Abstract
Typical metazoan core promoter elements, such as TATA boxes and Inr motifs, have yet to be identified in early-evolving eukaryotes, underscoring the extensive divergence of these organisms. Towards the identification of core promoters in protists, we have studied transcription of protein-encoding genes in one of the earliest-diverging lineages of Eukaryota, that represented by the parasitic protist Trichomonas vaginalis. A highly conserved element, comprised of a motif similar to a metazoan initiator (Inr) element, surrounds the start site of transcription in all examined T. vaginalis genes. In contrast, a metazoan-like TATA element appears to be absent in trichomonad promoters. We demonstrate that the conserved motif found in T. vaginalis protein-encoding genes is an Inr promoter element. This trichomonad Inr is essential for transcription, responsible for accurate start site selection, and interchangeable between genes, demonstrating its role as a core promoter element. The sequence requirements of the trichomonad Inr are similar to metazoan Inrs and can be replaced by a mammalian Inr. These studies show that the Inr is a ubiquitous, core promoter element for protein-encoding genes in an early-evolving eukaryote. Functional and structural similarities between this protist Inr and the metazoan Inr strongly indicate that the Inr promoter element evolved early in eukaryotic evolution.
Typical metazoan promoters use a TATA box, at −25 to −30 from the transcription start site, for accurate start site selection by RNA polymerase II (47). Some metazoan promoters, however, lack this TATA box and instead use a functionally analogous initiator element (Inr), with the consensus PyPyA+1NT/APyPy, to direct transcription initiation (3, 15, 27, 39, 40). The Inr is the only element in metazoan protein-encoding genes known to be a functional analog of the TATA box, in that it is sufficient for directing accurate transcription initiation in genes that lack TATA boxes (39). Genes of the Archaea, the ancestor of the eukaryotic cell (46), use TATA boxes to direct initiation but appear to lack metazoan-like Inr elements (43). Recent studies on the transcription of Archaea genes have shown that the minimal proteins required for accurate transcription initiation (TBP, TFIIB, and RNA polymerase) of eukaryotic and archaeal promoters are similar, indicating that they have a common ancestral transcription machinery (34).
Despite considerable effort, the quest to identify sequence-specific core promoter elements in early-diverging eukaryotic lineages has been largely unsuccessful. The lack of typical eukaryotic promoters is likely due to the immense divergence that has occurred since these organisms branched from the main line of eukaryotic evolution about a billion years ago. The most highly studied early-evolving eukaryotes are the parasitic protists, which are known for the evolution of unusual molecular mechanisms, such as RNA editing (37) and transcription of protein-encoding genes by RNA polymerase I (35), as found in kinetoplastids. Regulation of gene transcription in kinetoplastids has evolved in concert with ubiquitous trans-splicing, resulting in the apparent lack of sequence-specific promoters. Instead, genes are transcribed in large polycistronic units and formation of discrete mRNAs is regulated via trans-splicing (for reviews, see references 10 and 21). Although other protists have not evolved this elaborate mechanism for regulation of gene expression, the properties governing gene expression in this diverse group of eukaryotes are largely unknown.
An interest in the evolution and regulation of gene transcription in early-diverging eukaryotes led us to examine the promoter structure of protein-encoding genes of the parasitic protist Trichomonas vaginalis. This organism is a common human pathogen belonging to the phylum Parabasalia, one of the earliest-diverging eukaryotic lineages (8, 14). Trichomonads are characterized by a number of primitive cellular features, such as the absence of two classic eukaryotic organelles, peroxisomes and mitochondria (42). We have found that, without exception, T. vaginalis genes contain a highly conserved element that surrounds the start site of transcription (32). This element bears a remarkable resemblance to the metazoan initiator (Inr) (3, 15, 27, 39, 40). Using transfection assays to assess the effect of mutations in this conserved element on transcriptional activity, we demonstrate that it is essential for transcription, directs the selection of transcription start sites, and is exchangeable between trichomonad genes. Structural and functional analyses of the element indicate that it is a homologue of the metazoan Inr element. These studies identify the first cognate core promoter element found in both early-diverging and metazoan eukaryotes and indicate an early evolution of the Inr in Eukaryota.
MATERIALS AND METHODS
Cell culture.
T. vaginalis C1 (ATCC 3001) was grown in Diamond’s medium, supplemented with 10% (vol/vol) horse serum and iron as described previously (32).
Plasmid construction.
The αSCS-CAT (α-succinyl coenzyme A [CoA] synthetase-chloramphenicol acetyltransferase) construct has been described previously (11). αSCS-CAT Inr mutants were constructed by the unique restriction site elimination method of site-directed mutagenesis (Chameleon kit; Stratagene). Mutagenesis reactions to generate the Inr1 to Inr3, Inr5 to Inr13, Inr17, Inr20, Inr21, Inr33, Inr34, cHSP70 Inr, Fd Inr, and TdT Inr constructs contained the appropriate mutagenesis primer (Table 1), the pBS XhoI selection primer (Table 1), which changes the pBluescript XhoI site to a HpaI site, and the αSCS-CAT plasmid as a template. Mutagenesis reactions to generate the Inr22 and Inr24 to Inr32 constructs contained the appropriate mutagenesis primer (Table 1), the pBS HpaI selection primer (Table 1), which changes the HpaI site back to a XhoI site, and the Fd Inr plasmid as a template.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| αTub 1 | 5′-GGCTGGTACCGTGCTCAAGGCAGTAGAG-3′ |
| αTub 2 | 5′-GATCGGATCCATTCGCTAATTCCTAAAA-3′ |
| Neo 1 | 5′-GATCGGTACCATGATTGAACAAGATGGATTG-3′ |
| Neo 2 | 5′-GATCGGATCCTCAGAAGAACTCGTCAAGAAG-3′ |
| pBS XhoI | 5′-CCGGGCCCCCCGTTAACGTCGACGGTATC-3′ |
| pBS HpaI | 5′-CCGGGCCCCCCCTCGAGGTCGACGGTATCG-3′ |
| Inr1 | 5′-TAATGTGAAGCGAACAAAGCC-3′ |
| Inr2 | 5′-AGCATTAATGCGAAGTGAACA-3′ |
| Inr3 | 5′-GAGCATTAATGCGAAGCGAACAAAGC-3′ |
| Inr5 | 5′-GATGAAGAGAGCATACAAAGCCATTTTAG-3′ |
| Inr6 | 5′-TAATGTGAAGGGAACAAAGCC-3′ |
| Inr7 | 5′-AGCATTAATGGGAAGTGAACAAAGC-3′ |
| Inr8 | 5′-TAATGTGAAGAGAACAAAGCC-3′ |
| Inr9 | 5′-AGCATTAATGAGAAGTGAACAAAGC-3′ |
| Inr10 | 5′-AGCATTAATGGGAAGGGAACAAAGCC-3′ |
| Inr11 | 5′-AGCATTAATGAGAAGAGAACAAAGCC-3′ |
| Inr12 | 5′-GCATTAATGTCAAGTCAACAAAGCC-3′ |
| Inr13 | 5′-TGATGAAGAGAGCATATTAGAAGATATCACAAAGCCATTTTAGG-3′ |
| Inr17 | 5′-ATTAATGTGATGTGAACAAAG-3′ |
| Inr20 | 5′-CATTAATGTGCAGTGCACAAAGCCATTTTA-3′ |
| Inr21 | 5′-CATTAATGTGTAGTGTACAAAGCCATTTTA-3′ |
| Inr22 | 5′-TTAAAGAGAAGTGGACAAAGCCATTTTAG-3′ |
| Inr24 | 5′-TTAAAGAGAAGTAAACAAAGCCATTTTAG-3′ |
| Inr25 | 5′-GCATTAAAGAGAAGCGAACAAAGCC-3′ |
| Inr26 | 5′-GCATTAAAGAGAAGAGAACAAAGCC-3′ |
| Inr27 | 5′-GCATTAAAGAGAAGGGAACAAAGCC-3′ |
| Inr28 | 5′-TTAAAGAGAACTGAACAAAGCC-3′ |
| Inr29 | 5′-TTAAAGAGAATTGAACAAAGCC-3′ |
| Inr30 | 5′-CATTAAAGAGACGTGAACAAAGCC-3′ |
| Inr31 | 5′-CATTAAAGAGAGGTGAACAAAGCC-3′ |
| Inr32 | 5′-TGATGAAGAGAGCATCGCGTGGAAGTGAACAAAGCC-3′ |
| Inr33 | 5′-TTTAGGCTTACATTTATACCCCTTAATTG-3′ |
| Inr34 | 5′-ATTTTAGGCTTACAAGCGCACCCCTTAATTGAAG-3′ |
| cHSP70 Inr | 5′-TGATGAAGAGAGCATTATTAAAAAATGAACAAAGCCATTTTAG-3′ |
| Fd Inr | 5′-TGATGAAGAGAGCATTAAAAGAGAAGTGAACAAAGCCATTTTAG-3′ |
| TdT Inr | 5′-ATGAAGAGAGCATTACTCCAGAATGAGGGAAGCCATTTTAGGC-3′ |
| CAT-PE | 5′-TATATCAACGGTGGTATATCCAGTG-3′ |
| GS wt A | 5′-CTAAAATGGCTTTGTTCACTTCACATTAATGCTCTCT-3′ |
| GS wt B | 5′-AGAGAGCATTAATGTGAAGTGAACAAAGCCATTTTAG-3′ |
| GS Inr3 A | 5′-CTAAAATGGCTTTGTTCGCTTCGCATTAATGCTCTCT-3′ |
| GS Inr3 B | 5′-AGAGAGCATTAATGCGAAGCGAACAAAGCCATTTTAG-3′ |
| GS Inr12 A | 5′-CTAAAATGGCTTTGTTGACTTGACATTAATGCTCTCT-3′ |
| GS Inr12 B | 5′-AGAGAGCATTAATGTCAAGTCAACAAAGCCATTTTAG-3′ |
| GS Inr13 A | 5′-CTAAAATGGCTTTGTGATATCTTCTAATATGCTCTCT-3′ |
| GS Inr13 B | 5′-AGAGAGCATATTAGAAGATATCACAAAGCCATTTTAG-3′ |
| GS Inr20 A | 5′-CTAAAATGGCTTTGTGCACTGCACATTAATGCTCTCT-3′ |
| GS Inr20 B | 5′-AGAGAGCATTAATGTGCAGTGCACAAAGCCATTTTAG-3′ |
To generate the αTUB-Neo (α-tubulin–neomycin phosphotransferase) construct, primers αTub 1 and αTub 2 (Table 1), which correspond to the 5′ and 3′ ends of the α-tubulin (αTUB) gene and contain KpnI and BamHI restriction sites, respectively, were used in an inverse PCR with a genomic clone of the αTUB gene to generate the 1-kb 5′ and 1.3-kb 3′ αTUB untranslated regions (UTRs) and the pBluescript vector. The coding region of the neomycin (Neo) gene was generated by PCR with primers Neo 1 and Neo 2 (Table 1), which also contain KpnI (5′ end) and BamHI (3′ end) sites, and the pKm2 plasmid (4). The PCR products were then gel purified, digested with KpnI and BamHI, and ligated to each other to form αTUB-Neo. Constructs for the selectable-transfection experiments were generated by digesting the αTUB-Neo and αSCS-CAT Inr constructs with SacI and EcoRV and ligating them together to form the αSCS-CAT/αTUB-Neo constructs with the chloramphenicol acetyltransferase (CAT) and Neo genes in opposite transcriptional orientations.
Plasmid DNAs were purified by column chromatography (Qiagen). Constructs and Inr mutations were verified by sequencing with the Sequenase kit (Amersham).
Transient and selectable transfections.
Transient and selectable transfections of T. vaginalis C1 (ATCC 30001) were performed as described previously (11), except that cells were electroporated in Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4-KH2PO4 [pH 7.6], 25 mM HEPES [pH 7.6], 2 mM EGTA, 5 mM MgCl2, 0.375% glycerol) (44). DNA constructs are maintained as episomes in both transient and selectable transfectants (11).
CAT assay.
Cell extracts were prepared from transfected cells as described previously (11) and assayed for CAT activity by the phase extraction method of Seed and Sheen (36). Each 100-μl CAT assay mixture contained 50 μl of cell extract, 250 mM Tris-HCl (pH 8.0), 0.2 μCi of [14C]chloramphenicol (Amersham; 50 to 60 mCi/mmol) and 300 μM butyryl-CoA (Sigma). Reaction mixtures were incubated at 37°C for 2 to 4 h and then extracted with 200 μl of a 2:1 tetramethyl-p-phenylenediamine–xylene mixture. One hundred fifty microliters of the organic phase was removed and added to 5 ml of scintillation fluid, and the activity was determined in a liquid scintillation counter. Protein concentrations of the cell extracts were determined by the Bradford assay (Bio-Rad). CAT activities were normalized based on the protein concentration of the extracts and presented as the percentage of that of the wild-type control.
Primer extension analysis.
Total RNA from selectable transformants was obtained by a LiCl-urea method (32), and poly(A)+ RNA was isolated with an oligo(dT) column (Pharmacia). Each primer extension reaction mixture contained 2.5 μg of poly(A)+ RNA, except for Inr16, which had 15 μg of poly(A)+ RNA. Primer extension reactions using poly(A)+ RNA and the CAT-PE primer (Table 1) were performed as described before (32), except that hybridization was carried out at 42°C for 4 h and the reverse transcriptase reactions were performed at 42°C.
Mobility shift assay.
Preparation of T. vaginalis nuclear extracts was based on the method of Dignam et al. (12). T. vaginalis cells were harvested by centrifugation at 1,500 × g for 10 min at 4°C. Cell pellets were washed twice with phosphate-buffered saline and resuspended in 5 packed cell volumes of buffer A (20 mM HEPES-KOH [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 10 μg of leupeptin per ml, 50 μg of TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) per ml, 1 mM phenylmethylsulfonyl fluoride, 0.25% Nonidet P-40). The cell suspension was transferred to a glass Dounce homogenizer (Wheaton), and the cells were lysed with 10 to 20 strokes of a type A pestle. The cell lysate was then spun at 3,000 × g for 10 min at 4°C. The nuclear pellet was resuspended in 4 volumes of buffer C (20 mM HEPES-KOH [pH 7.6], 420 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 10 μg of leupeptin per ml, 50 μg of TLCK per ml, 1 mM phenylmethylsulfonyl fluoride, 20% glycerol) and incubated at 4°C while being stirred for 30 min. The extract was spun at 100,000 × g for 1 h at 4°C, and the supernatant was dialyzed against 1 liter of buffer D (20 mM HEPES-KOH [pH 7.6], 100 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 20% glycerol) overnight at 4°C.
A probe corresponding to −15 to +15 of the wild-type αSCS Inr was prepared by annealing two 37-mer oligonucleotides (GS wt A and GS wt B) (Table 1) and end labeling with [γ-32P]ATP and T4 polynucleotide kinase. Cold, mutant primers (GS Inr3 A and B, GS Inr12 A and B, Inr13 A and B, and Inr20 A and B) (Table 1) also corresponding to nucleotides −15 to +15 were annealed and used in competition assays. The labeled probe was purified on an 8% polyacrylamide gel. Each binding reaction mixture contained 5,000 cpm of labeled probe, 20 μg of nuclear extract, 500 ng of poly(dI-dC), 10 mM HEPES-KOH (pH 7.6), 40 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.015% Nonidet P-40, and 10% glycerol. Reaction mixtures were incubated for 30 min on ice and run on a prerun 8% 0.5× Tris-borate-EDTA polyacrylamide gel for 100 min at 15 mA.
RESULTS
Identification of a highly conserved core promoter element that is essential for transcription in an early-evolving eukaryote.
Very little is known about transcription control in early-evolving eukaryotes. Analyses of eukaryotic promoters and their interacting proteins have been confined, in large part, to the crown group of eukaryotes composed of animals, plants, and fungi. As illustrated previously (16), these organisms are quite similar in evolutionary terms and, in fact, represent only a fraction of the diversity exhibited by eukaryotic cells. By comparison, single-celled eukaryotes, such as those represented by trichomonads, kinetoplastids, and entamoebids, comprise a larger, more diverse group, and yet little is known regarding the mechanisms used to regulate gene transcription in these organisms. To address this problem, we have analyzed the 5′ UTRs of protein-encoding genes from one of the earliest-evolving eukaryotes studied to date, T. vaginalis, to identify promoter elements and to gain insight into the evolution of regulated gene transcription in eukaryotes.
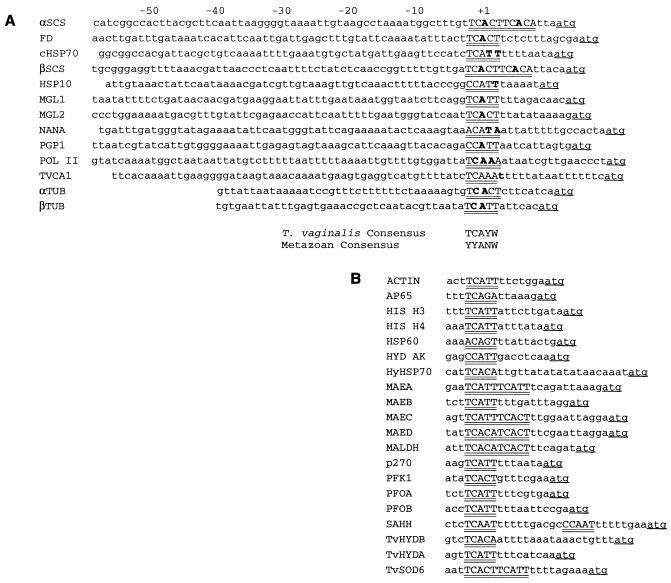
An alignment of the 5′ UTRs of all available T. vaginalis protein-encoding genes reveals the presence of a conserved motif positioned 6 to 20 nucleotides upstream from the translation initiation ATG codon (Fig. 1) (32). This motif, with the consensus sequence TCA+1T/CT/A, surrounds the start site of transcription of all genes for which the 5′ end of the mRNA has been determined (Fig. 1A). It also resembles the metazoan initiator (Inr) element that has been shown to be responsible for directing the start of transcription of metazoan genes that lack TATA boxes (39). The ubiquitous conservation of this element at the transcription start site and its similarity to metazoan Inr elements led us to question whether this apparent Inr element acts as a core promoter element. To address this, we have designed constructs that contain the α-succinyl CoA synthetase (αSCS) gene promoter with either wild-type or mutant versions of the element cloned directly upstream of the CAT gene. These constructs were then used to transiently transfect T. vaginalis cells, and the resulting CAT activities were measured. Using this approach, we show that deletion of the apparent Inr element completely abolishes detectable CAT activity (Fig. 2A, Inr5). Given the pyrimidine-rich nature of the sequences surrounding the start site of transcription of T. vaginalis genes (Fig. 1), we tested whether this element could be replaced with a random, pyrimidine-rich sequence and found that this severely reduces CAT activity, to only 6% ± 5% (mean ± standard deviation) (Fig. 2A, Inr13). These data demonstrate that this element is required for T. vaginalis promoter activity and that it cannot be replaced by random pyrimidine-rich sequences. Although we cannot rule out the presence of other, as-yet-unidentified T. vaginalis core promoter elements, these data also show that they cannot function without the conserved TCA+1CT/A element.
FIG. 1.
Alignment of sequences surrounding the start site of transcription of trichomonad genes. Highly conserved motifs are in uppercase letters and are double underlined. The longer motifs result from two conserved sequences in tandem. Transcription start sites, which were determined by primer extension analyses, except for those for MGL1 and MGL2 which were determined by 5′ rapid amplification of cDNA ends, are in bold type, and ATG start codons are underlined. (A) The 5′ flanking DNA sequences of the following T. vaginalis genes with start sites mapped to the conserved element are aligned: αSCS (accession no. L31930), FD (M33717), cHSP70 (U07783), and βSCS (M97553) (32); HSP10 (U26965) (7a); MGL1 (AJ000486) and MGL2 (AJ000487) (25); NANA (U35878) (28), PGP1 (U07785) (32), POL II (U20501) (33), TVCA1 (U65066) (26); αTUB (U07780) and βTUB (U07782) (32). The consensus sequence for the T. vaginalis conserved element is shown and compared to the consensus sequence for metazoan Inrs. For the T. vaginalis consensus, all 13 genes contain a C at −1, an A at +1, and a W (T or A) at +3; 10 of 13 contain a T at −2, and 11 of 13 contain a Y at +2. (B) Alignment of conserved sequence elements upstream of the ATG start codon of other available T. vaginalis genes where the transcription start site has not been mapped. Given that for genes where the start site has been determined, this invariably maps at <15 nucleotides upstream from the ATG (see panel A), it is likely that transcription commences in the underlined conserved motifs. Sequences are shown for the following genes: actin (accession no. U63122) and AP65 (U18347) (1); HIS H3 (X98016) and HIS H4 (X98017) (23); HSP60 (U26966) (6), Hyd AK (U07203) (20), and HyHSP70 (U27231) (6); MAEA (U16836), MAEB (U16837), MAEC (U16838), and MAED (U16839) (18); MALDH (U38692) (24), p270 (AF004357) (29), and PFK1 (AF044973); PFOA (U16822) and PFOB (U16823) (17); SAHH (U40872) (2); TvHYDA (U19897) and TvHYDB (U26964) (7); and TvSOD6 (AF022423) (45).
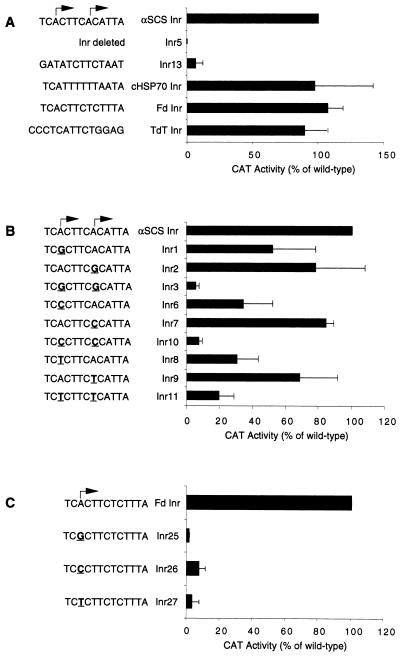
FIG. 2.
The T. vaginalis conserved element (Inr) is an essential core promoter element and requires an adenosine residue at the +1 position. CAT activities of T. vaginalis cells transfected with αSCS-CAT constructs containing the indicated Inr mutations are shown. All sequences of the Inr mutant constructs are identical to the wild-type αSCSB-CAT construct sequences except for the mutations shown. Results are reported as a percentage of the activity of the wild-type element and are the mean ± standard deviation of at least three independent experiments. (A) Mutant constructs in which the αSCS Inr was deleted or replaced with the sequence indicated to the left. (B) Mutant constructs with mutations in the transcription start site of the αSCS Inr. Mutations are shown in bold and are underlined. (C) Mutant constructs with mutations in the transcription start site of the Fd Inr shown in bold and underlined.
To determine whether this trichomonad Inr-like element is a core promoter element, we have tested whether these conserved motifs are interchangeable between T. vaginalis genes. The conserved sequences surrounding the start sites of the genes encoding cytosolic heat shock protein 70 (cHSP70; TCATTTTTTAATA) and ferredoxin (Fd; TCACTTCTCTTTA) (see Fig. 1) were substituted for the TCA+1CTTCA+1CATTA motif in the αSCS promoter. Swapping these sequences had virtually no effect on CAT activity, since essentially wild-type levels of CAT activity resulted (Fig. 2A). Moreover, replacement of the nucleotide sequence of the apparent trichomonad αSCS Inr element with that of a metazoan Inr, in particular the murine terminal deoxynucleotidyltransferase (TdT) gene Inr (CCCTCATTCTGGAG) (40), only slightly reduced activity, to 89% ± 17% of that of the wild type. These data show that the T. vaginalis element is not a gene-specific element and establish its identity as a core promoter element. Furthermore, they show that this element in T. vaginalis can be replaced in vivo by a metazoan Inr, strongly suggesting that the two elements perform homologous functions. Henceforth, we will refer to the trichomonad element as an Inr. Additional analyses, described below, confirm that this element is both structurally and functionally an Inr.
The T. vaginalis Inr requires an adenosine residue at the +1 position.
With rare exception, the start site of transcription of T. vaginalis protein-encoding genes has been mapped to adenosine residues within the Inr element (Fig. 1). Given the importance of this residue in the selection of transcription start sites in metazoan genes that require an Inr for activity, we tested whether conserved adenosines in T. vaginalis Inrs play a critical role in transcription. Our previous analyses have shown that the αSCS Inr has two TCA+1CT/A motifs in tandem, resulting in strong transcription start sites at both adenosines (32). To investigate the requirement for an adenosine residue at the +1 position, we have mutated the A’s at +1 in this element to either a G, C, or T (Fig. 2B). When only one of the two start sites is mutated individually to a G, C, or T, there is a modest reduction in CAT activity, to 30 to 84% of that of the wild type (Fig. 2B, Inr1, Inr2, and Inr6 to Inr9). This is presumably due to the nonmutated start site being used (see below for further discussion). However, when both the 5′ and 3′ start sites are mutated to a G or C, CAT activity is greatly reduced, to 5% ± 2% and 7% ± 2%, respectively (Fig. 2B, Inr3 and Inr10). Unexpectedly, when both start sites are mutated to T, CAT activity is only reduced to 19% ± 9% (Fig. 2B, Inr11). This is due to the creation of a new TCA+1NT/A motif (discussed in detail below). The effect of mutating the +1 position was confirmed by using another Inr element, the Fd Inr, which has only one transcription start site (Fig. 1) (32). Mutation of the +1 A residue to either a G, C, or T in the Fd Inr nearly eliminates CAT activity, to 1 to 7% of that of the wild type (Fig. 2C, Inr25 to Inr27). These results demonstrate the requirement of an adenosine residue at the +1 position of the T. vaginalis Inr element, since replacement of this nucleotide with any other nucleotide in two different Inrs severely reduces transcription.
The sequence requirements for a functional trichomonad Inr resemble those for metazoan Inrs.
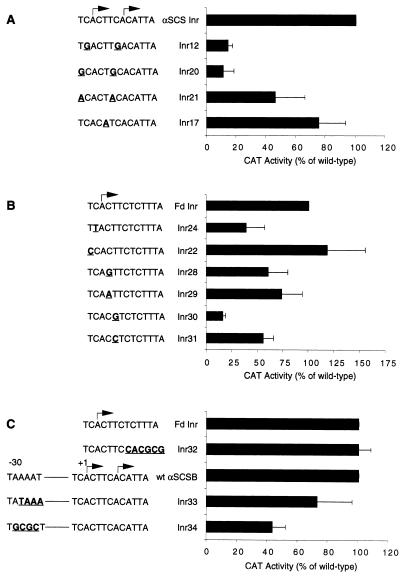
Functional analysis of the mammalian Inr has revealed a loose consensus of YYA+1NT/AYY for Inr activity, with the most critical residues for determining the strength of an Inr being the A at +1, the T or A at +3, and the pyrimidine at −1 (19, 22). The consensus sequence of the T. vaginalis Inr (TCA+1YT/A) looks very similar to the metazoan Inr consensus sequence, differing primarily in that the T. vaginalis Inr consensus sequence is more highly conserved (Fig. 1). To determine whether these conserved sequences play an essential functional role and whether the critical residues in the trichomonad and metazoan Inrs are similar, we carried out a detailed mutational analysis of the conserved residues of both the αSCS and Fd Inrs. First, we mutated the C at −1 to a G in the αSCS Inr and to a T in the Fd Inr. These mutations reduced CAT activity to 14% ± 3% and 38% ± 18%, respectively, of that of the wild type (Fig. 3A, Inr12; Fig. 3B, Inr24), indicating that a C is strongly preferred at this position while another pyrimidine is somewhat tolerated, similar to that observed for the mammalian Inr (19, 22). Next, we mutated the T at −2 to a G or A in the αSCS Inr construct and a C in the Fd Inr construct. These results show that a pyrimidine at this position allows for full CAT activity, that an A is tolerated (CAT activity = 46% ± 20%), but that a G is very detrimental, reducing CAT activity to 11% ± 7% (Fig. 3A, Inr20 and Inr21; Fig. 3B, Inr22). Mutation of the C at the +2 position in the Fd Inr to a G or an A had only a modest effect on CAT activity (activity reduced to 60% ± 19% or 73% ± 20%, respectively) (Fig. 3B, Inr28 and Inr29). Finally, when we mutated the +3 T to either an A or a C, the CAT activity was reduced to 75% ± 18% or 55% ± 10%, respectively, whereas a G mutation at this position severely reduced activity to only 15% ± 3% (Fig. 3A, Inr17; Fig. 3B, Inr30 and Inr31). These results show a preference for either a T or A at the +3 position. Taken together, our mutational analyses indicate that the sequence requirements for Inr activity in T. vaginalis are an A at +1, a C at −1, a Y at −2, and a T or an A at +3. These requirements are qualitatively the same as those for mammalian Inr activity (19, 22) but differ quantitatively since mutations at these four nucleotides are more deleterious for trichomonad Inr activity.
FIG. 3.
Sequence requirements of the T. vaginalis Inr. CAT activities of T. vaginalis cells transfected with αSCSB-CAT constructs containing the indicated Inr or promoter mutations are listed. Mutations are shown in bold and are underlined. All sequences of the Inr mutant constructs are identical to those of the wild-type αSCSB-CAT constructs except for the mutations shown. Results are reported as a percentage of the activity of the wild-type Inr control and are the mean ± standard deviation of at least three independent experiments. (A) Constructs with mutations at positions −1, −2, and +3 in the αSCSB Inr. (B) Constructs with mutations at positions −1, −2, +2, and +3 in the Fd Inr. (C) Constructs with mutations of +6 to +11 nucleotides downstream of the Fd Inr sequence and the AT-rich sequence at −30 in the αSCS promoter.
A 5-nucleotide core motif is sufficient for Inr activity, and an AT-rich sequence at −30 is not required.
Alignment of the sequences surrounding the mapped start sites of T. vaginalis genes (Fig. 1) suggests that the Inr may extend 3′ of the core Inr motif, TCA+1YT/A, whereas the metazoan Inr has been shown to be comprised of only a 5-nucleotide core sequence. To determine the core sequences required for trichomonad Inrs, we have tested whether the nucleotides immediately 3′ of the TCA+1YT/A consensus motif are important for T. vaginalis Inr activity. To do this, +6 to +11 of the Fd Inr was mutated by replacing this pyrimidine-rich sequence with a GC-rich sequence (Fig. 3C, Inr32). This mutation had no effect on CAT activity. This result and the fact that the TdT Inr, which has virtually no sequence similarity in this 3′ region to trichomonad Inrs, can replace the T. vaginalis Inr indicate that the core trichomonad Inr motif is composed of 5 nucleotides.
Examination of an alignment of the upstream regions of all available T. vaginalis genes fails to reveal a conserved TATA-like element at −30, suggesting that T. vaginalis promoters may be analogous to TATA-less metazoan promoters that rely entirely on an Inr element to select the start site of transcription. Although TATA-like elements are not typically found at −30 in T. vaginalis genes, the αSCS promoter does, in fact, have an AT-rich sequence, TAAAAT, at this position (Fig. 1). To examine the function of this sequence, we replaced it with a standard TATAAA sequence, to determine whether this change would augment transcription. To the contrary, a standard TATAAA box at this position results in a slight reduction in CAT activity, to 73% ± 23% (Fig. 3C, Inr33). To test whether the AT-rich motif at −30 can be replaced by a GC-rich element, we inserted a GC-rich sequence, TGCGCT, at this site. Although this reduces CAT activity to 43% ± 9% (Fig. 3C, Inr34), indicating that an AT-rich sequence at −30 is favored for full promoter activity, these data demonstrate that such an element is not required. These data indicate that a classic TATA box is not needed at −30 for efficient transcription of T. vaginalis genes.
The Inr selects the start site of transcription of T. vaginalis protein-encoding genes.
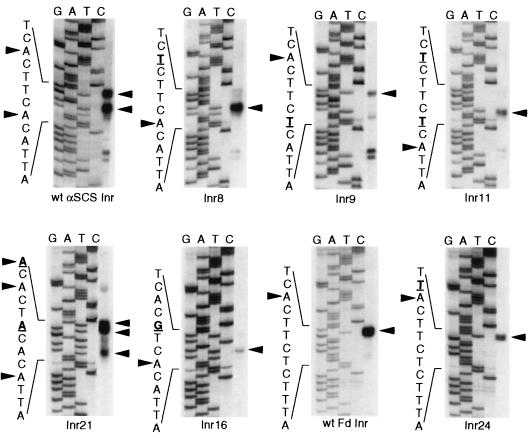
Our structural analyses of the trichomonad Inr show that mutation of specific nucleotides within this motif may severely inhibit or abolish transcriptional activity. To determine whether these mutations have a direct effect on transcription start site selection, as would be expected if this element performs the role of a metazoan Inr, we have analyzed selectable transfectants expressing different Inr mutations. T. vaginalis cells were transfected with αSCS-CAT/αTUB-Neo constructs that contain various Inr mutations as well as the neomycin phosphotransferase selectable marker under the control of a promoter with a wild-type Inr. Transfectants were selected, poly(A)+ RNA was prepared, and transcription start sites were determined by primer extension analysis. The CAT construct containing the wild-type αSCS Inr had two strong primer extension products at each TCA+1YT/A motif (Fig. 4), initiating at the adenosine start sites previously mapped for the endogeneous αSCS mRNA. When the 5′ A at +1 in this construct was mutated to a T (Inr8), an extension product was seen only at the 3′ start site. Conversely, when the 3′ start site was mutated to a T (Inr9), the principal start site used was the 5′ A; however, a minor extension product mapped to a T residue of a new TCATT sequence created by this mutation. Mutating both start site A’s in the αSCS Inr to T’s abolished the two wild-type TCATT/A motifs but created a new Inr with the sequence TCATT (discussed above) (Fig. 2B). Analysis of this mutant reveals an extension product only at the A of the newly created TCATT sequence (Fig. 4, Inr11). These results show that a TCA+1YT/A motif selects the transcription start site at A+1 and indicate that the position of the Inr relative to other sequence elements may be important in selecting the start site nucleotide.
FIG. 4.
The T. vaginalis Inr selects the start site of transcription. Primer extensions were performed with poly(A)+ RNA from T. vaginalis selectable transfectants containing αSCSB-CAT/αTUB-Neo constructs with the indicated Inr mutations. The complementary sequences are shown to the left of the sequencing ladder, where mutated nucleotides in the αSCS Inr construct are shown in bold and are underlined. Extension products are indicated by arrows. Note that the faster-migrating doublets observed in some primer extension reactions are observed in primer-alone controls and are not a product of the extension reaction.
We also have examined the effect of mutating both T’s at −2 in the αSCS Inr (Fig. 4, Inr21). In this case, accurate transcription initiation is lost, since three extra start sites are found while only one of the wild-type sites is used. Next, we mutated the 5′ T at +3 to a G in the αSCS Inr (Fig. 4, Inr16). This results in an extension product only at the 3′ TCA+1YT/A motif, indicating that the T/A at +3 is required for start site selection. Mutation of the C at −1 to a T in the Fd Inr (Fig. 4, Inr24) did not change the transcription start site, confirming that a pyrimidine can function at this position to accurately select the start site. It is noteworthy that nearly all alternative start sites observed upon mutation of the Inr map to adenosine residues, demonstrating a strong preference to initiate transcription at an A.
A nuclear protein specifically recognizes a functional Inr but not mutated, nonfunctional Inrs.
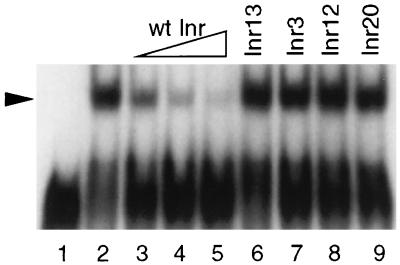
Analyses of the metazoan Inr indicate the presence of nuclear factors which may be involved in recognition of the Inr (31, 39). As a step toward determining whether T. vaginalis nuclear proteins directly interact with the Inr, we have identified an Inr-specific binding activity in T. vaginalis nuclear extracts. By using an electrophoretic mobility shift assay with a 37-bp double-stranded DNA probe containing the αSCS Inr, a binding activity that is specific for the αSCS Inr is detected (Fig. 5). Probes containing either a pyrimidine-rich, non-Inr sequence (Inr13) or Inr mutations at positions +1, −1, or −2 (Inr3, Inr12, and Inr20) do not allow binding (data not shown). In addition, the binding activity obtained with the wild-type αSCS Inr can be competed away with the wild-type αSCS Inr but not with a non-Inr sequence (Inr13) or with Inrs that have mutations at +1 (Inr3), −1 (Inr12), or −2 (Inr20) (Fig. 5, lanes 3 to 9). These data illustrate the presence of a protein(s) that can specifically recognize Inr sequences and that is able to distinguish between wild-type and mutant Inrs. Since Inr mutations that affect activity disrupt this binding activity, the protein or proteins responsible are likely to be involved in recognition of the T. vaginalis Inr by the transcriptional machinery.
FIG. 5.
Initiator specific binding activity in T. vaginalis nuclear extracts. A 37-bp 32P-labeled probe, corresponding to −15 to +15 of the αSCS Inr, was used in electrophoretic mobility shift assays with 20 μg of T. vaginalis crude nuclear extract. Competition assays were performed by using wild-type and mutant αSCS Inr probes. Lane 1, no extract; lane 2, with extract; lanes 3 to 5, competition with 20×, 50×, and 100× molar ratios of cold, wild-type αSCS Inr, respectively; lane 6, competition with 100× molar ratio of cold Inr13; lane 7, 100× molar ratio of cold Inr3; lane 8, 100× molar ratio of cold Inr12; lane 9, 100× molar ratio of cold Inr20.
DISCUSSION
Examination of the 5′ UTRs of all available protein-encoding genes from one of the earliest-diverging eukaryote studied to date, T. vaginalis (8, 14), has revealed a highly conserved TCA+1YT/A motif surrounding the start site of transcription (32). The functional analysis of this motif, presented here, shows that this element is essential for transcription, is interchangeable between trichomonad genes, and can be replaced by a mammalian initiator (Inr) element. We demonstrate that this motif is a promoter element which is both structurally and functionally similar to metazoan Inrs (3, 15, 27, 39, 40). Specific, conserved nucleotides comprising the core of this promoter element are shown to be necessary for accurate selection of the start of transcription of trichomonad genes, demonstrating its function as a bona fide Inr. These studies show that the Inr acts as a ubiquitous core promoter element in trichomonads.
It is remarkable that this early-diverging eukaryote appears not to use TATA elements to direct transcription initiation but instead invariably uses an Inr with strong similarity to metazoan Inrs. The structural and functional similarities between trichomonad and metazoan Inrs are particularly striking, since this is the first cognate promoter shown to be used by both protist and metazoan genes. There are, however, interesting differences between this protist Inr and its metazoan homologue. As revealed by our transcription analyses of mutant trichomonad Inrs, this Inr appears to have stricter sequence requirements than those observed for metazoan Inrs (19, 22). This is also reflected in a stronger consensus sequence for trichomonad Inrs. It is noteworthy that the Inr is found in all T. vaginalis genes, indicating that it is essential for transcription of all protein-encoding genes. This differs from the situation with metazoan Inrs, where only a small subset of genes have been shown to rely on the Inr for transcriptional activity (39). The trichomonad Inr also differs from its metazoan counterpart in its close proximity to the ATG translation initiation codon (see Fig. 1). The Inr is invariably located within 20 nucleotides of the ATG and may be as close as 6 nucleotides, resulting in unusually short 5′ UTRs for trichomonad mRNAs. The selection for a strict spatial conservation between the Inr and the ATG initiation codon is likely due to requirements for efficient translation of the mRNAs; however, since nothing is known about the translational machinery of trichomonads, this remains speculative.
Studies showing that transcription of protein-encoding genes in T. vaginalis is insensitive to the fungal toxin α-amanitin, an inhibitor of RNA polymerase II, have raised the question whether these genes are transcribed by this polymerase (33). The fact that the homologous Inr in metazoa is an RNA polymerase II promoter (39) indicates that RNA polymerase II transcribes these genes in T. vaginalis as well. RNA polymerase II promoters in other protists appear not to use either metazoan-like Inrs or TATA boxes, with the possible exception of the apicomplexan genus Toxoplasma (5, 9, 21, 30, 38, 41). The recent observation that a sequence similar to that of the Inr described here is required for the transcription of a Toxoplasma gene (30) raises the possibility that the Inr plays a more general role in the transcription of protist genes. However, it should be noted that this study did not test whether this sequence element actually functions as an Inr. Since so little is currently known about core promoter elements in protists, future studies on genes from this diverse group of eukaryotes will be necessary to determine whether the Inr, or elements which are functionally homologous to the Inr, is a common, essential feature of protists promoters.
The presence of a highly conserved ubiquitous Inr element that is indispensable for transcription initiation of T. vaginalis protein-encoding genes strongly indicates that the Inr evolved early during eukaryotic evolution. Although this promoter element is not generally conserved in all eukaryotes, little divergence has occurred between trichomonad and metazoan Inrs, supporting a common origin. Nevertheless, we cannot rule out the possibility that the metazoan and trichomonad Inrs evolved independently of one other. However, it seems more likely that the lack of conservation of the Inr in previously examined, early-evolving eukaryotes reflects either the divergence of these organisms or the limited number of sequence-specific protist promoters that have been identified. Indeed, conserved elements do surround and contain the transcription start site of genes in the entamoebid Entamoeba histolytica (38) and the diplomonad Giardia lamblia (13); however, these elements have no sequence similarity to each other or to trichomonad or metazoan Inrs. Finally, evidence of proteins that interact with metazoan Inrs exists (31, 39), but sequence-specific, Inr-binding proteins have been difficult to identify. Purification of the trichomonad nuclear protein that specifically recognizes a functional, but not a nonfunctional, Inr should advance our understanding of transcription in eukaryotes, since this protein will likely be a homologue of metazoan Inr-binding proteins.
ACKNOWLEDGMENTS
We thank Arnie Berk and Steve Smale for helpful comments on the manuscript, Doris Quon for assistance with primer extension analyses, Maria Delgadillo for assistance with site-directed mutagenesis, Kayvan Niazi for preparing the αTUB-Neo construct, and members of our laboratory for helpful advice and discussion.
This work was supported by an NIH grant (AI30537) to P.J.J. and a USPHS predoctoral training award (GM07185) to D.R.L. P.J.J. is the recipient of a Burroughs-Wellcome Fund Scholar Award.
REFERENCES
- 1.Alderete J F, O’Brien J L, Arroyo R, Engbring J A, Musatovova O, Lopez O, Lauriano C, Nguyen J. Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol Microbiol. 1995;17:69–83. doi: 10.1111/j.1365-2958.1995.mmi_17010069.x. [DOI] [PubMed] [Google Scholar]
- 2.Bagnara A S, Tucker V E, Minotto L, Howes E R, Ko G A, Edwards M R, Dawes I W. Molecular characterisation of adenosylhomocysteinase from Trichomonas vaginalis. Mol Biochem Parasitol. 1996;81:1–11. doi: 10.1016/0166-6851(96)02683-7. [DOI] [PubMed] [Google Scholar]
- 3.Beaupain D, Eleouet J F, Romeo P H. Initiation of transcription of the erythroid promoter of the porphobilinogen deaminase gene is regulated by a cis-acting sequence around the cap site. Nucleic Acids Res. 1990;18:6509–6515. doi: 10.1093/nar/18.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 5.Bruderer T, Wehrli C, Kohler P. Cloning and characterization of the gene encoding pyruvate phosphate dikinase from Giardia duodenalis. Mol Biochem Parasitol. 1996;77:225–233. doi: 10.1016/0166-6851(96)02605-9. [DOI] [PubMed] [Google Scholar]
- 6.Bui E T, Bradley P J, Johnson P J. A common evolutionary origin for mitochondria and hydrogenosomes. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui E T, Johnson P J. Identification and characterization of [Fe]-hydrogenases in the hydrogenosome of Trichomonas vaginalis. Mol Biochem Parasitol. 1996;76:305–310. doi: 10.1016/0166-6851(96)02567-4. [DOI] [PubMed] [Google Scholar]
- 7a.Bui, E. T., and P. Johnson. Unpublished data.
- 8.Cavalier-Smith T. Kingdom Protozoa and its 18 phyla. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabb B S, Cowman A F. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc Natl Acad Sci USA. 1996;93:7289–7294. doi: 10.1073/pnas.93.14.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross G A. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- 11.Delgadillo M G, Liston D R, Niazi K, Johnson P J. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc Natl Acad Sci USA. 1997;94:4716–4720. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillin F D, Reiner D S, McCaffery J M. Cell biology of the primitive eukaryote Giardia lamblia. Annu Rev Microbiol. 1996;50:679–705. doi: 10.1146/annurev.micro.50.1.679. [DOI] [PubMed] [Google Scholar]
- 14.Gunderson J, Hinkle G, Leipe D, Morrison H G, Stickel S K, Odelson D A, Breznak J A, Nerad T A, Muller M, Sogin M L. Phylogeny of trichomonads inferred from small-subunit rRNA sequences. J Eukaryot Microbiol. 1995;42:411–415. doi: 10.1111/j.1550-7408.1995.tb01604.x. [DOI] [PubMed] [Google Scholar]
- 15.Hariharan N, Perry R P. Functional dissection of a mouse ribosomal protein promoter: significance of the polypyrimidine initiator and an element in the TATA-box region. Proc Natl Acad Sci USA. 1990;87:1526–1530. doi: 10.1073/pnas.87.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinkle G, Leipe D D, Nerad T A, Sogin M L. The unusually long small subunit ribosomal RNA of Phreatamoeba balamuthi. Nucleic Acids Res. 1994;22:465–469. doi: 10.1093/nar/22.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrdy I, Muller M. Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J Mol Evol. 1995;41:388–396. [PubMed] [Google Scholar]
- 18.Hrdy I, Muller M. Primary structure of the hydrogenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J Eukaryot Microbiol. 1995;42:593–603. doi: 10.1111/j.1550-7408.1995.tb05913.x. [DOI] [PubMed] [Google Scholar]
- 19.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange S, Rozario C, Muller M. Primary structure of the hydrogenosomal adenylate kinase of Trichomonas vaginalis and its phylogenetic relationships. Mol Biochem Parasitol. 1994;66:297–308. doi: 10.1016/0166-6851(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee M G, Van der Ploeg L H. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu Rev Microbiol. 1997;51:463–489. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- 22.Lo K, Smale S T. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 23.Marinets A, Muller M, Johnson P J, Kulda J, Scheiner O, Wiedermann G, Duchene M. The sequence and organization of the core histone H3 and H4 genes in the early branching amitochondriate protist Trichomonas vaginalis. J Mol Evol. 1996;43:563–571. doi: 10.1007/BF02202104. [DOI] [PubMed] [Google Scholar]
- 24.Markos A, Morris A, Rozario C, Muller M. Primary structure of a cytosolic malate dehydrogenase of the amitochondriate eukaryote, Trichomonas vaginalis. FEMS Microbiol Lett. 1996;135:259–264. doi: 10.1111/j.1574-6968.1996.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 25.McKie A E, Edlind T, Walker J, Mottram J C, Coombs G H. The primitive protozoon Trichomonas vaginalis contains two methionine gamma-lyase genes that encode members of the gamma-family of pyridoxal 5′-phosphate-dependent enzymes. J Biol Chem. 1998;273:5549–5556. doi: 10.1074/jbc.273.10.5549. [DOI] [PubMed] [Google Scholar]
- 26.Meade J C, Li C, Moate M E, Davis-Hayman S R, Lushbaugh W B, Finley R W. Molecular characterization of a sarcoplasmic-endoplasmic reticulum Ca2+ ATPase gene from Trichomonas vaginalis. J Eukaryot Microbiol. 1997;44:480–486. doi: 10.1111/j.1550-7408.1997.tb05727.x. [DOI] [PubMed] [Google Scholar]
- 27.Means A L, Farnham P J. Transcription initiation from the dihydrofolate reductase promoter is positioned by HIP1 binding at the initiation site. Mol Cell Biol. 1990;10:653–661. doi: 10.1128/mcb.10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meysick K C, Dimock K, Garber G E. Molecular characterization and expression of a N-acetylneuraminate lyase gene from Trichomonas vaginalis. Mol Biochem Parasitol. 1996;76:289–292. doi: 10.1016/0166-6851(95)02544-8. [DOI] [PubMed] [Google Scholar]
- 29.Musatovova O, Alderete J F. Molecular analysis of the gene encoding the immunodominant phenotypically varying P270 protein of Trichomonas vaginalis. Microb Pathog. 1998;24:223–239. doi: 10.1006/mpat.1997.0199. [DOI] [PubMed] [Google Scholar]
- 30.Nakaar V, Bermudes D, Kyong R P, Joiner K A. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol Biochem Parasitol. 1998;92:229–239. doi: 10.1016/s0166-6851(97)00220-x. [DOI] [PubMed] [Google Scholar]
- 31.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 32.Quon D V, Delgadillo M G, Khachi A, Smale S T, Johnson P J. Similarity between a ubiquitous promoter element in an ancient eukaryote and mammalian initiator elements. Proc Natl Acad Sci USA. 1994;91:4579–4583. doi: 10.1073/pnas.91.10.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quon D V K, Delgadillo M G, Johnson P J. Transcription in the early diverging eukaryote Trichomonas vaginalis: an unusual RNA polymerase II and alpha-amanitin-resistant transcription of protein-coding genes. J Mol Evol. 1996;43:253–262. doi: 10.1007/BF02338833. [DOI] [PubMed] [Google Scholar]
- 34.Qureshi S A, Bell S D, Jackson S P. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudenko G, Blundell P A, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 36.Seed B, Sheen J Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 37.Simpson L, Maslov D A. RNA editing and the evolution of parasites. Science. 1994;264:1870–1871. doi: 10.1126/science.8009214. [DOI] [PubMed] [Google Scholar]
- 38.Singh U, Rogers J B, Mann B J, Petri W A., Jr Transcription initiation is controlled by three core promoter elements in the hgl5 gene of the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci USA. 1997;94:8812–8817. doi: 10.1073/pnas.94.16.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 40.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 41.Soldati D, Boothroyd J C. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer J M, Bradley P J, Wang C C, Johnson P J. Biogenesis of specialized organelles: glycosomes and hydrogenosomes. In: Smith D F, Parsons M, editors. Molecular biology of parasitic protozoa. Vol. 13. New York, N.Y: Oxford Press; 1996. pp. 159–180. [Google Scholar]
- 43.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 44.van den Hoff M J, Moorman A F, Lamers W H. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viscogliosi E, Delgado-Viscogliosi P, Gerbod D, Dauchez M, Gratepanche S, Alix A J, Dive D. Cloning and expression of an iron-containing superoxide dismutase in the parasitic protist, Trichomonas vaginalis. FEMS Microbiol Lett. 1998;161:115–123. doi: 10.1111/j.1574-6968.1998.tb12936.x. [DOI] [PubMed] [Google Scholar]
- 46.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]