Abstract
Pak1 protein kinase of Schizosaccharomyces pombe, a member of the p21-GTPase-activated protein kinase (PAK) family, participates in signaling pathways including sexual differentiation and morphogenesis. The regulatory domain of PAK proteins is thought to inhibit the kinase catalytic domain, as truncation of this region renders kinases more active. Here we report the detection in the two-hybrid system of the interaction between Pak1 regulatory domain and the kinase catalytic domain. Pak1 catalytic domain binds to the same highly conserved region on the regulatory domain that binds Cdc42, a GTPase protein capable of activating Pak1. Two-hybrid, mutant, and genetic analyses indicated that this intramolecular interaction rendered the kinase in a closed and inactive configuration. We show that Cdc42 can induce an open configuration of Pak1. We propose that Cdc42 interaction disrupts the intramolecular interactions of Pak1, thereby releasing the kinase from autoinhibition.
The p21-GTPase-activated protein kinase (PAK) family is present in all eukaryotes. Genetic evidence suggests that STE20, one of three Saccharomyces cerevisiae homologs of PAK, mediates signaling of pheromone response from receptor-coupled heterotrimeric G proteins to the mitogen-activated protein kinase (MAPK) cascade, which includes STE11, STE7, and the pair FUS3 and KSS1 (13, 14, 28). STE20 can phosphorylate STE11 in vitro (25, 36). Another homolog, CLA4, appears to regulate normal localization of cell growth and cytokinesis (7), and a third, SKM1, has broad functions in morphogenesis and growth (20). In the fission yeast, Schizosaccharomyces pombe, Pak1 (also known as Shk1) seems to be involved in both sexual differentiation and morphogenesis (17) and has a structural and functional homolog, Shk2 (26, 37). Pak1 has been shown to release the intramolecular and, presumably, autoinhibitory interactions of Byr2, the S. pombe homolog of STE11 (31). Mammalian PAK proteins have three major isoforms, and they appear to be mediators of signaling from members of the p21-GTPase family such as Rac1 and Cdc42 to the MAPKs including Jun kinase and p38 MAPKs (1, 3, 6, 11, 23, 27, 38).
All PAKs have an N-terminal regulatory domain and a conserved C-terminal kinase catalytic domain. The regulatory domains are poorly conserved except for a 70-amino-acid stretch, named CRIB (Cdc42-Rac interactive binding) domain, which is known to bind the small Rho-family GTPases (4). Cdc42 can activate PAK proteins in vitro, inducing a PAK autophosphorylation event (16). Two mechanistic models are consistent with the in vitro biochemical data: Cdc42-Rac directly induces an active conformation of the catalytic region, or the GTPases antagonize an autoinhibitory mechanism.
We have been utilizing genetic analysis and the two-hybrid system of Fields and Song (8) to probe the regulatory mechanisms of kinases in the RAS signaling pathways of yeast and mammalian systems (2, 5, 17, 18, 31, 32, 35). Byr2, one of the S. pombe Ras1 effectors that is required for sexual differentiation, has been analyzed in this way (31). The regulatory domain of Byr2 was found to bind to the kinase catalytic domain, and mutants in the regulatory domain that abolish this interaction were activating. Two-hybrid analysis has shown that this autoinhibitory intramolecular interaction also keeps the kinase in a closed configuration. With further analysis, we demonstrated that dominant activated Pak1 induced the open configuration of Byr2. Previous studies had strongly suggested a role for Pak1 in the integrity of the sexual differentiation pathways (17).
Using methods similar to those we have described previously, we have discovered an intramolecular interaction between the regulatory and catalytic domains of Pak1. The catalytic domain binds to the same highly conserved region on the regulatory domain that also binds Cdc42, and we have shown that wild-type Pak1 exists in a closed configuration with the kinase catalytic domain masked. We used these observations to isolate Pak1 mutants that are in an open configuration, with an accessible catalytic domain. Binding analysis of the regulatory domains of these Pak1 mutants has shown that they all have lost the ability to bind the catalytic domain. These results demonstrate that the intramolecular interaction keeps the kinase in a closed configuration. Moreover, in three different genetic assays, we have shown that most of these Pak1 mutants are more active than the wild-type kinase. Therefore, an autoinhibitory role for the intramolecular interaction is strongly suggested. Consistent with the in vitro result that Cdc42 induces PAK autophosphorylation (16), we have found that Cdc42 can induce the open configuration of Pak1 in vivo. Based on the conservation among PAK proteins, we propose that kinase autoinhibition and Cdc42 release of autoinhibition are general regulatory mechanisms for these protein kinases.
MATERIALS AND METHODS
Yeast, media, and genetic manipulations.
S. cerevisiae L40, a lexA-based two-hybrid reporter strain with both HIS3 and lacZ as reporter genes (33), was used to study two-hybrid interactions. AN43-5A has a FUS1-lacZ reporter system and was used to measure the activity of the S. cerevisiae mating signaling pathway (17). S. cerevisiae cultures were grown in YPD (2% peptone, 1% yeast extract, 2% glucose) or in dropout (DO) synthetic minimal medium (0.67% yeast nitrogen base without amino acids, 2% glucose) with appropriate auxotrophic supplements. The lithium acetate protocol was used for yeast transformation (12).
Generating Pak1 and Cdc42 clones.
PCR (24) was used to generate all constructs. Pak1-Cat, the kinase catalytic domain of Pak1 that encodes the C-terminal 385 amino acids, was made previously (31). Pak1-Reg, which encodes the N-terminal 284 amino acids, was made with the following pair of oligonucleotides (boldfacing indicates restriction enzyme sites): AAGGATCCGATGGAAAGAGGGACTTTACAA, which contains a BamHI site, and GGGGGGTTGTCGACTAGCATTAGAGGTAGTAGTTTTAAC, which contains a SalI site. The PCR product was digested with BamHI and SalI and cloned into pGAD and pLBD vectors. Full-length Pak1 was made by fusing Pak1-Reg to the C-terminal 375 amino acids of Pak1, which was generated by PCR with the following pair of oligonucleotides: CCCCCCAGTCGACAACCTTCTCCATTAGTTTCCAGCAAG and AAGGATCCCTGCACGTATTTACCAGAATGATGTATGGA. The 658-amino-acid full-length Pak1 clone thus had a new SalI site but was identical to wild-type full-length Pak1 at the amino acid level. Cdc42wt and Cdc42V12 were made by PCR with the primer pair GGGGATCCGATGCCCACCATTAAGTGTGTCGTAGTA, which contains a BamHI site, and CCCTTGGGTCGACTGCAGTTACAGTACCAAACACTTTGACTTTTT, which contains an overlapping SalI site and a PstI site. The templates for the PCRs were pREP-Cdc42wt and pREP-Cdc42V12 (kindly provided by Doug Johnson, University of Vermont). Cdc42 sequences were cloned into pGAD and pLBD. Cdc42 clones with a C189S mutation were made by PCR with the primer pair GGGGATCCGATGCCCACCATTAAGTGTGTCGTAGTA, which contains a BamHI site, and CCCCGTCGACAGTACCAAAGACTTTGACTTTTTCTTGTGAGGAAC, which contains the C189S mutation and a SalI site. Cdc42C189S sequences were cloned into pGAD, pLBD, and pLS104.
Detection of protein complex formation by the yeast two-hybrid system.
To determine if GAD fusions interact with LBD fusions in the two-hybrid system, the two fusions were transformed into L40 by the standard lithium acetate yeast transformation procedure. Cells were plated onto synthetic medium lacking leucine and tryptophan (DO-LT). Transformants were patched out on fresh DO-LT plates and examined for histidine prototrophy and β-galactosidase synthesis, since two-hybrid interactions result in transactivation of lexA-HIS3 and lexA-lacZ. Histidine prototrophy was tested by replicating patches onto medium lacking leucine, tryptophan, and histidine (DO-LTH) and was evident by growth on the His− plates. β-Galactosidase filter assay and liquid assay were conducted as previously described (32). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used as the substrate in the β-galactosidase filter assay. o-Nitrophenyl-β-d-galactopyranoside (ONPG) was used as the substrate in the β-galactosidase liquid assay for quantitative measurement.
Making Pak1 regulatory segment fusions by PCR.
Four Pak1 regulatory segments, each about 70 amino acids long, were made by PCR. The DNA fragment that encodes the first 70 amino acids of Pak1 was made by PCR with the oligonucleotide pair of AAGGATCCGATGGAAAGAGGGACTTTACAA and GGGGGTCGACTAGGATTGAGATAAAGGGAAACCGGA; the second 70-amino-acid fragment was made with the oligonucleotide pair of GTGGATCCAATGCGTACAACTGTATCTAGGGTTTCA and GGGGGTCGACTAGCTGGCAGAGCCTGACCCATAGGA; the third 70-amino-acid fragment was made with the oligonucleotide pair of GTGGATCCAATGCCTCGCAAATCGACTGTCATCTCT and GGGGGTCGACTAAAGATATTTCTTGGATTGGGAATA; and the last fragment, which is 74 amino acids long, was made with the oligonucleotide pair of GTGGATCCAATGGAGGAGGGAGCAAAGCCACCCTTT and GGGGGGTTGTCGACTAGCATTAGAGGTAGTAGTTTTAAC. The fragments were excised with BamHI and SalI and cloned into pGAD.
To map more precisely the domains mediating Cdc42 and Pak1-Cat interaction, we generated further segment fusions within the stretch of amino acids 141 to 210. Segments starting from amino acids 149, 153, 157, and 161 were made with the oligonucleotides GTGGATCCAATGTCTCCATTTGATCCGAAGCATGTC, GTGGATCCAATGCCGAAGCATGTCACTCACGTTGGT, GTGGATCCAATGACTCACGTTGGTTTTAATTATGAT, and GTGGATCCAATGTTTAATTATGATACTGGGGAATTT, respectively. The segments ending at amino acids 194, 198, 202, and 206 were made with the oligonucleotides GGGGGTCGACTACTGTGGAGTTTGTACTTGTTCCGA, GGGGGTCGACTAGTCCAAAACGGCCTGTGGATGTTG, GGGGGTCGACTAAAAAGCCATAGCGTCCAAAACGGC, and GGGGGTCGACTAGGATTGGGAATAAAAAGCCATAGC, respectively. The PCR products were excised with BamHI and SalI and cloned into the pGAD vector.
Creating and screening two-hybrid mutant libraries.
We constructed a library of Pak1 regulatory mutants by PCR mutagenesis of this region (40). We used the oligonucleotide pair AAGGATCCGATGGAAAGAGGGACTTTACAA and GGGGGGTTGTCGACTAGCATTAGAGGTAGTAGTTTTAAC, described above, to amplify and mutagenize wild-type Pak1 template. The PCR product was gel purified and digested with BamHI and SalI, and full-length Pak1 was reconstructed by ligation of the PCR products into the LBD fusion vector containing the C-terminal 375 amino acids of Pak1, as we described above. This mutant library had a complexity of over 104.
For screening, the pLBD-Pak1 mutant library was transformed into L40 containing pGAD-Pak1-Reg. Cells were plated onto DO-LTH to select for interacting pairs. A total of 3 × 104 clones were screened, and His+ transformants were patched out on fresh DO-LT for β-galactosidase filter assays. Twenty-five independent clones were both His+ and LacZ+. pLBD fusion plasmids were recovered, amplified, and tested individually with GAD-Pak1-Reg and GAD for binding specificity and reproducibility. Nineteen independent clones were found to bind Pak1-Reg specifically.
Recovery and amplification of plasmids from yeast cells.
To recover plasmids from yeast cells of interest, the yeast cells were collected and resuspended in 200 μl of lysis buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 0.1 M NaCl, 0.01 M Tris [pH 8], 0.001 M EDTA) and vortexed with equal volumes of glass beads and phenol-chloroform-isoamyl alcohol (25/24/1 [vol/vol/vol]) at 4°C for 5 min. After vortexing, cell extracts were centrifuged for 10 min, and the supernatants were used for electroporation into Escherichia coli. Plasmids were extracted from E. coli by standard DNA preparation procedures (Qiagen).
RESULTS
A conserved region of the Pak1 regulatory domain interacts with the catalytic domain.
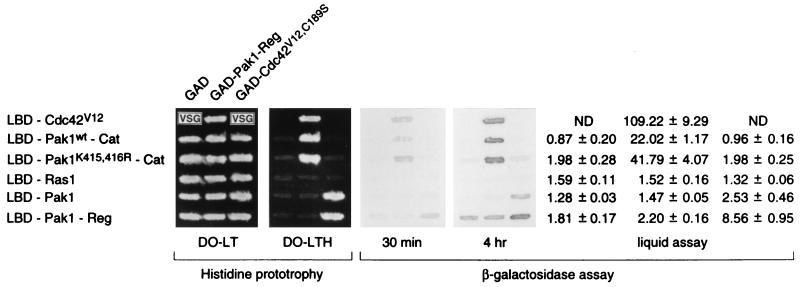
Many protein kinases have a regulatory domain that binds to and inhibits the kinase catalytic domain (29, 31), and we tested if Pak1 has domains capable of such intramolecular interaction, detectable by two-hybrid interaction. Pak1-Reg, the regulatory domain of Pak1, was fused to GAD (GAL4 transcription activation domain). The fusion was tested for interaction with LBD-Pak1-Cat, which is an LBD (lexA DNA binding domain) fusion of the kinase catalytic domain of Pak1. LBD-Cdc42V12, which had been shown elsewhere to bind GAD-Pak1-Reg (17, 26), was used as a positive control. GAD and LBD-Ras1 were employed as negative controls. The two-hybrid interaction was determined by histidine prototrophy and β-galactosidase production (see Materials and Methods). As shown in Fig. 1, GAD-Pak1-Reg was able to bind LBD-Cdc42 and LBD-Pak1-Cat, but not LBD-Ras1, while LBD-Pak1-Cat failed to bind GAD. This result established the specific binding between Pak1-Reg and Pak1-Cat. In keeping with this conclusion, we also tested and found that GAD-Pak1-Cat can bind LBD-Pak1-Reg faithfully as well (data not shown). We note in passing that the regulatory domain can even bind to a mutant, inactive catalytic domain.
FIG. 1.
Binding between the separated regulatory and catalytic domains of Pak1. L40 was transformed with either pGAD, pGAD-Pak1-Reg, or pGAD-Cdc42V12,C189S and either pLBD-Cdc42V12, pLBD-Pak1-Cat, pLBD-Pak1K415,416R-Cat, pLBD-Ras1, pLBD-Pak1, or pLBD-Pak1-Reg. Transformants were tested for growth on medium lacking histidine (DO-LTH) and assayed for β-galactosidase production. VSG, very slow growth. Values shown are relative levels (means ± standard deviations). DO-LT is the medium lacking leucine and tryptophan. ND, not determined.
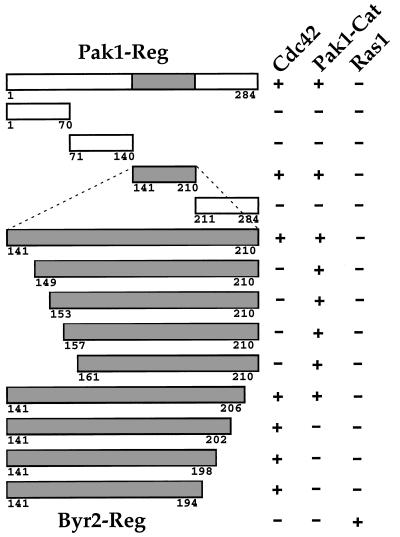
To identify the region on Pak1-Reg that is responsible for binding Pak1-Cat, we generated several Pak1-Reg deletion mutants by PCR and tested their ability to bind Pak1-Cat (see Materials and Methods). We found that a 70-amino-acid stretch from residues 141 to 210 is able to bind both Cdc42 and Pak1-Cat specifically (see Fig. 2). This region contains CRIB (Cdc42-Rac1 interactive binding) domain, the most conserved region on PAK proteins outside the kinase catalytic domain. Thus, Pak1-Cat binds to the same region on Pak1-Reg known to bind Cdc42.
FIG. 2.
Regions on Pak1-Reg mediating the interaction with the kinase catalytic domain. Pak1-Reg deletion mutants were made by PCR and fused to GAD. The GAD fusion to the regulatory domain of Byr2 was included as a control (last row). These fusions were assayed for interactions with LBD fused to Cdc42, Pak1-Cat, or Ras1 as a negative control. A plus sign represents a two-hybrid interaction; a minus sign represents no detectable two-hybrid interaction. The positive interactions were all of about similar intensities. The amino acid positions of the peptide sequences expressed as GAD fusions are shown. The conserved region of the Pak1 regulatory domain is shown in gray.
To map more precisely the regions on Pak1-Reg that mediate Cdc42 and Pak1-Cat interactions, several more deletion mutants within Pak1141–210 were made by PCR (see Materials and Methods). These deletion mutants were then tested for binding Cdc42V12, Pak1-Cat, and Ras1, the negative control. The two-hybrid binding results are also presented in Fig. 2. We found that truncation from the N-terminal portion of Pak1141–210 abolished binding to Cdc42 before affecting binding to Pak1-Cat, whereas truncation from the C terminus abolished binding to Pak1-Cat before binding to Cdc42V12. These experiments suggest that, in theory, Pak1160–206 should be the shortest peptide that can bind Pak1-Cat specifically. The experiments described below use slightly larger fragments that do not bind Cdc42.
Pak1 regulatory domains block truncated and activated Pak1 in vivo.
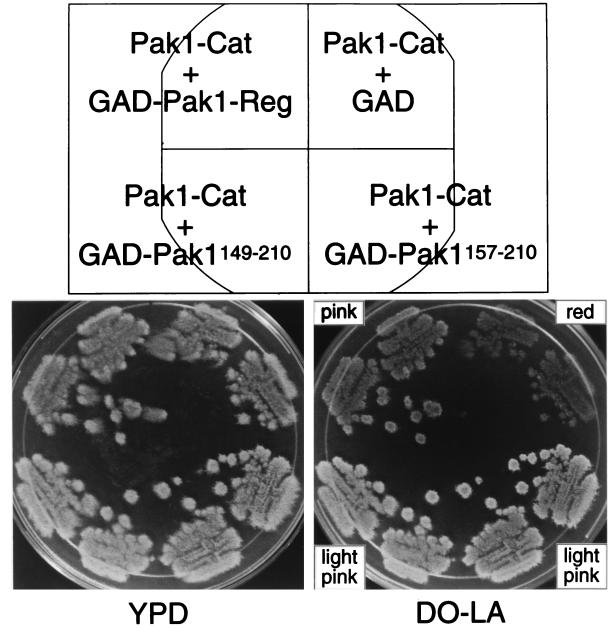
The standard autoinhibition model for protein kinases predicts that the regulatory domain inhibits the catalytic activity, and for Pak1, this is supported by the truncation experiments that have been performed and reported by others (17, 28). If our two-hybrid data correctly identifies the region of the regulatory molecule that binds to the catalytic domain, and the truncated Pak1 is activated because of the loss of the inhibitory influence of the regulatory domain, then expression of that domain should inhibit the activity of the truncated Pak1 when it is expressed in trans. To test this prediction, we exploited the observation that expression of the truncated Pak1 is somewhat toxic to S. cerevisiae. We thus performed an expression toxicity assay. L40 was transformed with either GAD-Pak1-Reg, GAD-Pak1149–210, GAD-Pak1157–210, or GAD alone, all carrying the LEU2 marker, and either pLS104-Pak1-Cat or pLS104 vector alone, each carrying ADE2. Cells were plated on medium lacking leucine and adenine (DO-LA), and transformants were patched out on fresh DO-LA plates. The patches were then replica plated and grown for several days on the nonselective medium, YPD, before being replica plated back on the selective medium. Cells expressing toxic ADE2 plasmids will tend to lose the same, which we can assay in two ways: by failure to thrive on the selective plates and by the red color characteristic of cells lacking ADE2. Cells with Pak1-Cat and GAD alone failed to grow effectively on the selective medium, and the patches displayed a red color. However, those with Pak1-Cat with either GAD-Pak1-Reg, GAD-Pak1149–210, or GAD-Pak1157–210 grew more effectively, and the patches displayed a pink to white color (Fig. 3). These studies confirm that the region we have identified not only binds to the catalytic domain but also inhibits it, even when expressed in trans.
FIG. 3.
Effect of expressing the Pak1 regulatory domain on the toxicity of Pak1-Cat. L40 was transformed with pLS104-Pak1-Cat and either pGAD, pGAD-Pak1-Reg, pGAD-Pak1149–210, or pGAD-Pak1157–210. Transformants were initially plated and streaked on medium lacking leucine and adenine (DO-LA). The Leu+ and Ade+ cells were then grown for several days in the nonselective medium, YPD, before being replica plated on the selective medium, DO-LA. Pictures were taken of the patches, and the color of the patches was noted.
Regulatory and catalytic interaction keeps wild-type, full-length Pak1 in a closed configuration.
Since Pak1, like Byr2, contains a regulatory domain capable of interacting with its catalytic domain, we suspected that full-length Pak1, like full-length Byr2, would exist in a closed configuration in which the catalytic domain is occupied by the regulatory domain. In support of this hypothesis, we found that although we could readily detect binding between Pak1-Reg and Pak1-Cat, we could not detect the binding of Pak1-Reg to full-length Pak1, even though the latter was perfectly capable of binding Cdc42V12,C189S (Fig. 1). (Note: in these experiments, the Cdc42V12,C189S protein, lacking the farnesylation site, was used because the combined expression of Pak1 and Cdc42V12 is toxic.) These results suggest that an intramolecular interaction exists between the regulatory and catalytic domains in full-length Pak1. This hypothesis is further strengthened by the experiments, described below, in which we searched for, found, and analyzed mutants of Pak1 that were in an open configuration.
If we correctly surmise that wild-type Pak1 failed to bind Pak1-Reg because of intramolecular interactions, we should be able to readily isolate Pak1 mutants that gain the ability to bind Pak1-Reg, and such mutants should have regulatory and catalytic domains that are no longer able to interact.
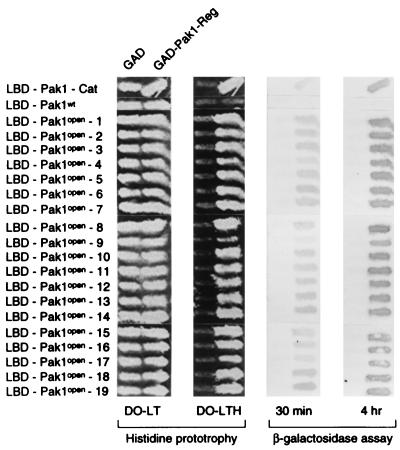
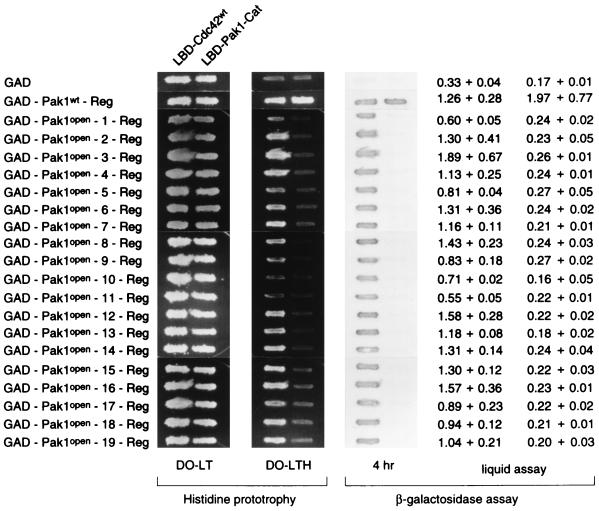
Pak1-Reg was randomly mutagenized by PCR and fused to Pak1-Cat in to form a library of LBD fusions of full-length, mutagenized Pak1. The DNA of this mutant library was transformed into L40 together with GAD-Pak1-Reg, and cells were plated in the absence of histidine to select for mutant, full-length Pak1 capable of interacting with the isolated regulatory domain. Colonies that grew on the His− plates were patched out and tested for the production of β-galactosidase. Twenty-five colonies that were both His+ and LacZ+ were isolated, and the LBD plasmids from these cells were recovered, amplified, and transformed back into L40 together with GAD-Pak1-Reg or GAD. Nineteen of the 25 LBD-Pak1 plasmids interacted with GAD-Pak1-Reg but not with GAD. Figure 4 shows the two-hybrid interactions of the 19 LBD-Pak1 mutants with GAD-Pak1-Reg and with the negative control. Since these Pak1 mutants can bind Pak1-Reg, we call them Pak1open mutants hereafter.
FIG. 4.
Binding of the separated regulatory domain to Pak1open mutants. L40 was transformed individually with either pGAD or pGAD-Pak1-Reg and either pLBD-Pak1-Cat, pLBD-Pak1, or the 19 pLBD-Pak1open mutants. Transformants were tested for growth on medium lacking histidine (DO-LTH) and assayed for β-galactosidase production. DO-LT is the medium lacking leucine and tryptophan.
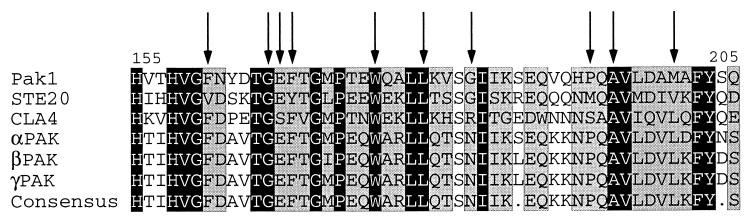
The regulatory domains of the 19 Pak1open mutants were sequenced. All of them contain a single mutation between residues 161 and 200, that is, within the CRIB domain, the highly conserved region that binds both Cdc42 and Pak1-Cat (Table 1). Several mutations were encountered more than once, and the mutants fell into 13 groups. All mutations except M200T and M200R were mapped to residues conserved among PAK proteins. Figure 5 shows the multiple alignments of this conserved region with representative homologs, with the sites of mutation indicated.
TABLE 1.
Mutations in Pak1open mutants
| Mutant group | Mutation | Member(s) |
|---|---|---|
| I | F161S | Pak1open-1 |
| II | G166W | Pak1open-9 |
| III | E167G | Pak1open-17 |
| IV | F168S | Pak1open-11 |
| V | W175R | Pak1open-5, Pak1open-10 |
| VI | L179P | Pak1open-6, Pak1open-18 |
| VII | I184T | Pak1open-3, Pak1open-12, Pak1open-13, Pak1open-15 |
| VIII | P193S | Pak1open-2 |
| IX | P193Q | Pak1open-19 |
| X | A195V | Pak1open-4 |
| XI | A195T | Pak1open-7, Pak1open-8 |
| XII | M200T | Pak1open-14 |
| XIII | M200R | Pak1open-16 |
FIG. 5.
Location of the altered amino acid residues of the Pak1open mutants in the highly conserved region of PAK proteins. The highly conserved regions on Pak1 (17, 26), STE20 (28), CLA4 (7), and three major mammalian PAK isoforms (3, 16, 21) are aligned, with identical residues in black boxes, conserved residues in grey boxes, and the residues altered in the Pak1open mutants indicated by arrows.
As the first step towards characterizing these Pak1open mutants, we tested the binding of the Pak1-Reg of these mutants to Pak1-Cat and Cdc42. The Pak1-Regs were excised and fused to GAD, and the GAD fusions were tested with LBD-Pak1-Cat and LBD-Cdc42, individually. GAD-Pak1wt-Reg and GAD were tested alongside. The two-hybrid results are presented in Fig. 6. Significantly, but not surprisingly, the regulatory domains of all of the Pak1open mutants failed to bind LBD-Pak1-Cat, while all still bound Cdc42 to varying degrees.
FIG. 6.
Failure of the separated Pak1open regulatory domains to bind the catalytic domain. L40 was transformed individually with either pLBD-Cdc42 or pLBD-Pak1-Cat and either pGAD, pGAD-Pak1-Reg, or 19 pGAD-Pak1open-Reg. Transformants were tested for growth on medium lacking histidine (DO-LTH) and assayed for β-galactosidase production. DO-LT is the medium lacking leucine and tryptophan.
These results demonstrate that all 19 Pak1open mutants contain mutations in the CRIB domain that abolish binding to the catalytic domain Pak1 and argue strongly that the loss of intramolecular interaction is the cause for the open configuration.
Genetic characterizations of Pak1open mutants.
If the disruption of the regulatory-catalytic interactions were sufficient to activate Pak1, we would expect the Pak1open mutants to be activated. To test this, we examined the activity of Pak1open mutants in comparison with that of the wild-type Pak1. Three phenotypes associated with the dominant activated Pak1, Pak1-Cat, were assayed: toxicity (references 17 and 26 and as described above), activation of the FUS1-lacZ reporter system (17), and the induction of the open configuration of Byr2 (31).
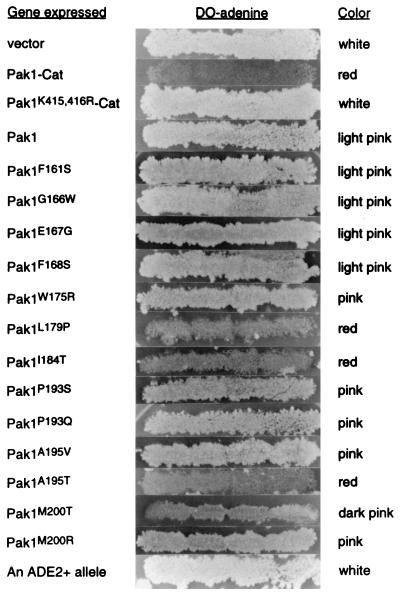
The format for expression toxicity assays was described above. Pak1, Pak1open mutants, Pak1-Cat, and Pak1K415,416R-Cat were cloned under an alcohol dehydrogenase promoter into an ADE2-based 2 μm plasmid, pLS104. L40 cells transformed with various pLS104 clones were plated on Ade− plates. Transformants were then patched onto fresh Ade− plates and replica plated on nonselective medium and then again on Ade− plates. Cells that grow well while carrying the pLS104 derivatives grow robustly as white patches on Ade− plates, while cells with pLS104 derivatives that cause toxicity or slower growth will not grow as well on Ade− plates and have a red to pink color, indicative of defective adenine biosynthesis. The patches of cells expressing Pak1-Cat, Pak1K415,416R-Cat, Pak1, or Pak1open mutants are shown in Fig. 7. As expected, cells containing pLS104-Pak1-Cat failed to grow efficiently on Ade− plates after being replica plated, and the patches displayed a red color; those containing pLS104 or pLS104-Pak1K415,416R-Cat grew perfectly well in the absence of adenine, and the patches displayed a white color. Cells containing pLS104-Pak1 grew well in the absence of adenine, and patches were light pink, suggesting a very mild toxicity resulting from the expression of wild-type Pak1. Cells expressing various Pak1open mutants displayed varying ranges between these two extremes. Those containing Pak1F161S, Pak1G166W, Pak1E167G, or Pak1F168S grew well, and their patches exhibited a light pink color, much like cells with wild-type Pak1; cells with Pak1W175R, Pak1P193S, Pak1P193Q, Pak1A195V, or Pak1M200R grew, but their patches were pink; cells with Pak1M200T were dark pink; and cells with Pak1L179P, Pak1I184T, or Pak1A195T grew poorly after being replica plated back to the Ade− plates, and the patches had a red color, much like those with dominant activated Pak1. From this, we conclude that the majority of Pak1open mutants have higher activity than wild-type Pak1, and the results support the model that Pak1 intramolecular interaction is responsible for autoinhibition.
FIG. 7.
Expression toxicity assay with Pak1open mutants. L40 was transformed with either pLS104, pLS104-Pak1-Cat, pLS104-Pak1K415,416R-Cat, pLS104-Pak1, or 13 pLS104-Pak1open mutants. Transformants were initially plated and then patched in groups of four on medium lacking adenine. The Ade+ cells were then replica plated on the nonselective medium, YPD, for several days, before being replica plated on medium lacking adenine (DO-adenine). Pictures were taken of the patches on DO-adenine, and the color of the patches was noted. The wild-type ADE2 allele was included for comparison.
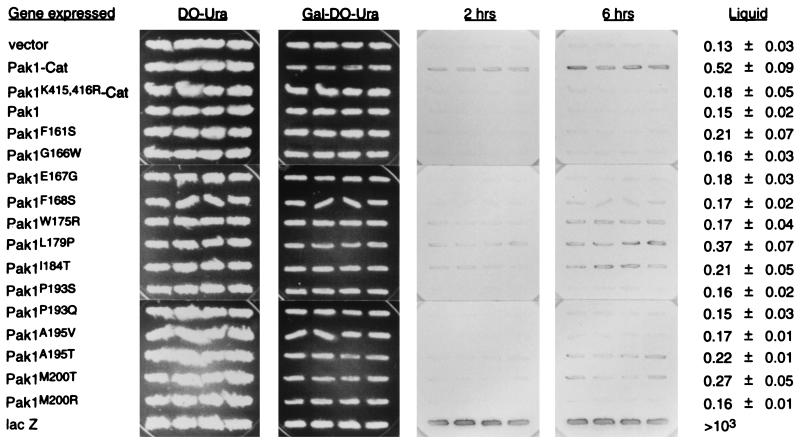
The FUS1-lacZ reporter system provides an indicator for the activity of the S. cerevisiae mating signaling pathway (10). We have shown previously that dominant activated forms of STE20 and Pak1 can activate this pathway in an STE11-dependent manner (17). It was noted then that full-length wild-type Pak1 failed to activate the pathway. Therefore, we asked if any of the Pak1open mutants could activate the S. cerevisiae mating pathway and stimulate β-galactosidase production by the reporter system. Pak1open mutants were cloned under a galactose-inducible GAL1 promoter, as described in Materials and Methods, to avoid the potential complications due to the toxicity of the expressed gene. Cells containing the plasmids were patched on medium rich in glucose and then replica plated to medium depleted of glucose but rich in galactose (2%). The amount of β-galactosidase in these cells was monitored by the conversion of X-Gal, and the results are presented in Fig. 8. As expected, cells with Pak1-Cat expressed produced more β-galactosidase than did cells with the vector alone or cells with Pak1K415,416R-Cat, the kinase-defective Pak1-Cat. While cells containing wild-type Pak1 were unable to activate the reporter system detectably, several of the strains carrying Pak1open mutants were able. In fact, some produced β-galactosidase as well as did cells carrying Pak1-Cat. These results demonstrate that some Pak1open mutants are more active than wild-type Pak1 and further confirm that intramolecular interaction is autoinhibitory.
FIG. 8.
Activity of the Pak1open mutants in the FUS1-lacZ induction assay. AN43-5A was transformed with either pYX113 (the empty vector with the GAL1 promoter), pYX113-Pak1-Cat, pYX113-Pak1K415,416R-Cat, pYX113-Pak1, 13 of the pYX113-Pak1open mutants, or pYX113-lacZ. Transformants were subjected to a 20-h galactose induction before being assayed for β-galactosidase production from the FUS1 promoter. Overlay filters were incubated with X-Gal for 2 to 6 h, and results of quadruplicate transformants are shown. Cultures were also harvested for β-galactosidase liquid assays, performed on four independent transformants. The assay results, ± standard deviations, are shown at right.
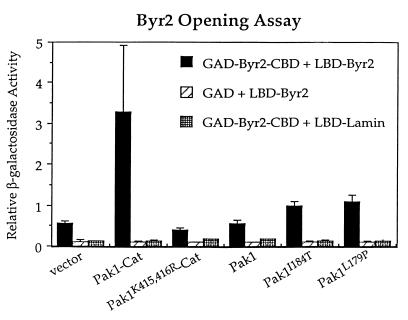
The third assay for Pak1 activation was based on its ability to induce the open configuration of Byr2. We have previously shown that expression of the dominant activated Pak1, Pak1-Cat, but not the wild-type full-length kinase, induced the two-hybrid interaction between GAD-Byr2-CBD (the GAD fusion to the smallest subregion of the regulatory domain of the Byr2 kinase sufficient to bind to its catalytic domain) and LBD-Byr2 (31). We therefore tested if Pak1open mutants were more effective than wild-type Pak1 at inducing this interaction. L40 was transformed with pGAD-Byr2-CBD, pLBD-Byr2, and either pLS104-Pak1-Cat, pLS104-Pak1K415,416R-Cat, pLS104-Pak1, pLS104-Pak1I184T, pLS104-Pak1L179P (the two Pak1 mutants that were most active in the previous assays), or just pLS104 vector alone. Transformants were tested quantitatively for the production of β-galactosidase. The results are presented in Fig. 9. As expected, Pak1wt-Cat induced the interaction between GAD-Byr2-CBD and LBD-Byr2 to about six times above the background level, while kinase-defective Pak1-Cat, Pak1K415,416R-Cat, failed to enhance this interaction. Wild-type full-length Pak1 also failed to increase this interaction, but both Pak1open mutants were able to induce levels twofold over the background level. These results once again confirm that the intramolecular interaction is autoinhibitory.
FIG. 9.
Induction of the open configuration of Byr2 by the overexpression of Pak1open mutants. L40 was transformed with either pGAD-Byr2-CBD or pGAD; either pLBD-Byr2 or pLBD-Lamin; and either pLS104-Pak1-Cat, pLS104-Pak1K415,416R-Cat, pLS104-Pak1wt, pLS104-Pak1I184T, pLS104-Pak1L179P, or just the pLS104 vector alone. Transformants were tested quantitatively for β-galactosidase production. Values shown are relative levels. Standard deviations from at least four independent transformants are shown by error bars.
Cdc42 promotes the open configuration of Pak1.
It has been shown in vitro, with a gel overlay assay, that purified Rac-Rho-Cdc42 can induce an autophosphorylation and activation event of Pak1 (16). Cdc42 is now known to be an upstream activator of Pak1 in vivo (17, 26), although the activation mechanism remains unknown. We have shown that Cdc42 and Pak1-Cat interact with a tightly overlapping region on Pak1-Reg. Moreover, we failed to find evidence for a trimeric complex among Cdc42V12, Pak1-Reg, and Pak1-Cat, detectable by the two-hybrid system, suggesting that the three-way interaction is sterically forbidden (data not shown). Therefore, we speculated that Cdc42 activates Pak1 by directly relieving Pak1 of the autoinhibition that results from the intramolecular binding of the regulatory and catalytic domains. This speculation led us to predict and test whether Cdc42 could induce the open configuration of Pak1.
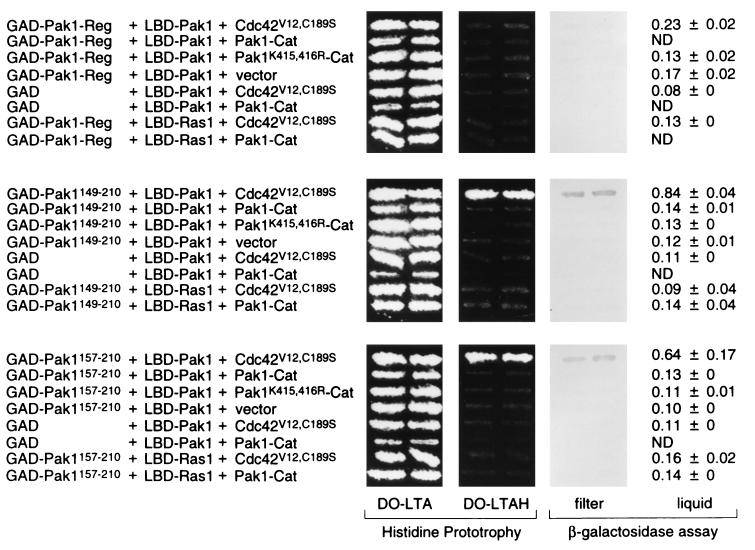
We have successfully used the two-hybrid system to identify signaling components that can induce the open configuration of Byr2 (31), and we applied the same principles to Pak1. The Pak1 opening assay was performed in the following fashion: L40 was transformed with (i) either GAD-Pak1-Reg, GAD-Pak1149–210, GAD-Pak1157–210, or GAD alone; (ii) either LBD-Pak1 or LBD-Ras1 as a control; and (iii) either pLS104-Cdc42V12,C189S, pLS104-Pak1-Cat, pLS104-Pak1K415,416R-Cat, or pLS104. Cells were patched on medium lacking leucine, tryptophan, and adenine (for pLS104 plasmid selection) (DO-LTA). Patches were replica plated on medium lacking histidine to test the transactivation of the HIS3 reporter gene. Transformants were also tested for lacZ expression, by both filter overlay and liquid assays. As shown in Fig. 10, only two kinds of cells displayed an interaction between GAD and LBD fusions: those expressing GAD-Pak1149–210, LBD-Pak1, and Cdc42V12,C189S and those expressing GAD-Pak1157–210, LBD-Pak1, and Cdc42V12,C189S. All other cells failed to display two-hybrid interactions. These results demonstrate that Cdc42V12,C189S can effectively and specifically induce the open configuration of Pak1.
FIG. 10.
Induction of the open configuration of Pak1 by the overexpression of Cdc42. L40 was transformed with either pGAD-Pak1-Reg, pGAD-Pak1149–210, pGAD-Pak1157–210, or pGAD vector alone; either pLBD-Pak1 or pLBD-Ras1; and either pLS104-Cdc42V12,C189S, pLS104-Pak1-Cat, pLS104-Pak1K415,416R-Cat, or pLS104 vector alone. Transformants were tested for growth on the medium lacking histidine (DO-LTAH) and assayed for β-galactosidase production. DO-LTA is the medium lacking leucine, tryptophan, and adenine. Values shown are relative levels (means ± standard deviations). ND, not determined.
Cells expressing GAD-Pak1-Reg, LBD-Pak1, and Cdc42V12,C189S did not yield a positive interaction. We attribute this to the fact that Pak1-Reg, unlike GAD-Pak1149–210 or GAD-Pak1157–210, also binds Cdc42V12,C189S and thus competes for its binding.
These experiments suggest that Cdc42 opens the configuration of Pak1 through its interaction with the regulatory domain. A more direct demonstration of this mechanism was obtained as follows. We screened for and identified two single-base-pair mutants of the regulatory domain of Pak1 that failed to bind Cdc42 yet still were capable of full-strength binding to the catalytic domain, as judged by two-hybrid interactions. The mutations, S148A and H155A, were each independently introduced into the full-length Pak1. We then tested if Cdc42V12,C189S could induce the open configuration of either Pak1S14A or Pak1H155A. It could not, indicating that the opening of Pak1 is the consequence of the direct binding of Cdc42 to the regulatory domain.
DISCUSSION
Previous studies showed that the intramolecular interaction between Byr2 regulatory and kinase catalytic domains keeps that kinase in a closed configuration and establishes autoinhibition (31). In this report, we first describe a similar potential for intramolecular interaction within Pak1, the fission yeast homolog of PAK. Expression of segment fusions indicated that the highly conserved region of the regulatory domain of Pak1 (Pak1-Reg), to which Cdc42 also binds, was capable of binding to the catalytic domain. This mapping was later confirmed by point mutation analysis.
Since this potential Pak1 intramolecular interaction resembles that found in Byr2, we incorporated the insights gained from Byr2 to guide us in further studies. In particular, we next demonstrated that Pak1 can exist in the wild-type closed configuration and a mutant open configuration, which differ in their ability to bind a free regulatory domain. Mutants with the open configuration have mutations in the conserved regulatory domain, and these mutant domains are unable to bind separated catalytic domains. These studies strongly support the existence of intramolecular interaction between the regulatory and catalytic domains of wild-type Pak1.
In the case of Byr2, the intramolecular interaction causes autoinhibition, and its release is associated with kinase activation. The same appears to be true for Pak1. First, the expression of the smallest regulatory region of Pak1 capable of binding the catalytic region, a region that does not bind to Cdc42, inhibits the toxicity resulting from expression of the free catalytic domain. Second, the majority of Pak1open mutants are more active than wild-type Pak1, and some of them behaved similarly to the activated Pak1 lacking its regulatory domain. Third, Cdc42, a known activator of Pak1, both in vivo and in vitro, induces the open configuration, as discussed below.
Our genetic results indicate that not all Pak1open mutants are equally activated, and none are as active as the construct which lacks the entire regulatory region. There are many possible explanations for this. First, these proteins may be expressed at different levels. Second, although we cannot detect intramolecular interaction in the mutants by two-hybrid analysis, the mutants may nevertheless have a closed configuration in vivo. Third, there may be other features of the regulatory domain that are inhibitory for full biological activity. Indeed, other proteins that bind to the regulatory domain of Pak1 have recently been identified (9). Our studies are not designed to resolve these questions.
In our previous studies, we found that activated Pak1 could induce the open configuration of Byr2. We suspected that Cdc42 might do the same to Pak1. First, it was known that Cdc42 was an activator. Second, Cdc42 and the catalytic domain bind to overlapping regions of the regulatory domain. Third, we could not observe a stable trimeric complex among Cdc42, Pak1-Reg, and Pak1-Cat. We thus tested if Cdc42 could open Pak1. We used three different molecular probes for the open configuration of Pak1: GAD-Pak1-Reg, GAD-Pak1149–210, and GAD-Pak1157–210. None are able to bind full-length Pak1, all three are able to bind the isolated catalytic domain, and only the first is also able to bind Cdc42. Indeed, when Cdc42 was overexpressed, the release of the kinase catalytic domain of full-length Pak1 to bind GAD-Pak1149–210 and GAD-Pak1157–210 was clear. Moreover, opening by Cdc42 could not be observed on mutant Pak1 proteins that do not bind Cdc42.
Although Cdc42 does activate Pak1, binds to Pak1, and opens its conformation and the open-conformation mutants are more active than wild type, these experiments do not rule out additional functions for Cdc42 in the activation of Pak1. For example, Cdc42 may facilitate the localization of Pak1 or the binding of other activating proteins.
It may be useful to draw a parallel between the interactions of Cdc42 and those of Ras1 with their respective protein kinase targets. Many of the same relations are retained: Ras1 is an in vivo regulator of Byr2, it binds directly to Byr2, and its domain of interaction overlaps with the site where the catalytic subunit also binds (22, 31, 32). Yet we were unable to demonstrate the opening of Byr2 by Ras1. In fact, no direct in vitro activation of Byr2 by Ras1 (or of Raf by H-ras) has been observed, and we have observed a stable complex between Ras1 and the Byr2 catalytic domain bridged by a mutant regulatory domain of Byr2 with enhanced affinity for the catalytic domain (30a). Thus, the mechanisms of action of these two very similar GTPases on two similar protein kinases are likely to be very different.
The region of the Pak1 regulatory domain that can bind to both Cdc42 and the catalytic domain is highly conserved among all members of the PAK family. Hence, this intramolecular interaction is highly likely to be conserved among them as well. Indeed, during the preparation of this paper, Zhao et al. reported the identification of a conserved negative regulatory region in αPAK (39). The authors showed that mutations on residues 101 to 137 of αPAK render that kinase constitutively active. They further provided evidence that αPAK83–149, a 67-amino-acid peptide, can block PAK activation by Cdc42 in vitro and suppresses PAK functions in vivo. This conserved negative regulatory region on αPAK corresponds to the Pak1 autoinhibitory region reported here. Our results are exactly complementary.
It may be proper to think of four kinases comprising the prototypic MAPK module: MAPK, MEK, MEKK, and PAK. MAPKs and MEKs have limited regions outside of the kinase catalytic domain and need to be phosphorylated at conserved residues in the catalytic domain to gain maximum kinase activities (34) (reviewed in reference 19). Thus, MEKs and MAPKs are predominantly regulated by dynamic phosphorylation and dephosphorylation and perhaps do not display autoregulation. MEKKs, such as Mekks, STE11, Byr2, and Raf, have long regulatory domains, which may bind and mask the kinase catalytic domains, and thus are kept in inactive form. MEKK autoregulation can be antagonized by PAK phosphorylation. PAKs, like MEKKs, also utilize regulatory and catalytic interaction to exert kinase autoregulation. Both PAKs and MEKKs can be regulated by p21 GTPases. However, while PAK regulation by Rho-family GTPases may be caused in part by direct release from autoinhibition, the regulation of MEKKs by GTPases may be more indirect (15, 30).
ACKNOWLEDGMENTS
We thank Ken Chang, Doug Johnson, and Aaron Neiman for providing DNA and yeast strains; Mike Riggs for DNA sequencing; Terry Vale, Hong Ma, Peter Gergen, and Marion Carlson for helpful discussion; the Cold Spring Harbor Laboratory Art Department for artwork; and Patricia Bird for secretarial assistance.
This work was supported by grants from the American Cancer Society and the National Cancer Institute (NIH) to M.W. M.W. is an American Cancer Society Professor.
REFERENCES
- 1.Bagrodia S, Derijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signaling leads to JNK and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:1–4. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 2.Barr M M, Tu H, Van Aelst L, Wigler M. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:5597–5603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J, Stowers L, Baer M, Trejo A, Coughlin S, Chant J. Human STE20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 4.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 5.Chang E C, Barr M, Wang Y, Jung V, Xu H-P, Wigler M. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 6.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 7.Cvrckova F, De Virgilio C, Manser E, Pringle J, Nasmyth K. Set20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeasts. Genes Dev. 1995;6:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 8.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 9.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. The highly conserved skb1 gene encodes a protein that interacts with SHK1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA. 1998;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen D, McCaffrey G, Sprague G. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 12.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leberer E, Dignar D, Harcus D, Thomas D, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein β subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leberer E, Dignard D, Hougan L, Thomas D Y, Whiteway M. Dominant-negative mutants of yeast G-protein β subunit identify two functional regions involved in pheromone signaling. EMBO J. 1992;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–418. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 16.Manser E, Leung T, Salihuddin H, Zhao Z S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 17.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65pak protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast, Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the yeast pheromone-responsive MAP kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall C. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 20.Martin H, Mendoza A, Rodriguez-Pachon J M, Molina M, Nombela C. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol Microbiol. 1997;23:431–444. doi: 10.1046/j.1365-2958.1997.d01-1870.x. [DOI] [PubMed] [Google Scholar]
- 21.Maser E, Chong C, Zhao Z S, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J Biol Chem. 1998;2740:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- 22.Masuda T, Kariya K-C, Shinkai M, Okada T, Kataoka T. Protein kinase Byr2 is a target of Ras1 in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1995;270:1979–1982. doi: 10.1074/jbc.270.5.1979. [DOI] [PubMed] [Google Scholar]
- 23.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 24.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 25.Neiman A, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEKK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottilie S, Miller P J, Johnson D I, Creasy C L, Sells M A, Bagrodia S, Forsburg S L, Chernoff J. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polverino A, Frost J, Yang P, Hutchison M, Neiman A, Cobb M, Marcus S. Activation of mitogen-activated protein kinase cascade by p21-activated protein kinase in cell-free extracts of Xenopus oocytes. J Biol Chem. 1995;270:26067–26070. doi: 10.1074/jbc.270.44.26067. [DOI] [PubMed] [Google Scholar]
- 28.Ramer S W, Davis R W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soderling T R. Protein kinases. J Biol Chem. 1990;265:1823–1826. [PubMed] [Google Scholar]
- 30.Stokoe D, MacDonald S G, Caddwallader K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 30a.Tu, H. Unpublished findings.
- 31.Tu H, Barr M, Dong D L, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vojtek A, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 34.Ward Y, Gupta S, Jensen P, Wartmann M, Davis R J, Kelly K. Control of MAP kinase activation by the mitogen-induced threonine/tyrosine phosphatase PAC1. Nature. 1994;367:651–654. doi: 10.1038/367651a0. [DOI] [PubMed] [Google Scholar]
- 35.White M, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M. Multiple RAS functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 36.Wu C, Whiteway M, Thomas D Y, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 37.Yang P, Kansra S, Pimental R A, Gibreth M, Marcus S. Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18481–18489. doi: 10.1074/jbc.273.29.18481. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, Han J, Sells M, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z S, Manser E, Chen X Q, Chong C, Leung T, Lim L. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with TAQ DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]