Abstract
A mutation in NMD3 was found to be lethal in the absence of XRN1, which encodes the major cytoplasmic exoribonuclease responsible for mRNA turnover. Molecular genetic analysis of NMD3 revealed that it is an essential gene required for stable 60S ribosomal subunits. Cells bearing a temperature-sensitive allele of NMD3 had decreased levels of 60S subunits at the nonpermissive temperature which resulted in the formation of half-mer polysomes. Pulse-chase analysis of rRNA biogenesis indicated that 25S rRNA was made and processed with kinetics similar to wild-type kinetics. However, the mature RNA was rapidly degraded, with a half-life of 4 min. Nmd3p fractionated as a cytoplasmic protein and sedimented in the position of free 60S subunits in sucrose gradients. These results suggest that Nmd3p is a cytoplasmic factor required for a late cytoplasmic assembly step of the 60S subunit but is not a ribosomal protein. Putative orthologs of Nmd3p exist in Drosophila, in nematodes, and in archaebacteria but not in eubacteria. The Nmd3 protein sequence does not contain readily recognizable motifs of known function. However, these proteins all have an amino-terminal domain containing four repeats of Cx2C, reminiscent of zinc-binding proteins, implicated in nucleic acid binding or protein oligomerization.
A screen for mutations that are lethal in yeast cells lacking the major cytoplasmic exoribonuclease, Xrn1p, identified mutations in SKI2 and SKI3 and one additional complementation group (27). ski6 and ski8 mutations are also synthetic lethal with xrn1Δ (1, 9). Although the superkiller (SKI) genes were initially described as host antiviral genes that repress expression of killer toxin encoded by an endogenous yeast RNA virus (58; reviewed in reference 63), they have more recently been shown to play a general role in the cell (27), to repress translation of poly(A)− RNA (7, 43, 64), and to be required for normal mRNA 3′-exonucleolytic degradation (1). SKI6 encodes an essential 3′→5′ exonuclease that is a component of the exosome (45), and ski6 mutants have defects in assembly of 60S ribosomal subunits (7). We have now cloned the wild-type gene of the third complementation group identified in the previous XRN1 synthetic lethal screen (27) and have shown that it is NMD3.
NMD3 (nonsense-mediated decay) was previously identified from a two-hybrid screen for proteins that interact with the nonsense-mediated decay factor Upf1p (22). Nonsense-mediated decay in yeast is a cytoplasmic pathway for the rapid elimination of aberrant transcripts containing premature stop codons (reviewed in reference 25). Homologous pathways are found in nematodes (49) and mammalian cells (5; reviewed in reference 41). Nonsense-mediated decay involves the recognition of a premature stop codon by a translating ribosome which is then thought to activate a scanning complex that recognizes downstream sequence elements, leading to rapid decapping of the transcript (12, 50). In yeast, the pathway depends on UPF1, UPF2/NMD2, and UPF3 (10, 22, 36–38). Deletion of any of these genes prevents nonsense-mediated decay, thus stabilizing otherwise unstable mRNA containing premature nonsense codons. However, such mutants display few growth defects, indicating that in yeast this pathway is dispensable for normal growth.
Dominant mutants of NMD3 (also referred to as SRC5) (30) have also been identified as suppressors of the growth defect of a temperature-sensitive mutation in QSR1/GRC5 (31), encoding the large ribosomal subunit protein identified as L10 in the current ribosomal protein nomenclature of Mager et al. (40) or as L7 in the nomenclature of Zinker and Warner (68). L10 is thought to be an exchangeable ribosomal protein (34) that may be added to the large subunit in a late cytoplasmic assembly step (13). The genetic interaction between NMD3 and QSR1 suggests a role for NMD3 in ribosome function or synthesis.
Eukaryotic ribosome biogenesis is a complex process occurring largely in the nucleolus, where rRNA is transcribed, modified, and processed during assembly with approximately 80 ribosomal proteins into mature ribosomal subunits. In Saccharomyces cerevisiae, three (25S, 5.8S, and 18S) of the four rRNAs are transcribed by RNA polymerase I as a single 35S precursor in the nucleolus, whereas the fourth rRNA, 5S rRNA, is transcribed independently by RNA polymerase III (66). The pre-rRNA then undergoes sequential cleavages and trimming to generate the mature rRNAs. Most of the ribosomal proteins assemble into subunits while in the nucleolus, although a few proteins, including L10, are exchangeable and may be added after transport of subunits to the cytoplasm.
rRNA processing and assembly of ribosomal proteins onto the rRNAs are intimately linked processes. Thus, mutations in ribosomal proteins or in factors required for assembly typically lead to the production of defective subunits and potentially alter the specificity of translation. Such defects may lead to the production of unstable subunits, resulting in an imbalance between 40S and 60S (51, 67), or directly affect ribosome function without resulting in a subunit imbalance (14). In this paper, we describe the identification and functional analysis of NMD3, an essential gene from S. cerevisiae. While our initial identification of a nmd3 mutant from a synthetic lethal screen with a xrn1 mutant and its previous implication in nonsense-mediated decay suggested a role in mRNA turnover, our results suggest that NMD3 is required for a late cytoplasmic assembly step of 60S ribosomal subunit biogenesis.
MATERIALS AND METHODS
Strains, media, and genetic methods.
Strains used in this study are listed in Table 1. Rich medium (YPD), 5-fluoroorotic acid (5FOA), synthetic complete (SC) dropout media, and standard yeast manipulations were as described elsewhere (28). Yeast transformations were carried out by the lithium acetate method as previously described (19) except that cells to be transformed were grown as light lawns on plates.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Plasmid | Reference or source |
|---|---|---|---|
| AJY362 | MATa/α leu2/leu2 ura3/ura3 trp1/trp1 L-A-o/L-A-o | ||
| AJY384 | MATa/α leu2/leu2 ura3/ura3 trp1/trp1 L-A-o/L-A-o NMD3/nmd3::TRP1 | ||
| AJY404 | MATa ade2 ade3 leu2 his3 ura3-52 xrn1Δ NMD3-LEU2-NMD3 | ||
| AJY483 | MATα ade2 ade3 his3Δ leu2 lys2-801 trp1Δ63 ura3-52 | ||
| AJY529 | MATα his3Δ leu2 trp1 ura3-52 nmd3::TRP1 | pAJ112 | |
| AJY543 | MATa ade2 ade3 leu2 lys2-801 ura3-52 nmd3-1 | 27 | |
| AJY590 | MATα his3 leu2 trp1 ura3-52 nmd3::TRP1 | pAJ124 | |
| AJY592 | MATα his3 leu2 trp1 ura3-52 nmd3::TRP1 | pAJ123 | |
| AJY596 | MATα his3 leu2 trp1 ura3-52 nmd3::TRP1 | pAJ129 | |
| AJY717 | MATa ade2 ade3 leu2 lys2-801 ura3-52 nmd3-4 ρ− | ||
| AJY734 | MATa ade2 ade3 leu2 lys2-801 ura3-52 nmd3-4 | ||
| AJY735 | MATa ade2 ade3 leu2 lys2-801 ura3-52 | ||
| CH1305 | MATa ade2 ade3 leu2 lys2-801 ura3-52 | 32 | |
| RDKY1977 | MATa ade2 ade3 leu2 lys2-801 ura3-52 xrn1Δ | ||
| RDKY1978 | MATα ade2 ade3 leu2 his3 ura3-52 xrn1::URA3 | 27 | |
| RDKY1979 | MATα ade2 ade3 leu2 his3 ura3-52 xrn1Δ | 27 | |
| RDKY1997 | MATa leu2 ura3 trp1 L-A-o | 27 | |
| RDKY2037 | MATα leu2 ura3 trp1 L-A-o | ||
| RDKY2050 | MATa ade2 ade3 leu2 lys2-801 ura3-52 nmd3-1 | pRDK297 | 27 |
| YAS398 | MATa ura3 trp1 his3 leu2 spb2::LEU2 | A. Sachs |
Cloning of NMD3 and DNA manipulation.
Strain RDKY2050, identified in a previous screen (27), was transformed with a centromeric LEU2 library containing 9- to 12-kbp inserts. Leu+ transformants were replica plated to 5FOA plates, and 5FOA-resistant colonies were restruck on 5FOA plates. To identify and eliminate wild-type XRN1 clones, 5FOA-resistant clones were analyzed by PCR using primers to distinguish wild-type XRN1 from the genomic xrn1 deletion (xrn1Δ). Plasmid DNA from one complementing clone that was not XRN1 was subcloned as a Sau3AI partial digest into YEp351 as described previously (27). The insert in pAJ78, containing the smallest complementing subclone, was sequenced (Sequenase dideoxy sequencing kit; Amersham) and found to contain one single complete open reading frame encoding NMD3.
nmd3::TRP1 disruption.
The TRP1-containing BstUI fragment from pRS424 was ligated into pAJ78 that had been digested with MscI and BglII and blunt ended with T4 DNA polymerase. This gave a TRP1 disruption of NMD3 from nucleotides (nt) 248 to 1146 to create pAJ81. The L-A double-stranded RNA virus-deficient diploid, AJY362, was made by mating RDKY1997 and RDKY2037. The nmd3::TRP1-containing NheI-to-SpeI fragment of pAJ81 was transformed into AJY362. Trp+ transformants were analyzed by PCR for heterozygotes containing the correct integration of the nmd3::TRP1 allele at the NMD3 locus to give AJY384.
Genomic NMD3-LEU2.
An NMD3-containing SmaI-to-NheI fragment from pAJ78 was ligated into the LEU2-containing yeast integrating plasmid pRS305 digested with SmaI and XbaI. The resulting plasmid (pAJ92) was digested with BamHI, which cuts within NMD3 coding sequence, and integrated into the genomic NMD3 locus of RDKY1979 (xrn1Δ) by transformation to give strain AJY404. The resulting NMD3 duplication with an intervening LEU2 gene was confirmed by Southern blotting.
Plasmids.
Table 2 describes the plasmids used in this study. All nucleotide numbering is relative to the translation start, defined as +1 of the respective gene. pAJ123 was constructed by ligating the NMD3-containing SmaI-to-SalI fragment from pAJ78 into the respective sites in pRS315. pAJ112, expressing His6-tagged NMD3, was constructed as follows. Oligonucleotide primers AJO108 (5′-CTAGTCTAGACTCGAGAAAATGCATCATCATCATCATCATTCCATGGAATTCACACCTATAGATCC), encoding the His6 tag and containing an XhoI site, and AJO109 (5′-GCGCAAGCTTGAGTATATACTACTCTCC), containing a HindIII site, were used to PCR amplify NMD3. The XhoI- and HindIII-digested PCR product was then ligated into XhoI- and HindIII-digested pVT102U to give pAJ112.
TABLE 2.
Plasmids used in this study
| Plasmids | Relevant markers | Source or reference |
|---|---|---|
| pAJ78 | 2μm LEU2 NMD3 | |
| pAJ81 | 2μm LEU2 nmd3::TRP1 | |
| pAJ92 | LEU2 NMD3 integrating vector | |
| pAJ112 | 2μm His6-NMD3(PCR) URA3 | |
| pAJ113 | 2μm His6-NMD3(WT) URA3 | |
| pAJ118 | 2μm LEU2 GAL10::NMD3 | |
| pAJ123 | CEN LEU2 NMD3 | |
| pAJ124 | CEN LEU2 nmd3-2 | |
| pAJ129 | CEN LEU2 nmd3-4 | |
| pAJ150 | URA3 nmd3-4 integrating vector | |
| pAJ153 | CEN LEU2 c-Myc-NMD3 | |
| pAJ234 | 2μm LEU2 GAL10::c-Myc-NMD3 | |
| pRDK306 | YEp351 GAL10::XRN1 | |
| pRP485 | GAL1::MFA2 URA3 | R. Parker |
| pVT102U | 2μm URA3 | 60 |
Epitope-tagged NMD3.
pAJ234, containing a galactose-inducible c-Myc-tagged NMD3, was created as follows. The PCR-amplified NMD3 coding sequence of pAJ112 was replaced with the NMD3-containing EcoRI-to-HindIII fragment from pAJ78 to give pAJ113. The NMD3-containing NcoI-to-HindIII fragment from pAJ113 was ligated into the corresponding sites in pRDK306 (47) to give pAJ118. Finally, oligonucleotide AJO148 (5′-CATGGAACAAAAGCTTATTTCTGAAGAAGACTTGAA), encoding the c-Myc epitope, and its complementary oligonucleotide were ligated into the NcoI site at the start codon of NMD3 in pAJ118 to give pAJ234. pAJ153, encoding c-Myc-tagged NMD3 expressed from its own promoter, was constructed as follows. The wild-type NMD3 promoter and 5′ sequences were amplified from pAJ78 by using an M13 reverse sequencing primer and oligonucleotide AJO168 (5′-GGTAACGGTACCATGGTTTTGTCAAATTCCTCAACG). The resulting PCR product was digested with NcoI and SmaI and ligated to the c-Myc-NMD3-containing NcoI-to-BglII fragment from pAJ234 and the BglII-to-SmaI vector pAJ123. The resulting construct complemented the temperature sensitivity of an nmd3-4 mutant.
nmd3 mutants.
The genomic nmd3-1 mutation was rescued to plasmid pAJ123 by in vivo recombination by transforming strain AJY543 with pAJ123 digested with SnaBI and HindIII, and the resulting plasmid was sequenced. nmd3-1 sequence was confirmed by directly sequencing the genomic locus amplified by PCR.
PCR mutagenesis was used to create random temperature-sensitive mutations in NMD3. Oligonucleotide primers AJO108 and AJO109 (see above) were used for PCR amplification of NMD3. The PCR product was digested with NcoI and was cotransformed into AJY529 with pAJ123 digested by MscI and BglII to remove NMD3 coding sequence from nt 245 to 1143. Leu+ transformants were then replica plated to 5FOA plates at 37 and 26°C. From approximately 2,000 transformants, three temperature-sensitive mutations, nmd3-2, nmd3-3, and nmd3-4, were obtained. All three nmd3 temperature-sensitive mutants behaved similarly, and nmd3-4 was used for most temperature shift experiments. pAJ124 (bearing nmd3-2) and pAJ129 (bearing nmd3-4) were transformed into AJY529, replacing pAJ112 to give strains AJY590 and AJY596 respectively. pAJ150, containing nmd3-4 on a URA3 integrating plasmid, was created by ligating the nmd3-4-containing NcoI-to-SalI fragment from pAJ129 into the respective sites of pRS406. pAJ150 was digested with BglII and transformed into the wild-type strain CH1305 to integrate pAJ150 into the NMD3 genomic locus. Ura+ transformants were then patched onto 5FOA plates to select for the loss of the intervening URA3 sequence and one copy of the NMD3 gene. 5FOA-resistant clones were then scored for temperature sensitivity, and loss of the integrated plasmid was confirmed by PCR of the genomic NMD3 locus. Since all temperature-sensitive isolates were petite, this strain (AJY717) bearing nmd3-4 was crossed to a wild-type grande (AJY483). The resulting heterozygous diploid was sporulated, and the resulting tetrads were dissected. All spore clones from the cross were grandes. AJY734 (nmd3-4) and AJY735 (wild type) from the same tetrad were used for further study.
Northern blot analysis.
For CYH2 steady-state mRNA analysis, 10-ml cultures were grown in SC dropout medium to a density of approximately 107 cells/ml at 26°C and then shifted to 37°C for 2 h. Cells were harvested and RNA was prepared and analyzed by Northern blotting as previously described (26). For time course experiments with thiolutin, 40-ml cultures were grown in YPD at 26°C and then shifted to 37°C. After 1.75 h at 37°C, cells were collected by centrifugation and resuspended in 10 ml of YPD at 37°C. After another 30 min at 37°C, thiolutin was added to 3 μg/ml (final concentration). Immediately after addition of thiolutin (time zero) and at various times afterward, 1.5-ml aliquots were removed, cells were collected by centrifugation, and cell pellets were placed on dry ice. RNA was prepared and analyzed by Northern blotting. For MFA2 Northern analysis, 25-ml cultures were grown at 26°C in SC Ura− medium containing 2% raffinose to a density of approximately 107 cells/ml and then shifted to 37°C for 2 h. Transcription was induced by adding galactose to 2%. After 10 min, glucose was added to 2% to repress transcription. At various times, cells were collected by centrifugation and cell pellets were placed on dry ice. RNA was prepared and analyzed by Northern blotting using oligonucleotide AJO101 (5′-GATCAGGAATTCCCCCCCCCCCCCCCCCCAAATTCCTA), specific to a poly(G) insertion in MFA2 mRNA.
For Northern analysis of pre-rRNA steady-state levels, total RNAs from strains AJY734 and AJY735 were prepared after a shift to 37°C for 1, 3, and 6 h, then separated on a formaldehyde-agarose gel, and transferred to a Zeta-probe blotting membrane (Bio-Rad) as described above. Membranes were probed with the following 32P-radiolabeled oligonucleotide probes: AJO215 (5′-GGTCTCTCTGCTGCCGGAAATG), specific to the 5’ external transcribed spacer (5′ETS); AJO130 (5′-TCTTGCCCAGTAAAAGCTCTCATG), specific to internal transcribed spacer 1 (ITS1); AJO214 (5′-GTTCGCCTAGACGCTCTCTTC), specific to ITS2; AJO213 (5′-CCACTTAGAAAGAAATAAAAAACAAATCAG), complementary to 9 nt of the 3′ end of 25S plus 22 nt of 3′ETS; and AJO216 (5′-CCCGGATCATAGAATTCTTAAG), complementary to 3′ETS, downstream of the RNase III cleavage site.
Pulse-chase analysis of pre-rRNA processing.
Labeling with [methyl-3H]methionine was carried out as described previously (67). Briefly, 5-ml cultures were grown in SC Met− medium to a density of approximately 0.6 × 107 cells/ml at room temperature. Cells were harvested by centrifugation, resuspended in 1.5 ml of SC Met− medium prewarmed to 37°C, and grown at 37°C for 2 h. Cells were then labeled by the addition of 150 μCi of [methyl-3H]methionine. After labeling for 2 min, 150 μg of unlabeled methionine was added as a chase. At various times, samples were taken and quickly frozen in dry ice.
A similar procedure was followed for pulse-chase experiments using [3H]uracil except that strains used had been transformed with pRS316 to allow growth in SC Ura− medium. Cells were shifted to 37°C for 2 h in 3 ml of SC Ura− medium, pulse-labeled with 150 μCi of [3H]uracil for 3 min, and chased with 900 μg of unlabeled uracil. RNAs were prepared and analyzed by electrophoresis through formaldehyde-agarose gels (6% formaldehyde, 1.2% agarose) as described elsewhere (28) and by electrophoresis in urea-polyacrylamide gels (6% polyacrylamide, 8.2 M urea). To detect labeled RNAs, the gels were soaked in Enlightning (New England Nuclear), dried, and subjected to autoradiography.
Drug sensitivity tests.
Fresh single colonies from plates were resuspended in sterile distilled water and plated on YPD plates. After the cell suspension had soaked into the plate, 0.25-in. sterile filters (Schleicher & Schuell catalog no. 740-E) were placed onto the plates, and the indicated amount of antibiotic was spotted onto the filter. Plates were incubated at room temperature or at 33°C.
Sucrose density gradients.
For polysome preparation, yeast cultures were grown to a density of 0.6 × 107 cells/ml in 500 ml of YPD at 26°C. One half of the culture was removed and shifted to 37°C for 2 h. Just before the cells were harvested, cycloheximide was added to a final concentration of 100 μg/ml. The cells were harvested, washed once with 25 ml of ice-cold buffer C (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 30 mM MgCl2, cycloheximide [50 μg/ml], heparin [200 μg/ml]), and resuspended in 1 ml of ice-cold buffer C; 300 μl of cold glass beads (400-μm diameter; Sigma Chemical Co.) was added, and the suspension was vortexed six times, each consisting of 30 s of vortexing followed by 1 min of cooling on ice. Extracts were clarified by centrifugation, and 30 A260 units of cell lysate was loaded onto each 12-ml linear 7 to 47% sucrose gradient and centrifuged for 2.5 h at 40,000 rpm in a Beckman SW40 rotor at 4°C as described elsewhere (2).
Analysis of dissociated ribosomal subunits was as described above except that extracts were prepared in buffer C containing 30 mM EDTA and lacking MgCl2 and cycloheximide. In addition, gradients were prepared with 30 mM EDTA and no MgCl2, and centrifugation was for 5 h. Sucrose gradients were analyzed by continuous monitoring at A254 with a Pharmacia Uvicord monitor.
Yeast cell fractionation and Western blotting.
Yeast cells were fractionated into nuclear and cytoplasmic fractions as described previously (65). Proteins from cytoplasmic and nuclear fractions and from sucrose density gradients were analyzed by Western blotting as described previously (26), using anti-c-Myc monoclonal antibody at 1:5,000 as the primary antibody to detect c-Myc-tagged Nmd3p. Rabbit anti-topoisomerase II (Topo II) and rabbit anti-glucose-6-phosphate dehydrogenase (G6PDH) polyclonal antibodies were diluted 1:7,500 and used to detect nuclear and cytoplasmic protein markers, respectively. Rabbit polyclonal anti-Topo II antibody was a gift from J. Lindsley and J. Wang, anti-G6PDH antibody was obtained from Sigma, and mouse monoclonal anti-c-Myc was derived from 9E10.2 cells (American Type Culture Collection, Rockville, Md.).
RESULTS
Identification of nmd3-1 as synthetically lethal with xrn1 deletion.
A previous screen for mutations synthetically lethal with xrn1Δ identified recessive mutations in three complementation groups. The wild-type genes for two of these were cloned and identified as SKI2 and SKI3 (27). The wild-type gene for the third complementation group was cloned from a centromeric LEU2 plasmid library by complementation and shown to be NMD3. NMD3 was previously found in a two-hybrid screen for proteins that interact with Upf1p (22). Mutants in the UPF1 gene, originally isolated on the basis of their ability to enhance the suppression of a frameshift mutation that led to premature translational termination (11), selectively stabilize mRNAs containing early nonsense mutations.
To confirm that the mutation in the third complementation group was allelic with NMD3, genetic linkage was assayed. RDKY2050 (xrn1Δ and putatively nmd3-1) containing pRDK297 (XRN1 URA3) was mated with AJY404 (xrn1Δ NMD3::LEU2::NMD3), diploids were sporulated, and the resulting tetrads were dissected. All tetrads containing four viable spores (17 tetrads) showed 2:2 segregation of synthetic lethality, scored as 5FOA sensitive (data not shown). No 5FOA-sensitive colonies were Leu+, indicating no recombination between the putative nmd3-1 allele and the LEU2-marked NMD3 locus, confirming that the original mutation was in NMD3.
NMD3 is an essential gene.
A heterozygous diploid containing one nmd3::TRP1 allele was constructed in a genetic background lacking the double-stranded RNA virus, L-A. Upon sporulation and tetrad dissection, no Trp+ spore clones were obtained from 37 tetrads and each ascus contained at least two dead spores (data not shown), indicating that NMD3 is essential. To further confirm the essentiality of NMD3, a LEU2 NMD3 plasmid was transformed into the NMD3 heterozygous diploid before sporulation. This plasmid rescued the inviability of Trp+ spore clones and allowed the recovery of tetrads containing four viable spores (data not shown). All Trp+ spore clones were also Leu+. Because NMD3 is essential, the original recessive allele, nmd3-1, identified as synthetically lethal with xrn1Δ, must be a hypomorphic, or partially functional, allele.
Sequence analysis of wild-type and mutant NMD3.
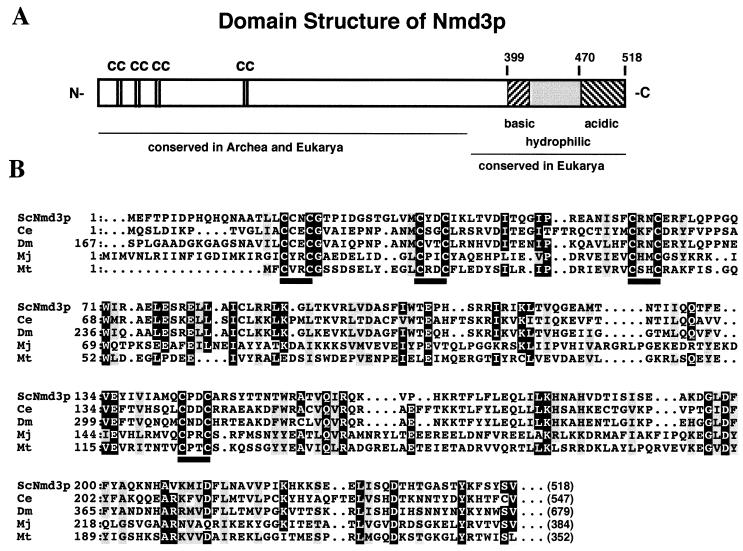
NMD3 encodes a 59.1-kDa protein that is homologous to hypothetical proteins from Caenorhabditis elegans and Drosophila melanogaster (Fig. 1) and from Schizosaccharomyces pombe (data not shown), suggesting conservation of NMD3 function throughout eukaryotes. In addition, apparent homologs, corresponding to the amino-terminal half of Nmd3p, are predicted from genome sequencing of the archaebacteria Methanococcus jannaschii and Methanobacterium thermoautotrophicum. Although this protein family does not contain readily identifiable protein motifs related to proteins of known function, it does contain a highly conserved cysteine repeat motif indicated in Fig. 1. These cysteine repeats are reminiscent of zinc-binding motifs of RING fingers, LIM domains (39), and type IV fingers (57). Determination of whether this motif is a novel zinc-binding motif will require further biochemical analysis.
FIG. 1.
Sequence analysis of Nmd3p. (A) Expected domain structure of Nmd3p based on sequence alignment and amino acid composition. Numbers indicate amino acid residues of Nmd3p. The acidic C terminus (residues 470 to 518) is lacking in the nmd3-1 mutant protein. (B) Multiple sequence alignment of the most highly conserved region of amino-terminal domain of Nmd3p with eukaryotic and archaeal homologs. Positions of ≥80% identity are shaded in black, and positions of ≥80% similarity are shaded in gray. Bars indicate the conserved Cx2C repeats. Numbers in parentheses indicate sizes of full-length proteins. Proteins and accession numbers: ScNmd3p, Nmd3p from S. cerevisiae; Ce, hypothetical protein from C. elegans (GenBank 1301731); Dm, hypothetical protein from D. melanogaster (GenBank 2661560); Mj, hypothetical protein from M. jannaschii (GenBank 150051); Mt, hypothetical protein from M. thermoautotrophicum (GenBank 2622898). Alignments were performed by using CLUSTALW at http://transfac.gbf-braunschweig.de/dresearch/clustalw.html. Shading was done with MacBoxshade, obtained from http://www.isrec.isb-sib.ch/sib-isrec/boxshade/.
NMD3 has also been identified from a screen for proteins that interact with Upf1p (6, 22). The domain of Nmd3p responsible for this interaction was the C-terminal 120 amino acids (6). This region is highly hydrophilic and is conserved among the eukaryotic homologs of Nmd3p but is not present in the archaeal proteins (Fig. 1A). Interestingly the nmd3-1 mutation is a single nucleotide insertion of an adenosine in a run of adenosines from nt 1399 to 1403. This insertion results in a frameshift in which the highly acidic C-terminal 51 amino acids (40% glutamate and aspartate) are truncated (Fig. 1A). This deletion is within the domain that interacts with Upf1p and thus may disrupt this interaction. We also sequenced three temperature-sensitive mutants generated by PCR (see Materials and Methods), all containing multiple mutations leading to multiple amino acid changes: nmd3-2 (Ile55Thr Ala122Thr Pro166Ser Leu296Pro), nmd3-3 (Thr105Ser Leu174Phe Asp195Gly Val258Asp), and nmd3-4 (Tyr136Asn Thr158Ala Leu214Trp Ser329Gly Arg441Cys Glu499Gly). Because of the number of changes and their distribution throughout the protein, they are not informative about protein structure and function. With the exception of synthetic lethality with xrn1Δ, the nmd3-1 and temperature-sensitive mutants behaved similarly in that all gave rise to reduced levels of 60S subunits resulting in half-mers, sensitivity to translation elongation inhibitors, and no obvious defects in mRNA turnover (see below; also data not shown).
nmd3 mutants do not display significant defects in mRNA turnover.
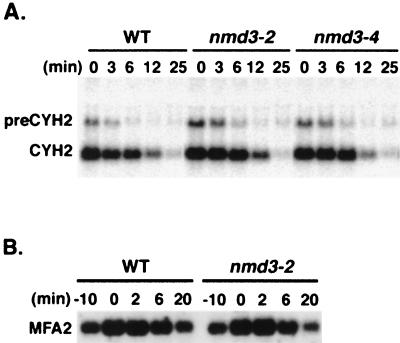
NMD3 was identified from two genetic screens for mRNA turnover factors: synthetic lethality with xrn1Δ and a two-hybrid screen with UPF1. Thus, it seemed likely that nmd3 mutants would display defects in mRNA turnover, especially for RNAs in the nonsense-mediated decay pathway. The transcript from the CYH2 gene is inefficiently spliced in yeast, and the pre-mRNA is rapidly degraded in the cytoplasm by nonsense-mediated decay. The effects of temperature-sensitive nmd3-2 and nmd3-4 mutants on pre-CYH2 degradation were assessed. Cultures were shifted to the nonpermissive temperature for 2 h, at which time thiolutin was added to inhibit RNA polymerase II transcription. In this experiment, the nmd3 mutant strains showed two- to fourfold-higher levels of pre-CYH2 at the time of thiolutin addition, suggesting that the steady-state levels of pre-CYH2 were slightly elevated (Fig. 2A). A similar elevation in the ratio of pre-CYH2 to mature CYH2 was also seen in steady-state experiments (data not shown). However, there was no significant difference in the rates of decay of pre-CYH2 or mature CYH2. The half-lives of pre-CYH2 in wild-type and mutant cells were about 4 and 5 min, respectively, and those of mature CYH2 cells were 9.5 and 10.5 min, respectively. In control experiments, an xrn1 null mutant showed significant stabilization of pre-CYH2 (data not shown). The degradation of MFA2 mRNA in a transcription pulse-chase experiment was also examined (Fig. 2B). After growth for 2 h at 37°C, transcription of a galactose-inducible MFA2 construct was transiently induced with galactose followed by inhibition of transcription by glucose. In this experiment, there was no discernible difference between mutant and wild type. Similar results were obtained for a galactose-inducible wild-type or nonsense codon-containing MATα1 transcript in the partially functional mutant nmd3-1 (data not shown). Thus, NMD3 does not appear to have a general role in mRNA degradation.
FIG. 2.
mRNA stability. (A) Strains containing wild-type (WT) NMD3 (AJY592) and the temperature-sensitive alleles nmd3-2 (AJY590) and nmd3-4 (AJY596) were grown at 26°C and shifted to 37°C for 2 h, after which time thiolutin was added. Cells were harvested at the indicated times after thiolutin addition, total RNA was prepared, and 10 μg of RNA was analyzed for each sample by Northern blotting for pre-CYH2 and mature CYH2. (B) NMD3 wild-type and temperature-sensitive strains AJY592 and AJY590 containing GAL1::MFA2 on a plasmid were grown at 26°C in SC Ura− medium containing 2% raffinose and shifted to 37°C for 2 h, after which time galactose was added to 2% (final concentration). After 10 min, glucose was added to 2%. Cells were harvested at the indicated times relative to the addition of glucose, total RNA was prepared, and 10 μg of RNA was analyzed for each sample by Northern blotting for MFA2.
Effects of translation inhibitory drugs on nmd3 mutants.
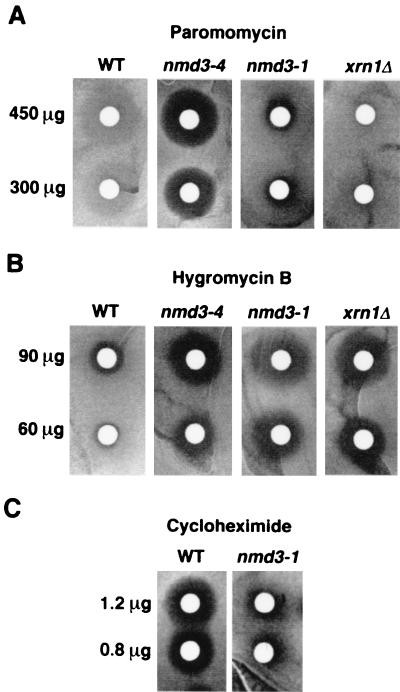
Since nmd3 mutants did not show significant defects in mRNA degradation, we examined their effects on translation, first by assaying for sensitivity to various antibiotics that inhibit translation. Figure 3 shows a disk diffusion assay for drug sensitivity on plates. In this assay, strains carrying both nmd3-1 and the temperature-sensitive allele nmd3-4 displayed sensitivity to paromomycin (Fig. 3A) and hypersensitivity to hygromycin B (Fig. 3B) relative to wild-type strains. The light halo seen for the wild type in the presence of paromomycin is a zone of slow growth distinct from the zone of total growth inhibition seen in the nmd3-4 strain. Interestingly, an xrn1Δ mutant showed hypersensitivity to hygromycin B but was not sensitive to paromomycin at the concentrations used (Fig. 3A and B). Paromomycin and hygromycin B increase the translational error rate during elongation in yeast cells, and mutations that affect translational fidelity often confer increased sensitivity to these drugs (21, 44, 53, 54). Cycloheximide also inhibits translation by direct interaction with the large ribosomal subunit, leading to the inhibition of translational elongation and the peptidyl transfer reaction (29). As shown in Fig. 3C, the nmd3-1 mutant displayed resistance to cycloheximide compared to that of wild-type cells, whereas xrn1 mutants were similar to wild-type cells in this assay (data not shown). The sensitivity of nmd3 mutants to paromomycin and hygromycin B and resistance to cycloheximide suggest a role for NMD3 in translation and ribosome function.
FIG. 3.
Drug diffusion assay. Filters containing the indicated amounts of antibiotic were placed on freshly spread lawns of cells on YPD plates and incubated at 33°C for 2 days. Strains used were CH1305 (wild type [WT]; NMD3), AJY734 (nmd3-4), AJY543 (nmd3-1), and RDKY1977 (xrn1Δ).
nmd3 mutants display decreased levels of 60S ribosomal subunits and half-mer polysomes.
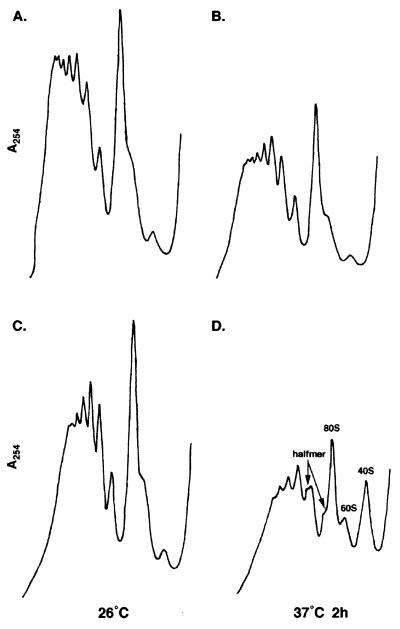
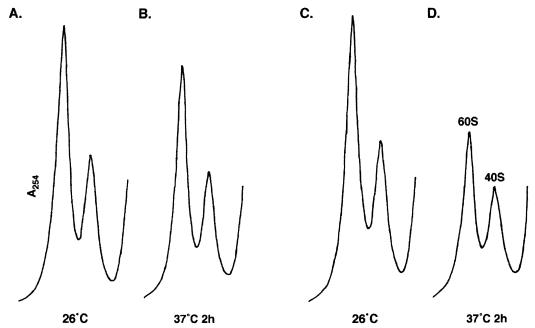
The role of NMD3 in translation was analyzed more directly by examining the effects of an nmd3 mutation on polysome profiles. Polysomes were prepared from wild-type and temperature-sensitive nmd3-4 mutant cells grown at 26 or 37°C and analyzed by ultracentrifugation on sucrose density gradients. At the permissive temperature, profiles for the nmd3-4 mutant (Fig. 4C) and the wild type (Fig. 4A) were similar, but after 2 h at 37°C, the mutant showed a deficit of free 60S relative to free 40S ribosomal subunits, an overall decrease in the average number of ribosomes in polysomes, and the appearance of half-mers (Fig. 4D). Similar results were obtained with the hypomorphic allele nmd3-1 grown at 30°C (data not shown). The presence of half-mers, which contain 43S initiation complexes stalled at the initiator AUG, is indicative of a defect in a late step of the translation initiation pathway (18).
FIG. 4.
Polyribosome profiles of nmd3-4 mutants at restrictive temperature. Extracts from strains AJY735 (NMD3) (A and B) and AJY734 (nmd3-4) (C and D) grown at room temperature (A and C) or shifted to 37°C for 2 h (B and D) were fractionated on 7 to 47% sucrose density gradients as described in Materials and Methods. Peaks representing free 40S, 60S, and 80S monoribosomes are labeled. Peaks representing half-mer polyribosomes are labeled with arrows.
The reduction in the amount of free 60S subunits and the presence of half-mers indicated an imbalance in the ratio of 40S to 60S subunits. This was confirmed by examining the levels of total 60S and 40S subunits under conditions that cause dissociation of subunits. Extracts were prepared in buffer lacking Mg2+ and with 30 mM EDTA and analyzed on sucrose gradients. Under these conditions, the nmd3-4 mutant showed significantly decreased total 60S levels relative to 40S after 2 h at the nonpermissive temperature (Fig. 5D) compared with the wild type (Fig. 5A and B) and the nmd3-4 mutant at 26°C (Fig. 5C). Quantitation of peaks in Fig. 5 yielded a 60S-to-40S ratio of 1.2 for the mutant at 37°C, compared to 1.7 for the mutant at 26°C and wild type at 37°C. The ratio of 40S in the mutant at 37°C versus the wild type at 37°C was 0.94. Thus, after 2 h at the nonpermissive temperature, there was a 25% reduction in total 60S subunits but little change in 40S levels. In a time course experiment, half-mers formed only after there was a measurable decrease in 60S subunits (data not shown). These data indicate that in an nmd3 mutant, subunit joining per se is not defective and the appearance of half-mers resulted from depletion of 60S levels.
FIG. 5.
nmd3-4 mutants display reduced 60S subunit levels at restrictive temperature. Extracts were prepared in the presence of EDTA and without Mg2+ to dissociate ribosomal subunits and sedimented through 7 to 47% sucrose density gradients. Strains AJY735 (NMD3; A and B) and AJY734 (nmd3-4; C and D) were grown at room temperature (A and C) or shifted to 37°C for 2 h (B and D).
Because nmd3 mutants have reduced levels of 60S subunits and nmd3-1 is synthetically lethal with xrn1Δ, it was possible that any mutation that resulted in reduction of 60S subunits would be synthetically lethal with xrn1Δ. This was tested genetically by crossing an spb2 mutant (YAS398) to an xrn1Δ mutant (RDKY1978) to determine if the double mutant was viable. SPB2 encodes ribosomal protein L46, and mutations in SPB2 lead to reduced levels of 60S subunits (51). The xrn1Δ spb2Δ double mutants were viable and did not grow appreciably more slowly than xrn1Δ single mutants (data not shown). Thus, nmd3-1 synthetic lethality with xrn1Δ may not be due simply to reduced 60S levels.
Nmd3p is required for stable 25S rRNA.
Defects in ribosomal subunit levels due to mutations in nonribosomal proteins typically suggest problems with rRNA processing and/or ribosomal subunit assembly. In yeast cells, the major portion of the rDNA repeat is transcribed by polymerase I and consists of a single transcription unit leading to the synthesis of a 35S rRNA precursor that is processed to form the 18S rRNA found in the 40S ribosomal subunit and the 25S and 5.8S rRNAs found in the 60S subunit. 5S rRNA, transcribed by polymerase III, also is found in 60S subunit (62, 66).
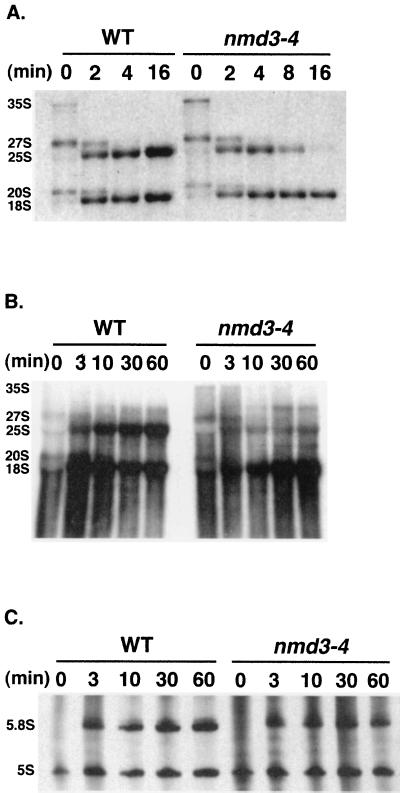
Since 25S and 18S rRNAs are highly methylated during processing, pre-rRNA processing is easily followed by metabolic labeling with [methyl-3H]methionine (61). RNA was pulse-chase labeled for 2 min at 37°C with [methyl-3H]methionine and then chased with unlabeled methionine. At the indicated times after the addition of unlabeled chase, total RNA was prepared and analyzed by formaldehyde-agarose gel electrophoresis. As shown in Fig. 6A, the nmd3-4 mutant showed a dramatic destabilization of 25S rRNA at 37°C. The half-life of 25S rRNA in the mutants was approximately 4 min, whereas in wild-type cells the label in 25S rRNA continued to accumulate throughout the chase (Fig. 6A). Although 25S rRNA failed to accumulate, the kinetics of processing of 18S and 25S rRNAs were normal in the nmd3-4 mutant at both room temperature and 37°C (Fig. 6A). This was seen by the transient appearance of mature 18S and 25S rRNAs at a 1:1 molar ratio at 2 min and the similar kinetics of disappearance of 27S and 20S precursor rRNAs, respectively. Similar results were obtained in separate experiments using different nmd3 mutant strains (data not shown). The slight increase in 35S precursor at time zero in the mutant was observed in separate experiments, whereas subsequent processing steps appeared to follow wild-type kinetics. It is not clear why the initial cleavage of 35S rRNA is slightly altered in the mutant; however, the localization of Nmd3p to the cytoplasm (see below) suggests that this is an indirect consequence of an nmd3 mutation.
FIG. 6.
Pulse-chase labeling analysis of rRNAs with [methyl-3H]methionine and [3H]uracil. (A) Log-phase cultures of AJY735 (wild type [WT]; NMD3) and AJY734 (nmd3-4) in SC Met− medium were shifted to 37°C for 2 h. Cells were labeled for 2 min with [methyl-3H]methionine, and then a saturating amount of unlabeled methionine for the chase was added. RNA was analyzed by electrophoresis in an formaldehyde-agarose gel (1.2% agarose, 6% formaldehyde). (B and C) Log-phase cultures of AJY735/pRS416 (NMD3) and AJY734/pRS416 (nmd3-4) in SC Ura− medium were shifted to 37°C for 2 h. Cells were labeled for 3 min with [3H]uracil and chased with a saturating amount of unlabeled uracil. RNA was prepared and analyzed by electrophoresis in a formaldehyde-agarose gel as described above (B) or urea-polyacrylamide gel (6% polyacrylamide, 8.2 M urea) (C) as described in Materials and Methods. Positions of the major pre-rRNAs and rRNAs are indicated.
To rule out the possibility that altered methylation was responsible for the apparent instability of 25S rRNA, pulse-chase labeling using [3H]uracil was carried out. Because [3H]uracil is utilized with slower kinetics than [methyl-3H]methionine and is more slowly chased by unlabeled uracil, it is not possible to compare directly the kinetics of processing using [3H]uracil and [methyl-3H]methionine. In addition, the time course of the [3H]uracil labeling experiment was longer than that in the [methyl-3H]methionine experiment. As seen in Fig. 6B, label accumulated normally in 18S rRNA in the mutant at 37°C but failed to accumulate in 25S rRNA. The lack of transient accumulation of processed 25S rRNA in this experiment was probably due to the rapid rate of degradation of 25S relative to the slow incorporation of label since [3H]uracil is chased with slower kinetics than [methyl-3H]methionine. The structure of the transient minor product migrating more slowly than 27S (Fig. 6B) is not known but was not observed by [methyl-3H]methionine labeling.
The processing and stability of 5S and 5.8S rRNAs were also examined by in vivo pulse-chase labeling with [3H]uracil (Fig. 6C). Total RNA was analyzed by electrophoresis on a denaturing urea-polyacrylamide gel. Quantitation of band intensities in Fig. 6C showed that after 10 min of chase, the ratio of 5.8S to 5S was 1.2 for both strains. At 30 and 60 min, the ratio increased to 1.3 for the wild type but decreased to 1.0 for the mutant. Thus, like 25S RNA, 5.8S appears to be synthesized with kinetics similar to wild-type kinetics, indicated by the similar ratios of 5.8S to 5S at 10 min, but fails to accumulate to wild-type levels. 5S RNA appears unaffected in the nmd3 mutant.
pre-rRNA processing was further examined by Northern blot analysis of total RNA using oligonucleotide probes specific for 5′ETS, ITSI, ITSII, 3′ETS and for 22 nt immediately downstream of the 3′ end of mature 25S RNA that are removed by 3′-exonucleolytic processing (see Materials and Methods). We observed no significant accumulation of pre-rRNA processing intermediates or aberrant processing products in nmd3-4 mutants at the nonpermissive temperature (data not shown). Thus, nmd3 mutants showed relatively normal processing of rRNAs but particular instability of 25S and 5.8S rRNAs. Since subunit assembly happens in a concerted fashion with rRNA processing, these results suggest that Nmd3p is necessary for a late step in ribosome assembly, after rRNA processing is completed.
Nmd3p cosediments with free 60S ribosomal subunits.
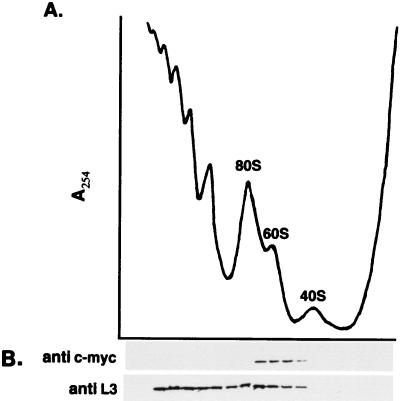
Destabilization of ribosomal subunits can result from mutations in ribosomal protein genes, factors required for rRNA processing and factors required for assembly. Since nmd3-4 mutants did not show defects in rRNA processing, we investigated the possible association of Nmd3p with ribosomes. The Nmd3 protein was epitope tagged with one copy of the c-Myc epitope at its amino terminus. This construct, expressed from a single-copy centromeric plasmid, complemented the temperature sensitivity of an nmd3 mutant. Extracts were prepared from cells expressing the c-Myc-Nmd3p and fractionated on sucrose density gradients. The A254 profile of the gradient was determined, and fractions were collected (Fig. 7A). The protein compositions of gradient fractions were analyzed by immunoblotting. Western blot analysis showed that the majority of Nmd3p migrated at the position of free 60S subunits on sucrose gradients and was not observed in the fractions containing polysomes or at the top of the gradient (Fig. 7B). The large ribosomal protein L3 was used as a marker for the sedimentation of 60S ribosomal proteins. L3 was found within the regions of the gradient containing 60S subunits, 80S monosomes, and polysomal ribosomes but was absent from fractions containing free 40S subunits (Fig. 7B). These results suggest that Nmd3p is associated with 60S subunits, but its absence from 80S ribosomes and polysomes indicates that Nmd3p is not an integral ribosomal protein.
FIG. 7.
Nmd3p cosediments with free 60S ribosomal subunits. (A) An extract from log-phase cells of strain CH1305 containing the c-Myc-tagged NMD3 on the centromeric plasmid pAJ153 was fractionated on a 7 to 47% sucrose density gradient. A254 peaks representing free 40S, 60S, and 80S monoribosomes are indicated. (B) Fractions from the gradient shown in panel A were collected and precipitated for immunoblotting. The Western blot was developed by using antibodies against the 60S subunit protein L3, used as a marker for positions of 60S subunits, and against the c-Myc epitope.
Cell fractionation.
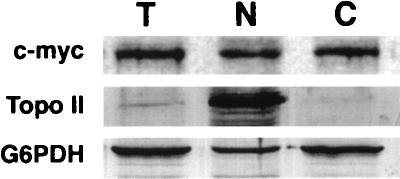
Since most ribosome assembly events occur in the nucleus, the apparent association of Nmd3p with free 60S subunits could have been explained by the cosedimentation of Nmd3p with nuclear pre-60S complexes. To address this possibility, cells expressing a functional c-Myc-tagged Nmd3p from a single-copy vector and expressed from its own promoter were fractionated into nuclear and cytoplasmic fractions. The proteins in arbitrary amounts of each fraction were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting. Topo II and G6PDH were used as nuclear and cytoplasmic markers, respectively. As seen in Fig. 8, Nmd3p fractionated similarly to G6PDH, the cytoplasmic marker protein. Topo II was highly enriched in the nuclear fraction lane, whereas Nmd3p and G6PDH were depleted. Quantitation of the band intensities in Fig. 8 revealed a 12-fold increased signal of Topo II in the nuclear fraction lane compared to total extract lane, whereas Nmd3p and G6PDH were depleted to 0.7- and 0.6-fold of their levels in the total extract lane. Thus, Nmd3p and G6PDH were present in the nuclear fraction at approximately 5% of the level expected if they were nuclear proteins. Although it is possible that a small amount of Nmd3p is present in the nucleus, the majority of the protein is cytoplasmic, supporting the notion that the apparent association with free 60S subunits reflects an association in the cytoplasm. The c-Myc cross-reactive protein was Nmd3p since no signal was observed from cells lacking c-Myc-tagged Nmd3p (data not shown).
FIG. 8.
Cell fractionation. Cells of strain CH1305 and containing a c-Myc-tagged NMD3 on the centromeric plasmid pAJ153 were fractionated into nuclear and cytoplasmic fractions. Protein from 10 μl each of total cell lysate (T), nuclear fraction (N), and cytoplasmic fraction (C) were separated by SDS-PAGE on 8% polyacrylamide gels. The separated proteins were transferred to nitrocellulose membrane, and Western blotting was performed with anti-c-Myc monoclonal antibody and anti-Topo II and G6PDH polyclonal antibodies.
DISCUSSION
We identified an allele of NMD3 from a screen for mutations synthetically lethal with a deletion of XRN1, which encodes the major cytoplasmic exoribonuclease responsible for mRNA degradation in yeast (27, 35, 55). NMD3 had previously been identified from a two-hybrid screen with UPF1 (22), a gene required for nonsense-mediated mRNA decay. In eukaryotes, premature stop codons within an mRNA stimulate rapid deadenylation-independent degradation of the mRNA, or nonsense-mediated decay (3, 25, 42, 48). In yeast, this pathway is not essential and depends on UPF1, UPF2/NMD2, and UPF3. Deletion of any one or all of these three genes prevents nonsense-mediated decay but confers few other apparent defects to the cell. NMD3, on the other hand, is an essential gene.
Dominant alleles of NMD3 (also referred to as SRC5) (30) have been identified from a screen for suppressors of a temperature-sensitive grc5 mutant (31). GRC5, also referred to as QSR1 (30), is an essential gene whose protein product, the large ribosomal subunit protein L10, is required for ribosomal subunit joining (13, 16). The loading of L10 to the 60S ribosomal subunit may be a late cytoplasmic event facilitated by the SQT1 gene product (15).
The essential nature of NMD3 along with the putative physical interaction of Nmd3p with Upf1p and the genetic interaction with xrn1 suggested to us that NMD3 may have a critical role in mRNA degradation. On the other hand, its genetic interaction with QSR1 implied a ribosomal function. Although these two functions are not mutually exclusive, our data support a primary role in ribosome biogenesis.
Examination of mRNA stability in nmd3 mutants indicated little defect in mRNA degradation. nmd3 mutants displayed a modest (two- to fourfold) increase in the abundance of pre-CYH2 mRNA, which is subject to nonsense-mediated decay. However, the increased levels of pre-CYH2 were not apparently due to impaired mRNA degradation since the half-life of pre-CYH2 was not significantly increased in the mutants. These modest mRNA stability phenotypes are in contrast to those observed for upf1 or xrn1 mutants, in which pre-CYH2 is highly stabilized. It is not clear why nmd3 mutants display elevated levels of pre-CYH2, but this could be due to a defect in export of pre-CYH2. Dominant alleles of nmd3 suppress grc5 mutations and restore the cytoskeletal defects of grc5 (31). Thus, it is conceivable that a defect in the cytoskeleton, caused by nmd3 mutation, affects mRNA export. Although we have observed no defects in mRNA turnover of deadenylation-dependent or of nonsense-mediated deadenylation-independent turnover in the nmd3-1 hypomorphic allele or in temperature-sensitive alleles, overexpression of a truncated form of Nmd3p, lacking the C-terminal 100 amino acids, leads to a dominant effect of general mRNA stabilization (6). Such stabilization appears to be a gain-of-function phenotype since it was not observed in our recessive alleles.
60S ribosomal subunits are unstable in a temperature-sensitive nmd3 mutant.
Several classes of mutations that affect ribosomal subunit stability in yeast have been identified. These include defects in integral ribosomal proteins (46, 51), defects in rRNA processing (4, 8, 17, 20, 24, 52, 56, 59, 67), and defects in ribosome assembly (15, 33). Our data suggest that nmd3 mutants fall into the latter class. The partially functional allele nmd3-1 and nmd3 temperature-sensitive mutations result in reduced levels of 60S subunits and half-mer polysomes. The half-mer polysomes appear to be a consequence of reduced 60S subunit levels and not specifically to a subunit joining defect since their appearance follows a time course similar to that of depletion of 60S subunits (unpublished observation). Pulse-chase analysis of rRNA processing indicated that 25S and 18S RNAs were made with normal kinetics in the temperature-sensitive mutant at nonpermissive temperature. However, the 25S rRNA had a half-life of only 4 min. Such destabilization of 25S rRNA has been observed previously in cells depleted of the 60S subunit protein L16 (46). Since rRNA processing and ribosome assembly are intimately linked processes, the apparently normal processing of 25S rRNA in nmd3 mutants suggest that the nmd3 defect is late in the assembly pathway.
Western blot analysis of a functional epitope-tagged Nmd3p suggested that Nmd3p was associated with free 60S subunits but not with polysomes. However, the finding that Nmd3p fractionated as a cytoplasmic protein supports the idea that the association is with free cytoplasmic 60S subunits. Although most 60S subunit proteins are loaded onto the particle in the nucleolus, at least four proteins are believed to be exchangeable, raising the possibility that they are added in the cytoplasm. One of these is the L10, encoded by QSR1 (13).
As noted above, NMD3 shows a genetic interaction with QSR1. A second gene, SQT1, was identified as a high-copy-number suppressor of a dominant negative phenotype conferred by overexpressing a truncated L10 (15). Sqt1p is thought to function in the cytoplasm in loading L10 onto the 60S subunit. Interestingly, depletion of L10 does not result in destabilization of 60S subunits, whereas depletion of Sqt1p does. Thus, in addition to loading L10 protein, Sqt1p must have an additional function that is required for subunit integrity. It has been suggested that Sqt1p may contribute to a novel mechanism of translational control by modulating ribosome function through exchanging protein L10 (13). That both SQT1 and NMD3 appear to be required for a late cytoplasmic assembly step and both have genetic interaction with QSR1 suggests that Sqt1p and Nmd3p may act together. However, in preliminary experiments we have not observed physical or genetic interaction between SQT1 and NMD3 (23). Additionally, whereas Sqt1p has been suggested to exchange L10 protein onto and off ribosomes, the depletion of 60S subunits in nmd3-4 mutants is observed only after several hours at the nonpermissive temperature (data not shown). This finding suggests that nmd3 mutations do not affect ribosomes already formed. Thus, NMD3 is most likely required for a maturation step and not for exchanging proteins on existing ribosomes. Such a maturation step could be protein loading, rRNA modification, or rearrangement of existing components. It is also possible that transport of the 60S subunit is affected.
Comparison of Nmd3p sequence with GenBank sequences indicated that the protein is conserved throughout eukaryotes. In addition, archaebacteria but not bacteria contain proteins of related sequence. Sequence analysis of Nmd3p revealed no readily recognizable protein motifs. Intriguingly, these proteins all contain Cx2Cx10–12Cx2Cx18–21Cx2Cx75–89Cx2C. This sequence is reminiscent of various cysteine-rich zinc-binding motifs including RING fingers, type IV zinc fingers, and LIM domains (39, 57), although the spacing of the Cx2C repeats and additional residues defining these motifs are not conserved in Nmd3p. Because Nmd3p is required for a ribosome assembly step, it is possible that this motif in Nmd3p is involved in either RNA binding or protein-protein interaction to facilitate the loading or rearrangement of a 60S ribosomal protein.
NMD3 and nonsense-mediated mRNA decay.
The identification of NMD3 from a two-hybrid screen with UPF1 suggests a role for NMD3 in nonsense-mediated decay. However, we were unable to observe a defect in nonsense mRNA decay in nmd3 mutants. Thus, NMD3 does not appear to be required for the nonsense-mediated decay pathway. It is intriguing that the nmd3-1 mutation results in truncation of the protein within the region identified as interacting with Upf1p. However, UPF1 is not an essential gene whereas NMD3 is, and nmd3-1 upf1 double mutants are viable (23). Thus, UPF1 is not required for NMD3 function. The finding that overexpression of a truncated Nmd3p gives rise to a dominant stabilization of mRNA suggests that Nmd3p may interact with mRNA turnover factors (6), although our work suggests that it is not required for normal mRNA turnover.
The basis for synthetic lethality between nmd3-1 and xrn1Δ.
At present we do not know why the nmd3-1 mutation is synthetic lethal with an xrn1 null mutation. nmd3 temperature-sensitive mutants are unable to assemble stable 60S subunits, manifest in a reduced pool of free and total 60S subunits. This in turn leads to a translation initiation defect. The partial function allele, nmd3-1, identified as synthetically lethal with xrn1, also displayed reduced 60S levels, suggesting that in an xrn1 mutant, normal levels of 60S subunits are essential. This idea suggests that other mutations leading to reduced 60S levels would also be synthetically lethal with an xrn1 deletion. However, an spb2 mutation, which leads to reduced 60S levels, is not synthetically lethal with xrn1. Thus, synthetic lethality is not due simply to reduced 60S levels, suggesting that there is a specific defect conferred by nmd3-1 on the 60S subunit. The identification of nmd3 suppressors should shed light on the specific function of NMD3 and the reason for its genetic interaction with XRN1.
ACKNOWLEDGMENTS
We thank Alan Sachs for providing strain YAS398, Janet Lindsley and Jim Wang for providing anti-Topo II antibodies, Jonathan Warner for anti-L3 antibodies, and Roy Parker for plasmid pRP485. We are grateful to Xianmei Yang for the spb2Δ cross with xrn1Δ and Justin Brown for c-Myc-NMD3 construct pAJ153. We are especially appreciative of John Woolford for critical reading of the manuscript and of Allan Jacobson for sharing results before publication. DNA sequence analysis was done by the Core Facility at the Institute for Cellular and Molecular Biology, University of Texas at Austin.
This work was supported by NIH grant GM056355 to A. W. Johnson.
ADDENDUM IN PROOF
Because we found that the c-Myc signal of epitope-tagged Nmd3p was weak, even when expressed from a galactose-inducible promoter, we have since tagged the genomic NMD3 with 13 tandem copies of c-Myc to verify the cytoplasmic localization. The multiply tagged protein was functional and by indirect immunofluorescence techniques it was found in the cytoplasm and excluded from the nucleus.
REFERENCES
- 1.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baim S B, Pietras D F, Eustice D C, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985;5:1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker G F, Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11:2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin-Baillien A, Tollervey D, Cullin C, Lacroute F. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol. 1997;17:5023–5032. doi: 10.1128/mcb.17.9.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belgrader P, Cheng J, Zhou X, Stephenson L S, Maquat L E. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belk, J., F. He, and A. Jacobson. 1998. Personal communication.
- 7.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs M W, Burkard K T, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. T., and A. W. Johnson. 1998. Unpublished data.
- 10.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 11.Culbertson M R, Underbrink K M, Fink G R. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick F A, Eisinger D P, Trumpower B L. Exchangeability of Qsr1p, a large ribosomal subunit protein required for subunit joining, suggests a novel translational regulatory mechanism. FEBS Lett. 1997;419:1–3. doi: 10.1016/s0014-5793(97)01402-6. [DOI] [PubMed] [Google Scholar]
- 14.Dinman J D, Wickner R B. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisinger D P, Dick F A, Denke E, Trumpower B L. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol Cell Biol. 1997;17:5146–5155. doi: 10.1128/mcb.17.9.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian G R, Hopper A K. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J Bacteriol. 1987;169:1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foiani M, Cigan A M, Paddon C J, Harashima S, Hinnebusch A G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gietz D, St. John A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez A, Jimenez A, Vazquez D, Davies J E, Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta. 1978;521:459–469. doi: 10.1016/0005-2787(78)90287-3. [DOI] [PubMed] [Google Scholar]
- 22.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 23.Ho, J. H.-N., and A. W. Johnson. Unpublished data.
- 24.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 26.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson A W, Kolodner R D. Synthetic lethality between SEP1/XRN1 and the antiviral genes SKI2 and SKI3 is independent of virus and suggests a general role in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 29.Kaufer N F, Fried H M, Schwindinger W F, Jasin M, Warner J R. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983;11:3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koller, L. Personal communication.
- 31.Koller, L., T. Karl, T. Klade, R. Kodzius, A. Thur, R. Andrejic, and M. Breitenbach. 1995. Personal communication.
- 32.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kressler D, de la Cruz J, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruiswijk T, Planta R J, Krop J M. The course of assembly of ribosomal subunits in yeast. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 35.Larimer F W, Hsu C L, Maupin M K, Stevens A. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 36.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 38.Leeds P, Wood J M, Lee B-S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackay J P, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 40.Mager W H, Planta R J, Ballesta J, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 42.Maquat L E, Kinneburgh A J, Rachmilewitz E A, Ross J. Unstable β-globin mRNA in mRNA deficient beta o thalassemia. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 43.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masurekar M, Palmer E, Ono B I, Wilhelm J M, Sherman F. Misreading of the ribosomal suppressor SUF46 due to an altered 40S subunit in yeast. J Mol Biol. 1981;147:381–390. doi: 10.1016/0022-2836(81)90490-3. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 46.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page A M, Davis K, Molineux C, Kolodner R D, Johnson A W. Mutational analysis of exoribonuclease I in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3707–3716. doi: 10.1093/nar/26.16.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peltz S W, He F, Welch E, Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–297. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 49.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 52.Sachs A B, Davis R W. Translation initiation and ribosome biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 53.Sandbaken M G, Culbertson M R. Mutations in elongation factor EF1-a affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics. 1988;120:923–934. doi: 10.1093/genetics/120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature (London) 1990;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 55.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 56.Sun C, Woolford J L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teakle G R, Gilmartin P M. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem Sci. 1998;23:100–102. doi: 10.1016/s0968-0004(98)01174-8. [DOI] [PubMed] [Google Scholar]
- 58.Toh-e A, Guerry P, Wickner R B. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 60.Vernet T, Dignard D, Thomas D Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 61.Warner J R. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–427. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 62.Warner J R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickner R B. Double-stranded viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Widner W R, Wickner R B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wise J A. Preparation and analysis of low molecular weight RNAs and small ribonucleoproteins. Methods Enzymol. 1991;194:405–415. doi: 10.1016/0076-6879(91)94031-7. [DOI] [PubMed] [Google Scholar]
- 66.Woolford J L Jr, Warner J R, editors. The ribosome and its synthesis. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- 67.Zanchin N I T, Roberts P, Desilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zinker S, Warner J R. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J Biol Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]