Abstract
Animal-microbe symbioses are often stable for millions of years. An example is the clade consisting of social corbiculate bees—honeybees, bumblebees, and stingless bees—in which a shared ancestor acquired specialized gut bacteria that subsequently diversified with hosts. This model may be incomplete, however, as few microbiomes have been characterized for stingless bees, which are diverse and ecologically dominant pollinators in the tropics. We surveyed gut microbiomes of Brazilian stingless bees, focusing on the genus Melipona, for which we sampled multiple species and biomes. Strikingly, Melipona lacks Snodgrassella and Gilliamella, bacterial symbionts ubiquitous in other social corbiculate bees. Instead, Melipona species harbor more environmental bacteria and bee-specific Starmerella yeasts. Loss of Snodgrassella and Gilliamella may stem from ecological shifts in Melipona or the acquisition of new symbionts as functional replacements. Our findings demonstrate the value of broadly sampling microbiome biodiversity and show that even ancient symbioses can be lost.
Subject terms: Symbiosis, Microbiome, Microbial ecology, Metagenomics, Microbial ecology
Introduction
Microbial symbionts have contributed to the diversification of many animal lineages. Host-microbe associations are often stable [1, 2]; however, ancient symbionts can be lost, and sometimes replaced by novel symbionts [3, 4]. These transitions may be driven by erosion of symbiont functionality [5] or shifts in host ecology [6]. Although symbiont turnover has been documented for intracellular symbionts, its prevalence among gut symbionts is unclear. Here we report the dissolution of a long-standing symbiosis between social bees and specialized gut bacteria.
The eusocial corbiculate bees (Apidae) contain distinctive gut bacterial symbionts that are restricted to bees and that have co-diversified with their hosts [7–9]. In Apis mellifera (honeybees) and Bombus impatiens and B. terrestris (bumblebees), Snodgrassella, Gilliamella and Lactobacillus have been observed to contribute to host health [10–13]. However, gut microbiomes of stingless bees (Meliponini), the most diverse clade of highly social bees and major pollinators in the tropics [14], are understudied, and seem to vary more than microbiomes of other social bees [8, 15–17]. Here, we characterize gut bacteria and fungi associated with the genus Melipona from multiple Brazilian biomes.
Methods and results
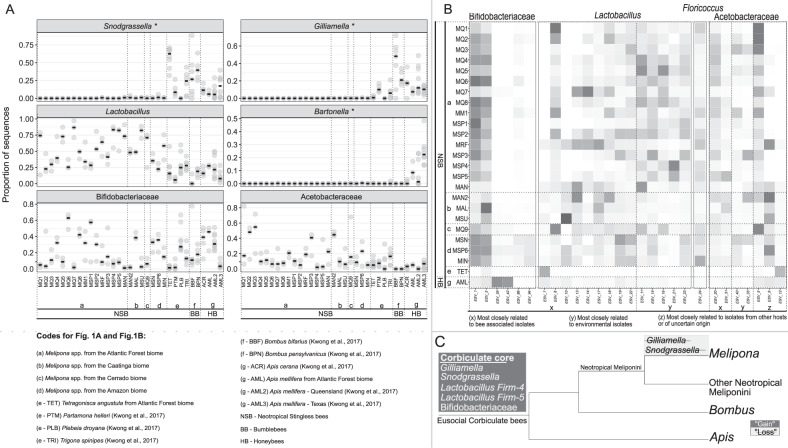
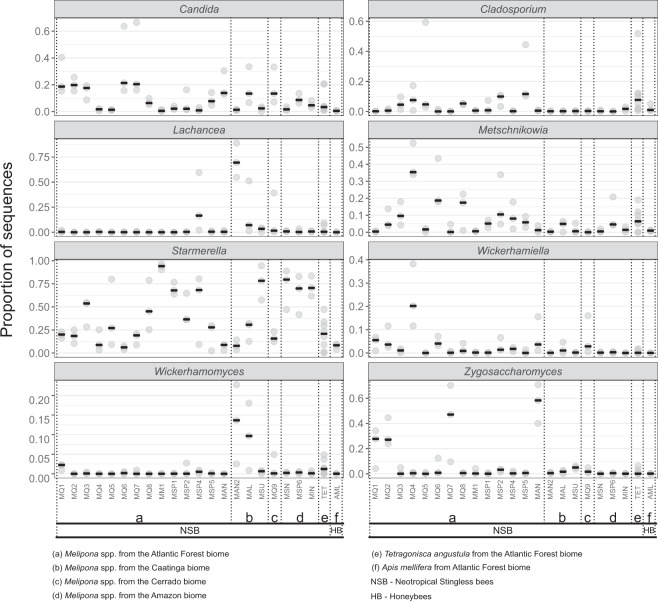
We sampled 3 colonies from each of 23 Melipona populations representing 4 Brazilian biomes (SISBIO/ICMBio authorization #69500-1). Three colonies of Apis mellifera and 8 of the stingless bee Tetragonisca angustula were also collected from one site. A pool of guts from five individuals/colony had regions of bacterial 16S rRNA and fungal ITS1 genes sequenced to characterize gut microbial community composition (Supplementary Table S1), and we analyzed these data (SRA accession #PRJNA678404) together with previously published data [8] (see Supplementary Methods). Our results show that Melipona lack Snodgrassella and Gilliamella, core gut symbionts of honeybees, bumblebees, and some other stingless bee clades (Fig. 1A). As expected, honeybee-specific Bartonella apis was also absent. These taxa were significantly lower in relative abundance in Melipona as compared with A. mellifera (Kruskal–Wallis p < 0.05) (Fig. 1A). The most abundant and consistent bacteria in Melipona belong to Lactobacillus, Bifidobacteriaceae, Acetobacteraceae, and Floricoccus (Fig. 1B). For the core Melipona bacterial taxa (i.e., taxa containing exact sequence variants [ESVs] in all Melipona samples), we constructed phylogenies using the most abundant ESVs and related sequences from GenBank to differentiate widespread environmental taxa from specialized associates of social bees. Melipona-associated Lactobacillus and Acetobacteraceae include putatively environmental ESVs as well as bee-specific ESVs (Fig. 1B, Supplementary Figs. S1, S2, and Table S2). Bifidobacteriaceae ESVs in Melipona are closely related to bee isolates (Fig. 1B, Supplementary Fig. S3 and Table S2). In contrast, the core Floricoccus ESV is closely related to environmental isolates (Fig. 1B, Supplementary Fig. S4 and Table S2). The fungal gut microbiome of Melipona is mainly composed of Starmerella, Lachancea, Zygosaccharomyces, and a few other yeasts (Fig. 2). Of these, Starmerella are noteworthy in that they form bee/beehive-specific clades (Supplementary Fig. S5), were present in all Melipona samples and comprised a higher proportion of gut microbiomes in Melipona than in other social bee species (Kruskal–Wallis p < 0.05) (Fig. 2). Apparent symbiont loss from Melipona (Fig. 1C) is not due to incomplete sampling, as rarefaction curves confirm sufficient characterization of bacterial communities (Supplementary Table S3 and Fig. S6).
Fig. 1. Gut bacterial community composition and a model of symbiont gain and loss in eusocial corbiculate bees.
A Distribution of six bee-specific gut bacterial symbionts across Neotropical stingless bees (NSB), bumblebees (BB) and honeybees (HB). Bee populations are on the x-axis (more detail in Supplementary Table S1). Previously published data [8] are included for comparison. The y-axis shows the relative abundance of each bacterial group as the proportion of sequences. Circles represent replicate bee samples, and black bars represent the median proportion for each bee population. Taxa with significantly higher relative abundances in Apis mellifera than Melipona are denoted with an asterisk. B Heatmap comprising the most abundant ESVs belonging to Melipona core bacterial taxa (i.e., taxa present in all samples of all Melipona populations). The x-axis represents ESVs labeled by the source from which close relatives were isolated (Supplementary Figs. S1–S4), and the y-axis represents each bee population sequenced. Darker squares correspond to higher log-transformed mean relative abundances for a given ESV in each bee population. C Phylogeny of eusocial corbiculate bees (based on [20]) showing the gains [7–9] and our hypothesized losses of core symbionts. A color version of this figure is available in the Supplementary Material.
Fig. 2. Distribution of eight dominant fungi across Melipona, Tetragonisca angustula and Apis mellifera.
The x-axis represents sequenced bee populations (Supplementary Table S1). The y-axis shows the relative abundance of each fungal genus as the proportion of sequences for each bee population. Circles represent replicate bee samples, and black bars represent the median proportion for each bee population. A color version of this figure is available in the Supplementary Material.
Discussion
Although present in our A. mellifera and T. angustula samples, Snodgrassella and Gilliamella were extremely rare or absent in samples of 23 Melipona populations from four biomes in Brazil (Fig. 1A). When present, the low abundance likely reflects artefacts such as cross-contamination. These findings, together with previous work [15, 17], suggest that Snodgrassella and Gilliamella have been lost entirely, or almost entirely, from the sampled species of Melipona. This shift is unlikely to have occurred in the common ancestor of stingless bees, because several other Neotropical stingless bees, including T. angustula and other species occurring in Brazil, do maintain these associations [8] (Fig. 1A and C). Furthermore, such losses are not geographically restricted as they were observed in Melipona from four Brazilian biomes (Fig. 1A) and from Central America [15]. While a few other stingless bee genera have been reported to lack Snodgrassella and Gilliamella, based on limited sampling [8, 15, 16], our finding of consistent absence from Melipona provides the strongest evidence of symbiont loss in a major social bee clade. Since Snodgrassella and Gilliamella can contribute to the health of honeybees and bumblebees [10–13], it is possible that Melipona has undergone ecological shifts that release them from dependence on symbiont-based nutrition or defense. Alternatively, either persisting members of the ancestral microbiome, or newly acquired symbionts, may compensate for their absence. Melipona retains bee-specialized strains of Lactobacillus (Supplementary Fig. S1), Acetobacteraceae (Supplementary Fig. S2), and Bifidobacteriaceae (Supplementary Fig. S3), which in theory could have gained new functions, such as metabolic and protective capabilities. Melipona has also gained new microbial associations, such as environmental strains of Lactobacillus, Acetobacteraceae, and Floricoccus. These bacteria are widespread in Melipona but only occasionally found in T. angustula and A. mellifera (Fig. 1B). Although phylogenetic evidence suggests they are acquired from the environment, the prevalence of these taxa points toward a stable, and possibly functional association with Melipona. Furthermore, certain fungi are also associated with Melipona (Fig. 2). Starmerella, the most abundant and widespread of these, has been found in stingless bee adults, pollen provisions and honey [18], suggesting specialization for the bee niche (Supplementary Fig. S5 and Table S2). In other insects, fungi have occasionally replaced degenerating bacterial symbionts [3], and dependence on a fungal symbiont is documented for another stingless bee, Scaptotrigona depilis [19]. Experimental work on Melipona and other stingless bees could give insights into the functional basis and dynamic nature of bee microbiomes.
Supplementary information
Acknowledgements
We acknowledge the UFV, the financial support from CNPq, CAPES – Finance Code 001 and FAPEMIG, a USDA NIFA postdoctoral fellowship (2018-08156) to TJH, and NIH award R35GM131738 to NAM. We thank Marina Cunha, Gil Viana, José Souza, Eduardo Ferreira, Leandro Campos, Marcelo Silva, Antônio Alves, Sidcley de Lucena, Flávio Yamamoto, Hilton Gomes, Kalhil França, Gilvan Santos and Helder Resende for providing bees, and Fernando da Silveira for Amazonian bees identification.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01000-1.
References
- 1.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 2.Bourguignon T, Lo N, Dietrich C, Roisin Y, Brune A, Evans TA, et al. Rampant host switching shaped the termite gut microbiome. Curr Biol. 2018;28:649–54. doi: 10.1016/j.cub.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 3.Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng X. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci USA. 2018;115:E5970–9. doi: 10.1073/pnas.1803245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong RA, Moran NA. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 2018;12:898–908. doi: 10.1038/s41396-017-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–76. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017;25:1–16. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–28. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 8.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, et al. Dynamic microbiome evolution in social bees. Sci Adv. 2017;3:1–17. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–84. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothman JA, Leger L, Graystock P, Russell K, McFrederick QS. The bumble bee microbiome increases survival of bees exposed to selenate toxicity. Environ Microbiol. 2019;21:3417–29. doi: 10.1111/1462-2920.14641. [DOI] [PubMed] [Google Scholar]
- 11.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108:19288–92. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA. 2017;114:4775–80. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mockler BK, Kwong WK, Moran NA, Koch H. Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl Environ Microbiol. 2018;84:1–11. doi: 10.1128/AEM.02335-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannini TCG, Boff S, Cordeiro GD, Cartonalo EA, Jr, Veiga AK, Imperatriz-Fonseca VL, et al. Crop pollinators in Brazil: a review of reported interactions. Apidologie. 2015;46:209–23. doi: 10.1007/s13592-014-0316-z. [DOI] [Google Scholar]
- 15.Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol. 2013;22:2028–44. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 16.Leonhardt SD, Kaltenpoth M. Microbial communities of three sympatric Australian stingless bee species. PLoS One. 2014;9:1–6. doi: 10.1371/journal.pone.0105718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díaz S, de Souza Urbano S, Caesar L, Blochtein B, Sattler A, Zuge V, et al. Report on the microbiota of Melipona quadrifasciata affected by a recurrent disease. J Invertebr Pathol. 2017;143:35–39. doi: 10.1016/j.jip.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Teixeira ACP, Marini MM, Nicoli JR, Antonini Y, Martins RP, Lachance M-A, et al. Starmerella meliponinorum sp. nov., a novel ascomycetous yeast species associated with stingless bees. Int J Syst Evol Microbiol. 2003;53:339–43. doi: 10.1099/ijs.0.02262-0. [DOI] [PubMed] [Google Scholar]
- 19.Paludo CR, Menezes C, Silva-Junior EA, Vollet-Neto A, Andrade-Dominguez A, Pishchany G, et al. Stingless bee larvae require fungal steroid to pupate. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-19583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramírez SR, Nieh JC, Quental TB, Roubik DW, Imperatriz-Fonseca VL, Pierce NE. A molecular phylogeny of the stingless bee genus Melipona (Hymenoptera: Apidae) Mol Phylogenet Evol. 2010;56:519–25. doi: 10.1016/j.ympev.2010.04.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.