Abstract
Transcription factors of the interferon regulatory factor (IRF) family bind to the type I interferon (IFN)-responsive element (ISRE) and activate transcription from IFN-inducible genes. To identify cofactors that associate with IRF proteins, DNA affinity binding assays were performed with nuclear extracts prepared from tissue culture cells. The results demonstrated that the endogenous IRFs bound to the ISRE are complexed with the histone acetylases, PCAF, GCN5, and p300/CREB binding protein and that histone acetylase activities are accumulated on the IRF-ISRE complexes. By testing recombinant proteins, we show that PCAF directly binds to some but not all members of the IRF family through distinct domains of the two proteins. This interaction was functionally significant, since transfection of PCAF strongly enhanced IRF-1- and IRF-2-dependent promoter activities. Further studies showed that expression of PCAF and other histone acetylases was markedly induced in U937 cells upon phorbol ester treatment, which led to increased recruitment of PCAF to the IRF-ISRE complexes. Coinciding with the induction of histone acetylases, phorbol ester markedly enhanced IFN-α-stimulated gene expression in U937 cells. Supporting the role for PCAF in conferring IFN responsiveness, transfection of PCAF into U937 cells led to a large increase in IFN-α-inducible promoter activity. These results demonstrate that PCAF is a phorbol ester-inducible coactivator of the IRF proteins which contributes to the establishment of type I IFN responsiveness.
Transcription factors belonging to the interferon (IFN) regulatory factor (IRF) family bind to the IFN-stimulated response element (ISRE) present in many type I alpha/beta IFN (IFN-α/β)-inducible promoters (37, 43). The N-terminal DNA binding domain of ∼110 amino acids conserved throughout the IRF family is responsible for binding to the ISRE. While some IRF members are activators of IFN-responsive genes, others act as repressors. For example, IRF-1 and ISGF3 activate transcription from promoters carrying the ISRE (24, 40, 51). The latter is a complex composed of the ISGF3γ and Stat proteins. IRF-3 (2), recently shown to complex with p300/CREB binding protein (CBP), is also an activator (53, 59). On the other hand, IRF-2, ICSBP, IRF-4/Pip/ICSAT, and IRF-7 repress transcription from a number of ISRE promoters (24, 35, 55, 60). However, these repressors function as an activator in other promoters (20, 50). There are additional members, such as IRF-5 and IRF-6, that belong to this IRF family whose functions are not well understood. In addition to playing a major role in eliciting IFNs’ broad biological activities, IRF proteins are also involved in controlling cell growth and apoptosis (37, 46).
Some members of the IRF family associate with other members, e.g., ICSBP is shown to interact with IRF-1 and IRF-2, and this interaction results in an increased binding activity for the ISRE (9, 42). Furthermore, IRF-4 and ICSBP both interact with PU.1, a member of the ETS family, and this interaction allows them to bind to the immunoglobulin light-chain enhancer λB (10, 20). Additionally, we have previously reported that some IRF members interact with a basal transcription factor, TFIIB, in a cell-type-specific manner (52).
However, despite extensive studies reported for the IRF’s diverse roles, the regulatory factors that interact with them have not been fully investigated. Such interactions are likely to be an important aspect of their function, since mobilization of the basal transcription machinery, as well as modification of the chromatin structure necessary for transcriptional activation, are dependent on interactions with other factors. Recently cloned enzymes that acetylate or deacetylate nucleosomal histones (11, 22, 46) are plausible candidates for factors that interact with IRF proteins.
There are at least three groups of histone acetylases (45). PCAF and GCN5 are the most-conserved acetylases that occur from yeasts to humans (12, 31, 57). These histone acetylases interact with the general, as well as the specific, transcription factors, the latter including the nuclear receptors, MyoD and NF-Y (8, 17, 21, 29, 39), although they also interact with basal transcription factors (21). In addition, the global coactivators p300 and CBP also have histone acetylase activity, although their substrate specificity in vitro differs from that of PCAF (3, 30). p300 and CBP interact with a wide variety of general and specific transcription factors, cell cycle regulators, and PCAF. They also interact with IRF-1 and IRF-3 (33, 53, 59). Further, specific coactivators, such as ACTR and SRC-1, that interact with nuclear receptors have also been shown to have histone acetylase activities (15, 44). These histone acetylases appear to generally enhance transcription, which is consistent with many previous observations indicating that transcriptionally active chromatin is composed of highly acetylated histones (49). The report that a component of TFIID, a TATA binding protein (TBP)-associated factor, is a histone acetylase (34) also supports their role in active transcription.
On the other hand, histone deacetylases, also highly conserved, are generally associated with transcriptional repression (22, 47, 54, 56). While expression of some of histone deacetylases is regulated during cell growth (5), little is known about the regulation of histone acetylases.
We show here that endogenous IRF proteins recruit multiple histone acetylases, including PCAF, GCN5, and CBP/p300, to bind to the ISRE. The functional importance of the recruitment is verified by enhanced transcription from an ISRE-carrying promoter after PCAF transfection, for which histone acetylase catalytic activity is required. More significantly, we show that phorbol ester treatment strongly induces expression of histone acetylases in human monocytic U937 cells. Our evidence indicates that this induction accounts for the acquisition of IFN responsiveness in U937 cells. Taken together, the expression of histone acetylases is dynamically regulated by external signals, and as such these acetylases play an integral role in IFN-mediated gene regulation.
MATERIALS AND METHODS
Cell cultures and nuclear extract preparations.
Human monocytic U937 cells and mouse “macrophage like” ANA-1 cells were maintained in RPMI 1640 and Dulbecco modified Eagle medium, respectively, each supplemented with 10% fetal bovine serum, 4 mM glutamine, and gentamicin (25 μg/ml). U937 cells were treated with 10 nM of O-tetradecanoylphorbol-13-acetate (TPA; Sigma) for the indicated periods of time. To prepare nuclear extracts, cells were suspended in 10 to 20 volumes of buffer A containing 10 mM KCl, 10 mM HEPES (pH 7.9), 1 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1% Nonidet P-40 (NP-40), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and homogenized and centrifuged at 10,000 rpm at 4°C for 5 min. Nuclear pellets were then suspended in buffer C containing 400 mM NaCl, 20 mM HEPES (pH 7.9), 15 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 25% glycerol, 1 mM PMSF, and 10 μg of leupeptin, 20 μg of pepstatin, and 10 μg of antipain per ml, incubated for 30 min at 4°C, and centrifuged at 14,000 rpm for 20 min. The supernatants were dialyzed against buffer D containing 100 mM NaCl, 20 mM HEPES (pH 7.9), 20% glycerol, 1 mM PMSF, and 1 mM DTT.
Cloning and purification of recombinant IRFs (rIRFs).
Full-length cDNAs for hIRF-1, mIRF-2, and mICSBP (24, 52) and full-length mPU.1 cDNA (20; a gift from H. Singh, University of Chicago) were cloned in the baculovirus vector, pAcSGHisNT (Pharmingen). Sf9 cells were infected with the recombinant viruses at an approximate multiplicity of infection of 1 for 3 days. Cells were disrupted by sonication in buffer containing 100 mM KCl, 40 mM Tris-HCl, 10% glycerol, 0.2 mM EDTA, and 1 mM DTT, and recombinant proteins were purified by affinity chromatography on the Ni-nitriloacetic acid resin (Qiagen) as described previously (52). Yields varied from 10 μg to 1 mg/108 infected cells.
Conjugation of ISRE to beads and binding assays.
Three tandem copies of the ISRE sequence (5′-GATCCTCGGGAAAGGGAAACCGAAACTGAAGCC-3′) from the ISG15 gene (9) and two copies of the λB sequence (5′-GAAAAAGAGAAATAAAAGGAAGTGAAACCAAG-3′) from the immunoglobulin λ chain gene (20) were cloned into the basic vector pGL-2-Ld40 (52). The biotinylated ISRE DNA (157 bp) and biotinylated λB DNA (124 bp) were synthesized by PCR from the above templates by using a biotinylated sense primer 5′-GAGGTACCGAGCTCTTACGCGTGC-3′ and an antisense primer 5′-TAACCAGCCTCCGCAGATCT-3′. Then, 2 to 4 pmol of biotinylated DNA was incubated with 100 to 200 μg of Dynabeads M-280 streptavidin (Dynal) in 200 μl of TE buffer containing 10 mM Tris-HCl (pH 8.0)–1 mM EDTA at room temperature for 30 min. More than 90% of the DNA was conjugated to the beads under these conditions. Unconjugated DNA was removed with a magnetic particle concentrator (Dynal). DNA-conjugated beads were then blocked by 0.5% bovine serum albumin in TGED buffer (20 mM HEPES [pH 7.9], 1 mM EDTA, 10% glycerol, 0.01% Triton X). Then, 2 to 12 pmol of rIRF-2 and rICSBP were incubated in 200 μl of beads conjugated to 2 to 4 pmol of ISRE DNA equilibrated in TGED buffer. Next, 2 to 8 pmol of rICSBP and rPU.1 were incubated with 4 pmol of the λB-conjugated beads. The beads were then washed once with TGED buffer, incubated with 500 μg of nuclear extracts from ANA-1 cells in 400 μl of TGED buffer supplemented with 100 mM NaCl for 2 to 4 h at 4°C, and washed three more times with TGED buffer. Bound materials were eluted in 20 μl of TGED buffer supplemented with 1 M NaCl. Oligomer competition assays were performed with a monomeric wild-type ISRE (9) or mutant ISG15 ISRE (5′-GATCCTCGGGAAAGatAAACatAAACTGAAGCC-3′; mutated sequences are indicated in lowercase letters).
Immunoblot assays.
Proteins eluted from the beads (∼20 μg of proteins) or nuclear extracts from U937 cells (∼30 μg of proteins) were resolved by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE); the gels were then transferred to a Millipore Immobilon P polyvinylidene difluoride membrane and blocked by 1% skim milk in phosphate-buffered saline containing 0.1% Tween 20. The membranes were incubated first with appropriate dilutions of primary antibodies and then with the secondary antibody, horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG (Amersham). The membranes were then developed with an Amersham ECL detection kit according to the instructions provided by the manufacturer. Rabbit antibodies to cyclin C, YY1, TFIIB, PU.1, and CBP (also reacting with p300) were obtained from Santa Cruz Biotechnology. Rabbit antibodies against PCAF, hGCN5, and hAda2 were raised against corresponding recombinant proteins (8, 57). Rabbit polyclonal antibodies to IRF-1, IRF-2, and ICSBP were as described previously (36). Monoclonal anti-flag M2 antibody was purchased from Kodak. Rabbit anti-HDAC1 and -HDAC2 antibodies were generous gifts from E. Sato (University of Florida) and G. Humphrey (Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, National Institutes of Health).
Histone acetylase activity.
Histone acetylase assays were performed as previously described (8, 38). First, 500 μg of nuclear extracts proteins or 100 to 400 ng of recombinant proteins were incubated with ISRE beads or λB beads (2 to 4 pmol each), and bound materials were then incubated with 2 μg of calf thymus type IIA histones (Sigma) for 30 min at 30°C in the presence of 0.1 μCi of 3H-labeled acetyl coenzyme A (Amersham) in 30 μl of reaction buffer. Radioactivity incorporated into acetylated histones was measured as described by Ogryzko et al. (38).
Flag and glutathione S-transferase (GST) pull-down assay.
Portions (1 μg) of flag-tagged rPCAFs produced in a baculovirus vector described by Yang et al. (57) were conjugated to 20 μl of M2 anti-flag antibody agarose beads (8) and were then incubated with in vitro-translated 35S-labeled IRFs in buffer containing 100 mM NaCl, 20 mM HEPES (pH 7.9), 10% glycerol, 0.5 mM EDTA, and 0.1% NP-40 for 30 min at 4°C, washed three times, eluted in SDS sample buffer, resolved by SDS–12% PAGE, and autoradiographed. A 330-bp HindIII and EcoRI fragment of the PCAF cDNA corresponding to the bromodomain (57) was subcloned into pGEX2T (Pharmacia). Then, 50 μl of glutathione-Sepharose beads (Pharmacia) was incubated with 500 μl of bacterial extracts containing approximately 10 μg of GST fusion proteins at 4°C for 30 min. The beads were washed four times in 1 ml of N100 containing 100 mM NaCl, 5 mM MgCl2, 20 mM Tris (pH 8.0), 10% glycerol, 0.05% NP-40, and 0.5 mM PMSF and suspended in 500 μl of N100. Afterward, 400 ng of rIRFs were incubated with the GST beads for 1 h at 4°C. The beads were then washed and incubated in 500 μl of N100 containing 20 mM glutathione (Sigma) for 15 min at room temperature. Eluted proteins were subjected to SDS–10% PAGE and analyzed by immunoblot assay.
Transfection assays.
The ISRE-Ld40 luciferase reporter containing three copies of the ISRE from the ISG15 gene was constructed from PGL2-Ld40 as described earlier (52). The H4 luciferase reporter was constructed by inserting three copies of the HiNF-M oligonucleotides (5′-CGCTTTCGCTTTTCAATCTGGTCGATAC-3′) from the H4 promoter (50) into PGL2-Ld40. The IRF-1 and IRF-2 expression vectors, pAct-1 and pAct-2, were a gift from T. Taniguchi (University of Tokyo, Tokyo, Japan). The indicated amounts of the reporter, pAct-1, or pAct-2 were cotransfected with the PCAF expression vectors pCXN2-PCAF or pCXN2-PCAFΔHAT-1 (8) into NIH 3T3 cells by using Lipofectamine (Life Technologies) or the Superfect reagent (Quiagen). Then, 107 U937 cells were transfected with 50 μg of pCXN2, pCXN2-PCAF, or pCXN2-PCAFΔHAT-1 by electroporation at 1,600 μF at 225 V for 2 s by using the cell porter (Life Technologies). One day after transfection, Geneticin (Life Technologies) was added at 400 μg/ml, and the cells were incubated for 10 to 14 days. Geneticin-resistant cells were pooled and used for reporter assays within a week. Pooled U937 cells (5 × 106 cells) were transfected with 15 μg of reporter by electroporation. After overnight incubation, cells were treated with 1,000 U of recombinant human IFN-α2b (Schering Plough, Tokyo, Japan) per ml for the indicated periods of time. For each pool of transfectants, the expression of transfected PCAF was confirmed by immunoblot analysis with anti-flag-M2 antibody.
RNA blot analysis.
Total RNA (20 μg) was prepared from U937 cells by using RNAzol B (Tel-Test) and was electrophoresed through a 1.2% formaldehyde agarose gel and blotted onto Hybond-N (Amersham) in 20× SSC (3 M NaCl plus 0.3 M sodium citrate, pH 7.0). The filters were hybridized with 32P-labeled probes at 42°C overnight and washed with 2× SSC containing 0.1% SDS at room temperature. The following probes were labeled with the random priming method by using the Prime-It RT kit (Stratagene): a 1.3-kb EcoRI fragment of oligo(A) synthetase (2′5′OAS [6]), a 2.2-kb HindIII fragment of hPKR (58), or a 477-bp NcoI-SalI fragment of ISG15 (18) genes.
Quantitative reverse transcriptase (RT)-PCR.
We used the method of Colle et al. (16) with a small modification. cDNAs were synthesized from total RNA from TPA-treated U937 cells by using Molony murine leukemia virus RT (Life Technologies) in a reaction mixture containing 75 mM KCl, 3 mM MgCl2 50 mM Tris-HCl (pH 8.3), 0.25 mM deoxynucleoside triphosphates, 0.8 U of RNasin, and random hexamer primers (Promega). Each PCR mixture contained 10 μl of cDNA, 50 mM KCl, 3 mM MgCl2 10 mM Tris-HCl (pH 9.0), 250 μM deoxynucleoside triphosphates, 1 U of Taq DNA polymerase (Promega), and 1 μg of sense and antisense primers in a total volume of 50 μl. A total of 25 cycles were completed for all samples. Each cycle consisted of denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min, and primer extension at 72°C for 1.5 min with HPRT (hypoxanthine phosphoribosyltransferase). RNA was used as a reference for equalization of cDNA input. RT-PCR products were separated on 1% agarose gel, and Southern blot hybridization was performed with γ-32P-labeled specific probes.
RESULTS
Recruitment of histone acetylases by the ISRE-IRF complex.
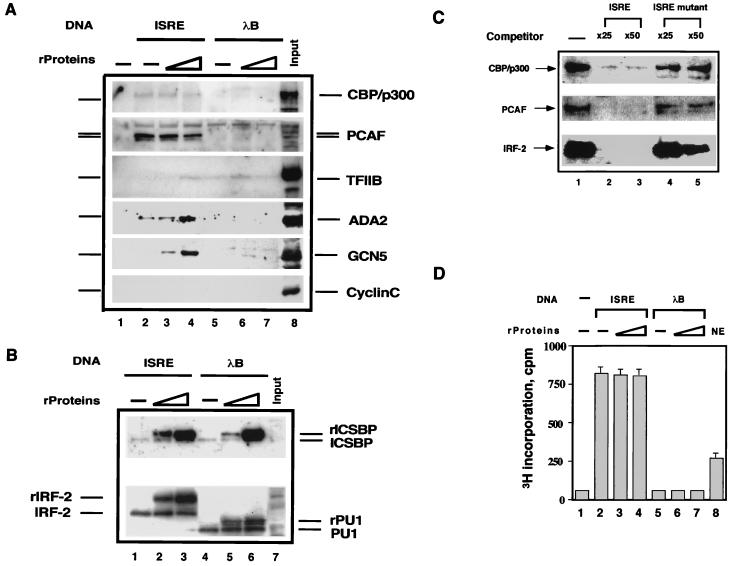
To identify nuclear factors that associate with ICSBP-IRF complexes, DNA affinity binding assays were performed. The biotinylated ISRE DNA (4 pmol) was conjugated to magnetic beads and bound to rICSBP and rIRF-2 (6 to 12 pmol). The beads were incubated with nuclear extracts from macrophage-like ANA-1 cells, and bound materials were then tested by immunoblot analysis. The ANA-1 cell line was chosen because our initial interest was to search for factors that bind to ICSBP, a member of the IRF family active in cells of the monocyte-macrophage lineage (26). rICSBP and rIRF-2 were used because they interact with each other to bind to the ISRE (9, 42). As a control, we tested beads conjugated to λB, an enhancer element of the immunoglobulin light-chain gene known to bind to ICSBP and PU.1 (10, 20). Reporter analysis indicated that the ISRE and λB can function as a positive regulatory element in these cells (data not shown). As presented in Fig. 1A, the histone acetylases, CBP, p300, PCAF, and GCN5 all bound to the ISRE-conjugated beads (ISRE beads), while cyclin C, tested as a control, did not (lanes 2 to 4). Significantly, the ISRE beads that were not bound to rIRFs also recruited PCAF, as well as CBP/p300, to a level similar to (or greater than) that of the beads bound to rIRFs (compare lane 2 to lanes 3 and 4). However, unconjugated beads (lane 1) did not recruit any of the acetylases. As presented below, the recruitment of these acetylases was attributed to the endogenous IRFs. Recruitment of another histone acetylase GCN5 was detected only with ISRE beads complexed with recombinant IRFs. Ada2, which has been shown to complex with GCN5 in yeast cells (13), was also found on the ISRE beads. Although weakly, TFIIB also bound to both ISRE and λB beads. In all cases PCAF was recruited much more efficiently than was CBP/p300 or GCN5: whereas a large fraction of PCAF in the nuclear extracts was recruited to the ISRE beads, only a small fraction of other histone acetylases was found on the beads (compare lanes 2 to 4 to lane 8). In contrast, none of the histone acetylases were recruited to the λB beads, with or without rICSBP and rPU.1 (lanes 5 to 7).
FIG. 1.
Recruitment of histone acetylases to the IRF-ISRE complexes. (A) Magnetic beads conjugated to no DNA (lane 1), ISRE (lanes 2 to 4), or λB (lanes 5 to 7) (4 pmol each) were incubated with 6 and 12 pmol of rIRF-2 plus rICSBP (lanes 3 and 4) or 4 and 8 pmol of rPU.1 plus rICSBP and then mixed with 500 μg of nuclear extracts from ANA-1 cells. Bound materials were analyzed by immunoblot assay. Lane 8 shows the total input of nuclear extracts (25 μg). (B) The membranes tested in panel A were stripped and reblotted with antibodies specific for ICSBP, IRF-2, and PU.1. The positions of the endogenous and recombinant proteins (rProteins) are marked. (C) Oligomer competition analysis. Four picomoles of ISRE conjugated to the beads was incubated with 500 μg of nuclear extracts without rIRFs in the presence of no oligomer (lane 1), excess wild-type ISRE oligomer (lanes 2 and 3), or mutant ISRE oligomer (lanes 4 and 5), and the bound materials were detected as in panel A. (D) Histone acetylase activity accumulated on the IRF-ISRE complexes. Four picomoles of ISRE or λB beads were incubated with 500 μg of nuclear extracts with or without rIRFs as in panel A. Bound materials were tested for histone acetylase activity. Lane 8 shows the histone acetylase activity by input nuclear extracts (25 μg). Values represent the average of triplicate determinations ± the standard deviation (SD).
The experiments shown in Fig. 1B examined the binding of endogenous IRFs to the ISRE and λB beads, in addition to the recombinant counterparts. Endogenous proteins were distinguished from recombinant counterparts by their different sizes, due to the added histidine tag. The endogenous IRF-2 and ICSBP bound to the ISRE beads, regardless of whether the beads had been incubated with the recombinant proteins. Similarly, the endogenous PU.1 and ICSBP bound the λB beads, irrespective of the binding of recombinant proteins (see lanes 4 to 6). These results show that the ISRE beads bind to the endogenous IRF proteins and recruit several histone acetylases.
PCAF and CBP/p300 are recruited to the ISRE beads by the endogenous IRFs: ISRE competition.
Histone acetylases recruited to the ISRE beads without bound recombinant proteins was likely to be attributed to the binding of endogenous IRFs to the ISRE, since these acetylases alone do not bind to the ISRE (Fig. 2A and data not shown). To ascertain whether the endogenous IRFs account for histone acetylase recruitment, oligonucleotide competition assays were performed. The results presented in Fig. 1C show that inclusion of wild-type ISRE oligomers in the binding reaction strongly inhibited the recruitment of PCAF and CBP/p300. However, the mutant ISRE oligomers that fail to bind to IRFs did not. These results show that the endogenous IRFs are complexed with PCAF and/or CBP/p300 to bind to the ISRE.
FIG. 2.
rIRF-1 and rIRF-2, but not rICSBP, directly bind to rPCAF. (A) IRFs (100 to 300 ng; 2 to 6 pmol) of rIRFs were incubated with 2 pmol of ISRE conjugated to beads and 400 ng of rPCAF or rhGCN5 in 400 μl of TGED buffer. Bound materials were detected by immunoblot assay with antibodies specific for PCAF, GCN5, and IRF. (B) rIRF-2 (6 pmol) was incubated with 2 pmol of ISRE immobilized to beads and with the indicated amounts of PCAF. Bound materials were analyzed as described above. (C) Histone acetylase activity recovered from the IRF-ISRE complexes. rIRFs (300 ng) were incubated with 2 pmol of ISRE conjugated to beads and with 400 ng of rPCAF. Bound materials were assayed for histone acetylase activity. Specific histone acetylase activity recovered from the bound rPCAF was estimated according to the value that 20 ng of free rPCAF gave 1,625 cpm of [3H]histone incorporation. Data are the average of triplicates ± SD.
Histone acetylase enzymatic activity accumulates on the IRF-ISRE complexes.
Materials bound to the ISRE beads were tested for histone acetylase activity. As shown in Fig. 1D, while the ISRE beads alone without extracts (lane 1) exhibited little enzymatic activity, the beads incubated with nuclear extracts showed high levels of histone acetylase activity, irrespective of whether they bound to the rIRF proteins. The enzymatic activity measured on the beads complexed with rIRFs was not significantly higher than on the beads without rIRFs. This is consistent with data presented in Fig. 1A, in which the levels of PCAF and CBP/p300 bound to the ISRE beads did not significantly change after addition of rIRFs. On the other hand, λB beads incubated with nuclear extracts exhibited little to no enzymatic activity, both with and without recombinant proteins (lane 5 to 7), a finding which is in agreement with the lack of recruitment of histone acetylases observed in Fig. 1A. Based on the histone acetylase activity of the input nuclear extracts (lane 8), approximately 10 to 12% of the total histone acetylases are estimated to bind to the ISRE beads under these conditions.
Recombinant IRF-1 and IRF-2, but not ICSBP, directly bind to PCAF and GCN5.
Since PCAF was recruited to the ISRE beads more efficiently than did the other histone acetylases, we sought to determine whether PCAF is capable of directly binding to IRFs. In Fig. 2A, the binding of rPCAF to the ISRE beads that had been complexed with rIRFs was tested by immunoblot assays. As expected, beads with and without ISRE, but without IRFs, did not bind to rPCAF (lanes 1 and 2). However, rPCAF avidly bound to the ISRE beads that had been complexed with rIRF-1 or rIRF-2. In contrast, beads complexed with rICSBP failed to bind to rPCAF. As with rPCAF, human rGCN5 (rhGCN5) bound to the ISRE beads complexed with rIRF-1 or rIRF-2, but not with rICSBP. It is of note that the rhGCN5 used in this work was a truncated protein which lacked the N-terminal portion but retained the conserved C-terminal domain. Figure 2B shows that the binding of PCAF to the ISRE beads complexed with rIRF-2 is PCAF dose dependent. A rough estimate suggests that approximately 50 ng (∼0.5 pmol) of rPCAF bound to 2 pmol of IRF-occupied ISRE under these conditions. These results show that IRF-1 and IRF-2, but not ICSBP, can directly interact with PCAF.
Figure 2C shows the histone acetylase activity of the rPCAF-bound to the IRF-ISRE complexes in Fig. 2A. Significant levels of enzymatic activity were found on the beads complexed with rIRF-1 or rIRF-2, while only a background level of enzymatic activity was seen on the beads with rICSBP (lane 5 and 6). The enzymatic activity was proportional to the amount of PCAF bound to the beads (see lanes 3 and 4 and lanes 7 and 8). The histone acetylase activity of the beads complexed with both rICSBP and rIRF-2 was similar to that with rIRF-2 alone (lanes 9 and 10). Further, levels of histone acetylase activities detected on the beads were similar to those measured after protein complexes were eluted from the beads (data not shown). Based on the measurement that 20 ng of rPCAF gave ∼1,500 cpm, approximately 50 to 75% of the bound PCAF gave acetylase activity in these assays. These results are in agreement with data in Fig. 1D and indicate that PCAF is a major component of the enzymatic activity associated with the IRF-ISRE complexes.
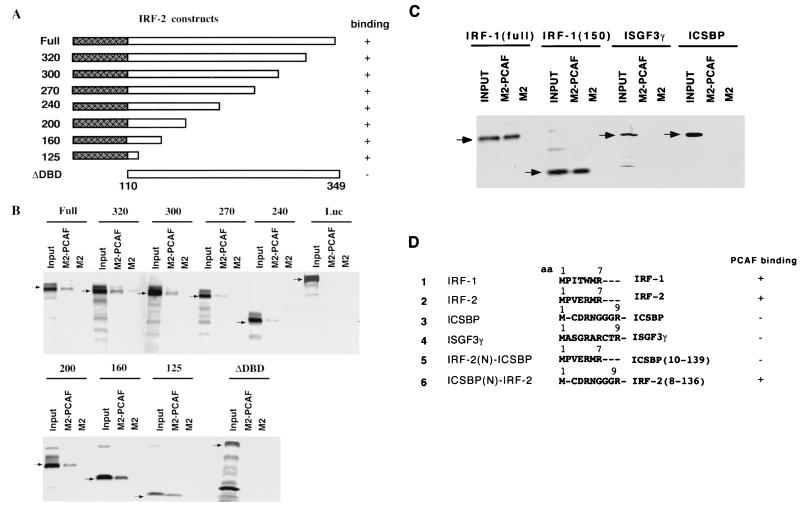
The DNA-binding domain (DBD) of IRFs binds to PCAF.
To determine a domain within an IRF protein that is involved in PCAF binding, successive C-terminal truncations of IRF-2 were tested for binding to rPCAF, which had been immobilized to agarose beads conjugated to anti-flag M2 antibody (Fig. 3A). Results are shown in Fig. 3B and summarized in Fig. 3A. All truncations, including IRF-2-125, which contained only the N-terminal 125 amino acids that correspond to the DBD, bound to PCAF. Conversely, the construct lacking the DBD (ΔDBD) completely failed to bind to PCAF. Similar binding assays were performed with IRF-1, ICSBP, and ISGF3γ (Fig. 3C). The full-length IRF-1 and the 150 N-terminal amino acids of IRF-1 both bound to PCAF. However, ICSBP and ISGF3γ failed to bind to PCAF. These results indicate that the DBD of IRF-1 and IRF-2 interacts with PCAF. Results also indicate that only some members of the IRF family are capable of binding to PCAF.
FIG. 3.
The DBD of IRFs binds to PCAF. (A) Diagram of IRF-2 deletion constructs. The shaded box in the N-terminal region represents the DBD. (B) Agarose beads conjugated to flag-tagged rPCAF by using anti-flag M2 antibody (M2-PCAF) or control anti-M2 agarose beads (M2) were incubated with 35S-labeled IRF-2 deletions (arrows). Bound materials were resolved on a 10 to 20% SDS-PAGE gradient and visualized by autoradiography. (C) Agarose beads conjugated to rPCAF as in panel B were incubated with 35S-labeled full-length IRF-1, an IRF-1 deletion mutant retaining 150 N-terminal amino acids, full-length ISGF3γ, and full-length ICSBP and then analyzed for binding as in panel B. (D) Analysis of chimeric ICSBP(N)-IRF2 and IRF-2(N)-ICSBP. Comparison of N-terminal amino acids in the DBD. In chimeric constructs, these amino acids were exchanged between IRF-2 and ICSBP. Numbers in parentheses indicate ICSBP or IRF-2 C-terminal amino acids, respectively. Chimeras were labeled with 35S and tested for binding to rPCAF immobilized to M-2 agarose. A summary of the binding results is shown on the right.
Since the DBD is the most-conserved region of the IRF family, it was of interest to assess which part of the DBD interacts with PCAF. Unlike the highly homologous internal region, the N-terminal 7 to 9 amino acids in the DBD show little homology among ICSBP, ISGF3γ, IRF-1, and IRF-2 (Fig. 3D). These amino acids are well conserved in IRF-1 and IRF-2. To investigated whether this N-terminal segment contributes to PCAF binding, we tested chimeric DBD constructs in which the N-terminal amino acids were exchanged between IRF-2 and ICSBP. While the ICSBP(N)-IRF2 chimera containing the ICSBP N-terminal amino acids and IRF-2 C-terminal amino acids bound to PCAF, the opposite chimera did not. These data indicate that the internal region of the DBD is responsible for PCAF binding.
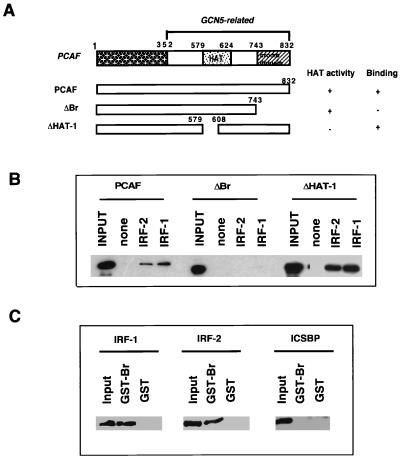
The bromodomain of PCAF is involved in IRF binding in vitro.
The C-terminal domain of PCAF is more conserved than the N-terminal domain, which contains the histone acetylase catalytic domain and a bromodomain (11, 57) (Fig. 4A). Although its function is still elusive, the bromodomain is present in many nuclear regulatory proteins, which may be involved in protein-protein interaction (4, 25). To assess a domain of PCAF involved in IRF binding, the deletion constructs depicted in Fig. 4A were tested for binding to rIRF bound to the ISRE beads. Full-length rPCAF, as well as rPCAF deletions without the catalytic domain (ΔHAT-1), bound to ISRE beads that had been complexed with rIRF-1 or rIRF-2. In contrast, rPCAF without the bromodomain failed to bind to the ISRE beads. To further investigate the significance of the bromodomain, GST pull-down assays were performed in which a GST fusion protein containing only the bromodomain of PCAF was tested for binding to rIRFs. As shown in Fig. 4C, IRF-1 and IRF-2 both bound to the GST-bromodomain fusion (GST-Br) but not to the control GST beads. In agreement with data presented in Fig. 2 and 3, ICSBP did not bind to GST-Br. These results show that the bromodomain of PCAF plays a role in IRF binding, at least in vitro.
FIG. 4.
Analysis of PCAF domains required for IRF binding. (A) Diagram of PCAF domains and deletion constructs tested for IRF binding. The N-terminal domain (1–352 [shaded box]), the histone acetylase catalytic domain (579–624 [dotted box]), and the bromodomain (743–832 [cross-hatched box]) in the C-terminal region are shaded. (B) Either 300 ng of rIRF-1 or 300 ng of rIRF-2 was incubated with 2 pmol of ISRE immobilized to beads and with 200 ng of truncated rPCAF. Bound rPCAF constructs were detected by immunoblot analysis with anti-flag M2 antibody. (C) Control GST (10 μg) or GST-PCAF-Br fusion proteins (10 μg) were incubated with 400 ng of rIRF-1, rIRF-2, or rICSBP. Bound materials were eluted with glutathione and analyzed by immunoblot with the corresponding antibodies. A summary of the binding is shown in panel A.
Ectopically expressed PCAF enhances IRF-mediated promoter activity.
PCAF is shown to enhance transcription mediated by specific transcription factors, such as nuclear hormone receptors, CREB, and MyoD (8, 29, 39). Similarly, GCN5 is reported to enhance NF-Y-dependent transcription (17). We studied whether PCAF affects the activity of promoters regulated by IRFs. To test IRF-1-dependent transcription, a luciferase reporter connected to the ISRE was transfected into NIH 3T3 cells along with IRF-1 and PCAF expression vectors, and the reporter activity was measured 24 h later (Fig. 5A). The ISRE used for the reporter was the same as that used in Fig. 1 and 2 and is known to be activated by IRF-1 (52). As expected, transfection of increasing amounts of IRF-1, without PCAF, increased the ISRE promoter activity (lanes 2 to 4). Cotransfection of IRF-1 and increasing amounts of the wild-type PCAF further increased the luciferase activity in a PCAF dose-dependent manner (lanes 6 to 8). The mutant PCAF without the histone acetylase domain (ΔHAT-1; see Fig. 4), however, did not increase luciferase activity but rather decreased the activity in a dose-dependent manner (lanes 10 to 12). Transfection of the wild-type or mutant PCAF alone did not increase promoter activity (lanes 5 and 9), as expected. These results indicate that PCAF acts as a coactivator of IRF-1.
FIG. 5.
PCAF enhances IRF-1 and IRF-2 mediated transcription. (A) IRF-1-dependent promoter activity. NIH 3T3 cells were transfected with 600 ng of the ISRE-Ld40 reporter and 10, 20, or 30 ng of IRF-1 (pACT-1) (lanes 2 to 4), 400 ng of PCAF or ΔHAT-1 alone (lanes 5 and 9), 10 ng of IRF-1 plus increasing amounts of PCAF (100, 200, and 400 ng) (lanes 6 to 8), or ΔHAT-1 (lanes 10 to 12). The total amounts of DNA transfected were adjusted with control pAct and pCXN vectors to be constant. Luciferase activity was measured 24 h after transfection. (B) IRF-2-dependent promoter activity. NIH 3T3 cells were transfected with 400 ng of H4-Ld40 reporter and 200 or 400 ng of PCAF or ΔHAT-1 alone (lanes 2 and 3 and lanes 9 and 10) or 1, 2, 5, 12.5, 25, or 50 ng of IRF-2 (pACT-2) alone (lanes 11 to 15), or 2 ng of IRF-2 plus 200 or 400 ng of PCAF (lanes 5 and 6) or ΔHAT-1. The luciferase activity was measured 24 h later. Values are the average of triplicate determinations ± SD.
IRF-2 represses transcription from a number of ISRE-containing promoters that are stimulated by IFNs or by IRF-1 (24, 32, 35). However, IRF-2 can stimulate some promoters containing an ISRE-like element including, for example, the histone H4 promoter (50). In experiments presented in Fig. 5B, the transcriptional activation by IRF-2 was tested with a luciferase reporter containing the binding site of the H4 gene (HiNF-M). Transfection of IRF-2 alone increased the luciferase activity in a dose-dependent manner (columns 11 to 15), as expected. Transfection of the wild-type PCAF and mutant ΔHAT-1 alone (columns 2 and 3 and columns 4 and 5) gave a modest increase, the basis of which has not been studied in detail. However, when IRF-2 and various doses of wild-type PCAF were cotransfected, luciferase activity was synergistically increased in a PCAF dose-dependent manner (columns 7 and 8). At the highest dose of PCAF, the level of luciferase activity by PCAF and IRF-2 greatly exceeded that by IRF-2 alone. However, when IRF-2 and mutant ΔHAT-1 were cotransfected, no increase in luciferase activity was detected (columns 9 and 10). Taken together, these results show that PCAF acts as a coactivator of both IRF-1 and IRF-2.
TPA induction of PCAF and CBP/p300 in U937 monocytic cells.
Human U937 cells undergo differentiation in response to the phorbol ester TPA and become more-mature monocytic cells (1, 28, 41, 58). Because untreated U937 cells do not respond to IFN-γ or IFN-α but do gain responsiveness after TPA treatment (reference 19; see also below), it was of interest to examine whether TPA treatment affects expression of histone acetylases and, if so, whether regulated expression of the enzymes influences IFN responsiveness in these cells.
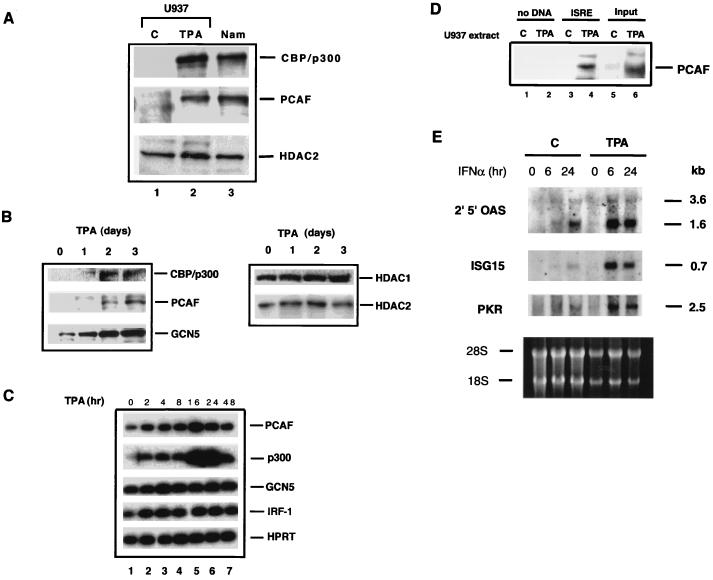
Figure 6A shows the immunoblot detection of several histone acetylases in U937 cells prior to and after TPA treatment. In untreated U937 cells the expression of PCAF and CBP/p300 was very low, almost at a background level, but their expression was dramatically induced after TPA treatment. The time course study in Fig. 6B shows that the induction of PCAF and CBP/p300 was detectable on day 1 and that the levels increased steadily, reaching a maximum on day 3. Expression of the smaller size of hGCN5 was also increased after TPA treatment. Interestingly, TPA treatment did not significantly change the levels of the histone deacetylases, HDAC1 and HDAC2, during this period (Fig. 6B).
FIG. 6.
Phorbol ester induction of histone acetylase expression in U937 cells. (A) Portions (30 μg) of nuclear extracts from untreated U937 cells (C) or from cells treated with 10 nM of TPA for 72 h (TPA) and untreated Namalwa B cells (Nam) were analyzed by immunoblotting. (B) Time course analysis of histone acetylase (left panel) and deacetylase (right panel) expression. Nuclear extracts from U937 cells treated with 10 nM TPA for 1, 2, and 3 days were analyzed by immunoblotting. (C) Quantitative RT-PCR analysis of histone acetylase mRNA induction in TPA-treated U937 cells. (D) Recruitment of TPA-induced PCAF to the ISRE complex. Nuclear extracts (500 μg) from U937 cells treated with TPA for 3 days were incubated with 4 pmol of ISRE-conjugated beads, and bound proteins were analyzed by immunoblot assay with anti-PCAF antibody as in Fig. 1A. (E) Enhanced expression of IFN-inducible genes in TPA-treated U937 cells. Untreated U937 cells (lanes 1 to 3) or cells treated with TPA for 3 days (lanes 4 to 6) were treated with rhIFN-α (1,000 U/ml) for indicated times and then analyzed in an RNA blot assay. 2′5′-OAS, 2′5′ oligo(A) synthetase; PKR, double-stranded RNA-dependent protein kinase. The size of each transcript (in kilobases) is indicated on the right. The bottom panel shows the rRNA staining to compare the levels of RNA loading.
The RNA levels of the histone acetylases were examined by quantitative RT-PCR (Fig. 6C). Both PCAF and CBP/p300 RNAs were increased within 2 h of TPA treatment. The RNA levels were further increased until 16 to 24 h, followed by a decrease by 48 h. These results show that TPA induction of PCAF and CBP/p300 occurs at the level of the RNA. GCN5 RNA levels, on the other hand, increased only slightly after treatment, suggesting the involvement of a posttranslational change. Similarly, the expression of IRF-1 RNA was not substantially increased after TPA treatment (Fig. 6C). Consistent with these results, IRF-1 protein levels, low in these cells, did not change after TPA treatment (results not shown). It is of note that these acetylases were constitutively expressed in several other lymphoid and/or myeloid cells tested, including Namalwa human B lymphocytes (Fig. 6A) and ANA-1 (data not shown). Levels of histone acetylases in U937 and other cells did not change after IFN treatment (data not shown).
By DNA affinity binding assays similar to those depicted in Fig. 1A, we investigated whether TPA-induced PCAF in U937 cells is recruited to the ISRE-IRF complex. Extracts from untreated U937 cells or cells treated with TPA for 3 days were incubated with ISRE beads (without rIRFs), and PCAF binding was tested by immunoblot assay. As seen in Fig. 6D, PCAF was efficiently recruited to the ISRE beads when extracts from TPA-treated cells were tested. In contrast, no recruitment was observed with extracts from untreated cells. Thus, histone acetylases are induced in U937 cells by phorbol ester treatment, and PCAF is then recruited to the ISRE.
Untreated U937 cells have been shown to respond poorly to type II IFN (IFN-γ) but to gain IFN responsiveness after TPA treatment (19). In light of TPA-induced PCAF recruitment to the ISRE, we felt it important to examine whether type I IFN-dependent gene expression is enhanced in these cells after TPA treatment. Figure 6E shows RNA blot analysis of several IFN-inducible genes. The 2,5-oligoadenylate synthetase (2′,5′-OAS), ISG15, and double-stranded RNA-dependent protein kinase (PKR) genes are stimulated by type I IFNs through the ISRE element in the promoters (6, 18, 30, 40). These genes play critical roles in IFN’s antiviral activity and in cytokine induction. Expression of these genes was only modestly induced by IFN-α in untreated U937 cells. In contrast, in TPA-treated cells the IFN-α treatment led to much greater levels of induction for all three genes. These results show that untreated U937 cells are a low responder to type I IFN, as they are to type II IFN, and that TPA confers increased competence in IFN responsiveness, a finding which coincides with the induction of histone acetylases.
U937 cells acquire type I IFN responsiveness after PCAF transfection.
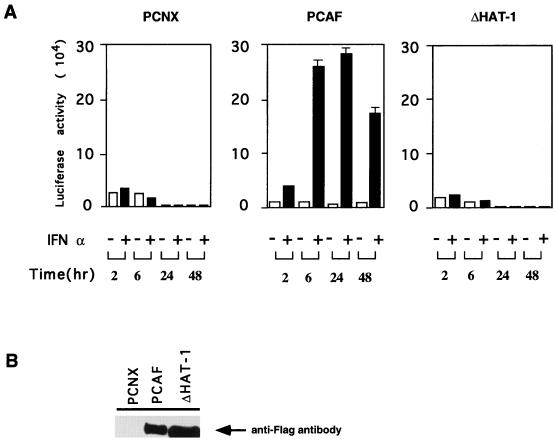
Results shown in Fig. 6E indicate that histone acetylases induced by TPA contribute to the acquisition of full IFN responsiveness in U937 cells. To further test the role for PCAF in type I IFN responsiveness, U937 cells that had been stably transfected with PCAF were tested for IFN-inducible ISRE promoter activity. Cells were first transfected with the wild-type PCAF or the ΔHAT-1 mutant (Fig. 4) and then selected by Geneticin treatment for about 10 to 14 days. Drug-resistant cells were pooled and then transiently transfected with the ISRE reporter (Fig. 5A). Luciferase activity was tested after treatment with IFN-α for various periods of time. As shown in Fig. 7, the activity of this reporter was not stimulated by IFN-α in U937 cells transfected with the control vector, which is consistent with the low IFN responsiveness seen in untreated U937 cells (Fig. 6E). In contrast, reporter activity was strongly induced (10- to 25-fold) by IFN-α treatment in cells transfected with the wild-type PCAF, whereas reporter activity was not stimulated in cells transfected with the ΔHAT mutant after IFN treatment. Immunoprecipitation results shown in the bottom panel of Fig. 7 show that pooled U937 cells expressed the exogenous PCAF or ΔHAT-1 at high levels, confirming the efficient transfection of the constructs. These data indicate that PCAF plays an important role in the acquisition of IFN responsiveness by U937 cells and that the histone acetylase catalytic domain is required for this activity.
FIG. 7.
Enhanced IFN-responsive promoter activity in PCAF-transfected U937 cells. A total of 5 × 106 pooled U937 cells transfected with pCNX (control), PCAF, or ΔHAT-1 were transiently transfected with 15 μg of the ISRE-Ld40 reporter. Cells were treated with 1,000 U of rhIFN-α per ml, and luciferase activity was measured at the indicated times. The values are the average of triplicates assays ± the SD. The bottom panel indicates the expression of flag-tagged PCAF in transfectants analyzed by immunoblot assay.
DISCUSSION
The ISRE affinity beads assay demonstrated that the endogenous IRF-1 and IRF-2 are complexed with multiple histone acetylases, PCAF, CBP, and p300 in vivo to bind to the ISRE. In addition, GCN5 was found on the ISRE beads when bound to the rIRFs, indicating that it, too, is recruited to the IFN-responsive promoters (Fig. 1A). These findings are in line with recent reports that histone acetylases interact with other transcription factors, including nuclear receptors, CREB, MyoD, and NF-Y (8, 14, 17, 29, 39), and support the view that they are recruited to transcriptionally active promoters through sequence-specific transcription factors. Consistent with our data, IRF-1 and IRF-3 have been shown to interact with CBP/p300 (33, 53, 59). In addition to type I IFN-responsive promoters, some histone acetylases are likely to be recruited to type II IFN-dependent promoters, since Stat1 and Stat2 have been shown to interact with p300 and/or CBP (7, 29, 61). In our recruitment assay (Fig. 1), PCAF and CBP/p300 were both found on the ISRE beads. It is possible that PCAF and CBP/p300 bind to IRF proteins independently of each other, as well as cooperatively, since PCAF and CBP/p300 interact with each other (57). Histone acetylases recruited to the IRF-ISRE complex may bring additional factors to the promoter, since acetylases appear to occur as a large complex in the cell (21).
It is of note that the λB DNA element, a powerful enhancer of the immunoglobulin light-chain gene (10, 20), did not recruit any of the histone acetylases tested here, even when bound to rPU.1 and rICSBP (Fig. 1). These results suggest that not all DNA binding activators are equipped to recruit histone acetylases and that transcriptional activation may not always involve histone acetylase recruitment.
In vitro analysis as presented in Fig. 2 and 3 show that IRF-1 and IRF-2 directly interact with rPCAF. In these analyses we also found that only some members of the IRF family interact with PCAF and that ICSBP and ISGF3γ fail to bind to PCAF. Further, the ability of IRF factors to bind to PCAF did not correlate with their known role as an activator or a repressor. Our preliminary data indicate that CBP/p300 also bind only to a subset of IRF proteins, suggesting that IRF family members may be classified into two groups based on their ability to interact with histone acetylases, which might be related to their ability to alter a chromatin structure of IFN-responsive promoters. The data in Fig. 3 show that although only some IRF members interact with PCAF, the region that contacts PCAF is the conserved DBD. Since PCAF binds to the IRF proteins that had been complexed with the ISRE, the DBD of IRF-1 and IRF-2 appears to have a dual binding activity in that it could interface ISRE DNA on one hand and PCAF on the other. In addition, we observed that 32P-labeled ISRE oligomers bound avidly to the IRF-2 DBD that had been bound to rPCAF on anti-M2 agarose beads (not shown), which lends credence to this possibility. Interestingly, a similar dual binding activity has been noted for the nuclear receptors, in that the PCAF binds to the DBD of the RXR-RAR heterodimer that had been complexed with the retinoid-responsive DNA element (8). Considering the lack of similarity between the IRFs and the nuclear receptors in terms of the structure and the relative locations of the DBD, PCAF binding to the DBD of IRF proteins may be of general significance and might be observed with other families of transcription factors. For example, by directly contacting the DBD of transcription factors, PCAF may affect their accessibility to chromatinized DNA.
As demonstrated by reporter assays in Fig. 5, PCAF is a coactivator, capable of enhancing transcription from the ISRE promoter stimulated by IRF-1. Similarly, PCAF enhanced transcription from the H4 promoter stimulated by IRF-2. This is in agreement with previous studies showing that PCAF serves as a coactivator for other families of transcription factors (8, 39). The transcriptional enhancement was dependent on the catalytic domain of PCAF in this and other studies, indicating that it is the histone acetylase activity that is responsible for enhanced transcription. PCAF may work by acetylating nucleosomal histones in or near the ISRE promoter. However, the possibility that PCAF targets the acetylation of nonhistone proteins important for transcription cannot be excluded at present (23, 27). It should be mentioned here that although the PCAF deletion lacking the bromodomain (ΔBr) failed to interact with IRF proteins in vitro (Fig. 4), ΔBr appeared to be capable of enhancing IRF-1-dependent reporter activity, albeit to a lower degree than the wild-type PCAF (data not shown), suggesting that interactions between PCAF and IRF proteins are complex and occur at multiple surfaces in vivo.
One of the most significant observations made here is the marked induction of multiple histone acetylases in U937 cells after TPA treatment. The induction of PCAF and CBP/p300 by TPA appeared well coordinated, since induction of these genes occurred at the level of RNA and the kinetics of induction were similar in these proteins. Considering the little knowledge available regarding the regulation of histone acetylases, our data are noteworthy, as they are one of the first examples showing a link between histone acetylase expression and signalling events. Since TPA affects cell growth and differentiation in U937 and other cells (1, 41), it is likely that histone acetylase expression is regulated by many other external signals that influence these processes. In this context it is interesting to note that TPA did not alter the expression of HDAC1 and HDAC2 in U937 cells, indicating that histone acetylases and deacetylases are regulated by distinct signals. We show that TPA induction of histone acetylases correlated with a marked enhancement in the expression of type I IFN-inducible genes. Consistent with the previous report that U937 cells are a low responder to type II IFN (19), our data show that these cells are a low responder to type I IFNs as well and that they are converted to a high responder after TPA treatment. In light of a marked increase in IFN-inducible ISRE reporter activity observed after transfection of PCAF (Fig. 7), it is most likely that increased histone acetylase expression plays a major role in conferring type I IFN responsiveness. Although detailed mechanisms of the enhanced IFN responsiveness induced by TPA are not known at present, PCAF may play a role in the activation of not only IRF-1 but also ISGF3 through Stat1 and Stat2 via CBP/p300.
In summary, this work identifies the histone acetylase PCAF as a powerful coactivator of the IRF family that affects cellular responsiveness to IFNs and whose expression is regulated by phorbol ester. Our work raises the possibility that histone acetylation may be an important parameter of the therapeutic efficacy of IFNs.
ACKNOWLEDGMENTS
We thank J. Blanco, V. Horn, and V. Orgryzko for helpful discussions; E. Sato and G. Humphrey for antibodies; H. Singh and T. Taniguchi for plasmids; and C. Hong, H. Fukazawa, and J. Boll for help in the preparation of the figures and manuscript.
REFERENCES
- 1.Asiedu C, Biggs J, Kraft A S. Complex regulation of CDK2 during phorbol ester-induced hematopoietic differentiation. Blood. 1997;90:3430–3437. [PubMed] [Google Scholar]
- 2.Au W C, Moore P A, Lowther W, Juang Y T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Barlev N A, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman J, Berger S L. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartl S, Tapliock J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benech P, Mory Y, Revel M, Chebath J. Structure of two forms of the interferon-induced (2′-5′) oligo A synthetase of human cells based on cDNAs and gene sequences. EMBO J. 1985;4:2249–2256. doi: 10.1002/j.1460-2075.1985.tb03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, Andrea A D, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-γ. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 8.Blanco J C G, Minucci S, Yang X-J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1641. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bovolenta, C., P. H. Driggers, M. S. Marks, J. A. Medin, A. D. Politis, S. N. Vogel, D. E. Levy, K. Sakaguchi, E. Appella, J. E. Coligan, and K. Ozato. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc. Natl. Acad. Sci. USA 91:5046–5050. [DOI] [PMC free article] [PubMed]
- 10.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;18:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 12.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p in linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 13.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of human proteins functionally conserved with the yeast putative adapters ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Lin R J, Schiltz R L, Chakravati D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:596–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 16.Colle J H, Falanga P B, Singer M, Hevin B, Milon G. Quantitation of messenger RNA by competitive RT-PCR. J Immunol Methods. 1997;210:175–184. doi: 10.1016/s0022-1759(97)00186-5. [DOI] [PubMed] [Google Scholar]
- 17.Currie R A. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 18.D’Cunha J, Knight E, Jr, Haas A L, Truitt R L, Borden E C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eilers A, Seegert D, Schindler C, Baccarini M, Decker T. The response of gamma interferon activation factor is under developmental control in cells of the macrophage lineage. Mol Cell Biol. 1993;13:3245–3254. doi: 10.1128/mcb.13.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 21.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 24.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 25.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;10:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch K P, Babriele L, Waring J P, Bachmann M F, Zinkernagel R M, Morse H C, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 27.Imhof A, Yang X-J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 28.Improta T, Pine R, Pfeffer L M. Interferon-gamma potentiates the antiviral activity and the expression of interferon-stimulated genes induced by interferon-alpha in U937 cells. J Interferon Res. 1992;2:87–94. doi: 10.1089/jir.1992.12.87. [DOI] [PubMed] [Google Scholar]
- 29.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 32.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 34.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 35.Nelson N, Marks M S, Driggers P H, Ozato K. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson N, Kanno Y, Hong C, Contursi C, Fujita T, Fowlkes B J, O’Connell E, Hu-Li J, Paul W E, Jankovic D, Sher A F, Coligan J E, Thornton A, Appella E, Yang Y, Ozato K. Expression of IFN regulatory factor family proteins in lymphocytes. Induction of Stat-1 and IFN consensus sequence binding protein expression by T cell activation. J Immunol. 1996;156:3711–3720. [PubMed] [Google Scholar]
- 37.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon factors. Cytokine Growth Factor Rev. 1997;4:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Puri P L, Sartorelli V, Yang X-J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y J, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 40.Schindler C, Fu X-Y, Improta R, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwantke N, Le Bouffant F, Doree M, Le Peuch C J. Protein kinase C: properties and possible role in cellular division and differentiation. Biochimie. 1985;67:1103–1110. doi: 10.1016/s0300-9084(85)80107-3. [DOI] [PubMed] [Google Scholar]
- 42.Sharf R, Azriel A, Lejbkowicz F, Winograd S S, Ehrlich R, Levi B-Z. Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J Biol Chem. 1995;270:13063–13069. doi: 10.1074/jbc.270.22.13063. [DOI] [PubMed] [Google Scholar]
- 43.Shuai K. Interferon-activated signal transduction to the nucleus. Curr Opin Cell Biol. 1994;6:253–259. doi: 10.1016/0955-0674(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 44.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna M J, Onate S A, Tsai S Y, Tsai M J, O’Malley B M. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 45.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi T, Harada H, Lamphier M S. Regulation of the interferon system and cell growth by the IRF transcription factors. J Cancer Res Clin Oncol. 1995;121:516–520. doi: 10.1007/BF01197763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 48.Thornton A M, Ogryzko V V, Dent A, Sharf R, Levi B-Z, Kanno Y, Staudt L M, Howard B H, Ozato K. A dominant negative mutant of an IFN regulatory factor family protein inhibits both type I and type II IFN-stimulated gene expression and antiproliferative activity of IFNs. J Immunol. 1996;157:5145–5154. [PubMed] [Google Scholar]
- 49.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 50.Vaughan P S, Aziz F, van Wijnen A J, Wu S, Harada H, Taniguchi T, Soprano K J, Stein J L, Stein G S. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 51.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang I-M, Blanco J C G, Tsai S Y, Tsai M-J, Ozato K. Interferon regulatory factors and TFIIB cooperatively regulate interferon-responsive promoter activity in vivo and in vitro. Mol Cell Biol. 1996;16:6313–6324. doi: 10.1128/mcb.16.11.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolffe A P, Wong J, Pruss D. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- 55.Yamagata T, Nishida J, Tanaka S, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 58.Yeung M C, Liu J, Lau A S. An essential role for the interferon-inducible, double-stranded RNA-activated protein kinase PKR in the tumor necrosis factor-induced apoptosis in U937 cells. Proc Natl Acad Sci USA. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Mishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p399. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]