In this study, Gao et al. demonstrate that the global loss of TDP-43 liquid–liquid phase separation perturbs brain function by modulating interactions of TDP-43 with PABPC4, RPS6, RPL7, and other translational factors to orchestrate protein synthesis.
Abstract
The in vivo physiological function of liquid–liquid phase separation (LLPS) that governs non–membrane-bound structures remains elusive. Among LLPS-prone proteins, TAR DNA-binding protein of 43 kD (TDP-43) is under intense investigation because of its close association with neurological disorders. Here, we generated mice expressing endogenous LLPS-deficient murine TDP-43. LLPS-deficient TDP-43 mice demonstrate impaired neuronal function and behavioral abnormalities specifically related to brain function. Brain neurons of these mice, however, did not show TDP-43 proteinopathy or neurodegeneration. Instead, the global rate of protein synthesis was found to be greatly enhanced by TDP-43 LLPS loss. Mechanistically, TDP-43 LLPS ablation increased its association with PABPC4, RPS6, RPL7, and other translational factors. The physical interactions between TDP-43 and translational factors relies on a motif, the deletion of which abolished the impact of LLPS-deficient TDP-43 on translation. Our findings show a specific physiological role for TDP-43 LLPS in the regulation of brain function and uncover an intriguing novel molecular mechanism of translational control by LLPS.
Introduction
Unlike membrane-bound organelles with physical separation, many subcellular compartments, such as the nucleolus, RNP granules, and stress granules, do not have membranes and exist as liquid-like droplets that carry out specialized biological processes within the nucleus and cytosol (Hyman et al., 2014). It is now well recognized that these non–membrane-bound compartments are formed by the condensation of proteins, nucleic acids, and other biomolecules via the process of liquid–liquid phase separation (LLPS; Ditlev et al., 2018). With the growing understanding of the biophysical properties and thermodynamic driving forces of LLPS and identification of new membraneless structures and proteins linked to LLPS, there has been a great desire to know whether the LLPS process observed and largely investigated in vitro is relevant to any specific biological process and, importantly, whether it has any physiological role in vivo.
Trans activation response (TAR) DNA-binding protein of 43 kD (TDP-43) is a highly and ubiquitously expressed heterogeneous nuclear RNP (hnRNP) composed of an N-terminal domain (NTD) containing NLS, two RNA recognition motifs (RRM1 and RRM2), and a C-terminal glycine-rich low-complexity domain (LCD). Despite its predominant nuclear localization, TDP-43 can also be present in the cytoplasm and has been implicated in the regulation of both nuclear and cytoplasmic functions, among which RNA alternative splicing is the most intensively studied function of TDP-43 (Humphrey et al., 2017; Jeong et al., 2017; Ling et al., 2015). Recent studies in vitro and in mammalian cells have repeatedly demonstrated that TDP-43 has a high propensity to undergo LLPS via its LCD (Conicella et al., 2016; Gasset-Rosa et al., 2019; Li et al., 2018b; Lim et al., 2016; Mann et al., 2019; Molliex et al., 2015). Because TDP-43 proteinopathy, manifested as aberrant mislocalization, aggregation, phosphorylation, ubiquitination, and cleavage of TDP-43, has been established as a prominent pathological feature of various major neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), the investigation of TDP-43 LLPS physiological function and its relationship with neuronal function in vivo appears to be an intriguing but unexplored area of interest. Here, we describe the first knock-in mouse model constitutively expressing endogenous LLPS-deficient murine TDP-43. In contrast to early established TDP-43 mouse models with manipulated TDP-43 expression in neurons (Igaz et al., 2011; Iguchi et al., 2013; Kraemer et al., 2010; Sephton et al., 2010; Wils et al., 2010; Wu et al., 2010; Wu et al., 2012; Xu et al., 2010), our mice with global TDP-43 LLPS deficiency showed characteristic robust behavioral deficits specifically related to brain dysfunction, thus providing a novel tool to dissect the in vivo function of TDP-43 and TDP-43 LLPS.
Results
Generation of TDP-43ΔCR mice
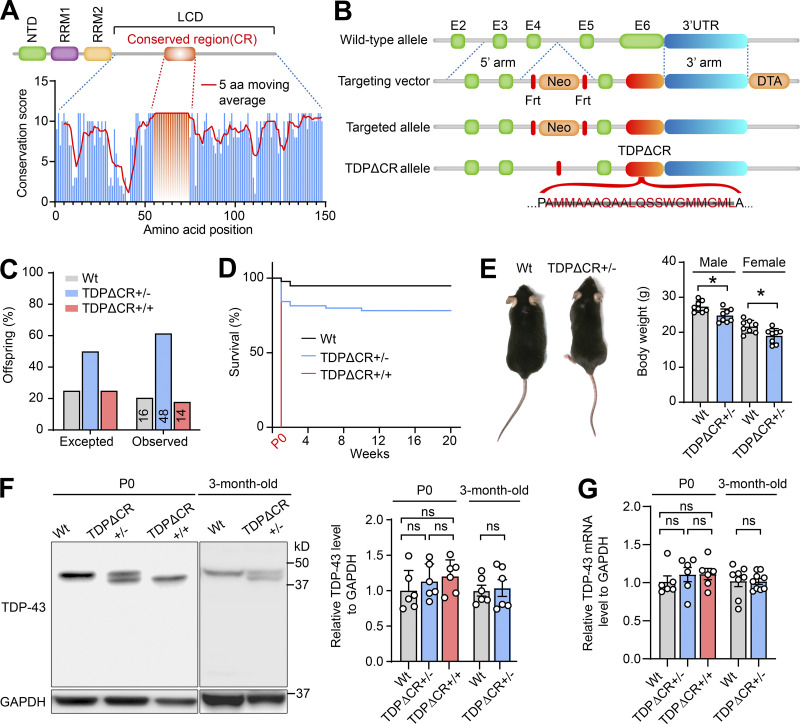
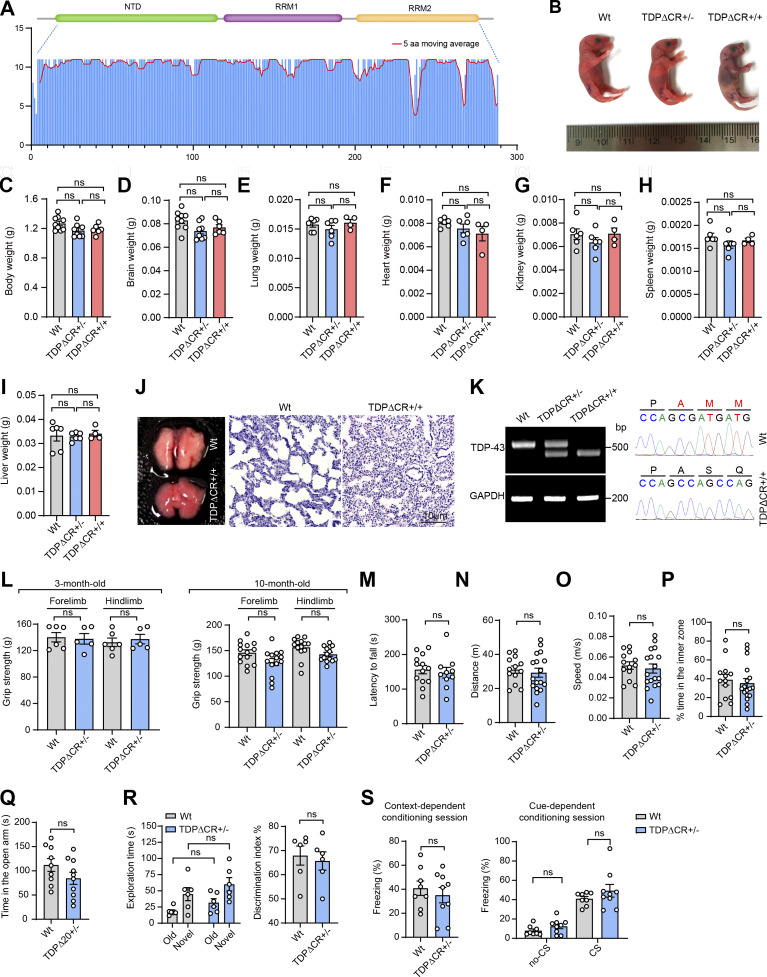
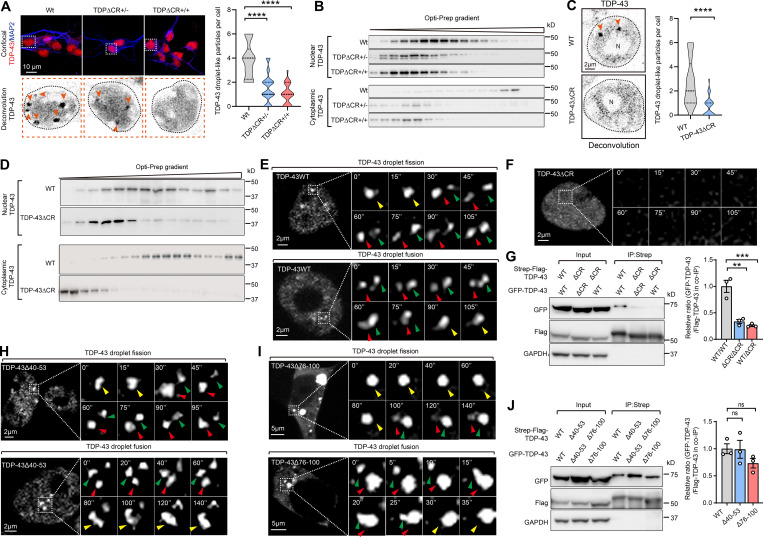
Recent studies have identified the α-helical region spanning residues from 321 to 340 within the LCD as the evolutionarily conserved region (CR) required for the LLPS of TDP-43 (Conicella et al., 2016; Li et al., 2018a; Li et al., 2018b). To understand the likely role of TDP-43 LLPS in the regulation its physiological function in vivo, we generated mutant mice with constitutive deletion of this motif encoded by exon 6 (i.e., TDPΔCR mice; Fig. 1, A and B; and Fig. S1 A). WT and TDPΔCR mice were born with expected Mendelian frequencies (Fig. 1 C). Newborn TDPΔCR mice appeared grossly comparable to WT littermates (Fig. S1 B). However, all homozygous TDPΔCR mice died within a few hours after birth, likely due to abnormalities in lung formation but without a change in weight of the lung or other organs, such as the brain, heart, kidney, spleen, and liver (Fig. 1 D; and Fig. S1, C–J). <20% heterozygous mice also died suddenly for unknown reasons during the first 2 wk (Fig. 1 D). Despite slightly lower body weight, surviving heterozygous TDPΔCR mice demonstrate no difference from age-matched WT littermates in feeding and survival until 12 mo old (Fig. 1 E). Targeted nucleotide deletion in newborn or adult TDPΔCR mice was confirmed by PCR amplifying the mutation site and was further verified by nucleotide sequencing of the PCR products and immunoblots showing the production of truncated TDP-43ΔCR protein (Fig. S1 K). Of note, quantitative analysis found that the deletion of the CR motif did not result in changed protein and mRNA levels of TDP-43 (Fig. 1, F and G).
Figure 1.
Generation of TDPΔCR mice. (A) Amino acid conservation score along an alignment of LCD of TDP-43 from 50 eukaryotic species. The histogram shows the conservation score for each position of the alignment; the red line shows the 5-aa moving average. (B) Generation of TDPΔCR mice by gene targeting using a targeting vector to replace the WT mouse TDP-43 allele with the targeted allele. Mice with the deletion of residues from 321 to 340 were produced after removal of the flippase recombinase target (Frt)-flanked selection cassette in the mutated TDP-43ΔCR allele. Diphtheria toxin A (DTA), a segment of the diphtheria toxin, was used for counter selection. (C) Genotype frequencies of WT mice (n = 16), TDPΔCR+/− mice (heterozygous; n = 48), and TDPΔCR+/+ mice (homozygous; n = 14) observed at postnatal day 0 (P0). (D) Kaplan-Meier survival curve of WT mice (n = 42), TDPΔCR+/− mice (n = 55), and TDPΔCR+/+ mice (n = 12). (E) Representative images and quantification of the body weight of WT mice (n = 16) and TDPΔCR+/− mice (n = 16) at 3 mo old. (F) Representative immunoblot and quantification of TDP-43 protein levels in the brains of WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 3–6 mice per group) at P0 or 3 mo old. (G) Relative mRNA levels of TDP-43 in the brains of WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 6–8 mice per group) at P0 or 3 mo old. Data were normalized to GAPDH. Data are mean ± SEM; two-tailed Student’s t test (E–G; 3-mo-old group) or one-way ANOVA followed by Tukey’s multiple comparison test (F and G; P0 group). *, P < 0.05.
Figure S1.
Characterization of TDP-43 LLPS-deficient knock-in mice. (A) Amino acid conservation score along an alignment of NTD and RRM1/2 of TDP-43 from 50 eukaryotic species. The histogram shows conservation scores for each position of the alignment; the red line shows the 5-aa moving average. (B) Representative images of WT, TDPΔCR+/−, and TDPΔCR+/+ mice at P0. (C–I) Quantification of the indicated organ weight of WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 4–9 mice per group) at P0. (J) Representative hematoxylin and eosin staining of lung tissues from WT and TDPΔCR+/+ mice at P0. (K) PCR and DNA-seq analysis of WT and mutant mouse TDP-43 alleles in WT, hemizygous (TDPΔCR+/−), or homozygous (TDPΔCR+/+) TDPΔCR mice. (L) Forelimb and hindlimb grip strength of WT and TDPΔCR+/− mice (n = 5–14 mice per group) at 3 mo old or 10 mo old. (M) Rotarod performance of WT mice (n = 13) and TDPΔCR+/− (n = 10 mice). (N–P) The distance traveled (N), velocity (O), and time spent in the center of apparatus (P) in the open field test. WT mice (n = 13) and TDPΔCR+/− (n = 17 mice). (Q) Time spent in the open arm in the elevated plus maze. WT mice (n = 9) and TDPΔCR+/− mice (n = 10). (R) Time spent with each object and preference ratio for an unfamiliar object in the two different sessions of the novel object recognition test. WT mice (n = 6) and TDPΔCR+/− mice (n = 6). (S) Freezing behavior of WT mice (n = 8) and TDPΔCR+/− mice (n = 9) in contextual fear learning and cue-dependent fear learning during the fear-conditioning test. CS, conditioned stimulus. Data are mean ± SEM; one-way ANOVA followed by Tukey’s multiple comparisons test (C–I), two-tailed Student’s t test (L–S).
TDP-43ΔCR mice exhibit characteristic behavioral abnormalities
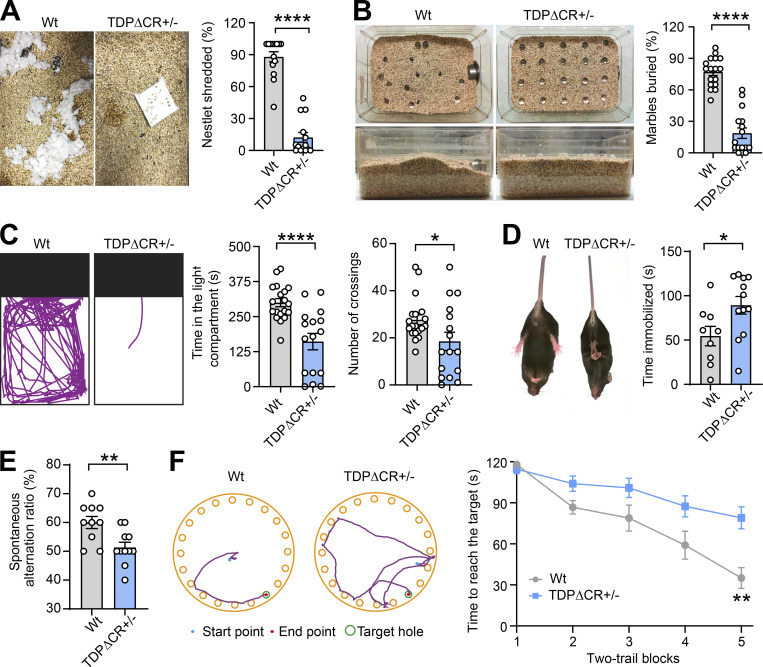
We next employed a battery of behavior tests to assess behavior changes in heterozygous TDPΔCR mice. No abnormalities were noted in grip strength, rotarod, or locomotor activity in the home cage in either adult or aged heterozygous TDPΔCR mice, indicative of undisturbed motor function or spontaneous behavior (Fig. S1, L–O). However, even young adult heterozygous TDPΔCR mice had already displayed remarkable deficits in shredding nestlets or burying marbles compared with their WT littermate controls (Fig. 2, A and B; and Video 1), suggesting that the repetitive, compulsion-like social behaviors are impaired in heterozygous TDPΔCR mice. Notably, despite unchanged locomotion, heterozygous TDPΔCR mice exhibited greatly reduced activity compared with WT animals in terms of the time spent in the light chamber in the light/dark exploration task (Fig. 2 C), further implying increased bright space anxiety-like behavior in heterozygous TDPΔCR mice. In contrast, open space anxiety-like behavior, as measured by both open field and elevated plus mazes, appeared to be unaltered (Fig. S1, P and Q). Heterozygous TDPΔCR mice also exhibited a depressive phenotype as evidenced by increased total time spent immobile during the tail suspension test (Fig. 2 D). Given that TDP-43 is associated with dementia, we further assessed the cognitive function in heterozygous TDPΔCR mice. While heterozygous TDPΔCR mice showed performance similar to that of control animals in the novel object recognition and fear-conditioning test, they were significantly impaired in the Y maze and Barnes maze tests (Fig. 2, E and F; and Fig. S2, R and S), revealing specifically impaired spatial learning and memory but not impaired nonspatial or emotional memory. Taken together, these data indicate that the global CR deletion specifically leads to brain-related behavioral deficits.
Figure 2.
TDPΔCR mice exhibit characteristic behavioral abnormalities. (A) Representative images and quantification of nestlet shredding of WT and TDPΔCR+/− mice (n = 14 mice per group). (B) Representative images and quantification of marble-burying activity of WT mice (n = 17) and TDPΔCR+/− mice (n = 16). (C) Representative track plots and quantification of WT mice (n = 19) and TDPΔCR+/− mice (n = 16) in the light/dark box task. (D) Representative images and quantification of WT mice (n = 9) and TDPΔCR +/− mice (n = 12) in the tail suspension test. (E) Performance of WT mice (n = 11) and TDPΔCR+/− mice (n = 12) in the Y-maze test. (F) Left: Representative track plots of WT and TDPΔCR mice in the second trial at day 5. Right: Quantification of the Barnes maze performance of WT mice (n = 12) and TDPΔCR+/− mice (n = 20) over 5 d of successive testing (two trials/day). Data are mean ± SEM; two-tailed Student’s t test (A–E) or two-way ANOVA followed by Bonferroni multiple comparisons test (F). *, P < 0.05; **, P < 0.001; and ****, P < 0.0001.
Video 1.
Representative 30-min movies showing the performance of 3-mo-old WT and TDPΔCR+/− mice in the marble-burying test. The movies are played at 8× normal speed at 30 frames per second and are related to Fig. 2 B.
Figure S2.
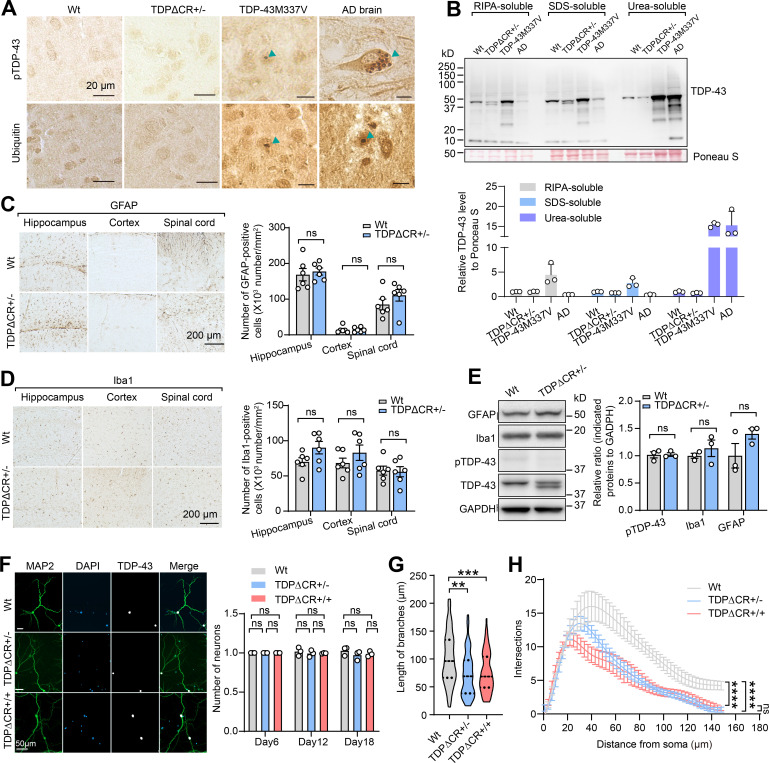
Absence of TDP-43 proteinopathy or neurodegeneration in TDPΔCR mice. (A) Representative immunohistochemistry of phosphorylated TDP-43 (pTDP-43) and ubiquitin in the brains of WT, TDPΔCR+/−, and TDP-43M337V mice at 3 mo old and in Alzheimer’s disease (AD) brains. (B) Representative immunoblot analysis of TDP-43 in SDS and urea soluble fraction from brain tissues of 3-mo-old WT or TDPΔCR+/− mice. Ponceau S staining was used for the loading control. (C and D) Representative immunohistochemistry and quantification of Iba1 (C) and glial fibrillary acidic protein (GFAP; D) in the hippocampus, cortex, and spinal cord of 3-mo-old WT and TDPΔCR+/− mice (n = 6 mice per group). (E) Representative immunoblot and quantification of Iba1, GFAP, pTDP-43, and TDP-43 in the brain lysates of WT and TDPΔCR+/− mice (n = 3 mice per group). GAPDH is included as the internal control. (F) Representative images of TDP-43 and MAP2 in primary cultured neurons from WT, TDPΔCR+/−, and TDPΔCR+/+ mice. Right: Quantification of neuronal cell numbers from WT, TDPΔCR+/−, and TDPΔCR+/+ mice. (G) Quantification of cultured hippocampal neurons from WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 8–10 neurons per genotype). (H) Sholl analysis of cultured hippocampal neurons from WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 8–10 neurons per genotype). Data are mean ± SEM; two-tailed Student’s t test (C–E), one-way ANOVA followed by Tukey’s multiple comparisons test (F and G), two-way ANOVA followed by Bonferroni multiple comparisons test (H). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TDP-43ΔCR mice show neuronal dysfunction but demonstrate no TDP-43 proteinopathy or neurodegeneration
In heterozygous TDP-43ΔCR mice, TDP-43 predominantly remained nuclear and did not aggregate or accumulate in the cytoplasm (Fig. 3 A). The post-translational modifications and solubility of TDP-43 also appeared to be similar between WT and heterozygous TDP-43ΔCR animals (Fig. S2, A and B), together implying the minimal effect of TDP-43 CR deletion on the development of its proteinopathy. Further histological and immunohistochemical analyses demonstrated no significant change in neuronal density in the brain and spinal cord of heterozygous TDPΔCR mice and no activation of astrocytes and microglia (Fig. 3 B; and Fig. S2, C–E). All TDPΔCR mouse behavior changes described above may be commonly linked to hippocampal activity (Deacon and Rawlins, 2005; Moser et al., 2015). Golgi-stained brain tissues of TDPΔCR mice were then subjected to morphological analysis of neuronal structures in the hippocampus. Adult heterozygous TDPΔCR mice exhibited significantly reduced basal dendrite branches, lengths, and branching complexity on hippocampal CA1 pyramidal neurons (Fig. 3, C and D), indicating impaired dendrite arborization, an essential component of neural network formation related to brain function. Consistently, a more detailed analysis under high magnification revealed a remarkable decrease in the dendritic spine density of hippocampal CA1 neurons of adult heterozygous TDPΔCR mice (Fig. 3 E). Impaired dendritic complexity was also noted in cultured neurons from both heterozygous and homozygous TDPΔCR mice (Fig. S2, F–H), together providing further histological evidence in support of deficits in brain function. To determine whether there were alterations in the neuronal function of heterozygous TDPΔCR mice, we measured synaptic activity and long-term potentiation (LTP) in the hippocampus by recording extracellular field excitatory post-synaptic potentials (fEPSPs) from the stratum radiatum of the CA1 area following stimulation of the Schaffer collateral fibers (Foster et al., 2019; Foster et al., 2018; Zhao et al., 2019). While LTP induced by modest θ-burst stimulation (TBS) appeared normal, the input–output (I/O) responses of TDPΔCR mice were largely deficient (Fig. 3, F and G), indicative of impaired synaptic strength, which is consistent with the reduction in dendrite complexity in the CA1 area. Little difference in the paired pulse ratio was also detected, except at 250 ms, which may indicate an issue with synaptic vesicle recycling (Fig. 3 H). Collectively, heterozygous TDPΔCR mice show the histological and physiological differences of neurons compared with their WT littermates, validating the important role of TDP-43 CR in the regulation of brain function.
Figure 3.
Neuronal dysfunction in TDPΔCR mice without TDP-43 proteinopathy or neurodegeneration. (A) Representative images of immunohistochemical staining of TDP-43 in the hippocampus, cortex, and spinal cord neuronal cells of WT and TDPΔCR+/− mice. (B) Representative images and quantification of immunohistochemical staining of NeuN in the hippocampus, cortex, and spinal cord of WT and TDPΔCR+/− mice (n = 6 mice per group). (C) Representative Golgi staining images and quantification of CA1 pyramidal neurons from WT mice (n = 14 neurons from 3 mice) and TDPΔCR+/− mice (n = 15 neurons from 3 mice). (D) Representative photomicrographs and Sholl analysis of CA1 pyramidal neurons from WT (n = 14 neurons from 3 mice) and TDPΔCR+/− mice (n = 15 neurons from 3 mice). (E) Representative images and quantification of apical dendritic spines from CA1 pyramidal neurons of WT and TDPΔCR+/− mice (n = 300–327 spines from 3 mice per group). (F) Slope of fEPSP after high-frequency stimulation of fEPSP in CA3–CA1 regions of WT (n = 22 slices from 7 mice) and TDPΔCR+/− mice (n = 16 slices from 7 mice), normalized by baseline. (G) I/O curve of fEPSP in CA3–CA1 regions of WT (n = 22 slices from 7 mice) and TDPΔCR+/− mice (n = 16 slices from 7 mice), normalized by fEPSP amplitude induced by minimum stimulation intensity. (H) Paired-pulse facilitation, elicited by application of two successive stimulation pulses to CA3 (interstimulus intervals between 50 and 250 ms), was measured as the amplitude ratio of the second fEPSP divided by the first fEPSP in a pair. Data are mean ± SEM; two-tailed Student’s t test (B, C, and E) or two-way ANOVA followed by Bonferroni multiple comparisons test (D, G, and H). *, P < 0.05; ***, P < 0.001; and ****, P < 0.0001.
Loss of CR disrupts TDP-43 LLPS in TDP-43ΔCR mice
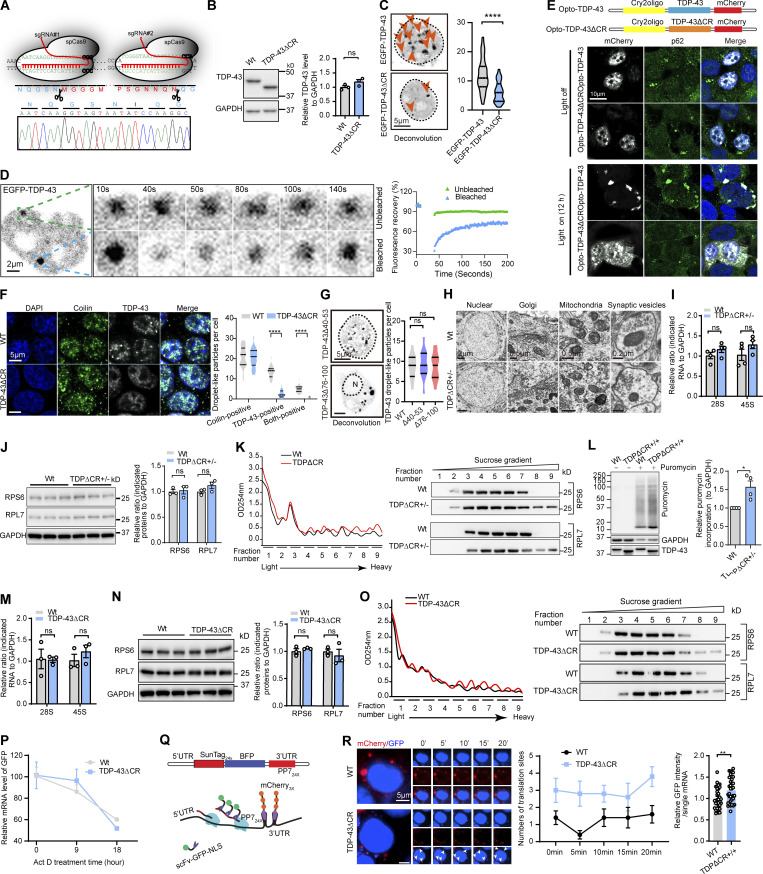
Previous studies implicated CR as the conserved motif required for TDP-43 LLPS (Conicella et al., 2016; Li et al., 2018a; Li et al., 2018b). Although the measurement of LLPS in mice in vivo is extremely challenging, high-resolution fluorescence images of in vitro cultured neurons have recently been used to determine the LLPS properties of intranuclear TDP-43 due to its predominant abundance in the nucleus (Gasset-Rosa et al., 2019; Mitrea et al., 2018). To obtain mechanistic insight into the brain dysfunction caused by CR loss, we then focused on the characterization of intranuclear TDP-43 LLPS in cultured brain neurons of TDPΔCR mice. Consistent with a previous study (Gasset-Rosa et al., 2019), under confocal microscopy, droplet-like round TDP-43 particles of 0.2–1-µm diameter were readily noted in the nucleus of neurons derived from WT mice (i.e., WT neurons; Fig. 4 A). As expected, the number of droplet-like intranuclear TDP-43 accumulations decreased to 40.5% of WT neurons in neurons from heterozygous TDPΔCR mice and further declined to the level of 26.4% of WT neurons in neurons from homozygous TDPΔCR mice, indicating impaired TDP-43 LLPS in TDPΔCR mice. Further OptiPrep gradient analysis consistently showed the disturbed subnuclear distribution of TDP-43 in the brains of newborn heterozygous and homozygous TDPΔCR mice (Fig. 4 B). Notably, although droplet-like TDP-43 structures in the cytoplasm were not detectable under confocal microscopy due to their low abundance, biochemical analysis also revealed disturbed subcytoplasmic distribution of TDP-43 in both heterozygous and homozygous TDPΔCR mice (Fig. 4 B). Consistent with these findings, TDP-43 in the HEK293 cell line expressing endogenous human TDP-43 lacking CR (TDPΔCR cells) demonstrated similarly suppressed formation of droplet-like particles and altered subnuclear or subcytoplasmic distribution (Fig. 4, C and D; and Fig. S3, A and B). Exogenously expressed GFP-tagged TDP-43 also exhibited greatly inhibited appearance of droplet-like structures after CR deletion by live imaging or FRAP assay (Fig. 4, E and F; and Fig. S3, C and D). Optogenetic approaches using an optogenetic model of TDP-43 proteinopathy (optoTDP43) have recently been developed to examine the role of LLPS in the formation of TDP-43 aggregates (Mann et al., 2019). Both WT optoTDP43 and optoTDP-43ΔCR were localized to nuclei in cells kept in darkness (Fig. S3 E). Upon blue light stimulation, WT optoTDP43 mislocalized to the cytoplasm and formed large p62-positive inclusions (Fig. S3 E). In contrast, optoTDP-43ΔCR failed to form large p62-positive aggregates in the cytoplasm after light stimulation, even though modest cytoplasmic mislocalization could be noted (Fig. S3 E), further indicating CR as the critical motif for LLPS-mediated optoTDP-43 aggregation. Cajal bodies are membraneless structures that exist as liquid droplets arising from LLPS (Gomes and Shorter, 2019). WT and TDP-43ΔCR cells were further costained with the widely used Cajal body marker coilin. Approximately 20% of TDP-43 puncta colocalized with Cajal bodies in WT cells, but they were barely observed in TDPΔCR cells (Fig. S3 F), suggesting that CR is also critical for TDP-43 LLPS in Cajal bodies. Notably, TDP-43 and coilin double-positive puncta only represented a small portion of Cajal bodies, and there was no difference in the number of total Cajal bodies between the WT and TDPΔCR animals, implying a specific impact of CR deletion on TDP-43 LLPS.
Figure 4.
Loss of CR causes abnormalities of TDP-43 LLPS. (A) Representative images and quantification of TDP-43 droplet-like particles in neurons from WT, TDPΔCR+/−, and TDPΔCR+/+ mice. Endogenous TDP-43 (red) and MAP2 (red) are shown. Lower panels, TDP-43 only (inverted in gray). Droplet-like particles are labeled by arrowheads. Cell nuclei are circled. (B) Representative immunoblot of nuclear and cytoplasmic TDP-43 from WT, TDPΔCR+/−, and TDPΔCR+/+ mouse brains separated by OptiPrep gradient ultracentrifugation. (C) Representative images and quantification of TDP-43 droplet-like particles in WT and TDP-43ΔCR HEK293 cells. TDP-43 droplet-like particles are labeled by arrowheads. (D) Representative immunoblot of nuclear and cytoplasmic TDP-43 from WT and TDP-43ΔCR HEK293 cells. (E and F) Representative images of dynamic changes of TDP-43 droplet-like particles in the nucleus of HEK293 cells expressing GFP-tagged TDP-43WT (E) or TDP-43ΔCR (F). Higher-magnification images at different time points are shown in the right panels. Green and red arrowheads point to two separate droplets before fusion, and yellow arrowheads point to the fused droplets. Note that TDP-43 is diffusive, and no droplet is noted in cells expressing TDP-43ΔCR. N, nuclear. (G) Representative immunoblot of coIP between transfected Strep-Flag–tagged TDP-43 (WT and ΔCR) and GFP-tagged TDP-43 (WT and ΔCR) in HEK293 cells. (H and I) Representative images of dynamic changes of TDP-43 droplet-like particles in the nucleus of HEK293 cells expressing GFP-tagged TDP-43Δ40-53 (G) and TDP-43Δ76-100 (H). Higher-magnification images at different time points are shown in the right panels. Green and red arrowheads point to two separate droplets before fusion, and yellow arrowheads point to the fused droplets. N, nuclear. (J) Representative immunoblot of coIP between transfected Strep-Flag–tagged TDP-43 (WT, Δ40-53, and Δ40-53) and GFP-tagged TDP-43 (WT, Δ40-53, and Δ40-53) in HEK293 cells. Data are mean ± SEM; one-way ANOVA followed by Tukey’s multiple comparisons test (A, G, and J), two-tailed Student’s t test (C). **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Figure S3.
Validation of LLPS loss by the deletion of the CR motif in TDP-43ΔCR cells and increased ribosomal assembly and protein translation in TDPΔCR mice and TDP-43ΔCR cells. (A) Schematic diagram of CRISPR/Cas9 genome editing of the TARDBP gene in HEK293 cells to obtain cells with deletion of the CR in human TDP-43 (TDP-43ΔCR cells). sgRNA, single-guide RNA. (B) Representative immunoblotting and quantification of TDP-43 in TDP-43ΔCR cells. GAPDH is used as the internal control. (C) Representative images and quantification of TDP-43 droplet-like particles formed in the nucleus of HEK293 cells expressing GFP-tagged TDP-43WT and TDP-43ΔCR. Cell nuclei are circled. (D) FRAP of GFP-tagged TDP-43 droplets in HEK293 cells after 24 h of expression. Higher magnification is shown in the right panel. The whole droplet was photobleached for 40 s. Mean fluorescence intensity at the bleached area was plotted over time, normalized to the average intensity of a droplet before photobleaching, and represented as the mean (from the recovery curves of five bleached droplets and four unbleached droplets). (E) optoTDP43 fusion protein and representative immunofluorescence images of optoTDP-43, optoTDP-43ΔCR, and p62 exposed to blue light stimulation (bottom) or darkness (top). (F) Representative immunofluorescence images and quantification of coilin and TDP-43 droplet-like particles in WT and TDP-43ΔCR HEK293 cells. (G) Representative images and quantification of TDP-43 droplet-like particles in the nucleus or cytoplasm of HEK293 cells expressing GFP-tagged TDP-43Δ40-53 or TDP-43Δ76-100, respectively. N, nuclear. Cell nuclei are circled. (H) EM analysis of nuclear, Golgi, mitochondrial, and synaptic vesicles in hippocampal neurons of 3-mo-old WT and TDPΔCR+/− mice. (I) Relative 28S and 45S ribosomal RNA levels in the brains of 3-mo-old WT and TDPΔCR+/− mice (n = 4 mice per group). Data were normalized to GAPDH. (J) Representative immunoblotting and quantification of RPS6 and RPL7 in the brain lysates of WT and TDPΔCR+/− mice (n = 3 mice per group). GAPDH is included as the internal control. (K) Representative immunoblots of RPS6 and RPL7 in the brains of WT and TDPΔCR+/− mice separated by sucrose gradient ultracentrifugation. UV absorbance profile at 254 nm of the extract from 3-mo-old WT and TDPΔCR+/− mice is shown at left. (L) Representative immunoblotting and quantification of newly synthesized polypeptide labeled by puromycin (10 µg/ml for 15 min) in primary cultured neurons from WT and TDPΔ+/+ mice at day in vitro 7. GAPDH was used as the internal control. (M) Relative 28S and 45S ribosomal RNA levels in WT and TDP-43ΔCR HEK293 cells. Data were normalized to GAPDH. (N) Representative immunoblotting and quantification of RPS6 and RPL7 in the cell lysates of WT and TDP-43ΔCR HEK293 cells. GAPDH was included as the internal control. (O) Representative immunoblots of RPS6 and RPL7 in WT and TDP-43ΔCR HEK293 cells separated by sucrose gradient ultracentrifugation. UV absorbance profile at 254 nm of the extract from WT and TDP-43ΔCR HEK293 cells is shown at left. (P) Relative level of the GFP mRNA in WT and TDP-43ΔCR HEK293 cells transfected with CMV-GFP plasmid. Cells were treated with 10 µM actinomycin D (Act D) for 0–18 h and then harvested at the time points indicated, followed by RNA isolation and RT-qPCR analysis. All the data were normalized to the GFP mRNA level before treatment. (Q) Schematic of nascent polypeptide labeling using the SunTag system and mRNA labeling using the PP7 system. (R) WT or TDP-43ΔCR cells expressing the SunTag24x-BFP-PP724x reporter was imaged by time-lapse microscopy. Active translation sites were quantified over time (n = 5 cells). Asterisks indicated active translation sites. Intensity of scFv-GFP at translational sites was measured in both WT and TDP-43ΔCR cells (n = 32–35 translation sites from 6–8 cells). Data are mean ± SEM; one-way ANOVA followed by Tukey’s multiple comparisons test (G) or two-tailed Student’s t test (B, C, F, I, J, L–N, and R). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
The protein’s self-association is crucial for TDP-43 LLPS (Conicella et al., 2016; Lim et al., 2016). Coimmunoprecipitation (coIP) assays were further performed to assess the self-interaction of WT TDP-43 and TDP-43ΔCR. The deletion of the CR was found to substantially reduce the interaction between WT TDP-43 and TDP-43ΔCR, or TDP-43ΔCR self-interaction (Fig. 4 G), further supporting CR as the critical motif required for TDP-43 LLPS. Notably, amino acid residues from 40 to 53 or from 76 to 100, two N-terminal motifs reported to mediate TDP-43 LLPS in vitro (McGurk et al., 2018; Wang et al., 2018), appear to have no effect on the formation of droplet-like TDP-43 particles or TDP-43 self-interaction in cells (Fig. 4, H–J; and Fig. S3 G). Collectively, these results indicated that CR is important for TDP-43 LLPS in vivo, suggesting, at least in part, a molecular mechanism whereby the loss of the CR affects brain function.
TDP-43ΔCR mice are associated with global translational alterations
To further explore aberrations that may be associated with TDP-43 CR loss, we analyzed TDPΔCR mouse brains by transmission EM. While hippocampal tissues from TDPΔCR animals showed healthy neuronal soma with intact nuclear envelope and normal characteristics of subcellular organelles, including the ER, Golgi, mitochondria, lysosomes, and even synaptic vesicles, they had much higher ribosomal density than those from WT animals either within the free cytoplasmic space or on the ER (Fig. 5 A and Fig. S3 H). Consistently, despite unchanged total RNA or ribosomal protein markers, ribosomal assembly was indeed found to be enhanced in TDP-43ΔCR mice compared with WT mice (Fig. S3, I–L). Considering the positive relationship between ribosomal density and translation rates (Plotkin and Kudla, 2011), we speculated that there might be a change in the rate of protein synthesis in TDPΔCR mice. To test this hypothesis, the surface sensing of translation (SUnSET) technique, based on the incorporation of puromycin to newly synthesized proteins, was employed to measure global protein synthesis in vivo (Fig. 5 B). Remarkably, puromycin-conjugated polypeptides were greatly increased in brains of TDPΔCR mice (Fig. 5 C), implying enhanced global protein synthesis by TDP-43 CR loss. Consistently, in vitro cultured neurons from TDPΔCR mice or HEK293 TDPΔCR cells also exhibited increased ribosomal assembly and protein synthesis without changes in total RNA or ribosomal protein markers (Fig. 5, D and E; and Fig. S3, M–O). Translation initiation can be either cap dependent or cap independent (Merrick, 2004). HEK293 cells were then transfected with reporters to express GFP either driven by the cytomegalovirus (CMV) promoter in a cap-dependent fashion or by the internal ribosome entry site (IRES) in a cap-independent manner. We found that both the cap-dependent and cap-independent translation of GFP was significantly increased in TDP-43ΔCR cells compared with WT HEK293 cells, while GFP mRNA stability remained unchanged as assessed by the mRNA stability assay using transcription inhibition by actinomycin D (Fig. 5 F and Fig. S3 P).
Figure 5.
Remarkable translational alterations in TDPΔCR mice and HEK293ΔCR cells. (A) Representative EM micrographs and quantification showing changed ribosomal densities in hippocampal neurons of WT and TDPΔCR+/− mice (n = 18–20 neurons from 3 mice per group). (B) Schematic diagram of in vivo labeling of newly synthesized polypeptides by SUnSET in WT and TDPΔCR+/− mice. Lower panel: Representative images of newly synthesized polypeptide labeled by puromycin in the brains of WT mice. Lv, lateral ventricle; III, third ventricle; IV, fourth ventricle. (C) Representative immunoblot and quantification of newly synthesized polypeptide labeled by puromycin in the hippocampal tissues from WT and TDPΔCR+/− mice (n = 4 mice per group). (D) Representative EM micrographs and quantification showing changed ribosomal densities in WT and TDP-43ΔCR HEK293 cells (n = 11–12 cells per group). (E) Representative immunoblot and quantification of newly synthesized proteins labeled by Click-iT AHA in WT and TDP-43ΔCR HEK293 cells (n = 3 independent experiments per group). (F) Representative immunoblot and quantification of EGFP expression driven by cap-dependent (CMV-EGFP) and cap-independent (IRES-EGFP) translation in WT and TDP-43ΔCR cells (n = 3 independent experiments per group). Data are mean ± SEM; two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001.
To validate these findings, we further imaged ongoing translation of single mRNAs in live cultured TDP-43ΔCR cells using the mCherry fused PP7 bacteriophage coat protein to label mRNA and the SunTag recognized by the GFP-tagged single-chain variable fragment (scFv-GFP) to label nascent proteins (Fig. S3 Q). In this SunTag system, mCherry puncta colocalizing with GFP correspond to mRNAs in translation, and scFv-GFP is fused to an NLS to sequester unbound antibody into the nucleus (Tanenbaum et al., 2014; Yan et al., 2016). Compared with WT cells, TDP-43ΔCR cells demonstrated more comigrated mCherry and GFP puncta and significantly brighter scFv-GFP fluorescence intensity (Fig. S3 R), indicating both increased active translation sites and ribosome loading on the reporter mRNA by ΔCR. Taken together, these results consistently imply that TDP-43 LLPS may have a general role in translational regulation.
TDP-43 CR deletion enhances its physical interactions with distinct translational factors
To investigate how translation changed as a function of TDP-43 CR loss, a mass spectrometry (MS)-based approach was used to monitor the dynamics of TDP-43–associated protein complexes in response to CR deletion using TDP-43-knockout HEK293 cells expressing Strep-tagged WT TDP-43 (TDP-43WT) or TDP-43ΔCR mutant (Fig. 6 A and Fig. S4 A). Because most translation occurs in the cytoplasm on intron-free mRNAs, our analysis focused on cytoplasmic interactomes. We identified 18 and 41 proteins associated with cytoplasmic TDP-43WT and TDP-43ΔCR, respectively, of which 9 proteins were commonly enriched in both TDP-43WT and TDP-43ΔCR precipitate samples for gene ontology terms largely related to translation (Fig. 6, B–D).
Figure 6.
Dependence of TDP-43 CR-mediated translation on physical interactions between TDP-43 and distinct translational factors. (A) Schematic overview of subcellular fraction and affinity purification strategies used in this study. After the removal of mitochondrial, nuclear, and unbroken cell pellets, the supernatant was subjected to the affinity purification of Strep-tagged TDP-43 by Strep-Tactin beads. The binding partners of TDP-43 were identified by LC-MS/MS. m/z, mass-to-charge ratio. (B) Volcano plots of TDP-43– and TDP-43ΔCR–interacting proteins detected by LC-MS/MS. The logarithmic ratios of average fold changes are shown on the x axis. The y axis data plot negative logarithmic false discovery rate (q) values from the t test performed on three biological replicates. Enriched proteins are denoted by red dots. (C) Venn diagram depicting the numbers of unique and shared interactors between TDP-43WT and TDP-43ΔCR. (D) Gene ontology (GO) enrichment analysis of unique and shared interactors in B according to categories based on biological processes. CRD, coding region instability determinant; SRP, signal recognition particle. (E) Representative immunoblot of coIP between transfected Strep-Flag–tagged TDP-43WT/ΔCR and EGFP-PABPC4, Myc-RPS6, or Myc-RPL7 in HEK293 cells. Cells transfected with empty vector were also included as controls. Relative intensity quantifications are shown in the middle of the blots. (F) Representative immunoblot and quantification of coIP between transfected Strep-Flag–tagged TDP-43WT or ΔCR and endogenous PABPC4, RPS6, or PRL7 in HEK293 cells. (G) Representative immunoblot and quantification of coIP between endogenous TDP-43WT or ΔCR and endogenous PABPC4, RPS6, or PRL7 in the brains of WT and TDPΔCR+/− mice. IP using IgG was included as a control. (H) Schematic of TDP-43 with the highlighted amino acids from 80 to 107. (I) Representative immunoblot of coIP between transfected Strep-Flag–tagged TDP-43 (WT, ΔCR, or ΔCRΔ99-105) and endogenous PABPC4, RPS6, or PRL7 in HEK293 cells. (J) Representative immunoblot and quantification of newly synthesized proteins labeled by Click-iT AHA in TDP-43–knockout HEK293 cells expressing TDP-43WT, ΔCR, or ΔCRΔ99-105; n = 3 independent experiments per group). (K) Representative immunoblot and quantification of GFP expression driven by cap-dependent (CMV-EGFP) and cap-independent (IRES-EGFP) translation in TDP-43–knockout HEK293 cells expressing TDP-43WT, ΔCR, or ΔCRΔ99-105 (n = 3 independent experiments per group). (L) A working model of TDP-43 LLPS-regulated protein translation. WT TDP-43 forms droplet-like particles, while TDP-43ΔCR disrupts the phase transition process. Diffusive TDP-43ΔCR in cytoplasm enhances its interaction with translation factors involved in multiple translational steps, together causing translational abnormalities. Data are mean ± SEM; two-tailed Student’s t test (F and G), one-way ANOVA followed by Tukey’s multiple comparisons test (J). *, P < 0.05; **, P < 0.001; ***, P < 0.001; and ****, P < 0.0001.
Figure S4.
Identifying TDP-43 interactome and mapping the binding site of TDP-43 for translational factors. (A) Schematic overview of subcellular fraction, affinity purification, and MS strategies used in this study. After the removal of mitochondrial, nuclear, and unbroken cells, the supernatant was subjected to affinity purification of Strep-tagged TDP-43 by Strep-Tactin beads. The purified proteins were digested, labeled by stable isotopes, and finally analyzed by quantitative MS. HEK293 cells with the knockout (KO) of TDP-43 were generated using CRISPR/Cas9 genome editing and used to express Strep-tagged TDP-43WT or TDP-43ΔCR by transient transfection. TDP-43 KO cells expressing TDP-43WT or TDP-43ΔCR without Strep tag were used as a control for IP. m/z, mass-to-charge ratio. (B) Representative immunoblot of coIP between exogenously expressed Strep-Flag–tagged PABPC4/PABPC1 and GFP-tagged TDP-43ΔCR in HEK293 cells. Cells transfected with empty vector were also included as controls. (C) Representative immunoblot of coIP between exogenously expressed GFP-tagged EEF1A1 and Strep-Flag–tagged TDP-43WT or Strep-Flag–tagged TDP-43ΔCR in HEK293 cells. Cells transfected with empty vector were also included as controls. (D) Representative immunoblot of coIP between exogenously expressed GFP-PABPC1 and Strep-Flag–tagged TDP-43WT or Strep-Flag–tagged TDP-43ΔCR in HEK293 cells. (E) Representative immunoblot of coIP between exogenously expressed GFP-tagged PARP1 and Strep-Flag–tagged TDP-43WT or Strep-Flag–tagged TDP-43ΔCR in HEK293 cells. (F) Schematic representation of TDP-43 deletion mutants used for mapping the binding site of TDP-43 for translational factors. (G–I) Representative immunoblot of coIP between transfected exogenously expressed GFP-tagged PABPC4 and the indicated Strep-Flag–tagged TDP-43ΔCR deletion mutants in HEK293 cells. (J) Representative immunoblot of coIP between exogenously expressed Strep-Flag–tagged TDP-43 WT or Δ99-105 and endogenous PABPC4, RPS6, and RPL7 in HEK293 cells. (K) Representative immunoblot of TDP-43 and mutations in exogenously expressed Flag-tagged TDP-43, Flag-tagged TDP-43ΔCR, and Flag-tagged TDP-43ΔCR/Δ99-105 in TDP-43 KO cells.
To validate and provide relative quantification of protein interactions, we performed coIP on HEK293 cells expressing TDP-43WT or TDP-43ΔCR. Either endogenously or exogenously expressed TDP-43ΔCR coprecipitated significantly more with poly(A)-binding protein cytoplasmic 4 (PABPC4) than did endogenous or exogenous TDP-43WT, although the levels of PABPC4 in input samples remained unchanged (Fig. 6, E and F). Consistently, a significantly higher amount of endogenous PABPC4 coprecipitated by TDP-43 was noted in TDPΔCR mouse brain extracts relative to WT control (Fig. 6 G). In addition to PABPC4, PABPC1 and some translation elongation factors and ribosomal subunits, such as EEF1A1, ribosomal protein S6 (RPS6), and ribosomal protein L7 (RPL7), could also be identified and validated as differentially enriched TDP-43ΔCR coprecipitating proteins (Fig. 6, E–G; and Fig. S4, B–D), thus implying that CR deletion may affect multiple steps in the translation process. There were some coprecipitating translation-unrelated proteins that instead showed decreased enrichment by TDP-43ΔCR (Fig. S4 E and Table S1), which may represent interacting proteins positively regulated by TDP-43 CR. Of note, TDP-43 coprecipitated PABPC4, RPS6, and RPL7 simultaneously (Fig. 6, F and G), further indicating the existence of TDP-43 in a protein complex composed of translation-related factors. Taken together, these findings provide further molecular evidence supporting the functional impact of CR loss on translational regulation.
The inhibition of TDP-43 and translational factor association blocks CR loss-induced translational dysregulation
To further investigate the importance of TDP-43 and translational factor association, we generated a series of deletion mutations in TDP-43ΔCR and analyzed their interaction with the identified major binding partner PABPC4 in HEK293 cells (Fig. 6 H and Fig. S4 F). Like the deletion of N-terminal 105 aa (ΔN1–105), the internal deletion of only seven residues from 99 to 105 (Δ99–105), which were immediately after the NLS, completely abrogated the ability of TDP-43ΔCR or TDP-43WT to coprecipitate exogenously expressed PABPC4 and other binding partners such as RPS6 and RPL7 (Fig. 6 I; and Fig. S4, G–J), suggesting them as CR-independent conserved amino acids required for interaction between TDP-43 and translational factors. The TDP-43ΔCR containing a part of this motif (ΔN1–102) still permitted a weaker interaction with PABPC4 (Fig. S4 I), indicating a likely length-dependent binding fashion. Notably, despite the predominant cytoplasmic localization (data not shown), the ΔCRΔN1–98 deletion showed interaction strength similar to that of the ΔCRΔN1–80 mutation or ΔCR that was largely localized in the nucleus (Fig. S4 H), implying that the interaction motif may be used in a protein expression–independent manner.
Based on the identification of the aa 99–105 motif required for interactions between TDP-43 and translational factors, transient transfection experiments were next performed in TDP-43–knockout HEK293 cells to validate their functional significance in translational regulation. After the reexpression of exogenous TDP-43, the expression level of transgene TDP-43 was very similar to the normal TDP-43 expression level in WT cells (Fig. S4 K). According to the L-azidohomoalanine (AHA) incorporation-based Click-iT assay, exogenous expression of TDP-43ΔCR significantly enhanced the rate of protein synthesis compared with TDP-43WT, which was abolished by the deletion of the binding motif (i.e., TDP-43ΔCRΔ99–105; Fig. 6 J). Consistently, TDP-43ΔCR–induced increases of cap-dependent and cap-independent translation, as measured by the above-mentioned translation reporters, were also completely suppressed by the binding motif deletion (Fig. 6 K). Overall, these data suggest that TDP-43 CR loss perturbs its association with various translational factors that are involved in multiple steps in the translation process, likely amounting to enhancement in global translation synergically (Fig. 6 I).
Discussion
Here, by generating a mouse model globally expressing endogenous murine TDP-43 lacking the conserved CR motif required for its LLPS, this study may identify a likely unexpected physiological function of TDP-43 LLPS in regulating protein translation by directly interfering physical interactions between TDP-43 and translational factors. This novel TDP-43ΔCR mouse model shows brain-related phenotypes, including social, emotional, and cognitive impairments in the absence of ALS-related muscle wasting, motor deficits, and paralysis, not only strongly supporting a likely important but specific role of TDP-43 LLPS in the maintenance of brain function but also implicating a likely ALS-irrelevant TDP-43 pathomechanism for Alzheimer’s disease and related brain disorders. Indeed, the previously reported TDP-43 proteinopathy, neurodegeneration, and aberrant RNA splicing were not observed in TDP-43ΔCR mice. Therefore, the behavior abnormalities of our TDP-43 mice are likely attributable to brain neuron impairment caused by dysregulation of TDP-43 LLPS-mediated global translation. Importantly, because our study demonstrates that the CR loss is sufficient to cause neuronal and brain dysfunction independent of protein mislocalization and aggregation, TDP-43 LLPS-mediated translational repression may represent an important mechanism contributing to the onset and progression of TDP-43–associated neurodegenerative diseases, which is worthy of further investigations.
Previously, TDP-ΔC knock-in mice with a deletion of the complete C-terminal domain (residues from 274 to 414) were generated (Nishino et al., 2019). Similar to TDP-ΔC mice as well as TDP-43–knockout mice, no homozygous TDP-43ΔCR mice were born, and heterozygous mice all developed normally. Yet, despite these very limited similarities, TDP-43ΔCR mice differ in many aspects. For example, in TDP-ΔC mice, the total expression level of endogenous WT TDP-43 plus TDP-43ΔC was increased, whereas in our TDP-43ΔCR mice, total TDP-43 remained unchanged. TDP-ΔC mice demonstrated motor dysfunction and differentially expressed Notch1 and genes related to the Akt signaling pathway, all of which are absent in our TDP-43ΔCR mice. TDP-ΔC protein is known to be predominantly localized in cytosol and to induce cytotoxicity. In TDP-ΔC mice, it is difficult to distinguish the impact of altered TDP-43 expression, subcellular localization, or C-terminal domain deletion. Therefore, due to altered cellular localization or the presence of neuronal loss and especially premature death (Igaz et al., 2011; Iguchi et al., 2013; Kraemer et al., 2010; Sephton et al., 2010; Wils et al., 2010; Wu et al., 2010; Wu et al., 2012; Xu et al., 2010), TDP-ΔC and other previously developed TDP-43 mice overexpressing or reducing TDP-43 specifically in neurons may not be ideal models in which to determine the in vivo physiological function of TDP-43, especially TDP-43 LLPS.
Heterozygous TDP-43ΔCR mice with constitutive genetic modifications showed specifically impaired brain and neuronal function without neuronal loss, altered survivorship, or noticeable changes of other tissues and organs until 15 mo of age (data not shown). Thus, we provide a unique animal model that enables molecular dissection of TDP-43 physiological function out of epiphenomena caused by neuron death or other irrelevant complications. Although TDP-43ΔCR homozygous mice died immediately after birth, they did not show gross and histopathological abnormalities in the brain and other tissues or organs. These results suggest that TDP-43 LLPS should not be involved in murine development. While TDP-43ΔCR heterozygous mice developed brain deficiency, they exhibited no typical ALS disease phenotypes reported in other TDP-43 animal models, such as muscle weakness, motor dysfunction, and mortality, further indicating that our TDP-43ΔCR mouse is a unique model for TDP-43 misregulation. Social, emotional, and cognitive function are all complex phenomena involving different brain regions and physiological processes. A series of behavioral tasks showed that these different brain functions were all affected in mice with TDP-43 CR loss, even though it seems paradoxical that they exhibited intact performance in other similar tests. Notably, the hallmark symptoms of FTD include progressive cognitive, behavioral, and socioemotional deficits (Bott et al., 2014). Because TDP-43 is both neuropathological and genetically connected to FTD, the presence of profound impairments in social, emotional, and cognitive function in TDP-43ΔCR mice may enable using them as a good model for the study of TDP-43 LLPS in the development of FTD-TDP. It is still unknown whether TDP-43 CR deletion differentially causes phenotypes correlating to regional neuronal dysfunction. Notably, the behavioral tests in which TDP-43ΔCR mice show deficits are all dependent on the hippocampus. Along this line, our TDP-43 mice presented significantly changed hippocampal neuron structure and function, while the number of hippocampal neurons remained unchanged. Therefore, these findings imply the likely specificity of the brain deficits that result from TDP-43 CR loss and pinpoint hippocampal dysfunction as a potential key contributor to brain defects. Reactive astrocytes or microglia were not noted in TDP-43ΔCR mice, probably because they are less vulnerable than neurons to the disturbance of translation. Cultured neurons from TDP-43ΔCR mice exhibited significantly impaired neurites. Consistently, our gene expression profiling of TDP-43ΔCR brains identified altered pathways related to neuronal dysfunction, but not to neuroinflammation (Fig. S5, A and B). Therefore, neuron-intrinsic mechanisms should be involved in synaptic dysfunction in TDP-43ΔCR mice. Nevertheless, translational dysregulation in inflammatory glia may still exert mild non–cell-autonomous effects that target dysfunctional neurons, and further detailed characterization of glia cells and brain neural circuits will be important to understand how TDP-43 LCDs are specifically involved in normal brain function.
Figure S5.
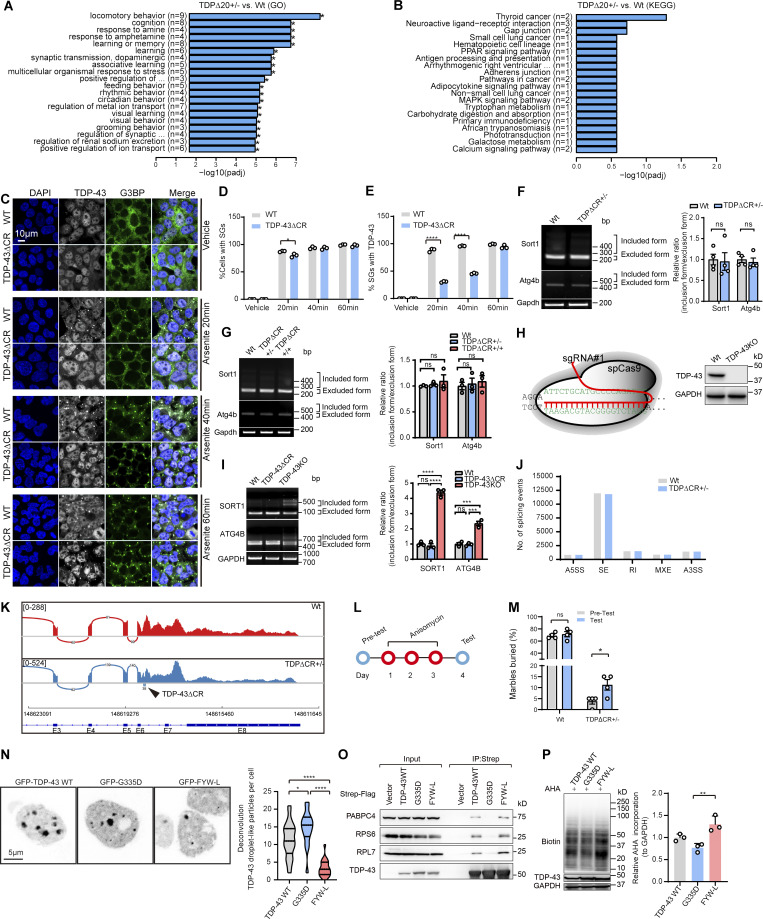
CR loss has no effect on RNA splicing activity and stress granule (SG) recruitment of TDP-43 and the effects of mutations of TDP-43 on protein translation. (A) Gene ontology (GO) enrichment analysis of altered expression of genes in RNA-seq data analysis. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of altered expression of genes in RNA-seq data analysis (n = 3 mice per group). (C) Representative immunofluorescence images of TDP-43 (white) and G3BP1 (green) in WT and TDP-43ΔCR HEK293 cells treated with sodium arsenite (500 µM) for the indicated times. DAPI was used to visualize nuclei. PPAR, peroxisome proliferator-activated receptor. (D and E) Quantification of SG-positive cells and TDP-43–positive SGs in WT and TDP-43ΔCR cells treated with sodium arsenite. n = 100–200 cells per group. (F) RT-PCR assay of Sort1 and Atg4b splicing in the brains of 3-mo-old WT and TDPΔCR mice (n = 4 per group). (G) RT-PCR assay of Sort1 and Atg4b splicing in the brains of newborn WT, TDPΔCR+/−, and TDPΔCR+/+ mice (n = 3 per group). (H) Schematic diagram of CRISPR/Cas9 genome editing of the TARDBP gene in HEK293 cells to obtain cells with the deletion of human TDP-43 (TDPKO cells). sgRNA, single-guide RNA. (I) RT-PCR assay of SORT1 and ATG4B splicing in WT, TDP-43ΔCR, and TDP-43KO HEK293 cells. (J) Classification of alternative splicing events in WT and TDPΔCR+/− mice (n = 3 mice per group). A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; MXE, mutually exclusive exons; RI, retained intron; events; SE, skipped exon. (K) Sashimi plots of Tardbp in WT and TDPΔCR+/− mouse brains. Arrowhead indicates the deletion of 60 nt coding 321–340 aa of TDP-43 in TDPΔCR+/− mice (n = 3 per group). (L) Schematic of anisomycin treatment in 3–4-mo-old WT and TDPΔCR+/− mice. (M) Quantification of the marble-burying activity of WT and TDPΔCR+/− mice before and after anisomycin treatment (10 mg/kg; n = 4 mice per group). (N) Representative images and quantification of TDP-43 droplet-like particles formed in the nucleus of HEK293 cells expressing GFP-tagged TDP-43WT, G335D, or FYW-L. (O) Representative immunoblot of coIP between exogenously expressed Strep-Flag–tagged TDP-43 WT, G335D, and FYW-L and endogenous PABPC4, RPS6, and RPL7 in HEK293 cells. (P) Representative immunoblot and quantification of newly synthesized proteins labeled by Click-iT AHA in TDP-43–knockout HEK293 cells expressing TDP-43WT, G335D, or FYW-L. Data are mean ± SEM; one-way ANOVA followed by Tukey’s multiple comparisons test (J, I, N, and P) and two-tailed Student’s t test (D, E, F, and M). *, P < 0.05; **, P < 0.01; and ****, P < 0.0001.
Previous in vitro studies of TDP-43 suggested that the disrupted phase separation ability by the deletion of the CR or mutagenesis of its essential amino acids may encourage the TDP-43 conversion to irreversible aggregates (Conicella et al., 2016; Schmidt and Rohatgi, 2016). In contrast, studies of the LLPS of other disease-associated proteins, such as hnRNPA1 and tau, propose that LLPS acts as an initial step toward pathological protein aggregation (Molliex et al., 2015; Wegmann et al., 2018). While these apparent discrepancies will need to be resolved, our study found no TDP-43 inclusion or related proteinopathy in TDP-43ΔCR mice, indicating that loss of the CR and likely LLPS should not induce TDP-43 aggregation in vivo. Time-course studies of arsenite-induced stress granules found that, compared with WT cells, the development of stress granule formation over the long term remained unchanged in TDP-43ΔCR cells (Fig. S5, C–E), excluding the possibility of TDP-43 inability to form protein aggregates. Notably, TDP-43ΔCR cells exhibited much slower appearance of TDP-43 condensates in response to arsenite stress (Fig. S5, C–E), further implying a likely role of TDP-43 LLPS in stress granule assembly during early stages. NLS is not located in the LCD. Also, TDP-43ΔCR mice are not associated with changed nuclear localization in vivo. While this phenotype together with the unchanged solubility and phosphorylation may imply that CR deletion does not affect TDP-43 subcellular localization and post-translational modification, it will still be interesting to test in further studies whether the enhanced TDP-43 self-interaction is able to induce proteinopathy. Nevertheless, the findings observed here provide an unrecognized possibility that the physiological function of TDP-43 may be regulated by its LLPS independent of protein aggregation, nucleocytoplasmic transport, or post-translational modification.
Splicing repression has been implicated as a major function of TDP-43 (Chen and Wong, 2019). However, TDP-43ΔCR mice did not show changed TDP-43 autoregulation or alternative splicing activity (Fig. S5, F–K). Consistently, the transcriptome also remained largely unaltered (Fig. S5, A and B; and Table S2). These findings strongly indicate that LLPS, at least in vivo, unlikely serves as an indispensable motif to regulate the RNA processing activity of TDP-43. On the basis of observations that global translation was greatly increased in TDP-43ΔCR mice or cells expressing TDP-43 lacking CR, we speculate that translational control may be the major molecular function for TDP-43 LLPS in the cytoplasm. In support of this notion, the short-term treatment of TDP-43ΔCR mice with anisomycin, a potent inhibitor of mRNA translation, can significantly improve their marble-burying performance (Fig. S5, L and M), strongly supporting an important role of translational dysregulation in brain deficits in our TDP-43 mice. Translational dysregulation has been increasingly implicated as a common feature across various major neurodegenerative diseases (Bosco, 2018). While early investigations largely focus on translational repression, whether aberrant translational enhancement by TDP-43 CR loss may contribute to the onset or development of disease at early stages appears to be an intriguing question for future studies.
Our proteomic analysis has identified a number of translation-related proteins that interact with TDP-43 in the cytoplasm. While the association between TDP-43 and most translational factors was found to be greatly increased by TDP-43 CR deletion, TDP-43 lacking CR also contrarily exhibited mildly reduced or unchanged interaction with some other translational factors. On top of these, the TDP-43 and translational factor association is dependent on a very short motif composed of only seven amino acids, which is located in the N-terminus and far away from the CR. Therefore, CR deletion unlikely affects protein interactions as simple as just losing this conserved motif. Instead, based on the facts that TDP-43 subcellular localization or splicing activity is not altered by CR loss and that the deletion of those seven N-terminal amino acids required for protein association is sufficient to completely abolish translational dysregulation caused by TDP-43 CR deletion, these results indicate that the impact of TDP-43 CR deletion on protein interaction and translation should be due to the altered function of LLPS. It is possible that LLPS acts to restrict the accessibility of TDP-43 to different subsets of translation-related proteins and that the loss of the CR increases the pool of TDP-43 available for binding motif recognition. Although it remains to be determined whether there are other cointeracting factors involved, these findings provide a molecular mechanism, at least in part, by which TDP-43 LLPS controls translation. Also, approaches to manipulate the interaction between TDP-43 and translational factors may be used to specifically counteract TDP-43–related translational dysregulation. Although total RNA or ribosomal protein markers remained unchanged, increased ribosomal density or assembly was noted in TDP-43ΔCR mice or cells. This is consistent with the observation that TDP-43 showed increased association with several ribosomal subunits, suggesting that ribosomal assembly may contribute to TDP-43 LLPS-mediated translational control. However, because TDP-43 also interacts with translation elongation and other factors, this is unlikely the only translational mechanism regulated by TDP-43 LLPS. It is highly possible that TDP-43 LLPS dynamically controls translation through protein complexes consisting of TDP-43 and various translational factors at multiple translational steps. Identifying the protein complexes that influence the dedicated translation machinery, specifically by TDP-43 LLPS, is a priority in future studies.
Previous and recent studies have reported severe phase separation defects in the TDP-43 FYW-L mutant but enhanced phase separation in the ALS-associated TDP-43 G335D mutant (Conicella et al., 2020; Jiang et al., 2016; Schmidt et al., 2019). Consistently, we found that, similar to TDPΔCR, the TDP-43 FYW-L mutant with impaired LLPS showed increased interaction with PABPC4, RPS6, and RPL7 and also promoted protein translation in cells. On the contrary, ALS-associated TDP-43 G335D mutant with enhanced LLPS exhibited reduced interaction with PABPC4, RPS6, and RPL7 and inhibited protein translation (Fig. S5, N–P). These findings collectively indicate that LLPS-mediated translational regulation may be a disease-relevant mechanism for TDP-43. However, because it is unlikely that G335D and other ALS-associated TDP-43 mutations only selectively affect its LLPS but no other functionality, further studies are needed to investigate the extent to which LLPS-mediated translational regulation contributes to TDP-43–induced neurotoxicity in disease models.
In conclusion, we have successfully generated TDP-43ΔCR mice, which demonstrate a robust phenotype and pathological changes specifically associated with the brain. In addition to dissecting the physiological function of TDP-43 or TDP-43 LLPS, this novel TDP-43 animal model without premature death or neuronal death makes it possible to cross with established disease models to elucidate potential relationships between TDP-43 and other neurodegenerative pathologies, such as amyloid, tau, and α-synuclein, providing an attractive tool for both physiological and pathological TDP-43 studies. TDP-43 LLPS may regulate its association with PABPC4, RPS6, RPL7, and other factors to influence translation. Therefore, our study also suggests a novel mechanism of translational regulation by TDP-43 LLPS, the dysregulation of which may contribute to disease onset and progression independently of functional motifs RRM1 and RRM2.
Materials and methods
Conservation analyses of TDP-43
Amino acid conservation scores were calculated by Jalview software with default parameters, and the results were plotted as a 5-aa moving average using GraphPad Prism. All the protein sequences from 50 eukaryotic species were automatically extracted from the UniProt database by Jalview software. All the information is listed in Table S3.
Generation of TDPΔCR mice
The targeting vector was generated by a series of PCR and DNA recombinations. In the targeting vector, the 5′ arm harboring exons 3–5 (4.5 kb), the 3′ arm harboring the 3′ UTR of the Tardbp gene (4.6 kb), and the neomycin resistance gene were inserted into the pDTA-down-ABC vector. The linearized targeting vector was electroporated into C57BL/6N embryonic stem cells, and targeted clones were screened by Southern blot analysis using 5′ and 3′ probes generated by PCR using mouse genomic DNA via primers 5′-CTGATACACCAAGAAATTCCAACC-3′, 5′-TAACTTTGATCTGCAAAATGGTCC-3′ and 5′-TAGTTTGACCCAGCAACAAGAC-3′, 5′-TAAATCATAGTTCCTGCTTTCCCAG-3′. Two independent targeted clones were injected into C57BL/6N mouse blastocysts to generate TDPΔCR chimeric mice. For genotyping of TDPΔCR mice, the following set of primers was used: 5′-GGCTGCTGGTTGGGTTTAAGTAAATT-3′ and 5′-GGCACCAGAATTAGAACCACTGTAG-3′. Mouse surgery and procedures were performed according to National Institutes of Health guidelines and were approved by the institutional animal care and use committee at Case Western Reserve University and the University of Nebraska Medical Center.
Behavior tests
3–4-mo-old TDPΔCR mice and age-matched littermates were subjected to behavior assessment batteries. Two separate testing batteries were performed with independent but age-matched cohort groups. One battery includes a rotarod (body coordination), grip strength (muscle strength), an open field (locomotor activity and anxiety), object recognition (short-term and long-term nonspatial memory), an elevated plus maze (anxiety), a light/dark box (anxiety-like behavior), and tail suspension (depression-like behavior). Individual mice were tested for these behavioral tasks on each test day in the following order: days 1–3 for the rotarod and grip strength tests, days 4–6 for the open field test and object recognition test, day 7 for the elevated plus maze, day 8 for the light/dark box test, and day 9 for the tail suspension test. The other battery contains tests involving marble burying (repetitive, compulsion-like behavior), nestlet shredding (repetitive, compulsion-like behavior), a Y-maze (working memory), a Barnes maze (long-term spatial memory), and fear conditioning (emotional memory). These behavioral tasks were performed in the following order: days 1–4 for the marble-burying and nestlet-shredding tests, day 5 for the Y-maze test, days 5–9 for the Barnes maze test, and days 10 and 11 for the fear-conditioning test. All tests were performed at the Case Western Reserve University Mouse Behavioral Phenotyping Core, with the investigator blinded to mouse genotype.
Rotarod test
Each mouse first underwent three trials per day for 3 d. During the training period, each mouse was placed on the rotarod, where cylinder speed was gradually increased from 4 rpm to 12 rpm for each trial. On the testing day, the rotarod was set to accelerating mode (4–40 rpm over 5 min), and maximal latency to fall-off was collected for statistical analysis.
Grip strength test
The muscular strength of mice was measured by a grip strength test meter (Bioseb). For the forelimb test, the two forepaws of a mouse were placed on a bar, and the mouse’s tail was pulled back. For the hindlimb test, the two forepaws were placed on a grid that was held by the examiner’s left hand, and the two hindpaws of the mouse were placed on a bar that was connected to the machine. The single best recorded value out of five trials was used for statistical analysis.
Open field test
The open field test consisted of a 50-cm-long square plastic apparatus closed with 50-cm-high walls, and activity was recorded using ANY-maze video tracking software (Stoelting). The field was digitally divided into an inner area (30 × 30 cm) and the periphery (10-cm-wide gallery) using ANY-maze software. Data were collected continually for 10 min, and the distance traveled (in meters), velocity (in meters per second), time spent immobile (>2 s of nonlocomotion), and time spent in the inner area were all recorded and scored automatically.
Object recognition test
The object recognition test apparatus was a 50-cm-long square plastic apparatus closed with 50-cm-high walls. Each mouse was placed in an apparatus for 10 min. Then, two identical objects were placed at the corner of the apparatus (T1). The mice were allowed to investigate these objects for 5 min. This session was followed by a 1.5-h delay during which the animals were returned to their home cages with their cagemates. After the delay, the animals performed a 5-min dissimilar stimulus session (T2). In this session, an object that was presented in T1 and another object that was unfamiliar were placed in the test cages. The objects were made of hard plastic and/or metal with apparently different shapes. The total amount of time spent to sniff and contact each object was recorded and scored using fully automated ANY-maze video tracking software.
Elevated plus maze test
The elevated plus maze test apparatus was made of stainless steel and consists of four arms (two open without walls and two enclosed by 15.25-cm-high walls) measuring 30 cm long and 5 cm wide. Each arm of the maze was attached to sturdy metal legs such that the maze was elevated 40 cm off the ground. There were two 32-W fluorescent lights on the top of the apparatus for consistent illumination. Animals were placed at the junction of the open and closed arms, facing the open arm. After 5 min of exploration in the maze, animals were removed from the elevated plus maze. The time spent on the open arm and the closed arm was recorded using fully automated ANY-maze video tracking software.
Light/dark box test
The light/dark box apparatus consisted of a box (42 × 21 × 25 cm) divided into a small (one-third) dark compartment and a large (two-thirds) illuminated compartment. A restricted opening 3 cm high by 4 cm wide connected the two chambers. The apparatus was placed in an isolated room away from any external interference and noise with a low-intensity white light source. The mice were allowed to move freely between the two chambers for 10 min. The time spent in the light and dark compartments was recorded using fully automated ANY-maze video tracking software.
Tail suspension test
Each mouse was suspended by its tail with adhesive tape for 6 min and was video recorded, which was later carefully scored by two independent observers for latency to immobility and total immobility time. Mice were considered immobile only when hung passively and motionless for at least 2 s.
Marble-burying test
The marble-burying test was performed in a standard polycarbonate mouse cage (25 × 16 × 13 cm) with fitted filter-top covers. Fresh mouse bedding materials were added to each cage to a depth of 5 cm. Then standard glass toy marbles (15-mm diameter, 5.2 g in weight) were gently placed on the surface of the bedding in five rows of four marbles. The testing animals were carefully placed into a corner of the cage containing marbles. After 30 min of exploration, mice were removed from the testing cages. Task scorers (two or three) blinded to the genotype of the mouse counted the number of marbles buried. A marble was scored as buried if two-thirds of its surface area was covered by bedding. Percentage scores (buried marbles/20) were calculated for each mouse.
Nestlet-shredding test
The cotton fiber nestlets (5 × 5 cm, 5 mm thick) were weighed on an analytical balance, and one nestlet was placed in each test cage. Animals were placed into a cage containing a single, preweighed nestlet to explore for 6 h. After testing, the intact nestlet materials were removed from the cage with forceps and allowed to dry overnight. The remaining unshredded nestlet was weighed, and this weight was divided by the starting weight to calculate the percentage of nestlet shredded.
Y-maze test
Mice were placed in a Plexiglass Y maze (with arms 60 cm in length) and allowed to explore the maze freely for 10 min. When put in the Y maze, the mice were recorded using the ANY-maze tracking system, and the time and frequency in a spontaneous alteration ratio were counted automatically.
Barnes maze test
The Barnes maze test apparatus consisted of a white acrylic circular disk 92 cm in diameter with 20 equally spaced holes (5 cm in diameter) located 2 cm from the edge of the disk. The maze was illuminated by two 60-W lamps to provide an aversive, bright disk surface. An acrylic escape box (7 × 7 × 5 cm) could be fitted under any of the holes in the maze. The maze was raised 30 cm from the floor and rested on a pedestal that enabled it to be rotated 360° on a horizontal plane. An acrylic start bin with 15-cm diameter and 15-cm height was used. Trials were recorded using a webcam and analyzed by automated ANY-maze video tracking software. Each trial began with the start bin positioned in the center of the maze with the mouse placed inside. Each mouse was allowed to explore the maze freely for 2 min. After the mouse entered the escape hole, the mouse was left in the escape box for 90 s before being returned to its home cage. If the mouse did not enter the escape box within 120 s, it was gently picked up by the experimenter and placed over the target hole and allowed to enter the escape box. After each trial, the maze and escape box were cleaned carefully with a 10% alcohol solution to dissipate odor cues and provide a standard olfactory context. Five training sessions consisting of two trials each were run on subsequent days, and escape latencies were measured.
Fear-conditioning test
All animals were placed in a conditioning box (Med Associates) and trained to associate a tone (white noise, 80 dB for 30 s, conditioned stimulus) with an electrical shock (0.5 mA for 1 s, unconditioned stimulus). This procedure was repeated four times with 120-s accumulation and a 60-s interstimulus interval. The tone and shock were coterminated. At the end of the trial, the animals were taken out and then placed back in the box 24 h later to evaluate their learned aversion for an environment associated with the shock (context-dependent fear). To this end, all animals were placed in the same box in which they were trained for the duration of 6 min, and freezing behavior in the absence of tone or aversive stimulus was measured. The animals were then removed, and the context was changed so that the animals could no longer recognize the chamber in which they had been trained. 2 h after the animals were tested for contextual fear conditioning, they were reintroduced into the now contextually altered box (shape, lighting, and odor [vanilla essence]), and freezing behavior was measured during the first 2 min to verify that the animals did not recognize the context. After 2 min (no-cue period), the tone (30 s, 5 kHz, 80 dB) was delivered 10 times without unconditional stimulus exposure in 60-s intertone interstimulus intervals, and freezing behavior was measured to determine cue-dependent fear conditioning.
Golgi staining and data analysis
Brains were harvested from 3-mo-old mice and stained using the FD Rapid GolgiStain kit (FD NeuroTechnologies). Briefly, brains were rinsed with double-distilled water and then immersed in a 1:1 mixture of FD Solution A:B for 2 wk at RT in the dark. Brains were then transferred to FD Solution C and kept in the dark at RT for 72 h. Solution C was replaced after the first 24 h. In preparation for freezing, individual brains were placed in Peel-A-Way disposable embedding molds (VWR) and immersed in tissue-freezing medium (Triangle Biomedical Sciences). Brains were kept at −80°C until sectioning. Cryosectioning was performed on a Leica CM 3050 S at −22°C. Coronal sections of 100-µm thickness were cut and transferred to gelatin-coated slides (Lab Scientific) on small drops of FD Solution C. After sections were allowed to dry at RT in the dark overnight, slides were then stained with a Solution D, Solution E, and distilled water mixture. Sections were rinsed in double-distilled water two times and dehydrated in 50%, 75%, 95%, and 100% ethanol. Permount (Thermo Fisher Scientific) was used for coverslipping. For each mouse, three independent coronal sections containing the hippocampus were imaged. Secondary and tertiary dendrites of these neurons were selected for analysis. Z-stacks of Golgi-stained dendrites were taken at low magnification or 63× magnification from each mouse.
Electrophysiology
Mouse brains were rapidly removed and placed in ice-cold cutting solution composed of 250 mM sucrose, 25 mM D-glucose, 2.5 mM KCl, 24 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM CaCl2, 1.5 mM MgSO4, and 1 mM kynurenic acid (pH 7.3–7.4). Coronal slices (400 µm) containing the hippocampus were prepared using a vibratome (Leica VT1200). Brain slices were transferred to a chamber containing artificial cerebrospinal fluid (aCSF; bubbled with 95% O2 and 5% CO2) composed of 124 mM NaCl, 3 mM KCl, 24 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM CaCl2, 1 mM MgSO4, and 10 mM glucose (pH 7.3–7.4) and allowed to recover for 30 min at 37°C followed by RT for at least 1 h. For recording, slices were maintained at 32°C with gravity-fed oxygenated aCSF (flow rate 2–3 ml/min). All data were acquired using an Axopatch 200B amplifier, Digidata 1440A, and pClamp 10.6 software (Molecular Devices). Local fEPSPs were recorded with borosilicate glass electrodes (1.5–5 MΩ, filled with aCSF containing 100 µM picrotoxin) from the stratum radiatum of CA1 and evoked by electrical stimulation (100-µs duration, every 20 s) of the Schaffer collateral fibers. I/O curves were generated for each slice with stimulation intensity ascending from 0 to 1.0 mA, with each step being 0.1 mA. Based on I/O curves, the stimulation intensity that evoked 50% of the maximum response (between 0.2 and 0.3 mA) was chosen to conduct experiments of synaptic plasticity. Synaptic field potentials were low-pass filtered at 1 kHz and digitally sampled at 50 kHz. LTP was induced by TBS consisting of five trains of four pulse bursts at 200 Hz separated by 200 ms, repeated six times with an intertrain interval of 10 s. The peak of the evoked fEPSPs was measured and normalized to the averaged baseline 5 min before TBS. Data were analyzed offline using Clampex 10.6 software. For statistical analysis, collected data were further processed in GraphPad Prism 8. Graphed data are displayed as mean ± SEM.
SUnSET
To measure the rate of protein synthesis, puromycin (Sigma-Aldrich) was stereotactically injected into the left ventricle of the mouse brain. For stereotactic injection, mice were anesthetized with isoflurane and immobilized using the stereotactic frame equipped with a heating blanket to maintain body temperature throughout the procedure (RWD Life Science). After hair removal and the cleaning of the shaved area with betadine and alcohol, mice were administered an injection of bupivacaine/lidocaine, and a small incision was made to expose the skull surface. One small hole was drilled into the skull (relative to bregma: anteroposterior −0.5 mm, medial lateral 0.9 mm) followed by injection of 2 µl puromycin (10 mg/ml) using Hamilton syringes into the left ventricle at a dorsal ventral position of 2.5 mm. The injection speed was pump controlled at 0.2 µl min−1. The needle was left in place for 5 min before it was slowly withdrawn. 1 h after injection, the brain was immediately removed, and hippocampal tissue was dissected for further immunoblot analysis or fixed in 10% formalin for immunofluorescence analysis. For the labeling of newly synthesized proteins in cultured primary neurons, puromycin (10 µg/ml) was added to the culture medium for 15 min. Cell lysates were subjected to immunoblot analysis.
Click-iT AHA protein synthesis assay
Click-iT AHA was used to label newly synthesized protein following the manufacturer’s protocol. After 12-h culture in 6-well plates, HEK293 cells were cultured in methionine-free DMEM for 60 min. A 50 µM concentration of Click-iT AHA was added into the culture medium and incubated at 37°C and 5% CO2 for 2 h. Cells were harvested by centrifugation at 400 ×g for 5 min at RT. After three washes with PBS, cells were lysed in 50 mM Tris-HCl, pH 8.0, with 1% SDS containing protease and phosphatase inhibitors. The lysate was sonicated with a probe sonicator to solubilize the proteins and disperse the DNA and centrifuged at 18,000 ×g at 4°C for 5 min. 100 µg protein sample in 50 mM Tris-HCl, pH 8.0, with 1% SDS was subjected to the click reaction as follows. Click reaction buffers were prepared by adding reagents in the following order with vortex mixing between the addition of each reagent: capture reagent (Biotin Alkyne 40 µM), Click-iT reaction buffer, CuSO4 (1 mM), Click-iT reaction buffer additive 1 solution, and Click-iT reaction buffer additive 2 solution. The mixture was rotated end over end for 20 min using a rotator. Subsequently, proteins were precipitated (chloroform/methanol, 0.25:1, relative to the sample volume). The precipitated proteins were pelleted by centrifugation at 14,000 rpm for 5 min, washed with methanol, and air dried for 10 min. The pellets were then resuspended in 1× SDS sample buffer and boiled for 10 min, and 20 µg of proteins were loaded onto 10% SDS-PAGE gels.
Generation of TDP-43ΔCR and TDP-43–knockout cells
TDP-43ΔCR cells were generated from HEK293 cells with a CRISPR/Cas9 system designed in our laboratory. pSpCas9(BB)-2A-Puro (PX459) version 2.0 was a gift from Zhang Feng at the Broad Institute of MIT and Harvard (Addgene; Ran et al., 2013). All single-guide RNA targeting sequences were predicted as first hits using the CRISPR guide design website (https://benchling.com). HEK293 cells were first transfected with two PX459 vectors, which targeted 5′-AATCAAGGTAGTAATATGGG-3′ and 5′-TCGGGTAATAACCAAAACCA-3′ of the TARDBP gene to delete the coding sequence of TDP-43 from aa 307 to 355. After puromycin selection, cells were diluted and seeded in 96-well plates at 1 cell/well to isolate monoclonal cells with truncated TDP-43ΔCR expression, as determined by PCR and Western blot analysis.
To generate TDP-43–knockout and rescued cell lines, HEK293 cells were first transfected with PX459 vectors, which targeted 5′-ATTCTGCATGCCCCAGATGC-3′ of the TARDBP gene. After puromycin selection, cells were diluted and seeded in 96-well plates at 1 cell/well to isolate monoclonal cells without TDP-43 expression, as determined by Western blot analysis. The coding sequences of TDP-43 WT or mutations with a resistance sequence to single-guide RNA 5′-ATTCTCCACGCTCCTGACGCAGGA-3′ (codon optimization by online tool at https://www.idtdna.com/pages) were cloned into the plenti6-LVP lentiviral vector (Thermo Fisher Scientific). Lentiviruses were generated and used to transduce TDP-43–knockout cells. Briefly, plenti6-LVP plasmids were cotransfected with lentiviral packaging mix into 293 cells using TransIT-293 Transfection Reagent. Supernatant containing lentivirus was harvested, and cellular debris was removed by centrifugation. The crude lentivirus mixture was added to the HEK293 cells. 3 d after transduction, single-cell clones were verified by Western blot analysis to validate the expression of exogenous TDP-43 and mutations.
Immunoblotting, immunocytochemistry, and immunofluorescence
Mouse brain tissues or HEK293 cells were homogenized or lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin; Cell Signaling Technology) containing 1 mM PMSF (EMD Millipore), protease inhibitor cocktail (Sigma-Aldrich), and phosphatase inhibitor cocktail (Sigma-Aldrich). The lysate was centrifuged at 14,000 ×g for 15 min at 4°C. 20 µg proteins were separated by 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Following blocking with 10% nonfat dry milk, primary and secondary antibodies were applied, and the blots were developed with Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore). Images were taken using a ChemiDoc Touch Imager (Bio-Rad Laboratories). Primary antibodies used in this study are listed in Table S3.
Immunocytochemistry was performed according to the peroxidase-antiperoxidase protocol (Wang et al., 2015). Briefly, paraffin-embedded brain tissue sections were first deparaffinized in xylene and rehydrated in graded ethanol and then incubated in Tris-buffered saline (50 mM Tris-HCl and 150 mM NaCl, pH 7.6) for 10 min before antigen retrieval in 1× antigen decloaker (Biocare Medical). Sections were rinsed with distilled water, incubated in Tris-buffered saline for 10 min, and blocked with 10% normal goat serum (NGS) in Tris-buffered saline at RT for 30 min. Tissue sections were further incubated with primary antibodies in Tris-buffered saline containing 1% NGS overnight at 4°C and immunostained by the peroxidase-antiperoxidase–based method.
For immunofluorescence staining, deparaffinized and rehydrated tissue sections or cells growing on the coverslips were washed three times with distilled water, and antigen retrieval was performed. The sections were then blocked with 10% NGS for 30 min at RT and incubated with primary antibodies in PBS containing 1% NGS overnight at 4°C. After three washes with PBS, the sections were incubated in 10% NGS for 10 min and then with Alexa Fluor–conjugated secondary antibody (Life Technologies; 1:300) for 2 h at RT in the dark. Finally, the sections were rinsed three times with PBS, stained with DAPI for nuclei, washed again with PBS three times, and mounted with Fluoromount-G mounting medium (SouthernBiotech).
Confocal microscopy, fluorescence microscopy, and EM
Confocal images were captured as serial sections along the Z-axis on a Zeiss LSM710 confocal microscope with a 63×/1.4 NA oil objective. The acquired images were processed with deconvolution (regularized inverse filter) by ZEN software. Then, the sharp regions from the individual sections of Z-stack images were combined to form extended depth of focus single images. The focus of all the z-positions were calculated to one extended depth of focus image, which enabled the display of a considerably larger depth of field than is possible with a single focus. For live-cell imaging using a Zeiss LSM710 confocal microscope (related to Fig. 4, E, G, and H), cells transfected with GFP-tagged TDP-43 or mutations were maintained on a stage-top incubator (37°C, 5% CO2). Fluorescent signaling was monitored at 5-s intervals. All images were processed using Zen software and Fiji (ImageJ).
Fluorescence images from brain sections, cultured neurons, and HEK293 cells were captured with a Zeiss Celldiscoverer 7 automated microscope (controlled by Zen software; Zeiss). For live-cell time-lapse imaging, HEK293 cells were seeded into a 35-mm glass-bottomed dish and transfected 24 h after seeding using Lipofectamine 2000 with 0.5 µg GFP-TDP-43, GFP-TDP-43ΔCR, GFP-TDP-43ΔNLS, and GFP-TDP-43ΔNLSΔCR constructs. 24 h after transfection, the culture dish was placed into the Celldiscoverer 7 system with a stage-top incubator (37°C, 5% CO2). All the images were captured by the water immersion objective at 100× magnification.
For transmission EM, samples were freshly dissected and processed. Small pieces of cortical tissue were fixed by immersion in triple-aldehyde–DMSO. After being rinsed in distilled water, they were postfixed in ferrocyanide-reduced osmium tetroxide. Another water rinse was followed by an overnight soak in acidified uranyl acetate. After being rinsed in distilled water again, the tissue blocks were dehydrated in ascending concentrations of ethanol, passed through propylene oxide, and embedded in Poly/Bed resin. Thin sections were sequentially stained with acidified uranyl acetate followed by a modification of Sato’s triple-lead stain and examined in an FEI Tecnai Spirit (T12) with a Gatan US4000 4k × 4k charge-coupled device. All EM images were obtained and quantified blindly and independently by investigators without knowledge of the samples.
Neuron, microglia, and astrocyte quantification
Digitized images acquired with all the parameters (contrast and brightness) kept constant were transformed into TIFF files and thresholded using Image-Pro Plus 6.0 software. Neuronal cell number counts of NeuN-positive cells in the cortices, hippocampi, and spinal cords were automatically performed using Image-Pro Plus 6.0 software, and cell numbers were calculated per mm2. Microglia and astrocytes were counted automatically after setting the brightness threshold. Quantitative comparisons were performed on sections processed at the same time with the same batch of solutions. Detailed mouse age and gender information for each experiment is presented in specific figure legends.
IP
pcDNA3.1+ (Invitrogen) plasmid was modified to express Strep-Flag–tagged proteins at their N termini containing the following sequence 5′-ATGAGATCTGACTACAAGGACGACGACGACAAGGGATCTGATTATAAAGATGACGATGATAAGGGATCTGACTACAAAGACGATGATGACAAAGGATCTGATTACAAGGATGATGACGATAAAGGATCTGCATGGAGCCATCCCCAGTTTGAGAAAGGTGGCGGATCAGGCGGGGGTTCTGGTGGATCTGCATGGAGCCATCCCCAGTTTGAGAAAGGTGGCGGATCAGGCGGGGGTTCTGGT-3′ (pcDNA-Strep-Flag). The cDNA of full-length or truncated human TDP-43 was inserted into pcDNA-Strep-Flag vectors. 10 µg plasmid was used to transfect one 10-cm dish of HEK293 cells with TransIT-293 Transfection Reagent (Mirus). Cells were collected at 24 h after transfection and lysed with lysis buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1% NP-40, pH 8.0) containing 1 mM PMSF (Millipore), protease inhibitor cocktail (Sigma-Aldrich), and phosphatase inhibitor cocktail (Sigma-Aldrich). The lysate was centrifuged at 14,000 ×g for 15 min at 4°C. Supernatant was incubated with MagStrep Type 3 XT beads (IBA Lifesciences) overnight at 4°C. Beads were washed three times with lysis buffer and eluted with BXT buffer (IBA Lifesciences) overnight at 4°C. The eluted proteins were subjected to Western blot or MS analysis.
For the endogenous TDP-43 IP from mouse brain lysates, anti–TDP-43 antibodies were conjugated to Dynabeads M-270 Epoxy following the manufacturer’s instructions. Briefly, lyophilized beads were resuspended in 0.1 M sodium phosphate buffer (pH 7.4) and vortexed for 30 s After being washed with sodium phosphate buffer three times, anti–TDP-43 antibodies and the same volume of 3 M ammonium sulfate were added to the beads. After 24-h incubation at 37°C with slow tilt rotation, the coated beads were washed four times with PBS, pH 7.4. Mouse brain lysates in lysis buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1% NP-40, pH 8.0) containing 1 mM PMSF (EMD Millipore), protease inhibitor cocktail (Sigma-Aldrich), and phosphatase inhibitor cocktail (Sigma-Aldrich) were added to coated beads and incubated for 12 h at 4°C. Then the beads were washed with PBS four times (5 min each time) at RT, followed by elution with 0.1 M citrate, pH 3.1. The eluted proteins were neutralized by adding the same volume of 0.25 M Tris buffer, pH 11.0, and subjected to Western blot analysis.
MS analysis
A proteomic approach was used to identify the specific binding partners of cytosolic TDP-43 WT and TDP-43ΔCR according to a previously published protocol (Kiser et al., 2009). Briefly, TDP-43 and TDP-43ΔCR without Strep-tag, Strep-TDP-43, and Strep-TDP-43ΔCR were transfected into TDP-43–knockout HEK293 cells with TransIT-293 Transfection Reagent (Mirus). After IP with MagStrep Type 3 XT beads, proteins were eluted with BXT buffer (IBA Lifesciences) and subjected to SDS-PAGE ∼1 cm into a 4–20% Mini-PROTEAN TGX precast protein gel (Bio-Rad Laboratories). The top 1 cm of the gel was then excised and in-gel digested by Lys-C protease (Shevchenko et al., 2006). Following digestion, liquid chromatography tandem MS (LC-MS/MS) was performed using the Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). The LC column used was an Acclaim PepMap rapid separation LC reverse-phase nanocolumn (75 µm × 15 cm, C18, 2 µm, 100 Å; Thermo Fisher Scientific), and peptides were chromatographed with a linear gradient of acetonitrile from 2% to 35% in aqueous 0.1% formic acid over 90 min at 300 nl/min. The eluent was directly introduced into the mass spectrometer operated in data-dependent MS to MS/MS switching mode, with the 15 most intense ions in each MS scan subjected to MS/MS. Full MS scanning was performed at 70,000 resolution between mass-to-charge ratios 380 and 1,300, and MS/MS spectra were collected at 17,500 resolution. High-energy collisional dissociation was performed at a normalized collision energy of 25%. The precursor ion masses were dynamically excluded from MS/MS analyses for a duration of 20 s. Ions with the charge state of 1 and >6 were excluded from MS/MS analyses. Proteins were identified by comparing all of the experimental peptide MS/MS spectra against the UniProt human database using the Andromeda search engine integrated into MaxQuant version 1.6.3.3 software (Cox and Mann, 2008; Cox et al., 2011). Carbamidomethylation of cysteine was set as a fixed modification, whereas variable modifications included oxidation of methionine to methionine sulfoxide and acetylation of N-terminal amino groups. For peptide/protein identification, strict Lys-C specificity was applied, the minimum peptide length was set to 7, the maximum missed cleavage was set to 2, and the cutoff false discovery rate was set to 0.01. Matching between runs (match time window, 0.7 min; alignment time window, 20 min) and label-free quantitation (LFQ) options were enabled. The LFQ minimum ratio count was set to 2. The remaining parameters were kept as the defaults. Protein quantitation was accomplished using Perseus (Tyanova et al., 2016). LFQ values were log2 transformed, and missing values were imputed using the “Replace missing value from normal distribution” function on the entire matrix using default parameters.
Sequential extraction of soluble and insoluble proteins from mouse brain
Fresh mouse brain tissues were solubilized in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin) with protease and phosphatase inhibitors on ice for 30 min, then centrifuged at 14,000 ×g, and the supernatants were collected as RIPA-soluble fractions. The pellet was washed with RIPA buffer twice and further solubilized in 1% SDS buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% SDS) with protease and phosphatase inhibitors on ice for 30 min. After centrifugation at 14,000 ×g, the supernatants were collected as SDS-soluble fractions. The resulting pellets were solubilized in urea buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, 5 mM magnesium acetate, pH 8.5) at RT. After centrifugation at 100,000 ×g for 1 h, the supernatant was collected as a urea-soluble fraction. The protein concentration was determined by bicinchoninic acid assay or Pierce 660-nm protein assay. Equal amounts (20 µg) of protein were separated by 10% SDS-PAGE.
RNA-sequencing (RNA-seq) and data analysis
For mRNA-seq analyses, whole-brain tissues prepared from WT and TDPΔCR+/− mice were submitted to Novogene for library construction and sequencing. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s instructions. Paired-end sequencing libraries were prepared by Novogene using the TruSeq Stranded Total Sample Preparation kit (Illumina), followed by quality control, cluster generation, and sequencing on the Illumina HiSeq 2000 platform. Sequence alignment was performed by STAR software to align for RNA-seq data analysis and for the junction reads against the UCSC mm10 assembly. Expression values were calculated with featureCounts version 1.4.6-p2, differential expression analysis was determined by DESeq2 (Love et al., 2014), and the downstream statistical analyses and plots were made in R (version 3.1.1; http://www.r-project.org/). Pathway overrepresentation analysis was performed using R package ReactomePA (Subramanian et al., 2005). Classifying and analyzing differential alternative splicing from RNA-seq data were performed by rMATS (replicate multivariate analysis of transcript splicing) and Integrative Genomics Viewer (version 2.6.0) software. rMATS analyzes skipped exon, alternative 5′ splice site, alternative 3′ splice site, mutually exclusive exons, and retained intron events. Every alternative splicing event corresponded to two isoforms: the exon inclusion isoform and the exon skipping isoform. Statistical analysis of the expression of quantity divided by the effective length can be performed to derive the adjusted expression quantity when calculating the ratio of the exon inclusion isoform and the two-isoform whole expression quantity in differential significance analysis. The threshold of the significant difference of alternative splicing analysis is set as false discovery rate <0.05.
RNA extraction and quantitative RT-PCR (RT-qPCR)
Total RNA was extracted from tissues and cells using the Qiagen All-Prep DNA/RNA/miRNA Universal kit (Qiagen) according to the manufacturer’s protocols. cDNA was synthesized by using High-Capacity cDNA Reverse Transcription kits (Applied Biosystems) with a random primer. qPCR was performed using SYBR Premix Ex Taq (Takara Bio) in a CFX96 real-time PCR system (Bio-Rad Laboratories) following the manufacturer’s protocol. Thermocycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. mRNA levels of targeted protein or ribosomal RNA were normalized to GAPDH using comparative cycle threshold analysis of qPCR data. The primers used in this study are shown in Table S3.
Embryonic primary hippocampal neuron and HEK293 cell culture and transfection
Primary hippocampal neurons were isolated from hippocampal tissue of embryonic day 18 mouse embryos from female TDPΔCR+/− mice mated with male TDPΔCR+/− mice. Briefly, mouse brains were dissected out in HBSS (Life Technologies) and stored in Hibernate E (BrainBits) supplemented with 2% B27 (Life Technologies). Under a dissecting microscope, the meninges were removed completely with fine forceps, and the hippocampus was dissected out. The hippocampus was then digested in 0.25% trypsin for 15 min at RT, followed by brief incubation in Opti-MEM (Life Technologies) supplemented with 10% FBS and 50 U/ml DNase I (Invitrogen). The digested hippocampus was further dissociated by gentle trituration with a pipette until the cell suspension was homogenous and no large pieces of tissue remained visible. Hippocampal neurons were finally collected and seeded onto poly-D-lysine– and laminin-coated coverslips or chamber slides (BD Biosciences) and cultured in neurobasal medium with 1% GlutaMAX (Gibco). Transfection of primary cultured neurons was performed with Lipofectamine 2000 following the manufacturer’s instructions.
HEK293 cells from the American Type Culture Collection were grown in DMEM (Sigma-Aldrich) supplemented with 10% FBS and 1% penicillin-streptomycin in 5% CO2 in a humid incubator at 37°C. HEK293 cells were tested and confirmed to be free of mycoplasma contamination. HEK293 cells were transfected with TransIT-293 Transfection Reagent according to the manufacturer’s protocol.
OptiPrep gradient ultracentrifuge
Mouse brain or HEK293 cells were resuspended in buffer A (0.25 M sucrose, 1 mM EDTA, 10 mM Hepes-NaOH, pH 7.4) with protease inhibitor cocktail and phosphatase inhibitor cocktail and homogenized by Dounce homogenization 20 times for buffer A and 20 times for buffer B. After centrifugation (700 ×g, 5 min, at 4°C), the pellets were collected as crude nuclear fractions. The crude nuclear fraction was further washed with buffer A three times. The same amounts of pellets resuspended in buffer A were separated by centrifugation using an Optima TLX ultracentrifuge (Beckman Coulter) with a swinging bucket rotor (SW50.1) at 200,000 ×g for 2.5 h in a 2.5–50% continuous gradient of OptiPrep (Sigma-Aldrich) in buffer A. After centrifugation, the OptiPrep gradient was fractionated in 25 fractions (150 µl/fraction) and subjected to Western blot analysis.
Anisomycin treatment
Anisomycin (Sigma-Aldrich) was used to impair protein synthesis. The drug powder was dissolved by adding several drops of 1 M HCl. The pH value was adjusted to 7.0 with 1 M NaOH. The final concentration was achieved by adding 0.9% saline. 3-mo-old WT or TDPΔCR+/− mice received anisomycin at a dose of 10 mg/kg in a volume of 100 µl by subcutaneous injection for 3 d (once per day). The behavior tests were performed before injection and 1 d after the last injection.
Ribosome profiling
Mouse brain tissues or HEK293 cells were lysed with lysis buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1 mM DTT, 100 µg/ml cycloheximide, 0.1% Triton X-100) with protease inhibitor cocktail and phosphatase inhibitor cocktail. Total lysate was clarified by centrifugation at 15,000 ×g for 5 min at 4°C, and 50 OD (at 254 nm) supernatant was loaded onto a continuous 25–35% sucrose gradient in 10 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2. After ultracentrifugation at 4°C in an SW50.1 Beckman rotor at 200,000 ×g for 2 h, the tubes were loaded into the tube holder of the gradient fractionator. After calibration of the UV monitor (Bio-Rad Laboratories), the fractions were collected and subjected to Western blot analysis.
SunTag fluorescence imaging
HEK293 WT and TDP-43ΔCR cells were seeded in a 35-mm glass-bottomed dish and transfected with SunTag labeling plasmids, including pcDNA4TO-24xGCN4_v4-BFP-24xPP7 (Addgene identifier 74929), pHR-scFv-GCN4-sfGFP-GB1-NLS (Addgene identifier 60906), and pHR-tdPP7-3xmCherry (Addgene identifier 74926). 24 h after transfection, the expression of the reporter gene was induced with 1 µg/ml doxycycline (Sigma-Aldrich) for 1 h. Then the culture dish was placed into the Celldiscoverer 7 system with a stage-top incubator (37°C, 5% CO2). Camera exposure times were set to 150 ms for both GFP and mCherry channels. Fluorescence was monitored at 5-s intervals for the times indicated in the figure legends.
FRAP analysis
FRAP experiments were performed on a Zeiss LSM710 confocal microscope with a 63×/1.4 NA oil objective. The intensity of the GFP signal is controlled in the detection range by changing the laser power, digital gain, and offset. Following five baseline images, bleaching was conducted by a 488-nm line from an argon laser at 100% intensity with 200 iterations. Fluorescence recovery was monitored at 1-s intervals for 5 min. During the experiment, cells were maintained in a stage-top incubator (37°C, 5% CO2). The images were analyzed by Zen software, and relative fluorescence intensity was plotted in GraphPad Prism 8 software.
Blue light illumination
After PCR amplification, Cry2oligo (Addgene identifier 60032) and TDP-43/TDP-43ΔCR were cloned into pcDNA3.1-cmCherry (generated in our laboratory) by Gibson assembly. HEK293 cells were seeded onto coverslips coated with poly-D-lysine (50 µg/ml; Sigma-Aldrich) in a 24-well plate and allowed to incubate overnight before transfection with 100 ng of DNA. 24 h after transfection, cells were exposed to blue light using light-emitting diode arrays designed to fit the plate dimensions. Individual light-emitting diodes were positioned ∼3.0 cm above the 24-well plate to provide 465-nm light stimulation to the transfected cells. All manipulations of cells expressing optogenetic constructs were performed under red lamp illumination to avoid ambient light exposure.
Statistical analysis
Statistical analyses were performed with Student’s t test, one-way ANOVA, and two-way ANOVA by GraphPad Prism 8 software (GraphPad Software). Detailed information about statistical analysis for each experiment is presented in specific figure legends. Data are presented as mean ± SEM, and n represents the number per experiment. P < 0.05 was considered to be statistically significant. Data distribution was assumed to be normal, but this was not formally tested.
Accession numbers
The raw data for the high-throughput sequencing results have been deposited in the National Center for Biotechnology Information Sequence Read Archive database with accession number PRJNA748406. MS raw files and MaxQuant output results have been deposited to the ProteomeXchange Consortium via the Proteomics Identifications Database partner repository with the dataset identifier PXD027494.
Online supplemental material
Fig. S1 (related to Figs. 1 and 2) shows the characterization of TDPΔCR mice. Fig. S2 (related to Fig. 3) shows the absence of TDP-43 proteinopathy, gliosis, or neurodegeneration in TDPΔCR mice. Fig. S3 (related to Figs. 4 and 5) shows the impact of ΔCR on TDP-43 LLPS, subcellular organelles, ribosome assembly, and protein translation. Fig. S4 (related to Fig. 6) shows the identification of TDP-43 binding sites for translational factors. Fig. S5 shows the impact of ΔCR on gene expression, arsenite-induced stress granule formation, and TDP-43–mediated RNA splicing. Fig. S5 also shows the rescue effect of anisomycin in TDPΔCR mice and the study of two TDP-43 mutants with altered LLPS. Video 1 shows the marble-burying performance of WT and TDPΔCR mice. Table S1 lists TDP-43WT– and TDP-43ΔCR–associated proteins identified by MS. Table S2 lists genes with significantly changed expression in TDPΔCR+/− mice. Table S3 lists protein identifiers used for TDP-43 sequence alignment, antibodies, and RT-PCR primers used in this study.
Supplementary Material
lists TDP-43WT– and TDP-43ΔCR–associated proteins identified by MS.
lists identification of genes with significantly changed expression in TDPΔCR+/− mice. The thresholds were set as P < 0.05 and Padj < 0.05.
lists protein identification used for TDP-43 sequence alignment, antibodies, and RT-PCR primers used in this study. IEM, immunoelectron microscopy; IF, immunofluorescence; IHC, immunohistochemistry; WB, Western blot.
Acknowledgments
This work was supported by the National Institutes of Health (1R01NS089604, R01NS097679, and RF1AG056320) and the Alzheimer’s Association (AARG-17-499682).
The authors declare no competing financial interests.
Author contributions: X. Wang designed and supervised the study; J. Gao, J. Liang, and X. Wang designed the experiments, analyzed the data, and wrote the manuscript; J. Gao, L. Wang, X. Ren, J.R. Dunn, and A. Peters performed experiments; and M. Miyagi, H. Fujioka, F. Zhao, and C. Askwith performed the experiments, analyzed the data, and wrote the manuscript.
References
- Bosco, D.A. 2018. Translation dysregulation in neurodegenerative disorders. Proc. Natl. Acad. Sci. USA. 115:12842–12844. 10.1073/pnas.1818493115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott, N.T., Radke A., Stephens M.L., and Kramer J.H.. 2014. Frontotemporal dementia: diagnosis, deficits and management. Neurodegener. Dis. Manag. 4:439–454. 10.2217/nmt.14.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., and Wong P.. 2019. Splicing repression is a major function of TDP-43 in motor neurons. Brain Pathol. 29:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella, A.E., Zerze G.H., Mittal J., and Fawzi N.L.. 2016. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 24:1537–1549. 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella, A.E., Dignon G.L., Zerze G.H., Schmidt H.B., D’Ordine A.M., Kim Y.C., Rohatgi R., Ayala Y.M., Mittal J., and Fawzi N.L.. 2020. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc. Natl. Acad. Sci. USA. 117:5883–5894. 10.1073/pnas.1912055117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox, J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Deacon, R.M., and Rawlins J.N.. 2005. Hippocampal lesions, species-typical behaviours and anxiety in mice. Behav. Brain Res. 156:241–249. 10.1016/j.bbr.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Ditlev, J.A., Case L.B., and Rosen M.K.. 2018. Who’s in and who’s out—compositional control of biomolecular condensates. J. Mol. Biol. 430:4666–4684. 10.1016/j.jmb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, J.B., Zhao F., Wang X., Xu Z., Lin K., Askwith C.C., Hodgetts K.J., and Lin C.G.. 2018. Pyridazine-derivatives enhance structural and functional plasticity of tripartite synapse via activation of local translation in astrocytic processes. Neuroscience. 388:224–238. 10.1016/j.neuroscience.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, J.B., Lashley R., Zhao F., Wang X., Kung N., Askwith C.C., Lin L., Shultis M.W., Hodgetts K.J., and Lin C.G.. 2019. Enhancement of tripartite synapses as a potential therapeutic strategy for Alzheimer’s disease: a preclinical study in rTg4510 mice. Alzheimers Res. Ther. 11:75. 10.1186/s13195-019-0530-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa, F., Lu S., Yu H., Chen C., Melamed Z., Guo L., Shorter J., Da Cruz S., and Cleveland D.W.. 2019. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 102:339–357.e7. 10.1016/j.neuron.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, E., and Shorter J.. 2019. The molecular language of membraneless organelles. J. Biol. Chem. 294:7115–7127. 10.1074/jbc.TM118.001192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, J., Emmett W., Fratta P., Isaacs A.M., and Plagnol V.. 2017. Quantitative analysis of cryptic splicing associated with TDP-43 depletion. BMC Med. Genomics. 10:38. 10.1186/s12920-017-0274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A., Weber C.A., and Jülicher F.. 2014. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30:39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Igaz, L.M., Kwong L.K., Lee E.B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M.J., Trojanowski J.Q., et al. 2011. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 121:726–738. 10.1172/JCI44867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi, Y., Katsuno M., Niwa J., Takagi S., Ishigaki S., Ikenaka K., Kawai K., Watanabe H., Yamanaka K., Takahashi R., et al. 2013. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 136:1371–1382. 10.1093/brain/awt029 [DOI] [PubMed] [Google Scholar]
- Jeong, Y.H., Ling J.P., Lin S.Z., Donde A.N., Braunstein K.E., Majounie E., Traynor B.J., LaClair K.D., Lloyd T.E., and Wong P.C.. 2017. Tdp-43 cryptic exons are highly variable between cell types. Mol. Neurodegener. 12:13. 10.1186/s13024-016-0144-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L.L., Zhao J., Yin X.F., He W.T., Yang H., Che M.X., and Hu H.Y.. 2016. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci. Rep. 6:23928. 10.1038/srep23928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser, J.Z., Post M., Wang B., and Miyagi M.. 2009. Streptomyces erythraeus trypsin for proteomics applications. J. Proteome Res. 8:1810–1817. 10.1021/pr8004919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer, B.C., Schuck T., Wheeler J.M., Robinson L.C., Trojanowski J.Q., Lee V.M., and Schellenberg G.D.. 2010. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 119:409–419. 10.1007/s00401-010-0659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.R., Chen T.C., Hsiao C.L., Shi L., Chou C.Y., and Huang J.R.. 2018a. The physical forces mediating self-association and phase-separation in the C-terminal domain of TDP-43. Biochim. Biophys. Acta. Proteins Proteomics. 1866:214–223. 10.1016/j.bbapap.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Li, H.R., Chiang W.C., Chou P.C., Wang W.J., and Huang J.R.. 2018b. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J. Biol. Chem. 293:6090–6098. 10.1074/jbc.AC117.001037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L., Wei Y., Lu Y., and Song J.. 2016. ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically disordered prion-like domain of TDP-43. PLoS Biol. 14:e1002338. 10.1371/journal.pbio.1002338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, J.P., Pletnikova O., Troncoso J.C., and Wong P.C.. 2015. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 349:650–655. 10.1126/science.aab0983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, J.R., Gleixner A.M., Mauna J.C., Gomes E., DeChellis-Marks M.R., Needham P.G., Copley K.E., Hurtle B., Portz B., Pyles N.J., et al. 2019. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 102:321–338.e8. 10.1016/j.neuron.2019.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk, L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R.G., Shorter J., and Bonini N.M.. 2018. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell. 71:703–717.e9. 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick, W.C. 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 332:1–11. 10.1016/j.gene.2004.02.051 [DOI] [PubMed] [Google Scholar]
- Mitrea, D.M., Chandra B., Ferrolino M.C., Gibbs E.B., Tolbert M., White M.R., and Kriwacki R.W.. 2018. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol. 430:4773–4805. 10.1016/j.jmb.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex, A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, M.B., Rowland D.C., and Moser E.I.. 2015. Place cells, grid cells, and memory. Cold Spring Harb. Perspect. Biol. 7:a021808. 10.1101/cshperspect.a021808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, K., Watanabe S., Shijie J., Murata Y., Oiwa K., Komine O., Endo F., Tsuiji H., Abe M., Sakimura K., et al. 2019. Mice deficient in the C-terminal domain of TAR DNA-binding protein 43 develop age-dependent motor dysfunction associated with impaired Notch1-Akt signaling pathway. Acta Neuropathol. Commun. 7:118. 10.1186/s40478-019-0776-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin, J.B., and Kudla G.. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12:32–42. 10.1038/nrg2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H.B., and Rohatgi R.. 2016. In vivo formation of vacuolated multi-phase compartments lacking membranes. Cell Rep. 16:1228–1236. 10.1016/j.celrep.2016.06.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H.B., Barreau A., and Rohatgi R.. 2019. Phase separation-deficient TDP43 remains functional in splicing. Nat. Commun. 10:4890. 10.1038/s41467-019-12740-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton, C.F., Good S.K., Atkin S., Dewey C.M., Mayer P. III, Herz J., and Yu G.. 2010. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285:6826–6834. 10.1074/jbc.M109.061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum, M.E., Gilbert L.A., Qi L.S., Weissman J.S., and Vale R.D.. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159:635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Wang, W., Zhang F., Li L., Tang F., Siedlak S.L., Fujioka H., Liu Y., Su B., Pi Y., and Wang X.. 2015. MFN2 couples glutamate excitotoxicity and mitochondrial dysfunction in motor neurons. J. Biol. Chem. 290:168–182. 10.1074/jbc.M114.617167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., Conicella A.E., Schmidt H.B., Martin E.W., Rhoads S.N., Reeb A.N., Nourse A., Ramirez Montero D., Ryan V.H., Rohatgi R., et al. 2018. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37:e97452. 10.15252/embj.201797452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann, S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., Laskowski P.R., MacKenzie D., Kamath T., Commins C., et al. 2018. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37:e98049. 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils, H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., and Kumar-Singh S.. 2010. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 107:3858–3863. 10.1073/pnas.0912417107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., and Shen C.K.J.. 2010. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 48:56–62. [DOI] [PubMed] [Google Scholar]
- Wu, L.S., Cheng W.C., and Shen C.K.. 2012. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J. Biol. Chem. 287:27335–27344. 10.1074/jbc.M112.359000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D’Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X., et al. 2010. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 30:10851–10859. 10.1523/JNEUROSCI.1630-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., Hoek T.A., Vale R.D., and Tanenbaum M.E.. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell. 165:976–989. 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F., Siu J.J., Huang W., Askwith C., and Cao L.. 2019. Insulin modulates excitatory synaptic transmission and synaptic plasticity in the mouse hippocampus. Neuroscience. 411:237–254. 10.1016/j.neuroscience.2019.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists TDP-43WT– and TDP-43ΔCR–associated proteins identified by MS.
lists identification of genes with significantly changed expression in TDPΔCR+/− mice. The thresholds were set as P < 0.05 and Padj < 0.05.
lists protein identification used for TDP-43 sequence alignment, antibodies, and RT-PCR primers used in this study. IEM, immunoelectron microscopy; IF, immunofluorescence; IHC, immunohistochemistry; WB, Western blot.