Abstract
Background
The most common B-cell cancers, chronic lymphocytic leukemia/lymphoma (CLL), follicular and diffuse large B-cell (FL, DLBCL) lymphomas, have distinct clinical courses, yet overlapping “cell-of-origin”. Dynamic changes to the epigenome are essential regulators of B-cell differentiation. Therefore, we reasoned that these distinct cancers may be driven by shared mechanisms of disruption in transcriptional circuitry.
Methods
We compared purified malignant B-cells from 52 patients with normal B-cell subsets (germinal center centrocytes and centroblasts, naïve and memory B-cells) from 36 donor tonsils using >325 high-resolution molecular profiling assays for histone modifications, open chromatin (ChIP-, FAIRE-seq), transcriptome (RNA-seq), transcription factor (TF) binding, and genome copy number (microarrays).
Findings
From the resulting data, we identified gains in active chromatin in enhancers/super-enhancers that likely promote unchecked B-cell receptor signaling, including one we validated near the immunoglobulin superfamily receptors FCMR and PIGR. More striking and pervasive was the profound loss of key B-cell identity TFs, tumor suppressors and their super-enhancers, including EBF1, OCT2(POU2F2), and RUNX3. Using a novel approach to identify transcriptional feedback, we showed that these core transcriptional circuitries are self-regulating. Their selective gain and loss form a complex, iterative, and interactive process that likely curbs B-cell maturation and spurs proliferation.
Interpretation
Our study is the first to map the transcriptional circuitry of the most common blood cancers. We demonstrate that a critical subset of B-cell TFs and their cognate enhancers form self-regulatory transcriptional feedback loops whose disruption is a shared mechanism underlying these diverse subtypes of B-cell lymphoma.
Funding
National Institute of Health, Siteman Cancer Center, Barnes-Jewish Hospital Foundation, Doris Duke Foundation.
Keywords: B-cell cancer, Lymphoma, Transcriptional regulation and feedback, Epigenetics, Super-enhancers
Research in context.
Evidence before this study
Chronic lymphocytic leukemia/lymphoma (CLL), follicular and diffuse large B-cell lymphomas (FL, DLBCL), are distinct in their presentation and clinical course, yet overlap in their “cell-of-origin” or normal counterpart B-cells. Previous studies showed that dynamic changes to the epigenome are essential regulators of normal B-cell differentiation. Notably, no studies have compared epigenetic and transcriptional regulation in these three lymphomas, which comprise the majority of blood cancers in the U.S.
Added value of this study
We reasoned that these distinct cancers may be driven by shared mechanisms of dysregulation of transcriptional circuitry, a question that has not been previously addressed to our knowledge. Our study is the first to map the transcriptional circuitry of the most common blood cancers. We report the results of our integrative analysis comparing epigenome, transcriptome, and genome copy number data from purified malignant B cells from 52 patients with sorted normal B-cell subsets from 36 donor tonsils, comprising 328 high resolution molecular profiling studies. Our studies identified selective gain and loss of self-regulating transcriptional regulators, including B-cell receptor signaling and identity factors, as well as tumor suppressors, that together form an iterative feed-forward process that curbs B-cell maturation and spurs proliferation.
Implications of all the available evidence
Our results suggest that diverse subtypes of B-cell cancer arise due to disruption of shared transcriptional circuitry involving core B-cell TFs and their cognate enhancers. We believe that our findings are novel and impactful in that they demonstrate a mechanism for perpetuating transcriptional and epigenetic dysregulation that is shared across diverse human B-cell cancers. These shared B-cell factors and their regulatory circuits represent previously unrecognized targets for future study and potential therapies for the most common blood cancers.
Alt-text: Unlabelled box
1. Introduction
Development of B lymphocytes from lymphoid progenitors is less a linear progression than a web of decision points in which response to internal and external cues drives phenotypic change and the next differentiation stage. Significant perturbations at developmental decision points should result in cell death, but rarely, cells escape this quality control system. Dynamic changes to the epigenome are essential regulators of B-cell differentiation, as demonstrated by the consistent alterations observed at B-cell specific promoters and enhancers during B-cell maturation [1,2] and the detrimental effects of epigenetic modifier knockouts [3], [4], [5], [6]. Several of these modifiers are also connected to the pathogenesis of human and murine B-cell cancers [7], [8], [9]. Widespread changes to DNA methylation landscapes have been reported in all major B-cell cancers, though unlike other blood cancers, mutations in DNA methylation genes are rare [10], [11], [12], [13], [14], [15].
The essential nature of histone modifications for epigenetic regulation of gene expression in B cells has been known for more than 50 years [16]. More recently, chromatin immunoprecipitation sequencing (ChIP-seq) studies across B-cell developmental stages have elucidated coordinated changes in histone acetylation and methylation linked to modulation of gene expression [1,2]. Effective B-cell maturation in the germinal center relies upon temporally regulating the activation or repression of proliferation and differentiation pathways, often via bivalent chromatin states marking key promoters [8,17]. Not surprisingly, mutation of essential chromatin modifiers and consequent alterations to histone landscapes is a hallmark of germinal center B-cell cancers, particularly follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL) [8,[18], [19], [20]]. Another mature B-cell cancer, chronic lymphocytic leukemia (CLL), harbors mutations in a different set of epigenetic modifier genes, albeit at much lower frequency [12,21]. Nevertheless, there are widespread changes to the chromatin landscape in CLL [22,23]. Notably, the chromatin landscapes of germinal center B-cell lymphomas (FL, DLBCL) and CLL have not been compared to identify unique and shared alterations that may illuminate pathways that are subtype-specific and shared, respectively, to the pathogenesis of each cancer type.
The process of B-cell differentiation generates a number of cellular states with distinct transcriptional and surface marker profiles, and importantly, different vulnerabilities for transformation to B-cell lymphoma/leukemia. Thus, identification of BCL-specific pathways requires comparison to the most similar normal B-cell subset, sometimes referred to as cell-of-origin [24], [25], [26]. Comparisons of whole transcriptomes, cell surface markers, and epigenome landscapes have demonstrated a close similarity between FL and centrocytes from the GC light zone [18,27]. The GCB subtype of DLBCL also resembles GC centrocytes, while the activated B-cell (ABC) subtype exhibits a profile more similar to centroblasts from the GC dark zone [27] or, more recently, has been compared to memory B cells [28]. The cell of origin for CLL is likewise dependent upon subtype, with unmutated IGHV CLL compared to pre-germinal center naïve B cells and mutated IGHV CLL compared to post-germinal center memory B cell [10,29,30]. Given the overlap in cell of origin, these distinct B cell cancers may be driven by dysregulation of shared epigenetic mechanisms.
Epigenetic modifications contribute to transcriptional regulation via a range of mechanisms, from structural genome organization to recruitment of transcriptional regulatory factors [31]. The genome is spatially organized into compartments defined by a shared chromatin state and organized into chromatin loops between enhancers and promoters of co-expressed genes within the domain. The domain boundaries and chromatin loops are anchored by CTCF and cohesin proteins [32], [33], [34], [35], [36]. Super enhancers (SEs) are clusters of highly active regulatory elements marked by high levels of active chromatin marks (e.g., H3K27ac), transcriptional regulatory proteins (e.g., Mediator), and master transcription factors (TFs) that regulate high expression of key cell identity genes [37], [38], [39]. These identity-driving factors, sometimes referred to as “core TFs”, form interconnected transcriptional loops that establish and maintain cell-type specific expression programs and disruption of this core transcriptional regulatory circuitry is likely oncogenic [40]. Given these characteristics, it is not surprising that SEs are also enriched in CTCF and cohesin binding to closely associate their transcriptional power with target genes for cell identity, while isolating them from other genes [41], [42], [43]. In many cancers, including BCL, altered genome copy number and/or epigenetic states create de novo SEs or increase the activity of existing SEs that then act to promote oncogenesis [44], [45], [46], [47], [48]. However, it is not known whether FL, DLBCL, and CLL harbor shared or disease-specific SE profiles and how each group contributes to pathogenesis.
To address these outstanding questions, we compared FL, DLBCL, and CLL and normal B lymphocytes to identify shared and distinct epigenetic perturbations that promote oncogenesis. We purified malignant B cells from lymph node and peripheral blood from 52 patients (18 FL, 11 DLBCL, 23 CLL) and isolated normal B-cell subsets (GC centrocytes and centroblasts, naïve and memory B cells) from 36 donor tonsils and subjected them to chromatin immunoprecipitation (ChIP) and sequencing for H3K4me1, H3K9/14ac, and H3K27ac, FAIRE-seq for open chromatin; gene expression and genome copy number were also profiled. These epigenetic studies identified novel SEs that likely promote aberrant BCR signaling and a striking loss of key B-cell transcription factors (TF) and tumor suppressors across BCL subtypes that we connected to the disruption of their linked SEs. Together, these results suggest that a critical subset of B-cell TFs and their cognate SEs form self-regulatory transcriptional feedback loops whose disruption is a shared mechanism driving mature B-cell leukemia/lymphoma.
2. Methods
2.1. Ethics
De-identified peripheral blood and lymph node biopsies were obtained from FL, DLBCL, and CLL patients, and excised tonsils obtained from otherwise healthy tonsillectomy patients (Table 1 and Fig. 1B) seen at Washington University School of Medicine who provided informed consent under IRB-approved protocols (IRB ID #: 201607102 and 201108251) in concordance with the Declaration of Helsinki.
Table 1.
Shows the patient characteristics (age, sex, diagnosis) for B-cell cancer patients in the WUSM study.
| Disease | Number | M/F | Age, years (Median/Range) |
|---|---|---|---|
| Chronic Lymphocytic Leukemia/Lymphoma (CLL) | 23 | 12/11 | 62 (38 - 86) |
| Diffuse Large B Cell Lymphoma (DLBCL) | 11 | 6/5 | 66 (50 - 77) |
| Activated B Cell (ABC) type | 6 | ||
| Germinal Center B cell (GCB) type | 5 | ||
| Follicular Lymphoma (FL) | 18 | 7/11 | 53 (35 - 78) |
| Stage 1-2 | 15 | ||
| Stage >2 | 3 |
Fig. 1.
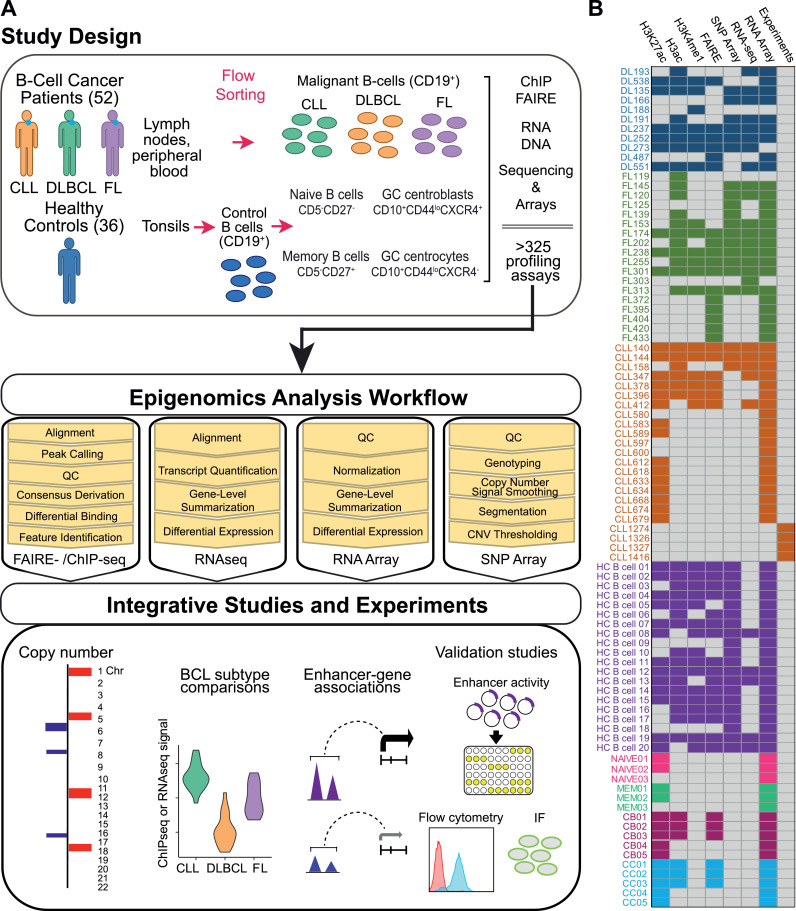
Study design, biospecimens, B-cell purification, molecular profiling, experiments and data analysis workflow. A) Lymph node biopsies and peripheral blood were collected from BCL patients (18 FL, 11 DLBCL, 23 CLL) and tonsils [36] from healthy donors. We purified CD19+ malignant B cells from lymph node biopsies (FL, CLL, DLBCL) or peripheral blood (CLL), and isolated CD19+ normal B-cells from tonsils [20]. From additional tonsils we sorted germinal center (GC) centrocytes (CD19+CD10+CD44loCXCR4−, 5), GC centroblasts (CD19+CD10+CD44loCXCR4+, 5), naive (CD19+CD5−CD27−, 3), and memory (CD19+CD5−CD27+, 3) control B-cell subsets. From these samples, we performed chromatin immunoprecipitation sequencing (ChIPseq, H3K27ac – 51, H3ac – 47, H3K4me1 – 35)), open chromatin profiling (FAIREseq - 45), RNA sequencing (RNAseq - 28) and microarrays [80], and whole genome copy number studies (SNP microarray - 42), totaling 328 high resolution molecular profiling studies. Our Integrative Analysis pipeline compared BCL subtypes to healthy control B-cells and identified copy number alterations, differentially bound enhancers, differentially expressed genes, and enhancer-gene associations. These studies also identified >1300 super-enhancers, many of which correlate with significantly altered expression of neighboring genes in BCL compared to control B-cells. We validated one of these, a novel super-enhancer with high levels of epigenetic activity and expression of two nearby genes, FCMR and PIGR, across BCL subtypes, using luciferase reporter assays and demonstrate high levels of FCMR and PIGR protein in primary BCL cells using flow cytometry and immunofluorescence (IF) staining. B) Heatmap shows assays performed for each sample: healthy control (HC) CD19+ B cells; NAIVE B cells; Memory B cells (MEM); Centroblast (CB); Centrocyte (CC); CLL; DLBCL; FL.
2.2. Human specimen processing
CD19+ B cells were purified from peripheral blood, lymph node or tonsil as in [18]. B-cell subsets isolated: centroblast CD10+CD44loCXCR4+ and centrocyte CD10+CD44loCXCR4− [18], naive CD27−CD5− and memory CD27+CD5− [49] populations. Specifically, CD19+ B cells were purified from lymph node by mechanical dissociation and flushing of the tissue with sort buffer (PBS, 1% FBS, 2 mM EDTA), followed by two rounds of red blood cell lysis (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 10 minutes each. Peripheral blood was subjected to at least four rounds of red blood cell lysis. CD19+ B cells were then purified from PBMCs via an EasySep CD19+ Positive Selection Kit (StemCell Technologies cat. 17854) or using CD19 MicroBeads (Miltenyi Biotec cat. 130-050-301) per the manufacturer's instructions to >90% purity as assessed by flow cytometry (CD19-APC, Miltenyi Biotec cat. 130-110-351). Tonsillar tissues were mechanically disrupted and subjected to red blood cell lysis. B cell subsets were isolated by CD19 MicroBeads and fluorescence activated cell sorting to collect CD10+CD44loCXCR4+ (Centroblast), CD10+CD44loCXCR4− (Centrocyte), CD27-CD5- (Naive), and CD27+CD5- (Memory) populations. Due to yield constraints, some samples were not subjected to all assays.
2.3. ChIP-seq
Crosslinking of 3-5 × 106 cells and chromatin immunoprecipitation for H3K9/K14ac (EMD-Millipore 06-599), H3K27ac (Abcam ab4729), and H3K4me1 (Abcam ab8895) was performed as described [7]. DNA was sequenced by the Washington University Genome Technology Access Center on an Illumina Hi-Seq 2000/2500/3000 to generate 42 or 50 bp single-end reads. Reads were aligned to hg19 with bowtie2 [8] (v2.2.5) using default settings. Reads in ENCODE blacklisted regions [9] were removed with samtools [10] (v1.9). Peaks were called with MACS25 (v2.1.0.20150420) with the parameters –nomodel –shiftsize=150 and input controls. RPKM normalized genome browser tracks were created with deepTools’(11) (v3.3.0) bamCoverage utility with settings –binSize 10 –extendReads 150 –normalizeUsing RPKM and visualized on the UCSC genome browser. ChIPQC [12] (v1.21.0) was used for quality control, and samples with fewer than 8.5% (H3ac), 4.5% (H3K27ac), 2.5% (FAIRE), or 7.5% (H3K4me1) reads in peaks were removed from subsequent analyses. ROSE [13] was used to call super enhancers for each sample with default settings and input controls, and a consensus set was derived using DiffBind [14] (v2.14.0). DiffBind was used to derive consensus peak sets for each histone mark and accessible chromatin, requiring peaks to overlap in at least three samples for each ChIP-seq or FAIRE-seq assay in order to be merged and retained. The consensus peaksets for each assay were then concatenated and merged to derive a list of putative regulatory elements. DiffBind was then used to determine differentially bound/accessible regions between sample groups for each assay. ChIPseeker [15] (v1.22.0) was used to annotate peaks with hg19 UCSC knownGene annotations. Averaged signal heatmaps were generated by merging all bam files for a group with samtools and subsequent visualization of RPKM-normalized reads in EaSeq [16] (v1.111). All bed file manipulations were performed with bedtools2 [17] (v2.29.2).
2.4. RNA-seq and expression microarrays
RNA was isolated from 1-2 × 106 cells from each sample with a Qiagen RNeasy kit (Qiagen cat. 74104). rRNA-depleted (Ribo-Zero, Epicentre, discontinued) libraries were prepared using TruSeq RNA Sample kits with indexed adaptors (Illumina) and subjected to 2 × 100 bp paired-end sequencing on an Illumina Hi-Seq 2000 by the Washington University Genome Technology Access Center. Reads were aligned to hg19 with Gencode v31 annotations using STAR [18] (v2.5.3a). RPKM normalized tracks were created with deepTools'(11) (v3.3.0) bamCoverage utility and visualized on the UCSC genome browser. Read quantification was performed by Salmon [19] (v0.14.1) using Gencode v31 annotations after generating a decoy-aware transcriptome index, and differential gene expression analyses performed with the DESeq2 [20] R package (v1.26.0). The pheatmap R package was used for heatmap generation and associated clustering. Manual gene curation was performed with Genotify (v1.2.2) [21]. For expression microarrays, RNA was purified, amplified, labeled and hybridized as in [18] on Affymetrix Human Gene 2.0ST arrays (cat. 902112) according to manufacturer instructions. Data was analyzed with Applied Biosystems Transcriptome Analysis Console (v4.0.2.15) using RMA normalization, HuGene2.0.na36 annotations, and default settings. Differential expression analysis was performed through the Transcriptome Analysis Console with limma [50]. Clonotyping was performed on raw RNA-seq reads with MiXCR (v3.0.12) [51] with the parameters -starting-material rna -only-productive -receptor-type bcr. The clonotypes were converted to VDJtools format with VDJtools (v1.2.1) [52] Convert functionality with the -S mixcr parameter. Gene usage was calculated with the CalcSegmentUsage utility and TCR segments were removed prior to visualization.
2.5. DNA SNP/copy number microarrays
Genomic DNA from 1 × 106 lymphoma or healthy control B cells isolated as above was purified via a Qiagen DNeasy Blood and Tissue kit (cat. 69504) and hybridized (0.5 ug) to Affymetrix Genome-Wide Human SNP6.0 Array GeneChip microarrays (ThermoFisher Scientific cat. 901150) according to the manufacturer's instructions. Arrays were processed using the Affymetrix Genotyping Console (v4.2) with hg19 na36 annotations. Samples were genotyped and copy number signal derived with regional GC correction and signal smoothing using default parameters and the HapMap reference. Resulting log2 ratios at each marker for each BCL array were concatenated and used as input for circular binary segmentation (CBS) [53] via the DNAcopy (v1.58.0) R package with min.width = 5. Additional visualizations were created with CNVkit (v0.9.3) [54] and the copynumber R package (v1.26.0) [55].
2.6. Pathway enrichment analyses
Gene pathway enrichment analyses were performed on the g:Profiler webserver [56] (version e98_eg45_p14_ce5b097), limiting terms to those with ≥5 and <1000 members and g:SCS multiple testing correction threshold of 0.05. For motif enrichment, term size limits were removed, and the lowest p-value was retained if there were multiple motifs for a given gene.
2.7. Luciferase assays
Putative regulatory elements were PCR-amplified from genomic DNA isolated from Raji (ATCC CCL-86; RRID:CVCL_0511) or LS180 cells (ATCC CCL-187; RRID:CVCL_0397) via phenol chloroform precipitation. Primer sequences are available in Table S6. PCR products were gel-extracted (Qiagen cat. 28706), digested with BamHI (New England BioLabs cat. R3136) and SalI (New England BioLabs cat. R3138), and ligated downstream of luciferase in the pGL3 Promoter Luciferase Reporter vector (Promega cat. E1761) overnight at 23 °C with T4 DNA ligase (New England BioLabs, cat. M0202S). Sanger sequencing confirmed successful cloning. 100 ng of each reporter plasmid and 15 ng of pRL renilla luciferase vector (Promega cat. E2231) were transfected into 20k LS180 cells in each well of a 96 well plate in triplicate with Lipofectamine 2000 (ThermoFisher Scientific cat. 11668019) per the manufacturer's instructions with 100 μL of MEM (ThermoFisher Scientific cat. 11095-080) supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum. 2.5 μg of each reporter plasmid and 50 ng of pRL renilla luciferase vector were nucleofected into 2 × 10^6 Raji cells in 2 mL RPMI-1640 (ThermoFisher Scientific cat. 11875119) in each well of a 6 well plate with a Nucleofector 2b (Lonza cat. AAB-1001) using the Human B Cell Nucleofector kit (Lonza cat. VAPA-1001) and the M-013 program per the manufacturer's instructions. After 24 h incubation at 37 °C with 5% CO2, LS180 media was replaced and luciferase activity read on a Cytation5 plate reader (BioTek cat. 12576) using the Dual-Glo Luciferase Assay System (Promega cat. E2920) according to the manufacturer's instructions. Raji cells were spun down, resuspended in 200 µL of media, and split into 3 wells of a 96 well plate prior to performing the assay. Assays were read in triplicate and performed at least twice. The average ratios between the firefly and renilla luciferase readings for each sample were compared to the average ratio for the empty pGL3-promoter vector to determine relative luciferase.
2.8. Immunofluorescence and confocal microscopy
For immunofluorescence and microscopy, 2 × 105 CLL, MEC1 (DSMZ cat. ACC 497; RRID:CVCL_1870), or HH (ATCC cat. CRL-2015; RRID:CVCL_1280) cells were placed on top of an 18 mm coverslip in one well of a 12 well plate in PBS for 30 min at 37 °C to adhere. LS180 cells were seeded on top of the slip and allowed to attach to the coverslip in typical culture conditions overnight. Primary antibody for PIGR (1:100, ThermoFisher Scientific cat. PA5-22096) was incubated with cells for 2 hours at room temperature. Secondary antibody (1:1000 goat anti-rabbit Alexa Fluor 594 ThermoFisher Scientific cat. A-11037) was incubated with cells for 1 hour at room temperature in the dark. Coverslips were mounted with ProLong Gold with DAPI (ThermoFisher Scientific cat. P36941) overnight before sealing with clear nail polish. Microscopy was performed on an Olympus fluorescence microscope (BX-53) using an ApoN 60X/1.49 NA oil immersion lens or a UPlanS-Apo 100X/1.4 oil immersion lens and cellSens Dimension software. Identical exposure times (DAPI - 10 ms; PIGR - 100 ms) were used for all cell types, and minor brightness adjustments were applied uniformly across images.
2.9. Flow cytometry
For flow cytometry, 2 × 105 freshly isolated PBMCs from peripheral blood CLL samples were blocked with FcX Trustain (BioLegend cat. 422301) for 5 min in Sort Buffer (PBS, 2% FBS, 10 mM EDTA) before incubation with 1 µg anti-FCMR antibody (Novus Biologicals cat. H00009214-M01) or 1 μg mouse IgG2b isotype control (abcam cat. ab170192) for 30 min at 25 °C. Cells were washed with sort buffer and stained with goat anti-mouse IgG BrilliantViolet421 (BioLegend cat. 405317) and anti-CD19 APC (Miltenyi Biotec cat. 130-114-168) for 30 min. After washing, flow cytometry was performed on a modified Becton Dickinson FACScan flow cytometer and analyzed using FlowJo (v9.9.5) software. At least 3000 events were recorded for each sample.
2.10. Statistics
Unless otherwise indicated, statistical testing was performed with GraphPad Prism (v8.3.0).
Role of the funding sources
The study sponsors had no role in the study design; collection, analysis, and interpretation of data; in writing of the manuscript; or in the decision to submit the paper for publication.
3. Results
3.1. Study overview and design
To generate a comprehensive map of the gene regulatory landscape in normal and malignant mature B-cell lymphoma/leukemia (BCL), we surveyed the active epigenomes and transcriptomes of primary biospecimens from FL, DLBCL, and CLL patients at the Washington University School of Medicine and compared them to normal B-cell subsets purified from donor tonsils (Fig. 1A). Clinical characteristics for BCL patients are summarized in Table 1. Briefly, we collected lymph node biopsies and peripheral blood from BCL patients (18 FL, 11 DLBCL, 23 CLL) and 36 tonsils from healthy donors. We purified CD19+ malignant B cells from lymph node biopsies (FL, CLL, DLBCL) or peripheral blood (CLL). We isolated CD19+ control B cells from tonsils and further sorted to obtain germinal center (GC) centrocytes (CD19+CD10+CD44loCXCR4−) and GC centroblasts (CD19+CD10+CD44loCXCR4+), naive (CD19+CD5−CD27−), and memory (CD19+CD5−CD27+) B-cell subsets [18,57]. Fig. 1B shows each sample and the assays performed. In total, we performed 328 high resolution molecular profiling studies of these samples.
We mapped the epigenomes of BCL and normal B cell samples using chromatin immunoprecipitation and sequencing (ChIP-seq) for H3K9/K14ac (H3ac) and H3K27ac, which mark active enhancers and actively transcribed genic regions, H3K4me1, which marks active and poised enhancers, and FAIRE-seq [58] to identify regions of open chromatin, which are located within active enhancers and promoters and indicate the binding of transcriptional regulatory factors. Whole transcriptome gene expression was profiled with RNA-seq and RNA microarray (Fig. 1). Immunoglobulin heavy and light chain clonotype analysis [52] of RNA-seq data revealed a single dominant clone in 87% (20/23 tested) of BCL samples compared to a polyclonal pattern in healthy control B cells, indicating substantial disease burden (Fig. S1 and Table S1). Clonotype analysis corroborated pathology review, which showed that all biopsies harbored >80% malignant B cells. We identified genome-wide copy number alterations in BCL samples using SNP microarray analysis. Integrative analysis of these ‘omics datasets compared BCL subtypes with healthy control B-cell subsets, mapped enhancer - gene associations, and identified a novel super-enhancer for experimental studies (Fig. 1). This extensive dataset enabled us to identify and correlate epigenetic, transcriptomic, and genetic differences and commonalities between three different mature B-cell neoplasms and normal B-cell subsets.
3.2. Epigenetic differences linked to gene expression changes distinguish BCL subtypes and healthy control B cells
To identify distinct chromatin and gene expression patterns for BCL subtypes, we developed an integrative bioinformatics pipeline to define putative regulatory elements across the genome and correlate changes in ChIP-seq signal or chromatin accessibility (FAIRE-seq) at these elements with expression of nearby genes (Fig. 1). We used MACS2 [59] to call peaks (q-value < 0.05) in all ChIP and FAIRE-seq datasets. Overlapping peaks called in at least three samples for each histone mark (ChIP-seq) or FAIRE-seq were merged. All ChIP-seq and FAIRE-seq datasets were then merged to derive a consensus peak set (Table S2). We used ChIPseeker [60] to annotate this consensus discovery set based on genomic location, segregating exon-overlapping peaks from putative regulatory elements (promoter, intronic, or intergenic peaks). Putative promoter and enhancer elements (intronic or intergenic peaks) were compared to the GeneHancer [61] database, a set of conserved regulatory elements generated from dozens of cell lines/tissues, including B-cell lines.
Our comparisons demonstrated that 76.8% of the putative B-cell enhancers and promoters (57,771/75,204) overlapped one or more GeneHancer regulatory elements (Fig. S2A). The remaining 17,433 comprise putative regulatory elements that are unique to primary mature B cells or to mature B-cell cancers, as these cell types are not represented in the GeneHancer database. We next compared the epigenetic landscapes in all BCL samples to healthy control CD19+ B cells to define differentially bound/accessible (DB) regulatory elements (FDR < 0.01; log2 fold-change > 2) using the DiffBind R package [62] for each histone mark (ChIP-seq) and for FAIRE-seq (Table S2). We evaluated the co-occurrence of DB peaks to identify unique and overlapping regions for these discrete gene regulatory modifications. As shown in Fig. 2A, H3K4me1 positive regions have the greatest proportional overlap with other histone marks and open chromatin (67%), as expected for this marker of both poised and active enhancers. Differentially accessible peaks (FAIREseq) had the greatest proportion of unique peaks, likely due to the fact that open or closed chromatin in these regions is often associated with reduced nucleosome density for the former and repressive chromatin for the latter, either of which would reduce the amount of H3K27ac, H3K9/14ac, and H3K4me1 in these regions [31]. The coincidence of active marks H3K27ac and H3K9/14ac in differentially bound peaks (1290) was high, as was the coincidence of H3K27ac, H3K9/14ac, and H3K4me1 (691), indicative of differentially active enhancers in BCL.
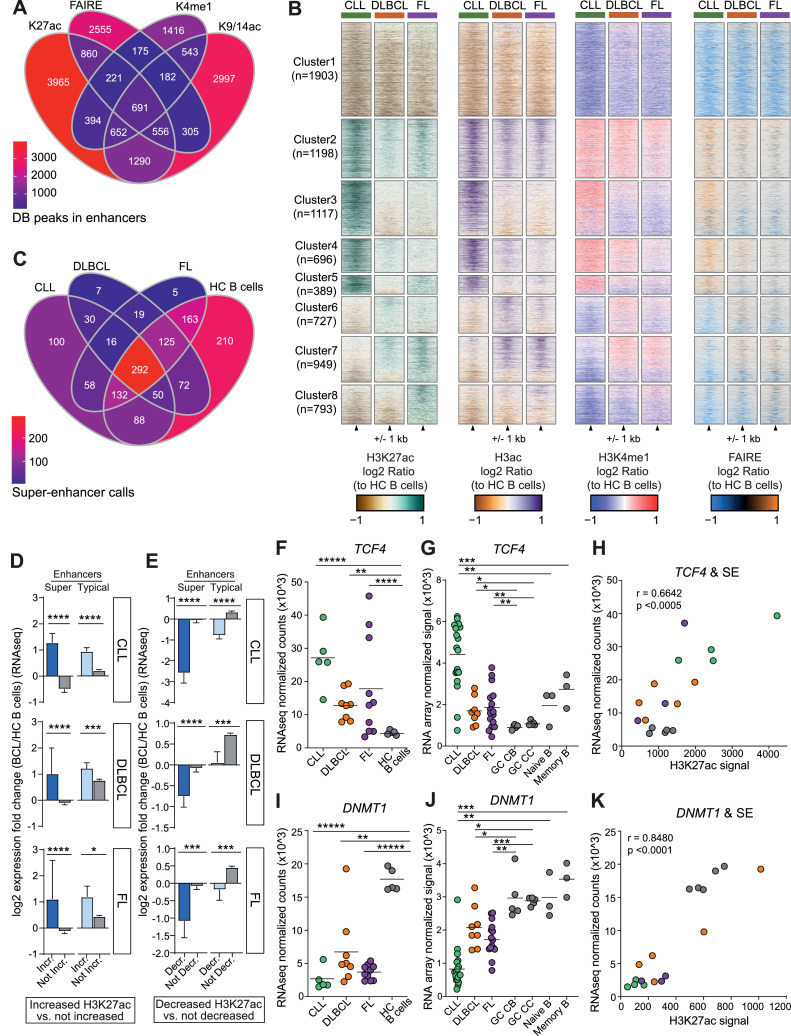
Fig. 2.
Super-enhancers are selectively gained and lost at key transcriptional regulators in B-cell cancers. A) Venn diagram shows the overlap of differentially accessible (FAIREseq) or differentially bound chromatin (H3K27ac-, H3ac-, H3K4me1- ChIPseq) (absolute log2 fold change > 2, FDR < 0.01) in WUSM BCL compared to healthy control CD19+ B-cells sorted from tonsils. B) Heatmaps show the log2 ratio of signal for H3K27ac-, H3ac-, H3K4me1-, and FAIRE-seq at differentially bound (as in A) H3K27ac peaks between all BCL subtypes and hc B-cells. C) Venn diagram shows the shared and unique super-enhancer (SE) calls in BCL subtypes and healthy control (HC) B-cells. D) Bar graphs show RNA-seq log2 fold changes of genes near enhancers and SE with increased H3K27ac signal (log2 fold change > 1, FDR < 0.01) between BCL subtypes and healthy control B-cells compared to genes near enhancers and SE that are not increased, grouped by BCL subtype. E) The same as (D), but for genes near enhancers and SE with decreased H3K27ac signal (log2 fold change < 2, FDR < 0.01). Mann Whitney test, ns not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. Mean with 95% confidence intervals shown. F-K) SE-associated genes with significantly increased (F-H) or decreased (I-K) expression and differentially bound SEs in BCL subtypes. Normalized RNA-seq TCF4 (F) or DNMT1 (I) gene counts grouped by BCL subtype and HC B-cells (DESeq2, ** adj p < 0.01, **** adj p < 0.0001, ***** adj p < 1e-5). Normalized RNA microarray signal for TCF4 (G) or DNMT1 (J) grouped by BCL subtype and healthy control germinal center (GC) centroblasts (CB), centrocytes (CC), naive and memory B-cells (limma * adj p < 0.05, ** adj p < 0.01, *** adj p < 0.001). Scatter plot shows significant correlation of TCF4 (H) or DNMT1 (K) expression (RNAseq) with H3K27ac levels at the associated SE (Spearman correlation).
To identify epigenetic regulatory elements with altered activity that is distinct or shared to each BCL subtype, we performed pairwise comparisons of each B-cell cancer type with healthy control B cells to define DB elements as before (FDR < 0.01; absolute log2 fold-change > 1). We focused on H3K27ac as the baseline set because this modification marks active enhancers and because we had H3K27ac ChIPseq data for the greatest number of BCL and healthy control B-cell subsets (Fig. S1). To ensure that the DB elements are cancer related, and not due to comparison with an inappropriate normal B-cell subset, we compared the chromatin profiles of each of the normal B-cell subsets: centrocytes [5], centroblasts [5], naïve [3], memory [3], and total CD19+ B cells [20] purified from tonsils. We found that 97% or greater of their profile peaks overlap, except for memory B cells (75%), and importantly, only 0.27% of the non-overlapping peaks (38/14168) were differentially bound/accessible compared to BCL samples. Thus, we concluded that, for identification of DB elements in BCL, the chromatin profiles of the healthy control CD19+ B cells effectively represent the epigenetic landscapes of the other B-cell subsets. Overall, chromatin accessibility and all other histone modification profiles exhibit similar signal patterns at H3K27ac DB elements in each of the BCL subtypes and are highly correlative (Pearson correlation r = 0.843-0.949, n = 7,772, Fig. S2B). Notably, DB elements for all histone modifications and chromatin accessibility occur more frequently than expected at enhancers (Chi-square test; ****, P < 0.0001; Fig. S2C).
CLL, FL and DLBCL subtypes exhibit distinct patterns of chromatin accessibility and histone modification compared to each other and to healthy control B-cell subsets. Log2 ratio heatmaps of histone marks and chromatin accessibility show eight distinct clusters of regulatory elements with similar patterns of epigenetic activity (Fig. 2B). The largest single cluster (1, n=1903) contains elements with decreased levels of all histone marks and decreased chromatin accessibility for all BCL subtypes. Cluster 2 is the next largest (n=1198) and contains elements with increased levels of chromatin modification and accessibility across BCL subtypes. Clusters 3–5 include elements increased in one or two subtypes, while elements in clusters 6–8 are decreased in one or two subtypes.
3.3. Key BCL driver genes are marked by super-enhancers
Super-enhancers (SE) are clusters of enhancers that control the expression of key cell identity and developmental genes [38] and arise during pathogenic events to drive oncogene expression [37,47,63,64]. We and others have shown that SEs contribute to B-cell cancers [18,44,47,65], which led us to further investigate SE dynamics in this dataset. We used the ROSE [37] tool to identify SEs from H3K27ac data for each BCL and HC B cell sample and derived a consensus set (n = 1367) by merging those that overlapped; nearly all (1254/1367, 91.7%) overlapped a previously described SE by at least 25% (dbSUPER [66] database). The majority of SEs were detected in multiple BCL subtypes and HC B cells; a minority (23.6%, 322/1367) were identified in only one. BCL subtypes were more similar to each other than to HC B cells, which harbored 210 SEs that were not detected in any BCL sample (Fig. 2C). Comparison of BCL to healthy control B cells revealed that 28.9% (395/1367) of SEs had significantly different levels of H3K27ac (absolute log2 fold change > 1; FDR < 0.01, DiffBind [62]; Table S3). To evaluate the effect of enhancer chromatin changes on gene expression, we divided total enhancers into DB increased or decreased SE and typical enhancer groups. We calculated the log2 fold change in gene expression (RNA-seq) for each BCL subtype relative to healthy control B cells and identified the gene most proximal to each DB element. As expected, genes near DB elements with increased histone acetylation in BCL groups exhibited a corresponding significant increase in expression fold change, with greater than two-fold (linear) increases in all comparisons. Increased SEs and typical enhancers had similar impacts on gene expression (Fig. 2D). However, we were surprised to find that decreased acetylation of SEs in BCL compared to healthy control B cells was associated with a much larger change in expression - up to 5-fold (linear) decrease (Fig. 2E). Taken together, these results demonstrate that BCL subtypes have distinct epigenetic changes compared to each other and to healthy control B cells, and that these differences correlate with expression changes in nearby genes. Most striking was that loss of SE acetylation had a greater impact on gene expression than increased acetylation. These findings suggest that loss of activity at key SEs may be an important contributor to BCL pathogenesis.
We next asked whether these DB SEs are associated with B-cell lymphoma oncogenes or tumor suppressor genes. Increased expression of TCF4 (E2-2), a basic helix-loop-helix transcription factor a critical for germinal center B and plasma cell development [67,68], has been reported in FL and DLBCL and likely contributes to lymphoma pathogenesis [69,70]. Here we found that TCF4 expression is significantly elevated (2.5- to 6-fold) in all BCL subtypes compared to all healthy control B-cell subsets (germinal center centrocytes and centroblasts, naïve and memory B cells) (Fig. 2F-G). Levels of K27ac within the TCF4 SEs are significantly correlated with expression level (r=0.6642, p<0.0005) (Fig. 2H). DNMT1 is a maintenance DNA methyltransferase whose downregulation has been implicated in EBV latency and is associated with worse outcomes in DLBCL [71], [72], [73]. We detected significantly lower (2- to 10-fold) DNMT1 expression in all BCL subtypes compared to all healthy control B-cell subsets (Fig. 2I-J). In addition, levels of H3K27ac within the DNMT1 SE significantly correlate with expression across all samples (r=0.8480, p<0.0001) (Fig. 2K). Together, these studies demonstrate that altered SE activity is recurrent across FL/DLBCL and CLL and these changes correlate with expression changes in neighboring genes with likely roles in lymphoma pathogenesis.
3.4. BCLs harbor a novel SE for immunoglobulin receptor genes FCMR and PIGR
We next used our unique approach for comparing gene regulation across multiple BCL subtypes to search for a novel BCL SE, reasoning that such an element may represent a shared oncogenic pathway in these disparate B-cell neoplasms. We identified an SE overlapping the immunoglobulin (Ig) receptor genes FCMR and PIGR that harbored several constituent elements with high levels of active and open chromatin (FAIRE/H3K27ac ChIP-seq) across all three BCL subtypes compared with healthy control B cells (Fig. 3A). A subset of BCLs harbored genome copy number gains in this region (2 FL, 2 DLBCL). In comparison, PIGR is normally expressed in intestinal mucosal epithelium and as expected, the PIGR gene body and flanking regions harbored active chromatin in small intestine (DNase/H3K27ac ChIP-seq, Roadmap Epigenomics Project17).
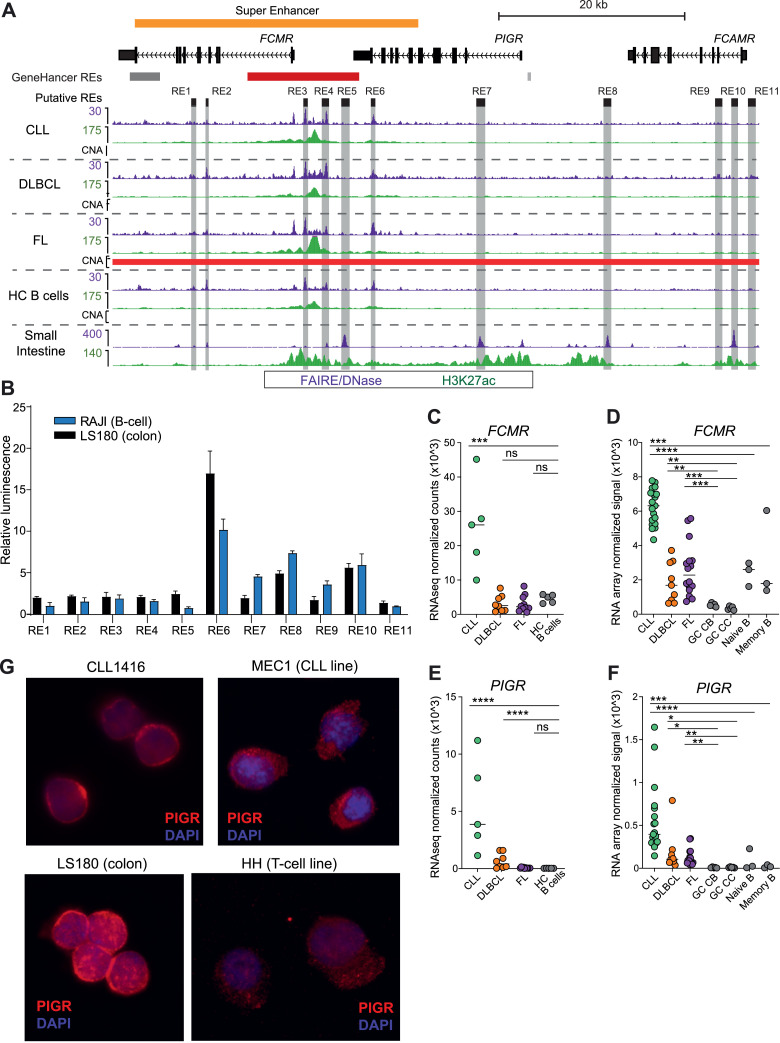
Fig. 3.
A novel super-enhancer associated with dysregulated high expression of PIGR and FCMR mRNA and protein. A) Genome browser screenshot shows signal for histone modifications (ChIPseq) and chromatin accessibility (FAIREseq - WUSM samples; DNase-seq - small intestine sample from Roadmap Epigenomic Project) for the FCMR/PIGR locus. Regulatory elements that were tested in luciferase assays are highlighted in gray. GeneHancer regulatory elements (red - promoter; gray - enhancer; darker color indicates higher confidence) and the identified super-enhancer are also shown. All tracks are RPKM normalized. Regions of copy number alteration (CNA) are shown by a red bar (amplifications identified in FL samples). B) Luciferase reporter assay results for all regulatory elements shown in (A). Assays were read in triplicate and performed at least twice. Mean with standard deviation shown. C-F) Expression of FCMR (C-D) and PIGR (E-F) determined by RNAseq (C&E) and RNA microarray (D&F) for BCL subtypes and healthy control B cells and B cell subsets (germinal center (GC) centroblasts (CB), centrocytes (CC), naive and memory B cells). RNAseq analyzed by DESeq2; microarrays analyzed by limma. * adj p > 0.05, ** adj p < 0.01, *** adj p < 0.001, **** adj p < 0.0001, ns not significant. G) Immunofluorescence microscopy for PIGR in purified primary CLL B-cells, MEC1 (CLL cell line), LS180 (colorectal cancer cell line), and HH (T-cell lymphoma cell line).
Because FCMR and PIGR expression is mutually exclusive in immune and mucosal epithelial cells, respectively, we reasoned that elements within the FCMR/PIGR SE may differentially regulate these genes in each cell type. We identified 6 putative component elements and 5 flanking potential regulatory elements based on overlapping accessible chromatin, H3K27ac, H3ac, and H3K4me1 in BCLs, healthy control B cells, and small intestine (Fig. 3A). To directly compare these elements in B cells and epithelial cells, we performed luciferase reporter assays in a B-cell line (Raji) and a colorectal cell line (LS180). Fig. 3B shows that four elements have robust enhancer activity in at least one cell type (REs 6, 7, 8, and 10). An element located within the super enhancer at the 3’ end of PIGR (RE6) has the greatest enhancer activity in both cell types, with 10- or 17-fold higher relative luminescence compared to the empty vector in B or colorectal cells, respectively. The relative luciferase activity induced by these elements in B cells versus colorectal cells did not always correlate with the relative levels of active or open chromatin detected FAIRE/DNase/ChIP-seq (e.g., RE6 versus RE7), suggesting that other factors that are absent from luciferase reporter plasmids, such as chromatin modifications and 3D genome organization [74], may be responsible for the tissue-specific expression of FCMR and PIGR. Indeed, there are several CTCF peaks flanking the FCMR promoter and the dense cluster of enhancers (RE2 - RE6) (Fig. S3A). CTCF is a DNA binding protein that forms chromatin loops that facilitate or inhibit promoter-enhancer or promoter-silencer interactions19. Moreover, high resolution Hi-C data from GM12878 B cells shows at least 6 chromatin interaction contact points in the region between RE1 in FCMR and RE6 in PIGR (Fig. S3A) [75,76]. Together, these data suggest that the strong enhancer cluster between FCMR and PIGR is tethered closely to the FCMR promoter in B cells. Overall, these results demonstrate that several enhancers throughout the FCMR/PIGR locus have robust regulatory activity in both cell types and suggest that 3D genome organization and chromatin modifications are likely responsible for the tissue-specific expression of FCMR and PIGR in non-malignant B cells.
In support of a functional role for this SE, the expression levels of FCMR were strikingly upregulated in CLL, and PIGR expression was significantly elevated in both CLL and DLBCL compared to healthy control B-cell subsets (Fig. 3C-F). FCMR is a high-affinity IgM receptor expressed by T and B lymphocytes, as well as NK cells. PIGR is a high-affinity IgA/lower-affinity IgM receptor normally expressed only in mucosal epithelial cells, where it transports Ig proteins produced by B cells from the basolateral surface across the cell to be secreted at the apical surface into the lumen of the gut or respiratory tract. FCMR overexpression in CLL has been previously reported [77,78], though its role in CLL pathogenicity remains unresolved. To our knowledge, PIGR expression has not been reported previously in normal or malignant human B cells, though it was expressed in a murine B-cell lymphoma line [79].
We validated and extended these findings in two additional cohorts. A recent study comparing RNA-seq of CLL to naive and memory B cells [22] shows similar upregulation of FCMR and PIGR as seen in our study (Fig. S3B-C). We also compared our dataset to a cohort that includes CLL, DLBCL, and FL, as well as mantle cell (MCL), mucosa-associated (MALT), and nodal marginal zone (NMZL) lymphomas [80]. Consistent with our study, FCMR expression is significantly increased in CLL and MCL relative to whole tonsil and lymph node samples (Fig. S3D). The expression in DLBCL samples is relatively lower, but this may be due to the ratio of GCB to ABC/non-GCB samples, which is not reported [80]. Our dataset contains an equal number of each DLBCL subtype and shows a trend toward a higher level of FCMR expression in ABC/non-GCB DLBCL samples (median ABC = 10.91, median GCB = 9.81, P = n.s.). Also consistent with our data, PIGR expression is significantly higher in CLL and DLBCL, as well as in MCL and MALT lymphomas (Fig. S3E). Together, these results demonstrate that elevated expression of FCMR and PIGR is shared across diverse mature B-cell malignancies and suggest a potential role for one or both proteins in BCL pathogenesis.
Having demonstrated significantly elevated FCMR and PIGR mRNA in BCL, we sought to validate this finding at the protein level. Using flow cytometry, we show high levels of FCMR surface expression in CD19+ B cells from three primary CLL samples (Fig. S3F), which is consistent with previous reports [77,78]. To evaluate PIGR, we performed immunofluorescence followed by confocal microscopy and demonstrated high levels of PIGR in CD19+ primary CLL cells and a CLL cell line (MEC1). As expected, PIGR expression was also high in a colorectal (mucosal epithelial) cell line (LS180), whereas expression was substantially lower in a T cell lymphoma line (HH) (Fig. 3G).
Taken together, these data demonstrate the complex interplay of epigenetic modification, genome conformation, and transcriptional regulators in the tissue-specific regulation of FCMR and PIGR. All of these control mechanisms are known to be disrupted in lymphoma [3,81] and may contribute to the deregulated expression observed in BCL.
3.5. A subset of genome copy number alterations (CNA) exhibit corresponding epigenetic changes
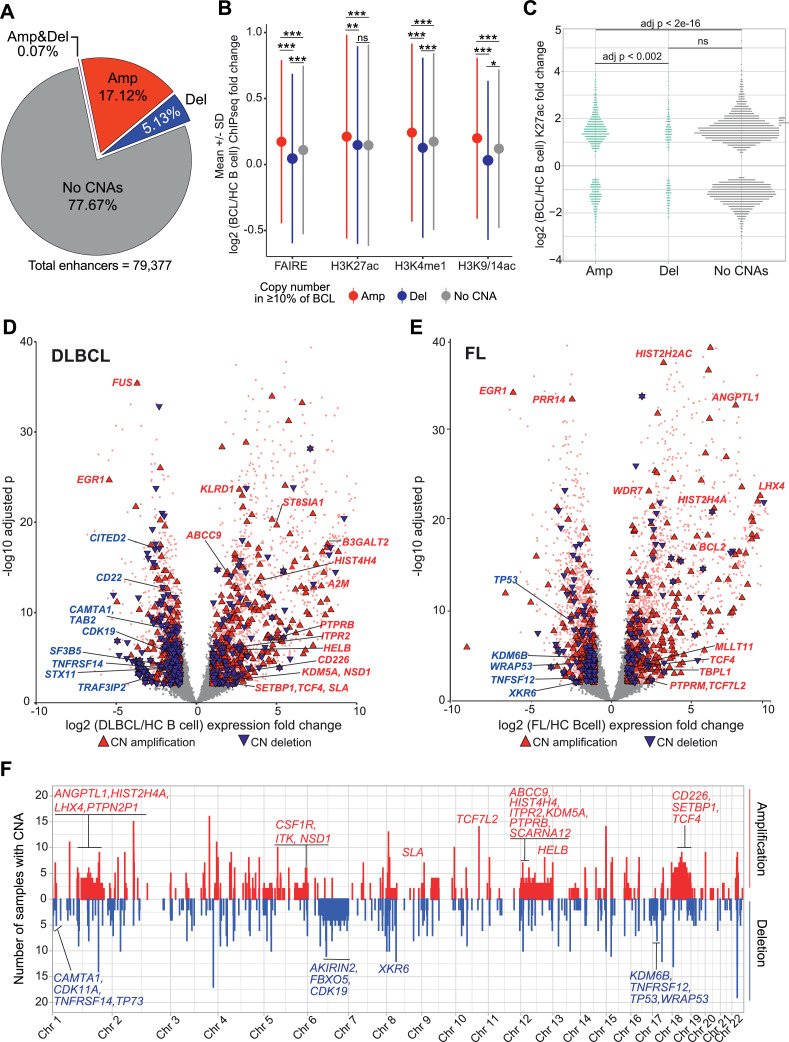
Alterations in genome copy number are a major driver of oncogenesis, including the amplification of oncogenes and deletion of tumor suppressor genes. We reasoned that gain or loss of genome regulatory regions could similarly impact the expression of key lymphoma pathogenesis genes. To identify copy number alterations, we compared purified B cells from DLBCL, FL, and CLL samples to healthy control B cells or a composite healthy control reference human genome using SNP microarrays. Using a log2 copy number fold change cutoff of +/− 0.2, we identified a range of 135 to 4907 (median 1443.5) regions of copy number alteration (CNA) in BCL samples (Table S4). We next evaluated the overlap of CNA with enhancers (Fig. 2) and found that 17.12% of enhancers overlap amplifications, 5.13% overlap deletions, 0.07% overlap deletions and amplifications in different samples, and the majority of enhancers, 77.67%, do not overlap a CNA (Fig. 4A). Percent overlaps were similar for CNAs with SEs (Fig. S4A). To determine if regulatory elements with CNA have altered chromatin activity that is consistent with the CNA type, we compared the log2 fold change of BCL versus healthy control B cells for elements overlapping amplifications, deletions, or no CNA, requiring that the CNA be detected in at least 10% of BCL samples. For each chromatin mark (H3K27ac, K3K4me1, K3K9ac) and open chromatin (FAIRE), those overlapping amplifications had significantly higher mean levels of activity and those overlapping deletions had significantly lower mean levels of activity compared to those without CNA overlap (Fig. 4B), with the exception of H3K27ac in elements overlapping deletions. The lack of significant difference could be due to the relatively lower number of DB elements overlapping deletions (Fig. 4C).
Fig. 4.
A subset of genome copy number alterations in BCL have corresponding epigenetic and expression changes. A) Pie chart shows the percentage of enhancers containing genome copy number alterations (CNA) in at least 10% of BCL samples. Amp - amplification, Del - deletion, Amp&Del - amplification or deletion detected in the same region in different samples. B) Mean +/− standard deviation (SD) of log2 fold change of BCL versus healthy control (HC) B cells for open chromatin (FAIREseq) and epigenetic marks (H3K27ac, H3ac, H3K4me1 ChIPseq) in enhancers containing CNA or not. Letters above SD lines indicate statistical comparator group (A - amplification, D - deletion, N - No CNA); superscript indicates adjusted p value (* p < 1e−04, ** p < 1e−5, *** p < 2e−16, ns not significant; one-way ANOVA with Tukey HSD). C) Plot shows the K27ac log2 fold change of BCL versus HC B cells of differentially bound (DB) enhancers with overlapping Amp, Del, or no CNA. Statistics as in (B). D) Volcano plot shows log2 fold change and -log10 adjusted p value for expression (RNAseq) in DLBCL versus HC B cells. Differentially expressed genes (absolute log2 fold change >1 and adjusted p < 0.01) are pink. Genes with differential expression and CNA are highlighted by text color and triangle shape and color: red/point up = amplification, blue/point down = deletion. Not differentially expressed and no CNA = grey. E) Volcano plot as in (D) for FL versus healthy control B-cells. F) Whole genome plot of number of BCL samples with copy number amplification (red) or deletion (blue) in 100kb bins. Differentially expressed genes (from D or E) with CNA in > 10% of BCL samples are labeled on the upper panel if they overlap an amplified region and on the lower panel for deleted regions. Gene label color indicates if expression is increased (red) or decreased (blue) in BCL relative to HC B cells.
Genome copy number alterations are sometimes associated with a corresponding change in the expression of genes within the altered region [82]. We evaluated this for genes within CNA present in at least 10% of samples, separated by BCL subtype. Using cutoffs for significantly different gene expression (adjust p < 0.01, absolute log2 fold-change >1, DEseq2), we determined that expression fold change corresponds to CNA direction (FC increase - amplification; FC decrease – deletion) for 50% to 61.5% of differentially expressed genes for each BCL subtype and CNA type. Simple cutoffs do not convey the magnitude of fold change expression difference, however. To evaluate this, we generated volcano plots and highlighted genes in CNAs with triangles with point direction and color indicating gain (up, red) or loss (down, blue). As shown in Figs. 4D-E and S4B genes overlapping genome amplifications had increased fold change and those with deletions had decreased fold change, though many genes with non-corresponding gene expression and CNA were also seen. Several genes with known roles in B-cell lymphoma demonstrate corresponding expression and copy number changes in the volcano plots, including BCL2, TNFRSF14, A2M, TCF4, and TP53. Multiple other transcriptional regulators, epigenetic factors, and DNA repair factors also show corresponding changes, such as KDM5A, KDM5B, SETBP1, NSD1, TRAF3IP2, TP73, WRAP53, and several histone genes. Many of these genes are in large regions of CNA, such as whole chromosome amplification (chr 12, chr 18) or deletion (chr 6), but some are in relatively small regions, such as NSD1 in chromosome 5 and TCF7L2 in chromosome 10 (Fig. 4F). These data demonstrate that genome copy number change is likely responsible for some amount of transcriptional dysregulation in B-cell lymphoma, but that other mechanisms play a substantial role in the altered expression of most genes.
3.6. Dysregulated transcription factors exhibit corresponding epigenetic changes in differentially bound enhancers
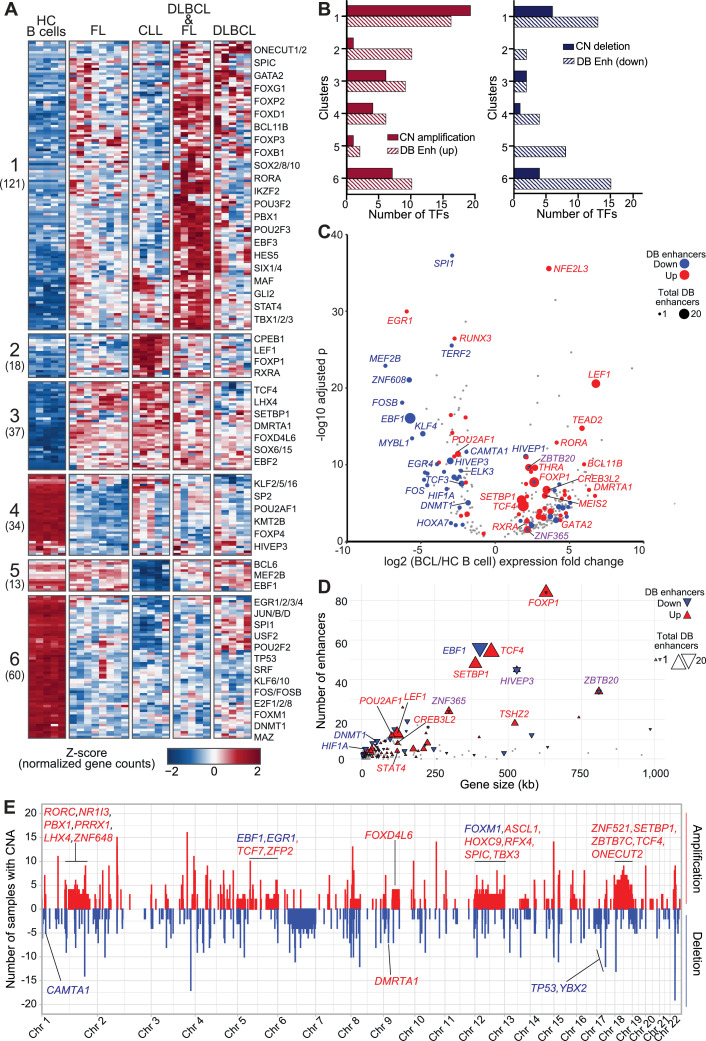
Transcriptional regulation is a carefully choreographed dance involving genome regulatory elements, chromatin landscapes, and a diverse set of protein modulators. Key among the last are certain transcription factors that are known to be oncogenic in hematopoietic cells [7,83]. With this in mind, we performed a comprehensive analysis of differentially expressed (DE) transcription factors (TF) to identify perturbed regulatory circuits that may contribute to lymphoma pathogenesis. Limiting our analysis to 1639 expertly curated TFs [84], we found 283 were differentially expressed (adjusted p-value < 0.01; absolute log2 fold change > 2) in at least one BCL subtype compared to healthy control B-cell subsets (DE TFs). Hierarchical clustering of RNA-seq data for these DE TFs defined 6 clusters of distinct expression profiles that separate BCL subtypes and healthy control B cells, with some overlap of FL and DLBCL samples. The two DLBCL groups did not separate GCB and non-GCB subtypes, which were confirmed by pathology review and by RNAseq (BCL6, IRF4/MUM1, CD10, LMO2), but rather had an equal number of non-GCB in each group (2 non-GCB, 3 GCB), suggesting that the differences in TF expression profiles transcend the GC/ABC designations (Fig. 5A and Table S5). The 176 DE TFs in Clusters 1-3 were more highly expressed in all BCL subtypes compared to healthy control B cells, with CLL and DLBCL groups exhibiting higher expression in Clusters 1 and 2, respectively. In contrast, the expression of 107 DE TF genes in Clusters 4–6 was significantly lower in one or more BCL subtypes compared to healthy control B cells, with Cluster 4 TFs more downregulated in FL and Cluster 5 more downregulated in CLL. Cluster 6 TFs were uniformly downregulated across BCL subtypes. This unbiased transcriptome analysis revealed genes that were also identified by epigenetic and CNA analyses above, such as SETBP1, TCF4, and TP53 (Figs. 2F-H, 4D-F). To evaluate the contribution of CNA and epigenetic regulation to the significant transcriptional changes observed in these DE TFs, we compared the number of TF genes in CNA with the number that overlap with differentially bound enhancers (adjusted p-value < 0.01; absolute log2 fold change > 1). Fig. 5B shows that, for each cluster, a greater number of TFs overlap differentially bound enhancers than overlap CNA.
Fig. 5.
BCL-altered transcription factor expression profiles associated with genome copy number alterations and differentially bound enhancers. A) Heatmap shows normalized gene count z-scores (RNA-seq) for transcription factors (TF) differentially expressed (adjusted p-value < 0.01, absolute log2 fold change > 1, n = 283) in at least one BCL subtype compared to healthy control (HC) B-cells. Six clusters of distinct expression profiles were detected; the number of genes is shown beneath each cluster label. B) Bar chart show the number of TFs with copy number alterations and differentially bound (DB) enhancers in each cluster. (Left) Copy number (CN) amplified, DB increased H3K27ac in BCL (DB up); (Right) CN deletion, DB decreased H3K27ac in BCL (DB down). C) Volcano plot shows log2 fold change and -log10 adjusted p value for expression of TFs (as in A) in BCL versus HC B cells. TF genes with overlapping DB enhancers are highlighted by color (red = increased, blue = decreased, grey = unchanged level of histone acetylation) and size (corresponding to the number of DB enhancers). Color of the gene label corresponds to the status of the overlapping DB enhancers (red = all increased, blue = all decreased, purple = both increased and decreased DB enhancers overlap). D) XY plot shows the number of total enhancers overlapping each TF (Y axis) by TF gene size in kilobases (kb) (X axis). Size and colors as in (C). E) Whole genome frequency plot of number of BCL samples with copy number amplification (red) or deletion (blue) (100kb bins). TFs (from A) with at least 3 samples with overlapping CN alterations (CNA) are labeled on the upper panel if they overlap an amplified region and on the lower panel for deleted regions. Gene label color indicates if expression is increased (red) or decreased (blue) in BCL relative to HC B cells.
There are many more enhancer elements throughout the genome than there are genes, and single genes are often controlled by multiple enhancers [85]. We reasoned that the large changes in expression of these DE TFs may be due to the transcriptional regulatory impact of multiple DB enhancers. The volcano plot in Fig. 5C demonstrates that many of the TFs overlap with up to 20 or more DB enhancers that largely correspond with the gene's expression change. Of those with increased expression and DB enhancers, 33 had increased enhancers, 12 had decreased, and 3 had both increased and decreased - though more increased than decreased. Of those with decreased expression and DB enhancers, 28 had decreased enhancers and 12 had increased. Because this analysis required overlap of enhancers with the TF genes, the largest genes had the greatest number of overlapping and DB enhancers, though some smaller genes also had similar numbers (Fig. 5D).
Pathway enrichment analyses revealed that Clusters 1-3, which exhibit significantly higher expression in BCL, are enriched in TFs involved in transcriptional regulation, WNT and beta-catenin signaling, and pathways downstream of key TFs such as TCF/LEF and RUNX families, suggesting that altered expression of multiple TFs within shared pathways reinforces aberrant regulation of pathway target genes. Clusters 4-6, with significantly lower expression in BCL, are enriched in TFs associated with cellular senescence and TP53-dependent apoptosis pathways. Cluster 6 in particular contains many known tumor suppressors and TFs crucial for B-cell development and differentiation, including TP53 [7,21], EGR1/2/3/4 [86], RUNX3 [87], and SPI1(PU.1) [88,89], among others (Fig. S5 and Table S6).
3.7. Lymphoma oncogenes and tumor-suppressors have self-targeting transcriptional feedback programs
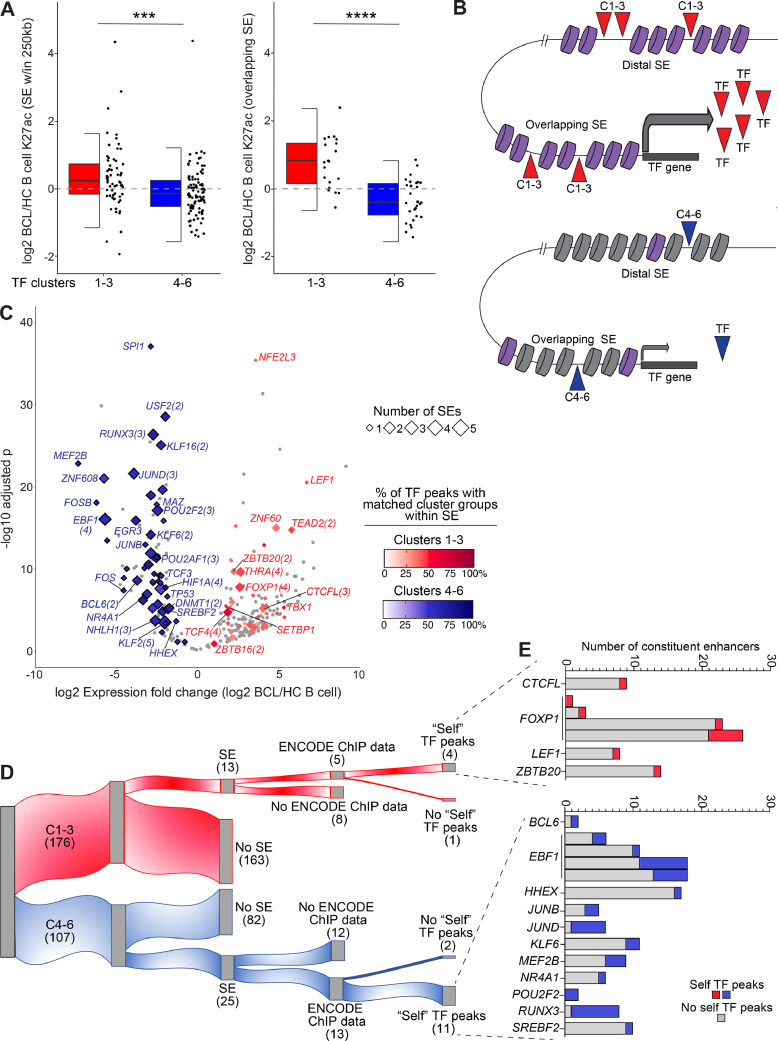
Having determined that many BCL-dysregulated TFs have multiple DB conventional enhancers overlapping the genes, we reasoned that SEs may also control their transcription. We allowed a distance of up to 250kb from genes to SEs, since SEs are often located many kilobases or even megabases away from their gene targets [37,38,47]. We identified 152 SEs located within 250kb of the 283 TFs in Clusters 1-6. Of these, 59 SE were associated with Cluster 1-3 TFs, which are highly expressed in BCL, and 86 SEs were associated with Cluster 4-6 TFs, which are significantly downregulated in BCL. Notably, there were fewer SEs associated with Cluster 1-3 TFs, even though there are a greater number of TFs in Clusters 1-3: 59 SEs and 40/176 Cluster 1-3 TFs vs 86 SEs and 54/107 Cluster 4-6 TFs). We next performed a similar analysis with SEs as we did with DB enhancers (Fig. 5). Compared to enhancers (Fig. 5D), the number of SEs was less concordant with gene size, likely due to the much larger size of SEs and the flanking distance allowed (Fig. S6A). However, the expression of DE TFs was generally concordant with the number and fold change direction (up/down) of DB SEs within 250kb of the TF gene's boundaries (Fig. S6B). Overall, SEs within 250kb of BCL-upregulated TFs in Clusters 1-3 harbored significantly higher levels of H3K27ac in BCL compared to healthy control B cells, while those associated with BCL-downregulated TFs in Clusters 4-6 exhibit significantly lower H3K27ac levels in BCL subtypes. The difference was even more striking for SEs that overlap DE TF genes (Fig. 6A).
Fig. 6.
BCL-altered transcription factors have super-enhancer powered transcriptional feedback loops. A) Half box/dot-plot shows the log2 fold change of K27ac in BCL versus healthy control (HC) B cells for super-enhancers (SE) within 250kb (left) or overlapping (right) the differentially expressed TF genes (red: clusters 1-3, upregulated in BCL, blue: clusters 4-6, downregulated in BCL, from Fig. 5). Wilcoxon test *** p < 0.001; **** p < 0.0001. B) Cartoon depicts distal and overlapping SEs with high (purple) or low (grey) levels of H3K27ac regulating the expression of a TF gene from one of clusters 1-3 (red) or 4-6 (blue). TFs from the same (matching) cluster groups (1-3/4-6) bind within the SEs and control TF expression. C) Volcano plot shows log2 fold change and -log10 adjusted p value for expression of TFs (from Fig. 5) in BCL versus HC B cells. TF genes with SE within 250kb are highlighted by diamonds, with size indicating number of SEs and color corresponding to the percentage of matching TF cluster group peaks within the SE(s), red = TF gene in Clusters 1-3, blue = TF gene in Clusters 4-6 (ENCODE 3 TF ChIPseq). Gene labels include the number of SEs if greater than one. D) Alluvial plot segregates TFs with overlapping SEs and ENCODE 3 ChIPseq data. “Self” TF peaks: ChIPseq binding peaks for a TF were found in the SE overlapping the gene encoding that TF, an indication of TF self-regulation. E) For 15 TFs with “self” TF peaks in overlapping super-enhancers from (D), bar graphs show the number of constituent enhancers with (red/blue) or without (grey) binding peaks of the “self” TF. Red = TF in Clusters 1-3, blue = TF in Clusters 4-6.
In addition to controlling transcription at promoters, TFs also function more broadly as epigenetic regulators by opening or maintaining open chromatin and by recruiting chromatin regulators and other transcription factors at promoters and enhancers [37,90,91]. We therefore reasoned that the DE TFs may act in this way on their neighboring SEs, with the upregulated TFs in Clusters 1-3 acting to increase active SE chromatin and the decreased levels of TFs in Clusters 4-6 resulting in decreased active SE chromatin (cartoon in Fig. 6B). This proposed mechanism of epigenetic regulation does not take into account the potential effects of transcriptional repressors or co-regulation with other factors but does evaluate a major mechanism of epigenetic regulation: that of activation. To test this hypothesis, we retrieved TF ChIP-seq peaks in SEs within 250kb of BCL-altered TFs from the ENCODE studies [31]. Factorbook [92] contains ChIP-seq data from 682 TFs from 142 human cell types; peaks from 665 of these TFs were found across all SEs in our dataset. Of 283 BCL-altered TFs, 89 had ChIP-seq data available in Factorbook. To evaluate the relative regulatory impact of TF cluster groups, we determined the number of TF peaks from Clusters 1-3 and 4-6 in each SE within 250kb of BCL-altered TFs, the total of which varied from 4 to 1500. Given this wide range, we expressed the relative regulatory impact of TF cluster groups as a percentage of the total TF peaks in any cluster. For simplicity, the percentage of “matching” TF cluster groups peaks is plotted in Fig. 6C (e.g., ChIP-seq peaks for a TF in Cluster 1-3 in an SE associated with a TF gene in Cluster 1-3). For SEs within 250kb of upregulated TFs (Clusters 1-3), a red color gradient (0–100%) indicates the percentage of TF peaks that are from Clusters 1-3 or “matching”; for Clusters 4-6, the color gradient for matching TF peaks is blue. We found that SEs within 250kb of Cluster 4-6 TFs have a majority of matching Cluster 4-6 TF peaks. In fact, 51/54 TFs had greater than 50% matching (Cluster 4-6) TF peaks in all SEs (median 60.2% for all SEs). In contrast, only 6/40 Cluster 1-3 TFs had greater than 50% matching (Cluster 1-3) TF peaks in all SEs (median 41.2% for all SEs). In addition, a greater number of Cluster 4-6 TFs have multiple SEs within 250kb of their gene (28/54 vs 13/40, size of diamond, Fig. 6C).
Having identified a correspondence of expression changes and TF peaks in SEs associated with DE TFs groups, we next focused our analysis on the potential role of TF self-regulation. To accomplish this more stringent analysis, we required that SEs overlap DE TF genes. Just as for SEs within 250kb, there were fewer Cluster 1-3 TFs with overlapping SEs [13] than cluster 4-6 TF [25] as shown in the alluvial plot in Fig. 6D. Of these, 18 TFs had ChIP-seq available from ENCODE (5 in Clusters 1-3, 13 in Clusters 4-6). We next evaluated the number of “self” TF ChIP-seq peaks within each SE, i.e., TF binding by the protein encoded by the SE-overlapping gene. All but three of the TFs had binding within their overlapping SEs (15/18). To quantify the relative regulatory impact of these “self” TF peaks we determined the number of constituent enhancers with self TF peaks within each SE. Fig. 6E demonstrates that these SEs harbor self TF peaks in a variable number of constituent enhancers, from 1 – 7 (4–100% of total constituent enhancers). TFs in Clusters 4-6 harbored self TF peaks in a higher percentage of constituent enhancers: median 33.3% vs 12.5% in Cluster 1-3 TF SEs. Together, these data suggest that BCL DE TFs act to promote and/or maintain epigenetic changes in their super-enhancers, forming self-regulating transcriptional feedback loops that may promote lymphoma pathogenesis.
4. Discussion
Disordered epigenetic landscapes and transcriptional regulation are known contributors to the pathogenesis of germinal center B-cell lymphomas (FL, DLBCL) and to chronic lymphocytic leukemia. However, the regulomes of these mature B-cell malignancies had not been previously evaluated together to identify shared and distinct pathways of oncogenesis. In this study, we profiled the chromatin landscapes of these three B-cell cancers, as well as their normal B-cell counterparts. We identified recurrent regulatory programs across BCL subtypes and a novel super enhancer locus driving tissue-specific expression of two immunoglobulin (Ig) receptor genes with aberrant expression in BCL. Most strikingly, we showed that BCL is marked by profound reductions in normal B-cell transcription factor expression, and that this may be mediated and reinforced by loss of self-regulatory feedback loops to the TF-associated SEs.
By comparing the regulomes of FL, DLBCL, and CLL, we identified a novel SE in each of the three subtypes that we linked to overexpression of its flanking genes. The SE locus is intriguing from both cancer and normal development perspectives. The flanking Ig receptor genes, FCMR and PIGR, are expressed in a mutually exclusive pattern in B lymphocytes and mucosal epithelial cells, respectively, but corresponding regulatory mechanisms had not previously been elucidated. Our studies suggest that expression is controlled by differential patterns of histone marks and chromatin accessibility as well as chromatin interactions via CTCF. Based on our studies, multiple CTCF-mediated chromatin interactions bring together a strong, active enhancer and the promoter of FCMR in B cells, while lack of CTCF and active chromatin marks “loop out” this enhancer away from the promoter in mucosal epithelial cells. Meanwhile, high levels of active chromatin marks in the flanking region overlapping PIGR likely drives expression of this gene in mucosal epithelial cells and not in B cells. The inappropriate expression of PIGR in some BCL samples is likely due to high levels of active histone marks and associated TFs binding to constituent SE elements overlapping the PIGR gene body promoting transcriptional activity. Gene activation via spreading of active histone marks, such as acetylation, is a known mechanism of enhancer driven transcriptional control that occurs during cellular development [93,94]. Studies in mice showed that FCMR regulates the surface expression of the B-cell receptor (BCR) and that B cells deficient in FCMR have enhanced tonic BCR signaling and increased differentiation [95]. Other reports connect FCMR to IgM binding and B-cell activation in human B cells and CLL [96]. Deregulated BCR signaling is known to play a role in the survival and growth of DLBCL and CLL [97,98]. To our knowledge, there is no previous report of PIGR expression in human B-cell neoplasms. We suggest that FCMR and PIGR overexpression may contribute to the aberrant BCR signaling that drives many B-cell cancers.
We identified SEs in all three subtypes of BCL and in healthy control B-cell subsets and compared their activity level. Strikingly, SEs with decreased activity in BCL were linked to the greatest changes in gene expression. Similarly, the most striking change in TF expression across BCL subtypes was downregulation of normal B-cell factors. These include known B lymphocyte differentiation factors and tumor suppressors, and some factors with no previously established role in B-cell cancer. Examples of the former group include SPI1 (PU.1), POU2F2 (OCT2), and TCF3 (E2A). PU.1 is an Ets family TF that is essential for the proper development of B cells and other lineages, though it has some redundancy with other factors [88,89,99]. Its loss via knockout or hypermethylation is associated with pre-B-cell leukemia or B-cell lymphoma, respectively [100,101]. Like PU.1, OCT2 cooperates with other essential factors to promote B-cell maturation [102,103]. OCT2 is amplified in DLBCL and required for survival of DLBCL cell lines [104]. In FL, recurrent mutations in the DNA binding domain led to loss of transactivational activity [105]. Our study revealed widespread loss of OCT2 expression associated with decreased SE activity, and in some cases, genomic copy number loss, suggesting that disparate mechanisms of POU2F2 disruption can be oncogenic. E2A is a E protein family helix-loop-helix (HLH) TF that plays a critical role in B and T lymphocyte development [106], is mutated in Burkitt's lymphoma, and is translocated in B-cell acute lymphocytic leukemia [107,108]. Like POU2F2/OCT2, TCF3/E2A expression and SE activity were significantly reduced and genomic deletions were detected in some samples. While MAZ is best known as a transactivator of MYC, its loss as seen in our study may promote BCL by loss of its repression of MYB, a master regulator of the germinal center reaction [109], [110], [111].
Genomic alterations are often considered a major driver of lymphomagenesis [7,112]. In our study, we found that a minority (<20%) of differentially bound enhancers and SEs in BCL were coincident with concordant copy number alterations (CNA), suggesting that epigenetic changes are independent of genome copy number. One caveat to our study is that we did not identify single nucleotide variants in our lymphoma samples. Chromatin modifier mutations are well-documented in FL and DLBCL [7], with a much lower frequency of mutations in these genes in CLL [12,21]. Nevertheless, chromatin modifier mutations are absent in 30-80% of BCL tumors, varying by subtype, yet we and others have found widespread changes to the chromatin landscape in all BCL tumors [8,[18], [19], [20],22,23]. Thus, epigenetic deregulation is common, and perhaps ubiquitous in BCL, and in a significant proportion of cases, is not connected to genomic mutations.
Our studies went a step further than simply identifying BCL-altered regulatory elements, TFs, and gene expression change; we connected these transcriptional control mechanisms to each other. Unexpectedly, our studies revealed that loss of core normal B-cell regulatory circuitry may play a larger role in lymphomagenesis than previously appreciated. First, we showed that changes in enhancer chromatin activity correlates with neighboring gene expression, and moreover that loss of acetylation in SEs was associated with a profound loss of expression. Further analysis demonstrated that TFs with differential expression in BCL were marked by enhancers and SEs with corresponding differences in acetylation levels. Some of these TF genes harbored up to 20 or more DB enhancers and up to 5 DB SE. Here again we observed a greater number of SEs with loss of active chromatin associated with downregulated TFs compared to SEs with increased active chromatin associated with upregulated TFs, even though there are nearly twice as many upregulated TFs. Further underscoring the role of loss of normal B cell development factors, we found that a majority of TF peaks in SEs near downregulated TFs (Clusters 4-6) are bound by TFs from these same groups. One caveat of the TF peak analyses is that a greater proportion of downregulated TFs in Clusters 4-6 had ChIP-seq data available compared to upregulated TFs in Clusters 1-3 (44/107 vs 45/176). To mitigate this and to evaluate self-regulation of TF expression, we analyzed “self” TF peak binding, i.e., binding of a TF within an SE overlapping the TF gene. This more stringent analysis identified four upregulated and 11 downregulated TFs with “self” SE binding peaks. In addition, these same SEs have significantly decreased active chromatin. While other studies have reported amplified signaling through core regulatory circuitries in cancer [40], our findings here demonstrate the less recognized but perhaps more impactful role of decommissioning of B-cell identity pathways in the genesis of lymphoma.
Many of the cluster group or self-regulated TFs are known BCL oncogenes or tumor suppressors, key B cell differentiation factors, or all of the above. For example, EBF1 is a pioneer TF and a B-cell lineage defining factor, acting to establish chromatin changes that enable activation of B cell-specific genes and repress genes associated with alternative cell fates. The loss of active chromatin in four SEs that overlap the EBF1 gene, coupled with a substantial number of EBF1 binding sites and loss of EBF1 expression in BCL, support a role for super-enhancer powered transcriptional feedback loops in the normal expression of such crucial B-cell TFs. RUNX3 is a tumor suppressor that has not been previously associated with BCL yet acts in pathways known to be lymphomagenic. Particularly relevant to this study, RUNX3 inhibits oncogenic Wnt signaling by interacting with TCF4/beta-catenin and preventing their binding and activation of MYC and CCND1 promoters [113], [114], [115]. The TCF4 gene, which codes for an E-box binding basic helix-loop-helix transcription factor, is located in a genomic amplification in activated B-cell type DLBCL, reported previously [69] and also detected in our study. TCF4 drives expression of IgM and MYC in these lymphomas [69]. The combined loss of RUNX3 and increased expression of TCF4 observed across BCL in our study suggests a synergistic feed-forward loop that promotes unchecked growth and proliferation. Compared to healthy B-cells, BCL have disruptions in key TFs and tumor suppressors and the epigenetic elements regulating these factors. Together, these findings suggest a complex, iterative, and interactive process that links TF and epigenetic activities together to repress B-cell maturation and promote proliferation, culminating in transformation.
In summary, we have taken an integrative approach to examine and compare the epigenomes and transcriptomes of three mature B-cell neoplasms with normal B-cell subsets. Our novel approach enabled us to define shared alterations in the regulomes of FL, DLBCL, and CLL and identify widespread disruption of SE-powered transcriptional feedback loops. Together, our findings implicate SEs as important hubs of tumor suppressing transcriptional feedback loops that maintain tumor suppression and developmental programs, and when perturbed, can drive lymphomagenesis.
Data sharing
Sequencing and array data was deposited in GEO: GSE145848. Additional datasets used: GSE32018, GSE119103, and GSE62246. UCSC session: http://genome.ucsc.edu/s/jepayton/Andrews_etal_2021. Additional documentation detailing the general RNA-seq and ChIP-seq analysis pipelines are available online and R code was wrapped into a high-level package composed of simple one-liner commands available at https://github.com/j-andrews7/OneLinerOmics. Original photomicrograph files are at zenodo.org. Flow cytometry data is available at: flowrepository.org, Experiment: FCMR_CLL_flow, ID: FR-FCM-Z3LM.
Contributors
Conception and design: J.M. Andrews, J.E. Payton
Development of methodology: J.M. Andrews, O.I. Koues, E.M. Oltz, J.E. Payton
Acquisition of data: J.M. Andrews, S.C. Pyfrom, J.A. Schmidt, O.I. Koues, R.A. Kowalewski, N.R. Grams, J.J. Sun, L.R. Berman, E.J. Duncavage, Y-S. Lee, A.F. Cashen, J.E. Payton
Analysis, interpretation of data, generation of figures: J.M. Andrews, J.E. Payton
Funding acquisition, writing, review, and/or revision of the manuscript: J.M. Andrews, E.M. Oltz, J.E. Payton
Study supervision: J.E. Payton.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgements
We thank the Washington University School of Medicine Lymphoma Banking Program of the Division of Medical Oncology in the Department of Medicine for support of the biopsy and banking program; the patients, their caregivers and families, and the patient coordinators who participated in this study. This work was supported by the National Institutes of Health (NIH) grants CA221012 (to J.M.A.) and CA188286 and CA156690 (to E.M.O. and J.E.P.). Additional funding from the Siteman Cancer Center, Barnes-Jewish Hospital Foundation, and the Doris Duke Foundation (to J.E.P.). Sequencing provided by the Genome Technology Access Center, which is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. None of these sources had any role in data collection, analysis, interpretation, trial design, patient recruitment, writing the manuscript, the decision to submit the manuscript, or any other aspect pertinent to the study. None of the authors was paid to write the article by a company or any other agency. The corresponding author (Jacqueline Payton) had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103559.
Appendix. Supplementary materials
References
- 1.Beekman R, Chapaprieta V, Russiñol N, Vilarrasa-Blasi R, Verdaguer-Dot N, Martens JHA. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018 Jun;24(6):868–880. doi: 10.1038/s41591-018-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015 Jul;47(7):746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews JM, Payton JE. Epigenetic dynamics in normal and malignant B cells: die a hero or live to become a villain. Curr Opin Immunol. 2019;57:15–22. doi: 10.1016/j.coi.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez PM, Ghamlouch H, Rosikiewicz W, Kumar P, Béguelin W, Fontán L. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discov. 2018;8(12):1632–1653. doi: 10.1158/2159-8290.CD-18-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Dominguez-Sola D, Hussein S, Lee J-E, Holmes AB, Bansal M. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015 Sep;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013 May;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasqualucci L. Molecular pathogenesis of germinal center-derived B cell lymphomas. Immunol Rev. 2019;288(1):240–261. doi: 10.1111/imr.12745. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Melnick A. The epigenetic basis of diffuse large B-cell lymphoma. Semin Hematol. 2015 Apr 1;52(2):86–96. doi: 10.1053/j.seminhematol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018 Apr 12;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012 Nov;44(11):1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 11.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011 Sep;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013 Feb;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011 Aug;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Riain C, O'Shea DM, Yang Y, Le Dieu R, Gribben JG, Summers K. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009 Oct;23(10):1858–1866. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaknovich R, Geng H, Johnson NA, Tsikitas L, Cerchietti L, Greally JM. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010 Nov 18;116(20):e81–e89. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pogo BG, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci. 1966 Apr 1;55(4):805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Good-Jacobson KL. Epigenetic regulation of B cell fate and function during an immune response. Immunol Rev. 2019;288(1):75–84. doi: 10.1111/imr.12733. [DOI] [PubMed] [Google Scholar]
- 18.Koues OI, Kowalewski RA, Chang L-WW, Pyfrom SC, Schmidt JA, Luo H. Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity. 2015 Jan;42(1):186–198. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gascoyne RD, Nadel B, Pasqualucci L, Fitzgibbon J, Payton JE, Melnick A. Follicular lymphoma: state-of-the-art ICML workshop in Lugano 2015. Hematol Oncol. 2017 Dec;35(4):397–407. doi: 10.1002/hon.2411. [DOI] [PubMed] [Google Scholar]
- 20.Koues OI, Oltz EM, Payton JE. Short-circuiting gene regulatory networks: origins of B cell lymphoma. Trends Genet. 2015;31(12):720–731. doi: 10.1016/j.tig.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015 Oct 22;526(7574):525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastore A, Gaiti F, Lu SX, Brand RM, Kulm S, Chaligne R. Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL. Nat Commun. 2019;10(1):1874. doi: 10.1038/s41467-019-09645-5. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018 Dec 10;34(6):982–995. doi: 10.1016/j.ccell.2018.11.001. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011 Oct;12(10):933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer AL, Young RM, Staudt LM. Shaffer 3rd AL. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012 Jan;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012 Sep;120(11):2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venturutti L, Melnick AM. The dangers of déjà vu: memory B cells as the cells of origin of ABC-DLBCLs. Blood. 2020 Nov 12;136(20):2263–2274. doi: 10.1182/blood.2020005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, Dürig J. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012 Nov 19;209(12):2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016 Mar;48(3):253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A. Integrative analysis of 111 reference human epigenomes. Nature. 2015 Feb;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569(7756):345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- 33.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014 Dec 18;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012 Apr 11;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016 Jun 2;62(5):668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015 Feb;518(7539):331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013 Apr 11;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Cairns MJ, Yan J. Super-enhancers in transcriptional regulation and genome organization. Nucl Acids Res. 2019 Nov 14 doi: 10.1093/nar/gkz1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Xu L, Lin RY-T, Müschen M, Koeffler HP. Core transcriptional regulatory circuitries in cancer. Oncogene. 2020 Oct;39(43):6633–6646. doi: 10.1038/s41388-020-01459-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H. CTCF binding polarity determines chromatin looping. Mol Cell. 2015 Oct;60(4):676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Vian L, Pękowska A, Rao SSP, Kieffer-Kwon K-R, Jung S, Baranello L. The energetics and physiological impact of cohesin extrusion. Cell. 2018;173(5):1165–1178. doi: 10.1016/j.cell.2018.03.072. 17e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan A, Mathelier A, Zhang X. Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers. Epigenetics. 2018;13(9):910–922. doi: 10.1080/15592294.2018.1514231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Koues OI, Zhao J, Liu R, Pyfrom SC, Payton JE. cis-regulatory circuits regulating NEK6 kinase overexpression in transformed B cells are super-enhancer independent. Cell Rep. 2017 Mar 21;18(12):2918–2931. doi: 10.1016/j.celrep.2017.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2015 Dec;48(2):176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon K-R, Resch W. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014 Dec 18;159(7):1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B Cell lymphoma. Cancer Cell. 2013 Dec 9;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kandaswamy R, Sava GP, Speedy HE, Beà S, Martín-Subero JI, Studd JB. Genetic predisposition to chronic lymphocytic leukemia is mediated by a BMF super-enhancer polymorphism. Cell Rep. 2016 23;16(8):2061–2067. doi: 10.1016/j.celrep.2016.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Andres M, Paiva B, Nieto WG, Caraux A, Schmitz A, Almeida J. Human peripheral blood B-cell compartments: a crossroad in B-cell traffic. Cytometry B Clin Cytom. 2010;78B(S1):S47–S60. doi: 10.1002/cyto.b.20547. [DOI] [PubMed] [Google Scholar]
- 50.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res. 2015 Apr 20;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015 May;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 52.Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV. VDJtools: unifying post-analysis of T cell receptor repertoires. PLOS Comput Biol. 2015 Nov 25;11(11) doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004 Oct 1;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 54.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput Biol. 2016 Apr 21;12(4) doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nilsen G, Liestøl K, Van Loo P, HK MV, Eide MB, Rueda OM. Copynumber: Efficient algorithms for single- and multi-track copy number segmentation. BMC Genom. 2012 Nov 4;13(1):591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucl Acids Res. 2019 Jul 2;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJP, Curti S. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015 Jan;16(3):318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007 Jun;17(6):877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012 Sep;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015 Jul 15;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 61.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;Jan 1(bax028) doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012 Jan;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015 Dec;528(7582):418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 Nov doi: 10.1126/science.1259037. science.1259037- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shull AY, Luo J, Pei L, Lee E-J, Liu J, Choi J. DNA hypomethylation within B-cell enhancers and super enhancers reveal a dependency on immune and metabolic mechanisms in chronic lymphocytic leukemia. Blood. 2016 Dec 2;128(22) 1049–1049. [Google Scholar]
- 66.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucl Acids Res. 2016 Jan 4;44(D1):D164–D171. doi: 10.1093/nar/gkv1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gloury R, Zotos D, Zuidscherwoude M, Masson F, Liao Y, Hasbold J. Dynamic changes in Id3 and E-protein activity orchestrate germinal center and plasma cell development. J Exp Med. 2016 May 23;213(6):1095–1111. doi: 10.1084/jem.20152003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wöhner M, Tagoh H, Bilic I, Jaritz M, Poliakova DK, Fischer M. Molecular functions of the transcription factors E2A and E2-2 in controlling germinal center B cell and plasma cell development. J Exp Med. 2016 Jun 3;213(7):1201–1221. doi: 10.1084/jem.20152002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain N, Hartert K, Tadros S, Fiskus W, Havranek O, Ma MCJ. Targetable genetic alterations of TCF4 (E2-2) drive immunoglobulin expression in diffuse large B cell lymphoma. Sci Transl Med. 2019 Jun 19;11(497) doi: 10.1126/scitranslmed.aav5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo S, Chan JKC, Iqbal J, McKeithan T, Fu K, Meng B. EZH2 mutations in follicular lymphoma from different ethnic groups and associated gene expression alterations. Clin Cancer Res Off J Am Assoc Cancer Res. 2014 Jun 15;20(12):3078–3086. doi: 10.1158/1078-0432.CCR-13-1597. [DOI] [PubMed] [Google Scholar]
- 71.Loo SK, Ch'ng ES, Lawrie CH, Muruzabal MA, Gaafar A, Pomposo MP. DNMT1 is predictive of survival and associated with Ki-67 expression in R-CHOP-treated diffuse large B-cell lymphomas. Pathology (Phila) 2017 Dec;49(7):731–739. doi: 10.1016/j.pathol.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Leonard S, Wei W, Anderton J, Vockerodt M, Rowe M, Murray PG. Epigenetic and transcriptional changes which follow Epstein-Barr virus infection of germinal center B cells and their relevance to the pathogenesis of Hodgkin's lymphoma. J Virol. 2011 Sep;85(18):9568–9577. doi: 10.1128/JVI.00468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo R, Zhang Y, Teng M, Jiang C, Schineller M, Zhao B. DNA methylation enzymes and PRC1 restrict B-cell Epstein-Barr virus oncoprotein expression. Nat Microbiol. 2020 Aug;5(8):1051–1063. doi: 10.1038/s41564-020-0724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaudhri VK, Dienger-Stambaugh K, Wu Z, Shrestha M, Singh H. Charting the cis-regulome of activated B cells by coupling structural and functional genomics. Nat Immunol. 2020 Feb;21(2):210–220. doi: 10.1038/s41590-019-0565-0. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Lee D, Dhiman V, Jiang P, Xu J, McGillivray P. An integrative ENCODE resource for cancer genomics. Nat Commun. 2020 Jul 29;11(1):3696. doi: 10.1038/s41467-020-14743-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013 May;10(5):375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pallasch CP, Schulz A, Kutsch N, Schwamb J, Hagist S, Kashkar H. Overexpression of TOSO in CLL is triggered by B-cell receptor signaling and associated with progressive disease. Blood. 2008 Nov 15;112(10):4213–4219. doi: 10.1182/blood-2008-05-157255. [DOI] [PubMed] [Google Scholar]
- 78.Proto-Siqueira R, Panepucci RA, Careta FP, Lee A, Clear A, Morris K. SAGE analysis demonstrates increased expression of TOSO contributing to Fas-mediated resistance in CLL. Blood. 2008 Jul 15;112(2):394–397. doi: 10.1182/blood-2007-11-124065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips-Quagliata JM, Patel S, Han J-K, Arakelov S, Rao TD, Shulman MJ. The IgA/IgM receptor expressed on a murine B cell lymphoma is poly-Ig receptor. J Immunol. 2000 Sep 1;165(5):2544–2555. doi: 10.4049/jimmunol.165.5.2544. [DOI] [PubMed] [Google Scholar]
- 80.Gómez-Abad C, Pisonero H, Blanco-Aparicio C, Roncador G, González-Menchén A, Martinez-Climent JA. PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood. 2011 Nov 17;118(20):5517–5527. doi: 10.1182/blood-2011-03-344374. [DOI] [PubMed] [Google Scholar]
- 81.Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Noia JMD. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat Rev Immunol. 2016 Mar;16(3):164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009 Aug 4;106(31):12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fabbri G. Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016 Mar;16(3):145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 84.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M. The human transcription factors. Cell. 2018 Feb 8;172(4):650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 85.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016 Aug;16(8):483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 86.Gururajan M, Simmons A, Dasu T, Spear BT, Calulot C, Robertson DA. Early growth response genes regulate B cell development, proliferation, and immune response. J Immunol Baltim Md. 2008 Oct 1;181(7):4590–4602. doi: 10.4049/jimmunol.181.7.4590. 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boto P, Csuth TI, Szatmari I. RUNX3-mediated immune cell development and maturation. Crit Rev Immunol. 2018;38(1):63–78. doi: 10.1615/CritRevImmunol.2018025488. [DOI] [PubMed] [Google Scholar]
- 88.Wang H, Jain S, Li P, Lin J-X, Oh J, Qi C. Transcription factors IRF8 and PU.1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc Natl Acad Sci USA. 2019 May 7;116(19):9511–9520. doi: 10.1073/pnas.1901258116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soodgupta D, White LS, Yang W, Johnston R, Andrews JM, Kohyama M. RAG-mediated DNA breaks attenuate PU.1 activity in early B cells through activation of a SPIC-BCLAF1 complex. Cell Rep. 2019 Oct 22;29(4):829–843. doi: 10.1016/j.celrep.2019.09.026. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014 Jun 5;54(5):844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015 Mar;521(7552):366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang J, Zhuang J, Iyer S, Lin X-Y, Greven MC, Kim B-H. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucl Acids Res. 2013 Jan;41:D171–D176. doi: 10.1093/nar/gks1221. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell. 2002 Dec 1;10(6):1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 94.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011 Feb 4;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen TTT, Kläsener K, Zürn C, Castillo PA, Brust-Mascher I, Imai DM. The FcμR limits tonic BCR signaling by regulating IgM-BCR expression. Nat Immunol. 2017 Mar;18(3):321–333. doi: 10.1038/ni.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vire B, David A, Wiestner A. TOSO, the Fcmicro receptor, is highly expressed on chronic lymphocytic leukemia B cells, internalizes upon IgM binding, shuttles to the lysosome, and is downregulated in response to TLR activation. J Immunol Baltim Md. 2011 Oct 15;187(8):4040–4050. doi: 10.4049/jimmunol.1100532. 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013 Dec;34(12):592–601. doi: 10.1016/j.it.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010 Nov;238(1):63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 100.Pang SHM, Minnich M, Gangatirkar P, Zheng Z, Ebert A, Song G. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia. 2016;30(6):1375–1387. doi: 10.1038/leu.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Özdemir İ, Pınarlı FG, Pınarlı FA, Aksakal FNB, Okur A, Uyar Göçün P. Epigenetic silencing of the tumor suppressor genes SPI1, PRDX2, KLF4, DLEC1, and DAPK1 in childhood and adolescent lymphomas. Pediatr Hematol Oncol. 2018 Mar;35(2):131–144. doi: 10.1080/08880018.2018.1467986. [DOI] [PubMed] [Google Scholar]
- 102.Corcoran LM, Karvelas M, Nossal GJ, Ye ZS, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993 Apr;7(4):570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 103.Corcoran L, Emslie D, Kratina T, Shi W, Hirsch S, Taubenheim N. Oct2 and Obf1 as facilitators of B:T cell collaboration during a humoral immune response. Front Immunol. 2014 Jan;5:108. doi: 10.3389/fimmu.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hodson DJ, Shaffer AL, Xiao W, Wright GW, Schmitz R, Phelan JD. Regulation of normal B-cell differentiation and malignant B-cell survival by OCT2. Proc Natl Acad Sci USA. 2016 Apr 5;113(14):E2039–E2046. doi: 10.1073/pnas.1600557113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014 Mar;123(10):1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quong MW, Romanow WJ. Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 107.Rohde M, Bonn BR, Zimmermann M, Lange J, Möricke A, Klapper W. Relevance of ID3-TCF3-CCND3 pathway mutations in pediatric aggressive B-cell lymphoma treated according to the non-Hodgkin Lymphoma Berlin-Frankfurt-Münster protocols. Haematologica. 2017;102(6):1091–1098. doi: 10.3324/haematol.2016.156885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mullighan CG. Genome sequencing of lymphoid malignancies. Blood. 2013 Dec;122(24):3899–3907. doi: 10.1182/blood-2013-08-460311. [DOI] [PubMed] [Google Scholar]
- 109.Álvaro-Blanco J, Urso K, Chiodo Y, Martín-Cortázar C, Kourani O, Arco PG-D. MAZ induces MYB expression during the exit from quiescence via the E2F site in the MYB promoter. Nucl Acids Res. 2017 Sep 29;45(17):9960–9975. doi: 10.1093/nar/gkx641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010 Jan 1;6(1):377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greig KT, de Graaf CA, Murphy JM, Carpinelli MR, Pang SHM, Frampton J. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010 Apr 8;115(14):2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gruber M, Wu CJ. Evolving understanding of the CLL genome. Semin Hematol. 2014 Jul;51(3):177–187. doi: 10.1053/j.seminhematol.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen F, Liu X, Bai J, Pei D, Zheng J. The emerging role of RUNX3 in cancer metastasis (Review) Oncol Rep. 2016 Mar 1;35(3):1227–1236. doi: 10.3892/or.2015.4515. [DOI] [PubMed] [Google Scholar]
- 114.Ito Y, Bae S-C, Chuang LSH. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015 Feb;15(2):81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 115.Otálora-Otálora BA, Henríquez B, López-Kleine L, Rojas A. RUNX family: oncogenes or tumor suppressors (Review) Oncol Rep. 2019 Jul;42(1):3–19. doi: 10.3892/or.2019.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.