Abstract
We generated genome-wide DNA data from four children buried roughly 8000 and 3000 years ago at Shum Laka (Cameroon), one of the earliest archaeological sites within the probable homeland of Bantu languages [1–11]. One individual carried the deeply divergent Y chromosome haplogroup A00, which is found today almost exclusively in the same region [12, 13]. However, all four individuals’ genome-wide ancestry profiles are most similar to West-Central African hunter-gatherers, implying that present-day populations in western Cameroon, as well as Bantu speakers across the continent, are not descended substantially from the population represented by these four people. We infer an Africa-wide phylogeny that features widespread admixture and three prominent radiations, including one giving rise to at least four major lineages deep in the history of modern humans.
The deposits at Shum Laka, a rockshelter located in the Grassfields region of western Cameroon, are among the most important archaeological sources for the study of Late Pleistocene and Holocene prehistory in West-Central Africa [1–4]. The oldest human-occupied layers at the site date to ∼30,000 calendar years before present (BP), but of special interest are a series of artifacts and skeletons from ∼8000–3000 BP, between the Later Stone Age (LSA) and the Iron Age (Extended Data Fig. 1; Supplementary Information section 1). This transitional period, sometimes referred to as the Stone to Metal Age (SMA), featured a gradual appearance of new stone tools as well as pottery [3–5]. Subsistence evidence in the rockshelter during the SMA points primarily to foraging, but with increasing use of fruits from Canarium schweinfurthii coinciding with developments in material culture and serving as a foundation for later agriculture [3] (Supplementary Information section 1; Supplementary Table 1). These cultural changes and their early appearance at Shum Laka are particularly intriguing because the Cameroon/Nigeria border area during the late Holocene was likely the cradle of Bantu languages, and of populations whose descendants would spread across much of the southern half of Africa between ∼3000–1500 BP, resulting in the vast range and diversity of the Bantu language family [6–11].
A total of 18 human skeletons have been discovered at Shum Laka, comprising two distinct burial phases (Supplementary Information section 1) [1–3]. We attempted to retrieve DNA from six petrous bone samples and obtained working data from two early SMA and two late SMA individuals (∼8000 and ∼3000 BP, respectively; Table 1, Supplementary Table 2). The two earlier individuals—a boy of 4±1 years (2/SE I) lying on top of the lower limbs of an adolescent male of 15±3 years (2/SE II) [2]—were recovered from a primary double burial, while the two later individuals—a boy of 8±2 years (4/A) and a girl of 4±1 years (5/B) [2]—were in adjacent single burials.
Table 1:
Details for the four ancient Shum Laka individuals in the study
| ID | Age at death (yrs) | Date (cal BP) | Radiocarbon date (uncal) | Sex | Mt hap | Y hap | Cov | SNPs | Mt/X contam (%) |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 2/SE I | 4±1 | 7920–7690 | 6985 ± 30 BP (PSUAMS-6307) | M | L0a2a1 | B | 0.70 | 564164 | 1.0/1.0 |
| 2/SE II | 15±3 | 7970–7800 | 7090 ± 35 BP (PSUAMS-6308) | M | L0a2a1 | A00 | 7.71 | 1082018 | 1.5/0.6 |
| 4/A | 8±2 | 3160–2970 | 2940 ± 20 BP (PSUAMS-6309) | M | L1c2a1b | B2b | 3.83 | 935777 | 0.3/0.5 |
| 5/B | 4±1 | 3210–3000 | 2970 ± 25 BP (PSUAMS-6310) | F | L1c2a1b | .. | 6.41 | 1014618 | 0.5/.. |
Calibrated direct radiocarbon dates are given as 95.4% CI (Methods). Age (mean ± SE) was determined from skeletal remains [2], and sex from genetic data. Mt/Y hap, mtDNA/Y-chromosome haplogroup; Cov, average sequencing coverage.; Mt/X contam, estimated contamination from mtDNA/X chromosome. See also Supplementary Table 2.
We extracted DNA from bone powder and prepared 2–4 libraries per individual for Illumina sequencing, enriching for ∼1.2 million target single-nucleotide polymorphisms (SNPs) across the genome (Methods; Supplementary Table 2). Final coverage ranged from 0.7–7.7× (0.56–1.08 million SNPs). Authenticity of the data was supported by the observed rate of apparent C-to-T substitutions in the final base of sequenced fragments (4–10%, within the expected range given our library preparation strategy [14]) and of heterozygosity for mitochondrial DNA (mtDNA) and for the X chromosome in males (estimated contamination 0.3–1.5%). We also generated whole-genome shotgun sequence data for individuals 2/SE II (∼18.5× coverage) and 4/A (∼3.9×), as well as genome-wide data (∼598,000 SNPs) for 63 individuals from five present-day Cameroonian populations (Extended Data Table 1; Supplementary Table 3).
Uniparental markers and kinship analysis
All of the mtDNA and Y chromosome haplogroups we observe at Shum Laka are associated today with sub-Saharan Africans. The two earlier individuals carry mtDNA haplogroup L0a (specifically L0a2a1), which is widespread in Africa, while the two later individuals carry L1c (specifically L1c2a1b), which is found among both farmers and hunter-gatherers in Central and West Africa [15, 16]. Individuals 2/SE I and 4/A have Y chromosomes from macrohaplogroup B, often found today in Central African hunter-gatherers [17], while 2/SE II has the rare Y chromosome haplogroup A00, which was discovered in 2013 and is present at appreciable frequencies only in Cameroon, in particular among the Mbo and Bangwa in the western part of the country [12, 13]. A00 is the oldest known branch of the modern human Y chromosome tree, with a split time of ∼300,000–200,000 BP [12, 18, 19]. At 1666 positions (from whole-genome sequence data; Supplementary Table 4) that differ between present-day A00 [18] and all other Y chromosomes, the Shum Laka A00 carries the non-reference allele at 1521, translating to a within-A00 split at ∼37,000–25,000 BP (95% CI; Methods; Fig. 1).
Figure 1: Y chromosome phylogeny.
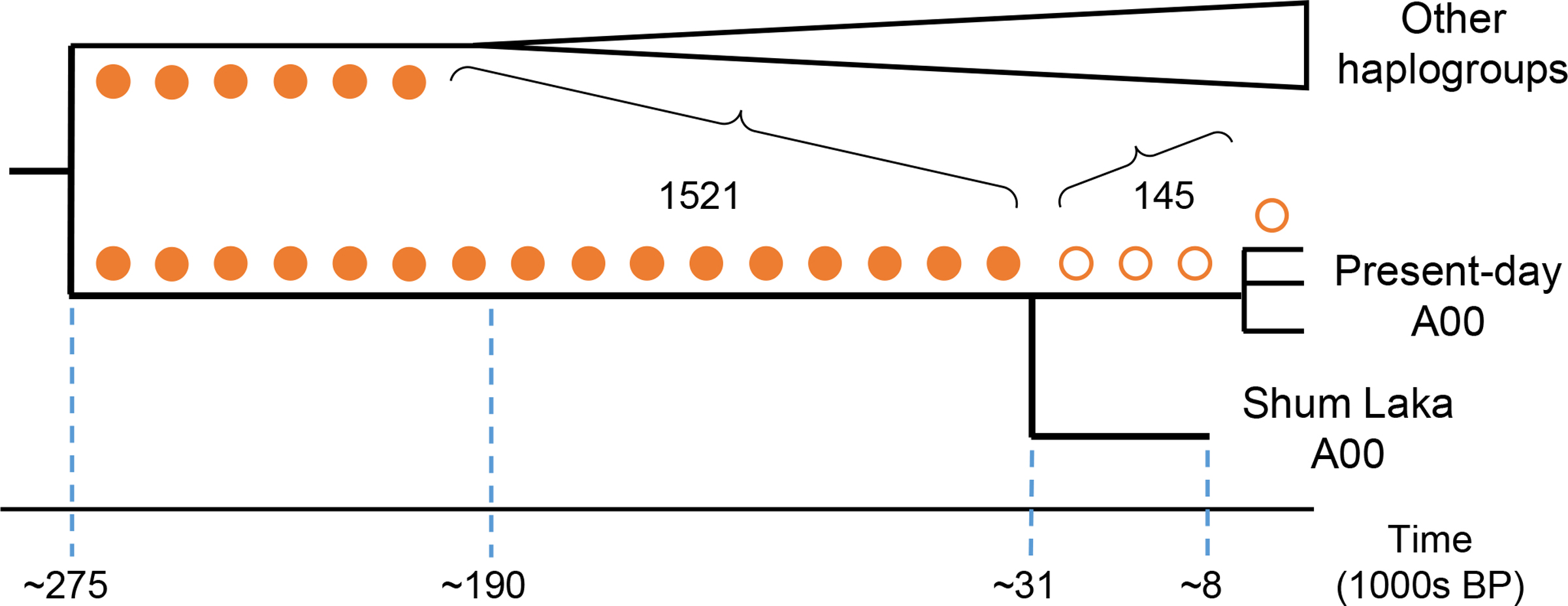
Circles represent mutations along the (unrooted) A00 lineage where we observe the alternative (filled) or reference (empty) allele in the Shum Laka A00.
Leveraging the effects of chromosomal segments shared identical by descent (IBD), we computed rates of allelic identity for each pair of individuals to infer degrees of relatedness. Both contemporaneous pairs display elevated identity, with 2/SE I and 2/SE II at the level of fourth-degree relatives and 4/A and 5/B at the level of second-degree relatives (either uncle and niece, aunt and nephew, or half-siblings; Extended Data Fig. 2), supporting archaeological interpretations that the rockshelter was used as an extended family cemetery during both burial phases [2]. We would expect more recent shared ancestry for the contemporaneous pairs even if they were not closely related, but we observe clear signatures of long IBD segments across the genome, confirming their close family relatedness (Supplementary Information section 2). All four individuals also show evidence of recent inbreeding (i.e., intra-individual IBD).
PCA and allele-sharing statistics
We visualized the genome-wide relationships between the Shum Laka individuals and diverse present-day and ancient sub-Saharan Africans (Extended Data Table 1) using principal component analysis (PCA). Initially, we computed axes using East and West Africans and southern and East-Central African hunter-gatherers (Fig. 2A). The Shum Laka individuals project to the right of Bantu speakers and related West African populations (Chewa, Mbo, and Mende), closest to present-day West-Central hunter-gatherers from Cameroon (Baka, Bakola, and Bedzan [20]) and the Central African Republic (Aka, often known as Biaka). We then carried out a second PCA using only West and East Africans and Aka to compute the axes, and again the Shum Laka individuals project in the direction of West-Central hunter-gatherers (Fig. 2B). By contrast, present-day Niger-Congo-speaking groups from western Cameroon cluster tightly with other West Africans (Fig. 2; Extended Data Fig. 3A). In both plots, the two earlier Shum Laka individuals fall slightly closer to West and East Africans, but based on their overall similarity, we grouped all four together for most subsequent analyses.
Figure 2: PCA results.
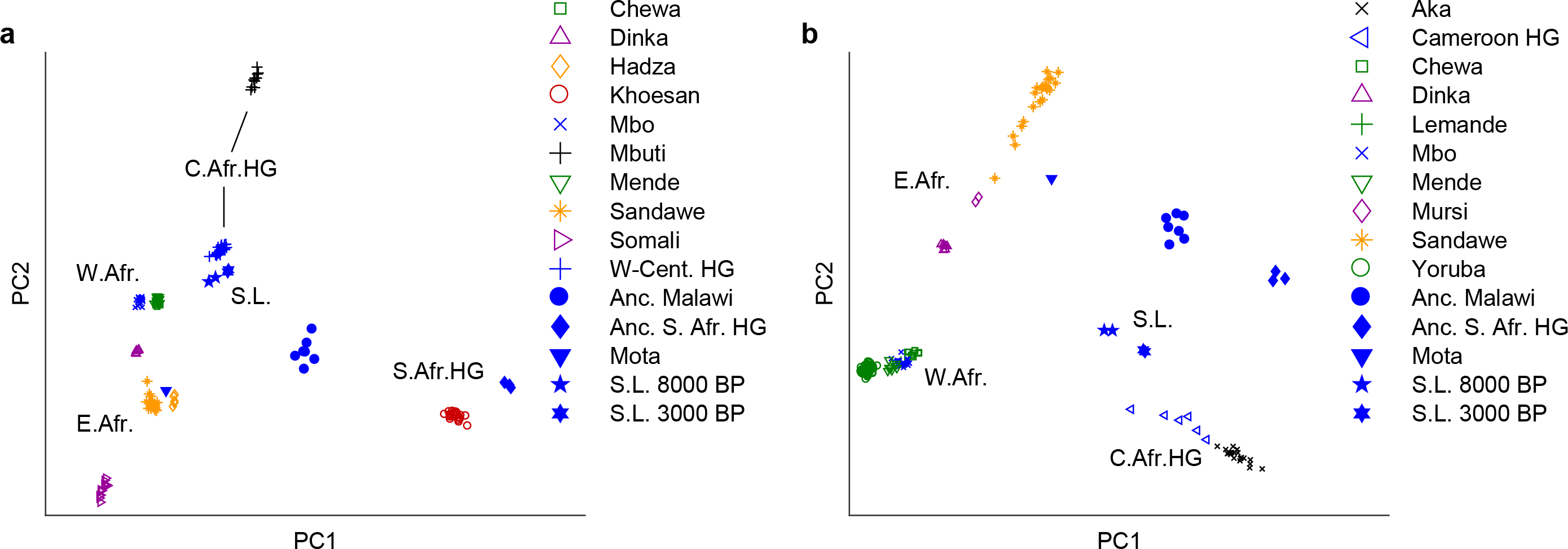
(A) Broad-scale analysis. (B) Narrow-scale analysis. Groups in blue (including ancient individuals, filled symbols) were projected onto axes computed using the other populations, using 593,124 SNPs (Methods). HG, hunter-gatherers; S.L., Shum Laka; W-Cent. HG, Aka plus Cameroon hunter-gatherers (Baka, Bakola, and Bedzan).
Using f-statistics (Fig. 3A), we investigated components of “deep ancestry” from sources diverging earlier than the split between non-Africans and most sub-Saharan Africans (above point (2) in Fig. 4A). We began with the statistic f4 (X, Mursi; South Africa HG, Han), which is expected to be increasingly positive for increasing deep ancestry in population X (via allele-sharing between X and ancient South African hunter-gatherers [21, 22]), with a baseline of zero set by Mursi, Nilotic-speaking pastoralists from western Ethiopia [20]. Shum Laka shows a large positive statistic, comparable to West-Central African hunter-gatherers (Fig. 3A, top), while other West Africans (e.g., Yoruba and Mende) yield smaller but significantly positive values, as do East African hunter-gatherers (Hadza from Tanzania and the ∼4500 BP Mota individual from Ethiopia [23]). We also obtained consistent results from analogous statistics with different reference groups (Extended Data Table 2).
Figure 3: Allele-sharing statistics.
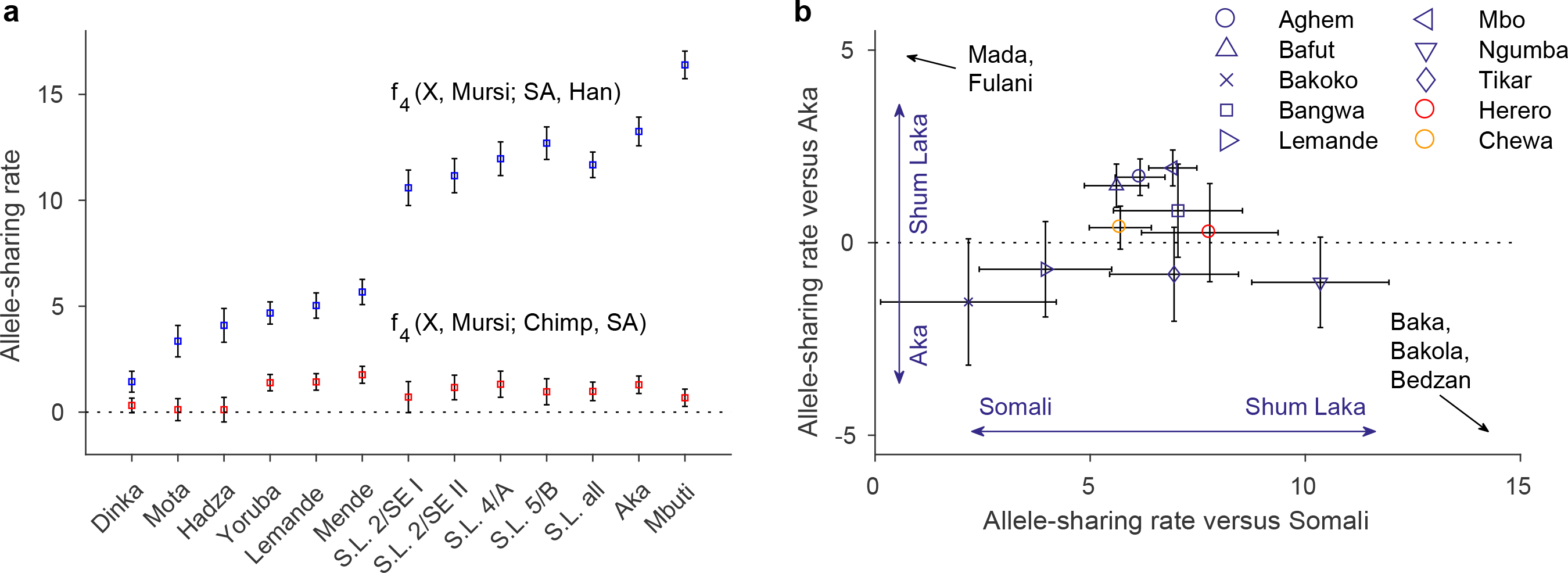
(A) Statistics sensitive to deep ancestry (mean ± 2SE, multiplied by 1000; blue, deeper than non-Africans; red, deeper than South African hunter-gatherers; computed on 1,121,119 SNPs). S.L., Shum Laka; SA, ancient South African hunter-gatherers. (B) Relative allele sharing (mean ± SE, multiplied by 10,000; computed on 538,133 SNPs) with Shum Laka versus East Africans (f4(X, Yoruba; Shum Laka, Somali); x-axis) and versus Aka (f4 (X, Yoruba; Shum Laka, Aka); y-axis) for present-day populations from Cameroon (blue) and southern (Herero, red) and eastern (Chewa, orange) Bantu speakers. See also Extended Data Fig. 3B.
Figure 4: Admixture graph results.
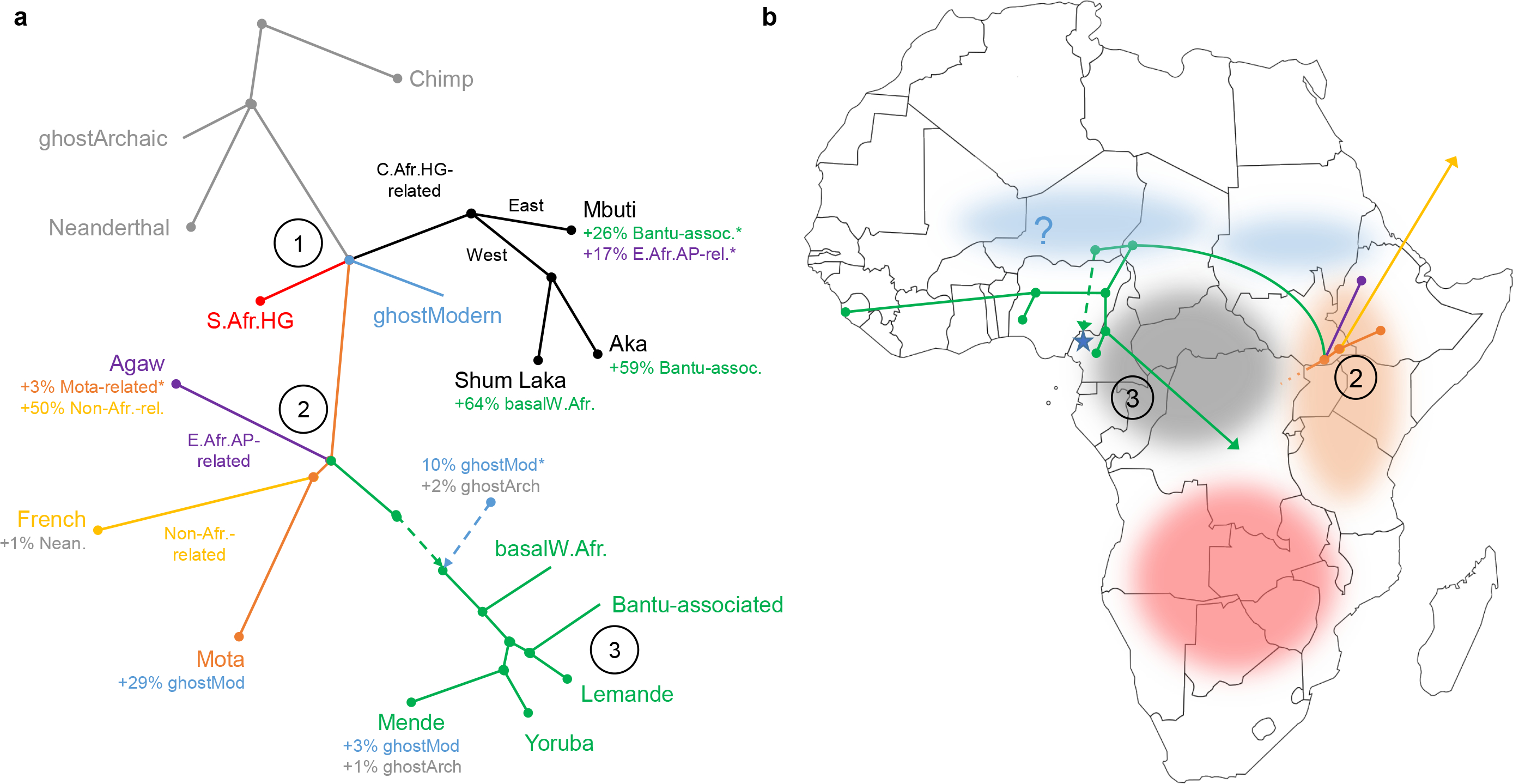
Points at which multiple lineages are shown diverging simultaneously indicate splits occurring in short succession (whose order we cannot confidently assess) but do not represent exact multifurcations. Key points are (1) early modern human split, (2) East African divergences, and (3) Bantu expansion. Branch lengths not drawn to scale. (A) Full model; see also Extended Data Fig. 4. HG, hunter-gatherer; AP, agro-pastoralist; *proportion not well constrained. (B) Geographical structure: shaded areas denote hypothesized historical locations of lineages descended from split point (1) in panel (A), and branching order is shown for populations descended from split point (2) (one ancestry component per population, with leaf nodes at sampling locations). The blue star represents Shum Laka (dashed line, possible direction of gene flow).
Next, we computed f4 (X, Mursi; Chimp, South Africa HG) (using chimpanzee as an outgroup symmetric to all human populations) to evaluate whether any of this deep ancestry is from sources diverging more deeply than southern African hunter-gatherers (the modern human lineage with the oldest known average split date [21, 24, 25]). Previous work has shown that southern African hunter-gatherers are not a symmetric outgroup relative to other sub-Saharan Africans, with West Africans (especially Mende) having excess affinity toward deeper outgroups [22]. Indeed, our test statistic is maximized in Mende and other West Africans (Fig. 3A, bottom). Hadza and Mota have values close to zero, and Shum Laka and Central African hunter-gatherers are intermediate. Some populations yield positive values for both f4-statistics (Fig. 3A), but the two sets are poorly correlated, implying that they in part reflect separate signals.
Combining our newly genotyped individuals with published data [20], we searched for differential allele-sharing between the Shum Laka individuals (compared to either East Africans [Somali] or Aka) and present-day Cameroonians (Fig. 3B, Extended Data Fig. 3B). We identified three distinct clusters: (a) Mada and Fulani, (b) hunter-gatherers, and (c) other Niger-Congo-speaking populations (in closeup in Fig. 3B). Within the third cluster are the only groups—Mbo, Aghem, and Bafut, all living close to Shum Laka today—with significantly Shum Laka-directed statistics in both dimensions, consistent with small proportions of Shum Laka-related admixture (maximum ∼7–8%; Supplementary Information section 3).
Admixture graph analysis
Finally, we built an admixture graph (Methods, Fig. 4A, Extended Data Fig. 4) co-modeling the ancient Shum Laka, Mota, and South African hunter-gatherer individuals; present-day Mbuti, Aka, Agaw (Afroasiatic speakers from Ethiopia [20]), Yoruba, Mende, and Lemande; non-Africans (French); and two outgroups (Altai Neanderthal and chimpanzee). We also fit versions of the model using alternative SNP ascertainments and additional populations (Hadza, Mbo, Herero, Chewa, Mursi, Baka, Bakola, Bedzan, Mada, Fulani, and ancient individuals from Taforalt in Morocco [26]) and obtained similar results (Extended Data Table 3; Supplementary Information section 3).
Among modern humans, the deepest-splitting branch is inferred to be the one leading to Central African hunter-gatherers, although four lineages diverge in a very short span: those contributing the primary ancestry to (a) Central African hunter-gatherers, (b) southern African hunter-gatherers, and (c) other modern human populations, along with (d) a “ghost” source contributing a minority of the ancestry in West Africans and the Mota individual. Central African hunter-gatherers separate into eastern (Mbuti) and western clades, with the latter then branching into components represented in Aka and Shum Laka. Next, a second cluster of divergences involves West Africans, two East African lineages (hunter-gatherer-associated and agro-pastoralist-associated), and non-Africans, the latter tentatively inferred to be a sister group to Mota but with no deep “ghost” ancestry. Within the West African clade, we identify Yoruba and Mende as sister groups, with Lemande as an outgroup, and most basally a separate West African-related lineage contributing to Shum Laka (64%). A Bantu-associated source (most closely related to Lemande) contributes 59% of the ancestry in Aka and 26% in Mbuti (who also harbor ancestry [17%] from an East African agro-pastoralist-related source). In a model separating the ∼8000 BP and ∼3000 BP Shum Laka pairs, the latter have ∼5% more Central African hunter-gatherer-related ancestry (as confirmed by the significantly positive statistic f4 (Shum Laka 8000 BP, Shum Laka 3000 BP; Yoruba, Aka) [Z=4.2]; Supplementary Information section 3).
We can also obtain a good fit for the Shum Laka individuals in a less-parsimonious alternative model using three components, replacing the basal West African source with a combination of ancestry from inside the clade defined by the other West African populations and from a source splitting between East and West Africans (near one lineage contributing to Taforalt; Extended Data Fig. 5, Supplementary Information section 3). However, two-component models for Shum Laka with the majority source splitting closer to other West or East Africans are rejected (Z=7.1 and Z=3.7, respectively).
The West African clade is distinguished by admixture from a deep source that can be modeled as a combination of modern human and archaic ancestry. The modern human component diverges at almost the same point as Central and southern African hunter-gatherers and is tentatively related to the deep source contributing ancestry to Mota, while the archaic component diverges close to the split between Neanderthals and modern humans (Supplementary Information section 3). The signals of deep ancestry in West African-related groups (Fig. 3A) can be explained by two admixture events: one along the ancestral West African lineage, and a second, smaller contribution (∼4%) to Mende from the same source (Fig. 4A). Accordingly, f4 -statistics testing for ancestry basal to southern African hunter-gatherers (Fig. 3A, bottom) are well correlated to inferred proportions of ancestry from the West African clade (Extended Data Fig. 6). We estimate the shared admixture to introduce 10% deep modern human and 2% archaic ancestry, although the first proportion is not well constrained (Extended Data Table 3). An alternative model with no archaic component, in which the West African clade receives deep ancestry from a single source [22] splitting before point (1) in Fig. 4A, also provides a reasonable fit to the data (Extended Data Fig. 5, Supplementary Information section 3), although it does not account for previous evidence of archaic ancestry in sub-Saharan Africans [27–31].
Shum Laka in genetic and archaeological context
Our analyses show that the four sampled children from Shum Laka can be modeled as admixed with ∼35% ancestry related to West-Central African hunter-gatherers and ∼65% from a basal West African-related source, or alternatively as a mixture of hunter-gatherer-related ancestry plus two additional components, one from inside the clade of present-day West Africans and one splitting between East and West Africans. The first component plausibly represents ancestry present in the area since at least the LSA, whereas the second component (third in the alternative model) may have originated farther to the north, given the geography and phylogeny of other sampled populations (Fig. 4B). The chronology of the archaeological record at Shum Laka suggests a possible northern influence on cultural developments during the SMA [3, 9]; these include changes in stone tools, which can be interpreted as a fusion of local LSA tool-making traditions with new macrolithic technologies introduced from the north [3], and the appearance of ceramics (four sherds found in the early SMA burial layer, and more abundant and distinct ceramics in later SMA deposits) potentially related to earlier pottery-working traditions in the Sahara and Sahel [3, 32]. Gene flow from the north before 8000 BP is also plausible due to a short period of Saharan and Sahelian aridification [3, 33]. Present-day groups in northern West Africa and the Sahel have substantial admixture connected to later migrations [34], so identifying the exact source area may await additional ancient DNA studies.
Although the scope of our sampling is limited to two individuals at either end of the SMA, the observed genetic similarity across a span of almost 5000 years—also consistent with skeletal morphometric analyses—suggests a long-term presence of related peoples who used the rockshelter for various activities, including burying their dead (Supplementary Information section 1). Today, however, most populations in Cameroon are more closely related to other West Africans than to the group represented by these individuals. Present-day hunter-gatherers in Cameroon are also not descended substantially from this specific group, as they lack the signal of basal West African ancestry (Supplementary Information section 3). We do observe elevated allele-sharing between the Shum Laka individuals and present-day Grassfields populations, so the genetic discontinuity is not absolute. Additionally, the adolescent male 2/SE II carried an A00 Y chromosome, suggesting that the concentration of this haplogroup in western Cameroon may have a long history, and moreover that A00 was formerly more diverse, given that the Shum Laka sequence falls outside of known present-day variation [12, 13]. The ∼300,000–200,000 BP divergence time of A00 from other modern human haplogroups [18, 19] could support its association either with the Central African hunter-gatherer-related ancestry component of the Shum Laka individuals or with the deep modern human portion of their West African-related ancestry.
Linguistic and genetic evidence points to western Cameroon as the most likely area for the development of Bantu languages and as the ultimate source of subsequent migrations of Bantu speakers, and while the regional mid-Holocene archaeological record is sparse, Shum Laka has been highlighted as possibly an important site in the early phase of this process [1–4, 6–11]. However, the genetic profiles of our four sampled individuals—even by ∼3000 BP, when the spreads of Bantu languages and of ancestry associated with Bantu-speaking populations was already underway—are very different from those of most Niger-Congo speakers today, implying that these individuals are not representative of the primary source population(s) ancestral to present-day Bantu speakers. These results neither support nor contradict a central role for the Grassfields area in the origins of Bantu-speaking peoples, and it may be that multiple, highly differentiated populations formerly lived in the region, with potentially either high or low levels of linguistic diversity. It would not be surprising if the Shum Laka site itself was used (either successively or concurrently) by multiple groups with different ancestry, cultural traditions, or languages [1], evidence of which may not be visible from the collection of remains as preserved today.
Implications for deep African population history
By analyzing data from Shum Laka and other ancient individuals in conjunction with present-day groups, we gain new insights into African population structure on multiple timescales. First, we infer a series of closely spaced population splits involving West African-related and two East African-related lineages, as well as non-Africans (point (2) in Fig. 4A). From the geography of the populations involved, the center of this radiation was plausibly in East Africa (Fig. 4B), with a date of ∼80,000–60,000 BP based on estimated divergences of African and non-African populations [24, 35]. Such an expansion is also consistent with mtDNA phylogeography—specifically the diversification of haplogroup L3, likely originating in East Africa ∼70,000 BP [36, 37]—and potentially with the origins of clade CT in the Y chromosome tree at a similar time depth [18, 38].
Second, we infer a phase of divergences involving at least four lineages early in the history of modern humans (point (1) in Fig. 4A). Recent consensus has been that southern African hunter-gatherers, who split from other populations ∼250,000–200,000 BP, represent the deepest sampled branch of modern human variation [21, 24, 25]. Our results suggest that Central African hunter-gatherers split at close to the same time (perhaps slightly earlier), and thus that both clades, as well as the lineage that would later diversify at point (2), originated as part of a large-scale African radiation.
In addition to the well-characterized deep lineages, we also detect at least one deep “ghost” source contributing to West Africans and East African hunter-gatherers. This signal corroborates previous evidence for Hadza and Sandawe [39] and for West Africans [22], although we find that the best fit is a source splitting near the same point as southern and Central African hunter-gatherers. Our results are also consistent with previous reports of archaic ancestry in African populations [27–31], specifically in West Africans. The presence of deep ancestry in the West African clade is notable in light of the Pleistocene archaeological record [5, 40], which includes Homo sapiens fossils dated to ∼300,000 BP in northwestern Africa [41], as well as an individual with archaic features buried ∼12,000 BP in southwestern Nigeria (the oldest known human fossil from West Africa proper) [42]. Middle Stone Age artifacts have also been found in parts of West Africa into the terminal Pleistocene [43], despite the development of LSA technologies elsewhere (e.g., Shum Laka). Thus, the available material and fossil evidence is concordant with our genetic results in indicating long-term African population structure and admixture [44, 45].
Further genetic studies may reveal additional complexities in deep human population history, while some early human groups will likely remain known only through fossils [44, 45]. Based on our current understanding, the presence of at least four modern human lineages that diversified ∼250,000–200,000 BP and are represented in people living today supports archaeological evidence that this was a pivotal period for human evolution in Africa.
Methods
Ancient DNA sample processing
We obtained bone powder from the Shum Laka skeletons (see Supplementary Information section 1 for more information on the site and burials) by drilling cochlear portions of petrous bone samples in a clean room facility at the Royal Belgian Institute of Natural Sciences. In dedicated clean rooms at Harvard Medical School, we extracted DNA using published protocols [46, 47]. From the extracts, we prepared barcoded double-stranded libraries treated with uracil-DNA glycosylase (UDG) to reduce the rate of characteristic ancient DNA damage [14, 48] in a modified partial UDG preparation including magnetic bead cleanups [14, 49]. For the SNP capture data, we used two rounds of in-solution target hybridization to enrich for sequences overlapping the mitochondrial genome and approximately 1.2 million genome-wide SNPs [50–54]. We then added 7-base-pair indexing barcodes to the adapters of each library [55] and sequenced on an Illumina NextSeq 500 machine with 76-base-pair paired-end reads. For individuals 2/SE II and 4/A, we also generated whole-genome shotgun data from the same libraries but without the target enrichment step. Sequencing was performed at the Broad Institute on an Illumina HiSeq X Ten machine, using 19 lanes for 2/SE II (yielding approximately 18.5× average coverage, including 1,216,658 sites covered from the set of target SNPs used in most analyses) and two lanes for 4/A (3.9× average coverage, 1,158,884 sites covered).
From the raw sequencing results, we retained reads with no more than one mismatch per read pair to the library-specific barcodes. Prior to alignment, we merged paired-end sequences based on forward and reverse mate overlaps and trimmed barcodes and adapters. Preprocessed reads were then mapped to both the mitochondrial reference genome RSRS [37] and the human reference genome (version hg19) using the “samse” command with default parameters in BWA (version 0.6.1) [56]. Duplicate molecules (having the same mapped start and end positions and strand orientation) were removed post-alignment. We filtered the mapped sequences (requiring mapping quality scores of at least 10 for targeted SNP capture and 30 for whole-genome shotgun data) and trimmed two terminal bases to eliminate (almost all) damage-induced errors.
For mitochondrial DNA, we called haplogroups using HaploGrep2 [57]. For nuclear DNA obtained from SNP capture and for the whole-genome shotgun data for individual 4/A, we selected one allele at random per site to create pseudo-haploid genotypes. For the whole-genome shotgun data for individual 2/SE II, we used a previously described reference-bias-free diploid genotype calling procedure [25], converting resulting genotypes into a fasta-like encoding allowing for extraction of data at specified sites via cascertain and cTools [25]. We determined the sex of each individual by examining the fractions of sequences mapping to the X and Y chromosomes [58], and we determined Y-chromosome haplogroups by comparing sequence-level SNP information to the tree established by the International Society of Genetic Genealogy (http://www.isogg.org).
To ensure authenticity, we computed the proportion of C-to-T deamination errors in terminal positions of sequenced molecules and evaluated possible contamination via heterozygosity at variable sites in haploid genome regions, using contamMix [50] and ANGSD [59] for mtDNA and the X chromosome (in males), respectively. Observed damage rates (4–10%) were relatively low but within the expected range after partial UDG treatment [14], and apparent heterozygosity rates for mtDNA (0.3–1.5% estimated contamination) and the X chromosome (0.5–1.0% estimated contamination) were minimal. The molecular preservation of the samples is impressive given the long-term warm and humid climate at Shum Laka [60] (supporting a mixed forest-savannah environment, at an elevation of ∼1650 meters above sea level).
Radiocarbon dates
At the Pennsylvania State University (PSU) Radiocarbon Laboratory, we generated new direct radiocarbon dates via accelerator mass spectrometry (AMS) for the four analyzed individuals, using fragments of the same temporal bone portions that were sampled for ancient DNA. We extracted and purified amino acids using a modified XAD process [61] and assessed sample quality via stable isotope analysis. C:N ratios for all four samples fell between 3.3 and 3.4, well within the nominal range of 2.9–3.6 indicating good collagen preservation [62]. The PSU dates were in good agreement with previously reported direct dates for different bones from individuals 2/SE II (8160–7790 cal BP, 7150 ± 70 BP, OxA-5203) and 4/A (3380–3010 cal BP, 3045 ± 60 BP, OxA-5205) [1, 2, 63, 64], but on the basis of a (modestly) aberrant date [65] from a rib of individual 2/SE I (Supplementary Table 5), we restricted our final reported results to the temporal bones. We performed calibrations using OxCal [66] version 4.3.2 with a mixture of the IntCal13 [67] and SHCal13 [68] curves, specifying “U(0,100)” to allow for a flexible combination [66, 69], and rounding final results to the nearest 10 years (see also Supplementary Information section 1).
New present-day data
We generated genome-wide SNP genotype data for 63 individuals from five present-day Cameroonian populations on the Human Origins array: Aghem (28), Bafut (11), Bakoko (1), Bangwa (2), and Mbo (21) (Extended Data Table 1; Supplementary Table 3). Samples were collected with informed consent, with collection and analysis approved by the UCL/UCLH Committee on the Ethics of Human Research, Committee A and Alpha.
A00 Y chromosome split time estimation
Present-day A00 Y chromosomes are classified into the subtypes A00a, A00b, and A00c, whose divergence times from each other have not been precisely estimated but are quite recent, perhaps only a few thousand years [12, 13]. To estimate the split time of the Shum Laka A00 Y chromosome from present-day A00, we called genotypes for individual 2/SE II (from our whole-genome sequence data) at a set of positions where sequences from two present-day individuals with haplogroup A00 [18] differ from all non-A00 individuals. (At every subtype-specific site for which we had coverage, the Shum Laka A00 carries the ancestral allele.) To avoid needing to determine the status of mutations as ancestral or derived, we considered the entire unrooted lineage specific to A00 (see Fig. 1). The total time span represented by this lineage is approximately 359,000 years, using published values of ∼275,000 BP for the divergence of the A00 lineage from other modern human haplogroups [19] and ∼191,000 BP for the next-oldest split within macrohaplogroup A [70]. With a requirement of at least 90% agreement among the reads at each site, we called 1521 positions as having the alternative allele (i.e., matching present-day A00 and differing from the human reference sequence) and 145 as having the reference allele (taking the average of 143 and 147 for the two present-day individuals). The fraction 145/(145+1521) then defines the position of the Shum Laka split along the (unrooted) A00 lineage. We note that split times computed either from all sites (relaxing the 90% threshold and using the majority allele), or from additionally requiring at least two reads per site, differ from our primary estimate by only a few hundred years. To produce a confidence interval, we used the variance in the published estimates and assumed an independent Poisson sampling error for the number of observed reference alleles. The final point estimate was ∼31,000 BP (95% CI: 37,000–25,000 BP), meaning that the Shum Laka A00 (with a sample date of ∼8000 BP) cannot be directly ancestral to the present-day subtypes.
PCA and allele-sharing statistics
We performed PCA using smartpca (with the “lsqproject” and “autoshrink” options) [71, 72] and computed f4-statistics using ADMIXTOOLS (with standard errors estimated via block jackknife over 5 cM chromosomal segments) [73]. We projected all ancient individuals in PCA rather than using them to compute axes in order to avoid artifacts caused by missing data. In each PCA, we also projected a subset of the present-day populations to allow controlled comparisons with ancient individuals. In most cases, reported f4-statistics are based on the approximately 1.15M autosomal SNPs from our target capture set. For PCA and for f4-statistics testing differential relatedness to Shum Laka, we used autosomal SNPs from the Human Origins array (a subset of the target capture set), with some populations in the analyses only genotyped on this subset (see Extended Data Table 1). For these latter f4-statistics, we excluded for all populations a set of roughly 40k SNPs having high missingness in the present-day Cameroon data.
Admixture graphs
We fit admixture graphs with the ADMIXTUREGRAPH (qpGraph) program in ADMIXTOOLS (with the options “outpop: NULL,” “lambdascale: 1,” “inbreed: YES,” and “diag: 0.0001”) [73–75], using the 1.15M autosomal SNPs from our target capture set by default, and other sets of SNPs in alternative model versions as specified. The program requires as input the branching order of the populations in the graph and a list of admixture events, and it then solves for the optimal parameters of the model (branch lengths and mixture proportions) via an objective function measuring the deviation between predicted and observed values of a basis set of f-statistics. From the inferred parameters, poorly fitting topologies (including positions of admixture sources) can be corrected by changing split orders at internal nodes that appear as trifurcations under the constraints enforced by the input (see Supplementary Information section 3).
To evaluate the fit quality of output models, we employed two metrics: first, a list of residual Z-scores for all f-statistics relating the populations in the graph, and second, a combined approximate log-likelihood score. The first metric is useful for identifying particularly poorly fitting models and the elements that are most responsible for the poor fits, while the second provides a means for comparing the overall fits of separate models (Supplementary Information section 3). In order to assess the degree of constraint on individual parameter inferences, we were guided primarily by the variability across different model versions (using different populations and SNP sets; see Extended Data Table 3 and Supplementary Information section 3), which reflects both statistical uncertainty and changes in model-specific assumptions. In our primary model, all f-statistics relating subsets of the populations are predicted to within 2.3 standard errors of their observed values.
Initially, we detected a slight but significant signal (max Z=2.5) of allele-sharing between Shum Laka and non-Africans, which we hypothesize is due to a small amount of DNA contamination. To prevent this effect from influencing our results, we included a “dummy” admixture of non-African ancestry into Shum Laka (inferred 1.1%, consistent with mtDNA- and X chromosome-based contamination estimates), although model parameters without the dummy admixture are also very similar (Extended Data Table 3, Supplementary Information section 3).
Data availability
The aligned sequences are available through the European Nucleotide Archive under accession number PRJEB32086. Genotype data used in analysis are available at https://reich.hms.harvard.edu/datasets.
Extended Data
Extended Data Figure 1: Overview of the site of Shum Laka.
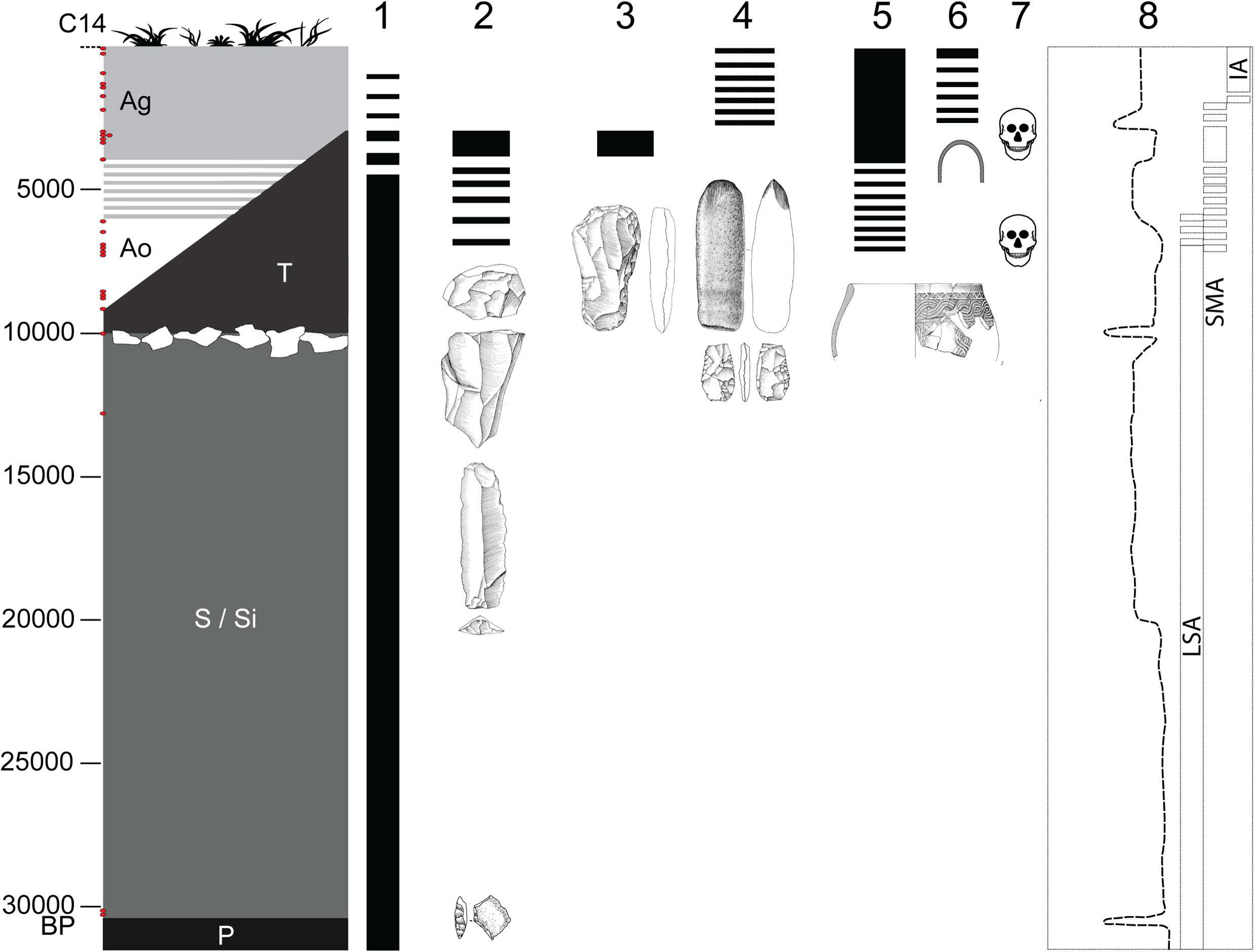
The left column represents generalized stratigraphy, with radiocarbon dates (uncalibrated) shown as red dots on the y-axis, and deposits indicated by their archaeological nomenclature (P, S/Si = Pleistocene; T, A = Holocene; Ao = Holocene ochre ashy layer; Ag = Holocene gray ashy layer; after ref. [76]). Columns 1–6 display chronological extents of technological traditions: 1, microlithic quartz industry; 2, macrolithic flake and blade industry on basalt; 3, bifaces of the axe-hoe type; 4, pecked grounded adze and arrow heads; 5, pottery; and 6, iron objects. Column 7 indicates the two Shum Laka burial phases. Column 8 shows climatic reconstructions based on carbon stable isotopes and pollen from organic matter extracted from sediment cores at Lake Barombi Mbo in western Cameroon (more arid conditions to the left and more humid conditions to the right [60, 76]), along with archaeological eras (LSA, Later Stone Age; SMA, Stone to Metal Age; IA, Iron Age). RMCA Collection; Drawings Y. Paquay, composition © RMCA, Tervuren; modified by E. Cornelissen [77].
Extended Data Figure 2: Kinship analysis.
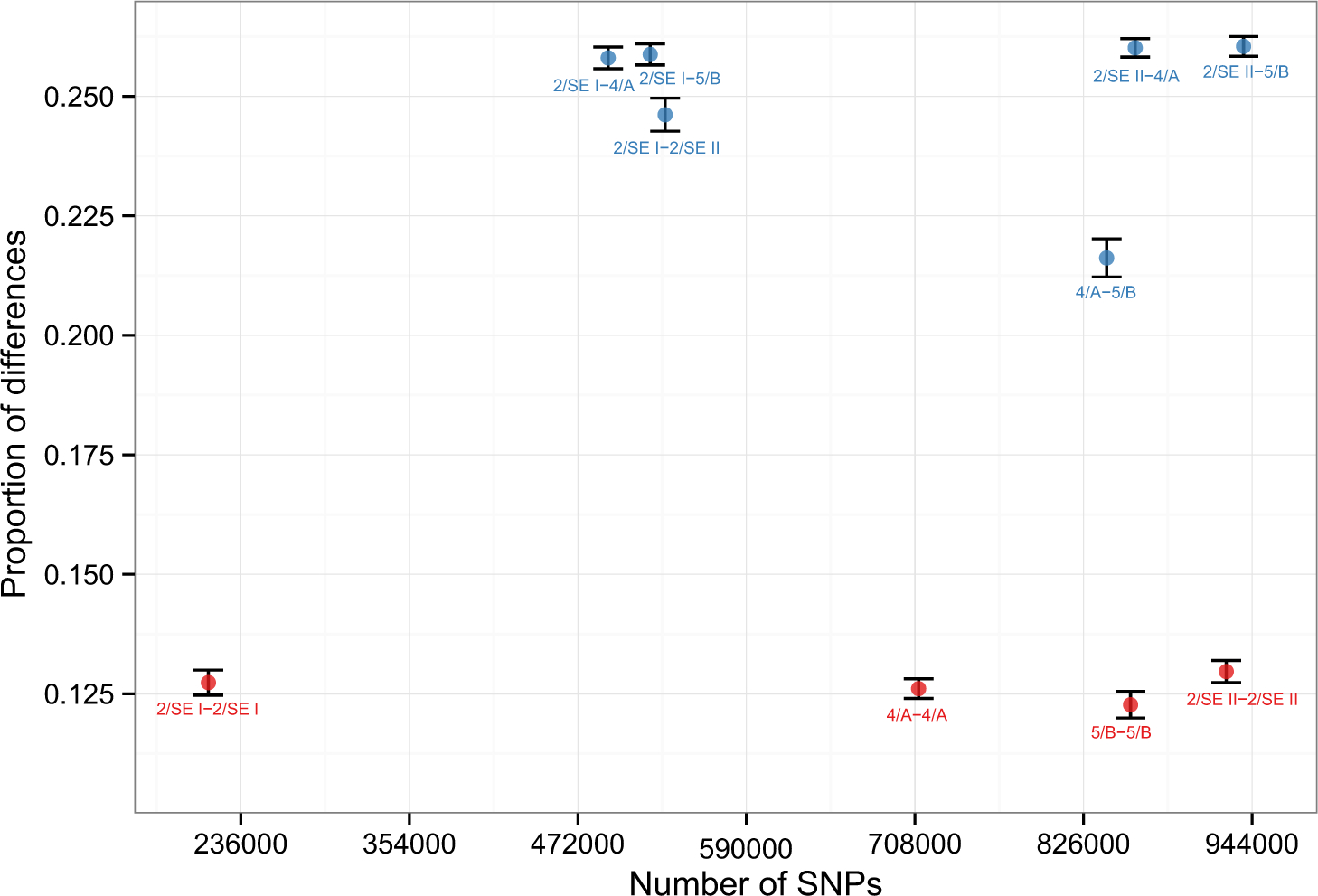
Shown are mean genome-wide allelic mismatch rates for each pair of individuals (blue), as well as intra-individual comparisons (red). We selected one read per individual at random at each targeted SNP (using all 1,233,013 targeted sites). Monozygotic twins (or intra-individual comparisons) are expected to have a value one-half as large as unrelated individuals; first-degree relatives, halfway between monozygotic twins and unrelated individuals; second-degree relatives, halfway between first-degree relatives and unrelated individuals; and so on. The presence of inbreeding also serves to reduce the rates of mismatches. For 4/A and 5/B, because both died as children, we can eliminate a grandparent-grandchild relationship, and the lack of long segments with both homologous chromosomes shared IBD implies that they are not double cousins (the few ostensible double-IBD stretches are likely a result of inbreeding; see Supplementary Information section 2). Thus, we can conclude that they were either uncle and niece (or aunt and nephew) or half-siblings. Bars show 99% confidence intervals (computed by block jackknife).
Extended Data Figure 3: Alternative PCA and allele-sharing analyses.
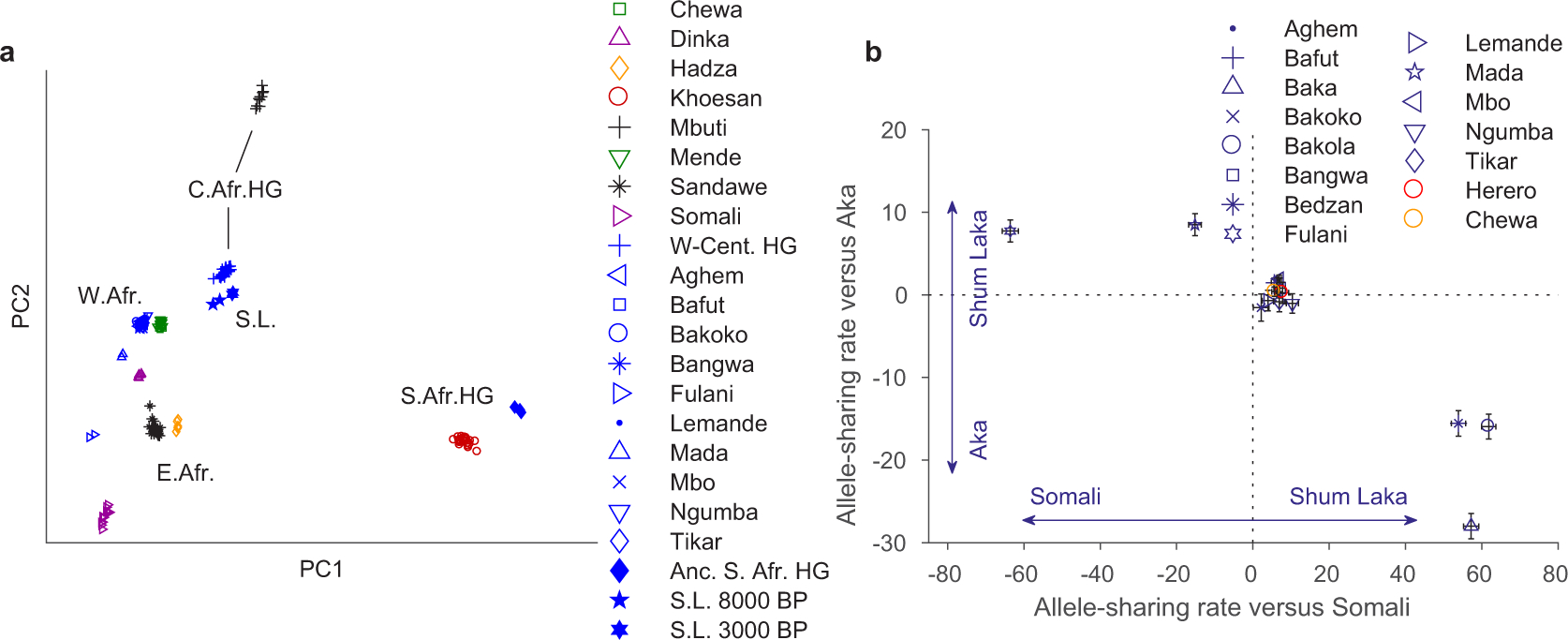
(A) Broad-scale PCA (differing from Fig. 2A by projecting all present-day Cameroon populations; again using 593,124 Human Origins SNPs). Groups shown in blue were projected onto axes computed using the other populations. HG, hunter-gatherers; S. L., Shum Laka. The W-Cent. HG grouping consists of Aka and Cameroon hunter-gatherers (Baka, Bakola, and Bedzan). The majority of the present-day Cameroon individuals fall in a tight cluster near other West Africans and Bantu speakers. (B) Relative allele sharing (mean ± SE, multiplied by 10,000, computed on 538,133 SNPs, as in Fig. 3B) with Shum Laka versus East Africans (f4 (X, Yoruba; Shum Laka, Somali); x-axis) and versus Aka (f4 (X, Yoruba; Shum Laka, Aka); y-axis) for present-day populations from Cameroon (blue points) and southern and eastern Bantu speakers (Herero in red and Chewa in orange). Mada and Fulani share more alleles with Shum Laka than with Aka, but this is likely a secondary consequence of admixture from East or North African sources (as reflected in greater allele sharing with Somali; see also Supplementary Information section 3). Bars show one standard error in each direction.
Extended Data Figure 4: Primary inferred admixture graph with full parameters.
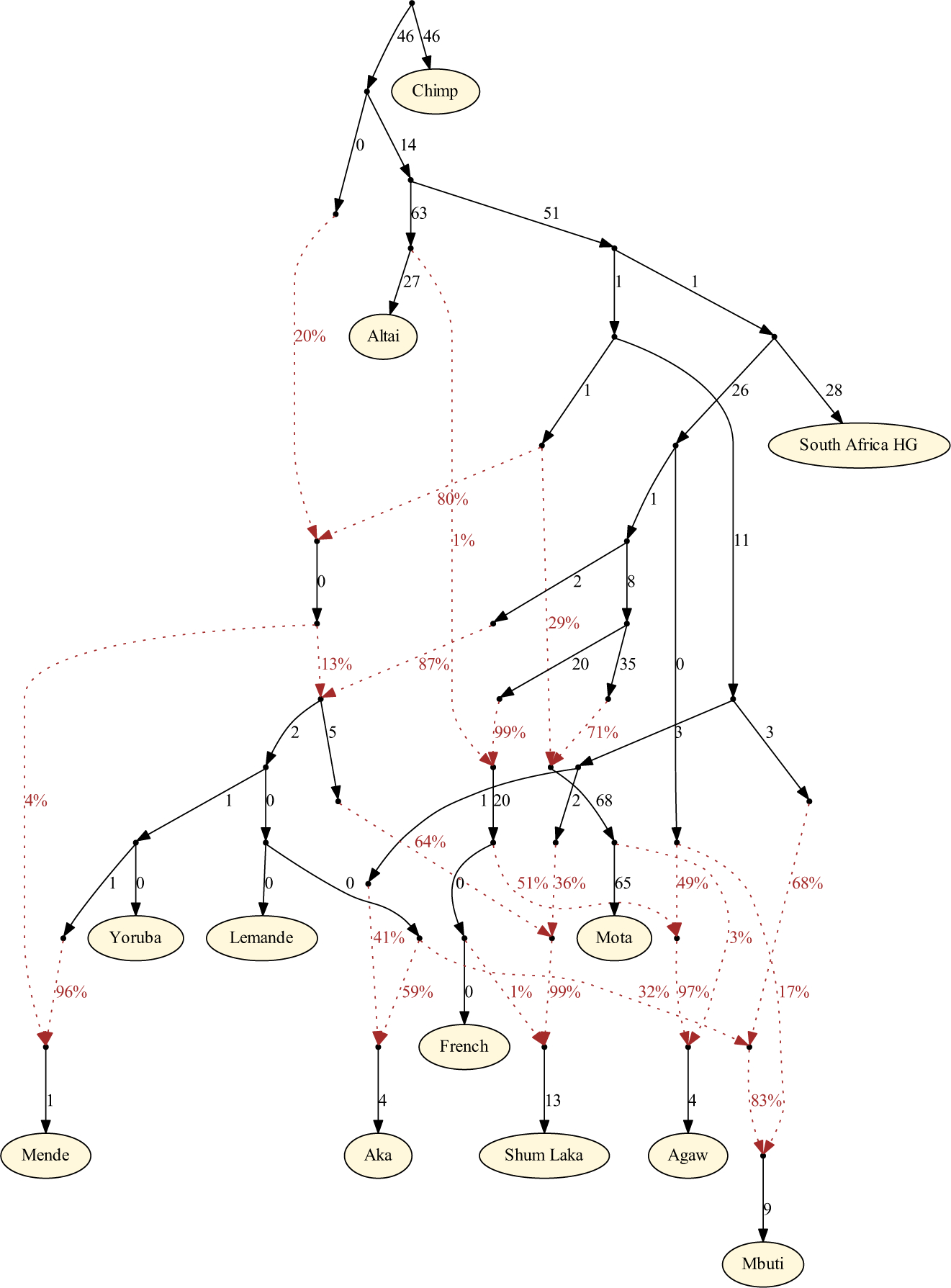
Of the ∼1.2M targeted SNPs, 932k are used for fitting (i.e., are covered by all populations in the model). Branch lengths (in units of squared allele frequency divergence) are rounded to the nearest integer. All f-statistics relating the populations are predicted to within 2.3 standard errors of their observed values.
Extended Data Figure 5: Schematic of first alternative admixture graph.
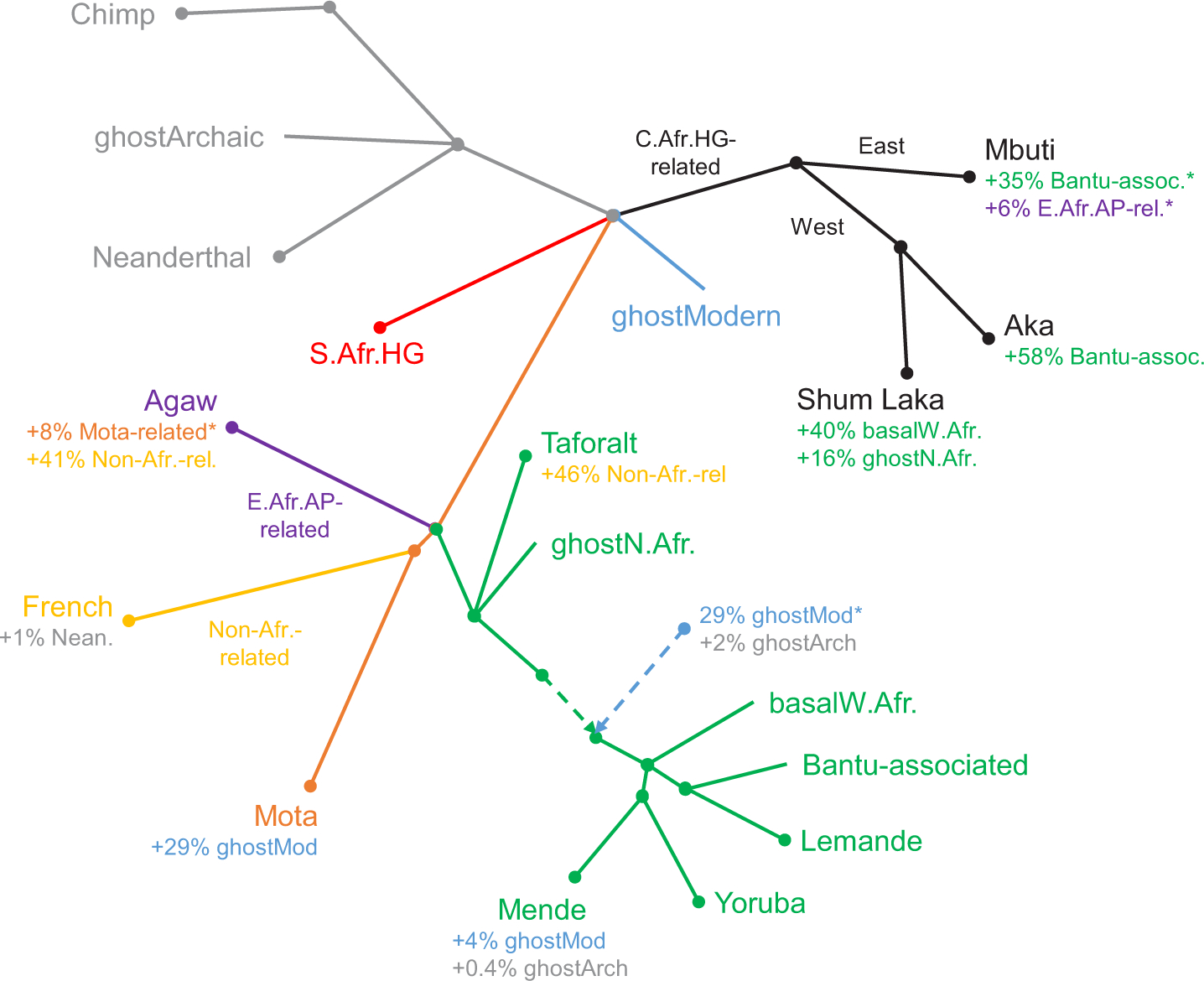
Results are shown including ancient individuals from Taforalt in Morocco associated with the Iberomaurusian culture, with the Shum Laka individuals modeled as having a mixture of hunter-gatherer-related ancestry plus two additional components: one from within the main portion of the West African clade, and one splitting at nearly the same point as one of the sources contributing ancestry to Taforalt. Branch lengths are not drawn to scale. Points at which multiple lineages are shown diverging simultaneously indicate splits occurring in short succession (whose order we cannot confidently assess) but are not meant to represent exact multifurcations. HG, hunter-gatherer; AP, agro-pastoralist. ∗ Proportion not well constrained (for Mbuti, the sum of the two indicated proportions is well constrained but not the separate values). See Supplementary Information section 3 for full inferred model parameters.
Extended Data Figure 6: Deep ancestry correlation from the West African clade.
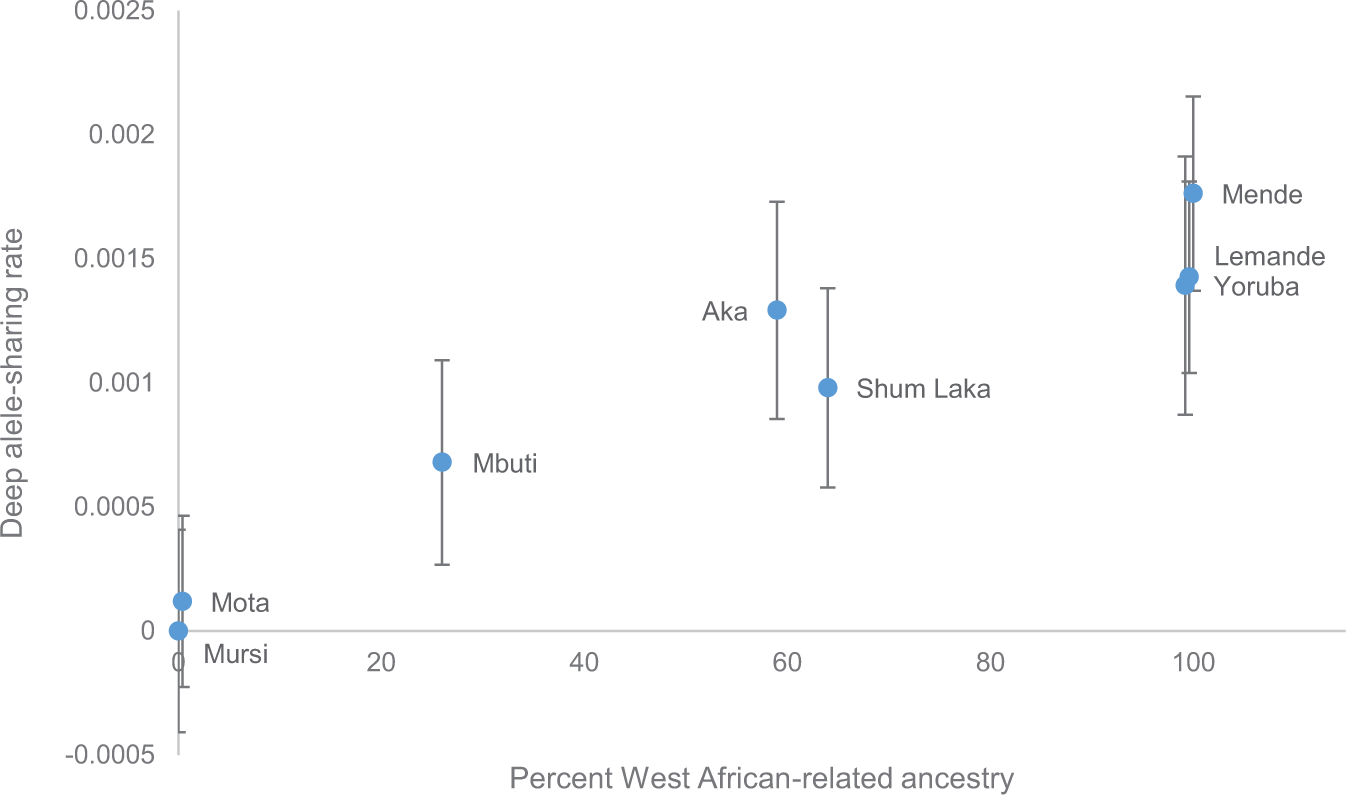
An allele-sharing statistic sensitive to ancestry splitting more deeply than South African hunter-gatherers (f4 (X, Mursi; Chimp, South Africa HG), mean ± 2SE from block jackknife, computed on 1,121,119 SNPs, as in Fig. 3A) is shown as a function of West African-related ancestry (from admixture graph results; Mota, Yoruba, and Lemande shifted slightly away from the boundaries for legibility). The (relative) allele-sharing rate for Mursi is identically zero according to the definition of the statistic.
Extended Data Figure 7: Schematic of second alternative admixture graph.
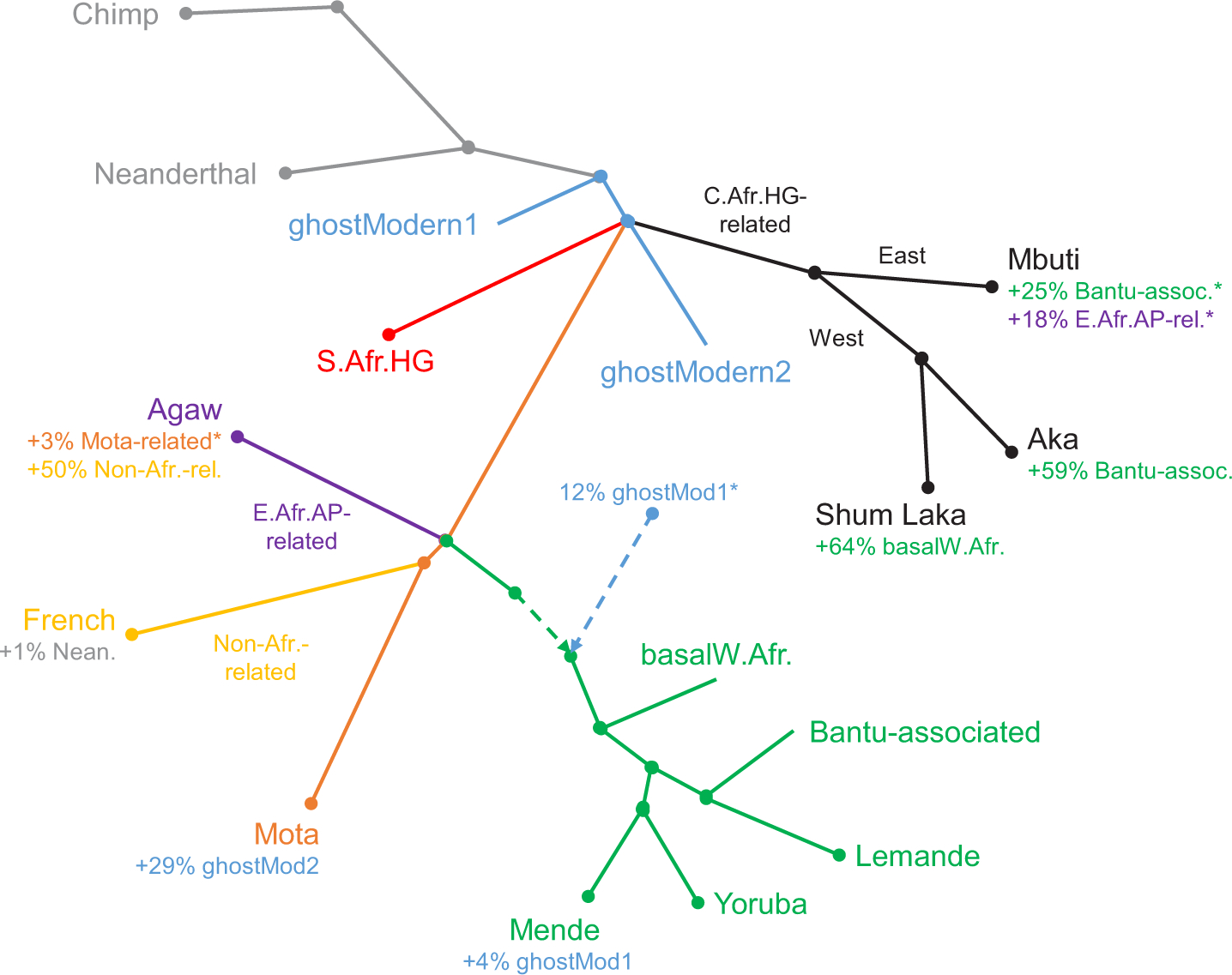
Results are shown with a single-component deep source for West Africans. Branch lengths are not drawn to scale. Points at which multiple lineages are shown diverging simultaneously indicate splits occurring in short succession (whose order we cannot confidently assess) but are not meant to represent exact multifurcations. HG, hunter-gatherer; AP, agro-pastoralist.*Proportion not well constrained (for Mbuti, the sum of the two indicated proportions is well constrained but not the separate values). See Supplementary Information section 3 for full inferred model parameters.
Extended Data Table 1:
Populations used in the study
| Population | Country | Language family | Date | Sample size | Data type | Reference |
|---|---|---|---|---|---|---|
|
| ||||||
| Shum Laka | Cameroon | ~8000–3000 BP | 4/1/1 | 1240k/DG/SG | This paper | |
| Ancient Malawi HG | Malawi | ~8100–2500 BP | 7* | 1240k | [22] | |
| Mota | Ethiopia | ~4500 BP | 1 | SG | [23] | |
| Ancient South African HG | South Africa | ~2000 BP | 3† | SG | [21,22] | |
| Taforalt | Morocco | ~15,000–14,000 BP | 6 | 1240k | [26] | |
| Altai Neanderthal | Russia | ~120,000 BP | 1 | DG | [78] | |
| Aghem | Cameroon | NC | Present | 28 | HO | This paper |
| Bafut | Cameroon | NC | Present | 11 | HO | This paper |
| Baka | Cameroon | NC | Present | 2 | DG | [20] |
| Bakoko | Cameroon | NC | Present | 1 | HO | This paper |
| Bakola | Cameroon | NC | Present | 2 | DG | [20] |
| Bangwa | Cameroon | NC | Present | 2 | HO | This paper |
| Bedzan | Cameroon | NC | Present | 2 | DG | [20] |
| Fulani | Cameroon | NC | Present | 2 | DG | [20] |
| Lemande | Cameroon | NC | Present | 2 | DG | [25] |
| Mada | Cameroon | AA | Present | 2 | DG | [20] |
| Mbo | Cameroon | NC | Present | 21 | HO | This paper |
| Ngumba | Cameroon | NC | Present | 2 | DG | [20] |
| Tikar | Cameroon | NC | Present | 2 | DG | [20] |
| Agaw | Ethiopia | AA | Present | 2 | DG | [20] |
| Aka (Biaka) | Central African Republic | NC | Present | 20/2 | HO/DG | [22,25] |
| Chewa | Malawi | NC | Present | 11 | HO | [22] |
| Dinka | Sudan | NS | Present | 7/4 | HO/DG | [22,25] |
| French | France | IE | Present | 3 | DG | [25] |
| Hadza | Tanzania | KS | Present | 5(2)/1 | HO/DG | [22,25] |
| Han | China | ST | Present | 4 | DG | [25] |
| Herero | Namibia | NC | Present | 2 | DG | [25] |
| Khoesan | Namibia | KS | Present | 22 | HO | [22] |
| Mbuti | DR Congo | NC, NS | Present | 10/4 | HO/DG | [22,25] |
| Mende | Sierra Leone | NC | Present | 8/2 | HO/DG | [22,25] |
| Mursi | Ethiopia | NS | Present | 2 | DG | [20] |
| Sandawe | Tanzania | KS | Present | 22 | HO | [22] |
| Somali | Kenya | AA | Present | 13 | HO | [22] |
| Yoruba | Nigeria | NC | Present | 70/3 | HO/DG | [22,25] |
List of populations used in analyses in the study. Data types are in-solution targeted SNP capture (1240k), whole-genome sequence with pseudo-haploid genotype calls (SG), high-coverage whole-genome sequence with diploid genotype calls (DG), and Human Origins SNP array (HO). For some populations, we used different sample sets for different analyses, indicated by slashes; Human Origins array genotyped individuals were used for PCA and for f-statistics testing differential relatedness to Shum Laka (Fig. 3B, Extended Data Fig. 3B). For Hadza, we used five individuals with Human Origins data for PCA and two of those five individuals for admixture graph modeling. HG, hunter-gatherers; AA, Afroasiatic; IE, Indo-European; KS, Khoesan; NC, Niger-Congo; NS, Nilo-Saharan; ST, Sino-Tibetan.
Individuals from Hora, Chencherere, and Fingira.
Individuals from Ballito Bay (A and B) and St. Helena Bay.
Extended Data Table 2:
Allele-sharing statistics for deep ancestry
| f 4 (X, Mursi; SA, Han) | f 4 (X, Mota; SA, Han) | f 4 (X, Han; SA, Mursi) | f 4 (X, Mota; SA, Mursi) | |||||
|
| ||||||||
| Test pop | Value | Z -score | Value | Z -score | Value | Z -score | Value | Z -score |
|
| ||||||||
| Dinka | 1.4 | 5.8 | −2.0 | −5.5 | 0.1 | 0.2 | −6.3 | −20.2 |
| Mota | 3.4 | 9.0 | 0 | 0 | 6.3 | 18.1 | 0 | 0 |
| Hadza | 4.1 | 10.3 | 0.8 | 1.7 | 7.3 | 21.2 | 1.0 | 2.7 |
| Yoruba | 4.7 | 17.8 | 1.3 | 3.8 | 5.2 | 18.2 | −1.1 | −3.5 |
| Lemande | 5.0 | 16.8 | 1.7 | 4.5 | 5.7 | 18.2 | −0.6 | −2.1 |
| Mende | 5.7 | 19.1 | 2.3 | 6.3 | 6.3 | 20.0 | 0 | 0 |
| Shum Laka | 11.7 | 38.7 | 8.3 | 22.6 | 12.7 | 40.8 | 6.4 | 20.5 |
| Aka | 13.3 | 39.1 | 9.9 | 25.2 | 13.6 | 40.4 | 7.3 | 22.0 |
| Mbuti | 16.4 | 50.4 | 13.0 | 34.9 | 16.4 | 49.9 | 10.0 | 31.8 |
| Mursi | 0 | 0 | −3.4 | −9.0 | .. | .. | .. | .. |
| Agaw | .. | .. | .. | .. | 0.1 | 0.3 | −6.2 | −18.9 |
| SA | .. | .. | .. | .. | .. | .. | .. | .. |
|
| ||||||||
| f 4 (X, Mursi; SA, Mota) | f 4 (X, Han; SA, Mota) | f 4 (X, Han; SA, Yor) | f 4 (X, Mursi; Chimp, Yor) | |||||
|
| ||||||||
| Test pop | Value | Z -score | Value | Z -score | Value | Z -score | Value | Z -score |
|
| ||||||||
| Dinka | 0.8 | 3.3 | 3.7 | 11.9 | −0.7 | −2.8 | −0.9 | −4.7 |
| Mota | .. | .. | .. | .. | 5.7 | 18.1 | 5.2 | 17.7 |
| Hadza | 4.1 | 11.5 | 7.0 | 17.7 | 4.8 | 15.2 | 3.4 | 11.4 |
| Yoruba | 4.1 | 15.7 | 7.1 | 21.6 | .. | .. | .. | .. |
| Lemande | 4.1 | 14.5 | 7.1 | 21.0 | .. | .. | .. | .. |
| Mende | 4.8 | 17.3 | 7.8 | 22.5 | .. | .. | .. | .. |
| Shum Laka | 9.1 | 29.8 | 12.0 | 33.7 | 8.0 | 28.7 | 8.3 | 31.9 |
| Aka | 10.3 | 33.4 | 13.2 | 35.5 | 7.8 | 24.8 | 8.5 | 30.1 |
| Mbuti | 12.5 | 41.8 | 15.5 | 44.1 | 11.6 | 40.8 | 11.8 | 46.3 |
| Mursi | 0 | 0 | 3.0 | 8.8 | 0.6 | 2.2 | 0 | 0 |
| Agaw | −2.4 | −7.7 | 0.6 | 1.8 | 0 | 0.2 | −0.2 | −0.9 |
| SA | .. | .. | .. | .. | .. | .. | 20.3 | 66.0 |
Variations of allele-sharing statistics (multiplied by 1000; computed on 1,121,119 SNPs) sensitive to ancestry in the test population X from a deeply-splitting lineage, along with Z-scores for difference from zero. We note that the zero level has a different meaning depending on which population is in the second position in the statistic. Blank entries are statistics that are confounded by specific relationships between the test population and one of the reference populations (in the third or fourth position; either duplication of the same group, Agaw with Han due to non-African-related ancestry, or Yoruba with other West Afrians). From the statistics f4 (Mursi/Agaw, Han; South Africa HG, Yoruba), we find minimal differences in deep ancestry proportions among Han, Mursi, and Agaw; from f4 (X, Mursi; Chimp, Yoruba), we obtain a value for South African hunter-gatherers that is roughly twice as large as for Central African hunter-gatherers. SA, ancient South African hunter-gatherers; Yor, Yoruba.
Extended Data Table 3:
Admixture graph parameter estimates
| Model version: | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||||
| Mixture proportions (%) | |||||||||||||||||||||||
|
| |||||||||||||||||||||||
| Shum Laka basal WA | 64 | 66 | 62 | 71 | 64 | 58 | 63 | 61 | 63 | 61 | 64 | 64 | 64 | 64 | 63 | 63 | 63 | 64 | 61 | 69 | 63 | 67/62* | |
| Aka Bantu-associated | 59 | 59 | 57 | 63 | 59 | 56 | 58 | 57 | 59 | 58 | 59 | 59 | 59 | 59 | 59 | 59 | 58 | 58 | 59 | 58 | 62 | 61 | 59 |
| Mbuti Bantu-associated | 26 | 24 | 33 | 19 | 28 | 27 | 26 | 12 | 28 | 30 | 32 | 25 | 24 | 26 | 29 | 28 | 35 | 35 | 25 | 35 | 23 | 36 | 27 |
| Mbuti East African-related | 17 | 19 | 10 | 27 | 14 | 9 | 16 | 23 | 15 | 13 | 11 | 19 | 20 | 18 | 13 | 14 | 6 | 6 | 18 | 9 | 23 | 8 | 16 |
| West African clade archaic | 2 | 2 | 4 | 4 | 3 | 3 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 3 | 3 | 2 | .. | .. | .. | .. | 2 |
| West African clade | 10 | 9 | 17 | 8 | 12 | 29 | 15 | 24 | 11 | 18 | 19 | 9 | 8 | 9 | 14 | 13 | 29 | 29 | .. | .. | .. | .. | 11 |
| deep modern human Mende deep | 4 | 4 | 4 | 3 | 4 | 3 | 4 | 6 | 5 | 5 | 5 | 4 | 4 | 4 | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 3 | 4 |
| ancestry Mota deep ancestry | 29 | 29 | 30 | 29 | 30 | 31 | 31 | 30 | 29 | 31 | 29 | 29 | 29 | 28 | 30 | 31 | 29 | 29 | 29 | 30 | 27 | 26 | 29 |
|
| |||||||||||||||||||||||
| Branch lengths | |||||||||||||||||||||||
|
| |||||||||||||||||||||||
| Basal WA split† | 2 | 3 | 3 | 3 | 3 | 1 | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 3 | .. | 2 | 3 | 3 | 1 | 3 |
| South African HG split‡ | 1 | 1 | 0 | 4 | 1 | −1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 4 | 0 | 1 |
| Ghost modern human split# | 1 | 1 | 1 | −3 | 1 | 1 | 0 | −2 | 1 | 0 | −1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | .. | .. | .. | .. | 2 |
Key admixture graph parameter estimates across different model versions (see Supplementary Information section 3 for full details): 1, primary model; 2, no “dummy” admixture; 3, African-ascertained SNPs; 4, transversion SNPs; 5, Shum Laka whole-genome sequence data; 6, outgroup-ascertained transversions; 7, Hadza added; 8, Mbo in place of Lemande; 9, Herero added; 10, Chewa added; 11, Mursi in place of Agaw; 12, Baka added; 13, Bakola added; 14, Bedzan added; 15, Mada added; 16, Fulani added; 17, Taforalt added; 18, alternative admixture for Shum Laka; 19, alternative deep source; 20, alternative deep source with African-ascertained SNPs; 21, alternative deep source with transversion SNPs; 22, alternative deep source with outgroup-ascertained transversions; 23, Shum Laka pairs fit separately. HG, hunter-gatherers.
Earlier pair/later pair
Units above the main West African clade
Units below the split of the Central African hunter-gather lineage (negative value indicates distance above)
Units along the Central African hunter-gather lineage (negative values indicate distances along an adjacent edge)
Supplementary Material
Acknowledgments
We thank Iosif Lazaridis, Vagheesh Narasimhan, and Kendra Sirak for discussions and comments; Monika Karmin for help with Y chromosome data; Laurie Eccles for help with radiocarbon dating; Brad Erkkila for help with isotopic analysis; Rebecca Bernardos, Matthew Mah, and Zhao Zhang for other technical assistance; Jean-Pierre Warnier for his role in locating the site of Shum Laka; and Otto Graf for proofreading, photo editing, and other figure assistance for the Supplementary Information. The Shum Laka excavations were supported by the Belgian Fund for Scientific Research (FNRS), the Université Libre de Bruxelles, the Royal Museum for Central Africa, and the Leakey Foundation. The collection of samples from present-day individuals in Cameroon was supported by Neil Bradman and the Melford Charitable Trust. I.R. was supported by a Université de Montréal exploration grant (2018–2020). M.G.T. was supported by Wellcome Trust Senior Investigator Award Grant 100719/Z/12/Z. G.H. was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant number 098386/Z/12/Z). C.L.F. was supported by Obra Social La Caixa 328, Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880), and a FEDER-MINECO grant (PGC2018-095931-B-100). Radiocarbon work was supported by the NSF Archaeometry program (grant BCS-1460369) to D.J.K. and B.J.C. M.E.P. was supported by a fellowship from the Radcliffe Institute for Advanced Study at Harvard University during the development of this project. D.R. was supported by the National Institutes of Health (NIGMS GM100233) and by an Allen Discovery Center grant, and is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- [1].de Maret P Shum Laka (Cameroon): human burials and general perspectives. In Pwiti G & Soper R (eds.) Aspects of African Archaeology: Papers from the 10th Congress of the Panafrican Association of Prehistory and Related Studies, 274–279 (University of Zimbabwe Publications, Harare, 1996). [Google Scholar]
- [2].Ribot I, Orban R & de Maret P The Prehistoric Burials of Shum Laka Rockshelter (North-West Cameroon). In Annales du Musée Royal de l’Afrique Centrale, vol. 164 (Musée Royal de l’Afrique Centrale, Tervuren, Belgium, 2001). [Google Scholar]
- [3].Lavachery P The Holocene archaeological sequence of Shum Laka rock shelter (Grassfields, western Cameroon). African Arch. Rev. 18, 213–247 (2001). [Google Scholar]
- [4].de Maret P Archaeologies of the Bantu expansion. In Mitchell P & Lane P (eds.) The Oxford Handbook of African Archaeology, 627–643 (Oxford University Press, 2013). [Google Scholar]
- [5].Cornelissen E Hunting and gathering in Africa’s tropical forests at the end of the Pleistocene and in the early Holocene. In Mitchell P & Lane P (eds.) The Oxford Handbook of African Archaeology, 403–417 (Oxford University Press, 2013). [Google Scholar]
- [6].Vansina J New linguistic evidence and ‘the Bantu expansion’. The Journal of African History 36, 173–195 (1995). [Google Scholar]
- [7].Tishkoff SA et al. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berniell-Lee G et al. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol. Biol. Evol. 26, 1581–1589 (2009). [DOI] [PubMed] [Google Scholar]
- [9].Bostoen K et al. Middle to late Holocene Paleoclimatic change and the early Bantu expansion in the rain forests of Western Central Africa. Curr. Anthropol. 56, 354–384 (2015). [Google Scholar]
- [10].Patin E et al. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science 356, 543–546 (2017). [DOI] [PubMed] [Google Scholar]
- [11].Bostoen K The Bantu Expansion. In Oxford Research Encyclopedia of African History (Oxford University Press, 2018). [Google Scholar]
- [12].Mendez FL et al. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am. J. Hum. Genet. 92, 454–459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Krahn T, Schrack B, Fomine FLM & Krahn A-M Searching for our most distant (paternal) cousins in Cameroon. Institute for Genetic Genealogy 2016 Conference, San Diego (2016). [Google Scholar]
- [14].Rohland N, Harney E, Mallick S, Nordenfelt S & Reich D Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Phil. Trans. R. Soc. B 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gonder MK, Mortensen HM, Reed FA, de Sousa A & Tishkoff SA Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol. 24, 757–768 (2006). [DOI] [PubMed] [Google Scholar]
- [16].Batini C et al. Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa. Mol. Phyl. Evol. 43, 635–644 (2007). [DOI] [PubMed] [Google Scholar]
- [17].Wood ET et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 13, 867 (2005). [DOI] [PubMed] [Google Scholar]
- [18].Karmin M et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 25, 459–466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mendez FL, Poznik GD, Castellano S & Bustamante CD The divergence of Neandertal and modern human Y chromosomes. Am. J. Hum. Genet. 98, 728–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fan S et al. African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations. Genome Biol. 20, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schlebusch CM et al. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358, 652–655 (2017). [DOI] [PubMed] [Google Scholar]
- [22].Skoglund P et al. Reconstructing prehistoric African population structure. Cell 171, 59–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gallego Llorente M et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820–822 (2015). [DOI] [PubMed] [Google Scholar]
- [24].Gronau I, Hubisz M, Gulko B, Danko C & Siepel A Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43, 1031–1034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mallick S et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van de Loosdrecht M et al. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- [27].Plagnol V & Wall JD Possible ancestral structure in human populations. PLoS Genet. 2, e105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hammer MF, Woerner AE, Mendez FL, Watkins JC & Wall JD Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. U. S. A. 108, 15123–15128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Durvasula A & Sankararaman S Recovering signals of ghost archaic admixture in the genomes of present-day Africans. bioRxiv preprint 285734 (2018). [Google Scholar]
- [30].Hey J et al. Phylogeny estimation by integration over isolation with migration models. Mol. Biol. Evol. 35, 2805–2818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ragsdale AP & Gravel S Models of archaic admixture and recent history from two-locus statistics. PLoS Genet. 15, e1008204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huysecom E et al. The emergence of pottery in Africa during the 10th millennium calBC: new evidence from Ounjougou (Mali). Antiquity 83, 905–917 (2009). [Google Scholar]
- [33].Gasse F Hydrological changes in the African tropics since the Last Glacial Maximum. Quat. Sci. Rev. 19, 189–211 (2000). [Google Scholar]
- [34].Triska P et al. Extensive admixture and selective pressure across the Sahel belt. Genome Biol. Evol. 7, 3484–3495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Laval G, Patin E, Barreiro L & Quintana-Murci L Formulating a historical and demographic model of recent human evolution based on resequencing data from noncoding regions. PLoS ONE 5, e10284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Soares P et al. The expansion of mtDNA haplogroup L3 within and out of Africa. Mol. Biol. Evol. 29, 915–927 (2012). [DOI] [PubMed] [Google Scholar]
- [37].Behar DM et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Poznik GD et al. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat. Genet. 48, 593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pickrell J et al. The genetic prehistory of southern Africa. Nat. Comm. 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Scerri E The Stone Age archaeology of West Africa. In Oxford Research Encyclopedia of African History (Oxford University Press, 2017). [Google Scholar]
- [41].Hublin J-J et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546, 289–292 (2017). [DOI] [PubMed] [Google Scholar]
- [42].Harvati K et al. The later Stone Age calvaria from Iwo Eleru, Nigeria: morphology and chronology. PLoS One 6, e24024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Scerri EM, Blinkhorn J, Niang K, Bateman MD & Groucutt HS Persistence of Middle Stone Age technology to the Pleistocene/Holocene transition supports a complex hominin evolutionary scenario in West Africa. J Archaeol. Sci. Rep. 11, 639–646 (2017). [Google Scholar]
- [44].Scerri EM et al. Did our species evolve in subdivided populations across Africa, and why does it matter? Trends Ecol. Evol. 33, 582–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Henn BM, Steele TE & Weaver TD Clarifying distinct models of modern human origins in Africa. Curr. Opinion Genet. Devel. 53, 148–156 (2018). [DOI] [PubMed] [Google Scholar]
Additional References
- [46].Dabney J et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U. S. A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Korlević P et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. BioTechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- [48].Briggs AW et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lipson M et al. Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fu Q et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U. S. A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Haak W et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fu Q et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mathieson I et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lazaridis I et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kircher M, Sawyer S & Meyer M Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucl. Acids Res. 40, e3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Weissensteiner H et al. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Skoglund P, Storå J, Götherström A & Jakobsson M Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol. Sci. 40, 4477–4482 (2013). [Google Scholar]
- [59].Korneliussen TS, Albrechtsen A & Nielsen R ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Giresse P, Maley J & Brenac P Late Quaternary palaeoenvironments in the Lake Barombi Mbo (West Cameroon) deduced from pollen and carbon isotopes of organic matter. Palaeogeography, Palaeoclimatology, Palaeoecology 107, 65–78 (1994). [Google Scholar]
- [61].Lohse JC, Culleton BJ, Black SL & Kennett DJ A precise chronology of middle to late Holocene bison exploitation in the far southern Great Plains. J. Texas Archeol. Hist. 1, 94–126 (2014). [Google Scholar]
- [62].Van Klinken GJ Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J Archaeol. Sci. 26, 687–695 (1999). [Google Scholar]
- [63].Lavachery P De la pierre au métal: archéologie des dépôts holocènes de l’abri de Shum Laka (Cameroun). Ph.D. thesis, Université libre de Bruxelles (1997). [Google Scholar]
- [64].Bronk Ramsey C, Higham TF, Owen D, Pike A & Hedges RE Radiocarbon dates from the Oxford AMS system: Archaeometry datelist 31. Archaeometry 44, 1–150 (2002). [Google Scholar]
- [65].Ward GK & Wilson SR Procedures for comparing and combining radiocarbon age determinations: a critique. Archaeometry 20, 19–31 (1978). [Google Scholar]
- [66].Ramsey CB & Lee S Recent and planned developments of the program OxCal. Radiocarbon 55, 720–730 (2013). [Google Scholar]
- [67].Reimer PJ et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon 55, 1869–1887 (2013). [Google Scholar]
- [68].Hogg AG et al. SHCal13 Southern Hemisphere calibration, 0–50,000 years cal BP. Radiocarbon 55, 1889–1903 (2013). [Google Scholar]
- [69].Marsh EJ et al. IntCal, SHCal, or a Mixed Curve? Choosing a 14C Calibration Curve for Archaeological and Paleoenvironmental Records from Tropical South America. Radiocarbon 60, 925–940 (2018). [Google Scholar]
- [70].Jobling MA & Tyler-Smith C Human Y-chromosome variation in the genome-sequencing era. Nat. Rev. Genet. 18, 485 (2017). [DOI] [PubMed] [Google Scholar]
- [71].Patterson N, Price A & Reich D Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu LT, Dobriban E & Singer A ePCA: High dimensional exponential family PCA. https://arxiv.org/abs/1611.05550 (2016).
- [73].Patterson N et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lipson M & Reich D A working model of the deep relationships of diverse modern human genetic lineages outside of Africa. Mol. Biol. Evol. 34, 889–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lipson M et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moeyersons J, Cornelissen E, Lavachery P & Doutrelepont H L’abri sous-roche de Shum Laka (Cameroun Occidental): Données climatologiques et occupation humaine depuis 30.000 ans. Géo-Eco-Trop 20, 39–60 (1996). [Google Scholar]
- [77].Cornelissen E A case study: Analyzing lithics from Shum Laka, NW Province, Cameroon. In Smith AL, Cornelissen E, Gosselain OP & MacEachern S (eds.) Field Manual for African Archaeology, 168–173 (Royal Museum for Central Africa, 2017). [Google Scholar]
- [78].Prüfer K et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The aligned sequences are available through the European Nucleotide Archive under accession number PRJEB32086. Genotype data used in analysis are available at https://reich.hms.harvard.edu/datasets.


