Abstract
In the present study, we investigated the function and the mechanism of action of RGS3, a member of a family of proteins called regulators of G protein signaling (RGS). Polyclonal antibodies against RGS3 were produced and characterized. An 80-kDa protein was identified as RGS3 by immunoprecipitation and immunoblotting with anti-RGS3 antibodies in a human mesangial cell line (HMC) stably transfected with RGS3 cDNA. Coimmunoprecipitation experiments in RGS3-overexpressing cell lysates revealed that RGS3 bound to aluminum fluoride-activated Gα11 and to a lesser extent to Gαi3 and that this binding was mediated by the RGS domain of RGS3. A role of RGS3 in postreceptor signaling was demonstrated by decreased calcium responses and mitogen-activated protein (MAP) kinase activity induced by endothelin-1 in HMC stably overexpressing RGS3. Moreover, depletion of endogenous RGS3 by transfection of antisense RGS3 cDNA in NIH 3T3 cells resulted in enhanced MAP kinase activation induced by endothelin-1. The study of intracellular distribution of RGS3 indicated its unique cytosolic localization. Activation of G proteins by AlF4−, NaF, or endothelin-1 resulted in redistribution of RGS3 from cytosol to the plasma membrane as determined by Western blotting of the cytosolic and particulate fractions with RGS3 antiserum as well as by immunofluorescence microscopy. Agonist-induced translocation of RGS3 occurred by a dual mechanism involving both C-terminal (RGS domain) and N-terminal regions of RGS3. Thus, coexpression of RGS3 with a constitutively active mutant of Gα11 (Gα11-QL) resulted in the binding of RGS3, but not of its N-terminal fragment, to the membrane fraction and in its interaction with Gα11-QL in vitro without any stimuli. However, both full-length RGS3 and its N-terminal domain translocated to the plasma membrane upon stimulation of intact cells with endothelin-1 as assayed by immunofluorescence microscopy. The effect of endothelin-1 was also mimicked by calcium ionophore A23187, suggesting the importance of Ca2+ in the mechanism of redistribution of RGS3. These data indicate that RGS3 inhibits G protein-coupled receptor signaling by a complex mechanism involving its translocation to the membrane in addition to its established function as a GTPase-activating protein.
A variety of hormones, neurotransmitters, and other stimuli elicit their cellular effects by binding to seven transmembrane-spanning receptors. A common feature of these receptors is coupling to and activation of heterotrimeric GTP-binding proteins (G proteins) in a ligand-dependent manner. The process of activation of G proteins involves an exchange of GDP for GTP, resulting in the dissociation of Gα from Gβγ subunits, which further transduce the signal to a variety of G protein effectors (5, 30). Gα is a diverse family of proteins subdivided by function and sequence homology into several groups, including αs, αi, αq/11, and α12, etc. (28). Among the most established functions of alpha subunits of G proteins are direct activation (αs) or inhibition (αi) of adenylyl cyclase, opening of Ca2+ channels (αs) and K+ channels (αi), activation of cGMP-phosphodiesterase (αt), and stimulation of phospholipase C (αq).
Recent genetic studies (11, 15, 22) have revealed the existence of proteins united by sequence homology and function into a family named regulators of G protein signaling (RGS). In vitro experiments demonstrated that RGS proteins bind directly to Gα in its active GTP-bound state and increase its GTPase activity, thus inactivating Gα (1, 2, 21, 35). In terms of specificity, RGS1, -4, and -10 and GAIP (G-α interacting protein) bound with much higher affinity to αi than to αq and did not bind to αs and α12 at all. However, the signaling studies demonstrated that both Gi- and Gq-mediated signal transduction were inhibited by RGS4 and GAIP (15, 19, 20, 37).
RGS3 is one of the less-studied members of the RGS family. In vitro experiments with purified glutathione S-transferase-fusion RGS3 suggested its ability to bind αq (25); however, αi was not investigated. The role of RGS3 in cell signaling is not clear either. Although data from two groups have shown its inhibitory effect on G protein-linked activation of mitogen-activated protein (MAP) kinase, they are contradictory in terms of the influence of RGS3 transient overexpression on Gq-mediated activation of phospholipase C (PLC) (9, 25). Therefore, the purpose of this work was to investigate the function and the mechanism of action of RGS3, with RGS3 antibodies as a tool and RGS3-overexpressing stable cell lines as an intact cellular experimental model.
The present work demonstrates that (i) RGS3 binds to the activated form of Gα11 (a member of the Gαq/11 family), which results in the inhibition of a G11-mediated increase in intracellular calcium concentration and MAP kinase phosphorylation; (ii) activation of G proteins (α11) induces translocation of RGS3 from the cytosol to the plasma membrane; and (iii) intracellular redistribution of RGS3 occurs by a calcium-induced and G protein-mediated mechanism.
MATERIALS AND METHODS
Materials.
Human RGS3 cDNA, previously cloned by Druey et al. (15), was excised from pRC/CMV and inserted into pcDNA3.1 (Invitrogen, San Diego, Calif.) in forward and reverse (antisense) orientations. RGS3(1-380) cDNA was amplified by PCR from the original RGS3 cDNA template and subcloned into pcDNA3.1. Mouse Gα11-WT and Gα11-Q209L cDNAs were kindly provided by Hiroshi Itoh (Yokohama, Japan). Anti-RGS10, anti-αq/α11, anti-αq, anti-α11, and anti-αi3 antibodies were from Santa Cruz Biotechnology (Santa Cruz, Calif.). Both anti-αq/α11 and anti-α11 but not anti-αq antibodies specifically recognized the same band corresponding to α11 in human mesangial cells (HMC). No αq was detected in HMC with either anti-αq/α11 or anti-αq antibodies. Guanine nucleotides and protease inhibitors were from Boehringer Mannheim (Indianapolis, Ind.). Endothelin-1 (ET-1) was from Calbiochem (Cambridge, Mass.).
Production and affinity purification of RGS3 antibodies.
The RGS3(40-52) peptide containing N-terminal cysteine was synthesized for antibody production based on the best immunogenic properties as determined by amino acid sequence analysis with the Wisconsin Sequence Analysis Package program (Madison, Wis.). The peptide was conjugated to maleimide-activated bovine serum albumin (Pierce, Rockford, Ill.) and purified from reaction compounds by dialysis. The conjugate (260 mg) was injected twice into a white male rabbit with a 2-week interval. The antiserum was tested by Western blotting with cell lysates from human embryonic kidney 293 (HEK293) cells transiently transfected with RGS3 cDNA (Fig. 1). Affinity purification of antibodies was performed on agarose beads coupled to cysteine-RGS3(40-52) by SulfoLink reagent (Pierce). RGS3-specific antibodies were washed and eluted from the column with ImmunoPure Gentle antigen/antibody binding and elution buffers (Pierce), respectively.
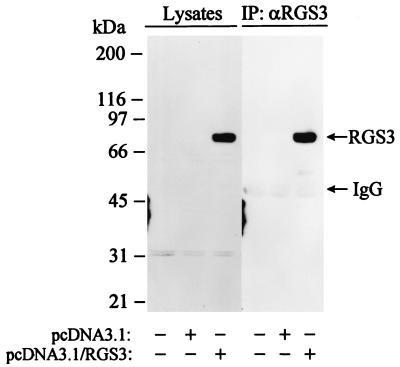
FIG. 1.
Stable transfection of RGS3 cDNA in glomerular mesangical cells. RIPA lysates from parental HMC stably transfected with pcDNA3.1 or pcDNA3.1/RGS3 were immunoprecipitated with RGS3 antiserum. Anti-RGS3 immunoprecipitates or total cell lysates were analyzed by Western blotting with RGS3 antiserum.
Cell culture and development of stable transfected cell line.
HEK293 cells and mouse NIH 3T3 fibroblasts (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium supplemented with 2 mM glutamine, 100 U of streptomycin per ml, 100 U of penicillin per ml, and 10% fetal bovine serum or 10% calf serum. HMC were kindly provided by Jean-Daniel Sraer (Paris, France) and cultured in RPMI supplemented with 5% fetal bovine serum, 10 mM HEPES, 2 mM glutamine, 100 U of streptomycin per ml, and 100 U of penicillin per ml, as described previously (31). The rat pulmonary arterial smooth muscle cell line PAC1 (27) was kindly provided by Abraham Rothman (San Diego, Calif.) and was maintained as HEK293 cells were. For transient overexpression of proteins, cells were transfected with 10 μg of DNA per 10-cm-diameter dish with LipofectAMINE reagent (Gibco BRL, Gaithersburg, Md.) or SuperFect reagent (Qiagen Inc., Valencia, Calif.). For development of stable cell lines, transfected HMC, PAC1, and NIH 3T3 cells were cloned in the presence of 0.4, 0.8, and 1 mg of geneticin G418 (Gibco BRL) per ml, respectively. Geneticin-resistant clones were tested for RGS3 expression by Western blotting with RGS3 antiserum.
Immunoprecipitation and Western blot analysis of RGS3.
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in the buffer, which contained 25 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM MgCl2, and protease inhibitors (1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride. The lysates were cleared from insoluble material by centrifugation at 14,000 rpm for 15 min and incubated with RGS3 antiserum (normally 1 to 2 μl of antiserum per 100 μg of protein) for 2 h at 4°C on a rotator, followed by incubation with protein A-Sepharose for another hour. The immunoprecipitates were washed three times with 1 ml of the same buffer, boiled in Laemmli buffer for 5 min, subjected to electrophoresis, and analyzed by Western blotting with a 1:1,000 dilution of RGS3 antiserum.
Measurement of calcium responses.
Intracellular Ca2+ responses were measured with fura-2 as described previously (18). Cells were maintained at 37°C throughout all incubations and experimental measurements. Serum-starved cells grown on glass coverslips were incubated with fura-2/AM (8 μM) for 1 h. After being washed with HEPES-buffered saline (pH 7.4), the cells were incubated with HEPES-buffered saline for an additional 30 min. Each coverslip was washed twice immediately before it was placed into the recording chamber of a spectrofluorimeter (Hitachi F2000). Excitation wavelength alternated between 340 and 380 nm during measurement of emission fluorescence at 510 nm. The ratio of emission fluorescence at an excitation wavelength of 340 nm versus emission fluorescence at an excitation wavelength of 380 nm (R340/380) was then calculated. After a stable baseline recording was obtained, ET-1 (10−7 M) was added, and the response was monitored in real time. All measurements were corrected for autofluorescence, with cells treated as described above except that fura-2 was omitted. Autofluorescence values for control and transfected cells were essentially identical.
Phosphorylation and activation of MAP kinase.
Serum-starved cells were stimulated with ET-1 (100 nM) or phorbol 13,14-myristate acetate (PMA; 100 nM) for 5 min at 37°C, washed twice with ice-cold PBS and lysed in Triton X100 buffer as described above. Cell lysates were directly analyzed by Western blotting with MAP kinase antibodies (electrophoretic mobility shift) or phospho-specific MAP kinase antibodies or subjected to immunoprecipitation with anti-p42 MAP kinase antibodies, followed by MAP kinase assay with myelin basic protein as a substrate, as described previously (34).
Binding of RGS3 to the membrane.
The crude membrane and cytosol fractions were prepared as described previously (17). Briefly, the cells were homogenized in the buffer containing 25 mM HEPES (pH 7.5), 250 mM sucrose, 1 mM EDTA, and protease inhibitors (as described above). Homogenates were cleared from the debris by centrifugation (2,000 × g, 5 min), and the membranes were separated from cytosol by centrifugation at 50,000 × g for 30 min. The supernatant (cytosol) was set aside, and the pellet (membranes) was washed once again and resuspended by sonication in the same buffer. After this procedure, the main RGS3 immunoreactivity was found in the cytosol (Fig. 7). Binding of RGS3 to the membrane was measured by incubation of equal amounts (per protein) of mixed membrane and cytosol fractions for 30 min at 4°C in the presence of the compounds indicated in Fig. 7, followed by membrane separation as described above. The membrane and cytosol fractions were then analyzed by Western blotting with anti-RGS3 or anti-G11 antibodies.
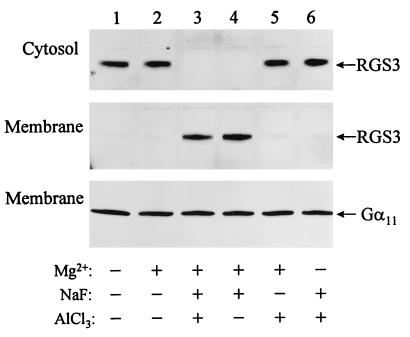
FIG. 7.
Redistribution of RGS3 from the cytosol to the membrane. The crude membrane and cytosol fractions were prepared from HMC/RGS3 as described in Materials and Methods. Equal amounts (per protein) of membranes and cytosol were mixed together in the presence of 10 μM GDP with or without 10 mM MgCl2, 10 mM NaF, and/or 30 μM AlCl3 as indicated. After incubation for 30 min at 4°C, the membranes were separated from the cytosol again by centrifugation, and the fractions were analyzed by Western blotting with anti-RGS3 or anti-αq/11 antibodies.
Indirect immunofluorescence microscopy.
The cells grown on glass chamber slides were fixed in Bouin’s solution containing 0.9% picric acid, 5% acetic acid, and 10% formaldehyde for 15 min at room temperature. The cells were then washed three times with 50% ethanol and twice with PBS, and the nonspecific binding was blocked for 30 min with 2% bovine serum albumin in PBS containing 2% normal goat serum. The cells were then incubated with affinity-purified polyclonal RGS3 antibodies (10 μg/ml) for 30 min, and washed four times with PBS, followed by incubation with 1:100 dilution of rhodamine-conjugated goat anti-rabbit immunoglobulin G (Pierce) for 30 min. The slides were additionally washed four times with PBS, and the coverslips were mounted with Gel/Mount aqueous mounting medium (Fisher, Pittsburgh, Pa.) and observed with a fluorescent microscope. Because the enhanced green fluorescent protein (EGFP) lost its fluorescence during the fixation procedure, visualization was achieved by immunofluorescence with monoclonal GFP antibodies (Clontech) in combination with fluorescein-conjugated goat anti-mouse antibodies (Pierce).
RESULTS
Stable overexpression and identification of RGS3.
In order to investigate the role of RGS3 in G protein-coupled receptor signaling and the mechanism of action of RGS3, we developed stable cell lines overexpressing RGS3. A human glomerular mesangial cell line (HMC), previously developed and characterized by Sraer et al. (31), was chosen as a host for two reasons. First, these cells do not express a detectable level of endogenous RGS3 as determined by immunoprecipitation followed by Western blotting with RGS3 antiserum (Fig. 1); second, HMC retained their characteristic endogenously expressed G protein-coupled ET-1 receptors, employed in the present work to evaluate the function of RGS3. Figure 1 shows an anti-RGS3 Western blot analysis of cell lysates from parental HMC, the cells stably transfected with pcDNA3.1 or pcDNA3.1/RGS3. Several bands were recognized by RGS3 antiserum in total cell lysates from parental and pcDNA3.1-transfected HMC. However, the major immunoreactive signal was produced by an 80-kDa protein from cells transfected with RGS3 cDNA. The identity of this band as RGS3 was confirmed by immunoprecipitation with RGS3 antiserum (Fig. 1), which was blocked by RGS3(40-52) peptide used as an antigen for antibody production (data not shown).
Coimmunoprecipitation of RGS3 with Gα subunits.
Several members of the RGS family have been reported to bind directly to an activated form of Gαi (10, 11, 18, 29) and to induce its GTPase activity (1, 2, 10, 19, 21, 35). GDP-AlF4−-, but not the GTP- or the GTPγS-bound form of αi was found to be preferable for binding RGS proteins in vitro (6, 35). RGS1, -4, and -10 and GAIP bound directly to αi but not to αs (21, 35) or αq (10, 11). Only one study has addressed this issue in respect to RGS3 and reported coprecipitation of purified RGS3 with αq in vitro (25). We investigated the ability of RGS3 to bind endogenous members of Gαi and Gαq/11 families by coimmunoprecipitation. Using the isoform-selective antibodies, we found that αi3 and α11 (but not αq) are the major isoforms of the Gαi and Gαq/11 families, respectively, endogenously expressed in HMC (data not shown). Therefore, the subsequent experiments were focused exclusively on these isoforms.
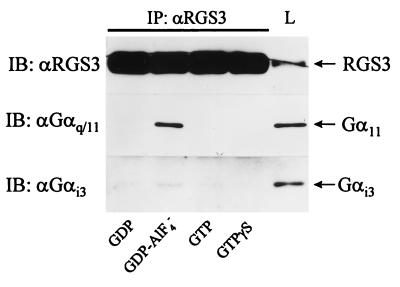
When G proteins were kept inactive in the presence of GDP alone, no significant coimmunoprecipitation of RGS3 with α11 and αi3 was detected (Fig. 2). The addition of AlF4− during immunoprecipitation of RGS3 resulted in a significant binding of RGS3 to α11 as determined by Western blotting of RGS3 immunoprecipitates with anti-αq/α11 antibodies. Consistent with binding characteristics of RGS1, RGS4, and GAIP to αi (1, 35), RGS3 interacted only with the GDP-AlF4−-bound form of endogenous α11 and failed to bind α11 in the presence of GTP or GTPγS (Fig. 2). Western blot analysis of the same RGS3 immunoprecipitates with anti-αi3 antibodies revealed a very weak but consistent αi3-immunoreactive signal only in the presence of AlF4− (Fig. 2). These data indicate that under these experimental conditions, activation of G proteins with AlF4− induces a physical association of RGS3 with endogenous α11 and, to a much lesser degree, with αi3.
FIG. 2.
Coimmunoprecipitation of RGS3 with Gα11 and Gαi3. HMC/RGS3 cells were lysed in Triton X-100 buffer as described in Materials and Methods, and the lysates were immunoprecipitated for 2 h with RGS3 antiserum in the presence of 10 μM GDP, 10 μM GDP plus 30 μM AlF4−, 10 μM GTP, or 10 μM GTPγS as indicated. Immunoprecipitates (IP; from 1 mg of protein) or total cell lysates (L; 50 μg of protein) were analyzed by immunoblotting with anti-RGS3, anti-αq/11, or anti-αi3 antibodies as indicated.
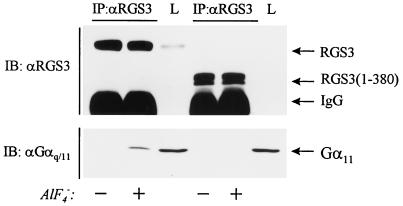
RGS3 contains a conserved region homologous within the family of RGS proteins (RGS domain) and a large N-terminal region which has no significant homology with any known protein (15). In order to validate the importance of the RGS domain of RGS3 in the binding of α11, we examined the ability of truncated RGS3(1-380), which lacks the RGS domain, to bind α11. As shown in Fig. 3, a band with an apparent molecular size of 70 kDa was identified as RGS(1-380) in human embryonic kidney 293 cells transiently transfected with RGS3(1-380) cDNA. Immunoprecipitation of cell lysates with RGS3 antiserum did not reveal any significant binding of RGS3(1-380) to α11 in the presence or absence of AlF4−. As was expected, under the same conditions AlF4− induced binding of the full-length RGS3 to α11 in the lysates from HEK293 cells transiently transfected with RGS3 cDNA (Fig. 3).
FIG. 3.
Truncated RGS3(1-380) does not bind α11. Human embryonic kidney 293 cells were transiently transfected with RGS3 cDNA (lanes 1 to 3) or with RGS3(1-380) cDNA (lanes 4 to 6) and lysed in Triton X-100 buffer 2 days after transfection. The cell lysates were immunoprecipitated for 2 h with RGS3 antiserum in the presence of 10 μM GDP with or without 30 μM AlF4−, as indicated. The RGS3 immunoprecipitates (IP) and total cell lysates (L) were analyzed by immunoblotting with anti-RGS3 or anti-αq/11 antibodies as indicated.
Taken together, these data indicate that in our experimental model, RGS3 binds preferably to AlF4−-activated α11 compared to αi3 and that this binding is mediated by the RGS domain of RGS3.
RGS3 inhibits ET-1-induced signaling.
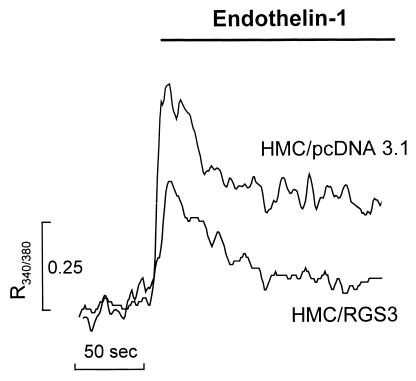
Gαq/11 were originally described as activators of phospholipase C (30). One of the downstream effects of PLC activation is inositol trisphosphate (IP3)-induced calcium mobilization followed by capacitative calcium entry (3, 4). Increasing evidence suggests that G11 is also responsible for the second phase (Ca2+ entry) of calcium response (24, 26). Therefore, in order to elucidate the physiological role of RGS3-G11 interaction, we first examined the effect of stable RGS3 overexpression on G protein-induced calcium rise in HMC cells. An increase of intracellular free-calcium concentration in response to ET-1 has been previously demonstrated in a variety of cells, including glomerular mesangial cells (29). In the present study, ET-1 elicited biphasic Ca2+ responses in HMC/pcDNA3.1 cells (Fig. 4), typical of those previously reported in glomerular mesangial cells. The biphasic profile consisted of an initial transient increase in cytosolic [Ca2+]i, determined as a fluorescent ratio (R340/380) equal to 0.63 ± 0.04 (n = 9), followed by a slow decline over time toward the resting level. By contrast, in HMC/RGS3 cells, the response of cytosolic [Ca2+] to ET-1 was significantly attenuated, having an initial rise in the R340/380 equal to 0.41 ± 0.06 (n = 8; P < 0.01 versus HMC/pcDNA3.1), with the second sustained phase not significantly different from the baseline (Fig. 4). This demonstrates a physiological role of RGS3 as an inhibitor of G protein signaling, presumably inhibiting both ET-1-induced activation of phospholipase C and calcium influx.
FIG. 4.
Inhibition of ET-1-induced Ca2+ responses by RGS3 overexpression. Serum-starved HMC/pcDNA3.1 or HMC/RGS3 cells were loaded with fura-2/AM, as outlined in Materials and Methods. The fluorescence ratio R340/380 was monitored as an index of cytosolic Ca2+ in the recording chamber of a spectrofluorimeter. The presence of ET-1 (10−7 M) is indicated by the bar above the traces. Shown are representative single traces from eight to nine individual experiments.
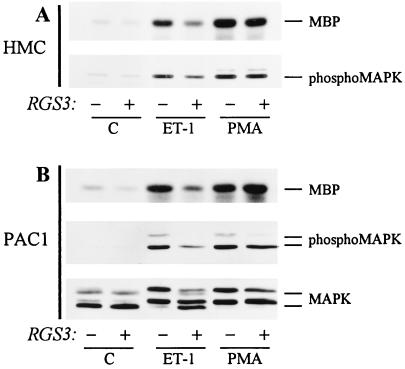
It was previously demonstrated that RGS3, as well as some other mammalian members of the RGS family, inhibited G protein-activated MAP kinase pathway in cells transiently cotransfected with RGS cDNA and receptor cDNA (9, 15). We examined whether stable RGS3 overexpression would influence MAP kinase activity in the HMC expressing a natural amount of receptors. As shown in Fig. 5a, ET-1 and PMA induced phosphorylation and activation of MAP kinase in HMC transfected with vector alone, as determined by Western blotting with phospho-specific MAP kinase antibodies and by myelin basic protein (MBP) phosphorylation assay of MAP kinase activity. A stable overexpression of RGS3 in HMC significantly attenuated phosphorylation and activation of MAP kinase induced by ET-1 by an average of 50%. By contrast, G protein-independent activation of MAP kinase by PMA in HMC/RGS3 was not significantly different from the one in HMC/pcDNA3.1 (Fig. 5A).
FIG. 5.
Inhibition of ET-1-induced MAP kinase activity by RGS3 overexpression. Serum-starved HMC (A) or PAC1 cells (B) stably transfected with pcDNA3.1 or pcDNA3.1/RGS3 were stimulated with 100 nM ET-1 or PMA for 5 min, as indicated. Cell lysates were subjected to the MAP kinase assay with MBP as a substrate or analyzed by Western blotting with anti-MAPK, or phospho-MAPK antibodies as indicated. Data represent the results of at least three experiments.
To ensure that the inhibitory effect of RGS3 overexpression on ET-1-induced MAP kinase activity was not a result of subcloning procedure, we performed similar experiments on pulmonary arterial smooth muscle cell line PAC1 (27), also stably expressing RGS3 (PAC1/RGS3). Consistently, the effect of ET1, but not PMA, on MAP kinase phosphorylation and activity, measured by MBP phosphorylation and by gel retardation of MAP kinase, was significantly smaller in PAC1/RGS3 than in PAC1/pcDNA3.1 (Fig. 5b).
Taken together, these data demonstrate a regulatory role of RGS3 in G protein-induced signaling in a cellular model with endogenous G proteins and receptors.
Regulatory role of endogenous RGS3 in ET-1-induced MAP kinase activity.
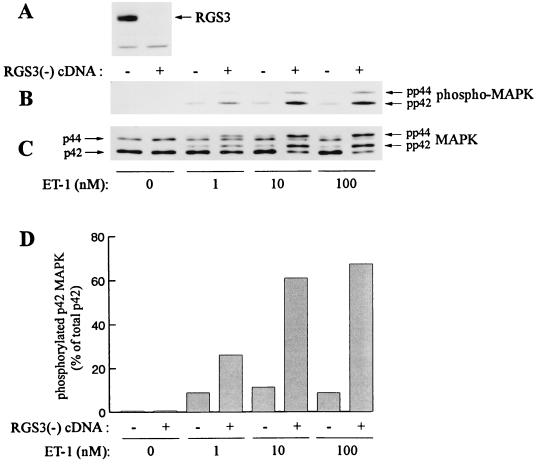
We have found that NIH 3T3 fibroblasts express significant amounts of RGS3 (Fig. 6A) and therefore could be used as a model for a study of endogenous RGS3. In our experiments, ET-1 at a concentration of 1 nM elicited a maximal effect on MAP kinase phosphorylation in NIH 3T3 cells as determined by immunoblotting of cell lysates with phospho-specific MAP kinase antibodies (Fig. 6B) and by gel retardation assay (Fig. 6C). Transfection of these cells with RGS3 cDNA in antisense orientation abolished the expression of endogenous RGS3 (Fig. 6A) and resulted in significant increase of MAP kinase phosphorylation induced by ET-1 at concentrations of 1 nM and even more dramatically at 10 and 100 nM (Fig. 6B and C). As assayed by densitometry of the shifted (phosphorylated) pp42 band over the total amount of MAP kinase [pp42/(p42 + pp42)], 60 versus 10% of total MAP kinase was phosphorylated in response to 10 nM ET-1 in RGS3-depleted versus control NIH 3T3 cells (Fig. 6D). This demonstrates a role of endogenous RGS3 in the regulation of Gq/11-coupled receptor signaling. Transfection of RGS3 antisense cDNA in NIH 3T3 cells did not affect the expression of a Gαi-specific member of the RGS family, RGS10 (21), as determined by immunoprecipitation and immunoblotting of cell lysates with anti-RGS10 antibodies (data not shown). Phosphorylation of MAP kinase induced by lysophosphatidic acid (Gi-linked), epidermal growth factor (G protein independent) and fetal bovine serum was not altered in RGS3-depleted cells (data not shown).
FIG. 6.
Effect of antisense RGS3 cDNA on ET-1-induced MAP kinase phosphorylation in NIH 3T3 cells. NIH 3T3 cells were stably transfected with the full-length RGS3 cDNA in reverse orientation [RGS3(-)] or with the vector alone, as indicated. Serum-starved cells were stimulated with different concentrations of ET-1 for 5 min. The total cell lysates were analyzed by immunoblotting with RGS3 antiserum (A), phospho-MAPK antibodies (B), or anti-p42/p44 MAPK antibodies (C). (D) The intensity of shifted (pp42) and unshifted (p42) MAPK (C) was analyzed by densitometry and represented as a percent of pp42 over the total (p42 plus pp42) MAPK. Data represent the results of two independent experiments.
Intracellular redistribution of RGS3 from the cytosol to the membrane fraction in vitro.
We further investigated the cellular distribution of RGS3 in HMC/RGS3 cells. Since RGS proteins interact with Gα subunit, it would be reasonable to assume a colocalization of RGS and Gα in the membrane. Surprisingly, after crude preparation of membranes from HMC/RGS3, we detected RGS3 immunoreactivity almost exclusively in the cytosol, while α11 was in the membrane fraction as expected (Fig. 7). However, activation of G proteins with AlF4− or NaF in the presence of Mg2+ resulted in redistribution of RGS3 from the cytosol to the membrane fraction (Fig. 7, lanes 3 and 4, respectively). Omission of Mg2+ or F− from the incubation mixture abolished redistribution of RGS3 (Fig. 7, lanes 5 and 6).
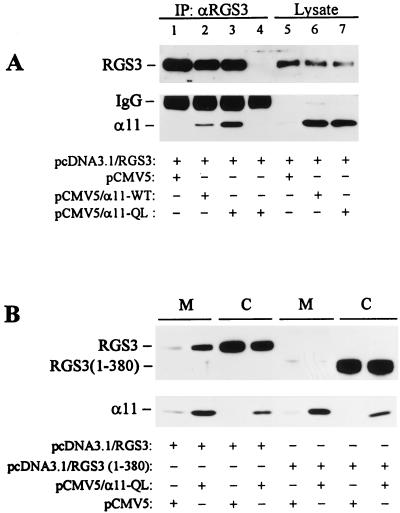
In order to demonstrate that redistribution of RGS3 reflects the binding of RGS3 to the activated α11, we determined whether RGS3 would bind to the membrane fraction in the presence of constitutively active α11 without the addition of AlF4−. Immunoprecipitation of RGS3 from the lysates of HEK293 cells transiently overexpressing RGS3 together with GTPase-deficient mutant of α11 (α11-QL) revealed a significant binding of α11-QL to RGS3 under the basal conditions (Fig. 8A). Interestingly, some coimmunoprecipitation of RGS3 with the wild-type α11-WT was also observed. This could reflect the presence of residual amounts of GTP-bound α11-WT unable to be inactivated by RGS3 because of its dramatic overexpression. On the other hand, RGS3 could interact with GDP-bound α11 with low affinity, detectable under these experimental conditions. Nevertheless, RGS3 bound to α11-QL with much higher efficiency than to α11-WT, and no binding was detected with endogenous α11 under the basal conditions (Fig. 8A). Overexpression of α11-QL in HEK293 cells also resulted in an increase of RGS3 immunoreactivity in the crude membrane fraction in the absence of AlF4− (Fig. 8B). No significant difference in membrane-bound RGS3 was detected between the cells transfected with the vector and α11-WT cDNA (data not shown). Moreover, consistent with the lack of interaction between RGS3(1-380) and α11 (Fig. 3), this RGS3 fragment also failed to bind to the membrane fraction in the presence of α11-QL (Fig. 8B).
FIG. 8.
Binding of RGS3 to the membrane as a result of interaction with constitutively active α11-QL. Human embryonic kidney 293 cells grown on 10-cm-diameter dishes were transiently cotransfected with 5 μg of pcDNA3.1/RGS3, or 5 μg of pcDNA3.1/RGS3(1-380) together with 5 μg of pCMV5, pCMV5/α11-WT, or pCMV5/α11-QL, as indicated. The cells were lysed in Triton X-100 buffer (A) or subjected to crude membrane and cytosol preparation (B) 2 days after transfection. The cell lysates were immunoprecipitated for 2 h with RGS3 antiserum (A, lanes 1 to 3) or preimmune serum (A, lane 4). The immunoprecipitates and total cell lysates (A) or crude membrane and cytosol fractions (B) were analyzed by immunoblotting with anti-RGS3 or anti-α11 antibodies, as indicated.
These data indicate that the binding of RGS3 to the membrane fraction in vitro is, at least in part, a result of interaction of RGS3 with GTP-bound α11. However, some observations suggested the possibility that other mechanisms are involved in intracellular redistribution of RGS3. First, a significant amount of RGS3 remained in the cytosolic fraction in the presence of α11-QL (Fig. 8B) compared to the effect of AlF4− (Fig. 7). Second, we were not able to detect the intracellular redistribution of RGS3 after stimulation of intact cells with physiological activators of α11 followed by crude membrane preparation. This might be explained by unstability of RGS3-α11 complex and/or by requirement of other factors necessary for RGS3 redistribution. To address this issue, we next used the immunofluorescence technique, which allowed us to preserve the intracellular structure and to view more precisely the distribution of RGS3 after stimulation with agonists.
Agonist-induced translocation of RGS3 from the cytosol to the plasma membrane in intact cells.
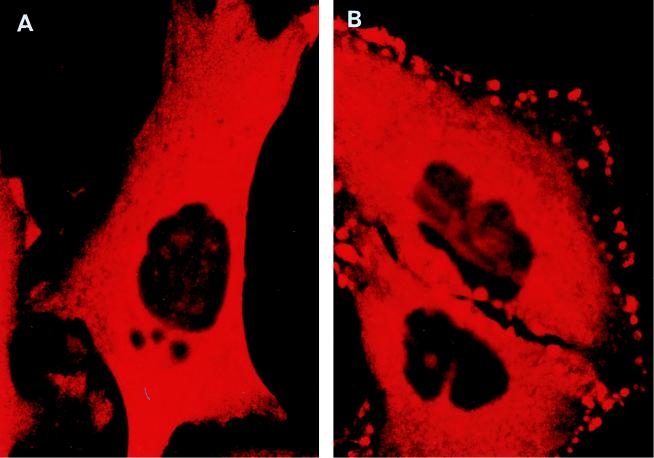
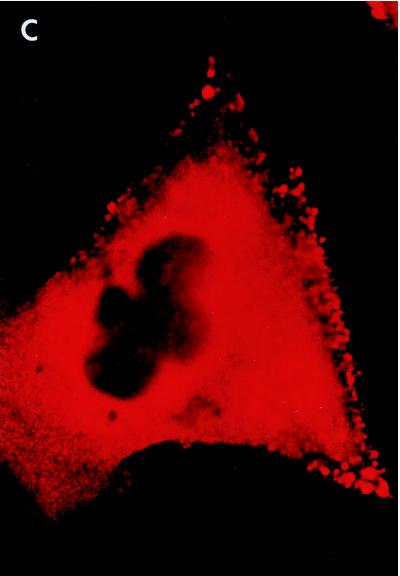
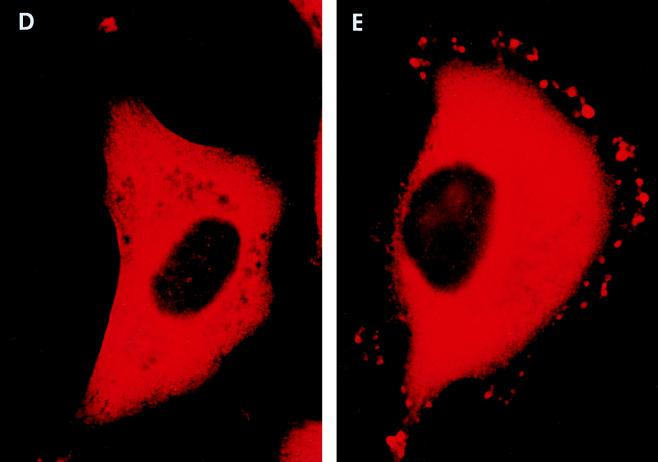
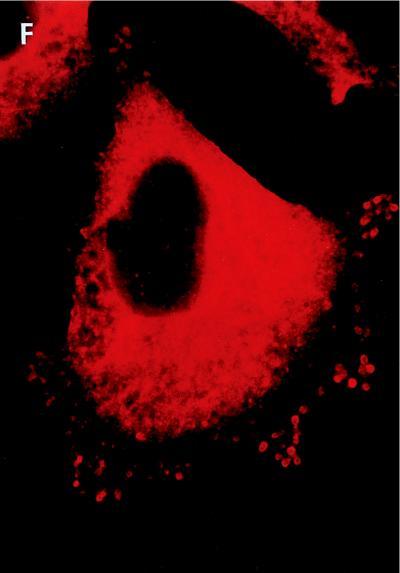
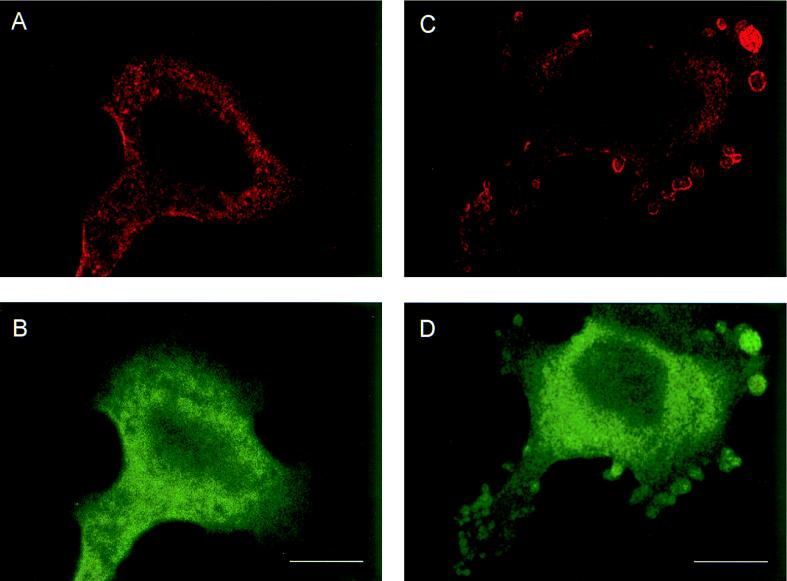
Whole-cell immunofluorescent microscopy demonstrated that RGS3 was diffusely localized predominantly in the cytoplasm of intact HMC/RGS3 cells (Fig. 9A). In some cells, occasional appearance of RGS3 was observed at the edge of the cell, probably due to the basal activity of G proteins. No significant immunofluorescence of HMC/pcDNA3.1 cells stained with RGS3 antibodies was observed (data not shown). Likewise, no labeling of HMC/RGS3 cells was detected when the primary antibodies were omitted during the staining procedure (data not shown). Incubation of HMC/RGS3 cells with ET-1 resulted in significant membrane ruffling viewed as small microprojections, different in size and amount per cell. These structures probably represent cytoplasmic processes or extensions common for glomerular mesangial cells in vivo (23) and have also been shown to be induced by angiotensin II (32). In ET-1-stimulated cells, RGS3 appeared to be concentrated in the membrane ruffles, although a significant amount still remained in the cytoplasm (Fig. 9B). Confocal immunofluorescent microscopy of ET-1-stimulated cells labeled with RGS3 antibodies showed significantly reduced cytoplasmic staining compared to whole-cell microscopy and indicated a distinctive membrane localization of RGS3 within the ruffles (Fig. 10C). By contrast, the green fluorescent protein variant (EGFP), coexpressed with RGS3 for control purposes, showed relatively diffuse staining in the cytoplasm and, if trapped in the ruffles, was not present in the membrane (Fig. 10B and D).
FIG. 9.
Immunofluorescent microscopy of RGS3 and RGS3(1-380) after stimulation of cells with ET-1 and A23187. HMC/RGS3 (A to C) or HMC/RGS3(1-380) (D to F) was incubated without (A and D) or with 300 nM ET-1 (B and E) or 10 μM A23187 (C and F) for 10 min, followed by indirect immunofluorescence microscopy with affinity-purified RGS3 antibodies as described in Materials and Methods. Magnification; × 100.
FIG. 10.
Confocal images of RGS3 and EGFP after stimulation of cells with ET-1. HMC were transiently cotransfected with RGS3 cDNA (A and C) and EGFP cDNA (B and D), serum starved, and incubated without (A and B) or with (C and D) 300 nM ET-1 for 10 min, followed by immunofluorescent staining as described in Materials and Methods. Shown are the RGS3 (red) and EGFP (green) images of the optical section at the bottom of the cells. Bars, 10 μm.
In order to determine whether redistribution of RGS3 was dependent exclusively on RGS3-α11 interaction, we performed similar immunofluorescence experiments on HMC stably overexpressing RGS3(1-380) fragment lacking the RGS domain, which did not interact with α11 (Fig. 3) and did not bind to the membrane fraction in the presence of α11-QL in vitro (Fig. 8B). Surprisingly, similar to the full-length RGS3, its N-terminal fragment also translocated to the membrane upon stimulation of HMC/RGS3(1-380) cells with ET-1 (Fig. 9E). This indicates that the N-terminal domain of RGS3 is important for its translocation, which could involve a mechanism(s) other than the interaction of RGS3 with α11.
IP3-induced increase of intracellular Ca2+ and diacyl glycerol-mediated activation of protein kinase C (PKC) are the most proximal to PLC second messengers involved in ET-1 signaling. Therefore, we investigated whether these pathways could mediate the ET-1-induced intracellular redistribution of RGS3. No significant translocation of RGS3 and RGS3(1-380) to the plasma membrane was observed after activation of PKC by PMA (data not shown). However, stimulation of cells with calcium ionophore A23187 resulted in increased membrane staining of RGS3, with a striking similarity to ET-1-induced redistribution of RGS3 (Fig. 9C). Moreover, A23187 elicited a similar effect on N-terminal fragment RGS3(1-380) (Fig. 9F).
Taken together, these data suggest that agonist-induced translocation of RGS3 is a complex process which occurs in a Ca2+- and G protein-dependent manner and involves both N-terminal and C-terminal domains of RGS3.
DISCUSSION
In the present study we describe three major findings. First, we demonstrate the physiological role of RGS3 as an inhibitor of Gαq/11 signaling, using intact cellular models with endogenous expression of receptors and G proteins as well as RGS3. Second, we describe a cytosolic localization of RGS3 and translocation of RGS3 to the plasma membrane after activation of G proteins. Finally, we show the importance of the N-terminal domain of RGS3, suggesting a dual role for RGS3 in cellular signaling.
RGS proteins were first described as inhibitors of Gi-mediated signaling (15, 22). In vitro experiments demonstrated that RGS1, -4, and -10 and GAIP function as GTPase-activating proteins for the alpha subunit of the Gi family (1, 2, 21, 35). AlF4−-activated αi/αo were found to bind directly to purified RGS proteins (1, 21, 35), while αq bound with much lower affinity (1), and αs did not bind at all (1, 21, 35). In terms of RGS3, only one study has addressed the issue of its interaction with G proteins and reported that RGS3 bound to αq applying purified proteins in vitro (25). Our study expanded this observation and demonstrated that RGS3 binds to endogenous G11 (Fig. 2), most likely by its conserved RGS domain (Fig. 3). Consistent with other members of the RGS family, RGS3 interacted only with activated α11, with a clear preference to the GDP-AlF4−-bound conformation (Fig. 2). The failure to coimmunoprecipitate α11 with RGS3 in the presence of GTP does not reflect a lack of interaction between RGS3 and GTP-bound α11 but, rather, may be a result of increased GTP hydrolysis and a subsequent release of RGS3 from inactivated α11. Supporting this possibility is the fact that RGS3 bound to constitutively active GTPase-deficient Q209L mutant of α11 in the absence of AlF4− (Fig. 8).
We further confirmed that binding of RGS3 to Gα11 results in inhibition of G protein-linked signaling. A biphasic increase of intracellular free Ca2+ concentration is a common response in Gq/11-coupled receptor signaling, and the effect of RGS proteins on this component of signaling has not been studied before. Since Gα11 is the major representative of the known members of Gαq/11 family in HMC, it is probably responsible for the ET-1-induced increase in [Ca2+]i in our system. In the present work, stable overexpression of RGS3 in HMC resulted in significant attenuation of both initial and the second phase of Ca2+ response to ET-1 (Fig. 4).
Inhibition of G protein-induced MAP kinase activity by RGS proteins has been established by transient cotransfection of RGS cDNA together with cDNA for specific receptor (9, 15). Using two independent stable cell lines, we demonstrate that RGS3 inhibits MAP kinase activity induced by endogenous Gq/11-coupled ET-1 receptors (Fig. 5). Moreover, depletion of endogenous RGS3 by antisense technique resulted in enhanced ET-1-induced phosphorylation of MAP kinase in the NIH 3T3 cellular model involving physiological amounts of RGS3 and ET-1 receptors (Fig. 6). Overexpression of antisense RGS3 cDNA did not alter phosphorylation of MAP kinase induced by lysophosphatidic acid (data not shown), which is known to activate this pathway via a Gi-dependent mechanism (33). This supports our coimmunoprecipitation data showing relatively weak binding of RGS3 to αi3 (Fig. 2).
The role of endogenous RGS proteins in mammalian cells has not been studied before. In our experiments, RGS3 depletion resulted in an increased maximal MAP kinase response rather than in sensitivity to ET-1 (Fig. 6). This is consistent with the effect of RGS4 overexpression on G protein signaling in both mammalian (20, 37), and yeast (15) systems, showing the reduction of maximal response without affecting the potency of the agonist. Moreover, gain-of-function mutations of the yeast RGS, Sst2p, had a similar effect on G protein-dependent pheromone response (13). However, Sst2 loss-of-function mutations (the model, somewhat similar to RGS3 depletion in our experiments) resulted in increased potency but not in increased efficacy of pheromone (12). This may suggest differences between the function of various RGSs or may reflect the differences in experimental models and the readout of final responses.
Perhaps the most intriguing and unexpected finding of the present work is the cytosolic localization of RGS3 (Fig. 7 and 8). The other mammalian RGS GAIP was reported to be membrane bound and behaved as integral membrane protein (10). The yeast RGS Sst2p was also found in the membrane and copurified with G proteins during cell fractionation (14). In fact, the membrane colocalization of RGS and G proteins would be a reasonable assumption, considering the function of RGS as a Gα-interacting protein. . However, our cell fractionation experiments (Fig. 7 and 8) as well as immunofluorescent microscopy (Fig. 9) clearly indicate that RGS3 is a predominantly cytosolic protein. One might speculate that RGS3 may serve some function in cytoplasm other than interaction with G proteins, suggesting a dual role of RGS3 in cellular signaling. Our additional experiments support this hypothesis by detecting a 28-kDa protein which coimmunoprecipitates with both full-length and N-terminal fragment of RGS3 from cell lysates of transfected HMC cells labeled with [35S]methionine (data not shown).
Agonist-induced translocation of RGS3 is another important aspect of its function which also involves both G protein-dependent and -independent mechanisms. In our experiments, in vitro binding of RGS3 to the crude membrane fraction was detectable only under conditions which provided a stable RGS3-G11 complex (Fig. 7 and 8). This binding was mediated by the RGS domain of RGS3 and probably reflected an interaction of RGS3 with the activated Gα subunit. Immunofluorescence microscopy permitted detection of RGS3 translocation induced by physiological agonist ET-1 (Fig. 9 and 10) but also indicated the importance of [Ca2+]i rise in the mechanism of translocation, which alternatively was mediated by N-terminal domain of RGS3 (Fig. 9). This may suggest that agonist-induced redistribution of RGS3 is a biphasic process consisting of (i) Ca2+-dependent, N-terminal domain-mediated translocation of RGS3 and (ii) RGS domain-mediated binding of RGS3 to G protein. On the other hand, in vitro binding of RGS3 to the membrane fraction induced by AlF4− (Fig. 6) was more efficient than the one mediated by constitutively active α11-QL (Fig. 8). Considering the ability of AlF4− to stimulate other GTP-binding proteins (7), one might suggest the possibility of other targets of RGS3 in the membrane. In this regard it is necessary to mention that constitutively active Gαi2(Q207L) induced translocation of RGS4 to the membrane by a mechanism independent of RGS4-G protein interaction (16). The fact that RGS4 consists almost entirely of the RGS domain may suggest that the mechanism of its recruitment to the membrane is different from that of RGS3, wherein G protein-independent component of RGS3 redistribution is mediated by its N-terminal domain. In addition, this may suggest that the RGS domain itself may have targets distinct from G proteins and function other than GAP for Gα subunits.
In conclusion, this work demonstrates that inhibition of G protein signaling by RGS3 is a complex process involving its translocation from the cytosol to the plasma membrane in addition to its established interaction with G proteins. Our data also suggest the possibility of cytosolic function of RGS3 distinct from the inactivation of G proteins.
ACKNOWLEDGMENTS
We thank Jean-Daniel Sraer for providing the human glomerular mesangial cell line, Abraham Rothman for providing the rat pulmonary arterial smooth muscle cell line, Hiroshi Itoh for providing α11-WT and α11-QL cDNA, and David J. Lacey for expert assistance in calcium measurements.
This work was supported by NIH grants HL 22563 and DK 41684 to M.J.D., a grant from the Milheim Foundation for Cancer Research to A.S., a grant-in-aid (96013570) from the National American Heart Association, and a grant from the Children’s Hospital of Wisconsin Foundation to S.J.E.
REFERENCES
- 1.Berman D M, Kozasa T, Gilman A G. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 2.Berman D M, Wilkie T M, Gilman A G. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 3.Berridge M J. Inositol trisphosphate and calcium signaling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 4.Berridge M J. Capacitative calcium entry. Biochem J. 1995;312:1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnbaumer L, Abramowitz J, Brown A M. Receptor-effector coupling by G proteins. Biochim Biophys Acta. 1990;1031:163–224. doi: 10.1016/0304-4157(90)90007-y. [DOI] [PubMed] [Google Scholar]
- 6.Carty D J, Iyengar R. Guanosine 5′-O-(γ-thio)triphosphate binding assay for solubilized G proteins. Methods Enzymol. 1994;273:38–44. doi: 10.1016/s0076-6879(94)37051-6. [DOI] [PubMed] [Google Scholar]
- 7.Chabre M. Aluminofluoride and beryllofluoride complexes: new phosphate analogs in enzymology. Trends Biochem Sci. 1990;15:6–10. doi: 10.1016/0968-0004(90)90117-t. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover A, Schwartz A L. The ubiquitin-mediated proteolytic pathway: mechanisms of recognition of proteolytic substrate and involvement in the degradation of native cellular proteins. FASEB J. 1994;8:182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- 9.Chattejee T K, Eapen A K, Fisher R A. A truncated form of RGS3 negatively regulates G protein-coupled receptor stimulation of adenylyl cyclase and phosphoinositide phospholipase C. J Biol Chem. 1997;272:15481–15487. doi: 10.1074/jbc.272.24.15481. [DOI] [PubMed] [Google Scholar]
- 10.De Vries L, Elenko E, Hubler L, Jones TLZ, Farquhar M G. GAIP is membrane-anchored by palmitoylation and interacts with the activated (GTP-bound) form of Gαi subunits. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vries L, Mousli M, Wurmser A, Farquhar M G. GAIP, a protein that specifically interacts with the trimeric G protein G alpha i3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietzel C, Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohlman H G, Apaniesk D, Chen Y, Song J, Nusskern D. Inhibition of G protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3635–3643. doi: 10.1128/mcb.15.7.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohlman H G, Song J, Ma D, Courschesne W E, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpal (the G protein α subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Druey K M, Blumer K J, Kang V H, Kehrl J U. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 16.Druey K M, Sullivan B M, Brown D, Fischer E R, Watson N, Blumer K J, Gerfen C R, Scheschonka A, Kehrl J H. Expression of GTPase-deficient Gia2 results in translocation of cytoplasmic RGS4 to the plasma membrane. J Biol Chem. 1998;273:18405–18410. doi: 10.1074/jbc.273.29.18405. [DOI] [PubMed] [Google Scholar]
- 17.Dulin N O, Madhun Z T, Chang C-H, Berti-Mattera L, Dickens D, Douglas J G. Angiotensin IV receptors and signaling in opossum kidney cells. Am J Physiol. 1995;269:F644–F652. doi: 10.1152/ajprenal.1995.269.5.F644. [DOI] [PubMed] [Google Scholar]
- 18.Elliott S J, Eskin S G, Schilling W P. Effect of t-butyl-hydroperoxide on bradykinin-stimulated changes in cytosolic calcium in vascular endothelial cells. J Biol Chem. 1989;264:3806–3810. [PubMed] [Google Scholar]
- 19.Hepler J R, Berman D M, Gilman A G, Kozasa T. RGS4 and GAIP are GTPase-activating proteins for Gqα and block activation of phospholipase Cβ by γ-thio-GTP-Gqa. Proc Natl Acad Sci USA. 1997;94:428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, Hepler J R, Gilman A G, Mumby S M. Attenuation of Gi- and Gq- mediated signaling by expression of RGS4 or GAIP in mammalian cells. Proc Natl Acad Sci USA. 1997;94:6159–6163. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt T W, Fields T A, Casey P J, Peralta E G. RGS10 is a selective activator of Gαi GTPase activity. Nature. 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- 22.Koelle M R, Horvitz H R. EGL-10 regulates G protein signaling in the C elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 23.Kriz W, Elger M, Lemley K, Sakai T. Structure of the glomerular mesangium: a biomechanical interpretation. Kidney Int. 1990;38:S2–S9. [PubMed] [Google Scholar]
- 24.Macrez-Lepretre N, Kalkbrenner F, Schultz G, Mironneau J. Distinct functions of Gq and G11 proteins in coupling α1-adrenoreceptors to Ca2+ release and Ca2+ entry in rat portal vein myocytes. J Biol Chem. 1997;272:5261–5268. doi: 10.1074/jbc.272.8.5261. [DOI] [PubMed] [Google Scholar]
- 25.Neill J D, Duck L W, Sellers J C, Musgrove L C, Scheschonka A, Druey K M, Kehrl J H. Potential role for a regulator of G protein signaling (RGS3) in gonadotropin-releasing hormone (GnRH) stimulated desensitization. Endocrinology. 1997;138:843–846. doi: 10.1210/endo.138.2.5034. [DOI] [PubMed] [Google Scholar]
- 26.Obukhov A G, Harteneck C, Zobel A, Harhammer R, Kalkbrenner F, Leopoldt D, Luckhoff A, Nurnberg B, Schultz G. Direct activation of trpl cation channels by Gα11 subunits. EMBO J. 1996;15:5833–5838. [PMC free article] [PubMed] [Google Scholar]
- 27.Rothman A, Kulik T J, Taubman M B, Berk B C, Smith C W J, Nadal-Ginard B. Development and characterization of a cloned rat pulmonary arterial smooth muscle cell line that maintains differentiated properties through multiple subcultures. Circulation. 1992;86:1977–1986. doi: 10.1161/01.cir.86.6.1977. [DOI] [PubMed] [Google Scholar]
- 28.Simon M I, Strathmann M P, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 29.Simonson M S, Wann S, Mene P, Dubyak G R, Kester M, Nakazato Y, Sedor J R, Dunn M J. Endothelin stimulates phospholipase C, Na+/H+ exchange, c-fos expression, and mitogenesis in rat mesangial cells. J Clin Investig. 1989;83:708–712. doi: 10.1172/JCI113935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiegel A M. Signal transduction by guanine nucleotide binding proteins. Mol Cell Endocrinol. 1987;49:1–6. doi: 10.1016/0303-7207(87)90058-x. [DOI] [PubMed] [Google Scholar]
- 31.Sraer J-D, Delarue F, Hagege J, Feunteun J, Pinet F, Nguyen G, Rondeau E. Stable cell lines of T-SV40 immortalized human glomerular mesangial cells. Kidney Int. 1996;49:267–270. doi: 10.1038/ki.1996.38. [DOI] [PubMed] [Google Scholar]
- 32.Takeda K, Meyer-Lehnert H, Kim J K, Schrier R W. Effect of angiotensin II on Ca2+ kinetics and contraction in cultured rat glomerular mesangial cells. Am J Physiol. 1988;254:F254–F266. doi: 10.1152/ajprenal.1988.254.2.F254. [DOI] [PubMed] [Google Scholar]
- 33.Van Biesen T, Hawes B E, Luttrell D K, Krueger K M, Touhara K, Porfiri E, Sakaue M, Luttrell L M, Lefkowitz R J. Receptor-tyrosine-kinase- and Gβγ-mediated MAP kinase activation by a common signalling pathway. Nature. 1995;376:781–784. doi: 10.1038/376781a0. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Simonson M S, Pouyssegur J, Dunn M J. Endothelin rapidly stimulates mitogen activated protein kinase activity in rat mesangial cells. Biochem J. 1992;287:589–594. doi: 10.1042/bj2870589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. RGS family members: GTPase-activating protein for heterotrimeric G protein α-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Lee C H, Rhee S G, Simon M I. Activation of phospholipase C by the α subunits of the Gq and G11 proteins in transfected Cos-7 cells. J Biol Chem. 1992;267:1811–1817. [PubMed] [Google Scholar]
- 37.Yan Y, Chi P P, Bourne H R. RGS4 inhibits Gq-mediated activation of mitogen-activated protein kinase and phosphoinositide synthesis. J Biol Chem. 1997;272:11924–11927. doi: 10.1074/jbc.272.18.11924. [DOI] [PubMed] [Google Scholar]