Abstract
Coronavirus disease 2019 (COVID-19) is raging worldwide. This potentially fatal infectious disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the complete mechanism of COVID-19 is not well understood. Therefore, we analyzed gene expression profiles of COVID-19 patients to identify disease-related genes through an innovative machine learning method that enables a data-driven strategy for gene selection from a data set with a small number of samples and many candidates. Principal-component-analysis-based unsupervised feature extraction (PCAUFE) was applied to the RNA expression profiles of 16 COVID-19 patients and 18 healthy control subjects. The results identified 123 genes as critical for COVID-19 progression from 60,683 candidate probes, including immune-related genes. The 123 genes were enriched in binding sites for transcription factors NFKB1 and RELA, which are involved in various biological phenomena such as immune response and cell survival: the primary mediator of canonical nuclear factor-kappa B (NF-κB) activity is the heterodimer RelA-p50. The genes were also enriched in histone modification H3K36me3, and they largely overlapped the target genes of NFKB1 and RELA. We found that the overlapping genes were downregulated in COVID-19 patients. These results suggest that canonical NF-κB activity was suppressed by H3K36me3 in COVID-19 patient blood.
Subject terms: Computational science, Machine learning
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was first identified in December 2019 in Wuhan, Hubei, China, and has resulted in an ongoing pandemic1–3. COVID-19 is a potential zoonotic disease with a moderate mortality rate (2–5%) and is primarily transmitted through droplets and direct contact with infected individuals or incubation carriers4. The large number of mild and asymptomatic cases is considered to be a feature of SARS-CoV-25–7. However, it can severely impact the lungs, and COVID-19 survivors can suffer long-term health effects. Although numerous studies on COVID-19 have been conducted, our understanding of it is still far from complete. Currently there are no clearly effective preventive or therapeutic remedies for COVID-19. Patients with COVID-19 have no choice but to receive supportive care to relieve symptoms8. Therefore, it is imperative to elucidate the mechanism of COVID-19 and find an effective treatment method.
The “silver bullet” approach requires analyzing RNA-Seq data containing RNA extracted from samples. By comparing the gene expressions of COVID-19 patients with those of non-patients, we can obtain more information about the infectious disease pathology. A data-driven approach using machine learning is an efficient strategy for predicting mechanisms that are difficult to elucidate through the application of conventional knowledge-based analysis in biology. Although it is not difficult to obtain various kinds of omics data for COVID-19, the data is difficult to analyze because they often include several tens of thousands of candidate genes and few samples.
Recently, an unsupervised feature extraction method based on principal component analysis (PCA) has been suggested for its utility in gene selection. This method, called PCAUFE9–27, enables analysis of data sets with a small number of samples and many variables. The algorithm, which is based on linear algebra, is computationally light and has been confirmed to work well for various gene selection problems. For example, an integrated analysis of the mRNA/miRNA expression associated with posttraumatic-stress-disorder- (PTSD-) mediated heart disease17 and various cancers12 identified a possible candidate gene associated with those diseases. More recently, an integrated gene expression analysis of blood from patients with dengue hemorrhagic fever by using PCAUFE identified 46 genes that are critical to the disease progression, whereas other methods of bioinformatic analysis were unable to obtain such results27. Furthermore, a theoretical justification for the PCAUFE methodology was already developed in previous studies (for more details, see27).
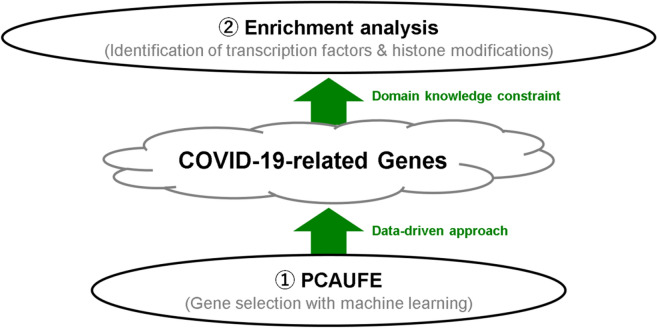
In this paper, we identify genes associated with COVID-19 by applying PCAUFE to the RNA expression profiles of COVID-19 patients and healthy control subjects. Figure 1 shows an outline of our study. We confirm the reliability of the identified genes from the viewpoints of both biology and machine learning. Furthermore, we use bioinformatics tools to identify the transcription factors and histone modifications that regulate the selected genes in the upper layers. The novelty in this manuscript lies our findings that the application of PCAUFE to the gene expression profiles provided the smallest number of genes which are reasonable to explain the COVID-19 development from both machine learning and biological perspectives by comparing to the other typical gene selection methods described in the following section.
Figure 1.
Outline of this study. First, we select genes related to the disease by using unsupervised machine learning; then, we enrich them by applying biological knowledge to identify the transcription factors and histone modifications of the selected genes.
Results
Application of PCAUFE to gene expression in COVID-19 patients
This section describes how we used PCAUFE to analyze the gene expression patterns of multiple COVID-19 patients.
The first example (data set 1, GSE152418) was obtained by Arunachalam et al28. It includes five severity categories: ICU patients (IP), severe patients (SP), moderate patients (MP), convalescent patients (CP), and healthy controls (HC). By investigating the principal component (PC) loadings that statistically differentiated the group of IP + SP + MP (16 patients) from the group of CP + HC (18 non-patients), we found the second and third PCs (PC2 and PC3). The P values computed with a t-test rejected the null hypothesizes that the mean loadings within the group of IP+SP+MP and within the group of CP+HC were identical: 9.69 × 10−5 for PC2 and 3.67 × 10−3 for PC3. Although the PC1 loadings were the significantly different between patients and non-patients, the P value (1.83 × 10−2) was larger than those of PCs 2 and 3. As also shown in Fig. S1, the 2nd and 3rd PCs more clearly separated samples into patients and non-patients than the first one. This is the reason why we chose PCs 2 and 3, but not 1. On this plane, we selected 141 probes embedded in the PC scores as outliers according to a χ2 test with the P values adjusted by the Benjamini and Hochberg (BH) criterion29. Table 1 lists all 123 genes associated with the 141 probes.
Table 1.
One hundred and twenty-three genes selected by PCAUFE. All of these genes were also selected by LIMMA.
| ACTB | ACTG1 | ADRBK1 | AHNAK | ALAS2 | ANXA1 |
|---|---|---|---|---|---|
| ANXA2 | APLP2 | ARL4C | B2M | BTG1 | BTG2 |
| C1orf63 | CCR7 | CD14 | CD163 | CD69 | CD74 |
| CD83 | CLU | COX1 | CTSB | CTSS | CXCR4 |
| CYFIP2 | DDX3X | DDX5 | DNAJB1 | DUSP1 | DUSP2 |
| EEF1A1 | EIF1 | EIF4G2 | ENO1 | F13A1 | FCN1 |
| FLNA | FOS | FOSB | FTL | GAPDH | GLUL |
| GPR183 | GRN | HBA1 | HBA2 | HBB | HLA-B |
| HLA-DPA1 | HLA-DRA | HLA-DRB1 | HLA-DRB5 | HLA-E | HMHA1 |
| HSP90B1 | HSPA5 | HSPA8 | IFI27 | IFITM3 | IGJ |
| IL10RA | IL1B | IRF1 | ISG15 | ITGA2B | ITGB2 |
| IVNS1ABP | JAK1 | JUNB | JUND | KLF2 | KLF6 |
| LCK | LCP1 | LOC100507709 | LOC100507714 | MAFB | MCL1 |
| MX1 | NFKBIA | NFKBIZ | NR4A1 | NR4A2 | PIK3IP1 |
| PKM2 | PLBD1 | PNRC1 | PPBP | PPP1R15A | PSAP |
| PTGER4 | PTPRC | RGS2 | RPL13 | RPL3 | RPS2 |
| S100A12 | S100A8 | S100A9 | SELL | SERPINA1 | SF3B1 |
| SH3BGRL3 | SLC2A3 | SORL1 | SPARC | SRSF5 | SRSF7 |
| SUN2 | TAGAP | TLN1 | TMEM66 | TNFAIP3 | TNFRSF1B |
| TSC22D3 | TUBA1A | TYMP | UBC | VCAN | YPEL5 |
| ZFP36 | ZFP36L2 | ZNF331 | |||
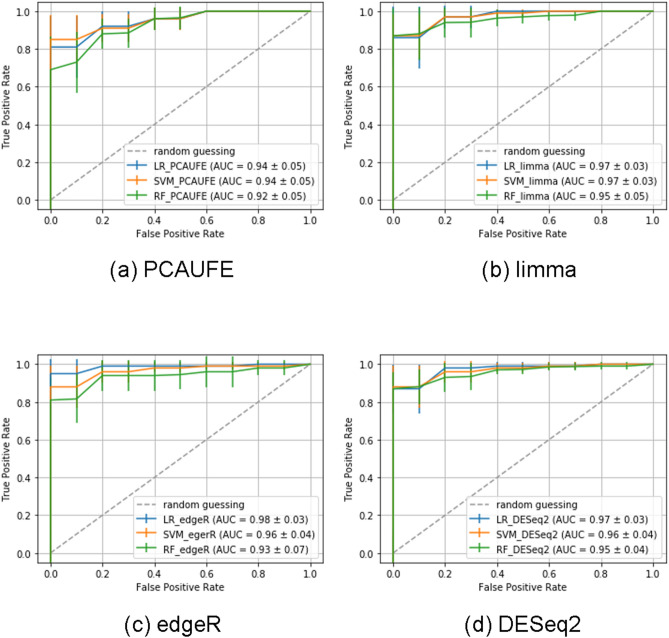
To confirm that we successfully selected critical genes representing the relationship between samples, we built the model to predict the COVID-19 patients or not from only the 123 genes selected with PCAUFE. We used data set 2, which consisted of 100 COVID-19 patients and 26 non-COVID-19 ones, to calculate the area under the curve (AUC)30. We used logistic regression (LR)31, support vector machine (SVM)32,33, and random forest (RF)34 as classification models. Table S1 shows each hyperparameter of the three models. We performed 5-fold cross-validation by randomly shuffling the samples of data set 2 for the three classifiers. Figure 2(a) shows the receiver operating characteristic (ROC) curves35,36 of each model. As shown in this figure, the average AUC for each model was derived to be above 0.9. From these results, we could use the 123 selected genes for the prediction of the COVID-19 outcome.
Figure 2.
ROC curves of each classification model to predict COVID-19 patients or not based on the probes selected by (a) PCAUFE, (b) LIMMA, (c) edgeR, and (d) DESeq2, respectively. Note that since the number of probes respectively selected by LIMMA, edgeR, and DESeq2 were all above 4000, too much more than the samples, the probes with the smallest adjusted. P values were further restricted to be almost the same number as in PCAUFE, 141, for the explanatory variables. Using each classification model, we respectively performed fivefold cross-validation and reported the averages and standard deviations of the 5 runs.
Comparison with other gene selection methodologies
To confirm the robustness of our results, we performed gene selection with two other classical methods: significance analysis of microarrays (SAM)37 and linear models for microarray data (LIMMA)38. By applying both SAM and LIMMA to data set 1 (GSE152418), we identified genes associated with adjusted P-values below 0.01. We confirmed that the majority of the genes selected by PCAUFE were included among those selected by SAM and LIMMA. Every time SAM was applied, the selected genes changed. Thus, the results of SAM are not shown in this report. As for LIMMA, the selected probes included all of the genes selected by PCAUFE. It was noteworthy that PCAUFE could limit the candidate genes to a much smaller number than the common gene expression analysis tools could; for example, 18,458 probes were selected by LIMMA. By further limiting the 18,458 probes to almost the same number as in PCAUFE from the smallest adjusted P-values, we also performed the patient/non-patient classification from the genes selected by LIMMA using data set 2. The results using LR, SVM and RF are shown in Fig. 2(b). We confirmed that the classification performances of each model were comparable to those in PCAUFE.
To increase the robustness of our results, we also selected genes using some more recent R packages: edgeR39 and DESeq240. As with SAM and LIMMA, by applying both edgeR and DESeq2 to data set 1, we identified genes associated with adjusted P-values below 0.01. The numbers of probes selected by edgeR and DESeq2 were 4452 and 5696, respectively. Thus, these methods selected much more genes than PCAUFE. The genes selected by edgeR and DESeq2 contained the 59 and 64 genes selected by PCAUFE, respectively. The common genes selected among the three methods were 57. For further comparison with PCAUFE, we conducted the classification analysis to predict whether the sample was the COVID-19 patient or not based on the genes selected by edgeR and DEseq2. In the classification analysis, we limited the same number of probes selected by each of edgeR and DEseq2 with the smallest adjusted P-values as in PCAUFE (i.e., 141), since both methods selected too many genes to use them for explanatory variables of the prediction models (i.e., 4452 and 5696). The numbers of genes associated with those probes were 111 for edgeR and 113 for DEseq2, respectively. Figures 2(c) and (d) shows the ROC curves of the patient-prediction models using the limited probes. Their AUCs were approximately equal to those in PCAUFE. As described above, we found that a smaller number of genes selected by PCAUFE than the other methods were significant in predicting the COVID-19 patients or not.
As the last part of this subsection, we conducted a weighted gene co-expression network analysis (WGCNA). Following the analytic procedure used in41 and42, we applied the WGCNA R package43 to data set 1. We here selected power of β = 7 as the soft threshold for constructing a scale-free network (Supplementary Fig. S2(A)). We then obtained 99 modules in the co-expression network as shown in Supplementary Fig. S2(B). These modules included 18882 probes, which were much more than those selected by PCAUFE (i.e., 141). Moreover, almost half (i.e., 58) of the genes selected by PCAUFE were contained in those by WGCNA. For reference, the protein-protein interaction (PPI) networks consisting of the genes that belonged to the top 3 modules with the smallest P values and the results of enrichment analyses for these genes are also shown in Supplementary Fig. S3 and Table S2, respectively.
As demonstrated above, we verified our approach, in which PCAUFE was adopted for gene selection, could narrow down the candidate genes more effectively than ordinary methods such as WGCNA, edgeR, and DEseq2.
Discussion
In this study, we first identified 123 genes related to COVID-19 patients using PCAUFE. We justified the use of the linear dimensional reduction method by the following supplementary analysis: To compare with PCAUFE, we also applied t- distributed stochastic neighborhood embedding (t-SNE)44 and uniform manifold approximation and projection (UMAP)45–47, two typical nonlinear dimension reduction methods, to dataset 1 for gene selection. In the same manner with the PCAUFE algorithm, we tried to search the probes which passed the χ2 test with the P value adjusted by the BH criterion. As shown in Fig. S4, however, we found no probes with the adjusted P values less than 0.01 on the planes. The first algorithm of PCAUFE searches for principal components (i.e., axes) whose loadings statistically separate two groups such as patients and non-patients. Therefore, the outliers through the χ2 test in the second algorithm of PCAUFE could be regarded as the probes that were abnormally up-or down-regulated in some patients compared to non-patients. On the other hand, t-SNE and UMAP emphasize preserving the similarity of the probes as a distance when reducing a high-dimensional data set to two dimensions, the axes in the low-dimensional space do not always correspond to the two groups. Thus, it is not suitable for gene selection to naively use these nonlinear dimensional reduction methods because it does not mean that the probes located far from the origin in the low-dimensional space can serve as the biomarkers for the diagnosis of COVID-19.
Furthermore, it should be noted that the use of state-of-the-art deep learning techniques is not always successful in the gene expression analysis of a new type of disease such as COVID-19: We also applied a recently published method called single-cell Decomposition using Hierarchical Autoencoder (scDHA)48, that was described as reliably extracting representative information of each cell, to data set 1 for comparison to PCAUFE. Supplementary Figure S5 displays the scatter plot of samples of data set 1 in the dimension reduction space of scDHA. As shown in this figure, scDHA could not separate the COVID-19 patients from non-patients based on gene expression profiles of PBMCs at all. The reason is probably that this data set has too few samples and too many variables for training the autoencoder. PCAUFE is computationally less expensive than other methods because it only requires one application of PCA to a gene expression matrix in a peculiar way. Therefore, it has been successfully used to tackle a variety of gene selection problems (for detail, see e.g.,49). As described above, we consequently demonstrated the usefulness of this non-novel but powerful gene selection method for the data sets of gene expression profiles from COVID-19 patients and proposed a novel mechanism underlying the COVID-19 development.
We second implemented three independent models to classify COVID-19 patients and non-patients based on the 123 genes selected by PCAUFE: LR, SVM, and RF, and confirmed that all the models archived the high AUCs of over 90%. The reason why we did not use the state-of-art deep learning techniques for the classification model was that the sample size in the dataset used for the cross-validation (i.e., 126) was not large enough for the number of explanatory variables (i.e., 123). For example, it may be possible to build the classification model with deep learning by virtually increasing the number of samples as in50. Liu et al.51 used convolutional neural networks (CNNs) to predict Alzheimer’s patients based on the fMRI images of their hippocampus, but we did not use them because the explanatory variables in our patient prediction model were gene expression levels, in which the similarities could not be assumed between elements close in location as in images. However, as mentioned above, the statistical relevance of the genes selected by PCAUFE is already guaranteed because we found the conventional machine learning models had sufficient prediction accuracies.
To show the further robustness of our results, we also perform gene selection using data set 2 and clustering analysis to validate the separability by the selected genes using data set 1. The numbers of genes selected from data set 2 by PCAUFE, LIMMA, edgeR, and DESeq2 were 145, 7360, 4809, and 5018, respectively. LIMMA, edgeR, and DESeq2 respectively included the 79, 82, and 82 of 145 genes selected by PCAUFE using data set 2. On the other hand, the number of genes overlapping between the 123 and 145 genes selected by PCAUFE was 38. For the clustering analysis, we adopted an unsupervised learning model, UMAP, since data set 1 included only 34 samples, too few to train and test the supervised learning models using it. The results shown in Supplementary Fig. S6 indicate that the genes selected by PCAUFE could classify the COVID-19 patients or not as well as LIMMA, edgeR, and DESeq2. In the end, we got similar results even if we switched the data sets for gene selection and patient/non-patient classification.
Because we successfully confirmed the robustness of our results, we next investigated the biological reliability of the 123 selected genes. First, we uploaded the 123 genes to three enrichment analysis servers, GeneSetDB52, Metascape53, and TargetMine54, to compensate for the bias introduced by each individual enrichment. Multiple immune-related enrichments were detected. For example, Gene Ontology (GO) biological process (BP) terms GO:0019221 (cytokine-mediated signaling pathway), GO:0060333 (interferon-gamma-mediated signaling pathway), and GO:0060337 (type I interferon-mediated signaling pathway) were identified by all three servers. GO cellular component (CC) term GO:0042613 (MHC class II protein complex) was identified by GeneSetDB and TargetMine. Reactome pathways R-HSA-877300 (interferon gamma signaling), R-HSA-6785807 (Interleukin-4 and Interleukin-13 signaling), R-HSA-449147 (signaling by interleukins), and R-HSA-1280218 (adaptive immune system) were identified by Metascape and TargetMine (for more details, see supplemental S1_File).
Second, we confirmed biological validation of the identified genes by examining the interactions between them. Tight relationships between the genes would indicate that the gene selection was reliable, because single proteins rarely function without collaboration with other proteins. Thus, we uploaded the 123 genes to the STRING server55, which detected 659 protein–protein interactions among the products of these genes. Therefore, the 123 genes were also enriched for protein–protein interactions because of the functional collaborations between their products. These enrichment analyses suggested that PCAUFE could successfully identify a biologically feasible set of genes related to COVID-19.
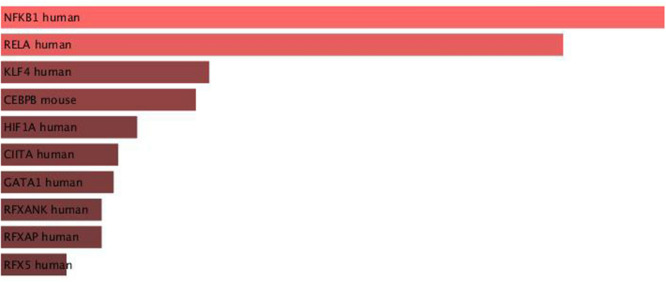
To investigate the upstream transcription factors (TFs) that regulate the 123 genes selected by PCAUFE, we also uploaded them to Enrichr56,57, a multi-functional enrichment analysis server. Among the results given by Enrichr, NFKB1 and RELA had smaller adjusted P-values for “TRRUST Transcription Factors 2019,” as shown in Fig. 3. We also noticed the three highest-ranked TF bindings for “ENCODE TF ChIP-seq 2015”: NELFE, RELA, and KAT2A (for more details, see Fig. S7 in the supplemental S2_File).
Figure 3.
Bar graph of TRRUST Transcription Factors 2019. The graph visualizes the top ten enriched transcription factors of the genes selected by PCAUFE. The bars are colored and sorted according to their P-values.
The 123 genes were also enriched for multiple histone modifications, and the results are listed in Table 2. Furthermore, as shown in Fig. 4, the genes associated with the histone modifications largely overlapped the TF target genes.
Table 2.
Enriched histone modifications detected by Enrichr (ENCODE Histone Modifications 2015) for the 123 selected genes. Only those with adjusted P-values below 0.05 are listed here.
| Rank | Histone modification | P value | adjusted P value | combined score |
|---|---|---|---|---|
| 1 | H3K36me3 Caco-2 hg19 | 1.93E−14 | 7.94 E−12 | 282.29 |
| 2 | H3K36me3 kidney epithelial cell hg19 | 7.73 E−10 | 1.59 E−07 | 206.28 |
| 3 | H3K36me3 bronchial epithelial cell hg19 | 1.26 E−09 | 1.73 E−07 | 65.42 |
| 4 | H3K36me3 splenic B cell mm9 | 7.07 E−09 | 5.83 E−07 | 53.40 |
| 5 | H3K36me3 thymus mm9 | 6.05 E−09 | 6.23 E−07 | 67.27 |
| 6 | H3K36me3 spleen mm9 | 2.57 E−08 | 1.76 E−06 | 48.31 |
| 7 | H3K36me3 GM06990 hg19 | 1.49 E−07 | 8.75 E−06 | 50.92 |
| 8 | H3K36me3 BJ hg19 | 2.98 E−07 | 1.53 E−05 | 46.92 |
| 9 | H3K36me3 kidney mm9 | 3.08 E−06 | 1.41 E−04 | 38.25 |
| 10 | H3K36me3 SK-N-SH hg19 | 7.04 E−06 | 2.90 E−04 | 43.66 |
| 11 | H4K20me1 skeletal muscle myoblast hg19 | 8.88 E−06 | 3.32 E−04 | 27.43 |
| 12 | H3K36me3 myocyte mm9 | 1.56 E−05 | 5.36 E−04 | 68.63 |
| 13 | H3K36me3 H7 hg19 | 1.76 E−05 | 5.57 E−04 | 25.97 |
| 14 | H3K36me3 MCF-7 hg19 | 3.85 E−05 | 1.13 E−03 | 26.07 |
| 15 | H3K36me3 C2C12 mm9 | 4.34 E−05 | 1.19 E−03 | 44.15 |
| 16 | H3K36me3 small intestine mm9 | 1.77 E−04 | 4.56 E−03 | 18.26 |
| 17 | H4K20me1 fibroblast of lung hg19 | 4.39 E−04 | 1.06 E−02 | 15.72 |
| 18 | H3K36me3 cardiac mesoderm hg19 | 5.37 E−04 | 1.23 E−02 | 14.12 |
| 19 | H3K36me3 CD14-positive monocyte hg19 | 1.04 E−03 | 2.14 E−02 | 13.41 |
| 20 | H4K20me1 GM12878 hg19 | 1.04 E−03 | 2.25 E−02 | 13.41 |
| 21 | H3K36me3 CH12.LX mm9 | 1.63 E−03 | 3.20 E−02 | 10.10 |
| 22 | H4K20me1 keratinocyte hg19 | 2.34 E−03 | 4.19 E−02 | 11.33 |
| 23 | H4K20me1 mammary epithelial cell hg19 | 2.34 E−03 | 4.38 E−02 | 11.33 |
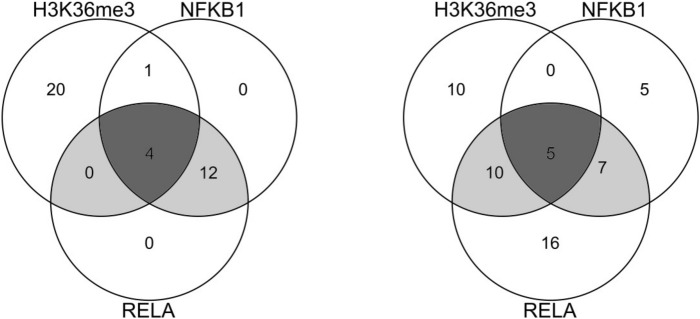
Figure 4.
Venn diagrams of the enrichment of TF-binding sites and histone modifications for the 123 genes, as identified by Enrichr. The numbers in each diagram indicate the numbers of genes selected by PCAUFE and regulated by the TFs. Left: NFKB1, RELA (TRRUST Transcription Factors 2019), and H3K36me3_GM06990_hg19 (ENCODE Histone Modifications 2015); right: NFKB1 (TRRUST Transcription Factors 2019), RELA_GM12892_hg19 (ENCODE TF ChIP-seq 2015), and H3K36me3_GM06990_hg19 (ENCODE Histone Modifications 2015).
The nuclear factor-kappa B (NF-κB) TFs play an evolutionarily conserved and critical role in the triggering and coordination of both innate and adaptive immune responses58. The NF-κB family of transcription factors consists of five members: p50, p52, p65 (RelA), c-Rel, and RelB, which are encoded by NFKB1, NFKB2, RELA, REL, and RELB, respectively59. The primary mediator of canonical NF-κB activity is the heterodimer RelA-p50, which consists of the RelA transcriptional activator and the nfkb1 protein p5060,61.
Nakshatri et al.62 suggested that NF-κB activity is suppressed by H3K36me3, which is consistent with the observed enrichment of NFKB1- and RELA-binding sites in these 123 genes. Many studies have also reported that the expression levels of genes associated with immune signaling are downregulated in naso/oropharyngeal swabs and peripheral blood mononuclear cells (PBMCs) in patients with COVID-19. Mick et al.63 showed that COVID-19 is characterized by a diminished innate immune response, with reduced expression of genes involved in toll-like receptor and interleukin signaling, chemokine binding, neutrophil degranulation, and interactions with lymphoid cells, as compared to other viral acute respiratory illnesses. Meckiff et al.64 showed that SARS-CoV-2-reactive CD4+ T cells express significantly lower levels of immune-related transcripts as compared to influenza-reactive cells. Ouyang et al.65 reported that the genes that are underexpressed in severe cases mainly involve Th17-cell differentiation, cytokine-mediated signaling pathways, and T-cell activation. Li et al.66 reported that proteins mediating T-cell receptor signaling are downregulated in severe COVID-19 PBMCs.
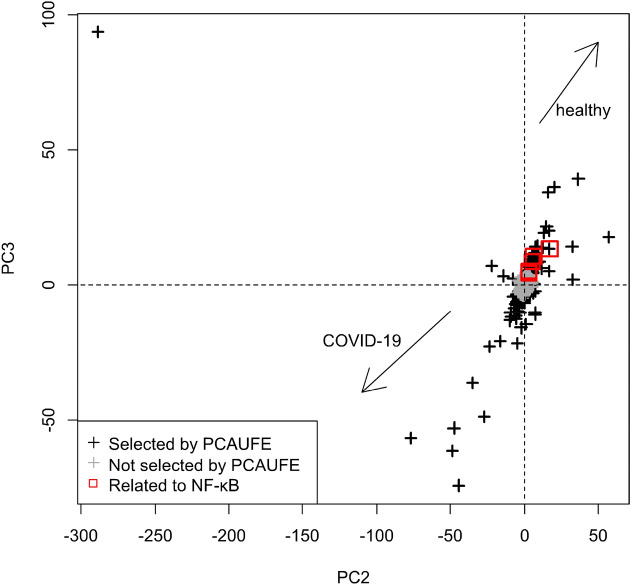
To investigate the expression variation of the five overlapping genes in Fig. 4, we confirmed the PC2 and PC3 scores for data set 1 via the scatter plot shown in Fig. 5. The probes that were negatively located for both PC2 and PC3 were mainly upregulated for COVID-19. On the other hand, the probes that were positively located for both PCs were mainly downregulated for COVID-19. As shown by the red squares in Fig. 5, the overlapping genes in Fig. 4 were positively located for both PCs. Therefore, those overlapping genes were downregulated for COVID-19. These analysis results are consistent with the above references62–66.
Figure 5.
Scatter plot of the PC2 and PC3 scores for data set 1. The black crosses represent the probes selected by PCAUFE, while the gray crosses represent unselected probes. The red squares are associated with the five overlapping genes in Fig. 4.
NF-κB is the subject of much active research among pharmaceutical companies as a target for anti-cancer therapy67. Abnormal expressions of NFKB1 and RELA are mediated through mRNA modifications68. The recent progress in N4- Acetylcytidine (N4A) on RNA expression is also playing key role on the cancer development69. Duan J et al.70 reported that N4A, a nucleoside metabolite, activated microglia and sustained NLRP3 inflammasome activation by inducing HMGB1 signaling. Released HMGB1 through N4A activated NF-κB and induced NLRP3 expression70. NLRP3 inflammasome appropriately activated and enabled to release mature IL-1β71–73. IL-1β is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis74,75.
Although we identified the 123 genes and the upstream TFs related to COVID-19 by using PCAUFE and enrichment analyses, we have yet assessed whether these genes have the potential causal effects on the COVID-19 development. Mendelian randomization (MR)76–80 approach has been widely used to investigate causality between genes and disease outcomes. For example, Zhang et al.79 investigated the causal relationships between PTSD and the depressive phenotypes using an MR approach. In another of their studies80, The results of MR analysis indicate that genetic variation mediates the causal influences of neuroticism on mental health and cardiovascular diseases. This method uses genetic polymorphism information as an operating variable, but unfortunately, the data sets we used do not include that information. Moreover, polymorphisms may have several phenotypic effects associated with the disease. Thus, we leave the application of the MR approach to the gene expression profiles from COVID-19 patients as future work.
In conclusion, we selected 123 COVID-19-related genes by applying PCAUFE to the gene expression levels of PBMCs from COVID-19 patients and healthy subjects. Then, by enrichment analysis, we identified the transcription factors and histone modifications that regulate the expression of these genes. Four transcription factors, NELFE, RELA, KAT2A and NFKB1, and a histone modification, H3K36me3, may be involved in the expression of the 123 genes. NFKB1, RELA, and H3K36me3 were found to overlap in the genes regulating expression. These two transcription factors are associated with NF-κB, and H3K36me3 may repress it. In fact, when we compared the expression levels of the genes duplicated in NFKB1, RELA, and H3K36me3 in GSE152418 between the COVID-19 patients and healthy subjects, we observed a decrease in expression levels in the COVID-19 patients. These results suggest that canonical NF-κB activity is suppressed by H3K36me3 in the PBMCs of COVID-19 patients.
Methods
Gene expression profiles
Two in vivo gene expression data sets, GSE15241828 and GSE15710381, were downloaded from Gene Expression Omnibus82. Hereafter, we denote these as data sets 1 and 2, respectively. PCAUFE was applied to data set 1, which described the expression level of each kind of mRNA in each subject’s PBMCs. The number of probes was 60,683. Data set 2 was then used to confirm the statistical validity of the genes selected by PCAUFE. This data set also described the expression level of each gene in each subject’s PBMCs. The data included both COVID-19 patients and non-COVID-19 patients who suffered from acute respiratory distress syndrome (ARDS) that was not associated with SARS-CoV-2. The number of genes was 19,472. The expression level of each gene i (= 1, 2,…, N) was standardized for PCAUFE, i.e., we set and . For the details of the samples included in these gene expression profiles, see Table S3 in the supplemental S2_File.
PCAUFE
The following briefly explains the PCAUFE procedure used in this study (for more details, see9,12,17,27,49). Let xi j be the expression of the i-th mRNA probe of the jth sample, and let and , where N is the number of m-RNA probes. First, we applied PCA to the dataset whose rows and columns were genes and samples, respectively. In contrast to the usual use of PCA, where samples are embedded, the genes were embedded in this implementation. By using a t-test, we specified two principal components (PCs) whose loadings statistically differentiated the patients from healthy control samples in order from the smallest P-value. Note that, unlike ordinary PCA, this operation does not guarantee that the first two PCs will be selected. Second, by using a χ2 test with the P values adjusted by the BH criterion29, we identified outlier PC scores (i.e., genes associated with the adjusted P-values less than 0.01) along with the specified PCs as candidates for the disease-related genes. Note that the PC scores in PCAUFE were associated with features (i.e., mRNA probes), not with samples, in contrast to the ordinary usage of PCA.
Patient/non-patient classification models
To verify the genes selected by PCAUFE were useful for the diagnosis of COVID-19 patients, we performed the patient/non- patient classification based on the selected genes using three standard prediction models: logistic regression (LR31), support vector machine (SVM32,33) and random forest (RF34). For the details of each hyperparameter of the three models, see Supplementary Table S1. The objective variable was given the value 0 or 1 for each sample depending on a non-COVID-19 or a COVID-19 patient, respectively. The explanatory variables were given the gene expressions of the probes associated with the genes selected by PCAUFE, edgeR, or DESeq2. We randomly allocated 80% of data set 2 to the training set and the remains to the test one. Receiver operating characteristic (ROC) curves of each model were drawn to calculate the area under the curves (AUCs).
Supplementary Information
Acknowledgements
This study was supported by Okinawa Prefecture’s Project to Promote the Use of Information Technology in the Health and Medical Industries (20G1000012).
Author contributions
K.F., Y.T., and R.M. designed the research; K.F., M.S., and R.M. wrote the manuscript; K.F. and M.S. performed the analysis; K.F., Y.T., S.I., and R.M. interpreted the results; S.I. and R.M. supervised the research; All of the authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kota Fujisawa, Email: fujisawa.k.ab@m.titech.ac.jp.
Ryota Miyata, Email: miyata26@tec.u-ryukyu.ac.jp.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95698-w.
References
- 1.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1101/2020.02.06.20020974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, et al. Temporal dynamics in viral shedding and transmissibility of covid-19. Nat. Med. 2020;26:672–675. doi: 10.1101/2020.03.15.20036707. [DOI] [PubMed] [Google Scholar]
- 6.Wei W, et al. Presymptomatic transmission of sars-cov-2—Singapore, January 23–March 16, 2020. MMWR Morb. Mortal Wkly. Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang R, Gui X, Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coron- avirus disease 2019 in Wuhan, China. JAMA Netw. Open. 2020;3:e2010182. doi: 10.1001/2Fjamanetworkopen.2020.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for disease control and prevention (2021, February 2). https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html.
- 9.Taguchi YH. Principal component analysis based unsupervised feature extraction applied to publicly available gene expression profiles provides new insights into the mechanisms of action of histone deacetylase inhibitors. Neuroepigenetcis. 2016;8:1–18. doi: 10.1016/j.nepig.2016.10.001. [DOI] [Google Scholar]
- 10.Taguchi Y-H, Iwadate M, Umeyama H. Principal component analysis based unsupervised feature extraction applied to budding yeast temporally periodic gene expression. BMC Med. Genomics. 2016;9:69–79. doi: 10.1186/s12920-016-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taguchi YH. Sfrp1 is a possible candidate for epigenetic therapy in non-small cell lung cancer. BioData Min. 2016;9:22. doi: 10.1186/s12920-016-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi YH. Identification of more feasible MicroRNA-mRNA interactions within multiple cancers using principal component analysis based unsupervised feature extraction. Int J Mol Sci. 2016;17(5):696. doi: 10.3390/ijms17050696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taguchi YH. Identification of aberrant gene expression associated with aberrant promoter methylation in primordial germ cells between E13 and E16 rat F3 generation vinclozolin lineage. BMC Bioinform. 2015;16:S16. doi: 10.1186/1471-2105-16-S18-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi, Y.-h. Integrative analysis of gene expression and promoter methylation during reprogramming of a non-small-cell lung cancer cell line using principal component analysis-based unsupervised feature extraction. In Huang, D.-S., Han, K. & Gromiha, M. (eds.) Intelligent Computing in Bioinformatics, vol. 8590 of LNCS, 445–455 (Springer International Publishing, Heidelberg, 2014).
- 15.Taguchi, Y.-h., Iwadate, M., Umeyama, H., Murakami, Y. & Okamoto, A. Heuristic principal component analysis-based unsupervised feature extraction and its application to bioinformatics. In Wang, B., Li, R. & Perrizo, W. (eds.) Big Data Analytics in Bioinformatics and Healthcare, 138–162 (IGI Global, 2015).
- 16.Taguchi, Y.-H., Iwadate, M. & Umeyama, H. Heuristic principal component analysis-based unsupervised feature extraction and its application to gene expression analysis of amyotrophic lateral sclerosis data sets. In Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 2015 IEEE Conference on, 1–10, 10.1109/CIBCB.2015.7300274 (2015).
- 17.Taguchi YH, Iwadate M, Umeyama H. Principal component analysis-based unsupervised feature extraction applied to in silico drug discovery for posttraumatic stress disorder-mediated heart disease. BMC Bioinform. 2015;16:139. doi: 10.1186/s12859-015-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umeyama H, Iwadate M, Taguchi YH. TINAGL1 and B3GALNT1 are potential therapy target genes to suppress metastasis in non-small cell lung cancer. BMC Genomics. 2014;15:S2. doi: 10.1186/1471-2164-15-s9-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami Y, et al. Comprehensive analysis of transcriptome and metabolome analysis in Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Sci. Rep. 2015;5:16294. doi: 10.1038/srep16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami Y, et al. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS ONE. 2014;9:e106314. doi: 10.1371/journal.pone.0106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami H, Toyoda Y, et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Li Q, Xu J, Zhang X, Zhang H, Xiang Y, Fang C, Wang T, Xia S, Zhang Q, Xing Q, He L, Wang L, Xu M, Zhao X. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci Rep. 2016;29(6):19910. doi: 10.1038/srep19910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taguchi YH, Murakami Y. Principal component analysis based feature extraction approach to identify circulating microRNA biomarkers. PLoS ONE. 2013;8:e66714. doi: 10.1371/journal.pone.0066714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita R, Iwadate M, Umeyama H, Taguchi YH. Genes associated with genotype-specific DNA methylation in squamous cell carcinoma as candidate drug targets. BMC Syst. Biol. 2014;8:S4. doi: 10.1186/1752-0509-8-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishida S, Umeyama H, Iwadate M, Taguchi YH. Bioinformatic screening of autoimmune disease genes and protein structure prediction with FAMS for drug discovery. Protein Pept. 2014;21:828–839. doi: 10.2174/09298665113209990052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi, Y.-h. & Okamoto, A. Principal component analysis for bacterial proteomic analysis. In Shibuya, T., Kashima, H., Sese, J. & Ahmad, S. (eds.) Pattern Recognition in Bioinformatics, vol. 7632 of LNCS, 141–152, 10.1109/BIBMW.2011.6112520 (Springer International Publishing, Heidelberg, 2012).
- 27.Taguchi Y-H. Principal components analysis based unsupervised feature extraction applied to gene expression analysis of blood from dengue haemorrhagic fever patients. Sci. Rep. 2017;7:44016. doi: 10.1038/srep44016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arunachalam PS, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 1995;57:289–300. [Google Scholar]
- 30.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145–1159. doi: 10.1016/S0031-3203(96)00142-2. [DOI] [Google Scholar]
- 31.Cox DR. The regression analysis of binary sequences (with discussion) J. R. Stat. Soc. Ser. B (Methodol.) 1958;20:215–232. doi: 10.1111/j.2517-6161.1958.tb00292.x. [DOI] [Google Scholar]
- 32.Vapnik V, Lerner A. Pattern recognition using generalized portrait method. Autom. Remote. Control. 1963;24:774–780. [Google Scholar]
- 33.Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. doi: 10.1007/2FBF00994018. [DOI] [Google Scholar]
- 34.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/2FA:1010933404324. [DOI] [Google Scholar]
- 35.Wang X, et al. Associations between maternal vitamin D status during three trimesters and cord blood 25(OH)D concentrations in newborns: a prospective Shanghai birth cohort study. Eur J Nutr. 2021 doi: 10.1007/s00394-021-02528-w. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, et al. LEPR hypomethylation is significantly associated with gastric cancer in males. Exp. Mol. Pathol. 2020 doi: 10.1016/j.yexmp.2020.104493. [DOI] [PubMed] [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. Co-expression network analysis identified hub genes critical to triglyceride and free fatty acid metabolism as key regulators of age-related vascular dysfunction in mice. AGING (Albany NY) 2019;11(18):7620–7638. doi: 10.18632/aging.102275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, et al. Genetic regulatory subnetworks and key regulating genes in rat hippocampus perturbed by prenatal malnutrition: implications for major brain disorders. AGING (Albany NY) 2020;12(9):8434–8458. doi: 10.18632/aging.103150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Der Maaten L, Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 45.McInnes L, Healy J, Saul N, Großberger L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018;3(29):861. doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 46.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 47.McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv https://arxiv.org/abs/1802.03426 (2020).
- 48.Tran HD, Nguyen TB, Vecchia LC, Luu NH, Nguyen T. Fast and precise single-cell data analysis using a hierarchical autoencoder. Nat. Commun. 2021;12(1):1029. doi: 10.1038/s41467-021-21312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taguchi Y-H. Unsupervised Feature Extraction Applied to Bioinformatics: A PCA Based and TD Based Approach. Springer International Publishing; 2019. [Google Scholar]
- 50.Feng C, et al. Gene expression data based deep learning model for accurate prediction of drug-induced liver injury in advance. J. Chem. Inform. Model. 2019;59:3240–3250. doi: 10.1021/acs.jcim.9b00143. [DOI] [PubMed] [Google Scholar]
- 51.Liu M, et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage. 2020 doi: 10.1016/j.neuroimage.2019.116459. [DOI] [PubMed] [Google Scholar]
- 52.Araki H, Knapp C, Tsai P, Print C. Genesetdb: A comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Openbio. 2012;2:76–82. doi: 10.1016/j.fob.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y-A, Tripathi L, Mizuguchi K. TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS ONE. 2011;6(3):e17844. doi: 10.1371/journal.pone.0017844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szklarczyk D, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen E, et al. Enrichr: interactive and collaborative html5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuleshov M, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 59.Hayden SM, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 60.Moorthy KA, et al. The 20S proteasome processes NF-κB1 p105 into p50 in a translation independent manner. EMBO J. 2006;25:1945–1956. doi: 10.1038/sj.emboj.7601081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basak S, Shih FV, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol. Cell Biol. 2008;28(10):3139–3150. doi: 10.1128/mcb.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakshatri H, et al. NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT. Cell Death Dis. 2014;6(1):e1608. doi: 10.1038/cddis.2014.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mick E, et al. Upper airway gene expression differentiates COVID-19 from other acute respiratory illnesses and reveals suppression of innate immune responses by SARS-CoV-2. medRxiv. 2020;4:e1608. doi: 10.1101/2F2020.05.18.20105171. [DOI] [Google Scholar]
- 64.Meckiff JB, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouyang Y, et al. Downregulated gene expression spectrum and immune responses changed during the disease progression in patients with COVID-19. Clin. Infect. Dis. 2020;ciaa462:1–9. doi: 10.1093/cid/ciaa462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med (N Y). 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Escárcega RO, Fuentes-Alexandro S, García-Carrasco M, Gatica A, Zamora A. The transcription factor nuclear factor-kappa B and cancer. Clin. Oncol. 2007;19(2):154–161. doi: 10.1016/j.clon.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 68.Ferrero-Andrés A, Panisello-Roselló A, Roselló-Catafau J, Folch-Puy E. NLRP3 inflammasome-mediated inflam- mation in acute pancreatitis. Int. J. Mol. Sci. 2020;21(15):5386. doi: 10.3390/ijms21155386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin G, Xu M, Zou M, Duan S. The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: a systematic review. Mol. Ther. Nucleic Acids. 2020;20:13–24. doi: 10.1016/j.omtn.2020.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan J, et al. N4-acetylcytidine is required for sustained NLRP3 inflammasome activation via HMGB1 pathway in microglia. Cell Signal. 2019;58:44–52. doi: 10.1016/j.cellsig.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 71.Mangan MSJ, et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 2018;17(9):688. doi: 10.1038/nrd.2018.149. [DOI] [PubMed] [Google Scholar]
- 72.Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. Trends Immunol. 2011;32(4):171–179. doi: 10.1038/nrd.2018.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng S, Zhao T, Yuan S, Yang L, Ding J, Cui L, Xu M. Immunodeficiency promotes adaptive alterations of host gut microbiome: an observational metagenomic study in mice. Front Microbiol. 2019;1(10):2415. doi: 10.3389/fmicb.2019.02415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tulotta C, Ottewell P. The role of IL-1B in breast cancer bone metastasis. Endocrine-Relat. Cancer. 2018;25(7):R421–R434. doi: 10.1530/2FERC-17-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan X, Zhao X, Li J, He L, Xu M. Effects of early-life malnutrition on neurodevelopment and neuropsychiatric disorders and the potential mechanisms. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;83:64–75. doi: 10.1016/j.pnpbp.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 76.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1989;327:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 77.Wu Y, Cao H, Baranova A, Huang H, Li S, Cai L, Rao S, Dai M, Xie M, Dou Y, Hao Q, Zhu L, Zhang X, Yao Y, Zhang F, Xu M, Wang Q. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl Psychiatry. 2020;10(1):209. doi: 10.1038/s41398-020-00902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, X., Fang, X., Zheng, W., Zhou, J., Song, Z., Xu, M., Min, J., & Wang, F. Genetic support of a causal relationship between iron status and type 2 diabetes: a Mendelian randomization study. J. Clin. Endocrinol. Metab. 2021. [DOI] [PMC free article] [PubMed]
- 79.Zhang F, et al. Causal influences of neuroticism on mental health and cardiovascular disease. Hum. Genet. 2021 doi: 10.1007/s00439-021-02288-x. [DOI] [PubMed] [Google Scholar]
- 80.Zhang F, et al. Genetic evidence suggests posttraumatic stress disorder as a subtype of major depressive disorder. J. Clin. Investig. 2021;27:145942. doi: 10.1172/jci145942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Overmyer KA, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2020;12:1–18. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar R, Domrachev M, Lash A. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.