Abstract
Four isoforms of human fibroblast growth factor 2 (FGF-2) result from alternative initiations of translation at three CUG start codons and one AUG start codon. Here we characterize a new 34-kDa FGF-2 isoform whose expression is initiated at a fifth initiation codon. This 34-kDa FGF-2 was identified in HeLa cells by using an N-terminal directed antibody. Its initiation codon was identified by site-directed mutagenesis as being a CUG codon located at 86 nucleotides (nt) from the FGF-2 mRNA 5′ end. Both in vitro translation and COS-7 cell transfection using bicistronic RNAs demonstrated that the 34-kDa FGF-2 was exclusively expressed in a cap-dependent manner. This contrasted with the expression of the other FGF-2 isoforms of 18, 22, 22.5, and 24 kDa, which is controlled by an internal ribosome entry site (IRES). Strikingly, expression of the other FGF-2 isoforms became partly cap dependent in vitro in the presence of the 5,823-nt-long 3′ untranslated region of FGF-2 mRNA. Thus, the FGF-2 mRNA can be translated both by cap-dependent and IRES-driven mechanisms, the balance between these two mechanisms modulating the ratio of the different FGF-2 isoforms. The function of the new FGF-2 was also investigated. We found that the 34-kDa FGF-2, in contrast to the other isoforms, permitted NIH 3T3 cell survival in low-serum conditions. A new arginine-rich nuclear localization sequence (NLS) in the N-terminal region of the 34-kDa FGF-2 was characterized and found to be similar to the NLS of human immunodeficiency virus type 1 Rev protein. These data suggest that the function of the 34-kDa FGF-2 is mediated by nuclear targets.
Fibroblast growth factor 2 (FGF-2) is a prototype of the FGF family of 17 genes coding for either mitogenic proteins, differentiating factors, or oncogenic proteins (22, 32, 35, 45, 52). FGF-2 is produced in many cell types and tissues, and its biological roles are pleiotropic. It is involved in embryogenesis and morphogenesis, especially in the nervous system and bone formation (11, 50). FGF-2 is a major angiogenic factor and thus a molecule of biological interest in cardiovascular disease therapeutics. However, this angiogenic effect also activates tumor neovascularization (25). In addition, the mitogenic and differentiating effects of FGF-2 confer on it oncogenic potential (13, 42). FGF-2 has also been described as playing a crucial role in wound healing (53).
The pleiotropic roles of FGF-2 can partly be explained by the different modes of action of this factor. On the one hand, it acts in a paracrine and autocrine manner, after being secreted by the producer cell. This mode of action is mediated by the recognition by FGF-2 of specific receptors, whose activation induces signal transduction cascades (54). This paracrine and autocrine effect may also be the result of nucleolar translocation of exogenous FGF-2 (2). On the other hand, FGF-2 also exhibits intracrine action, thereby allowing a direct effect on intracellular targets in the absence of secretion (6, 14).
The different modes of action of FGF-2 are in fact the direct consequence of a process of alternative initiation of translation on the FGF-2 mRNA. Four in-frame initiation codons, including three CUGs and one AUG, give rise to four FGF-2 isoforms with distinct features (15, 41). The CUG-initiated forms of 22, 22.5, and 24 kDa (HMW [high-molecular-weight] FGF-2) are localized in the nucleus, whereas the AUG-initiated form of 18 kDa is mostly cytosolic (9, 10). Constitutive expression of the 18-kDa form leads to transformation of adult bovine aortic endothelial cells, whereas expression of the HMW FGF-2 leads to immortalization of the same cells (13). The 18-kDa FGF-2 is also able to stimulate cell migration and to down-regulate its own receptor, which is not the case for the HMW FGF-2 (6, 33). These different features of the FGF-2 isoforms are correlated to their distinct modes of action: the 18-kDa isoform, secreted despite the absence of a signal sequence, is responsible for the paracrine and autocrine effects. In contrast, the nuclear HMW isoforms are not released from the cell and are responsible for the intracrine effect of FGF-2 (6, 14, 34).
We have known for a few years that FGF-2 expression is controlled at the translational level (40). Ninety percent of the 6,774-nucleotide (nt)-long human FGF-2 mRNA is composed of nontranslated regions, with a GC-rich leader of several hundred nucleotides and an AU-rich 3′ untranslated region (UTR) measuring almost 6,000 nt (41). Five regulatory elements have been identified in the leader region of the messenger, either in the 5′ UTR or in the alternatively translated region (40). One of these elements has been identified as an internal ribosome entry site (IRES) which enables the FGF-2 mRNA to be translated independently of the classical cap-dependent scanning mechanism (29, 47). A few cellular mRNAs have been shown to possess an IRES (5, 16, 30, 36, 46). This feature allows such mRNAs to be expressed in conditions of cap-dependent translation arrest, such as under stress (23).
A study of FGF-2 isoform expression in different human cell types has shown that the HMW isoforms are produced in various immortalized and transformed cell lines, while primary cells almost exclusively synthesize the 18-kDa form at confluence (48). Furthermore, the HMW isoforms are induced when normal cells are subjected to heat shock and oxidative stress, suggesting that the IRES, located just upstream from the CUG start codons, is activated by stress stimuli. In both transformed and stressed cells, CUG codon expression is correlated to the binding of specific factors to the FGF-2 mRNA leader (48).
In this study, we demonstrate the existence of a new FGF-2 isoform that is generated through an additional process of alternative initiation of translation in human FGF-2 mRNA. This fifth FGF-2, with a molecular mass of 34 kDa, is the result of a translation initiation at a CUG codon located 86 nt downstream from the mRNA 5′ end. The 34-kDa FGF-2, in contrast to the other FGF-2 isoforms, is exclusively synthesized by a cap-dependent, IRES-independent initiation mechanism. We also show that the 34-kDa FGF-2, in contrast to the other FGF-2 isoforms, permits NIH 3T3 cell survival in low-serum conditions. Furthermore, we have identified a new nuclear localization signal (NLS) sequence in the N-terminal region specific to this FGF-2 isoform.
MATERIALS AND METHODS
Plasmid construction.
The construct WT5′-FGF (plasmid pSCT-12V) contains the first 1,179 nt of FGF-2 cDNA (with a short 3′ UTR) cloned into the XbaI-SacI restriction sites of vector pSCT, under the control of T7 and cytomegalovirus promoters (40). The construct Δ1-257 (plasmid pSCT-27) corresponds to a deletion of nt 1 to 257 (SmaI site) of the FGF-2 cDNA in the above-mentioned vector pSCT-12V (40). To obtain the construct WT5′CAT, a PCR fragment containing nt 1 to 312 of the FGF-2 cDNA was synthesized, using oligonucleotide FGF5′, described previously (40), and oligonucleotide FLC, complementary to nt 293 to 312 of FGF-2 cDNA with an ApaI site (Table 1). This fragment was introduced between the XbaI and ApaI sites of plasmid pFC2 described previously (47), giving a fusion of nt 1 to 312 of the FGF-2 cDNA with the chloramphenicol acetyltransferase (CAT) coding sequence devoid of an AUG.
TABLE 1.
Sequences of oligonucleotides used for mutagenesis
| Name | Sequence |
|---|---|
| FLC | 5′-AAAATCGATTGGGCCCCCGCTCGGCCGCTCTTCTG-3′ |
| AUGm | 5′-CACCCGCCGGGCCGCGGCGTCAGATCTTCTAGATCTCCA-3′ |
| Sfi-S | 5′-GGCCCGGCGGGTGCCAGATTAGCGGCCGCGGTGGCCGCGGTTGCAACGGGATCCCGGGCGCTGCA-3′ |
| Sfi-AS | 5′-GCGCCCGGGATCCCGTTGCAACCGCGGCCACCGCGGCCGCTAATCTGGCACCCGCCGGGCCGC-3′ |
| ATG86-S | 5′-CGCGCAGGAGGGAGGAGAACTGGGGGCGCGGGATCATGACCGGTGTCGGGGGTGGAGAT-3′ |
| ATG86-AS | 5′-CTAGATCTCCACCCCCGACACCGGTTATGATCCCGCGCCCCCAGTTCTCCTCCCTCCTG-3′ |
| CCC86-S | 5′-GATCATAACGCGGGAGGCCCA-3′ |
| CCC86-AS | 5′-CCGGTGGGCCTCCCGCGTTAT-3′ |
| TAA89-S | 5′-GATCATCTAGAAGGCTGTAAA-3′ |
| TAA89-AS | 5′-CCGGTTTACAGCCTTCTAGAT-3′ |
| GCG122-S | 5′-GATCTAGAAGATGTAGCGCCGCGG-3′ |
| GCG122-AS | 5′-CCGGGCCGCGGCGCCTACATCTTCTA-3′ |
| CAT3′-S | 5′-ATGGCAATGAAAGACGGTGAG-3′ |
| UTR3′A1-AS | 5′-ACACATTTATTTTCTTTTACTCTC-3′ |
| bigATG-S | 5′-ACACTCTAGAGCGGGAGGATGGTGGGTGTCGGGGGT-3′ |
| FGF5′-AS | 5′-AGCCTCGAGCCGCTCGGCCGCTCTTCTGTC-3′ |
| DELNLS-5′ | 5′-GCTTGGGAGGCGGCTCTCCACCCATCCGTGAACCCCAGGTC-3′ |
| DELNLS-3′ | 5′-CCGGGACCTGGGGTTCACGGATGGGTGGAGAGCCGCCTCCCAAGCTGCA-3′ |
| SVNLS-5′ | 5′-AAAAAGCTTCCATGGCAAAAAAGAAGCGGAAAAAAGAGAAAAAAATCACTGGAGAT-3′ |
| CAT-rev | 5′-TTTGAGCTCAGATCTCATTACGCCCCGCCCTGCCA-3′ |
The wild-type FGF-CAT used for Fig. 5 corresponds to the previously described plasmid pFC1 (47); it contains the 539 5′ nt of the FGF-2 cDNA fused to the CAT sequence in the vector pSCT. Site-directed mutagenesis of CTG-86 and ACG-122 was obtained in two steps. The first step was the creation of new restriction sites. XbaI and BglII sites were introduced at positions 110 and 107 of FGF-2 cDNA by PCR mutagenesis using Pwo DNA polymerase (Boehringer) as follows. A PCR fragment was amplified from the template pFC1, using oligonucleotide FGF-5′ (see above) as the 5′ primer and oligonucleotide AUGm, containing the new BglII and XbaI restriction sites and the HgaI site at position 121, as the 3′ primer (Table 1; see Fig. 4A). The PCR fragment was cloned into the vector pBluescript KS+ EcoRV site. It was then subcloned from pBluescript, using enzymes HindIII and HgaI, into the vector pFC1; the new plasmid was called pFC1-XB. An SfiI site was introduced at position 152 by insertion of double-stranded oligonucleotide Sfi-S/Sfi-AS into plasmid pFC1, between positions 125 (SacII site) and 192 (PstI site) of the FGF-2 cDNA (Table 1); the new plasmid was called pFC1-Sfi. Both plasmids pFC1-XB and pFC1-Sfi gave the same FGF-2 isoform expression profile as pFC1 (not shown).
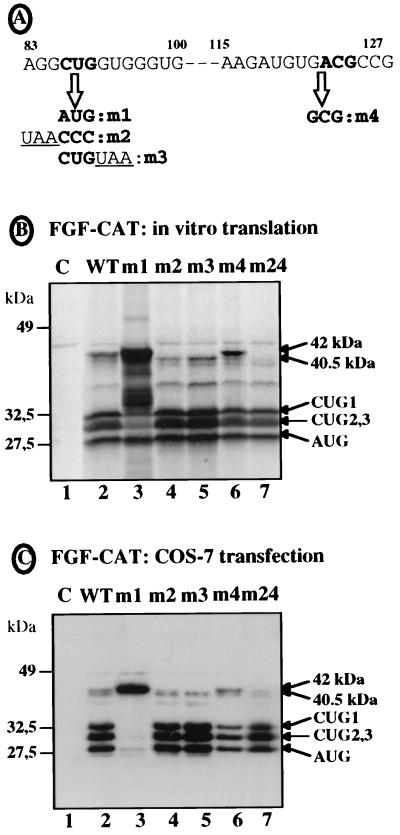
FIG. 5.
Identification of the start codon by site-directed mutagenesis. Several point mutations of CTG-86 or of ACG-122 were introduced into a chimeric FGF-CAT construct in which the first 539 nt of FGF-2 cDNA were fused to the CAT sequence (40) (see Materials and Methods). The mutant constructs were used either for in vitro transcription and translation in RRL or for COS-7 cell transfection followed by Western immunoblotting, as for Fig. 3. (A) Representation of nt 83 to 127 of the FGF-2 cDNA. CTG-86 and ACG-122 are represented in boldface; arrows indicate the mutations obtained. (B) Translation of the different in vitro-transcribed mRNAs in RRL. (C) Western immunoblotting of transfected COS-7 cell extracts with anti-CAT antibody. The names of the mutants are indicated above the lanes. WT corresponds to the wild-type FGF-CAT construct; the mutant m24 (lane 7) corresponds to a double mutant (m2 plus m4). Lane 1 corresponds to the control (C) assay without mRNA. Positions of migration of the size standards (left) and of FGF-CAT proteins (right) are indicated. The two bands corresponding to the largest FGF-CAT protein (42 and 40.5 kDa) are shown by arrows.
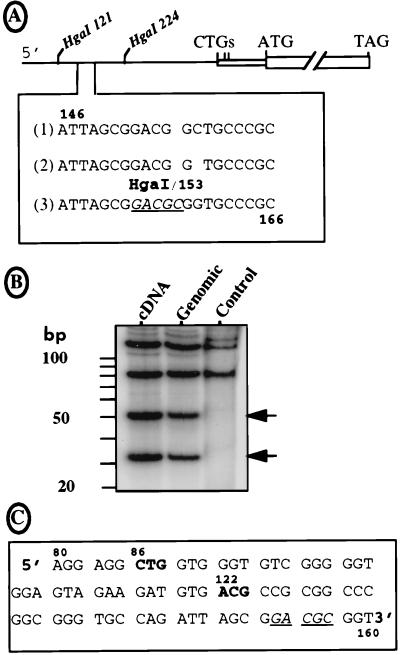
FIG. 4.
Sequence of the FGF-2 mRNA leader. (A) The sequence of the FGF-2 cDNA 5′ region was obtained by using a DNA analyzer (see Materials and Methods) and compared to the two published sequences. Under the schema of the FGF-2 cDNA indicating the two HgaI sites at positions 121 and 224 downstream from the 5′ end are represented the three sequences of the region from 146 to 166. Lines 1 and 2 correspond to the published sequences of the cDNA and genomic DNA, respectively (41, 44). Line 3 is the sequence obtained in this study for both DNAs with the DNA analyzer. The new HgaI site is underlined (position 153). (B) The pFC1 plasmid with the 5′ region of the FGF-2 cDNA, its homolog with the corresponding genomic sequence (kindly provided by R. Florkiewicz), and a pFC1-derived plasmid lacking the HgaI site at position 153 (point mutations change nt 153-GACGCGG to 153-GATGGC; published for its in-frame ATG-154 [47]) were BssHII digested. The 291-nt-long resulting fragments were dephosphorylated, kinase treated in the presence of [32P]ATP, and then digested with enzyme HgaI. Each step was followed by a G50 column purification. The restriction fragments were fractionated on a 15% polyacrylamide–Tris-borate-EDTA gel, which was dried and autoradiographed. Sizes of the expected fragments are (i) 121, 85, 51, and 34 nt in the presence of the new HgaI site and (ii) 121 nt plus a doublet at 85 nt in its absence. The DNA origin (cDNA or genomic) is indicated at the top; control corresponds to the plasmid lacking the HgaI site at position 153. (C) Representation of the new reading frame given by the DNA sequence shown in A (line 3). The sequence between nt 80 and 163 is shown. The two potential initiation codons are shown in boldface; the HgaI site (positions 153 to 157) is underlined.
The second step of mutagenesis involved use of the double-stranded oligonucleotide insertion strategy, starting either from plasmid pFC1-XB or from plasmid pFC1-Sfi. Plasmid pFC1-m1 (CTG-86 mutated to ATG) resulted from insertion of the double-stranded oligonucleotide ATG86-S/ATG86-AS into plasmid pFC1-XB between positions 50 (BssHII site) and 110 (XbaI site) (Table 1). The mutation introduced an AgeI site just downstream from the ATG-86 codon. Plasmid pFC1-m2 (CTG-86 mutated to CCC) resulted from insertion into plasmid pFC1-m1 of the double-stranded oligonucleotide CCC86-S/CCC86-AS between positions 1 (BamHI site in the polylinker) and 89 (AgeI site) of the FGF-2 leader sequence (Table 1). Plasmid pFC1-m3 (insertion of TAA downstream from CTG-86) was obtained in the same way as pFC1-m2, but using the double-stranded oligonucleotide TAA89-S/TAA89-AS (Table 1). Both constructs contained a deletion of the 5′ region (nt 1 to 76). Plasmid pFC1-m4 (ACG-122 mutated to GCG) was obtained by insertion of the double-stranded oligonucleotide GCG122-S/GCG122-AS into plasmid pFC1-XB between the BglII site (position 107) and the HgaI site (position 121) (Table 1). Plasmid pFC1-m24 (double mutant of CTG to CCC and of ACG to AAG) was obtained by ligation of a XbaI-SacI fragment from plasmid pFC1-m4 (containing the FGF-2 leader region downstream from position 107 and the CAT sequence) with an XbaI-SacI fragment from the plasmid pFC1-m2 (containing the vector sequence and the FGF-2 leader region in 5′ from position 107).
Plasmids p5′CAT-A0, p5′CAT-A1, and p5′CAT-A7 were derived from the plasmid pKSCAT-pA. pKSCAT-pA had been constructed by insertion of a 70-nt poly(A) fragment (5a) downstream from the CAT sequence, between the SpeI and BamHI sites of plasmid pKSCAT (containing the CAT sequence subcloned between the HindIII and BamHI sites of pBlueScript KS+, downstream from the T3 promoter). Plasmid p5′CAT-A0 resulted from introduction of a fragment of plasmid pFC1 (treated with BamHI, Klenow enzyme, and BspEI), containing the leader region of the FGF-2 cDNA, into plasmid pKSCAT-pA (digested by HincII and BspEI) upstream from the CAT sequence. To construct p5′CAT-A1, a fragment containing the CAT sequence and the shortest FGF-2 3′ UTR (90 nt long) was obtained by PCR from the template pSCT-DOG (this plasmid had been constructed by insertion of the complete FGF-2 3′ UTR downstream from the CAT sequence into the vector pSCT-CAT (42a), using oligonucleotides CAT3′-S and UTR3′A1-AS (Table 1) as 5′ and 3′ primers, respectively. This PCR fragment (digested by BstEI) was introduced into the BspEI and SmaI sites of p5′CAT-A0, giving rise to plasmid p5′CAT-A1, which has the 5′ end of FGF-2 fused to CAT, with a 90-nt FGF-2 3′ UTR and a poly(A) site. To construct plasmid p5′CAT-A7, a DNA fragment from pSCT-DOG treated with XbaI, Klenow enzyme, and BspEI, and containing the CAT and FGF-2 5,823-nt-long 3′ UTR sequences, was introduced into the BspEI and SmaI sites of p5′CAT-A0.
Plasmids used for transient transfection, pF18CAT and pF24CAT, expressing chimeric CAT proteins with the N-terminal parts of the 18- and 24-kDa FGF-2 isoforms, respectively, have been described in a previous report (40), as has plasmid pSCT-CAT. Plasmid pSV-CAT was obtained by introducing into the vector pSCT HindIII and SacI sites a PCR fragment, obtained with oligonucleotides SVNLS-5′ and CAT-rev (Table 1), containing the nucleotide sequence encoding the simian virus 40 (SV40) large-T-antigen NLS preceded by an ATG initiation codon, in frame with the CAT open reading frame (ORF). Plasmid pF34CAT was obtained by amplifying a PCR fragment with the 5′ primer bigATG-S (corresponding to nt 78 to 103 of FGF-2 cDNA but introducing an ATG start codon and preceded by a 5′ XbaI site) and the 3′ primer FGF5′-AS (complementary to nt 293 to 323 including the XhoI site [Table 1]). This 245-nt-long fragment was introduced between the XbaI and XhoI sites of plasmid pFC1 (see above). Plasmid pF34ΔX was obtained by introducing the double-stranded oligonucleotide DELNLS-5′/DELNLS-3′ between PstI and XmaI sites (positions 192 and 256, respectively); this resulted in deletion of nt 212 to 234.
All plasmids used for stable transfection except the plasmid expressing the 34-kDa FGF-2 from an ATG codon (pF34EN) have been described previously (31, 37). Each of these plasmids expresses distinct (or no, in the case of pEN) FGF-2 isoforms from the first cistron and the neomycin resistance gene (neo) from the second cistron controlled by the encephalomyocarditis virus (EMCV) IRES. pF34EN was obtained by introduction of the PCR fragment described in the previous paragraph between XbaI and XhoI sites of plasmid pFEN, which contains the wild-type FGF-2 cDNA (nt 1 to 1179) upstream from EMCV and neo; in addition, the start codon ATG-485 was changed to TTA by insertion of a PCR fragment in order to prevent expression of the 18-kDa FGF-2 from plasmid pF34EN (it was not necessary to mutate the CTGs, as they were extinguished by the upstream ATG start codon [see Fig. 8A]).
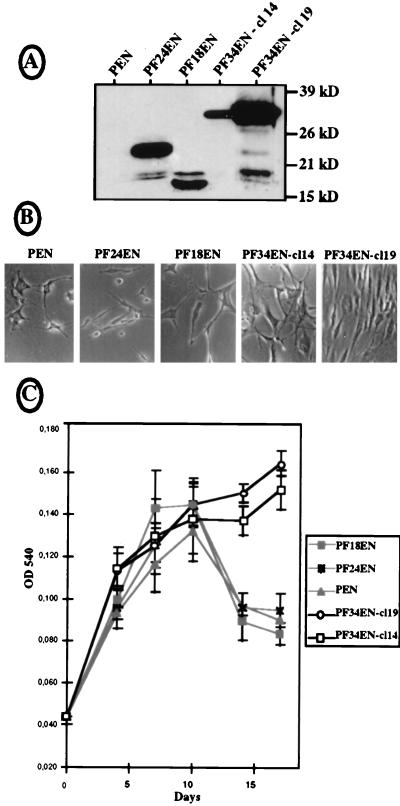
FIG. 8.
Expression of the different FGF-2 isoforms in permanently transfected NIH 3T3 cells. NIH 3T3 cells were transfected with bicistronic vectors expressing the 24-kDa (pF24EN), the 18-kDa (pF18EN), or the 34-kDa (pF34EN) FGF-2 or no protein (pEN) from their first cistrons and the neomycin resistance gene from their second cistrons; 10 to 12 G418-resistant clones were kept for each transfection, and analyses were performed with at least three clones of each origin. Two clones expressing two different levels of 34-kDa FGF-2 (cl14 and cl19), and one clone expressing a medium level of FGF-2, chosen for 24- and 18-kDa FGF-2 expression, are shown as representative clones. (A) Western immunoblotting was performed with cell extracts of the different clones, using anti-FGF-2 antibody as for Fig. 1. Positions of size standards are represented on the right. (B) Phase-contrast microscopy examination (magnification, ×100) of the different cell clones after 14 days of cultivation in six-well plates in 1% CS (see Materials and Methods). The panels correspond to representative parts of the culture dishes. (C) Proliferation curves obtained by the crystal violet method for cells seeded in 96-well plates and cultivated for 17 days (see Materials and Methods). In this procedure, cell growth is measured by determining the optical density at 540 nm (OD 540). Experiments were done in quadruplicate; they were also reproduced several times in triplicate with the classical cell counting procedure, which always gave similar results (not shown).
Anti-N23 antibody preparation.
The synthetic peptide NH2-VNPRSRAAGSPRTRGRRTEERPS-COOH, deduced from the nucleotide sequence of FGF-2 cDNA from positions 242 to 310 (41), was provided by Neosystem S.A. This peptide was coupled to keyhole limpet hemocyanin by using glutaraldehyde and carbodiimide, and the coupled product was used for rabbit immunization (20). Polyclonal antibodies were purified from rabbit serum by using an N23 peptide-coupled Affigel 10 column (Bio-Rad).
Cell transfection, fractionation, and immunocytolocalization.
COS-7 monkey cells were transfected with DNA (1 μg/ml) by the DEAE-dextran method (47). Cell lysates for Western blots were prepared 48 h later. For immunocytolocalization experiments, cells were seeded on lamella before transfection (Life Technologies); 48 h later, cells were washed four times with phosphate-buffered saline (PBS) and fixed for 7 min with 3% paraformaldehyde. Cells were washed again three times for 5 min each, once with PBS and twice with PBS plus 50 mM NH4Cl, permeabilized for 20 min at 37°C in PBS plus 0.025% saponin, and then incubated for 20 min at 37°C with nonimmune antibody (1/100 in PBS–0.025% saponin–0.5% bovine serum albumin). Cells were then incubated with anti-CAT antibody (1/500; Santa Cruz Biotechnology) for 1.5 h at 37°C, washed again three times as described above, incubated for 40 min at room temperature with fluorescein isothiocyanate-coupled anti-rabbit antibody (1/400; Tebu), and then washed again three times. Lamella were put on microscope slides in the presence of 8 μl of Citifluor (Pelco, Inc.).
SK-Hep-1 cells were transfected by using Lipofectin (Life Technologies). Cell fractionation to obtain cytosolic and nuclear extracts was performed as previously described (10, 37).
Stable NIH 3T3 clones were obtained by Lipofectin transfection with the different vectors of the pEN series (see above), and G418 at 1 mg/ml in Dulbecco modified Eagle medium (DMEM; Life Technologies) was added 72 h later. The cultures were maintained for 2 weeks in the presence of G418, and 10 to 12 clones from each transfection experiment were picked up and transferred into 24-well plates before cultivation in larger dishes.
Analysis of cell proliferation.
The proliferation curves were established with three stable clones from each transfection assay. Two procedures were used in parallel.
(i) Classical cell counting.
Six-well plates were seeded at 20,000 cells per well in DMEM plus 10% calf serum (CS). The next day, the complete medium was replaced by DMEM plus 1% CS. This medium was changed every 2 days. Cells were trypsinized every 3 days and counted in a Coulter counter (Coulter Electronics, Coultronics S.A., Andilly, France).
(ii) Crystal violet staining.
The staining procedure allows measurement of cell proliferation without the step of cell trypsination. Ninety-six-well plates were seeded with 750 cells/well in complete medium (DMEM plus 10% CS). The next day, the medium was replaced by DMEM plus 1% CS; it was then changed every 2 days. For the proliferation analysis, cells were fixed with 10% glutaraldehyde for 15 min under agitation, then abundantly washed extensively with double-distilled water, and dried for 1 h at room temperature. After addition of 100 μl of 0.1% crystal violet to each well, the plates were incubated for 30 min at room temperature under agitation. Cells were then washed with water three times and dried for 1 h at room temperature; 100 μl of 10% acetic acid (diluted in 200 mM formic acid [pH 6.7]) was added, and plates were shaken for 15 min. Optical density was read at 540 nm.
Western immunoblotting.
Total proteins were prepared, quantified, and analyzed by Western immunoblotting (5 μg of proteins from each cell lysate) as previously described (47). FGF-2 and FGF-CAT proteins were immunodetected with rabbit polyclonal anti-FGF-2 (Santa Cruz) and anti-CAT (homemade) antibodies, respectively. FGF-2 enrichment of an HeLa cell extract (from 107 cells) on heparin-Sepharose beads (see Fig. 2) was performed as previously described (13).
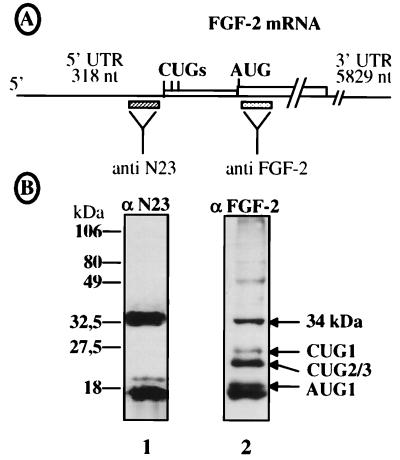
FIG. 2.
Identification of the 34-kDa FGF-2 with anti-N-terminal antibodies. (A) Schema representing the FGF-2 mRNA and the FGF-2 coding sequence, showing positions of the peptides used to prepare the anti-N23 and anti-FGF-2 polyclonal antibodies (see Materials and Methods). (B) Heparin-Sepharose-purified HeLa extract was analyzed by Western immunoblotting with either anti-N23 (αN23) or anti-FGF-2 (αFGF-2) antibodies (see Materials and Methods). Migration of size standards is indicated on the left; migration of FGF-2 isoforms is indicated on the right. The 16-kDa-migrating band revealed in lane 1 by anti-N23 probably corresponds to a cleavage product.
CAT activity analysis.
Cell pellets were resuspended in 100 mM Tris (pH 7.8)–2 mM MgCl2 and sonicated four times for 5 s; 35 μl of the cell extract was mixed with 5 μl of 1 M Tris (pH 7.8), 5 μl of butyryl coenzyme A at 5 mg/ml, and 5 μl of [14C]chloramphenicol and then incubated for 4 h at 37°C. Then 200 μl of 2,6,10,14-tetramethylpentadecane–xylene (2:1) was added, and the samples were vortexed for 30 s. Centrifugation was achieved for 3 min at 13,000 rpm, and 150 μl of the upper phase was counted in the presence of 3 ml of Ready Safe scintillating liquid in a scintillation counter (43).
In vitro transcription and translation.
DNAs were linearized and transcription was performed with T7 or T3 RNA polymerase, using the transcription kits provided by Ambion. RNA transcripts were quantitated by absorbance at 260 nm and ethidium bromide staining on agarose gels, and their integrity was verified. Translation was carried out in rabbit reticulocyte lysate (RRL) provided by Promega (47). The translation products were analyzed by electrophoresis on a 12.5% polyacrylamide gel (PAGE) followed by autoradiography and quantitation on a PhosphorImager (Molecular Dynamics).
RESULTS
Identification of a 34-kDa isoform of FGF-2 resulting from an upstream initiation of translation.
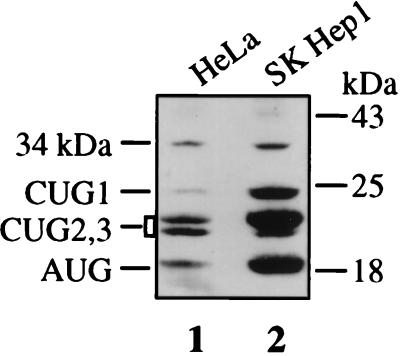
As we analyzed endogenous FGF-2 expression in different human cell types, it became apparent that a 34-kDa-migrating protein was recognized by anti-FGF-2 antibodies in various transformed cell lines (48). This protein was, for instance, clearly detectable by Western immunoblotting of HeLa and SK-Hep-1 cell extracts (Fig. 1). HeLa cell extracts were then run through a heparin-Sepharose column to see if this protein corresponded to FGF-2. As would be expected for an FGF-2 isoform, the 34-kDa protein was retained by heparin (Fig. 2B, lane 2).
FIG. 1.
Analysis of endogenous FGF-2 isoforms in HeLa and SK-Hep-1 cells. Aliquots (20 μg) of cell extracts from HeLa and SK-Hep-1 cells were analyzed by PAGE and transferred to nitrocellulose (see Materials and Methods). Immunoblotting was performed with anti-FGF-2 antibodies and chemiluminescence detection, immediately followed by autoradiography for 2 h. Positions of migration of size standards (right) and of the FGF-2 isoforms (left) are indicated.
The size of this potential new FGF-2 suggested that it could result from an additional initiation of translation at a codon located upstream from the known start codons. We tested this hypothesis by raising an antibody against a peptide (designated N23) deduced from the nucleic acid sequence of the FGF-2 cDNA upstream from the first CUG start codon (Fig. 2A). This antibody was used in a Western immunoblotting experiment with heparin-Sepharose-retained HeLa cell proteins. As shown in Fig. 2B (lane 1), the 34-kDa protein was recognized by the anti-N23 antibody, in contrast to the other FGF-2 isoforms. We also detected a 16-kDa-migrating band corresponding to a cleavage fragment of the 34-kDa protein (38).
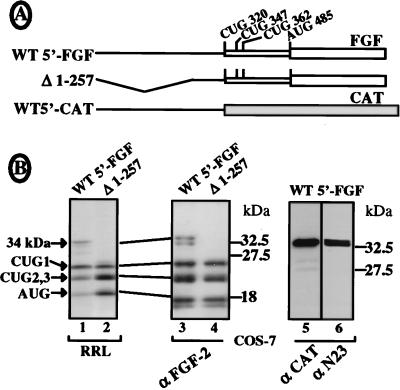
To check that the 5′ UTR of the FGF-2 cDNA was able to initiate translation of the 34-kDa protein, we then studied expression of the FGF-2 cDNA either with the complete leader or with a deletion of the 5′ 257 nt (Fig. 3A). FGF-2 isoform synthesis was analyzed either by in vitro translation in rabbit reticulocyte lysate (Fig. 3B, lanes 1 and 2) or by COS-7 cell transfection and Western immunoblotting with an anti-FGF-2 antibody (Fig. 3B, lanes 3 and 4). In both in vitro and in vivo experiments, an additional protein, sometimes migrating as a doublet, was detected at 34 kDa (Fig. 3B, lanes 1 and 3). This 34-kDa protein could not be detected with the 5′- deleted construct (Fig. 3B, lanes 2 and 4). Furthermore, an FGF-CAT chimeric construct containing the 5′ 312 nt of FGF-2 cDNA fused to the CAT coding sequence (Fig. 3A) was able to express a fusion protein, migrating at 33 kDa, which could be detected by both anti-CAT and anti-N23 antibodies (Fig. 3B, lanes 5 and 6).
FIG. 3.
Analysis of the ability of the FGF-2 mRNA 5′ UTR to promote translation initiation of the 34-kDa FGF-2. Expression of the 34-kDa FGF-2 was analyzed for three DNA constructs either by in vitro transcription and translation in RRL or by COS-7 cell transfection followed by Western immunoblotting (see Materials and Methods). (A) Schema of the mRNAs expressed from the different constructs (see Materials and Methods). WT5′-FGF corresponds to the FGF-2 mRNA with a complete 5′ leader; Δ1-257 corresponds to a deletion of nt 1 to 257 in the FGF-2 mRNA 5′ end; WT5′CAT corresponds to a fusion of FGF-2 cDNA nt 1 to 312 with the CAT sequence (devoid of an AUG start codon). (B) Analysis of mRNA expression either by in vitro translation (lanes 1 and 2) or by Western immunoblotting (lanes 3 to 6). Names of the antibodies (αFGF-2, αCAT, and αN23) are indicated at the bottom. Migration of size standards is indicated on the right; migration of FGF-2 isoforms is indicated on the left.
These results demonstrated the existence of a 34-kDa isoform of FGF-2 resulting from a process of alternative initiation of translation at a start codon located between nt 1 and 257 of FGF-2 mRNA.
The 34-kDa FGF-2 is initiated at a CUG codon located 86 nt from the mRNA 5′ end.
To identify the start codon that gave rise to the 34-kDa FGF-2, we looked for potential in-frame start codons in the published nucleotide sequence of the FGF-2 cDNA (41). The only potential initiation codon was an ACG at position 154 (Fig. 4A, line 1). However, examination of the published FGF-2 genomic DNA sequence (44) in this region revealed that a C was missing at position 159, setting the ACG-154 out of frame (Fig. 4A, line 2). Introduction of the genomic DNA fragment (kindly provided by R. Z. Florkiewicz) corresponding to nt 1 to 312 into our FGF cDNA construct revealed an expression profile identical to that obtained with the cDNA (not shown), suggesting an error in one of the published nucleotide sequences.
By sequencing this region of the FGF-2 cDNA with an automatic DNA sequencer (Applied Biosystems), we obtained the DNA sequence 153-GACGCGGT downstream from position 153, which still differed from the published sequences of cDNA and genomic DNA, i.e., 153-GACGGCT and 153-GACGGT, respectively (Fig. 4A, line 3). According to this new sequence, an HgaI restriction site (GACGC) had to be present at position 153 and two HgaI fragments of 34 and 51 nt should be generated following HgaI enzymatic digestion. As shown in Fig. 4B, such HgaI fragments were observed with both the cDNA and the genomic DNA (lanes 1 and 2) but not with a control DNA in which a point mutation removes the HgaI site (lane 3); this demonstrated that the new sequence shown in Fig. 4A, line 3, corresponded to the correct sequence effectively present in both the FGF-2 genomic DNA and cDNA. This sequence gave two possible in-frame start codons: a CTG codon at position 86 and an ACG codon at position 122 (Fig. 4C).
To determine which of the CUG-86 and ACG-122 codons was used as the start codon, point mutations of each or of both together were then introduced into a chimeric FGF-CAT construct in which the FGF-2 5′ 539 nt was fused to CAT (Fig. 5A). The effect of each mutation on translation initiation was analyzed either in vitro (in RRL [Fig. 5B]) or in vivo (in COS-7 cells [Fig. 5C]). Translation of the wild-type construct gave rise to a doublet at 42 and 40.5 kDa (consistent with the predicted size of the FGF-CAT protein derived from the 34-kDa FGF-2), the upper band being the most abundant in RRL (Fig. 5B and C, lanes 2). Whereas mutation of CUG-86 to AUG resulted in overexpression of the upper band (Fig. 5B and C, lanes 3), its mutation to CCC or the addition of a UAA codon downstream from it resulted in disappearance of the upper band and an increase of the lower band (lanes 4 and 5). In contrast, mutation of ACG-122 to GCG did not affect the upper band and resulted in strong decrease of the lower band (lane 6). The double mutation of CUG-86 and ACG-122 was, however, followed by the extinction of both bands (lane 7). These data clearly show that the FGF-2 34-kDa isoform was mostly initiated at CUG-86 (hereafter called CUG 0) in vitro and in vivo. The 40.5-kDa band of the doublet seemed to result in part from a secondary initiation at ACG-122 (lane 7), while the remaining 40.5-kDa protein expressed from the ACG-122 mutant could be attributed to the putative GUG start codon located just upstream from the ACG codon (Fig. 5A).
The 34-kDa isoform of FGF-2 is translated in a cap-dependent manner, independently of the IRES.
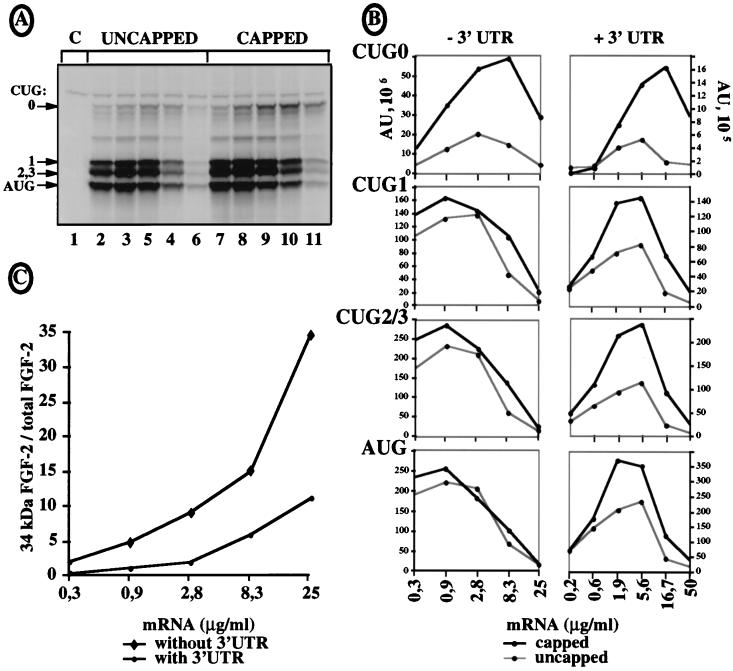
We have known for several years that the mechanism of synthesis of the FGF-2 isoforms is IRES driven (47). To find out whether the 34-kDa CUG 0-initiated isoform expression was cap dependent, in vitro translation experiments were performed with capped and uncapped mRNAs, using increasing concentrations of in vitro-synthesized polyadenylated FGF-2 mRNA with or without the 5,823-nt-long 3′ UTR (Fig. 6). When mRNAs devoid of a 3′ UTR or with a short (90-nt) 3′ UTR were used, expression of the isoforms initiated at CUG 1, 2, or 3 or AUG was, as expected, unaffected by the cap (Fig. 6A, lanes 2 to 11; Fig. 6B, left). In contrast, synthesis of the 34-kDa FGF-2 was clearly cap dependent (Fig. 6A and B, CUG 0): its expression at the optimal mRNA concentration increased three- to fourfold in the presence of the cap (Fig. 6B, CUG 0, left). Furthermore, this cap dependence was even enhanced in the presence of the 3′ UTR, with a fivefold increase of expression in the presence of the cap (Fig. 6B, CUG 0, right). Interestingly, the presence of the 3′ UTR also rendered the synthesis of the other isoforms partly cap dependent, with a twofold increase in the presence of the cap (Fig. 6B, right). mRNA stability was measured to check that the mRNAs were not degraded during translation both in the presence and in the absence of the 3′ UTR (not shown).
FIG. 6.
Cap dependence of 34-kDa FGF-2 expression. Capped and uncapped FGF-CAT mRNAs, with or without the FGF-2 mRNA 3′ UTR, were transcribed in vitro 2, and different dilutions of each mRNA were translated in RRL in the presence of [35S]methionine and analyzed as for Fig. 5. (A) Autoradiography after PAGE analysis of FGF-CAT mRNA expression. The mRNA was transcribed in vitro from plasmid p5′CAT-A1, in which the first 539 nt of FGF-2 cDNA were fused to the CAT sequence and the shortest FGF-2 3′ UTR (90 nt long). The results correspond to representative experiments that were repeated five times, either with a 90-nt-long 3′ UTR (p5′CAT-A1) or with no 3′ UTR (p5′CAT-A0). Migrations of the size standards and of FGF-CAT proteins are indicated (CUG 0 represents the largest isoform). C (lane 1) is the control without mRNA. FGF-CAT mRNA amounts used in each sample were 0.3, 0.9, 2.8, 8.3, and 25 μg/ml for lanes 2 to 6 (uncapped RNAs) and lanes 7 to 11 (capped RNAs), respectively. (B) FGF-CAT isoform expression using capped and uncapped mRNAs in which the 90-nt-long FGF-2 3′ UTR (−3′ UTR, corresponding to panel A) or the 5,823-nt-long FGF-2 3′ UTR (+3′ UTR) was quantified by PhosphorImager analysis (ImageQuant software). Expression of each isoform from capped or uncapped mRNA is shown in a separate plot. The data are from representative experiments that were repeated at least five times. (C) Ratio of 34-kDa FGF-2 isoform versus total FGF-2 expression in the presence or absence of a 3′ UTR, calculated for capped mRNAs from experiments shown in panel B (expressed as percentage of total FGF-2 expression).
The effect of the long FGF-2 3′ UTR on cap dependence of the different FGF-CAT isoforms suggested an effect on the ratio of the CUG 0-initiated isoform versus total FGF-CAT expression. This ratio was calculated for the capped RNA with or without the 3′ UTR (Fig. 6C). Interestingly, this ratio increased as a function of mRNA concentration. This increase was, however, much greater in the absence of the 3′ UTR, with the ratio of CUG 0 isoform versus total FGF-2 attaining 34.5% at the highest mRNA concentration. This ratio did not exceed 11% in the presence of the 3′ UTR, suggesting a negative regulation by the long FGF-2 mRNA 3′ UTR of the 34-kDa isoform in relation to the other FGF-2 isoforms.
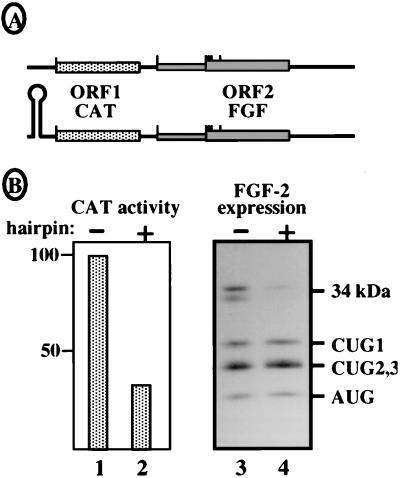
To confirm that the 34-kDa FGF-2 was exclusively expressed in a cap-dependent manner, we studied its expression in transfected COS-7 cells by using bicistronic vectors CAT-FGF, with or without a 5′ hairpin (Fig. 7A). These vectors enabled us to demonstrate the existence of the FGF-2 IRES in a previous report (47). As expected for IRES-driven translation, expression of the FGF-2 isoforms initiated at CUGs 1, 2, and 3 and AUG, analyzed by Western blotting, was not affected by the 5′ hairpin (Fig. 7B, lanes 3 and 4). In contrast, expression of the 34-kDa FGF-2 initiated at CUG 0 was detected in the absence of the hairpin but disappeared in its presence (lanes 3 and 4) following cap-dependent CAT expression (Fig. 7B, lanes 1 and 2). This finding indicated that the synthesis of the 34-kDa FGF-2 observed in the absence of a hairpin resulted from a cap-dependent reinitiation process, although the distance of 191 nt between the CAT stop codon and CUG-86 was longer than the optimal intercistronic length for reinitiation (28).
FIG. 7.
Expression of the 34-kDa FGF-2 from bicistronic vectors. Bicistronic vectors expressing CAT as first cistron (ORF1) and FGF-2 as second cistron (ORF2) were used to analyze the cap dependence of the 34-kDa FGF-2. (A) Representation of the two constructs with or without the presence of a 5′ hairpin. (B) Transfection of COS-7 cells with the bicistronic constructs. CAT expression was analyzed by measuring CAT activity (lanes 1 and 2). FGF-2 expression was analyzed by Western immunoblotting with anti-FGF-2 antibodies (lanes 3 and 4). The presence or absence of a hairpin is indicated by + or −, respectively. Migration of the different FGF-2 isoforms is shown on the right.
In conclusion, these results obtained for RRL and COS-7 transfected cells clearly showed that in contrast to the other isoforms, the new isoform of FGF-2 was exclusively synthesized according to a cap-dependent mechanism.
The 34-kDa FGF-2 permits NIH 3T3 cell survival in low-serum conditions.
To investigate the function of the 34-kDa FGF-2 and compare it to functions of the other FGF-2 isoforms, NIH 3T3 fibroblasts were permanently transfected with bicistronic vectors expressing separately the 24-kDa isoform, the 18-kDa isoform, or the 34-kDa isoform from the first cistron and the neomycin resistance gene from the second cistron (Fig. 8A, pF24EN, pF18EN, and pF34EN, respectively). As a control, NIH 3T3 cells were also transfected by the empty vector pEN. G418-resistant clones were derived from the different transfection experiments, and their FGF-2 production was checked by Western immunoblotting with anti-FGF-2 antibody (Fig. 8A).
The different clones were plated at low cell density and grown in the presence of 1% CS to analyze their ability to proliferate independently of serum. The cells were counted (by the crystal violet method; see Materials and Methods) during 17 days of growth, and their phenotypes were compared (Fig. 8B and C). Results showed that cells expressing either the empty vector pEN, the 24-kDa FGF-2, or the 18-kDa isoform were able to proliferate only during the first 9 days; thereafter all of them started to die, as shown by the drastic decrease in cell number in the dishes (Fig. 8C, pEN, pF24EN, and pF18EN). In contrast, the two clones expressing the 34-kDa FGF-2 did not stop growing and did not exhibit any cell death on day 17 (Fig. 8C, pF34EN-cl14 and -cl19). The proliferation rate of clone 19 was better than that of clone 14, in correlation with the different levels of 34-kDa FGF-2 production. The phenotype analysis performed also showed that the two cell clones expressing the 34-kDa FGF-2 were still growing in the end of the proliferation experiment and exhibited a fibroblast aspect, whereas the other three were sparse and had a very poor aspect (Fig. 8B).
These results led us to conclude that endogenous expression of 34 kDa FGF-2 permitted NIH 3T3 cell survival in conditions where expression of the other FGF-2 isoforms was unable to prevent cell death.
The 34-kDa FGF-2 contains a Rev-like NLS.
The survival role of the 34-kDa FGF-2 distinguished it from the other isoforms and thus raised the question of its mode of action. Our first attempt was to determine the intracellular localization of this new FGF-2.
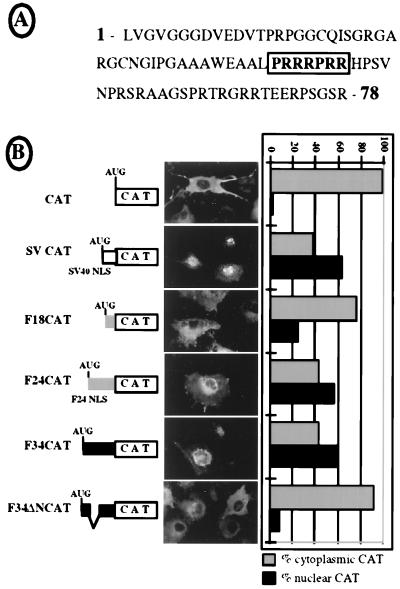
The 78-amino-acid-long sequence of the 34-kDa FGF-2 N-terminal region was examined and revealed an arginine-rich box, PRRRPRR, reminiscent of the NLS of the human immunodeficiency virus type 1 (HIV-1) Rev protein (Fig. 9A and reference 21). This N-terminal region, either wild type or with the putative NLS deleted, was fused to CAT protein (Fig. 9B, F34CAT or F34ΔNCAT). Plasmids expressing these two CAT fusions, as well as plasmids expressing either CAT alone or CAT fused to the SV40 NLS or to the 24-kDa or the 18-kDa FGF-2 N-terminal part (SV-, F24-, or F18CAT, respectively), were used for cell transient transfection (Fig. 9B). In the first experiment, COS-7 cell transfection was followed by CAT immunodetection in situ, using anti-CAT antibody (Fig. 9B, fluorescent staining). In a second experiment, SK-Hep-1 cell transfection by the different constructs was followed by cell fractionation and measurement of CAT activity in the cytoplasmic and nuclear fractions (Fig. 9B, histograms). As expected, the CAT and F18CAT controls were cytosolic with both procedures, while the SVCAT and F24CAT constructs were nuclear (Fig. 9B and references 10 and 24). The results obtained with the F34CAT fusion clearly indicated a nuclear localization, whereas F34ΔN-CAT was strictly cytosolic (Fig. 9B). These data show that the N-terminal portion of the 34-kDa FGF-2 contains an arginine-rich NLS involving the PRRRPRR box and suggest that the survival role of this protein is probably mediated by nuclear targets.
FIG. 9.
Characterization of an NLS in the N-terminal domain of the 34-kDa FGF-2. (A) Amino acid sequence of the NH2-terminal domain of the 34-kDa FGF-2 (78 amino acid residues). The boldface boxed characters represent the amino acids that were deleted in construct F34ΔNCAT. (B) Subcellular distribution of different chimeric CAT proteins was analyzed either by immunocytolocalization in transfected COS-7 cells (fluorescent staining micrographs in the middle) or by fractionation of transfected SK-Hep-1 cells into nuclear and cytosolic extracts (histograms on the right), as described in Materials and Methods. SVCAT fusion contains the minimal SV40 large-T antigen NLS (PKKKRKV [24]). F18- and F24CAT chimeras have N-terminal parts corresponding to the 20 and 75 N-terminal amino acid residues of 18- and 24-kDa FGF-2, respectively. The latter includes the 24-kDa FGF-2 NLS described previously (1). The F34CAT chimera contains the 78-amino-acid N-terminal domain of the 34-kDa FGF-2 shown in panel A. F34ΔNCAT has a deletion of the arginine-rich box shown in panel A, corresponding to residues 43 to 49. Results from immunocytolocalization and cell fractionation are presented at the right for each chimeric protein and the control (CAT). The subcellular distribution between nucleus and cytosol is represented as a percentage of the total CAT activity measured.
DISCUSSION
This work has revealed the existence of a new isoform of FGF-2 whose synthesis is initiated at a fifth alternative initiation codon, a CUG at position 86 downstream from the mRNA 5′ end. Expression of the 34-kDa FGF-2 was shown to be exclusively cap dependent, in contrast to the IRES-dependent expression of the other FGF-2 isoforms of 18, 22, 22.5, and 24 kDa. Furthermore, the long 3′ UTR of the FGF-2 mRNA seemed to influence the ratio of the CUG 0-initiated isoform versus total FGF-CAT expression in vitro. We have also demonstrated that the 34-kDa FGF-2 isoform, nuclearized by a novel arginine-rich NLS, has a specific survival function.
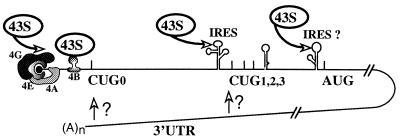
This makes the FGF-2 mRNA the most complex system of translational regulation described up to now. As schematized in Fig. 10, FGF-2 mRNA can be translated by two alternative mechanisms, depending on the activity of the cap binding protein eIF-4E. When translation occurs according to a cap-dependent mechanism, expression of the 34-kDa FGF-2 is favored. In contrast, the other isoforms are produced according to an IRES-driven mechanism, although their synthesis may also occur according to the cap-dependent mechanism in certain conditions (15a). The balance between these two mechanisms controls the use of the different initiation codons. This balance seems to be influenced, at least in vitro, by the long 3′ UTR of the FGF-2 mRNA (Fig. 6). The activities of the different regulatory cis elements are presumably controlled by trans-acting factors bound to the FGF-2 mRNA, whose identification is currently under investigation (48).
FIG. 10.
Model of translational regulation of FGF-2 expression. The FGF-2 mRNA can be translated by two alternative mechanisms (see the text): either a cap-dependent mechanism dependent on eIF-4E activity or an IRES-dependent mechanism that is independent of eIF-4E activity. The initiation factors 4A and 4G, forming with eIF-4E the complex eIF-4F, are represented. The 43S corresponds to the 40S ribosome subunit associated with eIF2 and with the initiator tRNA. The two putative IRESes and the 3′ UTR are schematized. The start codons of the FGF-2 isoforms are indicated.
From a mechanistic point of view, one could be surprised by the strong polar effect generated by introduction of an AUG in place of CUG 0, which completely extinguished expression of those isoforms initiated at CUGs 1, 2, and 3 (positions 320, 347, and 362) in vitro and in vivo (Fig. 5B and C, lanes 3). This interference of the AUG at position 86 with the downstream IRES can be explained by IRES masking and/or destruction due to the presence of the elongation machinery. Such a polar effect is not, however, observed with an AUG at position 156, which is in a different surrounding context and at a longer distance from the mRNA 5′ end (47). This IRES extinction observed in the case of an upstream efficient cap-dependent initiation favors the idea that the cap-dependent and IRES-driven mechanisms are exclusive and do not occur simultaneously on the same RNA molecule.
The extinction of downstream-initiated isoform synthesis resulting from the presence of an AUG codon in place of CUG 0 (Fig. 5) confers considerable significance on the presence of noncanonical codons as initiators in alternative translation initiation systems: if an AUG were naturally present at position 86, the 34-kDa FGF-2 would be predominant. This could be harmful to the function of FGF-2 in the cell as overexpression of the 34-kDa isoform results in maintenance of cell proliferation (Fig. 8). Consistent with this inference, we have detected it only in transformed cell lines (Fig. 1 and reference 48). The presence of a weakly efficient CUG codon to initiate 34-kDa FGF-2 synthesis is a clever way of maintaining a correct ratio of the 34-kDa protein in relation to the other isoforms. The crucial role of non-AUG codons in alternative initiation systems is not limited to FGF-2. Examples in the literature include the messengers of murine leukemia virus and of the proto-oncogene c-myc, both of which have an alternative CUG initiation codon (19, 39). In the first case, a cap-dependent CUG codon initiates the synthesis of a cell surface antigen important for viral dissemination within the animal, whereas an IRES-driven AUG codon controls synthesis of the viral capsid proteins; this allows the maintenance of a suitable ratio between the two synthesized proteins (4, 12, 49). As regards c-myc mRNA, the CUG- and AUG-initiated proteins c-Myc1 and c-Myc2, whose synthesis is also controlled by an IRES, have distinct targets as transcription factors and must also be present in a suitable ratio (18, 36).
This report suggests for the first time a role for the huge 3′ UTR of the FGF-2 mRNA (Fig. 6). This 5,823-nt-long 3′ UTR in fact corresponds to the longest of several species of the FGF-2 mRNA resulting from the use of alternative polyadenylation sites (3, 41, 51). The length of the 3′ UTR is in fact regulated according to cell density (7, 46a); this could result in a modulation of the ratio of 34-kDa FGF-2 to the other FGF-2 isoforms.
We have demonstrated here that the 34-kDa human FGF-2 does effectively exist in cells (Fig. 2). This protein is detected in various transformed cell types but not in primary cells (Fig. 1 and reference 48). Such an observation is in good agreement with previous reports that showed an up-regulation of eIF-4E in cancer cells (26). As the 34-kDa FGF-2 synthesis is cap dependent, it could be expected to follow the level of eIF-4E activity. This was supported in a study of translational enhancement of rat FGF-2 by eIF-4 factors which showed that translation of a rat 31-kDa FGF-2 isoform was induced when the eIF-4E level increased (27). Although the existence of the rat FGF-2 isoform has not been confirmed by specific antibodies, it could correspond to human 34-kDa FGF-2. In that same study, the 22- and 21-kDa rat isoforms were not enhanced by eIF-4E, suggesting that they corresponded to the CUG 1-, 2-, or 3-initiated isoforms of human FGF-2 whose synthesis is IRES driven. In conclusion, the complex mechanism of FGF-2 mRNA translation, with alternative initiations of translation regulated by the balance between cap-dependent and IRES-dependent initiation, seems to be evolutionarily conserved.
The important function of the 34-kDa FGF-2 evidenced in the present study could explain its conservation in evolution. Clearly, this FGF-2 isoform is involved in cell growth maintenance, by preventing cell death in low-serum conditions. Previous reports have shown that constitutive expression of the 24- or the 18-kDa FGF-2 leads to cell immortalization and/or transformation (13, 42). However in our experiments, where cells were seeded at low density, neither the 18-kDa nor the 24-kDa FGF-2 could prevent cell death. This survival effect of endogenous FGF-2 seems to be a specific function of the 34-kDa isoform.
As regards its mode of action, the presence of an NLS in the N-terminal part of the 34-kDa FGF-2 suggests that this protein has nuclear targets and that its effect may be intracrine, like that of the nuclear 24-kDa isoform. An Arg-rich NLS is also a novel feature for a cellular NLS-containing protein. This NLS resembles the NLS recently described for HIV-1 Rev (RQARRNRRRRWR [21]). Its Arg-rich NLS allows Rev to enter the nucleus through a nonclassical pathway involving its direct interaction via its NLS with the human nuclear import receptor, importin β (other NLS-containing proteins bind to importin α receptor, which itself interacts with importin β via an Arg-rich domain [17, 21]). Such an NLS is responsible in the case of HIV-1 Rev for regulation of importation, as the NLS overlaps with the RNA binding domain of the protein and is then masked when Rev binds RNA.
The existence of two different (and atypical) NLSs in the 34-kDa FGF-2 versus the 21-, 22-, and 24-kDa (HMW) FGF-2 isoforms offers the possibility of a differential nuclearization regulation. Furthermore, their different N-terminal domains may allow a differential targeting of FGF-2 isoforms within the nucleus. On the one hand, the HMW FGF-2 contains a glycine-arginine-rich domain (RGG), which might be involved in protein-protein interaction as shown for the RGG domain of nucleolin (8). On the other hand, the 34-kDa FGF-2 contains an Arg-rich N-terminal domain (Fig. 9A) that renders it more basic than the other isoforms, with a pI greater than 12. This suggests that the N-terminal domain of the 34-kDa FGF-2, in addition to its NLS function, could mediate direct interactions of the new FGF-2 with nucleic acids.
ACKNOWLEDGMENTS
Emmanuelle Arnaud and Christian Touriol contributed equally to this work.
We thank F. Bayard, B. Bugler, and B. Galy for helpful discussions, D. Villa for pictures, D. Warwick for English proofreading, and C. Zanibellato for technical assistance. We thank R. Z. Florkiewicz for the gift of the genomic DNA FGF-2 sequence. We also thank E. Bieth for the poly(A)-containing plasmid and A. Ramackers for plasmid pSCT-DOG.
This work was supported by grants from the Association pour la Recherche sur le Cancer, the Agence Nationale de Recherches sur le SIDA, the Ligue Nationale contre le Cancer, the Conseil Régional Midi-Pyrénées, and the European Community Biotechnology Program (subprogram Cell Factory, Actions de Recherches Concertées, contract 94/99-181). E. Arnaud received successive fellowships from the Association pour la Recherche sur le Cancer and from the Ligue Régionale contre le Cancer. C. Touriol received successive fellowships from the Ministère de l’Education Nationale et de la Recherche and from the Ligue Nationale contre le Cancer.
REFERENCES
- 1.Amalric F, Baldin V, Bosc-Bierne I, Bouche G, Bugler B, Couderc B, Prats H, Roman A M. Nuclear translocation of basic fibroblast growth factor. New York, N.Y: Academic Press; 1991. [DOI] [PubMed] [Google Scholar]
- 2.Baldin V, Roman A M, Bosc-Bierne I, Amalric F, Bouche G. Translocation of bFGF to the nucleus is G1 phase cell cycle specific in bovine aortic endothelial cells. EMBO J. 1990;9:1511–1517. doi: 10.1002/j.1460-2075.1990.tb08269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensaid M, Malecaze F, Prats H, Bayard F, Tauber J P. Autocrine regulation of bovine retinal capillary endothelial cells (BREC) proliferation by BREC derived fibroblast growth factor. Exp Eye Res. 1989;45:801–813. doi: 10.1016/0014-4835(89)90065-1. [DOI] [PubMed] [Google Scholar]
- 4.Berlioz C, Darlix J L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus Gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosome entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 5a.Bieth, E. Unpublished results.
- 6.Bikfalvi A, Klein S, Pintucci G, Quarto N, Mignatti P, Rifkin D B. Differential modulation of cell phenotype by different molecular weight forms of basic fibroblast growth factor: possible intracellular signaling by the high molecular weight forms. J Cell Biol. 1995;129:233–243. doi: 10.1083/jcb.129.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bost L M, Hjelmeland L M. Cell density regulates differential production of bFGF transcripts. Growth Factors. 1993;9:195–203. doi: 10.3109/08977199309010832. [DOI] [PubMed] [Google Scholar]
- 8.Bouvet P, Diaz J J, Kindbeiter K, Madjar J J, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 9.Brigstock D R, Sasse J, Klagsbrun M. Subcellular distribution of basic fibroblast growth factor in human hepatoma cells. Growth Factors. 1991;4:189–196. doi: 10.3109/08977199109104815. [DOI] [PubMed] [Google Scholar]
- 10.Bugler B, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin J D, Florkiewicz R Z, Neumann J, Mort-Hopkins T, Dorn II G W, Lightfoot P, German R, Howles P N, Kier A, O’Toole B A, et al. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6:1861–1973. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbin A, Prats A C, Darlix J L, Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol. 1994;68:3857–3867. doi: 10.1128/jvi.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couderc B, Prats H, Bayard F, Amalric F. Potential oncogenic effects of basic fibroblast growth factor requires cooperation between CUG and AUG-initiated forms. Cell Regul. 1991;2:709–718. doi: 10.1091/mbc.2.9.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis M G, Zhou M, Ali S, Coffin J D, Doetschman T, Dorn G W., II Intracrine and autocrine effects of basic fibroblast growth factor in vascular smooth muscle cells. J Mol Cell Cardiol. 1997;29:1061–1072. doi: 10.1006/jmcc.1997.0383. [DOI] [PubMed] [Google Scholar]
- 15.Florkiewicz R Z, Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Galy, B., et al. Submitted for publication.
- 16.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′ untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 17.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hann S R, Dixit M, Sears R C, Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 19.Hann S R, King M W, Bentley D L, Anderson C W, Eisenman R N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the rev nuclear localisation signal mediates specific binding to human importin β. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 22.Hoshikawa M, Ohbayashi N, Yonamine A, Konishi M, Ozaki K, Fukui S, Itoh N. Structure and expression of a novel fibroblast growth factor, FGF-17, preferentially expressed in the embryonic brain. Biochem Biophys Res Commun. 1998;244:187–191. doi: 10.1006/bbrc.1998.8239. [DOI] [PubMed] [Google Scholar]
- 23.Jackson R J. Initiation without an end. Nature (London) 1991;353:14–15. doi: 10.1038/353014a0. [DOI] [PubMed] [Google Scholar]
- 24.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 25.Kandel J, Bossy-Wetzel E, Radvanyi F, Klagsbrun M, Folkman J, Hanahan D. Neovascularisation is associated with a switch to the export of bFGF in the multistep development of fibrosarcoma. Cell. 1991;66:1095–1104. doi: 10.1016/0092-8674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- 26.Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, Benedetti A D. The proto-oncogene/translation factor eIF-4E: a survey of its expression in breast carcinomas. Int J Cancer. 1995;64:27–31. doi: 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- 27.Kevil C, Carter P, Hu B, DeBenedetti A. Translational enhancement of FGF-2 by eIF-4 factors, and alternate utilization of CUG and AUG codons for translation initiation. Oncogene. 1995;11:2239–2348. [PubMed] [Google Scholar]
- 28.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macejak D J, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature (London) 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 31.Maret A, Galy B, Arnaud E, Bayard F, Prats H. Inhibition of fibroblast growth factor 2 expression by antisense RNA induced a loss of the transformed phenotype in a human hepatoma cell line. Cancer Res. 1995;55:5075–5079. [PubMed] [Google Scholar]
- 32.Mason I J. The ins and outs of fibroblast growth factors. Cell. 1994;78:547–552. doi: 10.1016/0092-8674(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 33.Mignatti P, Morimoto T, Rifkin D B. Basic fibroblast growth factor (bFGF) released by single isolated cells stimulates their migration in an autocrine manner. Proc Natl Acad Sci USA. 1991;88:11007–11011. doi: 10.1073/pnas.88.24.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mignatti P, Morimoto T, Rifkin D B. Basic fibroblast growth factor, a protein devoid of secretory signal sequence, is released by cells via a pathway independent of the endoplasmic reticulum-Golgi complex. J Cell Physiol. 1992;151:81–93. doi: 10.1002/jcp.1041510113. [DOI] [PubMed] [Google Scholar]
- 35.Miyake A, Konishi M, Martin F, Hernday N, Ozaki K, Yamamoto S, Mikami T, Arakawa T, Itoh N. Structure and expression of a novel member, FGF-16, of the fibroblast growth factor family. Biochem Biophys Res Commun. 1998;243:148–152. doi: 10.1006/bbrc.1998.8073. [DOI] [PubMed] [Google Scholar]
- 36.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site (IRES) J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 37.Patry V, Arnaud E, Amalric F, Prats H. Involvement of basic fibroblast growth factor NH2 terminus in nuclear accumulation. Growth Factors. 1994;11:163–174. doi: 10.3109/08977199409046914. [DOI] [PubMed] [Google Scholar]
- 38.Patry V, Bugler B, Amalric F, Promé J C, Prats H. Purification and characterization of the 210-amino acid recombinant basic fibroblast growth factor form (FGF-2) FEBS Lett. 1994;349:23–28. doi: 10.1016/0014-5793(94)00633-4. [DOI] [PubMed] [Google Scholar]
- 39.Prats A C, De Billy G, Wang P, Darlix J L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 40.Prats A C, Vagner S, Prats H, Amalric F. cis-acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor. Mol Cell Biol. 1992;12:4796–4805. doi: 10.1128/mcb.12.10.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prats H, Kaghad M, Prats A C, Klagsbrun M, Lélias J M, Liauzun P, Chalon P, Tauber J P, Amalric F, Smith J A, Caput D. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quarto N, Talarico D, Florkiewicz R, Rifkin D B. Selective expression of high molecular weight basic fibroblast growth factor confers a unique phenotype to NIH 3T3 cells. Cell Regul. 1991;2:699–708. doi: 10.1091/mbc.2.9.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Ramackers, A. Unpublished results.
- 43.Seed B, Sheen J. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 44.Shibata F, Baird A, Florkiewicz R Z. Functional characterization of the human basic fibroblast growth factor gene promoter. Growth Factors. 1991;4:277–287. doi: 10.3109/08977199109043913. [DOI] [PubMed] [Google Scholar]
- 45.Smallwood P M, Munoz-Sanjuan I, Tong P, Macke J P, Hendry S H C, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family in nervous system development. Proc Natl Acad Sci USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teerink H, Voorma H O, Thomas A A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 46a.Touriol, C. Unpublished results.
- 47.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagner S, Touriol C, Galy B, Audigier S, Gensac M C, Amalric F, Bayard F, Prats H, Prats A C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 50.Wagner J A. The fibroblasts growth factors: an emerging family of neural growth factors. Curr Top Microbiol Immunol. 1991;165:95–118. doi: 10.1007/978-3-642-75747-1_6. [DOI] [PubMed] [Google Scholar]
- 51.Weich H A, Iberg N, Klagsbrun M, Folkman J. Expression of acidic and basic fibroblast growth factors in human and bovine vascular smooth muscle cells. Growth Factors. 1990;2:313–320. doi: 10.3109/08977199009167026. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi T P, Rossant J. Fibroblast growth factors in mammalian development. Curr Opin Genet Dev. 1995;5:485–491. doi: 10.1016/0959-437x(95)90053-j. [DOI] [PubMed] [Google Scholar]
- 53.Yanagisawa-Miwa A, Uchida Y, Nakamura F, Tomaru T, Kido H, Kamijo T, Sugimoto T, Kaji K, Utsuyama M, Kurashima C, Ito H. Salvage of infarcted myocardium by angiogenic action of basic fibroblast growth factor. Science. 1992;257:1401–1403. doi: 10.1126/science.1382313. [DOI] [PubMed] [Google Scholar]
- 54.Yayon A, Klagsbrun M, Esko J D, Leder P, Ornitz D M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]