Abstract
Fused in sarcoma (FUS) encodes an RNA-binding protein with diverse roles in transcriptional activation and RNA splicing. While oncogenic fusions of FUS and transcription factor DNA-binding domains are associated with soft tissue sarcomas, dominant mutations in FUS can cause amyotrophic lateral sclerosis. FUS has also been implicated in genome maintenance. However, the underlying mechanisms of its actions in genome stability are unknown. Here, we applied gene editing, functional reconstitution, and integrated proteomics and transcriptomics to illuminate roles for FUS in DNA replication and repair. Consistent with a supportive role in DNA double-strand break repair, FUS-deficient cells exhibited subtle alterations in the recruitment and retention of double-strand break–associated factors, including 53BP1 and BRCA1. FUS−/− cells also exhibited reduced proliferative potential that correlated with reduced speed of replication fork progression, diminished loading of prereplication complexes, enhanced micronucleus formation, and attenuated expression and splicing of S-phase–associated genes. Finally, FUS-deficient cells exhibited genome-wide alterations in DNA replication timing that were reversed upon re-expression of FUS complementary DNA. We also showed that FUS-dependent replication domains were enriched in transcriptionally active chromatin and that FUS was required for the timely replication of transcriptionally active DNA. These findings suggest that alterations in DNA replication kinetics and programming contribute to genome instability and functional defects in FUS-deficient cells.
Keywords: fused in sarcoma (FUS), DNA replication, DNA repair, replication timing, RNA binding protein, amyotrophic lateral sclerosis (ALS)
Abbreviations: ALS, amyotrophic lateral sclerosis; BrdU, 5-bromo-2′-deoxyuridine; BSA, bovine serum albumin; cDNA, complementary DNA; CF, chromatin fraction; CHOP, CCAAT/enhancer-binding protein homologous protein; CldU, 5-chloro-2′-deoxyuridine; CLM, calicheamicin γ1; CSK, cytoskeleton; DAPI, 4′,6-diamidino-2-phenylindole; DDR, DNA damage response; DSB, double-strand break; EdU, 5-ethynyl-2′-deoxyuridine; ERD, early replication domain; FET, FUS, EWSR1, TAF15; FTD, frontotemporal dementia; FUS, fused in sarcoma; GO, Gene Ontology; HDR, homology-directed repair; HEK293T, human embryonic kidney 293T; HU, hydroxyurea; IP, immunoprecipitation; iPOND, isolation of proteins on nascent DNA; LCD, low-complexity domain; LOF, loss-of-function; LRD, late replication domain; MMC, mitomycin C; MRD, mid replication domain; NHEJ, nonhomologous end joining; ORC, origin recognition complex; PAR, poly(ADP)-ribosyl; PARP, poly(ADP)-ribosyl polymerase; PCNA, proliferating cell nuclear antigen; PI, propidium iodide; pre-RC, prereplication complex; qPCR, quantitative PCR; RD, replication domain; RF, replication fork; RGG, arginine–glycine–glycine repeat; RIPA, radioimmunoprecipitation assay; RT, replication timing; SCAI, suppressor of cancer cell invasion; SSB, single-strand break
Fused in sarcoma (FUS, also referred to as translocated in liposarcoma) is a member of the FET (FUS, EWSR1, and TAF15) family of RNA- and DNA-binding proteins that play important roles in transcription and splicing (1, 2). Originally described as an oncogenic fusion to the CCAAT/enhancer-binding protein homologous protein (CHOP) transcription factor in myxoid liposarcoma (3, 4), FUS rose to prominence with the discovery that inherited, and de novo mutations in its ORF cause dominant forms of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (5, 6, 7). Although the underlying mechanisms are still unclear, the preponderance of ALS/FTD-associated mutations in FUS interferes with its nuclear import and folding, leading to the accumulation of cytosolic FUS aggregates that disrupt cellular function through loss-of-function (LOF) and gain-of function mechanisms impacting protein translation and nuclear transport among other processes (2, 8, 9, 10, 11).
FET proteins share a common domain structure that includes an N-terminal low-complexity domain (LCD), a Gly-rich region, one or more arginine–glycine—glycine repeat (RGG) domain, an RNA recognition motif with RNA- and DNA-binding activity, a zinc-finger domain, and a carboxyl-terminal PY-type nuclear localization signal that interacts with transportin nuclear import receptors that are essential for proper FUS folding (1, 8, 12, 13, 14). The LCD is also of particular interest as it exhibits strong transcriptional coactivation potential in vitro, and the fusion of this domain to the CHOP DNA-binding domain drives gene deregulation and oncogenesis in myxoid liposarcoma (1, 15). The LCD also mediates protein–protein interactions and participates in FUS oligomerization and liquid demixing (14, 16, 17, 18, 19) that may be central to its normal roles in transcription and splicing and pathologic roles in ALS/FTD (2).
In addition to their accepted roles in RNA processing, several lines of evidence support a role for the FET proteins in the cellular DNA damage response (DDR). FUS participation in the DDR was first inferred from chromosome instability and mild radiosensitive phenotypes of FUS−/− mice (20, 21, 22). FET proteins are capable of promoting invasion and pairing of a homologous ssDNA sequence with a dsDNA molecule in vitro (22, 23, 24), which suggests a possible role for FET proteins in the D-loop formation step of homology-directed repair (HDR) of DNA double-strand breaks (DSBs). Other studies showed that the FUS LCD is phosphorylated in response to DNA damage by DNA damage–activated protein kinases DNA-PKcs and ATM (17, 25), which are important regulators of the nonhomologous end joining (NHEJ) pathway of DSB repair. Consistent with a direct or an indirect role for FUS in DSB repair, we and others showed that shRNA-mediated depletion of FUS reduced the repair of HDR and NHEJ reporter substrates (26, 27, 28).
A role in the DDR is further suggested by poly(ADP)-ribosyl (PAR) polymerase (PARP)–dependent localization of FUS to sites of microirradiation-induced DNA damage (26, 27, 28). FUS is capable of interacting directly with PAR chains through its RGG domain (26), and the FET proteins are heavily PARylated in response to genotoxic stress (29). Mechanistically, it was reported that FUS mediates the recruitment of histone deacetylase 1, KU70, NBS1, and phosphorylated H2AX (γH2AX), and ATM at sites of DNA damage and that this recruitment pathway as well as FUS-dependent repair was compromised by ALS/FTD-associated mutations (27). It has also been proposed that FUS organizes DSBs in a PARP-dependent manner for their subsequent repair (30); while Wang et al. (31) reported that FUS recruits DNA ligase III downstream of PARP activation to repair single-strand breaks (SSBs) and that ALS-associated mutations in FUS disrupt SSB repair activity. Finally, it was recently reported that FUS regulates the response to transcription-associated recombinant DNA damage via association with topoisomerase 1 in the nucleolus (32). Despite these studies, the molecular mechanisms linking FUS to the different repair pathways in which it has been implicated remain unclear and the extent to which FUS-dependent RNA processing may contribute to reported DDR phenotypes in FUS-deficient cells is not known.
Here, we probed FUS-dependent genome protection using transcriptomic, proteomic, and functional analysis of FUS−/− cell lines reconstituted with FUS complementary DNAs (cDNAs). Our findings suggest that FUS plays particularly important roles in DNA replication where it contributes to replicon initiation and coordinates DNA replication timing (RT). These studies provide new insights into FUS-mediated genome protection in mitotically active cells.
Results
Generation and phenotypic characterization of FUS−/− cells
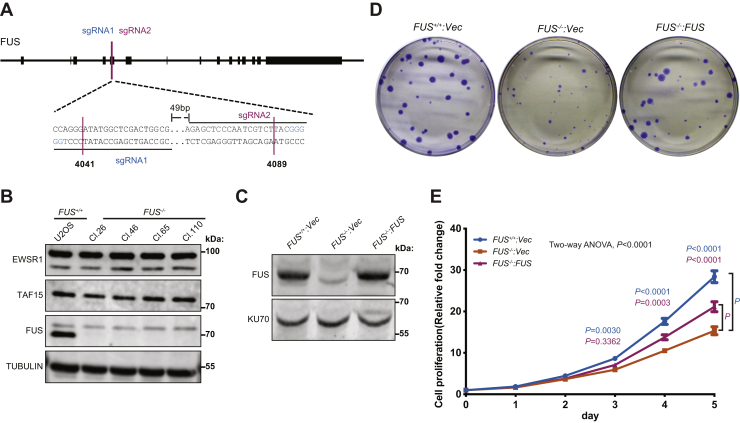
To discern roles of FUS in genome protection, we disrupted FUS gene loci in U-2 OS osteosarcoma cells using CRISPR–CAS9 followed by genetic reconstitution with a retroviral vector encoding the untagged FUS ORF (see Experimental procedures section). To ensure rigorous results, we studied multiple FUS−/− clones and selected a reconstituted FUS−/−:FUS line with physiological levels of FUS expression (Fig. 1, A–C). Notably, protein levels of TAF15 and EWSR1 were not upregulated in FUS−/− U-2 OS cells, diminishing concerns about functional compensation.
Figure 1.
FUS promotes cell proliferation.A, schematic of the FUS gene targeting. Two guide RNAs, sgRNA1 and sgRNA2, were used to target FUS exon 4 (see Experimental procedures section). B, expression of FET proteins (FUS, EWSR1, and TAF15) in FUS−/− clones. C, reconstitution of FUS−/− (Cl.110) with an untagged FUS retroviral vector. The same vector expressing β-glucuronidase (GUS) was introduced as a negative control into FUS−/− cells. D, FUS−/− cell colonies exhibited reduced growth relative to FUS+/+ and FUS−/−:FUS cells. E, cell proliferation rates of FUS+/+, FUS−/−, and FUS−/−: FUS U-2 OS cells. Three biological replicates were used. The bars represent mean ± SE. The two-way ANOVA test was performed, and the p values shown on plot are adjusted p values by Tukey's multiple comparisons test. FUS, fused in sarcoma.
FUS knockdown cells displayed mild IR sensitivity and modest defects in the repair of NHEJ and HDR reporter substrates (26), whereas a second study reported that FUS knockdown suppressed γH2AX and 53BP1 focus formation (27). We assessed time courses of γH2AX and 53BP1 accumulation and dissolution at IR-induced foci in FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells exposed to 2 Gy IR. The initial recruitment of 53BP1 to IR-induced nuclear foci was enhanced in FUS−/− cell lines, and 53BP1 foci persisted longer in FUS−/− cells relative to FUS+/+ cells (Fig. S1, A and C), and this was rescued in FUS−/−:FUS U-2 OS cells. The enhanced and prolonged accumulation of 53BP1 at IR-induced foci may reflect persistent DSBs. While no obvious γH2AX focus formation/dissociation defect was observed between FUS−/− and FUS+/+ cells, we observed reduced γH2AX focus formation in FUS−/−:FUS U-2 OS cells relative to FUS−/− cells (Fig. S1, B and C). Although the reason for discrepant findings between FUS+/+ and FUS−/−:FUS U-2 OS cells is unclear, it may reflect slightly increased FUS expression levels in FUS−/−:FUS U-2 OS cells relative to FUS+/+ cells (Fig. 1C).
We also investigated recruitment of the critical HDR factor, BRCA1. On a per-cell basis, the number of BRCA1 foci was comparable between FUS−/−, FUS+/+, and FUS−/−:FUS cells, suggesting FUS is not an essential component of the BRCA1 recruitment pathway (Fig. S2, A and C). On the other hand, the frequency of cells displaying IR-induced BRCA1 foci was significantly reduced in FUS−/− cells, and this was corrected by FUS reexpression (Fig. S2, B and C). Reduced BRCA1 focus formation was also seen in H460 cells stably transduced with FUS shRNA but not cells transduced with TAF15 or EWSR1 shRNA (Fig. S2, D and E), indicating a selective role for FUS. Because BRCA1 focus formation is largely restricted to S/G2 phase, these findings may indicate perturbed S-phase dynamics in FUS deficiency (see later). Despite the changes in 53BP1 and BRCA1 recruitment to IR-induced foci, FUS−/− cells did not exhibit significant hypersensitivity to mechanistically distinct genotoxins, including hydroxyurea (HU, replication stress), mitomycin C (MMC, DNA crosslinker), camptothecin (top1 inhibitor), and calicheamicin γ1 (CLM, radiomimetic). In fact, FUS−/−:FUS cells were slightly more resistant than FUS+/+ cells to MMC and CLM (Fig. S3). These findings suggest that FUS fulfills supportive rather than essential roles in DSB repair.
FUS−/− cells exhibit defects in DNA replication
FUS−/− U-2 OS cells exhibited reduced colony outgrowth and proliferative potential that was corrected by FUS reexpression (Fig. 1, D and E). Reduced proliferative capacity was observed in multiple FUS−/− U-2 OS clones as well as FUS-deficient NCI-H460 lung adenocarcinoma cells (Fig. S4, A and B). Finally, FUS−/− U-2 OS reconstituted with a FUS construct lacking the N-terminal LCD exhibited reduced colony growth rates relative to FUS−/−:FUS cells (Fig. S4, C and D). This finding implies that biochemical activities associated with the LCD, including transcriptional activation (15, 33) and phase separation/oligomerization (19, 34), contribute to its replication-associated functions.
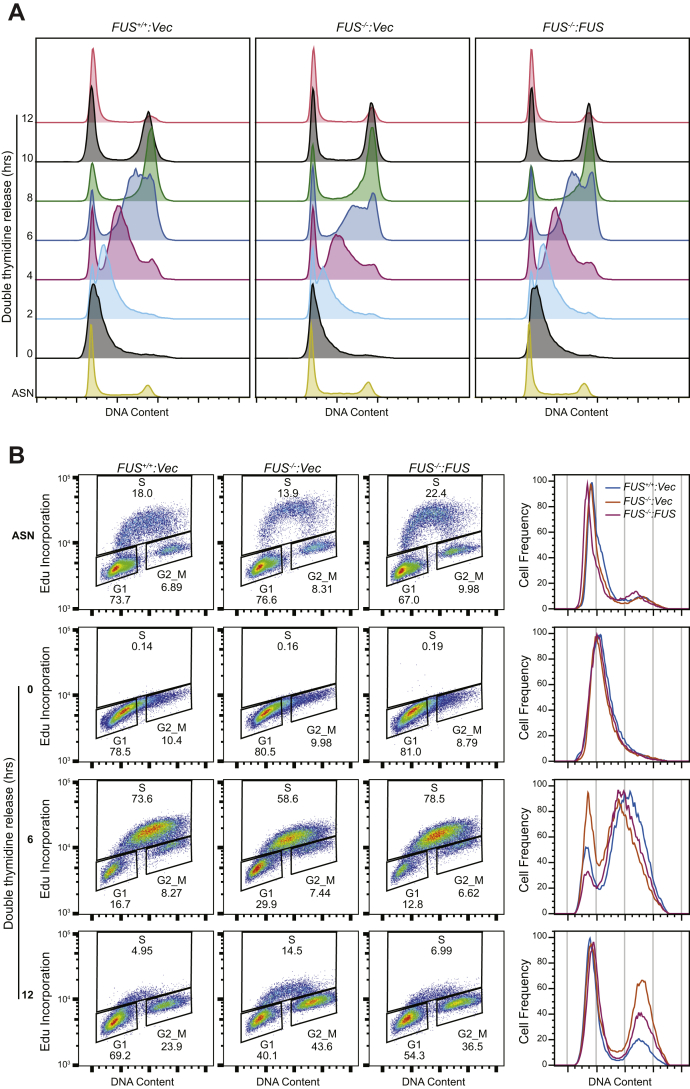
Following synchronous release from G1/S phase arrest, FUS−/− cells exhibited reduced reentry and progression through S phase, which was particularly pronounced at the 6 h time point (Fig. 2A, Fig. S5, A–C). The S-phase delay of FUS−/− cells was further revealed through 5-ethynyl-2′-deoxyuridine (EdU) incorporation experiments. Specifically, FUS−/− cells exhibited reduced S-phase entry 6 h following release from a double thymidine block and accumulated in G2/M to a lesser degree than FUS+/+ or FUS−/−:FUS cells 12 h following release (Fig. 2B; see Fig. S5D for additional time points). These experiments also revealed slightly reduced levels of EdU incorporation in asynchronously growing FUS−/− cells relative to FUS+/+ or FUS−/−:FUS cells (Fig. 2B).
Figure 2.
FUS is required for S-phase progression.A, DNA replication progression was analyzed by PI staining and flow cytometry. Cells were synchronized to G1/S boundary by double thymidine block and released into fresh growth medium for the indicated times and stained with PI for cell cycle analysis. B, DNA progression was monitored by EdU incorporation under the same conditions as in (A). Additional time points are presented in Fig. S5D. EdU, 5-ethynyl-2′-deoxyuridine; FUS, fused in sarcoma; PI, propidium iodide.
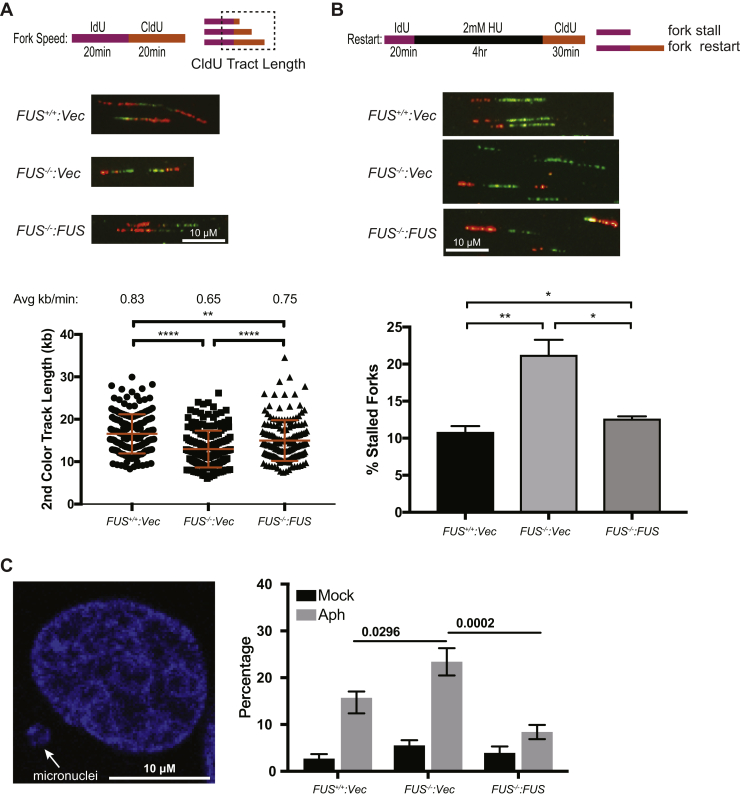
To ascertain impacts of FUS deficiency on replication fork (RF) dynamics, we performed DNA fiber analysis (35), on FUS−/−, FUS+/+, FUS−/−:FUS cells sequentially labeled with 5-iodo-2′-deoxyuridine and 5-chloro-2′-deoxyuridine (CldU). FUS−/− cells exhibited significant reductions in CldU track lengths indicative of reduced DNA replication rate (Fig. 3A). FUS−/− cells also showed delayed RF restart following release from a transient HU block (Fig. 3B). Both replication velocity and replication restart phenotypes were rescued by FUS reexpression.
Figure 3.
FUS deficiency leads to genomic instability and replication stress.A, replication fork speed is reduced in FUS−/− cells. The second pulse (CIdU) was used for measurement of track length, which was converted to micrometers using a 1 μm = 2.59 kb conversion factor. The average fork length was divided by 20 min to derive replication speed. B, replication fork restart was measured as shown in the schematic. Percentages of fork restart (percent of stalled forks) in HU-treated cells are shown. A and B, the representative DNA fiber images were included. Data are mean ± SD (n = 3). p Values were calculated using a t test with Welch's correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. C, FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells were treated with or without 0.2 μM aphidicolin (Aph) for 24 h, fixed, and stained with DAPI for micronucleus counting. p Values were calculated by two-way ANOVA test. Data are means ± SE (n = 3 biological replicates). More than 250 cells for each sample in each biological replicate were counted. CIdU, 5-chloro-2′-deoxyuridine; DAPI, 4′,6-diamidino-2-phenylindole; FUS, fused in sarcoma; HU, hydroxyurea; ns, no significance.
Because a reduced rate of DNA replication can lead to micronucleus formation and genomic instability (36), we measured micronuclei in FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells treated with a low dose of the DNA polymerase alpha inhibitor aphidicolin. FUS−/− cells exhibited increased rates of micronucleus formation relative to FUS+/+ and FUS−/−:FUS U-2 OS cells (Fig. 3C), suggesting that FUS enhances genome stability under replication stress.
Reduced S-phase gene expression in FUS-deficient cells
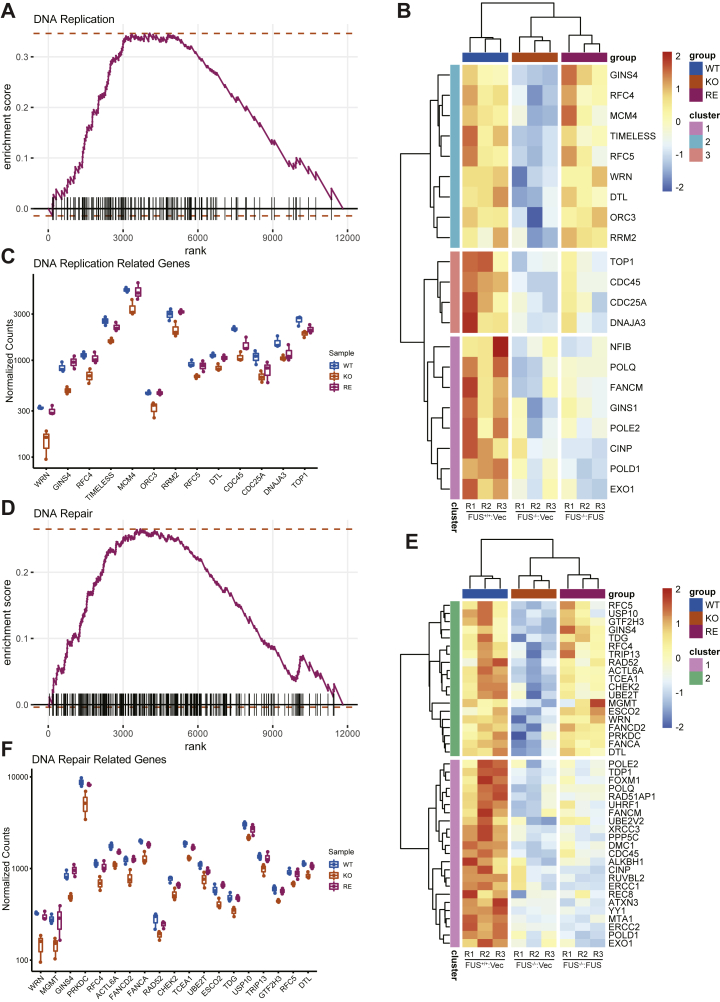
We performed RNA-Seq to establish gene expression correlates for DNA replication defects of FUS-deficient cells (Table S1). We identified 710 genes that were differentially expressed between FUS+/+ and FUS−/− cells that were corrected by FUS reexpression (Fig. S6, A and B and Table S3). Gene set enrichment analysis revealed that cell cycle, DNA repair, and DNA replication processes were downregulated, whereas immunomodulatory pathways were upregulated in FUS−/− cells (Fig. S6C and Fig. 4, A and D). DNA replication–associated genes that were downregulated in FUS−/− cells included GINS4, MCM4, RFC3, RCF4, and TIMELESS (Fig. 4, B and C). DNA repair–related genes, including WRN, PRKDC, FANCD2, FANCA, and RAD52, were also downregulated in FUS−/− cells (Fig. 4, E and F). Interestingly, the NHEJ factor 53BP1 was upregulated in FUS−/− cells (Fig. S6E). A subset of gene expression changes evident in RNA-Seq data were confirmed by quantitative PCR (qPCR; Fig. S6, D and E). Downregulation of S-phase genes may contribute to reduced proliferative potential of FUS−/− cells and/or may be a downstream consequence of DNA replication abnormalities.
Figure 4.
Reduced expression of replication-associated genes in FUS-deficient cells.A, enrichment plot of DNA replication pathway from GSEA using GO gene sets (biological process) in Table S2. B, heat map of differentially expressed DNA replication genes. Genes were clustered to three groups based on ward.D2 method. C, normalized RNA-Seq counts of cluster 2 genes involved in the DNA replication pathway. D, enrichment plot of DNA repair pathway from GSEA using GO gene sets (biological process) in Table S2. E, heat map of the leading gene list of DNA repair pathway showed significant change in all samples. Genes were clustered into two groups based on ward.D2 method. F, DNA repair–related gene expressions in cluster 1 were shown in normalized counts from RNA-Seq results. FUS, fused in sarcoma; GO, Gene Ontology; GSEA, gene set enrichment analysis.
We next considered the possibility that FUS regulates the alternative splicing of suites of genes involved in DNA replication and repair. We thus compared alternative splicing patterns between FUS+/+, FUS−/−, and FUS−/−:FUS cells. We identified 434 splicing events that differed between FUS−/− and FUS+/+ cells including alternative 5′ splice site selection, exon skipping events, altered 3′ splice site selection, and intron retention (Fig. S6F and Table S6). While DNA repair was not overrepresented in Gene Ontology (GO) terms, we nonetheless identified a handful of genes with annotated roles in DNA repair and replication that exhibited FUS-dependent splicing changes (Fig. S6, G and H). For instance, both origin recognition complex 3 (ORC3) and suppressor of cancer cell invasion (SCAI) saw increased inclusion of poison cassette exons predicted to terminate their respective ORFs and/or promote mRNA degradation via nonsense-mediated mRNA decay (Fig. S6, I–N). ORC3 is a component of the eukaryotic origin recognition complex, whereas SCAI is a negative regulator of the NHEJ factor RIF1 and has been implicated in restricting chromatin access to DSB factors (37, 38). Although functional implications are unclear, the inclusion of poison cassette exons may reduce ORC3 and/or SCAI gene dosage. We also identified an alternative cassette exon in the ubiquitin E3 ligase TRIP12 that was increased in FUS−/− U-2 OS cells relative to controls and rescued by FUS reexpression (Fig. S6, O–Q). TRIP12 has been implicated in the ubiquitylation of the p53 regulator ARF and RNF168 (39, 40), and its FUS-dependent alternative splicing may alter its activity toward ARF, RNF168, or other targets.
FUS regulates prereplication complex loading and associates with DNA replication factors
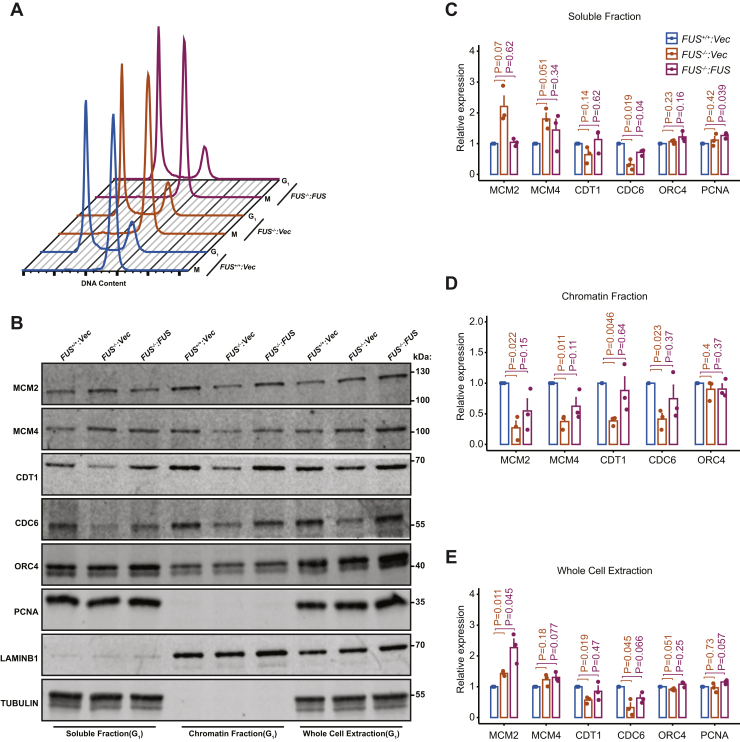
Given their reduced DNA replication rate, we investigated whether FUS−/− cells exhibited defects in the chromatin loading of replication licensing factors, including the ORC, CDC6, CDT1, and the MCM replicative helicase (41). Mitotically arrested FUS+/+, FUS−/−, and FUS−/−:FUS cells were released into early G1 phase, and soluble and chromatin fractions (CFs) were analyzed by immunoblotting. FUS-deficient cells showed normal cell progression from G2/M to G1 phase and unchanged ORC loading onto chromatin in G1 (Fig. 5, A–D). By contrast, both total abundance and chromatin loading of CDC6 and CDT1 was significantly decreased in FUS−/− cells and rescued by FUS reexpression (Fig. 5, B–E). There was a corresponding decrease in CDC6-dependent chromatin loading of MCM2 and MCM4 in FUS-deficient U-2 OS cells (Fig. 5, B–E). Collectively, these results revealed that FUS facilitates ORC-dependent recruitment of prereplication complex (pre-RC) factors to replication origins.
Figure 5.
FUS is required for efficient prereplication complex (pre-RC) loading.A, cell cycle profiles of FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells that were synchronized in early M phase with nocodazole (0.1 μg/ml for 16 h) and then harvested or released into G1 phase for 5 h. B, chromatin loading of ORC and pre-RC proteins in FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells. G1 fractions were immunoblotted with the indicated antibodies. C–E, quantification of Western blotting results for soluble fractions (SFs, panel C), chromatin fractions (CFs, panel D), and whole-cell extracts (WCE, panel E) shown in panel B. Three independent biological replicates were used for the quantification. Data are means ± SE (n = 3 biological replicates). p Values were calculated by Student's t test for comparison between two samples. The expression of proteins in CF was normalized to lamin B1, SF were normalized to tubulin, and WCE were normalized with mean of lamin B1 and tubulin. FUS, fused in sarcoma; ORC, origin recognition complex.
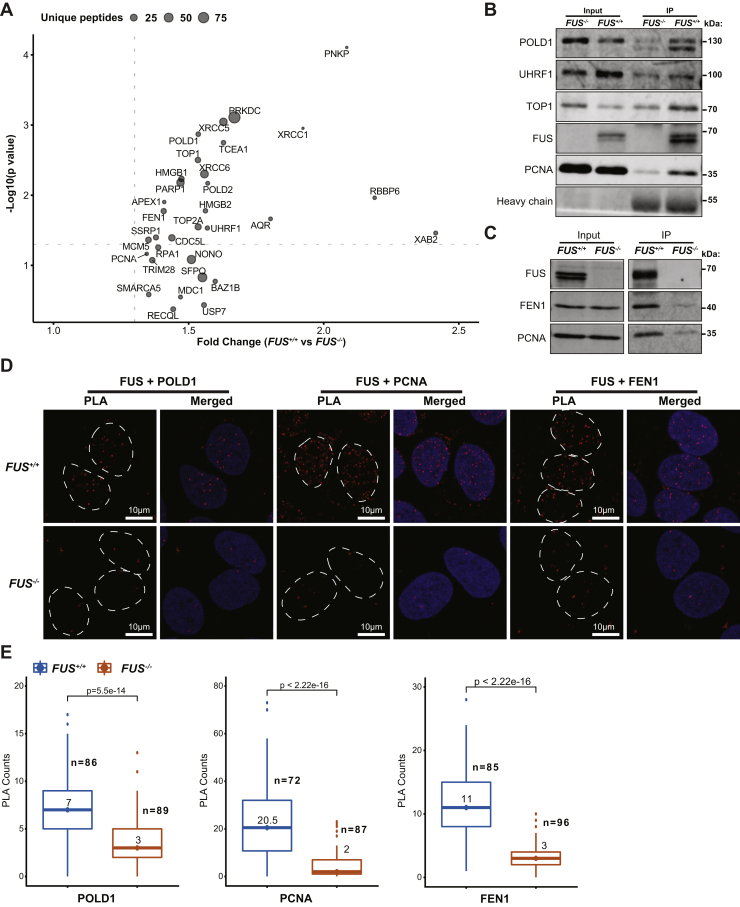
Reasoning that FUS may play direct roles in pre-RC loading and or DNA repair, we performed quantitative proteomic analysis of FUS complexes using a chromatin immunoprecipitation (IP) procedure in which endogenous FUS–chromatin complexes were digested with nuclease prior to IP with α-FUS antibodies and analysis by quantitative MS (42). The same chromatin IP procedure was carried out using FUS−/− cells as a negative control. Gene set enrichment analysis using all identified FUS interactants revealed RNA processing, DNA repair, and DNA replication as functional processes that were statistically overrepresented in the dataset of FUS-interacting proteins (Fig. S7A, Tables S4 and S5). The abundance of RNA-binding proteins in FUS complexes is consistent with other published studies (43, 44). Nucleotide excision repair and DNA strand elongation were among the most significantly enriched pathways within the DNA replication/repair gene sets (Fig. S7, B and C). We plotted those proteins within DNA repair and replication GO terms that showed a nominal 1.3-fold enrichment in IPs from FUS+/+ cells relative to FUS−/− cells (Fig. 6A). Proteins of interest include DSB repair factors (DNA-PK, Ku70, Ku80, and PNKP), single-strand break repair/base excision repair proteins (PARP1, FEN1, PNKP, and APEX1), DNA replication factors (DNA polymerase δ [POLδ or POLD1], proliferating cell nuclear antigen [PCNA], and UHRF1), and topoisomerases (TOP1 and TOP2α). The presence of single-strand break repair/base excision repair factors, including PARP, is consistent with the ability of FUS to bind to PAR chains (26), whereas the presence of POLδ but not POLε in FUS IPs is interesting given their participation in lagging strand and leading strand DNA synthesis, respectively (45). We carried out validation co-IP assays to confirm that endogenous FUS interacted with TOP1, PCNA, POLδ1, and FEN1 in unsynchronized (Fig. 6B) or synchronized S phase cells (Fig. 6C) and further validated association between FUS and POLδ1, PCNA, and FEN1 in proximity ligation assays (Fig. 6, D and E).
Figure 6.
FUS interacts with DNA repair and DNA replication factors.A, FUS-interacting proteins were identified by crosslinking chromatin immunoprecipitation (IP) and analyzed by MS. The results are combination of three biological replicates quantified by nonisotopic spectral peptide counting. The data shown are DNA repair and DNA replication pathway–related interactions based on GSEA (full list is shown in Fig. S6A). The unique peptides are summarized from the raw data of the three replicates. The gray dotted lines are 1.3 of fold change and 0.05 of p value. B, co-IP of FUS with POLD1, UHRF1, TOP1, and PCNA in unsynchronized cells. C, co-IP of FUS with FEN1 and PCNA in synchronized S-phase cells. D, in situ proximity ligation assay (PLA) was employed to verify the interactions between FUS and POLD1, PCNA, and FEN1. Nuclear regions were cycled by dashed lines in PLA red channel based on DAPI signal. E, quantification results of PLA signal in (D). The values are median of PLA foci in each sample. p Values were calculated by Wilcoxon test method. DAPI, 4′,6-diamidino-2-phenylindole; FUS, fused in sarcoma; GSEA, gene set enrichment analysis.
The replication defects in FUS−/− cells and interaction with DNA replication factors raised the possibility that FUS directly participates in DNA replication. To investigate this possibility, we carried out an isolation of proteins on nascent DNA (iPOND) assay that measures the association of proteins with nascently synthesized DNA (46). Human embryonic kidney 293T (HEK293T) cells were pulse labeled with EdU and then chased with thymidine in the absence or the presence of 2 mM HU prior to formaldehyde crosslinking and isolation of EdU–protein complexes. As expected, the abundance of the PCNA sliding clamp in EdU-labeled complexes decreased during the thymidine chase period as the replisome advanced beyond the region of nascent EdU-labeled DNA (Fig. S8). Although FUS was also observed in EdU-labeled DNA, its abundance was slightly increased following thymidine chase, as was histone H3 (Fig. S8). A similar iPOND labeling pattern has been described for DNA-binding proteins such as HMGA1 and LaminB1 that maintain high-order chromatin (47). This result suggests that FUS is proximal to replication factors on chromatin but does not translocate with the active replisome.
FUS regulates DNA RT
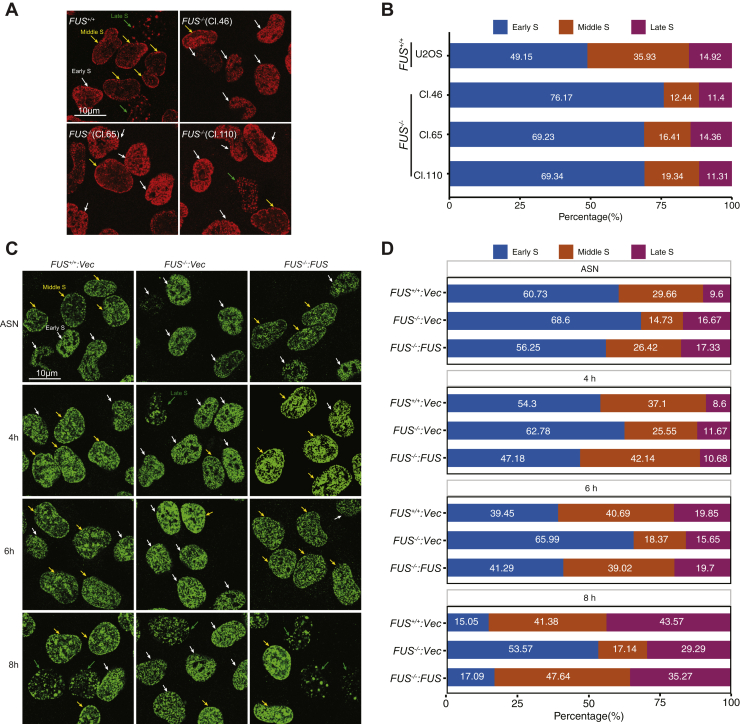
Chromosomal replication is stochastically initiated from origins that fire with characteristic heritable timing (41). RT can be qualitatively evaluated according to the pattern 5-bromo-2′-deoxyuridine (BrdU) or EdU incorporation following synchronized release from a double thymidine block (48). Early S-phase cells exhibit a uniform EdU incorporation pattern (Fig. 7A, white arrows); middle S-phase cells exhibit perinuclear and perinucleolar EdU incorporation (Fig. 7A, yellow arrows); and late S-phase cells exhibit large puncta of EdU incorporation (Fig. 7A, green arrows). Origins with shared firing kinetics are topologically organized into chromatin subdomains in a process that requires RIF1 (49, 50, 51, 52, 53, 54); however, few other timing regulators have been identified.
Figure 7.
FUS regulates DNA replication timing (RT).A, asynchronous U-2 OS cells and three FUS−/− clones (Cl. 46, Cl. 65, and Cl. 110) were pulse labeled with EdU for 20 min and scored for the presence of early, mid, or late EdU staining patterns. B, quantification analysis of cell numbers of each S-phase patterns in (A) and the percentages were calculated in each sample. C, cells were synchronized with double thymidine and then released into S phase for indicated times. Cells were then pulse labeled with BrdU, stained, and imaged by confocal microscopy. D, quantification results of samples using a minimum 100 cells per sample (C). BrdU, 5-bromo-2′-deoxyuridine; EdU, 5-ethynyl-2′-deoxyuridine; FUS, fused in sarcoma.
We noted that the frequency of mid-S phase staining patterns was reduced ∼50% in FUS−/− cells relative to FUS+/+ cells (Fig. 7, A and B), suggesting a potential RT defect. To rule out the apparent defect was not because of delayed S-phase entry of FUS−/− cells, we carried out a time course analysis of FUS+/+, FUS−/−, and FUS−/−:FUS cells released from thymidine block for 4, 6, or 8 h. FUS−/− cells exhibited reduced frequencies of the mid-S-phase staining pattern at all three time points, even though the frequency of late S-phase patterns more than doubled from 4 to 8 h (Fig. 7, C and D). Importantly, FUS reexpression largely reversed the mid-S phase RT defect of FUS−/− cells (Fig. 7, C and D). From this, we conclude that FUS-deficient cells harbor RT defects that cannot be solely attributed to reduced rates of replication.
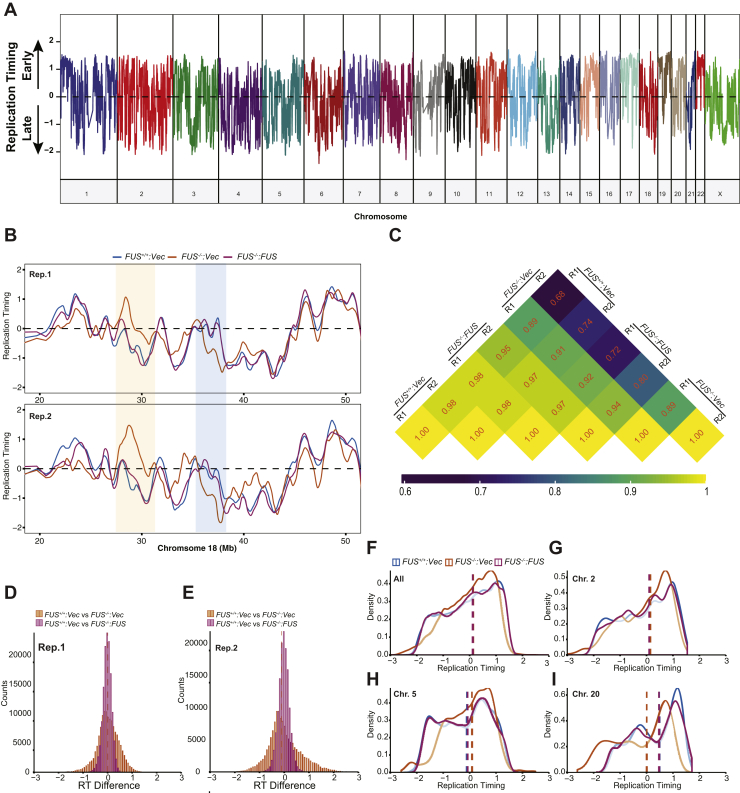
To follow up on the EdU labeling studies, we measured genome-wide RT in FUS+/+, FUS−/−, and FUS−/−:FUS U-2 OS cells using a Sort-Seq workflow (55) in which propidium iodide (PI)–stained FUS−/−, FUS+/+, and FUS−/−: FUS U-2 OS cells were sorted into G1- and S-phase fractions prior to genomic DNA isolation and deep sequencing (see Experimental procedures section). The read ratios between S- and G1-phase cells were used to establish relative DNA copy number between samples, with a higher ratio reflecting earlier replication (Fig. 8A). Using a fixed-window method of read binning, we found that FUS−/− cells exhibited widespread changes in RT relative to FUS+/+ and FUS−/−:FUS cells that was consistent across two biological replicates (Fig. 8, B and C). FUS deficiency impacted RT bidirectionally and was highly chromosome and position dependent. For example, within the same 30 Mb interval of Chr18, FUS−/− cells exhibited advanced RT (Fig. 8B, tan shading) and delayed RT (Fig. 8B, blue shading).
Figure 8.
FUS influences genome-wide RT.A, whole genome-wide replication timing profile of U-2 OS cells. The RT was calculated based on copy number variations between S- and G1-phase cells (S/G1 ratio). The signal was normalized by Z score and smoothed by Loess smoothing. B, representative RT profiles of FUS+/+, FUS−/−, and FUS−/−:FUS cells across two biological replicates. Regions of RT switching between FUS+/+ and FUS−/− are highlighted. C, correlation of RT between two biological replicates by Pearson's method. The smoothed RT values were used for the correlation matrix. D and E, genome-wide distribution of RT scores when comparing FUS+/+ to FUS−/− or FUS+/+versus FUS−/−:FUS in two biological replicates. The bin sizes are 50 and 100 for Replicate1 (Rep. 1) and Replicate2 (Rep. 2), respectively. F–I, the RT density distribution for Rep. 2 was analyzed across all chromosomes (F), Chr.2 (G), Chr.5 (H), and Chr.20 (I). The RT density distribution for Rep. 1 is shown in Fig. S9, A–D. The dashed lines are the median of each sample. The Loess smoothed data were used for analysis. FUS, fused in sarcoma; RT, replication timing.
Genome-wide bidirectional RT switches were further confirmed by RT distribution differences between FUS+/+ and FUS−/− cells in both biological replicates (Fig. 8, D and E). Although the RT distribution of FUS−/− cells skewed slightly earlier than FUS+/+ cells when examined across all chromosomes (Fig. 8F [replicate 2] and Fig. S9A [replicate 1]), RT directional changes were highly chromosome dependent. For example, while Chr2 did not show significant RT distribution differences between FUS+/+ and FUS−/− cells, the RT distributions of Chr5 an Chr20 skewed early and late, respectively, in FUS−/− cells relative to FUS+/+ cells (Fig. 8, G–I [replicate 2] and Fig. S9, B–D [replicate 1]). In summary, our data indicate that FUS influences genome-wide RT in a chromosomal context-dependent manner.
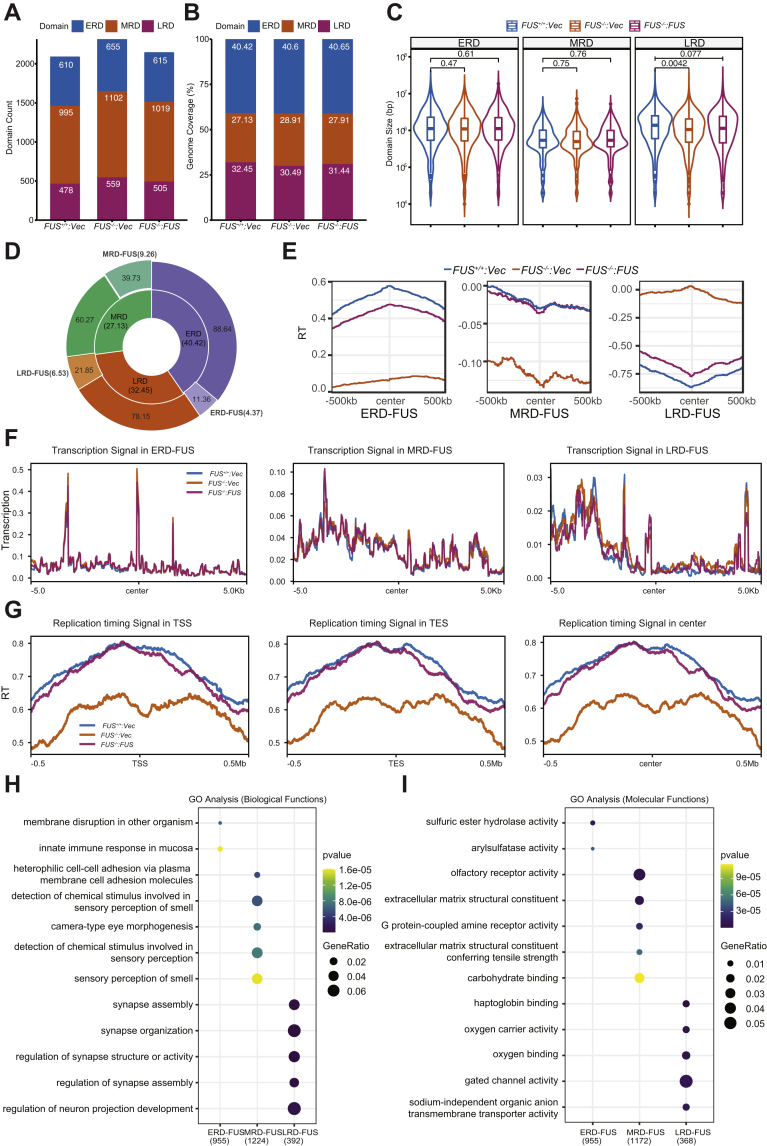
Characterization of FUS-dependent replication domains
We next used the unsupervised Segway deep learning tool (56, 57, 58) to de novo segment replication domains (RDs) in FUS+/+, FUS−/−, and FUS−/−:FUS cells (see Experimental procedures section). Three nonoverlapping contiguous segments were used to assign RT profiles into early replication domains (ERDs); late replication domains (LRDs), and mid replication domains (MRDs) spanning the transition between the early and late zone (Fig. 9A). Genomic coverage of all three types of RDs did not significantly change between FUS−/− cells relative to FUS+/+ cells (Fig. 9B). However, the average size of LRDs was significantly decreased in FUS−/− cells (Fig. 9C). To determine which fraction of ERDs, MRDs, and LRDs were dependent on FUS, overlapping RDs in FUS+/+ and FUS−/−:FUS cells were intersected and then subtracted from corresponding RDs in FUS−/− cells using bedtools. This analysis revealed that 11.36%, 39.73%, and 21.85% of total ERDs, MRDs, and LRDs, respectively, were FUS dependent (Fig. 9D), and we refer to these regions as ERD-FUS, MRD-FUS, and LRD-FUS (Fig. 9D). ERD-FUS, MRD-FUS, and LRD-FUS represented 4.37%, 9.26%, and 6.53% of whole genome sequence, respectively, and, in total, approximately 20% of the U-2 OS genome exhibited FUS-dependent RT. Finally, RT signals of FUS-associated RDs were centered, and the distribution and heat map analysis were performed and showed they were correctly identified (Fig. 9E and Fig. S9, E–G).
Figure 9.
Characterization of FUS-dependent replication domains (RDs).A, RT profiles were segmented into three states by nonsupervised package Segway as early RD (ERD), middle RD (MRD), and late RD (LRD). The domain numbers in each sample were plotted and labeled. The two biological replicates were merged for RD segmentation. B, percentages of genome coverage of RDs in each sample were calculated based on the segmentation. The values are percentages of each domain. C, the same RT domain sizes are compared among all the samples. The Student's t test was used for determination of significance. D, doughnut pie chart of FUS-dependent RD coverage. The percentage of each RD (ERD, MRD, and LRD; center pie) that is altered by FUS deficiency (FUS-dependent RDs) is shown in the outside layer, and the total percentage of each FUS-dependent RDs (ERD-FUS, MRD-FUS, and LRD-FUS) are calculated and shown in parentheses. The percentage was calculated based on the genome coverage. E, RT signal enrichment analysis of FUS-dependent replication domains in the samples. The average domain size is ∼106 bp C, and ∼0.5 × 106 bp flanking the midpoint was used for signal enrichment. Heat map results of RT signal enrichment of changing ERD, MRD, and LRD in all individual samples were shown in Fig. S9, E–G. F, transcription signal in the centered FUS-dependent RDs. Transcription signal was normalized with CPM by STAR. G, RT signal enrichment around TSS, TES, and center of FUS-regulated gene regions across a ±0.5 Mb window. RT signal was calculated by log2 ratio of S/G1 samples in 20 kb bin after CPM normalization and followed with Z score normalization. Only FUS-regulated genes (listed in Table S3) annotation was used. H, Gene Ontology (GO) enrichment in biological function level of FUS-dependent RDs. The FUS-dependent RDs were extended 3000 bases in both ends, and then, the gene list under the extended FUS-dependent RDs was extracted and used for GO analysis. I, GO analysis in molecular function level of extended FUS-dependent RDs. FUS, fused in sarcoma; RT, replication timing; TES, transcription end site; TSS, transcription start site.
Consistent with earlier studies (59, 60, 61), a positive correlation between gene activation and RT was found, with ERDs exhibiting active gene expression and LRDs exhibiting repressed gene expression (Fig. 9F and Fig. S9H). However, overall transcription signals were comparable between FUS−/− cells and FUS+/+ cells across ERD-FUS, MRD-FUS, and LRD-FUS domains (Fig. 9F). We next explored whether RT was changed proximal to genes showing FUS-dependent regulation by RNA-Seq. We found RT of annotated gene regions was delayed in FUS−/− cells relative to FUS+/+ and FUS−/−:FUS cells (Fig. 9G). This pattern of delayed timing was observed across the entirety of the gene, including the transcription start site, coding sequence, and termination site, and was observed for both upregulated and downregulated genes. To determine whether delayed RT was restricted to those genes regulated by FUS, we compared relative RT across all annotated genes. As shown in Fig. S9, I and J, a similar pattern of delayed RT was observed in FUS−/− cells relative to FUS+/+ and FUS−/−:FUS cells. These results imply that FUS plays a particularly important role in the early replication of transcriptionally active chromatin.
Finally, we examined FUS-dependent RDs for gene functional enrichment. Surprisingly, given the non-neuronal nature of U-2 OS cells, we found that LRD-FUS were highly enriched in nervous system development–related genes and, more specifically, genes encoding ion-gated channels (Fig. 9, H and I and Fig. S10, A and B). We further explored the transcription of genes in the top ten enriched GO terms in FUS-dependent LRDs (LRD-FUS) (Fig. S10, A and C). Despite being enriched in LRD-FUS, only ten of 55 genes showed significant expression differences between FUS+/+ and FUS−/− cells, and only a handful of these were rescued by FUS reexpression (Fig. S10, D–F). While the upregulation of repressed neuronal genes might be expected to occur in late-replicating chromatin, the neuron-related genes in LRD-FUS intervals exhibited both upregulation and downregulation in FUS−/− cells. These findings may be relevant to understanding FUS-dependent gene regulation in neurons.
Discussion
FUS DNA repair functions have been deduced from its PARP-dependent recruitment to sites of microirradiation; its co-IP with repair proteins; and the modest chromosome instability and DSB repair defects of FUS-deficient cells (21, 26, 27, 28, 30, 31, 32, 62). Despite these results, a unifying role for FUS in genome protection has yet to emerge. Using FUS−/− cells with and without reconstitution, we found that, while FUS may play a supporting role in DSB repair, it is more prominently required for timely DNA replication, which plausibly contributes to genome instability and DDR-related phenotypes ascribed to FUS-deficient cells.
FUS is among the first factors recruited to sites of microirradiation, which is driven through association of FUS RGG domains with PAR chains (26, 27, 28, 30, 63). FUS is also reportedly required for the assembly of IR-induced 53BP1 foci (27), despite the fact that FUS does not accumulate at these structures (26). Results in Fig. S1A clearly show that FUS is not required for 53BP1 focus formation. In fact, 53BP1 foci recruitment was more persistent in FUS−/− cells relative to FUS+/+ controls. This result appears to be congruent with the findings of Altmeyer et al. (63) who reported that overexpression of the EWSR LCD suppressed IR-induced 53BP1 focus formation. It is conceivable that FUS inhibits local assembly of 53BP1 complexes and/or limits their lateral spread along damaged chromatin. By contrast, FUS−/− cells exhibited reduced recruitment of BRCA1 (Fig. S2), which mediates HDR and mutually antagonistic to the 53BP1-RIF1 pathway (64). Despite these modest molecular defects, FUS deficiency does not confer sensitivity to mechanistically distinct genotoxins, including IR, camptothecin, MMC, or CLM (Figs. S3 and S4). Our findings are at odds with a recent report by Levone et al. (65) that FUS−/− HeLa cells are sensitive to DSB-inducing agents and that FUS is required for the recruitment of 53BP1, Ku80, and other DSB repair factors to DNA damage. The use of p53-wildtype (U-2 OS) versus p53-inactivated (HeLa) cells or different cell survival assays might underlie differences in genotoxin sensitivity between the studies, whereas discrepant 53BP1 recruitment findings may be related to the use of microirradiation, which induces supraphysiological levels of DSBs and SSBs (66).
FUS−/− cells exhibited reduced proliferative potential characterized by reduced RF speed (Fig. 3A), delayed RF restart (Fig. 3B), reduced expression of S-phase–associated genes (Fig. 4C), and reduced loading of pre-RC complexes (Fig. 5B). Participation of FUS in DNA replication was suggested by the presence of DNA replication factors in FUS–chromatin complexes (Fig. 6). The association of FUS with lagging strand synthesis factors POLδ1, PCNA, and FEN1, but not leading strand POLε, further suggested that FUS may play a role in the deposition or removal of RNA primers and/or the ligation of single-strand nicks on the lagging strand. It is worth noting that PARP, which was also present in FUS–chromatin complexes, contributes to the ligation of Okazaki fragments on the lagging strand (67). It is also conceivable that FUS plays a role in the postreplication repair of stalled RFs given established roles for POLδ in this process (68). A speculative model depicting a role for FUS in lagging strand synthesis is presented in Figure 10A. Alternatively, because FUS did not stably associate with translocating replisomes in the iPOND assay (Fig. S8), it is possible that FUS impacts DNA replication by influencing local chromatin structure or through splicing regulation. Indeed, we identified a handful of DNA replication/repair genes whose alternative splicing was altered in FUS deficiency (Fig. S6, F–Q). For instance, the inclusion of a poison cassette exon in ORC3 is upregulated in FUS−/− cells relative to FUS+/+ cells (Fig. S6K). Establishing a causal role for splicing changes in the growth and repair defects of FUS-deficient cells awaits future study.
Figure 10.
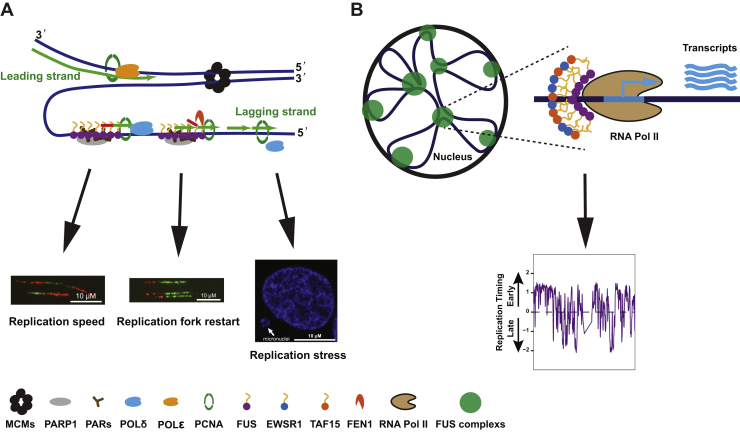
Working model of FUS in replication progression and replication timing.A, based on FUS chromatin proteomics, FUS specifically interacts with POLδ but not POLε. Many replication-coupled single-strand break (SSB) repair factors (PCNA, FEN1, and PARP1) were also enriched with FUS on chromatin. From this, we speculate that FUS facilitates Okazaki fragment processing and PARP-dependent repair of single-strand gaps on the lagging strand (67). Defects in this pathway may contribute to reduced RF speed, RF restart defects, and micronucleus formation. B, speculative model for FUS-dependent RT. FUS undergoes phase separation where it may interact transiently recruits RNA polymerase II, potentially in cooperation with EWSR1 and TAF15. Phase-separated FUS complexes (shown in green circles) organize chromatin into topologically distinct domains (ERD, MRD, and LRD) that are replicated during early, mid, and late S-phase, respectively. The DNA fiber and micronuclei images were reused from Figure 3 for illustration purpose only. ERD, early replication domain; FEN1, flap endonuclease-1; FUS, fused in sarcoma; LRD, late replication domain; MRD, mid replication domain; PARP1, poly(ADP)-ribosyl (PAR) polymerase 1; PCNA, proliferating cell nuclear antigen; POLδ, polymerase δ; RF, replication fork.
To our knowledge, this is the first study to implicate FUS in the control of RT, a stable cellular characteristic that is established in early G1 phase (48, 52, 69). Spatiotemporal control of RT is highly dependent on the master timing factor RIF1, a chromatin-bound factor that also plays important roles in RF stabilization and DSB repair pathway choice (64, 70, 71, 72, 73, 74). RIF1-deficient mammalian cells or yeast exhibit spatial changes in DNA replication that correlate with premature replication origin firing (53, 54, 75, 76, 77). RIF1 is thought to control RT by organizing chromatin domains with shared timing characteristics (78). Like RIF1-deficient cells, FUS−/− cells exhibited bidirectional RT changes; however, FUS and RIF1 are unlikely to act through the same pathway since they exhibited different intranuclear localization patterns (not shown) and did not detectably interact on chromatin (Fig. 6A). In addition, the fact that some chromosomal domains replicate earlier in FUS−/− cells relative to FUS+/+ cells suggests that the RT functions of FUS are at least partially independent from its positive contributions to DNA replication initiation.
Two plausible models may underlie participation of FUS in RT, with both models invoking the phase-separation characteristics of FUS as a central mechanistic feature (79). First, FUS may fulfill a chromatin-bundling function (53, 78). In this model, the DNA-binding and dynamic oligomerization properties of FUS promote the assembly of chromatin domains that are replicated with similar timing. This role would be conceptually similar to that proposed for RIF1. Nonexclusively, FUS-dependent RT may be linked to its roles in transcriptional activation. The LCDs of FUS, EWSR1, and TAF15 bind to the CTD of RNA pol II (19, 80, 81, 82) and function as potent transcriptional activators when fused to heterologous DNA-binding domains (15, 83, 84). Indeed, transcriptional deregulation is thought to drive malignant transformation in soft-tissue sarcomas harboring oncogenic fusions of FET genes with site-specific transcription factors, such as CHOP, FLI1, and CREB (4, 85, 86). A reciprocal relationship between RT and transcription is supported by studies showing that RT switches during embryonic development precede transcriptional changes of proximal genes (59, 87, 88) and work showing that transcriptional activation leads to RT advancement (89). Sima et al. (49) further demonstrated that cis-regulatory elements within an enhancer promoted early RT of the Dppa2/4 domain in mouse ESCs. While absolute levels of transcription were not significantly different, transcriptionally active genes showed delayed RT in FUS−/− cells relative to FUS+/+ cells (Fig. 9, F and G).
The LCD of FUS, in addition to intrinsically disordered regions of transcriptional coactivators, BRD4 and MED1, has been implicated in the assembly of phase-separated transcription “condensates” at gene enhancers (90, 91, 92). We speculate that FUS-dependent clustering of transcription complexes and chromatin looping may specify FUS-dependent RDs that undergo coordinate RT regulation (Fig. 10B). Chromatin capture approaches, such as ChIA-PET, will be needed to test this hypothesis.
It has been reported that ALS-associated mutations in FUS disrupt DSB repair (27, 65), SSB repair (31), and resolution of R-loops, which are RNA–DNA hybrids that can be processed into DSBs during transcription (93). While our present experiments did not evaluate the replication or repair functions of FUSALS mutants, the subtle DSB repair and genotoxin sensitivity phenotypes of FUS−/− cells makes it unlikely that such mutants drive genome stability through a purely LOF mechanism. On the other hand, FUS aggregation in end-stage ALS could confer LOF repair phenotypes or and/or sequester critical repair factors in the cytoplasm. Along these lines, FUS aggregation and nuclear LOF could impact the proliferative capacity, genome stability, and function of glia and other mitotic cell types in the central nervous system. The extent to which neuronal or glial genome instability would drive disease in ALS-FUS relative to other mechanisms, including translational deregulation, splicing deregulation, nuclear import/export deregulation, and proteostasis defects, remains to be determined.
Experimental procedures
Cell culture and gene editing
The U-2 OS, NCI-H460, and HEK293T cell lines were obtained from the American Type Culture Collection. U-2 OS and U-2 OS derivative cell lines were grown in McCoy's medium (Corning; 10-050-CV). HEK293T cells were grown in Dulbecco's modified Eagle's medium (Corning; 10-013-CV). NCI-H460 cells were maintained in RPMI1640 medium (Corning; 10-040-CV). All cell lines were grown in medium with 10% fetal bovine serum (Atlanta Biologicals, Inc) and 1% penicillin/streptomycin (Corning; 30-002-CI) and incubated at 37 °C in 5% CO2. For G1/S synchronization experiments, cells were treated with 2 mM thymidine for 19 h, released into thymidine-free growth media for 9 h, and then returned to thymidine-containing media for an additional 16 h. The cells were washed three times with PBS and then released into complete media for the indicated periods.
FUS−/− cells were generated by transient transfection of U-2 OS cells with pX459 vectors (v2, Addgene plasmid no. 62988) (94) expressing single guide RNAs (CGCCAGTCGAGCCATATCCC and AGAGCTCCCAATCGTCTTAC) targeting exon 4 using jetPRIME (Polyplus; 114-07). Twenty-four hours after transfection, cells were selected for 72 h with 1 μg/ml puromycin and then diluted into 96-well plates at an average density of one cell per well, and single clones were isolated and screened for FUS knockout by Western blotting. All clones were sequenced around the targeted sequence, and four clones were selected for further study. We reconstituted FUS−/− Cl.110 with a FUS CDS cloned into a pQCXIH CMV/TO DEST retroviral vector (Addgene; #17394) vector by gateway cloning. As a negative control, the GUS gene from vector pENTRGUS (Invitrogen) was also cloned into pQCXIH CMV/TO DEST vector. The FUSΔLCD construct was generated by PCR amplification of the FUS CDS beginning at codon 155 and recombination cloning into pQCXIH CMV/TO DEST as described previously. Retroviral plasmids were packaged with GP2-293 packaging cell line (Clonetech; 631458). Stably transduced cells were selected with 50 μg/ml hygromycin for 1 week, and single clones were isolated, expanded, and tested for FUS expression.
Lentivirus and retrovirus
The pLKO.1 system was used to package lentiviruses and deliver shRNA. The following shRNA target sequences were designed using the RNAi Consortium online tool (Broad Institute) and were cloned into pLKO.1-TRC (Addgene; #10878), according to the manufacturer's suggestions: EWSR1 5′-TGCATTGACTACCAGATTTAT-3′ and TAF15 5′-TGACATGATCCATAGTGAAAT-3′. The nontargeting shRNA (Addgene; #1864) and FUS sequences have been reported previously (26). For pSUPERIOR system, nontargeting 5′-TTCTCCGAACGTGTCACGT-3′, TAF15 5′-ACAGCGGAGATAGAAGTGG-3′. Lentiviral particles were produced by transient transfection of HEK293T cells with pLKO.1, psPAX2 (Addgene; #12260), and VSV-G (Addgene; #8454) in a ratio of 3:2:1. Retrovirus particles were produced as described previously using GP2-293 system.
EdU labeling, flow cytometry, microscopy, and DNA fiber analysis
For cell cycle progression experiments, U-2 OS cells were incubated with 20 μM EdU for 30 min before collection and then fixed with ice-cold 70% ethanol. EdU detection was performed using the Click-IT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies; C10634). PI was added to a concentration of 50 μg/ml. Flow cytometry was performed on Thermo Fisher Attune, and data were analyzed and organized using FlowJo software (FlowJo, LLC). For in situ EdU and BrdU staining, U-2 OS cells were pulse labeled for 30 min with 20 μM BrdU or EdU and fixed with 4% paraformaldehyde. For BrdU detection, cells were then incubated with 2 M HCl for 30 min and then permeabilized with 0.2% Triton-X100 for 15 min at room temperature, washed, and blocked in 3% bovine serum albumin (BSA). Cells were stained with BrdU primary antibody (Santa Cruz; sc-32323) in 3% BSA and incubated overnight at 4 °C, followed by washing in PBS with 0.02% Tween-20 and incubation with appropriate secondary antibodies in 3% BSA for 1 h at room temperature. EdU was detected by click chemistry and described previously. Samples were mounted in VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector; H-1200) before imaging. For general immunostaining experiments, cells were seeded into 12-well plate with glass coverslip (and transferred to a humidity chamber for immunostaining with appropriate antibodies). Nuclear DNA either stained with 0.5 μg/ml DAPI for 10 min at room temperature and then mounted with mounting medium for fluorescence (Vector; H-1000) or directly mounted in mounting medium with DAPI for fluorescence (Vector; H-1200) before imaging. Images were acquired using a Nikon A1RS Confocal Microscope under a 63× oil immersion objective. Images were organized using Fiji ImageJ software. Proximity ligation assay foci were counted in CellProfiler (version 3.1.5). DNA fibers were prepared and analyzed as described (35). In brief, cells were pulsed with 50 μM 5-iodo-2′-deoxyuridine and CldU for times indicated in each experiment. Cells were lyzed directly on glass slides, fixed, denatured, stained, and imaged with Keyence BZ-X710 microscope. Image analysis was done with ImageJ. A minimum of 150 fibers were measured for each independent experiment, and analysis shows mean of three independent experiments (biological replicates).
RNA-Seq and gene expression
Total RNA was isolated using the TRIzol reagent (Invitrogen; 15596018) following the manufacturer's protocol and treated with TURBO DNase (Invitrogen; AM2239). Then RNA samples were sent to Novogene (Novogene Co, Ltd) for nonstranded cDNA library building and sequencing at PE150 with NovoSeq 6000. Raw read adapters were trimmed by fastp (95) and then were mapped to human genome (GRCh38) by STAR. The number of RNA-Seq reads mapped to each transcript was summarized with featureCounts (96), and differential expression was called using DESeq2 (97). Three biological replicates were used for each sample. Splicing events were identified by MAJIQ2 (98)and filtered with an absolute dPSI ≥20%. GO analysis was performed on MetaScape Web site (99). Signal tracks were visualized by trackViewer (100). For qPCR analysis, total RNA was reverse transcribed into cDNA using SuperScript IV VILO Master Mix with ezDnase enzyme Kit (Invitrogen; 11766050). The primers were designed by Beacon Designer or National Center for Biotechnology Information primer-blast online tool. qPCR reaction was performed on Bio-Rad CFX RealTime PCR system using iTaq Universal SYBR Green Supermix (Bio-Rad; 1725125).
RT analysis
Cells were prepared and collected accordingly (55) with the following modifications. Approximately 10 million asynchronous cells were collected and fixed in 70% ethanol, washed with ice-cold PBS, and treated with Accutase (CORNING; 25-058-CI) for 20 min at room temperature. Cells were pelleted and resuspended in 2 ml PBS with 250 μl 10 mg/ml RNaseA and incubated at 37 °C for 30 min and stained with PI and then sorted to G1- and S-phase fractions by flow cytometry. DNA extracts from sorted cells were prepared using with DNeasy Blood and Tissue Kit (Qiagen; 69504) and single-end 100-base sequencing libraries prepared using TruSeq kit (Illumina), and deep sequencing was performed on HighSeq 2500. The analysis was carried out according to Marchal et al. (101). Briefly, reads were trimmed by fastp and mapped onto the human genome (GRCh38) using bowtie2. The RT (S/G1 ratio) was calculated in a fixed window size of 20 kb. Then RT raw data were used for quantile normalization and then smoothened with Loess smoothing. The RT signal and replication signal enrichment analysis were performed by deeptools (102). Two biological replicates were analyzed separately. RT domains were identified by unsupervised Segway deep learning tool (56, 57, 58) to de novo segment RDs in our samples with the setting: resolution = 1000, num-labels = 3. The running script can be found on GitHub (https://github.com/biofisherman/FusReplication).
Immunoblotting
For whole-cell extraction, cells were resuspended in high salt lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 0.5% Triton X-100, 2 mM MgCl2, 3 mM EDTA, 1% Protease Inhibitor Cocktail [Sigma, P8340-5 ml]) supplemented with benzonase (50 U/ml) and incubated on ice for 20 min followed by the addition of 4× SDS-loading buffer and heating at 95 °C for 15 min. For CF, cells were resuspended in cytoskeleton (CSK) buffer (20 mM Hepes–KOH [pH 7.4], 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 1% Protease Inhibitor Cocktail [Sigma; P8340-5 ml]) containing 0.5% Triton X-100, incubated on ice for 20 min, and centrifuged for 5 min at 5000g at 4 °C. The supernatant was transferred to a new tube and saved as soluble fraction, whereas the pellet/CF was washed twice in CSK buffer without detergent and resuspended in CSK buffer with benzonase (50 U/ml) for 20 min digestion at which time 4× SDS loading buffer was added and the lysates heated to 95 °C for 15 min. For immunoblotting, samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes and immunoblotted with primary antibodies and LI-COR IRDye secondary antibodies (IRDye 800CW goat anti-rabbit and IRDye 680RD goat antimouse) as described (103, 104). Signals were acquired using Odyssey bio-systems (LI-COR Biosciences). Immunoblotting results were analyzed and organized with ImageStudio Lite software (LI-COR Biosciences).
FUS purification and MS
Rapid IP MS of endogenous proteins assay of FUS was carried out as described (42) with the following modifications. Briefly, ∼20 million cells were counted and fixed with 20 ml 1% formaldehyde solution for 8 min at room temperature. Fixation was quenched by adding 0.12 M glycine. The soluble fraction was extracted in 10 ml of LB1 (50 mM Hepes–KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1% Protease Inhibitor Cocktail [Sigma; P8340-5 ml]) for 10 min with rotation at 4 °C. Cell nuclei were pelleted and washed once with 10 ml LB2 (10 mM Tris–HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 1% Protease Inhibitor Cocktail) and then resuspended in 500 μl LB3 (10 mM Tris–HCl [pH 8.0], 100 mM NaCl, 2.5 mM MgCl2, 0.1% [w/v] sodium deoxycholate, 0.5% Triton X-100, and 1% Protease Inhibitor Cocktail) with 500 U benzonase and incubated at room temperature for 30 min. Benzonase was deactivated with 2 mM EDTA and 1 mM EGTA. To this mixture was added 50 μl 10% Triton X-100, 37.5 μl of 4 M NaCl, and LB3 to bring the total lysate volume of each sample to 1 ml. Digested lysates were briefly sonicated using a 10 s/50 s on/off cycle for three times at 40% power and clarified by centrifugation at 20,000g for 10 min at 4 °C, and supernatants were incubated with 10 μg FUS antibody (Bethyl; A300-302A) overnight at 4 °C with rotation. Subsequently, 50 μl of prewashed Dynabeads protein G (Invitrogen; 10003D) was added to the lysates and incubated for additional 4 h at 4 °C. For Western blot, beads were washed sequentially with 1 ml LB3 and 1 ml radioimmunoprecipitation assay (RIPA) buffer (50 mM Hepes–KOH [pH 7.5], 0.5 M LiCl, 1 mM EDTA, 1% NP-40, 0.7% [w/v] sodium deoxycholate, and 1% Protease Inhibitor Cocktail) once and boiled in 100 μl 2× SDS buffer. For MS, beads were washed five times with 1 ml RIPA buffer and twice in 1 ml of cold fresh prepared 100 mM ammonium hydrogen carbonate (AMBIC) solution and processed as described (42).
FUS rapid IP MS of endogenous proteins from FUS+/+ and FUS−/− cells were subjected to tryptic digestion and orbitrap MS using the filter-aided sample preparation method (105). The tryptic digest solution was desalted/concentrated using an Omix 100 μl (80 μg capacity) C18 tip. The solution was pipetted over the C18 bed five times and rinsed three times with water and 0.1% TFA to desalt. The peptides were eluted from the C18 resin into 150 μl 70% acetonitrile, 0.1% TFA, and lyophilized. The peptides were resuspended in 95:5 H2O:acetonitrile, 0.2% formic acid, and analyzed in duplicate as described later. Samples were analyzed in duplicate (two technical replicates for each of the three biological replicates) by HPLC–electrospray ionization–MS/MS using a system consisting of a high-performance liquid chromatograph (nanoAcquity: Waters) connected to an electrospray ionization Orbitrap mass spectrometer (Q Exactive HF; Thermo Fisher Scientific). HPLC separation employed a 100 × 365 μm fused silica capillary microcolumn packed with 20 cm of 1.7 μm diameter, 130 Å pore size, C18 beads (Waters BEH), with an emitter tip pulled to approximately 1 μm using a laser puller (Sutter Instruments). Peptides were loaded on column at a flow rate of 400 nl/min for 30 min and then eluted over 120 min at a flow rate of 300 nl/min with a gradient of 5% to 35% acetonitrile, in 0.1% formic acid. Full-mass profile scans were performed in the FT orbitrap between 375 and 1500 m/z at a resolution of 120,000, followed by MS/MS HCD scans of the ten highest intensity parent ions at 30% relative collision energy and 15,000 resolution, with a mass range starting at 100 m/z. Dynamic exclusion was enabled with a repeat count of one over a duration of 30 s.
The data analysis was performed using MetaMorpheus, version 0.0.303 (106, 107). Peaks were read from the raw files, using ThermoRawFileReader for MS1 peak centroiding. The following search settings were used: protease = trypsin; maximum missed cleavages = 2; minimum peptide length = 7; maximum peptide length = unspecified; initiator methionine behavior = Variable; fixed modifications = Carbamidomethyl on C, Carbamidomethyl on U; variable modifications = Oxidation on M; max mods per peptide = 2; max modification isoforms = 1024; precursor mass tolerance = ±5 PPM; product mass tolerance = ±20 PPM. The search database (canonical human UniProt database downloaded 07/09/2017, appended with common Repository of Adventitious Proteins contaminants) contained 20,336 protein entries. Target peptides below 0.01 peptide spectrum match q value were quantified by label-free MS1 peak height with FlashLFQ (108, 109) (included in MetaMorpheus), where the q value was estimated from target-decoy competition with sequence-reversed decoys. The mean of two technical replicates of each biological replicate was used, and there were total three biological replicates for following analysis. The proteins with average PSMs in three biological replicates lower than 5 were filtered out for data analysis.
Cell proliferation and survival assays
For cell proliferation assay, 500 cells were plated in each well of 96-well plate, and each sample had six replicates and monitored for 6 days from day 0 to day 5 by CellTiter-Glo 2.0 Assay (Promega; G9242) according to the manufacturer's instructions. The luminescence was recorded by SpectraMax i3 (Molecular Devices). For cell viability assay, 1000 cells/well were plated in 96-well plate with drug-free medium, and varying amounts of drugs were added after 12 h in fresh medium. Cell survival was assayed as same as cell proliferation assay after 3 or 5 days as indicated in the legend to the figure. Data were analyzed and organized by Prism 8.
iPOND assay
The iPOND experiments were performed as described (35, 110) with minor modifications. Briefly, ∼108 HEK293T cells were pulse labeled with 20 μM EdU for 15 min followed by a 1 h chase with 20 μM thymidine. To induce replication stress, cells were treated with 2 mM HU after EdU labeling for 2 h, and then chased with 20 μM thymidine for 1 h. Each plate was crosslinked with 10 ml 1% formaldehyde in PBS for 20 min and quenched by adding 1 ml of 1.25 M glycine for 5 min. The conjugation of biotin to EdU was carried out by click chemistry reaction for 2 h at room temperature in click reaction buffer (10 μM biotin–azide, 10 mM sodium-l-ascorbate, 2 mM CuSO4, and 800 μM Tris(3-hydroxypropyltriazolylmethyl)amine in PBS) and followed by washing once in 0.5% BSA in PBS and once in PBS. Cells were resuspended in LB3 with 500 U benzonase (Santa Cruz; sc-202391) and incubated at room temperature for 30 min. Digested lysates were briefly sonicated using a 10 s/50 s on/off cycle for four times at 40% power and clarified by centrifugation at 8000g for 10 min at 4 °C, and supernatants were incubated overnight with 50 μl magnetic streptavidin beads (Dynabeads MyOne Streptavidin T1; 65601) at 4 °C with rotating. Beads were washed once in 1 ml washing buffer (20 mM Tris–HCl [pH 8.0], 500 mM NaCl, 2 mM EDTA, 0.1% [w/v] sodium deoxycholate, and 1% Triton X-100), once with 1 ml RIPA buffer (50 mM Hepes–KOH [pH 7.5], 0.5 M LiCl, 1 mM EDTA, 1% NP-40, 0.7% [w/v] sodium deoxycholate, and 1% Protease Inhibitor Cocktail) and twice in LB3 buffer. Proteins were eluted by boiling in 2× SDS buffer for 25 min.
Statistical processing
Statistical analysis information including individual replicates and biological replicates number, mean or median, and error bars are explained in the legends to the figures. The statistical tests and resulting p values are shown in the legends to the figures and/or figure panels.
Data availability
All the source data represented in the figures and bioinformatics analysis scripts are available on GitHub (https://github.com/biofisherman/FusReplication). Accession numbers: RT sequencing data have been deposited in the National Center for Biotechnology Information under accession code PRJNA615974. RNA-Seq data accession code is GSE147784 in Gene Expression Omnibus. The MS raw data have been deposited to MassIVE, and the access ID is MSV000087698.
Supporting information
This article contains supporting information (56, 96, 97, 98, 100, 102, 111, 112, 113, 114, 115, 116).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We gratefully thank Ammon Koren for the provided protocol for RT sample preparation and communication of RT analysis. We are grateful to Pfizer Compound Transfer Program for calicheamicin A. We thank Lance A. Rodenkirch in optical imaging core and Flow Cytometry Laboratory for assistance.
Author contributions
W. J., S. H. K., M. A. S., A. S. M., T. T. H., and R. S. T. conceptualization; W. J., S. H. K., M. A. S., P. T., R. J. M., L. L., A. S. M., and R. S. T. methodology; R. J. M. software; W. J. and W. M. G. validation; W. J., S. H. K., M. A. S., P. T., R. J. M., W. M. G., L. L., and A. S. M. formal analysis; W. J., R. J. M., and W. M. G. investigation; S. H. K., M. A. S., P. T., A. S. M., L. M. S., T. T. H., and R. S. T. resources; W. J., S. H. K., M. A. S., P. T., R. J. M., W. M. G., L. L., and A. S. M. data curation; W. J. and R. S. T. writing–original draft; W. J., M. A. S., and R. S. T. writing–review and editing; W. J. and L. L. visualization; L. M. S., T. T. H., and R. S. T. supervision; R. S. T. project administration; L. M. S., T. T. H., and R. S. T. funding acquisition.
Funding and additional information
This work was supported by the National Cancer Institute (R01CA180765-01 to R. S. T.); National Institute of Neurological Disorders and Stroke (1R21NS090313-01A1 to R. S. T.); National Institute of Environmental Health Sciences (R01ES025166 to T. T. H.); University of Wisconsin Carbone Cancer Center support grant (P30CA014520); National Institutes of Health Shared Instrumentation Grants (1S10RR025483-01); National Institute of General Medical Sciences (R35GM126914 to L. M. S.); National Human Genome Research Institute training grant to the Genomic Sciences Training Program (5T32HG002760 to R. J. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Patrick Sung
Supporting information
References
- 1.Tan A.Y., Manley J.L. The TET family of proteins: Functions and roles in disease. J. Mol. Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang Y., Huang E.J. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016;1647:65–78. doi: 10.1016/j.brainres.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabbitts T.H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 4.Crozat A., Aman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 5.Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., Valdmanis P., Rouleau G.A., Hosler B.A., Cortelli P., de Jong P.J. Mutations in the FUS/TLS on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 6.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K.L., Tripathi V., Al-Saraj S., Al-Chalabi A. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrado L., Del Bo R., Castellotti B., Ratti A., Cereda C., Penco S., Soraru G., Carlomagno Y., Ghezzi S., Pensato V., Colombrita C., Gagliardi S., Cozzi L., Orsetti V., Mancuso M. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2010;47:190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 8.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., Neumann M., Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Erauskin J., Tadokoro T., Baughn M.W., Myers B., McAlonis-Downes M., Chillon-Marinas C., Asiaban J.N., Artates J., Bui A.T., Vetto A.P., Lee S.K., Le A.V., Sun Y., Jambeau M., Boubaker J. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron. 2018;100:816–830.e817. doi: 10.1016/j.neuron.2018.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling S.C., Dastidar S.G., Tokunaga S., Ho W.Y., Lim K., Ilieva H., Parone P.A., Tyan S.H., Tse T.M., Chang J.C., Platoshyn O., Bui N.B., Bui A., Vetto A., Sun S. Overriding FUS autoregulation in mice triggers gain-of-toxic dysfunctions in RNA metabolism and autophagy-lysosome axis. Elife. 2019;8 doi: 10.7554/eLife.40811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamelgarn M., Chen J., Kuang L., Jin H., Kasarskis E.J., Zhu H. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11904–E11913. doi: 10.1073/pnas.1810413115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.D., Simons M., Niessing D., Madl T., Dormann D. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719.e713. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Yoshizawa T., Ali R., Jiou J., Fung H.Y.J., Burke K.A., Kim S.J., Lin Y., Peeples W.B., Saltzberg D., Soniat M., Baumhardt J.M., Oldenbourg R., Sali A., Fawzi N.L., Rosen M.K. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018;173:693–705.e622. doi: 10.1016/j.cell.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Strohl F., Meadows W., Ferry R., Dardov V.J., Tartaglia G.G., Farrer L.A. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–734.e715. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinszner H., Albalat R., Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z., Diaz Z., Fang X., Hart M.P., Chesi A., Shorter J., Gitler A.D. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S.L. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz J.C., Wang X., Podell E.R., Cech T.R. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., McKnight S.L. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., Ron D. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks G.G., Singh N., Nashabi A., Mai S., Bozek G., Klewes L., Arapovic D., White E.K., Koury M.J., Oltz E.M., Van Kaer L., Ruley H.E. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand P., Akhmedov A.T., Delacote F., Durrbach A., Lopez B.S. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: Both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–4521. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- 23.Baechtold H., Kuroda M., Sok J., Ron D., Lopez B.S., Akhmedov A.T. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 24.Guipaud O., Guillonneau F., Labas V., Praseuth D., Rossier J., Lopez B., Bertrand P. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- 25.Gardiner M., Toth R., Vandermoere F., Morrice N.A., Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- 26.Mastrocola A.S., Kim S.H., Trinh A.T., Rodenkirch L.A., Tibbetts R.S. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W.Y., Pan L., Su S.C., Quinn E.J., Sasaki M., Jimenez J.C., Mackenzie I.R., Huang E.J., Tsai L.H. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rulten S.L., Rotheray A., Green R.L., Grundy G.J., Moore D.A., Gomez-Herreros F., Hafezparast M., Caldecott K.W. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Singatulina A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I., Pastre D. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019;27:1809–1821.e1805. doi: 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Guo W., Mitra J., Hegde P.M., Vandoorne T., Eckelmann B.J., Mitra S., Tomkinson A.E., Van Den Bosch L., Hegde M.L. Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in amyotrophic lateral sclerosis. Nat. Commun. 2018;9:3683. doi: 10.1038/s41467-018-06111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Macias M.I., Moore D.A., Green R.L., Gomez-Herreros F., Naumann M., Hermann A., Van Damme P., Hafezparast M., Caldecott K.W. FUS (fused in sarcoma) is a component of the cellular response to topoisomerase I-induced DNA breakage and transcriptional stress. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201800222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Garcia I., Rabbitts T.H. Transcriptional activation by TAL1 and FUS-CHOP proteins expressed in acute malignancies as a result of chromosomal abnormalities. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7869–7873. doi: 10.1073/pnas.91.17.7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., Grishin N.V., Frantz D.E., Schneider J.W., Chen S., Li L. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonzi P., Yin Y., Lee C.W.T., Rothenberg E., Huang T.T. Translesion polymerase kappa-dependent DNA synthesis underlies replication fork recovery. Elife. 2018;7 doi: 10.7554/eLife.41426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffelder D.R., Luo L., Burke N.A., Watkins S.C., Gollin S.M., Saunders W.S. Resolution of anaphase bridges in cancer cells. Chromosoma. 2004;112:389–397. doi: 10.1007/s00412-004-0284-6. [DOI] [PubMed] [Google Scholar]
- 37.Hansen R.K., Mund A., Poulsen S.L., Sandoval M., Klement K., Tsouroula K., Tollenaere M.A., Raschle M., Soria R., Offermanns S., Worzfeld T., Grosse R., Brandt D.T., Rozell B., Mann M. SCAI promotes DNA double-strand break repair in distinct chromosomal contexts. Nat. Cell Biol. 2016;18:1357–1366. doi: 10.1038/ncb3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isobe S.Y., Nagao K., Nozaki N., Kimura H., Obuse C. Inhibition of RIF1 by SCAI allows BRCA1-mediated repair. Cell Rep. 2017;20:297–307. doi: 10.1016/j.celrep.2017.06.056. [DOI] [PubMed] [Google Scholar]
- 39.Chen D., Shan J., Zhu W.G., Qin J., Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S., Mailand N., Neumann B., Heriche J.K., Shearer R., Saunders D. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 41.Fragkos M., Ganier O., Coulombe P., Mechali M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 42.Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D'Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 43.Sun S., Ling S.C., Qiu J., Albuquerque C.P., Zhou Y., Tokunaga S., Li H., Qiu H., Bui A., Yeo G.W., Huang E.J., Eggan K., Zhou H., Fu X.D., Lagier-Tourenne C. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawaguchi T., Rollins M.G., Moinpour M., Morera A.A., Ebmeier C.C., Old W.M., Schwartz J.C. Changes to the TDP-43 and FUS interactomes induced by DNA damage. J. Proteome Res. 2020;19:360–370. doi: 10.1021/acs.jproteome.9b00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgers P.M.J., Kunkel T.A. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Contreras A.J., Ruppen I., Nieto-Soler M., Murga M., Rodriguez-Acebes S., Remeseiro S., Rodrigo-Perez S., Rojas A.M., Mendez J., Munoz J., Fernandez-Capetillo O. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013;3:1105–1116. doi: 10.1016/j.celrep.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimitrova D.S., Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 49.Sima J., Chakraborty A., Dileep V., Michalski M., Klein K.N., Holcomb N.P., Turner J.L., Paulsen M.T., Rivera-Mulia J.C., Trevilla-Garcia C., Bartlett D.A., Zhao P.A., Washburn B.K., Nora E.P., Kraft K. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell. 2019;176:816–830.e18. doi: 10.1016/j.cell.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattarocci S., Hafner L., Lezaja A., Shyian M., Shore D. Rif1: A conserved regulator of DNA replication and repair hijacked by telomeres in yeasts. Front. Genet. 2016;7:45. doi: 10.3389/fgene.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanoh Y., Matsumoto S., Fukatsu R., Kakusho N., Kono N., Renard-Guillet C., Masuda K., Iida K., Nagasawa K., Shirahige K., Masai H. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- 52.Dileep V., Ay F., Sima J., Vera D.L., Noble W.S., Gilbert D.M. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 2015;25:1104–1113. doi: 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamazaki S., Ishii A., Kanoh Y., Oda M., Nishito Y., Masai H. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012;31:3667–3677. doi: 10.1038/emboj.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornacchia D., Dileep V., Quivy J.P., Foti R., Tili F., Santarella-Mellwig R., Antony C., Almouzni G., Gilbert D.M., Buonomo S.B. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012;31:3678–3690. doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koren A., Polak P., Nemesh J., Michaelson J.J., Sebat J., Sunyaev S.R., McCarroll S.A. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan R.C.W., Libbrecht M.W., Roberts E.G., Bilmes J.A., Noble W.S., Hoffman M.M. Segway 2.0: Gaussian mixture models and minibatch training. Bioinformatics. 2018;34:669–671. doi: 10.1093/bioinformatics/btx603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F., Ren C., Li H., Zhou P., Bo X., Shu W. De novo identification of replication-timing domains in the human genome by deep learning. Bioinformatics. 2016;32:641–649. doi: 10.1093/bioinformatics/btv643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivera-Mulia J.C., Buckley Q., Sasaki T., Zimmerman J., Didier R.A., Nazor K., Loring J.F., Lian Z., Weissman S., Robins A.J., Schulz T.C., Menendez L., Kulik M.J., Dalton S., Gabr H. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S., Rathjen P.D., Gilbert D.M. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.W., Lyou Y., Townes T.M., Schubeler D., Gilbert D.M. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng Q., Holler C.J., Taylor G., Hudson K.F., Watkins W., Gearing M., Ito D., Murray M.E., Dickson D.W., Seyfried N.T., Kukar T. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M.-B.D., Streicher W., Jungmichel S., Nielsen M.L., Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., Xu D., Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 65.Levone B.R., Lenzken S.C., Antonaci M., Maiser A., Rapp A., Conte F., Reber S., Mechtersheimer J., Ronchi A.E., Muhlemann O., Leonhardt H., Cardoso M.C., Ruepp M.D., Barabino S.M.L. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202008030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Splinter J., Jakob B., Lang M., Yano K., Engelhardt J., Hell S.W., Chen D.J., Durante M., Taucher-Scholz G. Biological dose estimation of UVA laser microirradiation utilizing charged particle-induced protein foci. Mutagenesis. 2010;25:289–297. doi: 10.1093/mutage/geq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 2018;71:319–331.e313. doi: 10.1016/j.molcel.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao Y., Mutter-Rottmayer E., Zlatanou A., Vaziri C., Yang Y. Mechanisms of post-replication DNA repair. Genes (Basel) 2017;8:64. doi: 10.3390/genes8020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dimitrova D.S., Gilbert D.M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 70.Buonomo S.B.C. Rif1-dependent regulation of genome replication in mammals. Adv. Exp. Med. Biol. 2017;1042:259–272. doi: 10.1007/978-981-10-6955-0_12. [DOI] [PubMed] [Google Scholar]
- 71.Mirman Z., Lottersberger F., Takai H., Kibe T., Gong Y., Takai K., Bianchi A., Zimmermann M., Durocher D., de Lange T. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature. 2018;560:112–116. doi: 10.1038/s41586-018-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dev H., Chiang T.W., Lescale C., de Krijger I., Martin A.G., Pilger D., Coates J., Sczaniecka-Clift M., Wei W., Ostermaier M., Herzog M., Lam J., Shea A., Demir M., Wu Q. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 2018;20:954–965. doi: 10.1038/s41556-018-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A., Lukas J., Choudhary C. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018;173:972–988.e923. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Alvarez-Quilon A., Moatti N., Zimmermann M., Annunziato S., Krastev D.B., Song F., Brandsma I., Frankum J. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560:117–121. doi: 10.1038/s41586-018-0340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012;26:137–150. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silverman J., Takai H., Buonomo S.B., Eisenhaber F., de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu D., Muniandy P., Leo E., Yin J., Thangavel S., Shen X., Ii M., Agama K., Guo R., Fox D., 3rd, Meetei A.R., Wilson L., Nguyen H., Weng N.P., Brill S.J. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29:3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foti R., Gnan S., Cornacchia D., Dileep V., Bulut-Karslioglu A., Diehl S., Buness A., Klein F.A., Huber W., Johnstone E., Loos R., Bertone P., Gilbert D.M., Manke T., Jenuwein T. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol. Cell. 2016;61:260–273. doi: 10.1016/j.molcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bertolotti A., Melot T., Acker J., Vigneron M., Delattre O., Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwartz J.C., Ebmeier C.C., Podell E.R., Heimiller J., Taatjes D.J., Cech T.R. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burke K.A., Janke A.M., Rhine C.L., Fawzi N.L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.May W.A., Lessnick S.L., Braun B.S., Klemsz M., Lewis B.C., Lunsford L.B., Hromas R., Denny C.T. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 85.Bailly R.A., Bosselut R., Zucman J., Cormier F., Delattre O., Roussel M., Thomas G., Ghysdael J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Antonescu C.R., Nafa K., Segal N.H., Dal Cin P., Ladanyi M. EWS-CREB1: A recurrent variant fusion in clear cell sarcoma--association with gastrointestinal location and absence of melanocytic differentiation. Clin. Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- 87.Siefert J.C., Georgescu C., Wren J.D., Koren A., Sansam C.L. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 2017;27:1406–1416. doi: 10.1101/gr.218602.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaaij L.J.T., van der Weide R.H., Ketting R.F., de Wit E. Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep. 2018;24:1–10.e14. doi: 10.1016/j.celrep.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Therizols P., Illingworth R.S., Courilleau C., Boyle S., Wood A.J., Bickmore W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C., Li C.H., Guo Y.E., Day D.S., Schuijers J., Vasile E. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hill S.J., Mordes D.A., Cameron L.A., Neuberg D.S., Landini S., Eggan K., Livingston D.M. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 97.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vaquero-Garcia J., Barrera A., Gazzara M.R., Gonzalez-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5 doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ou J., Zhu L.J. trackViewer: A bioconductor package for interactive and integrative visualization of multi-omics data. Nat. Methods. 2019;16:453–454. doi: 10.1038/s41592-019-0430-y. [DOI] [PubMed] [Google Scholar]
- 101.Marchal C., Sasaki T., Vera D., Wilson K., Sima J., Rivera-Mulia J.C., Trevilla-Garcia C., Nogues C., Nafie E., Gilbert D.M. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 2018;13:819–839. doi: 10.1038/nprot.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim S.H., Stiles S.G., Feichtmeier J.M., Ramesh N., Zhan L., Scalf M.A., Smith L.M., Pandey U.B., Tibbetts R.S. Mutation-dependent aggregation and toxicity in a Drosophila model for UBQLN2-associated ALS. Hum. Mol. Genet. 2018;27:322–337. doi: 10.1093/hmg/ddx403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim S.H., Trinh A.T., Larsen M.C., Mastrocola A.S., Jefcoate C.R., Bushel P.R., Tibbetts R.S. Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism. Nucleic Acids Res. 2016;44:9667–9680. doi: 10.1093/nar/gkw643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 106.Shortreed M.R., Wenger C.D., Frey B.L., Sheynkman G.M., Scalf M., Keller M.P., Attie A.D., Smith L.M. Global identification of protein post-translational modifications in a single-pass database search. J. Proteome Res. 2015;14:4714–4720. doi: 10.1021/acs.jproteome.5b00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Solntsev S.K., Shortreed M.R., Frey B.L., Smith L.M. Enhanced global post-translational modification discovery with MetaMorpheus. J. Proteome Res. 2018;17:1844–1851. doi: 10.1021/acs.jproteome.7b00873. [DOI] [PubMed] [Google Scholar]
- 108.Millikin R.J., Solntsev S.K., Shortreed M.R., Smith L.M. Ultrafast peptide label-free quantification with FlashLFQ. J. Proteome Res. 2018;17:386–391. doi: 10.1021/acs.jproteome.7b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Millikin R.J., Shortreed M.R., Scalf M., Smith L.M. A Bayesian null interval hypothesis test controls false discovery rates and improves sensitivity in label-free quantitative proteomics. J. Proteome Res. 2020;19:1975–1981. doi: 10.1021/acs.jproteome.9b00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dungrawala H., Cortez D. Purification of proteins on newly synthesized DNA using iPOND. Methods Mol. Biol. 2015;1228:123–131. doi: 10.1007/978-1-4939-1680-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., Wiegraebe W., Singh S., Becker T., Caicedo J.C., Carpenter A.E. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sergushichev A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 [Google Scholar]
- 116.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the source data represented in the figures and bioinformatics analysis scripts are available on GitHub (https://github.com/biofisherman/FusReplication). Accession numbers: RT sequencing data have been deposited in the National Center for Biotechnology Information under accession code PRJNA615974. RNA-Seq data accession code is GSE147784 in Gene Expression Omnibus. The MS raw data have been deposited to MassIVE, and the access ID is MSV000087698.