Abstract
Axon degeneration is a prominent feature of the injured nervous system, occurs across neurological diseases, and drives functional loss in neural circuits. We have seen a paradigm shift in the last decade with the realization that injured axons are capable of actively driving their own destruction through the sterile-alpha and TIR motif containing 1 (SARM1) protein. Early studies of Wallerian degeneration highlighted a central role for NAD+ metabolites in axon survival, and this association has grown even stronger in recent years with a deeper understanding of SARM1 biology. Here we review our current knowledge of SARM1 function in vivo, and our evolving understanding of its complex architecture and regulation by injury-dependent changes in the local metabolic environment. The field is converging on a model whereby SARM1 acts as a sensor for metabolic changes that occur after injury, and then drives catastrophic NAD+ loss to promote degeneration. However, a number of observations suggest that SARM1 biology is more complicated, and there remains much to learn about how SARM1 governs nervous system responses to injury or disease.
Introduction
Axon degeneration occurs after neural injury and is a common feature of several acute and chronic, sporadic and familial neurological disorders including multiple sclerosis (MS)[1], spinal muscular atrophy (SMA) [2], amyotrophic lateral sclerosis (ALS), Parkinson’s disease (PD), Traumatic Brain Injury (TBI), stroke and myelin disorders [3]. It also occurs in peripheral neuropathies associated with chemotherapeutic regimens and in diabetes and genetic peripheral neuropathies (e.g. Charcot-Marie Tooth disease). Axonal degeneration drives the progressive loss of neurological function in patients suffering from neurodegenerative conditions [4–6], with functional loss in part resulting from the breakdown of circuit integrity. Despite its broad association with several diseases, we are only beginning to understand the molecular mechanisms that drive axon degeneration in any context. A comprehensive elucidation of molecules/pathways that drive axon degeneration, and, ultimately, therapeutic blockade of these pathways to preserve axon integrity in patients are central goals for the field.
The characterization of the Wallerian degeneration (WD) pathway as axon-intrinsic, injury activated molecular pathway has re-invigorated an interest in targeting axon degeneration in human disease. Central to the pathway is mammalian SARM1 (dSarm in Drosophila, TIR-1 in C. elegans), a primary regulator of axon auto-destruction [7•, 8•, 9•]. Significant progress has been made over the last decade in defining the phenotypic consequences of SARM1 loss, SARM1 enzymology and signaling, and how NAD+ metabolites regulate SARM1 activation. This review will discuss new roles for SARM1 in the injured nervous system, how new molecular knowledge about SARM1 enzymology and structure can be reconciled with in vivo function and highlight key questions for the future. The role of SARM1 in neurological diseases was recently reviewed [6], and will not be covered here.
How does Sarm1 signal in vivo?
Axotomy separates a distal axon stump from its cell body. After a latent phase, distal stumps undergo sudden and explosive fragmentation (WD). Two factors that likely drive WD are increases in axonal calcium [10] and depletion of NAD+/ATP [11]. In many experimental systems, axonal calcium levels increase dramatically immediately prior to degeneration, and blockade of calcium entry can significantly extend axon survival [12], but precise role(s) for calcium in driving axon degeneration remain elusive [13].
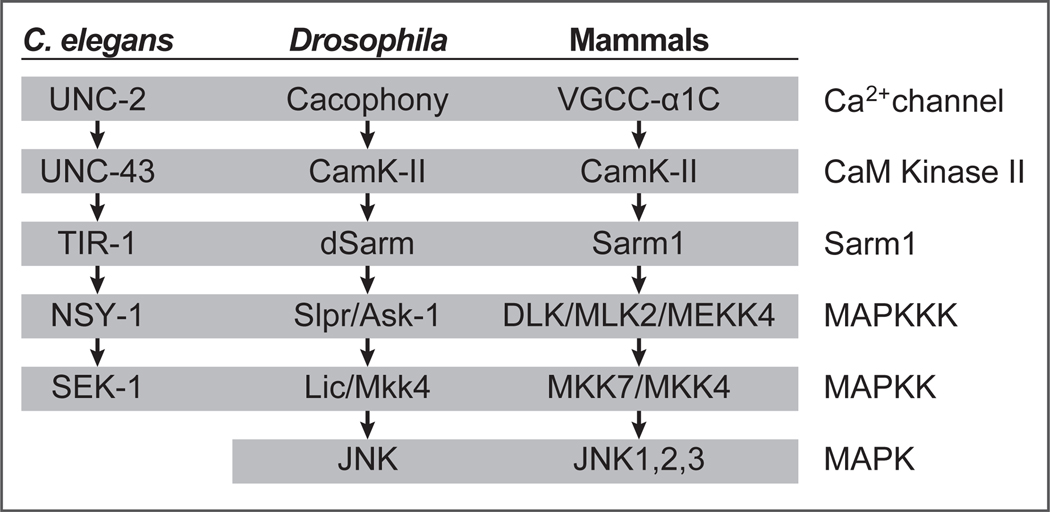
The role for SARM1 in axon degeneration was first discovered in a screen for Drosophila mutants that blocked WD [7•]: null alleles of Sarm1 blocked WD for the lifespan of the fly. Initial models for SARM1 signaling were built from elegant studies of TIR-1 (C. elegans Sarm1) [14]. TIR-1 (and dSarm/ SARM1) has three primary structural features: an N-terminal ARM, two SAM, and a C-terminal TIR domain. TIR-1-mediated signaling occurs via activation of the upstream UNC-2 Ca2+ channel, which leads to activated CamK-II binding the N-terminal auto-inhibitory ARM domain of TIR1, resulting in the re-localization of the TIR-1 to the post-synapse via its SAM domains, where the TIR domains promotes MAPK signaling (Figure 1).
Figure 1.
Components of the TIR-1 MAPK signaling cascade in worms, flies and mammals
The potent neuroprotective effects of Sarm1 null mutations are well conserved in mouse [7•,8•], and deletion of the auto-inhibitory ARM domain of SARM1 in flies [9•] or mice [8•] results in spontaneous axon degeneration, demonstrating that SARM1 is necessary and in some cases sufficient for axon destruction. In fact, all components of the TIR-1 pathway are well conserved from worms to mammals (Figure 1). Given the role of calcium in axon degeneration, it is intriguing that TIR-1 is genetically downstream of the UNC-2 voltage-gated calcium channel. However, in contrast to the TIR-1mediated signaling events studied in C. elegans (left-right asymmetry), null alleles of the Drosophila orthologs of TIR-1 signaling pathway components fail to block WD in vivo [7•,9,15••]. In mammals, blockade of MAPK signaling or downstream targets partially suppresses axon degeneration [16,17], but this level of protection does not approach that in SARM1 null animals, and WD phenotypes of other components of the TIR-1 pathway have not been reported.
The precise role of MAPK signaling in SARM1-mediated axon degeneration is not fully understood. Components of the MAPK signaling cascade (MKK4 and MKK7) are phosphorylated within 15 minutes after axotomy, and this is SARM1-dependent [16], demonstrating a role for SARM1 upstream of MAPK early in axonal responses to injury. MAPK phosphorylation then decreases a few hours after axotomy, and remains low, although axon degeneration occurs much later in the same cells [16]. Loss of MKK4 and MKK7 delays axotomy-induced axon degeneration [16] and depletion of NAD+ and ATP in axons [18]. However, while activation of a gain-of function SARM1 molecule led to phosphorylation of MAPK components [18,16], loss of MKK4 and MKK7 inhibited axon degeneration only partially [16] or not at all [18]. The latter observation has been used to argue that MAPK signaling is, instead, upstream of Sarm1.
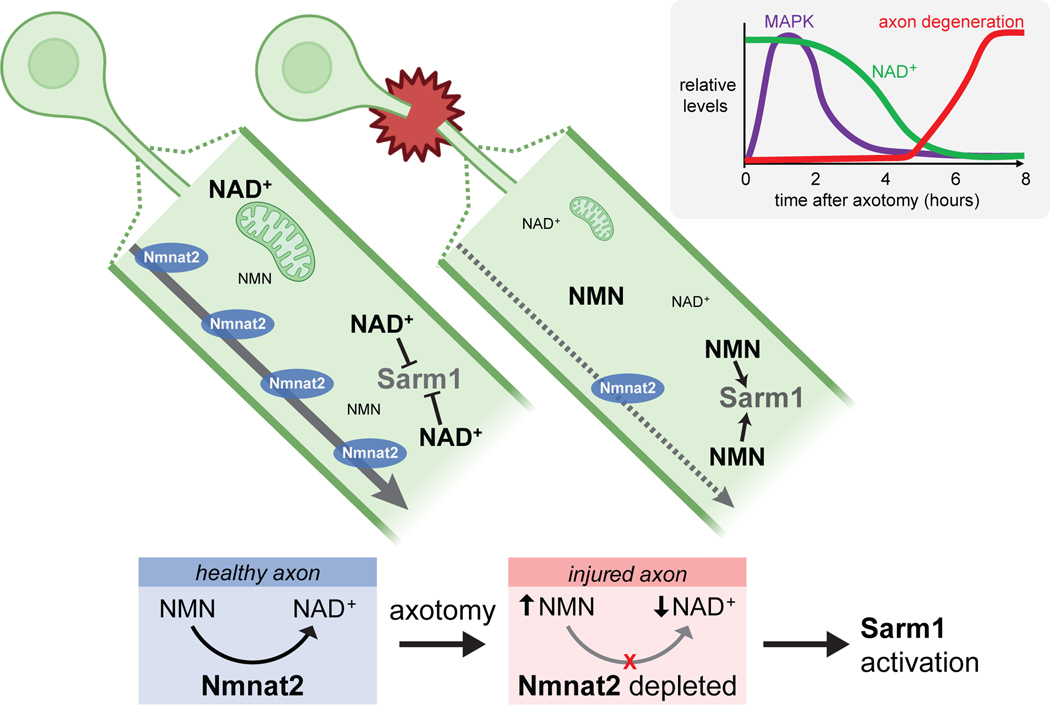
Significant progress has been made in exploring the upstream regulator of the WD pathway, Nmnat2, an NAD+ biosynthetic enzyme that is a survival factor normally transported down the axon (Figure 2). Nmnat2 generates NAD+ from NMN [19]. Nmnat2 is labile, so axotomy results in the eventual depletion of existing Nmnat2 pools in distal axon stumps— potentially explaining the latent phase between axotomy and degeneration. Nmnat2 loss results in both decreased axonal NAD+ and increased NMN [20]. NAD+ depletion is potentially energetically catastrophic for axons, and NMN is in fact toxic to axons [20,21]. The exciting recent discoveries that SARM1 itself can rapidly drive NAD+ depletion [22•,23••] through an intrinsic NAD+ hydrolase activity in its TIR domain [23••], revealed an unexpected and important direct link between SARM1 and NAD+ loss. Current models now propose SARM1 NADase activity drives an already sick (i.e. high NMN, low NAD+) axon over the edge to final auto-destruction (Figure 2). Additional central roles of NAD+, NMN, and Nmnat molecules in axon survival have been highlighted by extensive studies of the WldS molecule, and are discussed in detail elsewhere [6].
Figure 2. Nmnat2/NAD+ depletion model for axon degeneration.
Nmnat2 is a survival factor transported down axons from the cell body that generates NAD+ from NMN. After axotomy, the labile Nmnat2 molecule is depleted from distal severed axons, NAD+ drops, NMN rises, and Sarm1 is activated. In mammalian neurons MAPK signaling (based on MKK4/7 phosphorylation) in axons peaks early, NAD+ drops hours later, and axon fragmentation begins. Time reflects events in DRG primary cultures, in vivo times are much longer. (see text for details)
dSarm signals in two phases –early with MAPK and late with Axundead
Some clarity on the complex interaction between Sarm1 and MAPK signaling recently came from work in Drosophila [15••]. In the Drosophila L1 wing nerve it is possible to injure a subset of neurons, then examine the responses of both distal severed axon stumps and intact neighboring neurons (termed “bystanders”), with single cell/axon resolution. Within hours after injury of even a small number of axons, the transport of autophagosomes, lysosomes, and synaptic vesicles along axons are strongly suppressed in severed distal axons stumps. Surprisingly, transport is also blocked in neighboring, uninjured bystander neurons, as is their mechano- or chemosensory function. This observation revealed that partial nerve injury not only affects injured axons, but also neighboring intact bystander neurons. This raises an important point regarding how functional loss occurs in the nervous system. We generally envision functional loss to result from the breakdown of physical connections in circuits. This “bystander effect” indicates suppression of neuronal function after injury extends beyond injured neurons. If similar effects occur in neurodegenerative disease, this should force us to reconsider the cellular basis of functional loss in patients. This bystander effect can be observed in mammals after partial sciatic nerve injury [24] or mouse models of traumatic brain injury (TBI) [25]. Whether this occurs in Sarm1 knockout animals is an exciting, unanswered question. Interestingly, support for this idea comes from studies of TBI: while wild type mice show behavioral defects within hours after mild closed-head TBI, Sarm1 null animals behaved like pre-injury controls almost immediately after TBI [26].
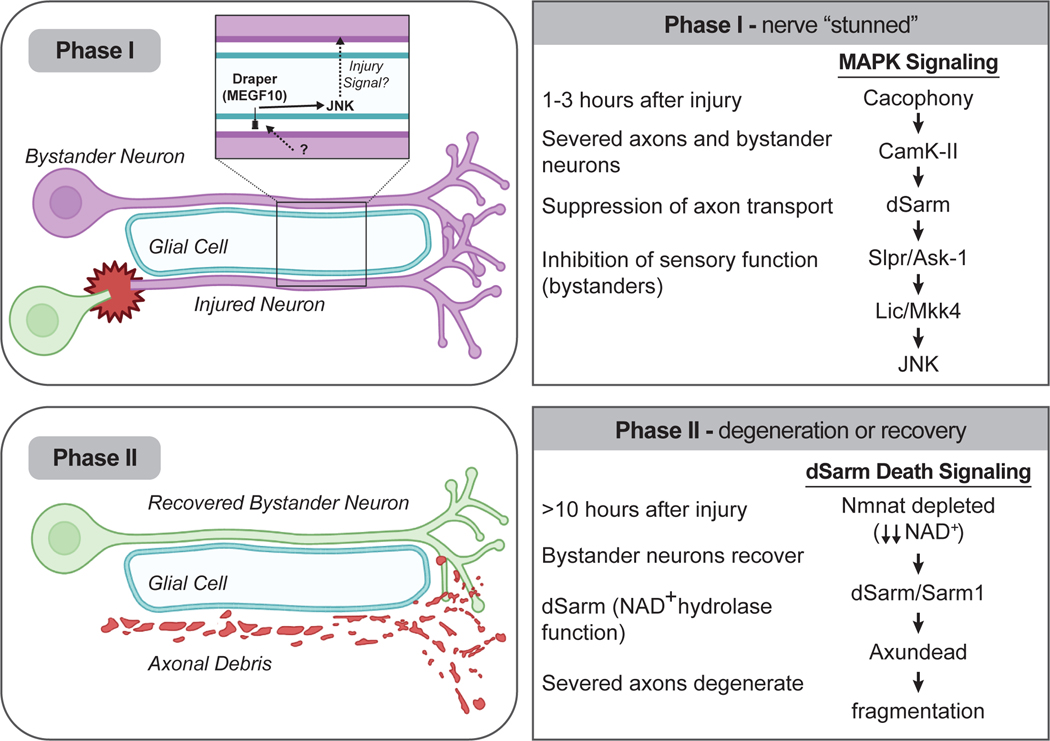
The rapid blockade of axon transport observed in Drosophila (termed “Phase I”) (Figure 3) is driven cell autonomously in all axons by dSarm: dsarm null mutants maintain axon transport in severed axon stumps and (unexpectedly) in intact bystanders. However, the BTB/Back domain molecule Axundead (Axed) is not involved, despite the absolute requirement later in dSarm-mediated axon degeneration (termed “Phase II”). Interestingly, components of the TIR-1-like MAPK signaling cascade are also required cell-autonomously for all blockade of neural function during Phase I. Given the fact null alleles of components of the TIR-1 MAPK cascade fail to block WD in Drosophila [7•,9•,15••], this argues that TIR-1-like MAPK signaling predominates early in axonal responses to injury, rather than driving axon death itself, and that dSarm drives axon degeneration later via NAD+ hydrolysis and Axundead [15••] (Figure 3).
Figure 3. SARM1 signals in two distinct phases.
Within 2 hours after axotomy the TIR-1-like MAPK cascade is activated in both injured axons and uninjured adjacent bystander neurons. Phase I leads to a broad suppression of neurophysiology in both severed and intact bystander neurons, with injury signals being spread to bystander neurons through glial Draper/MEGF10 signaling. Phase I does not require Axundead or dSarm NAD+ hydrolase function. During Phase II, bystanders recover functionally and severed axons activate dSarm/SARM1-mediated axon degeneration through dSarm NAD+ hydrolase activity and Axundead.
Insights into SARM1 activation from structural biology
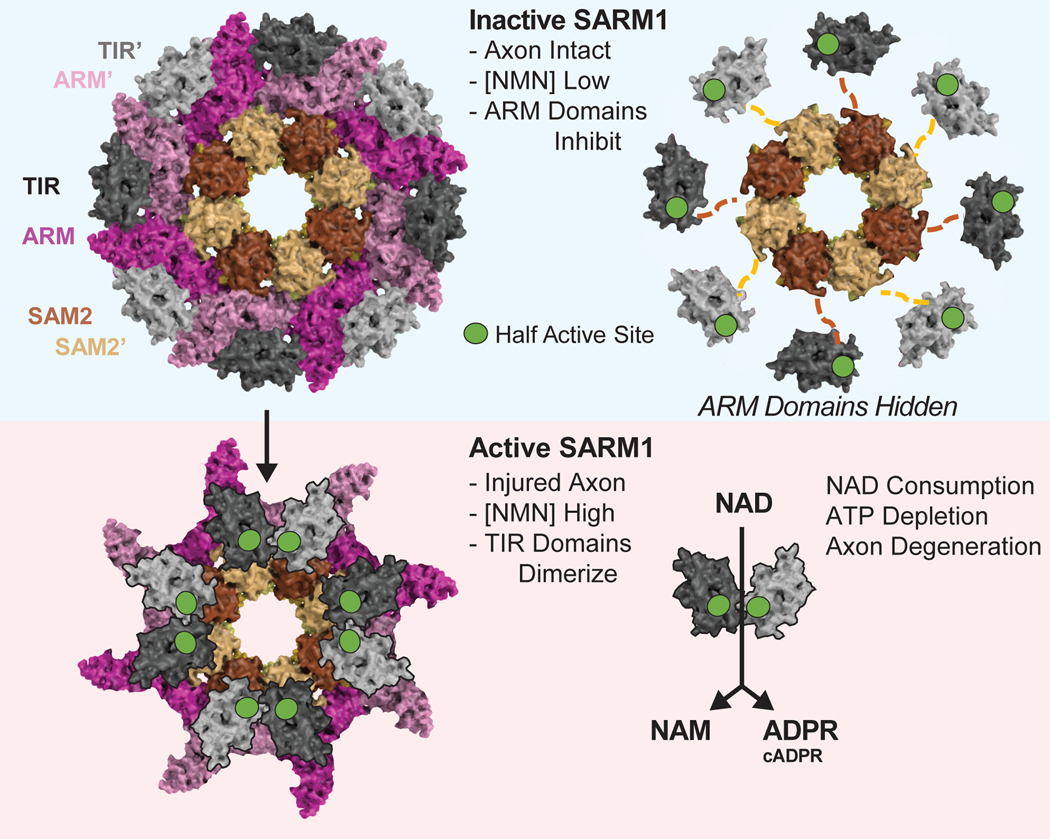
Emerging data on the structure, enzymatic function, and regulation of SARM1 are also growing our understanding of this complex metabolic sensor and axon death executioner. As discussed above, SARM1 is a structurally complex, multi-domain protein with an auto-inhibitory ARM domain, tandem oligomerization SAM domains and a catalytic TIR domain [8,14,27]. The multi-domain architecture of the full-length protein with flexible inter-domain interactions has made elucidation of high-resolution x-ray structures challenging. Crystal structures of single domains in isolation while important, have fallen short in informing the overall architecture of the inactive and active states of SARM1 and the molecular transitions from the inactive to active state [28•,29•]. Several lines of evidence strongly support the hypothesis that the oligomerization of the TIR domains is a pre-requisite for catalytic activity [8,22,23,30,31••]. In the absence of high-resolution structures of full-length SARM1, the initial hypothesis was that full-length SARM1 exists as a monomer in solution with the auto-inhibitory ARM domain preventing the oligomerization of the tandem SAM domains which in turn prevents the oligomerization of the catalytic TIR domain. The first high-resolution cryo-EM structure of full-length, inactive SARM1 revealed that to the contrary, full-length SARM1 is a pre-assembled octamer in its inactive state [31••]. As in the x-ray structure of the SAM domains alone [29•,32], the tandem SAM domains in the full-length structure also assume a closed ring architecture forming a do-octameric stacked core. Decorating the periphery of this complex are ARM-TIR units. This novel structure instantly clarified many aspects of SARM1 activity. While the assembly of the SAM domains is likely an important step en route to assembly of an active complex and disrupting the SAM domain assembly by mutating several key residues in the SAM-SAM and SAM-ARM interaction interfaces results in the loss of SARM1 catalytic activity [29•,31••,32], it is clearly not the rate-limiting step. Instead, the pre-assembled octameric inactive state ensures that enzyme is primed to rapidly transition from an inactive to active state upon injury. What then is the key driver of retaining SARM1 in an inactive state? The overall high-resolution of the cryo-EM structure led to the identification of a critical ARM-TIR lock that traps SARM1 in inactive state. The ARM-TIR lock ensures that the TIR domains stay spatially separate (~25Å apart), disfavoring the formation of productive active sites that likely require the TIR domains to come into proximity. The release of this lock by mutating key residues in the ARM domain is sufficient to allow this pre-assembled oligomeric complex to switch from inactive to catalytically active and cause extensive axon destruction. Thus, the first cryo-EM structure of full-length SARM1 explained some important aspects of SARM1 biology and identified important regulatory elements in the protein.
The identification of the ARM-TIR lock was followed by the identification of an allosteric pocket in SARM1 distant from the ARM-TIR lock site, but still in the ARM domain, that was shown to bind NAD+ and NMN [28•,33•]. While both NAD+ and NMN appear to bind in the same allosteric pocket, there are notable differences in the interactions between each of the metabolites and key residues in the ARM domain of SARM1. NMN has been previously identified as a potential endogenous activator of SARM1 [34]. The identification of the allosteric pocket that binds to both NAD+ and NMN leads to the logical hypothesis that under normal physiological conditions, when NAD+ is present at much higher levels compared to NMN, NAD+ binds to the allosteric pocket and stabilizes the inactive state. Upon injury, NMNAT2 depletion results in a reduction in NAD+ and a local/transient increase in NMN. This increase in the NMN/NAD+ ratio along with the higher affinity of NMN for SARM1 compared to NAD+ can result in replacement of the NAD+ with NMN in the ARM allosteric pocket and subsequent activation of enzymes by potentially breaking the ARM-TIR lock.
While the identification of the allosteric metabolite binding pocket is important, the exact conformational changes that result in the breaking of the ARM-TIR lock upon NMN binding remains unclear. Despite efforts by several groups, the structures of NMN-activated, full-length SARM1 have not been resolved to date. It also remains to be seen if the structural observations can be reconciled with the in vivo levels of these key metabolites in normal and disease states. For instance, are NAD+ levels in cells normally high enough to stabilize the ARM domain? Levels of NAD+ found in living cells range from 200 nM to 500 μM. If correct, it is unclear how the allosteric pocket is inhibited by NAD+ under these conditions. Likewise, NMN levels appear to be in the 10–20 nM range in axons even 6 hours after axotomy (just prior to degeneration)[35] while 2–6 μM of NMN is required for activation of Sarm1 NADase activity [28•,31••]. It is possible that local concentrations of NAD+ or NMN can reach levels necessary for inhibition or activation in cells and in vivo [11]. It is also conceivable that additional upstream signaling elements including the MAPK pathway might play an important role in activating SARM1. For instance, it is curious that NMN activation of full-length SARM1 in vitro results in lower hydrolase activity compared to ARM-deleted SARM1. Do other components in the MAPK pathway cause additional structural changes in SARM1 that are necessary for activity [36]? Does phosphorylation of SARM1 by the MAPK pathway in the first wave prime the enzyme for activity whereby when local NMN levels increase in the second wave, the enzyme may become fully active? In addition, mutations that release the ARM-TIR lock in full-length SARM1 are not as active as ARM-deleted, constitutively active SARM1, again suggesting that additional structural changes may be necessary to confer complete hydrolase activity.
The tight regulation of SARM1 by the local metabolic environment is further supported by the observation that high concentrations of Nicotinamide (NAM), one of the products of the NAD+ hydrolysis reaction, inhibits SARM1 [23,31••]. Product inhibition has been observed for several other NAD+ hydrolases including CD38, the NAD+ hydrolase most closely related to SARM1, as well as some members of the PARP and SIRTuin families [37–39]. Thus, SARM1 is a potent axon death factor whose death function is tightly regulated by changes in the local metabolite milieu. This now leads to the next question - is just a depletion of NAD+ adequate to drive catastrophic axon fragmentation or is the NAD+ hydrolysis merely the first event in the assembly of a larger death complex? Is cADPR, another product of the NAD+ hydrolysis reaction functioning as a second messenger triggering downstream signaling changes that contribute to greater calcium influx and activation of Phase II Sarm1 signaling, ultimately resulting in catastrophic axon destruction? Previous work has shown a drop in NAD+ and ATP beginning ~4 hours after axotomy in cultured mouse DRG neurons, with levels approaching zero by 6 hours, the time of axon degeneration [11]. This is also accompanied by an increase in cADPR [40]. This tight temporal association between axon degeneration and a drop in NAD+/ATP was identified almost two decades ago and implies that axon degeneration and energetic catastrophe are indeed tightly linked.
Despite significant advances in elucidating the inactive structure of SARM1 summarized above, the active state(s) of SARM1 have remained surprisingly elusive. If the active state of SARM1 results in the oligomerization of the TIR domains, one could argue that this may improve the likelihood of resolving cryo-EM structures of active, full-length SARM1. However, the opposite has been true. Cryo-EM structures of ARM-deleted SARM1 and NMN-activated SARM1 have revealed that while the SAM domains continue to be well-resolved, the TIR domains cannot be assigned unambiguously [31••]. This lack of resolvable high-quality density for TIRs in the active structure is surprising. Further, the active forms of the protein appear to remain octameric in solution. Hence, it is unclear if the TIRs come together in a higher-order oligomeric assembly upon removal of the auto-inhibitory ARM domain and if this were the case, would these interactions be transient and reversible/irreversible? The structure of the TIR domain alone has also not resulted in a better understanding of the active site or why an oligomeric state is active where a monomeric state is not [29•]. Finally, it is tempting to hypothesize that the TIR domains can both homo-oligomerize and hetero-oligomerize where the first order oligomerization involves homo-oligomerization resulting in the initial NAD+ hydrolysis event which then drives a second-order oligomerization event involving the re-arrangement of the complex and recruitment of other TIR containing proteins ultimately resulting in the formation of a death complex. Other mammalian TIRs such as Myd88 and TLR4 have been known to hetero-oligomerize and form larger functional complexes and signalosomes, impacting multiple signaling pathways including MAPK signaling. While little is known or understood about what happens following NAD+ hydrolysis leading up to catastrophic axon degeneration, what is clear is that the NAD+ hydrolysis aspect of SARM1 functioning is only one part of the SARM1 axon degeneration story and what happens downstream of the NAD+ hydrolysis event is yet to be uncovered.
Reconciling current models with structural and enzymology data, and in vivo biology?
It is not clear how SARM1 gets activated at each phase of signaling. In the context of axon degeneration, the substrates of both Nmnat (NMN) and Sarm1 (NAD+) have been proposed as regulators of SARM1. At least in vitro, NAD+ stabilizes the ARM domain to repress SARM1 NADase activity [32,33•], and NMN destabilizes the ARM domain to potentially promote it [28•,31••]. A simple model is that Nmnat turnover in severed axons increases NMN and decreases NAD+, and SARM1 is activated. This two-trigger model (high NMN, low NAD+) depending on Nmnat is consistent with the observations in multiple species that Nmnat loss drives SARM1-dependent axon degeneration [9•, 41], while NMN deamidase (which breaks down NMN) suppresses WD [15••,20,21]. If increases in NMN and a drop in NAD+ are required simultaneously, this might also explain why dramatically increasing NMN levels is not sufficient to induce axon degeneration [35].
There is a complex interaction between Nmnat, SARM1, and MAPK immediately after injury during Phase I. SARM1-mediated MAPK signaling is activated within 15 minutes after axotomy [16], although neither Nmnat depletion nor a drop in NAD+ have been observed by this early time point [15••][11,19]. So what is activating SARM1 and MAPK? In vivo Phase I signaling in Drosophila does not require dSarm NAD+ hydrolase activity or Axed [15••], both of which are absolutely essential for Phase II axon degeneration [15••,23••]. Instead, it requires the TIR-1-like MAPK signaling cascade, including transduction through the voltage gated calcium channel (VGCC) Cacophony [15••]. Based on timing and genetic data, it seems reasonable to assume that dSarm and SARM1 interact with MAPK components in Phase I responses to axotomy in a manner analogous to TIR-1 signaling in worms—the VGCC Cacophony signals through CamK-II to activate dSarm, and ultimately MAPK.
How do glia spread injury signals to bystander neurons? This requires glia sensing the injury, which somehow involves Draper/MEGF10 and JNK (Figure 3). This is puzzling as Draper primarily functions as an engulfment receptor [42,43], although non-phagocytic roles for Draper and MEGF10 have been described [44,45]. Drosophila dSarm can signal downstream of a Toll-like receptor to promote inflammatory-like signaling in glia [46], and such a signaling cascade could be utilized to activate early MAPK signaling in axons. More puzzling is the observation that glial cells can potently suppress axon transport and sensory function in uninjured bystanders—how does a glial cell tell a neuron to stop functioning? If dSarm NAD+ hydrolase activity is not driving early Sarm1-mediated MAPK signaling events, what is? (It remains to be determined if mammalian SARM1 promotes early suppression of neurophysiology, and if so, whether that depends on the NAD+ hydrolase function.) Could SARM1 act as a scaffold for MAPKs? To our knowledge direct binding of MAPK components to SARM1 has not been demonstrated in any system, so thus far it appears their interactions are likely indirect. The direct connection between SARM1 and MAPK signaling remains poorly defined. Finally, it is notable that dSarm signaling during Phase I is reversible. Bystander neurons, although they use dSarm to signal, recover eventually, so in vivo activation of dSarm is not necessarily a death sentence.
Does the NADase-mediated NAD+ depletion model explain SARM1 function satisfactorily? It seems that things must be more complicated at both early and late phases of SARM1 signaling. Several molecules that can potently block Phase II SARM1-mediated axon degeneration can also block early Phase I dSarm-MAPK interactions, including expression of dNmnat, WldS, or loss of Highwire/Phr1 [15••]. Our thinking has been that these keep Nmnat levels high enough to make NAD+ in axons to block degeneration, but how do we explain the protective effects of these molecules at Phase I where dNmnat and NAD+/ATP have not apparently dropped? In addition, dNmnat, unexpectedly, appears to be required for dSarm-mediated signaling during Phase I. Loss of dNmnat in fly neurons leads to axon death, but this can be suppressed by axed mutations, and Axed does not play a role early in Phase I signaling, which allowed for an exploration of the role of dNmnat in Phase I. In axed, dnmnat null mutants, Phase I signaling was blocked [15••]. Since Axed is not involved, this points to a positive role for dNmnat in promoting Phase I, dSarm-mediated signaling. In all other cases, loss of Nmnat activity activates SARM1. How would dNmnat promote SARM1 function? The dNmnat-MAPK relationship is emerging as complex in other contexts as well. In some cases, loss of Nmnat suppresses MAPK signaling [47], and in others, overexpression of Nmnat can potently suppress MAPK signaling [16]. It appears that dNmnat activity must be precisely tuned at each phase of Sarm1 signaling, as early it promotes Sarm1 signaling but later it inhibits SARM1. Finally, if SARM1 is the hydrolase driving NAD+ depletion, why can loss of Axed (a BTB and BACK domain molecule) completely suppress dSarm-mediated cell destruction [9]? Given that the NAD+ hydrolase function is intrinsic to SARM1/dSarm, it is hard to imagine how Axed could block its ability to degrade NAD+.
What role does calcium play in axon destruction, and is it upstream or downstream of SARM1? Beautiful imaging studies in zebrafish of the entire process of WD in vivo demonstrate that after axotomy calcium remains low, but immediately prior to degeneration it becomes pathologically high [13,48]. What is driving that increase, and what does the calcium increase do? Sarm1 NAD+ hydrolase activity leads to the production of cADPR, which can gate intracellular calcium stores [49]. Perhaps this helps drive the response, but entry of extracellular calcium is also important [12]. Part of the activity of increased calcium appears to be calpain activation, but blockade of these calcium activated proteases only has weak effects on axon degeneration [50]. This is the critical phase of axon degeneration that we understand very poorly from a cell biological perspective. Future studies aimed at identifying links between SARM1 signaling and pathological calcium rise will be essential.
In closing, while recent structural, biochemical and in vivo functional studies have propelled forward our understanding of SARM1 signaling mechanisms, there are still several questions to explore with respect to how SARM1 mediates neuronal responses to injury or disease. Determining precisely how SARM1 is activated in different contexts, and how it executes axon degeneration, top the list of most important questions for the future. The answers are likely to be complex and context dependent. The ability of SARM1 mutants to potently suppress a number of mouse models of neurological disease [6] highlight the promise of therapeutic blockade of SARM1 in saving axons in the context human disease.
Figure 4. Model for SARM1 activation.
SARM1 exists in an autoinhibited conformation where the ARM domains lock the TIR domains in an inactive conformation (top left). Hiding the ARM domains (top right) reveals that the TIR domains are connected to the SAM2 domains via a flexible (and unresolved) linker. A transient high concentration of NMN triggers a conformation change, releasing the ARM domains and likely causing the TIR domains to dimerize (possibly on top of the SAM2 domains), resulting in the formation of catalytically active TIRs (bottom). Note, this is a hypothetical model, and although active TIR domains are displayed as dimers, other oligomerization states are possible.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simkins TJ, Duncan GJ, Bourdette D: Chronic Demyelination and Axonal Degeneration in Multiple Sclerosis: Pathogenesis and Therapeutic Implications. Curr Neurol Neurosci 2021, 21:26. [DOI] [PubMed] [Google Scholar]
- 2.Kong L, Valdivia DO, Simon CM, Hassinan CW, Delestrée N, Ramos DM, Park JH, Pilato CM, Xu X, Crowder M, et al. : Impaired prenatal motor axon development necessitates early therapeutic intervention in severe SMA. Sci Transl Med 2021, 13:eabb6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauss R, Bosanac T, Devraj R, Engber T, Hughes RO: Axons Matter: The Promise of Treating Neurodegenerative Disorders by Targeting SARM1-Mediated Axonal Degeneration. Trends Pharmacol Sci 2020, 41:281–293. [DOI] [PubMed] [Google Scholar]
- 4.Coleman M: Axon degeneration mechanisms: commonality amid diversity. Nature reviews Neuroscience 2005, 6:889–898. [DOI] [PubMed] [Google Scholar]
- 5.Shy ME, Patzkó A: Axonal Charcot-Marie-Tooth disease. Current opinion in neurology 2011, 24:475–483. [DOI] [PubMed] [Google Scholar]
- 6.Coleman MP, Höke A: Programmed axon degeneration: from mouse to mechanism to medicine. Nat Rev Neurosci 2020, 21:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. : dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science (New York, NY) 2012, 337:481–484. •• Identification of dSarm in a Drosophila forward genetic screen for Wallerian degeneration mutants, and discovery of conservation of pro-degenerative function of SARM1 in mouse. dSarm/SARM1 mutations were the first loss of function mutations in and endogenous gene that protected axons at levels similar to WldS.
- 8. Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J: Sarm1-mediated axon degeneration requires both SAM and TIR interactions. The Journal of neuroscience: the official journal of the Society for Neuroscience 2013, 33:13569–13580. • SARM1 pro-degenerative function identified in siRNA screen of mammalian DRG neurons. Structure-function analysis demonstrated conserved roles for ARM inhibitory domain and TIR domain in axon degeneration, similar to functional roles in TIR-1 signaling in C. elegans.
- 9. Neukomm LJ, Burdett TC, Seeds AM, Hampel S, Coutinho-Budd JC, Farley JE, Wong J, Karadeniz YB, Osterloh JM, Sheehan AE, et al. : Axon Death Pathways Converge on Axundead to Promote Functional and Structural Axon Disassembly. Neuron 2017, 95:78–91.e5. • Identification of Axundead as downstream of dSarm and capable of fully blocking activated dSarm pro-degenerative function. Distal fibers of severed axons with axed or dSarm null mutations remain intact, integrated in circuits, and capable of driving complex behaviors for weeks. In the context of Nmnat loss, the protective effects of axed mutations far exceed those of dSarm.
- 10.Schlaepfer WW, Bunge RP: EFFECTS OF CALCIUM ION CONCENTRATION ON THE DEGENERATION OF AMPUTATED AXONS IN TISSUE CULTURE. J Cell Biology 1973, 59:456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, He Z: A local mechanism mediates NAD-dependent protection of axon degeneration. The Journal of cell biology 2005, 170:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George E, Glass J, Griffin J: Axotomy-induced axonal degeneration is mediated by calcium influx through ion-specific channels. J Neurosci 1995, 15:6445–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vargas ME, Yamagishi Y, Tessier-Lavigne M, Sagasti A: Live Imaging of Calcium Dynamics during Axon Degeneration Reveals Two Functionally Distinct Phases of Calcium Influx. The Journal of neuroscience: the official journal of the Society for Neuroscience 2015, 35:15026–15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang C-F, Bargmann CI: A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes & development 2005, 19:270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu J-M, Kang Y, Corty MM, Mathieson D, Peters OM, Freeman MR: Injury-Induced Inhibition of Bystander Neurons Requires dSarm and Signaling from Glia. Neuron 2021, 109:473–487.e5. •• Demonstrated a cell-autonomous role for dSarm in early suppression of neuronal physiology in severed axons and intact bystander neurons. dSarm signals in two phases, early with MAPK to suppress neurophysiology, and late with Axed to deplete NAD+ and drive axon degeneration. Spreading of injury signals to suppress bystander neuron function requires signaling through glia.
- 16.Yang J, Wu Z, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, Tessier-Lavigne M: Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 2015, 160:161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A: A dual leucine kinase–dependent axon self-destruction program promotes Wallerian degeneration. Nat Neurosci 2009, 12:387–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LJ, Summers DW, Sasaki Y, Brace EJ, Milbrandt J, DiAntonio A: MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife 2017, 6:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilley J, Coleman MP: Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS biology 2010, 8:e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefano MD, Nascimento-Ferreira I, Orsomando G, Mori V, Gilley J, Brown R, Janeckova L, Vargas ME, Worrell LA, Loreto A, et al. : A rise in NAD precursor nicotinamide mononucleotide (NMN) after injury promotes axon degeneration. Cell Death Differ 2015, 22:731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefano MD, Loreto A, Orsomando G, Mori V, Zamporlini F, Hulse RP, Webster J, Donaldson LF, Gering M, Raffaelli N, et al. : NMN Deamidase Delays Wallerian Degeneration and Rescues Axonal Defects Caused by NMNAT2 Deficiency In Vivo. Curr Biol 2017, 27:784–794. [DOI] [PubMed] [Google Scholar]
- 22. Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J: SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science (New York, NY) 2015, 348:453–457. • Dimerization of the SARM1 TIR domain leads to rapid depletion of NAD+ in axons and initiation of degeneration, and SARM1 acts in the axon after injury to drive degeneration. Supplementing NAD+ significantly rescued axons expressing activated SARM1, arguing NAD+ depletion was a key event in driving degeneration.
- 23. Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J: The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD+ Cleavage Activity that Promotes Pathological Axonal Degeneration. Neuron 2017, 93:1334–1343.e5. •• Made the surprising discovery that the TIR domain of SARM1 possesses an intrinsic NAD+ hydrolase activity that explains the rapid NAD+ drop after TIR domain dimerization [22] and is essential for SARM1 to drive axon degeneration.
- 24.Meyer RA, Ringkamp M: A role for uninjured afferents in neuropathic pain. Sheng Li Xue Bao Acta Physiologica Sinica 2008, 60:605–9. [PubMed] [Google Scholar]
- 25.Greer JE, Povlishock JT, Jacobs KM: Electrophysiological Abnormalities in Both Axotomized and Nonaxotomized Pyramidal Neurons following Mild Traumatic Brain Injury. J Neurosci 2012, 32:6682–6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henninger N, Bouley J, Sikoglu EM, An J, Moore CM, King JA, Bowser R, Freeman MR, Brown RH: Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain : a journal of neurology 2016, 139:1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essuman K, Summers DW, Sasaki Y, Mao X, Yim AKY, DiAntonio A, Milbrandt J: TIR Domain Proteins Are an Ancient Family of NAD+-Consuming Enzymes. Current biology : CB 2018, 28:421–430.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Figley MD, Gu W, Nanson JD, Shi Y, Sasaki Y, Cunnea K, Malde AK, Jia X, Luo Z, Saikot FK, et al. : SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration. Neuron 2021, 109:1118–1136.e11. • Crystal structure of the Drosophila ARM domain reveals that the endogenous activator NMN binds to a previously described NAD binding allosteric pocket in the auto-inhibitory ARM domain. Thus the endogenous activator NMN and the putative inhibitor NAD bind to the same allosteric pocket with some differences in interactions with specific residues in the ARM domain.
- 29. Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai J-S, Rank MX, et al. : NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 2019, 365:793–799. • The crystal structures of ribose and NADP+ (the oxidized form of nicotinamide adenine dinucleotide phosphate) complexes of SARM1 and plant NLR RUN1 TIR domains, respectively, reveal a conserved substrate binding site.
- 30.Summers DW, Gibson DA, DiAntonio A, Milbrandt J: SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc National Acad Sci 2016, 113:E6271–E6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bratkowski M, Xie T, Thayer DA, Lad S, Mathur P, Yang Y-S, Danko G, Burdett TC, Danao J, Cantor A, et al. : Structural and Mechanistic Regulation of the Pro-degenerative NAD Hydrolase SARM1. Cell Reports 2020, 32:107999. •• The high-resolution cryo-EM structure of full-length SARM1 reveals that it exists as a pre-assembled octameric auto-inhibited oligomer primed to transition rapidly to an active state. The molecular regulator of the inactive state is a critical “ARM-TIR” lock, an interaction interface between the auto-inhibitory N-terminal ARM domain and the catalytic C-terminal TIR domain that traps the catalytic TIR in an inactive conformation.
- 32.Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Shkolnisky Y, Mim C, Isupov MN, Zalk R, Hons M, et al. : The structural basis for SARM1 inhibition and activation under energetic stress. Elife 2020, 9:e62021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang Y, Liu T, Lee C-H, Chang Q, Yang J, Zhang Z: The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature 2020, 588:658–663. • Cryo-EM structure of full-length SARM1 that reveals an NAD binding allosteric pocket in the auto-inhibitory ARM domain. At high concentrations, NAD binds to the allosteric pocket on the auto-inhibitory ARM domain of SARM1 stabilizing the constitutively auto-inhibited state.
- 34.Zhao ZY, Xie XJ, Li WH, Liu J, Chen Z, Zhang B, Li T, Li SL, Lu JG, Zhang L, et al. : A Cell-Permeant Mimetic of NMN Activates SARM1 to Produce Cyclic ADP-Ribose and Induce Non-apoptotic Cell Death. Iscience 2019, 15:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki Y, Nakagawa T, Mao X, DiAntonio A, Milbrandt J: NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife 2016, 5:1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata H, Khine CC, Nishikawa A, Yamamoto K, Kinoshita R, Sakaguchi M: c-Jun N-terminal kinase (JNK)-mediated phosphorylation of SARM1 regulates NAD+ cleavage activity to inhibit mitochondrial respiration. J Biol Chem 2018, 293:18933–18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avalos JL, Bever KM, Wolberger C: Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD+ Cosubstrate Specificity of a Sir2 Enzyme . Mol Cell 2005, 17:855–868. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q: Structural Basis for the Mechanistic Understanding of Human CD38-controlled Multiple Catalysis*. J Biol Chem 2006, 281:32861–32869. [DOI] [PubMed] [Google Scholar]
- 39.Ruf A, de Murcia JM, de Murcia G, Schulz GE: Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc National Acad Sci 1996, 93:7481–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki Y, Kakita H, Kubota S, Sene A, Lee TJ, Ban N, Dong Z, Lin JB, Boye SL, DiAntonio A, et al. : SARM1 depletion rescues NMNAT1-dependent photoreceptor cell death and retinal degeneration. Elife 2020, 9:e62027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP: Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell reports 2015, 10:1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ: Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 2003, 38:567–580. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR: The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 2006, 50:869–881. [DOI] [PubMed] [Google Scholar]
- 44.McPhee CK, Logan MA, Freeman MR, Baehrecke EH: Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 2010, 465:1093–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kay JN, Chu MW, Sanes JR: MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 2012, 483:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaughlin CN, Perry-Richardson JJ, Coutinho-Budd JC, Broihier HT: Dying Neurons Utilize Innate Immune Signaling to Prime Glia for Phagocytosis during Development. Dev Cell 2019, 48:506–522.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou R-M, Shen Y, Yao J, Yang H, Shan K, Li X-M, Jiang Q, Yan B: Nmnat 1: a Security Guard of Retinal Ganglion Cells (RGCs) in Response to High Glucose Stress. Cell Physiol Biochem 2016, 38:2207–2218. [DOI] [PubMed] [Google Scholar]
- 48.Loreto A, Di Stefano M, Gering M, Conforti L: Wallerian degeneration is executed by an NMN-SARM1-Dependent late Ca(2+) influx but only modestly influenced by mitochondria. Cell Rep 2015, 13: 2539–2552. [DOI] [PubMed] [Google Scholar]
- 49.Galione A, Chuang K-T: Calcium Signaling. Adv Exp Med Biol 2019, 1131:371–394. [DOI] [PubMed] [Google Scholar]
- 50.Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier-Lavigne M: Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron 2013, 80:1175–1189. [DOI] [PubMed] [Google Scholar]