Abstract
Ultrasensitive, versatile sensors for molecular biomarkers are a critical component of disease diagnostics and personalized medicine as the COVID-19 pandemic has revealed in dramatic fashion. Integrated electrical nanopore sensors can fill this need via label-free, direct detection of individual biomolecules, but a fully functional device for clinical sample analysis has yet to be developed. Here, we report amplification-free detection of SARS-CoV-2 RNAs with single molecule sensitivity from clinical nasopharyngeal swab samples on an electro-optofluidic chip. The device relies on optically assisted delivery of target carrying microbeads to the nanopore for single RNA detection after release. A sensing rate enhancement of over 2,000x with favorable scaling towards lower concentrations is demonstrated. The combination of target specificity, chip-scale integration and rapid detection ensures the practicality of this approach for COVID-19 diagnosis over the entire clinically relevant concentration range from 104-109 copies/mL.
Keywords: SARS-CoV-2 RNAs, Solid-state nanopores, Nasopharyngeal swab samples, Single-molecule detection, Biosensors
1. Introduction
High performance molecular biosensors will form a core part of 21st century medicine. The ongoing COVID-19 pandemic has reinforced this notion in most dramatic fashion and, indeed, a recent survey has laid out the most desirable characteristics such a diagnostic tool should possess (Tong et al., 2021). These include excellent sensitivity and specificity, fast turnaround time, the ability to handle multiple diseases and multiple target types, and sufficiently low complexity to be suitable for point-of-care use. It has been challenging to implement all these traits in a single device due to their seeming incompatibility. This situation is reflected clearly in the COVID pandemic where relatively complex PCR based tests provide accurate and sensitive gold standard laboratory diagnostics while simpler antigen tests are used at the point of care, but at the price of lower sensitivity and reliability. Emerging approaches such as RNA detection via CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) offer high sensitivity but currently still require amplification-based processes to achieve this metric (Joung et al., 2020). The most promising approach to combining high sensitivity with low complexity would be the direct, label-free detection of individual molecular targets. Nanopore sensors are based on the electrical detection of single particles as they move through a nanoscopic opening in a membrane and have the potential to form the basis of such a long sought-after universal sensor. The current blockade sensing principle for molecular analysis was introduced in 1989 (Deamer et al., 2016), and the bulk of research and development was dedicated to the development of a next generation sequencing technology based on distinguishing the current signature produced by different nucleotides (Ashkenasy et al., 2005; Deamer and Akeson, 2000; Deamer and Branton, 2002; Kasianowicz et al., 1996). More recently, the potential of nanopores as single molecule detectors for a several diverse biomarker classes including nucleic acids, proteins, and small molecules has been recognized (Chien et al., 2019; Fologea et al., 2005; Garalde et al., 2018; Kowalczyk et al., 2010; Li et al., 2013; Liu et al., 2014; Rahman et al., 2019b; Shasha et al., 2014; Skinner et al., 2009; Wanunu et al., 2011; Wei et al., 2012; Yusko et al., 2017). However, the nanopore sensing paradigm poses its own set of challenges for creating a realistic diagnostic tool. The most prominent ones are ensuring specificity of the translocation signal for the desired target and efficient delivery of the targets to within the tiny (few micron) capture radius of the pore in which the electric field is strong enough to pull the targets through the opening (Chuah et al., 2019; Rahman et al., 2019a; Spitzberg et al., 2019). A few mitigation strategies such as concentration gradients and target delivery via magnetic nanoparticles have been demonstrated (Chuah et al., 2019; Wanunu et al., 2010), but none have been translated to complete diagnostic assays.
In order to create an integrated, label-free, amplification-free nanopore sensor that is capable of analyzing patient samples at clinically relevant concentrations, we use a new approach called optical trapping-assisted capture rate enhancement (TACRE (Rahman et al., 2019a)). It relies on a dual-beam optical trap of counterpropagating laser beams in liquid-core waveguides (Kühn et al., 2009) to position target-carrying microbeads within the capture radius of a nanopore. After applying heat to the nanopore chip, the bond between the pulldown sequence and the target nucleic acid is broken. The thermally released individual nucleic acids can be detected in rapid succession by the nanopore at an 80x increased rate compared to diffusion-limited capture from a bulk solution. Here, we use the TACRE approach to demonstrate the rapid, label-free, and amplification-free detection of SARS-CoV-2 RNAs with single molecule sensitivity on a nanopore electro-optofluidic chip. Specifically, RNAs were successfully detected from clinical nasopharyngeal swab samples, and a dynamic range covering the entire clinically relevant concentration range from 109 copies/mL to 104 copies/mL (or 17 aM) is reported. It is worth noting that the clinical viral load in posterior oropharyngeal saliva samples, which was ascertained by reverse transcriptase quantitative PCR (RT-qPCR), ranges from 104 to 106 copies/mL (To et al., 2020). The capture rate enhancement as a function of concentration is examined and shows favorable scaling with concentration, reaching values of over 2,000x. Consequently, this electro-optofluidic platform can serve as the basis of a new generation of true label-free, low complexity, high sensitivity diagnostic instruments.
2. Material and methods
2.1. Nanopore optofluidic device fabrication
The ARROW (Anti-Resonant Reflecting Optical Waveguide) optofluidic devices were fabricated on a 100 mm diameter, <100> oriented Si substrate. A sequence of alternating dielectric layers of SiO2 (n = 1.47) and Ta2O5 (n = 2.107) with respective thicknesses of 265 nm and 102 nm were created via sputtering over the substrate to form the ARROW layer. SU-8 photoresist was spun on top of the ARROW layer and then patterned to form the 6 μm × 12 μm (height x width) sacrificial layer for the microfluidic channel. The photoresist pattern was then hard-baked at 250 °C for 5 min. Afterwards, a self-aligning pedestal was formed by reactive ion etching (RIE) and a 6 μm thick low stress PECVD silicon dioxide layer was deposited on top of SU-8 pattern to form the wall of the microfluidic channel. The 5 μm tall solid-core waveguide intersecting the microfluidic channel was fabricated by reactive ion etching (RIE). The two ends of the microfluidic channel were opened by removing the oxide with wet etching, then a mixture of strong acids was used to remove the SU-8 to create the hollow microfluidic channel. The detailed step-by-step ARROW optofluidic platform fabrication process can be found in supplementary section 2 (Lunt et al., 2010).
To enable the fabrication of a nanopore in the microfluidic channel, a terraced microwell was first milled into the 6 μm thick top oxide using an FEI Quanta 3D FEG Dualbeam, as shown in Fig. 1 c and as described in detail in supplementary section 3. The nanopore was milled into the remaining thin oxide layer using a 30 kV, 1.6–10 pA ion beam controlled by the Nanometer Pattern Generation System (JC Nabity). The microwell and nanopore were milled as close as possible to the trapping region to ensure immediate capture of targets upon their release from the carrier beads.
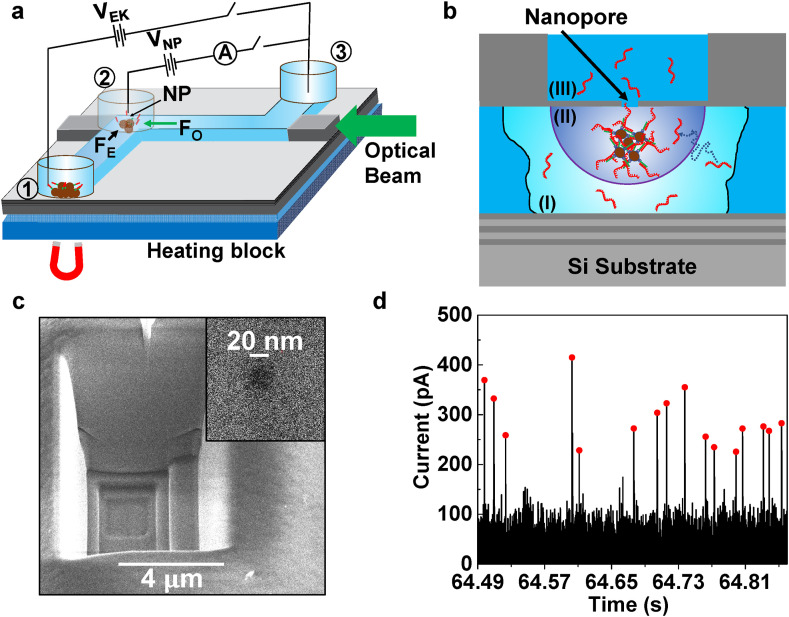
Fig. 1.
Optical trapping assisted nanopore capture rate enhancement (TACRE) platform. a, Optofluidic chip with connected solid-core (SC, gray) and liquid-core (LC, blue) waveguides. Reservoirs are attached over channel outlets and nanopore (NP) location. Voltages VEK and VNP are applied for electrokinetic delivery of bead-bound targets to the nanopore and translocation of released targets through nanopore, respectively. A light beam guided through the LC waveguide traps and collects carrier microbeads at the nanopore location. b, Closeup of TACRE process at NP location: targets are released from the beads while beads are trapped by the optical beam in the fluidic channel. Region (I): targets inside this region at the beginning of the experiment can diffuse to the nanopore capture volume within the duration of the experiment. Region (II): nanopore capture volume for particle translocations. c, SEM image of a terraced micro-well to create thin membrane for ion milling of 20 nm nanopore (inset). d, Baseline-corrected nanopore current for SARS-CoV-2 RNA segments translocating through the pore. Red dots mark individual RNA molecules identified by custom peak-finding algorithm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Electronic circuitry for thermal target release
A temperature controller (LDC 3724B, ILX Lightwave) was used to control the temperature of a Peltier heater (TES1 12703, Hebei I.T. (Shanghai) Co., Ltd). To implement the PID algorithm, a 10 kΩ thermistor as a feedback sensor was placed on the heater surface and connected with the controller circuit.
2.3. Nanopore current signal analysis
The nanopore current signal was processed by a sensitive current amplifier (Axopatch 200B, Molecular Device) with a low pass filter (cut off frequency of 10 KHz) and simultaneously recorded at 250KSa/s sampling rate with a data acquisition module (Digidata 1440A, Molecular Device). The current trace was later analyzed by a custom MATLAB program, which can distinguish translocation events from other ionic current fluctuations and electrical noise. First, the program calculates the standard deviation of the nanopore baseline current and then initializes the program with a user defined detection threshold, which is defined here as 4X the standard deviation of the background ionic current. In each cycle, a cluster of sampled data points are considered collectively to downsample the signal and the mean value of the cluster is considered as the new data point. This new data point is compared with the previously calculated reference data point, and if the difference does not cross the threshold then the reference point is updated with a proportional control algorithm. As soon as the threshold is crossed, the program tags this event as a possible translocation event and then calculates the peak height. When the current falls back below a predefined threshold within a user defined time (~10X longer than the expected translocation dwell time), the program saves the duration between this event and the threshold crossing event as the translocation dwell time, confirming it as a real translocation event. If the threshold is crossed by the nanopore current just because of a random baseline shifting event, the ionic current amplitude remains higher than the predefined threshold value within the user defined time. As a result, the program will discard the event as a timeout event.
2.4. Details of the SARS-CoV-2 samples and the pulldown sequence design
Synthetic RNA samples including fragments from ORF 1ab, Envelope, and Nucleocapsid region of the SARS-CoV-2 genome were purchased from American Type Culture Collection (ATCC: VR-3276SD). The stock concentration ranged from 1 × 105 to 1 × 106 copies/mL.
Biotinylated synthetic capture pulldown probes (14-mer:/5BiotinTEG/CATTTCGCTGATTT) were obtained from Integrated DNA Technologies, Inc. (IDT). They were designed to complement a part of the ORF 1ab region. (nt. 28,294-28,307; Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome, NCBI Reference Sequence: NC_045512.2). The simulated melting temperature of this pulldown segment in 50 mM Na+ salt solution is 35.1 °C (http://biotools.nubic.northwestern.edu/OligoCalc.html). The pulldown oligo ensures specificity of the sample preparation process and is designed here to enable thermal release of the target particles at moderate temperatures.
1-μm diameter streptavidin coated magnetic beads (4 mg/mL) were purchased from New England Biolabs, Inc. The binding capacity of this magnetic bead is more than 500 pico-moles of single-stranded 20bp biotinylated oligonucleotide per mg.
2.5. Experimental setup and TACRE implementation
Fig. 1a shows a schematic view of the nanopore sensor and the experimental setup. At its heart is an 8 × 8 mm silicon chip on which six layers of alternating SiO2 and Ta2O5 were deposited to enable optical waveguiding in low index layers based on the ARROW principle (Duguay et al., 1986). On top of these layers, solid-core (SC) ARROWs (shown in gray) and a 5 μm × 12 μm microfluidic channel (shown in blue) that also functions as a liquid-core (LC) ARROW waveguide are defined using SiO2 deposition and a sacrificial layer process (Measor et al., 2011; Yin et al., 2004). A nanopore with 20 nm diameter was milled with a focused ion beam into the SiO2 layer over the liquid channel right next to the SC-LC interface at the end of the horizontal channel section (see Fig. 1a). More details of the device fabrication are provided in the Methods section. Three fluid reservoirs were attached at the ends of the liquid channel (#1 and #3) and over the nanopore (#2), respectively. Reservoirs were attached to the device with a thin layer of thermal curing glue. They allow for introduction of liquids and particles into the channel and facilitate the insertion of Ag/AgCl electrodes (each electrode was vertically inserted into each reservoir) to apply an electrokinetic voltage VEK across the channel and a nanopore voltage VNP for inducing translocations, respectively.
The optical trapping-assisted capture rate enhancement (TACRE) approach is implemented by delivering microbeads that carry target molecules (here SARS-CoV-2 RNAs) from reservoir 1 into the channel and then optically trapping them directly under the nanopore location. In this way, all targets reside within the high-field capture radius of the nanopore at dramatically increased local concentration (region II in Fig. 1b) and can be drawn rapidly and efficiently through the nanopore upon thermal release. This is in stark contrast to detection of targets from bulk solution, where only a few targets that diffuse from region (I) into the capture volume are detected. In order to reach the attomolar concentration levels found in clinical SARS-CoV-2 samples, the process needs to be efficient in target delivery, trapping at the pore, and target release. For optimized carrier bead delivery to the trapping region, we use a magnet to attract the magnetic carrier beads to the bottom of the reservoir 1, where they can be drawn into the fluidic channel by the electrokinetic force applied via VEK. Electrophoretic transport of beads to the nanopore is faster and more controllable than purely pressure-based flow, and a detailed analysis is given in supplementary section 1. For efficient and simple trapping of the beads at the nanopore, we use a single optical beam to trap the carrier beads under the nanopore. As shown in Fig. 1a, a single-mode fiber is used to couple a laser beam (532 nm, Lighthouse Photonics) into the SC-waveguide at the edge of the chip with subsequent propagation in the adjoining section of the LC waveguide. The scattering force of the beam, F O, is the result of momentum transfer from photons that scatter off of the magnetic beads and overcomes the electrokinetic force, F E, and traps the beads against the channel wall. This approach provides a reproducible trapping location and a simplified optical setup compared to dual-beam or tweezer traps. Note that the optical force is only present in the horizontal channel section and, therefore, does not affect the continued electrokinetic delivery from reservoir 1 into this region. When a desired number of magnetic beads have been trapped under the nanopore, VEK is turned off and the beads remain immobilized due to the optical trapping force. Third, efficient capture of released target molecules by the nanopore is ensured by simultaneous application of heat on the heating block to release targets from the beads and the nanopore voltage VNP that initiates the sensing process. The absence of a delay between the release and sensing steps minimizes the possibility of target molecules escaping into the diffusion-limited region, making larger trapping enhancement factors possible as will be described below.
Using this approach, molecular targets are presented to and detected by the nanopore in label-free and amplification-free fashion as shown by the representative current trace in Fig. 1d where translocations from individual RNA targets are observed with good signal-to-noise ratio and marked with red dots. Events were identified by a custom threshold algorithm that is described in more detail in the Methods section. Before we turn to a detailed discussion of the results using clinical SARS-CoV-2 samples, we address the up-stream assay design that ensures specificity of the nanopore sensor.
2.6. SARS-CoV-2 assay design
When used as a molecular particle counter, nanopore sensors lack the specificity to reliably identify a particular target. In principle, different molecules produce different current blockade signatures, but in practice the variation in their signals is too large to ensure accurate analysis, especially in complex solutions such as bodily fluids. In order to preserve the label-free detection paradigm, a sample preparation process is required that ensures that the nanopore is only exposed to the target molecules. We use a modified version of a well-established magnetic bead-based solid phase extraction (SPE) method to provide the required specificity (Cai et al., 2016; Parks et al., 2014). Bead-based SPE is ideally suited for the TACRE assay as targets are selectively bound to the very microbeads that act as carriers to deliver them to the nanopore, and because SPE can be applied to different molecular target types, including nucleic acids, proteins, etc. (Meena et al., 2018). Fig. 2 illustrates the key elements of the SPE sample preparation process that was implemented for this work while a full step-by-step description is provided in the supplementary material. Streptavidin coated 1-μm magnetic beads (New England Biolabs, Inc.) were functionalized with 14-bp long biotinylated pulldown oligonucleotides specific to the ORF 1ab region of the SARS-CoV-2 genome (Fig. 2a). The specificity of this pulldown sequence to SARS-CoV-2 RNAs was verified by a negative control experiment with Zika virus nucleic acids and showed no false positives (Meena et al., 2021). A relatively short pulldown sequence was chosen for compatibility with a moderate target release temperature at the nanopore of 35.1 °C. To prepare the beads for the assay, a 5 μL aliquot of the beads at stock concentration (4 mg/mL) was washed 3x with pre-filtered 1X T50 (50 mM NaCl, 10 mM Tris, pH 6.5) buffer solution and then 0.5 μL of 100 μM biotinylated 14-bp pulldown sequence (purchased from IDT) was added with 6x molar excess to saturate the binding sites of each bead. The mixture was then kept on the rotary mixture at room temperature for 1 h. After the incubation process, unattached pulldown sequences were removed by washing and the magnetic beads were resuspended in 5 μL of 1X T50 solution.
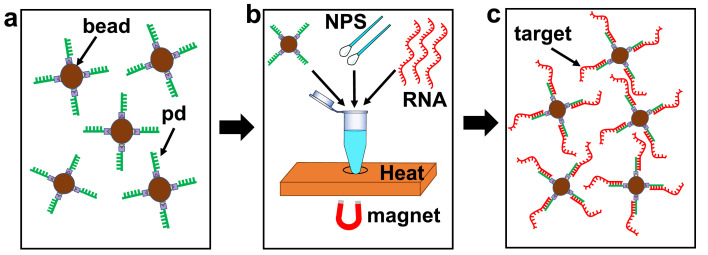
Fig. 2.
Flowchart of the magnetic-bead-based target extraction and preconcentration method. a, Magnetic carrier microbeads with 14 base pair long pulldown oligonucleotides. pd: pull-down sequence. b, SARS-CoV-2 negative human nasopharyngeal swab (NPS) solution and synthetic SARS-CoV-2 RNA solution is mixed with the magnetic carrier beads and processed using heating, magnetic pulldown, and washing steps to extract targets onto beads (details see main text). c, Target-functionalized magnetic carrier beads for detection on optofluidic nanopore chip.
To detect SARS-CoV-2 RNA in nasopharyngeal swab samples, human nasal swab samples that tested negatively for SARS-CoV-2 (confirmed by RT-qPCR) were collected by the UCSC Molecular Diagnostics Laboratory. A 10 μL aliquot of each sample was spiked with 10 μL of 1 × 109 copies/mL synthesized SARS-CoV-2 RNA fragments (ATCC) for a target concentration of 5 × 108 copies/mL (Fig. 2b). As mentioned above, the target RNA fragments are extracted sequence specifically and immobilized on the magnetic bead surface by a modified solid-phase extraction method (Cai et al., 2016; Parks et al., 2014). Secondary structures in the RNA were melted by incubating the solution on a heating block at 95 °C for 5 min, followed by the addition of 2.5 μL of 4 × 107 beads/mL of the functionalized magnetic bead solution to begin RNA hybridization. The number ratio between SARS-CoV-2 RNAs and magnetic beads corresponds to an average number of 100 RNAs per bead. While the exact extraction efficiency is not known, the results of our concentration dependent assay (see Fig. 3 d) suggest that this process worked with minimal loss. Since there are more than 105 binding sites on a bead, there is ample room for future optimization of the protocol, e.g for detection of higher concentrations, use of fewer beads for chip-based sample preparation, or to increase the throughput of the nanopore sensing step. To further increase the pulldown efficiency and accelerate annealing, the mixture was transferred to a 30 °C water bath for 1 min. Then the vial was gently flicked and centrifuged briefly, followed by 1-min heating in a dry bath at 95 °C. The water bath cooling and dry bath heating steps were repeated four times to maximize the binding possibility of the target particles to the magnetic beads. Then the solution was kept in an ice bath for 30 min. During the final bead washing process, the unattached target RNA molecules and nasal swab supernatant were discarded, followed by washing with 1x T50 buffer twice. The target-carrying magnetic beads (Fig. 2c) were resuspended in 1x T50 buffer with a final concentration of 3.3 × 106 beads/mL. An added 2% w/v sodium dodecyl sulfate (SDS) surfactant helps reduce bead clogging inside the LC channel. The release temperature is set to 10 °C above the melting temperature between the pulldown sequence and the target (35.1 °C), to account for heat transfer loss from the heater block to the ARROW chip. We reiterate that this bead based SPE method offers flexibility for adjusting the bead number for measurements at different target concentrations to reach a desired number of targets per bead. Save for the different starting matrix, the same protocol was used for measurements of targets in buffer solution.
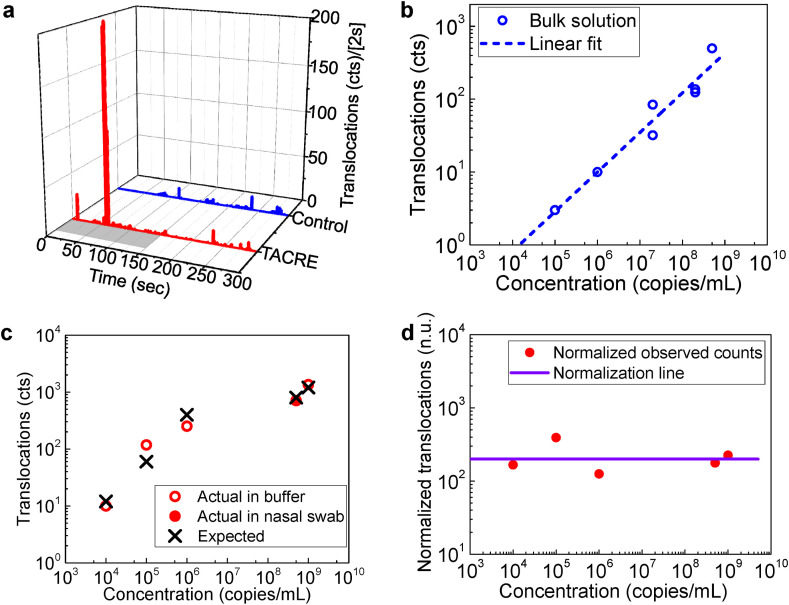
Fig. 3.
SARS-CoV-2 nanopore TACRE assay. a, Real-time translocations of SARS-CoV-2 RNAs from TACRE method and control experiment. The gray region in the TACRE trace represents the 150s heating period for target release. b, Number of translocations from SARS-CoV-2 RNAs in bulk solution at different concentrations within 360s detection time window. c, Detection of SARS-CoV-2 RNAs at different initial concentrations with TACRE method. (Circles: data; crosses: predicted number of events). d, Number of translocations (red solid circles) vs initial target concentration normalized to the case of 10 trapped beads and 20 RNAs/bead. Purple solid line: expected number of translocations in (c) after normalization, which is 200 for all the concentrations. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Results and discussion
Fig. 1d already showed the experimentally observed nanopore current traces obtained from the clinical NP swab samples using bead-based SPE and TACRE-based detection from eight trapped beads, demonstrating the feasibility of this approach for label-free, amplification-free diagnostics. In order to quantify the performance of the assay, we ran comparative assays with TACRE and without (i.e. detection from bulk solution), and over a large dynamic concentration range (104-109 copies/mL) that exceeds the clinically relevant range of 104-106 copies/mL (To et al., 2020). First, we assessed the speed of the TACRE-based assay. Fig. 3a shows a plot of real-time particle translocations versus running time for both TACRE and bulk control experiments at concentrations of 109 and 2 × 108 copies/mL, respectively. The control data (Fig. 3a top; blue bars) show stochastic, but continuous detection of small numbers of targets over the entire duration of the 5 min measurement due to the slow, diffusion-limited target delivery to the pore. The TACRE experiment shows a qualitatively and quantitatively different picture. Here, 90 beads holding an approximate average of 5 RNAs per bead were trapped at the nanopore. These target RNAs were released within the nanopore capture volume by increasing the temperature to 45 °C for the first 150 s (indicated by gray bar in Fig. 3a). After the chip reached the desired temperature (monitored by the nanopore baseline current), we observe a sharp rise in the translocation rate between t = 57s and t = 63s during which 75% of the total of 677 molecules were drawn through the nanopore and detected. This demonstrates the effectiveness of the TACRE process for a full clinical assay. We note that thermally released targets from the beads remaining in reservoir 1 could not contribute to the translocation events by diffusing to the nanopore capture volume within the duration of the experiment.
Next, we evaluated the suitability of this assay to cover the entire clinically relevant concentration range for COVID-19 (To et al., 2020). Fig. 3b shows the concentration of the control experiment with target RNAs spiked into buffer salt solution. Since this bulk solution test does not require bead pulldown, extraction of targets from nasal swabs was not necessary. We find that the number of translocations during the 360s detection window that represents a reasonable duration for a point of care test depends strongly on the concentration as expected for detection from bulk solution. While direct, label-free detection of single RNAs is indeed observed even in this arrangement, the assay has become unreliable at 105 copies/mL (2 ± 1 counts from three trials) and does not work at all at lower concentrations. Therefore, if we choose 10 translocation events with a standard deviation of less than 20% as the minimum number of events to positively identify the presence of SARS-CoV-2 RNAs, bulk detection has a limit of detection of 1 × 106 copies/mL and misses essentially the entire clinical concentration range. In stark contrast, the TACRE process does not suffer from these limitations and we can ensure that a sufficient number of carrier beads deliver at least 10 targets into the nanopore capture volume. Fig. 3c presents the concentration dependence of the TACRE assay. The solid circle marks the number of translocations (i.e. SARS-CoV-2 RNAs detected by the nanopore) for the clinical swab sample (c = 5 × 108 copies/mL). In this case, eight beads with an average 100 targets per bead were trapped and the measured number of detected RNAs is in excellent agreement with expectations (cross symbol). Another trial, with targets spiked into the same buffer solution that was used for the bulk experiments at a similar starting concentration (1 × 109 copies/mL), shows a very similar count (open circle) and again excellent agreement with the anticipated value. Clearly, the starting matrix (swab vs buffer) does not affect the outcome of the assay, and the remainder of the concentration series was carried out with RNA targets in buffer, analogous to the bulk series. We find that at least ten counts are observed for all concentrations down to the lower clinical limit of 104 copies/mL and that there is always good agreement between experiment and the number of counts predicted by multiplying the number of trapped microbeads with the average number of targets attached to each bead. This demonstrates unequivocally label-free and amplification-free detection of SARS-CoV-2 RNAs over the entire clinically relevant range. We emphasize that a linear dependence is not expected for this plot because the number of trapped beads and the average number of targets per bead varied between data points. On the other hand, given the agreement between data and predictions, it is possible to determine an unknown concentration from these data, i.e. from the number of translocations, number of trapped beads, and the concentration of magnetic beads used for extracting the targets.
In order to further clarify the favorable scaling of the TACRE method with concentration, we present normalized values of the detected counts as solid circles in Fig. 3d. The normalization is based on a standard trial of 10 trapped beads with 20 RNAs/bead, which would result in 200 counts independent of starting concentration and is represented by the solid purple line. For example, if only 5 beads were trapped with 8 RNAs/bead, the observed count number would be multiplied by a factor of (10/5)*(20/8) = 5 to obtain the normalized value. We find that these normalized values are close to 200 (within a factor of two) across the entire concentration range, which confirms that the TACRE assay is not limited at low concentrations in the same way as a bulk experiment. Indeed, the normalized counts at the lowest and highest concentration (167 and 225, respectively) are nearly identical to each other and the ideal value of 200. This shows that the capture process during the sample preparation phase was equally efficient at all concentrations and validates our earlier RNA-per-bead estimates. We can also conclude that the assay can readily be extended to even lower concentrations by increasing the number of trapped beads and/or mixing fewer capture beads with the sample to increase the number of targets per bead.
We now turn our attention to quantifying the benefits of the TACRE process in terms of the increased local concentration within the nanopore capture volume which is critical for reaching clinically relevant concentration levels as described above. To quantify the local target concentration near the nanopore during the TACRE experiments with SARS-CoV-2 RNAs, the capture volume is estimated from the bulk experiment as described in more detail in supplementary section 4. Using the calculated value of 1,309 μm3, the local target concentration for both the bulk control (solid blue line) and TACRE (red circles) assays of Fig. 3c is plotted versus starting concentration in Fig. 4 a. The bulk control curve shows a trivial linear dependence as the concentration at the nanopore is the same as in the rest of the channel. In the case of the TACRE assay, however, we observe local target concentrations that are orders of magnitude higher than the bulk value and can be maintained at picomolar levels even for initial concentrations in the attomolar range. This demonstrates that the TACRE approach is very efficient, even for a relatively small number of trapped carrier beads, and scales favorably to low concentrations. Indeed, the largest local concentration enhancement of a factor of 7.8 × 105 was observed for the lowest initial target concentration of 17 aM.
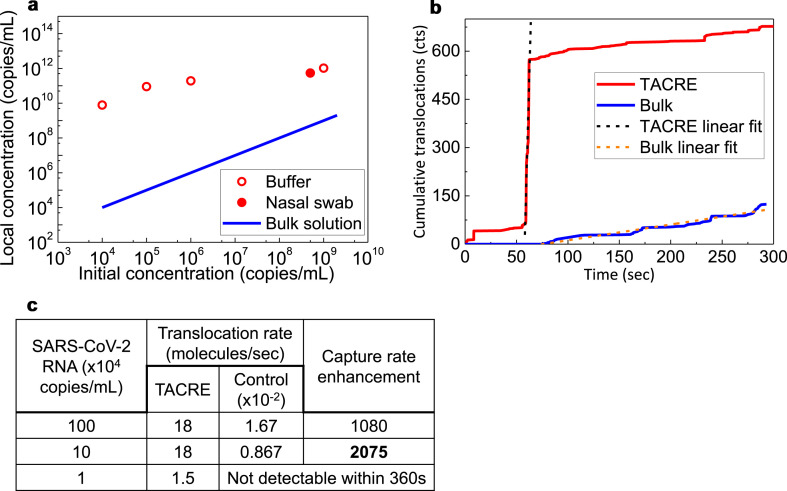
Fig. 4.
Target concentration enhancement and capture rate enhancement of TACRE method. a, Target concentration within the nanopore capture volume obtained by TACRE (open and closed circles) vs. initial target concentration in the bulk solution. Blue line: Target concentration at nanopore for control experiment (equal to starting concentration). b, Cumulative number of translocations of TACRE and control experiments over time. The slopes of the fitted lines represent the translocation rates. c, Capture rates and enhancements at different concentrations calculated from the respective fitted rates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Finally, we take a look at the improvements to the assay dynamics, i.e. the increase in assay detection speed enabled by the TACRE approach. To quantify this important benefit, we define a translocation rate enhancement as the ratio of peak translocation rate at each concentration of the TACRE assay and the rate of particle counts over the 360 s duration of the assay for the control case. Fig. 4b shows the cumulative target counts for the TACRE and bulk assays of Fig. 3a. The capture rates at different concentrations are then extracted from their respective linear fits and displayed in Fig. 4c along with the resulting enhancement factor for the clinically relevant concentration range of SARS-CoV-2 infection. Again, we find that the TACRE process becomes more powerful as the concentration decreases, and a record enhancement factor of 2,075x is observed at 1 × 105 copies/mL. This corresponds to an over 2,000 times reduction in the time needed to detect the same number of target molecules. Since no targets were detected in the control experiment at 104 copies/mL, the TACRE factor would nominally be infinite. With long enough assay times, the control experiment would produce counts and a finite enhancement factor, but it is safe to assume that this factor would significantly exceed the demonstrated value of 2,075x.
4. Conclusion
In summary, we have demonstrated a label-free and amplification-free assay for detection of SARS-CoV-2 RNAs in clinical samples on a nanopore-optofluidic chip. The use of target delivery to the nanopore via optical trapping of carrier microbeads at the pore solved the major issues of specificity and low detection rates commonly faced by nanopore sensors. The TACRE assay showed vastly superior performance over bulk solution-based analysis in terms of limit of detection, dynamic range, and detection speed with an improvement by more than three orders of magnitude. The assay was demonstrated with clinical nasopharyngeal swab samples and showed a dynamic range of 5 logs, covering the entire clinically relevant concentration range down to a concentration of 17 aM. The assay can be readily applied to detection of other infectious diseases that have comparable dynamic ranges (de Laval et al., 2017; Kuypers et al., 2006; Towner et al., 2004). It can also be improved, e.g. by increasing the number of trapped beads and their target loads, modifying the trapping process, or optimizing the target release method. These modifications can lead to even larger enhancement factors and lower limits of detection. Furthermore, the TACRE assay can be functionally expanded, e.g. by applying the assay to protein targets (Meena et al., 2018), by implementing the sample preparation step on a chip (Meena et al., 2018), implementing multi-channel, multiplex detection, or by applying advanced signal processing for real-time analysis of the nanopore signals. As a result, this integrated nanopore sensor has the potential to be used as a simple, ultrasensitive, and rapid molecular analysis tool with a diverse range of applications in disease diagnostics and biomedicine.
CRediT authorship contribution statement
Mohammad Julker Neyen Sampad: Conceptualization, Methodology, Software, Formal analysis, Investigation, Resources, Writing – review & editing, Visualization, contributed equally to this work. Han Zhang: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Writing – review & editing, Visualization, contributed equally to this work. Thomas D. Yuzvinsky: Resources, Writing – review & editing. Matthew A. Stott: Resources, Writing – review & editing. Aaron R. Hawkins: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition. Holger Schmidt: Conceptualization, Formal analysis, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Holger Schmidt and Aaron R. Hawkins have a financial interest in Fluxus, Inc. which commercializes optofluidic technology. The remaining authors declare no competing interests.
Acknowledgements
We thank M. Rahman for fruitful discussions. This work was supported by the W.M. Keck Center for Nanoscale Optofluidics at UC Santa Cruz, UC National Laboratory Fees Research Program (LFRP) block grant supporting COVID-19 research, and the National Institutes of Health [Grant No.1R01EB028608, and 3R01EB028608-02S1]. We also acknowledge the UCSC Molecular Diagnostics Laboratory for providing negative de-identified clinical samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113588.
Appendix B. Supplementary data
The following are the Supplementary data to this article:
References
- Ashkenasy N., Sánchez‐Quesada J., Bayley H., Ghadiri M.R. Angew. Chem. 2005;117(9):1425–1428. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Stott M.A., Ozcelik D., Parks J.W., Hawkins A.R., Schmidt H. Biomicrofluidics. 2016;10(6) doi: 10.1063/1.4968033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C.-C., Shekar S., Niedzwiecki D.J., Shepard K.L., Drndić M. ACS Nano. 2019;13(9):10545–10554. doi: 10.1021/acsnano.9b04626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah K., Wu Y., Vivekchand S., Gaus K., Reece P.J., Micolich A.P., Gooding J.J. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laval F., Matheus S., Labrousse T., Enfissi A., Rousset D., Briolant S. N. Engl. J. Med. 2017;377(7):697–699. doi: 10.1056/NEJMc1612600. [DOI] [PubMed] [Google Scholar]
- Deamer D., Akeson M., Branton D. Nat. Biotechnol. 2016;34(5):518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.W., Akeson M. Trends Biotechnol. 2000;18(4):147–151. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- Deamer D.W., Branton D. Acc. Chem. Res. 2002;35(10):817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- Duguay M., Kokubun Y., Koch T.L., Pfeiffer L. Appl. Phys. Lett. 1986;49(1):13–15. [Google Scholar]
- Fologea D., Gershow M., Ledden B., McNabb D.S., Golovchenko J.A., Li J. Nano Lett. 2005;5(10):1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A. Nat. Methods. 2018;15(3):201. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P., Ranu A., Macrae R.K., Faure G. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J.J., Brandin E., Branton D., Deamer D.W. Proc. Natl. Acad. Sci. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk S.W., Tuijtel M.W., Donkers S.P., Dekker C. Nano Lett. 2010;10(4):1414–1420. doi: 10.1021/nl100271c. [DOI] [PubMed] [Google Scholar]
- Kühn S., Measor P., Lunt E., Phillips B., Deamer D., Hawkins A., Schmidt H. Lab Chip. 2009;9(15):2212–2216. doi: 10.1039/b900555b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers J., Wright N., Ferrenberg J., Huang M.-L., Cent A., Corey L., Morrow R. J. Clin. Microbiol. 2006;44(7):2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Bell N.A., Hernández-Ainsa S., Thacker V.V., Thackray A.M., Bujdoso R., Keyser U.F. ACS Nano. 2013;7(5):4129–4134. doi: 10.1021/nn4004567. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhao Y., Parks J.W., Deamer D.W., Hawkins A.R., Schmidt H. Nano Lett. 2014;14(8):4816–4820. doi: 10.1021/nl502400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt E.J., Wu B., Keeley J.M., Measor P., Schmidt H., Hawkins A.R. IEEE Photon. Technol. Lett. 2010;22(15):1147–1149. doi: 10.1109/LPT.2010.2051145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measor P., Phillips B.S., Chen A., Hawkins A.R., Schmidt H. Lab Chip. 2011;11(5):899–904. doi: 10.1039/c0lc00496k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena G., Stambaugh A., Ganjalizadeh V., Stott M., Hawkins A., Schmidt H. APL Photonics. 2021;6(6) doi: 10.1063/5.0049735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena G.G., Jain A., Parks J.W., Stambaugh A., Patterson J.L., Hawkins A.R., Schmidt H. Lab Chip. 2018;18(23):3678–3686. doi: 10.1039/c8lc00966j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks J.W., Olson M.A., Kim J., Ozcelik D., Cai H., Carrion R., Jr., Patterson J., Mathies R., Hawkins A., Schmidt H. Biomicrofluidics. 2014;8(5) doi: 10.1063/1.4897226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Harrington M., Stott M., Li Y., Sampad M., Yuzvinsky T., Hawkins A., Schmidt H. Optica. 2019;6(9):1130–1131. doi: 10.1364/optica.6.001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Stott M., Harrington M., Li Y., Sampad M., Lancaster L., Yuzvinsky T., Noller H., Hawkins A., Schmidt H. Nat. Commun. 2019;10(1):1–7. doi: 10.1038/s41467-019-11723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shasha C., Henley R.Y., Stoloff D.H., Rynearson K.D., Hermann T., Wanunu M. ACS Nano. 2014;8(6):6425–6430. doi: 10.1021/nn501969r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner G.M., van den Hout M., Broekmans O., Dekker C., Dekker N.H. Nano Lett. 2009;9(8):2953–2960. doi: 10.1021/nl901370w. [DOI] [PubMed] [Google Scholar]
- Spitzberg J.D., Zrehen A., van Kooten X.F., Meller A. Adv. Mater. 2019;31(23) doi: 10.1002/adma.201900422. [DOI] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A., Sorrell T.C., Black A.J., Caillaud C., Chrzanowski W., Li E., Martinez-Martin D., McEwan A., Wang R., Motion A. Nat. Biotechnol. 2021;39(2):144–147. doi: 10.1038/s41587-021-00816-8. [DOI] [PubMed] [Google Scholar]
- Towner J.S., Rollin P.E., Bausch D.G., Sanchez A., Crary S.M., Vincent M., Lee W.F., Spiropoulou C.F., Ksiazek T.G., Lukwiya M. J. Virol. 2004;78(8):4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M., Bhattacharya S., Xie Y., Tor Y., Aksimentiev A., Drndic M. ACS Nano. 2011;5(12):9345–9353. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M., Morrison W., Rabin Y., Grosberg A.Y., Meller A. Nat. Nanotechnol. 2010;5(2):160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Gatterdam V., Wieneke R., Tampé R., Rant U. Nat. Nanotechnol. 2012;7(4):257–263. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- Yin D., Deamer D., Schmidt H., Barber J., Hawkins A. Appl. Phys. Lett. 2004;85(16):3477–3479. [Google Scholar]
- Yusko E.C., Bruhn B.R., Eggenberger O.M., Houghtaling J., Rollings R.C., Walsh N.C., Nandivada S., Pindrus M., Hall A.R., Sept D. Nat. Nanotechnol. 2017;12(4):360–367. doi: 10.1038/nnano.2016.267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.