Abstract
Diabetes disrupts mitochondrial function and often results in diabetic cardiomyopathy (DCM). Paeonol is a bioactive compound that has been reported to have pharmacological potential for cardiac and mitochondrial protection. This study aims to explore the effects of paeonol on mitochondrial disorderes in DCM and the underlying mechanisms. We showed that paeonol promoted Opa1-mediated mitochondrial fusion, inhibited mitochondrial oxidative stress, and preserved mitochondrial respiratory capacity and cardiac performance in DCM in vivo and in vitro. Knockdown of Opa1 blunted the above protective effects of paeonol in both diabetic hearts and high glucose-treated cardiomyocytes. Mechanistically, inhibitor screening, siRNA knockdown and chromatin immunoprecipitation experiments showed that paeonol-promoted Opa1-mediated mitochondrial fusion required the activation of Stat3, which directly bound to the promoter of Opa1 to upregulate its transcriptional expression. Moreover, pharmmapper screening and molecular docking studies revealed that CK2α served as a direct target of paeonol that interacted with Jak2 and induced the phosphorylation and activation of Jak2-Stat3. Knockdown of CK2α blunted the promoting effect of paeonol on Jak2-Stat3 phosphorylation and Opa1-mediated mitochondrial fusion. Collectively, we have demonstrated for the first time that paeonol is a novel mitochondrial fusion promoter in protecting against hyperglycemia-induced mitochondrial oxidative injury and DCM at least partially via an Opa1-mediated mechanism, a process in which paeonol interacts with CK2α and restores its kinase activity that subsequently increasing Jak2-Stat3 phosphorylation and enhancing the transcriptional level of Opa1. These findings suggest that paeonol or the promotion of mitochondrial fusion might be a promising strategy for the treatment of DCM.
Keywords: Diabetic cardiomyopathy, Paeonol, Mitochondrial fusion, CK2α, Stat3, Opa1
Graphical abstract
Highlights
-
•
Paeonol is identified as a novel mitochondrial fusion promoter against diabetic cardiomyopathy.
-
•
Paeonol promotes Opa1-mediated mitochondrial fusion in a Stat3-dependent transcriptional manner.
-
•
CK2α serves as a direct target of Paeonol that directly activates the Jak2–Stat3 signaling pathway.
1. Introduction
The incidence of diabetes has been rapidly increasing throughout the world. Cardiovascular complications are major causes of morbidity and mortality in diabetic patients [1]. Among these complications, diabetic cardiomyopathy (DCM) is a diabetes-associated heart disease that is independent of coronary artery disease and hypertension [2]. DCM is characterized by abnormalities in cardiac structure and function including myocardial hypertrophy, interstitial fibrosis, cardiomyocyte apoptosis, and diastolic and systolic dysfunction [3]. These characteristics of DCM lead to heart failure, contributing to increased morbidity and mortality of in patients with diabetes [4]. Currently, the availability of effective therapeutic interventions against DCM is limited.
The heart is an organ with high energy demand, largely dependent on the function of mitochondria, which provide approximately 90% energy and are a major source of reactive oxygen species (ROS) [5]. Increased mitochondrial ROS production and resultant mitochondrial dysfunction are generally recognized to play a pivotal role in the development of DCM [6,7]. Recently, it has been highlighted that imbalanced mitochondrial fusion/fission dynamics is an early component that increases mitochondrial ROS production and induces mitochondrial dysfunction [8,9]. Diabetic hearts display excessive mitochondrial fission and inhibited mitochondrial fusion. Our previous studies have found that correcting mitochondrial dynamics by moderately inhibiting mitochondrial fission or enhancing mitochondrial fusion inhibits mitochondria-derived ROS generation and mitigates mitochondrial dysfunction in diabetic hearts or high glucose-treated cardiomyocytes [10,11]. Of note, the study by Qin et al. has shown that promoting mitochondrial fusion by fusion protein Mfn2 overexpression does not impair cardiac mitochondrial function, whereas completely inhibiting Drp1-mediated mitochondrial fission impairs mitochondrial quality at baseline [12]. This suggests that mitochondrial fusion promotion is a more safe strategy than mitochondrial fission inhibition to maintain cardiac mitochondrial function. Hence, the discovery of safe and effective pharmacotherapy targeting mitochondrial fusion promotion is meaningful for the treatment of DCM.
Paeonol (Pae) is a bioactive compound extracted from the root of Paeonia albiflora and has a long history of clinical use. Currently, the dosage forms of Pae approved by the Food and Drug Administration of China for human use include tablets, injections and ointments. Oral and injection administrations of Pae are effective in relieving inflammation/pain-related diseases, such as rheumatoid arthritis, headache, and muscle pain [13]. Moreover, Pae has been reported to have great pharmacological potential for cardiac and mitochondrial protection. Indeed, it has been shown that Pae reduces ischemia-induced myocardial apoptosis by inhibiting oxidative stress [14,15] and attenuates glutamate-induced mitochondrial injury by restoring mitochondrial membrane potential and inhibiting cytochrome c release [16], suggesting Pae has a protective effect on the mitochondria. Given that the promotion of mitochondrial fusion is crucial in maintaining mitochondrial homeostasis and normal cardiac function, investigating whether Pae is an efficient mitochondrial fusion promoter in a DCM model is worthwhile. In the present study, we explored the effects of Pae on cardiac pathology and mitochondrial fusion/fission dynamics in diabetic hearts in vivo and in hyperglycemia-treated primary cardiomyocytes in vitro, with a specific focus on the molecular mechanisms of how Pae promotes mitochondrial fusion. Our results have revealed that Pae is a novel mitochondrial fusion promoter in protecting against DCM through the CK2α-Stat3-Opa1 signaling pathway.
2. Materials and methods
An extended “Materials and Methods” is provided in the Supplementary Materials and Methods.
2.1. Cardiomyocyte culture and treatment
Primary neonatal cardiomyocytes were isolated from 1-2-day-old neonatal Sprague-Dawley rats. The cardiomyocytes were subjected to normal glucose (5.5 mmol/L, NG) or high glucose (33 mmol/L, HG) challenge for 48 h with the vehicle or various concentrations of Pae supplement. Pae (catalog number: MB1762-S, purity > 98%) was obtained from Dalian Meilun Biology Technology (Dalian, China).
2.2. Assessment of mitochondrial morphology, ROS and ATP in cardiomyocytes
The mitochondria in the primary cardiomyocytes were labeled with MitoTracker Red CMXRos probe (100 mmol/L, Invitrogen, Carlsbad, USA) and imaged using a confocal laser-scanning microscope (Nikon A1R MP + Confocal Microscope). The number and volume of the mitochondria observed were analyzed and quantified as previously described [17]. After MitoTracker Red staining, flow cytometry was performed to analyze mitochondrial mass. MitoSOX staining (Invitrogen, Carlsbad, USA) was performed to evaluate mitochondrial ROS levels in cardiomyocytes. Intracellular ATP content was detected using an ATP bioluminescent assay kit (Biovision, California, USA) according to the manufacturer's protocol.
2.3. Animal experimental design and treatment
All animal experimental procedures were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals (8th Edition, 2011) and the study was approved by the Fourth Military Medical University Ethics Committee. Sixty-five male 6–8-week-old Sprague-Dawley rats were obtained and maintained under controlled conditions of temperature (22 ± 2 °C) and a 12-h light/dark cycle. Diabetes was induced by a single intraperitoneal injection of streptozotocin (STZ, Sigma-Aldrich, 65 mg/kg) dissolved in 0.1 mol/l citrate buffer (pH 4.5) [18]. Control animals received an intraperitoneal injection of vehicle (citrate buffer) only. One week after STZ injection, the rats with fasting glycemia levels ≧ 11.1 mM were considered diabetic. After confirmation of diabetes, the rats were administered with the vehicle or Pae (catalog number: MB1762, purity > 98%, Dalian Meilun Biology Technology, China) respectively for an additional 12 weeks. Pae was mixed with the vehicle (0.5% sodium carboxymethyl cellulose) and then administered to the animals by oral gastric gavage at 75, 150 or 300 mg/kg/day based on previous studies [19,20].
2.4. Prediction of potential targets of Pae and molecular docking
The SDF (Structure Data File) file of Pae was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/11092, accessed by October 15, 2020) and the effective targets were predicted using the PharmMapper Server (http://www.lilab-ecust.cn/pharmmapper/) by employing “All Targets” model [21,22]. The submission ID was recorded during the operation. Autodock Vina 1.1.2 was employed to investigate the binding affinity and binding sites between Pae and CK2α. Briefly, the SDF file of Pae was converted into PDBQT format by using AutoDock Tools. The 3D crystal structure of the CK2α kinase complex was obtained from the Protein Data Bank (PDB) (http://www.rcsb.org, PDB ID: 1M2P). The search grid of CK2 was identified as center_x: 23.331, center_y: 6.252 and center_z: 18.182 with dimensions size_x: 15, size_y: 15, and size_z: 15. The docking protocol was generated as described previously [23,24]. The Promega CK2α Kinase Assay kit was utilized to explore the effects of Pae on the activity of CK2α according to the manufacturer's instructions.
2.5. Western blot analysis
Total protein from rat hearts and the cardiomyocytes was extracted by using RIPA buffer containing protease inhibitor cocktail. The standard Western blotting method was used as previously described [25]. Detailed information for Western blotting is available in the Supplementary Materials and Methods section. The antibodies used were shown in Supplementary Table 1 in the Supplementary Materials and Methods.
2.6. Statistical analysis
All values were presented as the mean ± standard error of the mean (SEM). Data were analyzed by one-way ANOVA followed by Tukey's post-hoc test using GraphPad Prism software (version 8.0). Differences with P values less than 0.05 were considered to be statistically significant.
3. Results
3.1. Pae promoted mitochondrial fusion and improved mitochondrial function in high glucose-treated cardiomyocytes
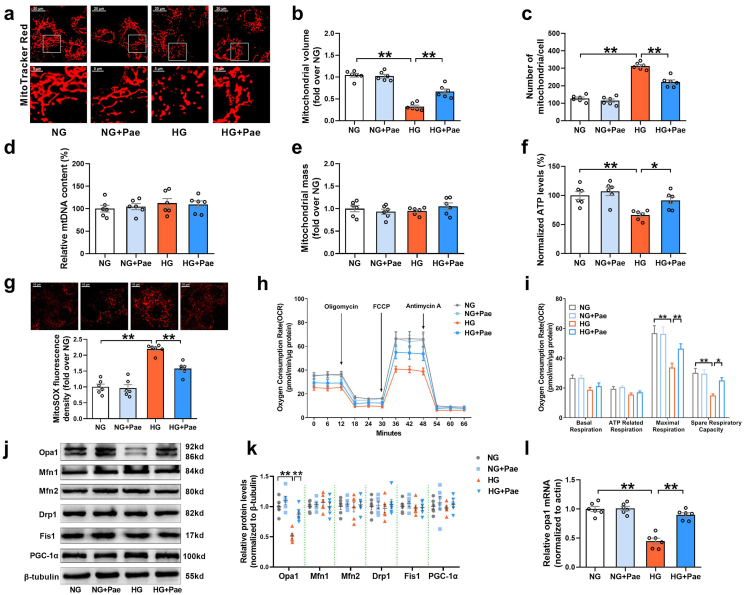
The cardiomyocytes were treated with Pae at 25, 50, 100 and 200 μmol/L for 48 h. As shown in Supplementary Fig. S1, the CCK8 assay showed that only 200 μmol/L Pae slightly inhibited cell viability, while the other concentrations of Pae had no significant effect on cell viability in the cardiomyocytes. Therefore, 100 μmol/L of Pae was considered to be the safest concentration. In light of the results from previous studies [26,27], this concentration was used to explore the effects of Pae on mitochondrial homeostasis including mitochondrial dynamics and biogenesis in primary cardiomyocytes. As shown in Fig. 1a, the mitochondria of the NG-cultured cardiomyocytes appeared as elongated tubules with highly interconnected networks. When subjected to HG medium, the mitochondria became spherical and shorter. Compared to NG-cultured cardiomyocytes, decreased mitochondrial volume and increased number of mitochondria were observed in the HG-cultured cardiomyocytes (Fig. 1a–c). Treatment with Pae resulted in a significant increase in the mitochondrial volume and a significant decrease in the number of mitochondria in the HG-cultured cardiomyocytes (Fig. 1a–c), indicating that Pae promoted mitochondrial fusion in hyperglycemia-treated primary cardiomyocytes. Mitochondrial biogenesis was determined by measuring the relative mtDNA content and the MitoTracker Red stained mitochondrial mass using quantitative real-time PCR and flow cytometry respectively. HG or Pae induced no changes in the mtDNA content and the mitochondrial mass in cardiomyocytes (Fig. 1d and e), suggesting that Pae did not enhance mitochondrial biogenesis. Moreover, compared to the NG-cultured cardiomyocytes, the HG-cultured cardiomyocytes exhibited elevated mitochondria-derived superoxide production (as detected by MitoSOX staining in Fig. 1g), reduced ATP production (Fig. 1f), and suppressed mitochondrial respiratory capacities including maximal respiration and spare respiratory capacity (Fig. 1h and i). Treatment with Pae significantly reduced mitochondria-derived superoxide production, increased ATP production, and recovered the mitochondrial respiratory capacity in the HG-treated cardiomyocytes (Fig. 1f–i).
Fig. 1.
Pae promoted Opa1-related mitochondrial fusion and enhanced mitochondrial respiratory capacity in high glucose (HG)-treated cardiomyocytes. (a) Representative confocal microscope images of mitochondrial morphology stained by MitoTracker Red. Original magnification × 600. (b) Quantification of mean mitochondrial volume. (c) Quantification of mitochondrial number per cell. (d) mtDNA content normalized to nuclear DNA content. (e) Quantification of mitochondrial mass. (f) Quantification of normalized ATP levels. (g) Representative images and quantitative analysis of MitoSOX-stained mitochondria-derived superoxide production. (h–i) Oxygen consumption rate (OCR) measured by Seahorse and quantitative statistical analysis of OCR. (j-k) Representative blots and quantitative analysis of the mitochondrial fission/fusion-related proteins including Opa1, Mfn1, Mfn2, Drp1 and Fis1. (l) Quantitative analysis of Opa1 mRNA expression determined by real-time PCR. Values are the means ± SEM. NG, normal glucose (5.5 mmol/L); HG, high glucose (33 mmol/L); Pae, paeonol (100 μmol/L). n = 6 in each group. All data are shown as means ± SEM. *P < 0.05, **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, the expression patterns of mitochondrial fission/fusion proteins and PGC-1α (a central regulator of mitochondrial biogenesis) were analyzed. Of the mitochondrial fission and fusion proteins (fission proteins: Drp1 and Fis1; fusion proteins: Mfn1, Mfn2 and Opa1), only the expression of Opa1 was significantly lower in the HG-treated cardiomyocytes than in the NG-cultured cardiomyocytes (Fig. 1j and k). Pae significantly reversed the decrease in Opa1 protein expression in the HG-cultured cardiomyocytes (Fig. 1j and k). No significant differences in PGC-1α levels were observed among the experimental groups (Fig. 1j and k). Meanwhile, Pae mitigated the HG-induced reduction in Opa1 mRNA expression (Figure 1l), suggesting that the expression of Opa1 may be regulated at the transcriptional level (mRNA level). Taken together, these results indicate that Pae had no significant effect on PGC-1α-related mitochondrial biogenesis in cardiomyocytes. Rather, Pae treatment enhanced the transcription of Opa1 and promoted mitochondrial fusion as well as mitochondrial function under hyperglycemic conditions in cardiomyocytes.
3.2. Pae-induced Opa1 upregulation was responsible for promoting mitochondrial fusion and function in HG-treated cardiomyocytes
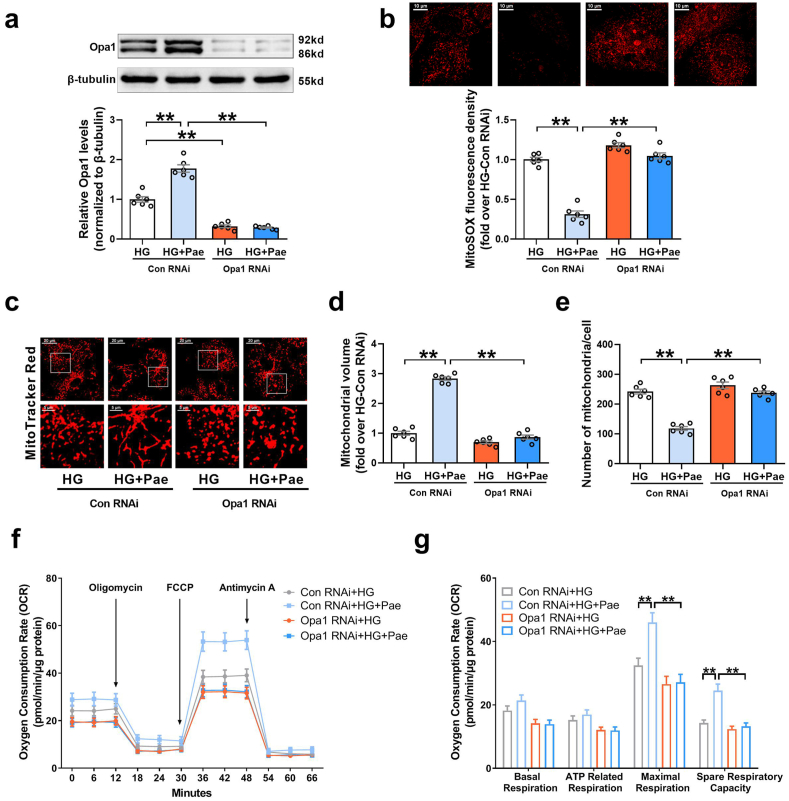
We subsequently explored whether the upregulation of Opa1 expression was responsible for the mitochondrial protective effects of Pae. As shown in Fig. 2, knockdown of Opa1 with small interfering (si)RNA blunted the inhibitory effects of Pae on mitochondrial oxidative stress (Fig. 2a–b) and largely abrogated the promoting effects of Pae on mitochondrial fusion (Fig. 2c–e) as well as mitochondrial respiratory capacity in HG-treated cardiomyocytes (Fig. 2f–g). On the other hand, upregulation of Opa1 by transfecting with the adenovirus encoding Opa1 (Ad-Opa1) alone efficiently inhibited mitochondrial oxidative stress and promoted mitochondrial fusion as well as mitochondrial respiratory capacity in HG-treated cardiomyocytes (Supplementary Fig. S2). These data indicated that the mitochondrial protective effects of Pae were largely dependent on the upregulation of Opa1.
Fig. 2.
Knockdown of Opa1 with siRNA blunted the inhibitory effects of Pae on mitochondrial oxidative stress and abrogated the promoting effects of Pae on mitochondrial fusion as well as mitochondrial respiratory capacity in high glucose (HG)-treated cardiomyocytes. (a) Representative blots and quantitative analysis of Opa1 expression. (b) Representative images and quantitative analysis of MitoSOX-stained mitochondria-derived superoxide production. (c) Representative confocal microscope images of mitochondrial morphology stained by MitoTracker Red. Original magnification × 600. (d) Quantification of mean mitochondrial volume. (e) Quantification of mitochondrial number per cell. (f–g) Oxygen consumption rate (OCR) measured by Seahorse and quantitative statistical analysis of OCR. HG, high glucose (33 mmol/L); Pae, paeonol (100 μmol/L). n = 6 in each group. All data are shown as means ± SEM. **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Pae alleviated diabetic cardiomyopathy in the rats
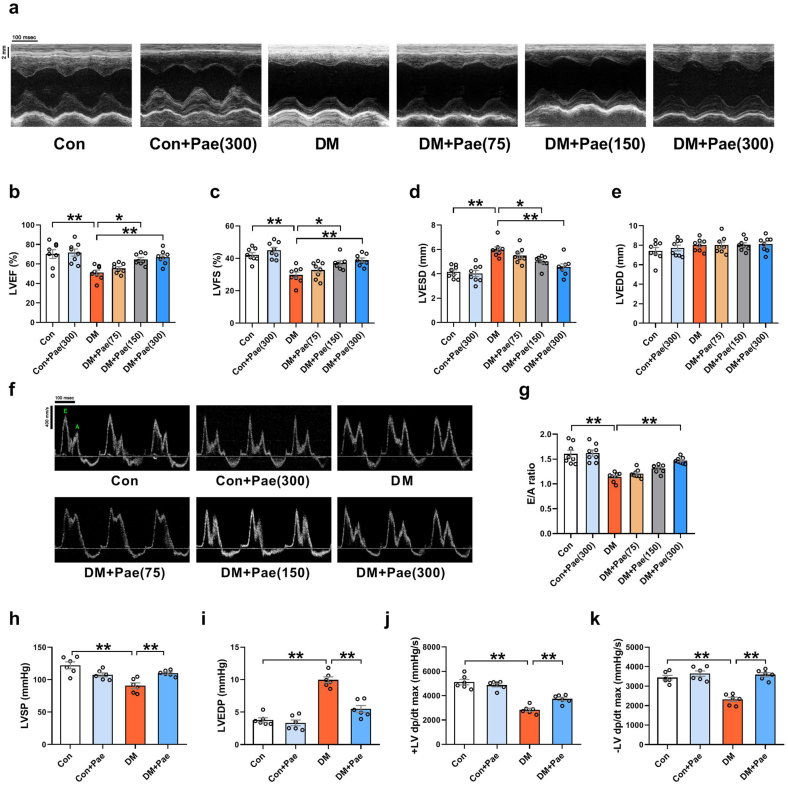
Since the data suggest that Pae was an efficient mitochondrial fusion promoter against hyperglycemia-induced mitochondrial oxidative injury in vitro, the effects of Pae on diabetic hearts were then explored in vivo. As shown in Table 1, compared with the control rats, the diabetic rats had decreased body weight as well as increased blood glucose and serum total cholesterol (TC) and triacylglycerol (TG) levels. All doses of Pae had no significant effects on body weight or blood glucose in the diabetic rats. Pae significantly reduced serum TC levels of diabetic rats at doses of 150 or 300 mg/kg/d. The diabetic rats showed decreased left ventricular ejection fraction (LVEF) and left ventricular fraction shortening (LVFS) and increased left ventricular end-systolic diameter (LVESD) (Fig. 3a–d). The diabetic rats that received Pae supplementation at doses of 150 or 300 mg/kg/d but not 75 mg/kg/d showed increased LVEF and LVFS and decreased LVESD (Fig. 3a–d). There were no significant differences in the left ventricular end-diastolic diameter (LVEDD) among all the groups (Fig. 3a and e). Moreover, the diabetic rats that received Pae treatment at a dose of 300 mg/kg/day showed improved diastolic function as evidenced by increased E/A ratio (Fig. 3f and g). A previous study has shown that Pae was detected at concentrations around 1 μg/ml (6 μmol/L) in plasma after oral administration of moutan cortex decoction (Moutan cortex is the root bark of Paeonia suffruticosa. Moutan cortex decoction is the water extraction of moutan cortex.) containing 20 mg/kg paeonol [28]. It is estimated that blood concentrations of 45 or 90 μmol/L Pae may be approximately reached after oral administration of 150 mg/kg or 300 mg/kg paeonol respectively. Since 100 μmol/L Pae was validated in our experiments on cardiomyocytes, 300 mg/kg/day Pae was considered to be the optimal dose for the animal experiments to yield similar action and was denoted as Con + Pae or DM + Pae group for subsequent experiments. Hemodynamic measurements indicated that the LVSP (left ventricular systolic pressure) and ±LV dP/dtmax (maximal and minimal first derivative of left ventricular pressure) were decreased, while the LVEDP (left ventricular end-diastolic pressure) was increased in the diabetic rats compared with the control rats (Fig. 3h–k). The diabetic rats that received Pae treatment at a dose of 300 mg/kg/day (DM + Pae) showed increased LVSP and ±LV dp/dtmax and decreased LVEDP (Fig. 3h–k).
Table 1.
Basic characteristics of animals.
| Groups | Con | Con + Pae (300 mg/kg) | DM | DM + Pae (75 mg/kg) | DM + Pae (150 mg/kg) | DM + Pae (300 mg/kg) |
|---|---|---|---|---|---|---|
| Body weight (g) | 553.4 ± 17.6 | 543.3 ± 13.9 | 299.5 ± 12.6** | 319.0 ± 11.5 | 317.8 ± 19.0 | 323.3 ± 19.5 |
| Blood glucose (mmol/L) | 5.01 ± 0.06 | 5.21 ± 0.14 | 21.3 ± 1.17** | 22.0 ± 0.46 | 22.3 ± 0.41 | 22.5 ± 0.44 |
| Serum TC (mmol/L) | 1.15 ± 0.06 | 1.31 ± 0.03 | 1.82 ± 0.08** | 1.77 ± 0.05 | 1.36 ± 0.09## | 1.16 ± 0.07## |
| Serum TG (mmol/L) | 0.40 ± 0.07 | 0.49 ± 0.07 | 1.56 ± 0.40** | 1.56 ± 0.12 | 1.37 ± 0.14 | 0.91 ± 0.13 |
Values presented are mean ± SEM. Con, control; DM, diabetes mellitus; TC, total cholesterol; TG, triacylglycerol. n = 8. **P < 0.01 vs. Con; ##P < 0.01 vs.DM.
Fig. 3.
Pae attenuated diabetes-induced cardiac functional abnormalities in rats. Control non-diabetic or diabetic rats were treated with Pae for 12 weeks and their cardiac function was analyzed. (a) Representative echocardiography images. (b-e) Quantitative analysis of echocardiography data including left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), left ventricular end-systolic diameter (LVESD) and Left ventricular end-diastolic diameter (LVEDD). (f-g) Representative pulse-wave Doppler images and quantitative analysis of the E/A ratio. DM, diabetes mellitus; Pae (75, 150, 300), paeonol at a dosage of 75, 150 or 300 mg/kg/day respectively; n = 8 in each group. (h-k) Quantitative analysis of hemodynamic data including LV systolic pressure (LVSP), LV end-diastolic pressure (LVEDP) and the maximal and minimal first derivative of LV pressure (±LV dP/dtmax). Pae, paeonol at a dosage of 300 mg/kg/day; n = 6 in each group. All data are shown as means ± SEM. *P < 0.05, **P < 0.01.
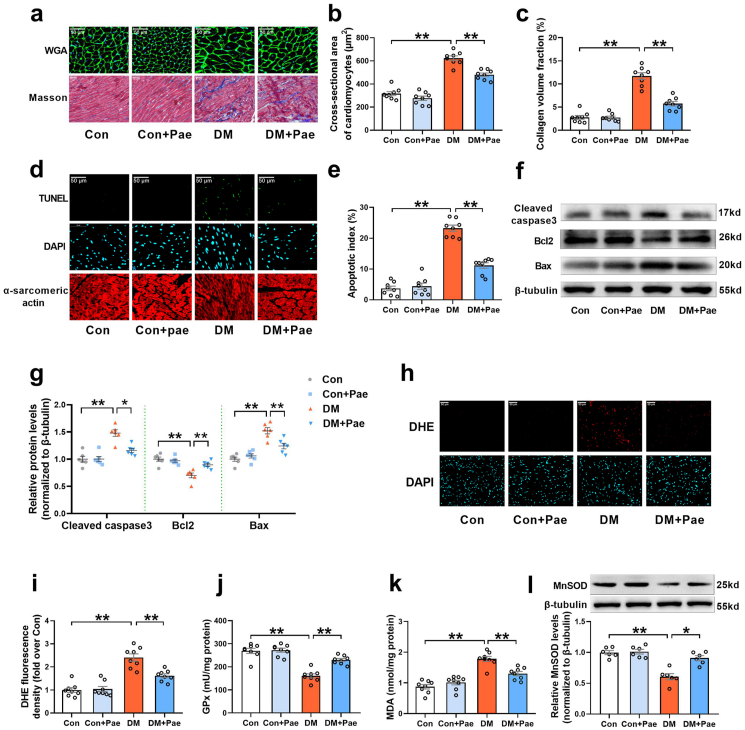
Cardiac hypertrophy and interstitial fibrosis are important characteristic changes in DCM. Compared with the control rats, the diabetic rats showed significant cardiac hypertrophy and interstitial fibrosis, as evidenced by increased cardiomyocyte cross-sectional area and collagen volume fraction (Fig. 4a–c). Treatment with Pae significantly inhibited cardiac hypertrophy and interstitial fibrosis in the diabetic rats, as evidenced by decreased cardiomyocyte cross-sectional area and reduced collagen volume fraction (Fig. 4a–c). Oxidative stress and cardiomyocyte apoptosis have been implicated in the pathogenesis of DCM. As expected, there were increased cardiomyocyte apoptosis (indicated by apoptosis index and the expression of apoptosis-related proteins including cleaved caspase 3, Bax and anti-apoptosis protein Bcl2 in Fig. 4d–g) and oxidative stress (indicated by DHE staining, MnSOD expression and the GPx and MDA content in Fig. 4h-l) in the diabetic hearts compared with the control hearts. Pae treatment significantly reduced cardiomyocyte apoptosis and inhibited myocardial oxidative stress in the diabetic hearts (Fig. 4d-l). These results indicated that Pae mitigated diabetes-induced oxidative stress and apoptosis and improved cardiac structure and function in diabetic animals.
Fig. 4.
Pae alleviated cardiac structural abnormalities and inhibited cardiomyocyte apoptosis and oxidative stress in diabetic hearts. (a) Representative images of cardiomyocyte size and interstitial fibrosis obtained using wheat germ agglutinin (WGA) staining and Masson trichrome staining respectively. Original magnification × 400. (b) Quantification of average cross-sectional area of cardiomyocytes. (c) Interstitial fibrosis was quantified as collagen volume fraction. (d) Representative images of TUNEL and DAPI staining of heart tissues. Original magnification × 800. (e) Percentage of apoptotic cells quantified as apoptotic index. (f, g) Representative blots and quantitative analysis of cleaved caspase 3, Bcl2, Bax and β-tubulin. (h-i) Representative images and quantitative analysis of DHE-stained heart tissues. Original magnification × 800. (j) Glutathione peroxidase (GPx) activity. (k) Malondialdehyde (MDA) content. (l) Representative blots and quantitative analysis of Mitochondrial manganese superoxide dismutase (MnSOD). DM, diabetes mellitus; Pae, paeonol at a dosage of 300 mg/kg/day. n = 8 for animal experiments (Figure a–e, h-k) and n = 6 for Western blot experiments (Figure f–g, l). All data are shown as means ± SEM. *P < 0.05, **P < 0.01.
3.4. Pae promoted mitochondrial fusion and the restoration of mitochondrial function in diabetic hearts
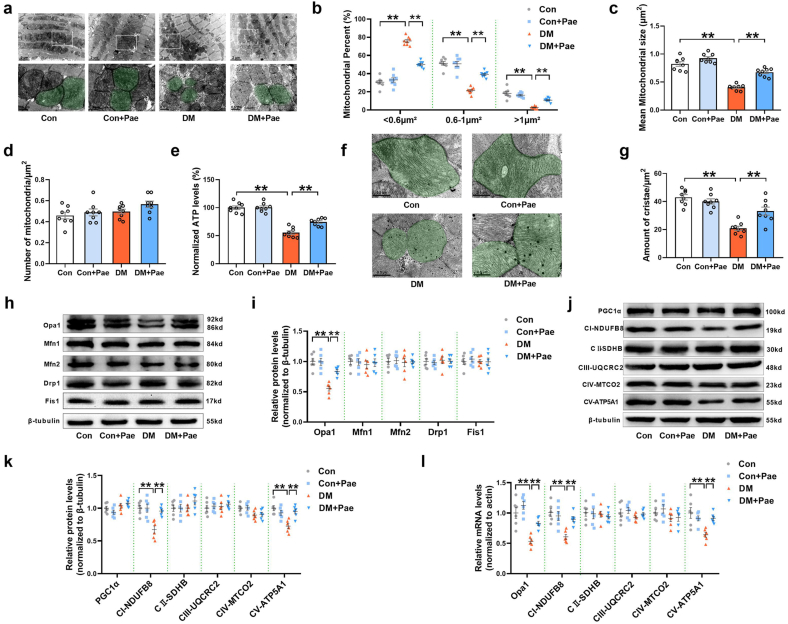
The effects of Pae on mitochondrial dynamics were further validated in vivo. Mitochondrial size and number were determined to evaluate the fusion of mitochondria. In the diabetic hearts, the percentage of mitochondria with sizes <0.6 μm2 was markedly higher, while the percentage of mitochondria with sizes of 0.6–1 μm2 or >1 μm2 was significantly lower than that in the control hearts (Fig. 5a and b). Moreover, the mean mitochondrial size was smaller in the diabetic hearts than in the control hearts (Fig. 5a and c). Pae treatment promoted mitochondrial fusion in the diabetic hearts, as reflected by the reduced percentage of mitochondria with sizes <0.6 μm2, an elevated percentage of mitochondria with sizes of 0.6–1 μm2 or >1 μm2, and increased mean mitochondrial size (Fig. 5a, b, and 5c). No significant differences in the number of mitochondria per μm2 were observed among the experimental groups (Fig. 5d). Pae treatment reversed the decrease in ATP levels in the diabetic hearts (Fig. 5e). Moreover, cristae density was significantly reduced in the diabetic hearts compared with that in the control hearts. Pae treatment increased the cristae density in the diabetic hearts (Fig. 5f–g).
Fig. 5.
Pae promoted Opa1-related mitochondrial fusion and restored mitochondrial function in diabetic hearts. (a) Representative images of mitochondrial morphology obtained by transmission electron microscope. Mitochondria are shown in green. Scale bars = 1 μm. Original magnification × 15,000. (b) Mitochondria were classified into three categories based on size (<0.6 μm2, 0.6–1 μm2, >1 μm2). (c) Quantification of mean mitochondrial size. (d) Quantification of mitochondrial number per μm2. (e) Quantification of normalized ATP levels. (f) Representative images of mitochondrial cristae obtained by transmission electron microscope. Mitochondria are shown in green. Scale bars = 0.5 μm. Original magnification × 40,000. (g) Quantification of cristae amount per mitochondrial area. (h-i) Representative blots and quantitative analysis of mitochondrial fission/fusion-related proteins including Opa1, Mfn1, Mfn2, Drp1 and Fis1. (j-k) Representative blots and quantitative analysis of PGC-1α and mitochondrial respiratory chain complex I–V (CI–V). (l) Quantitative analysis of Opa1 and CI–V mRNA expression determined by real-time PCR. DM, diabetes mellitus; Pae, paeonol at a dosage of 300 mg/kg/day. n = 8 for animal experiments (Figure a–g) and n = 6 for molecular biology experiments (Figure h–l). All data are shown as means ± SEM. **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, the expression patterns of mitochondrial fission/fusion proteins, PGC-1α and mitochondrial respiratory chain complex I–V were analyzed. Of the mitochondrial fission and fusion proteins, only the expression of fusion-related protein Opa1 was also significantly lower in the diabetic hearts than in the control hearts (Fig. 5h and i). Pae administration attenuated the decrease in Opa1 expression (Fig. 5h and i). No significant differences in PGC-1α and mitochondrial complexes II, III, IV (CII, CIII, CIV) were observed among the experimental groups (Fig. 5j and k). The expressions of mitochondrial complexes I and V were significantly reduced in the diabetic hearts, both of which were partially restored by Pae treatment (Fig. 5j and k). Pae also reversed the decrease in the mRNA expression of Opa1, complex I and V in the diabetic hearts (Figure 5l). Together, these data indicated that Pae promoted Opa1-related mitochondrial fusion and mitochondrial function in diabetic hearts.
3.5. Knockdown of Opa1 with AAV-Opa1 shRNA blunted the protective effects of Pae in diabetic hearts
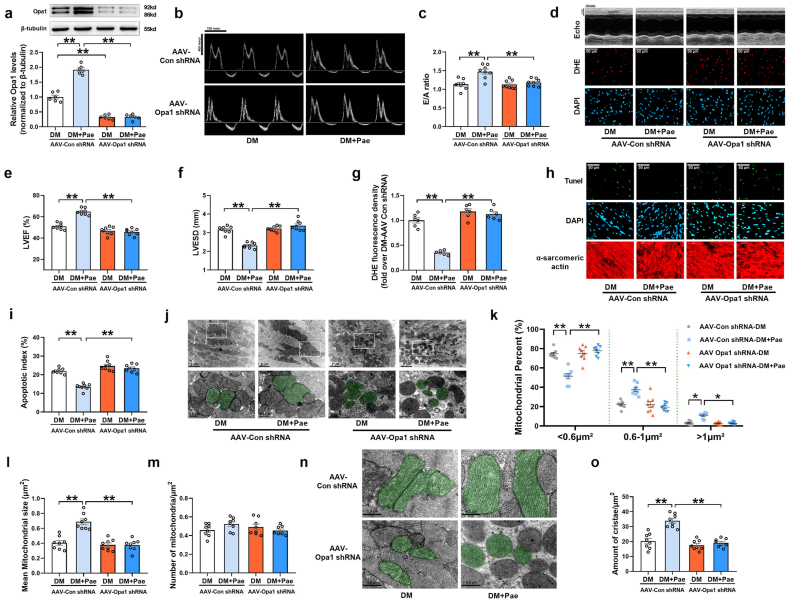
To explore whether Opa1 is essential for the cardiac protective effects of Pae in vivo, adeno-associated virus 9-harboring Opa1 miRNA backbone-based shRNA (AAV-Opa1 shRNA) construct was used to knockdown Opa1 in the diabetic hearts. Since virus transfection efficiency is relatively low in rat hearts, mouse hearts were then used for Opa1 gene silencing experiments. As shown in Fig. 6a, AAV-Opa1 shRNA significantly reduced Opa1 protein expression by ~70% in the diabetic hearts. Pae improved diastolic function as evidenced by the increased E/A ratio in the diabetic hearts receiving control shRNA (Fig. 6b and c). Moreover, Pae increased LVEF, decreased LVESD, inhibited myocardial oxidative stress and suppressed apoptosis in the diabetic hearts receiving Control shRNA (Fig. 6d–i), whereas all the effects were blunted in Opa1 knockdown hearts (Fig. 6b–i). Pae promoted mitochondrial fusion and increased cristae density in the diabetic hearts receiving control shRNA, as reflected by the reduced percentage of mitochondria with sizes <0.6 μm2, elevated percentage of mitochondria with sizes of 0.6–1 μm2 or >1 μm2, larger mean mitochondrial size and increased amount of cristae per mitochondrial area (Fig. 6j-o). Knockdown of Opa1 in the diabetic hearts largely abrogated the promoting effects of Pae on mitochondrial fusion and cristae density (Fig. 6j-o). These results indicated that Pae exerted its cardiac protective effects mainly via an Opa1-dependent way in the diabetic hearts.
Fig. 6.
Knockdown of Opa1 with AAV-Opa1 shRNA blunted the cardioprotective effects of Pae in diabetic hearts. (a) Representative blots and quantitative analysis of Opa1 expression. (b-c) Representative pulse-wave Doppler images and quantitative analysis of the E/A ratio. (d) Upper: Representative echocardiography images. Bottom: Representative images of DHE-stained heart tissues. Original magnification × 800. (e-f) Quantitative analysis of echocardiography data including left ventricular ejection fraction (LVEF) and left ventricular end-systolic diameter (LVESD). (g) Quantitative analysis of DHE staining. (h) Representative images of TUNEL and DAPI staining of the heart tissues. Original magnification × 800. (i) Percentage of apoptotic cells quantified as apoptotic index. (j) Representative images of mitochondrial morphology obtained by transmission electron microscope. Mitochondria are shown in green. Scale bars = 1 μm. Original magnification × 15,000. (k) Mitochondria were classified into three categories based on their size (<0.6 μm2, 0.6–1 μm2, >1 μm2). (l) Quantification of mean mitochondrial size. (m) Quantification of mitochondrial number per μm2. (n) Representative images of mitochondrial cristae obtained by transmission electron microscope. Mitochondria are shown in green. Scale bars = 0.5 μm. Original magnification × 40,000. (o) Quantification of cristae amount per mitochondrial area. DM, diabetes mellitus; Pae, paeonol at a dosage of 300 mg/kg/day. n = 6 for Western blot and DHE experiments (Figure a, g) and n = 8 for animal experiments (Figure b–o). All data are shown as means ± SEM. *P < 0.05, **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Pae upregulated the expression of Opa1 via Stat3-mediated transcription
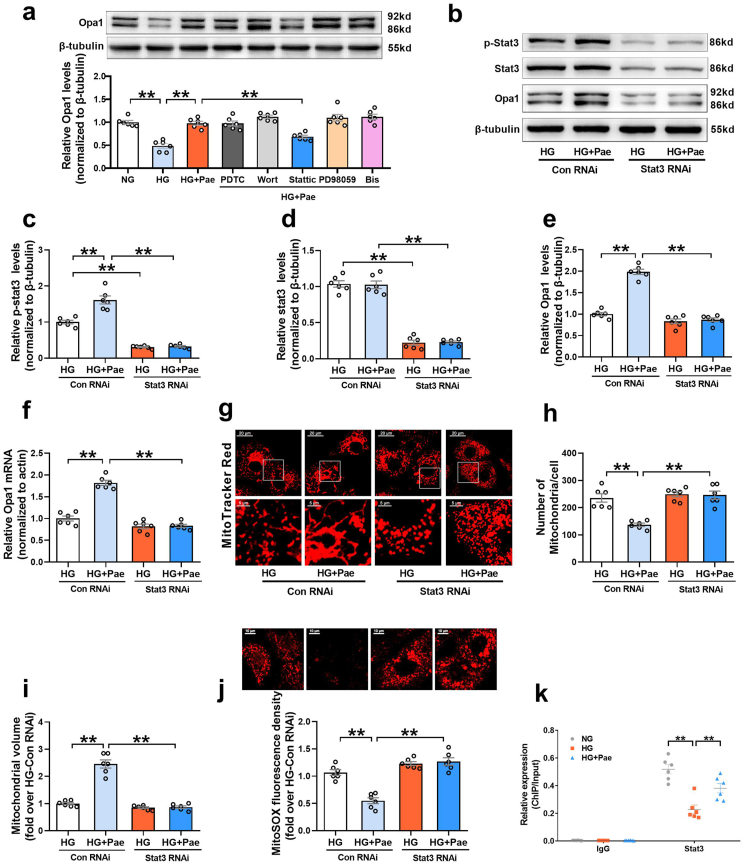
Li et al. has shown that Pae activates the PI3K signaling pathway in diabetic hearts [29]. To identify the signaling pathway that mediates the Pae-induced upregulation of Opa1, several specific inhibitors including wortmannin (Wort, a PI3K inhibitor) were used to screen for potential signaling pathways. As shown in Fig. 7a, the cardiomyocytes in the HG + Pae group were pretreated with the following agents: pyrrolidine dithiocarbamate (PDTC, an NF-κB inhibitor, 100 μmol/L, MedChem Express) [30], wortmannin (Wort, 0.1 μmol/L, MedChem Express) [31,32], Stattic (a Stat3 inhibitor, 10 μmol/L, MedChem Express) [33,34], PD98059 (a MEK inhibitor, 20 μmol/L, MedChem Express) [35], and bisindolylmaleimide I (Bis, a PKC inhibitor, 10 μmol/L, MedChem Express) [36,37]. None of these inhibitors affected the expression of Opa1 in the HG-cultured cardiomyocytes in the absence of Pae (Supplementary Fig. S3). Pae-induced upregulation of Opa1 in the HG-cultured cardiomyocytes was largely blunted by Stattic pretreatment, whereas it was unaffected by the other inhibitors, indicating that the activation of Stat3 may be involved in the upregulation of Opa1 expression induced by Pae.
Fig. 7.
Pae promoted Opa1-mediated mitochondrial fusion and inhibited mitochondrial oxidative stress in a Stat3-dependent way. (a) Several specific pharmacological inhibitors, including pyrrolidine dithiocarbamate (PDTC, an NF-κB inhibitor), wortmannin (Wort, a PI3K inhibitor), Stattic (a Stat3 inhibitor), PD98059 (a MEK inhibitor) and bisindolylmaleimide I (Bis, a PKC inhibitor), were pre-administered to the cardiomyocytes in the HG + Pae group and then the expression of Opa1 was quantified. (b-f) Stat3 was knocked down by siRNA, after which the cells were subjected to high glucose (HG) with or without paeonol (Pae). The protein expression of phosphorylated Stat3 (p-Stat3), total Stat3 and Opa1 were quantified in (b-e). The mRNA expression of Opa1 was quantified in (f). (g) Representative confocal microscope images of mitochondrial morphology stained by MitoTracker Red. Original magnification × 600. (h) Quantification of mitochondrial number per cell. (i) Quantification of mean mitochondrial volume. (j) Representative images and quantitative analysis of MitoSOX-stained mitochondria-derived superoxide production. (k) Chromatin immunoprecipitation (ChIP) and real-time PCR analysis for the binding of Stat3 to Opa1 promoter. n = 6 in each group. HG, high glucose (33 mmol/L); Pae, paeonol (100 μmol/L). All data are shown as means ± SEM. **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
SiRNA was used to further explore whether Stat3 was responsible for regulating Opa1-mediated mitochondrial fusion in response to Pae treatment. As shown in Fig. 7b–f, knockdown of Stat3 by siRNA blocked the upregulating effects of Pae on phosphorylated Stat3 (p-Stat3 Tyr705, an active form of Stat3), total Stat3, Opa1 protein and mRNA, implying that Stat3 modulates the expression of Opa1 at the level of transcription. Moreover, Stat3 siRNA blunted the promoting effects of Pae on mitochondrial fusion (Fig. 7g–i) and largely abrogated the inhibitory effects of Pae on mitochondrial oxidative stress (Fig. 7j). ChIP-PCR analysis revealed that Stat3 directly bound to the promoter of Opa1, while the binding was significantly decreased under HG conditions. Pae treatment significantly restored the binding of Stat3 to the Opa1 promoter in HG-treated cardiomyocytes (Fig. 7k). These data suggest that Pae promoted Opa1-mediated mitochondrial fusion and inhibited mitochondrial oxidative stress mainly in a Stat3-dependent transcriptional manner.
3.7. Pae increased Stat3 phosphorylation and promoted Opa1-mediated mitochondrial fusion via the CK2α-Jak2 signaling pathway
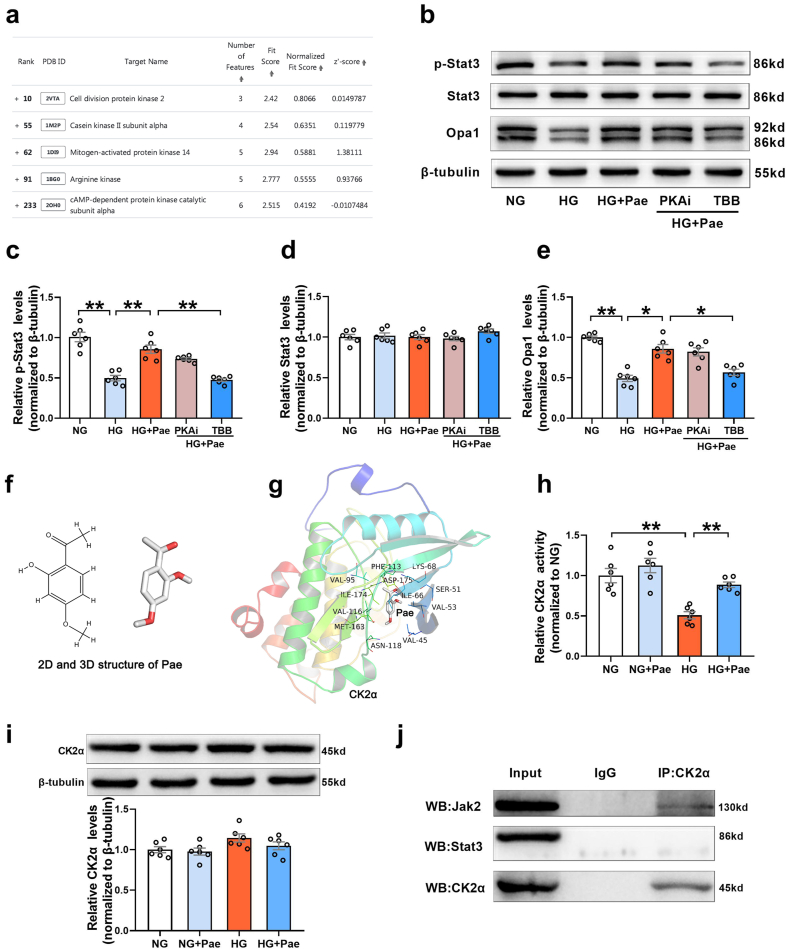
Pae is a highly liposoluble compound with a low molecular weight, which facilitates its easy transportation to the brain [38]. It is speculated that Pae can traverse the cell membrane and exert direct action on some adaptor molecules. To reveal the direct target of Pae, the SDF structure of Pae was submitted to the Pharmmapper selecting “All Targets” Model, and the top 300 potential targets ranked by the normalized fit score in descending order were listed (job ID: 201215135510). The detailed results were uploaded as a separate file (Table 2). Since Stat3 phosphorylation is usually regulated by kinases, the function annotations containing “Involved in kinase activity” were selected and five candidates were identified, as seen in Fig. 8a, which were “Cell division protein kinase 2” (Rank 10), “Casein kinase II subunit alpha” (Rank 55), “Mitogen-activated protein kinase 14” (Rank 62), “Arginine kinase” (Rank 91) and “cAMP-dependent protein kinase catalytic subunit alpha” (Rank 233). According to the information provided by Universal Protein Resource (UniProt), cell division protein kinase 2 is involved in the control of the cell cycle; Mitogen-activated protein kinase 14 is one of the four p38 MAPKs which have been implicated in apoptosis induction; Arginine kinase catalyzes the reversible phosphorylation between phosphoarginine and ADP. It seemed that the biological functions of these three kinases did not match the mitochondrial protective effects of Pae. Hence, CK2α (casein kinase II subunit alpha) and PKA Cα (cAMP-dependent protein kinase catalytic subunit alpha) were considered to be the most likely target candidates. A selective PKA inhibitor (PKA Inhibitor IV, 5 μmol/L, Santa Cruz) [39] and CK2α inhibitor (TBB, 10 μmol/L, MedChem Express) [40] were then used to identify whether CK2α or PKA Cα was involved in the upregulation of Stat3 phosphorylation and Opa1 expression. As shown in Fig. 8b–e, the Pae-induced increase in Stat3 phosphorylation and Opa1 expression was blunted by TBB but not PKA inhibitor (PKAi), suggesting that CK2α seemed to be the most likely target candidate of Pae in the activation of Stat3-Opa1 signaling.
Table 2.
Top 300 potential targets of Pae predicted by PharmMapper.
Fig. 8.
Pae interacted with CK2α and restored the activity of CK2α in high glucose (HG)-treated cardiomyocytes. (a) Listing of potential candidates involved in kinase activity. (b-e) PKA inhibitor (PKAi) or CK2α inhibitor (TBB) was pre-administered to the cardiomyocytes in the HG + Pae group and then the expression of phosphorylated Stat3 (p-Stat3), total Stat3 and Opa1 were quantified. (f) The 2D and 3D structure of Pae. (g) Binding mode between Pae and CK2α. CK2α is represented with the cartoon, and the representative binding residues are indicated by lines. (h) Relative CK2α activity. (i) Representative blots and quantitative analysis of CK2α expression. (j) Interaction between CK2α and Jak2 or Stat3 determined by co-immunoprecipitation. NG, normal glucose (5.5 mmol/L); HG, high glucose (33 mmol/L); Pae, paeonol (100 μmol/L). n = 6 in each group. All data are shown as means ± SEM. *P < 0.05, **P < 0.01.
The interaction between Pae and CK2α was further explored using computation docking. The 2D and 3D structure of Pae were exhibited in Fig. 8f. The AutoDock Vina binding test indicated that Pae might favorably bind to CK2α (maximum binding affinity −6.4 kcal/mol, Fig. 8g). The cellular experimental study showed that Pae reversed HG-induced inhibition of CK2α activity, although Pae did not affect the expression of CK2α in HG-treated cardiomyocytes (Fig. 8h and i). Pretreatment with the antioxidants NAC (N-acetyl-l-cysteine, 0.5 mmol/L) or Tempol (4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl, 10 μmol/L) increased the activity of CK2a in HG-cultured cardiomyocytes. Pae further elevated CK2α activity following pretreatment with the antioxidants (Supplementary Fig. S4). Furthermore, co-immunoprecipitation experiments indicated that Jak2, an upstream activator of Stat3, co-precipitated with endogenous CK2α, whereas Stat3 did not (Fig. 8j). This suggested that CK2α may directly interact with Jak2 and subsequently induce the activation of Jak2-Stat3 signaling.
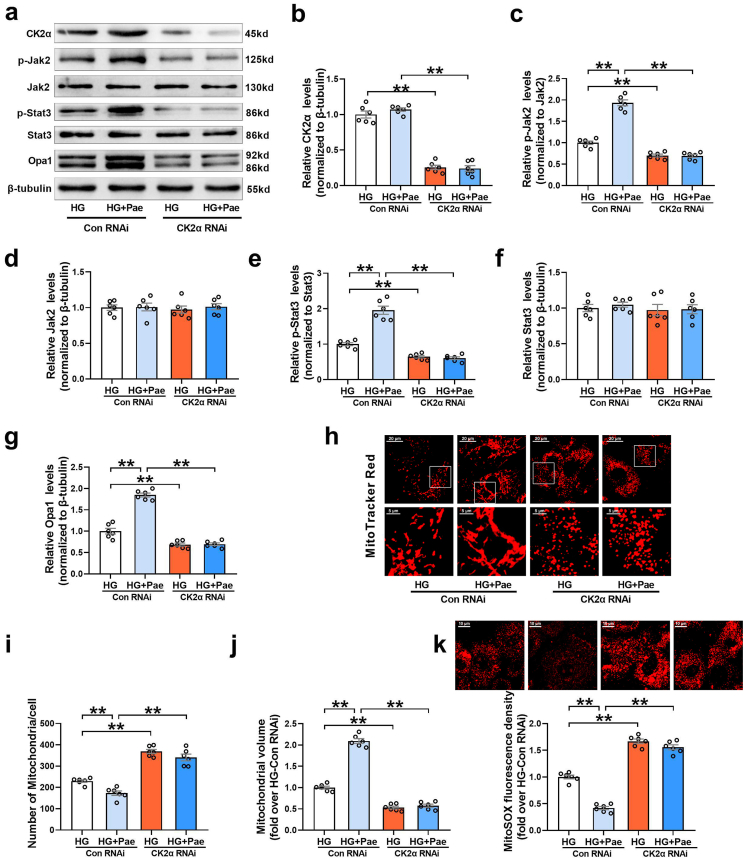
Finally, we investigated whether CK2α was responsible for enhanced Jak2-Stat3 signaling and Opa1-mediated mitochondrial fusion in response to Pae treatment. As shown in Fig. 9, knockdown of CK2α with siRNA further inhibited the phosphorylation of Jak2 and Stat3 (Fig. 9a–f), suppressed Opa1-mediated mitochondrial fusion (Fig. 9g–j) and increased mitochondrial oxidative stress (Fig. 9k) in HG-treated cardiomyocytes. Moreover, CK2α siRNA blunted the upregulating effects of Pae on Jak2 and Stat3 phosphorylation and Opa1-mediated mitochondrial fusion (Fig. 9a–j) as well as largely blocked the inhibitory effects of Pae on mitochondrial oxidative stress (Fig. 9k) in cardiomyocytes. The animal experimental study validated that Pae restored the activity of CK2α and increased Jak2 and Stat3 phosphorylation in the diabetic hearts (Supplementary Fig. S5). These results above indicated that CK2α served as a direct target of Pae that activated the Jak2–Stat3 signaling pathway and promoted Opa1-mediated mitochondrial fusion.
Fig. 9.
Knockdown of CK2α with siRNA blunted the promoting effect of Pae on Jak2 and Stat3 phosphorylation and Opa1-mediated mitochondrial fusion in high glucose (HG)-treated cardiomyocytes. (a-g) Representative blots and quantitative analysis of CK2α, phosphorylated Jak2 (p-Jak2), Jak2, phosphorylated Stat3 (p-Stat3), Stat3 and Opa1. (h) Representative confocal microscope images of mitochondrial morphology stained by MitoTracker Red. Original magnification × 600. (i) Quantification of mitochondrial number per cell. (j) Quantification of mean mitochondrial volume. (k) Representative images and quantitative analysis of MitoSOX-stained mitochondria-derived superoxide production. HG, high glucose (33 mmol/L); Pae, paeonol (100 μmol/L). n = 6 in each group. All data are shown as means ± SEM. **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
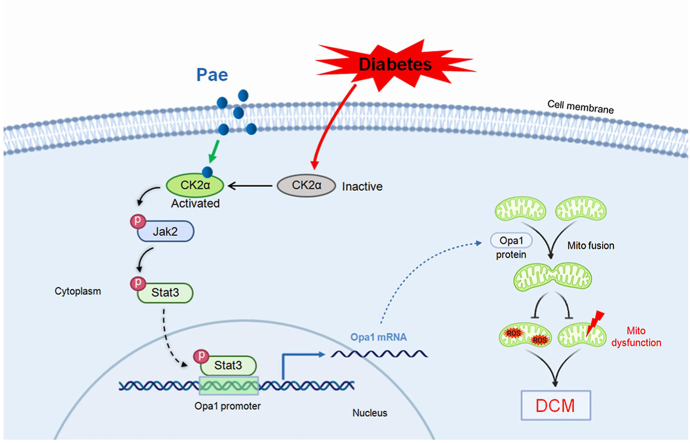
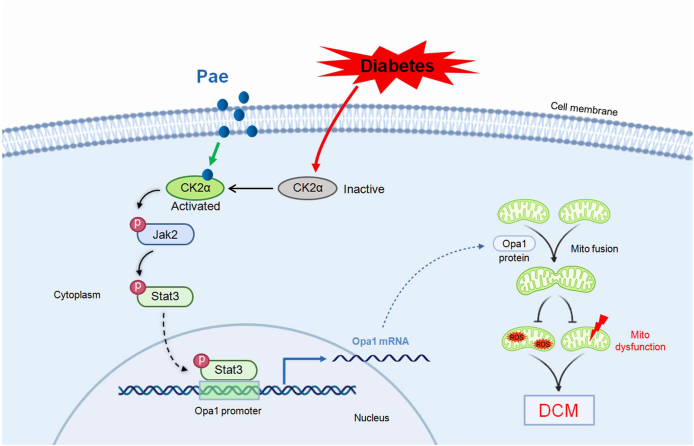
In this study, we demonstrated for the first time that Pae administration inhibits mitochondria-derived oxidative stress and preserves mitochondrial function as well as cardiac performance by promoting Opa1-mediated mitochondrial fusion in DCM in vivo and in vitro. Mechanistically, using in vitro (inhibitor screening and siRNA-based target confirmation) and in silico (pharmmapper screening and molecular docking) studies, it is verified that Pae binds to and activates CK2α that subsequently interacts with Jak2 and induces the phosphorylation and activation of the Jak2-Stat3 signaling, which then binds to the Opa1 promoter to promote Opa1-mediated mitochondrial fusion (Fig. 10). Thus, our study uncovers the detailed molecular mechanism on how Pae promotes mitochondrial fusion and inhibits mitochondrial oxidative stress under diabetic conditions.
Fig. 10.
Schematic figure illustrating that Pae alleviates diabetic cardiomyopathy by promoting Opa1-mediated mitochondrial fusion via activating the CK2α-Jak2-Stat3 pathway. Pae binds to diabetes-inactivated CK2α and restores its activity that subsequently interacts with Jak2 and induces the phosphorylation and activation of the Jak2-Stat3 signaling. Activated Stat3 binds to the Opa1 promoter to promote Opa1-mediated mitochondrial fusion. Afterward, Pae-induced upregulation of Opa1 promotes mitochondrial fusion, inhibits mitochondria-derived oxidative stress, preserves mitochondrial function and protects against diabetic cardiomyopathy. Pae, paeonol; Mito, mitochondrial; ROS, reactive oxygen species; DCM, diabetic cardiomyopathy.
Mitochondrial dysfunction associated with excessive ROS production has been recognized to significantly contribute to the development of DCM [41]. Recent studies from our lab and others have demonstrated that mitochondrial fusion is correlated with enhanced mitochondrial function and decreased mitochondrial ROS production [11,42,43]. Therefore, agents with the ability to promote mitochondrial fusion have great potential for inhibiting mitochondrial oxidative stress and protecting against the development of DCM. Previous reports have shown that Pae has potential as a protective agent in several cardiovascular diseases including atherosclerosis [44], hypertension [45] and myocardial ischemic injury [15], acting mainly via anti-inflammatory mechanisms. Few studies have focused on the role of mitochondria in the protective effects of Pae. Stimulating PGC1α-regulated mitochondrial biogenesis could increase mitochondrial content and also inhibit mitochondrial ROS production [46,47], which is similar with the protective effects of mitochondrial fusion promotion. Thus, when regarding the protective action of Pae on mitochondria, both mitochondrial biogenesis and mitochondrial fusion has been considered. Our data suggest that Pae inhibits mitochondrial ROS and improves mitochondrial function by promoting Opa1-mediated mitochondrial fusion rather than by stimulating PGC1α-regulated mitochondrial biogenesis. Moreover, Pae was shown to be effective in protecting against diabetic cardiac complications without significantly affecting blood glucose in the diabetic rats. A previous study by Liu et al. has shown that Pae has a weak glucose-lowering effect [48]. This discrepancy may be due to different animal model. In our study, the diabetic model was induced by relatively high dose of STZ (65 mg/kg). In the study by Liu et al. [48], the diabetic model was induced by high fat/high sucrose feed and relatively low dose of STZ (40 mg/kg). In general, Pae has not been considered as a potent glucose-lowering drug during long time of clinical practice.
In our previous article, M1 has been used as an agonist tool compound to promote mitochondrial fusion [11], whereas its chemical, physical and toxicological properties are so far largely unclear, which limits its application in practice. In contrast, Pae has the advantage that it is already approved and considered safe to use. One novelty of our study is that we have identified an approved drug Pae as a new mitochondrial fusion promoter to protect against diabetic cardiomyopathy. The promotion of mitochondrial fusion may represent a crucial mechanism by which Pae exerts its cardioprotective effects. Moreover, in our previous article [11], the complex mechanisms underlying inhibited Opa1-mediated mitochondrial fusion in diabetic hearts remain unclear. Another novelty of this study is that we have uncovered the detailed molecular mechanism on how Opa1-mediated mitochondrial fusion was inhibited by diabetes and restored by Pae after several times of screening and validation. Our results suggest that inactivated CK2α-Jak2-Stat3 signaling pathway is account for the reduced transcriptional level of Opa1 and inhibited mitochondrial fusion in diabetic hearts.
During the process of mitochondrial fusion, fusion of the outer mitochondrial membrane is followed by inner membrane fusion. Mfn1 and Mfn2 are required for the fusion of the outer mitochondrial membrane. Opa1 is localized to the mitochondrial inner membrane and is a key protein responsible for inner membrane fusion [49]. In the present study, we found that only the expression of Opa1 was markedly lower in the diabetic hearts and in the high glucose-treated cardiomyocytes, whereas the expression of other mitochondrial fusion proteins including Mfn1 and Mfn2 showed no significant change. This may be partly due to insulin deficiency in the STZ-induced diabetic model, since it has been demonstrated that insulin increases the expression of Opa-1 in cardiomyocytes without affecting the expression of Mfn1 and Mfn2 [50]. The level of Opa1 protein is regulated by transcriptional regulation and post-translational modifications. Long form and short form of Opa1 could be generated by proteolytic cleavage. The relative contributions of the two different isoforms of Opa1 are diverse. Long form of Opa1 (L-Opa1) is known as the functional variant for fusion between mitochondria, while short form of Opa1 (S-Opa1) is considered as an inactive variant [51,52]. Recently, Yoon and colleagues have interesting work showing that S-Opa1 protects against oxidant-induced cell death [53]. Our study suggests that Opa1 may not be proteolytically cleaved by hyperglycemia or Pae treatment, since long and short forms of Opa1 were not selectively affected by high glucose or Pae. In addition to promoting mitochondrial fusion, Opa1 has been demonstrated to participate in the regulation of mitochondrial cristae morphology, and that this facilitates electron transport and enhances mitochondrial oxidative phosphorylation in the respiratory chain [54,55]. Our study found that cristae density was significantly reduced in the diabetic hearts with decreased Opa1 expression. Knockdown of Opa1 in diabetic hearts blunted the upregulating effects of Pae on cristae density. These results suggest that the protective effects of Opa1 are mediated not only by the fusion of mitochondria but also by the remodeling of mitochondrial cristae.
Despite the known cardioprotective effects of Pae from previous studies [15,44,45], the mechanism of Pae's direct interaction with molecular targets is still largely unknown. An important observation is that we have shown for the first time that Pae binds to CK2α and then activates the Jak2–Stat3 pathway. As one catalytic subunit of the protein kinase CK2, CK2α bears most of the sequence and structural features common to holoenzymes [56]. CK2 does not require phosphorylation for its activation [57]. The expression and activity of CK2 are disturbed under many pathophysiological conditions. It has been reported that CK2 activity is decreased in hypertrophic hearts or cardiomyocytes, and oxidative modification results in a dramatic reduction in CK2 activity [58]. In our study, diabetes or high glucose inhibited the activity of CK2α rather than affecting its expression. Pretreatment with the antioxidants elevated the activity of CK2a in HG-cultured cardiomyocytes. Pae could further increase CK2α activity following pretreatment with the antioxidants. The results suggest that the impact of Paeonol on CK2a activity is not solely due to its antioxidant properties. The interaction between Pae and CK2α may help to increase its activity.
CK2 exerts anti-apoptotic and anti-hypertrophic effects in cardiomyocytes by phosphorylating the caspase-inhibiting protein ARC [58,59]. A recent study has shown that CK2 protects against oxidative stress by positively regulating NF-κB/RelA transcription activity in cardiomyoblasts [60]. Our study suggests that CK2α may promote mitochondrial fusion and exert myocardial protective effects by activating the Jak2-Stat3-Opa1 signaling pathway in the diabetic hearts. Nevertheless, it should be noted that there have been some contradictory results regarding the role of CK2α in the hearts. Zhou et al. have found that CK2α promotes the phosphorylation of Mff to facilitate mitochondrial fission and serves as a negative regulator of mitochondrial activity in cardiac ischemia-reperfusion injury [61,62]. This discrepancy may be due to distinct substrates phosphorylated and regulated by CK2 under different pathophysiological situations, since CK2 is a pleiotropic protein kinase acting on more than 300 substrates [56].
It is to be noted that our study has some limitations. First, Pae has been shown to be an efficient mitochondrial fusion promoter against mitochondrial oxidative injury and resultant DCM exclusively in an STZ-induced animal model of type 1 diabetes and high glucose-treated cardiomyocytes, which may not entirely represent the complex conditions caused by diabetes. Further investigation is needed to determine whether the mitochondrial fusion-promoting and cardioprotective effects of Pae could be applied to other diabetic models. Second, the role of CK2α-Stat3 signaling in the promoting effects of Pae on mitochondrial fusion was mainly explored in vitro using CK2α or Stat3 siRNA. Knockdown of CK2α or Stat3 in the heart in vivo will be helpful to verify the promoting action of Pae on Opa1-mediated mitochondrial fusion via a CK2α-Stat3-dependent way. Third, although Opa1 is required for the mitochondrial fusion-promoting effect of Pae in our study, it should be noted that Opa1 may not be the only key molecule involved in Pae-induced cardioprotection. A complex array of signaling pathways indeed participates in the therapeutic effects of Pae and future work will be needed to clarify this issue. Despite these limitations, we believe that our findings provide important novel insights for understanding the mitochondrial protective effects of Pae and the underlying mechanisms.
In summary, our study provides novel evidence indicating that Pae promotes mitochondrial fusion and protects against diabetic cardiomyopathy at least partially by upregulating Opa1 expression, a process in which Pae interacts with CK2α and restores its kinase activity that subsequently increasing Jak2-Stat3 phosphorylation and enhancing the transcriptional level of Opa1. These findings suggest that Pae or promotion of mitochondrial fusion might be a new potential option for the treatment of patients with DCM.
Fundings
This study was supported by the grants from National Natural Science Foundation of China (No. 82070387, No. 81970316, No. 82070051, No. 81770243, No. 82111530058), Key Research and Development Plan of Shaanxi (No. 2021SF-141), Innovation Capability Support Program of Shaanxi (No. 2019KJXX-084), the Fundamental Research Funds for the Central Universities (No. xzy012019115) and Russian Foundation of Basic Research Grant (No. 21-515-53003).
Declaration of competing interest
Authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102098.
Contributor Information
Jianming Pei, Email: jmpei8@fmmu.edu.cn.
Feng Fu, Email: fufeng048@126.com.
Mingge Ding, Email: dingmingge@xjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Knapp M., Tu X., Wu R. Vascular endothelial dysfunction, a major mediator in diabetic cardiomyopathy. Acta Pharmacol. Sin. 2019;40(1):1–8. doi: 10.1038/s41401-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chavali V., Tyagi S.C., Mishra P.K. Predictors and prevention of diabetic cardiomyopathy. Diabetes Metab Syndr Obes. 2013;6:151–160. doi: 10.2147/DMSO.S30968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huynh K., Bernardo B.C., McMullen J.R., Ritchie R.H. Diabetic cardiomyopathy: mechanisms and new treatment strategies targeting antioxidant signaling pathways. Pharmacol. Ther. 2014;142(3):375–415. doi: 10.1016/j.pharmthera.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Tribouilloy C., Rusinaru D., Mahjoub H., Tartiere J.M., Kesri-Tartiere L., Godard S., Peltier M. Prognostic impact of diabetes mellitus in patients with heart failure and preserved ejection fraction: a prospective five-year study. Heart. 2008;94(11):1450–1455. doi: 10.1136/hrt.2007.128769. [DOI] [PubMed] [Google Scholar]
- 5.Marin-Garcia J., Akhmedov A.T. Mitochondrial dynamics and cell death in heart failure. Heart Fail. Rev. 2016;21(2):123–136. doi: 10.1007/s10741-016-9530-2. [DOI] [PubMed] [Google Scholar]
- 6.Kaludercic N., Di Lisa F. Mitochondrial ROS formation in the pathogenesis of diabetic cardiomyopathy. Front Cardiovasc Med. 2020;7:12. doi: 10.3389/fcvm.2020.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jubaidi F.F., Zainalabidin S., Mariappan V., Budin S.B. Mitochondrial dysfunction in diabetic cardiomyopathy: the possible therapeutic roles of phenolic acids. Int. J. Mol. Sci. 2020;21(17):6043. doi: 10.3390/ijms21176043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu T., Sheu S.S., Robotham J.L., Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc. Res. 2008;79(2):341–351. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marin-Garcia J., Akhmedov A.T., Moe G.W. Mitochondria in heart failure: the emerging role of mitochondrial dynamics. Heart Fail. Rev. 2013;18(4):439–456. doi: 10.1007/s10741-012-9330-2. [DOI] [PubMed] [Google Scholar]
- 10.Ding M., Feng N., Tang D., Feng J., Li Z., Jia M., Liu Z., Gu X., Wang Y., Fu F., Pei J. Melatonin prevents Drp1-mediated mitochondrial fission in diabetic hearts through SIRT1-PGC1α pathway. J. Pineal Res. 2018;65(2) doi: 10.1111/jpi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding M., Liu C., Shi R., Yu M., Zeng K., Kang J., Fu F., Mi M. Mitochondrial fusion promoter restores mitochondrial dynamics balance and ameliorates diabetic cardiomyopathy in an optic atrophy 1-dependent way. Acta Physiol. 2020;229(1) doi: 10.1111/apha.13428. [DOI] [PubMed] [Google Scholar]
- 12.Qin Y., Li A., Liu B., Jiang W., Gao M., Tian X., Gong G. Mitochondrial fusion mediated by fusion promotion and fission inhibition directs adult mouse heart function toward a different direction. Faseb. J. 2020;34(1):663–675. doi: 10.1096/fj.201901671R. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Li D.C., Liu L.F. Paeonol: pharmacological effects and mechanisms of action. Int. Immunopharm. 2019;72:413–421. doi: 10.1016/j.intimp.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Nizamutdinova I.T., Jin Y.C., Kim J.S., Yean M.H., Kang S.S., Kim Y.S., Lee J.H., Seo H.G., Kim H.J., Chang K.C. Paeonol and paeoniflorin, the main active principles of Paeonia albiflora, protect the heart from myocardial ischemia/reperfusion injury in rats. Planta Med. 2008;74(1):14–18. doi: 10.1055/s-2007-993775. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Song F., Duan L.R., Sheng J.J., Xie Y.H., Yang Q., Chen Y., Dong Q.Q., Zhang B.L., Wang S.W. Paeonol and danshensu combination attenuates apoptosis in myocardial infarcted rats by inhibiting oxidative stress: roles of Nrf2/HO-1 and PI3K/Akt pathway. Sci. Rep. 2016;6:23693. doi: 10.1038/srep23693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Zhu G., Yang S., Wang X., Cheng H., Wang F., Li X., Li Q. Paeonol prevents excitotoxicity in rat pheochromocytoma PC12 cells via downregulation of ERK activation and inhibition of apoptosis. Planta Med. 2011;77(15):1695–1701. doi: 10.1055/s-0030-1271033. [DOI] [PubMed] [Google Scholar]
- 17.Ding M., Ning J., Feng N., Li Z., Liu Z., Wang Y., Wang Y., Li X., Huo C., Jia X., Xu R., Fu F., Wang X., Pei J. Dynamin-related protein 1-mediated mitochondrial fission contributes to post-traumatic cardiac dysfunction in rats and the protective effect of melatonin. J. Pineal Res. 2018;64(1) doi: 10.1111/jpi.12447. [DOI] [PubMed] [Google Scholar]
- 18.Ma Z.G., Yuan Y.P., Xu S.C., Wei W.Y., Xu C.R., Zhang X., Wu Q.Q., Liao H.H., Ni J., Tang Q.Z. CTRP3 attenuates cardiac dysfunction, inflammation, oxidative stress and cell death in diabetic cardiomyopathy in rats. Diabetologia. 2017;60(6):1126–1137. doi: 10.1007/s00125-017-4232-4. [DOI] [PubMed] [Google Scholar]
- 19.Xu F., Xiao H., Liu R., Yang Y., Zhang M., Chen L., Chen Z., Liu P., Huang H. Paeonol ameliorates glucose and lipid metabolism in experimental diabetes by activating Akt. Front. Pharmacol. 2019;10:261. doi: 10.3389/fphar.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Chen Z., Gong W., Zou Y., Xu F., Chen L., Huang H. Paeonol ameliorates diabetic renal fibrosis through promoting the activation of the Nrf2/ARE pathway via up-regulating Sirt1. Front. Pharmacol. 2018;9:512. doi: 10.3389/fphar.2018.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Shen Y., Wang S., Li S., Zhang W., Liu X., Lai L., Pei J., Li H. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):356–360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Ouyang S., Yu B., Liu Y., Huang K., Gong J., Zheng S., Li Z., Li H., Jiang H. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38(Web Server issue):609–614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., Wu X., Jia L., Li J., Zhang R., Tang H., Li Z., Bu H., Shen C. Design and synthesis of novel 2-arylbenzimidazoles as selective mutant isocitrate dehydrogenase 2 R140Q inhibitors. Bioorg. Med. Chem. Lett. 2020;30(9):127070. doi: 10.1016/j.bmcl.2020.127070. [DOI] [PubMed] [Google Scholar]
- 24.Li Z., Jia L., Tang H., Shen Y., Shen C. Synthesis and biological evaluation of geldanamycin–ferulic acid conjugate as a potent Hsp90 inhibitor. RSC Adv. 2019;9(72):42509–42515. doi: 10.1039/c9ra08665j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji L., Fu F., Zhang L., Liu W., Cai X., Zhang L., Zheng Q., Zhang H., Gao F. Insulin attenuates myocardial ischemia/reperfusion injury via reducing oxidative/nitrative stress. Am. J. Physiol. Endocrinol. Metab. 2010;298(4):E871–E880. doi: 10.1152/ajpendo.00623.2009. [DOI] [PubMed] [Google Scholar]
- 26.Liu N., Feng X., Wang W., Zhao X., Li X. Paeonol protects against TNF-alpha-induced proliferation and cytokine release of rheumatoid arthritis fibroblast-like synoviocytes by upregulating FOXO3 through inhibition of miR-155 expression. Inflamm. Res. 2017;66(7):603–610. doi: 10.1007/s00011-017-1041-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q., Xu X., Kang Z., Zhang Z., Li Y. Paeonol prevents IL-1beta-induced inflammatory response and degradation of type II collagen in human primary chondrocytes. Artif Cells Nanomed Biotechnol. 2019;47(1):2139–2145. doi: 10.1080/21691401.2019.1613418. [DOI] [PubMed] [Google Scholar]
- 28.Wu X., Chen H., Chen X., Hu Z. Determination of paeonol in rat plasma by high-performance liquid chromatography and its application to pharmacokinetic studies following oral administration of Moutan cortex decoction. Biomed. Chromatogr. 2003;17(8):504–508. doi: 10.1002/bmc.259. [DOI] [PubMed] [Google Scholar]
- 29.Li J., Xu Z.Q., Bao L., Li Z.C., Qu J.X. Protective effects of paeonol on cardiovascular complications in diabetes mellitus involves modulation of PI3K/Akt-GSK-3β signalling, regulation of protease-activated receptor-1 expressions and down-regulation of inflammatory mediators. Bangladesh J. Pharmacol. 2015;10(4):903–916. [Google Scholar]
- 30.Chang R.Q., Shao J., Meng Y.H., Wang J., Li D.J., Li M.Q. Decidual RANKL/RANK interaction promotes the residence and polarization of TGF-beta1-producing regulatory gammadelta T cells. Cell Death Dis. 2019;10(2):113. doi: 10.1038/s41419-019-1380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei S., Su W., Xia Z.Y., Wang Y., Zhou L., Qiao S., Zhao B., Xia Z., Irwin M.G. Hyperglycemia-induced oxidative stress abrogates remifentanil preconditioning-mediated cardioprotection in diabetic rats by impairing Caveolin-3-modulated PI3K/Akt and JAK2/STAT3 signaling. Oxid Med Cell Longev. 2019;(2019):9836302. doi: 10.1155/2019/9836302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florian M., Jankowski M., Gutkowska J. Oxytocin increases glucose uptake in neonatal rat cardiomyocytes. Endocrinology. 2010;151(2):482–491. doi: 10.1210/en.2009-0624. [DOI] [PubMed] [Google Scholar]
- 33.Leidgens V., Proske J., Rauer L., Moeckel S., Renner K., Bogdahn U., Riemenschneider M.J., Proescholdt M., Vollmann-Zwerenz A., Hau P., Seliger C. Stattic and metformin inhibit brain tumor initiating cells by reducing STAT3-phosphorylation. Oncotarget. 2017;8(5):8250–8263. doi: 10.18632/oncotarget.14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Zang W., Ji S., Cao J., Sun C. Three polymethoxyflavones purified from ougan (Citrus reticulata Cv. Suavissima) inhibited LPS-induced NO elevation in the Neuroglia BV-2 cell line via the JAK2/STAT3 pathway. Nutrients. 2019;11(4):791. doi: 10.3390/nu11040791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen D.A., Seki S.M., Fernandez-Castaneda A., Beiter R.M., Eccles J.D., Woodfolk J.A., Gaultier A. Modulation of the sigma-1 receptor-IRE1 pathway is beneficial in preclinical models of inflammation and sepsis. Sci. Transl. Med. 2019;11(478):5266. doi: 10.1126/scitranslmed.aau5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y.M., Park J.P., Lim K.T., Lee S.J. Intestinal epithelial cell apoptosis due to a hemolytic toxin from Vibrio vulnificus and protection by a 36kDa glycoprotein from Rhus verniciflua Stokes. Food Chem. Toxicol. 2019;125:46–54. doi: 10.1016/j.fct.2018.12.041. [DOI] [PubMed] [Google Scholar]
- 37.Kim J.Y., Lee Y.M., Kim D.W., Min T., Lee S.J. Nanosphere loaded with curcumin inhibits the gastrointestinal cell death signaling pathway induced by the foodborne pathogen vibrio vulnificus. Cells. 2020;9(3):631. doi: 10.3390/cells9030631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyawali A., Krol S., Kang Y.S. Involvement of a novel organic cation transporter in paeonol transport across the blood-brain barrier. Biomol Ther (Seoul). 2019;27(3):290–301. doi: 10.4062/biomolther.2019.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arakel E.C., Brandenburg S., Uchida K., Zhang H., Lin Y.W., Kohl T., Schrul B., Sulkin M.S., Efimov I.R., Nichols C.G., Lehnart S.E., Schwappach B. Tuning the electrical properties of the heart by differential trafficking of KATP ion channel complexes. J. Cell Sci. 2014;127(Pt 9):2106–2119. doi: 10.1242/jcs.141440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarno S., Reddy H., Meggio F., Ruzzene M., Davies S.P., Donella-Deana A., Shugar D., Pinna L.A. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 ('casein kinase-2') FEBS Lett. 2001;496(1):44–48. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- 41.Duncan J.G. Mitochondrial dysfunction in diabetic cardiomyopathy. Biochim. Biophys. Acta. 2011;1813(7):1351–1359. doi: 10.1016/j.bbamcr.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu L., Ding M., Tang D., Gao E., Li C., Wang K., Qi B., Qiu J., Zhao H., Chang P., Fu F., Li Y. Targeting mitochondrial dynamics by regulating Mfn2 for therapeutic intervention in diabetic cardiomyopathy. Theranostics. 2019;9(13):3687–3706. doi: 10.7150/thno.33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forte M., Schirone L., Ameri P., Basso C., Catalucci D., Modica J., Chimenti C., Crotti L., Frati G., Rubattu S., Schiattarella G.G., Torella D., Perrino C., Indolfi C., Sciarretta S. The role of mitochondrial dynamics in cardiovascular diseases. Br. J. Pharmacol. 2021;178(10):2060–2076. doi: 10.1111/bph.15068. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Dai M., Jia W. Paeonol attenuates high-fat-diet-induced atherosclerosis in rabbits by anti-inflammatory activity. Planta Med. 2009;75(1):7–11. doi: 10.1055/s-0028-1088332. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J.Y., Zhao L., Li Y.K., Weng W.L. [Effect of paeonol on blood pressure and blood flow in artery of spontaneously hypertensive rats and its mechanisms related on vasomotion] Zhongguo Zhongyao Zazhi. 2015;40(24):4903–4907. [PubMed] [Google Scholar]
- 46.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di W., Lv J., Jiang S., Lu C., Yang Z., Ma Z., Hu W., Yang Y., Xu B. PGC-1: the energetic regulator in cardiac metabolism. Curr. Issues Mol. Biol. 2018;28:29–46. doi: 10.21775/cimb.028.029. [DOI] [PubMed] [Google Scholar]
- 48.Liu J., Wang S., Feng L., Ma D., Fu Q., Song Y., Jia X., Ma S. Hypoglycemic and antioxidant activities of paeonol and its beneficial effect on diabetic encephalopathy in streptozotocin-induced diabetic rats. J. Med. Food. 2013;16(7):577–586. doi: 10.1089/jmf.2012.2654. [DOI] [PubMed] [Google Scholar]
- 49.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8(11):870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 50.Parra V., Verdejo H.E., Iglewski M., Del Campo A., Troncoso R., Jones D., Zhu Y., Kuzmicic J., Pennanen C., Lopez-Crisosto C., Jana F., Ferreira J., Noguera E., Chiong M., Bernlohr D.A., Klip A., Hill J.A., Rothermel B.A., Abel E.D., Zorzano A., Lavandero S. Insulin stimulates mitochondrial fusion and function in cardiomyocytes via the Akt-mTOR-NFkappaB-Opa-1 signaling pathway. Diabetes. 2014;63(1):75–88. doi: 10.2337/db13-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adaniya S.M., J O.U., Cypress M.W., Kusakari Y., Jhun B.S. Posttranslational modifications of mitochondrial fission and fusion proteins in cardiac physiology and pathophysiology. Am. J. Physiol. Cell Physiol. 2019;316(5):C583–C604. doi: 10.1152/ajpcell.00523.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishihara N., Fujita Y., Oka T., Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25(13):2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee H., Smith S.B., Sheu S.S., Yoon Y. The short variant of optic atrophy 1 (OPA1) improves cell survival under oxidative stress. J. Biol. Chem. 2020;295(19):6543–6560. doi: 10.1074/jbc.RA119.010983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., Derks C., Dejaegere T., Pellegrini L., D'Hooge R., Scorrano L., De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126(1):163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 55.Ong S.B., Kalkhoran S.B., Hernandez-Resendiz S., Samangouei P., Ong S.G., Hausenloy D.J. Mitochondrial-shaping proteins in cardiac health and disease - the long and the short of it! Cardiovasc. Drugs Ther. 2017;31(1):87–107. doi: 10.1007/s10557-016-6710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Battistutta R. Protein kinase CK2 in health and disease: structural bases of protein kinase CK2 inhibition. Cell. Mol. Life Sci. 2009;66(11–12):1868–1889. doi: 10.1007/s00018-009-9155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battistutta R., Lolli G. Structural and functional determinants of protein kinase CK2alpha: facts and open questions. Mol. Cell. Biochem. 2011;356(1–2):67–73. doi: 10.1007/s11010-011-0939-6. [DOI] [PubMed] [Google Scholar]
- 58.Murtaza I., Wang H.X., Feng X., Alenina N., Bader M., Prabhakar B.S., Li P.F. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J. Biol. Chem. 2008;283(10):5996–6004. doi: 10.1074/jbc.M706466200. [DOI] [PubMed] [Google Scholar]
- 59.Li P.F., Li J., Muller E.C., Otto A., Dietz R., von Harsdorf R. Phosphorylation by protein kinase CK2: a signaling switch for the caspase-inhibiting protein ARC. Mol Cell. 2002;10(2):247–258. doi: 10.1016/s1097-2765(02)00600-7. [DOI] [PubMed] [Google Scholar]
- 60.Schaefer S., Guerra B. Protein kinase CK2 regulates redox homeostasis through NF-kappaB and Bcl-xL in cardiomyoblasts. Mol. Cell. Biochem. 2017;436(1–2):137–150. doi: 10.1007/s11010-017-3085-y. [DOI] [PubMed] [Google Scholar]
- 61.Zhou H., Wang J., Zhu P., Zhu H., Toan S., Hu S., Ren J., Chen Y. NR4A1 aggravates the cardiac microvascular ischemia reperfusion injury through suppressing FUNDC1-mediated mitophagy and promoting Mff-required mitochondrial fission by CK2alpha. Basic Res. Cardiol. 2018;113(4):23. doi: 10.1007/s00395-018-0682-1. [DOI] [PubMed] [Google Scholar]
- 62.Zhou H., Zhu P., Wang J., Zhu H., Ren J., Chen Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2alpha-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ. 2018;25(6):1080–1093. doi: 10.1038/s41418-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.