Abstract
The chimeric transcription factor Pax3-FKHR, produced by the t(2;13)(q35;q14) chromosomal translocation in alveolar rhabdomyosarcoma, consists of the two Pax3 DNA binding domains (paired box and homeodomain) fused to the C-terminal forkhead (FKHR) sequences that contain a potent transcriptional activation domain. To determine which of these domains are required for cellular transformation, Pax3, Pax3-FKHR, and selected mutants were retrovirally expressed in NIH 3T3 cells and evaluated for their capacity to promote anchorage-independent cell growth. Mutational analysis revealed that both the third α-helix of the homeodomain and a small region of the FKHR transactivation domain are absolutely required for efficient transformation by the Pax3-FKHR fusion protein. Surprisingly, point mutations in the paired domain that abrogate sequence-specific DNA binding retained transformation potential equivalent to that of the wild-type protein. This finding suggests that DNA binding mediated through the Pax3 paired box is not required for transformation. Our results demonstrate that the integrity of the Pax3 homeodomain recognition helix and the FKHR transactivation domain is necessary for efficient cellular transformation by the Pax3-FKHR fusion protein.
Chromosomal translocations, which are hallmarks of both hematopoietic and solid malignancies (28), frequently involve the fusion of two independent genes to encode a novel chimeric protein with aberrant functions. Alveolar rhabdomyosarcoma, characterized by the t(2;13)(q35;q14) translocation (14, 31), is a malignant tumor of skeletal muscle and is one of the most common soft tissue sarcomas of childhood, accounting for more than half of such cases in pediatric patients (27). This translocation creates the Pax3-FKHR fusion gene. This gene, which encodes a protein of 837 amino acids, comprises the 5′ sequences of the Pax3 gene, a member of the paired class homeodomain family of transcription factors (32, 33), and the 3′ sequences of the FKHR gene, which encodes a member of the forkhead family of transcription factors (6). The fusion protein maintains the integrity of the Pax3 DNA binding motifs (paired domain and homeodomain), but the transactivation domain of Pax3 is replaced by the bisected FKHR DNA binding domain and transcriptional activation domain. As a result, the Pax3-FKHR fusion protein exhibits higher transcriptional activity than does Pax3 alone (13), suggesting that the oncogenic potential of Pax3-FKHR results from this gain-of-function mutation.
Many chromosomal translocations result in fusion proteins that involve transcription factors. A DNA binding-independent model has been described to explain the oncogenicity of the fusion protein E2a-Pbx1. In acute lymphoblastic leukemia, the t(1;19)(q23;p13) translocation fuses the N-terminal transcriptional activation domains of E2a to the C-terminal region of the homeodomain protein Pbx1 (21, 26). Transformation by the E2a-Pbx1 fusion protein is dependent on protein-protein interactions via the homeodomain cooperativity motif of Pbx1 (9) and is independent of DNA binding by the homeodomain of Pbx1 (23).
Pax3-FKHR has been demonstrated to transform chicken embryo fibroblasts in culture (30), but the structural requirements for its oncogenicity have not yet been defined. In the present study, we have analyzed the structure-function relationship of the Pax3-FKHR domains necessary for transcriptional activation and transformation. Our results show that paired domain-mediated DNA binding is not necessary for transformation. However, the integrity of the third α-helix of the Pax3 homeodomain, in conjunction with a region of the FKHR transactivation domain, is essential for Pax3-FKHR-mediated oncogenicity. These results suggest a model in which homeodomain-mediated DNA binding, with an additional role of homeodomain-mediated protein-protein interactions, is important for the oncogenic potential of Pax3-FKHR.
MATERIALS AND METHODS
Expression constructs and mutagenesis.
All expression plasmids were constructed in the mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, Calif.). The murine Pax3 cDNA, pBH3.2, kindly provided by Peter Gruss (Max Planck Institute, Göttingen, Germany), encodes a protein that has over 98% homology with human Pax3 and is completely identical in its DNA binding regions (15, 20). The isolation of the Pax3-FKHR fusion protein cDNA was previously reported (14, 31). The Pax3-FKHR point mutants Un-1, BU35, V265A, S268A, N269A, and S268N269A were created by standard PCR site-directed mutagenesis (18). The internal deletion mutants ΔPD-NH2, ΔHD-C, and ΔHD were constructed by overlap extension PCR (19), and the C-terminal deletion mutants Δ772, Δ728, and Δ672 were generated by PCR amplification and standard cloning techniques. All mutants were confirmed by sequence analysis. Further C-terminal truncation proteins (Δ503 and Δ606) were constructed by using restriction enzyme sites ScaI and NdeI at nucleotide positions 1497 and 1805, respectively (Fig. 1). The chloramphenicol acetyl transferase (CAT) reporter construct (pTK-CAT) containing six direct repeats of both the paired domain and homeodomain consensus binding sequences (PRS-9) [(PRS-9) PTKCAT] was a gift from Martyn Goulding (The Salk Institute, San Diego, Calif.).
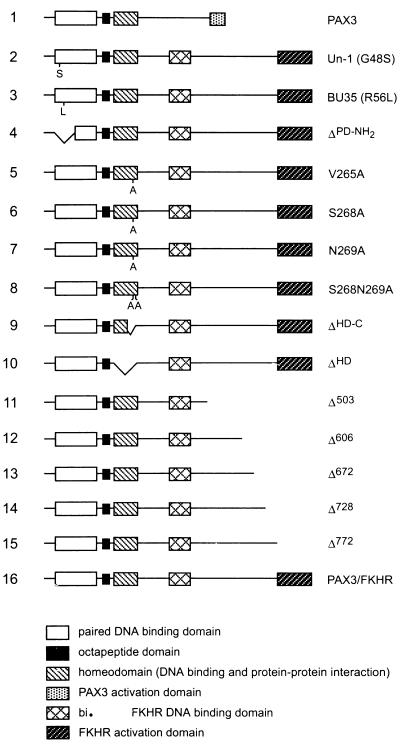
FIG. 1.
Schematic representation of the Pax3, Pax3-FKHR, and Pax3-FKHR mutant proteins. Two mutants containing a single amino acid change in the paired domain were constructed: Un-1 (G48S) and BU35 (R56L), which recapitulate naturally occurring mutations in Pax1 and Pax3, respectively (2, 20). ΔPD-NH2 contains an in-frame deletion of the paired-domain helices that are involved in DNA contacts (Asn53 to Thr93). S268A contains a single amino acid change in the homeodomain, which mediates Pax3 dimerization (12). V265A, N269A, and S268N269A contain single or double amino acid changes in the homeodomain recognition helix that have been shown to make direct DNA contacts (35). ΔHD-C and ΔHD contain in-frame deletions of the homeodomain DNA recognition helix (Glu269 to Lys276) and of the entire homeodomain (Gln219 to Gly279), respectively. Δ772, Δ728, Δ672, Δ606, and Δ503 contain progressive C-terminal deletions in the FKHR transactivation domain (see Materials and Methods. bi., bisected.
Electrophoretic mobility shift assay.
Protein-DNA binding reaction mixtures (20 μl) included 2 μl of in vitro-translated protein (Promega) and 3 μg of poly(dI-dC) in buffer containing 20 mM HEPES (pH 7.6), 50 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol, and 10% glycerol (13). Protein expression was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with detection by autoradiography of [35S]methionine-labeled protein. Unlabeled protein was incubated at room temperature for 20 min with 105 dpm of the oligonucleotide probe PRS-9, which contains both the paired domain and the homeodomain recognition sequences (7); this probe was labeled with [α-32P]dCTP (6,000 Ci/mmol; Amersham, Arlington, Ill.). Incubation of protein with competitor (1,500-fold molar excess of unlabeled PRS-9 probe) or with 1 μl of Pax3 rabbit antiserum was performed at room temperature for 10 min prior to the addition of labeled probe. DNA-protein complexes were resolved on a 6% native polyacrylamide gel, containing 2.5% glycerol in 25 mM Tris-HCl (pH 8.3) and 190 mM glycine, at 35 mA for 1.5 h. Gels were fixed in 10% acetic acid and dried, and labeled oligonucleotides were visualized by autoradiography.
Retroviral stocks and transformation assays.
The various forms of Pax3-FKHR cDNA were cloned into the EcoRI site of the vector pSRα(ΔHindIII)-tk-Neo (29), kindly provided by Charles Sawyers (UCLA Medical School, Los Angeles, Calif.). Retroviral stocks were generated by calcium phosphate cotransfection (16) of 293T cells, kindly provided by David Baltimore (California Institute of Technology, Pasadena, Calif.), with various constructs and an ecotropic packaging plasmid (ψ2), as described previously (24). Culture supernatants containing virus were collected between 36 and 72 h posttransfection, filtered, and subsequently used to infect NIH 3T3 cells, which were demonstrated to be transformed by EWS-FLI, kindly provided by Christopher Denny (University of California at Los Angeles). Soft agar assays were performed by resuspending 2 × 104 cells in 1 ml of 1× Iscove’s modified Dulbecco’s medium containing 15% fetal bovine serum and 0.3% Noble agar (Difco). The cell suspension was then layered over 2 ml of 0.6% bottom agar in 35-mm-diameter dishes. Colonies were scored 2 and 3 weeks after seeding.
Antibody production and immunoprecipitation.
The Pax3 antiserum was produced by immunization of rabbits with a bacterially expressed, His6 epitope-tagged Pax3 protein (a six-histidine-tagged Pax3 protein containing only the paired domain and the homeodomain). To confirm overexpression of the Pax3-FKHR constructs, NIH 3T3 cells infected with high-titer retroviruses encoding either Pax3-FKHR or the Pax3-FKHR mutant derivatives were metabolically labeled at 48 h after infection with [35S]methionine (0.5 mCi/ml; New England Nuclear, Beverly, Mass.) for 2 h, lysed in dissociation buffer (0.5% SDS, 1 mM dithiothreitol, 50 mM Tris-HCl [pH 7.4], 1 mM EDTA), and boiled for 5 min. Cell lysates were diluted fourfold with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% deoxycholic acid, 50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). The lysate supernatants were then centrifuged for 30 min at 4°C at 10,000 × g and incubated with the Pax3 rabbit polyclonal antiserum for 2 h, at 4°C, on a rotary shaker. Immune complexes were collected with protein A-Sepharose (Pharmacia, Piscataway, N.J.) and washed four times with radioimmunoprecipitation assay buffer containing 0.1% SDS. Pellets were resuspended in 30 μl of 2× Laemmli SDS-PAGE buffer, boiled for 5 min, and centrifuged to remove debris. Proteins were resolved on an SDS–10% PAGE gel. The gel was incubated in ENHANCE (New England Nuclear), dried, and autoradiographed at −80°C overnight.
Immunofluorescence.
NIH 3T3 cells were infected with retroviruses encoding either Pax3-FKHR or the Pax3-FKHR mutant derivatives. At 24 h postinfection, 2 × 104 cells were replated onto coverslips in 35-mm-diameter dishes and cultured for an additional 24 h. Cells were fixed with 2% paraformaldehyde for 15 min at room temperature, rinsed twice with phosphate-buffered saline (PBS), and permeabilized for 10 min in 2% paraformaldehyde containing 1% Triton X-100. The fixed cells were incubated for 1 h with a 1:200 dilution of the Pax3 rabbit polyclonal antiserum. After being washed with PBS, the cells were incubated with fluorescein isothiocyanate-conjugated donkey anti-rabbit antibody (1:1,000; Sigma Chemical Co., St. Louis, Mo.) for 30 min, washed with PBS, and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, Calif.). Slides were examined with an Olympus BH2 fluorescent microscope.
Transient transfections and transactivation assays.
NIH 3T3 cells (3 × 105) were plated in 60-mm-diameter dishes and transfected the following day, at approximately 70% confluency, by the Lipofectamine method (Gibco/BRL, Grand Island, N.Y.), in accordance with the manufacturer’s specifications. The Lipofectamine-DNA precipitate was formed in a total of 600 μl of serum-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM glutamine. The precipitate consisted of 1 μg of the (PRS-9)pTK-CAT reporter plasmid described above (7), 250 ng of expression plasmid (containing Pax3, Pax3-FKHR, or the mutant derivatives), and 1 μg of the secreted alkaline phosphatase (SEAP) control plasmid under control of the MAP1 promoter (5). The Lipofectamine-DNA precipitate was incubated on the cell monolayer for 5 h in 3.0 ml of DMEM supplemented with only 2 mM glutamine, after which the cell monolayer was fed with 5 ml of DMEM supplemented with 10% fetal calf serum and 2 mM glutamine. After 48 h, the medium was assayed for SEAP activity, as previously described (5), and the cells were harvested and assayed for CAT activity, as previously described (1). The percentage of [14C]chloramphenicol acetylation was quantitated from thin-layer chromatography plates by using a PhosphorImager (Molecular Dynamics). The transfection efficiency was normalized relative to the SEAP activity. Each experiment was repeated three times in duplicate plates.
RESULTS
Pax3-FKHR mutant constructs and DNA binding activity.
To define the Pax3-FKHR domains required for transformation, we generated a series of Pax3-FKHR mutants (Fig. 1). We made three mutants that disrupt DNA binding through the Pax3 paired domain: Un-1 contains a glycine-to-serine mutation at codon 48 (G48S), which is analogous to the mutation in the Pax1 paired domain of the undulated mouse mutant (2); BU35 contains an arginine-to-leucine mutation at codon 56 (R56L), which has been reported in a family with Waardenburg syndrome type 1 (20); and ΔPD-NH2 has a deletion in the N-terminal α-helices of the paired domain that are involved in making DNA contacts (35). We also constructed six mutants involving the homeodomain: S268A contains a serine-to-alanine mutation within the third α-helix of the homeodomain of Pax3-FKHR, which reduces Pax3 homodimerization upon binding to DNA (12); V265A, N269A, and S268N269A contain point mutations of amino acids within the homeodomain recognition helix that were demonstrated to be involved in making direct DNA contacts (35). ΔHD-C lacks the entire third α-helix from glutamic acid 260 to lysine 276 (ΔE260–K276) of the homeodomain. This helix, called the recognition helix, is believed to mediate homeodomain-DNA contacts, homeodomain-mediated protein-protein interactions, and Pax3 homodimerization on DNA (12, 34). ΔHD represents an in-frame deletion of the entire homeodomain from glutamine 219 to glycine 279 (ΔQ219–G279). Finally, we made progressive C-terminal deletions in the FKHR transactivation domain (Δ772 to Δ503 as shown in Fig. 1).
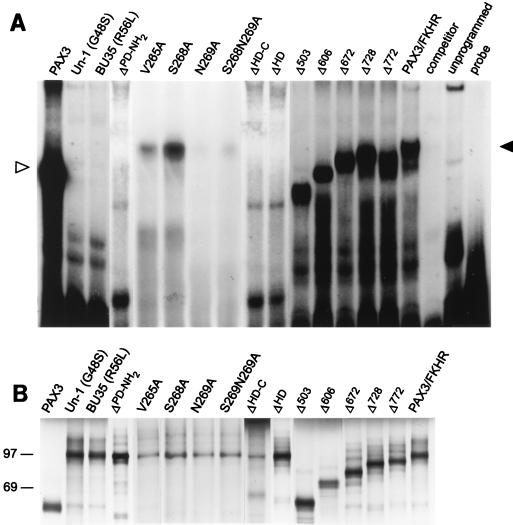
The DNA binding capabilities of the Pax3-FKHR mutant constructs were analyzed by gel retardation analysis. Deletions or point mutations which were introduced into either the DNA binding region of the paired domain (Un-1, BU35, and ΔPD-NH2) or the homeodomain (ΔHD-C and ΔHD) disrupted Pax3-FKHR binding to the PRS-9 probe. In the same manner, mutation of the homeodomain amino acids involved in making direct DNA contacts (V265A, N269A, and S268N269A) greatly reduced the binding capability of Pax3-FKHR to DNA. However, mutation of a single amino acid in the homeodomain (S268A) had no apparent negative effect on the ability of Pax3-FKHR to bind DNA. All mutants with deletions in the forkhead region (Δ503, Δ606, Δ672, Δ728, and Δ772) shifted the probe to a similar extent as wild-type Pax3-FKHR, consistent with the requirement of intact paired and homeobox domains for efficient DNA binding activity (Fig. 2A). Equivalent amounts of in vitro-translated protein were used for this analysis (Fig. 2B).
FIG. 2.
Gel retardation analysis with the paired recognition sequence (PRS-9) oligonucleotide probe, which contains both the paired domain and the homeodomain recognition sequences. (A) Only the Pax3-FKHR mutants containing an intact paired domain and homeodomain are able to efficiently bind to the probe. In vitro-translated Pax3, Pax3-FKHR, or Pax3-FKHR mutant proteins (2 μl) were each incubated with end-labeled PRS-9 oligonucleotide by using the binding conditions described in Materials and Methods. Protein-DNA complexes were separated on a 6% nondenaturing gel. The mobility of the Pax3 protein is indicated by an open arrowhead, and the mobility of the Pax3-FKHR, S268A, Δ772, Δ728 and Δ672 proteins is indicated by a closed arrowhead. Δ606 and Δ503 show a slightly lower mobility than that of Pax3-FKHR, as expected. The Pax3-FKHR and Pax3 protein-DNA complexes could be competed with a molar excess of unlabeled PRS-9 oligonucleotide probe. (B) Pilot in vitro translation reactions were performed with [35S]methionine-labeled proteins to confirm that comparable amounts of Pax3, Pax3-FKHR, and the Pax3-FKHR mutants were synthesized. The labeled proteins were separated by SDS–10% PAGE, and their molecular sizes were estimated by comparison with the rainbow protein molecular size markers (Amersham), whose positions are indicated to the left of the gel. Molecular sizes of the proteins are as follows: Pax3, 56 kDa; Pax3-FKHR, Un-1, BU35, V265A, S268A, N269A, and S268N269A, 97 kDa; ΔPD-NH2, 90 kDa; ΔHD-C, 94 kDa; ΔHD, 90 kDa; Δ503, 56 kDa; Δ606, 67 kDa; Δ672, 75 kDa; Δ728, 81 kDa; and Δ772, 86 kDa.
Oncogenic potential of Pax3-FKHR and its mutant derivatives.
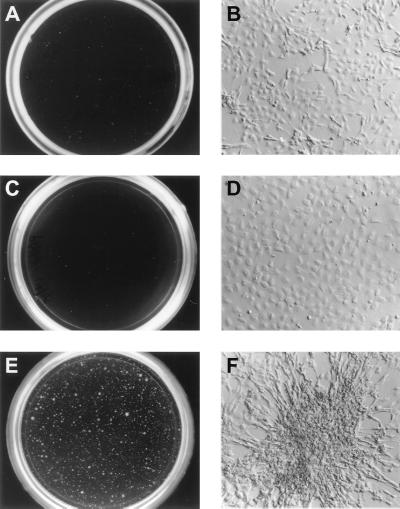
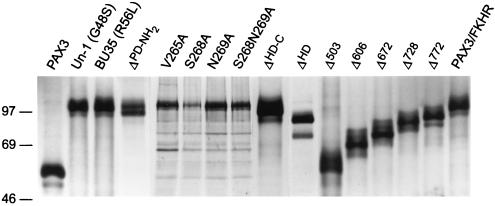
To examine the transforming properties of the Pax3-FKHR protein, wild-type Pax3-FKHR and its mutant derivatives were individually cloned into the retroviral vector pSRα(ΔHindIII)-tk-Neo (29). NIH 3T3 fibroblasts were infected with replication-deficient retroviral stocks and subsequently assayed for their ability to exhibit anchorage-independent growth in soft agar. Cells infected with the Pax3-FKHR virus displayed macroscopically visible colonies in soft agar (Fig. 3B and 4E) and possessed a highly refractile, spindle-shaped morphology under cell culture conditions (Fig. 4F). In contrast, cells infected with either the vector (Fig. 3A and 4A) or with Pax3 alone (Fig. 4C) showed no visible colony formation and displayed a flat morphology when grown on plastic (Fig. 4B and D, respectively). Immunoprecipitation with the Pax3 antiserum of Pax3-FKHR and the Pax3-FKHR mutant derivatives from metabolically labeled cell lysates confirmed that all cells expressed similar amounts of mutant proteins and that none of the mutations interfered with the expression of the Pax3-FKHR fusion (Fig. 5).
FIG. 3.
Soft agar assays to assess the tumorigenicity of Pax3-FKHR and the Pax3-FKHR mutants. NIH 3T3 fibroblasts were infected with high-titer retroviral stocks containing pSRα(ΔHindIII)-tk-Neo (A), Pax3-FKHR (B), Un-1 (C), ΔPD-NH2 (D), ΔHD-C (E), ΔHD (F), Δ728 (G), or Δ772 (H) and cloned in 0.3% soft agar. Colonies were visualized and counted 2 to 3 weeks after plating.
FIG. 4.
Cell morphology of NIH 3T3 cells infected with high-titer retroviral stocks containing pSRα(ΔHindIII)-tk-Neo (A and B), Pax3 (C and D), or Pax3-FKHR (E and F). Cells were grown on plastic, and colonies were visually scored 2 and 3 weeks after plating. Colonies shown are representative of the total population.
FIG. 5.
Overexpression of Pax3, Pax3-FKHR, and Pax3-FKHR deletion mutants in retrovirally infected NIH 3T3 fibroblasts. Retrovirally infected NIH 3T3 cells were metabolically labeled with [35S]methionine and lysed as described in Materials and Methods. Cell lysates were immunoprecipitated with Pax3 antiserum, separated on an SDS–10% PAGE gel, and visualized by autoradiography. Molecular sizes were estimated by comparison with protein molecular size markers (see legend to Fig. 2), which are indicated, in kilodaltons, to the left of the gel.
Surprisingly, cells that overexpressed mutant proteins that disrupt paired domain-mediated DNA binding (Un-1, BU35, and ΔPD-NH2) retained the ability to transform NIH 3T3 cells and did so with an efficiency similar to that of the wild-type Pax3-FKHR (Fig. 3C and D; see Fig. 7). To confirm that residual DNA binding activity from the bisected forkhead DNA-binding domain does not contribute to the transforming ability of the paired domain mutants, we constructed a recombinant retroviral vector containing only the forkhead portion of Pax3-FKHR. Cells expressing this bisected forkhead protein were not transformed (data not shown). This result suggested that Pax3 sequences outside of the paired domain were necessary for the oncogenic potential of Pax3-FKHR and that the transforming ability of the chimeric protein was independent of paired domain-mediated DNA binding.
FIG. 7.
Transformation and transcriptional activity of Pax3, Pax3-FKHR, and Pax3-FKHR mutants. Pax3, Pax3-FKHR, and Pax3-FKHR mutant proteins (as described in the legend to Fig. 1) are represented schematically (left column). To determine transformation (right column), NIH 3T3 fibroblasts were infected with high-titer retroviral stocks containing Pax3, Pax3-FKHR, or Pax3-FKHR mutant proteins and grown on 0.3% soft agar. Cell colonies were counted 2 to 3 weeks after plating. The values represent the number of colonies present on a 35-mm-diameter dish and an average of two independent determinations. To determine transactivation (Transact.) activity (right column), NIH 3T3 cells were transfected with 1 μg of the (PRS-9) pTK-CAT reporter construct, 1 μg of MAP1-SEAP, and 250 ng of an expression plasmid encoding either Pax3, Pax3-FKHR, or a Pax3-FKHR mutant protein. Quantification of the CAT activity was performed as described in Materials and Methods. Transfection efficiency was normalized relative to SEAP activity as described previously (5). The CAT activity in the presence of Pax3-FKHR was assigned a value of 100%, and all other activities are reported relative to this value. The error bars represent the standard deviation from the average value of three individual experiments performed in duplicate.
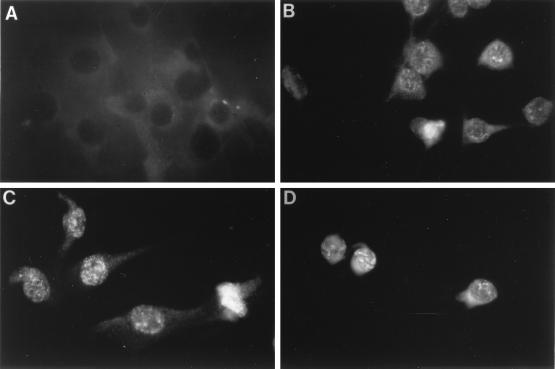
This conclusion is in keeping with the fact that cells overexpressing proteins with deletions in the homeodomain (ΔHD-C and ΔHD) were unable to transform NIH 3T3 cells (Fig. 3E and F and see Fig. 7). This finding suggests that deletion of all or part of the homeodomain removes an element required for efficient cellular transformation. Because deletion of the homeodomain removes the last two amino acids of the Pax3 nuclear localization signal (11), immunofluorescence was used to confirm that the ΔHD mutant protein was nuclear. Cells overexpressing the Pax3, ΔHD, and Pax3-FKHR proteins demonstrated strong nuclear staining consistent with proper localization of these proteins (Fig. 6).
FIG. 6.
Determination of the nuclear localization of Pax3, Pax3-FKHR, and ΔHD by immunofluorescence. NIH 3T3 cells (2 × 104) infected with either pSRα(ΔHindIII)-to-Neo (A), Pax3 (B), ΔHD (C), or Pax3-FKHR (D) were plated on glass coverslips. After 48 h, the cells were fixed, permeabilized, immunostained with Pax3 antiserum, and incubated with fluorescein isothiocyanate-conjugated donkey anti-rabbit antibody, as described in Materials and Methods.
The third α-helix of the homeodomain is involved in making both protein-DNA and protein-protein interactions (12, 34). To determine which of these interactions is important for transformation, cells overexpressing proteins with point mutations of homeodomain amino acids that are involved in making DNA contacts were tested for their oncogenic potential in soft agar assays. These point mutations either destroyed the transforming ability of Pax3-FKHR (N269A and S268N269A) (Fig. 7) or demonstrated a decreased transforming ability with a reduced colony size (V265A) (Fig. 7 and data not shown). The oncogenic potentials of these mutants appeared to be directly related to their respective DNA binding abilities (Fig. 2A), suggesting that homeodomain-mediated DNA binding is important for efficient transformation. However the S268A mutant demonstrated wild-type transforming ability, indicating that Pax3 homodimerization was not necessary for Pax3-FKHR-mediated oncogenicity (Fig. 7).
The FKHR deletion mutant Δ772 maintained near-wild-type transforming ability but produced colonies that were slightly smaller than those produced by cells retrovirally infected with Pax3-FKHR (Fig. 3H and 7). This transforming ability was completely lost, however, on further deletion of the FKHR protein (Fig. 3G and 7), demonstrating that the oncogenicity of Pax3-FKHR requires a region of the FKHR transactivation domain between amino acids 728 and 772.
Transcriptional activity of Pax3-FKHR mutants.
To relate transformation to transcriptional activity, wild-type Pax3-FKHR and its mutant derivatives were tested in transient-transfection assays using a Pax3-responsive reporter plasmid (see Materials and Methods). Pax3-FKHR demonstrated approximately ninefold greater transcriptional activity than Pax3 did (Fig. 7), consistent with previous reports (3, 13). Proteins mutated in the paired domain and deletion mutants of the homeodomain of Pax3 (Un-1, BU35, ΔPD-NH2, ΔHD, and ΔHD-C), which had no detectable DNA binding ability, were all transcriptionally inactive in NIH 3T3 cells (Fig. 7). Given that the S268A mutant retains its DNA binding potential (Fig. 2A), these results are consistent with the requirement of DNA binding for transcriptional activity. Surprisingly, proteins with point mutations in the homeodomain (V265A, N269A, and S268N269A), which retained only residual DNA binding capability, maintained nearly wild-type transcriptional activity. This finding suggests that in addition to affecting the DNA binding abilities of Pax3-FKHR, these point mutants may also interfere with the reported inhibitory action of the homeodomain (4) (see below).
The Δ772 FKHR mutant retained approximately 35% of wild-type transcriptional activity (Fig. 7) even though it lacked the reported transactivation domain (3). Thus, the complete transactivation domain may extend more N-terminally than was previously thought. Progressive FKHR deletion mutants showed even less transcriptional activity than Δ772, with Δ606 and Δ503 being transcriptionally inactive (Fig. 7), indicating that the FKHR transactivation domain extends N-terminally to a region between amino acids 606 and 672. These results demonstrate that both DNA binding and an intact FKHR transactivation domain are necessary for complete Pax3-FKHR transcriptional activity.
DISCUSSION
The oncogenic potential of Pax3-FKHR is independent of its transcriptional activity.
By creating a series of mutations targeting key structural regions of Pax3-FKHR, we have identified the domains necessary for transformation and transactivation by the fusion protein. We found that both the integrity of the third α-helix of the Pax3 homeodomain and a small region of the FKHR transactivation domain are absolutely required for effective transformation of NIH 3T3 mouse fibroblasts. However, the paired domain-mediated DNA binding is dispensable.
The observed oncogenic potential of Pax3-FKHR represents a gain-of-function mutation of Pax3 because neither Pax3 nor FKHR alone was transforming when overexpressed in rodent fibroblasts. The inability of Pax3 to transform NIH 3T3 cells in this study is inconsistent with previous reports in which Pax3 was found to be oncogenic (22). This apparent inconsistency may be attributable to differences between the two NIH 3T3 cell lines. Overexpression of the Pax3-FKHR fusion protein may be required to deregulate Pax3 target genes or to effectively compete for interactions between Pax3 and other proteins. This model would be consistent with a recent report that the t(2;13)(q35;q14) translocation is associated with overexpression of the fusion transcript (10). Such events might then establish an overall growth advantage for transformed cells.
We demonstrate that the majority of the transcriptional activity of Pax3-FKHR lies within the FKHR transactivation domain between amino acids 728 to 837, confirming a previous study that mapped the transactivation domain between amino acids 764 and 837 (3). However, the transcriptional activity of Pax3-FKHR does not appear to be directly responsible for the transforming capacity of the chimeric protein. First, there is no direct correlation between the oncogenic potentials of the FKHR deletion mutants and their respective transcriptional activities (Fig. 7, compare Δ772 and Δ728). Second, mutants of the paired box (UN-1, Bu35, and ΔPD-NH2) that are transcriptionally inactive exhibit a wild-type oncogenic potential. Finally, point mutant proteins of the homeodomain (N269A and S268N269A) that have lost their transforming ability retain nearly wild-type transcriptional activity. We therefore conclude that Pax3-FKHR transforming ability is largely independent of transcriptional activity.
Although it may seem unusual that homeodomain point mutants with minimal DNA binding (V265A, N269A, and S268N269A) transactivate with near-wild-type activity, this result is consistent with a previous report which identified a putative transcriptional inhibitory function associated with the Pax3 homeodomain (4). This inhibitory action is presumably mediated through protein-protein interactions. In the absence of structural data, the possible effect of the homeodomain point mutations on protein-protein interactions could not be determined. Therefore, in addition to decreasing the DNA binding ability of Pax3-FKHR, these point mutants may also disrupt protein-protein interactions, interfering with the reported inhibitory action of the homeodomain and resulting in near-wild-type transcriptional activity.
Pax3 homeodomain-mediated DNA binding is essential for transformation.
The two point mutants BU35 and Un-1 recapitulate naturally occurring mutations in the paired domains of Pax3 and Pax1, respectively (2, 20). These point mutations occur in the region of the paired domain that is involved in making DNA contacts (35) and prevent Pax3 and Pax1 from binding DNA (22). Introduction of these point mutations into the chimeric protein, as well as deletion of the region of the paired domain that is involved in making DNA contacts (ΔPD-NH2), destroyed the ability of Pax3-FKHR to bind DNA. Surprisingly, these mutants transformed cells as efficiently as did wild-type Pax3-FKHR. From this we conclude that Pax3 paired domain-mediated DNA binding is not required for the transformation by the fusion protein.
In contrast, removal of the third α-helix of the homeodomain, or removal of the entire homeodomain, prevents not only DNA binding and transcriptional activity but also transformation by Pax3-FKHR. In addition, mutation of key amino acids that are critical for making homeodomain-mediated DNA contacts greatly reduced DNA binding by Pax3-FKHR and either reduced (V265A) or destroyed (N269A and S268N269A) transformation by Pax3-FKHR. Therefore, homeodomain-mediated DNA binding is essential for the oncogenic potential of the chimeric protein.
Transformation requires the proximity of a small region of the FKHR transactivation domain.
Mutant proteins containing an intact FKHR transactivation domain and a mutated homeodomain (N269A, S268N269A, ΔHD-C, and ΔHD) or an intact homeodomain and a mutated FKHR transactivation domain (Δ728, Δ672, Δ606, and Δ503) were unable to transform NIH 3T3 cells, indicating that both the third α-helix of the Pax3 homeodomain and an element that lies between amino acids 728 and 772 of the FKHR transactivation domain must be present for efficient transformation by Pax3-FKHR. This gain-of-function mutation, in which transformation activity is critically dependent on the presence of two independent, nontransforming domains being brought into proximity in the fusion protein, is similar to the mechanism by which another homeodomain-containing fusion protein, E2a-Pbx1, induces transformation (26). E2a-Pbx1-mediated transformation is dependent on the homeodomain cooperativity motif, a small region proximal to the homeodomain in Pbx1 (9), and on two transcriptional activation domains in E2a being brought close to one another in the fusion protein (23).
The additional role of homeodomain-mediated protein-protein interactions.
In addition to making homeodomain-mediated DNA contacts, the Pax3 recognition helix also mediates protein-protein interactions (34). Several pieces of evidence support an additional role of protein-protein interactions in the oncogenic potential of Pax3-FKHR. First, Pax3-FKHR mutants that are unable to bind DNA (UN-1, BU35, and ΔPD-NH2) exhibit a wild-type oncogenic potential. These mutants contain the homeodomain structural components necessary for protein-protein interactions that would enable them to transform NIH 3T3 fibroblasts in the absence of DNA binding. Second, the reported inhibitory action of the homeodomain is no longer present in the nontransforming homeodomain point mutants N269A and S268N269A. This indicates that these point mutants disrupt homeodomain-mediated protein-protein interactions that are necessary for the inhibitory action of the homeodomain and which may be necessary for Pax3-FKHR’s oncogenic potential. Finally, protein-protein interactions have been implicated in the regulation of the biological activity of several homeodomain-containing proteins. For example, the homeodomain protein Msx-2 represses transcription by interacting with the basal transcriptional machinery in a DNA binding-independent manner (25). The Drosophila homeodomain protein fushi tarazu, through specific protein-protein interactions with the orphan nuclear receptor Ftz-F1, mediates the regulation of the genes engrailed and wingless (17). Finally, protein interactions between the homeodomain protein Pbx and several Hox proteins modulate the DNA binding specificity and transcriptional regulation of the Hox proteins (8). Therefore, protein-protein interactions, mediated through the homeodomain, may assist in the transforming ability of Pax3-FKHR.
Our results conclusively demonstrate the necessity of the integrity of the Pax3 homeodomain recognition helix and a small region of the FKHR transactivation domain for the efficient transformation by Pax3-FKHR. The oncogenic potential of Pax3-FKHR requires both homeodomain-mediated protein-DNA and protein-protein interactions and the proximity of the Pax3 homeodomain to the essential transforming element of the FKHR transactivation domain. We propose a model in which the Pax3 homeodomain recognition helix would be responsible for directing DNA contacts. The recognition helix would then recruit cellular proteins that, in conjunction with the FKHR transactivation domain, would improperly regulate Pax3 target genes. No binding partners for either Pax3 or FKHR have yet been identified, but the identification of such proteins will be essential to our understanding of how Pax3-FKHR transforms cells.
ACKNOWLEDGMENTS
We thank Peter Gruss, Max Planck Institute, for the murine Pax3 cDNA, pBH3.2; Charles Sawyers, UCLA Medical Center, for the pSRα(ΔHindIII)-tk-Neo retroviral vector; Martyn Goulding, the Salk Institute for Biological Studies, for the pTK-CAT vector; David Baltimore, California Institute of Technology, for the 293T cells; and Christopher Denny, University of California at Los Angeles, for the NIH 3T3 cells. We also thank Carol Bockhold, Craig McPherson and Rose Mathew for their expert technical assistance. Finally, we thank Suzanne Baker, Thomas Curran, Gerard Grosveld, and Susan Watson for their critical reading of the manuscript.
This work was supported, in part, by NIH grants CA-56819 and PO1-CA-71907 (M.F.R.), the Cancer Center (CORE) support grant CA21765, and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Boston, Mass: John Wiley & Sons; 1996. [Google Scholar]
- 2.Balling R, Deutsch U, Gruss P. Undulated, a mutation affecting the development of the mouse skeleton, has a point mutation in the paired box of Pax1. Cell. 1988;55:531–535. doi: 10.1016/0092-8674(88)90039-6. [DOI] [PubMed] [Google Scholar]
- 3.Bennicelli J L, Fredericks W J, Wilson R B, Rauscher III F J, Barr F G. Wild type Pax3 protein and the Pax3-FKHR fusion protein of alveolar rhabdomyosarcoma contain potent, structurally distinct transcriptional activation domains. Oncogene. 1995;11:119–130. [PubMed] [Google Scholar]
- 4.Bennicelli J L, Edwards R H, Barr F G. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1996;93:5455–5459. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braum R J, Hung D T, Martin P K, Schreiber S L, Crabtree G R. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan R G. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell. 1993;74:773–776. doi: 10.1016/0092-8674(93)90456-z. [DOI] [PubMed] [Google Scholar]
- 7.Chalepakis G, Fritsch R, Fickenscher H R, Deutsch U, Goulding M, Gruss P. The molecular basis of the undulated/Pax1 mutation. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- 8.Chang C-P, Brocchieri L, Shen W-F, Largman C, Cleary M L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C-P, de Vivo I, Cleary M L. The Hox cooperativity motif of the chimeric oncoprotein E2a-Pbx1 is necessary and sufficient for oncogenesis. Mol Cell Biol. 1997;17:81–88. doi: 10.1128/mcb.17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis R J, Barr F G. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall C, Laskey R. The nuclear membrane. Science. 1992;258:942–947. doi: 10.1126/science.1439805. [DOI] [PubMed] [Google Scholar]
- 12.Epstein J A, Lam P, Jepeal L, Maas R L, Shapiro D N. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 13.Fredericks W J, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr F G, Rauscher F J I., III The Pax3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcoma is a more potent transcriptional activator than Pax3. Mol Cell Biol. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher III F J, Emanuel B S, Rovera G, Barr F G. Fusion of a forkhead domain gene to Pax3 in the solid tumor alveolar rhabdomyosarcoma. Nature. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 15.Goulding M D, Chalepakis G, Deutsch U, Erselius J R, Gruss P. Pax3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 17.Guichet A, Copeland J W R, Erdelyi M, Hlousek D, Zavorsky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:552–555. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 18.Ho S, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 20.Hoth C F, Milunsky A, Lipsky N, Sheffer R, Clarren S K, Baldwin C T. Mutations in the paired domain of the human Pax3 gene cause Klein-Waardenburg Syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I) Am J Hum Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- 21.Kamps M P, Murre C, Sun X-H, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 22.Maulbecker C C, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12:2361–2367. doi: 10.1002/j.1460-2075.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monica K, LeBrun D P, Dedera D A, Brown R, Cleary M L. Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol Cell Biol. 1994;14:8304–8314. doi: 10.1128/mcb.14.12.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller A J, Young J C, Pendergast A-M, Pondel M, Landau N R, Littman D R, Witte O N. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive leukemias. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newberry E P, Latifi T, Battaile J T, Towler D A. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry. 1997;36:10451–10462. doi: 10.1021/bi971008x. [DOI] [PubMed] [Google Scholar]
- 26.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 27.Pappo A S, Shapiro D N, Crist W M, Maurer H M. Biology and therapy of pediatric rhabdomyosarcoma. J Clin Oncol. 1995;13:2123–2139. doi: 10.1200/JCO.1995.13.8.2123. [DOI] [PubMed] [Google Scholar]
- 28.Rabbitts T H. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 29.Roussel M F, Theodoras A M, Pagano M, Sherr C J. Rescue of defective mitogenic signaling by D-type cyclins. Proc Natl Acad Sci USA. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheidler S, Fredericks W J, Rauscher III F J, Barr F G, Vogt P K. The hybrid Pax3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci USA. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro D N, Sublett J E, Li B, Downing J R, Naeve C W. Fusion of Pax3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–5112. [PubMed] [Google Scholar]
- 32.Strachan T, Read A P. Pax genes. Curr Opin Gen Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 33.Stuart E T, Kioussi C, Gruss P. Mammalian Pax genes. Annu Rev Genet. 1993;27:219–236. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- 34.Wilson D S, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 35.Xu W, Rold M A, Jun S, Desplan C, Pabo C O. Crystal structure of a paired domain-DNA complex at 2.5 angstrom resolution reveals structural basis of Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]