Abstract
Over the past several decades, messenger RNA (mRNA) vaccines have progressed from a scepticism-inducing idea to clinical reality. In 2020, the COVID-19 pandemic catalysed the most rapid vaccine development in history, with mRNA vaccines at the forefront of those efforts. Although it is now clear that mRNA vaccines can rapidly and safely protect patients from infectious disease, additional research is required to optimize mRNA design, intracellular delivery and applications beyond SARS-CoV-2 prophylaxis. In this Review, we describe the technologies that underlie mRNA vaccines, with an emphasis on lipid nanoparticles and other non-viral delivery vehicles. We also overview the pipeline of mRNA vaccines against various infectious disease pathogens and discuss key questions for the future application of this breakthrough vaccine platform.
Subject terms: Nanoparticles, Nucleic-acid therapeutics, Infectious diseases, Drug delivery
The COVID-19 pandemic has established mRNA vaccines as a rapid, effective and safe approach for the protection of individuals from infectious disease. Here, Whitehead and colleagues review the principles of mRNA vaccine design, synthesis and delivery, assessing recent progress and key issues in the development of mRNA vaccines for a range of infectious diseases.
Introduction
Vaccination is the most effective public health intervention for preventing the spread of infectious diseases. Successful vaccination campaigns eradicated life-threatening diseases such as smallpox and nearly eradicated polio1, and the World Health Organization estimates that vaccines prevent 2–3 million deaths every year from tetanus, pertussis, influenza and measles (see Related links). Despite their evident success, however, conventional vaccines do not effectively tackle pathogens such as the malaria parasite Plasmodium falciparum, hepatitis C and human immunodeficiency virus (HIV), which evade immune surveillance2. Furthermore, they require regular modification to address rapidly mutating pathogens, such as the influenza virus.
Nucleic acid vaccines based on mRNA were conceived more than three decades ago in the hope of generating safe and versatile vaccines that are easy to produce3,4. In principle, mRNA vaccines have several advantages over conventional vaccines. Unlike some viral vaccines, mRNA does not integrate into the genome, obviating concerns about insertional mutagenesis5. mRNA vaccines can be manufactured in a cell-free manner, allowing rapid, scalable and cost-effective production. For example, a 5 litre bioreactor can produce almost a million mRNA vaccine doses in a single reaction6. Moreover, a single mRNA vaccine can encode multiple antigens, strengthening the immune response against resilient pathogens7 and enabling the targeting of multiple microbes or viral variants with a single formulation8. Initially, however, mRNA was not pursued as a therapeutic because of concerns about its stability, poor efficacy and excessive immunostimulation. Fortunately, a few tenacious researchers and companies persisted. And over the past decade, by determining mRNA pharmacology, developing effective delivery vehicles and controlling mRNA immunogenicity, interest in clinical applications of mRNA has renewed4.
During the SARS-CoV-2 pandemic, mRNA-based vaccines were shown to be highly effective against SARS-CoV-2 and were developed and administered at unprecedented speed to millions of people globally to combat COVID-19 (Box 1). These vaccines, initially developed by Pfizer–BioNTech and Moderna, have validated the platform and stimulated substantial interest in the application of mRNA for both prophylactic and therapeutic indications.
In this Review, we describe the technical basis of mRNA vaccines, including mRNA design and synthesis, as well as the enabling delivery technologies. The latter topic is a major focus of ongoing efforts for optimization of the platform and is thus emphasized here. We overview progress in developing mRNA vaccines for a wide range of infectious diseases such as influenza, Zika and respiratory syncytial virus (RSV), and discuss key issues facing the future of the platform, including safety, duration of response, application in specific populations and achieving global vaccine access.
Box 1 Developing mRNA vaccines at warp speed.
The sequencing of the SARS-CoV-2 genome in January 2020 kickstarted a race to develop potent vaccines against the novel coronavirus. At that time, it would have been optimistic to expect results within a couple of years. For example, MERS-CoV and SARS-CoV vaccines required about 2 years of development before entering phase I trials259 and have yet to be approved. Moreover, the mumps vaccine, which was then the fastest developed vaccine, required 4 years from conception to approval (see Related links). The Moderna and Pfizer–BioNTech vaccines, in contrast, were approved 11 months after initiation, 8 months of which were spent in clinical trials. Their lightning-fast development was facilitated by three factors: decades of previous research, large financial investments and the inordinate devotion of scientists, engineers, managers, medical personnel and regulatory officials working in concert and around the clock.
SARS-CoV-2 vaccines capitalized upon many years of research on other coronaviruses, small interfering RNA (siRNA) delivery systems and cancer vaccines. For example, research on MERS-CoV and SARS-CoV suggested that the antigen should be the spike protein and that its amino acid sequence should be slightly modified to lock the protein in its prefusion conformation, maximizing its immunogenicity127. These studies enabled Moderna to generate the mRNA-1273 sequence in 2 days and start phase I trials in 66 days127.
A colossal investment was also required for each vaccine candidate, with Pfizer–BioNTech and Moderna receiving $1.95 billion and $2.48 billion, respectively, from the US government through its Operation Warp Speed initiative (see Related links). Finally, none of this would have been possible without the extraordinary effort of the scientific and medical communities, the willingness of BioNTech, Pfizer and Moderna to take risks, and the flexibility of regulatory agencies to allow the simultaneous conduct of phase I, II and III trials.
Principles of mRNA design and synthesis
mRNA vaccines comprise synthetic mRNA molecules that direct the production of the antigen that will generate an immune response. In vitro-transcribed (IVT) mRNA mimics the structure of endogenous mRNA, with five sections, from 5ʹ to 3ʹ: 5ʹ cap, 5ʹ untranslated region (UTR), an open reading frame that encodes the antigen, 3ʹ UTR and a poly(A) tail (Fig. 1). A variation on IVT mRNA, self-amplifying mRNA, also contains replicase genes that encode RNA-dependent RNA polymerase. The virus-derived polymerase amplifies mRNA transcripts intracellularly, enabling the expression of large amounts of antigen with reduced mRNA doses9.
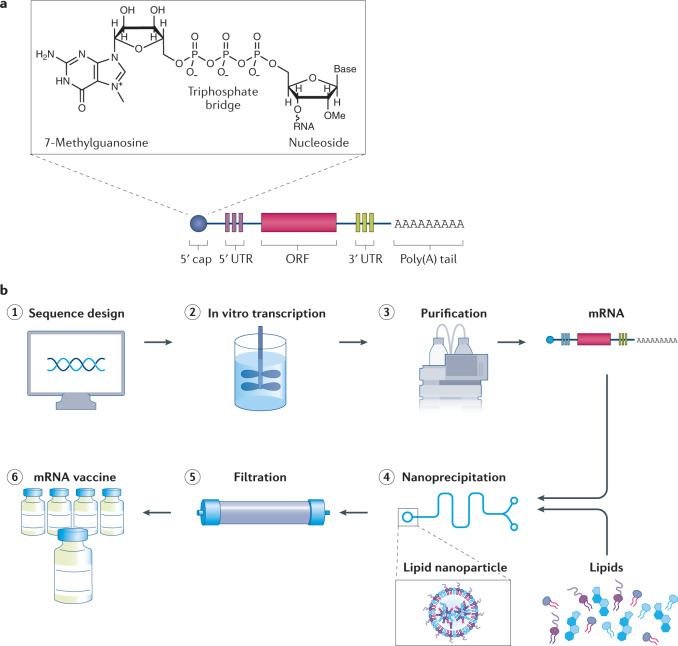
Fig. 1. IVT mRNA is formulated into lipid nanoparticle vaccines using a cell-free production pipeline.
a | In vitro-transcribed (IVT) mRNA contains five structural elements: a 5′ cap containing 7-methylguanosine linked through a triphosphate bridge to a 2′-O-methylated nucleoside, flanking 5′ and 3′ untranslated regions (UTRs), an open reading frame (ORF) and a poly(A) tail. b | The mRNA is synthetically produced and formulated into vaccines. (1) Once the genome of a pathogen has been sequenced, a sequence for the target antigen is designed and inserted into a plasmid DNA construct. (2) Plasmid DNA is transcribed into mRNA by bacteriophage polymerases in vitro and (3) mRNA transcripts are purified by high performance liquid chromatography (HPLC) to remove contaminants and reactants. (4) Purified mRNA is mixed with lipids in a microfluidic mixer to form lipid nanoparticles. Rapid mixing causes the lipids to encapsulate mRNA instantaneously and precipitate as self-assembled nanoparticles. (5) The nanoparticle solution is dialysed or filtered to remove non-aqueous solvents and any unencapsulated mRNA and (6) the filtered mRNA vaccine solution is stored in sterilized vials.
The 5′ cap structure, like that of natural eukaryotic mRNAs, contains a 7-methylguanosine nucleoside linked through a triphosphate bridge to the 5′ end of mRNA10. As in mammals, the first or second nucleotide from the 5′ end is methylated on the 2′ hydroxyl of the ribose (2′-O-methylation), which prevents recognition by cytosolic sensors of viral RNA10, and hence prevents unintended immune responses. Further, the 5′ cap protects the mRNA sterically from degradation by exonucleases, and it works synergistically with the poly(A) tail at the 3′ end, poly(A) binding proteins and translation initiation factor proteins to circularize mRNA and recruit ribosomes for initiating translation10,11. The length of the poly(A) tail indirectly regulates both mRNA translation and half-life. A sufficiently long tail (100–150 bp) is necessary to interact with poly(A) binding proteins that form complexes necessary for initiating translation11,12 and protecting the cap from degradation by decapping enzymes13.
The 5′ and 3′ UTRs flanking the coding region regulate mRNA translation, half-life and subcellular localization10,14. Naturally occurring UTRs from highly expressed genes, such as the α- and β-globin genes, are preferred for synthetic mRNAs15. However, because UTR performance can vary by cell type, alternative UTR sequences may be used that have been optimized for the desired application and intended cell target16–18. These engineered UTR sequences minimize mRNA degradation by excluding miRNA-binding sites11 and AU-rich regions in the 3′ UTR19. Furthermore, they minimize regions that prevent ribosomes from scanning the mRNA transcript, such as sequences with secondary and tertiary structure (for example, hairpins) in the 5′ UTR20.
The open reading frame of the mRNA vaccine is the most crucial component because it contains the coding sequence that is translated into protein. Although the open reading frame is not as malleable as the non-coding regions, it can be optimized to increase translation without altering the protein sequence by replacing rarely used codons with more frequently occurring codons that encode the same amino acid residue. For instance, the biopharmaceutical company CureVac AG discovered that human mRNA codons rarely have A or U at the third position and patented a strategy that replaces A or U at the third position in the open reading frame with G or C21. CureVac used this optimization strategy for its SARS-CoV-2 candidate CVnCoV, which is now in phase III trials (see the mRNA sequence in Related links). Although replacement of rare codons is an attractive optimization strategy, it must be used judiciously. This is because, in the case of some proteins, the slower translation rate of rare codons is necessary for proper protein folding22.
To maximize translation, the mRNA sequence typically incorporates modified nucleosides, such as pseudouridine, N1-methylpseudouridine or other nucleoside analogues23. Because all native mRNAs include modified nucleosides, the immune system has evolved to recognize unmodified single-stranded RNA, which is a hallmark of viral infection. Specifically, unmodified mRNA is recognized by pattern recognition receptors, such as Toll-like receptor 3 (TLR3), TLR7 and TLR8, and the retinoic acid-inducible gene I (RIGI) receptor. TLR7 and TLR8 receptors bind to guanosine- or uridine-rich regions in mRNA and trigger the production of type I interferons, such as IFNα, that can block mRNA translation24. The use of modified nucleosides, particularly modified uridine, prevents recognition by pattern recognition receptors, enabling sufficient levels of translation to produce prophylactic amounts of protein5. Both the Moderna and Pfizer–BioNTech SARS-CoV-2 vaccines, which produced >94% efficacy in phase III clinical trials25, contain nucleoside-modified mRNAs. Another strategy to avoid detection by pattern recognition receptors, pioneered by CureVac, uses sequence engineering and codon optimization to deplete uridines by boosting the GC content of the vaccine mRNA26.
In addition to improvements to the mRNA sequence, significant advances have also been made to streamline mRNA production. Clinically used synthetic mRNA is transcribed in vitro from a DNA plasmid by using the bacteriophage RNA polymerase T7 (T3 and SP6 polymerases can also be used). It is co-transcriptionally capped (CleanCap, developed by TriLink BioTechnologies) with a 2′-O-methylated cap27,28 and purified to remove double-stranded RNA (dsRNA) contaminants, reactants and incomplete transcripts. Other methods add the cap with a post-transcriptional reaction using capping and 2′-O-methyltransferase enzymes derived from the vaccinia virus. The poly(A) tail is encoded in the DNA template, which eliminates reaction steps and reduces overall production time and material loss29.
Incorporating the poly(A) tail in the DNA plasmid also overcomes the tail length variability that arises from enzymatic polyadenylation using poly(A) polymerase. Poly(A) tails of >100 bp are optimal for therapeutic mRNAs; however, the DNA sequences that encode these long poly(A) stretches can destabilize the DNA plasmids used for transcription. A solution to overcome this stability issue is to include a short UGC linker in the poly(A) tail30,31. The Pfizer–BioNTech vaccine BNT162b2 against SARS-CoV-2 uses this strategy and contains a 10 bp UGC linker to produce the sequence A30(10 bp UGC linker)A70 (see the mRNA sequence in Related links). Together, these innovations have overcome significant manufacturing bottlenecks and facilitated the development of a simple, cost-effective and scalable one-step mRNA synthesis process.
Delivery vehicles for mRNA vaccines
Because mRNA is large (104–106 Da) and negatively charged, it cannot pass through the anionic lipid bilayer of cell membranes. Moreover, inside the body, it is engulfed by cells of the innate immune system and degraded by nucleases. Various techniques, including electroporation, gene guns and ex vivo transfection can intracellularly deliver mRNA in a dish23. In vivo application, however, requires the use of mRNA delivery vehicles that transfect immune cells without causing toxicity or unwanted immunogenicity (Fig. 2). Fortunately, a number of innovative materials-based solutions have been developed for this purpose.
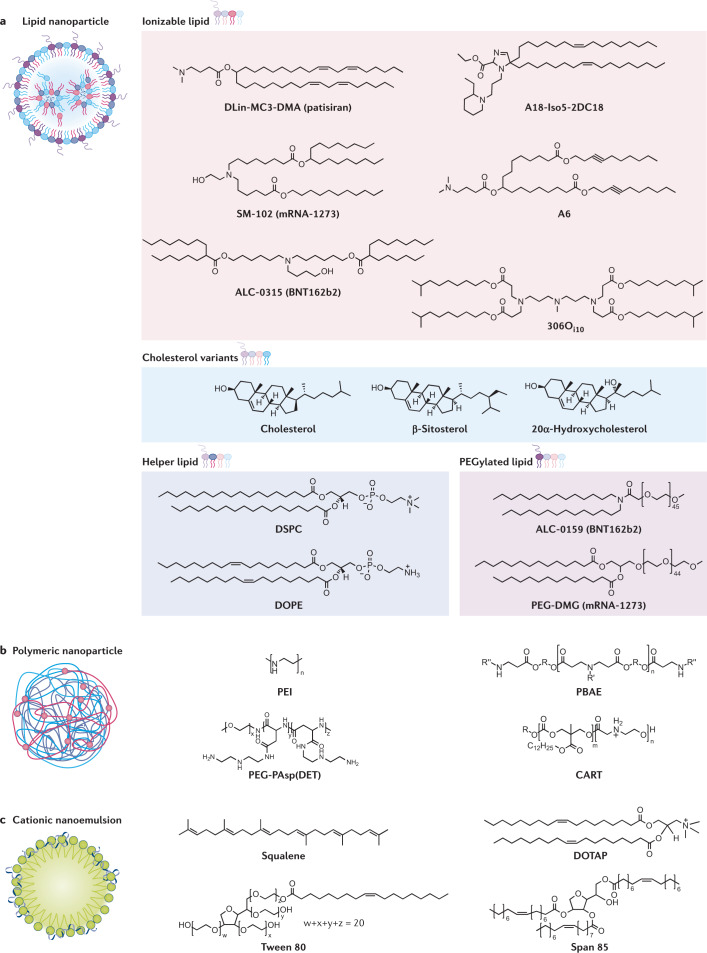
Fig. 2. All mRNA delivery vehicles contain cationic or ionizable molecules.
a | Lipid nanoparticles encapsulate mRNA in their core. They consist of four components: ionizable lipids, such as DLin-MC3-DMA42, SM-102 (ref.53), ALC-0315 (ref.54), A18-Iso5-2DC18 (ref.60), A6 (ref.57) and 306Oi10 (ref.46); cholesterol or its variants, β-sitosterol64 and 20α-hydroxycholesterol63; helper lipids, such as DSPC70 and DOPE69; and PEGylated lipids, such as ALC-0159 (ref.32) and PEG-DMG32. b | Polymers, such as PEI164, PBAE91, PEG-PAsp(DET)94 and CART99 form polymer–mRNA complexes. c | Cationic nanoemulsions contain a squalene core surrounded by an outer shell made of cationic lipid (for example, DOTAP) and surfactants, such as Tween 80 and Span 85. The mRNA adsorbs to the surface via electrostatic binding109.
Lipid-based nanoparticles
Lipid-based nanoparticles are the most clinically advanced of the mRNA delivery vehicles. All SARS-CoV-2 mRNA vaccines in development or approved for clinical use as of June 2021 employ lipid nanoparticles (LNPs). LNPs offer numerous benefits for mRNA delivery, including ease of formulation, modularity, biocompatibility and large mRNA payload capacity. Aside from the RNA drug, LNPs typically include four components (Box 2), each of which is described below: an ionizable lipid, cholesterol, a helper phospholipid and a PEGylated lipid, which together encapsulate and protect the fragile mRNA core32.
The cationic lipid DOTMA and its synthetic analogue DOTAP were the first lipids to deliver mRNA in 1989 (ref.33). Their positively charged amines facilitate the encapsulation of negatively charged RNA. Many other cationic lipids have been used for RNA delivery since then, including the commercially successful Lipofectamine34. Unfortunately, although cationic lipids are highly effective at mRNA delivery, they also trigger toxic pro-apoptotic and pro-inflammatory responses35,36.
Ionizable lipids were developed to overcome these safety issues. These lipids are neutral when injected into the bloodstream at physiological pH, which improves their safety and extends circulation time compared with cationic lipids37. Ionizable lipids are formulated with mRNA into nanoparticles in acidic buffer so that the lipids are positively charged and attract the RNA cargo. Moreover, they are positively charged in the acidic environment of the endosomes, which promotes their fusion with the endosomal membrane, releasing them into the cytoplasm38,39.
DODAP and DODMA were the first ionizable lipids used for RNA delivery40,41. Efforts to enhance the efficacy of DODMA through rational design led to the creation of DLinDMA41 and, ultimately, DLin-MC3-DMA42. The latter made history as the ionizable lipid in the first FDA-approved LNP formulation: the small interfering RNA (siRNA) drug, patisiran (Onpattro). In addition to potently and safely delivering siRNA, DLin-MC3-DMA has also been used for mRNA delivery43–46.
Although the rational lipid design approach described above has been successful in certain contexts, it is relatively slow. To accelerate materials discovery, many groups in academia and industry have used combinatorial reaction schemes to synthesize large libraries of potential delivery materials for testing. This approach has generated numerous potent lipids, including C12-200 (ref.47), 503O13 (ref.48), 306Oi10 (ref.46), OF-02 (ref.49), TT3 (ref.50), 5A2-SC8 (refs51,52), SM-102 (used in the Moderna vaccine mRNA-1273 against SARS-CoV-2)53 and ALC-0315 (used in the Pfizer–BioNTech vaccine BNT162b2)25,54. Also, structure–activity relationship datasets from these large screens have identified factors that predict efficacy, including lipid pKa48, surface charge at pH 5 (ref.55) and haemolytic activity at pH 5.5 (refs56,57), which can serve as design guides for future studies.
In addition to the search for improved efficacy, a growing interest in improving the specificity of delivery, particularly for vaccines and immunotherapies, has spurred efforts to target immune cells. Lipids containing polycyclic adamantane tails, such as 11-A-M58, or those containing cyclic imidazole heads, such as 93-O17S59, have been designed to target T cells in vivo. Although the mechanism is unclear, the cyclic groups of these lipids are crucial for targeting T cells. Furthermore, the cyclic amine head group of the lipid A18-Iso5-2DC18 has been shown to strongly bind to the stimulator of interferon genes (STING) protein, resulting in dendritic cell maturation and potent antitumour immunity60. It has also been shown that cyclic vitamin C-derived lipids delivered antimicrobial peptide and cathepsin B mRNA to macrophage lysosomes, eliminating multidrug-resistant bacteria and protecting mice from bacteria-induced sepsis61.
Although ionizable lipids are, arguably, the most important component of LNPs, the three other lipid components — cholesterol, helper lipid and PEGylated lipid — also promote nanoparticle formation and function. Cholesterol, a naturally occurring lipid, enhances the stability of the nanoparticles by filling gaps between lipids, and it aids fusion with the endosomal membrane during uptake into the cell62. Several studies have also shown improved efficacy with cholesterol analogues63–65, which likely reduce binding to Niemann–Pick type C1, a protein that traffics cholesterol and recycles ~70% of LNPs from late endosomes back into the extracellular fluid64,66.
Helper lipids modulate nanoparticle fluidity and enhance efficacy by promoting lipid phase transitions that aid membrane fusion with the endosome67,68. The choice of an optimal helper lipid depends on both the ionizable lipid material and the RNA cargo. For example, for lipidoid materials, saturated helper lipids (for example, DSPC) are best at delivering short RNAs (for example, siRNA)58, and unsaturated lipids (for example, DOPE) are best for mRNA delivery69,70. DSPC, however, has been incorporated into the FDA-approved SARS-CoV-2 vaccines mRNA-1273 and BNT162b2. It is possible that DSPC outperforms DOPE for these ionizable lipids, and it is also a convenient choice because it is a component of a previously FDA-approved LNP siRNA therapy, patisiran (Onpattro). Studying helper lipids has led to the design of potent unsaturated ionizable lipids, such as A6 (ref.57) and 4A3-Cit71 that enhance vesicle fusion, and zwitterionic lipids, such as 9A1P9, that improve endosomal escape by promoting phase transition72. In addition to enabling membrane fusion, helper lipids also influence target organ specificity: cationic lipids redirect liver-targeted formulations to the lungs, whereas anionic lipids redirect them to the spleen72,73.
The PEGylated lipid component of LNPs consists of polyethylene glycol (PEG) conjugated to an anchoring lipid such as DMPE or DMG. The hydrophilic PEG stabilizes the LNP, regulates nanoparticle size by limiting lipid fusion74 and increases nanoparticle half-life by reducing nonspecific interactions with macrophages75. Both the molecular weight of the PEG and the length of the lipid anchor can be tuned to alter efficacy, circulation time and immune cell uptake, depending on the application. The molecular weight of the PEG can vary from 350 to 3,000 Da76, and the lipid anchor’s tail length can vary from 10 to 18 carbons77,78. Larger molecular weights and longer lengths tend to result in nanoparticles with longer circulation times and reduced immune cell uptake. The mRNA-1273 and BNT162b2 SARS-CoV-2 vaccines contain PEGylated lipids of 2 kDa molecular weight and 13 and 14 carbon-long saturated lipid anchors, respectively32.
Box 2 Formulation and scale-up of lipid nanoparticles.
Lipid nanoparticles (LNPs) are formulated by precipitating lipids from an organic phase by mixing them with an aqueous phase. The lipids commonly used in mRNA vaccines — an ionizable lipid, a helper lipid, cholesterol and a PEGylated lipid — are dissolved in ethanol. Separately, mRNA is dissolved in an aqueous citrate or acetate buffer at pH 4. Mixing the two solutions protonates the ionizable lipid, causing electrostatic attraction between the protonated lipid and the anionic mRNA. This effect, when coupled with the hydrophobic interactions due to the poor aqueous solubility of the lipids, drives spontaneous self-assembly of LNPs with the mRNA encapsulated inside. LNPs are then dialysed or filtered to remove non-aqueous solvent and bring the solution to physiological pH.
Microfluidic mixers enable the formulation of small-sized nanoparticles with low dispersity and high encapsulation efficiency. Precision NanoSystems’ NanoAssemblr platform has been widely employed for LNP formulation in laboratory settings260. These systems use cartridges that have a staggered herringbone micromixer (SHM) architecture. The structure of SHMs enables the solvents to mix within microseconds. Because this timescale is smaller than the time necessary for lipid aggregation, SHMs produce small nanoparticles of uniform size260. The particle size distribution can be regulated by adjusting the total flow rate and the flow rate ratio between the aqueous and ethanol streams. A total flow rate of 12 ml min−1 and a flow rate ratio of 3:1 is commonly used to generate small monodisperse LNPs. Multiple SHMs can also be connected in parallel to increase throughput for large-scale production261.
Although SHMs have several advantages, their utility for good manufacturing practice (GMP) manufacturing is limited primarily by solvent incompatibility. SHMs are commonly fabricated using polydimethylsiloxane260, which can deform the mixer architecture upon long-term exposure to organic solvents such as ethanol. Although the cartridges are replaced after several runs in the lab, this is infeasible for continuous manufacturing at GMP scale. Instead, T mixers are employed for LNP scale-up25,135. They are compatible with organic solvents260, produce morphologically similar LNPs to SHMs74 and operate at the much higher flow rates (60–80 ml min−1) necessary for large-scale manufacturing260.
Polyplexes and polymeric nanoparticles
Although less clinically advanced than LNPs, polymers offer similar advantages to lipids and effectively deliver mRNA79. Cationic polymers condense nucleic acids into complexes called polyplexes that have various shapes and sizes and can be taken up into cells by endocytosis. The mechanisms by which polyplexes escape from endosomes are uncertain; one possible mechanism is that proton buffering by the polymer leads to osmotic swelling and rupture of the endosomes — the proton sponge hypothesis80.
Polyethylenimine is the most widely studied polymer for nucleic acid delivery. Although its efficacy is excellent, its application is limited by its toxicity81 owing to its high charge density82. Use of a low molecular weight form, incorporation of PEG into the formulation83, conjugation to cyclodextrin84–86 and disulfide linkage87 can mitigate the toxicity of polyethylenimine. Additionally, several alternative biodegradable polymers have been developed that are less toxic. Poly(β-amino ester)s, for example, excel at mRNA delivery, especially to the lung88–90. Because they are easily synthesized by the Michael reaction91, large poly(β-amino ester) libraries have been created that facilitate structure–function studies.
Similar to poly(β-amino ester)s, poly(amidoamine)s are biodegradable polymers that are synthesized by the Michael reaction and allow facile modifications to their core and periphery52. Poly(amidoamine)s form hyperbranched tree-like spherical dendrimers that efficiently form mRNA complexes owing to the high amine density on their periphery82. Although charge density is favourable for mRNA complexation, excessive charge can cause toxicity and serum aggregation82,92. Fortunately, these issues can be mitigated by introducing disulfide linkages or incorporating PEG in the dendrimer core82,92.
Like the ionizable lipids in LNPs, pH-responsive polymers have also been used for mRNA delivery. Poly(aspartamide)s conjugated to ionizable aminoethylene side chains are protonated at the acidic pH inside endosomes, facilitating RNA delivery. The hydrophobicity and length of the side chain influence poly(aspartamide) protonation and delivery efficacy93. For example, PEGylated poly(aspartamide) with an ethylenediamine side chain delivers mRNA to liver94, brain95, spinal cord96, knee joint97 and olfactory nerves98. In addition to poly(aspartamide)s, pH-responsive charge-altering releasable transporters have gained attention owing to their unique mRNA delivery mechanism. Instead of protonating inside endosomes, these charge-altering releasable transporters self-degrade into neutral, non-toxic by-products at cytosolic pH, leading to rapid release of the mRNA into the cytoplasm99,100.
Other delivery systems
In addition to lipid and polymer-based vehicles, peptides can also deliver mRNA into cells, thanks to the cationic or amphipathic amine groups (for example, arginine) in their backbone and side chains that electrostatically bind to mRNA and form nanocomplexes. For example, a fusogenic cell-penetrating peptide containing repetitive arginine-alanine-leucine-alanine (RALA) motifs changes conformation at endosomal pH, facilitating pore formation in the membrane and endosomal escape101,102. RALA delivers mRNA to dendritic cells (professional antigen-presenting cells of the immune system) to elicit T cell-mediated immunity103.
There is also a commercially available cell-penetrating peptide, PepFect14, that delivered mRNA to ovarian cancer cells in a mouse xenograft model104. Arginine-rich protamine peptides (of about 4 kDa), which are positively charged at neutral pH, can also condense mRNA and facilitate its delivery34. Protamine complexed with mRNA activates Toll-like receptor (TLR7, TLR8) pathways that recognize single-stranded mRNA105; thus, it can act as an adjuvant for vaccine or immunotherapy applications. CureVac AG is evaluating a protamine-containing delivery platform, RNActive, in clinical trials for melanoma106, prostate cancer107 and non-small-cell lung cancer108.
Finally, squalene-based cationic nanoemulsions also deliver mRNA. These nanoemulsions consist of an oily squalene core stabilized by a lipid shell that adsorbs mRNA onto its surface109. Some squalene formulations, such as Novartis’s MF59, act as adjuvants in FDA-approved influenza vaccines110. MF59 causes cells at the injection site to secrete chemokines, which recruits antigen-presenting cells, induces differentiation of monocytes into dendritic cells and enhances antigen uptake by antigen-presenting cells111–113. The mechanism by which squalene-based cationic nanoemulsions escape from endosomes to deliver mRNA into the cytoplasm remains unclear.
Progress with mRNA vaccines for infectious diseases
Vaccines for infectious diseases are currently the most advanced application for mRNA therapeutics. The majority of mRNA vaccines currently in preclinical trials and in clinical use are administered as a bolus injection into the skin, muscle or subcutaneous space, where they are taken up by immune or non-immune cells and translated into antigens that are displayed to T and B cells (Fig. 3). Both the mRNA and the delivery vehicle enhance the immunogenicity and efficacy of mRNA vaccines.
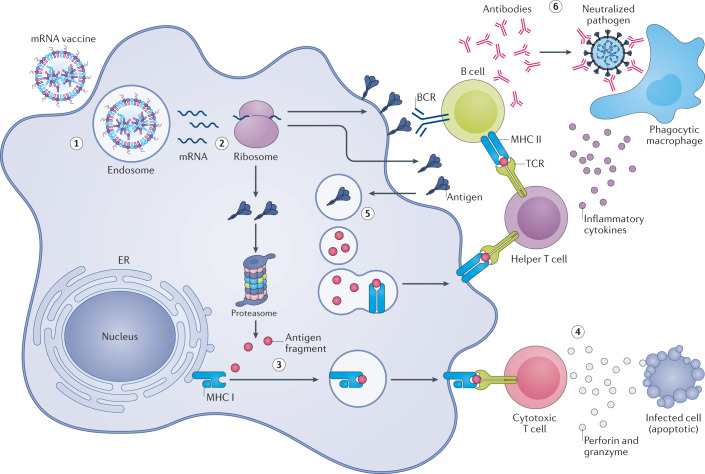
Fig. 3. Messenger RNA vaccines elicit immunity through transfection of antigen-presenting cells.
(1) Injected mRNA vaccines are endocytosed by antigen-presenting cells. (2) After escaping the endosome and entering the cytosol, mRNA is translated into protein by the ribosome. The translated antigenic protein can stimulate the immune system in several ways. (3) Intracellular antigen is broken down into smaller fragments by the proteasome complex, and the fragments are displayed on the cell surface to cytotoxic T cells by major histocompatibility complex (MHC) class I proteins. (4) Activated cytotoxic T cells kill infected cells by secreting cytolytic molecules, such as perforin and granzyme. (5) Additionally, secreted antigens can be taken up by cells, degraded inside endosomes and presented on the cell surface to helper T cells by MHC class II proteins. (6) Helper T cells facilitate the clearance of circulating pathogens by stimulating B cells to produce neutralizing antibodies, and by activating phagocytes, such as macrophages, through inflammatory cytokines. BCR, B cell receptor; ER, endoplasmic reticulum; TCR, T cell receptor.
By the end of 2019, 15 mRNA vaccine candidates against infectious diseases had entered clinical trials, with none in phase III trials79,114 (Table 1). At that time, it was thought that it would be at least another 5–6 years before an mRNA vaccine would obtain regulatory approval. These expectations were upended when the COVID-19 pandemic overtook the world in early 2020. Over the ensuing months, mRNA vaccine development, manufacturing and deployment were put to the ultimate test.
Table 1.
Clinical trials of mRNA vaccines against infectious diseases beyond COVID-19
| Funding source | Name | Target | Vaccine type | Route of administration | Clinical trial phase | Clinical trial identifier |
|---|---|---|---|---|---|---|
| Moderna | mRNA-1647 | CMV | Nucleoside-modified mRNA–LNP | Intramuscular | Phase II | NCT04232280, NCT03382405 |
| Moderna | mRNA-1443 | CMV | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03382405 |
| Moderna | mRNA-1893 | Zika | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT04064905 |
| Moderna | mRNA-1325 | Zika | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03014089 |
| Moderna | mRNA-1653 | hMPV/PIV3 | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT04144348, NCT03392389 |
| Moderna | mRNA-1345 | RSV | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT04528719 |
| Moderna, Merck | mRNA-1777 (V171) | RSV | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | Unregistered |
| Moderna, Merck | mRNA-1172 (V172) | RSV | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | Unregistered |
| Moderna | mRNA-1851 (VAL-339851) | Influenza A (H7N9) | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03345043 |
| Moderna | mRNA-1440 (VAL-506440) | Influenza A (H10N8) | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03076385 |
| Moderna | mRNA-1010 | Influenza A (H1N1, H3N2), influenza B (Yamagata lineage, Victoria lineage) | Unknown | Intramuscular | Phase I/II | NCT04956575 |
| Translate Bio, Sanofi | MRT5400 | Influenza A (H3N2) | Unknown | Intramuscular | Phase I | Unregistered |
| Translate Bio, Sanofi | MRT5401 | Influenza A (H3N2) | Unknown | Intramuscular | Phase I | Unregistered |
| Moderna | mRNA-1944 | Chikungunya | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03829384 |
| Moderna | mRNA-1388 (VAL-181388) | Chikungunya | Nucleoside-modified mRNA–LNP | Intramuscular | Phase I | NCT03325075 |
| CureVac | CV7201 | Rabies | Unmodified mRNA complexed in RNActive | Intradermal, intramuscular | Phase I | NCT02241135 |
| CureVac | CV7202 | Rabies | Unmodified mRNA–LNP | Intramuscular | Phase I | NCT03713086 |
| GSK | GSK3903133A | Rabies | Self-amplifying mRNA in cationic nanoemulsion | Intramuscular | Phase I | NCT04062669 |
CMV, cytomegalovirus; GSK, GlaxoSmithKline; HIV, human immunodeficiency virus; hMPV, human metapneumovirus; LNP, lipid nanoparticle; PIV3, parainfluenza virus type 3; RSV, respiratory syncytial virus.
SARS-CoV-2
At the start of 2021, a year after the first COVID-19 case was reported in China115, SARS-CoV-2 had infected more than 150 million people and claimed more than three million lives worldwide (see Related links). Although most SARS-CoV-2 infections are not life-threatening in younger people without pre-existing medical conditions, severe infection can trigger an unchecked immune response in the lungs that destroys epithelia and alveoli. This damage can cause pulmonary oedema, dangerous increases in vascular permeability and death116,117.
Most SARS-CoV-2 vaccine candidates induce an immune response to the spike protein on the virus surface (Fig. 4). The spike protein binds to its receptor on the host cell surface, angiotensin-converting enzyme 2 (ref.118). The attached spike protein is then cut open by the cell’s transmembrane serine protease 2, which induces a conformational change that exposes the spike protein’s fusion peptide and facilitates fusion with the cell or endosomal membrane118. Usually, the antigen encoded by the vaccine mRNA is either the full-length spike protein or the spike protein’s receptor-binding domain.
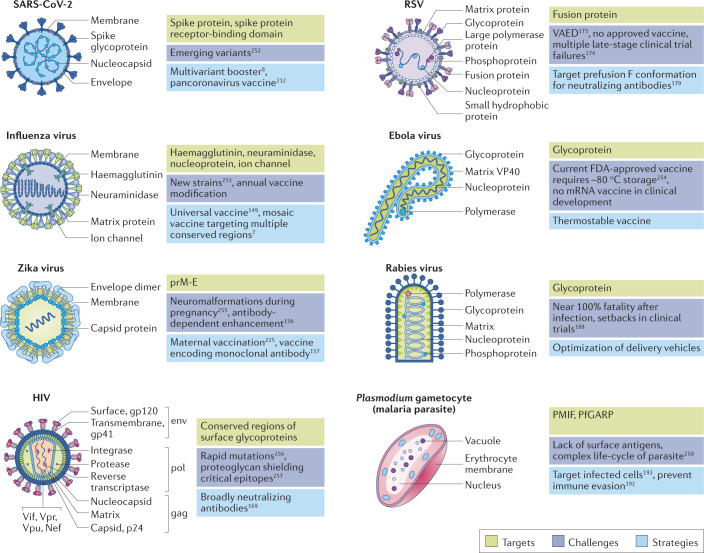
Fig. 4. mRNA vaccines in development protect against an array of common pathogens using disease-specific targeting strategies.
Surface proteins that enable cell entry are commonly used by mRNA vaccines to target viruses, for example, spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), haemagglutinin protein of influenza viruses, membrane and envelope protein (prM-E) of Zika virus, fusion protein of respiratory syncytial virus (RSV) and surface glycoproteins of human immunodeficiency virus (HIV), Ebola virus and rabies virus. Additionally, complex pathogens such as Plasmodium can be targeted using non-surface antigens such as Plasmodium macrophage migratory inhibiting factor (PMIF) or Plasmodium falciparum glutamic-acid-rich protein (PfGARP). Each pathogen poses a unique set of challenges, including high lethality, rapid mutations, immune evasion, new strains and variants252–258. Depending on the challenges, mRNA vaccines encoding conformation-specific proteins, conserved regions of antigens or monoclonal antibodies can be safely delivered to healthy adults, children, elderly people and pregnant people. VAED, vaccine-associated enhanced disease.
As of 18 June 2021, 185 COVID-19 vaccine candidates were in preclinical development and an additional 102 had entered clinical trials (see Related links). Of those in clinical trials, 19 were mRNA vaccines (Table 2). On 11 December 2020, the Pfizer–BioNTech vaccine BNT162b2 received emergency authorization from the FDA and became the first mRNA drug approved for use in humans. One week later, the Moderna vaccine mRNA-1273 was also authorized for use in the USA. Ultimately, they were the first SARS-CoV-2 vaccines to be authorized in the USA, the UK, Canada and several other countries.
Table 2.
Clinical trials of mRNA vaccines against SARS-CoV-2
| Name (INN)/funding source | mRNA type | Antigen | Phase | Clinical trial identifier (participants; location) | Clinical trial outcomes |
|---|---|---|---|---|---|
| BNT162b2 (Tozinameran)/BioNTech, Pfizer | Nucleoside-modified | Transmembrane prefusion spike | Phase III | NCT04805125 (431 participants; Switzerland) | EUA in several countries; >90% efficacy in real-world conditions in USA132 and Israel126; 95% overall efficacy in phase III trials125; 90–100% efficacy in phase III trials across subgroups defined by age, sex, race, ethnicity, baseline body mass index and the presence of coexisting conditions125; 1.7–4.6-fold higher neutralizing titres than convalescent serum in 18–55-year old participants, and 1.1–2.2-fold higher titres in 65–85-year old in phase I trials124 |
| NCT04816669 (610 participants; USA) | |||||
| NCT04800133 (900 participants; Hong Kong) | |||||
| NCT04713553 (1,530 participants; USA) | |||||
| Phase II/III | NCT04368728 (43,998 participants; Argentina, Brazil, Germany, South Africa, Turkey, USA) | ||||
| NCT04754594 (700 participants; Brazil, South Africa, Spain, UK, USA) | |||||
| Phase II | ISRCTN73765130 (2,886 participants; UK) | ||||
| NCT04894435 (1,200 participants; Canada) | |||||
| NCT04761822 (3,400 participants; USA) | |||||
| NCT04824638 (300 participants; France) | |||||
| NCT04860739 (676 participants; Spain) | |||||
| EUCTR2021-001978-37 (600 participants; Spain) | |||||
| NCT04649021 (950 participants; China) | |||||
| ISRCTN69254139 (820 participants; UK) | |||||
| NCT04907331 (3,000 participants; Austria) | |||||
| NCT04895982 (360 participants; Brazil, Germany, USA) | |||||
| Phase I/II | EUCTR2020-001038-36, NCT04380701 (476 participants; Germany) | ||||
| NCT04889209 (800 participants; USA) | |||||
| NCT04588480 (160 participants; Japan) | |||||
| Phase I | NCT04839315 (100 participants; USA) | ||||
| NCT04816643 (4,500 participants; Finland, Poland, Spain, USA) | |||||
| mRNA-1273/Moderna, NIAID, BARDA | Nucleoside-modified | Transmembrane prefusion spike | Phase III | NCT04811664 (37,500 participants; USA) | EUA in several countries;90% efficacy in real-world conditions in USA132; 94.1% overall efficacy in phase III trials131; dose-dependent increase in S2-P-binding antibodies and serum-neutralizing titres in phase I trials130 |
| NCT04470427 (30,420 participants; USA) | |||||
| NCT04860297 (240 participants; USA) | |||||
| NCT04806113 (220 participants; Canada) | |||||
| NCT04805125 (431 participants; Switzerland) | |||||
| Phase II/III | NCT04649151 (3,732 participants; USA) | ||||
| NCT04796896 (6,975 participants; USA) | |||||
| Phase II | ISRCTN73765130 (2,886 participants; UK) | ||||
| NCT04847050 (120 participants; USA) | |||||
| NCT04894435 (1,200 participants; Canada) | |||||
| NCT04748471 (180 participants; France) | |||||
| NCT04761822 (3,400 participants; USA) | |||||
| NCT04405076 (660 participants; USA) | |||||
| Phase I/II | NCT04889209 (800 participants; USA) | ||||
| Phase I | NCT04785144 (135 participants; USA) | ||||
| NCT04813796 (125 participants; USA) | |||||
| NCT04839315 (100 participants; USA) | |||||
| NCT04283461 (120 participants; USA) | |||||
| TAK-919/Takeda, Moderna | Nucleoside-modified | Transmembrane prefusion spike | Phase I/II | NCT04677660 (200 participants; Japan) | Approved in Japan based on positive interim phase I/II data (see Related links); same formulation as mRNA-1273 |
| CVnCoV (Zorecimeran)/CureVac | Unmodified | Transmembrane prefusion spike | Phase III | NCT04652102, EUCTR2020-003998-22 (39,693 participants; Argentina, Belgium, Colombia, Dominican Republic, Germany, Mexico, Netherlands, Panama, Peru, Spain) | 47% efficacy in phase III trials (see Related links); anti-spike IgG, anti-RBD IgG and serum-neutralization titres comparable to convalescent serum133 |
| EUCTR2020-004066-19, NCT04674189 (2,360 participants; Germany) | |||||
| NCT04860258 (1,200 participants; Belgium) | |||||
| NCT04848467 (1,000 participants; Argentina, Colombia, Peru) | |||||
| Phase II | ISRCTN73765130 (2,886 participants; UK) | ||||
| NCT04515147, PER-054-20 (674 participants; Panama, Peru) | |||||
| Phase I | NCT04449276 (280 participants; Belgium, Germany) | ||||
| ARCoV/Walvax Biotechnology, PLA | Unmodified | Secreted spike RBD | Phase III | NCT04847102 (28,000 participants; China, Mexico) | Unknown |
| Phase II | ChiCTR2100041855 (420 participants; China) | ||||
| Phase Ib | ChiCTR2000039212 (120 participants; China) | ||||
| Phase I | ChiCTR2000034112 (168 participants; China) | ||||
| BNT162b1 (Abdavomeran)/BioNTech, Pfizer | Nucleoside-modified | Secreted spike RBD | Phase II/III | NCT04368728 (43,998 participants; Argentina, Brazil, Germany, South Africa, Turkey, USA) | 8–50-fold higher anti-RBD IgG and 1.9–4.6-fold higher neutralization titres than convalescent serum27,123; higher frequency of fever, fatigue and chills in participants than BNT162b2 (ref.124) |
| Phase I/II | EudraCT 2020-001038-36, NCT04380701 (476 participants; Germany) | ||||
| Phase I | ChiCTR2000034825, NCT04523571 (144 participants; China) | ||||
| mRNA-1273.211/Moderna | Nucleoside-modified | Transmembrane prefusion spike | Phase II | NCT04405076 (660 participants; USA) | Unknown |
| ARCT-021/Arcturus | Self-amplifying | Transmembrane prefusion spike | Phase II | NCT04668339 (600 participants; Singapore, USA) | Unknown |
| NCT04728347 (106 participants; Singapore) | |||||
| Phase I/II | NCT04480957 (92 participants; Singapore) | ||||
| BNT162a1/BioNTech, Pfizer | Unmodified | Secreted spike RBD | Phase I/II | EudraCT 2020-001038-36, NCT04380701 (476 participants; Germany) | Unknown |
| BNT162b3 (Ganulameran)/BioNTech, Pfizer | Nucleoside-modified | Transmembrane spike RBD | Phase I/II | NCT04537949, EUCTR2020-003267-26-DE (96 participants; Germany) | Unknown |
| BNT162c2 (Pidacmeran)/BioNTech, Pfizer | Self-amplifying | Transmembrane prefusion spike | Phase I/II | EudraCT 2020-001038-36, NCT04380701 (476 participants; Germany) | Unknown |
| MRT5500/Sanofi, Translate Bio | Unmodified | Transmembrane prefusion spike | Phase I/II | NCT04798027 (333 participants; Honduras, USA) | Unknown |
| LNP-nCoVsaRNA/Imperial College London, Acuitas Therapeutics | Self-amplifying | Transmembrane prefusion spike | Phase I | ISRCTN17072692 (320 participants; UK) | Unknown |
| ChulaCov19/Chulalongkorn University | Nucleoside-modified | Transmembrane spike | Phase I/II | NCT04566276 (96 participants; Thailand) | Unknown |
| PTX-COVID19-B/Providence Therapeutics | Nucleoside-modified | Transmembrane spike | Phase I | NCT04765436 (60 participants; Canada) | Unknown |
| SAM-LNP-S/Gritstone Oncology, NIAID | Self-amplifying | Transmembrane spike | Phase I | NCT04776317 (150 participants; USA) | Unknown |
| mRNA-1273.351/Moderna | Nucleoside-modified | Transmembrane prefusion spike | Phase I | NCT04785144 (135 participants; USA) | Difference in serum neutralization between wild-type ancestral strain and B.1.351 reduced from 7.7-fold to 2.1-fold, 14 days after mRNA-1273.351 booster251 |
| mRNA-1283/Moderna | Nucleoside-modified | Transmembrane prefusion spike | Phase I | NCT04813796 (125 participants; USA) | Unknown |
| CoV2 SAM [LNP]/GSK | Self-amplifying | Transmembrane spike | Phase I | NCT04758962 (10 participants; USA) | Unknown |
All SARS-CoV-2 vaccine candidates in clinical trials are delivered intramuscularly. Clinical trials are regularly updated and the locations and the number of participants are subject to change. BARDA, Biomedical Advanced Research and Development Authority; EUA, emergency use authorization; GSK, GlaxoSmithKline; INN, international nonproprietary name; LNP, lipid nanoparticle; NIAID, National Institute of Allergy and Infectious Diseases; PLA, People’s Liberation Army; RBD, receptor-binding domain.
Pfizer and BioNTech co-developed five mRNA vaccine candidates that encode variants of the spike protein antigen119. The two lead candidates, BNT162b1 and BNT162b2, use LNPs that are formulated using Acuitas Therapeutics’ ionizable lipid ALC-0315 and a nucleoside-modified mRNA in which all uridines are replaced by N1-methylpseudouridine to enhance mRNA translation. BNT162b1 encodes a trimerized, secreted version of the spike protein’s receptor-binding domain, whereas BNT162b2 encodes the full-length SARS-CoV-2 spike glycoprotein with two proline substitutions in the S2 subunit, which lock the protein in its prefusion conformation120,121. In preclinical studies in rhesus macaques, two 100 μg doses of BNT162b2 delivered 21 days apart elicited neutralizing antibody titres 10.2–18.0 times higher than convalescent patient serum as well as strong CD4+ and CD8+ T cell responses122. In phase I trials of the two candidates, two 30 μg doses delivered 21 days apart induced high antibody and neutralizing titres and a robust CD4+ and CD8+ response with mild to moderate adverse events27,28,123,124. Both candidates were well tolerated and efficacious, but only the BNT162b2 vaccine advanced to phase II/III trials owing to its milder systemic and local adverse reactions124. In phase III trials involving 43,548 participants, BNT162b2 showed 95% overall efficacy at preventing COVID-19 and 90–100% efficacy across subgroups defined by age, sex, race, ethnicity, baseline body mass index and coexisting conditions125. In a mass vaccination campaign involving 3,159,136 participants in Israel, two doses of BNT162b2 were 94% effective at preventing symptomatic COVID-19, 87% effective at preventing hospitalizations and 92% effective at preventing the onset of severe COVID-19 (ref.126).
Moderna developed mRNA-1273 in collaboration with the National Institute for Allergy and Infectious Diseases at the National Institutes of Health and the Biomedical Advanced Research and Development Authority. The vaccine uses the ionizable lipid SM-102 to formulate LNPs that encapsulate an N1-methylpseudouridine-modified mRNA. The sequence encodes the SARS-CoV-2 spike protein with two proline substitutions that confer the prefusion conformation127. In a preclinical trial, two 1 μg doses of vaccine (a primer dose and a booster) injected into muscle induced robust virus-neutralizing activity as well as potent CD4+ and CD8+ T cell responses in mice127. It also protected the animals from upper and lower airway SARS-CoV-2 infection for at least 3 months after vaccination127. Rhesus macaques that received two 100 μg doses of the vaccine also developed potent humoral and cellular immune responses128. In comparison with serum from patients recovering from COVID-19, vaccinated macaque serum contained 15-fold higher titres of neutralizing antibodies, 348-fold more potent inhibitory activity for spike binding to the angiotensin-converting enzyme 2 and 12-fold higher virus-neutralizing activity128. Vaccinated macaques also elicited spike protein-specific IgG and IgA antibodies in nasal washes and bronchoalveolar lavages, suggesting that intramuscular vaccination with mRNA-1273 confers both serum and mucosal immunity128,129. Fortunately, these encouraging preclinical results extended to clinical trials in which mRNA-1273 was exceptionally efficacious and well tolerated130,131. In phase III trials involving 30,420 volunteers, two 100 μg vaccine doses were 94.1% effective at preventing the onset of COVID-19 (ref.131). Local pain at the site of injection was the most common side effect. After the second dose, half of the volunteers reported moderate-to-severe systemic side effects (for example, fatigue, muscle pain, joint pain) that resolved within 48 h131. In real-world conditions involving 3,950 healthcare personnel, first responders and other essential and frontline workers in the USA, two doses of mRNA-1273 or BNT162b2 were 90% effective against SARS-CoV-2 (ref.132).
Although the Pfizer–BioNTech and Moderna vaccines have demonstrated excellent efficacy and safety, their need for cold-chain storage poses logistical difficulties. mRNA-1273 can be stored at 4–8 °C for a month and 12 h at room temperature, while BNT162b2 requires storage at −60 °C. Thermostable vaccines that do not require refrigerators or freezers ease the distribution of vaccines considerably, particularly in warm countries or those without reliable cold chain storage. CureVac’s candidate vaccine CVnCoV is stable for 3 months at 5 °C and 24 h at room temperature (see Related links). CVnCoV uses LNPs formulated using an ionizable lipid from Acuitas Therapeutics (potentially ALC-0315) and an unmodified mRNA sequence encoding a full-length spike protein with the two proline substitutions133. In phase I clinical trials, primer and booster injections of a 12 μg dose of CVnCoV administered 28 days apart generated neutralizing antibodies similar to those in plasma from patients who were convalescing from COVID-19 and was well tolerated133. Unfortunately, in phase IIb/III trials involving 40,000 participants and spanning ten countries, CVnCoV demonstrated 47% efficacy against COVID-19 (see Related links). Interim analysis suggests that CVnCoV’s modest efficacy is attributable to emerging SARS-CoV-2 variants, as only one of the sequenced 124 cases was caused by the ancestral variant. Furthermore, 57% of the cases were caused by variants of concern: B.1.1.7, B.1.427, B.1.429, B.1.351, P.1 or B.1.617.2. At the time of writing, CureVac, in collaboration with GSK, is developing a second-generation candidate — CV2CoV — that has been optimized to enhance translation and immunogenicity relative to CVnCoV. CV2CoV uses a 5′ UTR from the human hydroxysteroid 17-β-dehydrogenase 4 gene and a 3′ UTR from the human proteasome 20S subunit β3 gene, whereas CVnCoV used a 3′ UTR from the human α-haemoglobin gene134. In preclinical studies, CV2CoV showed 1.8-fold higher protein expression than CVnCoV in vitro, and elicited high titres of cross-neutralizing antibodies against the B.1.1.7, B.1.1.298 and B.1.351 variants in rats134.
Another thermostable candidate vaccine, ARCoV, developed by the Academy of Military Science of the People’s Liberation Army in China in collaboration with Walvax Biotechnology, is stable at 25 °C for a week. ARCoV encodes the receptor-binding domain of the spike protein. In preclinical studies, two 10 μg doses in mice and two 100 μg doses in cynomolgus monkeys delivered 21 days apart elicited high SARS-CoV-2-specific IgG antibodies and strong virus neutralization titres135. Although the reasons behind the thermostability of CVnCoV and ARCoV are unknown to the public, the mRNA secondary structure, smaller mRNA size, GC content and the lipids may be important factors.
Several promising self-amplifying mRNA vaccine candidates are also in development. The mRNAs in these vaccines encode the antigen as well as RNA replication machinery that boosts the intracellular production of mRNA transcripts. The self-amplification supports the use of vaccine doses up to 100-fold lower than those used with standard mRNA. LNP-nCoVsaRNA, developed by Imperial College London and Acuitas Therapeutics, encodes the full-length spike protein and replication machinery from the genome of Venezuelan equine encephalitis virus. In mice, 10 ng primer and booster doses injected 28 days apart into muscle yielded a robust T cell response and IgG titres similar to those in plasma from patients who had recovered from COVID-19 (ref.136). At the time of writing, this candidate is being evaluated in a phase I trial using a 0.1–1 µg dose-escalation protocol (ISRCTN17072692), which is the lowest RNA dose of all mRNA vaccine candidates. Another self-amplifying mRNA vaccine candidate, ARCT-021 (also known as LUNAR-COV19), was developed by Arcturus Therapeutics with their proprietary LUNAR lipid delivery vehicle and self-transcribing and replicating RNA (STARR) platform. It encodes the full-length prefusion spike protein. In transgenic mice expressing human angiotensin-converting enzyme 2, doses of 2 µg and 10 µg produced strong T cell responses, high IgG titres and protected mice from SARS-CoV-2 infection137. Although the ARCT-021 dose is higher than the LNP-nCoVsaRNA dose, no booster is required, ensuring that recipients are fully vaccinated after a single injection.
Influenza viruses
Influenza viruses are responsible for an estimated 290,000–650,000 deaths annually worldwide138. Current vaccines target the virus haemagglutinin protein (Fig. 4), which facilitates viral entry. However, the virus mutates rapidly, causing antigenic drift that requires yearly review and modification of the haemagglutinin antigen component of the vaccines. Conventional influenza vaccines, which are inactivated influenza viruses grown in chicken eggs, are subject to long production times and purification difficulties. Furthermore, the viruses mutate for optimal growth in chicken eggs, sometimes rendering them ineffective in humans. For example, loss of a glycosylation site in egg-grown vaccines was associated with poor efficacy during the 2016–2017 season139. There is a real need, therefore, for alternative antigen targets and production methods. Synthetic mRNAs transcribed in vitro can meet this need and ensure rapid vaccine production in the event that an entirely new influenza strain emerges. In 2013, for example, a LNP (DLinDMA)-based self-amplifying mRNA vaccine was rapidly formulated within 8 days after the H7N9 outbreak in China140. Unfortunately, progress to a phase I trial was impossible given the unavailability of GMP facilities for mRNA manufacturing at that time.
There has also been work towards a universal influenza vaccine that would not require yearly modification. Such a vaccine would confer immunity against several influenza strains (heterologous immunity) and subtypes (heterosubtypic immunity). In the first demonstration of an effective mRNA vaccine against influenza in 2012, three intradermal injections of 80 µg RNActive-mRNA encoding haemagglutinin from the PR8 H1N1 strain induced homologous and heterologous immunity against H1N1 and H5N1 strains, respectively, and protected the mice against a lethal viral dose (10× LD50)141. Since then, several delivery vehicles (DLinDMA, DOTAP, polyethylenimine and cationic nanoemulsions), alternative mRNA technologies (nucleoside-modified mRNAs and self-amplifying mRNAs), and alternative antigen targets have been evaluated for mRNA-based influenza vaccines7,142–147.
Notably, the conserved stalk region of haemagglutinin, which is not prone to mutation, has recently emerged as a novel universal vaccine target148. A primer–booster regimen of a LNP-based mRNA vaccine (30 µg) encoding the conserved haemagglutinin stalk from the Cal09 H1N1 strain produced a stalk-specific antibody response in mice, ferrets and rabbits149. The broadly protective antibodies conferred homologous immunity against Cal09 H1N1, heterologous immunity against PR8 H1N1 and heterosubtypic immunity against H5N1 in mice and protected them against a lethal viral challenge149. Another study used LNPs to deliver a 50 ng dose of nucleoside-modified mRNA encoding three conserved influenza proteins: neuraminidase, nucleoprotein and matrix-2 ion channel protein (Fig. 4) in addition to the haemagglutinin stalk7. Incredibly, this minuscule mRNA dose generated broadly protective antibodies and protected mice from an extraordinarily large viral challenge (500× LD50).
In two separate phase I trials in 2016, Moderna evaluated two influenza candidates. These vaccines were dosed via two intramuscular injections of LNPs encapsulating nucleoside-modified mRNA expressing full-length haemagglutinin from H10N8 and H7N9 (Table 1). Both produced excellent seroconversion and seroprotection147. Adverse effects were limited to pain at the injection site, redness, muscle pain, joint pain, headache, fatigue and chills/common-cold-like symptoms, indicating that the vaccine is safe and well tolerated150.
Zika virus
Zika virus infection was first identified in 1947, and patients infected with Zika are often asymptomatic or experience mild symptoms such as fever, rash and muscle pain. However, Zika emerged as a global health crisis during the 2015–2016 epidemic in the Americas when the virus caused severe fetal neuromalformations and fetal death during pregnancy151. Fortunately, all Zika infections are caused by a single serotype, suggesting that vaccinating against an antigen from any strain could protect against all Zika strains152. The membrane and envelope protein (prM-E) is a common antigen choice for mRNA vaccines against Zika (Fig. 4), as neutralizing antibodies against prM-E can prevent viral fusion.
One study showed that a single 30 µg or 50 µg dose of a LNP-encapsulated nucleoside-modified prM-E mRNA protected mice and rhesus macaques, respectively, from a Zika challenge153. Of note, the neutralizing antibody titres generated by the mRNA vaccine were 50–100 times higher than those induced by purified inactivated virus and DNA vaccines in mice, and 50 times higher than a 1 mg DNA vaccine in macaques153.
Importantly, poorly designed Zika virus vaccines can increase the infectiousness of the dengue virus154. Dengue virus is from the same viral family as Zika, and their envelope proteins share 54–59% overlapping amino acid sequence155. Therefore, it is possible that the envelope protein antigen encoded by a Zika vaccine spurs the production of antibodies that are cross-reactive with the dengue envelope protein. In the event of a subsequent dengue virus infection, antibody-dependent enhancement can occur in which the suboptimal anti-Zika antibodies bind to the dengue virus. This binding enhances the entry of the virus into host cells and exacerbates dengue symptoms. Moderna collaborated with Washington University School of Medicine to deliver a modified prM-E mRNA containing a mutated fusion loop epitope in the E protein. Two 10 µg doses of the LNP-encapsulated modified mRNA delivered 21 days apart protected mice from a Zika challenge and diminished the production of dengue-enhancing antibodies156. These encouraging preclinical results prompted a phase I trial, and interim results suggest that the vaccine — mRNA-1893 (Table 1) — induces 94–100% seroconversion in 10 μg and 30 μg dose groups and is well tolerated (see Related links).
Another study used a passive immunization approach and delivered an mRNA encoding neutralizing ZIKV-117 monoclonal antibodies (mAbs) using a squalene-based nanocarrier157. A single 40 µg dose delivered either a day before or a day after viral inoculation protected immunocompromised mice from the lethal viral challenge157. Using a similar approach, Moderna successfully delivered mRNA encoding the mAb CHKV-24, 4 h after chikungunya virus inoculation and protected mice from chikungunya virus-induced arthritis158. Interestingly, these results suggest that mRNA therapeutics encoding neutralizing mAbs may have both prophylactic and therapeutic activity. Moreover, passive immunization is an appealing approach for vaccinating immunocompromised people who cannot synthesize their own antibodies owing to an impaired immune system.
HIV
Globally, HIV currently affects 38 million people and is projected to affect up to 42 million people by 2030 (ref.159). In 2020 alone, there were 1.5 million new infections and 680,000 deaths (see Related links). Despite 30 years of research, no effective vaccine has been developed, primarily owing to the remarkable antigenic diversity of the HIV envelope protein and the dense ‘glycan shield’ that conceals crucial envelope protein epitopes160. Several preclinical studies have delivered mRNA vaccines encoding HIV proteins using multiple delivery vehicles, including cationic nanoemulsions109,161, DOTAP/DOPE liposomes162, polymers84,163,164 and ionizable LNPs165,166, but they have had varied success. These studies suggest that novel antigens are necessary to effectively target HIV in addition to potent delivery vehicles.
A novel vaccination strategy against HIV is to isolate broadly neutralizing mAbs from infected individuals who neutralize several HIV strains. Notably, the broadly neutralizing mAbs, VRC01, have recently gained attention thanks to their ability to neutralize 98% of HIV strains167 and prevent transmission of antibody-sensitive strains with 75.4% efficacy168. In one study, a single 0.7 mg kg−1 intravenous injection of a LNP-encapsulated, nucleoside-modified mRNA expressing VRC01, produced similar antibody concentrations to those typically achieved by injecting a 10–20 mg kg−1 dose of mAb protein169. Importantly, a single dose protected mice from intravenous challenge with HIV-1.
Respiratory syncytial virus
Respiratory syncytial virus is the leading cause of acute lower respiratory infection globally. Annually, it is responsible for an estimated 60,000 deaths in children under the age of 5 (ref.170) and more than 14,000 deaths in people over 65 years of age171. Although the burden of RSV is well recognized, 40 years of vaccine development have not yet produced an approved RSV vaccine owing to numerous challenges. In 1968, for example, a formalin-inactivated RSV vaccine candidate caused vaccine-associated enhanced disease (VAED) in children. This response triggered excessive eosinophil and neutrophil infiltration in the lungs, resulting in severe bronchiolitis or pneumonia in 80% of the vaccinated children and two fatalities172. Although the exact mechanisms of VAED remain unclear, the formation of non-neutralizing antibodies and a T helper 2 (TH2)-skewed T cell response marked by upregulation of cytokines such as IL-4, IL-5 and IL-13 have been implicated173.
Current RSV vaccine candidates focus on targeting the highly conserved F protein, which facilitates viral fusion (Fig. 4). Although some candidates have failed clinical trials owing to insufficient neutralizing antibody titres174, newfound structural insights into F protein conformation have revealed that vaccinating against the prefusion conformation elicits superior neutralizing antibody response175–178. This discovery will hopefully improve future vaccine design. Fortunately, mRNA vaccines can be designed to encode stabilized F protein conformations by engineering the coding sequence179. In preclinical studies, mRNA vaccines encoding either the native RSV F protein or the stabilized prefusion conformation were successfully delivered using cationic nanoemulsions109 and LNPs179,180 without any observed instances of VAED.
Moderna is evaluating three single-dose vaccine candidates encoding the prefusion F protein (Table 1): mRNA-1172 (Merck’s proprietary LNPs) and mRNA-1777 (Moderna’s proprietary LNPs) for adults, as well as mRNA-1345 (Moderna’s proprietary LNPs) for children (see Related links). In phase I clinical trials, mRNA-1777 elicited a robust humoral response with RSV neutralizing antibodies, a CD4+ T cell response to RSV F peptides and no serious adverse events181. The sequence of mRNA-1345 has been further engineered and codon-optimized to enhance translation and immunogenicity relative to mRNA-1777. Interim phase I data suggest that a 100 μg dose of mRNA-1345 produces approximately eightfold higher neutralizing antibody titres than mRNA-1777 1 month after vaccination (see Related links). Ultimately, Moderna aims to integrate mRNA-1345 with its paediatric human metapneumovirus/parainfluenza virus type 3 (hMPV/PIV3) candidate mRNA-1653 and vaccinate children against three distinct pathogens with a single formulation (see Related links).
Ebola virus
Ebola virus (EBOV) was first identified in 1976 as the agent responsible for sporadic outbreaks of Ebola disease. This viral haemorrhagic fever is fatal in 50–90% of patients, depending on the strain; the 2014–2016 Ebola outbreak in West Africa claimed more than 11,000 lives182. In 2019, the FDA approved a recombinant vesicular stomatitis virus (VSV)-based Ebola vaccine (rVSV-EBOV). Although rVSV-EBOV is 97.5% effective at preventing Ebola transmission compared with no vaccination (see Related links), clinical trials note some safety concerns (for example, acute arthritis and skin rash at high doses)183. mRNA vaccines against EBOV are likely safer than this virus-based vaccine because they do not replicate inside the body. One mRNA vaccine has demonstrated efficacy in mice upon delivery of an unmodified, self-amplifying mRNA encoding the EBOV glycoprotein (Fig. 4) in a poly(amidoamine) dendrimer nanoparticle. The vaccine elicited glycoprotein-specific IgG antibodies and robust expression of IFNγ and IL-2 by CD8+ and CD4+ T cells, and two dosing regimens (two 4 μg doses or one 40 μg dose) protected mice from a lethal challenge184. Another study vaccinated guinea pigs with two 20 μg doses of a LNP-encapsulated, nucleoside-modified mRNA encoding the EBOV glycoprotein, inducing high antibody titres and protecting the animals from a lethal viral challenge185.
Rabies virus
Rabies is a zoonotic viral disease characterized by neurological symptoms resulting in near 100% fatality. Although vaccines are approved, more than 50,000 people succumb to rabies annually186, underscoring the need for more efficacious and affordable vaccines. To meet this need, CureVac has deployed its RNActive platform to deliver unmodified mRNA encoding the rabies virus glycoprotein in its rabies candidate, CV7201 (Table 1). In a preclinical study, two 80 µg vaccine doses delivered 21 days apart induced high neutralizing antibody titres and elicited antigen-specific CD4+ and CD8+ T cell responses in mice and pigs187. Resultant phase I trials examined both the route of delivery (intradermal or intramuscular) as well as the delivery device (standard needle-syringe or a needleless intradermal injector)188. Interestingly, although the delivery route did not affect the immune response, the delivery device did, with only the intradermal injector producing a short-lived humoral response188. This weak delivery efficacy, together with a high incidence of adverse events188 indicated the need for further optimization of the delivery platform. Subsequently, CureVac used proprietary LNPs made by Acuitas Therapeutics as the delivery vehicle for their newer rabies candidate CV7202 (ref.189). In a preclinical study, CV7202 delivered unmodified mRNA encoding the rabies virus glycoprotein and produced strong antibody and CD8+ and CD4+ T cell responses189. In non-human primates, two 100 µg doses spaced 28 days apart were well tolerated and provoked 20-fold higher antibody titres than a commercially licensed rabies vaccine189. Phase I results suggest that two 1 µg doses yield high neutralizing titres, strong adaptive immune responses, and were well tolerated190.
Plasmodium
Although the vast majority of mRNA vaccines under development are for protection from viruses, there are also efforts to prevent other infectious diseases. Malaria, which is caused by unicellular eukaryotic parasites of the genus Plasmodium, is at the top of that list owing to its incidence and lethality. Annually, malaria afflicts more than 200 million people and kills more than 400,000 patients worldwide (see Related links). Antimalarial vaccine production has been difficult owing to the lack of surface antigens and complex life cycle of Plasmodium. Fortunately, the interrogation of the body’s natural immune response to Plasmodium infection has identified potential non-surface antigen targets.
For example, the Plasmodium-secreted cytokine, macrophage migrating inhibitory factor (PMIF), has been shown to prevent T cells from developing long-term memory191. Following this discovery, a vaccine was created from a squalene-based cationic nanoemulsion loaded with self-amplifying mRNA encoding PMIF. Two 15 µg primer–booster doses improved helper T cell development and elicited anti-Plasmodium IgG antibodies and memory T cell responses192. Moreover, adoptive transfer of T cells from vaccinated mice protected unvaccinated mice from Plasmodium sporozoites192.
Another mechanistic study of malarial infection identified a protein, Plasmodium falciparum glutamic-acid-rich protein (PfGARP), as a potential mRNA vaccine target. PfGARP is expressed on the surface of infected erythrocytes and is recognized by antibodies from children who are relatively resistant to P. falciparum193. These antibodies bind to infected erythrocytes and induce programmed cell death. This discovery prompted the development of an mRNA vaccine comprising LNPs made using a proprietary lipid from Acuitas Therapeutics and encapsulating nucleoside-modified mRNA encoding PfGARP antigen. Three 50 μg doses were able to reduce parasitaemia in Aotus monkeys after a P. falciparum challenge193.
Key issues for the field
Duration of antibody response
After vaccination, translated antigens are produced or taken up by antigen-presenting cells and transported to lymph nodes, where interactions between B cells, antigen-presenting cells and follicular helper T cells (TFH cells) promote the formation of a germinal centre167. Within the germinal centre, B cells then proliferate, differentiate and mutate their antibody genes to produce high-affinity neutralizing antibodies against the offending antigen167. The germinal centre reaction and TFH cell induction are crucial for a durable antibody response that will protect the patient for months or years.
To enhance the first step of this immune response process, some delivery systems target antigen-presenting cells to translate the mRNA cargo. Several promising strategies that actively target antigen-presenting cells include conjugating mAbs to LNP surfaces194 and decorating LNP surfaces with dendritic cell-specific ligands145,146. Alternatively, modulation of physical properties of LNPs, such as surface charge195, has been used to improve cancer vaccines.
Additionally, altering vaccine pharmacokinetics by prolonging the translation of antigenic mRNA has emerged as an exciting tool to enhance antibody response196. Extending the availability of an intact antigen improves the affinity of neutralizing antibodies by diverting the efforts of the immune system away from hidden antigen epitopes and focusing them on accessible ones197,198. Sustained antigen availability during the germinal centre reaction has been shown to increase antibody production by approximately tenfold199. One study in mice showed that LNPs encapsulating nucleoside-modified mRNA circulated for longer and induced stronger TFH cell and germinal centre B cell responses than LNPs carrying unmodified mRNA200.
In preclinical studies, mRNA vaccines have elicited potent germinal centre reactions and TFH cell induction against SARS-CoV-2, HIV-1, Zika virus and influenza virus128,200–202. In humans, two doses of the Pfizer–BioNTech vaccine BNT162b2 induced strong germinal centre B cell responses for at least 12 weeks after immunization203. The antibody clones produced by the germinal centre B cells predominantly targeted the receptor-binding domain of the spike protein. Furthermore, the germinal centre responses after mRNA vaccination were superior to those seen after the seasonal influenza vaccination in humans204.
In clinical trials, two doses of mRNA-1273 also elicited durable antibody responses over a period of 6 months. Although antibody titres declined slightly over the duration of the study, high neutralizing ability was retained across all age groups205. These results are promising; however, the duration of antibody response is a complex phenomenon that will vary from antigen to antigen and will require longer-term data for a comprehensive understanding.
Vaccines against emerging viral variants
Mutations in the viral genome are common during replication. Although the majority of mutations have little or no effect on the functions of a virus, some mutations can enhance immune evasion, stymieing vaccine development. For example, rapid mutations in HIV have prevented the development of an effective vaccine for more than three decades, and the mutations in influenza viruses necessitate annual modification of vaccine formulations to target dominant strains. Novel strategies against these viruses involve delivering mRNA vaccines that target conserved regions such as the haemagglutinin stalk on influenza149, or encodeing broadly neutralizing antibodies such as VRC01 that bind to conserved regions of the CD4 binding site on the HIV gp120 protein206.
Emerging SARS-CoV-2 variants have also raised concerns about the cross-variant efficacy of mRNA vaccines. The B.1.351 and P.1 variants have a glutamate (E) to lysine (K) mutation at position 484 (E484K) in the receptor-binding domain of the spike protein, which promotes immune evasion207. Fortunately, the FDA-approved mRNA vaccines BNT162b2 and mRNA-1273 produce cross-neutralizing antibodies against B.1.351 and P.1, as well as against other variants, suggesting that they can provide protection against them. However, cross-neutralization efficacies have been significantly lower compared with the ancestral variant208–211. Furthermore, in phase IIb/III trials of CureVac’s candidate CVnCoV, 57% of the sequenced 124 COVID-19 cases were attributed to variants of concern, which include the B.1.351 and P.1 variants (see Related links).
If these variants of concern emerge as the dominant variant over time, variant-specific mRNA boosters may be required. At the time of this writing, Moderna is evaluating the original mRNA-1273 vaccine and updated versions of the vaccine as a third dose booster — mRNA-1273.351, which encodes the spike protein from the B.1.351 variant, and mRNA-1273.211, a multivariant vaccine that is a 1:1 mix of mRNA-1273 and mRNA-1273.351 (ref.8).
In the long term, a pancoronavirus vaccine that offers protection from SARS-CoV-2 and future coronavirus outbreaks would be a boon. One study has already demonstrated proof of concept. Specifically, Saunders and colleagues212 have shown that nucleoside-modified mRNA–LNP vaccines encoding the SARS-CoV-2 spike protein can elicit SARS-CoV-1 and batCoV cross-neutralizing antibodies. As with HIV and influenza, new structural insights are expected to facilitate the discovery of conserved sites across coronaviruses, accelerating antigen discovery and vaccine design.
Safety
Overall, mRNA vaccines have promising safety profiles, with only mild or moderate adverse events occurring in clinical trials. However, there have been scattered incidents that mandate further optimization of mRNA antigens and delivery vehicle components. For example, CureVac’s protamine-based rabies candidate CV7201 elicited severe adverse events in 78% of participants188, prompting CureVac to adopt LNPs as their preferred delivery platform for their subsequent rabies candidate CV7202 (ref.189). As with most drugs, adverse reactions to mRNA vaccines have often increased with dose. For example, in phase I trials of CV7202, a 5 μg dose had unacceptable reactogenicity, and 1 μg was the highest dose that was well tolerated. Furthermore, in phase I trials of the Moderna influenza H10N8 vaccine, severe adverse events from the 400 μg dose stopped further evaluation of that dose150. Trials including lower doses up to 100 μg continued.
Anaphylactic reactions have been observed in approximately 4.7 per million anti-COVID-19 vaccinations with the Pfizer–BioNTech vaccine and 2.5 per million vaccinations with the Moderna vaccine213, which is about two- to fourfold more than is typically seen with more traditional vaccines214. One hypothesis is that the allergic response is attributed to pre-existing antibodies against the PEGylated lipid used in LNPs. These antibodies are presumed to form in response to the presence of PEG in many consumer products (for example, toothpastes, shampoos and laxatives)215. Although PEG has long been regarded as safe, it is hypothesized to activate humoral immunity in some people in a T cell-independent manner by directly crosslinking the B cell receptor and initiating IgM production216. In preclinical studies, the presence of anti-PEG IgM in animal serum accelerated nanoparticle clearance and caused a complete loss of the efficacy of mRNA therapeutics217,218. Anti-PEG antibodies have been reported in 40% of the population, which might heighten the risk of allergic reactions in certain individuals and impede vaccine efficacy218. Currently, the CDC recommends that mRNA vaccines should not be given to individuals with a history of allergic response to any of the components of the Pfizer–BioNTech or Moderna vaccines (see Related links). It is clear that, as a field, we need a better understanding of how the mRNA vaccine formulations cause allergic reactions so that the formulations can be re-engineered for improved safety profiles.
Vaccination in specific populations
Most vaccines, whether traditional or mRNA, are developed with either children or healthy adults in mind. However, several populations may benefit from alternative vaccination strategies or respond to vaccination differently owing to differences in their immune system.
Maternal/neonatal vaccination
The dynamic nature of the immune system during pregnancy increases a person’s susceptibility to infectious diseases, and infection can have disastrous impacts on maternal health and fetal development219. Cytomegalovirus infection, for example, causes complications in up to 1% of pregnancies and can lead to congenital disabilities and neurological impairment in infants220. Zika virus can infect cortical neurons and glial cells in the fetus, resulting in cell death, neuroinflammation and severe congenital malformations221. Rare in utero transmission of SARS-CoV-2 has also been reported222,223; its implications on maternal, fetal and neonatal health are under active investigation.
To address these challenges, maternal vaccination has emerged as a tool to improve maternal health and reduce neonatal morbidity. Maternal IgG antibodies readily cross the placenta by binding to the neonatal crystallizable fragment (Fc) receptor, enter the fetal circulation and protect the fetus from pathogens224. In several studies, maternal vaccination with mRNA-loaded LNPs prevented fetal Zika virus transmission in pregnant mice225,226 and protected mouse neonates from herpesvirus227 and group A and group B streptococci228.
Although vertically transferred maternal antibodies can prevent neonatal infection, they can also impede the efficacy of vaccines that are administered to infants later in life. In a mouse model of H1N1 influenza infection in which influenza-specific maternal antibodies inhibited de novo vaccine responses in pups, a LNP-encapsulated, nucleoside-modified mRNA encoding haemagglutinin from the Cal09 H1N1 strain partially overcame this inhibition229. The mechanism remains unclear, but the authors of the report suggested that prolonged antigen availability compared with an FDA-approved inactivated virus vaccine promoted a stronger germinal centre reaction leading to robust infant immune responses in the presence of maternal antibodies.
mRNA vaccines against SARS-CoV-2 have also been shown to be immunogenic in pregnant and lactating people, and neutralizing antibodies have been detected in cord blood and human milk230. Preliminary data suggest that mRNA-1273 and BNT162b2 elicited similar adverse events in pregnant and non-pregnant people, and the vaccines did not increase the incidence of neonatal death or congenital anomalies231. However, further longitudinal studies are necessary to assess the impact of mRNA vaccines on maternal and neonatal health.
Elderly individuals
An increase in life expectancy accompanied by reduced birth rates has led to a shift in population demographics. By 2050, the proportion of the global population over 60 years old is expected to almost double from 12% to 22% (see Related links). Effective vaccines for this group are much needed, as many infectious diseases disproportionately affect the elderly. For example, 70–90% of influenza-related mortalities occur in people older than 65 years232, and COVID-19 is reported to be 62 times more fatal in patients older than 65 than it is in younger patients233.
Older populations are more difficult to vaccinate because ageing adversely affects both the innate and adaptive arms of the immune system. Reduced Toll-like receptor expression stalls cytokine and chemokine secretion by monocytes and macrophages and limits crosstalk with the adaptive immune system234. Adaptive immune responses during infection are often inadequate owing to impaired cytokine signalling and physiological and cellular changes. These changes can include an involuted thymus235, fewer naive B and T cells236, diminished T cell receptor diversity237, higher susceptibility to T cell apoptosis238 and reduced expression of crucial receptors such as CD28 on cytotoxic CD8+ T cells239.
Fortunately, evidence is mounting that mRNA vaccines might offer robust efficacy in all age groups. For instance, in phase III trials, the Pfizer–BioNTech vaccine candidate BNT162b2 elicited >93% efficacy across all treatment groups defined by age125. The Moderna vaccine candidate mRNA-1273 was also highly effective, and showed 86.4% efficacy in volunteers ≥65 years old, in comparison with 95.6% efficacy in 18–65-year-old volunteers131.
The design of delivery vehicles is important for improving vaccine efficacy in the elderly. mRNA delivery vehicles can act as inflammatory adjuvants and amplify vaccine response by enhancing antigen-presenting cell recruitment to the injection site. In a preclinical study, CureVac’s RNActive delivery platform activated TLR7 and generated durable immune responses against a lethal influenza dose in 18-month-old mice105. Novartis’s oil-in-water emulsion MF59, which has been used as an mRNA delivery vehicle, can also act as an adjuvant. MF59 amplifies the immunogenicity of influenza vaccines and has been approved for use in elderly adults110. In the elderly population, MF59-adjuvanted influenza vaccines enhance seroconversion and seroprotection rates compared with non-adjuvanted vaccines240 and induce broad serological protection against strains with antigenic drift241.
Access to vaccines
Access to vaccines is the greatest challenge in achieving widespread protection against infectious diseases, especially in low-income countries. Access is further limited by the cold storage requirements of the currently approved SARS-CoV-2 mRNA vaccines. During the 2014–2016 Ebola virus outbreak in West Africa, vaccines requiring −80 °C storage were supplied in the Democratic Republic of the Congo using portable and reusable Arktek freezers, allowing the vaccine to be administered to 400,000 people242. Such technologies are promising for the rapid deployment of a few million doses during new epidemics; however, vaccinating billions of people during pandemics such as COVID-19 will require thermostable vaccines. In preclinical studies, CureVac demonstrated that its sequence-optimized RABV-G vaccine against rabies could withstand exposure to temperatures ranging from −80 °C to +70 °C for several months243. In addition, two SARS-CoV-2 vaccine candidates are reported to be thermostable at room temperature135 (see Related links). If these thermostable candidates show promising results in clinical trials, they can potentially simplify global access to mRNA vaccines in the near future.
Vaccine acceptance
Vaccines are effective only if they are administered. Data supporting the safety and efficacy of vaccines are profuse, and it is undeniable that vaccines have eradicated several infectious diseases in portions of the world and saved countless lives. Nevertheless, public mistrust fuelled by misinformation and anti-vaccination movements threatens the maintenance of herd immunity and puts our most vulnerable populations at risk. Declining vaccination coverage can lead to the re-emergence of life-threatening diseases. For example, measles, which was eradicated from the USA in 2000, infected more than 1,200 people in 2019 owing to poor vaccine compliance in multiple communities (see Related links). For COVID-19, information from employers and government sources has been shown to improve vaccine acceptance rates, which range from 55% to 90% around the world244. In the USA, excellent efficacy data from mRNA vaccine trials have increased public confidence in these vaccines; however, current acceptance rates of 56–75%245 may be insufficient to reach at least 80–90% coverage246, the threshold thought to be necessary for herd immunity against SARS-CoV-2. Although much of the burden of improving vaccine coverage falls on governments and public health officials, the scientific community can help by improving mRNA vaccine efficacy and safety. Enhanced efficacy will lower the acceptance rates required for herd immunity, and improved safety will stem media reports of adverse events and, thus, decrease fear of vaccination.
Outlook
Decades of progress in mRNA design and nucleic acid delivery technology, together with the discovery of novel antigen targets have made mRNA vaccines an extraordinary tool for combating emerging pandemics and existing infectious diseases. The first two mRNA vaccines, which were developed at revolutionary speed to fight SARS-CoV-2, have exceeded expectations and offer hope that the COVID-19 pandemic will end. Furthermore, these vaccines have elevated LNPs and RNA therapy from small-market products for niche diseases to a prophylactic treatment deployed successfully in large swathes of the population. The resultant abundance of positive safety and efficacy data, together with a proven path to regulatory approval, leave us optimistic that mRNA therapeutics will transform modern medicine’s approach to vaccination1,5, cancer immunotherapy247–249, protein replacement therapy15,23 and beyond250.
Acknowledgements
The authors acknowledge support from the National Institutes of Health (NIH) award no. DP2-HD098860, Translate Bio and the Wadhwani Foundation.
Glossary
- Pattern recognition receptors
A family of membrane-bound and cytoplasmic proteins that are expressed by innate immune cells to detect microbial components, including bacterial carbohydrates such as lipopolysaccharide and mannose, viral and bacterial nucleic acids, bacterial peptides, peptidoglycans and lipoproteins.
- Nucleoside-modified
Chemically modified nucleosides have been introduced into in vitro-transcribed mRNA to reduce immunogenicity and increase translation.
- Endosome
A membrane-bound organelle that engulfs nutrients and macromolecules (including delivery vehicles) and transports them inside the cell.
- Adjuvant
An additive that boosts the immunogenicity of vaccines by triggering pattern recognition receptors, leading to the secretion of cytokines and chemokines.
- Neutralizing antibody
An antibody that binds to pathogens and prevents them from infecting cells.
- Variant
A subtype of a pathogen with a genetic sequence that differs from the reference pathogen.
- Strain
A variant of the pathogen with physical properties such as transmissibility, disease severity or immune response that are different from those of the reference pathogen.
Author contributions
N.C. and K.A.W. researched data for the article. N.C. and K.A.W. wrote the article. All authors reviewed and edited the manuscript before submission.
Competing interests
K.A.W. is an inventor on the following patents related to lipid nanoparticles: US Patent 8,450,298; 8,969,353; 9,556,110; 9,872,911; 9,227,917; 9,439,968; 10,189,802; and 10,844,028. K.A.W. is bound by confidential agreements that prevent her from disclosing related consulting relationships.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Ageing and health: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health
CureVac Provides Update on Phase 2b/3 Trial of First-Generation COVID-19 Vaccine Candidate, CVnCoV: https://www.curevac.com/en/2021/06/16/curevac-provides-update-on-phase-2b-3-trial-of-first-generation-covid-19-vaccine-candidate-cvncov/
CureVac’s COVID-19 Vaccine Candidate, CVnCoV, Suitable for Standard Fridge Temperature Logistics, Press Release, November 12, 2020: https://www.curevac.com/en/2020/11/12/curevacs-covid-19-vaccine-candidate-cvncov-suitable-for-standard-fridge-temperature-logistics/
Draft landscape of COVID-19 candidate vaccines: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
HIV & AIDS statistics — 2020 fact sheet: https://www.unaids.org/en/resources/fact-sheet
Impact of Malaria Worldwide: https://www.cdc.gov/malaria/malaria_worldwide/impact.html
Management of Anaphylaxis at COVID-19 Vaccination Sites: https://www.cdc.gov/vaccines/covid-19/clinical-considerations/managing-anaphylaxis.html
Measles Cases and Outbreaks: https://www.cdc.gov/measles/cases-outbreaks.html
Moderna Announces Clinical Progress from its Industry-Leading mRNA Vaccine Franchise and Continues Investments to Accelerate Pipeline Development: https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-clinical-progress-its-industry-leading-mrna
Moderna Announces Supply Agreement with U.S. Government for Initial 100 Million Doses of mRNA Vaccine Against COVID-19 (mRNA-1273): https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-supply-agreement-us-government-initial-100
Moderna Announces Updates on Respiratory Syncytial Virus (RSV) Vaccine Program: https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-updates-respiratory-syncytial-virus-rsv
Moderna Highlights Opportunity of mRNA Vaccines at its First Vaccines Day: https://investors.modernatx.com/news-releases/news-release-details/moderna-highlights-opportunity-mrna-vaccines-its-first-vaccines
Mumps: https://www.cdc.gov/vaccines/pubs/pinkbook/mumps.html
Pfizer and BioNTech Announce an Agreement with U.S. Government for up to 600 Million Doses of mRNA-based Vaccine Candidate Against SARS-CoV-2: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-announce-agreement-us-government-600
Preliminary results on the efficacy of rVSV-ZEBOV-GP Ebola vaccine using the ring vaccination strategy in the control of an Ebola outbreak in the Democratic Republic of the Congo: an example of integration of research into epidemic response: https://www.who.int/csr/resources/publications/ebola/ebola-ring-vaccination-results-12-april-2019.pdf
Takeda Announces Approval of Moderna’s COVID-19 Vaccine in Japan: https://www.takeda.com/newsroom/newsreleases/2021/takeda-announces-approval-of-modernas-covid-19-vaccine-in-japan/
Vaccines and immunization: https://www.who.int/health-topics/vaccines-and-immunization#tab=tab_1
WHO Coronavirus Disease (COVID-19) Dashboard: https://covid19.who.int/
11868 — Messenger RNA encoding full-length SARS-CoV-2 spike glycoprotein: https://extranet.who.int/soinn/pluginfile.php/1765/mod_folder/content/null/Structures/11868.doc
11889 — Messenger RNA encoding the full-length SARS-CoV-2 spike glycoprotein: https://extranet.who.int/soinn/pluginfile.php/1765/mod_folder/content/null/Structures/11889.doc
Change history
9/21/2021
A Correction to this paper has been published: 10.1038/s41573-021-00321-2
References
- 1.Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy RB, Ovsyannikova IG, Palese P, Poland GA. Current challenges in vaccinology. Front. Immunol. 2020;11:1181. doi: 10.3389/fimmu.2020.01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbeke R, Lentacker I, De Smedt SC, Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 4.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 5.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kis Z, Kontoravdi C, Dey AK, Shattock R, Shah N. Rapid development and deployment of high-volume vaccines for pandemic response. J. Adv. Manuf. Process. 2020;2:e10060. doi: 10.1002/amp2.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freyn AW, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, K. et al. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. bioRxivhttps://www.biorxiv.org/content/10.1101/2021.04.13.439482v1 (2021). [DOI] [PMC free article] [PubMed]
- 9.Bloom K, van den Berg F, Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadhwa A, Aljabbari A, Lokras A, Foged C, Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linares-Fernández S, Lacroix C, Exposito JY, Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Xiong Q, Lee GY, Ding J, Li W, Shi J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018;11:5281–5309. doi: 10.1007/s12274-018-2146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugridge JS, Coller J, Gross JD. Structural and molecular mechanisms for the control of eukaryotic 5′–3′ mRNA decay. Nat. Struct. Mol. Biol. 2018;25:1077–1085. doi: 10.1038/s41594-018-0164-z. [DOI] [PubMed] [Google Scholar]
- 14.Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng Y, et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 16.Sample PJ, et al. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019;37:803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlandini von Niessen AG, et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol. Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng C, et al. Leveraging mRNA sequences and nanoparticles to deliver SARS-CoV-2 antigens in vivo. Adv. Mater. 2020;32:2004452. doi: 10.1002/adma.202004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 20.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von der Mülbe, F., Hoerr, I. & Pascolo, S. Pharmaceutical composition containing a stabilised mRNA optimised for translation in its coding regions. US 2015/0104476 A1 (2015).
- 22.Spencer PS, Siller E, Anderson JF, Barral JM. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajj KA, Whitehead KA. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 24.Vaidyanathan S, et al. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschmann MD, et al. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thess A, et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahin U, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 28.Sahin U, et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 29.Pardi N, Hogan MJ, Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Stadler CR, et al. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 31.Eberle, F., Sahin, U., Kuhn, A., Vallazza, B. & Diken, M. Stabilization of poly(A) sequence encoding Dna sequences. US 2017/0166905 A1 (2017).
- 32.Kim J, Eygeris Y, Gupta M, Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc. Natl Acad. Sci. USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauffman KJ, Webber MJ, Anderson DG. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Rel. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 35.Cui S, et al. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. 2018;7:473–479. doi: 10.1039/C8TX00005K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonez C, Vandenbranden M, Ruysschaert JM. Cationic lipids activate intracellular signaling pathways. Adv. Drug Deliv. Rev. 2012;64:1749–1758. doi: 10.1016/j.addr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J. Control. Rel. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel S, et al. Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 2019;144:90–111. doi: 10.1016/j.addr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semple SC, et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta. 2001;1510:152–166. doi: 10.1016/S0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 41.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Rel. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 42.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaswamy S, et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veiga N, et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Sun J, Li M, Jin X. Modified mRNA-LNP vaccines confer protection against experimental DENV-2 infection in mice. Mol. Ther. Methods Clin. Dev. 2020;18:702–712. doi: 10.1016/j.omtm.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hajj KA, et al. A potent branched-tail lipid nanoparticle enables multiplexed mRNA delivery and gene editing in vivo. Nano Lett. 2020;20:5167–5175. doi: 10.1021/acs.nanolett.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci. USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehead KA, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenton OS, et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020;11:3232. doi: 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou K, et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl Acad. Sci. USA. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabnis S, et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hope, M. et al. Lipid nanoparticle formulations. WO 2018/081480 A1 (2018).
- 55.Hajj KA, et al. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15:1805097. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 56.Alabi CA, et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl Acad. Sci. USA. 2013;110:12881–12886. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miao L, et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lokugamage MP, Sago CD, Gan Z, Krupczak BR, Dahlman JE. Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv. Mater. 2019;31:1902251. doi: 10.1002/adma.201902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao X, et al. Imidazole-based synthetic lipidoids for in vivo mRNA delivery into primary T lymphocytes. Angew. Chem. Int. Ed. Engl. 2020;132:20258–20264. doi: 10.1002/ange.202008082. [DOI] [PubMed] [Google Scholar]
- 60.Miao L, et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 61.Hou X, et al. Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 2020;15:41–46. doi: 10.1038/s41565-019-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang ST, Kreutzberger AJB, Lee J, Kiessling V, Tamm LK. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paunovska K, et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater. 2019;31:1807748. doi: 10.1002/adma.201807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel S, et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim, J., Mukherjee, A., Nelson, D., Jozic, A. & Sahay, G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. bioRxivhttps://www.biorxiv.org/content/10.1101/2020.07.24.205583v1 (2020). [DOI] [PMC free article] [PubMed]
- 66.Sahay G, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 68.Koltover I, Salditt T, Rädler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 69.Kauffman KJ, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 70.Ball RL, Hajj KA, Vizelman J, Bajaj P, Whitehead KA. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett. 2018;18:3814–3822. doi: 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- 71.Lee SM, et al. A systematic study of unsaturation in lipid nanoparticles leads to improved mRNA transfection in vivo. Angew. Chem. Int. Ed. 2021;60:5848–5853. doi: 10.1002/anie.202013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu S, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng Q, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kulkarni JA, et al. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 75.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 76.Oberli MA, et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu X, et al. Surface de-PEGylation controls nanoparticle-mediated siRNA delivery in vitro and in vivo. Theranostics. 2017;7:1990–2002. doi: 10.7150/thno.18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akinc A, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol. Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bus T, Traeger A, Schubert US. The great escape: how cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B. 2018;6:6904–6918. doi: 10.1039/C8TB00967H. [DOI] [PubMed] [Google Scholar]
- 81.Moghimi SM, et al. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol. Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 82.Ulkoski D, Bak A, Wilson JT, Krishnamurthy VR. Recent advances in polymeric materials for the delivery of RNA therapeutics. Expert Opin. Drug. Deliv. 2019;16:1149–1167. doi: 10.1080/17425247.2019.1663822. [DOI] [PubMed] [Google Scholar]
- 83.Ke X, et al. Surface-functionalized PEGylated nanoparticles deliver messenger RNA to pulmonary immune cells. ACS Appl. Mater. Interfaces. 2020;12:35835–35844. doi: 10.1021/acsami.0c08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, et al. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Rel. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 85.Li M, et al. Engineering intranasal mRNA vaccines to enhance lymph node trafficking and immune responses. Acta Biomater. 2017;64:237–248. doi: 10.1016/j.actbio.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 86.Tan L, et al. Optimization of an mRNA vaccine assisted with cyclodextrin-polyethyleneimine conjugates. Drug Deliv. Transl. Res. 2020;10:678–689. doi: 10.1007/s13346-020-00725-4. [DOI] [PubMed] [Google Scholar]
- 87.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc. Natl Acad. Sci. USA. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaczmarek JC, et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 2018;18:6449–6454. doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaczmarek JC, et al. Polymer–lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew. Chem. — Int. Ed. 2016;55:13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patel AK, et al. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 2019;31:1805116. doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lynn DM, Langer R. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J. Am. Chem. Soc. 2000;122:10761–10768. doi: 10.1021/ja0015388. [DOI] [Google Scholar]
- 92.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem. Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 93.Kim HJ, et al. Fine-tuning of hydrophobicity in amphiphilic polyaspartamide derivatives for rapid and transient expression of messenger RNA directed toward genome engineering in brain. ACS Cent. Sci. 2019;5:1866–1875. doi: 10.1021/acscentsci.9b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matsui A, Uchida S, Ishii T, Itaka K, Kataoka K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci. Rep. 2015;5:15810. doi: 10.1038/srep15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin CY, et al. Messenger RNA-based therapeutics for brain diseases: an animal study for augmenting clearance of beta-amyloid by intracerebral administration of neprilysin mRNA loaded in polyplex nanomicelles. J. Control. Rel. 2016;235:268–275. doi: 10.1016/j.jconrel.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 96.Crowley ST, Fukushima Y, Uchida S, Kataoka K, Itaka K. Enhancement of motor function recovery after spinal cord injury in mice by delivery of brain-derived neurotrophic factor mRNA. Mol. Ther. Nucleic Acids. 2019;17:465–476. doi: 10.1016/j.omtn.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aini H, et al. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 2016;6:18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baba M, Itaka K, Kondo K, Yamasoba T, Kataoka K. Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles. J. Control. Rel. 2015;201:41–48. doi: 10.1016/j.jconrel.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 99.McKinlay CJ, et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc. Natl Acad. Sci. USA. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haabeth OAW, et al. mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice. Proc. Natl Acad. Sci. USA. 2018;115:E9153–E9161. doi: 10.1073/pnas.1810002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mccarthy HO, et al. Development and characterization of self-assembling nanoparticles using a bio-inspired amphipathic peptide for gene delivery. J. Control. Rel. 2014;189:141–149. doi: 10.1016/j.jconrel.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 102.Li W, Nicol F, Szoka FC. GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv. Drug Deliv. Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 103.Udhayakumar VK, et al. Arginine-rich peptide-based mRNA nanocomplexes efficiently instigate cytotoxic T cell immunity dependent on the amphipathic organization of the peptide. Adv. Healthc. Mater. 2017;6:1601412. doi: 10.1002/adhm.201601412. [DOI] [PubMed] [Google Scholar]
- 104.van den Brand D, et al. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019;141:180–190. doi: 10.1016/j.ejpb.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 105.Kallen KJ, et al. A novel, disruptive vaccination technology: self-adjuvanted RNActive® vaccines. Hum. Vaccines Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weide B, et al. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 107.Kübler H, et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer. 2015;3:26. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Papachristofilou A, et al. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J. Immunother. Cancer. 2019;7:38–38. doi: 10.1186/s40425-019-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brito LA, et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsai TF. Fluad®-MF59®-adjuvanted influenza vaccine in older adults. Infect. Chemother. 2013;45:159–174. doi: 10.3947/ic.2013.45.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O’Hagan DT, Ott GS, De Gregorio E, Seubert A. The mechanism of action of MF59 - an innately attractive adjuvant formulation. Vaccine. 2012;30:4341–4348. doi: 10.1016/j.vaccine.2011.09.061. [DOI] [PubMed] [Google Scholar]
- 112.Mosca F, et al. Molecular and cellular signatures of human vaccine adjuvants. Proc. Natl Acad. Sci. USA. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seubert A, Monaci E, Pizza M, O’Hagan DT, Wack A. The adjuvants aluminum hydroxide and MF59 induce monocyte and granulocyte chemoattractants and enhance monocyte differentiation toward dendritic cells. J. Immunol. 2008;180:5402–5412. doi: 10.4049/jimmunol.180.8.5402. [DOI] [PubMed] [Google Scholar]
- 114.Gómez-Aguado I, et al. Nanomedicines to deliver mRNA: state of the art and future perspectives. Nanomaterials. 2020;10:364. doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alon R, et al. Leukocyte trafficking to the lungs and beyond: lessons from influenza for COVID-19. Nat. Rev. Immunol. 2021;21:49–64. doi: 10.1038/s41577-020-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knezevic I, Liu MA, Peden K, Zhou T, Kang H-N. Development of mRNA vaccines: scientific and regulatory issues. Vaccines. 2021;9:81. doi: 10.3390/vaccines9020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vogel AB, et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 123.Mulligan MJ, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 124.Walsh EE, et al. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dagan N, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Corbett KS, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corbett KS, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Corbett, K. S. et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science eabj0299 (2021). [DOI] [PMC free article] [PubMed]
- 130.Jackson LA, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thompson MG, et al. Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers — eight U.S. locations, December 2020–March 2021. Morb. Mortal. Wkly Rep. 2021;70:495–500. doi: 10.15585/mmwr.mm7013e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kremsner, P. et al. Phase 1 assessment of the safety and immunogenicity of an mRNA- lipid nanoparticle vaccine candidate against SARS-CoV-2 in human volunteers. medRxivhttps://www.medrxiv.org/content/10.1101/2020.11.09.20228551v1 (2020). [DOI] [PMC free article] [PubMed]
- 134.Roth, N. et al. CV2CoV, an enhanced mRNA-based SARS-CoV-2 vaccine candidate, supports higher protein expression and improved immunogenicity in rats. bioRxiv, https://www.biorxiv.org/content/10.1101/2021.05.13.443734v1.full (2021).
- 135.Zhang NN, et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McKay PF, et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3–9. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.de Alwis R, et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021;29:1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iuliano A, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zost SJ, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl Acad. Sci. USA. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hekele A, et al. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Petsch B, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 142.Brazzoli M, et al. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2016;90:332–344. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Magini D, et al. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS ONE. 2016;11:e0161193. doi: 10.1371/journal.pone.0161193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vogel AB, et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goswami R, et al. Mannosylation of LNP results in improved potency for self-amplifying RNA (SAM) vaccines. ACS Infect. Dis. 2019;5:1546–1558. doi: 10.1021/acsinfecdis.9b00084. [DOI] [PubMed] [Google Scholar]
- 146.Zhuang X, et al. mRNA vaccines encoding the HA protein of influenza A H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines. 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bahl K, et al. Preclinical and clinical demonstration of immunogenicity by mrna vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nachbagauer R, et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines. 2017;2:26. doi: 10.1038/s41541-017-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pardi N, et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feldman RA, et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 151.Poland GA, et al. Development of vaccines against Zika virus. Lancet Infect. Dis. 2018;18:e211–e219. doi: 10.1016/S1473-3099(18)30063-X. [DOI] [PubMed] [Google Scholar]
- 152.Dowd KA, et al. Broadly neutralizing activity of zika virus-immune sera identifies a single viral serotype. Cell Rep. 2016;16:1485–1491. doi: 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pardi N, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wollner CJ, Richner JM. mRNA vaccines against flaviviruses. Vaccines. 2021;9:1–13. doi: 10.3390/vaccines9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Richner JM, et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Erasmus JH, et al. Intramuscular delivery of replicon RNA encoding ZIKV-117 human monoclonal antibody protects against Zika virus. Infect. Mol. Ther. Methods Clin. Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kose N, et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against Chikungunya infection. Sci. Immunol. 2019;4:6647. doi: 10.1126/sciimmunol.aaw6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dybul M, et al. The case for an HIV cure and how to get there. Lancet HIV. 2021;8:e51–e58. doi: 10.1016/S2352-3018(20)30232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mascola JR. The modern era of HIV-1 vaccine development. Science. 2015;349:139–140. doi: 10.1126/science.aac7800. [DOI] [PubMed] [Google Scholar]
- 161.Bogers WM, et al. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J. Infect. Dis. 2015;211:947–955. doi: 10.1093/infdis/jiu522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pollard C, et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Moyo N, et al. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol. Ther. Methods Clin. Dev. 2019;12:32–46. doi: 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao M, Li M, Zhang Z, Gong T, Sun X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016;23:2596–2607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 165.Blakney AK, McKay PF, Yus BI, Aldon Y, Shattock RJ. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pardi N, et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Singh A. Eliciting B cell immunity against infectious diseases using nanovaccines. Nat. Nanotechnol. 2021;16:16–24. doi: 10.1038/s41565-020-00790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Corey L, et al. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pardi N, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:6–13. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shi T, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 172.Kim HW, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 173.Haynes BF, et al. Prospects for a safe COVID-19 vaccine. Sci. Transl. Med. 2020;12:1–13. doi: 10.1126/scitranslmed.abe0948. [DOI] [PubMed] [Google Scholar]
- 174.Mazur NI, et al. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect. Dis. 2018;18:e295–e311. doi: 10.1016/S1473-3099(18)30292-5. [DOI] [PubMed] [Google Scholar]
- 175.Crank MC, et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science. 2019;365:505–509. doi: 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- 176.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Espeseth AS, et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines. 2020;5:1–14. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Geall AJ, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Aliprantis AO, et al. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum. Vaccines Immunother. 2021;17:1248–1261. doi: 10.1080/21645515.2020.1829899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaner J, Schaack S. Understanding Ebola: the 2014 epidemic. Glob. Heal. 2016;12:53. doi: 10.1186/s12992-016-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Agnandji ST, et al. Phase 1 trials of rVSV Ebola vaccine in Africa and Europe. N. Engl. J. Med. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chahal JS, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl Acad. Sci. USA. 2016;113:E5250. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meyer M, et al. Modified mRNA-based vaccines elicit robust immune responses and protect guinea pigs from Ebola virus disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hampson K, et al. Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 2015;9:e0003709. doi: 10.1371/journal.pntd.0003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schnee M, et al. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl. Trop. Dis. 2016;10:e0004746. doi: 10.1371/journal.pntd.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Alberer M, et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 189.Lutz J, et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Aldrich C, et al. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine. 2021;39:1310–1318. doi: 10.1016/j.vaccine.2020.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun T, et al. A Plasmodium-encoded cytokine suppresses T-cell immunity during malaria. Proc. Natl Acad. Sci. USA. 2012;109:E2117. doi: 10.1073/pnas.1206573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Baeza Garcia A, et al. Neutralization of the Plasmodium-encoded MIF ortholog confers protective immunity against malaria infection. Nat. Commun. 2018;9:2714. doi: 10.1038/s41467-018-05041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Raj DK, et al. Anti-PfGARP activates programmed cell death of parasites and reduces severe malaria. Nature. 2020;582:104–108. doi: 10.1038/s41586-020-2220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kedmi R, et al. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018;13:214–219. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 195.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 196.Irvine DJ, Aung A, Silva M. Controlling timing and location in vaccines. Adv. Drug Deliv. Rev. 2020;158:91–115. doi: 10.1016/j.addr.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cirelli KM, Crotty S. Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol. 2017;47:64–69. doi: 10.1016/j.coi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hu JK, et al. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J. Virol. 2015;89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tam HH, et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl Acad. Sci. USA. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pardi N, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Laczkó D, et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lederer K, et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Turner JS, et al. SARS-CoV-2 mRNA vaccines induce robust plasmablast and germinal centre responses in humans. Nature. 2021 doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Turner JS, et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature. 2020;586:127–132. doi: 10.1038/s41586-020-2711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Doria-Rose N, et al. Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Li Y, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yuan M, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021 doi: 10.1101/2021.02.16.430500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang Z, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu Y, et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wu K, et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pegu A, et al. Durability of mRNA-1273 vaccine–induced antibodies against SARS-CoV-2 variants. Science. 2021;12:eabj4176. doi: 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Saunders KO, et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shimabukuro TT, Cole M, Su JR. Reports of anaphylaxis after receipt of mRNA COVID-19 vaccines in the US — December 14, 2020-January 18, 2021. JAMA. 2021;325:1101–1102. doi: 10.1001/jama.2021.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McNeil MM, et al. Risk of anaphylaxis after vaccination in children and adults. J. Allergy Clin. Immunol. 2016;137:868–878. doi: 10.1016/j.jaci.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vrieze, J. Suspicions grow that nanoparticles in Pfizer’s COVID-19 vaccine trigger rare allergic reactions. Sciencehttps://www.sciencemag.org/news/2020/12/suspicions-grow-nanoparticles-pfizer-s-covid-19-vaccine-trigger-rare-allergic-reactions (2020).
- 216.Besin G, et al. Accelerated blood clearance of lipid nanoparticles entails a biphasic humoral response of B-1 followed by B-2 lymphocytes to distinct antigenic moieties. ImmunoHorizons. 2019;3:282–293. doi: 10.4049/immunohorizons.1900029. [DOI] [PubMed] [Google Scholar]
- 217.Abu Lila AS, Kiwada H, Ishida T. The accelerated blood clearance (ABC) phenomenon: clinical challenge and approaches to manage. J. Control. Rel. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 218.Kozma GT, Shimizu T, Ishida T, Szebeni J. Anti-PEG antibodies: properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Adv. Drug Deliv. Rev. 2020;154–155:163–175. doi: 10.1016/j.addr.2020.07.024. [DOI] [PubMed] [Google Scholar]
- 219.Vermillion MS, Klein SL. Pregnancy and infection: using disease pathogenesis to inform vaccine strategy. NPJ Vaccines. 2018;3:6. doi: 10.1038/s41541-017-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bialas KM, Swamy GK, Permar SR. Perinatal cytomegalovirus and varicella zoster virus infections: epidemiology, prevention, and treatment. Clin. Perinatol. 2015;42:61–75. doi: 10.1016/j.clp.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yockey LJ, Lucas C, Iwasaki A. Contributions of maternal and fetal antiviral immunity in congenital disease. Science. 2020;368:608–612. doi: 10.1126/science.aaz1960. [DOI] [PubMed] [Google Scholar]
- 222.Barrero-Castillero A, et al. COVID-19: neonatal-perinatal perspectives. J. Perinatol. 2021;41:940–951. doi: 10.1038/s41372-020-00874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Fenizia C, et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 2020;11:5128. doi: 10.1038/s41467-020-18933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 225.Richner JM, et al. Vaccine mediated protection against Zika virus-induced congenital disease. Cell. 2017;170:273–283.e12. doi: 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jagger BW, et al. Protective efficacy of nucleic acid vaccines against transmission of Zika virus during pregnancy in mice. J. Infect. Dis. 2019;220:1577–1588. doi: 10.1093/infdis/jiz338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.LaTourette PC, et al. Protection against herpes simplex virus type 2 infection in a neonatal murine model using a trivalent nucleoside-modified mRNA in lipid nanoparticle vaccine. Vaccine. 2020;38:7409–7413. doi: 10.1016/j.vaccine.2020.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maruggi G, et al. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35:361–368. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 229.Willis E, et al. Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Sci. Transl. Med. 2020;12:eaav5701. doi: 10.1126/scitranslmed.aav5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Collier AY, et al. Immunogenicity of COVID-19 mRNA vaccines in pregnant and lactating women. JAMA. 2021;325:2370–2380. doi: 10.1001/jama.2021.7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Shimabukuro TT, et al. Preliminary findings of mRNA Covid-19 vaccine safety in pregnant persons. N. Engl. J. Med. 2021;384:2273–2282. doi: 10.1056/NEJMoa2104983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Crooke SN, Ovsyannikova IG, Poland GA, Kennedy RB. Immunosenescence and human vaccine immune responses. Immun. Ageing. 2019;16:25. doi: 10.1186/s12979-019-0164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yanez ND, Weiss NS, Romand JA, Treggiari MM. COVID-19 mortality risk for older men and women. BMC Public Health. 2020;20:1742. doi: 10.1186/s12889-020-09826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Van Den Biggelaar AHJ, et al. Impaired innate immunity predicts frailty in old age. The Leiden 85-plus study. Exp. Gerontol. 2004;39:1407–1414. doi: 10.1016/j.exger.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 235.Palmer DB. The effect of age on thymic function. Front. Immunol. 2013;4:316. doi: 10.3389/fimmu.2013.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lazuardi L, et al. Age-related loss of naïve T cells and dysregulation of T-cell/B-cell interactions in human lymph nodes. Immunology. 2005;114:37–43. doi: 10.1111/j.1365-2567.2004.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl Acad. Sci. USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fang F, et al. Expression of CD39 on activated T cells impairs their survival in older individuals. Cell Rep. 2016;14:1218–1231. doi: 10.1016/j.celrep.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Boucher N, et al. CD28 expression in T cell aging and human longevity. Exp. Gerontol. 1998;33:267–282. doi: 10.1016/S0531-5565(97)00132-0. [DOI] [PubMed] [Google Scholar]
- 240.Yang J, et al. Effectiveness, immunogenicity, and safety of influenza vaccines with MF59 adjuvant in healthy people of different age groups. Medicine. 2020;99:e19095. doi: 10.1097/MD.0000000000019095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ansaldi F, et al. Cross-protection by MF59TM-adjuvanted influenza vaccine: neutralizing and haemagglutination-inhibiting antibody activity against A(H3N2) drifted influenza viruses. Vaccine. 2008;26:1525–1529. doi: 10.1016/j.vaccine.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 242.Kaiser, J. Temperature concerns could slow the rollout of new coronavirus vaccines. Sciencehttps://www.sciencemag.org/news/2020/11/temperature-concerns-could-slow-rollout-new-coronavirus-vaccines (2020).
- 243.Stitz L, et al. A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 2017;11:e0006108. doi: 10.1371/journal.pntd.0006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lazarus JV, et al. A global survey of potential acceptance of a COVID-19 vaccine. Nat. Med. 2020;27:225–228. doi: 10.1038/s41591-020-1124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sallam M. Covid-19 vaccine hesitancy worldwide: a concise systematic review of vaccine acceptance rates. Vaccines. 2021;9:1–15. doi: 10.3390/vaccines9020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lippi G, Henry BM. How will emerging SARS-CoV-2 variants impact herd immunity? Ann. Transl. Med. 2021;9:585–585. doi: 10.21037/atm-21-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Goldberg MS. Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer. 2019;19:587–602. doi: 10.1038/s41568-019-0186-9. [DOI] [PubMed] [Google Scholar]
- 249.Beck JD, et al. mRNA therapeutics in cancer immunotherapy. Mol. Cancer. 2021;20:69. doi: 10.1186/s12943-021-01348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Krienke C, et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 251.Wu, K. et al. Preliminary analysis of safety and immunogenicity of a SARS-CoV-2 variant vaccine booster. medRxivhttps://www.medrxiv.org/content/10.1101/2021.05.05.21256716v1.full (2021).
- 252.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Boni MF. Vaccination and antigenic drift in influenza. Vaccine. 2008;26:C8–C14. doi: 10.1016/j.vaccine.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Jusu MO, et al. Rapid establishment of a cold chain capacity of −60 °C or colder for the STRIVE Ebola vaccine trial during the Ebola outbreak in Sierra Leone. J. Infect. Dis. 2018;217:S48–S55. doi: 10.1093/infdis/jix336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Panchaud A, Stojanov M, Ammerdorffer A, Vouga M, Baud D. Emerging role of Zika virus in adverse fetal and neonatal outcomes. Clin. Microbiol. Rev. 2016;29:659–694. doi: 10.1128/CMR.00014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cuevas JM, Geller R, Garijo R, López-Aldeguer J, Sanjuán R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Berndsen ZT, et al. Visualization of the HIV-1 Env glycan shield across scales. Proc. Natl Acad. Sci. USA. 2020;117:28014–28025. doi: 10.1073/pnas.2000260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kirtane AR, et al. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021;16:369–384. doi: 10.1038/s41565-021-00866-8. [DOI] [PubMed] [Google Scholar]
- 259.Kim YC, Dema B, Reyes-Sandoval A. COVID-19 vaccines: breaking record times to first-in-human trials. NPJ Vaccines. 2020;5:34. doi: 10.1038/s41541-020-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Shepherd SJ, Issadore D, Mitchell MJ. Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials. 2021;274:120826. doi: 10.1016/j.biomaterials.2021.120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Belliveau NM, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]