Abstract
The ability to generate multi-omics data coupled with deeply characterizing the clinical phenotype of individual patients promises to improve understanding of complex cardiovascular pathobiology. There remains an important disconnection between the magnitude and granularity of these data and our ability to improve phenotype–genotype correlations for complex cardiovascular diseases. This shortcoming may be due to limitations associated with traditional reductionist analytical methods, which tend to emphasize a single molecular event in the pathogenesis of diseases more aptly characterized by crosstalk between overlapping molecular pathways. Network medicine is a rapidly growing discipline that considers diseases as the consequences of perturbed interactions between multiple interconnected biological components. This powerful integrative approach has enabled a number of important discoveries in complex disease mechanisms. In this review, we introduce the basic concepts of network medicine and highlight specific examples by which this approach has accelerated cardiovascular research. We also review how network medicine is well-positioned to promote rational drug design for patients with cardiovascular diseases, with particular emphasis on advancing precision medicine.
Keywords: Network medicine, Precision medicine, Cardiovascular disease, Pathobiology, Omics
1. Introduction
Our appreciation for the complex pathobiology of cardiovascular diseases has grown rapidly thanks to the wider availability of powerful diagnostic tools and large-scale data generation. This progress has been less dramatic with respect to our understanding of underlying disease mechanisms, however, particularly in terms of genotype–phenotype correlations. Despite exponential growth in ‘big data’ collected from patients by virtue of wearable devices, the electronic medical record, and clinical trials, important limitations persist for translating such valuable clinical patient data to explicate disease pathobiology. Accomplishing this end is likely to require strategies designed to interrogate complex biological systems as a complementary approach to conventional analytical methods.1
Cardiovascular diseases have been defined and classified traditionally using the ‘one-gene, one-disease’ reductionist approach; however, most clinical phenotypes are heterogeneous biologically, limiting the utility of single-gene disease hypotheses. Taking a broader view, complex diseases are often determined by a range of epigenetic, transcriptional, and post-transcriptional events in multiple cell types. Determining the mechanisms by which these events occur and interact is a necessary step towards understanding nuanced, but critical, differences between patients with the same disease diagnosis. Indeed, the precise molecular program driving phenotypic expression is known to differ for nearly all human diseases according to genetic context and acquired factors, ultimately causing variation in the pathogenic basis of a clinical disorder recognized at point-of-care. It follows that treatment responses often differ among patients with the same diagnosis, and in certain cases, this dilemma is a major obstacle to optimizing patient outcomes.
Network medicine combines the principles of network science and systems biology, and focuses on biological interactions as a collective. In this way, pathways regulating complex genotype–phenotype relationships can be determined, yielding novel mechanistic insights that may otherwise be difficult to assess from large-scale multi-omics datasets.2 In this review, we introduce the basic principles of network medicine and human disease networks, and review specific examples of various applications of network medicine in cardiovascular research, including (i) the identification of novel disease mechanisms,3,4,5 (ii) the redefinition of complex clinical entities, (iii) patient prognosis, (iv) biomarker discovery, and (v) drug discovery and repurposing.5,6 We also review the rising concept of reticulotypes (patient-specific networks) as an important development in the field and a key link to precision-based methods for diagnosis and treatment. For additional reviews focusing on how the network approach has facilitated cardiovascular research, we refer the reader to refs.7,8
2. Basic principles in network medicine
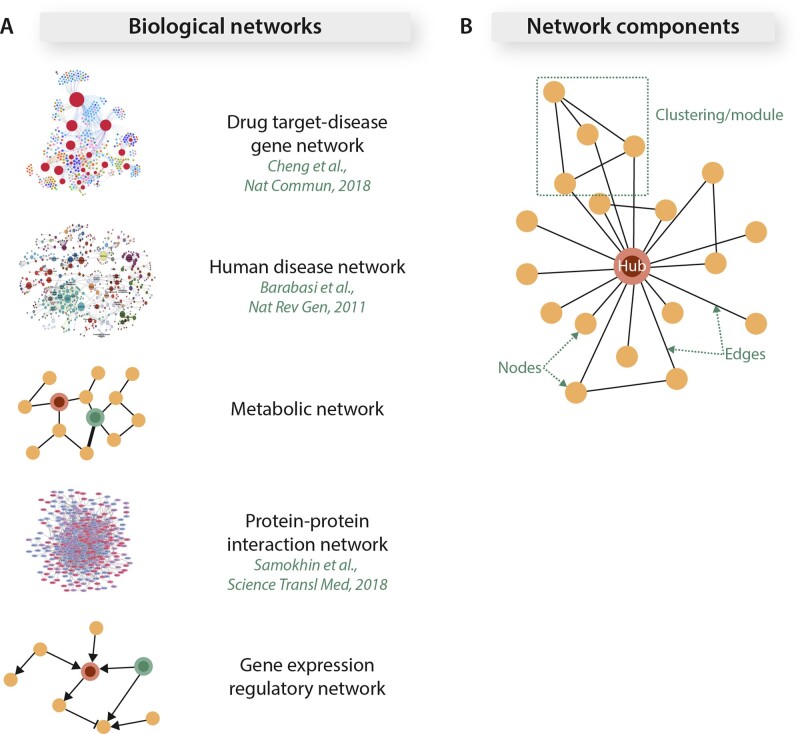
Networks can be used to represent a wide range of biological systems (Figure 1A). Notable examples of biological network types include, but are not limited to, protein–protein interaction networks, gene expression regulatory networks, Bayesian co-expression networks, and metabolic networks. Each network consists of individual biological entities (nodes), and their interactions are represented as interconnecting lines (edges) (Figure 1B).8 The number of edges connected to a node is referred to as the node’s degree. Topological clustering of interconnected neighbouring nodes forms a network subunit or module (Figure 1B).9 These network subunits tend to govern unique biological functions as demonstrated by the network containing multiple modules representing distinct biological aspects of the inflammatory response following myocardial infarction (MI).10
Figure 1.
Biological networks and basic network components. (A) Biological networks can represent a wide spectrum of biological dimensions. (B) In biological networks, distinct biological components, such as genes or metabolites, are represented as individual nodes. Edges connect two nodes and represent the interaction between them. Hubs are defined as nodes that are highly connected to other nodes. When a node and its neighbours are highly connected to one another, the resulting network neighbourhood is defined as a cluster or network submodule. Adapted from refs5–7 with permission.
In most biological networks, only a select few nodes are highly connected to a large number of other nodes (hub) (Figure 1B), while the majority of the nodes are sparsely connected with the overall network degree distribution following a power law. In these networks, the hubs tend to represent essential biological entities while the disease-associated genes tend to be located in the periphery of the network. Loss of a hub results in dissolution of the topological integrity of most biological networks.11
Biological networks are scale free,12 which means that their basic network organization properties (such as connectivity) remain invariant with changes in network size (or number of nodes) change. In these networks, the relationship between the logarithmic values of nodal degrees and their probabilities remain constant across varying network sizes. This property minimizes the rate of the (molecular) information flow across a network system and also allows understanding a network’s function when many of its components are missing or the network is incomplete (as is the case for most biological networks). These networks manifest emergent behaviour, which refers to the fact that the function of a network cannot be predicted based on what is understood about the function of an individual component studied in isolation.
The biological relationships represented by most networks are static (static networks), with ongoing efforts in play that incorporate dynamic processes evaluating network function (dynamic networks). The relative strengths of biological relationships between two nodes can be specified by differentially weighted edges (weighted networks). A network can consist of different types of nodes that represent different biological entities (bipartite networks, or in some cases, networks of networks). Examples include a drug target-disease gene network (Figure 1A) where the proximities between the nodes representing drug targets and those representing the genes associated with diseases may help predict drug efficacies and toxicities in silico.5 The computational approaches and technical details for constructing biological networks are outlined in ref.13
3. Human protein–protein interactome
Traditional molecular and biochemical inquiry over the last century has focused on analysing the function of individual molecules and their impact on cellular and physiological processes. However, this reductionist approach inherently does not capture the complexity that arises out of placing those individual molecules into a conglomerate, interactive biological system. An alternative approach is to begin with the premise that nearly all functions of a biomolecule require that biomolecule to interact physically with another entity. Biological molecules that interact with one another can then be grouped together and understood as a larger unit. Creation of an interaction map, or interactome, encompassing the totality of biomolecular interactions, therefore, allows both a more holistic and better organized approach to understanding cellular function than the traditional single-molecule or single-pathway approach.
The goal of creating such a human protein interactome is to represent protein–protein interactions (PPIs) among all proteins (or at least all of those that can be ascertained). It is important to note that ‘interactions’ in this context represent direct, specific physical binding of two proteins.14
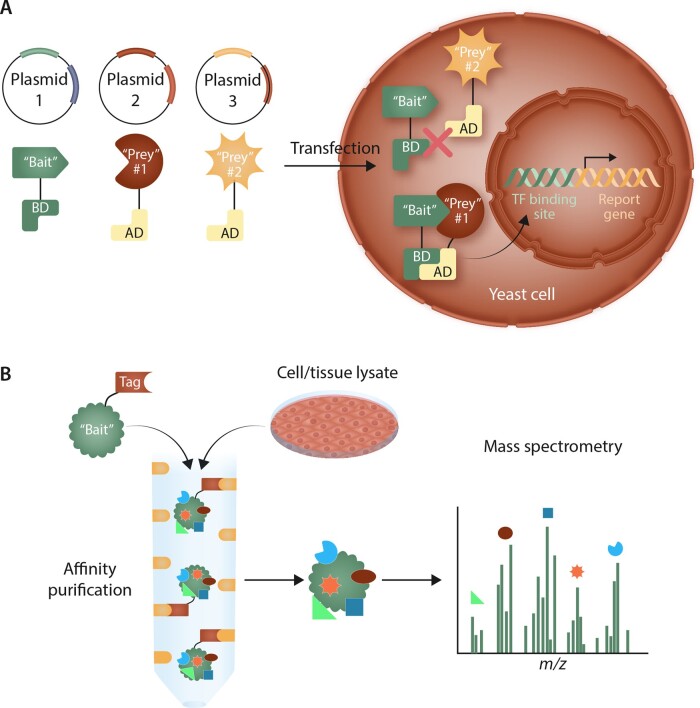
In order to elucidate these types of biologically relevant interactions in a large-scale and unbiased manner, methodologies have coalesced around two strategies: pairwise, or binary, discovery and co-complex discovery. While literature-curated PPIs also contribute to the construction of the interactome, this method does not lend itself to high-throughput evaluation of a large number of interactions and demonstrates bias towards better-studied proteins.15,16 Pair-wise discovery is based on observing interactions between two proteins; the most common high-throughput pair-wise strategy that has been utilized is the yeast two-hybrid (Y2H) system17–19 (Figure 2). In Y2H, a transcription factor necessary for expression of a reporter gene is separated into its constituent DNA-binding and activation domains. Two plasmids are created: one that expresses a particular ‘bait’ protein attached to the DNA-binding domain and a second that expresses a ‘prey’ protein (being investigated for its ability to interact with the ‘bait’) attached to the activation domain. If the two proteins physically interact, the binding and activation domains of the transcription factor will also be in close enough spatial proximity to mediate transcription of the reporter gene. In co-complex discovery methods, the ‘bait’ protein is expressed with a fused tag or is immunoprecipitated with a specific antibody. This ‘bait’ protein is then incubated with target cell or tissue lysates, after which the antibody or tag is used to isolate the ‘bait’ protein along with its associated ‘prey’ proteins. For high-throughput screening, co-complex discovery often takes the form of affinity purification with mass spectrometry (AP-MS), in which the tagged ‘bait’ protein is affinity-purified and associated ‘prey’ proteins are identified using MS20 (Figure 2). Beyond the classic techniques of Y2H and AP-MS, newer technologies are also being utilized to explore protein–protein interactions. These include protein-fragment complementation assays (such as biomolecular fluorescence complementation), in which the bait and prey proteins are attached to protein fragments that reconstitute to provide a direct signal or chemical reaction readout of the interaction21; surface plasmon resonance (in which changes in light refraction at a binding surface are used to determine binding and binding kinetics of bait and prey proteins); biolayer interferometry; mammalian protein–protein interaction traps22; and luminescence-based mammalian interactome.23 Other techniques, including nuclear magnetic resonance and isothermal titration calorimetry, are also used to identify and validate interactions between proteins.
Figure 2.
High-throughput methods for determining protein-protein interactions. (A) In the yeast two-hybrid system, a plasmid is created expressing a ‘bait’ protein linked to the DNA binding domain (BD) of a transcription factor (TF) required for expression of a reporter gene. Various ‘prey’ proteins are then linked to the activation domain (AD) of this TF in separate plasmids. Plasmids are co-transfected in pairs into yeast cells. If the ‘bait’ and ‘prey’ proteins interact, the BD and AD domains of the TF will be in close enough proximity to translocate to the nucleus and initiate reporter gene expression. If the ‘bait’ and ‘prey’ do not interact, the reporter gene will not be expressed. (B) In co-complex discovery, a ‘bait’ protein is linked to a tag and incubated with target cell or tissue lysates to allow ‘prey’ proteins to associate. The ‘bait’-‘prey’ complexes are isolated through affinity purification utilizing the linked tag. The ‘prey’ proteins can then be identified using mass spectrometry.
Current high-throughput methodological approaches have drawbacks that are important to understand in order to appreciate better the strengths and weakness of the generated PPIs. In the case of Y2H, false negative results may arise from the inability of the ‘bait’ and ‘prey’ proteins to interact due to conformational changes imposed by the linked transcription factor domains. The yeast expression system may lack the post-translational processing necessary for the interaction to occur. Additionally, complexes may fail to translocate to the nucleus and initiate transcription. False positives may occur from non-biological interaction of the ‘bait’ and ‘prey’ proteins arising from overexpression, or auto-activation of reporter gene expression. In order to overcome these shortcomings, validation of Y2H data is important to confirm that interactions observed are physiologically relevant. Two-hybrid technology has also been adapted to mammalian systems to overcome post-translational processing issues for human proteins expressed in non-mammalian cells.24,25
Co-complex discovery similarly takes place in the absence of a typical intracellular environment, which can alter the nature, occurrence, and properties of PPIs. One major potential limitation of co-complex discovery is detection of both direct and indirect interactions among proteins. Complexes are isolated in total, and those proteins that are part of the agglomeration, even if they are not in direct physical contact with the ‘bait’ protein, will nevertheless be identified as ‘prey’ proteins. The interactome, however, is a pair-wise representation of only direct interactions. In order to overcome this limitation and thereby represent co-complex data in a specific pair-wise fashion, protein interaction affinities can be computationally modelled. Several strategies to do so have been utilized, including a matrix model (which assumes interactions between all members of a complex), a spoke model (which directly pairs two proteins), and a socio-affinity model (a hybrid of spoke and matrix models).26,27 Optimization of these models is geared towards balancing stringency of affinity with inclusivity of all likely interactions in order to minimize false positive and false negative results.
Of note, the PPI also may not adequately capture context-specific effects on protein interactions, such as tissue type, developmental stage, or cellular environment. This limitation is, in part, due to the use of high-throughput screening methods and bulk-tissue sources that remove these context cues during PPI assessment. Bulk tissue sourcing limits the ability to understand spatial and temporal changes that may be induced by specific environmental stimuli or dynamic internal processes. Adding context information and creating subnetworks from these data can improve network predictions and provide better insights into cell-type- and disease-specific relationships.28–31 Single-cell level data are increasingly being investigated to give very detailed, high-resolution snapshots of protein interactions and their genetic regulators, as well as to generate cell atlases characterized by these interactions.32,33 By annotating more richly the PPI, these approaches hold the potential to elucidate better common interacting partners and critical hubs in the interactome.
4. Disease networks and disease–disease interactions
Development of the PPI lends itself to the question of whether higher-order phenotypes can similarly be represented as a network of inter-connected constituents. The intuitive logic of this approach is supported by several decades of research identifying shared mechanisms contributing to the pathobiology of disease processes. For example, perturbations in vascular homeostasis, cholesterol metabolism, and immune function contribute to the phenotype of MI. Using this rationale, different network spaces have been created to describe diseases and disease phenotypes, including a disease network, in which disease nodes are linked by shared genes; a disease-gene network, which links genes if they are associated with at least one common disease; and a symptom-disease network, in which two diseases are linked by common symptoms or by the likelihood of being co-morbid in patients.34–36 Disease networks can also be expanded further by incorporating additional layers of data, including microRNA associations and metabolic networks,37,38 creating networks of networks. For cardiovascular diseases, transcriptomic level networks39–41 and those incorporating long non-coding RNAs42,43 are being evaluated and generated for a variety of cardiovascular conditions to provide a richer representation of important interacting components.
The synthesis of data incorporating diseases, genes, and gene products into disease networks creates a unique representation of these inter-related components that can offer new insights into disease pathogenesis. These insights can arise from the topology of the network itself. Disease genes generally tend to populate the periphery of a network, rather than comprising central hubs34,44 (except for those that are associated with embryonic lethality).
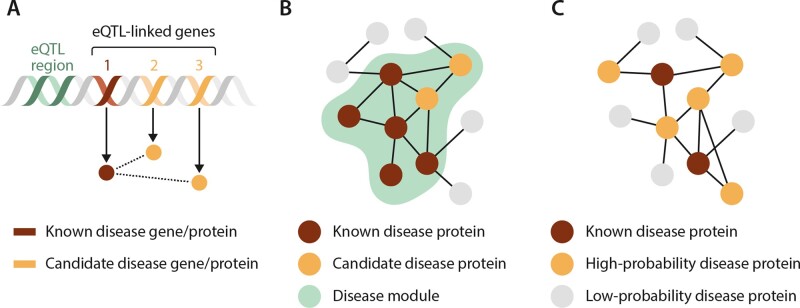
Analysis of disease networks has revealed that the genes associated with a particular disease demonstrate distinct network patterns. Disease-associated genes show a tendency to be expressed in the same tissue types and exhibit correlated expression compared to random control gene sets.34 Disease genes are more likely to interact with each other, and the products of these genes have overlapping PPIs.15,34,45 These similarities have helped to identify disease modules. A disease module can be defined as a network feature in which the constituents all contribute to a particular cellular function, disruption of which leads to a specific disease phenotype.6 Thus, on a network level, dysfunction in a disease module forms the underlying basis of disease (Figure 3).
Figure 3.
Network-based methods to predict disease genes. (A) Linked genetic elements, for example through shared expression quantitative trait loci (eQTLs) containing a known disease gene, have increased probability that other genes within the linkage interval contribute to the disease process. Linkage methods can be utilized to uncover these candidate disease genes and proteins. (B) Disease modules identify network regions that contribute to a disease. Proteins contained within these network elements can be investigated as candidate disease proteins through methods such as the seed connector algorithm. (C) Diffusion propagation methods start with known disease proteins and then assign probabilities to proteins in the interactome based on the likelihood they are associated with the disease. This analysis is based on frequency of candidate proteins’ interactions with and network distance from the known disease proteins. Adapted from ref.6 with permission.
The interconnectedness of disease modules can also help to explain how alterations in seemingly disparate genes may result in similar disease phenotypes. On a network level, components of a disease module share the same neighbourhood within the overall interactome. If two disease modules overlap, then perturbations in one will affect the other module, as well, leading to common (or convergent) phenotypes between the two disease states.13 Furthermore, the degree of overlap between two disease modules in the network space can be used as a surrogate for the degree of shared pathobiological links.
One example of how a network approach can help uncover new links is in the relationship between dementia and cardiovascular disease. While both entities share common physiological, metabolic, and environmental risk factors, including hypertension, obesity, and smoking, attempts to demonstrate positively-correlated clinical associations between cardiovascular disease and dementia have been mixed, and results can vary depending on study type and cohort.46 However, a network approach to this question yields a more robust relationship. Evaluation of disease modules in Alzheimer disease and cardiovascular disease demonstrates shared genes between the two phenotypes within a single (overlapping) module47 as well as shared regulatory elements.39 Utilizing a network-based definition of co-morbidity, Alzheimer disease and cardiovascular disease can further be shown to be highly co-morbid at a population level, elucidating a relationship that has been difficult to demonstrate based on clinical data alone.48
5. Uncovering novel disease mechanisms in cardiovascular diseases using network analyses
Network medicine emphasizes functional or physical associations between biological components (e.g., genes, transcripts, proteins, other intermediaries) as the principal step towards understanding complex pathophenotypes. In order to dissect complex mechanisms involved in coronary artery disease (CAD) and MI, for example, network construction and analysis using the existing inflammation- and MI-related PPIs (My-Inflamome) revealed 21 highly interconnected but distinct modules of unique biological properties and endophenotypes, including coagulation, cell death, wound healing, and immune responses.10
Overemphasizing the frequency of a genetic variant without proven knowledge of its functionality, or a single biochemical reaction without understanding the global effect of the pathway within which it operates, offers key challenges to reconciling the pathobiology of complex diseases.49 Data from genome-wide association studies (GWAS), for example, have yielded important gains such as the discovery that an inactivating mutation in NPC1L1 (in particular p.Arg406X) confers protection against coronary atherosclerotic disease via impaired cellular uptake of low-density lipoprotein cholesterol.50 Nonetheless, many false positives are described for this and other association-based methodologies, and a brief consideration to their limitations may be useful for understanding opportunities that network medicine aims to address.
5.1 Determining pathogenicity of gene variants
The true prevalence of ‘rare’ variant(s) in a disease population hinges on the adequacy of comparator datasets, which is critical to offset background (benign) genetic variation in the genome. In this regard, it is worth noting that much influential and highly cited early genomics-era data from population studies implicating monogenic cause(s) of complex cardiovascular diseases were based on limited access to sufficient control populations.51 For example, hypertrophic cardiomyopathy (HCM), which is characterized by left ventricular hypertrophy in the absence of a secondary cause, has long been regarded as a disease caused by a pathogenic gene variant coding one of several cardiomyocyte sarcomere proteins (reviewed in detail in ref.52). However, the phenotypic heterogeneity of HCM often involves cell-types that do not express cardiomyocyte sarcomere proteins, but does include endophenotypes beyond sarcomere-dependent hypercontractility (e.g., interstitial fibrosis, mitral valve leaflet elongation, and others).53 This finding suggested that using simple genetics to inform diagnosis could result in patient misclassification clinically.
Testing this hypothesis became possible only in the current era owing to newfound availability of large sequencing databases that include controls with pertinent demographic and clinical data. Manrai et al.54 showed that among N = 94 HCM-associated genetic variants, N = 5 [TNNT2 (K247R), OBSCN (R4344Q), TNNI3 (P82S), MYBPC3 (G278E), and JPH2 (G505S)] were classified as high-frequency variants, which accounted for 74% of the overall HCM genotype prevalence. Furthermore, African Americans harboured these variants disproportionately to a greater extent than white Americans (in part, because of a control population in which African Americans were severely under-represented), demonstrating the potential for misdiagnosis in specific ethnic or racial groups based on inappropriately attributing causality to genetic frequency alone. Similar observations have been made by other groups studying HCM,55 overall instilling an important cautionary message to efforts that reduce heterogeneous and complex cardiovascular disease phenotypes to a sentinel, easily measurable germline feature, such as a single germline gene mutation or biochemical reaction.53
5.2 Epigenetic and post-transcriptional mechanisms of cardiovascular disease
A wide range of functionally critical post-transcriptional events is recognized increasingly in the pathogenesis of cardiovascular disease, but may be under-represented in studies narrowing on genetic substrate. Indeed, complex diseases demonstrate a discordance between GWAS-identified genetic determinants and the relatively low odds ratios of, and percentage of, genetic variation explained by those GWAS signals.56 Data from intersecting network spaces, including epigenetic modifiers, transcriptional regulators, and post-translational effects, all influence the net effect of genetic variation on higher-order phenotypes. The contribution of epigenetics, microRNAs, and post-translational modifications, to name a few examples, has already been established in pulmonary vascular disease,57 MI,58 and essential systemic hypertension,59 among many other cardiovascular disorders. Transgenerational epigenetic imprinting has been documented in mice engineered to express a hypomorphic mutation in the gene encoding methionine synthase reductase (Mtrr),60 which regulates folate metabolism, and, thus, the bioavailability of pro-thrombotic and pro-atherosclerotic homocysteine. In pregnant rabbits fed a high cholesterol diet, an increase in predilection to atherosclerotic vascular diseases is noted in offspring through five successive generations.61 These examples provide mechanistic insight into observations linking perinatal events with adult-onset metabolic cardiovascular diseases, including diabetes mellitus, coronary heart disease, essential hypertension, and pulmonary arterial hypertension (PAH).62
Furthermore, ∼200 post-translational modifications are reported to regulate different cellular functions, some of which are already utilized daily in clinical practice (e.g., haemoglobin glycosylation for diagnosing diabetes mellitus). Although some modifications are well-established in the context of pre-programmed signalling, such as the case for the phosphorylation substrate of tyrosine kinases, many of these molecular responses function as a (mal)adaptive interface with the environment. For example, the amino acid cysteine is conserved within codons of higher-level eurkaryotic organisms to a lesser extent than is predicted computationally. This finding, in turn, suggests evolutionary pressure towards cysteine functionality.63 The sulfur of cysteine contains two pairs of free electrons, and, thus, in the setting of oxidant stress may react with oxygen free radicals (e.g., hydrogen peroxide, superoxide anion, others) to form higher oxidative intermediary species that alter protein structure and function. This mechanism is attributed to the development of numerous cardiovascular diseases,64 and articulates a critical interface between biochemistry and environmental cues, such as nutritional, toxin, or pollutant triggers, that may drive clinical phenotypes. Taken together, these points illustrate the additional importance of considering events after inborn coding that have the potential to affect cellular function and disease pathogenesis.
5.3 Analytical methods in network medicine for predicting disease genes
Determining pathogenic nodes or pathways involving a collection of nodes is important for translating data in silico to observations that may be tested empirically. To accomplish this end, several validated methods for interrogating network features have been developed (Figure 3).
5.4 Linkage methods
In this approach, information on important PPIs is derived from genetic loci that are known to influence the probability of developing a particular disease. Quantitative trait locus (eQTL) data are used increasingly to describe genes with strong local regulation within genetic loci.65 Using eQTL data in networks exploits the assumption that most such loci correspond to complex traits via programmed, integrated gene expression and downstream pathways.66 The integration of eQTL data with co-expression gene networks (in which a network edge is defined by correlative differences in expression between two genes) was used recently to decipher variability in lipid metabolism. Li et al.66 developed co-expression networks using publicly available liver genome-wide transcriptomic and proteomic data from a multitude of different mouse strains. The results were refined to modules that included cholesterol biosynthesis genes, captured using gene ontology datasets, and overall included N = 112 cholesterol/lipid metabolism genes that were conserved across all of the strains. From this approach, N = 25 novel genes were identified, and cross-referenced with validated GWAS databases enriched for patient populations with CAD (e.g. Million Veteran Program).67 The SESN1 gene, which codes for the protein sestrin, was linked with plasma lipid traits in the GWAS populations, and identified as a previously unrecognized cholesterol biosynthesis regulator in vivo. Focusing on eQTLs for pursuing the biological importance of genes has already been used to understand network features that may be important for predicting other common cardiovascular pathophenotypes, including the role of NFATC4 in type 2 diabetes mellitus.68 A similar approach was used recently to implicate SMAD3 in PAH incidence and clinical risk.69,70
This pathway-based approach to interpreting network data emphasizes the high probability that disease-associated proteins localize to discrete topological or functional segments, or modules, within a larger interactome. Several disease module strategies to determine gene pathogenicity have been reported,13,71 including the seed connector algorithm (SCA) approach.72 Seed proteins (or genes) are the specific proteins of interest with known disease association or other biological association. In SCA, linking proteins, which are first-order interactors (also denoted connector proteins) with seed proteins in the interactome, are added sequentially to networks derived from biological datasets; these connector proteins, only identified by knowledge of network topology, provide the ‘missing links’ among seed proteins, facilitating the identification of discrete disease modules within the PPI. This strategy is used to uncover potentially important PPIs that are not identified using seed proteins alone. The SCA approach aims to complement other established module-related methods, including DIAMOnD,73 prize-collecting Steiner tree,74 and GLADIATOR,75 which emphasize connectivity patterns of known disease proteins, network efficiency (relative to nodes and edge totals), and disease-disease phenotype similarities for pathogenic gene discovery.
5.5 Diffusion propagation
This methodology relies on the assumption that genes underlying like phenotypes are prone to interact together (or agglomerate in the PPI).13 Starting with a known or suspected (i.e., index) disease-causing gene, an iterative analysis is conducted to identify the outermost boundary of a potential, succinct subnetwork. This is accomplished by expanding the route of connectivity from the index node to additional nodes in a way that illustrates hierarchical connectivity.76 Capturing nodes that are not necessarily directly connected to the (actual or suspected) disease-causing gene is accomplished based on network topology, with consideration given to all potential pathways, and is, thus, a collective strength of this approach. Random walk and diffusion kernel methods are permutations of this approach that have been successful at identifying functionally essential proteins,77 as well as key molecular pathways underlying cardiovascular pathophenotypes, including heart failure.78
5.6 Specific Applications of network medicine to cardiovascular diseases
5.6.1. Pulmonary vascular disease
Plexigenic, hypertrophic, fibrotic, and proliferative remodelling of distal pulmonary arterioles is a cornerstone feature of PAH, which is a highly morbid cardiopulmonary disease associated with substantial disability and early mortality in most patients.79 Once perceived as primarily due to pulmonary arterial hypercontractility, in the contemporary era PAH is viewed as a complex disease driven by the interplay and cross-talk between numerous pathogenic signalling pathways.80 Deciphering the molecular underpinnings of PAH is needed, in turn, to identify novel therapeutic targets since at present all available pharmacotherapies modulate the same three signalling axes (the nitric oxide-, endothelin receptor-, and prostacyclin pathways).
Approximately 25% of presumed idiopathic PAH patients will harbour an established pathogenic variant (e.g. BMPR2, EIF2AK4, SOX17, others)81; however, most ‘PAH-causing’ genotypes are incompletely penetrant raising speculation that genetic co-regulation or integrated signalling pathways may be important in understanding PAH. Early reports utilizing network medicine in pulmonary vascular disease addressed post-transcriptional events related to BMPR-2 bioactivity, and observed that, indeed, hypoxia, inflammation, or genetic inhibition of BMPR-2 (all of which are reported in PAH patients) up-regulates miR-21 in pulmonary artery endothelial cells.82 Subsequently, numerous bioinformatics reports expanded the range of miR candidates involved in the development of various PAH endophenotypes, including miR-34a-angiopoietin-1 in neonatal lung injury83 and miR-133a in experimental right ventricular heart failure,84 among others.85 Zhang et al.86 reported recently on the regulation of abnormal adventitial fibroblast metabolism via miR-124/polypyrimidine tract binding protein 1/pyruvate kinase muscle signalling. Interesting, miR-124 was selected for analysis in that study based on its pathogenic role underlying dysregulated metabolism in carcinoma cells, reinforcing the importance of cell type-specific regulatory mechanisms in the pulmonary blood vessel despite the molecular promiscuity that is reported for many, if not most, miRs. This point has been clarified to some degree by reports focusing on cross-talk between heterologous pulmonary vascular cells, including data suggesting that miR-130/301-specific control of peroxisome proliferator-activated receptor-γ may regulate contractile signalling in pulmonary artery endothelial cells that affects contractility in pulmonary artery smooth muscle cells.87
Arterial fibrosis increases pulmonary vascular resistance in PAH, although effective anti-fibrotic therapies remain lacking. The conventional paradigm for understanding fibrosis invokes pro-fibrotic ‘master switch’ molecules, such as transforming growth factor (TGF)-88 This framework, however, overlooks collagen biofunctionality, differential sensing mechanism that regulate collagen transcription across cell types, and disease-specific molecular mechanisms that promote collagen deposition. For example, increased TGF- is implicated in dermal wound healing, a form of physiological fibrosis, but also in pathogenic and maladaptive fibrosis in numerous cardiovascular diseases such as HCM (interstitial fibrosis), MI (replacement fibrosis), and PAH (arterial fibrosis).89 Moreover, the functional consequences of TGF- activation differ across PAH subgroups: hereditary PAH due to a germline BMPR2 mutations and scleroderma-associated PAH are associated with proliferative- and fibrosis-dominant lesions, respectively, further underscoring the potential limitations of ascribing the totality of an entire endophenotype to a single point of molecular convergence.
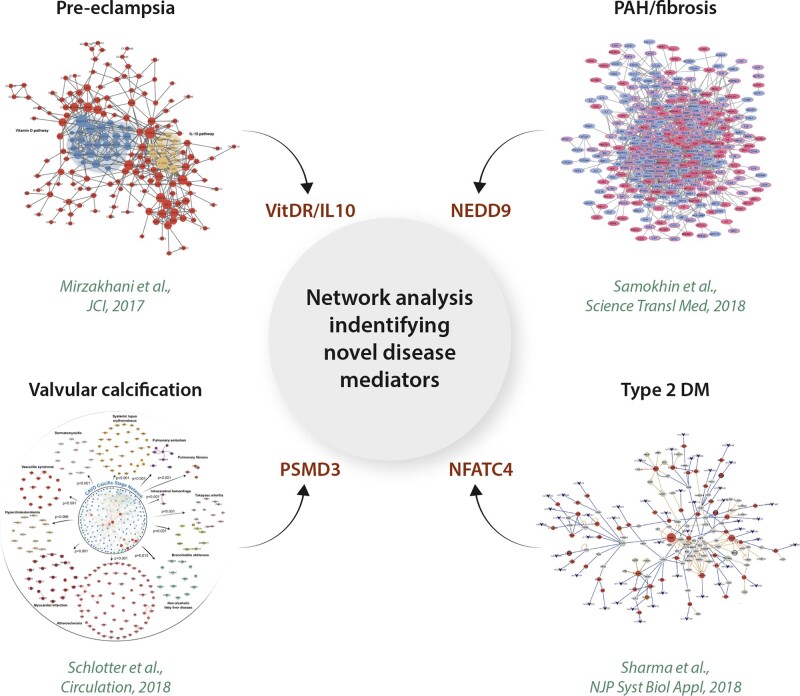
Our group addressed this issue recently by developing a PPI fibrosome, or fibrosis endophenotype module, which was constructed by considering differences in collagen biofunctionality among network genes. Specifically, fibrosis genes were collected from numerous validated gene ontology resources and designated as adaptive or pathogenic by virtue of their association with wound healing or vascular fibrosis, respectively.7 We focused on genes regulated by aldosterone (ALDO), which is increased in plasma of PAH patients and promotes fibrotic vascular remodelling in experimental disease models in vivo.82,90,91 The result was the identification of a novel network that included adaptive, pathogenic, ALDO-regulated, and overlapping genes (Figure 4). The betweenness centrality metric, which helps identify a key (or bottleneck) node within a network by measuring its relationship to other nodes’ interconnectivity, was used to rank order ALDO-regulated genes that were important in transitioning in silico between fibrosis phenotypes. From this approach, the Cas protein NEDD9 emerged as a potentially important pro-fibrotic intermediary in PAH, which, in cancer cell lines, targets the transcription factor SMAD3, itself previously implicated in BMPR-2-dependent pulmonary vascular remodelling.94 Oxidation of NEDD9 at Cys20 in pulmonary endothelial cells induced NEDD9-dependent fibrosis and led to the emergence of NEDD9 as a potential biomarker and therapeutic target in PAH.95
Figure 4.
Network analysis to identify novel disease mediators. A network approach has enabled a number of novel disease mechanism discoveries. Notable examples include the vitamin D receptor (VitDR) and interleukin (IL)-10 pathways in pre-eclampsia (upper left),92 NEDD9 in PAH (upper right),7 26S proteasome non-ATPase regulatory subunit 3 (PSMD3) in valvular calcification (lower left),93 and nuclear factor of activated T cells 4 (NFATC4) in type 2 diabetes (DM2) (lower right).68 (upper left) Network analysis of 348 differentially expressed vitamin D-associated genes in peripheral blood of pre-eclampsia patients identified the network modules specific to the changes in maternal immune responses. Of these, the IL-10 signalling pathway was noted to be closely interacting with the vitamin D signalling pathway with notable down-regulation of IL-10 signalling in pre-eclampsia patients. (upper right) Betweenness centrality analysis of a novel network consisting of ALDO-regulated genes associated with vascular fibrosis identified NEDD9 as a novel mediator of PAH pathogenesis. (lower left) Construction of the proteomics subnetworks involving the proteins overrepresented in the calcific stages of human calcific aortic valve disease in the fibrosa layers and betweenness centrality analysis identified fibronectin-1, PSMD3, and PSMA1 as the potential key disease mediators involved in valvular calcification. (lower right) Control centrality analysis of the human pancreatic islet tissue-specific gene regulatory network developed from diabetic and non-diabetic donors identified multiple biological pathways driving the type 2 diabetes disease network with NFATC4 as a key controller of these pathways. Adapted from refs7,68,92,93 with permission.
5.6.2. Pre-eclampsia
Pre-eclampsia is a major obstetrical complication that affects ∼6% of all pregnant women. The pathogenesis of pre-eclampsia is multifactorial and likely varies between patients according to genetic context, epigenetic mechanisms, age, and other acquired risk factors,96 but a unifying feature is trophoblastic vascular dysfunction.97 In one analysis, differentially expressed placental gene transcripts between pre-eclamptic and normal term controls identified N = 4 distinct network clusters, from which a high density of up-regulated and down-regulated placental genes were localized to two specific clusters. This approach yielded insight into trophoblastic differentiation via ZNF554, which is a member of the KRAB zinc finger family.98 Parallel efforts using PPI networks led to the discovery that proteins previously described as vasoactive in other circulatory beds may also be important in the pathogenesis of preeclampsia, including FLT1, VEGFA, FN1, LYN, and NDRG1.99 A recent study also linked maternal vitamin D level with asthma and pre-eclampsia92 and implicated vitamin D receptor and IL-10 signalling in the process (Figure 4). Further empiric data are needed, however, to clarify the precise circumstances by which these putative intermediaries generate or support pathogenic vascular responses in affected patients.
5.6.3. Calcific aortic valve disease
Calcific aortic valvular disease affects >25% of the population over the age of 65 years, and when clinically significant, is a high-risk condition that requires procedural intervention.100 Schlotter et al.93 characterized pathologic stage-specific differences in the aortic valvular proteome using a variety of methods, including in-tandem mass spectrometry. From a bioinformatics analysis, a total of N = 249 significantly enriched pathways were identified in highly calcific samples. When overexpressed proteins specific to calcific valvular remodelling were mapped to the human PPI, the derivative network could be interrogated by betweenness centrality to identify pathways that were critical to the network topology. This analysis identified PSMD3 and PSMA1, belonging to the TGF-β-, canonical wnt-, MAPK, and NOTCH-signalling pathways and important in osteoblast biology, as potential therapeutic targets to reverse or prevent valvular calcification (Figure 4).
5.6.4. Vitamin D receptor biology
The vitamin D receptor is expressed on several cardiovascular cell types, including cardiomyocytes, vascular smooth muscle cells, and endothelial cells.101 Several cross-sectional, cohort, and case-controlled studies have suggested a link between circulating vitamin D3 concentration and angina pectoris,102 coronary heart disease,103 and systemic hypertension,104 among other cardiovascular phenotypes. Vitamin D-dependent regulation of cardiovascular function is a complex process that lacks a dominant genetic-phenotype relationship. Indeed, differential expression of >1000 genes is reported in cells stimulated with 1,25(OH)2D3.105
Vukić et al.106 assessed serum 25(OH)2D3 (75 nM) from patients with insulin resistance in the VitDmet study, which measured the longitudinal effect of vitamin D3 supplementation on plasma levels.107 Focusing on N = 24 vitamin D receptor (VDR) genes (including N = 12 that were discovered in the study) derived from established VDR ChIP-seq data or the transcriptomic signature of B cell/THP-1 cells treated for 24 h with 1,25(OH)2D3, a correlation network was generated. In that network, Pearson correlation coefficients represented the relationship between changes in transcriptional data and 25(OH)D3 serum levels through the VitDmet study. The investigators identified involvement of STS, BCL6, ITGAM, LRRC25, LPGAT1, and TREM1, among others, in vitamin D3 responsiveness in vivo. The implications of these data to cardiovascular disease require further analysis, although several of these genes have been implicated previously in cardiomyocyte development108 and fibrosis.109
6. Network medicine and clinical risk stratification and disease classification reappraisal
Risk stratification tools in clinical cardiovascular medicine generally utilize probabilistic models. This process includes many point-of-care resources, such as Framingham110 and TIMI111 risk scores for predicting cardiovascular events and 30-day mortality in patients with suspected acute coronary syndrome. Although linear regression models are valid, there are several important limitations related to this approach. First, probabilistic modelling does not account for relationships between clinical parameters, and even multivariate calculations consider changes to risk based on the addition of individual inputs. Second, results may be driven disproportionately by extreme datapoints within the reference group. In the case of the REVEAL calculator in PAH, for example, maximized risk requires a pulmonary vascular resistance level >32 WU, which is extremely elevated and encountered rarely even in a referral centre, thereby affecting the wider utility of this scale in daily practice.112
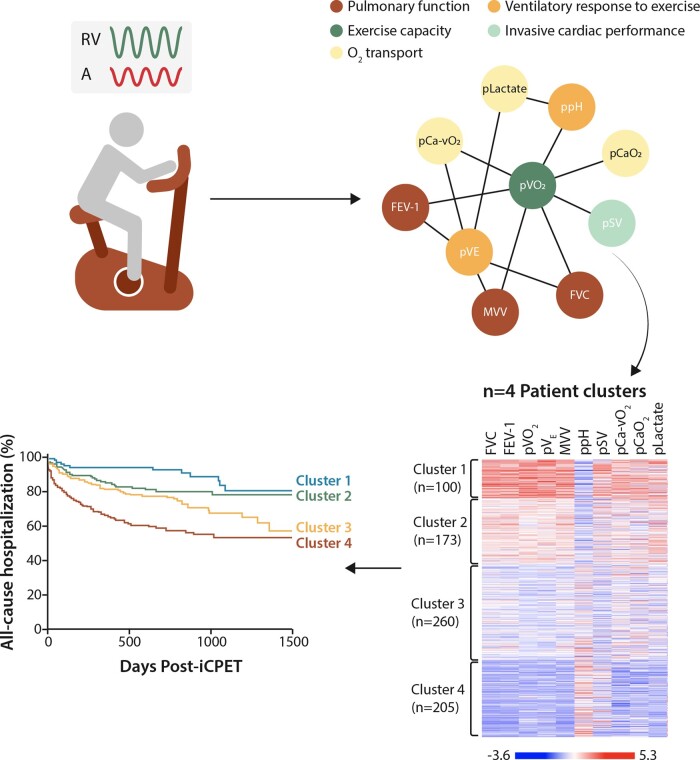
In contrast, network strategies focusing on clinical parameters that are functionally (e.g., pathophysiologically) related may lead to data on patient profile information not informed by binary, branch chain, or other reductionist methods. For example, we developed an exercise correlation network that was derived from clinical parameters collected during invasive cardiopulmonary exercise testing (iCPET) performed at point-of-care.113 Each of the 76 iCPET variables was assigned to one of eight physiological domains (Figure 5). We focused on a subnetwork that included 10 iCPET variables (inclusive of variables from five physiological parameters), which was used to identify four distinct patient clusters that varied by exercise profile and clinical outcome.114 These data were then used to prognosticate 3-year hospitalization rate; however, this tool was based on Euclidian principles rather than probabilistic modelling. In this way, the network approach was distinct from prior efforts classifying exercise profiles using artificial intelligence115 because in our method no single variable was used to drive prognostic estimates. Similar strategies have been proposed through ongoing deep-phenotyping efforts in patients with HCM53 and pulmonary hypertension,116 as well as comorbidities that affect cardiovascular function, including chronic obstructive lung disease.117
Figure 5.
Exercise correlation network from invasive cardiopulmonary exercise testing. Patients undergoing invasive cardiopulmonary exercise testing for unexplained exertional intolerance were used to construct an exercise network. Clinical measurements collected at the time of the study were used to construct this network with 39 nodes and 98 edges, grouped into 7 broader physiological domains. A subnetwork of 10 variables was used to identify 4 distinct patient clusters that differed in exercise profiles and clinical outcomes. Adapted from ref.114 with permission.
7. Network approach to biomarker discovery
Unbiased network analyses of large human datasets are accelerating novel biomarker discovery.Metabolomics data analysed for the strengths of association between a single or group of metabolites with disease have been most widely used for this purpose. Notable recent examples include a dynamic network analysis platform integrating patient plasma metabolite levels from multiple timepoints following alcohol septal ablation (‘planned myocardial injury’) that led to the identification of carnosine and glycocholic acid as novel metabolites associated with acute myocardial injury.118 NEDD9 in PAH is another important example of a novel biomarker discovery facilitated by the integrative network analysis of multiple perturbed biological pathways.7,95
At the same time, a paradigm shift that network medicine brings—approaching disease as a consequence of perturbed interactions among multiple biological entities rather than a single altered molecular pathway—highlights the current limitation of relying on a single molecule as a biomarker for diagnosing, prognosticating, or assessing therapeutic efficacy for a complex disease process (e.g., NT-proBNP in heart failure). While such biomarkers are unequivocally useful for our current clinical practice, our ability to collect large sets of patient data in multiple biological dimensions and growing capacity to integrate disease-specific data using network concepts suggest that a more comprehensive approach to biomarkers, such as a functionally rational subnetwork consisting of disease-associated molecules or pathways rather than the use of a single molecule, may become a reality in the near future.
8. Reticulotyping as a novel approach to precision medicine in cardiovascular health
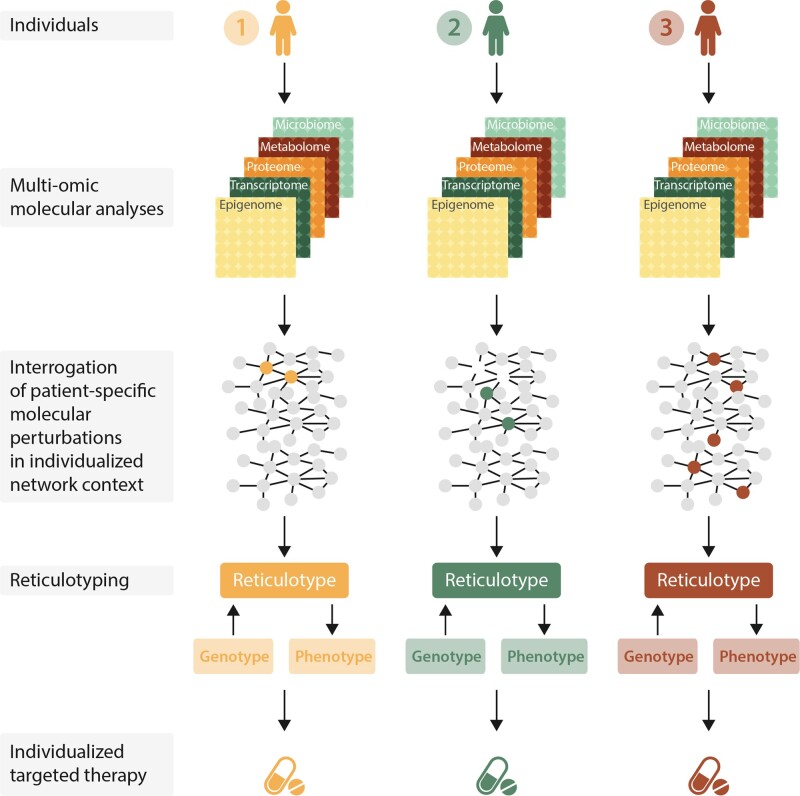
Network analysis can also accelerate our current endeavours towards precision medicine. Multi-omics data collected from an individual patient can be used to build a set of molecular networks across different biological domains (e.g., transcriptome, metabolome, proteome, microbiome). These networks then can be integrated into a network of networks that is unique to each patient’s biological makeup, including the interactions among multiple components; this personalized network is denoted the ‘reticulotype.’ (from reticulum or retiaculum, Lat.).119 An individual’s (molecular) reticulotype can be expanded and personalized further with his or her unique set of exposomes, such as environmental exposure, dietary or exercise habits, and psychosocial factors. Within this individualized network context established under homeostatic conditions, one can examine the effect of a specific molecular perturbation on the individual’s biological networks as a whole, especially how its effect alters interactions among the molecular constituents and their other interacting partners well beyond the select individual molecular targets of the perturbations.
As the baseline topological architecture and dynamic nature for each individual’s reticulotype likely differ greatly between any two individuals carrying the same disease diagnosis, it is not surprising that the net outcome (clinical phenotype) of a given pathogenic perturbant likely would differ, as well as their treatment responses. For example, the reticulotype for a 68-year-old male with heart failure with preserved left ventricular ejection fraction (HFpEF) and chronic obstructive pulmonary disease with tobacco exposure likely differs vastly from that of a 45-year-old premenopausal female with HFpEF with hypertension, obesity, and diabetes. The current therapeutic approach to these patients’ HFpEF remains mostly uniform (diuretics) despite the profound heterogeneity suspected among individuals’ disease mechanisms. The recent randomized controlled trial studying the efficacy of angiotensin receptor-neprilysin inhibitor in HFpEF patients (PARAGON-HF) has brought much needed attention to understanding the phenotypic heterogeneity underlying HFpEF. Although the overall study demonstrated no differences in the primary outcomes with sacubitril-valsartan vs. valsartan treatment alone, the prespecified subgroup analyses suggested possible benefit specific to female sex [hazard ratio 0.66 (0.49, 0.88)] and lower left ventricular ejection fraction (LVEF) [hazard ratio 0.76 (0.63, 0.92) per 10% decline in LVEF].120 While further mechanistic studies are needed to explain these findings, these observations are consistent with an individual’s baseline biological makeup as represented in his/her reticulotype and its interaction with (or perturbation by) disease as an important determinant of his or her treatment response, especially in complex cardiovascular disease settings such as HFpEF. In addition to facilitating an individual’s genotype–phenotype correlation, we propose that reticulotype analysis can guide identifying a therapeutic target specific to his or her disease network module while helping to avoid adverse off-target effects, benefiting from the precise knowledge of the topological relationship between the drug targets and disease- and non-disease genes within an individual’s biological network (Figure 6).
Figure 6.
Reticulotype analysis and precision medicine for cardiovascular patients. Complex sets of patient-specific molecular and clinical data can be integrated into a set of biological networks unique to each individual (reticulotype).119 The study of such network behaviour and the sequelae of its perturbation may facilitate genotype–phenotype correlation and predict treatment response in a patient-specific manner. Adapted from ref.2 with permission.
9. Network medicine and pharmacology
The prevailing paradigm of drug development and experimental pharmacology has relied heavily on single-drug/single-target investigations. This approach, however, does not take into account that most cardiovascular diseases arise from the interplay of genetic, metabolic, and environmental risk factors. In addition, off-target and side effects often become only after human trials or wide-spread use has begun.
Key areas in which a network pharmacological approach can yield new insights include drug target selection, repurposing of drugs for novel applications, and prediction of drug toxicities.121,122 The disease module hypothesis can serve as a framework for drug target selection. Drug efficacy, specificity, and toxicity can be assessed by mapping a drug’s known target(s) onto the PPI and assessing how close to this target (pathway) is to the intended disease module.
An initial step in this process is to understand the full array of drug targets and assess their interconnectedness. Yildirim et al.123 constructed a drug-target network composed of all FDA-approved drugs connected to their target proteins. From this analysis, the authors demonstrated that a large majority of drugs target more than one protein, and that there is a subset of proteins that is frequently targeted by many drugs. After mapping the drug-target network onto the PPI, the authors observed that most drug targets are on the periphery of disease modules and are less likely to include essential proteins of the module. This observation suggests that most drugs do not affect core underlying mechanisms of disease, and, perhaps, exert more effects on (disease non-specific) symptom control. Indeed, drug targets that demonstrate greater proximity to disease proteins in the PPI (assessed using network computational methods) demonstrate greater therapeutic effect, while drugs that have targets farther away from disease proteins are more palliative in nature.124
For cardiovascular diseases, one area to which this approach has been applied is the study of drugs used to treat MI. While the pathobiology of atherosclerosis and MI has been described in increasing detail and complexity, clinical treatment is still limited. Azuaje et al.125 constructed a network consisting of both approved and related drugs for the treatment of MI, the full set of drug-drug interactions of these drugs, the targets of both the drugs and their interacting partners, and the PPI of this broad set of targets. From this data set, they determined the interactome comprised 26 distinct modules, the majority of which showed strong associations with specific biological functions and pathways, showing that network modularity correlated with biologically relevant distinctions in function. Overlaying drug-target information, the authors observed that more than three-fourths of the total number of drugs are linked to two pathways: the complement-coagulation cascade and the calcium signalling pathway. Additionally, analysis of drug-drug interactions in this interactome revealed new interacting partners that were previously not listed in commercially and publicly available drug interaction listings.
Using the same starting point of approved and related MI drugs, another study similarly mapped drug-drug interactors, drug targets, and, uniquely, MI disease genes and gene products onto a comprehensive human interactome.126 Creating a network of the drug targets and MI disease genes/proteins and then overlaying the MI-related drugs and their interactors onto this network, the authors identified 12 distinct modules (termed drug, drug target, and disease protein or DTD modules). These DTD modules represented biologically relevant groupings with distinct and relatively cohesive higher-order function (assessed computationally by determining functional similarity of the protein pairs in the network). Additionally, drugs within the modules share similar therapeutic effects and similar side effects.
These characteristics of the DTD modules allow greater predictive power for understanding how the included non-MI drugs may be exerting cardiovascular effects. Drugs in a module are more likely to share mechanistic underpinnings with the other drugs in the module. Powerfully, this approach can begin to provide a rationale for drug repurposing. Non-MI drugs with targets that are heavily connected in these modules may be prime candidates for repurposing to a cardiovascular disease as we have recently shown.5 Another study utilized loci identified through GWAS for CAD and MI to identify druggable targets with low likelihood of systemic side effects; this strategy identified three drugs (adenosine triphosphate, pentolinium, and riociguat) that were targets for repurposing for MI/CAD.127 Yet another study also utilized GWAS-identified CAD loci to identify and rank druggable CAD disease modules to prioritize better drug discovery.128 It is useful to note that traditional cardiovascular therapeutics, including angiotensin converting enzyme inhibitors or P2Y12 receptor blockers, do not seem to be targeting highly central hubs in these modules, but, rather, more peripheral nodes.126 This finding is similar to what has been observed in larger drug-target networks.123 While network incompleteness could partly be an explanation, this phenomenon does highlight the importance of understanding better the connectedness and relationships between the peripheral nodes and central biological pathways so that drug repurposing predictions can be made accurately.
Network pharmacology is particularly suited for identifying more effective and rational approaches to polypharmacy, with a goal of optimizing efficacy and synergy of medications while minimizing toxicities. For example, one study demonstrated that multiple interventions in the nitric oxide-cyclic guanosine 3′,5′ monophosphate (cGMP) pathway were more effective at raising cGMP levels than a single intervention, providing an experimental basis for rational polypharmacy.129 Computational network approaches have been utilized to predict optimal drug combinations for cancer treatment and to evaluate drug combinations that will minimize perturbations in an inflammatory network of arachidonic acid metabolites.130,131 Network approaches have additionally provided insight into mechanisms of cardiovascular toxicities from cyclooxygenase 2 inhibitors.132 Looking forward to further applications of network pharmacology for cardiovascular diseases, one area that could particularly benefit from this approach is treatment of heart failure. Heart failure has adopted a multi-drug approach, largely driven by clinical data demonstrating the benefit of additive drug therapy; however, multi-drug therapy can also be associated with overlapping side effects and enhanced drug toxicities.133 A network pharmacology approach to this field could help to determine which combination of available therapies is most optimal for an individual.
10. Current limitations and future directions
Current challenges in network medicine are multiple. At the level of the protein–protein interactome, the current interactome remains incomplete albeit is expanding rapidly. Additionally, it is estimated that only 10% of genes are associated with human diseases.8 While this is also an advantage computationally as it reduces the network space needed to understand key disease mechanisms, it also suggests that some of the 90% of unassociated genes may become incorporated into disease modules as the interactome is completed. As the interactome has been established based on the curated PPIs, inspection bias is inherent in its construction. Additionally, PPIs curated in experimental settings may be substantially different from those involving endogenous proteins in the in vivo environment. The current interactome has thus far limited incorporation of more than one isoform or splice variant of a gene product134 or post-translational modifications,135 thereby omitting a major source of omic dimensionality.
At the network scope level, there is an increasing effort to incorporate tissue- and biological context-specificity in network constructions.136 Network approaches are also facilitating the studies of interactions between different organ systems.137 Additionally, going beyond the general concept of the exposome,138 there is a growing interest in classifying and defining different domains of an individual’s environment (e.g., personal, social,139 natural) in order to understand better how each domain and their interactions contribute to determining the clinical outcomes of complex cardiovascular diseases.140
At the computational levels, there needs to be more standardized approaches to large -omic data acquisition, quality control, normalization, and analysis pipelines, especially in studies involving proteomics and metabolomics.141 As none of the biological processes exist in isolation, studies assessing the interactions between the networks representing different biological dimensions (e.g., transcriptomics network and epigenetic modification network) are gaining increasing attention.142 Integration of multi-dimensional data also remains an important challenge (ref.143 as reviewed in refs.8,144). Notable integrative platforms include Pathway Representation and Analysis by Direct Reference on Graphical Models (PARADIGM), which integrates the multidimensional cancer patient data with biological pathways using a probabilistic graphical model and thereby infers pathways altered in glioblastoma patients.145 National Institute of Health-sponsored multicentre study Pulmonary Vascular Disease Phenomics (PVDOMICS), led by Leopold et al., is a superb example where complex and heterogeneous clinical entities, such as pulmonary hypertension, are being reappraised through the network integration of multi-omic patient data (transcriptomics, proteomics, metabolomics) with invasive hemodynamic and physiological data (cardiac catheterization, iCPET, 6-min walk test), and multi-modality clinical data (laboratory tests, body fat composition, echocardiogram, and cardiac magnetic resonance imaging).116
In summary, network medicine represents a powerful, integrative framework with which complex cardiovascular pathobiology can be dissected with greater precision at multiple biological levels. Ultimately, this discipline will facilitate complex phenotype–genotype correlation, improve drug discovery, and promote precision medicine in this multi-omics era.
Acknowledgements
We thank Stephanie Tribuna and Katherine Seropian for their expert technical assistance.
Conflict of interest: J.L. is the scientific cofounder of Scipher Medicine, Inc., which uses molecular interaction network analysis to identify disease biomarkers and drug targets. B.A.M. is a co-inventor on US patent 9,605,047, US pending patent PCT/US2019/059890, and provisional patent applications 62475955 and 029672; and is a member of the steering committee for a research grant supported by Actelion Pharmaceuticals. L.Y.L. and A.K.P. have none declared.
Funding
This work was supported, in part, by the National Institutes of Health [HG00760, HL108630, HL119145 to J.L., U01HL125215-01, 1R01HL139613-01, R01HL153502, R21HL134320, U54HL119145, to B.A.M., and T32 HL760434 to L.Y.L.]; American Heart Association [D700382, CV-19] to J.L.; the Cardiovascular Medical Research Education Foundation to B.A.M. and Boston Biomedical Innovation Center to B.A.M.
References
- 1.Leopold JA, Maron BA, Loscalzo J.. The application of big data to cardiovascular disease: paths to precision medicine. J Clin Invest 2020;130:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee LY, Loscalzo J.. Network medicine in pathobiology. Am J Pathol 2019;189:1311–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez D, Wheelock AM, Goto S, Haeggstrom JZ, Paulsson-Berne G, Hansson GK, Hedin U, Gabrielsen A, Wheelock CE.. The use of network analyses for elucidating mechanisms in cardiovascular disease. Mol Biosyst 2010;6:289–304. [DOI] [PubMed] [Google Scholar]
- 4.Chan SY, White K, Loscalzo J.. Deciphering the molecular basis of human cardiovascular disease through network biology. Curr Opin Cardiol 2012;27:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabási A-L, Loscalzo J.. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun 2018;9:2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samokhin AOStephens T, Wertheim BM, Wang RS, Vargas SO, Yung LM, Cao M, Brown M, Arons E, Dieffenbach PB, Fewell JG, Matar M, Bowman FP, Haley KJ, Alba GA, Marino SM, Kumar R, Rosas IO, Waxman AB, Oldham WM, Khanna D, Graham BB, Seo S, Gladyshev VN, Yu PB, Fredenburgh LE, Loscalzo J, Leopold JA, Maron BA.. NEDD9 targets COL3A1 to promote endothelial fibrosis and pulmonary arterial hypertension. Sci Transl Med 2018;10:eaap7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabasi ALGulbahce N, Loscalzo J.. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loscalzo J, Barabási A-L, Silverman EK.. Network Medicine: Complex Systems in Human Disease and Therapeutics. Cambridge, MA: Harvard University Press; 2017. [Google Scholar]
- 9.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL.. Hierarchical organization of modularity in metabolic networks. Science 2002;297:1551–1555. [DOI] [PubMed] [Google Scholar]
- 10.Azuaje FJ, Rodius S, Zhang L, Devaux Y, Wagner DR.. Information encoded in a network of inflammation proteins predicts clinical outcome after myocardial infarction. BMC Med Genomics 2011;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert R, Jeong H, Barabasi AL.. Error and attack tolerance of complex networks. Nature 2000;406:378–382. [DOI] [PubMed] [Google Scholar]
- 12.Barabasi AL, Albert R.. Emergence of scaling in random networks. Science 1999;286:509–512. [DOI] [PubMed] [Google Scholar]
- 13.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL.. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015;347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Las Rivas J, Fontanillo C.. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol 2010;6:e1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Li Y.. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics 2006;22:2800–2805. [DOI] [PubMed] [Google Scholar]
- 16.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M.. Literature-curated protein interaction datasets. Nat Methods 2009;6:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE.. A human protein-protein interaction network: a resource for annotating the proteome. Cell 2005;122:957–968. [DOI] [PubMed] [Google Scholar]
- 18.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M.. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005;437:1173–1178. [DOI] [PubMed] [Google Scholar]
- 19.Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis A-R, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere J-C, Vega K, Walsh J, Cusick ME, Xia Y, Barabási A-L, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M.. A proteome-scale map of the human interactome network. Cell 2014;159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gstaiger M, Aebersold R.. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet 2009;10:617–627. [DOI] [PubMed] [Google Scholar]
- 21.Li P, Wang L, Di LJ.. Applications of protein fragment complementation assays for analyzing biomolecular interactions and biochemical networks in living cells. J Proteome Res 2019;18:2987–2998. [DOI] [PubMed] [Google Scholar]
- 22.Vyncke L, Masschaele D, Tavernier J, Peelman F.. Straightforward protein-protein interaction interface mapping via random mutagenesis and mammalian protein protein interaction trap (MAPPIT). IJMS 2019;20:2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrios-Rodiles M, Ellis JD, Blencowe BJ, Wrana JL.. LUMIER: a discovery tool for mammalian protein interaction networks. Methods Mol Biol 2017;1550:137–148. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, Batalao A, Zhou H, Zhu L.. Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques 1997;22:350–352. [DOI] [PubMed] [Google Scholar]
- 25.Shioda T, Andriole S, Yahata T, Isselbacher KJ.. A green fluorescent protein-reporter mammalian two-hybrid system with extrachromosomal maintenance of a prey expression plasmid: application to interaction screening. Proc Natl Acad Sci USA 2000;97:5220–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader GD, Hogue CW.. Analyzing yeast protein-protein interaction data obtained from different sources. Nat Biotechnol 2002;20:991–917. [DOI] [PubMed] [Google Scholar]
- 27.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G.. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006;440:631–636. [DOI] [PubMed] [Google Scholar]
- 28.Lopes TJ, Schaefer M, Shoemaker J, Matsuoka Y, Fontaine JF, Neumann G, Andrade-Navarro MA, Kawaoka Y, Kitano H.. Tissue-specific subnetworks and characteristics of publicly available human protein interaction databases. Bioinformatics 2011;27:2414–2421. [DOI] [PubMed] [Google Scholar]
- 29.Schaefer MH, Lopes TJ, Mah N, Shoemaker JE, Matsuoka Y, Fontaine JF, Louis-Jeune C, Eisfeld AJ, Neumann G, Perez-Iratxeta C, Kawaoka Y, Kitano H, Andrade-Navarro MA.. Adding protein context to the human protein-protein interaction network to reveal meaningful interactions. PLoS Comput Biol 2013;9:e1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rachlin J, Cohen DD, Cantor C, Kasif S.. Biological context networks: a mosaic view of the interactome. Mol Syst Biol 2006;2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alanis-Lobato G, Andrade-Navarro MA, Schaefer MH.. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res 2017;45:D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi S, Davila-Velderrain J, Kellis M.. Reconstruction of cell-type-specific interactomes at single-cell resolution. Cell Syst 2019;9:559–568.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iacono G, Massoni-Badosa R, Heyn H.. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol 2019;20:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL.. The human disease network. Proc Natl Acad Sci USA 2007;104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Menche J, Barabasi AL, Sharma A.. Human symptoms-disease network. Nat Commun 2014;5:4212. [DOI] [PubMed] [Google Scholar]
- 36.Hidalgo CA, Blumm N, Barabasi AL, Christakis NA.. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol 2009;5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DS, Park J, Kay KA, Christakis NA, Oltvai ZN, Barabasi AL.. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci USA 2008;105:9880–9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q.. An analysis of human microRNA and disease associations. PLoS One 2008;3:e3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franzen O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, Foroughi-Asl H, Giambartolomei C, Fullard JF, Sukhavasi K, Koks S, Gan LM, Giannarelli C, Kovacic JC, Betsholtz C, Losic B, Michoel T, Hao K, Roussos P, Skogsberg J, Ruusalepp A, Schadt EE, Bjorkegren JL.. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 2016;353:827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng L, Talukdar HA, Koplev S, Giannarelli C, Ivert T, Gan LM, Ruusalepp A, Schadt EE, Kovacic JC, Lusis AJ, Michoel T, Schunkert H, Bjorkegren JLM.. Contribution of gene regulatory networks to heritability of coronary artery disease. J Am Coll Cardiol 2019;73:2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olin JW, Di Narzo AF, d’Escamard V, Kadian-Dodov D, Cheng H, Georges A, King A, Thomas A, Barwari T, Michelis KC, Bouchareb R, Bander E, Anyanwu A, Stelzer P, Filsoufi F, Florman S, Civelek M, Debette S, Jeunemaitre X, Björkegren JLM, Mayr M, Bouatia-Naji N, Hao K, Kovacic JC.. A plasma proteogenomic signature for fibromuscular dysplasia. Cardiovasc Res 2020;116:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, Balligand JL, Devaux Y, Cardiolinc N.. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids 2017;8:494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner AW, Wong D, Khan MD, Dreisbach CN, Palmore M, Miller CL.. Multi-omics approaches to study long non-coding RNA function in atherosclerosis. Front Cardiovasc Med 2019;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldman I, Rzhetsky A, Vitkup D.. Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci USA 2008;105:4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A.. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet 2006;38:285–293. [DOI] [PubMed] [Google Scholar]
- 46.Deckers K, Schievink SHJ, Rodriquez MMF, van Oostenbrugge RJ, van Boxtel MPJ, Verhey FRJ, Köhler S.. Coronary heart disease and risk for cognitive impairment or dementia: systematic review and meta-analysis. PLoS One 2017;12:e0184244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray M, Ruan J, Zhang W.. Variations in the transcriptome of Alzheimer's disease reveal molecular networks involved in cardiovascular diseases. Genome Biol 2008;9:R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J, Lee DS, Christakis NA, Barabasi AL.. The impact of cellular networks on disease comorbidity. Mol Syst Biol 2009;5:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB.. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet 2018;19:110–124. [DOI] [PubMed] [Google Scholar]
- 50.Myocardial Infarction Genetics Consortium InvestigatorsStitziel NO, Won H-H, Morrison AC, Peloso GM, Do R, Lange LA, Fontanillas P, Gupta N, Duga S, Goel A, Farrall M, Saleheen D, Ferrario P, König I, Asselta R, Merlini PA, Marziliano N, Notarangelo MF, Schick U, Auer P, Assimes TL, Reilly M, Wilensky R, Rader DJ, Hovingh GK, Meitinger T, Kessler T, Kastrati A, Laugwitz K-L, Siscovick D, Rotter JI, Hazen SL, Tracy R, Cresci S, Spertus J, Jackson R, Schwartz SM, Natarajan P, Crosby J, Muzny D, Ballantyne C, Rich SS, O'Donnell CJ, Abecasis G, Sunaev S, Nickerson DA, Buring JE, Ridker PM, Chasman DI, Austin E, Kullo IJ, Weeke PE, Shaffer CM, Bastarache LA, Denny JC, Roden DM, Palmer C, Deloukas P, Lin D-Y, Tang Z-Z, Erdmann J, Schunkert H, Danesh J, Marrugat J, Elosua R, Ardissino D, McPherson R, Watkins H, Reiner AP, Wilson JG, Altshuler D, Gibbs RA, Lander ES, Boerwinkle E, Gabriel S, Kathiresan S.. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med 2014;371:2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arad M, Penas-Lado M, Monserrat L, Maron BJ, Sherrid M, Ho CY, Barr S, Karim A, Olson TM, Kamisago M, Seidman JG, Seidman CE.. Gene mutations in apical hypertrophic cardiomyopathy. Circulation 2005;112:2805–2811. [DOI] [PubMed] [Google Scholar]
- 52.Maron BJ, Maron MS, Semsarian C.. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 2012;60:705–715. [DOI] [PubMed] [Google Scholar]
- 53.Maron BJ, Maron MS, Maron BA, Loscalzo J.. Moving beyond the sarcomere to explain heterogeneity in hypertrophic cardiomyopathy: JACC review topic of the week. J Am Coll Cardiol 2019;73:1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS.. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, Dougherty K, Harrison SM, McGlaughon J, Milko LV, Morales A, Seifert BA, Strande N, Thomson K, Peter van Tintelen J, Wallace K, Walsh R, Wells Q, Whiffin N, Witkowski L, Semsarian C, Ware JS, Hershberger RE, Funke B.. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med 2019;12:e002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverman EK, Loscalzo J.. Network medicine approaches to the genetics of complex diseases. Discov Med 2012;14:143–152. [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA, McLean DL, Park H, Comhair SA, Greif DM, Erzurum SC, Chun HJ.. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med 2013;19:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn TC, Knobel J, Burkert-Rettenmaier S, Li X, Meyer IS, Jungmann A, Sicklinger F, Backs J, Lasitschka F, Muller OJ, Katus HA, Krijgsveld J, Leuschner F.. Secretome analysis of cardiomyocytes identifies PCSK6 (Proprotein Convertase Subtilisin/Kexin Type 6) as a novel player in cardiac remodeling after myocardial infarction. Circulation 2020;141:1628–1644. [DOI] [PubMed] [Google Scholar]
- 59.Maron BA, Zhang YY, Handy DE, Beuve A, Tang SS, Loscalzo J, Leopold JA.. Aldosterone increases oxidant stress to impair guanylyl cyclase activity by cysteinyl thiol oxidation in vascular smooth muscle cells. J Biol Chem 2009;284:7665–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, Bieda MC, Snyder FF, Gravel RA, Cross JC, Watson ED.. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013;155:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Napoli C, Witztum JL, Calara F, de Nigris F, Palinski W.. Maternal hypercholesterolemia enhances atherogenesis in normocholesterolemic rabbits, which is inhibited by antioxidant or lipid-lowering intervention during pregnancy: an experimental model of atherogenic mechanisms in human fetuses. Circ Res 2000;87:946–952. [DOI] [PubMed] [Google Scholar]
- 62.Maron BA, Maron JL, Abman SH.. The case for bringing birthweight to adult cardiovascular medicine. Am J Cardiol 2020;127:191–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Go YM, Chandler JD, Jones DP.. The cysteine proteome. Free Radic Biol Med 2015;84:227–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maron BA, Zhang YY, White K, Chan SY, Handy DE, Mahoney CE, Loscalzo J, Leopold JA.. Aldosterone inactivates the endothelin-B receptor via a cysteinyl thiol redox switch to decrease pulmonary endothelial nitric oxide levels and modulate pulmonary arterial hypertension. Circulation 2012;126:963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Doring A, Elliott P, Epstein SE, Ingi Eyjolfsson G, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YI, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PEH, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BHR, Ordovas JM, Boerwinkle E, Palmer CNA, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR; Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Votava JA, Zajac GJM, Nguyen JN, Leyva Jaimes FB, Ly SM, Brinkman JA, De Giorgi M, Kaul S, Green CL, St. Clair SL, Belisle SL, Rios JM, Nelson DW, Sorci-Thomas MG, Lagor WR, Lamming DW, Eric Yen C-L, Parks BW.. Integrating mouse and human genetic data to move beyond GWAS and identify causal genes in cholesterol metabolism. Cell Metab 2020;31:741–754.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward RE, Cho K, Nguyen XT, Vassy JL, Ho YL, Quaden RM, Gagnon DR, Wilson PWF, Gaziano JM, Djousse L, Program VAMV.. Omega-3 supplement use, fish intake, and risk of non-fatal coronary artery disease and ischemic stroke in the Million Veteran Program. Clin Nutr 2020;39:574–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma A, Halu A, Decano JL, Padi M, Liu YY, Prasad RB, Fadista J, Santolini M, Menche J, Weiss ST, Vidal M, Silverman EK, Aikawa M, Barabasi AL, Groop L, Loscalzo J.. Controllability in an islet specific regulatory network identifies the transcriptional factor NFATC4, which regulates Type 2 Diabetes associated genes. NPJ Syst Biol Appl 2018;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rhodes CJ, Otero-Núñez P, Wharton J, Swietlik EM, Kariotis S, Harbaum L, Dunning MJ, Elinoff JM, Errington N, Thompson AAR, Iremonger J, Coghlan JG, Corris PA, Howard LS, Kiely DG, Church C, Pepke-Zaba J, Toshner M, Wort SJ, Desai AA, Humbert M, Nichols WC, Southgate L, Trégouët D-A, Trembath RC, Prokopenko I, Gräf S, Morrell NW, Wang D, Lawrie A, Wilkins MR; Consortium NB-RDP and the UKNPAHCSC. Whole blood RNA profiles associated with pulmonary arterial hypertension and clinical outcome. Am J Respir Crit Care Med 2020;202:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maron BA.Clarifying the pulmonary arterial hypertension molecular landscape using functional genetics. Am J Respir Crit Care Med 2020;202:488–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghiassian SD, Menche J, Chasman DI, Giulianini F, Wang R, Ricchiuto P, Aikawa M, Iwata H, Muller C, Zeller T, Sharma A, Wild P, Lackner K, Singh S, Ridker PM, Blankenberg S, Barabasi AL, Loscalzo J.. Endophenotype network models: common core of complex diseases. Sci Rep 2016;6:27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang RS, Loscalzo J.. Network-based disease module discovery by a novel seed connector algorithm with pathobiological implications. J MolBiol 2018;430:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghiassian SD, Menche J, Barabasi AL.. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput Biol 2015;11:e1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tuncbag N, Braunstein A, Pagnani A, Huang SS, Chayes J, Borgs C, Zecchina R, Fraenkel E.. Simultaneous reconstruction of multiple signaling pathways via the prize-collecting steiner forest problem. J Comput Biol 2013;20:124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silberberg Y, Kupiec M, Sharan R.. GLADIATOR: a global approach for elucidating disease modules. Genome Med 2017;9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cowen L, Ideker T, Raphael BJ, Sharan R.. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet 2017;18:551–562. [DOI] [PubMed] [Google Scholar]
- 77.Lee H, Tu Z, Deng M, Sun F, Chen T.. Diffusion kernel-based logistic regression models for protein function prediction. OMICS 2006;10:40–55. [DOI] [PubMed] [Google Scholar]
- 78.Fan Z, Gao S, Chen Y, Xu B, Yu C, Yue M, Tan X.. Integrative analysis of competing endogenous RNA networks reveals the functional lncRNAs in heart failure. J Cell Mol Med 2018;22:4818–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maron BA, Galie N.. Diagnosis, treatment, and clinical management of pulmonary arterial hypertension in the contemporary era: a review. JAMA Cardiol 2016;1:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thenappan T, Ormiston ML, Ryan JJ, Archer SL.. Pulmonary arterial hypertension: pathogenesis and clinical management. BMJ 2018;360:j5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morrell NW, Aldred MA, Chung WK, Elliott CG, Nichols WC, Soubrier F, Trembath RC, Loyd JE.. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J 2019;53:1801899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, Cottrill KA, Shaik RS, Waxman AB, Zhang YY, Maron BA, Hartner JC, Fujiwara Y, Orkin SH, Haley KJ, Barabasi AL, Loscalzo J, Chan SY.. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation 2012;125:1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Syed M, Das P, Pawar A, Aghai ZH, Kaskinen A, Zhuang ZW, Ambalavanan N, Pryhuber G, Andersson S, Bhandari V.. Hyperoxia causes miR-34a-mediated injury via angiopoietin-1 in neonatal lungs. Nat Commun 2017;8:1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Drake JI, Bogaard HJ, Mizuno S, Clifton B, Xie B, Gao Y, Dumur CI, Fawcett P, Voelkel NF, Natarajan R.. Molecular signature of a right heart failure program in chronic severe pulmonary hypertension. Am J Respir Cell Mol Biol 2011;45:1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Napoli C, Benincasa G, Loscalzo J.. Epigenetic inheritance underlying pulmonary arterial hypertension. Arterioscler Thromb Vasc Biol 2019;39:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, Wang D, Li M, Plecitá-Hlavatá L, D’Alessandro A, Tauber J, Riddle S, Kumar S, Flockton A, McKeon BA, Frid MG, Reisz JA, Caruso P, El Kasmi KC, Ježek P, Morrell NW, Hu C-J, Stenmark KR.. Metabolic and proliferative state of vascular adventitial fibroblasts in pulmonary hypertension is regulated through a microRNA-124/PTBP1 (Polypyrimidine Tract Binding Protein 1)/pyruvate kinase muscle axis. Circulation 2017;136:2468–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertero T, Cottrill K, Krauszman A, Lu Y, Annis S, Hale A, Bhat B, Waxman AB, Chau BN, Kuebler WM, Chan SY.. The microRNA-130/301 family controls vasoconstriction in pulmonary hypertension. J Biol Chem 2015;290:2069–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin M, Lefaix J, Delanian S.. TGF-beta1 and radiation fibrosis: a master switch and a specific therapeutic target? Int J Radiat Oncol Biol Phys 2000;47:277–290. [DOI] [PubMed] [Google Scholar]
- 89.Liu Q, Zhu LJ, Waaga-Gasser AM, Ding Y, Cao M, Jadhav SJ, Kirollos S, Shekar PS, Padera RF, Chang YC, Xu X, Zeisberg EM, Charytan DM, Hsiao LL.. The axis of local cardiac endogenous Klotho-TGF-beta1-Wnt signaling mediates cardiac fibrosis in human. J Mol Cell Cardiol 2019;136:113–124. [DOI] [PubMed] [Google Scholar]
- 90.Maron BA, Opotowsky AR, Landzberg MJ, Loscalzo J, Waxman AB, Leopold JA.. Plasma aldosterone levels are elevated in patients with pulmonary arterial hypertension in the absence of left ventricular heart failure: a pilot study. Eur J Heart Fail 2013;15:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calvier L, Legchenko E, Grimm L, Sallmon H, Hatch A, Plouffe BD, Schroeder C, Bauersachs J, Murthy SK, Hansmann G.. Galectin-3 and aldosterone as potential tandem biomarkers in pulmonary arterial hypertension. Heart 2016;102:390–396. [DOI] [PubMed] [Google Scholar]
- 92.Mirzakhani H, Litonjua AA, McElrath TF, O’Connor G, Lee-Parritz A, Iverson R, Macones G, Strunk RC, Bacharier LB, Zeiger R, Hollis BW, Handy DE, Sharma A, Laranjo N, Carey V, Qiu W, Santolini M, Liu S, Chhabra D, Enquobahrie DA, Williams MA, Loscalzo J, Weiss ST.. Early pregnancy vitamin D status and risk of preeclampsia. J Clin Invest 2016;126:4702–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schlotter F, Halu A, Goto S, Blaser MC, Body SC, Lee LH, Higashi H, DeLaughter DM, Hutcheson JD, Vyas P, Pham T, Rogers MA, Sharma A, Seidman CE, Loscalzo J, Seidman JG, Aikawa M, Singh SA, Aikawa E.. Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation 2018;138:377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Upton PD, Davies RJ, Tajsic T, Morrell NW.. Transforming growth factor-beta(1) represses bone morphogenetic protein-mediated Smad signaling in pulmonary artery smooth muscle cells via Smad3. Am J Respir Cell Mol Biol 2013;49:1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samokhin AO, Hsu S, Yu PB, Waxman AB, Alba GA, Wertheim BM, Hopkins CD, Bowman F, Channick RN, Nikolic I, Faria-Urbina M, Hassoun PM, Leopold JA, Tedford RJ, Ventetuolo CE, Leary PJ, Maron BA.. Circulating NEDD9 is increased in pulmonary arterial hypertension: a multicenter, retrospective analysis. J Heart Lung Transplant 2020;39:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL.. Epigenetics and the placenta. Hum Reprod Update 2011;17:397–417. [DOI] [PubMed] [Google Scholar]
- 97.Krause B, Sobrevia L, Casanello P.. Epigenetics: new concepts of old phenomena in vascular physiology. CVP 2009;7:513–520. [DOI] [PubMed] [Google Scholar]
- 98.Than NG, Romero R, Tarca AL, Kekesi KA, Xu Y, Xu Z, Juhasz K, Bhatti G, Leavitt RJ, Gelencser Z, Palhalmi J, Chung TH, Gyorffy BA, Orosz L, Demeter A, Szecsi A, Hunyadi-Gulyas E, Darula Z, Simor A, Eder K, Szabo S, Topping V, El-Azzamy H, LaJeunesse C, Balogh A, Szalai G, Land S, Torok O, Dong Z, Kovalszky I, Falus A, Meiri H, Draghici S, Hassan SS, Chaiworapongsa T, Krispin M, Knofler M, Erez O, Burton GJ, Kim CJ, Juhasz G, Papp Z.. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front Immunol 2018;9:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tejera E, Bernardes J, Rebelo I.. Preeclampsia: a bioinformatics approach through protein-protein interaction networks analysis. BMC Syst Biol 2012;6:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stewart BF, Siscovick D, Lind BK, Gardin JM, Gottdiener JS, Smith VE, Kitzman DW, Otto CM.. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol 1997;29:630–634. [DOI] [PubMed] [Google Scholar]
- 101.Merke J, Milde P, Lewicka S, Hugel U, Klaus G, Mangelsdorf DJ, Haussler MR, Rauterberg EW, Ritz E.. Identification and regulation of 1,25-dihydroxyvitamin D3 receptor activity and biosynthesis of 1,25-dihydroxyvitamin D3. Studies in cultured bovine aortic endothelial cells and human dermal capillaries. J Clin Invest 1989;83:1903–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kendrick J, Targher G, Smits G, Chonchol M.. 25-Hydroxyvitamin D deficiency is independently associated with cardiovascular disease in the Third National Health and Nutrition Examination Survey. Atherosclerosis 2009;205:255–260. [DOI] [PubMed] [Google Scholar]
- 103.Kim DH, Sabour S, Sagar UN, Adams S, Whellan DJ.. Prevalence of hypovitaminosis D in cardiovascular diseases (from the National Health and Nutrition Examination Survey 2001 to 2004). Am J Cardiol 2008;102:1540–1544. [DOI] [PubMed] [Google Scholar]
- 104.Giovannucci E, Liu Y, Hollis BW, Rimm EB.. 25-hydroxyvitamin D and risk of myocardial infarction in men: a prospective study. Arch Intern Med 2008;168:1174–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heikkinen S, Vaisanen S, Pehkonen P, Seuter S, Benes V, Carlberg C.. Nuclear hormone 1alpha, 25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res 2011;39:9181–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vukic M, Neme A, Seuter S, Saksa N, de Mello VD, Nurmi T, Uusitupa M, Tuomainen TP, Virtanen JK, Carlberg C.. Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS One 2015;10:e0124339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carlberg C, Seuter S, de Mello VD, Schwab U, Voutilainen S, Pulkki K, Nurmi T, Virtanen J, Tuomainen TP, Uusitupa M.. Primary vitamin D target genes allow a categorization of possible benefits of vitamin D(3) supplementation. PLoS One 2013;8:e71042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoshida T, Fukuda T, Hatano M, Koseki H, Okabe S, Ishibashi K, Kojima S, Arima M, Komuro I, Ishii G, Miki T, Hirosawa S, Miyasaka N, Taniguchi M, Ochiai T, Isono K, Tokuhisa T.. The role of Bcl6 in mature cardiac myocytes. Cardiovasc Res 1999;42:670–679. [DOI] [PubMed] [Google Scholar]
- 109.Zhang X, Zhang MC, Wang CT.. Loss of LRRC25 accelerates pathological cardiac hypertrophy through promoting fibrosis and inflammation regulated by TGF-beta1. Biochem Biophys Res Commun 2018;506:137–144. [DOI] [PubMed] [Google Scholar]
- 110.D’Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB.. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743–753. [DOI] [PubMed] [Google Scholar]
- 111.Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D, Braunwald E.. The TIMI risk score for unstable angina/non-ST elevation MI: a method for prognostication and therapeutic decision making. JAMA 2000;284:835–842. [DOI] [PubMed] [Google Scholar]
- 112.Benza RL, Gomberg-Maitland M, Miller DP, Frost A, Frantz RP, Foreman AJ, Badesch DB, McGoon MD.. The REVEAL Registry risk score calculator in patients newly diagnosed with pulmonary arterial hypertension. Chest 2012;141:354–362. [DOI] [PubMed] [Google Scholar]
- 113.Maron BA, Cockrill BA, Waxman AB, Systrom DM.. The invasive cardiopulmonary exercise test. Circulation 2013;127:1157–1164. [DOI] [PubMed] [Google Scholar]
- 114.Oldham WM, Oliveira RKF, Wang RS, Opotowsky AR, Rubins DM, Hainer J, Wertheim BM, Alba GA, Choudhary G, Tornyos A, MacRae CA, Loscalzo J, Leopold JA, Waxman AB, Olschewski H, Kovacs G, Systrom DM, Maron BA.. Network analysis to risk stratify patients with exercise intolerance. Circ Res 2018;122:864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arena R, Ozemek C, Laddu-Patel D, Myers J.. Refining the risk prediction of cardiorespiratory fitness with network analysis: a welcome and needed line of inquiry. Circ Res 2018;122:804–806. [DOI] [PubMed] [Google Scholar]
- 116.Hemnes AR, Beck GJ, Newman JH, Abidov A, Aldred MA, Barnard J, Berman Rosenzweig E, Borlaug BA, Chung WK, Comhair SAA, Erzurum SC, Frantz RP, Gray MP, Grunig G, Hassoun PM, Hill NS, Horn EM, Hu B, Lempel JK, Maron BA, Mathai SC, Olman MA, Rischard FP, Systrom DM, Tang WHW, Waxman AB, Xiao L, Yuan JX, Leopold JA, Group PS.. PVDOMICS: a multi-center study to improve understanding of pulmonary vascular disease through phenomics. Circ Res 2017;121:1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grosdidier S, Ferrer A, Faner R, Pinero J, Roca J, Cosio B, Agusti A, Gea J, Sanz F, Furlong LI.. Network medicine analysis of COPD multimorbidities. Respir Res 2014;15:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baumgartner C, Spath-Blass V, Niederkofler V, Bergmoser K, Langthaler S, Lassnig A, Rienmuller T, Baumgartner D, Asnani A, Gerszten RE.. A novel network-based approach for discovering dynamic metabolic biomarkers in cardiovascular disease. PLoS One 2018;13:e0208953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loscalzo J.Precision medicine: a new paradigm for diagnosis and management of hypertension. Circ Res 2019;124:987–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Solomon SD, McMurray JJV, Anand IS, Ge J, Lam CSP, Maggioni AP, Martinez F, Packer M, Pfeffer MA, Pieske B, Redfield MM, Rouleau JL, van Veldhuisen DJ, Zannad F, Zile MR, Desai AS, Claggett B, Jhund PS, Boytsov SA, Comin-Colet J, Cleland J, Düngen H-D, Goncalvesova E, Katova T, Kerr Saraiva JF, Lelonek M, Merkely B, Senni M, Shah SJ, Zhou J, Rizkala AR, Gong J, Shi VC, Lefkowitz MP.. Angiotensin–neprilysin inhibition in heart failure with preserved ejection fraction. N Engl J Med 2019;381:1609–1620. [DOI] [PubMed] [Google Scholar]
- 121.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL.. Predicting new molecular targets for known drugs. Nature 2009;462:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hopkins AL.Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008;4:682–690. [DOI] [PubMed] [Google Scholar]
- 123.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M.. Drug-target network. Nat Biotechnol 2007;25:1119–1126. [DOI] [PubMed] [Google Scholar]
- 124.Guney E, Menche J, Vidal M, Barabasi AL.. Network-based in silico drug efficacy screening. Nat Commun 2016;7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Azuaje FJ, Zhang L, Devaux Y, Wagner DR.. Drug-target network in myocardial infarction reveals multiple side effects of unrelated drugs. Sci Rep 2011;1:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang RS, Loscalzo J.. Illuminating drug action by network integration of disease genes: a case study of myocardial infarction. Mol Biosyst 2016;12:1653–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tragante V, Hemerich D, Alshabeeb M, Brænne I, Lempiäinen H, Patel RS, den Ruijter HM, Barnes MR, Moore JH, Schunkert H, Erdmann J, Asselbergs FW.. Druggability of coronary artery disease risk loci. Circ Genom Precis Med 2018;11:e001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lempiäinen H, Brænne I, Michoel T, Tragante V, Vilne B, Webb TR, Kyriakou T, Eichner J, Zeng L, Willenborg C, Franzen O, Ruusalepp A, Goel A, van der Laan SW, Biegert C, Hamby S, Talukdar HA, Foroughi Asl H, Pasterkamp G, Watkins H, Samani NJ, Wittenberger T, Erdmann J, Schunkert H, Asselbergs FW, Björkegren JLM.. Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci Rep 2018;8:3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garmaroudi FS, Handy DE, Liu YY, Loscalzo J.. Systems pharmacology and rational polypharmacy: nitric oxide-cyclic GMP signaling pathway as an illustrative example and derivation of the general case. PLoS Comput Biol 2016;12:e1004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang K, Bai H, Ouyang Q, Lai L, Tang C.. Finding multiple target optimal intervention in disease-related molecular network. Mol Syst Biol 2008;4:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vazquez A.Optimal drug combinations and minimal hitting sets. BMC Syst Biol 2009;3:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brænne I, Willenborg C, Tragante V, Kessler T, Zeng L, Reiz B, Kleinecke M, von Ameln S, Willer CJ, Laakso M, Wild PS, Zeller T, Wallentin L, Franks PW, Salomaa V, Dehghan A, Meitinger T, Samani NJ, Asselbergs FW, Erdmann J, Schunkert H.. A genomic exploration identifies mechanisms that may explain adverse cardiovascular effects of COX-2 inhibitors. Sci Rep 2017;7:10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tang WH, Francis GS.. Polypharmacy of heart failure. Creating a rational pharmacotherapeutic protocol. Cardiol Clin 2001;19:583–596. [DOI] [PubMed] [Google Scholar]
- 134.Talavera D, Robertson DL, Lovell SC.. Alternative splicing and protein interaction data sets. Nat Biotechnol 2013;31:292–293. [DOI] [PubMed] [Google Scholar]
- 135.Prabakaran S, Lippens G, Steen H, Gunawardena J.. Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wires Syst Biol Med 2012;4:565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jerby L, Shlomi T, Ruppin E.. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol 2010;6:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Westfall S, Lomis N, Kahouli I, Dia SY, Singh SP, Prakash S.. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell Mol Life Sci 2017;74:3769–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wild CP.The exposome: from concept to utility. Int J Epidemiol 2012;41:24–32. [DOI] [PubMed] [Google Scholar]
- 139.Luke DA, Stamatakis KA.. Systems science methods in public health: dynamics, networks, and agents. Annu Rev Public Health 2012;33:357–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Riggs DW, Yeager RA, Bhatnagar A.. Defining the human envirome: an omics approach for assessing the environmental risk of cardiovascular disease. Circ Res 2018;122:1259–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.McShane LM, Cavenagh MM, Lively TG, Eberhard DA, Bigbee WL, Williams PM, Mesirov JP, Polley MY, Kim KY, Tricoli JV, Taylor JM, Shuman DJ, Simon RM, Doroshow JH, Conley BA.. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration. BMC Med 2013;11:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leyva I, Sevilla-Escoboza R, Sendiña-Nadal I, Gutiérrez R, Buldú JM, Boccaletti S.. Inter-layer synchronization in non-identical multi-layer networks. Sci Rep 2017;7:45475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arakawa K, Tomita M.. Merging multiple omics datasets in silico: statistical analyses and data interpretation. Methods Mol Biol 2013;985:459–470. [DOI] [PubMed] [Google Scholar]
- 144.Sun YV, Hu YJ.. Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv Genet 2016;93:147–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM.. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 2010;26:i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]