Overview
Genetic model systems allow researchers to probe and decipher aspects of human disease, and animal models of disease are frequently specifically engineered and have been identified serendipitously as well. Animal models are useful for probing the etiology and pathophysiology of disease and are critical for effective discovery and development of novel therapeutics for rare diseases. Here we review the impact of animal model organism research in three examples of congenital metabolic disorders to highlight distinct advantages of model system research. First, we discuss phenylketonuria research where a wide variety of research fields and models came together to make impressive progress and where a nearly ideal mouse model has been central to therapeutic advancements. Second, we review advancements in Lesch-Nyhan Syndrome research to illustrate the role of models that do not perfectly recapitulate human disease as well as the need for multiple models of the same disease to fully investigate human disease aspects. Finally, we highlight research on the GM2 gangliosidoses Tay-Sachs and Sandhoff Disease to illustrate the important role of both engineered traditional laboratory animal models and serendipitously identified atypical models in congenital metabolic disorder research. We close with perspectives for the future for animal model research in congenital metabolic disorders.
Introduction
The history of eukaryotic cellular metabolism research is rich and the output has been fruitful. We understand core metabolism in significant detail; individual steps of metabolic pathways are elucidated and ordered, the genes encoding the required enzymatic activities are known, and those enzymatic reactions are frequently well-studied and understood biochemically and biophysically. Moreover, we have a systems-level understanding of how these core pathways operate within a vast network of highly interconnected activities. Links in this network remain unrecognized, and surprises are undoubtedly to be discovered. Nevertheless, our understanding of the vast complex of cellular metabolic reactions is extensive. In contrast, our knowledge of how perturbations in core metabolic pathways impact human development, function and health is much more limited.
Given that life is dependent on the extensive and complicated cellular metabolic network, it is not surprising that mutations in a vast number of genes that encode metabolic enzymes cause congenital metabolic disorders (CMDs), originally coined as inborn errors of metabolism (IEM) in 1908 (1). Recognition of the metabolic basis of disease also has a rich history. It has been almost 120 years since the first time a disease was noted to follow a Mendelian inheritance pattern (2). Identification of the heritable material was still 50 years away and the first genetic lesion causative of a disease of Mendelian inheritance was not identified for several more decades.
CMDs arise from the absence or low residual levels of enzymatic activity, leading to build-up of substrates and decreased production of important products, which each contribute to phenotypic outcome. CMDs are individually rare but collectively common, encompassing over 1400 types of diagnoses and affecting as many as 1 in 1000 newborns, with higher incidence in some populations that have reduced genetic diversity (3). Newborns are routinely screened for a selection of CMDs in technologically advanced countries, resulting in timely early diagnoses of most individuals in the United States with phenylketonuria (PKU) and a selection of other CMDs (4). In contrast other CMDs are ultra rare and are diagnosed only after genetic sequencing of patients with unique syndromes and thus, only in a few patients worldwide (5,6). Many CMDs are also thought to be underdiagnosed because they often present clinically as syndromes with a wide array of symptoms in a variety of tissue and organ systems making diagnoses difficult (7,8).
The study of congenital metabolic disease can be approached on a variety of levels. It is relatively straightforward to elucidate the effects of a mutation in an enzyme on its catalytic activity. Our mechanistic understanding of enzyme function is quite exhaustive in some cases, even permitting engineering of new activities. It is more difficult to predict network-wide metabolic effects from loss or reduction of activity in one reaction. However, advances in NMR and mass spectrometry techniques that allow simultaneous monitoring of both steady state and relative fluctuations of metabolites system-wide have made elucidation of such effects possible (9). It is even more difficult to explain or predict the effects of a mutation in a metabolic gene in terms of phenotypic outcome in a multicellular organism where certain tissues or processes may be hypersensitive to loss of activity or specifically sensitive to toxicity of a substrate that accumulates abnormally. In fact, metabolic mutations frequently result in syndromes with a suite of seemingly unrelated phenotypes. Further confounding diagnoses and study is that environmental factors, diet in particular, may have effects on outcomes and progression and presentation of symptoms.
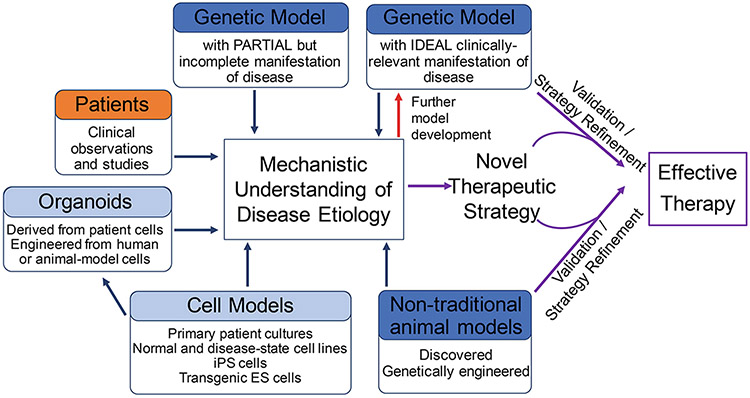
We have prepared a perspective to discuss examples of how the most complicated of models, both research model organisms such as mice and rats and less common models such as primates, and even domesticated animals, are contributing to the elucidation of the etiology of clinical phenotypes and to implementation of effective therapeutic interventions for CMDs (Figure 9.1).
Figure 9.1. The role of animal models in elucidating etiology and developing therapies of congenital metabolic disorders.
Animal models (highlighted in dark blue) are developed in different ways, including using traditional forward genetics, standard transgenic approaches and novel genome editing technologies. Genome editing is facilitating development of animal models in non-traditional laboratory animals. Some model animals also arise among animals outside of laboratory environments as a result of founder mutations and reduced genetic diversity stemming from human domestication. Animal models ideally mimic human disease, but often only partially recapitulate biochemical and /or phenotypic aspects of human disease. Each are useful for probing disease etiology. Research involving cell and in vitro models (highlighted in light blue) and human clinical studies (highlighted in orange) are combined with insights gained in animal models to both elucidate mechanisms disease etiology and enhance model development (red arrow). An ideal animal model is an extremely powerful tool (purple arrows) for advanced therapeutic development.
Phenylketonuria
We start with a discussion of Phenylketonuria (PKU, OMIM 261600). PKU is one of the most common CMDs (10). Left untreated, it results in profound and irreversible brain damage. Dietary intervention for PKU via restriction of phenylalanine in the diet was developed in 1954, and by 1966 routine newborn screening for PKU and dietary intervention for affected infants was being implemented (11,12). This has led to great success in reducing the impact of PKU on patient outcomes (13). While PKU dietary treatment is effective, it is a difficult regimen for patients and lack of compliance is common (14). Avoiding phenylalanine equates to avoiding protein, and patients must supplement with a phenylalanine-free protein formula to provide healthy nutritional requirements. Unfortunately, because many children and adults find the necessary formula supplement unpalatable and because dietary management is onerous for families, this treatment is less than ideal in practice.
We chose to highlight PKU because an excellent animal model was developed for PKU in the early 1990’s and this model has facilitated impressive progress in development of therapies to help patients manage PKU more effectively, even allowing lenience in dietary management. PKU research already represents a major success story for the contribution of animal models to congenital metabolic disease management. Moreover, additional therapeutic options that will continue to improve the life of patients are under development with the animal model in a central role. Another reason we chose to highlight PKU is to illustrate how models beyond animals contribute to human disease research in conjunction with animal models. From genetic screens in mice, to discovery of an alternative enzymatic pathway in plants, fungi and cyanobacteria, to investigations in non-traditional animal models, and finally, to treatments that employ prokaryotic systems, the PKU community has benefited from a wide swath of research models over many years with the mouse model as the key to many of the successes.
History and prevalence
PKU is an autosomal recessive metabolic disorder characterized by high plasma levels of phenylalanine. It arises as a result of mutations in the gene encoding the enzyme phenylalanine hydroxylase (PAH), which converts phenylalanine to tyrosine (15-17). PKU was first described by the Norwegian doctor Asbjørn Følling in 1934 (18). A mother reported that her two children had a musty urine odor. The doctor performed a ketone test, which requires acidification of a urine sample with ferric chloride and discovered that the children’s urine turned green instead of producing the typical reddish color. After further chemical analysis, he concluded that the green color was produced by phenylpyruvic acid, a phenylketone by-product derived from phenylalanine. PKU is found in 1 in 10,000 to 15,000 Caucasian births, but prevalence varies in other populations (19).
Clinical and molecular phenotypes and alleles
PKU can manifest across a wide spectrum of severity. However, untreated classical PKU results in symptoms affecting the nervous system, including severe cognitive disability, seizures, and neurobehavioral problems. The skin is also affected with reduced levels of pigmentation and susceptibility to eczema and other skin conditions (20).
Phenylalanine is an essential amino acid that comes from dietary intake. PAH functions in the liver to metabolize phenylalanine to tyrosine. In the absence of PAH, the excessive plasma phenylalanine is transported across the blood-brain barrier, where it is neurotoxic. Additionally, the inability to produce tyrosine from phenylalanine results in a deficit of other metabolites, such catecholamine neurotransmitters and melanin (15,21). Patients with PKU often have fair skin, blue eyes, and blond hair due to the inability to make melanin (21). High levels of transport of the phenylalanine across the blood-brain barrier can also hinder the transport of other important amino acids, which likely contributes to phenotypic outcome (22).
PKU patients may seem normal at birth. However, developmental delays become evident around 3-6 months of age if left untreated. Due to the prevalence of PKU and our ability to effect positive outcomes with a dietary intervention, newborns in the United States have been routinely screened since the 1960s. Long-term neurological consequences can be avoided with a strict low phenylalanine diet. Dietary adherence must be maintained for the lifetime to limit the irreversible brain damage and manage PKU. Phenylalanine application to neural cell cultures limits neurite outgrowth and high levels of phenylalanine in patients and model organisms is associated with decreased white matter and fewer synapses suggesting that phenylalanine is directly toxic to neurons (23,24).
Phenylalanine is a potent teratogen (25). Thus, control of phenylalanine levels is extremely important during pregnancy. Maternal PKU syndrome is caused by unmanaged elevated phenylalanine levels in pregnant PKU patients. Maternal PKU is associated with congenital heart disease, microcephaly, intrauterine growth retardation and spontaneous abortion, regardless of the fetus’ genetic PKU status (26).
The PAH gene was cloned in both humans (27) and mice (28) in the 1980s. More than 950 mutations have been associated with PKU and many categorized according to residual enzyme activity which negatively correlates with phenotypic severity (29). Mutations in the PAH gene include missense mutations (62%), deletions (13%), splicing defects (11%), silent polymorphisms (6%), nonsense mutations (5%), and insertions (2%) (Williams et al. 2008), and a frequent mechanism of reduced enzymatic function appears to be protein destabilization (30,31).
Animal Models
The first mouse model for PKU was purposefully generated in a forward genetic screen. After germline ethylnitrourea (ENU) mutagenesis in mice, animals were screened for hyperphenylalaninemia and three mutants were identified (32,33). The three alleles, Pahenu1, Pahenu2, and Pahenu3, were associated with varying degrees of hyperphenylalaninemia. Pahenu1 mice have a moderate elevation in serum phenylalanine concentration and urinary ketones, whereas Pahenu2 and Pahenu3 mice are severely elevated for both.
These animals were quickly appreciated as an ideal model in terms of their recapitulation of the molecular and pathological aspects of PKU. These mutants have provided an excellent model for unraveling differential effects of mutations in PAH on clinical outcome because the three alleles differ in severity of phenotype, amount of residual protein activity and type of molecular lesion in the gene (32-35). Moreover, as strong model for the human disease they have served as surrogates in investigating both etiology of disease and in testing of potential therapies. Here we focus on how the animal models have been useful for advancing therapeutic-focused research in conjunction with studies in multiple other systems from prokaryotes to plants.
Therapeutics
The concept of enzyme replacement therapy has heavily influenced PKU treatment research, even though significant challenges had to be overcome to make this a success story. PAH normally functions in the hepatocyte cytoplasm and requires the non-protein cofactor tetrahydrobiopterin (BH4) for activity. In fact, depletion of BH4, which can arise from mutations in enzymes in its biosynthetic pathway, reduces PAH activity and results in hyperphenylalaninemia and syndromes that overlap to some degree with PKU (36). The need for the BH4 co-factor and the liver localization of the normal protein complicates implementation of replacement enzyme therapy using PAH itself, as do issues with stability of the PAH enzyme (37). However, the same cofactor requirement attribute has been exploited to implement a distinct therapeutic for PKU.
Some patients with mild PKU benefit from administration of BH4 (Muntau et al. 2002). A synthetic form of BH4 called saptopterin dihydrochloride (tradename Kuvan™) was the first therapeutic approved for PKU treatment (38,39). Studies in the Pahenu1 model revealed the molecular mechanisms underlying BH4 function. BH4 stabilized the Pah protein in this model, acting as a chaperone to promote proper protein folding and prevent the aggregation and degradation that lead to low enzymatic activity (38). Patients who have destabilizing mutations in PAH can benefit from this therapy. Thus, the co-factor itself has led to an effective therapeutic, but attempts to bypass the co-factor entirely would also be beneficial for additional patients. It was research in a distinct field combined with academic scientists who communicated across disciplines and with industry researchers that led to a breakthrough on this front.
Researchers studying lignin biosynthesis in plants in the 1950s determined that metabolites of phenylalanine were important to lignin synthesis and then subsequently identified the enzyme phenylalanine ammonia lyase (PAL) as a key player in lignin biosynthesis (40). Phenylalanine ammonia lyase is found in plants as well as fungi and converts phenylalanine to trans-cinnamic acid and ammonia (40).
John Hoskins was an organic chemist who serendipitously shared lab space with a PKU diagnostics lab in the 1970s and became interested in the disease (41). His interest eventually led him to join forces with Dr. Henry Wade who knew that depletion of phenylalanine was important in treatment of PKU and had access to PAL enzyme. Together they implemented formulations to test the effect of PAL in humans, in both normal test subjects after a high protein meal and on the high levels of phenylalanine in PKU patients (41,42). Their work provided proof of principle for an effective therapeutic but years of research to develop a safe product lay ahead.
PAL is not a mammalian protein and its introduction in mammals leads to an immunogenic response, making it unsuitable as a therapeutic. Immunogenicity issues are not uncommon in molecular therapeutics, and PKU researchers employed a common technique to mask the immunogenic protein from the host (43). They used PEGylation, involving modification of the protein with polyethylene glycol, to try to limit the antigenicity and immunogenicity of the protein with some limited but insufficient success. In an academic / pharmaceutical collaborative effort in 2008, researchers turned to a selection of cyanobacteria, fungi and plants as sources of diverse PAL proteins to try to identify a suitably candidate for enzyme replacement therapy.
PEGylated PAL proteins from Anabaena variabilis, Nostoc punctiforme, Petroselinum crispum and Rhodospridium toruloides were tested for therapeutic effect in the Pahenu2 mice (44). High doses of protein followed by smaller, secondary doses decreased plasma and brain phenylalanine, with the Anabaena variabilis (Av) protein showing the most favorable results in terms of thermal stability, proteolytic resistance and optimal pH sensitivity. In addition to the decrease in phenylalanine levels, the characteristic hypopigmentation, due the lack of conversion of tyrosine to melanin, was also ameliorated (44). The effects of enzyme treatment were reversible once treatments were halted.
A modified and optimized version of PAL from Anabaena variabilis was chosen as a candidate for Phase I clinical trials conducted by BioMarin as a therapeutic treatment for classical PKU due to the success found in the Pahenu2 mice. During human clinical trials, 60.7% of patients had a blood phenylalanine level below the recommended guidelines within 24 months of starting treatment (45). Thus, the second therapeutic drug (trade name Palynziq™) and the first enzyme substitution therapy for PKU was approved by the FDA. This therapy no longer requires the onerous dietary restrictions associated with standard treatment.
Maternal PKU models
Avian, rodent and primate models for maternal PKU have been explored with emphasis on elucidating the etiology of the cardiac malformations associated with high phenylalanine during pregnancy (46-49). Again, the Pahenu2 mouse model is a relevant model for what is observed in humans. Pahenu2 females produce offspring with structural defects in the cardiovascular system, although the structural problems reported in humans and mouse differ (46). Gene expression analysis in chick and Pahenu2 reveal changes in genes relevant to cardiac development and function including cardiac troponin and myosin (47,50).
The Pahenu2 model and the future
PKU research benefits from the fact that it is less rare than many CMDs. As a result, many advances in therapeutics and disease etiology have come from insights gained in patients. Moreover, the mouse model for PKU is perhaps as close to ideal as can be expected between species. The future of PKU research will see Pahenu2 in a central role. Two recent advances illustrate the continued relevancy of this model for exciting future therapy development.
Our relatively recent realization of the importance of the gut microbiome and its interdependence with human metabolism has led to increased interest in probiotics as therapeutics (51). An engineered probiotic for PKU treatment is a reasonable approach given that oral administration of recombinant PAL enzyme was shown to lower serum phenylalanine levels in the Pahenu mouse models (52). A Lactobacillus strain engineered to express Anabaena variabilis-derived PAL was tested in the Pahenu2 model, resulting in a reduction in serum phenylalanine levels (53). Two other groups have engineered an E. coli Nissle strain to express phenylalanine-degrading enzymes and tested responses in the Pahenu2 model with positive outcomes (54,55). Isabella et al. produced an intricately engineered strain that expresses two phenylalanine degrading enzymes: cytoplasmic PAL and a membrane bound, periplasmic-localized L-amino acid deaminase (LAAD) that converts Phe to phenylpyruvate (55). Both enzymes are under inducible promoters to limit enzyme production until the final stages of manufacturing of the probiotic in the case of LAAD and until exposure to the microaerobic or anaerobic regions of the gut in the case of PAL. This strain was engineered with adherence to FDA guidelines regarding live biotherapeutic organisms with the goal of extending its use directly to clinical trials.
In the modern age of genome manipulation, it is now possible to consider editing of molecular lesions in patients. Again, Pahenu2 has provided the conditions for proof of principle for such experiments. In 2018, Villiger et al. reported use of a base editing CRISPR-CAS9 system (56,57) to revert the enu2 allele in cells of the mouse liver (58). The edited mice had reduced blood phenylalanine levels, increased PAH activity and a reversion of the mutant hypopigmentation.
In summary, twenty-five years of research with the Pah mouse model as a tool has already reaped benefits and is poised to promote additional progress for patients. This field is a success story. PKU is a great illustration where discoveries in animal models, combined with knowledge gleaned from multiple other systems from plants to bacteria led to the discovery of a therapy that efficiently clears the neurotoxic effects in people that suffer from this rare, metabolic disease.
Lesch-Nyhan Syndrome
While it may be tempting to define an ideal disease model, as we discussed with PKU, as one that recapitulates the entirety of the disorder seen in humans, e.g., genetically, biochemically, histopathologically, and phenotypically, models that demonstrate one facet of the disease have been useful in advancing our understanding of CMDs. Research into the inborn error of purine metabolism called Lesch-Nyhan Syndrome highlights the utility of establishing multiple animal models for a single CMD.
History and prevalence
Lesch-Nyhan Syndrome (LNS, OMIM 300322) is an X-linked recessive metabolic disorder of purine metabolism caused by mutations in the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene. It was first fully described by Michael Lesch and William Nyhan in 1964 (59). They reported on two young male patients, ages 3 and 5, at Johns Hopkins Hospital who presented with severe motor dysfunction, involuntary movements and muscle contractions, and crystals in the urine. Both patients also displayed compulsive self-injury involving biting of lips and digits. In 1967, Seegmiller et al. demonstrated that cells of patients with LNS lack hypoxanthine-guanine phosphoribosyltransferase (HPRT) activity (60). The HPRT gene was cloned and sequenced in 1983 (61,62). LNS prevalence is estimated at 1 in 380,000 live births (63). The recognition and description of LNS marked a milestone in that it was a first metabolic disturbance in purine metabolism linked to a neurobehavioral disease (59).
Clinical and molecular phenotypes and alleles
LNS patients have a diversity of clinical phenotypes, including hyperuricemia resulting in gouty arthritis and renal disease, aberrant motor function, reduced gastrointestinal motility and cognitive and behavioral problems (64-66). The syndrome presents on a continuum. Patients with the highest levels of residual enzyme activity present with gouty arthritis without neurological involvement. This milder clinical manifestation has been alternatively called Lesch-Nyhan variant and Kelley-Seegmiller syndrome (OMIM 300323). In contrast, patients with little to no residual enzyme activity are severely affected with motor dysfunction and the neurobehavioral issues in addition to the hyperuricemia. The most striking behavioral symptom is that these individuals are typically auto-aggressive, causing self-injury usually via biting of lips, tongues and digits.
As a result of a severe reduction or absence of HPRT enzymatic activity, individuals with LNS are unable to salvage purines to resynthesize nucleotides. Instead, the purines are degraded to uric acid, and purine biosynthesis is increased in a compensatory response, exacerbating the hyperuricemia (67). The excess uric acid crystalizes in various tissues and organs, causing acute arthritis in the joints and stones and renal disease in the kidneys (65,66,68). Uric acid production can by reduced in patients using drugs such as allopurinol, a xanthine oxidase inhibitor that prevents breakdown of hypoxanthine to uric acid, and this ameliorates the gouty aspects of the disease (65). However, allopurinol has no effect on the neurological and motor manifestations.
The self-injurious behavior has been a topic of intense study. Self-injuries are not due to the lack of sensation. They are believed to be associated with the dysfunction of dopaminergic pathways in the basal ganglia. Post-mortem tissue analyses revealed low HPRT enzymatic activity and a marked decrease in dopamine in the basal ganglia (Lloyd et al. 1981), as well as a decrease in markers of dopaminergic neurons (69,70).
Patients with this disorder usually require restraints to attempt to stop the neurobehavioral issues. Treatments such as mild tranquilizers (e.g., benzodiazepine) and anticonvulsants (e.g., carbamazepine) have been used to help with the behavioral issues (71,72). However, in extreme cases dental extractions are recommended(66,73). There are currently no treatments for motor deficits.
The official website of the Lesch-Nyhan Study Group lists 615 mutations in HPRT that are associated with LNS, in which 381 are single point mutations (61.9%). Previously, several mutational hot spots in the HPRT gene were found and the most predominant one included 12 unrelated individuals with a C to T mutation changing arginine to a stop codon in a CpG motif (74)(74).
Models
Cells
The first efforts to analyze the molecular etiology of LNS were carried out in cell models. The specific lack of HPRT activity in patients was demonstrated using erythrocytes and fibroblasts collected from patients (60). Such models have added to our understanding of the cellular biochemistry of purines as well. For example, these HPRT-deficient cell lines revealed alterations in de novo purine synthesis flux that occur upon loss of HPRT as well as changes in energetics as revealed by GTP:GDP ratios (75,76). Subsequently, cells engineered to lack HPRT activity, including human fibroblasts and dopaminergic neuroblastoma cell lines (77) and mouse dopaminergic neuroblastoma cell lines (78) have been used to address questions about the neurological biochemistry and functional ability of cells that lack HPRT activity. For example, recent application of genomics methods to examine gene expression in HPRT-deficient human fibroblasts and neuroblastoma cells reveals down regulation of Lmx1a and Engrailed 1, transcription factors required for development and function of dopaminergic neurons, dysregulation of canonical wnt signaling, aberrant presenilin expression (77) and disrupted miR181a expression, which subsequently regulates important dopaminergic cell developmental pathways (79). Each finding from these cell models generates new hypotheses about the possible link between HPRT and neural function and dysfunction. Moreover, cell lines have provided a platform to demonstrate proof of principle for enzyme replacement therapies at least in terms of correction of the biochemical deficits, if not the behavioral ones (80).
Rodents
LNS was the first human congenital disease for which a genetically engineered mouse model was created (81,82). There were high hopes for modeling the human disease in mice because the rarity of the syndrome presents added challenges beyond those already inherent in research involving patients. Hprt1-deficient mice lack HPRT enzymatic activity and were experimentally demonstrated to be unable to salvage purines, suggesting a strong biochemical correlate of LNS in this model (83). The brains of these mice also have reduced dopamine levels (84,85), similar to the findings in the post-mortem brains of LNS patients. Overall, the mouse model has been useful for validating and probing the neurochemical changes that arise when HPRT1 activity is compromised. However, the neurobehavioral aspects of the disease were not reproduced in the knock-out mice. There was one hopeful report that the self-injurious behaviors could be induced in the Hprt1-deficient mice upon inhibition of adenine phosphoribosyltransferase (86). However, this result has not been reproducible (87,88), and, thus, the mouse model is not suitable for investigation into causal relationships between the neurochemical signatures shared by mice and humans and the behavioral outputs observed only in humans.
Because rats often provide a model system physiologically and behaviorally more similar to humans than mice (89), rat models for LNS have also been explored. There were expectations of success in recapitulating the behavioral issues observed in patients via knock out of Hprt in rats because studies in rats, aimed at other purposes, serendipitously produced a model for the self-injurious behavior in LNS. Perinatal treatment of rats with 6-hydroxydopamine, which selectively kills dopaminergic and adrenergic neurons (90), results in a reduction of dopamine (91,92). Animals with this neural damage surprisingly exhibit self-mutilation of the limbs and abdomen when administered L-dopamine as adults (91,92). Similar outcomes are not observed when the 6-hydroxydopamine treatment is applied to adult animals. These studies lend support to the hypothesis that dopamine perturbations are causative of self-injurious behaviors and highlight the complicated etiology in terms of how changes in metabolite levels intersect with developmental events to produce the behaviors. Dopaminergic deficiencies have been directly observed using positron emission tomography in patients independent of age, again suggesting that the dopaminergic deficiencies arise from developmental as opposed to neurodegenerative processes (70).
The engineering of an Hprt knock out rat was reported in 2016 (93). The neurochemical changes observed in humans and mice are also recapitulated in these rats. However, the behavioral phenotype was once again not observed. Thus, while there is substantial evidence supporting the role of dopamine deficiencies in the neurobehavioral aspects of LNS, the link between loss of HPRT activity and the perturbations to dopaminergic signaling remain unclear.
While less than ideal for studying intriguing aspects of neurobehavior, the rodent models have been useful to shed light on the underlying etiology of other adverse symptoms seen in LNS patients, specifically gastrointestinal motility issues that result in vomiting, dysphagia, and constipation (94). The HPRT deficient model mice have an overall decrease in gastrointestinal motility that is correlated with dysfunction in dopaminergic neurons of the gastrointestinal tract (64). Moreover, gastrointestinal motility issues are not due to impairment of cholinergic or nitrergic systems (64). Thus, the mouse model did prove to be important for shedding light on the changes seen in gastrointestinal motility, illustrating significant benefit of a model that may be considered less than ideal on another front.
Back to cells and the future
The future of disease modeling is sure to include increased use of stem cells, including both induced pluripotent stem (iPS) cells and embryonic stem (ES) cells. Such modern models are being used for the study of HPRT activity in neural development and function. Knock down of HPRT expression in human iPS and ES cells results in lower expression of the P2Y1 purinergic signaling receptor and impaired downstream signaling (95). Gene and protein expression profiles across a timeline of dopaminergic neural development in mouse ES cells deficient for HPRT activity suggested that the cells are inhibited in neural differentiation in favor of glial differentiation (96). Moreover, dysregulation in a wide swath of cell functions was suggested by the gene expression changes observed, suggesting that HPRT may have roles in neural differentiation beyond what would be expected if considered only a purine biosynthetic mutant. In short, much biology remains to be uncovered.
Conclusions
LNS is a complex, multifaceted disease that is associated with varied pathophysiology. The rat and mouse models were first deemed only partial successes, due to the lack of the neurobehavioral issues seen in humans. However, the mouse model has effectively provided insight into the neurochemical and gastrointestinal issues. Given the success of the models in recapitulating the biochemical changes seen in LNS patients and the recent results of experiments examining neural development in these models, it is likely that the rodent models will still provide the clues needed to make a breakthrough in the neurobehavioral puzzle.
LNS is a particularly rare disease, making it more difficult to glean information from clinical studies. Because of the lack of a perfect model for assessing the most distressing clinical aspects of LNS, therapeutic research has received less emphasis relative to phenylketonuria as noted above. Instead a major thrust in LNS research has been efforts to develop the appropriate models and studying the etiology of the perturbations in the current models, both genetic and chemically induced, that will eventually advance therapeutic studies. This field highlights the usefulness of less than ideal models at the same time as illustrating the need for additional model development (Figure 9.1).
Tay-Sachs and Sandhoff Disease
In the examples above we discussed forward genetic and transgenic approaches to deliberately engineer genetic model systems. In this example, some of the engineered animal models do not perfectly reflect clinical aspects of the human disease. Researchers have instead made great use of serendipitous discoveries of useful models amongst domesticated animals. These animals are better suited to advance the types of effective therapeutic research we saw illustrated above for PKU.
History and frequency
Tay-Sachs Disease (TSD, OMIM 272800) and Sandhoff Disease (SD, OMIM 268800) are rare, autosomal recessive neurodegenerative disorders that develop as a result of mutations reducing hexosaminidase activities. They are clinically indistinguishable and manifest as mental and motor deficits that progressively worsen. Both are classified as lysosomal storage disorders and characterized by an accumulation of GM2 gangliosides in the lysosome.
Tay-Sachs was first described in a single patient by British doctor Waren Tay in 1881(97), who subsequently recognized the same syndrome in additional patients. In 1887, the American doctor Bernard Sachs, unaware of the work of William Tay, submitted a paper about a case in a girl of German descent (98), and in 1892 E.C. Kingdon reported a case and reviewed the reports of both Tay and Sachs. Kingdon subsequently identified additional patients and the syndrome became known as Tay-Sachs (99,100)
The American College of Medical Genetics and Genomics as well as American College of Obstetricians and Gynecologists recommend that people of Ashkenazi Jewish descent screen for TSD, as the carrier frequency in that population is 1 in 31 (3). The carrier frequency in eastern French Canadians is 1 in 14 (101). Other small founder populations include Louisiana Cajuns, Irish, and Brazilian populations also have an increased carrier frequency (100,102). Due to pre-screening for carrier status in the Ashkenazi Jewish population, TSD incidence decreased 90% between 1970 and 1993 in North America (100).
Sandhoff disease was described in 1968 (103) as a variant of Tay-Sachs with a distinct enzymatic activity profile (see below). Like TSD, SD is fatal, and the patient normally exhibits progressive neurodegeneration (Hadfield et al. 1977). SD prevalence is estimated to be 1 in 422,000, with a carrier rate of 1 in 310 (Meikle et al. 1999). Some isolated communities with high consanguinity have higher incidence rates. For example, northern Saskatchewan is home to isolated Métis communities where the population is mixed French Canadian and aboriginal and has a carrier frequency of 1 in 27 (Fitterer et al. 2014).
Clinical and molecular phenotypes and alleles
TSD and SD are classified clinically into multiple forms: infantile, juvenile and late-onset (100). The infantile form is typically associated with acute symptoms of mental and motor dysfunction and is usually diagnosed by 6 months of age (104). Catastrophic progressive neurodegeneration leads to hypotension, inability to sit or to hold up the head, eye abnormalities and dysphagia. Patients diagnosed with infantile forms typically do not survive past the age of 3 or 4 years (105).
The juvenile form is characteristically diagnosed between the ages of 3 and 10 years (104). There is more diversity in clinical manifestations in the adolescents with juvenile forms. However, patients usually exhibit ataxia, slowed and slurred speech (dysarthria), dysphagia, hypotension, and seizures. Patients diagnosed with juvenile forms typically do not survive past the age of 15 years due to the progressive nature of the disorder.
The late-onset form is often diagnosed in adolescence, but the symptoms can also appear as late as the 3rd decade of life. Recognition of previously undiagnosed mild neurodegenerative symptoms may be a characteristic of late-onset disease; patients in one study recalled previous clumsiness and motor skill issues prior to diagnoses (106). Furthermore, the same study showed that patients with the late-onset form did not get properly diagnosed, on average, for about 8 years from onset of symptoms. Clinical manifestations include a gradual reduction in motor, cerebral, and spinocerebellar activity.
SD is also associated with some distinct organ pathologies relative to TSD. Both hepatosplenomegaly and cardiac involvement have been described in SD patients (107-109).
TSD and Sandhoff are caused by mutations that inhibit β-N-acetylhexosaminidase (Hex) activity. Hex enzymes are homodimers or heterodimers of α and β subunits, encoded by the genes HEXA and HEXB, respectively. HexA enzyme is a heterodimer of an α and a β subunit. HexB enzyme is a homodimer of two β subunits, and HexS enzyme is a homodimer of two α subunits. Mutations in the HEXA gene disrupt HexA and HexS enzyme function and cause TSD. Mutations in the HEXB gene disrupt HexA and HexB enzyme function and cause SD. SD was first differentiated from TSD because of the observation that a patient lacked both HexA and HexB activity (103), whereas TSD patients retain HexB activity. Note that a third disorder with TSD and SD symptoms arises from mutations in a Hex enzyme activator (110).
The loss of HexA results in a defect in hydrolysis of GM2 gangliosides in the lysosome. The resulting accumulation of GM2 gangliosides inside the lysosomes of neurons causes neural toxicity. TSD is one of multiple pathologies dubbed GM2 gangliosidoses, as mutations in other enzymes in the pathway can lead to similar molecular and clinical phenotypes (111)
TSD severity is negatively correlated with HexA activity; patients with the infantile form have the most pronounced depletion of enzyme activity, whereas adult-onset patients may retain 5% to as much as 20% of normal enzyme activity (112). The wide diversity of clinical phenotypes is also reflective of the large number of specific disease-causing genetic lesions (111).
Models
Mouse models of TSD and SD were created via targeted deletion of the Hexa and Hexb genes (113-116). The Hexa mice recapitulate the biochemical aspects of TSD and histopathological aspects in some regions of the brain. However, clinical symptoms similar to TSD were not observed at first. As these animals age past 1 year, they do show clinical signs similar to late-onset TSD (117), but they offer a less than ideal model for studying TSD etiology or for exploring therapeutic approaches to the most severe forms of TSD. The difference between human and mouse phenotypes upon mutation of the Hexa gene appears to be due to sialidase activities in mice that can metabolize the GM2 gangliosides in combination with the residual HexB enzymatic activity when HexA enzymatic activity is absent (118,119). A HexA deficient mouse model in which NEU3 sialidase activity is also compromised displays a severe phenotype more characteristic of TSD (120).
In contrast, the Hexb mutant mice have progressive neurological phenotypes that parallel TSD and SD in humans, including a severely shortened life span (113). The Hexb mutant mice have an 8-week asymptomatic period following birth. This is followed by 8 weeks of rapid progression of the disease to the point of death (113). This model has been used effectively to study the etiology of the neurodegenerative events. During the asymptomatic period, accumulation of GM2 gangliosides damages neurons and impairs their survival. Subsequent inflammatory responses trigger reactive astrogliosis, further death and neurodegeneration (121,122). Defects in neurite outgrowth of hippocampal neurons from early embryos was observed in culture (123), and there are defects in differentiation that favor astrocytes at the expense of neurons in Hexb neural stem cells as well as Hexb mutant iPS cells (124).
Hexb deficiency has also been engineered in zebrafish using a CRISPR-Cas9 system to study the earliest stages of neural development (125). This model is very useful for the ability to image early development in live animals and Kuil et al. observed enlarged lysosomes and increased lysosomal numbers in glia as early as 3 days post-fertilization. The mutant animals also displayed reduced locomotor activity. Together, these results suggest early SD neurodegeneration is recapitulated in zebrafish, creating a powerful model for cell biological analysis of SD that will likely contribute significantly to the field. Interestingly, adult zebrafish are viable and do not show behavioral deficits (125), likely because zebrafish can make new neurons throughout its life (126).
TSD and SD have each been observed to arise spontaneously in domestic animal species (cats, dogs and pigs) and animal species kept in captivity (deer and flamingoes) (127-135). Feline colony models of SD have been maintained and studied in the laboratory for decades, and have been used to explore therapeutics for GM2 gangliosidosis (136-138). The size and complexity of the cat brain relative to the mouse provides a superior model for humans, and approaches using viral vectors to reintroduce missing gene activities in cats have demonstrated proof-of-principle for such approaches (136-138).
Intriguingly, a promising model that recapitulates human TSD phenotypically and offers an exciting model for therapeutics-focused investigations has been found in a rare breed of primitive sheep called Jacob sheep. Multiple lambs from a single flock in the United States displayed progressive neurological symptoms, and evaluation revealed histological changes in the nervous system, including enlarged neurons, a decrease in white matter and abnormalities in the retina. Hex enzyme activity was decreased in brain tissue and a mutation was found in the Hexa gene (139,140). The disease has also been identified in the population of British Jacob sheep from which the North American flock originated (141), and breeding populations have been established for research.
This model is being used to explore therapeutic approaches, and, again, because of brain size and complexity is an excellent model for exploring therapeutics to benefit children with Tay-Sachs. In 2018, Gray-Edwards et al. reported signficant success using intracranial administration of an adenovirus vector to introduce the Hexa gene or copies of both Hexa and Hexb. Experimental animals all showed delay of symptom onset (142).
These adenovirus-mediated gene therapies offer great promise as proof of principle for gene replacement therapies. However, adenovirus-mediated gene therapies have proven dangerous in humans and significant hurdles remain to be overcome to develop an efficacious strategy to reintroduce the missing enzymatic function in neural tissue. The cat and the sheep models offer great promise for testing engineered therapies to achieve the types of success achieved in PKU research.
Organoid Model – A step into patient-oriented models
While there is no substitute for animal models in the realm of probing neurobehavior, modern technologies offer options for modeling that will reap benefits for patients. Pluripotent cells can be both induced and then coaxed into formation of a variety of different organoids (143). These organoids are complex structures that recapitulate aspects of cell differentiation and organ development, morphology and function. Organoids formed from the cells of patients with specific genetic lesions is a step towards personalized medicine for rare CMDs. Cerebral organoids for Sandhoff Disease have been produced and examined (144). Fibroblasts were obtained from an infant with SD and used to produce iPS cells. The researchers also used a CRISPR-CAs9 system to edit the genetic lesion of the patient’s cells, correcting one copy of the HEXB gene and used the original and edited, isogenic control cells to generate cerebral organoids. The SD organoids were larger than the isogenic control organoids and produced GM2 gangliosides, similar to what was seen in the brain of SD patients (144). They were able to ameliorate GM2 accumulation using a gene replacement therapy approach. Whole transcriptome analysis of these organoids revealed the reduced expression of an intriguing set of genes related to neural development in the mutant organoids.
Conclusions
CMDs are rare diseases. Thus, the number of patients affected is low and gaining extensive information clinically is challenging. As a result, the development of animal models has been critical and will continue to play a role in CMD research and therapeutic advances for patients with CMDs. We have only discussed a handful of CMDs. However, these diseases illustrate two major principles in our research enterprise. Frist, a model that recapitulates human clinical symptoms is a tremendously powerful tool for facilitating therapeutic development. Second, models that do not recapitulate all clinical symptoms of a disease have significant value as well (Figure 9.1). They play a critical role in elucidating disease etiology which leads to new ideas and research pathways for therapeutic development as well as new ideas for further model development. Modeling rare disease is primarily aimed at helping patients. However, the impact is much broader because we have fundamental gaps in our understanding of the biology of disease phenotypic manifestation that will be addressed in these studies. For example, the catastrophic neural degeneration observed and investigated in Tay-Sachs models is likely to unravel aspects of immune responses in brain that are novel.
Use of animal models is necessary to generate the knowledge to ameliorate quality of life issues for patients and families. For disorders where great models have already been established, we expect the current era of genome manipulation to lead to human disease models more sophisticated than gene deletion. The Pahenu models, which are point mutations generated in forward screens, have been incredibly useful. As we move to the future, we also expect to see more frequent use of designer models engineered to carry specific human disease mutations. Advances in iPS and organoid technology will also be used to create patient-specific models more frequently. As model-making in atypical clinically-based research animals, such as pigs and cows, becomes possible because of advances in genome engineering (145), we also expect to see more complicated models for disorders. We are confident that each direction will result in eventual realization of truly designer treatments.
PKU is a rare disease, and LNS, TSD and SD are even rarer, yet all are heavily studied. There are many other CMDs that are ultra-rare, with fewer than 100 patients identified world-wide and these CMDs are understudied. While animal models can be engineered for such diseases, there is no a priori guarantee that the expense and research effort will produce a model for clinical aspects of disease and the research enterprise is simply not vast enough for an effective focus on each of these disorders. The community should also embrace genetic model systems such as zebrafish and invertebrates such as Drosophila and C. elegans for these diseases because these models are cheap and fast and bring the power of forward unbiased genetics to the table, which can be less expensive and yet quite effective in generating therapeutic ideas.
Abbreviations & Terminology
- CMD
congenital metabolic disorder
- ENU
ethylnitrosurea
- ES
embryonic stem cell
- GM2
G represents gangliosides, M indicates monosialic and 2 indicates 2nd monosialic ganglioside discovered
- HEXA, HEXA
human acetylhexosaminidase α subunit protein and gene, respectively. HEXA also refers to the enzymatic activity of the hexosaminidase α/ β dimer
- Hexa, Hexa
mouse acetylhexosaminidase α subunit protein and gene, respectively
- HEXB, HEXB
human acetylhexosaminidase β subunit protein and gene, respectively. HEXB also refers to the enzymatic activity of the hexosaminidase β dimers
- Hexb, Hexb
mouse acetylhexosaminidase β subunit protein and gene, respectively
- HEXS
refers to the enzymatic activity of the hexosaminidase α dimers
- HPRT, HPRT
human hypoxanthine-guanine phosphoribosyltransferase protein and gene, respectively
- Hprt, Hprt
rodent hypoxanthine-guanine phosphoribosyltransferase protein and gene, respectively
- IEM
Inborn Error of Metabolism
- iPS
induced pluripotent stem cells
- LNS
Lesch-Nyhan Syndrome
- PAH, PAH
human phenylalanine hydroxylase protein and gene, respectively
- Pah, Pah
rodent phenylalanine hydroxylase protein and gene, respectively
- PAL
phenylalanine ammonia lyase
- PEG
polyethylene glycol
- PKU
phenylketonuria
- SD
Sandhoff Disease
- THBD
tetrahydrobiopterin deficiency
- TSD
Tay-Sachs disease
- BH4
tetrahydrobiopterin
- Ataxia
is a lack of voluntary coordination of muscle movements
- Dysphagia
is difficulty swallowing
- Hyperuricemia
refers to elevated uric acid levels in the blood.
- Hyperphenylalaninemia
refers to elevated concentrations of the amino acid phenylalanine in the blood.
- PEGylated
refers to covalent and non-covalent modification of a protein with PEG.
References
- 1.Harthan AA. An Introduction to Pharmacotherapy for Inborn Errors of Metabolism. J Pediatr Pharmacol Ther. 2018;23(6):432–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. 1902. Mol Med [Internet]. 1996May [cited 2019 Sep 13];2(3):274–82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8784780 [PMC free article] [PubMed] [Google Scholar]
- 3.Kruszka P, Regier D. Inborn errors of metabolism: transition from childhood to adulthood. Am Fam Physician. American Family Physician; 2019;99(1):25–32. [PubMed] [Google Scholar]
- 4.Kanungo S, Patel DR, Neelakantan M, Ryali B. Newborn screening and changing face of inborn errors of metabolism in the United States. Ann Transl Med [Internet]. AME Publications; 2018December [cited 2019 Sep 13];6(24):468. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30740399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bick D, Jones M, Taylor SL, Taft RJ, Belmont J. Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J Med Genet [Internet]. 2019April25 [cited 2019 Sep 13];jmedgenet-2019-106111. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31023718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heimer G, Kerätär JM, Riley LG, Balasubramaniam S, Eyal E, Pietikäinen LP, et al. MECR Mutations Cause Childhood-Onset Dystonia and Optic Atrophy, a Mitochondrial Fatty Acid Synthesis Disorder. Am J Hum Genet. 2016;99(6):1229–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertheim-Tysarowska K, Gos M, Sykut-Cegielska J, Bal J. Genetic analysis in inherited metabolic disorders--from diagnosis to treatment. Own experience, current state of knowledge and perspectives. Dev period Med [Internet]. [cited 2019 Sep 13];19(4):413–31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26982749 [PubMed] [Google Scholar]
- 8.Ezgu F Inborn Errors of Metabolism. In: Advances in clinical chemistry [Internet]. 2016. [cited 2019 Sep 13]. p. 195–250. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26975974 [DOI] [PubMed] [Google Scholar]
- 9.Argmann CA, Houten SM, Zhu J, Schadt EE. A Next Generation Multiscale View of Inborn Errors of Metabolism. Cell Metab [Internet]. NIH Public Access; 2016January12 [cited 2019 Aug 1];23(1):13–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26712461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirrs S, Hollak C, Merkel M, Sechi A, Glamuzina E, Janssen MC, et al. The Frequencies of Different Inborn Errors of Metabolism in Adult Metabolic Centres: Report from the SSIEM Adult Metabolic Physicians Group. In: JIMD reports [Internet]. Wiley-Blackwell; 2015. [cited 2019 Sep 15]. p. 85–91. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26450566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.BICKEL H, GERRARD J, HICKMANS EM. The Influence of Phenylalanine Intake on the Chemistry and Behaviour of a Phenylketonuria Child. Acta Paediatr [Internet]. John Wiley & Sons, Ltd; (10.1111); 1954. January 1 [cited 2019 Sep 15];43(1):64–77. Available from: http://doi.wiley.com/10.1111/j.1651-2227.1954.tb04000.x [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharya K, Wotton T, Wiley V. The evolution of blood-spot newborn screening. Transl Pediatr [Internet]. AME Publications; 2014April [cited 2019 Sep 15];3(2):63–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26835325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burlina AP, Lachmann RH, Manara R, Cazzorla C, Celato A, van Spronsen FJ, et al. The neurological and psychological phenotype of adult patients with early-treated phenylketonuria: A systematic review. J Inherit Metab Dis [Internet]. John Wiley & Sons, Ltd; 2019March12 [cited 2019 Sep 15];42(2):209–19. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jimd.12065 [DOI] [PubMed] [Google Scholar]
- 14.Brown CS, Lichter-Konecki U. Phenylketonuria (PKU): A problem solved?Mol Genet Metab reports [Internet]. Elsevier; 2016March [cited 2019 Sep 15];6:8–12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27014571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams RA, Mamotte CDS, Burnett JR. Phenylketonuria: An Inborn Error of Phenylalanine Metabolism. Clin Biochem. 2008;49:31–41. [PMC free article] [PubMed] [Google Scholar]
- 16.Centerwall SA, Centerwall WR. The Discovery of Phenylketonuria: The Story of a Young Couple, Two Retarded Children, and a Scientist. Pediatrics. 2000;105(1):89–103. [DOI] [PubMed] [Google Scholar]
- 17.Blau N Genetics of Phenylketonuria: Then and Now. Hum Mutat. 2016;37(6):508–15. [DOI] [PubMed] [Google Scholar]
- 18.Christ SE. Asbjørn Følling and the Discovery of Phenylketonuria. J Hist Neurosci [Internet]. 2003March1 [cited 2019 Sep 7];12(1):44–54. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12785112 [DOI] [PubMed] [Google Scholar]
- 19.Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med [Internet]. Nature Publishing Group; 2011August6 [cited 2019 Sep 7];13(8):697–707. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00125817-201108000-00002 [DOI] [PubMed] [Google Scholar]
- 20.Nova MP, Kaufman M, Halperin A. Scleroderma-like skin indurations in a child with phenylketonuria: A clinicopathologic correlation and review of the literature. J Am Acad Dermatol [Internet]. Mosby; 1992February1 [cited 2019 Sep 15];26(2):329–33. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/019096229270048K?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 21.Van Vliet D, Van Wegberg AMJ, Ahring K, Bik-Multanowski M, Blau N, Bulut FD, et al. Can untreated PKU patients escape from intellectual disability? A systematic review. Orphanet J Rare Dis. Orphanet Journal of Rare Diseases; 2018;13(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Vliet D, Bruinenberg VM, Mazzola PN, Van Faassen MHJR, De Blaauw P, Pascucci T, et al. Therapeutic brain modulation with targeted large neutral amino acid supplements in the Pah-enu2 phenylketonuria mouse model. Am J Clin Nutr. 2016;104(5):1292–300. [DOI] [PubMed] [Google Scholar]
- 23.Pietz J Neurological aspects of adult phenylketonuria. Curr Opin Neurol. 1998;11(6):679–88. [DOI] [PubMed] [Google Scholar]
- 24.Anderson PJ, Wood SJ, Francis DE, Coleman L, Anderson V, Boneh A. Are neuropsychological impairments in children with early-treated phenylketonuria (PKU) related to white matter abnormalities or elevated phenylalanine levels? Dev Neuropsychol. 2007;32(2):645–68. [DOI] [PubMed] [Google Scholar]
- 25.Krishnamoorthy U, Dickson M. Maternal phenylketonuria in pregnancy. Obstet Gynaecol. 2005;7(1):28–33. [Google Scholar]
- 26.Lenke RR, Levy HL. Maternal Phenylketonuria and Hyperphenylalaninemia. N Engl J Med. 1980;303(21):1202–8. [DOI] [PubMed] [Google Scholar]
- 27.Woo SLC, Lidsky As, Güttler F, Chandra T, Robson KJH. Cloned human phenylalanine hydroxylase gene allows prenatal diagnosis and carrier detection of classical phenylketonuria. Nature. 1983;306:151–5. [DOI] [PubMed] [Google Scholar]
- 28.Kwok SCM, Ledley FD, DiLella AG, Robson KJH, Woo SLC. Nucleotide Sequence of a Full-Length Complementary DNA Clone and Amino Acid Sequence of Human Phenylalanine Hydroxylase. Biochemistry. 1985;24(3):556–61. [DOI] [PubMed] [Google Scholar]
- 29.Romani C, Palermo L, MacDonald A, Limback E, Hall SK, Geberhiwot T. The impact of phenylalanine levels on cognitive outcomes in adults with phenylketonuria: Effects across tasks and developmental stages. Neuropsychology. 2017;31(3):242–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Z, Sellers J, Moult J. Protein stability and in vivo concentration of missense mutations in phenylalanine hydroxylase. Proteins [Internet]. NIH Public Access; 2012January [cited 2019 Sep 11];80(1):61–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21953985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gersting SW, Kemter KF, Staudigl M, Messing DD, Danecka MK, Lagler FB, et al. Loss of Function in Phenylketonuria Is Caused by Impaired Molecular Motions and Conformational Instability. Am J Hum Genet [Internet]. Cell Press; 2008July11 [cited 2019 Sep 11];83(1):5–17. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/S0002929708003224?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shedlovsky A, McDonald JD, Symula D, Dove Wf. Mouse models of human phenylketonuria. Genetics. 1993;134(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mcdonald JDD, Bode VC, Dove WFF, Shedlovsky A, Vernon C, Dove WFF, et al. Pahhph-5: a mouse mutant deficient in phenylalanine hydroxylase. Proc Natl Acad Sci. 1990;87(5):1965–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haefele MJ, White G, McDonald JD. Characterization of the Mouse Phenylalanine Hydroxylase Mutation Pahenu3. Mol Genet Metab [Internet]. Academic Press; 2001January1 [cited 2019 Sep 11];72(1):27–30. Available from: https://www.sciencedirect.com/science/article/pii/S1096719200931044?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 35.McDonald JD, Charlton CK. Characterization of mutations at the mouse phenylalanine hydroxylase locus. Genomics. 1997;39(3):402–5. [DOI] [PubMed] [Google Scholar]
- 36.Blau N, Thony B, Heizmann CW, Dhondtt JL. Tetrahydrobiopterin deficiency: From phenotype to genotype. Pteridines. 1993;4(1):1–10. [Google Scholar]
- 37.Fitzpatrick PF. Allosteric Regulation of Phenylalanine Hydroxylase. Arch Biochem Biophys. 2012;519(2):194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gersting SW, Lagler FB, Eichinger A, Kemter KF, Danecka MK, Messing DD, et al. Pah enu1 is a mouse model for tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency and promotes analysis of the pharmacological chaperone mechanism in vivo. Hum Mol Genet [Internet]. Narnia; 2010May15 [cited 2019 Sep 11];19(10):2039–49. Available from: https://academic.oup.com/hmg/article-lookup/doi/10.1093/hmg/ddq085 [DOI] [PubMed] [Google Scholar]
- 39.Cederbaum S. Tetrahydrobiopterin and PKU: Into the future. J Pediatr. Mosby, Inc.; 2011;158(3):351–3. [DOI] [PubMed] [Google Scholar]
- 40.Koukol J, Conn EE. The Metabolism of Aromatic Compounds in Higher Plants. J Biol Chem. 1961;236(10):2692–8. [PubMed] [Google Scholar]
- 41.Levy HL, Sarkissian CN, Scriver CR. Phenylalanine ammonia lyase (PAL): From discovery to enzyme substitution therapy for phenylketonuria. Mol Genet Metab [Internet]. 2018August [cited 2019 Sep 16];124(4):223–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29941359 [DOI] [PubMed] [Google Scholar]
- 42.Hoskins JA, Jack G, Wade He, Peiris RJD, Wright EC, Starr DJT, et al. Enzymatic Control of Phenylalanine Intake in Phenylketonuria. Lancet. 1980;1(8165):392–4. [DOI] [PubMed] [Google Scholar]
- 43.Taipa MÂ, Fernandes P, de Carvalho CCCR. Production and Purification of Therapeutic Enzymes. In: Advances in experimental medicine and biology [Internet]. 2019. [cited 2019 Sep 16]. p. 1–24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31482492 [DOI] [PubMed] [Google Scholar]
- 44.Sarkissian CN, Gamez A, Wang L, Charbonneau M, Fitzpatrick P, Lemontt JF, et al. Preclinical evaluation of multiple species of PEGylated recombinant phenylalanine ammonia lyase for the treatment of phenylketonuria. Proc Natl Acad Sci. 2008;105(52):20894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahan kC, Gandhi MA, Anand S. Pegvaliase: a novel treatment option for adults with phenylketonuria. Curr Med Res Opin. 2019;35(4):647–51. [DOI] [PubMed] [Google Scholar]
- 46.McDonald JD, Dyer CA, Gailis L, Kirby ML. Cardiovascular Defects among the Progeny of Mouse Phenylketonuria Females. Pediatr Res [Internet]. 1997July [cited 2019 Sep 11];42(1):103–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9212044 [DOI] [PubMed] [Google Scholar]
- 47.Watson JN, Seagraves NJ. RNA-Seq analysis in an avian model of maternal phenylketonuria. Mol Genet Metab [Internet]. Academic Press; 2019January1 [cited 2019 Sep 1];126(1):23–9. Available from: https://www.sciencedirect.com/science/article/pii/S1096719218302567?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 48.Brass CA, Isaacs CE, McChesney R, Greencard O. The Effects og Hyperphenylalaninemia on Fetal Development: a new Animal Model of Maternal Phenylketonuria. Pediatr Res. 1982;16(5):388–94. [DOI] [PubMed] [Google Scholar]
- 49.Gr Kerr, CHamove AS, Harlow HF, Waisman HA. “Fetal PKU:” The Effect of Maternal Hyperphenylalaninemia During Pregnancy In The Rhesus Monkey (Macaca Mulatta). Pediatrics. 1968;42(1):27–36. [PubMed] [Google Scholar]
- 50.Matalon R, Surendran S, McDonald JDD, Okorodudu AOO, Tyring SKK, Michals-Matalon K, et al. Abnormal Expression of Genes Associated with Development and Inflammation in the Heart of Mouse Maternal Phenylketonuria Offspring. Int J Immunopathol Pharmacol [Internet]. SAGE PublicationsSage UK: London, England; 2005July24 [cited 2019 Sep 11];18(3):557–65. Available from: http://journals.sagepub.Com/doi/10.1177/039463200501800316 [DOI] [PubMed] [Google Scholar]
- 51.Le Barz M, Anhê FF, Varin TV, Desjardins Y, Levy E, Roy D, et al. Probiotics as Complementary Treatment for Metabolic Disorders. Diabetes Metab J [Internet]. Korean Diabetes Association; 2015August [cited 2019 Sep 16];39(4):291–303. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26301190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkissian CNN, Shao Z, Blain F, Peevers R, Su H, Heft R, et al. A different approach to treatment of phenylketonuria: Phenylalanine degradation with recombinant phenylalanine ammonia lyase. 1999March2 [cited 2019 Sep 7];96(5):2339–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10051643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durrer KE, Allen MS, Hunt von Herbing I. Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PkU. Wilson BA, editor. PLoS One [Internet]. 2017May17 [cited 2019 Sep 11];12(5):e0176286. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28520731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crook N, Ferreiro A, Gasparrini AJ, Pesesky MW, Gibson MK, Wang B, et al. Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut. Cell Host Microbe. Elsevier Inc.; 2019;25(4):499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol [Internet]. Nature Publishing Group; 2018October13 [cited 2019 Sep 10];36(9):857–64. Available from: http://www.nature.com/articles/nbt.4222 [DOI] [PubMed] [Google Scholar]
- 56.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. Nature Publishing Group; 2017;551(7681):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature [Internet]. Nature Publishing Group; 2016May20 [cited 2019 Sep 11];533(7603):420–4. Available from: http://www.nature.com/articles/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med [Internet]. Nature Publishing Group; 2018October8 [cited 2019 Sep 11];24(10):1519–25. Available from: http://www.nature.com/articles/s41591-018-0209-1 [DOI] [PubMed] [Google Scholar]
- 59.Lesch M, Nyhan WL. A familial disorder of uric acid metabolism and central nervous system function. Am J Med [Internet]. Elsevier; 1964April1 [cited 2019 Sep 2];36(4):561–70. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/0002934364901044?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 60.Seegmiller JE, Rosenbloom FM, Kelley WN. Enzyme defect associated with a sex-linked human neurological disorder and excessive purine synthesis. Science [Internet]. American Association for the Advancement of Science; 1967March31 [cited 2019 Sep 2];155(3770):1682–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/6020292 [DOI] [PubMed] [Google Scholar]
- 61.Jolly DJ, Okayama H, Berg P, Esty AC, Filpula D, Bohlen P, et al. Isolation and characterization of a full-length expressible cDNA for human hypoxanthine phosphoribosyl transferase. Proc Natl Acad Sci [Internet]. 1983January1 [cited 2019 Sep 10];80(2):477–81. Available from: http://www.ncbi.nlm.nih.gov/pubmed/6300847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nyhan Wl. Lesch-Nyhan disease. J Hist Neurosci. 2005;14:1–10. [DOI] [PubMed] [Google Scholar]
- 63.Crawhall JC, Henderson JF, Kelley WN. Diagnosis and Treatment of the Lesch-Nyhan Syndrome. Pediatr Res [Internet]. 1972May [cited 2019 Sep 2];6(5):504–13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/4558815 [DOI] [PubMed] [Google Scholar]
- 64.Zizzo MG, Frinchi M, Nuzzo D, Jinnah HA, Mudó G, Condorelli DF, et al. Altered gastrointestinal motility in an animal model of Lesch-Nyhan disease. Auton Neurosci [Internet]. Elsevier; 2018March1 [cited 2019 Sep 2];210:55–64. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/S1566070217302692?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres RJ, Puig JG. Hypoxanthine-guanine phosophoribosyltransferase (HPRT) deficiency: Lesch-Nyhan syndrome. Orphanet J Rare Dis [Internet]. 2007December8 [cited 2019 Sep 10];2(1). Available from: http://www.ncbi.nlm.nih.gov/pubmed/18067674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jinnah HA. Lesch-Nyhan disease: from mechanism to model and back again. Dis Model Mech [Internet]. 2009March1 [cited 2019 Sep 10];2(3–4):116–21. Available from: http://dmm.biologists.org/cgi/doi/10.1242/dmm.002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu R, Sutcliffe D, Zhao H, Huang X, Schretlen DJ, Benkovic S, et al. Clinical severity in Lesch–Nyhan disease: The role of residual enzyme and compensatory pathways. Mol Genet Metab [Internet]. Academic Press; 2015January1 [cited 2019 Sep 10];114(1):55–61. Available from: https://www.sciencedirect.com/science/article/pii/S1096719214003461?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bell S, Kolobova I, Crapper L, Ernst C. Lesch–Nyhan syndrome: Models, theories, and therapies. Mol Syndromol. 2016;7(6):302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ernst M, Zametkin AJ, Matochik jA, Pascualvaca D, Jons PH, Hardy K, et al. Presynaptic Dopaminergic Deficits in Lesch–Nyhan Disease. N Engl J Med [Internet]. Massachusetts Medical Society; ; 1996. June 13 [cited 2019 Sep 10];334(24):1568–72. Available from: http://www.nejm.org/doi/abs/10.1056/NEJM199606133342403 [DOI] [PubMed] [Google Scholar]
- 70.Wong DF, Harris JC, Naidu S, Yokoi F, Marenco S, Dannals RF, et al. Dopamine transporters are markedly reduced in Lesch-Nyhan disease in vivo. Proc Natl Acad Sci U S A [Internet]. National Academy of Sciences; 1996May28 [cited 2019 Sep 10];93(11):5539–43. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8643611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roach eS, Delgado M, Anderson L, Iannaccone ST, Bums DK. Carbamazepine Trial for Lesch-Nyhan Self-Mutilation. J Child Neurol [Internet]. 1996November2 [cited 2019 Sep 16];11(6):476–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9120227 [DOI] [PubMed] [Google Scholar]
- 72.Pozzi M, Piccinini L, Gallo M, Motta F, Radice S, Clementi E. Treatment of motor and behavioural symptoms in three Lesch-Nyhan patients with intrathecal baclofen. Orphanet J Rare Dis [Internet]. 2014December12 [cited 2019 Sep 16];9(1):208. Available from: http://ojrd.biomedcentral.com/articles/10.1186/s13023-014-0208-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodman eM, Torres RJ, Puig JG, Jinnah HA. Consequences of Delayed Dental Extraction in Lesch-Nyhan Disease. Mov Disord Clin Pract [Internet]. 2014September [cited 2019 Sep 10];1(3):225–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25419535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jinnah HA, De Gregorio L, Harris JC, Nyhan WL, O’Neill JP. The spectrum of inherited mutations causing HPRT deficiency: 75 new cases and a review of 196 previously reported cases. Mutat Res [Internet]. 2000October [cited 2019 Sep 10];463(3):309–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11018746 [DOI] [PubMed] [Google Scholar]
- 75.Zoref-Shani E, Bromberg Y, Brosh S, Sidi Y, Sperling O. Characterization of the Alterations in Purine Nucleotide Metabolism in Hypoxanthine-Guardne Phosphoribosyltransferase-Deficient Rat Neuroma Cell Line. J Neurochem [Internet]. John Wiley & Sons, Ltd; (10.1111); 2006. October 5 [cited 2019 Sep 2];61(2):457–63. Available from: http://doi.wiley.com/10.1111/j.1471-4159.1993.tb02146.x [DOI] [PubMed] [Google Scholar]
- 76.Shirley TL, Lewers JC, Egami K, Majumdar A, Kelly M, Ceballos-Picot I, et al. A human neuronal tissue culture model for Lesch-Nyhan disease. J Neurochem [Internet]. 2007May23 [cited 2015 Jun 14];101(3):841–53. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17448149 [DOI] [PubMed] [Google Scholar]
- 77.Kang TH, Guibinga G-H, Jinnah HA, Friedmann T. HPRT deficiency coordinately dysregulates canonical Wnt and presenilin-1 signaling: a neuro-developmental regulatory role for a housekeeping gene?PLoS One [Internet]. Public Library of Science; 2011January28 [cited 2019 Sep 10];6(1):e16572. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21305049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ceballos-Picot I, Mockel L, Potier M-C, Dauphinot L, Shirley TL, Torero-Ibad R, et al. Hypoxanthine-guanine phosphoribosyl transferase regulates early developmental programming of dopamine neurons: implications for Lesch-Nyhan disease pathogenesis. Hum Mol Genet [Internet]. Oxford University Press; 2009July1 [cited 2019 Sep 10];18(13):2317–27. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19342420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guibinga G-H, Hrustanovic G, Bouic K, Jinnah HA, Friedmann T. MicroRNA-mediated dysregulation of neural developmental genes in HPRT deficiency: clues for Lesch-Nyhan disease?Hum Mol Genet [Internet]. Oxford University Press; 2012February1 [cited 2019 Sep 10];21(3):609–22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22042773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wade-Martins R, White RE, Kimura H, Cook PR, James MR. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat Biotechnol [Internet]. 2000December [cited 2019 Sep 2];18(12):1311–4. Available from: http://www.nature.com/articles/nbt1200_1311 [DOI] [PubMed] [Google Scholar]
- 81.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch–Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature [Internet]. 1987March [cited 2019 Sep 2];326(6110):292–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3821905 [DOI] [PubMed] [Google Scholar]
- 82.Kuehn MR, Bradley A, Robertson EJ, Evans MJ. A potential animal model for Lesch–Nyhan syndrome through introduction of HPRT mutations into mice. Nature [Internet]. 1987March [cited 2019 Sep 2];326(6110):295–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3029599 [DOI] [PubMed] [Google Scholar]
- 83.Jinnah HA, Page T, Friedmann T. Brain Purines in a Genetic Mouse Model of Lesch-Nyhan Disease. J Neurochem. 1993;60(6):2036–45. [DOI] [PubMed] [Google Scholar]
- 84.Jinnah hA, Wojcik BE, Hunt M, Narang N, Lee KY, Goldstein M, et al. Dopamine deficiency in a genetic mouse model of Lesch-Nyhan disease. J Neurosci. 1994;14(3):1164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finger S, Heavens RP, Sirinathsinghji DJS, Kuehn MR, Dunnett SB. Behavioral and neurochemical evaluation of a transgenic mouse model of Lesch-Nyhan syndrome. J Neurol Sci. 1988;86:203–13. [DOI] [PubMed] [Google Scholar]
- 86.Wu C-L, Melton DW. Production of a model for Lesch–Nyhan syndrome in hypoxanthine phosphoribosyltransferase–deficient mice. Nat Genet [Internet]. Nature Publishing Group; 1993March [cited 2019 Sep 4];3(3):235–40. Available from: http://www.nature.com/articles/ng0393-235 [DOI] [PubMed] [Google Scholar]
- 87.Edamura K, Sasai H. No Self-Injurious Behavior Was Found in HPRT-Deficient Mice Treated With 9-Ethyladenine. Pharmacol Biochem Behav [Internet]. Elsevier; 1998October1 [cited 2019 Sep 4];61(2):175–9. Available from: https://www.sciencedirect.com/science/article/pii/S0091305798000951?via%3Dihub#BIB18 [DOI] [PubMed] [Google Scholar]
- 88.Engle S, Womer DE, Davies PM, Boivin G, Sahota A, Simmonds HA, et al. HPRT-APRT-deficient mice are not a model for lesch-nyhan syndrome. Hum Mol Genet [Internet]. Narnia; 1996October1 [cited 2019 Sep 4];5(10):1607–10. Available from: https://academic.oup.com/hmg/article-lookup/doi/10.1093/hmg/5.10.1607 [DOI] [PubMed] [Google Scholar]
- 89.Ellenbroek B, Youn J. Rodent models in neuroscience research: is it a rat race?Dis Model Mech [Internet]. Company of Biologists; 2016. [cited 2019 Sep 11];9(10):1079–87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27736744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.BREESE GR, TRAYLOR TD. Developmental characteristics of brain catecholamines and tyrosine hydroxylase in the rat: effects of 6-hydroxydopamine. Br J Pharmacol [Internet]. John Wiley & Sons, Ltd; (10.1111); 1972. February 1 [cited 2019 Sep 10];44(2):210–22. Available from: http://doi.wiley.com/10.1111/j.1476-5381.1972.tb07257.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breese GR, Baumeister AA, McCown TJ, Emerick SG, Frye GD, Mueller RA. Neonatal-6-hydroxydopamine treatment: Model of susceptibility for self-mutilation in the lesch-nyhan syndrome. Pharmacol Biochem Behav [Internet]. Elsevier; 1984September1 [cited 2019 Sep 10];21(3):459–61. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/S0091305784801100 [DOI] [PubMed] [Google Scholar]
- 92.Knapp DJ, Breese GR. The Use of Perinatal 6-Hydroxydopamine to Produce a Rodent Model of Lesch–Nyhan Disease. In: Current topics in behavioral neurosciences [Internet]. 2016. [cited 2019 Sep 2]. p. 265–77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27029809 [DOI] [PubMed] [Google Scholar]
- 93.Meek S, Thomson AJ, Sutherland L, Sharp MGF, Thomson J, Bishop V, et al. Reduced levels of dopamine and altered metabolism in brains of HPRT knock-out rats: a new rodent model of Lesch-Nyhan Disease. Sci Rep [Internet]. Nature Publishing Group; 2016. [cited 2019 Sep 2];6:25592. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27185277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Visser JE, Harris JC, Barabas G, Eddey GE, Jinnah HA. The Motor Disorder of Classic Lesch-Nyhan Disease. Nucleosides, Nucleotides and Nucleic Acids [Internet]. 2004December31 [cited 2019 Sep 10];23(8–9):1161–4. Available from: http://www.tandfonline.com/doi/abs/10.1081/NCN-200027432 [DOI] [PubMed] [Google Scholar]
- 95.Mastrangelo L, Kim J-E, Miyanohara A, Kang TH, Friedmann T. Purinergic signaling in human pluripotent stem cells is regulated by the housekeeping gene encoding hypoxanthine guanine phosphoribosyltransferase. Proc Natl Acad Sci U S A [Internet]. 2012February28 [cited 2019 Sep 10];109(9):3377–82. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1118067109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kang TH, Park Y, Bader JS, Friedmann T. The Housekeeping Gene Hypoxanthine Guanine Phosphoribosyltransferase (HPRT) Regulates Multiple Developmental and Metabolic Pathways of Murine Embryonic Stem Cell Neuronal Differentiation. Cooney AJ, editor. PLoS One [Internet]. Public Library of Science; 2013October9 [cited 2019 Sep 10];8(10):e74967. Available from: https://dx.plos.org/10.1371/journal.pone.0074967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tay W. Symmetrical Changes in the Region of the Yellow Spot in Each Eye of an Infant. Arch Neurol. 1969;20(1):104–5. [Google Scholar]
- 98.Sachs B Arrested Cerebral Development with Special Reference to its Cortical Pathology. J Nerv Ment Dis. 1887;14(9):541–3. [Google Scholar]
- 99.Kingdon EC, Russell JS. Infantile Cerebral Degeneration with Symmetrical Changes at the Macula. Med Chir Trans [Internet]. Royal Society of Medicine Press; 1897. [cited 2019 Sep 12];80:87–118.5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20896909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fernandes Filho JA, Shapiro BE. Tay-Sachs Disease. JAMA Neurol. 2004;61(9):1466–8. [DOI] [PubMed] [Google Scholar]
- 101.Maegawa GHB, Stockley T, Tropak M, Banwell B, Blaser S, Kok F, et al. The Natural History of Juvenile or Subacute GM2 Gangliosidosis: 21 New Cases and Literature Review of 134 Previously Reported. Pediatrics. 2006;118(5):e1550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferreira CR, Gahl WA. Lysosomal storage diseases. Metab Dis Found Clin Manag Genet Pathol. 2017;2:367–440. [Google Scholar]
- 103.Sandhoff K, Andreae U, Jatzkewitz H. Deficient hexosaminidase activity in an exceptional case of Tay-Sachs disease with additional storage of kidney globoside in visceral organs. Life Sci. 1968;7(6):283–8. [DOI] [PubMed] [Google Scholar]
- 104.Solovyeva VV, Shaimardanova AA, Chulpanova DS, Kitaeva KV., Chakrabarti L, Rizvanov AA. New approaches to Tay-Sachs disease therapy. Front Physiol. 2018;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bley AE, Giannikopoulos OA, Hayden D, Kubilus K, Tifft CJ, Eichler FS. Natural history of infantile G M2 gangliosidosis. Pediatrics. 2011;128(5):1233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neudorfer O, Pastores GM, Zeng BJ, Gianutsos J, Zaroff CM, Kolodny EH. Late-onset Tay-Sachs disease: Phenotypic characterization and genotypic correlation in 21 affected patients. Genet Med. 2005;7(2):119–23. [DOI] [PubMed] [Google Scholar]
- 107.Venugopalan P, Joshi S. Cardiac involvement in infantile Sandhoff disease. J Paediatr Child Health [Internet]. 2002February [cited 2019 Sep 16];38(1):98–100. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11869411 [DOI] [PubMed] [Google Scholar]
- 108.Sakpichaisakul K, Taeranawich P, Nitiapinyasakul A, Sirisopikun T. Identification of Sandhoff disease in a Thai family: clinical and biochemical characterization. J Med Assoc Thai [Internet]. 2010September [cited 2019 Sep 16];93(9):1088–92. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20873083 [PubMed] [Google Scholar]
- 109.Lee H-F, Chi C-S, Tsai C-R. Early cardiac involvement in an infantile Sandhoff disease case with novel mutations. Brain Dev [Internet]. 2017February [cited 2019 Sep 16];39(2):171–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27697305 [DOI] [PubMed] [Google Scholar]
- 110.Klima H, Tanaka A, Schnabel D, Nakano T, Schröder M, Suzuki K, et al. Characterization of full-length cDNAs and the gene coding for the human GM2 activator protein. FEBS Lett. 1991;289(2):260–4. [DOI] [PubMed] [Google Scholar]
- 111.Cachon-Gonzalez MB, Zaccariotto E, Cox TM. Genetics and Therapies for GM2 Gangliosidosis. Curr Gene Ther [Internet]. Bentham Science Publishers; 2018. [cited 2019 Sep 13];18(2):68–89. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29618308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lawson CA, Martin DR. Animal models of GM2 gangliosidosis: utility and limitations. Appl Clin Genet [Internet]. Dove Press; 2016. [cited 2019 Sep 1];9:111–20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27499644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Phaneuf D, Wakamatsu N, Huang JQ, Borowski A, Peterson AC, Fortunato SR, et al. Dramatically different phenotypes in mouse models of human Tay-Sachs and Sandhoff diseases. Hum Mol Genet. 1996;5(1):1–14. [DOI] [PubMed] [Google Scholar]
- 114.Yamanaka S, Johnson MD, Grinberg A, Westphal H, Crawley JN, Taniike M, et al. Targeted disruption of the Hexa gene results in mice with biochemical and pathologic features of Tay-Sachs disease. Proc Natl Acad Sci U S A. 1994;91(21):9975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sango K, Yamanaka S, Hoffmann A, Okuda Y, Sandhoff K, Suzuki K, et al. Mouse models of Tay-Sachs and neurologic phenotype and. Nature. 1995;11(2):170–6. [DOI] [PubMed] [Google Scholar]
- 116.Taniike M, Yamanaka S, Proia RLL, Langaman C, Bone-Turrentine T, Suzuki K. Neuropathology of mice with targeted disruption of Hexa gene, a model of Tay-Sachs disease. Acta Neuropathol [Internet]. 1995. [cited 2019 Sep 12];89(4):296–304. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7610760 [DOI] [PubMed] [Google Scholar]
- 117.Miklyaeva EI, Dong W, Bureau A, Fattahie R, Xu Y, Su M, et al. Late onset Tay–Sachs disease in mice with targeted disruption of the Hexa gene: behavioral changes and pathology of the central nervous system. Brain Res [Internet]. 2004March19 [cited 2019 Sep 12];1001(1–2):37–50. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14972652 [DOI] [PubMed] [Google Scholar]
- 118.Yuziuk JA, Bertoni C, Beccari T, Orlacchio A, Wu YY, Li SC, et al. Specificity of mouse GM2 activator protein and beta-N-acetylhexosaminidases A and B. Similarities and differences with their human counterparts in the catabolism of GM2. J Biol Chem [Internet]. 1998January2 [cited 2019 Sep 12];273(1):66–72. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9417048 [DOI] [PubMed] [Google Scholar]
- 119.Bertoni C, Li YT, Li SC. Catabolism of asialo-GM2 in man and mouse. Specificity of human/mouse chimeric GM2 activator proteins. J Biol Chem [Internet]. 1999October1 [cited 2019 Sep 12];274(40):28612–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10497228 [DOI] [PubMed] [Google Scholar]
- 120.Seyrantepe V, Demir SA, Timur ZK, Von Gerichten J, Marsching C, Erdemli E, et al. Murine Sialidase Neu3 facilitates GM2 degradation and bypass in mouse model of Tay-Sachs disease. Exp Neurol [Internet]. 2018January [cited 2019 Sep 12];299(Pt A):26–41. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28974375 [DOI] [PubMed] [Google Scholar]
- 121.Wada R, Tifft CJ, Proia RL. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. 2000September26 [cited 2019 Sep 13];97(20):10954–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11005868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jeyakumar M, Thomas R, Elliot-Smith E, Smith DA, van der Spoel AC, d’Azzo A, et al. Central nervous system inflammation is a hallmark of pathogenesis in mouse models of GM1 and GM2 gangliosidosis. Brain [Internet]. 2003April1 [cited 2019 Sep 13];126(4):974–87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12615653 [DOI] [PubMed] [Google Scholar]
- 123.Pelled D, Riebeling C, van Echten-Deckert G, Sandhoff K, Futerman AH. Reduced rates of axonal and dendritic growth in embryonic hippocampal neurones cultured from a mouse model of Sandhoff disease. Neuropathol Appl Neurobiol [Internet]. 2003August [cited 2019 Sep 13];29(4):341–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12887594 [DOI] [PubMed] [Google Scholar]
- 124.Ogawa Y, Kaizu K, Yanagi Y, Takada S, Sakuraba H, Oishi K. Abnormal differentiation of Sandhoff disease model mouse-derived multipotent stem cells toward a neural lineage. Kerkis I, editor. PLoS One [Internet]. 2017June2;12(6):e0178978. Available from: https://dx.plos.org/10.1371/journal.pone.0178978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.LEE Kuil, López Martí A, Carreras Mascaro A, van den Bosch JC, van den Berg P, van der Linde HC, et al. Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development. Glia [Internet]. John Wiley & Sons, Ltd; 2019May29 [cited 2019 Sep 11];67. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/glia.23641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker CG, Becker T. Adult zebrafish as a model for successful central nervous system regeneration. Restor Neurol Neurosci. 2008;26(2–3 AXONAL REGENERATO):71–80. [PubMed] [Google Scholar]
- 127.Cork L, Munnell J, Lorenz M, Murphy J, Baker H, Rattazzi M. GM2 ganglioside lysosomal storage disease in cats with beta-hexosaminidase deficiency. Science (80-) [Internet]. 1977May27 [cited 2019 Sep 13];196(4293):1014–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/404709 [DOI] [PubMed] [Google Scholar]
- 128.Kanae Y, Endoh D, Yamato O, Hayashi D, Matsunaga S, Ogawa H, et al. Nonsense mutation of feline β-hexosaminidase β-subunit (HEXB) gene causing Sandhoff disease in a family of Japanese domestic cats. Res Vet Sci [Internet]. 2007February [cited 2019 Sep 13];82(1):54–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16872651 [DOI] [PubMed] [Google Scholar]
- 129.Kolicheski A, Johnson GS, Villani NA, O’Brien DP, Mhlanga-Mutangadura T, Wenger DA, et al. GM2 Gangliosidosis in Shiba Inu Dogs with an In-Frame Deletion in HEXB. J Vet Intern Med [Internet]. 2017September [cited 2019 Sep 13];31(5):1520–6. Available from: http://doi.wiley.com/10.1111/jvim.14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin DR, Krum BK, Varadarajan GS, Hathcock TL, Smith BF, Baker HJ. An inversion of 25 base pairs causes feline G M2 gangliosidosis variant 0. Exp Neurol [Internet]. 2004May [cited 2019 Sep 13];187(1):30–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15081585 [DOI] [PubMed] [Google Scholar]
- 131.Rahman MM, Chang H-S, Mizukami K, Hossain MA, Yabuki A, Tamura S, et al. A frameshift mutation in the canine HEXB gene in toy poodles with GM2 gangliosidosis variant 0 (Sandhoff disease). Vet J [Internet]. 2012December [cited 2019 Sep 13];194(3):412–6. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1090023312002237 [DOI] [PubMed] [Google Scholar]
- 132.Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem [Internet]. 1998;273(22): 14037–45. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9593755 [DOI] [PubMed] [Google Scholar]
- 133.Zeng BJ, Torres PA, Viner TC, Wang ZH, Raghavan SS, Alroy J, et al. Spontaneous appearance of Tay–Sachs disease in an animal model. Mol Genet Metab [Internet]. Academic Press; 2008September1 [cited 2019 Sep 13];95(1–2):59–65. Available from: https://www-sciencedirect-com.ezaccess.libraries.psu.edu/science/article/pii/S1096719208001716?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 134.Fox J, Li YT, Dawson G, Alleman A, Johnsrude J, Schumacher J, et al. Naturally occurring GM2 gangliosidosis in two Muntjak deer with pathological and biochemical features of human classical Tay-Sachs disease (type B GM2 gangliosidosis). Acta Neuropathol [Internet]. 1999January1 [cited 2019 Sep 13];97(1):57–62. Available from: http://link.springer.com/10.1007/s004010050955 [DOI] [PubMed] [Google Scholar]
- 135.Kosanke SD, Pierce KR, Bay WW. Clinical and biochemical abnormalities in porcine GM2-gangliosidosis. Vet Pathol [Internet]. 1978November26 [cited 2019 Sep 13];15(6):685–99. Available from: http://journals.sagepub.com/doi/10.1177/030098587801500601 [DOI] [PubMed] [Google Scholar]
- 136.Rockwell HE, McCurdy VJ, Eaton SC, Wilson DU, Johnson AK, Randle AN, et al. AAV-mediated gene delivery in a feline model of Sandhoff disease corrects lysosomal storage in the central nervous system. ASN Neuro [Internet]. 2015March30 [cited 2019 Sep 13];7(2):175909141556990. Available from: http://journals.sagepub.com/doi/10.1177/1759091415569908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McCurdy VJ, Rockwell HE, Arthur JR, Bradbury AM, Johnson AK, Randle AN, et al. Widespread correction of central nervous system disease after intracranial gene therapy in a feline model of Sandhoff disease. Gene Ther [Internet]. 2015February4 [cited 2019 Sep 13];22(2):181–9. Available from: http://www.nature.com/articles/gt2014108 [DOI] [PubMed] [Google Scholar]
- 138.Bradbury AM, Cochran JN, McCurdy VJ, Johnson AK, Brunson BL, Gray-Edwards H, et al. Therapeutic Response in Feline Sandhoff Disease Despite Immunity to Intracranial Gene Therapy. Mol Ther [Internet]. 2013July [cited 2019 Sep 13];21(7):1306–15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23689599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Torres PA, Zeng BJ, Porter BF, Alroy J, Horak F, Horak J, et al. Tay-Sachs disease in Jacob sheep. Mol Genet Metab [Internet]. 2010December [cited 2019 Sep 12];101(4):357–63. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20817517 [DOI] [PubMed] [Google Scholar]
- 140.Porter BF, Lewis BC, Edwards JF, Alroy J, Zeng BJ, Torres PA, et al. Pathology of G M2 gangliosidosis in Jacob sheep. Vet Pathol. 2011;48(4):807–13. [DOI] [PubMed] [Google Scholar]
- 141.Wessels ME, Holmes JP, Jeffrey M, Jackson M, Mackintosh A, Kolodny EH, et al. GM2 Gangliosidosis in British Jacob Sheep. J Comp Pathol [Internet]. 2014February [cited 2019 Sep 13];150(2–3):253–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24309906 [DOI] [PubMed] [Google Scholar]
- 142.Gray-Edwards HL, Randle AN, Maitland SA, Benatti HR, Hubbard SM, Canning PF, et al. Adeno-Associated Virus Gene Therapy in a Sheep Model of Tay–Sachs Disease. Hum Gene Ther [Internet]. 2018March [cited 2019 Sep 13];29(3):312–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28922945 [DOI] [PubMed] [Google Scholar]
- 143.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov [Internet]. 2017February16 [cited 2019 Sep 16];16(2):115–30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27980341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Allende ML, Cook EK, Larman BC, Nugent A, Brady JM, Golebiowski D, et al. Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation. J Lipid Res [Internet]. 2018March;59(3):550–63. Available from: http://www.jlr.org/lookup/doi/10.1194/jlr.M081323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tu Z, Yang W, Yan S, Guo X, Li X-J. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener [Internet]. BioMed Central; 2015December4 [cited 2019 Sep 15];10(1):35. Available from: http://www.molecularneurodegeneration.com/content/10/1/35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hindle S, Hebbar S, Sweeney ST. Invertebrate models of lysosomal storage disease: what have we learned so far? Invertebr Neurosci [Internet]. 2011December25 [cited 2019 Sep 15];11(2):59–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22038288 [DOI] [PubMed] [Google Scholar]