Summary
Single particle cryo-EM has become a standard technique to determine protein structures at atomic resolution1–3. However, cryo-EM studies on protein-free RNA remain nascent. The Tetrahymena thermophila group I self-splicing intron was the first ribozyme discovered and has been a dominant model system for studying RNA catalysis and structure-function relationships4, but its full structure remains unknown. Here we report 3.1 Å cryo-EM structures of the full-length Tetrahymena ribozyme in substrate-free and bound states. Newly resolved peripheral regions form two coaxially stacked helices interconnected by two kissing loop pseudoknots wrapping around the catalytic core. The structures reveal two previously unforeseen tertiary interactions that embrace the catalytic core. The global architecture is nearly identical in both states, while the internal guide sequence and the guanosine binding site undergo a large conformational change and a localized shift, respectively, upon binding of RNA substrates. These results provide a long-sought structural view of a paradigmatic RNA enzyme and signal a new era for dissecting structure-function relationships in metalloribozymes guided by cryo-EM.
RNAs can fold into complex tertiary structures and participate in important biological processes in the absence of proteins, such as catalysis, transcriptional and translational regulation5,6. However, our understanding of RNA structure-function relationships remains limited due to the lack of RNA structural information. This paucity arises from the challenges posed to conventional X-ray crystallography and NMR by the intrinsic heterogeneity of RNAs7,8. Single particle cryogenic electron microscopy (cryo-EM) is an alternative structure determination method, but its application to RNA has been limited, similar to prior methods. There are currently fewer than ten protein-free RNA cryo-EM maps at better than 5 Å resolution, with the best resolved at 3.7 Å resolution, in the Electron Microscopy Data Bank9. Recently, we have developed an accelerated pipeline, Ribosolve, to determine protein-free RNA structures through sub-nanometer resolution cryo-EM maps, M2-seq secondary structure mapping, and Rosetta computational modeling. Using this pipeline, we determined 11 RNA cryo-EM structures10. A 6.8 Å structure of the full-length wild type Tetrahymena ribozyme was a highlight of this approach.
In 1982, the Tetrahymena group I self-splicing intron was discovered as the first example of a protein-free RNA catalyst and led to the coinage of the word “ribozyme”4. This ribozyme catalyzes two successive transesterification reactions to cleave the 5′ splice site and then ligate the 5′ and 3′ exons. Extensive studies indicated that the highly conserved core forms a compact structure11–15 (and references therein). Metal ions, especially Mg2+, are found essential to RNA structure stabilization and catalytic reactions12–14,16–22. The ribozyme’s peripheral regions have been predicted to form long-range interactions to stabilize the core23–27. Deletions and mutations of the ribozyme’s peripheral regions affect the folding pathways, perturb the stability of folded and misfolded states, and allosterically regulate catalysis23,24,26,28. The Tetrahymena ribozyme has also been used as a gene regulation tool in molecular therapeutics and RNA nanotechnology29.
Despite extensive prior efforts, the full-length Tetrahymena ribozyme at the resolution needed to guide structure-function studies remains unavailable. Westhof and colleagues predicted coordinates for the entire Tetrahymena ribozyme27. A groundbreaking 2.8 Å resolution crystal structure of the non-catalytic domain P4-P6 revealed important principles for RNA folding12,30 and crystallography15,31,32. Another crystal structure of the P3-P9 domain with 12 thermostabilizing mutations was resolved at 3.8 Å resolution13,14. However, such resolution and truncations/mutations precluded identification of all Mg2+ ions14. Determination of three-dimensional (3D) structures of the full-length ribozyme in different catalytic states are therefore needed to fully understand the tertiary structure fold and catalytic mechanism of the Tetrahymena ribozyme.
Here we determine structures of the Tetrahymena ribozyme in both apo and holo states at 3.1 Å resolution, the latter bound to two RNA substrates mimicking the state preceding the second step of splicing. The increased resolution and avoidance of extensive sequence truncations/mutations resolve new tertiary interactions and conformational changes of the internal guide sequence (IGS) and the catalytic site of the ribozyme upon substrate binding. These complete Tetrahymena ribozyme cryo-EM structures reveal a rich collection of novel structural and mechanistic insights into this classic RNA enzyme.
Apo L-21 ScaI ribozyme cryo-EM reconstruction
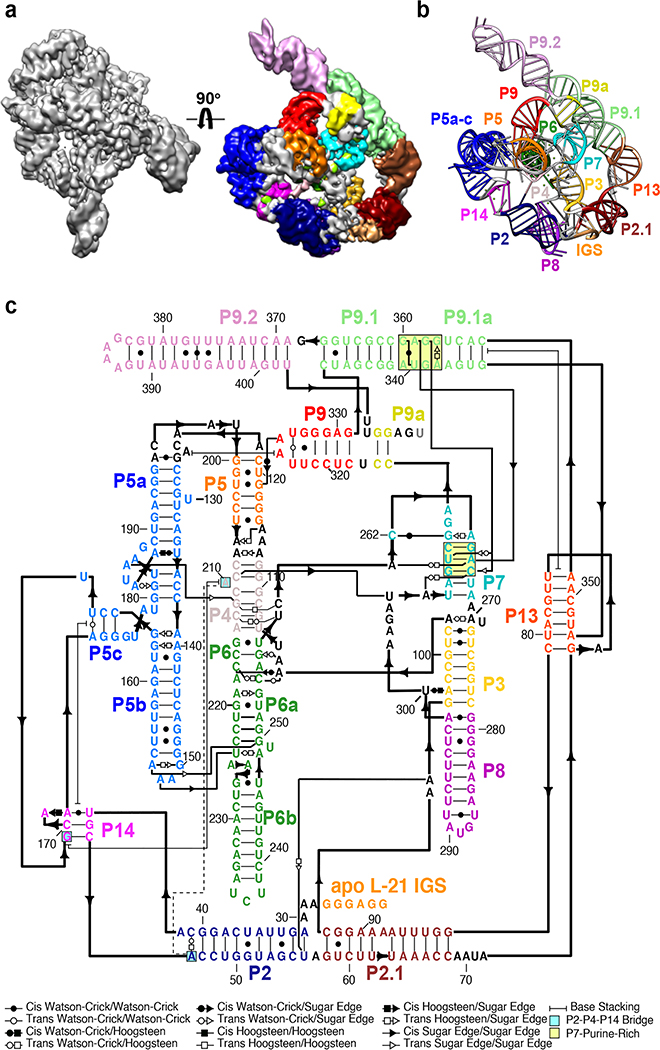
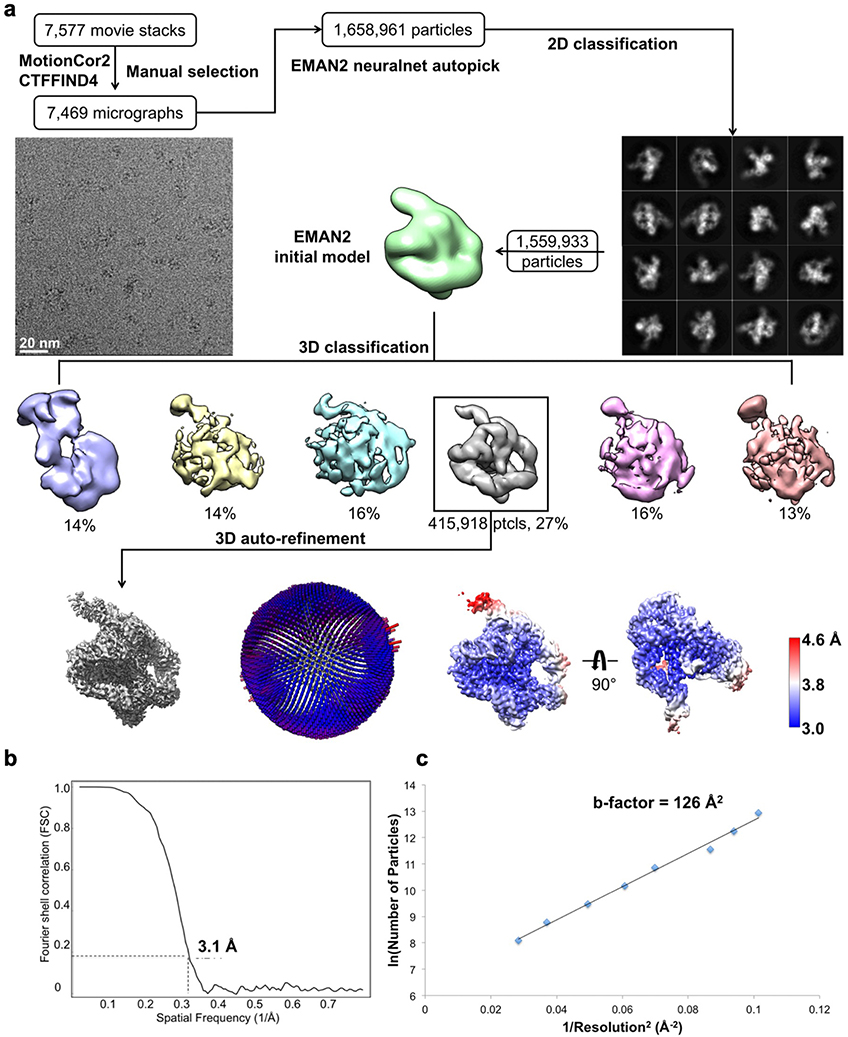
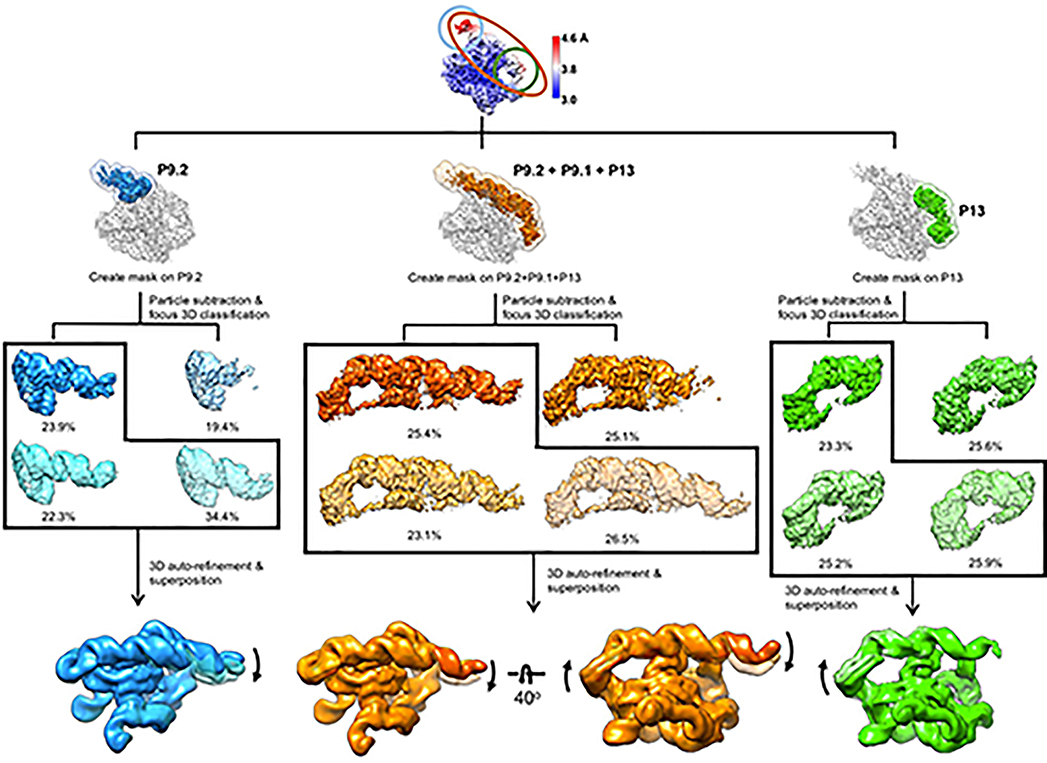
We applied cryo-EM to the apo L-21 ScaI Tetrahymena ribozyme, a linear form of the self-splicing intron without its first 21 nucleotides (nts 22–409), with transcription termination at a ScaI restriction endonuclease site in the DNA template. The ribozyme was readily visible in the cryo-EM images and yielded a 3.1 Å cryo-EM map (Figure 1a, Extended Data Figure 1, Extended Data Table 1). The local resolution map indicates that the catalytic core P3-P7 is better resolved to 3.0 Å, whereas flexible regions like stem P9.2 and P13 are resolved to 4.6 Å. Further 3D classifications that focus on P9.2 and/or P13 reveal modest conformational heterogeneity (Extended Data Figure 2).
Figure 1. Cryo-EM reconstruction of the apo L-21 ScaI ribozyme.
(a) The apo L-21 ScaI ribozyme cryo-EM reconstruction at 3.1 Å (left) and the segmented cryo-EM map (right) colored according to the secondary structure color scheme. (b) The cryo-EM model colored following the secondary structure color scheme. (c) The secondary structure of the apo L-21 ScaI ribozyme.
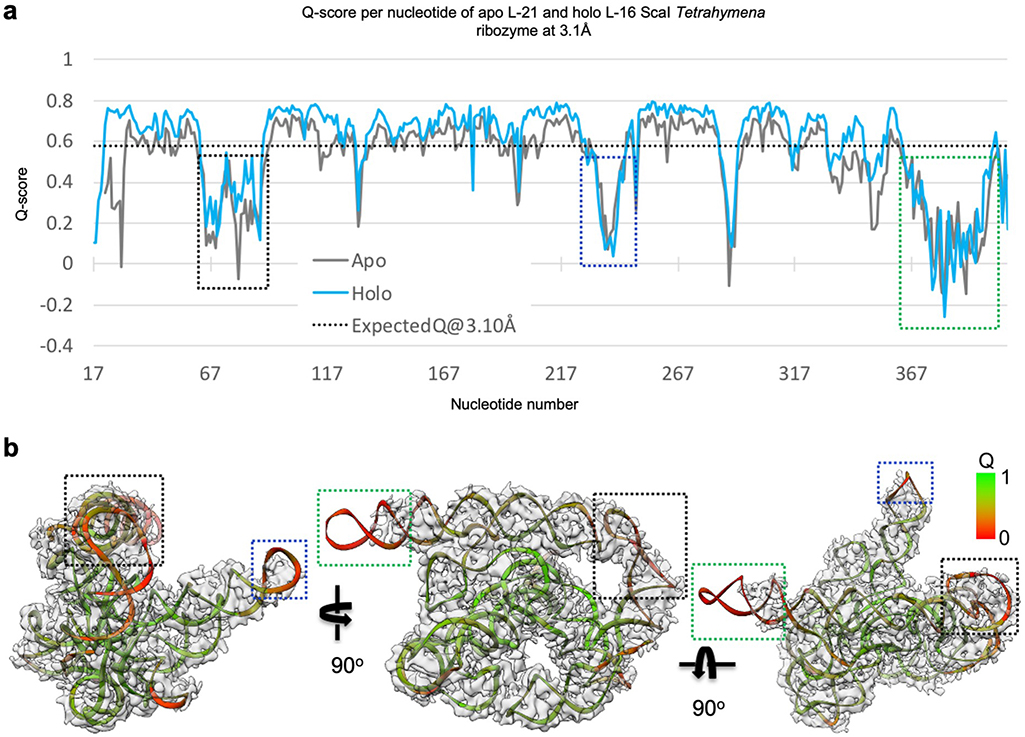
Most individual nucleotides are clearly resolved, allowing unambiguous modeling and an updated secondary structure layout based on the initial “Ribosolve” model (Figure 1b–c)10. The resolvability of the cryo-EM density is confirmed by Q-score analysis with an average Q-score of 0.53 per nucleotide, similar to those observed in other RNA-containing cryo-EM structures in this resolution range (Extended Data Figure 3)33.
Apo L-21 ScaI ribozyme architecture
The apo L-21 ScaI Tetrahymena ribozyme consists of the core P3-P9 domain previously captured by crystallography and the newly resolved peripheral regions including stems P2, P2.1, P9.1, P9.2, and two pseudoknots (PKs) P13 and P14 (Figure 1c). Tracing the ribozyme strand from 5′ to 3′ (Supplementary Video 1), the peripheral region starts from the 5′ single strand harboring the IGS that binds the oligonucleotide substrate, followed by the 11 base-paired (bp) P2, in which the loop (L2) forms a 3-bp PK P14 with L5c. After P2, the 11-bp P2.1 presents L2.1, which forms a 7-bp PK P13 with L9.1. This P13 connects to P2.1 via a 4-nt bulge. After P2.1, the ribozyme continues to residues 96–331, which presents the intricately pseudoknotted P3-P9 that forms binding sites for the substrates. The ribozyme strand continues on to P9.1, which exhibits a novel tertiary interaction with the P7 catalytic site and harbors L9.1 forming the other side of P13 with L2.1. P9.2 is a long stem subsequent to P9.1 that points away from the rest of the ribozyme. It is followed by a 2-bp P9a that returns the ribozyme’s 3′ end close to the catalytic site.
Cryo-EM confirms structural details in the P3-P9 domain
The P3-P9 region of the mutated Tetrahymena ribozyme was previously solved by X-ray crystallography12–15. The apo L-21 ScaI ribozyme cryo-EM structure largely agrees with those results and other group I ribozymes from Azoarcus (1U6B)17 and phage Twort (1Y0Q)18 (Extended Data Figure 4–6, Supplementary Table 1). Minor differences can be attributed to resolution and sequence differences13,14,17,18. Beyond this overall agreement in the core, the unique inclusion of the peripheral domains reveals novel structural and functional insights, described in the next sections.
Peripheral regions reveal unpredicted noncanonical interactions
The functional significance of the peripheral regions has been explored in previous work dissecting allosteric regulation of the catalytic process in the Tetrahymena ribozyme24,34. The apo L-21 ScaI ribozyme structure gives a clear view of tertiary interactions in the newly resolved peripheral regions that explain previous structural modeling and biochemical studies (Figure 2). At a domain level, Westhof and colleagues predicted the peripheral regions to stack coaxially and wrap around the catalytic core27. This generally agrees with our cryo-EM structure in that most peripheral helices are coaxially aligned, though there is an almost 90° bend involving the L2.1 loop between two long domains, P5c-P14-P2-P2.1 and P13-P9.1-P9.2 (Figure 2 center).
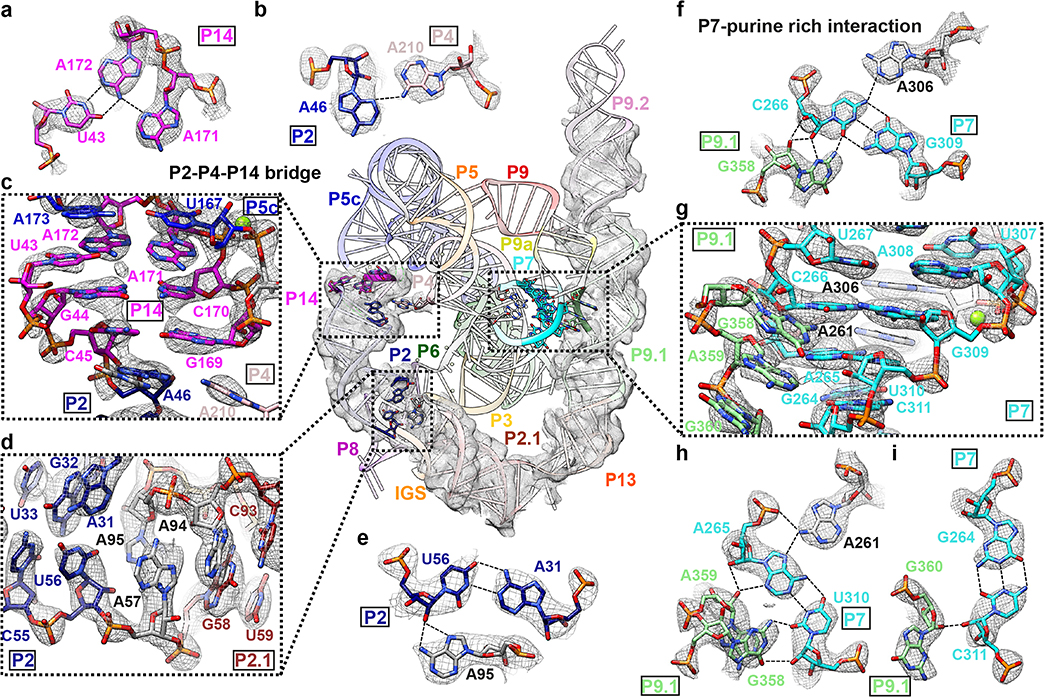
Figure 2. Novel structural insights in the peripheral regions of the apo L-21 ScaI ribozyme.
(a) Base triple consists of U43 and A171-A172 A-platform that connects P5c with P14. (b) A46 in P2 and A210 in P4 forms a ‘P2-P4-P14’ bridge (c) P14 that coaxially stacks with P5c and P2. (d) P2 forms a A31-U56-A95 base triple and connects with P2.1. (e) The A31-U56-A95 base triple. (f) G358 interacts with C266-G309-A306 to form a base quartet. (g) The bulge region in stem P9.1 stabilizes the catalytic site stem P7 via a purine-minor interaction. (h) G358 and A359 interacts with A265-U310-A261 to form base quartets. (i) G360 interacts with G264-C311 to form a base triple. Newly resolved peripheral region is presented with the cryo-EM map visualized at 3σ threshold. The cryo-EM local regions for all subpanels are visualized at 1σ threshold except for (c) at 2σ, (d and g) at 1.5σ. Dashed line indicates hydrogen bond.
At a nucleotide level, noncanonical pairings stack on Watson-Crick pairs to interconnect different peripheral helices (Supplementary Video 1). We resolve two interactions that involve base triples as examples of insights into RNA structure preorganization and impact on function. First, a seminal crystallographic analysis of the P4-P6 domain revealed a noncanonical “A-platform” motif in A171-A172 of L5c, potentially preorganizing this loop to interact with other parts of the ribozyme12,30. Our cryo-EM structure indeed reveals that A171-A172 retains the platform structure in the full-length ribozyme (Extended Data Figure 6e) and is involved in a U43-A171-A172 base triple that connects P5c and P2 via P14 (Figure 2a–c). Second, we observe A95 involved in a triple A31-U56-A95, positioned at the P2-P2.1 junction and stacked on both sides by other pairings (Figure 2d–e). This intricately folded junction is the ‘anchor point’ for the apo L-21 ScaI ribozyme’s IGS in the absence of substrates. Disruption of this junction would result in incorrect splice sites through changes in tertiary structure as was previously observed35.
Two new tertiary contacts with implications for catalysis and allosteric effects
The cryo-EM structure illuminates two new tertiary contacts not previously seen, both with potential significance for how peripheral domains impact catalysis. One contact, the ‘P2-P4-P14 bridge’, involves A210 flipping out of P4 and forming a hydrogen bond with A46 in P2 and a stack with G169 in P14 (Figure 2b–c). This interaction provides an explanation for the existence of bulged A210, which was found to be destabilizing in the context of the isolated P4-P6 domain but important for folding and function of the full-length Tetrahymena ribozyme15. Disruption of P14 formation led to decreased oligonucleotide substrate docking via an allosteric effect by P2 and the P2-P2.1 junction24,34, which is now explained directly by the new P4-P2-P14 bridge.
The second novel tertiary contact involves a larger interaction surface, the ‘P7-purine-rich interaction’. Previous observations indicated that P9.1-P9.2 played a role in the folding process to stabilize the catalytic site P3-P736. Our structure reveals that tertiary interactions between the P13-P9.1-P9.2 domain and the catalytic core are facilitated by a purine-rich internal loop in P9.1 (Figure 2f–i). This internal loop (nts 339–342, 357–360) forms a noncanonical G341-G357 pair that resembles an internal loop E structure of E. coli 5S rRNA (Extended Data Figure 6f)37. The loop’s other purines G358, A359 and G360 form noncanonical base pairs to the minor groove of the catalytic core helix P7 (Figure 2f, 2h, 2i). The P7 region harbors the ribozyme’s guanosine binding site and additionally forms base triples A265-U310-A261 and C266-G309-A306 at the catalytic site (Figure 2f, 2h, 2i)14. The P7-purine-rich interaction represents a tertiary contact that is closer in distance (about 17 Å) to the catalytic core than the previously proposed five tertiary contacts (about 23–48 Å), which may explain why mutations of P9.1-P9.2 lead to the largest effect of catalytic core folding stability24,34,36.
Rearrangement of the extended IGS in the holo L-16 ScaI ribozyme
To understand potential conformational changes through the Tetrahymena ribozyme functional cycle, we sought to capture the structure of more than just the apo state by cryo-EM. A classic crosslinking study indicated that the ribozyme’s substrate-binding IGS moves by 37 Å upon substrate binding, providing the earliest evidence that RNA can undergo large scale conformational changes and leading to the important hypothesis that such RNA flexibility is conserved in ancient machines like the ribosome and spliceosome38. However, structural evidence for the substantial conformational change of the IGS remains unavailable.
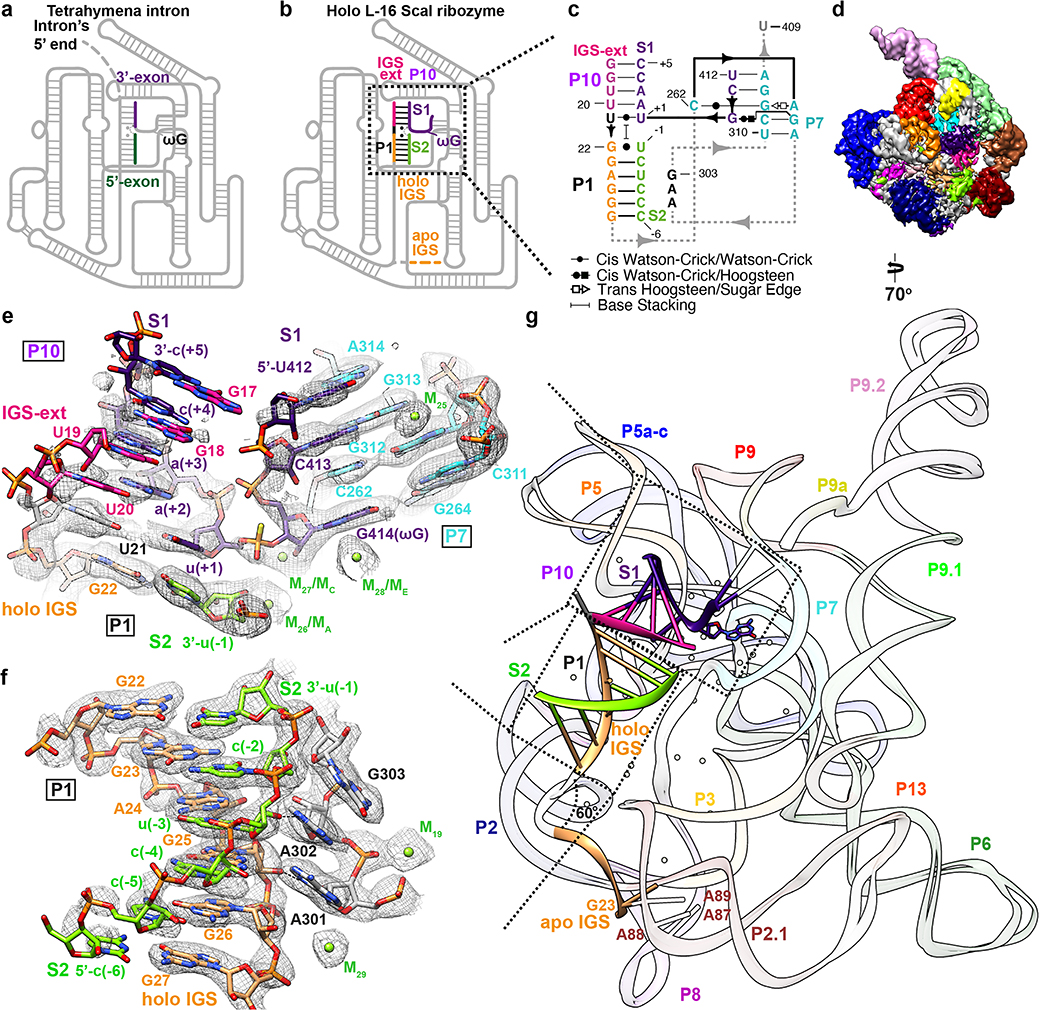
We determined a 3.1 Å cryo-EM map of the L-16 ScaI ribozyme (nts 17–409) bound to two RNA substrates. Substrate S1 contains the terminal G414 (ωG) of the intron and the 3′ splice site, while S2 comprises the cleaved 5′ exon. We introduced a phosphorothioate substitution at the scissile phosphate in S2 in order to isolate the docked complex mimicking the start of the second step of splicing (ligation of 5′- and 3′- exons)39, which is essentially the reverse reaction of the first step of splicing (Figure 3a–c)40,41. The holo L-16 ScaI ribozyme adopts an identical overall architecture with similar flexible regions compared to the apo L-21 ScaI ribozyme (Figure 3d, Extended Data Figure 7), except that the IGS (nts 17–27) undergoes a substantial conformational change to form a 4-bp stem P10 and a 6-bp stem P1 with a 8-nt substrate S1 and a 6-nt substrate S2 (also defined as P in previous studies41,42) (Figure 3c and 3e–g, Supplementary Video 2). The 5′ end of S1 (nts 412–414) mimics the Tetrahymena intron’s 3′ terminus that forms a 2-bp stem P7 extension, G313-C413 and A314-U412, in order to place the intron’s ωG in the guanosine binding site (Figure 3e). This is the same binding site as the exogenous guanosine that is the nucleophile for the first step of splicing13,14,17,18,20. The 3′ end of S1, from u(+1) to c(+5), mimics the 3′-exon (Figure 3a–b) and forms stem P10 with the 5-nt IGS extension at the 5′ end of L-16 ScaI ribozyme (nts 17–21) (Figure 3e). S2 mimics the 5′-exon of Tetrahymena (Figure 3a–b) and forms stem P1 with the 6-nt IGS (nts 22–27), which is stabilized by A301 and A302 in J8/7 (Figure 3f). Comparison between the apo and holo structures shows a ~60° conformational change of the IGS, moving its 5′ end from A87, A88 and A89, previously observed sites of crosslinking, into the catalytic core (Figure 3g, Supplementary Video 2).
Figure 3. Cryo-EM structure of the holo L-16 ScaI ribozyme reveals docked P1-P10 and a substantial conformational change of IGS that mimics the second step of splicing.
Schematic illustrations of (a) the Tetrahymena intron in the second step of splicing and (b) the holo L-16 ScaI ribozyme mimicking the reaction in (a). (c) Secondary structure of the docked P1-P10 in the catalytic site. (d) Cryo-EM map of the holo L-16 ScaI ribozyme colored following the secondary structure. (e) Substrate S1 (dark purple) forms P10 with the IGS extension (deep pink), P7 extension with G313 and A314 (cyan), and occupies the guanosine binding site, visualized at 0.9σ threshold. (f) Substrate S2 (chartreuse) and IGS (sandy brown) forms P1, visualized at 1σ threshold. (g) Superposition of the apo and holo structure reveals conformational change of IGS from a substrate-free site close to A87, A88, A89. Dashed arrows in (a) and (b) indicate nucleophilic attack direction of the second step of splicing, dashed line indicates hydrogen bond.
Localized shift in the guanosine binding site
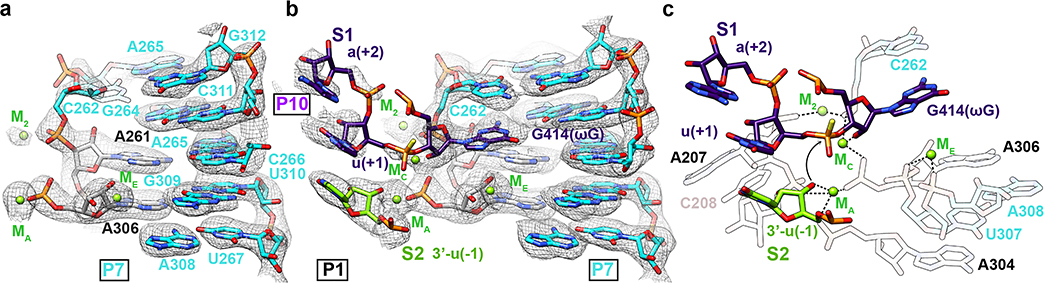
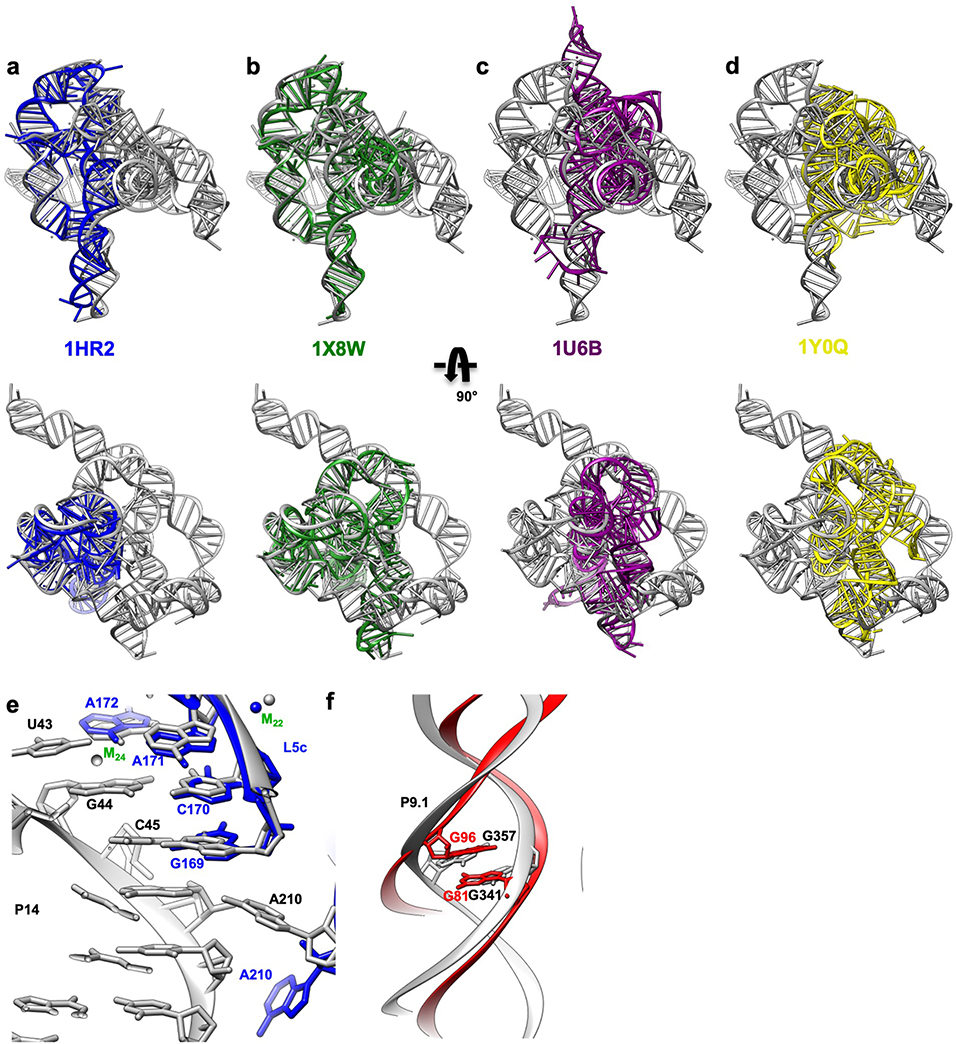
Previous studies based on pre-steady state enzymology40,42 have shown that the binding of the ribozyme’s guanosine substrate (here part of S1) is slow and may require conformational changes of the guanosine binding site, but such differences in apo and holo forms were not resolved. Comparison of our apo L-21 ScaI ribozyme with holo L-16 ScaI ribozyme and other group I introns shows that the guanosine binding site is largely preformed (Figure 4a–b, Extended Data Figure 8). However, our apo structure shows only weak density for the base of C262 (Figure 4a), in contrast to a well resolved C262 in the holo structure (Figure 4b). In addition, we observe that a peripheral metal ion (named M2 below) directly coordinates to the phosphate group of C262 in both structures. The M2 and C262 phosphate shift between apo and holo states to encapsulate the ribozyme’s guanosine substrate, providing a structural picture for fundamental functional observations (Extended Data Figure 8d).
Figure 4. Cryo-EM structures reveal conformational changes and mechanistic insights in the catalytic site.
(a) Cryo-EM map and model of the apo L-21 ScaI ribozyme with preorganized catalytic site. The density for C262 base is weak, likely due to the absence of ωG. The C262 phosphate backbone density is observed, likely stabilized by M2, visualized at 1σ threshold. (b) Cryo-EM map and model of the holo L-16 ScaI ribozyme catalytic site with docked substrate S1, which contains ωG and the 3′ splice site, visualized at 1σ threshold. (c) Mechanistic insights into the second catalytic reaction. The model illustrates one stereoisomer of phosphorothioate (yellow). Black dashed lines indicate inner-sphere coordination, coordination bond angles are shown. Black arrow indicates the direction of nucleophilic attack. See also Extended Data Figure 8.
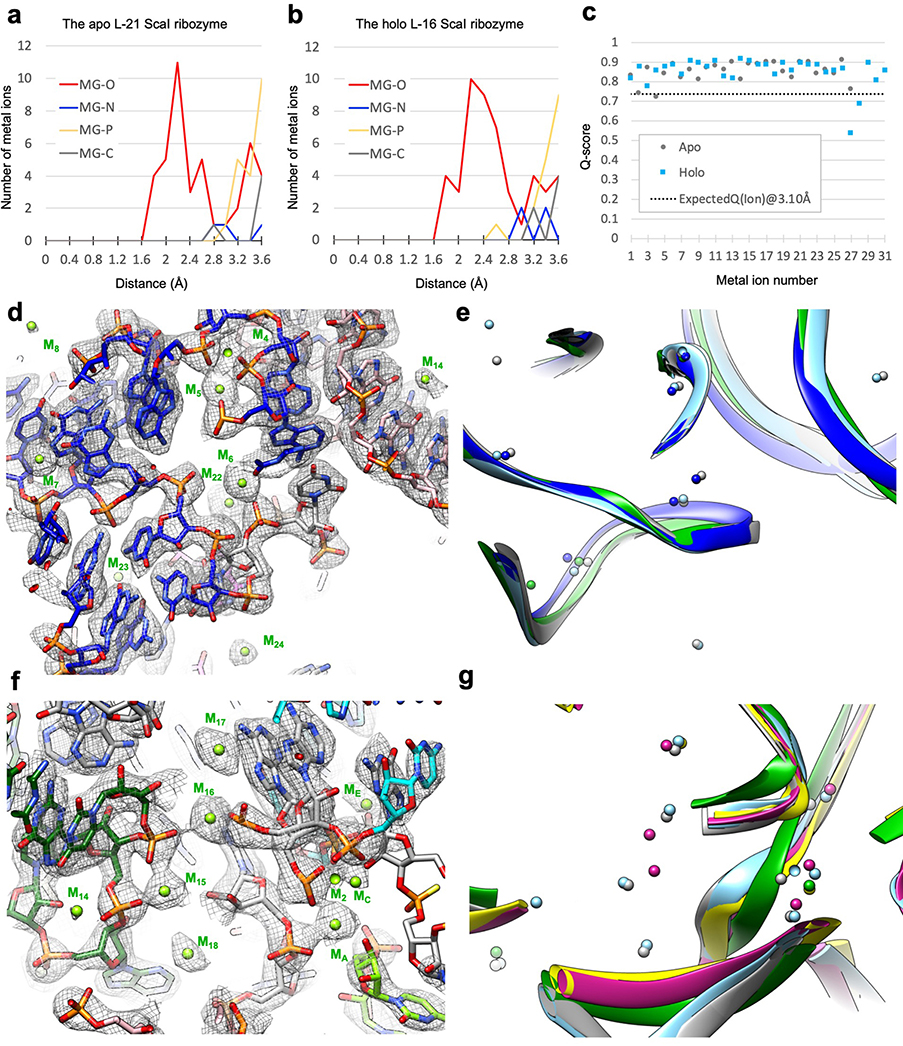
Metal ions in the Tetrahymena ribozyme
The Tetrahymena ribozyme is a metalloenzyme: metal ions are essential for structure stabilization and catalytic reactions in group I introns13–23. High resolution cryo-EM maps can resolve metal and solvent molecules33. A total of 31 metal ions (designated as M1 to M31) are identified in the holo L-16 ScaI ribozyme (Detailed in Methods), four of which are absent in the apo structure (M27, M29, M30 and M31; Supplementary Table 2). Most metal ions are in close proximity to oxygen atoms with Q-scores mostly greater than 0.8 (Extended Data Figure 9a–c), and more than half are consistent with previous crystal structures (Extended Data Figure 9d–g).
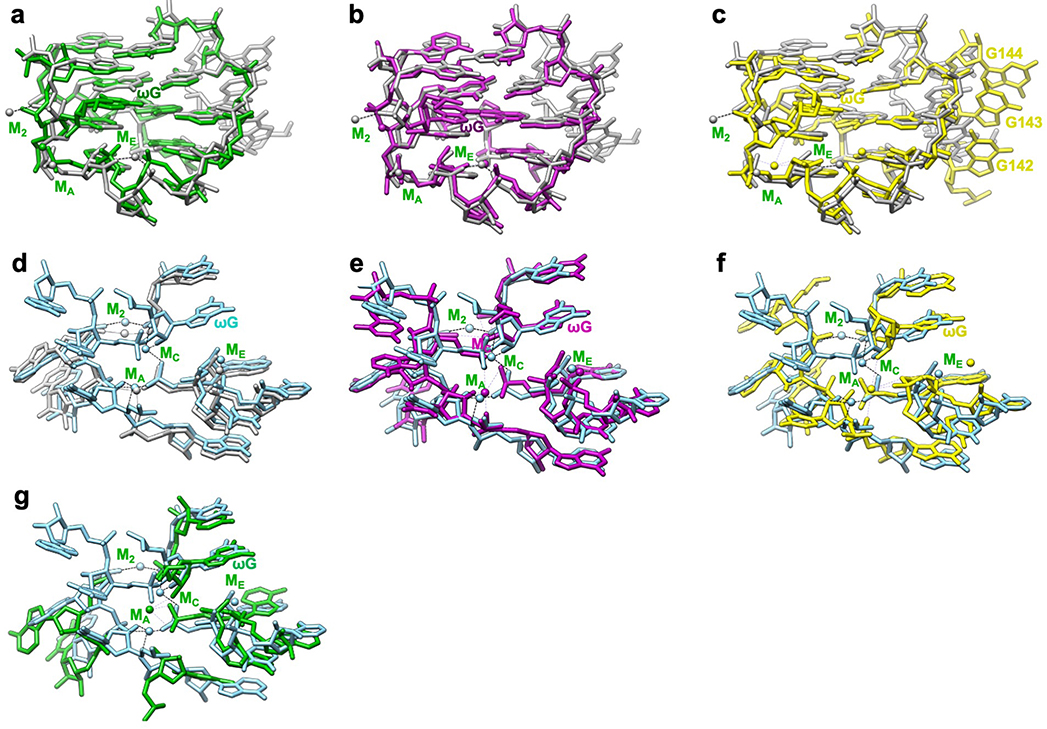
Some metal ions appear relevant for catalysis. Elegant metal ion rescue experiments along with crystallography on other group I ribozymes have identified five metal ions (MA-ME, in which MA, MC and ME correspond to M26, M27 and M28 in the cryo-EM structures16,22) as important for the ribozyme’s enzymatic reactions16,19,22,43–48. In the holo structure, we identified that M2, along with MA and MC, directly coordinate with multiple non-bridging phosphate oxygens of C208, C262, A304 and A306 (Figure 4a–b). Most importantly, MA directly coordinates with the nucleophilic 3′-OH oxygen of u(−1) of S2, whereas MC directly coordinates with the 2′-OH oxygen of ωG to establish the linear route for nucleophilic attack (Figure 4c). ME makes inner-sphere coordination with U307 and A308. MA and ME are in close overlap with those identified in Azoarcus ribozyme structure (0.74 and 1.06 Å deviations, respectively)17,20, whereas MC is shifted 2.67 Å in the Azoarcus ribozyme (Extended Data Figure 8e). The shift may be due to differences in sequence or chemical modifications between the ribozyme studies. Analogs of MB and MD are not observed in the cryo-EM structure (or in any previous crystal structures)17,18,20.
Discussion
RNA is considered inherently difficult for structural determination, particularly relatively large RNAs using conventional X-ray crystallography49. Cryo-EM is now routinely used to determine near-atomic resolution structures of proteins and RNA-protein complexes, without making mutations50. The Tetrahymena ribozyme is a very well-characterized catalytic RNA, probed in extensive biochemical and functional studies. In this study, we have determined 3.1 Å structures of the wild type full-length apo L-21 and holo L-16 ScaI ribozyme. At this resolution, most nucleotides and 31 metal ions are unambiguously resolved. Novel tertiary interactions revealed by cryo-EM structures suggest new explanations for allosteric effects connecting the catalytic core and the peripheral regions in previous biochemical and functional studies (Figure 2).
The apo L-21 ScaI ribozyme shows an overall preorganized guanosine binding site, whereas the holo L-16 ScaI ribozyme structure reveals the docked substrates P1 and P10, and four metal ions, M2, MA, MC and ME near the catalytic site. Conformational changes of the IGS and, at smaller scales, C262 and M2, occur upon substrate binding (Figure 3g, 4a–b). These results provide possible explanations of changes in cross-linking patterns during oligonucleotide substrate binding and slow binding of guanosine, which have been proposed to be important for the function and specificity during Tetrahymena group I intron self-splicing38,40,42.
In summary, our results show the stable tertiary structures formed by the Tetrahymena ribozyme in both apo and holo states, and demonstrate the capability and potential of cryo-EM in expanding our knowledge of RNA structure-function relationships. It now appears feasible to study this ribozyme with experimental structures illuminating all its aspects – substrate binding, metal ions, folding pathways – and to thereby complete a foundational RNA enzymology research program that began 40 years ago.
Methods
RNA preparation.
L-21 ScaI ribozyme was prepared as previously described10. Briefly, the DNA template was amplified from the pT7L-21 plasmid63, then RNA was prepared through in vitro transcription in a reaction containing 0.2 μM DNA template, 40 mM Tris·HCl, pH 8.1, 25 mM MgCl2, 3.5 mM spermidine, 0.01% TritonX-100, 40 mM DTT, 4% PEG 8000, 3 mM NTPs, and 7.5 U/μL T7 RNA polymerase (New England Biolabs). The transcription reaction was incubated at 37°C for 1 hour. The RNA was then isolated by ethanol precipitation, then purified on an 8% 29:1 acrylamide:bis, 7 M urea polyacrylamide gel. The gel was allowed to set overnight, then the precipitated RNA was mixed with loading buffer containing 95% formamide, 10mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue, and loaded on the gel. The gel was run at 25 W for 2 hours, then visualized briefly with a 254-nm UV lamp, held far from the gel to minimize RNA damage64. RNA was eluted from the gel overnight in RNase-free water at 4°C, then purified with Zymo RNA Clean and Concentrator columns (Zymo Research).
The DNA template of L-16 ScaI ribozyme was amplified from the pT7L-21 plasmid using forward primer 5′-TTCTAATACGACTCACTATAGGTTTGGAGGGAAAAGTTATCAGGCATGCACCTGGTAGC-3′ and reverse primer 5′-ACTCCAAAACTAATCAATATACTTTCGCATACAAATTAGTTCCCAGCGGCTCC-3′. RNA was prepared using the TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific) according to the manufacturer’s protocol, then purified with RNA Clean and Concentrator-25 columns (Zymo Research) and PAGE purified as described above for L-21 ScaI ribozyme. RNA was eluted from the gel using the ZR small-RNA PAGE Recovery Kit (Zymo Research) and then ethanol precipitated. RNA oligonucleotide substrate S1 (5′-UCG*UAACC) and S2 (5′-CCCUCU), where * indicates a phosphorothioate bond, were acquired from Integrated DNA Technologies. A phosphorothioate-substituted substrate was selected to capture the complex mimicking the second step of splicing based on previous work showing that a similar ligation reaction catalyzed by Tetrahymena ribozyme was significantly inhibited by substitution of phosphorothioate in the Rp isomeric form at the scissile phosphate (less than 5% product after 3 hours at 10 μM ribozyme concentration)65. Similarly, phosphorothioate substitution in the equivalent substrate for catalysis of the second step of splicing by the Azoarcus ribozyme reduced the reaction rate by a factor of >105 and ~14 for the Rp and Sp isomeric forms, respectively39.
Cryo-EM sample preparation.
To prepare L-21 and L-16 ScaI ribozyme samples for cryo-EM analysis, RNAs (20 μM or 15 μM final concentration, respectively) were denatured at 90°C for 3 min in 50 mM Na-HEPES, pH 8 and cooled to room temperature for 10 min. MgCl2 was added to a final concentration of 10 mM and the samples were incubated at 50°C for 30 min. Ribozyme samples were again cooled to room temperature for 10 min. At this point, L-21 ScaI ribozyme was kept on ice, while substrates S1 and S2 (75 μM final concentration each) were added to L-16 ScaI ribozyme and the sample was incubated at room temperature for 20 min to form the holoenzyme complex before being placed on ice. A total of 3 μl of the Tetrahymena ribozyme sample was applied onto glow-discharged (30 s) 200-mesh R2/1 Quantifoil Cu grids. The grids were blotted for 3 s in 100% humidity with no blotting offset and rapidly frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher).
Cryo-EM single particle data acquisition and data processing.
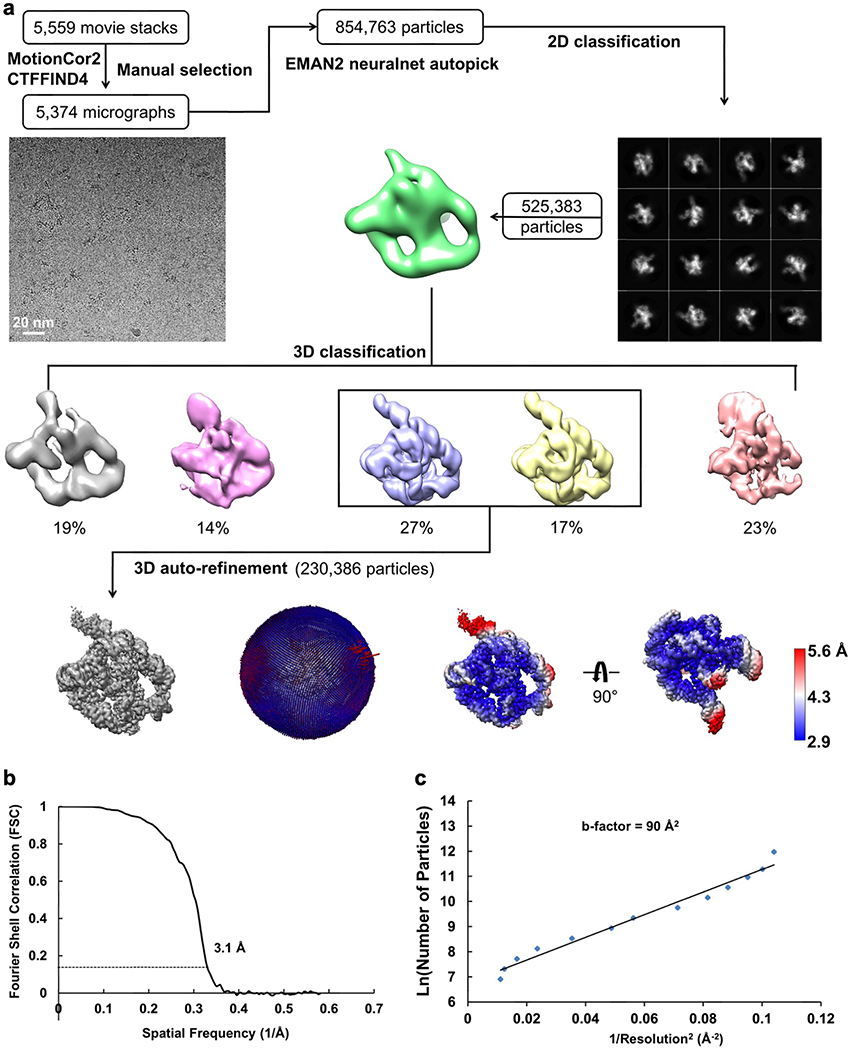
The frozen grids of apo L-21 ScaI ribozyme were loaded in Titan Krios (Thermo Fisher) operated at 300 kV, condenser lens aperture 50 μm, spot size 7, parallel beam with illuminated area of 0.85 μm in diameter. Microscope magnification was at 215,000× (corresponding to a calibrated sampling of 0.65 Å per physical pixel). Movie stacks were collected automatically using EPU software on a K2 direct electron camera equipped with a Bioquantum energy filter with an energy slit of 20 eV (Gatan), operating in counting mode at a recording rate of 5 raw frames per second and a total exposure time of 5 seconds, yielding 25 frames per stack, and a total dose of 75 e−/Å2. A total of 7,577 movie stacks were collected with defocus values ranging between −0.3 and −1.5 μm. These movie stacks were motion corrected using Motioncor266. After CTF correction by CTFFIND467, 7,469 micrographs were subjected to EMAN2.2 for neural network particle picking68. A total of 1,658,961 particles were extracted in Relion369 with the box size of 320 pixels. After two rounds of 2D classifications, the best classes by visual examination were subjected to EMAN2.2 to build the initial model, and a total of 1,559,933 particles were subjected to 3D classification in Relion3. The major class showing RNA features, including 415,918 particles, were subjected to auto refinement. The initial auto refinement result was subjected to Bayesian polishing followed by another round of auto refinement70. A sharpening B-factor of −14 Å2 was applied to the resulting cryo-EM map to yield the final sharpened map at 3.1 Å global resolution estimated by the 0.143 criterion of FSC curve.
Frozen grids of the holo L-16 ScaI ribozyme were loaded in Titan Krios (Thermo Fisher) operated at 300 kV, condenser lens aperture 70 μm, spot size 5, parallel beam with illuminated area of 1.1 μm in diameter. Microscope magnification was at 105,000× (corresponding to a calibrated sampling of 0.86 Å per physical pixel). Movie stacks were collected automatically using EPU software on a K3 direct electron camera equipped with a Bioquantum energy filter with an energy slit of 15 eV (Gatan), operating in counting mode and a total exposure time of 2.5 seconds, yielding 30 frames per stack with a total dose of 50 e−/Å2. A total of 5,559 movie stacks were collected with defocus values ranging between −0.8 and −2.0 μm. The data was processed using the above-mentioned protocol, and a total of 230,386 particles were subjected to auto refinement, Bayesian polishing and postprocessing with a sharpening B-factor of −30 Å2 to yield the final sharpened map at 3.1 Å global resolution estimated by the 0.143 criterion of FSC curve.
Both local resolution maps were determined in Relion3 and the final map was low-pass filtered accordingly and displayed in UCSF Chimera71.
Cryo-EM model building and refinement.
The initial Tetrahymena ribozyme models of both apo and holo cryo-EM structures were built with DRRAFTER72, then manually adjusted and rebuilt with Coot as needed73. The models were refined with Phenix.real_space_refine74, yielding an averaged model–map correlation coefficient (CCmask) of 0.79 and 0.82, respectively. The final model was validated by MolProbity75 and Q-score analysis33. Secondary structure diagrams were prepared with RiboDraw aided by manual adjustment (https://github.com/ribokit/RiboDraw).
Metal ion identification and validation in cryo-EM models.
Additional densities in our cryo-EM maps after fitting the RNA models are modeled as metal ions. These densities are also observed in both half maps of the reconstruction, which is another means of validating these metal ions. Several metal ions in the cryo-EM structures validate previous biochemical results: M4 and M5 have been previously identified to adopt inner-sphere coordination by metal-ion rescue experiments76; residues that make inner-sphere contacts with M2, M15, M16, M18 and M19 have been suggested as metal binding sites in a phosphorothioate interference assay77,78.
Extended Data
Extended Data Figure 1 related to Figure 1. Cryo-EM single particle reconstruction of the apo L-21 ScaI Tetrahymena ribozyme.
(a) Single particle pipeline yields the final cryo-EM reconstruction with the corresponding angular distribution and local resolution map. The local resolution map shows more flexibility and lower resolution in the 4-nt bulge that connects stem P2.1 and P13, and toward the end of stem P6 and P9.2. (b) FSC curve shows 3.1 Å resolution according to the 0.143 cutoff. (c) Cryo-EM B-factor51 that relates the number of particles to the map resolution attributed to cumulative experimental and computational factors affecting the final reconstruction.
Extended Data Figure 2. Focus 3D classification of apo L-21 ScaI ribozyme reveals local conformational dynamics.
The regions of low local resolution, P9.2 (blue), P9.2-P9.1-P13 (orange) and P13 (green), were extracted. Focus 3D classification were performed and different classes were superimposed to show rotational and translational motions on P9.2 (left), P9.2-P9.1-P13 (middle), P13 (right).
Extended Data Figure 3 related to Figure 1. Q-score analyses of cryo-EM maps and models of both the apo L-21 and holo L-16 Tetrahymena ribozymes.
(a) Q-score analyses per residue of the apo L-21 (grey) and holo L-16 (blue) Tetrahymena ribozyme cryo-EM models and maps, black dashed line indicates average Q score from nucleic acid cryo-EM models and maps at 3.1 Å resolution. (b) Colored cryo-EM model of apo L-21 ScaI ribozyme based on Q-score per residue. Dashed boxes (black, blue and green) correspond to different regions in the cryo-EM model with low Q-scores in panel (a).
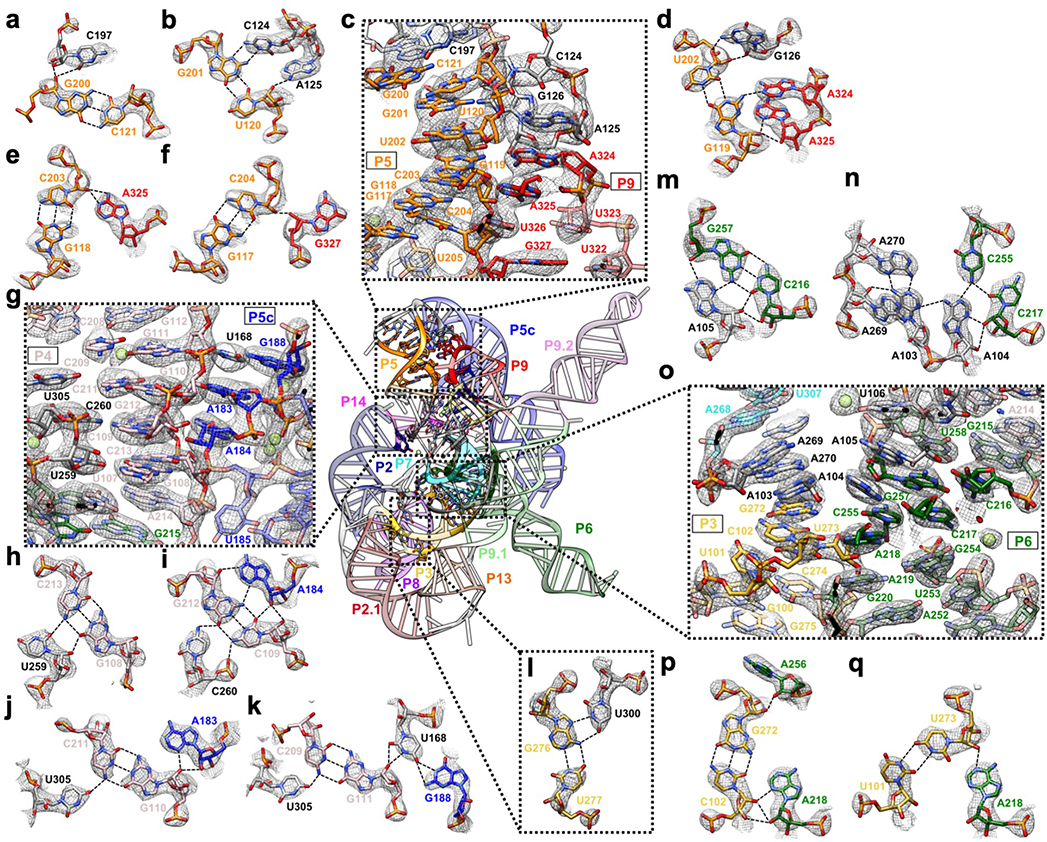
Extended Data Figure 4. Detailed tertiary interactions in the core region of the Tetrahymena ribozyme.
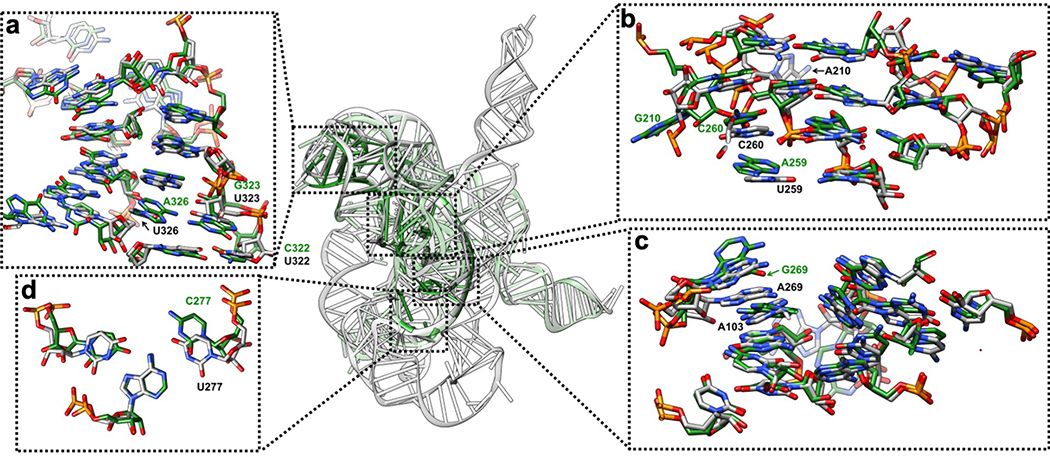
(a-f) The P5-J5/5a-L9 region has highly structured J5/5a junction in previous structures12,15. The cryo-EM structure shows tertiary interactions of (b) C124 and A125, (d) G126, (a) C197 and (d) A324, (e) A325, (f) G327 in the minor groove of P5. (g-k) Previous studies have identified A183 and A184 in A-rich bulge of the metal core, U259 and C260 from J6/7, U305 from J8/7 to be conserved and essential for catalytic site formation and splicing reactions52–58. The cryo-EM structure shows (h) U259, (i) C260, and (j-k) U305 stack continuously and interact with P4 base triples. (k) U168 from P5c stacks on the A-rich bulge and interacts with P4 in the minor groove, while pairing with G188 in P5a. (l) The Hoogsteen base triple U277-A97-U300 found to be essential for substrate helix recognition59. (m-q) In the P3-J3/4-P6-J7/3 region, A-rich J3/4 and J7/3 were previously found to interact with P6 in Azoarcus ribozyme17,60. Base triples formed by J3/4 were critical for catalysis52,57,58,61, and alterations in these regions result in RNA misfolding28,62. In the cryo-EM structure, (n) A104 and (m) A105 form A-minor interactions with P6, whereas (n) A103 and A104 join A269 and A270 from J7/3 to form an adenosine cluster. The proposed A103-U271 reverse-Hoogsteen pair is not found, instead we observed a noncanonical A103-A270 pair61. (p-q) The previously observed A-platform of A218-A219 is disrupted in the cryo-EM structure with P3 present12,30. A218 forms two A-minor interactions with C102-G272 and U273-U101 from P3, which also supports the conservation of this C-G pair in group Ib introns61. Black dashed line indicates hydrogen bond. The cryo-EM maps of all subpanels are visualized at 1σ threshold except for (c, g and o) at 1.5σ.
Extended Data Figure 5 related to Extended Data Figure 4. Comparison between the previous 3.8 Å crystal structure of the mutated Tetrahymena ribozyme catalytic core (green) and the cryo-EM structure of the wild type apo Tetrahymena ribozyme L-21 construct (grey) shows minor differences.
The overall RMSD for the catalytic core region (stem P3-P9) is 6.6 Å. (a) The same view of P5-J5/5a-P9 region as in Extended Data Figure 4a. The nucleotide conformations generally agree well between two models, three mutations U322C, U323G and U326A are not involved in tertiary interactions. The RMSD for this region is 4.9 Å. (b) The same view of P4-P5a-J6/7-J8/7 region as in Extended Data Figure 4b. In the crystal structure, U259A is slightly moved away from the G108-C213 base pair and disrupts this base triple interaction. A210G is moved far away from the wild type position of A210, because there is no A46 in stem P2 to interact with in the crystal structure. The very top base quartet is much more compact in the cryo-EM structure compared to the crystal structure, likely due to the presence of peripheral domain that wraps around the catalytic core to make it more compact. The RMSD for this region is 5.7 Å. (c) The same view of P3-J3/4-P6-J7/3 region as in Extended Data Figure 4c. The overall nucleotide conformations agree very well between two models, except for A269G and A270 in the crystal structure are completely moved away and disrupt their interactions with A103, which is observed in the cryo-EM structure. The RMSD for this region is 1.7 Å. (d) The same view as in Extended Data Figure 4d. U277C disrupts the U277-A97-U300 base triple. The RMSD for this base triple is 2.7 Å. See also Supplementary Table 1.
Extended Data Figure 6. Superposition of the apo L-21 ScaI ribozyme cryo-EM structure (grey) with previous crystal structures of the truncated and/or mutated Tetrahymena ribozyme, other group I introns and 5S rRNA loop E show global and local structural similarities.
Overlay of the cryo-EM structure (grey) with (a) The Tetrahymena ribozyme P4-P6 Delta C209 (blue, PDB 1HR2); (b) The mutated Tetrahymena ribozyme P3-P9 (green, PDB 1X8W); (c) Azoarcus ribozyme (violet, PDB 1U6B) and (d) phage Twort ribozyme (yellow, PDB 1Y0Q); (e) P5c region of the wild type P4-P6 crystal structure (blue, PDB 1GID); (f) 5S rRNA loop E crystal structure (red, PDB 354D).
Extended Data Figure 7 related to Figure 3. Cryo-EM single particle reconstruction of the holo L-16 ScaI Tetrahymena ribozyme.
(a) Single particle pipeline yields the final cryo-EM reconstruction with the corresponding angular distribution and local resolution map. The local resolution map shows more flexibility and lower resolution in the 4-nt bulge that connects stem P2.1 and P13, and toward the end of stem P6 and P9.2. (b) FSC curve shows 3.1 Å resolution according to the 0.143 cutoff. (c) Cryo-EM B-factor.
Extended Data Figure 8 related to Figure 4. Comparison of apo L-21 and holo L-16 ScaI ribozyme cryo-EM models with previous crystal structures show structural conservation and metal ion shifts in the guanosine binding site among group I introns.
The apo L-21 ScaI ribozyme adopts a preorganized guanosine binding site (grey) that superimposes with previous crystal structures of (a) mutated P3-P9 of the Tetrahymena ribozyme (green, PDB 1X8W), (b) Azoarcus ribozyme (violet, PDB 1U6B), and (c) phage Twort ribozyme (yellow, PDB 1y0q); The holo L-16 ScaI ribozyme (sky blue) superimposes with (d) apo L-21 ScaI ribozyme (grey), (e) Azoarcus ribozyme (violet), (f) mutated P3-P9 of the Tetrahymena ribozyme (green), and (g) phage Twort ribozyme (yellow). MC in the apo L-21 ScaI ribozyme is absent, whereas MC in Azoarcus ribozyme is shifted compared to the holo L-16 ScaI ribozyme. Dash line indicates metal ion coordination with surrounding atoms.
Extended Data Figure 9. Metal ion validations by distance and Q-score analysis, and illustrations in the apo L-21 and holo L-16 ScaI ribozyme cryo-EM structures compared with previous crystal structures.
(a) Distances between metal ions and other atoms in the apo L-21 ScaI ribozyme model. (b) Distances between metal ions and other atoms in the holo L-16 ScaI ribozyme model. (c) Q-score analysis per metal ion of the apo L-21 and holo L-16 ScaI Tetrahymena ribozyme cryo-EM models and maps. (d) Metal core region of the holo L-16 ScaI ribozyme, visualized at 1.1σ threshold. (e) Catalytic site of the holo L-16 ScaI ribozyme, visualized at 1.4σ threshold. (f) Comparisons of the apo L-21 (grey) and holo L-16 (sky blue) ScaI ribozyme cryo-EM models with P4-P6 Delta C209 (blue, PDB 1HR2) and mutated P3-P9 of the Tetrahymena ribozyme (green, PDB 1X8W) in the same view as (a). (d) Comparisons of the apo L-21 (grey) and holo L-16 (sky blue) ScaI ribozyme cryo-EM models with Azoarcus ribozyme (violet, PDB 1U6B), mutated P3-P9 of the Tetrahymena ribozyme (green, PDB 1X8W), and phage Twort ribozyme (yellow, PDB 1Y0Q) in the same view as (b). See also Supplementary Table 2.
Extended Data Table 1.
Cryo-EM data collection, processing, and model refinement statistics of the apo L-21 and holo L-16 ScaI ribozyme.
| Cryo-EM data collection and processing | The apo L-21 ScaI ribozyme | The holo L-16 ScaI ribozyme |
|---|---|---|
| Microscope | Titan Krios | Titan Krios |
| Voltage (kV) | 300 | 300 |
| GIF Quantum energy filter slit width (eV) | 20 | 15 |
| Detector | Gatan K2 | Gatan K3 |
| Nominal Magnification | 215,000 × | 105,000 × |
| Pixel size (Å) | 0.65 | 0.86 |
| Symmetry imposed | C1 | C1 |
| Defocus range (μm) | −0.3 – −1.5 | −0.8 – −2.0 |
| Electron exposure (e−/Å2) | 75 | 50 |
| Micrographs (acquired/used) | 7,577/7,469 | 5,559/5,374 |
| Number of extracted particles | 1,658,961 | 854,763 |
| Number of particles after 2D classifications | 1,559,933 | 525,373 |
| Number of particles going to 3D refinement | 415,918 | 230,386 |
| Map resolution at 0.143 FSC criterion (Å) | 3.1 | 3.06 |
| Local resolution range (Å) | 3.0–4.7 | 2.9–5.6 |
| Sharpening B-factor (Å2) | −14 | −30 |
| Model refinement | ||
| Atoms | 8,306 | 8,698 |
| Residues | 387 | 406 |
| CCmask | 0.79 | 0.82 |
| ResolutionFSC map vs. model @ 0.5 (Å) | 3.2 | 3.0 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.002 |
| Bond angles (°) | 0.547 | 0.523 |
| Clash score | 2.65 | 1.91 |
| MolProbity score | 2.20 | 2.10 |
Supplementary Material
Acknowledgements
The authors are grateful to insightful discussions with Dr. Raghuvir N. Sengupta, Prof. Daniel Herschlag and Ivan Zheludev. Cryo-EM data was collected at SLAC and Stanford, and processed at SLAC, Stanford and Duyu High Performance Computing Center in Sichuan University. This work was supported by National Institutes of Health (P41GM103832, R01GM079429, P01AI120943, and S10OD021600 to W.C.; R35GM112579 and R21AI145647 to R.D.); National Science Foundation (DGE-114747 to K.K. and DGE-1656518 to R.R.); Gabilan Stanford Graduate Fellowship to K.K.; Sichuan University start-up funding 20822041D4057 and Natural Science Foundation of China (NSFC82041016 and 32070049 to Z.S). We thank Nora Lawless for manuscript editing.
Footnotes
The authors declare no competing financial interests.
Data availability
The cryo-EM maps and associated atomic coordinate models of the apo L-21 and holo L-16 ScaI Tetrahymena ribozyme have been deposited in the wwPDB OneDep System under EMD accession code EMD-31385, EMD-31386 and PDB ID code 7EZ0, 7EZ2.
References
- 1.Nakane T et al. Single-particle cryo-EM at atomic resolution. Nature 587, 152–156, doi: 10.1038/s41586-020-2829-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yip KM, Fischer N, Paknia E, Chari A & Stark H Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161, doi: 10.1038/s41586-020-2833-4 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Zhang K, Pintilie GD, Li S, Schmid MF & Chiu W Resolving individual atoms of protein complex by cryo-electron microscopy. Cell Research 30, 1136–1139, doi: 10.1038/s41422-020-00432-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruger K et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157, doi: 10.1016/0092-8674(82)90414-7 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Cech TR The RNA worlds in context. Cold Spring Harb Perspect Biol 4, a006742, doi: 10.1101/cshperspect.a006742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cech TR & Steitz JA The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94, doi: 10.1016/j.cell.2014.03.008 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Zhang H & Keane SC Advances that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy. Wiley Interdiscip Rev RNA 10, e1541, doi: 10.1002/wrna.1541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J & Ferre-D’Amare AR New molecular engineering approaches for crystallographic studies of large RNAs. Curr Opin Struct Biol 26, 9–15, doi: 10.1016/j.sbi.2014.02.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K et al. Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 A resolution. Nat Commun 10, 5511, doi: 10.1038/s41467-019-13494-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kappel K et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat Methods 17, 699–707, doi: 10.1038/s41592-020-0878-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cech TR Ribozymes, the first 20 years. Biochemical Society transactions 30, 1162–1166, doi: 10.1042/bst0301162 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Cate JH et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685, doi: 10.1126/science.273.5282.1678 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Golden BL, Gooding AR, Podell ER & Cech TR A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282, 259–264, doi: 10.1126/science.282.5387.259 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Guo F, Gooding AR & Cech TR Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Molecular cell 16, 351–362, doi: 10.1016/j.molcel.2004.10.003 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Juneau K, Podell E, Harrington DJ & Cech TR Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure 9, 221–231, doi: 10.1016/s0969-2126(01)00579-2 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Piccirilli JA, Vyle JS, Caruthers MH & Cech TR Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature 361, 85–88, doi: 10.1038/361085a0 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Adams PL, Stahley MR, Kosek AB, Wang J & Strobel SA Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50, doi: 10.1038/nature02642 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Golden BL, Kim H & Chase E Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol 12, 82–89, doi: 10.1038/nsmb868 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Shan S, Yoshida A, Sun S, Piccirilli JA & Herschlag D Three metal ions at the active site of the Tetrahymena group I ribozyme. Proceedings of the National Academy of Sciences of the United States of America 96, 12299–12304, doi: 10.1073/pnas.96.22.12299 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahley MR & Strobel SA Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309, 1587–1590, doi: 10.1126/science.1114994 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Steitz TA & Steitz JA A general two-metal-ion mechanism for catalytic RNA. Proceedings of the National Academy of Sciences of the United States of America 90, 6498–6502, doi: 10.1073/pnas.90.14.6498 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forconi M, Piccirilli JA & Herschlag D Modulation of individual steps in group I intron catalysis by a peripheral metal ion. RNA 13, 1656–1667, doi: 10.1261/rna.632007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudry AA & Joyce GF Minimum secondary structure requirements for catalytic activity of a self-splicing group I intron. Biochemistry 29, 6534–6539, doi: 10.1021/bi00479a027 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Benz-Moy TL & Herschlag D Structure-function analysis from the outside in: long-range tertiary contacts in RNA exhibit distinct catalytic roles. Biochemistry 50, 8733–8755, doi: 10.1021/bi2008245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doherty EA & Doudna JA Ribozyme structures and mechanisms. Annual review of biophysics and biomolecular structure 30, 457–475, doi: 10.1146/annurev.biophys.30.1.457 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Joyce GF, van der Horst G & Inoue T Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic acids research 17, 7879–7889, doi: 10.1093/nar/17.19.7879 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehnert V, Jaeger L, Michel F & Westhof E New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chemistry & biology 3, 993–1009, doi: 10.1016/s1074-5521(96)90166-0 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Russell R et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. Journal of molecular biology 363, 531–544, doi: 10.1016/j.jmb.2006.08.024 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Kiyooka R et al. Catalytic RNA nano-objects formed by self-assembly of group I ribozyme dimers serving as unit structures. Journal of Bioscience and Bioengineering 130, 253–259, doi: 10.1016/j.jbiosc.2020.04.010 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Cate JH et al. RNA tertiary structure mediation by adenosine platforms. Science 273, 1696–1699, doi: 10.1126/science.273.5282.1696 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Cate JH, Hanna RL & Doudna JA A magnesium ion core at the heart of a ribozyme domain. Nature structural biology 4, 553–558, doi: 10.1038/nsb0797-553 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Shoffner GM, Wang R, Podell E, Cech TR & Guo F In Crystallo Selection to Establish New RNA Crystal Contacts. Structure 26, 1275–1283 e1273, doi: 10.1016/j.str.2018.05.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pintilie G et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nature methods, doi: 10.1038/s41592-020-0731-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi X et al. Roles of long-range tertiary interactions in limiting dynamics of the Tetrahymena group I ribozyme. J Am Chem Soc 136, 6643–6648, doi: 10.1021/ja413033d (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Downs WD & Cech TR A tertiary interaction in the Tetrahymena intron contributes to selection of the 5’ splice site. Genes & development 8, 1198–1211, doi: 10.1101/gad.8.10.1198 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Zarrinkar PP & Williamson JR The P9.1-P9.2 peripheral extension helps guide folding of the Tetrahymena ribozyme. Nucleic Acids Res 24, 854–858, doi: 10.1093/nar/24.5.854 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correll CC, Freeborn B, Moore PB & Steitz TA Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 91, 705–712, doi: 10.1016/s0092-8674(00)80457-2 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Wang JF, Downs WD & Cech TR Movement of the guide sequence during RNA catalysis by a group I ribozyme. Science 260, 504–508, doi: 10.1126/science.7682726 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Lipchock SV & Strobel SA A relaxed active site after exon ligation by the group I intron. Proceedings of the National Academy of Sciences 105, 5699–5704, doi: 10.1073/pnas.0712016105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karbstein K & Herschlag D Extraordinarily slow binding of guanosine to the Tetrahymena group I ribozyme: implications for RNA preorganization and function. Proc Natl Acad Sci U S A 100, 2300–2305, doi: 10.1073/pnas.252749799 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karbstein K, Lee J & Herschlag D Probing the role of a secondary structure element at the 5’- and 3’-splice sites in group I intron self-splicing: the tetrahymena L-16 ScaI ribozyme reveals a new role of the G.U pair in self-splicing. Biochemistry 46, 4861–4875, doi: 10.1021/bi062169g (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X, Mollova ET, Pljevaljcić G, Millar DP & Herschlag D Probing the dynamics of the P1 helix within the Tetrahymena group I intron. J Am Chem Soc 131, 9571–9578, doi: 10.1021/ja902797j (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan S, Kravchuk AV, Piccirilli JA & Herschlag D Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry 40, 5161–5171, doi: 10.1021/bi002887h (2001). [DOI] [PubMed] [Google Scholar]
- 44.Shan SO & Herschlag D An unconventional origin of metal-ion rescue and inhibition in the Tetrahymena group I ribozyme reaction. Rna 6, 795–813, doi: 10.1017/s1355838200000649 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sjogren AS, Pettersson E, Sjoberg BM & Stromberg R Metal ion interaction with cosubstrate in self-splicing of group I introns. Nucleic acids research 25, 648–653, doi: 10.1093/nar/25.3.648 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinstein LB, Jones BC, Cosstick R & Cech TR A second catalytic metal ion in group I ribozyme. Nature 388, 805–808, doi: 10.1038/42076 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Forconi M, Lee J, Lee JK, Piccirilli JA & Herschlag D Functional identification of ligands for a catalytic metal ion in group I introns. Biochemistry 47, 6883–6894, doi: 10.1021/bi800519a (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hougland JL, Kravchuk AV, Herschlag D & Piccirilli JA Functional Identification of Catalytic Metal Ion Binding Sites within RNA. PLOS Biology 3, e277, doi: 10.1371/journal.pbio.0030277 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reyes FE, Garst AD & Batey RT Strategies in RNA crystallography. Methods in enzymology 469, 119–139, doi: 10.1016/S0076-6879(09)69006-6 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Zhang K et al. Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM. Proc Natl Acad Sci U S A 117, 7176–7182, doi: 10.1073/pnas.1922638117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745, doi: 10.1016/j.jmb.2003.07.013 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Green R & Szostak JW In vitro genetic analysis of the hinge region between helical elements P5-P4-P6 and P7-P3-P8 in the sunY group I self-splicing intron. Journal of molecular biology 235, 140–155, doi: 10.1016/s0022-2836(05)80022-1 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Karbstein K, Tang KH & Herschlag D A base triple in the Tetrahymena group I core affects the reaction equilibrium via a threshold effect. Rna 10, 1730–1739, doi: 10.1261/rna.7118104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanner MA, Anderson EM, Gutell RR & Cech TR Mutagenesis and comparative sequence analysis of a base triple joining the two domains of group I ribozymes. Rna 3, 1037–1051 (1997). [PMC free article] [PubMed] [Google Scholar]
- 55.Tanner MA & Cech TR Joining the two domains of a group I ribozyme to form the catalytic core. Science 275, 847–849, doi: 10.1126/science.275.5301.847 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Ikawa Y, Yoshimura T, Hara H, Shiraishi H & Inoue T Two conserved structural components, A-rich bulge and P4 XJ6/7 base-triples, in activating the group I ribozymes. Genes to cells : devoted to molecular & cellular mechanisms 7, 1205–1215, doi: 10.1046/j.1365-2443.2002.00601.x (2002). [DOI] [PubMed] [Google Scholar]
- 57.Ikawa Y, Naito D, Shiraishi H & Inoue T Structure-function relationships of two closely related group IC3 intron ribozymes from Azoarcus and Synechococcus pre-tRNA. Nucleic acids research 28, 3269–3277, doi: 10.1093/nar/28.17.3269 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michel F, Ellington AD, Couture S & Szostak JW Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature 347, 578–580, doi: 10.1038/347578a0 (1990). [DOI] [PubMed] [Google Scholar]
- 59.Szewczak AA et al. An important base triple anchors the substrate helix recognition surface within the Tetrahymena ribozyme active site. Proceedings of the National Academy of Sciences of the United States of America 96, 11183–11188, doi: 10.1073/pnas.96.20.11183 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rangan P, Masquida B, Westhof E & Woodson SA Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proceedings of the National Academy of Sciences of the United States of America 100, 1574–1579, doi: 10.1073/pnas.0337743100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green R, Ellington AD & Szostak JW In vitro genetic analysis of the Tetrahymena self-splicing intron. Nature 347, 406–408, doi: 10.1038/347406a0 (1990). [DOI] [PubMed] [Google Scholar]
- 62.Mitchell D 3rd, Jarmoskaite I, Seval N, Seifert S & Russell R The long-range P3 helix of the Tetrahymena ribozyme is disrupted during folding between the native and misfolded conformations. Journal of molecular biology 425, 2670–2686, doi: 10.1016/j.jmb.2013.05.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaug AJ, Grosshans CA & Cech TR Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry 27, 8924–8931, doi: 10.1021/bi00425a008 (1988). [DOI] [PubMed] [Google Scholar]
- 64.Kladwang W, Hum J & Das R Ultraviolet shadowing of RNA can cause significant chemical damage in seconds. Scientific reports 2, 517, doi: 10.1038/srep00517 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajagopal J, Doudna JA & Szostak JW Stereochemical course of catalysis by the Tetrahymena ribozyme. Science 244, 692–694, doi: 10.1126/science.2470151 (1989). [DOI] [PubMed] [Google Scholar]
- 66.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen M et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nature methods 14, 983–985, doi: 10.1038/nmeth.4405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17, doi: 10.1107/S205225251801463X (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Kappel K et al. De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes. Nat Methods 15, 947–954, doi: 10.1038/s41592-018-0172-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544, doi: 10.1107/S2059798318006551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21, doi: 10.1107/S0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frederiksen JK, Li NS, Das R, Herschlag D & Piccirilli JA Metal-ion rescue revisited: biochemical detection of site-bound metal ions important for RNA folding. Rna 18, 1123–1141, doi: 10.1261/rna.028738.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strauss-Soukup JK & Strobel SA A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. Journal of molecular biology 302, 339–358, doi: 10.1006/jmbi.2000.4056 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Ortoleva-Donnelly L, Szewczak AA, Gutell RR & Strobel SA The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. Rna 4, 498–519, doi: 10.1017/S1355838298980086 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps and associated atomic coordinate models of the apo L-21 and holo L-16 ScaI Tetrahymena ribozyme have been deposited in the wwPDB OneDep System under EMD accession code EMD-31385, EMD-31386 and PDB ID code 7EZ0, 7EZ2.