Abstract
RSP5, an essential gene of Saccharomyces cerevisiae, encodes a hect domain E3 ubiquitin-protein ligase. Hect E3 proteins have been proposed to consist of two broad functional domains: a conserved catalytic carboxyl-terminal domain of approximately 350 amino acids (the hect domain) and a large, nonconserved amino-terminal domain containing determinants of substrate specificity. We report here the mapping of the minimal region of Rsp5 necessary for its essential in vivo function, the minimal region necessary to stably interact with a substrate of Rsp5 (Rpb1, the large subunit of RNA polymerase II), and the finding that the hect domain, by itself, is sufficient for formation of the ubiquitin-thioester intermediate. Mutations within the hect domain that affect either the ability to form a ubiquitin-thioester or to catalyze substrate ubiquitination abrogate in vivo function, strongly suggesting that the ubiquitin-protein ligase activity of Rsp5 is intrinsically linked to its essential function. The amino-terminal region of Rsp5 contains three WW domains and a C2 calcium-binding domain. Two of the three WW domains are required for the essential in vivo function, while the C2 domain is not, and requirements for Rpb1 binding and ubiquitination lie within the region required for in vivo function. Together, these results support the two-domain model for hect E3 function and indicate that the WW domains play a role in the recognition of at least some of the substrates of Rsp5, including those related to its essential function. In addition, we show that haploid yeast strains bearing complete disruptions of either of two other hect E3 genes of yeast, designated HUL4 (YJR036C) and HUL5 (YGL141W), are viable.
The number of proteins recognized as substrates for modification by ubiquitination has increased dramatically in recent years, yet we have a relatively poor understanding of how the components of the ubiquitin system function in defining and controlling substrate specificity. At least three classes of proteins are recognized that cooperate in catalyzing protein ubiquitination, two of which, the E1 ubiquitin-activating enzyme and the family of E2 ubiquitin-conjugating enzymes, are well characterized (37). The third group of activities, the E3 ubiquitin-protein ligases, are less well characterized, and the diversity of these activities, in terms of composition and mechanisms of action, is unknown. It is clear, however, that E3 activities play the major role in defining substrate specificity, since E1 and E2 activities, alone, are generally not sufficient to direct ubiquitination of biologically relevant substrates.
The largest known family of E3 proteins, the hect E3s, were discovered through characterization of human E6-AP (15, 35, 36). The interaction of E6-AP with the human papillomavirus (HPV) E6 protein of the cervical cancer-associated HPV types causes E6-AP to associate with and ubiquitinate the p53 tumor suppressor, and several lines of evidence suggest that this is an important component of the cell-immortalizing activity of the cancer-associated HPVs (13). While the natural (HPV E6-independent) substrates of E6-AP are not known, it has been proposed that lack of expression of the maternal allele of E6-AP (also known as UBE3A) in the human brain is the likely cause of Angelman syndrome, a severe neurologic disorder (20, 26). This suggests that E6-AP-dependent ubiquitination of one or more proteins in the brain, in particular, in the hippocampal and Purkinje neurons (1), is critical for normal brain function.
The family of E6-AP-related proteins is defined by a carboxyl-terminal hect domain (homologous to the E6-AP carboxyl terminus) of approximately 350 amino acids (14, 37). Hect E3s appear to be present in all eukaryotes, with exactly 5 encoded by the Saccharomyces cerevisiae genome and over 30 so far identified in mammalian species. Based on biochemical characterization of E6-AP, it has been proposed that an obligatory intermediate in the ubiquitination reaction catalyzed by hect E3s is a ubiquitin-thioester formed between the thiol of an absolutely conserved cysteine within the hect domain and the terminal carboxyl group of ubiquitin (36). The E3 becomes “charged” with ubiquitin via a cascade of ubiquitin-thioester transfers, in which ubiquitin is transferred from the active-site cysteine of the E1 enzyme to the active-site cysteine of an E2 and, finally, to the hect E3, which catalyzes isopeptide bond formation between ubiquitin and the substrate. The E3 is thought to be recharged with ubiquitin while bound to the substrate and can therefore catalyze ligation of multiple ubiquitin moieties to the substrate through conjugation either to other lysines on the substrate or to lysine residues on previously conjugated ubiquitin molecules. The resulting multiubiquitinated substrate is then recognized and degraded by the 26S proteasome.
The hect E3s range in mass from 92 to over 500 kDa and generally have only the hect domain in common. This pattern of similarity, along with structure-function analyses of human E6-AP (16), suggested that the large and highly variable amino-terminal domains of these proteins function in defining substrate specificity. Therefore, a simple model for the function of hect E3s is that they consist of two broad functional domains: a large amino-terminal domain that contains determinants of substrate specificity and the carboxyl-terminal hect domain, which catalyzes multiubiquitination of associated substrates. To test and further refine this model, we have explored the function and substrate specificity of the hect E3 encoded by the RSP5 gene of S. cerevisiae.
RSP5 (also known as NPI1 and MDP1) is an essential gene and has been isolated in at least three types of genetic screens, including as a suppressor of mutations in SPT3 (46b). SPT3 encodes a component of the SAGA (Spt/Ada/Gcn5/acetyltransferase) complex (8, 34), which has been proposed to affect a minimum of two aspects of RNA polymerase II (pol II) transcription: the function of the TATA binding protein (Spt15) and the activity of the Gcn5 histone acetyltransferase. We previously reported that the largest subunit of RNA polII, Rpb1, is a substrate of Rsp5 (17). While the biological relevance of Rpb1 ubiquitination is not clear, it may be related to the SPT3/RSP5 genetic interaction. In addition, another study showed that the large subunit of human polII (polII-LS) is subject to ubiquitination and degradation in response to UV irradiation, suggesting that ubiquitin-mediated degradation of the polII-LS is involved in the response to DNA damage (4, 32).
RSP5 has also been shown genetically to affect the activity of several plasma membrane-associated permeases, including Gap1 (general amino acid permease), Fur4 (uracil permease), maltose permease, and the plasma membrane H+-ATPase (6, 11, 23), and ubiquitination of Fur4 and Gap1 results in their down-regulation (9, 42). Mutations in RSP5 have also been shown to alter the mitochondrial-cytoplasmic protein distribution of the Mod5 protein, a tRNA modification enzyme that functions in both of these cellular compartments (49). Although the effect of Rsp5 on some of these pathways may be indirect, the broad range of cellular processes affected by RSP5 suggests that the Rsp5 protein has multiple diverse substrates.
The primary structure of Rsp5 reveals two types of domains within the amino-terminal region of the protein (see Fig. 1). The first is a C2 domain (amino acids 3 to 140), which is found in a variety of proteins, including protein kinase C, synaptotagmin, phospholipase C, and p120ras-GAP (33). C2 domains have been shown to interact with membrane phospholipids, inositol polyphosphates, and proteins. The interaction of C2 domains with their ligands is, in most cases, dependent on or regulated by Ca2+. Since several membrane-associated substrates of Rsp5 have been identified, it is conceivable that the C2 domain is involved in targeting these proteins, either by localizing Rsp5 to the plasma membrane or by directly mediating the interaction with membrane-associated substrates, such as Fur4 and Gap1.
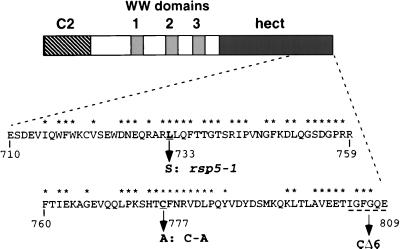
FIG. 1.
Schematic of the Rsp5 protein with the C2 domain, WW domains 1 to 3, and the hect domain indicated. The sequence of the carboxyl-terminal 100 amino acid residues of Rsp5 (amino acids 710 to 809) is shown. Amino acids marked with asterisks are residues highly conserved among the hect domain family of proteins. The rsp5-1 mutation (L to S, amino acid 733), the active-site mutation (C to A, amino acid 777), and the residues deleted by the CΔ6 mutation are indicated.
Rsp5 also contains three WW domains, spanning amino acids 231 to 418. WW domains are protein-protein interaction modules that have an affinity for proline-rich sequences, with the consensus binding site containing a PPxY sequence (PY motif) (7, 45). WW domains consist of 38 to 40 amino acids, have two absolutely conserved tryptophan residues, and form a three-stranded antiparallel β-sheet with a hydrophobic binding pocket for the PPxY ligand (24). Functionally, the WW domains are somewhat analogous to SH3 domains in that they both have affinities for proline-rich ligands, and in some cases, WW and SH3 domains can compete for binding to the same ligands in vitro (3, 44). Given their proposed function as protein-protein interaction modules, the WW domains are clearly strong candidates as sites of Rsp5 substrate interaction.
We have performed a structure-function analysis of Rsp5 in order to understand how the domain structure and in vitro biochemical activities of Rsp5 are related to its essential in vivo function. We have determined the minimal region and domains of Rsp5 necessary for complementation of the rsp5-1 temperature-sensitive mutant in vivo and the regions necessary for interaction and ubiquitination of Rpb1 in vitro. In addition, we show that the hect domain, by itself, is sufficient for formation of the ubiquitin-thioester intermediate.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strain FY56 (MATα his4-912 ΔR5 lys2-128Δ ura3-52) and isogenic rsp5-1 mutant strain FW1808 were obtained from Fred Winston (Harvard Medical School). The RSP5 and rsp5-1 open reading frames (ORFs) were amplified by PCR from the genomic DNAs of FY56 and FW1808, respectively, and cloned into the pGEM-1 vector. Three independent clones of each were sequenced and compared. The rsp5-1 ORF was subcloned into the pYES2 and pGEX-6p-1 vectors, along with the previously described wild-type (wt) RSP5 ORF, the active-site Cys-to-Ala (C-A) mutant (amino acid 777), and the CΔ6 mutant. Additional mutated RSP5 genes (see Fig. 3) were generated by PCR using gene-specific primers. 5′ PCR primers, in most cases, encoded the hemagglutinin (HA) epitope, and in other cases, PCR products were subcloned into a version of pYES2 with an initiating ATG and an HA epitope cloned upstream of the multicloning sites (pYES-HA). Mutants M, N, and O utilized primers encoding the indicated mutations, which introduced a SalI restriction site. Multiple clones of each construct were analyzed in both in vitro and in vivo assays, and the clones were partially sequenced to confirm the introduction of mutations and cloning junctions. All of the constructs shown in Fig. 3, in the pYES2 vector, were transformed into the FY56 and FW1808 strains. Plasmids encoding Rpb1 and the glutathione S-transferase (GST)-carboxy-terminal domain (CTD) fusion have been described previously (17).
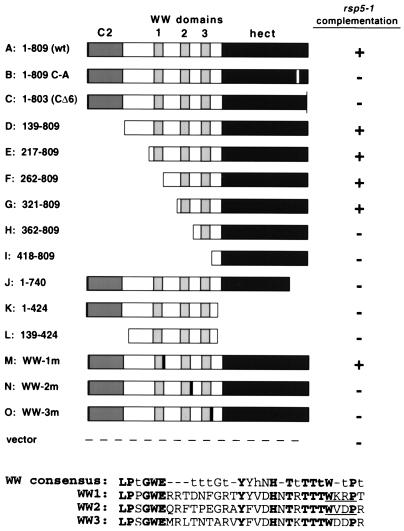
FIG. 3.
Schematic of mutated Rsp5 proteins (A to O; numbering corresponds to the amino acids retained in the mutated proteins) and summary of their abilities to complement the rsp5-1-encoded growth defect at 37°C on galactose. All of the proteins except E contained the HA epitope at the amino-terminal end. The WW consensus sequence (7) is shown relative to the sequences for WW domains 1, 2, and 3. The lowercase h in the consensus sequence represents hydrophobic residues, and a lowercase t represents a turn-like or polar position (45). Each of the WW domain mutant proteins (M, N, and O) contained the sequence FVDA in place of the WxxP sequence, as indicated.
The rsp5Δ::LEU2 mutant strains were derived from the W303 diploid strain (leu2-3112 ade2-1 his3-11 trp1-1 can1-100). A 4.5-kbp fragment of chromosome V spanning the RSP5 locus was amplified by PCR and cloned into the pBluescript II KS+ vector. Plasmid pBlueDR5 was generated by replacing the complete RSP5 ORF of this plasmid with the LEU2 gene. A linear 3.5-kbp disrupting fragment was isolated from pBlueDR5 by digestion with HindIII and NotI. This DNA fragment was transformed into diploid W303 cells, and Leu+ transformants were isolated. The deletion of one chromosomal copy of the RSP5 gene and the introduction of LEU2 at the RSP5 locus were confirmed by PCR. Sporulation of the heterozygous diploid (GW001) gave the expected 2:2 ratio of viable-to-inviable spores for an essential gene. GW001 was transformed with a centromere- and URA3-based plasmid, pRS416-RSP5, harboring the 4.5-kbp genomic RSP5 fragment, selecting for growth on plates lacking uracil and leucine. Haploid spores bearing both the rsp5Δ::LEU2 allele and the pRS416-RSP5 plasmid were isolated to yield strains GW003 (MATa rsp5Δ::LEU2 ade2-1 his3-11 trp1-1 can1-100[pRS416-RSP5]) and GW004, the equivalent MATα strain.
Centromere- and TRP1-based plasmids pRS414-Gal-A, -B, and -D were generated by subcloning the NgoM1-NotI fragment of pYES2-HA-Rsp5 (construct A; see Fig. 3), pYES2-HA-Rsp5C-A (construct B), or pYES2-HA-Rsp5ΔC2 (construct D) into the same sites of pRS414, yielding pRS414-Gal-A, -B, and -D. Expression of the respective Rsp5 proteins from these plasmids is under control of the GAL1 promoter. The pRS414-RSP5 plasmid contained the 4.5-kbp genomic RSP5 fragment, and expression of Rsp5 was therefore under control of the natural RSP5 promoter. Plasmids pRS414-Gal-A, pRS414-Gal-B, pRS414-Gal-D, and pRS414-RSP5 were transformed into yeast strains GW003 and GW004, followed by 5-fluoroorotic acid (5-FOA) counterselection against the pRS416-RSP5 plasmid. GW015 and GW016 are MATa and -α, respectively, containing the pRS414-Gal-A plasmid. GW017 and GW018 are MATa and -α, respectively, containing the pRS414-Gal-D plasmid, and GW013 and GW014 are MATa and -α, respectively, containing the pRS414-RSP5 plasmid. 5-FOA-resistant colonies could not be obtained from the pRS414-Gal-B (the C-A mutant) transformant.
The HUL4 (YJR036C) and HUL5 (YGL141W) ORFs, located on chromosomes X and VII, respectively, were isolated by PCR from FY56 genomic DNA and cloned into pBluescript. Heterozygous chromosomal disruptions of both ORFs were made individually in the W303 diploid background, as described above for RSP5. Haploid spores containing either the hul4Δ::LEU2 or hul5Δ::LEU2 allele were fully viable.
Protein expression and in vitro biochemical assays.
GST fusion proteins were expressed in Escherichia coli by standard methods and affinity purified on glutathione-Sepharose (Pharmacia). Rsp5 proteins expressed from the pGEX-6p-1 vector were cleaved from GST by using PreScission protease (Pharmacia) under manufacturer-recommended conditions, and these proteins were used in ubiquitin-thioester and ubiquitination reactions. Ubiquitin-thioester reactions were also performed by using in vitro-translated proteins (see Fig. 7), as described previously (14). E1 ubiquitin-activating enzyme and Arabidopsis thaliana Ubc8 protein were prepared as described previously (35). In vitro ubiquitin-thioester assays utilized 32P-labeled ubiquitin, prepared from pGEX-2TK-ubiquitin (35) and labeled in vitro with [γ-32P]ATP and heart muscle kinase (Sigma). The ubiquitin was cleaved from GST with thrombin, and the thrombin was then heat inactivated by incubation at 65°C for 10 min. In vitro translation reactions were carried out in coupled transcription-translation reactions by using Promega TNT reagents and [35S]methionine. Rpb1-ΔCTD was made by coupled in vitro transcription-translation by using as the template pYES2-Rbp1 plasmid DNA that had been treated with the BsiWI restriction endonuclease as described previously (17).
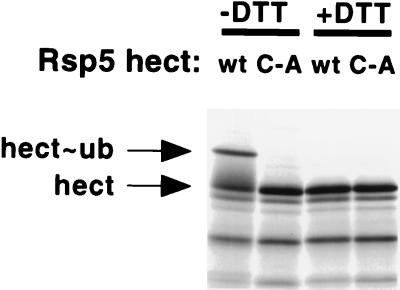
FIG. 7.
Ubiquitin-thioester assay. The wt and C-A mutant hect domains of Rsp5 (amino acids 456 to 809) were in vitro translated in a rabbit reticulocyte lysate and incubated with partially purified recombinant E1 (human) and E2 (A. thaliana UBC8) proteins. Products were subjected to SDS–10% PAGE by using loading buffer that either lacked or contained DTT.
Whole-cell yeast cell extracts for immunoblotting analyses (see Fig. 4A and B) were prepared by the method of Silver et al. (41). Primary antibodies were either an anti-HA rabbit polyclonal antibody (Santa Cruz Biotechnology) or an anti-Rsp5 mouse monoclonal antibody (17). Horseradish peroxidase-linked secondary antibodies and chemiluminescent reagents were obtained from Du Pont NEN.
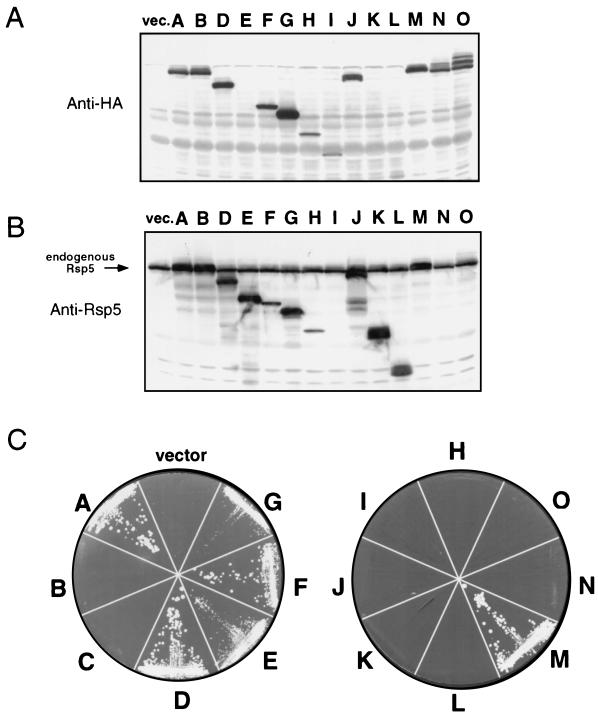
FIG. 4.
(A) Anti-HA immunoblot of total cell extracts made from the indicated FY56 transformants grown in galactose-containing medium. vec., vector. (B) Anti-Rsp5 immunoblot of the same extracts. The migration position of the endogenous Rsp5 protein is indicated. (C) Growth of the FW1808 transformants on galactose-containing agar plates at 37°C.
Rsp5/Rpb1 binding assays (see Fig. 6) were conducted as previously described (17). Briefly, each reaction mixture contained 125 μl of 25 mM Tris (pH 8.0), 125 mM NaCl, 0.1% Nonidet P-40, and 100 ng of GST fusion protein bound to 10 μl of glutathione-Sepharose. Five microliters of in vitro-translated protein was added to each reaction mixture. Reaction mixtures were rotated for 2 h at 4°C, and the Sepharose beads were washed three times with 500 μl of buffer containing 100 mM Tris, 100 mM NaCl, and 1% Nonidet P-40. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added directly to the beads and heated at 95°C for 5 min, and proteins were analyzed by SDS-PAGE and autoradiography.
FIG. 6.
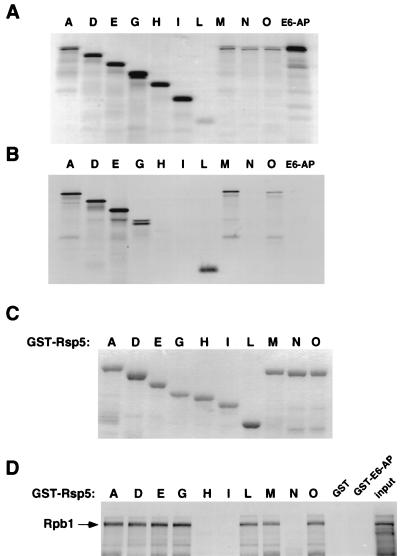
(A) 35S-labeled in vitro translation products of Rsp5 mutants, with designations corresponding to proteins depicted in Fig. 2, along with human E6-AP. (B) Translation products from panel A were assayed for binding to GST-CTD immobilized on glutathione-Sepharose. (C) Coomassie-stained SDS-PAGE of GST-Rsp5 fusion proteins, with designations corresponding to proteins depicted in Fig. 2. (D) GST-Rsp5 fusion proteins (100 ng in each lane) from panel C were assayed for binding to 35S-labeled, in vitro-translated Rpb1. GST (no fusion) and GST–E6-AP served as negative controls for binding. Twenty percent of the input amount of the translation reaction mixture used in each binding reaction is shown.
RESULTS
The protein encoded by the rsp5-1 allele is impaired in ubiquitin-thioester formation and catalysis of substrate ubiquitination.
The S. cerevisiae rsp5-1 mutant (strain FW1808), isolated by F. Winston and coworkers (cited in references 14 and 17), grows at 30°C in rich media with a doubling time only slightly longer than that of the isogenic RSP5 mutant strain (FY56) but is severely impaired for growth at elevated temperature. Characterization of the rsp5-1 allele and its protein product was undertaken in order to understand the biochemical basis of the mutant phenotype. The rsp5-1 and RSP5 ORFs were amplified by PCR from the genomic DNAs of the respective strains, and multiple independent clones were isolated and sequenced. The only difference between the rsp5-1 sequence and the wild-type RSP5 sequence was a T-to-C transition at nucleotide 2198 of the rsp5-1 ORF, resulting in a Leu-to-Ser alteration at amino acid 733 (Fig. 1). Leu733 is within the hect domain, amino terminal to the active-site cysteine at residue 777. Leu733 represents a conserved hydrophobic residue among hect domains, being either a leucine or a phenylalanine in nearly all cases.
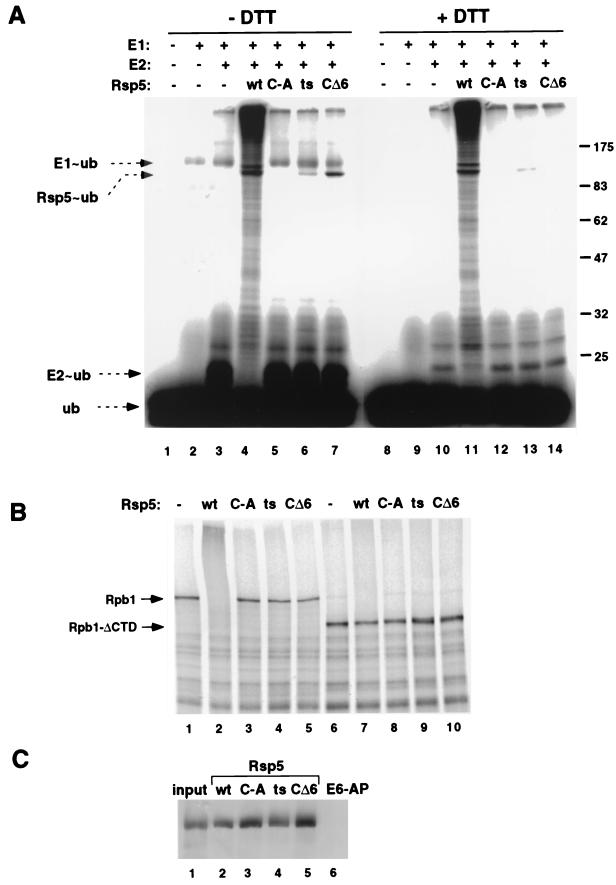
The protein encoded by the rsp5-1 allele was expressed as a GST fusion protein in the pGEX-6p vector, which allows for cleavage of the GST portion of the molecule with a highly specific protease (PreScission Protease; Pharmacia). Wt Rsp5, the active-site Cys-to-Ala (C-A) mutant, and the previously described CΔ6 mutant, in which the carboxyl-terminal six amino acids are truncated, were expressed in the same manner. Figure 2A shows the result of an in vitro ubiquitin-thioester assay utilizing these purified Rsp5 proteins along with partially purified recombinant E1 (human) and E2 (A. thaliana Ubc8) proteins and 32P-labeled ubiquitin. The basis of the assay is that thioester adducts formed between radiolabeled ubiquitin and enzyme E1, E2, or E3 will be stable when analyzed by SDS-PAGE performed in the absence of a reducing agent, but these adducts will be disrupted when analyzed under standard SDS-PAGE conditions (high reducing agent concentration). As reported previously (14), it is difficult to detect a ubiquitin-thioester intermediate with the wild-type Rsp5 protein because of the propensity of this protein to catalyze isopeptide-linked (dithiothreitol [DTT] insensitive) ubiquitination of itself via an intramolecular transfer from the cysteine to lysine residues elsewhere on the protein. While it is unclear if there is underlying biological relevance to the in vitro self-ubiquitination reaction, it is dependent on the active-site cysteine and, as shown below, correlates with the ability to catalyze intermolecular ubiquitination of substrates.
FIG. 2.
(A) Ubiquitin-thioester assay using 32P-labeled ubiquitin (ub) and purified Rsp5 proteins (the wt, the C-A mutant protein; the rsp5-1-encoded [ts] mutant protein, and the CΔ6 mutant protein). The presence or absence of recombinant E1 and E2 proteins is indicated, and the migration positions of the ubiquitin adducts are indicated. The samples on the left (lanes 1 to 7) were analyzed by SDS-PAGE using a loading buffer that lacked DTT, and the samples on the right were analyzed with a DTT-containing loading buffer (lanes 8 to 14). Migration positions of molecular mass standards (in kilodaltons) are indicated on the right. (B) Rpb1 ubiquitination assay using purified Rsp5 proteins and in vitro-translated, 35S-labeled Rpb1 (lanes 1 to 5) or Rpb1 with the CTD deleted (Rpb1-ΔCTD, lanes 6 to 10). (C) Binding of Rpb1, present in total yeast cell extract (prepared by glass bead lysis), to GST-Rsp5 fusion proteins (lanes 2 to 5) with GST–E6-AP as a negative control (lane 6). Lane 1 represents 20% of the input used in each binding reaction mixture. Rpb1 was detected by immunoblotting with an anti-CTD antibody.
The CΔ6 mutant forms a DTT-sensitive ubiquitin-thioester but, in contrast to the wild-type protein, does not catalyze isopeptide-linked self-ubiquitination, suggesting that the extreme carboxyl terminus of the protein is important for the ability to catalyze isopeptide-linked ubiquitin conjugates. This is supported by the observation that the analogous mutation in human E6-AP does not affect ubiquitin-thioester formation but abrogates ubiquitination of p53 (14). The rsp5-1-encoded protein was impaired in the formation of a ubiquitin-thioester intermediate relative to the CΔ6 mutant. A small amount of monoubiquitinated rsp5-1-encoded protein was detected after DTT and heat treatment, suggesting that it retains some capacity to catalyze isopeptide-linked ubiquitination. A temperature dependence of thioester formation was not observed for the rsp5-1-encoded protein in vitro, as it was impaired at all of the temperatures tested, from 4 to 37°C (data not shown).
We previously reported that the largest subunit of RNA polII, Rpb1, is a substrate of Rsp5 (17). The purified Rsp5 proteins were therefore assayed for the ability to catalyze ubiquitination of Rpb1 in vitro (Fig. 2B). The wt Rsp5 protein very efficiently ubiquitinated wt Rpb1 but did not ubiquitinate Rpb1 with the CTD, which is the binding site for Rsp5, deleted. The C-A protein, the CΔ6 protein, and the rsp5-1-encoded protein were all inactive in catalyzing ubiquitination of Rpb1. To determine if the inability of the mutant protein to ubiquitinate Rpb1 was due to an inability to bind to Rpb1, GST-Rsp5 fusion proteins were assayed for the ability to bind to Rpb1 present in crude yeast cell extracts. Figure 2C shows that all four GST fusion proteins (wt, C-A, CΔ6, and Rsp5-1) were equally capable of binding to Rpb1. Together, these results suggest that the biochemical basis of the rsp5-1-encoded phenotype is a defect in the ability of the enzyme to form a ubiquitin-thioester intermediate and to catalyze substrate ubiquitination, rather than a general defect in the recognition or binding of substrates.
The hect domain and WW domains 2 and 3 are required for the essential in vivo function of Rsp5.
In order to determine the minimal domain requirements for complementation of the rsp5-1 allele, a set of mutated Rsp5 proteins were expressed in rsp5-1 mutant yeast strain FW1808, as well as in isogenic RSP5 mutant strain FY56, under the control of the GAL1 promoter. Figure 3 illustrates the set of truncated and mutated Rsp5 proteins generated. All of the constructs encoded the HA antibody epitope at the 5′ end, except construct E. Mutations that disrupted individual WW domains altered the second conserved tryptophan and the nearby conserved proline residue from WxxP to FVDA, as shown in Fig. 3. These substitutions were originally suggested based on the nuclear magnetic resonance structure of the YAP WW domain, and both substitutions, individually, have been shown to render the domain completely inactive in terms of ligand (5, 24).
To confirm in vivo expression of the altered Rsp5 proteins, the FY56 transformants were grown in galactose-containing medium, and total cell extracts were analyzed for protein expression by immunoblotting by using either anti-HA or anti-Rsp5 antibodies (Fig. 4A and B). All proteins were expressed at detectable levels, and the level of expression of exogenous HA-tagged wt Rsp5 was roughly equivalent to the level of the endogenous Rsp5 protein (protein C, the CΔ6 mutant form, is not shown in the results of this experiment but was expressed at a level similar to that of wt Rsp5). Expression of protein E, which does not contain the HA epitope, was confirmed by using the anti-Rsp5 antibody (Fig. 4B). Proteins K and L were also only detected with the anti-Rsp5 antibody, even though the HA epitope was encoded in the ORF, suggesting that the epitope may have been removed by proteolysis posttranslationally. Protein I did not react with the anti-Rsp5 antibody, consistent with epitope mapping experiments that indicated that this monoclonal antibody recognizes an epitope spanning the region containing WW domains 2 and 3. The level of expression of the WW domain mutants M, N, and O was similar to that of wt Rsp5; however, proteins N and O consistently showed a pattern of higher-molecular-weight species which might represent ubiquitinated intermediates of the proteins. The expression pattern of the mutant proteins in the rsp5-1 transformants was identical to that seen in the RSP5 background (data not shown).
When expression of the exogenous RSP5 genes was repressed (by growth in dextrose), all of the FY56 (RSP5) and FW1808 (rsp5-1) transformants grew similarly to their respective untransformed strains; that is, the FY56 transformants grew at both 30 and 37°C, while the FW1808 transformants grew at 30°C but not at 37°C. When expression was induced (by growth in galactose-containing medium), all of the FY56 transformants, except K and L (Discussed below), grew similarly to the control strain (vector alone) at both 30 and 37°C. The ability of the various constructs to complement the rsp5-1 temperature-sensitive growth defect was tested by assaying for growth of the rsp5-1 transformants at 37°C on galactose-containing agar plates. Expression of the wild-type HA-tagged Rsp5 protein (A) fully complemented the temperature-sensitive growth defect, whereas the vector alone did not. Neither the active-site C-A mutant (B) nor the CΔ6 mutant (C) was able to complement the growth defect. These results strongly suggest that the ubiquitin-protein ligase activity of Rsp5 is intrinsically linked to its essential in vivo function.
Constructs D, E, F, and G, with deletions of increasing portions of the amino-terminal domain, all complemented the rsp5-1 mutation, although with variable efficiency. Protein D, with the entire C2 domain deleted, complemented the growth defect with an efficiency similar to that of full-length Rsp5; however, protein E, with sequences up to, but not including, WW domain 1 deleted grew significantly slower at 37°C. Truncation beyond WW domain 1 (F) restored nearly full complementation activity, while truncation of WW domain 1 and most of the spacer region between domains 1 and 2 supported growth with very reduced efficiency (G). Proteins with amino-terminal truncations which included WW domain 2 (proteins H and I) did not support growth at 37°C, nor did any of the carboxyl-terminally truncated proteins (C, J, K, or L). In addition, the WW domain 1 mutant fully complemented the rsp5-1 growth defect (protein M), whereas mutation of either WW domain 2 or 3 completely abrogated complementation activity (proteins N and O, respectively). These results indicate that the region of the amino-terminal domain that includes WW domains 2 and 3, along with a complete and functional carboxyl-terminal hect domain, are required for the essential function of Rsp5.
Interestingly, a subgroup of the noncomplementing mutant proteins actually inhibited the growth of rsp5-1 transformants on galactose-containing medium at the permissive temperature (data not shown) and may therefore be dominant-negative mutant proteins. This set included mutant proteins B, C, J, K, and L, which share two characteristics: a disrupted hect domain and intact WW domains. Expression of proteins K and L, which contain only amino-terminal domain sequences, inhibited growth most severely and also inhibited the growth of the RSP5 strain. We speculate that the growth-inhibitory effect of these mutant proteins is due to titration of substrates away from the endogenous rsp5-1-encoded protein, resulting in a lack of degradation of the substrate(s) related to the essential function of Rsp5. The fact that the region spanning the WW domains is required for both rsp5-1 complementation and the dominant-inhibitory effect is consistent with this hypothesis.
The C2 domain is not required for the essential in vivo function of Rsp5.
It was somewhat surprising that the C2 domain was dispensable for complementation of the rsp5-1 mutation, since several plasma membrane-associated substrates of Rsp5 have been identified. This led us to question whether the protein encoded by the rsp5-1 allele was playing a role in supporting the function of proteins with C2 deleted (proteins D, E, F, and G) in the complementation assay described above. For example, it was conceivable that at 37°C the temperature-inactivated rsp5-1 mutant protein, through heterodimerization with a protein with C2 deleted, might serve to localize the protein with C2 deleted to the plasma membrane, where it could then contact and ubiquitinate substrates. This idea was suggested by experiments that indicate that Rsp5, as well as human E6-AP, may form homodimers under certain conditions (2a). We therefore determined whether an Rsp5 protein with C2 deleted (protein D) could function as the sole source of Rsp5 protein in the cell.
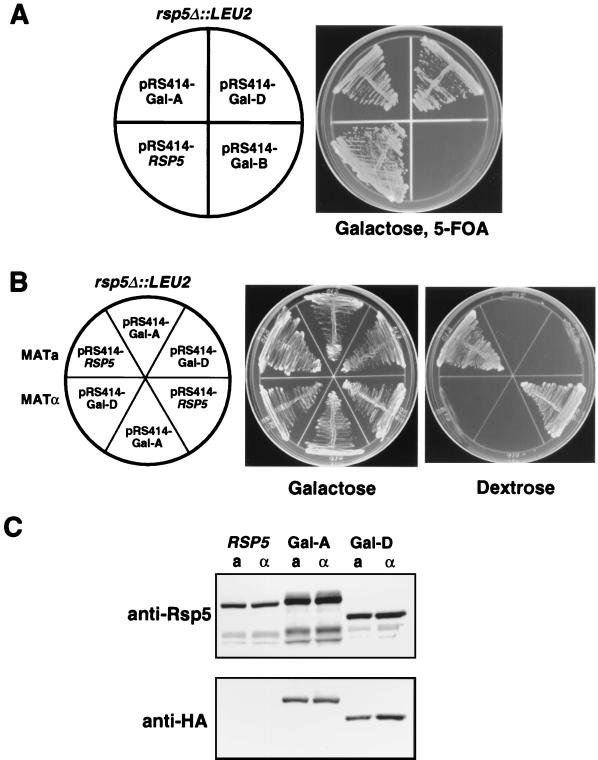
A chromosomal deletion of the complete RSP5 ORF was made in a diploid strain (W303). Tetrad dissections were consistent with RSP5 being an essential gene, as previously reported (48, 49). Viable rsp5Δ::LEU2 haploids were obtained by sporulation of the heterozygous diploid after transformation with a URA3-based centromere-containing plasmid expressing wild-type RSP5 from its own promoter. This strain was then transformed with a TRP1-based plasmid expressing either construct A (wt HA-Rsp5), B (the C-A mutant), or D (with the C2 domain deleted) under control of a galactose-inducible promoter or with wild-type RSP5 expressed from its own promoter. The URA3-based RSP5 plasmid was then selected against with 5-FOA. In accord with the rsp5-1 complementation results, 5-FOA-resistant transformants were obtained on galactose-containing plates with plasmids expressing protein A or D, as well as with the wild-type RSP5 genomic fragment, but not with construct B, the C-A mutant (Fig. 5A). As expected, the A and D transformants of either mating type could grow with galactose but not glucose as the carbon source (Fig. 5B). Immunoblotting confirmed that transformant D in the rsp5Δ background expresses only the form of Rsp5 with the C2 domain deleted (Fig. 5C). We therefore conclude that the C2 domain is not required for the essential Rsp5 function under standard growth conditions.
FIG. 5.
(A) Δrsp5::LEU2 haploid cells were transformed with a URA3-based plasmid expressing wt RSP5 and a TRP1-based plasmid expressing either HA-tagged wt Rsp5 (Gal-A), the C-A mutant protein (Gal-B), or a protein with the C2 domain deleted (Gal-D) under control of the GAL1 promoter or wt Rsp5 under the control of its own promoter (RSP5). The transformants were streaked onto galactose-containing agar plates containing uracil and 5-FOA to select for loss of the URA3 plasmid. All of the TRP1-based plasmids, except that expressing the C-A mutant protein (B), could support continued growth of the cells under these conditions. (B) Galactose and dextrose plates of the RSP5 and Gal-A and Gal-D transformants from panel A. The upper and lower sectors are a and α mating types, respectively. (C) Total cell extracts of the genomic RSP5 and the Gal-A and Gal-D transformants, grown in galactose, were analyzed by immunoblotting by using either an anti-Rsp5 (top) or an anti-HA (bottom) antibody. Both a and α mating types were analyzed.
WW domain 2 is essential for mediating the interaction of Rsp5 with Rpb1.
We previously showed that Rsp5 binds and ubiquitinates Rpb1, the largest subunit of RNA polII and that the CTD of Rpb1 is necessary and sufficient for stable binding to Rsp5 (17). To determine which regions of Rsp5 are critical for binding to Rpb1, a subset of the Rsp5 mutant proteins (A, D, E, G to I, and L to O) were translated in vitro in a rabbit reticulocyte lysate system (Fig. 6A) and assayed for the ability to bind to GST-CTD (Fig. 6B). Paralleling their importance in the in vivo rsp5-1 complementation assay, deletion of the C2 domain or WW domain 1 did not affect CTD binding (D, E, G, and L), while deletion of WW domain 2 or beyond (H and I) abolished CTD binding. Likewise, disruption of WW domain 1 by mutation did not affect CTD binding (M), while disruption of WW domain 2 completely abolished binding (N). Mutation of WW domain 3 (protein O) consistently reduced binding by approximately 50%. As shown previously (17), binding of Rsp5 to the CTD was independent of the hect domain, as complete deletion of the hect domain (L) resulted in binding to GST-CTD with efficiency similar to that of wild-type Rsp5 (A).
Binding of Rpb1 to Rsp5 was also analyzed by expressing the same subset of Rsp5 mutants as GST fusion proteins (Fig. 6C) and assaying for binding to in vitro-translated Rpb1 (Fig. 6D). The results were similar to those described above, except that in this assay, disruption of WW domain 3 did not significantly affect Rpb1 binding. The results were the same, regardless of whether Rpb1 was in vitro translated or present in crude yeast cell extracts (data not shown), consistent with our previous findings that Rsp5 can bind to Rpb1 whether it is free or present in the multisubunit polII enzyme. Together, these results indicate that the C2 domain, WW domain 1, and the hect domain are not required for Rpb1 binding, while WW domain 2 is absolutely required. WW domain 3 may also contribute to Rpb1 binding but is not required.
The hect domain, by itself, can form a ubiquitin-thioester intermediate.
To determine if the hect domain, by itself, is sufficient for recognition and activation by the E1 and E2 enzymes of the ubiquitin system, the isolated hect domain of Rsp5 (amino acids 456 to 809) was assayed for ubiquitin-thioester formation, along with the equivalent active-site (C-A) mutant. Figure 7 shows that the hect domain is sufficient for ubiquitin-thioester formation and that mutation of the conserved cysteine of the hect domain abolishes this activity. These results indicate that the hect domain is a separable functional domain and that determinants necessary for interaction with the upstream components of the ubiquitin-thioester cascade (the E1 and E2 enzymes) lie completely within the hect domain. The hect domain, unlike the full-length protein, did not catalyze isopeptide-linked self-ubiquitination, suggesting that the lysines that are the sites for self-ubiquitination of the full-length protein (Fig. 2A) lie outside of the hect domain.
Two other yeast hect E3 genes, HUL4 and HUL5, are nonessential genes.
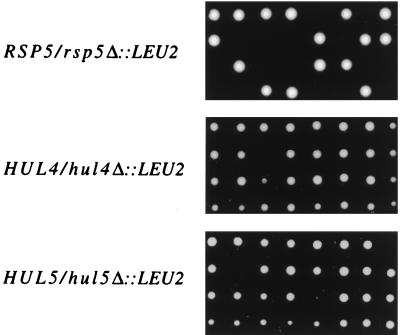
Database searches indicate that S. cerevisiae has five genes that encode apparent hect E3 proteins. These are RSP5, UFD4, TOM1, HUL4, and HUL5 (HUL stands for hect ubiquitin ligase). RSP5 is essential (49), UFD4 is not essential (19), and TOM1 (Trigger of Mitosis) is presumed to be essential (required for the G2/M cell cycle transition; unpublished data; see reference 46). To determine the phenotype of haploid strains bearing disruption of either of the previously uncharacterized hect E3 genes, here designated HUL4 (YJR036C) and HUL5 (YGL141W), we completely deleted each ORF individually in the W303 diploid background, creating the HUL4/hul4Δ::LEU2 and HUL5/hul5Δ::LEU2 heterozygous strains. We also created an RSP5/rsp5Δ::LEU2 diploid in the same manner. Tetrad analysis of the RSP5/rsp5Δ::LEU2 diploid showed the expected 2:2 ratio of viable-to-inviable spores for an essential gene, with the viable spores being leucine auxotrophs. In contrast, over 90% of the tetrads from both the HUL4/hul4Δ::LEU2 and HUL5/hul5Δ::LEU2 diploids yielded three or four viable spores (Fig. 8), with a 2:2 segregation of the LEU2 allele. Both haploid disruptants grew similarly to the parental strain at 37°C, as well. Therefore, HUL4 and HUL5 are not essential genes under standard growth conditions.
FIG. 8.
Tetrad analysis of RSP5/rsp5Δ::LEU2, HUL4/hul4Δ::LEU2, and HUL5/hul5Δ::LEU2 W303 diploids on leucine-containing agar plates.
DISCUSSION
The hect E3 proteins have been proposed to consist of two functional domains: a large amino-terminal domain that functions in defining substrate specificity and an approximately 40-kDa carboxyl-terminal domain that catalyzes ubiquitination of bound substrates. The results of our Rsp5 structure-function analysis support this two-domain model and point to those determinants within the amino-terminal domain that are critical for the essential in vivo function of Rsp5. In addition, our results strongly suggest that the ubiquitin-protein ligase activity of Rsp5 is intrinsically linked to its essential in vivo function. This is based on the finding that mutations within the hect domain that disrupt or impair formation of the ubiquitin-thioester and/or the ability of the protein to catalyze transfer of ubiquitin to substrates (the C777-to-A, rsp5-1, and CΔ6 mutations) abrogate the in vivo function of Rsp5. Several other rsp5 temperature-sensitive alleles were isolated by Zolladek et al. (49), and like rsp5-1, these mutations also lie within the hect domain and disrupt conserved amino acids. We speculate that these mutations also result in a defect in Rsp5 ubiquitination activity, rather than a general defect in binding of or association with substrates.
With respect to the specific determinants within the amino-terminal region of Rsp5, our results clearly indicate that the C2 domain is not required for the essential function of Rsp5. Several membrane-associated permeases have been shown to be regulated by ubiquitination in an Rsp5-dependent manner, including Fur4 (uracil permease) and Gap1 (general amino acid permease) (9, 42). Based on the fact that C2 domains have been shown to associate with membrane phospholipids or membrane-associated proteins, the C2 domain might be involved in targeting this group of substrates, either by affecting intracellular localization of Rsp5 or by directly contacting substrates. If this is indeed the case, then targeting of membrane-associated substrates must not be the essential function of Rsp5. Further experiments are necessary to determine the effect of specific Rsp5 mutations on ubiquitination of plasma membrane-associated substrates.
In contrast to the C2 domain, a subset of the WW domains are essential for Rsp5 function. Disruption of WW domain 2 or 3 abrogates Rsp5 function in vivo, while WW domain 1 can be disrupted with no significant effect on rsp5-1 complementation activity. WW domains have been proposed to be protein-protein interaction modules with an affinity for proline-rich sequences containing a PPxY motif (45). WW domains 2 and 3 are therefore likely to be involved in mediating the interaction with the substrate(s) of Rsp5 related to its essential function. WW domain 2 is absolutely required for Rpb1 binding, while WW domain 3 may also contribute, and the C2 domain and WW domain 1 can be disrupted without affecting Rpb1 binding. Thus, the determinants for Rpb1 binding and ubiquitination correspond to those required for rsp5-1 complementation, and Rpb1 is therefore a candidate for being at least one of the substrates related to the essential function of Rsp5.
The CTD of Rpb1, consisting of 26 copies of a heptapeptide repeat, is the binding site for Rsp5 (17). The CTD is essential for Rpb1 activity in vivo and is the site of interaction for many components of the transcriptional machinery. The consensus binding site for WW domains is a PPxY sequence, although certain substitutions in the first proline are permissible (5). The CTD heptapeptide sequence SPTSPSY may therefore be the direct recognition sequence for the WW domains of Rsp5. The function of Rpb1 ubiquitination with respect to polII transcription is not clear; however, human polII-LS is ubiquitinated in response to UV irradiation (32), suggesting a possible role in the response to DNA damage. Preliminary results have shown that DNA damage induces degradation of Rpb1 in yeast, as well, and that this is dependent on Rsp5 (2a). Apparent homologs of Rsp5 exist in mammalian cells (see below), and it is therefore possible that one or more of these mediate UV-dependent ubiquitination of human polII.
While the majority of hect E3s have unique amino-terminal domain sequences without recognizable or characterized sequence motifs, there are several hect E3s from both mammalian and nonmammalian species which, like Rsp5, contain multiple (two to four) WW domains (18, 21, 29, 30, 47). Although several of the cDNAs encoding these proteins remain to be sequenced in their entirety, all of the proteins appear to also contain a C2 domain at the extreme amino terminus. Schizosaccharomyces pombe encodes two hect E3 genes with three WW domains and one C2 domain, one of which is pub1, which targets the cdc25 protein phosphatase (27). The mouse-human Nedd4 protein (human Rpf1) is a C2/WW-hect E3 protein that has been proposed to down-regulate the epithelial Na+ channel by ubiquitinating the β and/or γ subunits (31, 43). Thus, the WW- or C2/WW-hect E3s appear to form a distinct subgroup of the hect E3s. The diversity of functions and substrates of the WW hect E3s suggests that these proteins may perform their functions in diverse intracellular locations. Studies on the mammalian C2/WW-hect E3, Nedd4, has shown that it is primarily cytoplasmic but also perinuclear and that calcium influx mediates localization to the plasma membrane (10, 21, 31). Preliminary localization studies with yeast suggest that Rsp5 may be both cytoplasmic and nuclear (46a).
Of the five yeast hect E3s, only Rsp5 contains WW domains, and neither Tom1, Ufd4, Hul4, nor Hul5 contains any previously identified protein motifs within its amino-terminal region. We speculate that each of the individual WW domains of Rsp5 is involved in mediating the interaction with one or more specific substrates. Five other yeast genes encode proteins with apparent WW domains (ESS1, PRP40, TIN1, YPR152C, and YFL010C), and all of these contain a single WW domain, except PRP40, which contains two. A subject for further investigation is whether any of the WW domain proteins that are not hect E3s can compete with Rsp5 for binding to its substrates, which could be a basis for regulated ubiquitination.
The hect domain of Rsp5, by itself, was found to be sufficient for formation of the ubiquitin-thioester intermediate, indicating that the hect domain has biochemical activity independent of the amino-terminal domain. Schwarz et al. recently reported similar results for human E6-AP (38). The hect domains of several E3s have been shown to interact with specific, in some cases multiple, E2 enzymes (10, 22, 28). The specific yeast Ubc proteins capable of activating Rsp5 are not known, but by homology to Arabidopsis Ubc8, which very efficiently activates Rsp5 in vitro, yeast Ubc4 and Ub5 are the best candidates, followed by Ubc1 and Ubc13. UBC1, UBC4, and UBC5 constitute an essential subset of UBC genes (39, 40). The fact that the hect domain possesses all of the determinants for ubiquitin-thioester formation supports the two-domain model of hect E3 function and suggests that hect domains, which, on average, share approximately 40% similarity, might be interchangeable between hect E3 proteins. This may not be generally the case, however, since Schwarz et al. showed that an E6-AP/hectH6 chimera, which was capable of forming a ubiquitin-thioester and binding the HPV E6 protein and p53, did not ubiquitinate p53 (38). In addition, structure-function analyses with E6-AP demonstrated that determinants for E6-dependent p53 binding lie partially within the hect domain (16). Thus, while our results obtained with Rsp5 support a simple two-domain model for hect E3 function, results obtained with E6-AP suggest a more complex picture of substrate recognition. Combined genetic and biochemical analyses of other yeast hect E3s should provide many opportunities to address the question of substrate recognition and specificity.
The results presented here support the idea that the ubiquitin-thioester intermediate is a requisite intermediate in ubiquitination reactions catalyzed by hect E3 proteins. Three other classes of E3 activities have been identified, which are represented by Ubr1 of yeast (2, 25), the multimeric anaphase-promoting complex (or cyclosome) (12), and the Cdc53/Cdc4/Skp1 complex (12). None of these types of E3 activities have been shown to function via a ubiquitin-thioester intermediate, and it is presumed, therefore, that E2 directly catalyzes substrate ubiquitination. These E3 activities may therefore function by providing a surface for simultaneous interaction of E2 and the substrate(s). Whether the mechanism of action for hect E3s is more the rule or the exception for E3 activities awaits further characterization of these and potentially other types of E3 activities and their substrates.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant CA72943-01 from the National Cancer Institute.
We thank Sylvie Beaudenon for many helpful discussions, Mike Kiledjian for critical reading of the manuscript, and Xin Li for assistance with tetrad dissections.
REFERENCES
- 1.Albrecht U, Sutcliffe J S, Cattanach B M, Beechey C V, Armstrong D, Eichele G, Beaudet A L. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 2.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Beaudenon, S., and J. M. Huibregtse. Unpublished data.
- 3.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H I, Einbond A, Kwak S-J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 6.de la Fuente N, Maldonado A M, Portillo F. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 1997;411:308–312. doi: 10.1016/s0014-5793(97)00721-7. [DOI] [PubMed] [Google Scholar]
- 7.Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 8.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 9.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama S, Jensen J P, Weissman A M. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 11.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoyt A M. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 13.Huibregtse J M, Maki C G, Howley P M. Ubiquitination of the p53 tumor suppressor. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Publishing Corp.; 1998. pp. 323–343. [Google Scholar]
- 14.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imhof M O, McDonnell D P. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson E S, Ma P C M, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 20.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Harvey K F, Kinoshita M, Copeland N G, Noda M, Jenkins N A. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics. 1997;40:435–443. doi: 10.1006/geno.1996.4582. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Kao W H, Howley P M. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 23.Lucero P, Lagunas R. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol Lett. 1997;147:273–277. doi: 10.1111/j.1574-6968.1997.tb10253.x. [DOI] [PubMed] [Google Scholar]
- 24.Macias M J, Hyvonen M, Baralk E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 25.Madura K, Varshavsky A. Degradation of Ga by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura T, Sutcliffe J S, Fang P, Galjaard R-J, Jiang Y-H, Benton C S, Rommens J M, Beaudet A L. De novo truncating mutations in the E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 27.Nefsky B, Beach D. Pub1 acts as an E6-AP-like protein ubiquitin ligase in the degradation of cdc25. EMBO J. 1996;15:1301–1312. [PMC free article] [PubMed] [Google Scholar]
- 28.Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J Biol Chem. 1996;271:2795–2800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- 29.Perry W L, Hustad C M, Swing D A, O’Sullivan T N, Jenkins N A, Copeland N G. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 30.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14666. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 31.Plant P J, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 32.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 33.Rizo J, Sudhof T C. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 34.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 36.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 37.Scheffner M, Smith S, Jentsch S. The ubiquitin-conjugation system. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Publishing Corp.; 1997. pp. 65–98. [Google Scholar]
- 38.Schwarz S E, Rosa J L, Scheffner M. Characterization of human hect domain family members and their interaction with UbcH5 and UbcH7. J Biol Chem. 1998;273:12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 39.Seufert W, Jentsch S. Ubiquitin conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seufert W, McGrath J P, Jentsch S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990;9:4535–4541. doi: 10.1002/j.1460-2075.1990.tb07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver P A, Chiang A, Sadler I. Mutations that alter both localization and production of a yeast nuclear protein. Genes Dev. 1988;2:707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- 42.Springael J Y, Andre B. Nitrogen-regulated ubiquitination of the gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 44.Sudol M. The WW module competes with the SH3 domain? Trends Biochem Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- 45.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 46.Ustugi T, Toh-e A, Kikuchi Y. A high dose of the STM1 gene suppresses the temperature sensitivity of the tom1 and htr1 mutants in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1263:285–288. doi: 10.1016/0167-4781(95)00123-x. [DOI] [PubMed] [Google Scholar]
- 46a.Wang, G., and J. M. Huibregtse. Unpublished data.
- 46b.Winston, F., et al. Unpublished data cited in references and .
- 47.Wood J D, Yuan J, Margolis R L, Colomer V, Duan K, Kushi J, Kaminsky Z, Kleiderlein J J, Sharp A H, Ross C A. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11:149–160. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]
- 48.Yashiroda H, Oguchi T, Yasuda Y, Akio T-E, Kikuchi Y. Bul1, a new protein that binds to the Rsp5 ubiquitin-protein ligase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3255–3263. doi: 10.1128/mcb.16.7.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zolladek T, Tobiasz A, Vaduva G, Boguta M, Martin N C, Hopper A K. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]