Abstract
Tf1 is a long terminal repeat (LTR)-containing retrotransposon that propagates within the fission yeast Schizosaccharomyces pombe. LTR-retrotransposons possess significant similarity to retroviruses and therefore serve as retrovirus models. To determine what features of the host cell are important for the proliferation of this class of retroelements, we screened for mutations in host genes that reduced the transposition activity of Tf1. We report here the isolation and characterization of pst1+, a gene required for Tf1 transposition. The predicted amino acid sequence of Pst1p possessed high sequence homology with the Sin3 family of proteins, known for their interaction with histone deacetylases. However, unlike the SIN3 gene of Saccharomyces cerevisiae, pst1+ is essential for cell viability. Immunofluorescence microscopy indicated that Pst1p was localized in the nucleus. Consistent with the critical role previously reported for Sin3 proteins in the histone acetylation process, we found that the growth of the strain with the pst1-1 allele was supersensitive to the specific histone deacetylase inhibitor trichostatin A. However, our analysis of strains with the pst1-1 mutation was unable to detect any changes in the acetylation of specific lysines of histones H3 and H4 as measured in bulk chromatin. Interestingly, the pst1-1 mutant strain produced wild-type levels of Tf1-encoded proteins and cDNA, indicating that the defect in transposition occurred after reverse transcription. The results of immunofluorescence microscopy showed that the nuclear localization of the Tf1 capsid protein was disrupted in the strain with the pst1-1 mutation, indicating an important role of pst1+ in modulating the nuclear import of Tf1 virus-like particles.
Retroviruses and retrovirus-like transposons belong to a widely distributed family of eukaryotic elements called long terminal repeat (LTR)-containing retroelements. Although these LTR-containing elements have been found in hosts as diverse as yeast and mammals, striking structural and functional conservation has been observed in the mechanisms responsible for their life cycles. The similarities include the proteolytic processing of precursor proteins, the assembly of particles, the reverse transcription of cDNA, and the integration of the cDNA into the host genome (10). Although retroviruses and retrotransposons possess a requirement for many components of host physiology, only a few host genes that control retroviral proliferation have been identified. One reason for this general lack of information is the difficulty of studying host organisms with high genetic complexity. One approach to understanding the propagation of retroelements is to study yeast retrotransposons, a family of elements that possess an extensive similarity to retroviruses and thus serve as a retrovirus model system (10, 63).
We study Tf1, an LTR-containing retrotransposon isolated from the fission yeast Schizosaccharomyces pombe. This retroelement has coding sequences for Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) proteins (40, 42). In vivo assays for transposition demonstrate that Tf1 is highly active and generates transposition frequencies varying from 2 to 20% (41). The results of sucrose gradient sedimentation revealed that Tf1 Gag, IN, mRNA, and reverse transcription products all assemble into virus-like particles (VLPs) that contain a 26-fold molar excess of Gag relative to IN (4, 38, 41).
In eukaryotic cells, genomic DNA is assembled with chromosomal proteins, particularly histones, to form a complex structure called chromatin. The level of chromatin condensation plays a key role in many cellular processes, including the regulation of transcription (68, 79), the replication of DNA (61, 72), the segregation of chromosomes (2, 19, 20, 25), and the integration of retroviral cDNA into host targets (56–59). In the last few years, factors capable of altering the structure of chromatin have been identified (35). One important class of modification is the acetylation of lysine residues in the core histones (13, 68, 80). Genetic and biochemical studies have revealed a correlation between histone acetylation and the transcriptional activity of promoters associated with acetylated histones (31, 44, 62, 79). Hyperacetylation of core histones destabilizes nucleosomes and correlates with gene activation, while hypoacetylation stabilizes nucleosomes and correlates with gene repression. Recently, factors containing histone acetylase and deacetylase activities have been identified (68, 79). Among these factors, the family of Sin3 proteins have been shown to play the role of scaffolding elements required for the formation and function of histone deacetylase complexes (54, 79). Indeed, each Sin3 protein contains four paired amphipathic helix (PAH) domains, which have been proposed to mediate protein-protein interactions (77). Sin3 proteins have been shown to use distinct domains to form histone deacetylase complexes and to interact with specific DNA-binding proteins (8, 79). Thus, Sin3p has been proposed to play an important role in recruiting the histone deacetylase activity to specific regions of chromosomes, causing targeted changes in chromatin structure and expression (8, 54, 79).
To identify genes of S. pombe that are required for Tf1 activity, we mutagenized cultures and screened for strains that were unable to support Tf1 transposition. In this paper, we report the isolation of the pst1+ (for pombe SIN three) gene in S. pombe that is a homologue of the SIN3 gene of Saccharomyces cerevisiae. The pst1+ gene is shown here to be important for Tf1 transposition and the ability of Tf1 cDNA to homologously recombine with Tf1 sequences contained in plasmids. Immunoblotting results indicated that the pst1-1 mutation did not cause changes in acetylation levels of key lysines in histones H3 or H4 when measured in bulk histone preparations. Nevertheless, the histone deacetylase inhibitor trichostatin A (TSA) was found to reduce the activity of the Tf1 element. Interestingly, strains with the pst1-1 allele exhibited pseudohyphal growth in the presence of TSA. Further analysis revealed that the pst1-1 mutation caused a block in the nuclear import of Tf1 Gag, and we propose that the mislocalization of Gag is responsible for the loss of Tf1 transposition.
MATERIALS AND METHODS
Media.
The S. pombe media were prepared as described previously (39, 46). Agar plates containing sporulation medium with malt extract (ME; Bio 101, La Jolla, Calif.) were prepared according to the manufacturer’s instructions. Phloxin B was used at 5 μg/ml to differentiate diploid cells from their haploid progeny. TSA (WAKO BioProducts) was dissolved in dimethyl sulfoxide and used at concentrations of 0.1 to 5 μg/ml. Ten micromolar vitamin B1 (thiamine) was added to minimal medium to repress the nmt1 promoter. 5-Fluoroorotic acid (5-FOA) (U.S. Biologicals, Swampscott, Mass.) was used at 1 mg/ml in Edinburgh minimal medium (EMM). Yeast extract plus supplements (YES) 5-FOA-G418 plates were made from YES medium containing 1 mg of 5-FOA/ml and 500 μg of Geneticin (Gibco)/ml (corrected for purity).
Plasmid construction.
Many plasmids used for this study were constructed by PCR cloning techniques. For this purpose, the plasmids were created in duplicate from independent PCRs and the properties of each plasmid were studied in parallel. The oligonucleotides and plasmids used in this study are listed in Table 1.
TABLE 1.
Description of plasmids and oligonucleotides used
| Plasmid or oligo-nucleotide | Description | Source |
|---|---|---|
| Plasmids | ||
| pAF1 | his3+ marker of S. pombe cloned into pBluescript II KS(+) | 52 |
| pSP1 | Replicative plasmid with ARS1 of S. pombe and a LEU2 marker of S. cerevisiae | 16 |
| pSP2 | Replicative plasmid with ARS1 of S. pombe and a URA3 marker of S. cerevisiae | 16 |
| pade6-M375-M26 | Replicative plasmid with ARS1 and ade6-M375-M26 allele of S. pombe and URA3 marker of S. cerevisiae | 55 |
| pHL449-1 | Tf1-neoAI assay plasmid | 38 |
| pHL481 | Integrative plasmid with a leu1+ gene of S. pombe | This study |
| pHL476-3 | Tf1-neoAI with frameshift in IN | 38 |
| pHL490-80 | Tf1-neoAI with frameshift in PR | 4 |
| pHL919-3 | Tf1-neoAI with frameshift in RT | 39 |
| pHL1277 | pHL449-1 with FLAG epitope inserted at the end of Tf1 Gag | This study |
| pHL1380 | pWH5 library plasmid containing pst1 and truncated gfa1 genes | This study |
| pHL1381 | 9.5-kb insert isolated from pHL1380 and cloned into integrative vector pHL481 | This study |
| pHL1383 | pHL481 with 5.6-kb Eco47III-NheI fragment containing the pst1 gene and 393 nt of the promoter from pHL1380 | This study |
| pHL1385-17 | Bluescript KS vector with Eco47III-NheI fragment isolated from pHL1383 | This study |
| pHL1386-1 | pSP1 vector carrying Eco47III-NheI fragment isolated from pHL1383 | This study |
| pHL1412-2 | pHL1385-17 with frameshift created at beginning of second exon of pst1 | This study |
| pHL1413-22 | pHL1386-1 with frameshift created at beginning of second exon of pst1 | This study |
| pHL1458-3 | pSP1 with PCR-generated 6.2-kb DNA fragment containing pst1+ gene and 997 nucleotides of promoter | This study |
| pHL1459-2 | Bluescript KS vector with 6.2-kb insert from pHL1458-3 and BamHI site created at ATG codon of pst1 | This study |
| pHL1460-2 | pHL1458-3 with BamHI site created at ATG codon of pst1 | This study |
| pHL1581-1 | pHL1459-2 with entire coding sequence of pst1+ replaced with a his3+ marker | This study |
| pHL1626-2 | pHL1458-3 with triple copy of HA epitope inserted upstream of stop codon of pst1 | This study |
| pHL1628-2 | pSP2 carrying 5.6-kb Eco47III-NheI isolated from pHL1383 | This study |
| Oligonucleotides | ||
| HL2 | 5′-GTAGGTGCTATTTTACTAGTCTAAGCTAATCAATAG-3′; bottom strand of artificial intron, used to detect neo mRNA | |
| HL220 | 5′-GACTACAAGGACGACGATGACAAG-3′; top-strand FLAG epitope | |
| HL221 | 5′-CTTGTCATCGTCGTCCTTGTAGTC-3′; FLAG epitope complementary strand | |
| HL335 | 5′-ATCCTTAACCCAATTGTATCGGATACAAACAGAGACT-3′; top-strand primer used to generate HA-tag in pst1 | |
| HL467 | 5′-GTATATATACCTTGGAAGCGTAGTCTGGGACATCGTATGGATATCCGGCGTAATCAGGCACATCATATGGGTAGGCAGCGTAGTCTGGAACGTCGTATGGGTACAGATCATCCTTTGAAGGCTCTTCTTCCAT-3′; bottom-strand primer used to generate triple HA-tag in pst1 | |
| HL426 | 5′-TAAGCATAATCTACTCGAGTCACTTTCATATCCAAGACGGC-3′; top-strand primer used to PCR amplify N-ter domain of pst1-1 | |
| HL326 | 5′-TTAGCATAATCTACTCGAGATTAACTGATACGTCAGGTAAA-3′; bottom-strand primer used to amplify N-ter domain of pst1-1 | |
| HL327 | 5′-ATCCTTAACCGAATTCGCCATTAATTTTGTTAATAATGTT-3′; top-strand primer used to PCR amplify central part of pst1-1 | |
| HL332 | 5′-TTAGCATAATTTACTCGAGCCCATTTTGATCAATTTCACC-3′; bottom-strand primer used to PCR amplify central part of pst1-1 | |
| HL333 | 5′-ATCCTTAACCCAATTGGAGCATGAGGAAATGACGA-3′; top-strand primer used to PCR amplify C-ter domain of pst1-1 | |
| HL336 | 5′-TTAGCATAATTTACTCGAGCTACAGATCATCCTTGGAAG-3′; bottom-strand primer used to amplify C-ter domain of pst1-1 | |
| HL440 | 5′-TTTGGCAGATCTCTTTCAACGTTT-3′; top-strand primer used to PCR amplify his3 marker from pAF1 | |
| HL441 | 5′-TTTGGCAGATCTCTTTCAACGTTT-3′; bottom-strand primer used to amplify his3 marker from pAF1 | |
| HL395 | 5′-TGATTCATTGGCAAGATATTGGC-3′; 3′-Flanking primer used to PCR create BamHI site at ATG of pst1 | |
| HL396 | 5′-TTAGCATAATCTACTCGAGGGCAGCTTTCTGCAGAACTTTG-3′; 5′-Flanking primer used to PCR create BamHI site at ATG of pst1 | |
| HL397 | 5′-CCAACAACTTTTGGGATCCGCAAAAGACTGGC-3′; top-strand fusion primer overlapping BamHI site at ATG of pst1 | |
| HL398 | 5′-GCCAGTCTTTTGCGGATCCCAAAAGTTGTTGG-3′; complementary oligonucleotide of primer HL397 |
The wild-type allele of the pst1 gene was isolated by transforming the pst1-1 mutant (YHL5018) with a genomic library (kindly provided by P. Young) and selecting for strains that showed wild-type levels of transposition and cDNA recombination. The library plasmid pHL1380 contained a 9-kb genomic insert with the entire coding sequence of the pst1+ gene. To create an integrative version of the pst1+ gene, an Eco47III-NheI (5.6-kb) fragment was isolated from pHL1380 and cloned into the integrative vector pHL481 at the SmaI and XbaI sites. The resulting plasmid was named pHL1383 and contained the entire open reading frame (ORF) of pst1+ and 393 nucleotides of the promoter. Vector pHL481 was created by removing from pJK148 the 742-nucleotide ClaI fragment (34). The NotI-ClaI fragment with pst1+ was isolated from pHL1383 and cloned into Bluescript KS(+) (Stratagene) at the NotI and ClaI sites to generate pHL1385-17. The insert with pst1+ from this plasmid was then transferred into the S. pombe plasmid pSP1 (16) at the unique NotI and XhoI sites to generate plasmid pHL1386-1.
To create a frameshift in the coding sequence of pst1+, the plasmid pHL1385-17 was digested with EcoNI, treated with Klenow enzyme (Boehringer), and recircularized with T4 ligase (United States Biochemical, Cleveland, Ohio). The presence of the newly created frameshift in the resulting plasmid pHL1412-2 was confirmed by sequencing. The 5.6-kb pst1 frameshift-containing insert of pHL1412-2 was isolated and cloned into the replicative vector pSP1 to generate pHL1413-22.
The Δpst1::his3 allele was constructed as follows. The putative translation start site of pst1+ (GGATGG) was first replaced by a unique BamHI site (GGATCC) by using the fusion PCR technique described previously (38). Primers HL395, HL396, HL397, and HL398 were used to generate from the template plasmid pHL1380 a 2.7-kb fusion PCR product that contained 997 nucleotides of the pst1+ promoter, a unique BamHI site at the ATG codon of pst1+, and 1.7 kb of its coding sequence. This PCR product was digested with SphI and XhoI and cloned into two plasmids, pHL1385-17 and pHL1386-1, at the SphI and XhoI sites to generate pHL1459-2 and pHL1460-2, respectively. A wild-type version of pHL1460-2, i.e., without the artificial BamHI site, was created and named pHL1458-3. The entire pst1+ coding sequence of pHL1459-2 was then removed by digesting it with BamHI-StyI and replaced by a 2-kb his3+ marker that was generated from the plasmid pAF1 (52) by PCR with the primers HL440 and HL441. The resulting plasmid was named pHL1581-1.
The HA-tagged allele of pst1+ included a triple copy of the HA epitope that was inserted at the C terminus of the pst1+ coding sequence in the plasmid pHL1458-3. For this purpose, the oligonucleotides HL335 and HL467 were used to generate a 1.1-kb PCR product that contained 1 kb of the pst1+ C terminus and a triple-HA epitope. HL335 annealed upstream of the unique EcoRI site located within the pst1+ coding sequence. HL467 annealed just upstream of the stop codon of pst1+, and its nucleotide sequence contained a triple-HA epitope and the last three amino acids of pst1+, including a StyI site. This EcoRI-StyI fragment with the HA sequence was used to replace the C-terminal EcoRI-StyI fragment of pst1+ carried by pHL1458-3. The resulting plasmid was named pHL1626-2. The junction of pst1+ and the HA epitope was sequenced, and the expression of the HA-tagged pst1+ was verified by immunoblot analysis with a monoclonal anti-HA antibody (Babco) (data not shown).
The FLAG-tagged Gag version of Tf1 was constructed by inserting a FLAG epitope (Eastman Kodak, New Haven, Conn.) at the C terminus of Tf1 Gag as described previously (6).
To determine the location of the mutation in the pst1-1 allele, we cloned the chromosomal copy of the pst1-1 allele by PCR and determined its nucleotide sequence. Genomic DNA of a yeast strain with the pst1-1 mutation (YHL5356) was extracted and used as a PCR template. The oligonucleotides used in this experiment are listed in Table 1. The sequence of the wild-type allele of pst1+ carried in the plasmid pHL1385-17 was also determined.
Yeast strain construction.
The yeast strains used in this study are listed in Table 2. To integrate the wild-type allele of pst1+ into its chromosomal locus, the integrative plasmid pHL1383 was linearized with SphI and used to transform the pst1-1 mutant YHL5356. Stable Leu+ transformants were selected, and DNA blot analysis was performed to verify the integration of a single copy of the plasmid pHL1383 at the original pst1+ locus (data not shown). The resulting integrant was YHL6211. The two strains YHL5761 and YHL5762 were from spores resulting from the mating of two haploid strains, YHL1101 and YHL5356. Strain YHL6403-2 was the diploid resulting from the mating of two haploid strains, YHL6381 and YHL6382 (gifts from R. Ohi and K. Gould). To construct the heterozygous diploid strain YHL6604, the 3.5-kb insert of the plasmid pHL1581-1 was isolated and used to transform the YHL6403-2 strain. His+ prototroph transformants were selected, and the replacement of one of the two wild-type pst1+ alleles by the Δpst1::his3 allele was checked by DNA blot analysis (data not shown). Plasmids were introduced into S. pombe strains by lithium acetate transformation (46).
TABLE 2.
Yeast strains used
| Strain | Relevant genotype or description | Parent strain/plasmid | Source |
| YHL912 | h− leu1-32 ura4-294 | No plasmid | J. Boeke, 21X5A |
| YHL1011 | h+ ade6-M210 leu1-32 ura4-D18 | No plasmid | S. Forsburg |
| YHL1282 | h− leu1-32 ura4-294 | YHL912/pHL449-1 | 38 |
| YHL965 | h− leu1-32 ura4-294 | YHL912/pSP1 | This study |
| YHL1554 | h− leu1-32 ura4-294 | YHL912/pHL476-3 | 38 |
| YHL1836 | h− leu1-32 ura4-294 | YHL912/pHL490-80 | 4 |
| YHL4988 | h− leu1-32 ura4-294 | YHL1282/pSP1 | This study |
| YHL4990 | h− leu1-32 ura4-294 | YHL1836/pSP1 | This study |
| YHL4992 | h− leu1-32 ura4-294 | YHL1554/pSP1 | This study |
| YHL4994 | h+ pst1-1 leu1-32 ura4-294 | YHL5018/pSP1 | This study |
| YHL5235 | h+ pst1-1 leu1-32 ura4-294 | No plasmid | This study |
| YHL5018 | h+ pst1-1 leu1-32 ura4-294 | YHL5235/pHL449-1 | This study |
| YHL5356 | h− pst1-1 leu1-32 ura4-294 (from third backcrosses) | No plasmid | This study |
| YHL5406 | h− pst1-1 leu1-32 ura4-294 | YHL5356/pHL449-1 | This study |
| YHL5392 | h+ pst1+ leu1-32 ura4-294 (after third backcrosses) | No plasmid | This study |
| YHL5426 | h+ pst1+ leu1-32 ura4-294 | YHL5392/pHL449-1 | This study |
| YHL5896 | h− leu1-32 ura4-294 | YHL912/pHL1277 | This study |
| YHL5761a | h+ ade6-M210 leu1-32 pst1-1 ura4− | No plasmid | This study |
| YHL5762a | h+ ade6-M210 leu1-32 pst1+ ura4− | No plasmid | This study |
| YHL5834a | h+ ade6-M210 leu1-32 pst1-1 ura4− | YHL5761/pade6-M375-M26 | This study |
| YHL5835a | h+ ade6-M210 leu1-32 pst1-1 ura4− | YHL5761/pade6-M375-M26 | This study |
| YHL5852a | h+ ade6-M210 leu1-32 pst1+ ura4− | YHL5762/pade6-M375-M26 | This study |
| YHL5834a | h+ ade6-M210 leu1-32 pst1+ ura4− | YHL5762/pade6-M375-M26 | This study |
| YHL6596 | h− leu1-32 ura4-294 | YHL912/pHL1626-2 | This study |
| YHL6211 | h− leu1-32 ura4-294 pst1-1-SphI::pst1+-leu1+ | No plasmid | This study |
| YHL6214 | h− leu1-32 ura4-294 pst1-1-SphI::pst1+-leu1+ | YHL6211/pHL449-1 | This study |
| YHL6221 | h− pst1-1 leu1-32 ura4-294 | YHL5406/pHLp1386-1 | This study |
| YHL6222 | h− pst1-1 leu1-32 ura4-294 | YHL5406/pHLp1413-22 | This study |
| YHL6381 | h+ ade6-M210 his3-D1 leu1-32 ura4-D18 | No plasmid | 52 |
| YHL6382 | h− ade6-M216 his3-D1 leu1-32 ura34-D18 | No plasmid | 52 |
| YHL6403-2 | h+/h− ade6-M210/ade6-M216 his3-D1/his3-D1 leu1-32/leu1-32 ura4-D18/ura4-D18 | No plasmid | This study |
| YHL6604 | h+/h− ade6-M210/ade6-M216 his3-D1/his3-D1 leu1-32/leu1-32 ura4-D18/ura4-D18 pst1+/Δpst1::his3+ | No plasmid | This study |
The exact allele of ura4 was not determined (either ura4-294 or ura4-D18).
Transposition and homologous recombination assays.
Tf1 transposition frequencies were determined as described previously (39). Briefly, Tf1 transposition was monitored by placing a neo-marked Tf1 element under the control of an inducible nmt1 promoter. The neo gene allowed cells to grow in the presence of 500 μg of G418/ml. S. pombe strains that contained a Tf1-neo plasmid were grown as patches on EMM-Ura dropout agar plates in the absence of thiamine to induce transcription of the nmt1-Tf1-neo fusion. After 4 days of 32°C incubation, these plates were then replica printed to medium containing 5-FOA to eliminate the URA3-Tf1-neo plasmid (11). Finally, 5-FOAr patches were printed to medium containing both 5-FOA and G418 and incubated at 32°C for 2 days to detect strains that became resistant to G418 as the result of insertions of the neo-marked Tf1 element into the genome. Wild-type Tf1 produced confluent G418r patches, whereas mutations in element-encoded components or host factors reduced the growth on the G418 plates. Quantitative measurements of transposition frequencies were performed as follows. Strains were grown as patches of cells on EMM-Ura dropout agar for 4 days at 32°C. These cells were then resuspended at an optical density at 600 nm (OD600) of 1.0 and diluted approximately 100-fold. Approximately 0.1 ml of the cells was then spread onto 5-FOA plates, and the resulting colonies (approximately 7,000/plate) were replica printed to YES plates containing FOA and 500 μg of G418/ml (41). The transposition frequency was the percentage of the FOA-resistant colonies that were also G418 resistant.
The presence of Tf1 cDNA in the nucleus was examined by cDNA recombination assays, which were conducted according to the method of Atwood et al. (3). This protocol is similar to the transposition assay in that strains with the neoAI-marked Tf1 plasmid were first grown as patches on agar plates that contained EMM (plus 10 μM thiamine and dropout powder) and then replica printed to similar EMM plates that lacked thiamine. After 4 days of 32°C incubation, the plates were replica printed directly to YES medium that contained 500 μg of G418/ml. Recombination between cDNA and cellular transposon sequences was scored on the G418 plates after 48 h of growth at 32°C.
Nucleic acid preparation and analysis.
cDNA preparation (4) and total RNA extraction (17) were performed as described previously. The protocols used for DNA and RNA blot analyses were adopted from previously published works (4, 17). To measure the expression level of the neoAI gene carried on the transposition assay plasmid pHL449-1, the RNA blot was hybridized with the oligonucleotide HL2 used as a strand-specific probe, which was 5′ radiolabelled with polynucleotide kinase (Boehringer). An EcoRI DNA fragment specific for Tf1 Gag (42) was used as a probe to detect Tf1 mRNA.
Protein extraction and immunoblot detection of Tf1 Gag and IN.
Total proteins were extracted from cells grown under inducing conditions (absence of thiamine) by a previously published protocol (4). Protein pellets were collected, and an equal volume of 2× sample buffer (4) was added. The mixture was boiled for 3 min, and 25 μg of total protein from each sample was loaded on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels for immunoblot analysis. Standard electrotransfer techniques were used (70) with Immobilon-P membranes (Millipore). The detection method used was the ECL system as described by the manufacturer (Amersham), except that the secondary antibody, horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin, was used at 1:10,000 dilution. The primary polyclonal antisera used for each filter were from production bleeds 660 (anti-Gag) and 657 (anti-IN) (42).
Indirect immunofluorescence. (i) Localization of the FLAG-tagged Gag.
The anti-FLAG M2 monoclonal antibody (Eastman Kodak), and fluorescein isothiocyanate (FITC)–Oregon green 488–goat anti-mouse immunoglobulin G (IgG) antibody (Molecular Probes) were used for immunofluorescence experiments. To visualize nuclear DNA, cells were stained with 1 μg of DAPI (4′,6′-diamino-2-phenylindole)/ml. Detection of FLAG-tagged Gag in intact cells was achieved by incubating mutant and wild-type cells containing the FLAG-tagged Gag under inducing conditions. Cells containing the Tf1 plasmid without FLAG, pHL449-1, were used as negative controls. Approximately 5 × 107 stationary-phase cells (OD600, 10 to 11) were harvested and fixed with 3.7% formaldehyde (F1268; Sigma) at 32°C for 40 min. The cells were next incubated in 0.1 M phosphate buffer, 1.2 M sorbitol, 0.01% β-mercaptoethanol, and 0.1 mg of 100T zymolase/ml at 32°C for 40 min. The resulting spheroplasts were washed five times in 1 ml of 0.1 M phosphate buffer–1.2 M sorbitol and then resuspended in 0.2 ml of the same buffer and transferred onto polylysine-coated slides. The cells were then immunostained with primary antibody (anti-FLAG M2; 1:1,000 dilution) and secondary antibody (FITC–Oregon green–anti-mouse IgG; 1:500 dilution), respectively. The cells were then mounted on glass slides with mounting solution (1 mg of p-phenylenediamine [P-1519; Sigma]/ml–1 μg DAPI/ml in 50% glycerol). The cells were examined by using a Zeiss Axioscope equipped with UV and FITC optics. Images were collected and imported into Adobe Photoshop version 4.0.1 for figure presentation.
(ii) Nuclear localization of HA-tagged Pst1p.
The yeast strain YHL912 was transformed with plasmids containing either the HA-tagged pst1+ (pHL1626-2) or the untagged pst1+ (pHL1458-3). Cells of the resulting transformants were grown to an OD600 of 0.3, and approximately 5 × 107 cells were harvested by centrifugation. Immunostaining of cells was performed by the same procedure as described above for FLAG-Gag localization. Primary and secondary antibodies were monoclonal anti-HA(Ab) (MMS-101P; Babco) (1:1,000 dilution) and FITC–Oregon green 488–anti-mouse IgG (Molecular Probes) (1:1,000 dilution), respectively.
Histone preparations and analysis.
Cells grown in rich YES medium were harvested at log phase (OD600 of 1). S. pombe histones were isolated from whole-cell extracts as described previously (20) with minor modifications. Histone samples (2 μg) were run on SDS–15% polyacrylamide gels and then electroblotted to nitrocellulose filters. Immunoblot analysis was performed to determine the acetylation levels of histone H4, using antisera raised against specific acetylated lysine residues in histone H4 (7, 71). Global acetylation levels of histones were also determined with rabbit polyclonal IgG antibodies against acetylated histones H3 and H4 (Upstate Biotechnology). Specific acetylation levels of the individual lysines AcK5-H4, AcK8-H4, AcK12-H4, and AcK16-H4 were determined with specific antibodies obtained from B. Turner (R41-K5, R232-K8, R101-K12, and R252-K16, respectively). Calf histones were used as molecular weight markers (Boehringer). Highly acetylated human histones, which were used as a positive control, were a gift from A. Wolffe. Rabbit IgG was detected by ECL (Amersham) with horseradish peroxidase-conjugated anti-rabbit IgG according to the manufacturer’s instructions.
RESULTS
Genetic screen to identify yeast strains deficient for Tf1 transposition.
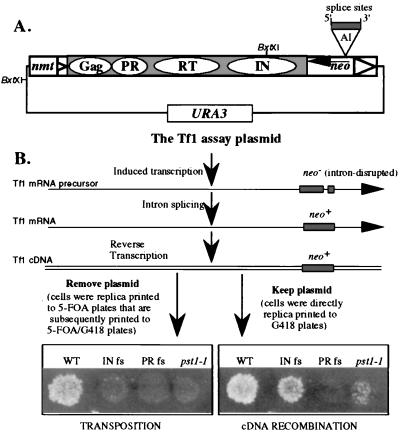
To identify genes in S. pombe that contribute to the propagation of the Tf1 element, we used ethyl methanesulfonate to mutagenize a strain that contained an active copy of Tf1 in a transposition assay plasmid (38). Mutagenized cells were screened for strains that showed reduced levels of transposition. Tf1 activity in individual colonies was monitored by using an assay that detected the resistance to G418 caused by the insertion of neo-marked Tf1 elements into the genome of S. pombe cells (41). The assay plasmid (pHL449-1) carried a copy of Tf1 (Tf1-neoAI) that was placed under the control of the inducible nmt1 promoter. The Tf1 element in the assay plasmid was marked with a bacterial neo gene that was inserted in the orientation opposite to Tf1 transcription (Fig. 1B). An artificial intron was inserted into the neo gene to block the translation of the neo mRNA. The orientation of the intron was opposite to that of neo transcription so that the intron could only be spliced out of the Tf1 transcript, and thus the neo gene could provide G418 resistance only after the Tf1 transcript was reverse transcribed. Cells harboring the plasmid with Tf1-neoAI were induced for transposition by activating the expression of the nmt1 promoter. Before the induced colonies were tested for resistance to G418, the plasmid with Tf1-neoAI was subjected to counterselection by replica printing cells to medium that contained 5-FOA. Thus, only cells that received a transposition event (conferring a G418r phenotype) and subsequently lost the assay plasmid (conferring a 5-FOAr phenotype) papillated on medium that contained G418 and 5-FOA. Figure 1B presents the results of a transposition assay and shows that a patch of cells that initially contained wild-type Tf1-neoAI produced confluent growth on a G418–5-FOA plate. A frameshift mutation in the N terminus of IN was shown to block IN expression and reduce the frequency of G418r by 34-fold (3). In addition, a frameshift in PR blocks the expression of RT and IN and produced no resistance to G418 (Fig. 1B).
FIG. 1.
Genetic screens used to identify strains with decreased Tf1 transposition and cDNA recombination. (A) The transposition assay plasmid, pHL449-1, contained the URA3 gene of S. cerevisiae and the Tf1 element fused to an inducible nmt1 promoter. The BxtXI restriction sites referred to in the text are shown. The shaded portion of the element represents the single ORF that encodes the Gag, PR, RT, and IN proteins. The triangles depict the LTRs. The Tf1 element was genetically marked with a bacterial neo gene, and the arrow indicates the orientation of neo transcription. An artificial intron (AI) was inserted into the neo sequence, and the 5′ and 3′ splice sites are shown. (B) Transposition was induced to high levels by growing cells on medium that activated the nmt1 promoter (the absence of thiamine) and was detected by the G418-resistant phenotype acquired when the neo-marked Tf1 element transposed. The extended arrows labeled with neo represent the Tf1 mRNA before and after the intron is spliced out. After splicing and reverse transcription, the neo gene carried on Tf1 cDNA became functional and was used to detect either the integration of the cDNA into genomic sequence (left panel) or its homologous recombination with other sources of transposon sequences (right panel). The manipulations required for the transposition and recombination assays are described in parentheses. The wild-type strain (YHL1282) and a strain with the pst1-1 mutation (YHL5406) were subjected to the transposition and cDNA recombination assays. These two strains contained the wild-type version of the nmt1-Tf1-neoAI assay plasmid (pHL449-1). The two control strains were the wild-type strain containing the nmt1-Tf1-neoAI assay plasmid with a frameshift (fs) created either in IN (YHL1554) or in PR (YHL1836) of Tf1.
Mutagenized strains that appeared to have significantly less transposition activity were then screened for trivial causes of reduced growth on the G418–5-FOA plates. The original strain we mutagenized contained an integrated copy of LacZ under the control of the nmt1 promoter so that each candidate suspected of possessing defects in transposition could be tested for reduced promoter function. We also retransformed each candidate with a fresh copy of the Tf1-neoAI plasmid to identify which strains were defective for transposition simply due to mutations in the assay plasmid. In addition, we tested strains with a version of Tf1 that contained the arg3+ gene as a transposition marker. In this way we could exclude candidates that showed low growth on the G418–5-FOA plates due to alterations specific to the metabolism of G418. A set of several strains with reduced transposition was identified after we screened a total 2,500 independent colonies. This report describes the analysis of a mutation in a gene that we named pst1+.
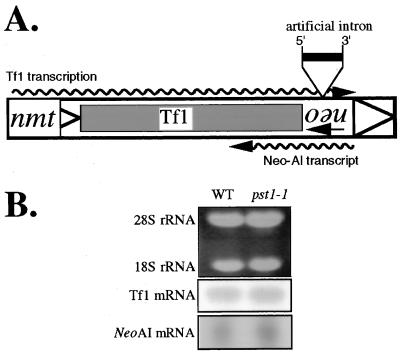
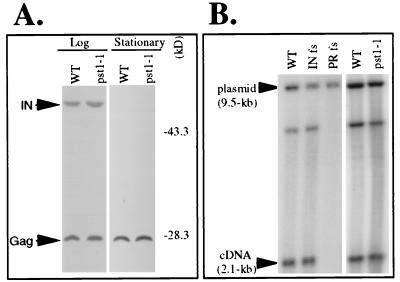
The strain with the pst1-1 mutation was backcrossed with a wild-type strain three times, and 20 tetrads were dissected. Of the 16 tetrads that produced four viable spores, all exhibited a 2:2 segregation of the transposition defect, indicating that the reduced Tf1 activity was due to a mutation in a single gene. Figure 1B shows the transposition defect of a pst1-1 mutant strain that was a product of the three backcrosses. We also found that in the context of diploid strains, this mutation was recessive for the reduced level of transposition (data not shown). After the three backcrosses, additional tests were conducted to determine whether the mutation in the pst1-1 allele lowered the transcription of Tf1 in the context of the assay plasmid. Total RNA was extracted from strains that were grown under inducing conditions (absence of thiamine) and was subjected to RNA blot analysis. The results presented in Fig. 2 showed that the pst1-1 mutation allowed normal levels of Tf1 transcripts to be expressed from the Tf1-neoAI plasmid. There was also the possibility that the pst1-1 mutation reduced the activity of the neo promoter so that even if transposition occurred normally, the insertion events would go undetected. The same RNA blot shown in Fig. 2 was hybridized with a neo-specific probe, and the mutation in the pst1-1 allele was found to have no effect on neo mRNA levels. Once we established that Tf1 mRNA levels were normal in these strains, we considered the possibility that the pst1-1 mutation could have indirectly reduced the level of one or all of the Tf1 proteins. Figure 3A is an immunoblot of proteins extracted from cultures harvested in both stationary and exponential phases. The levels of Gag and IN proteins that accumulated in cells with the pst1-1 mutation were indistinguishable from those of wild-type cells. Because IN is the last protein encoded by the single ORF of Tf1 (41), the normal levels of IN indicated that all Tf1 proteins were translated with wild-type efficiency. No IN is detected in stationary-phase cells because of a degradation mechanism that creates a significant molar excess of Gag relative to IN and RT (4).
FIG. 2.
RNA blot analysis of Tf1 and neoAI transcripts. (A) Schematic presentation of the two transcripts of interest. The largest rectangle represents the Tf1 element, and the triangles indicate the LTRs. The wavy arrows represent Tf1 and neo mRNAs and their transcription orientations. The 5′ and 3′ splice sites of the artificial intron (AI) inserted within the neo sequence are shown. The transcription of Tf1 was under control of the inducible nmt1 promoter, while neo transcription was driven by its own promoter. (B) Total RNA was extracted from cells that either did (YHL5406) or did not (YHL1282) (wild type [WT]) contain the pst1-1 mutation. The rRNAs (top panel), stained with ethidium bromide, were used to monitor equal loading of the lanes. Hybridizations were done with 32P-radiolabelled probes specific for either Tf1 mRNA (middle panel) or neo mRNA (bottom panel).
FIG. 3.
Effects of the pst1-1 mutation on the accumulation of Tf1 proteins and cDNA. (A) Levels of Tf1 Gag and IN accumulated in strains that were wild type (WT) (YHL1282) or contained the pst1-1 mutation (YHL5406). The immunoblots of S. pombe extracts were made from cells harvested either at log phase or at stationary phase. The filters were probed simultaneously with anti-Gag and anti-IN antisera. The positions of molecular mass standards are shown. The positions of Gag and IN are indicated by arrows. (B) DNA blot used to measure the levels of Tf1 cDNA produced in the strain with the pst1-1 mutation (YHL5406). The three control strains were the wild-type strain containing versions of Tf1 that either were wild-type (YHL1282) or contained a frameshift (fs) mutation in IN (YHL1554) or a frameshift mutation in PR (YHL1836). DNA was extracted from S. pombe strains induced for Tf1 expression. The DNA was digested with BstXI and probed with neo-specific sequence. The 2.1-kb band was generated by Tf1 cDNA, while the 9.5-kb band was produced by vector sequence and was used to monitor equal loading.
pst1-1 mutation caused a defect in a late step of the transposition pathway.
To measure the magnitude of the transposition defect caused by the pst1-1 mutation, we subjected strains to a previously developed quantitative assay (3). Patches of cells induced for Tf1 expression were resuspended in liquid and spread onto agar medium that contained 5-FOA. Approximately 5,000 to 10,000 5-FOA-resistant colonies per plate were then replica printed to plates with G418 and 5-FOA to determine what fraction of the cells induced for transposition received an insertion of Tf1-neo. The results of the quantitative transposition assay presented in Table 3 show that the strain with the pst1-1 mutation produced 10-fold fewer transposition events than did the wild-type strain.
TABLE 3.
Tf1 cDNA recombination and transposition frequencies of wild type and pst1-1 mutant strain
| Strain | Description | % Transposition (5-FOAr-G418r colonies/5-FOAr colonies) | % cDNA recombination (G418r colonies/YES colonies) |
|---|---|---|---|
| YHL1282 | Wild-type strain containing Tf1-neoAI | 2.1 (276/13,368) | 0.43 (1,734/398,200) |
| YHL1554 | Wild-type strain containing Tf1-neoAI with IN frameshift | 0.1 (10/11,640) | 0.13 (466/351,639) |
| YHL1836 | Wild-type strain containing Tf1-neoAI with PR frameshift | <0.01 (0/11,412) | <0.001 (0/347,096) |
| YHL5406 | pst1-1 mutant strain containing Tf1-neoAI | 0.2 (26/12,166) | 0.05 (186/402,450) |
The mutagenized strains were also screened with an assay that measured homologous recombination between copies of Tf1 cDNA and Tf1 plasmid sequences (3). Wild-type levels of this recombination indicate that normal levels of Tf1 cDNA are produced and that this cDNA is transported to the nucleus. The homologous recombination assay was performed with the same Tf1-neoAI plasmid shown in Fig. 1. As described above, the neo marker of the Tf1-neoAI plasmid was initially unable to provide resistance to G418 because the intron in neo could only be spliced from the Tf1 mRNA. The reverse transcripts of the spliced Tf1 mRNA, when present in the nucleus, homologously recombine with the Tf1-neoAI plasmid to generate a G418-resistant version of the assay plasmid. The difference between the transposition and recombination assays was that cells tested for recombination were printed from the induction medium directly to plates with G418 without first selecting against the presence of the plasmid on plates with 5-FOA. The recombination assay shown in Fig. 1B shows that a wild-type copy of Tf1-neoAI produced confluent growth on agar medium that contained G418. The confluent growth produced by a version of Tf1-neoAI that lacked IN and the lack of G418 resistance due to a frameshift just upstream of RT were demonstrations used to indicate that the homologous recombination assay detects products of reverse transcription even in the absence of IN activity (3). Figure 1B also shows that the pst1-1 mutation caused a reduction in the homologous recombination of Tf1 cDNA compared to the wild-type strain and the strain with the frameshift in IN. The magnitude of the recombination defect was determined by subjecting the strains to the quantitative version of the homologous recombination assay (3). Patches of yeast cells induced for Tf1 expression were resuspended in liquid and spread directly onto plates that contained G418. The number of colonies that grew on G418 was divided by the total number of viable cells plated to determine the fraction of cells that generated resistance to G418. Table 3 shows that the strain with the pst1-1 mutation produced eightfold-lower levels of cDNA recombination than the wild-type strain. However, the recombination defect in the pst1-1 mutant strain represented a threefold decrease when compared to the levels of recombination produced by a strain without IN (YHL1554 [Table 3]). This may be considered a more appropriate comparison, since the lack of IN generates a baseline level of resistance to G418 that does not include transposition.
The results of the recombination assays suggested that either the levels of Tf1 reverse transcripts were reduced by the pst1-1 mutation or that normal levels of cDNA were produced but they accumulated in a compartment that was inaccessible to the Tf1-neoAI plasmid. To investigate these possibilities, we used a previously published method of DNA blot analysis to measure the accumulated levels of Tf1 cDNA in cells that contained the pst1-1 mutation (3, 39). Liquid cultures of cells induced for Tf1 expression were extracted for total DNA that was then digested with BstXI and subjected to DNA blot analysis. The results presented in Fig. 3B show that the wild-type Tf1-neoAI produced a measurable amount of a 2.1-kb fragment of cDNA detected with a neo-specific probe. This BstXI fragment is derived from the terminal sequence of Tf1 that is the final region to be reverse transcribed and therefore represents the levels of full-length products. The DNA extracted from control strains showed that the frameshift in IN did not reduce the intensity of the 2.1-kb band, whereas the frameshift just upstream of RT blocked the accumulation of cDNA. In comparison, the strain with the pst1-1 mutation generated normal levels of the 2.1-kb fragment of cDNA. A 9.5-kb band resulted from the BstXI digestion of the Tf1-neoAI plasmid, and this species served as an internal control for levels of DNA loaded in each lane.
The fact that the strain with the pst1-1 mutation showed a defect in cDNA recombination despite its ability to produce wild-type levels of cDNA suggested that the general recombination machinery could be defective. To investigate this possibility, the frequencies of homologous recombination between two defective alleles of the ade6 gene were measured by testing for crossover events that generated adenine prototrophy. The plasmid pade6-M375-M26 (55) that contained an allele of ade6 with a double mutation at the beginning of the ORF was transformed into two yeast strains that both contained a chromosomal allele of ade6 with a point mutation near the end of the ORF, ade6-M210. One of the yeast strains contained the pst1-1 allele, while the other was pst1+. Two independent colonies of each strain were grown for 2 days in liquid EMM lacking uracil, and mitotic recombination frequencies were determined by plating the cells onto EMM agar lacking adenine. The number of Ade+ prototrophs compared to the number of cells viable on rich medium (YES) represented the frequency of recombination between the two point mutations at the ade6 locus. As shown in Table 4, the frequency of mitotic recombination was not significantly affected by the pst1-1 mutation.
TABLE 4.
Mitotic recombination in the isogenic wild-type and pst1-1 yeast strains
| Strain | Relevant genotype/plasmid | Recombination frequencya (per 106) |
|---|---|---|
| HL5834 | pst1-1 ade6-M210/pade6-M375-M26 | 25 |
| HL5835 | pst1-1 ade6-M210/pade6-M375-M26 | 40 |
| HL5852 | pst1+ ade6-M210/pade6-M375-M26 | 20 |
| HL5853 | pst1+ ade6-M210/pade6-M375-M26 | 30 |
Median mitotic recombination frequencies were determined as described previously (60).
Taken together, the accumulation of normal amounts of Tf1 cDNA, the low levels of cDNA recombination, and the wild-type recombination at the ade6+ locus indicated that the transposition defect caused by the pst1-1 mutation occurred in a late step in the retrotransposition pathway, i.e., after reverse transcription. To gain further information about the defect caused by the pst1-1 allele, we isolated the wild-type allele of the pst1+ gene from a library of genomic DNA.
pst1+ is a homologue of the SIN3 genes found in mammals and S. cerevisiae.
The reduced levels of transposition and cDNA recombination caused by the pst1-1 mutation served as the basis for cloning the wild-type copy of the pst1+ gene. For this purpose, we transformed a library of plasmids that contained S. pombe genomic DNA into a strain with the pst1-1 mutation and screened for complementation of the transposition defect. The plasmid library consisted of a multicopy vector containing genomic inserts that averaged 10 kb in length (kindly provided by P. Young). Out of a total of 130,000 colonies screened, only one plasmid was isolated for its ability to complement both the transposition and cDNA recombination defects caused by the pst1-1 mutation. Initial sequence data from this insert revealed that its sequence was available from the S. pombe genome project (accession no. Z54140).
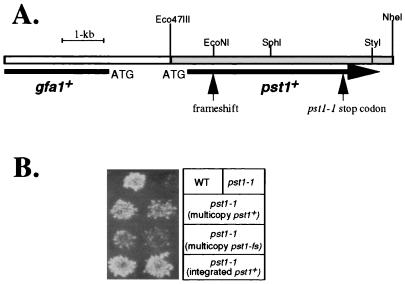
Figure 4A is a diagram of the isolated fragment with the significant ORFs and the restriction sites used for subsequent experiments. The sequence began with a truncated ORF whose predicted amino acid sequence had 58.6% identity to that of the product of the GFA1 gene of S. cerevisiae. The second part of the genomic fragment contains the entire ORF for a hypothetical protein with an amino acid sequence showing 36.6% identity to the ySin3p protein of S. cerevisiae (77). Subsequent subcloning studies showed that the 5.6-kb Eco47III-NheI fragment, which contained the homologue of SIN3, was responsible for the efficient complementation of the defects due to the pst1-1 mutation (Fig. 4). We also found that a frameshift generated at the beginning of the second exon of pst1+ severely reduced the complementation activity of the plasmid (Fig. 4). These data clearly indicated that the second ORF, which encodes a homologue of ySin3p, was the source of the complementation. To test whether this gene was allelic to pst1+, we subcloned it into an integrative vector that contained the leu1+ gene of S. pombe. The resulting plasmid was integrated into the genome of a haploid with the pst1-1 allele at the site of the complementing ORF. A single integrated copy of this genomic fragment was found to retain its ability to complement the pst1-1 transposition defect (Fig. 4B). The resulting strain was crossed with a wild-type pst1+ haploid strain, and 16 tetrads were dissected. All four spores of these tetrads possessed wild-type transposition and cDNA recombination activity (data not shown). Taken together, these data indicate that the homologue of SIN3 was allelic to pst1+. We named this gene pst1+ as an abbreviation for pombe SIN-three no. 1.
FIG. 4.
Isolation and characterization of the pst1+ gene of S. pombe. (A) Schematic representation of the original genomic fragment that complemented the transposition defect caused by the pst1-1 mutation. The ORF corresponding to the pst1+ allele is indicated by a large arrow, whereas the truncated ORF with high homology to the GFA1 gene of S. cerevisiae is also shown, with the location of its ATG codon. The restriction sites referred to in the text are shown. The Eco47III-NheI fragment responsible for the complementation of the pst1-1 transposition defect is indicated as a shaded region. The locations of the point mutation (stop codon) and the artificial frameshift in pst1 are also shown. (B) Transposition assay. Wild-type (WT; YHL4988) and pst1-1 (YHL4994) are the strains in the top row. The three lower rows are the pst1-1 strain that contained an additional allele of the pst1 gene. The name and copy number of each additional allele are indicated in parentheses. The second, third, and fourth rows are strains YHL6214, YHL6221, and YHL6222, respectively. The transposition assay was carried out as described in Materials and Methods.
To gain further information about the role of pst1+ in the propagation of Tf1, we cloned the chromosomal copy of the pst1-1 allele by PCR and determined its nucleotide sequence. Two independent PCR products of each region were cloned and sequenced (see Materials and Methods for details). The sequences of the cloned PCR products were compared to the wild-type pst1+ sequence retrieved from the S. pombe database as well as the sequence of the wild-type pst1+ clone isolated from the S. pombe library. A single G→A substitution was found in the sequence of the pst1-1 allele that converted codon 1305 from a tryptophan residue into a stop codon. The result of this nonsense mutation was predicted to stop translation of the pst1 mRNA, producing a truncated protein that lacked 217 amino acids from its C terminus (Fig. 4A and 5A).
FIG. 5.
Amino acid sequence for Pst1 protein. (A) The deduced amino acid sequence encoded by the pst1+ gene of S. pombe is shown. The ORF is interrupted by an intron in the gene whose position is indicated by the arrowhead. Each pair of boxes connected by underlining represents a PAH motif; all the pairs have been labeled PAH1 to PAH4. The solid circle denotes the position of the pst1-1 point mutation where a tryptophan residue was replaced by a stop codon. (B) Deduced amino acid sequences encoded by pst1+ and pst2+ of S. pombe are schematically shown aligned to that encoded by SIN3 of S. cerevisiae and mammalian mSIN3A and mSIN3B, using MACAW algorithms (65). The length of each protein is given. The solid boxes correspond to the four conserved PAH domains. The shaded boxes indicate the locations of HIDs, and the hatched boxes are conserved regions just downstream of PAH4. The scores of sequence similarity given in parentheses for each conserved domain represent the percentages of the amino acids in alignments that are similar within all five proteins. The open boxes represent nonconserved regions.
Putative splicing signals were predicted in the pst1+ ORF and reported in the S. pombe database (accession no. Z54140). A conserved splice branch and acceptor sequence TACTAATTATTTGATTAG (43) can be found in the pst1+ gene at nucleotides 249 to 266, relative to the ATG codon. The 5′ splice site (GTATGT; nucleotides 130 to 135) is also present. We demonstrated that this putative intron is spliced in vivo by determining the sequence of the pst1+ mRNA by a reverse-PCR technique (data not shown). The spliced message is predicted to encode a polypeptide of 1,522 amino acids (171 kDa) (Fig. 5A). As mentioned above, sequence comparisons showed that Pst1 protein (Pst1p) possessed significant similarity to ySin3p of S. cerevisiae and its mammalian mSin3A and mSin3B counterparts (5, 77). Furthermore, the examination of the S. pombe sequence database identified a second Sin3 homologue that we named pst2+ (accession no. Z98559). Figure 5A shows the predicted amino acid sequence of Pst1p and indicates the position of the four PAH domains. The PAH motif consists of two amphipathic helices separated by a loop of about 20 amino acids (77), which is similar to the amphipathic helical structure found in the tetratricopeptide repeat motif and the helix-loop-helix motif. It has been suggested that the amphipathic helices participate in protein dimerization or in the mediation of protein-protein interactions (23, 26, 29, 47, 66, 75). The sequences of Pst1p and Pst2p of S. pombe were aligned with the sequences of ySin3p, mSin3A, and mSin3B. A schematic representation of this alignment was generated by the program MACAW version 2.0.5 (Fig. 5B). Examination of the alignments revealed that the predicted amino acid sequence of S. pombe Pst1p and Pst2p displayed blocks of similarity to the S. cerevisiae and mammalian Sin3 proteins throughout their respective ORFs (Fig. 5B). The regions of highest similarity between the Pst and Sin3 proteins were centered around the four PAH domains, suggesting that these regions in the S. pombe proteins possess important functions. In addition, the histone deacetylase-interaction domain (HID) located between PAH3 and PAH4 (37) and the region just following PAH4 are highly conserved in all Sin3 and Pst proteins.
pst1+ is an essential gene.
Previous studies demonstrated that the budding yeast SIN3 is required for normal sporulation of diploid cells (50, 67, 73) but is not required for viability (77). To test whether pst1+ is required for sporulation or cell viability, we deleted just the ORF of pst1+ from one allele of a diploid strain and replaced it with the his3+ marker gene. Only 5 of 40 dissected tetrads produced two viable spores and two dead spores, while the rest of the tetrads (35) produced one or no viable spores. In addition, all of the germinating spores were auxotrophic for histidine, suggesting that the deletion of pst1+ and its replacement by the his3+ marker may inhibit spore germination and/or viability. The heterozygous diploid was subjected to random spore analysis, and none of the 500 germinating spores was prototrophic for histidine. Although these results suggested that the pst1+ gene was required for viability, we tested the possibility that the gene was only required for spore germination. A plasmid-shuffling technique was used for this purpose. From the diploid described above, we generated a haploid strain that contained the his3+ substitution of the pst1+ gene by introducing a wild-type copy of pst1+ carried on a LEU2 plasmid. The haploid was transformed with a second plasmid that contained pst1+ as well as the S. cerevisiae URA3 marker. This strain was first grown as a patch and then streaked on EMM that lacked uracil but contained leucine to allow for random loss of the LEU2 plasmid. These colonies were then replica printed to an agar plate that contained uracil and not leucine and to another agar plate that contained uracil, leucine, and 5-FOA. The 5-FOA selects against the presence of the URA3-expressing plasmid (11). The growth phenotypes were scored after a 2-day incubation either at 25, 32, or 37°C. After a second printing to plates that contained 5-FOA, the colonies that lacked the copy of pst1+ on the LEU2 plasmid did not grow on the 5-FOA plate. In comparison, the colonies that retained the LEU2 plasmid did form 5-FOA-resistant colonies. These results provided strong evidence that pst1+ was essential for vegetative growth.
Previous studies performed with Sin3 counterparts of other organisms strongly suggested a critical role for Sin3 proteins in histone deacetylation (54, 79). We therefore conducted a series of experiments to address whether pst1+ of S. pombe was also involved in these processes and whether alterations in histone deacetylation affected Tf1 transposition.
Cellular localization of HA-tagged Pst1 protein.
The role of Sin3 proteins in remodeling chromatin structure and regulating transcription, as shown in previous studies of mammalian and budding yeast Sin3 proteins, suggested that Sin3p is a nuclear protein. Indeed, budding yeast Sin3p and mammalian mSin3B proteins are located in the nucleus (1, 77). We found that Pst1p was also localized in the nucleus, by using indirect immunofluorescence microscopy. For this purpose, we created a yeast strain with a plasmid that contained pst1+ with a triple-HA epitope inserted at the C-terminal end. In this strain, HA-tagged pst1+ was expressed from its own promoter, and the presence of the HA epitope in the sequence of pst1+ was found not to alter its ability to complement the transposition and cDNA recombination defects (data not shown). S. pombe cells were grown to early log phase (OD600, 0.3), fixed with formaldehyde, prepared for immunofluorescence staining with an anti-HA monoclonal antibody (HA.11; Babco), and subjected to immunofluorescence microscopy. The results presented in Fig. 6 indicate that the FITC signals corresponding to HA-tagged Pst1p were distributed throughout the nucleus, as visualized by DAPI staining of chromosomal DNA (Fig. 6B). The control strain contained a plasmid with the pst1+ allele that lacked the HA tag. This strain produced no FITC signal (Fig. 6A), indicating that the signals generated by the experimental strain were specific for HA-tagged Pst1p. Nuclear localization of Pst1p was also observed when it was expressed from a single integrated copy of the HA-tagged pst1+ allele (data not shown). The fact that S. pombe Pst1p is a nuclear protein is consistent with its assumed role in the deacetylation of histones.
FIG. 6.
Cellular localization of Pst1p. Yeast cells with a plasmid copy of pst1+ that either did not (A) or did (B) contain an HA epitope were grown in YES medium to an OD600 of 0.3. The cells were harvested and subjected to immunofluorescence microscopy. The two right panels show the FITC signals obtained with the anti-HA antibody, and the two left panels show the locations of nuclei as indicated by DAPI counterstaining.
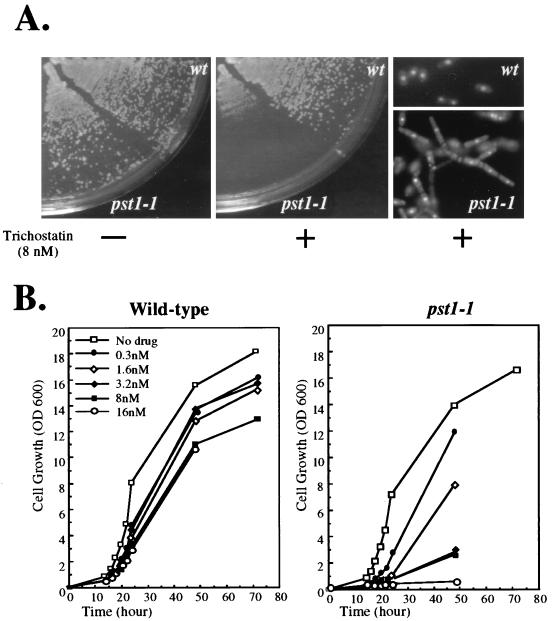
A strain with the pst1-1 mutation was highly sensitive to the histone deacetylase inhibitor TSA.
To investigate the possibility that Pst1p plays a role in the deacetylation of histones, we tested the ability of a strain with the pst1-1 mutation to grow in the presence of the histone deacetylase inhibitor TSA. As shown in Fig. 7A, a strain with the pst1+ allele was able to grow in the presence of 8 nM TSA whereas an isogenic strain with the pst1-1 allele grew much more slowly, such that no colonies were visible after 3 days of incubation at 32°C. We also measured the growth rates of the same strains in liquid cultures. Figure 7B shows the growth curves of the wild-type strain and the strain with the pst1-1 mutation in the presence of increasing TSA concentrations ranging from 0 to 16 nM. In the absence of TSA, the growth of the two strains was indistinguishable. When added at concentrations as high as 16 nM, TSA only caused small changes in the growth of the wild-type strain whereas the growth of the strain with the pst1-1 allele was strongly inhibited, and no growth was observed when TSA was used at 16 nM. The supersensitivity of the strain with the pst1-1 mutation to TSA was consistent with the proposed involvement of S. pombe Pst1p in the histone deacetylation process. Remarkably, when grown in liquid medium with 8 nM TSA, the strain with the pst1-1 mutation exhibited a morphological change by shifting from the unicellular growth mode to a filamentous growth mode based on the formation of pseudohyphal cells (Fig. 7A). Extensive filamentous growth led to the formation of mycelium-like structures that displayed pseudohyphae with branches and lateral buds. In comparison, exposure to TSA did not cause any apparent morphological change to the strain with the pst1+ allele (Fig. 7A).
FIG. 7.
Effect of the pst1-1 mutation on cell growth in the presence of TSA. (A) Supersensitivity of the strain with the pst1-1 mutation to TSA. Cells of strains that contained either pst1+ (YHL1282) or pst1-1 (YHL5406) allele were streaked on EMM-Ura. Growth in the presence (+) (middle panel) or absence (−) of 8 nM TSA (left panel) was scored after 3 days at 32°C. The pseudohyphal growth mode of the cells with the pst1-1 mutation when grown in liquid medium supplemented with 8 nM TSA is also shown (bottom right panel), while the wild-type cells maintained the yeast form (top right panel). (B) Growth rates of strains with pst1+ and pst1-1 allele in increasing concentrations of TSA. Cell growth in EMM-Ura was monitored by following OD600 as a function of time. The concentrations of TSA used for this experiment are also presented.
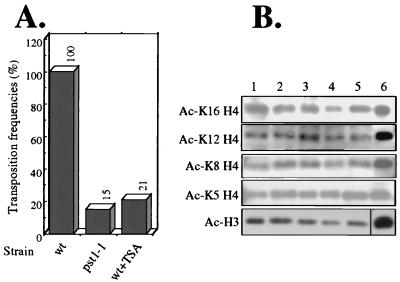
TSA inhibited Tf1 transposition.
If Pst1p of S. pombe is similar to the Sin3 proteins of other organisms it would work together with histone deacetylases to change the acetylation pattern of chromatin. If the mutation in the pst1-1 strain inhibited Tf1 transposition as the result of reduced deacetylase function then exposure to TSA could cause the reduction of Tf1 transposition by inhibiting the histone deacetylase activity associated with Pst1p. To test this prediction, we performed quantitative transposition assays on strains grown in the presence and absence of TSA. In these experiments, TSA was present at 8 nM in all steps of the transposition assay. As shown in Fig. 8A, TSA treatment during transposition significantly inhibited Tf1 activity in the strain with the pst1+ allele (wild type). The reduction in transposition caused by TSA treatment was similar in magnitude to the low transposition levels caused by the mutation in the pst1-1 strain. This finding suggests that the pst1-1 mutation may affect Tf1 transposition through the histone deacetylation pathway.
FIG. 8.
Effects of either the pst1-1 mutation or exposure to TSA on Tf1 transposition and histone acetylation of bulk chromatin. (A) Transposition activity of Tf1 was measured in wild-type (wt; YHL1282) and pst1-1 mutant (YHL5406) strains, using the quantitative transposition assay. If present, the concentration of TSA was 8 nM. The transposition level of the wild-type strain in the absence of TSA was defined as 100%. (B) Acetylation levels of bulk chromatin in wild-type (YHL1282) and pst1-1 mutant (YHL5406) strains. Whole-cell extracts of histones were isolated from cells grown in EMM-Ura medium and harvested at an OD600 of 1. Histone samples of two independent cultures of each strain, either wild-type (lanes 1 and 2) or the strain with the pst1-1 mutation (lanes 3 and 4), were separated on SDS-polyacrylamide gels, electroblotted onto nitrocellulose membranes, and probed with appropriate antibodies. Lane 5, histone extract of the wild-type strain grown in the presence of 8 nM TSA. Lane 6, highly acetylated human histones that were used as molecular weight markers. Coomassie staining was used to monitor equal loading (not shown). The blots were performed individually and assembled for the figure.
If the pst1-1 mutation reduced transposition due to changes in chromatin acetylation then large changes in the levels of acetylation may have occurred. To investigate this possibility, we measured the acetylation of specific lysines in bulk preparations of histones H3 and H4. In these experiments, cells with and without the pst1-1 mutation were harvested from cultures grown under the same conditions that were used for the quantitative transposition assay. Histones were extracted from whole cells by established methods (20), electrophoretically separated on SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with appropriate antibodies. Since histone H4 has been known to be mainly acetylated at the specific lysine residues K5, K8, K12, and K16 (13, 68, 80), we used antibodies that were raised against histone H4 with acetylation specifically at these lysines (7, 71). We also used antibodies against histone H3 acetylated at multiple lysines (Upstate Biotechnology). Data presented in Fig. 8B show that neither the pst1-1 mutation nor the treatment with 8 nM TSA significantly affected the global acetylation levels of lysines 5, 8, 12, and 16 of histone H4 or the bulk level of acetylation of lysines in histone H3. This suggests that the pst1-1 mutation did not cause a global change in the acetylation of histones. In contrast, it was shown previously that an extended exposure of S. pombe cells to very high concentrations of TSA, such as 80 nM, resulted in an overall increase in acetylation at lysine 5 of histone 4 (20).
Localization of the Tf1 complex is disrupted in the pst1-1 strain.
One interesting characteristic of cells with the pst1-1 mutation was the low levels of homologous recombination that occurred between Tf1 cDNA and cellular sequences of Tf1 (Fig. 1B). The fact that cells with the pst1-1 allele produced normal levels of cDNA led us to test the possibility that cDNA recombination was low because of a defect in nuclear import.
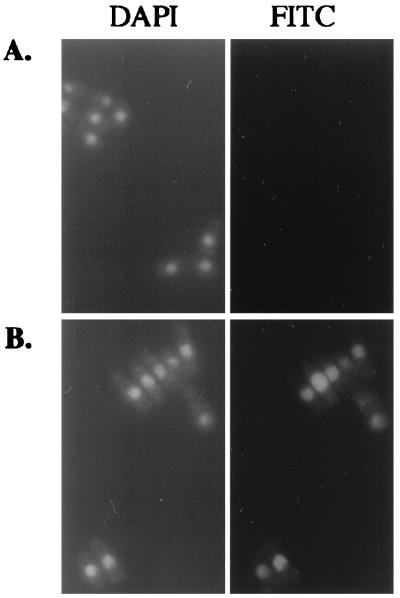
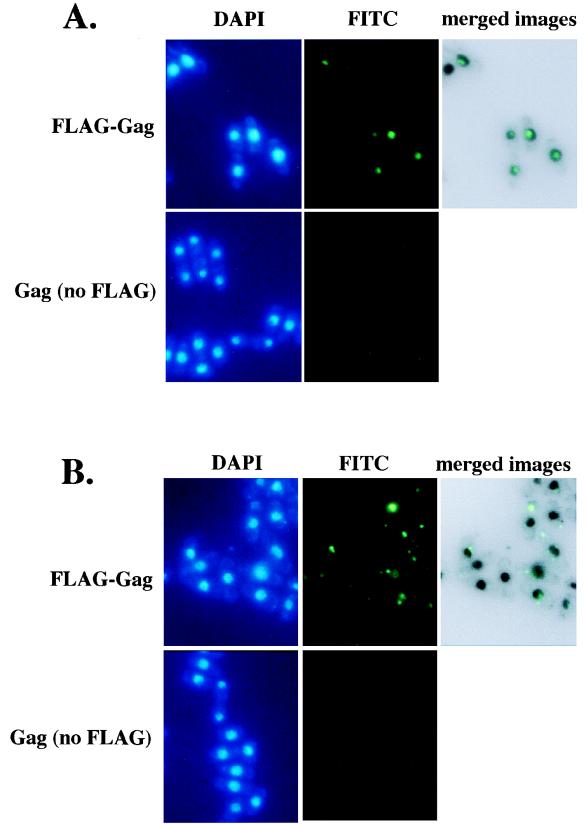
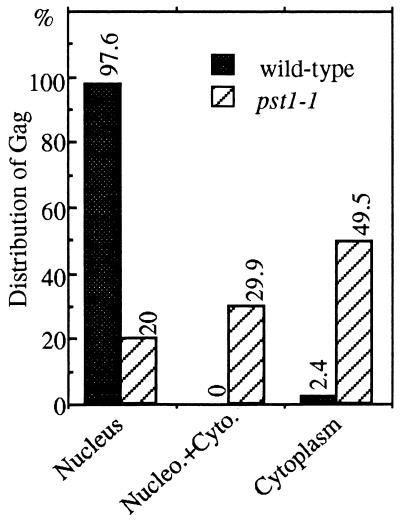
To investigate the possibility that the pst1-1 mutation reduced the nuclear localization of Tf1 material, we studied the localization of Tf1 Gag. This protein was chosen because it is present in high molar ratios in a large macromolecular complex with Tf1 cDNA and IN (41). We studied the localization of Gag in stationary-phase cells as expressed from the Tf1-neoAI plasmid. For this purpose, a FLAG epitope was inserted near the C terminus of Gag. The expression level of Gag was unaffected by the FLAG epitope, as determined by immunoblot analysis with Gag(Ab) and FLAG(Ab), and the resulting transposon, Tf1(FLAG)-neoAI, possessed wild-type levels of transposition and homologous recombination activity (data not shown). Wild-type cells that were induced for the expression of Tf1(FLAG)-neoAI were subjected to immunofluorescence staining with the M2 anti-FLAG monoclonal antibody and the Oregon green 488 goat anti-mouse IgG conjugate. We found that the majority of wild-type cells produced a single primary focus of Gag signal within the nucleus, as demonstrated by the colocalization of the anti-FLAG antibody and nuclear staining by DAPI (Fig. 9A). For this purpose, the fluorescence image produced by the anti-FLAG antibody was merged with an inverted black-and-white image of the nucleus generated by DAPI counterstaining. We found that no FLAG signal was observed from cells that were not induced for Tf1(FLAG)-neoAI expression (Fig. 9A), indicating that the anti-FLAG signals were specific for Tf1. A totally different scenario was observed in the strain with the pst1-1 mutation. FLAG-Gag appeared to form multiple aggregates spread quasirandomly throughout the cytoplasm (Fig. 9B). Indeed, very few nuclei appeared to contain Gag, suggesting that the nuclear localization of FLAG signals was disrupted in this strain. Figure 10 presents an analysis of 97 nuclei from cells with the pst1-1 mutation and 82 nuclei from wild-type cells. We categorized the cells into three classes reflecting the position of the Gag signal within the cells: (i) a single predominant signal that overlapped with and was thought to be located within the nucleus, (ii) multiple Gag signals observed predominantly in the cytoplasm but some Gag also overlapping with the DAPI staining, and (iii) single or multiple signals clearly positioned outside the nucleus. The data presented in Fig. 10 indicate that the pst1-1 mutation greatly reduced the nuclear localization of Gag.
FIG. 9.
Immunofluorescence analysis of Tf1 FLAG-Gag. (A) Immunofluorescence of Tf1 Gag in wild-type cells. The wild-type strain YHL5896 contained a FLAG-Gag version of the Tf1-neoAI plasmid (pHL1277). Cultures were grown under inducing conditions in EMM-Ura. In the top panels, the green FITC signals are specific for the FLAG-Gag protein and the blue signals indicate the locations of nuclei counterstained with DAPI. In the control experiments (bottom panels), the plasmid pHL449-1 was used in place of pHL1277 in the same wild-type strain. The upper right panel is a merge of the FLAG-Gag signals produced by YHL5895 with an inverted black-and-white image of its DAPI stain. The merge was generated with Adobe Photoshop version 4.0 with the screen function set at 65% opacity. (B) Same experiment shown in panel A, except the yeast strains contained the pst1-1 mutation (YHL5356). The top panel shows a strain with the FLAG-Gag version of the Tf1-neoAI plasmid (pHL1277), and the bottom panel shows a strain with the control plasmid pHL449-1.
FIG. 10.
Distribution of Gag in strains with pst1+ and pst1-1 alleles. Immunofluorescence-stained cells with the wild-type and pst1-1 alleles from the experiments described in the legend to Fig. 9 were analyzed for the localization of Gag relative to the locations of nuclei. The localization data are from approximately 100 cells. The cells were categorized into three classes. (i) Cytoplasm: all of the Gag signals were exclusively within the cytoplasm. (ii) Nucleus plus cytoplasm: the cells had significant amounts of Gag in both the cytoplasm and the nucleus. (iii) Nucleus: all the Gag in the cells was contained in the nucleus.
DISCUSSION
The application of a genetic assay for retrotransposition has allowed us to generate strains of S. pombe with host mutations that greatly reduce the function of Tf1. The yeast strain with the pst1-1 allele was found to have a mutation that lowered the frequency of transposition 10-fold and lowered homologous recombination of cDNA 8-fold. Despite this lack of transposition activity, we found that normal levels of element-encoded proteins and cDNA were produced, indicating that a defect occurred in a late step of transposition, i.e., after reverse transcription.
Chromatin remodeling factor Pst1p is required for retrotransposition.
The pst1+ gene was found to contain coding sequence for a protein of 1,522 amino acids that possessed four PAH motifs and high sequence similarity to the S. cerevisiae ySin3 protein as well as its counterparts in mammals (5, 77). In budding yeast, the single ySin3 protein was initially inferred to be a corepressor based on the observation that a LexA-Sin3 fusion can repress transcription when brought to a heterologous promoter (78). Indeed, ySin3p together with histone deacetylase Rpd3p negatively regulates a diverse set of yeast genes (74).
Recently, ySin3p, Rpd3p, and specific DNA-binding proteins have been found to form a large multiprotein complex (32, 33) that represses transcription by targeting histone deacetylase activity to specific regions of chromatin (31, 32, 62). Two Sin3 proteins, mSin3A and mSin3B, have been isolated from mice (5). Both mSin3A and mSin3B have been shown to function as corepressors that interact with proteins belonging to the Mad family of DNA-binding proteins. The resulting complex antagonizes the activation and transformation functions of the oncoprotein Myc (5, 64). In addition, mammalian Sin3 proteins have recently been shown to form large multiprotein complexes that include mammalian histone deacetylases (mRPD3, HDAC1, and HDAC2); DNA-binding repressors, such as Mad (27, 37), YY1 (81), and MeCP2 (30, 49); and transcriptional repressors for nuclear receptors, such as SMRT (48) and N-CoR (1, 28). The formation of complexes containing Sin3 proteins and histone deacetylases has also been proposed to be the mechanism used by mammalian cells to target histone deacetylase activity, thereby repressing transcription (24, 54, 68, 79).
Interestingly, BLAST-based alignments revealed that S. pombe also has two distinct members of the Sin3 gene family, pst1+ and a second gene we suggested be called pst2+. The presence of a second gene encoding a Sin3-like protein in S. pombe is in contrast to the single SIN3 gene of S. cerevisiae. In this respect, S. pombe is similar to mammalian cells. We found that both Pst1p and Pst2p displayed blocks of similarity to S. cerevisiae and mammalian Sin3 proteins throughout their respective ORFs. In particular, high levels of conservation exist in the regions around the four PAH domains and the HID located between PAH3 and PAH4 (37). This suggests that Pst1p and Pst2p are functionally related proteins. However, Pst1p and Pst2p possess significant sequence divergences. The most pronounced differences between Pst1p and Pst2p are at the N and C termini of the two proteins. In addition, Pst2p has a shorter amino-terminal region than Pst1p and has deletions relative to Pst1p in the regions between PAH1 and PAH2. Interestingly, similar types of sequence divergences and truncations in the mammalian mSin3B relative to mSin3A have also been reported (5). The obvious and interesting question is whether the two genes pst1+ and pst2+ in S. pombe play distinct roles in specific cellular functions and, if so, whether their roles are conserved from S. pombe to mammals. We demonstrated that the deletion of the chromosomal copy of pst1+ in a haploid cell caused cell death, indicating an essential role for pst1+ in vegetative growth. This result suggested that the two Pst proteins in S. pombe are not interchangeable.
pst1+ is an essential gene that contributes to histone deacetylation.
The fact that the single SIN3 gene of S. cerevisiae is not required for viability suggests that the chromatin remodeling due to Sin3p-mediated histone deacetylation does not play an essential role in the function of chromatin either at centromeres or at heavily transcribed regions of the genome. It was therefore surprising that the pst1+ gene of S. pombe was essential for growth. This finding raises the distinct possibility that Sin3 proteins of other organisms could possess vital functions. The presence of distinct homologues of Sin3 proteins in S. pombe and the similarity of these proteins to their mammalian counterparts suggest the possibility that Sin3 proteins of mammals may also be essential for viability.
Our observation that the growth of a strain with the pst1-1 mutation was severely inhibited by TSA was consistent with the possibility that TSA targeted the essential function of pst1+. Since TSA is an inhibitor of histone deacetylases, this result indicates that an essential function of pst1+ is its recruitment of deacetylases and its contribution to the deacetylation of histones. In addition, the supersensitivity of the pst1-1 strain to TSA suggests that the pst1-1 mutation resulted in reduced levels of histone deacetylase activity.
When TSA was added at sublethal concentrations, the strain with the pst1-1 mutation exhibited slow growth and an extensive filamentous morphology. This finding suggests that S. pombe required the corepressor Pst1p to maintain its yeast morphology. A similar situation has been reported for the pathogenic yeast Candida albicans, which requires another corepressor, the general transcription corepressor TUP1, to maintain its yeast morphology (12). When this repression is lifted under inducing environmental conditions, C. albicans shifts its growth mode from unicellular budding to growth that is pathogenic, invasive, and hyphal (51). In this respect, the TSA sensitivity of the pst1-1 strain could be used as a screen to identify mutations in deacetylation mechanisms. In addition, given the general lack of information about the mechanisms controlling pseudohyphal growth in S. pombe, the strain with the pst1-1 mutation could be used to identify mutations in genes responsible for initiating pseudohyphal growth.
Inhibition of Tf1 transposition by TSA.
Exposure of a wild-type strain of S. pombe to TSA significantly reduced the transposition frequency of Tf1. Intriguingly, the reduction in transposition caused by TSA treatment was similar in magnitude to the lowered transposition levels caused by the pst1-1 mutation. An interesting question was whether the mutation in pst1-1 affected transposition through histone deacetylation or through an independent mechanism. The sensitivity of Tf1 to TSA supported the idea that the low levels of transposition observed in a pst1-1 strain were due to a decrease in histone deacetylase activity. However, we cannot ignore the possibility that the defects in Tf1 function caused by TSA treatment were the indirect consequences of changes in other cellular functions (82).
It has recently been shown that in S. pombe, TSA treatment increased histone acetylation in centromeric chromatin, resulting in perturbations of centromere functions that included loss of centromere silencing, delocalization of centromere proteins, chromosome lagging in anaphase, and perturbation of chromosome segregation (20). Interestingly, the strain with the pst1-1 mutation also exhibited the same types of defects (19a), indicating that pst1+ may be required to maintain the underacetylated state at centromeres. To see whether strains with the pst1-1 mutation possessed a general change in histone deacetylation, we measured acetylation levels of specific lysines within bulk preparations of histones from wild-type and pst1-1 cells. Our results indicated that neither the pst1-1 mutation nor the exposure of cells to 8 nM TSA caused a measurable change in the acetylation levels of histones. This finding suggested that the pst1-1 mutation only affected histone acetylation at specific regions of chromatin. Because S. pombe has at least two histone deacetylases (53), the interesting question is which histone deacetylase activity works with pst1+ in regulating specific locations of chromatin.
Potential specificity of pst1+ versus pst2+ is suggested by the location of the pst1-1 mutation. The mutation in the pst1-1 allele created a stop codon in the C terminus such that the four conserved PAH domains were still encoded in the truncated product. In addition, sequence analysis of the C termini of Sin3 proteins did not identify any conserved motifs. One possibility is that the deletion of the C-terminal domain of Pst1p reduced its ability to bind a specific DNA-binding protein and this caused local changes in chromatin organization that could not be corrected by Pst2p.
We have reported here that the mutation in the pst1-1 allele disrupted the nuclear localization of the Tf1-encoded Gag. Indeed, we found that wild-type cells, when allowed to reach stationary phase, imported the Tf1 Gag protein into the nucleus, whereas a severe mislocalization of this protein was observed in cells with the pst1-1 mutation. The pst1-1 strain exhibited a 20.6-fold increase in the number of cells with multiple aggregates of Gag localized exclusively in the cytoplasm. The localization of Gag within the nucleus has been observed for some retroviruses. For example, the matrix protein of human immunodeficiency virus (HIV) is a component of the preintegration complex and possesses nuclear localization signal activity that contributes to the nuclear import of HIV protein in nondividing cells (14, 15, 21, 22). Although there is no direct evidence that the nuclear import of Gag is required for Tf1 transposition, several observations suggest the presence of Gag in the nucleus contributes to the propagation of Tf1. Previous analysis of Tf1 proteins in sucrose gradients indicated that in cells grown to stationary phase, the bulk of Gag and IN are coassembled into large macromolecular complexes called VLPs that also contain cDNA (41). Therefore, the results of the sucrose gradient fractionation and the immunolocalization of Gag indicate that these components are likely present together in the nucleus as VLPs. This colocalization suggests that Gag and cDNA (and likely RT and IN) were components of a complex that, in wild-type cells, was transported together as a unit. In addition, our finding that the pst1-1 mutation reduced the frequency of cDNA recombination with plasmid sequences supported the idea that Gag and cDNA were both excluded from the nucleus. In a study of the nuclear localization of Ty1 protein in S. cerevisiae, mutations in the nuclear localization signal of IN caused substantial defects in transposition but only modest reductions in cDNA recombination (45). The greater dependence of Ty1 transposition on IN transport may reflect differences in the import mechanisms of IN and cDNA. This is in contrast to the equivalent levels of defects in the transposition and cDNA recombination activities of Tf1 caused by the pst1-1 mutation.
We have recently shown that a mutation in a gene that encodes a component of the nucleopore complex, Nup124p, also inhibited the nuclear import of Tf1 Gag (6). In contrast to the phenotype of the strain with the pst1-1 mutation, in which Gag seemed to be distributed randomly throughout the cytoplasm, the mutation in the nup124+ gene caused large aggregates of Gag to adhere to the exterior of the nuclear envelope. The different phenotypes of the pst1-1 and nup124-1 mutations strongly suggests that there are multiple steps in the nuclear import mechanism of the capsid protein Gag.
We found that a mutation in the pst1 gene significantly inhibited the transposition activity of Tf1 and this lack of transposition correlated with a defect in the nuclear import of Tf1 Gag. One possible explanation for these phenotypes was that the modification in Pst1p resulted in the altered expression of a protein, thus far unidentified, that participates in the transport of Tf1 into the nucleus. This indirect contribution of pst1 to transposition is consistent with the ability of the SIN3 gene of S. cerevisiae to regulate the expression of many different genes (74). In addition, the localization of Pst1p to the nucleus limited the possible contributions that it could make to the import of Gag. However, an alternative hypothesis that may be just as likely as the indirect mechanism is that the mutation in the pst1 gene perturbed the function of the histone deacetylase complex and this in turn caused specific alterations in chromatin acetylation that directly reduced the integration efficiency of the Tf1 preintegration complex (PIC). The influence of chromatin structure on the integration of retroelement cDNA has already been reported for the integration of some yeast retrotransposons (18, 36, 83, 84) as well as HIV (56–59). Although it is reasonable to propose that altered histone acetylation could reduce Tf1 integration, it is more difficult to explain how a change in chromatin structure could change the localization of Gag to cytoplasmic. Since little is known about target site selection by Tf1, the possibility exists that, like Ty5 (84), Tf1 could avoid the disruption of host genes by specifically selecting regions of heterochromatin as targets of insertion. If nuclear pores contribute to the organization of heterochromatin, as has been suggested (69), and if heterochromatin mainly occupies the perinuclear areas (76), the insertion of cDNA into heterochromatin targets could be coupled to the nuclear localization of the PIC, as recently suggested by Boeke and Devine (9). In this way, a defect in the association of the Tf1 PIC with target sites that possess the proper chromatin structure could result in the accumulation of Tf1 proteins in the cytoplasm. An observation consistent with this model is the appearance of Gag fluorescence as single subnuclear signals adjacent to the nuclear envelope. An alternative model that is unrelated to the acetylation of histones is the possibility that direct acetylation and deacetylation of Tf1 proteins could be responsible for their import and activity. To reach a better understanding of how Pst1p contributes to Tf1 transposition, we will undertake a detailed analysis of target site selection as well as an examination of Tf1 nuclear import.
ACKNOWLEDGMENTS
We thank S. Forsburg, R. Ohi, K. Gould, and Gerald R. Smith for providing strains and plasmids. We also thank B. Turner, P. Young, and A. Wolffe for providing the S. pombe library and antibodies.
K.E. was supported by MFR project grant 11821.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 3.Atwood A, Choi J, Levin H L. The application of a homologous recombination assay revealed amino acid residues in an LTR-retrotransposon that were critical for integration. J Virol. 1998;72:1324–1333. doi: 10.1128/jvi.72.2.1324-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwood A, Lin J, Levin H. The retrotransposon Tf1 assembles virus-like particles with excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Balasundaram, D., M. Benedik, V.-D. Dang, and H. Levin. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that possesses a specialized function required for the nuclear import of the retrotransposon Tf1. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 7.Belyaev N D, Keohane A M, Turner B M. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp Cell Res. 1996;225:277–285. doi: 10.1006/excr.1996.0177. [DOI] [PubMed] [Google Scholar]
- 8.Bestor T H. Gene silencing. Methylation meets acetylation. Nature. 1998;393:311–312. doi: 10.1038/30613. [DOI] [PubMed] [Google Scholar]
- 9.Boeke J D, Devine S E. Yeast retrotransposon: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 10.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelement. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 11.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 12.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 13.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 14.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cottarel G, Beach D, Deuschle U. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr Genet. 1993;23:547–548. doi: 10.1007/BF00312650. [DOI] [PubMed] [Google Scholar]
- 17.Dang V D, Valens M, Bolotin-Fukuhara M, Daignan-Fornier B. A genetic screen to isolate genes regulated by the yeast CCAAT-box binding-protein Hap2p. Yeast. 1994;10:1273–1283. doi: 10.1002/yea.320101004. [DOI] [PubMed] [Google Scholar]
- 18.Devine S E, Boeke J D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 19.Ekwall K, Javerzat J P, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 19a.Ekwall, K., and R. C. Allshire. Unpublished data.
- 20.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 21.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 23.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 24.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 25.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 27.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 29.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 30.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 31.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 33.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeney J B, Boeke J D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 36.Kirchner J, Connolly C M, Sandmeyer S B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 37.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 38.Levin H L. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin H L. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin H L, Boeke J D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992;11:1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin H L, Weaver D C, Boeke J D. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. Embo J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. . (Erratum, 13:1494, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin H L, Weaver D C, Boeke J D. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mertins P, Gallwitz D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. Embo J. 1987;6:1757–1763. doi: 10.1002/j.1460-2075.1987.tb02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 47.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 48.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 49.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 50.Nasmyth K, Stillman D, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 51.Odds F C. Pathogenesis of Candida infections. J Am Acad Dermatol. 1994;31:S2–S5. doi: 10.1016/s0190-9622(08)81257-1. [DOI] [PubMed] [Google Scholar]
- 52.Ohi R, Feoktistova A, Gould K L. Construction of vectors and a genomic library for use with his3-deficient strains of Schizosaccharomyces pombe. Gene. 1996;174:315–318. doi: 10.1016/0378-1119(96)00085-6. [DOI] [PubMed] [Google Scholar]
- 53.Olsson T G S, Ekwall K, Allshire R C, Sunnerhagen P, Partridge J F, Richardson W A. Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res. 1998;26:3247–3254. doi: 10.1093/nar/26.13.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 55.Ponticelli A S, Smith G R. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc Natl Acad Sci USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruss D, Bushman F D, Wolffe A P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruss D, Reeves R, Bushman F D, Wolffe A P. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 58.Pryciak P M, Sil A, Varmus H E. Retroviral integration into minchromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pryciak P M, Varmus H E. Nucleosomes, DNA binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 60.Rattray A J, Symington L S. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 62.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 63.Sandmeyer S B. Yeast retrotransposons. Curr Opin Genet Dev. 1992;2:705–711. doi: 10.1016/s0959-437x(05)80130-3. [DOI] [PubMed] [Google Scholar]
- 64.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 65.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 66.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 67.Sternberg P W, Stern M J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple regulators. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 68.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 69.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 70.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 72.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 73.Vidal M, Buckley A M, Hilger F, Gaber R F. Direct selection for mutants with increased K+ transport in Saccharomyces cerevisiae. Genetics. 1990;125:313–320. doi: 10.1093/genetics/125.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wakimoto B T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 80.Wolffe A P, Pruss D, Turner B M, O’Neill L P. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 81.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 83.Zou S, Ke N, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 84.Zou S, Voytas D F. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc Natl Acad Sci USA. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]