Abstract
Mammalian SWI/SNF (mSWI/SNF) ATP-dependent chromatin remodelers modulate genomic architecture and gene expression and are frequently mutated in disease. However, the specific chromatin features that govern their nucleosome binding and remodeling activities remain unknown. Here, we subjected endogenously-purified mSWI/SNF complexes, and their constituent assembly modules to a diverse library of DNA-barcoded mononucleosomes, performing over 25,000 binding and remodeling measurements. We define histone modification-, variant-, and mutation-specific effects, alone and in combination, on mSWI/SNF activities and chromatin interactions. Further, we identify the combinatorial contributions of complex module components, reader domains, and nucleosome engagement properties to the localization of complexes to selectively permissive chromatin states. These findings uncover new principles that shape the genomic binding and activity of a major chromatin remodeler complex family.
One-Sentence Summary:
Integration of diverse chromatin landscape signals governs targeting and activity of SWI/SNF chromatin remodeling complexes.
Chromatin regulatory factors play critical roles in establishing and maintaining gene expression patterns, and their dysregulation is a common hallmark found in human disease (1). Whether single proteins or multimeric protein complexes with diverse functional roles, the local activity of these factors, dictated by structural features controlling their genomic targeting, must be tightly regulated in order to ensure fidelity in genomic processes. While transcription factors bind to well-defined stretches of DNA, the mechanisms by which chromatin regulatory proteins and protein complexes lacking DNA motif-specific domains localize and exert their activities genome-wide are multifactorial and remain less well-understood.
The megadalton-sized mammalian SWI/SNF (mSWI/SNF, or BAF) family of ATP-dependent chromatin remodeling complexes contain multiple histone recognition domains (readers) and generally sequence-non-specific DNA-binding domains, and thus have the potential for combinatorial readout of diverse epigenetic modifications and nucleosomal features (2). Further complicating matters, mSWI/SNF complexes are themselves diversified through altered subunit composition leading to three final-form assemblies, termed canonical BAF (cBAF), polybromo-associated BAF (PBAF), and non-canonical BAF (ncBAF) (3, 4), in which the number, type and possibly even accessibility of individual reader domains and nucleosome contact surfaces differ within a given structural framework. These three complexes have been shown to differentially localize on chromatin and exhibit differential functions and dependencies across human cancers (4, 5). Recent three-dimensional structural studies of yeast SWI/SNF and human BAF complexes bound to unmodified nucleosome substrates (6–8) have begun to resolve the multi-valent nature of chromatin engagement. In particular, studies on cBAF complexes have indicated the presence of bilateral nucleosome acidic patch recognition moieties on SMARCB1 and SMARCA4 subunits which “grip” the nucleosome in a “C-clamp” like shape and are recurrently mutated in human cancer (6, 7, 9). The overall complex architecture resolved in these structural studies agrees with earlier biochemical efforts which first highlighted the presence of both core and ATPase modules and their orders of assembly (3, 10).
While such studies have advanced our understanding of mSWI/SNF complex structure-function relationships, insights into the manner by which subunit composition and assembly cooperate to target these molecular machines in a specific manner and tune their chromatin remodeling activities as a function of nucleosome composition remain limited. Further, structural efforts performed on fully-formed SWI/SNF complexes to date have not resolved chromatin reader domains such as the bromodomain (BD) of SMARCA4 or the tandem PHD domains of DPF2, likely owing to the fact that complexes were solved on unmodified nucleosomes lacking cognate histone marks. As such, insights to date into mSWI/SNF targeting have come from (i) studies of individual subunit domains purified in isolation (11–13) and hence outside of their full complex contexts; (ii) studies examining the binding of yeast SWI/SNF or RSC complexes to acetylated nucleosomes and promoters (14–21) and (iii) genome-wide protein mapping technologies (e.g. ChIP-seq) used in cells (4, 10, 22–24). Genomic mapping analyses are inherently global, heterogeneous and correlative at best given the large numbers of cells required, and hence lack the ability to pinpoint specific nucleosomal features (e.g. PTMs) which directly interact with or modulate mSWI/SNF complex binding and chromatin remodeling activities. This is especially true given that specific nucleosomes that were once bound by active remodeling complexes may have been moved or ejected following binding, making it impossible to determine the features which initially facilitated complex engagement. Finally, no study to date has comprehensively evaluated the human mSWI/SNF complexes, including the most recently-discovered assembly, ncBAF, only present in higher eukaryotes (4). Taken together, there remain fundamental gaps in our understanding of the direct biochemical cues across the histone landscape that control mSWI/SNF complex function, gaps that if filled, could inform new strategies for site-specific modulation of remodeling activities.
Here we identify the features of nucleosome substrates which directly impact cBAF, PBAF, and ncBAF complex binding and remodeling activities. Using a high-throughput chemical biology approach, we define an extensive repertoire of histone PTMs, variants, mutations, and their combinations that confer mSWI/SNF complex stimulation or inhibition. The resulting functional ‘signatures’, validated with individually-synthesized nucleosomes, provide a mechanistic framework to explain genome-wide localization profiles, DNA accessibility data, complex roles in gene regulation, and the impact of disease-associated mutations on mSWI/SNF family complex targeting. By mapping the nucleosome landscape preferences through an ordered complex assembly pathway, we define the direct contributions of specific subunits and modules to the overall targeting and activity of this major chromatin remodeling entity.
Results
Final-form mSWI/SNF family complexes exhibit distinct remodeling activity signatures
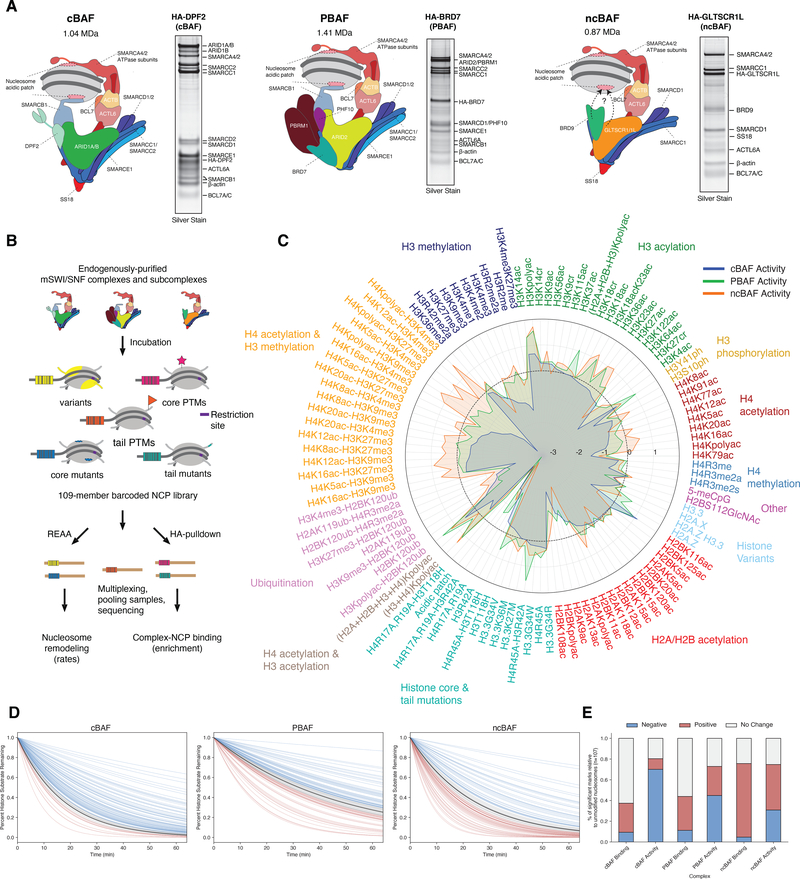
We began our investigations by isolating endogenous, fully-formed cBAF, PBAF, and ncBAF complexes from mammalian cells as previously described (3) (Fig. 1A). To understand how these complexes engage and respond to the chromatin landscape, we employed a previously described library of differentially modified, DNA-barcoded nucleosomes containing a diverse repertoire of histone modifications, histone variants and mutants, as well as combinations thereof (25). Two separate assays were performed on this library, each employing next generation sequencing (NGS) as a quantitative readout. The first assessed the chromatin remodeling activity of each complex using a restriction enzyme accessibility assay (REAA), and the second employed pulldown to determine complex binding preferences (Fig. 1B). Importantly, binding and activity experiments were highly reproducible across independent experiments (Fig. S1A) and remodeling activity was dependent on the presence of ATP (Fig. S1B). Further, in control experiments using GFP-NLS-HA in place of complexes purified using the same approaches, no activity was observed (Fig. S1C,D). Collectively, these initial datasets, representing >13,000 individual biochemical measurements, including replicates and controls, provide the first systematic view of nucleosome binding, activity, and their relationships for mSWI/SNF family remodelers, with the resulting ‘signatures’ revealing clear differences in the way individual complex subtypes interpret the epigenetic landscape (Fig. 1C, Fig. S1E–G, Table S1). Particularly striking and unexpected was the restrictive impact of many nucleosome modifications in the library on the remodeling activity of the canonical BAF complex, the most stoichiometrically abundant subtype within the mSWI/SNF family (3), in that 70% of the modified nucleosomes studied resulted in reduction in cBAF activity relative to activity on unmodified mononucleosomes (Fig. 1D–E). Notably, this inhibitory effect was not accounted for by binding alone, suggesting impacts on remodeling activity beyond nucleosome binding preferences or affinities. In contrast, activities of PBAF and ncBAF complexes were less constrained by the histone modifications and nucleosome variants in the library (at 45% and 30%, respectively), with ncBAF complexes exhibiting the highest degree of stimulation in both binding and activity across the collection of nucleosomes contained in the library (Fig. 1D–E).
Figure 1. Comprehensive profiling of nucleosome binding and remodeling activities of mSWI/SNF family complexes using DNA-barcoded nucleosome libraries.
A. Schematics and representative SDS-PAGE silver stain gel analyses of endogenous human mSWI/SNF family complexes from HEK-293T cells using HA-tagged DPF2, -BRD7, and -GLTSCR1L as baits for cBAF, PBAF, and ncBAF, respectively.
B. Strategy for high-throughput sequencing-based nucleosome binding and remodeling activity analyses of endogenous human mSWI/SNF complexes incubated with a DNA-barcoded mononucleosome library (n= 109 mononucleosomes).
C. Radar plots mapping the activity measurements of all three mSWI/SNF family complexes across the entire mononucleosome library, normalized to activity on unmodified substrates. Marks and variants are separated by color. Positive score indicates increased activity, negative score indicates decreased activity relative to complex activity on unmodified nucleosome substrates. Marks are sorted by cBAF remodeling activity within each mononucleosome subtype.
D. Activity curves for cBAF, PBAF, and ncBAF complexes across the n=109 mononucleosomes in the library using one-phase decay. Average of complex activity on unmodified (wild-type) mononucleosomes is shown as black line; mononuclesomes showing complex activity > 2 s.d. from WT are shown in red, < 2 s.d. from WT are shown in blue, remaining are shown in gray. See Methods for additional information.
E. Proportion of the unchanged and statistically significant positive and negative regulators of binding and activity measurements across cBAF, PBAF and ncBAF complexes. Positive and negative marks identified as those greater than +/− two standard deviations from the unmodified average. See Methods for additional information.
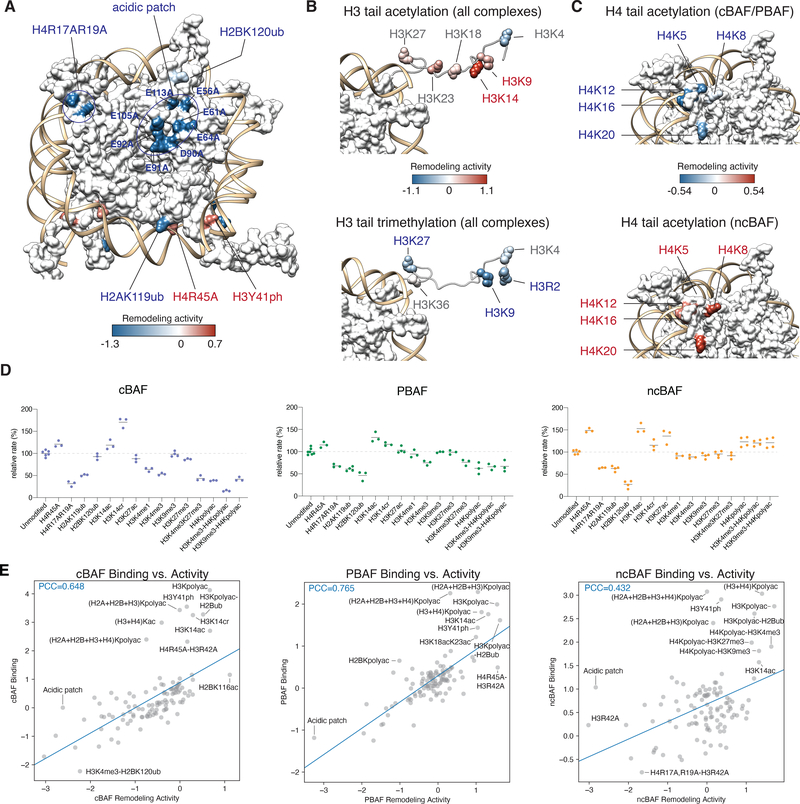
Mapping the results of our experiments on to the three-dimensional structure of the nucleosome revealed a number of modification hotspots that directly impact mSWI/SNF remodeling activities. As expected, screening results and validation experiments performed on individually-generated (non-DNA-barcoded) nucleosome substrates highlighted the H2A/H2B acidic patch as critical for the remodeling activity of all three mSWI/SNF complex types, in keeping with recent structural and functional studies (6, 25, 26) (Fig. 2A, Fig S2A–B). Likewise, modifications and mutants that map to histone-DNA interfaces such as H3Y41ph and H4R45A (a so-called SWI/SNF independent, or Sin-, mutant, (25, 27)) increased activity of all three mSWI/SNF complexes, a finding that is in line with recent results focused on ISWI complexes (25) and that suggests that histone PTMs and mutations that perturb DNA contacts broadly potentiate mSWI/SNF-mediated chromatin remodeling (Fig. 2A, Fig S2A). Library members containing mutations in the basic patch of the H4 tail, such as H4R17A and H4R19A, within a region known to be required for activation of the ATPase subunit (28), were poor substrates of all three mSWI/SNF complexes (Fig. 2A, Fig S2A). A strong inhibitory effect was also seen for ubiquitylation of H2A on lysine 119 (H2AK119ub), a mark associated with gene silencing (29, 30) (Fig. 2A, S2C).
Figure 2. Histone modification hotspots impact mSWI/SNF family complex nucleosome remodeling activities.
A. Modifications of all key residues in the acidic patch (H2AE56A, E61A, E64A, D90A, E91A, E92A and H2BE105A, E113A), the H4 tail basic patch (H4R17A, R19A), and H2AK119ub uniformly inhibit the remodeling activities of all three complex types (blue), while modifications mapping to histone-DNA interfaces (H3Y41ph and the H4R45A sin- mutant) promote the remodeling activity (red). All sites are colored according to the average of log2(fold-change vs. the unmodified nucleosome) values of the three complexes. Note, acidic patch sites have an average log2 value of −2.9, which is out of the color bar range, and is colored blue. (PDB: 1KX5).
B. Acetylation of the H3 tail predominantly promotes remodeling activity (top), while methylation marks tend to inhibit the remodeling activity of the three complexes (bottom). Modifications that consistently have negative, positive, or variable effects across all three complexes are indicated in blue, red, and gray, respectively. All sites are colored according to the average of log2(fold-change vs. the unmodified nucleosome) values of individual acetylation marks (top), and trimethylation marks (bottom) across the three complexes. (PDB: 1KX5).
C. Acetylation of the H4 tail predominantly inhibits the remodeling activity of cBAF and PBAF complexes (top, blue), while they selectively promote the remodeling activity of ncBAF complexes (bottom, red). Modifications that consistently have negative, and positive, or variable effects across all three complexes are written in red and blue, respectively. All sites are colored according to the average of log2(fold-change vs. the unmodified nucleosome) values of cBAF and PBAF (top), and ncBAF (bottom). (PDB: 1KX5).
D. Validation experiments using individual chemically-modified NCPs (lacking DNA barcodes; 10nM), performed on separately-purified cBAF, PBAF, and ncBAF complexes (5nM) across a selection of histone marks and variants from the screen (~15% of the library). n= 3–4 experimental replicates; dots highlight individual data points, black line represents mean value. See Methods for additional information.
E. Activity vs. binding scores for cBAF, PBAF and ncBAF complexes across all mononucleosomes profiled, normalized to unmodified nucleosomes. Pearson correlation coefficients (PCC) are reported for the simple linear regression using all marks (blue).
All three mSWI/SNF complexes were sensitive to marks on the histone H3 N-terminal tail (Fig. 2B). Poly-acetylation of this region, an activating epigenetic signature, significantly stimulated both binding and remodeling activity of all three complexes, an effect that was recapitulated with H3K14ac as a single mark (Fig. 2B, top, S2C,D). These results substantiate previous work showing that subunit domains (DPF2 tandem PHD domain and the SMARCA2/4 bromodomain) in isolation exhibit H3K14ac binding (11–13, 31, 32), as well as studies in yeast and Drosophila suggesting that RSC and SWI/SNF can be activated by histone H3 tail acetylation (18). In contrast to acetylation, the effects of H3 methylation varied as a function of position and BAF complex (Fig. 2B, bottom, Fig. S2E). In particular, H3K4 methylation selectively inhibited cBAF activity whilst having minimal impact on PBAF and ncBAF remodeling activities (Fig. S2E). A differential effect was also seen for acetylation of the H4 tail, particularly H4K20ac and K16ac, which selectively promoted the binding and activity of ncBAF complexes, while having negative effects on activity of cBAF and PBAF complexes (Fig. 2C, Figs. S2F,G).
We next validated key results from the library screen using individually synthesized nucleosomes assembled on identical DNA templates (but lacking DNA barcodes) (Fig. 2D, S2H–I). In vitro remodeling assays, employing separate batches of purified cBAF, PBAF, and ncBAF complexes, confirmed both the activating and inhibitory effects of selected histone PTMs across mSWI/SNF family subtypes. Notably, these experiments further highlighted the impact of dual H3K4me3 and H4 tail acetylation which has a large inhibitory effect on the cBAF complex while having a much smaller impact on PBAF and ncBAF complexes (in negative and positive directions, respectively) (Fig. 2D, S2I, Data S1–4). Importantly, these two marks co-localize to active promoters, at which the PBAF complex distribution is highest, whereas cBAF complexes primarily localize to distal enhancers at which their activities are required for enhancer maintenance (4, 22). Intriguingly, ubiquitylation of H2BK120 (H2BK120ub) was found to negatively impact the remodeling activity of ncBAF and PBAF, whilst having a minimal effect on cBAF. This result differs somewhat from the initial pooled library experiment in which the H2BK120ub mark inhibited all three complexes, albeit to variable extents (Fig. 1C, 2D).
Finally, in comparing the results of activity and binding measurements from the library experiments, we observed a moderately positive correlation between the binding ability and remodeling activity for cBAF and PBAF complexes (PCC= 0.65, 0.77, respectively) across all nucleosomes, although this was less pronounced for ncBAF (PCC= 0.43) (Fig. 2E, Figs. S2J–L). Taken together, these data present a comprehensive evaluation of the binding and activity signatures for the three human subtypes of the mSWI/SNF family across a large collection of diverse nucleosome substrates, revealing direct determinants of their localization and functions in cells.
Combined reader domain and complex architectural features underlie the ncBAF-specific binding signature
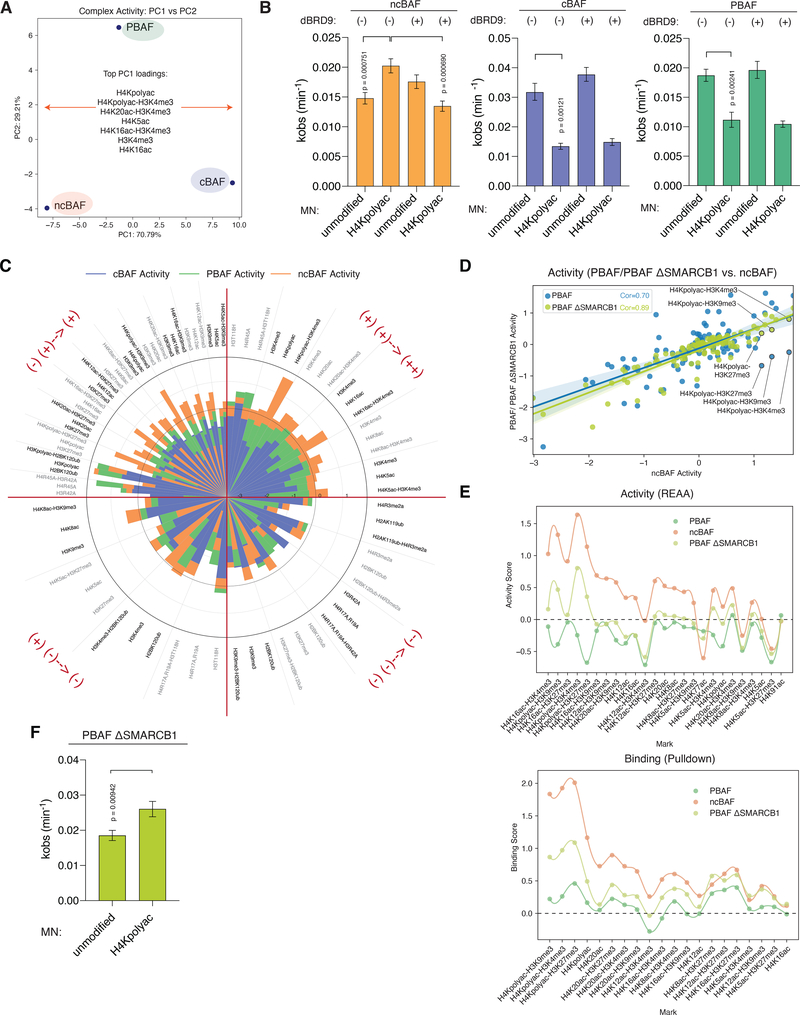
We next sought to better understand the behavior of the ncBAF complex, the most recently discovered member of the mSWI/SNF family whose function remains poorly understood (4, 33, 34). The binding and activity profiles of ncBAF complexes were the most distinct across the library, particularly owing to the strongly enriched binding to and remodeling of H4 acetylated substrates (Fig. 3A, 1C, S2F–GI, S3A), while such marks were inhibitory and strongly inhibitory for the remodeling of PBAF and cBAF complexes, respectively. Among the defining features of ncBAF is the presence of the complex-specific BRD9 subunit (Fig. 1A), the bromodomain of which has been shown to be capable of binding acetylated H3 and H4 peptides in solution (35, 36), potentially providing an explanation for the higher activity observed on substrates containing H4 tail acetylation marks. To test this, we generated individual unmodified or H4-poly-acetylated nucleosomes and performed remodeling assays with ncBAF complexes in the presence or absence of a highly selective BRD9-BD inhibitor, dBRD9 (4, 37). Consistent with our library data, we observed that in the absence of inhibitor (DMSO control), nucleosomal substrates containing the polyacetylated H4 tail significantly stimulated the remodeling activity of ncBAF (p=0.00075 (n=4), but inhibited the activity of cBAF and PBAF complexes (p=0.00121 (n=4–5), p=0.00241 (n=3), respectively) (Figs. 3B, Figs. S3B–D). Strikingly, addition of dBRD9 selectively reduced the remodeling activity of ncBAF on polyacetylated H4 substrates such that the rate of remodeling was closer to that seen on unmodified nucleosomes (p=0.00069 (n=3–4, while it had no effect on cBAF or PBAF activity on either nucleosomal substrate (Figs. 3B, S3B–D). Collectively, these data indicate that the presence of the BRD9 subunit within fully-formed ncBAF complexes is, at least in part, responsible for the sensitization of the complex toward substrates containing acetyl marks on the H4 tail. In contrast to ncBAF complexes, the remodeling activities of PBAF complexes are inhibited on H4ac-containing NCP substrates, despite binding at levels that are comparable or even slightly enhanced relative to binding on WT substrates (Fig. 1C, S1G, Fig. 2D). This is noteworthy as the PBAF-specific BRD7 BD-containing subunit, whose bromodomain is highly similar (~80%) to that of BRD9, is also capable of binding H4 acetylated tails in solution (38), suggesting that in the context of PBAF, BRD7 cannot engage H4ac marks, or, that if binding does occur, that it is not sufficient to overcome other inhibitory effects. In line with the latter possibility, acetylation of H4K16 and K20 within the basic patch of the H4 tail would be expected to disrupt the interaction with the ATPase subunit, as we observe with H4R17A, R19A mutations (Fig. 1C, 2A). As such, in the context of ncBAF, we presume that the inhibitory effect of H4 basic patch alterations is outweighed by the stimulation associated with BRD9 engagement, bearing in mind that the nucleosome contains two copies of H4. This result suggests that BAF complexes are able to integrate multiple biochemical inputs (either positive or negative) from the chromatin substrate, leading to a context-specific remodeling output. Indeed, there are several examples of this integrative behavior within our data in which a nucleosome decorated with two marks or variants result in either dominant effects for one or the other mark, or additive effects in positive or negative directions with respect to complex activity (Fig. 3C). For example, and as already noted, the combination of H4 polyacetylation and H3K4me3, each of which independently reduces cBAF activity, leads to a profound inhibition in cBAF remodeling (an additive effect) (Fig. 3C, 2D). These data suggest that combinatorial biochemical cues can play important roles in directing BAF complex activities, in this example, by possibly restricting cBAF activity to distal sites at which their activities are required for maintenance of enhancer accessibility (22, 39).
Figure 3. Preferential activity of ncBAF complexes on poly-acetylated histone H4 substrates is facilitated by BRD9 and the absence of the SMARCB1 subunit.
A. Principal component analysis (PCA) of cBAF, PBAF, and ncBAF complex activity measurements across the full n=109 nucleosome library; PC1: 70.79% and PC2: 29.21%. Top PC1 loadings are indicated.
B. Effect of H4polyac marks and dBRD9 (BRD9 inhibitor, 5 μM) on the remodeling activity (kinetics) of ncBAF, cBAF, and PBAF complexes. Graphs show the fit remodeling rates (kobs) for different conditions. Error bars represent 95% confidence interval. P-values of significant conditions are indicated (n = 3–5 replicates).
C. Radar plots containing stacked bar charts for cBAF, PBAF, and ncBAF complex activities, showing nucleosomes with sets of single and combination marks, distributed in quadrants showing additive positive and negative and dominant positive and negative combinations.
D. Correlation of pan-library activity scores for ncBAF with PBAF complexes (blue) or SMARCB1-deficient PBAF complexes (light green). Key H4ac marks are labeled.
E. Activity (REAA) (top) and binding (bottom) library screen results for PBAF, PBAF ΔSMARCB1, and ncBAF complexes over nucleosome substrates containing H4 tail acetylation. Curves representing smoothened activity and binding scores across the marks presented are shown.
F. SMARCB1-deficient PBAF remodeling activity on WT and H4polyac nucleosome substrates as measured by restriction enzyme accessibility assay (REAA). Graphs show the fit remodeling rates (kobs) for different conditions. Error bars represent 95% confidence interval. P-values of significant conditions are indicated (n = 3 replicates).
A second feature of ncBAF complexes is the absence of the evolutionarily conserved SMARCB1 (BAF47) subunit whose C-terminal domain directly engages the nucleosome acidic patch (6, 9) and which is required for binding of the DPF2 or PHF10 reader subunits in cBAF and PBAF complexes, respectively (6, 22). While cBAF (and by extension, PBAF) complexes are known to grip both nucleosome faces in a “C-clamp” type arrangement (6, 8), ncBAF is predicted to engage the acidic patch on just one face of the nucleosome via the ATPase module (which is present in all three complex forms) (Fig. 1A). To explore whether the SMARCB1-mediated architectural difference contributes to ncBAF complex nucleosome substrate specificities, especially in comparison to the BRD7-containing PBAF complexes, we purified PBAF complexes lacking SMARCB1 (PBAF ΔSMARCB1) using HA-SMARCD2 as bait in HEK-293T ΔSMARCB1 cells generated with CRISPR/Cas9-mediated gene editing (Fig. S3E), and performed full nucleosome library screens, comparing results to those obtained with full PBAF and ncBAF complexes. We observed that the library-wide binding pattern of PBAF ΔSMARCB1 has a high correlation with PBAF (PCC= 0.90), supporting the integrity of the subcomplex preparation (Fig. S3F). However, the remodeling activity signature of PBAF ΔSMARCB1 shifted towards that of ncBAF for a number of chromatin marks (Fig. 3D, Table S2). In particular, loss of SMARCB1 in PBAF resulted in increased (rather than inhibited) activity (and binding) on nucleosomes containing H4ac marks relative to unmodified nucleosomes, mirroring the ncBAF complex-specific activity preferences measured (Fig. 3E, Fig. S3G). This trend was also demonstrated using principal component (PC) analyses in which the top loadings driving separation of ncBAF and ΔSMARCB1 complexes from WT cBAF and PBAF complexes included histone H4ac marks such as H4 poly-acetylation and H4K16ac (Fig. S3H). These findings from the library data were recapitulated in experiments showing that individually-purified H4 polyacetylated nucleosomes stimulate the remodeling of PBAF ΔSMARCB1 complexes relative to unmodified nucleosomes (Fig. 3F, S3I). Finally, to evaluate whether the acidic patch binding function of SMARCB1 alone accounted for this affect, we purified either WT SMARCB1- or acidic patch-binding mutant SMARCB1 K364del-containing complexes (9) and subjected them to activity measurements on unmodified or H4polyAc-modified NCPs. Notably, while the SMARCB1 K364del acidic patch binding mutant complexes exhibited reduced activity overall relative to WT SMARCB1 complexes, as expected, the inhibitory impact of H4Ac polyacetylation was still observed (Fig. S3J). Taken together, these results indicate the combined requirements for an H4 acetylation binding subunit (i.e. BRD9) and the absence of the SMARCB1 subunit for H4 acetylation-induced stimulation of nucleosome remodeling activities. These data also provide insight into the biochemical basis underpinning the binding and activity signatures of mSWI/SNF family complexes on chromatin that are affected by the presence or absence of SMARCB1, as has been observed in cell-based genomic profiling efforts (4).
Modular deconstruction of cBAF complexes informs subunit- and domain-specific contributions to nucleosome remodeling
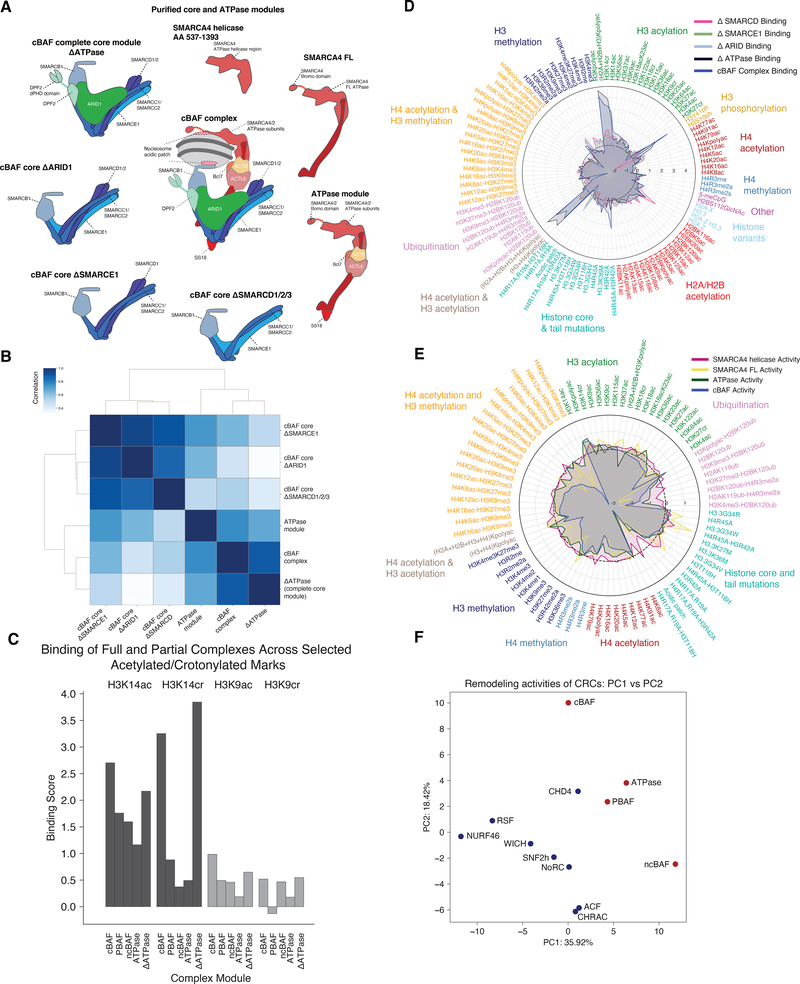
Next, we sought to deconstruct mSWI/SNF complexes into their component modules and subunits as a strategy to define the determinants of complex-nucleosome binding behavior, particularly, the highly selective chromatin preferences of canonical BAF complexes (Fig. 1C,D). Using a series of cell lines engineered to contain deletions of specific subunits (3), we isolated various stages of cBAF core module and ATPase module assembly (Fig. 4A, Fig. S4A). The ATPase module docks on to completed core modules to form final canonical BAF complexes (3). The complete cBAF complex core module (termed cBAF ΔATPase), as well as a series of purified partial modules lacking various core module subunits (i.e. those lacking ARID1A/B and DPF2 subunits [cBAF Core ΔARID1], lacking SMARCE1, ARID1/B and DPF2 subunits [cBAF Core ΔSMARCE1], or lacking SMARCD1/2, ARID1A/B and DPF2 subunits [cBAF Core ΔSMARCD1/2/3]), or variants of the SMARCA4 ATPase and ATPase module were then assayed for their ability to bind and remodel across the diverse mononucleosome library. Core module variants were not evaluated for remodeling activity given the absence of the ATPase and hence catalytic activity.
Figure 4. Epigenetic modification preferences of mSWI/SNF complexes are defined by module-specific histone binding properties.
A. Schematic summarizing the cBAF core and ATPase modules/subunits subjected to full library binding and activity experiments.
B. Correlation heatmap for pan-library binding profiles for all cBAF core modules, ATPase module, and full cBAF complexes.
C. Binding scores for cBAF, PBAF, ncBAF complexes as well as the full core module (Core, ΔATPase) and the ATPase module (ATPase) over H3 lysine acylation marks (H3K14ac, H3K14cr, H3K9ac, and H3K9cr).
D. Radar plots indicating the binding of cBAF cores (ΔARID, ΔSMARCD, ΔSMARCE1, ΔATPase) and full cBAF complexes across all mononucleosomes profiled in the library. Marks and variants are separated by color. The radar plots are sorted by cBAF full complex binding within each histone mark type.
E. Radar plots indicating the remodeling activities of the ATPase module, SMARCA4 FL, truncated SMARCA4 (aa 537–1393) and full cBAF complexes across all mononucleosomes profiled in the library. Marks and variants are separated by color. The radar plots are sorted by cBAF full complex binding within each histone mark type.
G. Principal component analysis (PCA) of mSWI/SNF, CHD4 and ISWI complex activities.
Analysis of nucleosome binding datasets for cBAF core and ATPase module variants revealed that the binding profile of the complete core module (ΔATPase) most closely resembled that of the final-form cBAF remodeler (PCC= 0.90), while the ATPase module in isolation exhibited a more moderately similar binding profile and the incomplete complex cores were far less sensitive (in both positive and negative directions) to the presence of nucleosome modifications contained in the library (Fig. 4B–C, Fig. S4B, Table S3). The complete cBAF core contains the tandem PHD domain-containing subunit, DPF2, whereas the other cores lack this subunit. Consequently, we found that binding to nucleosomes containing acetylated and crotonylated histone H3 tails (in particular H3K14) was enhanced in the full core module compared to any other core or ATPase module variant or the ncBAF and PBAF complexes lacking this subunit (Fig. 4C–D, Fig. S4C–D). These data implicate the core module, and specifically, DPF2, as a major determinant in the nucleosome binding specificity of fully-formed cBAF complexes. This observation is consistent with previous work which defines the double PHD domain of DPF2 as a preferential H3K14 crotonyl reader domain (11).
Finally, we sought to assess the activity of the isolated cBAF ATPase module and its constituents across the library of chromatin contexts. The complete module was purified from HEK-293T cells overexpressing an HA-tagged SS18 subunit (Fig. 4A, S4A). We then employed the barcoded-nucleosome library to profile the remodeling activity of the ATPase module as well as the full length SMARCA4 subunit and a truncated version thereof containing only the helicase region and SnAc/post-SnAc domain (residues 537–1393) that excludes the HSA and putative AT-hook binding regions (40) and is predicted to bind the nucleosome acidic patch (6) (Fig. 4E, Fig. S4A,C, Table S3). Remarkably, we found the restrictive remodeling behavior that is characteristic of the final-form cBAF complex was greatly relaxed in the ATPase module, especially in the magnitude of the inhibitory effects mediated by the majority of the library members (Fig. 4E). This more promiscuous behavior extended to the SMARCA4 subunit variants and was especially striking for the truncated form of SMARCA4, which was almost completely insensitive to the modifications in the library, including acidic patch mutations (Fig. 4E, S4E–F), despite the fact that this construct contains the region that has recently been implicated in acidic patch recognition (6, 8). These data highlight the requirement for the remaining regions of the SMARCA4 subunit, such as the C-terminal bromodomain and the rigid HSA domain which tethers the ARP module of ACTL6A/B and beta-actin subunits, in providing structural or biochemical recognition stability to facilitate its proper nucleosome engagement and acidic patch recognition. Collectively, these results are consistent with a model in which the chromatin landscape preferences of the cBAF complex become increasingly specific over the course of core module assembly. Given that the three final-from BAF complexes differ primarily in the subunit composition of the core modules (Fig. 1A), this model provides an attractive framework for understanding how functional specialization of distinct mSWI/SNF family complexes is acquired.
Discussion
The studies described herein provide a direct determination as to how variation in nucleosome structure impacts the recruitment and activity of mSWI/SNF family complexes, in either uniform or complex-specific manners. Our data indicate that mSWI/SNF complexes are able to respond to diverse features, or signals, present on the chromatin substrate, and that remodeling activity is an integrated response to these. This ability to contextualize chromatin landscape features is driven by a combination of module and overall complex architecture, acidic patch engagement, and inclusion of core module reader subunit componentry. One unexpected finding is that most histone marks present in our library had negative effects on the activity of canonical BAF and to a somewhat lesser extent, PBAF complexes, while ncBAF complexes displayed increases in binding and activity for many more nucleosomes across the library, suggesting their tolerance of a wider range of chromatin states. Intriguingly, marks that we found restricted the activity of cBAF complexes included some for which BAF has been positively correlated using genome-wide ChIP-seq-based mapping strategies and even suggested to serve as primary recruitment interactions, such as H3K4me1 and H3K27ac (41), underscoring the potential limitations in interpreting complex-histone mark co-occupancy using cellular genomic approaches. Our data indicates that the complex activities of the mSWI/SNF family differentiate from those of the ISWI and CHD families as revealed by PC and correlation analyses of both current and previously published (25) datasets (Fig. 4F, S4G–H). Moreover, our data suggest that architectural constraints imposed by the fully-formed modules and complexes play important roles in regulating activity, with the nature of nucleosome engagement and accessibility of reader domains emerging as potential determinants of mSWI/SNF binding (Fig. S4I). Our data also suggest that core module-mediated complex specificity is further tuned by the structural context imposed by the final docking of the long and highly interfaced ATPase module, in that full complexes further accentuate negative (i.e. H2BK120ub-H3K4me3-marked nucleosomes) and positive (i.e. H3R42A-mutant nucleosomes) effects (Fig. 4D).
Finally, findings from both pooled library experiments and those performed on individual nucleosomes highlight the concept of binding repulsion and inhibition of activity on certain nucleosome substrates as a mechanism that may contribute to the overall direction and distribution of mSWI/SNF complexes genome-wide. Indeed, such mechanisms may titrate appropriate levels of a given subcomplex at the sites at which their specific activities are needed, as exemplified by cBAF complexes avoiding H3K4me3, especially when combined with H4 acetylation, marks often found over active promoters. Taken together, these data suggest that a monolithic model in which the presence of chromatin marks only serve to recruit remodeling factors may not fully capture the significantly more nuanced nature of the input-output activity relationships at play, and, suggest a broadened mechanistic framework to include an avoidance paradigm in which certain epigenetic marks, or combinations thereof, can restrict the activity of remodelers, ultimately directing remodeling activities toward permissive chromatin states.
Supplementary Material
Acknowledgments:
We thank members of the Kadoch and Muir laboratories for helpful advice and discussions throughout this project. We thank N. Hananya, S. Daley and F. Wojcik for providing peptide materials.
Funding: This work was supported by Mark Foundation for Cancer Research- Emerging Leader Award (C.K.); National Institutes of Health Grant 1DP2CA195762-01 (C.K.); Pew-Stewart Scholars in Cancer Research Award (C.K.); American Cancer Society Research Scholar Award RSG-14-051-01-DMC (C.K.); National Institutes of Health Grant R37-GM086868 (T.W.M.); National Institutes of Health Grant PO1- CA196539 (T.W.M.); National Institutes of Health Grant K99/R00 K99CA237855 (N.M.); Jane Coffin Childs Memorial Fund Postdoctoral Fellowship Award (H.T.D.); National Institutes of Health Grant No. GM123659 (J.D.B.).
Footnotes
Competing interests: CK is the scientific founder, fiduciary Board of Directors member, Scientific Advisory Board Member, shareholder and consultant for Foghorn Therapeutics, Inc. (Cambridge, MA). The other authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the supplementary materials.
References and Notes
- 1.Bates SE, Epigenetic Therapies for Cancer. The New England journal of medicine 383, 650–663 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Kadoch C, Crabtree GR, Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science advances 1, e1500447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mashtalir N et al. , Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 175, 1272–1288.e1220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel BC et al. , A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nature cell biology 20, 1410–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan J et al. , Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell systems 6, 555–568.e557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashtalir N et al. , A Structural Model of the Endogenous Human BAF Complex Informs Disease Mechanisms. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Y, Reyes AA, Malik S, He Y, Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 579, 452–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He S et al. , Structure of nucleosome-bound human BAF complex. Science (New York, N.Y.) 367, 875–881 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Valencia AM et al. , Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell 179, 1342–1356 e1323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J et al. , The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nature genetics 51, 618–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong X et al. , Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nature chemical biology 12, 1111–1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng L et al. , Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen W et al. , Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 46, 2100–2110 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P, ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Molecular cell 6, 1049–1058 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Reinke H, Gregory PD, Horz W, A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Molecular cell 7, 529–538 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Agalioti T, Chen G, Thanos D, Deciphering the transcriptional histone acetylation code for a human gene. Cell 111, 381–392 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Charlop-Powers Z, Zeng L, Zhang Q, Zhou MM, Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell research 20, 529–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee N et al. , Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic acids research 39, 8378–8391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan AH, Neely KE, Workman JL, Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104, 817–827 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Hassan AH et al. , Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369–379 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Ferreira H, Flaus A, Owen-Hughes T, Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. Journal of molecular biology 374, 563–579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama RT et al. , SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nature genetics 49, 1613–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathur R et al. , ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nature genetics 49, 296–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X et al. , SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nature genetics 49, 289–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dann GP et al. , ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dao HT, Dul BE, Dann GP, Liszczak GP, Muir TW, A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nature chemical biology, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nacev BA et al. , The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567, 473–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Li M, Xia X, Li X, Chen Z, Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Tamburri S et al. , Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Molecular cell 77, 840–856 e845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H et al. , Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Huber FM et al. , Histone-binding of DPF2 mediates its repressive role in myeloid differentiation. Proceedings of the National Academy of Sciences of the United States of America 114, 6016–6021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange M et al. , Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes & development 22, 2370–2384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alpsoy A, Dykhuizen EC, Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. The Journal of biological chemistry 293, 3892–3903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X et al. , BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nature communications 10, 1881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn EM et al. , A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure 23, 1801–1814 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Filippakopoulos P et al. , Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remillard D et al. , Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angew Chem Int Ed Engl 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H et al. , Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun 358, 435–441 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Iurlaro M et al. , Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nature genetics 53, 279–287 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Singh M, D’Silva L, Holak TA, DNA-binding properties of the recombinant high-mobility-group-like AT-hook-containing region from human BRG1 protein. Biol Chem 387, 1469–1478 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Local A et al. , Identification of H3K4me1-associated proteins at mammalian enhancers. Nature genetics 50, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen UT et al. , Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nature methods 11, 834–840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ, Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of molecular biology 272, 301–311 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.