Abstract
The current taxonomy of the Lactiplantibacillus plantarum group comprises of 17 closely related species that are indistinguishable from each other by using commonly used 16S rRNA gene sequencing. In this study, a whole-genome-based analysis was carried out for exploring the highly distinguished target genes whose interspecific sequence identity is significantly less than those of 16S rRNA or conventional housekeeping genes. In silico analyses of 774 core genes by the cano-wgMLST_BacCompare analytics platform indicated that csbB, morA, murI, mutL, ntpJ, rutB, trmK, ydaF, and yhhX genes were the most promising candidates. Subsequently, the mutL gene was selected, and the discrimination power was further evaluated using Sanger sequencing. Among the type strains, mutL exhibited a clearly superior sequence identity (61.6–85.6%; average: 66.6%) to the 16S rRNA gene (96.7–100%; average: 98.4%) and the conventional phylogenetic marker genes (e.g., dnaJ, dnaK, pheS, recA, and rpoA), respectively, which could be used to separat tested strains into various species clusters. Consequently, species-specific primers were developed for fast and accurate identification of L. pentosus, L. argentoratensis, L. plantarum, and L. paraplantarum. During this study, one strain (BCRC 06B0048, L. pentosus) exhibited not only relatively low mutL sequence identities (97.0%) but also a low digital DNA–DNA hybridization value (78.1%) with the type strain DSM 20314T, signifying that it exhibits potential for reclassification as a novel subspecies. Our data demonstrate that mutL can be a genome-wide target for identifying and classifying the L. plantarum group species and for differentiating novel taxa from known species.
Keywords: comparative genome sequence analysis, genome-wide target, species-specific identification, Lactobacillus plantarum group, Lactiplantibacillus
1. Introduction
Following reclassification of Lactobacillus according to physiological criteria, clade-specific signature genes, core genome phylogeny, organism ecology, and pairwise average amino acid identity, this varied genus now comprises of 25 genera, with 23 of them being novel [1]. This change to the taxonomy has affected major probiotic Lactobacillus species; for example, Lactobacillus plantarum has become Lactiplantibacillus plantarum. Because the new genera suggested for this group all still begin with an “L”, the abbreviated genus and species names are unchanged. L. plantarum is a versatile lactic acid bacteria (LAB) species that one can isolate from different fermented foods and the human gastrointestinal tract [2,3]. Certain L. plantarum strains, some of which are recognized probiotics, are generally recognized as safe and have a qualified presumption of safety status [4,5], designations that are widely applied in food, agriculture, and industrial fermentation [6].
Relevant phylogenomic analyses have classified L. plantarum as a member of the L. plantarum group along with the following 16 species: L. argentoratensis, L. daoliensis, L. daowaiensis, L. dongliensis, L. fabifermentans, L. garii, L. herbarum, L. modestisalitolerans, L. mudanjiangensis, L. nangangensis, L. paraplantarum, L. pentosus, L. pingfangensis, L. plajomi, L. songbeiensis, and L. xiangfangensis [1,7,8]. Among these species, L. argentoratensis, L. paraplantarum, and L. pentosus are the most closely related to L. plantarum; they not only share extremely high sequence identities with full-length 16S rRNA genes (100%, 99.7%, and 99.9%, respectively) but also numerous common phenotypic characteristics [9]. Consequently, substantially conserved protein–coding housekeeping genes with more favorable discrimination power have been used to resolve the relationships among closely related bacterial species groups [10]. Researchers have mostly relied on sequence analysis of protein-coding housekeeping genes such as RNA polymerase alpha subunit (rpoA) and phenylalanyl t-RNA synthase alpha subunit (pheS) to differentiate LAB species, which are closely related, of the Lactobacillus, Leuconostoc, Weissella, Enterococcus, and Pediococcus genera [11,12,13,14,15,16,17,18,19,20,21]. A previously executed piece of research recommended the use of no less than two additional phylogenetic markers for accurate isolate identification, particularly for the description of novel species in the Lactobacillus genus [22]. In addition, higher-level taxonomy and phylogeny can be determined through the execution of multilocus sequence analysis (MLSA) involving the concatenation of several housekeeping genes (typically five or more; [23]). Nevertheless, MLSA is known to exhibit a few disadvantages, including the absence of a universal threshold for species definition, the lack of a common set of genes, and the excessive expense, labor, and time necessary when using various series of housekeeping genes [24,25,26,27,28]. In the current genomic era, prokaryotic taxonomy depends mainly on comparative genome sequence analysis [29,30,31,32]. As a minimum standard requirement, researchers have proposed species-level delineation through the calculation of overall genome-related indices, including average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH; [33,34,35]). Although whole-genome sequencing (WGS) constitutes a credible taxonomic information source, it can still be costly, time-consuming, and difficult to apply. Thus, developing a rapid and cost-effective identification and discrimination method that mainly relies on the most informative region of genome sequence data is reasonable and necessary.
In this study, we explored genome-wide taxonomic markers with high discrimination power for accurately identifying species in the L. plantarum group. We executed the direct sequencing of polymerase chain reaction (PCR) products through Sanger’s method for resolution validation by targeting the most distinguished phylogenetic marker of mutL. Moreover, a novel L. pentosus subspecies was differentiated according to genome-based and phenotypic characteristics.
2. Materials and Methods
2.1. Genome-Based Mining of Taxonomic Markers for Species-Level Discrimination within L. plantarum Group
The standalone version of the cano-wgMLST tool [36] was applied in this genome-wide analysis. Nine whole genomes belonging to L. argentoratensis, L. garii, L. herbarum, L. modestisalitolerans, L. nangangensis, L. paraplantarum, L. pentosus, L. plajomi, and L. xiangfangensis were annotated using Prokka [37] to identify the coding sequences. The pan genome analysis was performed using Roary [38] with a 75% minimum BLASTP percentage identity for the loci identification in species-level. BLAST [39] was then used for allele calling with a minimum identity of 90% and coverage greater than 90% for the locus assignment (presence/absence profile) and exact match for the allele assignment (allele profile). The loci with the largest allele size were selected as the candidate taxonomic markers for species-level discrimination, and their pairwise sequence identity matrices were calculated using Clustal Omega [40].
2.2. L. plantarum Group Strains and Culture Conditions
The Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan) served as the source of most of the reference strains employed in this study (Table 1). The bacterial strains were cultivated on lactobacilli MRS agar (Difco Laboratories, Detroit, MI, USA) anaerobically for 24 h at 37 °C. Genomic DNA was extracted using the DNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocols.
Table 1.
Strains used in this study and specificity tests with mutL-targeting PCR assays.
| No. | Species | Strain No. | Other Destination | Species-Specific PCR Assays | |||
|---|---|---|---|---|---|---|---|
| spLplan-F/R | spLarg-F/R | spLpara-F/R | spLpen-F/R | ||||
| 1 | L. plantarum | BCRC10069T | ATCC 14917T | + | − | − | − |
| 2 | BCRC 10357 | ATCC 8014 | + | − | − | − | |
| 3 | BCRC 12251 | ATCC 10241 | + | − | − | − | |
| 4 | BCRC 14059 | ATCC 10012 | + | − | − | − | |
| 5 | BCRC 15478 | NCDO 1193 | + | − | − | − | |
| 6 | BCRC 18858 | NRIC 1943 | + | − | − | − | |
| 7 | BCRC 80061 | − | + | − | − | − | |
| 8 | BCRC 80222 | CECT 5787 | + | − | − | − | |
| 9 | BCRC 80578 | NCDO 772 | + | − | − | − | |
| 10 | BCRC 80580 | CICC 6026 | + | − | − | − | |
| 11 | BCRC 80581 | CICC 20764 | + | − | − | − | |
| 12 | BCRC 06B0002 | − | + | − | − | − | |
| 13 | BCRC 06B0006 | − | + | − | − | − | |
| 14 | BCRC 06B0023 | − | + | − | − | − | |
| 15 | BCRC 06B0036 | − | + | − | − | − | |
| 16 | BCRC 11B0079 | − | + | − | − | − | |
| 17 | BCRC 11B0117 | − | + | − | − | − | |
| 18 | BCRC 11B0122 | − | + | − | − | − | |
| 19 | BCRC 11B0168 | − | + | − | − | − | |
| 20 | BCRC 11B0177 | − | + | − | − | − | |
| 21 | L. argentoratensis | BCRC17638T | DSM 16365T | − | + | − | − |
| 22 | BCRC 17639 | CCUG 50788 | − | + | − | − | |
| 23 | BCRC 17640 | CCUG 50789 | − | + | − | − | |
| 24 | L. paraplantarum | BCRC 17178T | DSM 10667T | − | − | + | − |
| 25 | BCRC 17970 | ATCC 10776 | − | − | + | − | |
| 26 | BCRC 17971 | ATCC 700210 | − | − | + | − | |
| 27 | BCRC 80221 | CECT 5783 | − | − | + | − | |
| 28 | L. pentosus | BCRC 11053T | ATCC 8041T | − | − | − | + |
| 29 | BCRC 17972 | LMG 9210 | − | − | − | + | |
| 30 | BCRC 80017 | JCM 8334 | − | − | − | + | |
| 31 | BCRC 80018 | JCM 8335 | − | − | − | + | |
| 32 | BCRC 06B0048 | − | − | − | − | + | |
| 33 | L. daoliensis | 116-1AT | NCIMB 15181 | − | − | − | − |
| 34 | L. pingfangensis | 382-1T | NCIMB 15187 | − | − | − | − |
| 35 | L. daowaiensis | 203-3T | NCIMB 15183 | − | − | − | − |
| 36 | L. nangangensis | 381-7T | NCIMB 15186 | − | − | − | − |
| 37 | L. herbarum | BCRC 80996T | DSM 100358T | − | − | − | − |
| 38 | L. fabifermentans | BCRC 18841T | LMG 24284T | − | − | − | − |
| 39 | L. plajomi | BCRC 80928T | NBRC 107233T | − | − | − | − |
| 40 | L. xiangfangensis | BCRC 80512T | LMG 26013T | − | − | − | − |
| 41 | L. modestisalitolerans | BCRC 80927T | NBRC 107235T | − | − | − | − |
| 42 | L. dongliensis | 218-3T | NCIMB 15184T | − | − | − | − |
| 43 | L. songbeiensis | 398-2T | NCIMB 15189T | − | − | − | − |
| 44 | L. mudanjiangensis | 11050T | LMG 27194T | − | − | − | − |
| 45 | L. acidophilus | BCRC 10695T | ATCC 4356T | − | − | − | − |
| 46 | L. casei | BCRC 10697T | ATCC 393T | − | − | − | − |
| 47 | L. crispatus | BCRC 14618T | ATCC 33820T | − | − | − | − |
| 48 | L. curvatus | BCRC 12189T | DSM 20019T | − | − | − | − |
| 49 | L. delbrueckii subsp. bulgaricus | BCRC 10696T | ATCC 11842T | − | − | − | − |
| 50 | L. gallinarum | BCRC 17266T | ATCC 33199T | − | − | − | − |
| 51 | L. gasseri | BCRC 14619T | ATCC 33323T | − | − | − | − |
| 52 | L. paracasei | BCRC 12248T | ATCC 25302T | − | − | − | − |
| 53 | L. rhamnosus | BCRC 10940T | ATCC 7469T | − | − | − | − |
| 54 | L. sakei | BCRC 14622T | ATCC 15521T | − | − | − | − |
| 55 | L. taiwanensis | BCRC 17755T | JCM 18086T | − | − | − | − |
| 56 | L. ultunensis | BCRC 17714T | DSM 16047T | − | − | − | − |
BCRC, Bioresource Collection and Research Center at Food Industry Research and Development Institute, Taiwan; +, PCR products with specific primer detected; −, PCR products with specific primer not detected.
2.3. Degenerate PCR Primer Design, Nucleotide Sequencing and Phylogenetic Analysis on mutL Gene
On comparison with the mutL gene sequence from the whole genome among the species of the L. plantarum group, the degenerate primers, LpmutL-F (5′-TSGAYGTSAAYGTKCAYCC-3′) and LpmutL-R (5′-ATGYGGRCARTTRAANGGAT-3′), were designed and targeted to the conserved region. PCRs were performed using 23 μL of sterile MilliQ water, 3 μL of 10× PCR buffer, 0.5 μL of denucleoside triphosphates (10 mM), 1.2 μL of forward primer (10 mM), 1.2 μL of reverse primer (10 mM), 1.5 U of Taq DNA polymerase (DreamTaq, Thermo Scientific, Waltham, MA, USA), and 1 μL of template DNA (100 ng/μL). The thermal protocol consisted of the following conditions: initial strand denaturation at 94 °C for 5 min, followed by 30 cycles at 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, with a final extension step at 72 °C for 7 min. The resulting amplicons were purified using a QIA quick PCR Purification Kit (Qiagen, Inc., Valencia, CA, USA) and sequenced using a BigDye Terminator v3.1 cycle-sequencing kit on a 3730 DNA Analyzer (Applied Biosystems and Hitachi, Foster City, CA, USA). All sequences were aligned using ClustalX version 1.8 [41], and the sequence identities were calculated using Clustal Omega. A phylogenetic tree was reconstructed using the software package mega version 7.0 [42]. The number of haplotypes was calculated using DnaSP version.5.1 [43].
2.4. Species-Specific Primer Design and Direct PCR-Based Identification
L. argentoratensis–, L. paraplantarum–, L. pentosus–, and L. plantarum–specific PCR primer sets were determined on the basis of mutL sequences using VectorNTI version 9.0 (Invitrogen, Carlsbad, CA, USA). Species-specific PCR testing was conducted on all L. plantarum group strains and 12 nontarget strains (Table 1). Regarding the steps constituting the thermal cycler protocol, initial strand denaturation was executed at 94 °C for 5 min; followed by 30 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min; and finally an extension step executed at 72 °C for 7 min.
Serial dilution and plating methods were used to isolate the LAB isolates from various sources (Supplementary Table S1), and were discriminated using a MALDI Microflex LT mass spectrometer (Bruker Daltonics, Bremen, Germany), as previously described [44]. This was followed by species-specific PCR and mutL sequencing.
2.5. WGS of BCRC 06B0048 and Calculation of dDDH and Phylogenomic Tree Analysis
Genomic DNA was extracted using the EasyPrepHY genomic DNA extraction kit (Biotools Co. Ltd., Taipei, Taiwan) in accordance with the manufacturer’s protocols. The draft genomes of strain BCRC 06B0048 was sequenced from an Illumina paired-end library with an average insert size of 350 bp by using an Illumina HiSeq4000 platform with the PE 150 strategy at Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The resulting raw reads were assembled de novo using CLC Genomics Workbench version 20.0 (QIAGEN). The dDDH value and the phylogenomic tree were estimated and reconstructed using the TYGS server [45].
2.6. Differentiation of Strain BCRC 06B0048 and BCRC 11053T Based on Biochemical and Chemotaxonomic Characteristics
Profiles of carbohydrate fermentation and enzymic activities were determined using the API 50 CH and API ZYM systems (bioMérieux, Marcy-l’Étoile, France), respectively, according to the manufacturer’s instructions. Whole-cell fatty acids were analyzed as fatty acid methyl esters (FAMEs) with the Sherlock Microbial Identification System (MIDI, Inc., Newark, NJ, USA), as described previously [46].
3. Results and Discussion
The gold standard marker for prokaryotic species identification has long been the 16S rRNA gene. Strains of a single species commonly exhibit 98.7% sequence identity with the 16S rRNA gene [47]. However, the 16S rRNA gene exhibits poor resolution in closely related species groups; for example, the Lentilactobacillus buchneri, Lacticaseibacillus casei, L. plantarum, and Latilactobacillus sakei groups [48,49,50,51,52]. Accordingly, researchers have employed comparative sequences of the housekeeping genes dnaJ, dnaK, hsp60, pheS, recA, rpoA, rpoB, and tuf to distinguish the members of the L. plantarum group [11,45,51,53,54]. Among them, recA exhibited the highest resolution at the interspecific level, along with relatively low sequence identities (75.6–93.0%; average: 80.6%; Supplementary Table S1). Moreover, researchers have established species-specific PCR primers that are targeted to the variable regions of the recA gene for species identification of L. argentoratensis, L. paraplantarum, L. pentosus, and L. plantarum [9,48]. However, because several phylogenetically closely related novel L. plantarum group species have been proposed in recent years [7,8], the usability and specificity of recA gene–based PCR and sequencing as well as species-specific primers should be reassessed through in silico methods. To identify highly distinguished taxonomic markers with deep-level phylogeny or species-specific genes according to the gain and loss of variable regions, the use of genome comparison methods have been applied to resolve the phylogenetically and phenotypically closely related species; this serves as an alternative to the conventional universal phylogenetic targets [55,56,57,58,59,60].
The genomic data of the L. plantarum group species exhibited sequences of high quality, as directly evidenced by the relatively small number of contigs (median, 48; Supplementary Table S2); we thus used the aforementioned data to execute our subsequent comparative genomic analyses. The pan genomic analysis of L. plantarum and its most closely related species was performed on the cano-wgMLST_BacCompare analytics platform; our derived results indicated that the pan genome contained 9382 genes, of which 13.12% (1231 genes) were present in all of the tested genomes. Subsequently, the 774 genes with the largest allele size were selected as the taxonomic markers, and target genes with high discrimination levels were developed by comparing their sequence identities, including maximum, minimum, and mean values within the L. plantarum group; the results revealed that csbB, morA, murI, mutL, ntpJ, rutB, trmK, ydaF, and yhhX genes were the most promising candidates (Supplementary Table S1). mutL encodes a DNA mismatch repair protein that has an essential role in the maintenance of genomic stability by correcting DNA replication error [61]. It is regarded as a crucial taxonomic marker for species- and strain-level differentiation in the L. casei group [44,62,63]. Thus, the discrimination power of mutL in the L. plantarum group merits further evaluation. A degenerate primer pair (LpmutL-F/R) was used to successfully amplify a PCR amplicon that comprised approximately of 1000 bp of mutL from L. plantarum group strains; this product was subsequently employed for sequencing. Among the type strains, the average sequence identity of mutL was 66.6% (61.6–85.6%), which was clearly superior to that of the 16S rRNA gene (96.7–100%; average: 98.4%) (Table 2) and conventional phylogenetic markers (Supplementary Table S1) and could be applied to effectively separate tested strains into various species clusters with a high bootstrap value in the phylogenetic tree (Figure 1). Consequently, species-specific primers for four species in the L. plantarum group were designed (Table 3); these primers could successfully generate a single specific amplicon (319, 115, 176, and 385 bp) when they were used in PCR processes with DNA from reference strains of L. argentoratensis, L. paraplantarum, L. plantarum, and L. pentosus (Table 1), with no cross-reaction against nontarget LAB species. Furthermore, 20 LAB isolates were isolated from various sources (animal feces, fermented foods, and silage), and MALDI-TOF MS indicated that 13 of them were related to L. plantarum; these LAB isolates were next identified to the species-level of L. plantarum by using mutL-based species-specific PCR and sequencing (Supplementary Table S3). On the other hand, all 33 L. plantarum strains could be assigned to a combination of 16 different haplotypes using mutL gene sequence, which was comparable with that of other conventional MLST targets to distinguishing between strains in L. plantarum species [64,65].
Table 2.
Identities of mutL and 16S rDNA sequences between the type strains of the L. plantarum group.
| No. | Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L. plantarum BCRC 10069T | 85.6 | 76.1 | 69.4 | 64.9 | 64.0 | 64.5 | 67.0 | 64.8 | 67.9 | 64.6 | 65.9 | 68.1 | 66.8 | 65.0 | 65.7 | 64.9 | |
| 2 | L. argentoratensis BCRC 17638T | 100 | 73.6 | 68.6 | 65.1 | 64.4 | 64.6 | 66.2 | 65.4 | 66.6 | 64.7 | 65.6 | 68.0 | 66.4 | 63.8 | 65.1 | 64.4 | |
| 3 | L. paraplantarum BCRC 17178T | 99.7 | 99.7 | 69.3 | 66.7 | 66.2 | 66.5 | 66.4 | 66.5 | 68.1 | 66.2 | 65.4 | 68.8 | 65.2 | 67.0 | 66.1 | 66.6 | |
| 4 | L. pentosus BCRC 11053T | 99.9 | 99.9 | 99.8 | 65.4 | 65.9 | 66.5 | 66.5 | 63.3 | 66.4 | 64.1 | 65.8 | 67.9 | 64.5 | 65.8 | 67.5 | 66.1 | |
| 5 | L. daoliensis 116-1AT | 99.0 | 99.0 | 99.0 | 99.1 | 71.6 | 65.6 | 65.9 | 73.8 | 63.7 | 66.4 | 64.5 | 64.3 | 65.1 | 66.0 | 66.3 | 65.9 | |
| 6 | L. pingfangensis 382-1T | 99.0 | 99.0 | 99.2 | 99.1 | 99.7 | 70.4 | 64.5 | 73.0 | 66.3 | 65.9 | 65.5 | 66.5 | 66.2 | 65.8 | 66.9 | 69.9 | |
| 7 | L. daowaiensis 203-3T | 98.9 | 98.9 | 98.8 | 98.9 | 98.5 | 98.5 | 66.0 | 66.1 | 64.3 | 68.2 | 62.7 | 66.1 | 67.0 | 70.2 | 69.5 | 71.1 | |
| 8 | L. garii FI11369T | 98.9 | 98.9 | 98.9 | 98.9 | 98.5 | 98.5 | 98.7 | 61.6 | 64.9 | 65.4 | 69.6 | 73.3 | 64.6 | 66.1 | 66.3 | 64.3 | |
| 9 | L. nangangensis 381-7T | 98.9 | 98.9 | 99.0 | 99.0 | 99.9 | 99.8 | 98.6 | 98.6 | 63.8 | 66.7 | 63.4 | 65.4 | 64.3 | 66.0 | 67.6 | 67.6 | |
| 10 | L. herbarum BCRC 80996T | 98.9 | 98.9 | 98.9 | 98.9 | 98.3 | 98.5 | 98.0 | 98.0 | 98.4 | 64.0 | 64.9 | 67.7 | 62.4 | 64.0 | 64.6 | 64.3 | |
| 11 | L. fabifermentans BCRC 18841T | 98.9 | 98.9 | 98.9 | 98.9 | 98.5 | 98.6 | 98.5 | 98.5 | 98.5 | 98.3 | 62.7 | 64.5 | 65.0 | 66.3 | 67.3 | 66.1 | |
| 12 | L. plajomi BCRC 80928T | 98.8 | 98.8 | 98.5 | 98.7 | 98.0 | 97.8 | 98.2 | 99.0 | 97.9 | 97.7 | 98.1 | 68.1 | 64.3 | 65.4 | 64.8 | 62.8 | |
| 13 | L. xiangfangensis BCRC 80512T | 98.7 | 98.7 | 98.7 | 98.8 | 99.0 | 99.0 | 98.4 | 99.4 | 99.1 | 98.0 | 98.5 | 98.7 | 65.6 | 68.0 | 70.5 | 68.6 | |
| 14 | L. modestisalitolerans BCRC 80927T | 98.5 | 98.5 | 98.4 | 98.5 | 97.8 | 97.7 | 97.8 | 98.7 | 97.8 | 97.5 | 97.5 | 98.9 | 98.4 | 67.0 | 67.7 | 66.0 | |
| 15 | L. dongliensis 218-3T | 98.0 | 98.0 | 97.8 | 97.9 | 97.9 | 98.1 | 98.3 | 97.6 | 98.0 | 97.3 | 97.5 | 96.9 | 97.9 | 96.8 | 80.2 | 69.9 | |
| 16 | L. songbeiensis 398-2T | 97.9 | 97.9 | 97.9 | 98.0 | 98.1 | 98.2 | 98.2 | 97.5 | 98.2 | 97.4 | 97.6 | 96.9 | 98.0 | 96.7 | 99.8 | 70.3 | |
| 17 | L. mudanjiangensis 11050T | 97.8 | 97.8 | 97.9 | 97.9 | 97.9 | 98.1 | 98.5 | 97.6 | 98.0 | 97.6 | 97.5 | 96.9 | 97.5 | 96.8 | 98.7 | 98.8 |
The values on the upper right are the similarities between mutL sequences, and the values on the lower left are the similarities between 16S rRNA sequences.
Figure 1.
Phylogenetic tree of 57 L. plantarum group strains based on mutL sequences. The tree was constructed with the neighbour-joining method. Genetic distances were computed by Kimura’s two-parameter model. L. casei was included as an outgroup. Only bootstrap percentages above 70% are shown (based on 1000 replications). The scale bar represents 0.1% sequence divergence.
Table 3.
Species-specific primers used in this study.
| No. | Primer | Target | Sequence (5′–3′) | Amplicon Size (bp) |
|---|---|---|---|---|
| 1 | spLarg-F | L. argentoratensis | CCTTTGGTGAACCCGCTGAA | 319 |
| spLarg-R | AGTTCGGCTAATAGTGGCAA | |||
| 2 | spLpara-F | L. paraplantarum | TCAGGTGGCGGATAAGACTAC | 115 |
| spLpara-R | GGTTGCCGAMGTGGCGTCA | |||
| 3 | spLpen-F | L. pentosus | CCTCCGCTGAACCAATCATG | 385 |
| spLpen-R | TTCAGGACATCACTGGTGGG | |||
| 4 | spLplan-F | L. plantarum | GCGRTTGTTCCGTCAGAAT | 176 |
| spLplan-R | CTTGCAGCCGTGCTGGTTT |
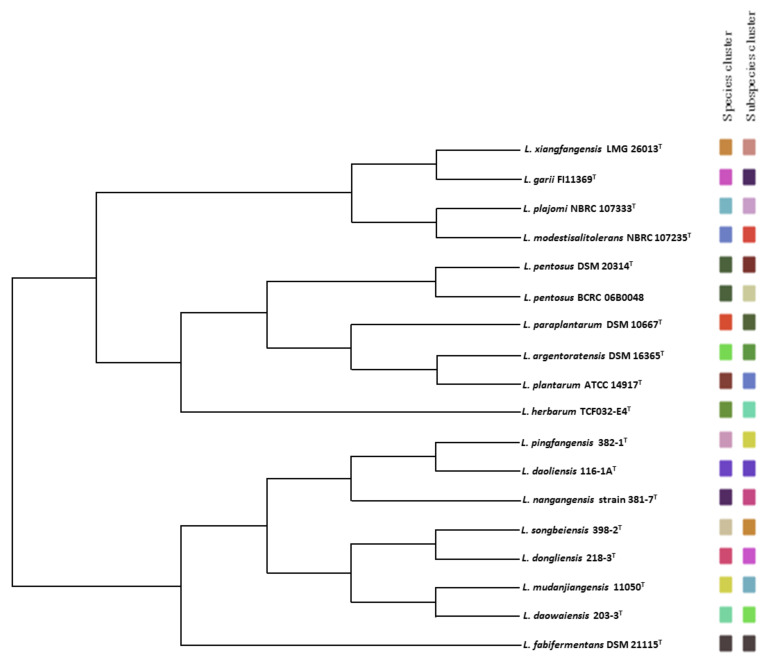
We note that the BCRC 06B0048 strain, which was originally identified as L. pentosus, had relatively low mutL sequence identities (97.0%) with the type strain DSM 20314T; moreover, the mutL-based phylogenetic tree indicates that the five strains of the species L. pentosus could be divided into two subclusters and that the nodes exhibited high bootstrap values (99%; Figure 1). Traditionally, subspecies-level differentiation mainly relied on the phenotypic and genotypic characteristics. Nevertheless, genome-based taxonomic designation for subspecies with the use of 79% dDDH has been suggested [66]. This approach has been successfully applied to propose and officially validate novel bacterial subspecies such as Bifidobacterium catenulatum, B. gallinarum B. thermacidophilum, and L. reuteri [67,68]. The draft genome size of the BCRC 06B0048 strain was 3.7 Mb (G+C content: 46.2 mol%), and it contained 3,293 protein-coding genes (Supplementary Table S2). The dDDH value between the BCRC 06B0048 and DSM 20314T strains was 78.1%, which is below the subspecies delineation threshold value. The phylogenomic tree revealed that the 17 type strains of the L. plantarum group were clearly differentiated into distinct species clusters, and the species L. pentosus could also be divided into two independent subclusters (Figure 2). In addition, strain BCRC 06B0048 has also shown several differential characteristics from the L. pentosus type strain BCRC 11053T (Supplementary Table S4). These results indicate that strain BCRC 06B0048 has the potential for reclassification as an independent subspecies of L. pentosus.
Figure 2.
Phylogenomic tree of the 18 L. plantarum group strains available on the TYGS database. The tree was inferred with FastME 2.1.6.1 from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The tree was rooted at the midpoint. Accession numbers of the genome sequences used for the reconstruction are shown in Supplementary Table S2.
4. Conclusions
In this study, we successfully applied a genome-based analysis method to assist in the selection of the promising target genes for a closely related species group by employing the cano-wgMLST_BacCompare analytics platform. The results suggest that the genome-wide taxonomic marker of mutL is an excellent phylogenetic target for precisely discriminating and identifying L. plantarum and related taxa through the use of direct sequencing as well as species-specific PCR assays. According to the genotypic and phenotypic characteristics, we deduced that strain BCRC 06B0048 may represent an undescribed subspecies of L. pentosus. We will endeavor to integrate polyphasic and combined with the genomic strategies to describe novel subspecies in the future.
Acknowledgments
We would like to thank Chii-Cherng Liao and Sung-Yuan Hsieh (Food Industry Research and Development Institute, Hsinchu, Taiwan) for their encouragement during the course of this research activity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081570/s1, Table S1: Interspecific analysis of the genome-based selection targets within the L. plantarum group. Table S2: Genomic characteristics of L. plantarum group type strains and BCRC 06B0048. Table S3: Identification of L. plantarum isolates from various sources using MALDI-TOF MS, species-specific PCR and mutL sequencing methods. Table S4: Differential characteristics between the L. pentosus BCRC 06B0048 and BCRC 11053T.
Author Contributions
Conceptualization, C.-H.H.; formal analysis, C.-H.H., C.-C.C., Y.-C.L., C.-H.C., A.-Y.L., J.-S.L. and C.-T.G.; funding acquisition, C.-H.H.; methodology, C.-H.H. and C.-C.C.; project administration, C.-H.H.; visualization, C.-H.H. and C.-C.C.; writing—original draft, C.-H.H.; writing—review and editing, C.-H.H. and L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Council of Agriculture, Taiwan, ROC (project no. COA 110AS-10.1.1-LI-L2) and Ministry of Economic Affairs, Taiwan, ROC (project no. 110-EC-17-A-22-0525).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequencing data has been uploaded to GenBank (KT591505~KT591231, MK473334~MK473363, and JAGHKR000000000).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harri H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 2.Tamang J.P., Holzapfel W.H., Watabane K. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016;7:377. doi: 10.3389/fmicb.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vries M.C., Vaughan E.E., Kleerebezem M., de Vos W.M. Lactobacillus plantarum-survival, functional and potential probiotic properties in the human gastrointestinal tract. Int. Dairy J. 2006;16:1018–1028. doi: 10.1016/j.idairyj.2005.09.003. [DOI] [Google Scholar]
- 4.Ray R.C., Joshi V.K. Fermented Foods: Past, present and future scenario. In: Ray R.C., Montet D., editors. Microorganisms and Fermentation of Traditional Foods. CRC Press; Boca Raton, FL, USA: 2015. pp. 1–36. [Google Scholar]
- 5.EFSA Panel on Biological Hazards (BIOHAZ) Ricci A., Allende A., Bolton D., Chemaly M., Davies R., Girones R., Koutsoumanis K., Herman L., Lindqvist R., et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: Suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 2020;18:6174. doi: 10.2903/j.efsa.2020.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venema K., Meijerink M. Lactobacilli as probiotics: Discovering new functional aspects and target sites. In: Venema K., do Carmo A.P., editors. Probiotics and Prebiotics: Current Research and Future Trends. Caister Academic Press; Norfolk, UK: 2015. pp. 29–42. [Google Scholar]
- 7.Diaz M., Sayavedra L., Atter A., Mayer M.J., Saha S., Amoa-Awua W., Narbad A. Lactobacillus garii sp. nov., isolated from a fermented cassava product. Int. J. Syst. Evol. Microbiol. 2020;70:3012–3017. doi: 10.1099/ijsem.0.004121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu D.D., Gu C.T. Lactobacillus pingfangensis sp. nov., Lactobacillus daoliensis sp. nov., Lactobacillus nangangensis sp. nov., Lactobacillus daowaiensis sp. nov., Lactobacillus dongliensis sp. nov., Lactobacillus songbeiensis sp. nov. and Lactobacillus kaifaensis sp. nov., isolated from traditional Chinese pickle. Int. J. Syst. Evol. Microbiol. 2019;69:3237–3247. doi: 10.1099/ijsem.0.003619. [DOI] [PubMed] [Google Scholar]
- 9.Bringel F., Castioni A., Olukoya D.K., Felis G.E., Torriani S., Dellaglio F. Lactobacillus plantarum subsp. argentoratensis subsp. nov., isolated from vegetable matrices. Int. J. Syst. Evol. Microbiol. 2005;55:1629–1634. doi: 10.1099/ijs.0.63333-0. [DOI] [PubMed] [Google Scholar]
- 10.Petti C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007;44:1108–1114. doi: 10.1086/512818. [DOI] [PubMed] [Google Scholar]
- 11.Naser S.M., Dawyndt P., Hoste B., Gevers D., Vandemeulebroecke K., Cleenwerck I., Vancanneyt M., Swings J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007;57:2777–2789. doi: 10.1099/ijs.0.64711-0. [DOI] [PubMed] [Google Scholar]
- 12.Naser S.M., Thompson F.L., Hoste B., Gevers D., Dawyndt P., Vancanneyt M., Swings J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology. 2005;151:2141–2150. doi: 10.1099/mic.0.27840-0. [DOI] [PubMed] [Google Scholar]
- 13.De Bruyne K., Camu N., De Vuyst L., Vandamme P. Weissella fabaria sp. nov., from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 2010;60:1999–2005. doi: 10.1099/ijs.0.019323-0. [DOI] [PubMed] [Google Scholar]
- 14.De Bruyne K., Franz C.M., Vancanneyt M., Schillinger U., Mozzi F., de Valdez G.F., de Vuyst L., Vandamme P. Pediococcus argentinicus sp. nov. from Argentinean fermented wheat flour and identification of Pediococcus species by pheS, rpoA and atpA sequence analysis. Int. J. Syst. Evol. Microbiol. 2008;58:2909–2916. doi: 10.1099/ijs.0.65833-0. [DOI] [PubMed] [Google Scholar]
- 15.De Bruyne K., Schillinger U., Caroline L., Boehringer B., Cleenwerck I., Vancanneyt M., De Vuyst L., Franz C.M.A.P., Vandamme P. Leuconostoc holzapfelii sp. nov., isolated from Ethiopian coffee fermentation and assessment of sequence analysis of housekeeping genes for delineation of Leuconostoc species. Int. J. Syst. Evol. Microbiol. 2007;57:2952–2959. doi: 10.1099/ijs.0.65292-0. [DOI] [PubMed] [Google Scholar]
- 16.Chao S.H., Huang H.Y., Kang Y.H., Watanabe K., Tsai Y.C. The diversity of lactic acid bacteria in a traditional Taiwanese millet alcoholic beverage during fermentation. LWT Food Sci. Technol. 2013;51:135–142. doi: 10.1016/j.lwt.2012.09.015. [DOI] [Google Scholar]
- 17.Chao S.H., Kudo Y., Tsai Y.C., Watanabe K. Lactobacillus futsaii sp. nov., isolated from traditional fermented mustard products of Taiwan, futsai and suan-tsai. Int. J. Syst. Evol. Microbiol. 2012;62:489–494. doi: 10.1099/ijs.0.030619-0. [DOI] [PubMed] [Google Scholar]
- 18.Chao S.H., Sasamoto M., Kudo Y., Fujimoto J., Tsai Y.C., Watanabe K. Lactobacillus odoratitofui sp. nov., isolated from stinky tofu brine. Int. J. Syst. Evol. Microbiol. 2010;60:2903–2907. doi: 10.1099/ijs.0.019307-0. [DOI] [PubMed] [Google Scholar]
- 19.Oki K., Kudo Y., Watanabe K. Lactobacillus saniviri sp. nov. and Lactobacillus senioris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012;62:601–607. doi: 10.1099/ijs.0.031658-0. [DOI] [PubMed] [Google Scholar]
- 20.Nyanzi R., Jooste P.J., Cameron M., Witthuhn C. Comparison of rpoA and pheS gene sequencing to 16S rRNA gene sequencing in identification and phylogenetic analysis of LAB from probiotic food products and supplements. Food Biotechnol. 2013;27:303–327. doi: 10.1080/08905436.2013.838783. [DOI] [Google Scholar]
- 21.Huang C.H., Liou J.S., Lee A.Y., Tseng M., Miyashita M., Huang L., Watanabe K. Polyphasic characterization of a novel species in the Lactobacillus casei group from cow manure of Taiwan: Description of L. chiayiensis sp. nov. Syst. Appl. Microbiol. 2018;41:270–278. doi: 10.1016/j.syapm.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Mattarelli P., Holzapfel W., Franz C.M., Endo A., Felis G.E., Hammes W., Pot B., Dicks L., Dellaglio F. Recommended minimal standards for description of new taxa of the genera Bifidobacterium, Lactobacillus and related genera. Int. J. Syst. Evol. Microbiol. 2014;64:1434–1451. doi: 10.1099/ijs.0.060046-0. [DOI] [PubMed] [Google Scholar]
- 23.Glaeser S.P., Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015;38:237–245. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Mulet M., Lalucat J., Valdes E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 25.Lai Q., Liu Y., Yuan J., Du J., Wang L., Sun F., Shao Z. Multilocus sequence analysis for assessment of phylogenetic diversity and biogeography in Thalassospira bacteria from diverse marine environments. PLoS ONE. 2014;9:e106353. doi: 10.1371/journal.pone.0106353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L., Wieme A., Spitaels F., Balzarini T., Nunes O.C., Manaia C.M., Landchoot A.V., De Vuyst L., Cleenwerck I., Vandamme P. Acetobacter sicerae sp. nov., isolated from cider and kefir, and identification of species of the genus Acetobacter by dnaK, groEL and rpoB sequence analysis. Int. J. Syst. Evol. Microbiol. 2014;64:2407–2415. doi: 10.1099/ijs.0.058354-0. [DOI] [PubMed] [Google Scholar]
- 27.Rosselló-Móra R., Amann R. Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 2015;38:209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Peeters C., Meier-Kolthoff J.P., Verheyde B., De Brandt E., Cooper V.S., Vandamme P. Phylogenomic study of Burkholderia glatheilike organisms, proposal of 13 novel Burkholderia species and emended descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front. Microbiol. 2016;7:877. doi: 10.3389/fmicb.2016.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun J., Rainey F.A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014;64:316–324. doi: 10.1099/ijs.0.054171-0. [DOI] [PubMed] [Google Scholar]
- 30.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E., Raoult D., Fournier P.E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int. J. Syst. Evol. Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme P., Peeters C. Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek. 2014;106:57–65. doi: 10.1007/s10482-014-0148-x. [DOI] [PubMed] [Google Scholar]
- 32.Zong Z. Genome-based Taxonomy for Bacteria: A Recent Advance. Trends Microbiol. 2020;28:871–874. doi: 10.1016/j.tim.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., Rooney A.P., Yi H., Xu X.W., De Meyer S., et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 34.Goris J., Konstantinidis T.K., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 35.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y.Y., Lin J.W., Chen C.C. cano-wgMLST_BacCompare: A Bacterial Genome Analysis Platform for Epidemiological Investigation and Comparative Genomic Analysis. Front. Microbiol. 2019;10:1687. doi: 10.3389/fmicb.2019.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W.Z., Lopez R., McWilliam H., Remmert M., Soding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 44.Huang C.H., Huang L. Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools. J. Dairy Sci. 2018;101:979–991. doi: 10.3168/jds.2017-13642. [DOI] [PubMed] [Google Scholar]
- 45.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of- the- art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chern L.L., Stackebrandt E., Lee S.F., Lee F.L., Chen J.K., Fu H.M. Chitinibacter tainanensis gen. nov., sp. nov., a chitin-degrading aerobe from soil in Taiwan. Int. J. Syst. Evol. Microbiol. 2004;54:1387–1391. doi: 10.1099/ijs.0.02834-0. [DOI] [PubMed] [Google Scholar]
- 47.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 48.Torriani S., Clementi F., Vancanneyt M., Hoste B., Dellaglio F., Kersters K. Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 2001;24:554–560. doi: 10.1078/0723-2020-00071. [DOI] [PubMed] [Google Scholar]
- 49.Koort J., Vandamme P., Schillinger U., Holzapfel W., Bjorkroth J. Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int. J. Syst. Evol. Microbiol. 2004;54:1621–1626. doi: 10.1099/ijs.0.63164-0. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe K., Fujimoto J., Tomii Y., Sasamoto M., Makino H., Kudo Y., Okada S. Lactobacillus kisonensis sp. nov., Lactobacillus otakiensis sp. nov. Lactobacillus rapi sp. nov. and Lactobacillus sunkii sp. nov., four novel heterofermentative species isolated from sunki, a Japanese traditional pickle. Int. J. Syst. Evol. Microbiol. 2009;59:754–760. doi: 10.1099/ijs.0.004689-0. [DOI] [PubMed] [Google Scholar]
- 51.Huang C.H., Lee F.L., Liou J.S. Rapid discrimination and classification of the Lactobacillus plantarum group based on a partial dnaK sequence and DNA fingerprinting techniques. Antonie Van Leeuwenhoek. 2010;97:289–296. doi: 10.1007/s10482-009-9409-5. [DOI] [PubMed] [Google Scholar]
- 52.Huang C.H., Lee F.L. The dnaK gene as a molecular marker for the classification and discrimination of the Lactobacillus casei group. Antonie Van Leeuwenhoek. 2011;99:319–327. doi: 10.1007/s10482-010-9493-6. [DOI] [PubMed] [Google Scholar]
- 53.Huang H., Huang L., Wu C.P., Chang M.T. Molecular discrimination of Lactobacillus plantarum group using comparative sequence analysis of the dnaJ gene and as a target for developing novel species-specific PCR primers. J. Chin. Soc. Anim. Sci. 2016;45:45–55. [Google Scholar]
- 54.Yu J., Sun Z., Liu W., Bao Q., Zhang J., Zhang H. Phylogenetic study of Lactobacillus acidophilus group, L. casei group and L. plantarum group based on partial hsp60, pheS and tuf gene sequences. Eur. Food Res. Technol. 2014;234:927–934. doi: 10.1007/s00217-012-1712-0. [DOI] [Google Scholar]
- 55.Conti A., Corte L., Pierantoni D.C., Robert V., Cardinali G. What Is the Best Lens? Comparing the Resolution Power of Genome-Derived Markers and Standard Barcodes. Microorganisms. 2021;2:299. doi: 10.3390/microorganisms9020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grana-Miraglia L., Arreguin-Perez C., Lopez-Leal G., Munoz A., Perez-Oseguera A., Miranda-Miranda E., Cossío-Bayúgar R., Castillo-Ramírez S. Phylogenomics picks out the par excellence markers for species phylogeny in the genus Staphylococcus. PeerJ. 2018;6:e5839. doi: 10.7717/peerj.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong W., Liu H., Xu C., Zuo Y., Chen Z., Zhou S. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genet. 2014;15:138. doi: 10.1186/s12863-014-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H.E., Kim E., Yang S.M., Lee S., Kim M.J., Kim H.Y. Development of Real-Time PCR Assay to Specifically Detect 22 Bifidobacterium Species and Subspecies Using Comparative Genomics. Front. Microbiol. 2020;11:2087. doi: 10.3389/fmicb.2020.02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim E., Yang S.M., Cho E.J., Kim H.Y. Novel real-time PCR assay for Lactobacillus casei group species using comparative genomics. Food Microbiol. 2020;90:103485. doi: 10.1016/j.fm.2020.103485. [DOI] [PubMed] [Google Scholar]
- 60.Kim E., Yang S.M., Lim B., Park S.H., Rackerby B., Kim H.Y. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 2020;20:96. doi: 10.1186/s12866-020-01781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 62.Bottari B., Felis G.E., Salvetti E., Castioni A., Campedelli I., Torriani S., Bernini V., Gatti M. Effective identification of Lactobacillus casei group species: Genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology. 2017;163:950–960. doi: 10.1099/mic.0.000497. [DOI] [PubMed] [Google Scholar]
- 63.Cai H., Rodriguez B.T., Zhang W., Broadbent J.R., Steele J.L. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology. 2007;153:2655–2665. doi: 10.1099/mic.0.2007/006452-0. [DOI] [PubMed] [Google Scholar]
- 64.de las Rivas B., Marcobal A., Muñoz R. Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiology. 2006;152:85–93. doi: 10.1099/mic.0.28482-0. [DOI] [PubMed] [Google Scholar]
- 65.Xu H., Liu W., Zhang W., Yu J., Song Y., Menhe B., Zhang F., Sun Z. Use of multilocus sequence typing to infer genetic diversity and population structure of Lactobacillus plantarum isolates from different sources. BMC Microbiol. 2015;15:241. doi: 10.1186/s12866-015-0584-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A., Rohde C., Rohde M., Fartmann B., Goodwin L.A., et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nouioui I., Carro L., Garcia-Lopez M., Meier-Kolthoff J.P., Woyke T., Kyrpides N.C., Pukall R., Klenk H.R., Goodfellow M., Göker M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018;9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li F., Cheng C.C., Zheng J., Liou J., Quevedo R.M., Li J., Roos S., Gänzle M.G., Walter J. Limosilactobacillus balticus sp. nov., Limosilactobacillus agrestis sp. nov., Limosilactobacillus albertensis sp. nov., Limosilactobacillus rudii sp. nov. and Limosilactobacillus fastidiosus sp. nov., five novel Limosilactobacillus species isolated from the vertebrate gastrointestinal tract, and proposal of six subspecies of Limosilactobacillus reuteri adapted to the gastrointestinal tract of specific vertebrate hosts. Int. J. Syst. Evol. Microbiol. 2021;71:004644. doi: 10.1099/ijsem.0.004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data has been uploaded to GenBank (KT591505~KT591231, MK473334~MK473363, and JAGHKR000000000).