Abstract
Formins generate unbranched actin filaments by a conserved, processive actin assembly mechanism. Most organisms express multiple formin isoforms that mediate distinct cellular processes and facilitate actin filament polymerization by significantly different rates, but how these actin assembly differences correlate to cellular activity is unclear. We used a computational model of fission yeast cytokinetic ring assembly to test the hypothesis that particular actin assembly properties help tailor formins for specific cellular roles. Simulations run in different actin filament nucleation and elongation conditions revealed that variations in formin’s nucleation efficiency critically impact both the probability and timing of contractile ring formation. To probe the physiological importance of nucleation efficiency, we engineered fission yeast formin chimera strains in which the FH1-FH2 actin assembly domains of full-length cytokinesis formin Cdc12 were replaced with the FH1-FH2 domains from functionally and evolutionarily diverse formins with significantly different actin assembly properties. Although Cdc12 chimeras generally support life in fission yeast, quantitative live-cell imaging revealed a range of cytokinesis defects from mild to severe. In agreement with the computational model, chimeras whose nucleation efficiencies are least similar to Cdc12 exhibit more severe cytokinesis defects, specifically in the rate of contractile ring assembly. Together, our computational and experimental results suggest that fission yeast cytokinesis is ideally mediated by a formin with properly tailored actin assembly parameters.
Significance
Cytokinesis, the physical separation of a mother cell into daughter cells, is a critical last step of the cell division cycle. Defects can negatively impact development or lead to the proliferation of cancer. The actin assembly factor formin facilitates cytokinetic contractile ring formation in eukaryotic cells by determining both how often and how fast actin filaments assemble. We investigated the importance of formin’s specific actin assembly properties on cytokinesis in fission yeast, a model for cell division. By altering formin properties using a computational model and by engineering mutant fission yeast strains, we conclude that efficient actin filament nucleation is the most critical factor for proper contractile ring formation.
Introduction
Formins (formin homology proteins) are a family of large, highly conserved proteins that nucleate actin filaments (F-actin) and remain processively associated with their barbed ends to significantly increase the elongation rate for a variety of fundamental cellular processes, including polarization, motility, division, and adhesion (1,2). Formins are characterized by two highly conserved formin homology (FH) domains that nucleate and elongate unbranched F-actin. The FH2 domain is a tethered, head-to-tail dimer that nucleates and then processively encircles the growing F-actin barbed end (3), whereas the unstructured FH1 domain binds profilin-actin via its multiple proline-rich regions and delivers it to the FH2-bound barbed end to facilitate rapid elongation (2,4, 5, 6, 7). The conserved FH1-FH2 domains are flanked by less-well-conserved regulatory domains that facilitate the activation of many formins at the correct time and place (1). Additionally, some formin isoforms carry out noncanonical functions, such as severing F-actin to promote depolymerization, generating F-actin bundles, and binding directly to microtubules to coordinate cross talk between the actin and microtubule cytoskeletons (2).
Most organisms express multiple formin isoforms, from two in budding yeast, three in fission yeast, seven in nematode worms, ∼15 in mammals, to more than 20 in plants (8, 9, 10). Despite having well-conserved structural folds and general actin assembly mechanisms, distinct formin isoforms are required for different cellular processes in many cell types. For example, each of the three fission yeast formins assembles actin filaments for a specific F-actin network (11): For3, polarizing actin cables (12,13); Cdc12, the contractile ring of dividing cells (14); Fus1, the fusion focus during mating (15,16). Although regulation via activation at the right time and place has been established to be critical for the functional specificity of formin isoforms (1), it is also possible that a formin’s particular actin assembly properties are also important (17). In vitro measurements reveal that many formins have significantly different actin assembly properties, such as F-actin nucleation efficiency, barbed-end elongation rate, and barbed-end dissociation rate (18,19). Consistent with this possibility, we previously determined that the actin assembly properties of the three fission yeast formins (For3, Cdc12, and Fus1) vary widely (Table 1; (20)), suggesting that a formin’s specific properties might also be tailored for its cellular role. However, the extent to which each property contributes to the assembly of particular F-actin networks in vivo is less clear.
Table 1.
In vitro actin assembly properties of formins used to generate formin chimeras
| Formin | Nucleation efficiency | Elongation rate (subunits s−1μM−1) | Dissociation rate (s−1) |
|---|---|---|---|
| fission yeast Cdc12 | one filament per two to three dimers (20,21) | 10–12 (19, 20, 21) | 4.7–7.0 × 10−5 (20,21) |
| fission yeast For3 | one filament per 170 dimers (20) | 10 (20) | 3.6 × 10−5 (20) |
| budding yeast Bni1 | one filament per ~20 dimers (4,22) | 20–25 (19,22) | 8 × 10−4 (4) |
| mouse Dia2 | one filament per ~4–6a dimers | 12 (19) | 1.3 × 10−4 (19) |
| worm CYK-1 | one filament per 25 dimers (21) | ~60 (21,23) | 3.9 × 10−3 (21) |
| fission yeast Fus1 | one filament per two dimers (20) | 5 (20) | 6.5 × 10−4 (20) |
Formin nucleation efficiency (filaments per dimer), barbed-end elongation rate (in the presence of profilin), and dissociation rate from the barbed end were determined previously with in vitro biochemical assays.
Data is as shown in Fig. S4C.
Cytokinesis, the physical separation of a mother cell into two daughter cells, is the final step of the cell cycle during which cells assemble a contractile ring of bundled, antiparallel actin filaments, and type II myosin (Myo2) motors. The molecular mechanism of cytokinesis and the role of formin Cdc12 are well established in fission yeast, providing an ideal system to investigate whether and how a formin’s particular actin assembly characteristics are suited for its cellular role (24). Fission yeast contractile ring assembly is accomplished via a primary mechanism known as search, capture, pull, and release (SCPR). In SCPR, several conserved cytokinesis proteins, including formin Cdc12 and Myo2, are associated with ∼140 prering cytokinesis nodes that are cortically distributed around the cell middle (Fig. 1 A; (25, 26, 27, 28)). Filaments nucleated and assembled by Cdc12, which is not thought to be canonically autoinhibited (29), are subsequently captured by Myo2 on an adjacent node (search and capture). Myo2 then exerts a pulling force on the Cdc12-assembled actin filament (pull), bringing the two nodes closer together. These node-node connections are lost via severing from cofilin (release), and the search phase starts again (Fig. 1 A). After multiple rounds of SCPR, actin filaments coalesce into a contractile ring (27,28). The SCPR model provides a solid framework for examining the importance of formin Cdc12’s specific actin assembly properties in the assembly of a contractile ring.
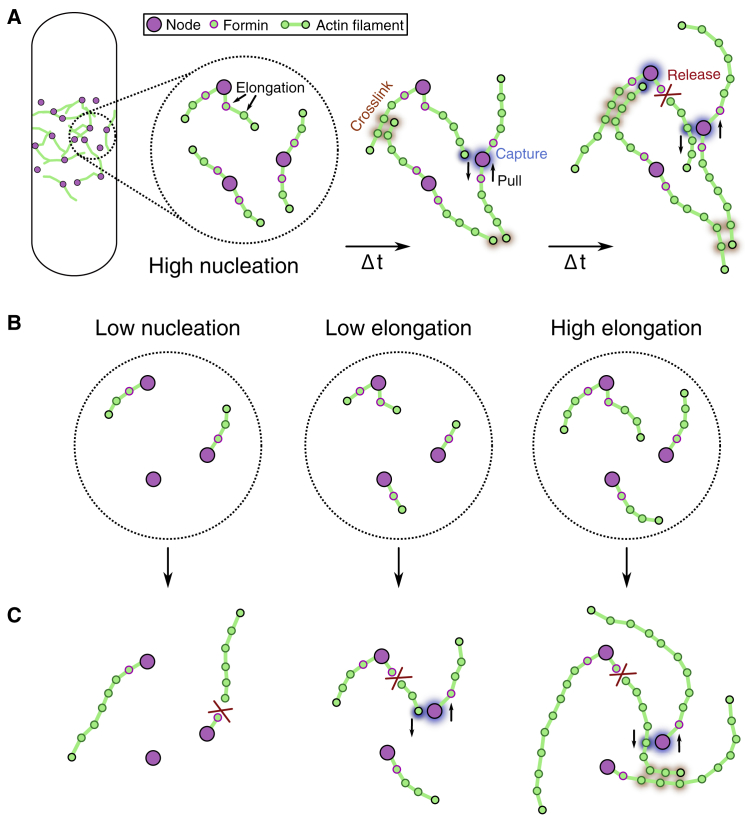
Figure 1.
Overview of 3D SCPR computational model. (A) Cartoon showing three time points of the SCPR mechanism used in the model. Under high nucleation wild-type conditions, approximately two actin filaments (string of green beads) elongate from formin beads connected to prering cytokinesis nodes (purple). The pointed end bead is highlighted in black. These elongating filaments may be captured by myosin on adjacent nodes, which exerts a pulling force that brings two nodes closer together. When actin filaments are severed, the connection between two nodes is broken. See Materials and methods for a detailed summary of the model. (B) We varied the rates of formin-mediated nucleation and elongation of actin filaments. Low nucleation, low elongation, and high elongation conditions are illustrated at early times in the simulation. (C) Illustrations of the dominant outcomes of non-wild-type formin-mediated nucleation and elongation properties. Low nucleation (left) results in few actin filaments that fail to be connect nodes. Low elongation (middle) produces short filaments that fail to form a continuous ring. High elongation (right) results in ectopic F-actin that fails to organize nodes favorably for ring formation. Simulations at various conditions can be seen animated in Video S2.
Here, we first used a computational three-dimensional (3D) SCPR model to characterize how formin-mediated actin filament nucleation efficiency and elongation rate affect cytokinetic ring formation. Our simulations predicted that both properties are important, but changes in formin nucleation efficiency have the largest impact on the timing and probability of contractile ring formation. We then tested the physiological importance of formin’s actin filament nucleation efficiency and elongation rate using in vivo experiments. We developed formin chimera strains that express engineered formin chimeras in which the FH1-FH2 domains of the cytokinesis formin Cdc12 were replaced with the FH1-FH2 domains of functionally, evolutionarily, and biochemically diverse formins from different organisms. Quantitative imaging and analysis of the formin chimera cells reveal that formin-mediated cytokinesis in fission yeast is robust, but supports the modeling predictions that Cdc12’s nucleation efficiency has the largest impact on contractile ring assembly rate.
Materials and methods
SCPR model of ring assembly
To test the individual and combined effects of formin’s actin assembly properties on fission yeast cytokinetic ring assembly, including actin filament nucleation and elongation rate, we extended a 3D model based on the SCPR mechanism (30). The model implements a Brownian dynamics algorithm within a domain that mimics shape, dimensions, and other physical properties of a fission yeast cell (Fig. 1). By integrating the Langevin equation of motion over time (31,32), the model reproduces the time evolution of actin filament nucleation, elongation, and severing of the SCPR mechanism, using explicit prering cytokinesis nodes containing myosin and formin (30).
The computational domain is a cylinder of radius 1.74 μm and length 13 μm, reproducing the dimensions of a fission yeast cell. Membrane-bound nodes are represented as explicit point particles that can move along the membrane. They are initially distributed according to a Gaussian distribution with the mean centered along the domain long axis and a standard deviation of 0.9 μm. Formins are represented as explicit point particles, bound to nodes, that nucleate and elongate actin filaments. The number of formins per node is selected depending on nucleation efficiency, which controls the total number of filaments, N. Formin-mediated actin elongation rate is assigned as a rate at which actin beads are added to the filament’s barbed end in contact with formin beads at the node. We simulated both conditions of fixed elongation rate, including formin mechanoinhibition reported in (33), whereby elongation rate was reduced to ∼30% of uninhibited values. Actin filaments are represented as series of beads and springs that form semiflexible polymers with persistence length 10 μm (34, 35, 36). Actin and formin beads experience an effective drag of ζb = 0.108 pN s/μm to account for cytoplasmic viscosity. When an actin bead comes within 0.1 μm of an adjacent node (other than the one it originates from), a harmonic interaction is formed between them (parameter values are reported in Table S1). Nodes then exert 4-pN tension on captured beads in the direction of the pointed end, mimicking the power-stroke of myosin heads. The nodes experience an equal and opposite force, which generates sliding motion of the node along the membrane. Node movement is constrained by a membrane drag ζnode = 400 pN s/μm, mimicking cell membrane friction.
Recent evidence suggests that there are 140 cytokinesis nodes in fission yeast (25). Previous versions of the SCPR model have varied the number of nodes and reported that values greater than ∼50 nodes all lead to comparable ring assembly kinetics (27). We performed a subset of our simulations at both 65 and 140 nodes and observed that our conclusions are reproduced using either 65 or 140 nodes (Fig. S1; Video S1). Therefore, we have used 65 nodes for simulations presented in the main text, which allowed a much larger exploration of parameter space because of the increased computational efficiency.
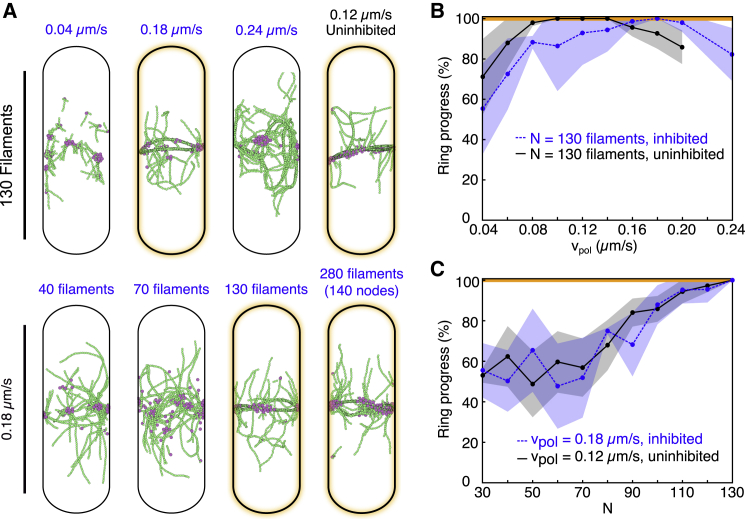
Representative simulations are animated next to graphs of corresponding time series data tracking the ring progress collective variable (see Materials and methods). Actin beads (green) elongate from formins bound to cytokinesis nodes (purple) at a rate of vpol. All runs include mechanoinhibition of formin. Raw data (gray) and 1-minute moving average (black) are shown. A red circle tracks this simulation time point on each graph. Under the same nucleation efficiency and elongation rate conditions, simulation outcomes are comparable whether 65 (left) or 140 nodes (right) are used. High and low values of vpol are compared in the top and bottom rows, respectively.
When actin beads of different filaments come within 0.1 μm of each other, a harmonic interaction is also formed between them. The rest length is 0.03 μm, and the stiffness is 1 pN/μm, mimicking the structural properties of the fission yeast contractile ring actin filament cross-linking protein Ain1 (37).
The positions of both filament beads and nodes, ri are updated at each time step of the simulation after the Langevin equation of motion (31,32) with inertia neglected:
where Fi is the total force acting on the particle and ζ is the drag coefficient. The force Fi acting on each particle is the 3D vector sum of a deterministic and stochastic force. In the case of filament beads, the deterministic force is the sum of cross-linking, bending, extension, myosin capturing, and myosin-pulling interactions. For the nodes, the deterministic force comes from filament elongation at the node-bound formin sites and node capturing and pulling. For both filament and formin beads, the stochastic force follows the fluctuation dissipation theorem:
where is the second-order unit tensor (32). A detailed description of the model parameter values, actin, formin and node interactions, and algorithm implementation scheme are reported in Supporting materials and methods and Tables S1 and S2.
Simulations were analyzed using a continuous collective variable that estimates how ring-like the actin and node geometry are by taking into account the node broad band and continuity of actin beads around the midzone. This “ring progress” collective variable is normalized between 0 and 100% such that 100% identifies fully formed rings. A complete description of this variable is provided in the Supporting materials and methods. Examples of simulations and the ring progress collective variable evolving side-by-side are shown in Videos S1 and S2. The ring progress value assigned to each simulation replicate was taken to be the maximal ring progress reached across all simulation frames. The first time point that reached 100% ring progress was recorded as the time to ring assembly.
Representative simulations are animated next to graphs of corresponding time series data tracking the ring progress collective variable (see Materials and methods). Actin beads (green) elongate from formins bound to cytokinesis nodes (purple) at a rate of vpol. The total number of filaments in each simulation, N, controls the nucleation efficiency. All runs come from 65 node simulations and include mechanoinhibition of formin. Raw data (gray) and 1-minute moving average (black) are shown. A red circle tracks this simulation time point on each graph. Left: At a constant elongation rate (vpol = 0.18 μm/s), ring formation occurs at high nucleation efficiency (top=130 filaments). As the nucleation efficiency decreases (middle=100 filaments, bottom=70 filaments), nodes coalesce progressively slower and ring assembly is compromised. Right: At high nucleation efficiency (N = 130), ring assembly is common across a range of values for vpol (top=0.2 μm/s, middle=0.16 μm/s, bottom=0.12 μm/s).
Formin chimera strain construction
The cdc12 N-terminal sequence was amplified by PCR (iProof; Bio-Rad Laboratories, Hercules, CA) from wild-type Schizosaccharomyces pombe genomic DNA and cloned into pBluescript II KS (−) (StrataGene, San Diego, CA) using restriction enzymes XhoI and BamHI. Overlap PCR was used to link formin (FH1-FH2) sequences from For3, Fus1, Bni1, mDia2, and CYK-1 to cdc12 C-terminal sequences, followed by homologous recombination using the In-Fusion Advantage PCR Cloning Kit (Clontech, Mountain View, CA) into pBluescript already containing cdc12 N-terminus, resulting in the cdc12 chimera fragment cdc12(N)::formin(FH1-FH2)::cdc12(C). PCR amplifications of the cdc12 promoter (1–700 bp upstream of the translation start site) from the wild-type S. pombe genome and monomeric GFP (mGFP) from the pSGP-572 vector were cloned into the S. pombe integration vector pJK210 (38) with restriction enzyme SacI. Cdc12 chimeras were also cloned into pJK210 by In-Fusion downstream of the cdc12 promoter and confirmed by sequencing.
Chimera formin constructs were integrated into the ura4 locus under control of the cdc12 promoter and tagged C-terminally with GFP to create pJK210-Pcdc12-cdc12(N-term)-formin(FH1-FH2)-cdc12(C-term)-GFP::ura+ (33). These strains, containing both integrated chimera formins and endogenous cdc12, were saved to perform the analysis seen in Fig. S6. For the remainder of the study, endogenous cdc12 was deleted through Kan cassette gene replacement (39), whereas markers for contractile rings (rlc1-tdTomato-natMX6) and spindle pole bodies (sad1-tdTomato-natMX6) were introduced to formin chimera strains through mating. Table S3 lists the fission yeast strains used in this study.
Cell imaging and growth
Differential interference contrast (DIC) and epifluorescence images were collected on an IX-81 microscope (Olympus, Tokyo, Japan) fitted with an Orca-ER camera (Hamamatsu, Bridgewater, NJ) and a 60×, 1.4 NA Plan Apo objective. Confocal images were acquired on a Zeiss Axiovert 200M microscope (ZEISS, Oberkochen, Germany) equipped with a Yokogawa CSU-10 spinning-disk unit (McBain, Simi Valley, CA) fitted with a Cascade 512B EM-CCD camera (Photometrics, Tuscon, AZ) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA) and illuminated with 50-mW, 473- and 561-nm, diode-pumped solid-state lasers (DPSS Lasers, Santa Clara, CA).
For live-cell DIC and epifluorescence imaging, cells were grown overnight in YE5S media at 25°C, subcultured into EMM5S minimal media without thiamine, and kept in log phase for 20–22 h. They were then imaged directly on glass slides using Z-stacks of 10 slices with a 0.5-μm step size.
Confocal spinning-disk images were obtained on an IX83-X1 microscope (Olympus) fitted with a Yokogawa CSU-X1 Spinning Disk Confocal Unit and ImagEM X2 EMCCD camera (Hamamatsu) with a 100×, 1.49 NA objective. Cells were grown as described above and imaged on gelatin pads using Z-stacks of 10 slices with a 0.5-μm step size.
Cell growth assay
Each strain was grown for 24–36 h in YE5S and then seeded in triplicate in a 96-well plate at initial OD600 readings of 0.03 and 0.06. OD600 readings were measured every 10 min for 24 h in a Tecan Infinite M200Pro (Tecan Systems, San Jose, CA) plate reader at 30°C with an orbital shaking amplitude of 4 mm.
BoDipy-phallicidin staining
Upon receipt, BoDipy-phallicidin stocks were prepared by resuspending 300 units BoDipy-phallicidin (Thermo Fisher Scientific, Waltham, MA) in 1.5 mL methanol. This solution was divided into 25 μL aliquots, vacuum dried, and stored at −20°C. Immediately before use, one dry BoDipy-phallicidin aliquot was resuspended in 10 μL PEM buffer (0.1 M NA PIPES (pH 6.8), 1 mM EGTA, and 1 mM MgCl2).
Cells were grown in YE5S for 36 h. 1 mL cells for each strain growing at 25°C and OD600 ∼0.4 were fixed with 333 μL 16% paraformaldehyde for 5 min. The fixed cells were washed three times at room temperature with PEM with 30-s spins at 7000 rpm in between. Cells were permeabilized in 1 mL PEM buffer with 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) at room temperature for 1 min. Cells were then washed three times as before and resuspended in 10 μL PEM and stained by adding 1 μL BoDipy-phallicidin solution for 30 min in the dark at room temperature. After staining, cells were washed once with PEM and spun for 30 s at 7000 rpm to obtain a pellet. The supernatant was removed, leaving a small amount of liquid. 4 μL of cells were imaged directly on glass slide as described above on an IX-81 microscope (Olympus).
4′,6-diamidino-2-phenylindole or calcofluor staining
Cells were grown in YE5S for 36 h, and DIC images were acquired directly on glass before fixation. 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor staining was conducted as described previously (40). Briefly, cells were grown in YE5S at 25°C for 36 h and then fixed with 100% cold methanol. For staining, cells were incubated in 300 μL 50 mM sodium citrate with 4 μL Calcofluor White Stain (Fluka Analytical; Sigma-Aldrich) for 5 min at 37°C. They were then washed with 1 mL 50 mM sodium citrate and resuspended in 15 μL sodium citrate and 4 μL DAPI stock (1 mg/mL in H2O; Life Technologies, Carlsbad, CA) and kept on ice until imaging. 2.5 μL of stained cells were placed on glass slide and imaged as described above on an IX-81 microscope (Olympus) with an Orca-ER camera (Hamamatsu) and 60×, 1.4 NA Plan Apo objective.
Quantification of DIC and DAPI and calcofluor images
DAPI-and-calcofluor-stained cells were scored for number of nuclei and appearance of septa, in which abnormal septa were those that were misplaced or misshapen. DIC images were used for quantification of cell length, cytokinesis defects, and morphology defects. Cell length was measured along the long axis of each cell. For cytokinesis defects, strains containing spindle pole body (SPB) marker Sad1-tdTomato were quantified for number of spindle pole bodies. For morphology defects, DIC images of each strain were quantified for three cell width measurements: 2 μm from each cell tip (m1 and m3) and in the cell midzone (m2). The standard deviation of these cell width measurements was then calculated for each cell.
Quantification of formin chimera expression
Z-stacks of strains expressing formin chimeras tagged with GFP and Rlc1-tdTomato were imaged on glass slides as described above, and sum slice projections were compiled. To measure formin chimera expression, ROIs were created of both contractile rings and the corresponding whole cell. The mean fluorescence of the formin-GFP was measured in the contractile ring and divided by the mean formin-GFP of the whole cell to control for background fluorescence. The average of this ratio was calculated for two replicates for each strain and normalized to the control.
Plasmid construction
A Cdc12 fragment containing the FH1-FH2 domains and a portion of the C-terminal tail (Cdc12 (882–1687)) was cloned by traditional restriction enzyme cloning into pET21a-MBP-TEV at BamHI-XhoI. The Bni1-Cdc12 (Bni1 (1228–1766)-Cdc12 (1391–1687)) and mDia2-Cdc12 (mDia2 (527–1022)-Cdc12 (1391–1687)) chimeras were amplified from plasmids constructed for building the formin chimera strains and were inserted via In-Fusion Cloning (Clontech) into pET21a-MBP-TEV at BamHI-NotI to form MBP-chimera-HIS(×6).
Protein purification
The Cdc12 fragment (Cdc12 (882–1687)) and the Bni1-Cdc12 and mDia2-Cdc12 chimeras were expressed in Escherichia coli strain BL21-Codon Plus (DE3)-RP (Agilent Technologies, Santa Clara, CA) with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 16 h at 16°C. Cells were lysed by sonication in extraction buffer (50 mM NaH2PO (anhydrous), 500 mM NaCl, 10% glycerol, 10 mM imidazole, and 10 mM β-mercaptoethanol (BME) (pH 8)) with EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Basel, Switzerland) and were clarified. The extract was incubated for 1 h at 4°C with Talon Resin (Clontech), loaded onto a column, washed with extraction buffer, and protein was eluted with 250 mM imidazole. The formin chimeras were dialyzed into buffer (50 mM HEPES (pH 7.0), 50 mM NaCl, 5% glycerol, 0.01% NaN3, and 1 mM dithiothreitol (DTT)) for cation exchange chromatography (GE Healthcare, Little Chalfont, UK). The cleanest fractions were pooled and dialyzed into SNAP buffer (20 mM HEPES (pH 7.4), 200 mM KCl, 0.01% NaN3, 10% glycerol, and 1 mM dithiothreitol). The mDia2-Cdc12 chimera required additional size-exclusion chromatography and was filtered on a Superdex 200 10/300 GL column (GE Healthcare). Aliquots of all formin proteins were flash frozen in liquid nitrogen and stored at −80°C. Chicken skeletal muscle actin was purified as described in (41). Fission yeast profilin Cdc3 was overexpressed and purified from E. coli using poly-L-proline affinity chromatography as described in (42).
Total internal reflection fluorescence microscopy and analysis
Total internal reflection fluorescence microscopy (TIRFM) was conducted with the formin chimeras as described previously (43). Briefly, time-lapse TIRFM videos were obtained with through-the-objective total internal reflection fluorescence illumination on an Olympus IX-71 microscope with an iXon EMCCD camera (Andor Technology) and a cell total internal reflection fluorescence four-line system (Olympus). 1 nM of either Cdc12, Bni1-Cdc12, or mDia2-Cdc12 formin chimeras was added to a polymerization mix (see (43)) along with 2.5 μM fission yeast profilin Cdc3, which was then added to Mg-ATP-actin (10% Alexa 488 labeled) to induce actin assembly. This mixture was added to a flow chamber and imaged at 5-s intervals at room temperature.
The nucleation activity was determined by counting the total number of actin filaments for each TIRFM video at frame 48, the same amount of time since the initiation of each actin assembly reaction. Each construct (actin only, Cdc12, Bni1-Cdc12, or mDia2-Cdc12) was counted in triplicate. To determine the actin filament elongation rate, 10 individual actin filaments from each TIRFM video were tracked over time, with their lengths measured every fifth frame for seven to nine total measurements. An average elongation rate was calculated for each filament, and then, those were averaged to obtain an average for each video. Each construct (actin only, Cdc12, or mDia2-Cdc12) was measured in triplicate with the exception of Bni1-Cdc12, which was only measured in duplicate because of a low number of Bni1-Cdc12-associated filaments in the third TIRFM video because of Bni1-Cdc12’s low nucleation activity.
Results
An SCPR model reveals the importance of nucleation efficiency on cytokinetic ring formation
We hypothesize that in addition to regulation of its activity in time and space, a formin’s ability to facilitate a particular cellular function also depends on its specific actin assembly properties (17). To begin to address this question, we investigated how the actin assembly properties of the fission yeast cytokinesis formin Cdc12 affect contractile ring assembly by extending a 3D SCPR model (30). Cdc12 nucleates F-actin very efficiently, generating one actin filament per 2.5 Cdc12 dimers, and elongates F-actin at a rate of 10–12 subunits/s/μM (Table 1). Thus, we systematically varied both the number of node-bound filaments, N, representing formin nucleation efficiency, and the formin-mediated processive actin filament elongation rate, vpol (Fig. 1). We varied N from 30 (∼one filament every two nodes) to 130 (two filaments per node) and vpol from 0.04 to 0.24 μm/s, capturing a relevant range for a wide array of formins (6,19, 20, 21, 22). Additionally, we performed simulations in which formin-mediated actin filament elongation was inhibited by forces exceeding 1 pN at the barbed end, mimicking the mechanoregulation of Cdc12 (33). We analyzed simulations using a continuous collective variable that tracks ring progress throughout the simulation by accounting for gradual changes in actin and node morphologies that are organized in more or less ring-like structures. 100% ring progress corresponds to a fully formed ring.
Our simulations reproduced cytokinesis ring assembly at nucleation efficiencies and unloaded elongation rates close to those measured for Cdc12 (N = 130 and vpol = 0.18 μm/s, respectively, with mechanoinhibition) (Fig. 2 A; Video S2). Keeping N constant at Cdc12’s nucleation efficiency of 130 filaments and varying the elongation rate, the model predicts that at low elongation rates (vpol = 0.04–0.10 μm/s), nodes form clumps with short actin filaments elongating radially from the nodes (Fig. 2 A, top row, left). However, at high actin elongation rates (vpol > 0.20 μm/s), nodes condense into a small number of clumps that elongate actin filament bundles along the domain’s long axis (Fig. 2 A, top row). Neither high nor low actin filament elongation rates facilitate reliable ring formation; however, a wide range of intermediate values supported ring assembly (Fig. 2 B). This held true for both mechanoinhibited and uninhibited formins, but shifted optimal ring-forming conditions toward lower values of vpol for uninhibited formins (Fig. 2 A, top row, right; Fig. 2, B and C). Mechanoinhibition also led to ring formation over a larger region of parameter space (Fig. S2), consistent with the idea that Cdc12’s activity is uniquely tailored for its role in cytokinesis.
Figure 2.
Ring assembly depends more strongly on formin nucleation efficiency than polymerization rate. (A) Snapshots at t = 800 s of simulations at different values of vpol and N. Upper: snapshots at a constant nucleation efficiency, N = 130, depict nonring morphologies resulting from too low or too high of polymerization rates, as well as proper ring formation for intermediate values (orange highlight). In the absence of formin mechanoinhibition, low polymerization rates (right) resemble higher elongation rates with inhibition. Lower: snapshots at a constant polymerization rate, vpol = 0.18 μm/s, depict a progression to more favorable conditions for ring assembly with increasing N. Outcomes of simulations using 65 nodes are comparable with those of equivalent simulations using 140 nodes (right, vpol = 0.2 μm/s). Video S2 shows simulations evolving in time. (B and C) Dependence of ring progress (see Materials and methods) on key parameters in which 100% corresponds to a fully formed ring (orange line). Data points are averages of three to seven independent runs and shaded regions represent standard deviations. Two-dimensional heat maps are shown in Fig. S2. (B) At a constant nucleation efficiency, N = 130, progress toward ring assembly exhibits a gradual dependence on polymerization rate, vpol, with many conditions supporting complete ring assembly. Peak values are shifted to higher vpol with formin mechanoinhibition. (C) At a constant polymerization rate that favors ring assembly, ring progress increases drastically at high nucleation efficiency for both inhibited and uninhibited formins.
Keeping the elongation rate constant at 0.18 μm/s, which maximizes ring assembly at Cdc12’s high nucleation efficiency (Fig. 2 B), and varying N, our simulations show that at low nucleation efficiency (N < 90), isolated clumps of nodes formed, but never assembled a ring (Fig. 2 A, bottom row). Only high nucleation efficiencies (N ≥ ∼110 filaments) reliably support complete ring formation (Fig. 2 C). Uninhibited formins follow the same trend at lower values of vpol (Fig. 2, B and C). Unlike high elongation rates, low N conditions were not noticeably different between mechanoinhibited and uninhibited formins (Fig. 2 C). Ring progress for the entire parameter space explored (i.e., varying vpol and N simultaneously) are reported as heat maps in the Fig. S2, A and B.
Together, these quantifications indicate that although both are important, the nucleation efficiency of formin is a more powerful determinant of ring formation than the formin-mediated actin filament elongation rate.
Fission yeast formin Cdc12 chimeras localize to the contractile ring and produce viable cells that complete cell division
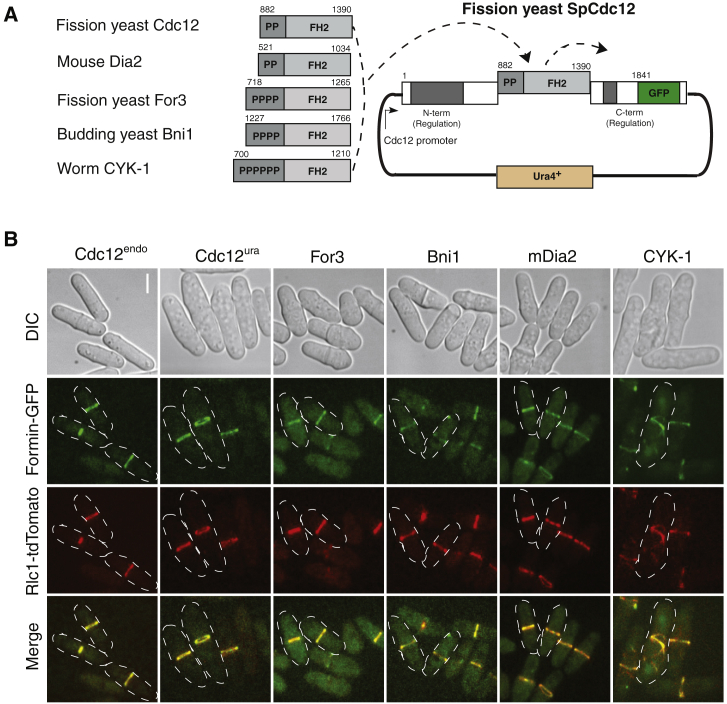
To test the results of our computational modeling in vivo, we aimed to alter the actin assembly properties of the contractile ring formin Cdc12 in fission yeast cells. Point mutations in formin’s actin assembly FH2 domain typically led to a nonfunctional protein because of the general importance of the FH2 domain in formin dimerization and barbed-end binding. Therefore, we instead engineered formin chimera strains in which the FH1-FH2 actin assembly domains of full-length Cdc12 were replaced with the FH1-FH2 domains from functionally and evolutionarily diverse formins with significantly different actin assembly properties: Fus1 (fission yeast), For3 (fission yeast), Bni1 (budding yeast), mDia2 (mouse), and CYK-1 (worm) (Fig. 3 A).
Figure 3.
Fission yeast formin Cdc12 chimeras localize to the contractile ring and produce viable cells that divide. (A) Schematic of fission yeast formin Cdc12 chimeras in which the FH1-FH2 actin assembly domains were replaced by FH1-FH2 domains from formins with diverse actin assembly properties. Each “P” indicates a single FH1 polyproline motif. Image not drawn to scale. (B) Representative micrographs of formin chimera strains showing morphology (DIC), the formin chimera (formin-GFP), the contractile ring (Rlc1-tdTomato), and the merge. Scale bar, 5 μm.
Given that we specifically replaced only the FH1-FH2 domains, we expected the regulation and localization of the formin chimeras to be maintained because Cdc12’s N- and C-terminal regions are thought to be primarily responsible for targeting it to the contractile ring (29). However, the formin chimeras should exhibit a wide range of actin assembly properties (nucleation efficiency, barbed-end elongation rate, barbed-end dissociation rate). Table 1 details a comparison of in vitro actin assembly properties for all the aforementioned formins relative to Cdc12. As described in the modeling section above, Cdc12 is an efficient nucleator (one filament per 2.5 dimers), but a comparatively slow elongator (10–12 subunits/s/μM) (19,20,44). If, as suggested by our computational modeling, formin’s actin filament nucleation rate is particularly critical for its role in cytokinesis, we predicted that the mDia2 chimera will compromise cytokinesis the least because its nucleation rates are most similar to Cdc12 (Table 1). However, if formin’s elongation rate is most important in vivo, then For3 and mDia2 chimeras will function best in fission yeast cytokinesis (Table 1).
One copy of each formin chimera was integrated into the ura4 locus under control of the endogenous cdc12 promoter and tagged at the C-terminus with GFP (Fig. 3 A). Endogenous cdc12 was deleted by replacement with a kanR cassette (39), and the contractile ring was observed by tagging the endogenous copy of the myosin II regulatory light chain Rlc1 with tdTomato. For controls, endogenous cdc12 was tagged C-terminally with GFP (Cdc12endo) and full-length Cdc12-GFP was integrated into the ura4 locus with endogenous cdc12 deleted (Cdc12ura). With the exception of fission yeast Fus1, which was not studied further (see Supporting materials and methods, for discussion), all of the formin chimera strains produced viable cells (Fig. 3 B), despite the diverse in vitro actin assembly parameters of the various formin FH1-FH2 domains (Table 1). Like control Cdc12 strains, the formin chimeras colocalize with Rlc1-tdTomato in the contractile ring or contractile ring-like material at the cell midzone (Fig. 3 B), suggesting that they are properly recruited to the cell division site. Furthermore, the formin chimera strains follow the same general growth curve over time as the control strains (Fig. S3 A), and staining of fixed cells with BoDipy-phallicidin did not reveal significant differences in their general actin cytoskeletal architectures (Fig. S3 B). Because purification of full-length Cdc12 is difficult, we validated that active fragments of purified formin mDia2-Cdc12 and Bni1-Cdc12 chimeras that contain the mDia2 or Bni1 FH1-FH2 domains and a 296-amino-acid Cdc12 tail exhibit similar in vitro actin assembly properties (nucleation activity and barbed-end elongation rate) as previously characterized for the isolated mDia2 and Bni1 FH1FH2 domains (Fig. S4; Table 1).
Formin Cdc12 chimera fission yeast strains display a range of cytokinesis and morphology defects
Although diverse formin chimeras localize to the contractile ring and generally support life in fission yeast, preliminary imaging suggested that some formin chimera strains may exhibit cytokinesis defects (for example, see CYK-1 in Fig. 3 B). Importantly, quantification of the total amount of formin-GFP fluorescence revealed that the expression levels of the formin chimeras are not statistically different from Cdc12endo (Fig. S3 C). Therefore, any observed cytokinesis defects are not likely due to differences in formin chimera expression.
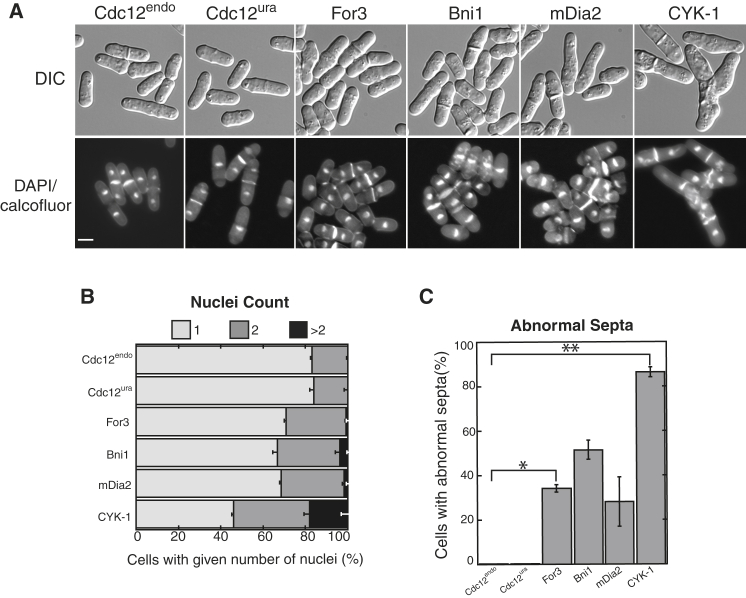
To characterize general cytokinesis defects in the formin chimera strains, we stained methanol-fixed cells with DAPI and calcofluor to quantify the number of nuclei and abnormal septa, respectively (Fig. 4). First, although only 15–20% of control Cdc12endo and Cdc12ura cells have two nuclei, 28% of For3, 30% of Bni1, 30% of mDia2, and 54% of CYK-1 cells have two or more nuclei (Fig. 4, A and B), suggesting cytokinesis defects. Second, whereas no Cdc12endo or Cdc12ura cells have abnormally placed and/or shaped septa, 34% of For3, 52% of Bni1, 28% of mDia2, and 86% of CYK-1 cells contain abnormal septa (Fig. 4, A and C). Formin chimera strains also exhibit varying degrees of morphological defects (Fig. S5). For example, CYK-1 chimera cells are abnormally long, growing to an average length of almost twice that of Cdc12endo cells (Fig. S5 A), presumably because of their failure to divide properly. By imaging the SPB marker Sad1-tdTomato, we also determined that formin chimera strains have at least twice as many cells in mitosis compared with control strains (Fig. S5 B). Finally, the formin chimera strains also exhibited abnormal bulging morphology defects (Fig. S5 C). Collectively, these data indicate that formin chimeras have a range of general division and morphology defects, with CYK-1 exhibiting extreme abnormalities and mDia2 appearing the most similar to control strains.
Figure 4.
Formin chimera strains exhibit a range of cytokinesis defects. (A) Representative micrographs of the indicated formin chimera strains; DIC (morphology) and methanol-fixed cells stained with DAPI (nuclei) and calcofluor (septa). Scale bar, 5 μm. (B and C) Quantification of general cytokinesis defects. (B) Percent of cells with one, two, or more than two nuclei. (C) Percent of cells with septa exhibiting abnormal septa (misplaced, misoriented, or abnormally broad). Shown are two-tailed t-tests for data sets with unequal variance yielded p-values ∗p = 0.030 and ∗∗p = 0.017. Error bars in (B and C) indicate standard error of two biological replicates of at least 400 cells each.
One possibility is that the formin chimeras are dominant negatives whose expression causes cytokinesis and morphology defects irrespective of their ability to functionally replace endogenous Cdc12 for division. Therefore, we also analyzed whether cells simultaneously expressing both the formin chimeras (integrated at the ura locus) and endogenous cdc12 have division defects (Fig. S6). Formin chimera strains exhibit dramatically less severe general cytokinesis defects (abnormal septa and multinucleated cells) in the presence of endogenous Cdc12 (Fig. S6). Therefore, the observed abnormalities in cells exclusively expressing formin chimeras result primarily from deficiencies in their ability to replace endogenous Cdc12 for cytokinesis rather than because of secondary dominant negative effects.
Formin chimera fission yeast cells progress through cytokinesis at different rates
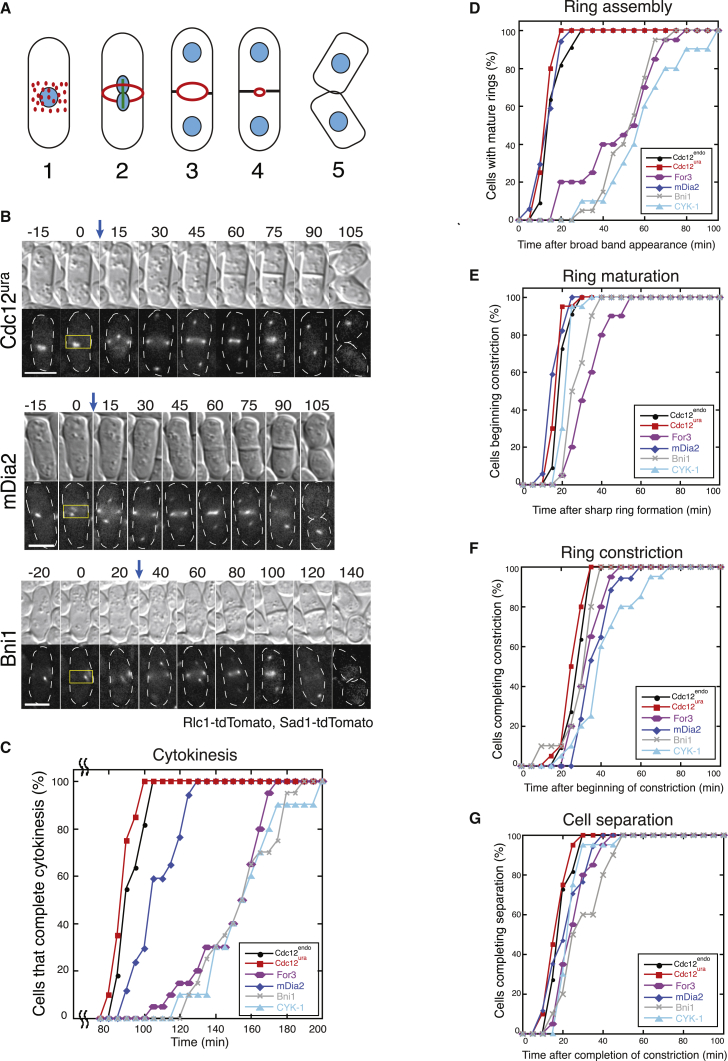
The computational model specifically revealed that inefficient actin filament nucleation by formin causes defects in the rate of contractile ring assembly (Figs. 2 and S2; Video S2). Because the formin chimera cells exhibit varying degrees of general division defects (Fig. 4; Fig. S5), we used quantitative imaging to determine which cytokinesis steps are specifically compromised in each strain. Fission yeast cells primarily assemble contractile rings from precursor cytokinesis nodes through a well-defined temporal pathway of events (Fig. 5 A): 1) formation of a broad band of precursor nodes, 2) contractile ring assembly, 3) initiation of constriction, 4) completion of constriction, and 5) cell separation (27,45). We imaged dividing cells expressing the contractile ring marker myosin II regulatory light chain Rlc1-tdTomato and the SPB marker Sad1-tdTomato by time-lapse fluorescence microscopy of a single plane to comparatively quantify the kinetics of cytokinesis for the formin chimera strains (Fig. 5, B and C).
Figure 5.
Formin chimeras with poor nucleation efficiency exhibit delays in ring assembly. (A) Schematic of fission yeast cytokinesis: 1) node assembly, 2) ring assembly, 3) ring maturation, 4) ring constriction, and 5) cell separation. (B) Representative time-lapse micrographs of the morphology (DIC) and contractile ring (Rlc1-tdTomato) of representative formin chimera cells. Time is indicated in minutes. Spindle pole bodies are labeled with Sad1-tdTomato. Blue arrows indicate time of SPB separation. Yellow boxes indicate broad band of precursor cytokinesis nodes. Scale bar, 5 μm. (C) Time required for at least 10 cells to complete the entire time course of cytokinesis for each strain. (D–G) Time required for at least 10 cells to complete the indicated step of cytokinesis for each strain. (D) Frequency of cells that assemble a sharp ring over time. (E) Frequency of cells that complete ring maturation. (F) Frequency of cells that complete ring constriction. (G) Frequency of cells that complete cell separation. The p-values for pairwise log-rank tests associated with plots in (C–G) are listed in Table S4.
We compared the entire cytokinesis time course by analyzing cells that completed cytokinesis from precursor node appearance to cell separation within the 3-h imaging window (Fig. 5, B and C). In control cells, SPBs separate around 10 min after Rlc1-labeled precursor nodes initially appear. However, the timing of SPB separation and the initiation of ring assembly is uncoupled in some formin chimera strains, where Rlc1 fluorescence appears at the midzone up to 1 h before SPB separation (Fig. 5 B, see Bni1 panel). Therefore, the initial appearance of the broad band of cytokinetic precursor nodes marks time zero in our experiments. 100% of Cdc12endo and Cdc12ura control cells complete cytokinesis with remarkably little variability in ∼90 min after the appearance of precursor nodes. Conversely, the formin chimera strains have distinct kinetic profiles of the progression of cytokinesis events, some with significant delays (Fig. 5 C). The mDia2 strain progresses through cytokinesis with only a slight delay as compared with control cells, with 100% of cells completing cell division within 120 min of precursor node appearance. However, For3, Bni1, and CYK-1 strains display substantial delays because 100% of cells have not finished cell division until after ∼160 min after precursor node appearance.
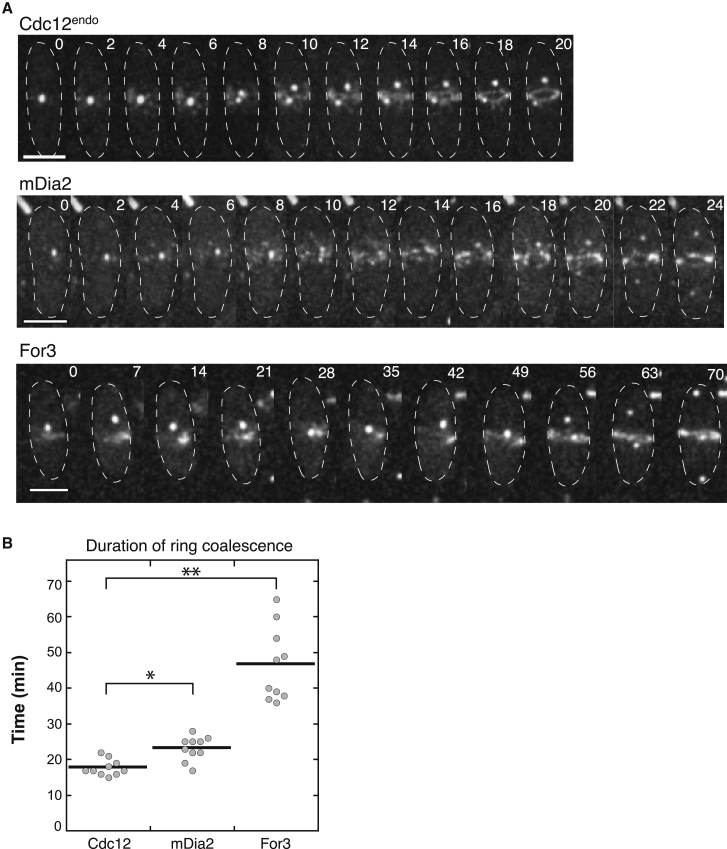
We next quantified the elapsed time for each well-defined cytokinetic event to determine which specific step(s) causes delays in formin chimera strain cytokinesis: ring assembly (Fig. 5 D), ring maturation (Fig. 5 E), ring constriction (Fig. 5 F), or cell separation (Fig. 5 G). For ring assembly, we measured the time for each cell to assemble a fully formed contractile ring after the initial appearance of precursor node fluorescence at the midzone (Fig. 5 D). Interestingly, two populations of ring assembly profiles emerged from this analysis. Similar to control strains, 100% of mDia2 cells assemble contractile rings within 25 min of broad brand appearance. Conversely, For3, Bni1, and CYK-1 exhibit a large lag in ring assembly, with 100% of cells finally forming contractile rings between 70 and 100 min after node appearance (Fig. 5 D). After the contractile ring was assembled, we quantified the ring maturation time, or the time elapsed before the onset of cell constriction (Fig. 5 E). The time course of ring maturation was similar for all formin chimeras, with most cells finishing ring maturation within 20 min of ring formation. Only For3 and Bni1 cells exhibit a brief lag, but are still much more similar to control strains than they are during ring assembly. Similarly, we found that the elapsed time for ring constriction (Fig. 5 F) and cell separation (Fig. 5 G) is similar for all of the formin chimera and control strains. To determine whether the observed lag in the rate of contractile ring assembly in For3 cells is due to slower ring coalescence, rather than the early appearance of static cytokinesis nodes, we performed higher resolution confocal microscopy (Fig. 6; Video S3) and quantified the duration of node coalescence from the first movement of cytokinesis nodes until the ring is fully formed (Fig. 6 B). As expected, the duration of dynamic cytokinesis node coalescence is longer for formin chimera cells because mDia2 coalescence takes 1.3-fold longer than Cdc12endo control cells, whereas For3 cells take 2.5-fold longer (Fig. 6; Video S3).
Figure 6.
Duration of contractile ring coalescence is longer in formin chimera cells. (A) Confocal time-lapse micrographs of representative cells from indicated strains during coalescence of the contractile ring. For comparison, time 0 is marked by the first appearance of Rlc1-tdTomato fluorescence at the midzone. Time is indicated in minutes. Scale bars, 5 μm. Also, see Video S3. (B) Duration of contractile ring coalescence from onset of cytokinesis node and spot movement. 10 cells were measured for each strain. Two-tailed t-tests for data sets with unequal variance yielded p-values ∗p = 6.09 × 10−3 and ∗∗p = 6.13 × 10−6.
Confocal time-lapse imaging of representative Cdc12endo, mDia2, and For3 formin chimera cells showing movement of cytokinesis nodes during assembly of the contractile ring (Rlc1-tdTomato). Scale bar, 5 μm. Time is indicated in minutes.
Our in vivo evidence reveals that different rates of contractile ring assembly are primarily responsible for major differences between the formin chimera strains in the overall time course of cytokinesis. This is consistent with formin-mediated actin assembly having a major role in generation of the contractile ring (14,24) and, because mDia2 has the most similar nucleation properties to Cdc12 (Fig. S4; Table 1), supports the conclusion from our quantitative modeling that efficient formin-mediated nucleation of actin filaments is particularly critical for contractile ring assembly in fission yeast (Fig. 1).
Discussion
Formins associate processively with the growing barbed end of actin filaments to rapidly assemble long, unbranched actin filaments that facilitate diverse cellular processes, such as cytokinesis and polarization. Although most formins assemble actin filaments by the same general nucleation and processive elongation mechanism, particular formins exhibit a wide range of specific rate constants. We previously determined that the quantitative actin assembly properties of the three fission yeast formin isoforms vary widely, indicating that in addition to activation at the right time and place, a formin’s specific properties might be tailored for a particular cellular role (17,20). Here, we investigated the effects of formin Cdc12’s actin assembly properties on cytokinetic ring assembly by extending a computational model that was previously developed and validated for reproducing fission yeast ring formation in 3D based on the SCPR mechanism (27,30,46). The model allowed us to independently control formin nucleation efficiency and elongation rate and to characterize their relative effects on ring assembly (Fig. 1). We also implemented a force-sensitive mechanism for actin filament elongation mimicking the mechanoregulation of formin Cdc12. Our simulations demonstrated that variations in formin nucleation efficiency sharply impact the probability of ring formation (Fig. 2, A and C; Video S2), which is consistent with our experimental characterization of formin chimera fission yeast cells. In contrast, changes in elongation rate had important effects at extreme values, but were less impactful overall (Fig. 2, A and B; Video S2).
To begin to test the roles of formin nucleation efficiency and elongation rate in vivo, we engineered formin chimera fission yeast strains in which the FH1-FH2 actin assembly domains of Cdc12 were replaced with the FH1-FH2 domains from a group of evolutionary and functionally distinct formins: Fus1 (fission yeast), For3 (fission yeast), Bni1 (budding yeast), mDia2 (mouse), and CYK-1 (worm) (Fig. 3 A). These formin chimeras retain Cdc12’s localization and regulation but demonstrate a wide range of actin assembly properties (Table 1).
We found that with the exception of fission yeast Fus1 (see further discussion in Supporting materials and methods), cells expressing formin chimeras instead of endogenous cdc12 are viable. More importantly, despite the proper localization of diverse formin chimeras to the division site, they exhibit a range of cytokinesis and morphological defects whose severity appears to be anticorrelated to the similarity of their nucleation efficiency to that of Cdc12. Specifically, the mDia2 formin chimera is generally less defective in cytokinesis than For3, Bni1, and CYK-1 formin chimeras (Figs. 4 and 5). Interestingly, an analysis of their in vitro actin assembly properties reveals that For3, Bni1, and CYK-1 elongate actin as fast or faster than Cdc12 (Table 1). In fact, both For3 and mDia2 elongate actin at the same rate as Cdc12 (Table 1). Thus, a formin’s elongation rate does not correlate well with its ability to successfully facilitate cytokinesis in fission yeast, at least over a ∼4- to 8-fold range. However, For3, Bni1, and CYK-1 have drastically lower nucleation efficiencies than Cdc12, at 1.5, 10, and 3% as efficient, respectively. Conversely, mDia2 nucleates actin filaments 25% as efficiently as Cdc12 and completes cytokinesis at similar rates as control strains (Fig. 5). Together, these data indicate that fission yeast cytokinesis is extremely robust, but formins with low nucleation efficiency are unable to facilitate the assembly of a mature contractile ring as effectively as Cdc12. Although our results indicate that contractile ring assembly in fission yeast relies on Cdc12’s particular actin assembly properties, whether the actin assembly properties of other formins are tailored for their cellular process remains to be determined (17).
Fission yeast cells use the SCPR mechanism for contractile ring assembly, which relies upon ∼140 prering cytokinesis nodes that contain formin Cdc12 and myosin, as well as other regulatory and scaffolding proteins (27,28). Results from simulations of 140 nodes confirm the data from simulations at 65 nodes (Fig. S1 B; Video S1). Node-bound Cdc12 is thought to nucleate and then elongate actin filaments that are captured and pulled by myosin on nearby nodes, bringing nodes closer together to assemble a continuous F-actin ring. These F-actin connections between nodes are severed by cofilin, and the process starts again until the ring is fully formed (27,28). It follows, then, that Cdc12’s efficient nucleation of F-actin (one filament for every 2.5 Cdc12 dimers; Table 1) is paramount in its role as the sole cytokinesis formin in fission yeast because the initial step of contractile ring assembly requires efficient formation of actin filaments. Conversely, we suspect that a formin’s processive run length (the number of actin monomers added) and dissociation rate contribute less to contractile ring assembly than its other actin assembly properties. Specifically, the F-actin-severing protein cofilin is thought to abolish node-node contacts before dissociation of formin from the barbed end (47). This is consistent with our experimental data because mDia2 dissociates from elongating actin filament barbed ends an order of magnitude faster than Cdc12 (Table 1) but still effectively completes cytokinesis in fission yeast.
It has been shown that each prering cytokinesis node contains only four Cdc12 dimers, which explains why the formin chimeras that are poor nucleators exhibit a long lag in ring assembly (25). For3, Bni1, and CYK-1 produce 10% or fewer actin filaments per formin dimer as Cdc12 (Table 1), apparently resulting in impaired node coalescence and slower contractile ring formation (Figs. 1, S1, and S2). It is also possible that because For3, Bni1, and CYK-1 are poor nucleators, some of these formin chimera cells use a distinct “spot” pathway for contractile ring assembly (Fig. S7; (48)).
Although a formin’s elongation rate is also presumably tailored for its cellular role, formin elongation rates can be modulated by external mechanical forces. Cdc12 is a mechanosensor, the elongation of which is inhibited by myosin-mediated pulling of Cdc12-bound filaments, which is required for optimally efficient contractile ring assembly (33). However, mDia2 is not mechanoinhibited like Cdc12, contributing to the modest delay in cytokinesis mediated by the mDia2 formin chimera (Fig. 5 C; (33)). Application of force to budding yeast Bni1 increases its F-actin elongation rate in the presence of profilin (49). The combination of low nucleation efficiency and absence of mechanoinhibition (such as in Bni1 chimera cells) might be particularly problematic for proper contractile ring assembly. Actin filaments, especially in contractile networks, often experience mechanical forces that could modify elongation by mechanosensitive formins to facilitate assembly of particular F-actin networks. Thus, formin chimera elongation rates might affect fission yeast cytokinesis less than their nucleation efficiencies.
Although our quantitative modeling and the associated in vivo experiments strongly support the importance of Cdc12’s tailored actin filament nucleation efficiency to facilitate contractile ring assembly in fission yeast, we cannot rule out that cytokinesis defects in the formin chimera strains are due to other differences between the formins such as 1) FH1 and/or FH2 domain association with accessory proteins, 2) recruitment of distinct actin-binding proteins to formin-mediated actin filaments, 3) posttranslational modifications of FH1 and/or FH2 domains and associated activities, or 4) interaction with microtubules (for further discussion of each of these possibilities, see Supporting materials and methods, Discussion). Finally, we cannot rule out the possibility that formin FH1-FH2 domains have evolved to function optimally in the presence of the formin’s native regulatory domains, such that swapping FH1-FH2 domains would not automatically confer the same actin assembly properties. Despite these potential caveats, our experimental and computational observations are consistent and provide evidence that Cdc12’s actin assembly properties are optimized for cytokinetic ring assembly, with its ability to efficiently nucleate actin filaments from cytokinesis nodes being a predominant factor.
Author contributions
K.E.H., E.M.N., and D.R.K. developed the project and the cell biological research, and K.E.H. and M.E.O. conducted cell biological experiments and analyzed the data. C.A.A. and M.E.O. conducted the biochemical experiments, and K.E.H. analyzed the data. V.Z. and T.C.B. developed the code, carried out the simulations, and analyzed the data. K.E.H., V.Z., and T.C.B. wrote the article, and D.R.K. and G.A.V. edited the article.
Acknowledgments
We thank Andy Bestul and Bonnie Scott for preliminary work on this project, as well as Michael Glotzer for helpful suggestions. Acknowledgment is also made to the computational resources provided by the Research Computing Center at The University of Chicago.
This work was supported by National Institutes of Health’s Molecular and Cellular Biology Training grant T32 GM007183 (to K.E.H.), National Institute for Health’s Grant RO1 GM079265 (to D.R.K.), Department of Defense Army Research Office’s Multidisciplinary University Research Initiative grant W911NF1410403 (to G.A.V. and D.R.K.), and by the University of Chicago Materials Research Science and Engineering Center, funded by National Science Foundation award DMR-1420709 (to G.A.V. and D.R.K.).
Editor: Dimitrios Vavylonis.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.06.023.
Contributor Information
Tamara C. Bidone, Email: tamarabidone@sci.utah.edu.
David R. Kovar, Email: drkovar@uchicago.edu.
Supporting citations
References (50, 51, 52, 53, 54) appear in the Supporting material.
Supporting material
References
- 1.Breitsprecher D., Goode B.L. Formins at a glance. J. Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courtemanche N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018;10:1553–1569. doi: 10.1007/s12551-018-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otomo T., Otomo C., Rosen M.K. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Kovar D.R., Pollard T.D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul A.S., Pollard T.D. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero S., Le Clainche C., Carlier M.-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 7.Vavylonis D., Kovar D.R., Pollard T.D. Model of formin-associated actin filament elongation. Mol. Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgs H.N., Peterson K.J. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivero F., Muramoto T., Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schönichen A., Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta. 2010;1803:152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Kovar D.R., Sirotkin V., Lord M. Three’s company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21:177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feierbach B., Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 13.Nakano K., Imai J., Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- 14.Chang F., Drubin D., Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen J., Nielsen O., Hagan I.M. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen J., Weilguny D., Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidali L., van Gisbergen P.A.C., Bezanilla M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA. 2009;106:13341–13346. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goode B.L., Eck M.J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 19.Kovar D.R., Harris E.S., Pollard T.D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Scott B.J., Neidt E.M., Kovar D.R. The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell. 2011;22:3826–3839. doi: 10.1091/mbc.E11-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidt E.M., Skau C.T., Kovar D.R. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moseley J.B., Goode B.L. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 23.Neidt E.M., Scott B.J., Kovar D.R. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- 24.Pollard T.D., Wu J.-Q. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante C., Huang F., Pollard T.D. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc. Natl. Acad. Sci. USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee I.-J., Coffman V.C., Wu J.-Q. Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton (Hoboken) 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vavylonis D., Wu J.-Q., Pollard T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 28.Wu J.-Q., Sirotkin V., Pollard T.D. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonetani A., Lustig R.J., Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol. Biol. Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidone T.C., Tang H., Vavylonis D. Dynamic network morphology and tension buildup in a 3D model of cytokinetic ring assembly. Biophys. J. 2014;107:2618–2628. doi: 10.1016/j.bpj.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nedelec F., Foethke D. Collective Langevin dynamics of flexible cytoskeletal fibers. New J. Phys. 2007;9:427. [Google Scholar]
- 32.Pasquali M., Shankar V., Morse D.C. Viscoelasticity of dilute solutions of semiflexible polymers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001;64:020802. doi: 10.1103/PhysRevE.64.020802. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann D., Homa K.E., Kovar D.R. Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat. Commun. 2017;8:703. doi: 10.1038/s41467-017-00445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberts J.B. Biophysically realistic filament bending dynamics in agent-based biological simulation. PLoS One. 2009;4:e4748. doi: 10.1371/journal.pone.0004748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isambert H., Venier P., Carlier M.F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 36.Tang H., Laporte D., Vavylonis D. Actin cable distribution and dynamics arising from cross-linking, motor pulling, and filament turnover. Mol. Biol. Cell. 2014;25:3006–3016. doi: 10.1091/mbc.E14-05-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laporte D., Ojkic N., Wu J.-Q. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell. 2012;23:3094–3110. doi: 10.1091/mbc.E12-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keeney J.B., Boeke J.D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bähler J., Wu J.Q., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Christensen J.R., Hocky G.M., Kovar D.R. Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. eLife. 2017;6:e23152. doi: 10.7554/eLife.23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 42.Lu J., Pollard T.D. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen J.R., Homa K.E., Kovar D.R. Cooperation between tropomyosin and α-actinin inhibits fimbrin association with actin filament networks in fission yeast. eLife. 2019;8:e47279. doi: 10.7554/eLife.47279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovar D.R., Kuhn J.R., Pollard T.D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J.-Q., Kuhn J.R., Pollard T.D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 46.Laporte D., Coffman V.C., Wu J.-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Q., Pollard T.D. Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Curr. Biol. 2013;23:1154–1162. doi: 10.1016/j.cub.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr. Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- 49.Courtemanche N., Lee J.Y., Greene E.C. Tension modulates actin filament polymerization mediated by formin and profilin. Proc. Natl. Acad. Sci. USA. 2013;110:9752–9757. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Christensen J.R., …, Kovar D.R. The F-actin bundler α-actinin Ain1 is tailored for ring assembly and constriction during cytokinesis in fission yeast. Mol. Biol. Cell. 2016;27:1821–1833. doi: 10.1091/mbc.E16-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris E.S., Rouiller I., …, Higgs H.N. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 52.Graziano B.R., DuPage A.G., …, Goode B.L. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol. Biol. Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin S.G., Rincón S.A., …, Chang F. Regulation of the formin for3p by cdc42p and bud6p. MBoC. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson M., East D.A., Mulvihill D.P. Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 2014;24:1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative simulations are animated next to graphs of corresponding time series data tracking the ring progress collective variable (see Materials and methods). Actin beads (green) elongate from formins bound to cytokinesis nodes (purple) at a rate of vpol. All runs include mechanoinhibition of formin. Raw data (gray) and 1-minute moving average (black) are shown. A red circle tracks this simulation time point on each graph. Under the same nucleation efficiency and elongation rate conditions, simulation outcomes are comparable whether 65 (left) or 140 nodes (right) are used. High and low values of vpol are compared in the top and bottom rows, respectively.
Representative simulations are animated next to graphs of corresponding time series data tracking the ring progress collective variable (see Materials and methods). Actin beads (green) elongate from formins bound to cytokinesis nodes (purple) at a rate of vpol. The total number of filaments in each simulation, N, controls the nucleation efficiency. All runs come from 65 node simulations and include mechanoinhibition of formin. Raw data (gray) and 1-minute moving average (black) are shown. A red circle tracks this simulation time point on each graph. Left: At a constant elongation rate (vpol = 0.18 μm/s), ring formation occurs at high nucleation efficiency (top=130 filaments). As the nucleation efficiency decreases (middle=100 filaments, bottom=70 filaments), nodes coalesce progressively slower and ring assembly is compromised. Right: At high nucleation efficiency (N = 130), ring assembly is common across a range of values for vpol (top=0.2 μm/s, middle=0.16 μm/s, bottom=0.12 μm/s).
Confocal time-lapse imaging of representative Cdc12endo, mDia2, and For3 formin chimera cells showing movement of cytokinesis nodes during assembly of the contractile ring (Rlc1-tdTomato). Scale bar, 5 μm. Time is indicated in minutes.