Abstract
Research on intranasal delivery of drugs, peptides, and proteins has grown over the past decade as an alternate way to deliver substrates to the brain. Recent work has shown intranasal (INL) delivery of insulin improves memory and cognition in healthy subjects as well as patients with Alzheimer’s disease (AD) and in AD mouse models. However, the molecular mechanism(s) for the beneficial effect of insulin on memory are still unclear. Using the SAMP8 mouse model of AD, we investigated the impact of INL insulin on protein and gene expression in brain regions including the olfactory bulb, hypothalamus, and hippocampus. We found genes and proteins in the insulin receptor signaling pathway were not activated by the doses tested. However, we did find the expression of genes present in the hippocampus involved in other pathways, especially those related to inflammation, were altered due to age and with a dose of INL insulin previously shown to improve cognition. These alternate pathways could be targets of insulin when delivered via the INL route to aid in memory improvement.
Keywords: Insulin, intranasal, hippocampus, cognition, RNA sequence analysis, Alzheimer’s disease
INTRODUCTION
Insulin plays a vital role in memory and cognition in the brain, distinct from the recognized role that it plays in blood glucose regulation. Central nervous system (CNS) insulin levels tend to decline with age [1] and with the development of Alzheimer’s disease (AD) [2]. When peripheral insulin levels are increased through the use of a euglycemic clamp, cognition is improved in AD patients [3] and can impact brain activity [4]. However, in those studies it was unclear if the memory improvements were due to increased CNS insulin levels.
The blood-brain barrier (BBB) limits the ability to deliver drugs and peptides to the brain. Intranasal (INL) delivery provides an alternative solution in accessing the brain [5, 6]. When delivered to the level of the cribriform plate, the substrate of interest, in this case insulin, is transported by various routes to distinct brain regions [7]. Some of the earlier studies reporting improvements in memory and cognition following INL insulin treatment [8–11] have been recapitulated by more recent independent studies [12–14]. These memory improvements in humans occur without affecting blood glucose and blood insulin levels [8].
Analogous mouse studies have shown that aged mice with an AD-like phenotype (SAMP8 mice) demonstrate memory improvements within 24 hrs after being administered a low dose of INL insulin [15]. The impact on memory is even greater with repeated injections over a two-week period. Pharmacokinetic studies investigating radioactive insulin uptake following INL administration show that insulin levels are highest throughout the brain 30 min later [16]. This transport is not different between young SAMP8 mice and aged, cognitively impaired SAMP8 mice suggesting the development of AD-like characteristics does not alter the access to insulin. SAMP8 mice are a non-transgenic model of AD [17] and useful for investigating the role of insulin in the brain [18]. Whereas the memory benefit has been shown consistently with INL insulin treatment, we wanted to investigate the molecular impact following treatment. We utilized young and aged SAMP8 mice treated with INL vehicle or a dose of insulin previously shown to improve cognition to investigate not only targeted changes in regional insulin receptor signaling but also an unbiased impact on hippocampal gene expression.
METHODS
Animal Use
Male SAMP8 mice were bred in-house at St. Louis University and aged to 2–4 month (young) and 12–14 month (aged) prior to transfer to the VA Puget Sound Health Care System. Male CD-1 mice (2 months) were purchased from Charles River Laboratory (Seattle, WA). Mice had ad libitum access to food and water and were kept on a 12/12 hour light/dark cycle. All animal protocols were approved by the local Institutional Animal Care and Use Committee (IACUC) and performed at an approved facility (Association for Assessment and Accreditation of Laboratory Animal Care International, AAALAC).
Administrations
INL: INL injections of a 1 μL volume directly at the cribriform plate were performed by using a 10 μL Multi-flex (Thermo Fisher Scientific, Waltham, MA) inserted 4 mm deep into the left nares. Mice were kept in the supine position for 30 seconds before turning to their side. For repeated INL administration, mice were anesthetized with 3.5% isofluorane. Mice received an INL administration of vehicle or insulin every other day for a total of 7 administrations. For terminal studies, mice were anesthetized with an intraperitoneal injection of 40% urethane solution (Sigma-Aldrich, St. Louis, MO). For dose response studies, mice were given vehicle, 0.1 μg, 1 μg, or 10 μg human insulin dissolved in 0.25 M phosphate buffer (PB).
ICV: Mice were anesthetized with an intraperitoneal injection of 40% urethane solution (Sigma-Aldrich, St. Louis, MO). ICV injections into the right lateral ventricle were performed by removing the scalp and drilling a hole 1 mm lateral and 0.5 mm posterior to the bregma, followed by injection of a 1 μL volume at a depth of 2.5 mm using a 26 g Hamilton syringe. For dose response studies, mice were given vehicle, 0.1 μg, 1 μg, or 10 μg human insulin dissolved in 0.25 M phosphate buffer (PB).
Sample collection and preparation
Brain regions were dissected on ice and snap frozen in liquid nitrogen. Briefly, tissue samples (olfactory bulb, hypothalamus, and hippocampus) were homogenized in RIPA buffer (150 mM NaCl, 0.5% deoxycholic acid, 0.1% SDS, 20 mM Tris HCl, 2 mM EDTA) plus freshly added 1/100 dilutions of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), 1mM sodium orthovanadate, and phosphatase inhibitor (1 tablet PhosSTOP per 10 mL buffer) (Roche). Samples were sonicated at 40% amplitude prior to centrifugation at 12,000 xg for 10 min at 4°C. Supernatants were collected and frozen at −80°C. Protein was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL).
Measurement of immunoactive human insulin
Samples processed for protein extraction were used to measure human insulin levels. Total immunoactive levels of human insulin were measured from 75 μg total protein in the olfactory bulb, hypothalamus, and hippocampus using the singleplex Meso Scale Discovery human insulin kit (MSD, Rockville, MD) according to the manufacturer’s directions.
Measurement of insulin receptor signaling
Tissue lysates prepared as described above were solubilized in NuPAGE sample buffer, warmed for 10 min at 70°C, and resolved on a 4–12% Bis-Tris gel (Invitrogen, Grand Island, NY). Protein was then transferred to nitrocellulose membranes using the iBlot transfer system (Invitrogen). Membranes were blocked with 5% BSA dissolved in TBS-T for 1 hour. Membranes were incubated in primary antibodies (see supplemental Table 1) specific to the phosphorylated form diluted in 5% BSA/TBS-T overnight at 4°C. Membranes were washed in TBS-T and then probed with respective secondary antibodies conjugated to horseradish peroxidase (Jackson Labs, West Grove, PA) for 1 hour at room temperature. Membranes were washed with TBS-T and then illuminated with chemiluminescent substrate (Amersham ECL Prime Western Blotting Detection Reagent, GE Life Sciences, Piscataway, NJ). Following band visualization on the ImageQuant LAS4000 CCD imaging system (GE Life Sciences, Piscataway, NJ), blots were stripped with Restore Western Blot Stripping Buffer (Thermo Scientific, Rockford, IL) and reprobed for total protein levels and β-actin. Densitometric analysis of bands was performed using IQTL software (GE Life Sciences, Piscataway, NJ). Band intensities of phosphorylated proteins were normalized to band intensities of corresponding total protein levels, and the ratios of phosphorylated/total protein were normalized to the young vehicle group. SYPRO Ruby (Invitrogen, Grand Island, NY) was also used to verify uniformity of protein loading where indicated [19].
INL treatment for RNA-Sequencing
Mice were transferred to the study space at least one hour prior to the administration to minimize stress. Mice were placed under 3.5% isofluorane prior to INL administration. Mice received an INL administration of vehicle or insulin as described above. Once recovered, mice were returned to their home cage. For the single INL administration (acute) study, the hippocampus was collected 4 hours later. For mice receiving repeated INL injections (chronic), home cages were returned to the animal facility after each administration. Mice received INL injections every other day for 7 injections. There was no effect of chronic INL insulin administration on body weight (data not shown). After the last injection, hippocampi were collected 4 hours later. Mice were sacrificed by cervical dislocation, in which the brain was extracted and the hippocampus immediately dissected and placed in 500 μL RNA later prior to placement in a −80 °C freezer.
RNA Extraction and processing of samples for RNA-Sequencing
The RNA from each hippocampus sample was extracted using the Qiagen miRNeasy mini kit (Qiagen, Germantown, MD) following the manufacturer’s recommended protocol. Integrity of RNA samples was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). RNA integrity was judged by observing distinct and sharp 18S and 28S ribosomal RNA peaks that were baseline separated. RNA quantity was determined by measuring OD260 with a Thermo Scientific NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.; Wilmington, DE). The NanoDrop instrument was also used to determine purity of RNA samples by measuring OD260/280 and OD260/230 ratios. Samples with both of these ratios >1.8 were considered pure. Only samples passing these stringent quality control criteria were used for RNA-Seq analysis. We pooled RNA from 2–3 hippocampi to generate a heterogeneous sample population within each experimental group (n=3/treatment). While each treatment group consists of three samples, 7–9 hippocampi are represented. cDNA libraries were prepared from 1 μg of total RNA in an automated, high-throughput format, following the manufacturer’s instructions for the TruSeq Stranded mRNA kit (Illumina, San Diego, CA) and the Sciclone NGSx Workstation (Perkin Elmer, Waltham, MA). During cDNA library construction, ribosomal RNA is removed by means of poly-A enrichment. Each library is then uniquely barcoded and subsequently amplified using a total of 13 cycles of PCR. Library concentrations are quantified using Qubit fluorometric quantitation (Life Technologies, Carlsbad, CA). Average fragment size and overall quality are evaluated with the DNA1000 assay on an Agilent 2100 Bioanalyzer. Each library is sequenced with paired-end 75bp reads to a minimum depth of 30 million reads on an Illumina HiSeq.
RNA-Sequencing Analysis
The results were aligned to mm10 [20]. The counts per million (CPM) were computed and filtered on CPM. A gene was only retained if it was expressed at a CPM above 0.5 in at least three samples. Normalization factors were calculated using the weighted trimmed mean of M-values (to the reference) as previously proposed [21]. Two generalized linear model functions [22], were used for the comparisons of the various experimental groups. Finally, multiple testing across genes and contrasts was performed. Genes that were significantly changed (p<0.05) from the comparison group and resulted in ≥ log 1.5 fold change (logFC) were used for pathway analysis. The data for all genes and contrasts that were computed is available in Supplemental Table 2.
Pathway Analysis
The genes that were significantly changed within each comparison (young vs aged, aged vehicle vs aged plus single INL insulin, aged vehicle vs aged plus repetitive INL insulin) were statistically analyzed for enrichment in functional categories such as Pathways using the online DAVID platform [23, 24]. Briefly, the Entrez Gene ID was entered into the database for each group comparison and the default criteria were used to generate the results. DAVID determines the p values of enrichment into pathways by Fisher exact test. A list of all pathways changed for each group comparison is presented in the Supplemental Data Table 3.
Statistical Analysis
Nonlinear regression analysis was performed using Prism 8.0 (GraphPad Inc, San Diego, CA). One-way ANOVAs were used to compare differences due to dose followed by Tukey’s multiple comparisons test post hoc analysis. Only p values ≤ 0.05 were considered significant. All data are reported as mean ± SEM. Sample sizes are listed in the figure legends.
RESULTS
Insulin levels in brain following INL and ICV delivery
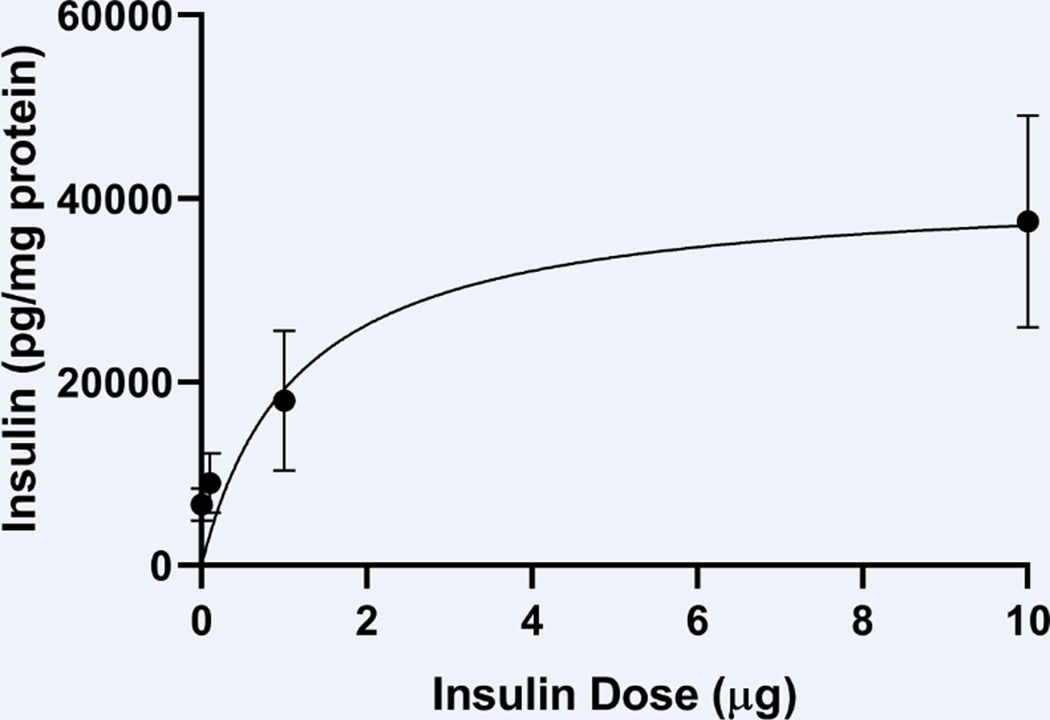
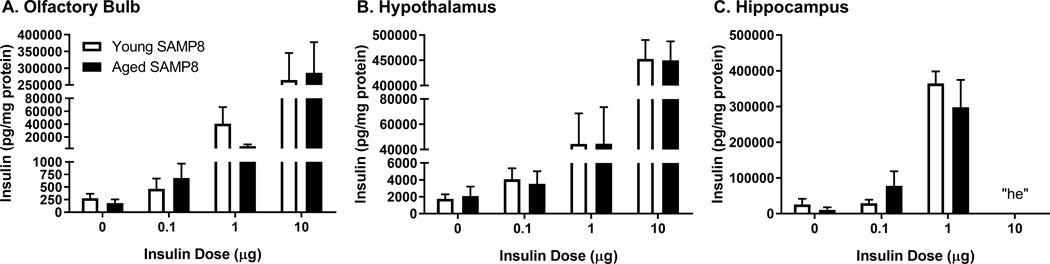
We have already shown radioactive insulin is distributed throughout the brain following INL delivery in SAMP8 mice [16]. There is no difference in distribution between young CD-1 mice and young or aged SAMP8 mice following INL delivery. Here, we measured the amount of immunoactive human insulin levels in the hippocampus 30 min following INL delivery of human insulin in CD-1 mice (Fig. 1). A nonlinear relationship existed, r(16) = 0.59, between the dose of insulin administered and the amount of insulin detected in the hippocampus, as expected, due to the saturable nature of insulin transport following INL delivery. We also measured human insulin levels in the olfactory bulb, hypothalamus, and hippocampus in young and aged SAMP8 male mice 10 min following ICV delivery (Fig. 2). We were able to detect a significant dose dependent increase in the amount of insulin present in the regions following ICV delivery (Fig 1A-C). In the hippocampus, the “hook effect” (he) interfered with immunoassy for the 10 μg insulin dose administration. That is, the analyte concentration was too high to accurately measure the levels and likely interfered with the capture antibody [25]. The hippocampus is nearest to the delivery site with ICV administration and thus, the 10 μg dose would deliver the most insulin to this region.
Figure 1.
Hippocampal insulin levels following INL insulin administration at increasing doses. Data are expressed as mean ± SEM; n=4–6/dose. Results were fitted to a nonlinear hyperbola model with r(16) = 0.59.
Figure 2.
Brain insulin levels following ICV insulin administration. One-way ANOVA revealed statistically significant differences between the doses (p<0.001). Data are expressed as mean ± SEM, n=4–5/group. “he” = hook effect
Insulin Receptor Signaling
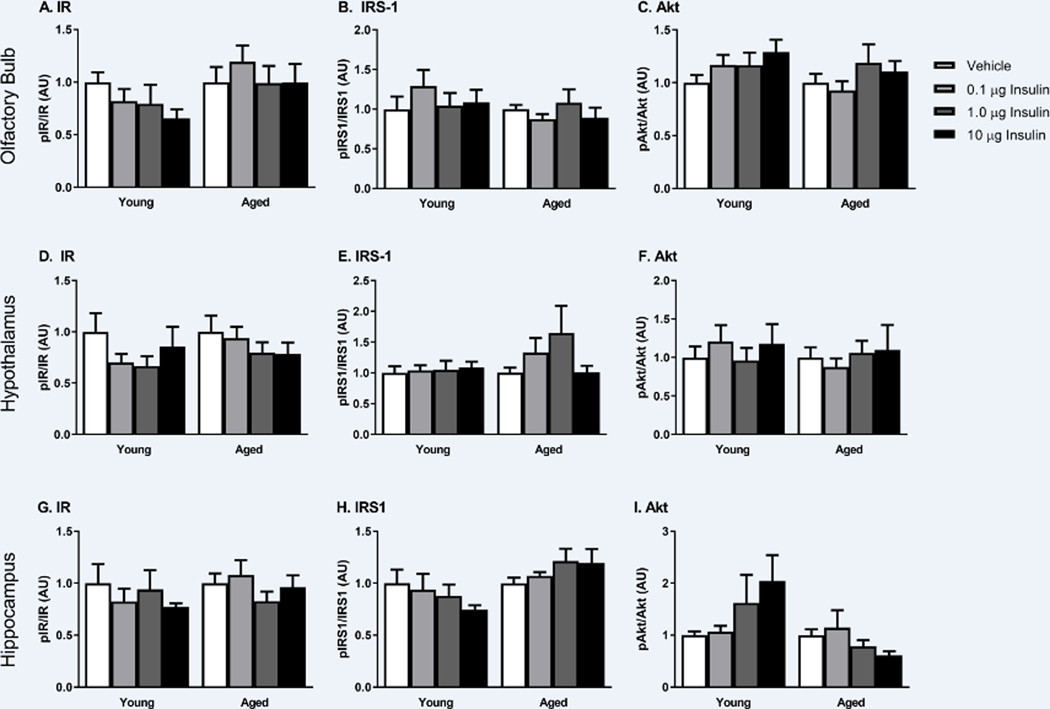
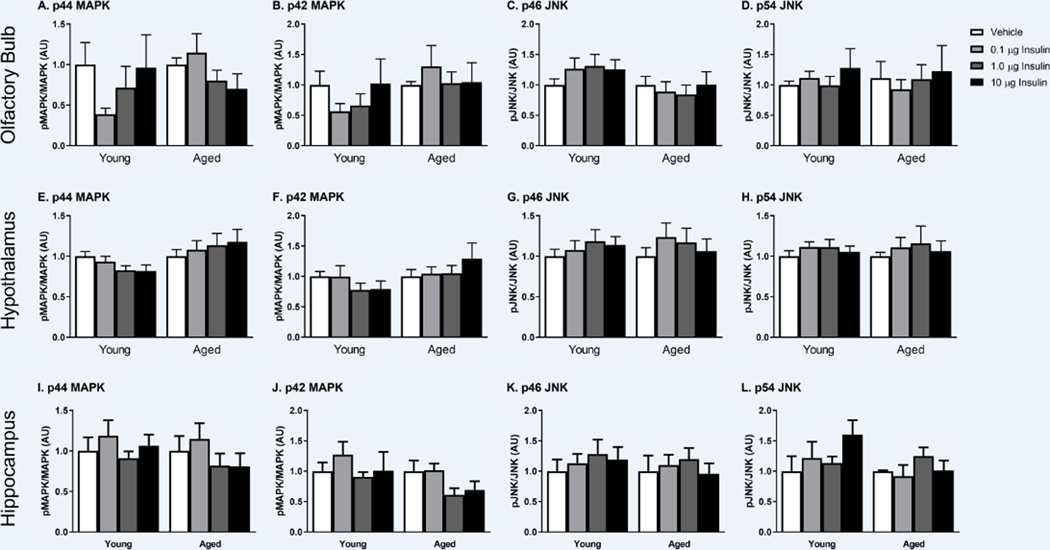
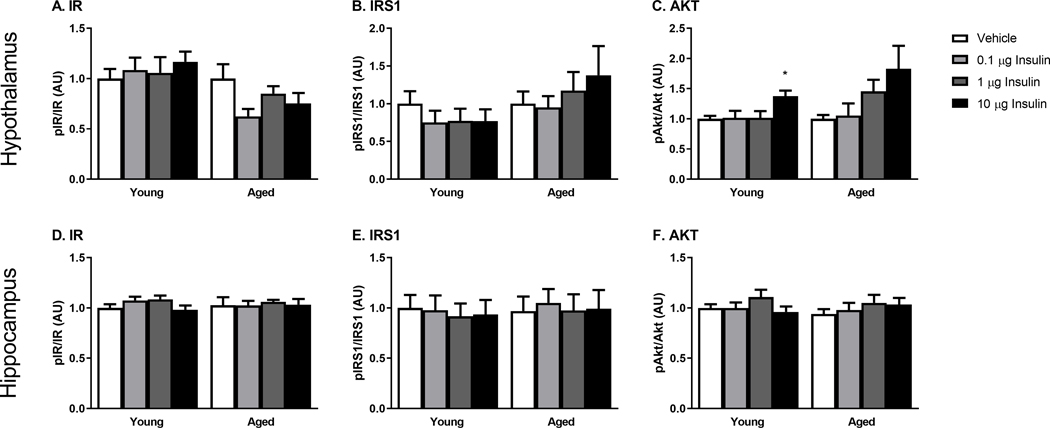
Changes in the phosphorylation of key insulin receptor signaling mediators were measured to determine changes in insulin receptor sensitivity in the young and SAMP8 mice. We investigated changes within the olfactory bulb (first site of delivery following INL administration), hypothalamus (site of high insulin levels) and the hippocampus (site of action) 30 min following INL administration. There were no differences between young and aged, vehicle treated groups. There was no dose-dependent change in the level of phosphorylated protein compared to total protein levels of key canonical insulin signaling proteins including the insulin receptor (IR), insulin receptor substrate 1 (IRS1), or Akt (Figure 3). Therefore, we also investigated the non-canonical insulin signaling pathway by looking at changes in phosphorylation of p44/42 MAPK (Erk1/2) and p46/54 SAPK/JNK (Figure 4). Again, with acute INL insulin treatment, there was no change in the levels of phosphorylated MAPK or JNK.
Figure 3.
Canonical insulin receptor signaling as measured by protein levels of phosphorylated IR, IRS1, and Akt is not affected by the tested doses of INL insulin administered 30 min after delivery in the olfactory bulb (A-C), hypothalamus (D-F), and hippocampus (G-I). Data are expressed as mean ± SEM, relative to vehicle. n=4–8/group
Figure 4.
Non-canonical insulin signaling as measured by phosphorylated MAPK and JNK following increasing doses of INL insulin administered 30 min after delivery in the olfactory bulb (A-D), hypothalamus (E-H), and hippocampus (I-L). Data are expressed as mean ± SEM, relative to vehicle, n=3–6/group.
In order to ensure insulin signaling was still intact in the young and aged SAMP8 mice, young and aged SAMP8 mice received increasing doses of insulin via ICV injection. We again measured changes in phosphorylation of IR, IRS1, and Akt 10 min following ICV administration (Figure 5). While we were not able to detect changes in phosphorylated IR or IRS1, we did see an insulin dose effect on phosphorylated Akt in the hypothalamus (Fig. 5C). We have previously detected this increase in phosphorylated Akt in the hypothalamus of young, CD-1 mice following ICV insulin [26].
Figure 5.
Canonical insulin receptor signaling following ICV insulin as measured by protein levels of phosphorylated IR, IRS1, and Akt in the hypothalamus (A-C) and hippocampus (D-F). Data are expressed as mean ± SEM, relative to vehicle, n=5–8/group; *p<0.05 vs Vehicle.
Due to the lack of protein expression changes in our targeted approach, we took an unbiased approach to investigate the effects of INL insulin on hippocampal gene regulation to determine what the molecular mechanisms of INL insulin might be. We used a dose of INL insulin previously shown to improve memory in the aged SAMP8 mouse [15].
Gene expression and pathway analysis
To investigate the genetic alterations taking place following INL delivery, we used multiple groups for comparison. We wanted to determine what hippocampal genes changed 1) with aging (Young vs Aged vehicle acute), 2) due to a single, acute INL insulin injection (Aged treated acute vs Aged vehicle acute) and 3) due to repeated, chronic injections of INL insulin (Aged treated chronic vs Aged vehicle chronic) (Table 1). Doses and regimens used were those that improve cognition in the aged SAMP8 mouse [15]. Repeated INL injections did not alter body weight or blood glucose in the current studies (data not shown).
Table 1.
RNA Sequencing Comparison Groups. The number of genes that were changed are listed to the right with the direction (increased or decreased) listed in regards to Group 2.
| Group 1 | Group 2 | Increased | Decreased | Total | |
|---|---|---|---|---|---|
| Young | vs | Aged Vehicle Acute | 835 | 202 | 1037 |
| Aged Vehicle Acute | vs | Aged Treated Acute | 138 | 178 | 316 |
| Aged Vehicle Chronic | vs | Aged Treated Chronic | 156 | 92 | 248 |
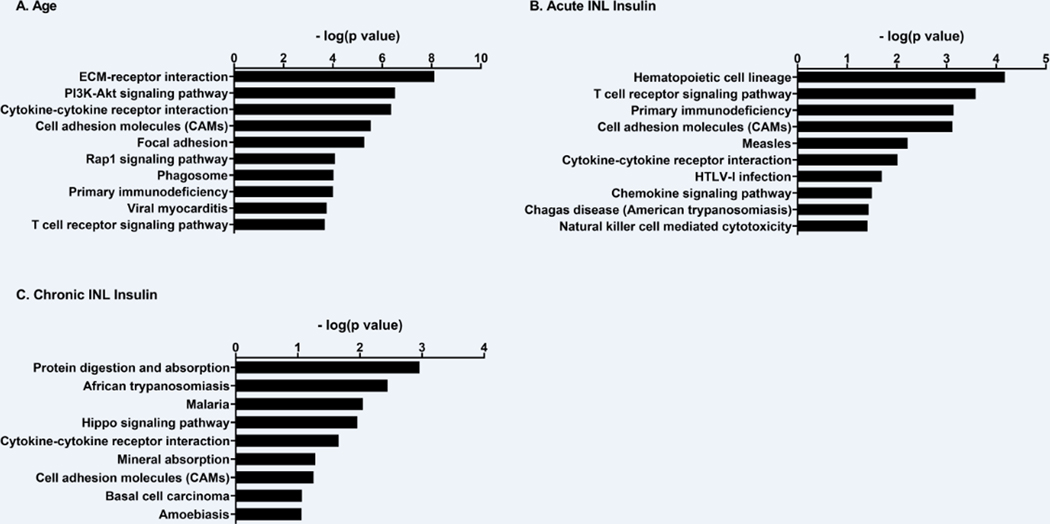
There were many genes that were differentially expressed in the hippocampus between these group comparisons. Supplemental Table 2 reports the log2 fold change, log2 counts per million, and p-value for each gene within each comparison. Age alone in SAMP8 mice resulted in a significant change in 1037 genes in the hippocampus (Table 1). Of these, 835 genes were increased with age while 202 genes were decreased. Acute INL insulin in aged SAMP8 mice resulted in a significant change in the expression of 316 genes: 138 increased with treatment and 178 decreased compared to vehicle. Chronic INL insulin in aged SAMP8 mice altered 248 genes: 156 increased with treatment and 92 decreased. Pathway analysis revealed significant changes in a number of KEGG pathways for each comparison (Supplemental Table 3). There were a total of 52 KEGG Pathways impacted due to age in the SAMP8 mice. The top 10 pathways changed due to age are presented in Figure 6 based on the minus log2(p value) along with the complete list of pathways impacted due to acute and chronic INL insulin treatment. There were a few pathways that were similarly changed within the comparisons (Table 2): cytokine-cytokine receptor interaction, cell adhesion molecule, and T cell receptor signaling pathway. Genes altered within the T cell receptor signaling pathway are listed in Table 3.
Figure 6.
KEGG pathway analysis of genes significantly altered within each comparison. Pathways are reported with the minus log(p value) values reported in response to A) age, a B) single (acute), or C) repeated (chronic) INL INS. Age resulted in a total of 52 pathways significantly altered. Due to space restraints, only the top 10 are listed here. See supplemental table 3 for a full list of pathways.
Table 2.
Common KEGG pathways altered between the comparisons. The p value is the modified Fisher Exact p-value reported from the DAVID platform.
| Common Pathways | Cytokine-Cytokine Receptor Interaction | Cell Adhesion Molecules | T cell receptor signaling pathway | |||
|---|---|---|---|---|---|---|
| Comparison | Genes Altered | P-value | Genes Altered | P-value | Genes Altered | P-value |
| Age | 31 | 4.30E-07 | 23 | 2.90E-06 | 15 | 2.10E-04 |
| Acute | 9 | 9.70E-03 | 9 | 7.60E-04 | 8 | 2.60E-04 |
| Chronic | 7 | 2.20E-0.02 | 5 | 5.60E-02 | “-” | ns |
Table 3.
T cell receptor signaling pathway. Genes altered within each comparison with corresponding logFC, p value, and effect of comparison.
| T Cell Receptor Signaling Pathway | Age | Acute | |||||
|---|---|---|---|---|---|---|---|
| Gene | Gene Symbol | LogFC | P Value | Effect of Age | LogFC | P Value | Effect of INL INS |
| CD3 antigen, delta polypeptide | Cd3d | 6.484 | 0.0261 | Up | |||
| CD3 antigen, gamma polypeptide | Cd3g | 5.449 | 0.0231 | Up | −4.812 | 0.0385 | Down |
| CD3 antigen, epsilon polypeptide | Cd3e | 4.378 | 0.038 | Up | −5.75 | 0.0139 | Down |
| CD247 antigen | Cd247 | 3.281 | 0.003 | Up | −2.825 | 0.0131 | Down |
| CD28 antigen | Cd28 | 3.075 | 0.0007 | Up | −3.377 | 0.0003 | Down |
| linker for activation of T cells | Lat | 1.787 | 0.0106 | Up | |||
| programmed cell death 1 | Pdcd1 | 1.425 | 0.0013 | Up | |||
| CD4 antigen | Cd4 | 1.365 | 0.0348 | Up | −1.73 | 0.0085 | Down |
| protein tyrosine phosphatase, receptor type, C | Ptprc | 1.167 | 0.0015 | Up | |||
| lymphocyte protein tyrosine kinase | Lck | 1.135 | 0.0282 | Up | −1.101 | 0.0346 | Down |
| inducible T cell co-stimulator | Icos | 0.7414 | 0.01309 | Up | |||
| caspase recruitment domain family, member 11 | Card11 | 0.697 | 0.042 | Up | |||
| IL2 inducible T cell kinase | Itk | 0.675 | 0.0368 | Up | −0.958 | 0.0037 | Down |
| mitogen-activated protein kinase 13 | Mapk13 | 0.646 | 0.0237 | Up | |||
| Casitas B-lineage lymphoma | Cbl | 0.609 | 0 | Up | |||
| vav 3 oncogene | Vav3 | 0.77 | 0.0013 | Up | |||
Genes altered with age and reversed with INL insulin
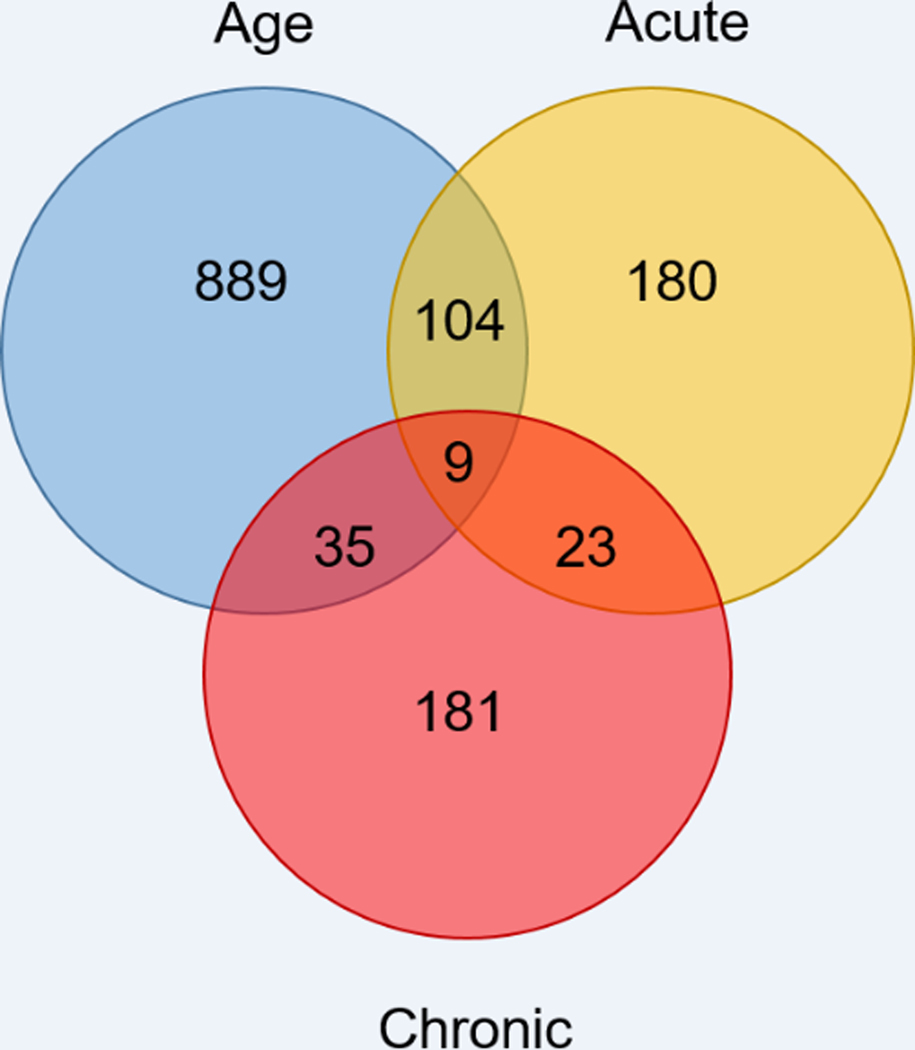
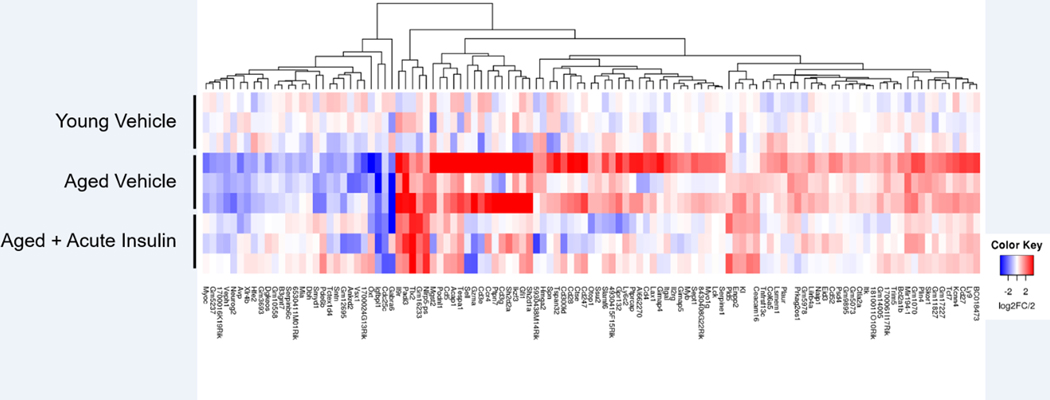
The effect of a single, acute INL insulin administration altered 113 genes that were commonly changed due to age (Young vs Aged vehicle acute) (Figure 7). The heat map shown in Figure 8 depicts the directional change of the genes and shows a reversal of most gene expression log2 fold change values with acute INL insulin treatment compared to the aged vehicle samples.
Figure 7.
Number of genes changed due to age, a single INL insulin injection (Acute), or repeated INL insulin injections (Chronic). The Venn diagram depicts the genes that were changed in each group and the genes commonly altered between comparisons.
Figure 8.
Heat map of 113 genes altered due to age that were also altered by a single INL insulin injection. Colors represent the log2 fold change (log2FC). Red indicates a downregulation while blue indicates an upregulation of gene expression. Each row represents a separate pooled hippocampal sample (n=3/comparison).
DISCUSSION
This study focuses on the molecular impact of INL insulin in an aged model of AD. Using male SAMP8 mice, we found hippocampal genes that were altered due to age in addition to a single and repetitive 1 μg INL insulin administration. We were unable to observe differences in protein expression level of various insulin receptor signaling mediators. However, the genetic data suggests there are likely alternative targets of insulin when administered via the INL route that could help mediate the improvements in memory.
To validate the radioactive insulin transport studies following INL administration performed previously [16], we wanted to measure the level of immunoactive human insulin following INL administration and ICV administration. We selected times based on previous data showing when insulin levels peak following INL delivery [16] and when insulin receptor signaling is activated following ICV delivery [26]. We saw a saturable transport effect with INL delivery as expected. In addition, we were able to detect a dose dependent increase in the amount of insulin when delivered ICV.
Others have reported protein changes in the insulin signaling pathway following chronic high-dose INL insulin in female APP/PS1 mice [27] or the 3xTg-AD mice [28]. However, the doses used in these studies were over 3 fold greater than our highest dose of insulin (10 μg) we delivered. Our dose was chosen based on a functional effect on cognition [15], rather than a dose known to activate the insulin receptor [26]. In our dosing studies following INL and ICV administration, we detected a 10 times greater amount of insulin present in the brain regions following ICV delivery compared to INL. This higher amount of insulin was necessary to increase phosphorylation levels of the insulin receptor signaling protein, Akt, in our study. The studies reported above detected a greater change in the phosphorylation of Akt, compared to the insulin receptor or IRS1, suggesting greater sensitivity in detecting changes in pAkt in response to INL insulin [27, 28]. It is possible the physiological response to lower doses of INL insulin is not strong enough to detect differences in phosphorylation by the technique used here. Future studies could be designed fully probe changes in phosphorylation of insulin signaling proteins, such as utilizing phosphoproteomics, which has previously elucidated abberant phosphorylation of proteins in AD brain tissue [29]. In this study, cytoskeletal proteins and proteins involved in the synapse were most altered in mouse AD brain tissue. The insulin receptor is present on the pre- and post-synaptic membranes of neurons [30] and therefore, localized studies investigating the phosphorylation of insulin receptor signaling proteins in this specific cellular location could reveal significant changes in the insulin receptor signaling pathway following INL insulin. Neurons expressing a mutant insulin receptor that cannot be activated have reduced synapse densities, further highlighting the role of the insulin receptor in this cellular location [31]. The direct role of the insulin receptor in memory and cognition has not been fully characterized as recently reviewed [32]. CNS-wide insulin receptor knock-out mice display intact cognition [33]. However, this is a life-long deficit in the insulin receptor so compensation could occur. In a hippocampal specific knock-down model, long-term memory is impaired [34]. The impact of INL insulin in CNS insulin receptor knock-out models remains to be determined. Further work as to the temporal and regional role of the insulin receptor in memory needs to be done. The goal of the current study was to investigate the molecular impact of a dose of insulin that elicits cognitive changes rather than a dose known to activate of the insulin receptor.
In order to investigate gene changes within the hippocampus due to age and due to INL insulin, we utilized an RNA-sequencing approach. We were able to detect multiple genes that changed due to age and due to INL insulin treatment. In an unbiased manner, we investigated these gene expression changes and the specific pathways impacted. With this approach, we determined unique pathways that INL insulin could act through in order to improve memory. A gene array profile has not been performed on the hippocampus in mice following INL insulin administration. However, other groups have investigated gene changes in the brain in aged SAMP8 mice compared to young SAMP8 mice using different methods [35–37]. Genes involved in long term potentiation, phosphatidylinositol signaling, and endocytosis pathways were significantly different in 12 month old mice compared to 4 month old mice [35, 37]. More specifically, in the frontal cortex, somatostatin is significantly decreased with age [36]. These results are similar to our findings and help validate our results in the young and aged SAMP8 mice in which we see a decrease in somatostatin with age and changes in MAPK signaling, PI3K/Akt signaling, and genes involved in long term potentiation.
To better investigate the impact of INL insulin, we focused on pathways similarly altered between the comparisons: effect of age and effect of INL insulin. Of importance, the T cell receptor signaling pathway was impacted not only due to age but also due to a single, acute INL injection of insulin. There is increasing interest in the role of inflammation in AD. Activation of the immune system is now thought to accompany AD pathology and contribute to the pathogenesis of this disease [38, 39]. Numerous anti-inflammatory agents have been successfully used to improve cognition in AD mouse models and improve AD pathology as recently reviewed [40]. T cells are able to enter the CNS by crossing the blood-CSF barrier, in addition to the blood-brain barrier (BBB) [41]. Indeed, there is increased infiltration of CD3+ cells into the cerebral cortex of the ArcAβ mouse model of AD [42]. In addition, T cells have been found in post-mortem brain tissues of patients with AD [43]. T cell infiltration can promote tau-triggered spatial memory deficits without a direct impact on tau protein deposition and phosphorylation [44]. In particular, it was found the chemoattractant, chemokine ligand 3 (CCL3), was increased in the hippocampus of mice overexpressing tau prior to T cell diapedesis. Our sequencing data detected an increase in the amount of CCL3 in the aged SAMP8 mice which could attract T cells to the hippocampus. T cells can directly affect neuronal function and plasticity by inhibiting neurite outgrowth [45]. Cytokines released from activated T cells could also contribute to interrupting neuronal connections. Activation of MAPKs can lead to secretion of cytokines and chemokines, contributing to the inflammation observed in AD [46]. CD3 cells accumulate in the brain with age [47]. Indeed, many of the CD3 antigens were increased with age in the SAMP8 mice in our study. Importantly, we found that acute INL insulin decreased these levels in the hippocampus of aged SAMP8 mice.
Previous studies have shown insulin can exert a vasodilatory effect of blood vessels and vasodilation reduces leukocyte adhesion to the endothelium and subsequent infiltration [48]. This neurovascular coupling role of insulin could be affecting the transport of various serum factors into the brain which can aid in the enhancement of memory due to INL insulin. Insulin can reduce activation of NFkB, downregulating soluble intercellular adhesion molecule 1 (ICAM1), which facilitates attachment of monocytes to endothelial cells [48]. The C-C chemokine receptor 4 (Ccr4) is increased with age and decreased with acute insulin in our sequencing data set. This protein is a receptor for monocyte chemoattractant protein 1 (MCP-1), which helps peripheral blood mononuclear cells transport into tissue. The cellular networks involved in the cognitively beneficial effect of INL insulin warrants further investigation.
Of note, there were no changes detected in the insulin signaling pathway with INL insulin, suggesting gene transcription of this pathway is not affected by INL insulin. The KEGG insulin signaling pathway involves 140 genes including the insulin receptor, IRS, and SHC, in addition to downstream mediators including Akt, MAPK, mTOR, and JNK [49]. Therefore, if insulin is affecting this pathway following INL insulin delivery, it would be in a post-translational manner (such as phosphorylation) or cellular localization that would require more sensitive techniques to observe changes in specific cell populations as discussed above.
The data presented here provides us with a better understanding of how INL insulin is acting within the hippocampus in aged SAMP8 mice and provides potential targets for AD. In addition, it suggests there is a low dose insulin-binding site with strong affinity in the CNS that could be responsible for eliciting a behavioral response without the activation of the insulin receptor signaling pathway. INL insulin can alter expression of genes involved in the cytokine-cytokine receptor interaction, cell adhesion molecules, and T cell receptor signaling pathways. Further investigations into the molecular impact of each of these pathways needs to be done to functionally characterize the impact of INL insulin.
Supplementary Material
Supplemental Table I. Antibodies used for western immunoblotting are listed with the catalog number. All antibodies were purchased from Cell Signaling Technology.
Supplemental Table III. (see file from Managing Editor) Significant KEGG pathways affected within each comparison are listed on separate tabs: Age, Acute INL insulin, Chronic INL insulin. KEGG terms are listed in column B. The number of genes involved in the term are listed in column C. The percentage of involved genes in the term compared to the total genes within each term are listed in column D. The modified Fisher Exact p-values are listed in column E. The –log(p-value) is listed in column F. This value was used to generate the graph in Figure 6.
Supplemental Table II. (see file from Managing Editor) Results from the RNA-sequencing analysis are listed. Gene names, symbols, and ENTREZ IDs are listed with the log2 fold change, log2 counts per million, and p-values for each comparison: young vs aged vehicle acute, aged treated acute vs aged vehicle acute, and aged treated chronic vs aged vehicle chronic.
ACKNOWLEDGEMENTS
Support for this project was provided by the NIH (RO1AG046619), T32AG000057 (EMR), VA Puget Sound Health Care System, and the University of Washington Nathan Shock Center. We would like to thank Ms. Sengkeo Srinouanprachan for isolation and QC of the RNA samples, and Dr. Nickerson’s lab at the University of Washington for processing samples for RNA-Sequence analysis and generating the RNA-Sequence data.
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
REFERENCES
- [1].Frolich L, Blum-Degen D, Bernstein HG, Engelsberger S, Humrich J, Laufer S, Muschner D, Thalheimer A, Turk A, Hoyer S, Zochling R, Boissl KW, Jellinger K, Riederer P (1998) Brain insulin and insulin receptors in aging and sporadic Alzheimer’s disease. J Neural Transm (Vienna) 105, 423–438. [DOI] [PubMed] [Google Scholar]
- [2].Craft S, Peskind E, Schwartz MW, Schellenberg GD, Raskind M, Porte D Jr. (1998) Cerebrospinal fluid and plasma insulin levels in Alzheimer’s disease: relationship to severity of dementia and apolipoprotein E genotype. Neurology 50, 164–168. [DOI] [PubMed] [Google Scholar]
- [3].Craft S, Newcomer J, Kanne S, Dagogo-Jack S, Cryer P, Sheline Y, Luby J, Dagogo-Jack A, Alderson A (1996) Memory improvement following induced hyperinsulinemia in Alzheimer’s disease. Neurobiol Aging 17, 123–130. [DOI] [PubMed] [Google Scholar]
- [4].Benedict L, Nelson CA, Schunk E, Sullwold K, Seaquist ER (2006) Effect of insulin on the brain activity obtained during visual and memory tasks in healthy human subjects. Neuroendocrinology 83, 20–26. [DOI] [PubMed] [Google Scholar]
- [5].Rhea EM, Salameh TS, Banks WA (2019) Routes for the delivery of insulin to the central nervous system: A comparative review. Exp Neurol 313, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Meredith ME, Salameh TS, Banks WA (2015) Intranasal Delivery of Proteins and Peptides in the Treatment of Neurodegenerative Diseases. Aaps j 17, 780–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lochhead JJ, Kellohen KL, Ronaldson PT, Davis TP (2019) Distribution of insulin in trigeminal nerve and brain after intranasal administration. Sci Rep 9, 2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Reger MA, Watson GS, Green PS, Baker LD, Cholerton B, Fishel MA, Plymate SR, Cherrier MM, Schellenberg GD, Frey WH, 2nd, Craft S (2008) Intranasal insulin administration dose-dependently modulates verbal memory and plasma amyloid-beta in memory-impaired older adults. J Alzheimers Dis 13, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reger MA, Watson GS, Green PS, Wilkinson CW, Baker LD, Cholerton B, Fishel MA, Plymate SR, Breitner JC, DeGroodt W, Mehta P, Craft S (2008) Intranasal insulin improves cognition and modulates beta-amyloid in early AD. Neurology 70, 440–448. [DOI] [PubMed] [Google Scholar]
- [10].Reger MA, Watson GS, Frey WH 2nd, Baker LD, Cholerton B, Keeling ML, Belongia DA, Fishel MA, Plymate SR, Schellenberg GD, Cherrier MM, Craft S (2006) Effects of intranasal insulin on cognition in memory-impaired older adults: modulation by APOE genotype. Neurobiol Aging 27, 451–458. [DOI] [PubMed] [Google Scholar]
- [11].Benedict C, Hallschmid M, Hatke A, Schultes B, Fehm HL, Born J, Kern W (2004) Intranasal insulin improves memory in humans. Psychoneuroendocrinology 29, 1326–1334. [DOI] [PubMed] [Google Scholar]
- [12].Claxton A, Baker LD, Hanson A, Trittschuh EH, Cholerton B, Morgan A, Callaghan M, Arbuckle M, Behl C, Craft S (2015) Long Acting Intranasal Insulin Detemir Improves Cognition for Adults with Mild Cognitive Impairment or Early-Stage Alzheimer’s Disease Dementia. J Alzheimers Dis 45, 1269–1270. [DOI] [PubMed] [Google Scholar]
- [13].Craft S, Claxton A, Baker LD, Hanson AJ, Cholerton B, Trittschuh EH, Dahl D, Caulder E, Neth B, Montine TJ, Jung Y, Maldjian J, Whitlow C, Friedman S (2017) Effects of Regular and Long-Acting Insulin on Cognition and Alzheimer’s Disease Biomarkers: A Pilot Clinical Trial. J Alzheimers Dis 57, 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feld GB, Wilhem I, Benedict C, Rudel B, Klameth C, Born J, Hallschmid M (2016) Central Nervous Insulin Signaling in Sleep-Associated Memory Formation and Neuroendocrine Regulation. Neuropsychopharmacology 41, 1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Salameh TS, Bullock KM, Hujoel IA, Niehoff ML, Wolden-Hanson T, Kim J, Morley JE, Farr SA, Banks WA (2015) Central Nervous System Delivery of Intranasal Insulin: Mechanisms of Uptake and Effects on Cognition. J Alzheimers Dis 47, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rhea EM, Humann SR, Nirkhe S, Farr SA, Morley JE, Banks WA (2017) Intranasal Insulin Transport is Preserved in Aged SAMP8 Mice and is Altered by Albumin and Insulin Receptor Inhibition. J Alzheimers Dis 57, 241–252. [DOI] [PubMed] [Google Scholar]
- [17].Morley JE, Farr SA, Kumar VB, Armbrecht HJ (2012) The SAMP8 mouse: a model to develop therapeutic interventions for Alzheimer’s disease. Curr Pharm Des 18, 1123–1130. [DOI] [PubMed] [Google Scholar]
- [18].Rhea EM, Banks WA (2017) The SAMP8 mouse for investigating memory and the role of insulin in the brain. Exp Gerontol 94, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hagiwara M, Kobayashi K, Tadokoro T, Yamamoto Y (2010) Application of SYPRO Ruby- and Flamingo-stained polyacrylamide gels to Western blot analysis. Anal Biochem 397, 262–264. [DOI] [PubMed] [Google Scholar]
- [20].Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- [25].Fernando SA, Wilson GS (1992) Studies of the ‘hook’ effect in the one-step sandwich immunoassay. J Immunol Methods 151, 47–66. [DOI] [PubMed] [Google Scholar]
- [26].Banks WA, Farr SA, Salameh TS, Niehoff ML, Rhea EM, Morley JE, Hanson AJ, Hansen KM, Craft S (2018) Triglycerides cross the blood-brain barrier and induce central leptin and insulin receptor resistance. Int J Obes (Lond) 42, 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mao YF, Guo Z, Zheng T, Jiang Y, Yan Y, Yin X, Chen Y, Zhang B (2016) Intranasal insulin alleviates cognitive deficits and amyloid pathology in young adult APPswe/PS1dE9 mice. Aging Cell 15, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen Y, Zhao Y, Dai CL, Liang Z, Run X, Iqbal K, Liu F, Gong CX (2014) Intranasal insulin restores insulin signaling, increases synaptic proteins, and reduces Abeta level and microglia activation in the brains of 3xTg-AD mice. Exp Neurol 261, 610–619. [DOI] [PubMed] [Google Scholar]
- [29].Butterfield DA (2019) Phosphoproteomics of Alzheimer disease brain: Insights into altered brain protein regulation of critical neuronal functions and their contributions to subsequent cognitive loss. Biochim Biophys Acta Mol Basis Dis 1865, 2031–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gralle M (2017) The neuronal insulin receptor in its environment. J Neurochem 140, 359–367. [DOI] [PubMed] [Google Scholar]
- [31].Chiu SL, Chen CM, Cline HT (2008) Insulin receptor signaling regulates synapse number, dendritic plasticity, and circuit function in vivo. Neuron 58, 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rhea EM, Banks WA (2019) Role of the Blood-Brain Barrier in Central Nervous System Insulin Resistance. Frontiers in Neuroscience 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schubert M, Gautam D, Surjo D, Ueki K, Baudler S, Schubert D, Kondo T, Alber J, Galldiks N, Kustermann E, Arndt S, Jacobs AH, Krone W, Kahn CR, Bruning JC (2004) Role for neuronal insulin resistance in neurodegenerative diseases. Proc Natl Acad Sci U S A 101, 3100–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Grillo CA, Piroli GG, Lawrence RC, Wrighten SA, Green AJ, Wilson SP, Sakai RR, Kelly SJ, Wilson MA, Mott DD, Reagan LP (2015) Hippocampal Insulin Resistance Impairs Spatial Learning and Synaptic Plasticity. Diabetes 64, 3927–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Armbrecht HJ, Siddiqui AM, Green M, Farr SA, Kumar VB, Banks WA, Patrick P, Shah GN, Morley JE (2014) SAMP8 mice have altered hippocampal gene expression in long term potentiation, phosphatidylinositol signaling, and endocytosis pathways. Neurobiol Aging 35, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen SC, Lu G, Chan CY, Chen Y, Wang H, Yew DT, Feng ZT, Kung HF (2010) Microarray profile of brain aging-related genes in the frontal cortex of SAMP8. J Mol Neurosci 41, 12–16. [DOI] [PubMed] [Google Scholar]
- [37].Currais A, Goldberg J, Farrokhi C, Chang M, Prior M, Dargusch R, Daugherty D, Armando A, Quehenberger O, Maher P, Schubert D (2015) A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging (Albany NY) 7, 937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Heppner FL, Ransohoff RM, Becher B (2015) Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16, 358–372. [DOI] [PubMed] [Google Scholar]
- [40].Martinez B, Peplow P (2019) Amelioration of Alzheimer’s disease pathology and cognitive deficits by immunomodulatory agents in animal models of Alzheimer’s disease. Neural Regeneration Research 14, 1158–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Engelhardt B (2008) Immune cell entry into the central nervous system: involvement of adhesion molecules and chemokines. J Neurol Sci 274, 23–26. [DOI] [PubMed] [Google Scholar]
- [42].Ferretti MT, Merlini M, Spani C, Gericke C, Schweizer N, Enzmann G, Engelhardt B, Kulic L, Suter T, Nitsch RM (2016) T-cell brain infiltration and immature antigen-presenting cells in transgenic models of Alzheimer’s disease-like cerebral amyloidosis. Brain Behav Immun 54, 211–225. [DOI] [PubMed] [Google Scholar]
- [43].Togo T, Akiyama H, Iseki E, Kondo H, Ikeda K, Kato M, Oda T, Tsuchiya K, Kosaka K (2002) Occurrence of T cells in the brain of Alzheimer’s disease and other neurological diseases. J Neuroimmunol 124, 83–92. [DOI] [PubMed] [Google Scholar]
- [44].Laurent C, Dorothee G, Hunot S, Martin E, Monnet Y, Duchamp M, Dong Y, Legeron FP, Leboucher A, Burnouf S, Faivre E, Carvalho K, Caillierez R, Zommer N, Demeyer D, Jouy N, Sazdovitch V, Schraen-Maschke S, Delarasse C, Buee L, Blum D (2017) Hippocampal T cell infiltration promotes neuroinflammation and cognitive decline in a mouse model of tauopathy. Brain 140, 184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liblau RS, Gonzalez-Dunia D, Wiendl H, Zipp F (2013) Neurons as targets for T cells in the nervous system. Trends Neurosci 36, 315–324. [DOI] [PubMed] [Google Scholar]
- [46].Ho GJ, Drego R, Hakimian E, Masliah E (2005) Mechanisms of cell signaling and inflammation in Alzheimer’s disease. Curr Drug Targets Inflamm Allergy 4, 247–256. [DOI] [PubMed] [Google Scholar]
- [47].Stichel CC, Luebbert H (2007) Inflammatory processes in the aging mouse brain: participation of dendritic cells and T-cells. Neurobiol Aging 28, 1507–1521. [DOI] [PubMed] [Google Scholar]
- [48].Sun Q, Li J, Gao F (2014) New insights into insulin: The anti-inflammatory effect and its clinical relevance. World J Diabetes 5, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table I. Antibodies used for western immunoblotting are listed with the catalog number. All antibodies were purchased from Cell Signaling Technology.
Supplemental Table III. (see file from Managing Editor) Significant KEGG pathways affected within each comparison are listed on separate tabs: Age, Acute INL insulin, Chronic INL insulin. KEGG terms are listed in column B. The number of genes involved in the term are listed in column C. The percentage of involved genes in the term compared to the total genes within each term are listed in column D. The modified Fisher Exact p-values are listed in column E. The –log(p-value) is listed in column F. This value was used to generate the graph in Figure 6.
Supplemental Table II. (see file from Managing Editor) Results from the RNA-sequencing analysis are listed. Gene names, symbols, and ENTREZ IDs are listed with the log2 fold change, log2 counts per million, and p-values for each comparison: young vs aged vehicle acute, aged treated acute vs aged vehicle acute, and aged treated chronic vs aged vehicle chronic.