.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu B.C., Hu J.Z., Wang J.N., Kong D.C. Direct visualization of RNA-DNA primer removal from okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J Biol Chem. 2017;292(12):4777. doi: 10.1074/jbc.M116.758599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Masuda-Sasa T., Imamura O., Campbell J.L. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34(6):1865. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bae S.H., Seo Y.S. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275(48):38022. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 101.Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412(6845):456. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 102.Zhou C., Pourmal S., Pavletich N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife. 2015;4 doi: 10.7554/eLife.09832. e09832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stewart J.A., Campbell J.L., Bambara R.A. Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J Biol Chem. 2009;284(13):8283. doi: 10.1074/jbc.M809189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stewart J.A., Campbell J.L., Bambara R.A. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J Biol Chem. 2006;281(50):38565. doi: 10.1074/jbc.M606884200. [DOI] [PubMed] [Google Scholar]
- 105.Qian L., Yuan F., Rodriguez-Tello P., Padgaonkar S., Zhang Y. Human Fanconi anemia complementation group a protein stimulates the 5' flap endonuclease activity of FEN1. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0082666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee M.Y.W.T., Wang X.X., Zhang S.F., Zhang Z.T., Lee E.Y.C. Regulation and Modulation of Human DNA Polymerase delta Activity and Function. Genes (Basel) 2017;8(7):190. doi: 10.3390/genes8070190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhagwat M., Nossal N.G. Bacteriophage T4 RNase H removes both RNA primers and adjacent DNA from the 5’ end of lagging strand fragments. J Biol Chem. 2001;276(30):28516. doi: 10.1074/jbc.M103914200. [DOI] [PubMed] [Google Scholar]
- 108.Stith C.M., Sterling J., Resnick M.A., Gordenin D.A., Burgers P.M. Flexibility of Eukaryotic Okazaki Fragment Maturation through Regulated Strand Displacement Synthesis. J Biol Chem. 2008;283(49):34129. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bruning J.B., Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12(12):2209. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 110.Rahmeh A.A., Zhou Y.J., Xie B., Li H., Lee E.Y.C., Lee M.Y.W.T. Phosphorylation of the p68 Subunit of Pol delta Acts as a Molecular Switch To Regulate Its Interaction with PCNA. Biochemistry. 2012;51(1):416. doi: 10.1021/bi201638e. [DOI] [PubMed] [Google Scholar]
- 111.Smith D.J., Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483(7390):434. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gulbis J.M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87(2):297. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.