ABSTRACT
Probiotics are consumed in fermented dairy products or as capsules for their putative health benefits. However, little research has been done to evaluate the effects of the delivery matrix on the health benefits of probiotics in humans. To examine the effects of delivering Bifidobacterium animalis subsp. lactis BB-12 (BB-12) (log10 10 ± 0.5 CFU/day) via a yogurt smoothie versus a capsule, we monitored the fecal microbiota, gut transit times (GTTs), and fecal excretion of short-chain fatty acids (SCFAs) in healthy adults. In a randomized, four-period, crossover study performed in a partially blind manner, 36 adults were recruited and randomly assigned to four treatments: control yogurt smoothie (YS), yogurt smoothie with BB-12 added prefermentation (PRE), yogurt smoothie with BB-12 added postfermentation (POST), and capsule containing BB-12 (CAP). Participants’ fecal microbiota was assessed using 16S rRNA sequencing, GTTs via SmartPill, and fecal SCFAs by gas chromatography (GC) before (baseline) and after each intervention. Participants had significantly higher percentage of Streptococcus after consuming YS versus CAP (P = 0.01). Bifidobacterium-specific terminal restriction fragment length polymorphism analysis revealed a significantly higher percentage of B. animalis after consuming PRE and POST compared to baseline, YS, CAP, and final washout (P < 0.0001). The predominant SCFAs were negatively correlated with GTTs. Consumption of BB-12 delivered in a yogurt smoothie or capsule did not significantly alter the composition of the gut microbiota, GTTs, or fecal SCFA concentration of the study cohort. However, daily consumption of BB-12 in yogurt smoothie may result in higher relative abundance of B. animalis in healthy adults. (This trial has been registered at ClinicalTrials.gov under identifier NCT01399996.)
IMPORTANCEBifidobacterium animalis subsp. lactis BB-12 is a probiotic strain that has been used worldwide since 1985. It has commonly been delivered in fermented dairy products for perceived benefits associated with gut health and enhanced immune function. In addition to fermented dairy products, many new probiotic-containing alternatives such as probiotic-containing juice, probiotic-containing chocolate, and capsules have been developed. While these products provide more options for people to access probiotics, little research has been done on the effect of delivery matrix (dairy versus nondairy) on their efficacy in humans. In addition, it was unclear how yogurt fermentation may influence the survival of BB-12 in the product or on its performance in vivo. The significance of our study is in simultaneously assessing the effect of BB-12, alone and in different delivery vehicles, on the gut transit time, fecal short-chain fatty acids, and the composition of the gut microbiota of the study cohort.
KEYWORDS: probiotics, BB-12, gut microbiota, short-chain fatty acids, gut transit time
INTRODUCTION
The human gastrointestinal tract (GIT) harbors a diverse and dynamic community of microorganisms collectively termed the gut microbiota that contributes to the homeostasis of the gut and the biology of the host (1). The gut microbiota is estimated to contain approximately 40 trillion microbes, including hundreds of species of facultative and obligate anaerobes (2). The balance and composition of the gut microbiota can be altered by several factors such as medical interventions, age, genetics, environment, diet, and human health (3). Disturbed gut microbiota, also referred to as “dysbiosis” (4), has been linked to diseases such as obesity (5–8), inflammatory bowel disease (IBD) (9), and antibiotic-associated diarrhea (AAD) (10). Probiotics may help restore the microbiota of a disrupted GIT. Studies have shown that probiotic interventions significantly reduced the incidence of AAD in infants and children (11, 12).
Whole gut transit time (WGTT) refers to the time it takes for food to move from the mouth to the anus. Gut transit time (GTT) varies markedly among individuals, as well as within individuals, and maintaining a regular WGTT is essential for health and general well-being (13). Fecal short-chain fatty acid (SCFA) production has been associated with changes in gut microbiota and WGTT as a result of consuming probiotics, prebiotics, or symbiotics, but the findings are inconsistent (14). To better understand their relationship, it is important to study fecal SCFAs together with WGTT and the gut microbiota.
Probiotics, defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (15, 16), can be ingested as supplements in powder, capsule, or liquid form. Such supplements have shown potential benefits in the treatment and prevention of numerous diseases, including diarrhea, asthma, necrotizing enterocolitis, and allergies (17–21). As an alternative to delivery in supplements, probiotic organisms are often included as ingredients in fermented dairy products to produce functional foods, that is, foods providing health benefits beyond their nutritional value (22). Yogurt, for example, is a fermented milk product often considered a functional food. According to a survey conducted by Monroe Mendelsohn Research in 2001, two-thirds of primary care physicians who counsel patients about nutrition recommend consuming yogurt containing live and active cultures for health benefits (23). However, it is not clear whether probiotics delivered via dairy- and non-dairy-based matrices are equally effective, or whether one matrix is better in terms of benefiting the host’s gut microbiota, gut transit time, and fecal SCFA production. It is also uncertain how the yogurt fermentation process may affect the performance of probiotic organisms in vivo.
Probiotics marketed as nutritional supplements and found in functional foods are predominantly members of the genera Bifidobacterium, Lactobacillus, Lactocaseibacillus, Ligilactobacillus, Lactiplantibacillus, and Limosilactobacillus. Bifidobacterium species can be found in the gastrointestinal tract (GIT) as both autochthonous and allochthonous residents (24). Bifidobacterium animalis subsp. lactis (BAL), BB-12 (BB-12 is a trademark of Chr. Hansen A/S), the primary focus of this study, has been shown to improve bowel function, to support a healthy gut microbiota, and to improve immune function (25). However, the nature of the delivery vehicle (food matrix, tablets, or capsules)—including parameters such as water activity, pH, level and types of macronutrients (fat, protein, and carbohydrates), presence of organic acids, oxygen level, and the presence of other functional ingredients—may be important in determining how BB-12 will behave in a product and when ingested by the consumer. In a review paper, Sanders and Marco (26) pointed out that, “little is known about the food matrix and product formulation impacts on probiotic functionality even though such information is essential to scientific understanding and regulatory substantiation of health benefits.”
The objective of the present study was to evaluate the efficacy of delivering strain BB-12 via dairy (yogurt smoothie) or nondairy (capsule) vehicles in humans. Moreover, we determined the effects of these delivery vehicles in terms of modifying the gut microbiota, modulating gut transit time, and increasing SCFA production. The primary outcome of the project was WGTT, and the focus of the present study was to survey the gut microbiota of participants before and after different BB-12 interventions.
RESULTS AND DISCUSSION
To test for the efficacy of delivery of B. animalis subsp. lactis BB-12, a randomized, four-period, crossover study performed in a partially blind manner was conducted. Thirty-six healthy adults were recruited and randomly assigned to four treatments: (i) control yogurt smoothie (YS), (ii) yogurt smoothie with strain BB-12 added prefermentation (PRE), (iii) yogurt smoothie with BB-12 added postfermentation (POST), and (iv) capsule containing BB-12 (CAP). Yogurt smoothies were manufactured using standard fermentation practices, and the concentration of BB-12 was measured through the shelf life of the products.
Interventions.
Viable counts of strain BB-12 in all yogurt smoothies were evaluated weekly following production over the 30 days. Results from 27 batches revealed a significant difference in the population of BB-12 between the PRE and POST fermentation treatments immediately following manufacture (week 0) (initial counts of log10 10.55 ± 0.12 CFU/serving and log10 10.50 ± 0.14 CFU/serving, respectively). However, we believe this difference is not of significance in terms of product performance since the amount of BB-12 was above log 10 CFU/serving in both treatments. As expected, the population of BB-12 declined throughout the shelf life of the products with a faster decrease in the population for the POST treatment than in the PRE treatment. This trend continued, and by the end of the shelf life, the BB-12 concentration decreased significantly in both PRE (log10 10.24 ± 0.13 CFU/serving) and POST (log10 9.54 ± 0.25 CFU/serving) after 4 weeks’ storage (Table 1). BB-12 survived significantly better in PRE than in POST (P < 0.001), indicating that BB-12 survives better when added before fermentation, possibly as a result of adaptation to the acidic environment (27). Overall, all BB-12 interventions remained at the targeted effective dose level (log10 10 ± 0.5 CFU/serving) during shelf life.
TABLE 1.
BB-12 concentration in yogurt smoothies during shelf life
| Treatmenta | BB-12 concentration (log10 CFU/serving)b at the following time: |
||||
|---|---|---|---|---|---|
| Week 0 | Week 1 | Week 2 | Week 3 | Week 4 | |
| PRE | 10.55 ± 0.12 Aa | 10.43 ± 0.13 Ba | 10.42 ± 0.13 Ba | 10.34 ± 0.13 Ca | 10.24 ± 0.13 Da |
| POST | 10.50 ± 0.14 Ab | 10.16 ± 0.15 Bb | 10.03 ± 0.20 Cb | 9.77 ± 0.20 Db | 9.54 ± 0.25 Eb |
PRE, yogurt smoothie with BB-12 added before fermentation; POST, yogurt smoothie with BB-12 added after fermentation.
Data are presented as means ± standard deviations (SD) from 27 batches. Values in a column without a common lowercase letter are significantly different (P < 0.05). Values in a row without a common uppercase letter are significantly different (P < 0.05).
Participant characteristics.
Baseline characteristics of the participants are shown in Table 2. Twenty-nine participants (18 females and 11 males) were included in the analyses, as they had completed at least one of the four intervention periods. Overall, participants were healthy young adults with a mean age of 28.1 ± 0.6 years. The average body mass index (BMI) was 24.1 ± 0.2 kg/m2: 17 (58.6%) participants were normal weight, 11 (37.9%) were overweight, and 1 (3.5%) was obese. Their blood pressure, waist circumference, fasting blood glucose, insulin, and C-reactive protein (CRP) levels were within the normal range (Table 2). Physical activity, as assessed from self-reported International Physical Activity Questionnaire (IPAQ) responses, indicated a median daily physical activity of 3.0 metabolic equivalents (METs) (range, 2.3 to 4.3 METs). The average daily total calorie intake of participants calculated from 3-day dietary recalls was estimated to be 2,241 ± 83 kcal. The daily intake of macronutrients, vitamins, minerals, and n-3 polyunsaturated fatty acids (PUFA), caffeine, and alcohol is also reported in Table 2.
TABLE 2.
Demographic characteristics of participants before treatment (baseline)a
| Characteristic | Valueb (n = 29) |
|---|---|
| Age (yr) | 28.1 ± 0.6 |
| Male, n (%) | 11 (37.9%) |
| Body mass index (kg/m2) | 24.1 ± 0.2 |
| ≤24.9 | 17 (58.6%) |
| 25.0−29.9 | 11 (37.9%) |
| ≥30 | 1 (3.5%) |
| Waist circumference (cm) | 85.1 ± 0.6 |
| Blood pressure (mm Hg) | |
| Systolic | 107.6 ± 0.8 |
| Diastolic | 72.6 ± 0.6 |
| Glucose (mg/dl) | 86.6 ± 0.8 |
| Insulin (mg/dl) | 5.3 ± 0.4 |
| hs-CRP (mg/liter) | 2.0 ± 0.5 |
| Physical activity (METs)c | 3.0 (2.3–4.3) |
| Dietary intakec | |
| Total calories (kcal/day) | 2,241 ± 83 |
| Carbohydrate (g/day) | 284.6 ± 10.9 |
| Protein (g/day) | 90.2 ± 3.7 |
| Fat (g/day) | 83.8 ± 3.4 |
| Vitamin C (mg/day) | 67.8 ± 4.8 |
| Vitamin D (IU/day) | 98.5 ± 11.2 |
| Vitamin E (mg/day) | 3.4 ± 0.3 |
| Iron (mg/day) | 14.4 ± 0.8 |
| Selenium (μg/day) | 46.0 ± 4.5 |
| Zinc (mg/day) | 5.7 ± 0.4 |
| n-3 PUFA (g/day) | 0.6 ± 0.1 |
| Caffeine (mg/day) | 75.3 ± 10.2 |
| Alcohol consumption (g/day) | 2.6 ± 0.9 |
Shared results with collaborators in the project.
Values are presented as means ± standard errors of the means (SEM) or n (%) or median (range).
Physical activity and dietary intake were assessed from self-reported responses to IPAQ and 3-day dietary recall records, respectively.
Compliance.
All stool DNA samples were tested for compliance using 16S rDNA-based subspecies-specific PCR. A total of 73 out of 78 samples were B. animalis subsp. lactis (BAL) positive after BB-12-containing interventions, while all the fecal samples were BAL negative after receiving the control intervention when the corresponding baselines were negative. Four participants were BAL positive before treatment (baseline) and throughout the study regardless of treatments, suggesting that BAL was autochthonous to these individuals.
Primary outcome.
Only 27 participants were included in primary outcome analysis because a few data points could not be retrieved from the data receiver due to technical difficulties. Results of the analysis indicated no treatment effect on gut transit times (data not shown). In the present study, participants had a wide range of WGTT (7.13 h to 128.58 h), colonic transit time (CTT) (0.5 h to 122.25 h), small bowel transit time (SBTT) (1 h to 19.02 h), and gastric emptying time (GET) (0.98 h to 18.83 h). Subjects with extremely short CTT (0.5 h) might have had diarrhea. The variability of responses among individuals made it difficult to detect a treatment effect, if there was any. In agreement with a previous study (28), males in the present cohort had shorter CTT (P = 0.0098), WGTT (P = 0.0036), and GET (P < 0.0001) than females but exhibited no difference in SBTT (P = 0.3). There was a significant correlation between the blue dye and SmartPill measurements (Spearman rho = 0.67, P < 0.0001), which suggests that the blue dye method remains a useful screening tool for GTT in healthy individuals. However, because the SmartPill is a more objective measure of GTT, the results for WGTT are likely better estimates of actual transit time.
In previous work, the effect of a BB-12 intervention (delivered in fermented milk, capsule, or fermented cereal) on host bowel movement was studied in both healthy subjects and subjects with functional bowel symptoms, and promising improvements were observed (29–31). A large clinical trial with 1,248 subjects performed in eight centers in Europe reported a treatment effect of strain BB-12 (delivered in capsule) on average defecation frequency (P = 0.0065) (32), despite the fact that the placebo group also had increased average defecation frequency compared to baseline. Several factors may explain the lack of treatment effect on GTT, as well as the discrepancy between our results and previous studies. First, the present study had a relatively small sample size, which makes it difficult to detect a small treatment effect within a highly variable data set. Second, the cohort studied is a generally healthy group of individuals, with whom there is only limited room for improvement in terms of bowel transit time. Finally, this study took direct measurements of the GTT using a wireless motility capsule in contrast to other studies that employed more subjective defecation frequency questionnaires.
Secondary outcomes. (i) Characteristics of the fecal microbiota of the participants.
Illumina sequencing of the 161 fecal samples generated over 2.6 million total reads. After removing samples with low quality and poor compliance, about 2.4 million sequences from 147 samples were used for data analyses. Overall, 10 phyla and 109 genera were identified in the participants. Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria accounted for >98% of the sequences at the phylum level. The predominant phyla and genera identified (represents > 0.1% of the sequences) among treatment groups are shown in Table 3. No difference at the phylum or genus level was detected among treatment groups, with the exception of a significantly higher percentage of Streptococcus in their fecal microbiota after the participants consumed control yogurt smoothie compared to consuming capsule (P = 0.01). All yogurt smoothies tended to have a higher percentage of Streptococcus compared to baseline, capsule, and final washout. This is likely due to the presence of the high level (log10 11.4 CFU/day) of S. thermophilus in the yogurt interventions.
TABLE 3.
Predominant fecal bacterial phyla and genera present in healthy adults before and after consuming BB-12-containing interventions in a crossover study
| Phylum and genus | % of sequencesa |
|||||||
|---|---|---|---|---|---|---|---|---|
| Baseline | YS | POST | PRE | CAP | Final washout | SEM | P value | |
| Firmicutes | 82 | 82 | 80 | 79 | 82 | 79 | 0.76 | 0.58 |
| Blautia | 8.4 | 8.3 | 7.9 | 6.8 | 8.2 | 7.3 | 0.35 | 0.77 |
| Faecalibacterium | 5.2 | 6.3 | 5.4 | 6.4 | 6.2 | 6.9 | 0.34 | 0.69 |
| Ruminococcus | 4.9 | 4.1 | 5.4 | 5.4 | 5.7 | 5.3 | 0.33 | 0.78 |
| Coprococcus | 2.7 | 3.3 | 3.3 | 2.8 | 3.1 | 3.1 | 0.13 | 0.74 |
| Roseburia | 1.7 | 1.8 | 1.8 | 1.4 | 2.0 | 2.6 | 0.19 | 0.62 |
| Lachnospira | 1.2 | 1.8 | 2.1 | 1.5 | 1.6 | 1.7 | 0.13 | 0.21 |
| Dialister | 1.0 | 1.1 | 1.1 | 1.3 | 1.4 | 0.8 | 0.12 | 0.77 |
| Dorea | 1.0 | 1.1 | 0.89 | 0.67 | 1.0 | 0.9 | 0.05 | 0.20 |
| Streptococcus | 0.59 AB | 1.04 A | 0.89 AB | 0.73 AB | 0.45 B | 0.38 B | 0.06 | 0.01 |
| Clostridium | 0.56 | 0.77 | 0.55 | 0.57 | 0.70 | 0.81 | 0.06 | 0.76 |
| Oscillospira | 0.83 | 0.51 | 0.61 | 0.69 | 0.57 | 0.56 | 0.05 | 0.46 |
| Enterococcus | 0.40 | 0.40 | 0.41 | 0.42 | 0.35 | 0.37 | 0.02 | 0.81 |
| Lactobacillus | 0.22 | 0.29 | 0.30 | 0.32 | 0.48 | 0.64 | 0.07 | 0.54 |
| Lachnobacterium | 0.29 | 0.28 | 0.08 | 0.47 | 0.19 | 0.33 | 0.07 | 0.72 |
| Anaerostipes | 0.23 | 0.18 | 0.22 | 0.23 | 0.31 | 0.20 | 0.02 | 0.65 |
| Turicibacter | 0.21 | 0.17 | 0.13 | 0.08 | 0.12 | 0.24 | 0.02 | 0.45 |
| Megasphaera | 0.08 | 0.07 | 0.10 | 0.21 | 0.10 | 0.13 | 0.03 | 0.79 |
| Bacteroidetes | 12 | 13 | 14 | 14 | 12 | 14 | 0.64 | 0.81 |
| Bacteroides | 8.3 | 8.9 | 9.9 | 9.2 | 7.9 | 10.3 | 0.53 | 0.81 |
| Parabacteroides | 0.92 | 1.0 | 1.1 | 1.0 | 1.0 | 1.1 | 0.08 | 0.96 |
| Prevotella | 0.44 | 0.46 | 0.38 | 0.81 | 0.28 | 0.28 | 0.14 | 0.91 |
| Actinobacteria | 3.7 | 2.8 | 3.6 | 4.4 | 3.7 | 3.7 | 0.36 | 0.72 |
| Bifidobacterium | 3.5 | 2.6 | 3.3 | 4.2 | 3.4 | 3.5 | 0.36 | 0.71 |
| Collinsella | 0.11 | 0.12 | 0.11 | 0.12 | 0.13 | 0.14 | 0.01 | 0.96 |
| Proteobacteria | 1.5 | 1.7 | 1.5 | 1.8 | 1.3 | 1.5 | 0.08 | 0.49 |
| Sutterella | 0.22 | 0.30 | 0.27 | 0.42 | 0.30 | 0.40 | 0.03 | 0.34 |
| Citrobacter | 0.22 | 0.20 | 0.21 | 0.37 | 0.22 | 0.24 | 0.03 | 0.32 |
| Haemophilus | 0.13 | 0.23 | 0.10 | 0.12 | 0.05 | 0.14 | 0.03 | 0.64 |
| Verrucomicrobia | 0.19 | 0.11 | 0.24 | 0.18 | 0.15 | 0.36 | 0.03 | 0.94 |
| Akkermansia | 0.19 | 0.11 | 0.24 | 0.18 | 0.15 | 0.36 | 0.00 | 0.41 |
Values are presented as means with pooled SEMs (n = 147). Values in a row without a common letter are significantly different (P < 0.05).
The Firmicutes/Bacteroidetes (F/B) ratio (median, 6.79) of the present study cohort is at the high end compared with the results of the Human Microbiome Project (HMP) (33). However, the results are comparable to other studies (6, 34, 35). The F/B ratio is known to vary dramatically among populations, as well as between age groups (36).
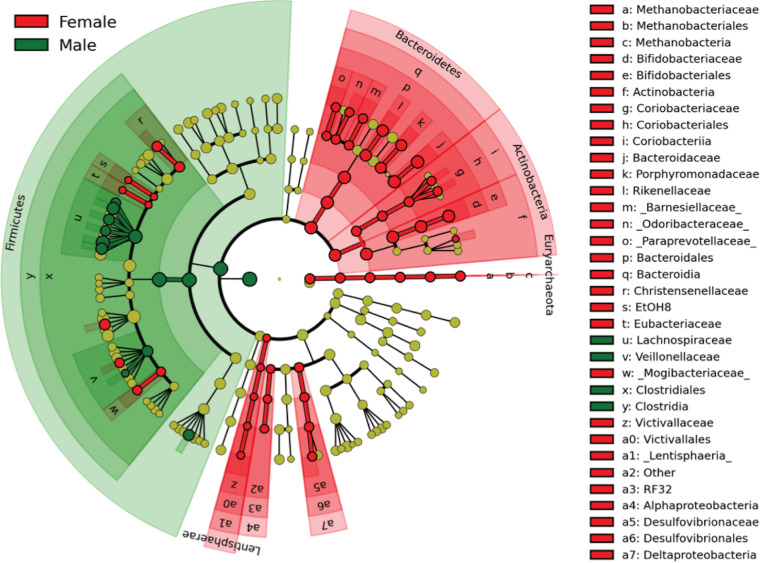
Comparisons at various taxonomic levels between each treatment period and baseline were performed using linear discriminant analysis effect size (LEfSe). Overall, there was little difference between any treatment and baseline, other than overrepresented Streptococcus in yogurt groups (data not shown). Interestingly, a gender difference was observed in the same data set (Fig. 1). Females had significantly greater abundance of Paraprevotella, Butyricimonas, Parabacteroides, Bacteroides, Collinsella, Bifidobacterium, Varibaculum, Methanobrevibacter, and Oscillospira, while males had significantly higher percentages of Anaerostipes, Blautia, Dorea, Lachnobacterium, and Roseburia. Gender differences in gut microbiota have also been observed in an animal study (37) and in other human studies (38–41), although some studies have reported no difference or a modest association (33). Gender differences in gut microbiota have been proposed to be influenced by sex hormone levels (40), gender-based differences in the immune system (41), and differences in dietary patterns (42). Coincidently, a gender difference in responsiveness of CD14+ HLA-DR+ cells to the interventions has been previously observed (43), where the percentage of CD14+ HLA-DR+ cells was increased only in the male participants following consumption of all yogurt-containing treatments compared with baseline.
FIG 1.
Taxonomic cladogram of LDA effect size comparing the relative abundance of taxa in males and females. Significantly discriminant taxon nodes are colored and branch areas are shaded according to the highest-ranked variety for that taxon. For each taxon detected, the corresponding node in the taxonomic cladogram is colored according to the highest-ranked group for that taxon. If the taxon is not significantly different between groups, the corresponding node is colored yellow (66).
In summary, only limited compositional changes were detected in participants’ fecal microbiota after consuming BB-12-containing capsules or yogurt smoothies compared to baseline. Yogurt interventions appeared to result in elevated relative abundance of Streptococcus in host fecal microbiota, especially in female participants, but gender appeared to be a more significant factor in shaping the gut microbiota than the treatments, at least in this study cohort.
(ii) Diversity of the participants’ fecal microbiota.
There were no significant differences in alpha diversity indices (Chao1 richness, Simpsons diversity, and Shannon evenness) between treatment groups or between genders (data not shown). A definitive stratification according to treatment group was not evident on UPGMA (unweighted pair group method with arithmetic mean) tree, but samples from each individual tended to cluster together (Fig. 2). Permutational multivariate analysis of variance (PERMANOVA) also confirmed the similarity between treatments (RANOSIM = −0.041, P = 1.0; R2ADONIS = 0.024, P = 1.0) and dissimilarity between individuals (RANOSIM = 0.925, P = 0.001; R2ADONIS = 0.433, P = 0.001). In this case, the results indicated that individual characteristics played a bigger role than treatments in shaping the host gut microbiota. This is supported by previous research on both animals (44) and humans (45). Specifically, Goodrich et al. studied over 1,000 stool samples from 416 pairs of twins (45). They found that fecal microbiota were more similar overall within individuals (resampled) than between unrelated individuals (P < 0.001) and were also more similar within twin pairs than unrelated individuals (P < 0.009). Moreover, monozygotic twin pairs had a more similar gut microbiota than dizygotic twin pairs (P = 0.032) (45).
FIG 2.
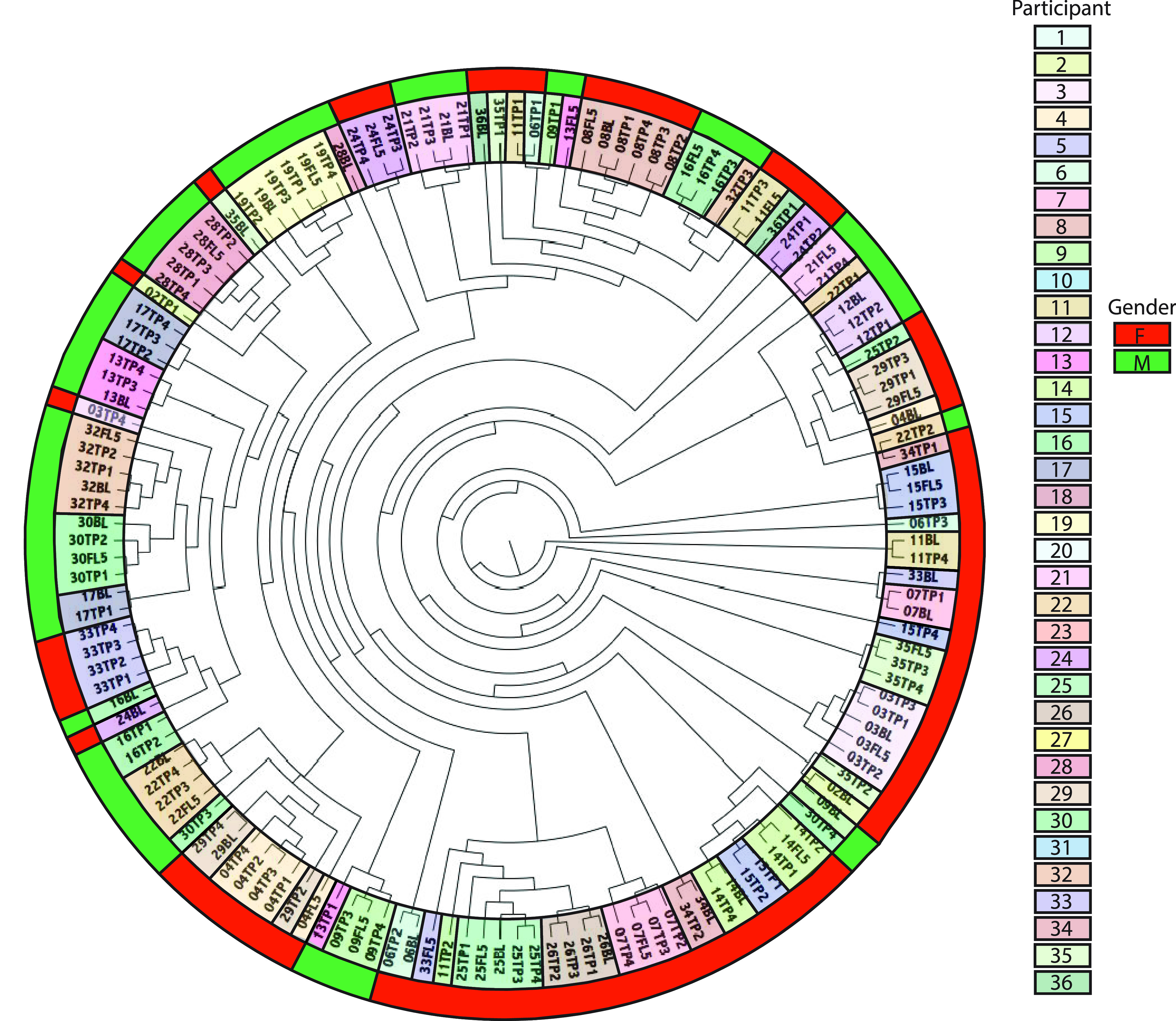
UPGMA tree based on weighted UniFrac distance (beta diversity) demonstrating the hierarchical relationships between the fecal samples. The code is participant’s study identifier (ID) followed by treatment period (i.e., TP1 is the first treatment period, BL is baseline, FL5 is final washout). The inside color bar indicates participant ID, and the outside color bar indicates the gender (female [F] or male [M]) of each participant. The data indicate that samples from each individual tend to cluster together regardless of treatment.
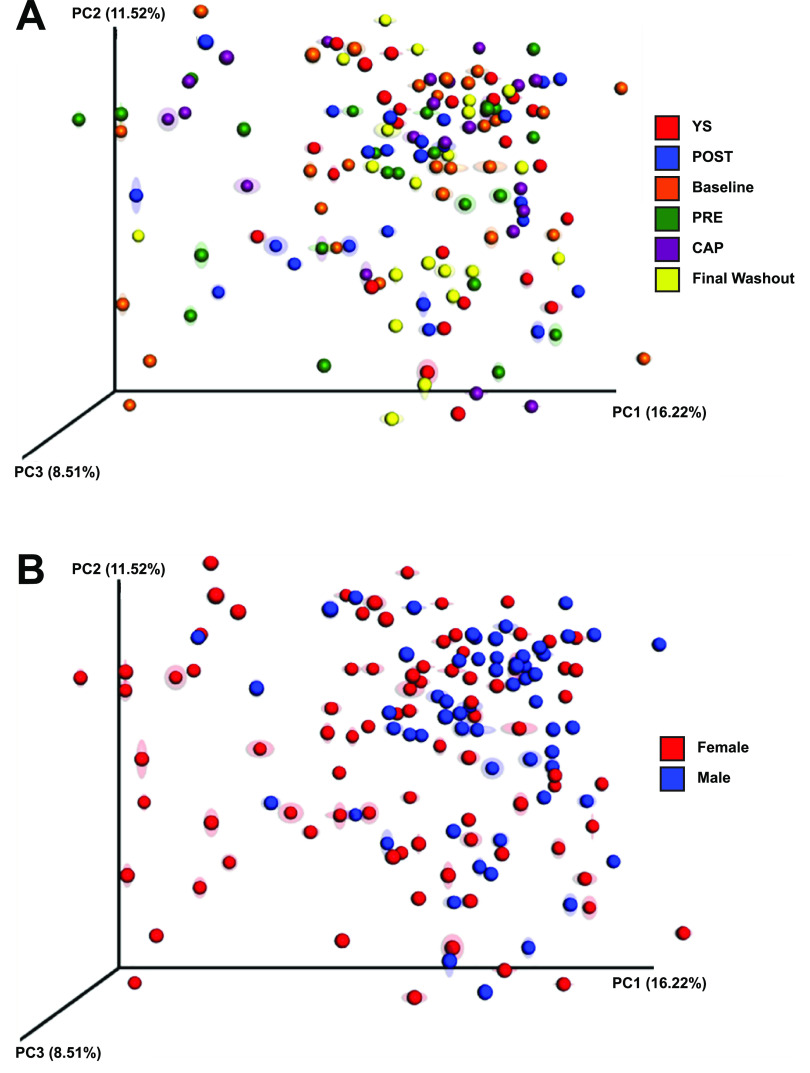
A visible difference in community structure between treatments was not noted on the principal coordinate analysis (PCoA) plot; however, samples tended to cluster when grouped by gender (Fig. 3). Statistical analyses revealed that community membership was different between males and females (RANOSIM = 0.096, P = 0.002; R2ADONIS = 0.021, P = 0.001), although only a small percentage of differences can be explained by the data set. Other metadata (i.e., glucose, high/low-density lipoprotein, triglycerides, C-reactive protein, tumor necrosis factor alpha, and interferon gamma) reported elsewhere (46, 47) were screened for possible associations with the host gut microbiota data. A number of statistically significant differences were found (see Table S1 in the supplemental material). However, further studies are needed to validate these relationships.
FIG 3.
Weighted UniFrac distance PCoA of bacterial communities with jackknife support grouped by treatment and gender. There were no patterns of clustering when samples were colored by treatments, while samples tend to cluster based on gender, suggesting a gender difference.
Statistical analyses of the weighted UniFrac distance (beta diversity) when grouped by the metabolic and immune parameters measured as described in references 46 and 47. Abbreviations: CRP, C-reactive protein; TC:HDL-C, total cholesterol (TC) to high-density lipoprotein cholesterol (HDL-C) ratio; WC, waist circumference; TG, triglycerides; LDL-C, low-density lipoprotein cholesterol; DCs, dendritic cells; IFN-γ, interferon gamma; TNF-α, tumor necrosis factor alpha; IL-2, interleukin-2. Download Table S1, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2021 Ba et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taken together, the diversity results are not completely surprising. Participants in the present study cohort were healthy individuals, and it is well understood that a healthy gut microbiota is stable and resilient (33, 48, 49). Although consuming a high-fat and low-fiber or low-fat and high-fiber diet for 10 days can induce statistically significant changes in the gut microbiota, these changes in species and gene content are small compared with baseline variations that occur between individuals (50).
In previous work, McNulty et al. (51) repeatedly sampled seven healthy adult female monozygotic twin pairs (aged 21 to 32 years, BMI of 20 to 25 kg/m3) 4 weeks before, 7 weeks during, and 4 weeks after consumption of a commercially available fermented milk product (8 oz/day) containing a consortium of BAL strain CNCM I-2494 (3.2 × 107 CFU/g), Lactobacillus delbrueckii subsp. bulgaricus strains CNCM I-1632 and I-1519 (6.3 × 107 CFU/g), Lactococcus lactis subsp. cremoris strain CNCM I-1631, and Streptococcus thermophilus strain CNCM I-1630. They found the species and gene content of the twins’ gut microbial communities remained stable and were not appreciably perturbed by consuming the intervention. However, another study of mice reported that introducing the fermented milk product strains resulted in marked changes in metabolic pathways related to carbohydrate processing, although the proportional representation of their gut microbiota acquired from their human donors remained the same (51). The authors suggested that analyses of the bacterial species and gene content of the gut microbiota/microbiome may not be informative biomarkers for understanding whether or how the interventions may have affected microbial community properties.
(iii) Fecal bifidobacterial distribution.
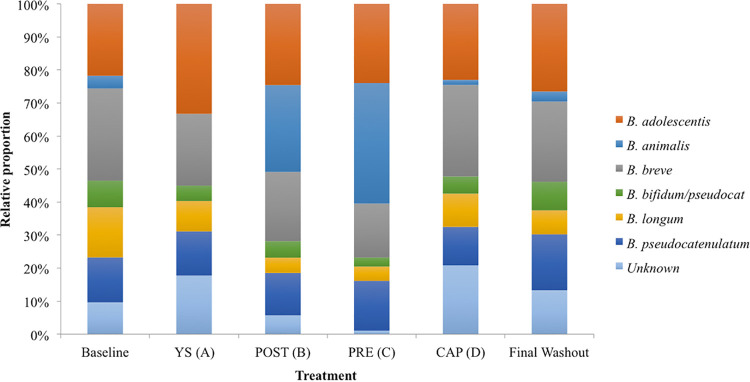
To further explore the effect of the study interventions on the host gut bifidobacteria at the species level, stool DNA samples were subjected to Bifidobacteria terminal restriction fragment length polymorphism (Bif-TRFLP) (52). Participants had a significantly higher percentage of B. animalis as a portion of the total bifidobacteria in their feces after consuming the two yogurt smoothies containing BB-12 (PRE and POST) compared to baseline, other interventions (YS or CAP), and final washout (P < 0.0001) (Fig. 4). There was no significant difference between PRE and POST yogurt smoothie treatments. It appears that the BB-12-containing yogurt smoothie resulted in higher relative abundance of B. animalis in the stool samples than capsules. This may be due to the buffering capacity of milk proteins that could protect strain BB-12 when passing through the acidic conditions of the stomach (53). Care should be taken when extrapolating this finding, because the results presented here are not absolute concentrations. Moreover, it is unclear what the physiological consequences are when the host has a higher level of B. animalis in their feces.
FIG 4.
Relative proportion of Bifidobacterium species in the stool DNA samples before treatment (baseline), after each treatment, and after final washout as determined by Bif-TRFLP (AluI).
The physiological effect of the delivery matrix on the performance of probiotics has been previously observed in an animal study. Lee et al. (54) fed dextran sulfate sodium (DSS)-induced ulcerative colitis mice, with a wild-type strain and two mutant (DltD− and RecA−) probiotic strains of Lacticaseibacillus casei (2 × 107 CFU/feeding) in milk or a nutrient-free buffer prior to and during administration of DSS for 15 consecutive days. Live Lb. casei cells in stool samples were recovered using selective De Man, Rogosa, and Sharpe (MRS) medium. A disease activity index (DAI) was calculated by percent total weight loss (before/after DSS treatment), histology score, the presence of blood in stools, and stool consistency. This study showed that mice fed with Lb. casei in milk had lower DAI than those fed with Lb. casei in nutrient-free buffer, milk only, and mutant in either milk or buffer, suggesting that milk might be the preferred delivery matrix for certain probiotic strains. Additional studies are needed to evaluate other strains, other delivery matrices, and different disease conditions. Moreover, the underlying mechanisms need to be further investigated.
(iv) Predominant fecal SCFAs correlate with GTTs.
Short-chain fatty acids are important for maintaining a healthy gastrointestinal (GI) environment as they promote the growth and differentiation of epithelial cells and provide energy to colonocytes (55). Fecal SCFA concentrations were measured before treatment (baseline), after each treatment, and after final washout. The most abundant SCFAs detected in human fecal samples were acetic acid (30.7%), propionic acid (21.3%), and butyric acid (31.8%), and there was no difference in SCFA levels among treatments. Previous studies have reported that the ratio of SCFAs in human fecal samples is 60:20:20 for acetate-butyrate-propionate (56, 57). The discrepancy between our study could be due to differences in analytical techniques (such as extraction or quantification), as the ratio was consistent throughout treatments and participants. In this study, the concentrations of SCFAs were significantly correlated (Table 4). Moreover, males had a higher butyric acid concentration than females in this study cohort (median of 908 μg/g versus 687 μg/g; P = 0.0233) in agreement with a previous study (58). High interindividual variations were observed in the content of fecal SCFAs (relative standard deviation, 52% to 118%).
TABLE 4.
Spearman correlations of GTT and fecal SCFAsa
| Variable or SCFA | Statistical parameterb | Spearman correlation or P value of variable and SCFA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WGTT | CTT | SBTT | GET | Acetic acid | Propionic acid | Isobutyric acid | Butyric acid | Isovaleric acid | Valeric acid | ||
| CTT | Sr | 0.97 | |||||||||
| P | <0.001 | ||||||||||
| SBTT | Sr | 0.26 | 0.12 | ||||||||
| P | 0.003 | 0.163 | |||||||||
| GET | Sr | 0.21 | 0.12 | −0.06 | |||||||
| P | 0.018 | 0.175 | 0.479 | ||||||||
| Acetic acid | Sr | −0.19 | −0.19 | 0.05 | −0.29 | ||||||
| P | 0.036 | 0.033 | 0.566 | 0.001 | |||||||
| Propionic acid | Sr | −0.25 | −0.25 | 0.03 | −0.27 | 0.68 | |||||
| P | 0.005 | 0.004 | 0.763 | 0.003 | <0.001 | ||||||
| Isobutyric acid | Sr | 0.04 | 0.06 | 0.03 | −0.16 | 0.11 | 0.29 | ||||
| P | 0.697 | 0.492 | 0.714 | 0.072 | 0.224 | 0.001 | |||||
| Butyric acid | Sr | −0.21 | −0.21 | 0.06 | −0.36 | 0.86 | 0.73 | 0.23 | |||
| P | 0.021 | 0.016 | 0.488 | <0.001 | <0.001 | <0.001 | 0.008 | ||||
| Isovaleric acid | Sr | 0.07 | 0.10 | 0.01 | −0.1 | −0.06 | 0.18 | 0.971 | 0.09 | ||
| P | 0.407 | 0.285 | 0.925 | 0.250 | 0.533 | 0.038 | <0.001 | 0.311 | |||
| Valeric acid | Sr | −0.1 | −0.12 | 0.20 | −0.12 | 0.49 | 0.54 | 0.64 | 0.51 | 0.55 | |
| P | 0.261 | 0.183 | 0.026 | 0.087 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| Caproic acid | Sr | 0.07 | 0.08 | 0.07 | −0.01 | 0.24 | −0.03 | 0.21 | 0.16 | 0.14 | 0.39 |
| P | 0.417 | 0.350 | 0.407 | 0.897 | 0.006 | 0.759 | 0.018 | 0.076 | 0.107 | <0.001 | |
Abbreviations: GTT, gut transit time; SCFAs, short-chain fatty acids.
Sr, Spearman rho value; P, P value.
Since fecal SCFAs only account for less than 5% of the total SCFAs produced in the colon (54, 57), the effectiveness of strain BB-12 in promoting SCFA production in the large intestine remains unclear. Previous studies reported inconsistent results. In an early study of 16 patients with ileal pouch-anal anastomosis for ulcerative colitis, no difference in SCFA content was observed after consumption of 500 ml of fermented milk (Cultura) containing >108 CFU/ml of both Lactobacillus acidophilus (LA-5) and BAL (BB-12) or heat-treated Cultura for 1 week compared to baseline or between groups (59). On the other hand, a more recent study demonstrated that consumption of yogurt containing log10 9.72 CFU of BAL LKM512 per day for 4 weeks tended to increase fecal butyrate concentration in patients with atopic dermatitis compared to baseline (P = 0.08) (60).
To further explore possible relationships between GTTs and fecal SCFA concentrations, correlation analyses were conducted. Gut transit times were negatively correlated with predominant SCFAs, but valeric acid was positively correlated with SBTT (Table 4). A previous study on the effect of GTT rate on fecal SCFA concentration showed negative correlations between GTT and fecal SCFAs, which was attributed to physiological factors of the participants (61).
Limitations.
One of the major limitations of the present study was the inability to recruit enough participants with significantly delayed WGTT (>60 h). As a result, volunteers with slightly delayed WGTT (>24 h) were recruited, leaving little room for the intervention to have a significant effect. Moreover, performing only one measurement before treatment (baseline) did not seem to reflect the true gut transit status, as each subject’s WGTT tended to vary significantly from one day to another. Finally, the sequencing technique employed had a low resolution that can only detect down to the genus level. This made it impossible to explore treatment effect at the species level, although supplemental methods were used to measure species of interest.
Conclusions.
The present study evaluated the effect of the probiotic BAL BB-12 alone (capsule) or when incorporated into yogurt smoothies before or after yogurt fermentation on the GTTs, fecal SCFA concentrations, the composition of the gut microbiota, and the bifidobacterial profile of young healthy adults. No significant treatment effects on the GTTs, fecal SCFAs, or the gut microbiota were detected due to the large interindividual and intraindividual variations observed. A significant gender effect was observed when comparing the gut microbiota of the cohort of the present study. Interestingly, the two BB-12-containing yogurt smoothies (PRE and POST) resulted in significantly higher percentage of B. animalis as a fraction of total bifidobacteria compared to baseline, to the BB-12-free yogurt smoothie (YS), the capsule (CAP), and final washout when analyzed by Bif-TRFLP. No difference was detected between PRE and POST addition of BB-12. Although Bif-TRFLP could not measure viability, the finding of the present study may shed light on the subtle effect of probiotic interventions on the gut microbiota at the species level. Further studies are warranted to study other bacterial groups at the species level, more importantly, to clarify the impact of these differences on human health.
MATERIALS AND METHODS
Study design.
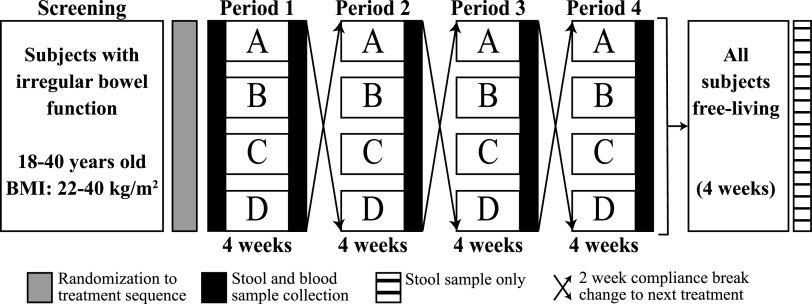
This clinical trial was a randomized, four-period, crossover study of free-living subjects performed in a partially blind manner. The study design scheme is shown in Fig. 5. The detailed clinical aspects of this study have been reported elsewhere (46, 47). Briefly, measurements taken during the baseline visit and after each treatment were anthropometric assessment (age and gender at the baseline visit only, BMI, waist circumference), biochemical parameters (fasting serum glucose, insulin, and high-sensitivity CRP [hs-CRP]), a physical activity questionnaire (self-reported IPAQ), and an immune endpoint assessment. Then each participant began the intervention phase as specified by the randomization order. The four treatments were YS (yogurt smoothie without BB-12) (treatment A), POST (yogurt smoothie with BB-12 added after fermentation) (treatment B), PRE (yogurt smoothie with BB-12 added before fermentation) (treatment C), and CAP (BB-12-containing capsule (treatment D). Each treatment period lasted 4 weeks, and a 2-week wash-out compliance break was scheduled between treatment periods.
FIG 5.
Schematic diagram for randomization design. The treatments are shown in boxes as follows: A, yogurt smoothie without BB-12 (YS); B, yogurt smoothie with BB-12 added postfermentation (POST); C, yogurt smoothie with BB-12 added prefermentation (PRE); D, BB-12-containing capsule (CAP).
Interventions.
The control and BB-12 (Chr. Hansen A/S, Hørsholm, Denmark) interventions were strawberry-flavored yogurt smoothies developed and manufactured at The Pennsylvania State University. The starter culture used was YF-L702 (Chr. Hansen A/S, Hørsholm, Denmark), a commercial blend of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus (59). Dry ingredients were mixed into the milk, pasteurized at 84.4°C for 41 s, and homogenized at 2,000 lb/in2 in the first stage and at 500 lb/in2 in the second stage. Yogurt mix was then heat treated at 85°C for 30 min, cooled to 43.3°C, inoculated with YF-L702, and split in two portions. One portion remained BB-12 free, while another portion was inoculated with BB-12 (PRE). Both yogurt mixes were fermented until a pH of 4.6 was reached. At this time, a prepared mixture containing strawberry, pectin, corn syrup solids, sugar, and water was added and blended into the yogurt until uniform. After addition of the slurry, the BB-12-free yogurt smoothie was further split into two parts; one part was inoculated with BB-12 (POST), while the other part remained BB-12-free (YS). Finally, each of the products was rehomogenized to produce a drinkable yogurt. The three products were identical in chemical and textural characteristics except for the addition of BB-12 and the timing of BB-12 addition.
To verify the viable count of Bifidobacterium animalis subsp. lactis (BAL) in the yogurt smoothies, both control and BB-12 products were analyzed immediately after manufacture and weekly during shelf life. To determine cell counts, suitable dilutions were pour plated on MRS-NNLP agar (62) followed by anaerobic incubation at 37°C for 72 h. Colonies counted as BAL were randomly picked and confirmed to be BAL by PCR using subspecies-specific primers (63).
During the yogurt smoothie treatment phases, participants consumed one 240-g serving of yogurt per day. Study interventions contained between log10 10 ± 0.5 × 109 or 3.16 × 109 and 3.16 × 1010 CFU of strain BB-12 per serving. During the CAP treatment phase, participants ingested one capsule per day. This CAP was specifically designed by Chr. Hansen A/S to deliver log10 10 ± 0.5 CFU of BB-12/capsule, which was confirmed by viable counting as described for the yogurt smoothies throughout the study. Participants were instructed to avoid consuming any other food or supplements containing probiotic bacteria, such as commercial yogurt, smoothies, and probiotic capsules or tablets, during each 4-week intervention phase. They were also asked not to change their habitual diets for the course of the study and to maintain their body weight.
Participants.
The primary outcome of the study, whole gut transit time (WGTT), was used for the sample size calculation. Specifically, the calculation was based on the data from a previous study (64) and indicated that 28 subjects were required to identify a mean difference of 6 h in WGTT. Considering the possibility of withdrawal from the study and potential lack of compliance, 36 healthy volunteers (18 to 40 years of age) with delayed WGTT (≥24 h) were recruited. Of these volunteers, 29 finished at least one treatment period. Detailed exclusion criteria are described elsewhere (46). All subjects were nonsmoking, normotensive, and not diagnosed with any chronic medical conditions. Prior to the start of the trial, all participants signed an informed consent form. The study was approved by the Institutional Review Board of The Pennsylvania State University (University Park, PA) under IRB 35111. This trial was registered at ClinicalTrials.gov as NCT01399996.
Primary outcome measures.
The primary outcome WGTT was measured using a modified blue food dye method (65) and using a wireless mobility capsule, SmartPill (Given Imaging, Duluth, GA) at baseline, and only SmartPill after each treatment. Unlike the blue dye method which only measures WGTT, the SmartPill measured regional gut transit times (GTTs) including gastric emptying time (GET), small bowel transit time (SBTT) and colonic transit time (CTT) along with pH, temperature, and pressure throughout the gastrointestinal tract (GIT).
Secondary outcome measures.
The secondary outcomes were to evaluate the effect of BB-12 interventions on the participants’ fecal microbiota (both composition and diversity) and to detect any shift in fecal SCFA concentrations.
(i) Stool DNA extraction.
Fecal samples were collected by participants and stored at home freezers until transported to the Penn State Clinical Research Center on ice before treatment (baseline), after each treatment, and after final washout. Stool DNA was extracted using a MOBIO PowerSoil DNA isolation kit (catalog no. 12888; Qiagen, Carlsbad, CA) according to the manufacturer’s protocol. Isolated genomic DNA was stored at −80°C prior to analysis.
(ii) Compliance.
Compliance was assessed by a BAL subspecies-specific PCR method (63). Detection of BAL in the feces of the BB-12 groups and the absence of BAL in the feces of the baseline group and the control smoothie group were taken as evidence of compliance. Results showing the presence of BAL in the control or baseline, or absence of BAL in the BB-12 groups, or missing fecal samples, were considered to indicate noncompliance.
(iii) 16S rDNA amplicon Illumina sequencing.
DNA samples were prepared as previously described (66) with the following modifications. Universal primers F515 (5′-NNNNNNNNGTGTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACHVGGGTWTCTAAT-3′), with the forward primer modified to contain an 8-nucleotide (nt) barcode [italicized poly(N) section of the primer above] and 2-nt linker sequence (boldface portion) at the 5′ end, were used to amplify the V4 region of the 16S rRNA gene. PCR mixtures contained 5.0 μl of 2 × GoTaq Green Master Mix (Promega, Madison, WI), 0.4 μl of 25 mM MgCl2, 2.4 μl of water, 0.2 μl of reverse primer (10 mM final concentration), 1.0 μl of forward primer (2 mM final concentration), and 1.0 μl of genomic DNA. Reactions were held at 94°C for 3 min to denature the DNA, with amplification proceeding for 25 cycles, with 1 cycle consisting of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s; a final extension of 10 min at 72°C was included to ensure complete amplification. The PCR products were purified using QIAquick PCR purification kit (catalog no. 28106; Qiagen, Valencia, CA). A sequencing library was created by combining equimolar ratios of amplicons from individual samples. The composite sample was sequenced at the DNA Technologies Core Facility of the University of California, Davis, on an Illumina Genome Analyzer II sequencing platform.
(iv) Bifidobacteria terminal restriction fragment length polymorphism assay.
The Bifidobacteria terminal restriction fragment length polymorphism (Bif-TRFLP) assay was performed based on the method described by Lewis et al. (52). Briefly, stool DNA samples were amplified for bifidobacterial 16S rRNA gene. The PCRs were carried out in a mixture (50 μl) that contained 1 μl of genomic DNA, 25 μl of 2× Promega GoTaq Green Master Mix (Promega, Madison, WI), 20 μl of nuclease-free water, 1 μl of each primer (NBIF389, 5′-[HEX]-GCCTTCGGGTTGTAAAC-3′, 10 μM; NBIF1018REV, 5′-GACCATGCACCACCTGTG-3′, 10 μM), and 2 μl of MgCl2 (25 mM). The reaction conditions were 95°C for 5 min, followed by 30 cycles, with 1 cycle consisting of 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min. A final extension at 72°C for 5 min was allowed following the cycles, and then the samples were stored at 4°C prior to analysis. PCR products were purified using the QIAquick PCR purification kit (catalog no. 28106; Qiagen, Valencia, CA).
A portion (8 μl) of the purified DNA was digested with two restriction enzymes (AluI [Thermo Fisher Scientific, Waltham, MA] and HaeIII [New England BioLabs Inc., Ipswich, MA]) in separate reactions, both of which involved 1 μl of enzyme (10 U/μl) in a 10-μl reaction mixture for 3 h at 37°C. Then, the enzymes were heat inactivated at 80°C for 20 min, and samples were stored at 4°C. Next, 1 μl of the digested mixture (diluted 1:20 in elution buffer) was submitted for fragment analysis on an ABI 3730 Capillary Electrophoresis Genetic Analyzer (Applied Biosystems, Grand Island, NY). The molecular size markers used were the ROX 50-500 size standards (Gel Company Inc., San Francisco, CA). The results were read using Peak Scanner software v1.0 (Applied Biosystems, Grand Island, NY). Detailed data processing is described in the original article (52).
(v) Microbiome sequence data analysis.
The data analysis pipeline used was modified from a previous study (67). Briefly, QIIME software package (68) was used to analyze the results of the Illumina sequencing run. Raw Illumina fastq files were first demultiplexed and quality filtered. Reads were truncated after a maximum number of three consecutive low-quality scores (<1e−5), and any read containing one or more ambiguous base calls was discarded. Reads with a minimum pairwise identity of 97% were clustered into operational taxonomic units (OTUs) using QIIME’s open-reference OTU-picking workflow, which was based on UCLUST (69) software. The Greengenes bacterial 16S rRNA database (13_8 release) was used for OTU picking (70). The most abundant sequence was chosen to represent each OTU. Taxonomy was assigned to each OTU using QIIME-based wrapper of the Ribosomal Database Project (RDP) classifier (71) against a representative subset of the Greengenes 16S rRNA database 13_8 release, using a 0.50 confidence threshold for taxonomic assignment. Bacterial 16S rRNA gene sequences were aligned using PyNAST (72) against a template alignment of the Greengenes core set filtered at 97% similarity. During the process, chimeras were identified and removed using the ChimeraSlayer (73) algorithm, and a phylogenetic tree was built from the filtered alignment using FastTree (74). Any OTU representing less than 0.001% of the total filtered sequences was removed to avoid erroneous reads that could lead to inflated estimates of diversity (75). After these quality-filtering steps, each sample was represented by less than 150 sequences, and the filtered OTU tables were ready for downstream analyses, such as diversity comparisons and biomarker discovery.
Alpha diversity and beta diversity were calculated within QIIME based on weighted UniFrac (76) distance between samples. Principal coordinates were calculated from the UniFrac distance matrices to decrease the dimensionality of the taxonomic data set into three-dimensional (3D) principal coordinate analysis (PCoA) plots, enabling visualization of sample relationships. To determine whether treatments caused differences in phylogenetic or species diversity, analysis of similarity (ANOSIM) (77) and permutational multivariate analysis of variance (PERMANOVA) (78) were used to test significant differences between sample groups based on weighted UniFrac.
Significant taxonomic differences between sample groups were also tested using the linear discriminant analysis (LDA) effect size (LEfSe) (79). LEfSe is an algorithm for high-dimensional biomarker discovery and identification of genomic features (genes, pathways, or taxa) that characterizes the differences between two or more classes/treatments. It first uses the nonparametric factorial Kruskal-Wallis (KW) rank sum test to detect taxa with significant differential abundances with respect to the class of interest (one-against-all strategy). Then LEfSe uses linear discriminant analysis to estimate the effect size of each differentially abundant feature.
(vi) Fecal SCFAs.
Short-chain fatty acids in the stool samples were analyzed using gas chromatography (GC) according to the method described in a previous study (80) with minor modifications. The GC system consisted of an Agilent 6890 gas chromatograph (Agilent Technologies, Palo Alto, CA), equipped with an automatic sampler (MPS) (Gerstel, Mülheim, Germany) and a flame ionization detector (FID). A high-polarity, polyethylene glycol (PFG), fused silica capillary column DB-WAXETR (30 m, 0.25-mm inner diameter [i.d.], 0.25-mm film thickness) (Agilent Technologies, Palo Alto, CA) was used for separation. Prior to sample analysis, a standard solution containing a mixture of standards (30 mM final concentration of acetic acid, propionic acid, isobutyric acid, butyric acid, isovaleric acid, valeric acid, and caproic acid) in ethyl acetate containing 1 mM heptanoic acid as internal standard (IS) was diluted to obtain a calibration curve ranging from 3 to 3,000 μM. Standard curves were constructed by plotting the concentration of each individual SCFA versus the ratio of SCFA peak area/IS peak area. Each point of the standard curves corresponds to the mean value from three independent injections. Three independent replicate extractions were performed per sample.
Statistical analysis.
Intention-to-treat (ITT) analyses were applied to treatment effect analyses (81). All data were first tested for normality using the Anderson-Darling test, and log transformation was applied when necessary. The analysis of treatment effect and period effect was performed using analysis of variance (ANOVA) or the Kruskal-Wallis test where appropriate. Mann-Whitney U test was used for gender comparison, and the Spearman rho test was employed for correlation analyses; a P value of <0.05 was considered significant. All statistical analyses were performed using Minitab 17.0 software (Minitab Inc., State College, PA).
Data availability.
Sequence data were deposited in NCBI SRA under BioProject PRJNA739252. Metadata and code used for microbiome analyses is available at https://github.com/LauRolon/BB12microbiome.
ACKNOWLEDGMENTS
We thank the participants for their engagement, the Penn State Berkey Creamery and its staff members for making the yogurt interventions, and the staff nurses at the Penn State Clinical Research Center for their collaboration.
This project was supported by a grant from Dairy Research Incorporated and by the Penn State Clinical & Translational Research Institute, Pennsylvania State University CTSA, NIH/NCATS grant UL1 TR000127. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or NCATS. This work was also supported by the USDA National Institute of Food and Agriculture and Hatch Appropriations under project PEN04708 and accession number 1019852. D.A.M. acknowledges National Institutes of Health awards AT008759 and AT007079.
Z. Ba, Y. Lee, H. Meng, P. M. Kris-Etherton, C. J. Rogers, Z. T. Lewis, E. J. Furumoto, M. L. Rolon, J. A. Fleming, and R. F. Roberts have no conflicts of interest. D. A. Mills is a cofounder of Evolve Biosystems, a company focused on diet-based manipulation of the gut microbiota, and BCD Biosciences, a company focused on bioactive glycans—neither of which were involved in this work.
Contributor Information
Robert F. Roberts, Email: rfr3@psu.edu.
Robert A. Britton, Baylor College of Medicine
REFERENCES
- 1.Tuohy K, Rouzaud G, Bruck W, Gibson G. 2005. Modulation of the human gut microflora towards improved health using prebiotics - assessment of efficacy. Curr Pharm Des 11:75–90. doi: 10.2174/1381612053382331. [DOI] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clausen MR, Bonnen H, Tvede M, Mortensen PB. 1991. Colonic fermentation to short-chain fatty acids is decreased in antibiotic-associated diarrhea. Gastroenterology 101:1497–1504. doi: 10.1016/0016-5085(91)90384-w. [DOI] [PubMed] [Google Scholar]
- 4.Tamboli CP, Neut C, Desreumaux P, Colombel JF. 2004. Dysbiosis in inflammatory bowel disease. Gut 53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Human gut microbes associated with obesity. Nature 444:1022–1028. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. 2012. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond) 36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso VR, Guarner F. 2013. Linking the gut microbiota to human health. Br J Nutr 109:S21–S27. doi: 10.1017/S0007114512005235. [DOI] [PubMed] [Google Scholar]
- 11.Engelbrektson AL, Korzenik JR, Sanders ME, Clement BG, Leyer G, Klaenhammer TR, Kitts CL. 2006. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol Ecol 57:239–250. doi: 10.1111/j.1574-6941.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 12.Corrêa NBO, Péret Filho LA, Penna FJ, Lima FMLS, Nicoli JR. 2005. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J Clin Gastroenterol 39:385–389. doi: 10.1097/01.mcg.0000159217.47419.5b. [DOI] [PubMed] [Google Scholar]
- 13.Raahave D. 2015. Faecal retention: a common cause in functional bowel disorders, appendicitis and haemorrhoids–with medical and surgical therapy. Dan Med J 62:B5031. [PubMed] [Google Scholar]
- 14.Wichmann A, Allahyar A, Greiner TU, Plovier H, Lundén GÖ, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Bäckhed F. 2013. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. 2002. Guidelines for the Evaluation of Probiotics in Food. Report of a joint FAO/WHO working group on drafting guidelines for evaluation of probiotics in food. Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food, London, Ontario, Canada.
- 16.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 17.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 18.Lin H, Su B, Chen A, Lin T, Tsai C, Yeh T, Oh H. 2005. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pedriatrics 115:1–4. doi: 10.1542/peds.2004-1463. [DOI] [PubMed] [Google Scholar]
- 19.Mastrandrea F, Coradduzza G, Sergio G, Minardi A, Manelli M, Ardito S, Murature L. 2004. Probiotics reduce the CD34+ hemopoietic precursor cell increased traffic in allergic subjects. Eur Ann Allergy Clin Immunol 36:118–122. [PubMed] [Google Scholar]
- 20.Rosenfeldt V, Michaelsen KF, Jakobsen M, Larsen CN, Møller PL, Pedersen P, Tvede M, Weyrehter H, Valerius NH, Pærregaard A. 2002. Effect of probiotic Lactobacillus strains in young children hospitalized with acute diarrhea. Pediatr Infect Dis J 21:411–416. doi: 10.1097/00006454-200205000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Vanderhoof JA, Whitney DB, Antonson DL, Hanner TL, Lupo JV, Young RJ. 1999. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J Pediatr 135:564–568. doi: 10.1016/S0022-3476(99)70053-3. [DOI] [PubMed] [Google Scholar]
- 22.Hasler CM, Brown AC, American Dietetic Association . 2009. Position of the American Dietetic Association: functional foods. J Am Diet Assoc 109:735–746. doi: 10.1016/j.jada.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Monroe Mendelsohn Research. 2001. Live active culture (LAC) yogurt survey. Monroe Mendelsohn Research, New York, NY.
- 24.Klijn A, Mercenier A, Arigoni F. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev 29:491–509. doi: 10.1016/j.femsre.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Jungersen M, Wind A, Johansen E, Christensen J, Stuer-Lauridsen B, Eskesen D. 2014. The science behind the probiotic strain Bifidobacterium animalis subsp. lactis BB-12. Microorganisms 2:92–110. doi: 10.3390/microorganisms2020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders ME, Marco ML. 2010. Food formats for effective delivery of probiotics. Annu Rev Food Sci Technol 1:65–85. doi: 10.1146/annurev.food.080708.100743. [DOI] [PubMed] [Google Scholar]
- 27.González-Rodríguez I, Ruiz L, Gueimonde M, Margolles A, Sánchez B. 2013. Factors involved in the colonization and survival of bifidobacteria in the gastrointestinal tract. FEMS Microbiol Lett 340:1–10. doi: 10.1111/1574-6968.12056. [DOI] [PubMed] [Google Scholar]
- 28.Degen L, Phillips S. 1996. Variability of gastrointestinal transit in healthy women and men. Gut 39:299–305. doi: 10.1136/gut.39.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen CN, Nielsen S, Kæstel P, Brockmann E, Bennedsen M, Christensen HR, Eskesen DC, Jacobsen BL, Michaelsen KF. 2006. Dose-response study of probiotic bacteria Bifidobacterium animalis subsp lactis BB-12 and Lactobacillus paracasei subsp paracasei CRL-341 in healthy young adults. Eur J Clin Nutr 60:1284–1293. doi: 10.1038/sj.ejcn.1602450. [DOI] [PubMed] [Google Scholar]
- 30.Ringel-Kulka T, Kotch JB, Jensen ET, Savage E, Weber DJ. 2015. Randomized, double-blind, placebo-controlled study of synbiotic yogurt effect on the health of children. J Pediatr 166:1475–1481.e3. doi: 10.1016/j.jpeds.2015.02.038. [DOI] [PubMed] [Google Scholar]
- 31.Pitkala K, Strandberg T, Finne-Soveri U, Ouwehand A, Poussa T, Salminen S. 2007. Fermented cereal with specific bifidobacteria normalizes bowel movements in elderly nursing home residents. A randomized, controlled trial. J Nutr Heal Aging 11:305–311. [PubMed] [Google Scholar]
- 32.Eskesen D, Jespersen L, Michelsen B, Whorwell PJ, Müller-Lissner S, Morberg CM. 2015. Effect of the probiotic strain Bifidobacterium animalis subsp. lactis, BB-12, on defecation frequency in healthy subjects with low defecation frequency and abdominal discomfort: a randomised, double-blind, placebo-controlled, parallel-group trial. Br J Nutr 114:1638–1646. doi: 10.1017/S0007114515003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Human Microbiome Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA, Singh U. 2013. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One 8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet J-P. 2009. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9:123–126. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity 39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli M, Clavel T, Koebnick C, Zunft H, Dore J, Blaut M. 2006. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solano-Aguilar G, Fernandez KP, Ets H, Molokin A, Vinyard B, Urban JF, Gutierrez MF. 2013. Characterization of fecal microbiota of children with diarrhea in 2 locations in Colombia. J Pediatr Gastroenterol Nutr 56:503–511. doi: 10.1097/MPG.0b013e318282aa12. [DOI] [PubMed] [Google Scholar]
- 40.Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Landa BB, Navas-Cortés JA, Tena-Sempere M, Clemente JC, López-Miranda J, Pérez-Jiménez F, Camargo A. 2016. Intestinal microbiota is influenced by gender and body mass index. PLoS One 11:e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, Savelkoul HF, de Jonge MI, Faas MM, Boekschoten MV, Smidt H, El Aidy S, de Vos P. 2017. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol 8:754. doi: 10.3389/fimmu.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos-Marcos JA, Haro C, Vega-Rojas A, Alcala-Diaz JF, Molina-Abril H, Leon-Acuña A, Lopez-Moreno J, Landa BB, Tena-Sempere M, Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F, Camargo A. 2019. Sex differences in the gut microbiota as potential determinants of gender predisposition to disease. Mol Nutr Food Res 63:e1800870. doi: 10.1002/mnfr.201800870. [DOI] [PubMed] [Google Scholar]
- 43.Meng H, Ba Z, Lee Y, Peng J, Lin J, Fleming JA, Furumoto EJ, Roberts RF, Kris-Etherton PM, Rogers CJ. 2017. Consumption of Bifidobacterium animalis subsp. lactis BB-12 in yogurt reduced expression of TLR-2 on peripheral blood-derived monocytes and pro-inflammatory cytokine secretion in young adults. Eur J Nutr 56:649–661. doi: 10.1007/s00394-015-1109-5. [DOI] [PubMed] [Google Scholar]
- 44.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng H, Lee Y, Ba Z, Fleming JA, Furumoto EJ, Roberts RF, Kris-Etherton PM, Rogers CJ. 2015. In vitro production of IL-6 and IFN-γ is influenced by dietary variables and predicts upper respiratory tract infection incidence and severity respectively in young adults. Front Immunol 66:94. doi: 10.3389/fimmu.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y, Ba Z, Roberts RF, Rogers CJ, Fleming JA, Meng H, Furumoto EJ, Kris-Etherton PM. 2017. Effects of Bifidobacterium animalis subsp. lactis BB-12 on the lipid/lipoprotein profile and short chain fatty acids in healthy young adults: a randomized controlled trial. Nutr J 16:39. doi: 10.1186/s12937-017-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coyte KZ, Schluter J, Foster KR. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 50.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis ZT, Bokulich NA, Kalanetra KM, Ruiz-Moyano S, Underwood MA, Mills DA. 2013. Use of bifidobacterial specific terminal restriction fragment length polymorphisms to complement next generation sequence profiling of infant gut communities. Anaerobe 19:62–69. doi: 10.1016/j.anaerobe.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziarno M, Zaręba D. 2015. Effects of milk components and food additives on survival of three bifidobacteria strains in fermented milk under simulated gastrointestinal tract conditions. Microb Ecol Heal Dis 26:27812. doi: 10.3402/mehd.v26.27812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee B, Yin X, Griffey SM, Marco ML. 2015. Attenuation of colitis by Lactobacillus casei BL23 is dependent on the dairy delivery matrix. Appl Environ Microbiol 81:6425–6435. doi: 10.1128/AEM.01360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy CC, Kien CL, Bouthillier L, Levy E. 2006. Short-chain fatty acids: ready for prime time? Nutr Clin Pract 21:351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 56.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 57.Primec M, Mičetić-Turk D, Langerholc T. 2017. Analysis of short-chain fatty acids in human feces: a scoping review. Anal Biochem 526:9–21. doi: 10.1016/j.ab.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 58.McOrist AL, Miller RB, Bird AR, Keogh JB, Noakes M, Topping DL, Conlon MA. 2011. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J Nutr 141:883–889. doi: 10.3945/jn.110.128504. [DOI] [PubMed] [Google Scholar]
- 59.Laake KO, Bjørneklett A, Bakka A, Midtvedt T, Norin KE, Eide TJ, Jacobsen MB, Lingaas E, Axelsen AK, Løtveit T, Vatn MH. 1999. Influence of fermented milk on clinical state, fecal bacterial counts and biochemical characteristics in patients with ileal- pouch- anal-anastomosis. Microb Ecol Health Dis 11:211–217. [Google Scholar]
- 60.Matsumoto M, Aranami A, Ishige A, Watanabe K, Benno Y. 2007. LKM512 yogurt consumption improves the intestinal environment and induces the T-helper type 1 cytokine in adult patients with intractable atopic dermatitis. Clin Exp Allergy 37:358–370. doi: 10.1111/j.1365-2222.2007.02642.x. [DOI] [PubMed] [Google Scholar]
- 61.Lewis S, Heaton K. 1997. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut 41:245–251. doi: 10.1136/gut.41.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laroia S, Martin J. 1991. Methods for enumerating and propagating bifidobacteria. Cult Dairy Prod J 26:32–34. [Google Scholar]
- 63.Ventura M, Reniero R, Zink R. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl Environ Microbiol 67:2760–2765. doi: 10.1128/AEM.67.6.2760-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouvier M, Meance S, Bouley C, Berta J-L, Grimaud J. 2001. Effects of consumption of a milk fermented by the probiotic strain Bifidobacterium animalis DN-173010 on colonic transit times in healthy humans. Biosci Microflora 20:43–48. doi: 10.12938/bifidus1996.20.43. [DOI] [Google Scholar]
- 65.Lu WZ, Song GH, Gwee KA, Ho KY. 2009. The effects of melatonin on colonic transit time in normal controls and IBS patients. Dig Dis Sci 54:1087–1093. doi: 10.1007/s10620-008-0463-z. [DOI] [PubMed] [Google Scholar]
- 66.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 70.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, The Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lozupone C, Hamady M, Knight R. 2006. UniFrac – an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371–314. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clarke KR. 1993. Non‐parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 78.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x. [DOI] [Google Scholar]
- 79.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.García-Villalba R, Giménez-Bastida JA, García-Conesa MT, Tomás-Barberán FA, Carlos Espín J, Larrosa M. 2012. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J Sep Sci 35:1906–1913. doi: 10.1002/jssc.201101121. [DOI] [PubMed] [Google Scholar]
- 81.Fisher LD, Dixon DO, Herson J, Frankowski RF, Hearon MS, Peace KE. 1990. Intention to treat in clinical trials, p 331–350. In Peace KE (ed), Statistical issues in drug research and development. Marcel Dekker, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analyses of the weighted UniFrac distance (beta diversity) when grouped by the metabolic and immune parameters measured as described in references 46 and 47. Abbreviations: CRP, C-reactive protein; TC:HDL-C, total cholesterol (TC) to high-density lipoprotein cholesterol (HDL-C) ratio; WC, waist circumference; TG, triglycerides; LDL-C, low-density lipoprotein cholesterol; DCs, dendritic cells; IFN-γ, interferon gamma; TNF-α, tumor necrosis factor alpha; IL-2, interleukin-2. Download Table S1, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2021 Ba et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequence data were deposited in NCBI SRA under BioProject PRJNA739252. Metadata and code used for microbiome analyses is available at https://github.com/LauRolon/BB12microbiome.