Abstract
Cell extrusion is a mechanism of cell elimination used by organisms as diverse as sponges, nematodes, insects and mammals1–3. During extrusion, a cell detaches from a layer of surrounding cells while maintaining the continuity of that layer4. Vertebrate epithelial tissues primarily eliminate cells by extrusion, and the dysregulation of cell extrusion has been linked to epithelial diseases, including cancer1,5. Mechanisms that drive cell extrusion remain incompletely understood. To analyze cell extrusion by C. elegans embryos3, we conducted a genome-wide RNAi screen, identified multiple cell-cycle genes with S-phase-specific function, and performed live-imaging experiments to establish how those genes control extrusion. We observed that extruding cells experience replication stress during S phase and activate a replication-stress response via ATR and Chk1 homologs. Preventing S-phase entry, inhibiting the replication-stress response, or allowing completion of the cell cycle blocked cell extrusion. Hydroxyurea-induced replication stress6,7 triggered ATR/Chk1- and p53-dependent cell extrusion from a mammalian epithelial monolayer. We conclude that replication-stress-induced cell extrusion is conserved among animals and propose that this extrusion process is a primordial mechanism of cell elimination with a tumor-suppressive function in mammals.
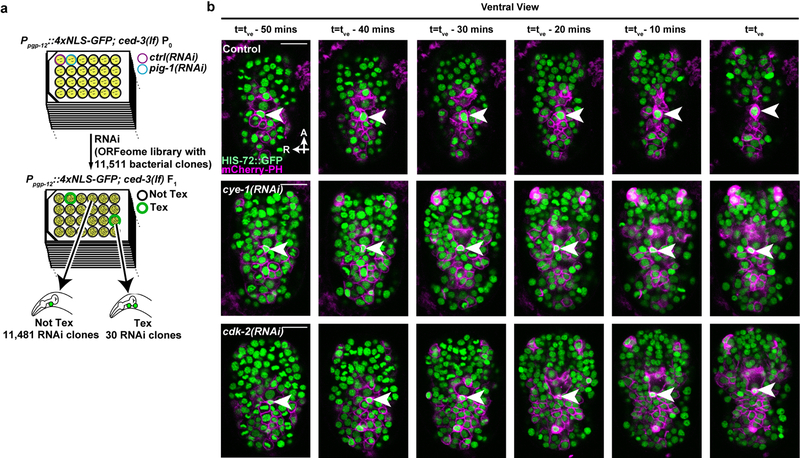
Mutants of C. elegans defective in caspase-mediated apoptosis, e.g. ced-3(lf) loss-of-function mutants, provide an excellent system for studies of cell extrusion3. Cell extrusion functions as a “backup” mechanism in ced-3(lf) embryos to eliminate certain cells otherwise eliminated by caspase-mediated apoptosis3. To identify genes that control cell extrusion, we screened the C. elegans ORFeome RNAi library of 11,511 bacterial clones for RNAi clones that in ced-3(lf) animals generated the two-excretory cell (Tex) phenotype3 (Figs. 1a–c, Extended Data Fig. 1a), which occurs when the cell ABplpappap fails to be extruded. From this screen, we identified 30 RNAi clones corresponding to 27 genes that produced a Tex phenotype (Extended Data Fig. 1a). Remarkably, 10 of the 27 genes identified were cell-cycle genes with functions mostly specific to the S phase8 (Figs. 1d, e). Additional RNAi screens identified an additional four such genes (Fig. 1d, Extended Data Table 1). Consistent with their functioning in cell extrusion, RNAi against 13 of the 14 identified cell-cycle genes produced a Tex phenotype only in the ced-3(lf) background and not in a ced-3(+) wild-type background (Extended Data Table 2).
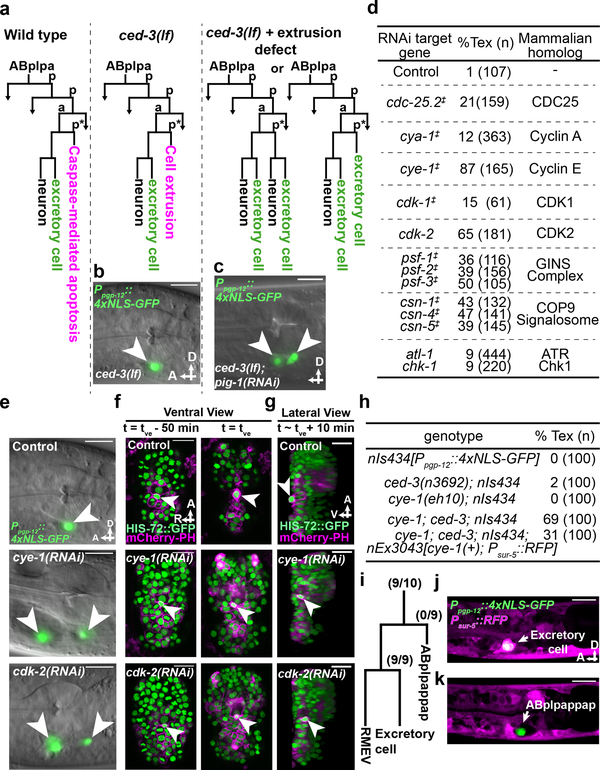
Figure 1. Cell-cycle genes control cell extrusion cell autonomously.
a, Sublineage diagram showing the fate of ABplpappap (*) in wild-type, ced-3(lf) and ced-3(lf)+extrusion-defective animals. b, c, Merged epifluorescence and Nomarski micrographs of the pharyngeal region of (b) nIs433[Ppgp-12::4xNLS-GFP]; ced-3(lf) and (c) extrusion-defective nIs433; ced-3(lf); pig-1(RNAi) animals (ref. 3). d, The percentages of animals with the Tex phenotype in ced-3(lf) animals after the indicated RNAi treatment. ‡, identified from screen shown in Extended Data Figure 1a; others identified from candidate RNAi screens. e, Merged epifluorescence and Nomarski micrographs of the pharyngeal region of nIs433; ced-3(lf) animals after the indicated RNAi treatment. f, g, Ventral and virtual lateral views of ced-3(lf); stIs10026[his-72::GFP]; nIs632[Pegl-1::mCherry::PH] embryos after the indicated RNAi treatment at the indicated time point. tve – time point of ventral enclosure. h, Percentage of animals of the indicated genotype displaying the Tex phenotype. i, Cell-lineage diagram showing number of animals with the Tex phenotype carrying the cye-1(+)-rescuing array nEx3043 in the indicated lineage or cell in 10 cye-1(lf); ced-3(lf); nIs434[Ppgp-12::4xNLS-GFP] animals. j, k, Two confocal fluorescence micrographs of one of 10 cye-1(lf); ced-3(lf); nIs434; nEx3043 genetic mosaic worms represented in (i). Arrowheads, excretory or ectopic excretory-like cells in (b, c, e); ABplpappap in (f, g). A, anterior; R, right; D, dorsal; V, ventral. Scale bars, 10 μm.
Cell-cycle genes cell-autonomously promote cell extrusion
To characterize the functional role of cell-cycle genes in cell extrusion, we used time-lapse confocal microscopy to observe ABplpappap in embryos treated with RNAi against cye-1 or cdk-2 or with an empty vector (“control embryos”). We tracked the fate of ABplpappap over a 50-min period ending in ventral enclosure, which normally coincides with cell extrusion3 (Fig. 1f, Extended Data Fig. 1b, Supplementary Videos 1–3). Virtual lateral sections of the embryos at the end of ventral enclosure confirmed that ABplpappap was extruded from control embryos (10 of 11 embryos; Fig. 1g). By contrast, ABplpappap was not extruded from cye-1(RNAi) (11 of 11 embryos; Fig. 1g) or cdk-2(RNAi) embryos (10 of 11 embryos; Fig. 1g), consistent with the highly penetrant Tex phenotype observed for cye-1(RNAi) and cdk-2(RNAi) larval animals (Figs. 1d, e). We conclude that cye-1 and cdk-2 are required for extrusion of the cell ABplpappap.
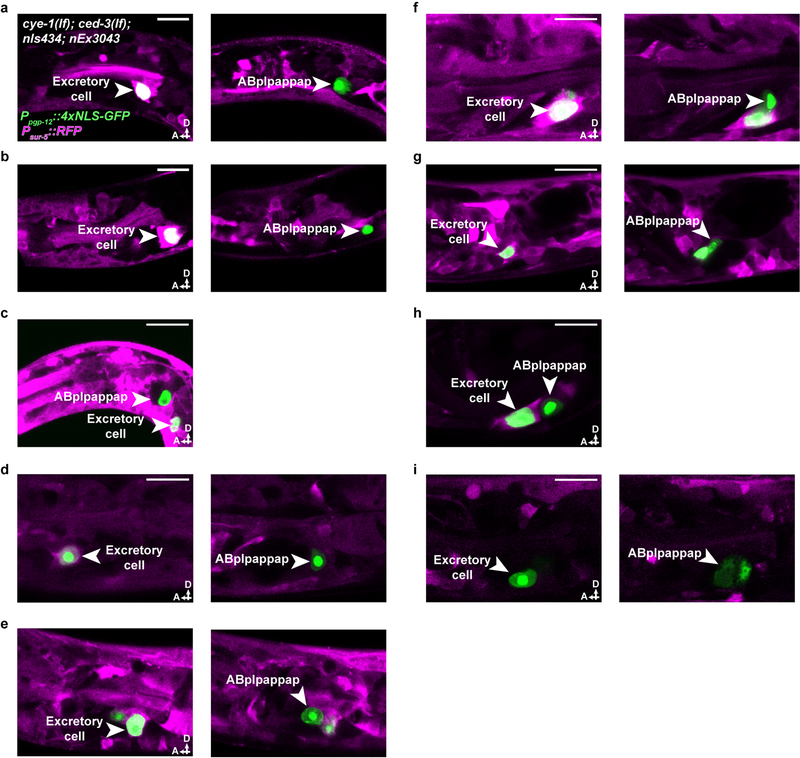
To identify the cellular site of cell-cycle gene function in ABplpappap cell extrusion, we performed genetic mosaic analyses. We examined cye-1(lf); ced-3(lf) double mutants and found that these animals displayed a Tex phenotype similar to that of ced-3(lf); cye-1(RNAi) animals (Figs. 1d, h), suggesting that extrusion is more sensitive to a reduction in cell-cycle gene levels than general embryonic development, for which maternal contribution of cye-1 is sufficient9. We generated an extrachromosomal array carrying the cell-autonomous RFP reporter Psur-5::RFP and a cye-1(+) transgene, which partially rescued the Tex phenotype of cye-1(lf); ced-3(lf) animals (Fig. 1h). Extrachromosomal arrays are mitotically unstable and randomly lost during development, which produces mosaic animals. We examined 10 mosaic animals that carried the cye-1(+)-rescuing array, as indicated by the presence of RFP in any set of cells or tissues, but were not rescued for the Tex phenotype (Fig. 1i). The RFP expression pattern indicated that the array was absent from ABplpappap in all 10 animals, despite being present in the ABplpappap niece (the excretory cell) in nine of the 10 animals (Figs. 1i–k, Extended Data Figs. 2a–i). We conclude that cye-1 and likely other cell-cycle genes function cell-autonomously for the extrusion of ABplpappap.
Cells arrest in S phase prior to extrusion
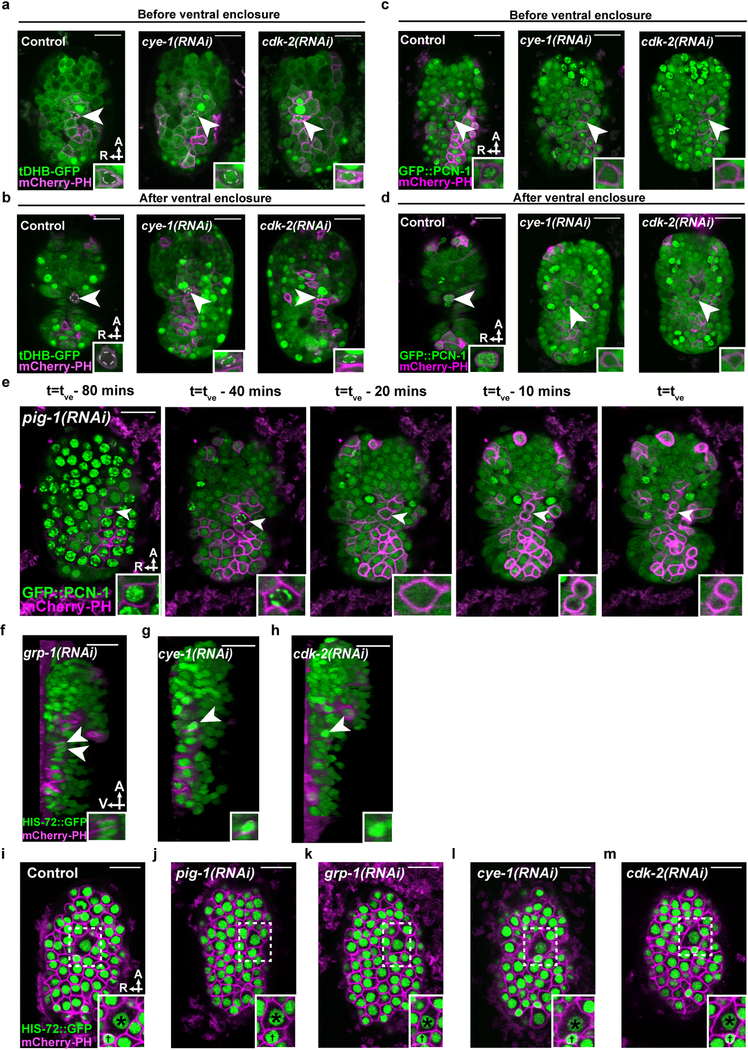
To characterize the cell-cycle phase of cells that are extruded, we used two cell-cycle reporters: (i) tDHB-GFP10, a truncated DNA Helicase B fragment fused to GFP, and (ii) GFP::PCN-1, an N-terminal translational fusion of GFP to the C. elegans homolog of the DNA replication processivity factor PCNA11. tDHB-GFP is enriched in the nuclei of quiescent, post-mitotic and G1-phase cells and transits to the cytoplasm for all other cell-cycle phases10 (Fig. 2a); GFP::PCN-1 exhibits a punctate sub-nuclear localization only during S phase in both mammalian and early C. elegans embryonic cells11,12 (Fig. 2b). In control embryos, tDHB-GFP reporter fluorescence was mostly absent from the ABplpappap nucleus (Fig. 2c) both before ventral enclosure (5 of 5 embryos; Extended Data Fig. 3a) and after extrusion (5 of 5 embryos; Extended Data Fig. 3b). Cells extruded from other sites of control or untreated embryos also displayed low levels of nuclear tDHB-GFP (Extended Data Figs. 4a–d). Therefore, ABplpappap and other extruded cells appear to enter but never complete the cell cycle during extrusion.
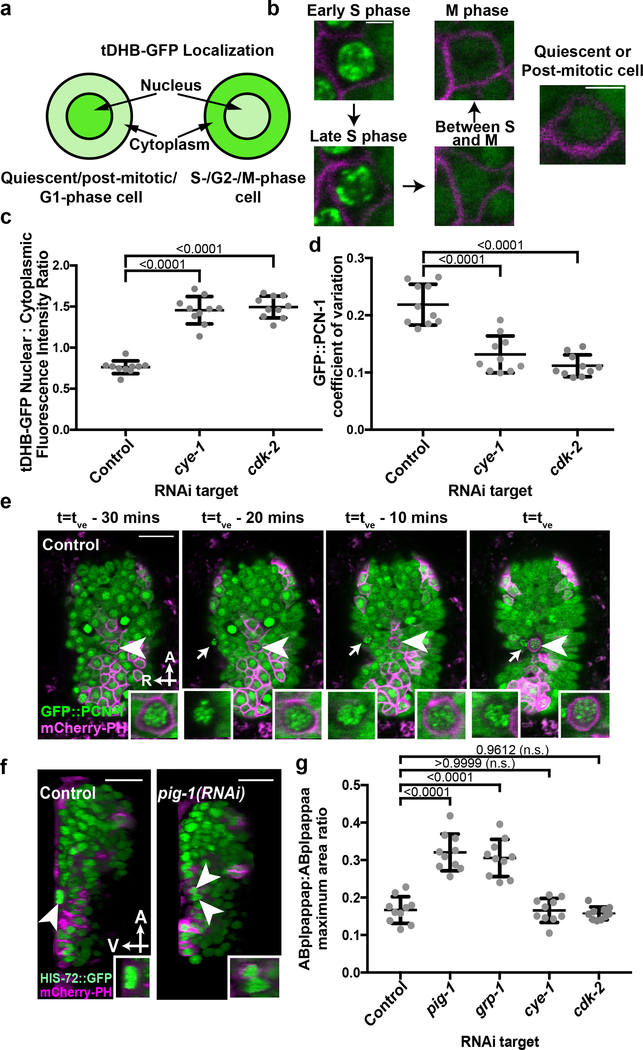
Figure 2. Cells undergoing extrusion arrest in S phase.
a, Relative nuclear/cytoplasmic localization of a tDHB-GFP fusion protein in the indicated cell-cycle phases10. b, localization pattern of GFP::PCN-1 in a typical C. elegans embryonic cell in the indicated cell-cycle phase. c, Nuclear:cytoplasmic ratio of tDHB-GFP fluorescence intensity in ABplpappap in heSi192[Peft-3::tDHB-GFP]; ced-3(lf); nIs861[Pegl-1::mCherry::PH] embryos after the indicated RNAi treatment. d, Quantification of the coefficient of variation of GFP::PCN-1 fluorescence intensity in ced-3(lf); isIs17[Ppie-1::GFP::pcn-1]; nIs861 embryos after indicated RNAi treatment. e, Time-lapse confocal fluorescence micrographs of GFP::PCN-1 fluorescence in ABplpappap (arrowhead) in ced-3(lf); isIs17; nIs861 embryos at the indicated times after treatment with control RNAi. Arrow, unidentified extruding cell. f, Micrographs of virtual lateral section of ced-3(lf); nIs861; stIs10026[his-72::GFP] embryos showing either ABplpappap (arrowhead) or its daughters (arrowheads) after indicated RNAi treatment. g, Quantification of the ratio of maximum area occupied by ABplpappap to that occupied by its sister, ABplpappaa, in ced-3(lf); ltIs44[Ppie-1::mCherry::PH]; stIs10026 embryos after the indicated RNAi treatment. Right inset, magnified view of ABplpappap or its daughters. Left inset in (e), magnified view of unidentified extruding cell. Scale bars, 10 μm in all micrographs except (b) and 2 μm in (b). A, anterior; R, right; V, ventral. n=10 embryos (biological replicates) for each RNAi treatment in (c, d, g). Data in (c, d, g) are represented as mean ± S.D. Ordinary one-way ANOVA with Dunnett’s correction for multiple comparisons (c, d, g). P values are indicated; n.s., not significant.
The GFP::PCN-1 reporter localized to bright sub-nuclear punctate foci in ABplpappap (Fig. 2d) both before extrusion (5 of 5 embryos; Extended Data Fig. 3c) and after extrusion (5 of 5 embryos; Extended Data Fig. 3d) from control embryos, indicating that ABplpappap entered but did not exit S phase. Time-lapse confocal microscopy of an embryo progressing towards ventral enclosure confirmed that GFP::PCN-1 localization did not change in either ABplpappap or a second, unidentified extruding cell indicating an arrested S-phase in both cells during the period ending in their extrusion (Fig. 2e, Supplementary Video 4). Nearly all cells extruded from ced-3(lf) embryos displayed bright sub-nuclear foci of GFP::PCN-1 (Extended Data Figs. 4e, f). We conclude that cells extruded by ced-3(lf) embryos enter the cell cycle before extrusion, arrest in S phase, and are then extruded.
The treatment of embryos with RNAi against cye-1 or cdk-2 dramatically altered the localization of tDHB-GFP and GFP::PCN-1 in ABplpappap. tDHB-GFP localized to the ABplpappap nucleus (Fig. 2c) and GFP::PCN-1 was diffusely nuclear in ABplpappap (Fig. 2d) in cye-1(RNAi) and cdk-2(RNAi) embryos both before (5 of 5 embryos each; Extended Data Figs. 3a, c) and after ventral enclosure (5 of 5 embryos each; Extended Data Figs. 3b, d). These observations show that ABplpappap fails to enter the cell cycle in embryos with reduced CYE-1 and CDK-2 and indicate that progression to S phase is required for cell extrusion.
Next we tested whether the previously identified cell-extrusion regulators3 pig-1 and grp-1 also promote S-phase arrest. Surprisingly, we found that ABplpappap in pig-1(RNAi) embryos completed the cell cycle and divided into daughter cells before ventral enclosure (Extended Data Fig. 3e, Supplementary Video 5); virtual lateral sections confirmed that the daughter cells were not extruded (5 of 6 embryos; Fig. 2f). ABplpappap similarly divided to generate surviving daughters in grp-1(RNAi) embryos (6 of 6 embryos; Extended Data Fig. 3f). Thus, failure either to initiate the cell cycle, as in cye-1(RNAi) or cdk-2(RNAi) embryos, or to arrest at S-phase, as in pig-1(RNAi) or grp-1(RNAi) embryos (Fig. 2f, Extended Data Figs. 3f–h), is associated with impaired cell extrusion.
Given that pig-1 and grp-1 regulate unequal cell divisions in multiple C. elegans cell lineages (e.g., the QR/L neuroblast lineages13), we tested whether they control the unequal division of ABplpappa, which produces the extruded cell ABplpappap and a sister cell almost 2.5-fold larger in diameter (Fig. 2g, Extended Data Fig. 3i). Both pig-1(RNAi) and grp-1(RNAi) generated an abnormally large ABplpappap cell (Fig. 2g, Extended Data Figs. 3i–k); by contrast, RNAi against cye-1 or cdk-2 did not affect the highly unequal ABplpappa division (Fig. 2g, Extended Data Figs. 3i, l, m). Thus, an unequal cell division precedes S-phase cell-cycle arrest in the small daughter fated for extrusion, whereas an abnormally large ABplpappap is competent to complete the cell cycle.
Extruding cells exhibit hallmarks of replication stress
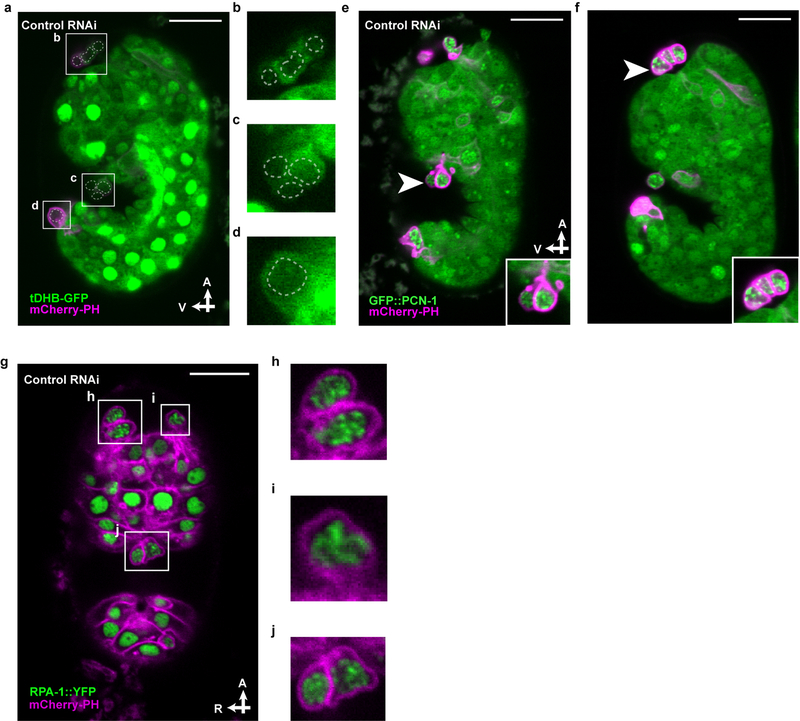
We speculated that ABplpappap enters S-phase arrest because it inherits from the unequal ABplpappa division a pool of resources insufficient to support DNA replication. We therefore examined ABplpappap for evidence of replication stress using a reporter of RPA-1, the C. elegans homolog of mammalian Replication Protein A. RPA-1 binds in distinct nuclear foci to single-stranded DNA segments generated during replication stress14. We observed that an RPA-1::YFP fusion protein localized to a small number of distinct foci in ABplpappap in control embryos (Figs. 3a, b), indicating this cell undergoes replication stress; other extruded cells exhibited a similar pattern of RPA-1::YFP foci (Extended Data Figs. 4g–j). By contrast, RPA-1::YFP was more diffuse in ABplpappap in pig-1(RNAi) or cdk-2(RNAi) embryos (Fig. 3b, Extended Data Figs. 5a, b), indicating, respectively, the absence of replication stress when ABplpappap is either abnormally large (with sufficient resources for DNA synthesis) or prevented from S-phase entry. In addition, most cells extruded from ced-3(lf) embryos displayed punctate TUNEL staining indicative of limited DNA damage15 (Figs. 3c, d) and unlike the diffuse staining of cells undergoing caspase-mediated apoptosis16, such as those visible in ced-5(lf) embryos (Figs. 3c, d). This observation suggests that a replication-stress response might limit TUNEL-reactive DNA damage17 in extruded cells. In short, cells extruded by ced-3(lf) embryos exhibited multiple hallmarks of replication stress.
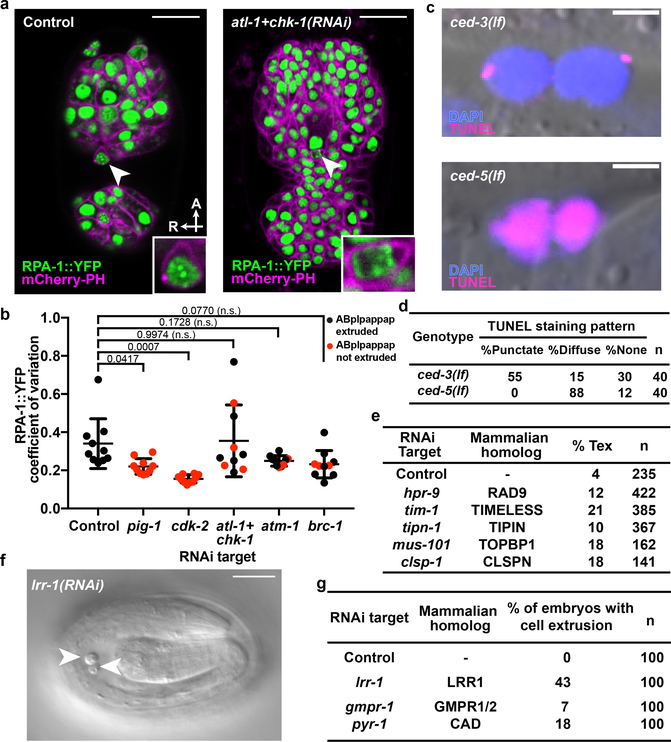
Figure 3. Replication stress is coincident with and promotes cell extrusion.
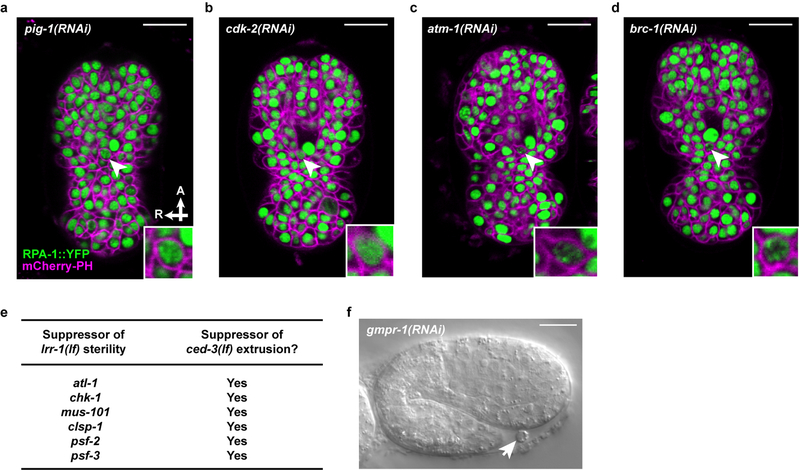
a, Confocal fluorescence micrographs showing the localization of RPA-1::YFP in ABplpappap (arrowhead) in ced-3(lf); ltIs44; opIs263[Prpa-1::rpa-1::YFP] embryos after the indicated RNAi treatment. Inset, magnified view of ABplpappap. A, anterior; R, right. Scale bars, 10 μm. b, Coefficient of variation of RPA-1::YFP fluorescence intensity in ABplpappap in ced-3(lf); ltIs44; opIs263 embryos after the indicated RNAi treatments. n=10 embryos (biological replicates) for each RNAi treatment. Black data point, ABplpappap extruded. Red data point, ABplpappap not extruded. All data are represented as Mean ± S.D. Ordinary one-way ANOVA with Dunnett’s correction for multiple comparison was performed. P values are indicated; n.s., not significant. c, Merged Nomarski and fluorescence micrographs showing DAPI and TUNEL staining in extruded cells in embryos of indicated genotype. Scale bars, 2 μm. d, Percentage of observed extruded cells in embryos of the indicated genotypes with the indicated TUNEL staining pattern. e, The percentages of animals with the Tex phenotype in ced-3(lf) animals after the indicated RNAi treatment along with mammalian homolog of the RNAi target. f, Nomarski micrograph showing cells extruded (arrowheads) from wild-type embryos after lrr-1(RNAi). Scale bar, 10 μm. g, Percentages of wild-type embryos exhibiting cell extrusion after the indicated RNAi treatments along with mammalian homologs of RNAi targets.
To determine if the replication-stress response directly promotes cell extrusion, we treated ced-3(lf) animals with RNAi against genes encoding critical replication-stress response proteins: TopBP1 (mus-101), Claspin (clsp-1), Timeless (tim-1), Tipin (tipn-1), Rad9 (hpr-9), ATR (atl-1) and Chk1 (chk-1)18. These RNAi treatments produced a Tex phenotype (Figs. 1e, 3e), indicating that the replication-stress response pathway is necessary for cell extrusion. Furthermore, RNAi against atm-1 (ATM homolog), brc-1 (BRCA1 homolog), or atl-1 (ATR homolog) and chk-1 (CHEK1 homolog) prevented ABplpappap extrusion in 3 to 4 of the 10 embryos examined after each treatment, despite the presence of RPA-1::YFP foci in the unextruded ABplpappap cell (Figs. 3a, b, Extended Data Figs. 5c, d). Thus, inhibition of the canonical replication-stress response signaling pathway prevented extrusion of cells undergoing replication stress.
We hypothesized that the source of replication stress in ABplpappap and other extruded cells is an insufficient pool of DNA replication proteins or nucleotides resulting from the unequal division of their mother cells, e.g., ABplpappa. We noted that genes previously identified from a screen for suppression of lrr-1(lf)-induced sterility19 also suppressed cell extrusion by ced-3(lf) animals (Extended Data Fig. 5e). lrr-1 encodes an adaptor protein for the CRL2LRR−1 E3 ubiquitin ligase and is required for the disassembly of terminated replisomes20,21. Loss of lrr-1 produces abnormalities in mitotic germline cells similar to those we observed in cells extruded from ced-3(lf) embryos: activation of the ATL-1/CHK-1-dependent replication-stress response, S-phase arrest, and DNA damage22. Additionally, we found that lrr-1(RNAi) causes ectopic extrusion of unidentified cells in a wild-type background (Figs. 3f, g). These findings indicate that LRR-1 insufficiency in the small ABplpappap cell can drive its extrusion from ced-3(lf) embryos, presumably by preventing the efficient removal of terminated replisomes. We also found that RNAi against a gene required for pyrimidine nucleotide synthesis (pyr-1) or a gene required for maintenance of purine nucleotide balance (gmpr-1) similarly causes ectopic extrusion of unidentified cells in wild-type animals (Fig. 3g, Extended Data Fig. 5f). pyr-1 and gmpr-1 encode the C. elegans homologs of mammalian CAD and GMP reductase enzymes, respectively. Thus, perturbing nucleotide levels is sufficient to drive cell extrusion. We propose that low levels of LRR-1 combined with insufficient nucleotide pools can result in replication stress and S-phase arrest in and ultimately cell extrusion of the small daughter cells of unequal cell divisions.
Replication stress is a conserved driver of cell extrusion
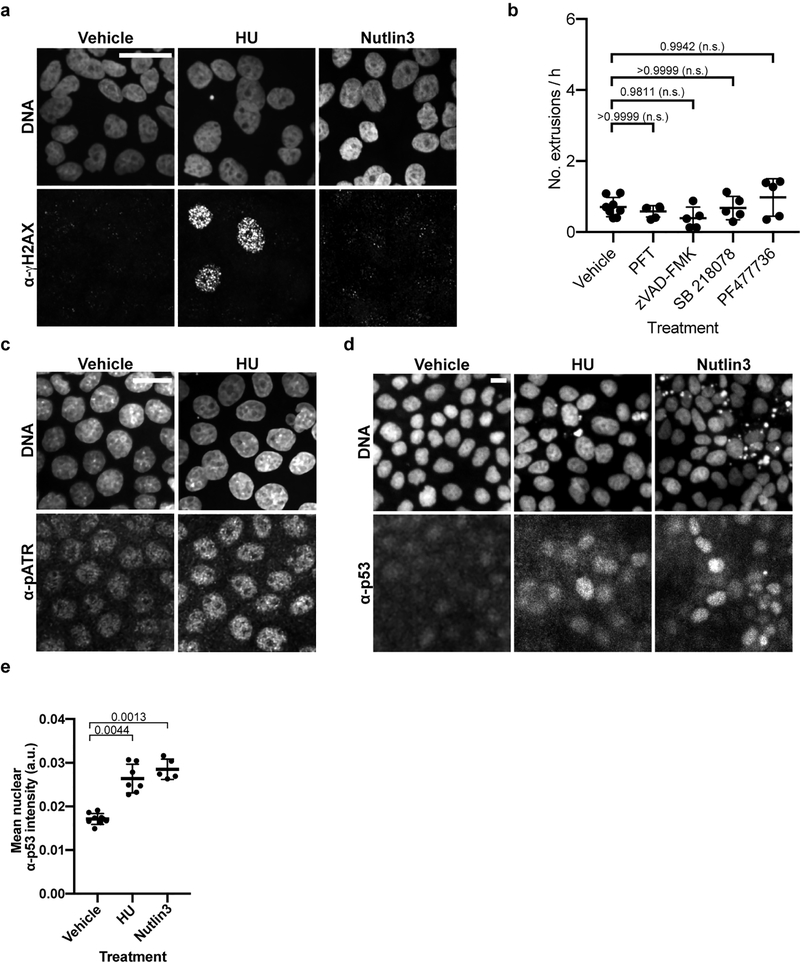
To test if replication stress-mediated cell extrusion is an evolutionarily conserved process, we examined monolayers of Madin-Darby Canine Kidney (MDCK-II) cells treated with 2 mM hydroxyurea (HU) to induce replication stress6,7 (Fig. 4a, Extended Data Fig. 6a) and measured the rate of apically directed cell extrusions via time-lapse microscopy. HU treatment increased the rate of cell extrusion from the MDCK-II monolayer by >3-fold over vehicle control (Figs. 4b, c; Supplementary Videos 6, 7). Using MDCK-Fucci cells, which produce a cell-cycle phase-specific fluorescence (G0/G1 - red; S/G2/M - green), we found that cells extruded stochastically from the monolayers (as observed with vehicle treatment) mostly exhibited red fluorescence indicative of the G0 or G1 phase (Fig. 4d). By contrast, most cells extruded from HU-treated monolayers displayed green fluorescence consistent with S-phase arrest (Fig. 4d).
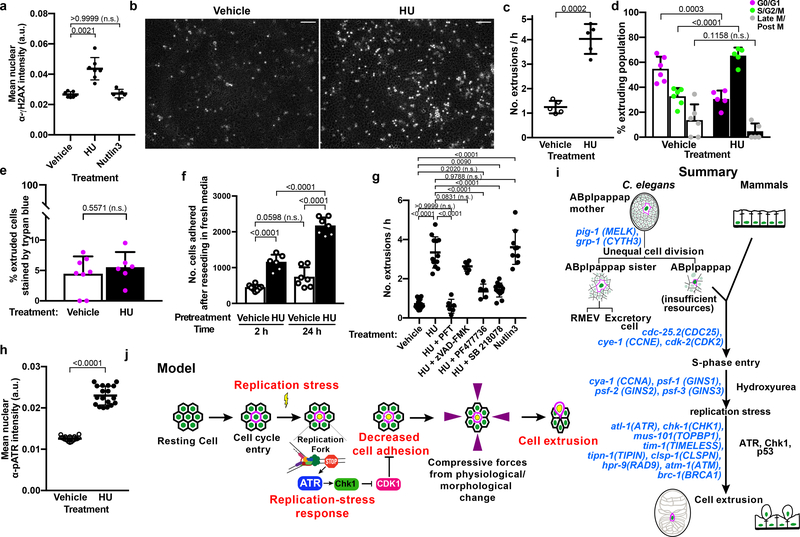
Figure 4. Replication stress promotes cell extrusion from a simple mammalian epithelial layer.
a, Quantification of α-γH2AX immunofluorescence signal in vehicle-control-, HU-, or Nutlin-3-treated MDCK-II cells. b, Representative micrographs of cells extruded (white rounded spots) from an MDCK-II monolayer after ~21 h of treatment with vehicle control (left) or HU (right). Scale bars, 100 μm. c, g, Quantification of extrusions per h after the indicated treatment. d, The cell-cycle phase of cells extruded after treatment with vehicle control or HU. e, Percentage of extruded cells staining with trypan blue after indicated treatment. f, Quantification of the number of HU-treated or vehicle-treated extruded cells that adhered at 2 h and 24 h after reseeding in fresh media. h, Quantification of α-pATR immunofluorescence signal in vehicle- or HU-treated MDCK-II cells, respectively. i, j, Summary and model of replication-stress induced cell extrusion. Each data point, separate experiment in (c-g); mean fluorescence intensity signal from one image of 100s of cells in (a, h). n = 7, 7 and 5 for vehicle, HU and Nutlin3, respectively, in (a); 5 each in (c); 6 for vehicle, 5 for HU in (d); 8 for vehicle, 6 for HU in (e); 8 for vehicle, 6 for HU at 2 h and 7 each at 24 h in (f); 13, 12, 6, 6, 5, 12 and 9 for vehicle, HU, HU+PFT, HU+zVAD-FMK, HU+PF477736, HU+SB218078, Nutlin-3, respectively in (g); 18 each in (h), all biologically independent. All data in (a, c-h) are represented as Mean ± S.D. Statistical analysis: Kruskal-Wallis one-way ANOVA followed by Dunn’s correction (a); two-tailed Welch’s t-test (c); Ordinary one-way ANOVA with Sidak’s correction (d, f, g); Mann-Whitney two-tailed test (e, h). P values are indicated; n.s., not significant.
Several observations indicate that cell death, e.g., apoptosis, did not cause the HU-mediated cell extrusions: (i) similar percentages of HU-induced and stochastically extruded cells showed trypan blue staining (Fig. 4e), (ii) cells extruded following HU treatment were viable and proliferated within 24 h of culturing in fresh media (Fig. 4f), and (iii) the caspase inhibitor zVAD-FMK did not prevent HU-induced cell extrusion (Fig. 4g, Extended Data Fig. 6b). These findings demonstrate that HU induces the extrusion of living cells.
Last, we tested the requirement for ATR and Chk1 in replication stress-mediated extrusions from MDCK-II cell layers. HU treatment significantly increased the level of phosphorylated ATR (pATR) in MDCK-II cells (Fig. 4h, Extended Data Fig. 6c). Also, the small molecule Chk1 inhibitors SB 218078 and PF477736 suppressed HU-induced cell extrusion (Fig. 4g, Extended Data Fig. 6b), indicating that HU-induced mammalian cell extrusion requires the ATR/Chk1 pathway. In mammals, the ATR/Chk1-mediated replication-stress response activates p5323. HU treatment increased p53 levels, as did treatment with Nutlin-3, a chemical activator of p53 (Extended Data Figs. 6d, e). Furthermore, the p53 inhibitor pifithrin-α (PFT) completely suppressed HU-induced cell extrusion (Fig. 4g, Extended Data Fig. 6b). Although Nutlin-3 increased extrusion rates of MDCK-II cells (Fig. 4g), it did not result in replication stress (Fig. 4a, Extended Data Fig. 6a), indicating that p53 activation is sufficient to induce cell extrusion in the absence of replication stress.
Discussion
Our findings are summarized in Fig. 4i and suggest the following four-step mechanistic model for replication-stress induced cell extrusions (Fig. 4j): (i) a cell in S phase experiences replication stress following a genotoxic insult (e.g., HU) or the inheritance of insufficient replicative resources from an unequal cell division; (ii) ATR and Chk1 (and p53 in mammals) mediate a replication-stress response; (iii) cell adhesion molecules are downregulated3, possibly through inhibition of CDK1, a regulator of cell adhesion during S phase24; and (iv) the cell is extruded as a consequence of this reduced adhesion25 and mechanical forces generated by neighboring cells (e.g., constrictive forces preceding ventral enclosure in developing C. elegans embryos26) or physiological crowding27.
Our finding that replication stress drives cell elimination by extrusion for both C. elegans and mammalian cells demonstrates that cell extrusion in ced-3(lf) embryos is not simply a result of the genetic perturbation of the caspase-mediated apoptosis pathway and indicates that this mechanism is evolutionarily conserved. The cell cycle is similarly conserved and is ancient. Since cell elimination caused by an inducible cell-cycle state does not require specialized molecular or cellular machinery (unlike caspase-mediated apoptosis and phagocytosis), we propose that replication-stress-driven cell extrusion is a primordial process of cell elimination. That cells targeted for caspase-dependent cell death extrude upon inactivating caspases suggests that extrusion as a primitive form of cell elimination has in some instances been supplanted by caspase-mediated apoptosis. We further suggest that a developmentally controlled replication-stress response might be utilized by metazoans for cell elimination via extrusion in a variety of biological contexts.
Replication stress and cell extrusion are key features of cancer biology1,28, but their relationship has not been explored. We propose that the extrusion of cells undergoing replication stress can be tumor-suppressive. Non-cancerous cells that fail to respond to replication stressors can accrue DNA damage, genomic rearrangements and ploidy defects associated with oncogenesis28. Pre-cancerous and cancerous cells experience persistent replication stress28 resulting from the over-expression, amplification or mutational activation of genes that drive uncontrolled proliferation. In such contexts, cell extrusion activated by replication stress could function as an early checkpoint to eliminate pre-cancerous and malignant cells.
In other contexts, cell extrusion induced by replication stress might be subverted to promote metastatic tumorigenesis. Indeed, the mouse p53 R172H mutation (equivalent to the R175H mutation common to human tumors) causes a high frequency of metastatic tumors29,30. Murine p53 R172H induces a replication-stress response by increasing both Chk1 expression and basal Chk1 phosphorylation31,32. We suggest that this ectopic replication-stress response triggers extrusion of cells harboring this mutation and that an oncogenic mutation in APC or KRas5, or perhaps p53 R172H itself reverses the direction of extrusion from apical (causing elimination) to basal (causing dissemination) to promote metastasis. Additionally, reports of chemotherapeutic inhibitors of DNA replication, such as doxorubicin and cyclophosphamide, promoting metastatic spread of tumor cells33 also support this notion.
In short, we identified a conserved mechanism that links replication stress to the process of cell extrusion. We suggest that cell extrusion driven by replication stress is a primordial mechanism of cell elimination common to all metazoa and that cell extrusion arising from replication stress regulates the survival and spread of tumor cells.
Methods
Strains
C. elegans hermaphrodite strains were maintained on Nematode Growth Medium (NGM) plates containing 3 g/L NaCl, 2.5 g/L peptone and 17 g/L agar supplemented with 1 mM CaCl2, 1 mM MgSO4, 1 mM KPO4 and 5 mg/L Cholesterol with E. coli OP50 as a source of food34. All strains were derived from Bristol N2. ced-3(lf) refers to the n3692 deletion allele of ced-33. cye-1(lf) refers to the eh10 allele of cye-135. ced-5(lf) refers to the n1812 allele of ced-536. C. elegans strains carrying the transgenes nIs861 and isIs17 were maintained at 25°C. All other strains were maintained at 22°C. The transgenes and mutations used are listed below:
LGI:nIs433[Ppgp-12::4xNLS::GFP::unc-54 3’UTR; p76–16B(unc-76(+))], cye-1(eh10)
LGII: heSi192[P eft-3 ::tDHB::eGFP::tbb-2 3’UTR + Cbr-unc119(+)]
LGIII: unc-119(ed3)
LGIV: ced-3(n3692), ced-5(n1812)
LGV: ltIs44[P pie-1 ::mCherry::PH(PLC1delta1) + unc-119(+)]
LGX: nIs434[P pgp-12 ::4xNLS::GFP::unc-54 3’UTR; p76–16B(unc-76(+))]
Unknown linkage:stIs10026[Phis-72::HIS-72::GFP], isIs17[pGZ295(Ppie-1::GFP::pcn-1(W03D2.4)), pDP#MM051 (unc-119(+))], nIs861[pDD111(Pegl-1::mCherry::PH::unc-54 3’UTR)], nIs632[pDD111(Pegl-1::mCherry::PH::unc-54 3’UTR), pML902 (dlg-1::GFP),p76–16B(unc-76(+))], opIs263[Prpa-1::rpa-1::YFP + unc-119(+)]
Extrachromosomal array: nEx3043[cye-1(+); Psur-5::RFP]
nIs632 and nIs861 express membrane-localized mCherry from the egl-1 promoter, which facilitated the identification of ABplpappap (an egl-1 expressing cell). nIs632 does not express dlg-1::GFP, presumably as a result of partial transgene silencing37–39. stIs1002640 ubiquitously expresses a GFP-tagged histone HIS-72 from its endogenous promoter, which produces fluorescence in the nuclei of all cells and facilitates in providing the context in which extrusion events are observed.
Plasmids and Fosmids
pDD111 - Pegl-1::mCherry::PH::unc-54 3’UTR was generated with the following steps: i) 6.8 Kb of the egl-1 promoter was amplified from genomic DNA with Phusion DNA polymerase using the primers 5’-CGCCTGCAGTTGAAATTTGGGGATATTTTGG-3’ and 5’- CGCGAGCTCCTGGAAATTAGTAAGGTTTTGAAGGGGG-3’; ii) the amplicon was digested with PstI and SacI (New England Biolabs) and ligated into pPD122.56, which encodes 4xNLS::GFP to generate Pegl-1::4xNLS::GFP::unc-54 3’UTR; iii) mCherry-PH (Pleckstrin Homology) sequence was amplified from pAA173 using primers 5’-CGCACCGGTCCAGATGGCTCAAACAAAGC-3’ and 5’- CGCGAATTCGGCACAAGTTCATTCACAGG-3’ and digested with EcoRI and AgeI (New England Biolabs) and ligated into the pDD122.56 - Pegl-1::4xNLS::GFP::unc-54 3’UTR, which generated the plasmid pDD122.56 - Pegl-1::4xNLS::mCherry::PH::unc-54 3’UTR; iv) the 4xNLS sequence was removed with the primers 5’- ggagctcAGAAAAAATGGTCTCAAAGGGTG-3’ and 5’- CACCCTTTGAGACCATTTTTTCTGAGCTCC-3’ using QuikChange Site-Directed Mutagenesis (Agilent) to generate pDD111 - Pegl-1::mCherry::PH::unc-54 3’UTR.
Sequences of all RNAi constructs that affected cell extrusion are provided in Supplementary Information. RNAi clones were constructed for atl-1, mat-2 and lin-15B. Genomic regions of about 1 kb length were amplified from wild-type genomic lysates using Q5 Hot Start high-fidelity polymerase (New England Biolabs) with the following primers:
atl-1
5’-TCGAATTCCTGCAGCTCCTCGAACCCATCATCCCT-3’
5’-TGACGCGTGGATCCCATGAAGCTGCGTGGTTGTTG-3’
mat-2
5’-TCGAATTCCTGCAGCCTGGAACTCATCCCATACGC-3’
5’-TGACGCGTGGATCCCCATTGGAACCTCCAGATGCT-3’
lin-15B
5’-TCGAATTCCTGCAGCGCTGACACAATTGCGAACAT-3’
5’-TGACGCGTGGATCCCCGTGTGCATAAAGACCAAGG-3’
atl-1+chk-1
atl-1 fragment 5’-TCGAATTCCTGCAGCTCCTCGAACCCATCATCCCT-3’
5’- ACACGACAGCGTCCGCAGAAATGAAGCTGCGTGGTTGTTG-3’
chk-1 fragment 5’-TTCTGCGGACGCTGTCGTGTCAAGCGGATCCGTGGTATCA-3’
5’- TGACGCGTGGATCCCCCGAGTGCTCCACATTGACT-3’
These inserts were cloned into the pL4440 vector linearized with XmaI (New England Biolabs) using the In-Fusion HD cloning kit (TaKaRa) according to manufacturer’s instructions. The cloned vector was then transformed into competent HT115 bacterial cells. Correct RNAi clones were identified by Sanger sequencing. Geneious 10.2.6 (Biomatters, Inc.) was used to guide all plasmid design and construction.
The fosmid WRM0637bF05 (Source Bioscience), which contains the genomic cye-1 sequence, was used for genomic rescue of the Tex phenotype in cye-1(eh10); ced-3(n3692); nIs434 animals. A plasmid containing the sequence for Psur-5::RFP, which expresses RFP cell-autonomously, was used as a fluorescent marker of cells carrying a mitotically-unstable extrachromosomal array for genetic mosaic analysis.
Germline transformation
Germline transformation experiments were performed as described41. To generate the cye-1-rescuing transgene nEx3043, the Fosmid WRM0637bF05, a plasmid containing the sequence for Psur-5::RFP and 1 kb plus DNA ladder (Invitrogen) were injected into hT2[qIs48]/cye-1(eh10); ced-3(n3692); nIs434 animals at 3 ng/μl, 20 ng/μl and 80 ng/ul, respectively.
RNAi treatments
Previously described feeding RNAi constructs and reagents were used to perform RNAi feeding experiments42,43. Briefly, HT115 Escherichia coli bacteria carrying RNAi clones in the pL4440 vector were grown for at least 12 h in Luria broth (LB) liquid media with 75 mg/L ampicillin at 37°C. These cultures were seeded onto 6 cm Petri plates with Nematode Growth Medium (NGM) containing 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Amresco) and 75 mg/L ampicillin and incubated for 24 h at 22°C. For imaging experiments using confocal microscopy, 10 L4 animals were added to each RNAi plate and imaging of progeny embryos was performed on the next day as described in Microscopy below. For excretory cell counts, five L4 animals were added to each RNAi plate and L3-L4 progeny were scored for number of excretory cells, as described in Excretory cell count below. In case a bacterial clone targeting a certain gene was not available in previously constructed libraries42,44, we generated our own RNAi clone as described in Molecular biology above. Each RNAi experiment included an empty pL4440 vector negative control.
Genome-wide RNAi screen
The ORFeome RNAi library was used to conduct a genome-wide RNAi screen42. For each day of the RNAi screen, all bacterial colonies from two 96-well plates were cultured for at least 12 h at 37°C in LB with 75 mg/L ampicillin. These cultures were then pre-incubated with 1 mM IPTG (Amresco) for 1 h to maximize induction of dsRNA production. 24-well plates with each well containing 2 mL NGM medium with 1 mM IPTG (Amresco) and 75 mg/L ampicillin were prepared in advance and stored at 4°C until needed; they were brought to room temperature a few hours before seeding. Each bacterial colony culture was then seeded onto an individual well of a 24-well plate and incubated for 24 h at 20°C. Three L4 animals were picked into a 10 μl drop of M9 medium, which facilitated their transfer into a well using a pipette. The progeny of these 3 animals were screened 3 days later. Each set of RNAi clones screened also included a pig-1 RNAi positive control and an empty pL4440 vector negative control. The scorer was blinded to the identity of the RNAi clones. Excretory cell counts were performed as described in Excretory cell counts below. Sanger sequencing was used to confirm the identity of RNAi clones that reproducibly generated a Tex phenotype for more than 10% of the animals scored.
Microscopy
All RNAi screens scoring excretory cells were performed using a Nikon SMZ18 fluorescent dissecting microscope.
DIC and epifluorescence images of L3-L4 larval stage animals carrying the transgene nIs433[Ppgp-12::4xNLS-GFP] or nIs434[Ppgp-12::4xNLS-GFP], which mark the excretory cell and ectopic excretory-like cells, were obtained using a 63x objective lens (Zeiss) on an AxioImager Z2 (Zeiss) compound microscope and Zen Blue software (Zeiss).
For confocal microscopy, embryos staged at the 200–300-cell stage were picked and mounted onto a glass slide (Corning) with a freshly prepared 2% agarose pad. Embryos with ventral surfaces facing the objective were selected for imaging. Confocal images of embryos were obtained using a 63x objective lens (Zeiss) on a Zeiss LSM800 confocal microscope.
For observing extrusion (or absence of extrusion), we focused particularly on the cell ABplpappap, the identification of which is facilitated by its central position on the ventral surface45. The fluorescent transgene nIs861[Pegl-1::mCherry::PH] or nIs632[Pegl-1::mCherry::PH; dlg-1::GFP], which express the Pleckstrin homology domain of PLC-δ fused to mCherry from the promoter of egl-1, was used to label the membrane of the ABplpappap cell, an egl-1 expressing cell3, to further facilitate cell identification. Another fluorescent transgene stIs10026[his-72::GFP], which expresses GFP-tagged HIS-72 histone protein, was used to label the nuclei of all cells to help define ABplpappap’s location within the embryo. Time-lapse confocal microscopy was used to monitor the location of ABplpappap in embryos, keeping the cell in view by refocusing on it every 30 sec. Confocal imaging during a period of about 50 min ending in ventral enclosure (the point at which hypodermal cells meet on the ventral surface of the embryo) occurs was sufficient to determine whether ABplpappap did or did not undergo extrusion.
For determining whether ABplpappap and other cells that are extruded entered the cell cycle, embryos carrying the transgene heSi192[Peft-3::tDHB::eGFP::tbb-2 3’UTR] expressing a codon-optimized (for C. elegans) C-terminal fragment of Human DNA Helicase B, which translocates from the nucleus to the cytoplasm in response to the activity of the cell cycle CDKs 1 and 210, were examined using confocal microscopy after various RNAi treatments. nIs861 labeled the membrane of ABplpappap with mCherry and facilitated cell identification in these embryos.
For determining the cell cycle phase of ABplpappap and other cells that are extruded, embryos carrying the transgene isIs17[Ppie-1::GFP::PCN-1] expressing GFP-tagged PCN-1 protein, which produces a phase-specific fluorescence intensity and localization pattern11, were examined using confocal microscopy after various RNAi treatments. nIs861 labeled the membrane of ABplpappap with mCherry and facilitated cell identification in these embryos.
For examining ABplpappap and other extruded cells for replication stress, embryos carrying the transgene opIs263[Prpa-1::rpa-1::YFP] expressing YFP-tagged RPA-1, which localizes to foci in conditions of replication stress14, were examined using confocal microscopy after various RNAi treatments. ltIs44[Ppie-1::mCherry::PH] labeled the membrane of all cells with mCherry and facilitated the identification of cell boundaries in these embryos.
Microscopy for the genetic mosaic analysis experiments are described in the ‘Mosaic analysis’ section below.
Images were processed with ImageJ software (NIH), Photoshop CC 2019 (Adobe) and Illustrator CC 2019 (Adobe) software. The Time Stamper function in the Stowers ImageJ plugin was used to mark elapsed time on time-lapse videos.
Excretory cell counts
Excretory cell counts were performed using a dissecting microscope equipped with fluorescence at a total magnification of 270x. For the genome-scale RNAi screen, roughly 50 animals were examined in each well of a 24-well plate and any well with more than 5 animals with two excretory cells was marked for confirmatory testing. Excretory cell counts in confirmatory RNAi experiments, candidate RNAi experiments and experiments with genetic mutants were conducted using 6 cm Petri plates with appropriate media. Animals were first immobilized by keeping the Petri plates on ice for 30 min. At least 100 animals at the L3-L4 larval stage were scored for each genotype or RNAi experiment unless there was extensive lethality or a growth defect, in which case a lower number or earlier-stage animals, respectively, were scored. A cell was scored as an excretory cell if it was located in the anterior half of the animal and its nucleus had strong GFP expression.
Mosaic analysis
To perform mosaic analysis, transgenic animals of the genotype hT2[qIs48]/cye-1(eh10); ced-3(n3692); nIs434; nEx3043[cye-1(+); Psur-5::RFP] were generated. Progeny of animals that lost the hT2 balancer and were of the genotype cye-1(eh10); ced-3(n3692); nIs434; nEx3043 were examined for the Tex phenotype. Ten such animals that displayed the Tex phenotype despite carrying the cye-1-rescuing array were examined using confocal microscopy with 63x (Zeiss) or 100x (Zeiss) objective lenses for the presence of the RFP-expressing extrachromosomal array in multiple cells, including the excretory cell (identified by the presence of distinctive canals extending from the cell body) and ABplpappap. Because a majority of these animals showed the presence of the rescuing array in the excretory cell, but not in ABplpappap, only the sublineage containing these cells was further analyzed.
Calculation of cell size
Confocal micrographs were obtained for multiple focal planes starting at the ventral surface and ending at the dorsal surface of the embryo, with each plane separated by a distance of 0.37 μm. The greatest area occupied by a cell in any plane was designated the “maximum area” of a cell.
To determine the ratio of diameters of ABplpappaa and ABplpappap, the square root of the mean of the area ratios was calculated and the two cells were assumed to be perfect spheres.
Generation of virtual lateral sections
Confocal micrographs were obtained for multiple focal planes starting at the ventral surface and ending at the dorsal surface of embryos that had completed epidermal ventral enclosure, with each plane separated by a distance of 0.37 μm.
Virtual lateral section for each Z-stack was generated at the plane bisecting ABplpappap along the anterior-posterior axis using the Orthogonal Views function of Fiji46.
Fluorescence signal quantifications
tDHB-GFP – The ABplpappap nuclear boundary, cell membrane boundary and the tDHB-GFP fluorescence signal were determined from DIC, mCherry and GFP channels, respectively, of confocal images of RNAi-treated ced-3(lf) embryos expressing the transgenes heSi192 and nIs861. Mean tDHB-GFP fluorescence intensities inside the nuclear region, entire cell and background were quantified using Fiji software46. Mean cytoplasmic tDHB-GFP fluorescence intensity was calculated by the formula Icytoplasm = ((Icell * cell area) – (Inucleus * nucleus area)) / (cell area – nucleus area), where Icytoplasm, Icell and Inucleus denote the mean tDHB fluorescence intensity in the cytoplasm, cell and nucleus, respectively. The ratio of nuclear-to-cytoplasmic tDHB fluorescence intensity in Figure 2c was adjusted for background fluorescence (measured from a random area outside the embryo boundaries), i.e., the background fluorescence intensity was subtracted from both nuclear and cytoplasmic fluorescence intensity values before calculating the ratios.
GFP::PCN-1 and RPA-1::YFP – The ABplpappap nuclear was determined from DIC channel of confocal images of RNAi-treated ced-3(lf) embryos expressing corresponding transgenes. Coefficient of variation (ratio of standard deviation to mean) of fluorescence intensity inside the nuclear region was used as a measure to differentiate between diffuse and punctate fluorescence signals and was quantified using Fiji software46. Briefly, highly localized fluorescence signals were expected to have pixel fluorescence intensity values that were either much higher or much lower than the mean fluorescence intensity of an area of interest, whereas a diffuse fluorescence signal were expected to have pixel intensity values that were closer to the mean fluorescence intensity of an area of interest. A high variation from the mean would produce a high variance and hence a higher standard deviation and vice versa. Since standard deviation is also influenced by fluorescence intensity, we used coefficient of variation as a measure of signal localization to remove any artifactual differences in fluorescence intensity among embryos.
TUNEL Staining
TUNEL staining was performed as described16 with the following modifications – after freezing embryos with liquid nitrogen, the embryos were fixed with 4% PFA/PBS, pH 7.4 for 15 min at room temperature and then permeabilized by incubation with 0.1% Triton X-100 in PBS for 10 min at room temperature. After washing, TUNEL staining was performed according to the manufacturer’s (Roche, Switzerland) instructions. Stained embryos were mounted using Fluoroshield with DAPI mounting solution (Sigma-Aldrich) to visualize the nuclei.
Cell culture
MDCK-II47 and MDCK-Fucci48 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a humidified incubator at 37ºC with 5% CO2.
Chemicals
2 mM HU (Millipore Sigma, Cat#H8627) was prepared in culture medium prior to each experiment. Nutlin-3 (Tocris, Cat#3984), Pifithrin-α (Focus Biomolecules, Cat#10–2480), SB 218078 (Tocris, Cat#2560), PF477736 (Tocris, Cat#4277) and zVAD-FMK (Promega, Cat# G7231) stocks were prepared in DMSO and kept at −20ºC until needed, when dilutions were prepared in culture medium for cell treatment for final concentrations of 10 μM, 10 μM, 30 nM, 5nM and 50 μM, respectively.
Mammalian cell imaging
These assays were performed using 6-well plastic plates. 20,000 MDCK-II cells were seeded in each well and grown to confluence for 72 h. The day of the experiment, cells were washed twice with PBS and treated with fresh medium or various chemicals in medium. After equilibration, plates were imaged at 15-min intervals for up to 24 h, using an Evos M7000 imaging system equipped with a humidified onstage incubator (37ºC, 5% CO2). Several positions per well were imaged in the phase contrast and green and red fluorescence channels (when applicable) available in this system.
Mammalian cell extrusion quantification
In time-lapse phase contrast images, extruding cells are easily identifiable as bright, white, rounded spots emerging from the epithelial plane. We counted the number of cells with these features for each condition using the Cell Counter plugin of Fiji46. Extrusions are reported as number of extruding cells/h for comparison between experiments of different duration.
Mammalian cell cycle phase determination
The Fucci system differentially labels the nuclei of cells in G1 (red) and S/G2/M (green)49. Images of MDCK-Fucci cells with HU or control treatment were obtained in the phase contrast, red and green fluorescence channels as per Mammalian cell imaging above. For each position, a multi-channel stack was built using Fiji46. After identifying an extruded cell in the phase contrast channel, the cell cycle phase was determined using the fluorescence channels.
Mammalian re-seeding experiments
At the end of an imaging experiment, supernatants were collected and centrifuged (1200 rpm, 5 min, room temperature). Pellets were re-suspended in 50 μL of PBS, and 10 μL of the suspension was used for cell counting with Trypan blue in a Neubauer chamber, allowing us to simultaneously calculate the number of cells being re-seeded and the fraction of cells that was dead. The remaining cells were seeded with 1 mL of fresh medium in a 24-well plate and grown in the cell culture incubator. Pictures were taken at 2 h and 24 h for cell counting.
Mammalian cell immunostaining
Cells were fixed in 4% PFA-PBS for 20 min at 37ºC, permeabilized with 0.5% Triton X-100-PBS for 5 min at room temperature and stained with primary antibodies (all 1:250 in 1%BSA-PBS: p53, abcam cat#ab26; phospho-ATR (Ser248), ThermoFisher cat# 720107; phospho-histone H2A.X(Ser139), CellSignaling cat# 9718) overnight at 4ºC, and secondary antibodies (with 1:500 AF647 phalloidin and 30 μg/mL Hoechst) for 1h at room temperature. 1x PBS was used for washing between steps and preparation of all solutions. Samples were mounted with Fluoromount-G Mounting Medium (Invitrogen cat# 00–4958-02).
Fixed mammalian cell imaging
Samples were imaged using 20x air and 60x oil objectives of a Nikon Eclipse Ti2-E microscope equipped with a Yokogawa CSU-W1 spinning disk system, an Andor DU-888 camera, and a Toptica multi-laser bed. All settings were kept constant between conditions.
Quantification of mammalian cell staining
All images were quantified with ad hoc Cell Profiler50 pipelines. Briefly, the IdentifyPrimaryObjects, MeasureObjectIntensity, and ExportToSpreadsheet modules were sequentially used for nuclear segmentation (Hoechst channel), nuclear intensity measurements (p53, pATR, or γH2AX channels) and data export.
Data Availability
Data supporting all figures are available within the paper and in the associated source data files. Raw microscopy data are available upon request to the corresponding author.
Statistics and Reproducibility
All experiments were repeated independently at least three times to ensure reproducibility. All representative micrographs are one example of five to ten biologically independent replicates of the same experiment, of which the remaining micrographs or quantification as a graph are provided in an associated figure panel, or have been described in the main text.
For calculation of statistical significance for fluorescence intensity and area ratios, the ratios were first transformed to logarithm values, with the assumption that logarithm of ratios produced a normal distribution of values. Normal distributions with unequal variances were assumed for coefficients of variance of GFP::PCN-1 and RPA-1::YFP signal fluorescence intensity. Normal distributions with unequal variances were assumed for rates of extrusion after different chemical treatments. No assumptions were made about the distributions of the rates of cell death under HU and vehicle treatments. Normal distribution with unequal variances were assumed for fraction of extruded cells in different phases of the cell cycle after HU and vehicle treatments. Normal distribution was assumed for numbers of cells reseeded in fresh media after pre-treatment in different conditions. Since each mammalian-cell staining image contained from 100s to 1000s of cells, per-cell statistical analysis would result in extremely low p-values. To avoid this sub-estimation, we first calculated descriptive statistics (median, average, SD, N) for each image and made no assumptions about the distributions of these values for subsequent statistical analysis. All statistical analysis was performed using Prism (GraphPad Software).
Extended Data
Extended Data Figure 1. A genome-wide RNAi screen for the Tex phenotype revealed control of cell extrusion by cye-1 and cdk-2.
a, Schematic representation of the genome-wide RNAi screen for the Tex phenotype. RNAi using pL4440 empty vector was used as negative control and pig-1(RNAi) was used as positive control3 b, Time-lapse confocal fluorescence micrographs of ced-3(lf); stIs10026[his-72::GFP]; nIs632[Pegl-1::mCherry::PH] embryos after indicated RNAi treatment at the indicated times. tve – time point of ventral enclosure. Arrowheads, ABplpappap. A, anterior; R, right. Scale bars, 10 μm.
Extended Data Figure 2. Genetically mosaic cye-1(lf); ced-3(lf) animals with the Tex phenotype lack a cye-1-rescuing transgene in ABplpappap.
a-i, Confocal micrographs showing the presence of the cye-1(+)-rescuing transgene in (a-h) the excretory cell but not in ABplpappap or in (i) neither the excretory cell nor ABplpappap of cye-1(eh10); ced-3(n3692); nIs434[Ppgp-12::4xNLS-GFP]; nEx3043[cye-1(+); Psur-5::RFP] animals with the Tex phenotype. A, anterior; D, dorsal. Scale bars, 10 μm.
Extended Data Figure 3. ABplpappap, which is generated by an unequal cell division, arrests in S phase and is extruded.
a, b, Confocal fluorescence micrographs of tDHB-GFP fluorescence in ABplpappap (arrowhead) (a) before and (b) after ventral enclosure in heSi192[Peft-3::tDHB-GFP]; ced-3(lf); nIs861[Pegl-1::mCherry::PH] embryos after the indicated RNAi treatment. Dotted line, ABplpappap nucleus, as identified by Nomarski optics. c, d, Confocal fluorescence micrographs of GFP::PCN-1 fluorescence in ABplpappap (arrowhead) (c) before and (d) after ventral enclosure in ced-3(lf); isIs17[Ppie-1::GFP::pcn-1]; nIs861 embryos after the indicated RNAi treatment. e, Time-lapse confocal fluorescence micrographs of GFP::PCN-1 fluorescence in ABplpappap (arrowhead) in a ced-3(lf); isIs17; nIs861; pig-1(RNAi) embryo at the indicated times. tve- time point of ventral enclosure. f-h, Micrographs of virtual lateral section of ced-3(lf); nIs861; stIs10026 embryos showing either ABplpappap (arrowhead) or its daughters (arrowheads) after indicated RNAi treatment. i-m, Confocal fluorescence micrographs of ced-3(lf); ltIs44[Ppie-1::mCherry::PH]; stIs10026 embryos showing the relative sizes of ABplpappap and its sister, ABplpappaa, in embryos after the indicated RNAi treatment. Insets, ABplpappap (a-d); ABplpappap or its daughters (e); magnified view of the region indicated, which includes ABplpappap (†) and ABplpappaa (∗) (i-m). A, anterior; R, right; V, ventral. Scale bars, 10 μm.
Extended Data Figure 4. All extruded cells display features of cell cycle entry, S-phase arrest, and replication stress.
a-d, tDHB-GFP fluorescence in unidentified extruded cells from (b) the anterior sensory depression, (c) the ventral pocket, and (d) the posterior tip of a comma stage embryo of the genotype heSi192[Peft-3::tDHB-GFP]; ced-3(lf); nIs861[Pegl-1::mCherry::PH] after RNAi against empty vector control. Nuclei of extruded cells, as identified by Nomarski optics, are marked by dotted lines. e, f, micrographs of GFP::PCN-1 fluorescence in unidentified extruded cells (arrowhead) at (e) the ventral pocket or (f) the anterior sensory depression from ced-3(lf); isIs17[Ppie-1::GFP::pcn-1]; nIs861 embryos after (e) RNAi against empty vector control or (f) no RNAi. Insets, extruded cells marked by arrowhead in micrograph. g-j, RPA-1::YFP fluorescence in unidentified extruded cells from (h, i) the anterior sensory depression and (j) ventral pocket from a in a ced-3(lf); ltIs44[Ppie-1::mCherry::PH]; opIs263[Prpa-1::rpa-1::YFP] embryos after RNAi against empty vector control. A, anterior; R, right; V, ventral. Scale bars, 10 μm.
Extended Data Figure 5. The replication-stress response, likely caused by lrr-1 and nucleotide insufficiency, promotes cell extrusion.
a-d, Confocal fluorescence micrographs showing the localization of RPA-1::YFP in ABplpappap (arrowhead) in ced-3(lf); ltIs44[Ppie-1::mCherry::PH]; opIs263[Prpa-1::rpa-1::YFP] embryos after the indicated RNAi treatment. Inset, magnified view of ABplpappap. A, anterior; R, right. e, Genes identified as suppressors of the sterility of lrr-1(lf) mutants (ref. 19) were tested for suppression of cell extrusion. f, Nomarski micrograph showing cells extruded (arrow) from a wild-type embryo after gmpr-1(RNAi) treatment. Scale bars, 10 μm.
Extended Data Figure 6. Inhibitors of HU-induced replication-stress response and pan-caspase inhibitors do not alter stochastic cell extrusion.
a, c, d, Representative micrographs of (a) α-γH2AX, (c) α-pATR and (d) α-p53 immunofluorescence signal in (c) vehicle- or HU-treated MDCK-II cells, and (a, d) vehicle-, HU- or Nutlin3-treated MDCK-II cells. DNA is stained with Hoechst. Scale bars, 20 μm. b, Quantification of extrusions per h after the indicated treatment. n = 10, 6, 5, 5 and 5 (biological replicates) each for control, PFT, zVAD-FMK, SB 218078 and PF477736 treatments, respectively. Each data point represents a separate experiment. These data were collected and analyzed for statistical significance with the data in Fig. 4g. P values are indicated; n.s., not significant. e, Quantification of α-p53 immunofluorescence signal in vehicle-control-, HU-, or Nutlin-3-treated MDCK-II cells. n = 9, 7 and 5 (biological replicates) for vehicle, HU and Nutlin3, respectively. Each data point represents mean fluorescence intensity signal from one image of 100s of cells. Kruskal-Wallis one-way ANOVA followed by Dunn’s correction was performed. P values are indicated. Data in (b, e) are represented as mean ± S.D.
Extended Data Table 1. Penetrances of the Tex phenotype produced by RNAi against cell cycle genes (and non-cell-cycle cyclins and CDKs) in ced-3(lf) animals.
Tex penetrance produced by indicated RNAi treatment, mammalian homolog of the RNAi target and whether or not the RNAi clone produced extensive lethality are shown. Genes corresponding to RNAi clones that produced more than 9% penetrance of the Tex phenotype are in bold. cyb-3 did not fit this criterion, as extensive lethality prevented the counting of sufficient number of animals to assign significance. Some cyclins and CDKs that function outside the cell cycle were included and served as negative controls.
| RNAi target | Mammalian Homolog | %Tex | n | extensive lethality? |
|---|---|---|---|---|
| empty vector | - | 1 | 159 | N |
| atl-1 | ATR | 10 | 509 | N |
| cdc-14 | CDC14 | 1 | 168 | N |
| cdc-25.1 | CDC25 | 0 | 111 | Y |
| cdc-25.2 | CDC25 | 12 | 159 | N |
| cdc-25.3 | CDC25 | 2 | 168 | N |
| cdc-25.4 | CDC25 | 0 | 183 | N |
| cdk-1 | CDK1 | 15 | 61 | Y |
| cdk-11.1 | CDK11 | 0 | 175 | N |
| cdk-11.2 | CDK11 | 1 | 132 | N |
| cdk-12 | CDK12 | 0 | 167 | N |
| cdk-2 | CDK2 | 65 | 181 | N |
| cdk-4 | CDK4 | 1 | 193 | N |
| cdk-5 | CDK5 | 0 | 155 | N |
| cdk-7 | CDK7 | 0 | 130 | Y |
| cdk-8 | CDK8 | 1 | 186 | N |
| cdk-9 | CDK9 | 1 | 155 | Y |
| cdt-1 | CDT1 | 1 | 147 | Y |
| chk-1 | CHEK1 | 10 | 164 | Y |
| cit-1.1 | CCNT1/2 | 0 | 105 | N |
| c it-1.2 | CCNT1/2 | 1 | 244 | N |
| cki-1 | CDKN1 | 0 | 125 | N |
| cki-2 | CDKN1 | 1 | 216 | N |
| clk-2 | TEL02 | 1 | 156 | N |
| cul-1 | CULI | 5 | 81 | Y |
| cul-2 | CUL2 | 0 | 21 | Y |
| cul-3 | CUL3 | 5 | 151 | Y |
| cul-4 | CUL4 | 0 | 141 | N |
| cya-1 | CCNA | 20 | 309 | N |
| cyb-1 | CCNB1 | 4 | 136 | N |
| cyb-2.1 | CCNB2 | 1 | 102 | N |
| cyb-2.2 | CCNB2 | 0 | 144 | N |
| cyb-3 | CCNB3 | 14 | 7 | Y |
| cic-1 | CCNC | 1 | 158 | N |
| cyd-1 | CCND | 0 | 111 | N |
| cye-1 | CCNE | 89 | 133 | Y |
| cyh-1 | CCNH | 0 | 119 | Y |
| cyl-1 | CCNL | 1 | 106 | Y |
| cyy-1 | CCNY | 1 | 108 | N |
| dpl-1 | TFDP1 | 0 | 167 | N |
| efl-1 | E2F | 0 | 141 | N |
| emb-27 | CDC16 | 0 | 107 | Y |
| emb-30 | ANAPC4 | 0 | 130 | Y |
| fzr-1 | FZR1 | 1 | 146 | N |
| fzy-1 | CDC20 | 0 | 130 | Y |
| hpr-17 | RAD 17 | 5 | 214 | N |
| hus-1 | HUS1 | 0 | 132 | N |
| lin-15 | - | 1 | 104 | N |
| lin-23 | βTrCP | 23 | 147 | N |
| lin-35 | Rb | 0 | 134 | N |
| lin-36 | - | 0 | 111 | N |
| lin-9 | LIN9 | 1 | 125 | N |
| mat-1 | CDC27 | 0 | 80 | Y |
| mat-2 | ANAPC1 | 1 | 163 | Y |
| mat-3 | CDC23 | 0 | 108 | Y |
| mdf-1 | MAD1L1 | 3 | 112 | N |
| mdf-2 | MAD2L1 | 1 | 111 | N |
| mrt-2 | RADI | 0 | 114 | N |
| rnr-1 | RRM1 | 5 | 103 | Y |
| san-1 | BUB1B | 1 | 145 | N |
| wee-1.1 | PKMYT1 | 1 | 159 | N |
| wee-1.3 | PKMYT1 | 0 | 60 | Y |
Extended Data Table 2. Penetrances of the Tex phenotype produced in wild-type animals by RNAi against cell-cycle genes with potential roles in cell extrusion.
The Tex penetrance produced in wild-type animals by RNAi clones against cell cycle genes that might be involved in cell extrusion (based on the corresponding Tex penetrance in ced-3(lf) animals) is provided. Bona fide candidates for cell extrusion regulation should not produce a Tex phenotype in wild-type animals, as cell extrusion does not occur in wild-type embryos. A Tex phenotype in wild-type animals could occur from other effects of RNAi against cell cycle genes, such as excessive proliferation leading to multiple excretory cells. Such proliferation is likely the case for lin-23, as RNAi against lin-23 has been previously described to cause excessive proliferation51. The 13 other genes are good candidates to be regulators of cell extrusion by the criterion of dependence of the Tex phenotype on the loss of function of ced-3.
| RNAi target | Mammalian Homolog | %Tex | n |
|---|---|---|---|
| empty vector | - | 0 | 127 |
| atl-1 | ATR | 0 | 198 |
| cdc-25.2 | CDC25 | 0 | 36 |
| cdk-1 | CDK1 | 0 | 51 |
| cdk-2 | CDK2 | 0 | 237 |
| chk-1 | CHK1 | 0 | 115 |
| csn-1 | GPS1 | 0 | 156 |
| csn-4 | COPS4 | 0 | 141 |
| csn-5 | COPS5 | 0 | 114 |
| cya-1 | CCNA | 0 | 167 |
| cye-1 | CCNE | 0 | 143 |
| lin-23 | βTrCP | 12 | 96 |
| psf-1 | GINS1 | 0 | 150 |
| psf-2 | GINS2 | 0 | 190 |
| psf-3 | GINS3 | 0 | 72 |
Supplementary Material
Acknowledgments
We thank S. van den Heuvel and the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing strains; L. Hufnagel and X. Trepat for providing MDCK-Fucci cells; G. van Meer and the ECACC for providing MDCK-II cells (ECACC cat # 62107); N. An for strain management; S. Luo, S.R. Sando, E.L.Q. Lee, A. Doi, and A. Corrionero and other members of the Horvitz laboratory for helpful discussions, and D. Ghosh, C.L. Pender, M.G. Vander Heiden, P.W. Reddien, and R.O. Hynes for suggestions regarding the manuscript. This work was supported by the Howard Hughes Medical Institute and by NIH grant R01GM024663. V.K.D. was a Howard Hughes Medical Institute International Student Research fellow. C.P.P. was the recipient of Human Frontiers Science Program postdoctoral fellowship LT000654/2019-L. J.N.K. was supported by NIH grant R01GM024663. N.T. was supported by NIH Pre-Doctoral Training Grant T32GM007287. D.P.D. was supported by postdoctoral fellowships from the Damon Runyon Cancer Research Foundation and from the Charles A. King Trust. J.R. and C.P.P. were funded by King’s College London startup funds. H.R.H. is the David H. Koch Professor of Biology at MIT and an Investigator at the Howard Hughes Medical Institute.
Footnotes
Competing Interests
The authors declare no competing interests.
Additional Information
Supplementary information is available for this paper.
Further information and resource sharing requests should be directed to and will be fulfilled by the lead contact, H. Robert Horvitz (horvitz@mit.edu).
The C. elegans gene F32D1.5 is referred to as gmpr-1 in this publication. A request to change the name of F32D1.5 to gmpr-1 has been approved by WormBase and this change will be reflected in a future WormBase release.
References:
- 1.Ohsawa S, Vaughen J & Igaki T. Cell Extrusion: A Stress-Responsive Force for Good or Evil in Epithelial Homeostasis. Dev. Cell 44, 284–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Goeij JM et al. Cell kinetics of the marine sponge Halisarca caerulea reveal rapid cell turnover and shedding. J. Exp. Biol 212, 3892–3900 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Denning DP, Hatch V & Horvitz HR. Programmed elimination of cells by caspase-independent cell extrusion in C. elegans. Nature 488, 226–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenblatt J, Raff MC & Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol 11, 1847–1857 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Gudipaty SA & Rosenblatt J. Epithelial cell extrusion: Pathways and pathologies. Semin. Cell Dev. Biol 67, 132–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timson J. Hydroxyurea. Mutat. Res 32, 115–132 (1975). [DOI] [PubMed] [Google Scholar]
- 7.Koç A, Wheeler LJ, Mathews CK & Merrill GF. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem 279, 223–230 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Takeda DY & Dutta A. DNA replication and progression through S phase. Oncogene 24, 2827–2843 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Fay DS & Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127, 4049–4060 (2000). [DOI] [PubMed] [Google Scholar]
- 10.van Rijnberk LM, van der Horst SEM, van den Heuvel S & Ruijtenberg S. A dual transcriptional reporter and CDK-activity sensor marks cell cycle entry and progression in C. elegans. PLOS ONE 12, e0171600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brauchle M, Baumer K & Gönczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol 13, 819–827 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Zerjatke T et al. Quantitative cell cycle analysis based on an endogenous all-in-one reporter for cell tracking and classification. Cell Rep. 19, 1953–1966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teuliere J & Garriga G. Size Matters: How C. elegans Asymmetric Divisions Regulate Apoptosis. Results Probl. Cell Differ. 61, 141–163 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Stergiou L, Eberhard R, Doukoumetzidis K & Hengartner MO. NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell Death Differ. 18, 897–906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinant C et al. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci 120, 2731–2740 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Wu YC, Stanfield GM & Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 14, 536–548 (2000). [PMC free article] [PubMed] [Google Scholar]
- 17.Toledo LI et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Stevens H, Williams AB & Michael WM. Cell-type specific responses to DNA replication stress in early C. elegans embryos. PloS One 11, e0164601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ossareh-Nazari B, Katsiarimpa A, Merlet J & Pintard L. RNAi-Based Suppressor Screens Reveal Genetic Interactions Between the CRL2LRR-1 E3-Ligase and the DNA Replication Machinery in Caenorhabditis elegans. G3 6, 3431–3442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonneville R et al. CUL-2LRR-1 and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis. Nat. Cell Biol 19, 468–479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dewar JM, Low E, Mann M, Räschle M & Walter JC. CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev. 31, 275–290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merlet J et al. The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137, 3857–3866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9, 714–723 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Jones MC, Askari JA, Humphries JD & Humphries MJ. Cell adhesion is regulated by CDK1 during the cell cycle. J. Cell Biol 217, 3203–3218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grieve AG & Rabouille C. Extracellular cleavage of E-cadherin promotes epithelial cell extrusion. J. Cell Sci 127, 3331–3346 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Wernike D, Chen Y, Mastronardi K, Makil N & Piekny A. Mechanical forces drive neuroblast morphogenesis and are required for epidermal closure. Dev. Biol 412, 261–277 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Eisenhoffer GT et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaillard H, García-Muse T & Aguilera A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Olive KP et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni Syndrome. Cell 119, 847–860 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Lang GA et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni Syndrome. Cell 119, 861–872 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Singh S et al. Mutant p53 establishes targetable tumor dependency by promoting unscheduled replication. J. Clin. Invest 127, 1839–1855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo CQX et al. p53 Maintains Genomic Stability by Preventing Interference between Transcription and Replication. Cell Rep. 15, 132–146 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Karagiannis GS et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci. Transl. Med 9, eaan0026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References:
- 34.Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodigan TM, Liu J. i, Park M, Kipreos ET & Krause M. Cyclin E expression during development in Caenorhabditis elegans. Dev. Biol 254, 102–115 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wu YC & Horvitz HRC. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392, 501–504 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Hsieh J et al. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13, 2958–2970 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishok A, Sinskey JL & Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19, 683–696 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer SEJ et al. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev. 27, 2678–2695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boeck ME et al. Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development. Dev. Biol 358, 345–355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mello CC, Kramer JM, Stinchcomb D & Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rual J-F et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser AG et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Kamath RS et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Sulston JE, Schierenberg E, White JG & Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 46.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansson GC, Simons K & van Meer G. Two strains of the Madin-Darby canine kidney (MDCK) cell line have distinct glycosphingolipid compositions. EMBO J. 5, 483–489 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streichan SJ, Hoerner CR, Schneidt T, Holzer D & Hufnagel L. Spatial constraints control cell proliferation in tissues. Proc. Natl. Acad. Sci. U. S. A 111, 5586–5591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakaue-Sawano A et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 50.McQuin C et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kipreos ET, Gohel SP & Hedgecock EM. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development 127, 5071–5082 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting all figures are available within the paper and in the associated source data files. Raw microscopy data are available upon request to the corresponding author.