Abstract
Gbp1p is a putative telomere-binding protein from Chlamydomonas reinhardtii that contains two RNA recognition motifs (RRMs) which are commonly found in heterogeneous nuclear ribonucleoproteins (hnRNPs). Previously we demonstrated that Gbp1p binds single-stranded DNA (ssDNA) containing the Chlamydomonas telomeric sequence but not the RNA containing the cognate sequence. Here we show that at lower protein concentrations Gbp1 can also bind an RNA containing the cognate sequence. We found that mutation of the two RRM motifs of Gbp1p to match the highly conserved region of hnRNP RRMs did not alter the affinity of Gbp1p for either RNA or DNA. The ability of Gbp1p to associate with either of these two nucleic acids is governed by the dimerization state of the protein. Monomeric Gbp1p associates with either ssDNA or RNA, showing a small binding preference for RNA. Dimeric Gbp1p has a strong preference for binding ssDNA and shows little affinity for RNA. To the best of our knowledge, this is the first example of a protein that qualitatively shifts its nucleic acid binding preference upon dimerization. The biological implications of a telomere-binding protein that is regulated by dimerization are discussed.
In most organisms, telomeric DNA is composed of simple short repeat sequences which show a strand bias such that there is a G-biased strand (G strand) and a complementary C-biased strand (C strand) (25). This double-stranded repeat sequence is bound in vivo by factors such as Rap1p in Saccharomyces cerevisiae (29) and TRF1 and TRF2 in humans (4, 47, 53), which are important for the regulation of telomere repeat length, the formation of telomeric chromatin, and the integrity of individual chromosome ends. Early sequencing of ciliate telomeres indicated that the G strand extends beyond the C strand, creating a single-stranded (ss) 3′ overhang of 12 to 16 bases which persists throughout the vegetative life cycle of the organism (18, 22). In contrast, in S. cerevisiae, longer G-strand overhangs that are more variable in length have been detected during late S phase, the cell cycle phase during which telomeres replicate (50). Even longer G-strand overhangs (200 ± 75 nucleotides) have been detected in humans (51). Thus, although they can be quite variable in length, the 3′ ss overhang structure at telomeres has been conserved in different eukaryotic kingdoms.
Unique factors associate with the 3′ ss telomeric overhang. Telomeric ss-G-strand binding proteins have been isolated and cloned from Oxytricha nova (42), Euplotes crassus (41), Tetrahymena thermophila (46), and Xenopus laevis (8). The Oxytricha and Euplotes ss-G-strand binding proteins remain bound to telomeric DNA in the presence of extremely high salt concentrations (up to 2 M NaCl) (41, 43). In contrast, telomere-binding proteins from other organisms are salt sensitive (46). Two yeast proteins, Est1p (48) and Cdc13p (16, 35), specifically associate with the yeast ss G strand in vitro, and mutations in these proteins affect telomere function in vivo. Heterogeneous nuclear ribonucleoproteins (hnRNPs) associate with RNA and play a variety of roles in RNA metabolism (reviewed in reference 49). hnRNPs A1, A2/B1, D, and E bind ss-G-strand overhang DNA in vitro, although they associate more tightly with RNA of the cognate sequence (20, 31). Recently, hnRNP A1 was shown to be essential for the maintenance of telomere length in a mouse cell line, and UP1, a truncated derivative of hnRNP A1, was shown to interact with telomerase in a cell extract (26), suggesting that hnRNP A1 acts both as a telomere-binding protein and as a splice site selection factor in vivo.
The G-strand binding protein of Chlamydomonas reinhardtii (Gbp1p) was identified and cloned on the basis of its ability to specifically associate with ssDNA containing the G-strand telomere repeat sequence (37, 38), and thus we have classified Gbp1p as a putative telomere-binding protein. RLF6 is the S. cerevisiae gene most similar to GBP1. Rlf6p binds ssDNA containing the Saccharomyces telomere repeat sequence in vitro (24, 28). Mutation of RLF6 alters the subnuclear localization of the double-stranded (ds) telomere-binding protein Rap1p but does not change other telomere-associated phenotypes (24). Gbp1p functionally complements rlf6 mutations: expression of this Chlamydomonas protein in rlf6 strains restores the subnuclear localization of Rap1p (24).
Gbp1p contains two RNA recognition motifs (RRMs) separated by a glycine-rich domain (38), an arrangement commonly found in human hnRNPs. RRMs are domains of approximately 80 to 100 amino acids (aa) that usually include at least one of two RNP consensus sequences (21), RNP-1 (K/R G F/Y G/A F V X F/Y) and RNP-2 (L/I F/Y V/I G/K N/G L/M) (5). The best-studied RRM-containing protein is hnRNP A1, an abundant nuclear protein which is important for splice site selection (7, 11). hnRNP A1 contains two RRM domains at its amino terminus and a glycine-rich domain at its carboxyl terminus. Each of the RRM domains independently binds either RNA or ssDNA but preferentially binds RNA (10, 32, 34). The conserved phenylalanines at position 5 in the RNP-1 consensus sequences of hnRNP A1 can be photochemically cross-linked to bound RNA (32), indicating a close association between this residue and the substrate. The glycine-rich domain of hnRNP A1 is sufficient for homodimerization or heterodimerization with other hnRNPs in vitro and in vivo (9).
Although most RRM-containing proteins clearly bind RNA with higher affinity than they bind DNA (49), at least one RRM-containing protein appears to preferentially bind DNA. Stage-specific activator protein 1 (SSAP-1) is a transcriptional activator of the sea urchin late histone H1 gene (12). The two RRM motifs of SSAP-1 are required for its association with either ss- or dsDNA but have little affinity for the cognate RNA molecule (12).
In this study, we investigated Gbp1p, a putative telomere end-binding protein from C. reinhardtii which contains two nonconsensus RRM domains and which binds ss telomeric DNA but not ds telomeric DNA (38). Despite the fact that a phenylalanine at position 5 in the RNP-1 consensus has been shown to have an important role in substrate binding, we found that mutation of the two RNP-1 motifs to match the conserved sequence had no measurable effect on the ability of Gbp1p to bind ssDNA or RNA or to discriminate between the two nucleic acids. We determined the specific sequence within the Chlamydomonas telomeric repeat sequence that is bound by Gbp1p. We found that monomeric Gbp1p binds either ss telomeric DNA or the cognate RNA sequence but has a small binding preference for RNA. In sharp contrast, dimeric Gbp1p associates with ssDNA with high affinity and binds RNA with much lower affinity. To the best of our knowledge, this is the first example of a protein that qualitatively changes its nucleic acid binding preference upon dimerization.
MATERIALS AND METHODS
Plasmids.
Plasmid pTrpE-Gbp1p (38) contains the Gbp1p coding sequence subcloned into pATH11 (23) so that it is fused with the TrpE open reading frame and is tryptophan inducible. This plasmid (also called pTrpE-IGIIC) was used as a starting point for construction of the deletion and point mutants of GBP1. pTrpE-IGII (aa 1 to 224) was made by subcloning the EcoRI-to-SacI fragment of pTrpE-GBP1 into pATH11. Similarly, pTrpE-IG (aa 1 to 144) contains the EcoRI-to-PstI fragment of pTrpE-GBP1 in pATH11, pTrpE-GIIC (aa 90 to 237) contains the Bpu1102I-to-EcoRI fragment of pTrpE-GBP1 in pATH11, and pTrpE-GII (aa 90 to 224) contains the Bpu1102I-to-SacI fragment of pTrpE-GBP1 in pATH11. pTrpE-IFGIIC and pTrpE-IGIIFC, which contain the mutations at positions 71 (substitution of phenylalanine for isoleucine) and 203 (substitution of phenylalanine for tyrosine), respectively, were constructed by using the oligonucleotides AAACTCCACGAAACCCCAGCC and GAACTTGACGAAGCCGTAGCC to mutagenize pTrpE-GBP1 essentially as described elsewhere (1). The double-mutant plasmid (pTrpE-IFGIIFC) was assembled from the two single-mutant plasmids by using the PstI site.
Protein preparation and analysis. (i) Extract preparation.
TrpE fusion proteins were expressed in Escherichia coli RR1 cells, extracts were prepared as described elsewhere (23) and then stored at −20°C. Most of the fusion protein was present in the insoluble fraction, but the soluble fusion protein was sufficient for purification and analysis. Chlamydomonas protein extracts were prepared essentially as described previously (38). Briefly, cells were disrupted by vortexing with glass beads at 4°C in a solution containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol (DTT), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and debris was removed by centrifugation. Gbp1p activity was stable at −20°C for at least 6 months and survived multiple freeze-thaw cycles. The total protein concentration was determined by the Bradford assay (Bio-Rad). Approximately 1 μg of total protein was used in each binding assay except where otherwise indicated.
(ii) Immunoaffinity purification.
A 500-μl portion of TrpE fusion protein extract or 500 μg of Chlamydomonas protein was mixed with 50 μg of polyclonal anti-Gbp1p in a solution consisting of 20 mM Tris-HCl (pH 8.0), 350 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40 (NP-40), and 0.1 mM PMSF. Complexes were allowed to form on ice for 1 h prior to the addition of 80 μl of protein A-agarose suspension (Santa Cruz Biotechnology). The suspension was mixed for 1 h at 4°C, and the beads were washed three times with 1 ml of chilled binding buffer. Gbp1p was eluted from the beads by two successive washed with 0.5 ml of a solution containing 100 mM glycine (pH 11.0), 2 M NaCl, 10 mM DTT, 1 mM EDTA, and 0.1 mM PMSF at room temperature (5 min each). Eluates were dialyzed against 200 ml of a buffer consisting of 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 50% glycerol, and 0.1 mM PMSF at 4°C overnight and then stored at −20°C.
(iii) DNA affinity purification.
A 500-μg portion of Chlamydomonas protein or 500 μl of TrpE fusion protein extract was mixed with 5 μg of the oligonucleotide (TTTTAGGG)4 with a biotin molecule covalently attached to the 5′ end (Integrated DNA Technologies, Inc.) in 1 ml of buffer (20 mM HEPES [pH 7.4], 50 mM NaCl, 1 mM EDTA, 5% glycerol, 0.05% NP-40). After incubation of the mixture on ice for 30 min, 150 μl of immobilized streptavidin (Boehringer Mannheim) was added, and the resulting suspension was shaken for 1 h at 4°C. The beads were washed three times with 1 ml of ice-cold binding buffer, and Gbp1p was eluted from the beads by two successive washes with 200 μl of a solution containing 20 mM Tris-HCl (pH 8.0), 1 M NaCl, 1 mM EDTA, 5% glycerol, and 0.05% NP-40 at room temperature (10 min each). Eluates were dialyzed against 200 ml of a buffer consisting of 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 50% glycerol, and 0.1 mM PMSF at 4°C overnight and then stored at −20°C. DNA affinity purification was found to yield more protein activity than immunoaffinity purification. To obtain higher concentrations of the fusion protein for the experiment shown in Fig. 5, the protocol was scaled up fivefold and the dialyzed protein was redialyzed against solid polyvinylpyrrolidone (molecular weight, 40,000) at 4°C for 2 h to further concentrate it and then stored at −20°C.
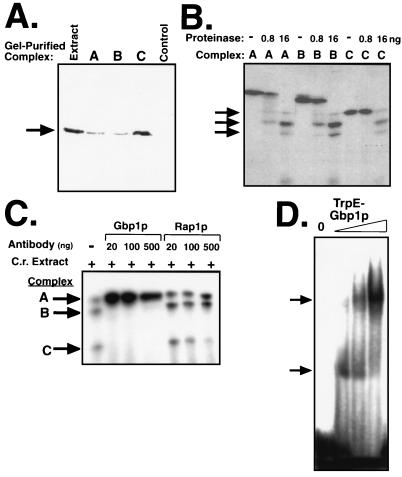
FIG. 5.
All three complexes contain Gbp1p and, likely, only Gbp1p. (A) Gbp1p is present in all three complexes. Complexes A, B, and C (or a region of the gel between B and C [Control]) were gel purified, the cross-linking was reversed, and the presence of Gbp1p (arrow) in the recovered proteins was assayed by immunoblotting with anti-Gbp1p serum. (B) Gbp1p is the only protein cross-linked to dCG3 in all three complexes. Complexes A, B, and C were gel purified, the cross-linked protein-DNA complexes were not (−) or were (+) subjected to limited proteolysis, and the resulting cross-linked fragments were resolved by SDS-PAGE on a 16% gel. The major proteolytic fragments are indicated by the arrows. (C) Anti-Gbp1p interferes with complexes B and C but not with complex A. Chlamydomonas extract (C.r. extract) was cross-linked to labeled dCG3 in the absence (−) or presence (+) of increasing amounts of anti-Gbp1p or anti-Rap1p serum as indicated. (D) Recombinant Gbp1p forms dimers. Full-length Gbp1p fused to TrpE was purified by DNA affinity chromatography and concentrated. Increasing amounts of the fusion protein (1, 3, or 10 μl) were assayed by EMSA with labeled dCG3 as a probe. The arrows indicate the two major TrpE-Gbp1p-dCG3 complexes.
(iv) Protein analysis.
Prior to being subjected to immunoblotting, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels, transferred to a polyvinylidene difluoride membrane, and blocked with 1% nonfat dry milk. Anti-Gbp1p serum at a dilution of 1:5,000 was used to probe the membrane, and protein was detected by enhanced chemiluminescence or chemofluorescence (Amersham). The anti-Gbp1p serum reacted with only one protein in the Chlamydomonas cell extract. Ammoniacal silver staining was performed as described elsewhere (1). Unstained Mark-12 protein markers (Novex) were used for the precise determination of molecular mass.
Nucleic acid binding assays. (i) Electrophoretic mobility shift assays (EMSAs).
Protein extracts or purified proteins were mixed with 1 ng of 5′ 32P-end-labeled oligonucleotide and competitors in 20 μl of buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1.25 mM MgCl2, 0.125 mM EDTA, 1 mM DTT, 6% glycerol, 0.125% NP-40, 2% Ficoll [final concentrations]). Inclusion of a nonspecific competitor [either E. coli DNA or poly (dI-dC)] had no affect on the assay when either purified protein or crude cell extract was used. Protein and nucleic acid preparations were incubated for 15 min on ice and then for 15 min at room temperature before being loaded on either a 4% or a 5% native polyacrylamide gel (30:1 acrylamide/bisacrylamide) in 0.5× Tris-borate-EDTA (TBE). Complexes were separated in 0.5× TBE at 4°C and 150 V for 2.5 h. The gels were dried and exposed to film or analyzed with a PhosphorImager (Storm 840; Molecular Dynamics).
(ii) Cross-linking.
Protein extract was mixed with 1 ng of 5′ 32P-end-labeled oligonucleotide and competitor in 10 μl of buffer (20 mM HEPES [pH 7.4], 50 mM NaCl, 1 mM EDTA, 5% glycerol, 0.05% NP-40 [final concentrations]). The mixture was incubated on ice for five min before the addition of 1 μl of 10% formaldehyde, and cross-linking was allowed to proceed at room temperature for 20 min. SDS loading buffer (5 μl) was added to each sample, and the complexes were resolved by SDS-PAGE through 10% polyacrylamide gels. The gels were fixed for 5 min in 10% acetic acid–10% methanol before being dried and exposed to film or analyzed with a PhosphorImager. Inclusion of a nonspecific competitor [either E. coli DNA or poly(dI-dC)] had no effect on the assay. To elute cross-linked bands from the gel, the reaction was scaled up 10-fold and the product was electrophoresed through a gel with wide lanes. The wet, unfixed gel was exposed to film overnight with orientation markers, and the three complexes were excised from the gel with a razor blade. Protein-DNA complexes were electroeluted from the gel slices into dialysis tubing in 0.5× TBE for 15 min at 100 V and 4°C and recovered by precipitation with 4 volumes of cold acetone and 10 μg of bovine serum albumin per sample as a carrier. The pellet was resuspended, and either the cross-linking was reversed by incubation at 65°C for 4 h followed by immunoblotting (see Fig. 5A) or the complexes were digested with protease (see Fig. 5B). Complexes were digested with 0, 0.8, or 16 ng of proteinase K (Boehringer Mannheim) in a solution containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM EDTA, and 5% glycerol at 37°C for 20 min. Immediately after digestion, the samples were electrophoresed through an SDS–16% polyacrylamide gel that was processed for autoradiography as described above.
RESULTS
Analysis of the RNP-1 octads of Gbp1p.
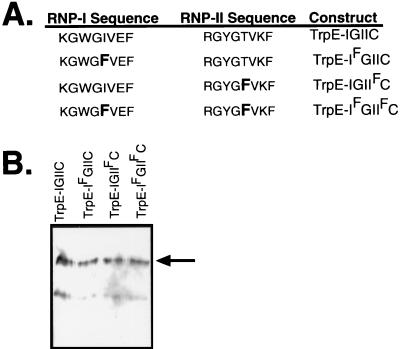
RNP-1 octads contain a highly conserved phenylalanine residue at position 5 (F5) (5) that has been photochemically cross-linked to bound RNA (32), suggesting that this residue either contacts or is very close to the RNA. One of the striking features of Gbp1p is that neither RRM-I nor RRM-II matches the consensus at this site (38). This difference from the consensus sequence also occurs in the Saccharomyces proteins Rlf6p, Nsr1p, and Tom34p, which suggests that these proteins can be grouped as a subfamily of RRM-containing proteins lacking F5 (24). Most RRM-containing proteins associate with RNA, but members of this subfamily lacking F5 have been shown to preferentially associate with ssDNA. Gbp1p in a whole-cell extract preferentially associates with telomeric G-strand ssDNA and not with the cognate RNA sequence (38). Rlf6p and Nsr1p bind telomeric G-strand ssDNA (24, 28). Thus, we hypothesized that the lack of F5 in RNP-1 might account for the unusual binding specificity of Gbp1p and perhaps of the subfamily in general. To test this hypothesis, GBP1 was subcloned into pATH11 to construct a plasmid that directs the production of a TrpE-Gbp1p fusion protein in E. coli (TrpE-IGIIC). We first confirmed that TrpE-IGIIC could be stably expressed and immunopurified from E. coli. TrpE-IGIIC specifically bound Chlamydomonas telomeric ssDNA (data not shown), demonstrating that no eukaryote-specific posttranslational modifications or auxillary proteins are required for this association. Additionally, we noted that TrpE-Gbp1p was remarkably heat stable (incubation at 65°C for 10 min resulted in less than a 20% reduction in binding activity [data not shown]) and that the TrpE-Gbp1p–ssDNA complex (unlike ciliate telomere-binding proteins [41, 43]) was highly sensitive to the NaCl concentration (diminishing greatly at NaCl concentrations above 200 mM [data not shown]). We constructed plasmids directing the expression of fusion proteins in which position 5 of the RNPs were mutated to phenylalanine in RRM-I (TrpE-IFGIIC), RRM-II (TrpE-IGIIFC), or in both RRMs (TrpE-IFGIIFC) (Fig. 1A). These fusion proteins were expressed in E. coli by tryptophan starvation and purified by immunoaffinity chromatography with a polyclonal anti-Gbp1p serum. All proteins were stable in E. coli and were analyzed qualitatively and quantitatively by immunoblotting with anti-Gbp1p serum (Fig. 1B).
FIG. 1.
Gbp1p with point mutations in one or both RNP-1 motifs. (A) Schematic representation of the two single mutants and one double mutant of Gbp1p as TrpE fusion proteins. The fifth residue in one or both of the RNP motifs was mutated to a phenylalanine to match the RNP-1 consensus sequence. (B) Immunoblot of the wild-type and point-mutant TrpE-Gbp1p proteins. The fusion proteins were produced in E. coli, purified by immunoaffinity chromatography, and detected with anti-Gbp1p serum. The upper band (arrow) is the predicted size for the TrpE-Gbp1p fusion; the lower band is likely a breakdown product, and its quantity differed among preparations (compare with Fig. 7B).
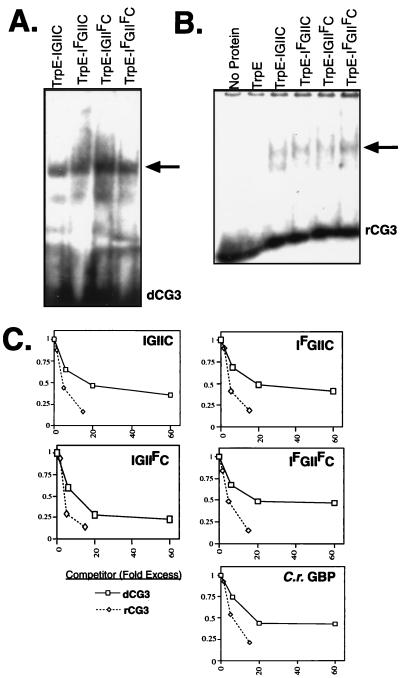
First, we asked whether the F5 mutations altered the affinity of Gbp1p for ssDNA. Approximately equal amounts of each of the proteins were assayed for activity in an EMSA using a radiolabeled ssDNA oligonucleotide that contains three repeats of the guanine-rich strand of the Chlamydomonas telomere sequence [(TTTTAGGG)3, hereafter referred to as dCG3]. All three mutant fusion proteins bound dCG3 equally well (Fig. 2A) and with an affinity similar to that of the nonmutated fusion protein. Thus, mutation of position 5 of the RNP consensus to phenylalanine does not have a major effect on the ability of Gbp1p to associate with ssDNA.
FIG. 2.
Point mutations in the RNP-1 motifs do not affect the association of Gbp1p with ssDNA or RNA either quantitatively or qualitatively. (A) Wild-type and point-mutant proteins shown in Fig. 1A were analyzed by EMSA with radiolabeled dCG3 as a probe. The arrow indicates the major protein-DNA complex. (B) Wild-type and point-mutant proteins were analyzed by EMSA with radiolabeled r(UUUUAGGG)3 rCG3 as a probe. The arrow indicates the major protein-RNA complex. (C) Wild-type and point-mutant TrpE-Gbp1p fusion protein or Gbp1p isolated from a Chlamydomonas extract by immunoaffinity chromatography (C.r. Gbp1p) was used in an EMSA with radiolabeled dCG3. Increasing amounts of unlabeled dCG3 (squares) or rCG3 (diamonds) were added to compete with the complexes. Complexes were quantitated by PhosphorImager analysis, and the data were normalized to the amount of complex present in the absence of competitor and plotted.
We next asked whether the F5 mutations that make Gbp1p fit the RNP-1 consensus sequence would alter the affinity of Gbp1p for RNA. We tested the fusion proteins in an EMSA using a radiolabeled RNA oligonucleotide (rCG3) of the cognate sequence of dCG3 [i.e., (UUUUAGGG)3]. All four fusion proteins bound rCG3 indistinguishably (Fig. 2B), indicating that a phenylalanine residue at position 5 of both of the RNP-1 motifs does not obviously alter the ability of Gbp1p to associate with RNA.
To determine if the RNP-1 F5 mutations might have subtler effects on the relative affinities of Gbp1p for DNA and RNA, EMSAs were performed with each of the fusion proteins, using radiolabeled dCG3 as a probe and including increasing amounts of unlabeled dCG3 or rCG3 as a competitor. The fraction of probe shifted in each experiment was determined by PhosphorImager analysis and normalized to the fraction shifted in the absence of competitor (Fig. 2C). Interestingly, rCG3 competed for binding to TrpE-Gbp1p, and mutation of RNP-I, RNP-II, or both RNP-I and RNP-II to F5 had no effect on the binding of Gbp1p to rCG3. Thus, the alteration of the RNP-1 motifs to F5 has no discernable effect on the ability of Gbp1p to bind either ssDNA or RNA or on the ability of Gbp1p to discriminate between the two nucleic acids, contradicting our original hypothesis.
Surprisingly, purified recombinant TrpE-Gbp1p fusion protein had a slight preference for RNA relative to ssDNA in an EMSA (Fig. 2C). This was unexpected because our laboratory previously observed that Gbp1p in a Chlamydomonas extract bound ssDNA much more efficiently than it bound the cognate RNA (38). A possible explanation for the difference in binding preference in the two experiments is that the recombinant TrpE-Gbp1p lacked a Chlamydomonas-specific modification or binding partner. Alternatively, the TrpE domain at the amino terminus altered the binding characteristics of Gbp1p. To address these possibilities, Gbp1p was isolated from a Chlamydomonas cell extract by immunoaffinity chromatography, using essentially the same methods that were used to purify the recombinant TrpE fusion proteins. This native Gbp1p had a binding profile indistinguishable from that of recombinant TrpE-Gbp1p; rCG3 competed slightly more effectively for binding than did dCG3 (Fig. 2C). Thus, Gbp1p purified from E. coli or from C. reinhardtii binds both ssDNA and RNA but shows a slight preference for RNA. This indicates that the lack of a Chlamydomonas-specific modification or binding partner is not the reason for the surprising binding preference of Gbp1p for RNA.
Gbp1p and ssDNA form three distinct complexes.
Another possible explanation for the difference between the results of Petracek et al. (38) and those shown in Fig. 2 is that different amounts of protein were used. To begin to address this possibility, EMSAs were performed with a range of crude Chlamydomonas extract concentrations (Fig. 3). As the protein concentration increased, three distinct complexes were observed, and the relative abundance of each complex changed with increasing protein-to-probe ratio. The relative abundance of the uppermost complex varied from experiment to experiment. At high protein concentrations, only one complex was observed. Because the original characterization of Gbp1p was performed at very high protein concentrations (>10 μg of total protein per assay) (38), it was the binding characteristics of this one complex that were reported.
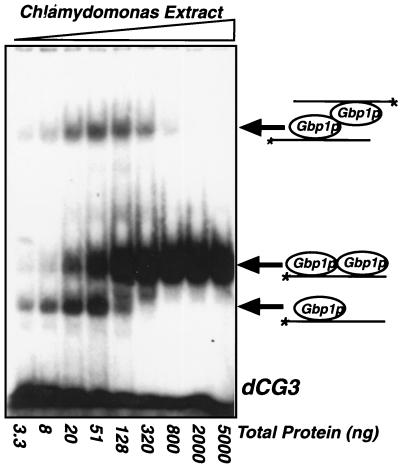
FIG. 3.
Three forms of Gbp1p-ssDNA complex exist within an extract. Increasing amounts of a Chlamydomonas whole-cell extract were incubated with 1 ng of radiolabeled dCG3 and analyzed by EMSA. The three complexes detected can each be alleviated via competition by unlabeled dCG3, and all contain Gbp1p as determined by an EMSA-immunoblot (data not shown). A model to explain the three complexes that is consistent with the observed concentration-dependent transitions between the complexes is presented on the right.
All three complexes evident in Fig. 3 were recognized by anti-Gbp1p serum in an EMSA-immunoblot analysis. Additionally, like the purified Gbp1p from C. reinhardtii and TrpE-Gbp1p, all three complexes were sensitive to treatment with 200 mM NaCl or proteinase K and were insensitive to treatment with micrococcal nuclease or to preincubation of the extract at 65°C for 10 min (data not shown). We considered the possibility that the multiple bands were a result of the probe being bound by differently phosphorylated Gbp1p isoforms. However, Gbp1p does not appear to be phosphorylated, since the protein as found in the extract ran as a single, discrete band on an immunoblot and its mobility was unaffected by treatment with calf alkaline phosphatase or potato acid phosphatase (data not shown). To determine whether nucleases were degrading the probe, leading to the generation of multiple bands, we examined the probe on a denaturing DNA gel after incubation with the extract and found that it had not been altered (data not shown). Thus, we conclude that all three complexes (Fig. 3) likely contain Gbp1p.
A model to explain the presence of three specific Gbp1p-DNA complexes is shown on the right side of Fig. 3. We hypothesize that the complex with the fastest electrophoretic mobility contains a Gbp1p monomer bound to a single dCG3 oligonucleotide, that complex with intermediate mobility contains a Gbp1p homodimer bound to a single oligonucleotide, and that the complex with the slowest mobility contains a Gbp1p homodimer bound to two oligonucleotides. hnRNP A1 is structurally similar to Gbp1p and is known to homodimerize through its glycine-rich domain (9). This model is consistent with the concentration-dependent transition between the three complexes; that is, the putative Gbp1p monomer-ssDNA complex occurs at low protein-to-probe ratios. As this ratio increases, the putative Gbp1p monomer-ssDNA complex disappears and the Gbp1p homodimer associated with one or two oligonucleotides appears. At high protein-to-probe ratios, the putative Gbp1p homodimer bound to a single oligonucleotide is the only complex detected.
We detected a specific Gbp1p-ssDNA putative monomeric complex with as little as 3.3 ng of total Chlamydomonas protein extract (Fig. 3), suggesting that either Gbp1p is an extremely abundant protein in C. reinhardtii or it has a very high affinity for this oligonucleotide, or both. To estimate the abundance of Gbp1p, we purified it from a Chlamydomonas extract by DNA affinity chromatography. By comparison with three known standards on a silver-stained gel, we estimated the amount of Gbp1p present in the sample, and then we used the sample as a standard to calibrate immunoblots of total Chlamydomonas extract. This analysis suggested that there was approximately 2 ng of Gbp1p per μg of total protein in our extract (data not shown). In other words, approximately 0.2% (by mass) of the extracted protein is Gbp1p; i.e., there are approximately 3.5 × 106 Gbp1p molecules per Chlamydomonas cell. It is apparent that Gbp1p is an extremely abundant protein.
Gbp1p dimers have a strong preference for ssDNA, and Gbp1p monomers have a weak preference for RNA.
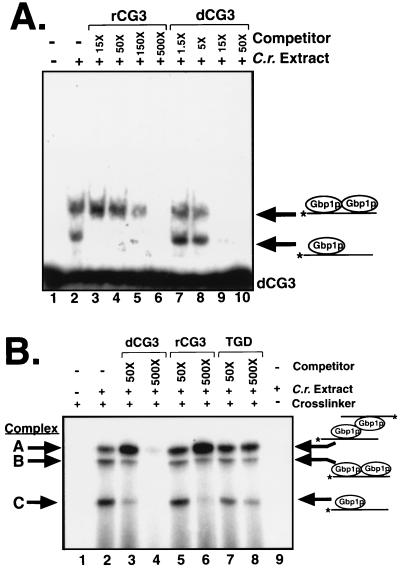
To investigate the possibility that the different isoforms of Gbp1p in Fig. 3 associate differently with RNA and ssDNA, we performed competition experiments with labeled dCG3 and either unlabeled dCG3 or unlabeled rCG3. We performed this experiment at a concentration of Chlamydomonas extract at which the putative Gbp1p monomer and Gbp1p dimer complexes were clearly visible in an EMSA (Fig. 4A). In this experiment, the most slowly migrating band (seen in Fig. 3) was not readily visible, although in other experiments this complex was observed and its binding affinity was similar to that of the intermediate complex. The putative Gbp1p monomer and dimer complexes responded similarly to the addition of the ssDNA competitor (Fig. 4A, lanes 7 to 10). Interestingly, these two complexes responded very differently to the RNA competitor (lanes 3 to 6). The putative Gbp1p dimer had a strong preference for ssDNA; a 15-fold excess of unlabeled dCG3 outcompeted the labeled dCG3 nearly completely, while a 15-fold excess of unlabeled rCG3 had essentially no effect (Fig. 4A, compare lanes 9 and 3, upper complexes). In contrast, the Gbp1p monomer had similar affinities for RNA and ssDNA, with a slight preference for RNA (Fig. 4A, compare lanes 9 and 3, lower complexes; also shown more clearly in Fig. 3C). Thus, the putative Gbp1p monomeric and dimeric isoforms have different binding preferences for ssDNA and RNA. Furthermore, the putative Gbp1p monomer had binding preferences like those observed for purified Gbp1p and TrpE-Gbp1p (Fig. 2), while the putative Gbp1p dimer exhibited binding preferences like those observed for Gbp1p from Chlamydomonas extracts (38).
FIG. 4.
Monomeric Gbp1p has a slight preference for binding RNA; dimeric Gbp1p has a strong preference for binding ssDNA. (A) EMSA using 0 (−) or 15 ng (+) of Chlamydomonas extract and radiolabeled dCG3. The molar fold excess of either rCG3 (C.r. extract) or dCG3 unlabeled competitor oligonucleotide that was added to each reaction is shown above the autoradiogram. (B) Three distinct DNA-protein complexes are identified in a cross-linking assay using radiolabeled dCG3 and Chlamydomonas extract. These complexes are named A, B, and C (from largest to smallest) and are all dependent on the presence of the extract and the cross-linker. The molar fold excess of either dCG3, rCG3, or TGD (G2T4)5 unlabeled competitor oligonucleotide that was added to each reaction is shown above the autoradiogram.
To measure the sizes of the Gbp1p-dCG3 complexes observed in the EMSA assays, we turned to a cross-linking assay. Chlamydomonas extract was mixed with labeled dCG3, the mixture was incubated on ice for 5 min, and the proteins and DNA were covalently cross-linked by the addition of formaldehyde. The resulting mixture was denatured, and the complexes were resolved by SDS-PAGE. Three specific complexes identified by this assay have been labeled (from largest to smallest) complexes A, B, and C (Fig. 4B, lane 2). This assay also had the advantages that all three complexes were always observed and the relative ratios of the three complexes were highly consistent between experiments, although it was less sensitive than the EMSA. The relative mobilities of complexes A to C in SDS-polyacrylamide gels corresponded to molecular masses of 49.6, 44.5, and 30.5 kDa, respectively. We consider it likely that the three complexes identified in this denaturing gel correspond to the three complexes identified in the native-gel EMSA assay. The formation of multiple covalent cross-links between protein and nucleic acid as well as between the two protein molecules leads to a highly cross-linked molecule whose migration in an SDS-PAGE system may be aberrant. Thus, the size estimates for these complexes (especially for the larger complexes) may not accurately reflect their true molecular masses.
The cross-linking assay was used to determine the relative affinities of the three complexes for ssDNA and RNA. Similar to the EMSA results (Fig. 4A), ssDNA competed with the two putative Gbp1p homodimer complexes (A and B) far more efficiently than did RNA, while ssDNA and RNA competed with the putative Gbp1p monomer complex (C) with similar efficiencies (Fig. 4B, compare lanes 4 and 6). In agreement with earlier data (38), an ssDNA oligonucleotide containing the sequence of the Tetrahymena telomere (GGTTTT)5 did not compete with any of the isoforms of Gbp1p. Thus, on the basis of both the EMSA and cross-linking data, we conclude that complexes A and B have strong preferences for associating with ssDNA while complex C has a weak preference for associating with RNA.
Previous data indicated that Gbp1p associated only with ssDNA and not with RNA (38). This conclusion was based on the results of EMSAs that utilized very large amounts of protein. In the present work with purified protein (at relatively low concentrations), we found that Gbp1p had a slight binding preference for RNA over ssDNA. We interpreted this to mean that the assays using purified protein were principally measuring binding by Gbp1p monomers, due to the relatively low protein concentration, and the previous EMSAs (38) were principally measuring binding by Gbp1p dimers, due to the high protein concentrations used.
All three complexes contain Gbp1p and only Gbp1p.
We next sought to test our model (presented in Fig. 3) which predicts that the only protein in all three complexes is Gbp1p. To determine which of the cross-linked complexes contain Gbp1p, the presence or absence of Gbp1p in each of the complexes was determined in a denaturing gel. The cross-linking assay was scaled up fourfold, and the positions of the three complexes were determined in an unfixed, wet gel by autoradiography. Each complex was excised and eluted from the gel before the cross-linking was reversed. The proteins (no longer cross-linked to DNA or to each other) were resolved by denaturing gel electrophoresis, and Gbp1p was detected by immunoblotting. Full-length Gbp1p was detected in all three complexes, but not in a gel slice excised from the region between complexes B and C (Fig. 5A). More Gbp1p was found in the gel slice representing complex C, probably because excess, unbound Gbp1p was not resolved from complex C. These results indicated that Gbp1p was intact in all of these complexes and that the multiple bands were not due to partial degradation of the protein. Therefore, we conclude that Gbp1p is found in all three complexes.
Although Gbp1p was determined to be present in all three complexes, it remained possible that Gbp1p heterodimerized with a second Chlamydomonas protein to generate complexes A and B. To investigate the nature of the protein(s) bound to the ssDNA in each complex, we analyzed the products of partial proteolysis of each complex. Each of the three cross-linked complexes was eluted from the gel, digested with a limiting amount of proteinase, and separated by SDS-PAGE. The three complexes had indistinguishable proteolytic profiles (Fig. 5B); complexes A and B generated a band that comigrated with complex C, and all three complexes produced identical bands corresponding to molecular masses of approximately 21 and 16 kDa. These data indicate that the proteins cross-linked to the oligonucleotide in all three complexes are indistinguishable. Therefore, we concluded that all three complexes contained Gbp1p and that Gbp1p was the only protein in the complexes that is close enough to the oligonucleotide probe to be chemically cross-linked to it. Thus, it is likely that Gbp1p is the only protein in each of the complexes.
To further test our model, we used our anti-Gbp1p serum as a nondenaturing probe to detect differences in conformation between the complexes, all of which contain Gbp1p. Depending on the location and availability of the epitope(s) in each complex, the antiserum could either supershift or ablate the complex or leave it unaffected. A polyclonal antiserum raised against Gbp1p or a control polyclonal antiserum raised against the S. cerevisiae protein Rap1p (13) was added to the Chlamydomonas extract, and after a 5-min incubation on ice, labeled dCG3 was added and the cross-linking protocol was continued. The anti-Gbp1p serum disrupted complexes B and C but had no effect on complex A, the putative Gbp1p homodimer bound to two molecules of dCG3. Anti-Rap1p serum had no effect on any of the complexes (Fig. 5C). The addition of the anti-Gbp1p serum increased the intensity of complex A, likely because disruption of the smaller complexes resulted in more Gbp1p and probe being available to form the larger complex. Because we know that complex A contains Gbp1p (Fig. 5A and B), we conclude that the Gbp1p epitope(s) is masked by the particular conformation of Gbp1p in complex A. In this experiment, the antiserum served as a structural probe, and the results indicate that the protein in complex A is in a conformation different from that of the proteins in complex B or C. Consistent with the model we have proposed, the presence of two oligonucleotides in complex A may limit the accessibility of the antibody to its epitope(s) on Gbp1p.
To definitively determine whether complex A is a Gbp1p homodimer, we next asked if it was possible for the recombinant Gbp1p to dimerize. In this system, Gbp1p is the only eukaryotic protein present, so if evidence of dimerization is seen it must be due to homodimerization and not heterodimerization. Full-length, wild-type TrpE-Gbp1p was purified by DNA affinity chromatography and concentrated before being used in the EMSA with dCG3 as a probe. Two distinct Gbp1p-dCG3 complexes were formed (Fig. 5D). As the protein concentration increased, the abundance of the larger complex increased at the expense of the smaller complex. This concentration-dependent transition with recombinant TrpE-Gbp1p is consistent with our model of Gbp1p homodimerization.
Definition of the Gbp1p-binding site.
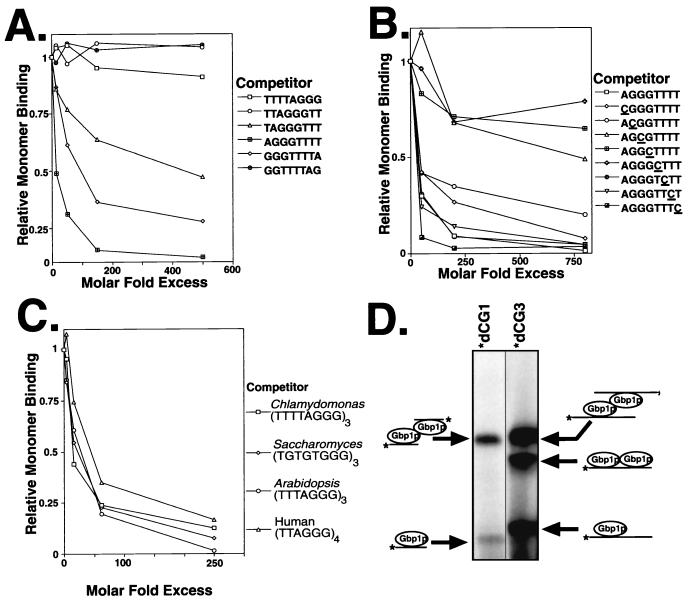
The probe used for both the EMSA and the cross-linking assay described above contains three copies of a TTTTAGGG repeat (dCG3). Depending on the exact sequence recognized by Gbp1p, this oligonucleotide likely contains multiple Gbp1p-binding sites. Our model predicts that a probe with only one binding site will not be able to form complex B (two Gbp1p molecules bound to two sites on one oligonucleotide) but will be able to form complexes A and C. To design a probe with only one Gbp1p-binding site, we first determined the minimal sequence necessary for Gbp1p binding. We tested six circularly permutated 8-mer oligonucleotides for their ability to compete with the Gbp1p monomer-dCG3 complex (complex C) in the cross-linking assay. Gbp1p was capable of binding three of the octamers (Fig. 6A). The sequence common to these three oligonucleotides was GGGTTT, indicating that this sequence of the Chlamydomonas ss-G-strand telomere is minimally sufficient for Gbp1p binding. Additionally, in the two cases in which GGGTTT (underlined) was located at the 5′ or 3′ end of the oligonucleotides (TAGGGTTT and GGGTTTTA), the binding affinity was lower than when the sequence was located in the middle (AGGGTTTT), suggesting that flanking DNA may assist in Gbp1p binding.
FIG. 6.
Definition of the Gbp1p binding site. (A) The minimal binding site for Gbp1p in the Chlamydomonas ss-G-strand telomere is GGGTTT. Chlamydomonas extract was covalently cross-linked to the radiolabeled dCG3 oligonucleotide in the presence of increasing amounts of the indicated unlabeled oligonucleotides. The resulting complexes were separated by denaturing gel electrophoresis, and the band corresponding to the Gbp1p monomer was quantitated by PhosphorImager analysis. The intensity of this band, relative to that in a reaction with no competitor, is plotted as a function of the molar fold excess of the competitor binding sites over the probe binding sites. (B) The GGT trinucleotide within the minimal binding site is critical for Gbp1p binding. Crucial nucleotides in the binding site were determined as for panel A, using the indicated oligonucleotides, in which each base was systematically changed to a cytosine (underlined). (C) Gbp1p also associates with telomere sequences from S. cerevisiae, Arabidopsis thaliana, and humans. The relative affinities of Gbp1p for ss-G-strand telomere sequences from other species were determined as for panel A by oligonucleotide competition. (D) Multiple binding sites per oligonucleotide are required for formation of complex B. Cross-linking was performed as described in the legend to Fig. 5B, using Chlamydomonas extract and either labeled dCG1 (AGGGTTTT) or labeled dCG3 (TTTTAGGG)3.
To more precisely determine the Gbp1p-binding site within the AGGGTTT octamer, we systematically altered each nucleotide to a cytosine and determined the effect of these mutations on binding. Increasing concentrations of the one wild-type and the eight mutant oligonucleotides were used as competitors for Gbp1p binding to labeled dCG3 in the cross-linking assay, and their effect on the Gbp1p monomer-dCG3 complex (complex C) was quantitated by PhosphorImager analysis. Five of the eight mutations had little effect on the ability to bind Gbp1p (Fig. 6B). Three oligonucleotides (AGCGTTTT, AGGCTTTT, and AGGGCTTT; mutated nucleotides are underlined) did not associate well with Gbp1p, thus identifying three nucleotides that are critical for Gbp1p binding. Mutation of the other nucleotides to cytosine had little effect on binding; it remains possible that mutation at these other sites to a different base would affect binding. These data identify the central GGT residues of the GGGTTT hexamer as being necessary for Gbp1p binding. However, the GGT trinucleotide alone is not sufficient for binding, since Gbp1p does not bind the Tetrahymena telomeric sequence (GGTTTT)5 (Fig. 4A) (38), which also includes GGT trinucleotides. It is likely that a T upstream of the core GGT sequence is not tolerated but a G or C can be tolerated.
Expression of GBP1 in S. cerevisiae is known to functionally complement the deletion of RLF6, the yeast homolog of GBP1 (24), which led us to predict that Gbp1p should associate with Saccharomyces ss-G-strand sequence. We tested this prediction in an oligonucleotide competition experiment similar to those described above. We compared the relative binding affinities of Gbp1p for Chlamydomonas (39), Saccharomyces (whose telomeres are highly degenerate, consisting of TG1–3 [45]), Arabidopsis (44), and human (33) ss telomere sequences. Gbp1p efficiently associated with telomeric sequences from these four organisms (Fig. 6C). Additionally, when the Saccharomyces oligonucleotide was labeled and used as a probe in the cross-linking assay, three complexes were identified whose mobilities were indistinguishable from the complexes formed on the labeled Chlamydomonas oligonucleotide (data not shown). These associations are consistent with the fact that all of these sequences contain more than one GGT core sequence, although there is some diversity in the nucleotides flanking this core. Additionally, this observation supports the idea that the expression of GBP1 in rlf6 mutant yeast cells complements the mutant phenotype through Gbp1p’s ability to associate with ss-G-strand telomeric DNA.
These binding studies indicate that the dCG3 oligonucleotide (TTTTAGGGTTTTAGGGTTTTAGGG) contains two Gbp1p-binding sites (underlined). If our interpretation of the three Gbp1p-dCG3 complexes is correct, an ssDNA molecule containing only one Gbp1p-binding site should be able to form complexes A and C but should be unable to form complex B (two Gbp1p molecules on one oligonucleotide). To test this aspect of our model, the minimal wild-type Gbp1p-binding sequence (AGGGTTTT [hereafter referred to as dCG1]) was radiolabeled and used as a probe in the cross-linking assay. As predicted, only two complexes were detected (Fig. 6D). Each of these complexes was slightly smaller than complexes A and C detected with dCG3. The slightly faster mobility of the two Gbp1p-dCG1 complexes relative to the Gbp1p-dCG3 A and C complexes is due to the smaller size of the dCG1 oligonucleotide. Even upon overexposure of the film, no complex B was detected when using dCG1 (data not shown), indicating that complex B cannot form on an oligonucleotide that contains only one Gbp1p-binding site. This result implies that at least two Gbp1p-binding sites on the oligonucleotide are required for the formation of complex B and is consistent with our interpretation of the nature of the three complexes.
Domain analysis of Gbp1p.
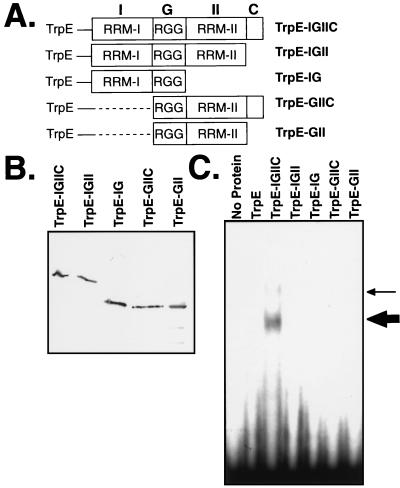
Sequence analysis of Gbp1p revealed the presence of two RRMs separated by an Arg-Gly-Gly domain, as well as a short carboxyl-terminal domain (38). The presence of two RRM domains suggests that a single Gbp1p molecule may be able to bind simultaneously to two nucleic acid molecules. To determine if the two RRM domains are able to associate independently with nucleic acids, we constructed deletion mutants of TrpE-Gbp1p. Four deletion variants of the pTrpE-IGIIC plasmid were created to eliminate the carboxyl-terminal domain and/or one of the two RRM domains (Fig. 7A). The fusion proteins were induced in E. coli and purified by immunoaffinity chromatography. Immunoblotting showed that all fusion proteins were produced and were stable (Fig. 7B). Approximately equal amounts of the proteins were assayed for activity in an EMSA using dCG3. TrpE-Gbp1p formed one major complex with the ssDNA; a second, minor complex was seen with some protein preparations (Fig. 7C). The major complex likely corresponds to the TrpE-Gbp1p monomer, and the minor complex likely corresponds to one of the dimer complexes.
FIG. 7.
RRM-I, RRM-II, and the carboxyl-terminal domain of Gbp1p are required for DNA binding. (A) Schematic representation of TrpE fusion proteins. Full-length Gbp1p (IGIIC, containing RRM-I, the glycine-rich domain, RRM-II, and the carboxyl-terminal domain) and various deletion derivatives were fused in frame with the TrpE protein. Dashed lines indicate deleted sequences. (B) Immunoblot of the five deletion variants of TrpE-Gbp1p. The fusion proteins were produced in E. coli, purified by immunoaffinity chromatography, and detected with anti-Gbp1p serum. (C) Only full-length Gbp1p binds DNA. The TrpE-Gbp1p deletion variants were assayed for DNA binding in an EMSA with radiolabeled (TTTTAGGG)3 (dCG3) being used as a probe. The thick arrow indicates the major TrpE-Gbp1p–ssDNA complex; the thin arrow a second, minor complex evident in some preparations.
Only the full-length Gbp1p bound dCG3. Deletion of either one of the RRMs abolished all binding of the protein to dCG3 (Fig. 7C). Removal of as few as the 13 carboxyl-terminal amino acids was also sufficient to abolish binding. Similarly, an 8-codon carboxyl-terminal deletion in RLF6, the S. cerevisiae homolog of GBP1, causes a loss of the Rap1p localization function of Rlf6p (20a). We conclude that the carboxyl-terminal domain and both RRM domains are required for Gbp1p to associate with ssDNA. Thus, unlike many RRM proteins, isolated RRM domains from Gbp1p cannot bind ssDNA independently, suggesting that a single Gbp1p molecule cannot simultaneously bind two nucleic acid molecules.
DISCUSSION
Characterization of Gbp1p.
Like many RRM-containing proteins, Gbp1p contains more than one RRM; however, both RRM domains are required for Gbp1p to associate with a single binding site on ssDNA. In contrast, for many proteins (including hnRNP A1 [32]), a single RRM is sufficient for RNA binding. Even small deletions in the carboxyl terminus of the protein ablate its ability to bind ssDNA. The phenylalanine at position 5 of the core RNP-1 sequence is one of the most highly conserved residues in RNP-1 (5), and it contacts the RNA associated with hnRNP A1 (32); yet Gbp1p, Rlf6p, and other proteins that bind G-strand telomeric DNA do not contain a phenylalanine at position 5 of any of their RNP-1 domains (24). This led us to ask whether F5 was important for the nucleic acid binding preference of Gbp1p. When we mutated position 5 of RRM-I and/or RRM-II to resemble the consensus motif, we found no effect on the ability to bind ssDNA or RNA or to discriminate between the two nucleic acids. Thus, the nonconsensus sequence of position 5 of the RNP does not control the discrimination by Gbp1p between ssDNA and RNA.
The telomeric repeat sequence in C. reinhardtii is (TTTTAGGG)n (39), and Gbp1p was previously identified as a protein that associated with ssDNA of this sequence (38). We precisely mapped the minimal Gbp1p-binding sequence to the GGGTTT hexamer within the Chlamydomonas telomere and determined that the central GGT nucleotides are crucial for binding. Additionally, Gbp1p binds to telomeric sequences from yeast (TG1–3)n, which is consistent with its ability to functionally complement a yeast strain bearing a deletion of its homolog, RLF6 (24). However, the sequence GGT is clearly not sufficient for binding, since the Tetrahymena telomeric sequence (GGTTTT)5 does not associate with Gbp1p (Fig. 4A) (38).
When assaying Gbp1p binding from a Chlamydomonas extract, three distinct complexes were identified by EMSA and in cross-linking assays (Fig. 3 and 4). The concentration-dependent transition that occurs between the three complexes is consistent with the model proposed in Fig. 3. Gbp1p is present in all three complexes (Fig. 5A), the Gbp1p in complex A is in a conformation different from that of the Gbp1p in complexes B and C (Fig. 5C), and recombinant Gbp1p is capable of homodimerization (Fig. 5D). Additionally, complex B does not form on an oligonucleotide with only one Gbp1p-binding site (Fig. 6D), which is consistent with our hypothesis that complex B is a Gbp1p homodimer associated with one oligonucleotide. All of these data are consistent with the idea that the three complexes consist of a Gbp1p monomer (complex C), a Gbp1p dimer with one oligonucleotide (complex B), or a Gbp1p dimer with two oligonucleotides (complex A), as depicted in Fig. 3.
Dimerization of Gbp1p changes the binding specificity.
Monomeric Gbp1p associates with either RNA or ssDNA, showing a slight binding preference for RNA. Yet dimeric Gbp1p, which is most readily identified in a Chlamydomonas cell extract due to its abundance, associates preferentially with ssDNA. This profound change in the binding characteristics of Gbp1p based on its dimerization state may have substantial physiological implications. The importance of dimerization of telomere-interacting proteins has been recently emphasized. In S. cerevisiae, telomerase is a complex that contains at least two functional telomerase RNAs (40), suggesting that the telomerase enzyme may need to dimerize to be functional. The Saccharomyces telomere-binding protein Rap1p associates with ds telomeric DNA as a dimer (10a). In humans, TRF1 (a structural homolog of Rap1p) also binds human ds telomeric DNA as a dimer (3). In vivo, telomeres are clustered both in S. cerevisiae (30) and in at least some mammalian cell types (52). Recently, Froelich-Ammon et al. (14) reported that the accessibility of Oxytricha telomere DNA to telomerase is regulated by the dimerization state of the telomere end-binding protein. When ss-G-strand DNA is bound by an α monomer, it is accessible to telomerase; in contrast, when it is bound by a homo-(α2) or heterodimer (αβ), it is inaccessible to telomerase (14). Dimerization is thus an emerging theme in telomere biology.
Gbp1p is not the only protein with RRM domains that associates with DNA. hnRNP A2/B1 associates specifically with ssDNA containing the human telomere repeat sequence, although it binds the cognate RNA sequence with a higher affinity (20, 31). Similarly, hnRNP A1, which is essential for telomere length control in mouse cells (26), preferentially binds to RNA but can associate with ss telomeric DNA (20). The RRM-containing protein human p54nrb and the mouse homolog NonO binds dsDNA to activate transcription but can also associate with RNA (2). The SSAP-1 transcription factor from the sea urchin, which contains two consensus RRM domains, associates with ssDNA and dsDNA. Like Gbp1p homodimers, the affinity of SSAP-1 for RNA is much weaker than its affinity for DNA (12).
Many proteins (such as hnRNPs A1, A2/B1, and K) are capable of associating with both ssDNA and RNA (2, 15, 20, 34). There are also many examples of proteins that undergo a quantitative change in affinity for their target sequence upon dimerization, such as Fos/Jun (6), lambda cro (36), and nuclear receptors (17). To our knowledge, Gbp1p is the first example of a protein that qualitatively alters its binding characteristics upon dimerization, changing from a monomeric protein that binds RNA slightly better than ssDNA to a homodimer that has a strong preference for ssDNA and very little affinity for RNA. Although we do not yet know if Gbp1p exists as a monomer, a homodimer, or a mixture of both in vivo, the abundance of Gbp1p (approximately 3.5 × 106 molecules per cell) suggests that the homodimer is likely to exist in vivo.
Is Gbp1p a telomere end-binding protein, and what might it be doing at the telomeres?
Gbp1p has been classified as a putative telomere-binding protein based on its ability to associate specifically with the ssDNA containing the Chlamydomonas telomere sequence (38) and its ability to functionally complement an rlf6 mutation that causes the mislocalization of the telomere-binding protein, Rap1p (24). Additionally, Gbp1p is similar to the mammalian protein hnRNP A1, which has recently been shown to affect telomere length control in cell culture and to bind telomerase in vitro (26). The facts that genes similar to GBP1 from S. cerevisiae and humans have telomeric phenotypes and that the binding specificity of Gbp1p matches the Chlamydomonas ss telomeric sequence are consistent with the idea that Gbp1p is a bona fide telomere-binding protein. However, the ability of monomeric Gbp1p to associate specifically with RNA and the abundance of Gbp1p (both of which are reminiscent of hnRNP A1) suggest that Gbp1p is not exclusively a telomere-binding protein. Regardless of whether it has a role at telomeres, it is likely that Gbp1p has other roles in the cell as well. Genetic studies in S. cerevisiae implicate proteins other than Rlf6p (e.g., Est1p and Cdc13p) as the principle in vivo telomere-regulating proteins (16, 27, 35, 48). Although the precise in vivo role(s) of Gbp1p remains to be determined, the previously unrealized ability of Gbp1p to modulate its nucleic acid discrimination by homodimerization gives this protein the ability to play a unique role in the cell.
Proteins that bind the ss-G-strand telomere DNA overhang have now been described for several organisms, and some patterns are emerging. In S. cerevisiae, several end-binding proteins have been described, including Cdc13p (27, 35), which physically interacts with Stn1p (19). Overexpression of either CDC13 or STN1 leads to shorter telomeres, while mutation of either gene leads to longer telomeres, suggesting that these two proteins work together to block telomerase action (19). In O. nova, telomeric DNA bound by an α monomer can serve as a telomerase substrate but telomeric DNA bound by either a homo-(α2) or heterodimer (αβ) cannot (14). In mammalian cells, hnRNP A1 is a telomere end-binding protein (20) that is needed for telomere length control (26). hnRNP A1 can physically interact with telomerase in vitro, but only when its glycine-rich domain (26), which mediates dimerization (9), is missing. This implies that hnRNP A1 monomers, but not dimers, may be able to interact with telomerase. These examples, from three different species, all point to a speculative model in which monomers of telomere end-binding proteins allow telomerase access to the telomere and dimers of telomere end-binding proteins block such access. Further work will be required to test this model in vitro and in vivo.
ACKNOWLEDGMENTS
We thank Craig Amundsen, Cathy Asleson, David Babcock, and Maryam Gerami-Nejad for excellent technical assistance and Cathy Asleson and Shinichiro Enomoto for helpful discussions and review of the manuscript. We also acknowledge Moffat Kable, Lisa Konkel, and Marie Petrack for preliminary studies of GBP.
S.D.J. was supported by a postdoctoral fellowship from the National Institute of General Medical Sciences (1 F32 GM19065-01). This work was supported by National Institutes of Health (NIH) grant GM38626.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 2.Basu A, Dong B, Krainer A R, Howe C C. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol. 1997;17:677–686. doi: 10.1128/mcb.17.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 5.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 6.Busch S J, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 7.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 8.Cardenas M, Bianchi A, de Lange T. A Xenopus egg factor with DNA-binding properties characteristic of terminus-specific telomeric proteins. Genes Dev. 1993;7:883–894. doi: 10.1101/gad.7.5.883. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 10.Casas-Finet J R, Smith J D, Kumar A, Kim J G, Wilson S H, Karpel R L. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains. J Mol Biol. 1993;229:873–889. doi: 10.1006/jmbi.1993.1093. [DOI] [PubMed] [Google Scholar]
- 10a.Cassidy-Stone, A., and S. Schultz. Personal communication.
- 11.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeAngelo D J, DeFalco J, Rybacki L, Childs G. The embryonic enhancer-binding protein SSAP contains a novel DNA-binding domain which has homology to several RNA-binding proteins. Mol Cell Biol. 1995;15:1254–1264. doi: 10.1128/mcb.15.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 14.Froelich-Ammon S J, Dickinson B A, Bevilacqua J M, Schultz S C, Cech T R. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard C, Cabannes E, Strauss F. Identity of the RNA-binding protein K of hnRNP particles with protein H16, a sequence-specific single strand DNA-binding protein. Nucleic Acids Res. 1994;22:4183–4186. doi: 10.1093/nar/22.20.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass C K. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 18.Gottschling D E, Zakian V A. DNA-protein interactions at telomeres in ciliated protozoans. Adv Cell Biol. 1988;2:291–307. [Google Scholar]
- 19.Grandin N, Reed S I, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Iyadurai, S., S. Enomoto, and J. Berman. Unpublished data.
- 21.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 22.Klobutcher L A, Swanton M T, Donini P, Prescott D M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koerner T J, Hill J E, Myers A M, Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 24.Konkel L M C, Enomoto S, Chamberlai E, McCune-Zierath P, Iyadura S J P, Berman J. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc Natl Acad Sci USA. 1995;92:5558–5562. doi: 10.1073/pnas.92.12.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurenova E V, Mason J M. Telomere functions. A review. Biochemistry. 1997;62:1242–1253. [PubMed] [Google Scholar]
- 26.LaBranche H, Dupuis S, Ben-David Y, Bani M-R, Wellinger R J, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 27.Lin J-J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J J, Zakian V A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. . (Erratum, 22:5516.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine M S, Wilson N M, Petracek M E, Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989;16:225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- 30.Lowell J E, Pillus L. Telomere tales: chromatin, telomerase and telomere function in Saccharomyces cerevisiae. Cell Mol Life Sci. 1998;54:32–49. doi: 10.1007/s000180050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay S J, Cooke H. hnRNP A2/B1 binds specifically to single-stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20:1387–1391. doi: 10.1093/nar/20.24.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrill B M, Stone K L, Cobianchi F, Wilson S H, Williams K R. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J Biol Chem. 1988;263:3307–3313. [PubMed] [Google Scholar]
- 33.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Raliff R L, Wu J-R. A highly conserved repetitive sequence (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadler S G, Merrill B M, Roberts W J, Keating K M, Lisbin M J, Barnett S F, Wilson S H, Williams K R. Interactions of the A1 heterogeneous nuclear ribonucleoprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry. 1991;30:2968–2976. doi: 10.1021/bi00225a034. [DOI] [PubMed] [Google Scholar]
- 35.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 36.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 37.Petracek M E, Berman J. Chlamydomonas reinhardtii telomere repeats form unstable structures involving guanine-guanine base pairs. Nucleic Acids Res. 1992;20:89–95. doi: 10.1093/nar/20.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petracek M E, Konkel L M C, Kable M L, Berman J. A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J. 1994;13:3648–3658. doi: 10.1002/j.1460-2075.1994.tb06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petracek M E, Lefebvre P A, Silflow C D, Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc Natl Acad Sci USA. 1990;87:8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price C M. Telomere structure in Euplotes crassus: characterization of DNA-protein interactions and isolation of a telomere-binding protein. Mol Cell Biol. 1990;10:3421–3431. doi: 10.1128/mcb.10.7.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price C M, Cech T R. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- 43.Price C M, Cech T R. Properties of the telomeric DNA-binding protein from Oxytricha nova. Biochemistry. 1989;28:769–774. doi: 10.1021/bi00428a053. [DOI] [PubMed] [Google Scholar]
- 44.Richards E J, Ausubel F M. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 45.Shampay J, Szostak J W, Blackburn E H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 46.Sheng H, Hou Z, Schierer T, Dobbs D L, Henderson E. Identification and characterization of a putative telomere end-binding protein from Tetrahymena thermophila. Mol Cell Biol. 1995;15:1144–1153. doi: 10.1128/mcb.15.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 48.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 49.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 50.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 51.Wright W E, Tesmer V M, Huffman K E, Levene S D, Shay J W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zalensky A O, Tomilin N V, Zalenskaya I A, Teplitz R L, Bradbury E M. Telomere-telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells. Exp Cell Res. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]
- 53.Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;12:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]