Abstract
Protein post-translational modification (PTM) is crucial to modulate protein interactions and activity in various biological processes. Emerging evidence has revealed PTM patterns participate in the pathology onset and progression of various diseases. Current PTM identification relies mainly on mass spectrometry-based approaches that limit the assessment to the entire protein population in question. Here we report a label-free method for the detection of the single peptide with only one amino acid modification via electronic fingerprinting using reengineered durable channel of phi29 DNA packaging motor, which bears the deletion of 25-amino acids (AA) at the C-terminus or 17-AA at the internal loop of the channel. The mutant channels were used to detect propionylation modification via single-molecule fingerprinting in either the traditional patch-clamp or the modern portable MinION™ Flow Cell system. Up to 2000 channels are available in the MinION™ Flow Cells. The current signatures and dwell time of individual channels were identified. Peptides with only one propionylation were differentiated. Excitingly, identification of single or multiple modifications on the MinION™ system was achieved. The successful application of PTM differentiation on the MinION™ system represents a significant advance towards developing a label-free and high-throughput detection platform utilizing nanopores for clinical diagnosis based on PTM.
Keywords: Engineered channels, Protein post-translational modifications, Lysine propionylation, DNA-Packaging Nanomotor, Nanopore sensing, MinION™ Flow Cell
1. Introduction:
Protein post-translational modification (PTM) represents a fundamental strategy to diversify protein primary structure via covalent attachment of functional groups, proteolytic cleavage, and degradation [1–3]. Currently, more than 200 characterized PTMs account for the vast proteome’s complexity and functionality in vivo, including acetylation, methylation, phosphorylation, and ubiquitination, among others [4–6]. PTMs can enhance protein stability, mediate protein-protein interactions, transduce cellular signaling, and serve as tags for spatial localization [7]. Therefore, PTMs are well-known key players in gene regulatory networks, cell cycle, disease pathogenesis, tumor progression, and drug resistance [8–11]. Given the importance and ubiquity of PTMs in cellular contexts, there is an increasing interest in high-throughput approaches to identify PTMs comprehensively. However, up-to-date techniques remain costly and time-consuming [12].
To date, PTM characterization depends mainly on antibody-based techniques such as immunoprecipitation (IP) [13], in combination with western blotting (WB) or mass spectrometry (MS). MS analysis of proteins is the measurement of the mass-to-charge ratio of ions to identify and quantify proteins. However, PTMS of eukaryotic protein is a dynamic process. Due to the conceivable combinations in the protein population in question, the identification of protein PTMs in such heterogeneous mixtures is challenging for conventional MS [14]. Mass can detect a single amino acid modification, but the outcome of the analysis is based on the protein population. If the population contains different PTMs that are not homogeneously modified, individual protein molecules can not be differentiated. Thus, it will be difficult to distinguish the specific modification of individual protein molecules using MS. In contrast, nanopore sensing applying single-molecule technology that can address this limitation [15]. Nanopore sensing offers the sensing capability at the single-molecule level [16]. In this case, the same type of proteins with variable PTMs in the population can be identified. Single pore sensing provides improved sensitivity and response time [17]. Currently, the single pore sensing technique mainly focuses on the characterization of nucleic acids [18–30]. For instance, the Oxford MinION™ nanopore sequencing technology has proven its application for DNA/RNA sequencing. The insertion of the Phi29 channel into the MinION™ Flow Cell and the testing of peptide translocation through the inserted channel have also been reported, revealing the potential of applying a Phi29 motor channel for high-throughput peptide sensing [31].
Additionally, biological and solid-state nanopores have shown great potential for peptide and protein sensing [32–39]. For example, differentiation among peptides with different numbers of arginine residues or peptides composed of a mixture of acidic amino acids have been reported using T7 nanopore technology [40, 41]. The single pore sensing technique has been applied to detect xenobiotics and cancer-specific antibody signals [42–44]. Characterization of different peptide mass shifts can be achieved by solid nanopore [15, 45–49]. A single pore sensing technique has recently reported detection of large PTMs such as N-glycosylation and ubiquitination [50, 51]. However, detection of small PTMs (about 50 Da) has remained a challenge [17, 35].
Among all 20 native proteinogenic amino acids, lysine (Lys, K) is the most diverse in terms of number and type of PTMs incorporated. The positively charged side chain plays an essential role in protein folding and catalysis. Neutralization of this charge often has a drastic impact on protein functionality. Its amino group shows a unique nucleophilicity that allows Lys to be selectively modified with several alkylation types with a large variety of small molecules and proteins [52]. Recent studies of protein PTMs, in both eukaryotic and bacterial proteins have revealed various kinds of lysine acylations, including formylation, butyrylation and propionylation [53–55]. In the case of lysine propionylation, the expected mass shift of one propyl group addition is 56 Da. This small change requires a nanopore with a smaller pore size and higher sensitivity [56]. The Phi29 connector protein contains three flexible loops, a N-terminal loop, followed by an internal channel loop, and a C-terminal loop at the end, which were not included in the crystal structure [57]. Our previous study found that removing the internal loop segment of the channel could reduce the channel size to 60%. This smaller channel was able to detect the translocation of single-stranded nucleic acids at a single-molecule level [58]. However, whether reengineered connector and flexible loop deletion can distinguish PTMs is still unexplored.
Here we report a label-free method for the detection of single peptide with only one propionylation via electronic fingerprinting using reengineered durable channel of phi29 DNA packaging motor. Up to 2000 channels are available in the MinION™ Flow Cells. The current signatures and dwell time of individual channels were identified. Excitingly, identification of single or multiple modifications was achieved using the 4-inch long USB-powered device MinION™ system. The successful application of PTM differentiation on the MinION™ system represents a significant advance towards label-free high-throughput detection.
2. Materials and methods
2.1. Materials
All chemicals were purchased from either Fisher Scientific or Sigma with purities >99.0%. 1, 2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) was purchased from Avanti Polar Lipids. The unmodified peptides in this study are synthesized by Genescript.
2.2. Peptide modification
Peptides (20 μg) were brought up to the same volume (30 μL) with 100 mM ammonium bicarbonate (ABC). Propionic reagent (3:1 ACN: propionic anhydride) was added (1:4, 10 μL) to each peptide and incubated for 15 min at room temperature (RT). This process was repeated one time and peptides were dried down in a speed-vac concentrator. Peptides were resuspended in the same volume of 100 mM ABC, and two additional rounds of propionylation were performed as before. Peptides were centrifuged in a speed-vac concentrator to dryness and resuspended in ddH2O prior to nanopore channel experiments.
2.3. Cloning, expression, and purification of loop deleted Phi29 connectors
Three mutant versions of the connector subunit protein were used in this research: a connector subunit with C-terminus 6×His tag and C-terminus 25 amino acids (AA287-309) deleted (C-Δ25 GP10), the connector subunit with the C-terminus 6×His tag and the flexible internal loop removed (AA229-246, Δloop GP10), and the connector subunit with the N-terminus 6×His tag and the flexible loop removed (AA1-14, N-Δ14 GP10). The cloning, expression, and purification of these connector protein subunits have been previously reported [57, 59].
2.4. Incorporation of wild type and mutant Phi29 motor channel into liposomes
The incorporation process contains dehydration and hydration steps[40]. Connector-reconstituted liposomes were prepared using a dehydration rehydration method. Briefly, DPhPC in chloroform (100 μl, 10 mg/ml) was dried for 4 min under a vacuum by an evaporator (Buchi). These four connectors (final concentration 300-500 μg/ml) and liposome buffer (3 M KC1, 250 mM sucrose, 5 mM HEPES, pH 7.4) were added and vortexed thoroughly to form giant unilamellar vesicles (GUVs). To prepare homogenous proteoliposomes, the GUVs were then filtered through a 0.4 μm polycarbonate membrane 25-30 times using an extruder (AvantiPolar Lipids).
2.5. Insertion of engineered channels into the copolymer membrane of MinION™ Flow Cell
The MinION™ was connected to the computer, and a blank Flow Cell without biological nanopore was inserted into the MinION™. The MinKNOW software, developed and provided by Oxford Nanopore Technologies Ltd., was used for pore insertion and data collection.
The Membrane Quality Check (MQC) script was run first to check the membrane’s qualityas determined through analysis via Traceview software. Air bubbles in the system were removed by 1000 μL pipette. To insert the channel into the Flow Cell membrane, proteoliposomes (20 μL) were mixed with C13 buffer (25 mM potassium phosphate, 150 mM potassium ferrocyanide, 150 mM potassium ferricyanide, pH 8) (280 μL) and loaded into the priming port. A voltage profile for the facilitation of channel insertion (−150 mV to −350 mV) was applied by MinKNOW™. C13 buffer (500 μL) was added from the priming port to flush away excess pores. The number of inserted channels was evaluated by Platform Quality Check (PQC) script. All data were visualized by custom software provided by Oxford Nanopore Technologies Ltd.
2.6. Peptide translocation experiments
For K6, K10, and K14 translocation experiments, three peptides (1 mg/ml) were added to both chambers in turn after single-channel insertion. For K10, proK10, K14, and proK14 detection experiments using C-Δ25 GP10 channel, the four peptides (1 mg/ml) were added to both chambers in turn after single-channel insertion. The electrical signals were recorded under ±50 mV. All nanopore experiments for peptide translocation were repeated several times and performed in conductance buffer (0.15 M KC1, 5 Mm HEPES, pH7.4). The final concentration of tested peptides in this study was 50 nM. The current trace was recorded by Bilayer Clamp Amplifier BC535 (Warner Instruments) system with an Axon DigiData 1440 A analog-digital converter (Molecular Devices). Data were recorded at 1 KHz bandwidth with a sampling frequency 20 KHz. The Clampex 10 (Molecular Devices), Clampfit 10 (Molecular Devices), MOSAIC, and PRISM were used to collect and analyze data.
On the MinION™ system, peptide solution (2 μL, 1 mg/ml) was mixed with C13 buffer (298 μL) and then loaded into the Flow Cell via priming port after the pore insertion. MinKNOW™ was used to observe peptide translocation under ±100 mV. After data collecting, the Flow Cell was flushed and washed with Flow cell buffer (1-2 ml), allowing for reuse.
3. Results and discussion
3.1. Construction of mutant channels of phi29 DNA packaging motor by removing each of the three flexible loops.
The WT Phi29 connector channel has a truncated cone structure (Fig. 1A), and it is composed of twelve protein GP10 subunits. Three flexible sequences are clearly differentiated: the N-terminus (AA1-14), the internal channel loop (Δloop, AA229-246), and the C-terminus (AA287-309, 25aa) (Fig. 1B). The E. coli host HMS174(DE3) was used to overexpress the mutant GP10 protein, which was self-assembled into the dodecamer connector. The C-ʔ25 (Fig. 1C, lane 1), and C-Δloop (Fig. 1C, lane 3) connectors were purified by His-tag affinity column into homogeneity and evaluated with 10% SDS-PAGE using WT connector (Fig. 1C, lane 4) and N-Δ14 (Fig. 1C, lane 2) as control proteins.
Figure 1. Illustration and characterization of the channel of wild type and mutant phi29 bacterial virus DNA packaging motor.
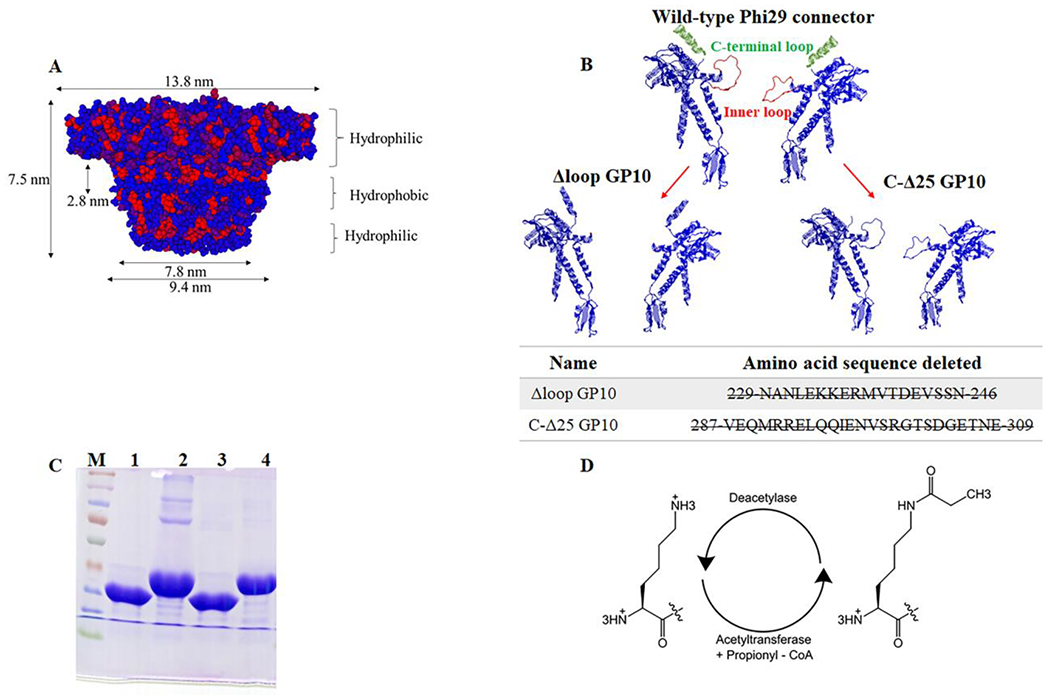
(A) Side view of the phi29 connector, red hydrophobic; blue hydrophilic. (B) Cross-section structure of two protein subunits of phi29 connector showing the inner loops and the C-terminus and N-terminus. (C) Molecular weight of purified C-Δ25 GP10 (lane 1), N-Δ14 GP10 (lane 2), Δloop GP10 (lane 3) and wild type GP10 (lane 4) on 10% SDS-PAGE; (D) Schematic diagram of lysine propionylation modification.
To explore the potential of engineered channels of the Phi29 DNA-packaging nanomotor for the identification and differentiation of PTMs, lysine propionylation was used as a model system. For lysine propionylation, peptides with lysine were incubated with ammonium bicarbonate in the presence of propionic reagent (3:1 ACN: propionic anhydride) (Fig. 1D).
3.2. Use of C-His Δloop to translocate peptides RK4 and proRK4 and identify one propionylation modification on lysine on chamber patch clamps system.
We first tested whether the mutant GP10 channel could detect only one propionylation modification by distinguishing the difference between unmodified peptide RRRKRRRRRRRR (RK4) and propionylated peptide RRRKproRRRRRRRR (proRK4). The blockage and dwell time were used to characterize the peptides. The blockage percentage was calculated using the equation (Io-Ib)/Io × 100%, in which Io is the channel current in the absence of peptide translocation (open channel current), and Ib is the current during peptide translocation (block current).
Robust peptides translocation signals were observed when either RK4 or a mixture of RK4/proRK4 peptides were added to the Δloop GP10 channel (Fig. 2A and D). Interestingly, the channel was capable of detecting the simultaneous passage of two RK4 peptides as well (Fig. 2D, yellow). Passage of RK4 (red) and proRK4 (green) through the channel generated significantly different current trace signals (Fig. 2A and D). The current blockage caused by RK4 was 32% (Fig. 2B, Table 1). Adding proRK4 peptides to the Δloop GP10 channel in the presence of RK4 resulted in one additional peak with a current blockage of 15% (Fig. 2E, Table 1). The scatter plot demonstrates that the Δloop GP10 channel can distinguish RK4 from proRK4 (Fig. 2C and F).
Figure 2. Δloop GP10 to translocate RK4 and proRK4 and distinguishion of only one lysine propionylation modification.
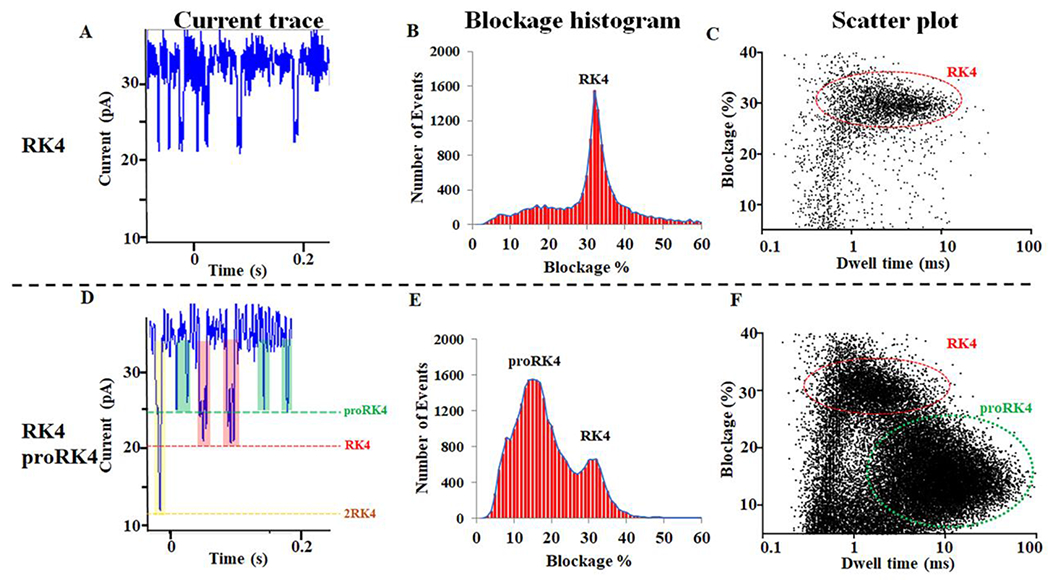
(A, D) The current trace shows the appearance of translocation signals after the addition of RK4 or RK4/proRK4 to the Δloop GP10 channel. (B, E) The current blockage caused by the addition of RK4 or RK4/proRK4. (C, F) The scatter plot caused by the addition of RK4 or RK4/proRK4 to the Δloop GP10 channel.
Table 1:
Statistical results of current blockage of different peptides.
| Name | Sequence | Current blockage(%) |
|---|---|---|
| RK4 | RRRKRRRRRRRR | 32 |
| proRK4 | RRRKproRRRRRRRR | 15 |
| K10 | KKKKKKKKKK | 45 |
| proK10 | proKproKproKproKproKproKproKproKproKproK | 26 |
| K14 | KKKKKKKKKKKKKK | 52 |
| proK14 | proKproKproKproKproKproKproKproKproKproKproKproKproKproK | 35 |
3.3. Use of C-Δ25 GP10 to translocate and differentiate peptide K6, K10 and K14 on chamber patch clamps system.
Protein modification plays important roles in diverse cellular processes, such as apoptosis, metabolism, transcription, and stress response [60, 61]. Additionally, multiple lysine propionylation sites have been discovered in single proteins often associated with cancer, neurodegenerative disorders, and cardiovascular diseases. For example, 2, 21, 9 and 12 lysine propionylation sites were identified in tumorigenesis-related protein p53, histone H4, p300, and CBP, respectively [54]. Therefore, it is necessary to analyze further whether the engineered channels can distinguish the propionylation modification at multiple lysine sites.
To do this, three unmodified peptides KKKKKK (K6), KKKKKKKKKK (K10) and KKKKKKKKKKKKKK (K14) were adopted to test their translocation on C-Δ25 Phi29 motor channels. We firstly investigated three different peptide lengths consisting of 6 (K6; FigS1. A–D), 10 (K10; Fig. 3A–D), and 14 (K14; Fig. 3E–H) lysines, using a C-Δ25 GP10 channel. As shown in Figure S1A–D, K6 peptide translocation did not affect current blockage, suggesting this peptide is too small to be detected with this technology. Obvious peptide translocation signals were observed after the addition of K10 or K14 to the channel (Fig. 3B and F), compared to the control (Fig. 3A and E). Current blockages of K10 and K14 were 45%, and 52%, respectively (Fig. 3C, G, Table 1). The electrical signal resulting from the peptide passage through the channel is related to the length of the narrow region of the channel. As shown in Figure 3D and H, the longer the peptide, the larger blockage percentages and dwell time.
Figure 3. C-Δ25 GP10 to differentiate polylysine (K10 and K14).
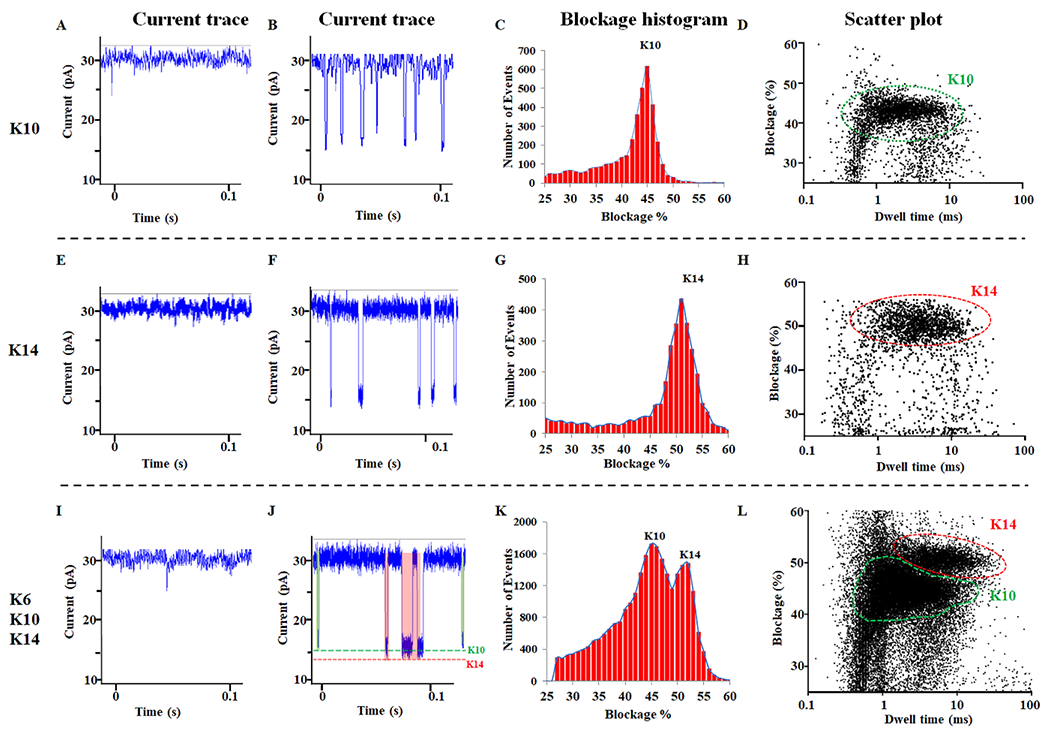
(A, E, I) The current trace without the presence of peptide. (B, F, J) The current trace of the peptide in conductance buffer. (C,G, K) The current blockage resulted from the addition of K10, or K14, or K6/10/14 mixture. (D,H, L) The scatter plot resulted from the addition of K10, or K14, or K6/10/14 mixture to the C-Δ25 GP10 connector.
The capability of the C-Δ25 GP10 channel to differentiate peptides in mixtures of biological samples was investigated. On the one hand, two distinct peaks (K10 and K14) of blockages were clearly observed as a result of fitting the data by Gaussian distributions (Fig. 3K). These peaks matched the blockage parameters of individual peptides measured separately. Furthermore, signals from K10 and K14 were observed in the current trace (Fig. 3J) and scatter plot (Fig. 3L), which were also in agreement with the blockage parameters of individual peptides measured separately.
3.4. Use of C-Δ25 GP10 channel to translocate peptide K10, K14 and proK10, proK14 and identify multiple propionylation modifications on lysine on chamber patch clamps system.
Since the Δloop GP10 channel could distinguish the incorporation of one lysine propionylation, and C-Δ25 GP10 channel could distinguish polylysine, we wanted to examine whether the C-Δ25 GP10 channel could detect differences between unmodified polylysine peptide (K10 and K14) and polylysine peptide containing multiple lysine propionylations (proK10, mass shift +560.26 Da and proK14, mass shift +784.27 Da). First, we tested K10 peptides on the C-Δ25 GP10 channel, without peptide current trace results as control (Fig. S2). After the addition of K10 to the C-Δ25 GP10 channel, one population (K10) was observed in the scatter plot (Fig. 4C) with a current blockage of 45% (Fig. 4B, Table 1). The resulting current trace further supported the presence of one population with uniform current signals (Fig. 4A). Next, the proK10 was added to the conductance buffer in the presence of K10. The current trace suggested the appearance of proK10 signals by the presence of a lower current blockage (Fig. 4D, green). The population of proK10 was apparent by the additional current blockage of 26% (Fig. 4E, Table 1) and the resulting scatter plot (Fig. 4F). After that, K14 was added to the conductance buffer in the presence of K10 and proK10. The current trace resulted in K14 translocation signals with the longest current blockage signals (Fig. 4G). One additional peak of the current blockage, 51% was observed in detection results of the K6, K10, and K14 peptide mixture (Fig. 4H, Table 1). The appearance of the K14 population was further evident in the scatter plot results (Fig. 4I, Fig.S3). Finally, the proK14 peptides were added to the conductance buffer. The current trace (Fig. 4J) and blockage results (Fig. 4K, Table 1) clearly confirmed the proK14 translocation signals. However, the proK10 and proK14 groups could not be distinguished from one another, as shown in the scatter plot (Fig. 4L, Fig.S4).
Figure 4. C-Δ25 GP10 to differentiate K10, proK10, K14, and proK14.
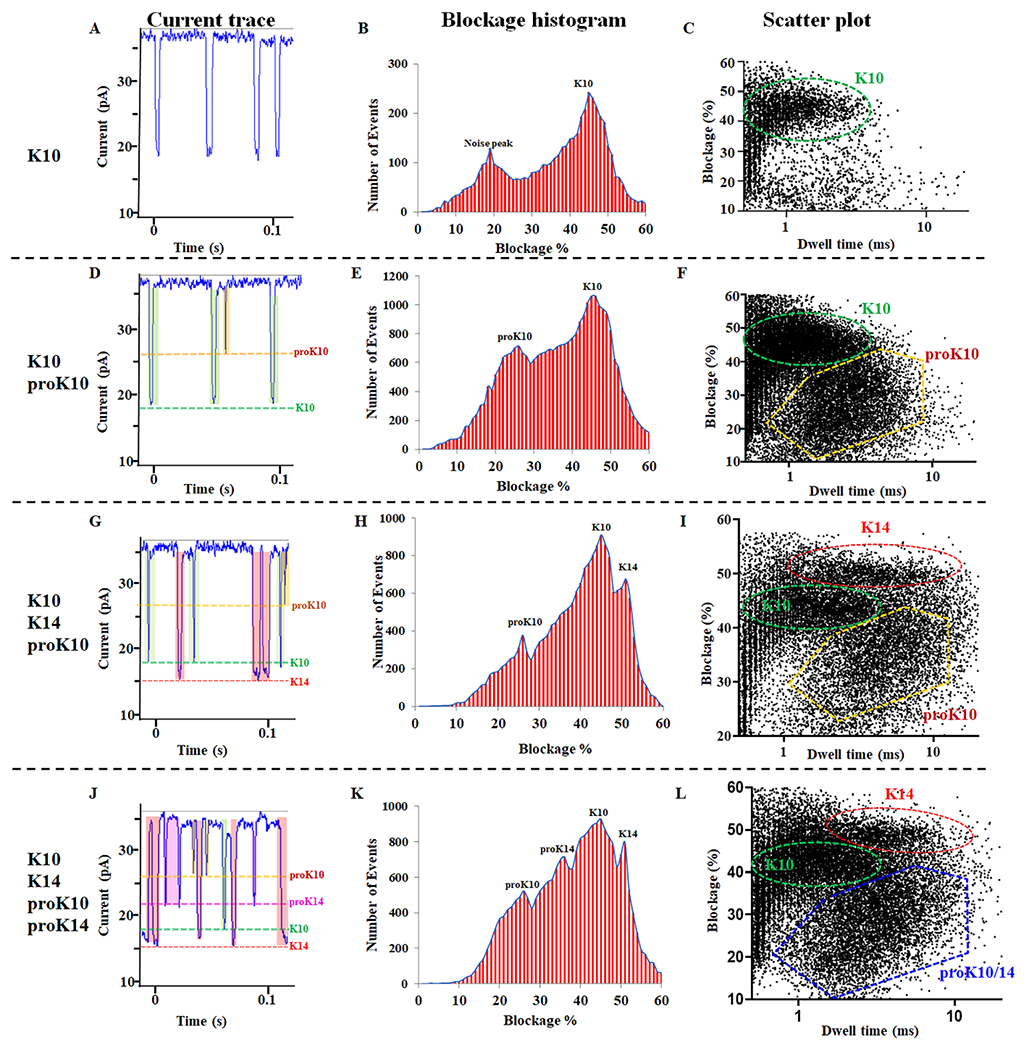
(A, D, G and J) The current trace showing the appearance of translocation signals after the addition of K10, K10/proK10, K10/proK10/K14, and K10/proK10/K14/proK14. (B, E, H, and K) The current blockage resulted from the addition of K10, K10/proK10, K10/proK10/K14, and K10/proK10/K14/proK14. (C, F, I and L) The scatter plot resulted from the addition of K10, K10/proK10, K10/proK10/K14, and K10/proK10/K14/proK14.
3.5. Use of C-Δ25 GP10 channel to translocate peptide RK4 and proRK4 and identify one propionylation modification on lysine on MinION™ Flow Cell system.
A feasible clinical application requires the translation of the chamber patch clamps system into a portable, real-time system for diagnosis and treatment based on PTMs. Oxford Nanopore Technologies Ltd. provides a new type of single-molecule sequencer (MinION™) for the third-generation sequencing technology by using a biological nanopore to provides direct DNA and RNA sequencing. It can plug directly into a standard USB3 port on a computer with low hardware requirement and simple configuration. Compared to DNA or RNA sequencing, proteins and peptides offer complexity from their size and structure, making them challenging for direct detection by protein-based nanopores [62, 63]. However, it is still unknown whether the MinION™ Flow Cell system can detect PTMs.
To develop a high-throughput sensing platform for disease screening and treatment based on PTMs, we tested modified and unmodified peptides on the MinION™ Flow Cell system using the C-Δ25 GP10 channel. About 100 single channels of C-Δ25 GP10 connector could be inserted into the MinION Flow Cell at one time, which enables itself for becoming a high-throughput sensing platform. Peptides were suspended in Flow Cell buffer (C13 buffer) and loaded into the Flow Cell via a priming port after confirming that the Phi29 connector channels have successfully directly inserted into the membrane. The peptide translocation was evaluated by Platform QC under ±100 mV (blue line, V). The proRK4 produced signals (events, red line) with a short dwell time and blockage (Fig. 5A and C, green arrow). After the addition of RK4, a second signal with a longer dwell time and a higher current blockage rate was detected (Fig. 5B and D, red arrow). The channel without peptide results as control (Fig. S5A). Two current blockage peaks with different blockage rates were observed after the addition of RK4 and proRK4 into the MinION™ Flow Cell (Fig.S6). The proRK4 (green arrow) and RK4 (red arrow) were distinguished from a premixture system with significantly different current blockage and/or dwell time (Fig. 5E and F). Interestingly, we also observed some noise events, characterized by no dwell time (black arrow).
Figure 5. C-Δ25 GP10 channel to translocate RK4 and proRK4 and to show the detection of only one lysine propionylation modification on the MinION™ Flow Cell system.
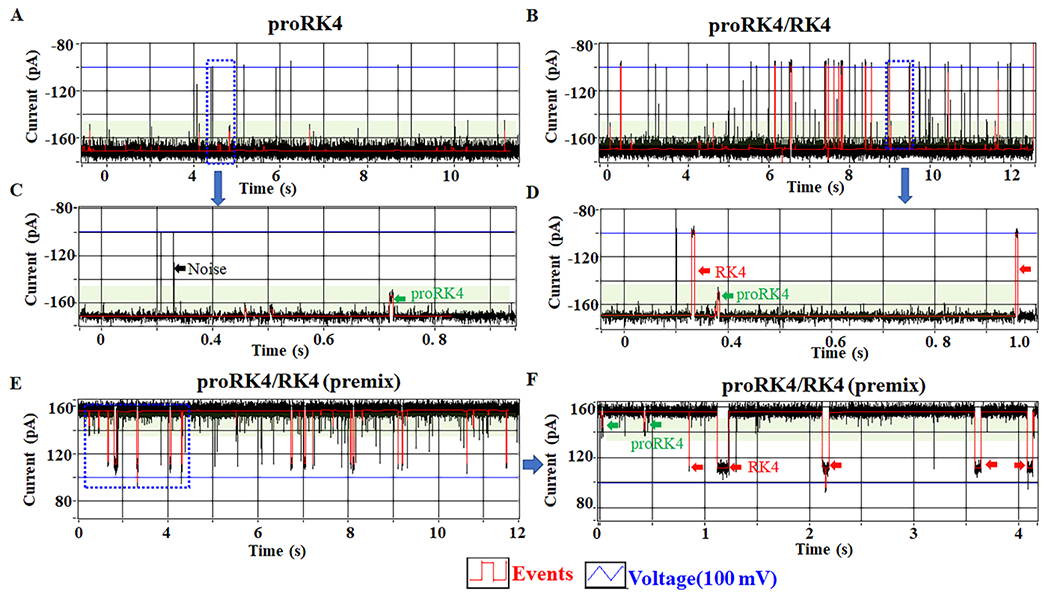
(A, C) The current trace showing the appearance of proRK4 translocation signals after the addition of proRK4 to MinION™ Flow Cell (green arrow). (B, D) The current trace showing the appearance of RK4 translocation signals (red arrow) after the addition of RK4 to MinION™ Flow Cell in the presence of proRK4. (E, F) Differentiating proRK4 (green arrow) from RK4 (red arrow) using MinION™ Flow Cell via premixing.
3.6. Use of C-Δ25 GP10 channel to translocate peptide K10 and proK10 and identify multiple propionylation modifications on lysine on MinION™ Flow Cell system.
We further analyzed the ability of the C-Δ25 GP10 channel to distinguish between K10 and proK10 on the MinION™ Flow Cell. Similar results for proK10 signals were obtained as before, with a shorter dwell time and current blockage rate (Fig. 6A and C, green arrow). In comparison, another significantly different signal with a longer dwell time, and higher current blockage rate was recorded after adding K10 to MinION™ Flow Cell in the presence of proK10 (Fig. 6B and D, red arrow). The channel without peptide results as control (Fig. S5B). Two current blockage peaks with different blockage rates were observed after the addition of K10 and proK10 into the MinION™ Flow Cell (Fig.S7). The current blockage of K10 and proK10 in the C13 buffer was 35% and 16%, respectively.
Figure 6. C-Δ25 GP10 channel to distinguish K10 and proK10 on the MinION™ Flow Cell system.
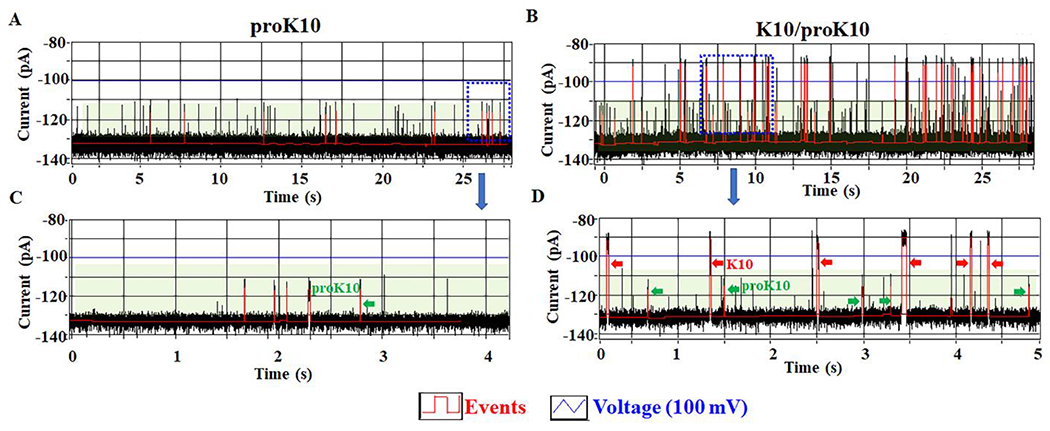
(A, C) The current trace showing the appearance of proRK10 translocation signals (green arrow). (B, D) The current trace showing the appearance of K10 translocation signals (red arrow) after the addition of K10 to MinION™ Flow Cell in the presence of proRK10 (green arrow).
Here, both the patch clamps and MinIONTM Flow Cell system allow for the identification of one or multiple propionylation modifications on lysine. However, the MinIONTM system holds two advantages comparing to the patch-clamp system: the robust property and the high through capacity. One of the important areas in single pore sensing is the translation of this technology into clinical application. That is, how to adapt and incoordinate the biological pore into the industrial instrument. The fragility of the lipid bilayer (BLM) is the weakness of the patch clamps system [64]. Protein pore in the patch-clamp system remains challenging to translate into clinical applications. Additionally, the lipid membrane has a large area and capacitance, leading to increased noise[65, 66]. Oxford Nanopore Technologies Ltd. provides the portable single-molecule sequencer (MinION™) for high-throughput applications. There are 2048 channels in the two-inch Flow Cell. Each channel can display current blockage with an independent current assessment. Compared with the lipid bilayer, the MinION™ Flow Cell polymeric membrane is mechanically stable, resistant to high voltage, higher capacity, thus making it a promising platform for PTMs detection.
4. Conclusion
The engineered Phi29 connectors with both the C-terminal 25-AA and the internal 17AA-loop deletion were used for single pore sensing to discriminate peptides with or without propionylation modifications. This was achieved on both the traditional patch-clamp and the modern MinION™ Flow Cell system. On the clamp system, the engineered loop-deleted Phi29 connector distinguished single or numerous propionylation modifications. PTMs were also detected on the MinION™ Flow Cell system, revealing the potential of applying the Phi29 connector for a label-free, high throughput nanopore detection platform.
Supplementary Material
Acknowledgements
The research was supported by NIH grant R01 EB012135, and Oxford Nanopore Technologies Ltd.
Peixuan Guo reports financial support was provided by Oxford Nanopore Technologies Ltd. Long Zhang reports financial support was provided by Oxford Nanopore Technologies Ltd. Peixuan Guo reports a relationship with Oxford Nanopore Technologies Ltd that includes: consulting or advisory. Peixuan Guo reports a relationship with Shenzhen P&Z Biomedical Co. Ltd that includes:. Peixuan Guo reports a relationship with ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC that includes: board membership. Michael A. Freitas reports a relationship with MassMatrix Inc. that includes: employment. P.G. is the consultant and licensor of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd., as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC.
Michael A. Freitas is the co-owner, co-founder, and Chief Scientific Officer of MassMatrix Inc., a for profit biotech company focusing on the development of bioinformatics software for biopharma. The work in this paper is not a product of MassMatrix Inc. The IP are properties of OSU as outlined in the OSU property rights agreements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
P.G. is the consultant and licensor of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd., as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC. Michael A. Freitas is the co-owner, co-founder, and Chief Scientific Officer of MassMatrix Inc., a for profit biotech company focusing on the development of bioinformatics software for biopharma. The work in this paper is not a product of MassMatrix Inc. The IP are properties of OSU as outlined in the OSU property rights agreements.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
The raw data required for these findings are available upon request by email to guo.1091@osu.edu.
Reference
- [1].Yang XJ, Seto E, Lysine acetylation: codified crosstalk with other posttranslational modifications, Mol Cell 31(4) (2008) 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuriakose A, Chirmule N, Nair P, Immunogenicity of Biotherapeutics: Causes and Association with Posttranslational Modifications, J Immunol Res 2016 (2016) 1298473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Darling AL, Uversky VN, Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter, Front Genet 9 (2018) 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Marko-Varga G, Fehniger TE, Proteomics and disease--the challenges for technology and discovery, J Proteome Res 3(2) (2004) 167–78. [DOI] [PubMed] [Google Scholar]
- [5].Audagnotto M, Dal Peraro M, Protein post-translational modifications: In silico prediction tools and molecular modeling, Comput Struct Biotechnol J 15 (2017) 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J, Regulation of autophagy by protein post-translational modification, Lab Invest 95(1) (2015) 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Seo J, Lee KJ, Post-translational modifications and their biological functions: proteomic analysis and systematic approaches, J Biochem Mol Biol 37(1) (2004) 35–44. [DOI] [PubMed] [Google Scholar]
- [8].Yan K, Wang K, Li P, The role of post-translational modifications in cardiac hypertrophy, J Cell Mol Med 23(6) (2019) 3795–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smith LE, White MY, The role of post-translational modifications in acute and chronic cardiovascular disease, Proteomics Clin Appl 8(7-8) (2014) 506–521. [DOI] [PubMed] [Google Scholar]
- [10].Gao J, Shao K, Chen X, Li Z, Liu Z, Yu Z, Aung LHH, Wang Y, Li P, The involvement of post-translational modifications in cardiovascular pathologies: Focus on SUMOylation, neddylation, succinylation, and prenylation, J Mol Cell Cardiol 138 (2020) 49–58. [DOI] [PubMed] [Google Scholar]
- [11].Lu H, Li G, Zhou C, Jin W, Qian X, Wang Z, Pan H, Jin H, Wang X, Regulation and role of post-translational modifications of enhancer of zeste homologue 2 in cancer development, Am J Cancer Res 6(12) (2016) 2737–2754. [PMC free article] [PubMed] [Google Scholar]
- [12].Smith LE, Rogowska-Wrzesinska A, The challenge of detecting modifications on proteins, Essays Biochem 64(1) (2020) 135–153. [DOI] [PubMed] [Google Scholar]
- [13].Fuchs SM, Strahl BD, Antibody recognition of histone post-translational modifications: emerging issues and future prospects, Epigenomics 3(3) (2011) 247–249. [DOI] [PubMed] [Google Scholar]
- [14].Restrepo-Perez L, Joo C, Dekker C, Paving the way to single-molecule protein sequencing, Nat Nanotechnol 13(9) (2018) 786–796. [DOI] [PubMed] [Google Scholar]
- [15].Huang G, Voet A, Maglia G, FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 dalton resolution, Nat Commun 10(1) (2019) 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Varongchayakul N, Song J, Meller A, Grinstaff MW, Single-molecule protein sensing in a nanopore: a tutorial, Chem Soc Rev 47(23) (2018) 8512–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Restrepo-Perez L, Wong CH, Maglia G, Dekker C, Joo C, Label-Free Detection of Post-translational Modifications with a Nanopore, Nano Lett 19(11) (2019) 7957–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li T, Liu L, Li Y, Xie J, Wu HC, A universal strategy for aptamer-based nanopore sensing through host-guest interactions inside alpha-hemolysin, Angew Chem Int Ed Engl 54(26) (2015) 7568–7571. [DOI] [PubMed] [Google Scholar]
- [19].Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P, Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores, Nat Nanotechnol 4(11) (2009) 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H, Continuous base identification for single-molecule nanopore DNA sequencing, Nat Nanotechnol 4(4) (2009) 265–270. [DOI] [PubMed] [Google Scholar]
- [21].Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH, Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase, Nat Biotechnol 30(4) (2012) 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Larkin J, Henley RY, Jadhav V, Korlach J, Wanunu M, Length-independent DNA packing into nanopore zero-mode waveguides for low-input DNA sequencing, Nat Nanotechnol 12(12) (2017) 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wen S, Zeng T, Liu L, Zhao K, Zhao Y, Liu X, Wu HC, Highly sensitive and selective DNA-based detection of mercury(II) with alpha-hemolysin nanopore, J Am Chem Soc 133(45) (2011) 18312–18317. [DOI] [PubMed] [Google Scholar]
- [24].Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A, Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient, Nat Nanotechnol 5(2) (2010) 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Venkatesan BM, Bashir R, Nanopore sensors for nucleic acid analysis, Nat Nanotechnol 6(10) (2011) 615–624. [DOI] [PubMed] [Google Scholar]
- [26].Iqbal SM, Akin D, Bashir R, Solid-state nanopore channels with DNA selectivity, Nat Nanotechnol 2(4) (2007) 243–248. [DOI] [PubMed] [Google Scholar]
- [27].Zhou Z, Hu Y, Wang H, Xu Z, Wang W, Bai X, Shan X, Lu X, DNA translocation through hydrophilic nanopore in hexagonal boron nitride, Sci Rep 3 (2013) 3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kowalczyk SW, Grosberg AY, Rabin Y, Dekker C, Modeling the conductance and DNA blockade of solid-state nanopores, Nanotechnology 22(31) (2011) 315101. [DOI] [PubMed] [Google Scholar]
- [29].Schneider GF, Dekker C, DNA sequencing with nanopores, Nat Biotechnol 30(4) (2012) 326–328. [DOI] [PubMed] [Google Scholar]
- [30].Verschueren DV, Pud S, Shi X, De Angelis L, Kuipers L, Dekker C, Label-Free Optical Detection of DNA Translocations through Plasmonic Nanopores, ACS Nano 13(1) (2019) 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ji Z, Jordan M, Jayasinghe L, Guo P, Insertion of channel of phi29 DNA packaging motor into polymer membrane for high-throughput sensing, Nanomedicine 25 (2020) 102170. [DOI] [PubMed] [Google Scholar]
- [32].Huang G, Willems K, Soskine M, Wloka C, Maglia G, Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores, Nat Commun 8(1) (2017) 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nivala J, Mulroney L, Li G, Schreiber J, Akeson M, Discrimination among protein variants using an unfoldase-coupled nanopore, ACS Nano 8(12) (2014) 12365–12375. [DOI] [PubMed] [Google Scholar]
- [34].Cressiot B, Braselmann E, Oukhaled A, Elcock AH, Pelta J, Clark PL, Dynamics and Energy Contributions for Transport of Unfolded Pertactin through a Protein Nanopore, ACS Nano 9(9) (2015) 9050–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rosen CB, Rodriguez-Larrea D, Bayley H, Single-molecule site-specific detection of protein phosphorylation with a nanopore, Nat Biotechnol 32(2) (2014) 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim JW, Tung S, Bio-Hybrid Micro/Nanodevices Powered by Flagellar Motor: Challenges and Strategies, Front Bioeng Biotechnol 3 (2015) 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA, The potential and challenges of nanopore sequencing, Nat Biotechnol 26(10) (2008) 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cressiot B, Greive SJ, Mojtabavi M, Antson AA, Wanunu M, Thermostable virus portal proteins as reprogrammable adapters for solid-state nanopore sensors, Nat Commun 9(1) (2018) 4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C, Fabrication of solid-state nanopores with single-nanometre precision, Nat Mater 2(8) (2003) 537–540. [DOI] [PubMed] [Google Scholar]
- [40].Ji Z, Kang X, Wang S, Guo P, Nano-channel of viral DNA packaging motor as single pore to differentiate peptides with single amino acid difference, Biomaterials 182 (2018) 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ji Z, Guo P, Channel from bacterial virus T7 DNA packaging motor for the differentiation of peptides composed of a mixture of acidic and basic amino acids, Biomaterials 214 (2019) 119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang S, Zhao Z, Haque F, Guo P, Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis, Curr Opin Biotechnol 51 (2018) 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Haque F, Li J, Wu HC, Liang XJ, Guo P, Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA, Nano Today 8(1) (2013) 56–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang S, Haque F, Rychahou PG, Evers BM, Guo P, Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum, ACS Nano 7(11)(2013) 9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Restrepo-Perez L, Huang G, Bohlander PR, Worp N, Eelkema R, Maglia G, Joo C, Dekker C, Resolving Chemical Modifications to a Single Amino Acid within a Peptide Using a Biological Nanopore, ACS Nano 13(12) (2019) 13668–13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J, Oukhaled A, Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore, Nat Commun 9(1) (2018) 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chavis AE, Brady KT, Hatmaker GA, Angevine CE, Kothalawala N, Dass A, Robertson JWF, Reiner JE, Single Molecule Nanopore Spectrometry for Peptide Detection, ACS Sens 2(9) (2017) 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM, Effect of a post-translational modification mimic on protein translocation through a nanopore, Nanoscale 12(20) (2020) 11070–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Robertson JW, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ, Single-molecule mass spectrometry in solution using a solitary nanopore, Proc Natl Acad Sci U S A 104(20) (2007) 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fahie MA, Chen M, Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms, J Phys Chem B 119(32) (2015) 10198–10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wloka C, Van Meervelt V, van Gelder D, Danda N, Jager N, Williams CP, Maglia G, Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore, ACS Nano 11(5) (2017) 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tamura T, Hamachi I, Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems, J Am Chem Soc 141(7) (2019) 2782–2799. [DOI] [PubMed] [Google Scholar]
- [53].Okanishi H, Kim K, Masui R, Kuramitsu S, Lysine propionylation is a prevalent post-translational modification in Thermus thermophilus, Mol Cell Proteomics 13(9) (2014) 2382–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y, Lysine propionylation and butyrylation are novel post-translational modifications in histones, Mol Cell Proteomics 6(5) (2007) 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang T, Li XM, Bao X, Fung YM, Li XD, Photo-lysine captures proteins that bind lysine post-translational modifications, Nat Chem Biol 12(2) (2016) 70–72. [DOI] [PubMed] [Google Scholar]
- [56].Zhang K, Chen Y, Zhang Z, Zhao Y, Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software, J Proteome Res 8(2) (2009) 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Geng J, Fang H, Haque F, Zhang L, Guo P, Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor, Biomaterials 32(32) (2011) 8234–8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Geng J, Wang S, Fang H, Guo P, Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids, ACS Nano 7(4) (2013) 3315–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cai Y, Xiao F, Guo P, The effect of N- or C-terminal alterations of the connector of bacteriophage phi29 DNA packaging motor on procapsid assembly, pRNA binding, and DNA packaging, Nanomedicine 4(1) (2008) 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Blander G, Guarente L, The Sir2 family of protein deacetylases, Annu Rev Biochem 73 (2004) 417–435. [DOI] [PubMed] [Google Scholar]
- [61].Kouzarides T, Acetylation: a regulatory modification to rival phosphorylation?, EMBO J 19(6) (2000) 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ying YL, Cao C, Long YT, Single molecule analysis by biological nanopore sensors, Analyst 139(16) (2014) 3826–3835. [DOI] [PubMed] [Google Scholar]
- [63].Kasianowicz JJ, Balijepalli AK, Ettedgui J, Forstater JH, Wang H, Zhang H, Robertson JW, Analytical applications for pore-forming proteins, Biochim Biophys Acta 1858(3) (2016) 593–606. [DOI] [PubMed] [Google Scholar]
- [64].Huang S, Romero-Ruiz M, Castell OK, Bayley H, Wallace MI, High-throughput optical sensing of nucleic acids in a nanopore array, Nat Nanotechnol 10(11) (2015) 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fragasso A, Schmid S, Dekker C, Comparing Current Noise in Biological and Solid-State Nanopores, ACS Nano 14(2) (2020) 1338–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Stranges PB, Palla M, Kalachikov S, Nivala J, Dorwart M, Trans A, Kumar S, Porel M, Chien M, Tao C, Morozova I, Li Z, Shi S, Aberra A, Arnold C, Yang A, Aguirre A, Harada ET, Korenblum D, Pollard J, Bhat A, Gremyachinskiy D, Bibillo A, Chen R, Davis R, Russo JJ, Fuller CW, Roever S, Ju J, Church GM, Design and characterization of a nanopore-coupled polymerase for single-molecule DNA sequencing by synthesis on an electrode array, Proc Natl Acad Sci U S A 113(44) (2016) E6749–E6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data required for these findings are available upon request by email to guo.1091@osu.edu.


