Abstract
Background
HIV infection and normal aging share immune and inflammatory changes that result in premature aging and non-communicable diseases (NCDs), but the exact pathophysiology is not yet uncovered. We identified the common metabolic pathways underlying various NCDs in treated HIV infection.
Methods
We performed untargeted metabolomics including 87 HIV-negative (–) normal controls (NCs), 87 HIV-positive (+) NCs, and 148 HIV+ subjects with only one type of NCDs, namely, subclinical carotid atherosclerosis, neurocognitive impairment (NCI), liver fibrosis (LF) and renal impairment. All HIV+ subjects were virally suppressed.
Results
HIV+ patients presented widespread alterations in cellular metabolism compared to HIV– NCs. Glycerophospholipid (GPL) metabolism was the only one disturbed pathway presented in comparisons including HIV– NCs across age groups, HIV+ NCs across age groups, HIV+ NCs vs HIV– NCs and each of HIV+ NCDs vs HIV+ NCs. D-glutamine and D-glutamate metabolism and alanine-aspartate-glutamate metabolism were presented in comparisons between HIV+ NCs vs HIV– NCs, HIV+ LF or HIV+ NCI vs HIV+ NCs. Consistently, subsequent analysis identified a metabolomic fingerprint specific for HIV+ NCDs, containing 42 metabolites whose relative abundance showed either an upward (mainly GPL-derived lipid mediators) or a downward trend (mainly plasmalogen phosphatidylcholines, plasmalogen phosphatidylethanolamines, and glutamine) from HIV– NCs to HIV+ NCs and then HIV+ NCDs, reflecting a trend of increased oxidative stress.
Interpretation
GPL metabolism emerges as the common metabolic disturbance linking HIV to NCDs, followed by glutamine and glutamate metabolism. Together, our data point to the aforementioned metabolisms and related metabolites as potential key targets in studying pathophysiology of NCDs in HIV infection and developing therapeutic interventions.
Funding
China National Science and Technology Major Projects on Infectious Diseases, National Natural Science Foundation of China, Yi-wu Institute of Fudan University, and Shanghai Municipal Health and Family Planning Commission.
Keywords: HIV, Aging, Metabolomics, Non-communicable diseases
Research in context.
Evidence before this study
HIV infection and normal aging share immune and inflammatory changes that result in premature aging of HIV-positive (+) individuals by promoting metabolic and cellular changes, which in turn increase their susceptibility to non-communicable diseases (NCDs); yet the exact pathophysiology is still being uncovered. We searched PubMed with any combination of each item from (1) “HIV”, or “human immunodeficiency virus” and (2) “metabolites”, “metabolic” “metabolism”, “metabolomic”, metabolomics”, “lipid”, or “lipidomics” for studies published up to Mar 1, 2021, to identify studies that applied the metabolomics to address the NCDs in HIV+ individuals. We found that all published studies focused on one type of NCDs, rendering the identification of common metabolic disturbances linking HIV infection to various NCDs impossible.
Added value of this study
Glycerophospholipid (GPL) metabolism emerges as the common metabolic disturbance in the normal aging process of both HIV+ and HIV– individuals as well as linking HIV to NCDs. D-glutamine and D-glutamate metabolism and alanine-aspartate-glutamate metabolism emerge as the common metabolic disturbance link HIV to both NCI and LF. Subsequent analysis in individual metabolites corroborated aforementioned findings and identified a metabolomic fingerprint containing 42 metabolites whose relative abundance showed either an upward (mainly GPL-derived lipid mediators) or a downward trend (mainly plasmalogen PCs and PEs, glutamine) from HIV– NCs to HIV+ NCs and then HIV+ NCDs, reflecting a trend of increased inflammation and oxidative stress across groups.
Implication of all the available evidence
Our data provide valuable information on the pathophysiology of NCDs in treated HIV infection, pointing to GPL, glutamine and glutamate metabolisms and related metabolites (plasmalogens, lipid mediators and glutamine) as potential key targets to study the molecular mechanisms linking HIV to NCDs via inflammation and oxidative stress, and further develop new therapeutic interventions and risk prediction models. Further research is needed to validate and extend these findings using longitudinal multi-omics.
Alt-text: Unlabelled box
1. Introduction
Despite the success of combination antiretroviral therapy (ART) in achieving viral suppression, HIV-positive (+) individuals experience early onset and elevated risk of chronic non-communicable diseases (NCDs) associated with normal aging in the general population, including atherosclerotic cardiovascular diseases, neurocognitive disorders, liver diseases, renal diseases and others [1]. Evidence suggests that HIV infection and normal aging share immune and inflammatory changes that result in the premature aging of HIV+ individuals by promoting metabolic and cellular changes [2], which in turn increase their susceptibility to these NCDs [3]; yet the exact pathophysiology is still being uncovered [4]. The challenge for the research on this topic is largely due to the complex nature of the aging process and development of NCDs as well as interconnected nature of causal pathways.
Metabolomics is a high-throughput analytical technology that measures a wide range of small molecule metabolites. Because small metabolites are intermediates and end products of all regulatory pathways, metabolic alterations may capture the complex physiological changes during the normal aging process and various NCDs, yielding clues to the molecular mechanisms [5]. HIV infection has profound effects on the cell metabolism which in turn plays a critical role in HIV pathogenesis and disease progression, pointing to targeting metabolism as a promising approach to prevent or treat HIV infection and associated NCDs [6,7]. Metabolic pathways associated with NCDs have been reported among HIV+ individuals [8], but all of these studies focused on one type of NCDs, such as left ventricular diastolic dysfunction [9], neurocognitive impairment (NCI) [10], subclinical carotid atherosclerosis (SCA) [11], and frailty [12], and thus are unable to identify common metabolic pathways linking HIV to various NCDs. A study including multiple NCDs may provide new promising avenue to elucidate the common molecular mechanisms and lead to new therapeutic interventions in treated HIV infection [13].
Here, we performed untargeted metabolomics of plasma from HIV-negative (–) normal controls (NCs), HIV+ NCs and HIV+ subjects with one and only one of the four different NCDs including SCA, NCI, liver fibrosis (LF) and renal impairment (RI), which are common diseases that could affect vital organs (Fig. 1). As they tend to co-occur [14], a design like this may help deconvolution of a disease-specific metabolomic profile from that of other diseases. The primary aims of this study were to characterize the key metabolites and metabolic pathways associated with HIV infection and four aforementioned major NCDs in treated HIV infection, and with a focus to identify the common metabolic pathways underlying these NCDs related to HIV infection. Furthermore, assuming it shares metabolic changes with the normal aging process, the metabolic pathways associated with age in both HIV– NCs and HIV+ NCs were investigated for comparison.
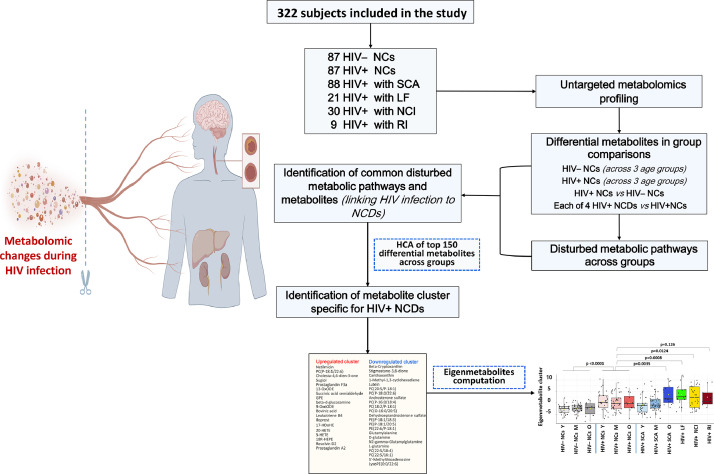
Fig. 1.
Flow chart of the study overview, subjects, and analysis.
2. Methods
2.1. Ethics
This study was performed in accordance with the Declaration of Helsinki, approved by the Institutional Review Board of Fudan University School of Public Health, China (IRB#2017-07-0629), and all subjects gave informed consent to participation.
2.2. Design and subjects
Plasma samples were drawn from the participants enrolled in the baseline evaluation (Years 2017–2018) of the CHART study, a prospective cohort of HIV and aging-related NCDs among HIV+ and HIV– participants. A detailed description of study design and participants recruitment has been previously published [15].
A flow chart depicting the study overview, subjects, and analysis are illustrated in Fig. 1. We profiled the metabolome of HIV– NCs, HIV+ NCs, and four groups of HIV+ NCD. Each group only had one specific type of the designated NCDs, namely SCA, NCI, LF and RI. Specifically, HIV+ SCA was defined as carotid artery intima-media thickness (IMT) ≥ 0·78 mm for 18–39 years and >1·00 mm for 40–65 years) [15,16]; HIV+ NCI was defined by Chinese version of Mini-mental State Examination [17]; HIV+ LF was defined as fibrosis-4 (FIB-4) > 3·25 [18]; HIV+ RI was estimated glomerular filtration rate (eGFR) < 60 mL/min/1·73 m2; HIV+ NCs and HIV– NCs were defined as carotid IMT < 0·78 mm, no NCI, no liver fibrosis (FIB-4 < 1·45 for HIV+ NCs, alanine aminotransferase [ALT] < 35 IU/L and aspartate aminotransferase [AST] < 35 IU/L and AST/ALT ratio [AAR] < 1·5 for HIV– NCs as platelet count [PLT] was not available), and no RI (eGFR ≥ 90 mL/min/1·73 m2) [19]. Furthermore, all subjects should: age at 18–65 years and sufficient volume of blood samples as inclusion criteria, and heavy alcohol use, prior clinical diagnosis of cancer and severe psychiatric disorders as exclusion criteria. All HIV+ subjects received ART and had current HIV RNA < 200 copies/mL. Additional details were presented in Supplementary methods: part A and B.
By this definition, eligible HIV+ SCA (n = 88), HIV+ LF (n = 21), NCI (n = 30), and RI (n = 9) subjects were all included for the present study due to small samples. HIV+ NCs (n = 87) and HIV– NCs (n = 87) were randomly selected from the eligible participants and frequency matched with HIV+ SCA at age (5-year category) and sex, and additionally Framingham risk score (< 10%, 10–20%, > 20%) distributions if possible (Supplementary methods: part C). In final, a total of 322 subjects were included.
2.3. Data collection and measurements
A standardized structured questionnaire was administered face-to-face by trained health staff to collect information on age, sex, education, and smoking habits, etc. HIV-related variables were extracted from the national HIV/AIDS Comprehensive Response Information Management System (CRIMS), including CD4 cell counts, HIV RNA, ART regimens, etc.
Physical examinations of height, weight and blood pressures (BP) were carried out. BP was measured twice after at least 5 min of rest, with at least 1 min between readings. The average of the two readings were recorded. Resting heart rate was measured by standard 12-lead electrocardiogram performed on all subjects at supine position after 5 min rest using standardized procedures. IMT of the left common carotid artery was measured by trained sonographers using a high-resolution B-mode ultrasound imager (LOGIQ P5 pro, GE, Indianapolis, USA), in accordance with standard procedures. Briefly, IMT image was obtained on about 10 mm of longitudinal carotid length which are free of plaque with a clearly identified double-line pattern. Fasting blood was collected for classical lipid profile, ALT, AST, PLT and creatinine.
2.4. Metabolomics profiling and data preprocessing
Fasting plasma samples were collected at the survey and were immediately stored at -80 °C. Processed plasma samples were analyzed by ultrahigh performance liquid chromatography-mass spectrometry (UPLC-MS) using UHPLC system (1290, Agilent Technologies) with UPLC BEH Amide column coupled to TripleTOF 6600 (Q-TOF, AB Sciex) and gas chromatography-mass spectrometry (GC-TOF-MS) using Agilent 7890 gas chromatograph coupled with a time-of-flight mass spectrometer. Details about metabolite extraction are presented in Supplementary methods: part D. During acquisition, one quality control (QC) sample was run after every 10 samples. The LC-MS data was converted from raw to mzXML file using the ProteoWizard software (Pala Alto, California, USA), and then were processed for peak detection, extraction, alignment, and integration by XCMS package (La Jolla, California, USA). In-house MS2 database (BiotreeDB) was applied in metabolite annotation. For GC-TOF-MS data, MS-DIAL software and Fiehn Binbase database were used for raw peaks exacting, the data baselines filtering and calibration of the baseline, peak alignment, deconvolution analysis, peak identification and integration of the peak area. Both of mass spectrum match and retention index match were considered in metabolites identification. The cut-off for annotation was set at 0·4.
Missing values were filled up by half of the minimum value. Metabolites with a relative standard deviation > 25% in QC samples deemed not suitably reproducible and were removed from further analysis. The normalization of each metabolite peak area was done by dividing peak area of metabolite by the peak area of the internal standards. This is a commonly applied technique for untargeted metabolomics. We identified 6091 metabolite features (613 named and 5478 unnamed). The preprocessing results generated a data matrix that consisted of retention time, mass to-charge ratio values, and normalized peak intensity was subjected to further analysis.
2.5. Statistical analysis
Data were expressed as the mean ± (standard deviation [SD]) for median (interquartile range [IQR]) for continuous variables and the number (percent) for categorical variables. In univariate statistical comparisons, chi-square test or Fisher's exact test was used for categorical variables, student's t test or ANOVA for normally distributed continuous variables, Wilcoxon signed rank test or Kruskal Wallis test for not normally distributed continuous variables.
Peak intensity for metabolites were scaled and logarithmic transformed to minimize the impact of both noise and high variance of the variables. After these transformations, first, principal component analysis (PCA), an unsupervised analysis that reduces the dimension of the data, was carried out to visualize the distribution and the grouping of the samples. Then, in order to visualize group separation, partial least squares-discriminant analysis (PLS-DA) was performed. To find significantly changed metabolites between group comparisons, orthogonal projections to latent structures discriminant analysis (OPLS-DA) was performed. The metabolites with variable importance in projection (VIP) > 1 was included for further analysis. The Benjamini-Hochberg procedure was applied to control the false discovery rate (FDR) due to multiple testing [20]. The p value < 0·05 (t test or Kruskal-Wallis test) and FDR-corrected p value (q value) ≤ 0·2 were considered as significantly changed metabolites (or differential). The named differential metabolites for each comparison are shown in Datafile S1. Such selection criteria were set so the biologically valuable metabolites were not missed given the exploratory and hypothesis generating nature of this study.
MetaboAnalyst (http://www.metabanalyst.ca/) was used for pathway analysis, hierarchical clustering analysis (HCA) and correlation analysis. The differential metabolites (Datafile S1) were mapped into their biochemical pathway. Pathway significance was based on the total number of metabolites that map to a pathway and their respective significances; a pathway was considered perturbed if the number of significant metabolites (i.e., hits) was ≥ 2 and the impact value was ≥ 0·10 and raw p < 0·10. Raw p values were determined on the basis of the number of hits and total number of compounds in the pathway. An impact value ≥ 0·10 indicates that the altered pathway has a clear impact. Such lenient criteria were used because the sample size for each comparison group was small. The FDR-corrected p values were presented in Datafile S2. Selected differential metabolites involved in the major disturbed metabolic pathways were compared between groups using student t-test. Bonferroni correction was applied to correct for multiple-testing. A corrected p value ≤ 0·05 was considered statistically significant. For each metabolite, multivariable linear model was performed to adjust for age, sex and body mass index (BMI) using R 4.0.5 packages. Furthermore, HCA was performed among top 150 of 315 differential metabolites with super class category in the Human Metabolome Database (HMDB) (https://hmdb.ca/) and using the “complete” method and the “Euclidean” distance measure. Eigenmetabolite is a single value summarizing the information contained in the first principal component of PCA [21]. Finally, to evaluation the relationships between eigenmetabolite, altered metabolites, clinical parameters and classical lipids among all participants, correlation matrix was performed.
2.6. Role of the funding sponsors
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Participant characteristics
Of 322 participants, the HIV– NCs, HIV+ NCs, and HIV+ SCA were comparable in age and sex. Due to limited number of young HIV+ patients with LF, NCI or RI, the majority of them were aged over 40 years. Characteristics by study groups are shown in Table 1.
Table 1.
Characteristics of the study participants
| HIV– NCs (n = 87) | HIV+ NCs (n = 87) | HIV+ SCA (n = 88) | HIV+ NCI (n = 30) | HIV+ LF (n = 21) | HIV+ RI (n = 9) | p valuea | p valueb | |
|---|---|---|---|---|---|---|---|---|
| Demographic and clinical variables | ||||||||
| Age, years | 0·91 | <0·001 | ||||||
| 18–29 | 30 (34·5%) | 30 (34·5%) | 30 (34·1%) | 2 (6·7%) | 0 (0%) | 1 (11·1%) | ||
| 30–44 | 43 (49·4%) | 45 (51·7%) | 43 (48.9%) | 14 (46·7%) | 6 (28·6%) | 4 (44·4%) | ||
| 45–65 | 14 (16·1%) | 12 (13·8%) | 15 (17·0%) | 14 (46·7%) | 15 (71·4%) | 4 (44·4%) | ||
| mean±SD | 35·2±9·6 | 34·9±9·9 | 35·5 ±10·0 | 45·1±10·3 | 50·7±8.4 | 42·7±8·8 | 0·81 | <0·001 |
| Male | 64 (73·6%) | 65 (74·7) | 65 (66·7) | 22 (73·3) | 13 (61·9%) | 6 (66·7%) | 0·86 | 0·72 |
| Years of education | 12 (9–15) | 9 (8–13) | 9 (9–12) | 6 (5–6) | 6 (5–8) | 9 (8–9) | 0·032 | <0·001 |
| Current smoker | 33 (37.9%) | 23 (26·4%) | 21 (23·9%) | 8 (26·7%) | 6 (28·6%) | 2 (22·2%) | 0·11 | 0·96 |
| BMI, kg/m2 | 23·8±3·3 | 21·7±2·9 | 22·2±2·9 | 21·4±2·4 | 21·4±1·9 | 23·1±3·0 | <0·001 | 0·35 |
| Waist to hip circumference ratio | 0·87±0·06 | 0·88±0·06 | 0·89±0·07 | 0·90±0·09 | 0·90±0·08 | 0·92±0·03 | 0·38 | 0·27 |
| Resting heart rate (n = 243), beats/min | 71·6±10·0 | 79·4±14·5 | 81·8±14·6 | 77·7±22·9 | 81·4±13·6 | 70·3±9·7 | <0·001 | 0·33 |
| SBP, mmHg | 122·2±15·6 | 121±14·2 | 124·6±18·3 | 121·5±13·4 | 126·1±11·1 | 130·2±20·3 | 0·82 | 0·38 |
| DBP, mmHg | 76·1±11·4 | 75·5±10·2 | 78·0±11·3 | 73·2±8·0 | 78·9±8·8 | 79·5±11·6 | 0·73 | 0·11 |
| Framingham risk score, % | 3·0 (1·3–7·5) | 2·7 (1·1–5·7) | 2·3 (1·2–6·4) | 7.1 (3·3–16·0) | 11·1 (4·1–13·9) | 6·2 (4·1–12·0) | 0·28 | <0·001 |
| Carotid IMT, mm | 0·54±0·11 | 0·55±0·11 | 0·94±0·21 | 0·56±0·12 | 0·68±0·14 | 0·67±0·15 | 0·53 | <0·001 |
| HDL cholesterol, mmol/L | 1·1±0·2 | 1·1±0·3 | 1·1±0·3 | 1·1±0·3 | 1·3±0·4 | 1·0±0·3 | 0·15 | 0·13 |
| LDL cholesterol, mmol/L | 2·8±0·7 | 2·4±0·6 | 2·4±0·8 | 2·4±0·7 | 2·7±0·8 | 2·6±0·8 | <0·001 | 0·51 |
| TG, mmol/L | 2·3±1·5 | 2·3±2·9 | 2·6±2·2 | 2·2±2·1 | 1·4±0·7 | 3·3±1·6 | 0·90 | 0·21 |
| TC, mmol/L | 5·0±0·9 | 4·8±1·1 | 4·9±1·1 | 5·1±1·1 | 4·9±1·0 | 5·2±1·2 | 0·10 | 0·62 |
| ALT, IU/L | 20·2±7·1 | 22·8±14·2 | 23·0±17·2 | 19·6±11·1 | 19·9±17.0 | 18·7±14·1 | 0·13 | 0·70 |
| AST, IU/L | 20·6±5·5 | 26·0±8·8 | 27·4±13·1 | 28·8±8·9 | 47·8±24·0 | 27.3±11·1 | <0·001 | <0·001 |
| AAR | 1·08±0·24 | 1·37±0·56 | 1·45±0·60 | 1·84±0·89 | 3·11±1·37 | 1·87±1·02 | <0·001 | <0·001 |
| FIB-4 | <0·001 | |||||||
| < 1·45 (Class 1) | NA | 87 (100%) | 88 (100%) | 10 (35·7%) | 0 (0%) | 4 (44·4%) | ||
| 1·45–<3·25 (Class 2) | NA | 0 (0%) | 0 (0%) | 18 (64·3%) | 0 (0%) | 5 (55·6%) | ||
| ≥ 3·25 (Class 3) | NA | 0 (0%) | 0 (0%) | 0 (0%) | 21 (100%) | 0 (0%) | ||
| eGFR, mL/min/1·73 m2 | 1·000 | <0·001 | ||||||
| ≥ 90 | 87 (100%) | 87 (100%) | 88 (100%) | 17 (56·7%) | 21 (100%) | 0 (0%) | ||
| 60–<90 | 0 (0%) | 0 (0%) | 0 (0%) | 13 (43·3%) | 0 (0%) | 0 (0%) | ||
| < 60 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 9 (100%) | ||
| MMSE score | 29·0±1·4 | 28·7±1·8 | 28·5±2·0 | 18·4±3·1 | 27·5±2·4 | 27.1±2·1 | 0·33 | <0·001 |
| HIV-specific variables | ||||||||
| Years since HIV diagnosis | NA | 3·7±2·6 | 4·2±2·6 | 3·6±3·2 | 4·1±3·6 | 5·9±3·4 | 0·21 | |
| Years since ART initiation | NA | 2·9±2·1 | 3·5±2·3 | 2·7±2·5 | 3·5±2·7 | 4·3±2·8 | 0·20 | |
| Current CD4 count, cells/μL | NA | 526 (388–642) | 539 (422–714) | 496 (393–584) | 331 (264–570) | 484 (418–569) | ||
| Current ARV regimens | NA | <·001 | ||||||
| EFV/3TC/AZT | NA | 35 (41·2%) | 43 (48·9%) | 10 (33·3%) | 9 (42·9%) | 3 (33·3%) | ||
| NVP/3TC/AZT | NA | 15 (17·7%) | 21 (23·9%) | 2 (6·7%) | 6 (28·6%) | 1 (11·1%) | ||
| EFV/3TC/TDF | NA | 26 (30·6%) | 17 (19·2%) | 13 (43·3%) | 5 (23·8%) | 1(11·1%) | ||
| NVP/3TC/TDF | NA | 4 (4·7%) | 3 (3·4%) | 2 (6·7%) | 1 (4·8%) | 0 (0%) | ||
| TDF/3TC/LPV/r | NA | 3 (3·5%) | 1 (1·1%) | 2 (6·7%) | 0 (0%) | 3 (33·3%) | ||
| AZT/3TC/LPV/r | NA | 1 (1·2%) | 3 (3·4%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| NVP/TDF/AZT | NA | 1 (1·2%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Current HIV viral suppression | NA | 87 (100%) | 88 (100%) | 30 (100%) | 21 (100%) | 9 (100%) | 1·000 |
Values are mean ± SD, median (interquartile range) or n (%).
Abbreviations: AAR=ALT to AST ratio; ALT=alanine aminotransferase; AST=aspartate aminotransferase; AZT=lamivudine; BMI=body mass index; carotid IMT=carotid artery intima-media thickness; DBP=diastolic blood pressure; EFV=efavirenz; eGFR=estimated glomerular filtration rate; FIB-4=fibrosis 4; NC=normal controls; HDL=high-density lipoprotein; LD=low-density lipoprotein; LF=liver fibrosis; LPV/r=ritonavir boosted lopinavir; MMSE=Mini-mental State Examination; NVP=nevirapine; NA=not available/applicable; NCI=neurocognitive impairment; RI=renal impairment; SBP=systolic blood pressure; SCA=subclinical carotid atherosclerosis; TC=total cholesterol; TDF=tenofovir disoproxil fumarate; TG=triglyceride.
p values represent chi-square (categorical), Student's t or Mann-Whitney U (continuous) comparisons between HIV+ NCs and HIV‒ NCs.
p values represent chi-square test (categorical), ANOVA or Kruskal-Wallis (continuous) comparisons across HIV+ NCs and four HIV+ NCDs.
3.2. Altered metabolites and metabolic pathways across HIV– NCs, HIV+ NCs and HIV+ NCDs
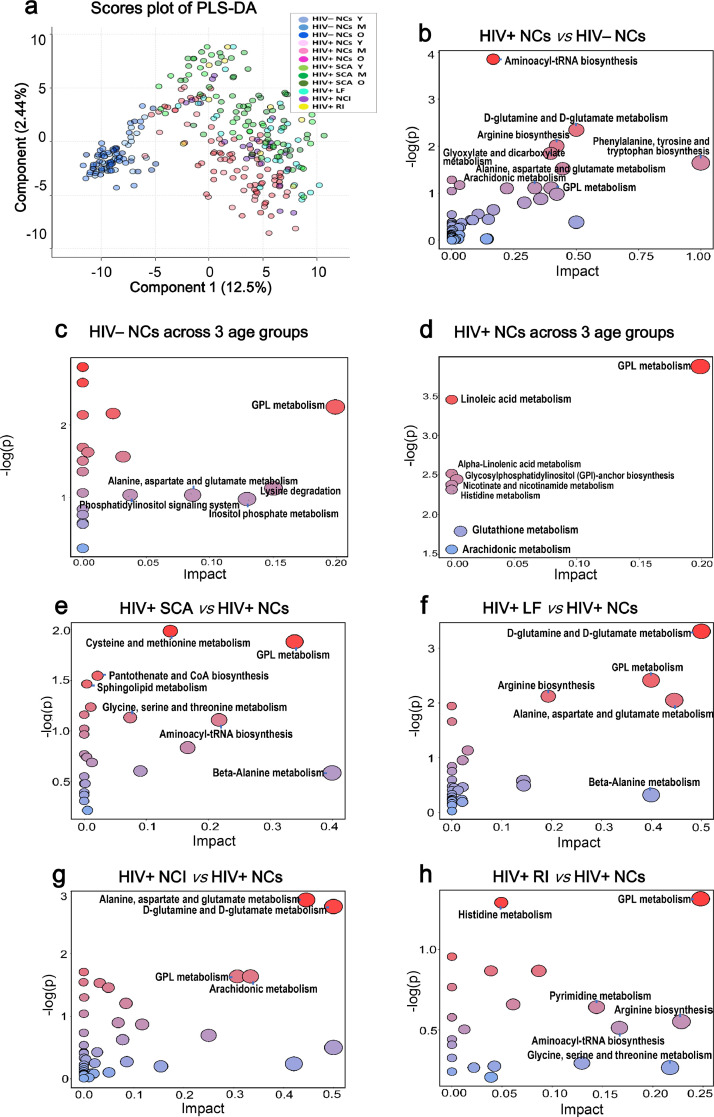
PCA was first performed for visualizing the metabolic separation across HIV– NCs, HIV+ NCs and HIV+ NCDs (Fig. S1). Then, PLS-DA results across all groups were analyzed and showed that significant separations were observed between HIV+ NCs vs HIV– NCs, and between four HIV+ NCDs vs HIV+ NCs (Fig. 2a). The OPLS-DA model were subsequently used to characterize the metabolic disturbances. The PCA score plots, OPLS-DA score plots and volcano plots are shown in Fig. S2. Here, stepwise comparisons were conducted for identifying the common metabolic pathways underlying these NCDs related to HIV infection. First, HIV– NCs and HIV+ NCs across 3 age groups were performed to identify metabolic pathways related to the normal aging process in those with and without HIV infection; second, HIV+ NCs vs HIV– NCs was to identify those related to HIV infection; third, each of HIV+ NCDs vs HIV+ NCs was to identify those related to HIV+ NCDs.
Fig. 2.
PLS-DA score plots and disturbed metabolic pathways across groups. (a) PLS-DA score plot across HIV– NCs, HIV+ NCs and HIV+ NCDs. The disturbed metabolic pathways showed various metabolism changes when comparing (b) HIV– NCs across age groups; (c) HIV– NCs across age groups; (d) HIV+ NCs vs HIV– NCs; (e) HIV+ SCA vs HIV+ NCs; (f) HIV+ LF vs HIV+ NCs; (g) HIV+ NCI vs HIV+ NCs; (h) HIV+ RI vs HIV+ NCs. Y indicates age range of 18–29 years; M indicates 30–44 years; O indicates 45–65 years.
The perturbed metabolic pathways are presented in Fig. 2b–h and Data file S2. For HIV– NCs across age groups, metabolism changed for glycerophospholipid (GPL); for HIV+ NCs across age groups, GPL, for HIV+ NCs vs HIV– NCs, phenylalanine-tyrosine-tryptophan biosynthesis, D-glutamine and D-glutamate, alanine-aspartate-glutamate, arginine biosynthesis, GPL, glyoxylate-dicarboxylate, arachidonic acid, histidine, and aminoacyl-tRNA biosynthesis; for HIV+ SCA vs HIV+ NCs, GPL, cysteine-methionine, and glycine-serine-threonine; for HIV+ LF vs HIV+ NCs, D-glutamine and D-glutamate, alanine-aspartate-glutamate, GPL, and arginine biosynthesis; for HIV+ NCI vs HIV+ NCs, D-glutamine and D-glutamate, alanine-aspartate-glutamate, arachidonic acid, and GPL; for HIV+ RI vs HIV+ NCs, GPL. Notably, GPL metabolism was the only disturbed pathway presented in all above comparisons, and thus emerged as the common metabolic disturbance in the normal aging process and linking HIV infection to NCDs. Furthermore, D-glutamine and D-glutamate metabolism, and alanine-aspartate-glutamate metabolism were the two disturbed pathways also presented in comparisons between HIV+ NCs vs HIV– NCs as well as HIV+ LF and HIV+ NCI vs HIV+ NCs (Fig. 2).
A sensitivity analysis comparing HIV+ NCI vs HIV+ NCs by excluding NCI with moderate fibrosis was conducted, the metabolism changed for the arachidonic acid, D-glutamine and D-glutamate, and alanine, aspartate and glutamate (Fig. S3 and Data file S2). Of note, GPL metabolism did not meet the criteria for disturbed metabolic pathway, possibly due to the small sample size.
3.3. Relative changes in individual metabolites across HIV– NCs, HIV+ NCs and HIV+ NCDs
To visualize relative changes across groups in individual metabolites involved in the major common disturbed pathways, autoscaled values for selected metabolites are presented in Fig. 3 and grouped into 3 categories: top 20 differential metabolites in GPL (Fig. 3a); 18 differential GPL-derived lipid mediators (Fig. 3b); 8 differential metabolites in D-glutamine and D-glutamate and alanine-aspartate-glutamate (Fig. 3c).
Fig. 3.
Relative changes in selected individual metabolites across HIV– NCs, HIV+ NCs and HIV+ NCDs. (a) Top 20 differential metabolites involved in GPL metabolism based on PLS-DA VIP values. PLS-DA was performed among all selected significantly changed GPLs across groups (as in Fig. 1a); (b) Differential PUFAs and PUFA-derived lipid mediators. Color coding on the left represents each biosynthetic precursor. (c) Differential metabolites involved in D-glutamine and D-glutamate metabolism, and alanine-aspartate-glutamate metabolism. Data are normalized values via log-transformation and autoscaling and presented as box that show the median and the 25th and 75th percentiles as the boxes and the mean values as the horizontal line. Corrected p value after Bonferroni correction for multiple testing was calculated (Table S1). A corrected p value < 0•05 was considered statistically significant. †p < 0•1, *p < 0•05, ⁎⁎p < 0•01, and ⁎⁎⁎p < 0•001. Age-, sex-, BMI- adjusted p values are presented in Table S2. Abbreviations: AA, arachidonic acid; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; EpOME, epoxyoctadecenoic acid; GPE, glycerophosphoethanolamine; GPL, glycerophospholipid; HDoHE, hydroxydocosahexaenoic acid; HEPE, hydroxyeicosapentaenoic acid; HETE, hydroxyeicosatetraenoic acid; HODE, hydroxyoctadecadienoic acid; HOTrE, hydroxyoctadecatrienoic acid; LA, linoleic acid; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine; OxoODE, oxo-octadecadienoic acid; PAF, platelet-activating factor; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PUFA, polyunsaturated fatty acid. O- indicates that the sn-1 position is ether-linked; P- indicates a plasmalogen form (sn-1 or sn-2 vinyl ether linkage).
Among changing metabolites, the major metabolites that relative abundance showed a decreasing trend from HIV– NCs to HIV+ NCs and then to HIV+ NCDs were mostly plasmalogen phosphatidylcholines (PCs) and phosphatidylethanolamines (PEs), arachidonic acid (AA), D-glutamine, L-glutamine and L-asparagine. The major metabolites that relative abundance showed an increasing trend from HIV– NCs to HIV+ NCs and then to HIV+ NCDs were platelet-activating factor (PAF), glycerophosphoethanolamine (GPE), LysoPC, LysoPE, ester PCs, and lipid mediators derived from arachidonic acid, linoleic acid (LA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) as well as L-glutamic acid and N-acetyl-L-aspartic acid. Notably, above changes in HIV+ SCA was mainly observed for age 45–65 years only (Fig. 3 and Table S1). Similar results were found with additional adjustment for age, sex, and BMI (Table S2).
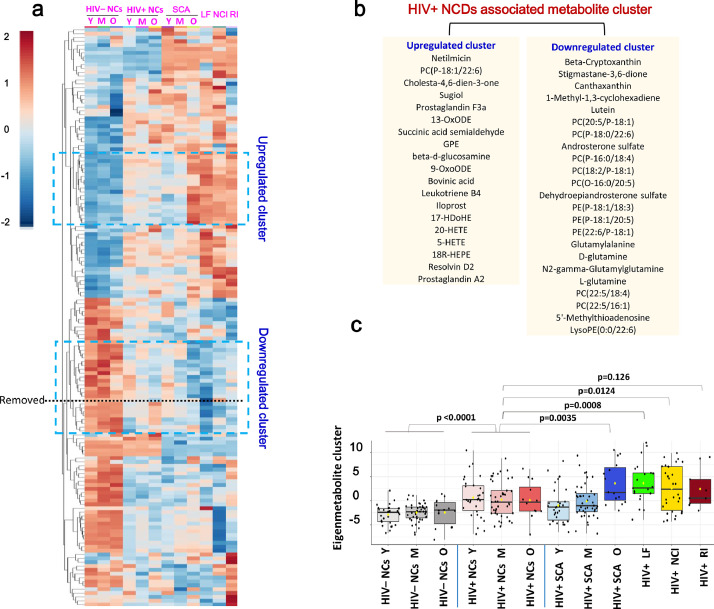
3.4. Blood metabolite signature for NCDs in HIV infection
We wondered whether any combination(s) of blood metabolites could reduce the dimension of the dataset and serve as a signature of NCDs in HIV infection. HCA based on top 150 differential metabolites provided intuitive visualizations of trends across groups in metabolites (Fig. 4a). We focused on the clusters which showed either an upward or a downward trend from HIV– NCs to HIV+ NCs and then to HIV+ NCDs, and belongs to organic acids and derivatives or lipids and lipid-like molecules. Two clusters emerged: the first composing of 19 metabolites (mainly PUFA-derived lipid mediators) showed an increasing trend, whereas the second composing of 23 metabolites (mainly plasmalogen PCs and PEs, D-glutamine, L-glutamine) showed a decreasing trend (Fig. 4b). Most of these metabolites were overlapped with that listed in Fig. 3. Then, we computed an eigenmetabolite (i.e., the eigenvector of the first principal component of the 42 metabolites) and presented the eigenmetabolite values progressively changed from HIV– NCs to HIV+ NCs and then to HIV+ NCDs (Table S3 and Fig. 4c). The crude and age-, sex- and BMI- adjusted p values were presented in Table S4. We further conducted the sensitivity analysis by using the combinations of different clusters, consisting of 38 upregulated, 34 downregulated, and 72 both upregulated and downregulated metabolites, respectively (Fig. S4). The results were not changed much, but the eigenmetabolite values of 42 metabolites and 72 metabolites changed similarly and more progressively across groups compared to the other two combinations (Tables S3 and S4).
Fig. 4.
Identification of a unique HIV+ NCD-associated blood metabolite signature. (a) Hierarchical clustering analysis of top 150 of 315 differential metabolites according to PLS-DA VIP values. PLS-DA was performed among all selected significantly changed (differential) metabolites with super class category in the Human Metabolome Database (HMDB) (https://hmdb.ca/). Red is increasing, blue is decreasing across each row. Here, the definition for differential metabolite was same (as in Fig. 2); (b) corresponding metabolites in upregulated cluster and downregulated cluster circled in (a), and 3 metabolites marked in red color are possibly drugs or food additives; (c) The eigenmetabolite values of 42 metabolites across groups. The eignemetabolite values were compared using student's t test.
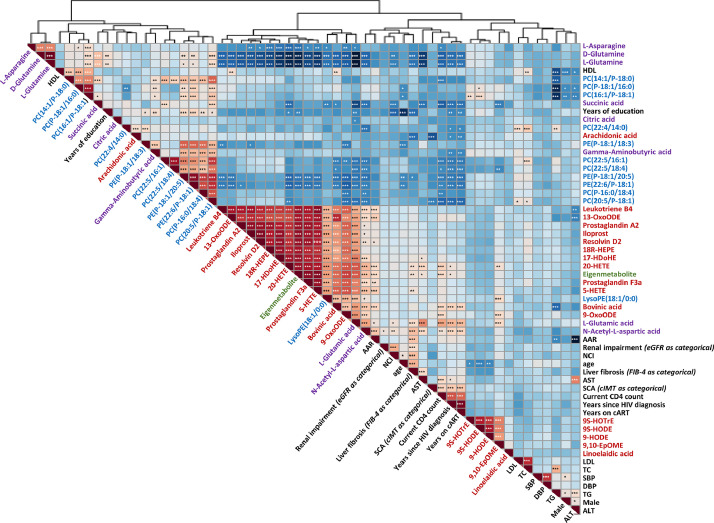
3.5. Correlation between eigenmetabolite, metabolites, clinical parameters and classical lipids
We conducted the correlation matrix between eigenmetabolite, altered metabolites, clinical parameters and classical lipids among all participants. Metabolites that were increased or decreased across groups tended to cluster together in the correlation matrix (Fig. 5). Eigenmetabolite, L-glutamic acid, and PUFA-derived lipid mediators were clustered together. High-density lipoprotein (HDL) cholesterol was clustered together with the downward metabolites such as plasmalogen PCs, plasmalogen PEs, L-glutamine and D-glutamine. Total cholesterol, triglyceride, low-density lipoprotein (LDL) cholesterol, systolic blood pressure and diastolic blood pressure were clustered together with LA-derived lipid mediators such as 9(S)-hydroxyoctadecatrienoic acid (HOTrE), 9(S)-hydroxyoctadecadienoic acid (HODE), 9-HODE and 9(10)-epoxyoctadecenoic acid (EpOME). In addition, age, years since HIV diagnosis, years being on ART, and current CD4 count were clustered together with N-acetyl-L-aspartic acid.
Fig. 5.
Spearman correlation matrix between eigenmetabolite, altered metabolites, classical lipids, and clinical parameters in all participants. Shades of blue indicate increasing positive correlation coefficient; shades of red indicate increasing negative correlation coefficient. Variables were ordered using hierarchical clustering analysis. The threshold after bonferroni correction for 1653 tests at *p < 0•00003025 (0•05/1,653), ⁎⁎p < 0•0000605 (0•01/1,653), and ⁎⁎⁎p < 0•000006 (0•001/1,653).
4. Discussion
This article reports the first comprehensive metabolomics study using plasma from HIV– NCs, HIV+ NCs and four types of HIV+ NCDs. HIV+ patients presented remarkable and profound alterations in cellular metabolism compared to HIV– individuals. GPL metabolism emerged as the common metabolic disturbance both in the normal aging process and linking HIV infection to NCDs, followed by glutamine/glutamate metabolism. Consistently, we identified a metabolomic fingerprint specific for HIV+ NCDs that reflecting elevated oxidative stress, containing 42-metabolites whose relative abundance showed either an upward (mainly lipid mediators) or a downward trend (mainly plasmalogen PCs and PEs, glutamine) from HIV– NCs to HIV+ NCs and then HIV+ NCDs. These are further supported by the correlation matrix indicating that metabolites that were increased or decreased across groups tended to cluster together, be related to each other and clinical parameters in expected direction.
A growing body of literature shows that disturbed GPL metabolism and altered homeostasis of GPL species play critical roles both in the normal aging process and in the etiology of NCDs [22], [23], [24]. HIV infection and subsequent ART have been both associated with changes in lipid profiles [25]. Recent studies have linked disturbed GPL metabolism to left ventricular diastolic dysfunction [9], abnormal lipidomic profiles to progression of SCA [11] and frailty [12] in HIV infection. Extending these prior reports, we included four types of HIV+ NCDs in comparison to HIV+ NCs and HIV– NCs. This offers an opportunity to identify the common metabolic pathways underlying the aging process and linking HIV to these NCDs. Beyond indicating that GPL metabolism was disturbed in HIV– NCs and HIV+ NCs across age groups and between HIV+ NCs vs HIV– NCs, we found that GPL metabolism was disturbed in each of the four major HIV+ NCDs vs HIV+ NCs, suggesting its key role in the aging process and pathophysiology of NCDs in treated HIV infection.
PCs and PEs are the most abundant GPLs in mammals which provide the majority of membrane lipids within cells. Our data further showed that the key metabolites showing significant differences in relative abundance between HIV+ NCs vs HIV– NCs, and between each of HIV+ NCDs vs HIV– NCs were PC and PE species. In our study, a number of plasmalogen PCs and PEs, such as PC (P-16:0/18:4), PE (P-18:1/20:5) that have been associated with healthy aging [22] and negatively associated with age-related diseases [26] were downregulated in HIV+ NCs and particularly in HIV+ NCDs. Plasmalogens are naturally occurring ether phospholipid species that are primarily presented as PC and PE species [27]. They contain a vinyl-ether bond at the sn-1 position of glycerol backbone and acyl-linked bond at the sn-2 position. Plasmalogens play a crucial role as endogenous antioxidants, protecting other phospholipids, lipid and lipoprotein particles from oxidative stress [24]. This is due to the fact that the vinyl ether bond is particularly susceptible to oxidation by reactive oxygen species (ROS) and thus protect the PUFA present in the sn-2 oxidation position [27]. Notable, we also identified several lipids species, such as PC (18:2/P-18:1) and PC (20:5/P-18:1), that contain vinyl-ether bond at the sn-2 position of glycerol backbone but have been rarely reported, showed similar changing trend across groups as classical plasmalogens, which requires further investigation. Plasmalogen levels decrease with aging, positively correlate with HDL and negatively correlate with LDL [27], as our data showed. In contrast, PAF, GPE, PC (18:0/P-16:0), LysoPC (20:0), LysoPE (18:1/0:0) and ester PCs that have been linked to age-related diseases [26,28]were upregulated in HIV+ NCs and particularly in HIV+ NCDs. It has been reported that lysoPCs (20:0) containing saturated fatty acids are pro-inflammatory, whereas PUFA-containing LysoPCs are anti-inflammatory [29]. These data reflect an elevated oxidative stress in HIV infection especially those with HIV+ NCDs.
Notably, GPLs are also a source of physiologically active compounds. GPL-derived PUFAs such as AA, LA, EPA and DHA are used to produce lipid mediators. In contrast to EPA and DHA giving rise to anti-inflammatory mediators such as resolvins and protectins and their precursors 18R-HEPE and 17-HDoHE [30], AA- and LA-derived pro-inflammatory eicosanoids such as leukotrienes and prostaglandins are well-known mediators of inflammation and oxidative stress [31], which have been recognized to play critical roles in the pathogenesis of aging and NCDs [32]. Interests in lipid mediators have recently grown rapidly and have been linked to various NCDs. A notable finding was that the relative abundance of most AA- and LA-derived lipid mediators, along with 18R-HEPE and 17-HDoHE, were higher among HIV+ NC vs HIV– NCs, and HIV+ NCDs vs HIV+ NCs. Although some anti-inflammatory lipid mediators were also elevated, the presence of pro-inflammatory lipid mediators in greater numbers may still reflect the inflammation and oxidative stress experienced in HIV+ patients especially those with NCDs. Stability of these mediators are better further evaluated in longitudinal design. Interestingly, for HIV+ SCA, above elevations in lipid mediators or depressions in ether lipids were observed at ages 45–60 years only, whereas 9-HODE and 9(S)-HOTrE were elevated at ages18–29 years only. HODEs are stable oxidation products, usually increased when oxidative stress is increased [33], and has been linked to atherosclerosis in the general population [34]. Thus, there is a possibility that HODEs may play a more important role in early onset of atherosclerosis. Additional disturbed metabolic pathways related to HIV infection and further NCDs in HIV infection were D-glutamine and D-glutamate metabolism and alanine-aspartate-glutamate metabolism. Both were disturbed between HIV+ NCs vs HIV– NCs, and between HIV+ NCI and HIV+ LF vs HIV– NCs. HIV infection causes long-lasting activation of the immune system along with increasing energy expenditure, resulting in widespread changes in cell metabolism and amino acid levels [6,35,36].
We observed an increase in relative abundance of glutamic acid and aspartic acid but a decrease in glutamine among HIV+ NCs vs HIV– NCs, consistent with previous literature [37]. Glutamate, the key excitatory neurotransmitter, is responsible for maintaining cognitive function and neuronal plasticity. Glutamate metabolism has been linked to HIV-associated NCI [38]. Liver is the site of protein and amino acid metabolism. Consistent with research in HIV-negative population [39], we observed that D-glutamine and D-glutamate metabolism, and alanine-aspartate-glutamate metabolism were disturbed in HIV+ LR vs HIV+ NCs. Interestingly, glutamine is a precursor for glutathione, and thus a decrease in glutamine may also indicate an elevated oxidative stress.
Emerging evidence shows that HIV+ patients, despite virally suppressed, experience chronic inflammation, immune activation, and elevated oxidative stress compared to the general population, resulting in accelerated aging and increasing risk of NCDs [2,3]. In aggregate, above findings point to GPLs, GPL-derived lipid mediators and glutamine/glutamate as the potential key targets to study the molecular mechanisms in the aging process and linking HIV infection to NCDs via oxidative stress and inflammation. Targeting the modulation of lipid specifies are active areas of study [25,27], some agents such as statin having being successfully applied in clinically settings for decades. Of note, glutamine supplementation has also been applied to maintain high levels of glutathione and to avoid oxidative stress. A therapy like this or as a part of multipronged therapeutic approach may have a promise to prevent or treat the NCDs in HIV infection as well as in the general population.
Several limitations should be considered. First, from statistical perspective, group sizes for HIV+ LF and RI are relatively small. Second, NCI was defined according to MMSE rather than a full battery of neurocognitive tests or clinical diagnosis. Third, using the cut-off points for signifying SCA cases vs controls might introduce non-differential misclassifications which could bias the association toward the null, especially in the youngest age strata of cases vs controls (≥ 0·78 vs < 0·78 mm), but should not be a major issue in the oldest age strata of cases vs controls (> 1·00 vs < 0·78 mm). Forth, we cannot exclude the possibility that the results may be affected by confounding that we cannot account for. For example, diet composition can affect plasma metabolite profiles but such data were not assessed. In addition, our findings on changed metabolites and identified signature for HIV-associated NCDs needs to be better confirmed using the longitudinal design.
In summary, our data highlights several metabolic shifts progressively changes from HIV– NCs to HIV+ NCs and then HIV+ NCDs. GPL metabolism emerges as the common metabolic disturbance in the normal aging process as well as linking HIV to NCDs, and the latter is also closely linked to glutamine and glutamate metabolism. Subsequent analysis in individual metabolites corroborate these findings and identify a metabolomic fingerprint containing metabolites whose relative abundance shows either an upward (mainly GPL-derived lipid mediators) or a downward trend (mainly plasmalogen PCs and PEs, glutamine) from HIV– NCs to HIV+ NCs and then NCDs, reflecting an increased inflammation and oxidative stress state across groups. In aggregate, our data point to GPL, glutamine and glutamate metabolisms and related metabolites including plasmalogens, lipid mediators and glutamine as the potential key targets to study the common molecular mechanisms linking HIV to NCDs via inflammation and oxidative stress, and further uncover new avenues of therapeutic opportunities. Further research is needed to validate and extend these findings using longitudinal multi-omics.
Contributors
NH contributed to study concept. YD contributed to data analysis, manuscript draft, and revision. HL and XC contributed to supervising subject enrollment and sample collection. XC, BZ, XX, and XX contributed to data collection. SW and MG contributed to performing the experiment. NH contributed to review and editing. YD and NH verified the underlying data.
All authors critically reviewed and edited the manuscript and consented to final publication. YD and NH had full access to all the data and had final responsibility for the decision to submit for publication.
Data sharing statement
All the data of this study are free to obtain in the paper or in the supplementary materials. Raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of Competing Interest
We declared no competing interests.
Acknowledgments
This study was supported by the China National Science and Technology Major Projects on Infectious Diseases (2018ZX10721102-004), the National Natural Science Foundation of China (81872671, 81773485), and partially supported by Yi-wu Institute of Fudan University and Shanghai Municipal Health Commission (GWV-10.1-XK16). We thank Zhonghui Ma (Fudan) for the sketch in central illustration; Shanghai Biotree Biotech Co., Ltd for LC-MS/GC-TOF-MS analysis; Chunyan He (Fudan) for assisting with boxplot illustration.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103548.
Appendix. Supplementary materials
References
- 1.Rasmussen L.D., May M.T., Kronborg G. Time trends for risk of severe age-related diseases in individuals with and without HIV infection in Denmark: a nation-wide population-based cohort study. Lancet HIV. 2015;2:e288–e298. doi: 10.1016/S2352-3018(15)00077-6. [DOI] [PubMed] [Google Scholar]
- 2.De Francesco D., Wit F.W., Burkle A. The Co-mor-Bidity in relation to AC. Do people living with HIV experience greater age advancement than their HIV-negative counterparts? AIDS. 2019;33:259–268. doi: 10.1097/QAD.0000000000002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furman D., Campisi J., Verdin E. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabuzda D., Jamieson B.D., Collman R.G. Pathogenesis of aging and age-related comorbidities in people with HIV: highlights from the HIV ACTION Workshop. Pathog Immun. 2020;5:143–174. doi: 10.20411/pai.v5i1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sáez-Cirión A., Sereti I. Immunometabolism and HIV-1 pathogenesis: food for thought. Nat Rev Immunol. 2021;21:5–19. doi: 10.1038/s41577-020-0381-7. [DOI] [PubMed] [Google Scholar]
- 7.Palmer C.S., Henstridge D.C., Yu D. Emerging role and characterization of immunometabolism: relevance to HIV pathogenesis, serious non-AIDS events, and a cure. J Immunol. 2016;196:4437–4444. doi: 10.4049/jimmunol.1600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams A.A., Sitole L.J., Meyer D. HIV/HAART-associated oxidative stress is detectable by metabolomics. Mol Biosyst. 2017;13:2202–2217. doi: 10.1039/c7mb00336f. [DOI] [PubMed] [Google Scholar]
- 9.Bravo C.A., Hua S., Deik A. Metabolomic profiling of left ventricular diastolic dysfunction in women with or at risk for HIV infection: the Women's interagency HIV study. J Am Heart Assoc. 2020;9 doi: 10.1161/JAHA.119.013522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassol E., Misra V., Dutta A., Morgello S., Gabuzda D. Cerebrospinal fluid metabolomics reveals altered waste clearance and accelerated aging in HIV patients with neurocognitive impairment. AIDS. 2014;28:1579–1591. doi: 10.1097/QAD.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai J.C., Deik A.A., Hua S. Association of lipidomic profiles with progression of carotid artery atherosclerosis in HIV infection. JAMA Cardiol. 2019;4:1239–1249. doi: 10.1001/jamacardio.2019.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeoh H.L., Cheng A.C., Cherry C.L. Immunometabolic and lipidomic markers associated with the frailty index and quality of life in aging HIV+ men on antiretroviral therapy. EBioMedicine. 2017;22:112–121. doi: 10.1016/j.ebiom.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Justice A.C., Erlandson K.M., Hunt P.W., Landay A., Miotti P., Tracy R.P. Can biomarkers advance HIV research and care in the antiretroviral therapy era? J Infect Dis. 2018;217:521–528. doi: 10.1093/infdis/jix586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licher S., Heshmatollah A., van der Willik K.D. Lifetime risk and multimorbidity of non-communicable diseases and disease-free life expectancy in the general population: a population-based cohort study. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002741. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H., Ding Y., Ning C. Age-specific associations between HIV infection and carotid artery intima-media thickness in China: a cross-sectional evaluation of baseline data from the CHART cohort. Lancet HIV. 2019;6:e860–e868. doi: 10.1016/S2352-3018(19)30263-2. [DOI] [PubMed] [Google Scholar]
- 16.de Groot E., Hovingh G.K., Wiegman A. Measurement of arterial wall thickness as a surrogate marker for atherosclerosis. Circulation. 2004;109:III33–III38. doi: 10.1161/01.CIR.0000131516.65699.ba. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y., Lin H., Shen W., Wu Q., Gao M., He N. Interaction effects between HIV and aging on selective neurocognitive impairment. J Neuroimmune Pharmacol. 2017;12:661–669. doi: 10.1007/s11481-017-9748-3. [DOI] [PubMed] [Google Scholar]
- 18.Sterling R.K., Lissen E., Clumeck N. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/NCV co-infection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y.C., Zuo L., Chen J.H. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol. 2006;17:2937–2944. doi: 10.1681/ASN.2006040368. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 21.Scott Chialvo C.H., Che R., Reif D., Motsinger-Reif A., Reed L.K. Eigenvector metabolite analysis reveals dietary effects on the association among metabolite correlation patterns, gene expression, and phenotypes. Metabolomics. 2016;12:167. doi: 10.1007/s11306-016-1117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Covarrubias V., Beekman M., Uh H.W. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meikle P.J., Summers S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat Rev Endocrinol. 2017;13:79–91. doi: 10.1038/nrendo.2016.169. [DOI] [PubMed] [Google Scholar]
- 24.Papsdorf K., Brunet A. Linking lipid metabolism to chromatin regulation in aging. Trends Cell Biol. 2019;29:97–116. doi: 10.1016/j.tcb.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowman E., Funderburg N.T. Lipidome abnormalities and cardiovascular disease risk in HIV infection. Curr HIV/AIDS Rep. 2019;16:214–223. doi: 10.1007/s11904-019-00442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Yu N., Zhao J. Advances in the biosynthetic pathways and application potential of plasmalogens in medicine. Front Cell Dev Biol. 2020;8:765. doi: 10.3389/fcell.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorninger F., Forss-Petter S., Wimmer I., Berger J. Plasmalogens, platelet-activating factor and beyond - ether lipids in signaling and neurodegeneration. Neurobiol Dis. 2020;145 doi: 10.1016/j.nbd.2020.105061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akerele O.A., Cheema S.K. Fatty acyl composition of lysophosphatidylcholine is important in atherosclerosis. Med Hypothese. 2015;85:754–760. doi: 10.1016/j.mehy.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis E.A., Orris P.C. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrasekaran A., Idelchik M.D., Andrés Melendez J. Redox control of senescence and age-related disease. Redox Biol. 2017;11:91–102. doi: 10.1016/j.redox.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umeno A., Sakashita M., Sugino S. Comprehensive analysis of PPARγ agonist activities of stereo-, regio-, and enantio-isomers of hydroxyoctadecadienoic acids. Biosci Rep. 2020;40 doi: 10.1042/BSR20193767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vangaveti V., Baune B.T., Kennedy R.L. Hydroxyoctadecadienoic acids: novel regulators of macrophage differentiation and atherogenesis. Ther Adv Endocrinol Metab. 2010;1:51–60. doi: 10.1177/2042018810375656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clerc I., Moussa D.A., Vahlas Z. Entry of glucose- and glutamine-derived carbons into the citric acid cycle supports early steps of HIV-1 infection in CD4 T cells. Nat Metab. 2019;1:717–730. doi: 10.1038/s42255-019-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michalek R.D., Gerriets V.A., Jacobs S.R. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitole L.J., Tugizimana F., Meyer D. Multi-platform metabonomics unravel amino acids as markers of HIV/combination antiretroviral therapy-induced oxidative stress. J Pharm Biomed Anal. 2019;176 doi: 10.1016/j.jpba.2019.112796. [DOI] [PubMed] [Google Scholar]
- 38.Vázquez-Santiago F.J., Noel R.J., Porter J.T., Rivera-Amill V. Glutamate metabolism and HIV-associated neurocognitive disorders. J Neurovirol. 2015;20:315–331. doi: 10.1007/s13365-014-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaggini M., Carli F., Rosso C. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology. 2018;67:145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.