Abstract
The nucleotide excision repair (NER) pathway is able to remove a wide variety of structurally unrelated lesions from DNA. NER operates throughout the genome, but the efficiencies of lesion removal are not the same for different genomic regions. Even within a single gene or DNA strand repair rates vary, and this intragenic heterogeneity is of considerable interest with respect to the mutagenic potential of carcinogens. In this study, we have analyzed the removal of the two major types of genotoxic DNA adducts induced by UV light, i.e., the pyrimidine (6-4)-pyrimidone photoproduct (6-4PP) and the cyclobutane pyrimidine dimer (CPD), from the Saccharomyces cerevisiae URA3 gene at nucleotide resolution. In contrast to the fast and uniform removal of CPDs from the transcribed strand, removal of lesions from the nontranscribed strand is generally less efficient and is modulated by the chromatin environment of the damage. Removal of 6-4PPs from nontranscribed sequences is also profoundly influenced by positioned nucleosomes, but this type of lesion is repaired at a much higher rate. Still, the transcribed strand is repaired preferentially, indicating that, as in the removal of CPDs, transcription-coupled repair predominates in the removal of 6-4PPs from transcribed DNA. The hypothesis that transcription machinery operates as the rate-determining damage recognition entity in transcription-coupled repair is supported by the observation that this pathway removes both types of UV photoproducts at equal rates without being profoundly influenced by the sequence or chromatin context.
UV light induces two major classes of genotoxic lesions in DNA, i.e., cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4)-pyrimidone photoproducts (6-4PPs). Both lesions are repaired by the nucleotide excision repair (NER) pathway, in which incision of the damaged strand on both sides of the lesion is followed by resynthesis of excised DNA with the undamaged strand as a template (reviewed in reference 2). Although the molecular mechanism of the NER reaction has become increasingly clear as it has been reconstituted in vitro by using purified components of either yeast or human origin (1, 4, 15), the mechanism of damage recognition in the nucleus where DNA is folded into chromatin with different levels of complexity is largely unknown. One possible way by which cells sense DNA damage is lesion interference with essential cellular DNA metabolic processes, like transcription, replication, or even recombination. This is exemplified by the intimate link found for the process of NER and mRNA transcription: UV-induced CPDs introduced in sequences transcribed by RNA polymerase or RNA polymerase II, respectively, in pro- and eukaryotes, are repaired preferentially to CPDs induced in nontranscribed DNA (13, 14). The molecular basis for this enhanced strand-specific repair, more commonly termed transcription-coupled repair (TCR) since it is dependent upon ongoing transcription (10, 18), is thought to originate from efficient recruitment of repair proteins towards RNA polymerase stalled at sites of base damage (7, 16). As a result of this coupling, DNA lesions that are located in transcribed DNA and constitute a block to RNA polymerase II transcription are repaired efficiently. Lesions in nontranscribed DNA are obviously not a target for TCR but nevertheless are removed by NER. This mode of repair, called global genome repair, has not been linked to any other DNA metabolic process, and the question of how lesions are located by the NER machinery in genomic DNA is still largely unanswered. For Saccharomyces cerevisiae, genetic and biochemical data suggest that a complex consisting of the RAD7 and RAD16 gene products is involved in damage recognition in global genome repair: a repair deficiency specifically of nontranscribed DNA is observed in rad7 and rad16 knockout mutants (27), while purified Rad7-Rad16 binds preferentially to UV-irradiated DNA (5).
Most of our current understanding of the organization of NER inside living cells has come from repair analysis of UV-induced CPDs. For this type of lesion, variations in repair rates are not confined to different DNA strands, as profound heterogeneity was observed when individual dinucleotide sequences within a single DNA strand were compared (3, 9, 17, 21, 22, 25). The level of repair at a specific sequence might very well constitute an important parameter for the mutation frequency at that position upon exposure to UV light. This also holds true for 6-4PPs. Albeit less frequently induced, this type of lesion contributes significantly to UV-induced mutagenesis in Escherichia coli, yeast, and mammalian cells (reference 2, and references therein). So far, high-resolution repair analysis of UV photoproducts has been confined to CPDs, mainly because technical limitations have hampered measurements of 6-4PP. We recently have succeeded in establishing a method to determine frequencies of 6-4PPs at nucleotide resolution in cells irradiated with a relatively low UV dose (24). Here, we have used this method to monitor the removal of 6-4PPs and CPDs from the S. cerevisiae URA3 gene. We chose this gene as a repair target for three reasons. (i) CPDs are removed from this RNA-polymerase-II-transcribed gene in a strand-specific manner (17, 23). Hence, by comparing NER-proficient cells with rad7 mutants, we can determine the relative contributions of TCR and global genome repair in the overall removal of both UV photoproducts. (ii) The URA3 gene contains several positioned nucleosomes, which have recently been determined at high resolution (19). Therefore, for both types of lesions, the efficiency of NER can be compared to the dipyrimidine’s chromatin environment. (iii) Because of the possibility of positive and negative selection, this gene can be used in a forward mutational assay in order to judge causality in the relation between induction and repair of DNA lesions and the induction of mutations.
In this paper, we show that although 6-4PPs are removed much faster from nontranscribed DNA than CPDs, NER of both types of UV-induced lesions is affected by chromatin. In contrast, the removal from transcribed DNA is predominated by TCR, which overrides chromatin-mediated repair modulation. Furthermore, we postulate that the similar rates with which structurally different lesions are removed from transcribed DNA result from processive RNA polymerase II serving as a DNA damage sensor.
MATERIALS AND METHODS
Strains and UV irradiation.
The S. cerevisiae NER-proficient (RAD+) strain used for this study is W303-1B (genotype: MATα ho can1-100 ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1) that was rendered URA3 by transformation of a linear PCR fragment and checked by Sanger sequencing for proper recombination at its chromosomal position. Subsequently, rad7, rad16, and rad14 disruptions were introduced into this background by one-step gene replacement. Strains were maintained on selective YNB (0.67% yeast nitrogen base, 2% glucose, 2% Bacto agar) supplemented with the appropriate markers. Cells were grown in complete medium (YEPD: 1% yeast extract, 2% Bacto peptone, 2% glucose) at 28°C under vigorous shaking. Cells diluted in chilled phosphate-buffered saline were irradiated with 254-nm UV light (Philips TUV; 30 W) at a rate of 3.5 J/m2 per s, collected by centrifugation, resuspended in complete medium, and incubated for various times in the dark at 28°C before DNA isolation. DNA samples were purified on CsCl gradients.
Detection of UV-induced CPDs and 6-4PPs at nucleotide resolution.
For a detailed protocol the reader is directed to references 22 and 24. For CPD analysis, DNA samples (25 μg) were digested with appropriate endonucleases and precipitated, and URA3 fragments were isolated and end-labelled by using fragment-specific oligonucleotides as described previously. After inactivation of SUPER Taq polymerase with 4 μl of 0.5 M EDTA, the labelled single-stranded DNA molecules were rehybridized by addition of a 50-fold molar excess of complementary strand (synthesized by linear amplification) followed by 3-min incubation at 93°C and gradual cooling to room temperature. DNA samples were treated or mock treated with T4endoV, subjected to spin column chromatography (Sephadex G-50), and lyophilized to small volumes. Portions with approximately equal counts per minute were loaded on denaturing 6% acrylamide gels.
For 6-4PP analysis, DNA samples (50 μg) were digested with appropriate endonucleases and precipitated. Anacystis nidulans photoreactivating enzyme (gift of A. Eker) was added to the DNA redissolved in 100 μl of the following reaction buffer: 10 mM KH2PO4-K2HPO4 (pH 7.0), 100 mM NaCl, 5 mM β-mercaptoethanol, 0.1-mg/ml bovine serum albumin. Then, samples were exposed at room temperature for 30 min to 425-nm light (Philips TLDK; 30 W) to completely convert CPDs to their native dipyrimidine sequences. The URA3 fragments were isolated from bulk DNA, end-labelled, and rehybridized as described above. Subsequently, these samples were subjected to a phenol-chloroform extraction, spin column chromatography, and lyophilization. Pellets were dissolved in 100 μl of UV dimer endonuclease (UVDE) reaction buffer (50 mM Tris-Cl [pH 8.0], 100 mM NaCl, 20 mM MgCl2, 1 mM dithiothreitol, and 1-μg/ml bovine serum albumin) followed by the addition of 1 μl (at 0.1 U/μl) of UVDE (reference 31; gift of A. Yasui) and incubation for 2 h at 37°C. Finally, the samples were again subjected to spin column chromatography and lyophilized to small volumes. Portions with approximately equal counts per minute were loaded on denaturing 6% acrylamide gels. After drying, autoradiograms were prepared from the gels.
Quantification of repair rates.
Multiple autoradiograms were obtained with different exposure times to allow signal determination within the linear range of Kodak X-OMAT-AR scientific imaging films for each individual photoproduct. Autoradiograms were scanned (UMAX; Astra 1200S) at 600 dots per in. and analyzed using ImageMaster software (Pharmacia). Background levels were subtracted, and gel band intensities were corrected for loading variations. Optical density values were plotted against repair time for lesions that gave sufficiently high signal-to-background ratios. Values for repair half-times (t1/2), defined as the time at which 50% of the initial damage (signal at t of 0) was removed, were derived from these plots. Quantification data were obtained from at least three independent experiments.
RESULTS
Detection of UV-induced photoproducts.
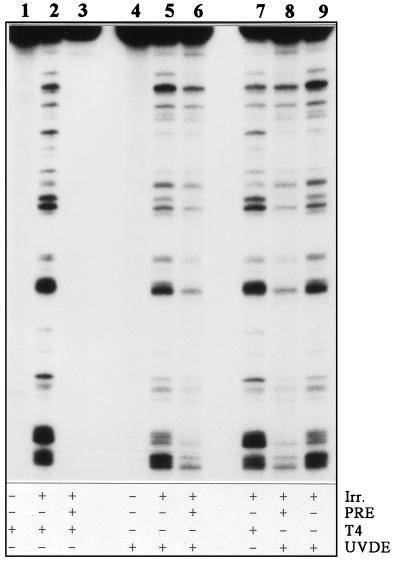
To determine the frequencies of UV-induced CPDs and 6-4PPs separately at any point after irradiation, we used an enzymatic approach (24). Figure 1 illustrates that both photoproducts can be detected separately and at nucleotide resolution with this procedure. CPDs were detected by using the phage enzyme T4 endonuclease V, which recognizes this damage specifically and incises the phosphodiester bond between the dimerized pyrimidines (lane 2). For detection of 6-4PPs no specific enzyme is available; therefore, samples were first subjected to photoreactivation to remove all CPDs from the DNA (lane 3), and subsequently the Neurospora crassa enzyme UVDE was used to incise DNA strands at sites of 6-4PPs (lane 6). The latter enzyme recognizes both CPDs and 6-4PPs and cuts the DNA strand 5′ of the damage (31). The obtained sensitivity allows us to analyze repair of both types of UV photoproducts induced at an identical dose in any S. cerevisiae target sequence.
FIG. 1.
Enzymatic detection of UV-induced photoproducts at nucleotide resolution. DNA (25 μg) isolated from cells exposed to either 0 (lanes 1 and 4) or 140 J/m2 (lanes 2, 3, 5, and 6–9) was mock treated (lanes 2, 5, 7, and 9) or treated with photoreactivating enzyme (PRE) (lanes 3, 6, and 8) and subsequently treated with either T4 endonuclease V (lanes 1–3 and 7) or UVDE (lanes 4–6, 8, and 9). Lanes 2 and 7 show CPD-specific incision, and lanes 6 and 8 show 6-4PP-specific incision, while in lanes 5 and 9 the combined distribution pattern is observed. Irr., irradiation; T4, phage enzyme T4 endonuclease V.
Repair of CPDs from the S. cerevisiae URA3 locus at nucleotide resolution.
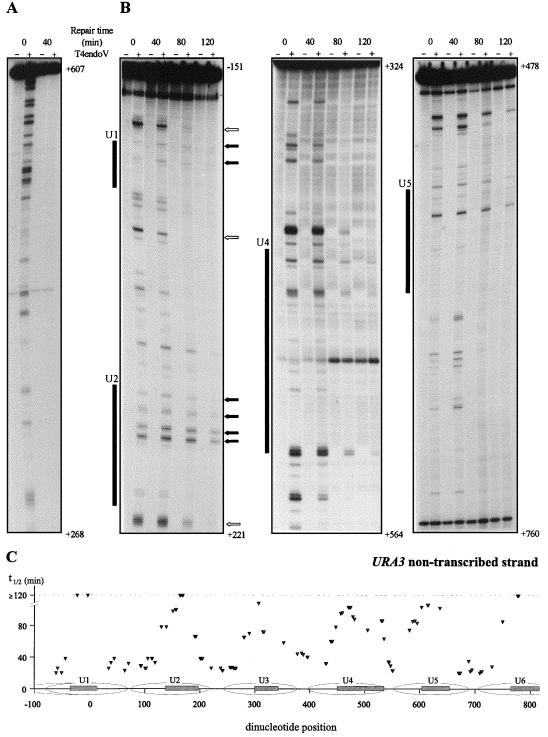
First, we analyzed repair of UV-induced CPDs along the transcribed strand and nontranscribed strand of the URA3 gene. S. cerevisiae cells were exposed to 70 J/m2 and incubated in growth medium to allow repair. At various time points samples were taken, DNA was isolated, and the damage distribution pattern was determined as described above. As shown in Fig. 2A, CPD removal from the transcribed strand was fast without apparent rate heterogeneity even when shorter intervals were chosen for analysis (data not shown; see also reference 23). In contrast, a profound degree of heterogeneity is observed when different dinucleotide positions in the URA3 nontranscribed strand are compared (Fig. 2B). As an example, some CPDs persisted even after 2 h of repair whereas some were repaired very fast, as hardly any signal could be detected after 80 min of repair despite the relatively high CPD induction frequency. Recently, the chromatin structure of the chromosomal URA3 locus was resolved at high resolution and six positioned nucleosomes flanked by nuclease-sensitive regions were identified (19). To allow a visual inspection the protected regions of nucleosomes U1, U2, U4, and U5 are schematically indicated along the repair plots shown in Fig. 2. By comparing the heterogeneity in CPD repair with the chromatin architecture of this locus, we find that regions where CPDs are slowly removed match with the internal protected regions of individual nucleosomes. This correlation between the chromatin environment of the damage and its rate of removal is evident throughout the nontranscribed strand (Fig. 2C), although a detailed analysis of the repair at certain nucleosomal regions is hampered by a relatively low amount of putative dimer sites in their protected DNA sequence.
FIG. 2.
Repair of UV-induced CPDs at single nucleotide resolution along (A) the transcribed strand, nt 268 to 607 (all positions are relative to the start codon, ATG, designated +1), and (B) the nontranscribed strand, nt −151 to 221, nt 324 to 518, and nt 478 to 760. Cells were irradiated with 70 J/m2, and repair was monitored at 0, 40, 80, and 120 min after irradiation. Samples that were mock treated or treated with the CPD-specific enzyme T4endoV are denoted by − and +, respectively. Shaded boxes indicate the internal protected regions of nucleosomes U1, U2, U4, and U5 positioned along the URA3 locus (19). Dark arrows mark CPDs that persisted after 2 h of repair, and open arrows mark some positions that were repaired very fast. (C) Graphic representation of quantified CPD repair rates along the nontranscribed strand of the URA3 locus. Repair t1/2 values, determined as the time at which 50% of the initial CPD signal was removed, were calculated for each individual CPD position with a sufficient signal-to-noise ratio and are plotted above their corresponding dipyrimidine positions. The internal protected regions are represented by the shaded boxes inside nucleosomes U1 through U6 (19).
Repair of (6-4)PPs from the S. cerevisiae URA3 locus at nucleotide resolution.
Because 6-4 photoproducts are less frequently induced by UV than CPDs we have analyzed repair of this type of lesion primarily at 140 J/m2. Although repair analysis could be performed at 70 J/m2, the signal-to-noise ratio of individual dimer sites was low and would therefore restrict the analysis to a smaller subset of highly induced dinucleotide positions.
As a control, we first repeated some of the CPD repair analysis at 140 J/m2 (data not shown), which revealed that although both strands are repaired less efficiently at this dose, the previously observed repair characteristics have not changed: (i) the transcribed strand is repaired faster than the nontranscribed strand; (ii) CPDs are removed from the transcribed strand with uniform kinetics, irrespective of the position of the damage; and (iii) repair of the nontranscribed strand displays a high degree of heterogeneity, with slow repair at the core of the nucleosomes and more efficient repair in between these regions.
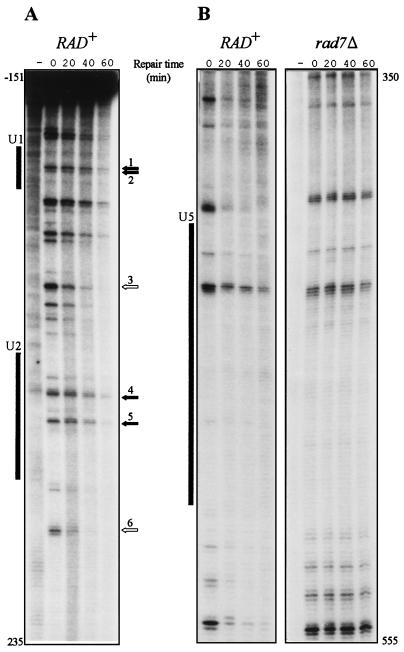
We first monitored repair of the URA3 nontranscribed strand to investigate whether repair of 6-4PPs, like that of CPDs, is influenced by the chromatin organization of the DNA around the lesion. Analysis at identical intervals revealed that 6-4PPs are removed from the URA3 nontranscribed strand much faster than CPDs, at both 140 and 70 J/m2. We therefore shifted to shorter intervals. Figure 3 shows the level of 6-4PPs in the nontranscribed strand (nucleotide [nt] −151 to 235 and nt 350 to 555) at 0, 20, 40, and 60 min after irradiation. Clearly, repair heterogeneity is observed and, as for CPD repair, dinucleotide sequences within the core of the nucleosome are repaired less efficiently compared to neighboring sequences. For instance, lesions at 5′-TC-3′ (nt −2 and −1), 5′-TC-3′ (nt 4 and 5), 5′-TCCC-3′ (nt 156 through 159), and 5′-CT-3′ (nt 172 and 173) are removed with t1/2 of 59, 59, 68, and 53 min, respectively, whereas sequences at 5′-CTTC-3′ (nt 101 through 104) and 5′-TTCC (nt 204 through 207) are repaired with t1/2 of 21 and 26 min, calculated from three independent experiments. As evident from Fig. 3, repair analysis is limited to a small number of sufficiently strong 6-4PP bands. For instance, the region occupied by nucleosome U5 harbors only two 6-4PP adducts with a yield sufficient to allow repair calculations. However, repair analysis throughout the URA3 gene (nucleosomes U1 through U6) provided an alternating pattern of slow repair at nucleosomal core sequences and relatively fast repair in between.
FIG. 3.
(A) Repair of UV-induced 6-4PPs along the nontranscribed strand of the URA3 gene. Numbering of arrows is as follows: 1, 5′-TC-3′ (nt −2 and −1; 2, 5′-TC-3′ (nt 4 and 5); 3, 5′-CTTC-3′ (nt 101 to 104); 4, 5′-TCCC-3′ (nt 156 to 159); 5, 5′-CT-3′ (nt 172 and 173); and 6, 5′-TTCC (nt 204 to 207). (B) Repair-proficient (RAD+) cells are compared with isogenic rad7 mutant cells. Cells were irradiated with 140 J/m2, and repair was monitored at 0, 20, 40, and 60 min after irradiation. To account for non-dimer-specific incision nonirradiated DNA was also assayed (indicated by a minus sign). Shaded boxes indicate the internal protected regions of nucleosomes U1, U2, and U5 positioned along the URA3 locus (19).
We have previously shown that removal of CPDs from the nontranscribed strand completely depends on the Rad7 and Rad16 gene products (22, 27), leading to the hypothesis that these gene products are required for global genome repair, defined as repair of lesions that is not mediated by the TCR pathway. Here, we demonstrate that this requirement also pertains to 6-4PPs as no repair of nontranscribed DNA was observed at all for this type of lesion in rad7 and rad16 mutants, either in the URA3 nontranscribed strand (Fig. 3B), in the upstream promoter region of this locus (see also below), or in the nontranscribed strand of the RPB2 locus (data not shown). Repair of all 6-4PPs was absent in an isogenic rad14Δ strain, confirming that removal of this type of lesion is accomplished by NER (as is the case for CPDs).
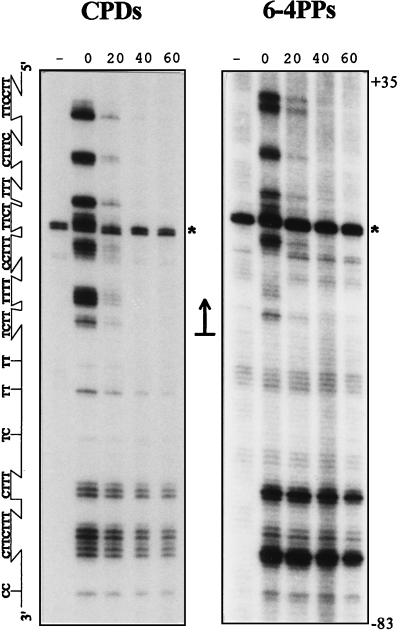
TCR removes 6-4PPs equally efficiently as CPDs.
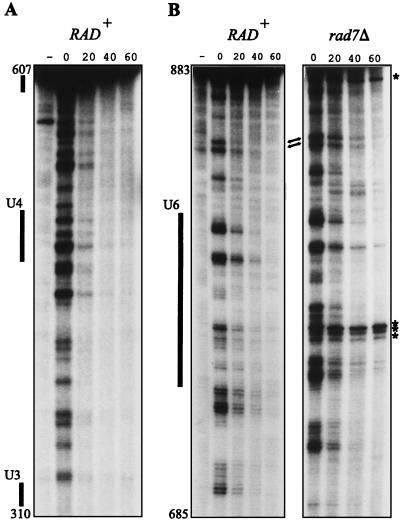
For CPDs, the rate of TCR greatly exceeds the rate of global genome repair, and as a result enhanced repair of the transcribed strand is observed. In fact, no contribution of global genome repair was observed at all, as repair kinetics of the transcribed strand in repair proficient cells were indistinguishable from those in rad7 or rad16 cells (22, 27). For 6-4PPs, on the other hand, global genome repair is more efficient, and at some sequences the t1/2, 20 min, even approximates the rate of TCR of CPDs (which is 17 ± 2 min at a UV dose of 140 J/m2). This suggests that (unlike for removal of CPDs) global genome repair can contribute to removal of 6-4PPs from transcribed DNA. To investigate this, we studied repair of 6-4PPs in the URA3 transcribed strand first in repair-proficient (RAD+) cells. Figure 4A shows that repair of transcribed DNA does not display the degree of heterogeneity characteristic of that of nontranscribed DNA, and in addition, removal from the transcribed strand exceeds that from the nontranscribed strand when dinucleotide repair rates are averaged. These data suggest that 6-4PP repair in the transcribed strand is dominated by TCR, as was observed for CPDs. However, at a few positions, exemplified in Fig. 4B, the repair rate is elevated with respect to the general repair level. Because these sequences are located outside core nucleosome regions—the removal of 6-4PPs from the nontranscribed strand was most efficient at such positions—their elevated repair level could result from a contribution from global genome repair. To test this hypothesis, we repeated the transcribed strand analysis in rad7 cells. Being defective in global genome repair, these cells allow the study of TCR exclusively. Indeed, as Fig. 4B illustrates, a deficiency in rad7 does not influence the repair rate of most 6-4PPs along transcribed DNA (supporting the notion that TCR is predominant), while the more efficiently repaired lesions in a RAD+ genetic background are repaired with kinetics similar to those for their neighboring sequences.
FIG. 4.
Repair of UV-induced 6-4PPs along the URA3 transcribed strand in repair-proficient (RAD+) cells (A) and in isogenic rad7 mutant cells (B). Cells were irradiated with 140 J/m2, and repair was monitored at 0, 20, 40, and 60 min. The minus signs indicate nonirradiated DNA assayed with UVDE. Several strong UV-independent incision products appearing at nondinucleotide sequences are indicated (asterisks); these were left out of the analysis. Arrows point to positions where the repair rate is elevated with respect to the general repair rate.
We previously hypothesized that the phenomenon of uniform rates of CPD removal from transcribed DNA results from an identical rate-determining recognition step for each individual dimer (22), namely, stalled RNA polymerase II elongation complexes at the site of base damage that might serve as a signal to initiate repair, as suggested by others (e.g., reference 7). Since transcription operates in a processive way, a 6-4PP will be encountered with equal probability to a CPD at any given dinucleotide sequence in the transcribed strand, which therefore predicts the operation of an identical rate-determining recognition process in the removal of both lesions. To test whether both photoproducts are indeed removed with equal kinetics by the TCR pathway, CPD removal and 6-4PP removal from the transcribed strand were monitored in rad7Δ cells, which are proficient only for TCR. Figure 5 shows the results for removal of both types of lesions around the transcription-initiation site on the template strand. Fast repair was observed for both types of lesions at sequences starting 2 nt downstream of transcription initiation (which corresponds to nt −33 with respect to the ATG at +1 [11]) and onwards into the transcription unit. Furthermore, the rates with which TCR operates are the same at individual dipyrimidine sequences for both types of lesions (t1/2 was 8 ± 1 min for UV dose of 70 J/m2 and 17 ± 2 min for dose of 140 J/m2) irrespective of the lesion’s sequence or chromatin context. Importantly, CPDs and 6-4PPs are removed from identical sequences in the transcribed strand with equal rates, suggesting that the rate of NER is determined by the rate of damage recognition. This mode of repair displays homogeneous repair, unlike repair in nontranscribed DNA, demonstrating that TCR is insensitive to chromatin-structure repair modulation in RNA-polymerase-II-transcribed genes.
FIG. 5.
Repair of UV-induced CPDs (A) and 6-4PPs (B) along the URA3 template strand in a rad7 strain. Repair was monitored at 0, 20, 40, and 60 min after irradiation (70 J/m2). The large arrow indicates the major transcription-initiation site (+1) and the direction of transcription. To account for background levels, nonirradiated DNA was also assayed (indicated by a minus sign). Due to the lower induction frequency of 6-4PPs than of CPDs, twice the amount of DNA was assayed in the 6-4PP analysis and different exposure times were used to allow visual inspection. As calculated from short exposures of the undamaged full-length fragment (not indicated), the autoradiograms display a 3- to 3.5-fold amplification of the actual 6-4PP signal relative to that of the CPDs. The asterisk indicates a UV-independent background signal.
DISCUSSION
Repair rates of UV-induced photoproducts were studied at nucleotide resolution by treating isolated and end-labelled genomic DNA fragments with damage-specific incision enzymes. This methodology provides a level of sensitivity sufficient to monitor the removal of both major classes of UV-induced DNA damage, i.e., CPDs and 6-4PPs, separately and individually from any target sequence in the yeast genome. Here, we have analyzed removal of these structurally different UV lesions from the entire chromosomal URA3 gene, which is well characterized with respect to its transcriptional and chromatin organization.
For CPDs, the rate of TCR generally exceeds the rate of global genome repair, and as a consequence enhanced repair of the transcribed strand is observed. Aside from this strand bias in NER, another level of intragenic variation is observed in the URA3 nontranscribed strand: “slowspots” of CPD repair coincide with the cores of positioned nucleosomes and are interspersed with regions that are quite efficiently repaired. In agreement with our data, a recent report has shown modulation of NER in the URA3 gene on a plasmid minichromosome (30). The method applied in that study does not discriminate between the two primary UV photoproducts, but it is likely that foremost CPD removal was determined because upon irradiation with 254-nm UV light the 6-4PP incidence level is on average about four- to fivefold lower than that of CPDs, which renders the former lesions difficult to detect in a distribution pattern which combines both photoproducts. Here we show that also for 6-4PPs NER is profoundly influenced by the chromatin environment of the damage when individual dimer sites are compared. Albeit more efficiently repaired than CPDs, the 6-4PPs also show repair patterns that include alternating slowly and quickly repaired regions along the URA3 nontranscribed strand.
Little is known about the mechanism by which NER locates DNA damage in chromatin and thus about the molecular basis for the differences in repair characteristics for the two UV photoproducts. The observation that the rate of 6-4PP removal from nontranscribed DNA exceeds the rate of CPD removal suggests a higher affinity of DNA-recognizing proteins towards this type of lesion that distorts the helical structure of the DNA duplex more profoundly: adjacent pyrimidine rings in cis-syn CPDs are believed to be nearly coplanar (2), whereas the pyrimidine planes within the 6-4PP are almost perpendicular, and for them a more-pronounced bending angle (44°) of the DNA has been observed (8, 20). Alternatively, induction of 6-4PPs might lead to an enhanced destabilization of nucleosomes (12), thereby rendering lesions more accessible to repair proteins. One can envisage that translocation of a damage-recognition component of NER along the DNA (in search of DNA damage) is hindered by chromatin components, and consequently sequences that are wrapped around histone octamers are less efficiently located and thus less efficiently repaired.
Recently, it was suggested that in S. cerevisiae the Rad7-Rad16 protein complex functions as the damage-recognition entity in global genome repair. This hypothesis is based on the finding that the encoded proteins, as a complex, bind UV-damaged DNA preferentially (5) and on the observation that repair of CPDs from nontranscribed DNA depends completely on RAD7 and RAD16 (22, 27), as is the case for the structurally different 6-4PP adduct (this study). In a reconstituted system, however, the Rad7-Rad16 complex only stimulates NER without being essential for damage-dependent incision (5), which indicates that in vitro other NER components are able to locate damage on naked DNA independent of Rad7 or Rad16. Interestingly, the Rad7-Rad16 protein complex displays double-stranded-DNA-stimulated ATPase activity that is inhibited when irradiated DNA is used (6). The latter feature could suggest that ATP hydrolysis is utilized to track along DNA and is subsequently blocked upon encountering a DNA lesion. Although not conflicting with a tracking mechanism, certainly the present in vivo data limit the possibilities for how such a mechanism might operate in the genomic DNA (see also reference 30). Since linker DNA is repaired efficiently, damage recognition in these sequences cannot depend on translocation of Rad7-Rad16 through nearby nucleosomal regions that are repaired less efficiently. This is in contrast to transcription elongation—a processive unidirectional “tracking” process starting at a defined position—which leads to identical repair rates in TCR for both types of lesions throughout the transcribed strand irrespective of the chromatin context. Clearly, the mechanism by which NER locates DNA damage in chromatin is unresolved at the present time and awaits additional biochemical data acquired with purified NER proteins operating on reconstituted chromatin. Our in vivo repair analysis of both major types of UV damage can provide a framework in which such data should be interpreted.
Although global genome repair can operate on UV-induced lesions in transcribed DNA (28), this pathway does not seem to contribute importantly to the removal of either CPDs (23) or 6-4PPs (this study) from the URA3 transcribed strand in repair-proficient S. cerevisiae cells. TCR of CPDs has been described extensively, but this study provides the first example of strand-specific repair of 6-4PPs. Although this type of lesion is more efficiently removed from nontranscribed DNA than CPDs, resulting in specific nontranscribed dipyrimidine sequences at repair rates comparable to those with TCR, the latter pathway still predominates in the removal of 6-4PPs from the URA3 transcribed strand. This observation contrasts with earlier studies on NER-proficient human and hamster cells (26, 29) which, possibly because of technical limitations inherent within the gene-specific repair assay, did not reveal any strand preference for 6-4PP repair. As the authors of these studies have indicated, the high UV dose needed to measure 6-4PPs in human genes, resulting in a total of about eight sites of damage per transcription unit, complicates the issue because CPDs are more frequently induced than 6-4PPs. Under the assumption that TCR operates in a sequential way, this process must repair at least one CPD before the elongating RNA polymerase complex (if not released from the template during NER) encounters a 6-4PP. On the other hand, global genome repair recognizes 6-4PP preferentially to CPDs and thus will be less affected by an increased CPD load.
Apart from being more efficient, removal of UV photolesions from the transcribed strand does not display a profound variation in repair rate when individual dimer sites are compared. This observation suggests that the mechanisms by which TCR and global genome repair detect DNA damage are fundamentally different. Three conclusions drawn from repair analysis in rad7 mutant cells, which are proficient only in TCR, support the idea that uniform repair kinetics result from RNA polymerase II acting as the DNA damage sensor in TCR. Firstly, both types of UV-induced lesions are repaired by TCR immediately downstream of transcription initiation and along the complete transcribed strand. Secondly, differently positioned DNA damage sites in the transcribed strand are repaired with similar kinetics, suggesting that all lesions are recognized with equal probability once RNA polymerase II transcription has been initiated and that subsequent steps of NER are not profoundly influenced by the sequence or chromatin context of the lesion. Thirdly, the kinetics of CPD removal at each position in the transcribed strand is similar to that of 6-4PP removal at that position. The latter observation agrees well with observations in human cells lacking functional global genome repair, which showed that the average rate of 6-4PP removal from the transcribed strand of the ADA gene was similar to that of CPDs, suggesting that TCR operates on both lesions to the same extent (26).
Finally, we believe that repair analyses at nucleotide resolution, such as those described in this report, are fundamental in the interpretation of UV-induced mutation spectra. The nonsymmetrical removal of the primary UV-induced lesions from the individual strands of active genes as well as the rate heterogeneity observed along the nontranscribed strand might constitute an important parameter that affects the probability that a mutation is induced at a particular position upon exposure to UV. We are currently analyzing this relation by comparing the incidence levels and the repair rates of CPDs and 6-4PPs quantitatively, with mutation spectra in repair-proficient and repair-deficient cellular backgrounds, using the URA3 gene as a mutational target.
ACKNOWLEDGEMENTS
We thank Richard Verhage for valuable discussion and critical reading of the manuscript, Esther Verhoeven for experimental assistance, and Akira Yasui and Andre Eker for generously providing N. crassa UV-dimer endonuclease and A. nidulans photoreactivating enzyme, respectively.
This work was supported by grants from the J. A. Cohen Institute for Radiopathology and Radiation Protection.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Produst V N, Protić M, Hübscher U, Egly J-M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 3.Gao S, Drouin R, Holmquist G P. DNA repair rates mapped along the human PGK1 gene at nucleotide resolution. Science. 1994;263:1438–1440. doi: 10.1126/science.8128226. [DOI] [PubMed] [Google Scholar]
- 4.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 5.Guzder S N, Sung P, Prakash L, Prakash S. Yeast Rad7-Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J Biol Chem. 1997;272:21665–21668. doi: 10.1074/jbc.272.35.21665. [DOI] [PubMed] [Google Scholar]
- 6.Guzder S N, Sung P, Prakash L, Prakash S. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J Biol Chem. 1998;273:6292–6296. doi: 10.1074/jbc.273.11.6292. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt P C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 8.Kim J-K, Choi B. The solution structure of DNA duplex-decamer containing the (6-4) photoproduct of thymidylyl(3′→5′)thymidine by NMR and relaxation matrix refinement. Eur J Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 9.Kunala S, Brash D E. Excision repair at individual bases of the Escherichia coli lacI gene: relation to mutation hot spots and transcription coupling activity. Proc Natl Acad Sci USA. 1992;89:11031–11035. doi: 10.1073/pnas.89.22.11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leadon S A, Lawrence D A. Strand-selective repair of DNA damage in the yeast GAL7 gene requires RNA polymerase II. J Biol Chem. 1992;267:23175–23182. [PubMed] [Google Scholar]
- 11.Losson R, Fuchs R P P, Lacroute F. Yeast promoters URA1 and URA3. Examples of positive control. J Mol Biol. 1985;185:65–81. doi: 10.1016/0022-2836(85)90183-4. [DOI] [PubMed] [Google Scholar]
- 12.Mann D B, Springer D L, Smerdon M J. DNA damage can alter the stability of nucleosomes: effects are dependent on damage type. Proc Natl Acad Sci USA. 1997;94:2215–2220. doi: 10.1073/pnas.94.6.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellon I, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Nature. 1989;342:95–98. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 14.Mellon I M, Spivak G S, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 15.Mu D, Park C-H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 16.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 17.Smerdon M J, Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 18.Sweder K S, Hanawalt P C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka S, Livingstone-Zatchej M, Thoma F. Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J Mol Biol. 1996;257:919–934. doi: 10.1006/jmbi.1996.0212. [DOI] [PubMed] [Google Scholar]
- 20.Taylor J-S, Garrett D S, Cohrs M P. Solution-state structure of the Dewar pyrimidinone photoproduct of thymidylyl-(3′-5′)-thymidine. Biochemistry. 1988;27:7206–7215. doi: 10.1021/bi00419a007. [DOI] [PubMed] [Google Scholar]
- 21.Teng Y, Li S, Waters R, Reed S H. Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J Mol Biol. 1997;267:324–337. doi: 10.1006/jmbi.1996.0908. [DOI] [PubMed] [Google Scholar]
- 22.Tijsterman M, Tasseron-de Jong J, van de Putte P, Brouwer J. Transcription-coupled and global genome repair in the Saccharomyces cerevisiae RPB2 gene at nucleotide resolution. Nucleic Acids Res. 1996;24:3499–3506. doi: 10.1093/nar/24.18.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tijsterman M, Verhage R A, van de Putte P, Tasseron-de Jong J G, Brouwer J. Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:8027–8032. doi: 10.1073/pnas.94.15.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tijsterman M, Verhoeven E E A, Tasseron-de Jong J G, Brouwer J. Enzymatic detection of ultraviolet-induced pyrimidine (6-4) pyrimidone photoproducts at nucleotide resolution in Saccharomyces cerevisiae. Anal Biochem. 1998;260:110–113. doi: 10.1006/abio.1998.2662. [DOI] [PubMed] [Google Scholar]
- 25.Tornaletti S, Pfeifer G P. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin cancer. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- 26.van Hoffen A, Venema J, Meschini R, van Zeeland A A, Mullenders L H F. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhage R, Zeeman A-M, de Groot N, Gleig F, Bang D D, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhage R A, van Gool A J, de Groot N, Hoeijmakers J H J, van de Putte P, Brouwer J. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol Cell Biol. 1996;16:496–502. doi: 10.1128/mcb.16.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vreeswijk M P, van Hoffen A, Westland B E, Vrieling H, van Zeeland A A, Mullenders L H F. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J Biol Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 30.Wellinger R E, Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;15:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yajima H, Takao M, Yasuhira S, Zhao J H, Ishii C, Inoue H, Yasui A. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damage by ultraviolet light. EMBO J. 1995;14:2393–2399. doi: 10.1002/j.1460-2075.1995.tb07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]