Abstract
With the gradual increase in the COVID-19 mortality rate, there is an urgent need for an effective drug/vaccine. Several drugs like Remdesivir, Azithromycin, Favirapir, Ritonavir, Darunavir, etc., are put under evaluation in more than 300 clinical trials to treat COVID-19. On the other hand, several vaccines like Pfizer-BioNTech, Moderna, Johnson & Johnson’s Janssen, Sputnik V, Covishield, Covaxin, etc., also evolved from the research study. While few of them already gets approved, others show encouraging results and are still under assessment. In parallel, there are also significant developments in new drug development. But, since the approval of new molecules takes substantial time, drug repurposing studies have also gained considerable momentum. The primary agent of the disease progression of COVID-19 is SARS-CoV2/nCoV, which is believed to have ~89% genetic resemblance with SARS-CoV, a coronavirus responsible for the massive outbreak in 2003. With this hypothesis, Human-SARS-CoV protein interactions are used to develop an in-silico Human-nCoV network by identifying potential COVID-19 human spreader proteins by applying the SIS model and fuzzy thresholding by a possible COVID-19 FDA drugs target-based validation. At first, the complete list of FDA drugs is identified for the level-1 and level-2 spreader proteins in this network, followed by applying a drug consensus scoring strategy. The same consensus strategy is involved in the second analysis but on a curated overlapping set of key genes/proteins identified from COVID-19 symptoms. Validation using subsequent docking study has also been performed on COVID-19 potential drugs with the available major COVID-19 crystal structures whose PDB IDs are: 6LU7, 6M2Q, 6W9C, 6M0J, 6M71 and 6VXX. Our computational study and docking results suggest that Fostamatinib (R406 as its active promoiety) may also be considered as one of the potential candidates for further clinical trials in pursuit to counter the spread of COVID-19.
Keywords: COVID-19, Drug repurposing, Fostamatinib/R406, nCoV-Human PPIN, COVID-19 symptom analysis, Molecular docking
1. Introduction
The world has witnessed several severe epidemics like Spanish flu, ebola, cholera etc. Now we are in front of the most life-threatening viral outburst with COVID-19. The feature that makes this new coronavirus, nCoV, unique is its ability to quickly transmit through an infected COVID patient [1]. The virus causing COVID-19 is an assimilation of accessory, non-structural and structural proteins [2]. According to World Health Organization (WHO) coronavirus disease dashboard [3], 162,704,139 confirmed cases of COVID-19, including 3,374,052 deaths, have been reported as of 1:39 pm CEST, 17 May 2021. Based on the prior knowledge of major outbreaks of Ebola, cholera etc., treatments with different antiviral drugs are considered and implemented to terminate COVID-19 based on previous knowledge of significant attacks. A literature survey [4] is recently carried out through a refined computational search in various online repositories like Google Scholar, Science Direct, PubMed, etc., to enlist various COVID-19 drug-related research articles since the onset of this pandemic. Almost 22 most relevant COVID-19 related drug articles [4] have been filtered out from the search results. All these significant researches and some others have been extensively studied, the details of which have been listed in the supplementary Table S1.
It is noted from Table S1 that the most recommended drugs are azithromycin, lopinavir, ritonavir, remdesivir, and favipiravir. It also appears that the amount of data accessible for these drugs is insufficient to recommend any one of them as a treatment for COVID-19 until and unless the necessary amount of appropriate clinical trials are executed. Relative data comparison is missing in almost all human-related studies about COVID-19. So, it is uncertain whether the COVID infected patient recovers due to applying the suggested drug or recover due to extensive clinical care and isolation. However, some of the in vitro studies have shown favourable results for these drugs. Still, these are all preliminary data, which need much more evidence before putting it in clinical trial.
COVID-19 vaccines are also in the same race as COVID-19 drugs. According to a current report [5] by the Centers for Disease Control and Prevention (CDC), Pfizer-BioNTech, Moderna, and Johnson & Johnson’s Janssen are the recommended and authorized vaccines in the United States COVID-19. In addition, another vaccine Sputnik V has been developed by Gamaleya National Center in Russia. Though it is found effective in initial trials, it has been recommended only for emergency use by the Technical Advisory Group (TAG) of WHO [6]. In contrast, Covishield [7] and Covaxin [8] are the recommended vaccines in India. All these vaccines might have specific side effects which need further analysis and research. But the two significant areas of concern are: 1) it is still unknown whether these vaccines are effective against all COVID-19 strains and 2) “getting vaccinated” does not guarantee that COVID-19 will not happen again. Still, it says it could save somebody’s life by refraining from getting seriously ill if they get infected with COVID-19. So, vaccines do not provide any herd immunity.
Due to the daily increase in deaths [9], there is an urgent need to identify a potential vaccine/drug that will eventually help eradicate COVID-19. So, with no other alternatives left, clinical trials have been started by WHO [10] on all the suggested drugs, including the ones mentioned in Table S1, which are somehow proved to be substantially beneficial in case studies of COVID-19. Drug design needs a proper understanding of disease transmission mechanisms that can be effectively done by analysing Host-pathogen protein–protein interaction networks (PPIN) [10]. Pathogen facilitates disease progression as it has the potential to transform itself by mutation. Infection of pathogen gets broadcasted through the connecting edge of interaction between host and pathogen. Thus, it is essential to explore target proteins and their interactions in Host-pathogen protein–protein interaction [11] networks for potential drug discovery [12]. However, the only recognized in vitro Human-nCoV PPIN available to date is in the work of Gordon et al. [13]. But UniProt reviewed nCoV proteins cannot be mapped through this in vitro generated PPIN. So, all this lead to the development of an in silico Human-nCoV PPIN through the SIS model [14] and fuzzy thresholding. Further study of protein targets of potential Food and Drug Administration (FDA) drugs [15] of COVID-19 in the formed Human-nCoV PPIN network also shows that FDA approved drug, Fostamatinib (R406 as its active promoiety) [16], can be a potential drug for COVID-19 treatment. Rigel Pharmaceuticals, Inc. [17] got approval for TAVALISSE (Fostamatinib disodium hexahydrate) for the treatment of Chronic Immune Thrombocytopenia [18] from the FDA on 17/04/2018.
The major contribution of the proposed work is described as follows: 1) It uses an in silico model which has been developed to identify potential spreader proteins in a Human-nCoV interaction network in the work of Saha et al. [19], [20], which was validated using proteins which are the targets of potential FDA drugs [15] for COVID-19 treatment. 2) A two-way analysis: a) Human-nCoV interaction network analysis b) COVID-19 symptom [21] based analysis (including “loss of smell”), have been implemented to detect the potential candidates in the list of FDA drugs for COVID-19. 3) In both the analyses, Fostamatinib/R406 [16], an FDA approved drug and commonly used for the treatment of chronic immune thrombocytopenia (ITP) [22], ranks at the top having a maximum overlap of target proteins in the Human-nCoV interaction network. 4) Fostamatinib/R406 is used for thrombocytopenia [23] which is also associated with severe coronavirus disease 2019 (COVID-19) infections [24]. 5) Molecular docking has also been performed on Fostamatinib/R406 and other potential FDA drugs [15] with the available major COVID-19 crystal structures having PDB IDs: 6LU7 [25], 6M2Q [26], 6W9C [27], 6M0J [28], 6M71 [29] and 6VXX [30]. While Fostamatinib registers the highest score for 6LU7 and 6M2Q, it obtains a second position than the other COVID-19 structures. 6) The active promoiety of Fostamatinib, i.e., R406, generates the highest docking scores compared to all other active metabolites. The detailed analysis of this entire methodology has been discussed in the subsequent sections and supplementary document.
2. Methods
The proposed methodology involves 4 datasets: 1) Human PPIN [31], [32] 2) SARS-CoV PPIN [33] 3) SARS-CoV-Human PPIN [33] and 4) SARS-CoV2 proteins [34]. The overall dataset statistics are highlighted in Table 1 . The entire proposed methodology of drug repurposing can be categorized into four major sections.
Table 1.
Description and details of the datasets.
| Database Name | Description | Nodes | Interactions/Edges |
|---|---|---|---|
| Human PPIN | Human-human protein interactions | 21,557 | 342,353 |
| SARS-CoV PPIN | SARS-CoV-SARS-CoV protein interactions | 7 | ---- |
| SARS-CoV-Human PPIN | SARS-CoV-Human protein interactions | 120 | 118 |
| SARS-CoV2 proteins | UniProt collected reviewed COVID19 proteins | 14 | ---- |
2.1. Detection of spreader nodes in Human-nCoV PPIN
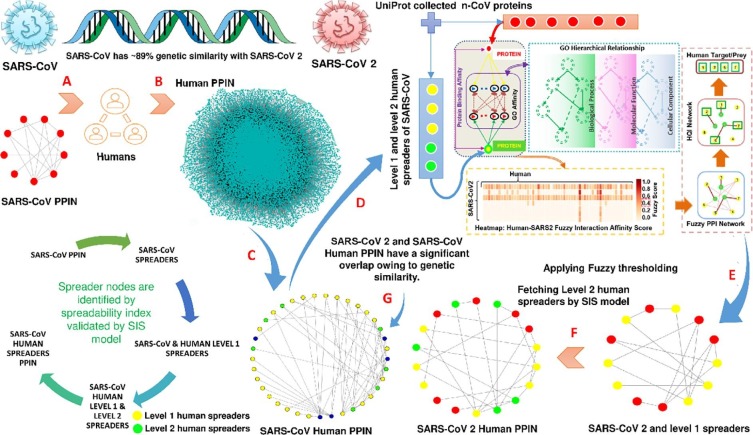
The only recognized in vitro Human-nCoV PPIN available to date is in the work of Gordon et al. [13]. But UniProt reviewed nCoV proteins cannot be mapped through this in vitro generated PPIN. So, an attempt has been made to construct a Human-nCoV PPIN based on the available PPIN information of SARS-CoV, which have ~89% similarity [35], [36] with SARS-CoV2. Not every protein in a PPIN is a spreader protein/node. Spreader proteins are considered to be those specific proteins that have a unique fast capability of transmitting infection in their neighbourhood in a short time [19]. They are identified through spreadability index computed by the combination of three terminologies: 1) Edge ratio [37], 2) Neighbourhood density [37] and 3) node weight [38]. Proteins having a high spreadability index are spreader proteins (for more details, please see supplementary). Identification of spreader proteins is conducted initially in the SARS-CoV PPIN dataset. Corresponding connected human proteins, i.e., level 1 and level 2 of selected SARS-CoV spreader proteins, are chosen from SARS-CoV-Human PPIN and Human PPIN datasets. Hence, the spreadability index detects spreader proteins in level 1 and 2 human proteins of SARS-CoV. The selected spreader nodes are also validated by Susceptible-Infected-Susceptible (SIS) model [14] (see Fig. 1 ). This results in forming a PPIN consisting of 7 SARS-CoV, 24 level 1 and 111 level 2 human spreader proteins, respectively, under a low threshold [19]. The potential Human-nCoV interactions have been identified using developed in silico fuzzy PPI model [39]. In this model, SARS-COV spreader (level-1 and level-2) proteins in humans are considered the candidate set of interactors for nCoV [20]. The nCoV-Human pair-wise relationships are quantified using the semantic similarity of their annotated GO pairs. A hybrid approach has been applied to assess the semantic similarity between GO target pairs using the topological properties of three GO subgraphs (BP: Biological Process, MF: Molecular Function and CC: Cellular Component) [40]. These GO-level assessment scores are incorporated to obtain the fuzzy interaction affinity (score ranges [0, 1]) between the target Human and nCoV protein pair and results (see Fig. 1). The high specificity (99.9%) has been achieved on a threshold of 0.4 fuzzy interaction affinity score on a benchmark Human PPI dataset. Finally, with the high specificity threshold, potential interactions are identified between nCoV bait and human prey [20].
Fig. 1.
The schematic diagram for the construction of COVID-19-Human PPIN. A. Formation of SARS-CoV PPIN (Red nodes) B. Formation of Human PPIN C. Spreader nodes identification in SARS-CoV-Human PPIN which is formed by the application of spreadability index and SIS model (Blue nodes represent SARS-CoV spreaders, while yellow and green denote level-1 and level-2 human spreaders) D. UniProt collected n-CoV proteins along with level-1 and level-2 human spreader nodes in SARS-CoV-Human PPIN are provided as an input to fuzzy thresholding E. SARS-CoV2, and level-1 Human spreaders are identified based on Fuzzy score (Red nodes represent SARS-CoV2 spreaders, while yellow denote level-1 human spreaders) F. Level-2 Human spreaders are identified by the application of the SIS model (Green nodes represent level-2 human spreaders) G. SARS-CoV-Human PPIN and SARS-CoV2-Human PPIN have a significant overlap owing to its genetic similarity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Identification of potential candidate FDA drugs with respect to COVID19 spreader nodes using Human-nCoV interaction network analysis
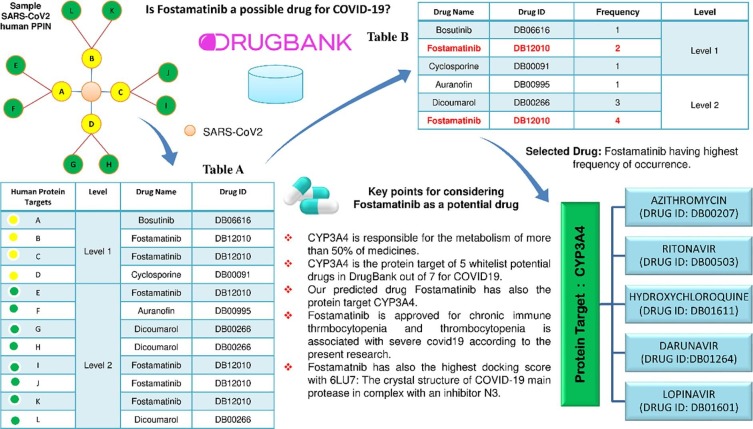
Once the COVID19-human PPIN is formed, all the level 1 and level 2 human proteins of COVID19 are mapped with their corresponding drugs from DrugBank [41]. DrugBank is an online repository [42] that contains comprehensive data about drugs, drug-protein targets and information about drug metabolism. Due to the high-quality annotation in DrugBank, it becomes the most used database in almost all in silico methodologies involved in drug design, docking of drugs, and drug interaction prediction. It contains about 60% and 10% of FDA approved and experimental drugs, respectively [41]. On proper analysis, it has been observed that various spreader nodes in COVID19-human PPIN are the protein targets of potential COVID19 FDA listed drugs [15]: hydroxychloroquine [43], [44], azithromycin [43], lopinavir [45], ritonavir [46], remdesivir [47], [48], [49], and favipiravir [50], [51]. The details of significant overlap between spreader nodes and drug-protein targets are highlighted in Table 2 [20]. It is observed from Table 2 that hydroxychloroquine has the highest hit/overlap, i.e. four, while each of azithromycin, lopinavir, ritonavir and darunavir has two hits [20]. Remdesivir and favipiravir have one impact individually [20]. Remdesivir is the only drug that acts directly on COVID19 protein R1AB_SARS2. Significant overlapping drug targets and spreader nodes in Table 2 motivate us to analyse further and develop a consensus strategy to identify a potential drug for COVID19 treatment. The consensus strategy is described in Algorithm 1 (PDS_CS). Drug consensus score (DCS) is used in PDS_CS, defined as the frequency of occurrences of a drug at a particular level of PPIN. Execution of the PDS_CS algorithm is also highlighted in Fig. 2 by considering a sample (randomly generated COVID19-Human PPIN). In this PPIN, corresponding linked drugs are mapped with each human protein (marked as green) in level-1 and level-2, as shown in Table A in Fig. 2. Hence the DCS, i.e., frequency of each drug, is computed and highlighted in Table B in Fig. 2. Since Fostamatinib has the highest DCS in both levels, it is considered the potential drug for the target nCoV protein in the randomly generated COVID19-Human PPIN (marked as red). Algorithm 1 is not only implemented in the in silico generated Human-nCoV PPIN [20] but also in the host targets of in vitro generated Human-nCoV PPIN of Gordon et al. [13] (for details, please see Section 3.6).
Table 2.
Details of overlap of spreader nodes and potential COVID19 FDA listed drugs.
| Sl. No. | COVID19 FDA listed Drugs | DrugBank ID | Drug Protein targets/Spreader nodes | No. of hits |
|---|---|---|---|---|
| 1 | Hydroxychloroquine | DB01611 | TLR9, ACE2, CYP3A4, ABCB1 | 4 |
| 2 | Azithromycin | DB00207 | CYP3A4, ABCB1 | 2 |
| 3 | Lopinavir | DB01601 | CYP3A4, ABCB1 | 2 |
| 4 | Ritonavir | DB00503 | CYP3A4, ABCB1 | 2 |
| 5 | Remdesivir | DB14761 | R1AB_SARS2 | 1 |
| 6 | Favipiravir | DB12466 | ABCB1 | 1 |
| 7 | Darunavir | DB01264 | CYP3A4, ABCB1 | 2 |
Fig. 2.
A drug consensus score was adopted to choose Fostamatinib/R406 as a potential COVID19 drug. Other connecting biological links for selecting the same have also been highlighted.
2.3. Identification of potential candidate FDA drugs with respect to COVID19 spreader nodes using COVID19 symptoms, risk factors and clinical outcome-based analysis
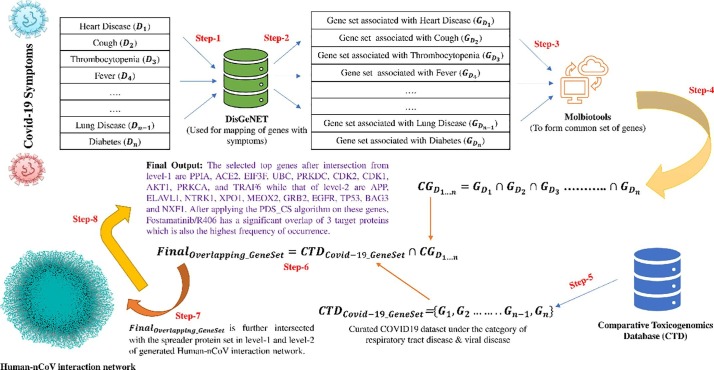
COVID19 is associated with specific health symptoms like cough, fever, breathing difficulty, loss of smell etc. Usually, the symptom “loss of smell” plays a higher significant role in comparison to the other existing symptoms [52], [53], [54] (for more details, please see the supplementary document). These symptoms are linked with specific human gene sets chosen as the bait's possible targets (preys), i.e., nCoV. The same is also true for other risk factors, clinical outcomes of COVID-19. So, all these genes under the mentioned categorization are grouped [46] from the disease-gene dataset available from DisGeNET. DisGeNET [55] is considered one of the significant resources covering all the relevant information about various diseases. These multiple gene sets are compared with each other [21] using molbiotools. The resultant gene set is again compared [21] with the curated COVID19 dataset available in Comparative Toxicogenomics Database [56] under respiratory tract disease & viral disease to obtain an overlapping gene set. CTD is yet another significant resource that collects, organizes and stores scientific data which describes the interrelationship between proteins, pathways, interactions, drugs etc. The overlapping gene set is further intersected with the spreader protein set in level-1 and level-2 of generated Human-nCoV interaction network [20]. The top 10 key genes are selected from the resultant intersection in each level based on the fuzzy score and spreadability index score in level-1 and level-2. These genes are considered the most significant ones that play an essential role in COVID19 transmission [57], [58], [59], [60], [61] and prevention [62], [63], [64], [65], [66] in the Human-nCoV interaction network. Potential FDA drugs having these key genes/spreader proteins as known targets are identified from DrugBank data [41], [42]. Then PDS_CS algorithm is executed to determine the most potential candidate FDA drug for COVID19. The entire process of the symptom-based analysis is highlighted in Fig. 3 .
Fig. 3.
Covid19 Symptoms based analysis. The analysis consists of the following steps. Step-1: Symptoms are searched in the DisGeNET database. Step-2: Symptom associated gene sets are fetched from the DisGeNET database. Step-3: All gene sets are provided as an input to Molbiotools online. Step-4: Common overlapping set of genes are obtained from Molbiotools. Step-5: Curated Covid19 dataset is extracted from the Comparative Toxicogenomics Database (CTD) under respiratory tract disease & viral disease. Step-6: These sets of genes intersect with the common overlapping set of genes obtained from Molbiotools to form a key set of Covid19 related genes. Step-7: This key set of Covid19 related genes, after mapping to their corresponding protein IDs, are compared with the spreader proteins in Human-nCoV PPIN. Step-8: After comparison, top genes are selected in both level-1 and level-2 of Human-nCoV PPIN, which are finally used for the PDS_CS algorithm to determine the most potential candidate FDA drug for COVID19. Here, Fostamatinib/R406 has the highest DCS score.
2.4. Computational docking of potential drugs with respect to COVID19 protein structures
The earlier sections discuss how several SARS-CoV2 proteins like R1AB_SARS2, SPIKE_SARS2, R1A_SARS2 etc., react with the human level-1 and level-2 spreaders to form SARS-CoV2-Human PPIN. Hence, a drug repurposing study is done based on network and symptom-based analysis. It reveals Fostamatinib/R406 might be a potential drug for COVID-19. However, a docking study is required to light this further, stating how well Fostamatinib/R406 binds with the SARS-CoV2 proteins. One of the most powerful approaches for structure-based drug discovery is molecular docking. It is defined as analysing how more than one molecular structure (drug and protein or enzyme) gets attached [67]. In other words, docking can be interpreted as molecular modelling methodology, which is implemented to anticipate how small molecules, i.e., ligands, interrelate with protein, i.e., enzyme. But to do docking, proteins structures of both SARS-CoV2 proteins and Fostamatinib/R406 are required. So, protein–ligand docking is executed by using Molegro Virtual Docker (version: 6.0) on potential COVID19 FDA listed drugs, Fostamatinib and R406, with all the so far available protein structures on nCoV having PDB IDs: 6LU7 [25], 6M2Q [26], 6W9C [27], 6M0J [28], 6M71 [29], 6VXX [30]. Grid-based cavity prediction is used to identify the potential binding sites. Models involving flexible ligands are taken into consideration. Orientation of ligands usually differs, and ranking for each ligand is based on the energy scores. The entire algorithm is implemented at 1500 iterations with a simplex evolution size of 10 runs. Compounds that take the lowest binding energy in comparison to others are considered to be the best. The molecules of the potential COVID19 FDA listed drugs are downloaded from DrugBank [41] in Structure data file (SDF/PDB) format. The docking returns two types of scores: 1) Moldock scores and 2) Rerank scores (for more details, please see supplementary) [68]. These scores assist in the identification of the best molecules docked in the selected target site. All the molecules are sorted based on these scores, representing the lowest energy required to get tied up with amino acid (AA) components.
3. Results and discussion
Computational study and results of associated drugs with human proteins in Human-nCoV PPIN shows that there is a probability that Fostamatinib/R406 may act as one of the potential candidates for COVID-19 treatment.
3.1. Drug-consensus results for COVID-19 spreader nodes using Human-nCoV interaction network analysis
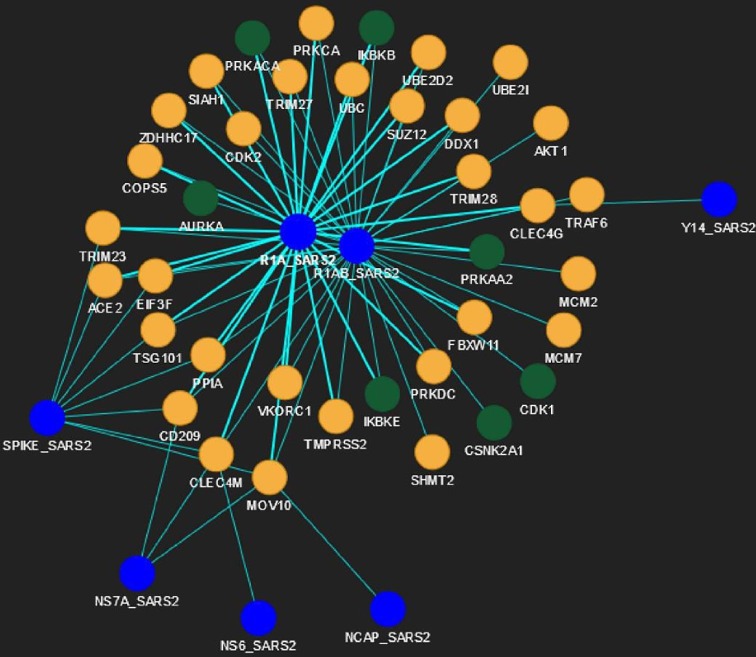
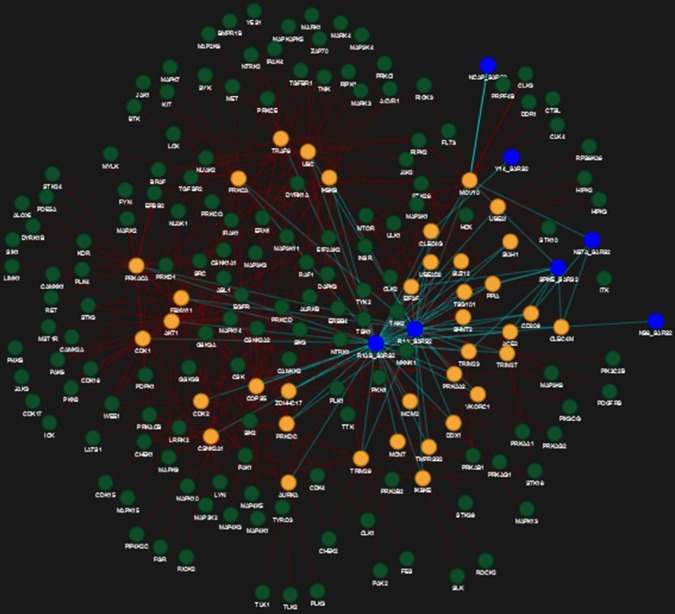
Drugs and their corresponding IDs are mapped with all human spreader proteins in Human-nCoV PPIN by matching the related drug-protein targets with spreader nodes. It is observed after applying the PDS_CS algorithm that Fostamatinib/R406 has a significant overlap of 155 target proteins in Human-nCoV PPIN, which is also the highest frequency of occurrence in the entire PPIN when compared to the remaining human protein associated drugs (i.e., highest Drug Consensus Score or DCS score as described earlier in the methodology section). It has a DCS score of 7 (i.e., count of level-1 protein targets of Fostamatinib as shown in Table 3 ) and 148 (i.e., count of level-2 protein targets of Fostamatinib as shown in Table 4 ) in level-1 and level-2 human spreader proteins. This establishes that the algorithm has succeeded in detecting the appropriate drug molecules with the highest protein targets in both levels. Protein targets corresponding to the DCS score of Fostamatinib in level-1 is highlighted in Fig. 4 , while that of level-2 is shown in Fig. 5 . In Fig. 4, green nodes represent level-1 protein targets of Fostamatinib, while blue and yellow nodes denote COVID-19 and other level-1 human proteins, respectively. In Fig. 5, green nodes represent level-2 protein targets of Fostamatinib, while blue and yellow nodes denote COVID-19 and other level-1 spreader human proteins, respectively. Other level-2 human spreaders in Fig. 5 are not shown to avoid visual complexity. The highest frequency of Fostamatinib/R406 is observed when the PDS_CS algorithm is implemented on Human-nCoV PPIN of Gordon et al. [13] (for more details, please see section 3.6).
Table 3.
Detailed analysis of DCS score at level 1 (Top 5 DCS have been shown).
| Drug | Drug ID | DCS (Level 1) |
|---|---|---|
| Fostamatinib/R406 | DB12010 | 7 |
| Arsenic trioxide | DB01169 | 2 |
| Acetylsalicylic acid | DB00945 | 2 |
| Resveratrol | DB02709 | 2 |
| Tamoxifen | DB00675 | 1 |
Table 4.
Detailed analysis of DCS score at level 2 (Top 5 DCS have been shown).
| Drug | Drug ID | DCS (Level 2) |
|---|---|---|
| Fostamatinib/R406 | DB12010 | 148 |
| Copper | DB09130 | 88 |
| Zinc acetate | DB14487 | 57 |
| Zinc | DB01593 | 57 |
| Zinc chloride | DB14533 | 57 |
Fig. 4.
Application of PDS_CS algorithm in Human-CoV PPIN (level-1). DCS score of Fostamatinib/R406 in level-1 is 7 (green nodes). Blue and yellow nodes denote COVID-19 and other level-1 proteins, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Application of PDS_CS algorithm in Human-CoV PPIN (level-2). DCS score of Fostamatinib/R406 in level-2 is 148 (green nodes). Blue and yellow nodes denote COVID-19 and other level-1 spreader proteins, respectively. Other level-2 spreaders are not shown to avoid visual complexity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Drug-consensus results for COVID-19 spreader nodes using COVID-19 symptoms, risk factors and clinical outcome-based analysis
Grouping genes based on various categories of COVID-19 symptoms, risk factors, and clinical outcomes [21] is done using DisGeNET [55]. The numerical statistics of the result is highlighted in supplementary Table S2. Mobiotools [69] are used to compare these gene sets to obtain 4931 unique genes. These genes are further compared with the curated COVID-19 dataset of CTD [56], containing 12,672 genes. The comparison generates an overlapping gene set containing 3525 genes. When used for validation against the spreader proteins in the Human-nCoV interaction network, these genes produce a significant overlap of 1448 genes in both level-1 and level-2. This highlights the fact that 1448 out of 3525 genes are selected as spreader nodes in the network. Hence, the top 10 key genes are selected from 1448 in each level based on the fuzzy and spreadability index scores in level-1 and level-2. The selected top genes from level-1 are PPIA, ACE2, EIF3F, UBC, PRKDC, CDK2, CDK1, AKT1, PRKCA, and TRAF6 level-2 are APP, ELAVL1, NTRK1, XPO1, MEOX2, GRB2, EGFR, TP53, BAG3 and NXF1. Potential FDA drugs having these key genes/spreader proteins as known targets are identified from DrugBank data (see the supplementary Table S3 and Table S4). It is also observed that after applying the PDS_CS algorithm on the obtained result in Table S3 and Table S4, Fostamatinib/R406 has a significant overlap of 3 target proteins which is also the highest frequency of occurrence. Similarly, for the symptom “loss of smell”, 12 overlapping genes are detected, mapping which with known drug targets in DrugBank has also been done (see supplementary Table S5). After applying the PDS_CS algorithm, only Fostamatinib/R406 and copper emerge based on their frequency of occurrence.
3.3. Docking results for potential COVID-19 drugs with respect to COVID-19 protein structures
Molecular docking is used in the proposed methodology to measure the binding capability of the potential COVID-19 drugs on 6LU7, 6M2Q, 6W9C, 6M0J, 6M71 and 6VXX. The detailed procedure of execution has been already discussed in the methodology section. In this work, SDF/PDB format is used for the docking of all the COVID-19 drugs. The results of docking with 6LU7 are highlighted in the supplementary Table S6. At the same time, docking results with others are shown in Table 5 and Table 6 . It is observed from the results that while Fostamatinib/R406 registers the highest score for 6LU7 and 6M2Q, it obtains the second position in comparison to the other COVID-19 structures.
Table 5.
Docking scores (Moldock Score) of drugs for 6VXX, 6M71, 6M2Q, 6W9C, 6M0J.
| Drugs | Drug ID | Moldock score (6VXX) | Moldock score (6M71) | Moldock score (6M2Q) | Moldock score (6W9C) | Moldock score (6M0J) |
|---|---|---|---|---|---|---|
| Ritonavir | DB00503 | −228.318 | −201.076 | −121.515 | −230.852 | −197.609 |
| Fostamatinib | DB12010 | −172.859 | −179.834 | −120.397 | −185.156 | −176.483 |
| Lopinavir | DB01601 | −166.217 | −166.388 | −114.563 | −181.474 | −170.136 |
| Remdesivir | DB14761 | −152.678 | −160.739 | −112.938 | −172.778 | −165.157 |
| Darunavir | DB01264 | −144.579 | −152.622 | −90.9735 | −156.453 | −152.907 |
| Hydroxychloroquine | DB01611 | −127.194 | −110.183 | −80.4403 | −132.054 | −122.721 |
| Azithromycin | DB00207 | −120.155 | −105.759 | −66.3967 | −127.711 | −115.321 |
| Favipiravir | DB12466 | −76.41 | −73.7678 | −14.436 | −72.6467 | −70.8629 |
Table 6.
Docking scores (Rerank Score) of drugs with for 6VXX, 6M71, 6M2Q, 6W9C, 6M0J.
| Drugs | Drug ID | Rerank Score (6VXX) | Rerank Score (6 M71) | Rerank Score (6M2Q) | Rerank Score (6W9C) | Rerank Score (6M0J) |
|---|---|---|---|---|---|---|
| Ritonavir | DB00503 | −154.226 | −140.533 | −92.2327 | −160.902 | −130.098 |
| Fostamatinib | DB12010 | −57.5164 | −110.545 | −87.5182 | −130.02 | −130.509 |
| Lopinavir | DB01601 | −81.6619 | −138.449 | 12.9996 | −147.683 | −125.165 |
| Remdesivir | DB14761 | −101.76 | −124.925 | −84.1368 | −108.703 | −119.626 |
| Darunavir | DB01264 | −103.522 | −122.378 | −66.1872 | −115.516 | −73.643 |
| Hydroxychloroquine | DB01611 | −100.612 | −88.6121 | 18.5798 | −102.419 | −97.8097 |
| Azithromycin | DB00207 | −88.4657 | −72.8345 | −54.8591 | −100.182 | −31.7329 |
| Favipiravir | DB12466 | −62.055 | −52.7567 | 793.471 | −59.1369 | −57.8456 |
3.4. Docking results for active metabolites/promoieties of COVID-19 Prodrugs with respect to COVID-19 protein structures
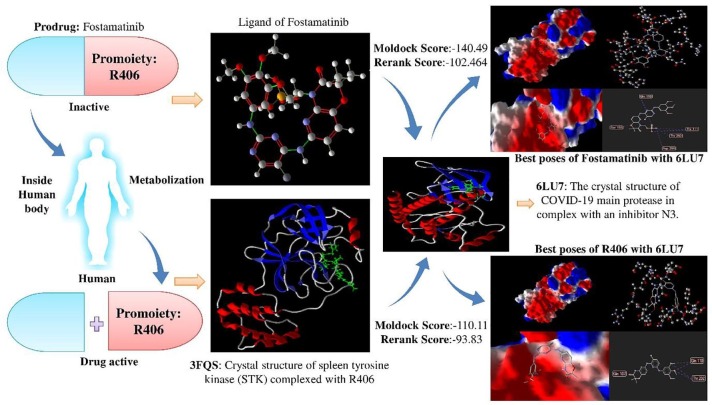
Several drug molecules consist of pharmacologically inactive compounds, which are known as Prodrugs [70]. These drugs get metabolized after entering the human body to liberate the active drug. On careful observation, it has been observed that Fostamatinib is also a prodrug. Fostamatinib (R788) is considered to be an orally induced prodrug in humans that releases active metabolite/promoiety R940406 (R406) [71]. R406 is a spleen tyrosine kinase (SYK) inhibitor responsible for treating rheumatoid arthritis [71]. Similar instances have also been observed in the case of remdesivir and favipiravir. So, the binding capability of these active metabolites/promoieties must be validated against 6LU7, 6M2Q, 6W9C, 6M0J, 6M71 and 6VXX by molecular docking for consideration of any prodrug as a COVID-19 drug. The results of this docking with 6LU7 are highlighted in the supplementary Table S7. At the same time, docking results with others are shown in Table 7 and Table 8 . The result draws the reference that R406 also shows high binding affinity scores compared to the others, which promotes the fact that Fostamatinib/R406 can be a potential COVID-19 drug. Molecular docking results of Fostamatinib and its corresponding promoiety, R406, have also been highlighted in Fig. 6 .
Table 7.
Docking scores (Moldock Score) of Prodrugs for 6VXX, 6M71, 6M2Q, 6W9C, 6M0J.
| Prodrugs | Drug id | Active promoieties | Moldock Score (6VXX) | Moldock Score (6M71) | Moldock Score (6M2Q) | Moldock Score (6W9C) | Moldock Score (6M0J) |
|---|---|---|---|---|---|---|---|
| Fostamatinib | DB12010 | RP406 (using 3FQS) | −143.34 | −131.064 | −115.229 | −150.184 | −134.057 |
| Remdesivir | DB14761 | GS-441524 | −115.54 | −106.624 | −106.108 | −107.333 | −120.417 |
| Favipiravir | DB12466 | RdRp complex (6K32) | −92.5709 | −100.143 | −54.3239 | −93.9083 | −71.9692 |
Table 8.
Docking scores (Rerank Score) of Prodrugs for 6VXX, 6M71, 6M2Q, 6W9C.
| Prodrugs | Drug id | Active promoieties | Rerank Score (6VXX) | Rerank Score (6 M71) | Rerank Score (6M2Q) | Rerank Score (6W9C) | Rerank Score (6M0J) |
|---|---|---|---|---|---|---|---|
| Fostamatinib | DB12010 | RP406 (using 3FQS) | −115.106 | −109.152 | −76.3059 | −111.853 | −111.887 |
| Remdesivir | DB14761 | GS-441524 | −91.7647 | −80.5349 | −70.1108 | −87.910 | −94.3608 |
| Favipiravir | DB12466 | RdRp complex (6 K32) | −66.4667 | −84.9671 | −49.3377 | −81.8392 | −57.9054 |
Fig. 6.
Molecular docking results of prodrug Fostamatinib and its corresponding promoiety, R406. Fostamatinib and R406 both have high binding affinity scores in comparison to the other potential COVID-19 drugs.
3.5. Analysis of 3 key target genes of Fostamatinib/R406 in Human-nCoV interaction network in symptom-based analysis
The three key target genes of Fostamatinib/R406, as identified in the supplementary Table S3 and Table S4, are CDK1 (level-1) and NTRK1, EGFR (level-2). It is noted that these three genes are related to the most significant COVID-19 symptoms, risk factors and clinical outcomes, which are highlighted in Table 9 . Moreover, these three genes also play an essential role in response to viral infections which has been highlighted in Table 10 . All these depict the fact that Fostamatinib/R406 might be a potential drug treatment to COVID-19 treatment.
Table 9.
Mapping of CDK1 and NTRK1, EGFR with COVID-19 symptoms, risk factors and clinical outcomes.
| Drug | Key Target Genes | Level | COVID-19 symptoms | Clinical outcome (Severe case) | Risk Factor |
|---|---|---|---|---|---|
| Fostamatinib | CDK1 | Level-1 | pneumonia | – | diabetes |
| cancer | |||||
| NTRK1 | Level-2 | fever | – | kidney disease | |
| cancer | |||||
| hypertension | |||||
| pneumonia | lung disease | ||||
| diabetes | |||||
| EGFR | Level-2 | pneumonia | neutrophilia | heart disease | |
| hypertension | |||||
| cancer | |||||
| dyspnea | kidney disease | ||||
| fever | kidney injury | lung disease | |||
| cough | thrombocytopenia | diabetes |
Table 10.
Role of CDK1 and NTRK1, EGFR in viral infections.
| Drug | Key Target Genes | Level | Role in viral infections |
|---|---|---|---|
| Fostamatinib | CDK1 | Level-1 | Viruses can express some oncoproteins. Genome replication gets induced inside the hosts' cells due to some signals generated due to the interference of these proteins with CDK and CIKs function [70]. |
| NTRK1 | Level-2 | NTRK1 has an active role in the immune response against viral infection [71]. | |
| EGFR | Level-2 | Hindrance of EGFR signalling might prevent an excessive fibrotic response to SARS-CoV. |
3.6. Application of algorithm S1 (PDS_CS) on in the host targets of in vitro generated Human-nCoV PPIN of Gordon et al. [13]
Gordon et al. [13] cloned, tagged, and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identified the human proteins that are physically associated with each of the SARS-CoV-2 proteins by affinity-purification mass spectrometry. As a result, 332 high-confidence protein–protein interactions between SARS-CoV-2 and human proteins are identified. These 332 host targets are collected, and Algorithm 1 (PDS_CS) is implemented on the same. It is observed from the implementation that Fostamatinib/R406 has a significant overlap of 10 target proteins (i.e., DCS score of 10) in Human-nCoV PPIN, which is also the highest frequency of occurrence in the entire PPIN when compared to the remaining human protein associated drugs. The result is highlighted in Table 11 .
Table 11.
Detailed analysis of DCS score (Top 6 DCS have been shown).
| Drug | Drug ID | DCS (Level 1) |
|---|---|---|
| Fostamatinib | DB12010 | 10 |
| NADH | DB00157 | 5 |
| Flavin adenine dinucleotide | DB03147 | 5 |
| Romidepsin | DB06176 | 2 |
| Glutamic acid | DB00142 | 2 |
| Atorvastatin | DB01076 | 1 |
The ten host target genes associated with Fostamatinib/R406 in Table 11 are TBK1, CIT, NEK9, RIPK1, COQ8B, CSNK2A2, MARK1, MARK3, MARK2 and PRKACA. In addition, on careful observation, it has been noted that these genes also play an essential role in response to viral infections, which has been discussed below:
-
1)
TBK1: TBK1 (TANK-binding kinase 1) plays a highly significant role in developing natural immunity against antiviral activities. It activates IRF (interferon regulatory factor) 3, which in turn induce type I interferon (IFNs) (IFN-α/β) proteins regulating immune activity [72].
-
2)
CIT: Encoded serine/threonine protein kinases are unique features in a specific set of giant DNA viruses. However, their role in the replication of virus varies. But different viral serine/ CIT (Citron Rho-Interacting Serine/Threonine Kinase) has the potentiality to act as the targets of antiviral drugs [17].
-
3)
NEK9: Nek9 exhaustion leads to the reduction of virus replication centres within which it remains confined. However, Nek9 overexpression will increase the number of viral genomes in the infected cell [73].
-
4)
RIPK1: Enhancement of plasma pro-inflammatory cytokines and lymphopenia is considered to be one of the significant predictors in increasing COVID-19 severity. Activating RIPK1 promotes the growth of these cytokines. In addition, it leads to the exhaustion of T cell populations (lymphopenia) in patients who get infected with HIV, which might pave the way for the entrance of SARS-CoV-2 in them [74].
-
5)
COQ8B: Mitochondrial metabolism is executed as a part of the metabolic pathway through the interaction of SARS-CoV-2′s M protein and COQ8B [75].
-
6)
CSNK2A2: CSNK2A2 is involved in the regulation of primary cellular processes as well as viral infection [76].
-
7)
MARK1: MARK1 plays an active role in viral responses [77].
-
8)
MARK2: MARK2 is engaged in stimulating FEZ1 (Fasciculation And Elongation Protein Zeta 1) phosphorylation on the central cores of viruses [80].
-
9)
PRKACA: PRKACA also plays a similar role in cardiovascular disease as that of MARK3 [78], [81].
4. Conclusion
In this computational study, we have analyzed the Human-nCoV PPIN and attempted to identify the candidate drugs for the level-1 and level-2 spreader proteins. Our study identifies Fostamatinib/R406, an FDA approved drug, as the most promising drug with the best chances to target the COVID-19 spreader proteins. The work relies on the hypothesis that SARS-CoV2/nCoV has ~89% genetic resemblance with SARS-CoV. Based on this, Human-nCoV PPIN has been developed, and its spreader nodes have been identified using the SIS model and fuzzy thresholding. Furthermore, a consensus strategy by a two-way analysis has been utilized to analyze drugs based on the overlap of spreader proteins and drug-protein targets. The consensus scores for Fostamatinib/R406 are highest in analysing the candidate drugs for COVID-19 spreader proteins. Besides, Fostamatinib/R406 also generates satisfactory results in molecular docking with the available COVID-19 protein structures. It also targets CAYP34A [23], [82], a common target for almost all the FDA approved drugs [15] for COVID-19. Moreover, recent studies also suggest that it is used for thrombocytopenia [23] which is also associated with severe coronavirus disease 2019 (COVID-19) infections [24].
A clinical test is needed as the FDA approves Fostamatinib/R406 in immune thrombocytopenia [83] and to determine its efficacy against SARS-CoV2. Rigel pharmaceuticals have already started the clinical trials of Fostamatinib/R406 [29]. The results obtained are quite encouraging and positive regarding reports published to date [27], [83]. According to the reports [27], [83], Fostamatinib meets the “primary endpoint of Safety in Phase 2 Clinical Trial” conducted in hospitalized patients affected with COVID-19. In addition to this, they have also enrolled themselves for Phase 3 clinical trial of fostamatinib/R406 to treat the same. But arriving at a specific conclusion needs time and more research analysis. In a nutshell, our computational research evidence discovers that Fostamatinib/R406 may be considered one of the strong contenders for COVID-19 treatment.
CRediT authorship contribution statement
Sovan Saha: Conceptualization, Data curation, Methodology, Writing - original draft, Software. Anup Kumar Halder: Conceptualization, Data curation, Methodology, Writing - original draft, Software. Soumyendu Sekhar Bandyopadhyay: Data curation, Writing - original draft, Visualization. Piyali Chatterjee: Supervision, Investigation, Formal analysis, Writing - review & editing. Mita Nasipuri: Supervision, Project administration, Investigation, Formal analysis, Writing - review & editing. Debdas Bose: Formal analysis, Validation. Subhadip Basu: Supervision, Project administration, Investigation, Formal analysis, Data curation, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors are thankful to the CMATER research laboratory of the Computer Science and Engineering Department, Jadavpur University, India, to provide infrastructure facilities during the work. The authors also express their sincere gratitude to Dr. Toufigh Gordi and the entire clinical team of Rigel Pharmaceuticals for their continuous help and motivation. Dr. Toufigh Gordi is a Senior Director Clinical Pharmacology at Rigel Pharmaceuticals based in South San Francisco, California.
Funding
This work is partially supported by the CMATER research laboratory of the Computer Science and Engineering Department, Jadavpur University, India, and Department of Biotechnology project (BT/PR16356/BID/7/596/2016), Ministry of Science and Technology, Government of India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2021.08.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations, 2021. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations. (Accessed 28-02-2021 Access 2021).
- 2.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discovery. 2019;19(2020):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 3.WHO Coronavirus Disease (COVID-19) Dashboard, 2021. https://covid19.who.int/. (Accessed 17-05-2021 Access 2021).
- 4.Rabby M.I.I. Current drugs with potential for treatment of covid-19: a literature review. J. Pharmacy Pharm. Sci. 2020;23(1):58–64. doi: 10.18433/jpps31002. [DOI] [PubMed] [Google Scholar]
- 5.Different COVID-19 Vaccines, 2021. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines.html.
- 6.WHO news updates – WHO | World Health Organization, 2021. https://www.who.int/news-room/news-updates.
- 7.RECOMMENDATION FOR AN EMERGENCY USE LISTING OF ON COVISHIELD™ SUBMITTED BY SIIPL, 2021. https://extranet.who.int/pqweb/sites/default/files/documents/COVISHIELD_TAG_REPORT_EULvaccine.pdf. (Accessed 17-05-2021 Access 2021).
- 8.Covaxin, 2021. https://www.bharatbiotech.com/covaxin.html.
- 9.COVID-19 Studies from the World Health Organization Database - ClinicalTrials.gov, 2021. https://clinicaltrials.gov/ct2/who_table. (Accessed 28-02-2021 Access 2021).
- 10.Saha S., Sengupta K., Chatterjee P., Basu S., Nasipuri M. Analysis of protein targets in pathogen–host interaction in infectious diseases: a case study on Plasmodium falciparum and Homo sapiens interaction network. Brief. Funct. Genomics. 2017;17(6):441–450. doi: 10.1093/bfgp/elx024. [DOI] [PubMed] [Google Scholar]
- 11.Halder A.K., Dutta P., Kundu M., Basu S., Nasipuri M. Review of computational methods for virus-host protein interaction prediction: a case study on novel Ebola-human interactions. Brief. Funct. Genomics. 2018;17(6):381–391. doi: 10.1093/bfgp/elx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhami M., Sadeghi B., Rezapour A., Haghdoost A.A., MotieGhader H. Repurposing novel therapeutic candidate drugs for coronavirus disease-19 based on protein-protein interaction network analysis. BMC Biotech. 2021;21(1):22. doi: 10.1186/s12896-021-00680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.N.T.J. Bailey, The mathematical theory of infectious diseases and its applications. 2nd edition, The mathematical theory of infectious diseases and its applications. 2nd edition. (1975).
- 15.J.C. Lucy Chin, Safiya Esmail, Mark Franklin, Diana Le., COVID-19 : Finding the Right Fit Identifying Potential Treatments Using a Data-Driven Approach, Drugbank White Paper (2020).
- 16.Fostamatinib - DrugBank, 2021. https://www.drugbank.ca/drugs/DB12010. (Accessed 28-02-2021 Access 2021).
- 17.T. Jacob, C. Van den Broeke, H.W. Favoreel, Viral Serine/Threonine Protein Kinases, Journal of Virology 85(3) (2011) 1158 LP-1173. [DOI] [PMC free article] [PubMed]
- 18.Drug Approval Package: TAVALISSE (fostamatinib disodium hexahydrate), 2021. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/209299Orig1s000TOC.cfm. (Accessed 28-02-2021 Access 2021).
- 19.S. Saha, P. Chatterjee, S. Basu, M. Nasipuri, Detection of spreader nodes and ranking of interacting edges in Human-SARS-CoV protein interaction network, PeerJ 9 (2021) e12117, 10.7717/peerj.12117. [DOI] [PMC free article] [PubMed]
- 20.S. Saha, A.K. Halder, S.S. Bandyopadhyay, P. Chatterjee, M. Nasipuri, S. Basu, Computational modeling of Human-nCoV protein-protein interaction network, arxiv (2020), arXiv:2005.04108v1. [DOI] [PMC free article] [PubMed]
- 21.Kumar S. COVID-19: A drug repurposing and biomarker identification by using comprehensive gene-disease associations through protein-protein interaction network analysis. Preprints. 2020 doi: 10.20944/preprints202003.0440.v1. [DOI] [Google Scholar]
- 22.FDA approves fostamatinib tablets for ITP | FDA, 2021. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-fostamatinib-tablets-itp. (Accessed 28-02-2021 Access 2021).
- 23.McKeage K., Lyseng-Williamson K.A. Fostamatinib in chronic immune thrombocytopenia: a profile of its use in the USA. Drugs Therapy Perspectives. 2018;34(10):451–456. doi: 10.1007/s40267-018-0551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin. Chim. Acta. 2019;506(2020):145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020:1–9. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 26.RCSB PDB - 6M2Q: SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21), 2021. http://www.rcsb.org/structure/6M2Q. (Accessed 28-02-2021 Access 2021).
- 27.RCSB PDB - 6W9C: The crystal structure of papain-like protease of SARS CoV-2, 2021. http://www.rcsb.org/structure/6W9C. (Accessed 28-02-2021 Access 2021).
- 28.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Agrawal, M. Zitnik, J. Leskovec, Large-Scale Analysis of Disease Pathways in the Human Interactome, bioRxiv (2017) 189787–189787. [PMC free article] [PubMed]
- 32.BioSNAP: Network datasets: Human protein-protein interaction network, 2021. https://snap.stanford.edu/biodata/datasets/10000/10000-PP-Pathways.html. (Accessed 28-02-2021 Access 2021).
- 33.Pfefferle S., Schöpf J., Kögl M., Friedel C.C., Müller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Züst R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Weßels C., Pöhlmann S., Haas J., Drosten C., von Brunn A. The SARS-Coronavirus-host interactome: identification of cyclophilins as target for pan-Coronavirus inhibitors. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bateman A., Martin M.J., O'Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye-Ajee H., Cowley A., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Castro L.G., Figueira L., Garmiri P., Georghiou G., Gonzalez D., Hatton-Ellis E., Li W., Liu W., Lopez R., Luo J., Lussi Y., MacDougall A., Nightingale A., Palka B., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Speretta E., Turner E., Tyagi N., Volynkin V., Wardell T., Warner K., Watkins X., Zaru R., Zellner H., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., ArgoudPuy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.China releases genetic data on new coronavirus, now deadly | CIDRAP, 2021. https://www.cidrap.umn.edu/news-perspective/2020/01/china-releases-genetic-data-new-coronavirus-now-deadly. (Accessed 28-02-2021 Access 2021).
- 37.Samadi N., Bouyer A. Identifying influential spreaders based on edge ratio and neighborhood diversity measures in complex networks. Computing. 2019;101(8):1147–1175. [Google Scholar]
- 38.Wang S., Wu F. Detecting overlapping protein complexes in PPI networks based on robustness. Proteome Science. 2013;11(Suppl 1):1–8. doi: 10.1186/1477-5956-11-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dutta P., Basu S., Kundu M. Assessment of semantic similarity between proteins using information content and topological properties of the gene ontology graph. IEEE/ACM Trans. Comput. Biol. Bioinf. 2018;15(3):839–849. doi: 10.1109/TCBB.2017.2689762. [DOI] [PubMed] [Google Scholar]
- 40.Consortium G.O., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(suppl 1):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D.S. Wishart, C. Knox, A.C. Guo, D. Cheng, S. Shrivastava, D. Tzur, B. Gautam, M. Hassanali, DrugBank: a knowledgebase for drugs, drug actions and drug targets, Nucleic acids research 36(Database issue) (2008) D901-D906. [DOI] [PMC free article] [PubMed]
- 42.DrugBank, 2021. https://www.drugbank.ca/. (Accessed 28-02-2021 Access 2021).
- 43.P. Gautret, J.-C. Lagier, P. Parola, V.T. Hoang, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V.E. Vieira, H.T. Dupont, S. Honoré, P. Colson, E. Chabrière, B. La Scola, J.-M. Rolain, P. Brouqui, D. Raoult, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int. J. Antimicrobial Agents (2020) 105949–105949. [DOI] [PMC free article] [PubMed]
- 44.Emergency Use Authorization | FDA, 2021. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization. (Accessed 28-02-2021 Access 2021).
- 45.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 46.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. PNAS. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emergency Access to Remdesivir Outside of Clinical Trials, 2021. https://www.gilead.com/purpose/advancing-global-health/covid-19/emergency-access-to-remdesivir-outside-of-clinical-trials. (Accessed 28-02-2021 Access 2021).
- 49.Remdesivir Clinical Trials, 2021. https://www.gilead.com/purpose/advancing-global-health/covid-19/remdesivir-clinical-trials. (Accessed 28-02-2021 Access 2021).
- 50.China approves antiviral favilavir to treat coronavirus - UPI.com, 2021. https://www.upi.com/Health_News/2020/02/17/China-approves-antiviral-favilavir-to-treat-coronavirus/5291581953892/. (Accessed 28-02-2021 Access 2021).
- 51.Taiwan synthesizes anti-viral drug favilavir for COVID-19 patients – Focus Taiwan, 2021. https://focustaiwan.tw/sci-tech/202003020012. (Accessed 28-02-2021 Access 2021).
- 52.Xydakis M.S., Dehgani-Mobaraki P., Holbrook E.H., Geisthoff U.W., Bauer C., Hautefort C., Herman P., Manley G.T., Lyon D.M., Hopkins C. Smell and taste dysfunction in patients with COVID-19. Lancet. Infect. Dis. 2020;20(9):1015–1016. doi: 10.1016/S1473-3099(20)30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menni C., Sudre C.H., Steves C.J., Ourselin S., Spector T.D. Quantifying additional COVID-19 symptoms will save lives. The Lancet. 2020;395(10241):e107–e108. doi: 10.1016/S0140-6736(20)31281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., Ganesh S., Varsavsky T., Cardoso M.J., El-Sayed Moustafa J.S., Visconti A., Hysi P., Bowyer R.C.E., Mangino M., Falchi M., Wolf J., Ourselin S., Chan A.T., Steves C.J., Spector T.D. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020;26(7):1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Piñero, N. Queralt-Rosinach, À. Bravo, J. Deu-Pons, A. Bauer-Mehren, M. Baron, F. Sanz, L.I. Furlong, DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes, Database : the journal of biological databases and curation 2015 (2015) bav028-bav028. [DOI] [PMC free article] [PubMed]
- 56.C.J. Mattingly, M.C. Rosenstein, G.T. Colby, J.N. Forrest, J.L. Boyer, The Comparative Toxicogenomics Database (CTD): A resource for comparative toxicological studies, 305 689-692. [DOI] [PMC free article] [PubMed]
- 57.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.E. Ciaglia, C. Vecchione, A.A. Puca, COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children, Front. Pediatrics 8 (2020) 206-206. [DOI] [PMC free article] [PubMed]
- 59.Xiao H., Xu L.H., Yamada Y., Liu D.X. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE. 2008;3(1) doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Müller M.A., Deng W., Lei J., Meyer B., Kusov Y., Von Brunn B., Bairad D.R., Hünten S., Drosten C., Hermeking H., Leonhardt H., Mann M., Hilgenfeld R., Von Brunn A. P53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U.S.A. 2016;113(35):E5192–E5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.L. Wendt, J. Brandt, B.S. Bodmer, S. Reiche, M.L. Schmidt, S. Traeger, T. Hoenen, The Ebola Virus Nucleoprotein Recruits the Nuclear RNA Export Factor NXF1 into Inclusion Bodies to Facilitate Viral Protein Expression, Cells 9(1) (2020) 187-187. [DOI] [PMC free article] [PubMed]
- 62.Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C., Sun T., He P., Chen J., Shen J., Luo X., Li Y., Liu H., Bai D., Shen J., Yang Y., Li F., Zuo J., Hilgenfeld R., Pei G., Chen K., Shen X., Jiang H. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321(3):557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.P. Nigro, G. Pompilio, M.C. Capogrossi, Cyclophilin A: A key player for human disease, Nature Publishing Group, 2013, pp. e888-e888. Access 2013). [DOI] [PMC free article] [PubMed]
- 64.Chai Q., Jovasevic V., Malikov V., Sabo Y., Morham S., Walsh D., Naghavi M.H. HIV-1 counteracts an innate restriction by amyloid precursor protein resulting in neurodegeneration. Nat. Commun. 2017;8(1) doi: 10.1038/s41467-017-01795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z., Kolokoltsov A.A., Wang J., Adhikary S., Lorinczi M., Elferink L.A., Davey R.A. GRB2 interaction with the ecotropic murine leukemia virus receptor, mCAT-1, controls virus entry and is stimulated by virus binding. J. Virol. 2012;86(3):1421–1432. doi: 10.1128/JVI.05993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venkataraman T., Frieman M.B. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus-induced pulmonary fibrosis. Antiviral Res. 2017;143:142–150. doi: 10.1016/j.antiviral.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.X.-Y. Meng, H.-X. Zhang, M. Mezei, M. Cui, Molecular Docking: A powerful approach for structure-based drug discovery, Curr. Computer-aided drug design 7(2) (2011) 146-146. [DOI] [PMC free article] [PubMed]
- 68.R. Thomsen, M.H. Christensen, MolDock: a new technique for high-accuracy molecular docking, Journal of Medicinal Chemistry (0022-2623 (Print)). [DOI] [PubMed]
- 69.MULTIPLE LIST COMPARATOR – A free online tool to find list overlaps and draw Venn diagrams, 2021. http://molbiotools.com/listcompare.html. (Accessed 28-02-2021 Access 2021).
- 70.Stella V.J., Charman W.N.A., Naringrekar V.H. Prodrugs. Drugs. 1985;29(5):455–473. doi: 10.2165/00003495-198529050-00002. [DOI] [PubMed] [Google Scholar]
- 71.Baluom M., Grossbard E.B., Mant T., Lau D.T.W. Pharmacokinetics of fostamatinib, a spleen tyrosine kinase (SYK) inhibitor, in healthy human subjects following single and multiple oral dosing in three phase I studies. Br. J. Clin. Pharmacol. 2013;76(1):78–88. doi: 10.1111/bcp.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 2013;587(6):542–548. doi: 10.1016/j.febslet.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R. Jung, S. Radko, P. Pelka, The dual nature of nek9 in adenovirus replication, J. Virol. 90(4) (2016) 1931 LP–1943. [DOI] [PMC free article] [PubMed]
- 74.Mifflin L., Ofengeim D., Yuan J. Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discovery. 2020;19(8):553–571. doi: 10.1038/s41573-020-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol.-Cell Physiol. 2020;319(2):C258–C267. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.CSNK2A2 protein expression summary – The Human Protein Atlas, 2021. https://www.proteinatlas.org/ENSG00000070770-CSNK2A2. (Accessed 28-02-2021 Access 2021).
- 77.ZFIN Gene: mark1, 2021. https://zfin.org/ZDB-GENE-070626-2. (Accessed 28-02-2021 Access 2021).
- 78.89 diseases associated with MARK3 – Open Targets Platform, 2021. https://www.targetvalidation.org/target/ENSG00000075413/associations. (Accessed 28-02-2021 Access 2021).
- 80.Malikov V., Naghavi M.H. Localized phosphorylation of a kinesin-1 adaptor by a capsid-associated kinase regulates HIV-1 motility and uncoating. Cell Reports. 2017;20(12):2792–2799. doi: 10.1016/j.celrep.2017.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turnham R.E., Scott J.D. Protein kinase A catalytic subunit isoform PRKACA; History, function and physiology. Gene. 2016;577(2):101–108. doi: 10.1016/j.gene.2015.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujita K.I. Food-drug interactions via human cytochrome P450 3A (CYP3A) Drug Metab. Drug Interact. 2004;20:195–217. doi: 10.1515/dmdi.2004.20.4.195. [DOI] [PubMed] [Google Scholar]
- 83.Positive Topline Data Shows Fostamatinib Meets Primary Endpoint of Safety in Phase 2 Clinical Trial in Hospitalized Patients with COVID-19, 2021. https://www.rigel.com/investors/news-events/press-releases/detail/312/positive-topline-data-shows-fostamatinib-meets-primary.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.