Abstract
The brain is now recognized as an insulin-sensitive tissue; however, the role of changing insulin concentrations in the peripheral circulation in gene expression in the brain is largely unknown. Here, we performed a hyperinsulinemic-euglycemic clamp on 3-month-old male C57BL/6 mice for 3 h. We show that, in comparison with results in saline-infused controls, increases in peripheral insulin within the physiological range regulate expression of a broad network of genes in the brain. Insulin regulates distinct pathways in the hypothalamus (HTM), hippocampus, and nucleus accumbens. Insulin shows its most robust effect in the HTM and regulates multiple genes involved in neurotransmission, including upregulating expression of multiple subunits of GABA-A receptors, Na+ and K+ channels, and SNARE proteins; differentially modulating glutamate receptors; and suppressing multiple neuropeptides. Insulin also strongly modulates metabolic genes in the HTM, suppressing genes in the glycolysis and pentose phosphate pathways, while increasing expression of genes regulating pyruvate dehydrogenase and long-chain fatty acyl-CoA and cholesterol biosynthesis, thereby rerouting of carbon substrates from glucose metabolism to lipid metabolism required for the biogenesis of membranes for neuronal and glial function and synaptic remodeling. Furthermore, based on the transcriptional signatures, these changes in gene expression involve neurons, astrocytes, oligodendrocytes, microglia, and endothelial cells. Thus, peripheral insulin acutely and potently regulates expression of a broad network of genes involved in neurotransmission and brain metabolism. Dysregulation of these pathways could have dramatic effects in normal physiology and diabetes.
Introduction
Insulin is central to the regulation of systemic metabolism, control of blood glucose, and energy homeostasis. Circulating insulin levels are low in fasting states but rise rapidly upon feeding. In the periphery, insulin acts on various insulin-sensitive tissues like liver, muscle, and adipose tissues by activating the insulin receptor tyrosine kinase, eliciting a network of signaling pathways and triggering robust transcriptional regulation to orchestrate the balance between energy storage and expenditure (1,2). Many of these actions are impaired in insulin resistance conditions like obesity and type 2 diabetes (3). It is now clear that insulin also has profound effects on the brain. Thus, circulating insulin can cross the blood-brain barrier, at least in part, through insulin receptor–mediated transcytosis, and act on insulin receptors, which are widely distributed throughout the brain (4–6). Although cerebrospinal fluid insulin levels are lower than circulating peripheral insulin levels, they are positively correlated over a range of physiological and pathophysiological conditions (7,8). For instance, insulin levels in the cerebrospinal fluid increase in parallel with circulating insulin levels after feeding or following intravenous insulin infusion (7,9). In addition, intracerebroventricular insulin injections suppress appetite in both rodents and primates, thereby reducing food intake and body weight (10,11).
The brain may also exhibit insulin resistance. Thus, in the context of obesity and type 2 diabetes, there is impaired insulin signaling in the brain despite peripheral hyperinsulinemia (8,12,13), supporting the concept that brain function may be altered in these insulin resistance states. Loss of insulin signaling in the brain in animals results in numerous metabolic and behavioral defects, including hyperphagia, hypothalamic hypogonadism, impaired hepatic glucose output control, systemic insulin resistance, impaired response to hypoglycemia, and depressive-like behavior (14–17). Using genetic tools to inactivate insulin receptors in distinct cell types in the brain, we and others have shown that insulin acts on both neuronal and nonneuronal cell types to regulate these responses (18–21). Insulin signaling in hypothalamic neurons, such as AgRP/NPY and POMC neurons, is important for the effects on appetite and systemic metabolism (22–24), whereas insulin signaling in astrocytes is particularly important for mood control and fertility (20,21). Insulin’s effects in the brain can also produce effects on synaptic plasticity (25), changes in cellular metabolism, changes in membrane potential (26), and release of glial transmitters (21). However, little is known about the effects of insulin on the regulation of gene transcription in the brain.
In the current study, we have used the euglycemic clamp to deliver intravenous insulin at physiological concentrations to mice while maintaining blood glucose in the normal range. Under these conditions, we find that insulin has robust and distinct effects on gene expression in different brain regions, with the most robust changes in the hypothalamus (HTM) followed by the hippocampus (HCA) and nucleus accumbens (NAC). Pathway analyses reveal that insulin regulates distinct pathways in different brain regions. In the HTM, in particular, many of the transcriptional targets of insulin are involved in neurotransmission, glucose metabolism, and lipid metabolism. In addition, computational analyses show that insulin regulates cell type–specific genes and pathways, which are important for distinct cellular functions. Together, these transcriptional effects of insulin in the brain can modulate neurotransmission and reroute the carbon source from glucose to plasma membrane regeneration important for synaptic plasticity and integrity.
Research Design and Methods
Animals
All animal studies were conducted in compliance with the regulations and ethics guidelines of the National Institutes of Health (NIH) and were approved by the Institutional Animal Care and Use Committee of the Joslin Diabetes Center and University of Massachusetts Medical School.
Hyperinsulinemic-Euglycemic Clamp Studies
The insulin clamp studies were conducted at the National Mouse Metabolic Phenotyping Center at University of Massachusetts Medical School similarly to how investigators conducted previous studies (27). Briefly, 3-month-old male C57BL/6J mice (N = 6 per condition) had an indwelling catheter placed in the internal jugular vein. After recovery, mice were fasted overnight (∼16 h) and placed in rat-sized restrainers. After an acclimation period, mice received an insulin bolus (16 or 48 mU/kg) followed by a continuous infusion of 4 or 12 mU/kg/min insulin or saline as control. Blood samples were collected from tail vein at 10- to 30-min intervals during the clamp, and a 20% glucose solution was infused for maintenance of blood glucose levels at euglycemia (110–150 mg/dL range). After 180 min, mice were euthanized by pentobarbital injection, and the HTM, HCA, and NAC were harvested, snap frozen in liquid nitrogen, and kept at −80°C until analysis.
RNA Isolation and Sequencing
Total RNA was extracted from the HTM, HCA, and NAC using QIAzol reagent (QIAGEN). RNA quality and quantification were verified with the Agilent 2100 Bioanalyzer. Total RNA samples (2 μg, RNA integrity score >7) were submitted to the Biopolymers Core at Harvard Medical School for library preparation and next-generation sequencing. Stranded cDNA libraries with unique index tags for each sample (48 multiplexed) were generated with a directional RNA-sequencing (RNA-seq) kit (WaferGen Bio-systems). Sequencing was performed on an Illumina HiSeq 2500 run on rapid mode (2 × 50).
Bioinformatics Analysis
Bioinformatics analysis was carried out with R software (www.r-project.org) by the Bioinformatics Core at Joslin Diabetes Center. Mouse genome (GRCm38.p6) and gene annotation files were downloaded from GENCODE (www.gencodegenes.org). Reads were aligned to the reference genome with STAR aligner, mapped to genes, and counted with Subread featureCounts. Genes that did not have at least one count per million in at least three samples were filtered out. Normalization was performed with the weighted trimmed mean of M values method (28). For single-cell data, classification of cell cycle phase and calculation of gene count normalization factors were done with use of the R package scran. Cells belong to G1 phase were selected, and the normalization factors were obtained by deconvolving size factors from cell pools. Cell type–enriched genes were selected when t score was >2 in comparisons with any other cell types. The voom method (29) was used to transform read counts to log2-counts per million (LogCPM), estimate the mean-variance relationship, and compute appropriate observational-level weights and sample quality weights. One sample of HTM from high-dose insulin infusion groups (TRA00038406) was of low quality and was excluded from further analysis. Principal components analysis (PCA) was applied to the LogCPM data with R function prcomp and was plotted with R package ggplot2. Differential expression was calculated with the limma package (30). Pathway analysis was performed with use of the mroast function in the limma package (31). Gene sets were from the Reactome and Gene Ontology (GO) databases. Functional annotation analysis was performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (32).
Insulin-Deficient Mouse Model by Streptozotocin
Three-month-old male C57BL/6J mice were purchased from The Jackson Laboratory and subjected to a single intraperitoneal injection of streptozotocin (STZ) (150 mg/kg i.p.; Sigma-Aldrich) in citrate buffer (pH 4.5). Control groups were injected with citrate buffer (pH 4.5) alone. At 8 days post–STZ injection, one-half of the mice were subjected to subcutaneous implantation of insulin pellet (LINBIT) according to the manufacturer’s instructions, while the other half of the STZ-injected mice remained untreated. At 14 days post–STZ injection, brain tissues and serum from these mice were collected. Serum insulin levels were measured with the Mouse Insulin ELISA (Ultra Sensitive) assay kit (Crystal Chem).
Primary Neuron and Astrocyte Cultures
Primary cortical neuron and astrocyte cultures were prepared from wild-type C57BL/6J newborn pups as previously described (16). Briefly, cortices were surgically dissected and digested in Hibernate-A Medium supplemented with 4 mg/mL papain (Sigma-Aldrich), 33 units/mL DNase I (Sigma-Aldrich), 0.05 mmol/L (2R)-amino-5-phosphonovaleric acid, AP-V (Tocris Bioscience), and 0.8 mmol/L kynurenic acid, pH 8.0 (Sigma-Aldrich), on a shaker at 37°C for 30 min. For primary neuron cultures, dissociated cells were cultured in Neurobasal-A Medium (Gibco) supplemented with 0.5 mmol/L glutamine (Gibco), B27 (Gibco), and penicillin (100 units/mL)-streptomycin (0.1 mg/mL) (Gibco) (complete neurobasal medium) on poly-d-lysine–coated sixwell plates. On the second day, 2 μmol/L AraC was added to prevent the proliferation of nonneuronal cells. Thereafter, the cells were refed with complete Neurobasal-A Medium by replacement of half the medium every 3–4 days. For primary astrocyte culture, dissociated cells were cultured in DMEM/F12 plus 10% FBS and 1× penicillin-streptomycin. Contaminating nonastrocytes were depleted by vigorous shaking. For insulin stimulation experiments, the neurons were starved in neural basal without B27 for 3 h followed by 100 nmol/L insulin treatment for 3 or 6 h. Primary astrocytes were serum starved for 6 h followed by 100 nmol/L insulin treatment for 6 h.
De Novo Lipogenesis of Primary Cultured Astrocytes
Primary astrocytes were serum starved for 5 h, followed by 1, 10, or 100 nmol/L insulin treatment overnight. Cell were then incubated with KRBH buffer supplemented with 10 mmol/L glucose, 0.1 μCi/mL 14C-labeled sodium acetate, and different concentrations of insulin for 2 h, followed by trypsinization and resuspension. Total lipid content was extracted from the cell suspension by chloroform:methanol (2:1). The incorporated 14C was measured by scintillation counting and normalized by protein concentration.
Total RNA Extraction, RT-PCR, and Quantitative Real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) from cell cultures or mouse brain tissues following the manufacturer’s standard protocol. RNA (500 ng) was reverse transcribed with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real-time PCR was performed with the SYBR Green PCR master mix (Bio-Rad Laboratories) and analyzed in a Bio-Rad Real-Time PCR analyzer (Bio-Rad Laboratories). All of the primer sequences used for this study are listed in Supplementary Table 4.
Statistical Analysis
Data are presented as means ± SEM and analyzed by Student t test or one-way ANOVA followed by Tukey post hoc test, as indicated. Significance level was set at P < 0.05.
Data and Resource Availability
RNA-sequencing (RNA-seq) data for brain samples are deposited in Gene Expression Omnibus (GEO) under accession no. GSE132352. RNA-seq data for liver and muscle samples from the same mice are deposited in GEO under accession no. GSE117741. All other original data and original codes used in this study are available from the corresponding author on request.
Results
Physiological Levels of Insulin Regulate Gene Expression in the Brain
To define the effects of physiological levels of insulin on transcriptional regulation in the brain, we performed euglycemic clamps on male C57BL/6J mice using two doses of insulin (4 mU/kg/min or 12 mU/kg/min) (Supplementary Fig. 1A). Despite some initial differences in insulin-infused versus saline-infused animals, glucose levels of all groups of mice reached steady state within 60 min and were maintained at euglycemic levels for the rest of the study (Supplementary Fig. 1B). During the initial 30–50 min of the clamp, the glucose infusion rates needed to maintain euglycemia of low-dose insulin animals were significantly lower than those of high-dose insulin–infused animals, while glucose infusion rates required to maintain euglycemia were comparable between groups during the last 2 h (Supplementary Fig. 1C), such that there was no significant difference in the total amount of glucose infused into the animals with low- versus high-dose insulin infusion (Supplementary Fig. 1D). As predicted, the low- and high-dose insulin infusions elevated plasma insulin levels to ∼2 ng/mL and 8 ng/mL, comparable with low and high postprandial levels, while plasma insulin levels of the saline-infused control mice were barely detectable (Supplementary Fig. 1E). Plasma insulin levels during the low-dose clamp (range 1.09–1.59 ng/mL, average 1.4 ng/mL) were similar to those observed after intraperitoneal glucose injection (Supplementary Fig. 1F), so we focused analysis on data from the low-dose clamp.
After 3 h of clamps, we collected the HTM, the key region to control appetite and systemic metabolism; the HCA, which plays critical roles in cognition; and the NAC, an integrating center for mood and reward control (Supplementary Fig. 1A). RNAs from these brain samples were isolated, enriched for poly-A tailed RNAs and subjected to next-generation RNA sequencing at a depth of ∼6 × 106 reads per sample. PCA of gene expression patterns in the saline-infused group showed three distinct clusters representing the three different regions of the brain, indicating the functional differences of the different brain regions (Fig. 1A). After 3 h of low-dose insulin infusion under euglycemic conditions, the HTM displayed the most dramatic transcriptional response, with 1,617 genes significantly upregulated and 1,772 genes significantly downregulated (P < 0.01 [Fig. 1B]). Of these, 930 and 926 genes were up- or downregulated by at least 50% (P < 0.01, |FC| > 1.5, where FC is fold change) (Fig. 1B, solid segment of bars, and Supplementary Table 1). The number of regulated genes in the HTM was almost twice that observed in classical peripheral targets of insulin action, like liver and muscle, under the same conditions (33) (Fig. 1B). While the responses to insulin in the HCA and NAC were less dramatic, there was still broad regulation, with 327 genes significantly up or downregulated (P < 0.01) in the HCA and 185 in the NAC at 3 h by low-dose insulin infusion (Fig. 1B and Supplementary Table 1). In contrast with results in peripheral tissues, there was no further increase of response in the HCA and NAC at the higher insulin concentration and a significant reduction in the number of regulated genes at higher insulin levels in the HTM (proportion test: P < 0.01) (Fig. 1B), suggesting either saturation of insulin delivery through blood-brain barrier or desensitization of the brain response at the higher insulin concentration.
Figure 1.
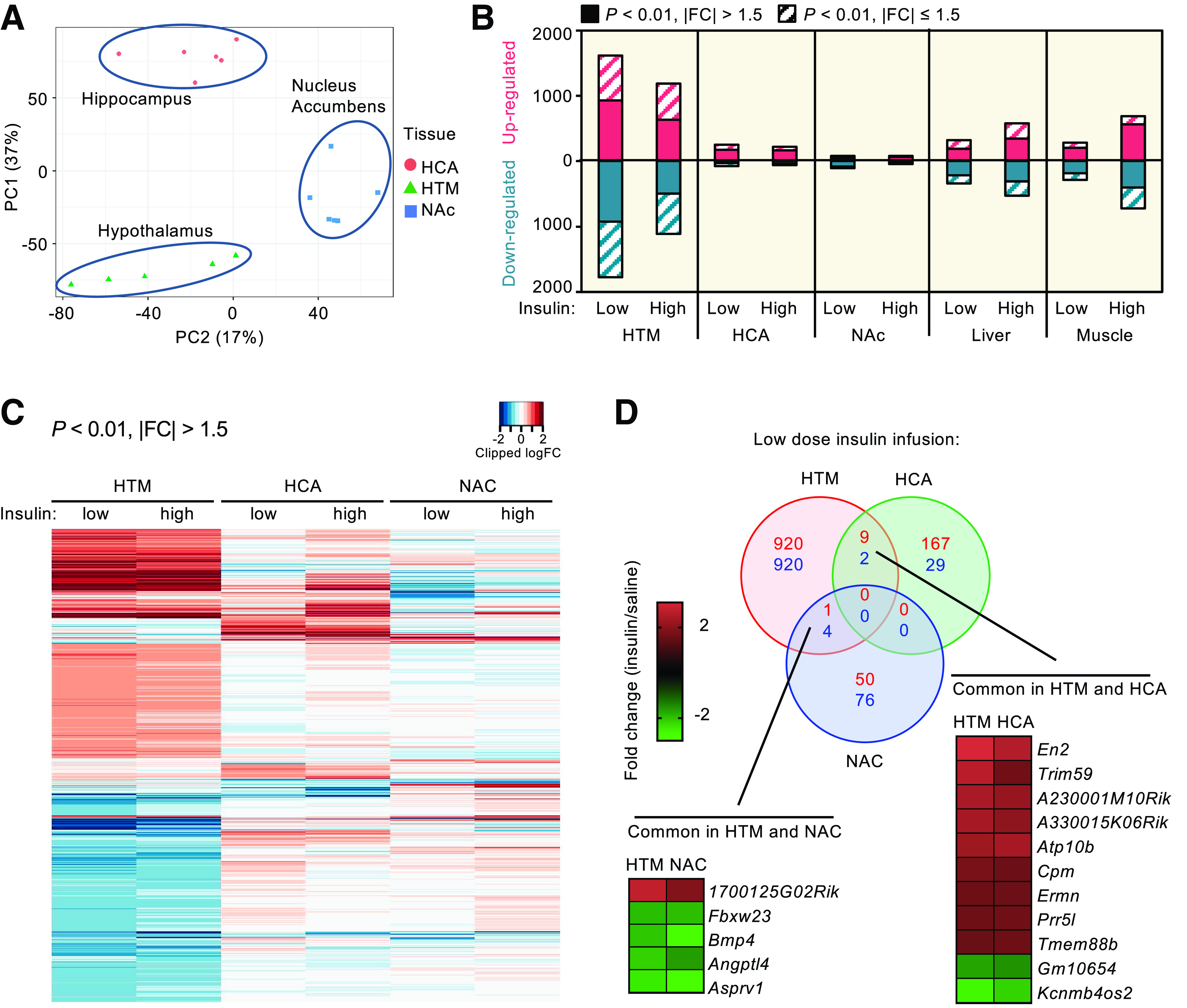
Insulin regulates robust gene expression in the brain. A: PCA of transcriptomes from HTM, HCA, and NAC of C57BL/6 mice in the fasted saline-infused group. B: Number of genes significantly (P < 0.01) up- or downregulated by low- and high-insulin infusion in the HTM, HCA, NAC, liver, and quadriceps muscle. The data of liver and muscle are from Batista et al. (33). Solid fractions of each bar represent the number of genes significantly regulated that changed by >50%. C: Heat map of all significantly (P < 0.01) regulated genes in brain regions by low-dose insulin with |FC| > 1.5. D: Venn diagram showing overlapping and unique genes significantly up- or downregulated by low-dose insulin (P < 0.01, |FC| > 1.5). The average fold changes of genes that were regulated in both HTM and NAC or HTM and HCA are shown in the inserted heat maps. PC1, principal component 1; PC2, principal component 2.
Insulin Differentially Regulates Gene Expression in Different Regions of the Brain
Analysis of the significantly (P < 0.01, |FC| > 1.5) regulated genes illustrates that each region shows a pattern of uniquely regulated genes (Fig. 1C). Interestingly, of the combined 2,178 significantly up- or downregulated genes in these three brain regions, there were no regulated genes common to all regions (Fig. 1D), and only 11 commonly regulated genes between the HTM and HCA, and five commonly regulated genes between HTM and NAC (Fig. 1D). Among the nine genes upregulated by insulin in both HTM and HCA, several are known to be involved in neural development and remodeling [engrailed 2 (En2), carboxypeptidase M (Cpm), and ERM-like protein (Ermn)], as well as two long noncoding (lnc)RNAs. The two genes suppressed by insulin in both HTM and HCA were Kcnmb4os2 (an antisense lncRNA) and Gm10654 (a predicted protein-coding gene of unknown function). Of the five genes commonly regulated in the HTM and the NAC (Fig. 1D), only one was induced by insulin (1700125G02Rik, an antisense lncRNA), while the other four, including angiopoietin-like 4 (Angptl4) and bone morphogenetic protein 4 (Bmp4), were suppressed by insulin.
Community network analysis of the significantly regulated genes identified different biological processes regulated by insulin in each of these brain regions. The large number of genes that were significantly regulated in the HTM clustered into several major interactive GO pathways (Fig. 2A). The top significant pathways include signal transduction, mRNA processing, extracellular structure remodeling, and several biosynthetic pathways. The HCA and the NAC showed distinct patterns of clustering in comparison with those of HTM. Thus, in the HCA, insulin regulated genes involved in the small GTPase signaling and phospholipid metabolic processing pathways (Fig. 2B), while in the NAC, insulin regulated genes involved in the steroid metabolic processing and cell cycle–related pathways (Fig. 2C). Notably, there were several regulated pathways common to all three brain regions, including G-protein–coupled receptor signaling, mRNA processing, and extracellular structure organization, indicating certain common roles of insulin in different brain regions despite the distinct responses at the individual gene level.
Figure 2.
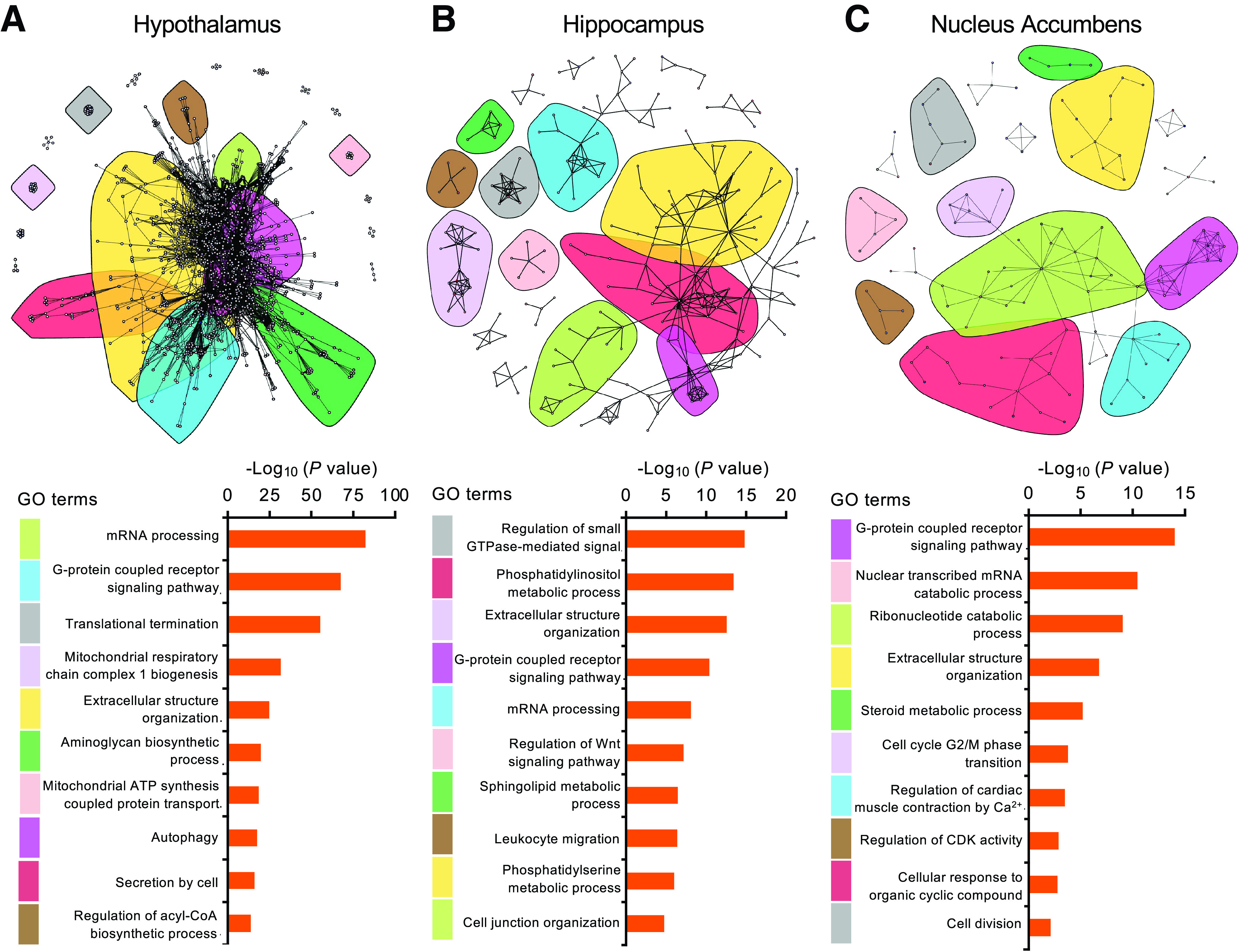
Insulin elicits distinct patterns of gene expression regulation in different brain regions. Community network analysis of significantly regulated genes by insulin in the HTM (A), HCA (B), and NAC (C) (P < 0.05). The top-enriched GO term pathways for the 10 major color-coded clusters are ranked by P value at the bottom.
Physiological Insulin Triggers Robust Responses in Both Protein-Coding and Noncoding RNA Expression in the HTM
Due to the robust response and the important role of the HTM in control of systemic metabolism, we conducted a more in-depth analysis of the 1,856 genes regulated by low-dose insulin in the HTM. Of these, 88.4% were protein-coding RNAs, 8.4% were noncoding RNAs, and 3.2% were transcripts of unknown function (Supplementary Fig. 2A). Of 158 noncoding RNAs regulated by insulin in the HTM (Supplementary Table 2), the majority were upregulated—some, such as Snord49a, as much as 19-fold (Supplementary Fig. 2A). More than half of the upregulated ncRNAs were lncRNAs (Supplementary Fig. 2A and B), including Malat1, an lncRNA that has been reported to regulate synapse formation (34) and play antiapoptotic and anti-inflammatory roles in brain microvasculature and reduce damage in ischemic stroke (35).
Among the protein-coding genes regulated by insulin by ≥|1.5|-fold, the top upregulated genes included mediators of cholesterol and phospholipid metabolism, such as insulin-induced gene 1 (Insig1) and lysophosphatidylglycerol acyltransferase 1 (Lgat1); intracellular signaling molecules, such as PI3K catalytic subunit p110a (Pik3ca) and Erb-B2 receptor tyrosine kinase-4 (Erbb4); and ion channels, including potassium voltage-gated channel subfamily A member 2 (Kcna2) (Supplementary Fig. 2C). The top downregulated genes were exemplified by small secreted peptides like agouti-related protein (Agrp) and angiopoietin-like 4 (Angptl4) (Supplementary Fig. 2C).
Unbiased Reactome pathway analysis revealed that insulin dramatically upregulated several metabolic pathways, including regulation of components of the pyruvate dehydrogenase (PDH) complex and biosynthesis pathways for multiple lipids, such as cholesterol, glycosylphosphatidylinositols, very-long-chain fatty acyl-CoAs, and fatty acyl-CoAs (Fig. 3A). In addition, insulin upregulated pathways involved in neurotransmission, including increased expression of 56–64% of genes in the GABA-A receptor activation pathway and the ligand-gated ion channel transport pathway (Fig. 3A). Insulin also increased expression of 48% of the genes in the BMP signaling pathway (Fig. 3A), which is important in brain development (36). On the other hand, insulin suppressed multiple genes involved in the nuclear factor-κB signaling, surface receptor regulation, and DNA repair (Fig. 3A). Thus, in the HTM, insulin is a potent regulator at the transcriptional level of pyruvate metabolism, lipid and cholesterol biosynthesis, neurotransmission, inflammation, and DNA repair.
Figure 3.
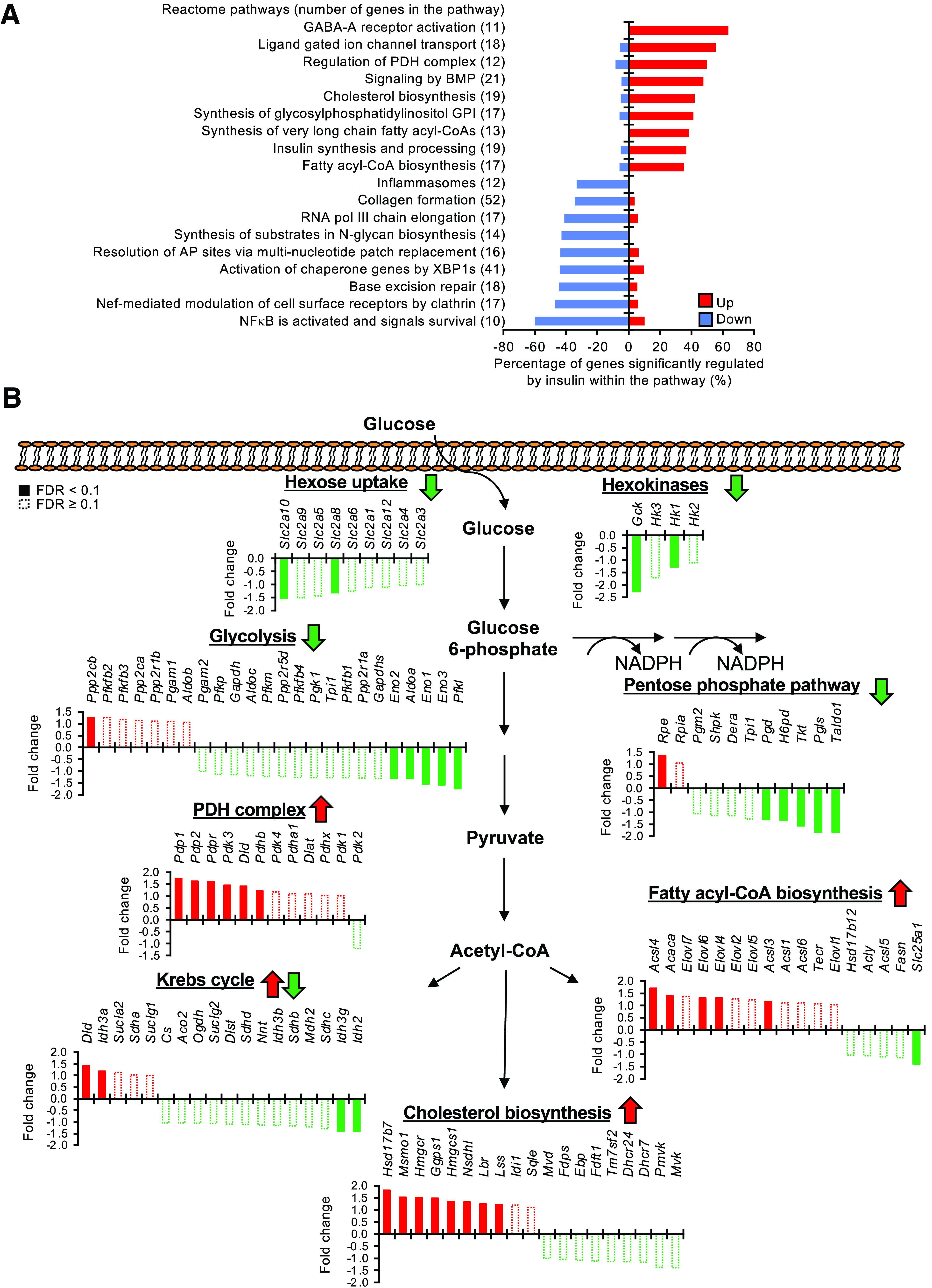
Insulin acutely reprograms glucose metabolism in the HTM. A: Pathway analysis showing top upregulated (Up) and downregulated (Down) pathways by insulin infusion. Percentages of genes significantly regulated by insulin (P < 0.05) within the pathway are shown. Total number of genes within the pathway are indicated in the parentheses. B: Regulation of genes involved in hexose uptake, hexokinases, glycolysis, pentose phosphate pathway, PDH complex regulation, Krebs cycle, fatty acyl-CoA biosynthesis, and cholesterol biosynthesis by low-dose insulin infusion. Upregulation of genes is represented by fold change (insulin/saline). Suppression of genes is represented by – fold change (saline/insulin). Solid bars indicate FDR <0.1; open dashed bars indicate FDR ≥0.1. The overall direction of the regulation of each pathway by insulin is indicated by arrows. pol, polymerase.
Insulin Regulates Genes of Glucose and Lipid Metabolism in the HTM
Although brain glucose uptake is traditionally considered insulin independent, expression analysis revealed novel aspects of insulin action on genes involved in glucose uptake and metabolism in the brain. Thus, insulin infusion significantly suppressed expression of the facilitated hexose transporters Slc2a8 and -10 (Glut-8 and -10) with a trend toward suppression of Slc2a1 (Glut-1) in the HTM (Fig. 3B). Likewise, among the four hexokinases critical for the initial step in intracellular glucose metabolism, glucokinase (Gck) and hexokinase 1 (Hk1) were suppressed by insulin (Fig. 3B). After phosphorylation, glucose-6-phosphate can flux into either the glycolysis pathway for production of ATP and pyruvate or the pentose phosphate pathway for NADPH generation, both of which were predominantly inhibited by insulin (Fig. 3B). Indeed, the most significantly suppressed glycolytic gene by insulin was Pfkl, the rate-limiting enzyme in glycolysis (Fig. 3B). Together, these data show that insulin suppresses the transcription of genes involved in glucose uptake/metabolism in the HTM.
Pyruvate can be converted to acetyl-CoA by PDH, oxaloacetate by pyruvate carboxylase (Pcx), and lactate by lactate dehydrogenase (Ldh). During the clamp, insulin upregulated the PDH phosphatase subunits Pdp1, Pdp2, and Pdpr (Fig. 3B) and suppressed Ldha and Ldhd (Supplementary Fig. 3). These changes would promote the conversion of pyruvate to acetyl-CoA rather than lactate. Along this line, insulin also significantly enhanced the expression of astrocyte-specific monocarboxylate transporter-1 (Mct-1 or Slc16a1), as well as neuron-specific Mct-2 (Slc16a7), in the HTM (Supplementary Fig. 3), suggesting that insulin would promote the shuttling of lactate between neurons and astrocytes.
Acetyl-CoA can enter the Krebs cycle for complete oxidation or serve as a building block for lipid biosynthesis. While insulin showed minor effects on the expression of genes in the Krebs cycle (Fig. 3B), it strongly stimulated the expression of multiple enzymes involved in the biosynthesis of long-chain fatty acyl-CoA and cholesterol. Thus, the expressions of long-chain fatty acyl-CoA synthetases (Acsl3, Acsl4, and Acsl6), very-long-chain fatty acid elongases (Elovl4 and Elovl6), and acetyl-CoA carboxylase 1 (Acaca) were all significantly upregulated by insulin (Fig. 3B). Insulin showed no significant transcriptional regulation of lipoprotein lipase (LPL), important for free fatty acid and lipoprotein sensing in the HTM and regulation of energy balance (37). Insulin also robustly enhanced the expression of 8 of the 19 enzymes in the cholesterol biosynthesis pathway, including Hmgcr, the rate-limiting enzyme in this pathway, which was increased by >50% (Fig. 3B). This is consistent with our previous finding that a major effect of insulin on the brain is to regulate cholesterol biosynthesis (38,39). Collectively, these changes indicate that insulin tunes down glycolysis and pentose shunt activity and reroutes carbons to synthesis of cholesterol and long-chain fatty acyl groups, which could eventually be incorporated into phospholipids important for plasma membrane remodeling.
Insulin Transcriptionally Modulates Neurotransmission in the HTM
In addition to neurometabolism, insulin significantly regulates the expression of genes involved in neurotransmission. Thus, insulin showed strong regulatory effects on both ionotropic and metabotropic glutamate receptors, with 8 of 23 genes regulated (false discovery rate [FDR] < 0.1) (Fig. 4). Notably, insulin’s effects on ionotropic glutamate receptors are restricted to N-methyl-d-aspartate (NMDA) receptors (induced, Grin2a and Grin2b; suppressed, Grin2d) and kainite receptors (induced, Grik2; suppressed, Grik3 and Grik5) but not AMPA receptors (Fig. 4). In contrast to the mixed regulation in glutamate receptors, insulin predominantly upregulated the expression of subunits of GABA receptors (Fig. 4). Insulin significantly upregulated 6 of the 17 GABA receptor subunits detected (35%), in some cases by as much as threefold, while expression was not significantly suppressed of any subunit (FDR < 0.1) (Fig. 4).
Figure 4.
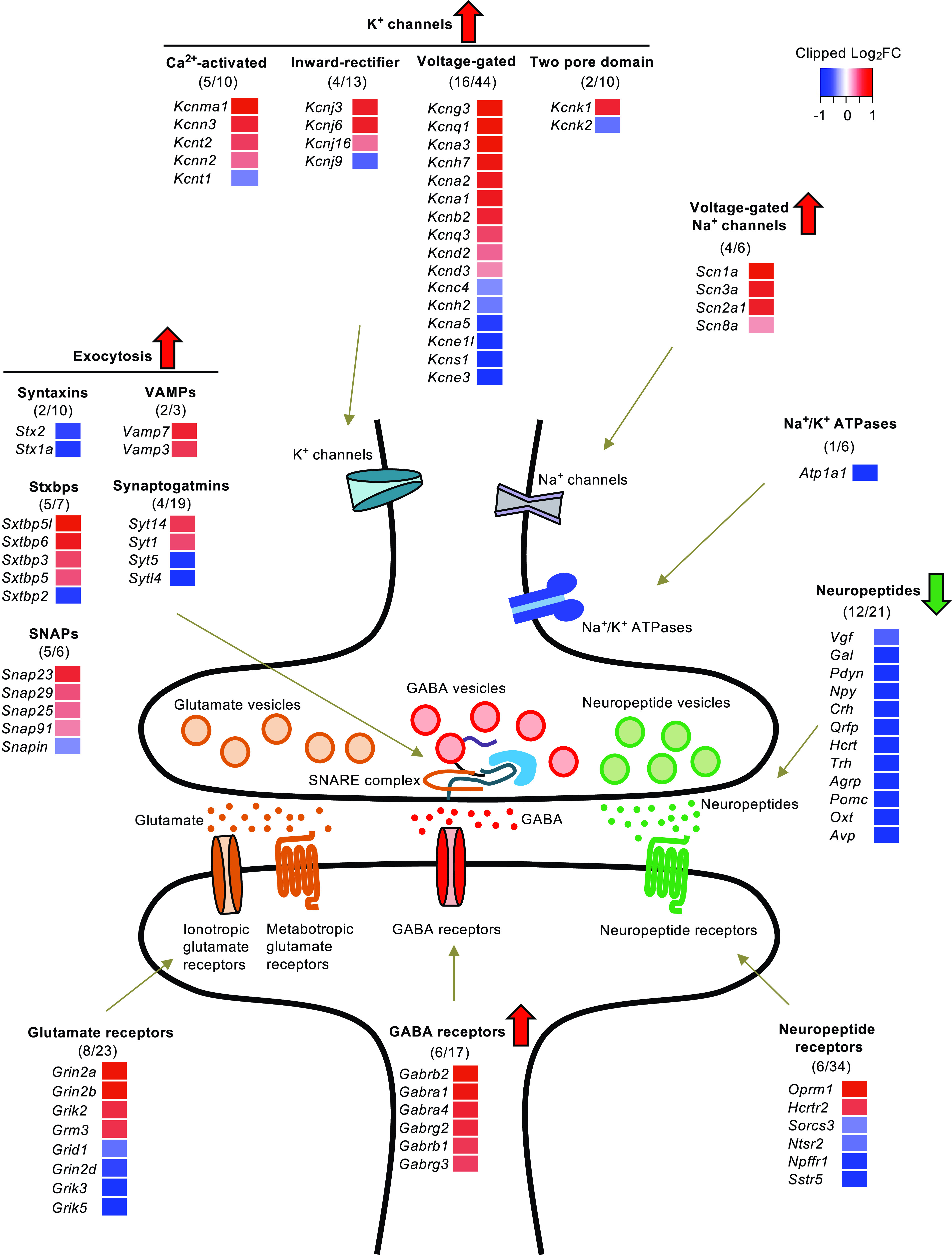
Insulin regulates multiple pathways of neurotransmission and neuromodulation in the HTM. Regulation of the subunits of the glutamate receptors, GABA receptors, neuropeptides and their receptors, components in exocytosis machinery, Na+ and K+ channels, and Na+/K+ ATPases by low-dose insulin. The number of genes significantly regulated by insulin (FDR <0.1) over the total number of genes within the group is indicated in the parentheses. The magnitude of regulation is represented in the heat map by the Log2(fold change).
The HTM is enriched for neurons expressing various neuropeptides known for the control of various types of behaviors and systemic metabolism. Among the 21 neuropeptides identified in the RNA-seq, 12 were significantly suppressed by insulin, whereas none were significantly upregulated (Fig. 4). The degree of the suppression of these neuropeptides during the low-dose insulin infusion was dramatic. For instance, oxytocin (Oxt) and arginine vasopressin (Avp) were suppressed by >90% within the 3 h insulin infusion, and proopiomelancortin-α (Pomc), agouti-related neuropeptide (Agrp), thyrotropin-releasing hormone (Trh), and several others were suppressed by >70% (Fig. 4). While the regulation of the receptors for neuropeptides by insulin was more moderate, some neuropeptide receptors showed more dramatic changes. For example, the opioid receptor mu 1 (Oprm1), which has high affinity for enkephalins and β-endorphin, was upregulated by low-dose insulin by ∼fourfold, while the somatostatin receptor 5 (Sstr5) and neuropeptide FF receptor 1 (Npffr1/Grp147) were suppressed by up to 50% (Fig. 4).
In addition to these neurotransmission-related pathways, insulin showed robust regulatory effects on pathways involved in neurotransmitter release and action potential generation and propagation. Insulin significantly regulated multiple components of the exocytosis machinery, including 5 of 7 syntaxin-binding proteins, 5 of 6 SNAPs, 2 of 3 VAMPs, 2 of 10 syntaxins, and 4 of 19 synaptotagmins (Fig. 4). Of these regulated genes, >66% were significantly upregulated, indicating increased capacity for neurotransmitter and neuropeptide release. Insulin also significantly induced four of the six voltage-gated sodium channel subunits and suppressed none (Fig. 4). Likewise, among the four types of potassium channels, insulin upregulated expression of 23.3% of the 77 subunits detected, while suppressing only 11.7% (Fig. 4). On the other hand, insulin had little effect on the expression of Na+/K+ ATPases (Fig. 4) involved in maintaining resting membrane potential. Taken together, these data demonstrate that changes in peripheral insulin levels can have dramatic and acute effects on expression of gene involved in neurotransmission and neuromodulation, indicating an important regulatory role of insulin on hypothalamic gene expression in control of systemic metabolism and behavior.
The Transcriptional Effects of Insulin Involve Multiple Cell Types in the HTM
Considering the high degree of cellular heterogeneity of the brain, we next sought to explore the role of insulin in different cell types within the HTM. Using the single-cell profiling data (40), we identified gene sets enriched for each cell type and analyzed the insulin-dependent regulation in these cell type–specific gene sets (Fig. 5 and Supplementary Table 3). Among the 164 genes that could be defined as neuron specific, 88 were significantly regulated by insulin. These insulin-regulated, neuron-specific genes were enriched in pathways related to the modulation of synaptic transmission and plasticity, cytoskeletal remodeling, and neuron projection development (Fig. 5A). By comparison, the main effects of insulin on astrocytes were related to the regulation of genes involved in cellular metabolism and communication. Thus, 60 of 181 astrocyte-specific genes were significantly regulated by insulin, and these were enriched in pathways of ion transport and carboxylic acid and lipid biosynthesis (Fig. 5B). Insulin also significantly regulated 52 of 137 oligodendrocyte-enriched genes, including genes involved in myelination, cytoskeletal organization, and long-chain fatty acid biosynthesis (Fig. 5C). Among the 252 microglia-specific genes, 77 were significantly regulated by insulin, including genes involved in protein translation and the biogenesis of ribosomes (Fig. 5D). Finally, of the 53 endothelial specific genes, insulin significantly regulated the expression of 17; these were highly enriched in processes of lipid metabolism, transmembrane transport, and other homeostatic processes (Fig. 5E), suggesting a potential role of insulin in blood-brain barrier function.
Figure 5.
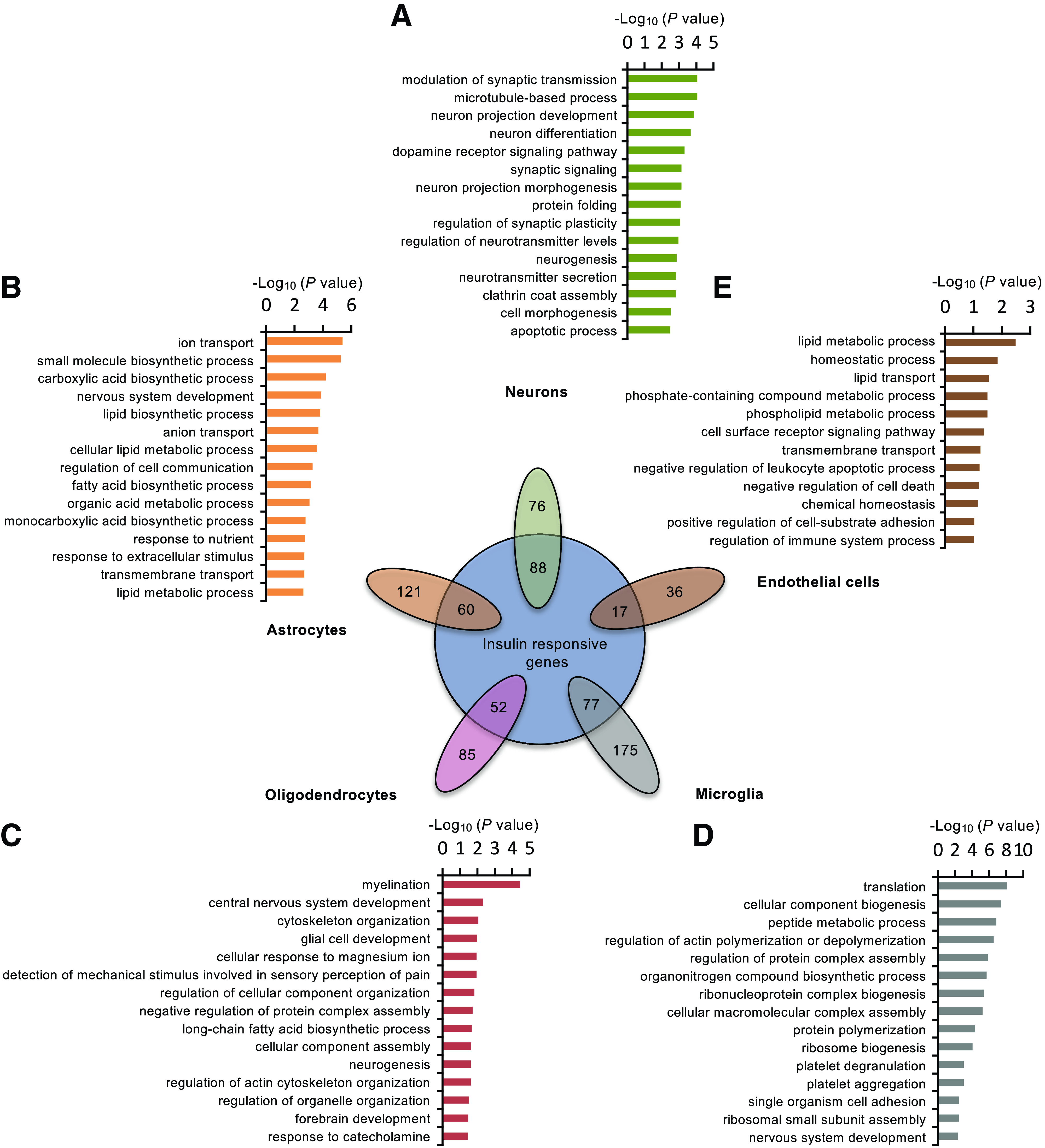
Insulin displays distinct transcriptional effects on populations of neuronal and nonneuronal cells. Venn diagram showing the proportion of cell type–specific genes based on previously performed single-cell RNA-seq (40) that were significantly regulated by low-dose insulin (P < 0.05). The top-enriched GO term FAT pathways from low-dose insulin regulated genes in neurons (A), astrocytes (B), oligodendrocytes (C), microglia (D), and endothelial cells (E) are shown.
Chronic Insulin Deficiency Impairs the Expression of Genes Involved in the De Novo Lipid Synthesis in the HTM
The insulin clamp reflects relatively acute effects of insulin action on the HTM under euglycemic conditions. To compare this with more chronic insulin actions in the brain, we induced insulin deficiency in 3-month-old male C57BL/6J mice by injection of the β-cell toxin STZ. These mice showed an almost complete absence of insulin in the serum (Supplementary Fig. 4A), which was accompanied by marked hyperglycemia (Supplementary Fig. 4B). Both the insulin deficiency and hyperglycemia could be rescued by insulin treatment of the STZ mice (Supplementary Fig. 4A and B).
In a manner opposite that of the insulin clamp, in the STZ mouse, the majority (∼83%) of the genes involved in the long-chain fatty acyl-CoA and cholesterol biosynthesis were dramatically downregulated in the HTM of STZ mice compared with controls (Fig. 6A and B). Furthermore, this reduction was largely normalized by insulin treatment of the STZ mice (Fig. 6A and B). Not all of the changes observed in the clamp are mirrored in STZ diabetes, since there are also secondary effects of hyperglycemia and the altered metabolic state. Thus, genes involved in glucose uptake and glycolysis were significantly suppressed, not increased, in the HTM of STZ mice (Supplementary Fig. 4C), likely reflecting the effects of chronic hyperglycemia (41). In addition, chronic insulin deficiency and hyperglycemia resulted in a pattern of regulation of genes involved in neurotransmission different from that of the clamp. Thus, loss of systemic insulin leads to dramatic increases of Agrp and Npy, and reduction of Pomc and Cart (Fig. 6C), reflecting the caloric status of the STZ mice. The changes in the expression of glutamate and GABA receptors were moderate in the chronic insulin deficiency model (Supplementary Fig. 4D). These studies demonstrate that insulin action in the brain has a complex kinetics and may be modified by many other metabolic signals, such as glucose levels and nutritional status. Importantly, the anabolic effects of insulin to promote long-chain fatty acyl-CoA and cholesterol biosynthesis, which are important for membrane and synapse formation, are observed in both the acute and chronic effects of insulin action.
Figure 6.
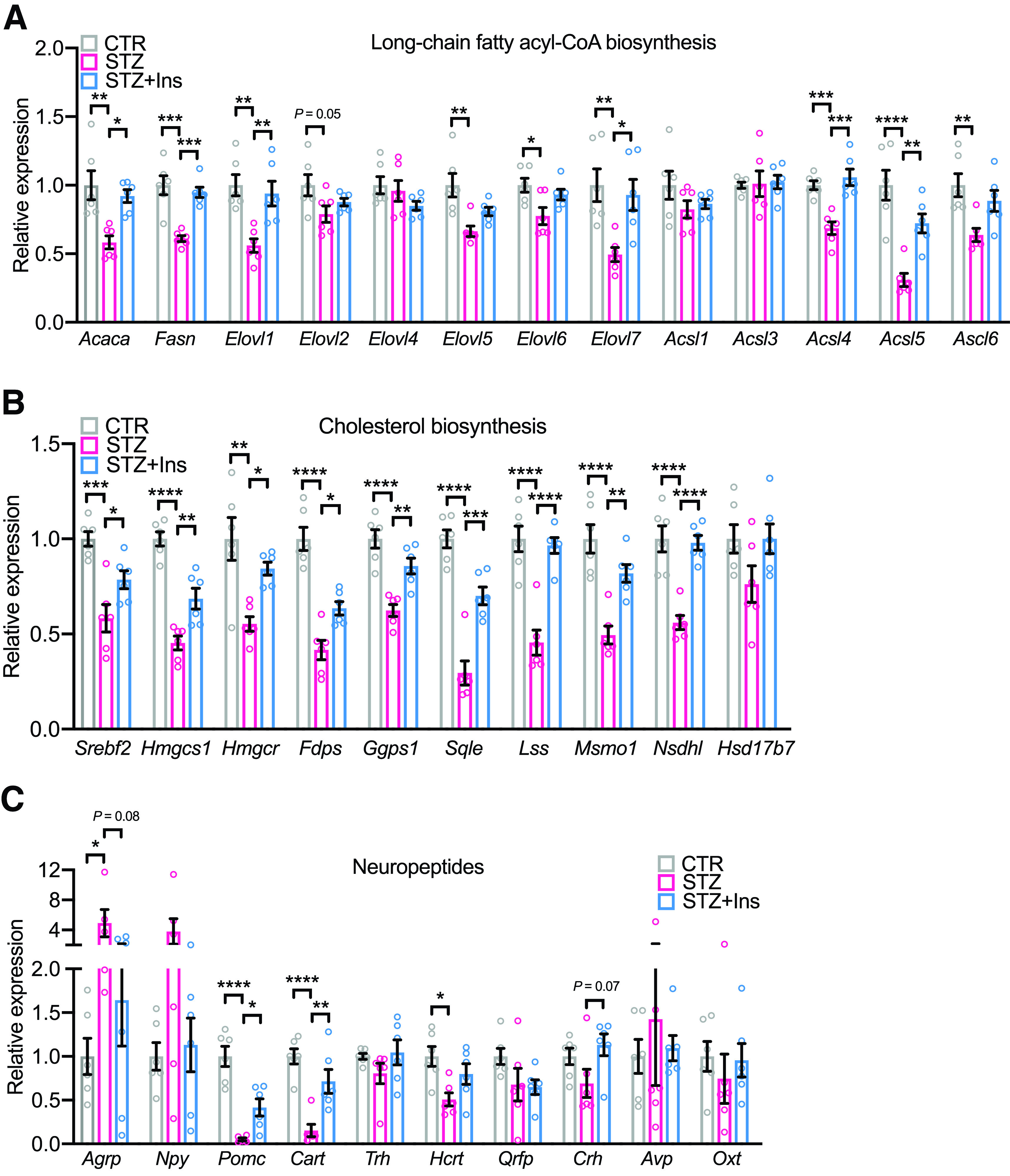
Insulin deficiency reduces the expression of genes involved in de novo lipid biosynthesis in the HTM. Relative mRNA expression of genes involved in long-chain fatty acyl-CoA biosynthesis (A), cholesterol biosynthesis (B), and neuropeptides (C) in the HTM of control (CTR), STZ, and STZ + insulin (STZ+Ins) groups. Tbp was used as internal control. N = 6. One-way ANOVA followed by Tukey multiple comparison test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are shown as means ± SEM.
Cell-Autonomous Effects of Insulin on Gene Transcription in Neurons and Astrocytes
To better dissect the primary effects of insulin versus the secondary effects due to systemic metabolic responses to insulin, we performed insulin stimulation of primary cultured neurons and astrocytes, the two most abundant cells in the brain. Indeed, acute insulin stimulation was sufficient to induce the expression of NMDA receptor subunits Grin2a and Grin2b in primary cultured neurons (Supplementary Fig. 5A), consistent with the insulin clamp data. On the other hand, insulin showed opposing regulatory effects on genes involved in glucose metabolism in neurons and astrocytes. Thus, insulin enhanced the expression of most glycolytic genes in primary neurons (Supplementary Fig. 5B), while suppressing expression of many of these genes in primary astrocytes (Supplementary Fig. 5C). Finally, insulin stimulation enhanced expression of many of the genes involved in long-chain fatty acyl-CoA and cholesterol biosynthesis in both primary neurons and astrocytes (Fig. 7A and B and Supplementary Fig. 6), consistent with the effects observed in vivo. Given the critical role of astrocytes as a source of supply of phospholipid and cholesterol to neurons and synapses (39,42), we assessed the effect of insulin on de novo lipogenesis in astrocytes using 14C-labeled sodium acetate. As shown in Fig. 7C, insulin induced de novo lipogenesis in primary astrocytes in a dose-dependent manner. Therefore, this important anabolic effect of insulin in the brain for phospholipid and cholesterol biosynthesis is observed in both acute and chronic in vivo models, as well as in primary cultured neurons and astrocytes.
Figure 7.
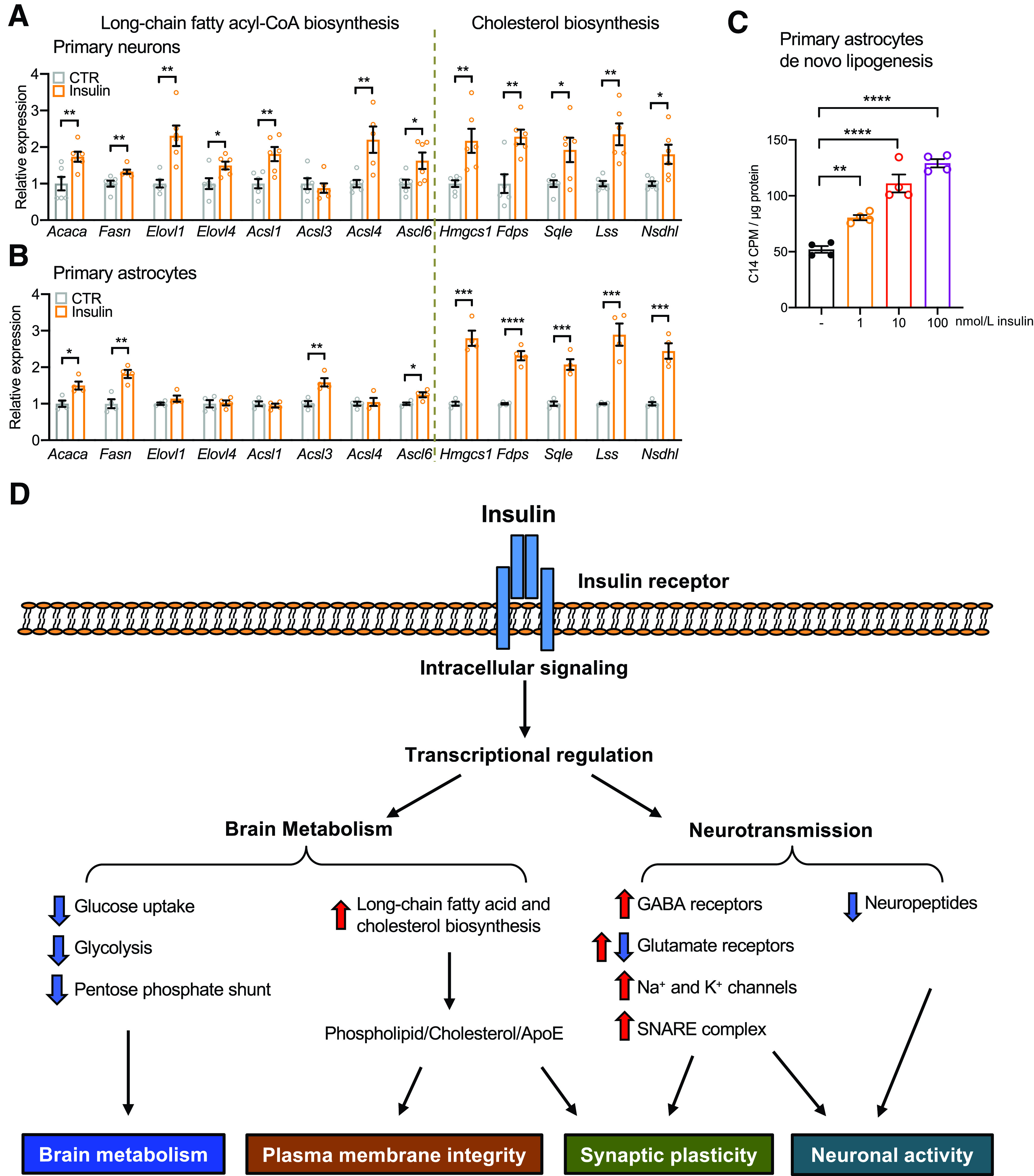
Insulin promotes long-chain fatty acyl-CoA and cholesterol biosynthesis in both neurons and astrocytes. A: Relative mRNA expression of key genes involved in the long-chain fatty acyl-CoA and cholesterol biosynthesis in the primary cultured neurons following 100 nmol/L insulin stimulation for 3 h. Tbp was used as internal control. N = 6. Student t test. *P < 0.05; **P < 0.01. B: Relative mRNA expression of key genes involved in the long-chain fatty acyl-CoA and cholesterol biosynthesis in the primary cultured astrocytes following 100 nmol/L insulin stimulation for 6 h. Tbp was used as internal control. N = 4. Student t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. C: 14C incorporation into lipids following 1, 10, or 100 nmol/L insulin stimulation in primary astrocytes in a de novo lipogenesis assay using 14C-labeled sodium acetate as substrates. N = 4. One-way ANOVA followed by Tukey multiple comparison test. **P < 0.01; ****P < 0.0001. Data are shown as mean ± SEM. D: Physiological levels of insulin regulate gene expression in the brain, especially in the HTM. As a result, insulin reduces glucose metabolism and reroutes carbons to biogenesis of long-chain fatty acid and cholesterol, both of which are important for plasma membrane integrity and synaptic plasticity. Insulin also directly upregulates GABA receptors, Na+ and K+ channels, and SNARE complex components; regulates many glutamate receptor subunits; and suppresses many neuropeptides in the HTM. Collectively, these changes modulate brain metabolism and neurotransmission. CTR, control.
In summary, our data show that relatively acute increases of insulin within the physiological range trigger robust transcriptional responses in the brain, which are most remarkable in the HTM. In general, insulin downregulates expression of genes involved in glucose metabolism and reroutes carbons to biogenesis of cholesterol and phospholipids, reducing oxidative stress and enhancing synthesis of plasma membranes and synaptic remodeling. In addition, insulin regulates the expression of glutamate receptors; induces GABA receptors, Na+ and K+ channels, and SNARE complex components; and suppresses multiple neuropeptides, all of which contribute to synaptic plasticity and neuronal activity (Fig. 7D). Taken together, these data reveal how changes in peripheral insulin can regulate a broad and complex network of genes critical for brain metabolism and neurotransmission.
Discussion
Insulin is central to the regulation of systemic metabolism, control of blood glucose, and energy homeostasis. This involves effects of insulin on multiple peripheral tissues, especially liver, muscle, and fat (3). Although it is well-documented that brain expresses insulin receptors, insulin action in the central nervous system at a molecular level is less well studied. In the current study, we performed gene expression analysis in three different brain regions in mice using a hyperinsulinemic-euglycemic clamp, which allowed us to assess the effects of physiological levels of peripheral insulin on the brain under euglycemia.
The most robust transcriptional regulation by peripheral insulin infusion is observed in the HTM followed by the HCA and the NAC. Indeed, the number of regulated genes in the HTM exceeds the number in two classical target tissues of insulin action (liver and muscle) combined (33). This may reflect the complexity of the cellular composition of the HTM compared with liver and muscle, as well as the fact that within the brain the HTM has a more permeable blood-brain barrier compared with HCA and NAC (43). Previous studies have shown that all areas of the brain respond to peripheral insulin with stimulation of insulin receptor and downstream phosphorylation events (5), indicating that there must be differences in intrinsic insulin responsiveness in each region. Furthermore, this occurs despite the fact that the concentrations of insulin in the cerebrospinal fluid lag those in the circulation and average out to be only 10–20% of peripheral insulin levels (8). These findings indicate that the brain, and especially the HTM, is at least as insulin responsive as classical peripheral target tissues, despite its exposure to lower insulin concentrations than peripheral tissues. Interestingly, in the case of the HTM, more genes were regulated by the low-dose insulin infusion than the high-dose infusion. This reversed dose-dependent response to insulin in the HTM in terms of gene regulation further indicates that for this brain region the higher-dose insulin infusion produces some downregulation, either in the insulin transport process or insulin signaling process. The exact cause remains to be determined.
Numerous loss-of-function studies have shown that insulin signaling in the brain acts on both neuronal and nonneuronal cells and is required for neural control of systemic metabolism, as well as mood and behavior (6,14–16,19–24). Insulin activates the PI3K/Akt signaling cascade to regulate intracellular ATP levels and KATP channel opening, thus inhibiting AgRP neurons and appetite (18). Insulin has been shown to regulate phosphorylation of subunits and membrane localization of NMDA and AMPA receptors in excitatory hippocampal neurons to facilitate opening of these cation channels and neuronal activation (44,45). In addition, we have shown that insulin controls ATP release from astrocytes by regulating Munc18c phosphorylation and SNARE-dependent exocytosis, thus modulating dopaminergic signaling and mood (21). The current study reveals that insulin also acutely regulates distinct sets of genes and pathways in neurons, astrocytes, oligodendrocytes, microglia, and endothelial cells and that these fall into distinct pathways and contribute to the different functions of these cells. Thus, insulin mainly controls transcription of genes related to synaptic function and plasticity in neurons, while it regulates the transcription of genes involved in metabolic, biosynthetic, and transport processes in non-neuronal cells, including astrocytes, oligodendrocytes, and endothelial cells. Thus, through both transcriptional and posttranscriptional effects in the brain, insulin can contribute to a wide range of neural and nonneuronal functions.
Brain is the most energy-demanding organ in the body and primarily relies on glucose as its energy source. The roles of insulin action on brain glucose uptake and metabolism are complex. While brain glucose uptake is primarily dependent on the insulin-independent Glut-1 or Glut-3 or other transporters (46), systemic insulin signaling and insulin resistance have been shown to affect brain glucose metabolism in different brain regions. Positron emission tomography–computed tomography studies have shown that peripheral infusion of insulin acutely increases glucose uptake in healthy humans (47). In a cohort with a wide range of systemic insulin sensitivity, however, brain glucose uptake under euglycemic-hyperinsulinemic conditions was negatively associated with insulin sensitivity (48), indicating a suppressive effect of insulin on brain glucose uptake. The transcriptional changes observed in the current study indicate novel aspects of insulin action on glucose metabolism in the brain. Our study indicates that insulin transcriptionally regulates brain glucose uptake and metabolism at multiple levels. Thus, genes controlling glucose phosphorylation, glycolysis, and the pentose phosphate pathway are markedly suppressed in the HTM in response to physiological changes in insulin, predicting a reduced protein expression of the enzymes for brain glucose uptake and metabolism. In this way, in the postprandial state, when insulin and glucose levels are high, the HTM can balance glucose uptake and oxidation rates and avoid excessive reactive oxygen species and ATP generation, which could be toxic to neurons. In addition, despite predominant suppression of glycolytic enzymes by insulin in the clamp study, insulin elicits opposing effects on primary cultured astrocytes and neurons. Thus, insulin primarily induced the expression of many genes involved in glycolysis in cultured neurons, while suppressing many of the same genes in astrocytes, highlighting differential effects of insulin response in different cell types in the brain.
On the other hand, insulin upregulates enzymes to use extra glucose that may enter the cell for lipid synthesis, such as cholesterol and long-chain fatty acyl-CoA biosynthesis, both of which are essential building blocks for plasma membranes and synaptic vesicles and important for synaptic remodeling and plasticity. The effect on cholesterol biosynthesis is consistent with the findings of our previous studies, which showed that decrease in insulin levels in an STZ mouse model of type 1 diabetes results in decreased cholesterol synthesis in the brain and is associated with impaired synaptic function (38). This may be one of the key mechanisms of the potential beneficial effects of insulin in the brain in neurodegenerative disorders, since the brain must synthesize almost all of its own cholesterol (49) and impairment of biosynthesis of cholesterol and long-chain fatty acyl groups in the brain has been shown to cause impaired neuronal activity, abnormal mood behavior, and cognitive decline (39,50).
A more unexpected and interesting finding is that insulin exhibits a potent regulatory role for multiple neurotransmission systems in the HTM. Insulin dramatically upregulates the expression of >75% of GABA receptor subunits. As the major inhibitory neurotransmitter system in the central nervous system, GABA and its receptors are essential “brakes” to prevent constant and excessive excitatory activity, which could lead to excitotoxicity and neurodegeneration (51). Indeed, defects in the GABA system are evident in many types of neurological disorders, including depression and Alzheimer disease (52,53). Additional studies will be needed to explore the role of insulin as a neuron-protective agent by enhancing GABA neurotransmission signaling.
Insulin also has strong effects on glutamate receptor signaling. While AMPA and NMDA receptors are two most important ionotropic channels gated by glutamate binding, insulin only shows robust transcriptional regulation on NMDA receptors, which are composed of two glutamate-binding NR2 subunits (Grin2a, Grin2b, Grin2c, or Grin2d) and two glycine/d-serine binding subunits. The upregulation of Grin2a and Grin2b and downregulation of Grin2d by insulin would change the composition and thus the kinetic properties of NMDA receptors (54). How these insulin-regulated changes in the expression levels of different subunits shift the stoichiometry and the electrophysiological properties of the NMDA receptors deserves further investigation. Although acute insulin infusion has only a moderate effect on the expression of the AMPA receptor subunits, chronic deletion of insulin signaling in the brain leads to decreased protein expression of GluA1 subunit (55) and thus associated decreases in GluA1/GluA2-containing channels important for long-term potentiation (44,56). Hence, instead of transcriptional regulation, insulin regulates AMPA receptors at the posttranscriptional level to modulate synaptic plasticity.
One major function of the HTM is regulation of appetite and energy balance. Insulin has been shown to have an anorexigenic effect when administered intracerebroventricularly (i.c.v.) (10,11). Indeed, both acute elevation of peripheral insulin (the current study) and chronic i.c.v. injections of insulin (57) strongly suppress the expression of the orexigenic neuropeptides Agrp and Npy, leading to suppression of appetite. Somewhat surprisingly, we found that insulin infusion during the clamp also inhibits the expression of anorexigenic neuropeptide Pomc. This is contrary to the effects of chronic insulin administration (58). This paradoxical suppression of Pomc suggests that there are different long- and short-term effects of insulin in these neurons or that the long-term effects could reflect secondary changes that can occur in a chronic setting. Supporting this, Könner et al. (23) have shown that insulin acutely suppresses neuronal activity of both AgRP and POMC neurons. An independent study has shown that insulin can excite POMC neurons (18), implying a potential heterogeneity of POMC neurons different on insulin response. The question of how the rapid transcriptional regulation of Agrp and Pomc contributes to behavior in mice needs further investigation. While the opposing regulation of the expression of Agrp and Pomc are evident at both fast and fed conditions, several studies suggest that the rapid appetite-controlling actions of the HTM may depend more on glutamate and GABA transmission (59,60). The excitability of AgRP and POMC neurons, on the other hand, are dynamically controlled by not only nutritional status (61) but also anticipation of feeding when the food is presented (62,63). In addition, insulin regulation of Agrp and Pomc expression in the HTM may contribute to some behavioral modulation in addition to food intake.
Due to the complex interplay of insulin, glucose, and many counterregulatory mechanisms, it is extremely challenging to dissect isolated brain insulin action under physiological conditions in a conscious animal or a human. The current study takes advantage of the best available experimental technique, hyperinsulinemic-euglycemic clamp, to tackle this question in mice under tight control of circulating blood glucose and minimized counterregulation (27). However, there are some technical considerations. First, although glucose levels were maintained between 110 and 170 mg/dL in all groups of mice, there were some glycemic fluctuations in the initial stage (∼60 min) of the clamp, which could potentially have some long-term effects on the transcriptional profiles at 3 h. While challenging, in future studies it would be interesting to dissect potential glucose effects on gene expression and compare and contrast these with the insulin effects. Second, we analyzed the global transcriptome of each brain regions to ensure in-depth coverage. While we were able to assess insulin responses on marker genes for different cell types using a bioinformatics approach, future studies using single-cell or single-nucleus RNA-seq will allow a more in-depth analysis of insulin response at the cellular level in important regions such as the HTM. Finally, future studies are needed to investigate the sex-dependent insulin responses in the brain, which have been demonstrated in several rodent models of insulin receptor deletion in the brain (14,16,21).
In summary, changes of peripheral levels of insulin within the physiological range elicit robust, acute transcriptional regulation in the brain, inducing changes that can regulate brain metabolism and neurotransmission. In addition to previously reported insulin-regulated genes such as Insig1, AgRP, and the genes of cholesterol synthesis, insulin regulates multiple genes involved in glucose and lipid metabolism and neurotransmission, as well as many noncoding RNAs. Together, these create a complex of interacting networks of important biological processes. These findings provide new insights into not only ways that insulin can modulate brain function but also how insulin resistance in brain can contribute to altered mood, behavior, cognition, and neurodegeneration.
Article Information
Funding. This work was supported by NIH grants R37 DK031036 and R01 DK033201 (to C.R.K.) and 5U2C-DK093000 (to J.K.K.) and the Mary K. Iacocca Professorship (to C.R.K.). W.C. was supported by NIH grants K01 DK120740 and P30 DK057521, A.K.R. was supported by NIH grant T32 DK007260-37, T.M.B was partially supported by a grant from Sao Paulo Research Foundation (2014/25370-8), R.G.-M. was supported by a Deutsche Forschungsgemeinschaft fellowship, and S.S. was supported by NIH grant P30 GM127211 and NASPGHAN Foundation Young Investigator Award. G.W. was supported by an American Diabetes Association postdoctoral fellowship grant (1-18-PDF-171), and B.T.O. was supported by NIH grant K08 DK100543. The authors thank Jonathan M. Dreyfuss and Hui Pan from Joslin Diabetes Center DRC Genomics and Bioinformatics Core (P30 DK036836) for assistance with data analysis.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. W.C. designed research, performed experiments, analyzed the data, and wrote the manuscript. X.Z., T.M.B., R.G.-M., S.S., G.W., A.K.R., M.K., and B.T.O. helped with experiments and data analysis and reviewed the manuscript. J.H.K. performed the hyperinsulinemic-euglycemic clamps. J.K.K. supervised clamp studies. C.R.K designed research, wrote the manuscript, and supervised the project. C.R.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.14568204.
References
- 1.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 2006;7:85–96 [DOI] [PubMed] [Google Scholar]
- 2.Haeusler RA, McGraw TE, Accili D. Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol 2018;19:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen MC, Shulman GI. Mechanisms of insulin action and insulin resistance. Physiol Rev 2018;98:2133–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray SM, Aylor KW, Barrett EJ. Unravelling the regulation of insulin transport across the brain endothelial cell. Diabetologia 2017;60:1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konishi M, Sakaguchi M, Lockhart SM, et al. Endothelial insulin receptors differentially control insulin signaling kinetics in peripheral tissues and brain of mice. Proc Natl Acad Sci U S A 2017;114:E8478–E8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinridders A, Ferris HA, Cai W, Kahn CR. Insulin action in brain regulates systemic metabolism and brain function. Diabetes 2014;63:2232–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strubbe JH, Porte D Jr, Woods SC. Insulin responses and glucose levels in plasma and cerebrospinal fluid during fasting and refeeding in the rat. Physiol Behav 1988;44:205–208 [DOI] [PubMed] [Google Scholar]
- 8.Kern W, Benedict C, Schultes B, et al. Low cerebrospinal fluid insulin levels in obese humans. Diabetologia 2006;49:2790–2792 [DOI] [PubMed] [Google Scholar]
- 9.Beard JC, Ward WK, Halter JB, Wallum BJ, Porte D Jr. Relationship of islet function to insulin action in human obesity. J Clin Endocrinol Metab 1987;65:59–64 [DOI] [PubMed] [Google Scholar]
- 10.Woods SC, Lotter EC, McKay LD, Porte D Jr. Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature 1979;282:503–505 [DOI] [PubMed] [Google Scholar]
- 11.Florant GL, Singer L, Scheurink AJ, Park CR, Richardson RD, Woods SC. Intraventricular insulin reduces food intake and body weight of marmots during the summer feeding period. Physiol Behav 1991;49:335–338 [DOI] [PubMed] [Google Scholar]
- 12.Soto M, Herzog C, Pacheco JA, et al. Gut microbiota modulate neurobehavior through changes in brain insulin sensitivity and metabolism. Mol Psychiatry 2018;23:2287–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinridders A, Lauritzen HP, Ussar S, et al. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. J Clin Invest 2013;123:4667–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brüning JC, Gautam D, Burks DJ, et al. Role of brain insulin receptor in control of body weight and reproduction. Science 2000;289:2122–2125 [DOI] [PubMed] [Google Scholar]
- 15.Fisher SJ, Brüning JC, Lannon S, Kahn CR. Insulin signaling in the central nervous system is critical for the normal sympathoadrenal response to hypoglycemia. Diabetes 2005;54:1447–1451 [DOI] [PubMed] [Google Scholar]
- 16.Kleinridders A, Cai W, Cappellucci L, et al. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc Natl Acad Sci U S A 2015;112:3463–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obici S, Feng Z, Karkanias G, Baskin DG, Rossetti L. Decreasing hypothalamic insulin receptors causes hyperphagia and insulin resistance in rats. Nat Neurosci 2002;5:566–572 [DOI] [PubMed] [Google Scholar]
- 18.Qiu J, Zhang C, Borgquist A, et al. Insulin excites anorexigenic proopiomelanocortin neurons via activation of canonical transient receptor potential channels. Cell Metab 2014;19:682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Cáceres C, Quarta C, Varela L, et al. Astrocytic insulin signaling couples brain glucose uptake with nutrient availability. Cell 2016;166:867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manaserh IH, Chikkamenahalli L, Ravi S, Dube PR, Park JJ, Hill JW. Ablating astrocyte insulin receptors leads to delayed puberty and hypogonadism in mice. PLoS Biol 2019;17:e3000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W, Xue C, Sakaguchi M, et al. Insulin regulates astrocyte gliotransmission and modulates behavior. J Clin Invest 2018;128:2914–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh K, Zhang L, Brandon A, et al. Insulin controls food intake and energy balance via NPY neurons. Mol Metab 2017;6:574–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Könner AC, Janoschek R, Plum L, et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab 2007;5:438–449 [DOI] [PubMed] [Google Scholar]
- 24.Shin AC, Filatova N, Lindtner C, et al. Insulin receptor signaling in POMC, but not AgRP, neurons controls adipose tissue insulin action. Diabetes 2017;66:1560–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skeberdis VA, Lan J, Zheng X, Zukin RS, Bennett MV. Insulin promotes rapid delivery of N-methyl-D- aspartate receptors to the cell surface by exocytosis. Proc Natl Acad Sci U S A 2001;98:3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spanswick D, Smith MA, Mirshamsi S, Routh VH, Ashford ML. Insulin activates ATP-sensitive K+ channels in hypothalamic neurons of lean, but not obese rats. Nat Neurosci 2000;3:757–758 [DOI] [PubMed] [Google Scholar]
- 27.Kim JK. Hyperinsulinemic-euglycemic clamp to assess insulin sensitivity in vivo. Methods Mol Biol 2009;560:221–238 [DOI] [PubMed] [Google Scholar]
- 28.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 2010;26:2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 33.Batista TM, Garcia-Martin R, Cai W, et al. Multi-dimensional transcriptional remodeling by physiological insulin in vivo. Cell Rep 2019;26:3429–3443.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 2010;29:3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ, Long Noncoding RNA. Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci 2017;37:1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat Rev Neurosci 2005;6:945–954 [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Astarita G, Taussig MD, et al. Deficiency of lipoprotein lipase in neurons modifies the regulation of energy balance and leads to obesity. Cell Metab 2011;13:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki R, Lee K, Jing E, et al. Diabetes and insulin in regulation of brain cholesterol metabolism. Cell Metab 2010;12:567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris HA, Perry RJ, Moreira GV, Shulman GI, Horton JD, Kahn CR. Loss of astrocyte cholesterol synthesis disrupts neuronal function and alters whole-body metabolism. Proc Natl Acad Sci U S A 2017;114:1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell JN, Macosko EZ, Fenselau H, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 2017;20: 484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang JJ, Jiang L, Sanchez Rangel E, et al. Glycemic variability and brain glucose levels in type 1 diabetes. Diabetes 2019;68:163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauch DH, Nägler K, Schumacher S, et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science 2001;294:1354–1357 [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez EM, Blázquez JL, Guerra M. The design of barriers in the hypothalamus allows the median eminence and the arcuate nucleus to enjoy private milieus: the former opens to the portal blood and the latter to the cerebrospinal fluid. Peptides 2010;31:757–776 [DOI] [PubMed] [Google Scholar]
- 44.Passafaro M, Piëch V, Sheng M. Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat Neurosci 2001;4:917–926 [DOI] [PubMed] [Google Scholar]
- 45.Christie JM, Wenthold RJ, Monaghan DT. Insulin causes a transient tyrosine phosphorylation of NR2A and NR2B NMDA receptor subunits in rat hippocampus. J Neurochem 1999;72:1523–1528 [DOI] [PubMed] [Google Scholar]
- 46.Koepsell H. Glucose transporters in brain in health and disease. Pflugers Arch 2020;472:1299–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bingham EM, Hopkins D, Smith D, et al. The role of insulin in human brain glucose metabolism: an 18fluoro-deoxyglucose positron emission tomography study. Diabetes 2002;51:3384–3390 [DOI] [PubMed] [Google Scholar]
- 48.Rebelos E, Bucci M, Karjalainen T, et al. Insulin resistance is associated with enhanced brain glucose uptake during euglycemic hyperinsulinemia: a large-scale PET cohort. Diabetes Care 2021;44:788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Björkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol 2004;24:806–815 [DOI] [PubMed] [Google Scholar]
- 50.Suzuki R, Ferris HA, Chee MJ, Maratos-Flier E, Kahn CR. Reduction of the cholesterol sensor SCAP in the brains of mice causes impaired synaptic transmission and altered cognitive function. PLoS Biol 2013;11:e1001532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer’s disease. Neurochem Int 2004;45:583–595 [DOI] [PubMed] [Google Scholar]
- 52.Villette V, Dutar P. GABAergic microcircuits in Alzheimer’s disease models. Curr Alzheimer Res 2017;14:30–39 [DOI] [PubMed] [Google Scholar]
- 53.Kendell SF, Krystal JH, Sanacora G. GABA and glutamate systems as therapeutic targets in depression and mood disorders. Expert Opin Ther Targets 2005;9:153–168 [DOI] [PubMed] [Google Scholar]
- 54.McBain CJ, Mayer ML. N-methyl-D-aspartic acid receptor structure and function. Physiol Rev 1994;74:723–760 [DOI] [PubMed] [Google Scholar]
- 55.Soto M, Cai W, Konishi M, Kahn CR. Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior. Proc Natl Acad Sci U S A 2019;116:6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science 2004;305:1972–1975 [DOI] [PubMed] [Google Scholar]
- 57.Schwartz MW, Sipols AJ, Marks JL, et al. Inhibition of hypothalamic neuropeptide Y gene expression by insulin. Endocrinology 1992;130:3608–3616 [DOI] [PubMed] [Google Scholar]
- 58.Benoit SC, Air EL, Coolen LM, et al. The catabolic action of insulin in the brain is mediated by melanocortins. J Neurosci 2002;22:9048–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci 2008;11:998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fenselau H, Campbell JN, Verstegen AMJ, et al. Corrigendum: a rapidly acting glutamatergic ARC→PVH satiety circuit postsynaptically regulated by α-MSH. Nat Neurosci 2017;20:1189. [DOI] [PubMed] [Google Scholar]
- 61.Williams KW, Elmquist JK. From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat Neurosci 2012;15:1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Lin YC, Kuo TW, Knight ZA. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell 2015;160:829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garfield AS, Shah BP, Burgess CR, et al. Dynamic GABAergic afferent modulation of AgRP neurons. Nat Neurosci 2016;19:1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]


