To reach the goal of curing currently incurable hematologic malignancies, we need to go beyond focusing on single gene mutatons and gain deeper understanding of the consequences of genetic alterations on gene-regulatory pathways. Edited by John Crispino, these 5 cutting-edge reviews from leaders in the their fields not only summarize our current understanding of key pathways that contribute to myeloid malignancies, but also discuss new therapeutic avenues related to them. They provide a springboard for further groundbreaking basic and clinical advances in hematologic malignancies.
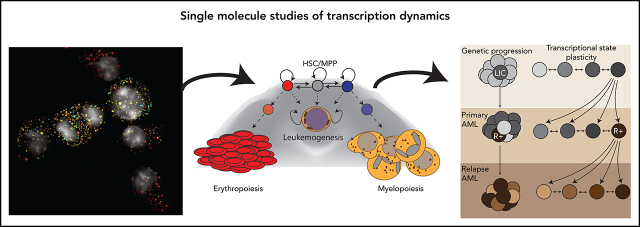
Visual Abstract
Abstract
Nongenetic heterogeneity, or gene expression stochasticity, is an important source of variability in biological systems. With the advent and improvement of single molecule resolution technologies, it has been shown that transcription dynamics and resultant transcript number fluctuations generate significant cell-to-cell variability that has important biological effects and may contribute substantially to both tissue homeostasis and disease. In this respect, the pathophysiology of stem cell-derived malignancies such as acute myeloid leukemia and myelodysplastic syndromes, which has historically been studied at the ensemble level, may require reevaluation. To that end, it is our aim in this review to highlight the results of recent single-molecule, biophysical, and systems studies of gene expression dynamics, with the explicit purpose of demonstrating how the insights from these basic science studies may help inform and progress the field of leukemia biology and, ultimately, research into novel therapies.
Introduction
Cellular identity is encoded by the repertoire of genes expressed and repressed by a cell. These baseline patterns of gene expression, as well as the functional modules executed by the cell, are organized in networks. The performance of all transcription networks is contingent on the selective participation of the genome in a spatiotemporally coherent manner. In that regard, transcription factors (TF) are essential players in the maintenance of cellular identity through the specific and regulated licensing of loci in the genome. Although developing a quantitative understanding of TF dynamics and their impact on transcriptional networks is an area of obvious interest to developmental biology, aberrant networks are also a defining and ubiquitous feature of oncogenesis. In particular, dysregulation of normal differentiation networks and maintenance of a progenitor-like state seems to be prevalent in acute leukemogenesis.1-9 Indeed, as the high degree of genetic heterogeneity found both within and between patients with myelodysplastic syndromes and acute leukemias continues to be appreciated,10-13 deregulation of essential hematopoietic TF such as CEBPA, RUNX1, MEIS1, HOXA9, GATA2, and PU.1, may be the unifying pathological hallmark of these notoriously recalcitrant tumors rather than any particular genetic event.14-22 Elucidating the operational rules of normal hematopoietic differentiation networks will therefore affect our understanding of tumor pathogenesis and may inform novel therapeutic strategies.
Over the past few decades, technological developments have facilitated the study of gene expression at increasingly finer resolution. The forefront of these techniques has enabled the investigation of single cell behaviors, and at times even the study of single messenger RNA (mRNA) and protein molecules in living cells. When combined with powerful computational tools, machine learning algorithms, and theoretical insights from systems biology, these studies have provided an unprecedented and rapidly evolving understanding of life within the cell. This insight has, in many cases, demonstrated substantial intrinsic heterogeneity, or stochasticity, in the behavior of genetically identical cells in homogeneous conditions. Although these results are largely congruent with predictions made many decades ago by theoretical biologists and physicists,23-25 how to reconcile the complicated, even counterintuitive, picture emerging of life at the single-cell level with our notion of robust steady-state hematopoiesis is a challenge. Similarly, these studies present a major hurdle for the field in understanding the pathogenesis and the therapeutic targeting of malignancies such as myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).
In this review, we will highlight the results of recent in situ, single-molecule, biophysical, and systems studies of transcription dynamics. The goal is to demonstrate how the results of these studies may require a reformatting of our understanding of leukemogenesis, particularly at the single-cell level. Indeed, as the oncogenicity imputed to TF “misexpression” is based on the premise that TF regulate their target genes in a concentration-dependent fashion, a quantitative model of TF expression and subsequent gene regulation is needed to ultimately understand AML pathogenesis.
Deregulation of transcription factors in AML
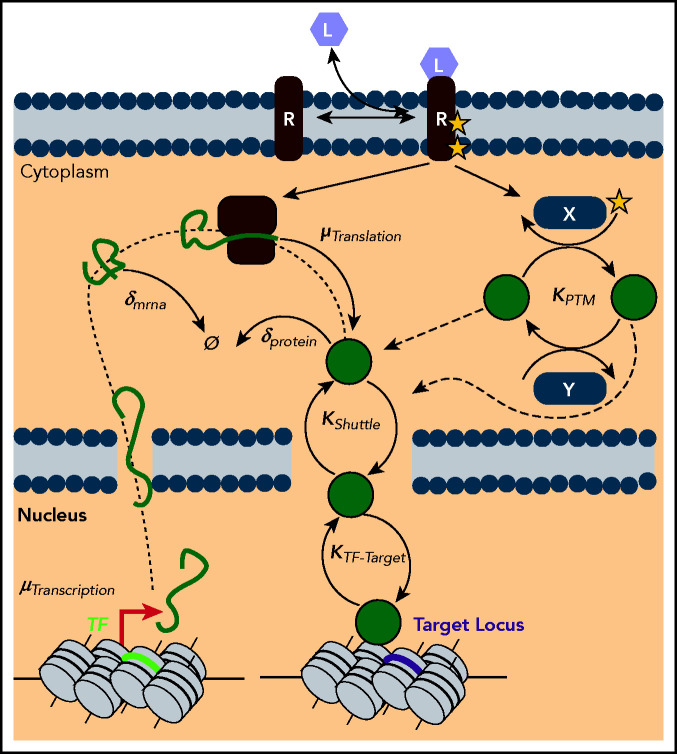
The complex cascade of reactions involved in “gene expression” offers multiple avenues by which TF levels and/or activity may be manipulated (Figure 1). Before delving into the major findings from single-cell gene expression studies, we will present a nonexhaustive survey of how hematopoietic transcription factors are known to be deregulated in AML.
Figure 1.
Gene expression as a multilevel reaction cascade. Schematic of the complex reaction cascade of gene expression. Deregulation of TF levels or activity can putatively occur along any step in the pathway, from synthesis steps such as transcription (µTranscription) and translation (µTranslation), to degradation steps such as the decay of mRNA (δmrna) and protein (δprotein), to dynamic processes such as posttranslational modifications by ligand activated pathways (KPTM), to shuttling dynamics (KShuttle), and finally to target locus search and binding (KTF-Target).
Transcriptional deregulation
The classic example of transcriptional deregulation of TF comes from one of the earliest identified AML oncogenes, the onco-fusion protein PML-RARα. In a series of seminal studies, it was determined that this fusion gene, the putative driver of acute promyelocytic leukemia, was actually the fusion of 2 TFs. As such, its pathogenicity is presumptively derived from its ability to deregulate the transcription network of the promyelocyte genome.3,26,27 More recently, the oncogenicity of another well-studied class of onco-fusion proteins, the MLL rearrangements, was demonstrated to be due to the mislocalization of the activating histone methyltransferase DOT1L, leading to increased RNA polymerase processivity genome wide and the overexpression of genes typically silenced during terminal myelopoiesis.28-34 In particular, MLL-AF9–mediated leukemogenesis in mouse models is thought to be dependent on the ability of this fusion to drive the aberrant expression of the HOX-related TF genes, Meis1 and Hoxa9, leading to a more primitive, blast-like state known as a leukemic granulocyte macrophage progenitor.35 It was also demonstrated that retroviral overexpression of these TF alone is sufficient to transform normal human hematopoietic stem and progenitor cells (HSPCs).36 Within human patient samples, one of the most striking examples of TF transcriptional deregulation is the aberrant expression of the EVI1 oncogene in inv(3) AML. This inversion event, which has 1 of the worst prognoses of all recurrent AML genetic events,37 reorients the highly active GATA2 enhancer toward the stem cell TF EVI1, leading to significant misexpression of the latter TF.6,9 EVI1 overexpression subsequently leads to installation and enforcement of a stem cell transcriptional network by physically antagonizing the transactivation of lineage specifying TF such as PU.1, GATA1, and RUNX1. Furthermore, the subsequent downregulation of GATA2 potentiates the EVI1 phenotype in human AML cells,9 a finding consistent with studies showing that haploinsufficiency of Gata2 accelerates Evi-1 driven AML pathogenesis in mice.38
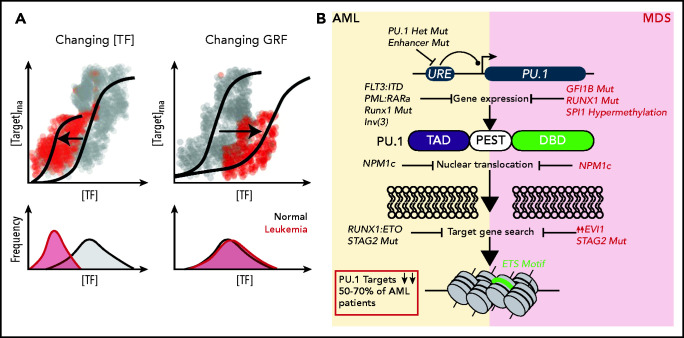
Direct transcriptional repression of TF is also a hallmark of many AML etiologies. For instance, the onco-fusion RUNX1-ETO, generated by the t(8;21) translocation, is thought to induce leukemia in part by the repression of CEBPA transcription and PU.1-dependent transcriptional activation,39 leading to a block in terminal granulocyte differentiation.40 RUNX1-ETO has also been shown to directly repress the expression of the tumor suppressor TF, p14(ARF).41 Another powerful example of how direct transcription deregulation of TF can precipitate AML includes the myriad of ways in which the master regulatory myeloid TF PU.1 is affected during leukemogenesis. This deregulation occurs at both the level of changing PU.1 concentration, as well as through direct uncoupling of the PU.1 gene regulatory function at transcriptional targets (Figure 2A). One example of the former is the murine model of spontaneous AML generated by loss of the −14 kb upstream regulatory element of the Spi1 (PU.1) gene.8,42 Loss of this upstream regulatory element leads to a roughly 80% reduction in the mRNA and protein level of PU.1 in hematopoietic stem cell and myeloid progenitors, ultimately precipitating a fulminant AML with virtually 100% penetrance. Moreover, heterozygous loss of this enhancer on a mutagenic, mismatch repair-deficient background also generates MDS and progression to AML in ∼70% of mice.43 This finding is paralleled in human MDS and AML pathogenesis, with roughly 50% to 70% of tumors downregulating this key TF via a variety of mechanisms, including mutation of upstream regulators such as GFI1b and RUNX1, NPM1c mutations that inhibit PU.1 shuttling into the nucleus, or through sequestration and physical blocking of PU.1 protein by oncoproteins such as RUNX1-ETO.44-51 Additionally, it appears that mutations in the cohesin complex, particularly Stag2, limits PU.1 ability to access and activate genes required for terminal differentiation.52 Finally, PU.1 heterozygous mutations are found in a subset of MLL rearranged leukemias, with subsequent downregulation of PU.1 target genes46 (Figure 2B).
Figure 2.
Multiple pathways of TF deregulation in AML and MDS. (A) TFs modify the output of target genes by influencing the reaction propensities of various steps in transcription in a manner proportional to some function of TF concentration. These functions can take on a variety of shapes, depending on the number of binding sites and the higher order complexes that a given TF is involved in at a target locus. Cooperativity at a locus produces gene regulatory functions (GRF) that are sigmoidal in shape. Although not mutually exclusive phenomena, in a simplistic sense deregulation can occur either through reducing the amount of TF produced or by changing the GRF shape through changing the degree of cooperativity. In the case of gene activation, this causes different effects at the level of single cells. In the former case (left), lower TF concentrations lead to less transcription of a target at the single-cell level. In the latter case (right), the amount of TF needed to sufficiently activate a target is increased, thereby reducing the number of cells which achieve the “threshold” concentration of TF. (B) Specific etiologies whereby the master myeloid TF PU.1 is deregulated in AML and MDS. Citations found within the main text. DBD, DNA-binding domain; PEST, Pro-Glu-Ser-Thr rich domain; TAD, transactivation domain.
Posttranscriptional deregulation
Although typically studied at the level of transcription, posttranscriptional deregulation is also known to occur in AML. This has been best described for the master granulocyte TF, CEBPA. CEBPA is translated as 2 major polypeptides, p30 and p42, where p30 can inhibit p42-mediated gene activation by competing for binding sites in the genome.53 Importantly, the selection of isoform expression is sensitive to eIF2 and eIF4e levels and consequently, mTOR signaling. Intriguingly, although frameshift mutations in CEBPA leading to exclusive p30 isoform expression have been described in ∼9% to 10% of AML cases, mTOR pathway perturbation has been demonstrated to be a hallmark feature seen in essentially all patients with AML.54-56
Beyond the level of a single TF, changes in global rate parameters of posttranscriptional reactions are also known to occur during leukemogenesis. One active area of research is focused on understanding how the variety of mutations in splicing factors57,58 frequently seen in both AML and MDS lead to pathology. These mutations, including in SRSF1 and U2AF1, are known to change pre-mRNA processing and alternative exon selection across many genes in the genome.57,58 Similarly, mutations in ribosomal genes can lead to a spectrum of ribosomopathies, with some such as dyskeratosis congenita, Shwachman-Diamond syndrome, and 5q syndrome (owing to consequent haploinsufficiency of the RPS14 gene) having an increased propensity for leukemia.59 Furthermore, mutations in ribosomal subunits have been described across various cancer subtypes.60 Finally, deregulation of mRNAs posttranscriptionally by either base editing or through the altered expression of RNA binding proteins has been recently recognized as having a critical impact on gene expression61-67 (this emerging field in leukemia research is addressed in a companion review). Although systems-level analyses will be required to fully understand how these aberrations promote leukemogenesis, given the typically short mRNA and protein half-lives of TF genes,68 it is anticipated that posttranscriptional deregulation will have an enormous impact on the concentration and stoichiometry of these factors.
Finally, 1 important mechanism for TF deregulation in leukemogenesis is through changes in the cellular distribution of TF molecules. For instance, NPM1 mutations, which occur in ∼35% of primary AML patients,69 are thought to confer pathogenicity through the cytoplasmic sequestration of myeloid regulatory factors, including PU.1.70 Our group recently described a role for the overexpression of MDMX in human AML, which subsequently leads to p53 sequestration.71 Additionally, the signaling pathways frequently found to be deregulated in leukemia communicate their aberrant signals through nuclear translocation of responsive TF such as ERK and STAT proteins (see previous work72 for a comprehensive review of this topic). Finally, 1 well-known mechanism of leukemogenesis, particularly in core-binding factor mutant AML, is the direct antagonism of lineage specifying TF,39,40,73 thereby leading to an uncoupling of TF levels and target gene responses.
Quite clearly, a number of distinct mechanisms have been described for how leukemia-initiating events generate and maintain a leukemic state. Although the exact details may vary, a hallmark seen in essentially every cytogenetic or mutational subgroup is the deregulation of TF behavior and/or expression. In that sense, TF deregulation may represent a unifying principle of leukemogenesis. As highly powerful analytical techniques and technologies with molecular resolution begin to be used in the study of leukemia, our understanding of how this process plays out at the molecular level in single cells is going to be a reality in the coming years. In the next section, we provide an overview of some of the basic findings derived from these types of technologies and what their impact may be on the future of leukemia research.
Gene expression at the molecular level
Although the studies listed have been foundational to our understanding of leukemogenesis, ensemble assays of gene expression inherently blur nongenetic variability between cells. This can be problematic when studying the pathogenesis of clonal diseases such as AML and MDS that arise from subpopulations of HSPCs,10,11 particularly given the lack of fully specific and sensitive markers to define leukemic stem cells. For one, the signal from the pathogenic process in question is diluted by signal from normal HSPCs. Arguably more important, however, is that even for identical cells in identical conditions, heterogeneity is a natural and unavoidable consequence of gene expression that has been documented at every level of the evolutionary tree.74-81 This heterogeneity, or stochasticity, derives from the physical nature of gene expression: the reactions constituting “gene expression” are a series of punctuated steps involving low copy numbers of reactants, in poorly mixed, partitioned reaction chambers within the cell. Moreover, capturing this variability is not simply an academic exercise. Indeed, this stochasticity provides critical information about the dynamics and potential regulatory systems of the process under study.82,83 As such, quantitatively studying gene-expression stochasticity, particularly in context of TF dynamics, is an essential “next step” in the field’s investigation of leukemia pathogenesis and treatment. To that end, in the following sections we focus on how molecular randomness and biophysical forces influence the behavior of TF in the cell.
Transcriptional bursting
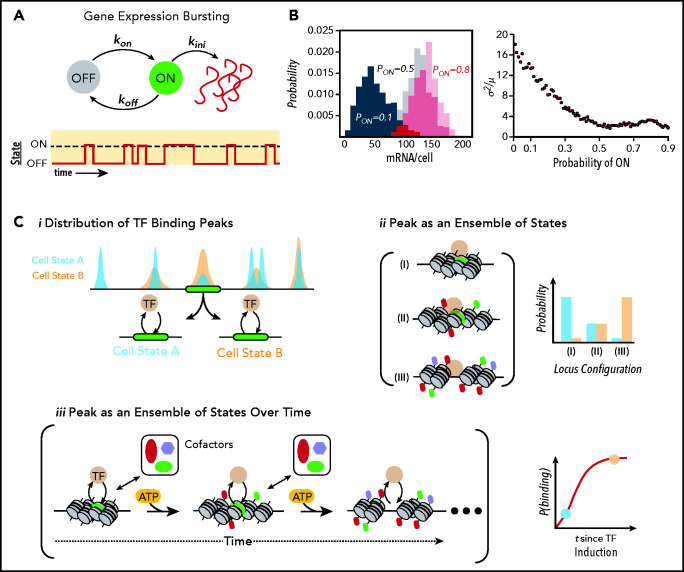
As the initial step in gene expression, the reaction kinetics of transcription have been a focus of intense investigation. The advent of in situ single-molecule imaging tools has provided the necessary resolution for the quantitative analysis of those kinetics. The first of these tools was single-molecule mRNA fluorescence in situ hybridization (smRNA-FISH), developed by Femino and colleagues in Rob Singer’s group in 1998.84 This transformative technology allows for the detection of individual molecules of mRNA while maintaining spatial information within the cell. This key property of smRNA-FISH enables one to not only accurately capture the probability distribution of a gene across a cell population, but also to facilitate direct observation of nascent transcription. When combined with computational modeling approaches, this has allowed for the inference of the underlying transcriptional rate parameters of a gene. The first of these efforts was performed in a seminal study by Arjun Raj and colleagues, who studied the transcriptional kinetics of a tetracycline inducible minigene using smRNA-FISH and a theoretical framework known as the random telegraph model.85,86 This work provided the first inference of key rate constants of an endogenous gene, including the rate of gene activation, gene inactivation, and polymerase initiation rates. Similar approaches have been subsequently used to study the transcription of genes from a diverse repertoire of species and situations, including bacteria,87 yeast,78 Drosophila,88 and mammalian cells,89-92 including recently in primary HSPCs from mice.93 Irrespective of the system studied, the common feature found is that transcription is a discontinuous (ie, bursting, phenomenon), whereby individual loci undergo cycling between intervals of productive transcription followed by longer periods of inactivity (Figure 3A). For instance, we have recently found that although the majority of primary HSPCs express the key TF PU.1, Gata1, and Gata2 genes at the mRNA level, active transcription is surprisingly rare, with only 10% to 20% of cells expressing at any given moment.93 Even in unipotent populations of cells typified by high levels of either Gata TF or PU.1 (ie, megakaryocyte-erythroid progenitor and granulocyte macrophage progenitor cells), these frequencies only increased to ∼40% and 20%, respectively. For low-copy-number bursts or mRNAs with shorter half-lives, features typical of many TF mRNAs,93,94 these temporal fluctuations in transcriptional activity can lead to markedly dispersed single-cell mRNA probability distributions.91,95 Furthermore, these observations with smRNA-FISH have been validated with live cell imaging studies of nascent transcription using the MS2 mRNA tagging system.96 In this technique, an array of stem loop sequences derived from the MS2 bacteriophage RNA genome are incorporated into the 3′ untranslated region of a gene of interest. By coexpressing a fluorescently tagged MS2 viral coat protein, which binds these stem loops, in the cell, single puncta corresponding to single mRNA are readily visualized. Using this approach, live cell dynamics of transcriptional bursting can be directly measured.97 Consistent with the data derived from smRNA-FISH, MS2-tagged endogenous alleles undergo cyclic bursting interspersed between long “OFF” periods in gene activity.71,97-105 For instance, a recent landmark paper from Dan Larson’s group demonstrated that bursts in the expression of the estrogen response gene TFF1 can be separated by inactivate periods lasting on the order of days, even after induction with estradiol.103
Figure 3.
Gene expression at the molecular level. (A) Schematic of a simple stochastic switch whereby a molecule of interest (ie, a gene, mRNA, or protein) undergoes cyclical activation and deactivation reactions, thereby rendering the molecule capable or refractory to an additional reaction. In the example of promoter cycling and transcription, this leads to probabilistically distributed bursts of transcription followed by periods of quiescence. (B) Simulated data showing how for a simple 1-species system produced in bursts and undergoing first-order decay, the probability of firing is directly related to the heterogeneity within a population of identical cells. Histograms on the left show the probability distributions for a gene with a probability of being “ON” (Pon) of 0.1 (black), 0.5 (gray), and 0.8 (red). Plot on the right demonstrates how population noise (captured with the Fano Factor, or the variance/mean) changes as a function of (Pon). A Poissonian distribution (Fano = 1) is the minimal noise observed within biological systems.83 For many TFs, Pon is ∼0.1 to 0.3.93 (C) Relationship of ensemble assays of TF-binding activity to actual behavior of molecules. (i) From the binding distributions derived from ensemble studies such as chromatin immunoprecipitation sequencing, one can conclude that at the green locus, cells in state B immunoprecipitated the locus more frequently than in state A, where states could be defined by a variety of measures including cell surface proteins, cell-cycle position, or metabolic profiling. However, the ensemble studies cannot determine whether this is due to changes in the configurations of chromatin states (ii), each with different affinities for the TF, or from differences in the temporal evolution of the locus (iii), or some combination. Of note, as indicated in the bottom right of iii, state A and state B may lie on a temporal continuum rather than representing discrete entities. This evolution in time is based on energy-dependent modification of the locus by epigenetic enzymes licensed to the site by the TF. Importantly, this implies that cells with identical concentrations of TF could have very different binding patterns (and therefore target gene activity) solely because of each cell’s position on the time curve. This ambiguity obviously complicates the interpretation of ensemble studies of TF binding and how such binding influences target locus expression.
Finally, these bursting behaviors are not restricted to mRNAs. Indeed, live cell imaging of translation has indicated that these reactions also occur in discrete bursts. In a series of seminal, concurrent papers, 4 groups demonstrated that translation of an mRNA is also an infrequent event, with the majority of mRNAs not translating at any given moment.106-109 For instance, it was demonstrated that for the Actb gene in murine neurons, only 10% to 30% of mRNAs were translating at any given moment in the cell. Furthermore, once an mRNA was engaged by the polysome, roughly 4.5 nascent peptides were produced before translation termination.110 Although these techniques are still relatively new and require further study, when taken together with results at the transcriptional level, there is a clear demonstration that bursting phenomena are intrinsic properties of multiple levels of the central dogma, effectively guaranteeing nongenetic, population heterogeneity in the level of any biochemical species of interest in the cell (Figure 3B).
Subcellular TF dynamics
Although it is evident that the intracellular concentration of a TF influences transcriptional networks, live cell protein imaging has demonstrated significant complexities in the processes of TF localization, target search, and locus binding. These dynamic processes impact target gene expression and transcriptional networks in ways that are only just now starting to be appreciated. Nevertheless, there is growing evidence that the spatiotemporal behavior of TF proteins is an important component of gene regulation and an additional layer of nongenetic heterogeneity in single cells.
TF shuttling between the nucleus and cytoplasm is currently the best studied dynamic behavior. In both yeast and mammalian cells, it appears that the rate and amplitude of these subcellular translocations impart meaningful information to the transcriptome not accounted for by concentration alone.111,112 For instance, differences in shuttling rates of stress TF in yeast can lead to differential output of coregulated genes,113 and differential p53 responses to γ-irradiation and ultraviolet genotoxic stress are encoded by differences in shuttling behaviors rather than changes p53 concentration.114-116 Similar shuttling behaviors have also been described for NF-κB,117 ERK2,118,119 and SMAD4.120 These studies and others demonstrate mechanistically that subcellular protein dynamics may provide critical information to the transcriptome that is independent of the TF concentration at a given moment. Of note, similar effects have recently been reported to be a key step in the oncogenesis of NPM1c mutations, with changes in the shuttling capacity of the master transcription factor PU.1.70 As such, this relatively unexplored area of gene deregulation may have important roles in AML pathogenesis.
Another area of TF biology markedly advanced by single-molecule resolution imaging studies is our understanding of how TF find and bind to target genes. Indeed, the facilitated diffusion models derived from bacterial studies,121 where TF passively explore 3-dimensional space followed by constrained 1-dimensional sliding on DNA, physically cannot work in eukaryotes given the enormity of the genome as well as the chromatin-derived energy barriers to sliding.122 In recent years, the application of single protein imaging in live cells has facilitated direct observation of TF search paths through the mammalian nucleus. The results have been striking and at times counterintuitive. For instance, although the general transcriptional amplifier MYC123 and the transcriptional elongation complex P-TEFb124 are postulated to work in concert, they explore the nuclear space with decidedly different modes: MYC globally surveys the entire volume of the nucleus whereas P-TEFb undergoes constrained diffusion.125 Similarly, CTCF and Rad21, 2 integral and cooperating proteins controlling the topology of the eukaryotic genome, also have markedly different search dynamics: although a diffusing CTCF typically searches for and binds to a cognate site within 1 minute, Rad21 requires on average 33 minutes of diffusion before rebinding.126
One somewhat surprising finding from the studies listed here is not just the differences in search strategies used by different regulatory proteins, but also the instability of binding once a target has been identified. Indeed, CTCF was found to bind to its cognate binding site for only 1 minute on average, whereas Rad21 bound for 22 minutes.126 These data indicate that the stable looping structures assumed through -omics approaches are actually maintained through a flux of protein. Even more remarkably, it appears that for many nuclear proteins, including SOX2,127,128 the Mediator complex, RNA polymerase II129 and BRD4,130 binding times are on the order of seconds, rather than minutes, indicating that most regulatory complexes are transient hubs maintained through dynamic binding and unbinding of factors to the genome. Although preliminary, our group has also recently discovered similar properties in the hematopoietic TF PU.1 and Gata1, with each having stable binding occurring in only 5% of all nuclear TF molecules at a given time and with residence times of only several seconds during each binding event.
Quantitative gene regulation
Another consideration that is exceedingly nontrivial in light of the preceding section is determining how a TF actually regulates a target locus. In principle, mammalian TF are known to either activate or repress target genes through the recruitment of enzymes to chromatin. This recruitment then changes the local energy landscape through covalent modification of histones and/or DNA, ultimately facilitating or hindering subsequent transcription131,132 (see the companion review for more on this topic). Although this model is typically assumed, implicit in this description is the consumption of energy, which means that the system out of thermodynamic equilibrium. This has at least 2 important implications for the leukemia field.
First, whereas the interaction of a TF and a target locus are typically viewed from the perspective of a static DNA substrate being bound by protein (Figure 3Ci), the microscopic reality of this interaction is actually much more nuanced. Indeed, the inclusion of epigenetic reactions actually means that the DNA-chromatin template could either be in a number of energetically different configurations within the same “cell state,” with consequently different binding activities by the TF that are stable properties for the locus over the cell’s lifespan, or it may reflect changes in the locus over time within the same cell (Figure 3Cii-iii).133,134 Although these phenomena are clearly not mutually exclusively, in the restricted case of simple temporal evolution, the binding affinities between a TF and a target locus will necessarily change over time. Given the probabilistic nature of binding and unbinding, as well as the short time scales over which these interactions take place, it is therefore evident that identical cells with similar TF concentrations could demonstrate markedly different behaviors over time through chance alone. Although this may seem to be a purely academic distinction, this directly affects the validity of the derived gene regulatory function87,133 linking the concentration of a TF to target gene expression. From a practical perspective, it indicates that contemporaneous transcriptional activity of a gene may be better correlated with the temporal integration of TF-binding events, rather than the current TF-binding peaks. This is likely to be particularly relevant for TF that are expressed at low, but fluctuating, levels beginning in early stem cells such as PU.1, Runx1, and Gata2. A second, related issue comes from the observation that many TFs appear to undergo stochastic state transitions between low and high expression states over the course of many cell divisions during differentiation.90,91,93,135,136 The number of these transitions, the time spent in each state, and the half-lives of the covalent modifications made at the TF’s target loci all have direct effect on the transcriptional network of the cell.
Taken together, these 2 effects of nonequilibrium dynamics necessitate the study of transcriptional network evolution over time, rather than through single snap shot analyses.134 At a technical level, this consideration ultimately requires that the root, leukemia-initiating cell populations are defined and studied. Recent advances in single-cell technologies, single-molecule imaging, and clonal tracing will be critical tools in this regard.90,93,137-141 Furthermore, advances in information theory,142,143 noise-control theory,144-147 and multitalk evolution theory148,149 will be of particular importance in making analytical sense of these complex data sets. Ultimately, these tools, both technical and analytical, should help formulate a more comprehensive, unified understanding of MDS and AML (and other cancers) and point the way toward novel therapeutic options.
Defining the leukemic state and therapeutic resistance
The aforementioned studies significantly complicate our understanding of transcriptional networks, how they emerge, and how TF regulate them. For one, transcriptional stochasticity leads to probability distributions that are significantly dispersed and typically dominated by low molecular copy numbers on the order of tens of mRNAs and hundreds to a few thousand protein molecules per cell. Second, even after gene expression reactions are completed, the behavior of these TF protein molecules is complex, relying heavily on protein–protein interactions and dynamic posttranslational modifications, in addition to the canonical interactions they make with the genome. Finally, all of these fluctuating sources of regulatory information ultimately must be “read out” and culminate in a stereotypical pattern of gene expression known as the transcription network. Undoubtedly, determining how these processes reliably transmit information to support robust tissue homeostasis is one of the major outstanding questions of molecular and developmental biology. By extension, how this putatively noisy flow of information can be further corrupted to generate a leukemic state or leukemic network also remains to be fully defined.
One very possible scenario in light of this realization is that a defined leukemic state may not be an appropriate view of disease pathogenesis in AML and MDS. This viewpoint is built from the large body of literature that has carefully collected and cataloged the litany of disease-associated mutational variants or deregulated genes found within these tumors. From those mutations, the assumption has been that a network must be operating to maintain fitness of leukemic clones. However, this model has not be able to explain why so many mutations both within10,11 and across leukemia patients37 lead to largely phenotypically similar conditions. Indeed, a “unified” theory of leukemogenesis is still needed.
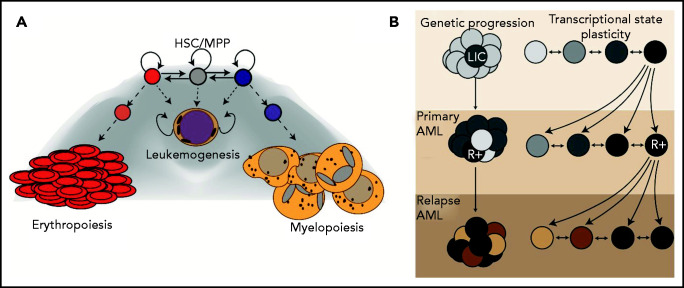
One alternative hypothesis is that leukemic cells or the leukemic network may be better understood as a set of “trapped” cellular states on the differentiation landscape normally supporting multilineage, hematopoietic differentiation, akin to a molecular purgatory from which the clone cannot escape (Figure 4A). In such a model, the leukemic state would effectively be a default state cells “fall into” when ≥1 critical network components are deregulated outside of their evolutionarily set operating window. Leukemogenicity would, in this situation, not be defined by a unique oncogenic program, but rather as the set of programs conducive to continued cell survival despite failure to progress along a differentiation trajectory. Importantly, the only requirement of such a default state is that the state is viable; it could comprise an innumerable number of substates driven through stochasticity of gene expression. Quite obviously, there will be a pressing need to concurrently define the boundaries within which “normal” hematopoietic network dynamics can operate to understand where in transcriptional space leukemic networks reside.
Figure 4.
The leukemic state and disease progression. (A) Topology of normal hematopoietic differentiation with the “leukemic state” as a trapped basin in gene expression space. (B) Two models of therapeutic resistance in AML. In the genetic scenario, acquisition of resistance mutations (denoted with R+) in a polyclonal malignancy creates a selective pressure that leads to AML relapse. In the transcriptional state plasticity scenario, cells can occupy a diversity of cellular microstates within the “leukemic state” that make them more or less susceptible to clearance by chemotherapy. If these states are based on the expression of particular TF, then selected states could be inherited in relapse tumors. Importantly, these mechanisms are likely not mutually exclusive and are intrinsically dependent on one another.
Beyond disease initiation, this view of leukemia gene expression states may also have an important role in therapy resistance. Indeed, network plasticity derived from nongenetic heterogeneity has been demonstrated in other tumors to generate population fitness. In both neuroblastoma150 and melanoma,138,151 stochastic transitions in the tumor’s gene expression network facilitate the escape of a subpopulation of tumor cells when challenged with chemotherapy. Foundational studies in metastases also have shown that heterogeneity within the primary tumor is central to the metastatic capacity of the tumor.152 Determining whether such mechanisms, as opposed to purely genetic mechanisms, are operating during leukemia relapse will be a critical step in the coming years (Figure 4B).
Summary/conclusion
In conclusion, recent studies at the level of single molecules and in single cells have shed new insight into the complexities of gene expression. In light of those studies, we believe that it will be particularly important to consider the molecular, spatial, and temporal resolution needed to study gene expression changes during leukemogenesis, particularly when the time scales (eg, binding times of TF, bursting frequencies) and the molecular thresholds (eg, the number of TF molecules needed to influence a network) operating in these systems remain poorly defined. We believe the technologies listed here will be critical in this regard, particularly when the results of those studies are used in parallel to the exciting advances coming from whole genome level, including single-cell RNA-sequencing,137,140,153-156 assay for transposase-accessible chromation sequencing,157,158 chromatin capture techniques,52 and chromatin immunoprecipitation seqencing.73,159 Moreover, they can be used to test the predictions made via elegant network analyses of hematopoiesis and leukemia.160-163 For one, tools such as single-molecule FISH will be vitally important toward understanding both the magnitude and source of gene expression stochasticity in primary hematopoietic cells. Live cell studies of transcription could also be a useful tool for investigating how different epigenetic aberrations seen in AML lead to changes in transcriptional kinetics. Single-protein imaging experiments in live cells could provide critical measurements of actual search times and binding times of nuclear proteins such as TF, transcriptional machinery, and cohesin complex components, which will help shape our understanding of chromatin dynamics, how combinatorial control of loci plays out in live cells, and how these dynamics are altered in leukemia. As with all technical approaches, it will be important to consider the biological and physical plausibility of models derived from these studies a priori,133,146,147,164,165 particularly in light of the wide range of single-cell behaviors that are evidently tolerated during normal hematopoiesis. Finally, it is also clearly evident that any of these tools (either alone or in combination) should also play a pivotal role in the next generation of high-throughput screening technologies in the search for novel therapeutics or therapeutic strategies. Indeed, the combination of high-resolution, single-cell technologies with systems analyses has recently yielded fundamental insights into how pharmacology can affect dynamic signaling processes in the cell.119 It behooves the leukemia community to begin applying similar experimental approaches in the quest for novel antileukemic agents. A particularly alluring prospect would be to use these tools to revisit previously developed compounds that, although not necessarily efficacious as single agents, limit the gene expression state space leukemia cells can occupy and possibly increase the potency of current standard of care regimes.
In summary, the advent of single-molecule gene expression studies has been a watershed moment for our understanding of life inside the cell. Those studies have demonstrated that stochasticity imparts an enormous influence on the process of gene expression. It therefore follows that defining “gene deregulation” may be much more a complicated process than initially anticipated. Nevertheless, by integrating these tools in parallel to the critical whole-genome approaches being used in the field, it is the opinion of these authors that there will be significant progress in our understanding of leukemia pathogenesis in the coming years. This progress should hopefully open new therapeutic avenues into the treatment of these recalcitrant malignancies.
Acknowledgments
J.C.W. is supported by grants from the National Institutes of Health, National Institute of General Medical Sciences (F30GM122308 and T32GM007288). U.S. is supported by grants from the National Institutes of Health, National Cancer Institute (R01CA217092 and P30CA013330).
Authorship
Contribution: J.C.W. and U.S. wrote the paper.
Conflict-of-interest disclosure: U.S. has received research funding from GlaxoSmithKline, Bayer Healthcare, Aileron Therapeutics, and Novartis; has received compensation for consultancy services and for serving on scientific advisory boards from GlaxoSmithKline, Bayer Healthcare, Celgene, Aileron Therapeutics, Stelexis Therapeutics, and Pieris Pharmaceuticals; and has equity ownership in and is serving on the board of directors of Stelexis Therapeutics. J.C.W. declares no competing financial interests.
Correspondence: Ulrich Steidl, Albert Einstein College of Medicine, Chanin Building, Rm. 601-605, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: ulrich.steidl@einsteinmed.org.
REFERENCES
- 1.Zhang Y, Wang J, Wheat J, et al. AML1-ETO mediates hematopoietic self-renewal and leukemogenesis through a COX/β-catenin signaling pathway. Blood. 2013;121(24):4906-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL.. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005; 65(24):11367-11374. [DOI] [PubMed] [Google Scholar]
- 3.Kakizuka A, Miller WH Jr, Umesono K, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66(4):663-674. [DOI] [PubMed] [Google Scholar]
- 4.Thirant C, Ignacimouttou C, Lopez CK, et al. ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell. 2017;31(3):452-465. [DOI] [PubMed] [Google Scholar]
- 5.Miyamoto T, Weissman IL, Akashi K.. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci USA. 2000;97(13):7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki H, Suzuki M, Otsuki A, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25(4):415-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbauer F, Owens BM, Yu L, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38(1):27-37. [DOI] [PubMed] [Google Scholar]
- 8.Steidl U, Rosenbauer F, Verhaak RGW, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38(11):1269-1277. [DOI] [PubMed] [Google Scholar]
- 9.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369-381. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Kao Y-R, Sun D, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level [published correction appears in Nat Med. 2019;25(3):529]. Nat Med. 2019;25(1):103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shlush LI, Mitchell A, Heisler L, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017; 547(7661):104-108. [DOI] [PubMed] [Google Scholar]
- 13.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crispino JD, Horwitz MS.. GATA factor mutations in hematologic disease. Blood. 2017;129(15):2103-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsumura KR, Bresnick EH; GATA Factor Mechanisms Group. The GATA factor revolution in hematology. Blood. 2017; 129(15):2092-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman AD.C/EBPα in normal and malignant myelopoiesis. Int J Hematol. 2015;101(4):330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porcher C, Chagraoui H, Kristiansen MS.. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood. 2017; 129(15):2051-2060. [DOI] [PubMed] [Google Scholar]
- 18.Sood R, Kamikubo Y, Liu P.. Role of RUNX1 in hematological malignancies [published correction appears in Blood. 2017;129(15):2070–2082]. Blood. 2017; 129(15):2070-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avellino R, Delwel R.. Expression and regulation of C/EBPα in normal myelopoiesis and in malignant transformation. Blood. 2017;129(15):2083-2091. [DOI] [PubMed] [Google Scholar]
- 20.Koschmieder S, Rosenbauer F, Steidl U, Owens BM, Tenen DG.. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J Hematol. 2005;81(5):368-377. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbauer F, Koschmieder S, Steidl U, Tenen DG.. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106(5):1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Zhou B, Mao F, et al. HOXA9 reprograms the enhancer landscape to promote leukemogenesis. Cancer Cell. 2018;34(4):643-658.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroedinger E.What is Life?: With Mind and Matter and Autobiographical Sketches. Cambridge: Cambridge University Press; 2012. [Google Scholar]
- 24.Berg OG.A model for the statistical fluctuations of protein numbers in a microbial population. J Theor Biol. 1978;71(4):587-603. [DOI] [PubMed] [Google Scholar]
- 25.Novick A, Weiner M.. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43(7):553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Thé H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A.. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343(6254):177-180. [DOI] [PubMed] [Google Scholar]
- 27.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A.. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66(4):675-684. [DOI] [PubMed] [Google Scholar]
- 28.Steger DJ, Lefterova MI, Ying L, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuntimaddi A, Achille NJ, Thorpe J, et al. Degree of recruitment of DOT1L to MLL-AF9 defines level of H3K79 Di- and tri-methylation on target genes and transformation potential. Cell Rep. 2015;11(5):808-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C, Smith ER, Takahashi H, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37(3):429-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas D, Milne TA, Basrur V, et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci USA. 2011;108(38):15751-15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Kumar AR, Hudson WA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13(5):432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernt KM, Zhu N, Sinha AU, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith E, Lin C, Shilatifard A.. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25(7):661-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818-822. [DOI] [PubMed] [Google Scholar]
- 36.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G.. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katayama S, Suzuki M, Yamaoka A, et al. GATA2 haploinsufficiency accelerates EVI1-driven leukemogenesis. Blood. 2017;130(7):908-919. [DOI] [PubMed] [Google Scholar]
- 39.Vangala RK, Heiss-Neumann MS, Rangatia JS, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101(1):270-277. [DOI] [PubMed] [Google Scholar]
- 40.Pabst T, Mueller BU, Harakawa N, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7(4):444-451. [DOI] [PubMed] [Google Scholar]
- 41.Linggi B, Müller-Tidow C, van de Locht L, et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8(7):743-750. [DOI] [PubMed] [Google Scholar]
- 42.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624-630. [DOI] [PubMed] [Google Scholar]
- 43.Will B, Vogler TO, Narayanagari S, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21(10):1172-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sive JI, Basilico S, Hannah R, Kinston SJ, Calero-Nieto FJ, Göttgens B.. Genome-scale definition of the transcriptional programme associated with compromised PU.1 activity in acute myeloid leukaemia. Leukemia. 2016; 30(1):14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164-3173. [DOI] [PubMed] [Google Scholar]
- 46.Lavallée V-P, Baccelli I, Krosl J, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47(9):1030-1037. [DOI] [PubMed] [Google Scholar]
- 47.Laricchia-Robbio L, Premanand K, Rinaldi CR, Nucifora G.. EVI1 impairs myelopoiesis by deregulation of PU.1 function. Cancer Res. 2009;69(4):1633-1642. [DOI] [PubMed] [Google Scholar]
- 48.Curik N, Burda P, Vargova K, et al. 5-azacitidine in aggressive myelodysplastic syndromes regulates chromatin structure at PU.1 gene and cell differentiation capacity. Leukemia. 2012;26(8):1804-1811. [DOI] [PubMed] [Google Scholar]
- 49.Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis [published correction appears in Nat Genet. 2008;40(2):255]. Nat Genet. 2008;40(1):51-60. [DOI] [PubMed] [Google Scholar]
- 50.Cheng JX, Anastasi J, Watanabe K, et al. Genome-wide profiling reveals epigenetic inactivation of the PU.1 pathway by histone H3 lysine 27 trimethylation in cytogenetically normal myelodysplastic syndrome. Leukemia. 2013;27(6):1291-1300. [DOI] [PubMed] [Google Scholar]
- 51.Anguita E, Gupta R, Olariu V, et al. A somatic mutation of GFI1B identified in leukemia alters cell fate via a SPI1 (PU.1) centered genetic regulatory network. Dev Biol. 2016;411(2):277-286. [DOI] [PubMed] [Google Scholar]
- 52.Viny AD, Bowman RL, Liu Y, et al. Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell. 2019;25(5):682-696.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calkhoven CF, Müller C, Leutz A.. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14(15):1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 54.Park S, Chapuis N, Tamburini J, et al. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica. 2010;95(5):819-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C.. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19(3):329-334. [DOI] [PubMed] [Google Scholar]
- 56.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299-310. [DOI] [PubMed] [Google Scholar]
- 57.Graubert TA, Shen D, Ding L, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogawa S.Splicing factor mutations in myelodysplasia. Int J Hematol. 2012;96(4):438-442. [DOI] [PubMed] [Google Scholar]
- 59.Narla A, Ebert BL.. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlachos A.Acquired ribosomopathies in leukemia and solid tumors. Hematology (Am Soc Hematol Educ Program). 2017;2017(1):716-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2013;110(3):1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kharas MG, Lengner CJ, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22(2):191-205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen C, Sheng Y, Zhu AC, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27(1):64-80.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vu LP, Cheng Y, Kharas MG.. The biology of m6A RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 2019;9(1):25-33. [DOI] [PubMed] [Google Scholar]
- 67.Vu LP, Prieto C, Amin EM, et al. Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat Genet. 2017;49(6):866-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control [published correction appears in Nature. 2013;495(7439):126-127]. Nature. 2011;473(7347):337-342. [DOI] [PubMed] [Google Scholar]
- 69.Falini B, Mecucci C, Tiacci E, et al. ; GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254-266. [DOI] [PubMed] [Google Scholar]
- 70.Gu X, Ebrahem Q, Mahfouz RZ, et al. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J Clin Invest. 2018;128(10):4260-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvajal LA, Neriah DB, Senecal A, et al. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10(436):eaao3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemmati S, Haque T, Gritsman K.. Inflammatory signaling pathways in preleukemic and leukemic stem cells. Front Oncol. 2017;7:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ptasinska A, Assi SA, Martinez-Soria N, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8(6):1974-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levsky JM, Singer RH.. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;13(1):4-6. [DOI] [PubMed] [Google Scholar]
- 75.Levsky JM, Shenoy SM, Pezo RC, Singer RH.. Single-cell gene expression profiling. Science. 2002;297(5582):836-840. [DOI] [PubMed] [Google Scholar]
- 76.Elowitz MB, Levine AJ, Siggia ED, Swain PS.. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183-1186. [DOI] [PubMed] [Google Scholar]
- 77.Raser JM, O’Shea EK.. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gandhi SJ, Zenklusen D, Lionnet T, Singer RH.. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18(1):27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar RM, Cahan P, Shalek AK, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells [published correction appears in Nature. 2015;519:118]. Nature. 2014;516(7529):56-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S.. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Balázsi G, van Oudenaarden A, Collins JJ.. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011; 144(6):910-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paulsson J.Summing up the noise in gene networks. Nature. 2004;427(6973):415-418. [DOI] [PubMed] [Google Scholar]
- 83.Paulsson J.Models of stochastic gene expression. Phys Life Rev. 2005;2(2):157-175. [Google Scholar]
- 84.Femino AM, Fay FS, Fogarty K, Singer RH.. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585-590. [DOI] [PubMed] [Google Scholar]
- 85.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S.. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peccoud J, Ycart B.. Markovian modeling of gene-product synthesis. Theor Popul Biol. 1995;48(2):222-234. [Google Scholar]
- 87.Sepúlveda LA, Xu H, Zhang J, Wang M, Golding I.. Measurement of gene regulation in individual cells reveals rapid switching between promoter states. Science. 2016; 351(6278):1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zoller B, Little SC, Gregor T.. Diverse spatial expression patterns emerge from unified kinetics of transcriptional bursting. Cell. 2018;175(3):835-847.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bartman CR, Hsu SC, Hsiung CC-S, Raj A, Blobel GA.. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell. 2016;62(2):237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hormoz S, Singer ZS, Linton JM, Antebi YE, Shraiman BI, Elowitz MB.. Inferring cell-state transition dynamics from lineage trees and endpoint single-cell measurements. Cell Syst. 2016;3(5):419-433.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singer ZS, Yong J, Tischler J, et al. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell. 2014;55(2):319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Senecal A, Munsky B, Proux F, et al. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014;8(1):75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wheat JC, Sella Y, Willcockson M, et al. Single-molecule imaging of transcription dynamics in somatic stem cells. Nature. 2020;583(7816):431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trcek T, Larson DR, Moldón A, Query CC, Singer RH.. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011;147(7):1484-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skinner SO, Xu H, Nagarkar-Jaiswal S, Freire PR, Zwaka TP, Golding I.. Single-cell analysis of transcription kinetics across the cell cycle. eLife. 2016;5:e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM.. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437-445. [DOI] [PubMed] [Google Scholar]
- 97.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH.. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011; 332(6028):475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Das S, Moon HC, Singer RH, Park HY.. A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci Adv. 2018;4(6):eaar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fukaya T, Lim B, Levine M.. Enhancer control of transcriptional bursting. Cell. 2016;166(2):358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larson DR, Fritzsch C, Sun L, Meng X, Lawrence DS, Singer RH.. Direct observation of frequency modulated transcription in single cells using light activation. eLife. 2013;2:e00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lenstra TL, Coulon A, Chow CC, Larson DR.. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol Cell. 2015;60(4):597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim B, Fukaya T, Heist T, Levine M.. Temporal dynamics of pair-rule stripes in living Drosophila embryos. Proc Natl Acad Sci USA. 2018;115(33):8376-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodriguez J, Ren G, Day CR, Zhao K, Chow CC, Larson DR.. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019;176(1-2):213-226.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Golding I, Paulsson J, Zawilski SM, Cox EC.. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025-1036. [DOI] [PubMed] [Google Scholar]
- 105.Chubb JR, Trcek T, Shenoy SM, Singer RH.. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16(10):1018-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morisaki T, Lyon K, DeLuca KF, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016; 352(6292):1425-1429. [DOI] [PubMed] [Google Scholar]
- 107.Yan X, Hoek TA, Vale RD, Tanenbaum ME.. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165(4):976-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Halstead JM, Lionnet T, Wilbertz JH, et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347(6228):1367-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang C, Han B, Zhou R, Zhuang X.. Real-time imaging of translation on single mRNA transcripts in live cells. Cell. 2016;165(4):990-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu B, Eliscovich C, Yoon YJ, Singer RH.. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352(6292):1430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Purvis JE, Lahav G.. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152(5):945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Levine JH, Lin Y, Elowitz MB.. Functional roles of pulsing in genetic circuits. Science. 2013;342(6163):1193-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin Y, Sohn CH, Dalal CK, Cai L, Elowitz MB.. Combinatorial gene regulation by modulation of relative pulse timing. Nature. 2015;527(7576):54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G.. p53 dynamics control cell fate. Science. 2012;336(6087):1440-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hafner A, Reyes J, Stewart-Ornstein J, Tsabar M, Jambhekar A, Lahav G.. Quantifying the central dogma in the p53 pathway in live single cells. Cell Syst. 2020;10(6):495-505.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lahav G, Rosenfeld N, Sigal A, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36(2):147-150. [DOI] [PubMed] [Google Scholar]
- 117.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW.. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466(7303):267-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U.. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol Cell. 2009;36(5):885-893. [DOI] [PubMed] [Google Scholar]
- 119.Goglia AG, Wilson MZ, Jena SG, et al. A live-cell screen for altered Erk dynamics reveals principles of proliferative control. Cell Syst. 2020;10(3):240-253.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Warmflash A, Zhang Q, Sorre B, Vonica A, Siggia ED, Brivanlou AH.. Dynamics of TGF-β signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc Natl Acad Sci USA. 2012;109(28):E1947-E1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bauer M, Metzler R.. Generalized facilitated diffusion model for DNA-binding proteins with search and recognition states. Biophys J. 2012;102(10):2321-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hansen AS, Amitai A, Cattoglio C, Tjian R, Darzacq X.. Guided nuclear exploration increases CTCF target search efficiency. Nat Chem Biol. 2020;16(3):257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Q, Li T, Price DH.. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81(1):119-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Izeddin I, Récamier V, Bosanac L, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:e02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X.. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife. 2017;6:e25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Z, Legant WR, Chen B-C, et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife. 2014;3:e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen J, Zhang Z, Li L, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156(6):1274-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cho W-K, Spille J-H, Hecht M, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361(6400):412-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sabari BR, Dall’Agnese A, Boija A, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361(6400):eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boettiger AN, Bintu B, Moffitt JR, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016; 529(7586):418-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bintu L, Yong J, Antebi YE, et al. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351(6274):720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Estrada J, Wong F, DePace A, Gunawardena J.. Information integration and energy expenditure in gene regulation. Cell. 2016;166(1):234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coulon A, Chow CC, Singer RH, Larson DR.. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14(8):572-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.MacArthur BD, Lemischka IR.. Statistical mechanics of pluripotency. Cell. 2013; 154(3):484-489. [DOI] [PubMed] [Google Scholar]
- 136.Stumpf PS, Smith RCG, Lenz M, et al. Stem cell differentiation as a non-Markov stochastic process. Cell Syst. 2017;5(3):268-282.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, et al. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018; 553(7687):212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shaffer SM, Emert BL, Reyes Hueros RA, et al. Memory sequencing reveals heritable single-cell gene expression programs associated with distinct cellular behaviors. Cell. 2020;182(4):947-959.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frieda KL, Linton JM, Hormoz S, et al. Synthetic recording and in situ readout of lineage information in single cells. Nature. 2017;541(7635):107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rodriguez-Fraticelli AE, Weinreb C, Wang S-W, et al. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature. 2020;583(7817):585-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bowling S, Sritharan D, Osorio FG, et al. An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells [published correction appears in Cell. 2020;181(7):1693-1694]. Cell. 2020;181(6):1410-1422.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A.. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334(6054):354-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hansen AS, O’Shea EK.. Limits on information transduction through amplitude and frequency regulation of transcription factor activity. eLife. 2015;4:e06559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gupta A, Hepp B, Khammash M.. Noise induces the population-level entrainment of incoherent, uncoupled intracellular oscillators. Cell Syst. 2016;3(6):521-531.e13. [DOI] [PubMed] [Google Scholar]
- 145.Briat C, Gupta A, Khammash M.. Antithetic integral feedback ensures robust perfect adaptation in noisy biomolecular networks. Cell Syst. 2016;2(1):15-26. [DOI] [PubMed] [Google Scholar]
- 146.Hilfinger A, Norman TM, Paulsson J.. Exploiting natural fluctuations to identify kinetic mechanisms in sparsely characterized systems. Cell Syst. 2016;2(4):251-259. [DOI] [PubMed] [Google Scholar]
- 147.Lestas I, Vinnicombe G, Paulsson J.. Fundamental limits on the suppression of molecular fluctuations. Nature. 2010; 467(7312):174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hausser J, Alon U.. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat Rev Cancer. 2020;20(4): 247-257. [DOI] [PubMed] [Google Scholar]
- 149.Hausser J, Szekely P, Bar N, et al. Tumor diversity and the trade-off between universal cancer tasks. Nat Commun. 2019;10(1):5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ryl T, Kuchen EE, Bell E, et al. Cell-cycle position of single MYC-driven cancer cells dictates their susceptibility to a chemotherapeutic drug. Cell Syst. 2017;5(3):237-250.e8. [DOI] [PubMed] [Google Scholar]
- 151.Shaffer SM, Dunagin MC, Torborg SR, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance [published correction appears in Nature. 2018;555(7695):274]. Nature. 2017;546(7658):431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fidler IJ, Kripke ML.. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197(4306):893-895. [DOI] [PubMed] [Google Scholar]
- 153.Nestorowa S, Hamey FK, Pijuan Sala B, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128(8):e20-e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olsson A, Venkatasubramanian M, Chaudhri VK, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice [published correction appears in Nature. 2019;569:E3]. Nature. 2016;537(7622):698-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu Z, Gao S, Diamond C, et al. Sequencing of RNA in single cells reveals a distinct transcriptome signature of hematopoiesis in GATA2 deficiency. Blood Adv. 2020;4(12):2656-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van Galen P, Hovestadt V, Wadsworth Ii MH, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176(6):1265-1281.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors [published correction appears in Cell. 2016;164(1):325]. Cell. 2015;163(7):1663-1677. [DOI] [PubMed] [Google Scholar]
- 158.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532-544. [DOI] [PubMed] [Google Scholar]
- 160.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.May G, Soneji S, Tipping AJ, et al. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 2013;13(6):754-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tusi BK, Wolock SL, Weinreb C, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 2018;555(7694):54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weinreb C, Wolock S, Tusi BK, Socolovsky M, Klein AM.. Fundamental limits on dynamic inference from single-cell snapshots. Proc Natl Acad Sci USA. 2018;115(10):E2467-E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gunawardena J.Models in biology: ‘accurate descriptions of our pathetic thinking’. BMC Biol. 2014;12(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Biddle JW, Nguyen M, Gunawardena J.. Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA. eLife. 2019;8:e41017. [DOI] [PMC free article] [PubMed] [Google Scholar]