Abstract
The tumor suppressor p53 plays a key role in inducing G1 arrest and apoptosis following DNA damage. The double-stranded-RNA-activated protein PKR is a serine/threonine interferon (IFN)-inducible kinase which plays an important role in regulation of gene expression at both transcriptional and translational levels. Since a cross talk between IFN-inducible proteins and p53 had already been established, we investigated whether and how p53 function was modulated by PKR. We analyzed p53 function in several cell lines derived from PKR+/+ and PKR−/− mouse embryonic fibroblasts (MEFs) after transfection with the temperature-sensitive (ts) mutant of mouse p53 [p53(Val135)]. Here we report that transactivation of transcription by p53 and G0/G1 arrest were impaired in PKR−/− cells upon conditions that ts p53 acquired a wild-type conformation. Phosphorylation of mouse p53 on Ser18 was defective in PKR−/− cells, consistent with an impaired transcriptional induction of the p53-inducible genes encoding p21WAF/Cip1 and Mdm2. In addition, Ser18 phosphorylation and transcriptional activation by mouse p53 were diminished in PKR−/− cells after DNA damage induced by the anticancer drug adriamycin or γ radiation but not by UV radiation. Furthermore, the specific phosphatidylinositol-3 (PI-3) kinase inhibitor LY294002 inhibited the induction of phosphorylation of Ser18 of p53 by adriamycin to a higher degree in PKR+/+ cells than in PKR−/− cells. These novel findings suggest that PKR enhances p53 transcriptional function and implicate PKR in cell signaling elicited by a specific type of DNA damage that leads to p53 phosphorylation, possibly through a PI-3 kinase pathway.
The tumor suppressor p53 protein is critical for regulation of the cell cycle upon genotoxic insult. When DNA is damaged by radiation or chemicals, cells respond rapidly by arresting the cell cycle and/or inducing apoptosis (reviewed in references 39, 47, and 72). G1 arrest requires the activity of wild-type p53; this presumably allows the damaged cell to carry out effective DNA repair, and thereby genomic integrity is maintained. In other cases, either when DNA repair cannot be completed successfully or when the cell is not programmed to respond by a viable growth arrest, p53 activation may destine the cell for apoptosis.
p53 functions as a transcription factor, and major downstream targets, including p21WAF1/Cip1, an inhibitor of the cyclin-dependent kinases, have been identified (39). Although the p53 transcriptional function is important for cell cycle arrest, it may not be essential for every case of p53-dependent apoptosis (72). Growth suppression by p53, through the induction of either growth arrest or apoptosis, can be modulated by several viral and cellular proteins (39, 47).
The double-stranded-RNA (dsRNA)-dependent protein kinase PKR is an interferon (IFN)-inducible protein (reviewed in reference 11). PKR, a polypeptide of 68 kDa in humans and 65 kDa in mice, is a serine/threonine kinase which is activated by autophosphorylation upon binding to dsRNA (11). Activated PKR then phosphorylates the α subunit of the translation initiation factor eIF-2, a modification that causes inhibition of protein synthesis (reviewed in reference 29). PKR exhibits antiviral (reviewed in reference 35) and antiproliferative (3, 8, 40, 51) activities. Consistent with an antiproliferative action, PKR has been shown to induce cell death by apoptosis (15, 45, 46, 66, 75). The molecular mechanisms by which PKR regulates cell growth are not fully understood. One possibility is the regulation of protein synthesis through eIF-2α phosphorylation (3, 16); another is the modulation of signaling pathways leading to gene transcriptional activation (11).
As proteins capable of regulating cell growth, PKR and p53 exhibit some similar properties. For example, (i) induction of both proteins results in inhibition of cell growth in mammalian (11, 47) or yeast (8, 56) cells, (ii) PKR and p53 are able to regulate gene expression at both the transcriptional and translational levels (11, 19, 39, 47), (iii) mutant forms of both PKR and p53 can transform cells in culture (3, 39, 40, 51), (iv) mutant forms of both proteins can block κ-chain transcription and pre-B-cell differentiation in vitro (39, 41), and (v) PKR and p53 can cooperate with the IFN regulatory factor 1 (IRF-1) in gene transcriptional activation (36, 43, 67).
However, many properties of these proteins are distinct. Specifically, (i) p53 is predominantly a nuclear protein (39), whereas only a small fraction (∼20%) of PKR exhibits nuclear localization (32); (ii) PKR is required for antiviral responses (35, 74), but p53 is not; (iii) p53 is essential for inducing cell cycle arrest upon DNA damage (39), but it is not known whether PKR is implicated in this process; (iv) depletion of p53 induces spontaneous tumors in mice (39), whereas depletion of PKR is not tumorigenic (1, 74); and (v) mutations in the p53 gene have been detected in a broad range of human cancers (57), but so far the involvement of PKR in human cancer has not been extensively studied, except in the case of some hematopoietic neoplasms (25).
It has been shown that the ability of some IFN-inducible proteins to modulate the function of proteins that play a key role in cell signaling and proliferation may explain, at least in part, the antiproliferative effects of IFNs (9, 10, 14, 53, 73). We have recently shown that PKR physically associates with p53 and modulates phosphorylation of human p53 on Ser392 in vitro (12). Here we further extend our observations on PKR and p53 by providing evidence for a role of PKR in p53-mediated transcriptional transactivation. We show that transcriptional induction of p21 and mdm2 genes is reduced in PKR−/− cells expressing the temperature-sensitive (ts) mouse p53(Val135) mutant compared to PKR+/+ ts p53-expressing cells at the permissive temperature of 32°C, where p53 acquires a wild-type conformation. In addition, basal phosphorylation levels of mouse ts p53 on Ser18 (the homologue of Ser15 on human p53) are lower in PKR−/− cells than in PKR+/+ cells. Moreover, induction of Ser18 phosphorylation of either ts p53 or endogenous p53 was impaired in PKR−/− cells upon DNA damage. These novel findings support the notion that PKR, in addition to its interaction with p53 (12), is able to modulate p53 transcriptional function by participating in signaling pathways leading to p53 activation.
MATERIALS AND METHODS
Cell culture, transfections, and production of antibody.
Mouse embryonic fibroblasts (MEFs), 3T3-like immortalized fibroblasts, and BALB/c(10)1 cells (generous gift from A. Levine) were grown in Dulbecco modified Eagle medium (DMEM; Life Technologies Inc.) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, and penicillin-streptomycin (100 U/ml).
Cells expressing the ts p53(Val135) mutant were generated by cotransfection of immortalized polyclonal populations of MEFs from PKR+/+ and PKR−/− mice (74) based on a 3T3-like protocol (69) with pLTRp53(val135) DNA (generous gift from B. Vogelstein) and pcDNA3.1/zeo vector (Invitrogen) followed by selection in zeocin (200 μg/ml; Invitrogen). All stable transfections were done by the calcium phosphate method (60), whereas transient transfections of BALB/c(10)1 cells were performed with Lipofectamine Plus (Gibco BRL) according to the manufacturer’s specifications.
The anti-P-Ser15 mouse monoclonal antibody (MAb) was developed by using a phosphopeptide containing P-Ser15 of human p53, SVEPPLpS15QETFSDC, which was chemically synthesized as described previously (38). A mouse was immunized with this phosphopeptide after conjugation with keyhole limpet hemocyanin. A hybridoma to produce a MAb was obtained as described by Harlow and Lane (26). MAb clone 5D7 was purified from the culture supernatant by column chromatography using Sepharose CL-4B linked with the same phosphopeptide, followed by passage through a column of Sepharose CL-4B linked with a corresponding unphosphorylated peptide, SVEPPLSQETFSD. Purified antibodies recognized phosphopeptide but not unphosphorylated peptide in an enzyme-linked immunosorbent assay (data not shown).
Induction of DNA damage.
PKR+/+ and PKR−/− cells (5 × 105 per 100-mm-diameter dish) were irradiated with a 60Co source and received 7 Gy of γ radiation, were treated with UV irradiation at 50 J/m2/s, or were treated with adriamycin (Sigma) at a final concentration of 1 μM as described elsewhere (63, 64, 67). Where indicated, the proteasome inhibitors ALLN (Sigma) and MG-132 (Biomol Research Laboratories) were added to the cells at a final concentration 20 μM each. The phosphatidylinositol-3 (PI-3) kinase inhibitor LY294002 (Calbiochem) was added to a final concentration 10 μM. Cells were harvested at indicated times after DNA damage.
Cell extraction, immunoprecipitation, and immunoblot analysis.
Cells were washed with ice-cold 1× phosphate-buffered saline (PBS; 140 mM NaCl, 15 mM KH2PO4 [pH 7.2], 2.7 mM KCl) and lysed with 10 volumes of lysis buffer consisting of 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.1 mM EDTA, 0.5% Nonidet P-40, 10% glycerol, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and aprotinin (3 μg/ml), pepstatin (1 μg/ml), leupeptin (1 μg/ml), 1 mM Na3VO4, and 10 mM NaF. The lysates were centrifuged at 10,000 × g for 10 min, and the supernatant (S10) was stored at −80°C.
For immunoprecipitation, 1 μg of affinity-purified anti-p53 rabbit polyclonal antibody SC-6243 (Santa Cruz Biotechnology) or 1 μg of anti-p53 MAb PAb421 (Calbiochem) was added to the lysates. The reactions were incubated on ice at 4°C for 2 h, protein A-Sepharose (Pharmacia) was added, and lysates were further incubated at 4°C for 1 h with rocking. The beads were collected by centrifugation and washed three times with ice-cold lysis buffer. The beads were boiled in sodium dodecyl sulfate (SDS) sample buffer, and immunoprecipitated p53 was subjected to SDS-polyacrylamide gel electrophoresis (PAGE).
For immunoblot analysis, immunoprecipitates or whole-cell extracts (∼50 μg of protein) were subjected to electrophoresis on SDS–10% polyacrylamide gels and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore) in 25 mM Tris-Cl (pH 7.5)–190 mM glycine–20% (vol/vol) methanol for 1 h at 300 mA. The filter was first blocked with 5% (wt/vol) nonfat dried skimmed milk powder in 1× PBS for 1 h at room temperature and then incubated for 1 h at room temperature with 25% fetal bovine serum and 0.5% (vol/vol) Triton X-100 in 1× PBS containing the following antibodies, each at 1 μg/ml: mouse anti-P-Ser15 p53 MAb clone 5D7, mouse anti-p53 MAb PAb421 (Calbiochem), mouse antiactin MAb clone C4 (ICN), a mix of mouse anti-Mdm2 MAbs (clones 2A10 and 4B11; generous gift from A. Levine), and rabbit antiserum to p53 (CM-1) or rabbit antiserum to p21 (C-19; Santa Cruz). Blots were washed three times with 1× PBS–0.5% Triton X-100 and incubated with horseradish peroxidase (HRP)-conjugated sheep anti-mouse or donkey anti-rabbit immunoglobulin (Ig) (Amersham) for 1 h at room temperature. The blots were washed three times with 1× PBS–0.5% Triton X-100, and the proteins were visualized by enhanced chemiluminescence as specified by the manufacturer (Amersham).
Electrophoretic mobility shift assays (EMSAs).
For gel mobility shift assays, cell extracts were prepared as previously described (73). To measure p53 DNA binding, 10 μg of protein extracts was added to [α-32P]dGTP-labeled oligonucleotide (0.5 to 2.0 ng) containing ∼2 × 105 cpm. The dsDNA oligonucleotide used was 5′-GATCCCGGCATGTCCGGGCATGTCCGGGCATGT-3′, which contains two tandem repeats (underlined) of the nearly palindromic sequence GGCATGTCC shown to be bound by p53 with high affinity (24). Binding reactions were contained in a buffer with 10 mM Tris-Cl (pH 8), 0.1 mM zinc acetate, 1 mM dithiothreitol, 5% glycerol, and poly(dI-dC) (250 ng/ml). Protein-DNA complexes formed during a 30-min incubation at room temperature were subsequently electrophoresed on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA at 400 V at 4°C. To induce p53-DNA complex formation, 100 ng of affinity-purified PAb421 (Calbiochem) was added to the binding reactions (24). To ensure the specificity of interactions, a 200-fold excess of unlabeled dsDNA oligonucleotide was used in cold competition reactions. DNA-protein complexes were visualized by autoradiography.
Northern blot analysis.
Total RNA was isolated by the guanidium thiocyanate method (7). RNA (10 μg) was denatured with glyoxal and dimethyl sulfoxide and subjected to electrophoresis on a 1% agarose gel in 10 mM sodium phosphate buffer (pH 7.0) (60). RNA was transferred onto a nylon membrane (BioTrans; ICN). Hybridization was performed at 65°C for 16 h with [α-32P]dATP-labeled random-primed cDNA probes (20) (5 × 106 cpm/ml) consisting of the EcoRV/StuI fragment of mouse p53 cDNA, the entire sequences of both mouse p21 and mdm2 cDNAs, or the entire sequence of mouse β-actin cDNA. After hybridization, the filters were washed with 0.1× SSC (150 mM NaCl, 15 mM sodium citrate [pH 7.0]) plus 0.1% SDS for 1 h at 50°C. The filters were exposed to X-ray film for 16 h.
FACS analysis.
Cells (3 × 106) were preplated in 150-mm-diameter petri dishes for 24 h in DMEM–10% FCS and were kept in DMEM–0.5% FCS for 48 h. Serum-deprived cells were then stimulated with 10% FCS and incubated at 32°C for various periods of time. At the appropriate time, cells were collected with PBS plus 1 mM EDTA and stained for DNA content with propidium iodide (Sigma) as previously described (70). Cell fractions were then subjected to fluorescence-activated cell sorting (FACS) analysis on a Becton Dickinson FACScan.
RESULTS
Generation of PKR+/+ and PKR−/− cell lines expressing a ts mutant of p53.
To investigate a role of PKR in p53 function, we used the mouse ts mutant p53(Val135) (52). It has been shown that in cells expressing p53(Val135), p53 is maintained in a mutant conformation at the restrictive temperature of 37°C, whereas at the permissive temperature of 32°C, p53 assumes a wild-type conformation (52).
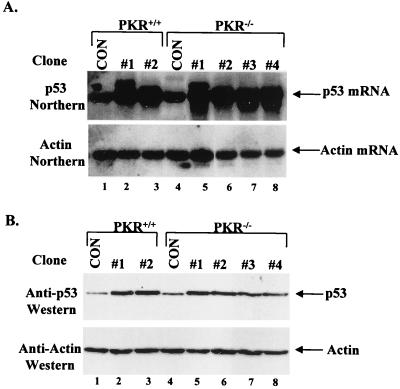
To generate cell lines expressing p53(Val135), immortalized polyclonal populations of PKR+/+ and PKR−/− MEFs (74) were transfected with the ts p53 gene and a zeocin resistance marker. Twenty drug-resistant clones from each cell type were screened for ts p53 expression by Northern blotting. Among them, two and four independent PKR+/+ and PKR−/− clones, respectively, were found to express ∼5-fold-higher levels of mouse p53 RNA compared to zeocin-resistant control clones (Fig. 1A, top panel). Similar levels of overexpression of the ts p53 protein were observed in all transfected clones after immunoblotting with anti-p53 antibody PAb421 (Fig. 1B, top panel) and normalization with an antiactin antibody (Fig. 1B, bottom panel).
FIG. 1.
Generation of PKR+/+ and PKR−/− cell lines expressing the ts p53. (A) Northern blot analysis. Total RNA (10 μg) from cell lines transfected with the ts p53 cDNA was subjected to Northern blotting using as a probe (top panel) the 32P-labeled mouse p53 cDNA, which recognizes both endogenous mouse p53 and ts mouse p53 RNAs. For normalization, the membrane was stripped and reprobed with 32P-labeled actin cDNA (bottom panel). (B) Immunodetection of p53 in PKR+/+ and PKR−/− cells transfected with p53(Val135). Lysates (50 μg of protein) from each clone were resolved by SDS-PAGE (10% gel) and then electrophoretically transferred to a PVDF membrane. p53 protein levels were detected by immunoblotting with PAb421 (top panel), which cross-reacts with both the endogenous mouse p53 and transfected ts p53. The membrane was stripped and reprobed with an antiactin antibody to ensure equal protein loading of the samples (bottom panel). (A and B) Lanes 1 and 4, PKR+/+ and PKR−/− cells transfected with the zeocin resistance gene only (control [CON] clones); lanes 2 and 3, PKR+/+ clones transfected with the ts p53 gene; lanes 5 to 8, PKR−/− clones transfected with ts p53 gene.
PKR is implicated in p53-mediated induction of p21WAF1/Cip1 and Mdm2 and in G0/G1 arrest.
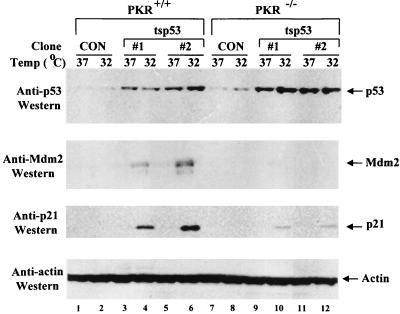
To test for an effect of PKR on p53, we examined the levels of p21 and Mdm2 proteins in the p53(Val135) clones. PKR+/+ and PKR−/− cells were either maintained for 12 h at 37°C, where p53 is mainly in a mutant conformation, or shifted to 32°C, where p53 acquires a wild-type form (Fig. 2). Immunoblot analysis of cell extracts prepared at the two different temperatures with anti-Mdm2 antibodies (Fig. 2, second panel) or an anti-p21 antibody (Fig. 2, third panel) showed that expression of either protein was more highly induced at 32°C in the two independent PKR+/+ cell lines (lanes 4 and 6) compared to the two independent PKR−/− cell lines (lanes 10 and 12). Similar data were obtained when the other two independent PKR−/− clones overexpressing p53(Val135) were examined (data not shown). These data implicate PKR in the induction of p21 and Mdm2 protein expression by ts p53.
FIG. 2.
Impaired expression of p21WAF1/Cip1 and Mdm2 in PKR−/− cells expressing p53(Val135). PKR+/+ (lanes 1 to 6) and PKR−/− clones (lanes 7 to 12) expressing p53(Val135) (lanes 3 to 6 and 9 to 12, respectively) were maintained for 12 h in conditions such that p53 acquires a mutant (37°C; lanes 1, 3, 5, 7, 9, and 11) or wild-type (32°C; lanes 2, 4, 6, 8, 10, and 12) form. Protein extracts (50 μg) were analyzed for p53, p21, and Mdm2 protein expression by immunoblot analysis using anti-p53 MAb PAb 421 (top panel), a mix of anti-Mdm2 MAbs 2A10 and 4B11 (second panel from top), rabbit antisera to p21 (third panel from top), or antiactin MAb (bottom panel). As controls (CON), PKR+/+ (lanes 1 and 2) and PKR−/− (lanes 7 and 8) clones expressing the zeocin resistance gene only were used. To avoid redundancy, results for two of the four PKR−/− clones overexpressing p53(Val135) are shown.
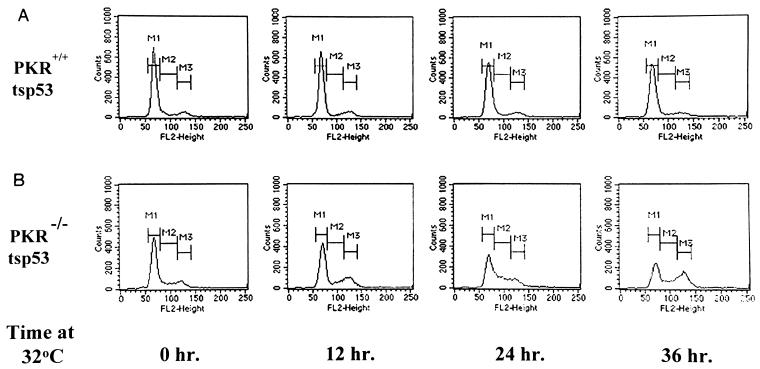
All cell lines expressing p53(Val135) displayed similar growth rates and morphology when maintained at 37°C. However, after being shifted to 32°C, PKR+/+ cells grew less well than PKR−/− cells (data not shown). We reasoned that the growth variations of PKR+/+ and PKR−/− cells were most likely due to the differences in p21 expression and cell cycle progression as result of p53 activation. In fact, cell cycle analysis of PKR+/+ and PKR−/− cells expressing ts p53 revealed some differences in cell cycle progression. As shown in Fig. 3A, ts p53-expressing PKR+/+ cells arrested at G0/G1 by serum deprivation failed to reenter cell cycle when stimulated with serum and maintained at 32°C for various periods of time. In contrast, PKR−/− cells expressing ts p53 bypassed the G0/G1 arrest after serum stimulation (Fig. 3B), although a fraction of these cells accumulated at G2+M when maintained at 32°C for prolonged time periods (see Discussion).
FIG. 3.
PKR is implicated in G0/G1 arrest by ts p53. Shown is cell cycle analysis of PKR+/+ and PKR−/− clones expressing ts p53. Cells were incubated in DMEM–0.5% FCS for 48 h, refed with 10% FCS, and incubated at 32°C for the indicated periods of time. Cells were then fixed, stained with propidium iodide, and subjected to cell cycle analysis as described in Materials and Methods. M1, fraction of cells in G0/G1 phase; M2, cells in S phase; M3, cells in G2+M.
PKR modulates p53 function at the transcriptional level.
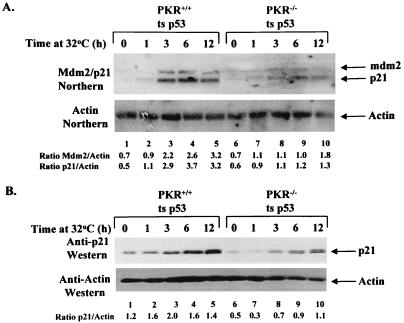
We then wished to determine the level at which PKR exerts its effect on p53. The regulation of p21 and Mdm2 protein expression in PKR+/+ and PKR−/− cells expressing ts p53 could have taken place at the transcriptional level and/or a posttranscriptional level. To distinguish between the two possibilities, p21 and mdm2 RNA levels were assessed by Northern blot analysis (Fig. 4). PKR+/+ and PKR−/− cell lines expressing ts p53 were either maintained at 37°C (lanes 1 and 6) or switched to 32°C for 1 (lanes 2 and 7), 3 (lanes 3 and 8), 6 (lanes 4 and 9), and 12 (lanes 5 and 10) h. RNA and protein extracts were isolated in parallel and subjected either to Northern blotting for mdm2 and p21 RNA expression (Fig. 4A) or immunoblotting for p21 protein expression (Fig. 4B). Induction of p21 and mdm2 RNAs took place with similar kinetics in both PKR+/+ and PKR−/− cells in conditions such that ts p53 acquired a wild-type conformation (Fig. 4A, top panel). However, mdm2 and p21 RNA levels (Fig. 4A, top panel) normalized to actin RNA (bottom panel) were 2.0- to 2.5-fold higher in PKR+/+ cells than in PKR−/− cells (compare lanes 2 to 5 with lanes 7 to 10). In parallel, immunoblot analysis with an anti-p21 antibody revealed that induction of p21 protein had kinetics similar to the kinetics of induction of p21 RNA and was ∼2.5-fold higher in PKR+/+ cells than in PKR−/− cells (Fig. 4B). The higher basal p21 protein levels for PKR+/+ cells (lane 1) than for PKR−/− cells (lane 6) may be explained by a higher fraction of ts p53 being in a wild-type conformation when cells are maintained at 37°C. The double band of p21 protein (Fig. 4B, top panel) might be due to expression of a p21 isoform (faster-migrating band) as reported previously (58). In this regard, it is worth mentioning that experiments with actinomycin D showed no differences in p21 and mdm2 RNA stability between PKR+/+ and PKR−/− ts p53-expressing cells in conditions such that ts p53 acquired a wild-type conformation (data not shown). The above findings suggest that PKR exerts an effect on p53 activity at the transcriptional level.
FIG. 4.
Transcriptional activation of p21 and mdm2 genes by p53 is enhanced by PKR. (A and B) PKR+/+ and PKR−/− cells expressing ts p53 were either maintained at 37°C (lanes 1 and 6) or switched to 32°C for 1 (lanes 2 and 7), 3 (lanes 3 and 8), 6 (lanes 4 and 9), and 12 (lanes 5 and 10) h. (A) Northern blot analysis. Total RNA (10 μg) was subjected to Northern blot analysis using a mix of 32P-labeled mdm2 and mouse p21 cDNAs as a probe (top panel). The levels of mdm2 and p21 RNA expression were normalized to actin RNA (bottom panel). (B) Immunodetection of p21 levels in PKR+/+ and PKR−/− cells expressing ts p53. Cell extracts (50 μg of protein) were subjected to SDS-PAGE (12% gel), electrotransferred to a PVDF membrane, and probed with an anti-p21 antibody. The blot was stripped and reprobed with an antiactin antibody for normalization of protein extracts. (A and B) Quantification of bands was performed by scanning autoradiograms in the linear range of exposure with a Chemiimager 4000 imaging system and analyzing with spot densitometry software (Alpha Innotech Corporation).
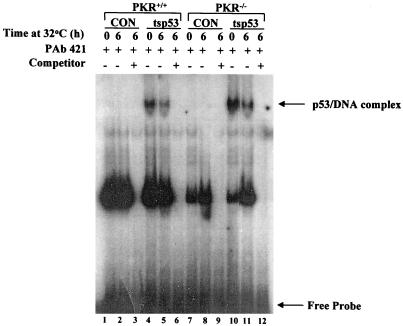
The DNA-binding activity of ts p53 is not modulated by PKR.
Given that the regulation of p53 transcriptional function by PKR could have been exerted at the DNA-binding and/or transactivation levels, we examined whether p53 DNA binding was affected by PKR. To this end, we analyzed the DNA-binding capacity of ts p53 in PKR+/+ and PKR−/− cells by EMSAs (Fig. 5). Control (zeocin-resistant) cells (lanes 1 to 3 and 7 to 9) or ts p53-expressing cells (lanes 4 to 6 and 10 to 12) were maintained either at 37°C or at 32°C, and extracts were isolated and incubated with a 32P-labeled dsDNA oligonucleotide containing a consensus sequence that is bound by p53 with high affinity (24). All reactions included the anti-p53 MAb PAb421 which binds to the carboxy terminus of p53 and enhances p53 sequence-specific DNA binding (24). The p53-DNA complex was readily detected in all cells expressing ts p53 (lanes 4, 5, 10, and 11) but not in control cells (lanes 1, 2, 7, and 8). This difference may be explained by the differences in p53 protein levels between control cells and cells expressing ts p53 (Fig. 1B). However, the amount and mobility of the p53-DNA complex did not significantly vary between PKR+/+ and PKR−/− cells expressing ts p53 (compare lanes 4 and 5 with lanes 10 and 11), suggesting that PKR most likely does not affect DNA binding of ts p53. In these experiments, we also noticed that there were no significant differences in DNA binding of ts p53 when cells were maintained either at 37°C (lanes 4 and 10) or at 32°C (lanes 5 and 11). Although this result appears to be inconsistent with the presence of an active form of ts p53 at 32°C, such a DNA-binding pattern of ts p53 has already been reported and explained by the possibility that a minor fraction of wild-type p53 suffices to promote growth suppression at 32°C (31).
FIG. 5.
DNA binding of ts p53 is not affected by PKR. EMSA was performed with cell extracts from control (CON) zeocin-resistant PKR+/+ and PKR−/− cells (lanes 1 to 3 and 7 to 9) or cells expressing ts p53 (lanes 4 to 6 and 10 to 12) which were incubated at either 37°C (lanes 1, 4, 7, and 10) or 32°C (lanes 2, 3, 5, 6, 8, 9, 11, and 12). Cell extracts (10 μg of protein) were incubated with a 32P-labeled dsDNA oligonucleotide which contains the consensus DNA-binding site of p53. Binding reaction mixtures contained 100 ng of affinity-purified anti-p53 MAb PAb421, which induces the sequence-specific DNA binding of p53. A 200-fold excess of unlabeled dsDNA oligonucleotide was added in cold competition reactions (lanes 3, 6, 9, and 12).
Transcriptional activation of p53-inducible genes is enhanced by PKR upon DNA damage.
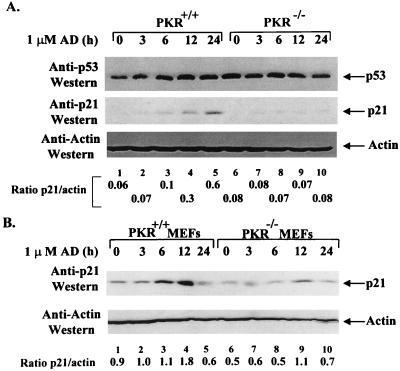
Since there were no detectable differences in DNA binding of ts p53 between PKR+/+ and PKR−/− cells, we sought to examine whether PKR might modulate p53 transactivation capacity. It has been shown that the transactivation capacity of p53 is highly induced in response to DNA damage (64). We reasoned that if PKR modulated p53 transactivation, then induction of p53-inducible genes should be defective in PKR−/− cells after DNA damage (17, 18, 34). This possibility was first examined in PKR+/+ and PKR−/− cells derived from spontaneously immortalized MEFs (74). As a means of inducing DNA damage, we used the anticancer drug adriamycin, which has been previously shown to activate p53 (48), whereas we examined the expression levels of p21 protein as a marker for p53-mediated transactivation (18). Adriamycin treatment resulted in induction of p21 protein levels in both PKR+/+ and PKR−/− cells (Fig. 6A, middle panel). However, expression of p21 protein normalized to actin levels was ∼3-fold higher in PKR+/+ cells than in PKR−/− cells and reached its maximal level at 24 h after treatment (Fig. 6A; compare lanes 2 to 5 with 7 to 10). This effect was not due to variations of p53 protein levels since immunoblot analysis with an anti-p53 antibody showed similar amounts of p53 in both PKR+/+ and PKR−/− cells (Fig. 6A, top panel). We also checked p21 levels in primary MEFs derived from PKR+/+ and PKR−/− mice (1). Similar to results for immortalized polyclonal populations (Fig. 6A), p21 protein levels normalized to actin were ∼2-fold higher in PKR+/+ MEFs than in PKR−/− MEFs (Fig. 6B; compare lanes 2 to 5 with lanes 7 to 10), reaching maxima 12 h after adriamycin treatment (compare lane 4 with lane 9). The downregulation of p21 protein in both PKR+/+ (lane 5) and PKR−/− (lane 10) MEFs could be due to cell death observed for both cell types 24 h after adriamycin treatment. In addition, FACS analysis of adriamycin-treated cells showed an impaired G1 arrest of PKR−/− cells compared to PKR+/+ cells but no significant differences in apoptosis (data not shown). These data are consistent with a role for PKR in p21 induction and cell cycle arrest in response to DNA damage and can be explained by a diminished p53 transactivation capacity in PKR−/− cells.
FIG. 6.
PKR enhances the induction of p21WAF1 expression upon DNA damage. (A and B) Induction of p21 in PKR+/+ and PKR−/− cells in response to adriamycin (AD). Exponentially grown cells (∼3 × 106/150-mm-diameter dish) were left untreated (lanes 1 and 6) or treated with 1 μM adriamycin for 3 (lanes 2 and 7), 6 (lanes 3 and 8), 12 (lanes 4 and 9), and 24 (lanes 5 and 10) h. Protein extracts (∼250 μg) were subjected to immunoprecipitation with anti-p53 antibody PAb421, transferred onto a PVDF membrane, and immunoblotted with the rabbit antiserum to p53 (CM-1; A, top panel). Cell extracts (50 μg of protein) were resolved by SDS-PAGE (12% gel) and subjected to immunoblot analysis first with anti-p21 antibody (A, middle panel; B, top panel) and then with antiactin antibody (bottom panels). (A) Spontaneously immortalized polyclonal populations derived from PKR+/+ and PKR−/− MEFs. (B) Primary PKR+/+ and PKR−/− MEFs grown between passages 2 and 3. (A and B) Quantification of bands performed by scanning autoradiograms in the linear range of exposure with a Chemiimager 4000 imaging system and analyzing the results with spot densitometry software (Alpha Innotech Corporation).
Phosphorylation of mouse p53 on Ser18 is impaired in PKR−/− cells.
Several lines of evidence indicate that p53 transactivation capacity is affected by phosphorylation (65). For example, phosphorylation of p53 within its amino-terminal domain has been suggested to be required for its maximal transcriptional activity (50). Of the seven serine residues present within the first 60 amino acids of human p53, Ser15 is the most highly conserved among all p53 species (21). Phosphorylation of Ser15 of human p53 has been shown to play a role in cell cycle progression (21) and be induced by a variety of genotoxic stimuli (63, 64).
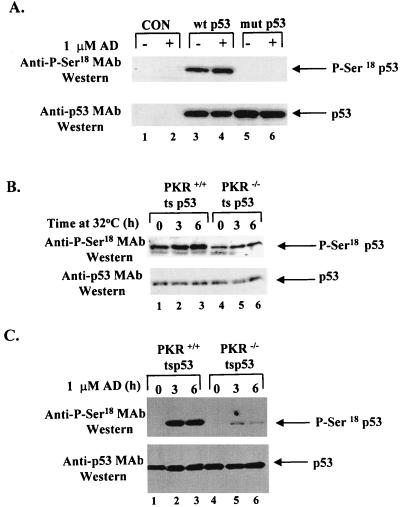
Detection of site-specific p53 phosphorylation is now possible with the development of antibodies that specifically recognize phosphorylated epitopes of p53 (2, 5, 33, 49, 63, 64, 71). To examine an effect of phosphorylation on p53 transactivation, we used a mouse MAb that specifically recognizes phosphorylation of human p53 on Ser15 (Ser18 in mouse p53) (2, 5). First, we tested the suitability of the P-Ser15 antibody for mouse p53. Wild-type mouse p53 cDNA or a Ser18-Ala mutant mouse p53 cDNA was expressed in the p53-negative BALB/c (10)1 cell line (27) by transient transfection (Fig. 7A). Cell extracts were subjected to immunoblot analysis first with the anti-P-Ser15 human p53 antibody (Fig. 7A, top panel) and then with the mouse anti-p53 PAb421 (Fig. 7A, bottom panel) before (lanes 1, 3, and 5) or after (lanes 2, 4, and 6) adriamycin treatment. Phosphorylation of p53 was detected with the wild-type mouse p53 (lanes 3) in the absence of DNA damage but not with the Ser18-Ala mutant of mouse p53 (lane 5). DNA damage with adriamycin slightly induced phosphorylation of wild-type mouse p53 (lane 4) but had no effect on mutant p53 (lane 6). These data demonstrate the suitability of the anti-P-Ser15 human p53 MAb for detection of phosphorylation of the mouse protein. The high levels of Ser18 phosphorylation of wild-type mouse p53 in the absence of DNA damage (lane 3) could be due to p53 activation by stress induced during the transient transfection.
FIG. 7.
PKR enhances phosphorylation of ts p53 on Ser18. (A) The anti-P-Ser15 human p53 antibody specifically recognizes P-Ser18 of mouse p53. BALB/c(10)1 cells lacking endogenous p53 were transiently transfected either with a wild-type mouse p53 cDNA (lanes 3 and 4) or a Ser18-Ala mutant (mut) of mouse p53 cDNA (lanes 5 and 6). Thirty-six hours after transfection, cells were treated with adriamycin (AD) for 1 h (lanes 2, 4, and 6). Cell extracts (50 μg of protein) were subjected to SDS-PAGE (12% gel), electrotransferred to a PVDF membrane, and probed first with the anti-P-Ser15 human p53 antibody (top panel) and second with mouse anti-p53 antibody PAb421 (bottom panel). Lanes 1 and 2, mock-transfected cells before (lane 1) and after (lane 2) adriamycin treatment. (B) Immunodetection of ts p53 phosphorylation in PKR+/+ and PKR−/− cells, using a mouse MAb specific for P-Ser15 of human p53. PKR+/+ and PKR−/− cells expressing ts p53 were either maintained at 37°C (lanes 1 and 4) or shifted to 32°C for 3 (lanes 2 and 5) and 6 (lanes 3 and 6) h. (C) For DNA damage, cells were shifted to 32°C for 6 h (lanes 1 and 4) and preincubated with the proteasome inhibitors ALLN and MG-132 for 1 h (lanes 1 to 6) before treatment with adriamycin for 3 (lanes 2 and 5) or 6 (lanes 3 and 6) h. Cell extracts (50 μg of protein) were subjected to SDS-PAGE (10% gel) and immunoblot analysis first with the mouse anti-P-Ser15 MAb (B and C, top panels) and then with mouse anti-p53 MAb PAb421 (bottom panels).
Then we examined the basal levels of p53 phosphorylation in PKR+/+ and PKR−/− cells expressing ts p53. Immunoblot analysis with the anti-P-Ser15 MAb showed that ts p53 is constitutively phosphorylated in both PKR+/+ and PKR−/− cells (Fig. 7B, top panel). We also observed that phosphorylation of p53 was detectably induced in both cell types in conditions such that ts p53 acquired a wild-type form (Fig. 7B, top panel; compare lane 1 with lanes 2 and 3, and lane 4 with lanes 5 and 6). However, the overall phosphorylation levels of ts p53 were ∼50% higher in PKR+/+ cells than in PKR−/− cells (Fig. 7B, top panel; compare lanes 1 to 3 with lanes 4 to 6) whereas p53 protein levels were the same in both cell types, as judged by immunoblot analysis with the anti-p53 antibody PAb421 (Fig. 7B, bottom panel).
Next we measured Ser18 phosphorylation of the ts p53 in PKR+/+ and PKR−/− cells in response to DNA damage induced by adriamycin (Fig. 7C, top panel). In this experiment, cells were first shifted to 32°C for 6 h (lanes 1 and 4) and then incubated in the presence of 1 μM adriamycin for additional 3 h (lanes 2 and 5) or 6 h (lanes 3 and 6). Immunoblot analysis with the anti-P-Ser15 MAb revealed a ∼5-fold higher level of p53 phosphorylation in PKR+/+ cells than in PKR−/− cells (Fig. 7C, top panel; compare lanes 2 and 3 with lanes 5 and 6). This effect was not due to the differences in ts p53 levels since immunoblot analysis with PAb421 showed equal levels of p53 protein in all samples (Fig. 7C, bottom panel). Detection of basal levels of ts p53 phosphorylation (Fig. 7C, top panel, lanes 1 and 4) was possible after long exposure (data not shown).
PKR enhances Ser18 phosphorylation of mouse p53 in response to DNA damage.
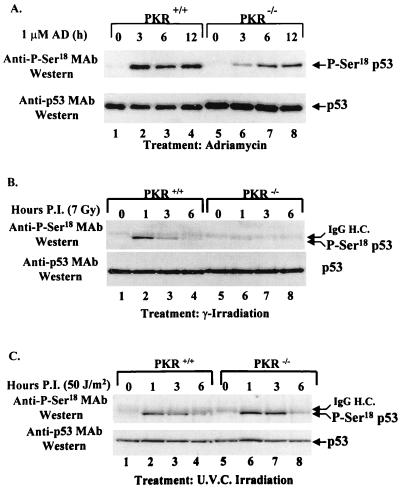
The anti-P-Ser15 MAb was subsequently used to examine the phosphorylation status of p53 in PKR+/+ and PKR−/− cells in response to a variety of genotoxic stimuli. Immortalized PKR+/+ and PKR−/− cells were subjected to DNA damage with adriamycin (Fig. 8A), γ radiation (Fig. 8B), or UV radiation (Fig. 8C) for various periods of time. Cell extracts were subjected to immunoprecipitation with rabbit antisera to p53 followed by immunoblot analysis with either the anti-P-Ser15 MAb (Fig. 8, top panels) or the mouse anti-p53 MAb PAb421 (Fig. 8, bottom panels). All treatments induced phosphorylation of endogenous mouse p53 on Ser18 in both PKR+/+ and PKR−/− fibroblasts. However, phosphorylation of p53 normalized to p53 protein levels was ∼5-fold higher in PKR+/+ cells than in PKR−/− cells after adriamycin or γ-radiation treatment (Fig. 8A and B; compare lanes 2 and 6, 3 and 7, and 4 and 8). In contrast, no differences in p53 phosphorylation were observed when cells were treated with UV (Fig. 8C). Similar data were obtained with the presence of ALLN and MG-132 upon adriamycin treatment (data not shown), suggesting that proteasome inhibitors have no pleiotropic effects leading to Ser18 p53 phosphorylation in response to DNA damage. These findings demonstrate that phosphorylation of Ser18 of mouse p53 is defective in PKR−/− cells upon certain types of DNA damage.
FIG. 8.
PKR enhances p53 phosphorylation on Ser18 in response to DNA damage. Cell extracts from PKR+/+ and PKR−/− cells were prepared after treatment with adriamycin (AD) (A), γ radiation (7 Gy), (B), or UV radiation (50 J/m2) (C) for the indicated periods of time postinfection (P.I.). Protein extracts (∼250 μg) were subjected to immunoprecipitation with a rabbit anti-p53 polyclonal antibody, transferred onto a PVDF membrane, and immunoblotted with the mouse anti-P-Ser15 MAb (upper panels). The membranes were stripped and reprobed with the mouse anti-p53 antibody PAb421 (lower panels). The faint slowly migrating band above p53 present in all lanes after immunoblotting with the anti-P-Ser18 antibody (B and C, top panels) is a fraction of the heavy chain of rabbit IgG (IgG H.C.) recognized by the secondary HRP-conjugated sheep anti-mouse IgG and was visible after long exposures only.
PKR is possibly implicated in a PI-3 kinase like pathway leading to activation of p53 by phosphorylation.
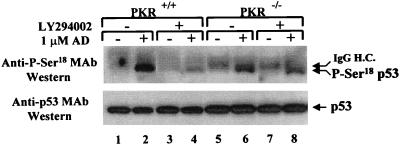
ATM (mutated in ataxia telangiectasia), DNA-PK (dsDNA-activated protein kinase), and possibly other members of the PI-3 kinase family have been implicated in the phosphorylation of human p53 on Ser15 (2, 5, 63, 64). Based on this finding, we wished to examine whether phosphorylation of mouse p53 on Ser18 in PKR+/+ and PKR−/− cells was affected by the potent and specific PI-3 kinase inhibitor LY294002 (Fig. 9). PKR+/+ and PKR−/− cells were incubated with the proteasome inhibitors ALLN and MG-132 in the absence or presence of LY294002 before or after treatment with adriamycin. Immunoprecipitation with rabbit antisera to p53 and immunoblot analysis with the anti-P-Ser15 MAb (Fig. 9, top panel) showed that adriamycin-induced p53 phosphorylation (lanes 2 and 6) was significantly reduced in PKR+/+ cells in the presence of LY294002 (lane 4) but not in PKR−/− cells (lane 8). Consequent immunoblot analysis with PAb421 (Fig. 9, bottom panel) showed that these differences were not due to variations of the immunoprecipitated p53. Similar results were obtained when another PI-3 kinase inhibitor, wortmannin, was used (data not shown). This finding indicates that PKR possibly participates in a PI-3 kinase signaling pathway(s) that leads to phosphorylation of p53.
FIG. 9.
PKR is implicated in PI-3 kinase pathway(s) leading to p53 phosphorylation. PKR+/+ and PKR−/− cells were treated with 10 μM specific PI-3 kinase inhibitor LY294002 and 1 μM adriamycin (AD) in the presence of ALLN and MG-132. To avoid a possible degradation of LY294002 during the treatment, adriamycin was added for 1 h only. Protein extracts (∼250 μg) were subjected to immunoprecipitation with a rabbit anti-p53 polyclonal antibody and resolved by SDS-PAGE (10% gel) followed by immunoblotting with the mouse anti-P-Ser15 MAb (upper panels). The membranes were stripped, and p53 protein levels were visualized by immunoblot analysis using mouse anti-p53 antibody PAb421 (lower panels). The faint slowly migrating band above p53 (top panel, lanes 5 to 8) is a fraction of the heavy chain of rabbit IgG (IgG H.C.) recognized by the secondary HRP-conjugated sheep anti-mouse IgG.
DISCUSSION
The IFN-inducible protein kinase PKR has been suggested to play an important role in viral infection by regulating protein synthesis through phosphorylation of eIF-2α (11). In addition to translational control, several reports have implicated PKR in signaling pathways leading to transcriptional activation (11). In this report, we have extended the observations on the transcriptional role of PKR by providing strong evidence for its function in the transcriptional activation by the tumor suppressor p53.
The approach that we took to address a possible role of PKR in p53 function was to generate mouse cell lines with either a PKR+/+ or a PKR−/− background expressing the ts p53 mutant p53(Val135). Using these cell lines, we have demonstrated that p53 transcriptional capacity is diminished in PKR−/− cells in conditions such that ts p53 acquires a wild-type form (Fig. 2 and 4). We have also shown that ts p53-expressing PKR−/− cells can bypass G0/G1 arrest imposed by serum deprivation when stimulated with serum and maintained at the permissive temperature of 32°C (Fig. 3, top panel). In contrast, ts p53-expressing PKR+/+ cells failed to reenter the cell cycle in the same conditions (Fig. 3, bottom panel). These differences may be explained, at least in part, by the lower levels of p21 expression in ts p53 PKR−/− cells than in PKR+/+ cells at 32°C (Fig. 2 and 4). In these experiments, we also noticed an accumulation of the ts p53 PKR−/− cells in G2+M after serum stimulation and incubation at 32°C for 24 or 36 h (Fig. 3, bottom panel). At the present there is no clear explanation for this event. Our data indicate that PKR may be required for progression through the G2/M phase. Since p53 activation is also implicated in the G2/M checkpoint (39, 47), such a property of PKR may be independent of p53 since G2/M accumulation cannot be explained by an impaired function of ts p53 in PKR−/− cells.
Having eliminated any changes in p53 DNA binding as a means of explaining how PKR regulates p53-mediated transcriptional activation, we turned our attention to the phosphorylation state of p53. p53 is phosphorylated on multiple sites by several kinases such as DNA-PK, protein kinase C, mitogen-activated protein kinase, casein kinase I (CKI), and CKII (reviewed in reference 65). In this regard, we have recently shown that PKR physically associates with p53 and phosphorylates human p53 on Ser392 in vitro (12). Although the role of Ser392 phosphorylation in p53-mediated transcriptional activation is not well understood (65), phosphorylation of human p53 on serine residues within the amino-terminal domain, including Ser15, has been suggested to play a role in p53 transcriptional function (63, 64) and p53-mediated inhibition of cell cycle progression (21). To detect p53 phosphorylation, a mouse MAb that specifically recognizes P-Ser15 of human p53 was used (2, 5). Experiments with wild-type and Ser18-Ala mutant forms of mouse p53 confirmed the suitability of this antibody for mouse p53 (Fig. 7A).
We initially observed that the basal levels of Ser18 phosphorylation of ts p53 were higher in PKR+/+ cells than in PKR−/− cells (Fig. 7B). Since PKR interacts with p53 (12), we examined whether phosphorylation of p53 within the amino terminus was mediated by PKR. We were unable to detect specific phosphorylation of either glutathione S-transferase–full-length human p53 or a glutathione S-transferase–human p53 fusion protein consisting of amino acids 1 to 50 by activated PKR in vitro (data not shown), indicating that PKR is unlikely to directly mediate phosphorylation of mouse p53 on Ser18 in vivo. The differences in the basal levels of Ser18 phosphorylation suggest that in the absence of a stimulus, PKR plays a role in regulating the activity of a kinase(s) that directly phosphorylate mouse p53 on Ser18. It is also conceivable that PKR could modulate the activity of a phosphatase(s) that dephosphorylates P-Ser18 of mouse p53, although treatment with the phosphatase inhibitor okadaic acid did not affect p53 phosphorylation in either PKR+/+ or PKR−/− cells (data not shown).
We extended our studies to examine the induction of Ser18 phosphorylation of mouse p53 in response to DNA damage. We observed a higher amount of Ser18 phosphorylation in PKR+/+ cells than in PKR−/− cells in response to the anticancer drug adriamycin (Fig. 7C and 8A) or γ radiation (Fig. 8B) but not UV radiation (Fig. 8C). In addition, we saw that the presence the specific PI-3 kinase inhibitor LY294002 caused a higher inhibition of adriamycin-induced Ser18 phosphorylation of p53 in PKR+/+ cells than in PKR−/− cells (Fig. 9). These data clearly implicate PKR in p53 activation by phosphorylation upon a certain type of DNA damage and indicate that PKR may regulate the activity of a PI-3 kinase that mediates p53 phosphorylation either directly or indirectly.
An explanation for the ability of PKR to enhance Ser18 phosphorylation of mouse p53 in response to DNA lesions caused by genotoxic stress may be twofold. One lies in the type of DNA damage and consequently the DNA repair enzymes activated by DNA damage. UV radiation causes cyclobutane pyrimidine dimers, and in this case an excision pathway is activated (49). On the other hand, γ radiation and adriamycin cause dsDNA breaks, leading to ligation and recombination repair (49, 68). Thus, it appears that distinct DNA damage detectors act to identify different types of DNA damage and signal to different parts of the p53 protein, resulting in p53 transcriptional activation.
A second explanation lies in the nature of the kinases that have been shown to play a role in the phosphorylation of p53 upon DNA damage. One is the ATM protein, a PI-3 like kinase (62) which activates p53 in response to γ radiation (6, 76). Transcriptional activation of human p53 and phosphorylation on Ser15 are defective in AT cells upon γ radiation but not UV treatment (64), which is similar to our observations in PKR+/+ and PKR−/− cells (Fig. 8). ATM acts upstream of p53 in the cellular response to γ radiation (64) and has been recently shown to phosphorylate human p53 on Ser15 (2, 5). Another kinase is DNA-PK, which also belongs to the PI-3 kinase family and has been implicated in phosphorylation of human p53 on Ser15 (63, 64). In addition to being activated by dsDNA breaks, DNA-PK is involved in V(D)J recombination (13). However, cells from scid mice, which are defective in DNA-PK, are still able to activate p53 in response to γ radiation (4, 23, 30, 55, 59), making this kinase an unlikely candidate to be modulated by PKR. Finally, CKI has been shown to directly phosphorylate murine p53 on multiple sites within the amino terminus (65), and CKI is activated when Drosophila melanogaster embryos are exposed to γ radiation (61). Our data indicate that the kinase regulated by PKR may be a member of the PI-3 kinase family (Fig. 9). How PKR mediates this effect is not immediately clear. Perhaps PKR binds to p53 in a complex with a PI-3 kinase and modulates PI-3 kinase activity by phosphorylation. Whether this is ATM or another PI-3 kinase is a possibility that remains to be examined.
It should be noted, however, that modulation of Ser18 p53 phosphorylation may be necessary but not sufficient to explain the PKR-mediated transcriptional activation of p53. It is possible that PKR mediates phosphorylation of the amino terminus of p53 on more than one site. For example, simultaneous mutations of Ser9, Ser18, and Ser37 of mouse p53 significantly reduced p53 transactivation, whereas point mutations of either of these serine residues had no effect (50). These data suggest that maximal transcriptional activity of p53 may require more than one phosphorylation site within the transactivation domain. Nevertheless, phosphorylation of human p53 on Ser15 has been shown to be crucial for its biological function. That is, overexpression of human p53 Ser15-Ala mutant protein in human cells resulted in partial failure of the mutant protein to inhibit cell cycle progression compared with cells that overexpress either wild-type p53 or the Ser37-Ala mutant protein (21). This is reminiscent of our data showing that the ts p53-expressing PKR−/− cells, which contain lower basal levels of p53 phosphorylation on Ser18 than the ts p53 PKR+/+ cells, progress through the cell cycle upon serum stimulation and activation of p53 (Fig. 3). In addition to the amino terminus of p53, modulation of p53 phosphorylation within the carboxy terminus has been recently implicated in p53 activation. That is, the ATM-dependent activation of a phosphatase that targets P-Ser376 of human p53 allows 14-3-3 proteins to bind to the carboxy terminus of p53, thus activating DNA binding and transcription by p53 (71). Considering the interaction of PKR with the carboxy terminus of p53 (12), a possible modulation of Ser376 dephosphorylation and 14-3-3 binding to p53 might provide a tentative, as yet unidentified link between PKR and p53 activation.
In response to DNA damage, p53 phosphorylation and p53-dependent transcription are increased, leading to p53-mediated induction of cell cycle arrest and/or apoptosis (22, 44, 72). One model for p53 activation involves its interaction with Mdm2, which results in inhibition of p53 transcriptional activation (54, 77), most likely as a result of rapid degradation of p53 (28, 42). Disruption of p53-Mdm2 interaction in normal cells results in the accumulation of p53 and activation of p53-dependent transcription. In this regard, phosphorylation of human p53 within the amino terminus including Ser15 has been shown to result in p53-Mdm2 dissociation, providing a possible link between p53 phosphorylation and protein stability upon DNA damage (63). However, our data show that p53 protein levels did not significantly vary between PKR+/+ and PKR−/− cells before or after DNA damage despite the fact that phosphorylation of p53 on Ser18 was diminished in PKR−/− cells after treatment with adriamycin or γ radiation (Fig. 8). In addition, the lower levels of p53 Ser18 phosphorylation in the ts p53-expressing PKR−/− cells (Fig. 7B) did not affect p53-Mdm2 interaction, nor was p53-Mdm2 interaction disrupted in either PKR+/+ or PKR−/− cells in response to DNA damage (data not shown). This finding suggests that phosphorylation of mouse p53 on Ser18 may not be essential for disruption of p53-Mdm2 interaction and p53 stabilization. In human cells, however, phosphorylation of Ser15 and Ser37 upon DNA damage result in inhibition of p53-Mdm2 interaction (63). One possible explanation for these contradictory findings may lie in the type and/or species of the cells used in these experiments. In this regard, it is not known whether phosphorylation within the amino terminus of mouse p53 on a site(s) other than Ser18 (i.e., Ser9 and/or Ser37) affects p53-Mdm2 interaction in mouse cells and, if so, whether phosphorylation on such a site(s) is induced in mouse cells upon DNA damage. Further experiments are required to clarify this issue.
In regard to possible alternate mechanisms by which PKR modulates p53-mediated transcription, the role of PKR in IRF-1 activation should be considered (36, 43). Interestingly, IRF-1 cooperates with p53 in transcriptional activation of the p21 gene and inhibition of cell growth (67). Considering the synergistic effect of IRF-1 with either p53 (67) or PKR (36, 43), it is reasonable to speculate that the decreased activity of p53 in PKR−/− cells could reflect a decrease in IRF-1 activity. However, the following observations do not favor this possibility. First, the interaction between PKR and p53 (12) and the regulation of expression of two different p53-dependent genes (i.e., p21 and mdm2) by PKR (Fig. 2 and 4) provide strong evidence that PKR and p53 function on the same pathway. In contrast to PKR, IRF-1 and p53 function in parallel pathways, and it is not known whether IRF-1 regulates the expression of p53-dependent genes other than p21 (67). Second, induction of p21 contributes significantly in cell growth inhibition by IRF-1 (67). However, activation of IRF-1 results in the same degree of cell growth inhibition in both PKR+/+ and PKR−/− cells (37), arguing against a role of PKR in IRF-1-mediated induction of p21.
In summary, phosphorylation of p53 represents an indispensable component of the DNA damage-induced pathways that modulate the multiple functions of p53. The identification of the specific kinase(s) that modulate p53 phosphorylation on Ser18 in vivo and are regulated by PKR may prove essential for understanding the mechanisms of cell growth regulation and signaling by both p53 and PKR.
ACKNOWLEDGMENTS
We thank Y.-L. Yang and C. Weissmann for PKR+/+ and PKR−/− MEFs; B. Vogelstein for the ts mutant p53(Val135) and mouse p21WAF1/Cip1 cDNAs; A. Levine for BALB/c (10)1 cells, mdm2 cDNA, and anti-Mdm2 MAbs 2A10 and 4B11; C. Midgley and D. Lane for wild-type mouse p53 and mouse p53 Ser18-Ala cDNAs; K. McDonald for flow cytometry analysis; S. Lehnert for assistance with the γ-irradiation experiments; S. Kirchhoff and H. Hauser for communicating results before publication; and J. Th’ng and C. Couture for helpful discussions and suggestions.
This work was supported by research grants from the National Cancer Institute of Canada (NCIC), Medical Research Council (MRC) of Canada, and The Cancer Research Society (CRS) Inc. to A.E.K. A.R.C. is a recipient of a CRS studentship, and A.H.-T.W. is a recipient of a NCIC Terry Fox research award. A.E.K. is a member of the Terry Fox Group in Molecular Oncology and an MRC Scientist awardee.
REFERENCES
- 1.Abraham N, Stojdl D F, Duncan P I, Methot N, Ishii T, Dube M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, Koromilas A E, Brown E G, Sonenberg N, Bell J C. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 2.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 3.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogue M A, Zhu C, Aguilar-Cordova E, Donehower L A, Roth D B. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- 5.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appela E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 6.Canman C E, Wolff A C, Chen C, Fornace A J, Kastan M B. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method for RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Chong K, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R G. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choubey D, Lengyel P. Binding of an interferon-inducible protein (p202) to the retinoblastoma protein. J Biol Chem. 1995;270:6134–6140. doi: 10.1074/jbc.270.11.6134. [DOI] [PubMed] [Google Scholar]
- 10.Choubey D, Li S J, Datta B, Gutterman J U, Lengyel P. Inhibition of E2F-mediated transcription by p202. EMBO J. 1996;15:5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 12.Cuddihy, A. R., A. H.-T. Wong, N. W. N. Tam, S. Li, and A. E. Koromilas. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 and phosphorylates human p53 on serine392 in vitro. Oncogene, in press. [DOI] [PubMed]
- 13.Danska J S, Holland D P, Mariathasan S, Willimans K M, Guidos C J. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem. 1996;271:27544–27555. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- 15.Der S D, Yang Y-L, Weissmann C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donze O, Jagus R, Koromilas A E, Hershey J W B, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 18.El-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 19.Ewen M E, Miller S J. p53 and translational control. Biochim Biophys Acta. 1996;1242:181–184. doi: 10.1016/0304-419x(95)00010-d. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Apella E. Mutation of the serine-15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 22.Giaccia A J, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 23.Guidos C J, Williams C J, Grandal I, Knowles G, Huang M T F. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 24.Halazonetis T D, Davis L J, Kandil A N. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 1993;12:1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanash S M, Beretta L, Barcroft C L, Sheldon S, Glover T W, Ungar D, Sonenberg N. Mapping of the gene for the interferon-inducible dsRNA-dependent protein kinase to chromosome region 2p21-22: a site of rearrangements in myeloproliferative disorders. Genes Chromosomes Cancer. 1993;8:34–37. doi: 10.1002/gcc.2870080107. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 27.Harvey D M, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Mdm-2 promotes the rapid degradation of p53. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Hershey J W B. Protein phosphorylation controls translation rates. J Biol Chem. 1989;264:20823–20826. [PubMed] [Google Scholar]
- 30.Huang L, Clarkin K C, Wahl G M. p53-dependent cell cycle arrests are preserved in DNA-activated protein kinase-deficient mouse fibroblasts. Cancer Res. 1996;56:2940–2944. [PubMed] [Google Scholar]
- 31.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 32.Jeffrey I W, Kadereit S, Meurs E F, Metzger T, Bachmann M, Schwemmle M, Hovanessian A G, Clemens M J. Nuclear localization of the interferon-inducible protein kinase PKR in human and transfected mouse cells. Exp Cell Res. 1995;218:17–27. doi: 10.1006/excr.1995.1126. [DOI] [PubMed] [Google Scholar]
- 33.Kapoor M, Lozano G. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 35.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 36.Kirchhoff S, Koromilas A E, Schaper F, Grashoff M, Sonenberg N, Hauser H. IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene. 1995;11:439–445. [PubMed] [Google Scholar]
- 37.Kirchhoff, S., F. Schaper, Y.-L. Yang, C. Weissmann, A. E. Koromilas, and H. Hauser. Unpublished data.
- 38.Kitagawa M, Higashi H, Jung H, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-cdk4 is different from that for phosphorylation by cyclin A/E-cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 39.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 40.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 41.Koromilas A E, Cantin C, Craig A W B, Jagus R, Hiscott J, Sonenberg N. The interferon-inducible protein kinase PKR modulates transcriptional activation of the immunoglobulin κ gene. J Biol Chem. 1995;270:25426–25434. doi: 10.1074/jbc.270.43.25426. [DOI] [PubMed] [Google Scholar]
- 42.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane D. Awakening angels. Nature (London) 1998;394:616–617. doi: 10.1038/29166. [DOI] [PubMed] [Google Scholar]
- 45.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 46.Lee S B, Rodriguez D, Rodriguez J R, Esteban M. The apoptosis pathway triggered by the interferon-induced protein kinase PKR requires the third basic domain, initiates upstream of bcl-2, and involves ICE-like proteases. Virology. 1997;231:81–88. doi: 10.1006/viro.1997.8494. [DOI] [PubMed] [Google Scholar]
- 47.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 48.Lowe S W, Ruley H E, Jacks T, Housman D E. p53-Dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 49.Lu H, Taya Y, Ikeda M, Levine A J. Ultraviolet radiation, but not γ radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayr G A, Reed M, Wang P, Wang Y, Schwedes J F, Tegtmeyer P. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 51.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michailovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 53.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-κB, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Momand J, Zambetti G P, Olson D C, George D L, Levine A J. The mdm2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated trans-activation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 55.Nacht M, Strasser A, Chan Y R, Harris A W, Schlissel M, Bronson R T, Jacks T. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 1996;10:2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 56.Nigro J M, Sikorski R, Reed S I, Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1357–1365. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeifer G P, Holmquist G P. Mutagenesis in the p53 gene. Biochim Biophys Acta. 1997;1333:M1–M8. doi: 10.1016/s0304-419x(97)00004-8. [DOI] [PubMed] [Google Scholar]
- 58.Poon R Y, Hunter T. Expression of a novel form of p21Cip1/WAF1 in UV-irradiated and transformed cells. Oncogene. 1998;16:1333–1343. doi: 10.1038/sj.onc.1201897. [DOI] [PubMed] [Google Scholar]
- 59.Rathmell W K, Kaufman W K, Hurt J C, Byrd L L, Chu G. DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer Res. 1997;57:68–74. [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 61.Santos J A, Logarinho E, Tapia C, Allende C C, Allende J E, Sunkel C E. The casein kinase 1α gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J Cell Sci. 1996;109:1847–1856. doi: 10.1242/jcs.109.7.1847. [DOI] [PubMed] [Google Scholar]
- 62.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali S R, Simsons A, Clines G A, Sartiel A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G J, Taylor A M R, Arlett C F, Miki T, Weissman S M, Lovett M, Collins F S, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 63.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 64.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steegenga W T, van der Eb A J, Jochemsen A G. How phosphorylation regulates the activity of p53. J Mol Biol. 1996;263:103–113. doi: 10.1006/jmbi.1996.0560. [DOI] [PubMed] [Google Scholar]
- 66.Takizawa T, Ohashi K, Nakanishi Y. Possible involvement of double-stranded RNA-activated protein kinase in cell death by influenza virus infection. J Virol. 1996;70:8128–8132. doi: 10.1128/jvi.70.11.8128-8132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka N, Ishihara M, Lamphier M S, Nozawa H, Matsuyama T, Mak T W, Aizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumor suppressor IRF-1 and p53 in response to DNA damage. Nature (London) 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 68.Tewey K M, Rowe T C, Yang L, Halligan B D, Liu L F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 69.Todaro G J, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vindelov L L, Christensen I J, Nissen N I. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry. 1983;3:323–327. doi: 10.1002/cyto.990030503. [DOI] [PubMed] [Google Scholar]
- 71.Waterman M J, Stavridi E S, Waterman J L, Halazonetis T D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 72.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 73.Wong A H-T, Tam N W N, Yang Y-L, Cuddihy A R, Li S, Kirchhoff S, Hauser H, Decker T, Koromilas A E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y L, Reis L F L, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeung M, Liu J, Lau A S. An essential role for interferon-inducible, double-stranded RNA-activated protein kinase PKR in the tumor necrosis factor-induced apoptosis in U937 cells. Proc Natl Acad Sci USA. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 77.Zauberman A, Barak Y, Raginov N, Levy N, Oren M. Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]