ABSTRACT
Human metapneumovirus (hMPV) is an important cause of acute viral respiratory infection. As the only target of neutralizing antibodies, the hMPV fusion (F) protein has been a major focus for vaccine development and targeting by drugs and monoclonal antibodies (MAbs). While X-ray structures of trimeric prefusion and postfusion hMPV F proteins from genotype A, and monomeric prefusion hMPV F protein from genotype B have been determined, structural data for the postfusion conformation for genotype B is lacking. We determined the crystal structure of this protein and compared the structural differences of postfusion hMPV F between hMPV A and B genotypes. We also assessed the receptor binding properties of the hMPV F protein to heparin and heparan sulfate (HS). A library of HS oligomers was used to verify the HS binding activity of hMPV F, and several compounds showed binding to predominantly prefusion hMPV F, but had limited binding to postfusion hMPV F. Furthermore, MAbs to antigenic sites III and the 66-87 intratrimeric epitope block heparin binding. In addition, we evaluated the efficacy of postfusion hMPV B2 F protein as a vaccine candidate in BALB/c mice. Mice immunized with hMPV B2 postfusion F protein showed a balanced Th1/Th2 immune response and generated neutralizing antibodies against both subgroup A2 and B2 hMPV strains, which protected the mice from hMPV challenge. Antibody competition analysis revealed the antibodies generated by immunization target two known antigenic sites (III and IV) on the hMPV F protein. Overall, this study provides new characteristics of the hMPV F protein, which may be informative for vaccine and therapy development.
IMPORTANCE Human metapneumovirus (hMPV) is an important cause of viral respiratory disease. In this paper, we report the X-ray crystal structure of the hMPV fusion (F) protein in the postfusion conformation from genotype B. We also assessed binding of the hMPV F protein to heparin and heparan sulfate, a previously reported receptor for the hMPV F protein. Furthermore, we determined the immunogenicity and protective efficacy of postfusion hMPV B2 F protein, which is the first study using a homogenous conformation of the protein. Antibodies generated in response to vaccination give a balanced Th1/Th2 response and target two previously discovered neutralizing epitopes.
KEYWORDS: crystal structure, fusion protein, heparan sulfate, heparin, human metapneumovirus, receptor binding
INTRODUCTION
Human metapneumovirus (hMPV) is a negative-sense single-stranded enveloped RNA virus in the family Pneumoviridae. There are two circulating genotypes of hMPV (A and B), which are further divided into four subgroups—A1, A2, B1, and B2—based on the sequence variability of the surface proteins (1). hMPV is one of the major causes of respiratory infections affecting infants and children under 5 years of age, and accounts for 6 to 40% cases of acute respiratory infections in hospitalized and outpatient children (2). Serological studies have shown that almost all people are exposed to hMPV by age 5 (3–5), and reinfections can happen throughout life. Premature infants, the elderly and immunocompromised patients are at high risk of severe disease caused by hMPV infection (2, 6). However, there are no licensed vaccines or specific treatments available for hMPV infection.
The fusion (F) protein of hMPV is highly conserved among hMPV subgroups, and it shares similar structural topology and approximately 30% amino acid sequence homology with the respiratory syncytial virus (RSV) F protein. In addition, the hMPV F protein, as with all studied pneumovirus and paramyxovirus F proteins, plays an indispensable role in viral infection. hMPV F belongs to the family of class I viral fusion proteins that mediate the fusion of viral envelope and cell membrane during infection. hMPV F is first synthesized as a polypeptide precursor, F0, and is then cleaved by an unknown enzyme to generate a F1-F2 heterodimer connected by disulfide bonds, which form the mature trimeric prefusion structure. The prefusion conformation of hMPV F is meta-stable and undergoes conformational rearrangement to the postfusion state during the process of membrane fusion (7). In addition, hMPV F is involved in virus attachment and receptor binding. Heparan sulfate (HS), a glycosaminoglycan that is ubiquitously expressed on the membrane surface of all animal tissues, has been hypothesized to be a receptor for the hMPV F protein (8). HS has been shown to block hMPV from infecting human lung cells and airway tissues in vitro (9). Integrin α5β1 is another potential cellular receptor for hMPV F, and the hMPV F protein has an Arg-Gly-Asp (RGD) binding motif (10, 11). Function-blocking monoclonal antibodies (MAbs) targeting α5β1 integrin, siRNA targeting α5 or β1, and EDTA all disrupt hMPV infection (12). Mutagenesis of the RGD motif inhibits cell-cell fusion, and mutant viruses have impaired growth in vitro and in vivo (13). However, there is still no evidence to show direct interactions between the hMPV F protein and these potential host receptors, and it is unclear whether hMPV F-specific MAbs can block receptor binding of hMPV F.
As the only target of neutralizing antibodies (14), hMPV F has been stabilized in both prefusion and postfusion conformations to facilitate recombinant expression and vaccine development (15, 16). The majority of hMPV F-specific human antibodies bind hMPV F in both prefusion and postfusion conformations (15, 17), while prefusion RSV F is preferred by neutralizing human antibodies (18). Like formalin-inactivated (FI)-RSV vaccines that induced aberrant immune responses and lead to enhanced respiratory disease in children after natural RSV reinfection (19–21), FI-hMPV and heat-inactivated hMPV vaccines also caused enhanced disease following viral infection in mice, cotton rats, and macaques potentially due to an abnormal Th2 immune response that leads to increased cytokine levels and lung inflammation (22, 23). Other forms of hMPV vaccines have also been explored in recent years. A recombinant live attenuated hMPV vaccine was tested in a phase I clinical trial in adults and children, but the vaccine was over attenuated and failed to efficiently infect hMPV-seronegative children (24). Several viral vector-based or virus-like particle-based hMPV vaccine candidates have also been evaluated in animal models and preclinical studies (14, 25, 26), which showed encouraging results. A bivalent fusion protein-based hMPV/PIV3 mRNA vaccine is currently under phase I clinical trials. With specific adjuvants, hMPV F-based subunit vaccines can induce protective immunity without enhancement of disease in cotton rats and nonhuman primates (27, 28), indicating hMPV F is a promising vaccine candidate. The crystal structure of postfusion hMPV A1 F has been solved and it can induce neutralizing antibodies after one immunization in concert with CpG adjuvant in mice (16). However, there remain several uncertainties regarding hMPV F vaccination, including the potential of postfusion hMPV F immunization to prevent viral replication, and the antigenic epitopes on the hMPV F protein targeted by B cells.
To further explore the structural, immunological, and receptor binding properties of hMPV F, we determined the X-ray crystal structure of the postfusion hMPV F protein from a genotype B strain. In addition, we determined the capacity of hMPV F to bind heparin and HS, and determined that neutralizing hMPV F MAbs can block heparin binding. Furthermore, the immunological properties of the protein were assessed by vaccination and challenge studies.
RESULTS
X-ray crystal structure of postfusion hMPV B2 F protein.
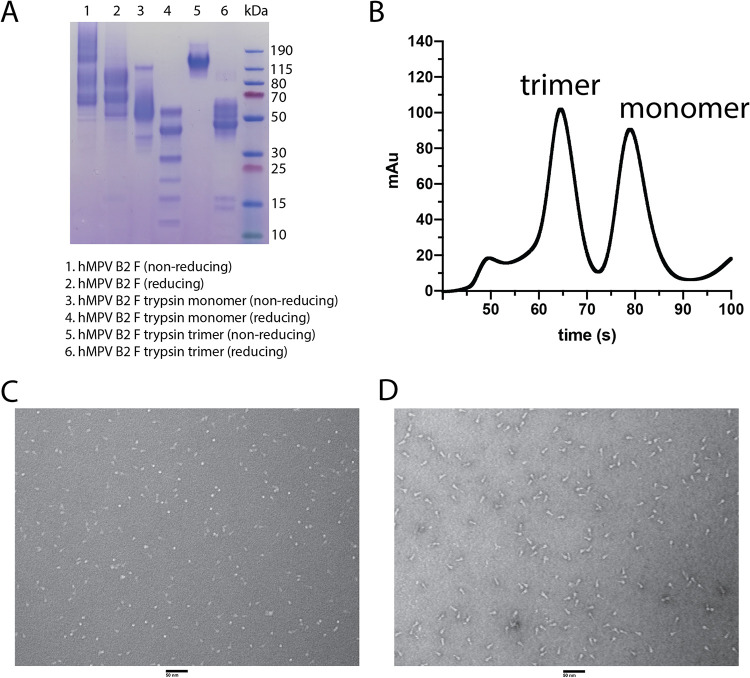
Stabilized postfusion hMPV A1 F protein has previously been generated by expression in CV-1 cells by removal of the hMPV F fusion peptide (residues 103 to 111) to prevent aggregation, replacing the transmembrane domain with the fibritin trimerization domain (Foldon) from T4 bacteriophage, and altering the cleavage site with the second furin-cleavage site of hRSV F (16). We previously reported a postfusion hMPV B2 F protein by incorporating similar genetic modifications, but this protein was expressed in HEK293F cells (29, 30). Here, we utilized this protein construct to determine the X-ray crystal structure of postfusion hMPV F from subgroup B2. Recombinantly expressed hMPV B2 F protein was subjected to trypsin digestion to induce cleavage, and both monomeric and trimeric hMPV F proteins were isolated by size exclusion chromatography (Fig. 1A and B). Limited cleavage was observed in a reducing SDS-PAGE without trypsin cleavage, however, the addition of trypsin results in multiple bands for the hMPV F monomer and a shift from the aggregated postfusion hMPV F to monomeric hMPV F for the trimeric fraction (Fig. 1A). We previously reported that the monomeric hMPV B2 F fraction is in the prefusion conformation based on the X-ray crystal structure of hMPV B2 F with the neutralizing MAb MPV458 (30). This trimeric fraction contains a mixture of elongated postfusion-like and spherical prefusion-like particles (Fig. 1C). To obtain homogeneous trimeric postfusion hMPV F, the protein was heated at 55°C to convert all particles to the postfusion conformation (Fig. 1D). This protein was crystallized and the X-ray crystal structure of this protein was determined to 3.1 Å (Fig. 2 and Table 1). Three N-linked glycans were observed at residues 57, 172 and 353 on each protomer, and residues that are different from A1 F are labeled in red (Fig. 2A). The overall structure of postfusion hMPV B2 F protein highly resembles the reported structure of postfusion hMPV F A1 F with an RMSD of 0.541 Å (Fig. 2B). Approximately 15 amino acid residues on the N terminus of the F1 domain are missing in the structure, possibly due to the cleavage by trypsin at site Arg129.
FIG 1.
Analysis of the hMPV B2 F protein. (A) SDS-PAGE of hMPV B2 F protein before and after trypsinization. Both monomeric and trimeric fractions were also isolated and analyzed. (B) Size exclusion chromatography curve of trypsinized hMPV B2 F protein. (C and D) Negative-stain electron micrograph of trypsinized hMPV B2 F trimeric protein before (C) and after (D) heating to 55°C. Scale bar, 50 nm.
FIG 2.

X-ray crystal structure of the postfusion hMPV B2 F protein. (A) The postfusion hMPV B2 F protein is displayed. Different residues between hMPV B2 F and A1 F are labeled in blue. (B) Overlay of postfusion hMPV A1 F (blue, PDB 5L1X) and B2 F (orange, PDB 7M0I).
TABLE 1.
Data collection and refinement statisticsa
| Parameter | Postfusion hMPV B2 F | After anisotropy correction |
|---|---|---|
| Data collection | ||
| Wavelength | 1 Å | |
| Space group | P 21 2 21 | |
| Cell dimensions | ||
| a, b, c (Å) | 114.5, 128.1, 431.3 | |
| α, β, γ (°) | 90, 90, 90 | |
| Resolution range (Å) | 49.63–2.811 (2.911–2.811) | a = 2.8, b = 2.9, c = 3.5 |
| Total reflections | 1,142,521 (110412) | |
| No. of unique reflections | 155,208 (1,266) | |
| Multiplicity | 7.4 (7.2) | |
| Rmerge | 0.535 (3.55) | 0.308 (0.716) |
| Mean I/σ〈I〉 | 5.6 (0.6) | 6.8 (2.1) |
| Completeness (%) | 99.7 (99.9) | 75.5 (8.2) |
| Refinement | ||
| Rwork/Rfree | 0.2542/0.2906 | |
| No. of atoms | 19,549 | |
| Protein | 19,279 | |
| Ligands | 252 | |
| Solvent | 18 | |
| RMSD | ||
| Bond lengths (Å) | 0.013 | |
| Bond angles (°) | 1.79 | |
| Ramachandran statistics | ||
| Favored regions (%) | 96.8 | |
| Allowed regions (%) | 3.0 | |
| Outliers (%) | 0.16 | |
| Rotamer outliers (%) | 1.34 | |
| Avg B-factor | 39.57 | |
| Macromolecules | 39.23 | |
| Ligands | 67.96 | |
| Solvent | 12.14 |
Statistics for the highest-resolution shell are shown in parentheses. RMSD, root mean square deviation.
Conformation dependence and epitope specificity of heparan sulfate binding by hMPV F.
We next sought to probe the receptor binding properties of this protein. Both heparan sulfate and integrin α5β1 have been identified as receptors for the hMPV F protein (8, 11). Since it is possible that receptor binding epitopes are not present in the postfusion conformation, we recombinantly expressed a previously described prefusion-like hMPV F protein containing the A185P stabilizing mutation, hMPV 130-BV (15). This protein contains the native hMPV F cleavage site and would not be proteolytically cleaved in HEK293F cells. To determine the conformation of the hMPV 130-BV F protein, we measured the binding of two prefusion-specific human MAbs, MPE8 (31), and the recently described MPV465 (30) by enzyme-linked immunosorbent assay (ELISA). The epitope for MPE8 on the RSV F protein spans two protomers at antigenic site III (32), and such an epitope would also be expected for hMPV F since this MAb neutralizes both viruses. In contrast, MPV465 binds monomeric hMPV F in the prefusion conformation (30). MPE8 had weak binding to monomeric and postfusion hMPV B2 F, while having 30-fold higher binding to the hMPV 130-BV F protein compared to postfusion hMPV B2 F (Fig. 3A). MPV465 had weak binding to postfusion hMPV B2 F, yet showed approximately 25-fold and 70-fold higher binding to hMPV 130-BV F and hMPV B2 F monomer, respectively. hMPV 130-BV F has a mixture of prefusion and postfusion-like particles in negative-stain electron micrographs (Fig. 3B). These data suggest the hMPV 130-BV F protein at least partially mimics the prefusion conformation of hMPV F. Binding of heparin to the hMPV B2 F protein was examined by surface plasmon resonance (SPR), and limited binding over the phosphate-buffered saline (PBS) control was observed for both monomeric (prefusion) and trimeric (postfusion) proteins (Fig. 3C). In contrast, heparin binding to hMPV F 130-BV showed an ∼60-fold improvement, suggesting a trimeric prefusion conformation is required for optimal heparin recognition (Fig. 3C). A hexahistidine peptide (Fig. 3D) was also tested as a control, to ensure that the his-tag has no contribution to heparin or HS saccharide binding by the protein. Heparin bound to hMPV F 130-BV with a Kd of 2.1 nM (Fig. 3E), and the binding can be inhibited by unfractionated heparin in a dose-dependent manner (Fig. 3F). To determine whether heparin binding is localized to a single epitope or binds to multiple sites, we assessed binding of heparin to hMPV F 130-BV in the presence of multiple concentrations of MAbs MPE8 and MPV458 (30). In both cases, the binding signal decreased as the concentration of MAb increased, with MPE8 demonstrating more inhibition than MPV458 (Fig. 3G and H). We also tested competition with MAbs 101F and DS7; however, these two MAbs bound nonspecifically to heparin (data not shown). Overall, these data suggest heparin binding to hMPV F occurs optimally to the trimeric prefusion conformation, and that binding occurs at or near the MPE8 and MPV458 epitopes.
FIG 3.
Recombinant hMPV F protein binds to heparin. (A) ELISA binding curves of MPE8 and MPV465 against hMPV B2 F monomer, hMPV B2 F postfusion trimer, and hMPV 130-BV F. (B) Negative-stain electron micrograph of hMPV 130-BV F protein. (C) Surface plasmon resonance (SPR) curves hMPV B2 F monomer, hMPV B2 F postfusion trimer, and hMPV 130-BV F binding to immobilized heparin. (D) Binding kinetics of 6×His-peptide to immobilized heparin. (E) Binding kinetics of hMPV 130-BV F at different concentrations (1.95 to 250 nM) to immobilized heparin. (F to G) Concentration-dependent inhibition of hMPV 130-BV F binding to immobilized heparin with unfractionated heparin (F), MPE8 (G), and MPV458 (H). The values of the data points in panels A and B are the averages of four replicates, and the error bars represent standard deviations. The data shown in panels E to H are from one experiment with at least two replicates.
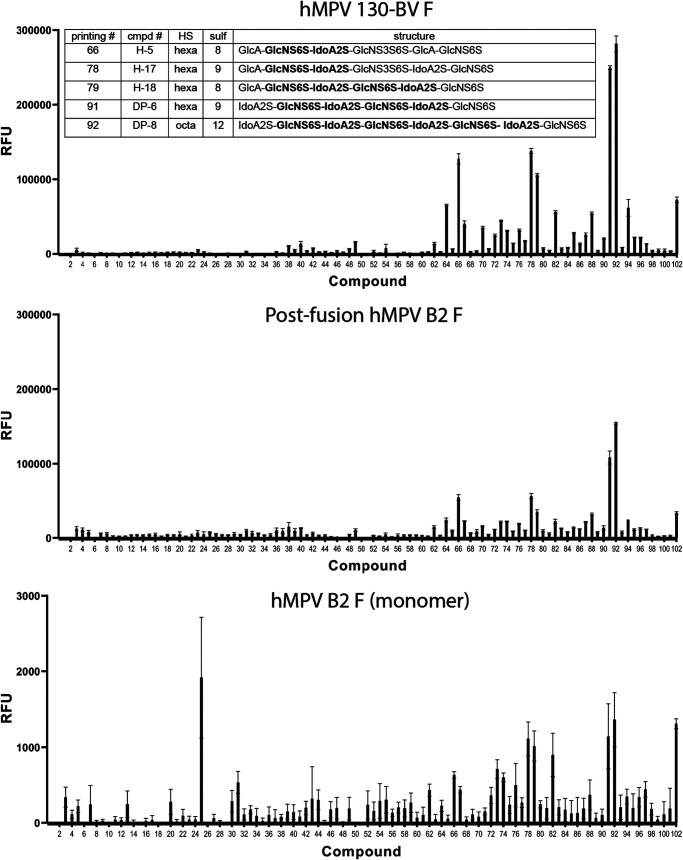
To further determine the motif on HS that is required for hMPV F binding, we tested binding of hMPV F to a panel of oligomers of HS (33, 34). Similar to heparin, hMPV 130-BV showed overall stronger binding to the HS oligomers than monomeric or postfusion hMPV B2 F. The microarray data showed hMPV F has a strong preference for specific HS oligosaccharides (compounds 66, 78, 79, 91, and 92) (Fig. 4; see also Table S1 in the supplemental material). All five compounds share the GlcNS6S-IdoA2S motif, indicating this unit potentially mediates the binding of HS to hMPV F. Since α5β1 integrin has also been shown to be a receptor for hMPV F (11, 12), we tested binding by ELISA and biolayer interferometry of multiple hMPV F constructs to recombinantly expressed integrin α5β1 using a stabilized construct as previously described (35), but we observed no binding of this protein to any recombinant hMPV F protein (data not shown). The lack of hMPV F binding to integrin α5β1 could be due to weak interactions, or the recombinant protein constructs may not be optimal for effective binding.
FIG 4.
Heparan sulfate binding to hMPV F proteins by microarray. Binding of synthetic heparan sulfate oligosaccharides to hMPV 130-BV, postfusion and monomeric hMPV B2 F proteins. The structures of the top five compounds for binding to hMPV 130-BV F protein are listed in the table. The value shown by each column is the average of six replicates, and the error bars indicate standard deviations.
Vaccination with postfusion hMPV B2 F induces a balanced immune response targeting two major epitopes.
While the prefusion conformation of RSV F elicits more robust neutralizing antibody titers due to the presence of prefusion-specific antigenic sites (18), the majority of antigenic sites on the hMPV F protein are present in both prefusion and postfusion conformations (15), suggesting the more stable postfusion F protein may be a viable vaccine candidate. However, there have been no studies to determine the protective efficacy of homogeneous postfusion hMPV F protein. hMPV challenge studies following hMPV F protein vaccination have recently been reported; however, all constructs in that study were mixtures of prefusion and postfusion conformations (17). To determine the immune response to vaccination with postfusion hMPV F, mice were immunized with postfusion hMPV F protein or PBS using a prime-boost-boost regimen as shown in Fig. 5A using TiterMax Gold adjuvant (Sigma-Aldrich). The serum from postfusion hMPV B2 F protein immunized mice showed increasing hMPV F-specific IgG levels after each immunization compared to prevaccination titers and the PBS+adjuvant-immunized group (Fig. 5B). Immunized mouse serum showed potent neutralization against both subgroups A2 (strain hMPV CAN/97-83) and B2 (strain hMPV TN/93-32) (Fig. 5C). In addition, a balanced Th1/Th2 immune response was observed since both robust IgG1 and IgG2a/2b were generated after the second boost (Fig. 5D). In all analyses, no major differences were observed between male and female groups of mice.
FIG 5.
Mouse vaccination study with postfusion hMPV B2 F. (A) Regimen of the vaccination study. (B) Serum hMPV F-specific IgG titers before (day −1) and after (days 14, 28, and 41) vaccinations. (C) Percent neutralization of 1/150 diluted endpoint serum against hMPV A2 (CAN/97-83) and B2 (TN/93-32). (D) Endpoint IgG subclasses titers against hMPV B2 F. Each data point represents a pool of serum samples from five mice in the same group. The values of the data points in panels B to D are the averages of four replicates.
To identify which antigenic sites were primarily targeted by postfusion hMPV F immunization, MAbs MPE8 (site III), 101F (site IV), DS7, and MPV458 (amino acids 66 to 87) were used in competition ELISAs with mouse serum against both monomeric (prefusion) and trimeric (postfusion) hMPV F. Anti-human secondary antibody was used to detect MAb binding to recombinant proteins, while anti-mouse secondary antibody was used to detect the binding of serum mouse antibodies. To confirm the conformation of the proteins, the prefusion-specific MAb MPE8 was used, and this MAb showed binding to the monomeric (prefusion) F protein but had limited binding to the trimeric (postfusion) hMPV F protein. In both male (groups 1 and 2) and female (groups 3 and 4) mice, MAb competition with mouse serum was observed for MAbs MPE8 and 101F, and no competition was observed for MAbs DS7 and 458 (Fig. 6A). These data suggest that antigenic sites III and IV are predominantly targeted by mouse B cells in response to postfusion hMPV F vaccination (Fig. 6B and C).
FIG 6.

Competition ELISA of postfusion hMPV B2 F vaccinated serum against human hMPV F MAbs. (A) Serial dilutions of vaccinated endpoint serum pooled from five mice in the same group competing with four human MAbs (MPE8-orange, 101F-blue, DS7-red, and MPV458-magenta). Mouse IgG was shown in dashed lines with solid symbols, while human IgG was shown in solid lines and empty symbols. Male mice are groups 1 and 2, and female mice are groups 3 and 4. The values of the data points are the averages of four replicates, and the error bars represent standard deviations. (B and C) The MAb binding sites of MPE8, 101F, DS7, and MPV458 are displayed on the surface of prefusion (B) and postfusion (C) hMPV B2 F.
Vaccination with postfusion hMPV B2 limited viral replication.
Since vaccination with postfusion hMPV B2 F elicited a robust and neutralizing IgG response, we sought to determine whether such vaccination can protect against viral replication. In this study, mice were primed and boosted with postfusion hMPV B2 F/PBS+TiterMax Gold adjuvant, then intranasally challenged with 5 × 105 PFU hMPV B2 TN/93-32 2 weeks after the boost (Fig. 7A). Vaccination with postfusion hMPV F limited viral replication below the detection limit for all mice, as no virus was detected by the plaque assay in the lung homogenates (Fig. 7B). Vaccine-enhanced disease is a potential concern with both RSV and hMPV, since formalin inactivation has previously been shown to exacerbate pulmonary pathological changes after challenge. We sectioned the lungs of two mice from each group and scored peribronchiolitis, perivasculitis, interstitial pneumonitis, and alveolitis as previously described (23). Mild to moderate pathological changes were observed in hMPV F- and PBS-vaccinated groups compared to uninfected mice (Fig. 7D).
FIG 7.
Mouse vaccination and challenge study with postfusion hMPV B2 F. (A) Regimen of the vaccination and challenge study. (B) Endpoint lung viral loads were quantified by PFU after immunostaining. Significant differences between unvaccinated and vaccinated groups are calculated by one-way analysis of variance (***, P = 0.002 via unpaired t test). The values of the data points are the averages of three replicates, and the error bars represent standard deviations. (C) Represented figures show the lung histopathology of untreated mice and vaccinated mice at ×2 magnification (left panel scale bar indicates 500 μm) and ×10 magnification (right panel scale bar, 100 μm). (D) Pulmonary pathology changes were quantified by scores of peribronchiolitis, perivasculitis, interstitial pneumonitis, and alveolitis.
DISCUSSION
In this study, we determined the X-ray crystal structure of postfusion hMPV F from genotype B and determined the receptor binding properties, immunogenicity, and protective efficacy of this homogeneous recombinant protein. The X-ray structure of postfusion hMPV B2 F closely aligns with the previously reported structure of postfusion hMPV A1 F (16). While the hMPV A1 F protein was expressed in CV-1 cells using a vaccinia virus expression system (16), the hMPV B2 F could be expressed in milligram quantities from HEK293F cells. It is worth noting that the Jardetzky group has previously demonstrated the hMPV B2 F protein can be observed in the postfusion conformation from HEK293F cells, although this protein contained a native cleavage and no high-resolution structure of the protein in the postfusion conformation was obtained (36). Overall, this expression protocol is reproducible and results in homogenous postfusion hMPV F particles.
Both integrin α5β1 and heparan sulfate have been previously reported to be potential receptors for the hMPV F protein (8, 9, 12, 13). We demonstrated binding of heparin to both postfusion hMPV B2 F and the prefusion-like protein hMPV 130-BV F. The prefusion conformation had much greater affinity for heparin than the postfusion conformation. Furthermore, we determined the hMPV 130-BV binding to heparin is dependent on the concentration of F protein. We also localized heparin binding to antigenic sites III and 66-87 using hMPV F neutralizing MAbs MPE8 and MPV458 (30, 31). Finally, we determined the optimal heparan sulfate oligomers that bind to the hMPV F protein.
It has previously been reported that the majority of neutralizing human antibodies in serum target both prefusion and postfusion conformations of the hMPV F protein, which suggests the postfusion hMPV F protein may contain the majority of neutralizing antigenic sites (15). Based on these findings, we hypothesized that the immunodominant epitopes on hMPV F are conserved in both prefusion and postfusion conformations. Therefore, immunization with postfusion F would induce antibodies that bind to prefusion F, which are capable of neutralizing the virus before fusion to host cells. To test this hypothesis, we vaccinated mice with postfusion hMPV B2 F protein and determined that such vaccination elicits neutralizing antibodies that primarily target antigenic sites III and IV. Antigenic site IV is conformationally conserved in both prefusion and postfusion conformations of hMPV F (37, 38). Antigenic site III elicits both prefusion-specific antibodies, such as MPE8, as well as antibodies that bind both prefusion and postfusion conformations (29, 32).
Previous studies have implicated multiple factors that can be attributed to the FI-RSV vaccine-enhanced disease. The postfusion conformation of the RSV F protein is dominant on FI-RSV particles (19), and such a vaccine cannot induce neutralizing antibodies to prefusion specific antigenic sites on the RSV F protein (39, 40). FI-RSV induces a Th2-skewed immune response that leads to eosinophil infiltration in the lungs, which may also contribute to the enhancement of the disease (41, 42). Similarly, FI-hMPV also boosts Th2 cytokines; however, neutralizing antibodies can still be induced to limit viral replication after challenge (22, 23). In this study, we found that postfusion hMPV B2 F elicits a balanced Th1/Th2 immune response when used with TiterMax adjuvant, which possibly alleviates the severity of pathological changes in the lungs. We then tested the protective efficacy of the hMPV B2 F protein and showed that vaccination completely protected mice from lung viral replication, consistent with result of the previous vaccination-challenge study using hMPV F proteins subtype A, which contained mixtures of prefusion and postfusion conformations (17). While lung viral titers were limited in the vaccinated groups, an increase in the total pathological score was observed for this group compared to the PBS-vaccinated group. While these groups were not large enough to assess statistical significance, assessing inflammation for hMPV vaccines is key to ensuring the safety of such vaccines as they move toward clinical trials. We did not directly compare these groups to a FI-inactivated hMPV vaccine group; therefore, we cannot make conclusions regarding potential enhanced disease, or whether the inflammation is a result of the particular mouse strain/model or the hMPV infection.
In summary, this work confirms the binding of hMPV F with a potential host receptor, heparan sulfate. The immunization and challenge studies bolster the potential of postfusion hMPV F to become an effective vaccine candidate. These findings will shed light on the development of novel drugs and vaccines against hMPV.
MATERIALS AND METHODS
Ethics statement.
All animal studies were approved by the University of Georgia Institutional Animal Care and Use Committee.
Production and purification of recombinant hMPV F proteins.
Plasmids encoding cDNA for hMPV B2 F and hMPV 130-BV F proteins were synthesized (GenScript) and cloned into the pcDNA3.1+ vector as previously described (29, 30). hMPV B2 F protein was derived from strain NL/1/94 and included residues 1 to 101, followed by the cleavage site KKRKRR and then residues 112 to 489. The sequence SGRENLYFQGGGGGSGYIPEAPRDQAYVRKDGEWVLLSTFLGGTEGRHHHHHH was appended to the C terminus and included, in order, a TEV cleavage site, Foldon trimerization domain, Xa cleavage site, and hexahistidine tag. The hMPV 130-BV protein was from strain NL/1/00 and included residues 1 to 485 with an A185P mutation, and the sequence SAIGGYIPEAPRDGQAYVRKDGEWVLLSTFLGGLVPRGSHHHHHH was appended to the C terminus and included a Foldon trimerization domain followed by a thrombin cleavage site and hexahistidine tag. The plasmids were expanded by transformation into Escherichia coli DH5α cells with 100 μg/ml of ampicillin (Thermo Fisher Scientific) for selection. Plasmids were purified using the E.Z.N.A. plasmid maxi kit (Omega BioTek), according to the manufacturer’s protocol. The stable cell line that expresses the hMPV B2 F protein was generated as previously described (30). For protein expression and purification, the stable cell lines were expanded in 500 ml of Freestyle293 medium supplemented with G418 at 1 × 106 cells/ml. After 5 to 7 days, recombinant protein was purified from the filtered culture supernatant using HisTrap Excel columns (GE Healthcare Life Sciences). Each column was stored in 20% ethanol and washed with 5 column volumes (CV) of wash buffer (20 mM Tris [pH 7.5], 500 mM NaCl, and 20 mM imidazole) before the samples were loaded onto the column. After sample application, columns were washed with 10 CV of wash buffer. Proteins were eluted from the column with 6 CV of elution buffer (20 mM Tris [pH 7.5], 500 mM NaCl, and 250 mM imidazole). Proteins were concentrated and buffer exchanged into PBS using Amicon Ultra-15 centrifugal filter units with a 30-kDa cutoff (Millipore Sigma).
Trypsinization of hMPV F.
In order to generate homogeneous cleaved trimeric hMPV F protein, trypsin-tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK; Thermo Scientific) was dissolved in double-distilled water (ddH2O) at 2 mg/ml. Purified hMPV B2 F was incubated with 5 TAME (p-toluene-sulfonyl-l-arginine methyl ester) U/mg of trypsin-TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone) for 1 h at 37°C. Trimeric hMPV B2 F protein was purified from the digestion reaction mixture by size exclusion chromatography on a Superdex S200, 16/600 column (GE Healthcare Life Sciences) in column buffer (50 mM Tris [pH 7.5], 100 mM NaCl). The trimeric hMPV F protein was identified by a shift in the elution profile from monomeric hMPV B2 F protein. The fractions containing the trimers and monomers were concentrated using 30 kDa Spin-X UF concentrators (Corning). To obtain homogenous postfusion hMPV F, trimeric trypsinized hMPV F protein was heated at 55°C for 20 min to induce conversion of remaining prefusion hMPV F protein to the postfusion conformation.
Negative-stain electron microscopy.
All samples were purified by size exclusion chromatography on a Superdex S200, 16/600 column (GE Healthcare Life Sciences) in column buffer before they were applied on grids. Carbon-coated copper grids (Electron Microscopy Sciences) were overlaid with 5 μl of protein solutions (10 μg/ml) for 3 min. The grid was washed in water twice and then stained with 0.75% uranyl formate for 1 min. Negative-stain electron micrographs were acquired using a JEOL JEM1011 transmission electron microscope equipped with a high-contrast 2K-by-2K AMT midmount digital camera.
Human MAb production and purification.
For recombinant MAbs, plasmids encoding cDNAs for heavy- and light-chain sequences of 101F (43), MPE8 (43), and DS7 (44) were synthesized (GenScript), and were cloned into vectors encoding human IgG1 and lambda or kappa light-chain constant regions, respectively. MAbs were obtained by transfection of plasmids into Freestyle HEK293F cells as described previously (30). For hybridoma-derived MAbs, hybridoma cell lines were expanded in serum-free medium (Hybridoma-SFM; Thermo Fisher Scientific). Recombinant cultures from transfection were stopped after 5 to 7 days, and hybridoma cultures were stopped after 30 days. Culture supernatants from both approaches were filtered and MAbs were purified from culture supernatants using HiTrap protein G columns (GE Healthcare Life Sciences) according to the manufacturer’s protocol.
Crystallization and structure determination of the trimeric postfusion hMPV B2 F.
Purified trypsinized postfusion hMPV B2 F protein was subjected to size exclusion chromatography (S200, 16/300, GE Healthcare Life Sciences) in 50 mM Tris (pH 7.5) and 100 mM NaCl. The fractions containing the trimeric hMPV F protein were concentrated to 12 mg/ml, and crystallization trials were prepared on a TTP LabTech Mosquito Robot in sitting-drop MRC-2 plates (Hampton Research) using several commercially available crystallization screens. Crystals were obtained in the Index HT (Hampton Research) in condition A6 (0.1 M Tris [pH 8.5], 2.0 M Ammonium sulfate). Crystals were harvested and cryo-protected with 30% glycerol in the mother liquor before being flash frozen in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source SER-CAT beamLine 21-ID-D. Data were indexed and scaled using XDS (45) and were significantly anisotropic. The data were submitted to the diffraction anisotropy server, and the data were truncated to 2.8 Å along the a* axis, 2.9 Å along the b* axes, and 3.5 Å along the c* axes (46, 47). A molecular replacement solution was obtained in Phaser (48) using the postfusion hMPV A1 F structure (PDB 5L1X). The crystal structure was completed by manually building in COOT (49), followed by subsequent rounds of manual rebuilding and refinement in Phenix. The data collection and refinement statistics are shown in Table 1.
HS microarray printing and screening.
All compounds were printed on NHS-ester activated glass slides (NEXTERION Slide H; Schott, Inc.) using a Scienion sciFLEXARRAYER S3 noncontact microarray equipped with a Scienion PDC80 nozzle (Scienion, Inc.). Individual samples were dissolved in sodium phosphate buffer (50 μl, 0.225 M [pH 8.5]) at a concentration of 100 μM and were printed in replicates of six with a spot volume of ∼400 pl at 20°C and 50% humidity. Each slide has 24 subarrays in a 3 × 8 layout. After printing, slides were incubated in a humidity chamber for 8 h and then blocked for 30 min with 5 mM ethanolamine in a Tris buffer (pH 9.0, 50 mM) at 40°C. Blocked slides were rinsed with deionized water (DI), spun dry, and kept in a desiccator at room temperature for future use. Screening was performed by incubating the slides with a protein solution for 1 h, followed by washing and drying. The buffers used in screening are TSM buffer (TSM; 20 mM Tris-Cl [pH 7.4], 150 mM NaCl, 2 mM CaCl2, and 2 mM MgCl2), TSM binding buffer (TSMBB; TSM buffer with 0.05% Tween 20 and 1% bovine serum albumin), and TSM washing buffer (TSMWB; TSM buffer with 0.05% Tween 20). A typical washing procedure includes sequentially dipping the glass slide in TSM wash buffer (2 min, containing 0.05% Tween 20), TSM buffer (2 min), and water (2 × 2 min), followed by spin dry.
The slides were incubated with His-tagged proteins diluted in TSMBB at different concentrations for 1 h, followed by washing and incubation with a solution of Alexa Fluor 647-conjugated anti-His antibody (BioLegend, 652513; 10 μg/ml). After washing and drying, the slides were scanned using a GenePix 4000B microarray scanner (Molecular Devices) at 635 nm with a resolution of 5 μM. Various gains and PMT values were employed for the scanning to ensure that all the signals were within the linear range of the scanner’s detector and there was no saturation of signals. The images were analyzed using GenePix Pro 7 software (version 7.2.29.2; Molecular Devices). The data were analyzed with a home written Excel macro. The highest and lowest values of the total fluorescence intensity of the replicate spots were removed, and the remaining values were used to provide the mean values and standard deviations (33, 34).
Heparin binding assay (SPR).
For the preparation of a heparin sensor chip, a CM5 chip was first coated with streptavidin by standard amine coupling using an amine coupling kit (Biacore; GE Healthcare), followed by immobilization of biotin-heparin (50). Briefly, the surface was activated using freshly mixed N-hydroxysuccinimide (NHS; 100 mM) and 1-(3-dimethylaminopropyl)-ethylcarbodiimide (EDC; 391 mM) (1/1, vol/vol) in water. Next, streptavidin (50 μg/ml; Invitrogen) in aqueous sodium acetate (10 mM [pH 4.5]) was passed over the chip surface until a ligand density of ∼2,000 RU was achieved. The remaining NHS-activated esters were quenched by aqueous ethanolamine (1.0 M [pH 8.5]). Next, biotin-heparin (50 μg/ml) was passed over one of the flow channels at a flow rate of 10 μl/min for 30 s resulting in a response of 83 RU. Next, the reference and modified flow cells were washed with three consecutive injections of 60 s with 1.0 M NaCl. Phosphate-buffered saline (PBS [pH 7.4]) was used as the running buffer for the immobilization, kinetic studies, and inhibition studies. Analytes were dissolved in running buffer, and a flow rate of 30 μl/min was employed for association (180 s) and dissociation (600 s) at a constant temperature of 25°C. A 30-s injection of 1.0 M NaCl at a flow rate of 30 μl/min was used for regeneration and to achieve prior baseline status. Using Biacore T100 evaluation software, the response curves of various analyte concentrations were globally fitted to a two-state binding model.
Growth of hMPV.
hMPV B2 strain TN/93-32 and hMPV A2 strain CAN/97-83 viruses were grown in LLC-MK2 cells (ATCC) as previously described (30). Briefly, cells were grown to 80% confluence in 225 cm2 flasks in Opti-MEM supplemented with 2% fetal bovine serum. For virus infection, cells were washed twice with Dulbecco phosphate-buffered saline (DPBS; Corning) and then infected with hMPV at 1:100 MOI, supplemented with 5 μg/ml trypsin-EDTA and 100 μg/ml CaCl2. Cells were incubated for 5 days, the medium was removed from the flask, and 5 ml of cold 25% (wt/vol) sterile-filtered sucrose was added to the flask. The flask was transferred to −80°C until the solution was frozen and then moved to thaw at 4°C, followed by another freeze-thaw cycle. Cell lysates were scraped and transferred to a sterile tube and centrifuged at 1,100 rpm for 5 min to remove cell debris. The clarified supernatant containing hMPV was flash frozen and titers were determined for later use.
Mice immunization and hMPV challenge.
BALB/c mice (6 to 8 weeks old; Charles River Laboratories) were immunized in a prime-boost-boost regimen with postfusion hMPV B2 F protein (50 μg protein/mouse) in a water in oil emulsion with TiterMax Gold adjuvant via the subcutaneous route into the loose skin over the neck, while mice in control groups were immunized with PBS+TiterMax Gold adjuvant emulsion. At 2 weeks after prime, the mice were boosted with the same amount of the emulsion, and a second boost without adjuvant was performed 2 weeks after the first boost. For the challenge study, BALB/c mice (6 to 8 weeks old; The Jackson Laboratory) were vaccinated with the same dosage as above, but only boosted once with adjuvant 4 weeks after the prime. At 2 weeks after the boost, mice were intranasally challenged with hMPV TN/93-32 (∼5 × 105 PFU/mouse). Mice were sacrificed 5 days postchallenge, and lungs were collected for virus titration and histological analysis.
Immunostaining of hMPV plaques.
For serum neutralization assays, heat-inactivated mouse serum was serially diluted (starting at 1:50, followed by 3-fold dilutions) and incubated 1:1 with a suspension of hMPV for 1 h at room temperature. LLC-MK2 cells in 24-well plates were then inoculated with the serum-virus mixture (50 μl/well) for 1 h and rocked at room temperature. Cells were then overlaid with 1 ml of 0.75% methylcellulose dissolved in Opti-MEM supplemented with 5 μg/ml trypsin-EDTA and 100 μg/ml CaCl2. Cells were incubated for 4 days, and then the cells were fixed with 10% neutral buffered formalin. The cell monolayers were then blocked with blocking buffer (2% Blotting-Grade Blocker [Bio-Rad] supplemented with 2% goat serum [Gibco] in PBS-Tween) for 1 h. The plates were washed with water, and 200 μl of MPV364 (29) primary antibody (1 μg/ml diluted in blocking buffer) was added to each well, and the plates were incubated for 1 h. The plates were then washed three times with water, and then 200 μl of goat anti-human IgG-horseradish peroxidase secondary antibody (catalog number 5220-0286; SeraCare) diluted 1:2,000 in blocking buffer was added to each well for 1 h. Plates were then washed five times with water, and 200 μl of TrueBlue peroxidase substrate (SeraCare) was added to each well. Plates were incubated until plaques were clearly visible. Plaques were counted by hand under a stereomicroscope and compared to a virus-only control, and the data were analyzed in GraphPad Prism using a nonlinear regression curve fit and the log(inhibitor)-versus-response function. To determine the viral load in lungs of challenged mice, the lungs were collected after euthanasia and homogenized with gentleMACS M Tubes (Miltenyi Biotec) in 1 ml of Opti-MEM, followed by centrifugation at 300 × g for 10 min to pellet the tissue debris. The supernatant was aliquoted and serial diluted with Opti-MEM, then plated on LLC-MK2 cells in 24-well plates (100 μl per well). After a 1 h incubation at 37°C, the cells were overlaid with 1 ml of 0.75% methylcellulose dissolved in Opti-MEM supplemented with 5 μg/ml trypsin-EDTA, 100 μg/ml CaCl2, and 1× antibiotic-antimycotic (Gibco). After 5 to 7 days, the plaques were stained in the same way described above.
ELISA for binding to hMPV F proteins.
For recombinant protein capture ELISAs, 384-well plates (Greiner Bio-One) were coated with 2 μg/ml of antigen in PBS overnight at 4°C. The plates were then washed once with water before blocking for 1 h with blocking buffer. Primary MAbs or serial dilutions of mouse serum (diluted 1:50, followed by 3-fold dilutions) were applied to wells for 1 h after three washes with water. Plates were washed with water three times before applying 25 μl of secondary antibody (goat anti-mouse IgG Fc; Southern Biotech, 1030-04) at a dilution of 1:4,000 in blocking solution. After incubation for 1 h, the plates were washed five times with PBS-Tween, and 25 μl of a PNPP (p-nitrophenyl phosphate) substrate solution (1 mg/ml PNPP in 1 M Tris base) was added to each well. The plates were incubated at room temperature for 1 h before reading the optical density at 405 nm (OD405) on a BioTek plate reader. Data were analyzed in GraphPad Prism using a nonlinear regression curve fit and the log(agonist)-versus-response function to calculate the binding EC50 values. Antibody titers were calculated from the highest dilution of a serum sample that produced OD readings of >0.3 above the background readings and were shown in a log10 scale as previously described (51). For competition ELISAs, 384-well plates were coated with 2 μg/ml of trypsinized hMPV B2 F monomer and trimer in PBS overnight at 4°C and then blocked for 1 h. Serial dilutions of mouse serum (diluted 1:100, followed by 3-fold dilutions) were premixed with equal volume of competing human MAbs (1 μg/ml) in blocking buffer and then applied to wells as primary antibodies for 1 h. Mouse IgG and human IgG were detected with separate secondary antibodies (goat anti-human IgG [Southern Biotech, 2014-14] and goat anti-mouse IgG Fc [Southern Biotech, 1030-04]) at a dilution of 1:4,000 in blocking buffer and PNPP as described above.
Pulmonary histopathological analysis.
After euthanasia, the lungs were collected, expanded through the trachea with 10% neutral buffered formalin (NBF) and immersion-fixed with 10% NBF. Fixed lungs were embedded in paraffin, sectioned at 4.0-μm thickness, mounted on positively charged glass slides, stained with hematoxylin-eosin, and coverslipped. Histological sections were evaluated by a board-certified veterinary pathologist. Histopathological scoring was performed according to previously established histopathologic criteria (23). Briefly, peribronchiolitis, perivasculitis, interstitial pneumonitis, and alveolitis were reviewed and scored on a scale of 0 to 4.
Data availability.
The structure factors and structure coordinates were deposited in the Protein Data Bank under accession code 7M0I.
ACKNOWLEDGMENTS
X-ray data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions (see www.ser-cat.org/members.htmL) and equipment grants (S10_RR25528 and S10_RR028976) from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank John Williams (University of Pittsburgh) for helpful discussions on the mouse challenge viruses. We thank Georgia Electron Microscopy at the University of Georgia for assistance with negative-stain electron microscopy.
These studies were supported by National Institutes of Health grants 1R01AI143865 (J.J.M.), 1K01OD026569 (J.J.M.), and 5R01HL151617 (G.-J.B.). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Jarrod J. Mousa, Email: jarrod.mousa@uga.edu.
Stacey Schultz-Cherry, St. Jude Children’s Research Hospital.
REFERENCES
- 1.den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, De Swart RL, Osterhaus ADME, Fouchier RAM. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis 10:658–666. 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafagati N, Williams J. 2018. Human metapneumovirus: what we know now. F1000Res 7:135. 10.12688/f1000research.12625.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung J, Esper F, Weibel C, Kahn JS. 2005. Seroepidemiology of human metapneumovirus (hMPV) on the basis of a novel enzyme-linked immunosorbent assay utilizing hMPV fusion protein expressed in recombinant vesicular stomatitis virus. J Clin Microbiol 43:1213–1219. 10.1128/JCM.43.3.1213-1219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavlin JA, Hickey AC, Ulbrandt N, Chan Y-P, Endy TP, Boukhvalova MS, Chunsuttiwat S, Nisalak A, Libraty DH, Green S, Rothman AL, Ennis FA, Jarman R, Gibbons RV, Broder CC, others. 2008. Human metapneumovirus reinfection among children in Thailand determined by ELISA using purified soluble fusion protein. J Infect Dis 198:836–842. 10.1086/591186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn SR, Ryder AB, Tollefson SJ, Xu M, Saville BR, Williams JV. 2013. Seroepidemiologies of human metapneumovirus and respiratory syncytial virus in young children, determined with a new recombinant fusion protein enzyme-linked immunosorbent assay. Clin Vaccine Immunol 20:1654–1656. 10.1128/CVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RAM, Osterhaus ADME. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang A, Masante C, Buchholz UJ, Dutch RE. 2012. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol 86:3230–3243. 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimyte EM, Smith SE, Oreste P, Lembo D, Dutch RE. 2016. Inhibition of human metapneumovirus binding to heparan sulfate blocks infection in human lung cells and airway tissues. J Virol 90:9237–9250. 10.1128/JVI.01362-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapp TG, Rechenmacher F, Neubauer S, Maltsev OV, Cavalcanti-Adam EA, Zarka R, Reuning U, Notni J, Wester H-J, Mas-Moruno C, Spatz J, Geiger B, Kessler H, others. 2017. A comprehensive evaluation of the activity and selectivity profile of ligands for RGD-binding integrins. Sci Rep 7:39805. 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox RG, Livesay SB, Johnson M, Ohi MD, Williams JV. 2012. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J Virol 86:12148–12160. 10.1128/JVI.01133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, Williams JV. 2009. Integrin αvβ1 promotes infection by human metapneumovirus. Proc Natl Acad Sci U S A 106:1566–1571. 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y, Zhang Y, Cai H, Mirza AM, Iorio RM, Peeples ME, Niewiesk S, Li J. 2014. Roles of the putative integrin-binding motif of the human metapneumovirus fusion (F) protein in cell-cell fusion, viral infectivity, and pathogenesis. J Virol 88:4338–4352. 10.1128/JVI.03491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skiadopoulos MH, Biacchesi S, Buchholz UJ, Amaro-Carambot E, Surman SR, Collins PL, Murphy BR. 2006. Individual contributions of the human metapneumovirus F, G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology 345:492–501. 10.1016/j.virol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Battles MB, Más V, Olmedillas E, Cano O, Vázquez M, Rodríguez L, Melero JA, McLellan JS. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8:1–11. 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Más V, Rodriguez L, Olmedillas E, Cano O, Palomo C, Terrón MC, Luque D, Melero JA, McLellan JS. 2016. Engineering, structure and immunogenicity of the human metapneumovirus F protein in the postfusion conformation. PLoS Pathog 12:e1005859. 10.1371/journal.ppat.1005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilaev M, Shen Y, Carbonneau J, Venable M-C, Rhéaume C, Lavigne S, Couture C, Guarné A, Hamelin M-È, Boivin G. 2020. Evaluation of pre-and post-fusion human metapneumovirus F proteins as subunit vaccine candidates in mice. Vaccine 38:2122–2127. 10.1016/j.vaccine.2020.01.047. [DOI] [PubMed] [Google Scholar]
- 18.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang G-Y, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS, others. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162–309ra162. 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killikelly AM, Kanekiyo M, Graham BS. 2016. Pre-fusion F is absent on the surface of formalin-inactivated respiratory syncytial virus. Sci Rep 6:34108–34107. 10.1038/srep34108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 21.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 22.Hamelin M-È, Couture C, Sackett MK, Boivin G. 2007. Enhanced lung disease and Th2 response following human metapneumovirus infection in mice immunized with the inactivated virus. J Gen Virol 88:3391–3400. 10.1099/vir.0.83250-0. [DOI] [PubMed] [Google Scholar]
- 23.Yim KC, Cragin RP, Boukhvalova MS, Blanco JCG, Hamlin M-È, Boivin G, Porter DD, Prince GA. 2007. Human metapneumovirus: enhanced pulmonary disease in cotton rats immunized with formalin-inactivated virus vaccine and challenged. Vaccine 25:5034–5040. 10.1016/j.vaccine.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karron RA, San Mateo J, Wanionek K, Collins PL, Buchholz UJ. 2018. Evaluation of a live attenuated human metapneumovirus vaccine in adults and children. J Pediatric Infect Dis Soc 7:86–89. 10.1093/jpids/pix006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lévy C, Aerts L, Hamelin M-È, Granier C, Szécsi J, Lavillette D, Boivin G, Cosset F-L. 2013. Virus-like particle vaccine induces cross-protection against human metapneumovirus infections in mice. Vaccine 31:2778–2785. 10.1016/j.vaccine.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 26.Russell CJ, Jones BG, Sealy RE, Surman SL, Mason JN, Hayden RT, Tripp RA, Takimoto T, Hurwitz JL. 2017. A Sendai virus recombinant vaccine expressing a gene for truncated human metapneumovirus (hMPV) fusion protein protects cotton rats from hMPV challenge. Virology 509:60–66. 10.1016/j.virol.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cseke G, Wright DW, Tollefson SJ, Johnson JE, Crowe JE, Williams JV. 2007. Human metapneumovirus fusion protein vaccines that are immunogenic and protective in cotton rats. J Virol 81:698–707. 10.1128/JVI.00844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herfst S, Schrauwen EJA, de Graaf M, van Amerongen G, van den Hoogen BG, de Swart RL, Osterhaus ADME, Fouchier RAM. 2008. Immunogenicity and efficacy of two candidate human metapneumovirus vaccines in cynomolgus macaques. Vaccine 26:4224–4230. 10.1016/j.vaccine.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Peled Y, Diaz D, Pena-Briseno A, Murray J, Huang J, Tripp RA, Mousa JJ. 2019. A potent neutralizing site III-specific human antibody neutralizes human metapneumovirus in vivo. J Virol 93. 10.1128/JVI.00342-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Diaz D, Mousa JJ. 2020. Antibody recognition of the pneumovirus fusion protein trimer interface. PLoS Pathog 16:e1008942. 10.1371/journal.ppat.1008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, Piralla A, Percivalle E, Sallusto F, Baldanti F, Lanzavecchia A, others. 2013. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501:439–443. 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 32.Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE, Jardetzky TS. 2017. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol 2:1–7. 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong C, Venot A, Li X, Lu W, Xiao W, Wilkes J-SL, Salanga CL, Handel TM, Wang L, Wolfert MA, Boons G-J, et al. 2017. Heparan sulfate microarray reveals that heparan sulfate–protein binding exhibits different ligand requirements. J Am Chem Soc 139:9534–9543. 10.1021/jacs.7b01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chopra P, Joshi A, Wu J, Lu W, Yadavalli T, Wolfert MA, Shukla D, Zaia J, Boons G-J. 2021. The 3-O-sulfation of heparan sulfate modulates protein binding and lyase degradation. Proc Natl Acad Sci U S A 118:e2012935118. 10.1073/pnas.2012935118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagae M, Re S, Mihara E, Nogi T, Sugita Y, Takagi J. 2012. Crystal structure of $α$5$β$1 integrin ectodomain: atomic details of the fibronectin receptor. J Cell Biol 197:131–140. 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, CroweJE, Jr, Jardetzky TS. 2012. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol 19:461–463. 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Diaz D, Mousa JJ. 2019. Antibody epitopes of pneumovirus fusion proteins. Front Immunol 10:2778. 10.3389/fimmu.2019.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mousa JJ, Binshtein E, Human S, Fong RH, Alvarado G, Doranz BJ, Moore ML, Ohi MD, Crowe JE. 2018. Human antibody recognition of antigenic site IV on Pneumovirus fusion proteins. PLoS Pathog 14:e1006837. 10.1371/journal.ppat.1006837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy BR, Walsh EE. 1988. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J Clin Microbiol 26:1595–1597. 10.1128/jcm.26.8.1595-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boelen A, Andeweg A, Kwakkel J, Lokhorst W, Bestebroer T, Dormans J, Kimman T. 2000. Both immunization with a formalin-inactivated respiratory syncytial virus (RSV) vaccine and a mock antigen vaccine induce severe lung pathology and a Th2 cytokine profile in RSV-challenged mice. Vaccine 19:982–991. 10.1016/S0264-410X(00)00213-9. [DOI] [PubMed] [Google Scholar]
- 42.Connors M, Giese NA, Kulkarni AB, Firestone C-Y, Morse HC, Murphy BR. 1994. Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J Virol 68:5321–5325. 10.1128/JVI.68.8.5321-5325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLellan JS, Chen M, Chang J-S, Yang Y, Kim A, Graham BS, Kwong PD. 2010. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol 84:12236–12244. 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams JV, Chen Z, Cseke G, Wright DW, Keefer CJ, Tollefson SJ, Hessell A, Podsiad A, Shepherd BE, Sanna PP, Burton DR, Crowe JE, Williamson RA, others. 2007. A recombinant human monoclonal antibody to human metapneumovirus fusion protein that neutralizes virus in vitro and is effective therapeutically in vivo. J Virol 81:8315–8324. 10.1128/JVI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:8060–8065. 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE. 2017. A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol 2:1–8. 10.1038/nmicrobiol.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Chopra P, Li X, Wolfert MA, Tompkins SM, Boons G-J. 2020. SARS-CoV-2 spike protein binds heparan sulfate in a length-and sequence-dependent manner. BioRxiv.
- 51.Ruan X, Sack DA, Zhang W. 2015. Genetic fusions of a CFA/I/II/IV MEFA (multiepitope fusion antigen) and a toxoid fusion of heat-stable toxin (STa) and heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) retain broad anti-CFA and antitoxin antigenicity. PLoS One 10:e0121623. 10.1371/journal.pone.0121623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1<br>. Download JVI.00593-21-s0001.pdf, PDF file, 0.09 MB (90.2KB, pdf)
Data Availability Statement
The structure factors and structure coordinates were deposited in the Protein Data Bank under accession code 7M0I.